Abstract
Purpose:
To develop a steady-state saturation with radial readout CEST method (starCEST) for acquiring CEST images at 3T. The polynomial Lorentzian line-shape fitting (PLOF) approach was further developed for extracting amideCEST intensities at this field.
Method:
StarCEST MRI using PROPELLER-based spatial sampling was implemented to acquire Z-spectra that are robust to brain motion. Multi-linear singular value decomposition (MLSVD) post-processing was applied to enhance the CEST SNR. The eggwhite phantom studies were performed at 3T to reveal the contributions to the 3.5 ppm CEST signal. Based on the phantom validation, the amideCEST peak was quantified using the PLOF, which exploits the inverse relationship between Z-spectral intensity and the longitudinal relaxation rate in the rotating frame. The 3D Turbo Spin Echo (TSE) CEST was also performed to compare with the starCEST method.
Results:
The amideCEST peak showed a negligible peak B1 dependence between 1.2 μT and 2.4 μT. The amideCEST images acquired with starCEST showed much improved image quality, SNR and motion robustness compared to the conventional 3D TSE CEST method with the same scan time. The amideCEST contrast extracted by the PLOF method trended towards a stronger gray matter (GM) signal (1.32% ± 0.30%) than white matter (WM) (0.92% ± 0.08%; p=0.02, n=5). When calculating the MTC and T1 corrected rotating frame relaxation rate maps, amideCEST again was not significantly different for WM and GM.
Conclusion:
Rapid multi-slice amideCEST mapping can be achieved by the starCEST method (< 5 minutes) at 3T by combing with the PLOF method.
Keywords: chemical exchange saturation transfer (CEST), magnetization transfer contrast (MTC), steady-state pulsed CEST/MT, steady-state radial (star), CEST, MultiVane, Periodically Rotated Overlapping ParallEL Lines with Enhanced Reconstruction (PROPELLER)
INTRODUCTION
Chemical exchange saturation transfer (CEST) MRI is a molecular imaging technique that can detect proteins and metabolites through the water signal by making use of the exchangeable protons in these molecules (1–4). Among endogenous CEST contrasts, amide proton transfer (APT or amideCEST), in mobile proteins has been intensively studied (5–9) due to its several favorable properties, including high concentration, large chemical shift, and optimal exchange rate (<400 Hz) for the NMR exchange regime at high and ultra-high field strengths (3T −7T) (5,6). The successful acquisition of amideCEST signal requires sufficient signal-to-noise ratio (SNR) and image stability as both contrasts are only a few percent of the water signal and need to compete with stronger background signals from direct water saturation and especially conventional semi-solid magnetization transfer contrast (MTC). Many efforts have been devoted to optimizing saturation parameters (e.g., saturation power and length) to increase CEST contrasts in tissues (10–13), but improvements have been limited. Reducing the noise caused by oscillations in the Z-spectrum due to subject physiological and physical motions can also improve CEST SNR. This has been shown using motion insensitive self-gated radial sampling CEST (14,15), navigators to correct frequency fluctuations (16–19) and/or some post-processing methods by exploiting the redundancy of CEST images between different saturation offsets (20–24). Although radial CEST MRI has been successfully demonstrated for acquiring CEST and relayed nuclear Overhauser effect (rNOE) images on animals at high magnetic fields (14,15), its application is hindered by the long acquisition time on human scanners. In this work, we combined the fast radial acquisition method of periodically rotated overlapping parallel lines with enhanced reconstruction (PROPELLER) (25,26), with steady state saturation for amideCEST imaging on the human brain at 3T (starCEST). PROPELLER is called MultiVane on the Philips system. To further improve the starCEST SNR, multilinear singular value decomposition (MLSVD) (27) was applied. Recently, a radial sampling CEST method combining PROPELLER readout was proposed on rat brain (28). However, due to the use of a conventional labeling scheme, that is, continuous-wave (CW) labeling followed by PROPELLER readout, this method was unable to reduce the total scan time.
The extraction of amideCEST signals from in vivo Z-spectra is another challenging issue for 3T MRI. The conventional CEST quantification method uses magnetization transfer ratio asymmetry analysis, that is, subtracting the labeling and control water saturation images acquired at the two symmetric offsets with respect to water resonance. However, amideCEST and aliphatic-rNOE resonances essentially are centered at opposite sides of the water frequency and therefore in part compensate each other when performing asymmetry analyses (5,29–32). In order to distinguish between rNOE signals, we name them as aromatic-rNOE and aliphatic-rNOE in the current study. Furthermore, in white matter the MTC effect is known to be asymmetric with respect to water frequency (33,34) and also contributes. One popular strategy in the CEST field has been to acquire a full Z-spectrum with low saturation field strength B1 and fit it by assuming a Lorentzian line-shape for each contributing signal, such as direct saturation (DS), guanidinium (Guan), amide, amine, and rNOE peaks (8,35–38). This method can remove the semi-solid tissue and direct saturation (DS) components, and it works well in terms of extracting the rNOE signal. However, it remains challenging to extract the amideCEST signal due to the difficulty of removing interference from aromatic-rNOE and other CEST peaks, such as the fast-exchanging amine and hydroxyl protons, to the CEST signal between 0 and 5 ppm. A similar strategy, named extrapolated semisolid magnetization transfer reference (EMR), was proposed for amideCEST/aliphatic-rNOE mapping with high saturation strengths; however, it still faces an issue similar to the Lorentzian line-shape fitting method when using low saturation strengths (39–41). Recently, more evidence has suggested that the amide peak is discernible even at 3T MR fields when using low B1 strengths (42,43). Hence, due to the smooth merging of background signals, the polynomial and Lorentzian line-shape Fitting (PLOF) approach (13,44,45) that was proposed to extract and quantify CEST signals with discernible peaks should be well-suited for amideCEST quantification imaging at 3T. In this work, we demonstrate that amideCEST imaging of the human brain at 3T is possible by combining the radial readout MRI with MLSVD in a clinically feasible acquisition time.
METHODS
Phantom preparation
Egg white is an appropriate model to demonstrate the pH sensitivity of the amide, guanidium, and amine protons from mobile proteins. To demonstrate the possibility to detect pH-dependent amideCEST and rNOE signals with starCEST MRI, a set of egg white phantoms (100% liquid egg white) was prepared in a group of NMR tubes and titrated to pH values of 6, 6.5, 7, and 7.5 during slow stirring to avoid denaturation. In order to demonstrate the CEST contributions at 3.5 ppm, another set of egg white phantom was prepared by mixing egg white protein powder (NOW Sports Nutrition) with pure water (0%D2O), 35% and 70% D2O solution, respectively. The egg white protein was fixed at 20% w/w with respect to the non-deuterated water in the solutions.
Human studies
Human studies were approved by the Johns Hopkins Medicine IRB and performed on nine healthy subjects (23–46 years old), all of whom provided informed consent. All human scans were acquired on a 3T Philips Elition system (Philips Healthcare, Best, The Netherlands) using a quadrature transmit body coil and a 32-channel phased array receive head coil (Philips Healthcare). Two pads were placed on both side of the head close to the temporal lobes to minimize the head movement.
MRI experiments
The eggwhite phantom studies were performed to investigate the 3.5 pp CEST contributions and to justify the proposed PLOF method for extracting the amideCEST at 3T. Considering several practical advantages over the human 3T, such as the high MRI image quality of the phantoms, the excellent B0/B1 homogeneity as well as the better temperature control with an air heater that is not available on the human scanners, the MRI experiments on the egg white phantoms were performed on a horizontal bore 3T Bruker Biospec system (Bruker, Ettlingen, Germany). During MRI experiments, the phantom was maintained at 37°C by an air heater. A 40-mm volume transceiver coil was used. The Z-spectrum was collected with an ultra-short echo (UTE)-CEST sequence detailed previously (14,15,46,47). The duration of the Gaussian saturation pulses was set to 50 ms with a peak B1 of 0.4–1.6 μT unless specified, and the repetition time (TR) was 60 ms. The excitation pulse for the 2D-UTE radial readout was 10°. In order to make the peak B1 comparable to the B1 values used in the conventional continuous wave CEST (CWCEST), the equivalent block pulse B1 values (B1ave) were also reported by B1ave = Aarea × B1, where Aarea is the area ratio between one sinc-Gauss (Aarea = 0.54) or Gaussian (Aarea = 0.42) pulse and one block pulse with the same peak B1. Here, the averaged B1ave value was only for each saturation pulse (length τp) and was not the averaged value for the total period τp + τmix that determines the steady state. The effective echo time (TE) was 0.3 ms. In the current study, single slice CEST images were collected with a matrix size of 96 × 96 and a slice thickness of 5 mm. The total imaging time for each offset was 12 s. The saturation offset was swept from −12 to 12 ppm using a 0.2-ppm increment between −8 and 8 ppm and a 0.5-ppm increment for other regions.
Due to the limited acquisition speed of the UTE-CEST based radial readout method (47,48), we applied the PROPELLER based method for the acquisition of multi-slice CEST on the 3T clinical scanner. Figure 1a depicts the starCEST sequence based on the MultiVane Philips version of this sequence. Notice that we plotted five blade lines and echoes for illustration in Fig. 1, but we acquired 12 blade lines in practice. Every TR contains a sinc-Gauss saturation pulse (50 ms) followed by a train of 12 gradient echoes for detection (TR/TE/flip angle = 4.42 ms/2.08 ms/10°), which generates the 12 blade lines in one blade. Each blade was rotated and shifted following the motion correction method proposed in the literature (26), and then the images were reconstructed by gridding them to a Cartesian data set. With fixed TR, the number of lines per blade determines the mixing time between saturation pulses, following τmix = TR × (#lines per blade). Previous pulsed CEST optimization has already demonstrated that one pulse width larger than 40 ms would achieve enough frequency selectivity at 3T (49). Hence, the pulse width of the sinc-gauss saturation pulses was set to 50 ms. For the human brain study, images were acquired at 5 × 3 × 3 mm3 resolution using 15 slices and a FOV of 220 × 220 mm2. The image matrix was resized to 96 × 96 during reconstruction. The parallel imaging SENSE factor was 2 in the AP direction for the lines in each blade. The oversampling rate was 150% for MultiVane, and 12 blade lines in one blade were collected unless specified. Hence, nine blades were collected for each slice. The time for the multi-slice acquisition was 7.6 s per irradiation frequency. Furthermore, 37 saturation frequency offsets from 1 to 7.8 ppm with 0.2-ppm increment were acquired for amideCEST imaging with a total acquisition time of 4min40s. The nonsaturated reference signals were acquired using the same sequence with a saturation offset of 200 ppm. The second-order pencil beam (PB)-volume shimming method was applied on the image slices before all the CEST experiments.
Figure 1:
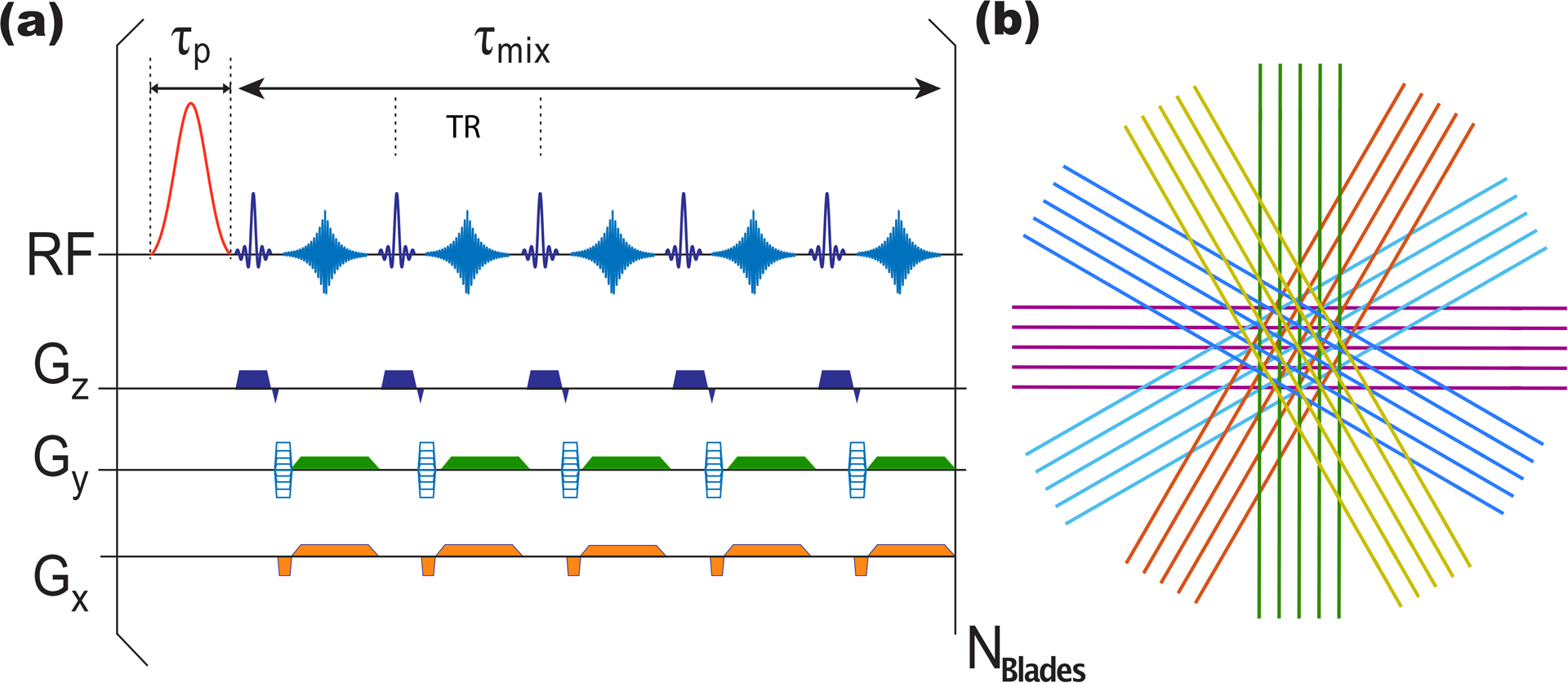
(a) Timing diagram of the starCEST sequence based on PROPELLER. τp is the duration of the saturation pulses; τmix is the mixing time for saturation transfer between successive saturation pulses; TR is the repetition time between two k-line acquisitions, determined by the number of phase encodes per k-line. (b) illustration of the sampling of k-space in starCEST using a set of radially directed blades. In this example, for demonstration purposes only, each blade is composed of five parallel phase-encoded lines that are collected using gradient echoes. However, 12 phase-encoded lines were acquired per blade in the current paper.
The conventional 3D Turbo Spin-Echo (TSE) CEST method (39,50,51) was performed on two subjects together with the starCEST method to compare the image quality, SNR and motion robustness. The CEST preparation module was identical to the saturation scheme in the starCEST, i.e. 50 ms sinc-Gauss saturation pulses with a peak B1 of 1.4 μT and 53 ms mixing times. 32 pulses and mixing times were implemented, which leaded to a total saturation time of 3.3 s and were considered as a steady-state saturation. The matrix and geometry sizes were identical to those used in the starCEST. The TSE factor was 174 with a 120-degree refocused angle. TR=7.2 s, TE=12 ms and the compressive sensing (CS) factor=4. The offsets and the total experimental time were identical to those of starCEST. In order to demonstrate the impact of brain motions on the amideCEST maps, starCEST and TSE experiments were performed sequentially with the two pads for fixing the brain removed.
T1 maps were acquired with a look-locker sequence with geometry identical to the CEST scan and were used for the CEST quantification in the PLOF method (52). Furthermore, 38 images were acquired with TR = 157 ms (TI = 35–5870 ms) and a readout flip angle of 4°. Pixel-by-pixel fitting was performed to a three-parameter model to obtain T1 (as T1=(B/A–1)T1*) (52). B0 maps were obtained using a dual-echo sequence with TR = 10 ms, TE = 4.6 ms, and a flip angle of 30°. B1 maps were obtained using the dual refocusing echo acquisition mode (DREAM) technique with a stimulated echo acquisition mode (STEAM) at a flip angle of 60° (53).
Data Analysis
All MRI images were processed using custom-written MATLAB scripts (MathWorks, www.mathworks.com). The amideCEST signal from both starCEST and 3D TSE CEST method was extracted using the PLOF method, which has been described previously (13,44,45). Briefly, the steady-state normalized Z-spectral intensity Zss has an inverse relationship to the longitudinal relaxation rate in the rotating frame, R1ρ,i.e. the rotating-frame relaxation spectrum (R-spectrum) (54–57):
| (1) |
R1 = 1/T1 is the longitudinal relaxation rate of water, and θ is the tilt angle of the effective magnetization with respect to the Z-axis. R1ρ is the water relaxation rate during saturation, which includes contributions from the effective water relaxation rate Reff, the rotating frame rate from the CEST signal of interest (Rexch), and a term Rback that accounts for the rotating frame rates of all other magnetization/exchange transfer processes in tissue (56):
| (2) |
where Reff is the longitudinal relaxation rate of water in the rotating frame without additional solution components. The observed CEST signal ΔZ is extracted by the following:
| (3) |
The amideCEST peak (R) and broad background (Rback) in the R-spectrum can be represented by a Lorentzian function and a mixed polynomial and Lorentzian function, respectively:
| (4) |
| (5) |
Where w is the peak full-width-at-half-maximum (FWHM) of the Lorentzian line-shape. Δω is the offset relative to the water proton resonance. Ramide is the intensity of the amideCEST peak in the R-spectrum. Δωamide is the chemical shift offset of the CEST peak relative to the water signal, that is, 3.5 ppm; The background function was modified from the original polynomial function (13,44,45) to improve the PLOF background fitting at 3T. The first Lorentzian function in Eq. 5 was included to account for the water direct saturation, while the polynomial function (C2 + C3 ·Δω) was implemented to fit the MTC and other exchanging protons such as amine/hydroxyl CEST. In the current study, we found a linear function can perfectly fit the combined effect with MTC and other CEST between 1–7.6 ppm. For a wider fitting range, a higher order approximation can be used. The Z-spectral range of [2.5,4.5] (ppm) was excluded from the background fitting (fitting range = 1.0–7.6 ppm) for amideCEST. When performing the PLOF calculation on the brain, the measured B0 map was used to shift the Z-spectrum for field homogeneity correction in each pixel. The MLSVD method was applied to enhance the SNR for amideCEST maps (27). The truncation numbers were 48, 48, and 10, and they were used for the two image dimensions and Z-spectral dimension, respectively. The SNR of the images were calculated by the mean values in the signal regions divided by the standard derivation of the noise regions at the four image corners. The intersubject reproducibility was quantified by coefficient of variance (COV; COV = standard derivation/mean×100%). The intraclass correlation coefficient (ICC) was used to measure the intrasubject agreement between two different scans. (58) The COV and ICC for the amideCEST (ΔZamide) were calculated based on the averaged values in the whole brain GM and WM regions.
RESULTS
Phantoms
We first studied the spectral contributions and pH dependence of the egg white Z-spectrum using UTE-CEST as a function of pH values. Figure 2 displays the Z-spectra of egg white as a function of saturation strength and pH. At low peak B1 values (<0.8 μT or B1ave < 0.43 μT), the amide, guanidinium, and aliphatic peaks show structure, whereas those peaks merge with the direct saturation lineshape at higher saturation field strengths (>1 μT). The results also demonstrate the frequency range of the protein Z-spectrum, which widens as a function of saturation field strength (43), due to the widening of the CEST peaks and larger bandwidths of the pulses. At 3 Tesla, the protein solution Z-spectrum range is approximately ±9 ppm for peak B1 = 0.8 μT (B1ave = 0.43 μT). For the amideCEST peak around 3.5 ppm, decreased effect size was found for lower pH values (Figure 2b), but relative to the estimated DS a strong residual saturation transfer background signal remained over a broad frequency range even at pH 6 (purple arrow in Fig. 2b). As expected for exchange relayed NOEs and shown previously in phantoms (59,60), the aliphatic rNOE at the opposite frequency range also demonstrated pH dependence. The guanidinium effect size first increased when lowering pH from 7.5 to 7 and then stabilized and decreased with further reduction of pH, which was consistent with previous observations (61). The contributions of CEST and rNOE signals in protein were examined by varying the hydrogen/deuterium ratio of water in egg white solutions and the results at a peak B1 of 0.8 μT (B1ave = 0.43 μT) are plotted in Fig. 2c. As expected, the aliphatic-rNOE showed only a small reduction, while the amide peak and part of the broad background in the exchangeable proton range were clearly reduced for the 35% D2O solution, and even more for the 70% D2O solution. For 70% D2O, the CEST effect between 0 to 6 ppm decreased dramatically but still a residual signal (about 56% in the 20% egg white solutions) was observable (purple arrow in Fig. 2c).
Figure 2:

Z-spectra of egg white as a function of peak B1 (a) and pH (b), acquired using UTE radial CEST. (a) Z-spectrum of egg white (pH 7.0) as a function of peak B1. A simulated DS spectrum for peak B1 = 0.4 μT (B1ave = 0.17 μT) is also shown (black curve). (b) Z-spectrum of egg white as a function of pH with peak B1 = 0.8 μT (B1ave = 0.34 μT). The exchangeable protons downfield, amide and guanidinium (Guan), and aliphaticNOE signals are indicated. The simulated DS spectrum with peak B1 = 0.8 μT (B1ave = 0.34 μT) is also plotted. (c) The Z-spectra of egg white solution with 0% (green), 35% (red) and 70% (blue) D2O acquired with UTECEST with peak B1 = 0.8 μT (B1ave = 0.34 μT). A residual signal is still observable in the exchangeable proton range for 70% D2O (purple arrow). The changes in the Z-spectral broad background at 3.5 ppm between 0% and 70% D2O (green arrow) are attributed to CEST effects of amine, and other fast exchangeable protons (e.g. hydroxyl in myoinositol) that are partially merged with the water signal due to being in the intermediate to fast exchange regime at 3T. The simulated DS spectrum for egg white in 0% D2O is also plotted.
Human brain
Examples of starCEST Z-spectra between 1.6 and 6.7 ppm for the human brain recorded as a function of saturation strength are depicted in Figs. 3a,b for gray matter (GM) and white matter (WM), respectively. Spectra are from regions of interest drawn on the brain in the mid-axial slice (8th slice) for which the T1-based mask for separating WM and GM is shown in Fig. 3c. A discernable amide peak at 3.5 ppm can be observed for peak B1 strengths below 2 μT (B1ave < 1.1 μT) for both GM and WM and the peak merges with the background above 2 μT. The Z-spectra acquired with a peak B1 of 1.4 μT from Figs. 3a,b are shown in Figs. 3d,e and demonstrate obvious differences in the amide peak for GM and WM, i.e. the amide peak in GM is more pronounced than that in WM. For example, the amide peak in GM is more pronounced than that in WM. The MTC background of WM is stronger than in GM, i.e. GM absolute Z-values are higher than those of WM, which is consistent with conventional MTC studies (62). In order to illustrate the process of extracting the amideCEST signal with the PLOF method, the PLOF fitting curves for the background Zback with the improved background function and the full Z-spectrum are plotted in the same figures (Figs. 3d,e). The background can be well fitted with four parameters in Eq. 5 for the whole 1–7.5 ppm, which allowed us fully utilizing all acquired offsets. The resulting B1-dependent amideCEST signal is shown in Fig. 3f. and did not show clear peak B1 dependence between 1.2–2.4 μT (B1ave between 0.65 and 1.3 μT) for both WM and GM. In order to make the sequence robust to the B1 variation in the brain, the peak B1 was therefore set to 1.4 μT (B1ave = 0.76 μT) in the following amideCEST studies.
Figure 3:

Typical Z-spectra of human brain acquired with starCEST on a 3T clinical scanner as a function of peak B1 for gray matter (GM) (a) and white matter (WM) (b), respectively. The regions of interest (GM and WM) were extracted from a mid-axial slice (8th slice) using a mask (c) based on the T1 map with a cut-off value of 0.8 s. (d) A GM Z-spectrum of the mid-axial slice recorded with peak B1 =1.4 μT (B1ave = 0.76 μT) and the PLOF fitting results with Eqs. 1–5. The method to extract ΔZamide signal with Eqs. 1–5 is indicated using a red arrow. The PLOF background fitted with the conventional 3th degree polynomial function (13,44,45) was plotted for comparison (red dashed line). The amide proton range and the whole fitting range were identical for the two background functions (Eq.5 and the 3th degree polynomial function). (e) A WM Z-spectrum of the mid-axial slice recorded with peak B1 =1.4 μT (B1ave = 0.76 μT) together with the PLOF fitting results. (f) B1 dependence of the amideCEST signal for the WM and GM in the mid-axial slices (n=4). Error bars reflect the standard deviation over different subjects. The signal was extracted with the PLOF method illustrated in (d) and (e).
Figure 4 depicts B0 and B1 maps of the brain of a typical subject. The B0 and B1 standard derivation (STD) for each slice are also plotted in Figs. 4b,d, respectively. The B0 inhomogeneity over the whole brain was in the range of −0.7–0.7 ppm. The major inhomogeneity areas (STD>0.2) appeared at the beginning of the slice package (slices 1–3). The B1 map was quite homogeneous across the brain (STD<0.11), except for the typical B1 hyperintensity in the midbrain region (slices 1–3). Figure 5 depicts starCEST, T1, ΔZamide and Ramide maps of the brain of a typical subject. The ΔZamide and Ramide maps were calculated with the PLOF method by including the B0 map and the measured T1 maps. Discernable WM and GM amide CEST effects can be observed in the ΔZamide maps and the GM amideCEST (ΔZamide) is higher that of WM, consistent with the observation from typical WM and GM Z-spectra (Figs. 3d&e). Correction for the MTC/DS background and T1 impacts the rotating frame relaxation method (Eqs.1–5), and the WM and GM contrast in the Ramide maps diminished significantly.
Figure 4:

(a) Typical multi-slice B0 maps acquired with dual-echo sequence on the human brain at 3T. The averaged standard derivation (n=3) for each slice of the B0 maps. (c) Typical multi-slice B1 ratio maps obtained with DREAM method on the same subject. (d) The averaged standard derivation (n=3) for each slice of the B1 maps.
Figure 5:

Typical multi-slice human brain images acquired at 3T. (a) starCEST images; (b) T1 maps by look-locker sequence. The amideCEST maps (ΔZamide) and the corresponding apparent relaxation rate (Ramide) maps extracted with the PLOF method by including the B0 and T1 maps are shown in (c) and (d), respectively.
The whole brain images acquired with the starCEST and 3D TSE CEST are plotted in Fig. 6 together with the corresponding amideCEST maps. The starCEST images show much better image quality than the TSE CEST images as judged from the clear contrast between WM/GM (Fig. 6a). On the contrary, the images obtained by the TSE method are blurry due to the long TSE acquisition time (2.1s) and it is challenging to differentiate WM and GM regions. Strong CSF signal was observed in the TSE images (Fig. 6c). The starCEST SNR values in the center slice were much higher than those of TSE CEST images (starCEST GM vs TSE GM: 96±19 vs 54±16; starCEST WM vs TSE WM: 34±6 vs 18±6;). The amideCEST maps obtained by starCEST (Fig.6b) did not show any obvious motion introduced artifacts in both GM and WM regions, but hyperintensities were present in some of the CSF regions. For the amideCEST maps obtained by the TSE CEST method, serious hypointensities were observed in the most of slices, tentatively attributed to the brain motions (Fig. 6d).
Figure 6:
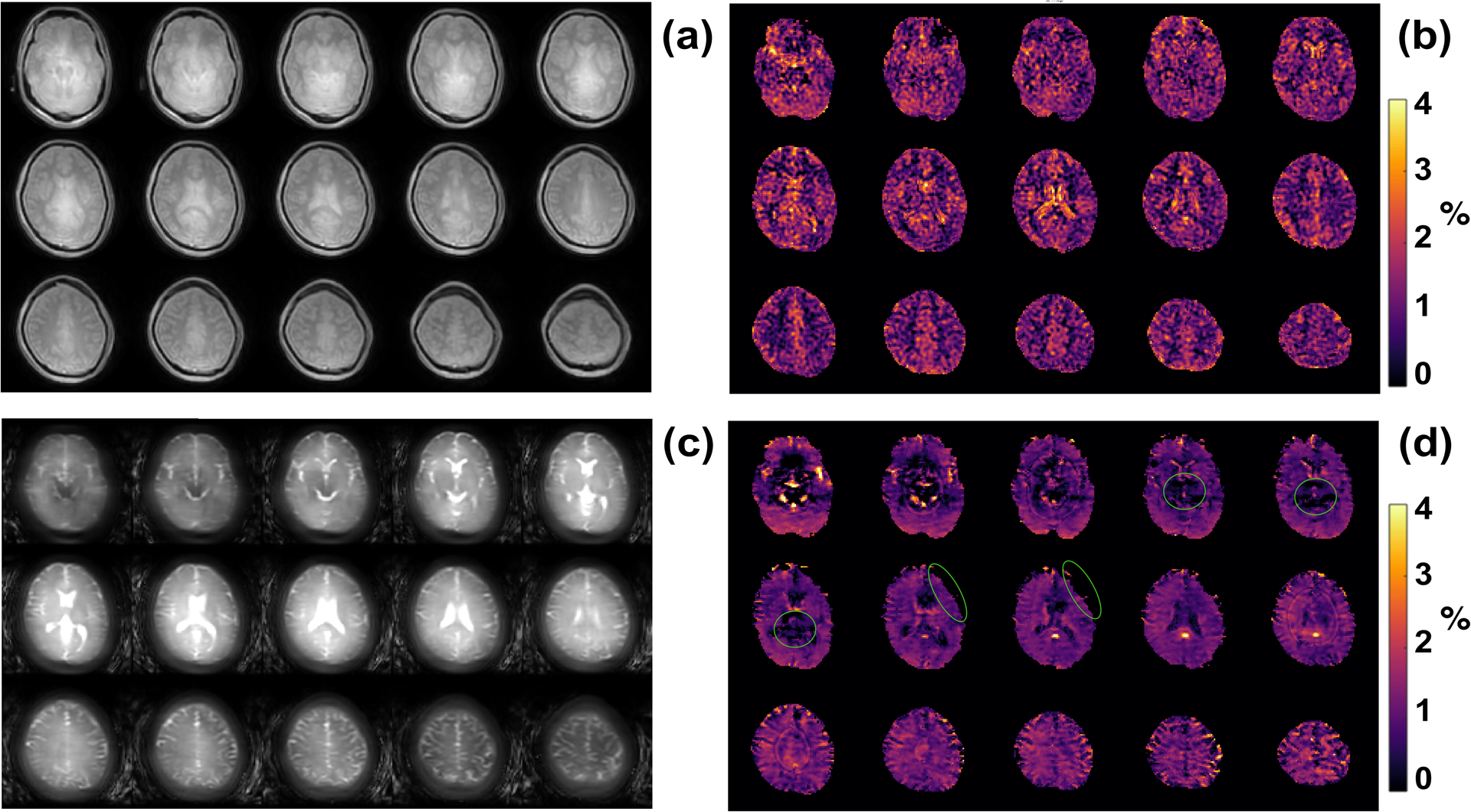
Comparison of the human brain images acquired at 3T with the multi-slice starCEST (a) and 3D TSE CEST sequence (c). The amideCEST maps extracted with the PLOF method from the multi-slice starCEST (c) and 3D TSE CEST (d) images. Hypointensity regions in the amideCEST maps due to the brain motions are indicated with green circles.
In order to quantify the amideCEST values, Figure 7 shows histograms of the amideCEST difference and rotating frame relaxation values in WM and GM for all subjects (n=5). The regions of interest for GM and WM were extracted from the T1 maps with a cut-off value of 0.8 s. The mean and standard deviation of the amideCEST signals were 1.32% ± 0.30% (GM) and 0.92% ± 0.08% (WM) (p=0.02, n=5) for ΔZamide, respectively (Fig. 7a). In the Ramide images, the averages also showed no significant difference (GM:17 ± 4 × 10−3 s−1; WM:18 ± 2 × 10−3 s−1; p=0.4, n=5, Fig. 7b), consistent with the low contrast in the Ramide maps (Fig. 5d). Reproducibility was investigated for the same protocol across different subjects (n=5) as well as the same subject (n=3) at two different sessions, and the results are plotted in Figs. 7c–d. The intersubject COV were 22% (GM) and 9% (WM) (n = 5) for amideCEST (Fig. 7c). Excellent intrasubject agreement were found in the amideCEST measurements from two scans with ICC=0.98 (GM) and 0.99 (WM) (n=3; Fig. 7e).
Figure 7:

(a) GM and WM distributions of fitted amideCEST signal differences (ΔZamide) as well as the corresponding apparent relaxation rates (b: Ramide) for all subjects (n=5). GM and WM regions show very similar distribution of ΔZamide. After correction with the rotating frame relaxation method, WM regions show slightly higher contrast in the rotating frame relaxation rate Ramide. Comparison of fitted amideCEST (c: ΔZamide) and the corresponding rotating frame relaxation rates (d: Ramide) in GM and WM for five healthy subjects. Comparison of amideCEST (e:ΔZamide) and the rotating frame relaxation rates (f: Ramide) in GM and WM for the same subject (n=3, subject 2, 4 and 5) at two different sessions i.e., scan1(S1) and scan 2 (S2) over two days. The error bar is the standard derivation for each subject.
Discussion
We applied radial CEST acquisition, post-processing, and quantification methods to isolate the amideCEST signal in human brain at 3T. We demonstrated that the amideCEST signal can be extracted with the updated PLOF method at 3T. The amideCEST images acquired with starCEST showed much improved image quality and SNR compared to the conventional 3D TSE CEST method with the same scan time. In order to achieve rapid acquisition for the TSE CEST, high TSE and CS factors had to be used. The amideCEST maps acquired by the starCEST were robust to the brain movements (Fig. 6), while motion corrections are required for the CEST maps with the TSE readout.
Contrary to a recent creatine validation study (45,63) using a knockout mouse model, validation of the amideCEST signal in the brain Z-spectrum cannot be easily achieved by removing the mobile proteins in tissue. However, we could exploit the property that CEST signals from amide and guanidinium protons are strongly pH-dependent, whereas rNOEs have weak pH dependence and MTC is insensitive to pH (60,64) in the physiological range. As demonstrated by the egg white model system (Fig. 2), the amide CEST signal is visible at 3T with a peak around 3.5 ppm. However, there is still a large residual signal at 3.5 ppm (purple arrow in Fig. 2b) that is not pH-dependent over the physiological range of pH 6.0 −7.5 (Fig. 2b). This conclusion is consistent with previous studies with animal stroke models at both high (27,61,64) and low MRI fields (65). The source of the strong residual background signal (purple arrow in Fig. 2b) in addition to amideCEST was further investigated with hydrogen/deuterium (H/D) exchange studies on egg white solutions. As seen from Fig. 2c, a large portion of this broad background signal between 0 and 6 ppm disappeared for 70% D2O, which we tentatively attribute to faster exchanging protons from amine and hydroxyl groups, signals of which are partially merged with water due to being in the intermediate to fast exchange regime at 3T. Due to this exchange regime, this portion of the CEST signal only has a weak pH dependence in the physiological range (6–7.5). In addition to this, the background (purple arrow in Fig. 2b) may have contributions from rNOEs, e.g. amide-rNOE and aromatic-rNOE. The existence of NOE components at 3.5 ppm has been suggested by hydrogen/deuterium (H/D) exchange studies (66,67), which have long been used to probe protein structures. The exchange rates of the amide protons depend on solvent accessibility and hydrogen bonding. The amide protons that are part of an intramolecular hydrogen bond or buried from the water will exchange slowly or do not exchange at all, whereas an amide on the surface of protein and side chains will exchange rapidly with water. Hence, NOE signals at approximately 3.5 ppm may be attributable to non-exchanging amide protons or amide proton with extremely slow exchange rates, but they will still be able to transfer magnetization to water following a two-step relayed NOE process with faster exchanging neighboring protons (68). However, this hypothesis and the contribution from faster exchanging amine and OH groups to the pH-independent background for the 3T exchange regime still needs to be further validated and is beyond the scope of the current study. The D2O study seems to indicate the contribution of the latter may be substantial. One recent study in which the myelin lipids in brain were removed also suggests that the rNOE signal from lipids may contribute to the positive side of the Z-spectrum (69). From the egg white study, the background CEST signal at 3.5 ppm, extracted using either Lorentzian fitting (8,35–38) or asymmetry analysis (70,71), includes many components such as amineCEST, hydroxylCEST, and rNOE signals. All these effects, including both CEST and rNOE effects, but not exclusively, also apply to mobile proteins in vivo. Hence, the signal at 3.5 ppm extracted by the conventional Lorentzian fitting or asymmetry analysis can still be applied for the protein profiling in tissues such as tumor. When probing pH changes in tissue at the physiological range, the PLOF method can thus provide a quite sensitive contrast by extracting the major pH-dependent component at physiological pH, i.e. amideCEST, or at higher B1, amineCEST, as shown by Ellingson et al. (72)
In the starCEST method, the steady-state saturation is a consequence of both the saturation pulses and the MRI readouts during the mixing period. As previous steady-state CEST optimization demonstrated, higher flip angles of the MRI readout pulses reduce the CEST contrast, while lower flip angles lead to low SNR for the MRI images. (73,74) In the current study, a flip angle of 10 degrees was selected as a good compromise. The time to reach steady-state in starCEST when switching saturation offset is determined by the water rotating frame relaxation time, which is close to water T1 at low saturation powers.(13) As the simulations in the Supporting Information Figure S1 show, the non-steady-state character of the saturation when switching irradiation frequency impacts mainly the initial offsets for slices 1–3. This can be easily solved by removing the first offset from the PLOF fitting.
Here, we demonstrated that, with same scan time, the starCEST approach can generate much better image quality than the 3D TSE CEST. The acquisition speed for the 3D MRI readout can be further improved with a 3D gradient- and spin-echo (GRASE) readout. Previously, a scan time of 20 s for each offset has been applied for the 3D GRASE CEST to achieve reasonable image quality (75). This is much slower than the starCEST method and, similar to the TSE CEST approach has the issue of not being robust to motion. Recently, several other whole brain CEST methods have been developed to reach acquisition speeds comparable or faster than the starCEST method, including steady-state 3D EPI (11s per offset) (76), snapCEST (6.9 s per offset) (42,77), and 3D EPI with CAIPIRINHA acceleration (4.3 s per offset) (38). It is known that segmented 3D MRI readouts are very sensitive to motion as suggested by arterial spin labeling MRI studies. (78) Thus, motion correction is still required for those 3D CEST MRI techniques, while starCEST is a multi-slice method and is inherently robust to motions. One advantage of the snapCEST and 3D EPI CEST methods over the starCEST and steady-state 3D EPI methods is more flexibility in the CEST preparation module. Therefore, both snapCEST and 3D EPI CEST methods are more suitable for the CEST experiments with fast-exchanging protons.
In the current study, the B0 maps were used for inhomogeneity correction in the amideCEST quantification. When the B0 shift is small (<0.3 ppm), the PLOF fitting method can find the amide peak and correct the B0 shift as demonstrated by Supporting Information Figure S2. It is still challenging for the PLOF method to correct the offsets with higher B0 shift values (>0.3 ppm). The pulsed CEST method is robust to B1 inhomogeneity compared with CWCEST, as 20% in the peak B1 value change is only equivalent to an 10.8% change in CWCEST (0.54 × 20%). As demonstrated in the B1 optimization for amideCEST (Fig. 3f), the amideCEST variation is less than 0.15% for GM and is less than 0.06% for WM (1.1–1.7 μT) with 25% B1 changes, which is the whole brain B1 inhomogeneity range (Fig. 4c).
The amideCEST maps closely resemble to the maps obtained by suppressing MTC with variable delay multi-pulsed method (79), in which strong GM signal was observed in the APT map. Although a clear difference in GM and WM was observed in the ΔZamide maps (Fig. 5c) as well as the Z-spectra in Figs. 3d,e, the GM/WM contrast was highly variable between different subjects (Fig. 7c). The intrasubject reproducibility study (Fig. 7e) showed consistent GM/WM contrast. Further large-scale studies will necessary to reveal the origin of the highly variable GM/WM contrast among different subjects. The amideCEST signal extracted by the PLOF method (<2%) was much lower than the total signal at 3.5 ppm extracted with the Lorentzian fitting method. (38,42,43) Hence, the MLSVD method was used to enhance the overall SNR as illustrated by the comparison study in Supporting Information Figure S3. Due to the low amideCEST signal, the background fitting in the PLOF method is critical for extracting reliable amideCEST values. Use of a third-degree polynomial function to fit the brain Z-spectrum at 3T was challenging due to the strong water direct saturation as demonstrated in Fig. 3d. As a consequence, a reduced fitting range (<5.1 ppm) had to be used for the PLOF method with the polynomial function, which underestimated the amideCEST values. The current fitting function (Eq. 5) provided a much-improved background fitting over a wider frequency range (6.5 ppm) (Fig. 3d), which lead to reliable amidCEST values as demonstrated by the intrasubject reproducibility study (Fig. 7e). The proposed starCEST method not only works for the brain but is also expected to be well-suited for study on body tissues, such as the liver and kidney, where motion is a serious issue as seen in our recent liver study (15).
CONCLUSIONS
We have demonstrated that a multi-slice starCEST method can be used for the imaging of amideCEST effect at clinical field strength. The PLOF method was implemented for quantifying the pH-sensitive amideCEST signal. The MLSVD post-processing method was applied to achieve enough SNR for the amideCEST maps, which trended towards slightly higher contrast in GM. This trend disappeared when calculating MTC- and T1-corrected rotating frame relaxation rates. This study paves the way for amideCEST contrast quantification with a clinically feasible scan time.
Supplementary Material
Supporting Information Figure S1: Simulation of the Z-spectrum for each slice in the starCEST to demonstrate the impact of the non-steady-state character of the saturation. Only the first four slices are shown for clarity. In the simulation, the 50 ms since-Gauss saturation pulses and 53 ms mixing times were applied. The frequency was swept from 6 ppm to 1 ppm. T1=1s, T2=80 ms were set for the water pool, while T1=1s, T2=80 ms, exchange rate 160 Hz, offset 3.5 ppm and concentration 500 mM were assumed for the amide proton pool. The non-steady-state saturation significantly impacts the Z values in the slices 1–3 at the initial offset (6 ppm) and also shift the Z-spectrum of the first slice by 0.03 ppm.
Supporting Information Figure S2: (a) B0 map recorded with a dual-echo sequence. Comparison between the amideCEST maps extracted using the PLOF method without (b) and with (c) B0 correction. Hyper/hypo intensity regions in the amideCEST maps are indicated with green circles and are due to the high B0 shifts (> 0.3 ppm).
Supporting Information Figure S3: Comparison of the amideCEST maps without and with MLSVD post-processing. (a) The original amide CEST maps extracted using PLOF without MLSVD denoising. (b) MLSVD denoising applied with truncation numbers 48, 48 and 10.
ACKNOWLEDGEMENTS
Grant support from NIH: P41EB015909, R01HL149742, R21NS118079, R21AG065794, and S10OD021648. We thank Caigui Lin for performing some pilot studies. Dr. van Zijl obtains research support from Philips, has technology licensed to Philips, and is a paid lecturer for Philips. This has been approved by the Johns Hopkins University Committee on Conflict of Interest.
Footnotes
Data Availability Statement
The code that supports the findings of this study is openly available at https://github.com/jiadixu/AmideCEST-aliphaticNOE.
REFERENCES:
- 1.Ward K, Aletras A, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST). J Magn Reson 2000;143(1):79–87. [DOI] [PubMed] [Google Scholar]
- 2.Van Zijl PC, Yadav NN. Chemical exchange saturation transfer (CEST): what is in a name and what isn’t? Magn Reson Med 2011;65(4):927–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woessner DE, Zhang S, Merritt ME, Sherry AD. Numerical solution of the Bloch equations provides insights into the optimum design of PARACEST agents for MRI. Magn Reson Med 2005;53(4):790–799. [DOI] [PubMed] [Google Scholar]
- 4.Liu G, Song X, Chan KW, McMahon MT. Nuts and bolts of chemical exchange saturation transfer MRI. NMR Biomed 2013;26(7):810–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu X, Yadav NN, Zeng H, Jones CK, Zhou J, van Zijl PC, Xu J. Magnetization transfer contrast–suppressed imaging of amide proton transfer and relayed nuclear overhauser enhancement chemical exchange saturation transfer effects in the human brain at 7T. Magn Reson Med 2016;75(1):88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones CK, Huang A, Xu J, Edden RA, Schär M, Hua J, Oskolkov N, Zacà D, Zhou J, McMahon MT. Nuclear Overhauser enhancement (NOE) imaging in the human brain at 7 T. NeuroImage 2013;77:114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desmond KL, Moosvi F, Stanisz GJ. Mapping of amide, amine, and aliphatic peaks in the CEST spectra of murine xenografts at 7 T. Magn Reson Med 2014;71(5):1841–1853. [DOI] [PubMed] [Google Scholar]
- 8.Zhou IY, Wang E, Cheung JS, Zhang X, Fulci G, Sun PZ. Quantitative chemical exchange saturation transfer (CEST) MRI of glioma using Image Downsampling Expedited Adaptive Least-squares (IDEAL) fitting. Scientific Reports 2017;7(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goerke S, Breitling J, Zaiss M, Windschuh J, Kunz P, Schuenke P, Paech D, Longo DL, Klika KD, Ladd ME. Dual‐frequency irradiation CEST‐MRI of endogenous bulk mobile proteins. NMR Biomed 2018;31(6):e3920. [DOI] [PubMed] [Google Scholar]
- 10.Sun PZ, Wang E, Cheung JS, Zhang X, Benner T, Sorensen AG. Simulation and optimization of pulsed radio frequency irradiation scheme for chemical exchange saturation transfer (CEST) MRI-demonstration of pH-weighted pulsed-amide proton CEST MRI in an animal model of acute cerebral ischemia. Magn Reson Med 2011;66(4):1042–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun PZ, Benner T, Kumar A, Sorensen AG. Investigation of optimizing and translating pH-sensitive pulsed-chemical exchange saturation transfer (CEST) imaging to a 3T clinical scanner. Magn Reson Med 2008;60(4):834–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zu Z, Li K, Janve VA, Does MD, Gochberg DF. Optimizing pulsed-chemical exchange saturation transfer imaging sequences. Magn Reson Med 2011;66(4):1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L, Wei Z, Cai S, Li Y, Liu G, Lu H, Weiss RG, van Zijl PCM, Xu J. High-resolution creatine mapping of mouse brain at 11.7 T using non-steady-state chemical exchange saturation transfer. NMR Biomed 2019;32(11):e4168. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Wei Z, Chan KWY, Cai S, Liu G, Lu H, Wong PC, van Zijl PCM, Li T, Xu J. Protein aggregation linked to Alzheimer’s disease revealed by saturation transfer MRI. Neuroimage 2018;188:380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y, van Zijl PCM, Xu X, Xu J, Li Y, Chen L, Yadav NN. Magnetic resonance imaging of glycogen using its magnetic coupling with water. Proc Natl Acad Sci U S A 2020;117(6):3144–3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu R, Zhang H, Niu W, Lai C, Ding Q, Chen W, Liang S, Zhou J, Wu D, Zhang Y. Improved chemical exchange saturation transfer imaging with real-time frequency drift correction. Magn Reson Med 2019;81(5):2915–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Windschuh J, Zaiss M, Ehses P, Lee JS, Jerschow A, Regatte RR. Assessment of frequency drift on CEST MRI and dynamic correction: application to gagCEST at 7 T. Magn Reson Med 2019;81(1):573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simegn GL, Van der Kouwe AJW, Robertson FC, Meintjes EM, Alhamud A. Real-time simultaneous shim and motion measurement and correction in glycoCEST MRI using double volumetric navigators (DvNavs). Magn Reson Med 2019;81(4):2600–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simegn GL, Alhamud A, van der Kouwe AJW, Meintjes E, Robertson F. Repeatability and reproducibility of prospective motion- and shim corrected 2D glycoCEST MRI. Quant Imaging Med Surg 2019;9(10):1674–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breitling J, Deshmane A, Goerke S, Korzowski A, Herz K, Ladd ME, Scheffler K, Bachert P, Zaiss M. Adaptive denoising for chemical exchange saturation transfer MR imaging. NMR Biomed 2019;32(11):e4133. [DOI] [PubMed] [Google Scholar]
- 21.Bie C, Liang Y, Zhang L, Zhao Y, Chen Y, Zhang X, He X, Song X. Motion correction of chemical exchange saturation transfer MRI series using robust principal component analysis (RPCA) and PCA. Quant Imaging Med Surg 2019;9(10):1697–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breitling J, Deshmane A, Goerke S, Korzowski A, Herz K, Ladd ME, Scheffler K, Bachert P, Zaiss M. Adaptive denoising for chemical exchange saturation transfer MR imaging. NMR Biomed 2019:e4133. [DOI] [PubMed] [Google Scholar]
- 23.Wech T, Kostler H. Robust motion correction in CEST imaging exploiting low-rank approximation of the z-spectrum. Magn Reson Med 2018;80(5):1979–1988. [DOI] [PubMed] [Google Scholar]
- 24.Dopfert J, Witte C, Kunth M, Schroder L. Sensitivity enhancement of (Hyper-)CEST image series by exploiting redundancies in the spectral domain. Contrast Media Mol Imaging 2014;9(1):100–107. [DOI] [PubMed] [Google Scholar]
- 25.Pipe JG. Motion correction with PROPELLER MRI: application to head motion and free-breathing cardiac imaging. Magn Reson Med 1999;42(5):963–969. [DOI] [PubMed] [Google Scholar]
- 26.Pipe JG, Gibbs WN, Li Z, Karis JP, Schar M, Zwart NR. Revised motion estimation algorithm for PROPELLER MRI. Magn Reson Med 2014;72(2):430–437. [DOI] [PubMed] [Google Scholar]
- 27.Chen L, Cao S, Koehler RC, van Zijl PCM, Xu J. High-sensitivity CEST mapping using a spatiotemporal correlation-enhanced method. Magn Reson Med 2020: 10.1002/mrm.28380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo C, Wu J, Rosenberg JT, Roussel T, Cai S, Cai C. Fast chemical exchange saturation transfer imaging based on PROPELLER acquisition and deep neural network reconstruction. Magn Reson Med 2020. [DOI] [PubMed] [Google Scholar]
- 29.Xu J, Yadav NN, Bar‐Shir A, Jones CK, Chan KW, Zhang J, Walczak P, McMahon MT, Van Zijl PC. Variable delay multi‐pulse train for fast chemical exchange saturation transfer and relayed‐nuclear overhauser enhancement MRI. Magn Reson Med 2014;71(5):1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou J, Hong X, Zhao X, Gao JH, Yuan J. APT‐weighted and NOE‐weighted image contrasts in glioma with different RF saturation powers based on magnetization transfer ratio asymmetry analyses. Magn Reson Med 2013;70(2):320–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heo HY, Zhang Y, Lee DH, Hong X, Zhou J. Quantitative assessment of amide proton transfer (APT) and nuclear Overhauser enhancement (NOE) imaging with extrapolated semi‐solid magnetization transfer reference (EMR) signals: application to a rat glioma model at 4.7 Tesla. Magn Reson Med 2016;75(1):137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heo HY, Zhang Y, Burton TM, Jiang S, Zhao Y, van Zijl PC, Leigh R, Zhou J. Improving the detection sensitivity of p H‐weighted amide proton transfer MRI in acute stroke patients using extrapolated semisolid magnetization transfer reference signals. Magn Reson Med 2017;78(3):871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pekar J, Jezzard P, Roberts DA, Leigh JS, Frank JA, McLaughlin AC. Perfusion imaging with compensation for asymmetric magnetization transfer effects. Magn Reson Med 1996;35(1):70–79. [DOI] [PubMed] [Google Scholar]
- 34.Hua J, Jones CK, Blakeley J, Smith SA, van Zijl PC, Zhou J. Quantitative description of the asymmetry in magnetization transfer effects around the water resonance in the human brain. Magn Reson Med 2007;58(4):786–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai K, Singh A, Poptani H, Li W, Yang S, Lu Y, Hariharan H, Zhou XJ, Reddy R. CEST signal at 2ppm (CEST@2ppm) from Z-spectral fitting correlates with creatine distribution in brain tumor. NMR Biomed 2015;28(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desmond KL, Moosvi F, Stanisz GJ. Mapping of amide, amine, and aliphatic peaks in the CEST spectra of murine xenografts at 7 T. Magn Reson Med 2014;71(5):1841–1853. [DOI] [PubMed] [Google Scholar]
- 37.Zaiss M, Windschuh J, Goerke S, Paech D, Meissner JE, Burth S, Kickingereder P, Wick W, Bendszus M, Schlemmer HP, Ladd ME, Bachert P, Radbruch A. Downfield-NOE-suppressed amide-CEST-MRI at 7 Tesla provides a unique contrast in human glioblastoma. Magn Reson Med 2016. [DOI] [PubMed] [Google Scholar]
- 38.Mueller S, Stirnberg R, Akbey S, Ehses P, Scheffler K, Stocker T, Zaiss M. Whole brain snapshot CEST at 3T using 3D-EPI: Aiming for speed, volume, and homogeneity. Magn Reson Med 2020. [DOI] [PubMed] [Google Scholar]
- 39.Heo HY, Zhang Y, Burton TM, Jiang S, Zhao Y, van Zijl PCM, Leigh R, Zhou J. Improving the detection sensitivity of pH-weighted amide proton transfer MRI in acute stroke patients using extrapolated semisolid magnetization transfer reference signals. Magn Reson Med 2017;78(3):871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heo HY, Zhang Y, Jiang S, Lee DH, Zhou J. Quantitative assessment of amide proton transfer (APT) and nuclear overhauser enhancement (NOE) imaging with extrapolated semisolid magnetization transfer reference (EMR) signals: II. Comparison of three EMR models and application to human brain glioma at 3 Tesla. Magn Reson Med 2016;75(4):1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heo HY, Zhang Y, Lee DH, Hong X, Zhou J. Quantitative assessment of amide proton transfer (APT) and nuclear overhauser enhancement (NOE) imaging with extrapolated semi-solid magnetization transfer reference (EMR) signals: Application to a rat glioma model at 4.7 Tesla. Magn Reson Med 2016;75(1):137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deshmane A, Zaiss M, Lindig T, Herz K, Schuppert M, Gandhi C, Bender B, Ernemann U, Scheffler K. 3D gradient echo snapshot CEST MRI with low power saturation for human studies at 3T. Magn Reson Med 2019;81(4):2412–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goerke S, Soehngen Y, Deshmane A, Zaiss M, Breitling J, Boyd PS, Herz K, Zimmermann F, Klika KD, Schlemmer HP, Paech D, Ladd ME, Bachert P. Relaxation-compensated APT and rNOE CEST-MRI of human brain tumors at 3 T. Magn Reson Med 2019;82(2):622–632. [DOI] [PubMed] [Google Scholar]
- 44.Chen L, Barker PB, Weiss RG, van Zijl PCM, Xu J. Creatine and phosphocreatine mapping of mouse skeletal muscle by a polynomial and Lorentzian line-shape fitting CEST method. Magn Reson Med 2019;81(1):69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L, Zeng H, Xu X, Yadav NN, Cai S, Puts NA, Barker PB, Li T, Weiss RG, van Zijl PCM, Xu J. Investigation of the contribution of total creatine to the CEST Z-spectrum of brain using a knockout mouse model. NMR Biomed 2017;30(12):e3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dixon WT, Hancu I, Ratnakar SJ, Sherry AD, Lenkinski RE, Alsop DC. A multislice gradient echo pulse sequence for CEST imaging. Magn Reson Med 2010;63(1):253–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du J, Takahashi AM, Bydder M, Chung CB, Bydder GM. Ultrashort TE imaging with off-resonance saturation contrast (UTE-OSC). Magn Reson Med 2009;62(2):527–531. [DOI] [PubMed] [Google Scholar]
- 48.Du J, Ma G, Li S, Carl M, Szeverenyi NM, VandenBerg S, Corey-Bloom J, Bydder GM. Ultrashort echo time (UTE) magnetic resonance imaging of the short T2 components in white matter of the brain using a clinical 3T scanner. Neuroimage 2014;87:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang J, Han X, Chen L, Xu X, Xu J, Chan KWY. Relayed nuclear Overhauser enhancement imaging with magnetization transfer contrast suppression at 3 T. Magn Reson Med 2020. [DOI] [PubMed] [Google Scholar]
- 50.Jiang S, Zou T, Eberhart CG, Villalobos MAV, Heo HY, Zhang Y, Wang Y, Wang X, Yu H, Du Y, van Zijl PCM, Wen Z, Zhou J. Predicting IDH mutation status in grade II gliomas using amide proton transfer-weighted (APTw) MRI. Magn Reson Med 2017;78(3):1100–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Togao O, Keupp J, Hiwatashi A, Yamashita K, Kikuchi K, Yoneyama M, Honda H. Amide proton transfer imaging of brain tumors using a self-corrected 3D fast spin-echo dixon method: Comparison With separate B0 correction. Magn Reson Med 2017;77(6):2272–2279. [DOI] [PubMed] [Google Scholar]
- 52.Gai N, Turkbey EB, Nazarian S, van der Geest RJ, Liu CY, Lima JA, Bluemke DA. T1 mapping of the gadolinium-enhanced myocardium: adjustment for factors affecting interpatient comparison. Magn Reson Med 2011;65(5):1407–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nehrke K, Bornert P. DREAM--a novel approach for robust, ultrafast, multislice B(1) mapping. Magn Reson Med 2012;68(5):1517–1526. [DOI] [PubMed] [Google Scholar]
- 54.Trott O, Palmer AG. R 1ρ relaxation outside of the fast-exchange limit. J Magn Reson 2002;154(1):157–160. [DOI] [PubMed] [Google Scholar]
- 55.Jin T, Autio J, Obata T, Kim SG. Spin-locking versus chemical exchange saturation transfer MRI for investigating chemical exchange process between water and labile metabolite protons. Magn Reson Med 2011;65(5):1448–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zaiss M, Bachert P. Chemical exchange saturation transfer (CEST) and MR Z -spectroscopy in vivo : a review of theoretical approaches and methods. Phys Med Biol 2013;58(22):R221. [DOI] [PubMed] [Google Scholar]
- 57.Zaiss M, Bachert P. Exchange-dependent relaxation in the rotating frame for slow and intermediate exchange -- modeling off-resonant spin-lock and chemical exchange saturation transfer. NMR Biomed 2013;26(5):507–518. [DOI] [PubMed] [Google Scholar]
- 58.McGraw KO, Wong SP. Forming inferences about some intraclass correlation coefficients. Psychological Methods 1996;1(1):30–46. [Google Scholar]
- 59.Jones CK, Huang A, Xu J, Edden RA, Schar M, Hua J, Oskolkov N, Zaca D, Zhou J, McMahon MT, Pillai JJ, van Zijl PC. Nuclear Overhauser enhancement (NOE) imaging in the human brain at 7T. NeuroImage 2013;77C:114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou Y, van Zijl PCM, Xu J, Yadav NN. Mechanism and quantitative assessment of saturation transfer for water-based detection of the aliphatic protons in carbohydrate polymers. Magn Reson Med 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jin T, Wang P, Hitchens TK, Kim SG. Enhancing sensitivity of pH-weighted MRI with combination of amide and guanidyl CEST. Neuroimage 2017;157:341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stanisz GJ, Kecojevic A, Bronskill MJ, Henkelman RM. Characterizing white matter with magnetization transfer and T(2). Magn Reson Med 1999;42(6):1128–1136. [DOI] [PubMed] [Google Scholar]
- 63.Zhang XY, Xie J, Wang F, Lin EC, Xu J, Gochberg DF, Gore JC, Zu Z. Assignment of the molecular origins of CEST signals at 2 ppm in rat brain. Magn Reson Med 2017;78(3):881–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin T, Wang P, Zong X, Kim S-G. MR imaging of the amide-proton transfer effect and the pH-insensitive nuclear overhauser effect at 9.4 T. Magn Reson Med 2013;69(3):760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou IY, Lu D, Ji Y, Wu L, Wang E, Cheung JS, Zhang XA, Sun PZ. Determination of multipool contributions to endogenous amide proton transfer effects in global ischemia with high spectral resolution in vivo chemical exchange saturation transfer MRI. Magn Reson Med 2019;81(1):645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krishna MM, Hoang L, Lin Y, Englander SW. Hydrogen exchange methods to study protein folding. Methods 2004;34(1):51–64. [DOI] [PubMed] [Google Scholar]
- 67.Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys 1983;16(4):521–655. [DOI] [PubMed] [Google Scholar]
- 68.Xu J, Yadav NN, Bar-Shir A, Jones CK, Chan KW, Zhang J, Walczak P, McMahon MT, van Zijl PC. Variable delay multi-pulse train for fast chemical exchange saturation transfer and relayed-nuclear overhauser enhancement MRI. Magn Reson Med 2014;71(5):1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Orzylowska A, Slowik T, Chudzik A, Pankowska A, Lam W, Stanisz G. The CLARITY procedure of lipid removal from brain tissue sample reveals the lipid-origin of MT contrast in CEST imaging experiment. 2020; Proceedings of the 28th Annual Meeting of ISMRM, Virtual Meeting. p 0497 [Google Scholar]
- 70.Zhou J, Payen JF, Wilson DA, Traystman RJ, van Zijl PC. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nat Med 2003;9(8):1085–1090. [DOI] [PubMed] [Google Scholar]
- 71.Zhou J, Tryggestad E, Wen Z, Lal B, Zhou T, Grossman R, Wang S, Yan K, Fu DX, Ford E, Tyler B, Blakeley J, Laterra J, van Zijl PC. Differentiation between glioma and radiation necrosis using molecular magnetic resonance imaging of endogenous proteins and peptides. Nat Med 2011;17(1):130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harris RJ, Cloughesy TF, Liau LM, Nghiemphu PL, Lai A, Pope WB, Ellingson BM. Simulation, phantom validation, and clinical evaluation of fast pH-weighted molecular imaging using amine chemical exchange saturation transfer echo planar imaging (CEST-EPI) in glioma at 3 T. NMR Biomed 2016;29(11):1563–1576. [DOI] [PubMed] [Google Scholar]
- 73.Kim B, So S, Park H. Optimization of steady-state pulsed CEST imaging for amide proton transfer at 3T MRI. Magn Reson Med 2019;81(6):3616–3627. [DOI] [PubMed] [Google Scholar]
- 74.Song X, Xu J, Xia S, Yadav NN, Lal B, Laterra J, Bulte JW, van Zijl PC, McMahon MT. Multi-echo length and offset VARied saturation (MeLOVARS) method for improved CEST imaging. Magn Reson Med 2015;73(2):488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu H, Jones CK, van Zijl PC, Barker PB, Zhou J. Fast 3D chemical exchange saturation transfer (CEST) imaging of the human brain. Magn Reson Med 2010;64(3):638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones CK, Polders D, Hua J, Zhu H, Hoogduin HJ, Zhou J, Luijten P, van Zijl PC. In vivo three-dimensional whole-brain pulsed steady-state chemical exchange saturation transfer at 7 T. Magn Reson Med 2012;67(6):1579–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zaiss M, Ehses P, Scheffler K. Snapshot-CEST: Optimizing spiral-centric-reordered gradient echo acquisition for fast and robust 3D CEST MRI at 9.4 T. NMR Biomed 2018;31(4):e3879. [DOI] [PubMed] [Google Scholar]
- 78.Alsop DC, Detre JA, Golay X, Gunther M, Hendrikse J, Hernandez-Garcia L, Lu H, Macintosh BJ, Parkes LM, Smits M, van Osch MJ, Wang DJ, Wong EC, Zaharchuk G. Recommended implementation of arterial spin-labeled perfusion MRI for clinical applications: A consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia. Magn Reson Med 2014; 10.1002/mrm.25197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu X, Yadav NN, Zeng HF, Jones CK, Zhou JY, van Zijl PCM, Xu JD. Magnetization Transfer Contrast-Suppressed Imaging of Amide Proton Transfer and Relayed Nuclear Overhauser Enhancement Chemical Exchange Saturation Transfer Effects in the Human Brain at 7T. Magn Reson Med 2016;75(1):88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1: Simulation of the Z-spectrum for each slice in the starCEST to demonstrate the impact of the non-steady-state character of the saturation. Only the first four slices are shown for clarity. In the simulation, the 50 ms since-Gauss saturation pulses and 53 ms mixing times were applied. The frequency was swept from 6 ppm to 1 ppm. T1=1s, T2=80 ms were set for the water pool, while T1=1s, T2=80 ms, exchange rate 160 Hz, offset 3.5 ppm and concentration 500 mM were assumed for the amide proton pool. The non-steady-state saturation significantly impacts the Z values in the slices 1–3 at the initial offset (6 ppm) and also shift the Z-spectrum of the first slice by 0.03 ppm.
Supporting Information Figure S2: (a) B0 map recorded with a dual-echo sequence. Comparison between the amideCEST maps extracted using the PLOF method without (b) and with (c) B0 correction. Hyper/hypo intensity regions in the amideCEST maps are indicated with green circles and are due to the high B0 shifts (> 0.3 ppm).
Supporting Information Figure S3: Comparison of the amideCEST maps without and with MLSVD post-processing. (a) The original amide CEST maps extracted using PLOF without MLSVD denoising. (b) MLSVD denoising applied with truncation numbers 48, 48 and 10.


