Abstract
Purpose
Endometrial cancer (EC) is often the sentinel cancer in women with Lynch syndrome (LS). However, efforts to implement universal LS screening in EC patients have been hampered by a lack of evidence detailing the proportion of EC patients that would be expected to screen positive for LS.
Methods
Studies were identified by electronic searches of Medline, Embase, Cochrane CENTRAL and Web of Science. Proportions of test positivity were calculated by random and fixed-effects meta-analysis models. I2 score was used to assess heterogeneity across studies.
Results
Fifty-three studies, including 12,633 EC patients, met the inclusion criteria. The overall proportion of endometrial tumors with microsatellite instability or mismatch repair (MMR) deficiency by immunohistochemistry (IHC) was 0.27 (95% confidence interval [CI] 0.25–0.28, I2: 71%) and 0.26 (95% CI 0.25–0.27, I2: 88%), respectively. Of those women with abnormal tumor testing, 0.29 (95% CI 0.25–0.33, I2: 83%) had LS-associated pathogenic variants on germline testing; therefore around 3% of ECs can be attributed to LS. Preselection of EC cases did increase the proportion of germline LS diagnoses.
Conclusion
The current study suggests that prevalence of LS in EC patients is approximately 3%, similar to that of colorectal cancer patients; therefore our data support the implementation of universal EC screening for LS.
Keywords: systematic review, Lynch syndrome, endometrial cancer, mismatch repair (MMR) immunohistochemistry, microsatellite instability (MSI)
INTRODUCTION
Lynch syndrome (LS) is an autosomal dominant cancer predisposition syndrome. Those affected most commonly inherit an inactivating variant in one of the four mismatch repair system (MMR) genes: MLH1, MSH2, MSH6, or PMS2. This highly conserved system is responsible for correcting insertion and deletion errors that occur during genomic replication.1 Loss of MMR functioning, termed MMR deficiency (MMRd), leads to microsatellite instability (MSI),2 a hypermutated phenotype, and increased cancer susceptibility. LS patients are at an increased risk for a number of different malignancies, but most commonly develop colorectal and endometrial cancer.3,4 As a result, patients with LS have a decreased life expectancy compared with nonaffected individuals.5
Diagnosing LS in endometrial cancer (EC) patients is an important step in clinical management. It allows for cascade testing to diagnose family members who may also have the disease.6 Furthermore, timely LS diagnosis allows for the initiation of lifestyle modification such as weight loss, chemoprophylaxis, and cancer site surveillance to prevent the development of further LS-related malignancies, most notably colorectal cancer (CRC).4,7 Annual colonoscopy has been shown to improve overall survival in LS patients through the detection and removal of adenomatous polyps.4 There is a growing drive for universal screening of CRC patients for LS.8–10 Indeed, the National Institute of Health and Care Excellence (NICE) in the United Kingdom has recently introduced a LS screening pathway for all CRC patients, alongside numerous institutions in the United States.11 LS screening pathways utilize tumor-based testing (immunohistochemistry [IHC] for MMR protein loss, MSI testing or MLH1 promoter methylation testing) to triage cases to undergo germline testing to identify a pathogenic variant in one of the MMR genes.
Universal screening of EC patients for LS has been recommended by numerous experts and specialist societies.12 Such practice has already been adopted in several cancer centers across the world.13–15 Proponents suggest a similar proportion of EC is related to LS as seen in CRC. Furthermore, there is evidence that EC is often the sentinel cancer in women with LS.16 Therefore, a diagnosis of LS at the time of EC diagnosis could afford earlier CRC surveillance and achieve greater survival benefit. However, the true proportion of EC associated with LS remains unclear. Published proportions vary greatly, with estimates ranging from 1% to around 10%.17,18 Such variation in estimates is in part due to variable testing strategies employed across different studies.
In this systematic review we sought to provide accurate data estimating the outcomes of testing for LS in EC patients. Specifically, we asked what proportion of EC patients would be expected to be put forward for definitive germline testing following initial tumor-based tests (namely IHC, MSI with or without MLH1 promoter methylation analysis), and secondly, what proportion of these would be confirmed Lynch syndrome by next-generation sequencing (NGS). The results of this study may be of benefit in informing the planning and implementation of universal LS screening in EC patients.
MATERIALS AND METHODS
Search strategy and study identification
A systematic literature search devised by a specialist librarian, following PRISMA guidelines,19 was undertaken. Medline, Embase, Cochrane CENTRAL, and Web of Science were searched. The gray literature and nonelectronic literature were not included. Search terms were “colorectal neoplasms, hereditary nonpolyposis” and “endometrial cancer” with associated Medical Subject Headings (MeSH). In addition, a secondary search was conducted using “Lynch syndrome” as a multipurpose term and “endometrial cancer” as a MeSH term. The search included all studies from source commencement to the end of July 2018. Citation searching was utilized to augment the initial results.
Studies found to have inconsistent results were excluded after unanimous review and agreement between all authors. Assessment of bias analysis was conducted by three reviewers (N.A.J.R., D.B., and M.C.D.) independently using Review Manager (RevMan) (Version 5.3. Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2014). Disagreements were resolved by either unanimous agreement after rereview or by the decision of the senior author (E.J.C.).
Selection criteria
The protocol for this systematic review was preregistered with the PROSPERO database registration (ID: CRD42017081707) and has been published.20 Only studies investigating LS in an EC population were included. Initial searches were limited by English language, human adults (>18 years), and female subjects. Only studies that used either direct germline analysis for pathogenic variants of MMR genes or proxy tumor-based molecular diagnostic methods (IHC, MSI with or without MLH1 promoter hypermethylation), or any combination of these were included. Microsatellite instability-high (MSI-H) was defined, where possible, as involving ≥30% of the included microsatellite markers. An IHC positive result was taken as loss of expression of one of the MMR proteins. Pathogenic variants of MMR genes were defined as per the authors’ analysis. To avoid double counting data, authors of more than one study were contacted for clarification and/or registry analysis was crosschecked. Where there was overlapping data, the larger study was included and the smaller excluded. Only articles that contributed at least 15 participants were included.
Data extraction
The results from the initial search were combined. The titles and abstracts were collated in a spreadsheet template downloaded from http://libguides.sph.uth.tmc.edu/excel_workbook_home. This is available from the authors on request. Duplicates were removed with the use of Endnote X7 (Thompson Reuters, New York, NY). All titles and abstracts where initially screened independently by three authors (N.A.J.R., D.B., and M.C.D.). Conflicts were resolved by unanimous agreement between the three observers. Where unanimous agreement could not be reached a senior author (E.J.C.) made the final decision. Those studies identified as meeting the inclusion criteria underwent full article review and data extraction. Those excluded at full manuscript review are detailed in supplementary materials appendix 1.
Baseline data extracted included type of study, selection criteria, number of participants, country of origin, demographic data, type of initial screening method for LS, gold standard test, pathogenic variant distribution, and pathology distribution.
Statistical analysis
The primary outcome measure was the proportion of EC patients who were identified as being likely LS (aberrant MMR IHC expression, MSI-H with or without MLH1 promoter hypermethylation) or as carrying a germline MMR pathogenic variant. The Freeman–Tukey (a double arcsine transformation) transformed proportions of LS positive EC patients were pooled using the inverse variance heterogeneity model.21 To aid interpretation, all results were presented after back transformation to natural proportions. A quantification of heterogeneity across studies was presented as an I2 score (with I2 score of 25%, 50%, and 75% representing low, moderate, and high levels of heterogeneity respectively).22 All statistical analyses were performed R, Version 3.3.1 (https://cran.r-project.org), using the package “meta.”
RESULTS
Search results
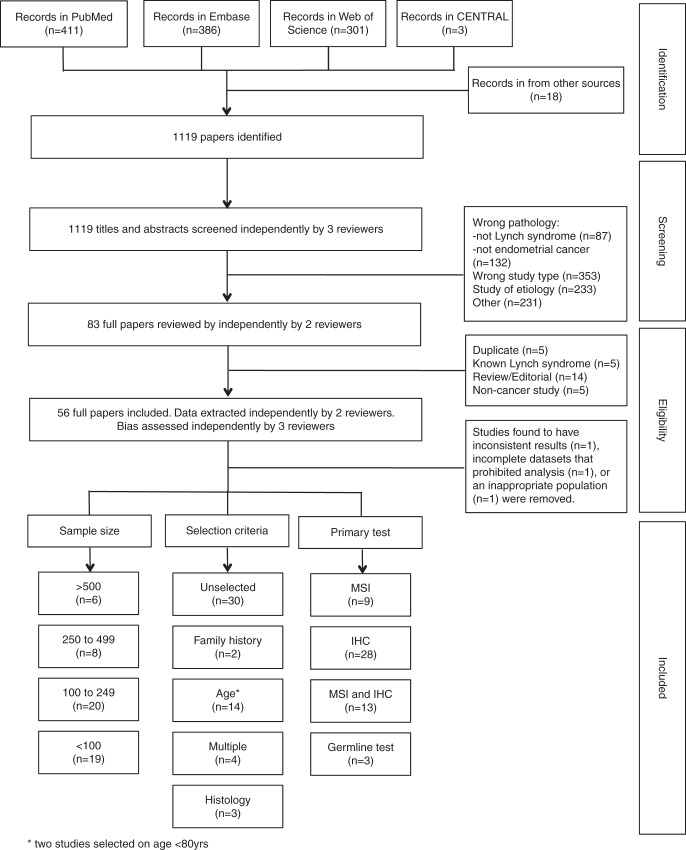
The combined search terms yielded 1119 articles. Primary review of titles and abstracts identified 83 articles that warranted full manuscript review. Of these, 56 studies met the inclusion criteria. At data extraction and quality assessment, three studies were removed due to incomplete data (n = 1) (ref. 23), inconsistent presentation of results (n = 1) (ref. 24), or an inappropriate population (n = 1) (ref. 25) (Fig. 1). Bias scores for each of the studies are outlined in supplementary appendix 2.
Fig. 1.
Flowchart detailing study identification, study selection, and characteristics of included studies. IHC immunohistochemistry, MSI microsatellite instability.
Fifty-three papers were included for the final analyses.13–15,17,18,26–73 These studies included 12,633 participants with EC. The majority of studies were conducted in North America (n = 33), with relatively small numbers carried out in Europe (n = 6), Southeast Asia (n = 7), Australasia (n = 4), and South America (n = 3). Twenty-three (43%) populations were preselected by age, family history, or other clinical parameters before analysis. Primary testing included MSI (n = 9), IHC (n = 28), MSI and IHC (n = 13), or germline testing (n = 3). Studies are summarized in Table 1.
Table 1.
Summary of included studies
| Author | Study year | Country | Selection | Initial tumor screen | Number of participants | Proportion of positive IHC | Proportion of positive MSI | Proportion of negative methylation after positive tumor triage | Proportion of positive germline samples after positive tumor-based triage | Comments |
|---|---|---|---|---|---|---|---|---|---|---|
| Backes et al.26 | 2009 | USA | None | IHC | 385 | 0.12 | NA | NA | 0.37 | IHC results not described by gene; MSI not clearly described |
| Batte et al.14 | 2014 | USA | None | IHC | 579 | 0.07 | NA | NA | 0.8 | Only 36 tumors had MSH6 IHC testing; PMS2 not tested; 20 tumors were MSI-H; 37 were MSI-L |
| Berends et al.27 | 2003 | USA | <50 years | IHC and MSI | 58 | 0.49 | 0.35 | NA | 0.09 | None |
| Bruegl et al.28 | 2014 | USA | None | IHC | 412 | 0.29 | NA | 0.11 | NA | Only one MS region investigated; only MLH1 and MSH2 IHC carried out; results quoted in the paper are not consistent |
| Buchanan et al.29 | 2013 | Australia | None | IHC | 702 | 0.24 | NA | 0.09 | 0.14 | Not clear how IHC was applied |
| Buttin et al.30 | 2004 | USA | None | MSI | 413 | NA | 0.27 | 0.19 | NA | None |
| Catasus et al.31 | 1998 | Spain | None | MSI | 42 | NA | 0.29 | NA | NA | PMS2 germline testing not performed; all MSI had GL |
| Chadwick et al.32 | 2001 | USA | None | MSI | 74 | NA | 0.23 | NA | 0.12 | None |
| Cook et al.33 | 2013 | Canada | <80 years | MSI | 480 | NA | 0.27 | NA | NA | Analysis includes 2 known LS carriers |
| Cossio et al.34 | 2010 | Brazil | <50 years of strong FHx | IHC and MSI | 30 | 0.33 | 0.23 | NA | NA | None |
| Dillon et al.13 | 2017 | USA | None | IHC | 233 | 0.26 | NA | 0.05 | 0.45 | None |
| Djordjevic et al.35 | 2013 | USA | None | IHC | 154 | 0.30 | NA | 0.14 | NA | None |
| Egoavil et al.36 | 2013 | Spain | None | IHC and MSI | 173 | 0.34 | 0.27 | 0.14 | 0.42 | 89 had unselected GL analysis; 4 GL results not available; those MSI +ve or MLH1 deficient without FHx did not have GL analysis; numbers of IHC MLH1 loss not clear |
| Ferguson et al.37 | 2014 | Canada | None | IHC and MSI | 119 | 0.29 | 0.23 | NA | 0.08 | A pre universal screening cohort, described in this paper, was not included in this analysis; those who underwent testing not as a result of universal screening were also excluded |
| Frolova et al.15 | 2015 | USA | None | IHC | 234 | 0.22 | NA | 0.09 | 0.31 | None |
| Garg et al.38 | 2009 | USA | <50 years or LS morphology | IHC | 71 | 0.45 | NA | NA | NA | PMS2 not tested |
| González et al.39 | 2012 | Puerto Rico | None | IHC | 20 | 0.25 | NA | NA | NA | None |
| Goodfellow et al.40 | 2015 | USA | Endometrioid only | IHC and MSI | 1002 | 0.36 | 0.30 | 0.11 | 0.4 | Not all variants detected could be classified as pathogenic |
| Hampel et al.41 | 2006 | USA | None | MSI | 543 | 0.34 | 0.22 | 0.06 | 0.02 | Germline pathogenic variants are not described; the PMS2 deficient case on IHC also lacked MSH6 expression |
| Hartnett et al.42 | 2015 | USA | None | IHC | 205 | 0.21 | NA | 0.03 | 0.01 | Of the 24 tumors tested 5 did not have sufficient material for any MSI; a further 4 had sufficient material for limited analysis |
| Hewitt et al.43 | 2006 | UK | FHx | MSI | 17 | 0.18 | NA | NA | None | |
| Joehlin-Price et al.44 | 2014 | USA | None | IHC | 1054 | 0.22 | NA | NA | NA | None |
| Kato et al.45 | 2016 | Japan | None | IHC | 360 | 0.03 | NA | 0.02 | 0.25 | Only PMS2 results clearly reported |
| Kost et al.66 | 2016 | USA | <50 years | IHC | 83 | 0.24 | NA | 0.16 | None | |
| Lee et al.46 | 2018 | Singapore | <50 years | IHC | 315 | 0.21 | NA | NA | NA | None |
| Leenen et al.47 | 2012 | Netherlands | <70 years | IHC and MSI | 179 | 0.23 | 0.23 | 0.06 | 0.7 | Only MLH1 and MSH2 on IHC |
| Lim et al.48 | 1996 | USA | None | MSI | 28 | NA | 0.32 | NA | NA | Two additional patients with previously known Lynch syndrome added to final results; only 3 germline results are clearly defined |
| Lin et al.49 | 2016 | USA | Mixed | IHC | 76 | 0.22 | NA | 0.08 | 0.6 | 21 of 41 showed MSH2/MSH6 and 10 of 41 showed MLH1/PMS2; however authors also report individual protein loss consisting of 72 in total |
| Long et al.66 | 2014 | China | None | IHC | 173 | 0.24 | NA | NA | NA | PMS2 not tested |
| Lu et al.50 | 2007 | USA | <50 years | GL | 100 | 0.34 | 0.33 | 0.22 | 0.09 | None |
| Chu et al.68 | 2015 | Hong Kong | <45 years | IHC | 67 | 0.33 | 0.23 | NA | None | |
| Mas-Moya et al.17 | 2015 | USA | None | IHC | 215 | 0.33 | NA | 0.17 | 0.65 | Only those with IHC loss had MSI |
| Matthews et al.51 | 2008 | USA | <50 years | IHC | 61 | 0.34 | 0.34 | NA | NA | MMR IHC done on TMA |
| McConechy et al.52 | 2015 | Canada | None | MSI | 89 | NA | 0.26 | NA | NA | MSI results are not clear |
| Mills et al.53 | 2014 | USA | None | IHC and MSI | 604 | 0.25 | NA | 0.09 | 0.81 | Three phases of testing using different tests and referral criteria; 4 had multiple gene loss on IHC; methylation testing not universally applied and therefore cannot make any sensible deductions from it |
| Moline et al.54 | 2013 | USA | Mixed | IHC or MSI or IHC with Methylation | 245 | 0.25 | 0.13 | NA | 0.24 | None |
| Najdawi et al.69 | 2017 | Australia | None | IHC | 124 | 0.24 | NA | NA | 0.33 | Only 9 of 11 of the Lynch-like tumors had germline testing |
| Parc et al.55 | 2000 | USA | <52 years | IHC and MSI | 62 | 0.24 | 0.34 | NA | NA | MSH2 and MSH6 IHC outcomes not reported |
| Pecorino et al.56 | 2017 | Italy | <50 years | IHC | 41 | 0.31 | 0.42 | NA | NA | None |
| Pennington et al.70 | 2013 | USA | Serous only | GL | 151 | NA | NA | NA | 0.00 | One subject with < 10% serous histology had a MSH6 pathogenic variant |
| Rabban et al.18 | 2014 | USA | >50 years | IHC | 273 | 0.15 | NA | 0.04 | 0.29 | None |
| Resnick et al.57 | 2009 | USA | None | IHC | 477 | 0.28 | NA | NA | NA | None |
| Riggi et al.58 | 2016 | Argentina | None | IHC | 84 | 0.33 | NA | NA | NA | None |
| Ring et al.71 | 2013 | USA | <50 years | IHC | 111 | 0.26 | NA | NA | NA | None |
| Ring et al.59 | 2016 | USA | None | IHC or MSI or IHC with methylation | 381 | NK | NK | NK | 0.06 | Tumor-based molecular triage was used but the results are not detailed |
| Rubio et al.60 | 2016 | Spain | None | IHC and MSI | 94 | 0.33 | 0.27 | NA | 0.15 | GL result not clear |
| Sugawara et al.61 | 2015 | Japan | AFP criteria | IHC | 182 | 0.30 | NA | 0.22 | NA | Of all samples 55% not tested |
| Tan et al.65 | 2013 | Australia | <80 years | IHC | 246 | 0.24 | NA | NA | NA | None |
| Walsh et al.62 | 2008 | Australia | <50 years | IHC and MSI | 146 | 0.26 | 0.35 | 0.18 | NA | None |
| Watkins et al.73 | 2016 | USA | None | IHC | 242 | 0.20 | NA | 0.05 | 0.4 | None |
| Woo et al.72 | 2014 | Malaya | Endometrioid only | IHC | 77 | 0.19 | NA | NA | NA | PMS2 not tested |
| Yoon et al.63 | 2007 | Korea | None | IHC and MSI | 113 | 0.23 | 0.44 | 0.17 | 0.04 | None |
| Zauber et al.64 | 2010 | USA | None | MSI | 213 | NA | 0.26 | 0.09 | NA | None |
FHx family history, GL germline, IHC immunohistochemistry, LS Lynch syndrome, MSI microsatellite instability, MSI-H microsatellite instability high, MSI-L microsatellite instability low. TMA tissue microarray, NA not applicable, NK not known, AFP age, family and personal history of cancer criteria.
All studies originated from specialist tertiary referral centers or their biobanks. Histological features were reported in 20 papers. Type 1/endometrioid tumors constituted 79.3% of tumors, consistent with the literature.74 There were insufficient data to describe the histological breakdown of tumors diagnosed in women who were found to have abnormal tumor triage or LS. From the studies that included age data (n = 43), the median age of subjects tested was 59.5 years (interquartile range [IQR:] 53–62).
Immunohistochemical analysis
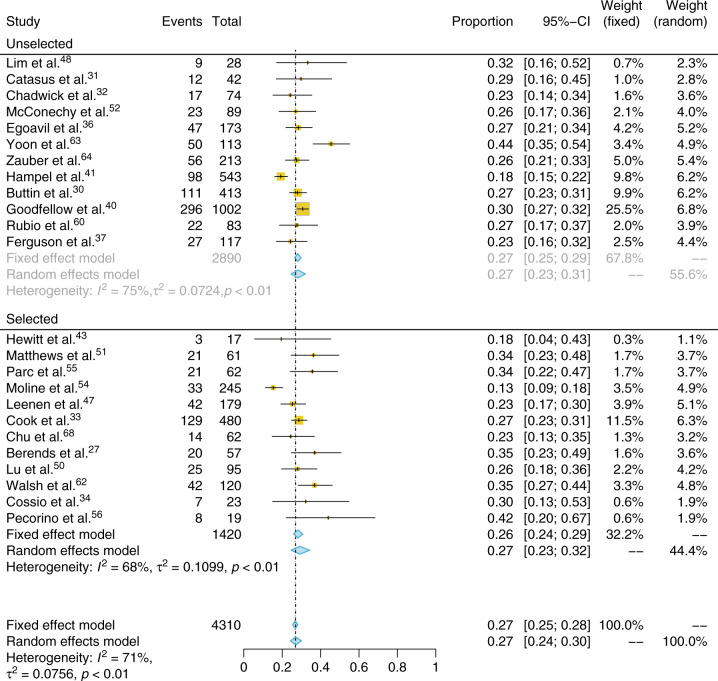
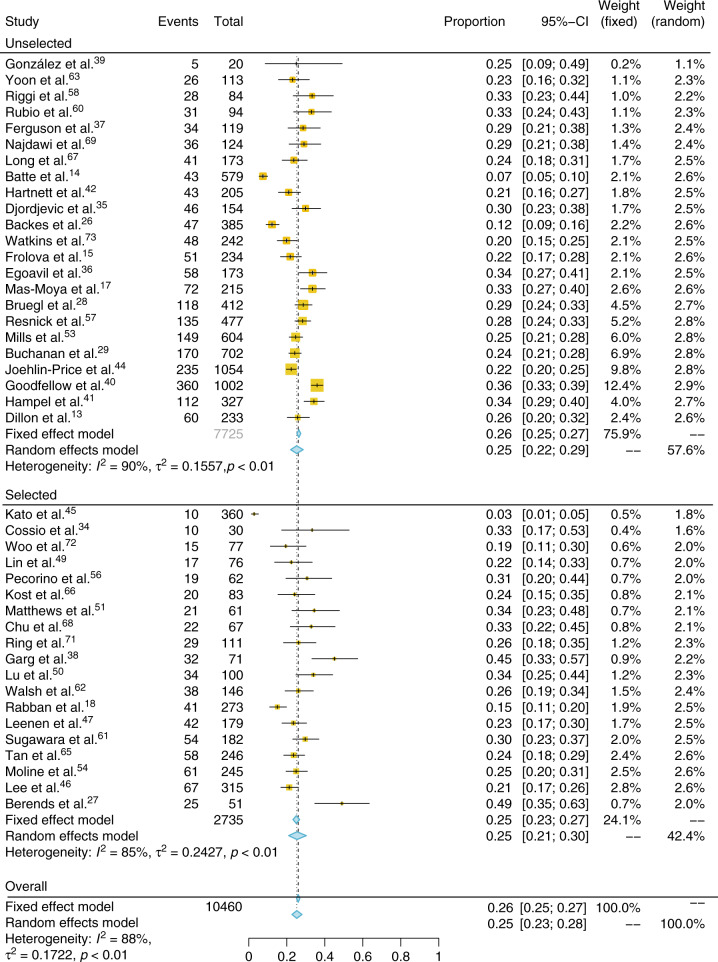
In total, 42 papers reported the outcome of IHC analysis. These studies include 10,683 participants, 10,460 with completed IHC analysis. Of these, 2563 (25%) were found to have absent expression of at least one of the MMR proteins on IHC. This represents an overall proportion of 0.26 (confidence interval [CI] 95% 0.25–0.27, I2: 88%). When preselected populations (cohorts restricted by clinical criteria such as age at EC diagnosis) were excluded, a total of 7725 tumors underwent MMR IHC, of which 1948 (25%) were MMR deficient (Fig. 3). Therefore, the proportion of unselected ECs with MMR deficiency is 0.26 (95% CI 0.25–0.27, I2: 90%) and 0.25 (95% CI 0.23–0.27, I2: 85%) in selected EC populations (Fig. 2).
Fig. 3.
Forest plot and meta-analysis of the proportion of endometrial tumors showing microsatellite instability, including those that did and did not preselect tumors for testing. CI confidence interval.
Fig. 2.
Forest plot and meta-analysis of the proportion of endometrial tumors with mismatch repair deficiency by immunohistochemistry, including those that did and did not preselect tumors for testing. CI confidence interval.
Of note, only a proportion of studies tested for loss in all four of the MMR proteins (23/42, 55%). MLH1-specific outcome data were available for a total of 9306 study participants. Of these, 1635 (18%) were found to be MLH1 protein deficient. When studies that preselected their populations were excluded, the total number of subjects was 7176, of whom 1343 (19%) were MLH1 deficient. Full protein specific data were available for 7385 subjects. Excluding MLH1, deficiencies in MSH6 were the most commonly recorded (n = 247/3%), followed by MSH2 (n = 211/3%) and finally PMS2 (n = 153/2%). As MMR proteins are dimeric, concurrent loss of both proteins within MutSα (MSH2/MSH6) and MutLα (MLH1/PMS2) would be expected. For MSH2, concurrent MSH6 loss was always seen across all studies. For MLH1, PMS2 loss was also seen, with the exception of one case. By contrast, isolated MSH6 was seen in 112 cases and isolated PMS2 loss in 13 cases. In studies with no preselection and with complete protein analysis data there were a total of 6104 study participants. Here the most prevalent protein deficiency was in MSH6 (n = 165/3%) followed by MSH2 (n = 153/3%) and PMS2 (n = 84/1%).
Of 5594 EC with MLH1 IHC and methylation analysis data, 1098 (20%) cases had MLH1 IHC loss. Of these, 963 (88%) were found to be due to somatic hypermethylation of the MLH1 promoter region. In unselected EC populations with methylation analysis data, there were 4525 participants, the tumors of whom showed MLH1 IHC loss in 960 (21%) cases. Eight hundred forty-three (88%) were as a result of somatic hypermethylation of the MLH1 promoter region. Therefore the proportion of MLH1 IHC loss not attributed to somatic hypermethylation of MLH1 was 0.11 (95% CI 0.10–0.13, I2: 78%) (supplementary appendix 4). From these data, there is no significant difference between the proportion of ECs with IHC MLH1 loss and normal methylation analysis according to cohort selection.
Microsatellite instability analysis
MSI data were available for 4310 tumors, 2580 of which were also tested by IHC. Of these, 1133 (26.3%) were MSI-H. When studies that used preselective criteria were excluded, the total number of participants was 2890, of whom 768 were positive (26.6%) The overall proportion of MSI-H ECs was 0.27 (CI 95% 0.25–0.28, I2: 71%). This was 0.27 (CI 95% 0.25–0.29, I2: 75%) and 0.26 (CI 95% 0.24–0.29, I2: 68%) for unselected and preselected populations, respectively (Fig. 3). The results of MLH1 methylation analysis in conjunction with MSI testing are shown in supplementary appendix 4.
There was no significant difference between the proportion of positive test results if IHC or MSI was used as the initial tumor triage (0.25 vs. 0.27 p value = 0.5 [Student's t test]). Analysis of MSI proportions pre-2011 vs. post-2011 did not find a significant difference (t test p value = 0.11).
Germline analysis
In total, 23 studies used some form of germline analysis to establish a diagnosis of LS. There was a wide degree of variation in the completeness of germline testing, with few studies (n = 9) testing all those they intended to. Furthermore, 12 studies that used a tumor-based triage test did not perform MLH1 promoter methylation testing. Two studies failed to report the outcome of their MLH1 methylation testing.14,54 The majority of these studies used another means of excluding a proportion of their MLH1 IHC positive results, for example a negative family history. Therefore, the population that went on to have definitive germline analysis is heterogeneous.
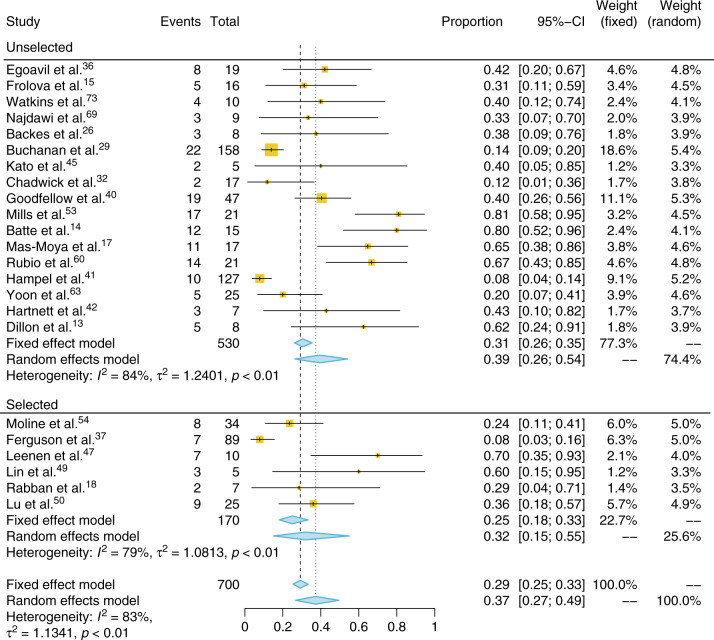
In total, 14,770 tumors underwent tumor-based triage with IHC (n = 10,460) and/or MSI (n = 4310). Concurrent testing with both IHC and MSI was sufficiently reported in ten studies16,27,34,36,37,40,47,51,63,68 and enabled removal of duplicates with positive concordant IHC and MSI data. Of the remaining 14,293 tumors, 1133 were MSI-H and 2563 had aberrant IHC. Studies that reported MLH1-specific IHC and MLH1 promoter methylation tumor outcomes (n = 5594) allowed further triage by removing tumors with likely somatic MLH1 loss. In total, 1005 women were eligible for and 700 women underwent germline testing for Lynch syndrome following tumor-based triage. A total of 181 (26%) were positive. When studies that preselected their population were excluded, the combined population was 5882, of whom, upon removal of methylated results, 821 (14%) were Lynch-like on the basis of their tumor analyses and therefore should have undergone germline testing. A total of 511 (62%) underwent testing and 150 (29%) were positive. This represents an overall proportion of LS-related pathogenic variants in those with positive tumor triage of 0.29 (CI 95% 0.25–0.33, I2: 83%). The same proportion in unselected cases was 0.31 (CI 95% 0.26–0.35, I2: 84%) and for selected 0.25 (CI 95% 0.18–0.33, I2: 79%) (Fig. 4). Therefore, around 3% of ECs can be attributed to LS. The gene breakdown from NGS is shown in supplementary appendix 3.
Fig. 4.
Forest plot and meta-analysis of the proportion of pathogenic variants in mismatch repairs (MMR) genes found in women with endometrial tumors showing mismatch repair deficiency and/or microsatellite instability, including studies that did and did not preselect women for testing. CI confidence interval.
Four studies, which examined unselected populations of EC, had complete germline testing of cases suggestive of LS on the basis of their tumor analysis. Focusing on these studies, 180 tumors were suggestive of LS of which 32 were found to have a pathological variant in one of the MMR genes. This represents a proportion of 0.21 (CI 95% 0.15–0.30, I2: 91%). This is significantly fewer than reported in studies with incomplete germline testing (p = 0.001) and suggests an overreporting of pathological variants in studies that fail to analyze all samples suggestive of LS from tumor triage. Only three studies did direct germline analysis without tumor triage. These studies were all highly selective, with Pennington and colleagues testing only tumors of a serous histology, which is not commonly associated with LS.75 Of 590 cancers tested in these three studies, 27 (4.6%) were found to have a germline pathological variant. If the data from Pennington are removed, 6% of ECs were found to have pathogenic variants associated with LS. These data are shown in supplementary appendix 5. Subgroup analysis of pre-2011 vs. post-2011 proportions of germline pathogenic variants found after tumor triage was not significant (t test p value = 0.14).
Further subgroup analysis was carried out to explore the relationship between potential confounding factors; these are shown in supplementary appendix 5. Of note, those studies that did not use a priori tumor-based triage, using instead direct germline sequencing of all samples, found a higher proportion of LS carriers (0.06); however, the number of studies is limited (n = 3). In addition, limiting testing to individuals less than 50 years yielded higher levels of MSI (0.31) and aberrant IHC (0.28).
DISCUSSION
Here we present a systematic review and meta-analysis to define the proportion of EC patients who test positive for LS. This work includes data from 53 studies and 12,633 participants with EC who underwent IHC, MSI, methylation, or germline analysis to diagnose LS. From these data, of 100 unselected cases of EC, approximately 3 people are estimated to have LS, consistent with current literature.76,77 While this is a modest percentage of positive tests, each diagnosis allows for cascade testing of family members. It is estimated that 80% of living first-degree relatives will accept LS screening.78 Newly diagnosed LS patients are able to benefit from interventions to prevent the development of other LS cancers, most notably CRC. The results of this meta-analysis are summarized in supplementary figure S12 showing the estimated outcome from each stage of the LS diagnostic pathway.
Our results do not support the use of a particular tumor triage method. The proportion of positive test results if IHC or MSI was used as the initial tumor test (0.26 vs. 0.27 p value = 0.5 [Student's t test]) was similar. This small difference could be explained by the commonly used Bethesda panel for the detection of MSI, which has only been validated in CRC and not EC.79 In addition, MSH6 deficient EC can be microsatellite low or stable.80 That said, IHC does enable a more targeted application of MLH1 promoter hypermethylation testing, given that it need only be applied to MLH1 deficient tumors. In addition, germline analysis could be limited to the gene(s) that corresponds to the protein lost; this has potential cost saving implications.
Preselecting EC populations by age or clinical criteria did not significantly change the proportion of positive IHC or MSI results, although we did find higher proportions in these subgroups. This is somewhat surprising, as preselected populations would be expected to harbor more women with LS. This may be partly explained by the later age of onset seen with MSH6 pathological variants and truncating variants.81 In other words, the application of age cutoffs reduces specificity without a significant increase in sensitivity. However, universal testing does seem to decrease the yield of pathogenic variant carriers; this may arise from somatic events that lead to false positives at the tumor triage stage. MLH1 methylation is associated with increased age and so is more common in older (unselected) populations.82 Subgroup analysis of studies that did not use a tumor triage stage, instead using direct germline sequencing, found a higher proportion of LS carriers (0.06). This could suggest that tumor triage itself misses potentially 50% of LS carriers. However, there were only three studies in the subgroup analysis. In addition, one of the studies preselected those who had germline testing on the basis of age (<50 years). However, this finding should encourage debate as to the application of NGS without tumor-based triage in EC populations; even more so given the decreasing cost of this technology.
Our work has several key strengths. Our conclusions are based on the results of over 50 studies and 12,600 participants; the search criteria were purposely broad as to capture the maximum number of studies. During the screening phase, three independent reviewers ensured the accuracy of study selection and data capture. Therefore, the foundations of our meta-analysis were robust. In addition, we have estimated the proportions of positive results seen in IHC, MSI, MLH1 methylation, and germline testing with a high degree of precision, as reflected in the narrow 95% confidence intervals in our meta-analysis.
The heterogeneity across the studies included in our review was high, and limits the strength of our conclusions. This is a reflection of the varying quality and rigor of the included studies, some of which had small numbers of participants, and were subject to bias. The majority of studies used retrospective cohorts. Furthermore, many studies failed to complete the indicated testing in their cohorts, leading to ambiguity in their conclusions. This is evidenced by the lower proportion of LS pathological variants in those studies with complete germline analysis of Lynch-like tumors versus those with partial germline analysis (0.21 vs. 0.29). To allow for the pooling of such heterogeneous data, studies were grouped according to selection and diagnostic method. All studies that were grouped reported the same endpoint.
Another potential weakness of this study is the evolution in diagnostic technology over time. The included studies were published between 1996 and 2017. During this time diagnostic technology and guidelines have evolved significantly. Although IHC based diagnostics has remained relatively constant, MSI diagnostics has been informed by the adoption of the Bethesda panel in 1998 and the development of the more modern panels such as the pentaplex and hexaplex panels, which became widely applied to clinical practice after 2011.83 Even so, analysis of MSI testing results pre- vs. post-2011 did not find a significant difference (t test p value = 0.11). Only one study predates the Bethesda guidelines. The area of germline diagnostics remains innovative, but again analysis of pre- vs. post-2011 proportions of germline pathogenic variants found after tumor triage was not significant (t test p value = 0.14). Generalizability is limited by the predominance of North American and European populations in our study. Most took place in insurance-based health-care systems, which impact negatively on the uptake of genetic testing.84 Therefore, it could be that the proportions are an underestimate due to reduced uptake of testing, especially in high-risk groups such as the young or those with a strong family history.
In summary, ours is the first meta-analysis to examine the proportion of EC cases that are associated with LS. Different tumor triage methods did not affect estimates of the proportion of EC associated with LS, which remained constant at around 3%. Our findings suggest that a similar proportion of EC patients will test positive for LS as seen in CRC LS screening. This supports the move toward the introduction of universal screening for LS in EC.
Supplementary information
Acknowledgements
N.A.J.R. was supported by a Medical Research Council (MRC) Doctoral Research Fellowship (MR/M018431/1). M.A.G. was supported by a Wellcome Trust Clinical Training Fellowship. E.J.C. was funded by a National Institute for Health Research (NIHR) Clinician Scientist Fellowship (NIHR-CS-012–009), and D.G.E. is an NIHR Senior Investigator (NF-SI-0513–10076). This article presents independent research funded by the MRC, the Wellcome Trust, and the NIHR, and supported by the NIHR Manchester Biomedical Research Centre (IS-BRC-1215–20007). The views expressed are those of the authors and not necessarily those of the funders or the Department of Health.
Disclosure
The authors declare no conflicts of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41436-019-0536-8) contains supplementary material, which is available to authorized users.
References
- 1.Lynch HT, Snyder CL, Shaw TG, Heinen CD, Hitchins MP. Milestones of Lynch syndrome: 1895-2015. Nat Rev Cancer. 2015;15:181–194. doi: 10.1038/nrc3878. [DOI] [PubMed] [Google Scholar]
- 2.Ahadova A, Gallon R, Gebert J, et al. Three molecular pathways model colorectal carcinogenesis in Lynch syndrome. Int J Cancer. 2018;138:2044. doi: 10.1002/ijc.31300. [DOI] [PubMed] [Google Scholar]
- 3.Barrow E, Robinson L, Alduaij W, et al. Cumulative lifetime incidence of extracolonic cancers in Lynch syndrome: a report of 121 families with proven mutations. Clin Genet. 2009;75:141–149. doi: 10.1111/j.1399-0004.2008.01125.x. [DOI] [PubMed] [Google Scholar]
- 4.Moller P, Seppälä T, Bernstein I, et al. Cancer incidence and survival in Lynch syndrome patients receiving colonoscopic and gynaecological surveillance: first report from the prospective Lynch syndrome database. Gut. 2017;66:464–472. doi: 10.1136/gutjnl-2015-309675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans DGR, Ingham SL. Reduced life expectancy seen in hereditary diseases which predispose to early-onset tumors. Appl Clin Genet. 2013;6:53–61. doi: 10.2147/TACG.S35605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hampel H. Genetic counseling and cascade genetic testing in Lynch syndrome. Fam Cancer. 2016;15:423–427. doi: 10.1007/s10689-016-9893-5. [DOI] [PubMed] [Google Scholar]
- 7.Movahedi M, Bishop DT, Macrae F, et al. Obesity, aspirin, and risk of colorectal cancer in carriers of hereditary colorectal cancer: a prospective investigation in the CAPP2 Study. J Clin Oncol. 2015;33:3591–3597. doi: 10.1200/JCO.2014.58.9952. [DOI] [PubMed] [Google Scholar]
- 8.Vasen HFA, Blanco I, Aktan-Collan K, et al. Revised guidelines for the clinical management of Lynch syndrome (HNPCC): recommendations by a group of European experts. Gut. 2013;62:812–823. doi: 10.1136/gutjnl-2012-304356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Provenzale D, Gupta S, Ahnen DJ, et al. Genetic/familial high-risk assessment: colorectal version 1.2016, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2016;14:1010–1030. doi: 10.6004/jnccn.2016.0108. [DOI] [PubMed] [Google Scholar]
- 10.Rubenstein JH, Enns R, Heidelbaugh J, et al. American Gastroenterological Association Institute guideline on the diagnosis and management of Lynch syndrome. Gastroenterology. 2015;149:777–782. doi: 10.1053/j.gastro.2015.07.036. [DOI] [PubMed] [Google Scholar]
- 11.The National Institute for Health and Care Excellence. Molecular testing strategies for Lynch syndrome in people with colorectal cancer. Diagnostic guideline. 2017;27:1–37. https://www.nice.org.uk/guidance/dg27. Accessed 10 May 2019.
- 12.Crosbie EJ, Ryan NAJ, Arends M, et al. (2019). The Manchester International Consensus Group Recommendations for the Management of Gynecological Cancers in Lynch Syndrome. Genet Med. 2019. 10.1038/s41436-019-0489-y. Accessed 10 May 2019. [DOI] [PMC free article] [PubMed]
- 13.Dillon JL, Gonzalez JL, DeMars L, Bloch KJ, Tafe LJ. Universal screening for Lynch syndrome in endometrial cancers: frequency of germline mutations and identification of patients with Lynch-like syndrome. Hum Pathol. 2017;70:121–128. doi: 10.1016/j.humpath.2017.10.022. [DOI] [PubMed] [Google Scholar]
- 14.Batte BAL, Bruegl AS, Daniels MS, et al. Consequences of universal MSI/IHC in screening endometrial cancer patients for Lynch syndrome. Gynecol Oncol. 2014;134:319–325. doi: 10.1016/j.ygyno.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frolova AI, Babb SA, Zantow E, et al. Impact of an immunohistochemistry-based universal screening protocol for Lynch syndrome in endometrial cancer on genetic counseling and testing. Gynecol Oncol. 2015;137:7–13. doi: 10.1016/j.ygyno.2015.01.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu KH, Dinh M, Kohlmann W, et al. Gynecologic cancer as a “sentinel cancer” for women with hereditary nonpolyposis colorectal cancer syndrome. Obstet Gynecol. 2005;105:569–574. doi: 10.1097/01.AOG.0000154885.44002.ae. [DOI] [PubMed] [Google Scholar]
- 17.Mas-Moya J, Dudley B, Brand RE, et al. Clinicopathological comparison of colorectal and endometrial carcinomas in patients with Lynch-like syndrome versus patients with Lynch syndrome. Hum Pathol. 2015;46:1616–1625. doi: 10.1016/j.humpath.2015.06.022. [DOI] [PubMed] [Google Scholar]
- 18.Rabban JT, Calkins SM, Karnezis AN, et al. Association of tumor morphology with mismatch-repair protein status in older endometrial cancer patients: implications for universal versus selective screening strategies for Lynch syndrome. Am J Surg Pathol. 2014;38:793–800. doi: 10.1097/PAS.0000000000000177. [DOI] [PubMed] [Google Scholar]
- 19.Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151:264–9–W64. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 20.Ryan NAJ, Blake D, Cabrera-Dandy M, Glaire MA, Evans DG, Crosbie EJ. The prevalence of Lynch syndrome in women with endometrial cancer: a systematic review protocol. Syst Rev. 2018;7:121. doi: 10.1186/s13643-018-0792-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doi SAR, Barendregt JJ, Khan S, Thalib L, Williams GM. Advances in the meta-analysis of heterogeneous clinical trials I: the inverse variance heterogeneity model. Contemp Clin Trials. 2015;45(Pt A):130–138. doi: 10.1016/j.cct.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 23.Nugent W, Durie N, Palaia T, Ragolia L, Hanna I, Staszewski H. Retrospective review of endometrial cancer (EC) specimens in individuals younger than age 50 for microsatellite instability (MSI) testing and DNA mismatch repair enzyme (MMR) expression. J Clin Oncol. 2011;29(:15 suppl):e15565. [Google Scholar]
- 24.Buchynska LG, Brieieva OV, Nekrasov KA, Nespryadko SV. The study of mismatch repair in endometrial cancer patients with a family history of cancer. Exp Oncol. 2015;37:272–276. [PubMed] [Google Scholar]
- 25.Thodi G, Fostira F, Sandaltzopoulos R, et al. Screening of the DNA mismatch repair genes MLH1, MSH2 and MSH6 in a Greek cohort of Lynch syndrome suspected families. BMC Cancer. 2010;10:544. doi: 10.1186/1471-2407-10-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Backes FJ, Leon ME, Ivanov I, et al. Prospective evaluation of DNA mismatch repair protein expression in primary endometrial cancer. Gynecol Oncol. 2009;114:486–490. doi: 10.1016/j.ygyno.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 27.Berends MJW, Wu Y, Sijmons RH, et al. Toward new strategies to select young endometrial cancer patients for mismatch repair gene mutation analysis. J Clin Oncol. 2003;21:4364–4370. doi: 10.1200/JCO.2003.04.094. [DOI] [PubMed] [Google Scholar]
- 28.Bruegl AS, Djordjevic B, Batte B, et al. Evaluation of clinical criteria for the identification of Lynch syndrome among unselected patients with endometrial cancer. Cancer Prev Res (Phila) 2014;7:686–697. doi: 10.1158/1940-6207.CAPR-13-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchanan DD, Tan YY, Walsh MD, et al. Tumor mismatch repair immunohistochemistry and DNA MLH1 methylation testing of patients with endometrial cancer diagnosed at age younger than 60 years optimizes triage for population-level germline mismatch repair gene mutation testing. J Clin Oncol. 2013;32:2129.. doi: 10.1200/JCO.2013.51.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buttin BM, Powell MA, Mutch DG, et al. Increased risk for hereditary nonpolyposis colorectal cancer-associated synchronous and metachronous malignancies in patients with microsatellite instability-positive endometrial carcinoma lacking MLH1 promoter methylation. Clin Cancer Res. 2004;10:481–490. doi: 10.1158/1078-0432.ccr-1110-03. [DOI] [PubMed] [Google Scholar]
- 31.Catasus L, Machin P, Matias-Guiu X, Prat J. Microsatellite instability in endometrial carcinomas: clinicopathologic correlations in a series of 42 cases. Hum Pathol. 1998;29:1160–1164. doi: 10.1016/s0046-8177(98)90430-0. [DOI] [PubMed] [Google Scholar]
- 32.Chadwick RB, Pyatt RE, Niemann TH, et al. Hereditary and somatic DNA mismatch repair gene mutations in sporadic endometrial carcinoma. J Med Genet. 2001;38:461–466. doi: 10.1136/jmg.38.7.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cook LS, Nelson HE, Stidley CA, et al. Endometrial cancer and a family history of cancer. Gynecol Oncol. 2013;130:334–339. doi: 10.1016/j.ygyno.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cossio SL, Koehler-Santos P, Pessini SA, et al. Clinical and histomolecular endometrial tumor characterization of patients at-risk for Lynch syndrome in South of Brazil. Fam Cancer. 2010;9:131–139. doi: 10.1007/s10689-009-9297-x. [DOI] [PubMed] [Google Scholar]
- 35.Djordjevic B, Barkoh BA, Luthra R, Broaddus RR. Relationship between PTEN, DNA mismatch repair, and tumor histotype in endometrial carcinoma: retained positive expression of PTEN preferentially identifies sporadic non-endometrioid carcinomas. Mod Pathol. 2013;26:1401–1412. doi: 10.1038/modpathol.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Egoavil C, Alenda C, Castillejo A, et al. Prevalence of Lynch syndrome among patients with newly diagnosed endometrial cancers. PLoS ONE. 2013;8:e79737.. doi: 10.1371/journal.pone.0079737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferguson SE, Aronson M, Pollett A, et al. Performance characteristics of screening strategies for Lynch syndrome in unselected women with newly diagnosed endometrial cancer who have undergone universal germline mutation testing. Cancer. 2014;120:3932–3939. doi: 10.1002/cncr.28933. [DOI] [PubMed] [Google Scholar]
- 38.Garg K, Soslow RA. Lynch syndrome (hereditary non-polyposis colorectal cancer) and endometrial carcinoma. J Clin Pathol. 2009;62:679–684. doi: 10.1136/jcp.2009.064949. [DOI] [PubMed] [Google Scholar]
- 39.González L, Ortiz AP, Suárez EL, et al. Case-case study of factors associated to hMLH1, hMSH2, and hMSH6 protein expression among endometrial cancer patients of the University District Hospital of San Juan, Puerto Rico. Int J Gynecol Cancer. 2012;22:826–829. doi: 10.1097/IGC.0b013e31825104de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodfellow PJ, Billingsley CC, Lankes HA, et al. Combined microsatellite instability, MLH1 methylation analysis, and immunohistochemistry for Lynch syndrome screening in endometrial cancers from GOG210: an NRG Oncology and Gynecologic Oncology Group Study. J Clin Oncol. 2015;33:4301–4308. doi: 10.1200/JCO.2015.63.9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hampel H, Frankel W, Panescu J, et al. Screening for Lynch syndrome (hereditary nonpolyposis colorectal cancer) among endometrial cancer patients. Cancer Res. 2006;66:7810–7817. doi: 10.1158/0008-5472.CAN-06-1114. [DOI] [PubMed] [Google Scholar]
- 42.Hartnett E, Stuckey A, Danilack V, McCourt C. Evaluation of universal immunohistochemistry screening for diagnosing Lynch syndrome in endometrial cancer patients at a tertiary care center. Gynecol Oncol. 2015;139:599. [Google Scholar]
- 43.Hewitt MJ, Wood N, Quinton ND, et al. The detection of microsatellite instability in blind endometrial samples—a potential novel screening tool for endometrial cancer in women from hereditary nonpolyposis colorectal cancer families? Int J Gynecol Cancer. 2006;16:1393–1400. doi: 10.1111/j.1525-1438.2006.00604.x. [DOI] [PubMed] [Google Scholar]
- 44.Joehlin-Price AS, Perrino CM, Stephens J, et al. Mismatch repair protein expression in 1049 endometrial carcinomas, associations with body mass index, and other clinicopathologic variables. Gynecol Oncol. 2014;133:43–47. doi: 10.1016/j.ygyno.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kato A, Sato N, Sugawara T, et al. Isolated loss of PMS2 immunohistochemical expression is frequently caused by heterogenous MLH1 promoter hypermethylation in Lynch syndrome screening for endometrial cancer patients. Am J Surg Pathol. 2016;40:770–776. doi: 10.1097/PAS.0000000000000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JCS, Andres J, Durai S, et al. Incidence of DNA mismatch repair (MMR) protein expression in young Asian women with endometrial cancer. 2018. https://epostersonline.com/rcog2018/node/2802. Accessed April 2018.
- 47.Leenen CHM, van Lier MGF, van Doorn HC, et al. Prospective evaluation of molecular screening for Lynch syndrome in patients with endometrial cancer ≤70 years. Gynecol Oncol. 2012;125:414–420. doi: 10.1016/j.ygyno.2012.01.049. [DOI] [PubMed] [Google Scholar]
- 48.Lim PC, Tester D, Cliby W, et al. Absence of mutations in DNA mismatch repair genes in sporadic endometrial tumors with microsatellite instability. Clin Cancer Res. 1996;2:1907–1911. [PubMed] [Google Scholar]
- 49.Lin DI, Hecht JL. Targeted screening with combined age- and morphology-based criteria enriches detection of Lynch syndrome in endometrial cancer. Int J Surg Pathol. 2016;24:297–305. doi: 10.1177/1066896916629782. [DOI] [PubMed] [Google Scholar]
- 50.Lu KH, Schorge JO, Rodabaugh KJ, et al. Prospective determination of prevalence of Lynch syndrome in young women with endometrial cancer. J Clin Oncol. 2007;25:5158–5164. doi: 10.1200/JCO.2007.10.8597. [DOI] [PubMed] [Google Scholar]
- 51.Matthews KS, Estes JM, Conner MG, et al. Lynch syndrome in women less than 50 years of age with endometrial cancer. Obstet Gynecol. 2008;111:1161–1166. doi: 10.1097/AOG.0b013e31817051d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McConechy MK, Talhouk A, Li-Chang HH, et al. Detection of DNA mismatch repair (MMR) deficiencies by immunohistochemistry can effectively diagnose the microsatellite instability (MSI) phenotype in endometrial carcinomas. Gynecol Oncol. 2015;137:306–310. doi: 10.1016/j.ygyno.2015.01.541. [DOI] [PubMed] [Google Scholar]
- 53.Mills AM, Liou S, Ford JM, Berek JS, Pai RK, Longacre TA. Lynch syndrome screening should be considered for all patients with newly diagnosed endometrial cancer. Am J Surgical Pathol. 2014;38:1501–1509. doi: 10.1097/PAS.0000000000000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moline J, Mahdi H, Yang B, et al. Implementation of tumor testing for Lynch syndrome in endometrial cancers at a large academic medical center. Gynecol Oncol. 2013;130:121–126. doi: 10.1016/j.ygyno.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 55.Parc YR, Halling KC, Burgart LJ, et al. Microsatellite instability and hMLH1/hMSH2 expression in young endometrial carcinoma patients: associations with family history and histopathology. Int J Cancer. 2000;86:60–66. doi: 10.1002/(sici)1097-0215(20000401)86:1<60::aid-ijc9>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 56.Pecorino B, Rubino C, Guardalà VF, Galia A, Scollo P. Genetic screening in young women diagnosed with endometrial cancer. J Gynecol Oncol. 2017;28:e4. 10.3802/jgo.2017.28.e4. Accessed 10 May 2019. [DOI] [PMC free article] [PubMed]
- 57.Resnick KE, Hampel H, Fishel R, Cohn DE. Current and emerging trends in Lynch syndrome identification in women with endometrial cancer. Gynecol Oncol. 2009;114:128–134. doi: 10.1016/j.ygyno.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riggi MC, Wernicke A, Salvo G, et al. Endometrial cancer and Lynch syndrome: immunohistochemical characterization of endometrial cancer associated with changes in mismatch repair protein expression. Gynecol Oncol. 2016;141:155–159. [Google Scholar]
- 59.Ring KL, Bruegl AS, Allen BA, et al. Germline multi-gene hereditary cancer panel testing in an unselected endometrial cancer cohort. Mod Pathol. 2016;29:1381–1389. doi: 10.1038/modpathol.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rubio I, Ibáñez-Feijoo E, Andrés L, et al. Analysis of Lynch syndrome mismatch repair genes in women with endometrial cancer. Oncology. 2016;91:171–176. doi: 10.1159/000447972. [DOI] [PubMed] [Google Scholar]
- 61.Sugawara T, Sato N, Shimizu D, et al. Efficient screening strategy for Lynch syndrome in Japanese endometrial cancer. Tohoku J Exp Med. 2015;235:117–125. doi: 10.1620/tjem.235.117. [DOI] [PubMed] [Google Scholar]
- 62.Walsh MD, Cummings MC, Buchanan DD, et al. Molecular, pathologic, and clinical features of early-onset endometrial cancer: identifying presumptive Lynch syndrome patients. Clin Cancer Res. 2008;14:1692–1700. doi: 10.1158/1078-0432.CCR-07-1849. [DOI] [PubMed] [Google Scholar]
- 63.Yoon SN, Ku JL, Shin YK, et al. Hereditary nonpolyposis colorectal cancer in endometrial cancer patients. Int J Cancer. 2007;122:1077–1081. doi: 10.1002/ijc.22986. [DOI] [PubMed] [Google Scholar]
- 64.Zauber NP, Denehy TR, Taylor RR, et al. Microsatellite instability and DNA methylation of endometrial tumors and clinical features in young women compared with older women. Int J Gynecol Cancer. 2010;20:1549–1556. [PubMed] [Google Scholar]
- 65.Tan YY, McGaughran J, Ferguson K, et al. Improving identification of Lynch syndrome patients: a comparison of research data with clinical records. Int J Cancer. 2013;132:2876–2883. doi: 10.1002/ijc.27978. [DOI] [PubMed] [Google Scholar]
- 66.Kost ER, Valente PT, Lynch BA, et al. Clinical and pathologic features of Hispanic endometrial cancer patients with loss of mismatch repair expression. Int J Gynecol Cancer. 2016;26:1129–1136. doi: 10.1097/IGC.0000000000000713. [DOI] [PubMed] [Google Scholar]
- 67.Long Q, Peng Y, Tang Z, Wu C. Role of endometrial cancer abnormal MMR protein in screening Lynch-syndrome families. Int J Clin Exp Pathol. 2014;7:7297–7303. [PMC free article] [PubMed] [Google Scholar]
- 68.Chu MMY, Liu SS, Tam KF, Ip PPC, Cheung ANY, Ngan HYS. The significance of mismatch repair deficiency in young patients with endometrial cancer. Int J Gynecol Pathol. 2015;34:403–410. doi: 10.1097/PGP.0000000000000174. [DOI] [PubMed] [Google Scholar]
- 69.Najdawi F, Crook A, Maidens J, et al. Lessons learnt from implementation of a Lynch syndrome screening program for patients with gynaecological malignancy. Pathology. 2017;49:457–464. doi: 10.1016/j.pathol.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 70.Pennington KP, Walsh T, Lee M, et al. BRCA1, TP53, and CHEK2 germline mutations in uterine serous carcinoma. Cancer. 2013;119:332–338. doi: 10.1002/cncr.27720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ring KL, Connor EV, Atkins KA, Ricketts W, Kashlan B, Modesitt SC. Women 50 years or younger with endometrial cancer: the argument for universal mismatch repair screening and potential for targeted therapeutics. Int J Gynecol Cancer. 2013;23:853–860. doi: 10.1097/IGC.0b013e31828eed9c. [DOI] [PubMed] [Google Scholar]
- 72.Woo YL, Cheah PL, Shahruddin SI, Omar SZ, Arends M. The immunohistochemistry signature of mismatch repair (MMR) proteins in a multiethnic Asian cohort with endometrial carcinoma. Int J Gynecol Pathol. 2014;33:554–559. doi: 10.1097/PGP.0000000000000099. [DOI] [PubMed] [Google Scholar]
- 73.Watkins Jaclyn C., Yang Eric J., Muto Michael G., Feltmate Colleen M., Berkowitz Ross S., Horowitz Neil S., Syngal Sapna, Yurgelun Matthew B., Chittenden Anu, Hornick Jason L., Crum Christopher P., Sholl Lynette M., Howitt Brooke E. Universal Screening for Mismatch-Repair Deficiency in Endometrial Cancers to Identify Patients With Lynch Syndrome and Lynch-like Syndrome. International Journal of Gynecological Pathology. 2017;36(2):115–127. doi: 10.1097/PGP.0000000000000312. [DOI] [PubMed] [Google Scholar]
- 74.Morice P, Leary A, Creutzberg C, Abu-Rustum N, Daraï E. Endometrial cancer. Lancet. 2016;387:1094–1108. doi: 10.1016/S0140-6736(15)00130-0. [DOI] [PubMed] [Google Scholar]
- 75.Meyer LA, Broaddus RR, Lu KH. Endometrial cancer and Lynch syndrome: clinical and pathologic considerations. Cancer Control. 2009;16:14–dju380. doi: 10.1177/107327480901600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 77.Moreira L, Balaguer F, Lindor N, et al. Identification of Lynch syndrome among patients with colorectal cancer. JAMA. 2012;308:1555–1565. doi: 10.1001/jama.2012.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barrow P., Green K., Clancy T., Lalloo F., Hill J., Evans D.G. Improving the uptake of predictive testing and colorectal screening in Lynch syndrome: a regional primary care survey. Clinical Genetics. 2015;87(6):517–524. doi: 10.1111/cge.12559. [DOI] [PubMed] [Google Scholar]
- 79.Wong YF, Cheung TH, Lo KWK, et al. Detection of microsatellite instability in endometrial cancer: advantages of a panel of five mononucleotide repeats over the National Cancer Institute panel of markers. Carcinogenesis. 2006;27:951–955. doi: 10.1093/carcin/bgi333. [DOI] [PubMed] [Google Scholar]
- 80.Zhao YS, Hu FL, Wang F, et al. Meta-analysis of MSH6 gene mutation frequency in colorectal and endometrial cancers. J Toxicol Environ Health A. 2009;72:690–697. doi: 10.1080/15287390902841003. [DOI] [PubMed] [Google Scholar]
- 81.Ryan NAJ, Morris J, Green K, et al. Association of mismatch repair mutation with age at cancer onset in Lynch syndrome: implications for stratified surveillance strategies. JAMA Oncol. 2017. [DOI] [PMC free article] [PubMed]
- 82.Levine AJ, Phipps AI, Baron JA, et al. Clinicopathologic risk factor distributions for MLH1 promoter region methylation in CIMP-positive tumors. Cancer Epidemiol Biomarkers Prev. 2016;25:68–75. doi: 10.1158/1055-9965.EPI-15-0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pagin A, Zerimech F, Leclerc J, et al. Evaluation of a new panel of six mononucleotide repeat markers for the detection of DNA mismatch repair-deficient tumours. British J Cancer. 2013;108:2079–2087. doi: 10.1038/bjc.2013.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Armstrong K, Calzone K, Stopfer J, Fitzgerald G, Coyne J, Weber B. Factors associated with decisions about clinical BRCA1/2 testing. Cancer Epidemiol Biomarkers Prev. 2000;9:1251–1254. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.