Abstract
Study Design:
Narrative review.
Objectives:
We aim to describe current progress in the application of artificial intelligence and machine learning technology to provide automated analysis of imaging in patients with spinal disorders.
Methods:
A literature search utilizing the PubMed database was performed. Relevant studies from all the evidence levels have been included.
Results:
Within spine surgery, artificial intelligence and machine learning technologies have achieved near-human performance in narrow image classification tasks on specific datasets in spinal degenerative disease, spinal deformity, spine trauma, and spine oncology.
Conclusion:
Although substantial challenges remain to be overcome it is clear that artificial intelligence and machine learning technology will influence the practice of spine surgery in the future.
Keywords: cervical, lumbar, thoracic, MRI, degenerative
Introduction
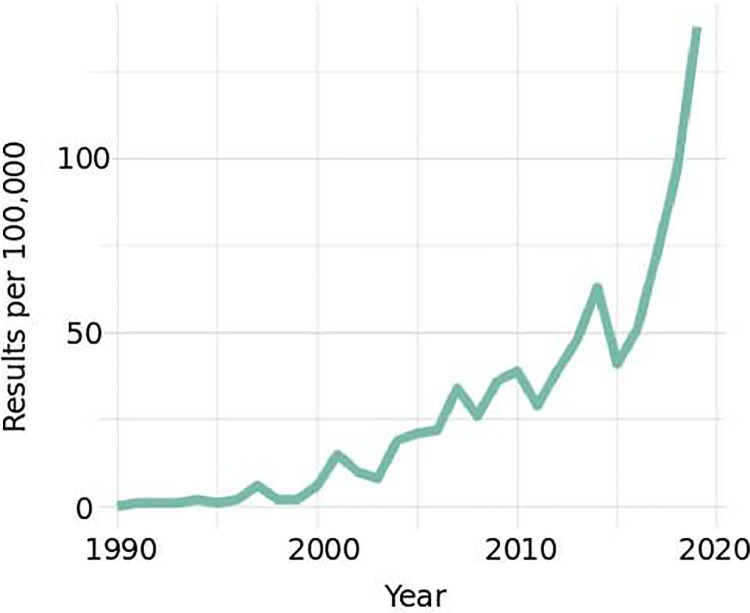
Medical imaging plays a central role in the diagnosis and management of spinal disorders.1 The combination of a growing burden of spinal disease associated with an aging population, and the greater availability of magnetic resonance imaging (MRI) and computed tomography (CT), has resulted in a dramatic increase in spine related imaging over the past few decades. Recent years have also seen major advances in machine learning (ML) and artificial intelligence (AI) technology, fueling a dramatic rise in research related to computer-aided interpretation of spinal imaging (Figure 1). While primarily a topic of research interest at present, it is possible that computer-aided interpretation of medical imaging will come to play a greater role in clinical medicine, particularly as it pertains to the diagnosis and management of spinal disorders, in the coming years.2
Figure 1.
PubMed indexed publications with the MeSH (medical subject heading) terms “Spine Surgery” AND “Medical Imaging” AND (“Artificial Intelligence” OR “Machine Learning”) since 1990.
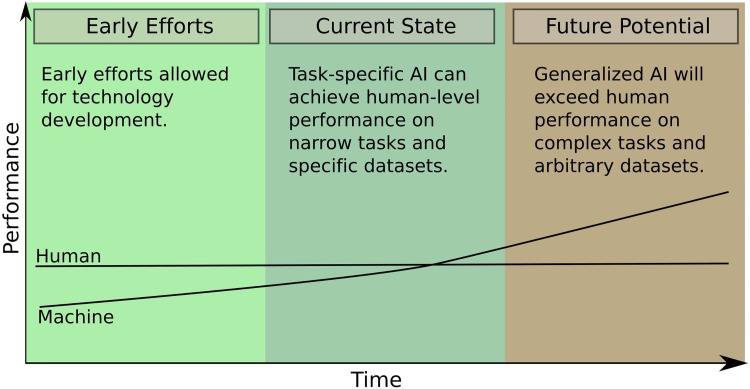
ML is a branch of AI that makes use of optimization algorithms to allow computer programs to improve through experience and exposure to data. Over the past decade, ML techniques have been increasingly used to inform clinical decisions, for example, in automated electrocardiography interpretation.3 The application of ML to automated interpretation of medical imaging, however, is a more complicated task, which is actively being developed.4 As the field of ML has progressed, specialized software has been able to achieve human-level performance in narrow image classification tasks.5,6 It is predicted that in the future, this technology will achieve generalized capabilities and be able to provide rapid interpretation of volumetric imaging and streamline care (Figure 2). Many such technologies are now being developed for a variety of clinical indications such as interpretation of CT scans of the brain and detection of pulmonary nodules.7-9
Figure 2.
Overview of artificial intelligence (AI) and human performance at image classification tasks over time. Early AI research achieved sub-human performance but allowed technologies to be developed. Currently, task-specific AI can achieve human performance on narrow image classification tasks and specific datasets in medical imaging. In the future, more generalized AI will affect medicine by providing rapid interpretation of volumetric medical imaging data.
There has been significant research into applying ML technology to the clinical management of patients with spinal disorders, but significant challenges remain. In this narrative literature review, we aim to describe current progress in the application of ML technology to imaging gathered in the care of patients with spine trauma, degenerative spinal disorders, spinal deformity, and spinal oncology. We have identified the trends and challenges that exist within this body of work and have attempted to identify areas that may see progress in future years.
Applications of Machine Learning in Spine Surgery
Degenerative Disease
Lumbar degenerative disease is highly prevalent and computer-aided interpretation of MRI scans could streamline care for these patients. A system for segmentation and classification of degenerated intervertebral discs on lumbar MRI T2-weighted images was first published in 2009.10 This system made use of classic imaging processing techniques and was validated on a small homogenous dataset. Similarly, Alomari et al11 published a system for binary classification of lumbar discs as degenerated or normal making use of a probabilistic computer vision model.
While early work tended to use probabilistic computer vision methods, subsequent work tended to make use of ML techniques. Hao et al12 made use of a support vector machine (SVM), which relied on disc segmentation followed by automated extraction of shape, intensity, and texture information. Building on this work, Ruiz-España et al,13 developed a classifier that classified lumbar discs into 1 of 5 classes as defined by Pfirrman.14 This approach made use of a segmentation algorithm that extracted features relating to the intensity, shape, and texture of the disc, which were passed to a custom classifier that made use of a classical image processing algorithm. The first published report to make use of an artificial neural network (ANN) was the study by Costro-Mateos et al,15 which used automated segmentation to extract disc features from a mid-sagittal T2-weighted image and trained an ANN based on these features.
The reports discussed up to this point all made use of an automated segmentation algorithm and relied on a relatively small number of training images, which limited their external validity. The use of a convolutional neural network (CNN) negates the need for a separate segmentation step prior to classification. The study by Jamaludin et al16 was innovative for its use of a relatively high number of MRIs when compared with previous studies. This study made use of 12 018 discs from 2009 patients and also made use of a CNN, which negated the need for a separate segmentation step, and allowed automatic extraction of features. This report achieved near human performance at grading lumbar disc degeneration, disc narrowing, upper/lower endplate defects, upper/lower marrow changes, spondylolisthesis, and central canal stenosis. Building on this work, the DeepSPINE framework used a large dataset of 22 796 lumbar disc levels extracted from 4075 patients and trained a CNN to perform classification of central canal stenosis and foraminal stenosis.17 These researchers achieved accuracy of 84.5% at grading lumbar spinal stenosis and 89.0% at grading lumbar foraminal stenosis, which exceeded that of any other published studies. In addition, the DeepSPINE framework performed similarly to human raters at detection and grading of lumbar spinal stenosis and foraminal stenosis.
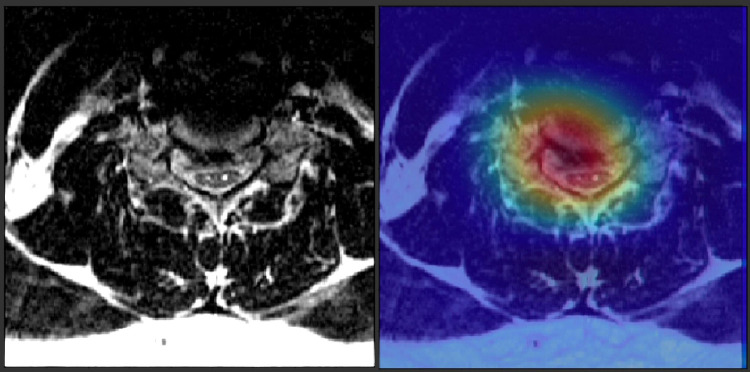
While most work on automated analysis of degenerative spinal imaging has focused on lumbar degenerative disease, some published reports have looked at cervical degenerative disease (Figure 3). Jin et al18 made use of diffusion tensor imaging (DTI) metrics in a population of patients undergoing surgery for degenerative cervical myelopathy. They employed ML methods to predict surgical outcome using these DTI metrics, but their predictive performance was limited by a small patient population. Weber et al19 made use of a CNN to quantify fatty infiltration in paraspinal musculature and showed clinical correlates of fat infiltration. These authors hypothesized that their metric could be used in the workup of patients with degenerative cervical myelopathy.
Figure 3.
Depiction of an attention map generated by a convolutional neural network (CNN) that has been trained to detect spinal cord compression on a cervical spine magnetic resonance imaging (MRI) scan. The CNN has identified the herniated intervertebral disc as being important to its classification decision.
Due to the aging population, the prevalence of spinal degenerative disease is increasing.20,21 The studies summarized here show the potential for ML technology to streamline patient care by providing rapid automated analysis of MRI scans, thereby reducing radiologist workloads. These studies also demonstrate that ML models can rapidly generate quantitative parameters from imaging data, which are time consuming for a radiologist to produce. Future ML models may be able to combine quantitative parameters from a patient’s imaging data, with clinical information such as patient demographics and neurologic exam and provide decision-making support to clinicians. In the future this type of decision-making tool making use of ML technology may be able to identify patients who will benefit most from surgery and may provide assistance with surgical planning.
Spinal Deformity
ML methods have also been applied to automated image analysis in spinal deformity. Initial work in the area focused on applying ML methods to data gathered from non-invasive surface topography scans to classify scoliotic curves. Ramirez et al22 made use of surface topography data to classify scoliotic patients into 3 categories: mild, moderate, and severe curves. This analysis found that a SVM achieved 85% accuracy in predicting scoliotic curve severity when using radiographs interpreted by radiologists as a gold standard. Seoud et al23 used data gathered from trunk surface topography to predict the Lenke class on 97 patients with scoliosis. These researchers achieved an overall accuracy of 72.7% when compared to radiologist classifications derived from radiographs.
More recently, research groups have attempted to automatically extract spinal parameters from spinal radiographs in the setting of scoliosis. Sun et al24 made use of coronal radiographs and trained an SVM to predict Cobb angles. When compared with human-derived measures, this classifier performed well with a high accuracy of 92.76%. The model had a root mean squared error of 21.6%, which indicates a good model fit to the training data. In a similar vein, Zhang et al25 used coronal radiographs to train an ANN to estimate Cobb angles. When compared with human-derived measures they achieved absolute errors of 3 degrees.
A comprehensive automated platform for assessment of idiopathic scoliosis was developed by Wu et al.26 This system, called MVC-Net, makes use of biplanar radiographs to identify each vertebral level and estimates Cobb angle on an anteroposterior and lateral view. This system used 526 images for training and validation and achieved high accuracy when compared with radiographs that were manually annotated by radiologists with a mean absolute error of 4° across the testing samples. Similarly, Galbusera et al27 developed a system for automated estimation of spinal parameters based on 2-planar radiographs. Their system involved a CNN trained on 493 patients and was able to extract the spatial locations of various landmarks such as endplate centers, hip joint centers, and the S1 endplate angle. From this they were able to derive spinal parameters such as T4-T12 kyphosis, L1-L5 lordosis, Cobb angle of scoliosis, pelvic incidence, sacral slope, and pelvic tilt. While the model-generated parameters showed good correlation to radiologist-derived values, the standard errors of the estimated parameters ranged from 2.7° for pelvic tilt to 11.5° for L1-5 lordosis.
Research to date has focused on automatically generating quantitative spinal parameters such as T4-T12 kyphosis, pelvic incidence, and pelvic tilt from radiographs. Spinal parameters are time consuming to manually annotate and interrater reliability can vary. The above studies demonstrate that a ML learning model could potentially facilitate more rapid and consistent interpretation of spinal radiographs. Future work may make use of larger datasets to compare spinal parameters generated by an ML model to those manually annotated by human raters. In addition, future research may seek to automatically classify adult spinal deformity according to the validated Scoliosis Research Society (SRS)–Schwab system. Ultimately, this line of research may lead to a model that makes use of automatically generated spinal parameters to provide decision-making support to clinicians by predicting patient response to surgery and optimal surgical approach.
Spine Trauma
The clinical management of patients with spine trauma requires rapid and accurate interpretation of volumetric imaging. For this reason, automated image analysis has a special potential to streamline the care of patient with spine trauma.
The majority of work in applying ML methods to automated image analysis in spine trauma has focused on thoracolumbar trauma. Yao et al28 first developed a fully automated system for detection of thoracolumbar fractures. This system used a segmentation method, followed by a transformation of the 3-dimensional (3D) vertebral body shape onto a 2D space. These researchers trained a committee of SVMs to recognize fracture patterns on the 2D representation. While they made use of a sophisticated method this initial report was limited by a low number of training samples. The same research group expanded this work by applying the same model to a larger dataset of 104 patients.29 In this dataset, they achieved a sensitivity of 0.81 with a false positive rate of 2.7. This research group later applied their model to detect and classify thoracolumbar osteoporotic fractures based on the Genant classification system.30 They assembled a cohort of 75 patients with thoracolumbar fractures and 75 matched patients with normal CT scans. They achieved a sensitivity for detection of compression fractures at the patient level of 98.7% and specificity of 77.3%. In addition, for each fracture within their imaging data their model attempted to classify the fracture into 1 of 4 Genant classes. They achieved an accuracy for classification by Genant type of 68%.
Some researchers have attempted to apply a CNN to detect thoracolumbar osteoporotic fractures. Nicolaes et al31 used a training database of 90 CT scans, which were labeled with localizing information by 2 raters. This group made use of a 3D CNN and achieved accuracy similar to previously published reports. Similarly, Baum et al32 developed a system for automatic detection of osteoporotic fractures in thoracolumbar imaging, which made use of a 2D CNN. The report by Tomita et al33 made use of the largest dataset, which included 1432 CT scans with 713 positive and 719 negative images of osteoporotic compression fractures. These researchers labeled mid sagittal images in a location agnostic manner and trained a CNN on their training dataset. This model achieved an accuracy of 89.2% and F1 score of 90.8% on the holdout dataset.
Studies to date have focused on specific fracture types, such as osteoporotic compression fractures and have used smaller datasets. This line of research is in its infancy and no published studies have been able to achieve human-level performance. However, a large volume of CT scans are gathered in an emergency setting and future work could leverage this large volume of data. The goal of this line of research is to produce a machine learning model that is general enough to detect any type of spinal fracture with better-than human-level performance. Such a model, if developed, could reduce workload for radiologists and increase sensitivity for subtle findings.
Spine Oncology
Compared with other aspects of spine surgery, fewer researchers have applied machine learning technology to interpretation of imaging in spinal oncology. Hammon et al34 made use of images from 114 patients and developed an SVM model to detect spinal metastases on CT scans. Similar work was undertaken by O’Connor et al,35 but both of these studies were limited by small imaging datasets. A more recent study made use of an innovate approach, which used a segmentation algorithm that identified regions of the image with similar signal characteristics.36 An SVM was then used to classify the segmented regions to identify areas suspicious for tumor. Again, the algorithm was limited by a small training sample of 49 patients and had a relatively high false positive rate. Wang et al37 made use of an CNN trained with MRI scans from 26 patients to detect metastatic lesions. In this case, a separate testing sample of images was not available to determine model performance.
Future research in spinal oncology may go beyond tumor detection and focus on automatically generating clinically meaningful parameters from MRI scans such as Bilsky grade and an estimate of spinal instability. In addition, the parameters generated by ML models may be combined with clinical information such as patient demographics, degree of impairment, and oncologic information, to assist clinicians with identifying patients who would benefit from surgery.
Future Directions
The increasing usage of ML technology has the potential to directly affect the care of patients with spinal disease by streamlining the interpretation of medical imaging and supporting diagnoses and clinical decisions. Despite the progress made in the research setting, there has thus far been limited usage of these technologies in a clinical setting, which highlights the challenges that remain to be overcome. The challenges that need to be overcome prior to wider clinical usage of ML technology can be broadly grouped into technological challenges and cultural challenges.
Improved accessibility of powerful workstations with graphical processing units have been largely responsible for the recent progress in computer vision and image processing. Medical imaging data, however, pose unique technological challenges.38 While most medical imaging data used within spine surgery is volumetric, many machine learning models are developed to accept 2D inputs.39,40 The majority of researchers have attempted to overcome this challenge by representing 3D spinal imaging in a 2D form. More recently, some research groups have attempted to use 3D convolutional neural networks to directly analyze 3D medical imaging data. While 3D CNNs can offer better performance, they are computationally expensive and medical imaging data must often be downsampled to accommodate currently available hardware limitations.41 Further advances in computer hardware and innovative data processing solutions are likely required to develop more robust ML models with human-level performance.
Perhaps as significant as the technological challenges are cultural challenges, which refer to systemic barriers in assembling large datasets due to data privacy concerns and the challenge of integrating computer-aided diagnostics into a traditional clinical workflow. The development of robust diagnostic tools relying on ML technology requires the assembly of large annotated datasets. The most successful CNNs, which have achieved widespread usage outside of medical imaging, such as Google’s Inception network, required millions of images during the training phase.42 It is recognized that to train an ML model for a specific clinical task would likely require tens of thousands of training images.4 Such datasets may need to be assembled from multiple institutions. Despite standard practices of data anonymization, medical imaging data in all jurisdictions are subject to strict regulations regarding storage, transmission, and usage, which creates challenges in assembling a large dataset.43 These challenges are more pronounced in certain jurisdictions. In the European Union, for example, the recent introduction of the General Data Protection Regulation, adopted an explicit opt-in policy for the usage of patient data for research purposes.44 Assemblage of sufficiently sized imaging datasets within spine surgery will require collaborations between institutions and a keen awareness of applicable data privacy regulations.
Another broad cultural challenge is that of accountability for medical decisions. Most ML models appear as a “black box” to an external user and the process by which the model arrives at a clinical diagnosis cannot be easily understood or interpreted by a human observer. If a hypothetical ML model were to fail, for example, to detect a cervical spinal fracture it is at this time unclear if the responsibility would fall on the clinician using the ML system or the manufacturer of the device. The high acuity of spinal disorders and the ramifications of a misdiagnosis within spinal surgery make this especially relevent.45,46 As diagnostic ML tools achieve better performance within spine surgery and clinicians come to more heavily rely on automated diagnoses this ethical issue will need to be debated further.
Conclusion
In recent years, ML technology has reached a substantial level of development. Within spine surgery, ML technologies have achieved near human performance in narrow classification tasks on specific datasets. Prior to more widespread clinical usage, however, ML technologies will need to demonstrate human-level performance on larger datasets and more generalized classification tasks. The goal of this field of research is to produce a generalized solution capable of providing comprehensive automated analysis of volumetric imaging. In addition, the coupling of clinical data with image analysis may one day permit improved treatment decision making, with an overall aim of improving surgical care and patient outcomes. Although substantial challenges remain to be overcome, ML technology has improved rapidly in the past few years and progress is expected to continue.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This supplement was supported by a grant from AO Spine North America.
ORCID iD: Zamir A. Merali, MD  https://orcid.org/0000-0002-6667-3035
https://orcid.org/0000-0002-6667-3035
References
- 1. Splawinski J, Fox R, Hall H, Fisher CG, Dvorak M. Imaging for spinal surgery. Can J Surg. 2006;49:311. [PMC free article] [PubMed] [Google Scholar]
- 2. Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys. 2019;29:102–127. doi:10.1016/j.zemedi.2018.11.002 [DOI] [PubMed] [Google Scholar]
- 3. Bortolan G, Degani R, Willems JL. ECG classification with neural networks and cluster analysis. In: Proceedings of Computers in Cardiology. IEEE; 1992:177–180. doi:10.1109/cic.1991.169074 [Google Scholar]
- 4. Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500–510. doi:10.1038/s41568-018-0016-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang S, Summers RM. Machine learning and radiology. Med Image Analysis. 2012;16:933–951. doi:10.1016/j.media.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan S, Siegel EL. Will machine learning end the viability of radiology as a thriving medical specialty? Br J Radiol. 2019;92:20180416. doi:10.1259/bjr.20180416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. US Food and Drug Administration. Proposed regulatory framework for modifications to artificial intelligence/machine learning (AI/ML)-based Software as a Medical Device (SaMD)—discussion paper and request for feedback. Accessed September 9, 2020. https://www.fda.gov/files/medical%20devices/published/US-FDA-Artificial-Intelligence-and-Machine-Learning-Discussion-Paper.pdf
- 8. Setio AAA, Ciompi F, Litjens G, et al. Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans Med Imaging. 2016;35:1160–1169. doi:10.1109/TMI.2016.2536809 [DOI] [PubMed] [Google Scholar]
- 9. Chilamkurthy S, Ghosh R, Tanamala S, et al. Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet. 2018;392:2388–2396. doi:10.1016/S0140-6736(18)31645-3 [DOI] [PubMed] [Google Scholar]
- 10. Michopoulou SK, Costaridou L, Panagiotopoulos E, Speller R, Panayiotakis G, Todd-Pokropek A. Atlas-based segmentation of degenerated lumbar intervertebral discs from MR images of the spine. IEEE Trans Biomed Eng. 2009;56:2225–2231. doi:10.1109/TBME.2009.2019765 [DOI] [PubMed] [Google Scholar]
- 11. Alomari RS, Corso JJ, Chaudhary V, Dhillon G. Desiccation diagnosis in lumbar discs from clinical MRI with a probabilistic model. In: Proceedings of the 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. ISBI; 2009:546–549. doi:10.1109/ISBI.2009.5193105 [Google Scholar]
- 12. Hao S, Jiang J, Guo Y, Li H. Active learning based intervertebral disk classification combining shape and texture similarities. Neurocomputing. 2013;101:252–257. doi:10.1016/j.neucom.2012.08.008 [Google Scholar]
- 13. Ruiz-España S, Arana E, Moratal D. Semiautomatic computer-aided classification of degenerative lumbar spine disease in magnetic resonance imaging. Comput Biol Med. 2015;62:196–205. doi:10.1016/j.compbiomed.2015.04.028 [DOI] [PubMed] [Google Scholar]
- 14. Urrutia J, Besa P, Campos M, et al. The Pfirrmann classification of lumbar intervertebral disc degeneration: an independent inter- and intra-observer agreement assessment. Eur Spine J. 2016;25:2728–2733. doi:10.1007/s00586-016-4438-z [DOI] [PubMed] [Google Scholar]
- 15. Castro-Mateos I, Pozo JM, Lazary A, Frangi AF. 2D segmentation of intervertebral discs and its degree of degeneration from T2-weighted magnetic resonance images. In: SPIE 9035 Medical Imaging; 2014: Computer-Aided Diagnosis. Vol. 9035. SPIE; 2014:903517. doi:10.1117/12.2043755 [Google Scholar]
- 16. Jamaludin A, Kadir T, Zisserman A. SpineNet: automated classification and evidence visualization in spinal MRIs. Med Image Analysis. 2017;41:63–73. doi:10.1016/j.media.2017.07.002 [DOI] [PubMed] [Google Scholar]
- 17. Lu JT, Pedemonte S, Bizzo B, et al. DeepSPINE: automated lumbar vertebral segmentation, disc-level designation, and spinal stenosis grading using deep learning. Published July 26, 2018. Accessed May 5, 2020. http://arxiv.org/abs/1807.10215
- 18. Jin R, Luk KD, Cheung J, Hu Y. A machine learning based prognostic prediction of cervical myelopathy using diffusion tensor imaging. Paper presented at: 2016 IEEE International Conference on Computational Intelligence and Virtual Environments for Measurement Systems and Applications, CIVEMSA; June 27-28, 2016; Budapest, Hungary. doi:10.1109/CIVEMSA.2016.7524318
- 19. Weber KA, Smith AC, Wasielewski M, et al. Deep learning convolutional neural networks for the automatic quantification of muscle fat infiltration following whiplash injury. Sci Rep. 2019;9:7973. doi:10.1038/s41598-019-44416-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nouri A, Tetreault L, Singh A, Karadimas SK, Fehlings MG. Degenerative cervical myelopathy: Epidemiology, genetics, and pathogenesis. Spine (Phila Pa 1976). 2015;40:E675–E693. doi:10.1097/BRS.0000000000000913 [DOI] [PubMed] [Google Scholar]
- 21. Karadimas SK, Erwin WM, Ely CG, Dettori JR, Fehlings MG. Pathophysiology and natural history of cervical spondylotic myelopathy. Spine (Phila Pa 1976). 2013;38(22 suppl 1):S21–S36. doi:10.1097/BRS.0b013e3182a7f2c3 [DOI] [PubMed] [Google Scholar]
- 22. Ramirez L, Durdle NG, Raso VJ, Hill DL. A support vector machines classifier to assess the severity of idiopathic scoliosis from surface topography. IEEE Trans Inf Technol Biomed. 2006;10:84–91. doi:10.1109/TITB.2005.855526 [DOI] [PubMed] [Google Scholar]
- 23. Seoud L, Adankon MM, Labelle H, Dansereau J, Cheriet F. Prediction of scoliosis curve type based on the analysis of trunk surface topography. In: Proceedings of 7th IEEE International Symposium on Biomedical Imaging: From Nano to Macro. ISBI; 2010:408–411. doi:10.1109/ISBI.2010.5490322 [Google Scholar]
- 24. Sun H, Zhen X, Bailey C, Rasoulinejad P, Yin Y, Li S. Direct estimation of spinal cobb angles by structured multi-output regression. In: Niethammer M, Styner M, Aylward S, et al. eds. Information Processing in Medical Imaging. IPMI 2017. Lecture Notes in Computer Science, Vol 10265. Springer; 2017. doi:10.1007/978-3-319-59050-9 [Google Scholar]
- 25. Zhang J, Lou E, Shi X, et al. A computer-aided cobb angle measurement method and its reliability. J Spinal Disord Tech. 2010;23:383–387. doi:10.1097/BSD.0b013e3181bb9a3c [DOI] [PubMed] [Google Scholar]
- 26. Wu H, Bailey C, Rasoulinejad P, Li S. Automated comprehensive Adolescent Idiopathic Scoliosis assessment using MVC-Net. Med Image Anal. 2018;48:1–11. doi:10.1016/j.media.2018.05.005 [DOI] [PubMed] [Google Scholar]
- 27. Galbusera F, Niemeyer F, Wilke HJ, et al. Fully automated radiological analysis of spinal disorders and deformities: a deep learning approach. Eur Spine J. 2019;28:951–960. doi:10.1007/s00586-019-05944-z [DOI] [PubMed] [Google Scholar]
- 28. Yao J, Burns JE, Munoz H, Summers RM. Detection of vertebral body fractures based on cortical shell unwrapping. In: In: Ayache N, Delingette H, Golland P, Mori K, eds. Medical Image Computing and Computer-Assisted Intervention—MICCAI 2012. MICCAI 2012. Lecture Notes in Computer Science. Vol. 7512. Springer Verlag; 2012:509–516. doi:10.1007/978-3-642-33454-2_63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burns JE, Yao J, Muñoz H, Summers RM. Automated detection, localization, and classification of traumatic vertebral body fractures in the thoracic and lumbar spine at CT. Radiology. 2016;278:64–73. doi:10.1148/radiol.2015142346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Burns JE, Yao J, Summers RM. Vertebral body compression fractures and bone density: Automated detection and classification on CT Images. Radiology. 2017;284:788–797. doi:10.1148/radiol.2017162100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nicolaes J, Raeymaeckers S, Robben D, et al. Detection of vertebral fractures in CT using 3D convolutional neural networks. Accessed May 7, 2020. http://arxiv.org/abs/1911.01816
- 32. Baum T, Bauer JS, Klinder T, et al. Automatic detection of osteoporotic vertebral fractures in routine thoracic and abdominal MDCT. Eur Radiol. 2014;24:872–880. doi:10.1007/s00330-013-3089-2 [DOI] [PubMed] [Google Scholar]
- 33. Tomita N, Cheung YY, Hassanpour S. Deep neural networks for automatic detection of osteoporotic vertebral fractures on CT scans. Comput Biol Med. 2018;98:8–15. doi:10.1016/j.compbiomed.2018.05.011 [DOI] [PubMed] [Google Scholar]
- 34. Hammon M, Dankerl P, Tsymbal A, et al. Automatic detection of lytic and blastic thoracolumbar spine metastases on computed tomography. Eur Radiol. 2013;23:1862–1870. doi:10.1007/s00330-013-2774-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. O’Connor SD, Yao J, Summers RM. Lytic metastases in thoracolumbar spine: Computer-aided detection at CT—preliminary study. Radiology. 2007;242:811–816. doi:10.1148/radiol.2423060260 [DOI] [PubMed] [Google Scholar]
- 36. Burns JE, Yao J, Wiese TS, Munoz HE, Jones EC, Summers RM. Automated detection of sclerotic metastases in the thoracolumbar spine at CT. [Erratum appears in Radiology. 2013;269(1):311]. Radiology. 2013;268:69–78. doi:10.1148/radiol.13121351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang J, Fang Z, Lang N, Yuan H, Su MY, Baldi P. A multi-resolution approach for spinal metastasis detection using deep Siamese neural networks. Comput Biol Med. 2017;84:137–146. doi:10.1016/j.compbiomed.2017.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Singh SP, Wang L, Gupta S, Goli H, Padmanabhan P, Gulyás B. 3D deep learning on medical images: a review. Accessed September 9, 2020. https://arxiv.org/abs/2004.00218 [DOI] [PMC free article] [PubMed]
- 39. Razzak MI, Naz S, Zaib A. Deep learning for medical image processing: overview, challenges and future. Accessed September 9, 2020. https://arxiv.org/abs/1704.06825
- 40. Kim M, Yun J, Cho Y, et al. Deep learning in medical imaging. Neurospine. 2019;16:657–668. doi:10.14245/ns.1938396.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deniz CM, Xiang S, Hallyburton RS, et al. Segmentation of the proximal femur from MR images using deep convolutional neural networks. Sci Rep. 2018;8:16785. doi:10.1038/s41598-018-34817-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. Vol. 07-12-June-2015. IEEE Computer Society; 2015:1–9. doi:10.1109/CVPR.2015.7298594 [Google Scholar]
- 43. Pesapane F, Volonté C, Codari M, Sardanelli F. Artificial intelligence as a medical device in radiology: ethical and regulatory issues in Europe and the United States. Insights Imaging. 2018;9:745–753. doi:10.1007/s13244-018-0645-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Char DS, Shah NH, Magnus D. Implementing machine learning in health care—addressing ethical challenges. N Engl J Med. 2018;378:981–983. doi:10.1056/NEJMp1714229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thierer A, O’sullivan AC, Russell R. Artificial Intelligence and Public Policy. Mercatus Center at George Mason; 2017. [Google Scholar]
- 46. Winfield AFT, Jirotka M. Ethical governance is essential to building trust in robotics and artificial intelligence systems. Philos Trans A Math Phys Eng Sci. 2018;376:20180085. doi:10.1098/rsta.2018.0085 [DOI] [PMC free article] [PubMed] [Google Scholar]