Abstract
There has been a recent increase in the exploration of cold-active β-galactosidases, as it offers new alternatives for the dairy industry, mainly in response to the current needs of lactose-intolerant consumers. Since extremophilic microbial compounds might have unique physical and chemical properties, this research aimed to study the capacity of Antarctic bacterial strains to produce cold-active β-galactosidases. A screening revealed 81 out of 304 strains with β-galactosidase activity. The strain Se8.10.12 showed the highest enzymatic activity. Morphological, biochemical, and molecular characterization based on whole-genome sequencing confirmed it as the first Rahnella inusitata isolate from the Antarctic, which retained 41–62% of its β-galactosidase activity in the cold (4 °C–15 °C). Three β-galactosidases genes were found in the R. inusitata genome, which belong to the glycoside hydrolase families GH2 (LacZ and EbgA) and GH42 (BglY). Based on molecular docking, some of these enzymes exhibited higher lactose predicted affinity than the commercial control enzyme from Aspergillus oryzae. Hence, this work reports a new Rahnella inusitata strain from the Antarctic continent as a prominent cold-active β-galactosidase producer.
Keywords: β-galactosidase, Antarctica, lactose, cold-adapted bacteria, extremozymes
1. Introduction
Antarctica is possibly one of the most hostile habitats on Earth. It can be defined as a polyextreme environment, given the multiple harsh conditions for life, such as low temperatures, low humidity, limited bioavailability of water, high solar radiation intervals, extended periods of darkness, and freeze–thaw cycles [1]. However, various organisms, mainly microorganisms, have adapted to colonize and proliferate under these extreme conditions and are of great interest as a source of novel bioactive biotechnological compounds [2,3]. In recent years, there has been a great interest in discovering new enzymes from extremophilic microorganisms, also called extremozymes, for industrial purposes [4]. Such enzymes belong to microorganisms adapted to multiple adverse conditions that might give them different biochemical and biophysical properties compared to those in mesophilic environments. Cold-adapted enzymes are a prime example, produced by organisms inhabiting low-temperature habitats, which are valuable in several industrial applications (e.g., food, processing, cleaning agents, leather processing, and degradation of xenobiotic agents in the cold.) [5]. During the last decades, Antarctic strains capable of producing lipases, proteases, amylases, cellulases, and β-galactosidases have been described [6].
β-galactosidases (lactases) are used in the food industry to produce probiotic galacto-oligosaccharides and reduce the lactose content in dairy products for lactose-intolerant consumers [7]. Lactose intolerance appears when human adults (and most adult mammals) downregulate intestinal lactase production, a necessary enzyme for the digestion of lactose into glucose and galactose [8]. Hence, lactose-intolerant people suffer from bloating, flatulence, cramps, and nausea after milk consumption. It is estimated that 65% of the world’s population suffers from lactose intolerance [8], ranging from 50 to nearly 100%, depending on the country [9].
Industrial lactose hydrolysis in milk is chiefly carried out using thermostable enzymes at high temperatures (optimum activity at ~50 °C) [10]. However, low-temperature processing is preferred to improve the hygienic conditions and the milk properties’ conservation, avoiding spoilage and flavor changes. Then an ideal enzyme for treating milk should retain a significant activity at 4 °C–8 °C. The activity of the major commercial β-galactosidases (e.g., Lactozym Pure® from Novozymes and Maxilact® from DSM Food Specialties) is severely diminished at 4 °C, with optimal activity at nearly 37 °C [11]. Thus, optional thermostable enzymes with activity ranging from dairy processing conditions (i.e., 4 °C) to moderate temperatures (i.e., 37 °C) are needed to increase the process efficiency [11,12].
In recent years, several low-temperature active β-galactosidases have been reported from different psychrophilic bacteria, including Rahnella sp. R3 [13], Halobacterium lacusprofundi, Arthrobacter sp. [7,14], Pseudoalteromonas sp. [15,16], Planococcus sp. [17], Paracoccus sp. [18], Enterobacter ludwigii [19], Halomonas sp. [20], Alkalilactibacillus ikkense [21], and Marinomonas sp. [22]. These enzymes have variable optimum temperatures for their activity, a characteristic that might have been driven by different evolutionary pathways [22]. However, most of the β-galactosidases identified so far display a relatively low activity in cold (~4 °C), lost their stability at moderate-high temperatures (>30 °C), or are produced in low amounts [21]. Since β-galactosidases are widespread (present in animals, plants, fungi, yeasts, bacteria, and Archaea), a great diversity of newly cold-active β-galactosidases is expected to be discovered in untapped environments. In this work, we aimed to characterize the enzymatic activity of the cold-active β-galactosidases produced by Antarctic bacteria. Our work described the strain Rahnella inusitata Se8.10.12, a species non previously reported in the Antarctic continent, with high β-galactosidase activity at low temperatures (4–15 °C).
2. Results and Discussion
2.1. A Rahnella Strain Exhibited the Highest Enzymatic Activity among Multiple Antarctic Strains with β-Galactosidase Activity
β-galactosidase enzymes cleave the disaccharide lactose to produce galactose and glucose to enter glycolysis. Microorganisms, vegetables, and animals produce β-galactosidases; however, bacterial sources usually have higher productivity [23]. Here we screened β-galactosidase activity of Antarctic bacteria since new cold-active enzymes need to be explored to process lactose-free products. Our results evidenced 81 (27%) out of 304 Antarctic strains with positive enzymatic activity according to the detection by the β-Bluo-Gal indicator on agar (Table S1, Figure S1). Similarly, other authors have explored the potential of polar bacterial strains for the production of β-galactosidase enzymes. Screenings based on cleaved X-Gal in agar plates revealed that 56% of Arctic bacteria were capable of milk degradation, and particularly 40% of the Pseudoalteromonas sp. strains showed β-galactosidase activity [24]. Meanwhile, from a collection of 107 Antarctic bacterial strains, 41 (38%) were positive for this enzymatic activity [16]. Both studies suggest that Arctic and Antarctic cultures are a relatively rich source of β-galactosidase producers.
Other β-galactosidases with potential for the agro-food industry have been described in different bacterial species. Strains, such as Arthrobacter psychrolactophilus, Thermotoga maritima, Lactobacillus acidophilus, Sulfolobus solfataricus, Enterobacter agglomerans, Alicyclobacillus acidocaldarius, and Bacillus licheniformis [25] have been characterized. Among other lactic acid bacteria, such as L. acidophilus, L. bulgaricus, and Streptococcus thermophilus [26,27,28], psychrotolerant bacteria of the genus Arthrobacter have been described as a producer of thermally stable and cold-active β-galactosidase with biotechnological potential [7,14]. Likewise, in our work, most strains with positive β-galactosidase activity were identified as Arthrobacter species (Table S1), along with other taxa previously reported in the Antarctic continent, including Stenotrophomonas sp., Sphingomonas sp., Sporosarcina sp., Serratia sp., and Streptomonas sp. [29,30].
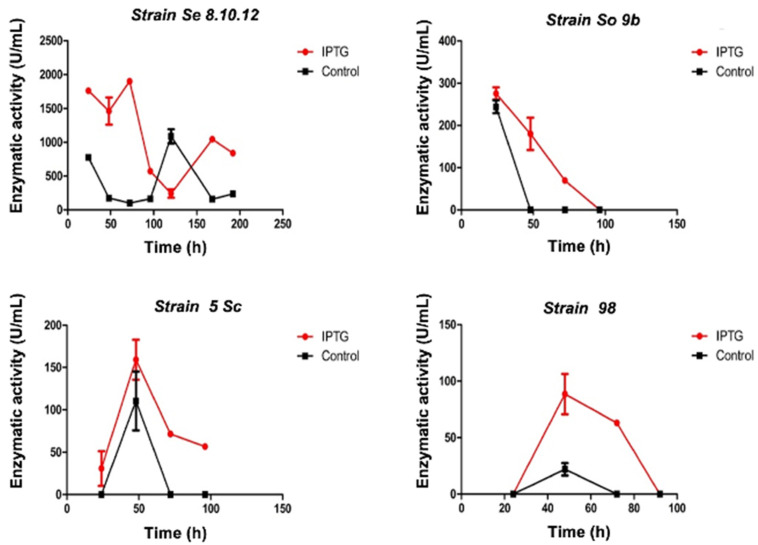
Quantification of enzymatic activity was performed from a set of the screened strains with higher activity (revealed by a more intense color change). The strain Se8.10.12 showed color diffusion on agar (Figure S1), which might indicate extracellular production of β-galactosidases. The potential of this strain (Se8.10.12) as the most significant β-galactosidase producer was confirmed since it produced the highest quantified activity in liquid culture (>2000 U/mL) compared to other selected strains (<500 U/mL) (Figure 1, Table S1) both in the presence and absence of the IPTG inducer (isopropyl-β-d-1-thiogalactopyranoside). IPTG triggered a higher enzymatic activity in all the assayed strains. Moreover, a faster response was observed on strain Se8.10.12, whose maximum activity was measured after five days of culture (120 h), changing to the third day (72 h) after IPTG was added. Based on the results described above, Se8.10.12 (Rahnella sp.) was selected to further characterize its β-galactosidase activity. Other studies have previously described species from the Rahnella genus as potential cold-active β-galactosidase producers [11,13,31], which specific enzymatic activity is discussed and compared in Section 2.3 and Section 2.4. However, none of them were obtained from Antarctic isolates.
Figure 1.
β-galactosidase activity quantified from enzyme extract of four Antarctic strains (specified in each graph title) at 15 °C reaction in the presence (red lines) and absence (black lines) of IPTG inducer. Error bars display the standard deviation of the data.
2.2. Antarctic Strain Se8.10.12 Showed Cold-Stable β-galactosidase Activity
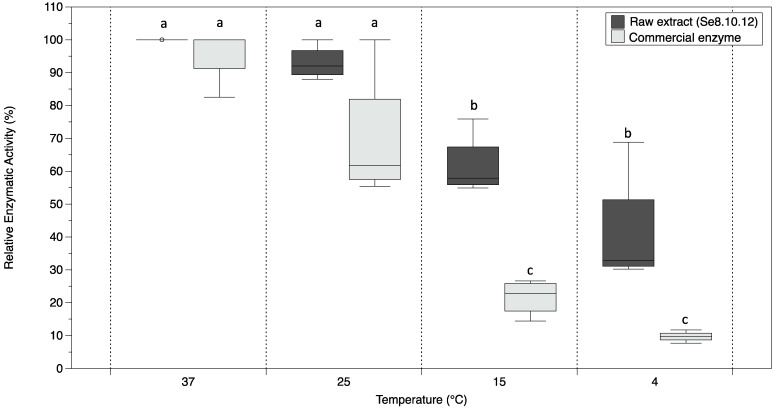
Among four promising β-galactosidase producer Antarctic strains, Rahnella sp. Se8.10.12 exhibited higher activity in the broth. Hence, we tested the stability of its lactose degradation activity at low temperatures. Using HPLC, we determined the lactose concentration (g/L) after 12 h in a reaction containing milk and the enzymatic extract or milk and commercial A. oryzae β-galactosidase. Relative activities were evaluated at 4 °C, 15 °C, 25 °C, and 37 °C. Higher concentrations of lactose were detected in the samples as the temperature decreased. Consequently, the enzymatic activity was reduced for both commercial enzyme and crude extract at temperatures below 15 °C (Figure 2). When comparing the lactose concentration for each reaction, it was observed that the crude enzymatic extract of the strain Se8.10.12 showed lower activity than the commercial enzyme at the same temperature and concentration equivalence (minimum concentration reached: 12.82 vs. 0.52 g/L, respectively). However, this behavior is expected for unpurified enzymes. On the other hand, the relative activity of the commercial enzyme of Aspergillus oryzae (maximum at 37 °C) is significantly decreased by temperatures lower than 15 °C, losing up to 90% of its activity at 4 °C (Figure 2). In contrast, the raw enzymatic extract of the Se8.10.12 strain showed significantly higher relative activity at lower temperatures than the commercial enzyme. Hence, the Antarctic strain extract showed more enzymatic efficiency in cold reaction, keeping up to 62% of its activity at 15 °C without significant differences below this temperature (i.e., 41% at 4 °C), but significantly higher than the commercial enzyme (Figure 2).
Figure 2.
Boxplot representing the β-galactosidase relative activity from Antarctic strain Se8.10.12 crude extract (Rahnella sp.) and commercial enzyme (Aspergillus oryzae) determined at 37 °C, 25 °C, 15 °C, and 4 °C after 12 h reaction with milk. The horizontal line indicates the mean values, and the length of each whisker indicates the interquartile range (IQR); n = 4. Different letters represent statistically significant differences (one-way ANOVA within temperatures, p < 0.005; t-student between enzymes, p < 0.01).
Cold-adapted galactosidases are a promising alternative for the current commercial enzymes used in the food industry because they hydrolyze lactose at low temperatures, thus preventing the degradation of other nutrients in the milk. Most of the recent studies describing robust cold-active β-galactosidases showed enzymes retaining nearly 20% of its highest activity at 4 °C (Antarctic Marinomonas sp. [22], Rahnella aquatilis from Xin-Jiang, China soil [32], Planoccocus sp. [33] from Daqing oil field in China and Enterobacter ludwigii from an Arctic Fjord [19]). Other few enzymes showed higher efficiencies in cold, among them, Rahnella sp. showed up to 27% of the maximum activity with ONPG as the substrate and nearly 80% with lactose at 4 °C [13], Halomonas sp. retains up to 25% of its activity at 0 °C [20], and for Alkalilactibacillus ikkense 60% of its maximal activity at 0 °C, and more than 70% at 10 °C was retained [21]. Similar results were reports for Arthrobacter psychrolactophilus strains isolated from Hokkaido, Japan, which showed high activity in cold; however, it significantly decreases above 15 °C [14], being less attractive because of its insufficient activity in moderate temperatures. This issue might be due to the optimal temperature activity, which is low (<15 °C) for most of the latter cases; thus, the enzymes lost activity in moderate temperatures. Our results suggest a highly efficient enzyme in cold, with optimal activity at moderate temperatures (opt T: 37 °C). The wide range of activity of our Antarctic β-galactosidase properly fits the necessities of the agro-food industry.
On the other hand, Antarctic bacteria have also been tested for potential cold-active β-galactosidase enzymes. Among them, the Antarctic strain Arthrobacter sp. C2-2 retained 20%, of its maximum activity in cold (~4 °C) [7]; Pseudoalteromonas sp. maintain up to 28% of activity [15]; a purified enzyme from bgaS gene of the Antarctic Arthrobacter sp. (strain SB) showed 50% of its activity at 0 ° C [34]; the purified enzyme of Pseudoalteromonas sp. isolated from Antarctic krill displayed 11–35% of maximum activity at 0 °C–20 °C [16], and a Planococcus sp. strain isolated from Ice Shelf in Antarctica keeps less than 20% activity below 10 °C [35]. Hence, these results indicate that our Rahnella sp. Se8.10.12 Antarctic strain has high activity in cold (retaining 41% of its maximum activity at 4 °C), which might help process liquid milk and generate low lactose products, tolerating moderate temperature (25 °C–30 °C). Thus, further molecular identification, genome sequencing, and analysis of this strain were performed.
2.3. Characterization of Strain Se8.10.12 Revealed an Antarctic Rahnella inusitata with Unique Metal-Resistance Related Genes among the Rahnella Species
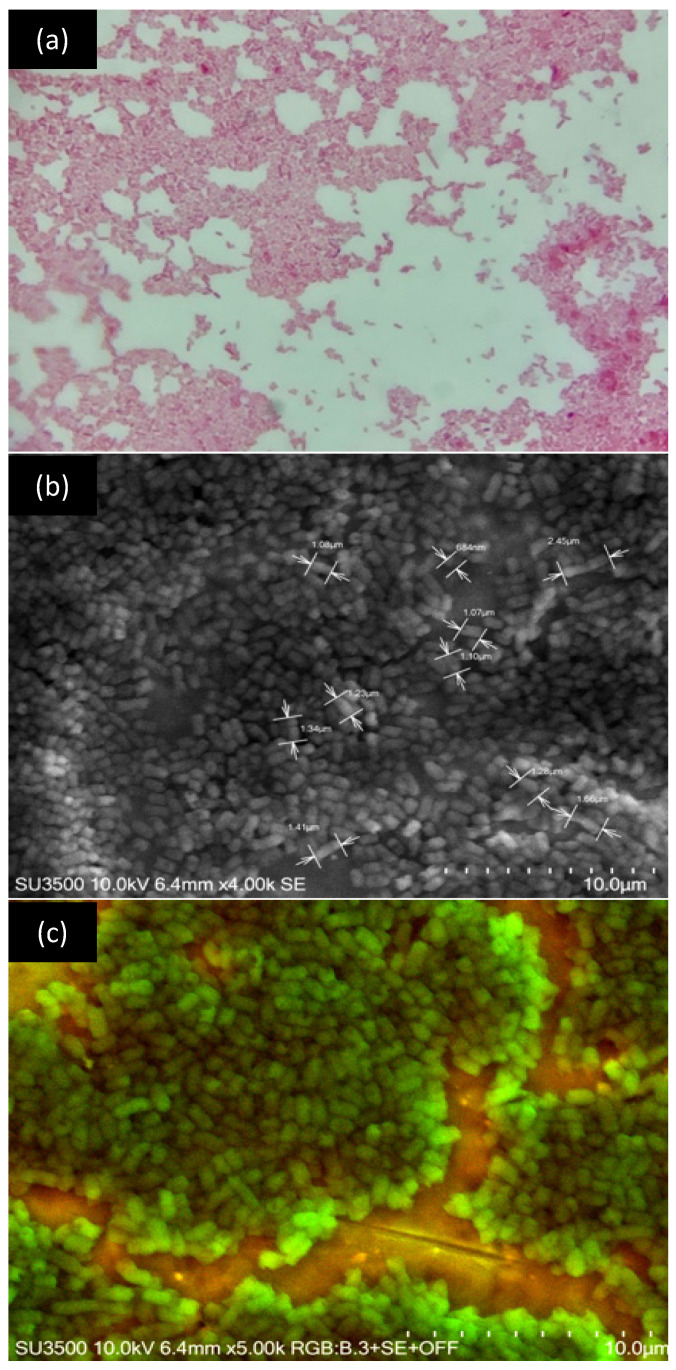
Strain Se8.10.12, showing higher potential as a β-galactosidase producer among 304 Antarctic strains, was characterized as a Gram-negative bacterium with a short bacillus or coccobacillus structure (Figure 3a). This bacterium lacks flagella and has a size of 1.33 µm approximately (Figure 3b, c). According to the biochemical test results, this strain can degrade sugars, as is expected from a lactase-producing strain. This bacterium also showed a lack of enzymes for lysine decarboxylation, ornithine decarboxylation, and urea hydrolysis, and it was negative for Citrate as the exclusive carbon pathway.
Figure 3.
Microscopic visualization of the strain Se8.10.12. (a) Gram staining at 100× magnification; (b,c) are images obtained by scanning electronic microscopy showing details of size and morphology, respectively.
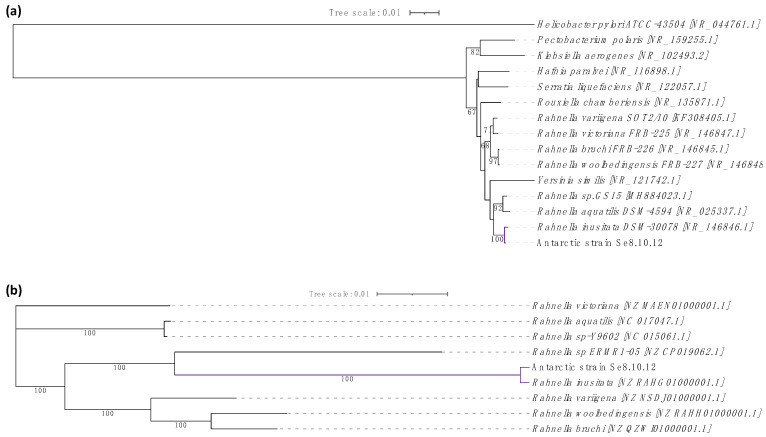
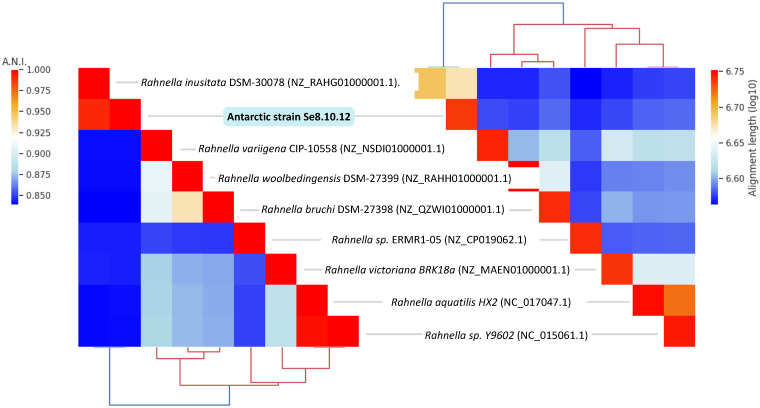
Regarding the 16S rRNA gene-based identity, this was associated with at least three Rahnella inusitata strains showing 99.93–100% identity by BLASTn comparison and 99.93% identity with Serratia sp. strains. The genus Rahnella belongs to the Enterobacteriaceae family; therefore, we evaluated the phylogenetic distances within the Rahnella genus (R. bruchi, R. woolbedingensis, R. variigena, R. victoriana, R. aquatilis, and R. inusitata), and other close members of the Enterobacteriaceae family (Yersinia similis, Yersinia bercovieri, Pectobacterium polaris, Serratia liquefaciens, Hafnia paralvei, Klebsiella aerogenes, and Rouxiella chamberiensis) [36,37]. The phylogenetic distance between the strain Se8.10.12 and the strain Rahnella inusitata relate them as the most proximate taxonomic group (Figure 4a). This taxonomic relatedness was also observed when analyzing the core-proteome-based phylogenetic history, showing a single and distant clade for Rahnella inusitata and our Antarctic strain (Figure 4b). This identity was further confirmed by Average Nucleotide Identity (ANI) after genome-wide analysis, where ANI values calculated across other eight Rahnella species reference genomes ranged from 84.09% to 99.04%, with alignment lengths from 3.51 Mb to 4.19 Mb (Figure 5 and Table S2). These results indicate that strains Se8.10.12 belong to genus Rahnella, crossing the species cut-off of a >96% ANI with Rahnella inusitata. Rahnella sp. are Gram-negative, bacillus-shaped bacteria described for the first time as Rahnella aquatilis [38]. These data agree with the results obtained by our morphological and biochemical characterization through Gram staining and sugars degradation capability, as expected for a β-galactosidase producing bacteria. Although some studies have described the lactase enzymatic activity for other Rahnella species [11,13,32], no reports described a β-galactosidase activity from Rahnella inusitata.
Figure 4.
Evolutionary history construction of the Antarctic strain Se.8.10.12 based on phylogeny. (a) Phylogenetic analysis of the 16S rRNA gene constructed with Bio-NJ method based on the Kimura two-parameter distance estimation model (Helicobacter pylori was used as an outgroup). (b) Maximum likelihood phylogenetic analysis of core-proteome based on the concatenated amino acid sequences of orthologous genes present in all Rahnella species using RAxML. GenBank accession numbers are provided in brackets. Branch lengths represent the number of substitutions per site (Bar: 0.01 substitutions per site), and bootstrap percentages of 1000 replicates are shown in the branches.
Figure 5.
Hierarchically clustered heatmap of Average Nucleotide Identity (ANI) calculated using BLASTn method (left), and alignment length (right) between nine Rahnella species genomes including the Antarctic isolate Se8.10.12. GenBank accession numbers are provided in brackets.
Rahnella inusitata is an uncommon species since little is known about its origin. It was recently described in damaged oak tissues in the United Kingdom [37], which contrasts with the Antarctic sediment from which strain Se8.10.12 was isolated. This is the first report of this species on the Antarctic continent to the best of our knowledge. The Se8.10.12 strain de novo genome assembly resulted in a 4.7 Mb chromosome in one complete linear contig with 134× coverage, 52.9 GC content, and showing 100% genome completeness based on 105 bacterial gene markers [39]. A total of 4312 coding sequences (CDS), including 22 rRNA, 78 tRNA, and two circular plasmids, were determined by Prokka annotation. CDS were assigned with eggNOG mapper to 4158 protein families.
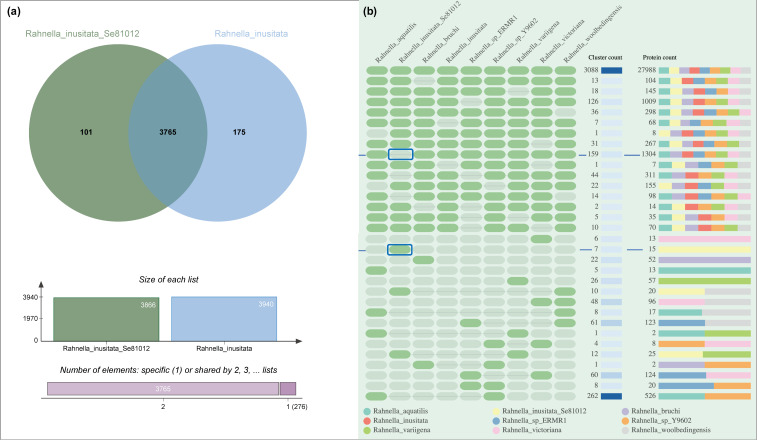
Comparative analysis of orthologous clusters revealed the presence of 101 protein families on our Antarctic strains that are absent in the reference R. inusitata (Figure 6a), most of them are primary biological functions shared with other Rahnella species, and at least seven clusters were unique of the Antarctic R. inusitata Se8.10.12 (Figure 6b). The functional assigned unique clusters are mostly related to heavy metal resistance, including copper, silver, cadmium, and cobalt (Table 1). On the other hand, based on gene ontology, the absent genes on our Antarctic strain showed two enriched biological processes: sporulation/formation of a cellular spore and response to zinc ion (p < 0.001). Interestingly, zinc response clusters were enriched in the Antarctic strain (R. inusitata Se8.10.12), R. aquatilis, and Rahnella sp. (Table 1), while absent in the reference R. inusitata. These data might suggest a gene loss of R. inusitata species related to zinc resistance, which may have been kept in the Antarctica strain by selective pressure due to heavy metals in the environment. These results might evidence the adaptation of this strain to its Antarctic habitat since it was isolated from sediment near a garbage disposal container at the Antarctic Chilean Base Julio Escudero. This environment could be considered contaminated compared to the environments where other Rahnella species have been found, which might influence the variances on metal-resistance-related genes gain/loss on the Antarctic R. inusitata. Besides, it has been extensively reported that contamination with heavy metals (e.g., cadmium, vanadium, arsenic, mercury, chromium, and gold) is usually found in Antarctic environments [3,40,41,42]. Moreover, a diversity of metal-resistance genes, including novel genes, has been described in the Antarctic bacteria, which occurrence are likely to be influenced by heavy metals at high concentrations (either by Anthropogenic or natural biogeochemical origin), shaping new taxonomic and functional diversities of soil microbial communities [43].
Figure 6.
Comparative analysis of orthologous gene cluster across Rahnella species genomes. (a) Venn diagram summarizing the distribution of shared orthologous clusters between two Rahnella inusitata genomes, including the Antarctic isolated strain Se8.10.12. (b) The occurrence table of main orthologous clusters between Rahnella species showing the 16 first cluster count in descending and ascending order. The presence or absence of a cluster group in the related species is represented by green or gray cells, respectively. The absent and unique clusters on the Antarctic strain Se8.10.12 are marked inside blue boxes.
Table 1.
Summary of main unique clusters found in the Antarctic Rahnella inusitata Se8.10.12 strain by gene ontology comparative analysis, including annotated clusters related to metal resistance.
| Cluster-ID | SwissProt ID: Protein, Function | GO Annotation | Genome|Protein |
|---|---|---|---|
| 5124 | P0CE53: transposase InsH, involved in the transposition of the insertion sequence IS5 | GO:0006313; P:transposition, DNA-mediated |
Rahnella_inusitata_Se81012|AJMHLFKD_00752 Rahnella_inusitata_Se81012|AJMHLFKD_00297 Rahnella_inusitata_Se81012|AJMHLFKD_03605 |
| 6148 | Q47459: protein PcoE, Required for the copper-inducible expression of copper resistance | GO:0046872; F: metal ion binding |
Rahnella_inusitata_Se81012|AJMHLFKD_00787 Rahnella_inusitata_Se81012|AJMHLFKD_00806 |
| 6151 | Q9ZHD4: sensor kinase SilS, component of the soil cation-efflux system that confers resistance to silver | GO:0000155; F:phospho-relay sensor kinase activity |
Rahnella_inusitata_Se81012|AJMHLFKD_00805 Rahnella_inusitata_Se81012|AJMHLFKD_00788 |
| 6149 | N/A | N/A |
Rahnella_inusitata_Se81012|AJMHLFKD_02409 Rahnella_inusitata_Se81012|AJMHLFKD_03172 |
| 6147 | N/A | N/A |
Rahnella_inusitata_Se81012|AJMHLFKD_02421 Rahnella_inusitata_Se81012|AJMHLFKD_03161 |
| 6146 | P76514: uncharacterized protein YfdR, probable cobalt ion binding | GO:0050897; F: cobalt ion binding |
Rahnella_inusitata_Se81012|AJMHLFKD_03184 Rahnella_inusitata_Se81012|AJMHLFKD_02399 |
| 6150 | Q9XJJ6: Spanin inner membrane subunit, participates in cell lysis during virus exit | GO:0019076; P: viral release from the host cell |
Rahnella_inusitata_Se81012|AJMHLFKD_03641 Rahnella_inusitata_Se81012|AJMHLFKD_03102 |
| 5653 | P41059: uncharacterized protein in gpA, probable zinc ion binding | GO:0008270; F: zinc ion binding |
Rahnella_sp_ERMR1|LLFBIOOH_04925 Rahnella_inusitata_Se81012|AJMHLFKD_02768 |
| 4804 | P74068: uncharacterized transporter sll1263, cadmium, iron, and zinc transmembrane transport | GO:0015341; F: zinc efflux active transmembrane transporter activity |
Rahnella_aquatilis|AEJLJCIE_02970 Rahnella_inusitata_Se81012AJMHLFKD_00295 Rahnella_sp_Y9602|CHBNGHAC_02899 |
2.4. Rahnella inusitata Se8.10.12 Genome Harbors Three β-Galactosidase Genes, In Silico Molecular Docking Showed That Two of Them Have a Higher Predicted Affinity for Lactose Than the Commercial Enzyme
The genomic and functional annotation results through Prokka and eggNOG delivered three genes associated with the production of β-galactosidases, including bglY, lacZ, and ebgA. BglY belongs to the family GH42 of β-galactosidases (EC 3.2.1.23) according to the CAZy database [44]. This protein has been reported as a cold-active enzyme in other species, including BglY from Alicyclobacillus acidocaldarius [45], and a β-galactosidases enzyme in the Rahnella genus [22]. LacZ is a GH2 β-galactosidase, which is a widely distributed family in bacteria (more than 22,000 GH2-protein entries in databases) [44]. LacZ has been described to have higher activity, at the optimum temperature, than other cold-active enzymes [46]. However, it is known as a mesophilic enzyme with little to no activity at 0 °C, regarding the LacZ of Escherichia coli [46]. Likewise, EbgA is also part of the GH2 family, known as an evolved ineffective LacZ paralog [47], since wild type E.coli was reported to have no lactase activity; however, single point mutations showed a drastically improved activity [48,49].
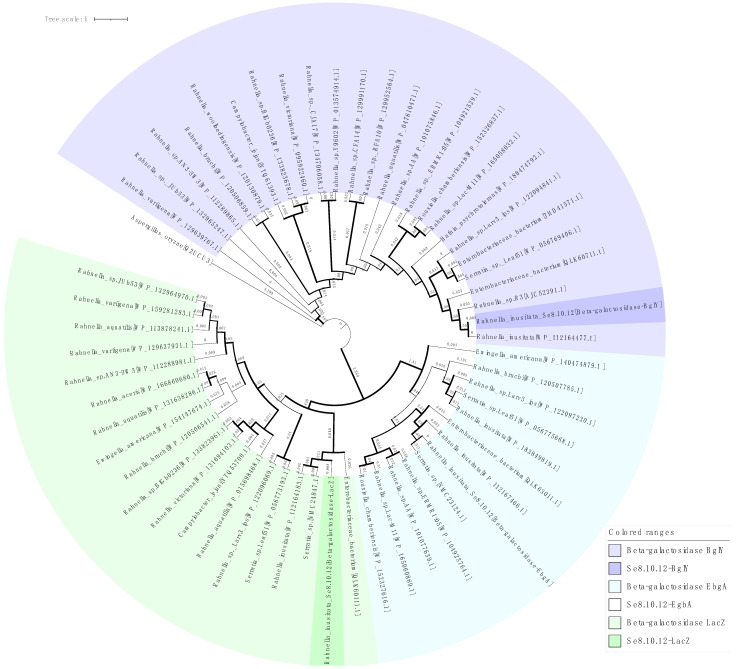
These bioinformatic analyses complement our experimental results, evidencing the existence of three genes of β-galactosidase in the Se8.10.12 strain genome. Despite its similarity with known β-galactosidases based on BLASP comparison, the phylogenetic analysis showed that these sequences are distant from most of the similar proteins and share a separate clade with enzymes from Rahnella inusitata, Serratia sp., and an Enterobacteriaceae strain (Figure 7), which might be related to their evolutionary history adapting to the specific environments. Moreover, two well-defined clades can be observed in the phylogenetic tree, which groups the proteins into the known glycoside hydrolases families GH2 and GH42 (clade 1: LacZ + EbgA, clade 2: BglY).
Figure 7.
Maximum likelihood phylogram based on three β-galactosidase amino acid sequences of the strain Se8.10.12 (highlighted color) and proteins on nucleotide database with >85% similarity (light color). Aspergillus oryzae β-galactosidase LacA was used as an outgroup. Branch lengths are shown in each branch and represent the number of nucleotide substitutions per site. Bootstrap support based on 1000 replicates ultrafast method is represented as branch width (1 px line = 47% to 10 px line = 100%). GenBank/EMBL/DDBJ accession numbers are provided in brackets.
To further study the enzymes’ specificity regarding their Antarctic nature, a presence/absence analysis of genes and determination of syntenic patterns were performed using PIRATE pangenome toolbox. Results showed that the genes lacZ and bglY were present in the eight of the available genomes of Rahnella strains (6 species), while gene ebgA was absent in the species R. vitoriana and R. varigena. Moreover, syntenic gene cluster was found for LacZ (including the genes: argO, Arginine exporter protein; mscS, small-conductance mechanosensitive channel; lacY, Lactose permease; and lacI Lactose operon repressor), and for EbgA (including ebgC, Evolved β-galactosidase subunit β; ygjK, Glucosidase; melR HTH-type transcriptional repressor; and ygjI inner membrane transporter). No syntenic blocks were found associated with gene bglY. These data suggest the presence of complete putative gene operons in the genome of the Antarctic Rahnella strain, which could effectively produce functional β-galactosidases. Moreover, it suggests that this is a highly conserved function in the Rahnella genus, while the EbgA gene might have been acquired by horizontal gene transference only in some species, as previously described [47].
On the other hand, it was established that Rahnella sp. R3 exhibited much higher catalytic activity (27 U/mg) at low temperatures than other β-galactosidases from Rahnella species (Rahnella sp. BS 1, Rahnella sp. HS-39 [31] and Rahnella aquatilis 14-1 [32]) [13]. Interestingly, Rahnella sp. R3 was isolated from a Glacier (Xinjiang, China), suggesting that, even though all the Rahnella species share orthologous β-galactosidase genes, they might evolve according to their specific niches. Our results support this idea, showing an Antarctic Rahnella with higher β-galactosidase activity at 4 °C than other Rahnella species previously reported, except for the R3 cold-adapted isolate. Moreover, the BglY β-galactosidase of our Antarctic R. inusitata might have acquired the same cold adaptability as the enzyme from Rahnella sp. R3, which can be deduced from its structural similarity (99% similarity to Rahnella sp. R3 protein) (Figure 8) and phylogenetic distance being more distant from other BglY Rahnella enzymes (Figure 7).
Figure 8.
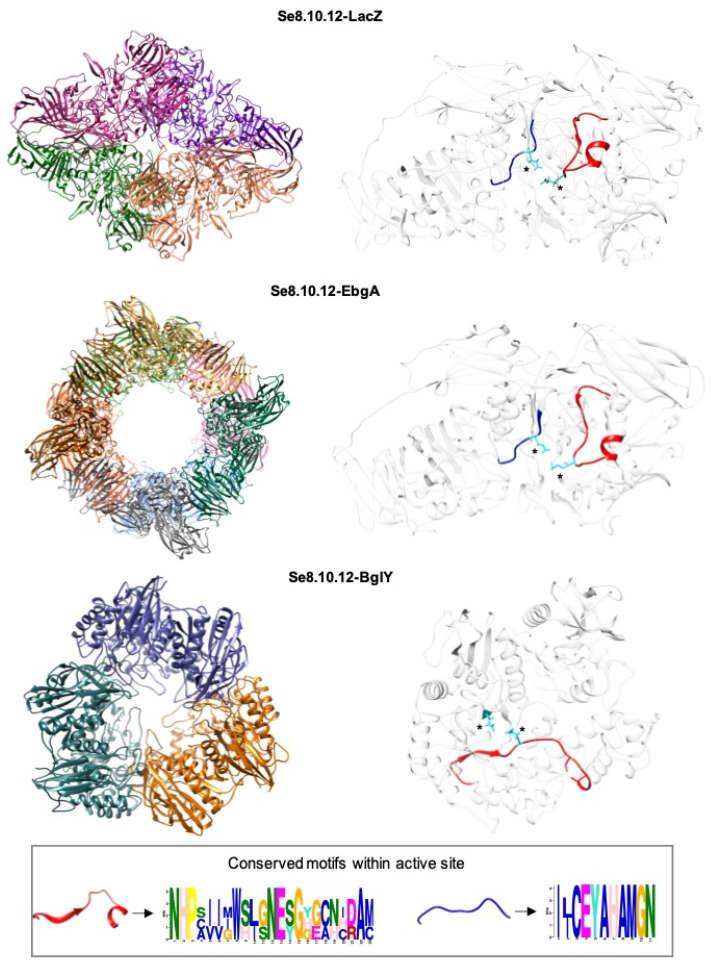
Overall predicted structure of the Rahnella inusitata Se8.10.12 β-galactosidases. The three-dimensional structure reconstructed by homology modeling is presented as a ribbon model. The reconstruction shows quaternary structure (left) and monomeric (right) views. In the monomers, the catalytic motifs are colored in red and blue, while asterisks indicate the glutamic acid amino acid in the active site.
The in silico prediction of the biochemical properties of R. inusitata Se8.10.12 β-galactosidases revealed that all sequences have similar characteristics to other β-gal proteins (Table 2), with complete sequences between 687 and 1032 amino acids, molecular masses of 77 to 116 kDa and pI between 5.2 and 5.8. The local alignment (BLASTp) against different β-gal from the PDB database showed that Se8.10.12 β-gal exhibited 30–40% of self-identity and 31–99% identity with other β-gal. BLASTp analysis of Se8.10.12 β-galactosidase proteins showed a variable degree of sequence similarity with different X-ray crystal structures from the PDB database. The crystal structure of Rahnella sp. R3 shares 99% sequence similarity with the Se8.10.12-BglY sequence. Likewise, high sequence similarity (68.9%) was found between LacZ from Escherichia coli and Se8.10.12-LacZ. In contrast, the sequence Se8.10.12-EbgA displayed the lowest identity percentage out of the three, only 37.24%, with a β-galactosidase structure from Thermotoga maritima (6S6Z_A). Nevertheless, all identity percentages met the recommended threshold for homology modeling using the PDB structures as template.
Table 2.
Biochemical properties of Rahnella inusitata Se.8.10.12 β-galactosidases and other β-gal enzymes from PDB database.
| Organism | Protein | Sequence | Molecular Mass (Da) | pI | Glycoside Hydrolase Family | PDB Number | % Identity | ||
|---|---|---|---|---|---|---|---|---|---|
| Lac-Z | BglY | EbgA | |||||||
| Rahnella inusitata Se.8.10.12 | Lac-Z | 1032 | 116,913.82 | 5.30 | GH 2 | - | 100 | 29.79 | 34.10 |
| Rahnella inusitata Se.8.10.12 | BglY | 687 | 77,230.21 | 5.84 | GH 42 | - | 29.79 | 100 | 40.91 |
| Rahnella inusitata Se.8.10.12 | EbgA | 1023 | 116,917.20 | 5.20 | GH 2 | - | 34.10 | 40.91 | 100 |
| Rahnella sp. R3 | β-gal | 712 | 80,188.46 | 5.98 | GH 42 | 5E9A_1 | 31.82 | 99.13 | 36.84 |
| Escherichia coli | Lac-Z | 1023 | 116,385.64 | 5.28 | GH 2 | 1jz2.1.D | 68.9 | 36.54 | 34.51 |
| Thermotoga maritima | β-gal | 1083 | 127,476.78 | 5.52 | GH 2 | 6S6Z_A | 36.39 | 29.27 | 37.24 |
Three-dimensional models of Se8.10.12 β-galactosidases (Figure 8) were reconstructed to compare the enzymes’ structures using homology modeling. The models got Qualitative Model Energy Analysis (QMEAN) Z-score quality values higher than −4.0 (Supplementary Table S3), indicating a good agreement between the model structure and experimental structures of similar size [50]. The SWISS-MODEL outputs were submitted to the YASARA energy minimization server to improve the quality and the accuracy of the predicted models. The refinement process vastly improved the raw β-galactosidases models’ accuracy, obtaining QMEAN Z-scores around zero, indicators of good quality. Ramachandran plots were also constructed for the three modeled enzymes using the PROCHECK server. Based on Ramachandran plot analysis, all models share a high overall structural quality, with more than 87–89% of residues in most favored regions, and only a few residues were in unfavorable conformations.
Three-dimensional models of the R. inusitata Se8.10.12 β-galactosidases showed noteworthy differences in their predicted quaternary structures. While BgIY was reconstructed as a homo-trimer, LacZ was modeled as a homo-tetramer and a homo-octamer for EbgA. For BgIY, the predicted homo-trimer structure could be associated with enzymatic activity at low temperatures, based on its similarity with the β-galactosidase from Rahnella sp. R3, which showed structural characteristics of cold adaptation [13]. Furthermore, a recently characterized β-galactosidase from the Antarctic archaea Halorubrum lacusprofundi, with a homo-trimeric quaternary structure, also showed cold adaptation and tolerance for high concentrations of NaCl [51]. However, no other associations have been made regarding a homo-trimeric conformation and cold tolerance. As these differences in conformation are likely to affect its molecular properties, it is important to consider that these assumptions are made by a prediction based on the highest template similarity with each homologous protein. However, other quaternary structures could occur in nature. Due to this, we included modeling for both the quaternary and monomeric structures for each enzyme.
For Se8.10.12 LacZ, the homo-tetrameric assembly shares structural properties with the quaternary structure of LacZ β-galactosidase from E. coli (comparable subunit mass, amino sequence identity, conservation of amino acid residues involved in catalysis, pI) [52]. The in silico analysis showed that both are tetramer of four identical polypeptide chains, each of around 1000 amino acids. Within each monomer, the amino acids form five well-defined structural domains. The central domain possesses the active site and is called (β/α)8-barrel [53]. Other β-galactosidases from psychrophile bacteria with homo-tetrameric structure have proven to be cold-adapted, including the cold-active β-galactosidases from the Antarctic bacteria Pseudoalteromonas haloplanktis [54], Pseudoalteromonas sp. 22b [55], and Alteromonas sp. ML117 [56]. Then, the predicted structure of Se8.10.12 LacZ could be associated with a cold-adapted enzyme. Unfortunately, the crystallized studies of those enzymes are not available for deeper structural comparisons.
Regarding the Se8.10.12 EbgA enzyme, the octameric predicted reconstruction of its three-dimensional structure is unusual. The octamer architecture has only been previously reported for one β-galactosidase from Thermotoga maritima, a hyperthermophilic organism isolated from a geothermal, marine volcano [57]. This enzyme has a characteristic TmLac domain, absent in other GH2 proteins, which determines its oligomeric arrangement. Moreover, despite the high conservation in the catalytic site, the enzyme has different features of the catalytic pocket and the regions surrounding it that might be important for interactions with large substrates and transglycosylation reactions [58].
Despite the marked differences in their quaternary structure’s prediction, which might or might not occur in nature regarding the physicochemical conditions, the monomers of the three Se8.10.12 β-galactosidases share characteristic conserved motifs within their structure. Figure 8 shows that the red motif is present in all three enzymes, while the blue motif is only present in LacZ and EbgA. These motifs are also shared by other β -galactosidases, such as LacZ from E. coli, and correspond, by the frequency of amino acids to highly conserved residues in the active site of each monomer. Two highly conserved glutamate residues for lactose hydrolysis were observed among both motifs, which are known to serve as proton donor and nucleophile, respectively, during catalysis [59].
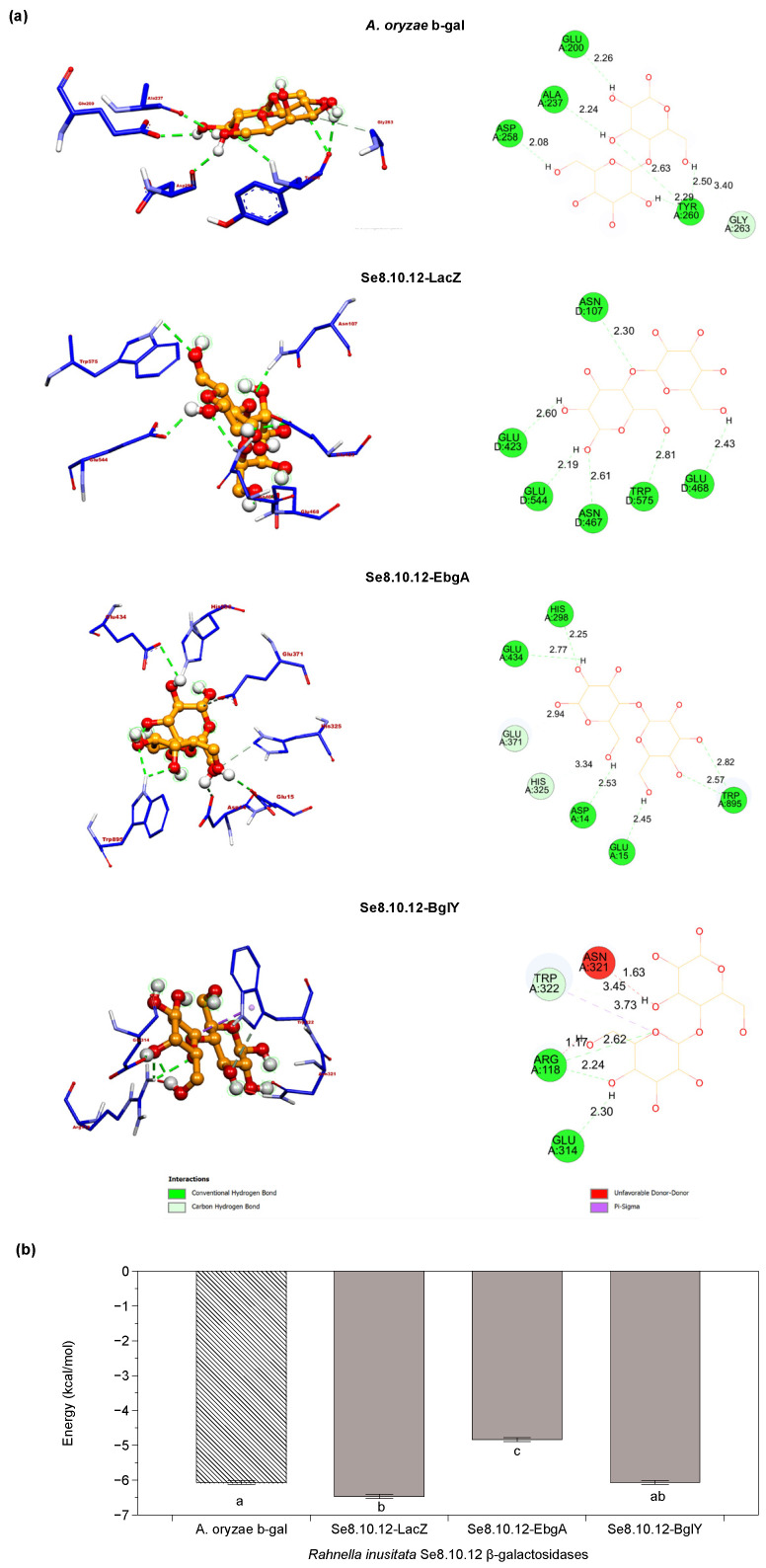
The refined Se8.10.12 β-galactosidases 3D models were used to predict the most likely interaction between the active site and lactose through molecular docking (Figure 9a). Molecular docking was performed to investigate whether the experimental results observed in Figure 2 would also be supported by in silico analysis. The docking score was the predicted binding affinity in kcal/mol, and the results are presented in Figure 9b. As a comparative parameter, the industrial–commercial Aspergillus oryzae β-galactosidase’s crystal structure was subjected to the same docking protocol. The calculated interaction energies between the lactose and the Se8.10.12 β-galactosidases models varied between −4.8 and −6.5 kcal mol−1, versus the −6.1 kcal mol−1 assigned for the interaction between A. oryzae β-galactosidase with lactose. The predicted binding affinity for lactose in Se8.10.12 LacZ was significantly higher than the binding affinity in A. oryzae for this molecule and comparably to the BglY enzyme. In contrast, the docking score in EbgA was significantly lower compared to the other enzymes. Thus, the in silico findings suggest that some β-galactosidases predicted from the Antarctic R. inusitata Se8.10.12 genome might have an effective lactose binding in the catalytic domain, comparable with the commercial control enzyme.
Figure 9.
Molecular docking of the Rahnella inusitata Se8.10.12 β-galactosidases and its substrate lactose. (a) Molecular docking simulation of lactose binding to Aspergillus oryzae β-galactosidase (4IUG) and Rahnella inusitata Se8.10.12 β-galactosidases LacZ, EbgA, and BglY (homology modeling). Schematic representation showing enzyme/ligand interactions (left image) and two-dimensional (2D) ligand-protein interaction diagram (right). The hydrogen bonds are represented dotted green lines (Å). (b) Docking scores (AutoDock Vina) for lactose against the selected β-galactosidases. Error bars denote SE of mean; n = 5. Different letters represent statistically significant differences between models (Kruskal–Wallis test; p < 0.05).
A. oryzae β-galactosidase, used as a control in our experimental and in silico analysis, belongs to the GH35 galactosidase family. While the β-galactosidases from R. inusitata are from galactosidase families 2 and 42. However, they all share the catalytic domain (β/α)8-barrel, with conserved motifs among them. It has even been proposed that all these families evolved from a common ancestor and shared a similar catalytic process. These types of enzymes share a common arrangement of two catalytically critical Glu residues at the end of the (β/α)8 barrel [60,61]. Hence, the binding affinities’ differences could be associated with details such as hydrophobic amino acids in the pocket, the number of hydrogen bonds, and the distance between them. Our data showed that the four enzymes’ active sites are highly hydrophilic (Figure S2), facilitating binding with the ligand since lactose is a hydrophilic molecule by itself [62]. However, slight tendencies towards neutrality or hydrophobicity can be seen in the active sites of A. oryzae and EbgA β-galactosidases, which matches with the lower binding affinity observed for those two enzymes in the molecular docking.
Docking results also showed that glutamate residues in both predicted active sites (E468/E544) of Se8.10.12 LacZ were in a conformation capable of forming hydrogen bonds with lactose; both close to the site of lactose cleavage (Figure 9a). The distance of these H-bonds was less than 3 Å, which agrees with the observed hydrogen bridge distances for LacZ from E. coli and β-gal from the Antarctic bacteria Alteromonas sp. and lactose [62]. As shown in Figure 9a, the residues ASN-107, ASN-467, GLU-423, and TRP-575 also form hydrogen bonds with lactose, respectively. In contrast, one glutamate residue in the active site of EbgA, BgIY, and A. oryzae (E-371, E-157, and E-298, respectively) does not appear capable of forming hydrogen bonds with lactose under the simulated tested conditions, which could explain why their affinity energies were lower than LacZ. Nevertheless, these models also form other hydrogen bonds with different amino acids in the active site, including tryptophan (TRP). TRP engages in-ring stacking with the glucosyl moiety of lactose and keeps the substrate inside the active pocket [63]. Besides, those H-bonds also were found at a suitable distance for catalysis.
Although hydrogen bonds may vary in an in vivo situation, the in silico analysis allowed the understanding of the potential catalytic efficiency, together with the binding affinity scores predicted for the enzymes [63]. Therefore, based on the docking score and hydrogen bond formation capability, the results suggest that Se8.10.12 LacZ has the highest affinity for lactose. In contrast, EbgA has the lowest apparent affinity, in agreement with previous data describing this as an inefficient enzyme. Accordingly, it could be suggested that the β-galactosidase activity observed in vitro could be given to a greater extent by LacZ and/or BgIY. Meanwhile, activity in cold might be expected from both LacZ and BglY based on the structural and sequence similarity to other cold-active enzymes; however, this must be confirmed for each different purified protein from the Antarctic R. inusitata.
3. Materials and Methods
3.1. Biological Material
The Antarctic bacteria included in this work (×304 strains) were previously isolated from soil and sediment samples from the Antarctic Peninsula and belong to the Chilean Type Culture Collection of the University of La Frontera (CCCT). The strains were reconstituted from the bacterial collection (−80 °C) both in nutritious broth and in nutritive agar and incubated at room temperature for 1 to 5 days until suitable growth.
3.2. Screening of β-Galactosidase Activity in Antarctic Bacteria
Determination of the β-galactosidase activity was performed at 15 °C in lactose agar (meat extract 0.3%, peptone 0.5%, lactose 0.5%, agar 1.8% and isopropyl-β-d-1-thiogalactopyranoside (IPTG) at 1 mM (Sigma-Aldrich, Gillingham, UK), using 5-Bromo-3-indolyl-β-d-galactopyranoside (β-Bluo-Gal) at 300 µg/mL (Thermo Fisher Scientific, Waltham, MA, USA) as an indicator of the β-galactosidase activity. One stripe of each strain (×304) was cultured in 150 × 15 mm plates in quadrants, allowing the simultaneous assay of 25 strains per plate. The positive β-galactosidase activity was determined after 48, 72, and 96 h of incubation when the color of the colonies changed to blue. A more intense color was considered indicative of a more bioactive strain.
3.3. Molecular Identification of β-Galactosidase Producing Antarctic Strains
DNA of positive β-galactosidase producers was extracted using the UltraClean® Microbial DNA Isolation Kit (MoBio Laboratories, Carlsbad, USA). The complete 16S rRNA gene was selectively amplified using the universal primers 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-GGTTACCTTGTTACGACTT-3′). PCR amplification was performed in a Multigene Optimal Thermal Cycler (Labnet, Edison, NJ, USA) in 50 μL reaction containing 25 μL SapphireAmp Fast PCR Master Mix, (Takara Bio Inc., Shiga, Japan), 22 μL ultra-pure water, 1 μL of each primer (10 μM), and 1 μL of genomic DNA. The temperature and cycling conditions were as follows: preheating at 94 °C for 2 min; 30 cycles at 94 °C for 1 min; 55 °C for 1 min; 72 °C for 1.5 min; and incubation at 72 °C for 10 min. Sequencing was performed with a Dye Terminator Cycle Sequencing Kit and an ABI 3730XL DNA Sequencer (Applied BiosystemsTM Thermo Fisher Scientific, Waltham, USA) by Macrogen Inc. (Korea). The nearest taxonomic group was identified by 16S rDNA nucleotide sequence BLASTN (http://www.ncbi.nlm.nih.gov/blast, accessed on 7 June 2020) using DNA Data Bank of Japan (DDBJ)/European Molecular Biology Laboratory (EMBL)/GenBank nucleotide sequence databases.
3.4. β-Galactosidase Activity Quantification
The most promising strains based on the β-galactosidase activity screening (showing the highest intensity of color change) were selected for enzyme activity quantification. The selected Antarctic strains (×4) were cultivated in lactose broth (meat extract 0.3%, peptone 0.5%, lactose 0.5%) at 15 °C for 48 h; then, cells were lysed by sonication. The cell lysate was centrifuged at 4000 rpm for 30 min at 4 °C, and the supernatant was used as the enzymatic extract [24]. 20 µL of the enzymatic extract was added to a 22 mM solution of o-nitrophenyl-β-d-galactopyranoside (ONPG) (Sigma-Aldrich, Gillingham, UK). The reaction was carried out for 15 min at 15 °C and pH 6.5. The amount of released o-nitrophenol (ONP) after ONPG hydrolysis was measured at 420 nm. One unit of β-galactosidase activity (U) was defined as the amount of enzyme required to release 1 µmol of ONP per minute from ONPG under the experimental conditions described above. Additionally, differences in β-galactosidase activity were evaluated in the presence and absence of IPTG (Sigma-Aldrich, Gillingham, UK). All determinations were performed in triplicates.
3.5. Morphological and Biochemical Characterization of Strain Se8.10.12
Strain Se8.10.12 was morphologically characterized by Gram staining and scanning electron microscopy in a SU-3500 Hitachi-Japan electron microscopy. Gram staining was performed based on the description in Gram (1884). For electronic microscopy characterization, the strain was incubated in nutrient broth for 24 h. Bacterial cells were harvested by centrifugation (5000 rpm, 5 min), followed by consecutive washings with sterile distilled water to eliminate traces of the culture broth media. After three washing-centrifugation steps, the supernatant was eliminated, and the strain was fixed with 500 µL of glutaraldehyde for 48 h. After fixation and before visualization by electronic microscopy, the sample was washed three times by centrifugation (10,000 rpm, 5 min) to eliminate the fixative excess. Once the supernatant was removed, the sample was resuspended in water and ethanol and visualized in the electronic microscope. Besides, to obtain a biochemical characterization of the strain, a battery of bacterial typing biochemical test tubes was used, composed of Triple Sugar Iron Agar (TSI), Motility Indole Ornithine (MIO), Simmons Citrate, Lysine Iron Agar (LIA), and Urea. The strain was inoculated into each media and incubated at room temperature for 24 h before evaluation.
3.6. Determination of the Effect of Temperature on the β-Galactosidase Relative Activity of Strain Se8.10.12
A raw enzymatic extract from Se8.10.12 strain was obtained as described in Section 3.4. The enzymatic reaction was performed with a 15 U/mL of a commercial enzyme for each mL of reaction (β-Galactosidase from Aspergillus oryzae CAS-9031-11-2; Sigma-Aldrich, Gillingham, UK), and the equivalent U/mL of the strain Se8.10.12 raw extract in 10 mL total reaction volume with commercial liquid whole milk (COLUN, Los Rios, Chile) at four different temperatures (4 °C, 15 °C, 25 °C, and 37 °C) for both enzymes. All samples were incubated for 12 h under constant agitation at 80 rpm. After 12 h, each sample was filtered with 0.22 μm filters and heated to 100 °C for 5 min to stop the enzymatic reaction. The samples were kept cold until the determination of the lactose content by HPLC (Shimadzu Prominence LC-20AT, Japan) using an Aminex HPX-87H ion exclusion column (300 × 7.8 mm, 9 µm particle size, 8% cross-linkage and pH range of 1–3; Bio-Rad, Hercules, CA, USA). Aliquots of 20 μL (injection volume) were separated at 0.5 mL min−1 (flow rate), at a constant 60 °C column temperature, with 0.01 N aqueous sulfuric acid elution phase. Detection of lactose was accomplished in a refractive index detector compared to a 40 mg/mL lactose commercial standard (Sigma-Aldrich, Saint Louis, MO, USA). Since a non-purified extract was used (targeting psychrophilic enzymes), the enzymatic activity was expressed as relative activity based on the maximum lactose degradation. All measurements were conducted with four replicates.
3.7. Whole-Genome Sequencing and Assembly
Strain Se8.10.12 was sequenced using the Illumina MiSeq (Illumina, San Diego, CA, USA) and Oxford Nanopore MinION (Oxford Nanopore Technologies, Oxford, UK) platforms. Briefly, 1 mL of bacterial culture (24 h growth in nutrient broth) was used for genomic DNA extraction using the UltraClean Microbial DNA Extraction Kit (MoBio Laboratories, Carlsbad, CA, USA). DNA quality was assessed by fluorometry using a QuantiFluor® ONE dsDNA System (Promega Corporation, Fitchburg, WI, USA) and the determination of purity determining ratios 260/280 and 260/230. A library was prepared with the rapid sequencing kits SQK-RAD004 (Oxford Nanopore Technologies, Littlemore, Oxford, UK), and sequencing was performed in a MinION platform (Oxford Nanopore Technologies (ONT)) using the software MinKNOW v2.3.5. DNA was also sequenced on an Illumina MiSeq X Ten sequencing platform using a paired-end library with an average insert size of 350-bp followed by 2 × 150-bp sequencing. The Illumina paired-end sequencing data were evaluated using FastQC v0.11.9 and then adjusted using the Trimmomatic v0.39 program by filtering out adapters and reads of less than a Phred score of 30. In parallel, the Oxford Nanopore data were evaluated using the NanoPlot v1.32.1 software and then adjusted using the NanoFilt v2.8.0 and Porechop v0.2.4 programs, filtering out reads of less than Q10, a read length fewer than 1000 bases, and removing the adapters, respectively. Finally, the hybrid assembly was performed using the Unicycler v0.4.8 program; additional finalization of the assembly was done using the tool FinisherSC v2.0 [64] and quickmerge v0.3 [65] to close the genome before and after Pilon v1.23 polishing [66]. The quality of the assembly was evaluated using the Quast v5.0.2 program [67], while completeness/contamination was determined with CheckM v1.1.3 [68] and miComplete v1.1.0 [39]. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the bioproject accessions PRJNA605861, BioSample SAMN16814261, which includes the genome CP065024 and plasmids CP065025 and CP065026
3.8. Genome-Wide Analysis for Taxonomic Identification and Characterization
Taxonomic affiliation of the strains Se8.10.12 was performed by 16S rRNA gene comparison using BLASTn [69]. Phylogenetic distances among the nearest taxonomic groups were also determined by a tree construction using the rRNA 16S complete gene sequence and core proteins of reference Rahnella sp. genomes. Core proteins were obtained with M1CR0B1AL1Z3R web server (https://microbializer.tau.ac.il/, accessed on 20 October 2020) [70] using the following cut-offs: 0.01 maximal e-value, 80.0% identity minimal percent, and 100.0% minimal percentage for the core. rRNA 16S gene phylogenetic tree construction was performed using Mafft alignment version 737 L-INS-i method [71] and SeaView version 5.0.4. with the Bio-NJ algorithm based on the Kimura-2-parameter model selection on 1000 bootstrapped data sets [72]. Core-proteome phylogenetic construction was conducted with a maximum-likelihood algorithm using RAxML [73] based on the inferred core-proteome alignment. Phylogenetic tree visualization was done using the iTOL v3 tool [74]. A genome-wide analysis was performed to confirm the strain identity by Pairwise Average Nucleotide Identity (ANI) values. Those were calculated among the available reference genomes representative of the genus Rahnella using the software Pyani v0.2.10 in mode ANIb [75]. The output was visualized using the software DiMHepy (available at https://github.com/lucaTriboli/DiMHepy, accessed on 12 August 2020).
Genomic annotation was performed using Prokka v1.14.5 [76] to determine the Antarctic strain’s genomic profile and potential genes associated with the production of β-galactosidases. The protein sequences from Prokka generated data were analyzed with the OrthoVenn2 web server (https://orthovenn2.bioinfotoolkits.net, accessed on 15 December 2020) for identification and comparison of orthologous clusters [77]. Briefly, OrthoVenn2 uses the OrthoMCL [78] clustering algorithm to annotate and compare ortholog groups. All-against-all protein similarity was obtained by DIAMOND v0.9.24 alignment [79] (0.05 e-value cut-off) from which putative orthology is identified. Disjoint clusters of closely related proteins are generated with the Markov Clustering Algorithm (MCL) [80] with a 1.5 inflation value. The Gene Ontology (GO) was assigned to the corresponding orthologous cluster by sequence similarity to the UniProt database [81].
3.9. Characterization of β-Galactosidase Enzymes of the Strain Se8.10.12 Based on Genomic Data
For the analyses focused on the protein sequences attributed to β-galactosidase genes, multiple alignments were carried out with MAFFT v7 [71] between the sequences obtained from strain Se8.10.12 and sequences deposited in NCBI with sequence similarity above 85% after BLASTp comparison. A phylogram was also constructed to determine the genetic distance between the aminoacidic sequences. The alignment file was used to build a Maximum Likelihood (ML) phylogenetic tree as described above by estimating the most accurate substitution model by Model Finder [82] with the Akaike Information Criterion in the IQ-TREE web server v1.4.2 [83]. The tree visualization was done with the iTOL v3 tool [74]. Gene presence/absence and association with syntenic clusters were studied using the pangenome toolbox PIRATE v1.0.4 (default settings used; 60, 70, 80, 90, 95% amino acid identity threshold) [84].
Moreover, in silico structural analyses of Se.8.10.12 β-galactosidase proteins were performed. Three-dimensional models of the β-galactosidases were reconstructed using a homology modeling approach. NCBI BLASTp against the PDB database was performed to identify β-galactosidase structure templates with the highest sequence similarity to the β-galactosidase sequences of Se.8.10.12. Only PDB structures with sufficient sequence similarity (>30%) were selected as modeling templates. A monomer and quaternary structure of each β-galactosidase, excluding any signal peptide sequence, was constructed using the SWISS-MODEL Workspace platform [85]. The crystal structures of Escherichia coli lacZ (PDB: 1jz2.1.D, 2.1 Å), Rahnella sp. R3 β-galactosidase (PDB: 5e9a.1.A, 2.6 Å), and Thermotoga maritima β-galactosidase (PDB: 6S6Z, 2.0 Å) were used as templates for Se8.10.12 LacZ, BglY, and EbgA model reconstruction, respectively. Based on the highest scores (Global Model Quality Estimation (GMQE) and the Qualitative Model Energy Analysis (QMEAN)) among all possible structural templates, the best models were submitted to refinement in the YASARA (Yet Another Scientific Artificial Reality Application) Energy Minimization Server [86]. The refinement outputs were reanalyzed by QMEAN [50] and SAVES v6.0 [87] servers. All three-dimensional models were visualized using UCSF Chimera software v1.14 [88].
Multiple sequence alignment using MultAlin online platform [89] was used to identify active sites in the β-galactosidases three-dimensional (3D) models. Binding conformations between lactose and the β-galactosidases from R. inusitata were performed using the Docking Algorithm of AutoDock Vina v1.1.2 [90]. The ligand structure was downloaded from the PubChem database (Compound ID (CID): 6134), and the energy minimizations were carried out with the PRODRG2 Server [91]. Gasteiger charges and polar hydrogen atoms were assigned in the protein structures and the ligand using AutoDockTools v4.2.6 [92]. The rotatable bonds were set up, and structure data files were converted into the Protein Data Bank, Partial Charge (Q), and Atom Type (T) format (PDBQT). Flexible molecular docking was applied to the center of the β-galactosidases catalytic site. An exhaustiveness value = 80 and 500 number modes were used. Conformations were docked, clustered, and ranked according to the binding free energy. Docking was conducted using β-galactosidase monomers since only residues from individual monomers interact directly with a substrate. Since the in vitro analysis was performed compared with an A. oryzae enzyme, a β-galactosidase from A. oryzae (PDB 4IUG) was included in the analysis as a reference from commercial lactase enzyme [93]. Discovery Studio Visualizer v4.5 software [94] was used to analyze, summarize, and cluster the AutoDock Vina results.
3.10. Statistic Data Analysis
Mean comparison was made with one-way analysis of variance (ANOVA) and Independent samples T-Test at a 95% level of significance (p < 0.05) using JASP software v0.14.1 [95]. The assumption of data normality and homoscedasticity were tested with the Shapiro–Wilk and Levene’s test. If data failed the parametric inference, Mann–Whitney U and Kruskal–Wallis H test was used instead. Stander Tukey or Dunn Post Hoc Test was carried out in those cases where ANOVA or Kruskal–Wallis H test was significant.
4. Conclusions
New β-galactosidase alternatives with activity in cold and wide-range thermostability are currently needed in the glycoside hydrolases industry, particularly for the preparation of lactose-free dairy products. Our work highlights Antarctic microorganisms’ bioprospection for extremozymes discovery, especially cold-active enzymes, since this is still an underexplored source of new molecules. Moreover, further efforts for commercial production of a new diversity of β-galactosidases may promote its regular use in other areas such as dairy whey to produce additional valuable bioproducts (i.e., ethanol and sweet syrup) [23].
Here we report for the first time the potential of an Antarctic Rahnella inusitata strain as a cold-active β-galactosidase producer. This strain produced an enzymatic extract with higher efficiency in cold than commercial enzyme from Aspergillus oryzae, regarding its remaining activity in cold compared with the activity at optimal temperature. The Antarctic strain harbors three β-galactosidase genes and its description will make possible future recombinant production of these potential cold-active enzymes to confirm its activity as a purified product and for commercial industrialization in GRAS platforms (e.g., Bifidobacterium bifidum and Bacillus subtilis). Despite the sequence and structure similarity of R. inusitata β-galactosidases with known enzymes, it is expected that adaptation to the Antarctic conditions shaped evolutive differences directly affecting their activity in cold. However, further studies are needed to understand each specific gene’s structure and functioning and possible evolutive variants at the catalytic domains related to thermostability and activity in cold, which is not entirely understood yet.
Acknowledgments
We acknowledge Carolina Shene deVidts and Allison Leyton from the Center for Food Biotechnology and Bioseparations for performing the Chromatographic Analyses, as well as Karina Godoy for her help in the microscopic analyses, both from the Scientific and Technological Bioresource Nucleus (BIOREN) of Universidad de La Frontera, Temuco, Chile.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22084144/s1, Table S1: Antarctic strains with positive detection of β-galactosidase activity based on β-Bluo-Gal reduction detection, Table S2: Alignment lengths and percentage of identity between nine Rahnella species genomes including the Antarctic isolate Se8.10.12. by Average Nucleotide Identity (ANI) calculation using BLASTn method, Table S3: Quality evaluation of the three-dimensional models of Rahnella inusitata Se8.10.12 β-galactosidases, Figure S1: Example of β-galactosidase producing Antarctic strains determined in Lactose Agar using β-Bluo-Gal as an indicator of the β-galactosidase activity, Figure S2: The hydrophobicity of the active site in docking analysis of β-galactosidases.
Author Contributions
Conceptualization, R.S., A.S., S.F., F.S. and L.B.D.; methodology, O.G.-E., K.N.-M., R.S., C.T., S.F., D.M., C.H. and A.S.; validation, K.N.-M., R.S., A.S.; formal analysis, K.N.-M., R.S., A.S. and O.G.-E.; investigation, K.N.-M., R.S., A.S., F.S. and L.B.D.; resources, L.B.D., K.N.-M., F.S.; data curation, O.G.-E. and A.S.; writing—original draft preparation, R.S. and K.N.-M.; writing—review and editing, K.N.-M., R.S., A.S., C.T., D.M., C.H., F.S. and L.B.D.; visualization, O.G.-E., K.N.-M., R.S. and A.S.; supervision, L.B.D.; project administration, L.B.D. and F.S; funding acquisition, L.B.D. and F.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the INSTITUTO ANTÁRTICO CHILENO (INACH), grant INACH RT_14-12, and INACH DG_01-19; UFRO/UANDES, grant INI4 from; UNIVERSIDAD DE LA FRONTERA, grant DI19-0079 and grant DI20-2018, NETWORK FOR EXTREME ENVIRONMENTS RESEARCH (NEXER), grant NXR17-0003; and CONICYT, grants CONICYT‒PFCHA/Doctorado Nacional/2017-21170263 for K.N.-M., CONICYT‒PFCHA/Doctorado Nacional/2017-21171392 to A.S., and CONICYT-PFCHA/Doctorado Nacional/2017-21170265 to O.G.-E. F. Scott gratefully acknowledges financial support by the National Agency for Research and Development (ANID Chile) (Fondecyt Iniciación 11170081).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found at https://www.ncbi.nlm.nih.gov under the accession number provided for each genome or nucleotide sequence.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vásquez-Ponce F., Higuera-Llantén S., Pavlov M.S., Marshall S.H., Olivares-Pacheco J. Phylogenetic MLSA and phenotypic analysis identification of three probable novel Pseudomonas species isolated on King George Island, South Shetland, Antarctica. Braz. J. Microbiol. 2018;49:695–702. doi: 10.1016/j.bjm.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cid F.P., Maruyama F., Murase K., Graether S.P., Larama G., Bravo L.A., Jorquera M.A. Draft genome sequences of bacteria isolated from the Deschampsia antarctica phyllosphere. Extremophiles. 2018;22:537–552. doi: 10.1007/s00792-018-1015-x. [DOI] [PubMed] [Google Scholar]
- 3.Núñez-Montero K., Barrientos L. Advances in Antarctic Research for Antimicrobial Discovery: A Comprehensive Narrative Review of Bacteria from Antarctic Environments as Potential Sources of Novel Antibiotic Compounds Against Human Pathogens and Microorganisms of Industrial Importance. Antibiotics. 2018;7:90. doi: 10.3390/antibiotics7040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrer M., Golyshina O., Beloqui A., Golyshin P.N. Mining enzymes from extreme environments. Curr. Opin. Microbiol. 2007;10:207–214. doi: 10.1016/j.mib.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Nichols D., Bowman J., Sanderson K., Nichols C.M., Lewis T., McMeekin T., Nichols P.D. Developments with Antarctic microorganisms: Culture collections, bioactivity screening, taxonomy, PUFA production and cold-adapted enzymes. Curr. Opin. Biotechnol. 1999;10:240–246. doi: 10.1016/S0958-1669(99)80042-1. [DOI] [PubMed] [Google Scholar]
- 6.Antony R., Sanyal A., Kapse N., Dhakephalkar P.K., Thamban M., Nair S. Microbial communities associated with Antarctic snow pack and their biogeochemical implications. Microbiol. Res. 2016;192:192–202. doi: 10.1016/j.micres.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Karasová-Lipovová P., Strnad H., Spiwok V., Malá Š., Králová B., Russell N.J. The cloning, purification and characterisation of a cold-active β-galactosidase from the psychrotolerant Antarctic bacterium Arthrobacter sp. C2-2. Enzyme Microb. Technol. 2003;6:836–844. doi: 10.1016/S0141-0229(03)00211-4. [DOI] [Google Scholar]
- 8.Itan Y., Jones B.L., Ingram C.J., Swallow D.M., Thomas M.G. A worldwide correlation of lactase persistence phenotype and genotypes. BMC Evol. Biol. 2010;10:36. doi: 10.1186/1471-2148-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vesa T.H., Korpela R., Marteau P. Lactose Intolerance. J. Am. Coll. Nutr. 2000;9:165–175. doi: 10.1080/07315724.2000.10718086. [DOI] [PubMed] [Google Scholar]
- 10.Hildebrandt P., Wanarska M., Kur J. A new cold-adapted β-D-galactosidase from the Antarctic Arthrobacter sp. 32c—Gene cloning, overexpression, purification and properties. BMC Microbiol. 2009;9:151. doi: 10.1186/1471-2180-9-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan Y., Yi J., Hua X., Zhang Y., Yang R. Preparation and characterization of gellan gum microspheres containing a cold-adapted β-galactosidase from Rahnella sp. R3. Carbohydr. Polym. 2017;162:10–15. doi: 10.1016/j.carbpol.2017.01.033. [DOI] [PubMed] [Google Scholar]
- 12.Margesin R., Schinner F., editors. Cold-Adapted Organisms: Ecology, Physiology, Enzymology. Springer; Berlin/Heidelberg, Germany: 1999. [Google Scholar]
- 13.Fan Y., Hua X., Zhang Y., Feng Y., Shen Q., Dong J., Zhao W., Zhang W., Jin Z., Yang R. Cloning, expression and structural stability of a cold-adapted β-galactosidase from Rahnella sp. R3. Protein Expr. Purif. 2015;115:158–164. doi: 10.1016/j.pep.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Nakagawa T., Fujimoto Y., Uchino M., Miyaji T., Takano K., Tomizuka N. Isolation and characterization of psychrophiles producing cold-active -galactosidase. Lett. Appl. Microbiol. 2003;37:154–157. doi: 10.1046/j.1472-765X.2003.01369.x. [DOI] [PubMed] [Google Scholar]
- 15.Fernandes S., Geueke B., Delgado O., Coleman J., Hatti-Kaul R. β-Galactosidase from a cold-adapted bacterium: Purification, characterization and application for lactose hydrolysis. Appl. Microbiol. Biotechnol. 2002;58:313–321. doi: 10.1007/s00253-001-0905-4. [DOI] [PubMed] [Google Scholar]
- 16.Turkiewicz M., Kur J., Białkowska A., Cieśliński H., Kalinowska H., Bielecki S. Antarctic marine bacterium Pseudoalteromonas sp. 22b as a source of cold-adapted β-galactosidase. Biomol. Eng. 2003;20:317–324. doi: 10.1016/S1389-0344(03)00039-X. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L., Wang K., Mo Z., Liu Y., Hu X. Crystallization and preliminary X-ray analysis of a cold-active β-galactosidase from the psychrotrophic and halotolerant Planococcus sp. L4. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011;67:911–913. doi: 10.1107/S1744309111022627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wierzbicka-Woś A., Cieśliński H., Wanarska M., Kozłowska-Tylingo K., Hildebrandt P., Kur J. A novel cold-active β-D-galactosidase from the Paracoccus sp. 32d—gene cloning, purification and characterization. Microb. Cell Fact. 2011;10:1–12. doi: 10.1186/1475-2859-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alikunju A.P., Joy S., Salam J.A., Silvester R., Antony A.C., Rahiman K.M.M., Krishnan K.P., Hatha A.A.M. Functional Characterization of a New Cold-Adapted β-Galactosidase from an Arctic Fjord Sediment Bacteria Enterobacter ludwigii MCC 3423. Catal. Lett. 2018;148:3223–3235. doi: 10.1007/s10562-018-2504-3. [DOI] [Google Scholar]
- 20.Wang G.X., Gao Y., Hu B., Lu X.L., Liu X.Y., Jiao B. hua A novel cold-adapted β-galactosidase isolated from Halomonas sp. S62: Gene cloning, purification and enzymatic characterization. World J. Microbiol. Biotechnol. 2013;29:1473–1480. doi: 10.1007/s11274-013-1311-7. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt M., Stougaard P. Identification, cloning and expression of a cold-active β-galactosidase from a novel Arctic bacterium, Alkalilactibacillus ikkense. Environ. Technol. 2010;31:1107–1114. doi: 10.1080/09593331003677872. [DOI] [PubMed] [Google Scholar]
- 22.Mangiagalli M., Lapi M., Maione S., Orlando M., Brocca S., Pesce A., Barbiroli A., Camilloni C., Pucciarelli S., Lotti M., et al. The co-existence of cold activity and thermal stability in an Antarctic GH42 β-galactosidase relies on its hexameric quaternary arrangement. FEBS J. 2020;288:546–565. doi: 10.1111/febs.15354. [DOI] [PubMed] [Google Scholar]
- 23.Saqib S., Akram A., Halim S.A., Tassaduq R. Sources of β-galactosidase and its applications in food industry. 3 Biotech. 2017;7:79. doi: 10.1007/s13205-017-0645-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groudieva T., Kambourova M., Yusef H., Royter M., Grote R., Trinks H., Antranikian G. Diversity and cold-active hydrolytic enzymes of culturable bacteria associated with Arctic sea ice, Spitzbergen. Extremophiles. 2004;8:475–488. doi: 10.1007/s00792-004-0409-0. [DOI] [PubMed] [Google Scholar]
- 25.Vincent V., Aghajari N., Pollet N., Boisson A., Boudebbouze S., Haser R., Maguin E., Rhimi M. The acid tolerant and cold-active β-galactosidase from Lactococcus lactis strain is an attractive biocatalyst for lactose hydrolysis. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2013;103:701–712. doi: 10.1007/s10482-012-9852-6. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen T.-H., Splechtna B., Krasteva S., Kneifel W., Kulbe K.D., Divne C., Haltrich D. Characterization and molecular cloning of a heterodimeric Î2-galactosidase from the probiotic strain Lactobacillus acidophilus R22. FEMS Microbiol. Lett. 2007;269:136–144. doi: 10.1111/j.1574-6968.2006.00614.x. [DOI] [PubMed] [Google Scholar]
- 27.Rhimi M., Boisson A., Dejob M., Boudebouze S., Maguin E., Haser R., Aghajari N. Efficient bioconversion of lactose in milk and whey: Immobilization and biochemical characterization of a β-galactosidase from the dairy Streptococcus thermophilus LMD9 strain. Res. Microbiol. 2010;161:515–525. doi: 10.1016/j.resmic.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 28.Rhimi M., Aghajari N., Jaouadi B., Juy M., Boudebbouze S., Maguin E., Haser R., Bejar S. Exploring the acidotolerance of β-galactosidase from Lactobacillus delbrueckii subsp. bulgaricus: An attractive enzyme for lactose bioconversion. Res. Microbiol. 2009;160:775–784. doi: 10.1016/j.resmic.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 29.Peeters K., Verleyen E., Hodgson D.A., Convey P., Ertz D., Vyverman W., Willems A. Heterotrophic bacterial diversity in aquatic microbial mat communities from Antarctica. Polar Biol. 2012;35:543–554. doi: 10.1007/s00300-011-1100-4. [DOI] [Google Scholar]
- 30.Tashyrev O., Romanovskaya V., Rokitko P., Tashyreva H., Prytula I., Suslova O., Govorukha V., Prekrasna I., Gladka G. Autecology and Taxonomy of Bacteria Isolated from Extreme Environments. Mikrobiol. Zh. 2017;79:100–113. doi: 10.15407/microbiolj79.01.100. [DOI] [Google Scholar]
- 31.Park J.W., Oh Y.S., Lim J.Y., Roh D.H. Isolation and characterization of cold-adapted strains producing β-galactosidase. J. Microbiol. 2006;44:396–402. [PubMed] [Google Scholar]
- 32.Liu W.Y., Shi Y.W., Wang X.Q., Wang Y., Wei C.Q., Lou K. Isolation and identification of a strain producing cold-adapted β-galactosidase, and purification and characterisation of the enzyme. Czech J. Food Sci. 2008;26:284–290. doi: 10.17221/31/2008-CJFS. [DOI] [Google Scholar]
- 33.Mahdian S.M.A., Karimi E., Tanipour M.H., Parizadeh S.M.R., Ghayour-Mobarhan M., Bazaz M.M., Mashkani B. Expression of a functional cold active β-galactosidase from Planococcus sp-L4 in Pichia pastoris. Protein Expr. Purif. 2016;125:19–25. doi: 10.1016/j.pep.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Coker J.A., Sheridan P.P., Loveland-Curtze J., Gutshall K.R., Auman A.J., Brenchley J.E. Biochemical characterization of a β-galactosidase with a low temperature optimum obtained from an Antarctic Arthrobacter isolate. J. Bacteriol. 2003;185:5473–5482. doi: 10.1128/JB.185.18.5473-5482.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheridan P.P., Brenchley J.E. Characterization of a salt-tolerant family 42 β-galactosidase from a psychrophilic antarctic Planococcus isolate. Appl. Environ. Microbiol. 2000;66:2438–2444. doi: 10.1128/AEM.66.6.2438-2444.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adeolu M., Alnajar S., Naushad S., Gupta R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016;66:5575–5599. doi: 10.1099/ijsem.0.001485. [DOI] [PubMed] [Google Scholar]
- 37.Brady C., Hunter G., Kirk S., Arnold D., Denman S. Rahnella victoriana sp. nov., Rahnella bruchi sp. nov., Rahnella woolbedingensis sp. nov., classification of Rahnella genomospecies 2 and 3 as Rahnella variigena sp. nov. and Rahnella inusitata sp. nov., respectively and emended description of the genus Rahnella. Syst. Appl. Microbiol. 2014;37:545–552. doi: 10.1016/j.syapm.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 38.Izard D., Gavini F., Trinel P.A., Leclerc H. “Rahnella aquatilis”, a new member of the “Enterobacteriaceae”. Ann. Microbiol. (Paris) 1979;130:163–177. [PubMed] [Google Scholar]
- 39.Hugoson E., Lam W.T., Guy L. miComplete: Weighted quality evaluation of assembled microbial genomes. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos I.R., Silva-Filho E.V., Schaefer C.E.G.R., Albuquerque-Filho M.R., Campos L.S. Heavy metal contamination in coastal sediments and soils near the Brazilian Antarctic Station, King George Island. Mar. Pollut. Bull. 2005;50:185–194. doi: 10.1016/j.marpolbul.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 41.Chu W.L., Dang N.L., Kok Y.Y., Ivan Yap K.S., Phang S.M., Convey P. Heavy metal pollution in Antarctica and its potential impacts on algae. Polar Sci. 2019;20:75–83. doi: 10.1016/j.polar.2018.10.004. [DOI] [Google Scholar]
- 42.Marina-Montes C., Pérez-Arribas L.V., Escudero M., Anzano J., Cáceres J.O. Heavy metal transport and evolution of atmospheric aerosols in the Antarctic region. Sci. Total Environ. 2020;721:137702. doi: 10.1016/j.scitotenv.2020.137702. [DOI] [PubMed] [Google Scholar]
- 43.Romaniuk K., Ciok A., Decewicz P., Uhrynowski W., Budzik K., Nieckarz M., Pawlowska J., Zdanowski M.K., Bartosik D., Dziewit L. Insight into heavy metal resistome of soil psychrotolerant bacteria originating from King George Island (Antarctica) Polar Biol. 2018;41:1319–1333. doi: 10.1007/s00300-018-2287-4. [DOI] [Google Scholar]
- 44.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan T., Yang P., Wang Y., Meng K., Luo H., Zhang W., Wu N., Fan Y., Yao B. Heterologous expression of a gene encoding a thermostable β-galactosidase from Alicyclobacillus acidocaldarius. Biotechnol. Lett. 2008;30:343–348. doi: 10.1007/s10529-007-9551-y. [DOI] [PubMed] [Google Scholar]
- 46.Coker J.A., Brenchley J.E. Protein engineering of a cold-active β-galactosidase from Arthrobacter sp. SB to increase lactose hydrolysis reveals new sites affecting low temperature activity. Extremophiles. 2006;10:515–524. doi: 10.1007/s00792-006-0526-z. [DOI] [PubMed] [Google Scholar]
- 47.Molina F., Simancas A., Tabla R., Roa I., Rebollodo E. Microbes in the Spotlight: Recent Progress in the Understanding of Beneficial and Harmful Microorganisms. BrownWalker Press; Boca Raton, FL, USA: 2016. Horizontal gene transfer and the evolving definition of coliforms; p. 496. [Google Scholar]
- 48.Stokes H.W., Betts P.W., Hall B.G. Sequence of the ebgA gene of Escherichia coli: Comparison with the lacZ gene. Mol. Biol. Evol. 1985;2:469–477. doi: 10.1093/oxfordjournals.molbev.a040372. [DOI] [PubMed] [Google Scholar]
- 49.Hall B.G. Evolutionary potential of the ebgA gene. Mol. Biol. Evol. 1995;12:514–517. doi: 10.1093/oxfordjournals.molbev.a040225. [DOI] [PubMed] [Google Scholar]
- 50.Benkert P., Biasini M., Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karan R., Mathew S., Muhammad R., Bautista D.B., Vogler M., Eppinger J., Oliva R., Cavallo L., Arold S.T., Rueping M. Understanding high-salt and cold adaptation of a polyextremophilic enzyme. Microorganisms. 2020;8:1594. doi: 10.3390/microorganisms8101594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Juers D.H., Heightman T.D., Vasella A., McCarter J.D., Mackenzie L., Withers S.G., Matthews B.W. A structural view of the action of Escherichia coli (lacZ) β-galactosidase. Biochemistry. 2001;40:14781–14794. doi: 10.1021/bi011727i. [DOI] [PubMed] [Google Scholar]
- 53.Juers D.H., Matthews B.W., Huber R.E. LacZ β-galactosidase: Structure and function of an enzyme of historical and molecular biological importance. Protein Sci. 2012;21:1792–1807. doi: 10.1002/pro.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoyoux A., Jennes I., Dubois P., Genicot S., Dubail F., François J.M., Baise E., Feller G., Gerday C. Cold-Adapted β-Galactosidase from the Antarctic Psychrophile Pseudoalteromonas haloplanktis. Appl. Environ. Microbiol. 2001;67:1529–1535. doi: 10.1128/AEM.67.4.1529-1535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cieśliński H., Kur J., Białkowska A., Baran I., Makowski K., Turkiewicz M. Cloning, expression, and purification of a recombinant cold-adapted β-galactosidase from antarctic bacterium Pseudoalteromonas sp. 22b. Protein Expr. Purif. 2005;39:27–34. doi: 10.1016/j.pep.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 56.Yao C., Sun J., Wang W., Zhuang Z., Liu J., Hao J. A novel cold-adapted β-galactosidase from Alteromonas sp. ML117 cleaves milk lactose effectively at low temperature. Process Biochem. 2019;82:94–101. doi: 10.1016/j.procbio.2019.04.016. [DOI] [Google Scholar]
- 57.Douillard F.P., Kant R., Ritari J., Paulin L., Palva A., De Vos W.M. Comparative genome analysis of Lactobacillus casei strains isolated from Actimel and Yakult products reveals marked similarities and points to a common origin. Microb. Biotechnol. 2013;6:576–587. doi: 10.1111/1751-7915.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Míguez Amil S., Jiménez-Ortega E., Ramírez-Escudero M., Talens-Perales D., Marín-Navarro J., Polaina J., Sanz-Aparicio J., Fernandez-Leiro R. The cryo-EM Structure of Thermotoga maritima β-Galactosidase: Quaternary Structure Guides Protein Engineering. ACS Chem. Biol. 2020;15:179–188. doi: 10.1021/acschembio.9b00752. [DOI] [PubMed] [Google Scholar]
- 59.Vukić V., Hrnjez D., Milanović S., Iličić M., Kanurić K., Petri E. Comparative Molecular Modeling and Docking Analysis of β-galactosidase Enzymes from Commercially Important Starter Cultures Used in the Dairy Industry. Food Biotechnol. 2015;28:248–262. doi: 10.1080/08905436.2015.1059766. [DOI] [Google Scholar]
- 60.Naumoff D.G. Hierarchical classification of glycoside hydrolases. Biochem. 2011;76:622–635. doi: 10.1134/S0006297911060022. [DOI] [PubMed] [Google Scholar]
- 61.Davies G., Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 62.Li S., Zhu X., Xing M. A new β-galactosidase from the antarctic bacterium Alteromonas sp. ANT48 and its potential in formation of prebiotic galacto-oligosaccharides. Mar. Drugs. 2019;17:599. doi: 10.3390/md17110599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu J., Sun J., Tang Y.J., Xie J., Wei D. Expression, characterization and structural profile of a heterodimeric β-galactosidase from the novel strain Lactobacillus curieae M2011381. Process Biochem. 2020;97:87–95. doi: 10.1016/j.procbio.2020.06.025. [DOI] [Google Scholar]
- 64.Lam K.K., Labutti K., Khalak A., Tse D. FinisherSC: A repeat-aware tool for upgrading de novo assembly using long reads. Bioinformatics. 2015;31:3207–3209. doi: 10.1093/bioinformatics/btv280. [DOI] [PubMed] [Google Scholar]
- 65.Chakraborty M., Baldwin-Brown J.G., Long A.D., Emerson J.J. Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nucleic Acids Res. 2016;44:e147. doi: 10.1093/nar/gkw654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J., Young S.K., et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 70.Avram O., Rapoport D., Portugez S., Pupko T. M1CR0B1AL1Z3R—A user-friendly web server for the analysis of large-scale microbial genomics data. Nucleic Acids Res. 2019;47:W88–W92. doi: 10.1093/nar/gkz423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017 doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gouy M., Guindon S., Gascuel O. SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 73.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Letunic I., Bork P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pritchard L., Glover R.H., Humphris S., Elphinstone J.G., Toth I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods. 2016;8:12–24. doi: 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 76.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 77.Xu L., Dong Z., Fang L., Luo Y., Wei Z., Guo H., Zhang G., Gu Y.Q., Coleman-Derr D., Xia Q., et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019;47:W52–W58. doi: 10.1093/nar/gkz333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li L., Stoeckert C.J., Roos D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2014;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 80.Dongen S. Van A cluster algorithm for graphs. Inf. Syst. [INS] 2000;R0010:40. [Google Scholar]
- 81.Apweiler R., Bairoch A., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004;32:2699. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., Von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nguyen L.T., Schmidt H.A., Von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bayliss S., Thorpe H., Coyle N., Sheppard S., Feil E. PIRATE: A fast and scalable pangenomics toolbox for clustering diverged orthologues in bacteria. bioRxiv. 2019 doi: 10.1093/gigascience/giz119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., De Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krieger E., Joo K., Lee J., Lee J., Raman S., Thompson J., Tyka M., Baker D., Karplus K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins Struct. Funct. Bioinform. 2009;77(Suppl. 9):114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Laskowski R.A., Rullmann J.A.C., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 88.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;15:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 89.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Trott O., Olson A. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schüttelkopf A.W., Van Aalten D.M.F. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;D60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 92.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009;16:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dekker P.J.T., Koenders D., Bruins M.J. Lactose-free dairy products: Market developments, production, nutrition and health benefits. Nutrients. 2019;11:551. doi: 10.3390/nu11030551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Biovia D.S., Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E., Darden T., et al. Dassault Systèmes BIOVIA, Discovery Studio Visualizer, v.17.2, San Diego: Dassault Systèmes, 2016. J. Chem. Phys. 2000;10:21–9991. doi: 10.1016/0021-9991(74)90010-2. [DOI] [Google Scholar]
- 95.JASP Team . JASP. University of Amsterdam; Amsterdam, The Netherlands: 2020. version 0.13.1; Computer software. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found at https://www.ncbi.nlm.nih.gov under the accession number provided for each genome or nucleotide sequence.