ABSTRACT
Reinfection by the severe acute respiratory syndrome coronavirus type 2 (SARS-COV-2) has been reported in many countries, suggesting that the virus may continue to circulate among humans despite the possibility of local herd immunity due to massive previous infections. The emergence of variants of concern (VOC) that are more transmissible than the previous circulating ones has raised particular concerns on the vaccines effectiveness and reinfection rates. The P.1 lineage was first identified in December 2020 in Manaus city and is now globally spread. We report the first case of reinfection of SARS-CoV-2 caused by the P.1 variant outside of Manaus. The potential of these new variants to escape naturally and vaccine- induced immunity highlights the need for a global vigilance.
KEYWORDS: SARS-CoV-2, COVID-19, Reinfection, Variants of concern, P.1 lineage
INTRODUCTION
As of March 2021, the coronavirus disease 2019 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) has resulted in over 118 million cases and 2.7 million deaths worldwide 1 . The absence of pre-existent immunity against SARS-CoV-2 could be one of the factors associated with its rapid global spread. Previous studies suggest that immunoglobulin G (IgG) antibodies against SARS-CoV-2 generated post-infection are associated with around 80% protection from reinfection for at least six months 2 – 4 . Since the beginning of the pandemic, concerns on the possibility of reinfection have been raised. However, detection of reinfection is challenging and so far few reinfection cases caused by phylogenetically distinct lineages have been described 5 , suggesting that reinfection is a rare event. More recently, the emergence of new variants of concern (VOC), such as B.1.1.7 (UK), B.1.351 (South Africa) and P.1 (Brazil) that rapidly spread and may escape from neutralizing antibodies 6 , 7 complicates matters further. Here we describe a symptomatic case of SARS-CoV-2 reinfection caused by the P.1 lineage.
CASE DESCRIPTION
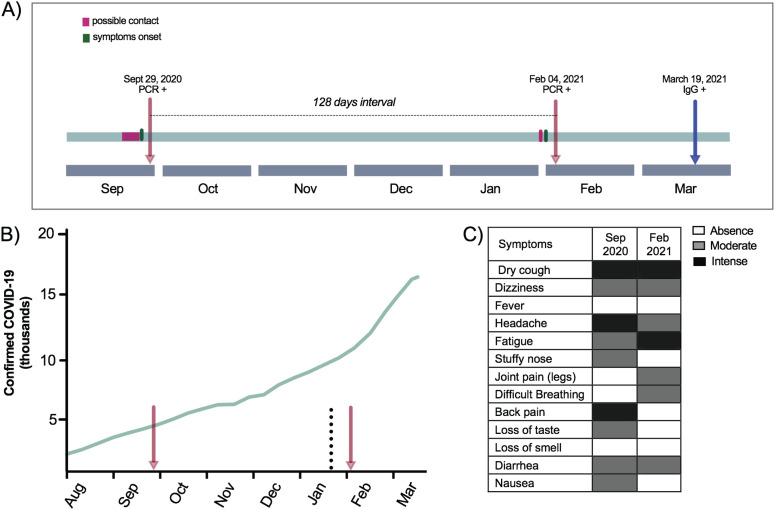
We present a case of SARS-CoV-2 reinfection in a 26-year-old woman, resident of Araraquara city, Sao Paulo State, Brazil. The patient reported having rheumatism but had never received any specific treatment. In the first infection episode, the patient sought the public health system for COVID-19 testing on September 29, 2020 (Figure 1A). The patient reported symptoms consistent with COVID-19 (Figure 1A and 1C) since September 25, 2020, and reported that her husband tested positive for SARS-CoV-2 by real-time quantitative reverse transcription-polymerase chain reaction (RT-qPCR) six days before. After 24h, her COVID-19 diagnosis was confirmed by RT-qPCR. The mild-COVID-19 was completely resolved during the isolation period, and no hospitalization was required.
Figure 1. The context of the two episodes of SARS-CoV-2 infection: A) Time line of the COVID-19 first and second episodes; B) Cumulative number of cases in Araraquara city from August, 2020 to March, 2021. Red arrows correspond to the patient's COVID-19 episodes. The dotted line highlights the first detection of the P.1 lineage in Araraquara city; C) Description of the symptoms reported by the patient during the first and the second episodes.
Then, in early February, 2021, she reported similar symptoms described in the COVID-19 episode in September 2020 with joint pain in the right leg in addition to difficulty breathing (Figure 1C). Two days later, her co-worker was also diagnosed with COVID-19, and all employees of the company were required to perform the RT-qPCR. On February 4, 2021, her reinfection was confirmed by means of SARS-CoV-2 RNA detection in a nasopharyngeal swab sample by RT-qPCR (Figure 1A). Similar to the previous SARS-CoV-2 episode, she presented only with mild symptoms and hospitalization was again not required. However, this time the patient reported persisting symptoms even two weeks after the second episode, including tiredness, dizziness and fatigue.
Samples from her two SARS-CoV-2 episodes were sent to the Institute of Tropical Medicine at the University of Sao Paulo to confirm the molecular diagnosis and to perform the whole virus genome sequencing. Samples were submitted to a RT-qPCR protocol that is able to differentiate between VOC and non-VOC viruses 8 . The results indicated that the first infection in September 2020 was caused by a non-VOC virus. This is expected as the two currently circulating VOCs in Brazil (B.1.1.7 and P.1) had not emerged yet 6 , 9 . However, the RT-qPCR results suggested that the second episode was caused by a VOC, that could be either B.1.1.7 or P.1. The cycle threshold (CT) values of the VOC RT-qPCR were 21 and 24 for the samples from the first and second episode, respectively.
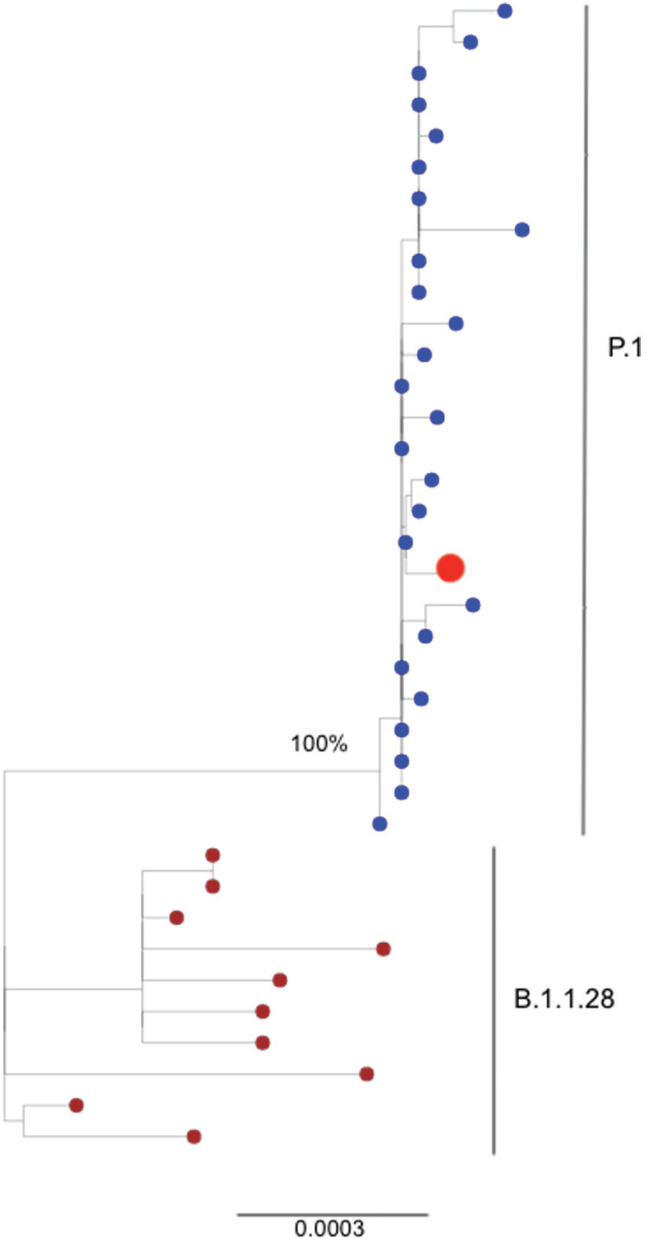
RNA from the second episode was submitted to SARS-CoV-2 complete genome sequencing using the MinION sequencing platform (Oxford Nanopore Technologies, ONT, UK) as previously described 10 . Unfortunately, the first sample was exhausted during RT-PCR protocols. The SARS-CoV-2 sequencing covered 83% of the whole virus genome (20-fold coverage) and according to Pangolin it was classified as P.1 lineage 11 . The SARS-CoV-2 complete genome generated in this study is available at GISAID under ID EPI_ISL_1004238. To contextualize the virus isolated from the second infection, a maximum likelihood (ML) phylogenetic tree was constructed with PhyML v.3.0 12 using as reference viruses from P.1 lineage (Manaus, Araraquara and Sao Paulo city) and the parent lineage B.1.1.28. According to the ML tree, the virus from the second infection of this case report clustered within a monophyletic cluster of P.1 viruses (Figure 2). Finally, the patient's serum was collected in March 19, 2021 and a serological assay for anti-SARS-COV-2 IgG detection was performed (Euroimmun Anti-SARS-CoV-2 ELISA Assay, Lübeck, Germany). The result was positive and the antibody detected was of high avidity as determined by the urea 6M treatment performed right after the ELISA test.
Figure 2. Maximum Likelihood (ML) phylogenetic tree of 38 complete genomes from P.1 and B.1.1.28 SARS-CoV-2. The isolate of the patient from Araraquara city corresponding to the second infection (red dot) clustered within the well supported clade of P.1 viruses from Manaus, Araraquara and Sao Paulo (blue dots) retrieved from GISAID. Viruses belonging to the B.1.1.28 parent lineage are represented by dark red dots.
DISCUSSION
We report here the first case of SARS-CoV-2 reinfection caused by the P.1 lineage outside of Manaus 13 . The P.1 lineage emerged in November, 2020 in Manaus city, and spread fast, causing the prevalence to increase from 25% to almost 90% in one month 6 . Later on, the P.1 lineage was detected in several Brazilian States and 45 countries around the world 14 . In Sao Paulo State, the P.1 lineage was first described in Araraquara city, in late January, 2021, during a dramatic increase in the number of cases and hospitalizations (Figure 1B). One month later, Araraquara's health system reached its limits and the P.1 lineage was responsible for >95% of all positive samples in the municipality (data not shown). Our finding supports that a natural SARS-CoV-2 infection may not necessarily prevent a secondary infection. As previous reported 15 , 16 , the secondary SARS-CoV-2 infection in the female patient did not present with increased severity of symptoms, except that in this case, some symptoms have persisted for more than 14 days.
Based on in vitro studies, antibodies binding to the viral spike induced by vaccine or from human convalescent sera present with decreased neutralization capacity against the emerging SARS-CoV-2 variants 7 , 17 . Furthermore, detailed data on SARS-CoV-2 seroprevalence in Manaus city suggested that P.1 has a 25-61% capacity to evade the immunity elicited by a previous infection caused by non-P.1 viruses 6 . However, it is too premature to attribute this particular reinfection case to the ability of the new variants to subvert the immune system. To the best of our knowledge, the 31 SARS-CoV-2 reinfection episodes described so far (with the exception of Naveca et al. 13 and Graham et al. 18 ) were caused by non-VOC viruses 5 , indicating that reinfection with P.1 evading immunity induced by the infection of previously circulating lineages is not a particularity of VOC.
Reinfection is well known characteristic of coronaviruses, such as NL63 and 229E, for which the protection elicited by antibodies may last only six months to one year 19 . According to Bauer et al. 20 , the avidity of IgG towards SARS-CoV-2 antigens is low for the majority of patients they tested, even several months after the disease. Here, the patient presented with high avidity IgG anti-SARS-CoV-2 a month after the second infection. Following this reasoning, we expected the patient to present with high avidity antibodies four months after the first episode, nevertheless, these antibodies were not able to prevent the second episode caused by the P.1 lineage.
The strategies used by SARS-CoV-2 to evade the immune responses remain unclear, and a better understanding on the quality and durability of cellular and humoral immunity elicited by natural infections with SARS-CoV-2 or vaccines are crucial. The emergence of variants with a repertoire of mutations in the spike protein highlights the need of quantitative assessments on the frequency of specific reinfection rates caused by SARS-CoV-2 variants.
ACKNOWLEDGMENTS
This work was supported by FAPESP grant Nº 2019/03859-9; by Medical Research Council and CADDE partnership award (MR/S0195/1) and FAPESP grant Nº 2018/14389-0; and a John Fell Research Fund (grant Nº 005166). NRF is supported by a Wellcome Trust and Royal Society Sir Henry Dale Fellowship (204311/Z/16/Z). WMS is supported by FAPESP Nº 2017/13981-0, 2019/24251-9 and CNPq Nº 408338/2018-0.
REFERENCES
- 1.World Health Organization WHO Coronavirus (COVID-19) dashboard. [cited 2021 Apr 6]. Available from: https://covid19.who.int.
- 2.Hansen CH, Michlmayr D, Gubbels SM, Molbak K, Etheberg S. Assessment of protection against reinfection with SARS-CoV-2 among 4 million PCR-tested individuals in Denmark in 2020: a population-level observational study. Lancet. 2021;397:1204–1212. doi: 10.1016/S0140-6736(21)00575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lumley SF, O’Donnell D, Stoesser NE, Matthews PC, Howarth A, Hatch SB, et al. Antibody status and incidence of SARS-CoV-2 infection in health care workers. N Engl J Med. 2021;384:533–540. doi: 10.1056/NEJMoa2034545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall V, Foulkes S, Charlett A, Atti A, Monk EJ, Simmons R, et al. Do antibody positive healthcare workers have lower SARS-CoV-2 infection rates than antibody negative healthcare workers? Large multi-centre prospective cohort study (the SIREN study), England June to Nov 2020. medRxiv. 2021 In Press. [Google Scholar]
- 5.Stokel-Walker C. What we know about covid-19 reinfection so far. BMJ. 2021;372:n99–n99. doi: 10.1136/bmj.n99. [DOI] [PubMed] [Google Scholar]
- 6.Faria NR, Mellan TA, Whittaker C, Claro IM, Candido DS, Mishra S, et al. Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. medRxiv. 2021 doi: 10.1126/science.abh2644. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen RE, Zhang X, Case JB, Winkler ES, Liu Y, VanBlargan LA, et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med. 2021 doi: 10.1038/s41591-021-01294-w. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romano CM, de Jesus JG, Felix AC, de Paula AV, Andrade PS, de Oliveira FM, et al. Real-Time PCR protocol to screen for SARS-COV-2 variants of concern (B.1.1.7, P.1 and B.1.1.35) [cited 2021 Apr 6]. Protocols.io. Available at: https://www.protocols.io/view/real-time-pcr-protocol-to-screen-for-sars-cov-2-va-bszbnf2n.
- 9.Claro IM, Sales FC, Ramundo MS, Candido DS, Silva CA, de Jesus JG, et al. Local transmission of SARS-CoV-2 Lineage B.1.1.7, Brazil, December 2020. Emerg Infect Dis. 2021;27:970–972. doi: 10.3201/eid2703.210038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jesus JG, Sacchi C, Candido CA, Claro IM, Sales FC, Manuli ER, et al. Importation and early local transmission of COVID-19 in Brazil, 2020. Rev Inst Med Trop Sao Paulo. 2020;62:e30. doi: 10.1590/S1678-9946202062030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centre for Genomic Pathogens Surveillance Pangolin COVID-19 lineage assignment. [cited 2021 Apr 6]. Available from: https://pangolin.cog-uk.io/
- 12.Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online: a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naveca F, da Costa C, Nascimento V, Souza V, Corado A, Nascimento F, et al. SARS-CoV-2 reinfection by the new Variant of Concern (VOC) P.1 in Amazonas, Brazil. Virol Org. 2021 In Press. [Google Scholar]
- 14.World Health Organization COVID-19 weekly epidemiological update. [2021 Apr 6]. Available at: https://reliefweb.int/sites/reliefweb.int/files/resources/20210330_Weekly_Epi_Update_33.pdf.
- 15.Van Elslande J, Vermeersch P, Vandervoort K, Wawina-Bokalanga T, Vanmechelen B, Wollants E, et al. Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1330. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.To KK, Hung IF, Ip JD, Chu AW, Chan WM, Tam AR, et al. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1275. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Beltran WF, Lam EC, St Denis K, Nitido AD, Garcia ZH, Hauser BM, et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021 doi: 10.1016/j.cell.2021.04.006. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham MS, Sudre CH, May A, Antonelli M, Murray B, Varsavsky T, et al. Changes in symptomatology, re-infection and transmissibility associated with SARS-CoV-2 variant B.1.1.7: an ecological study. medRxiv. 2021 doi: 10.1016/S2468-2667(21)00055-4. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edridge AW, Kaczorowska J, Hoste AC, Bakker M, Klein M, Loens K, et al. Seasonal coronavirus protective immunity is short-lasting. Nat Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- 20.Bauer G, Struck F, Schreiner P, Staschik E, Soutschek E, Motz M. The serological response to SARS corona virus-2 is characterized by frequent incomplete maturation of functional affinity (avidity) Sci Rep. 2021 In Press. [Google Scholar]