Abstract
Clinical judgment studies are an integral part of drug safety surveillance and pharmacovigilance frameworks. They help quantify the causal relationship between medication and its adverse drug reactions (ADRs). To conduct such studies, physicians need to review patients’ charts manually to answer Naranjo questionnaire1. In this paper, we propose a methodology to automatically infer causal relations from patients’ discharge summaries by combining the capabilities of deep learning and statistical learning models. We use Bidirectional Encoder Representations from Transformers (BERT)2 to extract relevant paragraphs for each Naranjo question and then use a statistical learning model such as logistic regression to predict the Naranjo score and the causal relation between the medication and an ADR. Our methodology achieves a macro-averaged f1-score of 0.50 and weighted f1-score of 0.63.
Introduction
Pharmacovigilance is an important research area in medical informatics and aims at evaluating the safety of medication usage and thereby improving patient safety. Clinical judgement studies that extract causal relations between drugs and adverse drug reactions (ADRs) are essential parts of Pharamcovigilance. An ADR can be loosely defined as any noxious, unintended or undesired effect of a medicine after doses used in humans for prophylaxis, diagnosis or therapy1. ADRs are the single largest contributor to hospital-related complications in inpatient settings3 and occur commonly at a rate of 2.4-5.2 per 100 hospitalized adult patients4–6. Anticoagulants are one of the most common drug classes that cause numerous ADRs, accounting for approximately 1 in every 10 of all drug-related adverse outcomes (specifically bleeding events)7 and one-third of all ADRs among hospitalized Medicare patients8.
Prior research work in this domain has been mainly focused on extracting drug and ADR entities9,10 or identifying relations between them from electronic health records11. However extracting relation between a drug and an ADR is quite different from inferring causality between the medication and ADRs. The relation also does not provide the level of causality which can be inferred from causality scores such as doubtful, possible, probable or definite. As described by the authors of Naranjo questionnaire1, the suspected medication is usually confounded with other causes, and the adverse reaction cannot be easily distinguished from the manifestations of the disease making it significantly harder to extract the accurate relation between the drug and ADRs. Hence, there is a need of formulating the problem of causal relation extraction in a different way to automate such clinical studies.
Due to the lack of an established methodology for clinical studies, Naranjo scale was developed to standardize the causality assessment of ADRs1. Naranjo scale is frequently used by physicians to conduct causality assessment studies between a medication and ADRs12,13. It comprises of 10 questions and a subset of these questions is shown in Table 1. A causality scale (e.g., doubtful or probable) is assessed based on the answers to those questions. Naranjo scale has shown a marked improvement in within-raters agreement, reproducibility, reliability as compared to other approaches1. One strength of the Naranjo scale is that it can handle missing values: the scale is valid even with the answers to some of the questions are missing. Therefore, Naranjo scale has been widely used as a standard in clinical domain.
Table 1:
Naranjo Scale Questionnaire.
| # | Naranjo Questions | Yes | No | Do not know |
| 1. | Are there previous conclusive reports on this reaction? | 1 | 0 | 0 |
| 2. | Did the adverse event occur after the suspected drug was administered? |
2 | -1 | 0 |
| 3. | Did the adverse reaction improve when the drug was discontinued or a specific antagonist was administered? |
1 | 0 | 0 |
| 4. | Did the adverse reaction reappear when the drug was readministered? |
2 | -1 | 0 |
| 5. | Are there alternative causes (other than the drug) that could have on their own cause the reaction? |
-1 | 2 | 0 |
| 6. | Did the reaction reappear when a placebo was given? | -1 | 1 | 0 |
| 7. | Was the drug detected in the blood (or other fluids) in concentrations known to be toxic? |
1 | 0 | 0 |
| 8. | Was the reaction more severe when the dose was increased or less severe when the dose was decreased? |
1 | 0 | 0 |
| 9. | Did the patient have a similar reaction to the same or similar drugs in any previous exposure? |
1 | 0 | 0 |
| 10. | Was the adverse event confirmed by any objective evidence? |
1 | 0 | 0 |
Previous clinical judgement studies have solely been conducted on time-consuming manual chart reviews of electronic health records (EHRs) which require significant manual efforts by experienced physicians. As such, these studies are usually conducted only on a subset of clinical notes due to multiple time constraints. To facilitate clinical judgement studies, we propose an end-to-end methodology which employs a deep learning model, BERT2, with statistical models to predict the causal relation between a medication and its ADRs. In this study, we investigate the causal relation between anticoagulants and bleeding events.
Our contributions are mainly three-folds:
We propose a methodology to predict the causal relation between a medication and its ADRs by accessing the Naranjo score based on Naranjo questionnaire. Our work may be a significant contribution to drug safety surveillance and pharmacovigilance, as the current practice relies on the labour-intensive process of domain-experts who manually chart-review the EHRs.
By effectively integrating deep learning and statistical modeling, our model provides a decent macro-averaged f-score of 0.50 and a weighted f1-score of 0.63.
To the best of our knowledge, our model is the first attempt at predicting the causal relation directly from the EHRs using Naranjo questions. Our work could be used as a strong baseline for further related research.
Naranjo Scale and Dataset
Naranjo Scale
The Naranjo Scale Questionnaire consists of 10 questions which are administered for each patient’s clinical note. Each question can be answered as “Yes”, “No” or “Do not know” , where “Do not know” is marked when the quality of the data does not allow an affirmative (yes) or negative (no) answer.
A score of {−1, 0, 1, 2} is assigned to each question as shown in Table 1. The Naranjo scale assigns a causality score, which is the sum of the scores of all questions, that falls into one of four causality types: doubtful (≤ 0), possible (1 − 4), probable (5 − 8), and definite (≥ 9). In clinical settings, it is typically rare to find answers for all 10 Naranjo questions. The Naranjo scale is designed such that it is valid even if the answers for only a subset of the Naranjo questionnaire are provided.
Cohort Selection
We built an expert annotated EHR cohort to be used for training and evaluation of our proposed model. We selected the clinical notes of patients who were administered one of these six anticoagulants: Apixaban, Clopidogrel, Dibigatran, Enoxaparin, Rivaroxaban and Warfarin. To increase the chance that the notes also contain ADRs, we focused on the patients who had any signs of internal bleeding such as gastrointestinal bleeding, blood clots or black tarry stools as these are the most common ADRs of anticoagulants. Physician annotators manually examined those notes and provided answers for each Naranjo question. The physicians provided granular information by annotating the relevant sentence in the EHR and then the answer of the related Naranjo question as one of the three answers: ‘Yes’, ‘No’ and ‘Do not know’. Experts provided two levels of annotation: the relevant sentences and answer for questions in the Naranjo questionnaire. Not all questions can be answered for an EHR since there may be no relevant information regarding some questions such as question 6 of Naranjo questionnaire as not all patients are provided with placebo during their treatment.
Dataset
Our dataset consists of discharge summaries of 991 unique patients. Since some of the patients were admitted more than once, there are 1385 discharge summaries in total. Four physicians, supervised by a senior physician, annotated the Naranjo scale questionnaire for each of these discharge summaries. Each discharge summary was annotated by one of the four physician independently. Reconciliation was done by the senior physician who examined every annotation and discussed the differences with other physicians. Each discharge summary could have multiple ADRs, each of which could have a different Naranjo questionnaire. Our model attempts to detect all of the ADRs and their corresponding questionnaires and answers. The distribution of unique patients and discharge summaries across six anticoagulants: Apixaban, Clopidogrel, Dibigatran, Enoxaparin, Rivaroxaban and Warfarin is shown in Table 2.
Table 2:
Distribution of unique patients and their discharge summaries across different anticoagulants.
| Anitcoagulant | # Unique Patients | # Discharge Summaries |
| Dabigtran | 38 | 48 |
| Apixaban | 82 | 121 |
| Rivaroxaban | 85 | 116 |
| Enoxaparin | 141 | 181 |
| Clopidogrel | 169 | 212 |
| Warfarin | 476 | 707 |
Since we are only interested in the questions that can be answered from the information provided in the discharge summary, we omitted the first question from our study. Similarly, question 6 was also eliminated as most of the patients are not provided placebo during their treatment. All the remaining questions were answered by the physicians. Questions 2, 3, 5, 7 and 10 were most frequently answered by the experts. Most of the answers (90% or more), for 4 questions, out of the remaining 9, were “Do not know”. Thus, major contribution in the Naranjo score is from the remaining 5 questions: 2, 3, 5, 7 and 10 which are shown in Table 1. As described earlier, the imbalanced answer distribution is typical for Naranjo scale assessment and it would still be clinically meaningful even if only a subset of the Naranjo questions could be answered.
Calculating Naranjo Score
The Naranjo score for each question is calculated according to the Table 1. If an answer is not annotated for any Naranjo question because of a lack of information provided in the EHR, it is considered as “Do not know”. The final Naranjo score (Nscore) is calculated by summing the scores of all questions. According to the total Naranjo score, a label for the causal relation, according to the conditions: doubtful (≤ 0), possible (1 − 4), probable (5 − 8), and definite (≥ 9), is assigned to each discharge summary. The condition as well as distribution for each causal relation is shown in Table 3
Table 3:
Distribution and condition for each causal relation between the medication and its ADRs.
| Causal Relation | Condition | # Discharge Summaries |
| Doubtful | Nscore ≤ 0 | 183 |
| Possible | 1 ≤ Nscore ≤ 4 | 916 |
| Probable | 5 ≤ Nscore ≤ 8 | 283 |
| Definite | 9 ≤ Nscore | 3 |
Methodology
In this section we discuss the problem formulation and briefly explain BERT2, which we use to extract relevant paragraphs from the EHRs, and then explain our methodology to predict the final Naranjo score.
Problem Formulation
As mentioned in the previous section, our annotators went through each of the clinical note meticulously and annotated all the ADRs with their corresponding Naranjo question-answers. The annotation resulted in two levels of information: relevant sentence for which the Naranjo question has been answered and answer (“Yes”, “No”, and “Do not Know”) for the specific Naranjo question. For example, the sentence “In ED, she was found to have a hgb of 9, INR 3.6, and rectal exam in ED revealed maroon stool” as shown in Figure 1 was annotated as a relevant sentence to answer the Naranjo question 2 for the ADR ”maroon stool” (the answer is “yes”). The sentences around the relevant sentence is also quite important as it provides context to that sentence. We consider this group of contiguous sentences as a paragraph. If a paragraph has even one relevant sentence then it is considered as a relevant paragraph otherwise non-relevant. For constructing a paragraph, we kept the length of contiguous sentences as variable, between 15 − 20, to make our model more robust.
Figure 1:
A synthetic example showing a paragraph, naranjo question and its answer. The relevant sentence of the paragraph is highlighted in green.
Given a discharge summary, a model should be able to identify the relevant paragraphs according to each Naranjo question. A paragraph could be relevant with respect to one Naranjo question but non-relevant with respect to another question. The relevant paragraphs should be used to predict either the causal relation or the final Naranjo score (Nscore) for the discharge summaries. Since our final aim is to predict the causal relation, the Nscore is used to get the causal relation according to the conditions enumerated in Table 3.
Bidirectional Encoder Representations from Transformers (BERT)
BERT2 uses multi-layer bidirectional Transformer14 networks to encode contextualised language representations. BERT representations are learned from two tasks: masked language modeling15 and next sentence prediction task. We chose BERT model as pre-trained BERT models, since fine-tuned pre-trained BERT models have achieved state-of-the-art results for a wide range of tasks such as question answering and multiple language inference tasks2. We utilised clinicalBERT16 for our experiments as it yielded superior performance on clinical-related NLP tasks such as i2b2 named entity recognition (NER) challenges17. clinicalBERT is created by further fine-tuning of BERT base2 with biomedical and clinical corpus (MIMIC-III)18. clinicalBERT is further fine-tuned for the classification task where it predicts whether the paragraph is relevant or not-relevant, given a question and the paragraph itself.
Proposed Methodology
Our proposed methodology consists of different parts: relevant paragraph selection, feature extraction and then causal relation prediction using the features extracted from the relevant paragraphs.
Relevant Paragraph Prediction
As explained in the Problem Formulation section, a paragraph is created from a contiguous set of sentences. If the paragraph has even one sentence which has been marked as relevant for a specific question, it is considered as relevant with respect to that question. We used clinicalBERT16 to create the classification model. clinicalBERT takes both question and the paragraph as input and predicts whether the paragraph is relevant or not. The paragraph is paired with each Naranjo question because it might have relevant information with regards to one question but not the other. In our model, we first identify relevant paragraphs. The relevant paragraphs are then passed for feature extraction before passing to statistical models.
The question and the paragraph are appended to each other with a separator in between [SEP] so that BERT can differentiate between the question and the paragraph. Another token [CLS] is appended in the starting of the sequence resulting in final sequence = [CLS]+ question +[SEP]+ paragraph. This final sequence is passed through BERT, which consists of multiple attention layers and attention heads14, to provide a contextual representation for each token. The sequence representation is provided by the output representation of [CLS] token. This [CLS] representation is passed through a softmax layer to provide the output probabilities for relevant and non-relevant label.
Feature Extraction
All relevant paragraphs are required for predicting the final causal relation between the medication and the ADR. There are two set of features that are extracted from the set of relevant paragraphs: n-grams and tf-idf.
n-g rams: An n-gram is a contiguous sequence of n words from the text corpus. They have shown to improve the performance of the model in several natural language processing tasks19. The n-grams extracted from the training corpus act as the features for training the statistical model. The n-grams capture important information. For example, the bi-gram ‘severe bleeding’ could capture the information which uni-grams ‘severe’ and ‘bleeding’ may not capture independently and ‘severe’ may be used in the corpus in non-relevant context such as ‘severe back pain’. We extract uni-grams, bi-grams, tri-grams and quad-grams for our model training.
tf-idf: Term frequency-inverse document frequency20 (tf-idf) reflects on the importance of a word to a document in the whole corpus. The tf-idf value for a word increases proportionally to the frequency of the word in the document and is offset by the number of documents in the corpus that contain the word. The offset helps in adjusting for the fact that some words appear more frequently in general such as ‘the’ and ‘in’. We used tf-idf for all n-grams: uni-grams, bi-grams, tri-grams and quad-grams. The tf-idf features convey the importance of each n-gram for a document.
Causal Relation Predication
The model could either directly predict the causal relation between the medication and its ADRs or predict the Naranjo score (Nscore) which could be used to infer the causal relation according to the conditions mentioned in Table 3. The first approach can be referred to as a classification approach and latter can be referred to as regression approach. We use multiple models for each approach.
Classification approach: For classification, we used multinomial naive bayes21, logistic regression22 and support vector machine23 (SVM). All these three supervised machine learning models have been widely used for statistical prediction modeling. We fine-tuned all the models for their different hyper-parameters before using them for predicting causal relation label (doubtful, probable, possible and definite) on testing corpora.
Regression approach: For this approach, we used linear regression24, ridge regression25 and support vector regression26 (SVR). All these models were also fine-tuned over their hyper-parameters before using for Nscore prediction on testing corpora.
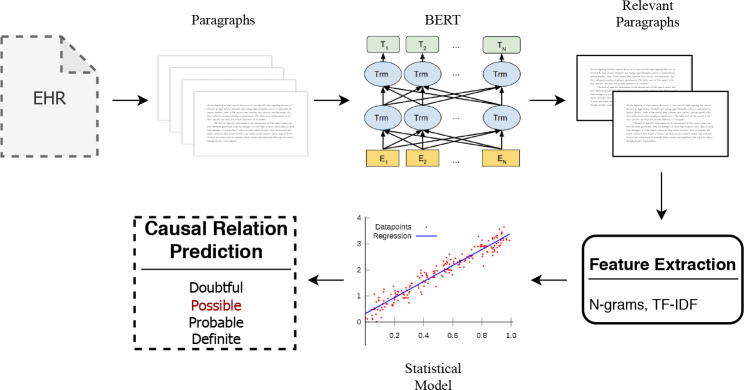
The final proposed model is illustrated in Fig. 2
Figure 2:
Proposed Model: Each electronic health record (EHR) is split up into multiple paragraphs which are passed through the BERT Model. BERT predicts the relevant paragraphs which are used to extract textual features such as n-grams and TF-IDF. These features are then passed through a statistical model such as logistic regression to predict the causal relation label.
Results and Discussion
We report the results by each model using the unseen test corpus. In our experiments, the dataset was pre-divided into training, validation and testing corpus in the ratio of 60 : 10 : 30. The evaluation metrics that compare the models against each other are explained below.
Evaluation Metrics: We evaluated our models on precision, recall and f-score metrics as the final causal relation prediction is limited to four classes: definite, probable, possible and definite. We report both weighted and macro-averaged precision recall and f-score for all models in Table 5. Macro-averaged metrics are calculated by averaging the performance across the labels and thus provide better insight on models’ performance across different labels. Whereas weighted metrics are calculated by averaging the weighted performance of each label according to the labels’ frequency making it more biased towards the most frequent label, which is possible in our case.
Table 5:
Macro-averaged and weighted precision, recall and f1-score for all models. The best performing scores are highlighted.
| Macro | Weighted | ||||||
| Model | precision | recall | f1-score | precision | recall | f1-score | |
| Multi-NB | 0.50 | 0.50 | 0.50 | 0.61 | 0.62 | 0.61 | |
| Classification | Logistic | 0.51 | 0.48 | 0.49 | 0.61 | 0.62 | 0.61 |
| SVM | 0.57 | 0.44 | 0.45 | 0.63 | 0.66 | 0.64 | |
| Linear | 0.36 | 0.38 | 0.36 | 0.52 | 0.64 | 0.56 | |
| Regression | Ridge | 0.36 | 0.38 | 0.36 | 0.52 | 0.64 | 0.56 |
| SVR | 0.39 | 0.43 | 0.40 | 0.54 | 0.65 | 0.59 | |
| Ensemble | 0.53 | 0.49 | 0.50 | 0.63 | 0.62 | 0.62 | |
Results: The relevant paragraph prediction results for clinicalBERT are provided in Table 4 and overall causal relation prediction results are provided in Table 5. The mutinomial naive bayes model is referred to as Multi-NB, similarly, logistic regression is referred to as Logistic, support vector machine as SVM, linear regression as Linear, ridge regression as Ridge and support vector regression as SVR. The hyper-parameters for the best performing models are provided in Appendix A in Table 6.
Table 4:
Macro-averaged precision, recall and f1-score for relevant paragraph prediction by clinicalBERT.
| Macro-averaged | |||
| Label | precision | recall | f1-score |
| Relevant | 0.73 | 0.92 | 0.81 |
| Non-Relevant | 0.96 | 0.87 | 0.91 |
| Overall | 0.85 | 0.89 | 0.86 |
clinicalBERT achieves a macro-weighted f1-score of 0.86 which suggests that the model is able to categorize the relevant and non-relevant paragraphs quite efficiently. clinicalBERT has low precision for relevant label which results in decreasing the overall performance of the model. The recall of clinicalBERT is quite high (0.90) and it is desired as well because it would result in extracting most of the relevant paragraphs from the EHR.
Multi-NB achieved the best macro-averaged f1-score of 0.50 amongst all the models. It also achieved the highest macro-weighted recall of 0.50 which suggests that it was able to correctly predict the most number of causal relations across labels. Though SVR was able to achieve the highest precision of 0.57, it under-performed in terms of recall resulting in lower f1-score as compared to Multi-NB. Overall, the classification models performed better than regression models which suggests that it is easier for the statistical learning model to create boundaries across causal relation labels than over the naranjo scores (Nscore).
SVM achieved the best weighted f1-score of 0.64 along with highest weighted precision and weighted recall of 0.63 and 0.66. Multi-NB and Logistic achieved a weighted f1-score of 0.61 which is quite close to the performance of SVM. The weighted evaluation metrics are higher as compared to macro-averaged evaluation metrics suggesting that the models perform better for the label with the highest frequency, which is possible in our dataset. For weighted evaluation metrics, as well, the classification models performed better than the regression models. These results suggest that the causal relation, between a medication and its ADRs, can be predicted with the help of deep learning and statistical models.
We also created multiple ensemble using our classification and regression model, the best ensemble consisted of Multi-NB and SVM which achieved the same macro-averaged f1-score as Multi-NB but improved the weighted f1-score of the model to 0.62. The precision of the ensemble model improved by 0.03 but reduction in recall resulted in the same f1-score as Multi-NB. Such ensemble model could be used for situations where higher precision model is desired for a clinical judgement study.
Conclusion
In this paper, we demonstrate that the causal relation, between a medication such as an anticoagulant and its ADRs such as bleeding, can be predicted with the help of our proposed methodology using Naranjo questionnaire. We show that the deep learning models could be used to extract the relevant paragraphs from the EHRs according to each Naranjo questionnaire1. These relevant paragraphs could then be used to extract textual features and predict the causal relation directly or Naranjo score using statistical learnning models. Our proposed methodology achieves a macro-averaged f1-score of 0.50 and weighted f1-score of 0.64 and provides a strong baseline for future research in this direction. To the best of our knowledge, this is the first study to automate the clinical judgement study by directly predicting the causal relation, over an EHR, between a medication and its ADRs using Naranjo questionnaire.
Acknowledgement
We would like to thank Drs. Steve Belknap, William Temps, and Edgard Granillo and Ms. Nadya Frid for annotating the discharge summaries regarding Naranjo Questionnaire and William Temps for also extracting the raw EHR data for experiments.
Appendix A
Table 6:
Hyper-parameters for the best performing models.
| Model | n-grams | Kernel | alpha | epsilon | use IDF |
| Multi-NB | bi-grams | - | - | - | TRUE |
| Logistic | tri-grams | - | - | - | TRUE |
| SVM | tri-grams | Linear | 1.00E-04 | - | TRUE |
| Linear Reg | bi-grams | - | - | - | TRUE |
| Ridge Reg | bi-grams | - | - | - | TRUE |
| SVR | bi-grams | Linear | - | 0.3 | TRUE |
Figures & Table
References
- 1.Naranjo Cla´udio A, Busto Usoa, Sellers Edward M, Sandor P, Ruiz I, Roberts EA, Janecek E, Domecq C, Greenblatt DJ. A method for estimating the probability of adverse drug reactions. Clinical Pharmacology & Therapeutics. 1981;30(2):239–245. doi: 10.1038/clpt.1981.154. [DOI] [PubMed] [Google Scholar]
- 2.Devlin Jacob, Chang Ming-Wei, Lee Kenton, Toutanova Kristina. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv. 2018;1810.04805 [Google Scholar]
- 3.Classen David C, Resar Roger, Griffin Frances, Federico Frank, Frankel Terri, Kimmel Nancy, Whittington John C, Frankel Allan, Seger Andrew, James Brent C. ‘global trigger tool’shows that adverse events in hospitals may be ten times greater than previously measured. Health affairs. 2011;30(4):581–589. doi: 10.1377/hlthaff.2011.0190. [DOI] [PubMed] [Google Scholar]
- 4.Bates David W, Spell Nathan, Cullen David J, Burdick Elisabeth, Laird Nan, Petersen Laura A, Small Stephen D, Sweitzer Bobbie J, Leape Lucian L. The costs of adverse drug events in hospitalized patients. Jama. 1997;277(4):307–311. [PubMed] [Google Scholar]
- 5.Classen David C, Pestotnik Stanley L, Scott Evans R, Lloyd James F, Burke John P. Adverse drug events in hospitalized patients: excess length of stay, extra costs, and attributable mortality. Jama. 1997;277(4):301–306. [PubMed] [Google Scholar]
- 6.Nebeker Jonathan R, Hoffman Jennifer M, Weir Charlene R, Bennett Charles L, Hurdle John F. High rates of adverse drug events in a highly computerized hospital. Archives of internal medicine. 2005;165(10):1111–1116. doi: 10.1001/archinte.165.10.1111. [DOI] [PubMed] [Google Scholar]
- 7.Lucado Jennifer, Paez Kathryn, Elixhauser A. Medication-related adverse outcomes in us hospitals and emergency departments, 2008. statistical brief#. 2006;109 [PubMed] [Google Scholar]
- 8.Levinson Daniel R, Inspector General Adverse events in hospitals: national incidence among medicare beneficiaries. Department of Health and Human Services Office of the Inspector General. 2010.
- 9.Jagannatha Abhyuday N, Yu Hong. Bidirectional rnn for medical event detection in electronic health records. Proceedings of the conference. Association for Computational Linguistics. North American Chapter. Meeting. volume 2016:473. doi: 10.18653/v1/n16-1056. NIH Public Access, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aramaki Eiji, Miura Yasuhide, Tonoike Masatsugu, Ohkuma Tomoko, Masuichi Hiroshi, Waki Kayo, Ohe Kazuhiko. Extraction of adverse drug effects from clinical records. MedInfo. 2010;160:739–743. [PubMed] [Google Scholar]
- 11.Munkhdalai Tsendsuren, Liu Feifan, Yu Hong. Clinical relation extraction toward drug safety surveillance using electronic health record narratives: Classical learning versus deep learning. JMIR public health and surveil- lance. 2018;4(2) doi: 10.2196/publichealth.9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma Rohini, Dogra Devraj, Dogra Naina. A study of cutaneous adverse drug reactions at a tertiary center in jammu, india. Indian dermatology online journal. 2015;6(3):168. doi: 10.4103/2229-5178.156384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shamna M, Dilip C, Ajmal M, Linu Mohan P, Shinu C, Jafer CP, Mohammed Yahiya. A prospective study on adverse drug reactions of antibiotics in a tertiary care hospital. Saudi pharmaceutical journal. 2014;22(4):303–308. doi: 10.1016/j.jsps.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaswani Ashish, Shazeer Noam, Parmar Niki, Uszkoreit Jakob, Jones Llion, Gomez Aidan N, Kaiser Łukasz, Polosukhin Illia. Attention is all you need. Advances in Neural Information Processing Systems. 2017. pp. 5998–6008.
- 15.Taylor Wilson L. “cloze procedure”: A new tool for measuring readability. Journalism quarterly. 1953;30(4):415–433. [Google Scholar]
- 16.Alsentzer Emily, Murphy John R, Boag Willie, Weng Wei-Hung, Jin Di, Naumann Tristan, McDermott Matthew. Publicly available clinical bert embeddings. arXiv preprint arXiv:1904.03323. 2019.
- 17.Uzuner O¨ zlem, South Brett R, Shen Shuying, DuVall Scott L. 2010 i2b2/va challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association. 2011;18(5):552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson Alistair EW, Pollard Tom J, Shen Lu, Li-wei H Lehman, Feng Mengling, Ghassemi Mohammad, Moody Benjamin, Szolovits Peter, Celi Leo Anthony, Mark Roger G. Mimic-iii, a freely accessible critical care database. Scientific data. 2016;3:160035. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sidorov Grigori, Velasquez Francisco, Stamatatos Efstathios, Gelbukh Alexander, Chanona-Herna´ndez Liliana. Syntactic n-grams as machine learning features for natural language processing. Expert Systems with Applications. 2014;41(3):853–860. [Google Scholar]
- 20.Juan Ramos, et al. Using tf-idf to determine word relevance in document queries; Proceedings of the first instructional conference on machine learning; Piscataway, NJ. 2003. pp. 133–142. [Google Scholar]
- 21.Kibriya Ashraf M, Eibe Frank, Bernhard Pfahringer, Geoffrey Holmes. Multinomial naive bayes for text categorization revisited; Australasian Joint Conference on Artificial Intelligence; Springer. 2004. pp. 488–499. [Google Scholar]
- 22.Scott Menard. Applied logistic regression analysis. Vol. 106. Sage; 2002. [Google Scholar]
- 23.Ingo Steinwart, Andreas Christmann. Support vector machines. Springer Science & Business Media; 2008. [Google Scholar]
- 24.Sanford Weisberg. Applied linear regression. Vol. 528. John Wiley & Sons; 2005. [Google Scholar]
- 25.Marquardt Donald W, Snee Ronald D. Ridge regression in practice. The American Statistician. 1975;29(1):3–20. [Google Scholar]
- 26.Mariette Awad, Rahul Khanna. Efficient Learning Machines. Springer; 2015. Support vector regression; pp. 67–80. [Google Scholar]