Abstract
Background
Bacterial urinary tract infection (BUTI) is the commonest urinary tract infection among people living with human immunodeficiency virus (PLHIV). It causes significant morbidity in this vulnerable group. Immunosuppression due to HIV can mask the signs and symptoms of infection leading to asymptomatic disease. There is limited evidence in Tanzania regarding BUTI and PLHIV. This study aimed to determine the prevalence, etiology, risk factors and susceptibility pattern of isolates causing asymptomatic UTI in PLHIV at Kilimanjaro Christian Medical Centre (KCMC).
Patients and Methods
This cross-sectional study was conducted from July to September 2020 at Kilimanjaro Christian Medical Centre (KCMC) hospital. A questionnaire was used to collect social demographic data from patients' files together with necessary information required by this study. Urine samples were obtained from participants for urinalysis and urine culture and sensitivity. Data from 300 adults aged ≥18 years were analyzed using Statistical Package for Social Science (SPSS) version 22. Mean or median with their measure of dispersion were calculated for continuous variables. Logistic regression was used to determine associations between variables, where P-value <0.05 was considered to be significant.
Results
Bacteriuria was prevalent in 37 (12.3%) of PLHIV. Positive nitrite in urine was associated with bacteriuria (P = 0.01). Gram-positive bacteria were the most common 25 (67.6%) isolates with high sensitivity to nitrofurantoin (88.2%) followed by gentamycin (69.2%). However, resistance was high against ciprofloxacin (77.8%). Multidrug resistance (MDR) was noted in 17 (45.9%).
Conclusion
One in eight of PLHIV attending CTC at KCMC referral hospital in Northern zone of Tanzania have BUTI. Presence of nitrites in urine is an important biomarker associated with BUTI. About two third of the isolates were Gram-positive bacteria, and nearly half of all isolates showed MDR to commonly used antibiotics.
Keywords: bacteriuria, bacteria urinary tract infection, people living with human immunodeficiency virus, multidrug resistance, Tanzania
Background
Bacterial urinary tract infection (BUTI) describes one of the common infections with a variety of clinical presentations ranging from asymptomatic, symptomatic to severe infection of the kidney. The clinical significance of asymptomatic bacteriuria (ABU) is its high risk of becoming symptomatic BUTI.1
Risk factors for BUTI include age, immunosuppressive conditions including diabetes mellitus and human immune virus (HIV), presence of underlying urological abnormality, multiple sclerosis and female gender. Vulnerable individuals, apart from being at risk of developing the condition, they are also at risk for recurrences, require long treatment duration and may develop more complex infection like pyelonephritis.2,3
The prevalence of bacteriuria in people living with HIV (PLHIV) has been documented to be high, and vary from one place to another ranging from 4–25.3%.3–6 In addition, PLHIV in Croatia had higher prevalence of bacteriuria than the control group of non-HIV-infected individuals.7 In Tanzania, the prevalence of UTI in asymptomatic pregnant women was 39 (16.6%).8 Risk factors for having bacteriuria in PLHIV include sex, and lower CD4 cell count.3,5,6,9
PLHIV have a diverse and broad spectrum of microbes causing BUTI. The most prevalent bacteria isolates causing bacteriuria in PLHIV were previously noted to be E. coli, E. faecalis, and S. aureus.3,6,9 Others have found Enterococcus spp. as the most common isolates in HIV-infected subjects while control group had E. coli.7
In Tanzania, E. coli was found to be the most common bacteria isolates in a study of both HIV positive and negative patients who were symptomatic for UTI.10 While in asymptomatic pregnant women, the common bacterial isolates were E. coli and K. pneumoniae.8
In this group of PLHIV, they have uropathogens which demonstrate moderate to high resistance against commonly prescribed antibiotics.5,11 Multidrug Resistance (MDR) has been demonstrated in 58.3% and in up to 78.4% of isolates causing ABU in PLHIV.4,9 MDR is the public health problem globally particularly in developing countries.12
This high prevalence of MDR isolates is a threat and therefore routine screening for bacteriuria has been recommended for early identification and treatment in order to prevent spread but also for betterment of patients.3–5,9
In our country, Tanzania, it was found that 75% of bacteria isolates from PLHIV were resistant to trimethoprim-sulfamethoxazole.10 Another study noted that E. coli were 90%, 16.7%, 10% and 13.3% resistant to trimethoprim-sulfamethoxazole, nitrofurantoin, gentamicin and ceftriaxone, respectively. The resistance rates for K. pneumoniae were 72.7%, 33.3% and 0% for trimethoprim-sulfamethoxazole, nitrofurantoin and ceftriaxone respectively.8 The prevalence of asymptomatic UTI and its resistance pattern in non-pregnant PLHIV in this context is not well known.
According to the Tanzanian standard treatment guideline of 2017, UTI in the general population is treated by ciprofloxacin with the exception of pregnant women and adolescents who are treated with amoxicillin-clavulanic acid. Referral to a high level of health facility is recommended for patients with acute pyelonephritis and the following characteristics: a) presence of vomiting, sepsis and known diabetes mellitus or b) pregnant women, women beyond reproductive age and in men. Before referral these patients receive ceftriaxone 50–80 mg/kg/dose as a single dose.13
There is no published study in Tanzania that has looked at the extent of asymptomatic UTI and the extent of multidrug resistance among adults living with HIV in this context. Additionally, the antibiotics recommended for empirical treatment in the general population are the ones used in PLHIV whenever empirical treatment is needed. However, the effectiveness of these antibiotics among PLHIV is uncertain. Therefore, this study aimed to determine the prevalence, etiology, risk factor and susceptibility pattern of isolates causing bacteriuria among people living with HIV attending Kilimanjaro Christian Medical Center (KCMC) care and treatment center (CTC).
Patients and Methods
Study Area and Population
This hospital based cross-sectional study was carried out from July to September 2020 at KCMC, Moshi, Tanzania. This zonal referral hospital in the Northern zone of Tanzania has a more than 600 bed capacity in a catchment area of approximately 15.7 million people. The hospital runs an outpatient CTC clinic three times a week, whereby an average of 100 patients are attended in each clinic day as per the local data from the clinic registry book.
Inclusion and Exclusion Criteria
The study included all PLHIV aged 18 years and above without sign and symptoms of UTI who consented to the study. A total of 311 participants were approached and consented to the study, four of them were exposed to other antibiotics other than trimethoprim-sulfamethoxazole for the past 2 weeks before attending our clinic, one was suspected to be pregnant and six males were on medical treatment for benign prostatic enlargement and therefore they were excluded from the study. The remaining 300 participants were interviewed, their files were reviewed for extraction of some of the information necessary for the study and mid stream clean-catch urine was collected from them for analysis.
Sample Size and Sampling Technique
The minimum sample size was calculated using Lesley Kish formulae. The assumptions included a standard deviation of 1.96, prevalence of ABU of 25.3% (Inyang-Etoh et al5) and the standard tolerable error of 0.05. The minimum sample size was therefore 290. To account for missing variables, we were set to recruit 300 PLHIV. The non-probability convenient sampling technique was employed to recruit study participants.
Sample Collection and Processing
The structured data questionnaire was used to collect the required data for the study. The questionnaire was pre-tested on five patients and errors noticed were corrected prior to the commencement of the study. These five patients were not included in this study. Social demography together with other variables were obtained from patient’s files and recorded on the questionnaire.
Two mid-stream clean-catch urine samples (each not less that five milliliters) from all eligible study participants were collected using two wide mouth screw-capped leak proof sterile containers by taking all precautions to avoid contamination. The two samples were taken to KCMC clinical laboratory within 30 minutes of collection. One specimen in each patient was cultured on blood agar (HiMedia laboratory Pvt Ltd, Mumbai, India) and cysteine lactose electrolyte deficient (CLED) agar (HiMedia laboratory Pvt Ltd, Mumbai, India). One-microliter disposable loop was used for nucleation on culture media plates then the plates were incubated at 37 °C for 24 hours. In case of delay in processing the sample, the samples were kept at 2–8 °C in a well-monitored refrigerator and cultured within 6 hours.
Urine cultures that grew a bacterial colony count of greater than or equal to 105 colony-forming units (CFU) per microliter of a typical urinary tract organism were regarded as significant bacterial growth, then a gram stain was performed from the colony for gram reaction identification. Gram negative bacteria identification test was performed by using the conventional method of identification which includes Kligler Iron Agar (KIA) (HiMedia laboratory Pvt Ltd, Mumbai-India), Sulphur Indole Motility (SIM) (HiMedia laboratory Pvt Ltd, Mumbai, India), Citrate (HiMedia laboratory Pvt Ltd, Mumbai, India), Urea (HiMedia laboratory Pvt Ltd, Mumbai, India) and Oxidase (Remel Europe Ltd, Dartford, UK) for suspected Pseudomonas spp., while gram positive identification tests was performed by using catalase (Remel Europe Ltd, Dartford, UK), coagulase (Remel Europe Ltd, Dartford, UK) for staphylococcus spp. and Bile esculin test (HiMedia laboratory Pvt Ltd, Mumbai, India) for Enterococcus spp. Bacitracin and optochin (all were from Oxoid Ltd, Basingstoke, England) test were used for identification of Streptococcus spp.
Five similar well isolated colonies of organisms from 24 hours pure culture were selected, the top of each colony was successively touched by using a one microliter loop and bacteria were transferred to a tube containing 5 mL of Mueller-Hinton broth (HiMedia laboratory Pvt Ltd, Mumbai, India), then the tube was incubated at 37 °C long enough (8hrs) to produce an organism suspension with slightly to moderate cloudiness. Then the broth was diluted with sterile saline to obtain turbidity equivalent to that of 0.5 McFarland standard. Then a sterile cotton swab was dipped into adjusted inoculum and the swab lifted above the fluid level and rotated against the inside of the tube to remove as much excess inoculum as possible, the entire agar (Mueller-Hinton Agar) was streaked with the swab evenly in three directions and the inoculum were left to dry for 5 minutes with the plate closed. Lastly antibiotics discs were placed on the agar with a sterile dispenser and the plates were incubated at 37 °C for 24 hrs.
Antimicrobial susceptibility tests were done for commonly prescribed antibiotics in our set up and zones of inhibition were interpreted using Clinical Laboratory Standard Institute guideline of year 2020 (CLSI).14 However, owing to the challenges in the availability of antibiotic discs, we did not conduct even number of drug sensitivity tests.
The following antibiotics discs (Basingstoke, England (UK)) were tested; gentamicin 10 µg, piperacillin 100 µg, ceftriaxone 30 µg, amoxicillin-clavulanic acid 20/10 µg, amikacin 30 µg, clindamycin 2 µg, nitrofurantoin 300 µg, chloramphenicol 30 µg, vancomicin 30 µg, trimethoprim-sulfamethoxazole 1.25/23.75 µg, ciprofloxacin 5 µg, cefotaxime 30 µg, penicillin 10 µg and erythromycin 15 µg. In this study, bacteria showing resistance to three or more categories of antibiotics tested was marked as multidrug resistant bacteria.12
The second sample of urine was analyzed using Cybow urinalysis test strips (DFI Co, Ltd, Gyeongsangnam-do, Korea) and recorded appearance, status of leucocytes and nitrites. The result of both urine culture and urinalysis were recorded in the questionnaire.
Quality Control
During the whole process of urine collection and analysis, standard operating procedures (SOP) were highly observed. These include making sure that the urine test strips, culture medias, antimicrobial identification tests and antibiotic discs have not expired and quality control parameters are followed as per CLSI 2020. Thorough inspection of the antibiotic disc and petri dishes to rule out cracks, presence of contaminated sample, bubbles and freezing was done. The quality of media was checked for each new lot by testing the S. aureus ATCC29213 for gram positive bacteria and E. coli ATCC 25922 for gram negative bacteria standard control strains.
Laboratory Analysis
KCMC clinical laboratory performs both CD4 count and viral load together with other tests including urinalysis and urine culture as part of its daily routine. It also does these tests together with other tests as part of research activity conducted at KCMC. The lab is equipped with Cobas 4800, Cobas Ampliprep/Cobas Taqman 96, Hologic Panther for HIV viral load testing, BD FACSPresto, BD FACSCalibur for CD4 count. The laboratory participates in external quality assurance with public health agency of Canada (QASI-LI), is also internationally recognized and accredited with ISO number 15189.
Data Management and Analysis
Data was analyzed by Statistical Package for Social Science (SPSS) version 22. Mean with standard diversion or median with interquartile ranges were calculate for continuous variables. Data was summarized in tables and figures. Logistic regression was used to determine association between dependent and independent variables, where variables with P value less than 0.09 in the univariate model were subjected to a multivariate logistic regression model. Odds ratio (OR) and 95% confidence interval (CI) were computed and P < 0.05 was considered to be statistically significant.
Ethical Consideration
The ethical clearance for this research was obtained from Kilimanjaro Christian Medical University Research and Ethical Review Committee (EC No.2420). Permission to conduct the study at KCMC CTC and use of KCMC main clinical laboratory was obtained from executive director of KCMC hospital. A written informed consent was obtained from each participant before interview and sample collection. All the data were kept confidential. For treatment purposes, the laboratory results were disclosed to attending physician. This study complied with the declaration of Helsinki with regard to ethical issues.
Results
Characteristics of the Study Participants
This study included a total of 300 participants who were attending KCMC CTC clinic. The mean (SD) age of the study participants was 46.1 (11.9) years. The majority 197 (65.7) were aged >40 years, more than two third; 233 (77.7%) were females. The median (range) CD4 count was 677.5 (4.0–3189) cells/mm3, 9 (3%) had a CD4 count < 200 cells/mm3, 23 (7.7%) had leucocytes in urine and 5 (1.7%) had nitrites in their urine (Table 1).
Table 1.
Characteristics of 300 Study Participants Living with HIV and Attending CTC Clinic at KCMC from July to September 2020
| Total | |
|---|---|
| Characteristics | n (%) |
| Age (years) (mean (SD) | 46.1 (11.9) |
| Age (years) | |
| 18–40 | 103 (34.3) |
| >40 | 197 (65.7) |
| Sex | |
| Male | 67 (22.3) |
| Female | 233 (77.7) |
| Marital status | |
| Single | 76 (25.3) |
| Married | 175 (58.3) |
| Widow | 39 (13.0) |
| Divorce/separated | 10 (3.3) |
| Known diabetic | |
| No | 285 (95.0) |
| Yes | 15 (5.0) |
| CD4 count(cells/mm3) | |
| < 200 | 9 (3.0) |
| 200–500 | 80 (26.7) |
| > 500 | 211 (70.3) |
| Use cotrimoxazole | |
| No | 263 (87.7) |
| Yes | 37 (12.3) |
| Urine appearance | |
| Cloud | 26(8.7) |
| Clear | 274 (91.3) |
| Leucocytes | |
| Negative | 277 (92.3) |
| Positive | 23 (7.7) |
| Nitrites | |
| Negative | 295 (98.3) |
| Positive | 5 (1.7) |
| Viral load | |
| < 20 | 218 (72.7) |
| 20–1000 | 73 (24.3) |
| >1000 | 9 (3.0) |
Prevalence and Risk Factors of UTI
The overall prevalence of bacteriuria was 37 (12.3%). The magnitude was high among females (14.2%) compared to males (6.0%), and in those who had a CD4 count of <200 cell/mm3, 2(22.2%), positive leucocytes 5 (21.7%) and four positive nitrites 4 (80%) compared to their counterparts. The presence of nitrites in urine was 21 times higher among people with ABU compared to their counterparts with no ABU (AOR = 21.18, 95% CI: 2.05; 218.86, p = 0.01). The risk of ABU was more than doubled among females and those having leucocytes in urine, however, such association did not reach a statistically significant level (Table 2).
Table 2.
Prevalence and Risk Factors for UTI Among 300 Patients Living with HIV Attending CTC Clinic at KCMC from July to September 2020
| Factors | Urine Culture Result | ABU | ||||
|---|---|---|---|---|---|---|
| Negative | Positive | |||||
| n (%) | n (%) | |||||
| 263 (87.7) | 37(12.3) | COR (95% CI) | p-value | AOR (95% CI) | p-value | |
| Age (years) | ||||||
| 18–40 | 88 (85.4) | 15(14.6) | 1 | |||
| >40 | 175 (88.8) | 22(11.2) | 0.74(0.36; 1.49) | 0.397 | ||
| Sex | ||||||
| Male | 63 (94.0) | 4 (6.0) | 1 | 1 | ||
| Female | 200 (85.8) | 33 (14.2) | 2.59 (0.89; 7.62) | 0.082 | 2.35 (0.74; 7.54) | 0.148 |
| Known diabetic | ||||||
| No | 250 (87.7) | 35 (12.2) | 1 | |||
| Yes | 13 (86.7) | 2 (13.3 | 1.09 (0.23; 5.07) | 0.904 | ||
| CD4 count (cells/mm3) | ||||||
| < 200 | 7 (77.8) | 2 (22.2) | 1 | |||
| 200–500 | 69 (86.3) | 11 (13.7) | 0.56 (0.10; 3.04) | 0.511 | ||
| > 500 | 187 (86.6) | 24 (11.4) | 0.45 (0.09; 2.29) | 0.335 | ||
| Use Cotrimoxazole | ||||||
| No | 229 (87.1) | 34 (12.9) | 1 | |||
| Yes | 34 (91.9) | 3 (8.1) | 0.59 (0.17; 2.04) | 0.409 | ||
| Urine appearance | ||||||
| Clear | 243 (88.7) | 31 (11.3) | 1 | 1 | ||
| Cloud | 20 (76.9) | 6 (23.1) | 2.35 (0.88; 6.30) | 0.089 | 1.58 (0.51; 4.89) | 0.427 |
| Leucocytes | ||||||
| Negative | 245 (88.4) | 32 (11.6) | 1 | |||
| Positive | 18 (78.3) | 5 (21.7) | 2.39 (0.82; 6.97) | 0.111 | ||
| Nitrites | ||||||
| Negative | 262 (88.8) | 33 (11.1) | 1 | 1 | ||
| Positive | 1 (20) | 4 (80) | 23.82 (2.41; 27.69) | 0.007 | 21.18(2.05; 218.86) | 0.01 |
| Viral load (c/mL) | ||||||
| <20 | 191 (87.6) | 27 (12.4) | 1 | |||
| 20–1000 | 64 (87.7) | 9 (12.3) | 0.99(0.44; 2.23 | 0.991 | ||
| >1000 | 8 (88.9) | 1 (11.1) | 0.88(0.11; 7.35) | 0.909 | ||
Abbreviations: COR, crude odds ratio; AOR, adjusted odds ratio; ABU, asymptomatic bacteriuria.
Microbiological Profile of the Isolates
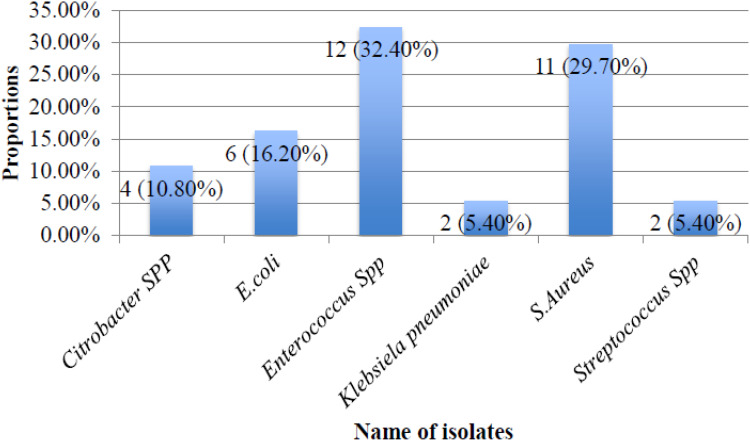
Among the 37 participants who had ABU, none of them had more than one type of bacterial isolate. The majority of the isolates were gram positive bacteria 25 (67.6%). Of the 37 isolates, the most frequent isolates were Enterococcus spp. 12 (32.4%), S. aureus 11 (29.7%), E. coli 6 (16.2%) and Citrobacter spp. 4 (10.8%) (Figure 1).
Figure 1.
Microbiological profile of 37 isolates among 300 participants living with HIV attending CTC clinic at KCMC from July to September 2020.
Antimicrobial Susceptibility Profile
The majority of the gram-positive bacteria isolates were tested for nitrofurantoin, vancomycin, ciprofloxacin, and erythromycin and most of them (88.2%) were sensitive to nitrofurantoin while they demonstrated high resistance against ciprofloxacin (77.8%) and erythromycin (60%). On the other hand, most of the gram negative bacteria were subjected to amoxicillin-clavulanic acid, ceftriaxone, gentamycin, trimethoprim-sulfamethoxazole and nitrofurantoin and they were 100%, 91%, 80% and 66.7% sensitive to amoxicillin-clavulanic acid, ceftriaxone, gentamycin and nitrofurantoin respectively. However, the resistances were high against trimethoprim-sulfamethoxazole (50%) (Table 3).
Table 3.
Antimicrobial Susceptibility Pattern of 37 Bacterial Isolates Among 300 Participants Living with HIV Attending CTC Clinic at KCMC from July to September 2020
| Isolated Bacteria | Gen | Pip | Ceft | Amoxc | Amik | Clin | Nitr | Chlor | Vanc | TX | Cipro | Cefo | Pen | Ery |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |
| Gram negative (n = 12) | ||||||||||||||
| E. coli (n=6) | NA | NA | NA | NA | NA | |||||||||
| Sensitive | 2 (50) | 5 (100) | 6 (100.0) | 1 (100) | 3 (100 | 4 (66.7) | 4 (66.7) | |||||||
| Resistant | 2 (50) | 0(0) | 0(0) | 0(0) | 0(0) | 2 (33.3) | 2 (33.3) | |||||||
| Citrobacter spp. (n=4) | NA | NA | NA | NA | ||||||||||
| Sensitive | 4 (100.0) | 1 (33.3) | 4 (100.0) | 4 (100.0) | 3 (100) | 1 (25.0) | 1 (100) | 1 (33.3) | ||||||
| Resistant | 0(0) | 2 (66.7) | 0(0) | 0(0) | 0(0) | 3 (75.0) | 2 (66.7) | 2 (100) | ||||||
| Klebsiella (n=2) | NA | NA | NA | NA | ||||||||||
| Sensitive | 2 (100.0) | 1 (50.0) | 2 (100.0) | 2(100) | 2 (100.0) | |||||||||
| Resistant | 0(0) | 1 (50.0) | 0(0) | 0(0) | 1 (100) | 0(0) | 1 (100) | |||||||
| Total n(%) | ||||||||||||||
| S | 8(80) | 1(33.3) | 10(90.9) | 12(100) | 4(100) | NA | 6(66.7) | 1(100) | NA | 5(50) | 6(60) | NA | 1(100) | NA |
| R | 2(20) | 2(66.7) | 1(9.1) | 0(0) | 0(0) | NA | 3(33.3) | 0(0) | NA | 5(50) | 4(40) | NA | 0(0) | NA |
| Gram positive (n = 25) | ||||||||||||||
| S. aureus (n=11) | NA | NA | NA | |||||||||||
| Sensitive | 5 (100) | 4 (44.4 | 1 (16.7) | 6 (100) | 6 (54.5) | 3 (50) | 1 (50) | 2 (22.2) | ||||||
| Intermediate | 0(0) | 0(0) | 1 (16.7) | 1 (9.1) | ||||||||||
| Resistant | 0(0) | 5 (55.6) | 4 (66.7) | 1 (100) | 3 (27.3) | 3 (100) | 3 (50) | 1 (50) | 4 (100) | 7 (77.8) | ||||
| Streptococcus spp. (n=2) | NA | NA | NA | NA | NA | |||||||||
| Sensitive | 1 (100) | 1 (50.0) | 1 (100) | |||||||||||
| Intermediate | 1 (100) | |||||||||||||
| Resistant | 1 (50.0) | |||||||||||||
| Enterococcus spp. (n=12) | NA | |||||||||||||
| Sensitive | 3 (42.9) | 4 (66.7) | 1 (100) | 9 (81.8) | 4 (100) | 9 (75.0) | 1 (8.3) | 6 (54.5) | 3 (30) | |||||
| Intermediate | 1 (14.3 | 1 (100) | 1 (9.1) | 1 (8.3) | 2 (20) | |||||||||
| Resistant | 3 (42.9) | 2 (33.3) | 1 (100) | 1 (9.1) | 2 (16.7) | 2 (16.7) | 11 (91.7) | 1 (100) | 5 (45.5) | 5 (50) | ||||
| Total n(%) | NA | |||||||||||||
| S | 9(69.2) | 8(53.3) | 1(100) | 1(14.3) | 15(88.) | 4(80) | 15(68.2) | 1(14.3) | 4(22.2) | 1(33.3) | 6(37.5) | 6(30) | ||
| I | 1(7.7) | 0(0) | 0(0) | 1(14.3) | 1(5.9) | 0(0) | 2(9.1) | 0(0) | 0(0) | 0(0) | 1(6.3) | 2(10) | ||
| R | 3(23.1) | 7(46.7) | 0(0) | 5(71.4) | 1(5.9) | 1(20) | 5(22.7) | 6(85.7) | 14(77.8) | 2(66.7) | 9(56.2) | 12(60) |
Abbreviations: Gen, gentamycin; Pip, piperacillin; Ceft, ceftriaxone; amoxc, amoxicillin-clavulanic acid; amik, amikacin; clind, clindamycin; Nitr, nitrofurantoin; chlor, chloramphenicol; vanc, vancomycin; TX, trimethoprim-sulfamethoxazole; cipro, ciprofloxacin; cefo, cefotaxime; pen, penicillin; ery, erythromycin.
The majority of Enterococcus spp. were 81.8% sensitive to nitrofurantoin and 75% vancomycin while they were 91.7% resistant to ciprofloxacin. Five out of eleven S. aureus were subjected to gentamycin and six were subjected to nitrofurantoin and they were all sensitive to these drugs. However the resistance was high against erythromycin (77.8%), followed by clindamycin (66.7%), ceftriaxone (55.6%) and ciprofloxacin (50%) (Table 3).
Multidrug Resistance
Among 37 culture positive samples, 17 of them showed resistance to three or more groups of commonly used antibiotics giving an overall MDR rate of 17 (45.9%) with Gram negative and positive bacteria having an MDR rate of 3 (25%) and 14 (56%) respectively. This is shown in Table 4.
Table 4.
Multidrug Resistance Pattern Among Bacterial 37 Isolates from 300 People Living with HIV Attending CTC Clinic at KCMC from July to September 2020
| Total | MDR | |||||||
|---|---|---|---|---|---|---|---|---|
| Isolated Bacteria | n (%) | R0 | R1 | R2 | R3 | R4 | R5 | n (%) |
| Gram negative | 12 (32.4) | 4 | 1 | 4 | 2 | 1 | 3 (25.0) | |
| E. coli (n=6) | 6 (50.0) | 3 | 1 | 1 | 1 | 2 (16.7) | ||
| Citrobacter spp. (n=4) | 4 (33.3) | 1 | 3 | 0 (0.0) | ||||
| K. pneumoniae (n=2) | 2 (16.7) | 1 | 1 | 1 (8.3) | ||||
| Gram positive | 25 (67.6) | 2 | 3 | 6 | 9 | 4 | 1 | 14 (56.0) |
| S. aureus (n=11) | 11 (44.0) | 1 | 3 | 4 | 2 | 1 | 7 (28.0) | |
| Streptococcus spp. (n=2) | 2 (8.0) | 1 | 1 | 1 (4.0) | ||||
| Enterococcus spp. (n=12) | 12 (48.0) | 2 | 2 | 2 | 4 | 2 | 6 (24.0) | |
| Total | 37 (100.0) | 6 (16.2) | 4 (10.8) | 10 (27.1) | 11 (29.7) | 5 (13.5) | 1 (2.7) | 17 (45.9) |
Abbreviations: R0, no resistant drug reported; R1, resistance to one group of antibiotic; R2, resistance to two groups of antibiotics; R3, resistance to three groups of antibiotics; R4, resistance to four groups of antibiotics; R5, resistance to five groups of antibiotics; MDR, resistance to more than two groups of antibiotics.
Discussion
The overall prevalence of bacteriuria (12.3%) in our study was relatively similar to two studies done in Ethiopia that documented prevalence of 10.3% and 11.3%.9,15 On the other hand, our finding was higher than that of India (4%)6 but lower than Uganda (32.2%),16 Ethiopia (18%)17 and Nigeria (19.5%, 26%)18,19. The difference might be due to differences in geographical location and social factors that may affect perinea hygiene.
Females 33 (14.2%) had a higher prevalence of bacteriuria compared to males although the difference did not reach a statistically significant level. This finding concurs with several other studies including the studies from India,6 Uganda16 and Kenya.20 The reason for having high prevalence in females could be related to the close proximity of the urethral to the anus but also the short and wide urethra. However, our study finding was different from a Nigerian study that found males to have a higher prevalence of 28.6% compared to 23.8% for female.5
In the current study, presence of nitrites in urine was significantly associated with bacteriuria (P = 0.01). Some of the uropathogenic bacteria converts nitrates in urine to nitrites, Therefore, PLHIV should undergo routine urinalysis screening for nitrites and those with positive nitrites should be investigated further by urine culture and sensitivity in order to have culture guided treatment results. However, other factors including CD4 count, age, diabetes status, and presence of leukocytes in urine were not statistically associated with bacteriuria. Although we had a small number of persons with CD4 count <200 cells/mm3, our study is in line with the findings of a Nigerian study which showed there is no association between CD4 count and bacteriuria.18 This is contrary to a study done in Ethiopia15 which showed female sex and having a CD4 count <200 cells/mm3 were significantly associated with bacteriuria. Other studies done elsewhere showed age to be a significant predictor of UTI.16,17 The differences could be due to the differences in sample size and study participants.
Most common bacterial isolates in our study were gram positive which is contradictory with other investigations which reported gram negative bacteria to be the most frequent isolate.9,15,21
Enterococcus spp. is now among the 3rd and 4th leading cause of nosocomial infection globally with a tendency to persist in a hospital environment. In Africa, the prevalence of this bacterium in different samples ranges from 5.7–88.9%. People living with HIV and a history of previous hospitalization are among the risk group for acquiring Enterococcus spp.22 Therefore, it is not surprising for this study to have Enterococcus spp. (32.4%) as the dominant isolate that resembles those found in nosocomial infection. A similar finding was reported in Croatia with the prevalence of 14 (26%).7 The reason for a high preponderance of this organism could be due to frequent hospitalization and HIV status of this patient as the risk factor for contacting nosocomial infection. However, a majority of other studies done in African countries like Nigeria19 and Ethiopia9 even in India6 and Europe3 showed the most prevalent organism was E. coli.
The high proportion of Enterococcus spp. in our study may be of public health concern, because evidence suggests that they have a tendency to form resistance to currently available antibiotics through various pathways and resistant populations proliferate very rapidly.22 This is an alarming sign in our environment and Africa at large because of the limited number of effective antibiotics in our environment.
In the current study, S. aureus was the second most common isolate, accounting for 29.70%. This is contrary to another study done in Tanzania where the proportions of S. aureus was 3.9% and was the fourth most common isolate.8 This might be due to differences in the study population. In the present study, we excluded pregnant women who were studied in the previous study. This is a different group that might have different characteristics including the type of isolates. Another study was done in Tanzania by Lyamuya et al, in his study on bacteriuria in diabetic women he found S. aureus to be the fourth most common isolate with a prevalence of 14.65%.23 Again this difference could be due to differences in the study population as in Lyamuya’s study involved diabetic women while the current study involved all patients, regardless of their sex and diabetic status.
The finding of the current study is similar to what is reported in India (25%)24 and Uganda (31.4%).16 However, other studies done in India and Ethiopia reported S. aureus to be the second most common isolate but with a lower proportion at 17.5%25 and 19.6%9 respectively. The differences in the proportions could be explained by differences in geographical location, and study populations.
Most of the gram positives bacteria were sensitive to nitrofurantoin (88.2%) while demonstrating resistance to ciprofloxacin and erythromycin by 77.8% and 60% respectively. This is in contradiction with one of the Ethiopian studies that found a very high sensitivity (100%) to nitrofurantoin and erythromycin.26 The differences could be due to the fact that in the previous study the proportion of gram positive bacteria (12.5%) was low as compared to ours (67.6%). Therefore, this could lead into an overestimation of the sensitivity and resistant patterns in their study.
In this study the majority of the gram negative bacteria were sensitive to amoxicillin-clavulanic acid (100%), ceftriaxone (91%), gentamycin (80%) and nitrofurantoin (66.7%). On the other hand, they were 50% resistant to trimethoprim-sulfamethoxazole. Contrarily, others had gram negative bacteria that were highly sensitive to nitrofurantoin (95%) and relatively similar sensitivity to ceftriaxone of 92.5% while they were highly resistant to trimethoprim-sulfamethoxazole (87.5%).26 In addition, Fenta G et al found that most gram negative bacteria were highly sensitive to nitrofurantoin by 100% and highly resistant to trimethoprim-sulfamethoxazole (71%).9 In Ethiopia, gram negative bacteria were highly sensitive to nitrofurantoin by 80% and similarly sensitivity (80%) to gentamycin, whereas resistance to trimethoprim-sulfamethoxazole was 57.1%.15 The differences in sensitivity and resistant patterns could be due to over-distribution of sensitivity and resistant strains of bacteria in these areas but also could be related to overuse and misuse of antimicrobials as they are easily available in the pharmacies.
In the current study, a large proportion of Enterococcus spp. was sensitive to nitrofurantoin (81.8%) and vancomycin (75%) while demonstrating high resistance to ciprofloxacin (91.7%). This is against the study by Fenta G et al, who found a very high sensitivity of 100% for nitrofurantoin and vancomycin.9 This difference could be contributed to by differences in the characteristics of the participants whereby in the previous study the proportion of gram positive isolates where only 39.3% as opposed to 67.6% in the current study, it may also be due to over-distribution of a vancomycin resistant strain of the Enterococcus (VRE) in our population.
The MDR rate of 45.9% is in line with one of the Indian studies that reported a rate of 48.5%25 but lower than other studies with a higher rate of 58.2%, 78% and 95%.4,9,27 The differences could be related to the extent of irrational use of antibiotics, with areas having a large degree of irrational use having high prevalence of MDR.
Of the 25 gram-positive bacteria, 14 (56.0%) were resistant to three or more groups of antibiotics (MDR), S. aureus accounted for 7 (28.0%) and Enterococcus spp. 6 (24.0%). On the other hand, only 3 (25%) gram-negative bacteria demonstrated MDR character, E. coli accounted for 2 (16.7%). Contrarily, in Ethiopia they found a prevalence of 16 (76.2%) among gram negative bacteria isolates and of those 57.1% were due to E. coli.15 Additionally, others had a MDR of 87% and 65% among gram negative and positive bacteria respectively.9
Study Limitations
Results of this study should be discussed in the light of the following limitations. First, this hospital based cross sectional study was conducted in a tertiary zonal and referral hospital. Therefore, results may not be generalizable to all health facilities, but to health facilities with similar contexts. Second, owing to the challenges in available antibiotic discs, we could not conduct an even number of sensitivity tests across all bacteria. Despite this limitation, our findings remained comparable to some other studies.
Conclusion
About one in eight of PLHIV in our setting have BUTI, of this, two thirds (67.6%) are gram-positive bacteria. Moreover, Enterococcus spp. accounts for the majority followed by S. aureus.
The presence of nitrites in urine was statistically associated with a positive urine culture. Therefore, routine urinalysis as a screening test for this group of people is recommended and for those with positive nitrites should be investigated further by urine culture and sensitivity in order to have culture-guided treatment.
About two thirds (67.6%) of the isolates were gram positive bacteria and they were sensitive to nitrofurantoin (88.2%), gentamycin (69.2%) and vancomycin (68.2%) while demonstrating resistance against ciprofloxacin (77.8%), erythromycin (60%), penicillin (56.2%) and ceftriaxone (46.7%). Hence we recommend use of either nitrofurantoin or gentamicin or vancomycin but never ciprofloxacin whenever empirical treatment for UTI is needed in PLHIV.
Nearly half of the isolates showed MDR to commonly used antibiotics. The alarming prevalence of MDR bacteria among PLHIV calls for measures to ensure judicious use of antibiotics, including culture-guided treatment.
Acknowledgments
Authors would like to extend their sincere thanks to the THET project for funding this research (award number R25TW011227) and Kilimanjaro Christian Medical University College for granting ethical clearance for this work. They also want to express their deep gratitude to all staff members of Kilimanjaro Christian Medical Centre, including the administrators, for their tireless support. Furthermore, their appreciation goes to all study participants because without their willingness this study wouldn’t have been possible.
Disclosure
All authors listed in this article declare that they have no conflicts of interest for this work.
References
- 1.Essien UC, Iheukwumere CC, Davou GI, et al. Prevalence and predictors of asymptomatic urinary tract infection among HIV positive patients in Jos, North central Nigeria. Int J Curr Microbiol Appl Sci. 2015;4(9):454–462. [Google Scholar]
- 2.Olowe OA, Ojo-Johnson B, Makanjuola OB, Olowe RA, Mabayoje VO. Detection of bacteriuria among human immunodeficiency virus seropositive individuals in Osogbo, south-western Nigeria. Eur J Microbiol Immunol. 2015;5(1):126–130. doi: 10.1556/eujmi-d-14-00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skrzat-klapaczy A, Matłosz B, Bednarska A, et al. Factors associated with urinary tract infections among HIV-1 infected patients. PLoS One. 2018;13(1):1–10. doi: 10.1371/journal.pone.0190564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murugesh K, Deepa S, Ravindranath C, Venkatesha D. Multidrug resistant uropathogens in HIV: are they a threat to community? Int J Sci Study. 2014;2(3):38–42. [Google Scholar]
- 5.Inyang-Etoh PC. Asymptomatic bacteriuria in patients on antiretroviral drug therapy in calabar” PC. Inyang-Etoh,” GC Udofa,” AA A. Alaribe and” NE Udonwa. J Med Sci. 2009;9(6):270–275. [Google Scholar]
- 6.Banu A, Ramachandrian J. Asymptomatic bacteriuria in HIV positive individuals in a tertiary care hospital. J HIV Hum Reprod. 2013;1(2):2–5. doi: 10.4103/2321-9157.126630 [DOI] [Google Scholar]
- 7.Schönwald S, Begovac J, Skerk V. Urinary tract infections in HIV disease. Int J Antimicrob Agents. 1999;11(3–4):309–311. doi: 10.1016/S0924-8579(99)00036-9 [DOI] [PubMed] [Google Scholar]
- 8.Chaula T, Seni J, Ng’walida N, et al. Urinary tract infections among HIV-positive pregnant women in Mwanza city, Tanzania, are high and predicted by low CD4+ count. Int J Microbiol. 2017;2017:1–7. doi: 10.1155/2017/4042686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenta GM, Legesse MH, Weldearegay GM. Bacteriuria and their antibiotic susceptibility patterns among people living with HIV attending Tikur Anbessa Specialized and Zewditu Memorial Hospital ART clinics, Addis Ababa, Ethiopia. J Bacteriol Parasitol. 2016;07(05). doi: 10.4172/2155-9597.1000292 [DOI] [Google Scholar]
- 10.Marwa KJ, Mushi MF, Konje E, Alele PE, Kidola J, Mirambo MM. Resistance to cotrimoxazole and other antimicrobials among isolates from HIV/AIDS and non-HIV/AIDS patients at Bugando Medical Centre, Mwanza, Tanzania. AIDS Res Treat. 2015;2015. doi: 10.1155/2015/103874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klasinc R, Rieger A, Presterl E, Wrba T, Diab- M. Epidemiology of urinary tract infections in HIV-positive patients at a tertiary care hospital in Central Europe (2011–2016). Infect Disord Targets. 2017;18(3):199–206. doi: 10.2174/1871526518666180405153616 [DOI] [PubMed] [Google Scholar]
- 12.Kemajou TS, Ajugwo AO, Oshoma CE, Oi E. Antibiotic resistance of bacterial isolates from HIV positive patients with Urinary Tract Infection (UTI) in Portharcourt, Nigeria. J AIDS Clin Res. 2016;7(8):8–11. doi: 10.4172/2155-6113.1000594 [DOI] [Google Scholar]
- 13.Tanzania Ministry of Health, Community Development, Gender E and C. Standard Treatment Guidelines and National Essential Medicines Lists. Vol. 39. Tanzania: National Drug Policy, Essential Drugs; 2018. Available from: http://www.tzdpg.or.tz/fileadmin/documents/dpg_internal/dpg_working_groups_clusters/cluster_2/health/Key_Sector_Documents/Tanzania_Key_Health_Documents/STANDARD_TREATMENT_GUIDELINES__CORRECT_FINAL_USE_THIS-1.pdf. Accessed March29, 2021. [Google Scholar]
- 14.CLSI. M100 Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. Wayne, PA; 2020. [Google Scholar]
- 15.Tessema NN, Ali MM, Zenebe MH. Bacterial associated urinary tract infection, risk factors, and drug susceptibility profile among adult people living with HIV at Haswassa University Comprehensive Specialized Hospital, Hawassa, Southern Esthiopia. Sci Rep. 2020;0123456789:1–9. doi: 10.1038/s41598-020-67840-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odoki M, Aliero AA, Tibyangye J, et al. Prevalence of bacterial urinary tract infections and associated factors among patients attending hospitals in Bushenyi District, Uganda. Int J Microbiol. 2019;2019:1–8. doi: 10.1155/2019/4246780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marami D, Balakrishnan S, Seyoum B. Prevalence, antimicrobial susceptibility pattern of bacterial isolates, and associated factors of urinary tract infections among HIV-positive patients at Hiwot Fana Specialized University Hospital, Eastern Ethiopia. Can J Infect Dis Med Microbiol. 2019;2019. doi: 10.1155/2019/6780354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danjuma ZA, Asuquo AE, Pokam T, Ibeneme E. Bacteriuric profiles and CD4 counts of Human Immunodeficient Virus (HIV) seropositve individuals seeking care in a Nigerian health facility sample collection and processing methods study subjects analysis of whole blood cells ethical clearance statistic. Int J Biomed Sci. 2019;8(1):1–9. [Google Scholar]
- 19.Adegoke AA, Akpan AA, Bassey N. Prevalence of significant asymptomatic bacteriuria in HIV-positive individuals under antiretroviral therapy. World J Appl Sci Technol. 2019;11(1):91–98. [Google Scholar]
- 20.Kayima JK, Otieno LS, Twahir A, Njenga E. Asymptomatic bacteriuria among diabetics attending Kenyatta National Hospital. East Afr Med J. 1996;73(8):524–526. [PubMed] [Google Scholar]
- 21.Olowe OA, Makanjuola OB, Olowe RA, Mabayoje VO. Detection of bacteriuria among human immunodeficiency virus seropositive individuals in Osogbo, south-western Nigeria. Eur J Microbiol Immunol. 2015;5:1–5. doi: 10.1556/EuJMI-D-14-00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ali S, Alemayehu M, Dagnew M, Gebrecherkos T. Vancomycin-resistant enterococci and its associated risk factors among HIV-positive and -negative clients attending Dessie Referral Hospital, Northeast Ethiopia. Int J Microbiol. 2018;2018:1–9. doi: 10.1155/2018/4753460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyamuya EF, Moyo SJ, Komba EV, Haule M. Prevalence, antimicrobial resistance and associated risk factors for bacteriuria in diabetic women in Dar es Salaam, Tanzania. Afr J Microbiol Res. 2011;5(6):683–689. [Google Scholar]
- 24.Banu A, Jyothi R. Asymptomatic bacteriuria in HIV positive individuals in a tertiary care hospital. J HIV Hum Reprod. 2013;1(2):54. doi: 10.4103/2321-9157.126630 [DOI] [Google Scholar]
- 25.Yadhav K, Samreen S. Asymptomatic urinary tract infection with multidrug resistant pathogens in retro positive patients on ART. Int J Med Res Rev. 2017;5(6):610–615. doi: 10.17511/ijmrr.2017.i06.10 [DOI] [Google Scholar]
- 26.Debalke S, Cheneke W, Tassew H, Awol M. Urinary tract infection among antiretroviral therapy users and nonusers in Jimma University Specialized Hospital, Jimma, Ethiopia. Int J Microbiol. 2014;2014:6. doi: 10.1155/2014/968716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alemu A, Dagnew M, Alem M, Gizachew M. Uropathogenic bacterial isolates and their antimicrobial susceptibility patterns among HIV/AIDS patients attending Gondar University Specialized Hospital Gondar, Northwest Ethiopia. J Microbiol Res Rev. 2013;1(4):42–51. [Google Scholar]