Abstract
Incompleteness of ontologies affects the quality of downstream ontology-based applications. In this paper, we introduce a novel lexical-based approach to automatically detect potentially missing hierarchical IS-A relations in SNOMED CT. We model each concept with an enriched set of lexical features, by leveraging words and noun phrases in the name of the concept itself and the concept’s ancestors. Then we perform subset inclusion checking to suggest potentially missing IS-A relations between concepts. We applied our approach to the September 2017 release of SNOMED CT (US edition) which suggested a total of 38,615 potentially missing IS-A relations. For evaluation, a domain expert reviewed a random sample of 100 missing IS-A relations selected from the “Clinical finding” sub-hierarchy, and confirmed 90 are valid (a precision of 90%). Additional review of invalid suggestions further revealed incorrect existing IS-A relations. Our results demonstrate that systematic analysis of the enriched lexical features of concepts is an effective approach to identify potentially missing hierarchical IS-A relations in SNOMED CT.
Introduction
Ontologies and terminologies have been increasingly used in biomedical research and applications, since they provide domain knowledge to facilitate biomedical data annotation, data integration and exchange, information retrieval, natural language processing (NLP), and clinical decision support1,2. For instance, SNOMED CT facilitates the exchange of healthcare information among healthcare providers and electronic health records (EHRs), leading to higher quality, consistency and safety in healthcare delivery3,4.
Given such important roles, the quality of biomedical ontologies directly impacts the quality of their downstream applications5. In particular, incompleteness of ontologies affects the quality of downstream applications such as leading to valid conclusions being missed6. For instance, in ontology-based search engines for patient cohort identification, incomplete ontology hierarchy impacts the quality of query results. As an example, value sets of SNOMED CT (consisting of subsets of SNOMED CT concepts) have been widely used for EHR decision support, quality reporting, and cohort selection. A value set can be defined as a list of concepts sharing some common features, e.g., all descendants of “Carcinoma of larynx”. However, “Primary adenosquamous cell carcinoma of larynx” is currently not listed as one of its descendants (i.e., a missing hierarchical IS-A relation), thus patients with “Primary adenosquamous cell carcinoma of larynx” would not be selected for a cohort of patients with “Carcinoma of larynx”.
Recently, lexical-based methods have shown great potential to automatically detect missing hierarchical IS-A relations for improving the completeness and quality of biomedical ontologies including SNOMED CT7-10, NCI Thesaurus11-14, and Gene Ontology15. For instance, in previous work10, we leveraged lexical features of concepts in non-lattice subgraphs of SNOMED CT to identify potentially missing IS-A relations, where the non-lattice subgraphs are potential problematic substructures which may reveal quality defects such as missing IS-A relations and missing concepts9.
In this paper, we introduce a novel lexical-based approach without relying on non-lattice substructures for automatic and exhaustive detection of potentially missing IS-A relations in SNOMED CT. We model each concept with an enriched set of lexical features, which leverages words and noun phrases not only in the name of the concept itself but also in the names of the concept’s ancestors. Then we perform subset inclusion checking for concept pairs that are not present in the existing inferred IS-A hierarchy. Each subset inclusion relation identified is a potentially missing IS-A relation detected. A random sample of detected missing IS-A relations is reviewed by a domain expert to evaluate the effectiveness of our approach.
1. Background
1.1. Auditing completeness of biomedical ontologies
Recent work has paid special attention to audit the completeness of biomedical ontologies. Chen et al. presented a recursive method to locate missing hierarchical relations in the Metathesaurus of the Unified Medical Language System (UMLS)16. Ochs et al. presented tribal-based17 and subject-based18 abstraction network methods to audit SNOMED CT for uncovering quality issues including missing parents. Bodenreider considered words in concept names as logical definitions and used a Description Logic (DL) classifier to automatically derive hierarchical IS-A relations among concepts in SNOMED CT; and then compared the DL-derived hierarchy with the original SNOMED CT hierarchy to detect missing IS-A relations7. Quesada-Martínez et al. analyzed concept names in SNOMED CT to identify lexical regularities and suggest missing relations (including missing IS-A relations)8. Cui et al. introduced a hybrid structural-lexical method by mining lexical patterns in non-lattice subgraphs to suggest missing IS-A relations and missing concepts in SNOMED CT9. Abeysinghe et al. extended Cui et al.’s structural-lexical approach9 to detect missing IS-A relations in NCI Thesaurus11 and proposed additional lexical patterns. Cui et al. further enriched lexical features of concepts in non-lattice subgraphs to identify missing IS-A relations in SNOMED CT without relying on predefined lexical patterns10. Keloth et al. leveraged horizontal density differences of concepts in different ontologies to identify missing child concepts19. Previously, we introduced a lexical-based inference approach to derive hierarchical inconsistencies and uncover missing IS-A relations in SNOMED CT, Gene Ontology and NCI Thesaurus15. After detecting missing IS-A relations, we leveraged external ontologies in the UMLS to identify supporting evidence which could potentially relieve the manual review effort of domain experts.
These approaches reveal different kinds of missing hierarchical relations in a given terminology. Even for the same terminology (e.g., SNOMED CT), each approach uncovers a certain portion of potentially missing hierarchical relations differently. In this paper, we introduce another lexical-based approach, to identify previously undiscovered missing hierarchical relations in SNOMED CT.
1.2. SNOMED CT
SNOMED CT20 is the most comprehensive clinical healthcare terminology used worldwide, containing more than 330,000 concepts and 19 sub-hierarchies. Each concept in SNOMED CT has a unique numeric concept identifier (e.g., 363504005) and a fully specified name (FSN). The FSN provides a unique, unambiguous description of a concept’s meaning. For instance, concept 363504005’s FSN is “Malignant tumor of lower limb (disorder)” with a semantic tag “disorder” in parentheses at the end.
Concepts in SNOMED CT are organized from the general to the more detailed using hierarchical IS-A relations (i.e., the detailed is a subtype of the general). Therefore, a concept is more detailed than its ancestors and is more general than its descendants. For example, concept 363504005 – “Malignant tumor of lower limb (disorder)” is more detailed than its parent concept 126655004 – “Neoplasm of lower limb (disorder).”
2. Methods
In this work, we use the September 2017 release of SNOMED CT (US edition). We perform three main steps for exhaustive detection of potentially missing IS-A relations in SNOMED CT: (1) identify a list of stop words/phrases and antonym pairs which may lead to incorrect suggestion of missing IS-A relations; (2) construct a set of lexical features for each concept by leveraging the words and noun phrases in the concept itself as well as in its ancestors; and (3) check the subset inclusions between each candidate pair of concepts to suggest potentially missing IS-A relations.
2.1. Identifying stop words/phrases and antonym pairs
Stop words/phrases may result in wrongly suggested missing IS-A relations (or false positives). Take concepts “Velopharyngeal incompetence due to cleft palate (disorder)” and “Cleft palate (disorder)” as an example, even though the set of lexical features of the former concept contains that of the latter concept, there should not be an IS-A relation between these two concepts. Words/phrases such as “due to” are highly likely to suggest false positives and thus are considered as stop words/phrases. In this work, we leveraged a list of stop words/phrases used in previous work10, including: “and”, “or”, “no”, “not”, “without”, “due to”, “secondary to”, “except”, “by”, “after”, “co-occurrent”, “bilateral”, “examination”, “able”, “amputation”, “removal”, “replacement”, “resection”, “excision”, “reaction to”, “unable”, “failure”, “failed”, “abnormal”, “excluding”, “non”, and “pre”.
Similarly, concept pairs whose lexical features contain antonym pairs are likely to generate erroneous suggestions. For instance, considering concepts “Secondary malignant neoplasm of right upper lobe of lung (disorder)” and “Neoplasm of right lower lobe of lung (disorder)”, apparently there should not be an IS-A relation between these two concepts, since the former concept is related to “right upper lobe of lung” while the latter concept is related to “right lower lobe of lung”. However, if the former concept inherited a lexical feature “lower” from one of its ancestors “Malignant neoplasm of lower respiratory tract (disorder)”, then the lexical feature set of the former would subsume that of the latter, as a result of which an incorrect IS-A between the former and latter would be suggested. To collect such potential antonym pairs, we adopted a list of adjective antonym pairs from WordNet21, including (“open”, “closed”), (“acute”, “chronic”), (“right”, “left”), etc. We also identified additional antonym pairs which are not included in WordNet, such as (“upper”, “lower”).
2.2. Constructing lexical features for concepts
Most existing lexical-based methods for identification of missing IS-A relations use words in concept names as the lexical features of concepts. In this work, we model concepts not only using words, but also utilizing noun phrases. For each concept, we first preprocess its FSN and identify an initial set of lexical features consisting of words and noun phrases in the concept’s FSN. Then we enrich the set with more lexical features inherited from the concept’s ancestors.
2.2.1. Preprocessing FSNs of concepts
We first preprocess the FSNs of concepts before the initialization of lexical feature sets. For each concept, we split its FSN (by space) into words sequentially and remove its semantic tag (e.g., “(disorder)”). The semantic tag will be leveraged while suggesting potentially missing IS-A relations. We further process special symbols in FSNs such as removing parentheses and square brackets, and replacing backslash with “or” if the FSN does not contain numbers (e.g., “Sickness/injury care” will result in “Sickness or injury care”, while “5 mg/ml” will remain intact).
2.2.2. Initializing lexical feature sets with noun phrases and words
In this work, instead of purely using the bag-of-words model, we consider noun phrases as meaning features of concepts to facilitate the identification of missing IS-A relations. Take two concepts “Acute sensitivity to pain (finding)” and “Acute pain (finding)” as an example, if we simply used the bag-of-words model, their lexical feature sets would be {acute, sensitivity, to, pain} and {acute, pain} respectively, where {acute, sensitivity, to, pain} is a superset of {acute, pain} (i.e., more detailed), and thus “Acute sensitivity to pain (finding)” would be suggested as a subtype of “Acute pain (finding)”. However, this suggestion is incorrect since “Acute sensitivity to pain” is a finding of pain threshold, while “Acute pain” is a finding of pattern of pain; and there should not be any subsumption relations between these two concepts. The reason for this incorrect suggestion is that the adjective “acute” is the modifier for two different nouns (“sensitivity” and “pain”) in these two concepts. To avoid such situations, we model a concept’s name as a set of noun phrases and words, where a noun phrase groups the modifier(s) and the corresponding noun as a single feature. Hence in the above example, the two concepts’ lexical features will become {acute, sensitivity, to, pain, acute sensitivity} and {acute, pain, acute pain}, which do not have any subset-superset relation.
We use Stanford CoreNLP Parser22 to identify noun phrases. Note that the parser may recognize noun phrases in different levels of granularity. For instance, for concept “Anesthesia for procedure on veins of lower leg (procedure)”, there is a base level noun phrase “lower leg” which is a component of a higher level noun phrase “veins of lower leg”. In this work, we only consider the base level noun phrases. That is, we model a concept’s FSN initially as a set of individual word(s) and base level noun phrase(s). In this example, the initial set of lexical features for the concept is {anesthesia, for, procedure, on, veins, of, lower, leg, lower leg}.
2.2.3. Enriching lexical feature sets
We enrich concepts’ lexical features in two steps. In the first step, for each concept c, we check if its FSN contains noun phrase(s) identified in the initial feature sets of other concepts which are not hierarchically linked with c; and if yes, we add such noun phrase(s) into c’s initial lexical feature set. We denote concept c’s set of lexical features obtained after the first-step enrichment process as E1c. In the second step, for each concept, we further enrich its set of lexical features with its ancestors’ sets of lexical features. It is intuitive that if concept x is a subtype of concept y, then the lexical features or attributes of concept y are also considered to be true for concept x (i.e., x inherits y’s attributes). In this work, we maintain a directed graph which is constructed using all the inferred hierarchical IS-A relations in SNOMED CT, compute its transitive closure, and obtain the ancestors of concepts using the breadth-first search. While performing the second-step enrichment process for a concept c, if an ancestor a contains stop word(s)/phrase(s), then we do not add a’s set of lexical features to c’s. More formally, we have
where E2c denotes concept c’s set of lexical features after the second-step enrichment process, and Ac is the set of c’s ancestors.
Table 1 shows an example of the initial and enriched sets of lexical features for a concept c: 371977004 – “Primary malignant neoplasm of cecum (disorder).” The noun phrase identified in the initial set of lexical features is “primary malignant neoplasm” (underlined). After the first-step enrichment (E1c), a new noun phrase “malignant neoplasm” is identified from concepts which are not hierarchically linked with c. After the second-step enrichment (E2c), more noun phrases and words are inherited from c’s ancestors. For instance, noun phrase “large intestine” is inherited from the initial lexical feature set of c’s parent - “Primary malignant neoplasm of large intestine (disorder)” and noun phrase “malignant tumor” is inherited from c’s other parent - “Malignant tumor of cecum (disorder)”
Table 1:
The initial and enriched sets of lexical features of an example concept c: 371977004 – Primary malignant neoplasm of cecum (disorder). Noun phrases are underlined.
| c's FSN | Primary malignant neoplasm of cecum (disorder) |
| Initial set | {primary, malignant, neoplasm, of, cecum, primary malignant neoplasm} |
| Enriched set E1c | {primary, malignant, neoplasm, of, cecum, primary malignant neoplasm, malignant neoplasm} |
| Enriched set E2c | {primary, malignant, neoplasm, of, cecum, primary malignant neoplasm, malignant neoplasm, abdominal, mass, abdominal mass, disorder, digestive, structure, digestive structure, finding, large, intestine, large intestine, neoplastic, disease, neoplastic disease, malignant neoplastic disease, viscus, structure finding, body, region, body region, trunk, trunk structure, abdomen, tumor, malignant tumor, organ, digestive organ, gastrointestinal, tract, gastrointestinal tract, system, digestive system, intraabdominal, intraabdominal organ, bowel, bowel finding, lower, lower gastrointestinal tract, body system, abdominal organ finding, abdominal organ, gastrointestinal tract finding, segment, abdominal segment, intestinal, intestinal tract, digestive system finding, system finding, body structure, digestive tract, cecal, cecal mass} |
2.3. Identifying potentially missing IS-A relations
To automatically suggest potentially missing IS-A relations, we first produce candidate pairs of concepts (say x and y) which meet the following conditions:
concepts x and y are within the same sub-hierarchy (we assume that concepts in different sub-hierarchies do not have hierarchical IS-A relations since sub-hierarchies in SNOMED CT do not share common concepts);
x and y are not hierarchically linked through existing IS-A relations;
x and y share the same semantic tag;
neither x nor y contains any stop word/phrase; and
the enriched sets of lexical features E2x and E2y do not contain antonym pairs.
Then for each candidate pair of concepts (x, y), we systematically compare their enriched sets of lexical features E2x and E2y as follows: if E2x is a superset of E2y, then “concept x IS-A concept y” will be suggested as a potentially missing IS-A relation; if E2x is a subset of E2y, then “concept y IS-A concept x” will be suggested as a potentially missing IS-A relation; otherwise, nothing will be suggested.
Since our suggestion of missing IS-A relations is in an exhaustive way, it may result in redundant missing IS-A relations. More specifically, the suggested missing IS-A relations may include cases like “concept x IS-A concept y” and “concept x IS-A concept z” while z is an ancestor of y. Here, “concept x IS-A concept z” is considered redundant, since it can be implied by that x is a subtype of y and y is a subtype of z. Similarly, there might be cases like “concept a IS-A concept c” and “concept b IS-A concept c” while b is an ancestor of a. In such cases, “concept a IS-A concept c” is considered redundant, since it can be implied by that a is a subtype of b and b is a subtype of c. To avoid unnecessary analyses, we remove redundant relations and only keep those suggested missing IS-A relations (say “concept x IS-A concept y”) satisfying the following two conditions:
there does not exist a concept z such that “concept x IS-A concept z” is a suggested missing relation and y is an ancestor of z; and
there does not exist a concept s such that “concept s IS-A concept y” is a suggested missing relation and s is an ancestor of x.
3. Results
In this paper, we applied our method to all the sub-hierarchies of SNOMED CT except “SNOMED CT Model Component (metadata)” (e.g., definition status) and “Special concept (special concept)” (e.g., inactive concept). A total of 38,615 potentially missing hierarchical IS-A relations were suggested. Table 2 shows the number of potentially missing IS-A relations in each sub-hierarchy. For instance, 6,946 potentially missing IS-A relations were identified from the “Clinical finding” sub-hierarchy.
Table 2:
Numbers of missing hierarchical IS-A relations detected in terms of the sub-hierarchies.
| Sub-hierarchy | # of Potentially Missing IS-A | Sub-hierarchy | # of Potentially Missing IS-A |
| Body structure | 26,161 | Situation with explicit context | 82 |
| Clinical finding | 6,946 | Staging and scales | 36 |
| Procedure | 3,861 | Social context | 33 |
| Substance | 390 | Specimen | 31 |
| Organism | 277 | Environment or geographical location | 26 |
| Observable entity | 242 | Pharmaceutical / biologic product | 22 |
| Physical object | 234 | Record artifact | 2 |
| Qualifier value | 185 | Physical force | 0 |
| Event | 87 |
3.1. Evaluation
To evaluate the effectiveness of our approach for detecting missing IS-A relations, we randomly selected a sample of 100 potentially missing IS-A relations from the “Clinical finding” sub-hierarchy. A domain expert (author JS) reviewed the sample and verified that 90 out of 100 missing IS-A relations are valid (or true positives), indicating that our approach achieved a precision of 90%. Table 3 lists 15 examples of missing IS-A relations in the “Clinical finding” sub-hierarchy verified by the domain expert, including “Open injury of diaphragm (disorder)” IS-A “Open wound of thorax (disorder)”, and “Primary malignant neoplasm of fibula (disorder)” IS-A “Malignant neoplasm of long bone of lower leg (disorder)”.
Table 3:
Examples of missing hierarchical IS-A relations in the “Clinical finding” sub-hierarchy confirmed by the domain expert.
| Subconcept | Superconcept |
| Primary adenosquamous cell carcinoma of larynx (disorder) | Carcinoma of larynx (disorder) |
| Strain of fascia of intrinsic muscle of thumb (disorder) | Injury of fascia of intrinsic muscle of thumb (disorder) |
| Pelvic muscular dystrophy (disorder) | Degenerative disorder of muscle (disorder) |
| Superficial injury of interscapular region with infection (disorder) | Superficial injury of trunk with infection (disorder) |
| Contracture of iliopsoas (disorder) | Disorder of soft tissue of trunk (disorder) |
| Carcinoma in situ of upper labial mucosa (disorder) | Tumor of upper labial mucosa (disorder) |
| Complete ankylosis of the spine (disorder) | Disorder of vertebra (disorder) |
| Plasmodium vivax malaria with rupture of spleen (disorder) | Infectious disease of abdomen (disorder) |
| Fracture subluxation of acromioclavicular joint (disorder) | Fracture subluxation of joint of upper limb (disorder) |
| Genital herpes simplex (disorder) | Infectious disease of genitourinary system (disorder) |
| Plasmodium vivax malaria with rupture of spleen (disorder) | Infectious disease of abdomen (disorder) |
| Open fracture of thoracic spine with spinal cord lesion (disorder) | Fracture of spine with spinal cord lesion (disorder) |
| Open injury of diaphragm (disorder) | Open wound of thorax (disorder) |
| Osteitis fibrosa cystica generalisata (disorder) | Degenerative disorder of bone (disorder) |
| Primary malignant neoplasm of fibula (disorder) | Malignant neoplasm of long bone of lower leg (disorder) |
For each false positive (i.e., invalid missing IS-A relation suggested), we provided the domain expert with the existing IS-A relation(s) which lead to the suggestion of the false positive. The domain expert further reviewed these existing IS-A relations and checked whether any of them is problematic.
3.2. Analysis of false positive cases
We manually examined the false positive cases for potential causes. For instance, our approach suggests a false positive: “Familial malignant neoplasm of pancreas (disorder)” IS-A “Malignant tumor of body of pancreas (disorder)”, since the former concept inherits a lexical feature “body” from its ancestor “Mass of body region (finding)”. However, the meaning of “body” in “Malignant tumor of body of pancreas (disorder)” is different than its meaning in “Mass of body region (finding)”. The former refers to the finding site of structure of body of pancreas, while the latter refers to the finding site of body region structure. Therefore, there should not be an IS-A relation between the two concepts. In this case, the false positive is due to the varied meanings of a word in different context.
Another cause of false positives is the incorrect existing IS-A relations in SNOMED CT that our approach leverages to suggest potentially missing IS-A relations. Table 4 shows seven examples of false positives generated by our approach due to the incorrect existing IS-A relations. For instance, our approach suggests “Encysted hydrocele of spermatic cord (disorder)” IS-A “Soft tissue lesion of pelvic region (disorder)”, which is incorrect since hydrocele refers to a small “bag of fluid” and is not considered as a soft tissue lesion. This incorrect suggestion is due to an existing relation: “Encysted hydrocele of spermatic cord (disorder)” IS-A “Soft tissue lesion (disorder)”. In addition, there are two false positives caused by the same existing relation: “Algodystrophy (disorder) IS-A “Degenerative disorder (disorder)” in the September 2017 release of SNOMED CT US edition that we used. It is worth noting that this relation is no longer existent in the current version of SNOMED CT, that is, this incorrect IS-A relation has been removed.
Table 4:
Examples of false positives caused by the incorrect existing hierarchical IS-A relations. *: indicates that the incorrect existing relation has been removed in the newer versions of SNOMED CT.
| False Positives | Reason: Incorrect Existing Relations |
| Disorder of left sacroiliac joint (disorder) IS-A Disorder of left lower extremity (disorder) | Disorder of pelvic girdle (disorder) IS-A Disorder of lower extremity (disorder) |
| Encysted hydrocele of spermatic cord (disorder) IS-A Soft tissue lesion of pelvic region (disorder) | Encysted hydrocele of spermatic cord (disorder) IS-A Soft tissue lesion (disorder) |
| Malignant neoplasm of sacral vertebra (disorder) IS-A Malignant neoplasm of bone of lower limb (disorder) | Neoplasm of sacrum (disorder) IS-A Neoplasm of lower limb (disorder) |
| Algodystrophy of foot (disorder) IS-A Degenerative disorder of extremity (disorder) | Algodystrophy (disorder) IS-A Degenerative disorder (disorder)* |
| Reflex sympathetic dystrophy of upper extremity (disorder) IS-A Degenerative disorder of extremity (disorder) | Algodystrophy (disorder) IS-A Degenerative disorder (disorder)* |
| Secondary malignant neoplasm of sacrum (disorder) ISA Secondary malignant neoplasm of bone of lower limb (disorder) | Neoplasm of sacrum (disorder) IS-A Neoplasm of lower limb (disorder) |
| Autosomal recessive popliteal pterygium syndrome (disorder) IS-A Dysplasia of limb (disorder) | Popliteal pterygium syndrome (disorder) IS-A Congenital anomaly of lower limb (disorder) |
4. Discussion
In this paper, we introduce a lexical approach for exhaustive detection of potentially missing hierarchical IS-A relations in SNOMED CT. It can be seen that our approach can not only detect intuitive/straightforward relations such as “Primary adenosquamous cell carcinoma of larynx (disorder)” IS-A “Carcinoma of larynx (disorder)”, but also uncover complicated cases such as “Genital herpes simplex (disorder)” IS-A “Infectious disease of genitourinary system (disorder)” and “Plasmodium vivax malaria with rupture of spleen (disorder)” IS-A “Infectious disease of abdomen (disorder)” (Table 3). Since our approach only requires the hierarchical IS-A structure and concept names as the input, it can be generally applied to other terminologies or ontologies.
4.1. Comparison with previous work
In previous work10, we introduced a structural-lexical approach for the detection of potentially missing IS-A relations in SNOMED CT, by leveraging the lexical attributes of concepts in non-lattice subgraphs. A pair of concepts is known as a non-lattice pair if they share more than one maximal common descendant. Non-lattice subgraphs derived from non-lattice pairs often reveal quality issues including missing IS-A relations. In this work, we perform exhaustive detection of potentially missing IS-A relations without limiting to the non-lattice substructures.
More importantly, this work identifies previously undiscovered missing IS-A relations. Among 38,615 potentially missing IS-A relations identified in this work, 36,534 (94.6%) are newly discovered compared with those in previous work10. Among 6,946 potentially missing IS-A relations from the “Clinical finding” sub-hierarchy in this work, 6,081 (87.5%) are newly identified compared with those in previous work10.
Since this work leverages the entire structure of SNOMED CT while the previous work focuses on non-lattice substructures, it is intuitive to further investigate the level differences of the subconcepts and superconcepts involved in the potentially missing ISA relations. Therefore, we computed the level of each concept in SNOMED CT (i.e., the number of concepts in the path from the root to the concept). For concepts with multiple paths from the root, we chose the number of the longest path. We considered the root’s level as 0. For instance, Figure 1 shows that the level of concept “Open injury of diaphragm (disorder)” is 12 and the level of concept “Open wound of thorax (disorder)” is 10. The level difference between these two concepts is 2.
Figure 1:
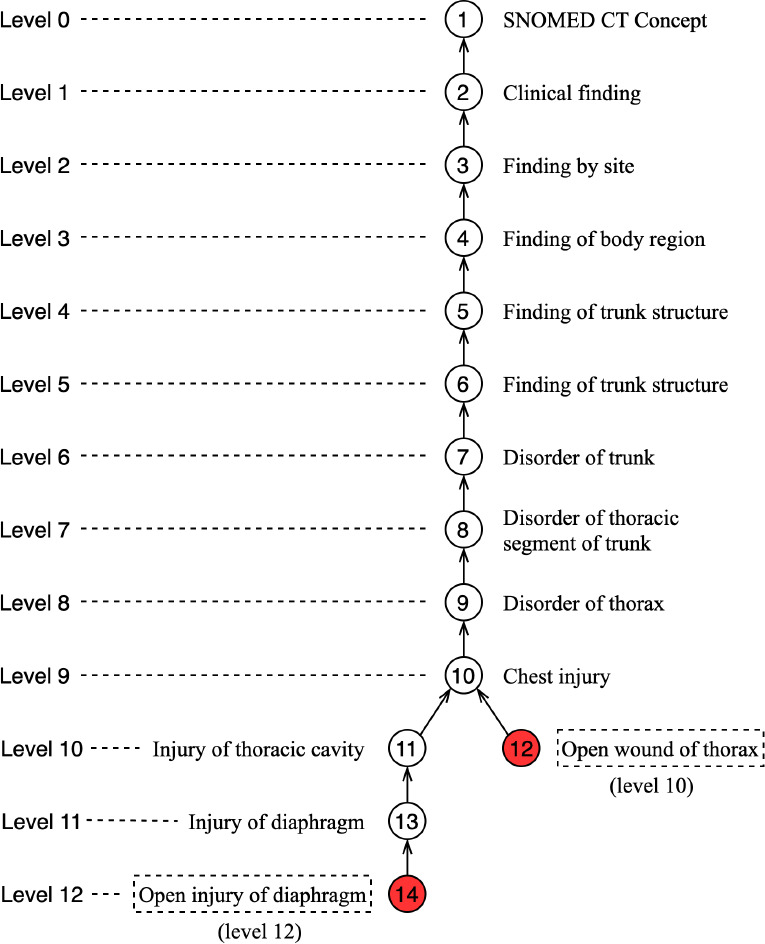
The levels of concepts involved in a missing IS-A relation: “Open injury of diaphragm (disorder)” IS-A “Open wound of thorax (disorder)”.
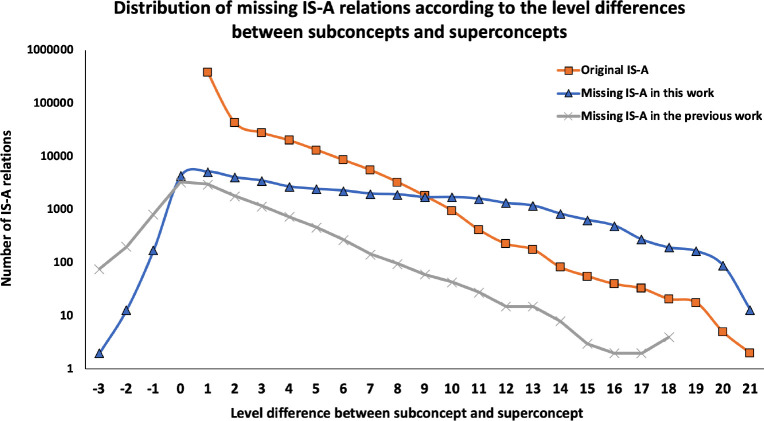
We compared the level difference of subconcepts and superconcepts for potentially missing IS-A relations identified in this work and previous work10. Figure 2 shows the number of potentially missing IS-A relations in terms of the level difference between the subconcept and superconcept. The level difference ranges from -3 to 21 in this work and -3 to 18 in previous work. A negative level difference indicates that the superconcept has a higher level than the subconcept does. For the previous work10, 6% of identified missing IS-A relations have a level difference that is greater than 5; while for this work, over 32% of the detected missing IS-A relations have a level difference that is greater than 5.
Figure 2:
Distribution of potentially missing IS-A relations detected in this work and previous work according to the level differences between subconcepts and superconcepts.
It can also be seen that for each of the non-negative level differences (0 to 21), this work consistently identifies more potentially missing IS-A relations than the previous work does; while for each of the negative level differences (-3 to -1), the previous work detects more potentially missing IS-A relations than this work does.
Another major distinction is regarding the construction of lexical features for concepts. In the previous work10, a concept in a non-lattice subgraph is modeled as a set of words in its FSN with enriched lexical features inherited from its ancestors within the nonlattice subgraph. For instance, Figure 3 shows a nonlattice subgraph identified in the previous work10, where concept 6, “Fracture subluxation of perilunate joint”, has a set of lexical features {fracture, subluxation, of, perilunate, joint, dislocation, lunate, wrist}. In this work, we model each concept as a set of words and noun phrases, with enriched lexical features inherited from all its ancestors in the entire SNOMED CT. Take the same concept “Fracture subluxation of perilunate joint” as an example, this work generates a set of lexical features for the concept as {fracture, subluxation, of, perilunate, joint, fracture subluxation, perilunate joint, traumatic dislocation, dislocation, traumatic, lunate, lunate bone, bone, wrist, limb structure, finding, structure, limb, upper limb, upper, wrist joint, disorder, fracture dislocation, lesion, musculoskeletal system, injury, system, musculoskeletal, arthropathy, wrist region, region, radiocarpal, radiocarpal joint, body region, body, extremity, upper extremity, traumatic injury, skeletal, skeletal system, connective tissue, tissue, joint injury, body system, bone finding, musculoskeletal finding, joint finding, carpal bone, disease, bone injury}. As can be seen, concepts have more enriched lexical features to represent their meanings in this work.
Figure 3:
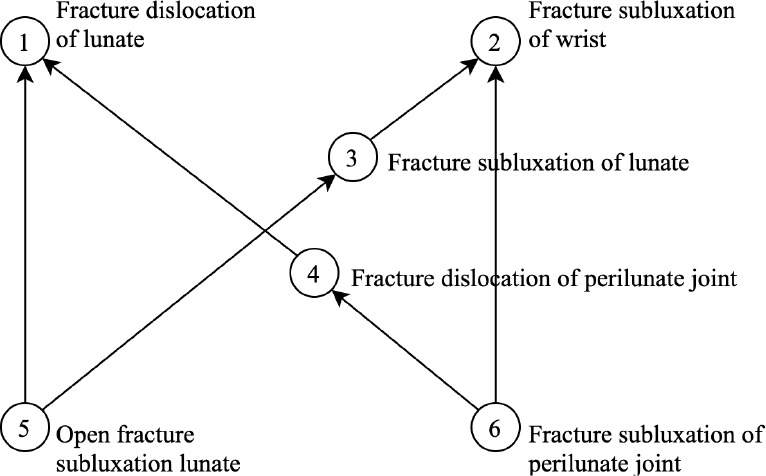
A non-lattice subgraph identified in the previous work10. This non-lattice subgraph suggests a missing IS-A relation between concepts 3 and 1: “Fracture subluxation of lunate” IS-A “Fracture dislocation of lunate”.
4.2. Limitations and future work
Although the results showed that our approach is effective in identifying missing hierarchical IS-A relations, there still remains several limitations.
Our evaluation is limited in that it only involved samples from the “Clinical Finding” sub-hierarchy. Manual review of the detected missing IS-A relations by domain experts is time-consuming and labor-intensive. In future work, we plan to investigate methods that leverage external knowledge (e.g., external ontologies, biomedical literature) to automatically identify supporting evidence for the detected missing IS-A relations to relieve the manual burden.
In addition, our approach relies on the Stanford CoreNLP Parser to identify noun phrases in the concept names. However, sometimes the Stanford Parser may not accurately recognize noun phrases. For instance, for concept “Gluthathione peroxidase deficiency (disorder)”, it recognizes “peroxidase deficiency” as a noun phrase. However, “Gluthathione peroxidase” is a more meaningful noun phrase to serve as a lexical feature for this concept.
Another limitation of this work is that the remediation of the suggested missing IS-A relations may not be simply adding them to the SNOMED CT hierarchy, since the IS-A hierarchy is inferred by a DL classifier based on the logical definitions of concepts. Further work is still needed to identify the potential causes of the missing IS-A relations from the logical definition point of view, so that missing IS-A relations can be properly inferred by modifying the logical definitions of concepts.
Regarding the performance evaluation of our approach, we mainly focused on measuring the precision. It is impracticable to report actual recall since there is no reference standard that contains false negatives for calculating the recall. However, one may use cumulative changes of IS-A relations over different versions of SNOMED CT as a surrogate standard to measure retrospective recall23. Due to the discovery nature of the task to identify missing IS-A relations, there may exist other auditing approaches that could propose missing IS-A relations in SNOMED CT that this approach did not uncover. We plan to apply a recent approach24 developed for suggesting missing IS-A relations in Gene Ontology to SNOMED CT, and compare the resulted missing IS-A relations with those identified in this work.
5. Conclusions
In this paper, we presented a lexical-based approach to exhaustively detect potentially missing hierarchical IS-A relations in SNOMED CT. We modeled each concept with a set of enriched lexical features consisting of words and noun phrases in the name of the concept itself and its ancestors. Pairwise comparison of the concepts’ lexical features automatically suggested potentially missing IS-A relations. The results showed that our approach is effective in detecting missing IS-A relations. Analysis of false positive cases further revealed incorrect existing IS-A relations in SNOMED CT.
Acknowledgment
This work was supported by the National Science Foundation (NSF) through grant IIS-1931134, and the National Institutes of Health (NIH) through grants R21CA231904 and R01NS116287. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NSF or NIH.
Footnotes
Corresponding author. Email: licong.cui@uth.tmc.edu
Figures & Table
References
- 1.Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearbook of medical informatics. 2008;17(01):67–79. [PMC free article] [PubMed] [Google Scholar]
- 2.Hoehndorf R, Schofield PN, Gkoutos GV. The role of ontologies in biological and biomedical research: a functional perspective. Briefings in bioinformatics. 2015;16(6):1069–1080. doi: 10.1093/bib/bbv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee D, de Keizer N, Lau F, Cornet R. Literature review of SNOMED CT use. Journal of the American Medical Informatics Association. 2013;21(e1):e11–e19. doi: 10.1136/amiajnl-2013-001636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winnenburg R, Bodenreider O. AMIA Annual Symposium Proceedings. Vol. 2013. American Medical Informatics Association; 2013. Metrics for assessing the quality of value sets in clinical quality measures; pp. 1497–1505. [PMC free article] [PubMed] [Google Scholar]
- 5.Cui L, Tao S, Zhang GQ. Biomedical ontology quality assurance using a big data approach. ACM Transactions on Knowledge Discovery from Data (TKDD) 2016;10(4):41. [Google Scholar]
- 6.Lambrix P, Wei-Kleiner F, Dragisic Z. Completing the is-a structure in light-weight ontologies. Journal of biomedical semantics. 2015;6(1):12. doi: 10.1186/s13326-015-0002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodenreider O. Identifying Missing Hierarchical Relations in SNOMED CT from Logical Definitions Based on the Lexical Features of Concept Names. ICBO/BioCreative. 2016. p. 2016. [PMC free article] [PubMed]
- 8.Quesada-Martínez M, Fernández-Breis JT, Karlsson D. Suggesting Missing Relations in Biomedical Ontologies Based on Lexical Regularities. MIE. 2016. pp. 384–388. [PubMed]
- 9.Cui L, Zhu W, Tao S, Case JT, Bodenreider O, Zhang GQ. Mining non-lattice subgraphs for detecting missing hierarchical relations and concepts in SNOMED CT. Journal of the American Medical Informatics Association. 2017;24(4):788–798. doi: 10.1093/jamia/ocw175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui L, Bodenreider O, Shi J, Zhang GQ. Auditing SNOMED CT hierarchical relations based on lexical features of concepts in non-lattice subgraphs. Journal of biomedical informatics. 2018;78:177–184. doi: 10.1016/j.jbi.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abeysinghe R, Brooks MA, Talbert J, Cui L. AMIA Annual Symposium Proceedings. Vol. 2017. American Medical Informatics Association; 2017. Quality assurance of NCI Thesaurus by mining structural-lexical patterns; pp. 364–373. [PMC free article] [PubMed] [Google Scholar]
- 12.Abeysinghe R, Brooks MA, Cui L. AMIA Annual Symposium Proceedings. Vol. 2019. American Medical Informatics Association; 2019. Leveraging Non-lattice Subgraphs to Audit Hierarchical Relations in NCI Thesaurus; pp. 982–991. [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng F, Abeysinghe R, Cui L. A Hybrid Method to Detect Missing Hierarchical Relations in NCI Thesaurus. 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). IEEE. 2019:1948–1953. [Google Scholar]
- 14.Abeysinghe R, Hinderer EW, Moseley HN, Cui L. Auditing subtype inconsistencies among gene ontology concepts. 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) IEEE. 2017:1242–1245. [Google Scholar]
- 15.Abeysinghe R, Zheng F, Hinderer EW, Moseley HN, Cui L. A Lexical Approach to Identifying Subtype Inconsistencies in Biomedical Terminologies. 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). IEEE. 2018:1982–1989. [Google Scholar]
- 16.Chen Y, Gu HH, Perl Y, Geller J. Structural group-based auditing of missing hierarchical relationships in UMLS. Journal of biomedical informatics. 2009;42(3):452–467. doi: 10.1016/j.jbi.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ochs C, Geller J, Perl Y, Chen Y, Agrawal A, Case JT, et al. A tribal abstraction network for SNOMED CT target hierarchies without attribute relationships. Journal of the American Medical Informatics Association. 2014;22(3):628–639. doi: 10.1136/amiajnl-2014-003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ochs C, Geller J, Perl Y, Chen Y, Xu J, Min H, et al. Scalable quality assurance for large SNOMED CT hierarchies using subject-based subtaxonomies. Journal of the American Medical Informatics Association. 2015;22(3):507–518. doi: 10.1136/amiajnl-2014-003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keloth VK, He Z, Chen Y, Geller J. AMIA Annual Symposium Proceedings. Vol. 2018. American Medical Informatics Association; 2018. Leveraging horizontal density differences between ontologies to identify missing child concepts: A proof of concept; pp. 644–653. [PMC free article] [PubMed] [Google Scholar]
- 20.SNOMED CT Starter Guide Available from: https://confluence.ihtsdotools.org/display/DOCSTART/SNOMED+CT+Starter+Guide .
- 21.Miller GA. WordNet: a lexical database for English. Communications of the ACM. 1995;38(11):39–41. [Google Scholar]
- 22.Manning C, Surdeanu M, Bauer J, Finkel J, Bethard S, McClosky D. The Stanford CoreNLP natural language processing toolkit. Proceedings of 52nd annual meeting of the association for computational linguistics: system demonstrations. 2014. pp. 55–60.
- 23.Zhang GQ, Yan H, Cui L. AMIA Annual Symposium Proceedings. Vol. 2017. American Medical Informatics Association; 2017. Can SNOMED CT changes be used as a surrogate standard for evaluating the performance of its auditing methods? pp. 1903–1912. [PMC free article] [PubMed] [Google Scholar]
- 24.Abeysinghe R. Hinderer III EW, Moseley HN, Cui L. SSIF: Subsumption-based Sub-term Inference Framework to audit Gene Ontology. Bioinformatics. 2020;36(10):3207–3214. doi: 10.1093/bioinformatics/btaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]