Abstract
Background and Aim:
Camels are a unique source of milk and meat, which helps recover from several diseases that affect humans worldwide. In Egypt, one of the great obstacles for this industry is tick-borne diseases. This study aimed to characterize blood parasite infections, such as Babesia (B.) bovis and Trypanosoma (T.) spp. in one-humped camel (Camelus dromedarius) (n=142) breeds in Halayeb and Shalateen, Egypt, through phylogenetic analysis.
Materials and Methods:
The prevalence of B. bovis and Trypanosoma spp. was identified in camels using polymerase chain reaction (PCR) assays targeting the Rhoptry-Associated Protein-1 and internal transcribed spacer 1 genes, respectively. A nested PCR technique was conducted to detect B. bovis. At the same time, KIN multispecies PCR assay was employed to diagnose and classify trypanosome DNA in camels.
Results:
B. bovis was detected in 4/142 camels with an infection rate of 2.81%. Sequencing and phylogenetic analyses revealed that the strain of B. bovis isolated from this population was closely related to strains isolated from Argentine, the United States, and Brazil. Moreover, Trypanosoma evansi was detected in 8/142 camels with an infection rate of 5.63%. Sequencing and phylogenetic analyses revealed that this isolated strain T. evansi was closely related to Trypanosoma theileri detected from cattle in Brazil.
Conclusion:
The obtained data indicated the existence of B. bovis and T. evansi in camels from two provinces of Egypt. The obtained findings have economic significance and reflect the importance of implementing effective prevention and control methods across Egypt to reduce the incidence of B. bovis and T. evansi in camels.
Keywords: Babesia bovis, camel, Egypt, epidemiology, Trypanosoma spp
Introduction
Global concerns about desertification and the resulting loss of biological productivity due to natural processes, climate change, or human activities have highlighted the important role of camels in socioeconomic aspects to protect human populations from health risks and promote species conservation. Thus, it is critical to implement a proper management system with appropriate disease control measures [1]. In several regions of the world, camels provide an important source of food and milk, which is considered a high protein source with substantial nutritive value. Moreover, the unique survival capability of camels in adverse environmental conditions permits breeders to adopt semi-intensive farming systems for camel production [2]. It was thought that camels are resistant to various pathogenic diseases [3]. Recently, different literatures have described the susceptibility of camels to large number of pathogenic agents, such as bacteria, fungi, parasites, and viruses [4].
In Egypt, vector-transmitted diseases cause various clinical manifestations in farm animals [3]. Members of piroplasms have been found in several species of equines, cattle, and camel in Egypt, where specific DNA fragments of those piroplasms were detected in the bloodstream of apparently healthy camels of Babesia and Theileria species [4]. One well-known piroplasm is Babesia spp., which infects humans through tick infestation [5]. Camel babesiosis is an acute to chronic infectious disease that is distributed all over the world [2]. The disease is responsible for deteriorative effects, high morbidity, and substantial economic losses due to transmission through tick-borne hemoparasitic protozoa [2,4]. Trypanosoma evansi is a well-known parasite that affects animals and is transmitted mechanically by tabanid species. T. evansi infections cause weight loss, anemia, and abortion in populations spanning several regions of the world, such as South America, Asia, and Africa [6]. In certain cases of trypanosomiasis, subclinical cases and other chronic infections can be found in hosts infected by low virulence strains [7].
Veterinary epidemiology offers the means to investigate disease outbreaks by tracking and monitoring diseases, identifying disease risk factors, implementing herd health programs, and developing biosecurity measures [8]. Accumulating epidemiological knowledge will lead to innovative strategies and clinical techniques that will result in the reduction, prevention, and eradication of important infectious diseases in animals as blood parasitic infections [9]. In this regard, a molecular technique using a polymerase chain reaction (PCR) assay with sequencing analysis can identify blood parasites in epidemiological studies with higher accuracy than other traditionally diagnostic methods, such as microscopic examination of Giemsa-stained blood smear and enzyme-linked immunoassay (ELISA) tests [3]. The latter two diagnostic tests (microscopy and ELISA) have low sensitivity against chronic and subclinical infections [10], with a lack of specificity in distinguishing between recent and old infections [7]. Most previous methods for epidemiological screening of blood parasite in camels that were performed in Egypt [11-14] used ELISA, the microscopy method, or PCR without performing sequencing and phylogenetic analysis for the diagnosis of parasites. Moreover, most of the previous studies screened only one blood parasite. For the last few years, most of the camels imported to Egypt came from Sudan. Identification and phylogenetic analysis of blood parasites in camels raised on the border between Egypt and Sudan are of great economic importance.
Therefore, this study aimed to use molecular diagnostic techniques to detect parasitic blood infections (Babesia bovis and Trypanosoma spp.) in camel breeds in Halayeb and Shalateen in Egypt followed by phylogenetic analysis to identify each parasite.
Materials and Methods
Ethical approval and informed consent
In this study, all experimental protocols were approved by the Animal Care and Use Committee, Faculty of Veterinary Medicine, Mansoura University, Egypt. According to the Egyptian Medical Research Ethics Committee (No. 14-126), all Institutional and National Guidelines for the care and use of animals were followed. All experiments were carried out under the authority of the Ministry of Education, Egypt, in compliance with the Fundamental Guidelines for Proper Conduct of Animal Experiment and Related Activities in Academic Research Institutions. Informed written consent was obtained from the owner.
Study period and location
The screened blood samples were collected during July 2017 from one-humped camels reared in Halayeb and Shalateen in Egypt, at the Sudan border. The samples were processed at Faculty of Veterinary Medicine, Mansoura University, Egypt.
Sampling
One hundred and forty two blood samples were collected from one-humped camels reared in Halayeb and Shalateen in Egypt, at the Sudan border (Figure-1). All obtained blood samples were tested for infection with B. bovis and Trypanosoma spp. During the sampling period, all animals were apparently healthy.
Figure-1.
Map of the study sites of the blood parasites molecular epidemiology study in Egypt. Blood samples were collected from animals reared in Halayeb and Shalateen as indicated by bullet points [Source: https://simplemaps.com/resources/svg-eg].
DNA extraction and PCR detection of hemoparasites
Blood samples were collected from the jugular veins of camels selected for our study. Approximately 2 mL whole blood was collected from each animal in a Vacutainer tube containing ethylenediaminetetraacetic acid (EDTA). The blood samples were labeled and stored at −20°C until DNA extractions were conducted. The DNA samples were extracted from 300 μL blood samples using a commercial kit (Promega, Madison, WI, USA), following the manufacturer’s instructions, and then stored at −20°C until further use. B. bovis and Trypanosoma spp. were detected in DNA samples using a previously described diagnostic PCR assay targeting Rhoptry-Associated Protein-1 (RAP-1) and internal transcribed spacer 1 (ITS1) genes, respectively [15-17]. Nested PCR assays were used for the detection of B. bovis. KIN multispecies PCR reaction was employed to detect and identify trypanosome DNA in camels. A KIN multispecies PCR amplifies ITS1 and simultaneously detects three major trypanosome species: T. evansi, Trypanosoma congolense, and Trypanosoma vivax [16]. Primer sequences and annealing temperatures are shown in Table-1. Enzyme activation and denaturation used for the amplification conditions for B. bovis and Trypanosoma spp. were 95°C for 3 min and 95°C for 30 s, 95°C for 5 min and 95°C for 30 s, and 94°C for 3 min and 94°C for 1 min, respectively [15-17]. After the product was chilled to 4°C, gel electrophoresis of the PCR products was performed on 1.5% agarose gel with Tris/Borate/EDTA buffer and was stained with ethidium bromide. The final PCR product was then visualized under ultraviolet light.
Table-1.
Primary and nested PCR primers used for PCR amplifications.
| Species | Assay | Primer sequence (5′ → 3′) | Annealing | Amplification cycles (No.) | Product size | Target gene | References |
|---|---|---|---|---|---|---|---|
| B. bovis | PCR | F-CACGAGCAAGGAACTACCGATGTTGA R- CCAAGGACCTTCAACGTACGAGGTCA |
55°C | 45 | 360 bp | RAP-1 | [15-17] |
| nPCR | F- TCAACAACGTACTCTATATGGCTACC R- CTACCGACCAGAACCTTCTTCACCAT |
55°C | 35 | 298 bp | |||
| Trypanosoma spp. | KIN-PCR | F- GCGTTCAAAGATTGGGCAAT R- CGCCCGAAAGTTCACC |
60°C | 40 | 540 bp | ITS1 | [16] |
B. bovis: Babesia bovis, PCR=Polymerase chain reaction, RAP-1=Rhoptry-Associated Protein-1, ITS1=Internal transcribed spacer 1
Cloning and sequencing of PCR products
Extraction of amplicons of PCR samples with high band intensities using a QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany) was conducted from agarose gels. The samples were then cloned into a plasmid vector (PCR 2.1-TOPO, Invitrogen, Carlsbad, CA, USA). A genetic analyzer AB1 PRISM 100 (Applied Biosystems, Foster City, CA, USA) was used to sequence two colonies for each amplicon.
Phylogenic analysis
The evolutionary history was predicted using the neighbor-joining method [18]. The optimal tree with a sum of 3-branch length = 56.08480007 is shown. The percentage of replicate trees in which associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches [19]. Evolutionary distances were computed using the maximum composite likelihood method [20] and are represented by the units of the number of base substitutions per site. The analysis involved 17 nucleotide sequences. The codon positions were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. There were 201 positions in the final dataset. Evolutionary analyses were conducted using MEGA6 [20].
Statistical analysis
The Open Epi program was used to detect the upper and lower limits of the confidence intervals of the positive rates for B. bovis and T. evansi parasites (http://www.openepi.com/v37/Proportion/Proportion.htm).
Results
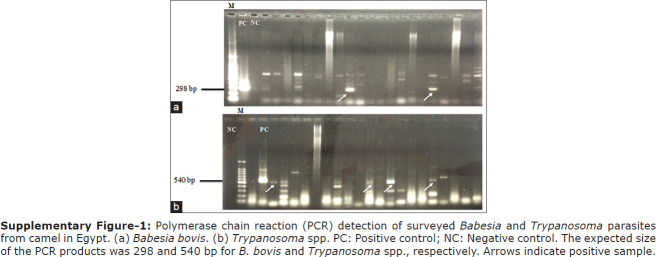
The genomic DNA sequences of the detected blood parasites in camel blood were subjected to PCR with universal primers to detect all possible strains of Babesia and Trypanosoma species to amplify small ribosomal subunits in those parasites using a molecular diagnostic approach. The infection rates for B. bovis and T. evansi were 2.81% and 5.63%, respectively (Table-2). Species-specific PCR assays detected B. bovis and KIN multispecies, while the PCR detected Trypanosoma species in the camel populations (Supplementary Figure-1). One sample harbored mixed infections from Babesia and Trypanosoma parasite species.
Table-2.
PCR detection of blood parasites in camels.
| Species | Positive No. | % CIa |
|---|---|---|
| B. bovis | 4 | 2.81 (1.10 7.01) |
| T. evansi | 8 | 5.63 (2.88 10.72) |
95% confidence interval, B. bovis: Babesia bovis, PCR=Polymerase chain reaction, T. evansi=Trypanosoma evansi
Supplementary Figure-1.
Polymerase chain reaction (PCR) detection of surveyed Babesia and Trypanosoma parasites from camel in Egypt. (a) Babesia bovis. (b) Trypanosoma spp. PC: Positive control; NC: Negative control. The expected size of the PCR products was 298 and 540 bp for B. bovis and Trypanosoma spp., respectively. Arrows indicate positive sample.
The nucleotide sequences of PCR amplified RAP-1 and ss-rRNA genes of Babesia species and the ITS1 gene of Trypanosoma species determined in this study have been registered and were assigned the following GenBank accession numbers: B. bovis (MF737083.1) and T. evansi (MF737081.1). For phylogenetic analysis, nucleotide sequences of the target genes from other species of Babesia and Trypanosoma were included for comparison (Tables-S1 and S2).
Table-S1.
Babesia and Trypanosoma species used for genetic analysis in this study.
| Babesia species | Origin | Accession number | |
|---|---|---|---|
| Geographic | Biological | ||
| B. bovis | Uruguay | Cattle | AF030061 |
| North Brazil | Cattle | FJ588011.1 | |
| China | Cattle | KT318580 | |
| USA | Cattle | AF030053.1 | |
| Argentina | Cattle | AF030055.1 | |
| Texas USA | Cattle | AF030059.1 | |
| Mexico | Cattle | AF027149.1 | |
| South Africa | Bovine | KC894392.1 | |
| Philippines | Cattle | LC006976.1 | |
| Colombia | Cattle | KX365053.1 | |
| Brazil | Bovine | JX177355.1 | |
| China | Cattle | KT312807.1 | |
| China | Cattle | KT318580.1 | |
| China | Cattle | KT312814.1 | |
| Cuba | Buffalo | JF279443.1 | |
| Sri Lanka | Buffalo | AB845432.1 | |
| Egypt | Cattle | AB917246.1 | |
| China | Cattle | KT312810.1 | |
| Guinea-Bissau | Cattle | KC312810.1 | |
| Brazil | Cattle | KC964615.1 | |
| Egypt | Camel | MF737083.1a | |
| Argentine | Cattle | AF030056.2 | |
| USA | Cattle | AF030054.1 | |
| Egypt | Cattle | BAP47555.1 | |
| China | Cattle | ALJ26466.1 | |
| USA | Cattle | AAB84269.1 | |
| USA | Cattle | AAB84264.1 | |
| Mexico | Cattle | AAC27387.1 | |
| USA | Cattle | AAB84263.1 | |
| China | Cattle | AKR16131.1 | |
| Philippines | Cattle | BAP81796.1 | |
| Argentine | Cattle | AAB84266.1 | |
| China | Cattle | AKR16127.1 | |
| Sri Lanka | Buffalo | BAO50704.1 | |
| China | Cattle | AKR16124.1 | |
| Brazil | Buffalo | AGT18676.1 | |
| Colombia | Cattle | APP91268.1 | |
| Argentine | Cattle | AAB84265.1 | |
| USA | Cattle | AAB84264.1 | |
| Portugal | Cattle | ACY76255.1 | |
| Sri Lanka | Cattle | BAW18785.1 | |
| South Africa | Bovine | AGU67927.1 | |
| Uruguay | Cattle | AAB84271.1 | |
| Brazil | Cattle | ACM44004.1 | |
| Cuba | Buffalo | AEI52891.1 | |
| B. bovis | Brazil | Bovine | AFQ30755.1 |
| Germany | Vole | AB085191.1 | |
GenBank accession numbers submitted by this study. B. bovis: Babesia bovis
Table-S2.
Trypanosoma species used for genetic analysis in this study.
| Trypanosoma species | Origin | Accession number | |
|---|---|---|---|
| Geographic | Geographic | ||
| Trypanosoma microti | Field vole | United Kingdom | AY043354.1 |
| Trypanosoma minasense | Squirrel-monkeys | Japan | AB362412.1 |
| Trypanosoma grosi | Wood mouse | United Kingdom | AY043355.1 |
| Trypanosoma evotomys | Bank vole | United Kingdom | AY043356.1 |
| Trypanosoma herthameyeri | Criolla frog | Brazil | KT765860.1 |
| Trypanosoma teixeirae | Red flying fox | Australia | KT907061.1 |
| Trypanosoma cruzi | ND | USA | LT220268.1 |
| Trypanosoma livingstonei | Roundleaf bat | Mozambique | KF192994.1 |
| Trypanosoma rangeli | bugs | Brazil | KT368799.1 |
| T. evansi | Camel | Egypt | Mf737081.1a |
| T. theileri | Cattle | Brazil | AY773713.1 |
| Trypanosoma dionisii | Free-tailed bat | Brazil | JN040980.1 |
| T. vivax | ND | Brazil | AY363165.1 |
| Trypanosoma equiperdum | Horse | South Africa | KY609969.1 |
GenBank accession numbers submitted by this study. T. evansi=Trypanosoma evansi, T. theileri=Trypanosoma theileri, T. vivax=Trypanosoma vivax
Detection of B. bovis in camel blood was determined using primer pairs flanking RAP-1. PCR of RAP-1 produced an amplicon size of 298 bp that was evaluated by GenBank for further confirmation of the determined sequence. The confirmed sequence was deposited in GenBank under accession number MF737083.1, where a similarity index test among other related B. bovis species was conducted using GenBank. A phylogenetic analysis was constructed using our obtained species and other related sequences of the RAP-1 gene in B. bovis. The phylogenetic analysis revealed a significant decrease in genetic distance between our obtained species and other species isolated from Argentina (AF030056), the United States (AF030054), and Brazil (KC964615) (Figure-2). Interspecies genetic distance analysis was conducted between our obtained RAP-1 gene sequences and sequences of related taxa, and a close genetic distance was detected between our isolated strain and other taxa isolated from cattle in Argentina (AF030056.2) and the United States (AF030054.1) (Supplementary Figure-2). For further confirmation, an amino acid sequence was deduced from our obtained sequence and was aligned with other related sequences of RAP-1 protein (Supplementary Figure-2). The results of the phylogenetic analysis of proteins showed a close genetic relationship with other strains from Mexico (AAC27387.1) (Figure-3). Pairwise genetic distance was performed between our obtained amino acid sequence and sequences of related species of RAP-1 genes, and close relationships with all aligned species except the isolated species from Brazilian bovine species (AFQ30755.1) were revealed (Supplementary Figure-3).
Figure-2.

Phylogenic tree of Babesia bovis Rhoptry-Associated Protein-1 gene. The nucleotide sequences determined in this study are shown in boldface type letters. Bootstrap values are provided at the beginning of each branch. The sequence identified from Egypt in this study is boxed in black.
Supplementary Figure-2.
Pairwise genetic distance based on Rhoptry-Associated Protein-1 gene sequences of Babesia bovis detected in camel in Egypt and compared with other related species on GenBank.
Figure-3.

Protein’s phylogenetic tree of Babesia bovis Rhoptry-Associated Protein-1 gene. The amino acids sequences determined in this study are shown in boldface type letters. Bootstrap values are provided at the beginning of each branch. The sequence identified from Egypt in this study is boxed in black.
Supplementary Figure-3.
Pairwise genetic distance based on Rhoptry-Associated Protein-1 amino acid deduced sequences of Babesia bovis detected in camel in Egypt and compared with other related species on GenBank.
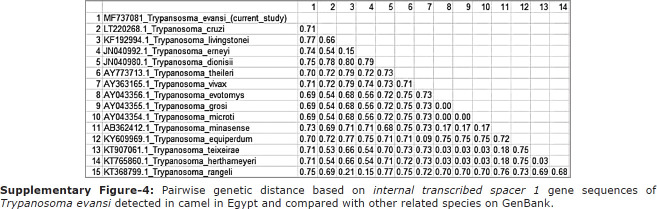
Internal transcript spacer 1 was used for simultaneous identification of three major trypanosome species (T. evansi, T. congolense, and T. vivax) in camels reared in Upper Egypt, with a product size of 540 bp using a KIN multispecies PCR assay. The sequencing results indicated that T. evansi is the causative agent of trypanosomosis in the camels under study. An accession number was deposited in GenBank for our obtained sequence (MF737081) (Figure-4). A phylogenetic tree was constructed depicting our isolated species of T. evansi and with related species where a higher similarity index was aligned with our sequence, and a lower genetic distance was observed between our isolated sequence and Trypanosoma theileri isolated from cattle in Brazil. The phylogenetic analysis revealed the presence of two clades (mammalian and non-mammalian), wherein T. evansi clustered with the mammalian group (Figure-4). Genetic distance analysis of our isolated T. evansi and other related trypanosome species revealed close genetic distance with AY0433156.1 from the United Kingdom in bank vole, AY043355.1 from the United Kingdom in wood mouse, and AY043354.1 from the United Kingdom in field vole with a 0.69 value in comparison with other included taxa (Supplementary Figure-4).
Figure-4.

Phylogenic tree of Trypanosoma evansi internal transcribed spacer 1 gene. The nucleotide sequences determined in this study are shown in boldface type letters. Bootstrap values are provided at the beginning of each branch. The sequence identified from Egypt in this study is boxed in black.
Supplementary Figure-4.
Pairwise genetic distance based on internal transcribed spacer 1 gene sequences of Trypanosoma evansi detected in camel in Egypt and compared with other related species on GenBank.
Discussion
A phylogenetic analysis was designed for our isolated sequence and other sequences with a high similarity index using the neighbor-joining method for statistical and phylogenetic analysis of 500 replicates. A close genetic relationship was observed between our strain and other strains from Argentina, the United States, and Brazil. A similar result was investigated through research conducted by Mtshali et al. [21] who found that a nested PCR technique to amplify RAP-1 revealed a close genetic relationship of a South African strain of B. bovis strains with other strains from Uruguay, Argentina, Brazil, and the United States. However, the presence of polymorphism can adequately discriminate between two species [22]. Similarly, the conservation of Babesia spp. sequence among species was observed among Brazilian strains of B. bovis (98-100%) [23].
Three main trypanosome species (T. evansi, T. congolense, and T. vivax) can be simultaneously detected in the circulating blood of camels using internal transcript spacer 1. A genetic relationship between our isolated species and other Trypanosoma species has been established through phylogenetic analysis. In this study, T. evansi was identified through PCR sequencing and was determined to be closely related to T. theileri detected from cattle in Brazil. Trypanosome was classified into several clades in a study in 1996 by Maslov et al. [24] which are as follows: Mammalian, bird group, and elasmobranch, vertebrates, and invertebrates parasites. Hughes and Piontkivska [25] classified trypanosome species according to the amino acid sequence of a small ribosomal subunit into 42 protein families. They also classified Trypanosoma families according to the host and location into American, and African. In Egypt, internal transcript spacer 1 was used to investigate the phylogenetic relationship among trypanosome species and revealed genetic diversity among those species, which can be used as a diagnostic tool for the identification of trypanosome species [26].
This study has some limitations that should be noted. The small sample size used in this study may not allow for a generalizable conclusion about the incidence rate of different hemoparasites included in the study. Moreover, this study was carried out in Upper Egypt, which so further investigation is needed to determine the presence blood parasites in other areas of Egypt.
Conclusion
The data provided here indicate the presence of B. bovis and T. evansi in camels from two provinces of Egypt. These findings are of economic significance and reflect the importance of adopting successful prevention and control methods across Egypt to reduce the incidence and spread of blood parasite infection in camels.
Authors’ Contributions
SAEE, MAR, MAE, and ME: Conceived and designed the experiments. SAEE, MAR, MAE, MOA, and ME: Performed the experiments. SAEE, MAE, MAO, and ME: Analyzed the data. SAEE, MAR, MAE, MA, ME, ME, and SSS: Contributed reagents/materials/analysis tools. SAEE, MAR, MAE, and ME: Wrote the manuscript. All authors reviewed the manuscript.
Data Availability
All data collected during this study are included in this article.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published map and institutional affiliation.
Acknowledgments
This study is supported by the Ministry of Higher Education of Egypt. The authors would like to thank the owners and staff of the study farms in Egypt.
References
- 1.Zarrin M, Riveros J.L, Ahmadpour A, de Almeida A.M, Konuspayeva G, Vargas-Bello-Pérez E, Faye B, Hernández-Castellano L.E. Camelids:New players in the international animal production context. Trop. Anim. Health Prod. 2020;52(3):903–913. doi: 10.1007/s11250-019-02197-2. [DOI] [PubMed] [Google Scholar]
- 2.Mihic T, Rainkie D, Wilby K.J, Pawluk S.A. The therapeutic effects of camel milk:A systematic review of animal and human trials. J. Evid. Based Complementary Altern. Med. 2016;21(4):NP110–Np126. doi: 10.1177/2156587216658846. [DOI] [PubMed] [Google Scholar]
- 3.Elsify A, Sivakumar T, Nayel M, Salama A, Elkhtam A, Rizk M, Mosaab O, Sultan K, Elsayed S, Igarashi I, Yokoyama N. An epidemiological survey of bovine Babesia and Theileria parasites in cattle, buffaloes, and sheep in Egypt. Parasitol. Int. 2015;64(1):79–85. doi: 10.1016/j.parint.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Qablan M.A, Sloboda M, Jirku M, Obornik M, Dwairi S, Amr Z.S, Horin P, Lukes J, Modry D. Quest for the piroplasms in camels:Identification of Theileria equi and Babesia caballi in Jordanian dromedaries by PCR. Vet. Parasitol. 2012;186(3-4):456–460. doi: 10.1016/j.vetpar.2011.11.070. [DOI] [PubMed] [Google Scholar]
- 5.Rizk M.A, El-Sayed S.A.E, Nassif M, Mosqueda J, Xuan X, Igarashi I. Assay methods for in vitro and in vivo anti-Babesia drug efficacy testing:Current progress, outlook, and challenges. Vet. Parasitol. 2020;279(3):109013. doi: 10.1016/j.vetpar.2019.109013. [DOI] [PubMed] [Google Scholar]
- 6.Desquesnes M, Dargantes A, Lai D.H, Lun Z.R, Holzmuller P, Jittapalapong S. Trypanosoma evansi and surra:A review and perspectives on transmission, epidemiology and control, impact, and zoonotic aspects. Biomed. Res. Int. 2013;2013(9):321237. doi: 10.1155/2013/321237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elhaig M.M, Youssef A.I, El-Gayar A.K. Molecular and parasitological detection of Trypanosoma evansi in Camels in Ismailia, Egypt. Vet. Parasitol. 2013;198(1-2):214–218. doi: 10.1016/j.vetpar.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Salman M.D. The role of veterinary epidemiology in combating infectious animal diseases on a global scale:The impact of training and outreach programs. Prev. Vet. Med. 2009;92(4):284–287. doi: 10.1016/j.prevetmed.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith R.S. Veterinary Clinical Epidemiology. CRC Press Boca Raton. 2005 [Google Scholar]
- 10.El-Sayed S, Rizk M.A, Terkawi M, Igarashi I. Cocktail Babesia bovis antigens for global detection of Babesia bovis infection in cattle. Exp. Parasitol. 2019;206(9):107758. doi: 10.1016/j.exppara.2019.107758. [DOI] [PubMed] [Google Scholar]
- 11.Abd-Elmaleck B.S, Abed G.H, Mandourt A.M. Some protozoan parasites infecting blood of camels (Camelus dromedarius) at Assiut Locality, Upper Egypt. J. Bacteriol. Parasitol. 2014;5(2):1–7. [Google Scholar]
- 12.Barghash,S.M Darwish, A.M. and El-Naga T.R.A. Molecular detection of pathogens in ticks infesting camels in Matrouh Governorate, Egypt. J. Bacteriol. Parasitol. 2016;7(2):1–7. [Google Scholar]
- 13.Haridy F.M, Khalil H.H, Morsy T.A. Trypanosoma evansi in dromedary camel:With a case report of zoonosis in greater Cairo, Egypt. J. Egypt Soc. Parasitol. 2011;41(1):65–76. [PubMed] [Google Scholar]
- 14.El-Naga T.R.A, Barghash S.M. Blood parasites in camels (Camelus dromedarius) in Northern West Coast of Egypt. J. Bacteriol. Parasitol. 2016;7(1):258. [Google Scholar]
- 15.Mtshali M.S, Mtshali P.S. Molecular diagnosis and phylogenetic analysis of Babesia bigemina and Babesia bovis hemoparasites from cattle in South Africa. BMC Vet. Res. 2013;9(8):154. doi: 10.1186/1746-6148-9-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mossaad E, Salim B, Suganuma K, Musinguzi P, Hassan M.A, Elamin E.A, Mohammed G.E, Bakhiet A.O, Xuan X, Satti R.A, Inoue N. Trypanosoma vivax is the second leading cause of camel trypanosomosis in Sudan after Trypanosoma evansi. Parasit. Vectors. 2017;10(1):176. doi: 10.1186/s13071-017-2117-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rizk M.A, AbouLaila M, El-Sayed S.A.E, Guswanto A, Yokoyama N, Igarashi I. Inhibitory effects of fluoroquinolone antibiotics on Babesia divergens and Babesia microti, blood parasites of veterinary and zoonotic importance. Infect. Drug Resist. 2018;2018(11):1605–1615. doi: 10.2147/IDR.S159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saitou N, Nei M. The neighbor-joining method:A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J. Confidence limits on phylogenies:An approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 20.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6:Molecular evolutionary genetics analysis Version 60. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mtshali P.S, Tsotetsi A.M, Thekisoe M.M, Mtshali M.S. Nested PCR detection and phylogenetic analysis of Babesia bovis and Babesia bigemina in cattle from Peri-urban localities in Gauteng Province, South Africa. J. Vet. Med. Sci. 2014;76(1):145–150. doi: 10.1292/jvms.13-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher T.G, McElwain T.F, Palmer G.H. Molecular basis for variable expression of merozoite surface antigen gp45 among American isolates of Babesia bigemina. Infect. Immun. 2001;69(6):3782–3790. doi: 10.1128/IAI.69.6.3782-3790.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramos C.A, Araújo F.R, Alves L.C, de Souza I.I.F, Guedes D.S, Jr, Soares C.O. Genetic conservation of potentially immunogenic proteins among Brazilian isolates of Babesia bovis. Vet. Parasitol. 2012;187(3-4):548–552. doi: 10.1016/j.vetpar.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 24.Maslov D.A, Lukes J, Jirku M, Simpson L. Phylogeny of trypanosomes as inferred from the small and large subunit rRNAs:Implications for the evolution of parasitism in the trypanosomatid protozoa. Mol. Biochem. Parasitol. 1996;75(2):197–205. doi: 10.1016/0166-6851(95)02526-x. [DOI] [PubMed] [Google Scholar]
- 25.Hughes A.L, Piontkivska H. Molecular phylogenetics of Trypanosomatidae:Contrasting results from 18S rRNA and protein phylogenies. Kinetoplastid Biol. Dis. 2003;2(1):15. doi: 10.1186/1475-9292-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amer S, Ryu O, Tada C, Fukuda Y, Inoue N, Nakai Y. Molecular identification and phylogenetic analysis of Trypanosoma evansi from dromedary camels (Camelus dromedarius) in Egypt, a pilot study. Acta Trop. 2011;117(1):39–46. doi: 10.1016/j.actatropica.2010.09.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data collected during this study are included in this article.