Abstract
Background and Aim:
Shewanella algae is ubiquitous in marine-associated environments and has been increasingly recognized as a significant human pathogen that can cause serious infections mainly associated with exposure to seawater and ingestion of raw seafood. This study aimed to isolate and characterize S. algae from ballast water of ships berthed at Port Klang, Malaysia.
Materials and Methods:
Ballast water was sampled from nine ships docked at Port Klang, Malaysia. The isolates were identified and characterized based on biochemical and enzymatic properties, 16S rRNA and gyrB sequencing, biofilm formation capability, and antibiotic susceptibility.
Results:
A total of four S. algae isolates were isolated from four ballast water samples tentatively name Sa-BW1, Sa-BW2, Sa-BW7, and Sa-BW8. All isolates showed positive reaction for cytochrome oxidase, catalase, high tolerance to NaCl (6% and 8%), ability to grow at 42°C, and on Salmonella-Shigella agar. The strains also exhibited b-hemolytic activity on sheep blood and human blood agar, positive reaction for lipase, protease, DNase and gelatinase, strong biofilm adherence capabilities and multiple antibiotic resistances against ampicillin, carbenicillin, cephalothin, colistin, novobiocin, oxacillin, penicillin, rifampicin, and tobramycin which suggested their potential pathogenicity.
Conclusion:
This study demonstrated the occurrence of putative pathogen S. algae in ballast water of ships docked at Malaysian port.
Keywords: ballast water, extracellular enzymes, putative pathogen, Shewanella algae
Introduction
Ships carry ballast water to control their stability and trim at the start of the voyage. However, the discharge of ballast water near ports risks the transport of invasive species, including pathogenic species from one continent to another. Should a novel genotype of pathogenic species arrived in ballast water, it may establish and persist in the local waters [1] which may lead to the emergence of new virulence strains in the existing population. The global distribution of pathogenic bacteria implicates a negative effect on the existing ecosystem as well as marine animal and human health. The concern about the transmission of potentially pathogenic bacteria through ballast water began in 1992, when the Centers for Disease Control and Prevention of the USA detected Vibrio cholerae in shellfish collected from ballast tanks of cargo ships that had come from South America [2]. Since then, the list of pathogenic species detected in ballast water has increased, including Listeria monocytogenes, Mycobacterium spp., several species of Aeromonas, Pseudomonas, Vibrio, Staphylococcus, the coral pathogens Serratia marcescens, and Sphingomonas spp. and some emerging opportunistic pathogen such as Stenotrophomonas maltophilia and Shewanella algae [3-5]. However, no discharge limits have been set for these pathogens.
Shewanella spp. is ubiquitous in marine-associated environments and widely spread in nature throughout the world. It is a facultative anaerobe, motile, and Gram-negative bacillus belonging to the family Shewanellaceae, order Alteromonadales. The physiological diversity and broad respiratory versatility of Shewanella spp. make it a highly adaptable organism that can survive in different environmental niches. In the previous studies, Shewanella spp. has been isolated from a wide range of environment including freshwater [6], estuary [7], deep sea [8], oil field [9], muddy sediment [10], fish [11], and marine sponge [12]. While members of this genus have been intensively studied for their role in bioremediation [13] and application in microbial fuel cells [14], several species have been reported as emerging pathogens in human and aquatic animals. Out of more than 60 known Shewanella species, Shewanella putrefaciens, Shewanella haliotis, Shewanella xiamenensis, and S. algae have been documented with pathogenicity in human beings [15-17]. Although human infection by these species is rare, increasing number of cases has been reported worldwide with >80% of clinical Shewanella isolates being attributed to the S. algae species. [18].
S. algae has been recognized as a conditionally pathogenic bacteria to human and aquatic animals [19,20]. The most common clinical symptoms described in human infections by S. algae are bacteremia, cellulitis, and chronic otitis media [18]. Rare cases of necrotizing soft-tissue infection [21], rupture of aortic aneurysm [22], peritonitis [23], and endocarditis was reported by Davidson et al. [24]. As for aquatic species, S. algae has been identified as a causative agent of ulcer disease for marine fish species, Scinenops ocellata and abalone, Haliotis diversicolor [11,25]. Other studies showed that S. algae can cause black spot disease to farmed freshwater shrimp, Penaeus vannamei and lesion in reared tonguefish, and Cynoglossus semilaevis [19,26]. The pathogenic potential of Shewanella has been controversial as most cases of human infection develop in people with underlying comorbidities and occurred as polymicrobial infections [18,27]. However, prior reports of monomicrobial infection with S. algae have suggested the organism as causal in many cases, confirming its ability to cause disease [28-30]. Although infection in healthy hosts is uncommon, rare cases have been reported in individuals with no underlying diseases due to massive exposure to marine environment and seafood consumption [31-33]. The increased in S. algae virulence was attributed to its hemolytic activity, enzymatic activity, and biofilm formation [18,34,35].
This study aimed to characterize S. algae strains isolated from ships’ ballast water incoming to Port Klang, Malaysia. The characterizations were carried out based on the biochemical and enzymatic properties, 16S rRNA and gyrB gene sequence analysis, biofilm formation ability, and susceptibility to antibiotics.
Materials and Methods
Ethical approval and Informed consent
No ethical approval was required in this study. The sheep blood for preparation of sheep blood agar was kindly provided by UKM Animal House. The handling and collection of sheep blood were conducted by an assistant veterinary officer. While for human blood agar preparation, human blood samples were collected from healthy volunteers by a medical laboratory technician of the UKM Health Center. Informed consent was obtained from the volunteers included in the study. There was no direct involvement during the blood withdrawal procedures and no further experiments were performed on the subjected animal and human.
Study period and location
This study was conducted from August 2016 to March 2017 in Port Klang, Malaysia, where ballast water samples were taken from several ships calling at Port Klang.
Sample collection
Ballast water samples were taken from nine ballast tanks of nine-unit ships docked at Port Klang, Malaysia. The samples were collected using 20 L bucket through the manhole or overflow pipe when manhole was inaccessible which later filled into 1 L sterilized Schott bottle. Seawater samples were also taken from four sampling points of surrounding port water using Niskin water sampler. The samples were then transferred back to the laboratory in an icebox for further microbiological analysis. Details of each ballast water samples are shown in Table-1.
Table-1.
Details of ballast water sampling.
| Sample No. | Type of vessel | BW source/Port of origin | Location of sampling |
|---|---|---|---|
| BW1 | Container ship | Singapore | Manhole |
| BW2 | Container ship | Yokkaichi | Manhole |
| BW3 | Container ship | Mundra | Sounding pipe |
| BW4 | Container ship | Hong Kong | Manhole |
| BW5 | Container ship | Ning Bo | Manhole |
| BW6 | Container ship | Mormugao | Manhole |
| BW7 | Container ship | Nhava Sheva | Manhole |
| BW8 | Bulk carrier | Malacca strait | Manhole |
| BW9 | Bulk carrier | Zhang Jia Gang | Manhole |
Isolation and selection of Shewanella spp.
The water samples were serially diluted up to 10−5 using 0.8% saline water and 0.1 mL of the diluted samples were inoculated on marine agar by spread plate technique. The plates were incubated at 30°C for 3 days. Different colonies were picked and purified by subculturing on marine agar for several times. Pure colonies with distinct morphologies were tested for the following key characteristics of Shewanella, according to the description by Holt and Bruun [18]: Gram stain, motility, cytochrome oxidase, catalase reaction (3% H2O2), and H2S production using sulfide indole motility medium. Colonies that fit the description of being Gram-negative bacillus, oxidase-catalase positive, and H2S producing were subjected to further tests.
Biochemical and enzymatic activities
All presumptive Shewanella isolates were tested for: Utilization of glucose, sucrose, lactose, maltose and citrate, IMViC test (indole, Methyl red, Voges–Proskauer and citrate) nitrate reduction, growth at 4°C and 42°C, growth in 6% and 8% NaCl, and growth on Salmonella-Shigella (SS) agar and MacConkey agar. The following enzymatic activities were also determined: Lipase, protease, DNase, and gelatinase which were performed on spirit blue agar, skim milk agar, DNase agar, and gelatine medium, respectively. Hemolytic activity was determined by streaking the strains on heart infusion agar supplemented with 5% washed erythrocytes of human and sheep blood. The incubation was performed at 30°C for up to 72 h and the results were recorded.
Confirmation of S. algae by 16S rRNA and gyrB gene sequencing
Total genomic DNA of the presumptive S. algae isolates was extracted using CTAB/NaCl method [36]. The 16S rRNA gene was amplified using universal primer, 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5′-TACGGCTACCTTGTTACGACTT-3′) [37]. A 25 mL polymerase chain reaction (PCR) mixture was prepared which consist of 1 mL genomic DNA, 12.5 mL 2 × PCR Master Dye Mix, 1 mL (0.1 mM) primer and 9.5 mL sterile distilled water. The PCR conditions were as follows: Initial denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, with a final extension at 72°C for 10 min.
Amplification of gyrB gene was carried out using two degenerated primers, UP1 (5’-GAA GTCATCATGACCGTTCTGCAYGC NGGNGGNAARTTY GA-3’) and UP2R (5’-AGC AGGGTACGATGTGCGAGCCRTCNACRTCN GCRTCNGTCAT-3’) with the following conditions: Initial denaturation at 94°C for 1 min, followed by 30 cycles of denaturation at 94°C for 1 min, primer annealing at 60°C for 1 min, primer extension at 72°C for 2 min, and final extension at 72°C for 7 min [38].
Sequencing of the 16S rRNA and gyrB amplicons was completed by a sequencing company (Apical Scientific Sdn Bhd, Selangor, Malaysia). The obtained sequence data were aligned and analyzed using BioEdit Version 7.0. Species identification was performed by Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Phylogenetic analysis
The phylogenetic relationship of S. algae was determined by comparing the obtained 16S rRNA and gyrB gene sequences with highly identical existing sequences in GenBank database using the BLAST algorithm. Sequences of other Shewanella species were also collected as comparison to the S. algae. The multiple alignment and construction of neighbour-joining (NJ) and maximum likelihood (ML) phylogenetic trees were performed using MEGA Version 6.0 [39]. The topology of the phylogenetic tree was evaluated by a bootstrap analysis through 1000 replications. Escherichia coli was used as an outgroup.
Biofilm formation potential test
The ability of S. algae isolates to form biofilm was tested based on microtiter dish biofilm formation assay adapted from O’Toole [40]. The biofilm formation by the isolates was quantified based on the absorbance (OD) reading at 595 nm. Negative control wells contained only marine broth were added in the assay, while suspension of Staphylococcus aureus from UKM Biotechnology and Marine Microbiology Laboratory culture collection was included as positive control. The adherence capabilities of the isolates were classified according to Stepanović et al. [41]. The cutoff OD (ODC) was defined as three standard deviations above the mean OD of the negative control. In this study, the OD mean of the negative control was 0.083±0.02. Hence, the cutoff OD (ODC) applied in this study was set as 0.143. Comparing the OD means of isolates well to ODc = 0.143, 2 × ODc = 0.286 and 4 × ODC = 0.332, the strains were then categorized as follows:
OD ≤ ODC = Non adherent
ODC < OD ≤ 2 × ODC = Weakly adherent
2 × ODC < OD ≤ 4 × ODC = Moderately adherent
4 × ODC < OD = Strongly adherent
Antibiotic susceptibility test
Susceptibility of S. algae isolates to antibiotics was determined by Kirby-Bauer disk-diffusion technique on Mueller-Hinton agar following protocol by Hudzicki [42] using broad spectrum antibiotics used to treat variety of bacterial infections. The disks used were ampicillin (20 μg), carbenicillin (100 μg), cephalothin (30 μg), chloramphenicol (30 μg), ciprofloxacin (10 μg), colistin (10 μg), gentamicin (10 μg), kanamycin (30 μg), novobiocin (30 μg), oxacillin (5 μg), oxytetracycline (30 μg), penicillin (10 μg), rifampicin (5 μg), streptomycin (25 μg), and tobramycin (10 μg). The diameter of inhibition zone around each antibiotic disk was measured to the nearest millimeter. The zone diameters of each drug were interpreted using the criteria published by the Clinical and Laboratory Standards Institute. The strains were classified as susceptible (S), resistant (R), and intermediate (I) depending on the size of zone inhibition. Multiple antibiotic resistance (MAR) index was calculated as ratio of the number of antibiotics to which the test isolate depicted resistance to the total number of antibiotics to which the test isolate had been evaluated for susceptibility [43]. The MAR index was used as a tool to assess the risk of the isolates coming from a region of high or low antibiotic use. A MAR index >0.2 indicates a “high-risk” source of contamination.
Results
Characterization and identification of S. algae
A total of 102 isolates were successfully isolated from ballast water. Four of the isolates were presumed as Shewanella species, namely, Sa-BW1, Sa-BW2, Sa-BW7, and Sa-BW8. As for seawater samples, out of 212 isolates that were isolated two of them were presumed as Shewanella, namely, Sa-SW2 and Sa-SW3. The presumptive Shewanella isolates presented as Gram-negative rods and displayed positive reaction for oxidase-catalase test and production of H2S. All isolates appeared as circular orange colonies on marine agar. Further biochemical tests revealed that all the presumptive Shewanella spp. isolates were tolerant to high NaCl concentration (6%, and 8%) and able to grow at 42°C but not at 4°C. Negative reactions were observed for indole, methyl-red, Voges-Proskauer, and citrate tests. All strains reduced nitrate to nitrite and produced acid from glucose but not from sucrose, maltose, and lactose. The isolates also grew well on MacConkey and SS agar. Positive enzymatic activities for lipase, protease, DNase, and gelatinase were detected from all strains. They also exhibited a clear zone of beta hemolysis on sheep blood and human blood agar after 48 h incubation.
Comparison of the biochemical and enzymatic profiles displayed by the presumptive Shewanella isolates with other Shewanella type strains (S. algae IAM 14159, S. putrefaciens ATCC 8071, and S. haliotis JCM 14758), as shown in Table-2, suggested all strains as S. algae. Further identification by 16S rRNA and gyrB gene sequencing confirmed the identity of the ballast water and seawater isolates. Nucleotide BLAST search of each strain showed >99% identity score to existing S. algae sequences in database. Sequence data of S. algae isolates of ballast water samples from this study were registered in GenBank under accession numbers: S. algae strain BW1 (16S; MN548355, gyrB; MN555561), S. algae strain BW2 (16S; MN548356, gyrB; MN555562), S. algae strain BW7 (16S; MN548357, gyrB; MN555563), and S. algae BW1 (16S; MN548358, gyrB; MN555564).
Table-2.
Biochemical and enzymatic profiles of S. algae strains from ballast water.
| Strain/Characteristics | S. algae (Sa-BW) | S. algae IAM 14159T | S. putrefaciens ATCC 8071T | S. haliotis JCM 14758T |
|---|---|---|---|---|
| Morphological characteristics | ||||
| Gram stain | − | − | − | − |
| Cell shape | rod | rod | rod | rod |
| Motility | + | + | + | + |
| Spores | − | − | − | − |
| Biochemical characteristics | ||||
| Oxidase | + | + | + | + |
| Catalase | + | NA | + | + |
| Voges–Proskauer | − | NA | NA | NA |
| Methyl red | − | NA | NA | NA |
| Indole production | − | NA | − | − |
| H2S production | + | + | + | + |
| Nitrate reduction | + | + | + | + |
| Utilization of glucose | + | + | + | − |
| Utilization of sucrose | − | − | − | − |
| Utilization of lactose | − | − | − | − |
| Utilization of maltose | − | − | − | − |
| Utilization of citrate | − | − | − | + |
| Growth on SS agar | + | + | − | NA |
| Growth on MacConkey | + | NA | NA | NA |
| Growth at 42°C | + | + | − | + |
| Growth at 4°C | − | − | + | − |
| Growth in 6% NaCl | + | + | − | + |
| Growth in 8% NaCl | + | NA | NA | + |
| Enzymatic characteristics: | ||||
| Lipase | + | + | + | − |
| Protease | + | NA | + | + |
| DNase | + | + | + | + |
| Gelatinase | + | + | NA | + |
| Hemolysis (sheep blood) | + | + | − | NA |
| Hemolysis (human blood) | + | NA | NA | NA |
=Type strain, NA=Not available, +=positive reaction, –=negative reaction. S. algae: Shewanella algae
Phylogenetic analysis
Approximately 1400 bp nucleotide sequences of 16S rRNA gene and 1100 bp nucleotide sequences of gyrB genes of S. algae ballast water isolates were used for phylogenetic analyses. GenBank nucleotide accession numbers for the 16S rRNA and gyrB gene sequences are shown in the phylogenetic tree. A NJ phylogenetic tree based on 16S rRNA (Figure-1a) indicated that the isolates fell within the clade comprising the members of genus Shewanella, forming a cluster with S. algae strain ATCC 51192 with sequences similarities of 99.4% (Sa-BW1), 99.9% (Sa-BW2), 99.8% (Sa-BW7), and 100% (Sa-BW8). The tree also revealed a very close phylogenetic relationship of the isolates with Shewanella upenei 20-23R with a same sequence similarities values as in S. algae ATCC 51192. The maximum-likelihood tree of 16S rRNA gene (Figure-1b) showed almost the same topology except for the location of the nearest neighbor, S. haliotis DW01 which being clustered together with ballast water isolates, S. algae and S. upenei. The isolates shared sequences similarities of 98.9% (SA-BW1) and 99.2% (Sa-BW2, Sa-BW7, and Sa-BW8), to S. haliotis DW01 and 92.8-98.7% similarities to the other Shewanella species used in the phylogenetic analysis.
Figure-1.
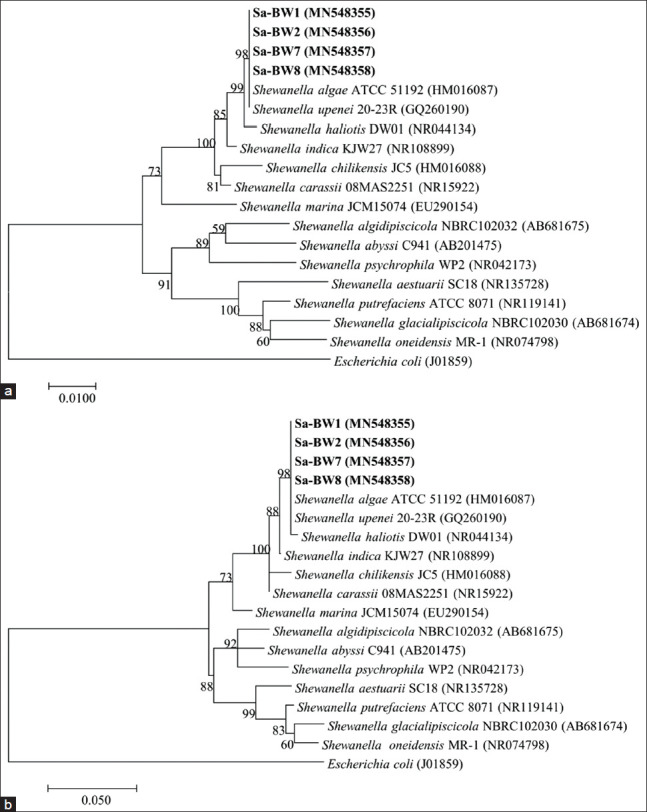
Phylogenetic tree based on 16S rRNA gene sequences of S. algae Sa-BW1, Sa-BW2, Sa-BW7, Sa-BW8, and some other related Shewanella species. (a) Neighbor-joining tree. (b) Maximum-likelihood tree. GenBank accession numbers are given in parentheses.
However, the topology of NJ tree based on the gyrB gene (Figure-2a) showed that these isolates clustered monophyletically, apart from S. upenei and S. haliotis. The strains showed gyrB gene sequence similarities of 97.7-98.9% to S. upenei, 97.5-97.9% to S. haliotis, and 73.1-95.2% to other Shewanella species. Almost the same topology was observed in ML tree of gyrB gene (Figure-2b). Isolates Sa-BW2 and Sa-BW7 formed an independent cluster next to Sa-BW1 indicating that these two isolates have an almost identical sequence to each other with similarity values of 99.5%. Meanwhile, strain Sa-BW8 formed a separated branch which suggests the greater number of sequence difference from the other isolates. Based on phylogenetic analysis and sequence similarity data, strains Sa-BW1, Sa-BW2, Sa-BW7, and Sa-BW8 were clearly regarded as S. algae.
Figure-2.
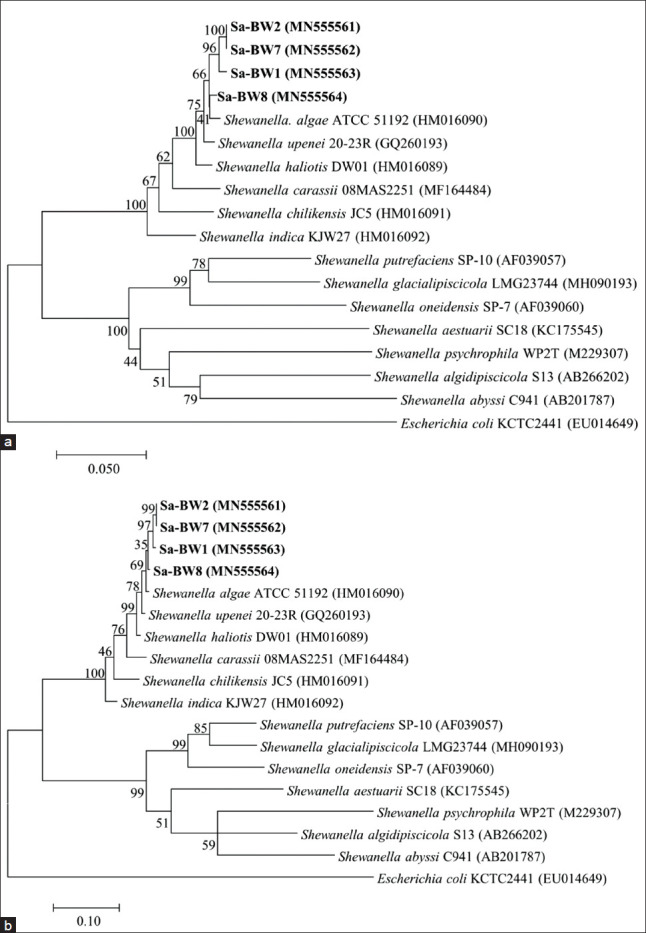
Phylogenetic tree based on gyrB gene sequences of S. algae Sa-BW1, Sa-BW2, Sa-BW7, Sa-BW8, and some other related Shewanella species. (a) Neighbor-joining tree. (b) Maximum-likelihood tree. GenBank accession numbers are given in parentheses.
Further phylogenetic analysis of S. algae gyrB sequences from ballast water isolates against several nearest S. algae sequences from GenBank database allows for reasonable interpretation in determining the most likely origin of each isolates from the ballast tank. However, it is impossible to determine the exact origin of the bacteria found in ballast water. Based on the ML tree of gyrB gene generated in Figure-3, isolate Sa-BW1 which ballast water source was from Singapore port was closely related to S. algae strains RQs-106 from China. Isolate Sa-BW2 and Sa-BW7 isolates with source of ballast water from Yokkaichi port and Nhava Sheva port, respectively, were clustered together with S. algae strain KC-Na-R1 from South Korea. Meanwhile, isolate Sa-BW8 which sourced from Malacca Straits formed a separated branch closely related to strain 2NE11 from Peru. This result suggests that ballast water could transport bacterial species from various regions around the globe contributing to the global spread of the species.
Figure-3.
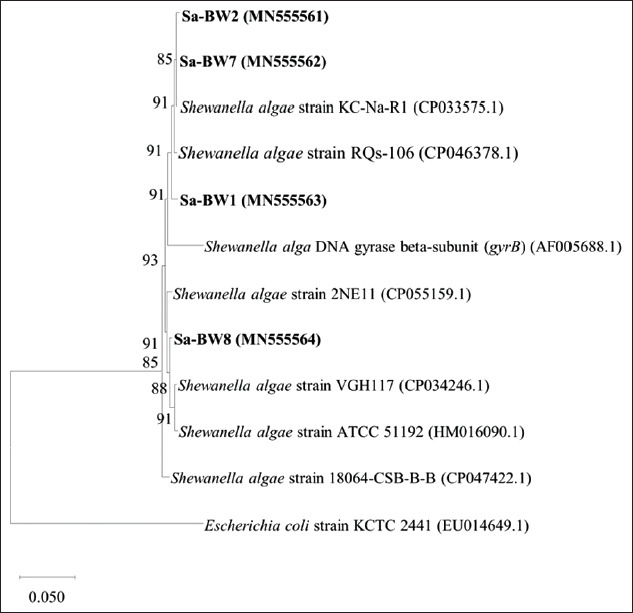
Maximum likelihood phylogenetic tree based on gyrB gene sequences of Shewanella algae Sa-BW1, Sa-BW2, Sa-BW7, Sa-BW8, and some other closely related S. algae strains. GenBank accession numbers are given in parentheses.
Biofilm formation capabilities
In this study, the cutoff ODC was set as 0.143. The isolates that showed OD value lesser than 0.143 were considered as non-biofilm former while isolates with OD value more than 4 × ODC were regarded as strong biofilm former. Comparing the OD means of each S. algae isolate to differentiation criterion; ODc = 0.143, 2 × ODc = 0.286, and 4 × ODC = 0.332, all strains were observed to be strongly adherent with OD value; 1.31±0.9 (Sa-BW1), 1.23±1.0 (Sa-BW2), 1.16±0.5 (Sa-BW7), 0.91±0.4 (Sa-BW8), 0.93±0.01 (Sa-SW2), and 0.89±0.6 (Sa-SW3), respectively. Positive control, S. aureus showed OD value 0.92±0.05 indicated strong adherence capabilities.
Antibiotic susceptibility
A similar pattern of antibiotic susceptibility was observed in all S. algae strains. As summarized in Table-3, multiple resistance was observed against oxacillin, ampicillin, carbenicillin, cephalothin, novobiocin, tobramycin, colistin, and rifampicin. Intermediate resistance to kanamycin, oxytetracycline, and streptomycin was also recorded. The isolates were only sensitive to ciprofloxacin, chloramphenicol and gentamicin. The MAR index recorded was >0.2 suggesting that the isolates were originated from a source that has antibiotic contamination [43].
Table-3.
Antibiotic resistance profile of S. algae strains from ballast water.
| Antibiotics | Disc content (μg) | Sensitivity |
|---|---|---|
| Ampicillin | 20 | R |
| Carbenicillin | 100 | R |
| Cephalothin | 30 | R |
| Ciprofloxacin | 10 | S |
| Chloramphenicol | 30 | S |
| Colistin | 10 | R |
| Gentamicin | 10 | S |
| Kanamycin | 30 | I |
| Novobiocin | 30 | R |
| Oxacillin | 5 | R |
| Oxytetracycline | 30 | I |
| Penicillin | 10 | R |
| Rifampicin | 5 | R |
| Streptomycin | 25 | I |
| Tobramycin | 10 | R |
| MAR index (a/b) | 0.6 |
S = Susceptible, I = Intermediate, R = Resistance, a/b = calculation ratio. S. algae: Shewanella algae
Discussion
In this study four S. algae was isolated from ballast water of ships docked in Port Klang, Malaysia. The isolates had key phenotypic characteristics of oxidase positive and sulfide production attributed to Shewanella spp. [18]. Additional phenotypic characteristics such as the ability to grow at 42°C, tolerance to high salt concentration (6-8%), and hemolysis on sheep blood agar distinguished the S. algae isolates from other species such as S. putrefaciens, S. haliotis and S. xiamenensis as described in the previous study [44]. These characteristics grouped the S. algae isolates into mesophilic and halophilic Shewanella strains as reported by several studies [45-47]. The biochemical characteristics of S. algae isolate from this study were similar to S. algae from the first Danish cases of S. algae bacteremia [48], shrimp (P. vannamei) [26], and type strain IAM 14159 [49]. Previous study by Altug et al. [4] also reported the presence of S. algae in several ballast water samples collected from ships berthed around Ambali Port, on the northern shores of Sea of Marmara.
The presence of S. algae in both ballast water and port water samples raises the possibility that some species may have been transported there through ballast water. Whether this species was introduced or is native and common in the local port water remains an open question. Since bacterial species may also be introduced to every marine environment in many ways and may be present in polluted environment, we cannot clearly conclude that the presence of S. algae in the local water is due to the release of ballast water. However, suppose the ballast water that contains the S. algae were released during cargo loading, the species will be transferred to local port water. The same goes to the species present in local water during unloading cargo process where ballasting procedure could transport the species into ballast tank to another port of call. These scenarios would validate the ballast water potential as a vector for global spread of microorganisms.
The phylogenetic analysis grouped the strains (Sa-BW1, Sa-BW2, Sa-BW7, and Sa-BW8) in the S. algae cluster, closest to S. algae ATCC 51192. Based on the comparative analysis of phylogenetic tree, the gyrB gene seems to be more reliable and useful than 16S rRNA for describing phylogenetic relationship at the species level. The S. algae isolates showed independent branching from S. haliotis and S. upenei based on gyrB gene with similarities values of 97.0-97.9% and 97.7-98.5%, respectively, which were much lower than in 16S rRNA (>99%). The lower occurrences of interspecies sequence similarities in gyrB gene compared to 16S rRNA described in this study implied a better resolution to distinguish S. algae isolates from its closely related species, S. haliotis and S. upenei. It is known that the rate of molecular evolution of gyrB sequences is faster than 16S rRNA which provides higher phylogenetic resolution. It was reported in several studies that gyrB gene has always been used as a discriminative detection for Shewanella species identification [50-52]. The similarities values of greater than 97% observed in both gyrB and 16S rRNA sequences of S. algae isolates with S. haliotis and S. upenei revealed a very close phylogenetic association between the three species. This might also indicate a potential taxonomic problem in the presumptive identification of this related strain. However, in recent whole-genome sequencing S. haliotis and S. upenei were proposed as a later heterotrophic synonym of S. algae [53-55]. The phylogenetic analysis of gyrB gene sequence between S. algae isolates from ballast water and nearest S. algae strains sequences from GenBank database revealed the phylogenetically relatedness of the ballast water strain with S. algae strains from various region around the globe. The fact that the S. algae isolates from this study itself were isolated from ballast tanks of different ballast water sources (port of origin) would validate the role of ballast water in transporting the bacterial species around the world, contributing to the global spread. It can be difficult and almost impossible to track the exact origin of the bacterial species as ballast water exchange can be made at different ports [3]. In addition, the ballast tank can contain mixture of water from different ports because there is always a small portion of unpumpable water that remains before taking on cargo.
The capability of all S. algae isolates in this study to produce extracellular enzymes such as lipase, protease, DNase, and hemolysin could be inferred as part of adaptive mechanism for the species to survive in their respective marine environment. The expression of this phenotype could be aimed at reducing surrounding microbial competition or degrading organic matter to gain access to their nutrients [20,56]. However, the adaptation allowing survival in marine environment could also allow for colonization of living hosts if the bacteria get the opportunity to enter the host. These enzymes which are known to have pathogenic potential are capable of enhancing bacterial virulence as they enable the bacteria to breach and invade host tissue contributing to a wide range of infections [57-59]. According to Edberg et al. [60], no single extracellular enzyme has been proved to be the sole factor responsible for bacterial pathogenicity. Thus, it is considered necessary for microbes to contain more than one extracellular enzyme to be virulent. In general, hemolytic activity has been considered as an important virulence marker for Shewanella spp. to predict potentially virulent strain [61-63]. In a mouse pathogenicity study performed by Khashe and Janda [34], S. algae was observed to be the more virulent species compared to S. putrefaciens, and it was speculated that hemolytic activity could play an important virulence factor. Thus, the ability of S. algae isolates in this study to exhibit beta-hemolytic activity on sheep blood and human blood agar suggested their potential as putative pathogen.
The ability of S. algae isolates to form a strong biofilm formation, as shown in this study suggested a significant advantage for the survival adaptation of this species in ballast tank. The production of exopolysaccharides matrix in the formation of biofilms could provide protection from mechanical or chemical treatment and from predatory protists [64]. The biofilm environment may also promote phenotypic modification as well as genetic exchanges among the communities of microorganisms within biofilms [65]. Although it has not been tested, antibiotic resistance or virulence in S. algae may be enhanced through horizontal gene transfer should a novel genotype or toxigenic species arrive in ballast water. This protective film may act as refuge for bacteria during transport allowing them to persist within the tank environment and proliferate. In term of pathogenicity, formation of biofilms provides protection against immune system and antibiotic treatment thus preventing access of certain antimicrobial agents from reaching the bacterial cells within the biofilm which could complicate the clinical treatment of S. algae [66].
Resistance to antibiotics has been inferred to encourage host pathogenesis allowing persistent or chronic diseases [67]. Studies on antibiotic susceptibility profiles of Shewanella spp. indicated that most species are susceptible to aminoglycosides, carbapenems, erythromycin, quinolones, extended-spectrum cephalosporins, and macrolides, but resistance to penicillin [68-70]. However, there has been an increase in the occurrence of multidrug resistance in S. algae strains [71-73] including the isolates from the current study. The most commonly reported antibiotic resistance is against beta-lactams, such as amoxicillin, ampicillin, and penicillin; and against cephalosporins such as cephalothin, cefazolin, and cefotaxime [74] in accordance with the resistance observed in this study. Notably, S. algae is frequently reported to be resistant to colistin [73]. Resistance to tobramycin, novobiocin, and rifampicin was also observed in several studies [24,26,73]. The multiple resistance profile observed in S. algae strains of ballast water are of concern as horizontal gene transfer of antibiotic resistance genes might occur due to the closed system and water retention time within ballast tanks [75].
Conclusion
In this study, four S. algae strains were isolated from ballast water samples taken from ships docked at Port Klang, Malaysia. The isolates were found to produce hemolytic activity on sheep blood agar, secreted several extracellular enzymes (lipase, protease, DNase, and gelatinase), performed a strongly adherent biofilm and demonstrated multiple resistances toward antibiotics. These characteristics may represent the putative pathogenic factor of the S. algae strains. The presence of putative pathogenic strains in ballast water suggested that ships carry a potential risk to local marine environment, should they release the pathogenic strains during ballasting operation. While S. algae is already present in Port Klang waters, further introduction of it could pose a risk to the local ecosystem. Hence, monitoring level of pathogenic species should be continued in incoming ballast water to protect the local environment from bacteriological risks and to guard the public on possible health risks in port environments.
Authors’ Contributions
NNNI, NMN, and FS designed the study. NNNI and NMN did laboratory analysis and collected data. FKS, FS, and AA reviewed the manuscript. FKS and FS were the supervisors for the study. All authors read and approved the final manuscript.
Acknowledgments
The authors thank the Universiti Kebangsaan Malaysia for providing equipment and infrastructures for the research. The study was financially supported by the Malaysian Ministry of Science, Technology and Innovation (MOSTI) Science Fund (Grant no.; 04-01-02-SF1244) and Ministry of Higher Education (MOHE) Fundamental Research Grant Scheme (Grant no. FRGS/1/2019/STG05/UKM/02/6).
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published institutional affiliation.
References
- 1.Ruiz G.M, Rawlings T.K, Dobbs F.C, Drake L.A, Mullady T, Huq A, Colwell R.R. Global spread of microorganisms by ships. Nature. 2000;408(6808):49–50. doi: 10.1038/35040695. [DOI] [PubMed] [Google Scholar]
- 2.Mccarthy S, McPhearson R.M, Guarino A, Gaines J. Toxigenic Vibrio cholerae01 and cargo ships entering Gulf of Mexico. Lancet. 1992;339(8793):624–625. doi: 10.1016/0140-6736(92)90918-s. [DOI] [PubMed] [Google Scholar]
- 3.Aguirre-Macedo M.L, Vidal-Martinez V.M, Herrera-Silveira J.A, Valdés-Lozano D.S, Herrera-Rodríguez M, Olvera-Novoa M.A. Ballast water as a vector of coral pathogens in the Gulf of Mexico:The case of the Cayo Arcas coral reef. Mar. Pollut. Bull. 2008;56(9):1570–1577. doi: 10.1016/j.marpolbul.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 4.Altug G, Gurun S, Cardak M, Ciftci P.S, Kalkan S. The occurrence of pathogenic bacteria in some ships'ballast water incoming from various marine regions to the Sea of Marmara, Turkey. Mar. Environ. Res. 2012;81(2012):35–42. doi: 10.1016/j.marenvres.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Brinkmeyer R. Diversity of bacteria in ships ballast water as revealed by next generation DNA sequencing. Mar. Pollut. Bull. 2016;107(1):277–285. doi: 10.1016/j.marpolbul.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 6.El-Barbary M.I. First recording of Shewanella putrefaciens in cultured Oreochromis niloticus and its identification by 16Sr RNA in Egypt. Egypt. J. Aquat. Res. 2017;43(1):101–107. [Google Scholar]
- 7.Skerratt J.H, Bowman J.P, Nichols P.D. Shewanella olleyana spp nov., a marine species isolated from a temperate estuary which produces high levels of polyunsaturated fatty acids. Int. J. Syst. Evol. Microbiol. 2002;52(6):2101–2106. doi: 10.1099/00207713-52-6-2101. [DOI] [PubMed] [Google Scholar]
- 8.Wang M, Sun L. Shewanella inventionis spp. nov., isolated from deep-sea sediment. Int. J. Syst. Evol. Microbiol. 2016;66(12):4947–4953. doi: 10.1099/ijsem.0.001450. [DOI] [PubMed] [Google Scholar]
- 9.Semple K.M, Westlake D.W.S. Characterization of iron-reducing Alteromonas putrefaciens strains from oil field fluids. Can. J. Microbiol. 1987;33(5):366–371. [Google Scholar]
- 10.Baaziz H, Lemaire O.N, Jourlin-Castelli C, Iobbi-Nivol C, Méjean V, Alatou R, Fons M. Draft genome sequence of Shewanella algidipiscicola H1, a highly chromate-resistant strain isolated from Mediterranean marine sediments. Microbiol. Resour. Announc. 2018;7(8):e00905–18. doi: 10.1128/MRA.00905-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang C, Chaoqun H, Xiaoyan C, Luping Z. Identification and characterization of Shewanella algae as a novel pathogen of ulcer disease of fish Scinenops ocellata. Oceanol. Limnol. Sin. 2003;34(1):1–8. [Google Scholar]
- 12.Yang S, Kwon K.K, Lee H, Kim S, Kim S. Shewanella spongiae sppnov., isolated from a marine sponge. Int. J. Syst. Evol. Microbiol. 2006;56(12):2879–2882. doi: 10.1099/ijs.0.64540-0. [DOI] [PubMed] [Google Scholar]
- 13.Fredrickson J.K, Romine M.F, Beliaev A.S, Jennifer M, Osterman L, Pinchuk G, Reed J.L, Rodionov D.A, Jorge L.M. Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 2008;6(8):592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 14.Li F, Li Y, Sun L, Li X, Yin C, An X, Chen X, Tian Y. Shewanella oneidensis enables xylose-fed microbial fuel cell. Biotechnol. Biofuels. 2017;10(196):1–10. doi: 10.1186/s13068-017-0881-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zong Z. Case report nosocomial peripancreatic infection associated with Shewanella xiamenensis. Med. Microbiol. 2011;60(9):1387–1390. doi: 10.1099/jmm.0.031625-0. [DOI] [PubMed] [Google Scholar]
- 16.Hochedez P, Vignier N, Barreau M, Olive C, Baubion E, Theodose R. Human infection with Shewanella putrefaciens and Salgae:Report of 16 cases in Martinique and review of the literature. Am. J. Trop. Med. Hyg. 2013;89(1):151–156. doi: 10.4269/ajtmh.13-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khurshid A, Shaukat S, Suleman R.M, Angez M. Shewanella haliotis associated with severe soft tissue infection, Thailand. Emerg. Infect. Dis. 2013;19(6):1019–1021. doi: 10.3201/eid1906.121607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holt H.M, Bruun B. Shewanella algae and Shewanella putrefaciens:Clinical and microbiological characteristics. Clin. Microbiol. Infect. 2005;11(5):347–352. doi: 10.1111/j.1469-0691.2005.01108.x. [DOI] [PubMed] [Google Scholar]
- 19.Han Z, Sun J, Lv A, Sung Y, Shi H, Hu X, Xing K. Isolation, identification and characterization of Shewanella algae from reared tongue sole, Cynoglossus semilaevis Günther. Aquaculture. 2017;468(2017):356–362. [Google Scholar]
- 20.Lemaire O.N, Méjean V, Iobbi-Nivol C. The Shewanella genus:Ubiquitous organisms sustaining and preserving aquatic ecosystems. FEMS Microbiol. Rev. 2020;44(2):155–170. doi: 10.1093/femsre/fuz031. [DOI] [PubMed] [Google Scholar]
- 21.Ananth A.L, Nassiri N, Pamoukian V.N. Case reports Shewanella algae:A rare cause of necrotizing fasciitis. Surg. Infect. 2014;15(3):336–338. doi: 10.1089/sur.2012.208. [DOI] [PubMed] [Google Scholar]
- 22.Paccalin M, Grollier G, Le Moal G, Rayeh F, Camiade C. Rupture of a primary aortic aneurysm infected with Shewanella alga. Scand. J. Infect. Dis. 2001;33(10):774–775. doi: 10.1080/003655401317074626. [DOI] [PubMed] [Google Scholar]
- 23.Shanmuganathan M, Goh B.L, Lim C, NorFadhlina Z, Fairol I. Shewanella algae peritonitis in patients on peritoneal dialysis. Perit. Dial. Int. 2016;36(5):574–575. doi: 10.3747/pdi.2015.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davidson N.L, Subedi S, Wilks K, Morgan J. A case of Shewanella algae endocarditis:An emerging pathogen with a diverse clinical spectrum. BMJ. Case. Rep. 2018;2018(2018):bcr2017223396. doi: 10.1136/bcr-2017-223396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai J, Thompson K.D. Isolation and identification of Shewanella alga and its pathogenic effects on post-larvae of abalone Haliotis diversicolor supertexta. J. Fish. Dis. 2006;29(8):505–508. doi: 10.1111/j.1365-2761.2006.00732.x. [DOI] [PubMed] [Google Scholar]
- 26.Cao H, Chen S, Lu L, An J. Shewanella algae:An emerging pathogen of black spot disease in freshwater-cultured white leg shrimp (Penaeus vannamei) Isr. J. Aquacult. Bamid. 2018;70(1):1–7. [Google Scholar]
- 27.Janda J.M, Abbott S.L. The genus Shewanella:From the briny depths below to human pathogen. Crit. Rev. Microbiol. 2012;40(4):293–312. doi: 10.3109/1040841X.2012.726209. [DOI] [PubMed] [Google Scholar]
- 28.Torri A, Bertini S, Schiavone P, Congestri F, Matteucci M, Sparacino M, Testa G, Pedna M.F, Sambri V. Shewanella algae infection in Italy:Report of 3 years'evaluation along coast of the Northern Adriatic sea. New Microbes New Infect. 2018;23(2018):39–43. doi: 10.1016/j.nmni.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dey S, Bhattacharya D, Roy S, Nadgir S.D, Patil A, Kholkute S.D. Shewanella algae in acute gastroenteritis. Indian J. Med. Microbiol. 2015;33(1):172–175. doi: 10.4103/0255-0857.148442. [DOI] [PubMed] [Google Scholar]
- 30.Takata T, Chikumi H, Morishita S, Hamada S, Hoi S, lyama T. Shewanella algae bacteremia in an end-stage renal disease patient:A case report and review of the literature. Intern. Med. 2017;56(6):729–732. doi: 10.2169/internalmedicine.56.7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baeur M.J, Stone-Garza K.K, Croom D, Andreoli C, Woodson P, Graf P.C.F, Maves R.C. Shewanella algae infections in United States naval special warfare trainees. Open Forum Infect. Dis. 2019;6(11):ofz442. doi: 10.1093/ofid/ofz442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin-Rodriguez A.J, Martin-Pujol O, Artiles-Campelo F, Bolanos-Rivero M, Romling U. Shewanella spp infections in Gran Canaria, Spain:Retrospective analysis of 31 cases and a literature review. JMM Case Rep. 2017;4(12):e005131. doi: 10.1099/jmmcr.0.005131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fluke E.C, Carayannopoulos N.L, Lindsey R.W. Pyogenic flexor tenosynovitis caused by Shewanella algae. J. Hand Surg. Am. 2016;41(7):1–4. doi: 10.1016/j.jhsa.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 34.Khashe S, Janda J.M. Biochemical and pathogenic properties of Shewanella alga and Shewanella putrefaciens. J. Clin. Microbiol. 1998;36(3):783–787. doi: 10.1128/jcm.36.3.783-787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srinivas J, Pillai M, Vinod V, Dinesh R.K. Skin and soft tissue infections due to Shewanella algae. An emerging pathogen. J. Clin. Diagn. Res. 2015;9(2):16–20. doi: 10.7860/JCDR/2015/12152.5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andreou L.V. Preparation of genomic DNA from bacteria. Methods. Enzymo. 2013;529:143–151. doi: 10.1016/B978-0-12-418687-3.00011-2. [DOI] [PubMed] [Google Scholar]
- 37.Weisburg W.G, Barns S.M, Pelletier D.A, Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto S, Harayama S. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl. Environ. Microbiol. 1995;61(3):1104–109. doi: 10.1128/aem.61.3.1104-1109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6:Molecular evolutionary genetics analysis version 60. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Toole G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2010;47(2011):10–11. doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stepanović S, Vuković D, Dakić I, Savić B, Švabić-Vlahović M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods. 2000;40(2):175–179. doi: 10.1016/s0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 42.Hudzicki J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol. 2016. Available from: https://www.asm.org/protocols/kirby-bauer-disk-diffusion-susceptibility-test-pro. Retrieved on 29-8-2018 .
- 43.Krumperman P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983;46(1):165–170. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Satomi M. The family Shewanellaceae. In: Rosenberg E, editor. The Prokaryotes:Gammaproteobacteria. Berlin, Heidilberg: Springer-Verlag; 2013. pp. 597–625. [Google Scholar]
- 45.DeFrank J.J, Beaudry W.T, Cheng T.C, Harvey S.P, Stroup A.N, Szafraniec L.L. Screening of halophilic bacteria and Alteromonas species for organophosphorus hydrolyzing enzyme activity. Chem. Biol. Interact. 1993;87(1-3):141–148. doi: 10.1016/0009-2797(93)90035-w. [DOI] [PubMed] [Google Scholar]
- 46.Gram L, Bundvad A, Melchiorsen J, Johansen C, Vogel B.F. Occurrence of Shewanella algae in Danish coastal water and effects of water temperature and culture conditions on its survival. Appl. Environ. Microbiol. 1999;65(9):3896–3900. doi: 10.1128/aem.65.9.3896-3900.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tseng S.Y, Liu P.Y, Lee Y.H, Wu Z.Y, Huang C.C, Cheng C.C, Tung K.C. The pathogenicity of Shewanella algae and ability to tolerate a wide range of temperatures and salinities. Can. J. Infect. Dis. Med. Microbiol. 2018;2018(2018):6976897. doi: 10.1155/2018/6976897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Domínguez H, Vogel B.F, Gram L, Hoffmann S, Schaebel S. Shewanella alga bacteremia in two patients with lower leg ulcers. Clin. Infect. Dis. 1996;22(6):1036–1039. doi: 10.1093/clinids/22.6.1036. [DOI] [PubMed] [Google Scholar]
- 49.Nozue H, Hayashi T, Hashimoto Y, Ezaki T, Hamasaki K, Ohwada K, Terawaki Y. Isolation and characterization of Shewanella alga from human clinical specimens and emendation of the description of Salga Simidu et al 1990. Int. J. Syst. Bacteriol. 1992;42(4):628–634. doi: 10.1099/00207713-42-4-628. [DOI] [PubMed] [Google Scholar]
- 50.Satomi M, Vogel B.F, Gram L, Venkateswaran K. Shewanella hafniensis spp Nov. and Shewanella morhuae spp. Nov., isolated from marine fish of the Baltic Sea. Int. J. Syst. Evol. Microbiol. 2006;56(1):243–249. doi: 10.1099/ijs.0.63931-0. [DOI] [PubMed] [Google Scholar]
- 51.Fang Y, Wang Y, Liu Z, Lu B, Dai H, Kan B, Wang D. Shewanella carassii spp Nov., isolated from surface swabs of crucian carp and faeces of a diarrhoea patient. Int. J. Syst. Evol. Microbiol. 2017;67(12):5284–5289. doi: 10.1099/ijsem.0.002511. [DOI] [PubMed] [Google Scholar]
- 52.Fang Y, Wang Y, Liu Z, Dai H, Cai H, Li Z, Du Z, Wang X, Jing H, Wei Q, Kan B, Wang D. Multilocus sequence analysis, a rapid and accurate tool for taxonomic classification, evolutionary relationship determination, and population biology studies of the genus Shewanella. Appl. Environ. Microbiol. 2019;85(11):1–13. doi: 10.1128/AEM.03126-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szeinbaum N, Kellum C.E, Glass J.B, Janda J.M, DiChristina T.J. Whole-genome sequencing reveals that Shewanella haliotis Kim et al 2018 2007 can be considered a later heterotypic synonym of Shewanella algae Simidu et al. Int. J. Syst. Evol. Microbiol. 1990;68(4):1356–1360. doi: 10.1099/ijsem.0.002678. [DOI] [PubMed] [Google Scholar]
- 54.Kim K.K, Kim Y.O, Park S, Kang S.J, Nam B.H, Kim D.N, Oh T.K, Yoon J.H. Shewanella upenei spp Nov., a lipolytic bacterium isolated from bensasi goatfish. Upeneus bensasi. J. Microbiol. 2011;49(3):381–386. doi: 10.1007/s12275-011-0175-5. [DOI] [PubMed] [Google Scholar]
- 55.Simidu U, Kita-Tsukamoto K, Yasumoto T, Yotsu M. Taxonomy of four marine bacterial strains that produce tetrodotoxin. Int. J. Syst. Evol. Microbiol. 1990;40(4):331–336. doi: 10.1099/00207713-40-4-331. [DOI] [PubMed] [Google Scholar]
- 56.Mogrovejo-Arias D.C, Brill F.H.H, Wagner D. Potentially pathogenic bacteria isolated from diverse habitats in Spitsbergen, Svalbard. Environ. Earth Sci. 2020;79(109):1–9. [Google Scholar]
- 57.Bottone E.J, Reitano M, Janda J.M, Troy K, Cuttner J. Pseudomonas maltophilia exoenzyme activity as correlate in pathogenesis of ecthyma gangrenosum. J. Clin. Microbiol. 1986;24(6):995–997. doi: 10.1128/jcm.24.6.995-997.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomas J.M. The main Aeromonas pathogenic factors. ISRN Microbiol. 2012;2012(2012):256261. doi: 10.5402/2012/256261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pavlov D, De Wet C.M, Grabow W.O, Ehlers M.M. Potentially pathogenic features of heterotrophic plate count bacteria isolated from treated and untreated drinking water. Int. J. Food. Microbiol. 2004;92(3):275–287. doi: 10.1016/j.ijfoodmicro.2003.08.018. [DOI] [PubMed] [Google Scholar]
- 60.Edberg S.C, Gallo P, Kontnick C. Analysis of the virulence characteristics of bacteria isolated from bottled, water cooler, and tap water. Microb. Ecol. Health Dis. 1996;9(2):67–77. [Google Scholar]
- 61.Myung D.S, Jung Y.S, Kang S.J, Song Y.A, Park K.H, Jung S.I, Kim S.H, Shin J.H. Primary Shewanella algae bacteremia mimicking Vibrio septicemia. J. Korean Med. Sci. 2009;24(6):1192–1194. doi: 10.3346/jkms.2009.24.6.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tseng S.Y, Liu P.Y, Lee Y.H, Wu Z.Y, Huang C.C, Cheng C.C, Tung K.C. The pathogenicity of Shewanella algae and ability to tolerate a wide range of temperatures and salinities. Can. J. Infect. Dis. Med. 2018;2018(2018):6976897. doi: 10.1155/2018/6976897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Richards G.P, Watson M.A, Crane E.J.M, Burt I.G, Bushek D. Shewanella and Photobacterium spp in oysters and seawater from the Delaware Bay. Appl. Environ. Microbiol. 2008;74(11):3323–3327. doi: 10.1128/AEM.00060-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Drake L.A, Doblin M.A, Dobbs F.C. Potential microbial invasion via ships'ballast water, sediment and biofilm. Mar. Pollut. Bull. 2007;55(7-9):333–341. doi: 10.1016/j.marpolbul.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 65.Drake L.A, Meyer A.E, Forsberg R.L, Baier R.E, Doblin M.A, Heinemann S, Johnson W.P, Koch M, Rublee P.A, Dobbs F.C. Potential invasion of microorganisms and pathogens via “interior hull fouling”:Biofilms inside ballast water tanks. Biol. Invasions. 2005;7(6):969–982. [Google Scholar]
- 66.Davies D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2003;2(2):114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 67.Schroeder M, Brooks B.D, Brooks A.E. The complex relationship between virulence and antibiotic resistance. Genes. 2017;8(1):39. doi: 10.3390/genes8010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holt H.M, Gahrn-Hansen B, Bruun B. Shewanella algae and Shewanella putrefaciens:Clinical and microbiological characteristics. Clin. Microbiol. Infect. 2005;11(5):347–352. doi: 10.1111/j.1469-0691.2005.01108.x. [DOI] [PubMed] [Google Scholar]
- 69.Janda J.M. Shewanella:A marine pathogen as an emerging cause of human disease. Clin. Microbiol. Newsl. 2014;36(4):25–29. [Google Scholar]
- 70.Martín-Rodríguez A.J, Martín-Pujol O, Artiles-Campelo F, Bolaños-Rivero M, Römling U. Shewanella spp infections in Gran Canaria, Spain:Retrospective analysis of 31 cases and a literature review. JMM Case Rep. 2017;4(12):1–9. doi: 10.1099/jmmcr.0.005131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhatawadekar S.M, Sharma J. Brevundimonas vesicularis bacteremia:A rare case report in a female infant. Indian J. Med. Microbiol. 2011;29(4):420–422. doi: 10.4103/0255-0857.90184. [DOI] [PubMed] [Google Scholar]
- 72.Jampala S, Meera P, Vivek V, Kavitha D.R. Skin and soft tissue infections due to Shewanella algae an emerging pathogen. J. Clin. Diagn. Res. 2015;9(2):16–20. doi: 10.7860/JCDR/2015/12152.5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cimmino T, Olaitan A.O, Rolain J.M. Whole-genome sequence to decipher the resistome of Shewanella algae, a multidrug-resistant bacterium responsible for pneumonia, Marseille, France. Expert. Rev. Antiinfect. Ther. 2016;14(2):269–275. doi: 10.1586/14787210.2016.1106936. [DOI] [PubMed] [Google Scholar]
- 74.Huang Y.T, Tang Y.Y, Cheng J.F, Wu Z.Y, Mao Y.C, Liu P.Y. Genome analysis of multidrug-resistant Shewanella algae isolated from human soft tissue sample. Front. Pharmacol. 2018;9(2018):419. doi: 10.3389/fphar.2018.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ng C, Le T.H, Goh S.G, Liang L, Kim Y, Rose J.B, Yew-Hoong K.G. A comparison of microbial water quality and diversity for ballast and tropical harbor waters. PLoS One. 2015;10(11):1–22. doi: 10.1371/journal.pone.0143123. [DOI] [PMC free article] [PubMed] [Google Scholar]


