Abstract
Opioid use disorder (OUD) represents a global public health crisis that challenges classic clinical decision making. As existing hospital screening methods are resource-intensive, patients with OUD are significantly under-detected. An automated and accurate approach is needed to improve OUD identification so that appropriate care can be provided to these patients in a timely fashion. In this study, we used a large-scale clinical database from Mass General Brigham (MGB; formerly Partners HealthCare) to develop an OUD patient identification algorithm, using multiple machine learning methods. Working closely with an addiction psychiatrist, we developed a set of hand-crafted rules for identifying information suggestive of OUD from free-text clinical notes. We implemented a natural language processing (NLP)-based classification algorithm within the Medical Text Extraction, Reasoning and Mapping System (MTERMS) tool suite to automatically label patients as positive or negative for OUD based on these rules. We further used the NLP output as features to build multiple machine learning and a neural classifier. Our methods yielded robust performance for classifying hospitalized patients as positive or negative for OUD, with the best performing feature set and model combination achieving an F1 score of 0.97. These results show promise for the future development of a real-time tool for quickly and accurately identifying patients with OUD in the hospital setting.
Introduction
The misuse of prescription opioids has become a major public health crisis.1 In 2017 alone, 47,600 people in the United States died from opioid overdoses, and a further 1.7 million people had prescription opioid-related substance use disorder.2 To combat this epidemic, the U.S. Department of Health and Human Services has made increasing access to addiction services the first of its five major priorities for fighting the opioid crisis.3 With 19% of patients who recognized a need for substance abuse treatment not receiving help due to a lack of information on where to go,4 it is clear that the current system is not always able to provide patients with the help they need to manage issues related to opioid misuse and abuse in an efficient and timely fashion.
One possible way to increase the number of people who get access to addiction services is to implement a system that helps health care providers know which patients should be referred to addiction services while independently alerting addiction services to potential patients. While hospital encounters provide an important opportunity for clinicians to provide effective interventions to patients, existing hospital screening methods are resource-intensive. Under most current workflows, including the one in place at our institution, a provider wishing to refer a patient must first reach out to addiction services, who must then reach back out to the provider after receiving the patient’s information. This process requires swift communication and coordination between multiple providers, making it easy for patients to inadvertently slip through the cracks. Patients’ detailed clinical and behavioral information, including their substance use, is often documented in clinical notes in the electronic health record (EHR). A new workflow that leverages natural language processing (NLP) techniques and machine learning models to automatically identify potential patients and alert the provider and addiction services simultaneously could have significant potential to facilitate an easier and more efficient referral process. Further, if this process is conducted in an inpatient setting, a patient’s care team can immediately present them with treatment options. In the present study, we used multiple natural language processing and machine learning methods to automatically classify patients as positive or negative for opioid use disorder (OUD) in an inpatient setting based on information contained in free-text notes.
Background
A number of prior studies have focused on the development of automated methods for classifying patients by their opioid use status. Many such studies relied on structured EHR data, such as diagnosis and billing codes, prescription data, procedure history, laboratory values, demographic information, and other coded variables.5-7 For example, a recent study involved the development of an automatic phenotyping system that was successfully able to identify emergency department patients with OUD based on clinician and billing codes, achieving a positive predictive value (PPV) of more than 95%.5 Another recent study used a combination of structured demographic information (e.g., age, sex, marital status) and clinical data (e.g., procedure history, laboratory values, medications) to predict sustained postoperative opioid prescription in patients status post lumbar disc surgery.6
While these and similar studies demonstrate the utility of structured EHR data for determining patients’ opioid use status, it is also the case that a substantial amount of relevant information is stored in free-text clinical notes, such as emergency department visit notes or inpatient progress notes, and is therefore not readily accessible for use in downstream informatics tasks. To address this challenge, multiple past studies have leveraged NLP techniques to identify various types of information related to patients’ opioid use from information contained in free-text notes.8-13 For example, a 2017 study used machine learning methods to identify patients with “aberrant” opioid use behavior based solely on information from free-text outpatient visit notes with more than 80% accuracy.9 In another recent study, the authors developed an NLP tool to identify multiple aspects of opioid-related overdose, such as intentionality and substance(s) involved, based on information documented in free-text with high specificity and good sensitivity.10
Currently, few studies have focused on developing methods for identifying OUD in an inpatient setting based on free-text clinical notes. The potential benefits of determining patients OUD status in this setting are manifold. Reliance on patient disclosure can be ineffective, as many patients are reluctant to disclose substance misuse for fear of judgement or mistreatment. Further, when OUD and other substance use disorders are documented, it is often mentioned in free-text clinical notes (e.g., emergency department visit notes and progress notes), and structured documentation formats such as ICD-9/10 billing codes have demonstrated high false negative rates for similar tasks like identifying opioid overdoses.14,15 To address these challenges, we describe an NLP and machine learning-based approach to identifying hospitalized patients with OUD based on the content of their free-text clinical notes with the goal of facilitating fast and efficient initiation of treatment and referral to ongoing outpatient treatment.
Methods
1. Clinical Setting and Data Collection
This study was conducted at Brigham and Women’s Hospital (BWH), a large academic hospital located in Boston, Massachusetts and a member of Mass General Brigham (MGB; formerly Partners HealthCare). The study’s protocol was reviewed and approved by the MGB Human Research Committee (IRB). We retrieved all emergency department visit notes, inpatient progress notes, and previous hospital discharge summaries (n = 846,302) for 22,626 patients admitted to BWH between July 1, 2017 and July 1, 2018.
2. Methods Overview
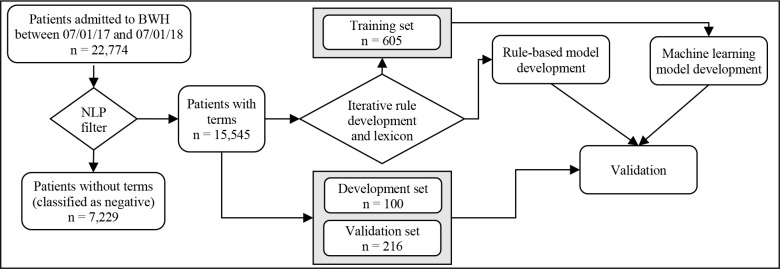
The study design is summarized in Figure 1 and consists of 4 steps. We first adapted a set of guidelines and a list of useful terms provided by an addiction psychiatrist into a rule-based NLP algorithm. We then iteratively improved the NLP algorithm and conducted manual chart review to determine the accuracy of the NLP system. Once the performance of the NLP algorithm was deemed sufficient (which we defined as achieving a precision and recall of at least 0.92), we compiled the manually labeled patients into a training dataset which we used with multiple machine learning algorithms. Finally, we tested both the NLP classification system and multiple machine learning classification algorithms against a randomly selected labeled subset of patients.
Figure 1.
Data flow and study design
3. Training, Development and Validation Sets
We manually labeled 915 randomly selected patients with their OUD status (positive, negative, or unclear) based on chart review. For the purposes of training and testing the models, patients whose OUD status was inconclusive based on the available data were excluded. The initial training set consisted of 599 patients that were manually labeled during the iterative rule development and lexicon refinement during the development of the NLP system (see sections 4.1-4.3). After excluding patients whose OUD status was unclear (n = 16), the final training set contained a total of 583 patients. The initial development set included 99 patients, with a roughly even distribution of positive and negative patients. Because of the rarity of OUD, we oversampled positively labeled patients to help ensure the model could identify different types of positive cases. After excluding 2 inconclusive patients, the final development set contained 97 patients. Finally, we created a validation set of all patients admitted between March 1 and March 14, 2018 (n = 217), excluding 1 inconclusive patient for a final total of 216 patients. Figure 1 shows the final training, development, and validation sets used in this study.
4. Case Identification Methods
We developed an NLP algorithm whose output was used both as the basis for the rule-based classifier’s decision-making process and as features for multiple machine learning classification methods. Notes were pre-processed using MTERMS (Medical Text Extraction, Reasoning and Mapping System), a suite of modular, multipurpose tools designed for use with clinical and biomedical text.16 MTERMS has been used to support a variety of clinical informatics tasks, including extracting medication and allergy information from narrative clinical text, identifying wounds and wound status from free-text notes, classifying smoking status, alcohol use, and other social and behavioral factors, and more.16-22 MTERMS has functionality to identify certain relevant contextual information, such as negation, and can distinguish between a patient’s personal and family history.23 The development of the NLP algorithm involved 3 steps: 1) logic development, 2) lexicon development, and 3) logic refinement.
4.1. Rule-based Classification
4.1.1. Logic Development
We developed a module within MTERMS specifically designed to identify patients with OUD. To develop the NLP algorithm, we adopted an empirical study design due to the rarity of patients with OUD, which has an estimated prevalence of 0.62-0.77%.24 To develop the NLP algorithm, we received an initial set of guidelines from a board-certified addiction psychiatrist (JS) at BWH for reviewing patients for potential opioid misuse, as well as a list of manually identified terms and phrases related to OUD. We then adapted these guidelines into a set of rules (Table 1) to capture the information needed in the clinical notes to automatically classify a patient’s OUD status.
Table 1.
Manually developed rules for classifying patients as positive for opioid use disorder
| Rule description | |
| 1 | Positive fentanyl urine test |
| 2 | At least 3 health issues related to intravenous drug use |
| 3a | Documented history of heroin use |
| 3b | Documented history of treatment for heroin addiction |
| 4a | Documented history of opioid misuse |
| 4b | Suspected history of opioid misuse |
| 5a | Opioid addiction without current treatment plan |
| 5b | Opioid addiction currently being treated |
| 6a | History of intravenous drug misuse with failed treatment attempts |
| 6b | Intravenous drug misuse being managed with treatment |
| 7a | Polysubstance misuse including opioids |
| 7b | Polysubstance misuse without mention of opioids |
| 8a | Nonstandard intake methods of opioids |
| 8b | Suspicion of nonstandard intake methods of opioids |
| 9 | History of medication-assisted treatment or other opioid addiction treatment |
| 10 | Patient administered Narcan prior to intake |
| 11a | Documented opioid seeking behavior |
| 11b | Possible or nonspecific drug seeking behavior |
4.1.2. Lexicon Development
In parallel to the rule development, we used word embeddings to expand the lexicon of key terms and phrases. Using the fastText library,25 we trained word embeddings on approximately 10.5 GB of clinical notes at MGB. We then compared the vector representations of the manually curated lexicon to other words in the embedding model and ranked them by similarity. We manually selected relevant terms from the ranked list to be added to the lexicon, including synonyms and misspellings of words already in the lexicon. This process was repeated iteratively until no new useful terms were identified. A subset of the lexicon is shown in Table 2.
Table 2.
Terms used by the NLP algorithm, including common synonyms and misspellings, grouped by category
| Term category | Terms |
| Drug (non-opioid) | Amphetamines; antidepressants; anxiolytics; benzo; benzodiazepine; benzos; cocaine; cocainse; ecstasy; ectasy; hallucinogens; hypnotic; illicits; recreational drug; sedative; sedative, hypnotic or anxiolytic abuse; sedatives |
| Drug misuse | Acute drug intoxication, acute intoxication, addict, addiction, alcohol-induce, amphetamines abuse, drug abuse, drug overdose, drug user, intravenous drug user, ivd, ivda, ivdu, long-term drug misuser, multi-drug misuser, overdose, poly-drug abuser, poly- drug misuser, polysub, poly-substance, polysubstance abuse, polysubstance drug abuse, polysubstance use, prescription drug abuse, psud, sedative abuse, seeking behavior, stoned, substance abuse, using substances, withdrawal |
| Heroin | Heroin, heroin addiction, heroin dependence, heroine, intravenous heroin, iv heroin |
| Narcotic | Narcotic, narcotic withdrawal, narcotics narcotism |
| Opioid | Avinza, codeine, dilaudid, fenanyl, fentanyl, fentantyl, fentayl, hydrocodone, morphine, opana, opiate, opiates, opioid, opioids, oxycontin, oxycodone, oxycone, oxycoodone, oxymorphone, percocet, roxycodone, sufentanyl, vicodin |
| Opioid misuse | Opiate addiction, opiate overdose, opiate use disorder, opiate withdrawal, opiod abuse, opiod addiction, opiod dependence, opiod overdose, opiod use disorder, opiod withdrawal, opioid abuse, opioid addiction, opioid dependence, opioid overdose, opioid use disorder, opioid withdrawal, oud |
| Opioid treatment | Addiction psych, addiction psychiatry, addiction services, addiction treatment, buprenorphine, mat, methadone, methadone clinic, methadone therapy, naltrexone, narcan, opiate maintenance, opioid maintenance, suboxone, suboxone clinic, suboxone therapy, vivitrol |
| Related illness | Abscess, bacteremia, cellulitis, cva, discitis, epidural abscess, hcv, hiv, osteomyelitis, septic emboli, spinal abscess, stroke |
4.1.3. Rule Logic Refinement
From there, we iteratively improved upon the rules and the lexicon, with regular manual chart reviews (JS, BM) to evaluate the OUD status of a random subset of the patients labeled by the algorithm. We then modified the rules and expanded the lexicon according to the feedback provided after the manual reviews. Once the NLP algorithm’s performance was deemed sufficient, we compiled the patients who were manually evaluated during this process into a training set for developing machine learning models. We then manually evaluated a final subset of patients labeled by the completed NLP algorithm to serve as a balanced development set for tuning the machine learning models.
The NLP algorithm is described in the following. Each note was first divided into sections using MTERMS’s sectioning module, which uses a combination of formatting features (e.g., multiple consecutive spaces or newline characters) and section header labels (e.g., “History of Present Illness”, “Discharge Instructions”) to identify different sections within a note, then further divided into tokenized sentences. All relevant terms were then extracted from the note, with MTERMS’s negation and family history modules applied to rule out irrelevant terms or negative test results. Any sections that did not contain a relevant term were automatically classified as negative and excluded from further analysis. All sentences in the remaining sections were then checked against the 11 manually developed rules to determine the patient’s opioid use status. Finally, note-level results were consolidated into patient-level classifications. All patients were assumed to be negative for opioid use disorder by default. Patients were labeled as positive if they had enough signs of opioid misuse that addiction services would want to consult the patient.
In addition to the rules, the presence of certain terms, such as those indicating current or prior treatment or signs of relapse, was used to determine if a patient was sober (and if so for how long) or relapsing on past sobriety. The rules were designed to distinguish between someone who is taking maintenance drugs (e.g., buprenorphine, methadone) but is not using illicit any opioids versus someone who is abstinent from all opioids, as well as whether or not the patient has shown signs of impending relapse, such as cravings. Unlike negated terms or terms associated with a patient’s family history, sobriety is determined based on all instances of a term rather than on a term-by-term basis and was not limited to a binary sober/not sober classification. This is because a patient’s sobriety status can change over time, and sobriety information is often not included with each mention of a term in a clinical note. For this project, we defined sobriety as a period of one year of no opioid use, both to follow the guidelines of the Diagnostic and Statistical Manual of Mental Disorders, 5th Edition (DSM-V) and because a patient who has not misused opioids in the last year and is not demonstrating signs of impending relapse is likely not currently in need of intervention or treatment.
Most rules did not individually assign a binary positive or negative label, but instead accumulated a score which was evaluated after all rules were processed. Each rule’s score is primarily a measure of its confidence in its classification but is also subject to rule-specific criteria, such as the combination of terms found or the length and extent of the patient’s sobriety. While developing the rule system, we found that the triggering of certain combinations of rules tended to correspond to a higher chance of opioid use issues than would be suggested by each of these rules individually. A weight of +1 was therefore applied to certain combinations of rules in order to label patients more accurately. Once all rules were evaluated, each rule’s score was combined to produce a singular positive or negative classification.
The following is an example of the application of the rule-based NLP algorithm for a hypothetical patient whose notes indicate 1) a positive fentanyl urine toxicology at some point during hospitalization and 2) a noted history of opioid use for which the patient is already being treated. For this patient, rule 1 will detect the positive urine toxicology test via the cooccurrence of indicative terms (e.g., “positive”, “utox”, and “fentanyl”) and flag the patient as a potential positive. Rules 2-5a do not apply to this patient and thus are not triggered. Rule 5b is triggered by the presence of terms indicative of opioid abuse (e.g., “OUD”, “withdrawal”) in conjunction with terms indicating treatment (e.g., “on suboxone”, “MAT”) also flagging the patient as a potential positive. Alone, each of these rules would be insufficient to assign a positive label. However, the presence of a positive fentanyl toxicology screening on top of treatment for known OUD suggests the treatment may be ineffective, indicating that this patient should be assigned a positive label.
4.2. Machine Learning-based Classification
We used Scikit-learn to implement support vector machines (SVM), logistic regression (LR), k-nearest neighbors (KNN) and random forest (RF) classifiers,26 using the default parameters for each model. The training set consisted of the 605 manually classified patient set created while building our NLP system. The same gold standard development and validation sets were also used to ensure the results were comparable to those of the NLP system. Patients who could not be classified into the binary positive or negative labels (e.g., because chart review proved inconclusive) were excluded from the data set. Each model was trained on multiple feature sets, including both features extracted by the NLP system, such as terms and rule-level results, and features independent of the NLP system, such as the full text of each of a patient’s notes in bag-of-words form. We used MTERMS to extract terms from the manually developed lexicon from each patient’s notes, the full text of the sentence in which the term was found, and the rule-level labels used by the NLP system. We additionally used a bag-of-words representation of the full text of a patient’s notes as features. Using both features extracted by the NLP system and features independent of the NLP system also allowed us to evaluate how successful the NLP system was at capturing useful information from clinical notes. By using the individual rule results as features, we were additionally able to rank the relative efficacy of each rule in the NLP system by examining the weights given by the machine learning models to each rule result.
We further used the NCRF++ toolkit for neural sequence labeling to develop a deep learning model for classification.27 We used the default model architecture, with a character level CNN layer feeding into a word level LSTM layer for input, as this structure has proven successful for neural NLP. Starting with a character-level CNN allowed the model to learn meaningful information from character combinations inside of full words, such as the root of a verb or a negation prefix like “un-”. It also allowed the model to be more robust when faced with unknown words, such as misspellings or less common medical terminology, since even if a particular word was previously unseen by the model, substrings of alphanumeric characters within the word will almost certainly have been seen previously. The model was trained using each patient’s clinical notes pooled chronologically, with the same training and validation data sets used for the other machine learning models.
5. Evaluation
Final evaluation was conducted using a held-out validation set of 216 patients admitted during the randomly selected two-week period of March 1, 2018 and March 14, 2018. To create the validation set, an NLP filter was applied to all patients admitted within this date range to exclude any patients whose notes contained no mentions of opioids or opioid-related diagnoses and were thus assumed to be negative for OUD (n = 7,229). Among the remaining 15,545 patients, 217 patients, that were not already in the training and development sets, were randomly selected for manual chart review. Of these, 13 were labeled as positive for OUD, 203 were labeled as negative for OUD, and 1 could not be conclusively labeled and was thus excluded, resulting in a total of 216 patients.
All models were evaluated in terms of precision (or positive predictive value [PPV]), recall (or sensitivity), and F1 score. Precision is defined as the number of cases correctly identified by the model as positive out of all the cases it classified as positive (Equation 1). Recall is defined as the number of cases correctly identified by the model as positive out of all known positive cases (Equation 2). The F1 score is the harmonic mean of precision and recall (Equation 3).

For the rule-based and statistical machine learning classifiers, overall accuracy (Equation 4) was also calculated.
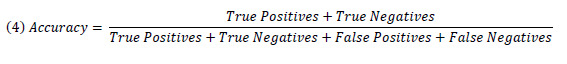
Results
1. Patient Cohort Description
The final training, development, and validation data sets had a roughly 65/10/25 split and are described in Table 3. Across all three datasets, patients were primarily white (69.8% overall), male (52.8%), and had one or more opioids on their medication list (76.1%) but did not have any opioid-related diagnoses on their problem list (92.5%). Unsurprisingly, the number of patients who had previously been prescribed opioids or received an opioid-related diagnosis was higher among patients classified as positive for OUD during manual review across all data sets.
Table 3.
Demographic information by data set and label for the subset of manually reviewed patients
| Characteristic | Training set | Development set | Validation set | Total | ||||||||
| Pos | Neg | Total | Pos | Neg | Total | Pos | Neg | Total | Pos | Neg | Total | |
| Patients | 153 | 430 | 583 | 46 | 51 | 97 | 13 | 203 | 216 | 212 | 684 | 896 |
| Age, mean (SD), yearsa | 46.6 (13.6) |
51.4 (23.1) |
50.1 (21.1) |
46.2 (12.0) |
50.6 (23.8) |
48.5 (19.2) |
39.2 (22.4) |
59.0 (21.1) |
57.7 (21.6) |
46.1 (14.0) |
53.5 (22.8) |
51.8 (21.3) |
| Sex, % | ||||||||||||
| Female | 37.9 | 50.2 | 47.0 | 37.0 | 54.9 | 46.4 | 23.1 | 51.3 | 49.5 | 36.8 | 50.9 | 47.5 |
| Male | 62.1 | 49.8 | 53.0 | 63.0 | 45.1 | 53.6 | 76.9 | 48.7 | 50.5 | 63.2 | 49.1 | 52.5 |
| Race, % | ||||||||||||
| White | 68.0 | 67.9 | 67.9 | 71.7 | 70.6 | 71.1 | 69.2 | 76.1 | 75.7 | 68.9 | 70.5 | 70.1 |
| Black | 17.6 | 10.2 | 12.2 | 15.2 | 7.8 | 11.3 | 0.0 | 10.7 | 10.0 | 16.0 | 10.2 | 11.6 |
| Asian | 1.3 | 3.0 | 2.6 | 0.0 | 0.0 | 0.0 | 7.7 | 3.6 | 3.8 | 1.4 | 2.9 | 2.6 |
| Others | 9.8 | 9.1 | 9.3 | 13.0 | 11.8 | 12.4 | 7.7 | 5.1 | 5.2 | 10.4 | 8.1 | 8.7 |
| Unknown | 3.3 | 9.8 | 8.1 | 0.0 | 9.8 | 5.2 | 15.4 | 4.6 | 5.2 | 3.3 | 8.3 | 7.1 |
| Ethnicity, % | ||||||||||||
| Non-Hispanic | 85.6 | 79.3 | 81.0 | 82.6 | 76.5 | 79.4 | 69.2 | 85.8 | 84.8 | 84.0 | 81.0 | 81.7 |
| Hispanic | 10.5 | 10.0 | 10.1 | 15.2 | 13.7 | 14.4 | 0.0 | 5.6 | 5.2 | 10.8 | 9.0 | 9.4 |
| Unknown | 3.9 | 10.7 | 8.9 | 2.2 | 9.8 | 6.2 | 30.8 | 8.6 | 10.0 | 5.2 | 10.0 | 8.9 |
| Opioid(s) on medication list, % | 71.2 | 66.6 | 67.8 | 64.4 | 60.8 | 62.5 | 53.8 | 70.1 | 69.0 | 68.7 | 67.2 | 67.5 |
| Opioid-related diagnosis on problem listb, % | 17.0 | 3.2 | 6.8 | 22.2 | 0.0 | 10.4 | 15.4 | 0.5 | 1.4 | 18.0 | 2.2 | 6.0 |
Age was calculated as the patient’s date of birth subtracted from the date of data collection (07/30/2018)
Includes: Opioid dependence with intoxication delirium (F11.221; ICD-10-CM); Opioid dependence with withdrawal (F11.23); Opioid dependence with opioid-induced mood disorder (F11.24); Opioid dependence with opioid-induced psychotic disorder with delusions (F11.250); Opioid dependence with other opioid-induced disorder (F11.288); Poisoning by heroin, accidental (unintentional), initial encounter (T40.1X1A); Poisoning by heroin, intentional self-harm, initial encounter (T40.1X2A); Poisoning by other opioids, accidental (unintentional), initial encounter (T40.2X1A); Adverse effect of other opioids, initial encounter (T40.2X5A); Underdosing of other opioids, initial encounter (T40.2X6A); Poisoning by analeptics and opioid receptor antagonists accidental (unintentional), initial encounter (T50.7X1A); Opioid Abuse And Dependence (All Patients Refined Diagnosis Related Groups [APR DRG] v30)
2. Model Performance
The performance of the different classification methods is summarized in Table 4. Hand-crafted rules, traditional machine learning, and deep neural networks all achieved high performance on the development set, with F1 scores ranging from 0.89 to 0.96. Performance on the validation set was more variable, with statistical machine learning models consistently outperforming both the rule-based and DNN models. The rule-based algorithm was comparable to the statistical machine learning models in terms of precision but achieved a notably poorer recall, indicating a higher rate of false negative classifications. On the other hand, the DNN model had somewhat higher recall than precision, although both measures were low compared to other methods.
Table 4.
Results by classification method and data set
| Rules | Machine learning models | DNN | |||||
| LR | SVM | KNN | RFC | ||||
| Development set | Precision | 0.9400 | 0.9601 | 0.9601 | 0.9033 | 0.9388 | 0.9167 |
| Recall | 0.9420 | 0.9592 | 0.9592 | 0.8980 | 0.9388 | 0.8684 | |
| F1 score | 0.9399 | 0.9592 | 0.9592 | 0.8972 | 0.9388 | 0.8919 | |
| Accuracy | 0.9495 | 0.9592 | 0.9592 | 0.8980 | 0.9388 | ||
| Validation set | Precision | 0.9324 | 0.9654 | 0.9730 | 0.9730 | 0.9730 | 0.5000 |
| Recall | 0.8052 | 0.9676 | 0.9722 | 0.9722 | 0.9722 | 0.6667 | |
| F1 score | 0.8563 | 0.9639 | 0.9683 | 0.9683 | 0.9683 | 0.5714 | |
| Accuracy | 0.9722 | 0.9676 | 0.9676 | 0.9676 | 0.9722 | ||
The most and least useful NLP-based features according to the machine learning models are listed in Tables 5 and 6. Ranks according to individual models were averaged together to determine the overall ranks used in Tables 5 and 6. Unsurprisingly, inconclusive NLP results generally received low weights, especially when they involved more complex rules, such as the presence of multiple diagnoses related to intravenous drug use, which received the lowest weight across all machine learning models. On the other hand, definitive results for rules specifically related to OUD were typically considered more important, such as a documented history of heroin use or opioid misuse, which were ranked most important and second most important, respectively.
Table 5.
The 5 most important NLP features by weights assigned during machine learning (where 1 is most important)
| Rank | Rule | Rule result via NLP |
| 1 | 3. Documented history of heroin use and/or addiction | Positive |
| 2 | 4. Documented positive or suspected history of opioid misuse | Positive |
| 3 | 8. Documented positive or suspected nonstandard intake methods of opioids | Positive |
| 4 | 7. Documented polysubstance misuse | Undetermined |
| 5 | 7. Documented polysubstance misuse | Positive |
Table 6.
The 5 least important features by weights assigned during machine learning (where 1 is least important)
| Rank | Rule | Rule result via NLP |
| 1 | 2. At least 3 health issues related to intravenous drug use | Undetermined |
| 2 | 5. Opioid addiction with or without current treatment plan | Undetermined |
| 3 | 8. Documented or suspected nonstandard intake method of opioids | Undetermined |
| 4 | 9. History of medication-assisted or other opioid addiction treatment | Undetermined |
| 5 | 11. Documented opioid or other drug seeking behavior | Undetermined |
Discussion
We developed a set of expert-created guidelines for identifying patients with opioid use disorder based on information from free-text clinical documents including emergency department visit notes, inpatient progress notes, and past hospital discharge summaries. Using MTERMS, we implemented an NLP algorithm to automatically identify the information captured by these guidelines for subsequent use in multiple types of classifiers, including a rule-based classifier, machine learning models and a deep neural network (DNN) using the NLP output as features. Our results suggest an NLP algorithm based on a physician-constructed ruleset can be used to accurately identify problematic opioid use based on information from free-text alone, especially when used as features for machine learning models.
While the rule-based model had strong results, using NLP-generated features for machine learning classification models proved more robust to sparse data, with comparable results on the development set, which had a relatively even representation of positive patients, and much stronger results on the validation set, which had a much lower proportion of positive patients. As the training set is more similar to the development set than the validation set in terms of the split between positive and negative patients, the stronger results on the validation set suggest the machine learning models are better at adapting to variations in dataset composition, which is particularly important when considering the viability of implementing the model within a hospital’s EHR system, where the features of the current subset of patients with problematic opioid use can vary over time based on unpredictable real world factors. However, due to this sparseness, evaluating the different machine learning models relative to each other is made more difficult, as the models shared most errors.
The use of the information captured by the NLP algorithm proved to be the performant set of features across all machine learning-based models, with the inclusion of all other tested features leading to worse performance. This also held true for the DNN model which, despite its comparatively low performance overall, achieved the best classification results when only the NLP-based features were used. However, since the data sets for machine learning were relatively small, the performance on other larger feature sets could be a result of a lack of similar training data to learn from, especially for feature sets using text from the clinical notes. Similarly, we suspect the DNN did not perform as well as the other models due to its greater dependency on large data sets for generating reliable results. The rule-based model performed best for cases where OUD or related risk factors were explicitly stated in a note’s text. Most incorrect classifications involved patients whose notes contained statements that were either ambiguous or inclusive statements (e.g., “may also be methadone withdrawal, though he is young for this”). Similarly, patients with multiple pieces of relevant information scattered throughout their notes were more likely to be misclassified. Because the rule-based and machine learning models both relied on the same NLP output, many cases were missed by all model types, suggesting a need for further development of the NLP algorithm to better handle ambiguous or uncertain information.
Although our positive label does not inherently represent an opioid-related diagnosis, an extreme number of patients that were manually determined to be of-interest to addiction services lacked any opioid-related diagnosis, including patients with problematic heroin usage or noted opioid misuse. This demonstrates the potential of our NLP surveillance system to help doctors better connect patients in need to addiction services. Patients with OUD, and substance use disorder (SUD) more broadly, are under-recognized in hospitals for a host of reasons. Among these are the stigma associated with SUD, making patients reluctant to disclose or discuss drug usage, as well as patients’ fears that they will be treated poorly if their SUD diagnosis was known, for example by having pain medication withheld due to concerns about “drug-seeking” behavior. Additionally, many providers also feel uncomfortable discussing SUD and, in some cases, lack sufficient training in how to ask patients questions related to SUD in the first place. In a majority of hospitals, the availability of SUD treatment in the hospital setting is limited. However, many hospitalized patients with SUD are willing to consider treatment when approached in the hospital, and these are critical opportunities to both initiate treatment and link patients to ongoing treatment as outpatients.
Limitations
Our models were trained and tested on data from a single institution and rely partially on hard-coded NLP rules. However, while the specific language used may vary across different institutions, the underlying rules are based on clinical guideline standards and are thus portable. To facilitate this, possible future work should aim to formalize and define the mapping from these clinical standards to NLP-based rules. Although the lexicon and rules must be manually updated as new terms or criteria are identified, a combined rule-based and machine learning approach can lead to improved performance. Deep learning proved infeasible due to the large quantity of patients that would need to be labeled given the rarity of OUD, but given sufficient resources, deep learning may be able to yield better performance.
In the present study, patients who could not be conclusively classified as either positive or negative during manual review were excluded from the training data set. In reality, however, such patients would still likely benefit from screening by a clinician, and the ability to accurately identify these patients for potential interventions is critical. Additionally, while we overall had a large set of patient notes, the effort required for manual chart reviews limited the number of patients in our manually evaluated data sets for machine learning. The validation set was created from a random sample; while we ensured a representative sample size for the evaluation by manually reviewing over 200 cases, the low prevalence of OUD among the population led to a relatively low number of positive cases.
Future Directions
A critical next step will be to evaluate our method on a larger data set better determine its accuracy and to assess its scalability. A larger sample size can provide a more generalizable representation of the patient population and will cover OUD patients with different presentations and levels of OUD severity, which can help facilitate more accurate model development. This will require further manual chart review to generate a sufficiently large set of labeled patients. However, recent similar work leveraging semi-supervised and reinforcement learning suggests such techniques can reduce the amount of manual required without sacrificing performance.28,29 From there, we aim to integrate our algorithm within our institution’s EHR system to automatically screen patients for potential OUD in real- or near real-time. In the event of a positive classification, we will implement system to automatically alert a specialist within addiction treatment services of that patient’s need and facilitate communication between addiction services and the patient’s care team. We are currently working with clinical and technical leadership at our institution to initiate this process.
Conclusion
Automatically identifying hospitalized patients with opioid use disorder based on information documented in free-text clinical notes has great potential to increase timely access to addiction treatment services for patients in need. Our results demonstrate the viability of leveraging NLP-generated features to train a machine learning model for automatically classifying patients by OUD status. Future work will focus on integrating our algorithm with the EHR for real-time patient screening.
Acknowledgement:
This work was supported by National Institutes of Health [grant number K23DA042326 (JS)]. We thank Sharmitha Yerneni, BS, for assisting with data collection and Tom Korach, MD, for assisting with lexicon expansion.
Figures & Table
References
- 1.Rose ME. Are Prescription Opioids Driving the Opioid Crisis? Assumptions vs Facts. Pain Med. 2018;19(4):793–807. doi: 10.1093/pm/pnx048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.National Institute on Drug Abuse (NIDA) https://www.drugabuse.gov/ . Accessed Oct. 24, 2019.
- 3.NIDA. Opioid Overdose Crisis https://www.drugabuse.gov/drugs-abuse/opioids/opioid-overdose-crisis . Published 2020. Accessed March 6, 2020.
- 4.2017 NSDUH Annual National Report https://www.samhsa.gov/data/report/2017-nsduh-annual-national- report . Accessed Oct. 24, 2019.
- 5.Chartash D, Paek H, Dziura JD, et al. Identifying Opioid Use Disorder in the Emergency Department: Multi-System Electronic Health Record-Based Computable Phenotype Derivation and Validation Study. JMIR medical informatics. 2019;7(4):e15794. doi: 10.2196/15794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karhade AV, Ogink PT, Thio Q, et al. Development of machine learning algorithms for prediction of prolonged opioid prescription after surgery for lumbar disc herniation. The spine journal : official journal of the North American Spine Society. 2019;19(11):1764–1771. doi: 10.1016/j.spinee.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Thompson HM, Hill K, Jadhav R, Webb TA, Pollack M, Karnik N. The Substance Use Intervention Team: A Preliminary Analysis of a Population-Level Strategy to Address the Opioid Crisis at an Academic Health Center. J Addict Med. 2019;13(6):460–463. doi: 10.1097/ADM.0000000000000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dligach D, Afshar M, Miller T. Toward a clinical text encoder: pretraining for clinical natural language processing with applications to substance misuse. J Am Med Inform Assoc. 2019;26(11):1272–1278. doi: 10.1093/jamia/ocz072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lingeman JM, Wang P, Becker W, Yu H. Detecting opioid-related aberrant behavior using natural language processing. AMIA Annual Symposium Proceedings. 2017. Paper presented at. [PMC free article] [PubMed]
- 10.Hazlehurst B, Green CA, Perrin NA, et al. Using natural language processing of clinical text to enhance identification of opioid-related overdoses in electronic health records data. Pharmacoepidemiology and drug safety. 2019;28(8):1143–1151. doi: 10.1002/pds.4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrell DS, Cronkite D, Palmer RE, et al. Using natural language processing to identify problem usage of prescription opioids. International journal of medical informatics. 2015;84(12):1057–1064. doi: 10.1016/j.ijmedinf.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Afshar M, Joyce C, Dligach D, et al. Subtypes in patients with opioid misuse: A prognostic enrichment strategy using electronic health record data in hospitalized patients. PloS one. 2019;14(7):e0219717. doi: 10.1371/journal.pone.0219717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Chen ES, Pakhomov S, et al. Automated Extraction of Substance Use Information from Clinical Texts. AMIA Annual Symposium proceedings AMIA Symposium. 2015;2015:2121–2130. [PMC free article] [PubMed] [Google Scholar]
- 14.Reardon JM, Harmon KJ, Schult GC, Staton CA, Waller AE. Use of diagnosis codes for detection of clinically significant opioid poisoning in the emergency department: A retrospective analysis of a surveillance case definition. BMC Emerg Med. 2016;16:11–11. doi: 10.1186/s12873-016-0075-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rowe C, Vittinghoff E, Santos GM, Behar E, Turner C, Coffin PO. Performance Measures of Diagnostic Codes for Detecting Opioid Overdose in the Emergency Department. Academic emergency medicine : official journal of the Society for Academic Emergency Medicine. 2017;24(4):475–483. doi: 10.1111/acem.13121. [DOI] [PubMed] [Google Scholar]
- 16.Zhou L, Plasek JM, Mahoney LM, et al. Using Medical Text Extraction, Reasoning and Mapping System (MTERMS) to process medication information in outpatient clinical notes. AMIA Annual Symposium proceedings AMIA Symposium. 2011;2011:1639–1648. [PMC free article] [PubMed] [Google Scholar]
- 17.Goss FR, Plasek JM, Lau JJ, Seger DL, Chang FY, Zhou L. An evaluation of a natural language processing tool for identifying and encoding allergy information in emergency department clinical notes. AMIA Annual Symposium proceedings AMIA Symposium. 2014;2014:580–588. [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou L, Baughman AW, Lei VJ, et al. Identifying Patients with Depression Using Free-Text Clinical Documents. Studies in health technology and informatics. 2015;216:629–633. [PubMed] [Google Scholar]
- 19.Topaz M, Radhakrishnan K, Blackley S, Lei V, Lai K, Zhou L. Studying Associations Between Heart Failure Self-Management and Rehospitalizations Using Natural Language Processing. Western journal of nursing research. 2016. [DOI] [PubMed]
- 20.Topaz M, Lai K, Dowding D, et al. Automated identification of wound information in clinical notes of patients with heart diseases: Developing and validating a natural language processing application. International journal of nursing studies. 2016;64:25–31. doi: 10.1016/j.ijnurstu.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 21.Navathe AS, Zhong F, Lei VJ, et al. Hospital Readmission and Social Risk Factors Identified from Physician Notes. Health services research. 2017. [DOI] [PMC free article] [PubMed]
- 22.Lin KJ, Singer DE, Glynn RJ, et al. Prediction Score for Anticoagulation Control Quality Among Older Adults. Journal of the American Heart Association. 2017;6(10) doi: 10.1161/JAHA.117.006814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou L, Lu Y, Vitale CJ, et al. Representation of information about family relatives as structured data in electronic health records. Applied clinical informatics. 2014;5(2):349–367. doi: 10.4338/ACI-2013-10-RA-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallow PJ, Sathe N, Topmiller M, et al. Estimating the Prevalence of Opioid use Disorder in the Cincinnati Region using Probabilistic Multiplier Methods and Model Averaging. 2019;6(2):61–69. doi: 10.36469/9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bojanowski P, Grave E, Joulin A. Mikolov TJTotAfCL. Enriching word vectors with subword information. 2017;5:135–146. [Google Scholar]
- 26.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-Learn. Machine learning in Python. 2011;12(Oct):2825–2830. [Google Scholar]
- 27.Yang J, Zhang YJapa. NCRF++: An open-source neural sequence labeling toolkit. 2018.
- 28.Halpern Y, Choi Y, Horng S, Sontag D. Using Anchors to Estimate Clinical State without Labeled Data. AMIA Annual Symposium proceedings AMIA Symposium. 2014;2014:606–615. [PMC free article] [PubMed] [Google Scholar]
- 29.Halpern Y, Horng S, Choi Y, Sontag D. Electronic medical record phenotyping using the anchor and learn framework. Journal of the American Medical Informatics Association. 2016;23(4):731–740. doi: 10.1093/jamia/ocw011. [DOI] [PMC free article] [PubMed] [Google Scholar]