Abstract
Focal segmental glomerulosclerosis (FSGS) is a podocytopathy leading to kidney failure, whose molecular cause frequently remains unresolved. Here, we describe a rare MYO9A loss of function nonsense heterozygous mutation (p.Arg701*) as a possible contributor to disease in a sibling pair with familial FSGS/proteinuria. MYO9A variants of uncertain significance were identified by whole exome sequencing in a cohort of 94 biopsy proven patients with FSGS. MYO9A is an unconventional myosin with a Rho-GAP domain that controls epithelial cell junction assembly, crosslinks and bundles actin and deactivates the small GTPase protein encoded by the RHOA gene. RhoA activity is associated with cytoskeleton regulation of actin stress fiber formation and actomyosin contractility. Myo9A was detected in mouse and human podocytes in vitro and in vivo. Knockin mice carrying the p.Arg701* MYO9A (Myo9AR701X) generated by gene editing develop proteinuria, podocyte effacement and FSGS. Kidneys and podocytes from Myo9AR701X/+ mutant mice revealed Myo9A haploinsufficiency, increased RhoA activity, decreased Myo9A-actin-calmodulin interaction, impaired podocyte attachment and migration. Our results indicate that Myo9A is a novel component of the podocyte cytoskeletal apparatus that regulates RhoA activity and podocyte function. Thus, Myo9AR701X/+ knock-in mice recapitulate the proband FSGS phenotype, demonstrate that p.R701X Myo9A is a FSGS-causing mutation in mice and suggest that heterozygous loss-of-function MYO9A mutations may cause a novel form of human autosomal dominant FSGS. Hence, identification of MYO9A pathogenic variants in additional individuals with familial or sporadic FSGS is needed to ascertain the gene contribution to disease.
Keywords: MYO9A variants, FSGS, RhoA activity, Myo9A-actin-calmodulin interaction
Graphical Abstract
INTRODUCTION
Focal segmental glomerulosclerosis (FSGS) is a histological lesion that can lead to end-stage renal disease.1 Genetic studies demonstrate that FSGS is largely a podocytopathy, as podocytes express most of the causative mutant genes.2,3 However, the molecular cause of FSGS is unknown in over half the cases.3 Thus, identification of additional FSGS-causing genes and their molecular pathogenesis is clinically relevant. Here we identify MYO9A as a novel candidate gene for human FSGS by whole exome sequencing (WES). This study overall goal is to determine if the rare MYO9A variant identified in patients alters podocyte function and causes FSGS in vivo.
MYO9A encodes an unconventional, non-muscle myosin not previously associated with human renal disease.4 MYO9A mutations were reported in arthrogryposis and congenital myasthenic syndrome patients.5,6 Myosins are a family of molecular motors that bind actin to generate force and movement and serve multiple cell functions, including motility and shape regulation, organelle trafficking and signaling.7,8,9 MyosinIIA (MY9H) and myosin1E (MYO1E) mutations have been reported to cause glomerular disease and autosomal recessive FSGS.10,11,12 Class-9 myosins comprise MYO9A and MYO9B that are expressed by epithelia and immune cells, respectively.9,13 Their distinctive features are a processive single head that moves on F-actin and a Rho-GAP tail domain that inactivates RhoA. Myo9A mRNA is expressed in brain ependymal epithelium, in testis, lung and kidney epithelia.4 Myo9A KO mice develop progressive hydrocephalus, mild hydronephrosis and tubulopathy.14,15 In vitro Myo9A controls epithelial cell junction assembly and collective cell migration.9 Importantly, Myo9A is sufficient to crosslink and bundle actin filaments inducing the formation of actin networks.16
RhoA modulates podocyte actin dynamics and maintains glomerular permeability.17 Perturbations of Rho-GTPase activity lead to glomerular disease (reviewed in18). Mutations of downstream effectors (INF2)19 and regulators of Rho-GTPases GDI (ARHGDIA)3 and RhoGAP (ARHGAP24)20 cause proteinuria and FSGS. Thus, fine-tuning of Rho-GTPase activity is critical to maintain a normal podocyte phenotype in vivo.18
Here we report that MYO9A is a novel podocyte protein, which when mutated in mice leads to proteinuria, foot process effacement and FSGS, replicating the human phenotype associated with p.Arg701* MYO9A variant. Our findings uncover Myo9A relevance in podocytes, demonstrate that Myo9A regulates podocyte RhoA and provide the first evidence that p.Arg701* Myo9A causes podocyte dysfunction by failing to negatively regulate RhoA and reducing Myo9A-actin-calmodulin interaction.
METHODS
WES
WES of DNA samples from two affected siblings and 94 FSGS-CT (ClinicalTrials.gov, NCT00135811) cohort subjects was performed at YCGA and Yale DNA Clinical lab on Illumina platforms.21–26 The FSGS-CT cohort WES data set was interrogated to identify MYO9A variants with minor allele frequency (MAF) cut off ≤0.01% for heterozygous variants and ≤0.1% for homozygous and compound heterozygous variants, assessed using bioinformatics metaSVM27, SIFT28 and CADD29 scores. MYO9A variants were confirmed by Sanger sequencing. A gene panel of 64 FSGS-causing genes based on OMIM database (omim.org) and literature associated with ‘FSGS’, ‘glomerular proteinuria’, ‘nephrotic syndrome’ were used to identify relevant variants.
Generation of knock-in Myo9AR701X mice
We generated Myo9R701X knock-in mice by CRISPR/Cas9-mediated gene editing.30 Complete methods are included in Supplementary Material. Myo9AR701X knock-in and wild type littermates, male and female, were used following Yale Animal Research Center Animal Care and Use Committee approved protocols.
Histology/ IF
Human kidney biopsy and mouse kidneys were processed for histology, IF and TEM as described.31,32 IF was performed in mouse kidney frozen sections and cultured podocytes. Primary antibodies were mouse anti-MYO9A (clone 4C11, Abnova), rabbit anti-podocin (P0372, Sigma), rabbit anti-laminin (L9393, Sigma), rabbit anti-aquaporin2 (PAS-78809, Invitrogen), rabbit anti-megalin33; secondary antibodies were Alexa Fluor-488 and 594-conjugated. Images were acquired using Olympus IX71 or Leica-SP5-Spectral confocal microscope.
Immunoblot/ Immunoprecipitation
Proteins were resolved by SDS-PAGE and immunoblotted as previously described31,32, with anti-MYO9A (clone 4C11), actin (A2066, Sigma), calmodulin (EP799Y, Abcam), GAPDH (6C5, Millipore), RhoA (67B9, Cell signaling) primary antibodies. Co-immunoprecipitation was performed as described34,35, with anti-MYO9A rabbit polyclonal antibody (A305–702A-M, Bethyl), followed by immunoblotting.
qPCR
Total RNA was reverse transcribed using iScript cDNA Synthesis kit (Bio-Rad), amplified with iQ SYBR Green Supermix (BioRad) and Myo9A PCR primers: 5’TCTGAGAAGTAGCAGGAATGC-3’ and 5’-AGCAACCACAGCTCTGAAA-3’ using CFX96-Touch-Real-Time-PCR (Bio-Rad). Myo9a mRNA was normalized to GAPDH mRNA with the 2-ΔΔCt method, as described.36
TEM/Immuno-Electron Microscopy
TEM was performed as described.32 For Immuno-gold EM wild type mouse kidney was fixed, ultrathin cryosections were quenched, blocked and incubated with mouse anti-MYO9A (clone 4C11) primary antibody. Secondary antibody was rabbit anti-mouse IgG, the gold conjugate was goat-anti-rabbit-10nm gold. Sections were post-fixed, stained and examined on FEI Tecnai Biotwin (LaB6, 80 kV) TEM.
Primary podocyte isolation and functional assays
Podocytes were isolated from wild type and Myo9AR701X/+ kidneys as described34 with few modifications.37,38 Adhesion assays were performed as described and quantified using crystal violet staining.37
Myo9A knockdown
Myo9A knockdown was induced in wild type immortalized mouse podocytes using Myo9A siRNA (L-006539-ON-TARGETplus, Dharmacon) following manufacturer’s instructions. Controls were untransfected podocytes and podocytes transfected with non-targeting siRNA (D-001810–10-ON-TARGETplus Non-Targeting-control, Dharmacon).
RhoA activity
Active RhoA was measured with Rho-activation pulldown assay (Millipore) following manufacturer’s instructions.39 Active RhoA was detected by immunoblotting.40
Statistical analysis
Data were analysed using GraphPad-Prism-8 software (San Diego, CA) with unpaired Student’s-t-test with Welch’s correction or Welch’s ANOVA, as appropriate. P<0.05 was deemed statistically significant.
RESULTS
Identification of MYO9A variants in patients with proteinuria and FSGS
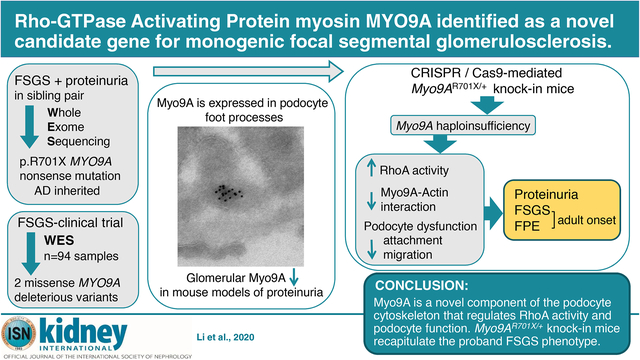
WES21 identified a rare truncating MYO9A mutation in two siblings diagnosed with FSGS/proteinuria at 15 and 14 years of age (Figure 1A), presenting with asymptomatic proteinuria ~1–2 g/day, normal blood pressure, normal renal ultrasounds, and eGFR of 72 and 96 ml/min/1.73m2, respectively. Renal biopsy performed on the index case showed FSGS, mild interstitial fibrosis and no tubular abnormalities (Figure 1B). The proband’s disease progressed over 3 years: eGFR decreased to 46 ml/min/1.73m2 and proteinuria >2g/day while his sister’s eGFR and proteinuria remained stable. Relevant family history includes a maternal uncle with chronic kidney disease of unknown etiology, mother with history of proteinuria and healthy father, and no consanguinity (Figure 1C). WES revealed a shared nonsense heterozygous MYO9A (NM_006901 c.2101C>T, p.Arg701*) in the siblings (Figure 1A). This rare MYO9A variant was identified in 1/113 632 alleles tested in the genome aggregation database (http://gnomad.broadinstitute.org, accessed November 7, 2020), allele frequency of 8.8e-6. The parents’ genotype revealed that only the mother is heterozygous for c.2101 C>T, p.Arg701* MYO9A variant, indicating autosomal dominant inheritance of the mutation (Figure 1C). Genotype from the maternal uncle with chronic kidney disease is not available. MYO9A p.Arg701* (p.R701X) is expected to result in loss-of-function due to nonsense-mediated mRNA decay (NMD) or to truncation in the loop 2 of MYO9A head domain (Figure 1D) that is required for Myo9A to crosslink actin.16 Table S1 shows all variants shared by the siblings. No mutations in FSGS or nephrotic syndrome-causing genes reported to date were detected in either sibling (Table S2). To our knowledge, this is the first report linking loss-of-function MYO9A mutation to a human renal disease.
Figure 1. Heterozygous nonsense mutation in MYO9A in a family with FSGS/proteinuria.
A) novel heterozygous single-bp mutation that leads to a stop codon in exon 14 of MYO9A c.C2101T: p.R701X (p.Arg701*) in a patient with proteinuria and FSGS. Nucleotide sequence traces show the C>T mutation shared by proband, sister and mother (top, indicated by asterisk) and wild type sequence from the father (bottom), based on human MYO9A reference sequence NM_006901.4. B) Kidney biopsy from the proband shows focal segmental glomerular sclerosis, mild interstitial infiltrate (PAS stain). C) Family pedigree shows genotype-phenotype segregation: individuals with proteinuria/FSGS and MYO9Ap.R701X variant (solid symbols), wild type (empty symbol), (#) indicates unknown MYO9A genotype. D) Wild type MYO9A (NP_008832.2) protein domains: motor domain (grey) includes loop 2 (yellow) and calmodulin binding site (red), neck domain (blue), tail domain (pink) including RhoGAP domain (green); domain boundaries are indicated by amino acid residue number; predicted p.R701X MYO9A terminates within calmodulin binding site in loop 2, lacking the neck and tail domains, asterisks indicate the location of missense MYO9A VUS identified. E) Evolutionary conservation of amino acid residues altered by MYO9A missense mutations identified in FSGS patients: MYO9A altered residues p.D156 (p.Asp156) and p.E765 (p.Glu765) (red arrows) are highly conserved through phylogeny. Accession numbers: H. sapiens (NP_008832.2), M. musculus (NP_766606.2), R. norvegicus (NP_599162.1), G. gallus (XP_015134553.1), D. rerio (E7EZG2.1), X. tropicalis (XP_002934596.2) were used for MYO9A multiple sequence alignment with Clustal Omega-EMBL-EBI (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Analysis of WES data from 94 FSGS patients (CT-FSGS, ClinicalTrials.gov, NCT00135811)41 identified 5 unrelated individuals with heterozygous MYO9A missense variants (Table 1) meeting a minor allele frequency (MAF) cutoff ≤ 0.01%; two of these individuals’ MYO9A variants, p.Asp156Gly and p.Glu765Asp (Figure 1D) are predicted to be deleterious by at least two bioinformatics scores: metaSVM, SIFT and CADD>18;27–29 the other three missense variants are deemed benign based on three bioinformatics prediction scores. The amino acids substituted in p.Asp156Gly and p.Glu765Asp variants are likely to have a relevant function since these residues are conserved through evolution (Figure 1E). We also identified one patient carrying a MYO9A variant (p.Arg2283His) that did not meet our MAF criteria previously reported in congenital myasthenia syndrome.6 No pathogenic mutations or deleterious variants in FSGS/NS/ proteinuria causing genes (Table S2) were identified in the 5 individuals with MYO9A variants.
Table 1:
Missense MYO9A variants identified in FSGS-CT patient cohort (NCT00135811)
| Sample | MYO9A variant | Pathogenicity prediction scores | gnomAD Freq. (> 240 000 alleles) | ||
|---|---|---|---|---|---|
| MetaSVM | SIFT | CADD | |||
| 1 | MYO9A:NM_006901:exon2:c.A467G:p.D156G | Deleterious | Damaging (0.003) | >18 | 0.0001 |
| 2 | MYO9A:NM_006901:exon15:c.G2295C:p.E765D | Not deleterious | Damaging (0.003) | >18 | 3.66E-05 |
| 3 | MYO9A:NM_006901:exon7:c.C1166T:p.T389M | Not deleterious | Benign | <10 | 1.22E-05 |
| 4 | MYO9A:NM_006901:exon26:c.C5110T:p.P1704S | Not deleterious | Benign | <10 | 8.94E-05 |
| 5 | MYO9A:NM_006901:exon41:c.A7150G:p.M2384V | Not deleterious | Benign | <10 | 0.0001 |
Variants filtered at minor allele frequency ≤ 0.01%, n=94 samples following ANNOVAR annotation; p.D156G = p.Asp156Gly; p.E765D = Glu765Asp; p.T389M = Thr389Met; p.P1704S = p.Pro1704Ser; p.M2384V = p.Met2384Val
MYO9A expression in podocytes and glomerular disease
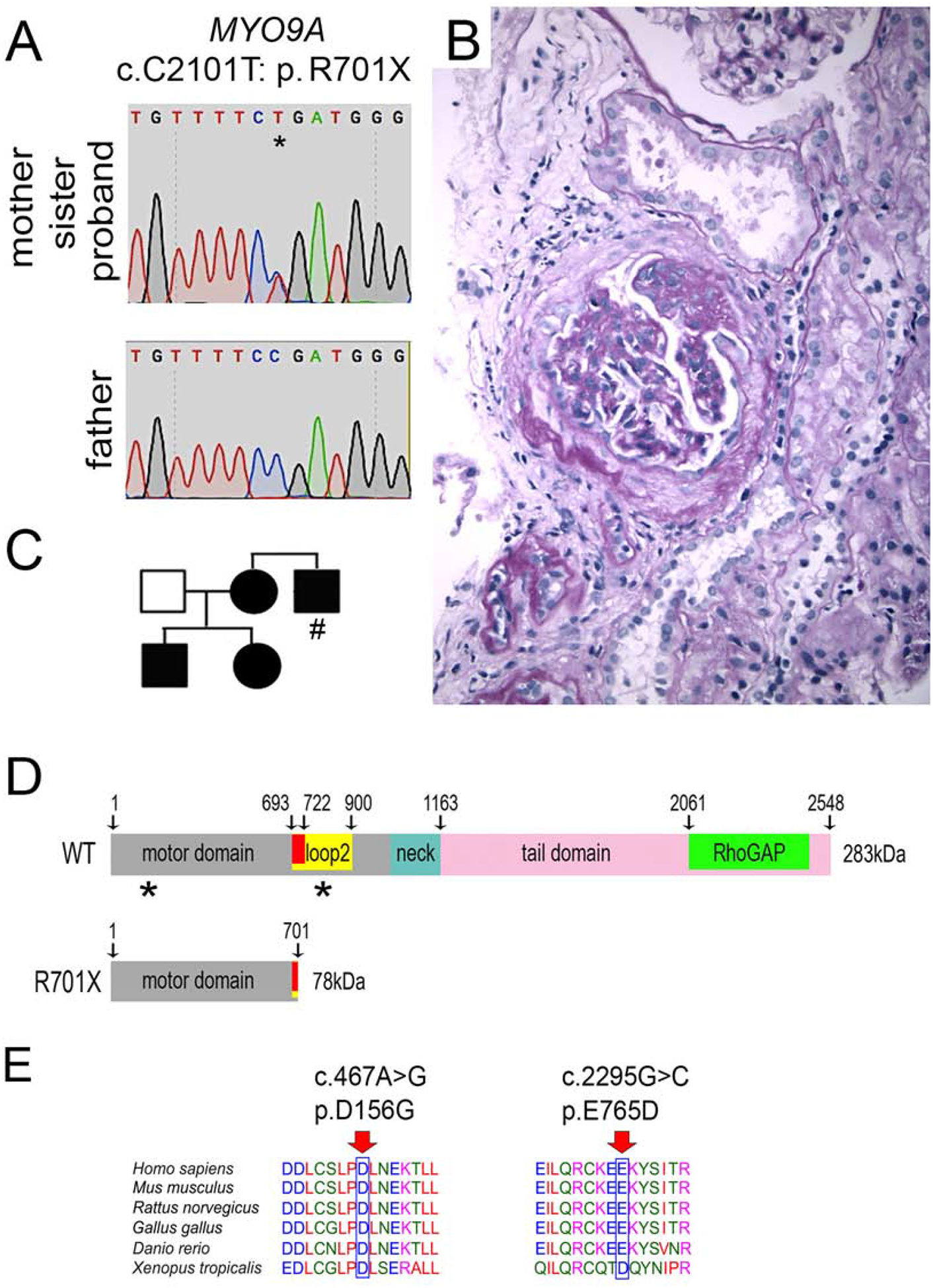
Myo9A protein expression was detected by immunoblotting in mouse kidneys, mouse and human podocytes (Figure 2A). Immunocytochemistry confirmed cytoplasmic expression of MYO9A in human podocytes (Figure 2B). Immunoreactive Myo9A is detected by fluorescence microscopy (IF) in mouse glomeruli, where it partially co-localizes with podocin (Figure 2C). Double labeling and confocal microscopy delineate Myo9A localization along the external aspect of the GBM tightly contiguous or co-localized with podocin (Figure 2D) and laminin (Figure 2E). Immuno-gold electron microscopy demonstrates Myo9A precise localization. Gold particles are distributed in the foot process cytoplasm and podocyte cell body (Figure 2F and G, Supplementary Figure S1B and S1D). Slit-diaphragms are uniformly unlabeled. Mesangial and endothelial cells show no significant labeling. Gold particles are also detected in proximal tubule cells (Supplementary Figure S1C).
Figure 2: Glomerular and podocyte Myo9A expression.
A) Immunoblots show Myo9A in mouse kidney, immortalized mouse podocytes (1), undifferentiated (2) and differentiated (3) human podocytes; B) Immuno-fluorescence microscopy (IF) shows MYO9A in the cytoplasm of undifferentiated and differentiated human podocytes (green), partially co-localized with F-actin (red) in differentiated podocytes (yellow), scale bar=10μm; C) IF shows Myo9A (red) and podocin (green) partially co-localized in mouse glomeruli (merge=yellow), scale bars=50μm; D) Dual immunolabeling and confocal microscopy show Myo9A staining (red) on podocytes tightly contiguous to podocin (green) on its GBM side (arrowheads), insets show higher magnification; scale bar = 25μm. E) Confocal microscopy shows Myo9A staining closely contiguous or co-localized with laminin (green) along the outer aspect of the GBM (insets and arrowhead); scale bar = 25μm. F an G) Immuno-EM localization of Myo9A in glomeruli of wild type mouse kidneys: gold particles (black arrows) are observed in the podocyte cell body (pod) and the foot processes cytoplasm (fp), some gold particles are seen at the fp base next to the GBM but not at slit diaphragms; scale bars=500 nm.
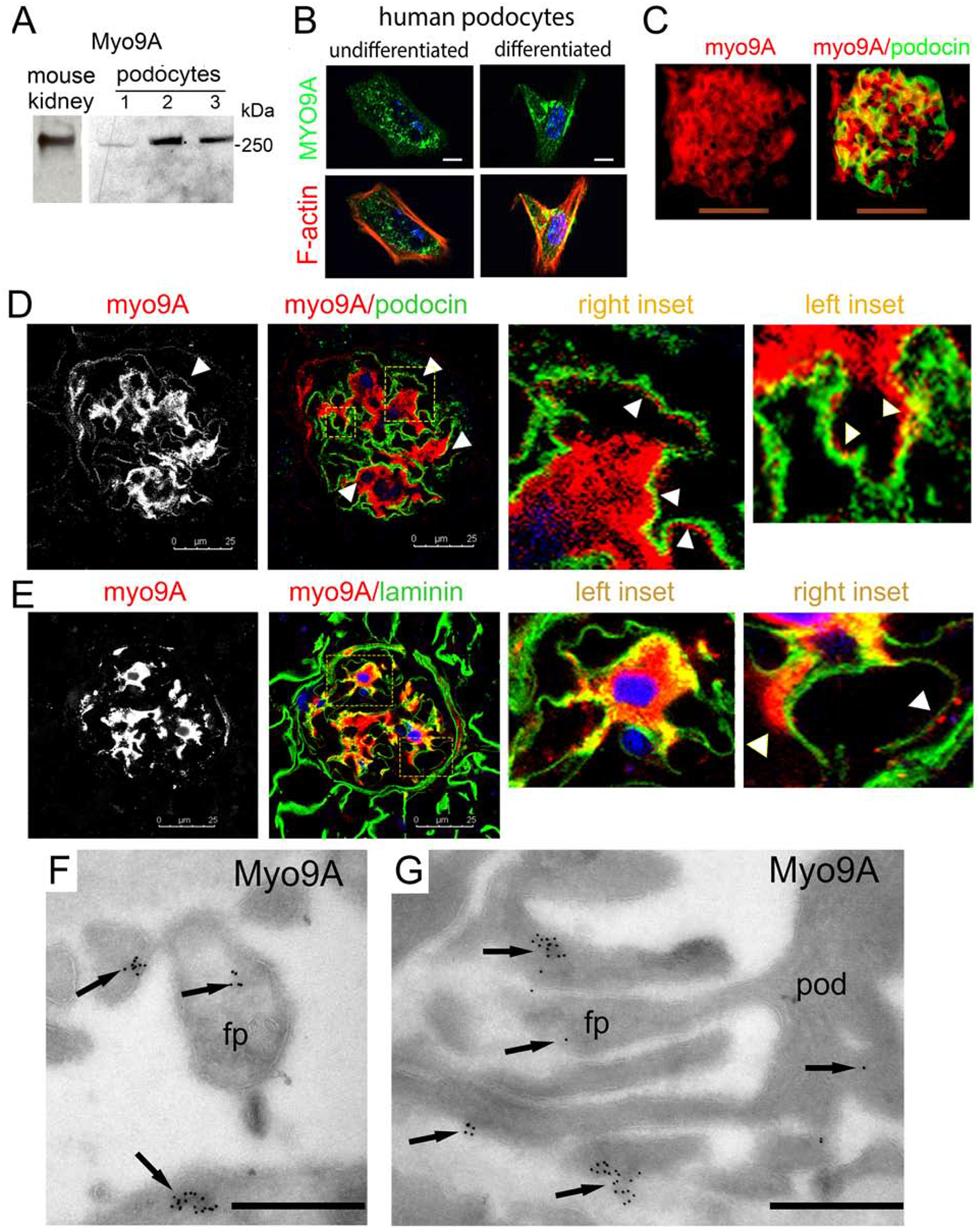
To evaluate whether Myo9A expression is disrupted in glomerular diseases we examined two mouse models associated to nephrotic range proteinuria.31,32 Developing mice with nephrotic syndrome induced by VEGF-A overexpression31 show decreased glomerular Myo9A (Figure 3A). Similarly, glomerular Myo9A is decreased in mice with streptozotocin-mediated advanced diabetic glomerulosclerosis induced by VEGF-A overexpression32, as compared to Myo9A detected in diabetic mice not overexpressing VEGF-A (Figure 3B) and non-diabetic mice (Figure 2C). Myo9A podocyte foot process localization and decreased glomerular Myo9A in both proteinuric disease models suggest that Myo9A may play a role in glomerular filtration barrier function.
Figure 3: Glomerular Myo9A expression in glomerular disease mouse models.
A) developmental nephrotic syndrome induced by VEGF-A overexpression:27 IF shows Myo9A (red) in glomeruli from normal, uninduced 2 week old mice and clearly decreased Myo9A expression in glomeruli from nephrotic mice overexpressing VEGF-A, while podocin (green) is unchanged. B) Streptozotocin-induced diabetes with or without VEGF-A overexpression:26 IF detects Myo9A (green) in glomeruli from uninduced diabetic kidneys (DM control) and markedly decreased immunoreactive Myo9A in glomeruli from diabetic mice overexpressing VEGF-A that develop diabetic glomerulosclerosis (advanced DN), Dapi (blue) shows nuclei from both sections with outlined glomeruli (hatched white line). Scale bars= 50μm.
Gene edited MYO9AR701X mice develop FSGS
To determine whether p.Arg701*(p.R701X) MYO9A nonsense mutation causes glomerular disease in mice we generated knock-in p.Arg701* (herein called Myo9AR701X) mice by CRISPR/Cas9-mediated gene editing.30 Heterozygous Myo9AR701X/+ mutant mice (Figure 4A) yield viable mutant pups in Mendelian fashion, demonstrating that the mutation is not embryonic lethal. Homozygous mutants develop progressive hydrocephalus (Supplementary Figure S2), ataxia and seizures and almost all die before 3 weeks of age, reminiscent of Myo9A-null mice.14 Heterozygous Myo9AR701X/+ mutants have no obvious phenotype at birth, are fertile and survive to adulthood.
Figure 4: Genotype and phenotype analysis of knock-in Myo9AR701X mice at 2 weeks of age.
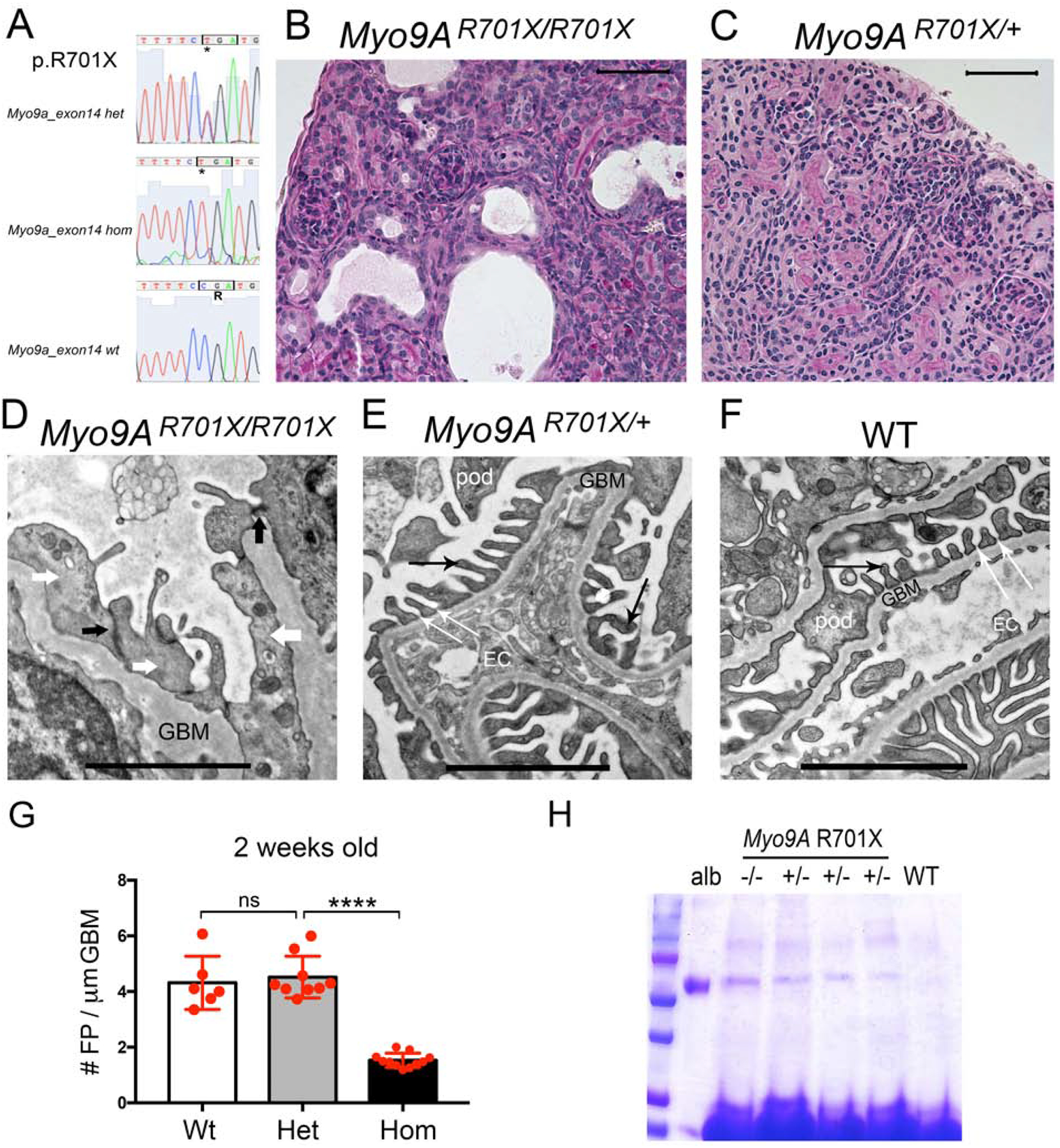
A) Sanger sequencing traces and deduced amino acid sequence from exon 14 are shown from mutant mice: heterozygous Myo9AR701X/+ (top), homozygous Myo9AR701X/R701X (middle) and wild type littermate (bottom); C>T mutation substitutes CGA encoding Arg (R) by TGA, a stop codon that predicts termination of translation at amino acid 701. B) Representative PAS stain from 2-week-old homozygous Myo9AR701X/R701X mutant kidney shows abnormal glomeruli, tubular dilatation and protein casts. C) Representative PAS stain from 2-week-old heterozygous Myo9AR701X/+ kidney shows normal histology. D) Representative TEM from 2-week-old Myo9AR701X/R701X glomerulus shows massive foot process effacement (white arrows), absence of slit-diaphragms, replaced by adherens junctions (black arrows) and thickened, irregular GBM. E) Representative TEM from 2-week-old heterozygous Myo9AR701X/+ glomerulus shows normal foot processes (black thin arrows), and normal slit-diaphragms (white thin arrows). F) TEM from 2-week-old wild type littermate (WT) shows normal glomerular filtration barrier: thin foot processes linked by slit-diaphragms, fenestrated endothelial cells and thin GBM. G) Quantitation of foot process effacement at 2 weeks of age: homozygous Myo9AR701X/R701X kidneys show less than half foot processes/μm GBM than willd type or heterozygous Myo9AR701X/+ littermates, indicating severe FPE , P<0.0001 (ANOVA). H) Coomassie blue stain of SDS-PAGE resolved urine samples shows albuminuria in both Myo9AR701X/R701X (−/−) and Myo9AR701X/+ (+/−), while WT urine is albumin free. BSA (10μg) serves as MW control. Scale bars: A and B= 20μm; C, D and E= 1μm.
All Myo9AR701X mutant mice develop foot process effacement (FPE) and proteinuria, albeit at different ages. Homozygous Myo9AR701X/R701X mutants show dysmorphic glomeruli, tubular dilatation and protein casts on light microscopy at 2 weeks of age (Figure 4B and Supplementary Figure S3A). Transmission electron microscopy (TEM) of 2-week-old Myo9AR701X/R701X mutant kidneys reveals extensive FPE, thick and irregular glomerular basement membrane (GBM) and lack of slit-diaphragms, replaced by adherens junctions (Figure 4D). In contrast, 2-week-old heterozygous Myo9AR701X/+ mutants show normal renal histology and glomerular filtration barrier ultrastructure (Figure 4C and E), indistinguishable from wild-type littermates (Figure 4F), as confirmed by quantitation of foot processes (FP/μm GBM, Figure 4G). Albuminuria is detected in urine samples from both heterozygous and homozygous Myo9AR701X mutant mice by SDS-PAGE (Figure 4H) at 2 weeks of age.
At 4 months of age heterozygous Myo9AR701X/+ mice show focal and segmental glomerulosclerosis (FSGS) on light microscopy (Figure 5A and G) and TEM reveals FPE (white arrows), abnormal slit-diaphragms (short black arrows), irregular GBM and mesangial expansion (Figure 5B). Quantitation of foot processes shows significant FPE in Myo9AR701X/+ vs. wild-type adult kidneys (Figure 5C). This glomerular phenotype is remarkably similar to that observed in the renal biopsy of the proband (Figure 1B). Adult heterozygous Myo9AR701X/+ mutant mice have significantly higher urine albumin:creatinine ratios than age-matched wild-type mice (Figure 5D) and show no evidence of polyuria or weight loss (Figure 5E and F). On PAS examination adult Myo9AR701X/+ kidneys show FSGS of various severity (arrows, Figure 5G) and absence of tubular atrophy and dilatation or interstitial infiltrates. Dual labeling and confocal microscopy detects Myo9A in proximal tubules, peripherally to megalin (arrowheads, Figure 5H and K), whereas Myo9A is not detected in distal nephron segments expressing aquaporin2 (Figure 5H and I). Labeling of both tubular markers in adult Myo9AR701X/+ is indistinguishable from wild-type kidneys by confocal microscopy (Figure 5H and K; I and L). In contrast, Myo9A staining in proximal tubules and in glomeruli is decreased in Myo9AR701X/+ (Figure 5H and I) vs. wild-type kidneys (Figure 5K and L). Together these findings demonstrate that p.Arg701* is a FSGS-causing Myo9A mutation in mice, which recapitulates the phenotype and autosomal dominant inheritance of the heterozygous MYO9A p.Arg701* variant identified in the proband.
Figure 5: Adult heterozygous Myo9AR701X/+ knock-in mice phenotype analysis.
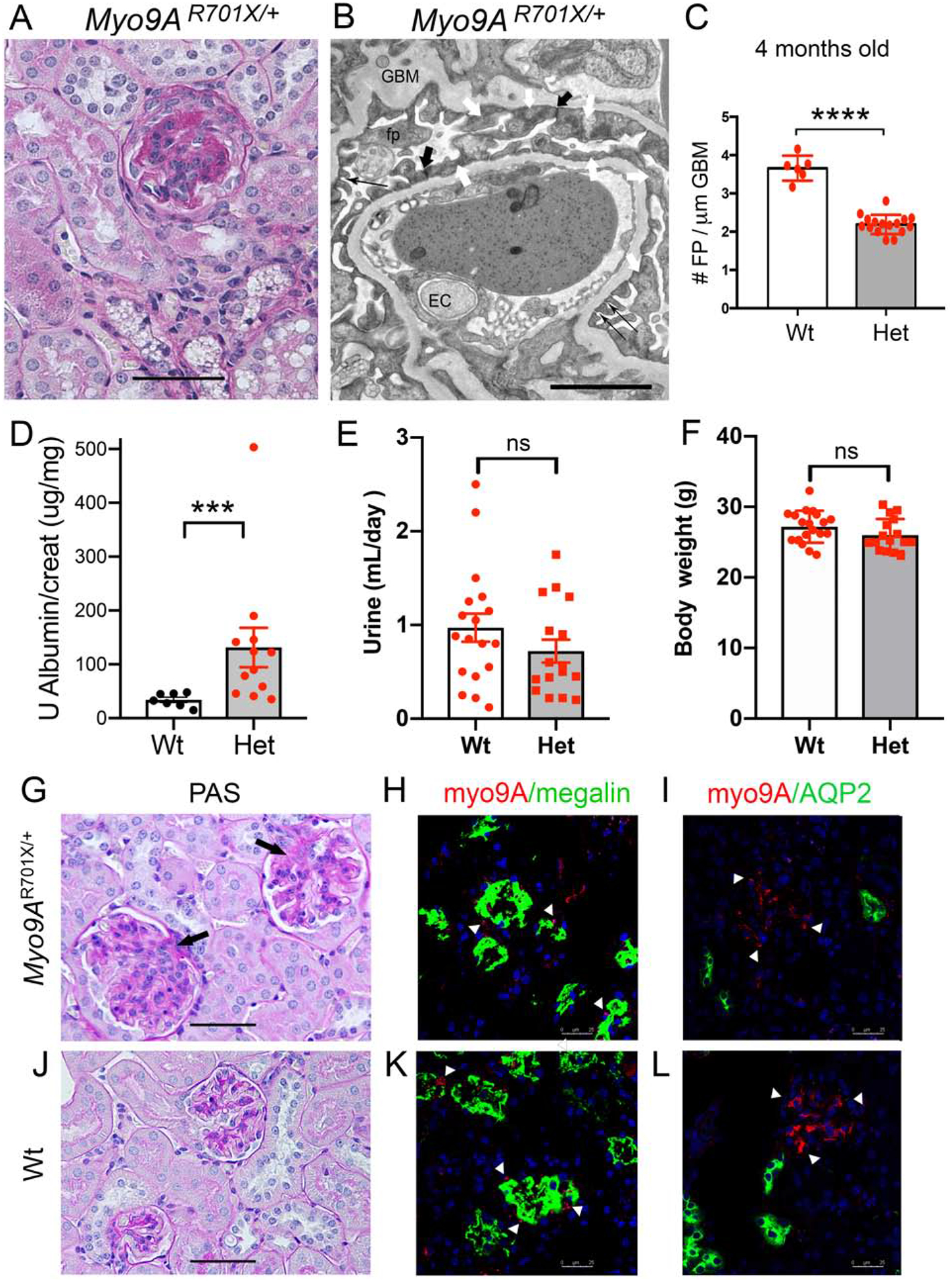
A) Representative PAS stain from 4 months old Myo9AR701X/+ kidney shows a glomerulus with extensive focal segmental glomerulosclerosis and mesangial proliferation. B) Representative TEM from 4 months old Myo9AR701X/+ kidney shows extensive foot process effacement (white arrows), partial replacement of slit-diaphragms (thin black arrows) by adherens junctions (thick black arrows). C) Quantitation of foot process effacement (FPE) in adult Myo9AR701X/+ mutant mice: 4 months of age heterozygous Myo9AR701X/+ show 2.2±0.06 FP/μm, as compared to wild type 3.7±0.13 FP/μm, **** indicates P<0.0001. D) Urine albumin/creatinine ratio at 4 months of age is higher in Myo9AR701X/+ (Het, red dots) than in wild type mice (Wt, black dots), *** indicates P>0.001. E) Urine volume (24 h) and F) body weight are not different in Wt (n=18) and Myo9AR701X/+ (Het, n=16) mice. G and J) Representative PAS stain from Myo9AR701X/+ (G) and Wt (J) kidneys: G) shows early focal sclerosis (arrows) and mesangial proliferation in 2 glomeruli, normal back to back tubules and no infiltrates; J) shows normal glomeruli and tubules in Wt kidney. H and K) Confocal microscopy of Myo9A/Megalin IF: H) Myo9AR701X/+ kidney shows limited Myo9A (red) signal (arrowheads) in proximal tubular cells labeled with megalin (green), proximal tubules (PT) are not dilated; K) Wt kidney shows more abundant Myo9A (arrowheads) contiguous to megalin in PT cells. I and L) Confocal microscopy of Myo9A/Aquaporin2 (AQP2) IF: I) Myo9AR701X/+ kidney shows limited Myo9A (red) in the glomerulus (arrowheads), Myo9A is not detected in distal nephron segments labeled with AQP2 (green), which are not dilated; L) Wt kidney shows Myo9A staining in glomerulus and AQP2 staining in distal nephron segments. Scale bars: A, G and J = 50μm; B= 2μm; H, I, K and L = 25μm.
How does p.Arg701* Myo9A mutation induce FSGS?
We used Myo9AR701X knock-in mice to examine the molecular pathogenesis of FSGS caused by p.Arg701* MYO9A mutation. Myo9A expression in knock-in Myo9AR701X kidneys significantly decreases in heterozygotes and is undetected in homozygotes by IF (Figure 6A) and immunoblotting (Figure 6B and C). We perfomed qPCR to assess whether nonsense mRNA decay (NMD)-mediated haploinsufficiency42 occurs in Myo9AR701X/+ mutants. Myo9A mRNA expression in Myo9AR701X/+ kidneys is ~50% lower than in wild type kidneys (48.3±5.6%, p<0.0001) (Figure 6D), consistent with haploinsuffciency.
Figure 6: Myo9A expression and Myo9A-actin-calmodulin interaction knock-in Myo9AR701X kidneys and podocytes.
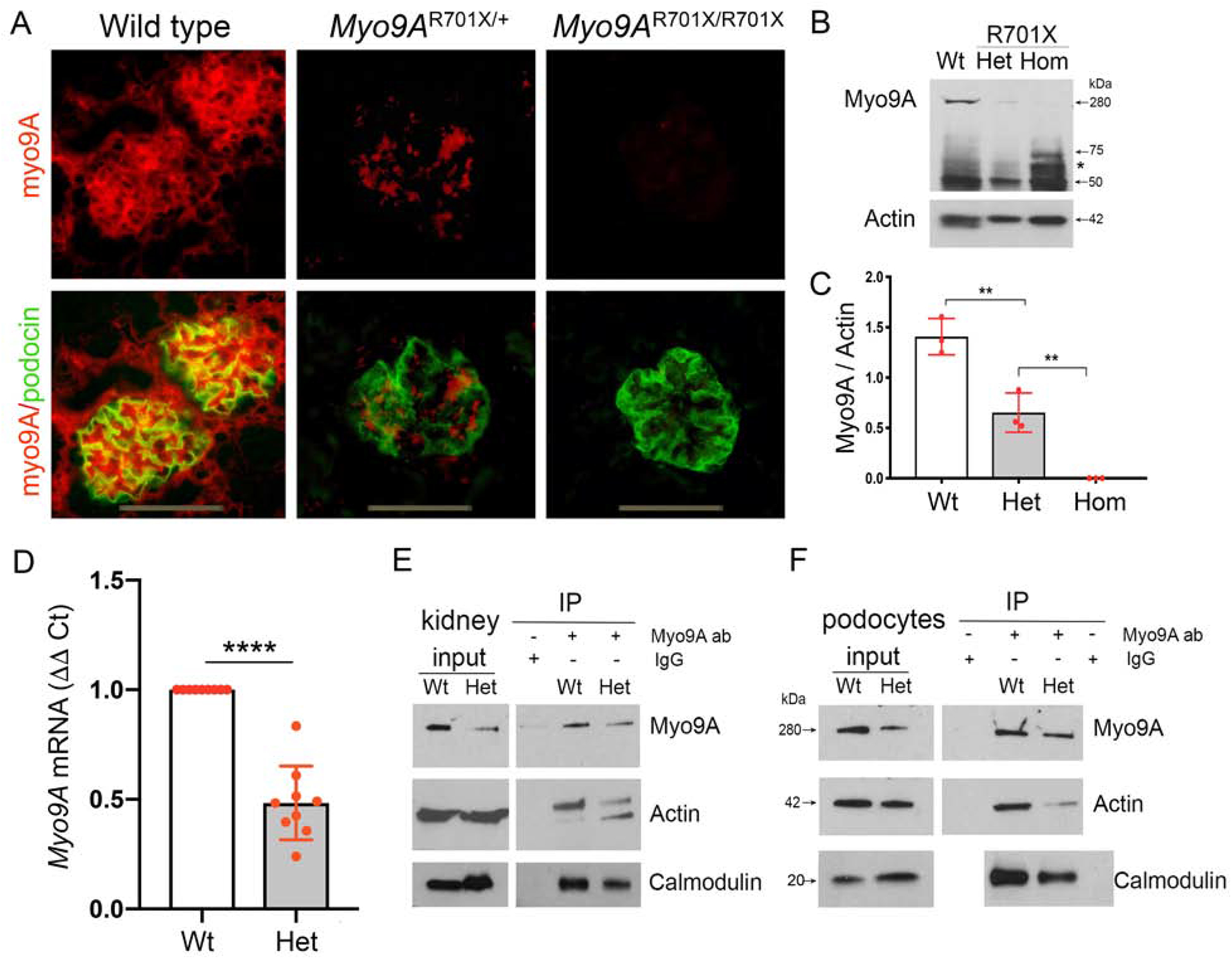
A) IF detects glomerular Myo9A and podocin: Myo9A glomerular immunostaining (red) is strong in wild type kidneys, decreases significantly in Myo9AR701X/+ glomeruli, and is not detected in Myo9AR701X/R701X glomeruli; podocin (green) localizes to podocytes in all mice; podocyte-Myo9A partial co-localization is shown by merge (yellow). B) Immunoblotting: Intact Myo9A (280kDa) expression decreases to ~ half in Myo9AR701X/+ (Het) kidneys and is not detected in Myo9AR701X/R701X (Hom) kidneys, smaller bands (<75kDa) are considered non-specific, ~50 kDa bands correspond to IgG heavy chain in all genotypes. C) Myo9A/actin quantification (mean±SD), n=3 independent experiments, pool from 3 mice each, ** indicates P<0.001. D) qPCR: Myo9A mRNA expression normalized to GAPDH mRNA, fold change (mean±SD), n=3 independent experiments, n=3 individual mice each, **** indicates P<0.0001. E-F) co-IP of kidney lysates (E) and primary podocytes (F) show decreased Myo9A-actin-calmodulin interaction in Myo9AR701X/+ (Het) proportionally to the reduced Myo9A expression (input), note input of actin and calmodulin is equal in both genotypes.
Decreased Myo9A diminishes its interaction with actin and calmodulin
We examined whether Myo9A haploinsufficiency alters Myo9A-actin-calmodulin interaction. Co-immunoprecipitation experiments comparing wild-type and Myo9AR701X/+ kidneys and podocytes confirm Myo9A interaction with actin and with calmodulin, and show that Myo9A interaction with both proteins decreases by ~50% in Myo9AR701X/+ mutant kidneys and podocytes (IP, Figure 6E and F), a reduction similar to that of full-length Myo9A. In contrast, actin and calmodulin expression levels remain intact (input, Figure 6E and F). To analyze Myo9A interactions in the setting of reduced expression, we induced ~50% Myo9A knockdown (Myo9AKD) in immortalized wild-type mouse podocytes using siRNA, assessed by qPCR and immunoblotting (Figure 7A and B), then performed co-IP experiments. Myo9A knockdown decreases Myo9A-actin-calmodulin interaction proportionally to Myo9A expression (Figure 7C). Together, co-IP findings indicate that heterozygous p.Arg701* Myo9A mutation and Myo9A knockdown reduce Myo9A abundance and thereby its interaction with actin and calmodulin. Since Myo9A crosslinks actin at 1:1 molar ratio,16 our data suggest that decreased Myo9A in heterozygous Myo9AR701X/+ podocytes may impair Myo9A-mediated actin crosslinking and bundling.
Figure 7: Myo9A knockdown decreases Myo9A-actin-calmodulin interaction in immortalized mouse podocytes.
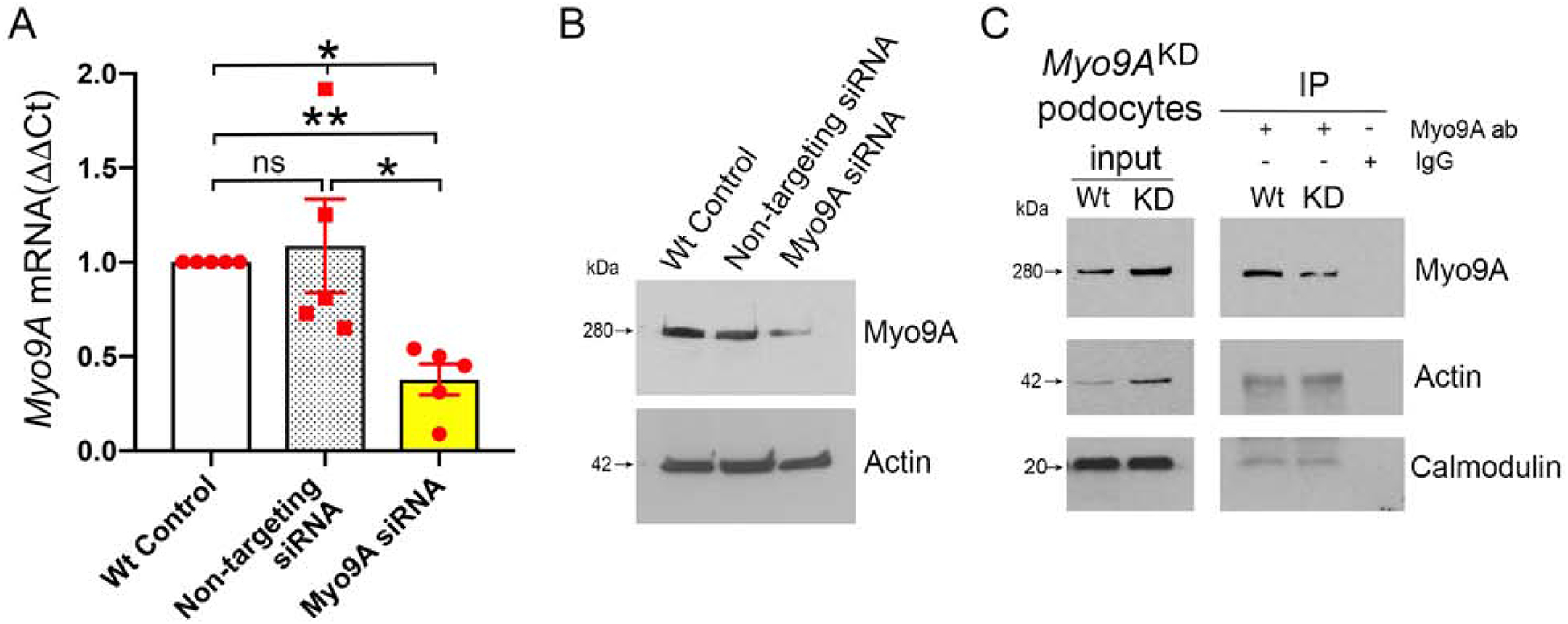
A) Myo9A siRNA [100nM] induces ~50% decrease in Myo9A mRNA, as compared to wild type control and non-targeting siRNA [100nM]. Myo9A mRNA normalized to GAPDH mRNA, expressed as fold change (mean±SD), n≥3 independent experiments,* indicates p<0.05. B) Representative IB shows that Myo9A siRNA induced ~50% reduction of Myo9A protein, as compared to Wt control and non-targeting siRNA; actin expression level was not altered by siRNAs; n≥3 independent experiments. C) Representative Myo9A IP shows decreased Myo9A-actin-calmodulin interaction in Myo9AKD podocytes (siRNA-treated=KD) as compared to wild type immortalized mouse podocytes (Wt), proportionally to the decrease in Myo9A immunoprecipitated; n≥3 independent experiments.
Myo9AR701X mutation increases Rho-A activity
We measured RhoA activity using a pull-down assay in Myo9AR701X mutant and wild-type mice, focusing on heterozygous Myo9AR701X/+ to provide mechanistic insight into the human p.Arg701* MYO9A phenotype. Active RhoA is significantly increased in Myo9AR701X/+ whole kidneys (Figure 8A), isolated glomeruli (Figure 8B) and primary podocytes (Figure 8C), as compared to the corresponding wild-type controls. The magnitude of GTP-RhoA increase in Myo9AR701X/+ samples ranges from ~60% in isolated glomeruli to ~2 fold in whole kidneys and podocytes (Figure 8A, B and C). Active RhoA is slightly higher in homozygous than heterozygous mutant kidneys but the difference is not significant (Figure 8A). Together, these experiments demonstrate that Myo9AR701X causes increased RhoA activity.
Figure 8: RhoA activity and podocyte function in Myo9AR701X kidneys and Myo9AKD podocytes.
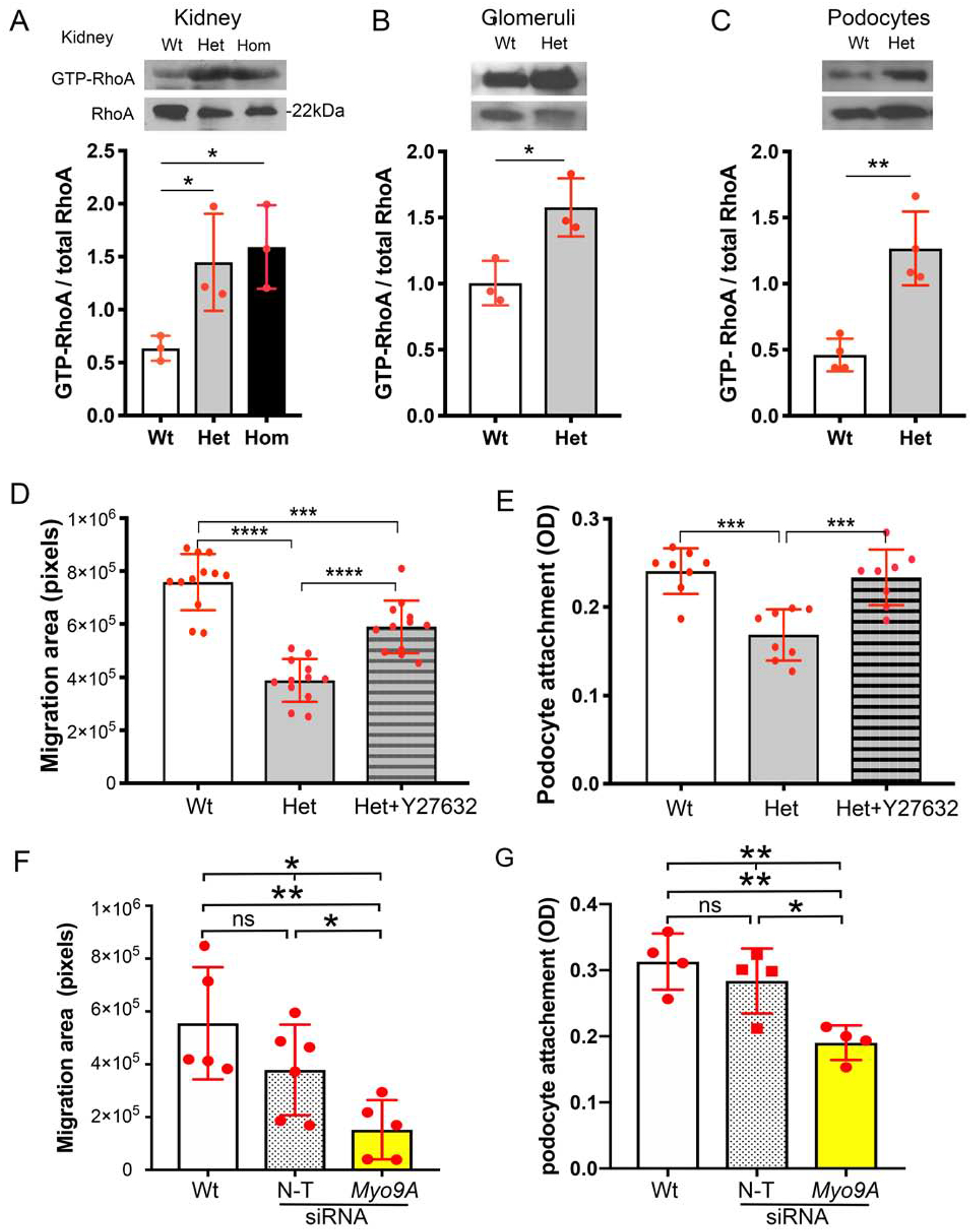
A-C) Active RhoA (GTP-RhoA) is increased in Myo9A R701X mutants vs. wild type: >2-fold in whole kidneys (A), ~60% in isolated glomeruli (B), >2-fold in primary podocytes (C), Western blots representative of n=3–4 independent experiments, lysates pooled from 3 mice each, quantitation (mean±SD), ANOVA (A) or unpaired t-test (B and C), * P<0.05, ** P<0.001. D) Podocyte migration assay (wound assay)57: quantification of the migration area shows that Myo9AR701X/+ (Het) podocyte migration is decreased; pre-incubation with Y26732 partially abrogates the migration defect, *** P<0.0005 (ANOVA),**** P<0.0001 (unpaired t-test). E) Podocyte attachment assay shows decreased attachment of Myo9AR701X/+ (Het) podocytes; pre-incubation with Y26732 corrects the attachment defect, *** P<0.0005 (ANOVA). D-E) Data are mean±SD, n=3–4 independent experiments, n=3 mice/genotype/experiment. F) Podocyte migration assay: siRNA-mediated Myo9A knockdown (Myo9AKD) decreases immortalized mouse podocytes migration (* P<0.05, ANOVA, ** P<0.01, unpaired t-test), whereas migration of non-targeting siRNA-treated podocytes (N-T) is not different from normal podocytes (Wt). G) Attachment assay: Myo9AKD decreases immortalized mouse podocytes attachment, as compared to podocytes treated with non-targeting siRNA (N-T) and untreated Wt podocytes. Myo9AKD data are mean±SD, n≥3 independent experiments. * indicates P<0.05, ** P<0.01, ns indicates no significant difference.
Myo9AR701X/+ mutation and Myo9A knockdown alter podocyte behavior
To assess the effect of impaired Myo9A-actin interaction and increased RhoA activity detected in Myo9AR701X/+ mutant podocytes on their function, we measured podocyte adhesion and motility using attachment and migration assays, comparing wild-type to mutant primary podocytes in control conditions and upon RhoA signaling inhibition with ROCK inhibitor Y27632. These experiments clearly demonstrate that Myo9AR701X/+ impairs podocyte motility (Figure 8D) and podocyte attachment (Figure 8E) in a RhoA activity-dependent manner. To directly evaluate the effect of Myo9A haploinsufficiency on podocyte function, we performed attachment and migration assays comparing wild-type mouse podocytes to Myo9A knockdown (Myo9AKD) podocytes expressing ~50% Myo9A mRNA and protein (Figure 7A and B). Non-targeting siRNA-treated podocytes served as additional control. Myo9AKD significantly decreases podocyte attachment and migration, the same defect observed in Myo9AR701X/+ podocytes (Figure 8F and 8G). Collectively, mechanistic data indicate that heterozygote p.Arg701* Myo9A mutation results in Myo9A haploinsufficiency leading to decreased Myo9A-actin-calmodulin interaction and increased active RhoA in the kidney, specifically in podocytes. We interpret that together, these mechanisms disrupt podocyte function, indicated by decreased attachment and motility in vitro and by the presence of proteinuria in vivo.
DISCUSSION
This study identifies Myo9A as a novel component of the podocyte cytoskeletal apparatus that influences podocyte function by negatively regulating RhoA activity. We describe a rare nonsense MYO9A mutation (p.Arg701*) associated to familial FSGS/proteinuria and demonstrate that this mutation causes FSGS-like lesions in mice, providing insight into the mechanism of disease.
Our findings support the notion that MYO9A is a FSGS candidate gene and that p.Arg701* MYO9A is a variant implicated in human FSGS, based on the American College of Medical Genetics (ACMG) guidelines to attribute causality using genetic prediction algorithms, allele frequencies, experimental animal models and functional assays.43,44 The proband siblings harboring p.Arg701* MYO9A have no mutations in FSGS-causing genes known to date and display no evidence suggesting secondary causes for FSGS. This variant co-segregates in a small kindred following an autosomal dominant pattern of inheritance with variable expressivity. MYO9A p.Arg701* is predicted to be a loss of function variant, is absent from control exomes and has an allele frequency <0.0001 in population databases, in compliance with ACMG standards to support pathogenicity and consistent with parameters used in a recent comprehensive report on genetic contributions to FSGS.43–45 Moreover, LOEUF (loss-of-function observed/expected upper bound fraction) for MYO9A is 0.3, suggestive of low tolerance to gene inactivation with a constraint similar to genes known to cause autosomal dominant FSGS (e.g. WT1, 0.25 or INF2, 0.3) (http://gnomad.broadinstitute.org, accessed 05/08/2020).19,45,46
WES screening of a large cohort of unrelated FSGS children and young adults41 identified two patients with rare heterozygous MYO9A missense variants (p.Asp156Gly and p.Glu765Asp). Both MYO9A missense variants involve highly conserved residues, have minor allele frequencies <0.01%, are predicted to be deleterious and have not previously been associated with human disease. Three additional MYO9A missense variants (Thr389Met; p.Pro1704Ser; p.Met2384Val) identified in this FSGS cohort were deemed benign by bioinformatics scores. These five individuals do not carry mutations in known FSGS-causing genes. For clinical purposes these missense variants should be considered variants of unknown significance (VUS) given the absence of experimental data evaluating how they impact MYO9A function.43,44 To our knowledge no MYO9A mutation has been proven to cause a specific human disease by ACMG guidelines.43,44 Association of compound heterozygote missense MYO9A variants (p.G2282E, and p.Y203C) with arthrogryposis was reported in one patient,5 and compound heterozygote or homozygote missense MYO9A variants (p.R1517H/p.R2283H and p.D1698G, respectively) were described in association with congenital myasthenia syndrome.6 A nonsense homozygous MYO9A (p.R513X) was associated with severe ventriculomegaly and fetal demise.47 Elegant expression studies support a role for MYO9A at the neuromuscular junction and its intriguing regulation of agrin secretion and actin dynamics.48
We demonstrate Myo9A protein expression in podocyte cell lines and mouse kidney glomeruli and Myo9A subcellular localization in podocytes. Dual immunolabeling and confocal microscopy localized Myo9A along the external aspect of the GBM stained by laminin, tightly contiguous to podocin. Immunogold electron microscopy determined precisely Myo9A localization to the cytoplasm of foot processes and podocyte cell bodies, leaving slit-diaphragms unlabeled. The apparent Myo9A mesangial localization in non-confocal IF images was not confirmed by immunogold-EM. Our findings are consistent with glomerular MYO9A immunohistochemical localization in human renal biopsies (www.proteinatlas.org/ENSG00000066933-MYO9A/tissue/kidney). Myo9A was also detected in proximal tubules by IF and immuno-EM, extending a previous report that did not detect Myo9A in glomeruli.15 The discrepancy in Myo9A tissue localization is probably due to different affinity of the primary antibodies used by Thelen et al.15 and in this study. Similar discrepancies in glomerular MYO9A staining of the same human renal biopsies were noticed using two affinity purified polyclonal antibodies, the higher specificity antibody by protein array resulting in stronger glomerular signal (https://www.proteinatlas.org/ENSG00000066933-MYO9A/antibody#protein_array). Localization of Myo9A mRNA in the kidney by in situ hybridization has also been discrepant,4,15 Myo9A developmental regulation might be involved. RNAseq data suggest Myo9A expression along most nephron segments in mice and humans (http://humphreyslab.com/SingleCell/displaycharts.php), raising the possibility that LOF mutations may lead to phenotypes involving several cell types. Interestingly, we observed a clear decrease in glomerular Myo9A expression in two mouse models of proteinuria: developmental nephrotic syndrome and advanced diabetic disease. The latter is consistent with a >2-fold decrease of Myo9A expression by DNA array reported in diabetic Zucker rats.49 Together, MyoA expression data suggest that Myo9A may play a role in the glomerular filtration barrier.
To understand the relevance of p.Arg701* MYO9A nonsense mutation in vivo we generated Myo9AR701X knock-in mice by gene editing. Homozygous Myo9AR701X/R701X mutants develop progressive hydrocephalus resulting in death by 3 weeks of age similar to Myo9A null mice.14 At 2 weeks of age homozygous Myo9AR701X/R701X kidneys reveal a severe glomerulo-tubular phenotype: extensive FPE, GBM abnormalities and paucity of slit-diaphragms and dilated tubules, associated with albuminuria. Adult Myo9A-null mice have shown proximal tubulopathy, polyuria, and low-molecular-weight proteinuria.15 Unfortunately, no glomerular ultrastructural data were included in the report15; surprisingly 3–6 months old Myo9A null mice did not succumb to progressive hydrocephalus as previously described.14 Because homozygous Myo9AR701X/R701X mutants do not survive to adulthood we can only ascertain the early glomerular phenotype described herein. Tubular dilatation observed in 2-week old Myo9AR701X/R701X mutants might represent early evidence of the tubulopathy described in adult Myo9A null mice.
A key finding of this study is that heterozygous Myo9AR701X/+ knock-in mice are phenotypically normal at birth but develop albuminuria, FSGS and podocyte foot process effacement at 4 months of age, demonstrating that p.Arg701* Myo9A is a FSGS-causing mutation in adult mice. Remarkably, Myo9AR701X/+ mutant kidneys histologic and ultrastructural changes recapitulate the diagnostic features of the proband’s renal biopsy. Moreover, Myo9AR701X/+ mutant mice develop significant albuminuria rather than massive albuminuria, reproducing the non-nephrotic proteinuria observed in the proband siblings and in many adult-onset FSGS patients.50 It has been suggested that Myo9A deletion may cause glomerular sclerosis secondary to a polyuric tubulopathy.15 Unconventional myosins have been implicated in albumin reabsorption by PT through their interactions with Dab2 and cubilin, but Myo9A involvement in this process has not been demonstrated.51 We did not detect any evidence of tubulopathy, such as polyuria, tubular dilatation, atrophy or abnormal distribution of proximal or distal tubular markers in adult heterozygous Myo9AR701X/+ mice. These data do not support the hypothesis that the FSGS observed in Myo9AR701X/+ mice result from obstruction or tubular defects.
Mechanistically, we determined that Myo9A mRNA and full-length Myo9A protein are reduced by ~50%, strongly suggesting that the mutation leads to Myo9A haploinsufficiency due to NMD.42 Since Myo9AR701X point mutation localizes to exon 14 out of 42, Myo9A mRNA regulation follows standard NMD rules for truncating mutations located upstream of the prior to last exon.42,52 Consistent with this, hydrocephalus and premature death observed in homozygous Myo9AR701X/R701X mutants mimic Myo9A null mice.14 We detemined that ~50% reduction of Myo9A protein expression in Myo9AR701X/+ mutant kidneys and primary podocytes as well as in Myo9AKD immortalized podocytes decrease proportionally the interaction between Myo9A, actin and calmodulin. These findings support the interpretation that Myo9A haploinsufficiency is the basis for the reduced Myo9A-actin-calmodulin interaction observed in Myo9AR701X/+ mutants.
A critical finding of this work is that p.Arg701* Myo9A induces loss of RhoGAP function resulting in increased active RhoA in Myo9AR701X whole kidneys, isolated glomeruli and primary podocytes. Moreover, Myo9AR701X/+ mutant podocytes demonstrate RhoA-dependent reduced attachment and motility. Myo9A-GAP signaling reduces active RhoA and has little effect on Rac1 and Cdc42.9,53 RhoGAP loss of function increases Rho-GTPase activity and leads to podocyte injury, proteinuria and FSGS.3,18,20 Thus, the abnormal behavior of Myo9AR701X/+ mutant podocytes may contribute to the development of foot process effacement and FSGS over time in Myo9AR701X/+ mice. Our data are consistent with the reported effect of RhoA disregulation on podocyte function in vivo and in vitro.54–56
Because Myo9A crosslinks actin and induces the formation of actin bundles in vitro at regular 36nm intervals by interacting cooperatively with actin and calmodulin, the stoichiometry of their interaction is thought to be crucial.16 Hence, Myo9A haploinsufficiency is likely to decrease Myo9A-mediated actin crosslinks and bundles, providing fewer sites along actin filaments where RhoA inactivation occurs, thereby altering actin dynamics in the immediate vicinity and podocyte behavior at the cellular level. ROCK inhibitor Y27632 partially restored the attachment and migration defect induced by Myo9AR701X/+, suggesting that additional downstream RhoA effectors, such as mDia1, are involved. Alternatively, decreased actin crosslinks in Myo9AR701X/+ may directly contribute to impair podocyte attachement and migration. Myo9A haploinsufficiency might also impact trafficking mechanisms.7–9 The precise molecular mechanisms whereby Myo9A regulates podocyte actin dynamics remain to be elucidated.
In summary, we identified Myo9A as a novel component of the podocyte cytoskeletal apparatus and a nonsense MYO9A mutation that implicates MYO9A as a candidate gene for human FSGS, by reproducing the human proteinuria/FSGS phenotype in Myo9AR701X/+ knock-in mice and by autosomal dominant co-segregation of the phenotype in a small kindred. Collectively, data strongly suggest that Myo9A haploinsufficiency is the molecular basis for the phenotype observed in adult Myo9AR701X/+ mice. We demonstrated that attachment and migration are impaired in knock-in mutant Myo9AR701X/+ podocytes, involving both increased RhoA activity and decreased Myo9A-actin-calmodulin interaction. Given that Myo9A induces actin crosslinking and bundling at 1:1 molar ratio16 and our observation that Myo9AR701X/+ leads to haploinsufficiency, podocyte effacement and FSGS overtime, we speculate that Myo9A may be critical for normal podocyte actin dynamics, akin to other actin-binding proteins (ACTN4, INF2) known to cause autosomal dominant FSGS in adults when mutated. Additional studies are warranted to elucidate how Myo9A interacts with podocyte RhoA, cytoskeletal and slit-diaphragm proteins, and whether Myo9A-mediated defects in actin crosslink and bundle formation directly alter podocyte cytoskeleton structure. Limitations of this study to establish causation include inability to extend the proband genotype pedigree and lack of additional kindreds with the same mutation. Further studies are necessary to validate our WES findings in additional familial or sporadic FSGS patients before p.Arg701* MYO9A variant can be considered disease-causing and this information can be used for clinical decision-making.
Supplementary Material
Figure S1: Immuno-EM localizes Myo9A expression to mouse glomerular podocytes and tubular epithelia.
Figure S2: Myo9AR701X/R701X causes neonatal progressive hydrocephalus.
Figure S3: Postnatal renal histology in Myo9AR701X mutant mice.
Figure S4: Podocyte migration assay (wound assay).
Table S1: List of all variants shared by proband siblings
Table S2: List of 64 FSGS-proteinuria-NS genes genes interrogated in the FSGS-CT cohort
TRANSLATIONAL STATEMENT.
This work identifies a myosin 9A (MYO9A) loss-of-function mutation in a family with proteinuria and focal segmental glomerulosclerosis (FSGS) and MYO9A variants of uncertain significance in a FSGS cohort. Myo9A knock-in mice harboring p.Arg701* mutation develop proteinuria and FSGS, reproducing the probands’ phenotype. This new animal model provides novel mechanistic insights on Myo9A functions in podocytes and enables to test experimental therapies for FSGS. Data suggest that heterozygous loss-of-function MYO9A mutations may contribute to FSGS pathogenesis. We propose including MYO9A in FSGS genetic testing panels.
ACKNOWLEDGEMENTS
We thank M. Saleem (U. Bristol, UK) for providing immortalized human podocytes, G. Moeckel (Yale School of Medicine, Dept. of Pathology) for providing biopsy images, S. Ishibe Lab (Yale School of Medicine, Dept. of Medicine/ Nephrology) for advice on podocyte isolation and adhesion assay, T. Ardito (Yale School of Medicine, Dept. of Pathology) and V. Kakade (Yale School of Medicine, Dept. of Medicine/ Nephrology) for contributions to IF and confocal imaging, FSGS patients and families and NIDDK repository (FSGS-CT, ClinicalTrials.gov NCT00135811) for providing DNA samples.
Sources of support: This work was supported by NIH-RO1DK109434 grant to A.T. and Yale Center for Mendelian Genomics (NIH-U54-HG006504).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURE
The authors have nothing to disclose.
SUPPLEMENTARY MATERIAL
Supplementary Methods
WES and bioinformatic data analysis, generation of knock-in Myo9AR701X mice and other detailed methods.
Supplementary References S1 to S7
Supplementary information is available at Kidney International’s website.
REFERENCES
- 1.Wiggins RC. The spectrum of podocytopathies: a unifying view of glomerular diseases. Kidney Int 2007;71:1205–1214. [DOI] [PubMed] [Google Scholar]
- 2.Sadowski CE, Lovric S, Ashraf S, et al. A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol 2015;26:1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gee HY, Saisawat P, Ashraf S, et al. ARHGDIA mutations cause nephrotic syndrome via defective RHO GTPase signaling. J Clin Invest 2013;123:3243–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorman SW, Haider NB, Grieshammer U, et al. The cloning and developmental expression of unconventional myosin IXA (MYO9A) a gene in the Bardet-Biedl syndrome (BBS4) region at chromosome 15q22-q23. Genomics 1999;59:150–160. [DOI] [PubMed] [Google Scholar]
- 5.Bayram Y, Karaca E, Coban Akdemir Z, et al. Molecular etiology of arthrogryposis in multiple families of mostly Turkish origin. J Clin Invest 2016;126:762–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Connor E, Töpf A, Müller JS, et al. Identification of mutations in the MYO9A gene in patients with congenital myasthenic syndrome. Brain 2016;139:2143–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masters TA, Kendrick-Jones J, Buss F. Myosins: domain organization, motor properties, physiological roles and cellular functions. Handb Exp Pharmacol 2017;235:77–122. [DOI] [PubMed] [Google Scholar]
- 8.Liu KC, Cheney RE: Myosins in cell junctions. Bioarchitecture 2012;2:158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Omelchenko T, Hall A: Myosin-IXA regulates collective epithelial cell migration by targeting RhoGAP activity to cell-cell junctions. Curr Biol 2012;22:278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekine T, Konno M, Sasaki S, et al. Patients with Epstein-Fechtner syndromes owing to MYH9 R702 mutations develop progressive proteinuric renal disease. Kidney Int 2010;78:207–214. [DOI] [PubMed] [Google Scholar]
- 11.Mele C, Iatropoulos P, Donadelli R, et al. MYO1E mutations and childhood familial focal segmental glomerulosclerosis. N Engl J Med 2011;365:295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krendel M, Kim SV, Willinger T, et al. Disruption of Myosin 1e promotes podocyte injury. J Am Soc Nephrol 2009;20:86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bähler M, Elfrink K, Hanley PJ, et al. Cellular functions of class IX myosins in epithelia and immune cells. Biochem Soc Trans 2011;39:1166–1168. [DOI] [PubMed] [Google Scholar]
- 14.Abouhamed M, Grobe K, San IV, et al. Myosin IXa regulates epithelial differentiation and its deficiency results in hydrocephalus. Mol Biol Cell 2009;20:5074–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thelen S, Abouhamed M, Ciarimboli G, et al. Rho GAP myosin IXa is a regulator of kidney tubule function. Am J Physiol Renal Physiol 2015;309:F501–F513. [DOI] [PubMed] [Google Scholar]
- 16.Saczko-Brack D, Warchol E, Rogez B, et al. Self-organization of actin networks by a monomeric myosin. Proc Natl Acad Med 2016;113:E8387–E8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asanuma K, Yanagida-Asanuma E, Faul C, et al. Synaptopodin orchestrates actin organization and cell motility via regulation of RhoA signalling. Nat Cell Biol 2006;8:485–491. [DOI] [PubMed] [Google Scholar]
- 18.Mouawad F, Tsui H, Takano T. Role of Rho-GTPases and their regulatory proteins in glomerular podocyte function. Can J Physiol Pharmacol 2013;91:773–782. [DOI] [PubMed] [Google Scholar]
- 19.Brown EJ, Schlöndorff JS, Becker DJ, et al. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet 2010;42:72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akilesh S, Suleiman H, Yu H, et al. Arhgap24 inactivates Rac1 in mouse podocytes, and a mutant form is associated with familial focal segmental glomerulosclerosis. J Clin Invest 2011;121:4127–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi M, Scholl I, Ji W, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA 2009;106:19096–190101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi M, Scholl UI, Yue P, et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science 2011;331:768–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bamshad MJ, Shendure JA, Valle D, et al. The Centers for Mendelian Genomics: a new large-scale initiative to identify the genes underlying rare Mendelian conditions. Am J Med Gen 2012;158A:1523–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc 2015;10:1556–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong C, Wei P, Jian X, et al. Comparison an integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet 2015;24:2125–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sim NL, Kumar P, Hu J, et al. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 2012;40(Web Server issue):W452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rentzsch P, Witten D, Cooper GM, et al. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 2019;47:D886–D894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H, Wang H, Jaenisch R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nature Protoc 2014;9:1956–1968. [DOI] [PubMed] [Google Scholar]
- 31.Veron D, Bertuccio CA, Marlier A, et al. Podocyte vascular endothelial growth factor (Vegf164) overexpression causes severe nodular glomerulosclerosis in a mouse model of type 1 diabetes. Diabetologia 2011;54:1227–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veron D, Reidy K, Marlier A, et al. Induction of podocyte VEGF164 overexpression at different stages of development causes congenital nephrosis or steroid-resistant nephrotic syndrome. Am J Pathol 2010;177:2225–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou Z, Chung B, Nguyen T, et al. Linking receptor-mediated endocytosis and cell signaling: evidence for regulated intramembrane proteolysis of megalin in proximal tubule. J Biol Chem 2004;279:34302–34310. [DOI] [PubMed] [Google Scholar]
- 34.Bertuccio C, Veron D, Aggarwal PK, et al. Vascular endothelial growth factor receptor 2 direct interaction with nephrin links VEGF-A signals to actin in kidney podocytes. J Biol Chem 2011;286: 39933–39944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veron D, Aggarwal PK, Velazquez H, et al. Podocyte-specific VEGF-a gain of function induces nodular glomerulosclerosis in eNOS null mice. J Am Soc Nephrol 2014;25:1814–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reidy KJ, Aggarwal PK, Jimenez JJ, et al. Excess podocyte semaphorin-3A leads to glomerular disease involving plexinA1-nephrin interaction. Am J Pathol 2013;183:1156–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian X, Kim JJ, Monkley SM, et al. Podocyte-associated talin1 is critical for glomerular filtration barrier maintenance. J Clin Invest 2014,124:1098–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ni L, Saleem M, Mathieson PW. Podocyte culture: tricks of the trade. Nephrology (Carlton) 2012;17:525–531. [DOI] [PubMed] [Google Scholar]
- 39.Ren XD, Kiosses WB, Schwartz MA. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J 1999;18:578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buvall L, Rashmi P, Lopez-Rivera E, et al. Proteasomal degradation of Nck1 but not Nck2 regulates RhoA activation and actin dynamics. Nat Commun 2013;4:2863, doi: 10.1038/ncomms3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kopp JB, Winkler CA, Zhao X, et al. Clinical Features and Histology of Apolipoprotein L1-Associated Nephropathy in the FSGS Clinical Trial. J Am Soc Nephrol 2015;26:1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurosaki T, Maquat LE. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci 2016;129:461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature 2014;508: 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology.Genet Med 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang M, Chun J, Genovese G, et al. Contributions of rare gene variants to familial and sporadic FSGS. J Am Soc Nephrol 2019. pii: ASN.2019020152. doi: 10.1681/ASN.2019020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karczewski KJ, Francioli LC, Tiao G et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maddirevula S, Alzahrani F, Al-Owain M, et al. Autozygome and high throughput confirmation of disease genes candidacy. Genet Med 2019. 21:736–742. doi: 10.1038/s41436-018-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Connor E, Phan V, Cordts I, et al. MYO9A deficiency in motor neurons is associated with reduced neuromuscular agrin secretion. Hum Mol Genet 2018;27:1434–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sárközy M, Zvara A, Gyémánt N, et al. Metabolic syndrome influences cardiac gene expression pattern at the transcript level in male ZDF rats. Cardiovasc Diabetol 2013;12:16, doi: 10.1186/1475-2840-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lepori N, Zand L, Sethi S, et al. Clinical and pathological phenotype of genetic causes of focal segmental glomerulosclerosis in adults. Clin Kidney J 2018;11:179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eshbach ML, Weisz OA. Receptor-mediated endocytosis in the proximal tubule. Annu Rev Physiol 2017, 79:425–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bouligand J, Delemer B, Hecart AC, et al. Familial glucocorticoid receptor haploinsufficiency by non-sense mediated mRNA decay, adrenal hyperplasia and apparent mineralocorticoid excess. PLoS One 2010;5: e13563, doi: 10.1371/journal.pone.0013563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yi F, Kong R, Ren J, et al. Noncanonical Myo9b-RhoGAP Accelerates RhoA GTP Hydrolysis by a Dual-Arginine-Finger Mechanism. J Mol Biol 2016;428:3043–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tian D, Jacobo SM, Billing D, et al. Antagonistic regulation of actin dynamics and cell motility by TRPC5 and TRPC6 channels. Sci Signal 2010;26;3(145):ra77 doi: 10.1126/scisignal.2001200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Ellis MJ, Gomez JA, et al. Mechanisms of the proteinuria induced by Rho GTPases. Kidney Int 2012;81:1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kistler AD, Altintas MM, Reiser J. Podocyte GTPases regulate kidney filter dynamics. Kidney Int 2012;81:1053–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cory G. Scratch-wound assay. Methods Mol Biol 2011;769:25–30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Immuno-EM localizes Myo9A expression to mouse glomerular podocytes and tubular epithelia.
Figure S2: Myo9AR701X/R701X causes neonatal progressive hydrocephalus.
Figure S3: Postnatal renal histology in Myo9AR701X mutant mice.
Figure S4: Podocyte migration assay (wound assay).
Table S1: List of all variants shared by proband siblings
Table S2: List of 64 FSGS-proteinuria-NS genes genes interrogated in the FSGS-CT cohort


