Abstract
Depletion of hepatocyte growth factor (HGF) or mesenchymal-epithelial transition factor (c-Met) in mice leads to fetal lethality and placental maldevelopment. However, the dynamic change pattern of HGF/c-Met signaling during placental development and its involvement in the early differentiation of trophoblasts remain to be elucidated. In this study, using in situ hybridization assay, we elaborately demonstrated the spatial-temporal expression of Hgf and c-Met in mouse placenta from E5.5, the very early stage after embryonic implantation, to E12.5, when the placental structure is well developed. The concentration of the soluble form of c-Met (sMet) in maternal circulation peaked at E10.5. By utilizing the induced differentiation model of mouse trophoblast stem cells (mTSCs), we found that HGF significantly promoted mTSC differentiation into syncytiotrophoblasts (STBs) and invasive parietal trophoblast giant cells (PTGCs). Interestingly, sMet efficiently reversed the effect of HGF on mTSC differentiation. These findings indicate that HGF/c-Met signaling participates in regulating placental trophoblast cell fate at the early differentiation stage and that sMet acts as an endogenous antagonist in this aspect.
Keywords: Hepatocyte growth factor (HGF), Mesenchymal-epithelial transition factor (c-Met), Placenta, Soluble form of c-Met (sMet), Syncytiotrophoblast cells, Trophoblast giant cells
Hepatocyte growth factor (HGF)/ mesenchymal-epithelial transition factor (c-Met) signaling is strongly associated with epithelial, mesenchymal, and hematological malignancies [1]. HGF acts as an essential cell mitogen, motogen, and morphogen and plays an important role in mesenchymal-epithelial transformation [2,3,4]. c-Met is a receptor tyrosine kinase that functions in cell growth and morphogenesis [5, 6] by activating multiple pathways after binding to its ligand HGF. The soluble form of c-Met (sMet) is hydrolyzed from c-Met, and it can competitively bind HGF and disrupt HGF/c-Met signaling [7].
During pregnancy, HGF homozygous mutant mice have been shown to exhibit an abnormal placental labyrinth region with a reduced number of trophoblast cells at embryonic day 11.5 (E11.5) and die after E13.5 [8]. Accordingly, the phenotype of c-Met homozygous mutant mice was similar to that of HGF homozygous mutant mice, which died around E12.5, and placental size was significantly reduced compared to the wild type [9]. In addition, conditional knockout of c-Met in placental trophoblast cells causes placental dysfunction [10], which indicates that HGF/c-Met signaling participates in embryonic and placental development.
The localization of HGF and c-Met has been analyzed in studies on human placenta. During the first and second trimesters of pregnancy, HGF is located in villous stromal cells, and c-Met is expressed in cytotrophoblast cells and decidual glands [11]. During the third trimester of pregnancy, HGF is found in extravillous trophoblast cells, mesenchymal cells, STBs, and vascular endothelial cells, while c-Met is strongly expressed in vascular endothelial cells and STBs [12, 13]. Although there has been a study that analyzed the localization of HGF or c-Met in mouse placenta, the dynamic change pattern of HGF/c-Met signaling during placental development and its involvement in early differentiation of trophoblasts are unclear.
From our data, we demonstrated the spatial-temporal expression of Hgf and c-Met in mouse placenta from E5.5, the very early stage after embryonic implantation, to E12.5, when the placental structure is well developed. Expression of Hgf and c-Met was observed in PTGCs at E5.5 and chorionic plate at E7.5, and the concentration of sMet in maternal circulation peaked at E10.5, which indicated that HGF/c-Met signaling participated in early differentiation of trophoblast cells. In addition, by utilizing the induced differentiation model of mouse trophoblast stem cells (mTSCs), we found that HGF promoted mTSC differentiation into STBs and invasive parietal trophoblast giant cells (PTGCs) and that sMet functionally blocked HGF’s role in mTSC differentiation. Therefore, HGF/c-Met signaling participates in regulating placental trophoblast cell fate at the early differentiation stage, and sMet acts as an endogenous antagonist.
Materials and Methods
Sample collection
Mouse placental tissues and plasma were obtained from pregnant female SPF CD1 mice at different pregnancy stages (Beijing SPF Biotechnology Co., Ltd., Beijing, China). Mice were sacrificed by cervical dislocation, and plasma samples were collected using EDTA-Na anticoagulant. The experimental procedure was approved by the Animal Welfare and Ethics Committee of the Institute of Zoology, Chinese Academy of Sciences.
In situ hybridization
Hgf and c-Met specific riboprobes were transcribed in vitro from CD1 mouse cDNA templates. Hgf riboprobes were designed using the forward primer: TCGGATAGGAGCCACAAGGA and reverse primer: GAAGGCCTTGCAAGTGAACG. c-Met riboprobes were designed using the forward primer: AGTGCCCGAAGTGTAAGTCC and reverse primer: AGACACAGCCAAAATGCCCT. Digoxigenin-labeled riboprobes were synthesized according to the manufacturer’s protocol (Roche, Indianapolis, IN, USA). Mouse placental tissues were embedded in O.C.T. compound immediately after sampling, and frozen sections (10 µm) were briefly fixed in 4% PFA and hybridized with the riboprobe overnight at 55○C. The slides were incubated with AP-conjugated anti-digoxigenin antibody (Roche, Indianapolis, IN, USA) and visualized with BCIP/NBT (Promega, Madison, WI, USA) as a substrate. Fast red (Ding Guo, Beijing, China) was stained for 10 min at room temperature to identify the nucleus. The images were recorded using a light microscope with a CCD (Olympus, Tokyo, Japan).
Hematoxylin and eosin staining
Frozen sections (10 µm) were briefly fixed in 4% PFA and washed with PBS. Subsequently, the sections were immersed in Hematoxylin (Ding Guo) for 3 min at room temperature. After dehydration, the sections were immersed in alcohol-soluble eosin (Baso, Zhuhai, China) for 1 min at room temperature. Thereafter, the sections were washed with 95% ethanol, dehydrated, and mounted. The images were recorded using a light microscope with a CCD (Olympus).
ELISA for sMet
Levels of sMet secretion in CD1 mouse peripheral blood were analyzed using a sandwich ELISA according to the manufacturer’s instructions (RD System, MN, USA). In brief, the plasma samples were 5-fold diluted and subjected to sandwich ELISA. The levels of sMet were determined using a standard curve.
Cell culture of mice trophoblast stem cells (mTSCs)
Mouse fibroblasts obtained from E13.5 fetal and mouse embryonic fibroblast-conditioned media (MEF-CM) were collected after mitomycin treatment. Trophoblast stem cell medium (TS medium) was comprised of RPMI-1640 (Gibco, New York, USA) supplemented with 20% FBS (Gibco), glutamine, sodium pyruvate, and 50 µM beta-mercaptoethanol. For normal culture and cell passage, mTSCs were cultured in mTSC complete medium containing 70% MEFs-CM, 30% TS medium, 25 ng/ml FGF4 (PeproTech, New Jersey, USA), and 1 μg/ml heparin (Sigma, St. Louis, USA). mTSCs were digested and passaged using 0.05% trypsin and 0.02% EDTA. For the induction of STB formation, mTSCs were seeded onto 35 mm dishes and cultured in TS medium without FGF4, heparin, and MEF-CM for two days, followed by pre-treatment with 3 μM CHIR99021 (Selleck, Houston, USA) for 2 days. Cells were treated with either 10 ng/ml HGF (Peprotech, New Jersey, USA) or 50 ng/ml sMet (SAB, Maryland, USA) for two days. To induce TGC formation, mTSCs were cultured in the TS medium with HGF/sMet-treatment for 48 h, and the experiments were withdrawn FGF4, heparin, and MEF-CM.
RNA preparation and quantitative real-time PCR
Total RNA from mTSCs was extracted with TRIzol reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). Reverse transcription was carried out using the Oligo Primer (TIANGEN, Beijing, China), and cDNA was used as a template for quantitative real-time PCR with the SYBR Premix Ex TaqTM kit (Takara, Dalian, China) according to the manufacturer’s instructions. The primer sequences are listed in Supplementary Table 1 (online only). After the PCR reaction, the relative expression of certain genes was normalized by GAPDH using the 2−ΔΔCT method.
Cell invasion assay
Matrigel (BD Biosciences, New Jersey, USA) was diluted in RPMI 1640 and added to a transwell insert with an 8 µm pore polycarbonate membrane (Corning, New York, USA). mTSCs were dissociated and resuspended in RPMI 1640 medium supplemented with 1% FBS. Approximately 6 × 104 cells cultured with 1% FBS were added to the transwell insert, and 600 μl RPMI 1640 medium containing 10% FBS was added to the 24-well plate below the cell. After 36 h of culture, invasive cells in the transwell insert were fixed in 0.25% glutaraldehyde and stained with hematoxylin solution for 15 min. The images were recorded using a light microscope with a CCD (Olympus).
Cell proliferation assay
Cell Counting Kit-8 (CCK-8) (Dojindo, Kamamoto, Japan) was used for cell proliferation analysis according to the manufacturer’s instructions. Briefly, 10 μl CCK-8 solution was added to each culture well, and after culturing for 4 h at 37oC, the absorbance at 450 nm was determined using a microplate reader.
Statistical analysis
Statistical analyses were performed using GraphPad Prism version 5.01 (GraphPad Software, San Diego, CA, USA). Data are shown as mean ± SD. Statistical comparisons between groups were performed by one-way ANOVA with post hoc multiple comparison analysis, and equal variances were assumed by Tukey-Kramer test according to at least three independent experiments. The P value of less than 0.05 was considered statistically significant.
Results
The localization of HGF and c-Met during early placental development in CD1 mice
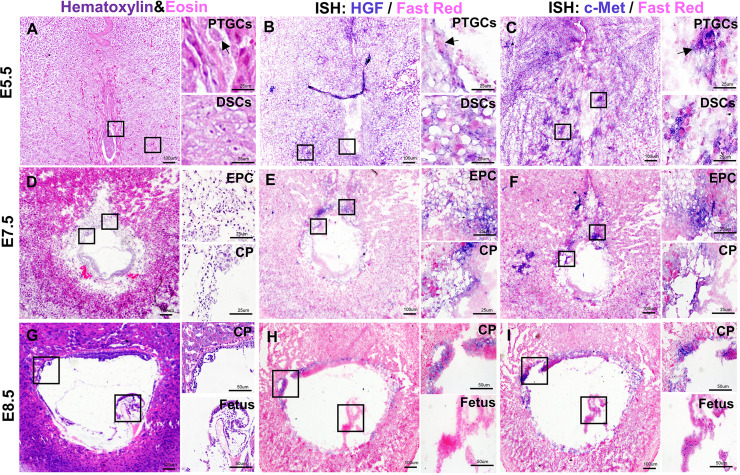
Embryo implantation occurs at E4.5 [14]. After implantation, the PTGCs begin to differentiate at E5.5. PTGCs invade the decidua and form an initial connection between the trophoblastic layer and the maternal vascular system [15]. At E5.5, Hgf was found to be expressed in PTGCs and decidual stromal cells (DSCs) (Fig. 1B). Most c-Met was also detected in PTGCs and DSCs, and c-Met+ DSCs were found to be more extensive than those expressing Hgf (Fig. 1C).
Fig. 1.
The localization of Hgf and c-Met during early placental development in CD1 mice. A: H&E staining of E5.5 placenta. B: In situ hybridization for Hgf on E5.5 placenta. C: In situ hybridization for c-Met on E5.5 placenta. D: H&E staining of E7.5 placenta. E: In situ hybridization for Hgf on E7.5 placenta. F: In situ hybridization for c-Met on E7.5 placenta. G: H&E staining of E8.5 placenta. H: In situ hybridization for Hgf on E8.5 placenta. I: In situ hybridization for c-Met on E8.5 placenta. PTGCs, parietal trophoblast giant cells; EPC, ectoplacental cone; CP, chorionic plate. Arrows indicate PTGCs. Scale bars are shown in panels.
At E7.5, the outer membrane of PTGCs closely connects with the decidua and forms an ectoplacental cone (EPC) [16]. Hgf was mainly distributed in the chorionic plate (CP) and EPC (Fig. 1E), and c-Met was also found in the CP and EPC (Fig. 1F).
At E8.5, PTGC continues to invade the mesenteric side of the uterus, and allantois extends to the CP and fuses together [17]. At this time, Hgf was mainly located in the CP (Fig. 1H), and c-Met was also distributed in the CP (Fig. 1I). Hematoxylin and eosin staining was performed in the early placenta to identify the structure (Figs. 1A, D and G).
The localization of HGF and c-Met during the development of mid-placenta in CD1 mice
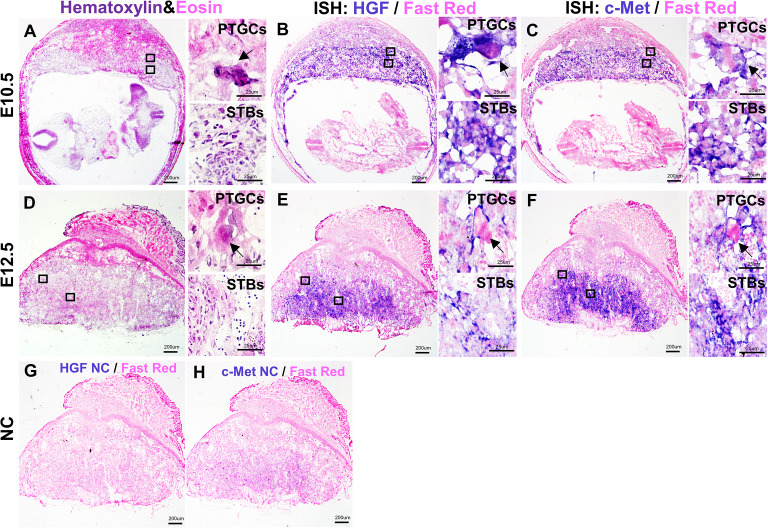
After the fusion of CP and allantois, the allantois mesoderm further penetrates into the CP and branches the villi, and the formation of the labyrinth layer begins [18, 19]. At E10.5, Hgf was mainly found in PTGCs and labyrinth STBs (Fig. 2B), and c-Met was also distributed in PTGCs and labyrinth STBs (Fig. 2C).
Fig. 2.
The localization of Hgf and c-Met during the development of mid-placenta in CD1 mice. A: H&E staining of E10.5 placenta. B: In situ hybridization for Hgf on E10.5 placenta. C: In situ hybridization for c-Met on E10.5 placenta. D: H&E staining of E12.5 placenta. E: In situ hybridization for Hgf on E12.5 placenta. F: In situ hybridization for c-Met on E12.5 placenta. G: In situ hybridization of negative controls for Hgf probes. H: In situ hybridization of negative controls for c-Met probes. STBs, syncytiotrophoblast cells; PTGCs, parietal trophoblast giant cells. Arrows indicate PTGCs. Scale bars are shown in panels.
At E12.5, a mature placenta was gradually built. The structures of the labyrinth, junctional zone, and decidua were well defined. At this time, Hgf was mainly located in PTGCs and STBs (Fig. 2E), and c-Met was also distributed in PTGCs and STBs (Fig. 2F). Negative controls of Hgf and c-Met probes were performed on E12.5 placenta (Figs. 2G–H) to determine probe specificity. Hematoxylin and eosin staining was performed at the mid-placenta to identify the structure (Figs. 2A and D).
The concentration of plasma sMet peaked at E10.5 in the majority of CD1 mice
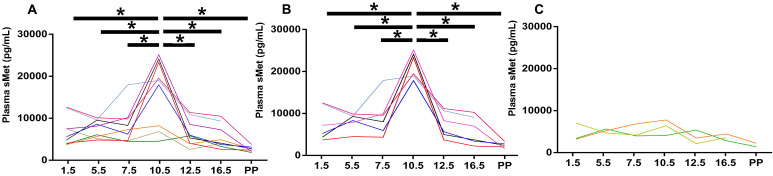
Met is the soluble form of c-Met, formed after metalloprotease hydrolysis, and it hinders HGF binding to c-Met to a certain extent [7]. Plasma sMet levels peak during the second trimester (21–26 weeks) in humans [20, 21], but the dynamic change of plasma sMet in pregnant mice is unclear. ELISA was performed to analyze the dynamic changes in plasma sMet levels at different time points throughout pregnancy in CD1 mice. Plasma samples obtained from 12 pregnant mice on E1.5, E4.5, E7.5, E10.5, E13.5, E16.5, and postpartum were analyzed for sMet concentration. We drew a line chart to show the dynamic changes in plasma sMet levels during pregnancy in each mouse. We found that although individual differences existed, sMet peaked at E10.5 (Fig. 3A). Since pregnant mice exhibited variation in the basal level of plasma sMet, we divided them into sMet-high and sMet-low groups at E10.5 to analyze the concentration of the plasma sMet more accurately. No difference in maternal weight at E13.5, and fetal weight on the first day postpartum was found between the two groups (Supplementary Fig. 1: online only). We found that plasma sMet also peaked at E10.5 in the sMet-high group (Fig. 3B). In the sMet-low group, no significant differences were found among the different gestational days (Fig. 3C). These data were similar to the results in most of the human plasma, which indicated that the concentration of sMet during midterm pregnancy may reach the highest level in both mice and humans.
Fig. 3.
ELISA for plasma sMet in CD1 pregnant mice. A: The concentration of plasma sMet during pregnancy. B: The concentration of plasma sMet in the sMet-high group. C: The concentration of plasma sMet in the sMet-low group. PP, postpartum. Comparison between groups was performed with one-way ANOVA multiple comparisons and analyzed with Tukey-Kramer test. * P < 0.05.
Met blocked HGF-induced differentiation of mice STB
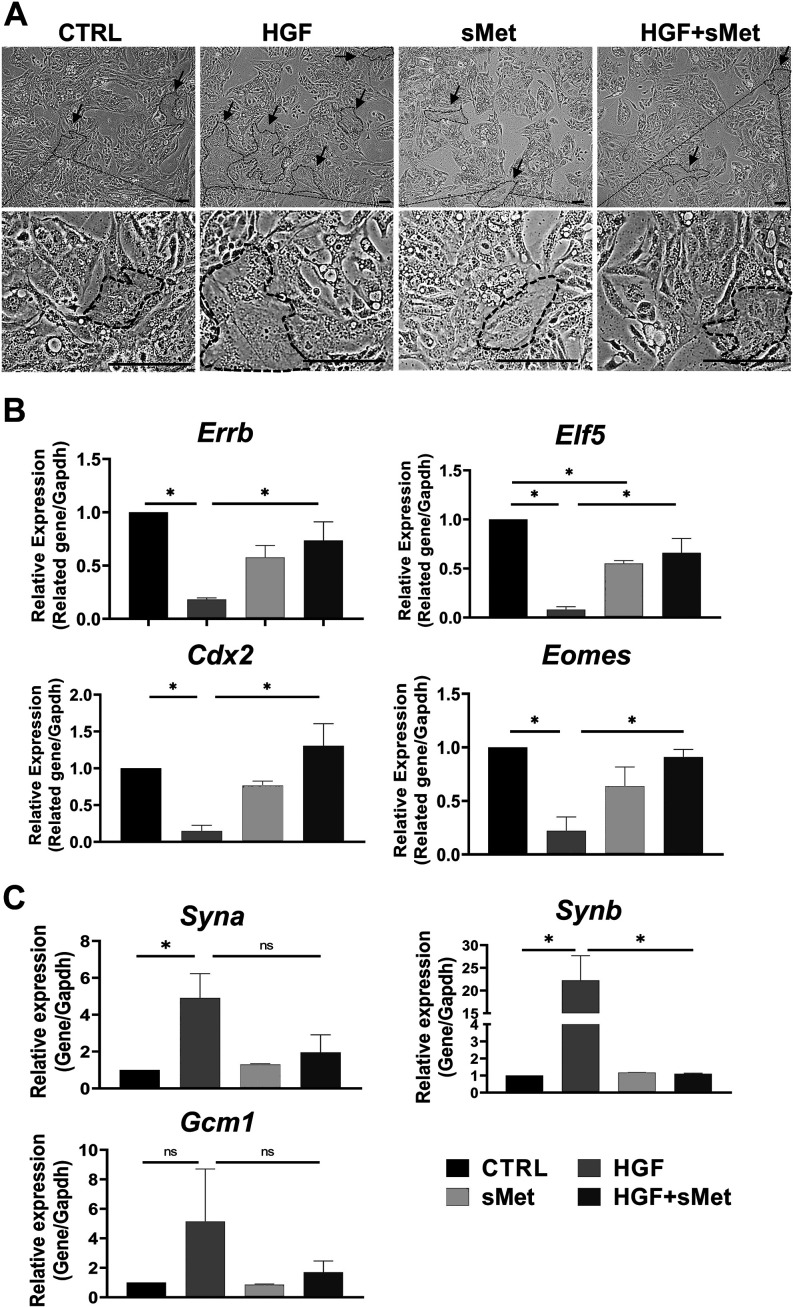
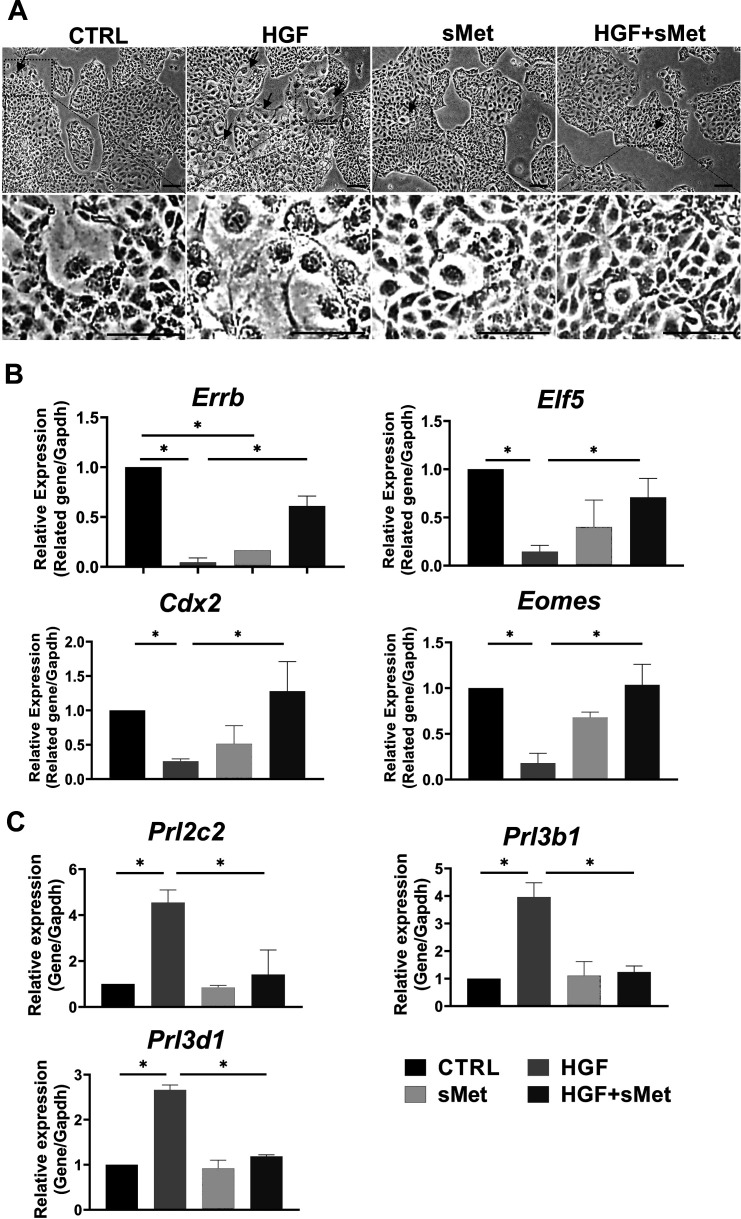
HGF/c-Met signaling participates in the formation of mouse SynT-II cells after activation of Wnt signaling [22]. According to our data, HGF and c-Met were both expressed in STBs during placental development (Fig. 2). In our experiments, mTSCs were cultured in TS medium without FGF4, heparin, and MEF-CM for two days and then pre-treated with 3 μM CHIR99021 for 48 h to activate Wnt signaling. After activation, in the control group, the pre-treated mTSCs were continuously cultured with TS medium for 48 h. In the HGF group, pre-treated mTSCs were cultured with TS medium and 10 ng/ml HGF for 48 h. In the sMet group, pre-treated mTSCs were cultured with TS medium and 50 ng/ml sMet for 48 h. In the HGF + sMet group, the pre-treated mTSCs were cultured with TS medium, 10 ng/ml HGF, and 50 ng/ml sMet for 48 h. In all these groups, differentiation was induced by withdrawal of FGF4, heparin, and MEF-CM. We used 25 ng/ml, 50 ng/ml, and 100 ng/ml sMet for mTSC differentiation and found that 25 ng/ml sMet had almost no impact on mTSC differentiation, while 50 ng/ml and 100 ng/ml sMet had similar effects on the differentiation of the pre-treated mTSCs. Therefore, we used 50 ng/ml of sMet in the pre-treated mTSC differentiation experiments (Supplementary Fig. 2: online only). The cell morphology under different treatments is shown in Fig. 4A. We could see that the syncytialized cells that gradually lost cell boundaries and fused together abundantly existed only in the HGF group, and only a few syncytialized cells were found in control, sMet, and HGF + sMet groups. We found that sMet functionally blocked the role of HGF in the differentiation of pre-treated mTSCs. The expression of stemness markers Errb, Elf5, Cdx2, and Eomes significantly decreased (Fig. 4B), and the STB markers like Syna and Synb were sharply upregulated compared with the control group; Gcm1 could have an upregulated tendency with no significance (Fig. 4C). When we added 50 ng/ml sMet to the HGF-induced system, sMet reversed the dysregulation of stemness markers, and STB markers were significantly downregulated compared with the HGF group (Figs. 4B–C). Thus, sMet functionally blocked the role of HGF in the differentiation of STBs, which is consistent with the antagonizing effect of sMet on HGF [7].
Fig. 4.
Met blocked HGF-induced differentiation of STBs. A: A typical result for morphological observation of the mTSCs among different groups. B: Relative mRNA expression of stemness markers after different treatments (n = 3). C: Relative mRNA expression of STB markers after different treatments. Arrows indicate STBs (n = 3). Scale bars: 100 μm. Data are presented as mean ± SD. Comparison between groups was performed with one-way ANOVA multiple comparisons and analyzed with Tukey-Kramer test. NS, no significance. * P < 0.05.
Met blocked HGF-induced differentiation of mice PTGCs
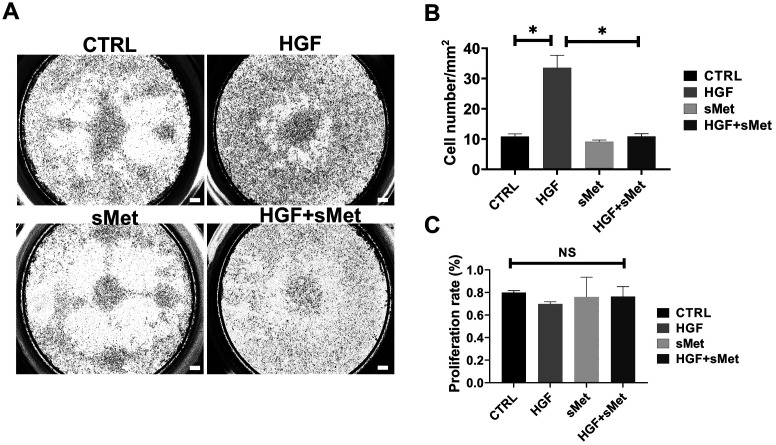
HGF is expressed in several types of trophoblast cells and promotes the invasion and development of human trophoblast cells [11,12,13, 23]. We found that Hgf was expressed in PTGCs during placental development in mice (Figs. 1 and 2), but whether HGF promoted the differentiation of mTSCs into invasive PTGCs is unknown. In the control group, mTSCs were obtained after 48 h of treatment with TS medium. In the HGF group, mTSCs were obtained after 48 h of treatment with TS medium and 10 ng/ml HGF. None of the groups was exposed to FGF4, heparin, or MEF-CM. We found that stemness markers such as Errb, Elf5, Cdx2, and Eomes were significantly decreased (Fig. 5B), and PTGC markers such as Prl2c2, Prl3b1, and Prl3d1 were sharply upregulated after HGF induction compared with the control group (Fig. 5C). Cell morphology observation indicated that HGF induced the differentiation of TGCs (Fig. 5A). In the cell invasion assay, the number of invasive cells in the unit area was statistically quantified, showing that HGF-induced cells had much stronger invasive ability than the control group (Figs. 6A–B). These results indicated that HGF promoted the differentiation of TGCs cultured in TS medium.
Fig. 5.
Met blocked HGF-induced differentiation of PTGCs. A: A typical result for morphological observation of the mTSCs among different groups. B: Relative mRNA expression of stemness markers after different treatments (n = 3). C: Relative mRNA expression of TGC markers after different treatments (n = 3). Arrows indicate TGCs. Scale bars: 100 μm. Data are presented as mean ± SD. Comparison between groups was performed with one-way ANOVA multiple comparisons and analyzed with Tukey-Kramer test. * P < 0.05.
Fig. 6.
Met blocked the cell invasion of HGF-induced PTGCs. A: Transwell assay among different groups. B: Statistical analysis of invasive cells among different groups (n = 3). C: Cell proliferation analysis among different groups (n = 3). Scale bars are shown in panels. Data are presented as mean ± SD. Comparison between groups was performed with one-way ANOVA multiple comparisons and analyzed with Tukey-Kramer test. * P < 0.05. NS, no significance. Scale bars: 100 μm.
We found that sMet functionally blocked the role of HGF in TGC differentiation. In the HGF-induced system, sMet reversed the expression levels of stemness-related genes, such as Errb, Elf5, Cdx2, and Eomes (Fig. 5B), and PTGC markers such as Prl2c2, Prl3b1, and Prl3d1 were also significantly reversed (Fig. 5C) compared with the HGF group, which is consistent with the antagonizing effect of sMet on HGF [7]. We observed that TGCs with large nuclei were abundant only in the HGF group, and only a few TGCs were found in control, sMet, and HGF + sMet groups (Fig. 5A). The transwell assay showed that sMet significantly blocked the HGF-induced invasion of PTGCs (Figs. 6A–B). Cell proliferation was excluded by the CCK8 assay, and we found no differences between the groups (Fig. 6C).
Discussion
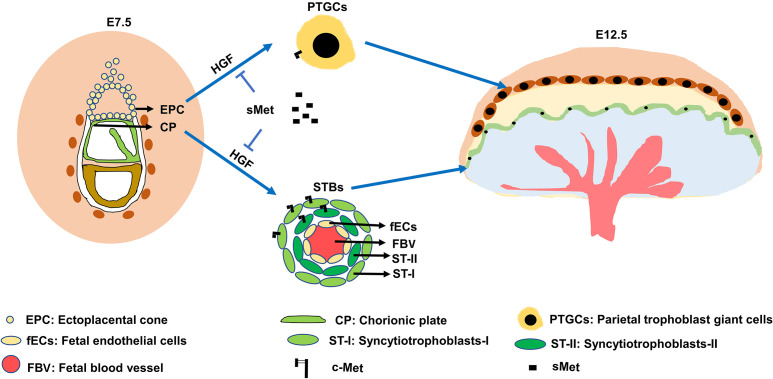
In CD1 pregnant mice, HGF and c-Met were expressed in EPC and CP at E7.5, and both were expressed in PTGCs and STBs at E12.5 (Figs. 1 and 2). PTGCs and STBs were differentiated from EPC and CP, respectively. In addition, sMet neutralized endogenous HGF (Figs. 4 and 5). These results suggest that HGF/c-Met signaling could regulate trophoblast differentiation in an autocrine manner. The proposed model scheme based on the results of this study is shown in Fig. 7. Because of sampling limitations, continuous sampling of the tissue from the human early placenta is not possible, and whether HGF/c-Met signaling, including sMet, is involved in the differentiation of trophoblast cells, is still unclear. Therefore, a suitable animal model should be developed. Our results showed that in CD1 pregnant mice, the expression patterns of Hgf and c-Met were similar to those in the human placenta. We analyzed the concentration of plasma sMet and found that plasma sMet peaked at E10.5 in most of the CD1 pregnant mice. Consistently, plasma sMet levels also peak during mid-term pregnancy in normal pregnant women [20, 21]. To some extent, our data suggest that HGF/c-Met signaling regulates placental cell functions in humans and mice in a similar manner and that CD1 pregnant mice could be a suitable animal model to study the function of HGF/c-Met signaling (Table 1). We also found that HGF significantly promoted the differentiation of pre-treated mTSCs into STBs and PTGCs, while sMet reversed this effect.
Fig. 7.
A scheme of the role of HGF/c-Met signal in trophoblast differentiation. Autocrine HGF/c-Met signaling pathway for the regulation of trophoblast differentiation.
Table 1. Comparison of the distribution and function of HGF/ c-Met signaling in human and mouse placentas.
| Pregnant women | CD1 pregnant mice | |
|---|---|---|
| HGF expression | Syncytiotrophoblasts | Syncytiotrophoblasts |
| Extravillous trophoblasts | PTGCs | |
| c-Met expression | Syncytiotrophoblasts | Syncytiotrophoblasts |
| Extravillous trophoblasts | PTGCs | |
| sMet expression | Peak at 21–26w | Peak at E10.5 |
| HGF/c-Met function | Trophoblast invasion | Invasive PTGC differentiation |
| Trophoblast development | Syncytiotrophoblast differentiation | |
| Conclusion | HGF/c-Met regulates placenta cell functions in human and mouse with similar manner | |
HGF/c-Met signaling regulates placental cell functions in human and mouse in a similar manner.
Depletion of HGF or c-Met in mice leads to fetal lethality and placental maldevelopment during early development [8,9,10]. For most trophoblast differentiation, PTGCs are differentiated from EPC, and STBs are differentiated from CP, which develops from the extraembryonic ectoderm [24]. Both the EPC and extraembryonic ectoderm are derived from the trophectoderm. HGF plays a role in equine trophectoderm proliferation [25], and HGF receptor is expressed in the ovine trophectoderm [26]. Our in vivo data showed that Hgf was strongly expressed in CP as well as the ectoplacental cone (Fig. 1), which represent the origins of STBs and PTGCs, respectively. Based on these results, it is reasonable to assume that HGF/c-Met signaling plays a role in trophoblast differentiation. Hgf and c-Met were expressed in PTGCs at E5.5, which indicated that HGF/c-Met participated in the early differentiation of PTGCs. CP eventually develops into STBs, and the molecules specifically expressed in CP can branch the fetal blood vessels and promote the formation of the labyrinth layer [27, 28]. We found that Hgf and c-Met were expressed in EPC and CP at E7.5, which indicated that HGF/c-Met signaling might play a role in the early differentiation of PTGCs and STBs. At E10.5, the basic structure of the mouse placenta is formed, different subtypes of TGCs exist in the placenta, the labyrinth layer is expanded, and Gcm1 is wildly expressed in the labyrinth layer [24]. We found that Hgf and c-Met were expressed in labyrinth STBs, and plasma sMet levels peaked at E10.5. Moreover, we found that HGF significantly promoted Gcm1 expression and that sMet blocked the function of HGF, indicating that HGF/c-Met signaling might participate in the expression of Gcm1, which is one of the most important transcription factors in STB differentiation and formation [27, 29]. At E10.5 and E12.5, Hgf and c-Met were both expressed in PTGCs and STBs; thus, HGF/c-Met signaling has the potential to regulate trophoblast cell differentiation.
Expression of Hgf and c-Met was observed in PTGCs and STBs (Figs. 1 and 2). Therefore, it is reasonable to assume that HGF functions in an autocrine or juxtacrine manner to regulate cell differentiation. However, when cells were treated with sMet alone, we found that sMet significantly reduced the expression levels of Elf5 or Errb compared with the control group (Figs. 4 and 5). If endogenous HGF exists, this result is in contrast to the neutralizing function of sMet to HGF. From our perspective, this result may be due to the absence of endogenous HGF in mTSCs, and the exogenous sMet acting as a stimulus and promoting cell differentiation. The supplementary data from several transcriptome analyses of mTSCs indicate extremely low HGF levels in mTSCs [30, 31]. Therefore, a better cell model is required for endogenous HGF analysis.
HGF/c-Met signaling was found to be dysfunctional in the preeclamptic placenta. HGF expression is significantly reduced in preeclamptic placenta compared to that in the normal placenta [12, 32, 33]. HGF functionally promotes invasion of placental trophoblast cells [34], and decreased HGF expression is related to the failure of trophoblast invasion, which is one of the essential causes of preeclampsia [35,36,37,38]. The placental sMet sheds from c-Met by ADAM10/ADAM17, and ADAM10/ADAM17 shed more placental sMet to inhibit trophoblast invasion in the preeclamptic placenta [39]. In a normal pregnancy, the concentration of peripheral sMet peaks in the second trimester, but in preeclampsia patients, peripheral sMet is sharply downregulated compared with normal pregnancies [20, 21]. Peripheral sMet reduction is accompanied by the upregulation of placental sMet. Thus, the distribution preference of sMet in the local placenta may cause dysregulation of peripheral sMet, which correlates with the pathogenesis of preeclampsia. Our data suggested that sMet could functionally block HGF’s role in trophoblast invasion; thus, an increased placental sMet in the preeclamptic placenta is related to inadequate trophoblast invasion and causes failure of spiral artery remodeling.
Nutrient demands dramatically increase during pregnancy, and several physiological changes occur in body organs to adapt to the pregnant environment. A healthy pregnancy depends on the coordination of all the maternal organs. Placental sMet perfuses into maternal peripheral blood along with maternal circulation and may play a role in the adaptation of other organs during pregnancy. In the gestational process, the maternal liver size is properly maintained to prevent physical damage to other organs. Hepatocyte size decreases at D10 during normal pregnancy in mice [40]. At the same time, peak levels of plasma sMet in CD1 pregnant mice were observed in our data. sMet may suppress the effect of HGF/c-Met signaling on hepatocyte growth to prevent the unrestrained growth of the maternal liver. Uncontrolled enlargement of the liver may endanger maternal and fetal health. Therefore, the peak concentration of plasma sMet in the second trimester of both humans and mice is likely to participate in the adaptive regulation of the liver to maintain pregnancy. Under pathological conditions, such as preeclampsia, the liver considerably increases in size [41], and dysregulation of plasma sMet cannot control liver size. Under physiological conditions, we found that the basal level of plasma sMet was different in pregnant mice, but the maternal weight at E13.5 and fetal weight at the first day postpartum showed no differences between the sMet-high and sMet-low groups (Supplementary Fig. 1), which was in accordance with other studies [42, 43]. In a normal pregnancy, individual differences exist in response to pregnancy. The sMet-high group mice may be prone to liver enlargement in response to pregnancy, so a high concentration of sMet may prevent the unrestrained growth of the liver. The sMet-low group mice may not be prone to liver enlargement, so a high concentration of plasma sMet is not required. Therefore, the liver size was normal in both the sMet-high and sMet-low groups of normal pregnancy. The adaptations during pregnancy not only include liver growth but also observed in the cardiovascular system [44], bone metabolism [45] and renal function [46]. HGF/c-Met signaling also participates in the cardiovascular function [47], bone development [48] and renal function [49]. Based on these findings, the role of sMet in pregnant adaptation may be a potential reason for normal maternal and fetal development in the sMet-low group of normal pregnancy.
In conclusion, our results demonstrated similar expression patterns of Hgf and c-Met in CD1 pregnant mice to those in humans; plasma sMet peaking in the second trimester was also consistent with that in normal human pregnancy. Therefore, CD1 pregnant mice can be a suitable animal model for HGF/c-Met signaling. Moreover, we showed that Hgf and c-Met are expressed in PTGCs, EPC, and CP in the early placenta, and PTGCs and STB in the mid-placenta. We also found that plasma sMet peaked at E10.5 during pregnancy. On analyzing the dynamic change pattern of HGF/c-Met signaling during placental development and its involvement in early trophoblast differentiation, we found that HGF/c-Met signaling participated in the differentiation of pre-treated mTSCs to STBs and invasive PTGCs, while sMet reversed these effects. In addition, reduced levels of plasma sMet in the second trimester in preeclampsia patients indicate that sMet can be a good candidate as a clinical biomarker. Further research might explore the roles of HGF/c-Met signaling and that of sMet in pregnancy-related diseases such as preeclampsia.
Conflict of interests
The authors declare no conflict of interest.
Supplementary
Acknowledgments
The technical support from Dr. Guangming Cao in the in situ hybridization experiments is appreciated.
This research was funded by the National Key Research and Development Program of China (grant number: 2016YFC1000200 and 2018YFC1004100) and the National Natural Science Foundation of China (grant number: 81730040).
References
- 1.Ma WW, Adjei AA. Novel agents on the horizon for cancer therapy. CA Cancer J Clin 2009; 59: 111–137. [DOI] [PubMed] [Google Scholar]
- 2.Bottaro DP, Rubin JS, Faletto DL, Chan AM, Kmiecik TE, Vande Woude GF, Aaronson SA. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science 1991; 251: 802–804. [DOI] [PubMed] [Google Scholar]
- 3.Stoker M, Gherardi E, Perryman M, Gray J. Scatter factor is a fibroblast-derived modulator of epithelial cell mobility. Nature 1987; 327: 239–242. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura T, Nishizawa T, Hagiya M, Seki T, Shimonishi M, Sugimura A, Tashiro K, Shimizu S. Molecular cloning and expression of human hepatocyte growth factor. Nature 1989; 342: 440–443. [DOI] [PubMed] [Google Scholar]
- 5.Jiang WG, Martin TA, Parr C, Davies G, Matsumoto K, Nakamura T. Hepatocyte growth factor, its receptor, and their potential value in cancer therapies. Crit Rev Oncol Hematol 2005; 53: 35–69. [DOI] [PubMed] [Google Scholar]
- 6.Hurle RA, Davies G, Parr C, Mason MD, Jenkins SA, Kynaston HG, Jiang WG. Hepatocyte growth factor/scatter factor and prostate cancer: a review. Histol Histopathol 2005; 20: 1339–1349. [DOI] [PubMed] [Google Scholar]
- 7.Wajih N, Walter J, Sane DC. Vascular origin of a soluble truncated form of the hepatocyte growth factor receptor (c-met). Circ Res 2002; 90: 46–52. [DOI] [PubMed] [Google Scholar]
- 8.Uehara Y, Minowa O, Mori C, Shiota K, Kuno J, Noda T, Kitamura N. Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature 1995; 373: 702–705. [DOI] [PubMed] [Google Scholar]
- 9.Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature 1995; 376: 768–771. [DOI] [PubMed] [Google Scholar]
- 10.Ueno M, Lee LK, Chhabra A, Kim YJ, Sasidharan R, Van Handel B, Wang Y, Kamata M, Kamran P, Sereti KI, Ardehali R, Jiang M, Mikkola HK. c-Met-dependent multipotent labyrinth trophoblast progenitors establish placental exchange interface. Dev Cell 2013; 27: 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark DE, Smith SK, Sharkey AM, Sowter HM, Charnock-Jones DS. Hepatocyte growth factor/scatter factor and its receptor c-met: localisation and expression in the human placenta throughout pregnancy. J Endocrinol 1996; 151: 459–467. [DOI] [PubMed] [Google Scholar]
- 12.Wolf HK, Zarnegar R, Oliver L, Michalopoulos GK. Hepatocyte growth factor in human placenta and trophoblastic disease. Am J Pathol 1991; 138: 1035–1043. [PMC free article] [PubMed] [Google Scholar]
- 13.Kilby MD, Afford S, Li XF, Strain AJ, Ahmed A, Whittle MJ. Localisation of hepatocyte growth factor and its receptor (c-met) protein and mRNA in human term placenta. Growth Factors 1996; 13: 133–139. [DOI] [PubMed] [Google Scholar]
- 14.Krupinski P, Chickarmane V, Peterson C. Simulating the mammalian blastocyst—molecular and mechanical interactions pattern the embryo. PLOS Comput Biol 2011; 7: e1001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemberger M, Hughes M, Cross JC. Trophoblast stem cells differentiate in vitro into invasive trophoblast giant cells. Dev Biol 2004; 271: 362–371. [DOI] [PubMed] [Google Scholar]
- 16.Corbel C, Heard E. Transcriptional analysis by nascent RNA FISH of in vivo trophoblast giant cells or in vitro short-term cultures of ectoplacental cone explants. J Vis Exp 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cross JC, Simmons DG, Watson ED. Chorioallantoic morphogenesis and formation of the placental villous tree. Ann NY Acad Sci 2003; 995: 84–93. [DOI] [PubMed] [Google Scholar]
- 18.Dupressoir A, Vernochet C, Harper F, Guégan J, Dessen P, Pierron G, Heidmann T. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc Natl Acad Sci USA 2011; 108: E1164–E1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou W, Jerome-Majewska LA. TMED2/emp24 is required in both the chorion and the allantois for placental labyrinth layer development. Dev Biol 2018; 444: 20–32. [DOI] [PubMed] [Google Scholar]
- 20.Naghshvar F, Torabizadeh Z, Moslemi Zadeh N, Mirbaha H, Gheshlaghi P. Investigating the relationship between serum level of s-Met (soluble hepatic growth factor receptor) and preeclampsia in the first and second trimesters of pregnancy. ISRN Obstet Gynecol 2013; 2013: 925062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng X, Sun Y, Yang HX, Li D, Li YX, Liao QP, Wang YL. Plasma level of soluble c-Met is tightly associated with the clinical risk of preeclampsia. Am J Obstet Gynecol 2009; 201: 618.e1–618.e7. [DOI] [PubMed] [Google Scholar]
- 22.Zhu D, Gong X, Miao L, Fang J, Zhang J. Efficient induction of syncytiotrophoblast layer ii cells from trophoblast stem cells by canonical Wnt signaling activation. Stem Cell Reports 2017; 9: 2034–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Somerset DA, Strain AJ, Afford S, Whittle MJ, Kilby MD. Hepatocyte growth factor activator (HGF-A) and its zymogen in human placenta. Placenta 2000; 21: 615–620. [DOI] [PubMed] [Google Scholar]
- 24.Cross JC, Nakano H, Natale DR, Simmons DG, Watson ED. Branching morphogenesis during development of placental villi. Differentiation 2006; 74: 393–401. [DOI] [PubMed] [Google Scholar]
- 25.Bonometti S, Menarim BC, Reinholt BM, Ealy AD, Johnson SE. Growth factor modulation of equine trophoblast mitosis and prostaglandin gene expression. J Anim Sci 2019; 97: 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C, Spencer TE, Bazer FW. Expression of hepatocyte growth factor and its receptor c-met in the ovine uterus. Biol Reprod 2000; 62: 1844–1850. [DOI] [PubMed] [Google Scholar]
- 27.Lu J, Zhang S, Nakano H, Simmons DG, Wang S, Kong S, Wang Q, Shen L, Tu Z, Wang W, Wang B, Wang H, Wang Y, van Es JH, Clevers H, Leone G, Cross JC, Wang H. A positive feedback loop involving Gcm1 and Fzd5 directs chorionic branching morphogenesis in the placenta. PLoS Biol 2013; 11: e1001536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anson-Cartwright L, Dawson K, Holmyard D, Fisher SJ, Lazzarini RA, Cross JC. The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat Genet 2000; 25: 311–314. [DOI] [PubMed] [Google Scholar]
- 29.Bainbridge SA, Minhas A, Whiteley KJ, Qu D, Sled JG, Kingdom JC, Adamson SL. Effects of reduced Gcm1 expression on trophoblast morphology, fetoplacental vascularity, and pregnancy outcomes in mice. Hypertension 2012; 59: 732–739. [DOI] [PubMed] [Google Scholar]
- 30.Latos PA, Sienerth AR, Murray A, Senner CE, Muto M, Ikawa M, Oxley D, Burge S, Cox BJ, Hemberger M. Elf5-centered transcription factor hub controls trophoblast stem cell self-renewal and differentiation through stoichiometry-sensitive shifts in target gene networks. Genes Dev 2015; 29: 2435–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishiuchi T, Ohishi H, Sato T, Kamimura S, Yorino M, Abe S, Suzuki A, Wakayama T, Suyama M, Sasaki H. Zfp281 shapes the transcriptome of trophoblast stem cells and is essential for placental development. Cell Reports 2019; 27: 1742–1754.e6. [DOI] [PubMed] [Google Scholar]
- 32.Kauma SW, Bae-Jump V, Walsh SW. Hepatocyte growth factor stimulates trophoblast invasion: a potential mechanism for abnormal placentation in preeclampsia. J Clin Endocrinol Metab 1999; 84: 4092–4096. [DOI] [PubMed] [Google Scholar]
- 33.Moon HB, Ahn HY, Shin JC. Expression of hepatocyte growth factor and its receptor in the placental basal plate in pre-eclamptic pregnancies. Int J Gynaecol Obstet 2003; 83: 203–206. [DOI] [PubMed] [Google Scholar]
- 34.Nasu K, Zhou Y, McMaster MT, Fisher SJ. Upregulation of human cytotrophoblast invasion by hepatocyte growth factor. J Reprod Fertil Suppl 2000; 55: 73–80. [PubMed] [Google Scholar]
- 35.Fisher SJ. Why is placentation abnormal in preeclampsia? Am J Obstet Gynecol 2015; 213(Suppl): S115–S122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pollheimer J, Vondra S, Baltayeva J, Beristain AG, Knöfler M. Regulation of placental extravillous trophoblasts by the maternal uterine environment. Front Immunol 2018; 9: 2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pijnenborg R, Vercruysse L, Hanssens M. Fetal-maternal conflict, trophoblast invasion, preeclampsia, and the red queen. Hypertens Pregnancy 2008; 27: 183–196. [DOI] [PubMed] [Google Scholar]
- 38.Huppertz B. The critical role of abnormal trophoblast development in the etiology of preeclampsia. Curr Pharm Biotechnol 2018; 19: 771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y, Wang Y, Zeng X, Ma XJ, Zhao Y, Qiao J, Cao B, Li YX, Ji L, Wang YL. Self-control of HGF regulation on human trophoblast cell invasion via enhancing c-Met receptor shedding by ADAM10 and ADAM17. J Clin Endocrinol Metab 2012; 97: E1390–E1401. [DOI] [PubMed] [Google Scholar]
- 40.Zou Y, Hu M, Bao Q, Chan JY, Dai G. Nrf2 participates in regulating maternal hepatic adaptations to pregnancy. J Cell Sci 2013; 126: 1618–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikhaleva LM, Gracheva NA, Biryukov AE. The clinical and anatomical aspects of preeclampsia: current features of its course. Arkh Patol 2018; 80: 11–17 (in Russian). [DOI] [PubMed] [Google Scholar]
- 42.Momoi N, Tinney JP, Liu LJ, Elshershari H, Hoffmann PJ, Ralphe JC, Keller BB, Tobita K. Modest maternal caffeine exposure affects developing embryonic cardiovascular function and growth. Am J Physiol Heart Circ Physiol 2008; 294: H2248–H2256. [DOI] [PubMed] [Google Scholar]
- 43.Eudaly JA, Tizzano JP, Higdon GL, Todd GC. Developmental toxicity of gemcitabine, an antimetabolite oncolytic, administered during gestation to CD-1 mice. Teratology 1993; 48: 365–381. [DOI] [PubMed] [Google Scholar]
- 44.Boeldt DS, Bird IM. Vascular adaptation in pregnancy and endothelial dysfunction in preeclampsia. J Endocrinol 2017; 232: R27–R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kovacs CS. Maternal mineral and bone metabolism during pregnancy, lactation, and post-weaning recovery. Physiol Rev 2016; 96: 449–547. [DOI] [PubMed] [Google Scholar]
- 46.de Souza AMA, West CA. Adaptive remodeling of renal Na+ and K+ transport during pregnancy. Curr Opin Nephrol Hypertens 2018; 27: 379–383. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y, Wang T, Yan J, Jiagbogu N, Heideman DAM, Canfield AE, Alexander MY. HGF/c-Met signalling promotes Notch3 activation and human vascular smooth muscle cell osteogenic differentiation in vitro. Atherosclerosis 2011; 219: 440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strømme O, Psonka-Antonczyk KM, Stokke BT, Sundan A, Arum CJ, Brede G. Myeloma-derived extracellular vesicles mediate HGF/c-Met signaling in osteoblast-like cells. Exp Cell Res 2019; 383: 111490. [DOI] [PubMed] [Google Scholar]
- 49.Dudkowska M, Stachurska A, Chmurzyska W, Grzelakowska-Sztabert B, Manteuffel-Cymborowska M. Cross-talk between steroid-receptor-mediated and cell-membrane-receptor-mediated signalling pathways results in the in vivo modulation of c-Met and ornithine decarboxylase gene expression in mouse kidney. Biochem J 2001; 353: 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.