Abstract
Nanoparticle carriers are effective drug delivery vehicles. Along with other design parameters including size, composition, and surface charge, particle shape strongly influences cellular uptake. How nanoparticle geometry affects targeted delivery under physiologically relevant conditions, however, is inconclusive. Here, we demonstrate that nanoconstruct core shape influences the dynamics of targeting ligand-receptor interactions on cancer cell membranes. By single-particle tracking of translational and rotational motion, we compared DNA aptamer AS1411 conjugated gold nanostars (AS1411-AuNS) and 50-nm gold spheres (AS1411-50NPs) on cells with and without targeted nucleolin membrane receptors. On nucleolin-expressing cells, AS1411-AuNS exhibited faster velocities under directed diffusion and translated over larger areas during restricted diffusion compared to AS1411-50NPs, despite their similar protein corona profiles. On nucleolin-inhibited cells, AS1411-AuNS showed faster rotation dynamics over smaller translational areas, while AS1411-50NPs did not display significant changes in translation. These differences in translational and rotational motions indicate that nanoparticle shape affects how targeting nanoconstructs bind to cell-membrane receptors.
Keywords: nanoparticle shape, single-particle dynamics, differential interference contrast microscopy, protein corona, nucleolin, targeting nanoconstructs
Graphical Abstract
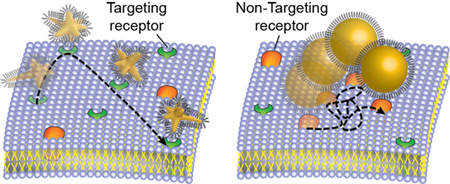
Nanoparticle (NP) shape is an important parameter that influences cellular uptake and intracellular trafficking.1–2 Nanoconstructs—NP cores surrounded by ligand shells—with different particle geometries can promote different interactions with cell membranes based on surface ligand presentation,3 including altered binding kinetics to receptors.3–5 Efforts to prepare nanoconstructs with different shapes have focused on various core materials, including noble metal NPs,6–7 metal organic frameworks,8–9 DNA nanostructures,10 and polymer NPs.11 In cellular environments, the non-specific adsorption of proteins on NP surfaces forms a so-called protein corona that can screen the engineered targeting moieties as well as change the physicochemical properties of NPs.12–13 Although the composition of the protein corona has been shown to depend on NP geometry14–15 and conformation of ligand shell,16 the impact on the subcellular fate of targeting nanoconstructs is unknown.
In situ monitoring of nanoconstruct-cell interactions using NPs as probes can deconvolute the effects of NP shape and protein corona in physiologically relevant conditions. However, because of the intrinsic low optical contrast compared to cells, ligand-functionalized organic NPs and DNA origamis cannot function as probes without conjugation to dyes or inorganic nanoparticles.17–19 In contrast, gold nanoparticles (AuNPs) can be tracked via optical methods because of their strong scattering properties20 and also have advantages as probes because they are biocompatible, can be synthesized into various shapes,21–22 and can be covalently functionalized with diverse ligands.23–25 Anisotropic AuNPs such as gold nanostars (AuNS) show angle-dependent patterns in differential interference contrast (DIC) microscopy26–28 with potential for 3D orientation tracking,29 which can provide information on ligand-receptor binding and endocytosis at the molecular level.
DNA aptamer AS1411, the first aptamer to enter clinical oncology trials,30 is a widely used tumor-targeting ligand that binds to nucleolin (NCL), an overexpressed and ubiquitous protein on the surface of various cancer cell types.31 Previous work showed that AS1411-conjugated AuNS (AS1411-AuNS) are internalized after binding to NCL and shuttled to the peri-nuclear region to induce cell apoptosis with in vitro efficacy higher than free AS1411 aptamer.23, 32 Recently, we found that AuNS nanoconstructs functionalized with targeting versus non-targeting ligands exhibit distinct translational and rotational behavior even with similar protein corona profiles;26 hence, the ligand shell properties are preserved on anisotropic NPs and are critical for NP-cell membrane interactions.24, 26, 32–33 The influence of nanoconstruct shape on the presentation of targeting ligands to receptors on cell membranes has received little attention.
Here we demonstrate that NP shape determines whether engineered nanoconstructs maintain their targeting abilities on cell membranes. By real-time single-particle tracking using DIC-epifluorescence imaging, we compared dynamics of AS1411-conjugated AuNS (AS1411-AuNS) and AuNP spheres (AS1411-50NPs) on NCL-expressing (NCL+) and NCL-inhibited (NCL−) MCF-7 cells. We confirmed AS1411-NCL binding by tracking AS1411-AuNS rotation on NCL+ and NCL− cells; limited angular rotation was observed on NCL+ cells but free, fast rotation on NCL− cells. We then compared translational dynamics of AS1411-AuNS and AS1411-50NPs. Despite similar protein corona profiles, AS1411-AuNS exhibited faster velocities under directed diffusion and translated over larger areas during restricted diffusion compared to AS1411-50NPs. On NCL− cells, AS1411-AuNS showed much shorter translational trajectories, while AS1411-50NPs showed translation similar to that on NCL+ cells, which demonstrates that the two nanoconstructs have different targeting specificities for NCL. Our results suggest that NP core shape should be considered an important design parameter to maintain the targeting properties of nanoconstructs, especially for drug-delivery carriers.
Scheme 1 depicts representative single-particle dynamics of targeting constructs on cancer cell membranes. We prepared AS1411-AuNS and AS1411-50NPs with the same surface ligand density (Table S1) to investigate effects of particle geometry on AS1411-NCL interactions. We compared the single-particle dynamics of the two nanoconstructs on both NCL+ and NCL− MCF-7 cells to confirm the specificity of AS1411-nanoconstructs towards NCL. NCL− cells were prepared by pre-incubating MCF-7 cells with a NCL inhibitor to determine how blocking the NCL receptor would change nanoconstruct motion (SI, Methods). 30-s video streams were acquired to track individual AS1411-AuNS and AS1411-50NPs motion on cell membranes with either expressing or blocked NCL.
Scheme 1.
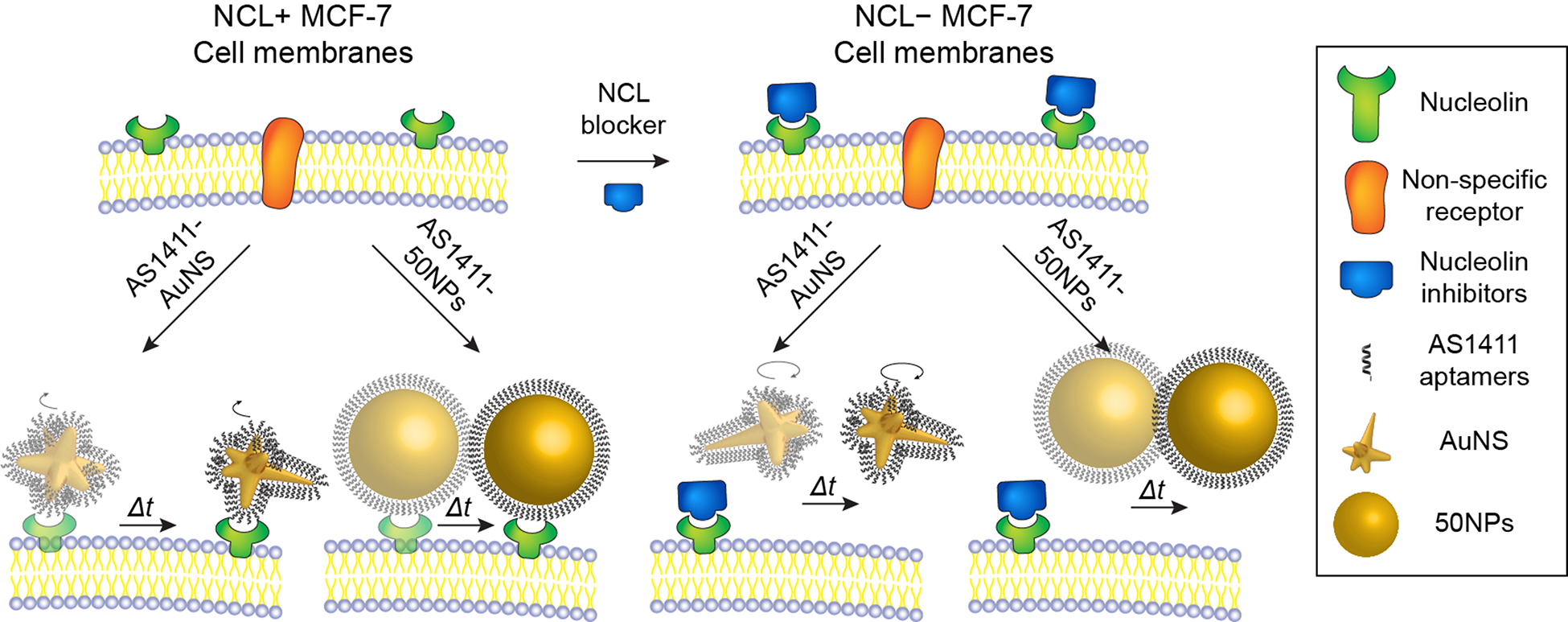
Live-cell imaging of AS1411-AuNS and AS1411-50NPs on (left) NCL+ and (right) NCL− cancer cell membranes.
Figure 1 illustrates how the single-particle dynamics of AS1411-AuNS and AS1411-50NPs on cancer cell membranes were characterized via multi-channel and multi-wavelength imaging. Figures 1a–b depict a series of DIC images of AS1411-nanoconstructs on a glass substrate under rotation over 180° in increments of 45°; AS1411-AuNS showed angle-dependent DIC image patterns because of their anisotropic structure. AS1411-50NPs did not show significant changes at different angles because of symmetry, and moreover, were difficult to distinguish from spherical cellular vesicles by DIC microscopy alone (Figure S1).
Figure 1. Multi-channel live-cell imaging of AS1411-nanoconstructs on cell membrane.
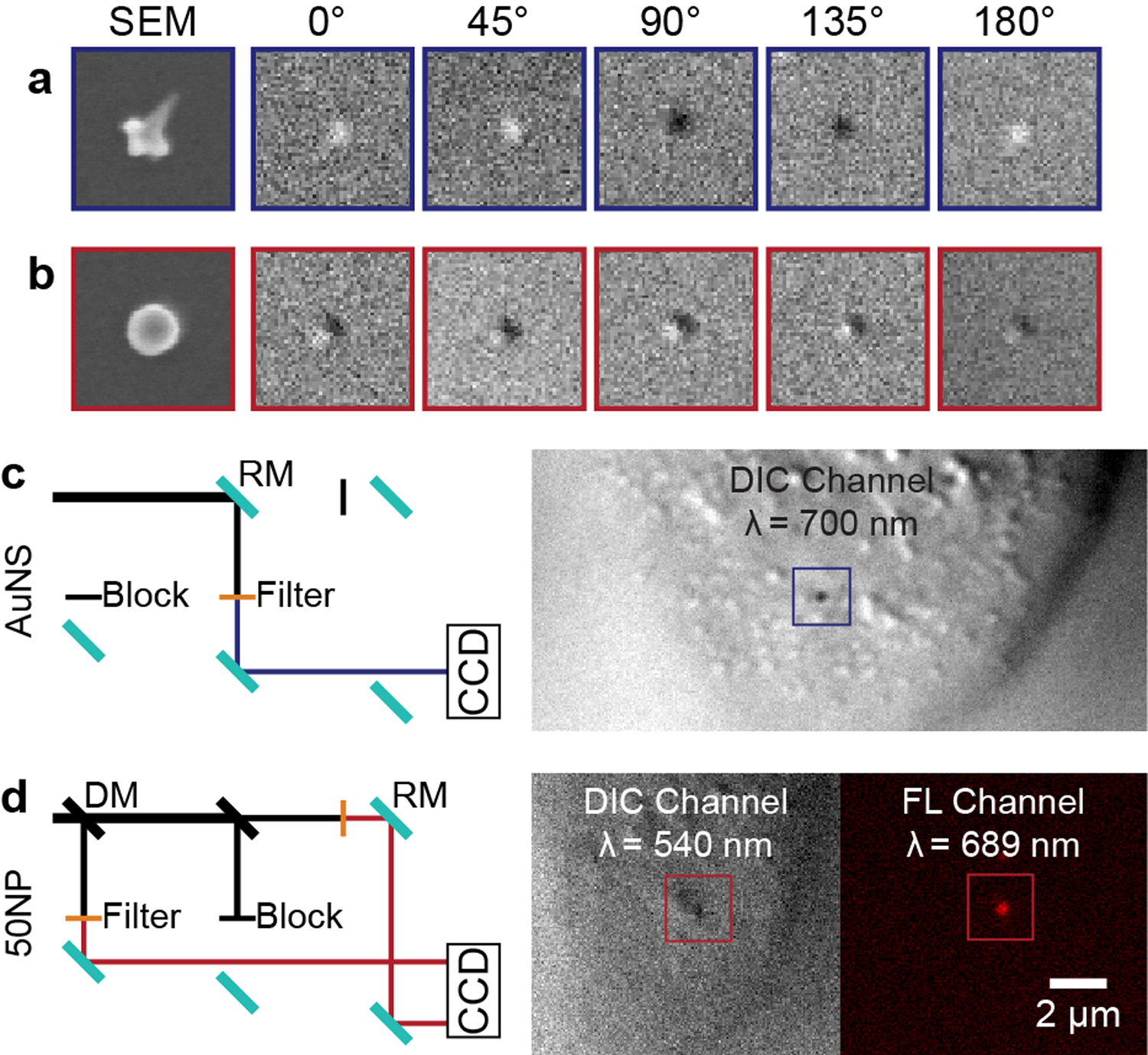
SEM and DIC images of (a) AuNS and (b) 50NP taken at 0°, 45°, 90°, 135°, and 180°. DIC images: 2 μm × 2 μm, SEM images: 200 nm × 200 nm. (c) DIC light path of AS1411-AuNS and an example DIC image. (d) DIC-epifluorescence light path for tracking AS1411-50NPs and representative DIC and epifluorescence image. DM: dichroic mirror, RM: reflecting mirror.
We carried out single-channel DIC imaging of AS1411-AuNS at 700 nm, near the localized surface plasmon resonance of the AuNS, which showed high signal-to-noise ratios for both cellular features and nanoconstruct probes (Figure 1c). We conjugated Cy5-labeled AS141134 to the 50-nm spheres and tracked particle motions with multi-channel DIC-epifluorescence imaging (Figure 1d). NP motion was tracked in the epifluorescence channel (λ = 689 nm) by the Cy5 signal; the DIC channel (λ = 543 nm)20 was used to locate the particles with respect to the cell membrane. We identified the image plane of the cell membrane by optical sectioning (Figure S2, S3).
We first evaluated rotation of the nanoconstructs because of their relevance to ligand/receptor binding and clustering.35 Figure 2 shows that AS1411-AuNS on NCL+ cells and on NCL− cells have distinct rotational behaviors, indicating that targeting specificity of AS1411-AuNS to NCL was maintained in vitro. AS1411-AuNS exhibited a constant DIC pattern over several seconds on NCL+ cells (Figure 2a and S4). In contrast, AS1411-AuNS on NCL− cells fluctuated rapidly over a similar time period (Figure 2b), indicating a higher degree of rotational freedom for AS1411-AuNS when NCL was unavailable for binding. We calculated the contrast for AuNS in each frame, which we define as the average intensity of the circular AuNS pattern normalized against the background after subtracting the background intensity, (I − Ibkg)/Ibkg (SI Methods).27–28 On NCL+ cells, the contrast remained mostly constant, either positive (bright) or negative (dark); on NCL− cells, fast changes from negative to positive were observed (Figure 2c–d).
Figure 2. Rotational Dynamics AS1411-AuNS on NCL+ and NCL− MCF-7 cells.

DIC images of AS1411-AuNS on (a) NCL+ and (b) NCL− cell membranes. (c, d) Plots of DIC contrast for the two particles in (a, b) over the whole 30-s stream. (e) Comparison of maximum duration for AS1411-AuNS. (NCL+: N = 56, NCL−: N = 38) Statistical significance: ***p < 0.001, student t-test.
To distinguish between large and small angular changes during construct rotation, we defined the maximum contrast duration instead of the previous rotational frequency analysis method.26 We quantified the longest time when the nanoconstruct contrast remained the same sign over the 30-s stream; on average, the maximum duration for AS1411-AuNS was ~1.8 times longer on NCL+ cells compared to NCL− cells (Figure 2e). We also confirmed that rotational dynamic variations between AuNS/NCL+ and AuNS/NCL− were not from cell behavior changes after NCL blocking by comparing rotations of non-targeting control aptamer-conjugated AuNS on NCL+ and NCL− cells; no statistical difference was observed between the two cases (p > 0.05) (Figure S5). This result supports that the faster rotation of AS1411-AuNS on NCL− cells may be attributed to the unavailability of NCL for binding. A viability test with increased inhibitor concentration also established that the concentration used to block NCL did not induce cytotoxicity (Figure S6).
To determine whether the protein corona affected single-particle motion, we characterized the composition for AS1411-AuNS and AS1411-50NPs (SI Methods).26 The protein corona profiles for both nanoconstructs were almost identical (Figure S7, Table S2), with serum albumin being the most abundant protein (23% for AS1411-AuNS, 18% for AS1411-50NPs). Although alpha-2-HS-glycoprotein, known to target scavenger receptor-A,36 was found in the protein corona of both nanoconstructs (7% for AS1411-AuNS, 6% for AS1411-50NPs), this receptor is not expressed on MCF-7 cell membranes.37 Therefore, the protein corona does not influence the nanoconstruct targeting abilities to NCL or corresponding single-particle dynamics of AS1411-nanoconstructs during live-cell imaging.
Since translation of nanoconstructs after NP-receptor binding can provide insight on the lateral diffusion of surface receptors,38–40 we carried out single-particle tracking of AS1411-AuNS and AS1411-50NPs to probe shape-dependent translational dynamics. We processed the tracked particles by mean square displacement (MSD) analysis categorized into four diffusion modes: directed diffusion (DD), simple diffusion (SD), restricted diffusion (RD), and stationary (ST).26 For each nanoconstruct, the diffusion mode with the best MSD fit was assigned (Figure 3a and S8). Generally, AS1411-AuNS showed a drastic decrease in trajectory length on NCL− cells for all three types of motion: DD, SD, RD, while the trajectory lengths of AS1411-50NPs did not change significantly (Figure 3b).
Figure 3. Categorizing diffusion modes via MSD fitting.
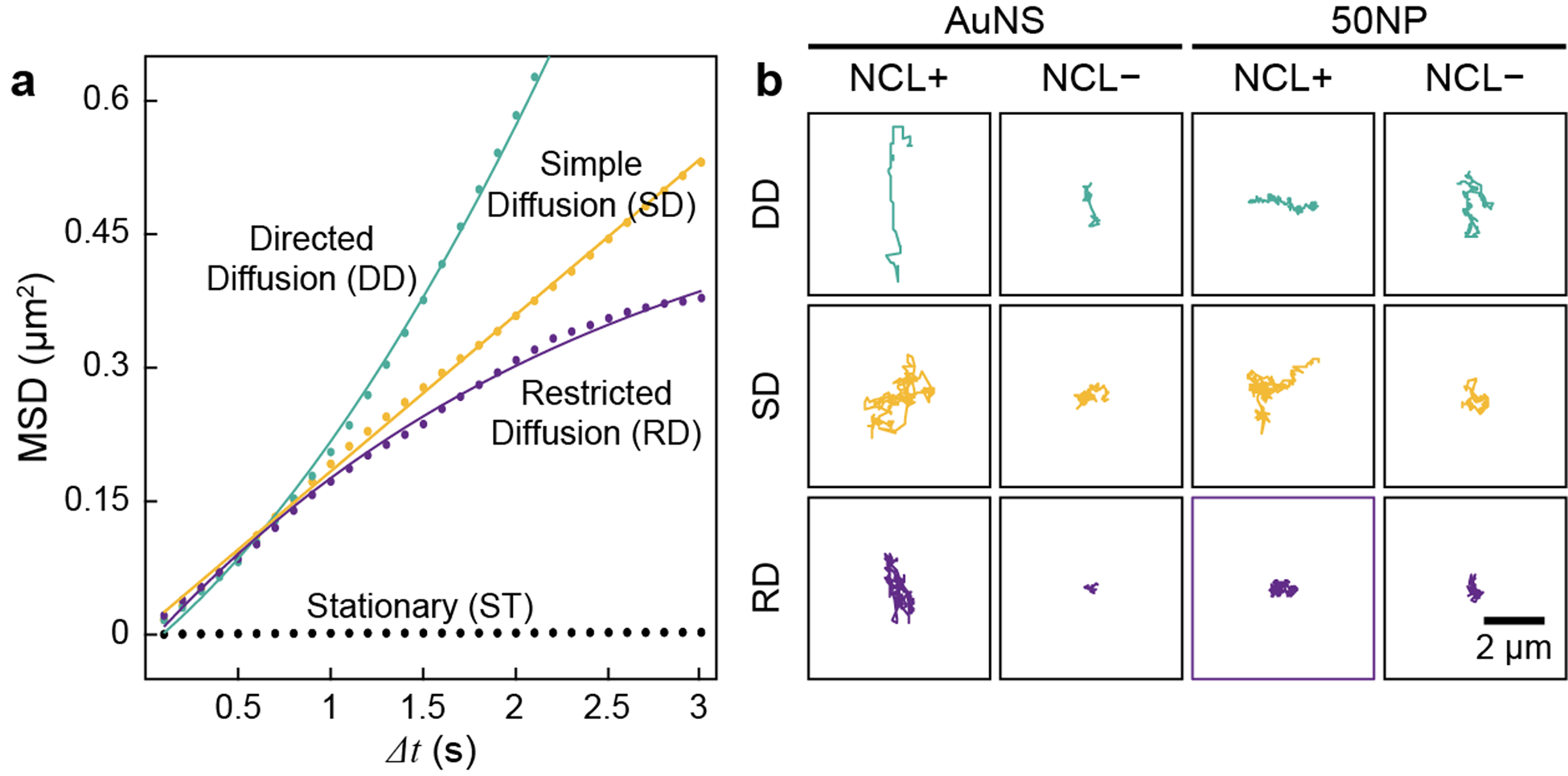
(a) Representative MSD fitting. Raw data (dots) were fit with corresponding equations (solid lines). (b) Representative trajectories for AS1411-AuNS/NCL+, AS1411-AuNS/NCL−, AS1411-50NPs/NCL+, and AS1411-50NPs/NCL−.
Furthermore, we quantitively compared the translational motion of the two differently shaped nanoconstructs. The proportion of AS1411-AuNS under DD decreased from 51.8% to 15.8% after NCL was blocked, while the percentage of AS1411-AuNS under RD increased from 35.7% to 60.5% (Figures 4a–b). In addition, AS1411-AuNS that were ST were only observed on NCL− cells. AS1411-50NPs showed a lower percentage decrease in DD after NCL blocking (37.8% to 20.6%) compared to AS1411-AuNS; however, RD accounted for the highest population on both NCL+ and NCL− cells (37.8% and 47.1%, respectively). Different from AS1411-AuNS, ST particles were observed for AS1411-50NPs on both NCL+ and NCL− cells (Figures 4c–d). The change in diffusion mode distribution after NCL inhibition confirms that AS1411-AuNS binds to NCL, consistent with rotational dynamics results (Figure 2). We attribute the high percentage of AS1411-AuNS going through DD on NCL+ cells to NCL clustering being highly dependent on the presence of intact actin filaments41 in the cytoskeleton, which are responsible for directed movement on the cell membrane.
Figure 4. Impact of NP shape on nanoconstruct translations.
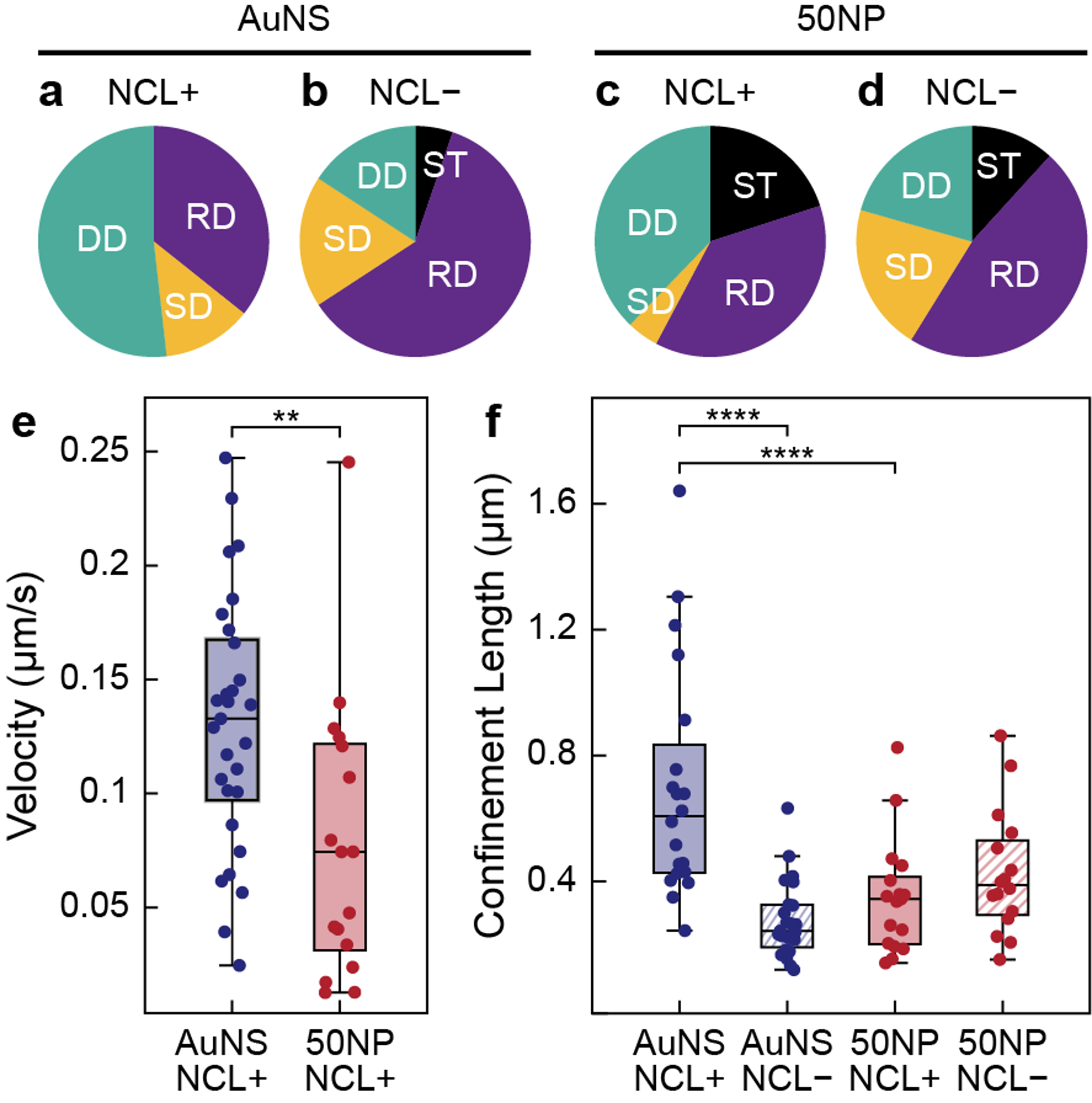
(a-d) Translational mode distribution for AS1411-AuNS/NCL+, AS1411-AuNS/NCL−, AS1411-50NPs/NCL+ (N = 45), and AS1411-50NPs/NCL− (N = 34). (e) Velocity for AS1411-AuNS and AS1411-50NPs under DD on NCL+ MCF-7 cell membranes. (f) Confinement lengths for AS1411-AuNS and AS1411-50NPs under RD on NCL+ and NCL− MCF-7 cells. Statistical significance: **p < 0.01, ****p < 0.0001, one-way ANOVA.
We further compared the diffusion parameters, including velocity and confinement length. For nanoconstructs under DD, AS1411-AuNS translated at ~1.7 times faster velocities compared to AS1411-50NPs (Figure 4e). For nanoconstructs with RD, AS1411-AuNS showed ~2 times higher confinement lengths compared to AS1411-50NPs on NCL+ cell membranes (Figure 4f). These differences in translational dynamics indicate that AS1411-AuNS and AS1411-50NPs bind to receptors with different diffusion dynamics. When NCL was blocked, the confinement lengths of AS1411-AuNS under RD decreased ~0.4 times, while spheres increased by ~1.2 times. The significant decrease in AS1411-AuNS confinement lengths on NCL− cells provides additional support that AS1411-AuNS bind specifically to NCL. For AS1411-50NPs, however, NCL inhibition did not significantly change the confinement lengths, which suggests that NCL was not the dominant receptor interacting with AS1411-50NPs. Therefore, the two nanoconstructs have different targeting selectivity towards NCL because of differences in their core shapes.
In summary, we demonstrated that nanoconstruct core shape governs single-particle dynamics during in vitro targeted ligand-receptor interactions on cancer cells. The differences in translation of AS1411-AuNS and AS1411-50NPs suggest that the two constructs show different targeting specificity towards NCL. We hypothesize that the branched structure of the AuNS core provides multiple areas of contact and ligand presentation and enables multivalent binding between AS1411 to the NCL receptor, while AS1411-50NPs only provides a single limited area of contact. Our study indicates that NP shape is a critical factor in determining ligand-receptor interactions that will affect downstream effects during targeted delivery.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Health (NIH) under awards 5 R01 GM131421-02 (P.C., T.L., T.W.O.) and 1R01GM115763. This work made use of the EPIC facility of Northwestern University’s NUANCE Center, which has received support from the SHyNE Resource (NSF ECCS-2025633), the IIN, and Northwestern’s MRSEC program (NSF DMR-1720139). Proteomics services were performed by the Northwestern Proteomics Core Facility, generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center, instrumentation award (S10OD025194) from NIH Office of Director, and the National Resource for Translational and Developmental Proteomics supported by P41 GM108569. Metal analysis was performed at the Northwestern University Quantitative Bio-element Imaging Center generously supported by NASA Ames Research Center NNA06CB93G. We are grateful for the support from Northwestern University High Throughput Analysis Laboratory.
Footnotes
Supporting Information available: The Supporting Information is available free of charge via the Internet at http://pubs.acs.org. Experimental methods; AS1411 nanoconstruct properties; differentiation of 50-nm spheres from cellular organelles; rotational motions of AS1411-AuNS on NCL+ and NCL− cells; effect of NCL inhibition on cAS1411-AuNS dynamics; the influence of endostatin on MCF-7 cell health; protein corona composition; determination of single-particle translational modes
The authors declare no competing financial interest.
References
- 1.Xie X; Liao J; Shao X; Li Q; Lin Y, The Effect of shape on Cellular Uptake of Gold Nanoparticles in the forms of Stars, Rods, and Triangles. Sci Rep 2017, 7 (1), 3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foroozandeh P; Aziz AA, Insight into Cellular Uptake and Intracellular Trafficking of Nanoparticles. Nanoscale Res Lett 2018, 13 (1), 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee K, Huang ZN, Mirkin CA, Odom TW, Endosomal Organization of CpG Constructs Correlates with Enhanced Immune Activation. Nano Letters 2020, 20 (8), 6170–6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah S, Liu Y, Hu W, Gao J, Modeling Particle Shape-Dependent Dynamics in Nanomedicine. J Nanosci Nanotechnol 2011, 11 (2), 919–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKenzie M, Ha SM, Rammohan A, Radhakrishnan R, Ramakrishnan N, Multivalent Binding of a Ligand-Coated Particle: Role of Shape, Size, and Ligand Heterogeneity. Biophysical Journal 2018, 114 (8), 1830–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheon JY; Kim SJ; Rhee YH; Kwon OH; Park WH, Shape-dependent antimicrobial activities of silver nanoparticles. Int J Nanomedicine 2019, 14, 2773–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Y; Xia Y, Shape-controlled synthesis of gold and silver nanoparticles. Science 2002, 298 (5601), 2176–9. [DOI] [PubMed] [Google Scholar]
- 8.Sarawade P; Tan H; Anjum D; Cha D; Polshettiwar V, Size- and shape-controlled synthesis of hexagonal bipyramidal crystals and hollow self-assembled Al-MOF spheres. ChemSusChem 2014, 7 (2), 529–35. [DOI] [PubMed] [Google Scholar]
- 9.Shang W; Kang X; Ning H; Zhang J; Zhang X; Wu Z; Mo G; Xing X; Han B, Shape and size controlled synthesis of MOF nanocrystals with the assistance of ionic liquid mircoemulsions. Langmuir 2013, 29 (43), 13168–74. [DOI] [PubMed] [Google Scholar]
- 10.Bastings MMC, Anastassacos FM, Ponnuswamy N, Leiger FG, Cuneo G, Lin C, Ingber DE, Ryu JH, Shih WM, Modulation of the Cellular Uptake of DNA Origami through Control over Mass and Shape. Nano Letters 2018, 18 (6), 3557–3564. [DOI] [PubMed] [Google Scholar]
- 11.Geng Y; Dalhaimer P; Cai S; Tsai R; Tewari M; Minko T; Discher DE, Shape effects of filaments versus spherical particles in flow and drug delivery. Nat Nanotechnol 2007, 2 (4), 249–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai Q, Yan Y, Ang C-S, Kempe K, Kamphuis MMJ, Dodds SJ, Caruso F, Monoclonal Antibody-Functionalized Multilayered Particles: Targeting Cancer Cells in the Presence of Protein Coronas. ACS Nano 2015, 9 (3), 2876–2885. [DOI] [PubMed] [Google Scholar]
- 13.Oh JY; Kim HS; Palanikumar L; Go EM; Jana B; Park SA; Kim HY; Kim K; Seo JK; Kwak SK; Kim C; Kang S; Ryu JH, Cloaking nanoparticles with protein corona shield for targeted drug delivery. Nat Commun 2018, 9 (1), 4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madathiparambil Visalakshan R; Gonzalez Garcia LE; Benzigar MR; Ghazaryan A; Simon J; Mierczynska-Vasilev A; Michl TD; Vinu A; Mailander V; Morsbach S; Landfester K; Vasilev K, The Influence of Nanoparticle Shape on Protein Corona Formation. Small 2020, 16 (25), e2000285. [DOI] [PubMed] [Google Scholar]
- 15.Wang G; Wang W; Shangguan E; Gao S; Liu Y, Effects of gold nanoparticle morphologies on interactions with proteins. Mater Sci Eng C Mater Biol Appl 2020, 111, 110830. [DOI] [PubMed] [Google Scholar]
- 16.Chinen AB; Guan CM; Mirkin CA, Spherical nucleic acid nanoparticle conjugates enhance G-quadruplex formation and increase serum protein interactions. Angew Chem Int Ed Engl 2015, 54 (2), 527–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K; He X; Yang X; Shi H, Functionalized silica nanoparticles: a platform for fluorescence imaging at the cell and small animal levels. Acc Chem Res 2013, 46 (7), 1367–76. [DOI] [PubMed] [Google Scholar]
- 18.Li K; Liu B, Polymer-encapsulated organic nanoparticles for fluorescence and photoacoustic imaging. Chem Soc Rev 2014, 43 (18), 6570–97. [DOI] [PubMed] [Google Scholar]
- 19.Wang P, Rahman MA, Zhao Z, Weiss K, Zhang C, Chen Zhenjia, Hurwitz SJ, Chen ZG, Shin DM, Ke Y, Visualization of Cellular Uptake and Trafficking of DNA Origami Nanostructures in Cancer Cells. JACS 2018, 140, 2478–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun W; Wang G; Fang N; Yeung ES, Wavelength-dependent differential interference contrast microscopy: selectively imaging nanoparticle probes in live cells. Anal Chem 2009, 81 (22), 9203–8. [DOI] [PubMed] [Google Scholar]
- 21.Sau TK; Murphy CJ, Room temperature, high-yield synthesis of multiple shapes of gold nanoparticles in aqueous solution. J Am Chem Soc 2004, 126 (28), 8648–9. [DOI] [PubMed] [Google Scholar]
- 22.Grzelczak M; Perez-Juste J; Mulvaney P; Liz-Marzan LM, Shape control in gold nanoparticle synthesis. Chem Soc Rev 2008, 37 (9), 1783–91. [DOI] [PubMed] [Google Scholar]
- 23.Dam DHM, Culver KSB, Odom TW,, Grafting Aptamers onto Gold Nanostars Increases in Vitro Efficacy in a Wide Range of Cancer Cell Types. Molecular Pharmaceutics 2014, 11 (2), 580–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H, Dam D, Ha J Yue J, Odom TW, Enhanced Human Epidermal Growth Factor Receptor 2 Degradation in Breast Cancer Cells by Lysosome-Targeting Gold Nanoconstructs. ACS Nano 2015, 9, 9589–9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rotz MW, Culver KSB, Parigi G, MacRenaris KW, Luchinat C, Odom TW, Meade TJ, High Relaxivity Gd(III)–DNA Gold Nanostars: Investigation of Shape Effects on Proton Relaxation. ACS Nano 2015, 9 (3), 3385–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhowmik D, Culver KSB, Liu T, Odom TW, Resolving Single-Nanoconstruct Dynamics during Targeting and Nontargeting Live-Cell Membrane Interactions. ACS Nano 2019, 13 (12), 13637–13644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Culver KSB; Liu T; Hryn AJ; Fang N; Odom TW, In Situ Identification of Nanoparticle Structural Information Using Optical Microscopy. The Journal of Physical Chemistry Letters 2018, 9 (11), 2886–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choo P; Hryn AJ; Culver KS; Bhowmik D; Hu J; Odom TW, Wavelength-Dependent Differential Interference Contrast Inversion of Anisotropic Gold Nanoparticles. The Journal of Physical Chemistry C 2018, 122 (47), 27024–27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu J; Liu T; Choo P; Wang S; Reese T; Sample AD; Odom TW, Single-Nanoparticle Orientation Sensing by Deep Learning. ACS Cent Sci 2020, 6 (12), 2339–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laber DA, Sharma VR, Bhupalam L, Taft B, Hendler FJ, Barnhart KM, Update on the first phase I study of AGRO100 in advanced cancer. Journal of Clinical Oncology 2006, 23 (16), 3064. [Google Scholar]
- 31.Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO, Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Experimental and Molecular Biology 2009, 86 (2), 151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dam DHM, Lee JH, Sisco PN, Co DT, Zhang M, Wasielewski MR Odom TW,, Direct Observation of Nanoparticle-Cancer Cell Nucleus Interactions. ACS Nano 2012, 6 (4), 3318–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dam DH; Culver KS; Kandela I; Lee RC; Chandra K; Lee H; Mantis C; Ugolkov A; Mazar AP; Odom TW, Biodistribution and in vivo toxicity of aptamer-loaded gold nanostars. Nanomedicine 2015, 11 (3), 671–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dam DHM; Lee RC; Odom TW, Improved in Vitro Efficacy of Gold Nanoconstructs by Increased Loading of G-quadruplex Aptamer. Nano Letters 2014, 14 (5), 2843–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao L; Wei L; Liu C; He Y; Yeung ES, Unsynchronized translational and rotational diffusion of nanocargo on a living cell membrane. Angew Chem Int Ed Engl 2012, 51 (17), 4181–4. [DOI] [PubMed] [Google Scholar]
- 36.Herrmann M; Schafer C; Heiss A; Graber S; Kinkeldey A; Buscher A; Schmitt MM; Bornemann J; Nimmerjahn F; Herrmann M; Helming L; Gordon S; Jahnen-Dechent W, Clearance of fetuin-A--containing calciprotein particles is mediated by scavenger receptor-A. Circ Res 2012, 111 (5), 575–84. [DOI] [PubMed] [Google Scholar]
- 37.Vetter S, Chapter Five - Glycated Serum Albumin and AGE Receptors. Adv Clin Chem 2015, 72, 205–275. [DOI] [PubMed] [Google Scholar]
- 38.Spillane KM; Ortega-Arroyo J; de Wit G; Eggeling C; Ewers H; Wallace MI; Kukura P, High-speed single-particle tracking of GM1 in model membranes reveals anomalous diffusion due to interleaflet coupling and molecular pinning. Nano Lett 2014, 14 (9), 5390–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H; Zhang C; Vincent J; Zala D; Benstaali C; Sainlos M; Grillo-Bosch D; Daburon S; Coussen F; Cho Y; David DJ; Saudou F; Humeau Y; Choquet D, Modulation of AMPA receptor surface diffusion restores hippocampal plasticity and memory in Huntington’s disease models. Nat Commun 2018, 9 (1), 4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lasne D; Blab GA; Berciaud S; Heine M; Groc L; Choquet D; Cognet L; Lounis B, Single nanoparticle photothermal tracking (SNaPT) of 5-nm gold beads in live cells. Biophys J 2006, 91 (12), 4598–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hovanessian AG; Puvion-Dutilleul F; Nisole S; Svab J; Perret E; Deng JS; Krust B, The cell-surface-expressed nucleolin is associated with the actin cytoskeleton. Exp Cell Res 2000, 261 (2), 312–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


