Summary
This protocol describes how to visualize surface protein-protein co-localization across a cell-cell interface between antigen-presenting γδ-T cells and CD4 T cells. By consolidating immunofluorescence assay, confocal microscopy and 3D imaging analysis, it enables assessment of interaction between cell surface proteins such as Δ42PD1 and TLR4 between co-cultured γδ-T and CD4 T cells. This protocol can be applied to study a surface protein of interest and its potential interaction with a target cell/protein at the cell-cell interface.
For complete details on the use and execution of this profile, please refer to Mo et al. (2020).
Subject areas: Cell culture, Cell isolation, Immunology, Microscopy
Graphical abstract

Highlights
-
•
Protocol to co-culture antigen-pulsed Vδ2 T cell and antigen-specific CD4 T cell
-
•
Use of IFA for visualizing colocalization of proteins across cell-cell interface
-
•
Detailed procedures to perform Z-stack and 3D imaging analysis
This protocol describes how to visualize surface protein-protein co-localization across a cell-cell interface between antigen-presenting γδ-T cells and CD4 T cells. By consolidating immunofluorescence assay, confocal microscopy, and 3D imaging analysis, it enables assessment of interaction between cell surface proteins such as Δ42PD1 and TLR4 between co-cultured γδ-T and CD4 T cells. This protocol can be applied to study a surface protein of interest and its potential interaction with a target cell/protein at the cell-cell interface.
Before you begin
Generally, in this protocol, pp65-specific CD4 T cell and pp65-pulsed γδ-T cell are co-cultured and visualized by confocal microscope. Figure 1 below displays the timeline for the whole protocol. All cells are to be incubated in 37°C/ 5% CO2 cell culture incubator.
Figure 1.
Timeline of the procedures described in this protocol
PBMC isolation
Timing: 2 h 30 min
-
1.Preparation before Gradient Centrifugation
-
a.Buffy coat blood from healthy donors (n=3) is collected under existing ethic approval protocols with local regulatory agencies.
-
b.Dilute buffy coat blood with sterile 1× phosphate buffered saline (PBS) at a ratio of 1:3.
-
c.Load 30 mL diluted buffy coat gently in a 50 mL sterile conical centrifuge tube underlaid with 15 mL Ficoll-Paque.
-
i.Add 15 mL Ficoll-Paque to the bottom of a 50 mL centrifuge tube first.Note: Temperature may affect Ficoll-Paque density, so it is important to keep Ficoll-Paque at room temperature (22°C~28°C) to ensure good performance on PBMC isolation.
-
ii.When gently loading 30 mL diluted blood onto Ficoll-Paque, the pipetting speed should be as slow as possible at the beginning and increase gradually, in order to avoid heavily disturbing the Ficoll-Paque surface.
-
i.
-
a.
-
2.Gradient Centrifugation and Cell Collection
-
a.Centrifuge at 652 g for 30 min.
 CRITICAL: To ensure good performance, this procedure should be done at room temperature (22°C~28°C) due to the suitable Ficoll-Paque density. It is also important to keep acceleration at a slow rate (usually at 5) and brake at exactly 0.
CRITICAL: To ensure good performance, this procedure should be done at room temperature (22°C~28°C) due to the suitable Ficoll-Paque density. It is also important to keep acceleration at a slow rate (usually at 5) and brake at exactly 0. -
b.After centrifuging, the peripheral blood mononuclear cell (PBMC) layer would appear as a white yellow thin interface layer between the middle clear Ficoll-Paque layer and the upper yellow plasma layer.
-
c.Use a Pasteur pipette to aspirate the PBMC layer and transfer into a new 50 mL sterile conical centrifuge tube.
-
d.Makeup the volume to 45 mL with 1× PBS and centrifuge the tube at 200 g for 10 min.
-
e.The supernatant should be disposed by pouring into a waste container gently to avoid the loss of the cell pellet.
-
f.Flick the bottom of the tubes to dislodge the pellet and resuspend the PBMC cell pellet with a total of 45 mL PBS and centrifuge again at 200 g for 10 min.
-
g.Repeat e-f.
-
h.Count the number of PBMC and check cell viability using 0.4% Trypan Blue and Hemacytometer under microscope.Note: If the counting step takes less than 5 min, PBMC can be kept at room temperature (22°C~28°C). Otherwise, it would be better to keep cells at 4°C or on ice before further experiments. Normally, cell viability should be greater than 90% under good practice during PBMC isolation.
-
i.Based on the cell count, remove 10 million PBMC for the immediate experiment (preparation of pp65-specific CD4 T cell) into a new tube.
-
j.The remaining PBMCs can be centrifuged for a cell pellet and resuspended in freezing medium before use. Prepare the following tubes: one vial (100 million PBMC in 1 mL freezing medium) for γδ-T cell isolation, one vial (10 million PBMC in 500 μL freezing medium) for CD4 T cell isolation, while one vial (10 million PBMC in 500μL freezing medium) for PBMC supplementation in cell culture. The residual PBMC can also be stored (around 100 million PBMC in 1 mL freezing medium) as back-up when applicable.
-
a.
Note: For short-term storage, cells can be stored at −80°C. For long-term storage, cells are suggested to be stored at −150°C.
Note: The protocol for isolating peripheral blood mononuclear cells is adapted from the recommended protocol from GE Healthcare, which is originally developed by Bøyum, A. (Böyum, 1968).
Preparation of pp65-specific CD4 T cell
Timing: 14 days
-
3.pp65 Stimulation on Fresh PBMC
-
a.On the day of PBMC isolation (Day 1), PBMC culture A (PBMCa) is prepared. In order to stimulate pp65-specific memory CD4 T cells, resuspend 10 million PBMC in 2 mL R10 medium and supplement with 10 μL PepTivator CMV pp65 (30 μM). Distribute 1 mL of the cell suspension into each well of a 12-well plate.Note: The expansion method of pp65-specific T cell is modified and adapted from the protocol published by other researchers (Loeth et al., 2012).Note: This step is to obtain pp65-specific T cell for further co-culture experiment. Unstimulated CD4 T cell is isolated from frozen PBMC in step 4(a), serving as a negative control in the further experiment.
-
b.On the next day (Day 2), add human IL-2 (working concentration at 100 U/mL) into the cell culture media.Note: Two days after pp65 stimulation (on Day 3), cell clumps should be formed. The purpose of addition of IL-2 is to maintain the proliferation of activated pp65-specific CD4 T cell.
 CRITICAL: If little or no clump is shown, it suggests that PBMCs from the donor might not be sensitive to pp65 or may not be CMV-seropositive, which should be excluded for the rest of the experiment.
CRITICAL: If little or no clump is shown, it suggests that PBMCs from the donor might not be sensitive to pp65 or may not be CMV-seropositive, which should be excluded for the rest of the experiment. -
c.Replace half of cell culture media with fresh R10 medium supplemented with 100 U/mL IL-2 every 2–3 days until step 3(f). We recommend to do it on Day 5.
-
d.On Day 7 (6 days after PBMCa being prepared), recover one vial of frozen 10 million frozen PBMC (the same donor) in R10 medium pre-warmed to 37°C. PBMC culture B (PBMCb) is prepared in this step.Note: To ensure that the recovered PBMC will be healthy, it should be cultured in R10 medium overnight (18~24 h) before any next steps.
-
e.On Day 8 [the next day after step 3(d)], resuspend 5 million PBMCb with 1 mL R10 medium and supplement 5 μL of PepTivator CMV pp65 in a 12-well plate.
-
f.On Day 9 [the next day after step 3(e)], for the purpose of restimulation, add PBMCb into PBMCa culture at the cell number ratio of 1:10. Split each well of PBMCa/PBMCb mixture into two wells and add equivalent volume of R10 medium with 100 U/mL IL-2.
-
g.Replace half of cell culture media with fresh R10 medium supplemented with 100 U/mL IL-2 every 2-3 days until step 3(h). We recommend to do it on Day 11 or Day 12.
-
h.On the day before pp65-specific CD4 T cell isolation (Day 14), change PBMC culture media with fresh R10 medium supplemented with IL-2 at 20 U/mL to let cells rest.
-
a.
-
4.Total CD4 T cell Isolation from Untreated PBMC and pp65-treated PBMC.
-
a.On Day 11, recover frozen PBMC (the vial with 10 million PBMC for CD4 T cell isolation) in 5 mL R10 medium at the cell concentration at around 2 million cells per mL pre-warmed to 37°C. On the next day (Day 12), isolate CD4 T cell from recovered PBMCs using human total CD4 T cell isolation kit (negative selection), strictly according to manufacturer’s instructions (https://www.miltenyibiotec.com/DE-en/shop/comMiltenyiDatasheet/product?productId=130). Purified unstimulated CD4 T cells should be cultured in R10 medium supplemented with 20 U/mL IL-2 at cell density of 5 million per mL for around 3 days before co-culture.
 CRITICAL: The purity of isolated CD4 T cell should be confirmed by FACS analysis using antibodies against CD3 and CD4.Note: The duration of IL-2 treatment on total CD4 T cell isolated from untreated PBMC is optional from 2 to 4 days. The purpose of IL-2 treatment here is to maintain survival to avoid unpredicted effects of IL-2 on surface markers of CD4 T cell which may affect the interaction between γδ-T cell and CD4 T cell. The cells here will serve as untreated control.
CRITICAL: The purity of isolated CD4 T cell should be confirmed by FACS analysis using antibodies against CD3 and CD4.Note: The duration of IL-2 treatment on total CD4 T cell isolated from untreated PBMC is optional from 2 to 4 days. The purpose of IL-2 treatment here is to maintain survival to avoid unpredicted effects of IL-2 on surface markers of CD4 T cell which may affect the interaction between γδ-T cell and CD4 T cell. The cells here will serve as untreated control. -
b.On the day of co-culture experiment (Day 15), pp65-treated PBMC can be harvested for total CD4 T cell isolation using the isolation kit mentioned in the above procedure (strictly according to manufacturer’s instructions).
-
a.
CRITICAL: The purity of isolated CD4 T cell should be confirmed by FACS analysis using antibodies against CD3 and CD4.
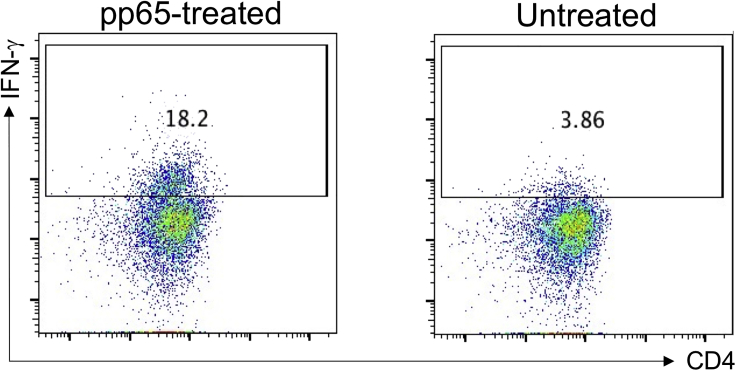
Note: According to the instruction of PepTivator CMV pp65, to purify IFNγ+CD4+ T cell by flow cytometer is recommended as a good way to purify pp65-specific CD4 T cell. However, the reason we could not do so is that we have to keep cells unlabeled from fluorescence-conjugated antibodies before confocal study. Technically, two-week culture with high dose of IL-2 may be good for activation and expansion of antigen-specific CD4 T cell. The percentage of IFNγ+ in CD4 T cell isolated from pp65-treated PBMC can be monitored by FACS analysis using antibodies against CD3, CD4, and intracellular stained IFN-γ. An example of such FACS data is shown in Figure 2.
Figure 2.
Examples for assessing IFN-γ-expression in CD4 T cell isolated from pp65-treated PBMC (left) and untreated PBMC (right) shown as dot plots by flow cytometry
Preparation of pp65-pulsed γδ-T cell
Timing: 4 days
-
5.γδ-T cell Isolation and Activation
-
a.On Day 10, recover frozen PBMC (same donor as the above procedure, the vial containing 100 million PBMC) in 10 mL pre-warmed R10 medium and culture them at the concentration of 10 million cells per mL R10 medium overnight (for 18~24 h).
-
b.On Day 11, isolate γδ-T cell from recovered PBMCs using human total γδ-T cell isolation kit (negative selection), strictly according to manufacturer’s instructions (https://www.miltenyibiotec.com/DE-en/shop/comMiltenyiDatasheet/product?productId=200). Purified γδ-T cell is left untreated or treated with IL-12 (4 ng/mL) and IL-15 (20 ng/mL) in the presence of 70 Gy γ-irradiated Raji B cell at the ratio of (T:B = 5:1) at a total cell concentration at 1~2 million cells per mL for 18–72 h (at least 18 h for activation) to express the surface proteins such as Δ42PD1 and HLA-DR.
-
a.
CRITICAL: The purity of isolated γδ-T cell (CD3+Vδ2+) and activation phenotype (expression of Δ42PD1 and HLA-DR) should be confirmed by FACS analysis.
Note: Similar to other studies that used irradiated Epstein-Barr virus (EBV)-transformed B cells or primary B cells (Brandes et al., 2005, Brandes et al., 2003) as feeder cells for CD40/CD40L interaction, these cells can induce an increased upregulation of co-stimulatory molecules such as HLA-DR, CD40, CD80 and CD86 on Vδ2+ cells during soluble antigen presentation process (Brandes et al., 2005, Himoudi et al., 2012). One type of EBV-transformed B cells, Daudi cells (Himoudi et al., 2012), could directly stimulate the Vδ2+ cells for additional functions. In contrast, Raji B cell line is not directly stimulatory to Vδ2+ cells (L'Faqihi et al., 1999, Fisch et al., 1990). In our experiment, we aim at investigating the antigen presenting role of Δ42PD1+Vδ2+ cells induced by IL-12 and IL-15, without additional stimulation by feeder cells (such as Daudi cells), so Raji B cells are the suitable EBV-transformed B cell line here. Importantly, in our experiments Raji cells were γ-irradiated prior to co-culture with antigen-pulsed γδ-T cells and therefore would not respond to stimulation or produce new cytokines to affect CD4 T cells in the co-culture.
-
6.pp65-pulsed antigen presenting γδ-T cells
-
a.Three days after IL-12/IL-15 treatment (on Day 14), pulse 5 million IL-12/IL-15 activated γδ-T cells with 5 μL of PepTivator CMV pp65 (30 μM) in 1 mL culture for 24–36 h before co-culture experiment (details of co-culture experiment are displayed in step-by-step method details).
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Δ42PD1 antibody (clone: CD34), mouse IgG2b (See note below) |
In-house | N/A |
| TCR V delta 2 Monoclonal Antibody (clone: 15D), mouse IgG1 | Thermo Fisher | Cat# TCR1732 RRID:AB_417090 |
| TLR4 Antibody (clone: HTA125), mouse IgG2a | Santa Cruz | Cat# sc-13593 RRID:AB_628366 |
| TLR4 Antibody (clone: 25), mouse IgG1 | Santa Cruz | Cat# sc-293072 RRID:AB_10611320 |
| Purified anti-human CD4 Antibody (clone: RPA-T4), mouse IgG1 | BioLegend | Cat# 300502 RRID:AB_314070 |
| HLA-DR Antibody (clone: L243), mouse IgG2a | Novus Biologicals | Cat# NB100-77855 RRID:AB_1085035 |
| Goat anti-Mouse IgG1 Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat# A-21121 RRID:AB_2535764 |
| Goat anti-Mouse IgG2a Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Invitrogen | Cat# A-21134 RRID:AB_2535773 |
| Goat anti-Mouse IgG2b Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Invitrogen | Cat# A-21242 RRIA:AB_2535811 |
| Biological samples | ||
| Buffy coat from healthy donors | Hong Kong Red Cross (See note below) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Human IL-12 | Miltenyi Biotec | Cat# 130-096-705 |
| Human IL-15 | Miltenyi Biotec | Cat# 130-095-765 |
| Recombinant Human IL-2 Protein | R&D Systems | Cat# 202-IL-050 |
| PepTivator CMV pp65, human | Miltenyi Biotec | Cat# 130-093-438 |
| Ficoll-Paque | GE Healthcare | Cat# 17-5442-03 |
| Fetal bovine serum | Gibco | Cat# 10270106 |
| PBS (10×), pH 7.4 | Gibco | Cat# 70011069 |
| Bovine serum albumin [Fraction V Bovine Albumin, pH 7] | Thermo Scientific | Cat# AAJ1085722 CAS: 9048-46-8 |
| DMSO | Sigma | Cat# D2650-100ML CAS: 67-68-5 |
| L-Glutamine (200 mM) | Gibco | Cat# 25030081 |
| Penicillin-streptomycin (10,000 U/mL) | Gibco | Cat# 15140122 CAS: 3810-74-0 (Streptomycin) |
| Paraformaldehyde | Sigma | Cat# 158127-100G CAS: 30525-89-4 |
| TritonTM X 100 | Sigma | Cat# X100-500ML CAS: 9002-03-1 |
| Trypan blue solution, 0.4% | Life Technologies | Cat# 15250061 CAS: 72-57-1 |
| UltraPureTM DNase/RNase-free Distilled Water | Invitrogen | Cat# 10977015 |
| RPMI 1640 Medium, no glutamine | Gibco | Cat# 21870092 |
| Critical commercial assays | ||
| CD4+ T Cell Isolation Kit, human | Miltenyi Biotec | Cat# 130-096-533 |
| TCRγ/δ+ T Cell Isolation Kit, human | Miltenyi Biotec | Cat# 130-092-892 |
| Experimental models: cell lines | ||
| Raji B cell | Kindly provided by Prof Honglin Chen (HKU). | N/A |
| Software and algorithms | ||
| ImageJ | (Schneider et al., 2012) | https://imagej.nih.gov/ij/ |
| JACoP plugin for ImageJ | (Bolte and Cordelières, 2006) | http://www.blackwell-synergy.com/doi/pdf/10.1111/j.1365-2818.2006.01706.x |
| ZEN Blue | Zeiss | v2.3 |
| Imaris | Oxford Instruments | v7.5.2 |
| Other | ||
| NuncTM Lab-TekTM II 8-well chambered coverglass | Thermo Fischer | Cat# 155409 |
| SlowFadeTM Gold antifade mounting medium with DAPI | Invitrogen | Cat# S36938 |
| Carl Zeiss LSM 700 | Zeiss | N/A |
| Carl Zeiss LSM 800 | Zeiss | N/A |
| Leica TCS SP5 II | Leica | N/A |
| Centrifuge 5810R | Eppendorf | N/A |
| BD FACSAriaTM III Cell Sorter | BD Biosciences | N/A |
| MDS Nordion Gammacell 3000 Elan | Best® Theratronics | N/A |
Note: Anti-Δ42PD1 antibody (clone: CD34, Mouse IgG2b) is available upon request.
Note: Buffy coat from healthy donors can also be obtained by other sources appropriate for the researchers’ country, districts or areas with ethics approval.
Note: The antibodies for FACS analysis in those critical points (assessing purity or activation) are optional and depend on the user, so they are not included here.
Optional: Other imaging processing software may be used for data analysis.
Materials and equipment
| R10 medium | Final concentration | Volume (mL) |
|---|---|---|
| RPMI 1640 medium | N/A | 440 |
| FBS | 10% | 50 |
| L-glutamine (200 mM) | 2 mM | 5 |
| Penicillin and streptomycin (10000 U/mL and 10 mg/mL, respectively) | 100 U/mL and 100 μg/mL, respectively | 5 |
| Total | 500 |
Once prepared, it should be stored at 4°C and used within 4 weeks.
| Freezing medium | Final concentration | Volume (mL) |
|---|---|---|
| FBS | 90% | 9 |
| DMSO | 10% | 1 |
| Total | 10 |
It should be freshly prepared before use.
| Stock of PepTivator CMV pp65 | Final concentration | Amount |
|---|---|---|
| PepTivator CMV pp65 | N/A | 6 nmol |
| Sterile UltraPureTM DNase/RNase-free Distilled Water | N/A | 0.2 mL |
| Total | 0.2 mL |
Once prepared, it should be aliquoted into 10 μL per tube and frozen at −80°C for one year. Repeated freeze-thaw cycles should be avoided.
| Perm/Block buffer | Final concentration | Amount |
|---|---|---|
| PBS | N/A | 9.45 mL |
| FBS | 5% | 0.5 mL |
| Bovine Serum Albumin (BSA) | 3% | 0.3 gram |
| Triton-X 100 | 0.5% | 0.05 mL |
| Total | 10 mL |
Once prepared, it should be stored at 4°C and used within 4 weeks.
| Staining Buffer | Final concentration | Amount |
|---|---|---|
| PBS | N/A | 49.95 mL |
| Bovine Serum Albumin (BSA) | 3% | 1.5 gram |
| Triton-X 100 | 0.1% | 0.05 mL |
| Total | 50 mL |
Once prepared, it should be stored at 4°C and used within 4 weeks.
Step-by-step method details
Co-culture of pp65-specific CD4 T cell and pp65-pulsed γδ T cell
Timing: 2 h
In this step, CD4 T cell isolated from pp65-stimulated PBMCs is co-cultured with pp65-pulsed γδ-T cell before proceeding to immunofluorescent assay. It is Day 1 for co-culture experiment, as well as Day 15 for the whole experimental procedure.
-
1.Co-culture
-
a.NuncTM Lab-TekTM II 8-well chambered coverglass is used for the co-culture experiment.Note: The NuncTM Lab-TekTM II 8-well chambered coverglass is made up of cell-culture treated glass bottom chambers, which allowed our cells to adhere to the bottom of chamber after overnight culture. Since we sought to analyze the cell-cell interaction, the situation that cells tightly adhere to the chamber bottom before cell-cell interaction should be avoided. Therefore, we do not recommend using coating reagents facilitating cell adherence, for instance, fibronectin and poly-L-lysine.
-
b.Co-culture CD4 T cell isolated from pp65-stimulated PBMCs or unstimulated PBMCs with pp65-pulsed activated or unactivated γδ-T cell, at a ratio of γδ-T:CD4 T = 1:1 (a total of 0.2 million cells in 200 μL R10 medium per well) overnight (around 18 h). Thus, eight groups are involved:
Group Cell culture Aim 1 Unactivated γδ-T cell Unactivated cell only control 2 pp65-pulsed activated γδ-T cell Antigen presenting cell only control 3 Unstimulated CD4 T cell Effector cell negative control 4 pp65-stimulated CD4 T cell Effector cell only control 5 Unactivated γδ-T cell plus unstimulated CD4 T cell Negative control 6 Unactivated γδ-T cell plus pp65-stimulated CD4 T cell Internal control 7 pp65-pulsed activated γδ-T cell plus unstimulated CD4 T cell Internal control 8 pp65-pulsed activated γδ-T cell plus pp65-stimulated CD4 T cell Test
-
a.
Note: The recommended culture volume in the NuncTM Lab-TekTM II 8-well chambered coverglass is 0.2–0.5 mL, which is capable of culturing 0.2 million to 0.5 million cells at the density of ~1 million cells per mL. To assist the cells to settle at the bottom of the well, culture volume lower than 0.4 mL would be better. Thus, the maximal cell number per well should be 0.4 million in 0.4 mL culture.
Note: In total, 24 wells (8 groups × 3 donors) are needed. Cells are cultured in a 37°C/5% CO2 incubator overnight (around 18 h).
CRITICAL: For the sake of seeing specific conjugates, co-culture of “unstimulated CD4 T cell and pp65-pulsed γδ-T cell” and co-culture of “pp65-specific CD4 T cell and unactivated γδ-T cell” should be included as control.
CRITICAL: Spare one or two more wells serving for the secondary antibody control in the immunofluorescence assay.
Immunofluorescence assay
Timing: 2 days
γδ-T cells and CD4 T cells are allowed to interact after 18-h co-culture, cells are then subjected to immunofluorescence assay (IFA) and evaluated with confocal microscopy (Day 15 overnight to Day 16, for the whole experimental procedure).
-
2.
Following overnight (~18 h) co-culture, cells are now ready for antibody staining.
-
3.
On Day 1 for immunofluorescence assay, using a 200 μL pipette, aspirate the media slowly and carefully from top-to-bottom at the bottom-right corner of each well.
CRITICAL: After overnight culture, these suspension cells should be settled down on the bottom of each well though they are not attached tightly, as mentioned above. In order to avoid lifting of the cells and to minimize cell loss, the procedure of moving the chamber, aspirating or adding any buffer should be done slowly and carefully along the sides of the wells. For consistency, the down-right corner of each well should be fixed for removing the media or PBS, while the upper-left corner of each well is used to add buffer, in a slow and gentle manner. This applies for the rest of the following steps.
-
4.
Add 200 μL PBS slowly and gently to each well at the upper-left corner.
-
5.
Aspirate the PBS slowly and carefully from top-to-bottom at the bottom-right corner of each well.
-
6.
Add 200 μL of 4% paraformaldehyde (PFA, dissolved in PBS, pH 7.4) to each well and keep it at room temperature (22°C~28°C) in the dark for 20 min to fix the cells.
-
7.
Aspirate the 4% PFA and wash with 200 μL PBS twice.
-
8.
Add 200 μL of Perm/Block buffer and keep the chamber at 4°C for 20 min.
-
9.
Wash with 200 μL Staining Buffer twice.
-
10.Incubate with primary antibodies in 200 μL Staining Buffer at 4°C overnight (~18 h). For example, in our experiment, we used the following primary antibodies:
-
a.Panel 1: anti-Δ42PD1 (CH34, Mouse IgG2b, 1:100), anti-Vδ2 (15D, Mouse IgG1, 1:20), and anti-TLR4 (HTA125, Mouse IgG2a, 1:20).
-
b.Panel 2: anti-Δ42PD1 (CH34, Mouse IgG2b, 1:100), anti-HLA-DR (L243, Mouse IgG2a, 1:250), anti-TLR4 (25, Mouse IgG1, 1:50)
-
a.
CRITICAL: For non-specific signal control, do not use primary antibodies and only add appropriate secondary antibodies.
-
11.
On the next day (Day 2 for immunofluorescence assay), wash with 200 μL Staining Buffer twice.
-
12.Incubate with secondary antibodies in 200 μL Staining Buffer at 4°C for 30 min. In our experiment, we used the following secondary antibodies for the above both panels:
-
a.Goat anti-mouse IgG1 AF488 (1:1000), goat anti-mouse IgG2a AF568 (1:1000) and goat anti-mouse IgG2b 647 (1:1000).
-
a.
-
13.
Wash with 200 μL Staining Buffer twice.
-
14.
Mount the cells by adding 200 μL SlowFadeTM Gold antifade reagent with DAPI, and incubate at room temperature (22°C~28°C) in the dark for 30 min.
CRITICAL: Unlike coverslip/microscope slides, the chamber is not needed to be turned upside-down for imaging, as it is imaged from the bottom of the chamber. No coverslip is needed here as well. However, sufficient volume of the mounting medium (200 μL) is important to fully cover the cells and to prevent evaporation.
Optional: If mounting with SlowFadeTM reagent without DAPI, cells can be stained with DAPI in 200 μL PBS at room temperature (22°C~28°C in the dark; incubation time depends on the instruction of different commercial products) and then washed with 200 μL PBS before applying mounting medium.
-
15.
Keep the chamber with the lid at 4°C before imaging under confocal microscope.
Note: It is better to perform imaging as soon as possible to get clearer signals without bleaching, within one month after IFA is done.
Pause point: The chamber can be kept in dark place at 4°C, placed in a sealed container or glad wrapped, for the purpose of protecting from light and from drying, if performing confocal microscopy at a later time.
3D imaging using confocal microscope
Timing: Depends on availability of the confocal microscope at your institution and familiarity of operation, it may take multiple sessions of several hours to obtain sufficient data.
In this step, IFA samples can be imaged using confocal microscopes such as LSM700, LSM800 and Leica SP 5 II. We mainly focused on the colocalization of the signal of Δ42PD1 on Vδ2+ cell and the signal of TLR4 on CD4 T cell, as well as the presence of HLA-DR on Vδ2+ cell. The protein expression on the cell only groups (Group 1, 2, 3 and 4) serve as controls.
-
16.
The pixel resolution for the different objective lens used will depend on the specific parameters and the confocal microscope being used.
Note: The details about the objectives we used are shown below:
| Magnification | Numerical aperture | Free working distance | Coverglass thickness | Immersion |
|---|---|---|---|---|
| 10× | 0.3 | 2 mm | 0.17 mm | Air |
| 40× | 1.4 | 0.13 mm | 0.17 mm | Oil |
| 63× | 1.4 | 0.19 mm | 0.17 mm | Oil |
To acquire images with high quality, we recommend using the optimal setting of ZEN or relevant acquisition software during image capturing.
-
17.
Scan at least 3 images for each group using 10× lens first.
Note: Scanning images using 10× lens first is helpful to have a general view of the distribution of signals and to identify the cells of interest, which is a preliminary step before locating the scanning position using 40× or 63× lens.
-
18.
For experimental group 5 (unactivated γδ-T cell plus unstimulated CD4 T cell), group 6 (unactivated γδ-T cell plus pp65-stimulated CD4 T cell), group 7 (pp65-pulsed activated γδ-T cell plus unstimulated CD4 T cell) and group 8 (pp65-pulsed activated γδ-T cell plus pp65-stimulated CD4 T cell), scan at least five images per chamber using 40× or 63× oil lens. Scan at least five Z-stack images for group 8.
Note: A resolution of at least 1024∗1024 is recommended but depends on the objective lens used. User should check the hardware parameters.
Note: Sufficient number of images is helpful in a robust data quantification and analysis afterwards.
Note: T-PMT should be turned off and the bright field should be excluded during Z-stack imaging. Setting the Z-position: decrease focus height until you cannot see the cell structure and set this as the first scanning position of the Z-position and increase focus height until you cannot see the cell structure and set as its last scanning position. Click “1 AU” hotkey to adjust pinhole size. Use the recommended parameters on frame size and resolution (click “Optimal” option) by the software. Use the scan speed recommended by the software.
Optional: The condition we set is that Z-stack interval is 0.506 μm with 25 images acquired. The interval should be smaller than one airy unit. Smaller interval leads to slower scanning but higher resolution along the Z-axis. It should be adjusted to the resolution limit of the objective lens used. The number of acquired images depends on both the length of Z-position and the interval.
Expected outcomes
At the end of this protocol the protein-protein co-localization on the interface between antigen-presenting γδ-T and CD4 T cells can be visualized by 3D imaging. Data generated using this protocol has recently been published (Mo et al., 2020), as shown in Figure 3.
Figure 3.
Example of data from using this protocol describing Δ42PD1-TLR4 interaction between Vδ2 and CD4+ T cells
(Figure reprinted with permission from Mo et al., 2020). pp65-specific CD4+ T cells were used to co-culture with autologous IL-12/IL-15 stimulated and pp65-pulsed γδ-T cells or untreated (Unt) γδ-T cells for around 18 h, before fixed and immunostained for Δ42PD1 (white), TLR4 (green), HLA-DR (red) and DAPI (blue). Co-culture of pp65-specific CD4+ T cells with (A) unstimulated γδ-T cells or (B) with IL-12/IL-15 stimulated and pp65-pulsed γδ-T cells. Pink and blue arrows point to Δ42PD1+HLA-DR+ and TLR4+ cells, respectively. Pixel size = 120.2 nm.
(C) Representative images showing colocalization of Δ42PD1 and HLA-DR expression signals on Unt (n = 65) and pp65-pulsed γδ-T cells (n=96) were analyzed, for the overlap and Manders’ coefficient values shown in the graph.
(D) Representative images showing the interaction between pp65-specific CD4+ T cell and pp65-pulsed γδ-T cells analyzed by the signal values for Δ42PD1, MHC-II and TLR4 along the yellow line transecting from bottom left to top right equivalent to left to right in the histogram. Values are indicated by the histogram of n=32 cell-cell pairs at ten pixels before and after interface between the cells set at x = 0.
(E) Orthogonal views over XY, XZ and YZ axes of a Z-stack of 78 images at 0.29 μm thickness each. Image at stack 72 is shown for the interaction between pp65-specific CD4+ T cell and pp65-pulsed γδ-T cells. Pixel size = 80.2 nm.
(F) Imaris analysis on a maximum projection of a Z-stack of 25 images each with a thickness of 0.506 μm (upper) to generate the 3D model (bottom). Pixel size = 112 nm. The rightmost image is a flipped version of the middle image to indicate interactions at two different angles. White bar represents a scale of 10 μm. Representative images from three independent experiments are shown. Data in the graphs represent mean±SEM.
Generally, the technique of image acquisition through Z-stack provides us important data of the cell-cell interface. Further data analysis through ImageJ provides valuable quantitative results on the co-localization coefficients values and signal profile across interface. Imaris analysis also constructs a 3D model for the interface of interest.
Quantification and statistical analysis
Determining co-localization coefficients
To determine whether two proteins are expressed at the same location, co-localization coefficients are calculated using the JACoP plugin in ImageJ.
-
1.
Choose an image taken using high magnification such as 630× and open using ImageJ.
-
2.
Identify the cells of that appears to be interacting and crop the image to the particular area. Save this image.
-
3.
Split the image into individual channels, such that one image only displays one parameter. Save the individual images.
Note: In our study, we aimed to determine the co-expression of Δ42PD1 and HLA-DR. Thus, the confocal image was split into individual images displaying Δ42PD1, HLA-DR, DAPI, and brightfield channels.
-
4.
Choose the two channels for analysis in JACoP, e.g., Δ42PD1 for Image A and HLA-DR for Image B. Save these images.
-
5.
In the analysis to perform, choose the ones appropriate to you. Our analysis used Overlap and Manders’ coefficients.
Note: Overlap coefficient is based on Pearson’s coefficient that is dependent on linear signals while Manders’ coefficient take into account of ratios of two signals on top of the compartment and vice versa. Having both coefficients will provide a more comprehensive co-localization analysis.
-
6.
In the Threshold tab, adjust the slider for channel A and B to the level of signals that can clearly differentiate between background without signal saturation. Press Analyze.
-
7.
In the data log window in ImageJ, record the Overlap coefficient and Manders’ coefficients (using threshold value, for both A overlapping B and B overlapping A).
-
8.
Using steps 3–7, analyze at least 50 cells per experimental group across multiple images.
-
9.
Comparisons can be made in a table or as a graph where statistics can be calculated.
Determining the co-localization between two surface proteins across a cell-cell interface
-
10.
Choose an image taken using high magnification such as 630× and open using ImageJ (Figure 4A).
-
11.
Identify the cells that are in close proximity and expressing the protein interaction of interest. Crop the image to the particular cell. Save this image (Figure 4A).
-
12.
Split the image into individual channels, such that one image only displays one parameter. Save the individual images (Figure 4B).
-
13.
Using the drawing tool, draw a line across the membrane of two cells beginning and ending in the respective cytoplasmic regions. Make sure the middle of the line (usually displayed as a square dot) is at the middle of cell-cell interface. Draw for one image first (Figure 4B).
-
14.
Go to Analyze\Tools\ROI Manager then click on “Add”, this will designate a number for the line. Click on the images showing the other individual channels and click the newly added line number in ROI Manager to apply the same line to the other images (Figure 4C).
-
15.
Click on the channels of interest with line drawn, then go to Analyze\Plot Profile to show the signals for the individual channels (Figure 4D).
-
16.
In one of the plots, click the “Data” button and choose “Add from Plot” for the other channels with appropriate colors (typed the color you desire, e.g., “green”, “red”) (Figure 4E).
Note: In our study, we wanted to examine the interaction between Δ42PD1/HLA-DR on pp65-pulsed activated γδ-T cell and TLR4 on pp65-stimulated CD4 T cell. Thus, the plot included signals for Δ42PD1, HLA-DR and TLR4 across the line drawn between the γδ-T cell and CD4 T cell.
-
17.
Repeat these steps to analyze additional cell-cell pairs.
-
18.
Data from the plots can be exported for the different signals from one cell-cell pair by clicking “Data” then “Copy all data” and paste into Excel worksheet or other softwares (Figure 4F).
Note: The data will depict the signal value of each pixel across the line that is drawn across the cell-cell interface. However, the length of the line can vary, and it is why in step 13, the middle square dot of the line should be placed in the middle of the interface. Then only the signal values of 10 pixels forward and backward from the middle square dot should be used across the different cell-cell pairs and different line lengths. Taking the example in Figure 1, the line is 0 to 52 pixels in length, so the middle square dot sits at pixel 26, and the data from 16 to 36 should be included for analysis.
Note: At least 30 cell-cell pairs are recommended for drawing the line, generating plot profiles, and for data analysis.
Figure 4.
Procedures to analyze pixel profiles across a cell-cell interface
(A) Example of choosing a cell-cell interface area of interest. (B) Drawing a line of interest across the interface where yellow arrow points to the middle of the line. (C) Adding the line of interest in the ROI Manager. (D) Profile plots of the different protein expression across the line. (E) Overlaying the profile plots. (F) Extracting the pixel data from the profile plots.
Examining the cell-cell interface in Z stack and orthogonal views
The purpose of viewing a Z-stack in an orthogonal view is to identify cell-cell and protein-protein co-localization at different angles. There could be interactions that occur in an up-down orientation that would be missed if only the XY-plane is considered. Further, any interactions found on one plane should be confirmed in another plane and the orthogonal views will enable this.
-
19.
Choose a Z-stack taken using high magnification such as 630× and open using ImageJ.
-
20.
Identify the cells that are in close proximity and expressing the protein interaction of interest. Crop the Z-stack to display the desired cell-cell interaction. Save this Z-stack.
-
21.
Using ImageJ, click on Image\Stacks\Orthogonal views. The YZ and XZ images will be displayed and can see to the cell-cell and protein-protein interaction at different angles. Save the displayed images.
-
22.
Move the crosshair to highlight the interaction at the different stack, which could be used for analysis as above.
Generating a 3D model for cell-cell and protein-protein interaction using Imaris
Imaris is a useful image analysis software on generating a visual 3D model for protein-protein interaction among cells.
-
23.
Import Z-stack files generated from LSM700 into Imaris software, as shown in Figure 5A.
-
24.
Select “Surpass” mode → check the channel that needs to be processed and uncheck others (Figure 5B) → tick “Light Source 1” and “Volume” → click “Add new surface” (Figure 5C) and define color for each channel [color can be set by experimenter in the “color” column in Figure 5D, and it is not limited to the default one.]→ set the optimal algorithm (using default parameters), source channel (Figure 5E), threshold (Figure 5F) and number of voxels (Figure 5G) → process and obtain the virtual image of one channel (Figure5H). → Process other channels using similar procedures.
Note: Although DAPI represents the cell nuclear, we still choose the “surface” mode for it, which may show the surface of nuclear.
Note: To make those surfaces better visualize, the optimal threshold and number of voxels should be adjusted. The adjustment is supposed to exclude noise signals but cover all the clear signals. Too high threshold leads to large noise (Figure 6A), while too low one leads to failure of covering signals (Figure 6B). Figure 6C shows the suitable threshold which covers most of bright signals. As for number of voxels, too high number of voxels (Figure 6D) fails in exclude noise signals, but too low number of voxels (Figure 6E) is not able to fully cover those clear signals. Figure 6F shows the optimal number of voxels that excludes noise and that makes a suitable coverage of bright signal.
Figure 5.
Procedures for how to process virtual image from one channel
(A) Opening the Z-stack file of interest in the working space of Imaris software. Different channels (B) and functional options (C) in the working space are shown in enlarged display. (D) General settings on parameters and color before proceeding to algorithm creation. Channel (E), threshold (F) and number of voxels (G) can be selected during algorithm creation. (H) Algorithm can be processed in order to obtain the virtual image of one specific channel.
Figure 6.
Comparison on results of high, low or optimal level of threshold and number of voxels
The results of high threshold (A), low threshold (B) and optimal threshold (C) are displayed. The results of high number of voxels (D), low number of voxels (E) and optimal number of voxels (F) are displayed.
-
25.
After virtual images of all channels are processed, check the processed surface programs and uncheck the raw signals, obtain one 3D model for the interface, as shown in Figure 7.
Figure 7.
Virtual 3D model for the interface by Imaris
Limitations
Although we showed an applicable protocol on displaying the protein co-localization on the cell-cell interface, an ideal model for cell-cell interaction is to do live cell imaging on the cell-cell co-culture system. However, since the biomarker antibody also serves as blocking antibody and the two clones of in-house antibodies have the same problem, it is difficult to image cell-cell interface. To counteract with this problem, more clones of antibody for the specific biomarker, not limited to the antibodies we used for our research, can be considered to develop for different experimental purpose separately. It should be addressed that the antibodies under investigation would not block the interaction between the proteins of interest and would be able to access between cells of interest, which is a long and difficult way to develop.
In terms of problems involving signal strength in the confocal study, conjugated primary antibodies may provide relatively weak signal. Labeling of the proteins using unconjugated primary antibody followed by conjugated secondary antibody allows amplification of signals. It will increase signal-to-noise ratio, while using conjugated primary antibodies may have reduced signal-to-noise ratio. Under this limitation, we chose the primary/secondary antibodies approach to ensure signal strength. We should also be careful about determining the optimal concentration of primary and secondary antibodies in the immunofluorescence assay.
Troubleshooting
Problem 1
Cell loss during IFA procedure (steps 1, 3, 4, and 17 in step-by-step method details).
Potential solutions
Solution 1. Total cell number seeded in each assay well should not be less than 0.2 million cells or larger than 0.4 million cells per well, given the appropriate culture volume of 0.2–0.4 mL. If experimenter can not find enough cells for imaging or experimenter suffer from cell loss during IFA procedure, total cell number can reach around 0.4 million. However, too high density of cell number may not be good for regular practice of cell culture for PBMC (appropriate density at around 1 million cells per mL).
Solution 2. Overnight culture helps these suspension cells settle down on the bottom.
Solution 3. In order to avoid blowing cells up from the bottom of chamber, it is a good performance to fix one corner for adding reagents and the opposite one for aspirating out reagents.
Solution 4. The procedure of moving the chamber and aspirating or adding any buffer should be as slowly as possible.
Problem 2
No cell clusters are observed in the co-culture of pp65-specific CD4 T cell and pp65-pulsed γδ T cell (step 3 and 5 in before you begin and step 17 in step-by-step method details).
Potential solutions
Solution 1. The main problem is that the cultured CD4 T cells stimulated with pp65 was not successful likely due to the donor cells being unresponsive due to CMV-seronegative status. As mentioned above, the PBMCs that showed clumps within 2 days should be used for subsequent experimental procedures. If not, cells from another donor should be sought.
Solution 2. Failure of activation on γδ T cell may also lead to little cell cluster in the co-culture system. The control group of co-culture of unactivated γδ T cell and unstimulated CD4 T cell should show little cell cluster.
Problem 3
A lot of cell debris shown under the microscope in brightfield (steps 4–15 and 17 in step-by-step method details).
Potential solutions
Solution 1. Cells might be left dried during experimental procedures of removing the different buffers. To avoid this, after removing the buffer such as PBS, PFA, Perm/Block Buffer and Staining Buffer, the subsequent buffer or solutions should be added into the wells in a timely manner and thoroughly. In our experience, this should be done after removing 3–5 wells.
Solution 2. To avoid cells getting dried in the well-prepared IFA chamber before confocal study, ensure the mounting reagent covers the cells all the time.
Problem 4
Signals are not sufficiently bright for capturing during confocal study (steps 10, 12, and 17 in step-by-step method details).
Potential solutions
Solution 1. Firstly, a preliminary experiment on testing the incubation concentration of primary antibodies is recommended. This should be based on the recommended range from the manufacturers’ instructions to find the optimal concentration for bright signals.
Solution 2. If the tested primary antibody does not work well at the highest concentration, try to increase the concentration of the secondary antibody until the signal is shown clearly.
Problem 5
Noise of images is too high (steps 8, 12, and 17 in step-by-step method details).
Potential solutions
Solution 1. Firstly, try to adjust the settings of confocal microscope before capturing, for instance, voltage of channels, time of exposure and so on. For instance, increasing laser power is advisable compared to Digital Gain.
Solution 2. If the adjustment on confocal microscope is not helpful, make sure that enough concentration of FBS and Bovine Serum Albumin (BSA) is added into Perm/Block Buffer and the Perm/Block procedure is performed at room temperature (22°C~28°C) in the dark for 20 min.
Solution 3. Reduce the concentration of secondary antibodies to avoid non-specific binding.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Allen Ka Loon Cheung (akcheung@hkbu.edu.hk).
Materials availability
No new materials were generated in this study. Note that the anti-Δ42PD1 monoclonal antibodies used were described by another publication (Cheng et al., 2015), which are available upon request.
Data and code availability
No dataset or code was generated or analyzed during this study.
Acknowledgments
We thank Jingjing Li and Dongyan Zhou for generating the anti-Δ42PD1 monoclonal antibodies that were used in this study. This work was supported by Hong Kong Research Grant Council TRS-T11-706/18-N, GRF17115818, GRF17104919, GRF17103514, and HKU5/CRF/13G, University Development Fund of the University of Hong Kong to AIDS Institute and the National Science and Technology Major Project 2018ZX10731-101-002-001 (to Z.C.), and Health and Medical Research Fund (HMRF) (15140732 and 18170032), Interdisciplinary Research Matching Scheme (RC-IRCs-1718-03), Faculty Research Grant (FRG2/17-18/066), Faculty Start-up Fund (SCI-17-18-01), Tier 2 Start-up Grant (RC-SGT2/18-19/SCI/007) (to A.K.L.C.).
Author contributions
Conceptualization, A.K.L.C. and Z.W.; methodology, A.K.L.C., Y.M., and Z.W.; investigation, A.K.L.C. and Y.M.; writing – original draft, A.K.L.C. and Y.M.; writing – review & editing, A.K.L.C., Y.M., and Z.C.; funding acquisition, A.K.L.C. and Z.C.; supervision, A.K.L.C. and Z.W.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Allen Ka Loon Cheung, Email: akcheung@hkbu.edu.hk.
Zhiwei Chen, Email: zchenai@hku.hk.
References
- Bolte S., Cordelières F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Böyum A. Isolation of mononuclear cells and granulocytes from human blood. Isolation of monuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand. J. Clin. Lab. Invest. Suppl. 1968;97:77–89. [PubMed] [Google Scholar]
- Brandes M., Willimann K., Lang A.B., Nam K.H., Jin C., Brenner M.B., Morita C.T., Moser B. Flexible migration program regulates gamma delta T-cell involvement in humoral immunity. Blood. 2003;102:3693–3701. doi: 10.1182/blood-2003-04-1016. [DOI] [PubMed] [Google Scholar]
- Brandes M., Willimann K., Moser B. Professional antigen-presentation function by human gammadelta T Cells. Science. 2005;309:264–268. doi: 10.1126/science.1110267. [DOI] [PubMed] [Google Scholar]
- Cheng L., Tang X., Liu L., Peng J., Nishiura K., Cheung A.K., Guo J., Wu X., Tang H.Y., An M. Monoclonal antibodies specific to human Δ42PD1: A novel immunoregulator potentially involved in HIV-1 and tumor pathogenesis. MAbs. 2015;7:620–629. doi: 10.1080/19420862.2015.1016695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch P., Malkovsky M., Kovats S., Sturm E., Braakman E., Klein B.S., Voss S.D., Morrissey L.W., Demars R., Welch W.J. Recognition by human V gamma 9/V delta 2 T cells of a GroEL homolog on Daudi Burkitt's lymphoma cells. Science. 1990;250:1269–1273. doi: 10.1126/science.1978758. [DOI] [PubMed] [Google Scholar]
- Himoudi N., Morgenstern D.A., Yan M., Vernay B., Saraiva L., Wu Y., Cohen C.J., Gustafsson K., Anderson J. Human γδ T lymphocytes are licensed for professional antigen presentation by interaction with opsonized target cells. J. Immunol. 2012;188:1708–1716. doi: 10.4049/jimmunol.1102654. [DOI] [PubMed] [Google Scholar]
- L'Faqihi F.E., Guiraud M., Dastugue N., Brousset P., Le Bouteiller P., Halary F., Bonneville M., Fournié J.J., Champagne E. Acquisition of a stimulatory activity for Vgamma9/Vdelta2 T cells by a Burkitt's lymphoma cell line without loss of HLA class I expression. Hum. Immunol. 1999;60:928–938. doi: 10.1016/s0198-8859(99)00076-2. [DOI] [PubMed] [Google Scholar]
- Loeth N., Assing K., Madsen H.O., Vindeløv L., Buus S., Stryhn A. Humoral and cellular CMV responses in healthy donors; identification of a frequent population of CMV-specific, CD4+ T cells in seronegative donors. PLoS One. 2012;7:e31420. doi: 10.1371/journal.pone.0031420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y., Cheung A.K.L., Liu Y., Liu L., Chen Z., Mo Y., Cheung A.K.L., Liu Y., Liu L., Chen Z. Δ42PD1-TLR4 augments γδ-T cell activation of the transitional memory subset of CD4(+) T cells. iScience. 2020;23:101620. doi: 10.1016/j.isci.2020.101620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No dataset or code was generated or analyzed during this study.