SUMMARY
Mouse embryonic stem cells (ESCs) sporadically express preimplantation two-cell-stage (2C) transcripts, including MERVL endogenous retrovirus and Zscan4 cluster genes. Such 2C-like cells (2CLCs) can contribute to both embryonic and extraembryonic tissues when reintroduced into early embryos, although the molecular mechanism underlying such an expanded 2CLC potency remains elusive. We examine global nucleosome occupancy and gene expression in 2CLCs and identified miR-344 as the noncoding molecule that positively controls 2CLC potency. We find that activation of endogenous MERVL or miR-344-2 alone is sufficient to induce 2CLCs with activation of 2C genes and an expanded potency. Mechanistically, miR-344 is activated by DUX and post-transcriptionally represses ZMYM2 and its partner LSD1, and ZMYM2 recruits LSD1/HDAC corepressor complex to MERVL LTR for transcriptional repression. Consistently, zygotic depletion of Zmym2 compromises the totipotency-to-pluripotency transition during early development. Our studies establish the previously unappreciated DUX-miR-344-Zmym2/Lsd1 axis that controls MERVL for expanded stem cell potency.
Keywords: Endogenous retrovirus, MERVL, miR-344, Zmym2, Lsd1, Dux, Gata2, 2C-like cells, totipotency
eTOC Blurb
Wang and colleagues demonstrate that expanded stem cell potency can be obtained by endogenous activation of MERVL or miR-344. Mechanistically, miR-344, a direct transcriptional target of DUX, activates endogenous MERVL via repressing downstream target Zmym2 that directly binds to MERVL LTRs and recruits HDAC corepressors for transcriptional repression.
Graphical Abstract
INTRODUCTION
Mouse embryonic stem cells (ESCs) are derived from the inner cell mass (ICM) of blastocy-ststage embryos and considered “pluripotent” owing to their ability to contribute to all three germ layers of the embryo, but rarely to the extraembryonic tissues. In contrast, totipotent cells, such as zygote and 2-cell-stage (2C) blastomeres in vivo, can generate both the embryo proper and the extraembryonic tissues from a single cell (Tarkowski, 1959). A small subset of ESCs, known as 2C-like cells (2CLCs), are also found to have an expanded potency (Macfarlan et al., 2012). Unlike pluripotent ESCs, 2CLCs arise spontaneously in ESC cultures (1~5%) at any given time (Dan et al., 2013; Macfarlan et al., 2012) and are characterized by activation of major satellites (Borsos and Torres-Padilla, 2016; Dang-Nguyen and Torres-Padilla, 2015) and endogenous retroviral (ERV) elements (Lu and Zhang, 2015). ERVs contribute a significant portion of the transcripts to promoting zygote genome activation (ZGA) (Gifford et al., 2013). Particularly, MERVL is actively transcribed exclusively in the 2C embryo together with a group of 2C-specific genes including the Zscan4 gene family, likely through epigenetic mechanisms such as DNA methylation and histone modifications involving DNMT (Eckersley-Maslin et al., 2016) and LSD1 (Ancelin et al., 2016; Wasson et al., 2016) (Macfarlan et al., 2011; Wang et al., 2009).
During maternal-to-zygotic transition, miRNAs have been reported to accelerate the deadenylation and decay of maternal mRNAs, facilitating ZGA and the establishment of novel cellular states in Xenopus, Drosophila and Zebrafish (Giraldez, 2010). In mouse, miR-34a was recently found to control pluripotency of ESCs through post-transcriptional repression of Gata2, a transcriptional activator of MERVL. miR-34a knockout ESCs upregulate MERVL and 2C genes with an expanded developmental potency in chimeric embryos (Choi et al., 2017). These findings establish the negative regulatory role of miR-34a in suppressing totipotency features in ESCs. Conversely, recent studies reveal a positive role of DUX (DUX4 in human) in activating mammalian embryonic genome chromatin landscape as well as 2C-specific genes and repeat elements (De Iaco et al., 2017; Hendrickson et al., 2017). The molecular events downstream of DUX and the potential crosstalk between miRNA and DUX pathways are not known.
In this study, we establish for the first time a causative role of MERVL activation in contributing to the expanded potency of 2CLCs. We discovered and established miR-344 as an important positive regulator of MERVL in 2CLCs and identified a previously unappreciated molecular axis of DUX→miR-344--|ZMYM2/LSD1--|MERVL invoking transcriptional and posttranscriptional mechanisms underlying MERVL control for expanded stem cell potency.
RESULTS
Endogenous MERVL activation induces 2C-like cells (2CLCs)
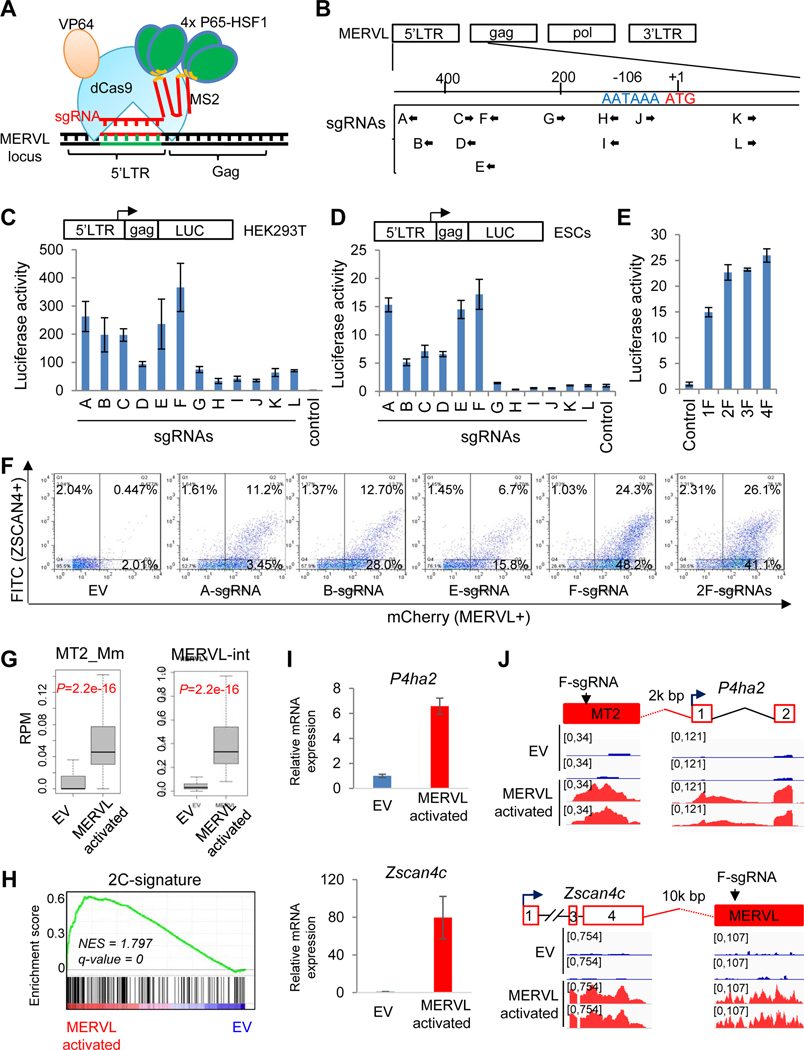
MERVL is a member of Class III ERVs and is present in more than 650 full-length copies in the mouse genome (Schoorlemmer et al., 2014). To address whether MERVL activation is a driver or a byproduct of the totipotent state in developing embryos or in 2CLCs, we employed the CRISPR synergistic activation mediator (SAM) (CRISPRSAM hereafter), composed of dCas9-VP64 and helper MS2-P65-HSF1 (Konermann et al., 2015) (Figure 1A) and sgRNAs targeting the 730-bp fragment that was previously reported to recapitulate MERVL expression (Macfarlan et al., 2011), to achieve the activation of MERVL repeats in ESC line co-expressing MERVL-tdTomato (Macfarlan et al., 2012) and pZscan4c-EGFP (Dan et al., 2013) fluorescent reporters (double reporters; DR) (Figures 1B–E and S1A-B) (see Star* Methods for details). We found single F- and double 2F-sgRNA treatments resulted in the highest percentages of MERVL+ (~70%) as well as double positive (DR+/+, ~25%) populations by fluorescence activated cell sorting (FACS) analysis, compared with an empty vector (EV) control or other sgRNAs treated cells (Figure 1F). These sgRNA-activated ESCs were maintained for three passages with high ratios of Zscan4c+ (Figure S1C) and MERVL+ populations (Figure S1D). F-sgRNA-activated ESCs were then FACS-sorted into DR+/+ and DR−/− populations and subsequently replated separately (Figure S1E). FACS-sorted DR+/+ and DR−/− cells fluctuate (Figure S1F), which is consistent with the fluctuating nature of 2CLC population in ESCs (Macfarlan et al., 2012; Zalzman et al., 2010). Remarkably, 65.9% of the sorted DR+/+ cells still maintained MERVL+ state, whereas 36.0% of the sorted DR−/− cells reached MERVL+ state in 3 days after FACS sorting (Figure S1G; the total of yellow and red bars), which is far more abundant than the typical 1~5% fluctuating 2CLCs observed in conventional ESCs.
Figure 1. MERVL activation is sufficient to induce 2C-like cells.
(A) Illustration of CRISPR/sgRNA-directed synergistic activation mediator (SAM) system (CRISPRSAM) for the activation of MERVL.
(B) Schematic depiction of the MERVL structure showing 12 potential sgRNA target sites (A-L) within 730 bp of the 5’ LTR and partial gag sequence.
(C-D) Effects of sgRNAs for MERVL activation by CRISPRSAM in HEK293T cells (C) and mESCs (D). Fold activation denotes relative luciferase activity normalized to an empty sgRNA expression vector. Data are presented as average ± SD.
(E) Multiplex expression of F sgRNAs increases MERVL activation in mESCs. Fold activation denotes relative luciferase activity normalized to an empty sgRNA expression vector. Data are presented as average ± SD.
(F) FACS profiles of MERVL+ (tdTomato) and Zscan4c+ (GFP) population after MERVL activation using different sgRNAs as indicated.
(G) Box plots showing expression (reads per million, RPM) of the MERVL solo LTR (left) and internal region (right) in empty vector (EV) and MERVL-activated ESCs.
(H) GSEA indicating that upregulated genes by F-sgRNA activation were highly enriched in the 2-cell embryo gene set. Red, upregulated genes; blue, downregulated genes.
(I) RT-qPCR of 2C-specific gene P4ha2 and Zscan4c expression in the empty vector (EV) and MERVL-activated ESCs. Data are presented as average ± SD.
(J) Examples of induced expression of MERVL-proximal genes P4ha2 and Zscan4c in MERVL-activated ESCs compared with empty vector (EV) treated cells.
To molecularly characterize these MERVL-activated ESCs, we profiled the transcriptomes of CRIPSPRSAM/2F-sgRNA-activated ESCs and empty vector (EV)-transfected ESCs using RNA sequencing (RNA-seq). Expression of total ERV elements was slightly downregulated (Figure S1H top). However, expression of ERVL class, including MERVL family solo LTR promoters (MT2_Mm) and internal regions (MERVL-int), was significantly upregulated in MERVL-activated ESCs (Figures 1G and S1H bottom). RT-qPCR confirmed that retrotransposon induction in ESCs was specific to the MERVL family but not other repetitive elements such as intracisternal A-particle (IAP), long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), or MMERGLN (Figure S1I). From the RNA-seq data, we identified 130 downregulated and 924 upregulated genes (fold-change>2, P<0.05) in F-sgRNA activated ESCs (Table S2), suggesting an overall effect of transcriptional activation by F-sgRNA. F-sgRNA activation also led to the enrichment of the geneset associated with 2-cell embryo development (Wu et al., 2016) (Figure 1H). MERVL LTRs can be co-opted as functional promoters or enhancers for nearby coding genes (Macfarlan et al., 2012). Indeed, we identified F-sgRNA binding sites at solo LTR (MT2) or entire MERVL regions, which may serve as regulatory elements for nearby genes, such as P4ha2, Zscan4c (Figures 1I–J), Zfp352, Prelid2, and Ddit4l (Figure S1J), leading to their transcriptional activation by CRISPRSAM.
Together, our data demonstrate that MERVL activation, which leads to the activation of neighboring genes like Zscan4, P4ha2, and Zfp352, as well as other 2C-specific genes, is causative to the induction of 2CLCs.
Discovery of miR-344 cluster miRNAs as totipotency-associated miRNAs
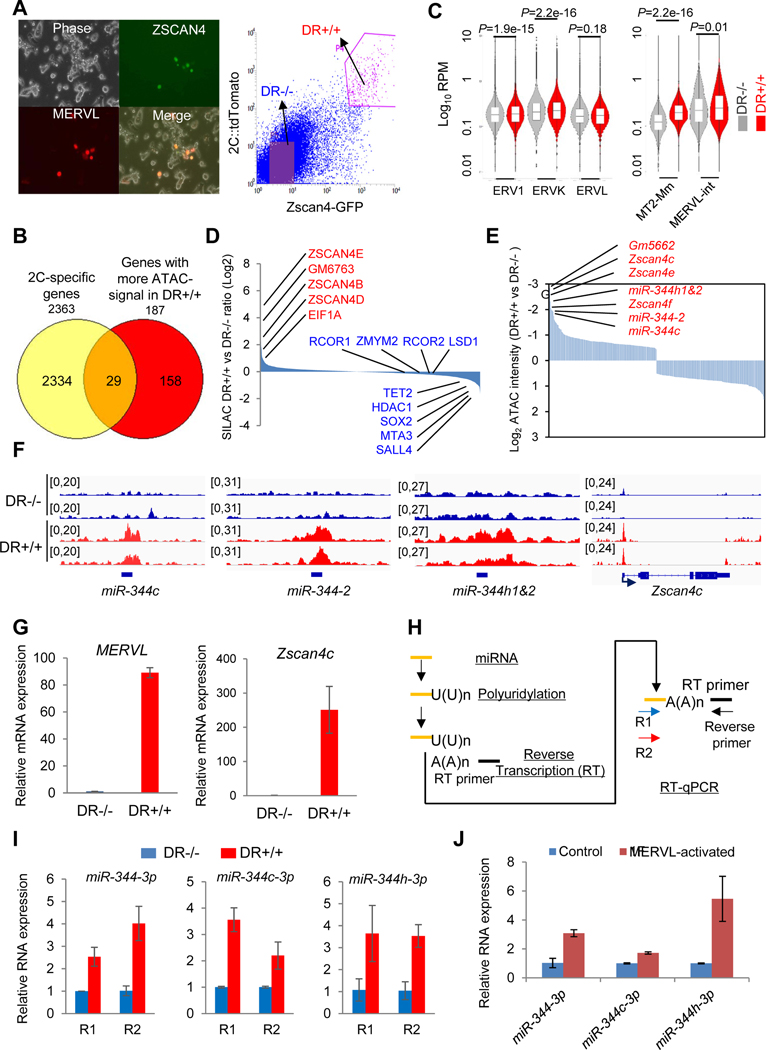
The causative role of MERVL activation in inducing 2CLCs in ESCs prompted us to search for endogenous regulators of MERVL activation underlying 2CLC expanded potency. To identify factors/genes that control 2CLCs in ESCs, we sorted DR+/+ and DR−/− cells from our double reporter line (Figure 2A) for profiling the genome-wide accessibility of open chromatin by ATAC-seq (Buenrostro et al., 2013; Buenrostro et al., 2015). We annotated 377 differentially enriched peaks (188 peaks enriched in DR+/+, 189 peaks enriched in DR−/−) in the mouse genome, identified 171 protein-coding genes and 16 ncRNAs (187 genes in total) that have more open-chromatin in DR+/+ cells, and 178 protein-coding genes and 9 ncRNAs that have more open-chromatin in DR−/− cells. For the 187 genes with more ATAC signaling in DR+/+ cells, 29 out of 187 (15.5%) genes are in the list of known 2C-specific genes (Figure 2B, Table S1). Gene ontology (GO) analysis reveals that genes with high peak intensities in DR+/+ are related to metabolism and RNA regulation, whereas those with low peak intensities in DR+/+ are mainly involved in organ development (Figure S2A). The enrichment of the GO term on metabolism in 2CLCs is consistent with the presence of vigorous metabolic activity triggering ZGA at the 2-cell stage (Zhang et al., 2018). We also observed an overall increase in chromatin accessibility in DR+/+ relative to DR−/− cells across different ERV classes, particularly notable for MERVL-LTR MT2_Mm (Figure 2C). As expected, the 2C genes like Zscan4 cluster genes are strongly enriched for ATAC signals in the DR+/+ cells (Figure S2B).
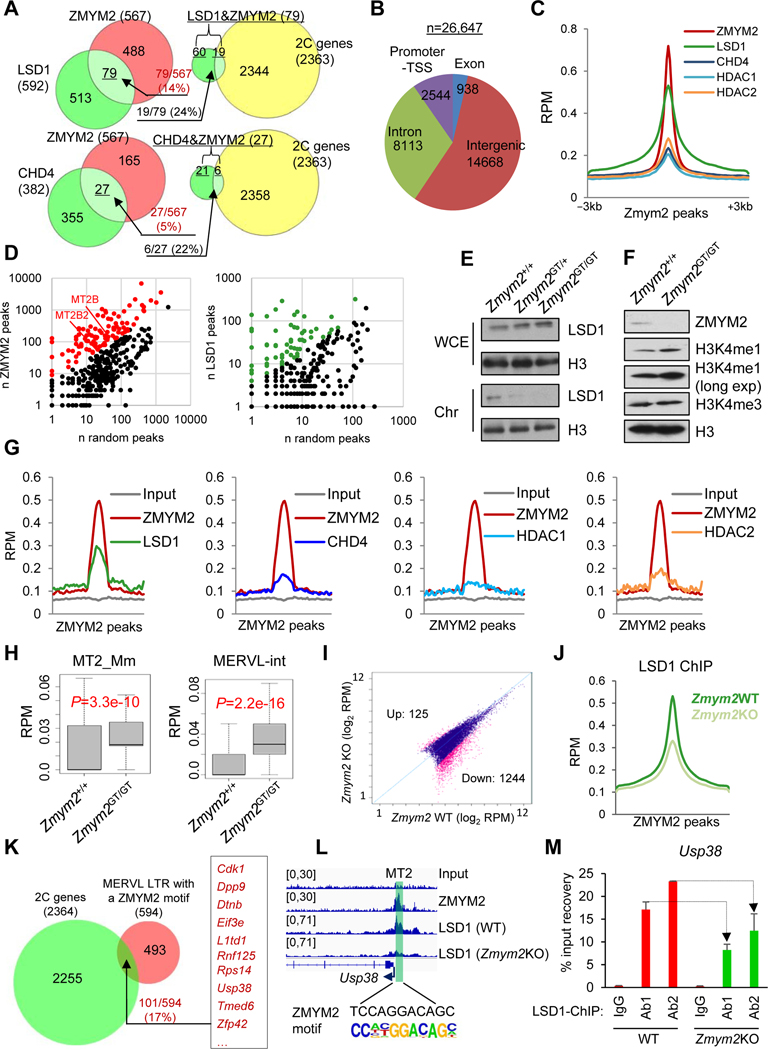
Figure 2. Discovery of miR-344 as a totipotency-associated miRNA.
(A) ESCs co-transfected with pZscan4c-GFP and 2C::tdTomato. (left) Phase contrast and fluorescence microscope images. (right) FACS plots of the pZscan4c-GFP and MERVL::tdTomato dual-reporter ESC line (DR+/+). Scale bars, 250 μm.
(B) Venn diagram for shared genes between 2C-specific genes (Macfarlan et al., 2012) and genes with significantly higher ATAC-signal in DR+/+ population.
(C) Chromatin accessibility of different genomic features determined by ATAC-seq analysis in DR−/− (gray) and DR+/+ (red) cells. Bars represent mean levels of accessibility. P-value is from Mann-Whitney test.
(D) Relative abundance of proteins in DR+/+ versus DR−/− cells identified by SILAC-MS.
(E) Open chromatin states of Zscan4 and miR-344 cluster gene loci in DR+/+ cells identified by ATAC-seq.
(F) Representative genomic regions (miR-344-2/c/h and Zscan4c) with higher ATAC-seq intensities (reads per million, RPM) in DR+/+ (red) cells.
(G) Relative expression of MERVL and Zscan4c in DR+/+ and DR−/− cells by RT-qPCR. Data are presented as average ± SD, relative to the levels in DR−/− cells.
(H) Strategy for mature miRNA measurement by RT-qPCR. Two miR-344-specific primers (R1/R2) and a common reverse primer are used to determine each miRNA expression.
(I) Relative expression levels of mature miR-344-3p, miR-344c-3p, and miR-344h-3p in DR+/+ versus DR−/− cells by RT-qPCR. Data are presented as average ± SD, relative to U6 control, and normalized to the levels in DR−/− cells.
(J) Relative expression levels of mature miR-344-3p, miR-344c-3p, and miR-344h-3p in control and MERVL-activated ESCs by F-sgRNA. Data are presented as average ± SD.
Next, we employed a stable isotope labeling by amino acids in cell culture (SILAC) method and liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) technique (Mann, 2006) to profile the relative protein abundance in DR+/+ and DR−/− cells (Figure S2C). A total of 2,484 proteins were quantified, with an FDR cutoff of 0.01 (Table S1). We found that 2C gene products such as ZSCAN4 family members and EIF1A are upregulated in DR+/+ cells, whereas pluripotency factors such as SALL4 and SOX2 are downregulated in DR+/+ cells (Figure 2D). We also found a higher expression level of TET2 in DR−/− population (Figure 2D), which is consistent with its role in repressing MERVL in ESCs through a novel post-transcriptional mechanism that we defined recently (Guallar et al., 2018). By combining the SILAC protein expression and ATAC-seq data, we found a positive correlation of higher expression of ZSCAN4 family proteins (Figure 2D) with more open chromatin signals (Figure 2E) in DR+/+ cells.
Interestingly, we also identified noncoding miR-344 cluster genes, including miR-344-h1&2, miR-344-2, and miR-344c, that are ranked as top ATAC signal enriched loci in DR+/+ cells, together with 2C genes Zscan4 and Gm5662 (Figures 2E–F and S2D), suggesting a positive role of this miRNA cluster in the regulation of 2CLCs. Similar to MERVL and other 2C genes (Figures 2G and S2E), mature miR-344-3p (miR-344, derived from both miR-344-1 and miR-344-2), miR-344c-3p (derived from miR-344c), and miR-344h-3p (derived from miR-344h1&2) miRNAs are all highly expressed in DR+/+ cells (Figures 2H–I) as well as in MERVL-activated ESCs (Figure 2J). From a published miRNA-array dataset in early embryo development (Liu et al., 2012), we found that miR-344 expression increased from pronucleus to 2C-8C stages and peaked at the 8C stage, and then downregulated in later stages (Figure S2F). In contrast, MERVL/MT2 similarly increased from pronucleus to 2C stages but downregulated immediately thereafter (Figure S2G) (Xue et al., 2013), suggesting a relatively tighter control of MERVL/MT2 during early development.
Together, these data establish a positively correlated dynamic control of both miR-344 and MERVL/MT2, i.e., upregulation in totipotent cells and downregulation in pluripotent cells, during the totipotency-to-pluripotency transition.
Activation of miR-344 promotes 2CLCs in ESCs with an in vivo expanded potency
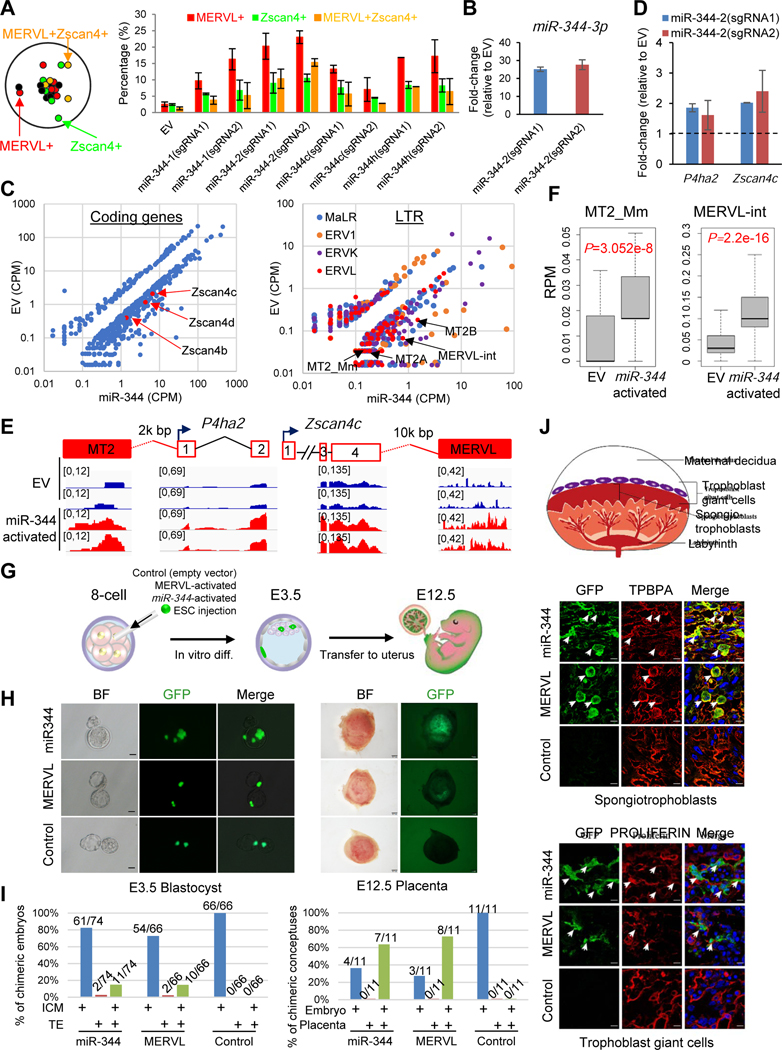
Given the high expression of miR-344 cluster in DR+/+ and MERVL-activated ESCs in culture as well as in totipotent 2C-to-8C cells in vivo (Figures 2J and S2F–G), we employed the same CRISPRSAM strategy to activate endogenous miR-344 genes in ESCs and test whether miR-344 activation could induce 2CLCs with an expanded potency. We designed 12 sgRNAs to activate 6 individual miR-344 genes (2 sgRNAs for each gene), including miR-344-1/2/c/h with enriched ATAC signal in DR+/+ cells (Figure 2E), and miR-344-d/f with no enrichment as negative controls (Figure S3A). Indeed, we found that only miR-344-1/2/c/h activation led to a dramatic upregulation of DR+/+ population (Figure S3B). Notably, we found a higher MERVL+ population than Zscan4c+ population by miR-344-1/2/c/h activation (Figure 3A), suggesting a more specific role of MERVL activation by these miRNAs. RT-qPCR also confirmed higher expression of mature miR-344-3p, miR-344c-3p, and miR-344h-3p in sgRNA-activated ESCs, indicating efficient activation and processing of these miRNAs upon CRISPRSAM (Figures 3B and S3C). Hereafter, we particularly studied effects of miR-344-2 activation (which produces mature miR-344) because of its highest activation of MERVL+ and DR+/+ populations (Figure 3A). By comparing the transcriptomes between miR-344-activated and EV-transfected ESCs, we found that many coding genes (Figure 3C, left panel, Table S2) and non-coding LTR elements (Figure 3C, right panel, Table S2) were differentially expressed, particularly upregulated, among which are Zscan4 family members and MERVL LTRs (MT2A/B), respectively. There are 182 genes (P=9.605e-143, Chi-square test) shared between the 344 and 924 up-regulated genes (fold-change>2, P<0.05) in miR-344-activated ESCs and F-sgRNA-MERVL-activated ESCs, respectively, among which 23 (12.6%) are in the list of 2C-specific genes and 46 (25.3%) genes are upregulated in Lsd1 knockout ESCs from the previous study (Figure S3D, Table S2) (Macfarlan et al., 2012). Higher expression levels of MERVL-nearby 2C genes P4ha2 and Zscan4c in miR-344-activated ESCs were further confirmed by RT-qPCR (Figure 3D). miR-344 activation significantly upregulates the expression of the total ERVs as well as the class of ERVL (Figure S3E), especially the solo LTR promoters (MT2_Mm) or internal regions (MERVL-int) of MERVL family (Figure 3F). As expected, a significant enrichment of the geneset associated with 2-cell embryo development was also observed in miR-344-activated ESCs (Figure S3F). Next, we examined the expression levels of the pluripotency and 2C genes in MERVL- or miR-344-activated ESCs compared to those in spontaneous 2CLCs or 2C embryos. We sorted the DR+/+ population from untreated ESCs, MERVL- or miR-344- activated ESCs, then performed RNA-seq analyses of both bulk and DR+/+ populations, as well as 2C embryos.
Figure 3. Endogenous miR-344 activation by CRISPRSAM induces 2C-like cells with expanded potency in vivo.
(A) FACS quantification of the proportion of MERVL+, Zscan4+ and MRVL+Zscan4+ (DR+/+) cells upon activation by sgRNAs targeting miR-344-1/2/c/h.
(B and D) RT-qPCR for relative expression of mature miR-344-3p (B) or P4ha2 and Zscan4c expression (D) in miR-344-2-activated ESCs. Data are presented as average ± SD, relative to U6 (B), and each normalized to the level in the empty vector.
(C) Scatter plots of transcript profiles of coding genes (left) and noncoding LTRs (right) in miR-344-2-activated ESCs relative to the empty vector (EV) control from RNA-seq data.
(E) Induced expression of MERVL-proximal genes P4ha2 and Zscan4c in miR-344-2-activated ESCs compared with empty vector (EV) treated cells. RNA-seq peaks were shown.
(F) Expression (RPM, reads per million) of the MERVL solo LTR (left) and internal region (right) in empty vector (EV) and miR-344-2-activated ESCs. P-value is from Mann-Whitney test.
(G) Procedure for injecting single GFP-labeled ESC into 8-cell embryo, followed by in vitro culture until the blastocyst stage (E3.5), then transferred back to female uterus till E12.5.
(H-J) Images of injected GFP-labeled MERVL-/miR-344-activated, or control EV transfected mESCs differentiating to the inner cell mass (ICM) and trophectoderm (TE) in E3.5 blastocysts (H; left, scale bars, 20 μm) and E12.5 placenta (H; right, scale bars, 1 mm) or to spongiotrophoblasts and trophoblast giant cells in the placentae with so-staining of TPBPA and PROLIFERIN (J; scale bars, 20 μm) in chimera assay. The total number of embryos and numbers of chimeric embryos with contributions to TE, ICM, and TE&ICM at E3.5, and numbers of chimeric conceptuses with contributions to embryonic tissue or placentas at E12.5 were summarized in I.
Expression of pluripotency genes Pou5f1 and Nanog were highly expressed in all samples except for 2C embryos. In contrast, 2C genes Zscan4c and Zfp352 were extremely low in bulk untreated ESCs, but highly expressed in DR+/+ population of untreated ESCs and in both bulk and DR+/+ populations of MERVL- and miR-344-activated ESCs (Figure S3G). Expression of Zfp352 in these 2CLCs was, however, much lower than that in 2C embryos (Figure S3G). Together, these data indicate that direct activation of miR-344 can promote 2CLCs in ESCs with activation of MERVL/2C genes, and also suggest there may exist intrinsic differences in certain 2C gene expression among variant 2CLCs and 2C embryos.
To conclusively address whether those miR-344- and MERVL-activated ESCs are indeed 2CLCs with an expanded developmental potency, we labeled miR-344-/MERVL-activated and control ESCs by GFP expression using a GFP-sgRNA-MS2 or a control GFP-(No-sgRNA)-MS2 vector (Figure S1A), respectively, and injected single (or up to 3) ESC into an 8-cell embryo to test their contribution to embryonic and extraembryonic tissues (Figure 3G). 14.9% (11/74) and 15.1% (10/66) of recovered blastocysts (E3.5) showed concomitant TE and ICM differentiation with injection of miR-344- and MERVL-activated ESCs, respectively, which is strikingly different from that 0% (0/66) of recovered blastocysts showed concomitant TE and ICM with injection of control ESCs (Figures 3H–I, left panels). To further examine the developmental potency of those 2CLCs, chimeric embryos were transferred to uteruses of pseudo-pregnant females and examined at E12.5. The control ESCs only gave rise to embryonic tissue, but not placenta, in chimeric conceptuses. On the other hand, the injected miR-344- and MERVL-activated ESCs differentiate to cells of both embryos and placentas at E12.5 (Figures 3H–I, right panels). Immunostaining of TPBPA and PROLIFERIN combined with GFP fluorescence revealed a clear regional distribution of spongiotrophoblasts and giant trophoblasts, respectively, in the placenta (Figure 3J) and that GFP signals were mostly present in the trophoblast lineages but not decidual region (Figure S3H), demonstrating the expanded developmental potency of miR-344- and MERVL-activated 2CLCs.
miR-344 directly represses Zmym2/Lsd1 to mediate MERVL induction
To understand how miR-344 regulates MERVL and 2C genes for the expanded potency, we hypothesize that miR-344 may mediate post-transcriptional repression of the repressors for MERVL/2C genes in 2CLCs. We evaluated the miR-344 putative targets identified by TargetScan (Agarwal et al., 2015), and were particularly interested in two targets of miR-344: ZMYM2 and LSD1 for the following reasons. First, from our RNA-seq data, Zmym2 is downregulated (ratio=0.579, P=0.0005, Table S2) upon miR-344-activation in ESCs. Second, ZMYM2 was reported to stabilize the HDAC-containing LSD1-CoREST (RCOR1) corepressor complex on the chromatin (Gocke and Yu, 2008) and both LSD1 and HDAC1 were identified as top RNAi hits in repressing MERVL 2C::tdTomato reporter (Li et al., 2017), although ZMYM2 itself was not present in the shRNA library of that study (Li et al., 2017). Third, another genome-wide RNAi study also identified ZMYM2 as an ERV silencer (Yang et al., 2015). However, the detailed molecular mechanism by which ZMYM2 controls ERVs, especially MERVL, remains undefined. Therefore, we focused our studies on dissecting the functional relationship between ZMYM2/LSD1-HDAC1 and miR-344 in MERVL regulation.
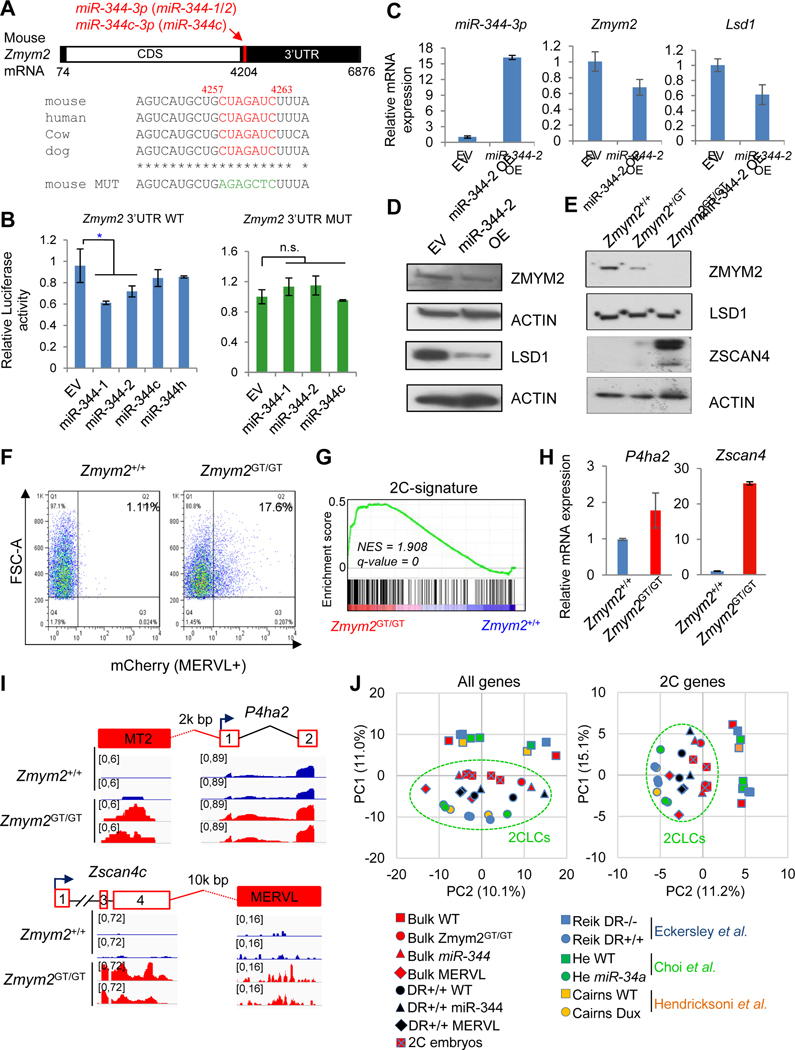
By examining sequence complementarities between miR-344 mature miRNAs and the 3’-UTRs of target genes, we found that both Zmym2 (Figure 4A) and Lsd1 (Figure S4A) 3’-UTRs contain conserved binding sites for miR-344 and miR-344c, suggesting direct post-transcriptional repression of Zmym2/Lsd1 by miR-344. We confirmed this by luciferase reporter assays in 293T cells, demonstrating that the luciferase reporters containing Zmym2 and Lsd1 3’-UTR with predicted miR-344 binding sites exhibited miR-344-1 and miR-344-2 dependent repression, respectively, when cotransfected with miR-344 gene expression vectors, while the repression was lost after mutating the predicted miR-344 binding sites (Figures 4A–B and S4A–B). When miR-344-2 was overexpressed in ESCs (Figure 4C), downregulation of Zmym2 and Lsd1 was observed at both mRNA (Figure 4C) and protein (Figure 4D) levels. Consistently, the percentage of MERVL+ cells was decreased in ZMYM2- or LSD1-overexpressed ESCs (Figure S4C). Lastly, our previous SILAC data have confirmed downregulation of both LSD1 and ZMYM2 proteins in DR+/+ cells (Figures 2D and S4D), where miR-344 is highly abundant (Figure 2E). Together, these data establish Zmym2 and Lsd1 as the direct targets of miR-344.
Figure 4. Zmym2 and Lsd1 are the targets of miR-344.
(A) Schematic layout of the Zmym2 mRNA with predicted miR-344 binding sites indicated. The evolutionarily conserved seed sequence and its mutated version are indicated.
(B) Luciferase reporter assay in ESCs co-transfected with reporter constructs containing the Zmym2 WT (left) or mutated (right) 3’UTRs and miR-344-1/2/c/h expression vectors or empty vector (EV). miR-344-h is a non-targeting control. Data are presented as average ± SD, P-value is from T-test, * P<0.05, n.s. non-significant.
(C) Relative expression levels of miR-344-3p, Zmym2, and Lsd1 upon overexpression (OE) of miR-344-2 or empty vector (EV). RT-qPCR data are presented as average ± SD.
(D) Protein expression of ZMYM2 and LSD1 in mESCs after overexpression (OE) of miR-344-2 or empty vector (EV). ACTIN is a loading control.
(E) Protein expression of ZMYM2, LSD1, and ZSCAN4 in Zmym2+/+, Zmym2+/GT, Zmym2GT/GT ESCs. ACTIN is a loading control.
(F) FACS profiles of 2C::tdTomato positive cells in Zmym2+/+ and Zmym2GT/GT ESCs.
(G) GSEA indicating that upregulated genes in Zmym2GT/GT ESCs were highly enriched in the 2-cell embryo gene set. Red, upregulated genes; blue, downregulated genes.
(H) RT-qPCR of 2C-specific P4ha2 and Zscan4c expression in Zmym2+/+ and Zmym2GT/GT ESCs. Data are presented as average ± SD.
(I) Examples of induced expression (shown as RNA-seq peaks) of MERVL-proximal P4ha2 and Zscan4c in Zmym2GT/GT compared with Zmym2+/+ cells.
(J) Principle-component analysis (PCA) of RNA-seq data from bulk or sorted DR+/+ population of miR-344-/MERVL-activated cells, Zmym2GT/GT ESCs, 2C embryos, and other published 2CLC cells. PCA was performed based on all genes (left) and only 2C-specific genes (Macfarlan et al., 2012) (right).
To further investigate the functional significance of ZMYM2 in regulating MERVL and 2CLCs in ESCs, we derived homozygous gene-trap Zmym2 (Zmym2GT/GT) mutant ESCs (Figure S4E) from the intercrosses of heterozygous (Zmym2GT/+) mice. The resulting Zmym2GT/GT mutant ESCs are null for ZMYM2 protein expression (Figure 4E), with little effect on LSD1 but a marked increase of ZSCAN4 expression (Figure 4E). In Zmym2GT/GT ESCs, only the expression of MERVL family transcripts was highly upregulated, compared with the other repetitive elements such as IAP, LINE1 and SINE (Figure S4F). To determine if ZMYM2 represses MERVL, we transfected Zmym2+/+ (wild-type, WT) and Zmym2GT/GT ESCs with a MERVL-Luc (luciferase) reporter containing the same MERVL fragment as shown in Figures 1C–D. As expected, luciferase activity elevated in Zmym2GT/GT ESCs relative to WT (Figure S4G). Using MERVL-containing 2C::tdTomato fluorescence reporter as a proxy for the MERVL+ population in Zmym2GT/GT ESCs, we found that the percentage of MERVL+ cells was increased in Zmym2GT/GT relative to Zmym2+/+ ESCs (Figures 4F and S4H). Next, we profiled the transcriptomes of Zmym2GT/GT and Zmym2+/+ ESCs by RNA-seq. We identified 1148 differentially expressed genes (581 down-/567 up-regulated genes, fold-change>2, P<0.05) in Zmym2GT/GT ESCs, which enrich significantly the geneset associated with 2-cell embryo development (Wu et al., 2016) (Figure 4G). Consistently, expression levels of Zscan4c and P4ha2 (Figures 4H–I), and notably the 2C gene activator Gata2 (Choi et al., 2017) (see more in Figure S7 and in Discussion), were all upregulated in Zmym2GT/GT ESCs. We then performed principal component analysis (PCA) to characterize the 2CLC populations from us including spontaneous DR+/+ population of ESCs, MERVL-activated, miR-344-activated, and Zmym2GT/GT ESCs, 2C embryos, together with 2CLC datasets from others including a similar DR+/+ population (Eckersley-Maslin et al., 2016), 2C::tdTomato-marked ESCs (Macfarlan et al., 2012), miR-34a−/− ESCs (Choi et al., 2017), and DUX-induced ESCs (De Iaco et al., 2017; Hendrickson et al., 2017; Whiddon et al., 2017), as well as the respective control ESCs. PCA revealed a similarity of 2C embryos with our 2CLCs and those established ones with expanded potency by clustering with both “all genes” (Figure 4J, left) and “2C genes” (Figure 4J, right).
Together, our data identify direct gene targets of miR-344 including Zmym2, whose posttranscriptional repression by miR-344 leads to derepression of MERVL and 2C genes, supporting a transcriptional repressor role of ZMYM2 in restricting MERVL/2C gene expression.
ZMYM2 recruits HDAC-containing complexes to directly bind to MERVL and repress MERVL expression
To understand how ZMYM2 represses MERVL and 2C gene expression, we investigated the ZMYM2 interactome in ESCs. We employed affinity purification followed by LC-MS/MS as described (Ding et al., 2012) to identify ZMYM2-interacting proteins. A total of 149 high-confidence ZMYM2-interacting partners were identified (Figure S5A and Table S3). GO analysis of the ZMYM2 partners revealed a significant enrichment of histone modification and transcription regulation (Figure S5B). Consistent with previous findings that ZMYM2 interacts with HDAC-containing LSD1-RCOR1/2 corepressor complex in ESCs (Yang et al., 2011) and HeLa cells (Gocke and Yu, 2008), we also found subunits of the LSD1-RCOR1/2 complex in our ZMYM2 interactome (Figure S5A and Table S3). Interestingly, the LSD1-NuRD (CHD4, GATAD2B, RBBP4, HDAC1/2) corepressor complex, required for enhancer decommissioning during ESC differentiation (Whyte et al., 2012), was also enriched in our ZMYM2 interactome (Figure S5A and Table S3). To understand how these partner proteins may contribute to ZMYM2 functions in repressing MERVL and 2C genes, we first confirmed the interactions of ZMYM2 with LSD1, CHD4, and HDAC1/2 by co-immunoprecipitation (coIP) (Figure S5C). We then studied the gene regulation by ZMYM2 and its partner proteins using our own (Figure 4G) and published (Macfarlan et al., 2012; Stevens et al., 2017) RNA-seq datasets (Table S2). We found that ZMYM2 shared 14% and 5% upregulated genes with LSD1 and CHD4, respectively, upon their depletion, 24% and 22% of which belong to 2C genes, respectively (Figure 5A).
Figure 5. ZMYM2 physically associates and functionally interacts with the LSD1-NuRD corepressor complex in controlling MERVL expression.
(A) Overlap of the common genes repressed by ZMYM2/LSD1 (top) or ZMYM2/CHD4 (bottom), which are compared with the 2C-specific genes (Macfarlan et al., 2012).
(B) Distribution of ZMYM2 ChIP-seq peaks.
(C, G and J) Average ChIP-seq density (reads per million, RPM) of factors indicated around the ZMYM2 peak center (−3 kb to 3 kb) (C and J) or at ZMYM2 peaks overlapped with MERVL LTR (MT2) regions (G).
(D) Relative enrichment of MERVL elements (MT2B and MT2B2) in ZMYM2 (left) but not in LSD1 (right) peaks regions over a random control.
(E-F) Reduced chromatin occupancy of LSD1 (E) and increased H3K4me1 expression (F) upon Zmym2 depletion. H3 is a loading control. WCE, whole cell extracts; Chr, chromatin-bound fractions.
(H) Expression (reads per million, RPM) of the MERVL solo LTR (MT2_Mm, left) and internal region (MERVL-int, right) in WT and Zmym2GT/GT ESCs. P-value is from Mann-Whitney test.
(I) LSD1 ChIP-seq peak intensity in WT versus Zmym2KO ESCs. Significantly decreased and increased numbers of peaks upon loss of ZMYM2 are indicated with FDR < 0.01.
(K) Overlap of genes activated in 2C embryos (n=2363) and MERVL-proximal genes with a ZMYM2 motif at LTR promoter (n=594).
(L) ZMYM2 and LSD1 cobind at Usp38 promoter with an MT2 region.
(M) LSD1 ChIP-qPCR with 2 different antibodies, at promoters of Usp38 in WT and Zmym2KO ESCs. Data are presented as average ± SD.
To further understand the functional specificity of ZMYM2 in MERVL repression, we identified global genomic targets of ZMYM2 by ChIP-seq. We created ZMYM2-3xFLAG (ZMYM23xFL) knockin ESC line (Figure S5D) to obviate the lack of ChIP-grade ZMYM2 antibody. A total of 26,647 ZMYM2 peaks were identified, revealing a broad range of ZMYM2 binding at different genomic loci such as promoters, introns, and intergenic regions (Figure 5B). We first analyzed ZMYM2 peaks at transcription start sites (TSSs) and enhancers, and confirmed the enrichment of ZMYM2 in those regions (Figure S5E). Consistent with their physical partnerships (Figure S5F), CHD4, HDAC1/2 and LSD1 were found to co-occupy the ZMYM2 peaks (Figure 5C).
Although LSD1 was known to be involved in epigenetic silencing of MERVL (Macfarlan et al., 2011), how LSD1 is recruited to the chromatin is unclear. We thus explored the potential function of ZMYM2, a sequence-specific DNA-binding transcription factor, in recruiting LSD1 for specific ERV repression by performing LSD1 ChIP-seq in both WT and Zmym2KO ESCs (independently created by a CRISPR/Cas9 strategy; see Method). Two biological replicates with different LSD1 antibodies were used for ChIP-seq. A high correlation was obtained within the two biological replicates indicative of high-quality datasets (Figure S5H). We uniquely mapped ZMYM2 and LSD1 ChIP-seq data with RepeatMasker annotation and calculated the proportion of peaks overlapping each repeat class. ZMYM2 peaks, but not LSD1 peaks, were significantly (P<0.05, Binomial test) enriched at MERVL LTR (MT2) regions such as MT2B and MT2B2 (Figure 5D, Table S4) and MT2_Mm (Figure S5G), supporting a ZMYM2-dependent targeting of LSD1 to MERVL LTR.
Given that ZMYM2 binds to MT2 LTR specifically, we examined if ZMYM2 recruits LSD1, CHD4, and HDAC1/2 specifically to the LTR regions. Supporting this, we found reduced chromatin occupancy of LSD1 (Figure 5E) and enhanced H3K4me1 level (Figure 5F) upon ZMYM2 depletion in the presence of unchanged total LSD1 (Figures 5E and S5F), consistent with the reported role of LSD1 in demethylating monomethyl and dimethyl histone H3 lysine 4 (Shi et al., 2003). To further understand how ZMYM2 cooperates with HDAC-containing LSD1-RCOR1/2-NuRD corepressor complex in restricting expanded potency in ESCs, we employed MT2-related ZMYM2 peaks to find the overlap with LSD1, CHD4 and HDAC1/2 enrichment. Confirmatively, ZMYM2 peaks showed a clear enrichment for LSD1, CHD4 and HDAC1/2 at MERVL LTR (MT2) regions (Figure 5G). Consequently, expression of both MERVL regions (MERVL-int) and its LTR (MT2_Mm) is upregulated significantly upon ZMYM2 loss in Zmym2GT/GT relative to WT cells (Figure 5H). We also examined enrichment of these proteins at other LTR elements, such as MaLR MTC and IAP, which have similar copy numbers compared to MT2. We found that MTC regions have no enrichment of ZMYM2 or LSD1 (Table S4), and that IAP has an enrichment of ZMYM2, but not LSD1/CHD4/HDAC1/2 (data not shown), which is consistent with IAPs being not upregulated in Zmym2GT/GT ESCs (Figure S4F). To understand how LSD1 chromatin-binding is globally affected by loss of ZMYM2, we compared LSD1 binding events in WT and Zmym2KO ESCs. A total of 12,776 of LSD1 peaks were identified from either WT or Zmym2KO ESCs, composed of 3012, 7904, and 1850 Zmym2WT-only, Zmym2WT/KO-common, Zmym2KO-only LSD1 peaks, respectively (Figure S5I, top). Although a large number of LSD1 binding peaks (7904) was observed regardless of Zmym2 status (Zmym2WT/KO-common), it is noteworthy that the LSD1 peaks of the Zmym2WT-only and Zmym2KO-only contain the highest (1906/3012 or 63.3%) and lowest (455/1850 or 24.6%) percentage of shared peaks with ZMYM2 binding, respectively (Figure S5I, bottom), supporting ZMYM2-dependent LSD1 binding. Further substantiating this, there are many more LSD1 peaks with significantly decreased intensity (Up 125, Down 1244, FDR<0.01) in Zmym2KO ESCs (Figure 5I), and the overall LSD1 ChIP-seq signal also decreased at ZMYM2 peak regions upon Zmym2 depletion (Figure 5J). Together, our data conclusively establish a ZMYM2-dependent LSD1 chromatin-binding, i.e., ZMYM2 recruits the LSD1 complex, to specific regions in the genome.
Next, we focused on the MERVL LTR regions where ZMYM2 may recruit LSD1-RCOR1/2-NuRD corepressor complex and thus affect expression of nearby 2C-specific genes. By searching all MERVL LTRs (MT2) with a ZMYM2 binding motif, and locating them to the nearest gene (< 50 kb) promoters that harbor such LTRs, we found 17% (101/594, P=0.007, Chi-square test, Table S5) of these genes are 2C-specific genes defined previously (Macfarlan et al., 2012) (Figure 5K). For example, both ZMYM2 and LSD1 bind at the promoters of 2C genes Usp38 and Rps14 in WT ESCs, and LSD1 ChIP intensity at the binding loci decreases in Zmym2KO ESCs compared to that in WT ESCs (Figures 5L–M and S5J–K), demonstrating ZMYM2-dependent binding of LSD1 to the chromatin harboring MERVL/LTR for 2C gene control. Consistent with the repressive roles of ZMYM2 and LSD1 in ERV silencing (Macfarlan et al., 2011; Yang et al., 2015) and the requirement of MERVL activation for expanded potency (Figures 3 and S3I), both transcripts (Figure S5L) and proteins (Figure 2D) of Zmym2 and Lsd1 were downregulated in totipotent 2C embryos and DR+/+ 2CLCs.
Together, our results establish a negative regulatory role of ZMYM2 in restricting 2CLCs in ESCs by recruiting LSD1-RCOR1/2-NuRD corepressor complex to MERVL LTRs for direct transcriptional repression of MERVL/2C genes (Figure S5M).
Zygotic depletion of ZMYM2 compromises the totipotency-to-pluripotency transition
To further appreciate the physiological relevance of our findings that Zmym2 maintains pluripotency by restricting totipotent 2CLCs in ESC culture, we asked how Zmym2 depletion would affect the totipotency-to-pluripotency transition in embryonic development. We first performed zygotic injections of mmu-miR-344-3p mimics (mimics) or miR non-targeting control (miNC) as shown in Figure S6A. After injection of miR-344-3p at zygote stage (Figure S6B), 7 out of 44 (16%) of embryos failed to develop to the blastocyst stage, compared with one embryo (2.7%, 1/36) failed to develop by miNC injection (Figures 6A–B, blue rectangles and lines; Table S6). While the compromised totipotency-to-pluripotency transition is readily appreciable, the difference is however statistically non-significant (n.s.), likely due to the general fine-tuning function of miRNAs and already highly abundant miR-344 at that stage. Nonetheless, totipotency markers MERVL and Zscan4 were upregulated in 8-cell embryos upon mimics’ treatment (Figure 6C). Zmym2 and Lsd1 expression levels were downregulated at 8-cell and blastocyst stages upon mimics treatment (Figures 6D–E), indicating a conserved role of miR-344 in repressing Zmym2/Lsd1 during early development. Moreover, we observed downregulation of a group of predicted miR-344 target genes, including Zmym2, measured by RNA-seq analysis of these embryos (Figure S6C).
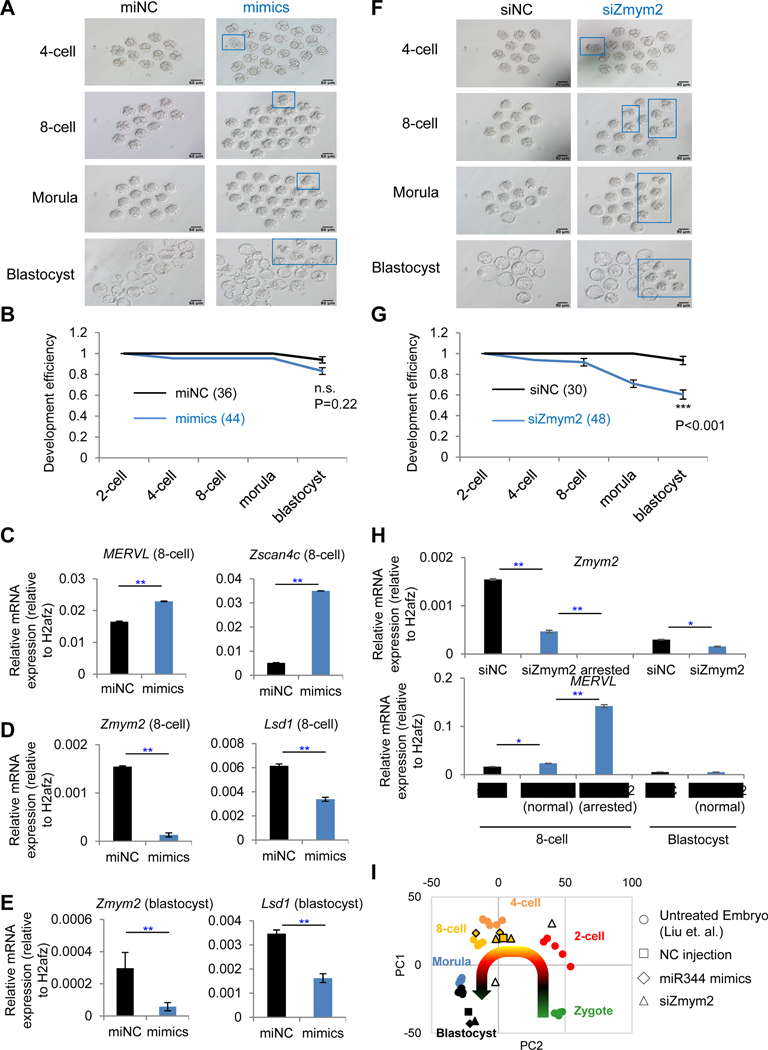
Figure 6. Zygotic depletion of Zmym2 arrests development at 2C-stage embryos.
(A and F) Morphology of embryos after zygotic injection with miR-344-3p mimics (mimics) and negative control (miNC) (A) or siRNA against Zmym2 (siZmym2) and non-targeting siRNA (siNC) (F) at each indicated developmental stage. The blue squares indicate aberrantly developed embryos. Scale bars, 50 μm.
(B and G) Statistics for the development efficiency (portion of normal embryos) at each developmental stage in Figure 6A (B) or Figure 6F (G). Data are presented as average percentage ± SD. P-value is from two-way ANOVA test; n.s. non-significant; *** P<0.001.
(C-E) RT-qPCR of MERVL/Zscan4 (C) and Zmym2/Lsd1 expression (D) in embryos with miR-344-3p mimics (mimics) or negative control (miNC) treatments, collected at the 8-cell stage or at the blastocyst stage (E). Data are presented as average ± SD. P-value is from T-test, ** P<0.01.
(H) RT-qPCR analysis of Zmym2 and MERVL expression in embryos with siZmym2 or siNC. Embryos were collected at the 8-cell and blastocyst stages, and siZmym2-treated embryos with normal or arrested morphology were collected separately. Data are presented as average ± SD. P-value is from T-test, * P<0.05, ** P<0.01.
(I) PCA of RNA-seq data of embryos with miR-344 mimics, siZmym2, and non-target control (NC) injections from this study, and of normal embryos at different embryonic stages from (Liu et al., 2012). Each sample represents a pool of embryos (~10) with identical treatment and developmental stage. Shapes of samples indicate the treatments, and colors of samples indicate the stage for RNA collection. The curved arrow indicates development progression from zygote to blastocyst. Two pools of embryos (clear triangles) are developmentally arrested upon siZmym2 treatment. RNA is collected at 8-cell/early morula stage.
Next, we addressed how Zmym2 loss would affect the totipotency-to-pluripotency transition during embryonic development by injecting siRNA against Zmym2 (siZmym2) or a non-targeting control (siNC) into mouse zygotes following the same strategy (Figure S6A). 19 out of 48 (40%) of embryos failed to develop to the blastocyst stage, compared with 2 embryos (6.7%, 2/30) failed to develop by siNC injection (P<0.001, Figures 6F–G, blue rectangles and lines; Table S6). The majority of abnormal embryos after injection with siZmym2 were arrested at 8C stage morphologically (Figure 6F). To elucidate the molecular difference between the morphologically normal and arrested embryos by siZmym2, we carried out RT-qPCR and found that a relatively higher level Zmym2 in normal embryos than arrested ones. More importantly, the lower levels of Zmym2 correspond to higher induction of MERVL in arrested embryos relative to normal ones at the 8-cell stage (Figure 6H), which was further confirmed by RNA-seq analysis of treated embryos (Figure S6D). Furthermore, by comparing the transcriptomes between morphologically normal and arrested embryos upon siZmym2, totipotency-related genes such as Zscan4c/d were highly expressed in the arrested embryos (Figure S6E). Genes upregulated in those arrested embryos were enriched with the GO terms “negative regulation of cell differentiation”, “positive regulation of cell proliferation”, “negative regulation of apoptotic process” and “nucleosome assembly” (Figure S6F). PCA of RNA-seq data suggests that embryos injected with siZmym2 or miR-344 mimics with normal developmental morphology have a similar transcriptional expression with the untreated embryos at PC1 (Figure 6I, orange triangles and diamonds with solid border versus orange circles). In contrast, the developmentally arrested embryos with siZmym2 treatment (collected at 8-cell/early morula stages) displayed a trend of moving toward the direction of 2-cell embryos (Figure 6I, the two clear triangles with solid border sit in between orange/4-cell and red/2-cell circles at PC2).
Together, these results conclusively establish the in vivo functional significance of miR-344 and its direct target Zmym2 in regulating developmental potency of early mouse embryos.
Transcriptional activation of miR-344 by DUX in MERVL+ cells
Having established the miR-344--|Zmym2--|MERVL regulatory axis for 2CLC control with direct in vivo relevance for expanded stem cell potency, we wondered how miR-344 expression is regulated in early embryos. Recently, several studies identified DUX as a positive regulator of cleavage-stage genes and MERVL LTR elements in early mouse embryos leading to transcriptional activation and chromatin opening (Hendrickson et al., 2017; Iturbide and Torres-Padilla, 2017; Whiddon et al., 2017). We also found that Dux has higher ATAC signals and expression levels in DR+/+ than DR−/− cell population (Figures 7A–B). As mouse DUX binds to conserved sites to activate genes associated with cleavage-stage embryos, including MERVL retrotransposon (Hendrickson et al., 2017; Iturbide and Torres-Padilla, 2017; Whiddon et al., 2017), we asked whether DUX activates miR-344. By analyzing the published ChIP-seq of DUX and performing ChIP-qPCR, we confirmed direct binding of DUX to totipotency-associated genes Zscan4c and Zscan4d (Figures S7A–B). More importantly, we also found mouse DUX directly occupied the loci of miR-344-2, miR-344c and miR-344h (Figure 7C), indicating a direct regulation of these genes. To establish the transcriptional regulation of miR-344 by DUX, we overexpressed DUX-3xFLAG in ESCs. ChIP-qPCR was then performed to analyze DUX binding to the miR-344 loci upon DUX overexpression, revealing a specific binding of DUX to miR-344-2, miR-344c and miR-344h genes (Figure 7D). We also found that mature miR-344-3p, miR-344c-3p, and miR-344h-3p (Figure 7E) and Zscan4/MERVL mRNAs (Figure S7C) were upregulated upon DUX overexpression. Conversely, Dux knockdown in sorted DR+/+ population of 2CLCs leads to downregulation of mature miR-344-3p, miR-344c-3p, miR-344h-3p (Figure 7F), and Zscan4/MERVL transcripts (Figure S7D). Moreover, when inserting the conserved DUX-binding motif fragment present in miR-344 (Figure 7G) into a luciferase reporter (Wu et al., 2006), we found that the activation of this reporter by DUX is dependent on intact DUX-binding motif (Figures 7H and S7E), and that luciferase activity decreased if one or both of the predicted DUX-binding sites were mutated (Figure 7I; m1 and m2). Together, these data establish DUX as an upstream transcription activator of miR-344 in 2CLCs.
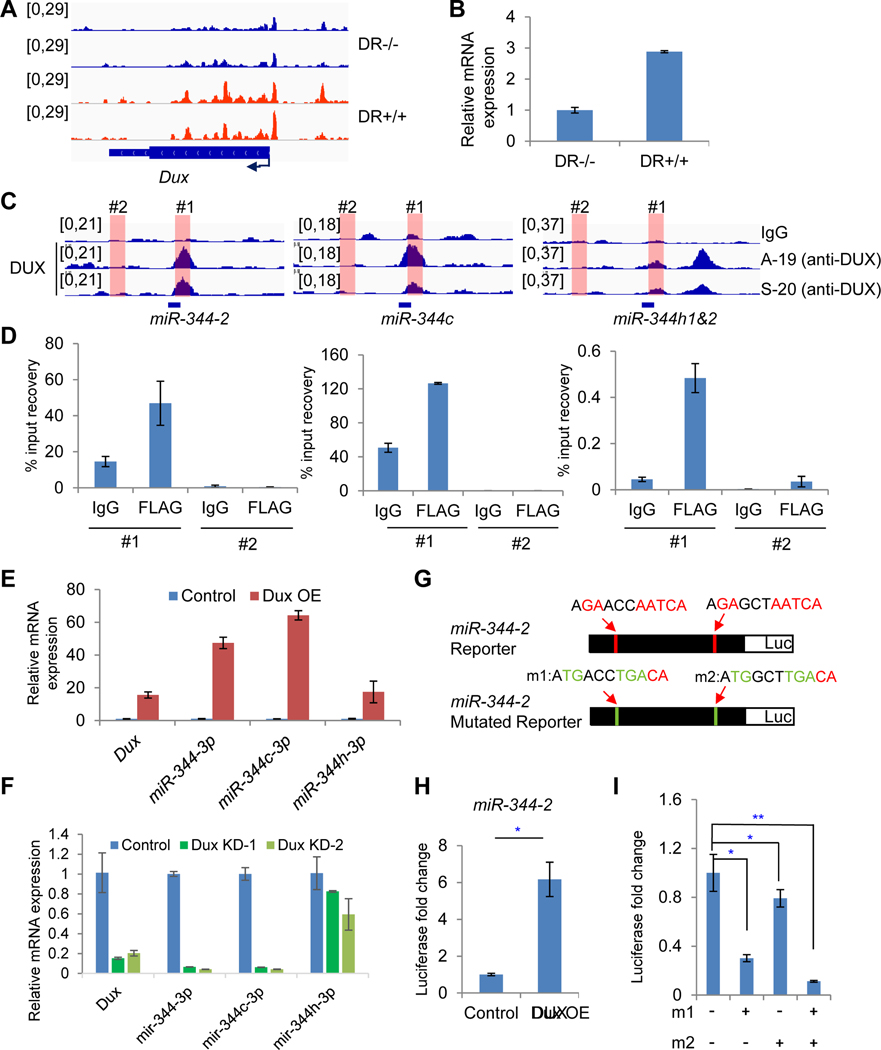
Figure 7. DUX binds to miR-344 promoter and activates miR-344 expression in MERVL+ cells.
(A) ATAC-seq tracks at Dux locus in DR+/+ (red) and DR−/− (blue) cells.
(B) RT-qPCR analysis of relative Dux expression in DR+/+ and DR−/− cells.
(C) ChIP-seq tracks of DUX (Whiddon et al., 2017) at miR-344-2, miR-344c, and miR-344h1&2 loci. Locations of primers for ChIP-qPCR analysis in panel D are indicated.
(D) ChIP-qPCR of DUX at miR-344-2 (left), miR-344c (middle), and miR-344h1&2 (right) loci. Region #1 is positive for DUX enrichment, whereas #2 is negative from DUX ChIP-seq data.
(E) RT-qPCR of Dux and mature miR-344-3p, miR-344c-3p, and miR-344h-3p expression upon Dux overexpression (OE) in mESCs.
(F) RT-qPCR of Dux and mature miR-344-3p, miR-344c-3p, and miR-344h-3p expression upon Dux knockdown in sorted MERVL+ mESCs. KD-1 and KD-2 are experiments with 2 independent shRNAs.
(G) Depiction of luciferase reporters containing miR-344-2 sequence with two DUX-binding motifs in wild type (top) or mutated (m1 and m2) version (bottom).
(H) Luciferase reporter assay in mESCs co-transfected with the miR-344-2 reporter and a DUX expression vector or a control. P-value is from T-test, *P < 0.05.
(I) Luciferase reporter assay in mESCs co-transfected with the miR-344-2 wildtype and mutated reporter and DUX expression vector. “m1 and m2” refer to the mutations at the two potential DUX motifs in panel G. P-value is from T-test, * P<0.05, ** P<0.01.
(B-H) The qPCR data are presented as average ± SD.
DISCUSSION
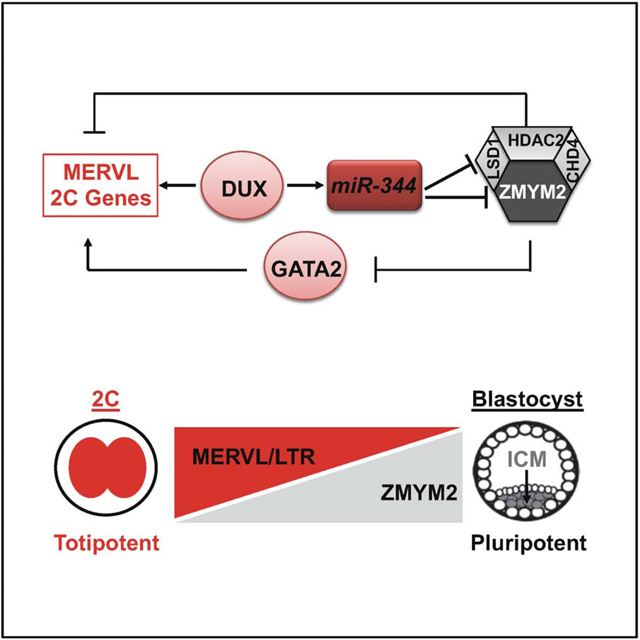
Our current understanding of the regulation of totipotent 2C state is largely limited in identifying and characterizing ESC-enriched coding and/or noncoding molecules that restrict the cell fate potential to a pluripotent state rather than activate it to the 2C state. For example, previous studies have revealed a number of key factors that restrict the 2C state and 2CLCs in mouse ESCs, including LSD1 (Macfarlan et al., 2012), CAF-1 (Ishiuchi et al., 2015), KAP1 (Rowe et al., 2013), G9A (Maksakova et al., 2013), miR-34a (Choi et al., 2017), PRC1.6 and EP400-TIP60 complexes (Rodriguez-Terrones et al., 2018), and PIAS4 SUMO E3 ligase (Yan et al., 2019). In contrast, DUX is the first transcription factor that was identified to be the activator of the 2C state and 2CLCs in heterogeneous ESCs (De Iaco et al., 2017; Hendrickson et al., 2017), and Yan et al. further identified DPPA2/4 as potential upstream regulators of DUX controlling zygotic transcriptional program (Yan et al., 2019). However, the detailed molecular events downstream of DUX regulating the 2C state are poorly defined. In this regard, our study delineates a previously unappreciated molecular axis downstream of DUX involving both transcriptional and post-transcriptional regulatory modes leading to activation of the 2C state and 2CLCs. Specifically, we identify a novel DUX→miR-344--|Zmym2/Lsd1--|MERVL regulatory pathway controlling 2CLC totipotency.
In our model, the open chromatin of the 2C state makes it more accessible for transcription factors like DUX to activate miR-344, which in turn activates MERVL through multiple layers of control. First, miR-344 post-transcriptionally represses ZMYM2 and LSD1, and the depletion of ZMYM2 leads to the loss of LSD1 binding on MERVL and 2C-specific genes. Second, LSD1 can act as a lysine-specific demethylase by specifically demethylating monomethyl and dimethyl histone H3 lysine 4 (H3K4me1 and H3K4me2), which are marks of active transcription state. The post-transcriptional repression of Lsd1 by miR-344 leads to activation of MERVL and 2C-specific genes. Third, DUX can also directly activate MERVL and associated 2C genes (De Iaco et al., 2017; Hendrickson et al., 2017; Whiddon et al., 2017). Finally, ZMYM2 can directly bind to and repress Gata2 (Figures S7F–H), a critical transcription factor with a demonstrated role in activating MERVL/MT2 and 2C genes (Choi et al., 2017). Our studies thus establish miR-344 as the first noncoding positive regulator for 2CLC expanded potency and preimplantation development, which is in stark contrast with the reported role of miR-34a in post-transcriptional restriction of the 2C state and 2CLCs (Choi et al., 2017), adding a new layer of complexity in molecular control of totipotency and its transition to pluripotency (Figure S7I).
ZMYM2 is a zinc finger protein containing a stretch of unique tandem zinc fingers called MYM (myeloproliferative and mental retardation) domains (Smedley et al., 1999) that are essential for the interaction of ZMYM2 with HDAC1 and for the binding of ZMYM2 to chromatin through its SUMO-interacting motifs (SIMs) (Aguilar-Martinez et al., 2015; Gocke and Yu, 2008). It is noteworthy that Zmym2 was identified from an RNAi screen as a hit for retroviral silencing together with Sumo2 as a potent inhibitor of ERVs including MERVL (Yang et al., 2015). Interestingly, besides SIMs, the MYM-type zinc fingers were also found to be legitimate SUMO-binding domains in ZMYM2 (Guzzo et al., 2014), and ZMYM2 itself could be a SUMOylated protein (Hendriks et al., 2018; Kunapuli et al., 2006). A recent study also discovered that the SUMO E3 ligase PIAS4 impairs 2C-like state by repressing DUX, MERVL, and 2C genes (Yan et al., 2019). Future studies are warranted to investigate whether ZMYM2 is a direct substrate of SUMO2 and/or functions as a binder to other Sumo2-modified proteins in pluripotent stem cells and how such a potential connection with the protein SUMOylation pathway may have endowed ZMYM2 with its unique roles in regulating the totipotency-to-pluripotency transition. Such studies in defining the molecular pathways underlying 2CLC totipotency will be significant in further understanding cellular plasticity and mammalian development.
STAR ★ METHODS
Detailed methods are provided in the online version of this paper and include the following:
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jianlong Wang (jw3925@cumc.columbia.edu). All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals and Collection of Mouse Embryos
The specific pathogen-free (SPF) grade mice were housed in the animal facility of Tongji University, Shanghai, China and Icahn School of Medicine at Mount Sinai. All animal maintenance and experimental procedures were performed according to Institutional Guides for the use of laboratory animals.
To get MII oocytes and pre-implantation embryos, B6D2F1 or C57BL/6 female mice (8~10-weeks old) were super-ovulated by injection with 7 IU each of pregnant mare serum gonadotropin (PMSG), followed by injection of 5 IU of human chorionic gonadotropin (hCG) (San-Sheng Pharmaceutical) 48 h later. The super-ovulated female mice were mated with B6D2F1 or DBA2 male mice. Then, the zygotes or 2-cell stage embryos were collected from the oviducts of female B6D2F1 mice. To obtain 4-cell, 8-cell, morula and blastocyst stage embryos, 2-cell stage embryos were cultured in G1 plus medium to reach the corresponding stage. MII oocytes were collected from the oviducts of unmated female mice.
Cell Culture
Feeder-free mouse embryonic stem cells (mESCs) were cultured on 0.1% gelatin-coated plates and in ESM medium: DMEM supplemented with 15% fetal bovine serum (FBS), 1000 units/mL recombinant leukemia inhibitory factor (LIF), 0.1 mM 2-mercaptoethanol, 2 mM L-glutamine, 0.1 mM MEM non-essential amino acids (NEAA), 1% nucleoside mix (100X stock, Sigma), and 50 U/mL Penicillin/Streptomycin).
METHOD DETAILS
Zygotic Injection of siRNA or miRNA and Embryo Development
B6D2F1 (BDF1) female mice (7–8 weeks old) were superovulated by intraperitoneally injecting with pregnant mare serum gonadotropin (PMSG) and human chorionic gonadotropin (hCG), and then mated with BDF1 male mice. The fertilized embryos (zygotes) were collected from oviducts. A mixture of Zmym2 siRNA (20 μM), scramble siRNA (20 μM), miR-344 (50 μM), or scramble miRNA (50 μM) was separately injected into the cytoplasm of fertilized eggs with visible pronuclei. The injected zygotes were then cultured in G1 plus medium (10128, Vitrolife), and 2-cell, 4-cell, 8-cell, morula and blastocyst embryos were obtained after culturing. In addition, the development potential was recorded at each stage during culturing. The siRNA and miRNA were synthesized by Genepharma (Shanghai, China), and their sequences are listed in Table S7.
Single-cell Microinjection and Chimeric Assay
Chimeric embryo generation was performed by single-cell injection in 8-cell stage embryo. To generate chimeric blastocysts by microinjection, a single cell (or up to three cells) from ESC line miR-344-2- activated mESCs, MERVL-activated mESCs and the empty vector control was separately injected into each 8-cell stage BDF1 recipient embryos. GFP+ve ESCs after sorting were seeded on plates and passaged once before they were used for chimera operation (microscopic exposure of these cells to light source was avoided to ensure their maximal viability). The injected embryos were cultured in G1 plus medium and chimeric blastocysts could then be obtained. Chimeric embryos were cultured for 1 day in a humidified incubator under 5% CO2 at 37°C. The chimeric blastocysts (E3.5) were dissected under an immunofluorescence stereomicroscope for detecting GFP+ cell localization. Then the chimeric blastocysts were transferred to uterine horns of 2.5-day post coitum pseudo-pregnant females.
The conceptuses were dissected at E12.5 and observed using an immunofluorescence stereomicroscope for detecting GFP+ve cell localization. The placenta was isolated from the E12.5 conceptuses, followed by embedding, freezing, slicing (5um thick) from the sagittal side and then, immunofluorescence staining of frozen sections.
Immunofluorescence Staining
For immunofluorescence staining, the placenta frozen sections were permeabilized with 0.5% Triton X-100 (Sigma) for 30 mins. The samples were blocked with 2.5% BSA in PBS for 1 hour at room temperature. Then, they were incubated overnight at 4°C with the primary antibodies against PROLIFERIN (1:200; Santa Cruz Biotechnology, sc-271891), TPBPA (1:100; Abcam, ab104401) or GFP (50430–2-AP/66002–1-Ig, Proteintech, 1:100). Next, the samples were washed three times with PBS and incubated for 1 hour at room temperature with secondary antibodies. The DNA was labeled with 4’,6-diamidino-2-phenylindole (DAPI) (Merk Millipore). The slides were analyzed by the Leica TCS sp8 microscope.
ATAC-seq and Data Analysis
The ATAC-seq libraries of mESCs were prepared as previously described (Buenrostro et al., 2015) with minor modifications. Briefly, samples were lysed in lysis buffer (10 mM Tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2 and 0.15% NP-40) for 10 min on ice to prepare the nuclei. Immediately after lysis, nuclei were spun down at 500g for 5 min to remove the supernatant, incubated with the Tn5 transposase and tagmentation buffer at 37 °C for 30 min (Vazyme Biotech). After the tagmentation, ATAC-seq library was prepared following a published protocol (Buenrostro et al., 2013), and sequenced by Illumina HiSeq2500 at New York University Genome Technology Center following a standard protocol. Paired-end 50 bp-length ATAC reads were produced. The ATAC-seq raw data were processed as previously described (Buenrostro et al., 2015). Briefly, sequencing reads were aligned to mouse genome (NCBI build 37, mm9) using the bowtie2 (v2.3.0) program, with parameters -X 2000 --no-mixed. Aligned reads were filtered by samtools (v0.1.19) program with parameters -F 0×04 -f 0×02 -q 20. ATAC-seq peaks were determined by the MACS program (v.2.0.10) with default settings. Peak intensity by reads per million (RPM) for each ATAC-seq peak was calculated by DiffBind (v1.16.3) program, with minimal overlap of two peaks between different samples. The ATAC-seq peaks with significantly enriched intensities in either DR+/+ and DR−/− cells were exported by DiffBind.
SILAC-MS Profiling of Relative Protein Levels
The SILAC-MS procedure was illustrated in Figure S2C. Briefly, ESCs were cultured in either SILAC Light (Lys0, Arg0) or Heavy (Lys8, Arg10) medium. DR+/+ and DR−/− cells were sorted from both culture conditions. The cell lysates of each population at different SILAC condition were equally mixed, resulting in 2 replicates with reciprocal labeling. Protein lysates were dissolved in 8M Urea buffer, and subjected to tryptic digestion, followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using an Obitrap-Velos mass spectrometer. Proteome Discoverer™ Software (Thermo) was used for protein quantification and identification.
Nuclear Extract Preparation and Affinity Purification
To identify ZMYM2-interacting partners, four large square dishes of Zmym2GT/GT and WT serum/LIF (SL) ESCs were prepared after culturing for 2 weeks in SILAC ESC medium supplemented with either light or heavy lysine and arginine as described above (Ding et al., 2015). Nuclear extracts from Zmym2GT/GT and WT SL ESCs were precleared with Protein G agarose beads rotating overnight at 4oC. The next day, ZMYM2 antibodies were incubated with pre-cleared nuclear extracts for 8 hours with gentle rotation. The immunoprecipitates were washed five times with buffer D (20 mM HEPES pH 7.9, 0.2 mM EDTA, 1.5 mM MgCl2, 100 mM KCl, 20% glycerol) containing 0.02% NP40, and eluted from the beads by using buffer D. Eluted protein was then concentrated, quantified, mixed in a 1:1 ratio for each sample, and subjected to SDS-PAGE. Finally, a whole lane was cut into 10 pieces and subjected to quantitative liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
Co-Immunoprecipitation (CoIP) and Western Blot
To test the interactions of ZMYM2 with LSD1 and CHD4, 2 × 15 cm dishes of confluent ESCs were harvested, and nuclear extracts were prepared as described (Ding et al., 2015). For immunoprecipitation, nuclear extracts were incubated with 4 μg ZMYM2 (Abcam, ab30783), LSD1 (Abcam, ab17721), or IgG (Millipore, PP64) antibodies and then incubated with protein G-Agarose beads (#11243233001, Roche) overnight at 4 °C. The immunoprecipitates were washed five times with wash buffer (50 mM HEPES, pH 7.9, 180 mM NaCl, 0.1% NP-40, 0.2 mM EDTA) containing 0.2 mM PMSF, protease inhibitor cocktail and 0.5 mM DTT. Proteins were eluted from the beads by boiling in wash buffer and 4X SDS loading buffer. Western blotting with the following primary antibodies were performed: ZMYM2 (Abcam, ab30783), LSD1 (Abcam, ab17721), CHD4 (Abcam, ab70469), HDAC1 (Bethyl, A300–713A), HDAC2 (Bethyl, A300–705A-1).
For FLAG IP, nuclear extracts were prepared from Zmym2-3xFLAG knockin ESCs and wildtype ESCs and incubated with 50 μl of α-FLAG-agarose beads (M2, Sigma) for 3 hrs. The immunoprecipitates were washed five times with wash buffer, eluted from the beads by boiling in wash buffer and 4X SDS loading buffer, and separated by SDS-PAGE. Western blotting was performed using the following primary antibodies: ZMYM2 (Abcam, ab30783), LSD1 (Abcam, ab17721), CHD4 (Abcam, ab70469) and HDAC2 (Bethyl, A300–705A-1).
Genome-Scale Transcriptional Activation by CRISPRSAM
Genome-scale transcriptional activation was achieved by using the CRISPR-Cas9 Synergistic Activation Mediator (SAM) system. Given that 2C::tdTomato Reporter contains hygromycin (which has been used to establish DR mESCs), we first replaced Hygro resistance gene with puromycin in MS2-P65-HSF1 following a similar protocol described (Konermann et al., 2015) with minor modifications. Briefly, lentivirus containing dCas9-VP64 and MS2-P65-HSF1 were prepared for infection of mESCs containing pZscan4c-GFP and 2C::tdTomato reporter (DR) followed by puromycin and blasticidin selection for 6 days. U6-MS2-Zeo containing specific sgRNA was used to infect these cells followed by zeocin selection.
To express multiplex sgRNA-containing MS2, we inserted two MS2 RNA aptamers at the tetraloop and stem-loop 2 by the cut-and-paste method in pmU6-gRNA (Addgene, # 53187), ph7SK-gRNA (Addgene, #53189), phH1-gRNA (Addgene, #53186), phU6-gRNA (Addgene, #53188). We also modified U6-MS2-Zeo by inserting the lacZ fragment containing two Bsmb I sites for golden gate assembly. Finally, these vectors were used to assemble four promotergRNA cassettes into lentiviral destination vector U6-MS2-Zeo (modified)/U6-MS2-GFP by golden gate assembly shown in Figure S1A. The multiplex sgRNA expression vector was combined with dCas9-VP64 and MS2-P65-HSF1 following the similar protocol described above for MERVL activation in DR mESCs. We designed and tested 12 sgRNAs covering the 730-bp fragment (Figure 1B) for MERVL activation by an engineered CRISPRSAM complex. These sgRNAs were screened in HEK293T cells with ectopic expression of the luciferase reporter driven by the 730-bp fragment sequence together with dCas9-VP64 and helper MS2-P65-HSF1. We found that all sgRNAs activated MERVL from 30-fold to 400-fold, and that sgRNAs from A to F upstream of the “ATG” start codon, in particular, yielded the most significant activation (Figure 1C). When this experiment was repeated in mouse ESCs, we observed a relatively lower luciferase activity but the same trend of activation (Figure 1D), likely due to the dilution of sgRNAs by over 650 copies of endogenous full-length MERVL and many thousands of MERVL-derived LTR elements present in the mouse genome (Schoorlemmer et al., 2014). We therefore attempted another strategy for stronger activation in mouse ESCs by using the sgRNA multiplex expression system (Figures S1A–B) (Kabadi et al., 2014). Indeed, we found an increased luciferase activity with increasing copy numbers of sgRNAs, with a maximum of ~25fold activation by expressing 2~4 copies of F-sgRNA (2F, 3F, and 4F) (Figure 1E).
3xFLAG Knock-in at Zmym2 Locus
The fragments for homology arms of Zmym2 and 3xFLAG-P2A-Neomycin cassette were PCR amplified, and assembled by Gibson Assembly® Master Mix (New England BioLabs, E2611S) to obtain 5’arm-3xFLAG-2A-Neo-3’arm (5a-3F2ANeo-3a) fragment. The 5a-3F2ANeo-3a fragment was subcloned into pCR™-Blunt II-TOPO® vector (Invitrogen) using Zero Blunt® TOPO® PCR Cloning Kit (Invitrogen, #45–0245) to obtain Topo-5a-3FNeo-3a. CRISPR gRNAs that were inserted into pSpCas9(BB)-2A-Puro (PX459) V2.0 were provided in Table S7.
To introduce 3xFLAG-P2A-Neomycine into the stop codon site of Zmym2, 5 × 105 mESCs were transfected using Lipofectamine 2000 (Invitrogen) with 2 μg linearized Topo-5a-3FNeo-3a and 2 μg sgRNA expression vector. Forty-eight hours after transfection, cells were seeded into 10 cm dishes supplied with 500 μg/ml G418 (Corning, 30–234-CR), and 1 μg/ml puromycin (Sigma, P9620–10ML). After selection for 72 h, cells were reseeded into a new 10 cm dishes with 500 μg/ml G418 (Corning, 30–234-CR) only. Single clones were picked and expanded for validation of homologous recombination by primers listed in Table S7, and the correctly targeted clones were further confirmed with anti-FLAG western blotting test.
CRISPR Knockout of Zmym2 in ESCs
A Zmym2KO ESC line was generated by CRISPR knockout technique. Briefly, ESCs were transfected with a puro-resistant pX330 vector with a guide RNA targeting the first exon of Zmym2. After transfection and drug selection, the ESCs were seeded as single clones, which were then picked up and expanded. All clones were examined for ZMYM2 protein expression by western blotting analysis, and the KO clones were further validated by Sanger sequencing.
shRNA Knockdown
Small hairpin RNAs (shRNAs) for Dux knockdown were synthesized and subcloned into pLKO.1 vector expressing a puromycin-resistant gene. The shRNA sequences used in this study are listed in Table S7.
Flow Cytometry
Single-cell suspensions were evaluated on an LSRII Flow Cytometer System (BD Biosciences). Cell viability was determined by 1 μM 4’−6-diamidino-2-phenylindole (DAPI, Molecular Probes) staining in unfixed cells, and data were analyzed with FlowJo software.
Luciferase Assay
To screen sgRNAs targeted to the MERVL LTR, 293T cells or mESCs were infected with dCas9-VP64, helper MS2-p65-HSF1 (Konermann et al., 2015), and transfected in triplicate using Lipofectamine 2000 with 10 ng pRL-TK, 200 ng of pGL3-MERVL (Macfarlan et al., 2011). Forty-eight hr after transfection, Luciferase and Renilla activity were determined using Dual-Glo Luciferase Assay kit (#E2920, Promega) following manufacturer’s instructions. All Luciferase activities were normalized to the Renilla activity in the same sample.
To investigate miR-344 regulation on Zmym2 and Lsd1 3’UTR, 293T cells were transfected in triplicate using Lipofectamine 2000 with 10 ng pRL-TK, 200 ng of MDH1-PGK-GFP-miR-344, and 200 ng psiCheck2 containing Zmym2 or Lsd1 3’UTR. Dual luciferase activity was determined as described above.
RT-qPCR
To quantify mature miR-344 expression, total RNA was extracted using Trizol. Polyuridylation was performed as described (Mei et al., 2012). Briefly, total RNA was polyuridylated with UTP by poly(U) (New England Biolabs, catalog no. M0337S) at 37 °C for 1 h in a 20 μL reaction volume. Reverse transcription was performed by using SuperScript® III First-Strand Synthesis System (Invitrogen, Cat# 18080–051) with specific primer SL-poly(A) “GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAAAAAAAAA AAAAAAAVN”. Relative expression levels were determined using LightCycler® 480 SYBR Green (Roche, 4729749001). Gene expression was normalized to U6.
To quantify mRNA expression, total RNA was extracted using the RNeasy kit (Qiagen). Reverse transcription was performed and cDNA was generated using qScript (Quanta, Cat# 95048). Gene expression was normalized to beta-Actin.
For embryo RT-qPCR analysis, RNA from embryos was extracted and purified by using Arcturus™ PicoPureTM RNA Isolation Kit (Applied Biosystems) and then reversely transcribed using 5×All-In-One RT Master Mix (Applied Biologic Materials, G492) according to manufacturer’s recommendations. Quantitative RT-PCR was performed using SYBR Premix Ex Taq II (Takara, RR820B), and signals were detected with an ABI7500 Real-Time PCR system (Applied BioSystems). Gene expression was normalized to H2afz. The primers for qPCR are provided in Table S7.
ChIP-seq and Data Analysis
ZMYM2 ChIP was performed as previously described (Ding et al., 2015) by using FLAG antibody (Sigma, F1804) based IP in Zmym2–3xFLAG knockin ESCs. We prepared 6 μl of 138 ng/μl and 9 μl of 232 ng/μl as input, 32 μl 0.184 ng/μl and 32 μl 0.266 ng/μl FLAG ChIPed DNA to prepare ZMYM2 ChIP-seq libraries. Massively parallel sequencing was performed with the Illumina HiSeq2500 according to the manufacturer’s protocol, and single-end 50 bp-length reads were produced. After sequencing, FastQC (v0.11.5) was used to check the sequencing quality. Reads from two biological replicates were combined, and aligned to the mouse genome (NCBI build 37, mm9) using the bowtie (v1.0.0) program, with parameters -M 1 --chunkmbs 200. The “-M 1” parameter ensures that the best match is randomly selected if more than one equivalent best alignments are found, which is important for alignments of reads at repetitive regions. Aligned reads were converted to a binary BAM file, sorted, PCR duplicates removed, and indexed with samtools (v0.1.19), followed by visualization using IGV software.
ZMYM2 ChIP-seq peaks were determined by the MACS program (v.2.0.10), using input ChIP-seq as the control data, and parameters -q 0.01 -m 5 50, other parameters followed the default settings. ZMYM2 binding motif was determined using the findMotifsGenome.pl script in HOMER tools, and the top de novo motif was used. Intensity heat-maps of ChIP-seq enrichment at ZMYM2-bound regions were obtained by ngsplot program (v2.61, available at https://code.google.com/p/ngsplot/). Public ChIP-seq data were downloaded (refer to Key Resource Table) and processed with the same settings.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-β-Actin, Clone AC-15 (human/mouse) | Sigma-Aldrich | Cat. A5441; RRID:AB_476744 |
| Anti-mouse-ZMYM2 antibody, clone F4 | Abcam | Cat. ab30783 (AP: 228894); RRID:AB_874057 |
| Anti-mouse/human LSD1, | Abcam | Cat. ab129195; RRID:AB_11145494 |
| Anti-mouse ZSCAN4 | Millipore | Cat. AB4340 |
| Anti-mouse/human CHD4 | Abcam | Cat. ab70469; RRID:AB_2229454 |
| Anti-mouse/human HDAC1 | Bethyl | Cat. A300-713A; RRID:AB_533395 |
| Anti-mouse/human HDAC2 | Bethyl | Cat. A300-705A; RRID:AB_533399 |
| Anti-Histone H3, monomethyl (Lys4) | Abcam | Cat. ab8895; RRID:AB_306847 |
| Anti-Histone H3, trimethyl (Lys4) | Abcam | Cat. ab8580; RRID:AB_306649 |
| Anti-Histone H3 | Abcam | Cat. ab1791; RRID:AB_302613 |
| Anti-FLAG | Sigma-Aldrich | Cat. A8592; RRID:AB_439702 |
| Anti-PROLIFERIN | Santa Cruz | Cat. Sc-271891; RRID: AB 10710396 |
| Anti-TPBPA | Abcam | Cat: ab104401; RRID: AB 10901888 |
| Anti-GFP | Proteintech Group | Cat. 50430-2-AP; RRID:AB_11042881 |
| Experimental Models: Cell & mouse Lines | ||
| C57BL/6(C57) mice | Beijing Vital River Laboratory Animal Technology | Stock No.: 213 |
| DBA2 mice | Beijing Vital River Laboratory Animal Technology | Stock No.: 214 |
| Mouse embryonic stem cell line J1 | ATCC | SCRC-1010 |
| Zmym2GT/GT mESCs | This study | N/A |
| Zmym23Flag mESCs | This study | N/A |
| Zmym2−/− Knock out mESCs | This study | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMEM | GIBCO | Cat. 11965-092 |
| Heat inactivated FBS | GIBCO | Cat. 35-010-cv |
| Penicillin-Streptomycin (5,000 U/mL) | GIBCO | Cat. 15070-063 |
| Glutamine | GIBCO | Cat. 25030-081 |
| Non-essential NEAA | GIBCO | Cat. 1140-050 |
| 2-Mercaptoethanol | Sigma-Aldrich | Cat. M6250 |
| Puromycin | Sigma-Aldrich | Cat. P9620-10ML |
| Blasticidin S HCl | Life Technologies | Cat. A11139-03 |
| Zeocin | Fisher Scientific | Cat. Z22100-0.25 |
| Oligonucleotides | ||
| mmu-miR-344-3p mimics and Zmym2 siRNAs | GenePharma (Shanghai, China) | Table S7 |
| Recombinant DNA | ||
| lenti sgRNA(MS2)_zeo backbone | Konermann et al., 2015 | RRID:Addgene_61427 |
| lenti MS2-P65-HSF1_Hygro | Konermann et al., 2015 | RRID:Addgene_61426 |
| lenti dCAS-VP64_Blast | Konermann et al., 2015 | RRID:Addgene_61425 |
| phH1-gRNA | Kabadi et al., 2014 | RRID:Addgene_53186 |
| pmU6-gRNA | Kabadi et al., 2014 | RRID:Addgene_53187 |
| phU6-gRNA | Kabadi et al., 2014 | RRID:Addgene_53188 |
| ph7SK-gRNA | Kabadi et al., 2014 | RRID:Addgene_53189 |
| pLV hUbC-dCas9-T2A-GFP | Kabadi et al., 2014 | RRID:Addgene_53191 |
| 2C::tdTomato Reporter | Macfarlan et al., 2012 | RRID:Addgene_40281 |
| pZscan4c-EGFP | Dan et al., 2013 | N/A |
| Critical Commercial Assays | ||
| Dual-Glo Luciferase Assay | Promega | Cat. E2920 |
| SimpleChIP Plus Enzymatic Chromatin IP Kit | Cell Signaling Tech. | Cat. #9005 |
| Deposited Data | ||
| RNA-seq on control (EV) and MERVL or miR-344-2 activated cells, raw and processed data | This study | NCBI GEO: GSE119819 |
| RNA-Seq on WT and Zmym2 deficient mESCs, raw and processed data | This study | NCBI GEO: GSE119819 |
| RNA-seq on 8-cell and blastocyst injected with NC, raw data and processed data | This study | NCBI GEO: GSE119819 |
| RNA-seq on 8-cell and blastocyst injected with siRNA against Zmym2, raw data and processed data | This study | NCBI GEO: GSE119819 |
| RNA-seq on 8-cell and blastocyst injected with miR-344 mimics, raw data and processed data | This study | NCBI GEO: GSE119819 |
| RNA-seq on control and MERVL or miR-344-2 activated cells after sorting of DR+/+ population, raw and processed data. | This study | NCBI GEO: GSE119819 |
| RNA-seq of 2-cell embryos. | This study | NCBI GEO: GSE119819 |
| ChIP-seq on 3Flag-Zmym2 in mESCs, raw and processed data | This study | NCBI GEO: GSE119819 |
| ChIP-seq of LSD1 in WT and Zmym2KO ESCs | This study | NCBI GEO: GSE119819 |
| ATAC-seq on DR+/+ and DR−/− mESCs population | This study | NCBI GEO: GSE119817 |
| Other Data | ||
| RNA-seq on WT and Chd4 deficient mESCs, raw data and processed data | Stevens et al., 2017 | NCBI GEO: GSE80280 |
| RNA-seq on DR−/− and DR+/+ mESCs, raw data and processed data | Macfarlan et al., 2012 | E-MTAB-5058 |
| RNA-seq on miR34a and miR34a deficient mESCs, raw data and processed data | Choi et al., 2017 | NCBI GEO: GSE69484 |
| RNA-seq on WT and DUX deficient mESCs, raw data and processed data | Hendrickson et al., 2017 | NCBI GEO: GSE85632 |
| RNA-seq on untreated 8-cell and blastocyst, raw data and processed data | Liu et al., 2016 | NCBI GEO: GSE70608 |
| ChIP-seq on LSD1 in mESCs | Whyte et al., 2012 | NCBI GEO: GSE27844 |
| ChIP-seq on DUX/DXU4 in mESCs | Whiddon et al., 2017 | NCBI GEO: GSE87279 |
| Software and Algorithms | Version | Source |
| FlowJo | 7.6.1 | https://www.flowjo.com/ |
| STAR | 2.5.3 | https://github.com/alexdobin/STAR |
| Tophat | 2.1.1 | http://ccb.jhu.edu/software/tophat/index.shtml |
| Cufflinks | 2.1.1 | http://cole-trapnell-lab.github.io/cufflinks/ |
| Diffbind | 1.16.3 | https://bioconductor.org/packages/release/bioc/html/DiffBind.html |
LSD1 ChIP-seq was performed with SimpleChIP Plus Enzymatic Chromatin IP Kit (Cell Signaling Tech. #9005) following the standard protocol. Briefly, 4 million cells were used for each ChIP experiment, with two antibodies of LSD1 (Cell Signaling Tech. #2184, clone C69G12; and Abcam, ab17721). Massively parallel sequencing was performed with the Illumina HiSeq4000 according to the manufacturer’s protocol, and paired-end 150 bp-length reads were produced. After sequencing, FastQC (v0.11.5) was used to check the sequencing quality. Reads were aligned to the mouse genome (NCBI build 37, mm9) using the bowtie2 (v2.3.4) program, with parameters -X 1000 --no-mixed --no-discordant. Aligned reads were converted to a binary BAM file, sorted, and indexed with samtools (v0.1.19), followed by visualization using IGV software.
LSD1 ChIP-seq peaks were determined by the MACS program (v.2.0.10), using input ChIP-seq as the control data, and parameters -q 0.01 -m 5 50, other parameters followed the default settings. All LSD1 peaks were imported by DiffBind (v1.16.3) program, with minimal overlap of two peaks between different samples. The ChIP-seq peaks with significantly enriched intensities (FDR < 0.01) in either WT or Zmym2KO ESCs were exported.
ChIP-qPCR
ChIP assays were performed as described (Ding et al., 2015) with minor modifications. The purified immunoprecipitated DNA was analyzed by qPCR using LightCycler® 480 SYBR Green (Roche, 4729749001) and a Roche LightCycler480 machine. The percentage of input recovery was calculated for each locus. The primary antibodies used for ChIP are as follows: ZMYM2 (Abcam, ab30783), FLAG (Sigma, F1804–5MG), and IgG (Millipore, PP64). The primers for qPCR are provided in Table S7.
RNA-seq and Data Analysis
For ESCs, total RNAs were extracted using the RNeasy kit (#74136, Qiagen) according to the manufacturer’s instructions. RNA quality was evaluated by Agilent 2100 BioAnalyzer. About 1 μg total RNA from each sample was taken for the preparation of PolyA RNA-seq libraries, and massively parallel sequencing was performed with the HiSeq4000 platform. Paired-end 150 bp-length reads were produced.
For embryos, full-length RNA-seq libraries of miR-344 mimics, Zmym2 siRNA, and non-target control RNA-injected embryos were prepared according to the Smart-seq2 protocol (Picelli et al., 2014) with minor modifications. A total of 10–20 embryos with the same treatment and embryonic stage were pooled for each reaction. In brief, injected embryos were harvested, washed several times in 0.5% BSA-PBS (Sigma) solution and subsequently picked and transferred into lysis buffer by a mouth pipette. Reverse transcription was performed using SuperScript II (Invitrogen). cDNA was pre-amplified (10 PCR cycles) and purified with Ampure XP Beads (Agencourt) at 0.8 beads/1 DNA (v/v). One microliter (1 μl) cDNA, diluted by 19 μl nuclease-free water, was used for Real-time PCR quality check. The amplified cDNA was fragmented using a Covaris sonicator (Covaris S220). To generate the sequence libraries, the KAPA Hyper Prep Kit was used following the manufacturer’s instructions. NOVA pair-end 150-bp sequencing was performed on a HiSeq 2500 or 2000 sequencer (Illumina) at Berry Genomics Corporation.
RNA-seq reads were aligned to the genome using STAR software (v2.5.3) with the default parameter settings. UCSC mm9 mouse genome, as well as the transcript annotation, was downloaded from the iGenomes site. Transcript assembly and differential expression analysis were performed using Cufflinks (v2.1.1). Assembling of novel transcripts was not allowed (-G), other parameters of Cufflinks followed the default setting. The summed RPKM (reads per kilobase per million mapped reads) of transcripts sharing each gene_id was calculated and exported by the Cuffdiff program. For expression of LTRs, a reference genome with all LTRs was created based on the RMSK database. RNA-seq intensity at each LTR region was counted by HTseq software (v0.6.1) with parameters -a 10 -m intersection-nonempty, and normalized to total mapped reads per million total reads (RPM).
Public ChIP-seq data were downloaded (refer to Key Resource Table), RNA-seq data of embryos at different stages in early embryonic development were from our previous study (Liu et al., 2016). All RNA-seq data were processed with the same settings.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Analysis
All statistical analysis was performed with GraphPad Prism (GraphPad Software, Inc.) or R (www.r-project.org/). Specific statistical method was performed as indicated in the manuscript or figure legends. For quantification of qPCR and luciferase data, unpaired t test was performed with two-tailed distribution. For testing the embryo development efficiency, a two-way analysis of variance (ANOVA) was performed assuming equal variances. For comparison of the expression of ERV elements, a non-parametric Mann-Whitney test was used. For all statistical tests, differences were considered significant at p < 0.05.
Principle component analysis (PCA) was performed for the RNA-seq expression (RPKM) data for all genes (global) or the 2C-specific genes (Macfarlan et al., 2012). In the RPKM data matrix, a minimal RPKM value of 0.1 was applied if the gene expression was less than this minimum value. Batch effects were adjusted by ComBat function implemented in the sva Bioconductor package (v.3.18.0). The expression data matrix was imported by Cluster 3.0 software (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) for PCA analysis.
Gene ontology analyses were performed using the DAVID gene ontology functional annotation tool (http://david.abcc.ncifcrf.gov/tools.jsp) with all NCBI Mus musculus genes as a reference list.
GSEA (v3.0, available at http://www.broadinstitute.org/gsea) was used to determine whether the set of 2C-signature genes was statistically enriched in F-sgRNA-activated versus EV, miR-344-activation versus EV, and Zmym2GT/GT versus WT ESCs RNA-seq data. The 2C-signature genes (n=254) were from a published RNA-seq dataset containing genes that are only activated in 2C embryos in early development (Wu et al., 2016). The normalized enrichment score (NES), and FDR q-value were indicated for each enrichment test.
DATA AND CODE AVAILABILITY
The accession number for the data reported in this paper is NCBI Gene Expression Omnibus (GEO): GSE119820.
Supplementary Material
Table S1. ATAC-seq and SILAC data comparing ESCs and 2CLCs. Related to Figure 2.
Table S3. ZMYM2 interactome in ESCs. Related to Figure 5.
Table S4. ZMYM2, LSD1 peaks versus random peaks. Related to Figure 5.
Table S5. Final 101 genes. Related to Figure 5.
Table S6. Developmental efficiency. Related to Figure 6.
Table S7. Primers. Related to STAR Methods.
HIGHLIGHTS.
Activation of endogenous MERVL or miR-344 induces 2CLCs with totipotency features
miR-344 directly silences Zmym2 and Lsd1 to activate MERVL and 2C-specific genes
Zmym2 zygotic depletion compromises embryo totipotency-to-pluripotency transition
DUX directly binds to the miR-344 cluster and activates its expression
ACKNOWLEDGMENTS
We thank Dr. Yanhong Shi (Beckman Research Institute/City of Hope) for the Lsd1 3’-UTR luciferase reporter construct, Dr. Lin Liu (Nankai University) for pZscan4c-EGFP1 reporter construct, Dr. Stephen Tapscott for the mouse Dux and human DUX4 expression constructs. This research was funded by grants from the National Institutes of Health (NIH) (1R01GM129157; 1R01HD095938; and 1R01HD097268) and New York State Stem Cell Fund (NYSTEM) (C32583GG and C32569GG) to J.W., and The National Key R&D Program of China (2016YFA0100400 and 2017YFA0102600), the National Natural Science Foundation of China (31721003; 31871446; and 81630035), the Shanghai Rising-Star Program (19QA1409600), the Shanghai Chenguang Program (16CG17), the key project of the Science and Technology Commission of Shanghai Municipality (19JC1415300), the Shanghai municipal medical and health discipline construction projects (2017ZZ02015), to S.G. and J.C.. J.W. is a recipient of Irma T. Hirschl and Weill-Caulier Trusts Career Scientist Award. F.Y. was a visiting student at Icahn School of Medicine at Mount Sinai sponsored by the China Scholarship Council.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at …
DECLARATION OF INTERESTS
All authors declare no competing interests.
SUPPORTING CITATIONS
The following references appear in the Supplemental Information: (Ding et al., 2015; Kabadi et al., 2014; Liu et al., 2016; Mei et al., 2012; Picelli et al., 2014; Yu et al., 2016).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agarwal V, Bell GW, Nam JW, and Bartel DP (2015). Predicting effective microRNA target sites in mammalian mRNAs. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Martinez E, Chen X, Webber A, Mould AP, Seifert A, Hay RT, and Sharrocks AD (2015). Screen for multi-SUMO-binding proteins reveals a multi-SIM-binding mechanism for recruitment of the transcriptional regulator ZMYM2 to chromatin. Proceedings of the National Academy of Sciences of the United States of America 112, E4854–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancelin K, Syx L, Borensztein M, Ranisavljevic N, Vassilev I, Briseno-Roa L, Liu T, Metzger E, Servant N, Barillot E, et al. (2016). Maternal LSD1/KDM1A is an essential regulator of chromatin and transcription landscapes during zygotic genome activation. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsos M, and Torres-Padilla ME (2016). Building up the nucleus: nuclear organization in the establishment of totipotency and pluripotency during mammalian development. Genes Dev 30, 611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Current protocols in molecular biology 109, 21 29 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Lin CP, Risso D, Chen S, Kim TA, Tan MH, Li JB, Wu Y, Chen C, Xuan Z, et al. (2017). Deficiency of microRNA miR-34a expands cell fate potential in pluripotent stem cells. Science 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J, Li M, Yang J, Li J, Okuka M, Ye X, and Liu L. (2013). Roles for Tbx3 in regulation of two-cell state and telomere elongation in mouse ES cells. Scientific reports 3, 3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang-Nguyen TQ, and Torres-Padilla ME (2015). How cells build totipotency and pluripotency: nuclear, chromatin and transcriptional architecture. Curr Opin Cell Biol 34, 9–15. [DOI] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, and Trono D. (2017). DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nature genetics 49, 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Huang X, Shao N, Zhou H, Lee DF, Faiola F, Fidalgo M, Guallar D, Saunders A, Shliaha PV, et al. (2015). Tex10 Coordinates Epigenetic Control of Super-Enhancer Activity in Pluripotency and Reprogramming. Cell stem cell 16, 653–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Xu H, Faiola F, Ma’ayan A, and Wang J. (2012). Oct4 links multiple epigenetic pathways to the pluripotency network. Cell research 22, 155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley-Maslin MA, Svensson V, Krueger C, Stubbs TM, Giehr P, Krueger F, Miragaia RJ, Kyriakopoulos C, Berrens RV, Milagre I, et al. (2016). MERVL/Zscan4 Network Activation Results in Transient Genome-wide DNA Demethylation of mESCs. Cell reports 17, 179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford WD, Pfaff SL, and Macfarlan TS (2013). Transposable elements as genetic regulatory substrates in early development. Trends in cell biology 23, 218–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ (2010). microRNAs, the cell’s Nepenthe: clearing the past during the maternal-to-zygotic transition and cellular reprogramming. Current opinion in genetics & development 20, 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocke CB, and Yu H. (2008). ZNF198 stabilizes the LSD1-CoREST-HDAC1 complex on chromatin through its MYM-type zinc fingers. PloS one 3, e3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guallar D, Bi X, Pardavila JA, Huang X, Saenz C, Shi X, Zhou H, Faiola F, Ding J, Haruehanroengra P, et al. (2018). RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat Genet 50, 443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzo CM, Ringel A, Cox E, Uzoma I, Zhu H, Blackshaw S, Wolberger C, and Matunis MJ (2014). Characterization of the SUMO-binding activity of the myeloproliferative and mental retardation (MYM)-type zinc fingers in ZNF261 and ZNF198. PloS one 9, e105271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson PG, Dorais JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. (2017). Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nature genetics 49, 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks IA, Lyon D, Su D, Skotte NH, Daniel JA, Jensen LJ, and Nielsen ML (2018). Site-specific characterization of endogenous SUMOylation across species and organs. Nature communications 9, 2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiuchi T, Enriquez-Gasca R, Mizutani E, Boskovic A, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas JM, and Torres-Padilla ME (2015). Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nature structural & molecular biology 22, 662–671. [DOI] [PubMed] [Google Scholar]
- Iturbide A, and Torres-Padilla ME (2017). Starting embryonic transcription for the first time. Nature genetics 49, 820–821. [DOI] [PubMed] [Google Scholar]
- Kabadi AM, Ousterout DG, Hilton IB, and Gersbach CA (2014). Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic acids research 42, e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunapuli P, Kasyapa CS, Chin SF, Caldas C, and Cowell JK (2006). ZNF198, a zinc finger protein rearranged in myeloproliferative disease, localizes to the PML nuclear bodies and interacts with SUMO-1 and PML. Experimental cell research 312, 3739–3751. [DOI] [PubMed] [Google Scholar]
- Li P, Wang L, Bennett BD, Wang J, Li J, Qin Y, Takaku M, Wade PA, Wong J, and Hu G. (2017). Rif1 promotes a repressive chromatin state to safeguard against endogenous retrovirus activation. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WM, Pang RT, Chiu PC, Wong BP, Lao K, Lee KF, and Yeung WS (2012). Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proceedings of the National Academy of Sciences of the United States of America 109, 490–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang C, Liu W, Li J, Li C, Kou X, Chen J, Zhao Y, Gao H, Wang H, et al. (2016). Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562. [DOI] [PubMed] [Google Scholar]
- Lu F, and Zhang Y. (2015). Cell totipotency: molecular features, induction, and maintenance. National science review 2, 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews SE, Franco L, Rosenfeld MG, Ren B, et al. (2011). Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes & development 25, 594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, and Pfaff SL (2012). Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, Thompson PJ, Goyal P, Jones SJ, Singh PB, Karimi MM, and Lorincz MC (2013). Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenetics & chromatin 6, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M. (2006). Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol 7, 952–958. [DOI] [PubMed] [Google Scholar]
- Mei Q, Li X, Meng Y, Wu Z, Guo M, Zhao Y, Fu X, and Han W. (2012). A facile and specific assay for quantifying microRNA by an optimized RT-qPCR approach. PloS one 7, e46890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, and Sandberg R. (2014). Full-length RNA-seq from single cells using Smart-seq2. Nature protocols 9, 171–181. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Terrones D, Gaume X, Ishiuchi T, Weiss A, Kopp A, Kruse K, Penning A, Vaquerizas JM, Brino L, and Torres-Padilla ME (2018). A molecular roadmap for the emergence of early-embryonic-like cells in culture. Nature genetics 50, 106–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, Viville S, Jakobsson J, Pfaff SL, and Trono D. (2013). TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome research 23, 452–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoorlemmer J, Perez-Palacios R, Climent M, Guallar D, and Muniesa P. (2014). Regulation of Mouse Retroelement MuERV-L/MERVL Expression by REX1 and Epigenetic Control of Stem Cell Potency. Frontiers in oncology 4, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, and Shi Y. (2003). Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422, 735–738. [DOI] [PubMed] [Google Scholar]
- Smedley D, Hamoudi R, Lu YJ, Cooper C, and Shipley J. (1999). Cloning and mapping of members of the MYM family. Genomics 60, 244–247. [DOI] [PubMed] [Google Scholar]
- Stevens TJ, Lando D, Basu S, Atkinson LP, Cao Y, Lee SF, Leeb M, Wohlfahrt KJ, Boucher W, O’Shaughnessy-Kirwan A, et al. (2017). 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkowski AK (1959). Experiments on the development of isolated blastomers of mouse eggs. Nature 184, 1286–1287. [DOI] [PubMed] [Google Scholar]
- Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, et al. (2009). The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nature genetics 41, 125–129. [DOI] [PubMed] [Google Scholar]
- Wasson JA, Simon AK, Myrick DA, Wolf G, Driscoll S, Pfaff SL, Macfarlan TS, and Katz DJ (2016). Maternally provided LSD1/KDM1A enables the maternal-to-zygotic transition and prevents defects that manifest postnatally. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiddon JL, Langford AT, Wong CJ, Zhong JW, and Tapscott SJ (2017). Conservation and innovation in the DUX4-family gene network. Nat Genet 49, 935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte W, Bilodeau S, Orlando D, Hoke H, Frampton G, Foster C, Cowley S, and Young R. (2012). Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, Zhang B, Liu B, Wang Q, Xia W, et al. (2016). The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657. [DOI] [PubMed] [Google Scholar]
- Wu Q, Chen X, Zhang J, Loh YH, Low TY, Zhang W, Zhang W, Sze SK, Lim B, and Ng HH (2006). Sall4 interacts with Nanog and co-occupies Nanog genomic sites in embryonic stem cells. The Journal of biological chemistry 281, 24090–24094. [DOI] [PubMed] [Google Scholar]
- Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, et al. (2013). Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500, 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan YL, Zhang C, Hao J, Wang XL, Ming J, Mi L, Na J, Hu X, and Wang Y. (2019). DPPA2/4 and SUMO E3 ligase PIAS4 opposingly regulate zygotic transcriptional program. PLoS biology 17, e3000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BX, El Farran CA, Guo HC, Yu T, Fang HT, Wang HF, Schlesinger S, Seah YF, Goh GY, Neo SP, et al. (2015). Systematic identification of factors for provirus silencing in embryonic stem cells. Cell 163, 230–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Wang Y, Chen J, Li H, Kang L, Zhang Y, Chen S, Zhu B, and Gao S. (2011). RCOR2 is a subunit of the LSD1 complex that regulates ESC property and substitutes for SOX2 in reprogramming somatic cells to pluripotency. Stem cells 29, 791–801. [DOI] [PubMed] [Google Scholar]
- Yu C, Ji SY, Sha QQ, Dang YJ, Zhou JJ, Zhang YL, Liu Y, Wang ZW, Hu BQ, Sun QY, et al. (2016). BTG4 is a meiotic cell cycle-coupled maternal-zygotic transition licensing factor in oocytes. Nature structural & molecular biology 23, 387–394. [DOI] [PubMed] [Google Scholar]
- Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, et al. (2010). Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 464, 858–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhao J, Dahan P, Lu V, Zhang C, Li H, and Teitell MA (2018). Metabolism in Pluripotent Stem Cells and Early Mammalian Development. Cell metabolism 27, 332–338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. ATAC-seq and SILAC data comparing ESCs and 2CLCs. Related to Figure 2.
Table S3. ZMYM2 interactome in ESCs. Related to Figure 5.
Table S4. ZMYM2, LSD1 peaks versus random peaks. Related to Figure 5.
Table S5. Final 101 genes. Related to Figure 5.
Table S6. Developmental efficiency. Related to Figure 6.
Table S7. Primers. Related to STAR Methods.
Data Availability Statement
The accession number for the data reported in this paper is NCBI Gene Expression Omnibus (GEO): GSE119820.