Abstract
Over the last decade, strong evidence has emerged that protein degradation mediated by the ubiquitin-proteasome system is critical for fear memory formation in the amygdala. However, this work has been done primarily in males, leaving unanswered questions about whether females also require protein degradation during fear memory formation. Here, we found that male and female rats differed in their engagement and regulation of, but not need for, protein degradation in the amygdala during fear memory formation. Male, but not female, rats had increased protein degradation in the nuclei of amygdala cells after fear conditioning. Conversely, females had elevated baseline levels of overall ubiquitin-proteasome activity in amygdala nuclei. Gene expression and DNA methylation analyses identified that females had increased baseline expression of the ubiquitin coding gene Uba52, which had increased DNA 5-hydroxymethylation (5hmc) in its promoter region, indicating a euchromatin state necessary for increased levels of ubiquitin in females. Consistent with this, persistent CRISPR-dCas9 mediated silencing of Uba52 and proteasome subunit Psmd14 in the amygdala reduced baseline protein degradation levels and impaired fear memory in male and female rats, while enhancing baseline protein degradation in the amygdala of both sexes promoted fear memory formation. These results suggest that while both males and females require protein degradation in the amygdala for fear memory formation, they differ in their baseline regulation and engagement of this process following learning. These results have important implications for understanding the etiology of sex-related differences in fear memory formation.
Keywords: Ubiquitin, Proteasome, Sex Differences, Consolidation, Amygdala
1. Introduction
The process of forming long-term memories is complex and requires the coordinated actions of hundreds of different transcription factors and intracellular signaling molecules, which are thought to lead to increased transcriptional regulation and de novo protein synthesis in a select population of cells, or engram, that ultimately store the memory (Bisaz, Travaglia, and Alberini, 2014; Josselyn and Tonegawa, 2020; Kandel, 2012; Kandel, Dudai, and Mayford, 2014). Much of this data comes from rodent Pavlovian fear conditioning tasks, where strong evidence suggests a role for new gene transcription and protein translation in fear memory formation, primarily in the amygdala (Hoeffer et al., 2011; Keeley et al., 2006; Mahan et al., 2012; Morey et al., 2012; Ou and Gean, 2007; Parsons, Gafford, and Helmstetter, 2006; Phillips and LeDoux, 1992; Shin, Rauch, and Pitman, 2006). However, much of the work on the molecular and cellular mechanisms of fear memory formation have focused primarily on male rodents, leaving unanswered questions about whether similar mechanisms govern fear memory formation in females. Importantly, numerous studies suggest that the molecular mechanisms involved in the formation of long-term memories, including fear memories, can differ between sexes (Frick, Kim, Tuscher, and Fortress, 2015; Gresack, Schafe, Orr, and Frick, 2009; Maren, De Oca, and Fanselow, 1994; Mendez-Lopez, Mendez, Lopez, and Arias, 2009; Mizuno et al., 2007; Mizuno and Giese, 2010), indicating that a more thorough examination of sex differences in the neurobiology of fear memory formation is needed.
Despite the traditional focus on increased transcriptional and translational regulation in fear memory formation, over the last decade strong evidence has emerged that protein degradation mediated by the ubiquitin-proteasome system (UPS) is also critically involved this process. In the UPS system, the small protein modifier ubiquitin targets substrates for degradation by the 26S proteasome complex (Jarome and Helmstetter, 2013). In most cases proteins that are going to be degraded acquire multiple ubiquitin modifications that are linked together at lysine 48 (K48), known as the K48 polyubiquitin tag (Musaus, Navabpour, and Jarome, 2020). Since its discovery, the UPS has been implicated in many neurological, neurodegenerative and psychiatric disorders (Ciechanover and Kwon, 2015; Dantuma and Bott, 2014; Minelli et al., 2015), as well as various forms of synaptic plasticity (Dong, Bach, Haynes, and Hegde, 2014; Ehlers, 2003; Fioravante, Liu, and Byrne, 2008; Fonseca et al., 2006). Recent work has shown a critical role for ubiquitin-proteasome mediated protein degradation in fear memory formation (Cullen, Ferrara, Pullins, and Helmstetter, 2017; Lee et al., 2008; Lopez-Salon et al., 2001; Reis, Jarome, and Helmstetter, 2013; Rosenberg, Elkobi, Dieterich, and Rosenblum, 2016; Rosenberg, Elkobi, and Rosenblum, 2016), especially within the amygdala (Jarome, Ferrara, Kwapis, and Helmstetter, 2016; Jarome, Kwapis, Ruenzel, and Helmstetter, 2013; Jarome, Werner, Kwapis, and Helmstetter, 2011; Orsi et al., 2019; Rodriguez-Ortiz et al., 2011). Additionally, recent evidence suggests a role for the immunoproteasome, in which elevated interferon levels induce incorporation of catalytic subunit variants into the proteasome core, in constraining fear memory formation (Gorny et al., 2019). However, this previous rodent work has been done exclusively in male rodents, so little is known about whether similar UPS mechanisms control the formation of fear memories in females. Importantly, we recently found that male and female rats differ in their regulator mechanisms of the proteasome in the amygdala and hippocampus (Devulapalli et al., 2019). Furthermore, recent evidence suggests that sexual dimorphism exists in proteasome function, with higher activity seen in females across a range of tissues (Jenkins et al., 2020). These data suggest sex differences likely exist in the role and regulation of the protein degradation process in the brain. However, the functional significance of this has never been examined.
In this study, we investigated sex differences in the role and regulation of protein degradation in the amygdala during fear memory formation. We found that while males engage protein degradation in the amygdala following fear conditioning, females do not. Instead, females have elevated baseline levels of protein degradation in the amygdala in comparison to male, which is associated with increased epigenetic regulation of a major ubiquitin coding gene. Importantly, genetic manipulations of protein degradation in the amygdala alter fear memory in both males and females in a similar manner, suggesting that while they differ in baseline regulation and activity-dependent engagement of this process, both sexes need protein degradation in the amygdala for fear memory formation. Collectively, these studies provide the first evidence of baseline differences in brain ubiquitin-proteasome signaling in males and females, which may have important implications for understanding the etiology of sex-related differences in fear memory formation.
2. Methods and Materials
2.1. Subjects
All experiments used 8-9 week old male and female Sprague Dawley rats obtained from Envigo (Frederick, MA). Animals were housed two per cage with free access to water and rat chow. Male and female animals were housed in separate rooms. The colony was maintained under a 12:12-hr light/dark cycle. All experiments took place during the light portion of the cycle. All procedures were approved by the Virginia Polytechnic Institute and State University Institutional Animal Care and Use Committee and conducted with the ethical guidelines of the National Institutes of Health
2.2. CRISPR plasmid preparation
CRISPR guide RNA (gRNA) targeting a 200bp region within the Uba52 and Psmd14 promoters were designed using CRISPOR. The sequences (PSMD14 gRNA: CCGCCAATTGGAATCCGCCG; UBA52 gRNA 1: AATCCGCGCCGCACGTCGTC; UBA52 gRNA 2: ACATGGCGCCCGGCGAAGCT) were inserted in the 293-T backbone, cloned into the CRISPR gRNA vector driven by the murine U6 promoter (gift from Stanley Qi; #44248, Addgene, Watertown, MA) and transformed into One Shot Top10 chemically compotent E. coli. Colonies were selected and sequenced to confirm proper insertion of the sgRNA into the vector (gRNA). Both the gRNA and dCas9m4-KRAB-MECP2 (gift from Alejandro Chavez and George Church; #110821, Addgene) or VP64-dCas9-VP64 (gift from Charles Gersbach; #59791, Addgene) cells were grown in LB Broth with ampicillin and purified with the Endotoxin-free Midi-Prep kit (Takara Bio, Mountain View, CA) according to the manufacturer’s instructions. For stereotaxic delivery, plasmids were mixed with In Vivo Jet-PEI (Polyplus Transfection, New York, NY) according the manufacturer’s instructions using a previously described procedure (Butler, Johnston, Kaur, and Lubin, 2019).
2.3. Surgeries
Rats underwent stereotaxic surgeries where CRISPR-dCas9 plasmids were injected into the basolateral amygdala (BLA) using coordinates relative to Bregma (A/P: −3.0, M/L: +/− 5.0, D/V: −7.7). Animals were anesthetized with 1.5-4% isoflurane and received bilateral injections into the amygdala using a 26-guage Hamilton syringe connected to an automated pump (Harvard Apparatus, Cambridge, MA) at a rate of 0.1μl per minute for a total of 0.5μl per hemisphere. Animals received a subcutaneous injection of carprofen (5mg/kg) and topical lidocaine on the day of surgery.
2.4. Apparatus
This project used two identical fear conditioning chambers. Each Habitest chamber consisted of a steel test cage with front and back Plexiglas walls and a grid shock floor above a plastic drop pan. The right wall of the chamber consisted of a house light in the top back corner, which remained on during the behavioral procedures, and an infrared light in the top middle, which was not illuminated during this project. The left wall of the chamber consisted of a high-bright light, which was not illuminated during this project. All remaining slots of both walls were filled with blank metal panels. A USB camera was mounted on a steel panel outside the back Plexiglas wall of the chamber, angled at ~45 degrees. The entire chamber was housed in an isolation cubicle with an acoustic liner and a house fan, which remained active during all behavioral procedures. Shock was delivered through the grid floor via a Precision Animal Shocker under the control of FreezeFrame 4 software, which also analyzed animal behavior in real-time. A freezing threshold of 2.0 was used as the scoring parameter for all animals. All video was recorded and stored for later analysis. The chamber walls were wiped with 70% isopropanol before use.
2.5. Behavioral Procedures
Rats underwent a contextual fear conditioning procedure as described previously (Devulapalli et al., 2019; Orsi et al., 2019) in a Habitest chamber. Animals were handled for 4 days prior to behavioral training; the first two days occurred in the animal housing room and the second two days occurred in an adjacent room where behavioral training were to occur. Following this, animals were placed into the fear conditioning apparatus and after a 1 min baseline, received 4 unsignaled footshock (1.0mA, 1 sec) presentations. After a 1 min post-shock period, the animals were returned to their homecages. The procedure for weak contextual fear conditioning was similar except that the shock intensity was lowered to 0.6mA, though the same number of shocks were given. For testing, which occurred 24 hrs after training, animals were placed back into the training context for 5 minutes in the absence of shock. Male and female animals underwent identical procedures and were ran at the same time in a counterbalanced manner so that they could be directly compared, except for the CRISPR-dCas9 experiments described below. When males and females were trained on the same day, the following procedure was used. Two animals of the sex were trained in adjacent chambers. Immediately following training, the chamber floor and drop pan were removed, washed in a sink and replaced. Chamber walls were cleaned with water and subsequently were wiped with 70% isopropanol before the next group of two rats were trained, which consisted of animals of the opposite sex from the first cohort. This pattern was repeated until all animals had completed the training procedure.
2.6. Specific Experimental Procedures
EXPERIMENT 1:
Male (n = 10) and female (n = 10) rats were trained to contextual fear conditioning and the following day re-exposed to the training context to assess memory retention. These animals were only used for behavioral comparison between sexes, as well as shock reactivity analysis.
EXPERIMENT 2:
Male and female rats (n = 8-10 per sex) were trained to contextual fear conditioning and brain tissue collected 1 hr later. Separate male and female rats (n = 8-10 per sex) that were not exposed to the training context had brain tissue collected from them during the training day served as naïve controls. Collected brain regions were split by hemisphere and one side used for protein assays, while the other was used for RNA and DNA analyses as described below. For protein assays, groups were compared for K48 polyubiquitination, total polyubiquitination and proteasome activity. For RNA assays, groups were compared for Ubb, Ubc, Uba52 and Rps27a expression. For DNA assays, groups were compared for DNA methylation analyses using methylated DNA immunoprecipitation and bisulfite sequencing as described below. Group sizes for RNA/DNA analyses (n = 7-8 per group) were lower than protein assays due to low RNA/DNA quantity being collected for some samples. Final group sizes are listed in figure legends.
EXPERIMENT 3:
One naïve male and one naïve female rat were trained to contextual fear conditioning and brain tissue collected 1 hr later for cyrosectioning. Non-quantitative immunofluorescence was performed on these slices as described below.
EXPERIMENT 4:
Female rats (n = 8) were trained to contextual fear conditioning and brain tissue collected 4 hrs later. Separate female rats (n = 8) that were not exposed to the training context and had brain tissue collected from them during the training day served as naïve controls. Collected brain regions were used for protein assays and were compared for K48 polyubiquitination and proteasome activity.
EXPERIMENT 5:
Female rats were stereotaxically injected with CRISPR gRNA+dCas9-KRAB-MECP2 (n = 10) or CRISPR gRNA alone (n = 9) into the basolateral amygdala. Two weeks later, animals were trained to contextual fear conditioning and the following day re-exposed to the training context to assess memory retention. Twenty-four hours after testing, basolateral amygdala tissue was collected, hemispheres split and processed for protein and RNA assays. Protein assays consisted of proteasome activity and K48 polyubiquitination. RNA assays consisted of qRT-PCR for Uba52 and Psmd14. Group sizes for RNA analysis (n = 8-9 per group) were lower than protein assays due to low RNA quantity being collected for two samples. Final group sizes are listed in figure legends. Behavioral and molecular analyses were performed on the same animals to allow a direct comparison between baseline regulation of the protein degradation process in the basolateral amygdala and behavioral performance.
EXPERIMENT 6:
Female rats were stereotaxically injected with CRISPR gRNA+VP64-dCas9-VP64 (n = 8) or CRISPR gRNA alone (n = 8) into the basolateral amygdala. Two weeks later, animals were trained to contextual fear conditioning and the following day re-exposed to the training context to assess memory retention. For this experiment the shock intensity was lowered to 0.6mA in order to produce a weaker memory in the controls.
EXPERIMENT 7:
Male rats were stereotaxically injected with CRISPR gRNA+dCas9-KRAB-MECP2 (n = 5) or CRISPR gRNA alone (n = 4) into the basolateral amygdala. Two weeks later, animals were trained to contextual fear conditioning and the following day re-exposed to the training context to assess memory retention.
EXPERIMENT 8:
Male rats were stereotaxically injected with CRISPR gRNA+VP64-dCas9-VP64 (n = 4) or CRISPR gRNA alone (n = 4) into the basolateral amygdala. Two weeks later, animals were trained to contextual fear conditioning and the following day re-exposed to the training context to assess memory retention. For this experiment the shock intensity was lowered to 0.6mA in order to produce a weaker memory in the controls.
2.7. Shock Reactivity Analysis
Shock reactivity analysis was performed as described previously (Jarome, Kwapis, Nye, and Helmstetter, 2010) by obtaining scores for each animal’s reaction to the four shocks given during the training session. For each shock, the time from when the shock terminated until the first instance of freezing behavior occurred was recorded. This period from shock termination until freezing behavior is exhibited is an “activity burst” that results as a response elicited from the unconditioned shock stimulus. The first instance of freezing behavior was identified by FreezeFrame 4 software using 2.0 as the freezing threshold and was scored offline from saved video files. Due to a system error, one video file was lost for the males and could not be included in the analysis.
2.8. Tissue Collection
Amygdala and dorsal hippocampus tissue were collected as described previously (Devulapalli et al., 2019). Rats were overdosed on isoflurane in a necrosis chamber and the brain rapidly removed and immediately frozen on dry ice. Tissue containing the basolateral amygdala and CA1 region of the dorsal hippocampus were then dissected out by blocking the brain in a rat brain matrix (Harvard Apparatus, Holliston, MA) incubated with dry ice. Amygdala and hippocampal hemispheres were split so that one was used for nuclear and cytoplasmic extractions and the other for RNA/DNA extractions, which were counterbalanced across extraction procedures to account for any possible laterality effects. All dissected tissue was frozen at −80°C until needed.
2.9. Nuclear and Cytoplasmic Extractions
Nuclear and cytoplasmic extracts were collected using our previously described protocol (Devulapalli et al., 2019; McFadden, Devulapalli, and Jarome, 2019; Orsi et al., 2019). The basolateral amygdala and dorsal CA1 tissue were homogenized with Teflon tissue grinders using 15 strokes in 500μl of lysis buffer (10mM HEPES, 1.5mM MgCl2, 10mM KCl, 0.5mM DTT, 0.5% IGEPAL, 1μl/ml protease inhibitor cocktail, and 1μl/ml phosphatase inhibitor cocktail) and incubated on ice for 30 min. The homogenized samples were then centrifuged for 10 min at 845 x g at 4 °C. The supernatant was collected as the cytoplasmic fractions. Pelleted nuclei were resuspended in 75μl of extraction buffer (20mM HEPES, 6.25% glycerol, 1.5mM MgCl2, 300mM NaCl, 0.25mM EDTA, 0.5mM DTT, 1μl/ml protease inhibitor cocktail, and 1μl/ml phosphatase inhibitor cocktail) and mixed by pipetting. The resuspended nuclei were incubated on ice for 30 min and then centrifuged for 20 min at 21,130 x g at 4°C. The resulting supernatant containing nuclear proteins was collected and protein concentration was determined by using the Bio-Rad (Hercules, CA) Dc protein assay.
2.10. Proteasome Activity Assay
Proteasome activity assays were performed as described previously (Devulapalli et al., 2019; McFadden et al., 2019; Orsi et al., 2019). Normalized samples (nuclear = 2μg; cytoplasmic = 20μg) were diluted in MilliQ H2O and mixed with reaction buffer (250mM HEPES, pH 7.5, 5mM EDTA, 0.5% NP-40, 0.01% SDS, 5mM ATP). Fluorogenic peptide Suc-leu-leu-val-thy-AMC (#BML-P802-0005, Enzo Life Sciences) was then added to the samples at a concentration of 10μM to assess proteasome chymotrypsin-like activity. The reaction was incubated at 37°C for 2 hrs and fluorescence monitored at 360 (excitation)/460 (emission) on a monochromatic plate reader (Synergy H1; Biotek, Winooski, VT). Protein free blanks were used and an AMC standard curve was produced. The scan with the peak level of AMC was used for statistical analyses. Data are presented as the percentage change in relative fluorescent units (RFU) relative to the Control group.
2.11. Antibodies
Rabbit monoclonal antibodies included K48 polyubiquitin (1:1000; #ab140601, Abcam, Cambridge, MA) and β-actin (1:1000; #4967, Cell Signaling, Danvers, MA). Mouse monoclonal antibodies included Ubiquitin (1:1000; #BML-PW8810, Enzo Life Sciences).
2.12. Western Blots
Western blots were performed as describe previously (Orsi et al., 2019). Samples (5-10μg) were loaded on 7% Acrylamide gels, ran through SDS-PAGE and transferred using a Turbo Transfer System (Biorad). Membranes were incubated in a 50:50 blocking buffer (50% Licor TBS blocking buffer and 50% TBS + 0.1% Tween-20) for 1 h at room temperature, followed by an overnight incubation in primary antibody in 50:50 blocking buffer at 4°C. Membranes were then washed 3 times for 10 min with TBS + 0.1% Tween-20 (TBSt) and incubated in secondary antibody (1:20,000; goat anti-rabbit 700CW or goat anti-mouse IgG1 800CW) in 50:50 blocking buffer for 45 min. After two 10 min washes in TBSt, the membranes were washed in TBS and imaged using the Odyssey Fc (LI-COR, Lincoln, NE). Visualized proteins were analyzed using Image Studio Ver 5.2. Between developments, membranes were stripped for 10 min with 0.2M NaOH followed by two 15 min washes in TBSt and blocking buffer for 1 hr. Samples were normalized to β-actin or Coomassie blue, which were used as loading controls. For Coomassie blue staining, membranes were stained for 10 sec, washed extensively in 50% methanol and imaged at 700CW using the Odyssey Fc.
2.13. Immunofluorescence
Immunofluorescence procedures were performed as described previously with a small-scale modification (Webb et al., 2017). Animals were overdosed on isoflurane and euthanized, after which their brains were removed and mounted in O.C.T. and flash frozen. Slices (20μM) were taken throughout the amygdala and mounted to slides. Slices were fixed with 4% paraformaldehyde for 10 min, followed by antigen retrieval in boiling citric acid buffer. Slices were washed in PBS, blocked for 1 hr (4% normal goat serum and 3% Triton-X in PBS) and incubated in primary antibody for K48 polyubiquitin (1:500) overnight at 4°C. The following day slices were rinsed with PBS and incubated in Alexa Fluor 488 (1:500, #715-025-150, Jackson ImmunoResearch, West Grove, PA) secondary antibody for 2 hrs, rinsed with PBS and coverslipped with VectaShield Mounting Media Hardset with DAPI. Images were taken on a Nikon Eclipse Ci-L microscope (Melville, NY) and adjusted with Image J.
2.14. RNA/DNA Extractions
RNA and DNA were extracted from BLA lysates using the Qiagen (Germantown, MD) Allprep kit. RNA concentration was measured on the Take3 (BioTek, Winooski, VT), normalized (200ng) and converted to cDNA using the iScript cDNA synthesis kit (Bio-rad). The resulting DNA was measured on the Take3 and used for meDIP and bisulfite sequencing as described below.
2.15. Quantitative Real-Time PCR
Real-time PCR amplifications of the cDNA were performed on the Bio-rad CFX96 Real-Time System using the following protocol: 95.0°C for 3 min, then 95.0°C for 10 s, followed by 60°C for 30 s (39 repeats), 55–95°C for 0.5°C/cycle, followed by a melt curve starting at 55.0°C for 10 s (81 repeats), and then held at 4.0°C. Primers were PSMD14 (F: AAACAGGAAGGCCCGAGATG; R: CACACCAGAAAGCCAACAGC), Rps27a (F: CTTCCTTTCCGATCCGCCAT; R:GAGCGTGATGGTCTTCCCTG), Uba52 (F: TTTCTCTTCAACGAGGCGGC; R: AAGAGTGATGGTCTTGCCCG), Ubb (F: GAGAGGCTTTGTCCGGTTCC; R: CGATGAAGGAACAAACCGCC) and Ubc (F: CTCGTACCTTTCTCACCACAGT; R: GACACCTCCCCATCAAACCC).
2.16. Methylated DNA Immunoprecipitation (meDIP)
Methylated DNA immunoprecipitations were performed as described previously with a small-scale modification (Jarome et al., 2018). Normalized amounts of genomic DNA (1 μg) were sheared to ~300 bp using the QSonica Q800R2 Sonicator with 50% amplitude and 20 sec pulse for 30 min, denatured at 95°C for 15 min, diluted with IP buffer, mixed with MagnaChip magnetic protein A or protein A/G beads (MilliporeSigma, St. Louis, MO), and immunoprecipitated at 4 °C overnight with primary antibody against 5-hydroxymethylcytosine (5hmC; Active Motif, Carlsbad, CA). Immune complexes were sequentially washed with low salt buffer, high salt buffer, LiCl immune complex buffer, and TE buffer and extracted in 1× TE containing 1% SDS and 100 μg of proteinase K. DNA was extracted by phenol/chloroform/isoamyl alcohol, ethanolprecipitated and subjected to quantitative real-time PCR. The cumulative fluorescence for each amplicon was taken as a percentage of the input fraction and taken as a fold change of the control group. Primers were Uba52 Promoter (F: GCTGTCACGTGCTGAAGGTA; R: CTGGCAGGAGAAGCCAAACT) and Uba52 Exon I (F: CCGCTGTCCTCTTTCTCTTCA; R: GACCCCTACAGTCCCCTCG).
2.17. Direct Bisulfite Sequencing
Genomic DNA (50ng) was bisulfite converted through the Qiagen EpiTect Bisulfite Kit and amplified using a semi-nested PCR protocol in which 0.4 μl of the forward and 0.4 μl of the reverse primers (5 μM) were mixed with 1 μl of bisulfite treated DNA, 3.2 μl of nuclease free water, and 5 μl of HotStarTaq Master Mix (#203443, Qiagen). Samples were run on a thermal cycler for target amplification using the methyl-specific primers for Uba52 (F: TAAATAAAAATTATGTTTGAAAGGTAATAT; R:AAAAAAAACCAAACTATCTAAAACC). PCR parameters were as follows: 95°C for 5 min, then 49 cycles of 95°C for 1 min, 60°C for 1 min and 72°C for 1 min, followed by a 5 min incubation step at 72°C. Then, 1 μl of the product from the first set of primers was used to repeat the experiment with the same parameters and mix using the same reverse primer with forward primer 2 (AATAAAAATTATGTTTGAAAGGTAATATAG). Product sizes were verified on a gel. PCR product was cleaned using ExoSAP-IT (Affymetrix, Santa Clara, CA) and sequenced by the Genomics Sequencing Center at the Fralin Life Science Institute of Virginia Tech using the reverse primer. Analysis was done using Chromas software to read the electropherogram and percent methylation of the CpG sites was determined by the ratio between peak values of guanine (G) and cytosine (C), C / (C +T) * 100.
2.18. Statistical Analyses
All data are presented as mean with standard error, with scatter plots to identify individual samples (except in line graphs). Molecular data with two group comparisons were analyzed with two-tailed t-tests. Molecular data with four group comparisons were analyzed with two-way ANOVA and Fisher LSD posthoc test. Molecular data that was nonparametric, as indicated by a significant Bartlett’s test, was LOG transformed prior to analysis with two-way ANOVA. All behavioral data were analyzed with two-way ANOVA and Fisher LSD posthoc tests, unless otherwise noted. Statistical outliers were defined as those samples that were two or more standard deviations from the mean and were determined by the outlier function in Prism.
3. Results
3.1. Males and females differ in the baseline regulation and learning-dependent recruitment of protein degradation in the amygdala
Several studies have indicated that male and female rats perform differently on some, but not all, fear conditioning tasks (Cossio, Carreira, Vasquez, and Britton, 2016; Graham, 2009; Wood and Shors, 1998). We first assessed whether male and female rats differed in performance on our foreground contextual fear conditioning task (Figure 1A). We used this task because it is the most commonly utilized paradigm for studying protein degradation in the amygdala and other brain regions (Devulapalli et al., 2019; Jarome et al., 2016; Jarome et al., 2011; Lee et al., 2008; Orsi et al., 2019) and behaviorally produces similar performance in male and female Sprague Dawley rats (Graham, 2009). During training, we found that while both sexes showed enhanced fear across time, males showed greater fear responses overall than females (two-way repeated measure ANOVA, Sex effect F1,18 = 4.573, p = 0.0464; Time effect F4,72 = 26.36, p < 0.0001; Interaction F4,72 = 1.574, p = 0.1905; Figure 1B). However, during test there were no significant differences between groups, indicating that both males and females showed similar retention for the task (two-way repeated measure ANOVA, Sex effect F1,18 = 1.899, p = 0.1851; Time effect F4,72 = 6.47, p = 0.0002; Interaction F4,72 = 1.129, p = 0.3496; Figure 1C). Additionally, shock reactivity analysis did not reveal and differences between sexes in response to the shock stimulus during the training session (two-way ANOVA, Sex: F(1,68) = 0.0005943, p = 0.9806; Shock Number: F(3,68) = 2.109, p = 0.1072; Interaction: F(3,68) = 0.07605, p = 0.9727; Figure 1D), suggesting that differences in freezing behavior during the training session where unrelated to differences in sensory processing. These results indicate that while male and female rats may vary some in performance during the training of the fear conditioning task, both sexes show similar retention for it when tested at later times.
Figure 1. Male and female rats both acquire context fear memories.
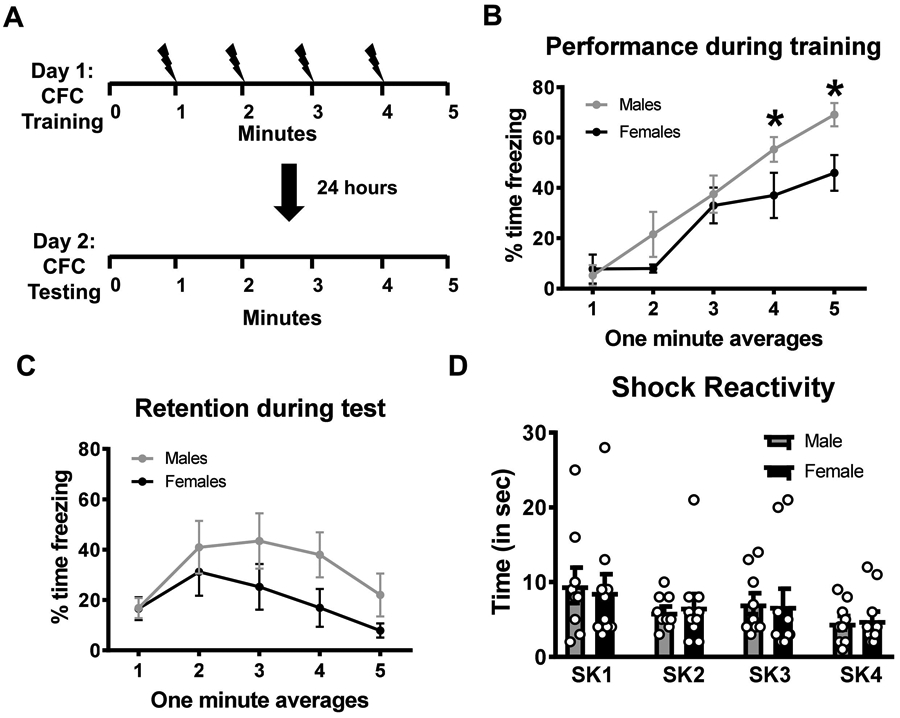
Young adult male and female rats (n = 10 per sex) were trained on a contextual fear conditioning paradigm and tested the following day for the retention to the training context. (A) Schematic of the training protocol in which 4 shocks were delivered over a 5 minute session. (B) Males showed higher freezing behavior than females during the training session, however, (C) both groups performed similar during the memory retention test. (D) Shock reactivity analysis did not reveal any differences between males and females in response to the shock stimulus during the training session. * p < 0.05 from Females.
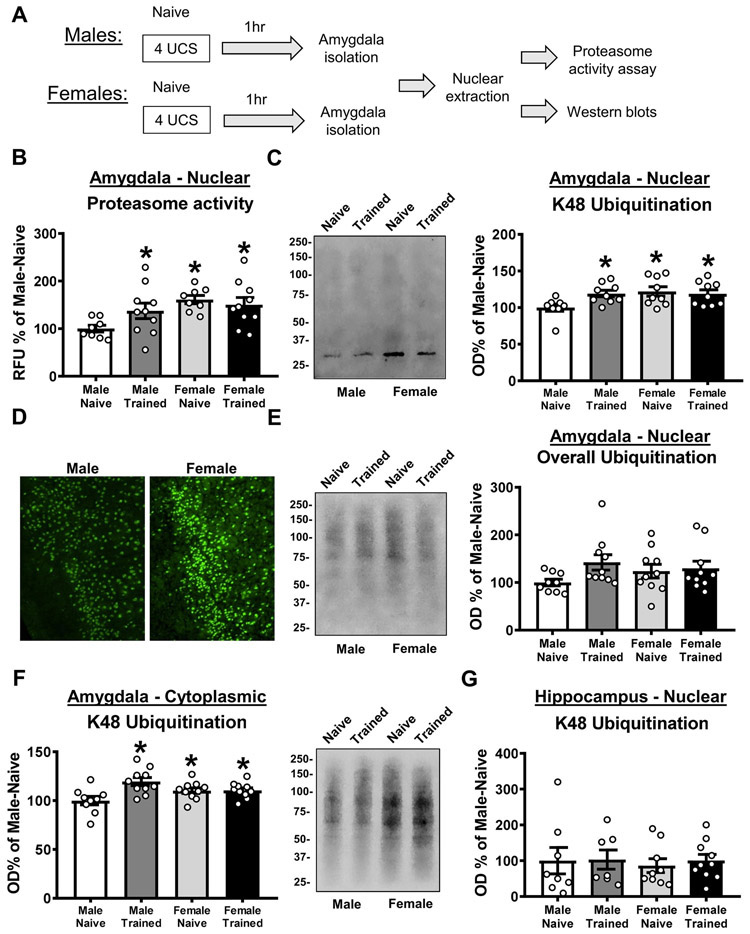
We next tested whether both male and female rats had increased protein degradation in the amygdala following fear conditioning (Figure 2A). For this we examined nuclear extracts of the basolateral amygdala (BLA) 1 hr after fear conditioning, as this is the timepoint and subcellular compartment in which we previously reported protein degradation changes in males (Orsi et al., 2019). For proteasome activity, we observed a significant main effect for sex (two-way ANOVA, F(1,32) = 7.509, p = 0.01; Figure 2B), but not Training (two-way ANOVA, F(1,32) = 0.9714, p = 0.3317), and there was not a significant interaction (two-way ANOVA, F(1,32) = 3.232, p = 0.0817). Posthoc analyses revealed that when comparing naïve and trained animals of the same sex, proteasome activity increased in males (p = 0.0578), but not females (p = 0.5698), following fear conditioning. Interestingly, both naïve (p = 0.0046) and trained (p = 0.0129) female animals had higher proteasome activity than naïve males, which was similar to that of trained males (Male-Trained vs Female-Naïve, p = 0.2237; Male-Trained vs Female-Trained, p = 0.4848), suggesting that females have higher resting levels of proteasome activity than males in the BLA. Consistent with this, we found a similar pattern for degradation-specific K48 polyubiquitination in the BLA where there was a significant interaction between sex and training (two-way ANOVA, F(1,31) = 4.298, p = 0.0466; Figure 2C) and a strong trend for a main effect of sex (two-way ANOVA, F(1,31) = 4.038, p = 0.0533). Posthoc analyses revealed that when comparing naïve and trained animals of the same sex, K48 levels increased in males (p = 0.0191), but not females (p = 0.6720), following fear conditioning. Again, both naïve (p = 0.0078) and trained (p = 0.0211) female animals had higher K48 levels than naïve males, which was similar to that of trained males (Male-Trained vs Female-Naïve, p = 7053; Male-Trained vs Female-Trained, p = 0.9638). This suggest that females have higher resting levels of K48 polyubiquitination than males in the BLA, which we confirmed with immunofluorescence, and applied to both the basal and lateral nuclei of the amygdala (Figure 2D). We did not see these learning-dependent or baseline differences in overall protein polyubiquitination levels in the BLA (two-way ANOVA, Sex: F(1,35) = 0.1557, p = 0.6956; Training: F(1,35) = 2.975, p = 0.0934; Interaction: F(1,35) = 1.721, p = 0.1981; Figure 2E). In the BLA cytoplasmic fraction we found a significant effect for training (two-way ANOVA, F(1,35) = 8.096, p = 0.0074) and a sex by training interaction (two-way ANOVA, F(1,35) = 7.57, p = 0.0093; Figure 2F). Importantly, while males showed increases in K48 polyubiquitination in BLA cytoplasmic fractions following learning (p = 0.0004), females did not (p = 0.9466), suggesting that the lack of change in the nucleus in females was not due to them engaging protein degradation in a different subcellular compartment than males. However, again naïve (p = 0.0485) and trained (p = 0.0421) females had elevated baseline levels of K48 polyubiquitination when compared with naïve males. Additionally, neither males nor females engaged protein degradation (indicated by K48 polyubiquitination) in nuclear fractions from the hippocampus following fear conditioning (two-way ANOVA, Sex: F(1,32) = 0.1631, p = 0.6890; Training: F(1,32) = 0.1445, p = 0.7063; Interaction: F(1,32) = 0.02466, p = 0.8762; Figure 2G) and no baseline differences were observed between sexes, suggesting that females have a general upregulation of protein degradation at rest in comparison to males that is selective to the amygdala. Collectively, these results suggest that while both males and females showed similar performance during the fear conditioning task, they differ in the baseline regulation of the protein degradation process in the amygdala and only males engage the ubiquitin-proteasome system in this region following fear conditioning.
Figure 2. Males and females differ in baseline and learning-dependent ubiquitin-proteasome activity in the amygdala.
(A) Schematic of the experimental design. Basolateral amygdala (BLA) nuclear fractions were collected from male and female rats 1 hr after training to a contextual fear conditioning procedure (n = 8-10 per group per sex). (B) Relative to naïve animals of the same sex, males (white bar vs dark grey bar), but not females (light grey bar vs black bar), had increased proteasome activity following fear conditioning. Conversely, females had elevated baseline levels of proteasome activity relative to males (white bar vs light grey bar). (C) Relative to naïve animals of the same sex, males (white bar vs dark grey bar), but not females (light grey bar vs black bar), had increased K48 polyubiquitination following fear conditioning. Conversely, females had elevated baseline levels of K48 polyubiquitination relative to males (white bar vs light grey bar). (D) Qualitative immunofluorescence analysis confirmed higher baseline level of K48 polyubiquitination in the BLA of naïve females relative to naïve males. (E) Relative to naïve animals of the same sex, neither males (white bar vs dark grey bar) or females (light grey bar vs black bar) had learning-dependent increases in overall protein polyubiquitination following fear conditioning. Males and females did not differ in baseline levels of overall protein polyubiquitination (white bar vs light grey bar). (F) Relative to the same sex naïve control, males (white bar vs dark grey bar), but not females (light grey bar vs black bar), have increased K48 polyubiquitination levels in the cytoplasm of basolateral amygdala (BLA) cells 1 hour after fear conditioning. (G) Males and females did not differ in learning-dependent or baseline K48 polyubiquitination levels in the dorsal hippocampus following fear conditioning. Representative western blot images were taken from the same gel in the order shown. * p < 0.05 from Naïve-Male.
3.2. Females do not have delayed increases in protein degradation in the amygdala following fear conditioning
In our previous work with male rats we found both early (1 hr) and delayed (4 hrs) increases in protein degradation in the amygdala following fear conditioning (Jarome et al., 2013; Jarome et al., 2011). Though in our present study females did not have increases in protein degradation soon after behavioral training, it is possible that they do at later timepoints in the consolidation process. In order to test this, we next examined changes in ubiquitin-proteasome activity in the BLA of female rats 4 hrs after fear conditioning (Figure 3A). Consistent with our first experiment, we found no changes in proteasome activity (t14 = 0.1455, p = 0.8864; Figure 3B) or K48 polyubiquitination (t14 = 0.2363, p = 0.8166; Figure 3C) in nuclear fractions from the amygdala.
Figure 3. Females do not have delayed increases in protein degradation in the amygdala following fear learning.
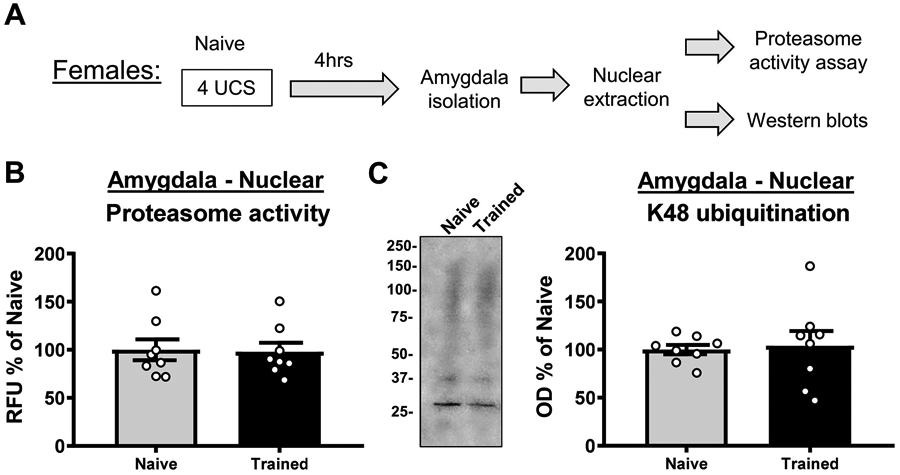
(A) Schematic of the experimental design. Basolateral amygdala (BLA) nuclear fractions were collected from female rats 4 hr after training to a contextual fear conditioning procedure (n = 8 per group). (B-C) Relative to naïve animals, fear learning did not change levels of proteasome activity (B) or K48 polyubiquitination (C) in the BLA.
3.3. Increased baseline protein degradation in the amygdala of females is due to increased epigenetic regulation of Uba52
Our results suggest that females do not show increases in protein degradation in the amygdala following fear learning, which could be due to their high resting level of ubiquitin-proteasome activity, an effect that has been observed in other peripheral tissues but not yet reported in the brain (Jenkins et al., 2020). One possible reason for why females have higher protein degradation at baseline is that they could be using ubiquitin more efficiently, tagging substrates at a higher/faster rate than males. However, when we examined free ubiquitin levels in the BLA we found that naïve females had elevated levels in comparison to naïve males (t test, t16 = 2.125, p = 0.0495; Figure 4A). These results suggest that females have higher levels of free and conjugated ubiquitin in the amygdala at baseline, which may lead to increased protein degradation at rest.
Figure 4. Females have increased baseline promoter 5-hydroxymethylation and expression of the ubiquitin coding gene Uba52 in the amygdala.
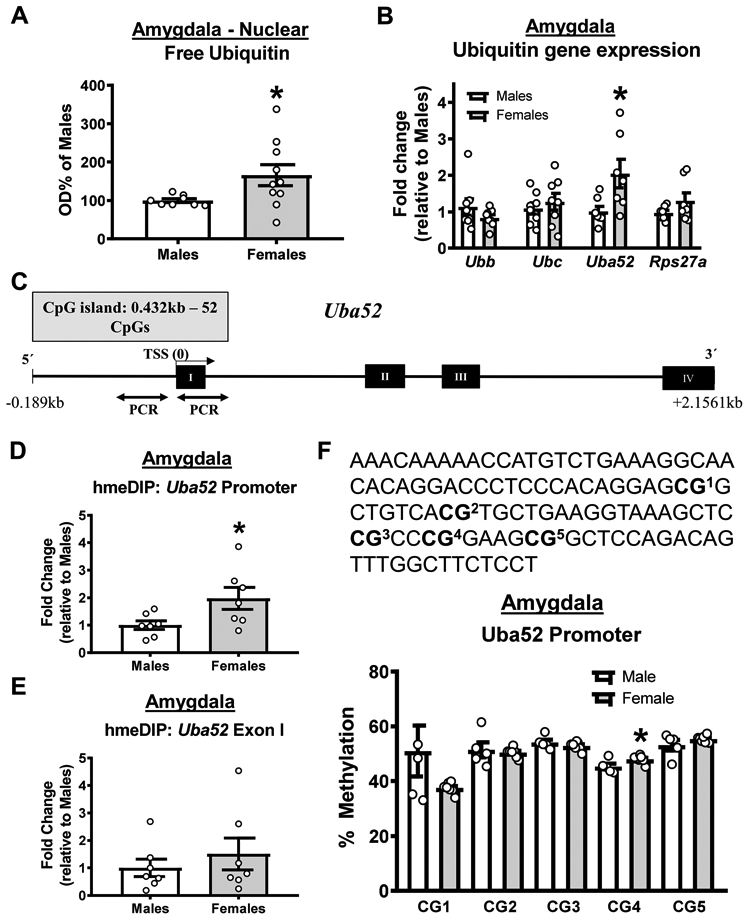
(A) Females have increased baseline levels of free ubiquitin in the basolateral amygdala (BLA; n = 8-10 per group) (B). qRT-PCR analysis of RNA collected from BLA of naïve male and female rats (n = 7-8 per group). Females had elevated baseline levels of Uba52, but not Ubb, Ubc or Rps27a, in the BLA. (C) Schematic of the Uba52 gene showing CpG island and site of methylated DNA immunoprecipitation (meDIP) PCR analysis. (D-E) DNA 5-hydroxymethylation was increased at the Uba52 promoter (D) but not coding (E) region in females (n = 7 per group). (F) Bisulfite sequencing of 5 CpG sites in the Uba52 promoter in naïve males and females revealed an increase in methylation at CpG4 in females (n = 5-7 per group). * p < 0.05 from Males.
We next examined why females have higher baseline levels of ubiquitin the amygdala. First, we quantified the expression of the four ubiquitin coding genes (Ubb, Ubc, Uba52, Rps27a), which all code for the same ubiquitin protein, in BLA tissue collected from naïve male and female rats. At baseline, females had higher expression of a single ubiquitin coding gene in the BLA, Uba52 (t test, Ubb t14 = 1.227, p = 0.2402; Ubc t14 = 0.6534, p = 0.5241; Uba52 t12 = 2.49, p = 0.0284; Rps27a t12 = 1.432, p = 0.1778; Figure 4B). This Uba52 dual-coding gene has a large CpG island in its promoter region that runs through the first exon (Figure 4C). These CpG islands are the primary site of DNA methylation in the genome, suggesting that the upregulation of Uba52 expression in females may be due to sex differences in epigenetic regulation of this gene. To test this, we performed methylated DNA immunoprecipitation assays targeting two CpGs in the Uba52 promoter region that were −152 and −157 base pairs from the transcriptional start site. While we were unable to detect the repressive 5-methylcytosine form of DNA methylation, which is consistent with the high transcription rate of this gene, we observed significantly higher levels of the transcriptional activator DNA 5-hydroxymethylation (5hmc) in the Uba52 promoter (t test, t12 = 2.285, p = 0.0413; Figure 4D), but not coding (t test, t12 = 0.7694, p = 0.4565; Figure 4E) region. Additionally, this was confirmed with bisulfite sequencing, which revealed an increase in DNA methylation of a CpG dinucleotide −31 base pair upstream of the Uba52 transcriptional start site (t test, t10 = 2.438, p = 0.0350; Figure 4F). Collectively, these results suggest that females likely have more ubiquitin in the amygdala at baseline due to increased epigenetic regulation of the Uba52 gene.
3.4. Baseline increases in amygdala protein degradation are critical for fear memory formation in females
Our data suggests that Uba52-dependent elevated baseline levels of protein degradation in the amygdala may be critical for fear memory formation in females. In order to test this, we used the CRISPR-dCas9-KRAB-MECP2 system to persistently repress endogenous expression of Uba52 in the BLA of females (Figure 5A top and middle). Additionally, in order to reduce proteasome activity, which was also elevated at baseline (Figure 2B), we multiplexed with a guide RNA against Psmd14; this subunit of the proteasome is essential for proteasome assembly and function and upregulation of Psmd14 in invertebrates has been shown to increase proteasome activity (Tonoki et al., 2009). This CRISPR-dCas9 method was chosen over pharmacological proteasome manipulations because 1) the dCas9-KRAB-MECP2 fusion is a robust transcriptional repressor that can result in sustained reduction of Uba52 and Psmd14 specifically, 2) it will allow us to reduce but not abolish functional proteasome activity, preventing a potentially toxic accumulation of ubiquitinated proteins and depletion of the free ubiquitin pool, and 3) it is capable of overcoming the euchromatin state that is associated with the increased DNA 5hmc levels we found at the Uba52 promoter. Two weeks after infusion of the CRISPR plasmids into the BLA, we observed significant reductions in Uba52 (t test, t17 = 2.728, p = 0.0143; Figure 5B) and Psmd14 (t test, t16 = 3.829, p = 0.0015; Figure 5B) levels, which correlated with reduced K48 polyubiquitination (t test, t15 = 2.38, p = 0.0310; Figure 5C) and proteasome activity (t test, t15 = 2.159, p = 0.0474; Figure 5D) in this region. The expression of the other gene ubiquitin coding genes was not altered by our manipulation of Uba52 (Figure 5B). This suggests that we were able to successfully reduce, but not abolish, baseline protein degradation in the amygdala of females using the CRISPR-dCas9 system. Importantly, while this manipulation had no effect on animal performance during the fear conditioning task (two-way repeated measure ANOVA, Group effect F1,17 = 1.269, p = 0.2756; Time effect F4,68 = 47.39, p < 0.0001; Interaction F4,68 = 0.6287, p = 0.6437; Figure 5E), it significantly impaired memory retention during test the following day (two-way repeated measure ANOVA, Group effect F1,17 = 10.37, p = 0.0050; Time effect F4,68 = 1.192, p = 0.3224; Interaction F4,68 = 0.7906, p = 0.5354; Figure 5F). Next, we artificially enhanced resting levels of ubiquitin-proteasome activity in the BLA of female rats. To do this, we used the same gRNAs from the previous experiment in combination with a double VP64-dCas9 fusion (Figure 5A top and bottom), a robust transcriptional activator, under weaker training conditions (lower intensity shock, 0.6mA). Two weeks after infusion of the CRISPR plasmids into the BLA, we found that this manipulation had no effect on performance during the fear conditioning task (two-way repeated measure ANOVA, Group effect F1,14 = 0.2145, p = 0.6504; Time effect F4,56 = 31.76, p < 0.0001; Interaction F4,56 = 0.07802, p = 0.9887; Figure 5G), nor did it enhance memory retention during the one minute intervals of the test session the following day (two-way repeated measure ANOVA, Group effect F1,14 = 3.04, p = 0.1032; Time effect F4,56 = 3.446, p = 0.0138; Interaction F4,56 = 0.5095, p = 0.7289; Figure 5H). However, this lack of a group effect was likely driven by the experimental group showing signs of extinction across the testing session, which was not evident in the control group that had low freezing levels throughout. Consistent with this, when we compared average overall freezing behavior across the entire test session we found that the CRISPR manipulation enhanced memory in females (U = 11, p = 0.0281). Collectively, these results suggest that females do need protein degradation in the amygdala for fear memory formation and that elevated baseline levels of this likely bypass the need for learning-dependent increases in ubiquitin-proteasome activity.
Figure 5. CRISPR-dCas9-KRAB-MECP2-mediated reductions in baseline protein degradation in the amygdala impairs fear memory in females.
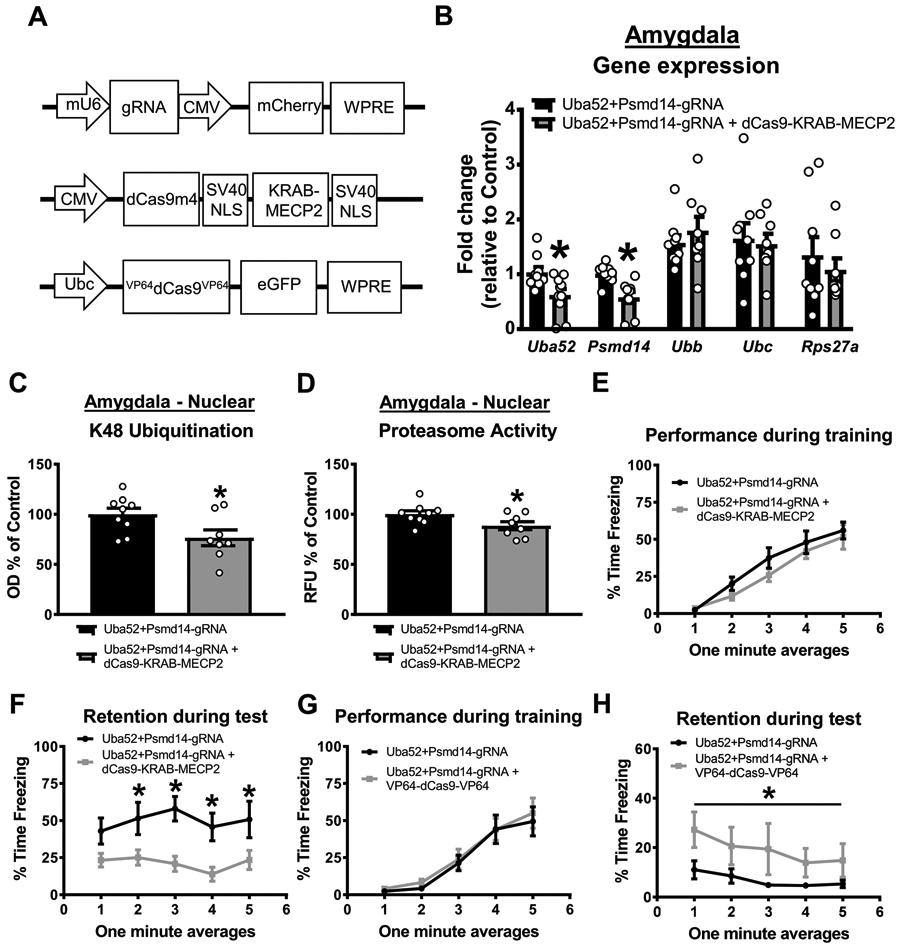
Female rats received basolateral amygdala (BLA) injections of CRISPR guide RNA (gRNA) targeting the ubiquitin coding gene Uba52 and proteasome subunit Psmd14 with or without dCas9-KRAB-MECP2 or VP64-dCas9-VP64 fusion. Two weeks later, animals were trained and tested on a contextual fear conditioning paradigm. (A) Schematic of the gRNA (bottom), dCas9-KRAB-MECP2 fusion (middle) or VP64-dCas9-VP64 fusion. (B) Two weeks after co-infusion of gRNA and dCas9-KRAB-MECP2 there was reduced expression of Uba52 and Psmd14, but not Ubb, Ubc or Rps27a, gene expression in the BLA (n = 9-10 per group). (C-D) This decreases in gene expression correlated with reduced K48 polyubiquitination (C) and proteasome activity (D) in the BLA (n = 8-9 per group). (E-F) Co-infusion of gRNA and dCas9-KRAB-MECP2 did not impair performance during the fear conditioning task (E) but significantly impaired long-term memory when tested the following day (F; n = 9-10 per group). (G-H) Female rats received basolateral amygdala (BLA) injections of CRISPR gRNA targeting the ubiquitin coding gene Uba52 and proteasome subunit Psmd14 with or without the VP64-dCas9-VP64 transcriptional activator fusion. Two weeks later, animals were trained and tested to a weak contextual fear conditioning paradigm in which the shock intensity was lowered to 0.6mA (still receiving 4 shocks). Co-infusion of gRNA and VP64-dCas9-VP64 did not alter performance during the fear conditioning task (G) but significantly enhanced long-term memory during the test session the following day (H; n = 8 per group). Line denotes that the average freezing across the entire testing session (all minutes summed) was significantly different between groups. * p < 0.05 from Uba52+Psmd14-gRNA.
3.5. Increasing baseline protein degradation in the amygdala of males enhances fear memory
To further test the importance of amygdala protein degradation to the consolidation of fear memory in females, we next tested whether manipulating baseline protein degradation in the amygdala of males could control fear memory formation. First, we reduced baseline protein degradation in males using the same CRISPR-dCas9-KRAB-MECP2 system described above targeting Uba52 and Psmd14 promoter regions. Two weeks after infusion of the CRISPR plasmids into the BLA, we found that while this manipulation had no effect on the animals performance during the fear conditioning task (two-way repeated measure ANOVA, Group effect F1,7 = 1.794, p = 0.2223; Time effect F4,28 = 14.64, p < 0.0001; Interaction F4,28 = 0.9723, p = 0.4383; Figure 6A), it significantly impaired memory retention during test the following day (two-way repeated measure ANOVA, Group effect F1,7 = 3.065, p = 0.1234; Time effect F4,28 = 4.300, p = 0.0078; Interaction F4,28 = 5.034, p = 0.0035; Figure 6B). Next, we artificially enhanced resting levels of ubiquitin-proteasome activity in the BLA of male rats using dCas9-VP64 under weaker training conditions (lower intensity shock, 0.6mA). Two weeks after infusion of the CRISPR plasmids into the BLA, we found that while this manipulation had no effect on performance during the fear conditioning task (two-way repeated measure ANOVA, Group effect F1,6 = 0.6193, p = 0.4613; Time effect F4,24 = 33.75, p < 0.0001; Interaction F4,24 = 2.066, p = 0.1169; Figure 6C), it significantly enhanced memory retention during test the following day (two-way repeated measure ANOVA, Group effect F1,6 = 32.04, p = 0.0013; Time effect F4,24 = 3.696, p = 0.0176; Interaction F4,24 = 1.839, p = 0.1543; Figure 6D). In combination with our previous experiments, these results suggest that behaviorally both males and females have a similar need for protein degradation in the amygdala following fear conditioning, though they differ significantly in how this process is engaged and regulated within in this brain region (Figure 7).
Figure 6. CRISPR-VP64-dCas9-VP64-mediated increases in baseline protein degradation in the amygdala enhances fear memory in males.
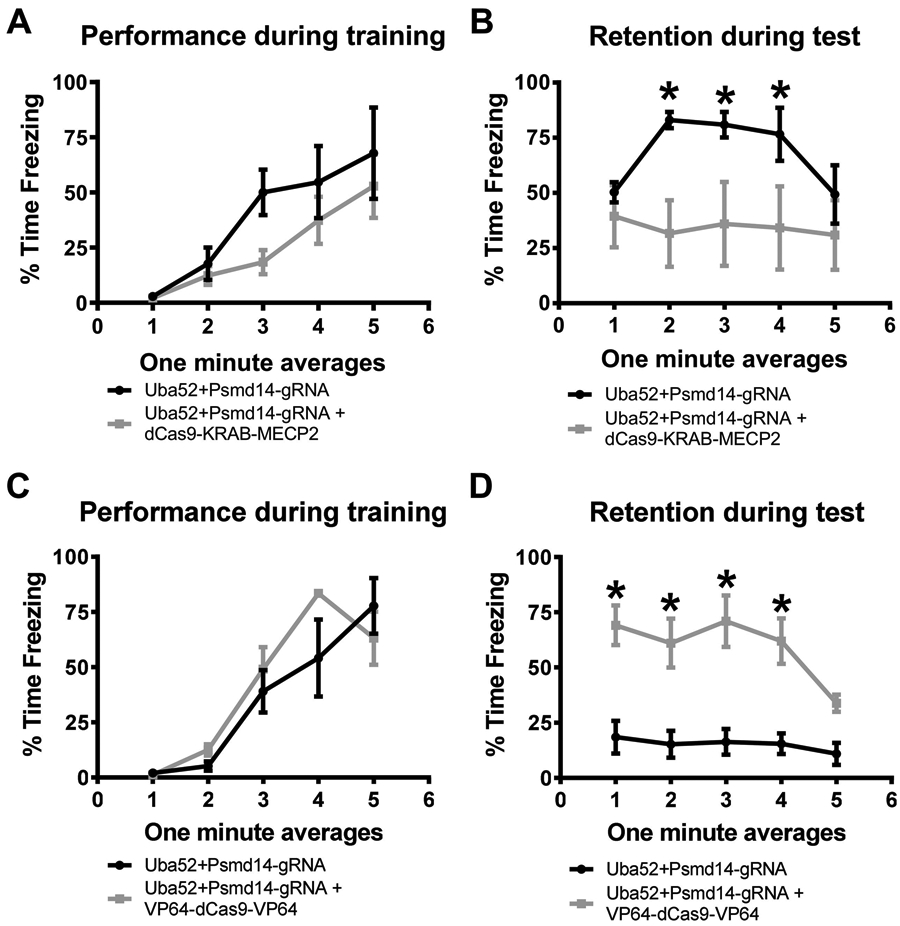
(A-B) Male rats received basolateral amygdala (BLA) injections of CRISPR guide RNA (gRNA) targeting the ubiquitin coding gene Uba52 and proteasome subunit Psmd14 with or without dCas9-KRAB-MECP2 fusion. Two weeks later, animals were trained and tested to a contextual fear conditioning paradigm. Co-infusion of gRNA and dCas9-KRAB-MECP2 did not alter performance during the fear conditioning task (A) but significantly impaired long-term memory when tested the following day (B; n = 4-5 per group). (C-D) Male rats received basolateral amygdala (BLA) injections of CRISPR gRNA targeting the ubiquitin coding gene Uba52 and proteasome subunit Psmd14 with or without the VP64-dCas9-VP64 transcriptional activator fusion. Two weeks later, animals were trained and tested to a weak contextual fear conditioning paradigm in which the shock intensity was lowered to 0.6mA (still receiving 4 shocks). Co-infusion of gRNA and VP64-dCas9-VP64 did not alter performance during the fear conditioning task (C) but significantly enhanced long-term memory when tested the following day (D; n = 4 per group). * p < 0.05 from Uba52+Psmd14-gRNA.
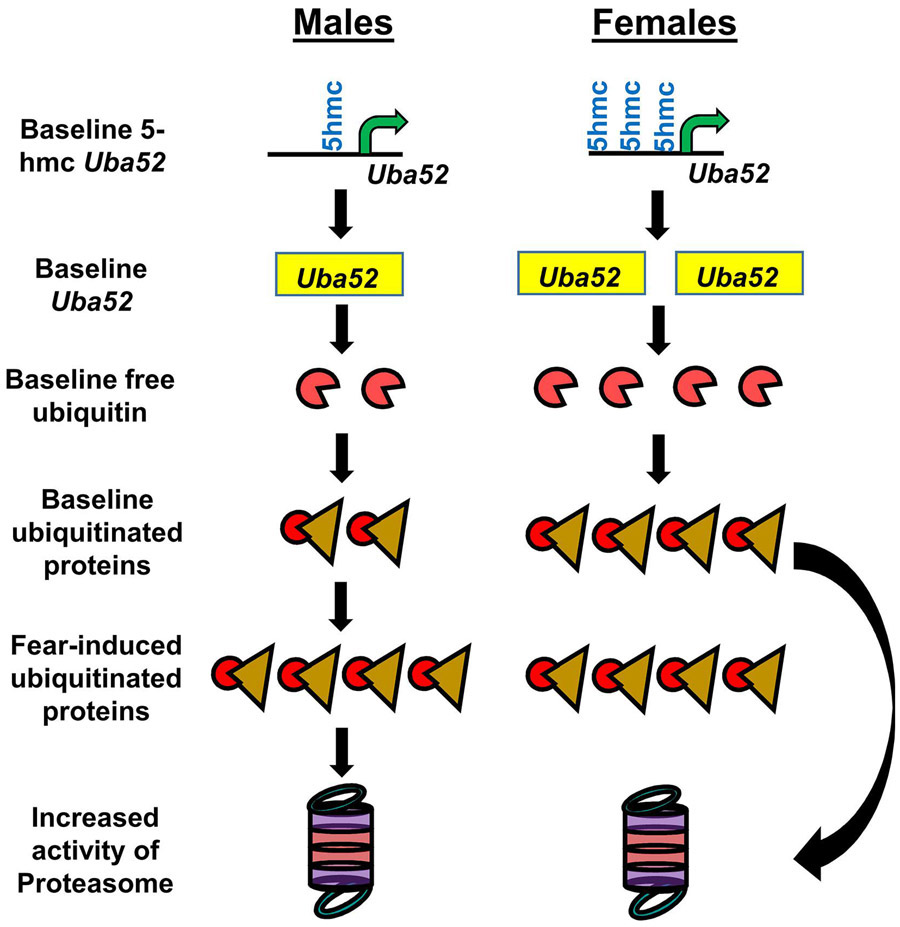
Figure 7. Model of how increased levels of baseline protein degradation in the amygdala may be sufficient for females to acquire fear memories.
At rest females have higher levels of DNA 5-hydroxymethylation (5hmc) of the Uba52 promoter than males. This leads to increased expression of Uba52, which results in greater free and conjugated ubiquitin levels in the female amygdala. This elevated state of ubiquitination causes a reciprocal increase in proteasome activity, resulting in levels of protein degradation typically seen in males only after fear conditioning.
4. Discussion
Protein degradation mediated by the ubiquitin-proteasome system has been widely reported to be involved in the process of fear memory formation (Jarome et al., 2016; Jarome and Helmstetter, 2013; Jarome et al., 2014; Jarome et al., 2011; Lee et al., 2008; Lopez-Salon et al., 2001; Orsi et al., 2019; Reis et al., 2013; Rodriguez-Ortiz et al., 2011; Yeh, Mao, Lin, and Gean, 2006), however, whether this requirement is shared in both males and females remained unknown. Here, we found that males, but not females, had increased levels of protein degradation in the amygdala following contextual fear conditioning. This learning-dependent sex difference was associated with elevated baseline levels of ubiquitin-proteasome activity in the amygdala, which was due to increased epigenetic-mediated expression of the ubiquitin coding gene Uba52. Furthermore, CRISPR-dCas9-mediated repression of baseline protein degradation levels in the amygdala impaired fear memory formation in both males and females while enhancing protein degradation promoted memory in both sexes. Collectively, these results suggest that both males and females require protein degradation in the amygdala for fear memory formation, though differ in how this system is regulated at baseline and engaged following learning. Importantly, females may be able to acquire fear memories through elevated baseline activity of the ubiquitin-proteasome system in the amygdala (Figure 7).
Our data strongly suggest that unlike males (Jarome et al., 2011), females do not need activity-dependent increases in protein degradation in the amygdala for fear memory formation, though other interpretations could exist. For example, it is possible that the protein targets of ubiquitin and the proteasome change in the amygdala following fear conditioning. Such a change in protein targets would allow increased degradation of a subset of proteins critical for fear memory formation without changing net levels of overall and degradation-specific polyubiquitination. However, this theory is particularly difficult to test as, to date, the protein targets of ubiquitin and the proteasome remain largely unidentified (Jarome et al., 2011; Lee et al., 2008). Furthermore, this possibility does not negate the significance of our data showing increased baseline levels of protein degradation in the amygdala. Additionally, while our CRISPR-dCas9 manipulation directly reduced baseline proteasome activity, it would have still been possible for the protein targets of ubiquitin to change in response to fear learning as ubiquitin ligase and deubiquitinating activity remained intact. If the above alternative interpretation was correct then, our manipulation should not have been able to impair fear memory. Thus, while we cannot completely rule out this possibility, our data strongly suggest that females do not need activity-dependent changes in protein degradation in the amygdala for fear memory formation.
The behavioral deficit we observed in males and females following sustained CRISPR-dCas9-KRAB-MECP2 reductions in amygdala ubiquitin levels and proteasome activity is similar to what we have previously reported in male animals following transient pharmacological inhibition of the proteasome (Jarome et al., 2011). This leads to the interpretation that fear learning in both males and females is sensitive to protein degradation inhibition in the amygdala. However, several important differences exist between these studies. First, in our previous work the pharmacological manipulation used renders proteasomes irreversibly nonfunctional, resulting in a potentially toxic accumulation of ubiquitinated proteins and depletion of the free ubiquitin pool (Jarome et al., 2011). Conversely, in the present study our CRISPR-dCas9-KRAB-MECP2 manipulation reduced but did not abolish proteasome activity, which occurred simultaneously with reductions in the levels of ubiquitinated proteins. This latter method then was better able to sustain a normal balance between ubiquitin protein tagging and proteasome activity, albeit at reduced levels from the normal physiological state. Additionally, our CRISPR-dCas9-KRAB-MECP2 manipulation is persistent and able to stably control protein degradation across time, as opposed to the transient nature of the pharmacological manipulation, and could achieve cell-type specificity if desired. Thus, while the pharmacological manipulations in our previous work and the CRISPR-dCas9-KRAB-MECP2 manipulation in our present study both resulted in behavioral deficits, it is important to note that the former method is better for activity-dependent changes while the latter is better suited for sustained manipulations. Additionally, in general, the CRISPR-dCas9-KRAB-MECP2 method we describe here is better at maintaining ubiquitin-proteasome homeostasis, which could alleviate confounding effects from toxicity that may occur with abnormal accumulation of ubiquitinated proteins following proteasome inhibition. Collectively, these results suggest that our CRISPR-dCas9 approach can achieve highly specific, bidirectional, sustained control over the protein degradation process, which could have significant implications for the study of ubiquitin-proteasome system in normal and abnormal disease states.
Contextual fear memories require both the amygdala and hippocampus for their long-term storage (Kim and Fanselow, 1992; Phillips and LeDoux, 1992). However, we found that baseline protein degradation was increased in the amygdala, but not hippocampus, of females. This suggests that in the brain these sex-dependent increases in baseline ubiquitin proteasome system may be unique to the amygdala. Importantly, this data adds to compelling recent evidence showing that females have higher resting activity of the proteasome across a variety of peripheral tissues (Jenkins et al., 2020). While the exact reasons for this brain region selective increase in resting protein degradation are unknown, one possibility is that it helps regulate estrogen signaling in this region. Estrogen receptors are abundant in the amygdala (Mitra et al., 2003) and the proteasome is heavily involved in regulating the expression and function of these receptors (Lonard, Nawaz, Smith, and O'Malley, 2000; Nawaz et al., 1999). It is possible then that increased baseline protein degradation may be needed in this region to regulate the activation and continued turnover of estrogen receptors. However, as stated above, the protein targets of the proteasome during baseline or learning-dependent synaptic plasticity remain unknown, making it difficult to determine the exact reasons for this elevation of resting protein degradation levels in the amygdala of females.
One surprising result from our study was that only one of the ubiquitin coding genes, Uba52, was increased at baseline in the amygdala of females. This result is especially significant because while Ubb and Ubc each code for multiple ubiquitin, Uba52 only codes for a single ubiquitin fused at the C-terminus to ribosomal protein L40 (Callis, 2014; Musaus et al., 2020). In the case of the latter, ubiquitin does not become functional until it is post-translationally cleaved from the ribosomal protein. Importantly, how each of the four ubiquitin coding genes serves as a source of ubiquitin for the free ubiquitin pool remains equivocal (Kobayashi et al., 2016; Park and Ryu, 2014). While the significance of only Uba52 being upregulated in the amygdala of females is unclear, our data strongly suggest that it is responsible for increased ubiquitin levels at baseline and that this upregulation is mediated by a DNA 5-hydroxymethylation-dependent process. Future studies should aim to determine what targets DNA 5-hydroxymethylation enzymes to the Uba52 in the amygdala, which could provide a better understanding of why this specific ubiquitin coding gene is selectively upregulated in the amygdala of females.
5. Conclusion
In conclusion, we found a novel sex difference in the role and regulation of ubiquitin-proteasome mediated protein degradation in the amygdala during fear memory formation. In addition, baseline increases in protein degradation in the amygdala of females may be sufficient for them to acquire fear memories. Collectively, these data suggest that while experience produces similar mnemonic and behavioral endpoints, males and females differ significantly in how they regulate and engage the ubiquitin-proteasome system in the amygdala. These results provide important information for understanding the etiology of sex-related differences in fear memory formation, which could have implications for understanding the molecular biology behind sex differences in the development of post-traumatic stress disorder
Highlights.
Males, but not females, have increased amygdala protein degradation following fear learning
Females have higher resting levels of protein degradation in the amygdala
These higher resting levels may be due to increased epigenetic regulation of Uba52
Genetic reduction in baseline protein degradation impairs memory in males and females
Genetic increase in baseline protein degradation enhances memory in males and females
Acknowledgements
This work was supported by National Institute of Health (NIH) grants MH120498, MH120569, MH122414 and MH123742 and startup funds from the College of Agricultural and Life Sciences and the School of Neuroscience at Virginia Tech (T.J.J.). T.M. is supported by the George Washington Carver Program at Virginia Tech. The authors declare no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- Bisaz R, Travaglia A, & Alberini CM (2014). The neurobiological bases of memory formation: from physiological conditions to psychopathology. Psychopathology, 47, 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler AA, Johnston DR, Kaur S, & Lubin FD (2019). Long noncoding RNA NEAT1 mediates neuronal histone methylation and age-related memory impairment. Sci Signal, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J (2014). The ubiquitination machinery of the ubiquitin system. Arabidopsis Book, 12, e0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, & Kwon YT (2015). Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp Mol Med, 47, e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossio R, Carreira MB, Vasquez CE, & Britton GB (2016). Sex differences and estrous cycle effects on foreground contextual fear conditioning. Physiol Behav, 163, 305–311. [DOI] [PubMed] [Google Scholar]
- Cullen PK, Ferrara NC, Pullins SE, & Helmstetter FJ (2017). Context memory formation requires activity-dependent protein degradation in the hippocampus. Learn Mem, 24, 589–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma NP, & Bott LC (2014). The ubiquitin-proteasome system in neurodegenerative diseases: precipitating factor, yet part of the solution. Front Mol Neurosci, 7, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devulapalli RK, Nelsen JL, Orsi SA, McFadden T, Navabpour S, Jones N, Martin K, O'Donnell M, McCoig EL, & Jarome TJ (2019). Males and Females Differ in the Subcellular and Brain Region Dependent Regulation of Proteasome Activity by CaMKII and Protein Kinase A. Neuroscience, 418, 1–14. [DOI] [PubMed] [Google Scholar]
- Dong C, Bach SV, Haynes KA, & Hegde AN (2014). Proteasome modulates positive and negative translational regulators in long-term synaptic plasticity. J Neurosci, 34, 3171–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD (2003). Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci, 6, 231–242. [DOI] [PubMed] [Google Scholar]
- Fioravante D, Liu RY, & Byrne JH (2008). The ubiquitin-proteasome system is necessary for long-term synaptic depression in Aplysia. J Neurosci, 28, 10245–10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca R, Vabulas RM, Hartl FU, Bonhoeffer T, & Nagerl UV (2006). A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron, 52, 239–245. [DOI] [PubMed] [Google Scholar]
- Frick KM, Kim J, Tuscher JJ, & Fortress AM (2015). Sex steroid hormones matter for learning and memory: estrogenic regulation of hippocampal function in male and female rodents. Learn Mem, 22, 472–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny X, Saring P, Bergado Acosta JR, Kahl E, Kolodziejczyk MH, Cammann C, Wernecke KEA, Mayer D, Landgraf P, Seifert U, Dieterich DC, & Fendt M (2019). Deficiency of the immunoproteasome subunit beta5i/LMP7 supports the anxiogenic effects of mild stress and facilitates cued fear memory in mice. Brain Behav Immun, 80, 35–43. [DOI] [PubMed] [Google Scholar]
- Graham LK, Yoon T, Lee HJ, Kim JJ (2009). Strain and sex differences in fear conditioning: 22 kHz ultrasonic vocalizations and freezing in rats. Psychology & Neuroscience, 2, 219–225. [Google Scholar]
- Gresack JE, Schafe GE, Orr PT, & Frick KM (2009). Sex differences in contextual fear conditioning are associated with differential ventral hippocampal extracellular signal-regulated kinase activation. Neuroscience, 159, 451–467. [DOI] [PubMed] [Google Scholar]
- Hoeffer CA, Cowansage KK, Arnold EC, Banko JL, Moerke NJ, Rodriguez R, Schmidt EK, Klosi E, Chorev M, Lloyd RE, Pierre P, Wagner G, LeDoux JE, & Klann E (2011). Inhibition of the interactions between eukaryotic initiation factors 4E and 4G impairs long-term associative memory consolidation but not reconsolidation. Proc Natl Acad Sci U S A, 108, 3383–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome TJ, Ferrara NC, Kwapis JL, & Helmstetter FJ (2016). CaMKII regulates proteasome phosphorylation and activity and promotes memory destabilization following retrieval. Neurobiol Learn Mem, 128, 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome TJ, & Helmstetter FJ (2013). The ubiquitin-proteasome system as a critical regulator of synaptic plasticity and long-term memory formation. Neurobiol Learn Mem, 105, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome TJ, Kwapis JL, Hallengren JJ, Wilson SM, & Helmstetter FJ (2014). The ubiquitin-specific protease 14 (USP14) is a critical regulator of long-term memory formation. Learn Mem, 21, 9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome TJ, Kwapis JL, Nye SH, & Helmstetter FJ (2010). Introgression of Brown Norway chromosome 1 onto the fawn hooded hypertensive background rescues long-term fear memory deficits. Behav Genet, 40, 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome TJ, Kwapis JL, Ruenzel WL, & Helmstetter FJ (2013). CaMKII, but not protein kinase A, regulates Rpt6 phosphorylation and proteasome activity during the formation of long-term memories. Front Behav Neurosci, 7, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome TJ, Perez GA, Hauser RM, Hatch KM, & Lubin FD (2018). EZH2 Methyltransferase Activity Controls Pten Expression and mTOR Signaling during Fear Memory Reconsolidation. J Neurosci, 38, 7635–7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome TJ, Werner CT, Kwapis JL, & Helmstetter FJ (2011). Activity dependent protein degradation is critical for the formation and stability of fear memory in the amygdala. PLoS One, 6, e24349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins EC, Shah N, Gomez M, Casalena G, Zhao D, Kenny TC, Guariglia SR, Manfredi G, & Germain D (2020). Proteasome mapping reveals sexual dimorphism in tissue-specific sensitivity to protein aggregations. EMBO Rep, 21, e48978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josselyn SA, & Tonegawa S (2020). Memory engrams: Recalling the past and imagining the future. Science, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER (2012). The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol Brain, 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER, Dudai Y, & Mayford MR (2014). The molecular and systems biology of memory. Cell, 157, 163–186. [DOI] [PubMed] [Google Scholar]
- Keeley MB, Wood MA, Isiegas C, Stein J, Hellman K, Hannenhalli S, & Abel T (2006). Differential transcriptional response to nonassociative and associative components of classical fear conditioning in the amygdala and hippocampus. Learn Mem, 13, 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, & Fanselow MS (1992). Modality-specific retrograde amnesia of fear. Science, 256, 675–677. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Oshima S, Maeyashiki C, Nibe Y, Otsubo K, Matsuzawa Y, Nemoto Y, Nagaishi T, Okamoto R, Tsuchiya K, Nakamura T, & Watanabe M (2016). The ubiquitin hybrid gene UBA52 regulates ubiquitination of ribosome and sustains embryonic development. Sci Rep, 6, 36780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Choi JH, Lee N, Lee HR, Kim JI, Yu NK, Choi SL, Lee SH, Kim H, & Kaang BK (2008). Synaptic protein degradation underlies destabilization of retrieved fear memory. Science, 319, 1253–1256. [DOI] [PubMed] [Google Scholar]
- Lonard DM, Nawaz Z, Smith CL, & O’Malley BW (2000). The 26S proteasome is required for estrogen receptor-alpha and coactivator turnover and for efficient estrogen receptor-alpha transactivation. Mol Cell, 5, 939–948. [DOI] [PubMed] [Google Scholar]
- Lopez-Salon M, Alonso M, Vianna MR, Viola H, Mello e Souza T, Izquierdo I, Pasquini JM, & Medina JH (2001). The ubiquitin-proteasome cascade is required for mammalian long-term memory formation. Eur J Neurosci, 14, 1820–1826. [DOI] [PubMed] [Google Scholar]
- Mahan AL, Mou L, Shah N, Hu JH, Worley PF, & Ressler KJ (2012). Epigenetic modulation of Homer1a transcription regulation in amygdala and hippocampus with pavlovian fear conditioning. J Neurosci, 32, 4651–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maren S, De Oca B, & Fanselow MS (1994). Sex differences in hippocampal long-term potentiation (LTP) and Pavlovian fear conditioning in rats: positive correlation between LTP and contextual learning. Brain Res, 661, 25–34. [DOI] [PubMed] [Google Scholar]
- McFadden T, Devulapalli RK, & Jarome TJ (2019). Quantifying Subcellular Ubiquitin-Proteasome Activity in the Rodent Brain. Journal of Visualized Experiments, e59695. [DOI] [PubMed] [Google Scholar]
- Mendez-Lopez M, Mendez M, Lopez L, & Arias JL (2009). Sexually dimorphic c-Fos expression following spatial working memory in young and adult rats. Physiol Behav, 98, 307–317. [DOI] [PubMed] [Google Scholar]
- Minelli A, Magri C, Barbon A, Bonvicini C, Segala M, Congiu C, Bignotti S, Milanesi E, Trabucchi L, Cattane N, Bortolomasi M, & Gennarelli M (2015). Proteasome system dysregulation and treatment resistance mechanisms in major depressive disorder. Transl Psychiatry, 5, e687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra SW, Hoskin E, Yudkovitz J, Pear L, Wilkinson HA, Hayashi S, Pfaff DW, Ogawa S, Rohrer SP, Schaeffer JM, McEwen BS, & Alves SE (2003). Immunolocalization of estrogen receptor beta in the mouse brain: comparison with estrogen receptor alpha. Endocrinology, 144, 2055–2067. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Antunes-Martins A, Ris L, Peters M, Godaux E, & Giese KP (2007). Calcium/calmodulin kinase kinase beta has a male-specific role in memory formation. Neuroscience, 145, 393–402. [DOI] [PubMed] [Google Scholar]
- Mizuno K, & Giese KP (2010). Towards a molecular understanding of sex differences in memory formation. Trends Neurosci, 33, 285–291. [DOI] [PubMed] [Google Scholar]
- Morey RA, Gold AL, LaBar KS, Beall SK, Brown VM, Haswell CC, Nasser JD, Wagner HR, McCarthy G, & Mid-Atlantic MW (2012). Amygdala volume changes in posttraumatic stress disorder in a large case-controlled veterans group. Arch Gen Psychiatry, 69, 1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musaus M, Navabpour S, & Jarome TJ (2020). The diversity of linkage-specific polyubiquitin chains and their role in synaptic plasticity and memory formation. Neurobiol Learn Mem, 174, 107286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz Z, Lonard DM, Dennis AP, Smith CL, & O'Malley BW (1999). Proteasome-dependent degradation of the human estrogen receptor. Proc Natl Acad Sci U S A, 96, 1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi SA, Devulapalli RK, Nelsen JL, McFadden T, Surineni R, & Jarome TJ (2019). Distinct subcellular changes in proteasome activity and linkage-specific protein polyubiquitination in the amygdala during the consolidation and reconsolidation of a fear memory. Neurobiol Learn Mem, 157, 1–11. [DOI] [PubMed] [Google Scholar]
- Ou LC, & Gean PW (2007). Transcriptional regulation of brain-derived neurotrophic factor in the amygdala during consolidation of fear memory. Mol Pharmacol, 72, 350–358. [DOI] [PubMed] [Google Scholar]
- Park CW, & Ryu KY (2014). Cellular ubiquitin pool dynamics and homeostasis. BMB Rep, 47, 475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons RG, Gafford GM, & Helmstetter FJ (2006). Translational control via the mammalian target of rapamycin pathway is critical for the formation and stability of long-term fear memory in amygdala neurons. J Neurosci, 26, 12977–12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips RG, & LeDoux JE (1992). Differential contribution of amygdala and hippocampus to cued and contextual fear conditioning. Behav Neurosci, 106, 274–285. [DOI] [PubMed] [Google Scholar]
- Reis DS, Jarome TJ, & Helmstetter FJ (2013). Memory formation for trace fear conditioning requires ubiquitin-proteasome mediated protein degradation in the prefrontal cortex. Front Behav Neurosci, 7, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Ortiz CJ, Balderas I, Saucedo-Alquicira F, Cruz-Castaneda P, & Bermudez-Rattoni F (2011). Long-term aversive taste memory requires insular and amygdala protein degradation. Neurobiol Learn Mem, 95, 311–315. [DOI] [PubMed] [Google Scholar]
- Rosenberg T, Elkobi A, Dieterich DC, & Rosenblum K (2016). NMDAR-dependent proteasome activity in the gustatory cortex is necessary for conditioned taste aversion. Neurobiol Learn Mem, 130, 7–16. [DOI] [PubMed] [Google Scholar]
- Rosenberg T, Elkobi A, & Rosenblum K (2016). mAChR-dependent decrease in proteasome activity in the gustatory cortex is necessary for novel taste learning. Neurobiol Learn Mem, 135, 115–124. [DOI] [PubMed] [Google Scholar]
- Shin LM, Rauch SL, & Pitman RK (2006). Amygdala, medial prefrontal cortex, and hippocampal function in PTSD. Ann N Y Acad Sci, 1071, 67–79. [DOI] [PubMed] [Google Scholar]
- Tonoki A, Kuranaga E, Tomioka T, Hamazaki J, Murata S, Tanaka K, & Miura M (2009). Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol Cell Biol, 29, 1095–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb WM, Sanchez RG, Perez G, Butler AA, Hauser RM, Rich MC, O'Bierne AL, Jarome TJ, & Lubin FD (2017). Dynamic association of epigenetic H3K4me3 and DNA 5hmC marks in the dorsal hippocampus and anterior cingulate cortex following reactivation of a fear memory. Neurobiol Learn Mem, 142, 66–78. [DOI] [PubMed] [Google Scholar]
- Wood GE, & Shors TJ (1998). Stress facilitates classical conditioning in males, but impairs classical conditioning in females through activational effects of ovarian hormones. Proc Natl Acad Sci U S A, 95, 4066–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh SH, Mao SC, Lin HC, & Gean PW (2006). Synaptic expression of glutamate receptor after encoding of fear memory in the rat amygdala. Mol Pharmacol, 69, 299–308. [DOI] [PubMed] [Google Scholar]