Abstract
The efficacy of early fluid treatment in patients with sepsis is unclear and may contribute to serious adverse events due to fluid non-responsiveness. The current method of deciding if patients are responsive to fluid administration is often subjective and requires manual intervention. This study utilizes MIMIC III and associated matched waveform datasets across the entire ICU stay duration of each patient to develop prediction models for assessing fluid responsiveness in sepsis patients. We developed a pipeline to extract high frequency continuous waveform data and included waveform features in the prediction models. Comparing across five machine learning models, random forest performed the best when no waveform information is added (AUC = 0.84), with mean arterial blood pressure and age identified as key factors. After incorporation of features from physiologic waveforms, logistic regression with L1 penalty provided consistent performance and high interpretability, achieving an accuracy of 0.89 and F1 score of 0.90.
Keywords: Sepsis, fluid responsiveness prediction, MIMIC III, waveform data, machine learning
Introduction
Sepsis is defined as “life-threatening organ dysfunction caused by a dysregulated host response to infection", and it is the leading cause of hospital mortality in the United States.1 Sepsis-related mortality risk factors vary significantly due to non-uniformity of disease development. Different rules have been proposed over time to define sepsis by combining different physiological and laboratory observations. While sepsis-1 and sepsis-2 defined sepsis as a combination of systemic inflammatory response syndrome (SIRS) response and infection, they have poor specificities and hence overlap with symptoms of ‘sepsis-mimics’.2 In this study, we have adopted the latest sepsis-3 definition as a more reliable and effective diagnostic criterion to identify sepsis patients.2,3
Patients in septic shock sometimes respond poorly to intravenous fluid administration.4 While fluid administration is a first-line strategy to combat sepsis onset, it is unclear if the patient will respond to the treatment positively. Aggressive fluid administration on unresponsive patients can lead to serious adverse events such as organ dysfunction, tissue edema, and tissue hypoxia.2-4 Many sepsis patients have prior heart problems, such as diastolic dysfunction or systolic dysfunction, and improper fluid administration for these patients will worsen the condition.5 Hence, the decision to administer fluids or not is critical for better patient outcomes.6
This study builds prediction models to determine which sepsis patients are likely to respond to bolus fluid treatment significant change in systolic blood pressure after fluid being administered) up to 3 hours prior to the intervention.7 We utilized both MIMIC III dataset and its high frequency continuous waveform data8,9 and validated the results across multiple time windows. We developed a pipeline to extract and convert high-resolution waveform data to readable numeric values from MIMIC-III Waveform Database Matched Subset and linked it to MIMIC-III structured data. The study also identified lead indicators (including vital signs and demographic information) to consider when administering fluids for sepsis patients.
To the best of our knowledge, this is the first study to incorporate high-resolution waveform matched dataset with MIMIC-III data for volume responsiveness prediction.
Related Work
The analysis of continuous data streams to predict sepsis has been of great interest to the research community.10-14 Studies have also used continuous data from bedside monitoring for improving sepsis prediction in models which primarily have used clinical data.15,16 While these studies have focused on improving early prediction of sepsis, others have proposed models that aid in the management of sepsis care.17 The focus on care management is unique since a multitude of alternative treatment plans exist to manage complex sepsis patients, the efficacies of which may be uncertain in time-critical decision making.18
To aid clinical decision support regarding appropriate interventions, a number of studies have demonstrated that volume responsiveness can be characterized earlier using various forms of clinical data, including echocardiography,19 using non-invasive stroke volume coupled with passive leg raising (PLR)20 and using end-tidal CO2 with PLR21. These findings support the notion that non-invasive measures, such as those derived from Arterial Blood Pressure (ABP) may predict intervention effectiveness.
In evaluating intervention effectiveness, there is limited evidence suggesting that machine learning methods can predict volume responsiveness and early initiation of vasopressors using EMR data.22-24 Table 1 highlights recent work that focus on modeling specific intervention efficacies for critically ill patients using public and private datasets. These findings reinforce the premise that salient characteristics can be captured earlier to characterize hypotensive patients who respond to treatment. However, these approaches use only the static clinical data in the MIMIC-III dataset and have not explored prediction performance within a septic cohort. Finally, to the best of our knowledge, no existing study has incorporated ‘physiomarkers’ derived from continuous physiologic waveform, along with clinical data from the EMR, to predict fluid responsiveness among septic patients.
Table 1.
Prior research on models to predict intervention efficacy among critically ill patients
| Title | Reference | Year | Method |
| The artificial intelligence clinician learns optimal treatment strategies for sepsis in intensive care | Komorowski et al.17 | 2018 | Reinforcement Learning |
| Predicting Blood Pressure Response to Fluid Bolus Therapy Using Neural Networks with Clinical Interpretability | Girkar et al.23 | 2019 | RNN (with attention) |
| Understanding vasopressor intervention and weaning: Risk prediction in a public heterogeneous clinical time series database | Wu et al.22 | 2017 | Switching-state Autoregressive Model |
| Improving Sepsis Treatment Strategies by Combining Deep and Kernel-Based Reinforcement Learning | Peng et al.24 | 2018 | Deep/Kernel Reinforcement Learning |
Methods
Data Pre-processing: MIMIC-III Data
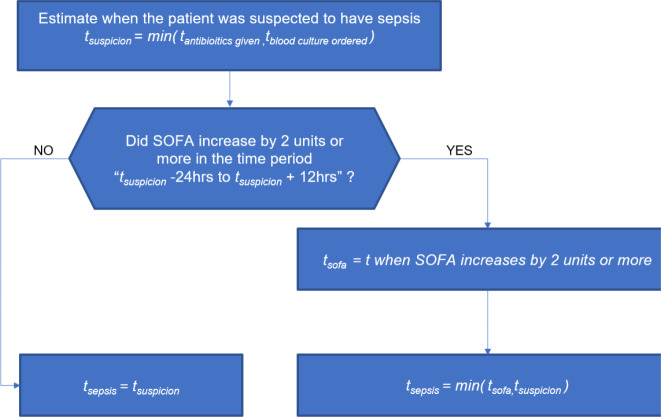
The study uses 61,532 unique ICU stay records extracted from publicly available Medical Information Mart for Intensive Care (MIMIC-III) dataset. It includes demographics, vital signs, laboratory results, medication information, and bolus/fluid events of ICU patients. We applied the sepsis-3 definition to all eligible patients by calculating time of sepsis (tSepsis) sequentially from admission until discharge. We calculated the sequential organ failure assessment (SOFA) scores as follows: tsuspicionis calculated as earlier timestamp of antibiotics and blood cultures within a specified duration (given in ICUStay table). tSOFAis identified as a 2-point deterioration in SOFA score within a 24-hour period. SOFA scores for every hour can be obtained from the SOFA table. Once we have tsuspicionand tSOFA, we obtain tsepsis = min (tsuspicion, tSOFA ) (Figure 1).
| (1) |
Figure 1.
Clinical criteria to identify time of sepsis onset
Similarly, building on Girkar et al.,22 we defined volume responsiveness as follows: First, we checked whether the bolus administered was greater than 500 ml/hr and record time of administration as tbolus. At tbolus, the change in mean blood pressure for the next three hours is noted. If any blood pressure value shows a change > 10%, the patient was labeled responsive (eqn 1)
We developed a master dataset of such patients with their demographic information, vital signs, and laboratory examination results.
While the MIMIC-III schema is rich with a variety of clinical concepts and relationships, extracting such information posed two major challenges: (1) understanding the relations among the entities in the complex schema, represented with multiple joins across a wide range of entities, to extract required relationships, and (2) generating SOFA scores for each patient for each hour of stay. Sepsis-3 event identification across an entire ICU stay of a patient requires tracking SOFA sequentially at an hourly level.
We addressed the first challenge by identifying a list of most relevant entities and untethered this entity subset from the complex schema for our master dataset creation, as well as key-value pair tables, such as D_Items and D_Labitems, that contained lookup maps required to map the above entities to the units, description and definition of a particular key used with other entities. We addressed the second challenge by creating our own custom subschema that builds on Johnson et al.25 to derive entities from querying the MIMIC-III database, and we roll out the tables at an hourly level. The simplified database contains five entities as follows:
ICUStay: Contains the basic demographic information of the patient. It also contains certain important flags such as metastatic cancer and diabetes. Attribute tsuspicion, the time of clinical suspicion as defined in26 present in this table, is essential for developing an accurate tSepsis timestamp.
VitalsInfo: Contains information about all the vital signs monitored and recorded for the patient. This table contains the change in blood pressure and certain important features like Glucose, systolic and diastolic blood pressure.
BolusInfo: Contains fluid bolus administered to the patient at a given chart time in milligram per hour. From the 53,432 adult patients considered, only 30,000 patients had bolus information.
Lab Values: Has Lab values for features that can improve the volume responsiveness prediction.
SOFA: Consists of hourly SOFA values for a patient admitted to the ICU, with the start-time and end-time associated with each interval.
Sepsis evaluation has routinely been performed on data using the first 24 hours of ICU stay, therefore potentially missing patients who go on to develop sepsis later during ICU stay.25 Attributes such as tsuspicionand tantibiotic, when the antibiotic is first administered, have an average value of more than one day across all the patients, presenting potential conflicting timestamps. Furthermore, a change of SOFA greater than 2 (the key criterion for sepsis-3 definition) is detected after the initial 24 hours in almost 25% of the cases.
Starting from 61,532 ICU IDs in the MIMIC-III database, 29,560 (49%) were identified to be sepsis-3 related using the rules over the entire length of ICU stay. Fluids and vital signs information were available for 23,540 ICU IDs.
Linking on both criteria resulted in a master dataset of 15,062 ICU IDs, of which 10,539 (~40%) were volume responsive. Demographic (age, gender, race), comorbidity (diabetes, metastatic cancer), and vitals (systolic, diastolic, and mean arterial blood pressure, respiration rate, and oxygen saturation) information were added to the master dataset, resulting in 49 variables. On volume responsiveness outcome (i.e., when given a bolus of 500ml or more, a 10% rise in blood pressure was observed), 70% were responsive.
Data Pre-processing: MIMIC-III Waveform Data
Independent of the MIMIC-III data collection, the MIMIC-III Waveform Database includes recorded physiological waveforms obtained from patient bedside monitors. The MIMIC-III Waveform Matched Subset is the intersection of MIMIC-III database and the waveform records, consisting of 22,317 physiologic signals (“waveforms”) and 22,247 vital signs time series. We identified 5,960 patients who developed sepsis-3 during their ICU stays and had high frequency waveforms, including electrocardiogram (EKG), arterial blood pressure (ABP) and plethysmography (PPG) within the matched database.
Once we aggregate the waveform data for each patient, we performed signal processing methods to identify signal quality and baseline drifts. We applied a signal quality index to derive 5-second segments of EKG with data quality > 80%. We derived peak detection using the Christov real-time QRS algorithm.30 Statistical features were derived from the R-R interval between neighboring QRS complex from each EKG. For the pulsatile signals, e.g. ABP and PPG, we applied peak detection algorithms by Lazaro et al.31 and Zong et al.32 Features were then derived from the peak-to-peak interval of these waveforms from time-frequency and signal entropy domains.
Linking All Relevant Tables
All relevant files are linked to create a master reference dictionary. This includes data from MIMIC-III database, a complete list of waveform record file and fluid events. There are two reasons for joining relevant tables to create a master reference dictionary. Fluid events are organized at the ICU stay level, while waveform records are recorded with patient identifiers. Thus, we need to map patient identifier to ICU stays. Secondly, boosting execution speed is critical. Iterating through waveform records to perform read-in and extractions, performing linear search for each incoming identifier for its corresponding ICU stay and then for waveforms is extremely inefficient. The time complexity is O(n*m*l), where n, m, and l denote the size of each file. Joining all relevant tables free up disk space as well as effectively shorten the time complexity to O(1) for search.
Timestamp Check & Read in Waveform Records
This step iterates through the list of waveform records, performs timestamp check and passes on those qualified waveform records for extraction. There are two check mechanisms involved in the step, one before read-in, and another before passing for extraction. The waveform read and feature extraction are both very expensive tasks that consume considerable disk space and program run time. In our test run, we observed that on average, reading in a waveform record with 11 hours of signals will take up approximately 5 to 10 minutes, and extracting features from the waveform can take anywhere from 3 to 90 minutes, depending on the size of its signal vector. Hence, we incorporated several heuristics into the pipeline to help improve performance.
When the pipeline iterates through the waveform record list, and before it reads in the record file, we compare the timestamp embedded in the filename with those of fluid events under the patient identifier. If the file timestamp is larger than all of the recorded fluid administration, we conclude that the waveform records physiologic signals after fluid is given. Since our primary focus is to predict volume responsiveness prior to fluid administration, those files will be discarded because they will not contain important information about the patient before fluid time. Similarly, we compare waveform start time and end time with fluid time. We are interested in the waveform information from eight hours prior to fluid time to two hours prior, or [t-8, t-2], where t denotes time of fluid administration, t-fluid. If there is no intersection between waveform record time and the six-hour window, [t-8, t-2], we excluded the waveform record.
Feature Extraction
Receiving the waveforms from previous steps, the third step is to perform statistical transformation. We identified more than 150 features that are highly relevant to our study. However, this comprehensive extraction takes, on average, 90 minutes on a 11-hour-long waveform record. Thus, we trimmed feature space down to 73 so that the entire pipeline is more efficient. The complete list of features extracted is available on our GitHub repository. The extraction pipeline takes about 60 GB disk space, 150+ storage space, and more than 90 hours of run time despite all the intelligent construction aforementioned. Nevertheless, we estimate that we obtained a 3X speed up with the check mechanism and join in place.
Waveform Data Imputation
Finally, we performed MICE (Multivariate Imputation by Chained Equations)27 to fill the missing values. It has many advantages over single imputation methods, such as replacing missing values by mean or median. The MICE algorithm works by running multiple regression models and each missing value is modeled conditionally, depending on the remaining variables in the dataset. We applied the IterativeImputer function from the sklearn python library to impute waveform related data points to handle missingness.
Integration with MIMIC-III
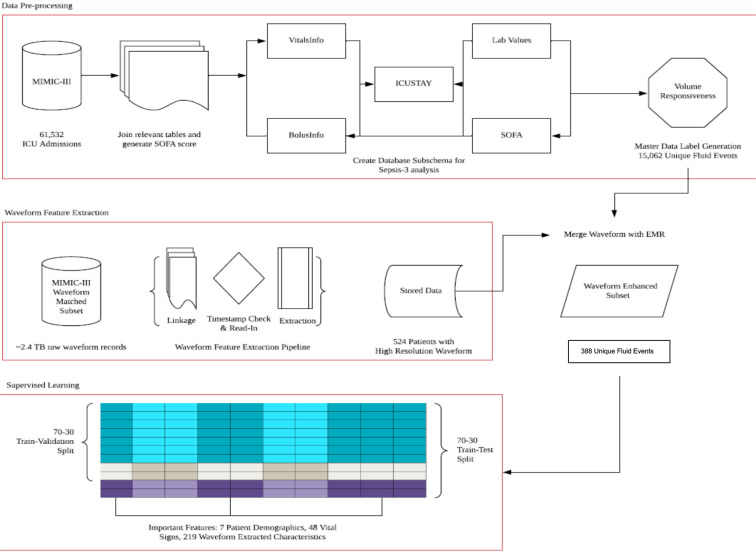
From the MIMIC-III sub-schema, we extracted data about sepsis-3 patients who received fluid treatment at infusion rate greater than 500 ml/hour and whose vitals were recorded. In this focused dataset, there were 15,062 unique fluid events and 10,539 events where patients were responsive to fluid volume. After matching waveform extraction results with MIMIC-III database, we obtain 388 unique fluid event observations with vital signs and waveform records present. The integrated dataset has 274 features, including 219 waveform extracted characteristics, 48 vital signs, and 7 patient demographics. Figure 2 summarizes all the steps in the data processing, extraction and integration schema.
Figure 2.
MIMIC-III Data Processing, Waveform Feature Extraction, and Prediction Steps
Models and Evaluation
We evaluated multiple machine learning models to predict volume responsiveness in sepsis-3 patients and identify key indicators. Due to the limited sample size and interpretability concerns, we excluded recurrent neural networks and other deep learning models. Accuracy, AUC, and F1 score were used as evaluation criteria, and performance robustness and clinical interpretability were considered in the discussions with clinicians.
MIMIC-III data: We applied logistic regression, random forest and support vector machines (SVM).28 15,062 patient data were split into 80% for training using 10-fold cross validation, and 20% for testing. The best versions of each were evaluated on the 20% holdout data. We eliminated SVM models based on poor AUC and further compared the logistic regression (LR) and random forest (RF) models systematically on reduced test datasets as follows. First, we removed one hour of data from all the patients in the test set, then predicted the outcome with the two models, LR and RF. Next, we removed another hour and compared the predictions. The aim was to assess the robustness of the two models with minimum data for early prediction and action.
Adding waveform data: We compared random forest (RF), XGBoost, standard Logistic Regression (LR), Logistic Regression with L1 penalty and sparsity (LR with L1), and Support Vector Machines (SVM) with all features normalized using standard scaler available in scikit learn and 70-30 train-hold out split, and conducted 10-fold cross validation on an extensive hyperparameter space.
Results
Table 2 summarizes the demographic, comorbidities and vital signs data on 15,062 ICU patients that were included in our final analysis.
Table 2.
Descriptive summary
| Category | Variable | Mean (SD) | Occurrence |
| Demographics | Sex = Male | 58% | |
| Age | 66 (16) | ||
| Race = White | 73% | ||
| Comorbidities | Metastatic cancer | 51% | |
| Diabetes | 29% | ||
| Vitals | Heart rate | 90 (19) | |
| Systolic BP | 106 (19) | ||
| Diastolic BP | 55 (12) | ||
| Mean BP | 70 (13) | ||
| Respiration rate | 19 (6) | ||
| SPO2 | 97 (4) |
MIMIC-III data: The most robust model for volume responsiveness prediction with MIMIC-III structured data was the Random Forest model, with an AUC of 0.84 and accuracy of 78% (Table 3). Among the demographic variables, Age is the most significant factor. The logistic regression coefficient for this was significant and negative, suggesting that older patients are less responsive to fluid administration. Gender, race and comorbidities were not significant in both models. However, the master dataset patients are predominantly white and have no comorbidities. Hence, we cannot reject the importance of race and comorbidity in volume responsiveness prediction. The most significant variable in Random Forest was mean blood pressure and the logistic regression coefficient for this was significant and negative, suggesting that a patient with low mean blood pressure is less likely to not respond to volume compared to one with a higher mean blood pressure.
Table 3.
Prediction Results with MIMIC-III and Waveform data
| Waveform | EMR | |||
| Model | Accuracy (%) | AUC | Accuracy (%) | AUC |
| Random Forest | 83 | 0.91 | 78 | 0.84 |
| XGBoost | 86 | 0.85 | 69 | 0.64 |
| Logistic Regression | 89 | 0.86 | 79 | 0.86 |
| Linear SVM | 81 | 0.57 | 79 | 0.72 |
| SVM Ploynomial 3 | 80 | 0.57 | 77 | 0.65 |
Integration of waveform data: Inclusion of features extracted from the waveform resulted in a better overall accuracy. With Logistic Regression using L1 sparsity feature selector, achieving an accuracy of 89% and an AUC of 0.86 (Table 3). This model included 25 features; 6 were vital signs related and 19 were extracted from waveform. On the given metrics, accuracy, AUC, and F1, there is no strictly dominant model that has a clear advantage on all three performance measures. We performed simple robustness check with different random data sets and train-test splits. RandomForest, XGBoost and LR with L1 displayed steady performance on the test data, showing strong performance metrics across all test scenarios. Using interpretability as the determining factor, LR with L1 has clear advantages as it provides widely used, understandable interpretation for clinicians. Figure 3 shows the ROC curve for the models.
Figure 3.
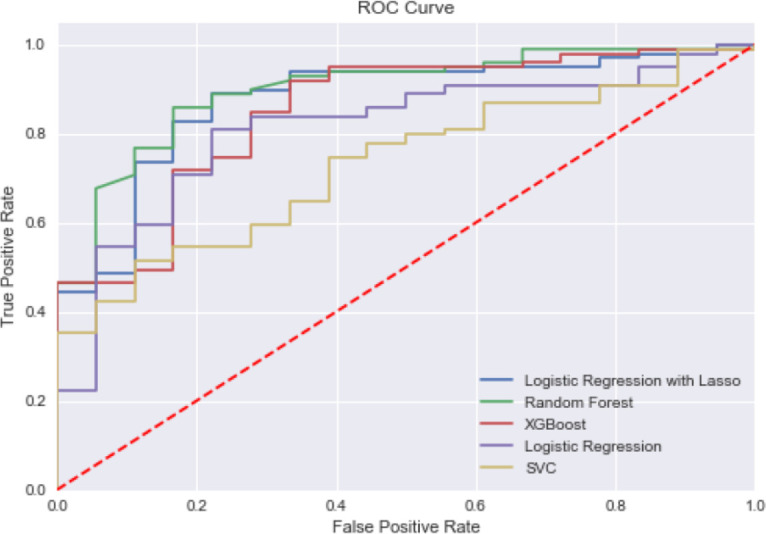
ROC curve for competing models of Volume Responsiveness Prediction with MIMIC-III+ Waveform Data
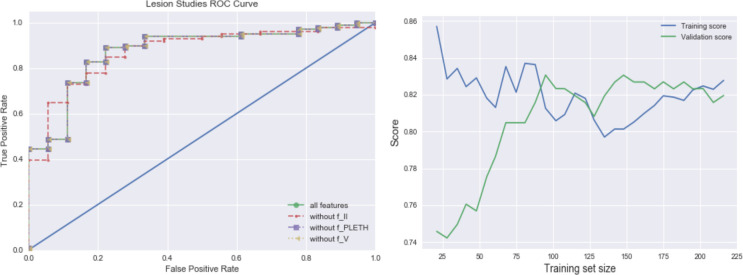
Lesion studies: To gain insight into what contributes to our model performance by lesioning components of it, we evaluated models with all but EKG lead II, plethysmography (PPG), or EKG lead V, respectively. The ROC curves in Figure 4 (a) show how performance is affected when different feature types are removed from the model. It is clear that without lead II information, the model performance deteriorates. Furthermore, among all three categories of waveform lead records, only lead II records have an impact on model performance. Removing lead PPG or lead V records provided incremental value for our scenario.
Figure 4.
(a) Lesion studies; (b) Learning curves with MIMIC-III+ Waveform Data
Learning Curve: From the learning curve of the model in Figure 4 (b), we observe a reasonable improvement as training sample size increases. The model construction is able to quickly improve performance from the additional variation it observes from a marginally increased sample size. With only ⅓ of the original training set size, the model is able to generate reasonably good scores; and with half of the training set size, it achieves similar scores compared to utilizing the entire training data.
Discussion and Conclusion
Patients who develop sepsis-3 are in a vulnerable condition that requires extreme care and clinicians often consider fluid infusion as the first-line treatment. Studies have indicated that inappropriate fluid strategy can lead to severe adverse events, and it is imperative to develop a reliable prediction mechanism to identify patients who are responsive to fluid administration from those who are unresponsive. Our study utilized the MIMIC-III dataset which was integrated high-resolution matched waveform data, and applied multiple machine learning models using patient demographics, comorbidity data, vital signs, and waveform information as predictors.
The top performing models in our study were those that included features from the waveform and are logistic regression and random forest, achieving AUC of 0.86 and 0.84, respectively. To determine a robust model, we considered potential clinical use cases, where clinicians may prefer a model with early prediction capability, easy interpretability and requiring less data points. For analysis with waveform data, a constrained logistic regression that uses L1 sparsity feature selector provides a robust, interpretable model with strong performance on accuracy and AUC. In terms of clinical interpretability, this model also provides clear indicators that waveform extracted features are beneficial in improving the prediction model, as there are 19 significant waveform features among the 25 features selected. These results demonstrate the benefits of integrating waveform records in assisting clinicians in determination of fluid management strategies for sepsis patients.
Significant information was found within the EKG lead II waveform, pertaining to energy of the signal, standard deviation and autocorrelation of R-R intervals. The PPG signal did not significantly contribute to the predictive power of the algorithm, this could be due to a number of reasons. First, we considered only the peak-to-peak interval, and therefore exclude more dynamic and rich information that may be derived from amplitude or phase shifts. Secondly, we perform basic statistical feature extraction of the time-frequency and information theory domains, hence there may be an opportunity to improve the performance through feeding the raw signal into deep neural networks.
This work has some limitations, hence additional studies are needed to better understand volume responsiveness, and better inform clinical practitioners. An important limitation is the inability to analyze the impact of race, comorbidity and laboratory results on sepsis-3 related volume responsiveness due to unavailability of the data in the data source. The MIMIC III sepsis-3 patients are disproportionately white with no reported comorbidities. Laboratory data was sparse and could not be included in the analysis. With access to a richer dataset, future research could build on this work and also explore interaction effects on volume responsiveness. All the relevant features for modeling different types of blood pressure, vital signs, demographic features, flags for metastatic cancer and diabetes within the schema that we developed are not restricted to the analysis of volume responsiveness prediction. It can also be applied to investigate and predict how body temperature and respiration rate changes within sepsis-3 prone patients admitted to ICU, tracking SOFA scores for patients suspected of sepsis-3 development, and analyzing the impact of laboratory values over the duration of ICU stay.
In conclusion, this study applied the sepsis-3 definition to identify target patients, whereas most of the existing research has focused on dated sepsis-1 or sepsis-2 definitions. A complete schema was developed to identify sepsis-3 patients in MIMIC III database that can be leveraged by future researchers. We expanded the analysis to include patients’ complete ICU stay while prior research has been limited to only the first 24 hours of ICU stay. We constructed a pipeline to extract and transform features from waveform information into readable, organized form, developed models to predict fluid responsiveness in the subset of sepsis-3 patients, and validated the results across multiple time windows. The study also identified lead indicators (including vitals and demographic information) to consider when clinicians administer fluid for sepsis-3 patients, and integrated key waveform features with vitals and demographic information to develop a high performance prediction model.
Assessing for volume status in a septic patient is both challenging and inexact at the point of initiation of a sepsis protocol and the current dose of 30cc/kg as a bolus requires further examination and validation as we acquire and try to merge mismatched and novel data streams. Developing a predictive capability to early identify poor responders would help clinicians confidently move from IV fluid boluses to the initiation of vasopressors to chemically assist blood vessels to provide oxygen to tissues. Future studies would also benefit from pre-hospital and emergency room administration of fluid boluses that are generally initiated prior to transfer to the ICU which would give a more complete picture for assessment. The integration of additional data points, including physiological10-17 and biomarkers29 may further improve the performance of the model.
Acknowledgements
We thank K. Silvas, MD, and J. O'Neill, MD, for insightful feedback during the development of this study.
Figures & Table
References
- 1.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent JL, Angus DC. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis- 3) JAMA. 2016 Feb 23;315(8):801–10. doi: 10.1001/jama.2016.0287. doi: 10.1001/jama.2016.0287. PMID: 26903338; PMCID: PMC4968574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gül F, Arslantaş MK, Cinel İ, Kumar A. Changing Definitions of Sepsis. Turk J Anaesthesiol Reanim. 2017 Jun;45(3):129–138. doi: 10.5152/TJAR.2017.93753. doi: 10.5152/TJAR.2017.93753. Epub 2017 Feb 1. PMID: 28752002; PMCID: PMC5512390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joynes E. More challenges around sepsis: definitions and diagnosis. Journal of Thoracic Disease. 2016 Nov;8(11):E1467–E1469. doi: 10.21037/jtd.2016.11.10. DOI: 10.21037/jtd.2016.11.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monnet X, Marik PE, Teboul JL. Prediction of fluid responsiveness: an update. Ann Intensive Care. 2016;6(1):111. doi: 10.1186/s13613-016-0216-7. doi:10.1186/s13613-016-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Micek ST, McEvoy C, McKenzie M, Hampton N, Doherty JA, Kollef MH. Fluid balance and cardiac function in septic shock as predictors of hospital mortality. Crit Care. 2013;17(5):R246. doi: 10.1186/cc13072. Published 2013 Oct 20. doi:10.1186/cc13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd JH, Forbes J, Nakada TA, Walley KR, Russell JA. Fluid resuscitation in septic shock: a positive fluid balance and elevated central venous pressure are associated with increased mortality. Crit Care Med. 2011;39(2):259–65. doi: 10.1097/CCM.0b013e3181feeb15. Epub 2010/10/27. 10.1097/CCM.0b013e3181feeb15. [DOI] [PubMed] [Google Scholar]
- 7.“CORE EM: Fluid Resuscitation” EmDOCs.net - Emergency Medicine Education, 26 Jan 2018 www.emdocs.net/core-em-fluid-resuscitation/
- 8.MIMIC-III, a freely accessible critical care database Johnson AEW, Pollard TJ, Shen L, Lehman L, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG. Scientific Data. 2016. DOI: 10.1038/sdata.2016.35. Available at: http://www.nature.com/articles/sdata201635 . [DOI] [PMC free article] [PubMed]
- 9.Goldberger AL, Amaral LAN, Glass L, Hausdorff JM. Ivanov PCh, Mark RG, Mietus JE, Moody GB, Peng C- K, Stanley HE. PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation. 2000 (June 13);101(23):e215–e220. doi: 10.1161/01.cir.101.23.e215. [Circulation Electronic Pages; http://circ.ahajournals.org/cgi/content/full/101/23/e215] [DOI] [PubMed] [Google Scholar]
- 10.Kamaleswaran R, Akbilgic O, Hallman MA, West AN, Davis RL, Shah SH. Applying Artificial Intelligence to Identify Physiomarkers Predicting Severe Sepsis in the PICU. Pediatr Crit Care Med. 2018;19:e495–e503. doi: 10.1097/PCC.0000000000001666. doi:10.1097/PCC.096_3417142000001666. [DOI] [PubMed] [Google Scholar]
- 11.Van Wyk F, Khojandi A, Kamaleswaran R, Akbilgic O, Nemati S, Davis RL. A Case Study in Sepsis Detection Using Deep Learning. NIH-IEEE; 2017. How Much Data Should We Collect? Special Topics Conference on Healthcare Innovations and Point of Care Technologies: Technology in Translation. 2017. [Google Scholar]
- 12.Van Wyk F, Khojandi A, Mohammad A, Begoli E, Davis RL, Kamaleswaran R. Physiomarkers in High Frequency Real-Time Physiological Data Streams Predict Adult Sepsis Onset Earlier. Int J Med Inform. 2018. In review. [DOI] [PubMed]
- 13.van Wyk F, Khojandi A, Kamaleswaran R. Improving Prediction Performance Using Hierarchical Analysis of Real-Time Data: A Sepsis Case Study. IEEE Journal of Biomedical and Health Informatics. 2019. doi:10.1109/jbhi.2019.2894570. [DOI] [PubMed]
- 14.Kamaleswaran R, Koo C, Helmick R, Mas V, Eason J, Maluf D. Predicting Early Post-Operative Sepsis in Liver Transplantation Applying Artificial Intelligence. International Liver Transplantation Society Annual Congress. 2019.
- 15.Ahmad S, Tejuja A, Newman KD, Zarychanski R, Seely AJE. Clinical review: A review and analysis of heart rate variability and the diagnosis and prognosis of infection. Crit Care. 2009;13:232. doi: 10.1186/cc8132. doi:10.1186/cc8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shashikumar SP, Stanley MD, Sadiq I, Li Q, Holder A, Clifford GD, et al. Early sepsis detection in critical care patients using multiscale blood pressure and heart rate dynamics. J Electrocardiol. 2017;50:739–743. doi: 10.1016/j.jelectrocard.2017.08.013. doi:10.1016/j.jelectrocard.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komorowski M., Celi L.A., Badawi O., Gordon A.C., Faisal A.A. The artificial intelligence clinician learns optimal treatment strategies for sepsis in intensive care. Nature medicine. 2018;24(11):1716–1720. doi: 10.1038/s41591-018-0213-5. [DOI] [PubMed] [Google Scholar]
- 18.Hunter J.G., Pritchett C., Pandya D., Cripps A., Langford R. Sim-Sepsis: improving sepsis treatment in the emergency department? BMJ Simulation and Technology Enhanced Learning. 2019;5(4):232–233. doi: 10.1136/bmjstel-2018-000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamia B, Ochagavia A, Monnet X, Chemla D, Richard C, Teboul J-L. Echocardiographic prediction of volume responsiveness in critically ill patients with spontaneously breathing activity. Intensive Care Med. 2007;33:1125–1132. doi: 10.1007/s00134-007-0646-7. doi:10.1007/s00134-007-0646-7. [DOI] [PubMed] [Google Scholar]
- 20.Thiel SW, Kollef MH, Isakow W. Non-Invasive stroke volume measurement and passive leg raising predict volume responsiveness in medical ICU patients: an observational cohort study. Crit Care. 2009;13:R111. doi: 10.1186/cc7955. doi:10.1186/cc7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monnet X, Bataille A, Magalhaes E, Barrois J, Le Corre M, Gosset C, et al. End-Tidal carbon dioxide is better than arterial pressure for predicting volume responsiveness by the passive leg raising test. Intensive Care Med. 2013;39:93–100. doi: 10.1007/s00134-012-2693-y. doi:10.1007/s00134-012-2693-y. [DOI] [PubMed] [Google Scholar]
- 22.Wu M, Ghassemi M, Feng M, Celi LA, Szolovits P, Doshi-Velez F. Understanding vasopressor intervention and weaning: Risk prediction in a public heterogeneous clinical time series database. J Am Med Inform Assoc. 2017. doi:10.1093/jamia/ocw138. [DOI] [PMC free article] [PubMed]
- 23.Girkar U, Uchimido R, Lehman L-WH, Szolovits P, Celi L, Weng W-H. Predicting Blood Pressure Response to Fluid Bolus Therapy Using Neural Networks with Clinical Interpretability. Circ Res. 2019;125:A448–A448. Available: https://www.ahajournals.org/doi/abs/10.1161/res.125.suppl_1.448 . [Google Scholar]
- 24.Peng X., Ding Y., Wihl D., Gottesman O., Komorowski M., Lehman L.W.H., Ross A., Faisal A., Doshi- Velez F. AMIA Annual Symposium Proceedings. 887) (Vol 2018. American Medical Informatics Association; 2018. Improving sepsis treatment strategies by combining deep and kernel-based reinforcement learning. [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson A.E., Aboab J., Raffa J.D., Pollard T.J., Deliberato R.O., Celi L.A., Stone D.J. A comparative analysis of sepsis identification methods in an electronic database. Critical care medicine. 2018;46(4):494. doi: 10.1097/CCM.0000000000002965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemati S., Holder A., Razmi F., Stanley M.D., Clifford G.D., Buchman T.G. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Critical care medicine. 2018;46(4):547–553. doi: 10.1097/CCM.0000000000002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azur MJ, Stuart EA, Frangakis C, Leaf PJ. Multiple imputation by chained equations: what is it and how does it work? Int J Methods Psychiatr Res. 2011 Mar;20(1):40–9. doi: 10.1002/mpr.329. doi: 10.1002/mpr.329. PMID: 21499542; PMCID: PMC3074241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hastie T, Tibshirani R, Friedman J. Data Mining, Inference and Prediction. 2nd Edition. Springer; 2008. [Google Scholar]
- 29.Mohammed A, Cui Y, Mas VR, Kamaleswaran R. Differential gene expression analysis reveals novel genes and pathways in pediatric septic shock patients. Scientific Reports. 2019;9(1):11270. doi: 10.1038/s41598-019-47703-6. doi:10.1038/s41598-019-47703-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christov Ivaylo I. “Real time electrocardiogram QRS detection using combined adaptive threshold”. BioMedical Engineering OnLine. 2004; vol.3(28):2004. doi: 10.1186/1475-925X-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lázaro J., Gil E., Vergara J.M., Laguna P. Pulse rate variability analysis for discrimination of sleep- apnea-related decreases in the amplitude fluctuations of pulse photoplethysmographic signal in children. IEEE journal of biomedical and health informatics. 2013;18(1):240–246. doi: 10.1109/JBHI.2013.2267096. [DOI] [PubMed] [Google Scholar]
- 32.Zong W, Heldt T, Moody GB, Mark RG. An open-source algorithm to detect onset of arterial blood pressure pulses. Computers in Cardiology. 2003;30:259–262. [Google Scholar]