Abstract
Aims:
This study aims to derive simple yet robust formula(s) for the calculation of cranial tumor volume using linear tumor dimensions in anterioposterior (AP), mediolateral (ML) and craniocaudal (CC) directions and also propose a reproducible methodology for tumor dimension measurements.
Materials and Methods:
Magnetic resonance images (MRI) of 337 patients planned for Gammaknife Stereotactic Radiosurgery for different types of brain tumors were analyzed using Leksell Gamma Plan (LGP) software. Tumor volume in three dimensional was outlined and maximum tumor diameters were measured in three orthogonal directions AP, ML, and CC on the MRI. Formulas were derived to calculate tumor volume from AP, ML, and CC diameters using linear regression technique. An agreement between the calculated volume and standard volume observed from LGP software was determined using Bland Altman (B-A) plot. A comparison was made between the volume calculated using traditionally used formula of ellipsoid, standard volume obtained from LGP software and volume calculated from formulas derived in the present study.
Results:
The tumors were divided into two categories based on their size for better volume prediction. The tumors having product of their diameters in the range 0–2.5cc were called “small tumors” and the formula proposed for their volume estimation (V = 1.513) × (AP × ML × CC) + 0.047 ) was found to predict the tumor volume with an average bias of 0.0005cc. For “large tumors,” having product of diameters in the range 2.5–36cc, the proposed formula (V = 0.444 × (AP × ML × CC) + 0.339 ) predicted the tumor volume with an average bias of 0.007cc.
Conclusions:
The two formulas proposed in the study are more accurate as compared to the commonly used formula that considers the tumors as ellipsoids. The methodology proposed in the study for measurement of linear tumor dimensions is simple and reproducible.
Keywords: Brain tumour, gamma knife radiosurgery, magnetic resonance images, tumour volume
INTRODUCTION
The volume of intra-cranial lesions is an important parameter, among others, for deciding the appropriate treatment strategy. Though surgery remains a principal treatment modality, stereotactic radiosurgery (SRS) has also emerged as an alternative mode of treatment for managing these lesions/tumors. SRS involves delivering ionizing radiation to a well identified target, generally in one sitting (fraction), with high precision (<1 mm), high dose conformality, and sharp dose gradient beyond the target (tumor) using three dimensional (3D) stereotactic localization of the tumor. In some literature, the SRS definition also includes radiation delivered in up to five fractions.[1] For dose delivery either linear accelerator based or cobalt-60 radioactive source based technologies are extensively in use nowadays. The 3D localization of the tumor is achieved using modern imaging techniques such as computed tomography (CT) and magnetic resonance imaging (MRI). Tumor size, its location, and relative position with respect to critical normal tissues play important roles in deciding the suitability of SRS and the number of fractions in SRS. There are well laid down international guidelines on deciding the suitability of SRS for brain lesions.[2,3,4] All these guidelines highlight the importance of tumor size/volume for the purpose of decision making. Further, tumor diameter and volume are important factors for studying the post-SRS responses, and can also be used to determine the index of malignancy.[5,6,7,8]
Diagnostic imaging modalities such as CT and MRI generally report the linear dimensions of a tumor. Estimating the tumor volume from initial diagnostic CT/MRI work-up of a patient helps the treatment team to choose the appropriate treatment modality and also to explain the expected outcome to the patients. Yet, despite tremendous technological advancements, no single standard protocol has been established for the measurement and reporting of linear tumor dimensions, and estimation of tumor volume from the latter. Different methods are used by radiologists or other physicians to report tumor diameters that represent their dimensions. Many authors have reported two largest perpendicular diameters on the axial image slice showing the largest lesion size among axial images as the two dimensions of tumor, and the maximum extent in cranio-caudal (CC) direction as the third dimension.[9] In such a situation the maximum diameter chosen for a tumor is dependent on the direction it is looked for. To overcome this subjectivity, the maximum diameter in the anterioposterior (AP) direction, and not in any random direction, may be chosen on the axial slice showing the largest lesion size. The maximum diameter in perpendicular direction, that is, medio-lateral (ML) direction, on the same axial image may then be chosen as the second dimension. However, choosing the axial slice with largest lesion size poses some difficulty as many tumors may be more extended in the AP direction while some others may be extended more in the ML direction. Shi et al. used dimensional extent in three largest orthogonal directions as the tumor dimensions which posed greater difficulty during actual measurements introducing a source of uncertainty in the measurements.[10]
In addition to the variations in lesion/tumor dimensions introduced due to the aforementioned variations in measurement methods, usage of different mathematical formulations for volume estimation from these linear dimensions further enhances the variations. None of these methods and formulations results in one unique estimated value for volume of each lesion. Therefore, there is no unanimity among SRS practitioners to rely on one specific formula for lesion volume estimation. The simplest among the various formulas is the one applicable for estimation of volume of a sphere. It relies on the measurement of a single diameter-the largest tumor dimension for volume estimation.[11] Formula for the volume of a cylinder has also been applied in which CC extent of the tumor is used as the height of tumour and the maximum diameter in an axial section with largest lesion size is taken as its radius. An equation that either uses a single diameter or two diameters in orthogonal directions is inadequate in estimating brain tumor volumes.[5] In general, a 3-D shape of a tumor is assumed to be hemi-ellipsoid whose volume is given by, 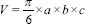 where a, b, c are the linear dimensions of the tumor.[12,13] However, its mathematical simplification
where a, b, c are the linear dimensions of the tumor.[12,13] However, its mathematical simplification 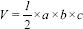 is predominantly used by many practitioners of SRS.[5] Davies et al. reported the use of a formula applicable for an ellipsoid in case of pituitary adenoma but they did not include the preoperated tumors in their study due to the complexity of shape of the latter.[14] Despite its good correlation with actual volume for many types of cranial tumors, this formula of ellipsoid overestimates the true lesion volume in case of acoustic schwannoma and meningioma, and underestimates the volume in case of arteriovenus malformations (S).[15,16] The variations in the methods used for the measurement of tumor linear dimensions and the subsequent volume estimation may not only introduce uncertainties in deciding the right treatment modality but also in the follow-up studies. This would be especially important for tumors whose volumes fall close to the upper limit (13 cc) generally agreed for SRS.[12]
is predominantly used by many practitioners of SRS.[5] Davies et al. reported the use of a formula applicable for an ellipsoid in case of pituitary adenoma but they did not include the preoperated tumors in their study due to the complexity of shape of the latter.[14] Despite its good correlation with actual volume for many types of cranial tumors, this formula of ellipsoid overestimates the true lesion volume in case of acoustic schwannoma and meningioma, and underestimates the volume in case of arteriovenus malformations (S).[15,16] The variations in the methods used for the measurement of tumor linear dimensions and the subsequent volume estimation may not only introduce uncertainties in deciding the right treatment modality but also in the follow-up studies. This would be especially important for tumors whose volumes fall close to the upper limit (13 cc) generally agreed for SRS.[12]
We retrospectively analyzed data of patients who underwent SRS on a Leksell Gamma Knife (LGK) system. The Leksell Gamma Plan (LGP) software application associated with LGK has provision for estimating linear tumor dimensions and volumes. Three hundred and thirty seven number of tumors of different types, shapes and sizes were included in this study. The objectives of the study were to propose (i) an easily reproducible method for measurement of tumor dimensions and (ii) a robust formalism for accurately calculating tumor volumes from these measurements irrespective of type, shape, and size of the tumors applying appropriate statistical techniques. The tumor volumes reported by the LGP system were used as a standard for comparison.
MATERIALS AND METHODS
Database
For this retrospective study cases treated with Gammaknife SRS between September 2016 and 2019 were included. Three hundred and thirty seven number of consecutive patients for whom tumor volumes had been contoured were included in this study excluding 65 patients with more than 1 lesion. The data included 116 cases of vestibular schwannoma (VS), 43 of AVM, 78 of meningioma, 50 of pituitary adenoma, 11 of metastases, 8 of cavernoma, 4 of craniopharyngioma, and 27 categorized as other benign tumors. Out of 337 tumors, 137 tumors were postoperated cases.
Target volume delineation
The Leksell Gamma Plan (LGP, Elekta Instruments AB, Stockholm, Sweden) versions 5.34 and 10.0 supplied with Leksell Gammaknife System model C (Elekta, Sweden) were used for linear dimension measurements and target delineation purposes. All patients for Gammaknife SRS (GKRS) underwent MRI after fixation of the stereotactic frame (Leksell coordinate frame G) on the patient head, which is MRI compatible. The MRI scans were performed on 1.5 Tesla Magnetom Avanto (Siemens, Germany) with the magnetic resonance localizer fitted on the G-frame. The most common MRI sequences were contrast-enhanced T1-weighted images and T2-weighted images obtained with Fast Spin Echo pulse sequence with axial slice thickness of 1 mm. For AVM cases an additional imaging in the form of a planar digital subtraction angiography (DSA) on Axiom Artis BA machine (Siemens, Germany) with the G-frame and special angio localizer was obtained. The MRI and the DSA images were transferred to the LGP system in DICOM format and defined in the 3D space with reference to the G-frame with the help of the fiducials from the localizers embedded on the images. The sagittal and coronal planes were reconstructed from the acquired axial images within the LGP system. Following this, the tumor/lesion was contoured on any one set of images, generally axial image set, by a trained GKRS team member. The help of other image sets and other orientations was taken to draw the contours as accurately as possible. No margin was given for clinical target volume or planning target volume. In the case of AVM, the nidus was first contoured on the DSA images. This helped generate projections on the MRI set. The nidus in 3D was then contoured on the MRIs within the box projected from the DSA images.
Data acquisition
The tumor volume, as contoured on the MRIs for GKRS planning, was provided by the LGP system. The volume was a sum of cross-sectional areas of consecutive slices multiplied by the section thickness. This volume was designated as the standard volume. The maximum AP, ML and CC diameters of the tumor were recorded from the MRIs. The AP diameter was obtained from the axial image with maximum spread in AP direction. Similarly, the ML diameter was noted from the axial slice with maximum expansion in the ML direction. The maximum CC diameter was obtained from the coronal image. The product of the three diameters obtained and standard volume provided by LGP software were used for further statistical investigations.
Statistical analysis
The statistical analysis was performed using origin 9.0 software (OriginLab Corporation, Northampton, MA, USA) and Microsoft Office Excel 2007. Distribution of standard volume and product of diameters was displayed using a box plot. Furthermore, a scatter plot was obtained between the standard volumes and the product of diameters. Pearson's correlation coefficient, r was calculated to determine the existence of any linear relationship between them and determine its strength.[17,18,19] The significance of the correlation coefficient was estimated using t-test.[20,21] A best fit curve was obtained using least square deviation method. Formula to calculate the tumor volume was obtained using the value of slope m and intercept c as obtained from the best fit line. The volume obtained using the derived relationship was termed as calculated volume.
Bland-Altman (B-A) plot was used to describe the agreement between standard and calculated volumes.[22,23,24,25] The differences between the two paired volumes were plotted against their averages. Linear regression analysis was performed to check for any relationship between the differences and the averages of paired volumes. In case of significant linear relationship between the differences and magnitudes of average volumes, the B-A plot with ratios of the two volumes plotted against their averages was obtained.[26,27,28,29] Kolmogorov–Smirnov test was applied to ascertain if the differences and the ratios between the two methods of estimating volumes were distributed normally.[30] The tumor volumes estimated from the formulas derived in the present study were compared with the standard volume obtained from LGP software and the traditionally used formula for an ellipsoid. The median percentage deviation between (i) the tumor volumes calculated using the formula for an ellipsoid (ellipsoid volumes) and volumes obtained from the LGP software (standard volumes), (ii) the ellipsoid volumes and the volumes estimated in the present study (calculated volumes), and (iii) the standard volumes and the calculated volumes were calculated and compared.
RESULTS
Small and large tumor volumes
Figure 1 shows the scatter plot of standard volumes plotted against the products of diameters. The Pearson correlation coefficient between the standard volumes and the product of diameters was 0.969 (P < 0.01). This indicated a strong linear relationship between the standard volume and the product of diameters validating our method to measure the linear tumor dimensions for estimation of tumor volumes. The box plot in Figure 2 represents the distribution of standard volume and the product of the diameters obtained. The standard volumes ranged from 0.045cc to 14.9cc, with a median of 3.15cc. The mean standard volume was 3.818cc ± 3.129cc. The product of diameters ranged from 0.071cc to 35.957cc with a median of 6.730cc. The mean of the product of diameters was 7.997cc ± 6.812cc. From Figure 2, it is evident that the product of the tumor diameters needs further correction to be in agreement with the standard volume. Linear regression analysis between standard volumes and product of diameters resulted into a best fit line with an intercept value of 0.256 and slope value of 0.445 with coefficient of determination as 94.0% [Figure 1]. Based on the obtained values of slope and intercept, the formula V = 0.445 × (AP × ML × CC) + 0.256 was devised to calculate the tumor volume.
Figure 1.
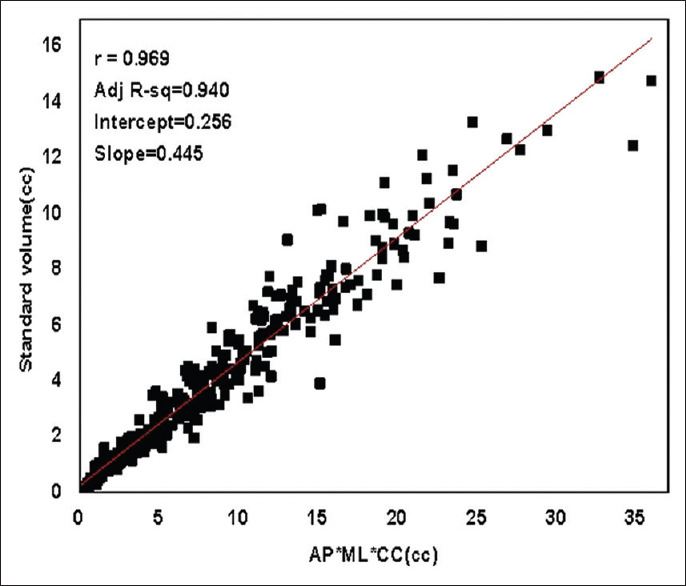
Scatter plot: Standard volumes obtained from Leksell Gamma Plan software plotted against the product of diameters
Figure 2.
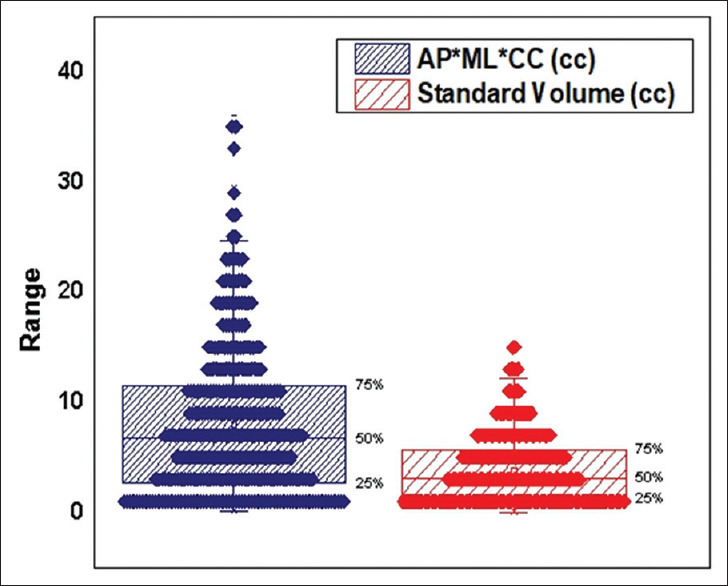
Box plot: The distribution of standard volumes and the product of diameters along with lower quartile, upper quartile and median
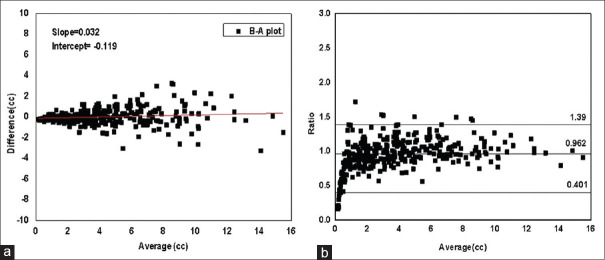
The B-A plot was generated to test the agreement between the standard and calculated volumes using two methods. In one method, the difference between the standard and the calculated volumes was plotted against their averages as shown in Figure 3a. It was observed that B-A plot started narrow and widened toward the right of the window with the increase in magnitude of averages which meant that the difference increased with volume. Linear regression analysis provided a slope value of 0.032 with an intercept value of −0.119 which depicted considerable variation of differences with volumes. Therefore, the ratios of the standard volumes to the calculated volumes were plotted against the averages of the volumes [Figure 3b]. Since the distribution of ratios was not observed to be normal, the statistical limits of agreement were found using the nonparametric method.
Figure 3.
(a) Bland-Altman plot: Differences between the standard and the calculated volume plotted against their average along with best fit line (b) Bland-Altman plot: Ratios of the standard volume to the calculated volume plotted against the averages of volumes
The median value of the ratios was observed to be 0.962 with agreement limits of 95% class interval (CI) at 0.401 and 1.39. The average bias of 0.038 was observed which implied that the standard volume differed from the calculated volume by 3.8% on an average. However, it was observed that the ratios were distributed closely around the line of equality for higher volumes, but there was considerable deviation from unity in case of relatively smaller volumes. It was therefore suggested that instead of using a common formula for all tumor sizes, two separate but simple formulas were required for the calculation of smaller and larger tumor volumes.
Tumor volume calculations
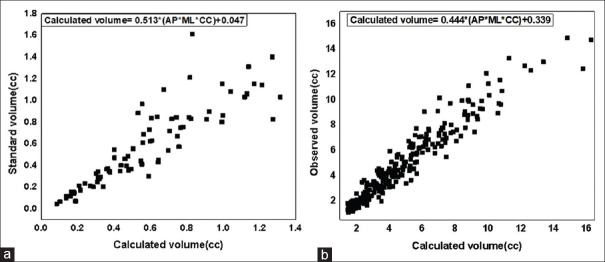
A trial and error method was used to divide the tumors into two groups based on the product of their diameters. Group 1 (small tumors) included tumors with product of diameters in the range 0–2.5cc. Pearson Correlation Coefficient between the standard volume and the product of diameters in Group 1 was calculated to be 0.883 (P < 0.01). A linear regression analysis for Group 1 provided the best fit line with a slope value of 0.513 and intercept value of 0.047 with coefficient of determination at 77.7%. The formula derived to calculate tumor volumes for Group 1 was V = 0.513 × (AP × ML × CC) + 0.047. Figure 4a shows the scatter plot of standard volume against the calculated volume. The Group 2 (large tumors) included tumors with values of the product of diameters in the range 2.5–36cc. The Pearson correlation coefficient between the standard volume and the calculated volume (product of diameters) for Group 2 was calculated to be 0.962 (P < 0.01). A linear regression analysis provided the best fit line with a slope value of 0.444 and intercept value of 0.339 with a coefficient of determination at 92.5% as shown in Figure 4b. The formula derived to calculate tumor volumes in Group 2 was V = 0.444 × (AP × ML × CC) + 0.339.
Figure 4.
(a) Scatter plot: The standard volumes plotted against the product of three diameters with best fit line displayed for product of diameters in the range 0–2.5 cc (Group 1) (b) Scatter plot: The standard volumes plotted against the product of three diameters with best fit line displayed for products of diameters in the range 2.5–36cc (Group-2)
Comparison between standard and calculated volumes
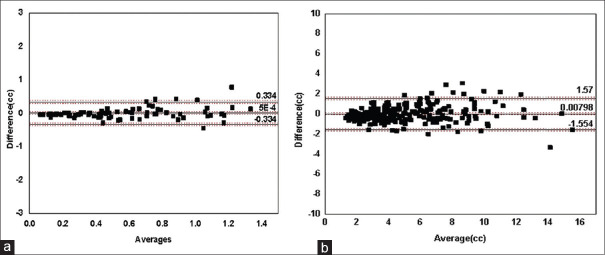
A B-A plot with a difference between standard and calculated volumes against the average of two for Group-1 is shown in Figure 5a. Since the differences were distributed normally, the agreement limits were found using parametric method. The mean difference was found to be 0.0005cc ± 0.171cc having a negligible average bias. This meant that the standard volume very closely matched the calculated volume on an average. The lower and upper limits of agreement were drawn at 0.334 and −0.334 for 95% CI. The 95% CI intervals for mean difference and agreement limits were drawn in the figure. A B-A plot with difference between the standard and the calculated volumes against the average of two for Group 2 (large tumors) with volumes in the range 2.5–36cc is shown in Figure 5b. The mean difference was found to be 0.00798cc ± 0.797 cc. This implied that the standard volume differed by 0.7% from the calculated volume on an average. The lower and upper limits of agreement were drawn at −1.554 and 1.57 for 95% CI.
Figure 5.
(a) Bland-Altman plot: Differences between the standard volumes and calculated volumes plotted against their averages for product of diameters in the range 0-2.5cc (Group 1). (b) Bland-Altman plot: Differences between the standard volume and calculated volume plotted against their averages for products of diameters in the range 2.5–36cc (Group-2)
For small tumors, mean values of standard, calculated and ellipsoid volumes were found to be 0.569cc ± 0.352cc, 0.573cc ± 0.322cc and 0.513cc ± 0.313cc, respectively, whereas for the large tumors, the corresponding means volumes were 4.792cc ± 2.921cc, 4.784cc ± 2.805cc, and 5.005cc ± 3.159cc, respectively. The calculated volumes using the formulas proposed in this study were found to closely approximate the standard volumes as compared to ellipsoid volumes for both small and large tumors. Table 1 compares the median percentage deviations between (1) the calculated and the standard volume (2) the standard and the ellipsoid volumes and (3) the calculated and the ellipsoid volumes along with their quartile values. For the small tumors, the ellipsoid volumes deviated from the standard tumor volume by 14.13%, while the volume calculated from our derived formula deviated by 11.95% from the standard volume. For the large tumors, the ellipsoid formula deviated from the standard tumor volume by 11.29% while the volume calculated by our derived formula differed only by 10.8%. However, in both the cases, larger discrepancy was observed in the case of smaller tumor volumes as compared to the larger ones.
Table 1.
Percentage deviation (median) between standard tumor volume (A), calculated volume (B) and ellipsoid volume (C)
| Percentage deviation | |(A- C)× 100/A| | |(A- B)× 100/A| | |(C-B)x 100/C| |
|---|---|---|---|
| Small tumors (%) | 14.13 (6-21) | 11.95 (5-25) | 12.19 (9-21) |
| Large tumors (%) | 11.29 (5-18) | 10.8 (5-18) | 5.41 (3-7.5) |
DISCUSSION
Figure 3a shows that the differences in tumor volumes estimated using initially derived formula in this study and the volumes observed from LGP software increased with the increasing tumor volumes. Similar increase in the differences between the volumes calculated using formula of an ellipsoid and perimeter method was observed with increasing tumor volumes in a study by Davies et al.[14] Lower values of absolute differences at small tumor volumes however may lead to misinterpretation regarding the agreement between the two techniques employed for tumor volume calculation as relative differences can be appreciable despite of small absolute differences. This necessitates separate analysis of differences in the case of small and the large size tumors. Moreover, a relatively lower value of Pearson Correlation Coefficient was obtained for small tumors. This could be because the same amount of absolute errors in linear dimension measurements would mean relatively higher errors for the small tumors as compared to the large tumors. It would magnify into still larger relative errors in tumor volume estimation for the small tumors. Similar results were also obtained by Bathla et al.[31] The measurement errors could be due to reasons such as displacement or slightly wrong placement of cursor at tumor edge during measurements. We realized that since the measurement errors are applicable to all cases, the method to calculate volumes should be different for small and large size tumors.
For Group 1 (small) tumors, only one data point was found to be outside the agreement limits with standard volume greater than the volume calculated by our formula. It corresponded to the case of a postoperated VS. As observed on its MRIs the lesion had highly irregular shape due to prior surgery. The small size and shape might have led to increased errors in measurement of linear tumor dimensions. Three major outliers were observed in Group 2. In two cases the calculated volumes overestimated the true volumes. These were residual pituitary adenoma tumors after at least two prior surgeries with highly irregular shapes. The third was a case of Meningioma for which the true volume was underestimated by the applied formula. This also was a residual tumor after surgery whose shape was almost like a flat disc or discoid. Thus, we observed that highly irregular or discoid shaped tumors continued to pose challenge for tumor volume estimation.
The median percentage deviations between the volumes calculated using the standard method and the formula of ellipsoid as reported in literature is 36% (quartiles 16%–46%) which is considerably larger than the deviations obtained by us.[11] This is due to the difference in the methodology adopted for the measurement of tumor dimensions.
Accurate measurement of tumor dimensions is a source of error as it depends on the personal judgment of the observer apart from many other factors. Sticking to the proposed protocol for measuring tumor dimensions may help in substantially mitigating the errors associated with measurement of linear dimensions on CT/MRIs.
CONCLUSION
Consistency and accuracy in measuring tumor dimensions and estimating tumor volumes from them are required for not only deciding the appropriate treatment strategy but also for assessing the posttreatment responses and for maintaining uniformity in reporting of the results. To improve reproducibility in the measurement of tumor linear dimensions on CT/MRIs, we have proposed a simple method. A strong correlation was found between the products of measured linear dimensions and the volume obtained from the LGP software validating our method for the measurement of tumor dimensions. One unique formula was not feasible for accurately calculating tumor volumes for the range and shape of tumor volumes encountered in our study. Therefore, we have derived two separate formulas-one for small tumors and the other for large tumors. The formulas derived in this study are simple enough to be used in clinical settings. Highly lobulated and flat/elongated tumors that are considered less amenable to SRS are also the tumors that pose challenge for tumor volume estimation. We believe that the proposed method for measurement of intra-cranial tumor dimensions and the formulas for their volume estimation will find acceptability for clinical decision-making in cranial SRS.
Financial Support and Sponsorship
Nil.
Conflicts of Interest
There are no conflicts of interest.
REFERENCES
- 1.Kirpatrick JP, Soltys SG, Lo SS, Beal K, Shrieve DC, Brown PD. The radiosurgery fractionation quandary: Single fraction or hypofractionation? Neuro Oncol. 2017;19(Supp 2):ii38–49. doi: 10.1093/neuonc/now301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seung SK, Larson DA, Galvin JM, Mehta MP, Potters L, Schultz CJ, et al. American college of radiology (ACR) and American society for radiation oncology (ASTRO) practice guideline for the performance of stereotactic radiosurgery (SRS) Am J Clin Oncol. 2013;36:310–5. doi: 10.1097/COC.0b013e31826e053d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahgal A, Kellett S, Ruschin M, Greenspoon J, Follwell M, Sinclair J, et al. Stereotactic radiosurgery for brain metastasis guideline development Group. A cancer care Ontario organizational guideline for the delivery of stereotactic radiosurgery for brain metastasis in Ontario, Canada. Pract Radiat Oncol. 2020;10:243–54. doi: 10.1016/j.prro.2019.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Potters L, Steinberg M, Rose C, Timmerman R, Ryu S, Hevezi JM, et al. American Society for therapeutic radiology and oncology and American College of Radiology practice guideline for the performance of stereotactic body radiation therapy. Int J Radiat Oncol Biol Phys. 2004;60:1026–32. doi: 10.1016/j.ijrobp.2004.07.701. [DOI] [PubMed] [Google Scholar]
- 5.Sreenivasan SA, Madhugiri VS, Sasidharan GM, Kumar RV. Measuring glioma volumes: A comparison of linear measurement based formulae with the manual image segmentation technique. J Cancer Res Ther. 2016;12:161–8. doi: 10.4103/0973-1482.153999. [DOI] [PubMed] [Google Scholar]
- 6.Wheatly JM, Rosenfield NS, Heller G, Feldstein D, LaQuaglia MP. Validation of a technique of computer-aided tumour volume determination. J Surg Res. 1995;59:621–6. doi: 10.1006/jsre.1995.1214. [DOI] [PubMed] [Google Scholar]
- 7.Ytre-Hauge S, Husby JA, Magnussen IJ, Werner HM, Salvesen ØO, Bjørge L, et al. Preoperative tumor size at MRI predicts deep myometrial invasion, lymph node metastases, and patient outcome in endometrial carcinomas. Int J Gynecol Cancer. 2015;25:459–66. doi: 10.1097/IGC.0000000000000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Routman DM, Bian SX, Diao K, Liu JL, Yu C, Ye J, et al. The growing importance of lesion volume as a prognostic factor in patients with multiple brain metastases treated with stereotactic radiosurgery. Cancer Med. 2018;7:757–64. doi: 10.1002/cam4.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pshak TJ, Cho DS, Hayes KL, Vemulakonda VM. Correlation between CT-estimated tumour volume, pathologic tumour volume, and final pathologic specimen weight in children with Wilms' tumour. J Pediatr Urol. 2014;10:148–54. doi: 10.1016/j.jpurol.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Sorensen AG, Patel S, Harmath C, Bridges S, Synnott J, Sievers A, et al. Comparison of diameter and perimeter methods for volume calculation. J Clin Oncol. 2001;19:551–7. doi: 10.1200/JCO.2001.19.2.551. [DOI] [PubMed] [Google Scholar]
- 11.Dachman AH, MacEneaney PM, Adedipe A, Carlin M, Schumm LP. tumor size on computed tomography scans: Is one measurement enough? Cancer. 2001;91:555–60. [PubMed] [Google Scholar]
- 12.Feldman JP, Goldwasser R, Mark S, Schwartz J, Orion I. A Mathematical model for tumour volume evaluation using two dimensions. J.Appl Quant Methods. 2009;4:455–62. [Google Scholar]
- 13.Mayr NA, Taoka T, Yuh WT, Denning LM, Zhen WK, Paulino AC, et al. Method and timing of tumor volume measurement for outcome prediction in cervical cancer using magnetic resonance imaging. Int J Radiat Oncol Biol Phys. 2002;52:14–22. doi: 10.1016/s0360-3016(01)01808-9. [DOI] [PubMed] [Google Scholar]
- 14.Davies BM, Carr E, Soh C, Gnanalingham KK. Assessing size of pituitary adenomas: A comparison of qualitative and quantitative methods on MR. Acta Neurochirurgica. 2016;158:677–83. doi: 10.1007/s00701-015-2699-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu YL, Lee MS, Juan CJ, Hueng DY. Calculating the tumor volume of acoustic neuromas: Comparison of ABC/2 formula with planimetry method. Clin Neurol Neurosurg. 2013;115:1371–4. doi: 10.1016/j.clineuro.2012.12.029. [DOI] [PubMed] [Google Scholar]
- 16.Ishi Y, Terasaka S, Yamaguchi S, Yoshida M, Endo S, Kobayashi H, et al. Reliability of the size evaluation method for meningiomas: Maximum Diameter, ABC/2 Formula, and Planimetry Method. World Neurosurg. 2016;94:80–8. doi: 10.1016/j.wneu.2016.06.108. [DOI] [PubMed] [Google Scholar]
- 17.Bewick V, Cheek L, Ball J. Statistics review 7: Correlation and regression. Crit Care. 2003;7:451–9. doi: 10.1186/cc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor R. Interpretation of the correlation coefficient: A basic review. J Diagn Med Sonogr. 1990;6:35–9. [Google Scholar]
- 19.Havlicek LL, Peterson NL. Robustness of the pearson correlation against violations of assumptions. Percept MotSkills. 1976;43:1319–34. [Google Scholar]
- 20.Courses Lumenlearning Testing the Significance of Correlation Coefficient. [[Last accessed on 2019 Dec 10]]. Available from: https://courses.lumenlearning.com/introstats1/chapter/ testing-the-significance-of-the-correlation-coefficient.
- 21.Asuero AG, Sayago A, González AG. The correlation coefficient: An overview. Crit Rev Anal Chem. 2006;36:41–59. [Google Scholar]
- 22.Giavarina D. Understanding bland Altman analysis. Biochem Med. 2015;25:141–51. doi: 10.11613/BM.2015.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–10. [PubMed] [Google Scholar]
- 24.Krouwer JS. Why bland-Altman plots should use X, not (Y + X)/2 when X is a reference method. Stat Med. 2008;27:778–80. doi: 10.1002/sim.3086. [DOI] [PubMed] [Google Scholar]
- 25.Bland JM, Altman DG. Comparing methods of measurement: Why plotting difference against standard method is misleading. Lancet. 1995;346:1085–7. doi: 10.1016/s0140-6736(95)91748-9. [DOI] [PubMed] [Google Scholar]
- 26.Chhapola V, Kanwal SK, Brar R. Reporting standards for Bland-Altman agreement analysis in laboratory research: A cross-sectional survey of current practice. Ann Clin Biochem. 2015;52:382–86. doi: 10.1177/0004563214553438. [DOI] [PubMed] [Google Scholar]
- 27.Abu AA, Jordan H, Drummond G. Reporting of method comparison studies: A review of advice, an assessment of current practice, and specific suggestions for future reports. Br J Anaesth. 2016;117:569–75. doi: 10.1093/bja/aew320. [DOI] [PubMed] [Google Scholar]
- 28.Medcalc. [[Last accessed on 2019 Dec 10]]. Available from: https://www.medcalc.org/manual/ blandaltman.php.
- 29.Robert JF, Andrew T, Ian DW, Robin W. Basic Laboratory operations. In: Flanagan RJ, Taylor A, Watson ID, Whelpton R, editors. John Wiley & Sons, Inc. Hoboken, NJ, USA: Fundamentals of Analytical Toxicology; 2008. pp. p.353–98. [Google Scholar]
- 30.Stephens MA. Use of the kolmogorov-smirnov, cramér-von mises and related statistics without extensive tables. J R Stat Soc B. 1970;32:115–22. [Google Scholar]
- 31.Bathla G, Policeni B, Hansen MR, Berbaum K. Calculating the Tumor Volumes in Vestibular Schwannomas: Are the ABC/2 and Volumetric Methods Comparable? Otol Neurotol. 2017;38:889–94. doi: 10.1097/MAO.0000000000001423. [DOI] [PubMed] [Google Scholar]