Abstract
Tripartite motif protein 21 (TRIM21) is an E3 ubiquitin ligase and cytosolic antibody receptor of the TRIM family. Previous reports have indicated that TRIM21 plays an important role during viral infection. This study aimed at examining the role of TRIM21 in the replication of porcine epidemic diarrhea virus (PEDV) and showed that TRIM21 inhibits PEDV proliferation by targeting and degrading the nucleocapsid (N) protein through the proteasomal pathway. Furthermore, the endogenous expression of TRIM21 was found to be downregulated by PEDV infection in Vero and LLC-PK1 cells. Overexpression of TRIM21 inhibited PEDV replication, whereas knockdown of TRIM21 increased viral titers and N protein levels. TRIM21 was found to interact and colocalize with the N protein, and the TRIM21-mediated antiviral effect was dependent on its ubiquitin ligase activity, which engages in polyubiquitination and degradation of the N protein in a proteasome-dependent manner. Taken together, these findings provide information about the role of TRIM21 in PEDV proliferation and increase our understanding of host-virus interactions.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00705-021-05080-4.
Introduction
Porcine epidemic diarrhea virus (PEDV) is one of the most devastating pathogens threatening the development of the pig industry. PEDV infection leads to severe watery diarrhea, dehydration, and even death in piglets, causing huge economic losses [1]. PEDV was first reported in Asian countries in the 1990s [2, 3]. Recently, porcine epidemic diarrhea (PED) outbreaks occurred in China, South Korea, and Japan, resulting from the emergence of new PEDV variant strain since 2010 [4–6]. Currently, vaccination is the most effective approach for preventing and controlling PED. However, widespread vaccination is likely to favor the generation of mutations, leading to a wide variety of genotypes, some of which may have increased virulence. Therefore, knowledge about host factors that restrict viral replication is needed for understanding host-virus interactions, and this may provide potential targets for antiviral therapies against viral infection.
PEDV, a single-stranded positive-sense RNA virus, is a member of the genus Alphacoronavirus of the family Coronaviridae. It has a 28-kb genome encoding two polyproteins (pp1a and pp1b), four structural proteins (spike, S; envelope, E; membrane, M; and nucleocapsid, N), and an accessory protein (ORF3) that is associated with virulence [7, 8]. The N protein, 55-58 kDa in size, is abundantly expressed and conserved among members of the genus Alphacoronavirus. It is predominantly located in the cytoplasm of the host cell and is involved in transcription and synthesis of the viral genome, viral particle assembly, host stress, and the immune response [9]. For example, the N protein can antagonize Sendai virus (SeV)-induced interferon-β (IFN-β) production by blocking the interaction between TANK binding kinase 1 (TBK1) and IFN regulatory factor 3 (IRF3) [10]. In addition, the N protein decreases IFN-λ production by inhibiting the translocation of nuclear factor-κB (NF-κB) [11]. Most recently, host viperin was found to bind to the N protein to inhibit PEDV proliferation [12]. These findings suggest that interaction of the N protein with host factors plays a significant role in viral infection.
The tripartite motif protein (TRIM) family is part of the E3 ubiquitin ligase family. To date, more than 100 different TRIM proteins have been discovered that may play key roles in cell proliferation, differentiation, and apoptosis [13]. These proteins contain one or two B-box and coiled-coil domains, a variable C-terminal domain (RBCC), and three conserved functional domains [14]. There is accumulating evidence that TRIM proteins play a regulatory role in innate immune responses by interacting with antiviral factors. For example, TRIM25 potentiates the innate immune response by targeting and ubiquitinating the viral RNA receptor retinoic acid-inducible gene I (RIG-I) [15]. TRIM31 interacts with mitochondrial antiviral signaling (MAVS) protein and catalyzes the K63-linked polyubiquitination of Lys10, Lys311, and Lys461 of MAVS, regulating antiviral signaling [16]. In addition, TRIM56 promotes the ubiquitination of cyclic GMP–AMP synthase (cGAS) to affect viral growth [17].
TRIM21, first identified as an antibody-bound protein that serves as an Fc receptor (FcR), recognizes pathogen-bound antibodies, and catalyzes K63-linked polyubiquitination to initiate cellular innate immune responses against viral infection [18]. During infection with antibody-opsonized virus, TRIM21 expedites the uncoating of the virus to expose the viral genome, promoting RIG-I and cGAS sensing and triggering an innate immune response [19]. In contrast, it has been reported that TRIM21 negatively regulates innate immunity by targeting and degrading the viral DNA sensor DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 (DDX41) [20]. Intriguingly, TRIM21 has been found to enhance antiviral effects on RNA virus infections by catalyzing K27-linked polyubiquitination of MAVS [21]. Additionally, although TRIM21 has no direct effect on enterovirus 71 (EV71) infection, it regulates sterile alpha motif and histidine–aspartic acid domain-containing protein 1 (SAMHD1) to affect viral growth [22]. These findings indicate that TRIM21 serves as a multifaceted regulator during viral infections. Recently, porcine TRIM21 was found to inhibit proliferation of foot-and-mouth disease virus (FMDV) by targeting and degrading Fc-fused viral protein 1 (VP1) [23]; however, the ability of porcine TRIM21 to regulate PEDV infection and the underlying mechanisms involved have not been fully investigated.
In this study, we investigated the role of TRIM21 in PEDV replication and found that TRIM21 negatively regulates PEDV replication through polyubiquitination and degradation of the viral N protein in a proteasome-dependent manner. These findings increase our understanding of host factors that modulate viral replication and provide new insights into the role of the N protein in PEDV pathogenesis.
Materials and methods
Cell lines and transfection
Human embryonic kidney cells (293T cells; CRL-11,268) and HeLa cells (CCL-2) were obtained from ATCC. 293T and HeLa cells were used for immunoprecipitation and confocal assay, respectively. African green monkey kidney cells (Vero cells, CCL-81) and porcine kidney cells (LLC-PK1 cells, CL-101) were obtained from ATCC. These cells were cultured using 10% fetal bovine serum (FBS; Gibco, 10,099,141) in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, 12,430,054) and maintained at 37 °C with 5% CO2. Lipofectamine 3000 (Invitrogen, L3000015) was used to transfect cells with the indicated plasmids. Lipofectamine RNAiMAX (Invitrogen, 13,778,150) was used to transfect cells with siRNA. The simian TRIM21 siRNA and the control siRNA were synthesized by GenePharma (Shanghai, China). The siRNA sequences are listed in Table 1.
Table 1.
Primer and siRNA sequences used in this study
| Purpose | Primer | Sequence (5′-3′) |
|---|---|---|
| Real-time PCR | PEDV N forward | GAGGGTGTTTTCTGGGTTG |
| PEDV N reverse | CGTGAAGTAGGAGGTGTGTTAG | |
| SUS TRIM21 forward | CCAGACTCCCCTCTACCCT | |
| SUS TRIM21 reverse | TTCCACCGTCATTGAAACC | |
| MACACA TRIM21 forward | CCTTCGTGGAGCCTGTGAGC | |
| MACACA TRIM21 reverse | GGCGGAGATTCCTGAGCAGA | |
| β-actin forward | TCCCTGGAGAAGAGCTACGA | |
| β-actin reverse | AGCACTGTGTTGGCGTACAG | |
| siRNA assay | si-mTRIM21 sense | GGAAUGCAUCUCUCAGGUUTT |
| si-mTRIM21antisense | AACCUGAGAGAUGCAUUCCTT | |
| NC sense | UUCUCCGAACGUGUCACGUTT | |
| NC antisense | ACGUGACACGUUCGGAGAATT |
Antibodies, plasmids, and reagents
Antibodies against the HA tag (26183) and DYKDDDDK tag (PA1-984B) were purchased from Invitrogen. The antibody against TRIM21 (ab4396) was obtained from Abcam. Alexa FluorTM 594 goat anti-mouse IgG (H+L) (A11032) and Alexa FluorTM 488 goat anti-rabbit IgG (H+L) (A11034) were purchased from Invitrogen. Antibodies against the GST tag (AF0174) and ACTB/β-actin (AF5001) were obtained from Beyotime Biotechnology. Horseradish peroxidase (HRP)-conjugated antibody against mouse IgG (SA00001-1) and HRP-conjugated antibody against rabbit IgG (SA00001-2) were obtained from the Proteintech Group. The antibody against ubiquitin (SC-8017) was obtained from Santa Cruz Biotechnology. The antibody against the PEDV N protein and the plasmids PCAGGS, pCold-GST, and pCold-TF were from our laboratory. Recombinant plasmids were generated by homologous recombination using a ClonExpress II One Step Cloning Kit (Vazyme Biotech, C112-02) following the manufacturer's protocols. Bafilomycin A1 (Baf A1, 54,645) was obtained from CST. MG132 (M7449), 3-methyladenine (3-MA, M9281), and chloroquine phosphate (CQ, PHR1258) were obtained from Sigma-Aldrich.
Virus and viral infection
PEDV strain JS-2013 (GenBank no. MH910099), maintained in our laboratory, was used in this study. Vero and LLC-PK1 cells were mock infected with DMEM or infected with PEDV at the indicated multiply of infection (MOI) with trypsin at a concentration of 5 ng/μL (Invitrogen, 15,050,065). After incubation at 37 °C for 1 h, the cells were washed three times with phosphate-buffered saline (PBS) to remove unattached viruses. The cells were then maintained in serum-free DMEM supplemented with 5 ng of trypsin per µl at 37 °C and harvested at different time points. Viral titers in the supernatants of infected cells were determined by the Kaerber method and expressed as 50% tissue culture infectious doses (TCID50) per milliliter.
Quantitative real-time PCR
Total RNA extraction was performed using TRIzol Reagent (15596-026, Thermo Fisher Scientific). For reverse transcription (RT)-qPCR analysis, cDNA was synthesized using a PrimeScript™ RT Reagent Kit (Takara, RRO47A) and quantitated by real-time PCR with specific primers, using the ΔΔCT method with a LightCycler system (Roche, Switzerland). The specific primer sequences are listed in Table 1.
Western blotting
Cells were washed twice with ice-cold PBS and then lysed on ice using RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific, 89,901) supplemented with Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, 78420) and Protease Inhibitor Cocktail (Thermo Fisher Scientific, 87786). The western blot assay was performed as described previously [24]. Briefly, the lysates were centrifuged and boiled in 5× SDS-PAGE loading buffer for 5 min. After that, the proteins were separated by SDS-PAGE and transferred to 0.2-μm nitrocellulose membranes (Thermo Fisher Scientific, STM2007). Next, the membranes were blocked with Tris-buffered saline (TBS) supplemented with 5% skim milk (Yeasen, 36120ES60) for 1 h and incubated with the primary antibodies for 2 h at room temperature. After washing for 30 min, the membranes were incubated with HRP anti-mouse IgG antibodies or HRP anti-rabbit IgG antibodies for 1 h. Then, the membranes were washed for 30 min and probed using chemiluminescence reagents. Finally, the protein bands were analyzed using ImageJ software.
Co-immunoprecipitation and confocal microscopy
293T or HeLa cells were transfected with the indicated plasmids. Co-IP and confocal microscopy were performed as described previously [25]. Briefly, for IP assay, 293T cell lysates were collected and centrifuged. The supernatants were then incubated with antibody-bound Dynabeads Protein G (Life Technologies, 10004D) for 20 min at room temperature. The Dynabeads were then washed with ice-cold PBS containing 0.1% Tween-20 (MACKLIN, T885238) and eluted in 1× SDS-PAGE sample buffer. Then proteins were analyzed by western blotting with the indicated antibodies. For confocal microscopy, HeLa cells were first fixed with PBS supplemented with 4% paraformaldehyde (Sigma-Aldrich, P6148) at room temperature for 15 min. The cells were then washed and permeabilized with PBS containing 0.1% Triton X-100 (Sigma-Aldrich, T9284) at 4 °C for 20 min. Next, cells were blocked with PBS supplemented with 5% bovine serum albumin (BSA; Yeasen, 36101ES25). After that, the cells were incubated with the primary antibodies (anti-Flag tag or anti-HA tag) at 37 °C for 1 h, washed five times with PBS, and incubated with the secondary antibodies at 37 °C for 1 h. After washing five times, the cells were examined using a confocal immunofluorescence microscope (Carl Zeiss, Oberkochen, Germany).
GST pull-down assay
The plasmids pCold-GST-N and pCold-TF-TRIM21 were used to express the recombinant proteins GST-N and TF-TRIM21, respectively, which were then purified. The proteins were processed using a GST Protein Interaction Pull-Down Kit (Thermo, 21,516) according to the manufacturer’s instructions. Briefly, the bait proteins (GST-N) and prey proteins (TF-TRIM21) were incubated together with GST glutathione agarose resin with gentle shaking overnight at 4 °C. Then, the proteins were washed five times with a washing buffer composed of pull-down lysis buffer and TBS. Finally, the proteins were eluted and boiled for 5 min in 1× SDS-PAGE sample buffer and detected by western blot with the indicated antibodies.
Statistical analysis
All experiments were performed at least three times independently, and statistical significance was analyzed using the two-tailed Student’s t-test. A P-value less than 0.05 was considered statistically significant.
Results
Endogenous expression of TRIM21 is downregulated by PEDV infection
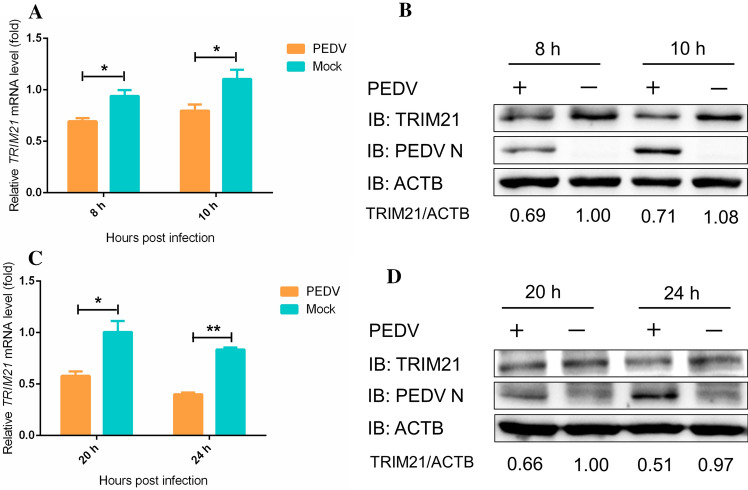
To investigate whether PEDV infection affects endogenous TRIM21 expression, LLC-PK1 and Vero cells were mock infected or infected with PEDV and processed at the indicated time points. The results showed that the mRNA level of TRIM21 in Vero cells decreased during viral infection (Fig. 1A), and the level of TRIM21 protein also decreased (Fig. 1B). Likewise, the mRNA and protein levels of TRIM21 in LLC-PK1 cells were reduced by PEDV infection (Fig. 1C and D). Collectively, these results suggest that PEDV infection downregulates endogenous expression of TRIM21 in Vero and LLC-PK1 cells.
Fig. 1.
PEDV infection reduces endogenous expression of TRIM21. Vero and LLC-PK1 cells were mock infected or infected with PEDV at an MOI of 1, and the cells were collected and processed at the indicated time points. (A) The mRNA level of TRIM21 in Vero cells was measured by real-time PCR. (B) The protein levels of TRIM21, N, and β-actin in Vero cells were measured by western blotting using the indicated antibodies. β-actin was used as the sample loading control. (C) The mRNA level of TRIM21 in LLC-PK1 cells was measured by real-time PCR. (D) TRIM21, N, and β-actin protein levels in LLC-PK1 cells were measured by western blot using the indicated antibodies. β-actin was used as the sample loading control. Data are the mean ± SD of triplicate samples. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (two-tailed Student’s t-test)
TRIM21 inhibits PEDV proliferation
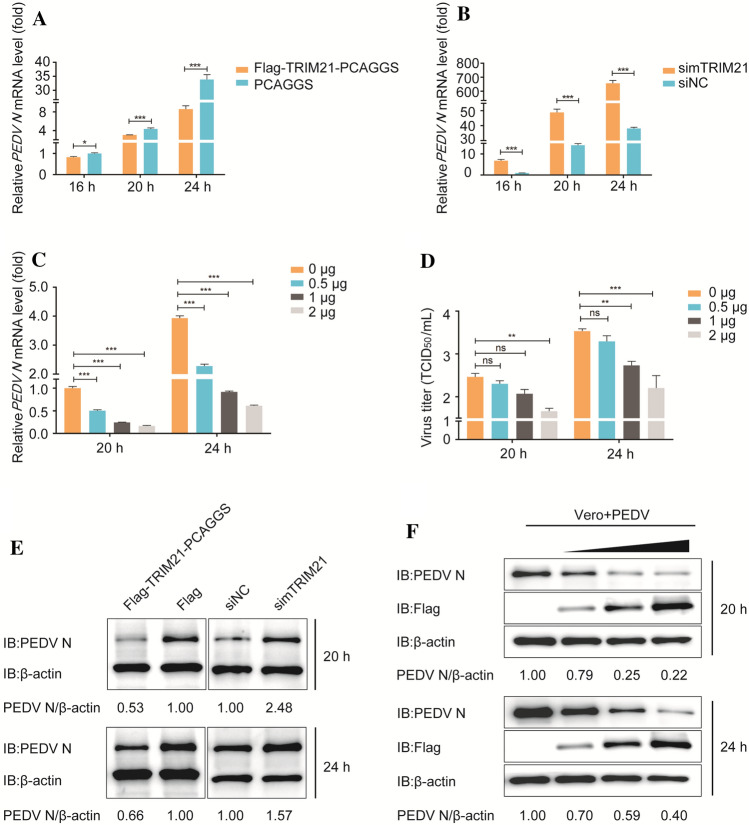
Next, to investigate their effect on PEDV replication, Flag-TRIM21 proteins were overexpressed in Vero cells. We found that the PEDV N mRNA level was lower in cells transfected with Flag-TRIM21-encoding plasmids than in cells transfected with the empty vector (Fig. 2A). In addition, the N mRNA level and viral titers were consistently reduced with increasing doses of Flag-TRIM21-encoding plasmids (Fig. 2C and D). Furthermore, western blot analysis showed that TRIM21 upregulation led to a strong reduction in the accumulation of N protein (Fig. 2E and F), confirming an antiviral role for TRIM21 against PEDV proliferation. We then confirmed the effect of TRIM21 using a knockdown assay. The knockdown efficiency is shown in Supplementary Fig. S1, and we selected siRNA-1. As shown in Fig. 2B, the N mRNA level was higher in cells transfected with simTRIM21 than in cells transfected with the control siRNA. Likewise, the level of N protein was also higher in cells transfected with simTRIM21 (Fig. 2E). Together, these data indicate that TRIM21 negatively regulates PEDV proliferation.
Fig. 2.
TRIM21 inhibits PEDV proliferation. (A) Vero cells were transfected with Flag-TRIM21-PCAGGS (1 μg) or the empty vector (1 μg), and infected 24 h later with PEDV at an MOI of 0.01. The cells were then collected and the PEDV N mRNA level was analyzed by real-time PCR. (B) Vero cells were transfected with simTRIM21 or siNC and infected 24 h later with PEDV at an MOI of 0.01. Cells were then harvested, and the PEDV N mRNA level was analyzed by real-time PCR. (C) Vero cells were transfected with increasing doses of Flag-TRIM21-PCAGGS or the empty vector, and the PEDV N mRNA level was determined by real-time PCR. (D) The amount of virus in the supernatants from the samples (C) was determined using the TCID50 method. (E) Vero cells treated as described above (A and B) were collected at the indicated time points, and the protein levels of N and β-actin were estimated by western blot using the primary antibody (anti-N antibody and anti-β-actin antibody) and the secondary antibody (HRP-conjugated antibody against mouse IgG). (F) Vero cells treated as described above (C) were processed, and the protein levels of N, Flag-TRIM21, and β-actin were estimated by western blot using the indicated antibodies. Data are the mean ± SD of triplicate samples, *p < 0.05; **, p < 0.01; ***, p < 0.001 (two-tailed Student’s t-test)
TRIM21 interacts with N protein
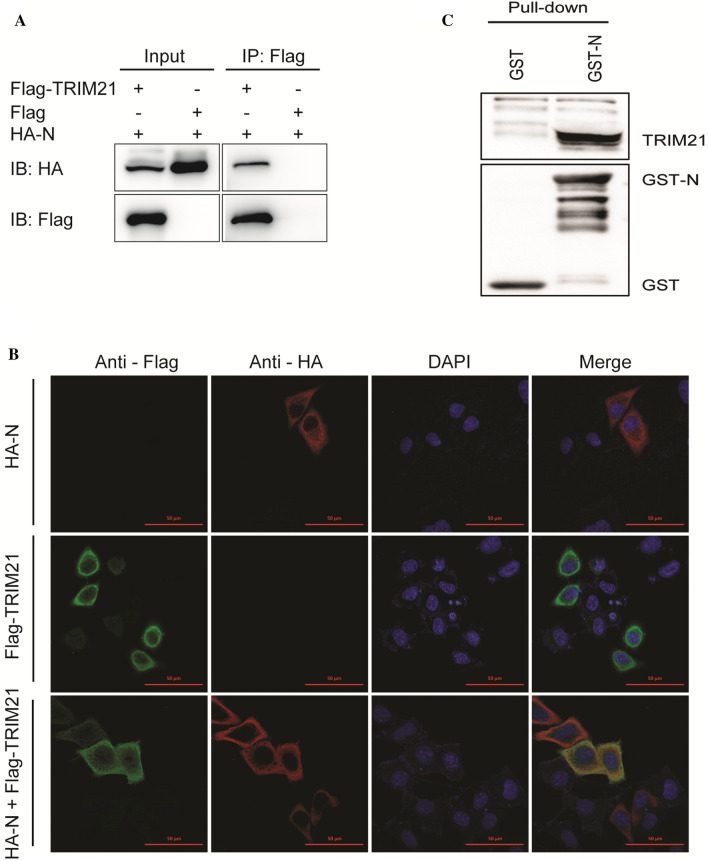
To investigate the mechanisms by which TRIM21 suppresses PEDV replication, a co-IP assay was performed. We found that TRIM21 could interact with the N protein (Fig. 3A). Additionally, a confocal immunofluorescence assay showed that TRIM21 colocalized with the N protein in the cytoplasm (Fig. 3B). Moreover, a GST pull-down assay was performed, and this confirmed the binding between TRIM21 and N protein. GST-N bound to TRIM21, but GST alone did not, suggesting a direct interaction with TRIM21 (Fig. 3C).
Fig. 3.
TRIM21 interacts with the N protein. (A) 293T cells were co-transfected with HA-N-PCAGGS (1 μg) and Flag-TRIM21-PCAGGS (1 μg) or PCAGGS (1 μg). At 24 hours post-transfection, cells were collected and processed for co-IP assay, followed by western blot with the indicated antibodies. (B) HeLa cells were transfected with HA-N-PCAGGS (1 μg) or Flag-TRIM21-PCAGGS (1 μg) or co-transfected with both and processed 24 h later for a confocal imaging assay with the indicated antibodies. (C) The recombinant proteins were expressed in E. coli strain BL21 and purified for the GST pull-down analysis. After washing, proteins eluted from beads were analyzed by western blot using the indicated antibodies. Data are the mean ± SD of triplicate samples. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (two-tailed Student’s t-test)
TRIM21 targets the N protein for proteasomal degradation
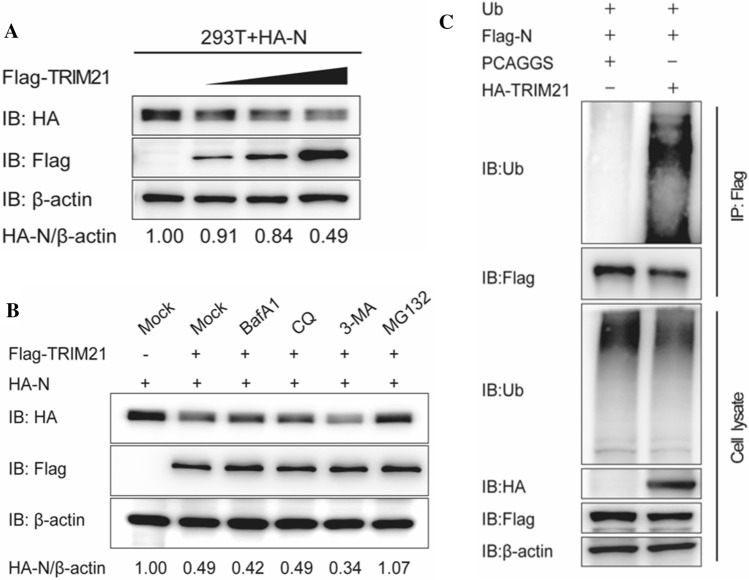
Given that TRIM21 is an E3 ubiquitin ligase, we investigated whether TRIM21 could induce degradation of the N protein. For this purpose, 293T cells were co-transfected with HA-N-encoding plasmids and increasing doses of Flag-TRIM21-encoding plasmids. As shown in Fig. 4A, TRIM21 overexpression reduced the N protein level in a dose-dependent manner. Next, to test whether the N protein undergoes proteasomal degradation or lysosomal proteolysis, we treated 293T cells with the proteasome inhibitor MG132 or the autophagy inhibitor BafA1, CQ, or 3-MA and found that TRIM21-mediated degradation of N was inhibited by MG132, but not by BafA1, CQ, or 3-MA (Fig. 4B). As ubiquitination is a key step in the proteasome-dependent pathway, we investigated whether N ubiquitination contributes to the antiviral role of TRIM21. To this end, we co-transfected 293T cells with the indicated plasmids, which led to increased N ubiquitination in the presence of TRIM21, suggesting that TRIM21 mediates N ubiquitination (Fig. 4C). Notably, other proteins might also interact with the PEDV N protein and cause its polyubiquitination. For example, the E3 ubiquitin ligase MARCHF8 interacts and colocalizes with the N protein. Furthermore, MARCHF8 overexpression promotes N polyubiquitination, whereas knockdown has the opposite effect [26]. Interestingly, the porcine deltacoronavirus (PDCoV) N protein inhibits K63-linked ubiquitination of RIG-I, thus suppressing RIG-I signaling [27]. These studies increase our understanding of host-virus interactions and the role of the coronavirus N protein in viral pathogenesis. We conclude that TRIM21 inhibits viral growth by degrading the N protein in a proteasome-dependent manner, which involves ubiquitination of the N protein.
Fig. 4.
TRIM21 degrades the N protein in a proteasome-dependent manner. (A) 293T cells were co-transfected with plasmids encoding HA-N (1 μg) and increasing doses of plasmids encoding Flag-TRIIM21 (0.5 μg, 1 μg, or 2 μg). After 24 h, the cells were collected and analyzed by western blot using the indicated antibodies. (B) 293T cells were co-transfected with the plasmid encoding HA-N (1 μg) and the plasmid encoding Flag-TRIM21 (1 μg) or the empty vector (1 μg) for 24 h and treated with BafA1 (0.1 mM), CQ (10 mM), 3-MA (0.5 mM), or MG132 (5 mM) for 8 h. The cells were then collected and analyzed by western blot using the indicated antibodies. (C) 293T cells were co-transfected with the plasmids encoding Ub (1 μg), Flag-N (1 μg), and HA-TRIM21 (1 μg) or PCAGGS (1 μg) for 24 h. Then cells were then collected and analyzed by co-IP assay followed by western blot using the indicated antibodies. Data are the mean ± SD of triplicate samples. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (two-tailed Student’s t-test)
Discussion
PEDV is one of the most devastating pathogens in the swine industry. Knowledge about host factors that modulate viral replication is needed for understanding host-virus interactions and identifying potential targets for antiviral therapies. In this study, we investigated the antiviral role of host TRIM21 in viral infection. We found that TRIM21 overexpression markedly suppressed viral replication in Vero cells, whereas TRIM21 knockdown had the opposite effect. We also confirmed these results using LLC-PK1 cells (data not shown). This indicates that TRIM21 may serve as a novel target to effectively control PEDV infection.
In previous studies, TRIM21 was shown to play a dual role in the defense against viral infection. For example, TRIM21 promotes EV71 infection by inducing proteasomal degradation of SAMHD1, which is a restriction factor for EV71 replication [22]. In contrast, TRIM21 inhibits infection of several viruses, including Newcastle disease virus (NDV), SeV, and vesicular stomatitis virus (VSV). This is because TRIM21 catalyzes the K27-linked polyubiquitination of MAVS, resulting in the recruitment of TBK1 to MAVS, which facilitates innate immune responses [21]. Differences between these viruses and the underlying mechanisms involved in the host antiviral responses may partly account for the observed differences. In the present study, we found that PEDV infection downregulated TRIM21 expression and that inhibition of TRIM21 expression facilitated viral replication, indicating an important role of TRIM21 during viral infection. These results suggest that PEDV seems to hijack the translation machinery of the host to reduce TRIM21 expression in response to infection, thus establishing a successful infection. However, the network of host-virus interactions is very complicated and will require further investigation.
TRIM21 also serves as a cytosolic antibody receptor, binding antibodies with high affinity [28]. Upon infection, TRIM21 detects the antibody-coated viruses in the cytosol. It then activates the cellular degradation machinery, leading to the destruction of viral particles and neutralization of infection. Intriguingly, TRIM21 also initiates innate signaling, resulting in the induction of cytokines and IFN-stimulated genes (ISGs) [29]. Recently, porcine TRIM21 was found to inhibit FMDV replication through an intracellular neutralization mechanism. TRIM21 first interacts with VP1 that is fused with the Fc fragment of porcine immunoglobulin G and then promotes the degradation of Fc-fused VP1 in a proteasome-dependent manner, thus leading to the inhibition of FMDV replication [23]. This provides new insights into the antiviral role of TRIM21, and will be of interest to determine whether TRIM21 exerts inhibitory effects on other viruses via an antibody-dependent mechanism.
In conclusion, we show here that porcine and simian TRIM21 are downregulated by PEDV infection in Vero and LLC-PK1 cells. The antiviral role of TRIM21 against PEDV was confirmed using transient overexpression and knockdown assays in Vero cells. We also found that TRIM21 could interact and colocalize with the N protein. Overexpression of TRIM21 reduced N protein levels in a dose-independent manner, which was inhibited by the addition of the proteasome inhibitor MG132. In addition, a co-IP assay and western blotting showed that the ubiquitination of N was mediated by TRIM21. These results indicate that TRIM21 targets the N protein for degradation in a proteasome-dependent manner, leading to the suppression of PEDV replication. Together, these results shed light on the critical role of TRIM21 in PEDV infection and increase our understanding of host-virus interactions.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was funded by the National Key Research and Development Programs of China (no. 2016YFD0500103), the Natural Science Foundation of Shanghai (no. 19ZR1469100), and the National Natural Science Foundation of China (no. 31872478).
Declarations
Conflict of interest
The authors have no conflicts of interest and have read the manuscript and approved its submission to Archives of Virology.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hua Wang and Xiaoyong Chen contributed equally to this work.
Contributor Information
Guangzhi Tong, Email: gztong@shvri.ac.cn.
Tongling Shan, Email: shantongling@shvri.ac.cn.
References
- 1.Choudhury B, Dastjerdi A, Doyle N, Frossard JP, Steinbach F. From the field to the lab—an European view on the global spread of PEDV. Virus Res. 2016;226:40–49. doi: 10.1016/j.virusres.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu Z, Zhang Y, Cao Y. The roles of apoptosis in swine response to viral infection and pathogenesis of swine enteropathogenic coronaviruses. Front Vet Sci. 2020;7:572425. doi: 10.3389/fvets.2020.572425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shibata I, Tsuda T, Mori M, Ono M, Sueyoshi M, Uruno K. Isolation of porcine epidemic diarrhea virus in porcine cell cultures and experimental infection of pigs of different ages. Vet Microbiol. 2000;72(3–4):173–182. doi: 10.1016/s0378-1135(99)00199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang DK, Kim HH, Lee SH, Yoon SS, Park JW, Cho IS. Isolation and characterization of a new porcine epidemic diarrhea virus variant that occurred in Korea in 2014. J Vet Sci. 2018;19(1):71–78. doi: 10.4142/jvs.2018.19.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan Y, Tian X, Li W, Zhou Q, Wang D, Bi Y, Chen F, Song Y. Isolation and characterization of a variant porcine epidemic diarrhea virus in China. Virol J. 2012;9:195. doi: 10.1186/1743-422x-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Diep N, Sueyoshi M, Norimine J, Hirai T, Myint O, Teh APP, Izzati UZ, Fuke N, Yamaguchi R. Molecular characterization of US-like and Asian non-S INDEL strains of porcine epidemic diarrhea virus (PEDV) that circulated in Japan during 2013–2016 and PEDVs collected from recurrent outbreaks. BMC Vet Res. 2018;14(1):96. doi: 10.1186/s12917-018-1409-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z, Ma Z, Li Y, Gao S, Xiao S. Porcine epidemic diarrhea virus: molecular mechanisms of attenuation and vaccines. Microb Pathog. 2020;149:104553. doi: 10.1016/j.micpath.2020.104553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q, Vlasova AN, Kenney SP, Saif LJ. Emerging and re-emerging coronaviruses in pigs. Curr Opin Virol. 2019;34:39–49. doi: 10.1016/j.coviro.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung K, Saif LJ. Porcine epidemic diarrhea virus infection: Etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet J (London, England: 1997) 2015;204(2):134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding Z, Fang L, Jing H, Zeng S, Wang D, Liu L, Zhang H, Luo R, Chen H, Xiao S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J Virol. 2014;88(16):8936–8945. doi: 10.1128/jvi.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shan Y, Liu ZQ, Li GW, Chen C, Luo H, Liu YJ, Zhuo XH, Shi XF, Fang WH, Li XL. Nucleocapsid protein from porcine epidemic diarrhea virus isolates can antagonize interferon-λ production by blocking the nuclear factor-κB nuclear translocation. J Zhejiang Univ Sci B. 2018;19(7):570–580. doi: 10.1631/jzus.B1700283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J, Chi H, Fu Y, Cao A, Shi J, Zhu M, Zhang L, Hua D, Huang J. The antiviral protein viperin interacts with the viral N protein to inhibit proliferation of porcine epidemic diarrhea virus. Adv Virol. 2020;165(10):2279–2289. doi: 10.1007/s00705-020-04747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Wu H, Wu W, Zhuo W, Liu W, Zhang Y, Cheng M, Chen YG, Gao N, Yu H, Wang L, Li W, Yang M. Structural insights into the TRIM family of ubiquitin E3 ligases. Cell Res. 2014;24(6):762–765. doi: 10.1038/cr.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajsbaum R, García-Sastre A, Versteeg GA. TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J Mol Biol. 2014;426(6):1265–1284. doi: 10.1016/j.jmb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, Liu S, Goraya MU, Maarouf M, Huang S, Chen JL. Host immune response to influenza A virus infection. Front Immunol. 2018;9:320. doi: 10.3389/fimmu.2018.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu B, Zhang M, Chu H, Zhang H, Wu H, Song G, Wang P, Zhao K, Hou J, Wang X, Zhang L, Gao C. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat Immunol. 2017;18(2):214–224. doi: 10.1038/ni.3641. [DOI] [PubMed] [Google Scholar]
- 17.Seo GJ, Kim C, Shin WJ, Sklan EH, Eoh H, Jung JU. TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat Commun. 2018;9(1):613. doi: 10.1038/s41467-018-02936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McEwan WA, Tam JC, Watkinson RE, Bidgood SR, Mallery DL, James LC. Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat Immunol. 2013;14(4):327–336. doi: 10.1038/ni.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watkinson RE, McEwan WA, Tam JC, Vaysburd M, James LC. TRIM21 promotes cGAS and RIG-I sensing of viral genomes during infection by antibody-opsonized virus. PLoS Pathog. 2015;11(10):e1005253. doi: 10.1371/journal.ppat.1005253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z, Bao M, Lu N, Weng L, Yuan B, Liu YJ. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat Immunol. 2013;14(2):172–178. doi: 10.1038/ni.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue B, Li H, Guo M, Wang J, Xu Y, Zou X, Deng R, Li G, Zhu H. TRIM21 promotes innate immune response to RNA viral infection through Lys27-linked polyubiquitination of MAVS. J Virol. 2018 doi: 10.1128/jvi.00321-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Huan C, Wang H, Liu Y, Liu X, Su X, Yu J, Zhao Z, Yu XF, Zheng B, Zhang W. TRIM21-mediated proteasomal degradation of SAMHD1 regulates its antiviral activity. EMBO Rep. 2020;21(1):e47528. doi: 10.15252/embr.201847528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan W, Zhang D, Qian P, Qian S, Wu M, Chen H, Li X. Swine TRIM21 restricts FMDV infection via an intracellular neutralization mechanism. Antiviral Res. 2016;127:32–40. doi: 10.1016/j.antiviral.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Ma Y, Chen X, Chen K, Zeng X, Yang S, Chang W, Tang Y, Chen X, Wang S, Chen JL. Identification and characterization of a distinct strain of beak and feather disease virus in Southeast China. Virol Sin. 2020;35(1):43–51. doi: 10.1007/s12250-019-00159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong N, Shan T, Wang H, Jiao Y, Zuo Y, Li L, Tong W, Yu L, Jiang Y, Zhou Y, Li G, Gao F, Yu H, Zheng H, Tong G. BST2 suppresses porcine epidemic diarrhea virus replication by targeting and degrading virus nucleocapsid protein with selective autophagy. Autophagy. 2020;16(10):1737–1752. doi: 10.1080/15548627.2019.1707487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong N, Shan T, Wang H, Jiao Y, Zuo Y, Li L, Tong W, Yu L, Jiang Y, Zhou Y, Li G, Gao F, Yu H, Zheng H, Tong G. BST2 suppresses porcine epidemic diarrhea virus replication by targeting and degrading virus nucleocapsid protein with selective autophagy. Autophagy. 2019 doi: 10.1080/15548627.2019.1707487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Likai J, Shasha L, Wenxian Z, Jingjiao M, Yaxian Y. Porcine deltacoronavirus nucleocapsid protein suppressed IFN-β production by interfering porcine RIG-I dsRNA-binding and K63-linked polyubiquitinationtable. Front Immunol. 2019;10:1024. doi: 10.3389/fimmu.2019.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc Natl Acad Sci USA. 2007;104(15):6200–6205. doi: 10.1073/pnas.0609174104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dickson C, Fletcher AJ, Vaysburd M, Yang JC, Mallery DL, Zeng J, Johnson CM, McLaughlin SH, Skehel M, Maslen S, Cruickshank J, Huguenin-Dezot N, Chin JW, Neuhaus D, James LC. Intracellular antibody signalling is regulated by phosphorylation of the Fc receptor TRIM21. Elife. 2018 doi: 10.7554/eLife.32660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.