Highlights
-
•
27 SOX (high-mobility group HMG-box) genes were identified in the C. carp genome.
-
•
SOX genes ranging from 3496 (SOX6) to 924bp (SOX17b) which coded with putative protein series from 307 to 509 amino acids.
-
•
Gene ontology revealed SOX proteins maximum involvement is in metabolic process 49.796 %.
-
•
Chromosomal location and synteny analysis display all SOX gene are located on different chromosomes.
Keywords: Common carp, Sex determination, SOX genes, Genome wide analysis, Fishes, Conserved domains, Protein motif, Syntenic analysis, Phylogenetic tree
Abstract
Common carp (Cyprinus carpio) is a commercial fish species valuable for nutritious components and plays a vital role in human healthy nutrition. The SOX (SRY-related genes systematically characterized by a high-mobility group HMG-box) encoded important gene regulatory proteins, a family of transcription factors found in a broad range of animal taxa and extensively known for its contribution in multiple developmental processes including contribution in sex determination across phyla. In our current study, we initially accomplished a genome-wide analysis to report the SOX gene family in common carp fish based on available genomic sequences of zebrafish retrieved from gene repository databases, we focused on the global identification of the Sox gene family in Common carp among wide range of vertebrates and teleosts based on bioinformatics tools and techniques and explore the evolutionary relationships. In our results, a total of 27 SOX (high-mobility group HMG-box) domain genes were identified in the C. carp genome. The full length sequences of SOX genes ranging from 3496 (SOX6) to 924bp (SOX17b) which coded with putative proteins series from 307 to 509 amino acids and all gene having exon number expect SOX9 and SOX13. All the SOX proteins contained at least one conserved DNA-binding HMG-box domain and two (SOX7 and SOX18) were found C terminal. The Gene ontology revealed SOX proteins maximum involvement is in metabolic process 49.796 %, average in biological regulation 45.188 %, biosynthetic process (19.992 %), regulation of cellular process 39.68, 45.508 % organic substance metabolic process, multicellular organismal process 23.23 %,developmental process 21.74 %, system development 16.59 %, gene expression 16.05 % and 14.337 % of RNA metabolic process. Chromosomal location and syntanic analysis show all SOX gene are located on different chromosomes and apparently does not fallow the unique pattern. The maximum linkage of chromosome is (2) on Unplaced Scaffold region. Finally, our results provide important genomic suggestion for upcoming studies of biochemical, physiological, and phylogenetic understanding on SOX genes among teleost.
1. Introduction
Sex determination (SD) and differentiation pattern are rudimentary developmental processes, which have been commonly seen with transcriptional factors between wide range diversity of vertebrates and invertebrates [1]. Different genetic and environmental factors are involved to facilitate the tools and technologies for investigation of sex determination mechanism and differentiation patterns. Among vertebrates, gonadal differentiation and determination are accomplished by studying the interaction between complex networks of transcription factors [2]. High diversity of SD mechanisms observed in fish is connected to the high turnover rate of their sex chromosomes, that’s why they are considered as young and labelled as homomorphic. Sex chromosomes are interrelated with gonadal differentiation and their turnover, which turn to appear with new master genes for sex determination [3]. Approximately half of the vertebrates are includes with fishes and shows great variation in sex determination process [3], [4]. In vertebrates including fish, genes are reported related to sex determination mechanisms including AMHR2, GDF6, DMRT genes having conserved DNA binding DM domains [5], and SOX gene family with HMG domain (high-mobility group) play important role in sex determination and differentiation.
The SOX gene family is a group of evolutionary transcriptional factors that involves in various developmental process, not only in sex determination and sex differentiation process but also the formation of multiple organs, endoderm development, eyes [6], [7] angiogenesis, gonad [8], [9], chondrogenesis, neurogenesis [10], [11], [12], cardiogenesis [13], [14], cartilage [15], [16] and pancreas [17], [18]. Consideration of SOX genes identification was initiative step with the discovery of Sry (testis determining factor) in mammals [19], [20] which brings out high mobility group HMG domain specifically binds with DNA sequences [21]. SOX genes having 80 % sequence identity with HMG domain and commonly conserved in SOX 1 to SOX 32. With the facility of whole-genome sequencing (WGS) and genome wide characterization (GWC), more than 40 members of SOX family and diverse varieties of proteins have been identified and analyzed in mammals, birds, amphibians, reptiles, insects and fishes [22], [23]. For instance, earlier researcher report the variability of SOX genes have been found in various organism like 8 SOX genes in drosophila, 19 SOX genes in Japanese medaka, over 20 SOX genes in mouse and human [24], and 27 SOX genes in Nile tilapia [25].
The SOX gene family is divide in different groups and subgroups among higher vertebrates and teleost based on similarity of DNA or protein sequences [23], [26], [28]. Earlier studies on growth and development of teleost fishes have explored the potential roles of SOX genes for example SOX1, SOX2, and SOX3 proteins sequence bind specifically with DNA via HMG domain and act as a transcription factor or involves in protein-protein interactions such as POU proteins binding with other proteins [29]. The SOX1 and SOX3 has a C-terminal region, which facilitate it to act as a transcriptional factor by developing protein-protein complex and sox1 expressed in development process for formation of neural plate [30]. The expression of Sox2 and SOX3 help in maintaining the identity of progenitor cell by inhibiting neurogenesis [29]. In mammals and fish sox6 act as transcription factor to controls the identity of skeletal muscle fiber [31]. The Sox8, sox9 and sox 10 involved in many developmental processes like testis development maintenance of male fertility, humans early developing sate of gonads, expression in somatic cells and sex determination with the help of DMRT1 gene [32], [33]. Sox9 is an essential transcription regulatory factor for the development of adult cartilage and activates the genes transcriptional factor for structural components [34]. In same contrast the many members of SOX gene families are identified and able to share some common biochemical properties and function overlapping in different biological contexts Sarkar et al., 2013. Among all available types of SOX genes, SOX9 is known to be most important transcription factor necessary for Sex differentiation in vertebrates and have two variant forms SOX9a and SOX 9b in teleost’s [2]. Documentation of comparative genomic involves in evaluation and analysis of gene sequences and regulatory regions between different species, and provides a novel approach to identify the Common carp SOX genes. Computational approach for whole genome analysis is becoming more common not only for biological science but also for aquatic biotechnology research topics.
Our study identifies the presence of SOX genes in the common carp (Cyprinus carpio) to indicate the difference between genes among different species and to provide genomic resources on the very best SOX genes for future work. Our aim of this present study were to recognize the sufficiency of the SOX gene family in common carp (Cyprinus carpio) fish, associated with the gene divergence between different species having diverse environmental factors and to provide genomic resources for future work on SOX genes. Here we utilized all the accessible gene resources from the model fish and reported the whole genome identification of SOX gene family in common carp (Cyprinus carpio) fish. Prediction of the structure of the sequences and functional domains of the SOX genes will be carried out, followed by a phylogenetic and structural analysis. Our systematic study of the SOX genes will also provide some basic genomic resources that may be remain available to better understand the evolutionary and physiological aspects of whole post-genome duplication (WGD) in common carp.
However, genome-wide analyses on SOX gene family are scarce in fish
2. Material and methods
The availability of all SOX genes in Zebrafish (Danio rerio) genome provide the facility to check the candidate genes downloaded from Ensembl (https://asia.ensembl.org/Danio_rerio/Info/Index) given in (Table 1). The Zebrafish genes were used as query sequences to search against the Common carp (Cyprinus carpio) genome accessible in NCBI genome database (https://www.ncbi.nlm.nih.gov/genome) for identifying candidate genes. We used different strategies on available genomic resources of Common Carp including whole genome sequences and retrieve cDNAs by BLAST searches with 1e-10 E values, according to the earlier reports [20], [35]. Then, reciprocal BLAST searches were conducted using the candidate common carp SOX genes as queries, to confirm the accuracy of the candidate genes. Furthermore, the coding sequences were confirmed by BLASTN searches against the NCBI non-redundant protein sequence database (nr). Subsequently, the conserved HMG-box (PDOC00305) domain was searched against the local database by BLASTp program [36], and manually deletes irrelevant sequences. The SOX proteins sequences from other organisms were retrieved from the NCBI (https://www.ncbi.nlm.nih.gov/) genome databases using BLAST program with an threshold of 1e-10 and mapped all protein genes on individual chromosomal locations. The Pfam (https://pfam.xfam.org/), SMART (http://smart.embl-heidelberg.de/), and other databases were utilized to check the candidate sequences that contained HMG-box domains [37].
Table 1.
Summary of SOX genes in Danio rerio: All available SOX genes of zebrafish (Danio rerio) downloaded from Ensembl (http://asia.ensembl.org/index.html) were used as query sequences to search against the Common carp (Cyprinus carpio). Table consistent of Accession number of genes, name of Sox genes, Genomic length(bp), CDS (bp), CDS of amino acids, No. of exons, Chromosomes, Genome location and Assembly of organism amiable for open access. In above mentioned table chromosomes 7 contain highest number of SOX gene like (SOX6, SOX7, SOX9b and SOX32), chromosome 3 consist of three SOX genes (SOX8b, SOX9b and SOX10), Chromosomes 11 have only two SOX genes (SOX12 and SOX12), and reaming all SOX genes are consist of individual chromosomes.
| Gene ID | Sox genes | Genomic length(bp) | CDS (bp) | CDS (aa) | CDS status | No. of exons | Chromosomes Number | Genome location | Assembly |
|---|---|---|---|---|---|---|---|---|---|
| ZDB-GENE-040718-186 | Sox1a | 1945 | 1010 | 336 | Complete | 1 | 9 | NC_007120.7 (21722734..21724661) | GRCz11 |
| ZDB-GENE-060322-5 | Sox1b | 1773 | 1022 | 340 | Complete | 1 | 1 | NC_007112.7 (46194333..46196106) | GRCz11 |
| ZDB-GENE-030909-1 | Sox2 | 2120 | 947 | 315 | Complete | 1 | 22 | NC_007133.7 (37347893..37349978) | GRCz11 |
| ZDB-GENE-980526-333 | Sox3 | 1781 | 902 | 300 | Complete | 1 | 14 | NC_007125.7 (32742701..32744464) | GRCz11 |
| ZDB-GENE-030131-8290 | Sox4a | 3350 | 1091 | 363 | Complete | 1 | 19 | NC_007130.7 (28786149..28789409) | GRCz11 |
| ZDB-GENE-040426-1274 | Sox4b | 3015 | 1028 | 342 | Complete | 2 | 16 | NC_007127.7 (68069..71110) | GRCz11 |
| ZDB-GENE-000607-13 | Sox5 | 2393 | 2279 | 759 | Complete | 30 | 4 | NC_007115.7 (16981414..17244201) | GRCz11 |
| ZDB-GENE-081120-6 | Sox6 | 1242 | 410 | 136 | Incomplete | 14 | 7 | NC_007118.7 (27320462..27449391) | GRCz11 |
| ZDB-GENE-040109-4 | Sox7 | 2246 | 1172 | 390 | Complete | 2 | 20 | NC_007131.7 (19075127..19078424) | GRCz11 |
| ZDB-GENE-130530-719 | Sox8a | 2000 | 1205 | 401 | Complete | 4 | 12 | NC_007123.7 (1072485..1085503) | GRCz11 |
| ZDB-GENE-031114-1 | Sox8b | 1934 | 1076 | 358 | Complete | 3 | 3 | NC_007114.7 (62397711..62403488) | GRCz11 |
| ZDB-GENE-001103-1 | Sox9a | 1790 | 1,388 | 462 | Complete | 3 | 12 | NC_007123.7 (1947593..1951233) | GRCz11 |
| ZDB-GENE-001103-2 | Sox9b | 1916 | 1232 | 407 | Complete | 3 | 3 | NC_007114.7 (62522542..62527667) | GRCz11 |
| ZDB-GENE-011207-1 | Sox10 | 3231 | 1457 | 485 | Complete | 4 | 3 | NC_007114.7 (1492174..1501252) | GRCz11 |
| ZDB-GENE-980526-395 | Sox11a | 3280 | 1064 | 354 | Complete | 1 | 17 | NC_007128.7 (35878547..35881807) | GRCz11 |
| ZDB-GENE-980526-466 | Sox11b | 2666 | 1106 | 368 | Complete | 1 | 20 | NC_007131.7 (30032636..30035292) | GRCz11 |
| ZDB-GENE-040724-33 | Sox12 | 1401 | 1067 | 355 | Complete | 2 | 11 | NC_007122.7 (24190164..24191928) | GRCz11 |
| ZDB-GENE-100519-1 | Sox13 | 5380 | 1790 | 596 | Complete | 14 | 11 | NC_007122.7 (37768409..37831781) | GRCz11 |
| ZDB-GENE-051113-268 | Sox14 | 1935 | 716 | 238 | Complete | 1 | 6 | NC_007117.7 (26557979..26559883) | GRCz11 |
| ZDB-GENE-991213-1 | Sox17 | 1705 | 1241 | 413 | Complete | 2 | 7 | NC_007118.7 (58773148..58776152) | GRCz11 |
| ZDB-GENE-080725-1 | Sox18 | 2545 | 1295 | 331 | Complete | 2 | 23 | NC_007134.7 (8797002..8800754) | GRCz11 |
| ZDB-GENE-980526-102 | Sox19a | 2181 | 893 | 297 | Complete | 2 | 5 | NC_007116.7 (24199180..24201444) | GRCz11 |
| ZDB-GENE-010111-1 | Sox19b | 2556 | 881 | 293 | Complete | 2 | 7 | NC_007118.7 (26494585..26497947) | GRCz11 |
| ZDB-GENE-990715-6 | Sox21a | 1098 | 719 | 239 | Complete | 1 | 6 | NC_007117.7 (7414273..7415343) | GRCz11 |
| ZDB-GENE-040429-1 | Sox21b | 1374 | 737 | 245 | Complete | 1 | 9 | NC_007120.7 (53276356..53277702) | GRCz11 |
| ZDB-GENE-011026-1 | Sox32 | 1222 | 923 | 307 | Complete | 2 | 7 | NC_007118.7 (58824526..58826178) | GRCz11 |
2.1. Gene characterization and structure
To characterize the common carp genes structures, compare them with their orthologs in zebrafish (Danio rerio) and human (Homo sapiens) genome, we first accomplished exon-intron structure analysis with online analysis tool Gene Structure Display Server 2.0 (http://gsds.cbi.pku.edu.cn/). The Multiple Motif Elicitation for Motif Elicitation (MEME) 5.05 (http://meme-suite.org/tools/meme) used to recognized conserved motif regions of protein sequences. MEME (Multiple Ex-pectation-Maximization for Motif Elicitation) is online suite containing with motif discovery and searching tools [38]. We use default setting of MEME tools for performing our motif analysis with maximum 10 different ranges of motif on individual sequence among 50 optimum widths for each motif.
2.2. Proteomics analysis of SOX gene family
The size of individual protein sequences, molecular weight (MW), extinction co-efficient and theoretical isoelectric point (pI) of each SOX protein were calculated by using the online tool ProtParam (http://www.protparam.net/index.html), the web based ExPASy Proteomics Server and sequence analysis performed on PSIPRED 4.0 (http://bioinf.cs.ucl.ac.uk/psipred/) server. Discovered MEME motifs were searched Expasy-Prosite database in ScanProsite (https://prosite.expasy.org/scanprosite/) tool linked with knowledge based database was used to predict the conserved domain (CD) based on sequence homology (SH), which was further confirm or analyzed by BLAST with N program (https://blast.ncbi.nlm.nih.gov/Blast).
2.3. Protein-protein and Gene Ontology (GO) of SOX genes
Here we obligate to carry out direct (Physical) or indirect (Functional) connotation of protein-protein network interactions (PPNIs) for all SOX genes identified in common carp genome was done with the help of online available databases like STRIG (Search Tool for the Retrieval of Interacting Genes/Proteins) (https://string-db.org/), Pfam (https://pfam.xfam.org/), and SMART (http://smart.embl-heidelberg.de/) etc. or interactively represent using genemania online available desktop based software and tools.
2.4. Chromosome mapping and synteny analysis
For physical map of individual chromosomes protocol is design to find the actual location, data of all SOX genes (Common carp, Zebrafish and Human) was downloaded from NCBI (https://www.ncbi.nlm.nih.gov/) and Ensemble (http://asia.ensembl.org/index.html) databases. For the graphical demonstration of linkage maps and quantitative trait locus (QTLs) used Mapchart 2.32 software to map onto the individual chromosomes of common carp. The syntenic analysis of SOX paralogs pairs were retrieve or identified by searching the gene duplication among all the species at NCBI and Ensembl database and Map individually with map chart software.
2.5. Sequence alignment and phylogenetic analysis
The DNA sequences for HMG box, all SOX genes of Common carp genes were aligned on Clustal Omega (https://www.ebi.ac.uk) by online available software T-Coffee (http://tcoffee.crg.cat) with default parameters were extracted base on the SMART analysis [37]. The amino acid sequences of protein from 10 species including Human, Mouse, Dog, and Chimpanzee, Nile tilapia, gold fish, Medaka, Rainbow trout, Guppy, and Atlantic salmon were aligned with ClustalX program [39]. For phylogenetic analysis neighbor-joining method was conducted with reference of SOX proteins from Common Carp to construct phylogenetic tree with a bootstrap test of 1000 replicates by using MEGA 7.0 program [40]. For interactive representation of both tree we used interactive Tree Of Life (iTOL) online available software (https://itol.embl.de) to display phylogenetic tree with different color coded ranges [41].
3. Results
3.1. Identification of SOX genes in common carp
We identified a total of 27 SOX genes in the common carp genome using all available genomic resources, including SOX1a, SOX1b, SOX2, SOX3, SOX4a, SOX4b, SOX5, SOX6, SOX7, SOX8a, SOX8b, SOX9, SOX9a, SOX9b, SOX10, SOX10a, SOX10b, SOX11a, SOX11b, SOX13, SOX14, SOX17b, SOX18, SOX19a, SOX19b, SOX21a and SOX21b from Zebrafish given in (Table 2). The Complete material and evidence on their corresponding genomic sequences, coding sequences (CDS) and total number of exons is précised in Table 2. The maximum genomic length of SOX6 was 3496bps and coding region of SOX9 and SOX18 were 1373bp or 1328bp which encodes 457 and 442 amino acids, in detail we categorized the coding sequence according to length of nucleotide sequences which included 1000pb genes are in cluster one SOX1a (1010), SOX1b (1007), SOX4a (1046), SOX11a (1061), SOX11b (1064), the genes start from 1100bp are in second cluster SOX10b (1151), SOX10a (1145), SOX13 (1187), SOX7 (1193), third cluster include with two genes having diverse length of coding sequence SOX8a (1265) and SOX9b (1316) and at the end fourth cluster start for minimum genomic length of coding sequences from 413 to maximum length 944. The SOX9 and SOX13 has no exon number, SOX1a, SOX2, SOX3, SOX4a, SOX4b, SOX10, SOX21a, SOX21b individual genes has one exons respectively, SOX7, SOX11a, SOX11b, SOX18, SOX19a, SOX19b having same exon number such as two, SOX8a, SOX8b, SOX9a, SOX9b, SOX10a, SOX10b, SOX14, SOX17b has three exons. The SOX6 has four exons and SOX5, SOX1b have same exon number i.e. 6. We observe that the genes of SOX families were located individual chromosome such as SOX 8a on chromosome number 5, SOX19a (9), SOX14 (10), SOX13 (22), SOX9a (23), SOX9b (30), SOX9 (31), SOX3 (35), SOX8b (41), SOX2 (44), and remaining all are located on Unplaced Scaffold region of common carp genome. The comparative analysis of common carp SOX genes with other higher vertebrates (Human, Mouse, Dog, and Chimpanzee) and fishes (Nile tilapia, gold fish, Medaka, Rainbow trout, Guppy, and Atlantic salmon) are given in (Table 3). The Presence of SOX genes on Chromosomal position of individual species of teleost and other vertebrate were identified and given in (Table 4). The chromosome with star (*) indicate the presence of highly conserved gene and similar variant present on chromosome place (Table 5).
Table 2.
Summery of all SOX genes in Cyprinus carpio: Resulted SOX genes in Common carp (Cyprinus carpio) based on query sequence of all available SOX genes of zebrafish (Danio rerio) in Ensembl database online available for public access. Table contain with information of Accession number of genes, name of Sox genes, Genomic length(bp), CDS (bp), CDS of amino acids, No. of exons, Chromosomes, Genome location and Assembly of organism amiable for open access. In which above mentioned table we observe that all SOX genes are consistent of induvial chromosomes and reaming all are on unplaced region.
| AN | Sox genes | Genomic length(bp) | CDS (bp) | CDS (aa) | No. of exons | Chromosomes Number | Genome location | Assembly |
|---|---|---|---|---|---|---|---|---|
| XM_019075156.1 | Sox1a | 2209 | 1010 | 336 | 1 | Unplaced | NW_017538026.1 (90488..92696) | GCF_000951615.1 |
| XM_019072343.1 | Sox1b | 1357 | 1028 | 342 | 1 | Unplaced | NW_017537908.1 (994268..995625, | GCF_000951615.1 |
| XM_019065499.1 | Sox2 | 2120 | 947 | 315 | 1 | 44 | NC_031740.1 (10639735..106418iiiii53) | GCF_000951615.1 |
| XM_019122086.1 | Sox3 | 1505 | 899 | 299 | 1 | 35 | NC_031731.1 (13437760..13439264) | GCF_000951615.1 |
| XM_019079512.1 | Sox4a | 1670 | 1046 | 348 | 1 | Unplaced | NW_017538201.1 (270388..272057) | GCF_000951615.1 |
| XM_019069830.1 | Sox4b | 1503 | 929 | 309 | 1 | Unplaced | NW_017537801.1 (274672..276174) | GCF_000951615.1 |
| XM_019089698.1 | Sox5 | 925 | 818 | 272 | 6 | Unplaced | NW_017540372.1 (84204..107152 | GCF_000951615.1 |
| XM_019083302.1 | Sox6 | 3496 | 509 | 169 | 4 | Unplaced | NW_017538301.1 (846107..850788, | GCF_000951615.1 |
| XM_019069299.1 | Sox7 | 1571 | 1193 | 397 | 2 | Unplaced | NW_017537780.1 (246443..249212) | GCF_000951615.1 |
| XM_019064819.1 | Sox8a | 2568 | 1265 | 421 | 3 | 5 | NC_031701.1 (904555..907715) | GCF_000951615.1 |
| XM_019063104.1 | Sox8b | 962 | 944 | 314 | 3 | 41 | NC_031737.1 (4710056..4713500) | GCF_000951615.1 |
| DQ201318.1 | Sox9 | 2107 | 1373 | 457 | 0 | 31 | NC_031727.1 (12689194..12693097 | (GCF_000951615.1) |
| XR_002019411.1 | Sox9a | 2906 | 637 | 457 | 3 | 23 | NC_031719.1 (41616..44965 | GCF_000951615.1 |
| XM_019117253.1 | Sox9b | 2534 | 1316 | 438 | 3 | 30 | NC_031726.1 (7371120..7374563) | GCF_000951615.1 |
| XM_019092904.1 | Sox10 | 523 | 443 | 147 | 1 | Unplaced | NW_017541515.1 (421..943) | GCF_000951615.1 |
| MF573939.1 | Sox10a | 3221 | 1145 | 481 | 3 | Unplaced | NW_017543385.1 (111150..116572 | (GCF_000951615.1) |
| MF538663.1 | Sox10b | 3167 | 1151 | 483 | 3 | Unplaced | NW_017543385.1 (111150..116572 | GCF_000951615.1 |
| XM_019096628.1 | Sox11a | 1271 | 1061 | 353 | 2 | Unplaced | NW_017542566.1 (213843..215409) | GCF_000951615.1 |
| XM_019096626.1 | Sox11b | 1728 | 1064 | 354 | 2 | Unplaced | NW_017542566.1 (357335..359362) | GCF_000951615.1 |
| XM_019083671.1 | Sox13 | 1341 | 1187 | 395 | – | 22 | NC_031718.1 (7903690..7921638) | GCF_000951615.1 |
| XM_019104301.1 | Sox14 | 2000 | 716 | 238 | 3 | 10 | NC_031706.1 (7744712..7748222 | GCF_000951615.1 |
| XM_019086643.1 | Sox17b | 924 | 923 | 307 | 3 | Unplaced | NW_017539346.1 (376..1868) | GCF_000951615.1 |
| XM_019069992.1 | Sox18 | 1609 | 1328 | 442 | 2 | Unplaced | NW_017537809.1 (403527..406325) | GCF_000951615.1 |
| XM_019097725.1 | Sox19a | 1345 | 890 | 296 | 2 | 9 | NC_031705.1 (5973083..5974515) | GCF_000951615.1 |
| XM_019075219.1 | Sox19b | 1881 | 869 | 289 | 2 | Unplaced | NW_017538029.1 (162955..165835 | GCF_000951615.1 |
| XM_019072662.1 | Sox21a | 1066 | 716 | 238 | 1 | Unplaced | NW_017537923.1 (191505..192570 | GCF_000951615.1 |
| XM_019095743.1 | Sox21b | 2310 | 728 | 242 | 1 | Unplaced | NW_017542346.1 (20482..22791 | GCF_000951615.1 |
Table 3.
Comparative analysis of SOX genes of “Cyprinus carpio” with vertebrate’s species: in our finding Star (*) indicate that highly conserved variant with gene having 90 to 100 similarity.
| Gene Name | Zebrafish | Common carp | Nile tilapia | Gold fish | Medaka | Rainbow trout | Guppy | Atlantic salmon | Chimpanzee | Human | Mouse | Dog |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sox-1a | 1 | 2 | 1 + 2* | 2 | 1 + 1* | 4 | 1 + 4* | 2 + 3* | 1 | 1* | 1* | 1* |
| Sox-1b | 1 | 2 | 1 + 2* | 2 | 1 + 1* | 4* | 1 + 2* | 3 + 2* | 1 | 1* | 1* | 1* |
| Sox2 | 1 | 2 | 1 | 1 + 1* | 1 | 1 + 1* | 1 + 1* | 1 + 1* | 1 | 1 | 1 | 1 |
| Sox3 | 1 | 1 | 1 | 1 + 1* | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 1 |
| Sox4a | 1 | 1 + 1* | 1 + 1* | 1 + 4* | 2* | 2 + 4* | 1 + 1* | 5* | 1 | 1* | 1 | 1* |
| Sox4b | 1 | 1 + 1* | 1 + 1* | 1 + 4* | 1 + 1* | 2 + 4* | 1 + 1* | 5* | 1 | 1* | 1 | 1* |
| Sox5 | 1 | 3 + 1* | 1 + 4* | 4 + 4* | 1 + 9* | 2 | 1 + 8* | 2 + 20* | 7* | 1 + 3* | 1 + 20* | 1 + 6* |
| Sox6 | 1 | 3 | 1 + 8* | 2 + 5* | 1 | 3 | 2 + 3* | 4 + 2* | 21* | 1 + 2* | 1 + 11* | 17* |
| Sox7 | 1 | 3 | 1 | 4 | 1 + 1* | 1 + 1* | 1 + 1* | 3 | 1 | 1 | 1 | 1 |
| Sox8a | 1 | 1 | 1 + 1* | 1 + 3* | 1 | 1 + 2* | 2* | 1 + 5* | 1 | 1* | 1* | 1* |
| Sox8b | 1 | 1 | 1 + 1* | 1 + 3* | 1* | 1 + 2* | 2* | 1 + 5* | 1 | 1* | 1* | 1* |
| Sox9 | 1 | 1 | 2 | 2 + 3* | 1 | 4 + 2* | 1 + 1* | 5* | 1 | 1 | 1 | 1 |
| Sox9a | 1 | 1 | 1 + 1* | 2 + 3* | 2* | 2* | 1 + 1* | 2 + 2* | 1 | 1* | 1* | 1* |
| Sox9b | 1 | 2 | 1 + 1* | 2 + 3* | 2* | 2* | 2* | 1 + 4* | 1 | 1* | 1* | 1* |
| Sox10 | 1 | 3 | 3 | 3 | 1 + 2* | 4 | 1 | 4 | 1 | 1 | 1 | 1 |
| Sox 10a | 1 | 1* | 1 + 1* | 1 + 2* | 1 + 2* | 4* | 2* | 4* | 1 | 1* | 1* | 1 |
| Sox 10b | 1 | 1* | 1 + 1* | 1 + 2* | 1 + 2* | 4* | 1 + 1* | 4* | 1 | 1* | 1* | 1 |
| Sox11a | 1 | 1* | 1 + 1* | 4 + 2* | 1 | 3* | 1 + 1* | 3* | 1 | 1* | 1* | 0 |
| Sox11b | 1 | 1* | 1 + 1* | 4 + 2* | 1 | 3* | 1 + 1* | 3* | 1 | 1* | 1* | 0 |
| Sox13 | 1 | 3 | 1 | 1 + 1* | 1 | 1 | 1 | 2 + 3* | 6* | 2 | 1 + 4* | 1 |
| Sox14 | 1 | 3 | 2 | 3 | 1 + 1* | 1 | 1 + 1* | 1 + 1*2 | 1 | 1 | 1 | 1 |
| Sox17b | 1 | 1 | 1 + 1* | 2* | 1* | 1* | 1 + 1* | 4* | 1 | 1* | 2* | 1 |
| Sox18 | 1 | 2 | 1 | 2 | 0 | 2 | 1 | 1 + 1* | 1 | 1 | 1 | 1 |
| Sox19a | 1 | 1 | 1 | 2 + 2* | 1* | 2 + 2* | 1* | 2 + 2* | 0 | 0 | 0 | 0 |
| Sox19b | 1 | 2 | 1 | 2 + 2* | 1 | 1 + 3* | 1 | 1 + 3* | 0 | 0 | 0 | 0 |
| Sox21a | 1 | 2 | 1 | 2 + 2* | 1* | 2* | 1* | 2* | 1 | 1* | 1 | 0 |
| Sox21b | 1 | 1* | 1 | 2 + 2* | 1 | 1 + 1* | 1 | 2 | 1 | 1* | 1 | 0 |
| Total | 27 | 41 | 31 | 53 | 18 | 35 | 22 | 35 | 21 | 11 | 14 | 12 |
Table 4.
Presence of SOX genes on Chromosomes of individual species: chromosome with star (*) indicate the presence of highly conserved and similar variant present on mentioned chromosome place.
| Gene Name | Zebrafish | Common carp | Nile tilapia | Gold fish | Medaka | Rainbow trout | Guppy | Atlantic salmon | Human | Mouse | Dog |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sox-1a | 9 | Unplaced | LG16 | 9 | 21 | 22 | LG2 | 16 | 13* | 8* | 0 |
| 18 | 25 | ||||||||||
| Sox-1b | 1 | Unplaced | LG23 | up | 2 | 3 | LG23* | 17 | 13* | 8* | 0 |
| 6 | 7 | ||||||||||
| Sox2 | 22 | 44 | LG17 | 47 | 4 | 4 | LG4 | 23 | 3 | 3 | 34 |
| Sox3 | 14 | 35 | 39 | 10 | 14 | LG10 | 9 | X | x | X | |
| 25 | 5 | ||||||||||
| Sox4a | 19 | Unplaced | LG22 | 19 | 16* | 14 | LG16 | 12* | 6* | 13* | 35 |
| 14* | |||||||||||
| Sox4b | 16 | Unplaced | LG11 | 16 | 16 | 18 | LG11 | 12* | 6* | 13* | 27 |
| 41* | |||||||||||
| Sox5 | 4 | Unplaced | LG17 | 4 | 23 | 21 | LG13 | 17 | 12 | 6 | 27 |
| 29* | 7 | ||||||||||
| Sox6 | 7 | Unplaced | LG 1 | 7 | 6 | 2 | LG3 | 11 | 11 | 7 | 21 |
| 30* | 3* | 4 | 26 | ||||||||
| Sox7 | 20 | Unplaced | LG 15 | 20 | 8 | LG21 | 6 | 8 | 14 | ||
| 45* | 15 | ||||||||||
| Sox8a | 12 | 5 | LG 8 | 28 | 8 | 13 | LG8* | 28 | 16* | 17* | 8* |
| 23 | |||||||||||
| Sox8b | 3 | 41 | LG 4 | LG37 | 20 | LG8* | 3 | 16* | 17* | 8* | |
| Sox9 | 31 | LG 4 | 28 | 8 | 13 | LG8 | 28* | 17 | 11 | 9 | |
| Sox9a | 12 | 23 | LG 4 | 12 | 8* | 13* | LG8 | 6 | 17* | 11* | 9* |
| 37* | |||||||||||
| Sox9b | 3 | 30 | LG 8 | 12* | 8* | 13* | LG8* | 19 | 17* | 11* | 9* |
| Sox10 | 3 | Unplaced | 3 | 8 | 13 | LG8 | 3 | 22 | 15 | 10 | |
| Sox 10a | 3 * | Unplaced | LG 6 | LG28* | 1 | 12* | LG8* | 12* | 22* | 15* | 10* |
| Sox 10b | 3* | Unplaced | LG 4 | LG28* | 8 | 17* | LG8* | 3* | 22* | 15* | 10* |
| Sox11a | 17 | Unplaced | LG 19 | 17* | 22 | 17* | LG22 | 1* | 2* | 0 | 38* |
| Sox11b | 20 | Unplaced | LG 15 | 45* | 22 | 19* | LG21 | 15* | 2* | 0 | 38* |
| Sox13 | 11 | 22 | LG 5 | 11 | 5 | 17 | LG5 | 22 | 1 | 1 | 38 |
| Unplaced | 36* | 12 | |||||||||
| Sox14 | 6 | 10 | LG 18* | 2 | 4 | 28 | LG4 | 3 | 3 | 9 | 23 |
| Unplaced | 27* | ||||||||||
| 31* | |||||||||||
| Sox17b | 7* | Unplaced | LG 9* | 20 | 15* | LG20 | 19 | 8* | 1* | 29* | |
| Sox18 | 23 | Unplaced | LG 20 | up | 7 | 16 | LG7 | 15 | 20 | 2 | 24 |
| Sox19a | 5 | 9 | 30 | 18* | 10 | 18* | 4 | 0 | 0 | 0 | |
| 2 | 10 | ||||||||||
| Sox19b | 7 | Unplaced | LG 3 | 32 | 18 | 21 | LG18 | 7 | 0 | 0 | 0 |
| Sox21a | 6 | Unplaced | LG 16* | 6 | 21* | 3 | LG21* | 25* | 13* | 14* | 0 |
| 31* | |||||||||||
| Sox21b | 9 | Unplaced | LG 16 | 9 | 21 | 22 | LG2 | 21 | 12* | 14* | 0 |
| 34* |
Table 5.
Detailed characteristics information of SOX genes in Cyprinus carpio. The molecular weight, Weight in Kilodaltons, theoretical PI, Extinction Coefficient, Number of Atom and N-glycosylation sites of individual SOX genes.
| Gene Name | Molecular weight | Weight in Kilodaltons (kDa) | PI (Isoelectric Point) | Extinction Coefficient | Total Number of Atom | N-glycosylation sites |
|---|---|---|---|---|---|---|
| Sox-1a | 36152.4881 | 36.16 | 9.70 | 37360 | 4956 | 2 |
| Sox-1b | 35946.3331 | 36.7 | 9.70 | 37360 | 4932 | 2 |
| Sox2 | 34542.9331 | 34.55 | 9.74 | 37360 | 4731 | 2 |
| Sox3 | 33326.7361 | 33.33 | 9.63 | 37360 | 4589 | 3 |
| Sox4a | 37999.9801 | 38 | 6.08 | 32430 | 5230 | 1 |
| Sox4b | 33945.7861 | 33.95 | 8.59 | 33920 | 4705 | 0 |
| Sox5 | 30049.8056 | 30.06 | 9.07 | 21430 | 4233 | 0 |
| Sox6 | 19299.6776 | 19.3 | 5.45 | 13410 | 2653 | 1 |
| Sox7 | 43535.2099 | 43.54 | 6.14 | 43320 | 5966 | 1 |
| Sox8a | 46255.0010 | 35.11 | 6.64 | 58790 | 6346 | 0 |
| Sox8b | 35105.9064 | 46.26 | 6.65 | 35410 | 4826 | 0 |
| Sox9 | 50794.7645 | 50.81 | 6.13 | 55810 | 6965 | 1 |
| Sox9a | 50794.7645 | 50.81 | 6.13 | 55810 | 6965 | 1 |
| Sox9b | 48461.2156 | 48.47 | 6.07 | 57300 | 6640 | 0 |
| Sox10 | 15735.5147 | 15.74 | 5.61 | 19480 | 2160 | 0 |
| Sox 10a | 51135.7732 | 51.14 | 6.27 | 54320 | 6985 | 1 |
| Sox 10b | 51292.9020 | 51.3 | 6.20 | 54320 | 7003 | 1 |
| Sox11a | 39908.2661 | 39.91 | 5.26 | 38390 | 5491 | 3 |
| Sox11b | 39892.2381 | 39.9 | 5.15 | 38390 | 5481 | 3 |
| Sox13 | 44313.0145 | 44.33 | 7.91 | 18450 | 6191 | 3 |
| Sox14 | 26673.4610 | 26.68 | 9.69 | 32890 | 3713 | 1 |
| Sox17b | 35357.9587 | 35.36 | 7.30 | 37360 | 4854 | 2 |
| Sox18 | 47999.5073 | 48.01 | 6.81 | 43890 | 6624 | 2 |
| Sox19a | 32763.9188 | 32.77 | 9.66 | 40340 | 4510 | 0 |
| Sox19b | 31966.1471 | 31.97 | 9.61 | 41830 | 4398 | 0 |
| Sox21a | 26512.5282 | 26.51 | 9.74 | 32890 | 3705 | 2 |
| Sox21b | 26662.8271 | 26.67 | 9.69 | 31400 | 3711 | 0 |
3.2. Physiochemical properties of SOX proteins
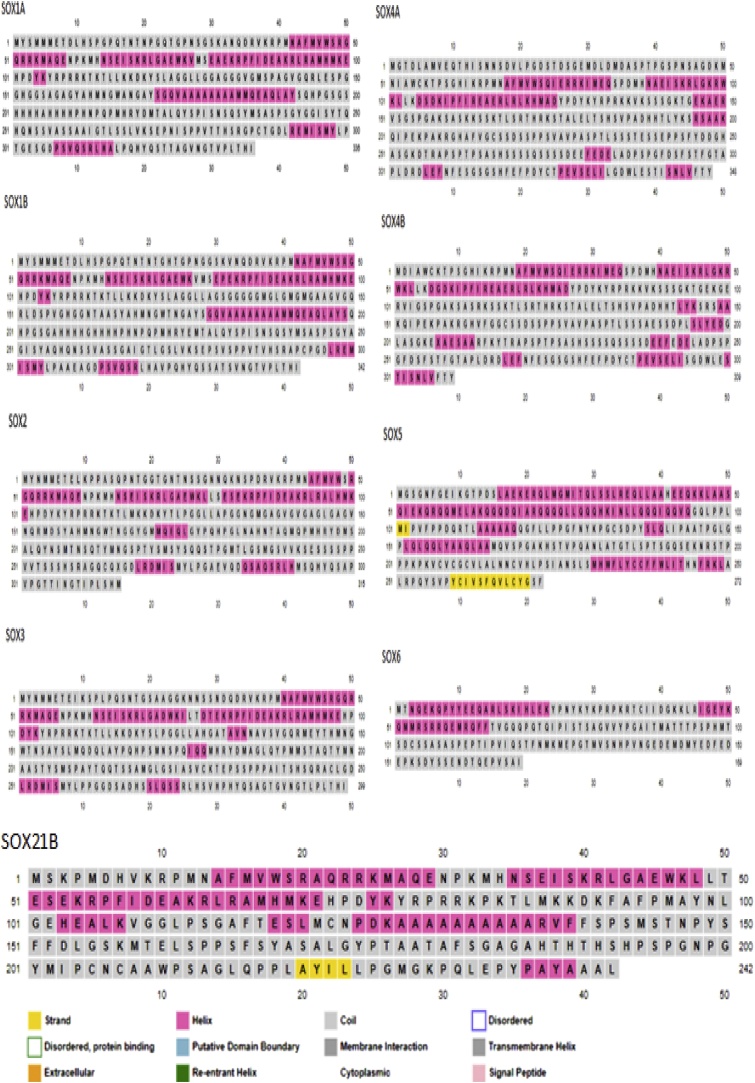
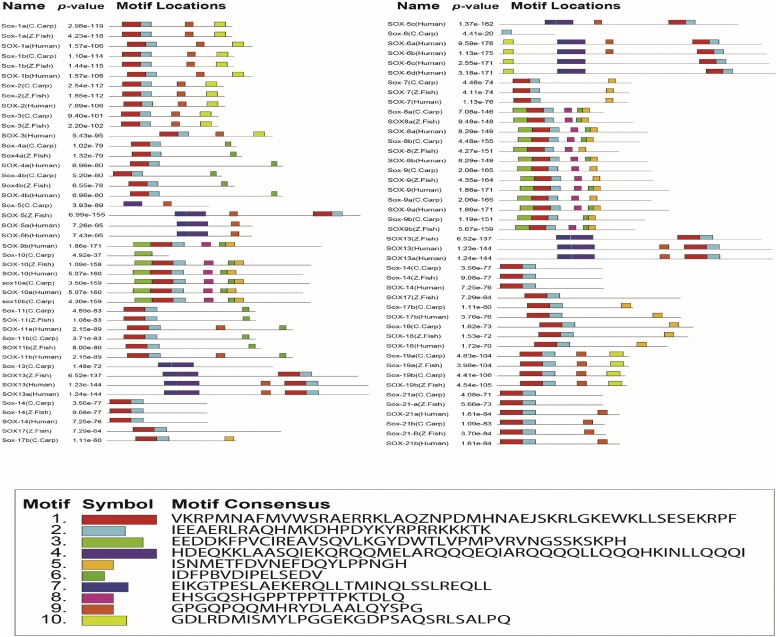
We check Physiochemical properties of individual SOX proteins for further understanding in involvement of biological functions, having different parametric details i.e analysis of functional domains, molecular mass, potential N-glycosylation sites, theoretical PI and the extinction coefficient values. The molecular weight of most SOX proteins ranged from 31.97 to 51.14 kDa, theoretical PI of most SOX proteins were within 7.9.6.5. The Extinction Coefficient values of individual proteins observed or set in Table 4 in which various proteins founded in 37360–55810 in range. The number of potential Nglycosylation sites varied in Cyprinus carpio SOX proteins, ranging from 3 to 1 but total of nine genes (SOX4b, SOX5, SOX8a, SOX8b, SOX9b, SOX10, SOX19a SOX19b and SOX21b) were 0 Nglycosylation sites mentioned in (Table 4). A secondary structure of all SOX proteins annotation consists of stand, Coils, helix, extracellular, putative domain boundary and signal peptide 43 % to 15 % beta sheet, transmembrane, cysteine residues signal Peptide are summarized in Fig. 1. The functional domains and motif of all SOX genes were foreseen based on their sequences, all SOX gene show closed relation based on distance based phylogenetic suggestion and individual gene have a HMG (High mobility group) domains which are family of chromosomal proteins comparatively low molecular weight and non-histone mechanisms that bind with DNA among low sequence specific identity. We also perceive the individual SOX genes encompassing with high mobility group A and B (DNA-binding domains) which mean HMG1-A also called HMG-T in fish and HMG2-B are related genes that have two distinguishing features: two HMG boxes (A and B), homologous folded domains of around 80 amino acid residues, and a long acidic tail containing 20–30 aspartic or glutamic acid residues, all common carp SOX genes in HMG group-1 and two genes SOX7 and SOX18 SOX having C-terminal domain were founded (Fig. 2). For motif (super secondary structure) analysis we compare the protein sequences of three organism (common carp, zebrafish and human) to find the overall ten motifs mentioned with different color code (motif number, motif symbol with diverse ten colors and motif consensus mentioned in motif legend), all SOX genes according to ascending order have at least one and maximum ten motifs on single sequence with cut of values (p value) and motif location are given in Fig. 3).
Fig. 1.
SOX protein annotation in Cyprinus carpio.
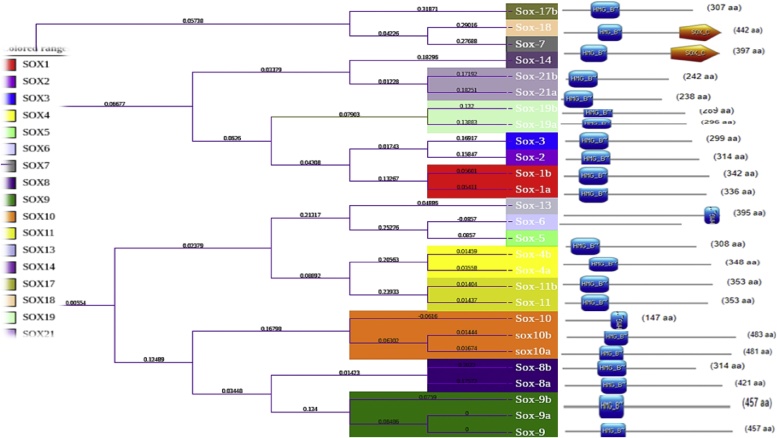
Fig. 2.
SOX genes Binding Domains: Relationship of SOX genes in common carp based on distance made phylogenetic tree. The each SOX genes have a HMG (High mobility group) domains or SOX7 and SOX18 having C-terminal domains, to display phylogenetic tree with different color coded ranges labeled with induvial gene names.
Fig. 3.
SOX genes in common carp and other vertebrates with their motif location and sequences.
3.3. Retrieval of all SOX protein network form five big protein databases
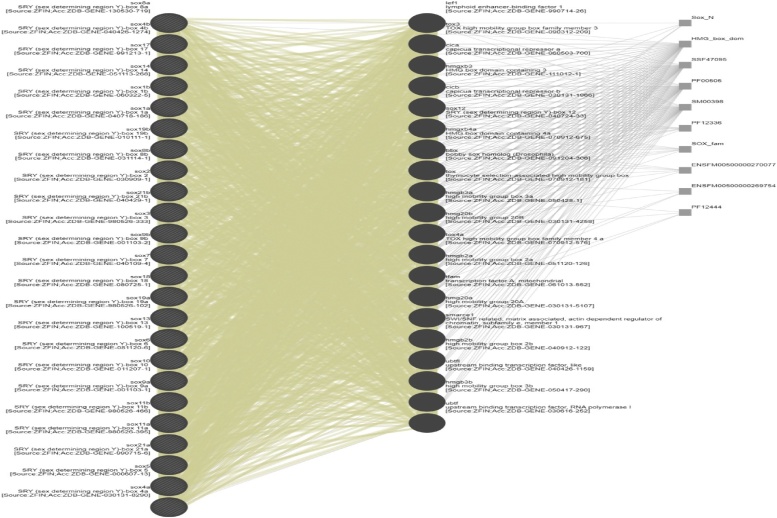
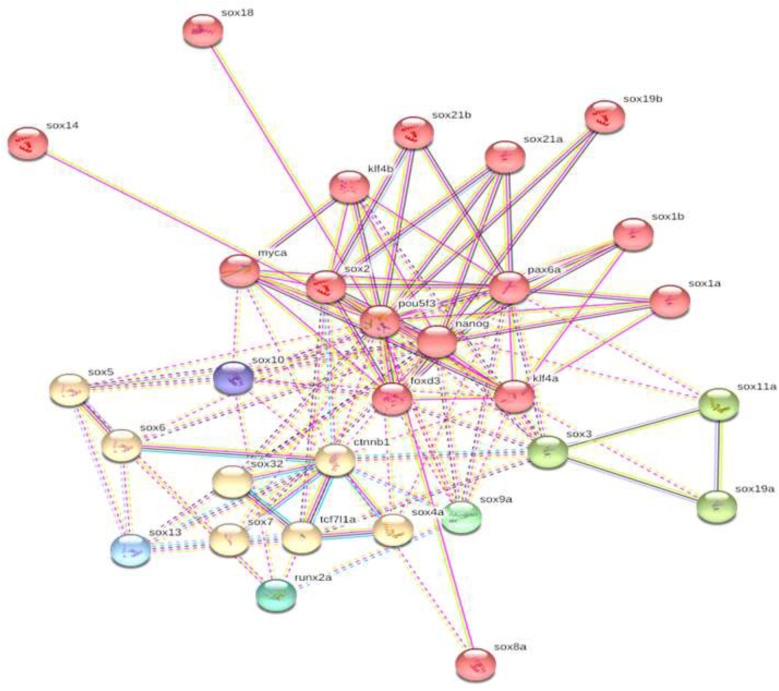
For retrieval of all SOX protein networks we use genemania to retrieve the interaction of individual protein from different protein databases such 22 query related protein interaction involved with 44 proteins among 5 wide range attributes linked with 2035 individual proteins. The network were further categorized according to weightage of interaction mentioned in color coded lines as like share protein domains was 30.93 %, interaction from interpro (26.45 %),PFAM 18.35 %, SMART 7.70 %, Ensesmble protein family 6.62 %, super family 5.44 %, co expression among all proteins (4.09 %), and physical interaction among whole network was o.42 % showing in (Fig. 4)
Fig. 4.
Retrieval of all Sox protein network form five big protein databases.
3.4. Protein-protein interaction and GO of all SOX proteins
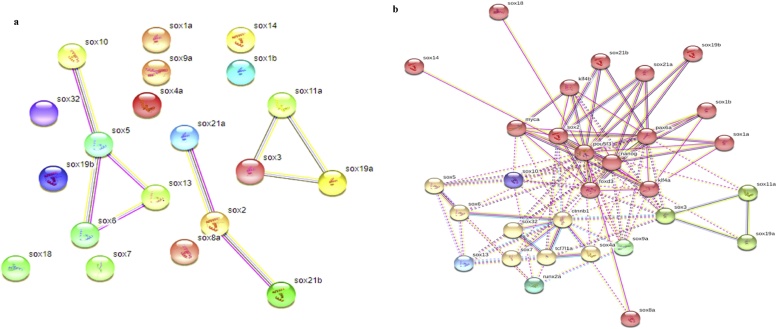
For Protein-protein interaction we use sting database to retrieve the network interaction with 0.417 average clustering coefficients and observed total 20 numbers of nodes with 0.9 average degrees and 9 numbers of edges with 1 expected edge, many SOX protein were remain individual and this will interact many other protein as showing in Fig. 5b, but some of SOX proteins showing strong interaction network cluster with each other such as network 1 consistent of (SOX19a, SOX11a and SOX3), network 2 (SOX21a, SOX21b and SOX2), ad network 3 were including (SOX13, SOX6, SOX19b, SOX5, SOX32, and SOX10 proteins) as showing in Fig. 5a. For better understanding we divided All SOX proteins into 9 clusters by using K-mean cluster. The cluster was composed of closely connected proteins interaction, in which queried proteins were used to predict total of 13 functional partner proteins with total number of amino acid like pou5f3 (472 aa), runx2a (467 aa), nanog (384 aa), pou5f3 (472 aa), klf4a (352 aa), pax6a (461 aa), myca (409 aa), ctnnb1 (780 aa), klf4b (409 aa), and foxd3(371 aa) with p-value: < 1.0e-16 and score between 0.981 to 0.958. The balls giving unspecified effects in the interaction network and arrows shows positive action effect and one is show negative effects. The networks were divided into different colors clusters shown in Fig. 5b. The GO probability revealed based on five set of p values labeled with different colors (orange< = 1e-10, yellow 1e-10 to 1e-0, green 1e-0 to 1e-6, light blue1e-6 to 1e-4, and gray >0.01) for all SOX genes, to identify the maximum involvement of these genes were in metabolic process 49.796 %. Average SOX genes involved in biological regulation 45.188 %, biosynthetic process (19.992 %), regulation of cellular process 39.68, 45.508 % organic substance metabolic process, multicellular organismal process 23.23 %,developmental process 21.74 %, system development 16.59 %, gene expression 16.05 % and 14.337 % of RNA metabolic process as given in Fig. 6.
Fig. 5.
a SOX Protein interaction (Protein-protein interaction) with other proteins. b SOX Protein interaction (Protein-protein interaction) with other proteins (K means clustering).
Fig. 6.
Gene ontology analysis of all SOX proteins.
3.5. Phylogenetic analysis of SOX genes
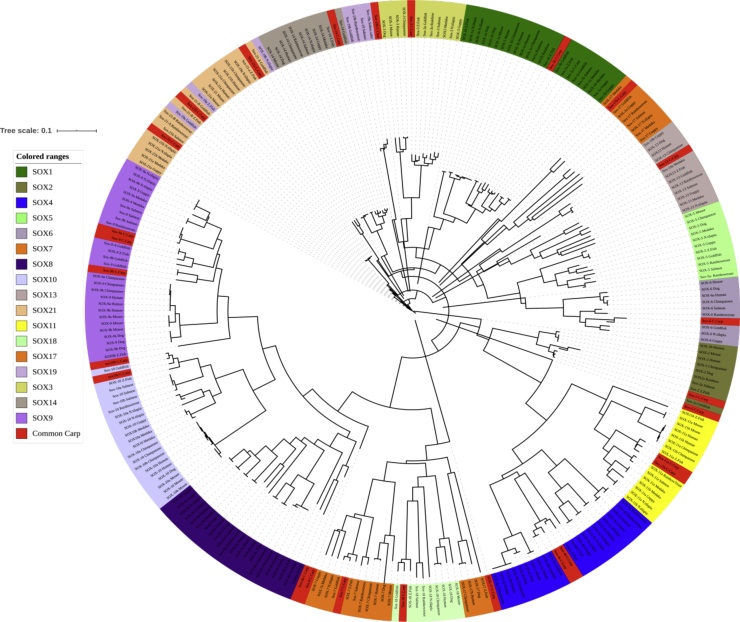
The phylogenetic analysis give extrapolation for all the SOX genes of common carp were grouped with their corresponding homologs from other higher vertebrates and fish species (Human (Homo sapiens), Mouse (Mus musculus), Dog(Canis lupus familiaris), Nile tilapia (Oreochromis niloticus), Gold fish (Carassius auratus), Chimpanzee (Pan troglodytes), Medaka (Oryzias latipes), Rainbow trout (Oncorhynchus mykiss), Guppy (Poecilia reticulata), and Atlantic salmon(Salmo salar), the indication of these groups are signifying that all protein sequences in SOX gene family are highly conserved except SOX19 of Common carp, Zebrafish, Nile tilapia and Goldfish or mix with SOX21. The specified higher vertebrates and teleost, SOX gene were assembled into individual clades shown with different color ranges, our candidate fish (Cyprinus carpio) indicate in phylogenetic tree with red color or found in all respective clads. Among the teleost cluster common carp close to Zebrafish(Danio rerio), Goldfish (Carassius auratus) and rainbow trout (Oncorhynchus mykiss) as given in Fig. 7. The multiple alignment of all SOX protein sequences of individual organism are given in supplementary file 1
Fig. 7.
The phylogenetic analysis of all SOX genes of common carp with their corresponding homologs from other higher vertebrates and fish species.
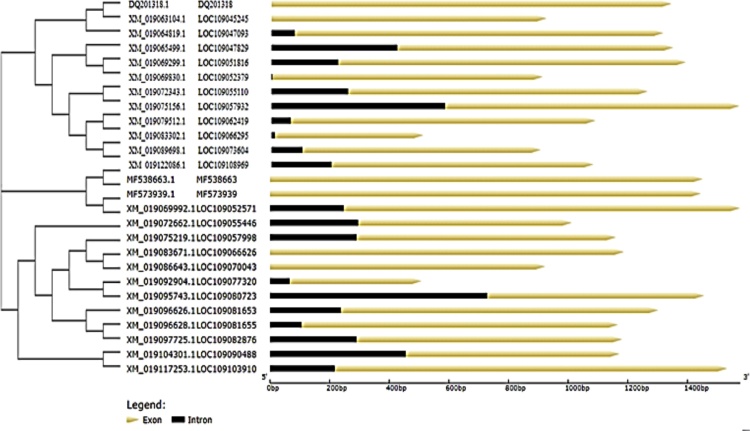
3.6. Synteny analysis of SOX genes
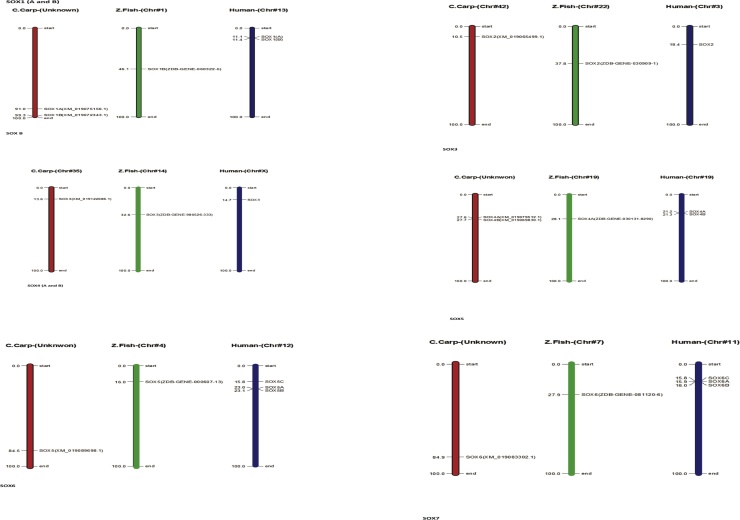
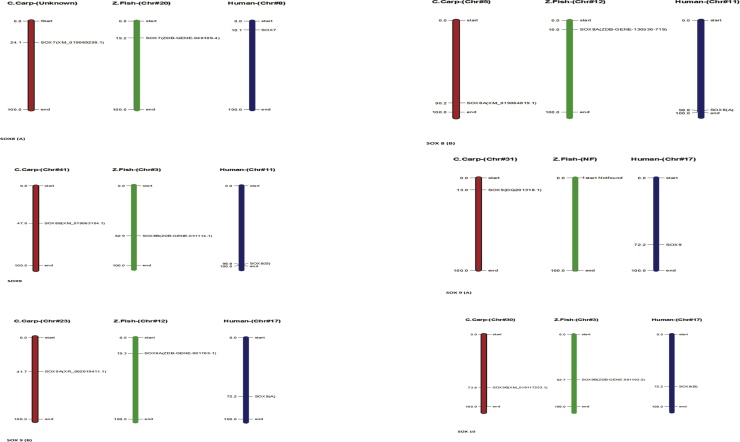
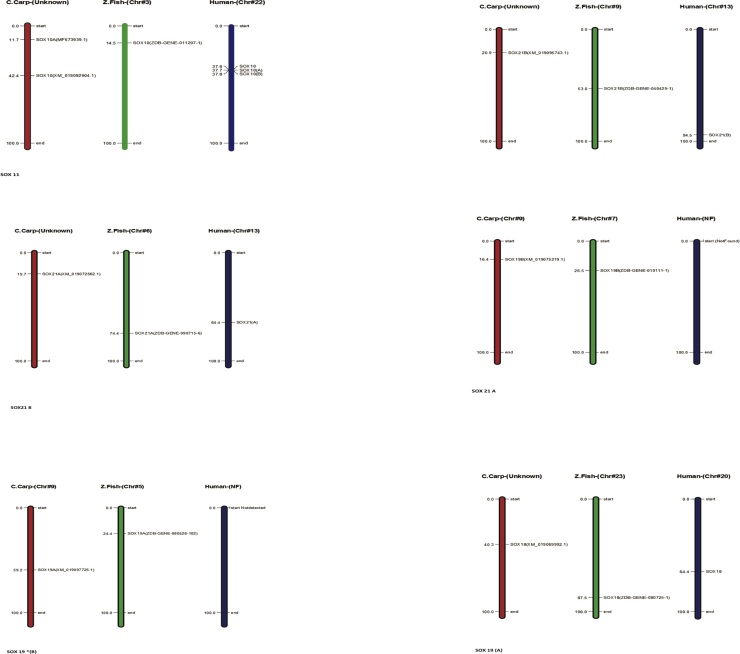
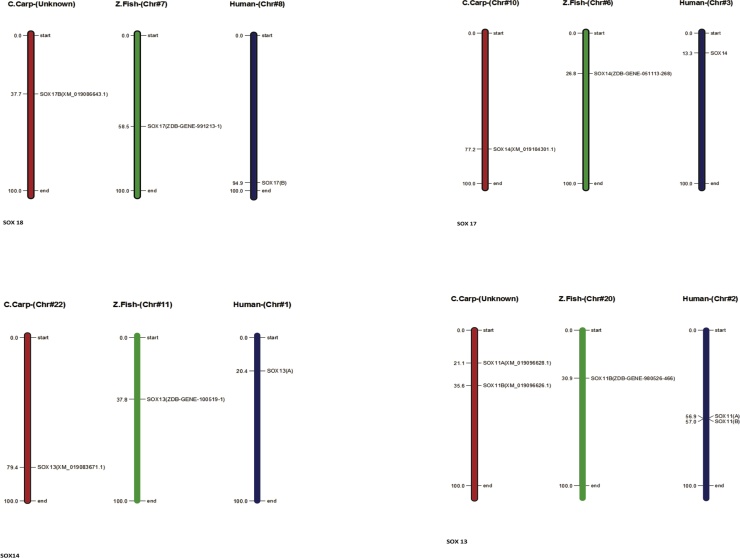
In Common carp SOX gene, we perceived all the gene for characterization ensuring close relation among each other with information of phylogenetic association and display intron and exon presence on individual gene excepting total of (6) genes does not have a intron structure such as (Acc. # DQ201318, XM_019063104.1) are present on first cluster in the start of the tree, two genes (Acc. # MF573939.1 and MF538663.1) present in middle of tree with same cluster weightage and in the latter two genes (Acc. # XM_019083671.1 and XM_019086643.1) are present in last of the tree and demonstrated in Fig. 8. All the SOX genes of common carp (Cyprinus carpio) were physically mapped among Zebrafish (Danio rerio) and Human (Homo sapiens) on the chromosomes level as per availability of acquired data from public resource databases and mentioned with different colors coded as like red identify common carp, green zebrafish and blue indicate human chromosomes. Among all chromosomes, the highest number of SOX genes (2) was found in common carp on unplaced region matched with many other chromosomes mentioned in zebrafish and human as like maximum (3) genes located on human chromosomes number 11, 12 and 22 and zebrafish contain at least one SOX gene, in detail SOX1B present on chromosomes number 1 in zebrafish and chromosome 13 in human, respectively all genes mapped according to their order and presence on respective chromosomes numbers mentioned in Fig. 9 except SOX9 in zebrafish and SOX19A and SOX19B in human because both they were absent in mentioned genome. The distribution pattern of SOX genes on chromosomes also identified with certain physical areas along with comparatively higher accumulation of SOX genes gene clusters.
Fig. 8.
Phylogenetic association and presence intron and exon on individual gene.
Fig. 9.
Common carp physically mapping of chromosomes among Zebrafish (Danio rerio) and Human (Homo sapiens).
4. Discussion
Our main goal this current study was to identify the genome wide identification of highly conserved SRY-related HMG-box (SOX) genes in common carp genome with confirmation of in silico parametric techniques for gene characterization and structural insight of protein sequence’s with verification of phylogenetic investigation and syntenic analysis. In higher vertebrates SOX genes show diverse distribution pattern among different gene subfamilies and fishes including SOX proteins, such as channel catfish [42], tilapia [23], pufferfish [43], zebrafish [44] and medaka [20], based on conserved structure of protein domains and intron exon regions SOX1A, SOX1B, SOX2, SOX3, SOX4A, SOX4B, SOX5, SOX6, SOX7, SOX8A, SOX8B, SOX9, SOX9A, SOX9B, SOX10, SOX10A, SOX10B, SOX11A, SOX11B, SOX13, SOX14, SOX17B, SOX18, SOX19A, SOX19B, SOX21A AND SOX21B [3], [45], [46].
All SOX proteins including SOX1 to SOX 32, are a class of transcriptional regulators related sex determining factor SRY of mammals and other vertebrates [1]. In Genome wide distribution pattern due to evolutionary full genome repetition of SOX genes, these are reported in different teleost species, like 27 in Nile tilapia, Zebrafish (27), Pufferfish(25), Medaka (19), Human(20), Florida Lancelet (10) and 18, 18 in Western Clawed and chicken [23], [46], and no sox genes like SOX12, SOX15, SOX16 could be identified in certain teleost species [46], like sox 12 genes in acanthopterygian genomes and sox 30 in other fish orthologues. Until now in vertebrate more than 20 SOX different genes families are discovered based on conservancy of HMG‐box and classified with evidence of phylogenetic analysis [22]. SOX1 is highly functionally conserved with SOX1A, SOX1B, SOX2 and SOX3, all are identified in the genome of Homo sapiens [47], Rattus norvegicus [48], Gallus gallus [49], Xenopus tropicalis [50]. SOX5 and SOX6 identified in human, mouse and zebrafish (16, 17). SOX7, SOX17 and SOX18 are three closely related SOX proteins and identified in many corresponding genome like mouse, human and teleost (19)
In the current study full length of SOX1A and SOX1B gene in common carp is 2209bp and 1357bp with 1010 bp or 1028bp open reading frame (ORF) which encoded in 336 and 342 amino acid (aa) sequences for protein. In Chinese sturgeon (Acipenser sinensis), full length of SOX1 is 2029 which encode with 343 amino acids [51]. In Nile tilapia SOX1A gene CDS length is 1533bp and protein encode with 344aa and SOXB gene encode with 354aa [23]. SOX1 has been already reported in broad range of animal taxa especially in teleost like Zebrafish, catfish, Medaka, Nile tilapia and rainbow trout [23], [46], [52]. In common carp SOX1A and SOX1B has 1, 1 exon, same in zebrafish but in the case of Nile tilapia SOX1A has only 1 and SOXB total have 7 exons [23]. The full-length cDNA of SOX2 gene is 997 encoded with 315aa having 1 exon, which is highly comparable with CDS of Indian major carps (Catla catla, Cirrhinus cirrhosis and Labeo rohita) and nile tilapia such as 936–997 nucleotides sequences with 315, 322 and 404aa [23], [53], [54]. Current work identified SOX such as SOX3 to SOX21 genes have diverse range CDS length like 716 to 1373bp, full-length in base pair is 523–3496 and putative protein sequences 147 to 483aa, pI>6, and Weight in Kilodaltons (kDa) ranges are 50, which are highly comparable with earlier reported SOX genes in fish species [23], [42], [53], [55].
All the SOX family encoded by protein sequences of common carp was analogous to other higher vertebrate species. It contains with highly conserved HMG domains and structural motif of the model zebrafish, have well conserved domains among the manifold species based on multiple protein sequences. Our present observations in all selected organisms are strongly aggregated with earlier reports confirming that the HMG domains of SOX gene family are highly conserved [53], [45]. The HMG (High mobility group) domains which are family of chromosomal proteins comparatively low molecular weight and non-histone mechanisms those bind with DNA among low sequence specific identity [56]. We also perceive that the individual SOX genes including with high mobility group A and B (DNA-binding domains) which mean HMG1-A also called HMG-T in fish and HMG2-B are related with other vertebrate genes, same observation in the report of structural features of HMG genes and proteins [57], which make double distinguishing features: two HMG boxes (A and B), homologous folded domains consist of approximately 80 amino acid residues, and a long acidic tail containing 20–30 aspartic or glutamic acid residues, all common carp SOX genes in HMG Group-I(SOX7) and Group-II (SOX18) have C-terminal domains. The SOX gene families of HMG BOX (PDOC00305) transcriptional factor encoded with sex determination factor on Y-chromosome play a vital role for embryonic development.In teleost, sex chromosomes are commonly monomorphic and feasibly evolutionary young [58]. With the help of bioinformatics analysis the SOX 18 combine with SOX7 and SOX17 or show C-termini among different higher vertebrates and teleost (Homo sapiens, Mus musculus, Canis lupus familiaris, Oreochromis niloticus, Carassius auratus, and Pan troglodytes), they have highly conserved aa residues and comprises with a strong trans activating domains [59], [60].
In Protein–protein interaction of SOX family are interact many other transcriptional factors like POU5F3, RUNX2A, NANOG, POU5F3, KLF4A, PAX6A, MYCA, CTNNB1, KLF4B, AND FOXD3 etc. All the SOX genes in common carp are mentioned in networks are directly or indirectly involved with sex determination mechanisms including Sry -related HMG box. This has previously been observed in other vertebrates including fish, genes are reported related to sex determination mechanisms including Sry -related HMG box (SOX genes), Anti-Mullerian hormone receptor type 2, Double-sex and mab-3 related transcription factors and Growth differentiation factor 6. In the same contrast SOX proteins showing strong interaction network cluster with each other such as network-1 consistent of (SOX19a, SOX11a and SOX3) same as earlier researcher reported in Zebrafish [61], network-II consistent of (SOX21a, SOX21b and SOX2) same as compared in Japanese rice fish [20], and in network 3 of common carp were including (SOX13, SOX6, SOX19b, SOX5, SOX32, and SOX10 proteins), as compared with network-III of common carp and same trend observe in study of SOX proteins for cell regulation and interaction in fish, SOX proteins make network of SOX13, SOX6, SOX5, SOX32, and SOX10 [26].
In Common carp for GO analysis SOX gene Group-I (SOX1, SOX2, SOX3, and SOX19) involved in positive regulation of DNA binding transcriptional factor activation, positive regulation of molecular function and involved in metabolic processes with different percentage given in results part or pathway relation shown in Fig. 4. The SOX1, SOX2, and SOX3 proteins sequence bind specifically with DNA via HMG domain and act as a transcription factor or involves in protein-protein interactions such as POU proteins binding with other proteins [29]. The SOX1 and SOX3 has a C-terminal region, which facilitate it to act as a transcriptional factor by developing protein-protein complex and sox1 expressed in development process for formation of neural plate [30]. The expression of Sox2 and SOX3 help in maintaining the identity of progenitor cell by inhibiting neurogenesis [29].
The Group-II (SOX5, SOX6, and SOX13) in common carp via gene ontology involved in negative regulation of transcription (DNA templated), gene expression and macromolecule metabolic process. The sox5 and sox6 have been identified in human, mouse and other organisms. Sox5 is expressed for developing CFu neurons and prevents premature differentiation [62], in mammals and fish sox6 act as transcription factor to controls the identity of skeletal muscle fiber [31]. The SOX13 expressed in different embryonic lineages and show development of central nervous system (CNS), visceral mesoderm of yolk sac, pancreas, kidney and liver [63].
In Common Carp Group-III (SOX8, SOX9, SOX10, and SOX21) involved in negative and positive regulation in transcription by RNA polymerase II. The Sox8, sox9 and sox 10 involved in many developmental processes like testis development maintenance of male fertility, humans early developing sate of gonads, expression in somatic cells and sex determination with the help of DMRT1 gene [32], [33]. Sox9 is an essential transcription regulatory factor for the development of adult cartilage and activates the genes transcriptional factor for structural components [34]. In the regulatory network of oligodendrocytes and schwann cells sox10 HMG domain show complex relation with sox8 and sox9, and include as important transcription factors for lineage progression, terminal differentiation and myelin induction [64].
In present work the chromosomal organization and synteny analysis of common carp between Zebrafish, and Human indicate that, all SOX a gene are located on different chromosomes and apparently does not fallow the unique pattern. The maximum linkage of chromosome is unknown in common carp and highest number of SOX genes (2) were found on unplaced region matched with many other chromosomes mentioned in zebrafish and human as like maximum (3) genes located on human chromosomes number 11, 12 and 22. Same pattern earlier reported in zebrafish on chromosome number 3, 12 and 22 [65]. The previously reported SOX2 gene was localized at 44 chromosome (2n) genome of Tilapiine species as like Nile tilapia, Mozambique tilapia, Oreochromis aureus and other teleost Haplochromis obliquidens, Kennyi cichlid, Scrapermouth mbuna, Pseudetroplus maculatus, which is analogous with current observation in common carp [65]. In our genome wide analysis the SOX3 gene is located on X chromosomes of Human, Dog and mouse which is more closely related with common carp 34 chromosomes sequence then to other teleost on the basis of HMG box region which are highly comparable earlier reports [66], [67]. The previous study also reported that SOX3 is X liked gene in multiple organisms [68], including human [69], mouse [70], dog, frog and some other fishes [71]. The numerous gene clusters of common carp is found on unknown region as same compared with recent study on genome wide identification of SOX genes in Nile tilapia [23], and Paralichthys olivaceus [72]. For evolution history of SOX gene family, phylogenetic analyses was done among higher vertebrates and teleost’s class of Actinopterygii, and detect all the genes of SOX families have been identified and clustered in respected clades such as Common carp shown closed phylogenic relation between Zebrafish and Goldfish. The same trend of phylogenic analysis of SOX gene family is observed in recent studies [53], [46]. The diversity of SOX protein sequences in common carp is consistent with other number of identified SOX genes in fishies such as Oryzias latipes, Nile tilapia, zebrafish, Goldfish, Nile tilapia, and channel catfish [23], [42]. The mammalian class (Human, Dog, mouse and Chimpanzee) SOX genes (12, 15, 16, 20) group have not been found in common carp genome which represented by single genes such as SOX15 and SRY. In our syntenic analysis SOX3 consider to link with mammalian Y‐chromosome, in previous research SRY evolve from the X‐linked gene SOX3 during the genesis [73]. The possible elucidation could be the wide-ranging soxgenes identified in diverse kind of fish genomes and other species [74]. The large number of novel genes could be derived from non‐conserved areas among the common carp and other teleosts [75], which unsuccessful to be annotate with functional informative proteins.
5. Conclusion
In conclusion, we identified expanded set of 27 SOX genes of Common carp. The total of 27 SOX genes can be divided SOX1A, SOX1B, SOX2, SOX3, SOX4A, SOX4B, SOX5, SOX6, SOX7, SOX8A, SOX8B, SOX9, SOX9A, SOX9B, SOX10, SOX10A, SOX10B, SOX11A, SOX11B, SOX13, SOX14, SOX17B, SOX18, SOX19A, SOX19B, SOX21A AND SOX21B. All SOX genes have conserved HMG domains in all higher vertebrates and teleosts. SOX gene in chromosomal mapping located on diverse number of chromosomes and mostly were found on unplaced regions. The current evidence will be supportive for further understanding of structural and functional properties of SOX gene family in fishes.
Funding’s
This work did not receive any fund form funding agencies in the public and commercial domains.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
The authors thankful to Dr. Mohd Ashraf Rather technical assistance, the authors are also thankful to Ensemble, NCBI and other genome centers for providing genomic resources in public domain for conducting Bioinformatics analysis.
References
- 1.Watanabe M., Kawasaki K., Kawasaki M., Portaveetus T., Oommen S., Blackburn J. Spatio-temporal expression of Sox genes in murine palatogenesis. Gene Expr. Patterns. 2016;21:111–118. doi: 10.1016/j.gep.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Bhat I.A., Rather M.A., Saha R., Pathakota G.B., Pavan-Kumar A., Sharma R. Expression analysis of Sox9 genes during annual reproductive cycles in gonads and after nanodelivery of LHRH in Clarias batrachus. Res. Vet. Sci. 2016:100–106. doi: 10.1016/j.rvsc.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 3.Pan Q., Anderson J., Bertho S., Herpin A., Wilson C., Postlethwait J.H. Vertebrate sex-determining genes play musical chairs. Comptes rendus biologies. 2016;339(7–8):258–262. doi: 10.1016/j.crvi.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kikuchi K., Hamaguchi S. Novel sex‐determining genes in fish and sex chromosome evolution. Dev. Dyn. 2013;242(Apr (4)):339–353. doi: 10.1002/dvdy.23927. [DOI] [PubMed] [Google Scholar]
- 5.Rather M.A., Dhandare B.C. Genome-wide identification of doublesex and Mab-3-related transcription factor (DMRT) genes in Nile Tilapia (Oreochromis niloticus) Biotechnol. Rep. 2019;24 doi: 10.1016/j.btre.2019.e00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soriano N.S., Russell S. The Drosophila SOX-domain protein Dichaete is required for the development of the central nervous system midline. Development. 1998;125:3989–3996. doi: 10.1242/dev.125.20.3989. [DOI] [PubMed] [Google Scholar]
- 7.Mukherjee A., Shan X., Mutsuddi M., Ma Y., Nambu J.R. The Drosophila sox gene, fish-hook, is required for postembryonic development. Dev. Biol. 2000;217:91–106. doi: 10.1006/dbio.1999.9506. [DOI] [PubMed] [Google Scholar]
- 8.Clarkson M.J., Harley V.R. Sex with two SOX on: SRY and SOX9 in testis development. Trends Endocrinol. Metab. 2002;13:106–111. doi: 10.1016/S1043-2760(01)00541-0. [DOI] [PubMed] [Google Scholar]
- 9.Barrionuevo F., Scherer G. SOX E genes: SOX9 and SOX8 in mammalian testis development. Int. J. Biochem. Cell Biol. 2010;42:433–436. doi: 10.1016/j.biocel.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 10.Jager M., Queinnec E., Houliston E., Manuel M. Expansion of the SOX gene family predated the emergence of the Bilateria. Mol. Phylogenet. Evol. 2006;39:468–477. doi: 10.1016/j.ympev.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki T., Sakai D., Osumi N., Wada H., Wakamatsu Y. Sox genes regulate type 2 collagen expression in avian neural crest cells. Dev. Growth Differ. 2006;48:477–486. doi: 10.1111/j.1440-169X.2006.00886. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L.H., Zhu T.Y., Lin D., Zhang Y., Zhang W.M. A second form of Sox11 homologue identified in the orange-spotted grouper Epinephelus coioides: analysis of sequence and mRNA expression patterns. Comp. Biochem. Phys. B. 2010;157:415–422. doi: 10.1016/j.cbpb.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Kashimada K., Koopman P. Sry: the master switch in mammalian sex determination. Development. 2010;137:3921–3930. doi: 10.1242/dev.048983. [DOI] [PubMed] [Google Scholar]
- 14.Jiang T., Hou C.C., She Z.Y., Yang W.X. The SOX gene family: function and regulation in testis determination and male fertility maintenance. Mol. Biol. Rep. 2012;40:2187–2194. doi: 10.1007/s11033-012-2279-3. [DOI] [PubMed] [Google Scholar]
- 15.Ikeda T., Kamekura S., Mabuchi A., Kou I., Seki S., Takato T., Nakamura K., Kawaguchi H., Ikegawa S., Chung U.I. The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum. 2004;50:3561–3573. doi: 10.1002/art.20611. [DOI] [PubMed] [Google Scholar]
- 16.Yan Y.L., Willoughby J., Liu D., Crump J.G., Wilson C., Miller C.T., Singer A., Kimmel C., Westerfield M., Postlethwait J.H. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132:1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
- 17.McDonald E., Krishnamurthy M., Goodyer C.G., Wang R. The emerging role of SOX transcription factors in pancreatic endocrine cell development and function. Stem Cells Dev. 2009;18:1379–1388. doi: 10.1089/scd.2009.0240. [DOI] [PubMed] [Google Scholar]
- 18.Gracz A.D., Magness S.T. Sry-box (Sox) transcription factors in gastrointestinal physiology and disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;300:G503–G515. doi: 10.1152/ajpgi.00489.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang Y.K., Srivastava Y., Hu C., Joyce A., Yang X., Zuo Z. Quantitative profiling of selective Sox/POU pairing on hundreds of sequences in parallel by Coop-seq. Nucleic Acids Res. 2017;45:832–845. doi: 10.1093/nar/gkw1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui J., Shen X., Zhao H., Nagahama Y. Genome-wide analysis of Sox genes in Medaka (Oryzias latipes) and their expression pattern in embryonic development. Cytogenet. Genome. Res. 2011;134:283–294. doi: 10.1159/000329480. [DOI] [PubMed] [Google Scholar]
- 21.Sarkar A., Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12(Jan (1)):15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.She Z.Y., Yang W.X. SOX family transcription factors involved in diverse cellular events during development. Eur. J. Cell Biol. 2015;94:547–563. doi: 10.1016/j.ejcb.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Wei L., Yang C., Tao W., Wang D. Genome-wide identification and transcriptome-based expression profiling of the Sox gene family in the nile tilapia (Oreochromis niloticus) Int. J. Mol. Sci. 2016;17:270. doi: 10.3390/ijms17030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schepers G.E., Teasdale R.D., Koopman P. Twenty pairs of Sox: extent, homology, and nomenclature of the mouse and human Sox transcription factor gene families. Dev. Cell. 2002;3:167–170. doi: 10.1016/s1534-5807(02)00223-x. [DOI] [PubMed] [Google Scholar]
- 25.Han F., Wang Z., Wu F., Liu Z., Huang B., Wang D. Characterization, phylogeny, alternative splicing and expression of Sox30 gene. BMC Mol. Biol. 2010;11:1–11. doi: 10.1186/1471-2199-11-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamachi Y., Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140(20):4129–4144. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- 28.Fu L., Shi Y.B. The Sox transcriptional factors: functions during intestinal development in vertebrates. Semin. Cell Dev. Biol. 2017;3:58–67. doi: 10.1016/j.semcdb.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kan L., Israsena N., Zhang Z., Hu M., Zhao L.R., Jalali A. Sox1 acts through multiple independent pathways to promote neurogenesis. Dev. Biol. 2004;269(2):580–594. doi: 10.1016/j.ydbio.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Gao J., Zhang W., Li P., Liu J., Song H., Wang X., Zhang Q. Identification, molecular characterization and gene expression analysis of sox1a and sox1b genes in Japanese flounder, Paralichthys olivaceus. Gene. 2015;574(2):225–234. doi: 10.1016/j.gene.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 31.Jackson H.E., Ono Y., Wang X., Elworthy S., Cunliffe V.T., Ingham P.W. The role of Sox6 in zebrafish muscle fiber type specification. Skelet. Muscle. 2015;5(1):2. doi: 10.1186/s13395-014-0026-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrionuevo F.J., Hurtado A., Kim G.J., Real F.M., Bakkali M., Kopp J.L. Sox9 and Sox8 protect the adult testis from male-to-female genetic reprogramming and complete degeneration. Elife. 2016;5 doi: 10.7554/eLife.15635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Portnoi M.F., Dumargne M.C., Rojo S., Witchel S.F., Duncan A.J., Eozenou C. Mutations involving the SRY-related gene SOX8 are associated with a spectrum of human reproductive anomalies. Hum. Mol. Genet. 2018;27(7):1228–1240. doi: 10.1093/hmg/ddy037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lefebvre V., Dvir-Ginzberg M. SOX9 and the many facets of its regulation in the chondrocyte lineage. Connect. Tissue Res. 2017;58(1):2–14. doi: 10.1080/03008207.2016.1183667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu P., Zhang X., Wang X., Li J., Liu G., Kuang Y., Xu J., Zheng X., Ren L., Wang G. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 2014;46:1212–1219. doi: 10.1038/ng.3098. [DOI] [PubMed] [Google Scholar]
- 36.McGinnis S., Madden T.L. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Letunic I., Doerks T., Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeanmougin F., Thompson J.D., Gouy M., Higgins D.G., Gibson T.J. Multiple sequence alignment with Clustal x. Trends Biochem. Sci. 1998;23:403–405. doi: 10.1016/S0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 40.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letunic I., Bork P. Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2006;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 42.Zhang S., Chen X., Wang M., Zhang W., Pan J., Qin Q. Genome‐wide identification, phylogeny and expressional profile of the sox family in channel catfish (Ictalurus punctatus) Comp. Biochem. Physiol. Part D. 2018;28:17–26. doi: 10.1016/j.cbd.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Koopman P., Schepers G., Brenner S., Venkatesh B. Origin and diversity of the SOX transcription factor gene family: genome-wide analysis in Fugu rubripes. Gene. 2004;328:177–186. doi: 10.1016/j.gene.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 44.Okuda Y., Yoda H., Uchikawa M., Furutani‐Seiki M., Takeda H., Kondoh H., Kamachi Y. Comparative genomic and expression analysis of group B1 sox genes in zebrafish indicates their diversification during vertebrate evolution. Dev. Dyn. 2006;235:811–825. doi: 10.1002/dvdy.20678. [DOI] [PubMed] [Google Scholar]
- 45.Godwin J., Luckenbach J.A., Borski R.J. Ecology meets endocrinology: environmental sex determination in fishes. Evol. Dev. 2003;5(1):40–49. doi: 10.1046/j.1525-142x.2003.03007.x. [DOI] [PubMed] [Google Scholar]
- 46.Voldoire E., Brunet F., Naville M., Volff J.N., Galiana D. Expansion by whole genome duplication and evolution of the sox gene family in teleost fish. PLoS One. 2017;12(7) doi: 10.1371/journal.pone.0180936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan M., Yao L., Abulizi G. Tumor-suppressor gene SOX1 is a methylation-specific expression gene in cervical adenocarcinoma. Medicine. 2019;98(38) doi: 10.1097/MD.0000000000017225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubio-Osornio C., Eguiluz-Meléndez A., Trejo-Solís C., Custodio V., Rubio-Osornio M., Rosiles-Abonce A. Decreased expression of Sox-1 in cerebellum of rat with generalized seizures induced by kindling model. CNS Neurol. Disord. Drug Targets (Formerly Curr. Drug Targets-CNS & Neurol. Disord.) 2016;15(6):723–729. doi: 10.2174/1871527315666160321105818. [DOI] [PubMed] [Google Scholar]
- 49.Kamachi Y., Uchikawa M., Collignon J., Lovell-Badge R., Kondoh H. Involvement of Sox1, 2 and 3 in the early and subsequent molecular events of lens induction. Development. 1998;125(13):2521–2532. doi: 10.1242/dev.125.13.2521. [DOI] [PubMed] [Google Scholar]
- 50.Klein S.L., Strausberg R.L., Wagner L., Pontius J., Clifton S.W., Richardson P. Genetic and genomic tools for Xenopus research: the NIH Xenopus initiative: a peer reviewed forum. Dev. Dyn. 2002;225(4):384–391. doi: 10.1002/dvdy.10174. [DOI] [PubMed] [Google Scholar]
- 51.Yang J., Hu Y., Han J., Xiao K., Liu X., Tan C. Genome-wide analysis of the Chinese sturgeon sox gene family: identification, characterisation and expression profiles of different tissues. J. Fish Biol. 2019;96(1):175–184. doi: 10.1111/jfb.14199. [DOI] [PubMed] [Google Scholar]
- 52.Zhong L., Yu X., Tong J. Sox genes in grass carp (Ctenopharyngodon idella) with their implications for genome duplication and evolution. Genet. Sel. Evol. 2006;38(6):673. doi: 10.1186/1297-9686-38-6-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang L., Bi D., Ding H., Wu X., Zhu R., Zeng J. Systematic identification and evolution analysis of sox genes in Coturnix japonica based on comparative genomics. Genes. 2019;10(4):314. doi: 10.3390/genes10040314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patra S.K., Chakrapani V., Panda R.P., Mohapatra C., Jayasankar P., Barman H.K. First evidence of molecular characterization of rohu carp Sox2 gene being expressed in proliferating spermatogonial cells. Theriogenology. 2015;84(2):268–276. doi: 10.1016/j.theriogenology.2015.03.017. [DOI] [PubMed] [Google Scholar]
- 55.Kyte Jack, Doolittle Russell F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 56.Malarkey C.S., Churchill M.E. The high mobility group box: the ultimate utility player of a cell. Trends Biochem. Sci. 2012;37(12):553–562. doi: 10.1016/j.tibs.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bustin M., Lehn D.A., Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim. Biophys. Acta (BBA)-Gene Struct. Expression. 1990;1049(3):231–243. doi: 10.1016/0167-4781(90)90092-g. [DOI] [PubMed] [Google Scholar]
- 58.Charlesworth D., Charlesworth B., Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- 59.Sandholzer J., Hoeth M., Piskacek M., Mayer H., de Martin R. A novel 9-amino-acid transactivation domain in the C-terminal part of Sox18. Biochem. Biophys. Res. Commun. 2007;360(2):370–374. doi: 10.1016/j.bbrc.2007.06.095. [DOI] [PubMed] [Google Scholar]
- 60.Grimm D., Bauer J., Wise P., Krüger M., Simonsen U., Wehland M. Seminars in Cancer Biology. Academic Press; 2019. The role of SOX family members in solid tumours and metastasis. March. [DOI] [PubMed] [Google Scholar]
- 61.Onichtchouk D., Geier F., Polok B., Messerschmidt D.M., Mössner R., Wendik B. Zebrafish Pou5f1‐dependent transcriptional networks in temporal control of early development. Mol. Syst. Biol. 2010;6(1) doi: 10.1038/msb.2010.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lai T., Jabaudon D., Molyneaux B.J., Azim E., Arlotta P., Menezes J.R., Macklis J.D. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57(2):232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 63.Wang Y., Ristevski S., Harley V.R. SOX13 exhibits a distinct spatial and temporal expression pattern during chondrogenesis, neurogenesis, and limb development. J. Histochem. Cytochem. 2006;54(12):1327–1333. doi: 10.1369/jhc.6A6923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turnescu T., Arter J., Reiprich S., Tamm E.R., Waisman A., Wegner M. Sox8 and Sox10 jointly maintain myelin gene expression in oligodendrocytes. Glia. 2018;66(2):279–294. doi: 10.1002/glia.23242. [DOI] [PubMed] [Google Scholar]
- 65.Mazzuchelli J., Yang F., Kocher T.D., Martins C. Comparative cytogenetic mapping of Sox2 and Sox14 in cichlid fishes and inferences on the genomic organization of both genes in vertebrates. Chromosome Res. 2011;19(5):657–667. doi: 10.1007/s10577-011-9225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ijiri S., Kaneko H., Kobayashi T., Wang D.S., Sakai F., Paul-Prasanth B. Sexual dimorphic expression of genes in gonads during early differentiation of a teleost fish, the Nile tilapia Oreochromis niloticus. Biol. Reprod. 2008;78(2):333–341. doi: 10.1095/biolreprod.107.064246. [DOI] [PubMed] [Google Scholar]
- 67.Howe K., Clark M.D., Torroja C.F., Torrance J., Berthelot C., Muffato M. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stevanovlć M., Lovell-Badge R., Collignon J.M., Goodfellow P.N. SOX3 is an X-linked gene related to SRY. Hum. Mol. Genet. 1993;2(12):2013–2018. doi: 10.1093/hmg/2.12.2013. [DOI] [PubMed] [Google Scholar]
- 69.Sinclair A.H., Berta P., Palmer M.S., Hawkins J.R., Griffiths B.L., Smith M.J. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346(6281):240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 70.Raverot G., Lejeune H., Kotlar T., Pugeat M., Jameson J.L. X-linked sex-determining region Y box 3 (SOX3) gene mutations are uncommon in men with idiopathic oligoazoospermic infertility. J. Clin. Endocrinol. Metab. 2004;89(8):4146–4148. doi: 10.1210/jc.2004-0191. [DOI] [PubMed] [Google Scholar]
- 71.Graves J.A.M. How to evolve new vertebrate sex determining genes. Dev. Dyn. 2013;242(4):354–359. doi: 10.1002/dvdy.23887. [DOI] [PubMed] [Google Scholar]
- 72.Yu H., Du X., Li X., Qu J., Zhu H., Zhang Q., Wang X. Genome-wide identification and transcriptome-based expression analysis of sox gene family in the Japanese flounder Paralichthys olivaceus. J. Oceanol. Limnol. 2018;36(5):1731–1745. [Google Scholar]
- 73.Foster J.W., Graves J.A. An SRY-related sequence on the marsupial X chromosome: implications for the evolution of the mammalian testis-determining gene. Proc. Natl. Acad. Sci. 1994;91(5):1927–1931. doi: 10.1073/pnas.91.5.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Freeling M., Thomas B.C. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16(7):805–814. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]
- 75.Yang J., Hu Y., Xiao K., Liu X., Tan C., Wang B., Du H. Transcriptome profiling reveals candidate cleft palate-related genes in cultured Chinese sturgeons (Acipenser sinensis) Gene. 2018;666:1–8. doi: 10.1016/j.gene.2018.05.016. [DOI] [PubMed] [Google Scholar]