Abstract
This year less than 200 National Library of Medicine indexers expect to index 1 million articles, and this would not be possible without the assistance of the Medical Text Indexer (MTI) system. MTI is an automated indexing system that provides MeSH main heading/subheading pair recommendations to assist indexers with their heavy workload. Over the years, a lot of research effort has focused on improving main heading prediction performance, but automated fine-grained indexing with main heading/subheading pairs has received much less attention. This work revisits the subheading attachment problem, and demonstrates very significant performance improvements using modern Convolutional Neural Network classifiers. The best performing method is shown to outperform the current MTI implementation with a 3.7% absolute improvement in precision, and a 27.6% absolute improvement in recall. We also conducted a manual review of false positive predictions, and 70% were found to be acceptable indexing.
Introduction
PubMed® is a free online resource provided by the National Library of Medicine (NLM) to support the search and retrieval of biomedical and life science literature. MEDLINE® is the indexed subset of PubMed, and it contains over 26 million citations indexed with Medical Subject Headings (MeSH®). The MeSH vocabulary contains over 29,000 main headings representing biomedical concepts (e.g. “Myocardial Infarction”, “Parkinson Disease”, or “Dopamine Agonists”) and 76 subheadings (e.g. “surgery”, “drug therapy”, or “therapeutic use”) that may be coordinated with main headings to index a more specific aspect of a concept. For example, an article discussing the surgical treatment of myocardial infarction may be indexed with “Myocardial Infarction/surgery”. Frequently, subheadings are used to represent relationships between concepts. For example, the treatment of Parkinson’s Disease with a dopamine agonist could be indexed as “Parkinson Disease/drug therapy; Dopamine Agonists/therapeutic use”.
As the size of the biomedical literature continues to grow, it is a constant challenge to keep MEDLINE up-to-date and relevant. The manual indexing of scientific articles is a highly specialized and time-consuming activity, and this year a team of less than 200 NLM indexers expect to index 1 million articles. To assist indexers with their heavy workload, the NLM has developed an automated indexing system called the Medical Text Indexer1 (MTI). MTI is a machine learning and rule-based system that processes an article title and abstract and returns a pick-list of recommended MeSH terms. The system was first introduced in 2002, and at this time it only made main heading recommendations. Later in 2008, it was updated to recommend main heading/subheading pairs (MeSH pairs). MTI uses statistical, dictionary lookup, rule-based, and machine learning methods for subheading attachment2, and a recent performance analysis has shown that it recommends MeSH pairs with relatively high precision and low recall.
NLM indexers are finding MTI increasingly useful1, but it is acknowledged that its subheading recommendations could be improved. This work revisits the subheading attachment problem, 12 years after the feature was first added to MTI. The paper focuses on 17 “critical” subheadings that are known to be particularly important to MEDLINE users, and shows that very significant performance improvements can be achieved using modern neural network classifiers. The paper also includes an indexer evaluation of the new method, focusing on particularly problematic false positive predictions. The results of the evaluation were encouraging, and we expect that the improved subheading recommendations will be very useful for NLM indexers.
Related Work
The automated indexing of biomedical articles with MeSH terms is a very challenging multi-label text classification problem. The key challenges are the large number of labels and their highly imbalanced distribution. In 2019 MeSH there are 631,568 allowed main heading/subheading combinations. Some of these MeSH pairs are assigned many thousands of times per year, whereas others may only have ever been assigned a few times. For example, the pair “MicroRNAs/genetics” was assigned 6,815 times in 2018, while the pair “Calcimycin/adverse effects” was only assigned once in the same year.
Many machine learning methods are not suitable for extremely large label spaces, and this is one reason why previous work has used a two-stage approach: first the main headings are predicted, and then subheadings are attached to these main headings. This is also the recommended way for human indexers to approach the problem. Most prior work has focused on the main heading prediction problem3–5, and in comparison the subheading attachment problem has received much less attention. The problem was last studied by Neveol et al.2 at the NLM in 2008, and MTI still uses the presented algorithms today.
In their paper, Neveol et al. assess 7 different methods for subheading attachment. Three of these methods (“Dictionary”, “Journal Descriptor Indexing”, and “MTI”) are described as “jigsaw puzzle” methods because the main headings and subheadings are extracted independently, and then combined in any allowed combination. The “Dictionary” method extracts subheadings based on the presence of key words or bi-grams in the title or abstract. The “Journal Descriptor Indexing” method generates word and subheading vectors based on statistical associations with a set of 120 main headings (called Journal Descriptors) that are manually assigned to MEDLINE journals. For a new article, the average cosine similarity between word and subheading vectors is used to generate a ranked list of subheadings. The final “jigsaw puzzle” method is the “MTI” method, and this is a rules-based approach that extracts subheadings based on the predicted main headings. “MTI” method rules are general and do not specify the type of main heading that an extracted subheading should be attached to.
In addition to “jigsaw puzzle” methods, the paper also evaluates three other rules-based methods that attach subheadings to specific main headings: “Natural Language Processing” rules were derived from relationships between concepts that are present in the Unified Medical Language System® Semantic Network, “Coordination” rules enforce the main heading/subheading coordination rules that are detailed in the NLM indexing manual, and “Post-processing” rules are other rules that are applied after main heading extraction. The final assessed method is a k-nearest neighbor approach based on the PubMed Related Citations6 (PRC) algorithm: any MeSH pair that occurs more than once in the 10 nearest neighbors is recommended.
The paper explores various different combination strategies and finds that the different methods are complementary: the Dictionary and PRC algorithms are found to have high recall and lower precision, whereas the rules-based methods are found to have high precision and lower recall. The best combination strategy achieves 48% precision and 30% recall on a large test set of 100,000 articles randomly selected from the MEDLINE 2006 baseline. These performance metrics are for all subheadings, and MeSH pairs containing main headings not in the manual indexing were filtered out. As such, they cannot be directly compared to the performance metrics reported in this paper.
Methods
Dataset
The dataset is comprised of citation data for manually indexed articles published from 2004 onwards. Fully and semi-automatically7 indexed articles were identified using the indexing method attribute of the PubMed XML format8, and excluded as their indexing may be biased towards MTI’s predictions. The dataset contains about 10 million articles: 20,000 articles published in 2018 were randomly selected for the validation set, and 38,903 articles indexed between 16th November 2017 and 31th January 2018 were randomly selected for the test set. All citation data was downloaded from the MEDLINE/PubMed 2019 baseline9. We have a record of MTI’s original recommendations for the test set articles; this is important because MTI uses nearest neighbor algorithms, and its performance will improve after articles have been indexed.
Convolutional Neural Network
This paper uses a type of deep neural network called a Convolutional Neural Network (CNN) for fine-grained automatic MeSH indexing. The CNN architecture was chosen because it is computationally efficient and because it has been shown to be effective for text classification problems with large numbers of labels10. The paper compares two different machine learning methods: an end-to-end method that predicts main heading/subheading pairs directly, and a chained method that predicts the main headings and then the subheadings. Three separate models were trained: a single model for the end-to-end method, and standalone main heading and subheading prediction models for the chained method. These three models share the same CNN architecture, and this is described in the next section.
CNN Architecture
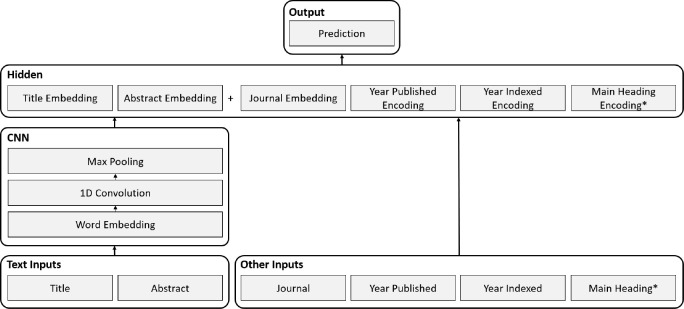
The neural network architecture used in this paper (Figure 1) is based on the CNN architecture presented by Kim11 for sentence classification. The architecture represents words as vectors and input text as the concatenation of word vectors. The network learns a set of convolutional filters that are convolved along the length of the input text to produce an activation map; filters learn to activate when they detect a specific type of feature (e.g. discriminative words or phrases) at some position in the text. The convolution operation is followed by a max pooling operation that keeps only the maximum activation of each filter. The result is a fixed length representation of the input text that is invariant to the position of the detected features.
Figure 1:
CNN architecture. *The main heading input is only used for the standalone subheading prediction model.
This paper applies a custom neural network architecture that uses a CNN to process text inputs. A similar architecture was previously used at the NLM for MEDLINE article selection12. The three different models share five common inputs: the article title, abstract, journal, publication year, and indexing year, and the standalone subheading prediction model requires an additional input for the main heading that subheadings are being predicted for. The network generates a fixed length representation of each input and then concatenates them to construct the input to the hidden layer. The final classification layer uses a sigmoid activation function and its size is equal to the number of labels. The output size of the end-to-end model is the number of allowable MeSH pairs for the 17 critical subheadings (122,542), and the output sizes for the standalone main heading and subheading prediction models are the number of main headings (29,351) and the number of critical subheadings (17) respectively.
Models use randomly initialized word vectors, dropout regularization, and batch normalization for the hidden and convolution layers. The title and abstract inputs are processed separately using the same word embeddings and convolutional filter weights. Standard max pooling is used for the title, whereas dynamic max pooling10 is used for the abstract. The journal is treated as a categorical input, and each journal is represented by a fixed length vector. Like the word embeddings, the journal embeddings are learned during training. The two year inputs are represented using the special encoding scheme previously described in Rae et al.12. The encoding is similar to one-hot encoding; however, positions for the year and preceding years are activated. The main heading input of the standalone subheading prediction model is one-hot encoded.
Configuration
The CNN models were implemented in Tensorflow version 1.12.0, and model hyperparameters are listed in Table 1. The three models share the same hyperparameters, except for the dropout rate, which was adjusted individually. Subword tokenization was performed using SentencePiece13 (byte-pair-encoding algorithm), and the SentencePiece model was trained on title and abstract sentences from training set articles published in 2015 or later. The Tensorflow code for the CNN models is available on GitHub at http://github.com/indexing-initiative/subheading_attachment.
Table 1:
Model hyperparameters.
| Hyperparameter | Value |
| Vocabulary size | 64,000 |
| Word embedding size | 300 |
| Title max words | 64 |
| Abstract max words | 448 |
| Number of convolution filters | 350 |
| Convolution filter sizes | 2, 5, 8 |
| Dynamic max pooling number of regions | 5 |
| Classification layer activation function | Sigmoid |
| Activation function for all other layers | Relu |
| Hidden layer size | 2048 |
| Journal embedding size | 50 |
| Dropout rate | 0.05-0.5 |
| Batch size | 128 |
| Learning rate | 0.001 |
Models were trained using the Adam optimizer and binary cross-entropy loss on a single 16GB NVIDIA V100 GPU. The end-to-end and standalone main heading prediction models were trained on the full training set, while the standalone subheading prediction model was trained on the 2.2 million training set articles published in 2015 or later. The standalone subheading prediction model was run for all manually indexed main headings, and for this reason a smaller training set was necessary to reduce the training time per epoch. Articles published in the last 5 years were chosen for the smaller training set as it is important for the model to learn how to index recent articles. During training, the learning rate was reduced by a factor of 3 if the validation set micro F1 score did not improve by more than 0.001 between epochs, and training was stopped early if the F1 score did not improve by more than 0.001 over two epochs. The total training time for the end-to-end and chained methods was 92 and 55 hours respectively.
Journal Statistics Baseline
It was not possible to use MTI as a baseline for standalone subheading prediction performance since we do not have a record of MTI’s original subheading recommendations for all manually indexed main headings in the test set. As an alternative, a simple “Journal Statistics” baseline was implemented based on the indexing statistics of training set articles published in 2015 or later. For each journal and main heading the probability of the 17 critical subheadings being indexed was recorded, and at test time, for every manually indexed main heading, critical subheading were randomly sampled according to these statistics. This is an interesting baseline because it does not consider the article text.
Indexing Consistency Study
It is useful to compare machine learning performance to human performance. Unfortunately, the most recent study of MEDLINE indexing consistency was published 37 years ago14, and the raw study data is not available for analysis. In this paper, we obtain a more recent estimate of indexing consistency by finding articles in the MEDLINE 2019 baseline that have been inadvertently double indexed.
Double indexed articles were found by searching for exactly matching titles, abstracts, and author last names. When PubMed curators become aware of duplicates, they may synchronize the assigned indexing. For this reason, article pairs annotated with the “Duplicate Publication” publication type, or certain relationships1, were excluded. Furthermore, it is unlikely for independently indexed articles to have exactly the same indexing, and so these pairs were excluded too. The remaining article pairs were reviewed manually, and some further pairs were excluded because they were not considered suitable for the study for various reasons. In total 1839 double indexed article pairs were identified for the indexing consistency study.
Indexer Evaluation
The precision, recall, and F1 score metrics used in this study provide a useful measure of performance, but they do have some limitations. One particularly relevant limitation is their treatment of false positives. From an indexer’s perspective, false positives predictions can either be acceptable alternative indexing, acceptable imperfect indexing, or unacceptable indexing that needs to be removed. The third case is particularly problematic because it is more time-consuming to remove incorrect indexing than to add missing indexing. The problem is that precision, recall, and F1 score metrics penalize all false positives equally, making it difficult to judge how useful the MeSH term recommendations will be in practice.
In order to better understand the strengths and weaknesses of the proposed neural network approach, we conducted an indexer evaluation of the best performing machine learning method. The evaluation focused on particularly problematic false positive main heading/subheading pair predictions, and indexers were asked to decide whether each false positive pair is acceptable or unacceptable (would need to be removed). There are three types of false positive of interest to this study, and these are: incorrect main heading and subheading, correct main heading and incorrect subheading, and incorrect main heading and correct subheading (attached to another indexed main heading).
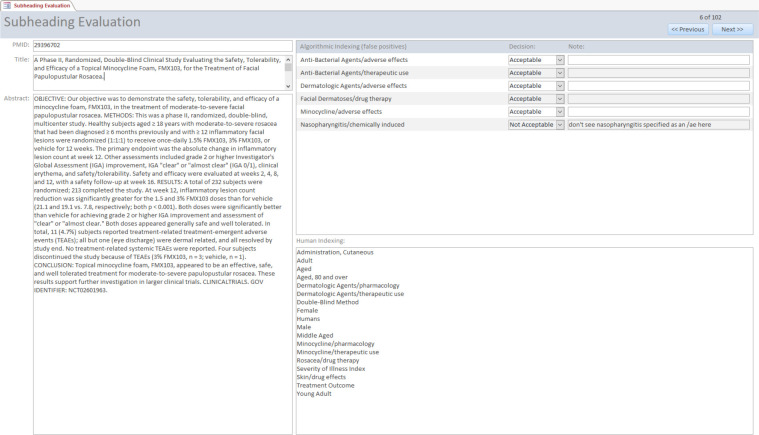
The evaluation set consisted of 180 articles from the test set, and these articles were selected to have a good balance of false positive types and critical subheadings. Two senior indexers participated in the study, and each indexer evaluated 90 articles using the Microsoft Access form shown in Figure 2. The form displays the article title, abstract, and existing manual indexing, and the indexers were required to select “Acceptable” or “Not Acceptable” in a drop down list for each false positive prediction. An additional set of 12 articles were evaluated by both indexers in order to understand the level of inter-indexer agreement for this task.
Figure 2:
Microsoft Access form used for the indexer evaluation.
Evaluation Metrics
For automated indexing evaluations main heading, subheading, or MeSH pair predictions were obtained after applying a single decision threshold to all model outputs, and then micro F1 score (MiF), micro precision (MiP), and micro recall (MiR) performance metrics were computed by comparing the manual indexing y to the model predictions :
where N and L are the number of examples and labels respectively. Optimum decision threshold values were determined by a linear search for maximum F1 score on the validation set. The evaluation metric for the indexer evaluation was the fraction of acceptable false positive predictions.
Results
Table 2 shows main heading/critical subheading pair prediction performance for the proposed neural network methods compared to the MTI baseline. The chained method has the best performance: outperforming MTI with a 3.7% absolute improvement in precision, and a 27.6% absolute improvement in recall. Note that the chained method has 2.7 times the recall of MTI. The two neural network methods are shown to have very similar performance: they have almost identical recall, but the chained method has slightly better precision.
Table 2:
Main heading/subheading pair prediction performance for critical subheadings.
| Method | Precision | Recall | F1 Score |
| MTI | 0.416 | 0.162 | 0.234 |
| End-to-end | 0.448 | 0.438 | 0.443 |
| Chained | 0.453 | 0.438 | 0.445 |
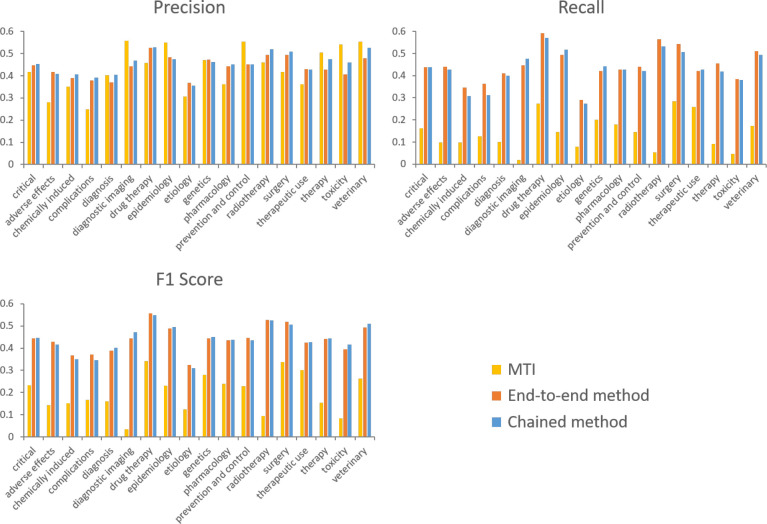
Figure 3 plots micro precision, recall, and F1 score for MeSH pair prediction by critical subheading. The plots highlight the fact that MTI has high precision for some subheadings (e.g. “prevention and control”), but consistently low recall compared to the neural network methods. The performance of the end-to-end and chained methods are close for all subheadings, and the largest absolute difference in F1 score is 2.8% for the “diagnostic imaging” subheading. Both neural network methods can be seen to have reasonably consistent performance across the different critical subheadings, and per-subheading precision and recall are also fairly balanced.
Figure 3:
Micro precision, recall, and F1 score for MeSH pair prediction by critical subheading.
Table 3 shows the standalone performance of the main and subheading prediction CNN models of the chained method. The subheading prediction CNN was evaluated for all manually indexed main headings in the test set. The main and subheading prediction models are shown to have standalone F1 scores of 0.638 and 0.664 respectively, and error propagation from main heading prediction explains the lower end-to-end MeSH pair prediction performance reported in Table 2.
Table 3:
Standalone performance of chained method models.
| Model | Precision | Recall | F1 Score |
| Main heading prediction CNN | 0.681 | 0.600 | 0.638 |
| Subheading prediction CNN | 0.634 | 0.697 | 0.664 |
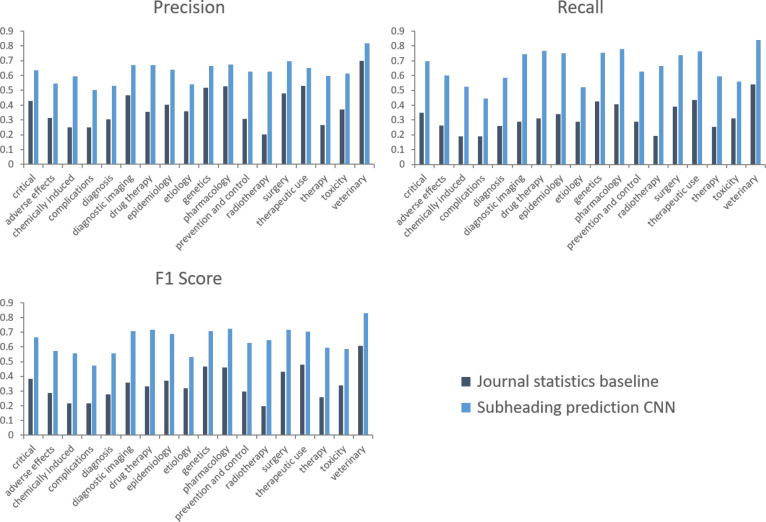
Figure 4 compares the performance of the subheading prediction CNN to the journal statistics baseline for manually indexed main headings. The CNN model is found to outperform the baseline by 28.0% points in terms of F1 score, and can be seen to have consistently higher precision and recall. Like for the end-to-end evaluation, the CNN subheading prediction performance is reasonably consistent across the different critical subheadings, and per-subheading precision and recall are also fairly balanced. Comparing Figures 3 and 4, it looks like the subheading prediction CNN is responsible for much of the per-subheading performance variation of the chained method.
Figure 4:
Micro precision, recall, and F1 score subheading prediction performance for manually indexed main headings.
Indexing Consistency Study Results
In total 1839 double indexed article pairs were identified in the MEDLINE 2019 baseline, and the study results are shown in Table 4. Micro precision, recall, and F1 score metrics were computed assuming that one article in the pair has the ground-truth indexing. Subheading assignment metrics were computed for MeSH pairs for which both indexers agreed on the main heading, and the reported errors were computed using bootstrapping with 10,000 samples.
Table 4:
Indexing consistency study results for critical subheadings.
| Study | Precision | Recall | F1 Score |
| Main heading/subheading pair assignment | 0.42±0.01 | 0.44±0.01 | 0.43±0.01 |
| Subheading assignment | 0.65±0.01 | 0.68±0.01 | 0.66±0.01 |
Comparing manual and automatic indexing consistency using F1 score, the best performing chained neural network method is found to exceed human indexing consistency for MeSH pair assignment by about 1.5% points. For the subtask of assigning subheadings to known main headings, the standalone subheading prediction model of the chained method is found to match manual indexing consistency.
Indexer Evaluation Results
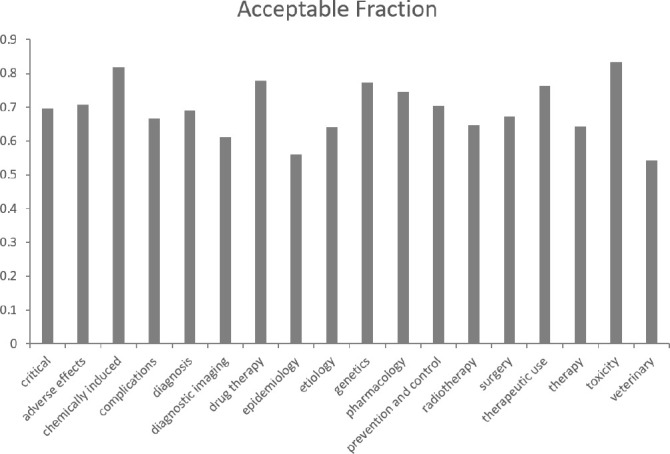
Indexers evaluated the false positive predictions of the chained method, and the evaluation results are shown in Figure 5. The figure plots the fraction of acceptable false positive predictions by critical subheading, and shows that on average 70% of false positives were considered to be acceptable. The results are fairly consistent for different critical subheadings: the highest acceptable fraction is 0.83 for “toxicity”, while the lowest acceptable fraction is 0.54 for “veterinary”.
The shared set of 12 articles contained 57 false positives, and the two indexers agreed on 95% of their determinations. The indexers have told us that they discussed the shared set false positive examples in order to calibrate their determinations for the main study, and 95% may therefore be an over estimate of their inter-indexer agreement.
Discussion
This paper has compared two neural network methods for fine-grained automatic MeSH indexing. Both methods were found to significantly outperform the existing MTI implementation: offering a small improvement in precision, and close to three times the recall of current system. The two methods were also found to exceed manual indexing consistency for MeSH pair assignment when compared using F1 score, and this is important because it suggests that future improvements in performance may be more limited and difficult to achieve.
The paper also includes an indexer evaluation of particularly problematic false positive predictions, and the evaluation results were encouraging because 70% of false positives were considered to be either acceptable alternative indexing, or imperfect indexing that does not need to be removed. This gives us confidence that the updated MeSH term recommendations will be well received by NLM indexers.
For many tasks, end-to-end approaches have been shown to outperform multi-step methods. However, in this paper the performance of end-to-end and multi-step methods are found to be similar: the multi-step chained method achieves a marginally better F1 score, even when trained on less data. The lower than expected performance of the end-to-end method may be explained by the sparsity of training data for individual MeSH pairs. The network may find it difficult to learn general rules that apply to individual subheadings, or groups of related main headings, because it is not provided with the information that certain MeSH pairs are closely related due to shared subheadings and/or similar main headings.
In comparison, the data sparsity problem is less severe for the chained method. The standalone subheading prediction model is explicitly told the subheading of each MeSH pair and is able to learn subheading prediction rules that generalize across main headings. The model does still suffer from a sparse input problem, and this is because the one-hot encoded main heading input does not provide any information about how main headings are related (e.g. by the MeSH hierarchy). Another potential weakness of the chained method is that it predicts subheading independently for each main heading, and it is therefore unable to coordinate its subheading predictions to describe two-way relationships between main headings (e.g.“Parkinson Disease/drug therapy; Dopamine Agonists/therapeutic use”).
Finally, there are also various advantages and disadvantages of the two methods relating to training and deployment. The main advantage of the chained method, compared to the end-to-end method, is that its component models have fewer parameters due to their smaller output sizes. The models therefore require less GPU memory, and the training time per example is lower. The main disadvantage of the chained method, compared to the end-to-end method, is the additional complexity that arises from managing two models and passing data between them.
Conclusion
This work has revisited the subheading attachment problem, 12 years after the feature was first added to MTI, and very significant performance improvements were demonstrated using a modern Convolutional Neural Network text classification architecture. The paper focused on 17 critical subheadings, and the performance of end-to-end and chained classifier methods were compared. The best performing chained classifier method was found to outperform the current MTI implementation with a 3.7% absolute improvement in precision, and close to three times the recall of the existing system. An indexer evaluation of false positive predictions was performed, and the results are encouraging, with 70% of false positives considered to be acceptable. In the future, we plan to integrate the chained classifier method into MTI, and to extend this work to provide recommendations for all 76 subheadings. We are also interested to see if pretrained transformer neural network architectures like BioBERT15 can provide additional performance improvements.
Acknowledgements
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM), and Lister Hill National Center for Biomedical Communications (LHNCBC).
We would like to thank Susan Schmidt and Deborah Whitman for completing the indexer evaluation and for their invaluable advice on MeSH indexing.
Footnotes
Excluded relationships: “Corrected and Republished”, “Reprint”, “Republished”, “Retracted and Republished”, “Update”.
Figures & Table
Figure 5:
Indexer evaluation results.
References
- 1.Mork J, Aronson A, Demner-Fushman D. 12 years on – is the NLM medical text indexer still useful and relevant? Journal of Biomedical Semantics. 2017;8(1):8. doi: 10.1186/s13326-017-0113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neveol A, Shooshan SE, Humphrey SM, Mork JG, Aronson AR. A recent advance in the automatic indexing of the biomedical literature. Journal of Biomedical Informatics. 2009;42(5):814–823. doi: 10.1016/j.jbi.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsatsaronis G, Balikas G, Malakasiotis P, Partalas I, Zschunke M, Alvers MR, et al. An overview of the BioASQ large-scale biomedical semantic indexing and question answering competition. BMC Bioinformatics. 2015;16(1):138. doi: 10.1186/s12859-015-0564-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng S, You R, Wang H, Zhai C, Mamitsuka H, Zhu S. DeepMeSH: deep semantic representation for improving large-scale MeSH indexing. Bioinformatics. 2016;32(12):i70–i79. doi: 10.1093/bioinformatics/btw294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xun G, Jha K, Yuan Y, Wang Y, Zhang A. MeSHProbeNet: a self-attentive probe net for MeSH indexing. Bioinformatics. 2019;35(19):3794–3802. doi: 10.1093/bioinformatics/btz142. [DOI] [PubMed] [Google Scholar]
- 6.Lin J, Wilbur WJ. PubMed related articles: a probabilistic topic-based model for content similarity. BMC Bioinformatics. 2007;8(1):423. doi: 10.1186/1471-2105-8-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.NLM medical text indexer first line indexing (MTIFL) and MTI review filtering (MTIR) [Internet] Bethesda (MD): National Library of Medicine (US); 2020 Feb 3. [cited 2020 July 2]. Available from: https://ii.nlm.nih.gov/MTI/MTIFL.shtml . [Google Scholar]
- 8.Incorporating values for indexing method in MEDLINE/PubMed XML. NLM Technical Bulletin. 2018 Jul- Aug;(423).
- 9.MEDLINE/PubMed 2019 baseline. 2019 Jan 2. Available from: https://mbr.nlm.nih.gov/ Download/Baselines/2019/
- 10.Liu J, Chang WC, Wu Y, Yang Y. Deep learning for extreme multi-label text classification. Proceedings of the 40th International ACM SIGIR Conference on Research and Development in Information Retrieval; Tokyo, Japan. ACM; 2017 Aug 7-11. pp. 115–124. 2017. [Google Scholar]
- 11.Kim Y. Convolutional neural networks for sentence classification. Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing; 2014 Oct 25-29; Doha, Qatar. ACL; 2014. pp. 1746–1751. [Google Scholar]
- 12.Rae AR, Savery ME, Mork JG, Demner-Fushman D. A high recall classifier for selecting articles for MEDLINE indexing. Proceedings of the 2019 AMIA Annual Symposium; 2019 Nov 14-16; Washington DC, USA. AMIA; 2019. pp. 727–734. [PMC free article] [PubMed] [Google Scholar]
- 13.Kudo T, Richardson J. SentencePiece: a simple and language independent subword tokenizer and detokenizer for neural text processing. Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing; 2018 Nov 2-4; Brussels, Belgium. ACL; 2018. pp. 66–71. [Google Scholar]
- 14.Funk ME, Reid CA. Indexing consistency in MEDLINE. Bulletin of the Medical Library Association. 1983;71(2):176. [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2019;36(4):1234–1240. doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]