Abstract
The intracellular penetration of darunavir, a second-generation HIV protease inhibitor, is limited by the activity of the efflux P-glycoprotein (ABCB1). ABCB1 expression and/or activity levels can vary between individuals due to genetic polymorphisms including the c.1199G>A, c.1236C>T, c.2677G>T and c.3435C>T variants, which could in part explain why the pharmacokinetics of darunavir are so variable from one individual to another. While a few clinical studies have failed to demonstrate an influence of these polymorphisms on darunavir pharmacokinetics, drug-drug interactions and methodological limitations may have prevented them from revealing the true influence of ABCB1 variants. In this work, we report on the intracellular accumulation of darunavir in recombinant HEK293 cell lines expressing wild-type ABCB1 or one of several variants: ABCB1 1199A, ABCB1 3435T, and ABCB1 1236T/2677T/3435T. We demonstrate that while ABCB1 expression limits intracellular accumulation of darunavir, there is no significant difference in efflux activity between cells expressing wild-type ABCB1 and those that express any of the studied variants.
Subject terms: Immunological deficiency syndromes, Genetics research, Pharmacogenetics
Introduction
Darunavir (DRV) is a second-generation protease inhibitor used as part of a multi-drug cocktail in HIV therapy1,2. Its pharmacokinetics (PK) are known to be highly heterogeneous, with the plasma AUC varying more than fivefold between individuals receiving the same dosing regimen3. Many individual factors could potentially contribute to this variability, including genetic variations in key biotransformation enzymes and transporters involved in the disposition of DRV. As far as transporters are concerned, DRV is a substrate of influx transporters of the SLCO family (SLCO1A2, SLCO1B1 and SLCO1B3) and efflux transporters of the ABC family (ABCB1, ABCC2)4–6. The ABCB1 gene, which encodes the P-glycoprotein, is of particular interest7. ABCB1 efflux limits drug penetration in enterocytes, therefore decreasing the oral bioavailability of DRV, which is low when administered alone8. This is one of the reasons why DRV is always coadministered with a PK booster such as ritonavir (RTV) or cobicistat (COB), which reduces first-pass metabolism and systemic clearance of DRV, mainly through inhibition of CYP metabolism and ABCB1 activity. ABCB1 also controls drug accumulation in lymphocytes, the active site of antiretrovirals, thereby directly limiting the ability of DRV to reach its target. It is also expressed in other organs, including the blood–brain barrier, meaning it could limit penetration in the central nervous system, which has been described as a sanctuary site for HIV. And while single nucleotide polymorphisms (SNPs) that alter the expression and/or activity of ABCB1 have been described and could potentially explain—at least in part—the important PK variability that characterizes DRV, their exact contribution remains unclear. Functional ABCB1 SNPs of interest include ABCB1 c.1199G>A, c.1236C>T, c.2677T>G/A and c.3435C>T (Table 1). Both ABCB1 c.1236C>T and c.2677T>G/A are in strong linkage disequilibrium with c.3435C>T, and the haplotype they define is considered to capture most of the genetic variability for ABCB19. There is some controversy over the impact of these variants on the PK of several drugs, perhaps due to differences in studied populations and experimental protocols, but also due to substrate-specific effects, which explains why observations made for one drug cannot be directly transposed to another, even if they are both known substrates of this efflux transporter. Moreover, even when in vitro associations are found, these do not necessarily translate into in vivo associations probably due to compensatory mechanisms, confounding factors or DDIs10.
Table 1.
ABCB1 polymorphisms of interest.
| Nucleotide change | rs number | Description | MAF | ||
|---|---|---|---|---|---|
| Euro (%) | Asian (%) | African (%) | |||
| c.1199G>A | rs2229109 | Missense variant: Ser400Asn. Effect are possibly substrate-dependent | 3 | < 1 | < 1 |
| c.1236C>T | rs1128503 | Synonymous variant. Controversial effects | 42 | 59–63 | 14 |
| c.2677T>G/A | rs2032582 | Missense variant: Ser893Ala/Thr. Controversial effects | 41 | 40–59 | 3 |
| c.3435C>T | rs1045642 | Synonymous variant. Alters the timing of protein folding and decreases mRNA stability. Effect are possibly substrate-dependent | 52 | 40–58 | 15 |
Euro European, MAF minor allele frequency. MAFs are from the 1000 Genomes Project11. Asian MAFs include South and East Asians.
The influence of some of these ABCB1 variants on DRV PK has been investigated in clinical studies3,12–14, but they did not appear to be good predictors of inter-individual variability. However, the inhibitory effect of RTV and COB on ABCB1 could limit the in vivo effect of genetic variants for this transporter towards DRV6. Combined with small sample sizes and other confounding factors, this makes it difficult to delineate the true contribution of ABCB1 variants. Therefore, we decided to study the intracellular accumulation of DRV in human embryonic kidney cells (HEK293) overexpressing the wild-type ABCB1 or one of several variants (ABCB1 1199A, ABCB1 3435T, and ABCB1 1236T/2677T/3435T) to clarify this point.
Methods
Chemicals and reagents
Dulbecco’s Modified Eagle Medium (DMEM), fetal bovine serum (FBS), penicillin/streptomycin and enzyme-free cell dissociation buffer were purchased from Gibco (Thermo Fisher); G418 from Roche; flow cytometry antibodies from BD Biosciences (FITC mouse anti-human CD243, clone 17F9, reference 557002; and FITC mouse IgG2b κ isotype control, clone 27–35, reference 555742); DRV, deuterated DRV (DRV-d9) and COB from Toronto Research Chemicals. All chemicals used in drug quantification were of analytical grade.
Characterization of cell lines
The generation and characterization of recombinant cell lines have been described in previous work conducted by our lab15,16. Briefly, HEK293 cells were transfected with plasmids carrying wild-type ABCB1 (1199G-1236C-2677G-3435C, HEKWT); variants 1199G-1236C-2677G-3435T (HEKCGT), 1199G-1236T-2677T-3435T (HEKTTT) or 1199A-1236C-2677G-3435C (HEK1199A) obtained using site-directed mutagenesis; or an empty vector (HEKcontrol). HEK293 cells feature low endogenous levels of expression of ABCB1, ensuring that all ABCB1 expression originates from the transfection process. This model has previously been extensively characterized using flow cytometry, Western blot, and fluorescence microscopy, and validated using reference substrates and inhibitors of ABCB115,16. Since the same cells were used, only flow cytometry was used to re-characterize them. After thawing, cells were grown in DMEM, FBS 10% and penicillin/streptomycin 1% at 37 °C in the presence of 5% CO2. After at least seven days of growth in the presence of the selection antibiotic G418 (1 g/l), ABCB1 expression was assessed by flow cytometry: 0.5 × 106 cells were collected by centrifugation and washed twice with ice-cold buffer (PBS, FBS 1%, EDTA 1 mM) then re-suspended in buffer supplemented with 10% anti-ABCB1 antibody, 10% isotype control or in buffer with no antibody, and left to incubate for 45 min on ice and in the dark. Finally, the cells were washed with buffer and resuspended before being analyzed using a BD FACSVerse flow cytometer (for characterization) or BD FACSAria III (for cell sorting) and the BD FACSuite software. Additional data analysis and plotting were carried out in FlowJo (version 10.6.1).
Intracellular accumulation experiments
0.35 × 106 cells were seeded on poly-l-lysine-coated 24-well plates and incubated overnight. The next day, DRV dilutions in DMEM were prepared from a stock solution and added in each well at a final concentration of 0.5, 1, 2.5, 5 or 10 mg/l (final volume: 500 µl). These values were chosen to cover the range of total plasma concentrations found in patients treated with COB- or RTV-boosted DRV. Cells were incubated in triplicate at 37 °C in the presence of 5% CO2 for 2 h. The plates were then centrifuged for 5 min at 450×g, 4 °C, then kept on ice for the remainder of the experiment to block drug efflux. Cells were washed twice with 500 µl of ice-cold PBS, then 400 µl of a mixture of methanol/water 60/40% (v/v) containing 20 ng/ml DRV-d9 was added and cells were detached by scratching the surface of the well. Cell suspensions were kept at − 20 °C until quantification.
Cell viability
The viability of HEK cells in the presence of increasing concentrations of DRV (1, 5 and 10 mg/l) was assessed using the WST-1 assay according to the manufacturer’s instructions. Cells were grown in 96-well plates and viability was assessed separately for the HEKcontrol and HEKWT groups. Absorbance was measured at 450 nm after 0.5, 1, 1.25, 1.5 and 1.75 h of incubation.
Drug quantification
DRV intracellular concentrations were determined using an adapted version of a previously published liquid chromatography-tandem mass spectrometry (LC–MS/MS) method17. Cell suspensions were vortex mixed, sonicated for 5 min, placed on an orbital shaker for 2 h and centrifuged for 10 min at 10,500×g. The supernatant was transferred to a vial for injection and the pellet was set aside for protein quantification. Calibrators ranging from 1.25 to 125 ng/ml were prepared in a similar fashion. Chromatographic separation was achieved on a Waters UPLC BEH C18 1.7 µm column (2.1 × 50 mm) maintained at 40 °C. The injection volume was 5 µl and the flow rate was 0.5 ml/min. The mobile phase consisted of a gradient of water/formic acid 0.1% (mobile phase A) and acetonitrile (mobile phase B), starting with 95% A and 5% B, ramping up to 80% B over 6 min, then returning to 5% B at 6.1 min and remaining at this ratio until the end of the run (total run time: 8 min). The MS system was a Xevo TQS-micro tandem quadrupole mass spectrometer (Waters). The following ion transitions were monitored: 548.2 > 392.3 for DRV and 557.3 > 113 for DRV-d9.
Protein quantification
Total proteins were quantified in pellets using the BCA kit (Thermofisher Scientific) according to the manufacturer’s instructions. Absolute DRV concentrations were normalized by total protein content.
Statistical analysis
Each set of experiments were performed thrice. Normalized intracellular concentrations at each dose level were compared using one-way ANOVA (α = 0.05) followed by Tukey’s HSD test. All statistical analyses were carried out in R (version 3.6.3)18.
Results
Flow cytometry
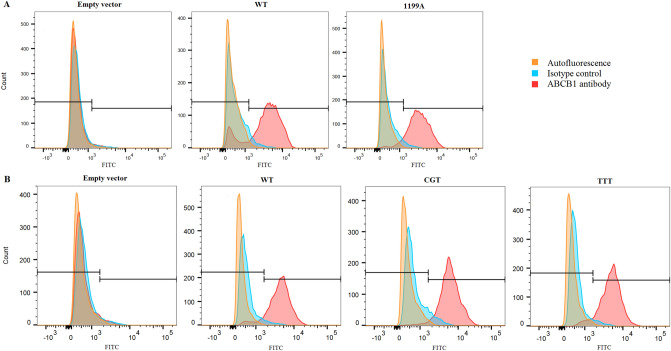
ABCB1 expression in our model was confirmed by flow cytometry analysis. After sorting, all cell lines expressed comparable levels of ABCB1 (> 80%) except for HEKcontrol, which, predictably, were characterized by low levels of expression (Fig. 1).
Figure 1.
ABCB1 expression assessed by flow cytometry for cells stained with ABCB1 antibody, isotype control and no staining (autofluorescence). (A) Histograms of HEKcontrol, HEKWT and HEK1199A. (B) Histograms of HEKcontrol, HEKWT, HEKCGT and HEKTTT. FITC Fluorescein isothiocyanate.
Intracellular accumulation
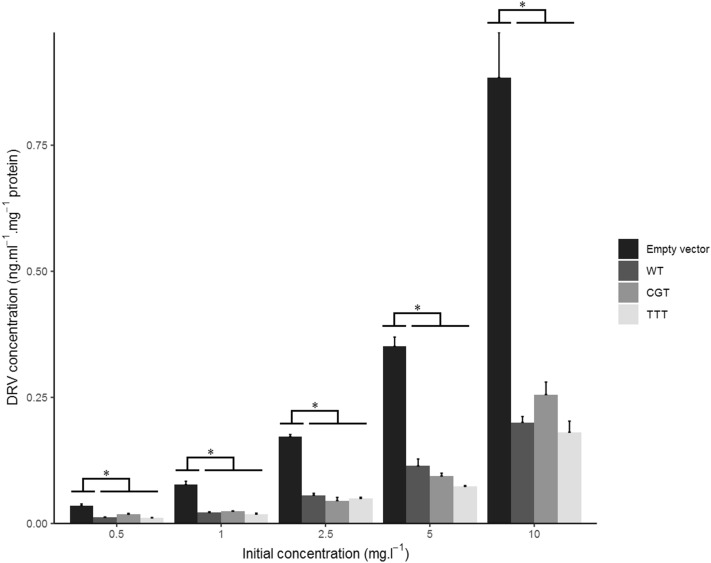
Intracellular concentrations of DRV, adjusted by the total protein content of the sample, markedly differed between cell lines: they were significantly lower in all ABCB1-expressing cells compared to control cells transfected with an empty plasmid in all experiments (Figs. 2 and 3), confirming that DRV is a substrate of ABCB1 and that drug efflux greatly limits cellular penetration in this model. There was no difference in intracellular concentrations between cells expressing the c.1199G or the c.1199A variant (p = 0.89, 0.98, 0.81, 0.38, 0.56 at the 10, 5, 2.5, 1 and 0.5 mg.l-1 dose levels, respectively) (Fig. 2). Likewise, concerning the ABCB1 haplotype defined by the c.1236C>T, c.2677G>T and c.3435C>T variants, intracellular concentrations did not significantly differ between CGC (wild-type) versus CGT cells (p = 0.85, 0.66, 0.37, 0.95, 0.20), CGC versus TTT (p = 0.99, 0.16, 0.74, 0.93, 0.98), or CGT versus TTT (p = 0.7, 0.64, 0.9, 0.68, 0.12) (Fig. 3).
Figure 2.
Intracellular protein-normalized DRV concentrations in HEKcontrol (empty vector), HEKWT and HEK1199A cells at several dose levels after 2 h of incubation. Results reported as mean + standard error (n = 3). *Denotes statistically significant difference (p < 0.05) between groups.
Figure 3.
Intracellular protein-normalized DRV concentrations in HEKcontrol (empty vector), HEKWT, HEKCGT and HEKTTT cells at several dose levels after 2 h of incubation. Results reported as mean + standard error (n = 3). *Denotes statistically significant difference (p < 0.05) between groups.
Cell viability
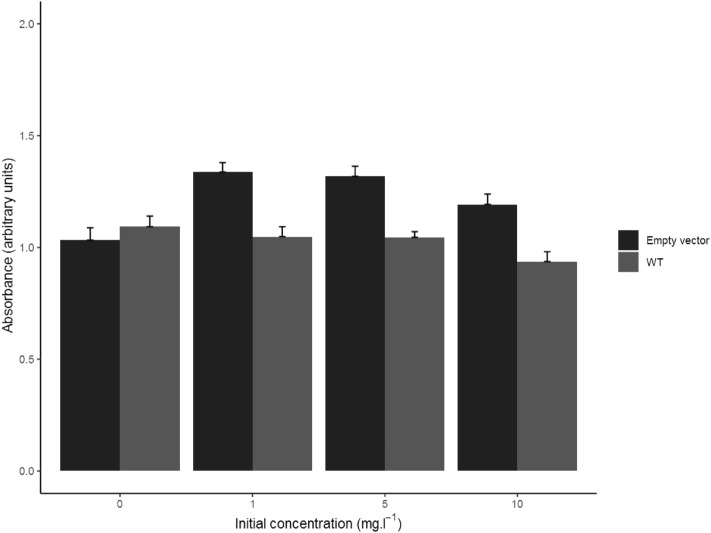
Cell viability was unaffected by DRV at concentration levels ranging from 1 to 10 mg/l. The results of these experiments are displayed in Fig. 4.
Figure 4.
WST-1 cell viability assay results in HEKcontrol (empty vector) and HEKWT cells at several dose levels (0 being the control condition) after 1.75 h of incubation. Results reported as mean absorbance + standard error (n = 6).
Discussion
Several pharmacogenetic studies have attempted to determine the influence of ABCB1 variants on DRV pharmacokinetics. Calcagno et al. studied the paired PK of DRV in plasma and cerebrospinal fluid (CSF) in 41 patients and showed that there was no influence of ABCB1 c.1236C>T, c.2677G>T or c.3435C>T on DRV penetration in the central nervous system, as assessed by the ratio of concentrations between the CSF and plasma14. Moltó et al. developed a population PK model of DRV on 75 subjects and assessed the influence of 148 SNPs in 15 genes, including ABCB1 c.1236C>T and c.2677G>T, neither of which showed a significant effect on PK parameters13. PK parameters were similarly unaffected by the c.1236C>T and c.3435C>T variants in a small group of 25 patients in another population study19. Moreover, Nagano et al. found that the ABCB1 c.3435C>T variant was not correlated with concentrations of DRV in either plasma or peripheral blood mononuclear cells (PBMCs) in a group of 19 patients12. Finally, we recently published a population PK model developed on 127 patients for which neither ABCB1 c.1199G>A nor ABCB1 c.3435C>T were correlated with DRV PK parameters3. Although these studies were not necessarily powered with a specific set of covariates in mind, they suggest that genetic polymorphisms in ABCB1 either do not alter protein activity toward this molecule, or that other, unidentified factors mask or neuter the effect of ABCB1 variants in vivo. RTV- or COB-based boosting likely has a role to play since both of these boosters are capable of inhibiting ABCB1 to some degree, but other factors may also contribute. Besides these considerations related to DRV based therapy, there have been conflicting results in the literature regarding the effect of these genetic variants not only in vivo but also in vitro. For instance, Woodahl et al. showed that ABCB1-mediated efflux for older protease inhibitors such as lopinavir, amprenavir, indinavir, saquinavir and RTV was increased in cells expressing the c.1199A variant compared to the wild-type protein20. Meanwhile, tumor resistance to anticancer agents was increased in c.1199A cells for certain agents such as vinblastine, vincristine, paclitaxel and etoposide, with fold-changes ranging from 1.9 to 11 depending on the drug, but was unchanged for doxorubicin21. Dessilly et al. also showed that the c.1199G>A variant modulated the intracellular accumulation of tacrolimus, but not that of ciclosporine15. Further, they showed that the 1236-2677-3435 haplotype had little to no effect on the accumulation and activity of several tyrosine kinase inhibitors (dasatinib, nilotinib and ponatinib), whereas the variant ABCB1 displayed lower activity toward imatinib16. Sennesael et al. also showed that there was no effect of any of the four aforementioned SNPs on the efflux of the anticoagulant drug rivaroxaban22. Because of these conflicting results, there is a need for case-by-case assessment of drug accumulation to determine the relevance of genetic variants in ABCB1. An existing model of intracellular accumulation was used to assess the influence of ABCB1 variants. Our experiments showed that the c.1199G>A, c.1236C>T, c.2677G>T and c.3435C>T variants have no effect on the ability of ABCB1 to transport DRV in this model, similarly to what was observed for other substrates, such as rivaroxaban. While it could be argued that HEK293 cells are not a physiologically representative model, they feature a low basal expression level of ABCB1, ensuring all ABCB1 activity in our model can be attributed to the transfection process15,16, unlike other cell lines like Caco-2, that would be more representative of the digestive tract but would also introduce a bias due to a combination of native and transfection-induced expression. Further, this in vitro model is characterized by supraphysiological levels of ABCB1 expression and these SNPs may exert slightly different effects with in vivo levels. In any event, variants in other influx and efflux transporters, as well as a range of genetic and non-genetic factors, could potentially explain the wide range of plasma and intracellular concentrations that have been reported for DRV. In conclusion, using a recombinant model of HEK293 cells, we showed that ABCB1 variants do not appear to modulate the ability of this efflux pump to transport DRV, and as such, they do not alter the intracellular accumulation of this antiretroviral.
Acknowledgments
The authors would like to thank Dr Nicolas Dauguet (Institut de Duve, Université catholique de Louvain) for his assistance in cell sorting.
Abbreviations
- COB
Cobicistat
- CSF
Cerebrospinal fluid
- DRV
Darunavir
- DRV-d9
Deuterated darunavir
- EDTA
Ethylenediaminetetraacetate
- FBS
Fetal bovine serum
- FITC
Fluorescein isothiocyanate
- HEK
Human embryonic kidney
- LC–MS/MS
Liquid chromatography coupled with tandem mass spectrometry
- MAF
Minor allele frequency
- PBS
Phosphate buffer saline
- PK
Pharmacokinetic
- RTV
Ritonavir
- SNP
Single nucleotide polymorphism
Author contributions
G.S., V.H. and L.E. designed the study. G.S., H.D. and N.P. performed accumulation and flow cytometry experiments. G.S., H.D. and K.A.D. performed LC–MS/MS measurements. G.S., H.D., H.E.H. and N.P. performed protein quantification, H.E.H. performed cell viability experiments. All authors contributed to data analysis and visualization. G.S. wrote the first draft of the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the Fonds pour la Formation à la Recherche dans l’Industrie et dans l’Agriculture (FRIA) under Grant Number FC16749 to GS, and by grants from the Fonds pour la Recherche scientifique [FRS-FNRS] (Grants F4509.19, J018317 and J006920).
Data availability
Data generated and analyzed during this study are available from the authors on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Vincent Haufroid and Laure Elens.
References
- 1.Guidelines Version 10.0, European AIDS Clinical Society (2019).
- 2.Saag MS, et al. Antiretroviral drugs for treatment and prevention of HIV infection in adults: 2018 recommendations of the international antiviral society–USA panel. JAMA. 2018;320:379–396. doi: 10.1001/jama.2018.8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stillemans G, et al. Exploration of reduced doses and short-cycle therapy for darunavir/cobicistat in patients with HIV using population pharmacokinetic modeling and simulations. Clin. Pharmacokinet. 2020;2:5. doi: 10.1007/s40262-020-00920-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.König SK, et al. Impact of drug transporters on cellular resistance towards saquinavir and darunavir. J. Antimicrob. Chemother. 2010;65:2319–2328. doi: 10.1093/jac/dkq324. [DOI] [PubMed] [Google Scholar]
- 5.Hartkoorn RC, et al. HIV protease inhibitors are substrates for OATP1A2, OATP1B1 and OATP1B3 and lopinavir plasma concentrations are influenced by SLCO1B1 polymorphisms. Pharmacogenet. Genomics. 2010;20:112–120. doi: 10.1097/FPC.0b013e328335b02d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lepist E-I, Phan TK, Roy A, Tong L, MacLennan K, Murray B, et al. Cobicistat boosts the intestinal absorption of transport substrates, including HIV protease inhibitors and GS-7340, in vitro. Antimicrob. Agents Chemother. 2012;56:5409–5413. doi: 10.1128/AAC.01089-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujimoto H, et al. P-glycoprotein mediates efflux transport of darunavir in human intestinal caco-2 and ABCB1 gene-transfected renal LLC-PK1 cell lines. Biol. Pharm. Bull. 2009;32:1588–1593. doi: 10.1248/bpb.32.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prezista—Résumé Des Caractéristiques Du Produit, Janssen-Cilag International NV (Turnhoutseweg 30, Beerse, Belgium, 2007).
- 9.Hodges LM, et al. Very important pharmacogene summary: ABCB1 (MDR1, P-glycoprotein) Pharmacogenet. Genomics. 2011;21:152–161. doi: 10.1097/FPC.0b013e3283385a1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolking S, Schaeffeler E, Lerche H, Schwab M, Nies AT. Impact of genetic polymorphisms of ABCB1 (MDR1, P-glycoprotein) on drug disposition and potential clinical implications: update of the literature. Clin Pharmacokinet. 2015;54:709–735. doi: 10.1007/s40262-015-0267-1. [DOI] [PubMed] [Google Scholar]
- 11.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagano D, et al. Darunavir concentration in PBMCs may be a better indicator of drug exposure in HIV patients. Eur. J. Clin. Pharmacol. 2018;74:1055–1060. doi: 10.1007/s00228-018-2464-y. [DOI] [PubMed] [Google Scholar]
- 13.Moltó J, et al. Simultaneous pharmacogenetics-based population pharmacokinetic analysis of darunavir and ritonavir in HIV-infected patients. Clin Pharmacokinet. 2013;52:543–553. doi: 10.1007/s40262-013-0057-6. [DOI] [PubMed] [Google Scholar]
- 14.Calcagno A, et al. Determinants of darunavir cerebrospinal fluid concentrations: impact of once-daily dosing and pharmacogenetics. AIDS. 2012;26:1529–1533. doi: 10.1097/QAD.0b013e3283553619. [DOI] [PubMed] [Google Scholar]
- 15.Dessilly G, et al. ABCB1 1199G>A genetic polymorphism (Rs2229109) influences the intracellular accumulation of tacrolimus in HEK293 and K562 recombinant cell lines. PLoS ONE. 2014;9:e91555. doi: 10.1371/journal.pone.0091555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dessilly G, Panin N, Elens L, Haufroid V, Demoulin J-B. Impact of ABCB1 1236C > T-2677G > T-3435C > T polymorphisms on the anti-proliferative activity of imatinib, nilotinib, dasatinib and ponatinib. Sci. Rep. 2016;5:2. doi: 10.1038/srep29559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belkhir L, et al. Quantification of darunavir and etravirine in human peripheral blood mononuclear cells using high performance liquid chromatography tandem mass spectrometry (LC–MS/MS), clinical application in a cohort of 110 HIV-1 infected patients and evidence of a potential drug–drug interaction. Clin. Biochem. 2016;49:580–586. doi: 10.1016/j.clinbiochem.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 18.R Core Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, 2014).
- 19.Arab-Alameddine M, et al. Population pharmacokinetic modelling and evaluation of different dosage regimens for darunavir and ritonavir in HIV-infected individuals. J. Antimicrob. Chemother. 2014;69:2489–2498. doi: 10.1093/jac/dku131. [DOI] [PubMed] [Google Scholar]
- 20.Woodahl EL, Yang Z, Bui T, Shen DD, Ho RJ. MDR1 G1199A polymorphism alters permeability of HIV protease inhibitors across P-glycoprotein-expressing epithelial cells. AIDS. 2005;19:1617–1625. doi: 10.1097/01.aids.0000183626.74299.77. [DOI] [PubMed] [Google Scholar]
- 21.Woodahl EL, Crouthamel MH, Bui T, Shen DD, Ho RJY. MDR1 (ABCB1) G1199A (Ser400Asn) polymorphism alters transepithelial permeability and sensitivity to anticancer agents. Cancer Chemother. Pharmacol. 2009;64:183–188. doi: 10.1007/s00280-008-0906-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sennesael A-L, et al. Effect of ABCB1 genetic polymorphisms on the transport of rivaroxaban in HEK293 recombinant cell lines. Sci. Rep. 2018;8:10514. doi: 10.1038/s41598-018-28622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data generated and analyzed during this study are available from the authors on reasonable request.