Abstract
While antimicrobial resistance (AMR) continues to be a major public health problem in Pakistan, data regarding trends of resistance among pathogenic bacteria remains scarce, with few studies presenting long-term trends in AMR. This study was therefore designed to analyze long-term AMR trends at a national level in Pakistan. We report here results of a comprehensive analysis of resistance, among pathogens isolated from blood and cerebrospinal fluid (CSF), between 2011 and 2015. Susceptibility data was obtained from a local laboratory with collection points all across Pakistan (Chughtai Laboratory). Resistance proportions to most commonly used antimicrobials were calculated for each pathogen over a period of five years. While Acinetobacter species demonstrated highest resistance rates to all tested antimicrobials, a sharp increase in carbapenem resistance was the most noticeable (50%-95%) between 2011–2015. Our results also highlight the presence of third and fourth generation cephalosporins resistance in Salmonella enterica serovar Typhi in Pakistan. Interestingly, where rise in AMR was being observed in some major invasive pathogens, decreasing resistance trends were observed in Staphylococcus aureus, against commonly used antimicrobials. Overall pathogens isolated from blood and CSF between 2011–2015, showed an increase in resistance towards commonly used antimicrobials.
Introduction
The use of broad-spectrum antimicrobials for treatment of invasive infections, defined as bloodstream and cerebrospinal fluid (CSF) infections, has resulted in an increase in antimicrobial resistance (AMR). As a consequence, the treatment of such infections is becoming increasingly difficult leading to treatment failures and increased mortality. Availability of over the counter drugs in developing countries such as Pakistan has led to the epidemic of AMR in these countries. Indiscriminate usage of antimicrobials exerts an increased selection pressure on the bacterial population resulting in accelerated emergence of AMR [1]. Recent estimates suggested that invasive infections, in particular antimicrobial resistant invasive infections, account for 5.3 million deaths around the world annually. A significant proportion of these deaths occurs in low to middle income countries (LMIC) such as Pakistan [2]. While developing countries are battling an accelerated spread of AMR, developed countries are also experiencing the same trend. In the United States between 1999 and 2012, 47.9% Acinetobacter baumanii were carbapenem resistant, 68.4% Staphylococcus epidermidis were ciprofloxacin resistant, and 13.7% Escherichia coli (E. coli) were β-lactam resistant [3–5]. Furthermore, a similar resistance landscape had emerged in Canada between 2007 and 2011, where 27% of E.coli were resistant to ciprofloxacin, 19.3% Staphylococcus aureus (S. aureus) were resistant to methicillin, and 16.8% Streptococcus pneumoniae (S. pneumoniae) were resistant to penicillin [6].
Unavailability of reliable data in the developing countries like Pakistan, makes it difficult to develop efficient methods to monitor and control AMR [1]. The limited number of studies undertaken to investigate resistance in Pakistan indicate that most of the pathogens are resistant to commonly used antibiotics. For instance, between 1997 and 2014, 91% of E. coli were reported to be resistant to amikacin, while 91.7% Salmonella enterica serovar Typhi (S. Typhi) were fluoroquinolone resistant, and 90.9% Acinetobacter species were imipenem resistant [7–9]. These studies indicate that AMR poses a burgeoning public health problem in Pakistan and highlights the need of an in-depth assessment of AMR situation.
Towards this goal, the present study examined AMR data from a large diagnostic lab to identify (i) pathogens that are most commonly isolated from blood and CSF cultures in Pakistan, (ii) the patterns of resistance amongst these pathogens, and (iii) their co-resistance trends.
Materials and methods
For this retrospective cross-sectional study, AMR data on pathogens isolated between 2011–2015 and their antimicrobial susceptibilities were obtained from Chughtai Lab (CL), a local diagnostic facility with over 180 collection centers in 60 cities of Pakistan. Standard biochemical tests and Analytical Profile Index (API) identification kits (BioMerieux) [10] were used to identify bacterial species at CL. Antimicrobial susceptibility testing was done using disc diffusion method following Clinical Laboratory & Standards Institute (CLSI) guidelines [11].
Data on positive cultures were obtained from an electronic database containing patients’ reports. Information obtained on each case included: (1) patient’s age, (2) patient’s sex, (3) year of sample collection, (4) specimen source, (5) city in which the sample was drawn, (6) isolated pathogen in the positive culture, and (7) susceptibility results (defined as susceptible, intermediate, or resistant).
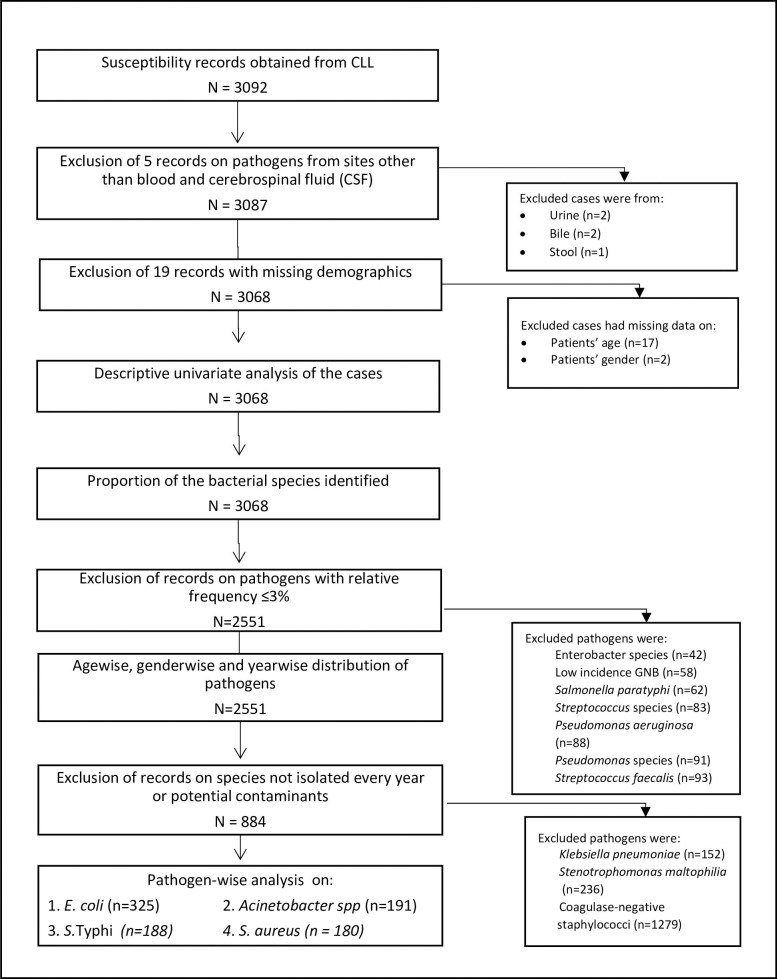
Of a total of 3092 cases, datasets with missing information on age (n = 17) and sex (n = 2) were excluded from the study. Cases with information on pathogens isolated from sites other than blood and CSF (n = 5) were also excluded from the dataset (Fig 1). Most commonly isolated microorganisms from blood and CSF cultures were identified and used for all later analysis. Microorganisms that demonstrated low frequency of isolation and were found in less than 100 samples were also eliminated from the analysis. Pathogens were stratified by year of sample collection and patients’ age (<5 years, 6–18 years, 19–45 years, 46–65 years, and 65< years), and patients’ sex. For each case, intermediate resistance was considered as resistant. Susceptibility for all tested antimicrobials was examined and reported in each species separately. The bacterial species analyzed included E. coli, Acinetobacter species, S. aureus, and S. Typhi.
Fig 1. Flow of cases.
Flow of the cases through the study has been shown.
Statistical analysis
Statistical analyses were performed using IBM SPSS statistics 22 (SPSS Inc., Chicago, Ill., USA). SAS university edition software (Cary, NC: SAS Institute Inc) was employed to perform Cochran Armitage test for trends. Figures were plotted using Circa software (www.omgenomics.com). P-value of <0.05 was considered significant for all statistical analyses. Frequency of cases was calculated by pathogens, by age and sex of patients, site of infection, and year of isolation using univariate analysis. Unadjusted resistance rates in pathogens were determined over age groups, sex, and year of isolation using univariate analysis. As the number of isolates varied annually throughout the study period, proportions of the resistant pathogens were evaluated to normalize the data. The proportions of resistant isolates were examined over age and sex respectively using Fisher’s exact test, with Bonferroni corrections. Cochran Armitage test for trends was used to investigate trends of AMR over time. Pairwise resistance (referred to as co-resistance) trends were examined using chi-square test of independence. For significantly associated pairwise resistance, binary logistic regression was used to calculate odds ratios, which outlined odd of resistance to one antimicrobial given resistance to another antimicrobial. Antimicrobials with an overall resistance above 1.5% were plotted in a Circos-based AMR map for each pathogen.
Results
Geographical, temporal, and demographical distribution of participants
Burden of invasive infections vary between different geographic regions as well as patients’ demographics [2, 12]. Hence, we wanted to profile the distribution of bacterial pathogens isolated from blood and CSF cultures over patients’ demographics, geographic location and year of isolation. Results of univariate analysis of demographic distribution of pathogens associated with invasive infections and the phenotypic characteristics of such pathogens, are presented in Table 1.
Table 1. Attributes of pathogens isolated from blood and cerebrospinal fluid between 2011–2015 in Pakistan.
| Number of bacterial pathogens (n = 3068) |
|
|---|---|
| Age (years) | |
| ≤5 | 1061 (34.6%) |
| 6–18 | 262 (8.5%) |
| 19–45 | 711 (23.2%) |
| 46–65 | 550 (17.9%) |
| >65 | 484 (15.8%) |
| Sex | |
| Female | 1245 (40.6%) |
| Male | 1823 (59.4%) |
| Site of Infection | |
| Blood | 2917 (95.1%) |
| CSF | 151 (4.9%) |
| Year of Isolation | |
| 2011 | 262 (8.5%) |
| 2012 | 296 (9.6%) |
| 2013 | 493 (16.1%) |
| 2014 | 750 (24.4%) |
| 2015 | 1267 (41.3%) |
| Location of sample collection | |
| Punjab | 2701 (87.9%) |
| KPK | 338 (11%) |
| Sindh | 25 (1%) |
| Baluchistan | 1 (0%) |
| No information | 3 (.1%) |
| Phenotypic characters/Gram Staining | |
| Gram-positive bacteria | 1433 (46.7%) |
| Gram-negative bacteria | 1635 (53.3%) |
A total of 3068 microorganisms were isolated from blood and CSF cultures between 2011 and 2015. These cases were reported from 44 cities, from all four provinces of Pakistan, however, majority of cases were reported from 13 cities, whereas the remaining 31 cities reported less than 11 cases. The median number of cases from any given city was 4 with an interquartile range of 2–15. Analysis by province indicated that highest number of cases were reported from Punjab (87.9%), followed by Khyber Pakhtunkhwa (11%), Sindh (1%) and Baluchistan (0.033%). Within Punjab most (60.5%) of reported cases were from the city of Lahore, which is the most populous city of Punjab followed by Faisalabad (18.3%) and Abbottabad (8.3%). Detailed distribution of cases over geographic location is given in the S1 Table.
Temporal and demographical distribution of pathogens frequently isolated from blood and CSF cultures
Distribution of isolates by patient characteristics (age and sex) and by year is given in Table 2. Previous studies have shown that the incidence rates of pathogen-specific invasive infections fluctuate on the basis of their temporal, spatial, and demographical characteristics [13]. Hence, the next logical step was to determine the distribution patterns of pathogens isolated from blood and CSF specimens. To investigate the temporal and demographical distribution of the bacterial species, univariate analyses were performed. Over 75% of these pathogens were from one of the seven most common bacterial species. These included CoNS (41.7%), E. coli (10.6%), Stenotrophomonas. maltophilia (S. maltophilia) (6.2%), Acinetobacter species (6.2%), S. Typhi (6.1%), S. aureus (5.9%), and Klebseilla pneumoniae (K. pneumoniae) (5%). Since data was collected from a diagnostic laboratory, it was difficult to obtain data on patient characteristics such as clinical manifestation as well as mortality and morbidity rates for CoNS. Therefore, samples containing CoNS were eliminated from the study (please see discussion for further information on CoNS in Pakistan).
Table 2. Profile of common bacterial species isolated from blood and cerebrospinal fluid (CSF) cultures with corresponding demographical and temporal distribution.
| Organism | Age (years) | Sex | Year | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <5 | 6–18 | 19–45 | 46–65 | >65 | Female | Male | 2011 | 2012 | 2013 | 2014 | 2015 | ||
| Coagulase-negative staphylococci | 444 (41.8%) | 74 (28.2%) | 297 (41.8%) | 255 (19.9%) | 209 (43.2%) | 557 (44.7%) | 722 (39.6%) | 101 (38.5%) | 111 (37.5%) | 198 (40.2%) | 337 (44.9%) | 532 (42%) | 1279 (41.7%) |
| Escherichia coli | 57 (5.4%) | 11 (4.2%) | 58 (8.2%) | 90 (27.7%) | 109 (22.5%) | 144 (11.6%) | 181 (9.9%) | 44 (16.8%) | 53 (17.9%) | 73 (14.8%) | 66 (8.8%) | 89 (7%) | 325 (10.6%) |
| Stenotrophomonas maltophilia | 215 (20.3%) | 3(1.15%) | 8 (1.1%) | 3(0.55%) | 7 (1.4%) | 76 (6.1%) | 160 (8.8%) | 0 | 0 | 5(1.01%) | 46 (6.1%) | 185 (14.6%) | 236 (7.7%) |
| Acinetobacter species | 85 (8%) | 9 (3.4%) | 41 (5.8%) | 35 (18.3%) | 21 (4.3%) | 81 (6.5%) | 110 (6%) | 15 (5.7%) | 16 (5.4%) | 26 (5.3%) | 44 (5.9%) | 90 (7.1%) | 191 (6.2%) |
| Salmonella enterica serovar Typhi | 16 (1.5%) | 82 (31.3%) | 86 (12.1%) | 3(0.55%) | 1(0.21%) | 81 (6.5%) | 107 (5.9%) | 13 (5%) | 20 (6.8%) | 33 (6.7%) | 58 (7.7%) | 64 (5.1%) | 188 (6.1%) |
| Staphylococcus aureus | 41 (3.9%) | 13 (5%) | 52 (7.3%) | 45 (25%) | 29 (6%) | 71 (5.7%) | 109 (6%) | 19 (7.3%) | 20 (6.8%) | 33 (6.7%) | 47 (6.3%) | 61 (4.8%) | 180 (5.9%) |
| Klebsiella pneumoniae | 66 (6.2%) | 8 (3.1%) | 26 (3.7%) | 29 (19.1%) | 23 (4.8%) | 53 (4.3%) | 99 (5.4%) | 1(0.38%) | 0 | 29 (5.9%) | 51 (6.8%) | 71 (5.6%) | 152 (5%) |
Data are n (% isolates in a column). Bacterial species with relative frequency above 5% are shown.in the table.
Antimicrobial resistance (AMR) trends in pathogens isolated from blood and CSF cultures
Variations in the treatment approaches of physicians within a geographical location, patients’ compliance, and the ability of pathogens to acquire and disseminate resistance, impact resistance trends in different pathogens [14]. The ability of resistant pathogens to cause infections is also dependent on host-related factors including age, gender, and co-morbidities [15]. Towards this, we determined temporal and demographical AMR trends in pathogens most frequently isolated from blood and CSF. Susceptibility data was not available for all years throughout the study period on S. maltophilia and K. pneumoniae, therefore these two pathogens were excluded from the analysis. Moreover, in the absence of clinical data on patients to support CoNS role in disease manifestation, CoNS was taken out of the downstream analysis. Each of the remaining four pathogens were analyzed separately. For each isolate, susceptibility data for all antimicrobials was not available. To account for missing values, available case approach was employed to analyze resistance trends for each antimicrobial. As a result, the number of data points (n) varied between analyses involving different antimicrobials in a pathogen. Tables 3–6 and Figs 2–5 demonstrate resistance trends in E. coli, Acinetobacter species, S. Typhi and S. aureus.
Table 3. Temporal, age-wise, and gender-wise prevalence of resistant Escherichia coli strains isolated from blood and cerebrospinal fluid (CSF) cultures in Pakistan (2011–2015).
| Antimicrobials | Antimicrobial group | Prevalence (%) of resistant organisms isolated from patients of different age-groups | Prevalence (%) of resistant organisms isolated from males and female patients | Year-wise prevalence (%) of resistant organisms | Total | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <5 Years | 6–18 Years | 19–45 Years | 46–65 Years | >65 Years | P for difference | Female | Male | P for difference | 2011 | 2012 | 2013 | 2014 | 2015 | P for trend | |||
| Cephalexin | Cephalosporins | 56/56 (100%) | 10/11 (90.9%) | 55/58 (94.8%) | 88/90 (97.8%) | 107/109 (98.2%) | NS | 139/144 (96.5%) | 177/180 (98.3%) | NS | 37/43 (86%) | 52/53 (98.1%) | 72/73 (98.6%) | 66/66 (100%) | 89/89 (100%) | < .0001 | 316/324 (97.5%) |
| Cephradine | Cephalosporins | 56/56 (100%) | 10/11 (90.9%) | 55/58 (94.8%) | 88/90 (97.8%) | 107/109 (98.2%) | NS | 139/144 (96.5%) | 177/180 (98.3%) | NS | 37/43 (86%) | 52/53 (98.1%) | 72/73 (98.6%) | 66/66 (100%) | 89/89 (100%) | < .0001 | 316/324 (97.5%) |
| Cefaclor | Cephalosporins | 53/57 (93%) | 10/11 (90.9%) | 54/58 (93.1%) | 82/90 (91.1%) | 102/109 (93.6%) | NS | 133/144 (92.4%) | 168/181 (92.8%) | NS | 40/44 (90.9%) | 52/53 (98.1%) | 68/73 (93.2%) | 56/66 (84.8%) | 85/89 (95.5%) | NS | 301/325 (92.6%) |
| Cefuroxime | Cephalosporins | 52/57 (91.2%) | 8/9 (88.9%) | 51/54 (94.4%) | 79/87 (90.8%) | 96/105 (91.4%) | NS | 124/136 (91.2%) | 162/176 (92%) | NS | 28/31 (90.3%) | 49/53 (92.5%) | 68/73 (93.2%) | 56/66 (84.8%) | 85/89 (95.5%) | NS | 286/312 (91.7%) |
| Ampicillin-Sulbactam | Penicillin-Inhibitor | 45/50 (90%) | 7/7 (100%) | 35/38 (92.1%) | 61/67 (91%) | 78/88 (88.6%) | NS | 91/101 (90.1%) | 135/149 (90.6%) | NS | 17/21 (81%) | 11/12 (91.7%) | 57/62 (91.9%) | 56/66 (84.8%) | 85/89 (95.5%) | NS | 226/250 (90.4%) |
| Amoxicillin-Clavulanicacid | Penicillin-Inhibitor | 51/57 (89.5%) | 10/11 (90.9%) | 54/58 (93.1%) | 82/90 (91.1%) | 95/109 (87.2%) | NS | 129/144 (89.6%) | 163/181 (90.1%) | NS | 33/44 (75%) | 51/53 (96.2%) | 67/73 (91.8%) | 56/66 (84.8%) | 85/89 (95.5%) | 0.0284 | 292/325 (89.8%) |
| Ceftriaxone | Cephalosporins | 51/57 (89.5%) | 8/11 (72.7%) | 53/58 (91.4%) | 76/90 (84.4%) | 93/109 (85.3%) | NS | 125/144 (86.8%) | 156/181 (86.2%) | NS | 37/44 (84.1%) | 47/53 (88.7%) | 63/73 (86.3%) | 52/66 (78.8%) | 82/89 (92.1%) | NS | 281/325 (86.5%) |
| Cefixime | Cephalosporins | 38/44 (86.4%) | 5/5 (100%) | 35/39 (89.7%) | 53/64 (82.8%) | 73/84 (86.9%) | NS | 81/93 (87.1%) | 123/143 (86%) | NS | 8/9 (88.9%) | 62/72 (86.1%) | 52/66 (78.8%) | 82/89 (92.1%) | NS | 204/236 (86.4%) | |
| Cefoperazone | Cephalosporins | 41/47 (87.2%) | 5/7 (71.4%) | 44/48 (91.7%) | 62/74 (83.8%) | 81/94 (86.2%) | NS | 97/111 (87.4%) | 136/159 (85.5%) | NS | 36/42 (85.7%) | 63/73 (86.3%) | 52/66 (78.8%) | 82/89 (92.1%) | NS | 233/270 (86.3%) | |
| Trime-Sulphamethoxazole | Sulphonamides | 45/53 (84.9%) | 10/11 (90.9%) | 50/55 (90.9%) | 76/86 (88.4%) | 84/102 (82.4%) | NS | 123/138 (89.1%) | 142/169 (84%) | NS | 42/44 (95.5%) | 47/52 (90.4%) | 58/67 (86.6%) | 47/64 (73.4%) | 71/80 (88.8%) | NS | 265/307 (86.3%) |
| Cefotaxime | Cephalosporins | 51/57 (89.5%) | 8/11 (72.7%) | 52/58 (89.7%) | 75/89 (84.3%) | 92/109 (84.4%) | NS | 123/144 (85.4%) | 155/180 (86.1%) | NS | 35/44 (79.5%) | 46/52 (88.5%) | 63/73 (86.3%) | 52/66 (78.8%) | 82/89 (92.1%) | NS | 278/324 (85.8%) |
| Aztreonam | Monobactams | 45/52 (86.5%) | 8/9 (88.9%) | 35/42 (83.3%) | 61/73 (83.6%) | 77/94 (81.9%) | NS | 98/117 (83.8%) | 128/153 (83.7%) | NS | 32/44 (72.7%) | 13/17 (76.5%) | 46/55 (83.6%) | 53/65 (81.5%) | 82/89 (92.1%) | 0.0045 | 226/270 (83.7%) |
| Ceftazidime | Cephalosporins | 49/56 (87.5%) | 6/11 (54.5%) | 51/58 (87.9%) | 76/90 (84.4%) | 89/109 (81.7%) | NS | 119/144 (82.6%) | 152/180 (84.4%) | NS | 28/43 (65.1%) | 46/53 (86.8%) | 63/73 (86.3%) | 52/66 (78.8%) | 82/89 (92.1%) | 0.0043 | 271/324 (83.6%) |
| Doxycycline | Tetracycline | 36/51 (70.6%) | 8/10 (80%) | 43/54 (79.6%) | 66/81 (81.5%) | 91/106 (85.8%) | NS | 114/134 (85.1%) | 130/168 (77.4%) | NS | 37/44 (84.1%) | 47/53 (88.7%) | 48/57 (84.2%) | 48/66 (72.7%) | 64/82 (78%) | NS | 244/302 (80.8%) |
| Cefepime | Cephalosporins | 44/57 (77.2%) | 7/11 (63.6%) | 47/58 (81%) | 71/90 (78.9%) | 82/109 (75.2%) | NS | 111/144 (77.1%) | 140/181 (77.3%) | NS | 29/44 (65.9%) | 38/53 (71.7%) | 55/73 (75.3%) | 50/66 (75.8%) | 79/89 (88.8%) | 0.0019 | 251/325 (77.2%) |
| Moxifloxacin | Fluoroquinolone | 27/55 (49.1%)* | 5/8 (62.5%) | 40/54 (74.1%) | 61/85 (71.8%) | 84/104 (80.8%)# | 0.001 | 93/134 (69.4%) | 124/172 (72.1%) | NS | 14/28 (50%) | 31/50 (62%) | 59/73 (80.8%) | 47/66 (71.2%) | 66/89 (74.2%) | 0.0328 | 217/306 (70.9%) |
| Ofloxacin | Fluoroquinolone | 29/57 (50.9%)* | 5/11 (45.5%) | 41/58 (70.7%) | 66/90 (73.3%) | 88/109 (80.7%)# | 0.001 | 98/144 (68.1%) | 131/181 (72.4%) | NS | 23/44 (52.3%) | 34/53 (64.2%) | 59/73 (80.8%) | 47/66 (71.2%) | 66/89 (74.2%) | 0.0198 | 229/325 (70.5%) |
| Tobramycin | Aminoglycosides | 45/56 (80.4%) | 8/11 (72.7%) | 41/57 (71.9%) | 59/90 (65.6%) | 73/107 (68.2%) | NS | 103/141 (73%) | 123/180 (68.3%) | NS | 33/43 (76.7%) | 34/53 (64.2%) | 59/73 (80.8%) | 44/65 (67.7%) | 56/87 (64.4%) | NS | 226/321 (70.4%) |
| Ciprofloxacin | Fluoroquinolone | 28/57 (49.1%)* | 5/11 (45.5%) | 43/58 (74.1%) | 65/90 (72.2%)# | 87/109 (79.8%)# | 0.001 | 98/144 (68.1%) | 130/181 (71.8%) | NS | 24/44 (54.5%) | 32/53 (60.4%) | 59/73 (80.8%) | 47/66 (71.2%) | 66/89 (74.2%) | 0.0188 | 228/325 (70.2%) |
| Levofloxacin | Fluoroquinolone | 29/57 (50.9%)* | 5/11 (45.5%) | 41/58 (70.7%) | 66/90 (73.3%) | 87/109 (79.8%)# | 0.001 | 97/144 (67.4%) | 131/181 (72.4%) | NS | 22/44 (50%) | 34/53 (64.2%) | 59/73 (80.8%) | 47/66 (71.2%) | 66/89 (74.2%) | 0.0115 | 228/325 (70.2%) |
| Gentamicin | Aminoglycosides | 40/57 (70.2%) | 6/11 (54.5%) | 34/58 (58.6%) | 52/89 (58.4%) | 63/109 (57.8%) | NS | 91/144 (63.2%) | 104/180 (57.8%) | NS | 29/44 (65.9%) | 32/53 (60.4%) | 51/72 (70.8%) | 38/66 (57.6%) | 45/89 (50.6%) | 0.0487 (-) | 195/324 (60.2%) |
| Amikacin | Aminoglycosides | 19/56 (33.9%)# | 4/11 (36.4%) | 10/57 (17.5%) | 12/90 (13.3%)* | 10/106 (9.4%)* | 0.001 | 22/142 (15.5%) | 33/178 (18.5%) | NS | 14/44 (31.8%) | 12/53 (22.6%) | 10/73 (13.7%) | 11/66 (16.7%) | 8/84 (9.5%) | 0.0018 | 55/320 (17.2%) |
| Piperacillin-Tazobactam | Penicillin-Inhibitor | 10/55 (18.2%)# | 3/11 (27.3%)# | 5/56 (8.9%) | 3/87 (3.4%)* | 6/106 (5.7%) | 0.005 | 8/140 (5.7%) | 19/175 (10.9%) | NS | 2/44 (4.5%) | 4/53 (7.5%) | 5/71 (7%) | 7/63 (11.1%) | 9/84 (10.7%) | NS | 27/315 (8.6%) |
| Cefoperazone-Sulbactam | Cephalosporins-Inhibitor | 13/57 (22.8%)# | 2/11 (18.2%) | 4/57 (7%) | 2/87 (2.3%)* | 6/108 (5.6%)* | 0.001 | 8/141 (5.7%) | 19/179 (10.6%) | NS | 2/44 (4.5%) | 3/53 (5.7%) | 7/69 (10.1%) | 7/65 (10.8%) | 8/89 (9%) | NS | 27/320 (8.4%) |
| Imipenem | Carbapenem | 4/56 (7.1%) | 3/11 (27.3%)# | 3/58 (5.2%) | 0/90 (0%)* | 2/109 (1.8%)* | 0.001 | 3/143 (2.1%) | 9/181 (5%) | NS | 2/43 (4.7%) | 3/53 (5.7%) | 0/73 (0%) | 3/66 (4.5%) | 4/89 (4.5%) | NS | 12/324 (3.7%) |
| Meropenem | Carbapenem | 4/56 (7.1%) | 3/11 (27.3%)# | 3/58 (5.2%) | 0/90 (0%)* | 2/109 (1.8%)* | 0.001 | 3/143 (2.1%) | 9/181 (5%) | NS | 2/43 (4.7%) | 3/53 (5.7%) | 0/73 (0%) | 3/66 (4.5%) | 4/89 (4.5%) | NS | 12/324 (3.7%) |
Data are number of resistant isolates/total number of isolates (%). Denominator varies across cells because all isolates were not tested against the complete set of antimicrobials listed here. Empty cells indicate no cases were tested against the antimicrobial. P-value for difference was calculated by Fisher’s exact test. Bonferroni correction for pairwise comparisons was used to detect differences in percentage of resistant isolates between different age groups.
* indicates age groups less susceptible to resistant infections as compared to the age groups labeled with
#. P-value for trends was calculated by Cochran Armitage test for trends. Two-sided p-value has been reported. P for trend of <0.05 indicate an increasing resistance trend unless P-value is followed by “(-)” which shows a decreasing resistance rates against the antimicrobial in Escherichia coli strains. NS: Non significant p-value (i.e. >0.05).
Table 6. Temporal, age-wise and gender-wise prevalence of resistant Staphylococcus aureus strains isolated from blood and cerebrospinal fluid (CSF) cultures in Pakistan (2011–2015).
| Antimicrobials | Antimicrobial group | Prevalence (%) of resistant organisms isolated from patients of different age-groups | Prevalence (%) of resistant organisms isolated from males and female patients | Year-wise prevalence (%) of resistant organisms | Total | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <5 | 6-18Y | 19-45Y | 46-65Y | >65Y | P for difference | Female | Male | P for difference | 2011 | 2012 | 2013 | 2014 | 2015 | P for Trend | |||
| Ampicillin | Penicillin | 40/41 (97.6%) | 12/13 (92.3%) | 48/52 (92.3%) | 41/45 (91.1%) | 28/29 (96.6%) | NS | 64/71 (90.1%) | 105/109 (96.3%) | NS | 16/19 (84.2%) | 17/20 (85%) | 33/33 (100%) | 46/47 (97.9%) | 57/61 (93.4%) | NS | 169/180 (93.9%) |
| Amoxicillin | Penicillin | 40/41 (97.6%) | 12/13 (92.3%) | 48/52 (92.3%) | 41/45 (91.1%) | 28/29 (96.6%) | NS | 64/71 (90.1%) | 105/109 (96.3%) | NS | 16/19 (84.2%) | 17/20 (85%) | 33/33 (100%) | 46/47 (97.9%) | 57/61 (93.4%) | NS | 169/180 (93.9%) |
| Tobramycin | Aminoglycosides | 19/30 (63.3%) | 8/12 (66.7%) | 24/39 (61.5%) | 22/39 (56.4%) | 12/24 (50%) | NS | 35/59 (59.3%) | 50/85 (58.8%) | NS | 10/19 (52.6%) | 5/7 (71.4%) | 11/16 (68.8%) | 29/44 (65.9%) | 30/58 (51.7%) | NS | 85/144 (59%) |
| Trimethoprim-Sulphamethoxazole | Sulphonamides | 19/38 (50%) | 6/12 (50%) | 31/50 (62%) | 20/40 (50%) | 15/27 (55.6%) | NS | 31/65 (47.7%) | 60/102 (58.8%) | NS | 18/19 (94.7%) | 15/20 (75%) | 17/27 (63%) | 23/47 (48.9%) | 18/54 (33.3%) | < .0001 (-) | 91/167 (54.5%) |
| Erythromycin | Macrolides | 22/38 (57.9%) | 6/13 (46.2%) | 22/51 (43.1%) | 21/44 (47.7%) | 17/29 (58.6%) | NS | 29/69 (42%) | 59/106 (55.7%) | NS | 11/18 (61.1%) | 12/19 (63.2%) | 13/33 (39.4%) | 24/44 (54.5%) | 28/61 (45.9%) | NS | 88/175 (50.3%) |
| Azithromycin | Macrolides | 23/39 (59%) | 6/13 (46.2%) | 22/51 (43.1%) | 20/44 (45.5%) | 17/29 (58.6%) | NS | 28/69 (40.6%) | 60/107 (56.1%) | 0.046 | 11/18 (61.1%) | 11/19 (57.9%) | 13/33 (39.4%) | 25/45 (55.6%) | 28/61 (45.9%) | NS | 88/176 (50%) |
| Levofloxacin | Fluoroquinolone | 12/36 (33.3%) | 3/12 (25%) | 25/48 (52.1%) | 23/42 (54.8%) | 16/28 (57.1%) | NS | 26/68 (38.2%) | 53/98 (54.1%) | NS | 10/19 (52.6%) | 8/20 (40%) | 8/19 (42.1%) | 23/47 (48.9%) | 30/61 (49.2%) | NS | 79/166 (47.6%) |
| Moxifloxacin | Fluoroquinolone | 12/36 (33.3%) | 4/12 (33.3%) | 21/40 (52.5%) | 21/38 (55.3%) | 11/21 (52.4%) | NS | 24/61 (39.3%) | 45/86 (52.3%) | NS | 0/1 (0%) |
9/20 (45%) | 8/19 (42.1%) | 22/46 (47.8%) | 30/61 (49.2%) | NS | 69/147 (46.9%) |
| Amoxicillin-Clavulanic acid | Penicillin-Inhibitor | 20/41 (48.8%) | 4/13 (30.8%) | 24/52 (46.2%) | 19/45 (42.2%) | 16/29 (55.2%) | NS | 29/71 (40.8%) | 54/109 (49.5%) | NS | 16/19 (84.2%) | 7/20 (35%) | 11/33 (33.3%) | 23/47 (48.9%) | 26/61 (42.6%) | NS | 83/180 (46.1%) |
| Ofloxacin | Fluoroquinolone | 13/40 (32.5%) | 3/13 (23.1%) | 25/52 (48.1%) | 24/45 (53.3%) | 17/29 (58.6%) | NS | 27/71 (38%) | 55/108 (50.9%) | NS | 10/19 (52.6%) | 8/20 (40%) | 12/33 (36.4%) | 23/47 (48.9%) | 29/60 (48.3%) | NS | 82/179 (45.8%) |
| Imipenem | Carbapenem | 20/41 (48.8%) | 4/13 (30.8%) | 24/52 (46.2%) | 19/45 (42.2%) | 15/29 (51.7%) | NS | 29/71 (40.8%) | 53/109 (48.6%) | NS | 16/19 (84.2%) | 7/20 (35%) | 11/33 (33.3%) | 23/47 (48.9%) | 25/61 (41%) | NS | 82/180 (45.6%) |
| Meropenem | Carbapenem | 20/41 (48.8%) | 4/13 (30.8%) | 24/52 (46.2%) | 19/45 (42.2%) | 15/29 (51.7%) | NS | 29/71 (40.8%) | 53/109 (48.6%) | NS | 16/19 (84.2%) | 7/20 (35%) | 11/33 (33.3%) | 23/47 (48.9%) | 25/61 (41%) | NS | 82/180 (45.6%) |
| Cefaclor | Cephalosporins | 20/41 (48.8%) | 4/13 (30.8%) | 24/52 (46.2%) | 19/45 (42.2%) | 15/29 (51.7%) | NS | 29/71 (40.8%) | 53/109 (48.6%) | NS | 16/19 (84.2%) | 7/20 (35%) | 11/33 (33.3%) | 23/47 (48.9%) | 25/61 (41%) | NS | 82/180 (45.6%) |
| Ceftazidime | Cephalosporins | 20/41 (48.8%) | 4/13 (30.8%) | 24/52 (46.2%) | 19/45 (42.2%) | 15/29 (51.7%) | NS | 29/71 (40.8%) | 53/109 (48.6%) | NS | 16/19 (84.2%) | 7/20 (35%) | 11/33 (33.3%) | 23/47 (48.9%) | 25/61 (41%) | NS | 82/180 (45.6%) |
| Ceftriaxone | Cephalosporins | 20/41 (48.8%) | 4/13 (30.8%) | 24/52 (46.2%) | 19/45 (42.2%) | 15/29 (51.7%) | NS | 29/71 (40.8%) | 53/109 (48.6%) | NS | 16/19 (84.2%) | 7/20 (35%) | 11/33 (33.3%) | 23/47 (48.9%) | 25/61 (41%) | NS | 82/180 (45.6%) |
| Cephalexin | Cephalosporins | 20/41 (48.8%) | 4/13 (30.8%) | 24/52 (46.2%) | 19/45 (42.2%) | 15/29 (51.7%) | NS | 29/71 (40.8%) | 53/109 (48.6%) | NS | 16/19 (84.2%) | 7/20 (35%) | 11/33 (33.3%) | 23/47 (48.9%) | 25/61 (41%) | NS | 82/180 (45.6%) |
| Ciprofloxacin | Fluoroquinolone | 14/41 (34.1%) | 3/13 (23.1%) | 25/52 (48.1%) | 23/45 (51.1%) | 17/29 (58.6%) | NS | 26/71 (36.6%) | 56/109 (51.4%) | NS | 10/19 (52.6%) | 7/20 (35%) | 12/33 (36.4%) | 23/47 (48.9%) | 30/61 (49.2%) | NS | 82/180 (45.6%) |
| Ampicillin-Sulbactam | Penicillin-Inhibitor | 20/39 (51.3%) | 4/13 (30.8%) | 16/40 (40%) | 17/42 (40.5%) | 9/20 (45%) | NS | 24/62 (38.7%) | 42/92 (45.7%) | NS | 0/1 (0%) |
7/14 (50%) | 10/31 (32.3%) | 23/47 (48.9%) | 26/61 (42.6%) | NS | 66/154 (42.9%) |
| Cefepime | Cephalosporins | 18/39 (46.2%) | 4/12 (33.3%) | 17/43 (39.5%) | 15/39 (38.5%) | 7/20 (35%) | NS | 20/60 (33.3%) | 41/93 (44.1%) | NS | 2/12 (16.7%) | 11/33 (33.3%) | 23/47 (48.9%) | 25/61 (41%) | NS | 61/153 (39.9%) | |
| Cefoperazone | Cephalosporins | 18/39 (46.2%) | 4/12 (33.3%) | 17/43 (39.5%) | 15/39 (38.5%) | 7/20 (35%) | NS | 20/60 (33.3%) | 41/93 (44.1%) | NS | 2/12 (16.7%) | 11/33 (33.3%) | 23/47 (48.9%) | 25/61 (41%) | NS | 61/153 (39.9%) | |
| Cefotaxime | Cephalosporins | 18/39 (46.2%) | 4/12 (33.3%) | 17/43 (39.5%) | 15/39 (38.5%) | 7/20 (35%) | NS | 20/60 (33.3%) | 41/93 (44.1%) | NS | 2/12 (16.7%) | 11/33 (33.3%) | 23/47 (48.9%) | 25/61 (41%) | NS | 61/153 (39.9%) | |
| Cephradine | Cephalosporins | 19/40 (47.5%) | 4/13 (30.8%) | 24/52 (46.2%) | 19/45 (42.2%) | 15/29 (51.7%) | NS | 29/71 (40.8%) | 52/108 (48.1%) | NS | 16/19 (84.2%) | 7/20 (35%) | 11/33 (33.3%) | 23/47 (48.9%) | 24/60 (40%) | NS | 81/179 (45.3%) |
| Cefixime | Cephalosporins | 18/37 (48.6%) | 4/12 (33.3%) | 15/37 (40.5%) | 14/37 (37.8%) | 7/19 (36.8%) | NS | 18/52 (34.6%) | 40/90 (44.4%) | NS | 0/2 (0%) | 11/33 (33.3%) | 22/46 (47.8%) | 25/61 (41%) | NS | 58/142 (40.8%) | |
| Gentamicin | Aminoglycosides | 16/40 (40%) | 4/13 (30.8%) | 23/51 (45.1%) | 17/44 (38.6%) | 11/29 (37.9%) | NS | 27/71 (38%) | 44/106 (41.5%) | NS | 11/19 (57.9%) | 8/20 (40%) | 10/31 (32.3%) | 19/47 (40.4%) | 23/60 (38.3%) | NS | 71/177 (40.1%) |
| Cefuroxime | Cephalosporins | 18/39 (46.2%) | 4/12 (33.3%) | 17/43 (39.5%) | 15/39 (38.5%) | 7/20 (35%) | NS | 20/60 (33.3%) | 41/93 (44.1%) | NS | 2/12 (16.7%) | 11/33 (33.3%) | 23/47 (48.9%) | 25/61 (41%) | NS | 61/153 (39.9%) | |
| Doxycycline | Tetracycline | 12/34 (35.3%) | 4/12 (33.3%) | 10/49 (20.4%) | 16/43 (37.2%) | 11/27 (40.7%) | NS | 18/68 (26.5%) | 35/97 (36.1%) | NS | 8/18 (44.4%) | 12/20 (60%) | 11/24 (45.8%) | 13/47 (27.7%) | 9/56 (16.1%) | 0.0002 (-) | 53/165 (32.1%) |
| Clindamycin | Lincosamide | 16/41 (39%) | 4/13 (30.8%) | 11/52 (21.2%) | 13/45 (28.9%) | 9/29 (31%) | NS | 16/71 (22.5%) | 37/109 (33.9%) | NS | 2/19 (10.5%) | 6/20 (30%) | 14/33 (42.4%) | 13/47 (27.7%) | 18/61 (29.5%) | NS | 53/180 (29.4%) |
| Amikacin | Aminoglycosides | 8/32 (25%) | 1/12 (8.3%) | 9/39 (23.1%) | 11/41 (26.8%) | 2/22 (9.1%) | NS | 13/58 (22.4%) | 18/88 (20.5%) | NS | 8/19 (42.1%) | 3/7 (42.9%) | 3/16 (18.8%) | 7/47 (14.9%) | 10/57 (17.5%) | 0.0126 (-) | 31/146 (21.2%) |
Data are number of resistant isolates/total number of isolates (%). Denominator varies across cells because all isolates were not tested against the complete set of antimicrobials listed here Empty cells indicate no cases were tested against the antimicrobial. P-value for difference was calculated by Fisher’s exact test. P-value for trends was calculated by Cochran Armitage test for trends. Two-sided p-value has been reported. P for trend of <0.05 indicate an increasing resistance trend unless P-value is followed by “(-)” which shows a decreasing resistance rates against the antimicrobial. NS: Non significant p-value (i.e. >0.05)
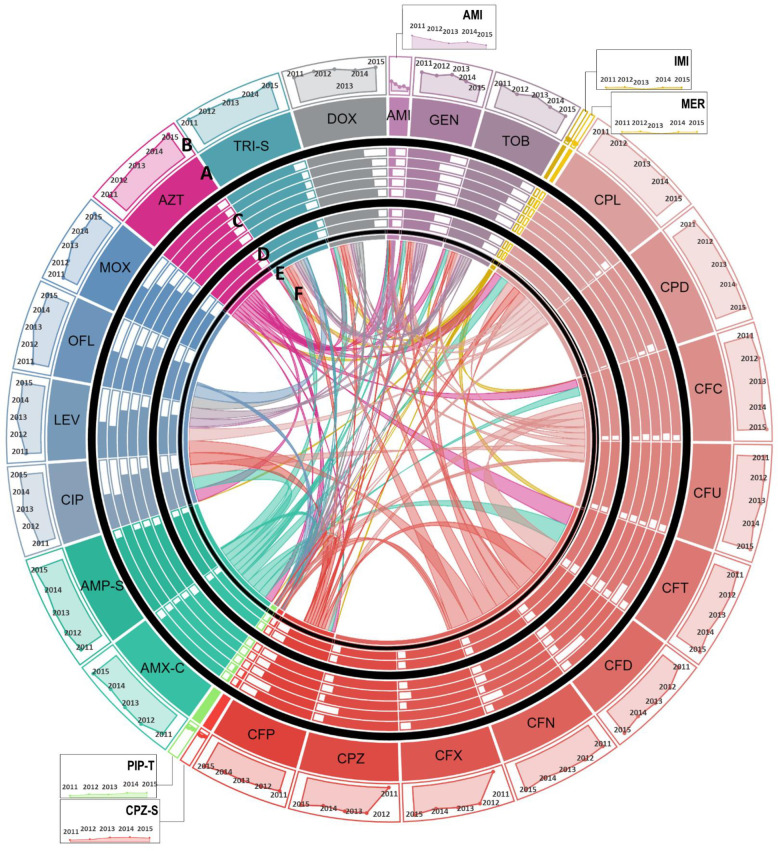
Fig 2. Antimicrobial resistance in Escherichia coli (E. coli).
A: Each section of the diagram represents the resistance observed in E. coli against the antibiotic. Size of each section is proportional to the proportion of E. coli resistant to the antibiotic over the study period. Antibiotics of the same class are shown in similar colors. B: Line graphs show temporal trends of proportion of resistant E. coli in a clockwise direction from 2011 to 2015. C: Bar charts show the comparison of susceptibility to resistant strains in patients of different age groups. Moving from out to inward, bars represent proportion of resistant E. coli reported in children <5 years of age, young adults between 6 to 18 years, middle aged 19 to 45 years old, 45 to 65 years old patients, and elderly over 65 years of age, respectively. D: Gender-wise comparison to susceptibility to resistant E. coli is shown in form bars. Outer circle and inner circle shows proportion of resistant E. coli isolated from women vs. men, respectively. E: For co-resistance analysis, antibiotics belonging to the same class with same susceptibility profile for all isolates of E. coli were merged into a single variable. F: Proportion of E. coli isolates resistant to one antimicrobial resistant to another antimicrobial are shown in the connections. The area covered by the connection on E is proportional to the level of co-resistance observed. Co-resistance proportions were scaled down to 1/15th of the actual overlap for visualization. Abbreviations: AMI: Amikacin, GEN: Gentamicin, TOB: Tobramycin, IMI: Imipenem, MER: Meropenem, CPL: Cephalexin, CPD: Cephradine, CFC: Cefaclor, CFU: Cefuroxime, CFT: Cefotaxime, CFD: Ceftazidime, CFN: Ceftriaxone, CFX: Cefixime, CPZ: Cefoperazone, CFP: Cefepime, CPZ-S: Cefoperazone-Sulbactam, PIP-T: Piperacillin-Tazobactam, AMX-C: Amoxicillin-Clavulanic acid, AMP-S: Ampicillin-Sulbactam, CIP: Ciprofloxacin, LEV: Levofloxacin, OFL: Ofloxacin, MOX: Moxifloxacin, AZT: Aztreonam, TRI-S: Trime-Sulphamethoxazole, and DOX: Doxycycline.
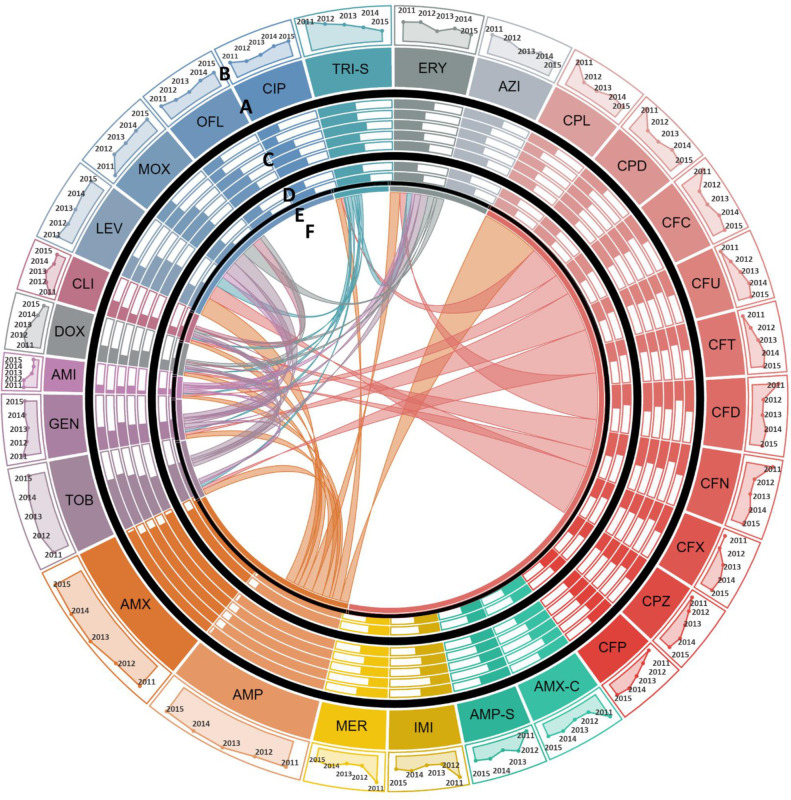
Fig 5. Antimicrobial resistance in Staphylococcus aureus (S. aureus).
A: Each section of the diagram represents the resistance observed in S. aureus against the antibiotic. Size of each section is proportional to the proportion of S. aureus resistant to the antibiotic over the study period. Antibiotics of the same class are shown in similar colors. B: Line graphs show temporal trends of proportion of resistant S. aureus in a clockwise direction from 2011 to 2015. C: Bar charts show the comparison of susceptibility to resistant strains in patients of different age groups. Moving from out to inward, bars represent proportion of resistant S. aureus reported children in <5 years of age, young adults between 6 to 18 years, middle aged 19 to 45 years old, 45 to 65 years old patients, and elderly over 65 years of age, respectively. D: Gender-wise comparison to susceptibility to resistant S. aureus is shown in form bars. Outer circle and inner circle show proportion of resistant S. aureus isolated from women vs. men, respectively. E: For co-resistance analysis, antibiotics belonging to the same class with same susceptibility profile for all isolates of S. aureus were merged into a single variable. F: Proportion of S. aureus isolates resistant to one antimicrobial resistant to another antimicrobial are shown in the connections. The area covered by the connection on E is proportional to the level of co-resistance observed. Co-resistance proportions were scaled down to 1/10th of the actual overlap for visualization. Abbreviations: AMI: Amikacin, GEN: Gentamicin, TOB: Tobramycin, AMP: Ampicillin, AMX: Amoxicillin, IMI: Imipenem, MER: Meropenem, CPL: Cephalexin, CPD: Cephradine, CFC: Cefaclor, CFU: Cefuroxime, CFT: Cefotaxime, CFD: Ceftazidime, CFN: Ceftriaxone, CFX: Cefixime, CPZ: Cefoperazone, CFP: Cefepime, AMX-C: Amoxicillin-Clavulanic acid, AMP-S: Ampicillin-Sulbactam, CIP: Ciprofloxacin, LEV: Levofloxacin, OFL: Ofloxacin, MOX: Moxifloxacin, CLI: Clindamycin, AZI: Azithromycin, ERY: Erythromycin, TRI-S: Trime-Sulphamethoxazole, and DOX: Doxycycline.
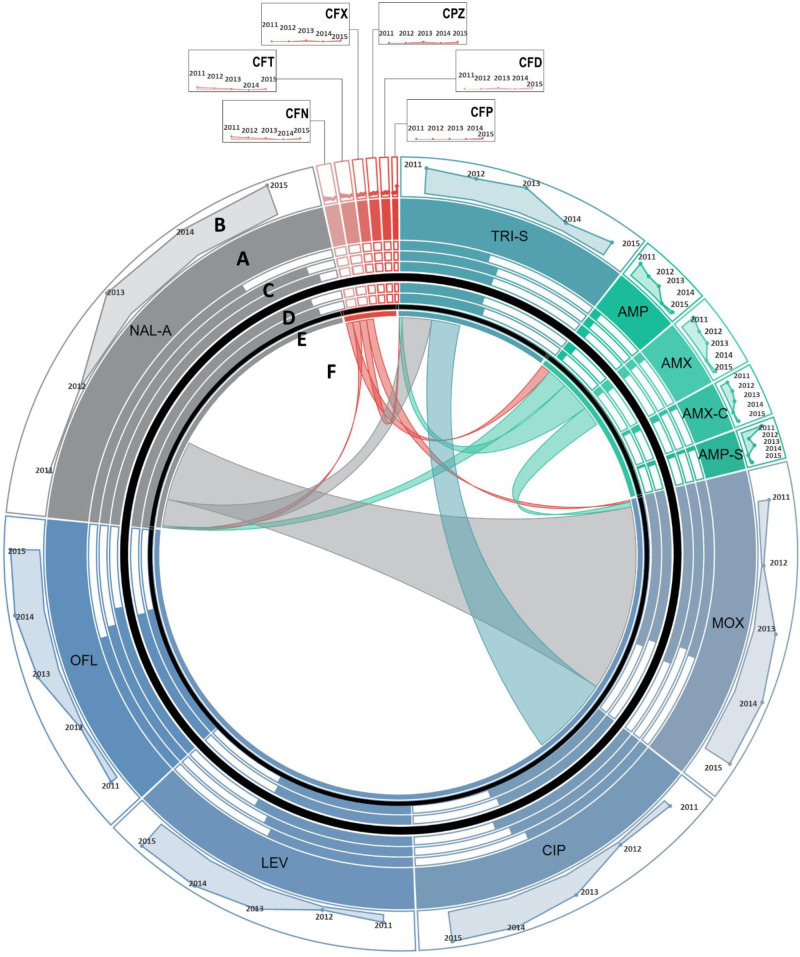
To determine the association between age and sex and trends of antimicrobial resistance, the frequency of isolation of resistance pathogen was calculated by age and sex. The sex-wise comparisons showed that the rate of isolation of resistant gram-negative pathogens were independent of patients’ sex. Evaluation of age-wise resistance trends highlighted that rate of isolation of resistant pathogens is age dependent in that children under 5 years of age had a higher rate of isolation of E. coli resistant to fluoroquinolones as compared to the elderly (Table 3 and Fig 2). Contrasting trend was observed for amikacin, piperacillin-tazobactam, cefoperazone-sulbactam and carbapenem resistance in E. coli where we observed that children and young adults had a higher rate of isolation of resistant strains compared to the elderly (Table 3 and Fig 2). Our results have shown a similar trend for cefoperazone-sulbactam resistance in Acinetobacter species–children under 5 years of age had higher rate of isolation of cefoperazone-sulbactam resistant Acinetobacter species strains as compared to young adults and elderly (Table 4 and Fig 3). We have also reported that S. Typhi isolated from adults between 18 to 45 years of age had a higher proportion of Nalidixic acid resistance as compared to those isolated from children under 5 years of age (Table 5 and Fig 4).
Table 4. Temporal, age-wise and gender-wise prevalence of resistant Acinetobacter species strains isolated from blood and cerebrospinal fluid (CSF) cultures in Pakistan (2011–2015).
| Antimicrobials | Antimicrobial group | Prevalence (%) of resistant organisms isolated from patients of different age-groups | Prevalence (%) of resistant organisms isolated from males and female patients | Year-wise prevalence (%) of resistant organisms | Total | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <5 Years | 6–18 Years | 19–45 Years | 46–65 Years | >65 Years | P for difference | Female | Male | P for difference | 2011 | 2012 | 2013 | 2014 | 2015 | P for trends | |||
| Ampicillin-Sulbactam | Penicillin-Inhibitor | 79/81 (97.5%) | 6/6 (100%) | 29/29 (100%) | 26/26 (100%) | 15/15 (100%) | NS | 59/59 (100%) | 96/98 (98%) | NS | 1/1 (100%) | 4/4 (100%) | 17/18 (94.4%) | 44/44 (100%) | 89/90 (98.9%) | NS | 155/157 (98.7%) |
| Cefixime | Cephalosporins | 79/83 (95.2%) | 6/7 (85.7%) | 32/32 (100%) | 27/27 (100%) | 15/15 (100%) | NS | 66/67 (98.5%) | 93/97 (95.9%) | NS | 4/5 (80%) | 24/25 (96%) | 42/44 (95.4%) | 89/90 (98.9%) | 0.0476 | 159/164 (96.9%) | |
| Cefoperazone | Cephalosporins | 80/84 (95.2%) | 6/7 (85.7%) | 37/38 (97.4%) | 33/33 (100%) | 18/18 (100%) | NS | 74/76 (97.4%) | 100/104 (96.2%) | NS | 5/6 (83.3%) | 14/15 (93.3%) | 24/25 (96%) | 42/44 (95.4%) | 89/90 (98.9%) | 0.0397 | 174/180 (96.7%) |
| Amoxicillin-Clavulanic acid | Penicillin-Inhibitor | 82/84 (97.6%) | 8/9 (88.9%) | 39/41 (95.1%) | 35/35 (100%) | 20/21 (95.2%) | NS | 79/81 (97.5%) | 105/109 (96.3%) | NS | 12/15 (80%) | 15/16 (93.7%) | 24/25 (96%) | 44/44 (100%) | 89/90 (98.9%) | 0.0006 | 184/190 (96.8%) |
| Ceftriaxone | Cephalosporins | 81/85 (95.3%) | 7/9 (77.8%) | 38/40 (95%) | 35/35 (100%) | 20/20 (100%) | NS | 78/81 (96.3%) | 103/108 (95.4%) | NS | 12/15 (80%) | 14/15 (93.3%) | 25/26 (96.1%) | 41/43 (95.3%) | 89/90 (98.9%) | 0.003 | 181/189 (95.8%) |
| Cefotaxime | Cephalosporins | 81/85 (95.3%) | 7/9 (77.8%) | 39/41 (95.1%) | 35/35 (100%) | 20/21 (95.2%) | NS | 78/81 (96.3%) | 104/110 (94.5%) | NS | 11/15 (73.3%) | 15/16 (93.7%) | 25/26 (96.1%) | 42/44 (95.4%) | 89/90 (98.9%) | 0.0004 | 182/191 (95.3%) |
| Ceftazidime | Cephalosporins | 81/85 (95.3%) | 7/9 (77.8%) | 39/41 (95.1%) | 33/34 (97.1%) | 20/21 (95.2%) | NS | 77/80 (96.2%) | 103/110 (93.6%) | NS | 11/15 (73.3%) | 14/16 (87.5%) | 24/25 (96%) | 42/44 (95.4%) | 89/90 (98.9%) | 0.0001 | 180/190 (94.7%) |
| Cefepime | Cephalosporins | 79/84 (94.0%) | 7/8 (87.5%) | 39/41 (95.1%) | 33/35 (94.3%) | 18/18 (100%) | NS | 76/80 (95%) | 100/106 (94.3%) | NS | 7/10 (70%) | 15/16 (93.7%) | 25/26 (96.1%) | 41/44 (93.2%) | 88/90 (97.8%) | 0.0064 | 176/186 (94.6%) |
| Gentamicin | Aminoglycosides | 76/84 (90.5%) | 7/9 (77.8%) | 35/39 (89.7%) | 32/34 (94.1%) | 19/21 (90.5%) | NS | 71/78 (91%) | 98/109 (89.9%) | NS | 9/14 (64.3%) | 14/16 (87.5%) | 24/26 (92.3%) | 39/44 (88.6%) | 83/87 (95.4%) | 0.0021 | 169/187 (90.4%) |
| Piperacillin-Tazobactam | Penicillin-Inhibitor | 73/78 (93.6%) | 6/7 (85.7%) | 33/38 (86.8%) | 27/32 (84.4%) | 17/20 (85%) | NS | 66/73 (90.4%) | 90/102 (88.2%) | NS | 6/13 (46.1%) | 12/16 (75%) | 22/25 (88%) | 34/37 (91.9%) | 82/84 (97.6%) | < .0001 | 156/175 (89.1%) |
| Imipenem | Carbapenem | 76/84 (90.5%) | 7/9 (77.8%) | 37/41 (90.2%) | 31/34 (91.2%) | 17/21 (80.9%) | NS | 74/80 (92.5%) | 94/109 (86.2%) | NS | 7/14 (50%) | 13/16 (81.2%) | 23/26 (88.5%) | 40/44 (90.9%) | 85/89 (95.5%) | < .0001 | 168/189 (88.9%) |
| Meropenem | Carbapenem | 76/84 (90.5%) | 7/9 (77.8%) | 36/40 (90%) | 31/34 (91.2%) | 17/21 (80.9%) | NS | 73/79 (92.4%) | 94/109 (86.2%) | NS | 7/14 (50%) | 12/15 (80%) | 23/26 (88.5%) | 40/44 (90.9%) | 85/89 (95.5%) | < .0001 | 167/188 (88.8%) |
| Moxifloxacin | Fluoroquinolone | 72/84 (85.7%) | 6/7 (85.7%) | 32/37 (86.5%) | 28/31 (90.3%) | 19/19 (100%) | NS | 67/75 (89.3%) | 90/103 (87.4%) | NS | 4/6 (66.7%) | 11/13 (84.6%) | 23/25 (92%) | 39/44 (88.6%) | 80/90 (88.9%) | NS | 157/178 (88.2%) |
| Ofloxacin | Fluoroquinolone | 72/85 (84.7%) | 7/9 (77.8%) | 35/40 (87.5%) | 33/35 (94.3%) | 19/21 (90.5%) | NS | 72/80 (90%) | 94/110 (85.4%) | NS | 9/14 (64.3%) | 14/16 (87.5%) | 24/26 (92.3%) | 39/44 (88.6%) | 80/90 (88.9%) | NS | 166/190 (87.4%) |
| Trimethoprim-Sulphamethoxazole | Sulphonamides | 70/80 (87.5%) | 7/9 (77.8%) | 35/40 (87.5%) | 26/31 (83.9%) | 19/20 (95%) | NS | 69/80 (86.3%) | 88/100 (88%) | NS | 13/15 (86.7%) | 14/16 (87.5%) | 18/21 (85.7%) | 33/43 (76.7%) | 79/85 (92.9%) | NS | 157/180 (87.2%) |
| Amikacin | Aminoglycosides | 74/81 (91.4%) | 6/9 (66.7%) | 32/39 (82%) | 29/33 (87.9%) | 18/21 (85.7%) | NS | 72/79 (91.1%) | 87/104 (83.6%) | NS | 11/15 (73.3%) | 12/16 (75%) | 21/26 (80.8%) | 39/44 (88.6%) | 76/82 (92.7%) | 0.0058 | 159/183 (86.9%) |
| Ciprofloxacin | Fluoroquinolone | 72/85 (84.7%) | 7/9 (77.8%) | 35/41 (85.4%) | 33/35 (94.3%) | 19/21 (90.5%) | NS | 72/81 (88.9%) | 94/110 (85.4%) | NS | 9/15 (60%) | 14/16 (87.5%) | 24/26 (92.3%) | 39/44 (88.6%) | 80/90 (88.9%) | 0.0396 | 166/191 (86.9%) |
| Levofloxacin | Fluoroquinolone | 72/85 (84.7%) | 7/9 (77.8%) | 35/41 (85.4%) | 33/35 (94.3%) | 19/21 (90.5%) | NS | 72/81 (88.9%) | 94/110 (85.4%) | NS | 9/15 (60%) | 14/16 (87.5%) | 24/26 (92.3%) | 39/44 (88.6%) | 80/90 (88.9%) | 0.0396 | 166/191 (86.9%) |
| Tobramycin | Aminoglycosides | 51/79 (64.6%) | 3/8 (37.5%) | 30/40 (75%) | 26/34 (76.5%) | 15/21 (71.4%) | NS | 57/75 (76%) | 68/107 (63.5%) | NS | 9/15 (60%) | 12/16 (75%) | 18/26 (69.2%) | 23/42 (54.8%) | 63/83 (75.9%) | NS | 125/182 (68.7%) |
| Doxycycline | Tetracycline | 37/80 (46.2%) | 3/8 (37.5%) | 20/35 (57.1%) | 16/31 (51.6%) | 14/21 (66.7%) | NS | 43/72 (59.7%) | 47/103 (45.6%) | NS | 6/14 (42.9%) | 10/15 (66.7%) | 11/17 (64.7%) | 18/43 (41.9%) | 45/86 (52.3%) | NS | 90/175 (51.4%) |
| Cefoperazone-Sulbactam | Cephalosporins-Inhibitor | 48/84 (57.1%)# | 1/9 (11.1%)* | 7/41 (17.1%)* | 12/34 (35.3%) | 3/20 (15%)* | < .0001 | 26/81 (32.1%) | 45/107 (42.1%) | NS | 3/15 (20%) | 0/16 (0%) | 1/25 (4%) | 22/43 (51.2%) | 45/89 (50.6%) | < .0001 | 71/188 (37.8%) |
Data are number of resistant isolates/total number of isolates (%). Denominator varies across cells because all isolates were not tested against the complete set of antimicrobials listed here Empty cells indicate no cases were tested against the antimicrobial. P-value for difference was calculated by Fisher’s exact test. Bonferroni correction for pairwise comparisons was used to detect differences in percentage of resistant isolates between different age groups.
* indicates age groups less susceptible to resistant strains as compared to the age-groups labeled with
#. P-value for trends was calculated by Cochran Armitage test for trends. Two-sided p-value has been reported. P for trend of <0.05 indicate an increasing resistance trend. NS: Non significant p-value (i.e. >0.05)
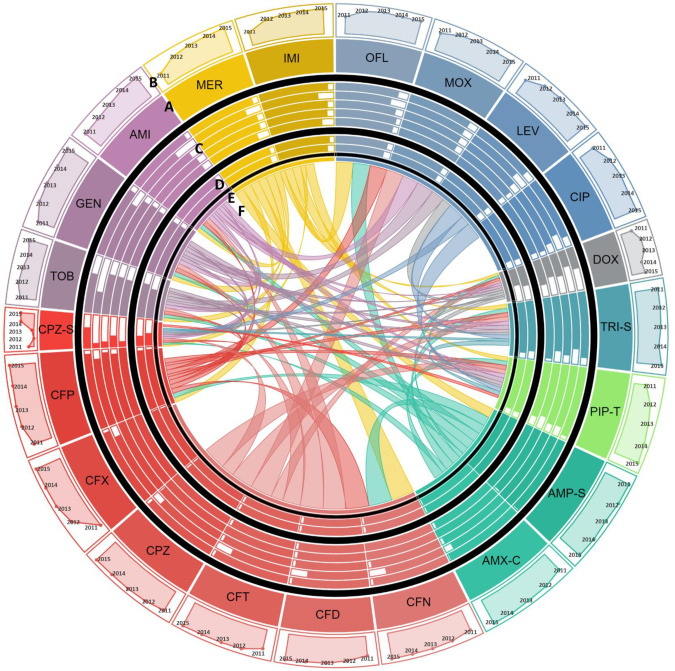
Fig 3. Antimicrobial resistance in Acinetobacter.
A: Each section of the diagram represents the resistance observed in Acinetobacter species against the antibiotic. Size of each section is proportional to the proportion of Acinetobacter species resistant to the antibiotic over the study period. Antibiotics of the same class are shown in similar colors. B: Line graphs show temporal trends of proportion of resistant Acinetobacter species in a clockwise direction from 2011 to 2015. C: Bar charts show the comparison of susceptibility to resistant isolates in patients of different age groups. Moving from out to inward, bars represent proportion of resistant Acinetobacter species reported in children <5 years of age, young adults between 6 to 18 years, middle aged 19 to 45 years old, 45 to 65 years old patients, and elderly over 65 years of age, respectively. D: Gender-wise comparison to susceptibility to resistant Acinetobacter species is shown in form bars. Outer circle and inner circle shows proportion of resistant Acinetobacter species isolated from women vs. men, respectively. E: For co-resistance analysis, antibiotics belonging to the same class with same susceptibility profile for all isolates of E. coli were merged into a single variable. F: Proportion Acinetobacter species isolates resistant to one antimicrobial resistant to another antimicrobial are shown in the connections. The area covered by the connection on E is proportional to the level of co-resistance observed. Co-resistance proportions were scaled down to 1/10th of the actual overlap for visualization. Abbreviations: DOX: Doxycycline, TRI-S: Trime-Sulphamethoxazole, PIP-T: Piperacillin-Tazobactam, AMP-S: Ampicillin-Sulbactam, AMX-C: Amoxicillin-Clavulanic acid, CIP: Ciprofloxacin, LEV: Levofloxacin, OFL: Ofloxacin, MOX: Moxifloxacin, CFN: Ceftriaxone, CFD: Ceftazidime, CFT: Cefotaxime, CPZ: Cefoperazone, CFX: Cefixime, CFP: Cefepime, CPZ-S: Cefoperazone-Sulbactam, IMI: Imipenem, MER: Meropenem, TOB: Tobramycin, GEN: Gentamicin, and AMI: Amikacin.
Table 5. Temporal, age-wise and gender-wise prevalence of resistant Salmonella enterica serovar Typhi strains isolated from blood and cerebrospinal fluid (CSF) cultures in Pakistan (2011–2015).
| Antimicrobials | Antimicrobial group | Prevalence (%) of resistant organisms isolated from patients of different age-groups | Prevalence (%) of resistant organisms isolated from males and female patients | Year-wise prevalence (%) of resistant organisms | Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <5 Years | 6–18 Years | 19–45 Years | P for difference | Female | Male | P for difference | 2011 | 2012 | 2013 | 2014 | 2015 | P for trend | |||
| Nalidixic acid | Fluoroquinolone | 12/16 (75%)* | 60/65 (92.3%) | 61/64 (95.3%) # | 0.03 | 57/63 (90.5%) | 78/85 (91.8%) | NS | 25/26 (96.2%) | 52/58 (89.7%) | 58/64 (90.6%) | NS | 135/148 (91.2%) | ||
| Moxifloxacin | Fluoroquinolone | 9/14 (64.3%) | 48/77 (62.3%) | 57/82 (69.5%) | NS | 49/77 (63.6%) | 67/100 (67%) | NS | 3/9 (33.3%) | 1/18 (5.6%) | 22/33 (66.7%) | 52/58 (89.7%) | 38/59 (64.4%) | < .0001 | 116/177 (65.5%) |
| Ciprofloxacin | Fluoroquinolone | 9/16 (56.3%) | 49/80 (61.3%) | 56/84 (66.7%) | NS | 50/81 (61.7%) | 66/103 (64.1%) | NS | 3/13 (23.1%) | 2/20 (10%) | 22/33 (66.7%) | 52/58 (89.7%) | 37/60 (61.7%) | < .0001 | 116/184 (63%) |
| Levofloxacin | Fluoroquinolone | 9/16 (56.3%) | 49/82 (59.8%) | 57/86 (66.3%) | NS | 50/81 (61.7%) | 67/107 (62.6%) | NS | 3/13 (23.1%) | 2/20 (10%) | 22/33 (66.7%) | 52/58 (89.7%) | 38/64 (59.4%) | < .0001 | 117/188 (62.2%) |
| Ofloxacin | Fluoroquinolone | 9/16 (56.3%) | 49/82 (59.8%) | 57/86 (66.3%) | NS | 50/81 (61.7%) | 67/107 (62.6%) | NS | 3/13 (23.1%) | 2/20 (10%) | 22/33 (66.7%) | 52/58 (89.7%) | 38/64 (59.4%) | < .0001 | 117/188 (62.2%) |
| Trimethoprim-Sulphamethoxazole | Sulphonamides | 7/16 (43.8%) | 38/77 (49.4%) | 39/80 (48.8%) | NS | 36/76 (47.4%) | 50/101 (49.5%) | NS | 10/13 (76.9%) | 13/20 (65%) | 20/26 (76.9%) | 15/57 (26.3%) | 28/61 (45.9%) | 0.0013 (-) | 86/177 (48.6%) |
| Ampicillin | Penicillin | 2/16 (12.5%) | 9/74 (12.2%) | 13/77 (16.9%) | NS | 13/75 (17.3%) | 11/95 (11.6%) | NS | 6/13 (46.2%) | 2/6 (33.3%) | 10/30 (33.3%) | 1/58 (1.7%) | 5/63 (7.9%) | < .0001 (-) | 24/170 (14.1%) |
| Amoxicillin | Penicillin | 2/16 (12.5%) | 9/74 (12.2%) | 13/77 (16.9%) | NS | 13/75 (17.3%) | 11/95 (11.6%) | NS | 6/13 (46.2%) | 2/6 (33.3%) | 10/30 (33.3%) | 1/58 (1.7%) | 5/63 (7.9%) | < .0001 (-) | 24/170 (14.1%) |
| Amoxicillin-Clavulanic acid | Penicillin-Inhibitor | 1/12 (8.3%) | 5/76 (6.6%) | 11/79 (13.9%) | NS | 10/72 (13.9%) | 7/98 (7.1%) | NS | 3/13 (23.1%) | 5/20 (25%) | 6/33 (18.2%) | 0/50 (0%) | 3/54 (5.6%) | 0.0009 (-) | 17/170 (10%) |
| Ampicillin-Sulbactam | Penicillin-Inhibitor | 1/11 (9.1%) | 4/60 (6.7%) | 5/54 (9.3%) | NS | 6/53 (11.3%) | 4/74 (5.4%) | NS | 2/3 (66.7%) | 1/6 (16.7%) | 4/14 (28.6%) | 0/50 (0%) | 3/54 (5.6%) | 0.0003 (-) | 10/127 (7.9%) |
| Ceftriaxone | Cephalosporins | 0/16 (0%) | 1/81 (1.2%) | 4/84 (4.8%) | NS | 4/81 (4.9%) | 1/104 (1%) | NS | 1/13 (7.7%) | 1/20 (5%) | 1/33 (3%) | 0/56 (0%) | 2/63 (3.2%) | NS | 5/185 (2.7%) |
| Cefotaxime | Cephalosporins | 0/16 (0%) | 1/81 (1.2%) | 4/85 (4.7%) | NS | 4/80 (5%) | 1/106 (0.9%) | NS | 1/13 (7.7%) | 1/20 (5%) | 1/32 (3.1%) | 0/57 (0%) | 2/64 (3.1%) | NS | 5/186 (2.7%) |
| Cefixime | Cephalosporins | 0/16 (0%) | 0/69 (0%) | 3/70 (4.3%) | NS | 3/67 (4.5%) | 0/91 (0%) | NS | 0/6 (0%) | 1/32 (3.1%) | 0/56 (0%) | 2/64 (3.1%) | NS | 3/158 (1.9%) | |
| Cefoperazone | Cephalosporins | 0/16 (0%) | 0/74 (0%) | 3/74 (4.1%) | NS | 3/69 (4.3%) | 0/98 (0%) | NS | 0/13 (0%) | 1/32 (3.1%) | 0/58 (0%) | 2/64 (3.1%) | NS | 3/167 (1.8%) | |
| Ceftazidime | Cephalosporins | 0/16 (0%) | 0/79 (0%) | 3/85 (3.5%) | NS | 3/81 (3.7%) | 0/103 (0%) | NS | 0/13 (0%) | 0/20 (0%) | 1/33 (3%) | 0/56 (0%) | 2/62 (3.2%) | NS | 3/184 (1.6%) |
| Cefepime | Cephalosporins | 0/7 (0%) | 0/53 (0%) | 1/40 (2.5%) | NS | 1/44 (2.3%) | 0/58 (0%) | NS | 0/11 (0%) | 0/20 (0%) | 0/4 (0%) | 0/17 (0%) | 1/50 (2%) | NS | 1/102 (1%) |
Data are number of resistant isolates/total number of isolates (%). Less than 5 isolates were isolated from patients above 45 year of age and these cases were excluded from the age-wise comparison for S. Typhi. Denominator varies across cells because all isolates were not tested against the complete set of antimicrobials listed here. Empty cells indicate no cases were tested against the antimicrobial. P-value for difference was calculated by Fisher’s exact test. Bonferroni correction for pairwise comparisons was used to detect differences in percentage of resistant isolates between different age groups.
* indicates age groups less susceptible to resistant infections as compared to the age groups labeled with
#. P-value for trends was calculated by Cochran Armitage test for trends. Two-sided p-value has been reported. P for trend of <0.05 indicate an increasing resistance trend unless P-value is followed by “(-)” which shows a decreasing resistance rates against the antimicrobial in Salmonella enterica serovar Typhi strains. NS: Non significant p-value (i.e. >0.05).
Fig 4. Antimicrobial resistance in Salmonella enterica serovar Typhi (S. Typhi).
A: Each section of the diagram represents the resistance observed in S. Typhi against the antibiotic. Size of each section is proportional to the proportion of S. Typhi resistant to the antibiotic over the study period. Antibiotics of the same class are shown in similar colors. B: Line graphs show temporal trends of proportion of resistant S. Typhi in a clockwise direction from 2011 to 2015. C: Bar charts show the comparison of susceptibility to resistant isolates in patients of different age groups. Moving from out to inward, bars represent proportion of resistant S. Typhi reported in children <5 years of age, young adults between 6 to 18 years, and middle aged 19 to 45 years old, respectively. D: Gender-wise comparison to susceptibility to resistant S. Typhi is shown in form bars. Outer circle and inner circle show proportion of resistant S. Typhi isolated from women vs. men, respectively. E: For co-resistance analysis, antibiotics belonging to the same class with same susceptibility profile for all isolates of E. coli were merged into a single variable. F: Proportion S. Typhi isolates resistant to one antimicrobial resistant to another antimicrobial are shown in the connections. The area covered by the connection on E is proportional to the level of co-resistance observed. Co-resistance proportions were scaled down to 1/10th of the actual overlap for visualization. Abbreviations: CFN: Ceftriaxone, CFD: Ceftazidime, CFT: Cefotaxime, CPZ: Cefoperazone, CFX: Cefixime, CFP: Cefepime, CIP: Ciprofloxacin, LEV: Levofloxacin, OFL: Ofloxacin, MOX: Moxifloxacin, NAL-A: Nalidixic acid, AMP-S: Ampicillin-Sulbactam, AMX-C: Amoxicillin-Clavulanic acid, AMP: Ampicillin, AMX: Amoxicillin, and TRI-S: Trime-Sulphamethoxazole.
We then assessed temporal antimicrobial resistance trends in these pathogens using Cochran Armitage test for trends. Our results show that among gram-negative organisms, E. coli, demonstrated increasing resistance trend to fluoroquinolones, in that a sharp increase from 50–74% was reported between 2011 and 2015 (Table 3 and Fig 2). Moreover, an increasing resistance trend against cefipime, a fourth-generation cephalosporin, was also observed in E. coli (Table 3 and Fig 2). While increasing resistance rates were observed for most of the tested antimicrobials, we found decreasing resistance trends against amikacin and gentamicin in E. coli (Table 3 and Fig 2). The most alarming trends were observed for Acinetobacter species (Table 4 and Fig 3), which was found to be resistant to almost all antibiotics. Among these resistance trends, the most notable was the steep increase in carbapenem resistance from 50% in 2011 to 95.5% in 2015 (Table 4 and Fig 3). Between 2011 and 2015 in S. Typhi, our results have demonstrated an increasing resistance trend against fluoroquinolones with resistance rates reaching up to 60% in 2015. Additionally, existing trends showed resistance to third and emergence of resistance to fourth generation cephalosporins in S. Typhi (Table 5 and Fig 4).
Surprisingly, we did not observe any increasing resistance trend against the tested antimicrobials in S. aureus. Unlike other pathogens, a decreasing resistance trend to amikacin, doxycycline, and trimethoprim-sulfamethoxazole was observed in S. aureus (Table 6 and Fig 5). Detailed AMR trends in S. aureus have been tabulated in Table 6 and shown in Fig 5.
Co-resistance trends in pathogens isolated from blood and CSF cultures
Multidrug resistance (MDR) has emerged as a major public health problem globally as well as in Pakistan. Co-resistance to multiple drugs emerges with the selection of strains which are resistant to multiple antimicrobials. Clonal expansion of MDR clones is faster as compared to strains resistant to a single antimicrobial [16]. Hence, it is important to identify those antimicrobials, which do not exhibit resistance with any other type of antimicrobials.
To investigate this, antimicrobials belonging to the same class and exhibiting identical resistance profiles in isolates of a given species were merged and entries with missing data were excluded. Chi-square test was used to identify patterns of co-resistance in each pathogen and these patterns are tabulated in Tables 7 and S2–S5. Evaluation of E. coli showed that resistance against all antimicrobials was significantly associated except β-lactams, aminoglycosides, fluoroquinolones, and tetracycline. Resistance against these four antimicrobials in E. coli was found to be independent of resistance against other antimicrobials (Table 7 and Fig 2). We then evaluated Acinetobacter species and our results showed that cefoperazone-sulbactam resistance is not significantly associated with resistance against aminoglycosides, trimethoprim-sulfamethoxazole and tetracycline (Table 7 and Fig 3). For S. Typhi, we only detected a significant co-resistance between nalidixic acid and fluoroquinolones (Table 7 and Fig 4). Analysis of S. aureus showed that penicillin resistance was not associated with resistance against any other antimicrobials. Furthermore, doxycycline resistance in S. aureus was found to be independent of resistance to all antimicrobials except macrolides and tobramycin (Table 7 and Fig 5). The co-resistance proportions, p-values, and associated odds ratios for all tested antimicrobials are provided in Tables 7 and S2–S5. The co-resistance trends for E. coli, Acinetobacter species, S. Typhi, and S. aureus are presented in Figs 2–5.
Table 7. Selected co-resistance trends in pathogens isolated from blood and cerebrospinal fluid (CSF) cultures in Pakistan (2011–2015).
| Pathogen | Antimicrobial 1 | Number of isolates resistant to antimicrobial 1 & 2/ Number of isolates resistant to antimicrobial 1 (%) | Number of isolates resistant to antimicrobial 1 & 2/ Number of isolates resistant to antimicrobial 2 (%) | P for difference | Odds ratio 95% CI) | Antimicrobial 2 |
|---|---|---|---|---|---|---|
| Acinetobacter | Cefoperazone-sulbactam | 54/ 54 (100) | 54/ 104 (51.9) | 0.118 | NA | Piperacillin-tazobactam |
| 52/ 54 (96.3) | 52/ 100 (52) | 0.2702 | 3.25 (0.63–16.88) | Amikacin | ||
| 42/ 54 (77.8) | 42/ 79 (53.2) | 0.2777 | 1.61 (0.68–3.8) | Tobramycin | ||
| 53/ 54 (98.1) | 53/ 103 (51.5) | 0.3632 | 4.24 (0.46–39.24) | Gentamicin | ||
| 29/ 54 (53.7) | 29/ 55 (52.7) | 0.5636 | 1.25 (0.59–2.66) | Doxycycline | ||
| 48/ 54 (88.9) | 48/ 96 (50) | 1 | 1 (0.3–3.32) | Trimethoprim-sulphamethoxazole | ||
| Cephalosporin | 79/ 106 (74.5) | 79/ 79 (100) | 0.0703 | NA | Tobramycin | |
| 103/ 106 (97.2) | 103/ 104 (99) | 0.073 | 34.33 (1.71–689.68) | Piperacillin-tazobactam | ||
| 99/ 106 (93.4) | 99/ 100 (99) | 0.1433 | 14.14 (0.8–250.9) | Amikacin | ||
| 55/ 106 (51.9) | 55/ 55 (100) | 0.2385 | NA | Doxycycline | ||
| 54/ 106 (50.9) | 54/ 54 (100) | 0.4953 | NA | Cefoperazone-sulbactam | ||
| Carbapenem | 78/ 104 (75) | 78/ 79 (98.7) | 0.0583 | 9 (0.9–90.33) | Tobramycin | |
| 103/ 104 (99) | 103/ 106 (97.2) | 0.073 | 34.33 (1.71–689.68) | Cephalosporin | ||
| 54/ 104 (51.9) | 54/ 54 (100) | 0.118 | NA | Cefoperazone-sulbactam | ||
| 54/ 104 (51.9) | 54/ 55 (98.2) | 0.3587 | 3.24 (0.33–32.18) | Doxycycline | ||
| Piperacillin-tazobactam | 78/ 104 (75) | 78/ 79 (98.7) | 0.0583 | 9 (0.9–90.33) | Tobramycin | |
| 54/ 104 (51.9) | 54/ 55 (98.2) | 0.3587 | 3.24 (0.33–32.18) | Doxycycline | ||
| Doxycycline | 51/ 55(92.7) | 51/ 96(53.1) | 0.196 | 2.27(0.64–8.03) | Trimethoprim-sulphamethoxazole | |
| Tobramycin | 72/ 79(91.1) | 72/ 96(75) | 0.2984 | 2.14(0.62–7.38) | Trimethoprim-sulphamethoxazole | |
| Escherichia coli | Carbapenem | 3/ 6 (50) | 3/ 133 (2.3) | 0.1118 | 0.25 (0.05–1.32) | Doxycycline |
| 6/ 6 (100) | 6/ 119 (5) | 0.181 | NA | Tobramycin | ||
| 5/ 6 (83.3) | 5/ 96 (5.2) | 0.2367 | 3.96 (0.45–34.62) | Gentamicin | ||
| 6/ 6 (100) | 6/ 137 (4.4) | 0.3604 | NA | 4th generation cephalosporin | ||
| 6/ 6 (100) | 6/ 139 (4.3) | 0.3732 | NA | Trimethoprim-sulphamethoxazole | ||
| 6/ 6 (100) | 6/ 144 (4.2) | 0.5935 | NA | 3rd generation cephalosporin | ||
| 6/ 6 (100) | 6/ 145 (4.1) | 0.5958 | NA | Aztreonam | ||
| 6/ 6 (100) | 6/ 145 (4.1) | 0.5958 | NA | Aztreonam | ||
| 6/ 6 (100) | 6/ 153 (3.9) | 0.6462 | NA | Penicillin | ||
| 6/ 6 (100) | 6/ 153 (3.9) | 0.6462 | NA | Penicillin & β-lactamase inhibitor | ||
| 6/ 6 (100) | 6/ 154 (3.9) | 0.6572 | NA | 2nd generation cephalosporin | ||
| 5/ 6 (83.3) | 5/ 128 (3.9) | 1 | 1.63 (0.18–14.33) | Fluoroquinolone | ||
| Cefoperazone-sulbactam | 12/ 15 (80) | 12/ 96 (12.5) | 0.0575 | 3.33 (0.9–12.28) | Gentamicin | |
| 15/ 15 (100) | 15/ 137 (10.9) | 0.0772 | 1 (17–120) | 4th generation cephalosporin | ||
| 15/ 15 (100) | 15/ 144 (10.4) | 0.1308 | NA | 3rd generation cephalosporin | ||
| 14/ 15 (93.3) | 14/ 139 (10.1) | 0.3139 | 3.25 (0.41–25.7) | Trimethoprim-sulphamethoxazole | ||
| 15/ 15 (100) | 15/ 153 (9.8) | 0.3657 | 1 (17–136) | Penicillin & β-lactamase inhibitor | ||
| 15/ 15 (100) | 15/ 154 (9.7) | 0.3665 | 1 (17–137) | 2nd generation cephalosporin | ||
| 13/ 15 (86.7) | 13/ 128 (10.2) | 0.3665 | 2.2 (0.48–10.2) | Fluoroquinolone | ||
| 14/ 15 (93.3) | 14/ 145 (9.7) | 0.4817 | 2.46 (0.31–19.61) | Aztreonam | ||
| 11/ 15 (73.3) | 11/ 133 (8.3) | 0.7408 | 0.72 (0.22–2.42) | Doxycycline | ||
| Piperacillin-tazobactam | 17/ 17 (100) | 17/ 144 (11.8) | 0.0795 | NA | 3rd generation cephalosporin | |
| 13/ 17 (76.5) | 13/ 96 (13.5) | 0.0843 | 2.7 (0.84–8.66) | Gentamicin | ||
| 17/ 17 (100) | 17/ 145 (11.7) | 0.1351 | 1 (152–1) | Aztreonam | ||
| 16/ 17 (94.1) | 16/ 139 (11.5) | 0.2094 | 3.77 (0.48–29.61) | Trimethoprim-sulphamethoxazole | ||
| 17/ 17 (100) | 17/ 153 (11.1) | 0.2284 | 1 (137–17) | Penicillin & β-lactamase inhibitor | ||
| 17/ 17 (100) | 17/ 154 (11) | 0.2374 | 1 (137–0) | 2nd generation cephalosporin | ||
| 15/ 17 (88.2) | 15/ 128 (11.7) | 0.2494 | 2.59 (0.57–11.83) | Fluoroquinolone | ||
| 14/ 17 (82.4) | 14/ 133 (10.5) | 0.7702 | 1.29 (0.35–4.77) | Doxycycline | ||
| Amikacin | 15/ 22(68.2) | 15/ 133(11.3) | 0.2613 | 0.53 (0.2–1.41) | Doxycycline | |
| 21/ 22(95.5) | 21/ 153(13.7) | 0.4866 | 2.39 (0.3–19.02) | Penicillin & β-lactamase inhibitor | ||
| 21/ 22(95.5) | 21/ 154(13.6) | 0.6957 | 2.21 (0.28–17.7) | 2nd generation cephalosporin | ||
| 16/ 22(72.7) | 16/ 128(12.5) | 0.7238 | 0.83 (0.3–2.29) | Fluoroquinolone | ||
| 17/ 22(77.3) | 17/ 137(12.4) | 0.7703 | 0.77 (0.26–2.25) | 4th generation cephalosporin | ||
| 18/ 22(81.8) | 18/ 139(12.9) | 1 | 0.97 (0.3–3.09) | Trimethoprim-sulphamethoxazole | ||
| 19/ 22(86.4) | 19/ 145(13.1) | 1 | 1.06 (0.29–3.88) | Aztreonam | ||
| 19/ 22(86.4) | 19/ 144(13.2) | 1 | 1.11 (0.3–4.09) | 3rd generation cephalosporin | ||
| Gentamicin | 78/ 96 (81.3) | 78/ 133 (58.6) | 0.3528 | 1.42 (0.68–2.97) | Doxycycline | |
| Salmonella enterica serovar Typhi | Penicillin | 4/5 (80) | 4/ 49 (8.1) | 0.172 | 5.24 (0.567–48.55) | Trimethoprim-sulphamethoxazole |
| 4/5 (80) | 4/ 100 (4) | 0.356 | 0.333 (.033–3.348) | Nalidixic acid | ||
| 4/5 (80) | 4/ 86 (4.65) | 1 | 1.073 (0.114–10.93) | Fluoroquinolone | ||
| Trimethoprim-sulphamethoxazole | 46/49 (93.8) | 46/100 (46) | 0.51 | 1.704 (0.403–7.195) | Nalidixic acid | |
| 39/49 (79.6) | 39/86 (45.3) | 1 | 1.079 (.427–2.727) | Fluoroquinolone | ||
| Staphylococcus aureus | Doxycycline | 7/ 24 (29.2) | 7/ 16 (43.8) | 0.0502 | 2.97 (0.97–9.14) | Amikacin |
| 11/ 24 (45.8) | 11/ 31 (35.5) | 0.0852 | 2.28 (0.88–5.92) | Clindamycin | ||
| 15/ 24 (62.5) | 15/ 48 (31.3) | 0.1273 | 2.07 (0.8–5.33) | Macrolides | ||
| 13/ 24 (54.2) | 13/ 44 (29.5) | 0.2935 | 1.64 (0.65–4.14) | β-lactam (except penicillin) | ||
| 11/ 24 (45.8) | 11/ 37 (29.7) | 0.3475 | 1.56 (0.61–3.98) | Gentamicin | ||
| 12/ 24 (50) | 12/ 47 (25.5) | 0.8179 | 1.11 (0.44–2.8) | Fluoroquinolone | ||
| Trimethoprim-sulphamethoxazole | 25/ 44 (56.8) | 25/ 48 (52.1) | 0.1612 | 1.77 (0.79–3.96) | Macrolides | |
| 8/ 44 (18.2) | 8/ 16 (50) | 0.6538 | 1.28 (0.44–3.74) | Amikacin | ||
| Penicillin | 58/ 95 (61.1) | 58/ 59 (98.3) | 0.5613 | 3.14 (0.27–35.81) | Tobramycin |
Data are number of isolates resistant to both antimicrobials / number of isolates resistant to either antimicrobial 1 (in the case of R1) or antimicrobial 2 (in the case of R1) (%). P-value for difference was calculated using Chi-square test. Odds-ratio was calculated using binary logistic regression and is listed with 95% confidence interval (95% CI). Two-sided p-value has been reported.
Discussion
Antimicrobial resistance (AMR) has emerged as a major public health concern in both developing and developed countries. Continuous surveillance of AMR has been recommended by World Health Organization (WHO) as a necessary step for controlling emergence of resistance as well as infections caused by resistant pathogens [17]. Despite this urgent need to investigate AMR trends, only a handful of studies till date have reported resistance trends in pathogens isolated from blood and CSF cultures in Pakistan [18, 19]. The current study stands to fill this gap in knowledge of AMR patterns and trends in these bacterial pathogens at a national scale. For that, we undertook a retrospective analysis of AMR of pathogens isolated from clinical specimens of blood and CSF. To the best of our knowledge, this study is the first of its kind in Pakistan providing both demographic and temporal AMR trends in major pathogens from blood and CSF cultures. Previously published results have been limited by the number of participants the sample size and analysis. Data presented in this report provides a more in-depth analysis of antimicrobial resistance. While most of the previous studies were cross-sectional presenting resistance at single time point, our long-term analysis presents alterations in resistance patterns over five years.
Our results show that CoNS was the most frequently isolated pathogen from invasive isolates which is consistent with previous studies in southeast Asia [20]. However, as we did not have information on the clinical manifestation of these cases and they may have been skin contaminants, we removed CoNS from the downstream analysis. Two of the most probable reasons for high rate of isolation of CoNS in Pakistan could be poor clinical sample management and presence of contaminants on surfaces [21].
In accordance with international reports, our data also reflects emergence of antimicrobial resistance in Pakistan, where pathogens are resistant to most commonly used antimicrobials [1, 3, 22]. Our analysis showed a wide distribution of resistant pathogen over 44 cities of Pakistan, however most of our observations were from the province of Punjab, which could be because the facility is primarily located in Punjab. No significant association was observed when comparisons were made by sex, indicating that both male and female are equally susceptible to infections caused by resistant pathogens. Similarly, no significant association between a sex and a specific pathogen was observed. However, we did observe a significant association between rates of isolation of resistant pathogens with age, younger children and elderly were at a higher risk of infections by drug resistant pathogens. This observation is in line with previous findings where infections with resistant pathogens were found to be associated with a children and elderly [23–25]. The most likely explanation for these observations is the increase rates of infections in both elderly and children. The observed increase susceptibility to infections in children and elderly is due to poor immune responses. As a result antimicrobials are frequently and empirically used in both children and elderly resulting in increased rates of infections by resistant pathogens.
Upper respiratory tract infections and gastrointestinal tract infections are the most frequent cause of doctors visit for children and elderly in Pakistan. Such infections are most often treated empirically by broad spectrum antibiotics without performing culture sensitivity, leading to development of resistance in both, the resident bacteria and infecting pathogen. Comparison of rates of isolation of different invasive pathogens indicated an increase in rate of isolation of S. maltophilia between 2013 and 2014. Stenotrophomonas maltophilia, previously known as Pseudomonas maltophilia primarily cause infections in immune compromised individuals. Together with Pseudomonas aeruginosa and Acinetobacter baumannii, S.maltophilia is an important cause of healthcare-associated infections. Moreover, A. baumannii and S. maltophilia have also been shown to co-exist, causing multi drug resistant polymicrobial infections in hospitalized patients. Most common source of these infections are contaminated endoscopes and dialysis machines where S. maltophilia and A. baumannii has been shown to form biofilms [21, 26–29].
Another salient observation of our study is the rapid rise of carbapenems resistance among Acinetobacter species (CRA) (from 50% to 95% between 2011 and 2015) in Pakistan. This is in line with earlier CRA trends in Southeast Asian countries [30, 31]. This increase is concomitant with the two-fold increase in carbapenem usage in Pakistan over the last decade [32, 33]. It is critical to note that carbapenems are one of the few last resort broad-spectrum antibiotics recommended for treatment of sepsis in Pakistan [34] and emergence of CRA has further limited the therapeutic options. Only doxycycline and cefoperazone-sulbactam remain effective against many of the Acinetobacter strains (Table 4). The persistent susceptibility to doxycycline may be attributed to its limited usage in Acinetobacter infections treatment due to its bacteriostatic nature [35]. Further, while resistance against cefoperazone-sulbactam was found to be on the rise (Table 4), a wider acquisition of resistance against it might require more time. Additionally, Acinetobacter is an environmental pathogen found in the soil, the acquisition of resistance therefore suggests inappropriate disposal of antimicrobials. To further explain this, it is likely that unused or expired antimicrobials are being disposed of in a way that they are available to environmental organisms resulting in emergence of resistance in such organism [36, 37]. This situation provides policymakers with an opportunity to legislate regulated usage of carbapenem, cefoperazone-sulbactam, and doxycycline in the country, and ensure proper disposal of such antimicrobials from hospital systems. Developed countries like the US and the UK have resorted to using colistin, polymixin B, and tigecycline for the treatment of CRA infections [38, 39]. Susceptibility data against these antibiotics were not accounted for in this study. Future studies should highlight the susceptibility patterns of Acinetobacter against colistin, polymixin B, and tigecycline.
Another important finding of this study is the presence of third and fourth generation cephalosporins resistance in S. Typhi. Recent studies have reported an incidence of third generation resistant S. Typhi infections in Pakistan and neighboring countries [40, 41]. These antimicrobials are the drugs of choice for the empirical treatment of these infections [42]. This suggests that treatment of these infections with cephalosporins will become ineffective. Case studies from Canada and the US have shown that these infections can successfully be treated with carbapenems and ceftriaxone-azithromycin combination [43]. However, it is highly likely that S. Typhi will acquire resistance against these antimicrobials in the short term. In the light of this, WHO has recommended large scale implementation of Typbar-TCV, a typhoid vaccine, for the containment of S. Typhi infections in high risk regions [42, 44]. Improved personal hygiene, handwashing, availability of clean drinking water, and well cooked food lowers the risk of S. Typhi infections by 20% [42, 45]. The government should actively work towards implementing these suggestions to limit the spread of drug resistant S. Typhi infections.
While AMR was rising in other major pathogens, we unexpectedly found decreasing resistance rates against few major antimicrobials in S. aureus. This is in line with the reports on incidence of antimicrobial resistant S. aureus infections worldwide [46, 47]. While the exact reason for this decrease is yet to be identified, studies have suggested that it may be due to improved infection control practices and improved antibiotic prescription guidelines [46]. The ability of S. aureus to excise resistance markers, out of its genome, in the absence of antimicrobials may have played a role in the decreasing resistance as well. Excision of resistance markers is associated with reduced metabolic cost and overall increase in the bacterial fitness [46, 48, 49]. While the current resistance levels in S. aureus are low, continuous surveillance is required to keep track of emerging resistance trends in this pathogen.
Study limitation
This study is limited in that two provinces, namely Baluchistan and Gilgit-Baltistan, were not represented. The combined population of these two provinces constitutes less than 10% of the population of and our sample was representative of two large provinces namely Punjab and Sindh where 90% of Pakistani population resides. Therefore we can comfortably state that this is the representative sample of the country. However, this does not exclude the need for performing similar studies in areas such as Gilgit-Baltistan and Baluchistan [50]. However, excessing data from these regions is difficult and adds to the limitations of our study. Next, while our results have shown CoNS to be the most prevalent bacterial pathogen isolated from the blood and CSF cultures, this can be explained by CoNS being a common skin contaminant. For the purpose of this study, we have treated CoNS as a contaminant and not reported resistance trends in CoNS. However, further studies need to be carried to determine the role of CoNS as a potential bacteremia pathogen. Due to the unavailability of the patient characteristics we were also limited in our analysis of the impact of comorbid conditions on the susceptibility to invasive infections by resistant bacteria. Data regarding antimicrobial prescription practices was also lacking, preventing us to determine the impact of prescription practices in a geographic region on emergence of resistance. Since a significant proportion of our patient population was from Punjab and KPK, our analysis by geographic region was limited by lack of power. Lastly, susceptibility data on last resort antibiotics, including colistin, polymixin B, and linezolid, was not available. Susceptibility patterns of invasive pathogens to these antibiotics still need to be carried out.
Conclusions
In this study we set out to determine resistance patterns in pathogens isolated from blood and CSF cultures. We found that resistance has been at rise for several of these pathogens. Highest resistance rates were observed in Acinetobacter species against all tested antimicrobials including carbapenems. Resistance against 3rd and 4th generation cephalosporins has been reported in S. Typhi during the study period. Policy makers should prioritize and expedite implementation of infection control practices and antimicrobial stewardship in the country to control the emerging threat of AMR to public health.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
Acknowledgments
We would like to thank the staff at Department of Microbiology at Chughtai Lab Lahore and Lahore University of Management Sciences for the core support and assistance. We would like to thank Nestor Mojica from The Center for Disease Dynamics, Economics & Policy (CDDEP) for assistance in data organization and processing. We also acknowledge Iqra Manzoor for technical support and assistance throughout the study.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the Government of Pakistan through National Research Program for Universities – Higher Education Commission [Grant: 5193 awarded to Dr. Shaper Mirza; Grant: 3629 awarded to Dr. Safee Ullah Chaudhary]. Partial support for this project was provided by Lahore University of Management Sciences through Startup Funds [Grant: STG071 awarded to Dr. Shaper Mirza] and Ignite Funds [Grant: ICTRDF/TR&D/2014/33 awarded to Dr. Safee Ullah Chaudhary]. The funders had no role in study design, data collection, data analysis, decision to publish or preparation of the manuscript.
References
- 1.Okeke IN, Laxminarayan R, Bhutta ZA, Duse AG, Jenkins P, O’Brien TF, et al. Antimicrobial resistance in developing countries. Part I: Recent trends and current status. Lancet Infectious Diseases. 2005. 10.1016/S1473-3099(05)70189-4 [DOI] [PubMed] [Google Scholar]
- 2.Fleischmann C, Scherag A, Adhikari NKJ, Hartog CS, Tsaganos T, Schlattmann P, et al. Assessment of global incidence and mortality of hospital-treated sepsis current estimates and limitations. Am J Respir Crit Care Med. 2016; 10.1164/rccm.201504-0781OC [DOI] [PubMed] [Google Scholar]
- 3.Zilberberg MD, Kollef MH, Shorr AF. Secular trends in Acinetobacter baumannii resistance in respiratory and blood stream specimens in the United States, 2003 to 2012: A survey study. J Hosp Med. 2016; 10.1002/jhm.2477 [DOI] [PubMed] [Google Scholar]
- 4.May L, Klein EY, Rothman RE, Laxminarayan R. Trends in antibiotic resistance in coagulase-negative staphylococci in the United States, 1999 to 2012. Antimicrob Agents Chemother. 2014; 10.1128/AAC.01908-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sader HS, Farrell DJ, Flamm RK, Jones RN. Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalized in intensive care units in United States and European hospitals (2009–2011). Diagn Microbiol Infect Dis. 2014; [DOI] [PubMed] [Google Scholar]
- 6.Lagacé-Wiens PRS, Adam HJ, Low DE, Blondeau JM, Baxter MR, Denisuik AJ, et al. Trends in antibiotic resistance over time among pathogens from Canadian hospitals: results of the CANWARD study 2007–11. J Antimicrob Chemother. 2013; [DOI] [PubMed] [Google Scholar]
- 7.Sohail M, Khurshid M, Murtaza Saleem HG, Javed H, Khan AA. Characteristics and antibiotic resistance of urinary tract pathogens isolated from Punjab, Pakistan. Jundishapur J Microbiol. 2015; 10.5812/jjm.19272v2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qamar FN, Azmatullah A, Kazi AM, Khan E, Zaidi AKM. A three-year review of antimicrobial resistance of Salmonella enterica serovars Typhi and Paratyphi A in Pakistan. J Infect Dev Ctries. 2014; 10.3855/jidc.3817 [DOI] [PubMed] [Google Scholar]
- 9.Sohail M, Rashid A, Aslam B, Waseem M, Shahid M, Akram M, et al. Antimicrobial susceptibility of acinetobacter clinical isolates and emerging antibiogram trends for nosocomial infection management. Rev Soc Bras Med Trop. 2016; 10.1590/0037-8682-0111-2016 [DOI] [PubMed] [Google Scholar]
- 10.Smith PB, Tomfohrde KM, Rhoden DL, Balows A. API system: a multitube micromethod for identification of Enterobacteriaceae. Appl Microbiol. 1972; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clsi. Performance Standards for Antimicrobial Susceptibility Testing; Clinical and Laboratory Standards Institute. 2013.
- 12.Jawad I, Lukšić I, Rafnsson SB. Assessing available information on the burden of sepsis: global estimates of incidence, prevalence and mortality. J Glob Health. 2012; 10.7189/jogh.02.010404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Den Hoogen A, Gerards LJ, Verboon-Maciolek MA, Fleer A, Krediet TG. Long-term trends in the epidemiology of neonatal sepsis and antibiotic susceptibility of causative agents. Neonatology. 2009; 10.1159/000226604 [DOI] [PubMed] [Google Scholar]
- 14.Franco BE, Martínez MA, Sánchez Rodríguez MA, Wertheimer AI. The determinants of the antibiotic resistance process. Infection and Drug Resistance. 2009. [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes AH, Moore LSP, Sundsfjord A, Steinbakk M, Regmi S, Karkey A, et al. Understanding the mechanisms and drivers of antimicrobial resistance. The Lancet. 2016. 10.1016/S0140-6736(15)00473-0 [DOI] [PubMed] [Google Scholar]
- 16.Cantón R, Ruiz-Garbajosa P. Co-resistance: An opportunity for the bacteria and resistance genes. Current Opinion in Pharmacology. 2011. 10.1016/j.coph.2011.07.007 [DOI] [PubMed] [Google Scholar]
- 17.World Health Organisation. Global action plan on antimicrobial resistance. World Health Organisation. 2015.
- 18.Taj Y, Abdullah FE, Kazmi SU. Current pattern of antibiotic resistance in Staphylococcus aureus clinical isolates and the emergence of vancomycin resistance. J Coll Physicians Surg Pakistan. 2010. [PubMed] [Google Scholar]
- 19.Idrees F, Jabeen K, Khan MS, Zafar A. Antimicrobial resistance profile of Methicillin Resistant Staphylococcal Aureus from skin and soft tissue isolates. J Pak Med Assoc. 2009. [PubMed] [Google Scholar]
- 20.Gandra S, Mojica N, Klein EY, Ashok A, Nerurkar V, Kumari M, et al. Trends in antibiotic resistance among major bacterial pathogens isolated from blood cultures tested at a large private laboratory network in India, 2008–2014. Int J Infect Dis. 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adegoke AA, Stenström TA, Okoh AI. Stenotrophomonas maltophilia as an emerging ubiquitous pathogen: Looking beyond contemporary antibiotic therapy. Frontiers in Microbiology. 2017. 10.3389/fmicb.2017.02276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmed D, Nahid MA, Sami AB, Halim F, Akter N, Sadique T, et al. Bacterial etiology of bloodstream infections and antimicrobial resistance in Dhaka, Bangladesh, 2005–2014. Antimicrob Resist Infect Control. 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia A, Delorme T, Nasr P. Patient age as a factor of antibiotic resistance in methicillinresistant staphylococcus aureus. J Med Microbiol. 2017; [DOI] [PubMed] [Google Scholar]
- 24.Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019; 10.1016/S1473-3099(18)30605-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medernach RL, Logan LK. The Growing Threat of Antibiotic Resistance in Children. Infectious Disease Clinics of North America. 2018. 10.1016/j.idc.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ali U, Abbasi SA, Kaleem F, Butt T, Raza S. Antimicrobial Resistance Pattern of Stenotrophomonas maltophilia Isolates from a Tertiary Care Setting in Rawalpindi, Pakistan. J Med Microbiol Infect Dis. 2016;4(3):83–7. [Google Scholar]
- 27.Tariq TM, Rasool E. Emerging Trends of Bloodstream Infections: A Six-Year Study at a Paediatric Tertiary Care Hospital in Kabul. J Coll Physicians Surg Pak. 2016; [PubMed] [Google Scholar]
- 28.Chang YT, Lin CY, Chen YH, Hsueh PR. Update on infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options. Frontiers in Microbiology. 2015. 10.3389/fmicb.2015.00893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gajdács M, Urbán E. Epidemiological Trends and Resistance Associated with Stenotrophomonas maltophilia Bacteremia: A 10-Year Retrospective Cohort Study in a Tertiary-Care Hospital in Hungary. Diseases. 2019;7(2):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Gu B, Mei Y, Wen Y, Xia W. Increasing resistance rate to carbapenem among blood culture isolates of Klebsiella pneumoniae, Acinetobacter baumannii and Pseudomonas aeruginosa in a university-affiliated Hospital in China, 2004–2011. J Antibiot (Tokyo). 2015; [DOI] [PubMed] [Google Scholar]
- 31.Kamolvit W, Sidjabat HE, Paterson DL. Molecular Epidemiology and Mechanisms of Carbapenem Resistance of Acinetobacter spp. in Asia and Oceania. Microb Drug Resist. 2015; 10.1089/mdr.2014.0234 [DOI] [PubMed] [Google Scholar]
- 32.Retail sales of carbapenem antibiotics to treat Gram-negative bacteria are increasing rapidly in India and Pakistan—Center for Disease Dynamics, Economics & Policy (CDDEP) [Internet]. [cited 2019 Jun 18]. Available from: https://cddep.org/tool/retail_sales_carbapenem_antibiotics_treat_gram_negative_bacteria_are_increasing_rapidly_india/
- 33.Khan EA. Situation Analysis Report on Antimicrobial Resistance in Pakistan-Findings and Recommendations for Antibiotic Use and Resistance. Internet] Glob Antibiot Resist Partnersh (GARP), Pakistan[cited 2018 Mar 19] Available from https//www cddep org/publications/garp-pakistan-situation-analysis. 2018;
- 34.Hussain E, Jamil B, Salahuddin N. Guidelines for the initial management of adults with sepsis/severe sepsis/septic shock: 2015. 2015; [Google Scholar]
- 35.Rodríguez-Hernández MJ, Pachón J, Pichardo C, Cuberos L, Ibáñez-Martínez J, García-Curiel A, et al. Imipenem, doxycycline and amikacin in monotherapy and in combination in Acinetobacter baumannii experimental pneumonia. J Antimicrob Chemother. 2000; 10.1093/jac/45.4.493 [DOI] [PubMed] [Google Scholar]
- 36.Harris SJ, Cormican M, Cummins E. Antimicrobial Residues and Antimicrobial-Resistant Bacteria: Impact on the Microbial Environment and Risk to Human Health-A Review. Hum Ecol Risk Assess. 2012; [Google Scholar]
- 37.Clark NM, Zhanel GG, Lynch JP. Emergence of antimicrobial resistance among Acinetobacter species: A global threat. Current Opinion in Critical Care. 2016. 10.1097/MCC.0000000000000337 [DOI] [PubMed] [Google Scholar]
- 38.Pogue JM, Mann T, Barber KE, Kaye KS. Carbapenem-resistant Acinetobacter baumannii: Epidemiology, surveillance and management. Expert Review of Anti-Infective Therapy. 2013. 10.1586/eri.13.14 [DOI] [PubMed] [Google Scholar]
- 39.Qureshi ZA, Hittle LE, O’Hara JA, Rivera JI, Syed A, Shields RK, et al. Colistin-resistant acinetobacter baumannii: Beyond carbapenem resistance. Clin Infect Dis. 2015; 10.1093/cid/civ048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanmoy AM, Westeel E, De Bruyne K, Goris J, Rajoharison A, Sajib MSI, et al. Salmonella enterica Serovar Typhi in Bangladesh: Exploration of Genomic Diversity and Antimicrobial Resistance. MBio. 2018; 10.1128/mBio.02112-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qamar FN, Yousafzai MT, Khalid M, Kazi AM, Lohana H, Karim S, et al. Outbreak investigation of ceftriaxone-resistant Salmonella enterica serotype Typhi and its risk factors among the general population in Hyderabad, Pakistan: a matched case-control study. Lancet Infect Dis. 2018; 10.1016/S1473-3099(18)30483-3 [DOI] [PubMed] [Google Scholar]
- 42.Stoesser N, Eyre D, Basnyat B, Parry C. Treatment of enteric fever (typhoid and paratyphoid fever) with third and fourth generation cephalosporins. Cochrane Database Syst Rev. 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kariuki S, Gordon MA, Feasey N, Parry CM. Antimicrobial resistance and management of invasive Salmonella disease. Vaccine. 2015; 10.1016/j.vaccine.2015.03.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.World Health Organization. Typhoid vaccines: WHO position paper, March 2018 –Recommendations. Vaccine. 2019. [DOI] [PubMed]
- 45.Levine MM, Simon R. The Gathering Storm: Is Untreatable Typhoid Fever on the Way? MBio. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perovic O, Iyaloo S, Kularatne R, Lowman W, Bosman N, Wadula J, et al. Prevalence and trends of staphylococcus aureus bacteraemia in hospitalized patients in South Africa, 2010 to 2012: Laboratory-based surveillance mapping of antimicrobial resistance and molecular epidemiology. PLoS One. 2015; 10.1371/journal.pone.0145429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lafaurie M, Porcher R, Donay JL, Touratier S, Molina JM. Reduction of fluoroquinolone use is associated with a decrease in methicillin-resistant Staphylococcus aureus and fluoroquinolone-resistant Pseudomonas aeruginosa isolation rates: A 10 year study. J Antimicrob Chemother. 2012; 10.1093/jac/dkr555 [DOI] [PubMed] [Google Scholar]
- 48.Boundy S, Zhao Q, Fairbanks C, Folgosa L, Climo M, Archer GL. Spontaneous staphylococcal cassette chromosome mec element excision in Staphylococcus aureus nasal carriers. J Clin Microbiol. 2012; 10.1128/JCM.01063-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Massey RC, Buckling A, Peacock SJ. Phenotypic switching of antibiotic resistance circumvents permanent costs in Staphylococcus aureus. Curr Biol. 2001; 10.1016/s0960-9822(01)00507-3 [DOI] [PubMed] [Google Scholar]
- 50.Pakistan Bureau of Statistics. Pakistan Bureau of Statistics | 6th Population and Housing Census. Pakistan Bureau of Statistics. 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.