Abstract
Background
Type 2 diabetes (T2D) is a common metabolic disease. Variants in human IGF2 mRNA binding protein 2 (IMP2/IGF2BP2) are associated with increased risk of T2D. IMP2 contributes to T2D susceptibility primarily through effects on insulin secretion. However, the underlying mechanism is not known.
Methods
To understand the role of IMP2 in insulin secretion and T2D pathophysiology, we generated Imp2 pancreatic β-cell specific knockout mice (βIMP2KO) by recombining the Imp2flox allele with Cre recombinase driven by the rat insulin 2 promoter. We further characterized metabolic phenotypes of βIMP2KO mice and assessed their β-cell functions.
Results
The deletion of IMP2 in pancreatic β-cells leads to reduced compensatory β-cell proliferation and function. Mechanically, IMP2 directly binds to Pdx1 mRNA and stimulates its translation in an m6A dependent manner. Moreover, IMP2 orchestrates IGF2-AKT-GSK3β-PDX1 signaling to stable PDX1 polypeptides. In human EndoC-βH1 cells, the over-expression of IMP2 is capable to enhance cell proliferation, PDX1 protein level and insulin secretion.
Conclusion
Our work therefore reveals IMP2 as a critical regulator of pancreatic β-cell proliferation and function; highlights the importance of posttranscriptional gene expression in T2D pathology.
Keywords: IMP2/IGF2BP2, T2D, Insulin secretion, m6A, Post-transcriptional gene expression regulation
Highlights
-
•
Specific deletion of IMP2 in mouse pancreatic β-cells leads to reduced compensatory β-cell proliferation and function.
-
•
IMP2 directly binds to Pdx1 mRNA and stimulates its translation in a m6A dependent manner.
-
•
IMP2 orchestrates IGF2-AKT-GSK3β-PDX1 axis to promote PDX1 polypeptide stability.
-
•
In human EndoC-βH1 cells, the expression of IMP2 enhances cell proliferation and insulin secretion.
1. Introduction
Human type 2 diabetes (T2D) is a common metabolic disease attributable to both environment and genetics [1]. In 2007, three independent genome-wide association studies (GWAS), WTCCC, Fusion, and DGI scans, reported a strong association between IMP2 genetic variances and T2D [[2], [3], [4]]. These GWAS each identified a cluster of single-nucleotide polymorphisms (SNPs) in the second intron of IMP2 gene as the basis for the T2D association [[2], [3], [4]]. To date, GWAS have identified ~100 SNPs in the second intron of the human IMP2 gene associated with increased risk for T2D [[5], [6], [7], [8], [9], [10]]. IMP2 contributes to T2D susceptibility primarily through effects on insulin secretion [[11], [12], [13]]. Any strong evidence for IMP2 association with insulin sensitivity, a primary component of T2D susceptibility, is strikingly absent [[11], [12], [13]]. In line with these discoveries, IMP2 mRNA and protein are readily detected in human islets and pancreatic β cells [14] (microarray data from the Diabetes Genome Anatomy Project), and IMP2 transcript levels are significantly altered in the islets of T2D patients [15]. Notably, in 2019, two independent studies provided evidence that the association of the IMP2 intronic SNPs with T2D is affected through modification of the expression of IMP2 itself instead of neighboring genes [16,17]. These human findings have further confirmed that IMP2 is a T2D causal gene.
IMP2 belongs to a small IMP protein family (IMP1-3) and is composed of six canonical RNA-binding domains: two RNA recognition motif (RRM) domains and four K homology (KH) domains [18]. IMPs have been recognized as “onco-fetal” proteins as they are highly expressed in development and carcinogenesis [[19], [20], [21], [22], [23], [24]]. Distinct from IMP1 and IMP3, which are expressed at very low levels in most adult tissues, IMP2 expression is broadly maintained postnatally [25,26]. IMPs play key roles in post-transcriptional regulation of RNAs, in a cell typespecific manner, through ribonucleoprotein complex (RNP) [27]. They participate in the life cycle control of a wide variety of RNAs, including long noncoding RNAs and mRNAs [[28], [29], [30], [31], [32], [33], [34]]. The conserved IMP binding sites are enriched in “GGAC”, which is highly overlapped with N6-methyladenosine (m6A) motif [[35], [36], [37]]. IMPs preferentially recognize m6A-modified mRNAs and regulate their stability and translation [36]. Recent studies have suggested that the m6 A modification plays a critical role in the regulation of T2D [38,39]. For example, the increased expression of m6A methylation upregulates the IGF1 –AKT-PDX1 pathway in human β-cells, which ultimately inhibits cell-cycle arrest and protects insulin secretion [40]. Therefore, m6 A modulators are potential therapeutic targets for maintaining glucose metabolism and insulin functions in T2D.
Nevertheless, nothing was known about the metabolic functions of IMP2 when it was first nominated as a human T2D-associated gene. To understand the role of IMP2 in T2D pathophysiology, we previously characterized global Imp2-deficient mice [26]. Imp2 null mice have decreased lean and fat body mass, increased energy expenditure, reduced insulin secretion coupled with improved insulin sensitivity, and glucose tolerance [26]. The complex metabolic effects of global Imp2 deficiency make it is impossible to clarify Imp2's function in each major metabolic tissue and to provide a foundation for patient care. Guided by human studies, we decided to further focus on the islet-specific function of IMP2 and its function in insulin secretion. Here, we report that IMP2 pancreatic β-cell-specific deletion mice (βIMP2KO) have reduced compensatory β-cell proliferation and insulin secretion in response to the increased insulin demand of high-fat diet (HFD) feeding. We identified IMP2-binding mRNAs in islets, uncovered alternations of mRNAs and polypeptides from βIMP2KO mice, confirmed the binding of IMP2 protein to Igf2, Pdx1and many other islet transcripts, demonstrated that IMP2 regulates PDX1 mRNA translation in an m6A dependent manner, and showed that Imp2 regulates insulin secretion by orchestrating Igf2-Akt-Gsk3-Pdx1 signaling. Moreover, expression of IMP2 in human EndoC-βH1 cells promotes cell proliferation, PDX1 expression and Igf2-Akt-Gsk3 signaling. Based on the above evidence, we conclude that the ablation of Imp2 in mouse pancreatic β cells leads to impaired insulin secretion recapitulating human T2D traits.
2. Results
2.1. Deletion of IMP2 in mouse pancreatic β-cells causes reduced compensatory β-cell proliferation and insulin secretion
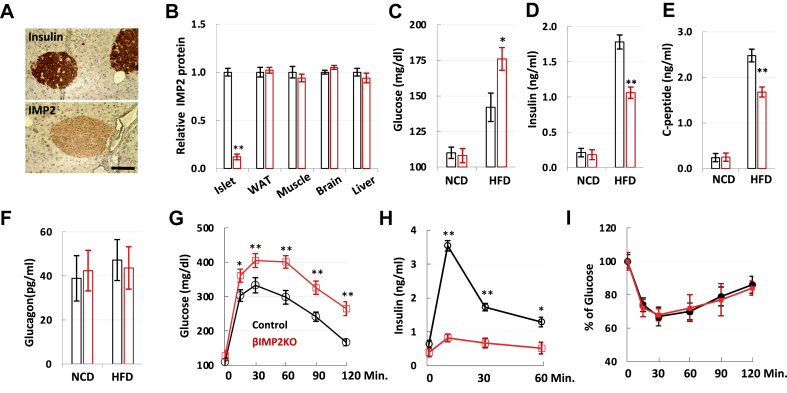
IMP2 is expressed at moderate levels in most mouse tissues, including liver, muscle, fat, and brain [25]. In the pancreas, IMP2 is highly expressed in islets, particularly in insulin-producing β cells (Figure 1A). As the causal T2D risk alleles at the IMP2 locus are correlated with reduced insulin secretion and lower islet IMP2 mRNA expression [16,17], we hypothesized that the decreased IMP2 activity in pancreatic β-cells contributes to the T2D pathophysiology. To test this hypothesis, we generated Imp2 pancreatic β-cell-specific knockout mice (βIMP2KO) under RIP2 promoter (Supplementary Figure 1a), which showed specific reduced expression of Imp2 in isolated islets (Figure 1B). βIMP2KO mice have normal body weight, body composition, physical activity, energy expenditure and serum lipid levels (Supplementary Table 1) as littermate controls. On normal calorie diet (NCD), βIMP2KO mice showed similar fasting blood glucose (Figure 1C) and serum insulin (Figure 1D) as controls. However, on HFD, blood glucose levels of βIMP2KO mice were elevated (Figure 1C) due to decreased insulin level (Figure 1D). We also observed decreased C-peptides (Figure 1E) with normal circulating glucagon levels (Figure 1F) from HFD-fed βIMP2KO mice. When challenged with an intraperitoneal glucose injection, βIMP2KO mice exhibited significantly higher glucose and lower insulin levels than RIP2-Cre control mice (Figure 1G and H). Importantly, this was not due to a difference in insulin sensitivity, as blood glucose levels after an intraperitoneal insulin injection were similar in RIP2-Cre and βIMP2KO mice (Figure 1I).
Figure 1.
The pancreatic β-cell specific deletion of IMP2 impairs insulin secretion on HFD feeding. A. Immunostaining of insulin and IMP2 in 8-week-old mouse pancreas. Scale bar: 80 μm B. Relative IMP2 protein abundance in control (black) and βIMP2KO (red) mice (n = 4 pairs). C–F. 6-hour fast blood glucose, serum insulin, C-peptide and glucagon from 12-week-old male control (black) and βIMP2KO (red) mice (n = 8 pairs). G–H. Glucose tolerance test of 14-week-old, HFD-fed male control (black) and βIMP2KO (red) mice (n = 8 pairs). g. blood glucose h. serum insulin. I. Insulin tolerance test of 16-week-old, HFD-fed male control (black) and βIMP2KO (red) mice (n = 8 pairs). Data are means ± SD. Two-way ANOVA test. ∗P < 0.05; ∗∗P < 0.01.
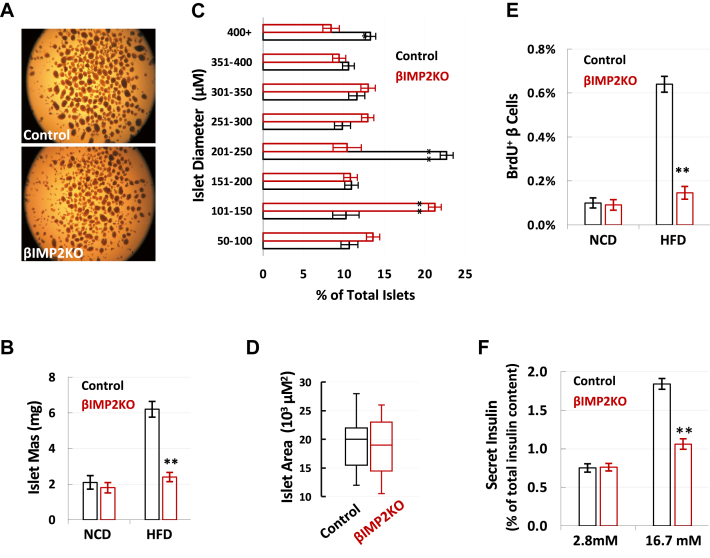
To determine whether Imp2 deficiency impairs β-cell function during HFD feeding, we isolated and measured islet mass (Figure 1A). Compared to NCD, HFD induced a 1.3-fold increase in islet mass of βIMP2KO mice (NCD: 1.84 ± 0.09 mg, HFD: 2.38 ± 0.07 mg), much less than a 3.0-fold increase in islet mass (NCD: 2.06 ± 0.12 mg, HFD: 6.23 ± 0.19 mg) in RIP2-Cre mice (Figure 1B). Although βIMP2KO mice have more small islets and less big islets (Figure 2C), the average islet diameter is not different due to the large variation (Figure 2D). This suggests that significantly decreased islet mass from HFD-fed βIMP2KO mice was primarily due to reduced β-cell number instead of cell size. As IMP2 is known to promote cell proliferation in various cell types, we next analyzed β-cell proliferation by BrdU incorporation and immune-fluorescence staining of 6-week-old, HFD-fed mice. The number of BrdU+ β-cells from βIMP2KO mice was reduced 70% compared to age-matched RIP2-Cre controls (Figure 1E), indicating that the loss of Imp2 led to impaired compensatory β-cell proliferation. Next, we performed glucose-stimulated insulin secretion (GSIS) using isolated islets. Although the size-matched islets from βIMP2KO and RIP2-Cre mice secrete similar insulin levels upon low glucose (2.8 mM) stimulation, βIMP2KO islets released approximately half the amount of insulin as controls after high glucose (16.7 mM) stimulation (Figure 1F). Therefore, deleting Imp2 in mouse β-cells impairs high glucose-stimulated insulin secretion. Based on all data above, we conclude that mouse pancreatic β-cell specific Imp2 is required for the full, compensatory β-cell proliferation and glucose stimulated insulin secretion in response to elevated insulin demand of HFD feeding.
Figure 2.
IMP2 deletion reduces β-cell proliferation and function. A. The representative images of isolated islets from HFD-fed, 16-weeks-old control and βIMP2KO mice (n = 6 pairs). B. The islet mass from 16-weeks-old NCD (n = 7 pairs) and HFD fed mice (n = 7 pairs). C–D. The distribution of islet size and area from HFD-fed control (black) and βIMP2KO (red) (n = 6 pairs). E. The percentage of BrdU labeled β-cells from 6-week-old mice (n = 8 pairs). F. the glucose stimulated insulin secretion of isolated 16-weeks-old, HFD-fed islets (n = 8 pairs). Data are means ± SD. Two-way ANOVA test. ∗P < 0.05 ∗∗P < 0.01.
2.2. Imp2 promotes insulin secretion through post-transcriptional regulation
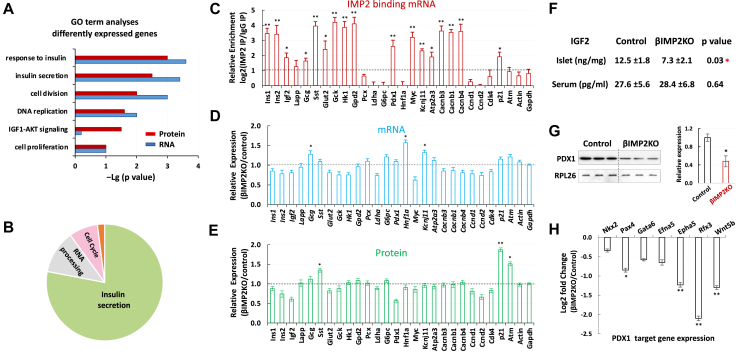
To understand the molecular mechanism through which Imp2 contributes to β-cell proliferation and function, we conducted RNA-seq and Imp2 RNA immunoprecipitation sequencing (RIP-seq) using islets from 8-week-old mice fed on HFD. RNA-seq identified total 30,636 transcripts and the expression levels of 1,291 transcripts were altered significantly more than 2 folds (Supplementary 2a). Consistent with reduced β-cell proliferation observed in βIMP2KO mice, RNA sequencing analyses showed downregulation of key cell-cycle genes (Ccnd1, Ccnd2, Cdk4) and increased expression of the cyclin-dependent kinase inhibitor (p21). Enriched pathway analyses of differentially expressed genes (FDR < 0.10) revealed response to insulin, cell proliferation/cell cycle, and insulin secretion as the most commonly affected biological events in βIMP2KO islets (Figure 3A). To identify Imp2-binding RNAs, each IP-associated RNA was corrected for abundance in the total extract and divided by the corrected IP value from βIMP2KO islets. This nominated ~1,600 islet mRNAs as Imp2 binding partners (Supplemetary Figure 2a–b) that are highly enriched for insulin secretion, RNA processing, and cell cycle (Figure 3B).
Figure 3.
IMP2 promotes β-cell proliferation and function. A. GO analyses of differentially expressed islet proteins (red) and RNAs (blue). B. Functions of IMP2 binding RNAs. C–E. Confirmation of abundances of selected IMP2 binding mRNAs (c), total mRNAs (d) and proteins (e) from 8-week-old, HFD fed control and βIMP2KO animals (n = 5 pairs). F. The islet and serum IGF2 levels from 12- weeks-old HFD-fed mice (n = 7 pairs). G. Western blot of PDX1 protein from isolated islets of 12-weeks-old, HFD-fed mice (n = 3 pairs) H. the relative expression of PDX1 transcriptionally regulated genes. RNA was isolated from 12-weeks-old, HFD-fed control and βIMP2KO mice (n = 5 pairs). Data are means ± SD. T-test ∗P < 0.05;∗∗P < 0.01.
Since IMP2 plays a key role in post-transcriptional gene expression regulation, we next performed liquid chromatography tandem mass spectrometry (LC-MS/MS)-based proteomics using islets from the same cohort of mice for RNA-seq and RIP-Seq. The analysis resulted in the confident identification of 29,021 different tryptic peptides covering 3,365 proteins (≥2 unique peptide identifications per protein). As expected, three major islet hormones (insulin, glucagon, and somatostatin) were detected, as well as various β-cell enriched secretory products, ion channels, and transcription factors. In addition, significant proteome coverage of islet metabolic enzymes and cellular pathways was observed. Quantitative measurement of relative polypeptide abundances showed significant differences of a total of 1,252 proteins. Pathway analyses of differentially expressed proteins revealed a highly significant enrichment of the insulin secretion, cell proliferation/cell cycle, and IGF1-AKT pathway (Figure 3A). Although the latter was not evident in the transcriptome analysis, the impairment of insulin/IGF–AKT signaling is a well-known mechanism associated with T2D [[40], [41], [42]].
Guided by the results of genome-wide research, we further validated the abundances of critical RNAs and polypeptides for β cell function (Figure 3C–E). Consistent with RNA-Seq and proteomic studies, 50% of validated Imp2 targets did not show significant alternation at either mRNA or protein level. As expected, Igf2 mRNA was confirmed as an Imp2 binding partner and reduced in βIMP2KO islets (Figure 3F). Notably, validated as one of Imp2 targeted mRNAs (Figure 3C), the Pdx1 transcriptional level is similar between βIMP2KO and control (Figure 3D). However, the Pdx1 polypeptides significantly decreased in βIMP2KO (Figure 3G), indicating Imp2 regulates Pdx1 mRNA translation and/or Pdx1 polypeptide stability. In line with reduced Pdx1 protein levels, the mRNA abundances of major Pdx1 transcriptionally regulated genes were reduced by 12–54% (Figure 3H).
2.3. IMP2 directly binds PDX1 mRNA and promotes its translation in an m6A-dependent manner
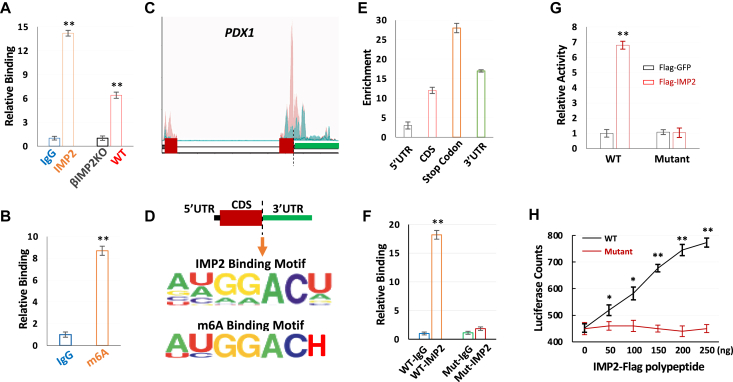
As IMPs preferentially bind m6A-modified mRNAs [36], we next hypothesized that IMP2 regulates PDX1 mRNA translation through m6A recognition. We first confirmed the direct binding of IMP2 with PDX1 mRNAs by IMP2 RIP-qPCR (Figure 4A), and demonstrated m6A modification of PDX1 mRNAs through PDX1-specific m6A assays using human islets (Figure 4B). Notably, the PDX1 stop codon containing region which showed dramatic hypo-methylation in T2D patients [40] has most abundant m6A modifications (Figure 4C) relative to other regions of PDX1 transcript (such as 5′UTR). As conserved IMP2 binding motifs significantly overlaps with m6A modification sites (Figure 4D), this locus is also enriched for IMP2 binding. To determine whether the stop codon-containing region is critical for IMP2-guided PDX1 mRNA translation, we inserted 200 nt PDX1 mRNA wild-type or mutant sequence (Supplementary Figure 3a) flanking stop codon into firefly luciferase reporters (Supplementary Figure 3b) into EndoC-βH1 cells [43] stably expressing Flag-IMP2 or Flag-GFP. IMP2 RIP-qPCR demonstrated a strong binding of IMP2 with wild-type PDX1 reporter but not with the mutant form (Figure 4F). Moreover, IMP2 dramatically enhanced luciferase activity, which was also dependent on the presence of wild-type m6A motifs surrounding the stop codon of PDX1 mRNA (Figure 4G) as the mutant m6A motif form abolished the IMP2-stimulated luciferase activity. We further confirmed the effect of IMP2 stimulated PDX1 mRNA translation in vitro using rabbit reticulate lysate as Flag-IMP2-induced a significant increase of PDX1 wild-type reporter activity in a dose-dependent manner (Figure 4H). This effect was greatly impaired by mutations in the m6A consensus sites/IMP2 binding sites in the PDX1 stop codon-containing region (Figure 4H). Taken together, these data demonstrate that IMP2 preferentially binds m6A-modified PDX1 mRNAs and promotes their translation.
Figure 4.
IMP2 directly binds PDX1 mRNA and promotes its translation in an m6A dependent manner in vivo and in vitro. A. IMP2 binds PDX1 mRNA. IPs were performed using human islets (left pair) with nonimmune IgG (blue) anti-IMP2 antibody (orange) or from the IMP2-IP using islet extracts of βIMP2KO and WT mice (right pair) IP, immunoprecipitation. B. the enrichment of PDX1 mRNA with nonimmune IgG (blue) and m6A antibody (orange). IPs were performed using human islets with nonimmune IgG (blue) or m6A antibody (orange). C. Schematic representation of human PDX1 gene and published m6A peaks from control (pink) and T2D patients (blue) [40]. D. the IMP2 and m6A binding motif around the stop codon of PDX1 gene. H: not G. E. the enrichment of IMP2 binding sites on different location of PDX1 mRNA. F. the relative binding of m6A WT and mutant to IMP2 antibody and IgG. G. IMP2 promotes WT luciferase activity in vitro. Flag construct and luciferase reporter were co-transfected into Endoc-βH1 cells. The luciferase activities were assayed 36 h post-transfection. H. IMP2 promotes m6A WT, but not mutant luciferase in vitro translation assay. Data are means ± SD. T-test ∗P < 0.05; ∗∗P < 0.01.
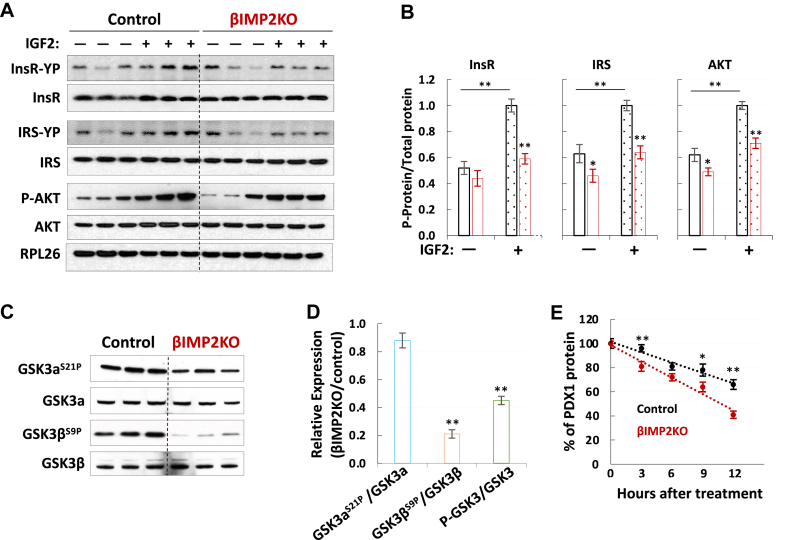
2.4. IMP2 stabilizes PDX1 polypeptides by enhancing IGF2–AKT–GSK3 signaling
In addition to reduced PDX1 mRNA translation, the decreased Pdx1 proteins in βIMP2KO mice could also be attributed to post-translational regulation. Indeed, the impairment of IGF2-AKT signaling promotes PDX1 protein degradation and decreases β-cell mass and function [44,45]. The nominated reduced IGF1–AKT signaling by proteomics studies promoted us to further explore the post-translational mechanism. We previously published that, in addition to its ability to promote IGF2 production, IMP2 strongly stimulates IGF2 action [46]. In mouse muscles, the deficiency of Imp2 results in impaired Igf2 signaling through diminishing the phosphorylation of insulin/Igf1 receptors, reducing Akt1 activation and disinhibiting Gsk3α [30]. Therefore, we hypothesized that IMP2 regulates IGF2 signaling, in a similar way, to promote insulin secretion in pancreatic β cells. Islets from HFD-fed βIMP2KO and control mice were stimulated with exogenous IGF2. Immunoblots of total InsR and Irs polypeptides showed no difference (Figure 5A–B); however, the phosphorylated forms of both proteins reduced by 35–40% (P < 0.05) (Figure 5A–B) in βIMP2KO islets. Similarly, the total Akt polypeptide levels are also not different. However, phos-Akts (Thr308 and Ser473) are both marked reduced in βIMP2KO islets (Figure 5A–B). We next examined Akt-catalyzed inhibitory phosphorylation of Gsk3. While Gsk3α (Ser21) phosphorylation is not dramatically different (Figure 5C–D), the corresponding phosphorylation of Gsk3β (Ser9) is reduced by ~50%, pointing to higher activity of Gsk3β in βIMP2KO islets (Figure 5C–D). Most importantly, the Gsk3β-catalyzed Pdx1 degradation is increased more than 3-fold in βIMP2KO islets over controls measured by the reduced half-life of Pdx1 protein in βIMP2KO islets (Figure 5E). Taken together, the above data show that the IMP2 deficiency in mouse islets led to impaired insulin secretion through the downregulation of the IGF2–AKT–GSK3β–PDX1 pathway.
Figure 5.
IMP2 promotes IGF2-AKT-GSK3-PDX1 signaling. A–B. Overall abundances and phosphorylation of InsR, IRS and AKT with/without IGF2 (100 nM) stimulation (n = 3 for each condition). C–D. The quantification of overall abundances and phosphorylation of GSK3 (n = 3). E. Cumulative distribution of half life of PDX1 protein (n = 5). All islets were isolated from 12-weekold, HFD-fed male mice. Data are means ± SD. T-test ∗P < 0.05; ∗∗P < 0.01.
2.5. Overexpression of IMP2 in EndoC-βH1 cells promotes insulin secretion
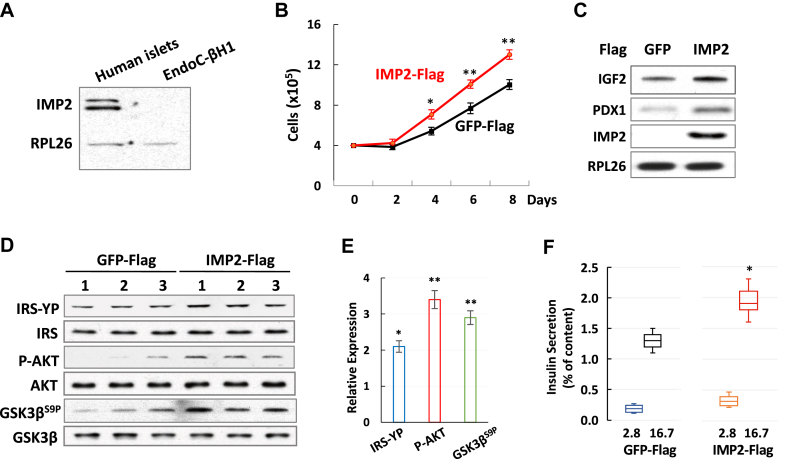
We found that IMP2 is highly expressed in human islets but cannot be detected by immunoblotting in EndoC-βH1 cells (Figure 6A), a commonly used human β cell line. In addition, EndoC-βH1cells have much lower capability of insulin secretion compared to human islets [43]. We therefore hypothesized that the expression of IMP2 is able to promote insulin secretion in EndoC-βH1 cells. Next, we transfected EndoC-βH1 cells with Flag-IMP2 or Flag-GFP, and selected stable cells. EndoC-βH1 cells stably expressing IMP2 showed accelerated cell proliferation (Figure 6B), which is consistent with IMP2's function as a promoter of cell growth. Moreover, the immunoblotting of IGF2 and PDX1 showed increased polypeptide levels in IMP2-expressing cells in comparison with GFP-expressing controls (Figure 6C). Furthermore, consistent with the result from βIMP2KO mice studies, IMP2 expression also improved the IGF2 signaling by promoting AKT phosphorylation and inhibiting GSK3β activity (Figure 6D–E). Finally, the results of GSIS demonstrated that the expression of IMP2 is capable to increase insulin secretion at high glucose (16.7 mM), but not at low glucose (2.8 mM), in EndoC-βH1cells (Figure 6F). Thus, these data demonstrate that the expression of IMP2 in EndoC-βH1 cells stimulates cell proliferation, enhances IGF2 and PDX1 expression, promotes IGF2-AKT-GSK3β signaling, and increases insulin secretion.
Figure 6.
IMP2 promotes insulin secretion in EndoC-βH1 cells. A. Western blot of IMP2 in human islets and EndoC-βH1 cells. B. Cell proliferation of IMP2 and GFP expressing cells C. Western blot of IGF2 and PDX1 in cells expressing IMP2 or GFP. D–E. western blot (D) and quantification (E) of IRS-YP, p-AKT and GSK3βS9P in IMP2 and GFP expressing cells. F. Relative insulin secretion from IMP2 or GFP expressing cells. Data are means ± SD. T-test ∗P < 0.05; ∗∗P < 0.01.
Collectively, our study revealed an unexpected function of Imp2 in pancreatic β-cells through promoting Pdx1 mRNA translation and enhancing Igf2–Akt–Gsk3β signaling, which impairs insulin secretion and β-cell proliferation upon the ablation of Imp2.
3. Discussion
Human T2D is widely considered as a chronic and progressive disease without cure [47]. As insulin-producing β-cell function progressively declines over time, blood glucose rises. This study focuses on the pancreatic β-cell specific function of IMP2. We provide experimental evidence for the molecular mechanisms of IMP2 in β-cell biology which highlights the significance of post-transcriptional gene expression regulation in T2D pathophysiology.
IMP2 was first nominated as a human T2D-associated gene in 2007 without any experimental evidence supporting its role in metabolic regulations [[2], [3], [4]]. To understand the function of IMP2 in T2D pathogenesis and provide a foundation for patient care, we generated and studied the Imp2 global knockout mice [26]. The global Imp2-null mice exhibit a normal birth size and a body weight similar to that of control littermates until weaning [26]. However, they gain less weight due to slower accumulation of both lean and fat body mass [26]. The lower fat mass of Imp2-null mice is especially marked on HFD and is accompanied by reduced levels of circulating lipids, less liver triglyceride accumulation and better glucose tolerance and insulin sensitivity [26]. However, it is unclear which of these favorable metabolic phenotypes exhibited by Imp2 null mice (smaller white fat depots, resistance to hepatic steatosis, superior insulin sensitivity, reduced insulin secretion) is relevant to its role in human T2D; it is also unclear which is a direct consequence of Imp2 deficiency in the target organ (adipocyte, liver, skeletal muscle, islet). Seeking to understand the tissue(s) in which Imp2 has its primary impact, we generated a series of mice lacking Imp2 in specific organs. The postnatal inactivation of Imp2 in skeletal muscle decreased accrual of muscle mass after weaning and reduced wheel-running activity [30]. However, their whole-body glucose metabolism and insulin sensitivity are unaltered [30], in contrast to the very pronounced improvements seen in global Imp2 deficiency [26]. The primary finding of elimination of the hepatocyte Imp2 is increased intrahepatic triglyceride deposition in HFD-fed mice, which developed slight hypertriglyceridemia and hyperglycemia by 30 weeks of age [31]. This indicates that the marked protection against fatty liver observed in global Imp2 deficiency mice is entirely due to their reduced adiposity. Although we have plausibly attributed the reduced insulin secretion of the global Imp2-null mice to their enhanced insulin sensitivity, the majority of human studies have identified impaired insulin secretion as the predominant defect of IMP2 T2D risk alleles [16,17]. In the current study, we showed that β-cell specific deletion of Imp2 in mice resulted in impaired compensatory β-cell proliferation and function due to the increased demand of insulin secretion by HFD. This study revealed that pancreatic β-cell intrinsic functions of Imp2 are completely obscured by the tremendous insulin sensitivity of the global Imp2-null mice caused by their dramatically reduced fat mass. While studying Imp2 murine models, we noticed that many phenotypes are diet-dependent. For example, the impaired insulin secretion and reduced β cell proliferation in βIMP2KO mice only showed on HFD but not on NCD. This phenomenon is likely due to IMP2 functioning as a post-transcriptional gene regulator, which is critical for stress conditions [48], such as HFD feeding. As Imp2 is not critical under normal conditions, the Imp2 deficiency has less/no impact on whole-body metabolism on NCD.
The data collected from all Imp2 murine models has shown that GWAS signals can be moved forward into biological understanding by embracing all the technologies at our disposal. To date, no deleterious mutations encoding IMP2 have been found in human populations; this knowledge would be informative to the future T2D research. In line with our data collected from murine models, the recent human T2D research grouped IMP2 into a “proinsulin cluster” featured with decreased proinsulin and reduced β-cell function [49]. The blood IMP2 mRNA levels were positively associated with fasting insulin from people without diabetes [50] indicating that IMP2 may be involved in the regulation of β-cell function before onset of T2D. Despite these emerging data, it remained unclear what these findings brought to our greater understanding of human T2D. The caveat is that, despite a large number of variants discovered by GWAS, the total of associated variants explains only a small proportion (10%) of the heritability of T2D.
Recently, mRNA m6A methylation has been reported to contribute significantly to the pathogenesis of T2D and is a better mark to segregate human T2D islets from controls [40]. IMPs are a newly identified family of m6A “reader” which can decode m6A methylation and generate functional signals [36]. Consistent with the downregulation of m6A methylation components in T2D patients [40], we observed that the deletion of Imp2 in β cells results in downregulation of key β-cell regulator Pdx1 and Igf2. PDX1 is one of most vital transcriptional factors that controls β-cell development, identity, proliferation, survival, and function [51]. The β cell Imp2-specific deficiency leads to decreased Pdx1 protein levels. We discovered that Imp2 directly binds to Pdx1 mRNA and promotes its translation in an m6A-dependent manner. In addition, Imp2 enhances Igf2-Akt-Gsk3β signaling to prevent Pdx1 polypeptide degradation. IGF2 is a potent growth factor from insulin/IGF family and involved in a spectrum of biological events. IGF2 is also a human T2D-associated gene. During mouse embryonic development, Igf2 is a major paracrine regulator of pancreatic growth and function (http://doi.org/10.1101/714121). In adult mice, the autocrine action of Igf2 is required for maintaining proper insulin secretion as well as β-cell functions in response to metabolic stress such as HFD feeding [52]. In this study, we demonstrated that Imp2 is required to promote Igf2–Akt–Gsk3β–Pdx1 signaling in response to HFD feeding. Taken together, we propose that reduced IMP2 expression in pancreatic β-cells contributes to the pathophysiology of human T2D. Therapeutic targeting of IMP2 in a β-cell-specific manner in combination with current therapeutic agents might be a new avenue to counter the decreased m6A levels in T2D islets and to promote β-cell proliferation and function.
4. Methods
4.1. Animal studies
All animal procedures were approved by the Institutional Animal Care and Use Committee of Massachusetts General Hospital and were performed in accordance with the National Research Council guidelines for laboratory animal care. RIP2-Cre transgenic mice were purchased from Jackson Lab (Stock Number #003573), which has a 668 bp fragment of the rat insulin II promoter. βIMP2KO and RIP2-Cre control mice were maintained on C57BL/6J background in a specific pathogen-free facility with 12:12 light:dark cycle and fed irradiated chow (Prolab 5P75 Isopro 3000; 5% crude fat; PMI nutrition international) or a HFD (D12492i; 60 kca% fat; Research Diets, Inc.).
4.2. Glucose and insulin tolerance tests
For glucose tolerance tests, mice were fasted overnight. Twenty percent D-glucose (Sigma) (1 g/kg body weight) was administered by intraperitoneal injection. At 0, 30, 60, 90, and 120 minutes after administration, blood was collected by tail vein bleeding. Glucose levels were measured by a One Touch Ultra AlphaTrak2 glucometer (Zoetis, Parsippany, NJ). For insulin tolerance tests, HFD-fed mice were fasted for 6 h. Human insulin (Eli Lily) at 0.75 U/kg was injected intraperitoneally. Blood was drawn from the tail vein at 0, 30, 60, 90, and 120 minutes after injection, and glucose levels were measured.
4.3. Serum analyses
Blood was collected into EDTA-coated tubes (Sarstedt, Newton, NC). Serum was separated by centrifugation at 4 °C, frozen in liquid nitrogen, and assayed at the Vanderbilt University Mouse Metabolic Phenotyping Center (MMPC). The blood Igf2 concentration was measured using a mouse Igf2 enzyme-linked immunosorbent assay (ELISA) kit (catalog number EK0381; Boster Biological Technology Co., Ltd., CA) according to the manufacturer's instructions.
4.4. Mouse islet isolation and quantification
Mouse islet isolations were performed at MGH islet transplant core. Briefly, mice were anesthetized and their pancreases were infused with liberase (Roche) and dissected. Following incubation at 37 °C, the digested pancreases were washed, filtered, and run on a sucrose gradient. The islet cell counter (ICC) used for this study (Biorep Technologies) is a machine vision system that includes both hardware and software designed specifically for automated islet counting [53]. For each sample, a single high-resolution image is taken to assess the amount and purity of the islet cell sample. ICC uses a fully automated counting method implemented in LabVIEW software [Laboratory Virtual Instrumentation Engineering Workbench (LabVIEW); National Instruments, Austin, TX, USA]. Islet samples were prepared and stained with diphenylthiocarbazone. The software uses pixel coloration to separate islets from acinar tissue and a segmentation-based algorithm to delineate each individual islet and calculate its area. The IEQ (mass) contribution of each islet is then obtained using the 50-μm increment group classification and standard IEQ group values. Estimated islet mass (total IEQ), number, purity, and size group distributions, as well as the high-resolution image, are saved in the final output as both Word document and Adobe pdf files.
For islet histomorphometry study, number of islet/mm2 of pancreatic area, mean area of islet and number of β–cell nuclei/mm2 of insulin-positive area were carried out for each group of animals.
4.5. Insulin secretion assay
After overnight culture in RPMI 1640 medium (11 mmol/L glucose and 10% FBS), triplicate samples of 10 equilibrated islets for each mouse placed in wells of a 24-well plate were sequentially incubated with 2.8 and 16.8 mmol/L glucose in Krebs–Ringer buffer (16 mmol/L HEPES and 0.1% BSA, pH 7.4) for 50 minutes. Supernatant fractions and cell lysates were frozen until assayed for insulin.
4.6. BrdU treatment
Mice were provided BrdU in drinking water (80 mg/ml) after weaning at 4-week-old and received intraperitoneal injections of BrdU (100 mg/kg body weight) every three days for two weeks. Mice were sacrificed at 6 weeks old and β-cell proliferation was assessed by co-immunostaining sections of each pancreas sample with BrdU, insulin, and DAPI. β-cell proliferation was assessed by analyzing images acquired from 30 islets per animal.
4.7. Pancreas immunostaining and analyses
Mouse pancreas was collected and fixed in 3.7% formaldehyde at 4 °C overnight, followed by paraffin embedding. Four micron-thick slides were cut and subjected to immunostaining. Slides were heated in 10 mM sodium citrate, followed by blocking with donkey serum and incubated with various primary antibodies (Supplementary Table 2). The specific signal was detected using fluorescence-conjugated secondary antibodies.
4.8. RNA-seq
RNA from 8-week-old, HFD-fed mouse islets was extracted using a Qiagen RNase kit. RNA-seq libraries were constructed from poly(A)-selected RNA sequenced on an Illumina HiSeq2500 instrument, resulting in approximately 30 million reads per sample on average. STAR aligner was used to map sequencing reads to transcripts in the mouse mm10 reference genome. Read counts for individual transcripts were produced with HTSeq-count, followed by the estimation of expression values and detection of differentially expressed transcripts using EdgeR.
4.9. Proteomic studies
Purified islets from 8-week-old, HFD-fed mice were processed and analyzed through the Thermo Fisher Scientific Center for Multiplexed Proteomics at Harvard Medical School. Peptides derived from digestion using LysC and trypsin were labeled with Tandem Mass Tag 8-plex reagents and fractionated. Multiplexed quantitative mass spectrometry data were collected on an Orbitrap Fusion mass spectrometer operating in a MS3 mode using synchronous precursor selection for the MS2 to MS3 fragmentation. MS data were searched against an Uniprot mouse database with both the forward and reverse sequences using the SEQUEST algorithm. Further data processing steps included controlling peptide and protein level false discovery rates, assembling proteins from peptides, and protein quantification from peptides.
4.10. IMP2 RIP-seq
Purified islets from 8-week-old, HFD-fed mice were extracted directly using a tissue lyser (Qiagen) in ice-cold lysis buffer (140 mM KCl, 1.5 mM MgCl2, 20 mM Tris–HCl at pH 7.4, 0.5% Nonidet P-40, 0.5 mM dithiothreitol, 1 U/μl RNase inhibitor, one complete EDTA-free protease inhibitor cocktail tablet) for 10 min. The lysates were centrifuged for 10 min at 14,000 rpm, and the supernatant was transferred to a fresh 1.5-ml tube. Total protein was measured by a Bradford assay, and 5 mg of the cytoplasmic lysate protein was subjected to immunoprecipitation. Lysates were incubated with 500 μl of protein A Dynabeads magnetic beads precoated with IMP2 antibody and incubated for 6 hours at 4 °C with rotation. Dyna beads were extensively washed with lysis buffer five times and digested with DNase I and protease K. RNA was extracted with phenol-chloroform and precipitated with ethanol. RNA sequencing was performed to examine RNAs associated with cytoplasmic IMP2 in islets.
4.11. Protein assays
Proteins were extracted using a tissue homogenizer (Qiagen). Purified islets from 8-week-old, HFD-fed mice were homogenized in ice-cold buffer (20 mM Tris [pH 7.5], 2.7 mM KCl, 1 mM MgCl2, 1% Triton X-100, 10% [wt/vol] glycerol, 1 mM EDTA, 1 mM dithiothreitol) supplemented with protease (Thermo Scientific) and a phosphatase inhibitor cocktails (Millipore). Samples were then centrifuged at 13,000 rpm for 10 min at 4 °C, and the supernatants were collected. The protein content of the supernatant was determined using a bicinchoninic acid (BCA) assay (Thermo Scientific). Aliquots of each extract containing 50 μg protein were loaded for Western blotting. Proteins were resolved on a 4–12% gradient SDS Bis-Tris gel (Invitrogen, Carlsbad, CA). Antibodies for immunoblotting are listed in Supplementary Table 2. For protein half-life assay, 250 mM cycloheximide was used to inhibit mRNA translation; protein half-life was estimated from immunoblot of extracts prepared before and at various times after cycloheximide addition.
4.12. Gene-specific m6A qPCR
m6A modifications of specific genes were determined using Magna MeRIP m6A Kit (Millipore, Billerica, MA). In short, 1 mg of total RNA was sheared to 100 nt, incubated with anti-m6A antibody (Synaptic Systems) or mouse IgG (Millipore) with RNase inhibitors at 4 °C. Methylated RNA was immunoprecipitated with beads, eluted by competition with free m6A, and recovered with an RNeasy kit (Qiagen). Relative enrichment of m6A was calculated by normalizing to input.
4.13. Human samples
Human islet samples were obtained from the Pancreatic Islet Isolation Core of Boston Area Diabetes Endocrinology Research Center (BADRC). The detailed donor information is in Supplementary Table 3.
4.14. EndoC-βH1 cell culture
Culture flasks were coated with DMEM (glucose 4.5 g/L, Gibco) containing PS (1%, Gibco), fibronectin (2 μg/ml; Gibco), and extracellular matrix (1% vol/vol, Sigma) and incubated for 2 hours in 5% CO2 at 37 °C before the cells were seeded. EndoC-βH1 cells were grown on Matrigel/fibronectin-coated culture flasks containing DMEM (glucose 1 g/L), BSA fraction V (2% wt/vol) (Roche), 2-mercaptoethanol (50 μM, Sigma), nicotinamide (10 mM, Sigma), transferrin (5.5 μg/ml Sigma), sodium selenite (6.7 ng/ml Sigma), PS (1%). For insulin secretion assays, EndoC-βH1 cells were starved for 2 h in Krebs ringer buffer (KRB) containing NaCl (115 mM), NaHCO3 (24 mM), KCl (5 mM), MgCl2 (1 mM), CaCl2 (1 mM), HEPES (10 mM), BSA (0.2% wt/vol), and 0.5 mM glucose. Static insulin secretion assays were then initiated by adding KRB containing 2.8 mM or 16.7 mM of glucose for 1 hour. Aliquots of supernatant were removed, centrifuged to discard dead cells, and stored for analysis in ice-cold acid ethanol that was added to extract insulin content from cells. Insulin secretion and content were measured by the human insulin ELISA (Mercodia) according to the manufacturer's instructions. For stable cell lines, EndoC-βH1 cells were transduced with lentiviral particles. Puromycin (1 μg/ml) was added to the culture media to select cells stably expressing Flag-IMP2 or Flag-GFP. Transient transfections were performed by mixing cells with Lipofectamine according to the manufacturers' instructions.
4.15. Dual luciferase reporter assay
Luciferase activity was examined by a dual luciferase reporter assay using the dual luciferase reporter assay kit (Promega) on a TD-20/20 luminometer (Turner Designs). To generate the PDX1 firefly luciferase reporter construct, DNA fragments of wild-type and mutant sequences were synthesized by Integrated DNA Technologies (Coralville, Iowa) and cloned into the pcDNA5/TO vector (Invitrogen). The EndoC-βH1 cells were transiently transfected with luciferase vector together with Renilla luciferase control (Promega). Luciferase activity was measured at 36 hours post-transfection.
4.16. Statistical analysis
Comparisons between the mean ± SD of two groups were calculated using two-way analysis of variance (ANOVA) or Student's unpaired two-tailed t test. P values of 0.05 or less were considered statistically significant.
Acknowledgments
This work was supported by NIH grants DK17776, DK057521, and MGH institutional funds. We thank the MGH islet transplant core for providing human islets and technical support; MGH microscopy core of the program in membrane biology for imaging; Dr. Susan Weir, Dr. Gordon Weir, Dr. Dario Jesus, and Dr. Rohit Kulkarni for advice.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101209.
Conflict of interest
All authors do not have conflict of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Neel J.V. Diabetes mellitus: a "thrifty" genotype rendered detrimental by "progress"? The American Journal of Human Genetics. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- 2.Zeggini E., Weedon M.N., Lindgren C.M., Frayling T.M., Elliott K.S., Lango H. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott L.J., Mohlke K.L., Bonnycastle L.L., Willer C.J., Li Y., Duren W.L. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saxena R., Voight B.F., Lyssenko V., Burtt N.P., Bakker P., Chen H. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 5.Votsi C., Toufexis C., Michailidou K., Antoniades A., Skordis N., Karaolis M. Type 2 diabetes susceptibility in the Greek-Cypriot population: replication of associations with TCF7L2, FTO, HHEX, SLC30A8 and IGF2BP2 polymorphisms. Genes. 2017;8(1) doi: 10.3390/genes8010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson D., Cordell H.J., Fakiola M., Francis R.W., Syn G., Scaman E.S. First genome-wide association study in an Australian aboriginal population provides insights into genetic risk factors for body mass index and type 2 diabetes. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0119333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phani N.M., Adhikari P., Nagri S.K., D’Souza S.C., Satyamoorthy K., Rai P.S. Replication and relevance of multiple susceptibility loci discovered from genome wide association studies for type 2 diabetes in an Indian population. PLoS One. 2016;11(6) doi: 10.1371/journal.pone.0157364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han L., Li Y., Tang L., Chen Z., Zhang T., Chen S. IGF2BP2 rs11705701 polymorphisms are associated with prediabetes in a Chinese population: a population-based case-control study. Experimental Therapeutics Medicine. 2016;12(3):1849–1856. doi: 10.3892/etm.2016.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng M.C., Shriner D., Chen B.H., Li J., Chen W., Guo X. Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLoS Genetics. 2014;10(8) doi: 10.1371/journal.pgen.1004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christiansen J., Kolte A.M., Hansen T.O., Nielsen F.C. IGF2 mRNA-binding protein 2: biological function and putative role in type 2 diabetes. Journal of Molecular Endocrinology. 2009;43(5):187–195. doi: 10.1677/JME-09-0016. [DOI] [PubMed] [Google Scholar]
- 11.Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nature Genetics. 2010;42(7):579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segrè A.V., Steinthorsdottir V. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nature Genetics. 2012;44(9):981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashcroft F.M., Rorsman P. Diabetes mellitus and the beta cell: the last ten years. Cell. 2012;148(6):1160–1171. doi: 10.1016/j.cell.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Bunt M., Fox J.E.M., Dai X., Barret A., Grey C., Li L. Transcript expression data from human islets links regulatory signals from genome-wide association studies for type 2 diabetes and glycemic traits to their downstream effectors. PLoS Genetics. 2015;11(12) doi: 10.1371/journal.pgen.1005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marselli L., Thorne J., Dahiya S., Sgroi D.C., Sharma A., Bonner-Weir S. Gene expression profiles of Beta-cell enriched tissue obtained by laser capture microdissection from subjects with type 2 diabetes. PLoS One. 2010;5(7):e11499. doi: 10.1371/journal.pone.0011499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bysani M., Agren R., Davegårdh C., Volkov P., Rönn T., Unneberg P. ATAC-seq reveals alterations in open chromatin in pancreatic islets from subjects with type 2 diabetes. Scientific Reports. 2019;9(1):7785. doi: 10.1038/s41598-019-44076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenwald W.W., Chiou J., Yan J., Qiu Y., Dai N., Wang A. Pancreatic islet chromatin accessibility and conformation reveals distal enhancer networks of type 2 diabetes risk. Nature Communications. 2019;10(1):2078. doi: 10.1038/s41467-019-09975-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen J., Christiansen J., Lykke-Andersen J., Johnsen A.H., Wewer U.M., Nielsen F.C. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Molecular and Cellular Biology. 1999;19(2):1262–1270. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yisraeli J.K. VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biologie Cellulaire. 2005;97(1):87–96. doi: 10.1042/BC20040151. [DOI] [PubMed] [Google Scholar]
- 20.Deshler J.O., Highett M.I., Schnapp B.J. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276(5315):1128–1131. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- 21.Yaniv K., Yisraeli J.K. The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene. 2002;287(1–2):49–54. doi: 10.1016/s0378-1119(01)00866-6. [DOI] [PubMed] [Google Scholar]
- 22.Hansen T.V., Hammer N.A., Nielsen J., Madsen M., Dalbaeck C., Wewer U.M. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Molecular and Cellular Biology. 2004;24(10):4448–4464. doi: 10.1128/MCB.24.10.4448-4464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janiszewska M., Suvà M.L., Riggi N., Houtkooper R.H., Auwerx J., Virginie Clément-Schatlo V. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes & Development. 2012;26(17):1926–1944. doi: 10.1101/gad.188292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z., Gilber J.A., Zhang Y., Zhang M., Qiu Q., Ramanujan K. An HMGA2-IGF2BP2 axis regulates myoblast proliferation and myogenesis. Developmental Cell. 2012;23(6):1176–1188. doi: 10.1016/j.devcel.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai N., Rapley J., Angel M., Yanik A.M., Blower M.D., Avruch J. mTOR phosphorylates IMP2 to promote IGF2 mRNA translation by internal ribosomal entry. Genes & Development. 2011;25(11):1159–1172. doi: 10.1101/gad.2042311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai N., Zhao L., Wrighting D., Krämer D., Majithia A., Wang Y. IGF2BP2/IMP2-Deficient mice resist obesity through enhanced translation of Ucp1 mRNA and Other mRNAs encoding mitochondrial proteins. Cell Metabolism. 2015;21(4):609–621. doi: 10.1016/j.cmet.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai N. The diverse functions of IMP2/IGF2BP2 in metabolism. Trends in Endocrinology and Metabolism. 2020;31(9):670–679. doi: 10.1016/j.tem.2020.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen F.C., Ostergaard L., Nielsen J., Christiansen J. Growth-dependent translation of IGF-II mRNA by a rapamycin-sensitive pathway. Nature. 1995;377(6547):358–362. doi: 10.1038/377358a0. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y., Lu J., Wu Q.N., Jin Y., Wang D.S., Chen Y.X. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Molecular Cancer. 2019;18(1):174. doi: 10.1186/s12943-019-1105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Regué L., Ji F., Flicker D., Kramer D., Pierce W., Davidoff T. IMP2 increases mouse skeletal muscle mass and voluntary activity by enhancing autocrine IGF2 production and optimizing muscle metabolism. Molecular and Cellular Biology. 2019 doi: 10.1128/MCB.00528-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regué L., Minichiello L., Avruch J., Dai N. Liver-specific deletion of IGF2 mRNA binding protein-2/IMP2 reduces hepatic fatty acid oxidation and increases hepatic triglyceride accumulation. Journal of Biological Chemistry. 2019;294(31):11944–11951. doi: 10.1074/jbc.RA119.008778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H.B., Muhammad T., Guo Y., Li M., Sha Q., Zhang C. RNA-binding protein IGF2BP2/IMP2 is a critical maternal activator in early zygotic genome activation. Advancement of Science. 2019;6(15):1900295. doi: 10.1002/advs.201900295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang S., Wu Z., Cheng Y., Wei W., Hao L. Insulin-like growth factor 2 mRNA binding protein 2 promotes aerobic glycolysis and cell proliferation in pancreatic ductal adenocarcinoma via stabilizing GLUT1 mRNA. Acta Biochimica et Biophysica Sinica. 2019;51(7):743–752. doi: 10.1093/abbs/gmz048. [DOI] [PubMed] [Google Scholar]
- 34.Gong C., Li Z., Ramanujan K., Clay L., Zhang Y., Lemire-Brachat S. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Developmental Cell. 2015;34(2):181–191. doi: 10.1016/j.devcel.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang H., Weng H., Sun W., Qin X., Shi H., Wu H. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nature Cell Biology. 2018;20(3):285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conway A.E., Nostrand E.L., Pratt G.A., Aigner S., Wilbert M.L., Sundararaman B. Enhanced CLIP uncovers IMP protein-RNA targets in human pluripotent stem cells important for cell adhesion and survival. Cell Reports. 2016;15(3):666–679. doi: 10.1016/j.celrep.2016.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y., Shen F., Huang W., Qin S., Huang J., Sergi C. Glucose is involved in the dynamic regulation of m6A in patients with type 2 diabetes. Journal of Clinical Endocrinology & Metabolism. 2019;104(3):665–673. doi: 10.1210/jc.2018-00619. [DOI] [PubMed] [Google Scholar]
- 39.Fischer J., Koch L., Emmerling C., Vierkotten J., Peters T., Brüning J.C. Inactivation of the Fto gene protects from obesity. Nature. 2009;458(7240):894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 40.De Jesus D.F., Zhang Z., Kahraman S., Brown N.K., Chen M., Hu J. m(6)A mRNA methylation regulates human beta-cell biology in physiological states and in type 2 diabetes. Nature Metabolism. 2019;1(8):765–774. doi: 10.1038/s42255-019-0089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fuchsberger C., Flannick J., Teslovich T.M., Mahajan A., Agarwala V., Gaulton K. The genetic architecture of type 2 diabetes. Nature. 2016;536(7614):41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manning B.D., Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169(3):381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ravassard P., Hazhouz Y., Pechberty S., Bricout-Neveu E., Armanet M., Czernichow P. A genetically engineered human pancreatic beta cell line exhibiting glucose-inducible insulin secretion. Journal of Clinical Investigation. 2011;121(9):3589–3597. doi: 10.1172/JCI58447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Modi H., Cornu M., Thorens B. Glutamine stimulates biosynthesis and secretion of insulin-like growth factor 2 (IGF2), an autocrine regulator of beta cell mass and function. Journal of Biological Chemistry. 2014;289(46):31972–31982. doi: 10.1074/jbc.M114.587733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sacco F., Seeling A., Humphrey S.J., Krahmaer N., Volta F., Reggio A. Phosphoproteomics reveals the GSK3-PDX1 Axis as a key pathogenic signaling node in diabetic islets. Cell Metabolism. 2019;29(6):1422–1432 e3. doi: 10.1016/j.cmet.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 46.Dai N., Ji F., Wright J., Minichiello L., Sadreyev R., Avruch IGF2 mRNA binding protein-2 is a tumor promoter that drives cancer proliferation through its client mRNAs IGF2 and HMGA1. Elife. 2017;6 doi: 10.7554/eLife.27155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng Y., Ley S.H., Hu F.B. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nature Reviews Endocrinology. 2018;14(2):88–98. doi: 10.1038/nrendo.2017.151. [DOI] [PubMed] [Google Scholar]
- 48.Zeng W.J., Lu C., Shi Y., Wu C., Chen X., Li C. Initiation of stress granule assembly by rapid clustering of IGF2BP proteins upon osmotic shock. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2020;1867(10):118795. doi: 10.1016/j.bbamcr.2020.118795. [DOI] [PubMed] [Google Scholar]
- 49.Udler M.S., Kim J., Grotthuss M.V., Bonàs-Guarch S., Cole J.B., Chiou J. Type 2 diabetes genetic loci informed by multi-trait associations point to disease mechanisms and subtypes: a soft clustering analysis. PLoS Medicine. 2018;15(9) doi: 10.1371/journal.pmed.1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen B.H., Hivert M., Peters M.J., Pilling L.C., Hogan J.D., Pham L.M. Peripheral blood transcriptomic signatures of fasting glucose and insulin concentrations. Diabetes. 2016;65(12):3794–3804. doi: 10.2337/db16-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujitani Y. Transcriptional regulation of pancreas development and beta-cell function. Endocrine Journal. 2017;64(5):477–486. doi: 10.1507/endocrj.EJ17-0098. [Review] [DOI] [PubMed] [Google Scholar]
- 52.Modi H., Jacovetti C., Tarussio D.T., Metref S., Madsen O., Zhang F.P. Autocrine action of IGF2 regulates adult beta-cell mass and function. Diabetes. 2015;64(12):4148–4157. doi: 10.2337/db14-1735. [DOI] [PubMed] [Google Scholar]
- 53.Buchwald P., Bernal A., Echeverri F., Tamayo-Garcia A., Linetsky E., Ricordi C. Fully automated islet cell counter (ICC) for the assessment of islet mass, purity, and size distribution by digital image analysis. Cell Transplantation. 2016;25(10):1747–1761. doi: 10.3727/096368916X691655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.