Abstract
The number of mitochondria in blastocysts is a potential marker of embryo quality. However, the molecular mechanisms governing the mitochondrial number in embryos are unclear. This study was conducted to investigate the effect of reduced mitochondrial reactive oxygen species (ROS) levels on mitochondrial biogenesis in porcine embryos. Oocytes were collected from gilt ovaries and activated to generate over 4 cell-stage embryos at day 2 after activation. These embryos were cultured in media containing either 0.1 μM MitoTEMPOL (MitoT), 0.5 μM Mitoquinol (MitoQ), or vehicle (ethanol) for 5 days to determine the rate of development to the blastocyst stage. The mitochondrial number in blastocysts was evaluated by real-time polymerase chain reaction (PCR). Five days after activation, the embryos (early morula stage) were subjected to immunostaining to determine the expression levels of NRF2 in the nucleus. In addition, the expression levels of PGC1α and TFAM in the embryos were examined by reverse transcription PCR. One day of incubation with the antioxidants reduced the ROS content in the embryos but did not affect the rate of development to the blastocyst stage. Blastocysts developed in medium containing MitoT had lower mitochondrial DNA copy numbers and ATP content, whereas MitoQ showed similar but insignificantly trends. Treatment of embryos with either MitoT or MitoQ decreased the expression levels of NRF2 in the nucleus and levels of PGC1α and TFAM. These findings indicate that reductions in mitochondrial ROS levels are associated with low mitochondrial biogenesis in embryos.
Keywords: Blastocyst, Embryo development, Mitochondrial biogenesis, Mitochondrial number, Reactive oxygen species
Technology for in vitro embryo production has been developed for humans and cows, and blastocysts developed in vitro are used for embryo transfer worldwide. Through these numerous efforts, the quality of in vitro-developed embryos has been improved but remains lower than that of in vivo-developed embryos. Mitochondria are important organelles for oocytes and embryos, and their number is thought to be an important marker of oocyte quality. The mitochondrial number in oocytes is notably greater than that observed in cells, and increases in mitochondrial number have been observed between early antral follicles (approximately 50,000) and antral follicles (200,000–350,000) in pigs [1,2,3,4]. A larger mitochondrial number in oocytes is closely associated with high oocyte maturation, fertilization, and developmental competence [5,6,7]. After fertilization, the mitochondrial number decreases and is maintained at a low level until the blastocyst stage because of the low mitochondrial biosynthesis activity [8]. In blastocysts, the mitochondrial number increases and mitochondrial morphology changes to meet the active proliferation requirement of trophoblasts [9, 10]. Therefore, a greater mitochondrial number is considered as an important factor underlying proper post-implantation development [11]. However, the relationship between mitochondrial number and reproductive potential in blastocysts is not completely clear. Accumulating evidence supports the quiet embryo hypothesis [12], which states that blastocysts with higher mitochondrial DNA copy numbers have low quality or low reproductive ability [13,14,15,16].
Mitochondrial biogenesis is regulated at the transcriptional, translational, and post-translational levels [17], and peroxisome proliferator-activated receptor γ co-activator 1α (PGC1α) is a key regulator of mitochondrial biogenesis [18]. In addition, NRF2, NRF1, and downstream TFAM are the main regulators of mitochondrial biosynthesis [19, 20].
However, the factors that stimulate mitochondrial biogenesis during implantation development are unclear. In an exercise model, it was suggested that reactive oxygen stress acts as a signal to increase mitochondrial biogenesis through the NRF2-PGC1a-TFAM pathway [20]. Accumulating evidence has shown that the expression levels of NRF-1, NRF-2, PGC1α, and PGC1β are upregulated by oxidative stress [21,22,23], indicating a close link between reactive oxygen species (ROS) and mitochondrial biogenesis [24]. Furthermore, ROS levels are considered as signaling molecules that regulate the redox regulation of cellular processes including proliferation, differentiation, and apoptosis [25, 26]. 2,2,6,6-Tetramethyl-4-[[5-(triphenylphosphonio)pentyl]oxy]-1-piperidinyloxy bromide (MitoTEMPOL: MitoT) and 10-(2,5-dihydroxy-3,4-dimethoxy-6-methylphenyl)decyl]triphenyl-phosphonium, monomethanesulfonate (Mitoquinol: MitoQ) are specific inhibitors of mitochondrial reactive oxygen that can reduce the generation of mitochondria-specific ROS or superoxide [27]. Therefore, we hypothesized that modulation of ROS levels in mitochondria affects mitochondrial biogenesis in embryos. To evaluate this hypothesis, we added mitochondrial antioxidants to the culture medium of porcine parthenogenetic-activated embryos and evaluated embryo development and mitochondrial DNA copy number.
Materials and Methods
Ethics statement
The animal ethics committee of the Tokyo University of Agriculture approved the use of oocytes from gilt ovaries collected from a slaughterhouse where ovaries are otherwise discarded.
Chemicals and media
All drugs used in this study were purchased from Nacalai Tesque (Kyoto, Japan) unless stated otherwise. The medium for in vitro maturation (IVM) of oocytes was TCM199 (Gibco, Grand Island, NY, USA) supplemented with 10% v/v porcine follicular fluid, 0.5 mM L-cysteine, 1 mM L-glutamine, 0.9 mM sodium pyruvate, 10 ng/ml epidermal growth factor (Sigma-Aldrich, St. Louis, MO, USA), 5% fetal calf serum, 10 IU/ml equine chorionic gonadotropin (ASKA Pharma Co., Ltd., Tokyo, Japan), and 10 IU/ml human chorionic gonadotropin (Fuji Pharma Co., Ltd., Tokyo, Japan). Porcine follicular fluid was aspirated from the antrum follicles (3–5 mm in diameter) of gilts, centrifuged (10,000 × g for 5 min), and stored at –20°C until use. The medium for in vitro culture (IVC) of embryos was porcine zygote medium 3 (PZM3) [19, 28]. IVM was performed under atmospheric conditions of 5% CO2 and 95% air at 38.5°C, and IVC was performed under atmospheric conditions of 5% O2, 5% CO2, and 90% N2 at 38.5°C. MitoTEMPOL (MitoT; 18796, Cayman Chemical, Ann Arbor, MI, USA) and Mitoquinol (MitoQ; 89950, Cayman Chemical) were diluted in water and ethanol by1000-fold to obtain the final concentrations (0.1 and 0.5 μM for MitoT and MitoQ, respectively). The concentrations of the antioxidants were determined in a previous study [29, 30]. In addition, in our preliminary experiments, we found that 0.1 μM MitoT and 0.5 μM MitoQ were the most effective among 0, 0.1, and 1 μM of MitoT and 0, 0.5, and 5 μM MitoQ, respectively. Control groups were prepared in the same volume of vehicle (1/1000 ethanol). Supplementation of 1/1000 ethanol to the culture medium did not affect developmental rate to blastocyst stage (Control 16.7 ± 3.6%, Ethanol 19.0 ± 2.4, P = 0.59).
Oocyte collection and in vitro maturation
Cumulus-oocyte complexes (COCs) were collected and used for IVM according to our previous method [31]. Briefly, ovaries were collected from gilts at the slaughter house, stored at 37°C in phosphate-buffered saline containing antibiotics, and transported to the laboratory within 1 h. The COCs were aspirated from antral follicles (3–5 mm in diameter) using a syringe with a 21-G needle (Terumo, Tokyo, Japan). The COCs were cultured in IVM medium for 44 h (10 COCs/100 μl drops) and then subjected to activation.
Parthenogenetic activation (PA) of oocytes and in vitro culture of embryos
After IVM, all oocytes were subjected to parthenogenetic activation. Polyspermy occurs frequently in porcine oocytes fertilized in vitro, and the rate of polyspermic fertilization varies to a great extent among ovary batches. In addition, the ploidy of the embryos affects mitochondrial numbers in the resultant blastocysts [31]. To avoid these differences, we conducted parthenogenetic activation rather than in vitro fertilization. The oocytes were denuded from surrounding cumulus cells and activated in a solution containing 280 mM mannitol, 0.05 mM CaCl2 and 0.1 mM MgSO4 using a single electrical pulse of 100 V for 0.1 ms and CUY500P1 electrode (NEPA21 (NepaGene Ltd., Chiba, Japan) followed by incubation in PZM3 containing 10 µM cytochalasin B and 10 µM cycloheximide for 4.5 h. The embryos were then cultured in IVC media (10 embryos/50 µl drops). After 2 days of PA, cleaved embryos with normal morphology (> 4-cell stage) were individually cultured in 6 µl of IVC media supplemented with mitochondrial antioxidants (0.1 μM of MitoT and 0.5 μM of MitoQ, or 1/1000 ethanol vehicle) for 5 days. Embryos at the > 4-cell stage after 2 days of PA were predicted to have good subsequent developmental ability. We used greater than 4-cell stage embryos for the experiments to obtain a uniform developmental background.
Evaluation of developmental competence of embryos
After 5 days of incubation (day 7 post-activation), the blastocyst rate was examined, and the total cell number of randomly selected blastocysts was counted after Hoechst 33342 staining under a fluorescence microscope (Olympus, Tokyo, Japan). Approximately 20–25 embryos (> 4-cell stage embryos) were cultured for each replicate and repeated 25 times, and the rate of embryos (day 2) reaching the blastocyst stage at day 7 was calculated. All blastocysts derived from each of the 6 trials were used for ATP (29 embryos were used for each experimental group, respectively) and mitochondrial DNA copy number measurement; 31, 24, and 32 embryos were used for control, MitoT, and MitoQ groups), and blastocysts derived from others (13 trials) were used to measure the total cell number of the blastocysts. To obtain cleaved embryos, 3,350 oocytes were used, and the total cleaved rate (over 4-cell stage embryos/oocytes at 48 h post-activation) was 60.0%. To measure the total cell number of the blastocysts, embryos were fixed in 4% paraformaldehyde overnight, followed by treatment with Triton-X 100 (0.25%) for 30 min and mounted on a slide glass with antifade reagent with DAPI (Invitrogen, Carlsbad, CA, USA). The number of nuclei was examined under a fluorescence microscope (DMI 6000B, Leica, Wetzlar, Germany).
Evaluation of mitochondrial function and number
Blastocysts obtained by IVC with or without antioxidants were used for the experiment. The ATP content of individual blastocysts was measured based on the luminescence generated in an ATP-dependent luciferin-luciferase reaction (ATP Assay Kit; TOYO B-Net., Ltd., Tokyo, Japan). Luminescence was measured immediately after the reaction using a luminometer (Spark 10M; Tecan, Männedorf, Switzerland). Blastocysts were individually transferred into 50 µl of distilled water and stored at −20°C until measurement. DNA from individual blastocysts was extracted by incubation in 30 µl of lysis buffer (final concentration: 20 mM Tris, 0.4 mg/ml proteinase K, 0.9% Nonidet P-40, and 0.9% Tween-20) at 55°C for 30 min, followed by incubation at 98°C for 10 min. The mitochondrial number was determined by PCR using a real-time system (Bio-Rad Laboratories Hercules, CA, USA) and KAPA SYBR Fast (Kapa Biosystem, Wilmington, MA, USA) as described previously [32,33,34,35]. Real-time PCR was performed at 95°C for 3 min, followed by 40 cycles at 98°C for 5 sec and 60°C for 11 sec. The primer set was designed using Primer-BLAST (NC_000845.1: forward, 5-atccaagcactatccatcacca-3 and reverse, 5-ccgatgattacgtgcaaccc-3; 10277-10431, 155 bp). A standard curve was generated for each assay using 10-fold serial dilutions, representing copies of the external standard, which was the PCR product of the corresponding gene cloned into a vector using the Zero Blunt TOPO PCR Cloning Kit (Invitrogen). To obtain the copy number of the standard DNA, the concentration of the standard DNA, molecular weight of the vector, and Avogadro’s number were used. Prior to its use, this product was sequenced for confirmation.
Measurement of mitochondrial ROS
We examined whether mitochondrial antioxidants reduced mitochondrial ROS. Embryos (> 4-cell stage embryos at day 2 post-activation) were cultured in medium containing vehicle (ethanol), MitoT (0.1 µM), or MitoQ (0.5 µM) for 24 h, and ROS in mitochondria were measured using MitoSOX (mitochondrial superoxide indicator, Invitrogen). Embryos were incubated with 5 µM MitoSox for 10 min, and then observed under a fluorescence microscope (Leica) to obtain images. Fluorescence intensity was calculated using ImageJ software (NIH https://imagej.nih.gov/ij/).
Reverse transcription PCR (RT-PCR)
Mitochondrial biogenesis starts from the morula stage, and when mitochondrial DNA copy number was compared between embryos, differential mitochondrial DNA copy number was detected from day 4 post-activation in pigs [31]. Therefore, embryos on day 5 (morula-stage 130 h after PA) were used for RT-PCR. Two days post-activation, cleaved embryos (> 4-cell stage) were transferred into medium containing MitoT, MitoQ, and vehicle (ethanol), and cultured for 3 days. Then, 30 embryos from each group were used for RNA extraction. RNA was extracted using an RNAqueous total RNA isolation kit (Invitrogen) and reverse-transcribed into cDNA using SuperScript IV Reverse Transcriptase and oligo(dT) primer. Next, cDNAs were diluted by 20-fold with DNAase and RNAase-free water. PCR was conducted with 8 μl of cDNA, 10 μl of KAPA SYBR® FAST qPCR (Kapa Biosystems), and 2 μl of primer set (final concentration: 0.2 µM). The primer sets are shown in Table 1. All PCRs were conducted at 95°C for 3 min, followed by 40 cycles of 98°C for 8 sec and 60°C for 10 sec. PCR products were examined by melting curve analysis and electrophoresis. Gene expression was determined by absolute quantification using a serially diluted standard plasmid into which each PCR product was cloned using the Zero Blunt™ TOPO™ PCR Cloning Kit. According to the minimal information for publication of Quantitative Real Time PCR Experiments guidelines [36], the expression levels of PGC1α and TFAM were normalized to that of B2M. The amplification efficiency was > 1.98. The experiment was repeated 4 times using differential embryo batches.
Table 1. Primer sequences used for reverse transcription PCR.
| Gene | Primer | Accession number | Amplicon size (bp) | |
|---|---|---|---|---|
| PGC1α | F | ttccgtatcaccacccaaat | NM_213963.2 | 137 |
| R | atctactgcctggggacctt | |||
| TFAM | F | ggcagactggcaggtgta | NM_001130211.1 | 165 |
| R | cgaggtctttttggttttcca | |||
| B2M | F | gatcagtatagctgccgcgt | NM_213978.1 | 87 |
| R | ctgtgatgccggttagtggt | |||
Expression levels of NRF2 in the nucleus
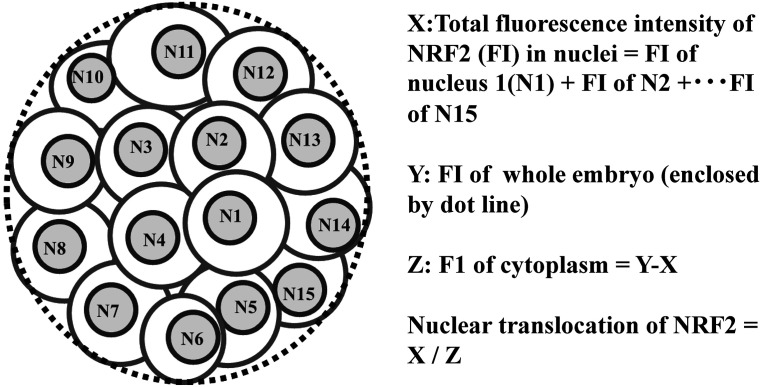
Embryos were immunoassayed against NRF2, and the levels of NRF2 in the nuclei of embryos were examined. Embryos (> 4-cell stage embryos at 2 days post-activation) were cultured in IVC medium containing MitoT, MitoQ, and vehicle (ethanol) for 3 days and subjected to immunostaining. Embryos were fixed in 4% paraformaldehyde overnight, followed by treatment with Triton-X 100 (0.25%) for 30 min, blocking in 5% bovine serum albumin-phosphate-buffered saline for 1 h, and immunostained as previously described [37]. The primary antibody was rabbit polyclonal anti-NRF2 (1:200, overnight, ab31163, Abcam), and the secondary antibody used was anti-rabbit IgG (H+L) F(ab')2 fragment (1:500, 1 h, Alexa Fluor® 555 Conjugate) (Cell Signaling Technology, Danvers, MA, USA), which were diluted in blocking solution, mounted onto glass slides using an anti-fade reagent containing DAPI, and observed under a Leica DMI 6000B microscope using LAS AF software (Leica). Nuclei in the blastomeres were visualized by DAPI staining. The fluorescence intensities of NRF2 in the nuclear region evidenced by DAPI and those of the whole embryo were captured to calculate the ratio of NRF2 nuclei expression level per cytoplasm for each embryo (Fig. 1). Next, 43, 40, and 43 embryos were examined for their NRF2 expression levels in the control (vehicle, ethanol), MitoT, and MitoQ groups, respectively. The fluorescence intensities were quantified using ImageJ software (NIH, Bethesda, MD, USA).
Fig. 1.
Evaluation of nuclear translocation of NRF2. Representative image of an embryo immune-stained against NRF2, and formulas to obtain fluorescence intensity of NRF2 in nuclei per fluorescence intensity of cytoplasm.
Statistical analysis
All data were analyzed by one-way analysis of variance followed by Fisher’s post hoc test. The percentage of embryo development was arcsine-transformed prior to analysis. P-values less than 0.05 were considered as statistically significant.
Results
Mitochondrial antioxidants decreased the levels of mitochondrial reactive oxygen.
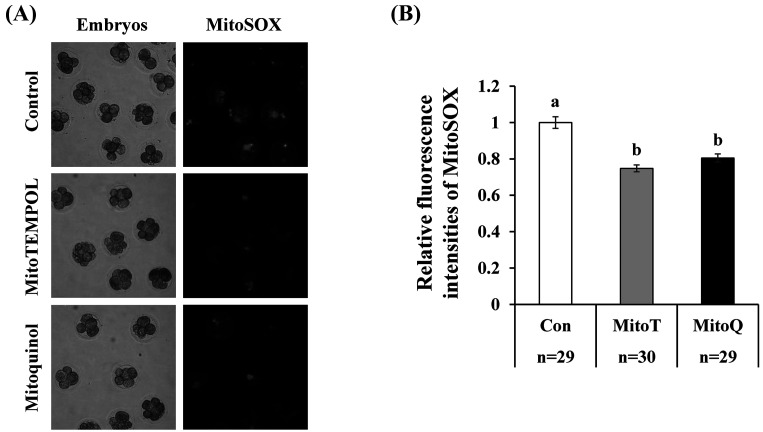
When early cleavage-stage embryos (> 4 cells at day 2 after activation) were cultured in a medium containing mitochondrial antioxidants (MitoT or MitoQ) for 24 h, the amount of mitochondrial ROS determined by MitoSOX significantly decreased compared to that in cleavage-stage embryos cultured with the vehicle (ethanol) (Fig. 2).
Fig. 2.
Effect of mitochondrial antioxidants on mitochondrial ROS levels in embryos. Cleaved embryos (> 4-cell stage at 2 days after parthenogenetic activation) were cultured in medium containing vehicle (ethanol 1:1000, Con) or mitochondrial antioxidants (0.1 µM MitoT or 0.5 µM MitoQ) for 24 h. Relative fluorescence intensities of embryos stained with MitoSOX were compared among the three groups. The fluorescence intensity of the control was normalized to 1.0. a–b, P < 0.05. Number of embryos (n) examined is stated at the bottom of the figure.
Developmental competence and mitochondrial function and quantity in blastocysts
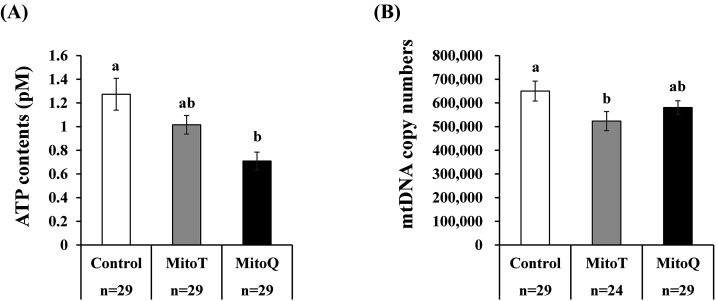
After culturing the embryos (> 4-cell stage embryos) in medium containing mitochondrial antioxidants, the developmental rate to the blastocyst stage and total cell number of the blastocysts did not change among the experimental groups (MitoT, MitoQ, and vehicle (ethanol)) (Table 2). Supplementation with MitoT and MitoQ led to decreased ATP and mitochondrial DNA contents in the blastocysts compared to in those developed with the vehicle, but the difference was not significant for MitoQ (Figs. 3A, 3B).
Table 2. Effect of mitochondrial antioxidants on developmental competence of porcine embryos.
| Group | No. of | No. of | No. of | Rate of | n= | TCN |
|---|---|---|---|---|---|---|
| replicates | embryos | blastocysts | blastocyst (%) | |||
| Control | 25 | 522 | 147 | 28.5 ± 3.2 | 89 | 37.8 ± 1.1 |
| MitoT | 25 | 526 | 158 | 30.5 ± 3.2 | 105 | 37.5 ± 1.0 |
| MitoQ | 25 | 534 | 171 | 32.2 ± 3.2 | 113 | 35.0 ± 1.3 |
At day 2 post-activation, > 4-cell stage embryos were cultured in medium containing ethanol (Con, 1:1000), and antioxidants (0.1 µM MitoT or 0.5 µM MitoQ) for 5 days, and the rate of blastocyst per embryos were examined. Total cell number (TCN) of the blastocysts and number of blastocyst (n) examined are shown. Data are presented as the mean ± SEM.
Fig. 3.
Effect of mitochondrial antioxidants on mitochondrial function and quantity in blastocysts. Cleaved embryos (> 4-cell stage embryos at 2 days after parthenogenetic activation) were cultured in medium containing vehicle (ethanol 1:1000, Con) or mitochondrial antioxidants (0.1 µM MitoT or 0.5 µM MitoQ) for 5 days. The ATP content (A) and mitochondrial DNA copy number (B) in blastocysts were compared among the three groups. a–b, P < 0.05. Number of embryos (n) examined is stated at the bottom of the figure.
Mitochondrial antioxidants decreased mitochondrial biogenesis
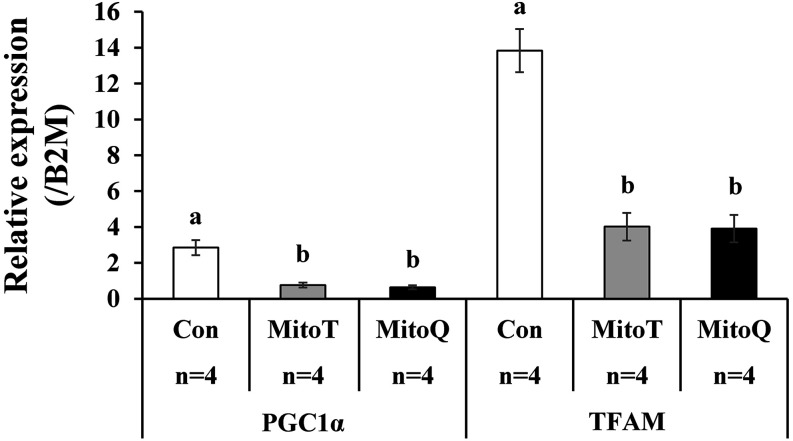
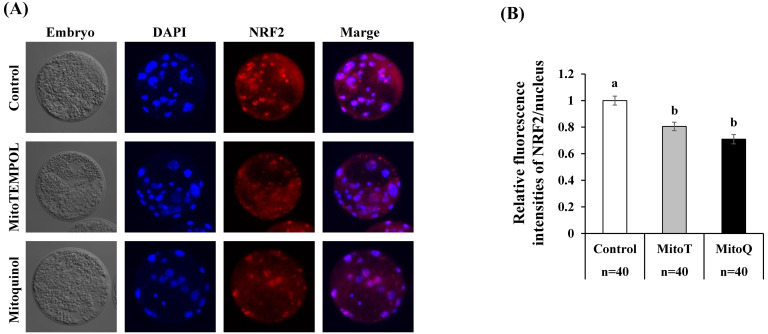
RT-PCR revealed that the expression levels of PGC1a and TFAM were significantly lower in embryos cultured with mitochondrial antioxidants than in their vehicle (ethanol)-treated counterparts (Fig. 4). Immunostaining revealed that the expression levels of NRF2 in the nucleus were significantly decreased in embryos cultured with mitochondrial antioxidants compared to in their vehicle-treated counterparts (Fig. 5).
Fig. 4.
Effect of mitochondrial antioxidants on expression levels of genes associated with mitochondrial biogenesis. Cleaved embryos (> 4-cell stage at 2 days after parthenogenetic activation) were cultured in medium containing vehicle (ethanol 1:1000, Con) or mitochondrial antioxidants (0.1 µM MitoT or 0.5 µM MitoQ) for 3 days. The expression levels of genes were normalized to that of beta-2-microglobulin (B2M) (A). a–b or c–d, P < 0.05. The experiment was repeated 4 times (n) using embryos derived from differential oocyte batches.
Fig. 5.
Effect of mitochondrial antioxidants on expression levels of NRF2 in the nucleus. Cleaved embryos (> 4-cell stage embryos at 2 days after parthenogenetic activation) were cultured in medium containing (ethanol 1:1000, Con) or mitochondrial antioxidants (0.1 µM MitoT or 0.5 µM MitoQ) for 3 days. A: Representative images of embryos stained to visualize NRF2 and nuclei (DAPI) to examine the nuclear translocation of NRF2. B: Relative fluorescence intensities of NRF2 in the nucleus were compared among the three groups. The fluorescence intensity of control embryos was normalized to 1.0. a–b, P < 0.05. Number of embryos (n) examined is stated at the bottom of the figure.
Discussion
MitoT and MitoQ mitochondrial superoxide scavengers reduced mitochondrial ROS levels in early cleaved embryos and reduced mitochondrial biogenesis during embryonic development. In addition, exposure to MitoT resulted in blastocysts with a low mitochondrial DNA copy number, while exposure to MitoQ resulted in blastocysts with low ATP content.
Mitochondria are crucial in the production of energy in cells. ROS are a common by-product of ATP generation in mitochondria. Although normal levels of ROS play a role in intercellular signaling, excessive ROS can cause mitochondrial dysfunction, damage cellular organelles, and induce apoptosis [38]. We confirmed that 24 h of incubation with MitoT and MitoQ reduced the levels of mitochondrial ROS in embryos. However, these antioxidants did not increase the rate of development to the blastocyst stage or total cell number of blastocysts compared with those treated with the vehicle. Yang et al. [29] reported that supplementation of the culture medium of porcine embryos with MitoT in an atmosphere of 5% CO2 in air improved embryonic development and increased cellular numbers. It is widely accepted that low oxygen tension is beneficial for embryonic development [39], and that high oxygen tension changes the morphology of mitochondria and decreases the development of mouse embryos, resulting in lower cell numbers and higher ATP content [40]. This suggests that under low oxygen tension, the amount of ROS in embryos was not sufficient to improve embryo development.
Interestingly, supplementation of the culture medium with MitoT decreased the mitochondrial DNA content in the blastocysts. In addition, these antioxidants induced inactive mitochondrial biogenesis, as indicated by decreased expression levels of PGC1α and TFAM expression as well as by low translocation of NRF2 into the nucleus. Mitochondrial biogenesis is well-regulated by crosstalk between mitochondria and the nucleus. Mitochondrial ROS are involved in the crosstalk, although their precise role remains unclear [41]. It has been reported that key factors regulating mitochondrial biogenesis, such as NRF1, NRF2, PGC1-1α, and PGC1-β, are upregulated by oxidative stress [21,22,23,24], and nuclear translocation of NRF2 was induced by oxidative stress [42, 43]. Therefore, mitochondrial ROS regulate mitochondrial biogenesis in embryos. One question remains as to why MitoT was more effective than MitoQ in reducing ATP and mitochondrial DNA copy number in embryos, even though both drugs reduced mitochondrial biogenesis and mitochondrial ROS. MitoT is a superoxide dismutase mimetic that reduces mitochondrial O2– to H2O2. MitoT combines an antioxidant moiety with the lipophilic cation triphenylphosphonium, which allows it to pass through lipid bilayers and accumulate in mitochondria [44]. MitoQ is a mitochondria-targeted antioxidant and a reduced form of mitoquinone [45]. Both drugs target mitochondrial ROS but exert their effects under different molecular mechanisms. Unraveling the differential extent of their effects is a target for future research.
In the present study, blastocysts that developed in the presence of mitochondrial antioxidants had low mitochondrial DNA copy numbers and ATP content. It is unclear whether these embryos have high developmental competence. However, we found a higher mitochondrial copy number in invitro-grown bovine blastocysts than in those grown in vivo [46]. It is widely accepted that in vitro culture conditions are closely related to ROS generation. In our previous report, granulosa cells cultured in vitro showed increased mitochondrial DNA content and ATP generation, whereas under low oxygen tension, both mitochondrial number and ATP were decreased [47]. In addition, it has been reported that low mitochondrial numbers in blastocyst-stage embryos are associated with the high implantation potential of the embryos [14]. In addition, embryos with abnormal ploidy have a large number of mitochondria in humans and pigs [35, 48]. In cells with abnormal ploidy, an imbalanced contribution of the mitochondrial genome and nuclear genomes induces an imbalance of mitochondrial proteins, which induces ER stress and generation of ROS [49]. In addition, dysfunction in the electron transport chain and increased misfolding of mitochondrial proteins induce increased ROS generation, which stimulates the biogenesis of mitochondrial DNA and thus an increased copy number [10]. Based on these reports and the results of the present study, ROS in embryos may be a key factor determining the mitochondrial number in blastocysts.
In conclusion, our results suggest that reducing the mitochondrial ROS level attenuates mitochondrial biogenesis during embryo development and results in blastocysts with low mitochondrial DNA copy number and function.
Conflicts of Interests
The authors declare that there are no conflicts of interest regarding this study.
Acknowledgments
We thank Kanagawa Meat Center for providing the ovaries.
References
- 1.Cotterill M, Harris SE, Collado Fernandez E, Lu J, Huntriss JD, Campbell BK, Picton HM. The activity and copy number of mitochondrial DNA in ovine oocytes throughout oogenesis in vivo and during oocyte maturation in vitro. Mol Hum Reprod 2013; 19: 444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Itami N, Shirasuna K, Kuwayama T, Iwata H. Resveratrol improves the quality of pig oocytes derived from early antral follicles through sirtuin 1 activation. Theriogenology 2015; 83: 1360–1367. [DOI] [PubMed] [Google Scholar]
- 3.Munakata Y, Ichinose T, Ogawa K, Itami N, Tasaki H, Shirasuna K, Kuwayama T, Iwata H. Relationship between the number of cells surrounding oocytes and energy states of oocytes. Theriogenology 2016; 86: 1789–1798.e1. [DOI] [PubMed] [Google Scholar]
- 4.Kansaku K, Munakata Y, Shirasuna K, Kuwayama T, Iwata H. Mitochondrial cell-free DNA secreted from porcine granulosa cells. Zygote 2019; 27: 272–278. [DOI] [PubMed] [Google Scholar]
- 5.Santos TA, El Shourbagy S, St John JC. Mitochondrial content reflects oocyte variability and fertilization outcome. Fertil Steril 2006; 85: 584–591. [DOI] [PubMed] [Google Scholar]
- 6.Reynier P, May-Panloup P, Chrétien MF, Morgan CJ, Jean M, Savagner F, Barrière P, Malthièry Y. Mitochondrial DNA content affects the fertilizability of human oocytes. Mol Hum Reprod 2001; 7: 425–429. [DOI] [PubMed] [Google Scholar]
- 7.Zeng HT, Ren Z, Yeung WSB, Shu YM, Xu YW, Zhuang GL, Liang XY. Low mitochondrial DNA and ATP contents contribute to the absence of birefringent spindle imaged with PolScope in in vitro matured human oocytes. Hum Reprod 2007; 22: 1681–1686. [DOI] [PubMed] [Google Scholar]
- 8.May-Panloup P, Vignon X, Chrétien MF, Heyman Y, Tamassia M, Malthièry Y, Reynier P. Increase of mitochondrial DNA content and transcripts in early bovine embryogenesis associated with upregulation of mtTFA and NRF1 transcription factors. Reprod Biol Endocrinol 2005; 3: 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendriks WK, Colleoni S, Galli C, Paris DBBP, Colenbrander B, Stout TAE. Mitochondrial DNA replication is initiated at blastocyst formation in equine embryos. Reprod Fertil Dev 2019; 31: 570–578. [DOI] [PubMed] [Google Scholar]
- 10.Seli E, Wang T, Horvath TL. Mitochondrial unfolded protein response: a stress response with implications for fertility and reproductive aging. Fertil Steril 2019; 111: 197–204. [DOI] [PubMed] [Google Scholar]
- 11.Wai T, Ao A, Zhang X, Cyr D, Dufort D, Shoubridge EA. The role of mitochondrial DNA copy number in mammalian fertility. Biol Reprod 2010; 83: 52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leese HJ. Quiet please, do not disturb: a hypothesis of embryo metabolism and viability. BioEssays 2002; 24: 845–849. [DOI] [PubMed] [Google Scholar]
- 13.Jing Y, Li L, Li YY, Ouyang YC, Sun QY, Zhang CL, Li R. Embryo quality, and not chromosome nondiploidy, affects mitochondrial DNA content in mouse blastocysts. J Cell Physiol 2019; 234: 10481–10488. [DOI] [PubMed] [Google Scholar]
- 14.Fragouli E, Spath K, Alfarawati S, Kaper F, Craig A, Michel CE, Kokocinski F, Cohen J, Munne S, Wells D. Altered levels of mitochondrial DNA are associated with female age, aneuploidy, and provide an independent measure of embryonic implantation potential. PLoS Genet 2015; 11: e1005241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shang W, Zhang Y, Shu M, Wang W, Ren L, Chen F, Shao L, Lu S, Bo S, Ma S, Gao Y. Comprehensive chromosomal and mitochondrial copy number profiling in human IVF embryos. Reprod Biomed Online 2018; 36: 67–74. [DOI] [PubMed] [Google Scholar]
- 16.Bayram A, De Munck N, Elkhatib I, Arnanz A, Liñán A, Lawrenz B, Fatemi HM. Cleavage stage mitochondrial DNA is correlated with preimplantation human embryo development and ploidy status. J Assist Reprod Genet 2019; 36: 1847–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Xu H. Translational regulation of mitochondrial biogenesis. Biochem Soc Trans 2016; 44: 1717–1724. [DOI] [PubMed] [Google Scholar]
- 18.Zhang GM, Guo YX, Deng MT, Wan YJ, Deng KP, Xiao SH, Meng FX, Wang F, Lei ZH. Effect of PPARGC1A on the development and metabolism of early rabbit embryos in vitro. Mol Reprod Dev 2019; 86: 1758–1770. [DOI] [PubMed] [Google Scholar]
- 19.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 2008; 88: 611–638. [DOI] [PubMed] [Google Scholar]
- 20.Gureev AP, Shaforostova EA, Popov VN. Regulation of mitochondrial biogenesis as a way for active longevity: interaction between the Nrf2 and PGC-1α signaling pathways. Front Genet 2019; 10: 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HC, Yin PH, Chi CW, Wei YH. Increase in mitochondrial mass in human fibroblasts under oxidative stress and during replicative cell senescence. J Biomed Sci 2002; 9: 517–526. [DOI] [PubMed] [Google Scholar]
- 22.Suliman HB, Carraway MS, Welty-Wolf KE, Whorton AR, Piantadosi CA. Lipopolysaccharide stimulates mitochondrial biogenesis via activation of nuclear respiratory factor-1. J Biol Chem 2003; 278: 41510–41518. [DOI] [PubMed] [Google Scholar]
- 23.St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jäger S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 2006; 127: 397–408. [DOI] [PubMed] [Google Scholar]
- 24.Yoboue ED, Devin A. Reactive oxygen species-mediated control of mitochondrial biogenesis. Int J Cell Biol 2012; 2012: 403870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finkel T. Signal transduction by reactive oxygen species. J Cell Biol 2011; 194: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rigoulet M, Yoboue ED, Devin A. Mitochondrial ROS generation and its regulation: mechanisms involved in H2O2 signaling. Antioxid Redox Signal 2011; 14: 459–468. [DOI] [PubMed] [Google Scholar]
- 27.Silva FS, Simoes RF, Couto R, Oliveira PJ. Targeting mitochondria in cardiovascular diseases. Curr Pharm Des 2016; 22: 5698–5717. [DOI] [PubMed] [Google Scholar]
- 28.Yoshioka K, Suzuki C, Tanaka A, Anas IM, Iwamura S. Birth of piglets derived from porcine zygotes cultured in a chemically defined medium. Biol Reprod 2002; 66: 112–119. [DOI] [PubMed] [Google Scholar]
- 29.Yang SG, Park HJ, Kim JW, Jung JM, Kim MJ, Jegal HG, Kim IS, Kang MJ, Wee G, Yang HY, Lee YH, Seo JH, Kim SU, Koo DB. Mito-TEMPO improves development competence by reducing superoxide in preimplantation porcine embryos. Sci Rep 2018; 8: 10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nierobisz LS, McFarland DC, Mozdziak PE. MitoQ10 induces adipogenesis and oxidative metabolism in myotube cultures. Comp Biochem Physiol B Biochem Mol Biol 2011; 158: 125–131. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi M, Ito J, Shirasuna K, Kuwayama T, Iwata H. Comparative analysis of cell-free DNA content in culture medium and mitochondrial DNA copy number in porcine parthenogenetically activated embryos. J Reprod Dev 2020; 66: 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato D, Itami N, Tasaki H, Takeo S, Kuwayama T, Iwata H. Relationship between mitochondrial DNA copy number and SIRT1 expression in porcine oocytes. PLoS One 2014; 9: e94488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Itami N, Shirasuna K, Kuwayama T, Iwata H. Short-term heat stress induces mitochondrial degradation and biogenesis and enhances mitochondrial quality in porcine oocytes. J Therm Biol 2018; 74: 256–263. [DOI] [PubMed] [Google Scholar]
- 34.Itami N, Shirasuna K, Kuwayama T, Iwata H. Palmitic acid induces ceramide accumulation, mitochondrial protein hyperacetylation, and mitochondrial dysfunction in porcine oocytes. Biol Reprod 2018; 98: 644–653. [DOI] [PubMed] [Google Scholar]
- 35.Ichikawa K, Shibahara H, Shirasuna K, Kuwayama T, Iwata H. Cell-free DNA content in follicular fluid: A marker for the developmental ability of porcine oocytes. Reprod Med Biol 2019; 19: 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, Hellemans J, Kubista M, Mueller RD, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT, Bustin SA. The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clin Chem 2013; 59: 892–902. [DOI] [PubMed] [Google Scholar]
- 37.Hara T, Kin A, Aoki S, Nakamura S, Shirasuna K, Kuwayama T, Iwata H. Resveratrol enhances the clearance of mitochondrial damage by vitrification and improves the development of vitrified-warmed bovine embryos. PLoS One 2018; 13: e0204571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ristow M, Schmeisser K. Mitohormesis: Promoting health and lifespan by increased levels of reactive oxygen species (ROS). Dose Response 2014; 12: 288–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arias ME, Sanchez R, Felmer R. Evaluation of different culture systems with low oxygen tension on the development, quality and oxidative stress-related genes of bovine embryos produced in vitro. Zygote 2012; 20: 209–217. [DOI] [PubMed] [Google Scholar]
- 40.Belli M, Zhang L, Liu X, Donjacour A, Ruggeri E, Palmerini MG, Nottola SA, Macchiarelli G, Rinaudo P. Oxygen concentration alters mitochondrial structure and function in in vitro fertilized preimplantation mouse embryos. Hum Reprod 2019; 34: 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouchez C, Devin A. Mitochondrial biogenesis and mitochondrial reactive oxygen species (ROS): a complex relationship regulated by the cAMP/PKA signaling pathway. Cells 2019; 8: E287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryoo IG, Kwak MK. Regulatory crosstalk between the oxidative stress-related transcription factor Nfe2l2/Nrf2 and mitochondria. Toxicol Appl Pharmacol 2018; 359: 24–33. [DOI] [PubMed] [Google Scholar]
- 43.Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R, Tang D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 2016; 63: 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dikalov S. Cross talk between mitochondria and NADPH oxidases. Free Radic Biol Med 2011; 51: 1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelso GF, Porteous CM, Coulter CV, Hughes G, Porteous WK, Ledgerwood EC, Smith RA, Murphy MP. Selective targeting of a redox-active ubiquinone to mitochondria within cells: antioxidant and antiapoptotic properties. J Biol Chem 2001; 276: 4588–4596. [DOI] [PubMed] [Google Scholar]
- 46.Noguchi T, Aizawa T, Munakata Y, Iwata H. Comparison of gene expression and mitochondria number between bovine blastocysts obtained in vitro and in vivo. J Reprod Dev 2020; 66: 35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagata S, Tatematsu K, Kansaku K, Inoue Y, Kobayashi M, Shirasuna K, Iwata H. Effect of aging on mitochondria and metabolism of bovine granulosa cells. J Reprod Dev 2020; 66: 547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee YX, Chen CH, Lin SY, Lin YH, Tzeng CR. Adjusted mitochondrial DNA quantification in human embryos may not be applicable as a biomarker of implantation potential. J Assist Reprod Genet 2019; 36: 1855–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newman DL, Thurgood LA, Gregory SL. The impact of aneuploidy on cellular homeostasis. Free Radic Res 2019; 53: 705–713. [DOI] [PubMed] [Google Scholar]