Abstract
Because they contain detailed individual-level data on various patient characteristics including their medical conditions and treatment histories, electronic health record (EHR) systems have been widely adopted as an efficient source for health research. Compared to data from a single health system, real-world data (RWD) from multiple clinical sites provide a larger and more generalizable population for accurate estimation, leading to better decision making for health care. However, due to concerns over protecting patient privacy, it is challenging to share individual patient-level data across sites in practice. To tackle this issue, many distributed algorithms have been developed to transfer summary-level statistics to derive accurate estimates. Nevertheless, many of these algorithms require multiple rounds of communication to exchange intermediate results across different sites. Among them, the One-shot Distributed Algorithm for Logistic regression (termed ODAL) was developed to reduce communication overhead while protecting patient privacy. In this paper, we applied the ODAL algorithm to RWD from a large clinical data research network—the OneFlorida Clinical Research Consortium and estimated the associations between risk factors and the diagnosis of opioid use disorder (OUD) among individuals who received at least one opioid prescription. The ODAL algorithm provided consistent findings of the associated risk factors and yielded better estimates than meta-analysis.
Introduction
Electronic health record (EHR) systems have increasingly been implemented around the world and across the United States (U.S.), providing an extensive data resource for the conduct of biomedical research and improvement of health care1-3. Many large clinical data consortia have been founded to provide platforms and tools to collect and integrate EHR data from multiple clinical sites to obtain more reliable and generalizable conclusions4-7. The Patient-Centered Clinical Research Network (PCORnet) is a large national network of networks covering more than 100 million patients through 348 health systems in the U.S5,6, funded by the Patient-Centered Outcomes Research Institute (PCORI), one of the prominent examples of large-scale, national research networks. The OneFlorida Clinical Research Consortium (OneFlorida CRC) is one of the 9 clinical data research networks (CDRNs) funded by the Patient-Centered Outcomes Research Institute (PCORI) that contribute to the national PCORnet to accelerate the translation of promising research findings into improved patient care. The OneFlorida network has collected robust longitudinal and linked patient-level real-world data (RWD) for ~15 million (>50%) Floridians, including data from Medicaid & Medicare claims, cancer registries, vital statistics, and EHRs from its clinical partners8.
The OneFlorida data repository integrates various data sources from contributing organizations, which current included 12 healthcare organizations: 1) four academic health centers (i.e., University of Florida Health [UFHealth], University of Miami Health System [UMHealth], Florida State University and regional campus practice partners, and University of South Florida [USF]), 2) seven healthcare systems including Tallahassee Memorial Healthcare (TMH affiliated with Florida State University), Orlando Health (ORH), Adventist Health (AH, formerly known as Florida Hospital), Nicklaus Children’s Hospital (NCH, formerly known as Miami Children’s Hospital), Bond Community Health (BCH), Capital Health Plan (CHP), and Tampa General Hospital (TGH affiliated with USF), and 3) CommunityHealth IT—a rural health network in Florida. In addition, the OneFlorida network has obtained claims data from the Florida Medicaid (FLM) program. As a network, the OneFlorida CRC provides care for more than 50% of Floridians through 4,100 physicians, 914 clinical practices, and 22 hospitals with a catchment area covering all of the 67 Florida counties9. The scale of the data in OneFlorida is ever-growing and as of December 2019 included longitudinal and robust patient-level records of approximately 14.4 million Floridians and over 561.1 million encounters, 1.16 billion diagnoses, 1 billion prescribing records, and 1.44 billion procedures.
The availability of EHR data allows opioid use disorder (OUD) to be studied using larger and more representative samples than previously was possible. OUD is defined in the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM–5) as a problematic pattern of opioid use that leads to clinically significant impairment or distress10. Currently, it is estimated that globally 27 million people have OUD11. In 2018, over 2 million people in the U.S. suffered from OUD and over 47,600 people died from opioid overdose12. In 2017, the U.S. Department of Health and Human Services (HHS) announced a public health emergency due to the increasing prevalence of OUD and the occurrence of opioid overdose and called for strategies to control the opioid epidemic13. Studies have shown that OUD is heterogeneous in terms of demographic and clinical characteristics and treatment outcomes. For example, OUD disproportionally affects non-Hispanic Whites and Native Americans, younger adults, and those with a history of mental health disorders14. Geographic variations have also been documented, where rural areas are affected most by OUD15. Analyses based on data from a single site within a small geographically constrained area and relatively homogenous population cannot capture the geographic variations and the heterogeneity of OUD patterns that limit the generalizability of the findings. Therefore, in this analysis, we aim to utilize diverse multicenter data from the different sites in OneFlorida CRC to account for the potential variation in the OUD population.
In multicenter studies, sharing data is a major challenge due to privacy concerns16. To circumvent the issue of sharing individual patient-level data, many distributed algorithms have been developed to jointly study multiple datasets by communicating only summary-level statistics17-20. Among them, Duan et al19-20 proposed a privacy-preserving One-shot Distributed Algorithm for Logistic regression (ODAL), which can be used to identify risk factors of a binary healthcare outcome of interest. Compared to existing methods, ODAL requires only one round of communication across sites and can achieve high accuracy as a pooled analysis in which a logistic regression is fitted on the combined dataset20. However, Duan et al.20 evaluated the performance of ODAL using simulated data and random partition of a real-world dataset from a single health system. Such a dataset may not be representative of the multisite scenarios in a real-world data research network, where the data from each site were collected at different locations and with different population characteristics.
In this paper, we evaluated the performance of the ODAL algorithm using real-world linked EHR and claims data from the OneFlorida network. We studied the association between OUD and several relevant clinical risk factors among individuals receiving at least one opioid prescription. Our results demonstrate that ODAL yields greater accuracy than meta-analysis, which is currently the most popular and preferred method for distributed analysis. From the analysis, the significant risk factors for OUD that we identified are consistent with those reported in previous studies.
Materials and Methods
Data Source and Study Population
OneFlorida contains robust longitudinal and linked patient-level RWD of ~15 million (>50%) Floridians, including data from Medicaid & Medicare claims, cancer registries, vital statistics, and EHRs from its clinical partners. OneFlorida is a HIPAA limited data set (i.e., dates are not shifted and patients’ 9-digit zip codes are available) that contains detailed patient and clinical variables, including demographics, encounters, diagnoses, procedures, vitals, medications, and labs, following the PCORnet Common Data Model (CDM)21. The OneFlorida data undergo rigorous quality checks at a data coordinating center (i.e., University of Florida [UF]), and a privacy-preserving record linkage process is used to deduplicate records of patients seen in different healthcare systems within the network22. Figure 1 shows the geographic locations of OneFlorida partners.
Figure 1.
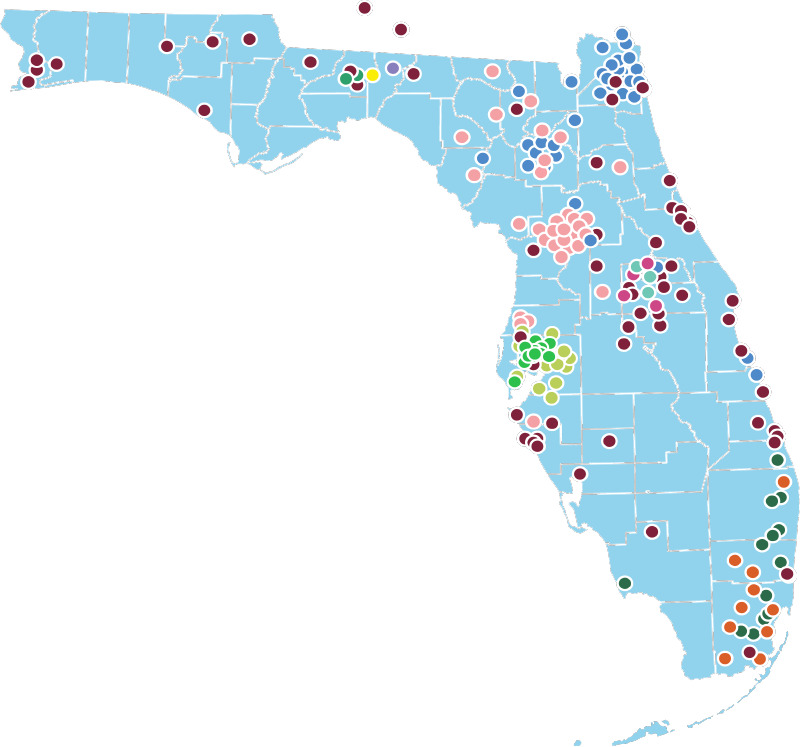
The OneFlorida Clinical Research Consortium.
Based on the OneFlorida data, individuals whose first opioid prescriptions were made between 01/01/2012 and 03/01/2019 were identified. The date of the first opioid prescription is set to be the index date. We considered a total of 9 most frequently used opioid medications, including codeine, fentanyl, hydromorphone, meperidine, methadone, morphine, and oxycodone23. These nine medications accounts for more than 90% of opioid prescriptions in the United States23-26. All brands and dosages of these medications were identified with RXCUI. The outcome of interest is the 12-month risk of OUD after patients’ first opioid prescription. Our primary outcome was the recorded diagnosis of OUD as a proxy for OUD, which was identified using the corresponding codes from the International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) (304.0, 305.5) and ICD-10-CM (F11.1*, F11.2*)27. Once eligible, all individuals remain in the cohort, regardless of whether they continue to receive opioid prescriptions until they are censored because of an OUD diagnosis or the end of the observation period. In addition, we excluded individuals with an OUD diagnosis prior to their first opioid prescription and those who had any cancer diagnosis. A total of 1,155,304 records were identified in OneFlorida. After the exclusion of patients with missing data, a total of 336,800 individuals were included in our final analysis from 5 sites. Table 1 displays the distribution of the included population across different sites.
Table 1.
Breakdown of study population by site.
| Site | OUD | At-risk population | OUD rate |
| Site 1 | 47 | 31,836 | 0.15% |
| Site 2 | 36 | 17,368 | 0.21% |
| Site 3 | 16 | 3,379 | 0.47% |
| Site 4 | 72 | 27,871 | 0.26% |
| Site 5 | 930 | 256,115 | 0.36% |
| All sources | 1,407 | 336,711 | 0.42% |
Risk Factors
Because the primary goal of this analysis is to show how ODAL performs compare to meta-analysis in RWD, we extracted only a set of demographic and clinical risk factors that were identified from the literature. All records 12 months before the first opioid prescription (i.e., the index date) and 3 months after the first opioid prescription were used. For patients who developed an OUD 3 month after the first opioid, their records before OUD diagnosis were used. We included demographic variables (age, race, gender, and insurance type), BMI, lipid panel results, smoking status, selected clinical diagnoses, and prescriptions. Because laboratory test results suffered from a high rate of missing values (>50% in the total study population), they were removed from the analyses. In addition, the clinical diagnoses that were made in <1% of the total study population were removed to minimize potential bias introduced by the small sample size. Finally, patients who had missing values for any risk factors were removed, as the current algorithms are unable to handle missing values and our sample size is sufficient to power the study even after removing patients with missing values. A total of 16 risk factors/predictors were included in the analysis. Table 2 displays the summary statistics for these predictors. Overall, patients diagnosed with OUD are younger and more likely to be male, non-Hispanic Whites, smokers, with Medicaid insurance, and have different clinical conditions that were included in the analysis as risk factors.
Table 2.
Characteristics of included risk factors between individuals developing incident of OUD diagnosis vs. those without OUD
| Without OUD (n=335,610) | With OUD (n=1,101) | Overall (n=336,711) | |
| Mean age, mean (SD) | 47.7 (20.8) | 43.9 (14.2) | 47.7 (20.8) |
| Gender | |||
| Female | 190,141 (56.7%) | 584 (53.0%) | 190,725 (56.6%) |
| Male | 145,469 (43.3%) | 517 (47.0%) | 145,986 (43.4%) |
| Race/ethnicity | |||
| Hispanic | 27,047 (8.1%) | 49 (4.5%) | 27,096 (8.0%) |
| Non-Hispanic black | 79,952 (23.8%) | 171 (15.5%) | 80,123 (23.8%) |
| Non-Hispanic White | 215,855 (64.3%) | 863 (78.4%) | 216718 (64.4%) |
| Other | 12,756 (3.8%) | 18 (1.6%) | 12,774 (3.8%) |
| Type of insurance | |||
| Medicaid | 64,633 (19.3%) | 339 (30.8%) | 64,972 (19.3%) |
| Medicare | 83,393 (24.8%) | 246 (22.3%) | 83,639 (24.8%) |
| Cash or no payment* | 24,984 (7.4%) | 213 (19.3%) | 25,197 (7.5%) |
| Other | 18,712 (5.6%) | 32 (2.9%) | 18,744 (5.6%) |
| Other governmental payment | 13,414 (4.0%) | 17 (1.5%) | 13,431 (4.0%) |
| Private | 130,474 (38.9%) | 254 (23.1%) | 130,728 (38.8%) |
| BMI, mean (SD) | 29.0 (8.07) | 27.5 (7.33) | 29.0 (8.10) |
| Smoking | |||
| Former smoker | 80,357 (23.9%) | 214 (19.4%) | 80,571 (23.9%) |
| Never smoker | 179438 (53.5%) | 244 (22.2%) | 179682 (53.4%) |
| Smoker | 75,815 (22.6%) | 643 (58.4%) | 76,458 (22.7%) |
| Alcohol use disorders (ICD-9: 291, 303; ICD-10: F10) | |||
| 1 | 9745 (2.9%) | 75 (6.8%) | 9820 (2.9%) |
| Depression (ICD-9: 311; ICD-10: F33, F32) | |||
| 1 | 35,928 (10.7%) | 260 (23.6%) | 36,188 (10.7%) |
| Anxiety (ICD-9: 300; ICD-10: F41) | |||
| 1 | 43,821 (13.1%) | 348 (31.6%) | 44,169 (13.1%) |
| Sleep disorders (ICD-9: 327; ICD-10: G47) | |||
| 1 | 27,585 (8.2%) | 74 (6.7%) | 27,659 (8.2%) |
| Rheumatoid arthritis (ICD-9: 714; ICD-10: M05, M06) | |||
| 1 | 5,030 (1.5%) | 23 (2.1%) | 5,053 (1.5%) |
| Other pain conditions (ICD-9: 338; ICD-10: G89, R52) | |||
| 1 | 53,399 (15.9%) | 378 (34.3%) | 53,777 (16.0%) |
| Cannabis-related disorders (ICD-9: 304.3, 305.2; ICD-10: F12) | |||
| 1 | 8,699 (2.6%) | 105 (9.5%) | 8,804 (2.6%) |
| Nicotine-related disorders (ICD-9: 305.1; ICD-10: F17) | |||
| 1 | 67,110 (20.0%) | 561 (51.0%) | 67,671 (20.1%) |
| Other psychoactive disorders (ICD-9: 305.9; ICD-10: F19) | |||
| 1 | 3,738 (1.1%) | 160 (14.5%) | 3,898 (1.2%) |
| Cocaine-related disorders (ICD-9: 304.2, 305.6; ICD-10: F14) | |||
| 1 | 4,013 (1.2%) | 99 (9.0%) | 4,112 (1.2%) |
no payment includes self-pay, nor charge, refusal to pay/bad debt, Hill-Burton free care, research/donor, and other.
Statistical Analysis
We assume that there is a total of K sites, with observations. Define logit. Let zij and yij to be the risk factors (as a vector) and the binary outcome (i.e., status of opioid use disorder) for the i-th patient in the j-th site. Denote , we aim to fit a logistic regression between and , i.e.,
and we are interested in estimating the regression coefficients (including the intercept), i.e., β.
Our statistical analysis is based on the second-order algorithm (i.e., ODAL2) proposed in Duan et al20, with slight modifications. The ODAL algorithms require one site in the research network to serve as a local site, which provides patient-level data and all the other sites share their summary-level statistics with the local site to estimate the results more accurately. Using data from OneFlorida, a collaborative environment, we modify the algorithm by allowing all sites to serve as the local site. By allowing each site to serve as the local site, the surrogate likelihood function is built in each site, and the surrogate estimates are obtained from each site. The final estimator is obtained through a weighted average, which reduces the impact of a specific local site (e.g., distorted local population, poor data quality in a specific local site) on the final estimate, and potentially improves the robustness and accuracy of the ODAL algorithm.
More specifically, we consider the following detailed procedure:
Step 1: In site j = 1, ...K, run logistic regression at the j-th site to obtain and its variance . Broadcast
Step 2: In site j = 1, ...K,
obtain
calculate and broadcast , and ;
- construct
, obtain and broadcast .
Step 3: Output
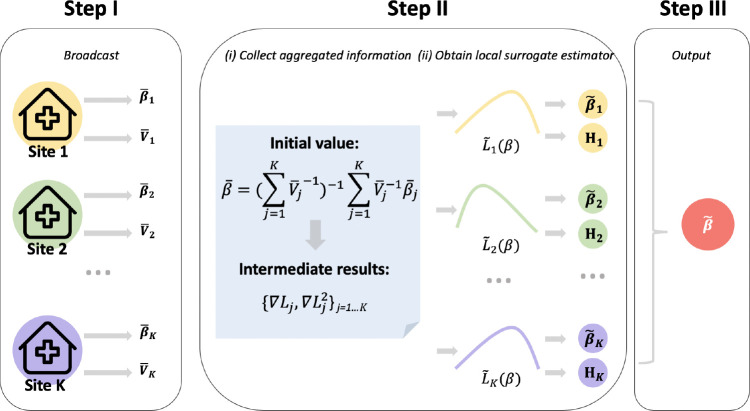
The following Figure 2 provides a schematic illustration of the above algorithm.
Figure 2.
Illustration of the ODAL algorithm. Step I: Use patient-level data within each site to obtain and its variance , then broadcast , where j = 1,...,K. Step II: Obtain the initial value with and calculate the intermediate terms and at at j-th site. With intermediate information, construct local surrogate likelihoods to obtain and broadcast and Hj. Step III: Synthesize the evidence to get output .
In practice, to utilize the ODAL algorithm within a clinical data network, the first step is to distribute the pre-written code to the collaborating sites. With the code, the initial values of parameters are calculated locally within each site. Then, these values are broadcasted by uploading them to a shared cloud folder. When all of the initial values are uploaded, the local or central site can calculate the initial value, , which is broadcasted to the rest of the sites for calculating the intermediate results (i.e., and ). These results are uploaded to the shared cloud folder and are synthesized into the final estimate . The procedure of applying ODAL algorithm to various data networks keeps the same.
With the data from OneFlorida, we compared the ODAL algorithm with the pooled analysis and meta-analysis. The pooled analysis is treated as the gold standard, which fits a unified logistic regression on the combined dataset. Results from the meta-analysis are obtained by fitting logistic regressions separately within each site and synthesizing the local estimates through a weighted average. Compared with meta-analysis, ODAL has better estimation accuracy for studying rare conditions, and similar performance when the outcome and exposures are common.
Results
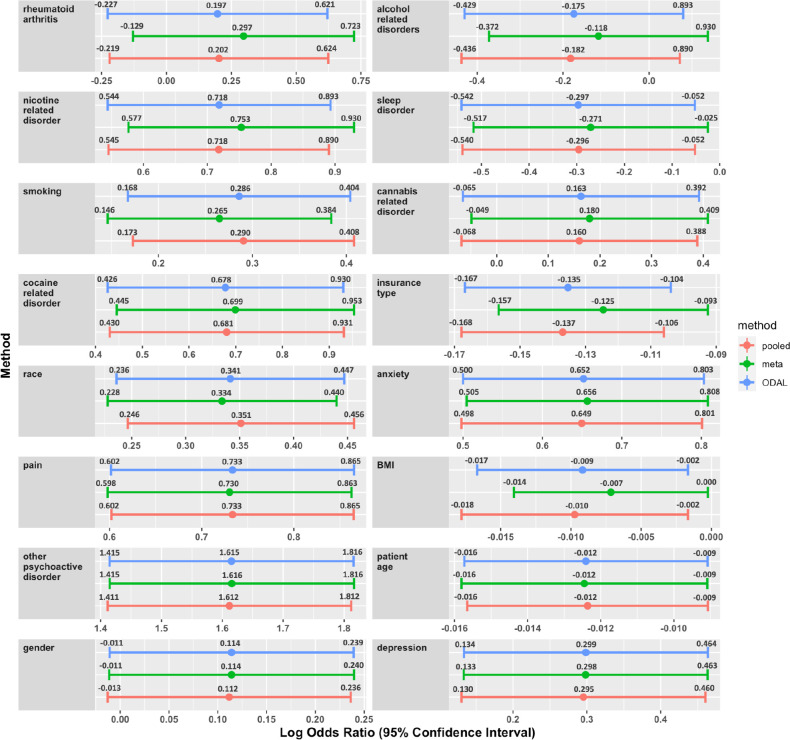
Figure 3 presents the comparison of the estimated log odds ratio and 95% confidence interval for each risk factor using three methods: pooled estimate (red), meta-analysis (green), and ODAL (blue). ODAL provides more accurate estimation results for most of the risk factors than meta-analysis, in that the estimates are closer to the analysis where all data are pooled together. Of the 16 risk factors, ODAL improved the estimation for 15 risk factors. For the one risk factor that was not improved (i.e., depression), the relative bias of ODAL compared to the pooled analysis is below 1%. For risk factors such as alcohol use disorder, insurance type, and cannabis-related disorders, the relative bias of meta-analysis is greater than 20%, which ODAL can reduce to below 3%.
In the analysis using the ODAL method, we identified 10 risk factors that were statistically significantly associated with OUD: anxiety, cocaine-related disorders, depression, insurance type, nicotine-related disorders, other psychoactive disorders, other pain conditions, age, race, and smoking. These findings are consistent with previous studies examining risk factors for OUD. For example, mental health conditions, such as anxiety, depression, and psychoactive substance use disorders have previously been associated with higher risks of OUD28-30. Smoking, the use of other substances, and polysubstance use were also associated with a higher risk of OUD among individuals who primarily use opioids31,32. Demographic variables, such as sex (being female), age (being a young adult), or ethnicity (being non-Hispanic White) were also associated with having OUD. Among these variables, although the finding of gender, which had a positive estimated log odds ratio, was not consistent with previous finding33, the observed gender effect was not statistically significant. The consistent findings from our analysis suggested the ODAL method can be used to identify clinically relevant risk factors for OUD using distributed real-world data. Moreover, due to the low event rates of OUD in the clinical sites, the estimates provided by the ODAL algorithm outperform those of meta-analysis, which is consistent with the findings in Duan et al20. ODAL may be especially valuable for studying rare outcomes or exposures in a multicenter analysis.
Figure 3 displays the estimated log odds ratio and 95% confidence interval for each risk factor using the three methods: pooled estimate (red), meta-analysis (green), and ODAL (blue). The risk factors are ranked by the performance of ODAL compared with meta-analysis starting from the best one (i.e., rheumatoid arthritis). The last risk factor (i.e., depression) is the only one whose estimated effect size is not improved by the ODAL algorithm among the 16 risk factors.
Conclusion and Discussion
In this paper, we evaluated the performance of the ODAL algorithm using real-world EHR data from the OneFlorida Clinical Research Consortium and investigated the association between OUD and 16 clinical risk factors. Compared with meta-analysis, the ODAL algorithm yielded a better estimate of 15 risk factors. Ten of the 16 factors showed statistically significant associations. Our findings are consistent with previous reports on risk factors for OUD, supporting the reliability of the real-world performance using the ODAL algorithm.
We modified the original ODAL algorithm by allowing each site to serve as the local site. For this modification, one extra step (i.e., broadcast the local estimate and variance obtained by logistic regression in each site) is required to obtain the initial value compared with the original ODAL algorithm. This additional step requires not much effort as it only requires the local estimates of the model parameters and their standard errors to be transferred from each site, and it can reduce the impact of a specific local site on the final estimate, and improve the robustness and accuracy of the ODAL algorithm.
The ODAL algorithm was developed using the pooled analysis as the gold standard method, which fits a unified logistic regression model on the combined dataset. Therefore, it requires that the data are homogeneously distributed across sites. However, when there exists heterogeneity in the distribution of data across sites, the pooled analysis may not be a gold standard and will require correcting the model to address the heterogeneity34. Taking this into consideration, we plan to extend our method to handle heterogeneity across clinical sites by allowing site-specific effects and covariates.
For future works, the use of both structured and unstructured data would also be of great interest when analyzing large data networks. The wide adoption of EHR systems has made large-scale, longitudinal clinical data available for research. The U.S. Food and Drug Administration (FDA) recently coined the term real-world data (RWD) to refer to information derived from sources outside research settings, including EHRs, claims data, and billing data, among others. EHRs contain important structured data, such as diagnoses and procedures, as well as unstructured clinical narratives such as physicians’ notes. More than 80% of the clinical information in the EHR is documented in clinical narratives35, which contain much detailed patient information. In this study, we used only the structured data from the OneFlorida network because other important risk factors (e.g., homelessness, social determinants of health, prescription, and utilization patterns) were not readily available and thus could not be included in our analysis. In future studies, we plan to use advanced natural language processing (NLP) methods to extract additional risk factors from clinical narratives. Furthermore, to avoid the bias caused by using a complete-case analysis, methods for handling missing data, such as multiple imputation, can be considered before applying the ODAL algorithm.
In addition to the OneFlorida network, there are a number of other large-scale national and international data research networks. The Observational Health Data Sciences and Informatics (OHDSI) network is another prominent example—an international network of observational health databases that covers more than half a billion patient records4. Novel distributed-learning methods, which are privacy-preserving, communication efficient, and accurate are needed to exploit these large data networks in the future.
Finally, the ODAL algorithm can be extended in several aspects. First, methods to integrate and analyze other types of outcomes can be considered36, such as count data or time-to-event outcomes. Second, we have been developing open-source and user-friendly software to implement the ODAL algorithm within research networks to facilitate data integration across health systems and promote research that can provide novel insights into important issues in healthcare.
Acknowledgment
This work was supported in part by NIH grants 1R01AI130460, 1R01LM012607, R01CA246418, R21AG061431 and UL1TR001427, PCORI grants ME-2018C3-14754, and the VISN4 Mental Illness Research, Education and Clinical Center of the U.S. Department of Veterans Affairs. The content is solely the responsibility of the authors and does not represent the official views of the NIH, PCORI, or the VA. Disclosure: Dr. Kranzler is an advisory board member for Dicerna Pharmaceuticals; a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was sponsored in the past three years by AbbVie, Alkermes, Amygdala Neurosciences, Arbor Pharmaceuticals, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, and Pfizer; and is named as an inventor on PCT patent application #15/878,640 entitled: “Genotype-guided dosing of opioid agonists,” filed January 24, 2018.
Footnotes
first three authors contributed equally
correspondence
Figures & Table
References
- 1.Center for Devices and Radiological Health Real-World Evidence to Support Regulatory Decision-Making for Devices. FDA Med Bull. www.fda.gov/regulatory-information/search-fda-guidance-documents/use-real-world-evidence-support-regulatory-decision-making-medical-devices .
- 2.Center for Drug Evaluation and Research Submitting Documents Using Real-World Data and Real-World Evidence. FDA Med Bull. www.fda.gov/regulatory-information/search-fda-guidance-documents/submitting-documents-using-real-world-data-and-real-world-evidence-fda-drugs-and-biologics-guidance .
- 3.Center for Drug Evaluation and Research Use of Electronic Health Record Data in Clinical Investigations. FDA Med Bull. www.fda.gov/regulatory-information/search-fda-guidance-documents/use-electronic-health-record-data-clinical-investigations-guidance-industry .
- 4.Hripcsak G, Duke JD, Shah NH, et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Stud Health Technol Inform. 2015;216:574–578. [PMC free article] [PubMed] [Google Scholar]
- 5.Collins FS, Hudson KL, Briggs JP, et al. PCORnet: turning a dream into reality. J Am Med Inform Assoc. 2014;21:576–577. doi: 10.1136/amiajnl-2014-002864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.PCORnet: National Patient-Centered Clinical Research Network https://pcornet.org/
- 7.Forrest CB, Margolis PA, Bailey LC, et al. PEDSnet: a National Pediatric Learning Health System. J Am Med Inform Assoc. 2014;21:602–606. doi: 10.1136/amiajnl-2014-002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shenkman E, Hurt M, Hogan W, et al. OneFlorida Clinical Research Consortium: Linking a Clinical and Translational Science Institute with a Community-Based Distributive Medical Education Model. Acad Med. 2018;93:451–455. doi: 10.1097/ACM.0000000000002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shenkman EA. OneFlorida Clinical Research Consortium. PCORI. 2019. https://www.pcori.org/research-results/2015/oneflorida-clinical-research-consortium .
- 10.CDC-Opioid Basics-Commonly Used Terms CDC. 2019. https://www.cdc.gov/drugoverdose/opioids/terms.html .
- 11.Information sheet on opioid overdose . WHO; 2018. https://www.who.int/substance_abuse/information-sheet/en/ [Google Scholar]
- 12.What is the U.S. Opioid Epidemic? United States Department of Health and Human Services; https ://www. hhs.gov/opioids/about-the-epidemic/index. html. [Google Scholar]
- 13.of Health USD, Services H, Others 5-Point strategy to combat the opioid crisis
- 14.Santoro TN, Santoro JD. Racial Bias in the US Opioid Epidemic: A Review of the History of Systemic Bias and Implications for Care. Cureus. 2018;10:e3733. doi: 10.7759/cureus.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haffajee RL, Lin LA, Bohnert ASB, et al. Characteristics of US Counties With High Opioid Overdose Mortality and Low Capacity to Deliver Medications for Opioid Use Disorder. JAMA Network Open. 2019;2:e196373. doi: 10.1001/jamanetworkopen.2019.6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tucker K, Branson J, Dilleen M, et al. Protecting patient privacy when sharing patient-level data from clinical trials. BMC Med Res Methodol. 2016;1(16 Suppl):77. doi: 10.1186/s12874-016-0169-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Jiang X, Kim J, et al. Grid Binary LOgistic REgression (GLORE): building shared models without sharing data. J Am Med Inform Assoc. 2012;19:758–764. doi: 10.1136/amiajnl-2012-000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu C-L, Wang S, Ji Z, et al. WebDISCO: a web service for distributed cox model learning without patient-level data sharing. J Am Med Inform Assoc. 2015;22:1212–1219. doi: 10.1093/jamia/ocv083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan R, Boland MR, Moore JH, et al. ODAL: A one-shot distributed algorithm to perform logistic regressions on electronic health records data from multiple clinical sites. Pac Symp Biocomput. 2019;24:30–41. [PMC free article] [PubMed] [Google Scholar]
- 20.Duan R, Boland MR, Liu Z, et al. Learning from electronic health records across multiple sites: A communication-efficient and privacy-preserving distributed algorithm. J Am Med Inform Assoc. 2020;27:376–385. doi: 10.1093/jamia/ocz199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Common Data Model (CDM) Specification https://pcornet.org/wp-content/uploads/2019/09/PCORnet-Common-Data-Model-v51-2019_09_12.pdf .
- 22.Bian J, Loiacono A, Sura A, et al. Implementing a hash-based privacy-preserving record linkage tool in the OneFlorida clinical research network. Jamia Open. 2019;2:562–569. doi: 10.1093/jamiaopen/ooz050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kenan K, Mack K, Paulozzi L. Trends in prescriptions for oxycodone and other commonly used opioids in the United States, 2000-2010. Open Medicine. 2012;6(2):e41. [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly JP, Cook SF, Kaufman DW, Anderson T, Rosenberg L, Mitchell AA. Prevalence and characteristics of opioid use in the US adult population. Pain. 2008 Sep 15;138(3):507–13. doi: 10.1016/j.pain.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Baker O, Schuur JD, Weiner SG. Effects of Rescheduling Hydrocodone on Opioid Prescribing in Ohio. Pain Medicine. 2019. Sep 10, [DOI] [PMC free article] [PubMed]
- 26.Cassidy TA, Oyedele N, Mickle TC, Guenther S, Budman SH. Patterns of abuse and routes of administration for immediate - release hydrocodone combination products. Pharmacoepidemiology and drug safety. 2017 Sep;26(9):1071–82. doi: 10.1002/pds.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DSM 5 Diagnostic Codes Related to Substance Use Disorders http://www.acbhcs.org/providers/qa/docs/training/DSM-IV_DSM-5_SUD_DX.pdf .
- 28.Carroll I, Barelka P, Wang CKM, et al. A pilot cohort study of the determinants of longitudinal opioid use after surgery. Anesth Analg. 2012;115:694–702. doi: 10.1213/ANE.0b013e31825c049f. [DOI] [PubMed] [Google Scholar]
- 29.Edlund MJ, Martin BC, Fan M-Y, et al. Risks for opioid abuse and dependence among recipients of chronic opioid therapy: results from the TROUP study. Drug Alcohol Depend. 2010;112:90–98. doi: 10.1016/j.drugalcdep.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katz C, El-Gabalawy R, Keyes KM, et al. Risk factors for incident nonmedical prescription opioid use and abuse and dependence: results from a longitudinal nationally representative sample. Drug Alcohol Depend. 2013;132:107–113. doi: 10.1016/j.drugalcdep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leeman RF, Sun Q, Bogart D, et al. Comparisons of Cocaine-Only, Opioid-Only, and Users of Both Substances in the National Epidemiologic Survey on Alcohol and Related Conditions (NESARC) Subst Use Misuse. 2016;51:553–564. doi: 10.3109/10826084.2015.1122063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Elliott AL, Serdarevic M, et al. A latent class analysis of the past-30-day substance use patterns among lifetime cocaine users: Findings from a community sample in North Central Florida. Addict Behav Rep. 2019;9:100170. doi: 10.1016/j.abrep.2019.100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davenport S, Katie Matthews AS. Milliman white paper. March; 2018. Opioid use disorder in the United States: Diagnosed prevalence by payer, age, sex, and state. [Google Scholar]
- 34.Tong J, Duan R, Li R, Scheuemie MJ, Moore JH, Chen Y. Robust-ODAL: Learning from heterogeneous health systems without sharing patient-level data. InPacific Symposium on Biocomputing. Pacific Symposium on Biocomputing. 2020 Jan 1;25:695. NIH Public Access. [PMC free article] [PubMed] [Google Scholar]
- 35.Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008:128–144. [PubMed] [Google Scholar]
- 36.Duan R, Luo C, Schuemie MJ, et al. Learning from local to global - an efficient distributed algorithm for modeling time-to-event data. J Am Med Inform Assoc. Epub ahead of print 6 July 2020. DOI: 10.1093/jamia/ocaa044. [DOI] [PMC free article] [PubMed]