Abstract
A longstanding issue with knowledge bases that discuss drug-drug interactions (DDIs) is that they are inconsistent with one another. Computerized support might help experts be more objective in assessing DDI evidence. A requirement for such systems is accurate automatic classification of evidence types. In this pilot study, we developed a hierarchical classifier to classify clinical DDI studies into formally defined evidence types. The area under the ROC curve for sub-classifiers in the ensemble ranged from 0.78 to 0.87. The entire system achieved an F1 of 0.83 and 0.63 on two held-out datasets, the latter consisting focused on completely novel drugs from what the system was trained on. The results suggest that it is feasible to accurately automate the classification of a sub-set of DDI evidence types and that the hierarchical approach shows promise. Future work will test more advanced feature engineering techniques while expanding the system to classify a more complex set of evidence types.
Introduction
Identifying drug combinations that could result in a clinically meaningful alteration to patient safety or therapeutic efficacy is an important patient care activity, especially given that clinicians are known to have incomplete knowledge about drug-drug interactions (DDIs)1,2. Computerized alerting systems can help clinicians by providing relevant reference information and suggestions, intelligently filtered and presented at appropriate times3. Unfortunately, the knowledge bases underlying these systems have long been known to be incomplete and inconsistent with one another. For example, a recent study by Fung et al. found that only 5% of 8.6 million unique interacting drug pairs were present in all 3 of the knowledge bases they included in the study4.
We refer to individuals who maintain knowledge bases used by clinicians for DDI clinical decision support as compendium editors. In our prior work, we established that the workflow of compendium editors generally involves topic identification, evidence search, evidence synthesis, and generating recommendations5. Focusing on the evidence synthesis step, compendium editors tend to evaluate evidence informally, with no dedicated support from information tools such as reference management software or databases. Although there exist systematic approaches to evaluate a collection of evidence relevant to establishing DDIs6,7, compendium editors generally reported using heuristic and subjective approaches to determine when sufficient evidence had been gathered to make a recommendation. Variation in evidence assessment suggests a potentially important factor underlying the lack of agreement that exists among different DDI knowledge base.
Evaluating study design is a critical component of evidence synthesis that can present challenges because the nuances of flawed studies are not always obvious. We previously reported on an experiment that found inter-rater agreement on evidence sufficiency among compendia editors to be poor8. Among the possible explanations for the finding is that experts tend to be subjective when assessing an evidence item’s type and study design. The degree of subjectivity might be related to an expert’s experience and knowledge of specific in vitro and in vivo pharmacology research methods.
We think that a particularly promising future research direction would be computerized support to help experts be more efficient and objective in assessing DDI evidence from biomedical literature. In this pilot study, we tested the feasibility of using machine learning to classify clinical DDI studies into the formally defined evidence types present in the DIDEO- the potential Drug-drug Interaction and potential Drug-drug Interaction Evidence Ontology9. The goal was to determine the feasibility of accurate automatic classification of study evidence types that could form the basis of a more efficient and objective approach to DDI evidence synthesis.
Methods
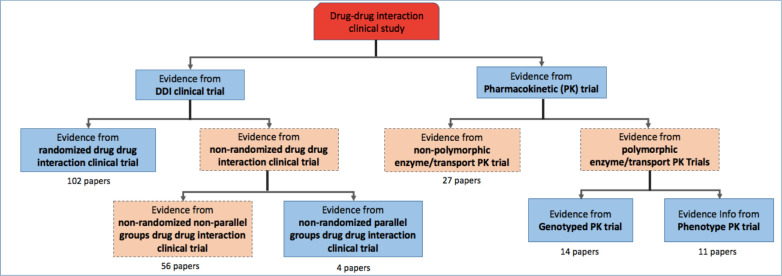
The DIDEO ontology is a foundational domain representation that contains 44 evidence types used in in vitro and in vivo pharmacokinetic DDI research10. DIDEO specifies the necessary and sufficient conditions for each evidence type using terms either defined in DIDEO or imported from other formal ontologies11. We set out to test machine learning classifiers that predict seven of the 44 specific types of DDI evidence being reported in a DDI paper (blue boxes in Figure 1). These seven evidence types were chosen because they are commonly used in in vivo study designs that we thought would be useful for showing proof-of-concept and identifying requirements for a larger scale study. During the development of the training corpus for this study, we found a need to create additional evidence types. For example, for those papers that were annotated as Pharmacokinetic (PK) Trial at the second level but were neither Genotype PK trial nor Phenotype PK trial at the third level, we labeled them as “non-polymorphic enzyme/transport PK Trial” which provides a novel alternative evidence type at the second level. Figure 1 also shows the novel evidence types that we added (in orange) and the number of papers in each category in our training set.
Figure 1.
Evidence types hierarchy that was used in the classification system. The blue boxes represent the portion of DIDEO’s evidence type hierarchy that we used in this study. The orange boxes were added to cover the full range of evidence types identified in the training corpus. Numbers of papers in the training set for each category are shown below each box.
The machine learning approach tested in this pilot study was an ensemble of hierarchical classifiers. This was chosen based on the observation that there exist multiple logical distinctions between the evidence types. For example, participant randomization is the major distinction between a Randomized DDI Clinical Trial and the two Non-Randomized DDI Clinical Trial types. Similarly, genetic genotyping distinguishes the two polymorphic enzyme/transport PK trial types. The hierarchical approach is a combination of five sub-classifiers—each of which was designed as a binary classifier that distinguished a specific pair of evidence types using models that operates at each level of the hierarchy. Our intuition was that each model in the hierarchical ensemble would pick up on the primary distinguishing features better than a single model that forms the non-hierarchical approach.
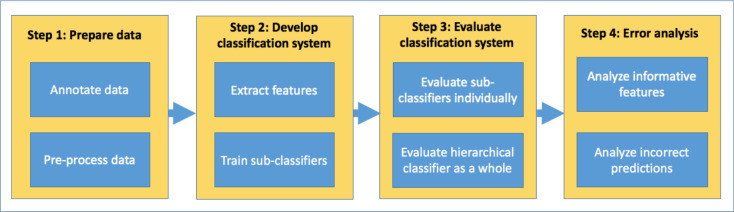
The implementation of the classification system consisted of four main steps which are shown in Figure 2 and described further below.
Figure 2.
Implementation steps used in this study to build a hierarchical evidence type classification system.
Prepare data
The training dataset for this study contained 214 unique papers about DDIs that were systematically collected by DDI experts during previous knowledge representation research involving 65 drugs12–14. The papers were manually annotated to assign evidence type labels from the DIDEO ontology using an annotation guideline15. One investigator (RB) was the primary annotator with a second (BS) independently annotating one-third of the dataset for quality assurance. The developer of the classification system (LH) also observed the annotation process in order to identify relevant text that could be used for training the classifier. The annotation process was repeated to add two additional data sets that were used for “hold out” testing of the classifiers. One data set contained 32 DDI studies involving the same 65 drugs as for the training set. The other data set contained 94 papers identified using the same search strategy used for the other two datasets but focusing on drugs other than the 65 that were the focus of prior research. More details about the search and screening strategies used to construct all three datasets can be found in a written protocol available on the project’s github15.
In the preprocessing step, we automatically collected the papers’ metadata, including titles, abstracts and PubMed publication types through the PubMed API16. We also manually collected full-text PDFs of these papers and converted them to plain text using the PDFMiner Python library17. For PDFs that were scanned images, we manually copied text from the PDFs and saved as plain text. We then standardized the plain text files by converting the text into lowercase and removing English stop words from the Natural Language Toolkit (NLTK) library version 3.4.518 run on Python version 3.7.
The features extracted from the papers included stemmed unigrams taken from the titles, abstracts, and the Methods sections of the papers. The Methods sections were included based on our observation during the annotation process that the section often contained information needed to determine the DDI evidence type that was not present in the title or abstract. Stemming was applied based on our observation that many words that experts use to distinguish evidence types have the same roots (e.g. genotyped, genotyping and genotype) and should not be treated by the classification system as different features. In order to avoid overfitting, we used MetaMap19 to remove all of the drug and enzyme names from the text as well as regular expression to eliminate numeric strings, including numbers tied with measurement units. After the final feature engineering process, our feature space contained 11325 features for the 214 instances, corresponding to the 214 papers in our training set.
Develop the classification system
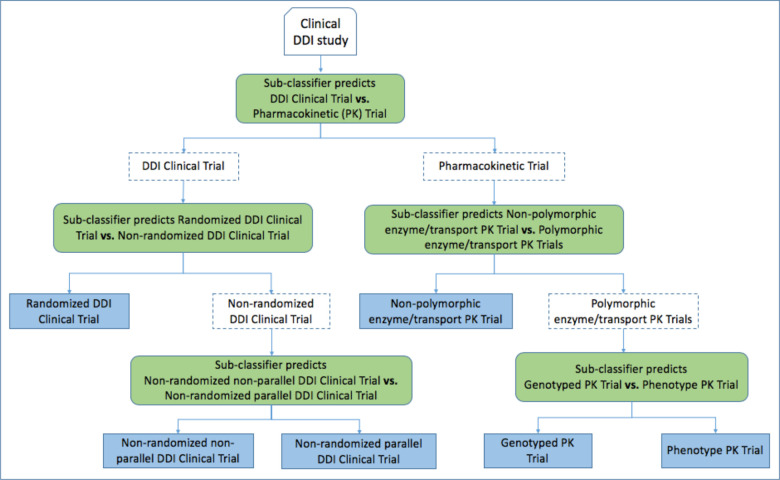
In the hierarchical classification system, a DDI clinical paper (one input instance) would be passed through a series of sub-classifiers until the paper reaches the lowest level of the evidence type hierarchy and is predicted with a specific evidence type. Corresponding to the hierarchy of evidence types shown in Figure 1, our final hierarchical classification system was a combination of five sub-classifiers, each designed as a binary classifier that distinguished a specific pair of evidence types, hierarchically divided into three levels (Figure 3). Each sub-classifier was trained using the support vector machine (SVM) algorithm (linear kernel, class weight balance applied).
Figure 3.
Design of the hierarchical classifier.
In order to prevent over-estimation of the classification system’s accuracy, we used cross-validation (5 folds) to randomly split the train and test sets to develop the sub-classifiers. More specifically, in each cross-validation iteration, data was randomly split into a training set and a testing set. All papers in the training dataset were used to train and test the top-level sub-classifier. A subset of the training set from the top-level classifier was passed down to and used to train the next lower level sub-classifiers following a particular path in the hierarchy. Similarly, a subset of the testing set from the top classifier was used to test the next level sub-classifiers. This process was repeated until the sub-classifiers at the lowest level were trained and tested.
Evaluation of the classification system
Precision, recall, balanced F measure (F1), and the area under the receiver operating characteristic curve (AUC ROC) were used to evaluate the classification performance during training. These metrics were calculated based on the predictions of each sub-classifier at each level against the actual labels of the papers at the same level. The four metrics were calculated for each sub-classifier for each cross-validation iteration. We then calculated the average of each metric, by dividing their sum by the number of cross-validation iterations (five). To take into account the hierarchical classifier structure, we supplemented these metrics with hierarchical precision, hierarchical recall, and F1 metrics for hierarchical classification systems20. We describe this in more detail in the next paragraph. The weighted average of the hierarchical classifier was calculated by dividing the sum of the hierarchical metrics by the number of cross validation (5 of them). After training the hierarchical classifier, we evaluated its performance on the two held-out datasets mentioned above.
The hierarchical metrics aggregate the predictions of all sub-classifiers for every single data point into their formula20. For example, suppose that an instance is classified into the label “Non RCT non parallel DDI Clinical trial” while it really belongs to label “Non RCT parallel DDI Clinical trial” (Figure 1). To calculate our hierarchical measure, we extend the set of real labels: {Actual Label} = {“Non RCT parallel DDI Clinical Trial”} with all its ancestors: {Actual Labels}’ = {“Non RCT parallel DDI Clinical Trial”, “Non RCT DDI Clinical Trial, “DDI Clinical Trial”}. We also extend the set of predicted labels: {Predicted Label} = {“Non RCT non parallel DDI Clinical Trial”} with all its ancestors: {Predicted Label}’ = {“Non RCT non parallel DDI Clinical Trial”, “Non RCT DDI Clinical Trial, “DDI Clinical Trial”}. Then, the hierarchical precision (hP), recall (hR) and F1 (hF) score were calculated based on the extended label sets as following:
According to these formulas, the number of correctly assigned labels for this instance from the extended set would be the union of the actual labels and the predicted labels, which is 2, instead of 0. This approach reduces the penalty for misclassification when the predicted label is “near” the actual label in the hierarchy.
Error Analysis
We obtained insight into the classification system’s behavior by examining the most informative features (unigrams) that were associated with each evidence type as ranked by a Pearson’s Chi-squared statistical test. We also looked at the papers that were given wrong predictions by the hierarchical classification system on the held-out datasets and analyzed the papers’ titles, abstracts and Method sections in order to identify the possible reasons for the wrong predictions.
Results
Classification Performance
Table 1 reports the sub-classifiers’ prediction performance on the training dataset. Table 2 shows the performance of the hierarchical classifier with the two held-out testing datasets.
Table 1.
Hierarchical classification system performance on the training dataset. Shown is the performance of each individual classifier at each level in the evidence hierarchy from Figure 1.
| Level | Sub-classifier |
Average AUC ROC (over 5 folds) |
Average Precision (over 5 folds) | Average Recall (over 5 folds) | Average F1 (over 5 folds) |
| 1 | DDI Clinical Trial vs. Pharmacokinetic Trial | 0.79 | 0.87 | 0.87 | 0.86 |
| 2 | Randomized DDI Clinical Trial vs. Non-randomized DDI Clinical Trial | 0.87 | 0.89 | 0.88 | 0.87 |
| 2 | Polymorphic enzyme/transport PK Trial vs. Non-polymorphic enzyme/transport PK Trial | 0.78 | 0.79 | 0.77 | 0.77 |
| 3 | Non-randomized parallel DDI Clinical Trial vs. Non-randomized non-parallel DDI Clinical Trial | 0.78 | 0.87 | 0.85 | 0.85 |
| 3 | Genotyped PK Trial vs. Phenotype PK Trial | 0.78 | 0.92 | 0.88 | 0.87 |
Table 2.
Classification performance of the hierarchical classifier on the two held-out datasets.
| Dataset | Hierarchical classifier | ||
| Hierarchical Precision | Hierarchical Recall | Hierarchical F1-score | |
| Performance on the held-out 32 papers about the same drugs as in the training set | 0.8 | 0.86 | 0.83 |
| Performance on the held-out 94 papers about entirely different drugs than in the training set | 0.81 | 0.51 | 0.63 |
Error Analysis
We printed out the most informative features (unigrams) associated with each evidence type, ranked by Chi-square scores. We found that evidence types that have terms strongly associated with them are easier to predict. For example, “Randomized DDI clinical trial” has terms related to this study design, including “random”, “double”, “blind”, while “Genotyped PK Trial” has terms related to drug metabolism and excretion, including “genotype” and “polymorphism”.
Another observation is that certain unigrams are highly correlated with some of the evidence types but not others, based on their Chi-square score ranking. For example, the “random” unigram and its variants (e.g. randomis, nonrandom) were given higher Chi-square scores and thus is more highly relevance ranked for RCT DDI Clinical Trial than any other types. In contrast, while the “genotyp” unigram was ranked as the most important unigram for Genotyped PK Trial evidence, it was ranked as less important for the others. Similarly, the “phenotyp” unigram is one of the top relevant unigrams for Phenotype PK Trial but not for Genotyped PK Trial. Table 3 shows the list of study design-associated features and their corresponding ranks in each evidence type.
Table 3.
Examples of the common unigrams between different evidence types and their rankings.
| Ancestor evidence type | Evidence type | Examples of features and their ranks (unigrams in stemmed format) |
| DDI Clinical Trial | Randomized DDI Clinical Trial | placebo (rank 7), crossov (rank 24), random (rank 26), doubl (rank 61), blind (rank 64), pharmacokinet (rank 1149) |
| Non-randomized (non-parallel) DDI Clinical Trial | placebo (rank 26), random (rank 84), blind (153), doubl (rank 317), crossov (rank 807), pharmacokinet (rank 3807) | |
| Non-randomized parallel DDI Clinical Trial |
crossov (rank 166), placebo (rank 579), blind (847), doubl (rank 1670), random (rank 3898), pharmacokinet (rank 7518) |
|
| PK Trial | Non-polymorphic enzyme/Transport PK Trial |
genotype (rank 84), phenotyp (rank 389), pharmacokinet (rank 580) |
| Genotyped PK Trial | genotype (rank 1), pharmacokinet (rank 398), phenotyp (rank 1218) | |
| Phenotype PK Trial | phenotyp (rank 49), pharmacokinet (rank 93), genotype (rank 1394) |
We also conducted an error analysis of incorrect predictions on the held-out 32 papers. Two examples are shown in Table 4. We found that the unigram features are not sufficient to assist the hierarchical classifier in making correct predictions in some cases where the study designs should be determined based on the whole context rather than single words. For example, in the first example of Table 4, the actual label is “Non RCT parallel group DDI trial”, however, there are no mentions of “parallel” in the text. Instead, the authors described the parallel design differently by using phrases such as “simultaneous and time-separated administration” and “in two treatment groups”. In the second example in Table 4, the classifier’s incorrect prediction of “RCT DDI Trial” was likely caused by the “doubl”, “blind” and “random” unigrams which are among the most informative features for the RCT DDI Trial evidence type. However, in this case, they occur in the context of describing a population pharmacokinetics study rather than a DDI study.
Table 4.
Examples of incorrect predictions of the hierarchical classifier on the held-out 32 papers.
| Example | Actual evidence type | Predicted evidence type | Sample text from the paper |
| 1 | Non RCT parallel DDI Trial | Non RCT non parallel DDI Trial |
Title: “Almorexant effects on CYP3A4 activity studied by its simultaneous and time-separated administration with simvastatin and atorvastatin.” Abstract: “…To characterise further the previously observed cytochrome P450 3A4 (CYP3A4) interaction of the dual orexin receptor antagonist almorexant. Pharmacokinetic interactions were investigated (n_=_14 healthy male subjects in two treatment groups) between almorexant at steady-state when administered either concomitantly…” |
| 2 | Non polymorphic enzyme transport PK Trial | RCT DDI Trial |
Title: “Population pharmacokinetics and pharmacodynamics of rivaroxaban--an oral, direct factor Xa inhibitor--in patients undergoing major orthopaedic surgery.” Abstract: “…This analysis was performed to characterize the population pharmacokinetics and pharmacodynamics of rivaroxaban in patients participating in two phase II, double-blind, randomized, active-comparator-controlled studies of twice-daily rivaroxaban for the prevention of venous thromboembolism after total hip- or knee-replacement surgery…” |
Discussion
The results suggest that it is feasible to accurately automate the classification of a sub-set of DDI evidence types. They also suggest that the hierarchical ensemble approach we tested based on the DIDEO evidence is a promising approach to build upon in future work. To our knowledge, this is the first study to test a hierarchical approach for classifying DDI clinical studies into highly specific evidence types. A 2020 study on extracting evidence of drug repurposing classified studies into more basic evidence types such as “Pre-clinical” (F1 = 0.96), “Clinical observational study” (F1 = 0.84), and “Clinical trial” (F1 = 0.80)21. Compared to that study, we target a more detailed set of evidence types. While further work will be necessary to improve the NLP performance and expand the classifiers to in vitro evidence types, this study is an important step towards more sophisticated computer support for DDI evidence synthesis.
Using existing knowledge about DDI evidence provided by the ontology was beneficial in several ways. First, the annotation process allowed us to identify evidence types that do not exist in the current ontology, resulting in novel evidence types that we plan to contribute to the ontology. Second, the ontology suggests an efficient design for the hierarchical classification system. Machine learning classifiers depend extensively on training data. Using the hierarchical structure of the evidence types suggested by the ontology helps to reduce the quantity of papers needed to train the classifiers successfully (especially in our case where the dataset is quite small to start with). This two-way feedback contributes to both the machine learning classifier development and can inform further development of the ontology. In future work, we plan to rigorously compare the performance of our hierarchical approach with alternative approaches to multi-class labeling.
The performance of the hierarchical classifier on two different held-out datasets did raise some possible concerns. While the precision scores are comparable between the two held-out datasets, recall and F1 on the second held-out set (different drugs) decrease significantly compared with the first held-out set (the same drugs as used in the training data). This result could be interpreted as: with “unseen” data (new drugs, different language and contexts), the hierarchical classifier did a good job picking up true positives but also produced a high number of false negatives. This could be an indication that the current features generated from the training set might favor some evidence type labels over others. In future work we plan to test if this is an issue with the classifier, feature engineering techniques, or both.
The error analysis results suggest improvements for the classification system in the future. We think that more sophisticated representation of key entities such as drugs and enzymes might be useful as features to enhance the prediction performance. In this pilot study, we implemented a preprocessing approach using Metamap and regular expressions to remove all of the drug entities and numeric strings from the text, which helped us to avoid potential overfitting problems. However, one of the strong textual indications to distinguish genotyped papers from phenotype papers is mention of genes or enzymes (like CYP2B6 or CYP2C19) along with certain symbolic/numeric strings. However, when we tested keeping these drug-related features as unigrams, we observed significant overfitting. More advanced methods such as term embedding (e.g., word2vec22) might overcome these issues by de-emphasizing term syntax and increasing the emphasis on term context. Also, we think that relationships between the drug entities should also be taken into account, such as information about which drugs play object vs. precipitant roles in the interactions. We learned from this study that evidence type associated words/phrases are important but that contextual features would be helpful to more accurately classify complex edge cases.
Our study has several potential limitations. The training and held out datasets were relatively small. To overcome this problem, we need to obtain and have experts annotate more data. Alternately, a computational approach to increasing the annotated data would be to semi-automatically collect and label the data using techniques such as rule-based distant supervision23. Another limitation is that our implementation used all features (unigrams) to train all of the sub-classifiers regardless of which labels they were predicting. In the future, developing different feature selection strategies tailored to different sub-classifiers could be helpful, because in the error analysis, we found that some specific words and phrases (especially the ones indicating study design) are more important for some labels than others.
Conclusion
We combined machine learning and knowledge representation in a pilot experiment classifying evidence types of from the DDI literature. Drawing on an existing ontology of evidence types, DIDEO, we built a hierarchical classifier, which combined a series of sub-classifiers to categorize a DDI study’s evidence type. In the future, such an automatic classification system could be a key component of a computerized system to help experts be more efficient and objective in DDI evidence assessment, ultimately assisting drug experts as they assess evidence items. Other promising applications of the technology would be to support automatic identification of new clinical DDI papers, and to help extend the DIDEO ontology to new evidence sub-types.
Acknowledgements
This study was funded in part by two grants from the National Library of Medicine: “Addressing gaps in clinically useful evidence on drug-drug interactions” (R01LM011838) and “Text Mining Pipeline to Accelerate Systematic Reviews in Evidence-based Medicine” (R01LM010817).
Figures & Table
References
- 1.van der Sijs H, Aarts J, van Gelder T, Berg M, Vulto A. Turning off frequently overridden drug alerts: limited opportunities for doing it safely. J Am Med Inform Assoc. 2008;15(4):439–448. doi: 10.1197/jamia.M2311. doi:10.1197/jamia.M2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheife RT, Hines LE, Boyce RD, Chung SP, Momper JD, Sommer CD, Abernethy DR, Horn JR, Sklar SJ, Wong SK, Jones G. Consensus recommendations for systematic evaluation of drug-drug interaction evidence for clinical decision support. Drug Saf. 2015;38(2):197–206. doi: 10.1007/s40264-014-0262-8. doi: 10.1007/s40264-014-0262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sirajuddin AM, Osheroff JA, Sittig DF, Chuo J, Velasco F, Collins DA. Implementation pearls from a new guidebook on improving medication use and outcomes with clinical decision support. Effective CDS is essential for addressing healthcare performance improvement imperatives. J Healthc Inf Manag. 2009 Fall;23(4):38–45. [PMC free article] [PubMed] [Google Scholar]
- 4.Fung KW, Kapusnik-Uner J, Cunningham J, Higby-Baker S, Bodenreider O. Comparison of three commercial knowledge bases for detection of drug-drug interactions in clinical decision support. J Am Med Inform Assoc. 2017;24(4):806–812. doi: 10.1093/jamia/ocx010. doi:10.1093/jamia/ocx010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romagnoli KM, Nelson SD, Hines L, Empey P, Boyce RD, Hochheiser H. Information needs for making clinical recommendations about potential drug-drug interactions: a synthesis of literature review and interviews. BMC Med Inform Decis Mak. 2017;17(1):21. doi: 10.1186/s12911-017-0419-3. doi:10.1186/s12911-017-0419-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bottiger Y, Laine K, Andersson ML, Korhonen T, Molin B, Ovesjo ML, Tirkkonen T, Rane A, Gustafsson LL, Eiermann B. SFINX—a drug-drug interaction database designed for clinical decision support systems. Eur J Clin Pharmacol. 2009;65(6):627–633. doi: 10.1007/s00228-008-0612-5. doi:10.1007/s00228-008-0612-5. [DOI] [PubMed] [Google Scholar]
- 7.Seden K, Gibbons S, Marzolini C, Schapiro JM, Burger DM, Back DJ, Khoo SH. Winston A, editor. Development of an evidence evaluation and synthesis system for drug-drug interactions, and its application to a systematic review of HIV and malaria co-infection. PLoS One. 2017;12(3):e0173509. doi: 10.1371/journal.pone.0173509. doi:10.1371/journal.pone.0173509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grizzle AJ, Hines LE, Malone DC, Kravchenko O, Hochheiser H, Boyce RD. Testing the face validity and inter-rater agreement of a simple approach to drug-drug interaction evidence assessment. J Biomed Inform. 2020;101:103355. doi: 10.1016/j.jbi.2019.103355. doi:10.1016/j.jbi.2019.103355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DIDEO - The Potential Drug-drug Interaction and Potential Drug-drug Interaction Evidence Ontology [Internet] [cited 2020 Mar 8]. Available from: http://www.ontobee.org/ontology/DIDEO .
- 10.Boyce R, Collins C, Horn J, Kalet I. Computing with evidence. J Biomed Inform. 2009;42(6):979–989. doi: 10.1016/j.jbi.2009.05.001. doi: 10.1016/j.jbi.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brochhausen M, Schneider J, Malone D, Empey PE, Hogan R, Boyce RD. Towards a foundational representation of potential drug- drug interaction knowledge. First International Workshop on Drug Interaction Knowledge Representation at ICBO. 2014. http://ceur-ws.org/Vol-1309/paper2.pdf . [PMC free article] [PubMed]
- 12.Boyce R, Collins C, Horn J, Kalet I. Computing with evidence: Part I: A drug-mechanism evidence taxonomy oriented toward confidence assignment. J Biomed Inform. 2009;42(6):979–989. doi: 10.1016/j.jbi.2009.05.001. doi:10.1016/j.jbi.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyce R, Collins C, Horn J, Kalet I. Modeling drug mechanism knowledge using evidence and truth maintenance. IEEE Trans Inf Technol Biomed. 2007;11(4):386–397. doi: 10.1109/titb.2007.890842. doi: 10.1109/TITB.2007.890842. [DOI] [PubMed] [Google Scholar]
- 14.Schneider J, Brochhausen M, Rosko S, Ciccarese P, Hogan WR, Malone DC, Ning Y, Clark T, Boyce RD. Formalizing knowledge and evidence about potential drug-drug interactions. Biomedical Data Mining, Modeling, and Semantic Integration at ISWC. 2015. http://ceur-ws.org/Vol-1428/BDM2I_2015_paper_10.pdf .
- 15.DDI_Evidence_Classification [Internet]. GitHub. 2020. [cited 2020 Mar 8]. Available from: https://github.com/infoqualitylab/DDI_Evidence_Classification .
- 16.APIs - Develop - NCBI [Internet]. National Center for Biotechnology Information. U.S: National Library of Medicine; [cited 2020 Mar 8]. Available from: https://www.ncbi.nlm.nih.gov/home/develop/api/ . [Google Scholar]
- 17.Pdfminer [Internet]. GitHub. 2020. [cited 2020 Mar 8]. Available from: https://github.com/pdfminer/pdfminer.six .
- 18.Natural Language Toolkit [Internet]. [cited 2020 Mar 8] Available from: https://www.nltk.org/
- 19.MetaMap - A Tool For Recognizing UMLS Concepts in Text [Internet] U.S. National Library of Medicine. National Institutes of Health; [cited 2020 Mar 8]. Available from: http://metamap.nlm.nih.gov/ . [Google Scholar]
- 20.Kiritchenko S, Matwin S, Famili AF. Proceedings of BioLINK SIG: Linking Literature, Information and Knowledge for Biology at ISMB-05. Michigan: 2005. Jun 24, Functional annotation of genes using hierarchical text categorization. [Google Scholar]
- 21.Subramanian S, Baldini I, Ravichandran S, Katz-Rogozhnikov DA, Ramamurthy KN, Sattigeri P, Varshney KR, Wang A, Mangalath P, Kleiman LB. Proceedings of The Thirty-Second Annual Conference on Innovative Applications of Artificial Intelligence IAAI-20. New York: 2020 Feb 9. A natural language processing system for extracting evidence of drug repurposing from scientific publications; 11 pp. [Google Scholar]
- 22.Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Proceedings of Advances in neural information processing systems NIPS. California: 2013 Dec 510. Distributed representations of words and phrases and their compositionality. [Google Scholar]
- 23.Mintz M, Bills S, Snow R, Jurafsky D. Distant supervision for relation extraction without labeled data. Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP; Singapore. Association for Computational Linguistics; 2009 August. pp. 1003–1011. doi: 10.3115/1690219.1690287. [Google Scholar]