Abstract
A bleeding event is a common adverse drug reaction amongst patients on anticoagulation and factors critically into a clinician’s decision to prescribe or continue anticoagulation for atrial fibrillation. However, bleeding events are not uniformly captured in the administrative data of electronic health records (EHR). As manual review is prohibitively expensive, we investigate the effectiveness of various natural language processing (NLP) methods for automatic extraction of bleeding events. Using our expert-annotated 1,079 de-identified EHR notes, we evaluated state-of-the-art NLP models such as biLSTM-CRF with language modeling, and different BERT variants for six entity types. On our dataset, the biLSTM-CRF surpassed other models resulting in a macro F1-score of 0.75 whereas the performance difference is negligible for sentence and document-level predictions with the best macro F1-scores of 0.84 and 0.96, respectively. Our error analyses suggest that the models’ incorrect predictions can be attributed to variability in entity spans, memorization, and missing negation signals.
Introduction
In the US, atrial fibrillation (AF) affects 5.2 million mostly older Americans, with 12 million projected by 2050, commensurate with an increasing number of Americans living longer despite being affected by AF risk factors1. Nearly 10% suffer a stroke within 5 years of an AF diagnosis2. AF therapy has also become more complex with the introduction of target specific oral anticoagulants (ACs)3. ACs help to reduce the risks of strokes but can have significant adverse effects, including major bleeding complications. Weighing stroke risk against the risk of bleeding from ACs is central to AF management for the current ~2.8 million Americans with AF who are eligible for AC therapy4,5. Available stroke and bleeding risk calculators were not developed to counter these challenges simultaneously4,6 and it remains challenging to advise older AF patients about AC since they are frequently at high risk for stroke and complications from AC, particularly bleeding7–9.
In clinical text, a bleeding event can be identified by a phrase or word (e.g., hemorrhage, maroon stools) indicative of the escape of blood from the circulatory system. Bleeding events factor critically into a clinician’s decision to prescribe or continue AC for AF10. However, they are not captured well in the structured data of electronic health records (EHR), many are documented in the unstructured EHR notes11. In addition, although there are bleeding-related ICD codes (9 and 10) and prediction scores, they lack details about severity (severe or mild), cause-related information (e.g., trauma, wrong dose, fall), and other characteristics (e.g., site – intracranial or gastrointestinal) that influence the decision to prescribe AC and choice of AC. Therefore, for patients with AF, we are evaluating different natural language processing (NLP) approaches on EHR notes to detect bleeding events and their complex attributes including anatomic site, alternative causes, medications, lab evaluations and severity. The new risk factors related to bleeding events extracted by such approaches would be helpful in improving the existing risk and benefit evaluation techniques for AC.
Robust NLP approaches can be used to extract bleeding related information such as bleeding events, their anatomic sites, bleeding lab evaluations mentioned in the EHR notes and suspected alternative causes for bleeding. This extracted information can help the clinicians in making decisions while prescribing ACs to different patients. Despite various works on entity recognition for clinical texts12–14, the focus has been mostly for adverse drug events, heart disease, smoking, etc. To the authors’ knowledge, there has been no previous work regarding bleeding named entity recognition (NER) from patient EHR notes. Previously, we developed a binary classifier which, given a sentence from EHR, can predict whether it contains any bleeding event or not 15. However, the study does not detect the bleeding named entity, its site, medication etc. Hence, we decided to annotate EHR notes for different patients to mark the aforementioned information related to bleeding. In this paper, we explored multiple state-of-the-art end-to-end deep learning models for automated bleeding related entity detection from clinical text. We evaluated the models and compared their performance both qualitatively and quantitatively. We also provide a detailed error analysis to understand the instances where different models fail to extract the information.
In summary, our contributions are threefold: (1) To our knowledge, this is the first work that explores various NLP approaches on the extraction of bleeding-related entities. (2) We compared three state-of-the-art machine learning architectures and reported thorough analyses. (3) Our detailed error analyses over different NLP techniques provide insights about factors critical to the development of efficient NLP models for any further research work in this area.
Materials and Methods Models
In this section, we will briefly explain the models used for this work. In particular, we chose three representative models based on their unique architectural differences and effectiveness for NER. These include the popular statistical modeling method for sequence labelling - CRF, a variation of widely used biLSTM-CRF architecture – LM-LST-CRF and transformer-based architectures such as BERT, BioBERT etc.
CRF
CRF (conditional random field)16 is a widely-used probabilistic graphical model for sequence labelling tasks such as NER, parts of speech tagging, chunking etc.17–19 So, we chose CRF as our baseline model. In CRF, each feature function, fj, takes a sentence s, the position i of a word in that sentence, the label li of the current word and the label li−1 of the previous word as inputs. The final probability distribution for the tokens over the labels in CRF is given by:
| (1) |
Here λj is the weight assigned to each feature function fj, n is the number of words in the current sentence and m is the number of features considered. The above-mentioned variant is a special case of linear-chain CRF.
LM-LSTM-CRF
Previous work suggests that bidirectional LSTM (biLSTM) outperforms CRF in the clinical domain12. Hence, we decided to use a powerful biLSTM architecture, namely LM-LSTM-CRF20, which incorporates language modeling (LM) with biLSTM layers and adds a CRF layer on top. The system includes highway units after the character level LSTM layers, analogous to the fully connected (fc) layers after the convolutional layers in computer vision tasks. It follows a multi-task strategy where both the language and sequence labeling models are trained simultaneously, and predictions are made at the word level. The sequence labeling model has the following loss function:
| (2) |
where, Zi = (zi,1,zi,2,...,zi,n) is the output of the word level LSTM for training instance (xi,ci,yi). Here xi is the i-th word, ci the list of all the characters of that word and yi the label for the word. On the other hand, the loss function for the language model is:
| (3) |
This is a reversed-order language model, where pf is the generation probability from left to right and pr is the generation probability from right to left. Combining equations 1 and 2, the final joint loss function becomes,
| (4) |
BERT
BERT21 (Bidirectional Encoder Representations from Transformers) is a pre-trained language model which learns using semi-supervised objectives from large text corpus. Since its inception, BERT-based models have been shown to outperform earlier state-of-the-art models in multiple NLP tasks, including many clinical NLP applications22. Given any input token, BERT can provide an unsupervised and rich contextual representation. Leveraging this power of BERT, it is possible to add an additional linear classification layer on top and fine-tune the whole system for any downstream NLP task. We used three variants of BERT, vanilla BERT - using BERT-base released by the authors21, BioBERT23 - fine-tuned on biomedical corpora, and Bio+Clinical BERT24 - BioBERT fine-tuned on clinical data. Additionally, we used RoBERTa25, which has the same model architecture with differences in the pre-training stage.
Dataset
With the approval from the Institutional Review Board (IRB) at the University of Massachusetts Medical School (UMMS) and a Memorandum of Understanding (MOU) between the UMMS and Northwestern University , we annotated 1,079 de-identified EHR notes for six entity types with the help of 5 expert annotators supervised by 2 senior physicians. These notes are de-identified discharge summaries from patients who received care at hospitals affiliated with Northwestern University. The cohort was made by selecting EHR notes for which an anticoagulant or bleeding event was mentioned in the EHR and at least one ICD-9 code related to cardiovascular diseases was mentioned in their structured tables. Each discharge summary was annotated by one of the 5 annotators and reconciliation was done by the senior physicians. Regular weekly meetings were held to maintain uniformity amongst the annotations. All six entity types and their relevant statistics are provided in Table 1 below. The ‘suspected alternative cause’ entity has the highest average number of words per annotation followed by ‘bleeding event’. Figure 1 shows definition and examples of each entity.
Table 1.
Data Statistics.
| Entity Type | Instances | Average Span Length (word/instance) |
Prevalence in EHR notes (%) |
Prevalence in Sentences (%) |
| Bleeding event | 10232 | 1.49 | 100.00 | 8.26 |
| Bleeding anatomic site | 3372 | 1.35 | 73.59 | 3.08 |
| Suspected alternative cause | 3622 | 1.59 | 79.98 | 3.32 |
| Severity | 3490 | 1.39 | 78.31 | 3.00 |
| Medication | 8726 | 1.01 | 99.81 | 6.52 |
| Bleeding lab evaluation | 3010 | 1.07 | 78.31 | 2.96 |
Figure 1.
A brief description of all the 6 entity types.
Evaluation Metrics
We use precision, recall and F1-score as evaluation metrics , which are commonly used in information extraction26,27, to compare the NER performance of our models. We report both micro and macro-averaged metrics. Micro-averaged metrics represent the performance of all instances whereas macro-averaged metrics help compare the performance by entity types. We consider both strict-matching (i.e., if both the span boundary and entity type match with that of the ground truth26), and relaxed-matching (i.e., if the span boundaries overlap and entity types match).
Data Processing
The dataset comprises a total of 1079 expert-annotated EHR notes. We used NLTK28 (Natural Language Toolkit) to split the EHR text into sentences and then tokenize them. We split the dataset into training (852 notes) and test (227 notes) sets with an approximate ratio of 79:21. The BIO tag scheme29 was followed to generate the final corpus.
Experimental Setup
At first, we chose 198 EHR notes (approx. 18% of the whole dataset) from the training set as development set for tuning the hyperparameters and the rest were considered for final training. This process was repeated 3 times ensuring that each time the development set is different. Three instances of each model were run independently on these 3 different train-development sets and the final reported results are averaged over the three runs. All the hyperparameters were tuned on the first development set only.
We used sklearn-crfsuite30 package for the CRF model whereas PyTorch was used for LM-LSTM-CRF and all BERT variants. For CRF, we have used syntactic features such as the last 2 and 3 characters of a token, its parts of speech tag, case, it is a digit or not and semantic features i.e. a concatenation of syntactic features of the previous two tokens. For LM-LSTM-CRF, after hyperparameter tuning, we chose stochastic gradient descent (SGD) as the optimizer with a final learning rate of 0.03. The dropout rate was 0.55 and the model was trained for 100 epochs. As pre-trained word embeddings we experimented with both GLOVE31 (100 dimensional) and biomedical (200 dimensional) word embeddings32. We also fine-tuned word embedding to analyze its effect on the model.
For all the transformer based architectures like BERT variants and RoBERTa, we used the popular PyTorch library - Transformers33. Maximum sequence length was 512 with a learning rate of 5e-5. For each of these models, we initialized the weights from pretrained models, added a linear classification layer on top and fine-tuned it for 15 epochs. Early stopping was used for selecting the final model for testing. All models were trained on Tesla V100 GPUs except CRF.
Results
Table 2 shows the results on the test set for CRF, the best LM-LSTM-CRF model and the best BERT variant (BioBERT). ‘Medication’ and ‘bleeding lab evaluation’ yielded the best scores irrespective of the model. We hypothesize this is due to the fact that on average, both entities have a single word per annotation (1.01 and 1.07 respectively, please refer to Table 1) and all the models do a good job at labelling single word entities. The third best performance was for 'bleeding event'. Though it does not have the least words per annotation number, it has a high number of annotations (10,220). Following the same trend, the worst performance was for the entity with the highest words per annotation count (1.6) – suspected alternative cause. Another reason behind this might be the challenges raised by the annotators for 'suspected alternative cause'. Even for human experts, it is difficult to properly identify the alternative causes for bleeding events. We will elaborate this in the “Error Analysis” section.
Table 2.
Model performance comparison on entity level. Each cell represents the tuple (precision, recall, F1 score). LM-LSTM-CRF achieves the best performance over the other two models.
| Entity Type | CRF | LM-LSTM-CRF | BioBERT |
| Bleeding event | 0.83, 0.77, 0.80 | 0.76, 0.78, 0.77 | 0.74, 0.76, 0.75 |
| Bleeding anatomic site | 0.71, 0.51, 0.60 | 0.74, 0.75, 0.74 | 0.72, 0.73, 0.72 |
| Suspected alternative cause | 0.59, 0.23, 0.33 | 0.56, 0.43, 0.48 | 0.50, 0.43, 0.46 |
| Severity | 0.71, 0.54, 0.62 | 0.67, 0.78, 0.72 | 0.66, 0.76, 0.71 |
| Medication | 0.93, 0.91, 0.92 | 0.92, 0.94, 0.93 | 0.93, 0.88, 0.91 |
| Bleeding lab evaluation | 0.83, 0.77, 0.80 | 0.80, 0.91, 0.85 | 0.82, 0.88, 0.85 |
| Micro | 0.79, 0.64, 0.71 | 0.78, 0.79, 0.78 | 0.76, 0.76, 0.76 |
| Macro | 0.75, 0.60, 0.66 | 0.74, 0.76, 0.75 | 0.73, 0.74, 0.73 |
A comparison of three models unfolds that both deep learning models outperformed CRF by a large margin (e.g. 9% by LM-LSTM-CRF and 7% by BioBERT in macro F1). The only metric CRF excels at is precision i.e. CRF is more accurate than any other model when it predicts an instance as positive. At the same time, CRF misses many positive labels (low recall), resulting low F1 scores. Despite the use of various syntactic features, this is expected as CRF is a simple machine learning algorithm whereas LM-LSTM-CRF and BioBERT are deep-learning models, capable to capture the context and pattern of any sequential data without explicit feature engineering. LM-LSTM-CRF also includes a CRF layer at the top, which lets it leverage the power of both CRF and LSTM networks. Of the two deep learning architectures, LM-LSTM-CRF outperformed BioBERT, which is surprising as most works have shown the opposite34. We believe this is in part because of the maximum sequence length limit for BERT variants. Because of this hard constraint, sentences longer than the maximum length were truncated, which not only disrupted the context but also removed many gold label entities in the process. For example, our gold test set had 6,435 labeled entities whereas with a maximum sequence length of 512, the processed test set for BERT got 6,409 entities. In addition, training a language model in parallel with the sequence labelling task also helped LM-LSTM-CRF.
Of all the BERT variants, BioBERT and RoBERTa have the best F1 scores (Table 3). However, BioBERT achieved slightly higher recall (Table 3), hence, we chose it for comparison with LM-LSTM-CRF. Despite being fine-tuned on the clinical data, Bio+Clinical BERT was underperformed by BioBERT. This is not surprising as Bio+Clinical BERT was fine-tuned only on one publicly available clinical domain (MIMIC III v 1.4) and this might limit its generalizability. On the contrary, BioBERT was fine-tuned on a wide range of biomedical domain corpora. Both Bio+Clinical BERT and vanilla BERT achieved similar F1 scores. For the remaining of the paper, we will detail the comparison of the best two models, i.e. LM-LSTM-CRF and BioBERT.
Table 3.
Entity level performance comparison for all BERT models. Each cell represents the tuple (precision, recall, F1 score). BioBERT and RoBERTa give the best performance.
| Entity Type | BERT | BioBERT | Bio+Clinical BERT | RoBERTa |
| Bleeding event | 0.72, 0.76, 0.74 | 0.74, 0.76, 0.75 | 0.72, 0.78, 0.75 | 0.75,0.76,0.75 |
| Bleeding anatomic site | 0.72, 0.71, 0.71 | 0.72, 0.73, 0.72 | 0.73, 0.72, 0.72 | 0.76,0.70,0.73 |
| Suspected alternative cause | 0.52, 0.40, 0.45 | 0.50, 0.43, 0.46 | 0.46, 0.41, 0.43 | 0.51,0.40,0.45 |
| Severity | 0.66, 0.74, 0.70 | 0.66, 0.76, 0.71 | 0.63, 0.74, 0.68 | 0.68,0.73,0.70 |
| Medication | 0.92, 0.89, 0.91 | 0.93, 0.88, 0.91 | 0.92, 0.89, 0.90 | 0.94.0.89.0.91 |
| Bleeding lab evaluation | 0.81, 0.87, 0.84 | 0.82, 0.88, 0.85 | 0.78, 0.91, 0.84 | 0.83,0.85,0.84 |
| Micro | 0.75, 0.75 0.75 | 0.76, 0.76, 0.76 | 0.74, 0.77, 0.75 | 0.77,0.75,0.76 |
| Macro | 0.73, 0.73, 0.72 | 0.73, 0.74, 0.73 | 0.71, 0.74, 0.72 | 0.75,0.72,0.73 |
We also evaluated how well the models perform on the sentence and document levels, as they are clinically more significant. It is more important to a physician to find out whether the patient has a bleeding event or not based on the entire EHR note, rather than caring about the exact span boundary of the extracted named entities. Here, we define a sentence as positive for an entity type if it contains at least one token of that type. Given such a sentence, if a model labels at-least one token within the sentence as that entity type, we consider it as a correct classification. Based on this criterion, we calculated all the metrics for both LM-LSTM-CRF and BioBERT. To facilitate better understanding, we have also added the majority baseline classifier. For each entity type, the majority baseline classifier labels all the sentences as that type. We also extended this definition to note level and calculated the F1 scores for all entity types. We report these results in Table 4. In both cases, LM-LSTM-CRF and BioBERT gave similar performance.
Table 4.
Sentence and note level performance comparison (F1 score).
| Entity Type | Sentence Level | Note Level | ||||
| LM-LSTM-CRF | BioBERT | Baseline | LM-LSTM-CRF | BioBERT | Baseline | |
| Bleeding event | 0.8785 | 0.8761 | 0.1492 | 0.9985 | 1.0 | 1.0 |
| Bleeding anatomic site | 0.8083 | 0.8043 | 0.0592 | 0.9454 | 0.9338 | 0.8478 |
| Suspected alternative cause | 0.6356 | 0.6371 | 0.0658 | 0.8838 | 0.8931 | 0.8734 |
| Severity | 0.8902 | 0.8973 | 0.0609 | 0.9574 | 0.9642 | 0.8506 |
| Medication | 0.9484 | 0.9388 | 0.1242 | 0.9985 | 0.9993 | 1.0 |
| Bleeding lab evaluation | 0.8934 | 0.8976 | 0.0593 | 0.9575 | 0.9620 | 0.8506 |
| Macro | 0.8424 | 0.8419 | 0.0864 | 0.9569 | 0.9587 | 0.9154 |
In addition to exact matching which is a strict evaluation, relaxed matching35 can also be used in entity recognition tasks to provide further insight. So, we decided to use relaxed matching criteria and compare the results with that of the exact matching. In particular we chose two variants - based on text overlaps36 and word based evaluation37 The results have been shown in Table 5 for LM-LSTM-CRF and BioBERT. Relaxed matching based on text overlaps gave significant improvements for both models. For example, the macro F1 score of LM-LSTM-CRF and BioBERT increased by 5.83% and 6.58% respectively. For word based relaxed matching, there was a 1.69% improvement in the macro F1 score of LM-LSTM-CRF and a 3.22% improvement for BioBERT. Since CRF calculates the joint probability over the labels of the whole sequence, it assists LM-LSTM-CRF to perform better than BioBERT for both relaxed matching evaluations.
Table 5.
Performance comparison for different matching criteria based on F1 scores. Here, Relaxed (O) refers to relaxed matching based on text overlaps while Relaxed (W) indicates word based relaxed evaluation. Both relaxed matching criteria increased all the scores. LM-LSTM-CRF still outperforms BioBERT.
| Entity Type | LM-LSTM-CRF | BioBERT | ||||
| Exact | Relaxed (O) | Relaxed (W) | Exact | Relaxed (O) | Relaxed (W) | |
| Bleeding event | 0.7701 | 0.8682 | 0.8240 | 0.7483 | 0.8517 | 0.8078 |
| Bleeding anatomic site | 0.7422 | 0.7803 | 0.7221 | 0.7226 | 0.7703 | 0.7390 |
| Suspected alternative cause | 0.4845 | 0.5795 | 0.5117 | 0.4595 | 0.5749 | 0.5057 |
| Severity | 0.7218 | 0.8363 | 0.7815 | 0.7052 | 0.8263 | 0.7820 |
| Medication | 0.9315 | 0.9337 | 0.9307 | 0.9057 | 0.9076 | 0.9052 |
| Bleeding lab evaluation | 0.8524 | 0.8539 | 0.8339 | 0.8495 | 0.8550 | 0.8440 |
| Micro | 0.7823 | 0.8420 | 0.7934 | 0.7584 | 0.8252 | 0.7822 |
| Macro | 0.7504 | 0.8087 | 0.7673 | 0.7318 | 0.7976 | 0.7640 |
Discussions Error Analysis
We manually analyzed cases where the models (LM-LSTM-CRF and BioBERT) made wrong predictions, we noticed four main patterns:
We noticed inconsistent annotations in terms of entity span boundaries. For example, given the phrase “acute bleeding”, the complete phrase was annotated as a bleeding event in one note whereas only the word “bleeding” was annotated in another note. Given the variability in the entity annotations, it is often difficult for the models to predict the exact spans. This is also confirmed by the improvements in the model performances when a relaxed text matching scheme was applied compared to the exact matching. In cases like these, the models mostly developed an idea of the entity type but were often struggling with the exact text span, leading to a low exact matching score.
The models memorized tokens which were prevalent in the training corpus. For example, in our dataset, ‘bleeding lab evaluation’ and ‘severity’ were annotated only if the corresponding lab test value is not within the normal range. Usually, for every ‘bleeding lab evaluation’ annotation, there is a ‘severity’ annotation within the same sentence. Due to memorization, the models labelled any mention of lab tests related to bleeding as ‘bleeding lab evaluation’. Moreover, any nearby token with numerical values was labelled as ‘severity’. All these generated many false positives.
One other aspect where the models failed was to handle the negations, possibly due to a small number of negated sentences in the training data. For example, the token ‘bleeding’ in ‘No source of bleeding’ was labelled as a ‘bleeding event’ by BioBERT. Similarly, ‘Coumadin’ in ‘Not a candidate for Coumadin’ was labelled as ‘medication’ by LM-LSTM-CRF. This is also partly due to memorization.
Finally, entity type ‘suspected alternative causes’ was the most difficult category for the models to learn. It might be attributed to a couple of factors. Firstly, it requires domain knowledge and contextual information, which makes it a challenging task even for the annotators. For example, portal hypertension (HTN) can cause esophageal varices (EV) which can cause GI bleeding, resulting in bloody stools. Similarly, thrombocytopenia (low platelet count) interferes with proper blood clotting, which can lead to bleeding events. Because these conditions are not the immediate reasons for bleeding, all annotators may not necessarily decide to associate them with the relevant bleeding event(s). Secondly, ‘suspected alternative causes’ has the highest entity span with an average span length of 1.59 (refer to Table 1), contrast to 1.26 for other entity types. This makes it difficult for the models to predict the correct spans.
Ablation with LM-LSTM-CRF
In this section, we experimented with different word embeddings for LM-LSTM-CRF. As can be seen from Table 5, language modeling (LM) improved the model performance (No LM refers to no language model) which proves the effectiveness of the joint learning setup in this architecture. Quite surprisingly, biomedical word embeddings did not help (row 1 and row 3) compared to GLOVE. However, models using biomedical embeddings had significantly higher precision scores implying these models were more confident at detecting the correct entities. But low recall scores resulted in an overall decrease in F1 scores. We also fine-tuned the biomedical word embeddings, but the result was almost the same as before (row 3 and row 4).
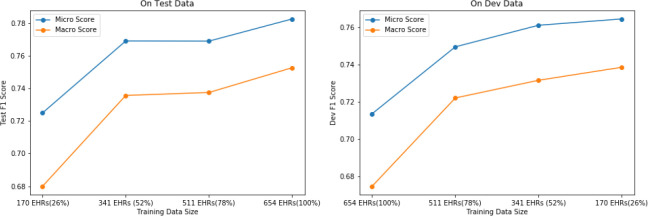
Model Performance with Data Size
Regardless of the task domain, in a supervised learning setup the training data size always plays a vital role in the performance of deep learning models. Even with transfer learning, model with a bigger dataset often outperforms itself when trained on a smaller dataset. So, in this experiment we tried to explore how the data size affects the NER task in the healthcare domain. Figure 2 shows the performance of LM-LSTM-CRF model with different training data sizes. As the amount of training data is increased (x axis on Figure 2), both the test and dev scores improve. This strongly suggests that the model’s performance has not reached the upper bound yet and with more training data it is possible to achieve a better performance. We also observed the same trend with the BERT models. This is an indication that the lack of sufficient labeled data is a crucial bottleneck in our task.
Figure 1.
Effect of data size on model performance.
Conclusion
In this work, we explored several popular architectures and evaluated their effectiveness for bleeding entity detection task on a novel EHR dataset. To the author’s knowledge, this is the first work on bleeding entity recognition from EHR notes. We found that on token-level BERT-based models performed worse than a biLSTM-CRF model with language modeling. Further experiments suggest that BERT models are more context-driven and hence work better on sentence and note level. Moreover, the scores can be significantly improved with an increase in the training data. Additional error analyses show that most of the deep learning models’ wrong predictions can be linked to inconsistent span boundary, memorization, negation, and higher span length.
In future, we hope to explore span-based approaches38,39 for entity recognition. We believe Recurrent Transformer40 and hierarchical BERT41 may solve the sequence length limitation of BERT. We will also try distant supervision which might be a solution to the scarcity of labelled data. Another direction worth exploring is to utilize additional domain specific contextual features.
Conflict of Interest
DDM receives sponsored research grant support from Bristol-Myers Squibb, Boehringher-Ingelheim, Pfizer, Flexcon, Fitbit, Philips Healthcare, Biotronik, and Apple and has received consultancy fees from Bristol-Myers Squibb, Pfizer, Flexcon, Fitbit, Boston Biomedical Associates, and Rose Consulting.
Acknowledgement
We would like to thank our annotators - Raelene Goodwin, Edgard Granillo, Nadiya Frid, Heather Keating and Brian Corner for annotating the discharge summaries. Additionally, we would like to thank our anonymous reviewers for their constructive comments on this work.
Footnotes
Equal contribution, alphabetically ordered.
Figures & Table
Table 6.
Effect of LM and Word Embedding on LM-LSTM-CRF
| Model | Macro Precision | Macro Recall | Macro F1 Score |
| LM+GLOVE | 0.7431 | 0.7638 | 0.7504 |
| No LM+GLOVE | 0.7371 | 0.7428 | 0.7383 |
| LM + Biomedical | 0.7486 | 0.7386 | 0.7399 |
| LM + Biomedical* | 0.7684 | 0.7209 | 0.7384 |
Word embeddings were fine-tuned
References
- 1.Colilla S, Crow A, Petkun W, Singer DE, Simon T, Liu X. Estimates of current and future incidence and prevalence of atrial fibrillation in the U.S. adult population. Am J Cardiol. 2013 Oct 15;112(8):1142–7. doi: 10.1016/j.amjcard.2013.05.063. [DOI] [PubMed] [Google Scholar]
- 2.Wolf PA, Abbott RD, Kannel WB. Atrial fibrillation as an independent risk factor for stroke: The framingham study. Stroke. 1991;22(8):983–8. doi: 10.1161/01.str.22.8.983. [DOI] [PubMed] [Google Scholar]
- 3.Giugliano RP, Ruff CT, Braunwald E, Murphy SA, Wiviott SD, Halperin JL, et al. Edoxaban versus warfarin in patients with atrial fibrillation. N Engl J Med. 2013;369(22):2093–104. doi: 10.1056/NEJMoa1310907. [DOI] [PubMed] [Google Scholar]
- 4.Lip GYH, Lane DA, Buller H, Apostolakis S. Development of a novel composite stroke and bleeding risk score in patients with atrial fibrillation: The AMADEUS study. Chest [Internet] 2013 Dec [cited 2020 Mar 24];144(6):1839–47. doi: 10.1378/chest.13-1635. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24009027 . [DOI] [PubMed] [Google Scholar]
- 5.Di Biase L, Burkhardt JD, Santangeli P, Mohanty P, Sanchez JE, Horton R, et al. Periprocedural stroke and bleeding complications in patients undergoing catheter ablation of atrial fibrillation with different anticoagulation nagement results from the role of coumadin in preventing thromboembolism in atrial fibrillation (AF) patients undergoing catheter ablation (COMPARE) randomized trial. Circulation. [Internet] 2014 Jun 24 [cited 2020 Mar 24];129(25):2638–44. doi: 10.1161/CIRCULATIONAHA.113.006426. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24744272 . [DOI] [PubMed] [Google Scholar]
- 6.Piccini JP, Hammill BG, Sinner MF, Hernandez AF, Walkey AJ, Benjamin EJ, et al. Clinical course of atrial fibrillation in older adults: the importance of cardiovascular events beyond stroke. Eur Heart J [Internet] 2014 Jan [cited 2020 Mar 24];35(4):250–6. doi: 10.1093/eurheartj/eht483. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24282186 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.F.D.R. H, A.K. R, G.Y.H. L, K. F, D.A. F, J. M, et al. Performance of stroke risk scores in older people with atrial fibrillation not taking warfarin: Comparative cohort study from BAFTA trial. BMJ [Internet] 2011;343(7815):d3653. doi: 10.1136/bmj.d3653. [cited 2020 Mar 24]; Available from: http://www.embase.com/search/results?subaction=viewrecord&from=export&id=L362135106 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hylek EM, Evans-Molina C, Shea C, Henault LE, Regan S. Major hemorrhage and tolerability of warfarin in the first year of therapy among elderly patients with atrial fibrillation. Circulation. [Internet] 2007 May 29 [cited 2020 Mar 24];115(21):2689–96. doi: 10.1161/CIRCULATIONAHA.106.653048. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17515465 . [DOI] [PubMed] [Google Scholar]
- 9.Mant J, Hobbs FR, Fletcher K, Roalfe A, Fitzmaurice D, Lip GY, et al. Warfarin versus aspirin for stroke prevention in an elderly community population with atrial fibrillation (the Birmingham Atrial Fibrillation Treatment of the Aged Study, BAFTA): a randomised controlled trial. Lancet [Internet] 2007 Aug 11 [cited 2020 Mar 24];370(9586):493–503. doi: 10.1016/S0140-6736(07)61233-1. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17693178 . [DOI] [PubMed] [Google Scholar]
- 10.Reynolds MR, Shah J, Essebag V, Olshansky B, Friedman PA, Hadjis T, et al. Patterns and predictors of Warfarin use in patients with new-onset atrial fibrillation from the FRACTAL registry. Am J Cardiol. 2006 Feb 15;97(4):538–43. doi: 10.1016/j.amjcard.2005.09.086. [DOI] [PubMed] [Google Scholar]
- 11.Turchin A, Shubina M, Breydo E, Pendergrass ML, Einbinder JS. Comparison of Information Content of Structured and Narrative Text Data Sources on the Example of Medication Intensification. J Am Med Informatics Assoc [Internet] 2009 May 1 [cited 2019 Dec 26];16(3):362–70. doi: 10.1197/jamia.M2777. Available from: https://academic.oup.com/jamia/article-lookup/doi/10.1197/jamia.M2777 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jagannatha AN, Yu H. Bidirectional RNN for medical event detection in electronic health records. 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, NAACL HLT 2016 - Proceedings of the Conference. Association for Computational Linguistics (ACL); 2016. pp. 473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F, Liu W, Yu H. Extraction of Information Related to Adverse Drug Events from Electronic Health Record Notes: Design of an End-to-End Model Based on Deep Learning. JMIR Med Informatics. 2018 Nov 26;6(4):e12159. doi: 10.2196/12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman AB, Peterson KS, Alba PR, DuVall SL, Patterson O V. Detecting Adverse Drug Events with Rapidly Trained Classification Models. Drug Saf. 2019 Jan 21;42(1):147–56. doi: 10.1007/s40264-018-0763-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li R, Hu B, Liu F, Liu W, Cunningham F, McManus DD, et al. Detection of bleeding events in electronic health record notes using convolutional neural network models enhanced with recurrent neural network autoencoders: Deep learning approach. J Med Internet Res. 2019 Feb 1;21(2) doi: 10.2196/10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lafferty J, McCallum A, Pereira F. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data. Dep Pap [Internet] 2001 Jun 28 [cited 2019 Dec 26] Available from: https://repository.upenn.edu/cis_papers/159 . [Google Scholar]
- 17.Sha F, Pereira F. Shallow parsing with conditional random fields. Proceedings of the 2003 Conference of the North American Chapter of the Association for Computational Linguistics on Human Language Technology - NAACL ’03 [Internet]; Morristown, NJ, USA. Association for Computational Linguistics (ACL). 2003. pp. 134–41. [cited 2020 Mar 2]. Available from: http://portal.acm.org/citation.cfm?doid=1073445.1073473 . [Google Scholar]
- 18.McCallum A, Li W. dl.acm.org. [Internet]; 2003. Early results for named entity recognition with conditional random fields, feature induction and web-enhanced lexicons; pp. 188–91. [cited 2020 Mar 24]. Available from: https://dl.acm.org/citation.cfm?id=1119206 . [Google Scholar]
- 19.Sarawagi S, Cohen WW. Advances in Neural Information Processing Systems [Internet] 2005. Semi-markov conditional random fields for information extraction. [cited 2020 Mar 24]. Available from: http://papers.nips.cc/paper/2648-semi-markov-conditional-random-fields-for-information-extraction.pdf . [Google Scholar]
- 20.Liu L, Shang J, Ren X, Xu FF, Gui H, Peng J, et al. Empower sequence labeling with task-aware neural language model. 32nd AAAI Conference on Artificial Intelligence, AAAI 2018 [Internet]; 2018. pp. 5253–60. [cited 2019 Dec 26]. Available from: www.aaai.org . [Google Scholar]
- 21.Devlin J, Chang M-W, Lee K, Toutanova K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. 2018 Oct 10 [cited 2019 Dec 26]. Available from: http://arxiv.org/abs/1810.04805 .
- 22.Li F, Jin Y, Liu W, Rawat BPS, Cai P, Yu H. Fine-tuning bidirectional encoder representations from transformers (BERT)–based models on large-scale electronic health record notes: An empirical study. J Med Internet Res [Internet] 2019 Sep 12 [cited 2020 Mar 24];21(9):e14830. doi: 10.2196/14830. Available from: http://www.ncbi.nlm.nih.gov/pubmed/31516126 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2019 Sep 10. [DOI] [PMC free article] [PubMed]
- 24.Alsentzer E, Murphy JR, Boag W, Weng W-H, Jin D, Naumann T, et al. Publicly Available Clinical BERT Embeddings. 2019 Apr 5 [cited 2019 Dec 26]. Available from: http://arxiv.org/abs/1904.03323 .
- 25.Liu Y, Ott M, Goyal N, Du J, Joshi M, Chen D, et al. RoBERTa: A Robustly Optimized BERT Pretraining Approach. 2019 Jul 26 [cited 2020 Jan 17]. Available from: http://arxiv.org/abs/1907.11692 .
- 26.Tjong EF, Sang K. Introduction to the CoNLL-2002 Shared Task: Language-Independent Named Entity Recognition. COLING-02 6th Conf Nat. Lang Learn. 2002:2002. [Google Scholar]
- 27.Lample G, Ballesteros M, Subramanian S, Kawakami K, Dyer C. Neural architectures for named entity recognition. 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, NAACL HLT 2016 - Proceedings of the Conference; 2016. pp. 260–70. [Google Scholar]
- 28.Loper E, Bird S. NLTK: The Natural Language Toolkit. [Internet]. arxiv.org. 2002. [cited 2020 Mar 24]. Available from: http://nltk.sf.net/
- 29.Ramshaw LA, Marcus MP. Text Chunking Using Transformation-Based Learning. 1999. pp. 157–76.
- 30.GitHub - TeamHG-Memex/sklearn-crfsuite: scikit-learn inspired API for CRFsuite [Internet] [cited 2019 Dec 26]. Available from: https://github.com/TeamHG-Memex/sklearn-crfsuite .
- 31.Pennington J, Socher R, Manning CD. GloVe: Global vectors for word representation. EMNLP 2014-2014 Conference on Empirical Methods in Natural Language Processing, Proceedings of the Conference; 2014. pp. 1532–43. [Google Scholar]
- 32.Pyysalo S, Ginter F, Moen H, Salakoski T, Ananiadou S. Distributional Semantics Resources for Biomedical Text Processing. [Internet]. [cited 2020 Mar 25]. Available from: https://github.com/spyysalo/nxml2txt .
- 33.Wolf T, Debut L, Sanh V, Chaumond J, Delangue C, Moi A, et al. HuggingFace’s Transformers: State-of- the-art Natural Language Processing. 2019 Oct 8 [cited 2019 Dec 26]. Available from: http://arxiv.org/abs/1910.03771 .
- 34.Li J, Sun A, Han J, Li C. A Survey on Deep Learning for Named Entity Recognition. 2018 Dec 21 [cited 2020 Mar 25]. Available from: http://arxiv.org/abs/1812.09449 .
- 35.Tsai RTH, Wu SH, Chou WC, Lin YC, He D, Hsiang J, et al. Various criteria in the evaluation of biomedical named entity recognition. BMC Bioinformatics. 2006 Feb 24;7:92. doi: 10.1186/1471-2105-7-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Z, Yang M, Wang X, Chen Q, Tang B, Wang Z, et al. Entity recognition from clinical texts via recurrent neural network. BMC Med Inform Decis Mak [Internet] 2017 Jul 5 [cited 2019 Dec 26];17(S2):67. doi: 10.1186/s12911-017-0468-7. Available from: http://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-017-0468-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jagannatha AN, Yu H. Structured prediction models for RNN based sequence labeling in clinical text. EMNLP 2016 - Conference on Empirical Methods in Natural Language Processing, Proceedings. [Internet]; 2016. pp. 856–65. [cited 2020 Feb 18]. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5167535/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wadden D, Wennberg U, Luan Y, Entity Hajishirzi H. Relation, and Event Extraction with Contextualized Span Representations. Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP). [Internet]. Association for Computational Linguistics; 2019. pp. 5784–9. [cited 2020 Mar 21]. Available from: https://www.aclweb.org/anthology/D19-1585 . [Google Scholar]
- 39.Eberts M, Ulges A. Span-based Joint Entity and Relation Extraction with Transformer Pre-training. 2019 Sep 17 [cited 2020 Mar 21]. Available from: http://arxiv.org/abs/1909.07755 .
- 40.Dai Z, Yang Z, Yang Y, Carbonell J, Le Q, Salakhutdinov R. Transformer-XL: Attentive Language Models beyond a Fixed-Length Context. 2019. pp. 2978–88. [cited 2019 Dec 26]. Available from: http://arxiv.org/abs/1901.02860 .
- 41.Zhang X, Wei F, Zhou M. HIBERT: Document Level Pre-training of Hierarchical Bidirectional Transformers for Document Summarization. Association for Computational Linguistics (ACL) 2019. pp. 5059–69.