Abstract
Background
Soluble receptor for advanced glycation end products (sRAGE) is a proposed emphysema and airflow obstruction biomarker; however, previous publications have shown inconsistent associations and only one study has investigate the association between sRAGE and emphysema. No cohorts have examined the association between sRAGE and progressive decline of lung function. There have also been no evaluation of assay compatibility, receiver operating characteristics, and little examination of the effect of genetic variability in non-white population. This manuscript addresses these deficiencies and introduces novel data from Pittsburgh COPD SCCOR and as well as novel work on airflow obstruction. A meta-analysis is used to quantify sRAGE associations with clinical phenotypes.
Methods
sRAGE was measured in four independent longitudinal cohorts on different analytic assays: COPDGene (n = 1443); SPIROMICS (n = 1623); ECLIPSE (n = 2349); Pittsburgh COPD SCCOR (n = 399). We constructed adjusted linear mixed models to determine associations of sRAGE with baseline and follow up forced expiratory volume at one second (FEV1) and emphysema by quantitative high-resolution CT lung density at the 15th percentile (adjusted for total lung capacity).
Results
Lower plasma or serum sRAGE values were associated with a COPD diagnosis (P < 0.001), reduced FEV1 (P < 0.001), and emphysema severity (P < 0.001). In an inverse-variance weighted meta-analysis, one SD lower log10-transformed sRAGE was associated with 105 ± 22 mL lower FEV1 and 4.14 ± 0.55 g/L lower adjusted lung density. After adjusting for covariates, lower sRAGE at baseline was associated with greater FEV1 decline and emphysema progression only in the ECLIPSE cohort. Non-Hispanic white subjects carrying the rs2070600 minor allele (A) and non-Hispanic African Americans carrying the rs2071288 minor allele (A) had lower sRAGE measurements compare to those with the major allele, but their emphysema-sRAGE regression slopes were similar.
Conclusions
Lower blood sRAGE is associated with more severe airflow obstruction and emphysema, but associations with progression are inconsistent in the cohorts analyzed. In these cohorts, genotype influenced sRAGE measurements and strengthened variance modelling. Thus, genotype should be included in sRAGE evaluations.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12931-021-01686-z.
Keywords: COPD, Emphysema, Biomarkers, Progression
Clinical relevance
The soluble receptor for advanced glycation end products (sRAGE) is an emerging blood protein biomarker for emphysema and chronic obstructive pulmonary disease (COPD).
This study uses four independent cohorts and four distinct sRAGE assay platforms to confirm that sRAGE is an independent blood biomarker for the presence and severity of both emphysema and COPD; however, the association between baseline sRAGE and emphysema progression or COPD progression is less consistent. Furthermore, although there is correlation among different sRAGE assay platforms, many platforms, but not all, have variable sRAGE detection dependent on a subject’s genotype, suggesting that the genetic background should be considered when interpreting sRAGE measurements.
Background
The Receptor for Advanced Glycation End Products (RAGE; UniProtKB—Q15109) is a 41-kD multi-ligand transmembrane receptor belonging to the immunoglobulin gene superfamily [1]. Cleavage of the extracellular domain of RAGE results in a 35 kD soluble RAGE (sRAGE), which can be measured in plasma or serum. Little is known about the exact functions of sRAGE; however, lower levels of sRAGE have been reported to be associated with increase risk of chronic diseases such as diabetes [2], atherosclerosis [3], coronary artery disease [2], diabetic retinopathy [4], and chronic obstructive pulmonary disease (COPD) [5]. Elevated sRAGE indicated alveolar epithelial cell injury in infection-related ARDS [6] and in diabetic nephropathy [7, 8]. sRAGE is positively associated with other proinflammatory advanced glycation end products (AGEs) [9] and negatively associated with other proinflammatory markers such as C-reactive protein (CRP), fibrinogen, and white blood cell counts [10].
More is known about transmembrane RAGE. Over-expression of transmembrane RAGE has been shown to have a protective role in experimental models including RSV infection in HEK293 cells [11]. Mice overexpressing AGER, the gene encoding RAGE, develop emphysema [12]. AGER knockout mice are resistant to tobacco smoke-induced lung disease [13] and are protected from LPS-induced lung injury [6]. Engagement of RAGE by AGEs activates inflammatory signalling pathways, including nuclear factor (NF)-kB [14] and several mitogen-activated protein kinases [15, 16]. This RAGE signalling may contribute to the sustained inflammation seen in COPD. When the extracellular portion of RAGE is cleaved, the protein becomes soluble (sRAGE) and can be measured in serum and plasma. sRAGE has been hypothesized to bind competitively to AGEs, thus reducing the transmembrane signalling of RAGE and of other pathogen-associated molecular patterns (PAMPs) or damage-associated molecular pattern (DAMPs) receptors through other pattern recognition receptors (PRRs) implicated in chronic lung inflammation [17].
In human studies, four large COPD cohorts (ECLIPSE, COPDGene, TESRA, SPIROMICS) [18–20] and several smaller studies [21–23] have reported that sRAGE is the biomarker showing the strongest known association with emphysema, even independent of airflow obstruction and other clinical covariates (age, sex, current smoking, pack-years, BMI, and prior exacerbation history). All studies demonstrated that lower levels of plasma or serum sRAGE were associated with more emphysema as measured by the 15th percentile density of lung density (PD15) or the low attenuation area at -950 Hounsfield units (LAA, the percent lung tissue voxels less than -950 HU). One study (ECLIPSE) was sufficiently powered to show that lower sRAGE was associated with more rapid progression of emphysema as measured by change in PD15 over time [20]. Although abundant evidence supports a cross-sectional association between plasma/serum sRAGE and emphysema/airflow obstruction, there are few reports of its association with COPD progression, its receiver operating characteristics, and how genetics simultaneously impacts protein level and disease associations. The goal of this study is to conduct all of these evaluations using 4 different COPD cohorts and summarize results using a meta analysis for sRAGE’s association with PD15adj. and FEV1 decline.
Methods
Cohorts
This analysis includes data from participants from four independent cohorts: Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE) [24]; Genetic Epidemiology of COPD (COPDGene) [25]; Subpopulations and Intermediate Outcome Measures in COPD Study (SPIROMICS) [26]; and Specialized Center for Clinically Oriented Research (SCCOR) in COPD at the University of Pittsburgh [27, 28]. Although all four cohorts enrolled predominantly older current and former smokers, there were some differences in study recruitment and the percentage of participants with COPD. COPDGene and SPIROMICS were multi-center U.S. cohorts of current and ex-smokers (> 10 and 20 pack-years respectively) with and without COPD. ECLIPSE was an international cohort from 12 countries that included predominantly moderate-very severe COPD subjects. The Pittsburgh COPD SCCOR was a single-center U.S. study. All participants signed a written informed consent. All studies were approved by the ethics and review boards at all participating centers. The current analyses include only the subset of subjects from those four cohorts who had at least one measurement of sRAGE and either spirometry or quantitative CT measurements of emphysema (Table 1).
Table 1.
Baseline clinical characteristics of subjects who have an sRAGE measurement by cohort
| COPDGene (n = 1443) | ECLIPSE (n = 2349) | SCCOR (n = 399) | SPIROMICS (n = 1623) | p-value | |
|---|---|---|---|---|---|
| Age (mean ± SD) | 61 ± 9 | 62 ± 8 | 65 ± 6 | 64 ± 9 | < 0.001 |
| Sex (male) (%) | 49% | 62% | 53% | 54% | < 0.001 |
| Race (%) | |||||
| Non-Hispanic White | 86% | 93.5% | 95% | 75% | < 0.001 |
| Non-Hispanic African American | 14% | 1.5% | 4% | 16% | |
| Other | 0% | 5% | 1% | 9% | |
| BMI (kg/m2) (mean ± SD) | 29 ± 6 | 27 ± 5 | 28 ± 4 | 28 ± 5 | < 0.001 |
| Never Smoker (%) | 2% | 9% | 0% | 8% | < 0.001 |
| Current Smoker (%) | 39% | 35% | 42% | 34% | |
| Pack-years median (5th and 95th percentile) | 38.4 (11.3; 90.0) | 39 (0; 95) | 46.0 (19.0; 118.0) | 42.0 (0; 96.0) | < 0.001 |
| COPD (%) | 41% | 78% | 48% | 62% | < 0.001 |
| PRISm (%) | 10% | 0.2% | 5% | 2% | < 0.001 |
| FEV1 (% predicted) (mean ± SD) | 81 ± 25 | 62 ± 30 | 83 ± 20 | 76 ± 26 | < 0.001 |
| FEV1 (L) (mean ± SD) | 2.36 ± 0.91 | 1.79 ± 1.00 | 2.39 ± 0.76 | 2.15 ± 0.91 | < 0.001 |
| FVC (L) (mean ± SD) | 3.44 ± 1.00 | 3.30 ± 1.03 | 3.54 ± 0.89 | 3.44 ± 1.02 | 0.006 |
| Emphysema (% LAA < − 950 HU) median (5th and 95th percentile) | 1.40 (0.08; 26.19) | 11.48 (0.49; 39.18) | 0.80 (0.10; 17.40) | 3.07 (0.29; 29.54) | < 0.001 |
| PD15adj (g/L) (mean ± SD) | 89 ± 24 | 61 ± 26 | 87 ± 21 | 83 ± 26 | < 0.001 |
| History of diabetes (%) | 11% | 9% | 8% | 13% | < 0.001 |
| History of heart attack (%) | 6% | 8% | 5% | 6% | 0.009 |
| History of coronary artery disease (%) | 7% | NC | 6% | 9% | 0.12 |
| History of stroke (%) | 2% | 3% | 3% | 4% | 0.11 |
| Follow-up (years) median (5th and 95th percentile) | 5.14 (0; 10.1) | 3.0 (1.5; 3.0) | 6.0 (2.0; 6.0) | 3.1 (0; 7.5) | < 0.001 |
| Percentage of visits with a spirometry per participant | |||||
| 0 | 0.1% | 0.1% | |||
| 1 | 39% | 0.04% | 12% | ||
| 2 | 44% | 0.04% | 39% | 12% | |
| 3 | 16% | 1% | 61% | 18% | |
| 4 | 4% | 34% | |||
| 5 | 4% | 24% | |||
| 6 | 5% | ||||
| 7 | 10% | ||||
| 8 | 76% | ||||
| Number of visits with CT scan per participant | |||||
| 0 | 1% | 7% | 0.1% | ||
| 1 | 41% | 11% | 13% | ||
| 2 | 58% | 21% | 39% | 50% | |
| 3 | 61% | 61% | 36% | ||
To evaluate differences between cohorts, analysis of variance (ANOVA) was used for normally distributed continuous variables and Kruskal–Wallis test for non-normaly distributed variables; and a Chi square/Fisher’s exact test for categorical
COPD chronic obstructive pulmonary disease, PRISm Preserved Ratio Impaired Spirometry, FEV1 forced expiratory volume in one second, FVC forced vital capacity, PD15adj HU of the 15th Percentile adjusted for total lung capacity, NC not collected
COPDGene (NCT02445183) enrolled 10,300 subjects ages 45–80. sRAGE was measured in a representative sample of 1,443 subjects at the baseline using fresh frozen plasma with an sRAGE assay by Quotient Bioresearch (QBR) as previously described [29]. Additional sRAGE assays were performed in 594 subjects using fresh frozen plasma, with an sRAGE assay by Myriad-Rules Based Medicine (Myriad-RBM) as previously described [30], in 509 subjects using liquid chromatography-mass spectrometry (LCMS) [31], and in 1243 subjects using an sRAGE specific aptameric assay (Sequence ID 4125_52_2) on the SOMAscan 1.3 K panel (Fig. 1). Spirometry and CT scans were obtained at baseline and Year 5, with spirometry data available for the first 2,088 returning Year 10 participants.
Fig. 1.
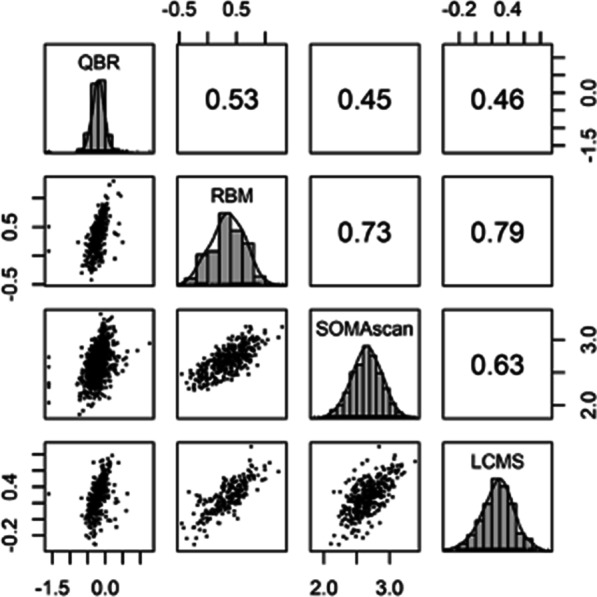
sRAGE correlation among different platforms: Quotient Bioresearch (QBR, n = 1448), Rules Based Medicine (RBM, n = 594), SOMAscan (aptamer 4125_52_2, n = 1248) and liquid chromatography/mass spectrometry (LCMS, n = 509). Axes are on a log10 scale. Units are ng/ml (QBR, RBM, LCMS), and scale free (SOMAscan). Data are from the COPDGene cohort. p-value < 0.001 for all correlations shown
SPIROMICS (NCT01969344) enrolled 2973 subjects. sRAGE was measured in 1623 subjects at baseline using fresh frozen plasma from EDTA (BD) tubes with the Myriad-RBM assay, as previously described [30]. Spirometry was measured at baseline (visit 1), visit 2–4 (Year 1–3), and visit 5 (mean 6 years after baseline), and; CT scans were obtained at baseline and visit 1 (Year 1) and visit 5 (mean 6 years after baseline).
ECLIPSE (NCT00292552) enrolled 2746 subjects. sRAGE was measured in serum from the Year 1 visit in 2349 subjects using a QBR assay as previously described [19]. CT scans were obtained at baseline, Year 1 and Year 3. There have been previous publications of the associations between emphysema and sRAGE [20]. Spirometry was measured at baseline, 3 and 6 months, and every 6 months, with the last measurement obtained at Year 3.
Pittsburgh SCCOR recruited subjects primarily from the Pittsburgh Lung Screening Study cohort, a tobacco-exposed cohort with only a subset of subjects having spirometrically confirmed obstructive lung disease. The complete description of subject recruitment and clinical evaluation were described in detail elsewhere [27, 28]. A total of 399 of the Pittsburgh SCCOR subjects with available follow-up study were used to analyze sRAGE levels using ELISA (DuoSet for human sRAGE, R & D Systems) and citrate plasma according to the manufacturer’s instructions. All samples were analyzed in duplicate. Spirometry and CTs were measured at baseline, Years 2 and 6.
Clinical phenotypes and their harmonization, sRAGE assays, genotyping, and statistical analyses plan are described in the Additional file 1: Methods.
Results
Correlation of different sRAGE assays
Among the four platforms used to measure the same samples in COPDGene, correlations were highest between RBM and LCMS (0.79), then RBM and SOMAscan (0.73), SOMAscan and LCMS (0.63), QBR and RBM (0.53), QBR and LCMS (0.46) and lowest for QBR and SOMAscan (0.45) (Fig. 1). Bland–Altman plots reveal that there were significant differences among the means, and also proportional bias, particularly when the RBM platform was a comparator (Additional file 1: Figure S1). For this reason, we chose to meta-analysis and recommend assay specific parameters be used.
Demographics
Baseline characteristics of the COPDGene, ECLIPSE, Pittsburgh SCCOR, and SPIROMICS cohorts are shown in Table 1. SPIROMICS and COPDGene had more than 10% minorities (mostly African Americans), but the ECLIPSE and SCCOR cohorts were almost exclusively white. The COPDGene subsets of subjects who had sRAGE measured on more than one biomarker platform were similar (Additional file 1: Table S1).
On cross-sectional analysis (Additional file 1: Table S2), higher sRAGE was significantly associated with more advanced age, female sex, and non-Hispanic white race (compared to non-Hispanic African American race). Current smoking was associated with significantly higher levels of sRAGE in ECLIPSE and SCCOR. These two cohort populations were predominantly non-Hispanic white, which is associated with higher levels of sRAGE compared to non-Hispanic African Americans. Controlling for race in the analysis of current smoking had minimal effect on the associations with current smoking in ECLIPSE (β = 0.031; p = 0.0004) or SCCOR (β = 0.059; p = 0.0082); but in COPDGene and SPIROMICS significantly higher levels of sRAGE were associated with current smoking, (β = 0.039; p = 0.001) and (β = 0.051, p = 0.0043) respectively. sRAGE was not associated with comorbidities such as diabetes, cardiovascular disease, or stroke (Additional file 1: Table S2).
sRAGE is strongly associated with severe airflow obstruction
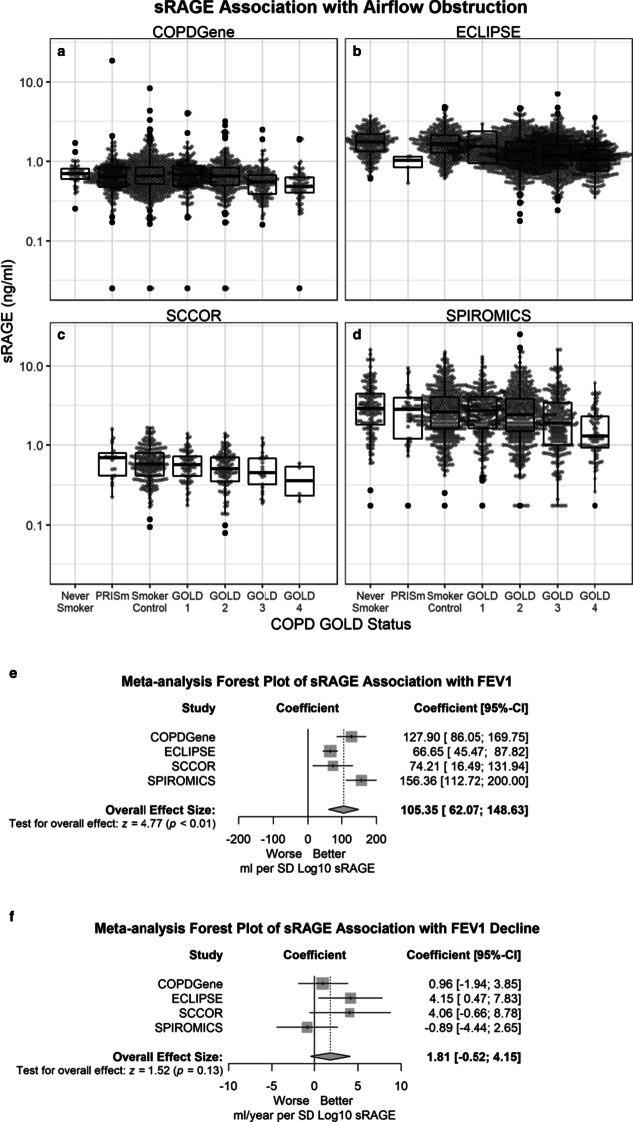
sRAGE was significantly lower in subjects with airflow obstruction compared to never smokers and current and former smokers without COPD (Fig. 2; P < 0.001 for all cohorts except SCCOR: P = 0.03) and with adjusted decrease in FEV1 at baseline (Table 2). After adjustment for covariates, one standard deviation lower log10 sRAGE was associated with a weighted average of 105.35 ml lower FEV1 (62.07; 148.63) (Fig. 2). After adjustment for covariates, sRAGE was not significantly associated with changes in FEV1 over time in 3 cohorts, except in ECLIPSE (Fig. 2).
Fig. 2.
More severe airflow obstruction is associated with lower plasma and serum sRAGE in multiple cohorts and with different assay platforms for sRAGE. sRAGE is shown on the log-scale y-axis. Shown are the QBR assays for COPDGene (n = 1437) (a) and ECLIPSE (n = 2342) (b), DuoSet for sRAGE assay for SCCOR (n = 399) (c), and RBM assay for R&D SPIROMICS (n = 1620) (d). Median, 25th percentile, 75th percentile, and whiskers (the minimum of 1.5 times interquartile range (IQR) or highest/lowest value) are shown in the box plots. e Forest plot of sRAGE effect size estimates for baseline FEV1 for each cohort (squares) as well as a weighted estimate of the meta-analysis (diamond). The shaded represents the interquartile range and the whiskers represent the 95% confidence interval. f Forest plot of sRAGE effect size estimates with FEV1 decline for each cohort (squares) as well as a weighted estimate of the meta-analysis (diamond). The shaded represents the interquartile range and the whiskers represent the 95% confidence interval
Table 2.
Results from the random coefficient models for change in FEV1, (coefficients per standard deviation of Log10 sRAGE)
| Cohort | Effect on baseline FEV1 (ml per SD Log10 sRAGE) | Effect on annual change in FEV1 (ml/year per SD Log10 sRAGE) | ||
|---|---|---|---|---|
| Coefficient (SE) | p-value | Coefficient (SE) | p-value | |
| COPDGene QBR (n = 1408) | 127.90 (21.35) | < 0.0001 | 0.96 (1.48) | 0.52 |
| ECLIPSE (n = 1847) | 66.65 (10.80) | < 0.0001 | 4.15 (1.88) | 0.0272 |
| SCCOR (n = 399) | 74.21 (29.45) | 0.0121 | 4.06 (2.41) | 0.09 |
| SPIROMICS (n = 1408) | 156.36 (22.27) | < 0.0001 | − 0.89 (1.81) | 0.62 |
sRAGE is associated with the presence and severity of emphysema, but not progression of emphysema
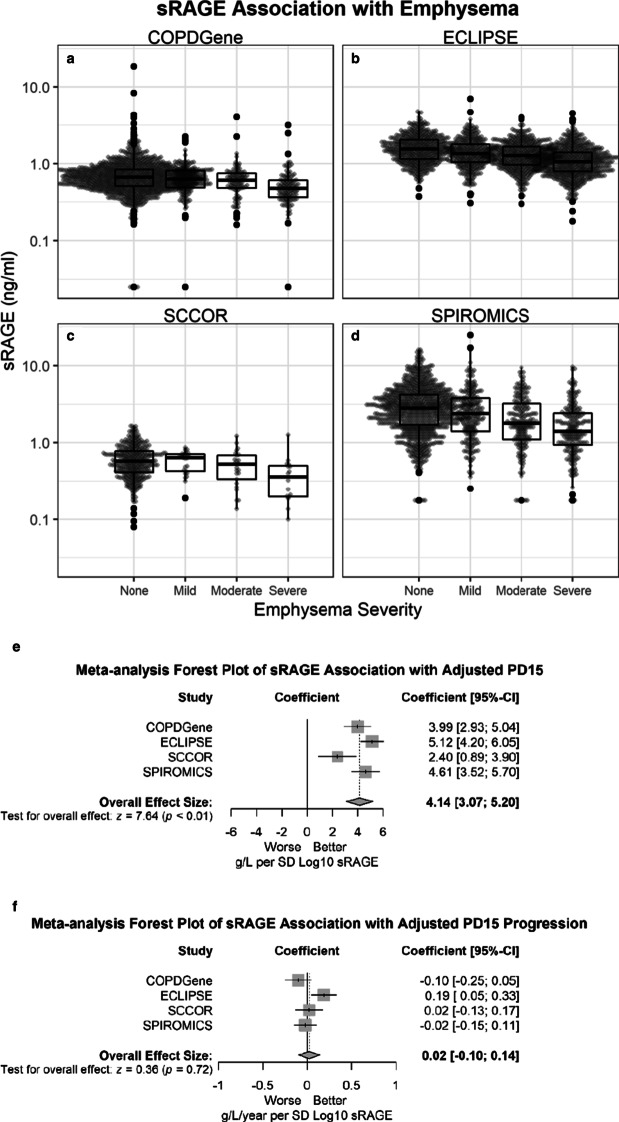
Emphysema and more severe emphysema were associated with lower plasma or serum sRAGE in all cohorts regardless of whether emphysema was assessed by lung attenuation area below − 950 HU (p < 0.001) (Fig. 3) or PD15adj (P < 0.001) (Additional file 1: Figure S2). Compared to no visual emphysema, sRAGE was significantly lower (p-value < 0.05) for moderate, confluent, and advanced destructive emphysema for COPDGene and ECLIPSE, and only for advanced destructive emphysema for SCCOR (Additional file 1: Figure S3). These associations were significant even after adjusting for important clinical predictors of emphysema, including age, sex, race, height, weight, smoking status, pack-years, exacerbations, and airflow limitation (GOLD group) (Table 3; Fig. 3). Lower sRAGE was associated with more emphysema progression in the ECLIPSE cohort, but not in COPDGene, SPIROMICS, or SCCOR (Fig. 3).
Fig. 3.
Severe emphysema is associated with lower sRAGE. sRAGE is shown on a log-scale y-axis. Each dot represents one subject. Overall p-value < 0.001 for all cohorts [COPDGene (n = 1372) (a), ECLIPSE (n = 1849) (b), SCCOR (n = 399) (c), and SPIROMICS (n = 1477) (d)]. Emphysema severity was defined as (LAA ≤ 5%), mild (LAA > 5 and ≤ 10%), moderate (LAA > 10 and ≤ 20%), or severe (LAA > 20%). Median, 25th percentile, 75th percentile, and whiskers (the minimum of 1.5 times IQR or highest/lowest value) are shown in the box plots. e Forest plot of sRAGE effect size estimates for baseline PD15adj for each cohort (squares) as well as a weighted estimate of the meta-analysis (diamond). The shaded represents the interquartile range and the whiskers represent the 95% confidence interval. f Forest plot of sRAGE effect size estimates for a change in PD15adj for each cohort (square) as well as a weighted estimate of the meta-analysis (diamond). The shaded represents the interquartile range and the whiskers represent the 95% confidence interval
Table 3.
Results from the random coefficient models for change in PD15adj., (coefficients per standard deviation of Log10 sRAGE)
| Cohort | Effect on baseline PD15adj (g/L per SD Log10 sRAGE) | Effect on annual change in PD15adj (g/L/year per SD Log10 sRAGE | ||
|---|---|---|---|---|
| Coefficient (SE) | p-value | Coefficient (SE) | p-value | |
| COPDGene QBR (n = 1402) | 3.99 (0.0.54) | < 0.0001 | − 0.10 (0.08) | 0.18 |
| ECLIPSE (n = 1699) | 5.12 (0.47) | < 0.0001 | 0.19 (0.07) | 0.009 |
| SCCOR (n = 399) | 2.40 (0.77) | 0.0343 | 0.02 (0.08) | 0.81 |
| SPIROMICS (n = 1406) | 4.61 (0.56) | < 0.0001 | − 0.02 (0.06) | 0.75 |
sRAGE receiver operating characteristic (ROC) curves for emphysema
We tested the sensitivity and specificity of sRAGE using ROC for both quantitative emphysema (Additional file 1: Figure S4) and qualitative emphysema (Additional file 1: Figure S5). The ROC area under the curve increased (COPDGene 0.68–0.73, ECLIPSE 0.67–0.68, SCCOR 0.60–0.78, SPIROMICS 0.64–0.69) as the emphysema cutoff increased from 5 to 25% (Additional file 1: Table S3). ROC estimates for sRAGE were slightly lower for visually assessed emphysema (present/absent) (COPDGene: LCMS 0.57, QBR 0.53, RBM 0.63, SOMAscan 0.56; ECLIPSE 0.54; SCCOR 0.52; SPIROMICS 0.58) (Additional file 1: Figure S5, Table S4); however, ROC were higher for DLco (Additional file 1: Figure S6, Table S5).
Effect of rs2070600 and rs2071288 genotypes on sRAGE measured levels and interaction with clinical phenotypes
Prior studies have reported that the AGER rs2070600 minor allele variant genotype (A versus common allele G) was associated with lower serum sRAGE [32]. This coding variant results in the substitution of a glycine-to-serine at amino acid position 82 (G82S). Using an RBM assay for sRAGE we have reported a similar association in both COPDGene (Additional file 1: Figure S7A) and SPIROMICS (Additional file 1: Figure S7B) [30]. Similar associations have been reported in the ECLIPSE and TESRA cohorts [19]. These used the same monoclonal antibody for capture. We also found the same association using the 4125_52_2 aptamer on the SOMAscan 1.3 k assay, which is an antibody free assay that uses aptamers for protein detection, (Additional file 1: Figure S7A); however, in COPDGene using a Quotient Bioresearch (QBR) assay, we found no association between the rs2070600 genotype and plasma sRAGE (Additional file 1: Figure S7A). Since the QBR assay correlates (⍴ = 0.53; Fig. 1) with the RBM assay in the COPDGene subjects, this suggests certain platforms may not be as sensitive to the presence of the G82S variant.
In African Americans, the minor allele (A) of the rs2071288 SNP in the AGER gene has been reported to be associated with lower circulating levels of sRAGE [33, 34]. This SNP is an intronic SNP located at a splice site in intron 9 and has been reported to be associated with sRAGE levels [33–35]. In the non-Hispanic African American population in COPDGene, the rs2070600 SNP was removed from the GWAS data during the QC process because of SNP frequency < 0.01; however, rs2071288 was kept in the African American population (minor allele frequency (MAF) = 0.11), but not in the non-Hispanic whites (MAF = 0.005). In the non-Hispanic African American population, the rs2071288 genotype was found to be significantly associated in the QBR assay (p = 0.0002) sRAGE levels but not with SOMAscan (p = 0.195) (Additional file 1: Figure S7C).
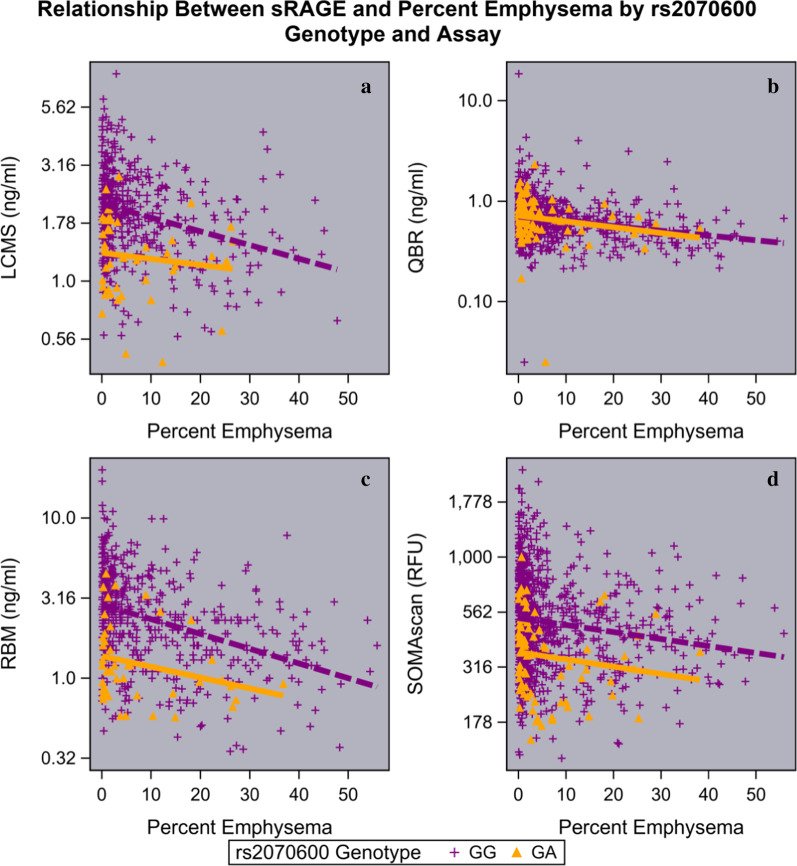
Although non-Hispanic whites who carry the rs2070600 variant have lower measurements of sRAGE, there is still an inverse relationship between emphysema severity and sRAGE levels regardless of genotype (Fig. 4); however, we found no significant interaction between the rs2070600 genotype and percent emphysema for sRAGE measured by either LCMS (p = 0.39), QBR assay (p = 0.70), RBM assay (p = 0.61), or SOMAscan (in regression models for emphysema (p = 0.96) for COPDGene (Fig. 4); or for ECLIPSE QBR (p = 0.65), SCCOR DuoSet (p = 0.61), and SPIROMICS RBM (p = 0.64) (Additional file 1: Figure S8A). A similar association was found with rs2071288 in non-Hispanic African Americans for QBR with an inverse relationship with a non-significant interaction between rs2071288 and percent emphysema (p = 0.51), but with the SOMAscan platform there was no inverse relationship with percent emphysema (Additional file 1: Figure S8B) and the relationship did not differ by genotype (p = 0.50).
Fig. 4.
Scatter plots showing the inverse relationship between emphysema and plasma sRAGE by rs2070600 genotype showing that the slopes are not different (genotype × percent emphysema interaction) even if the intercept is lower for subjects carrying the minor allele. COPDGene subjects with sRAGE measured using a LCMS ( n= 491); b QBR (n = 1166); c RBM (n = 569); d SOMAscan (n = 998). The minor allele homozygotes are not shown because of small numbers
Discussion
This is the first report of sRAGE-COPD associations in SPIROMICS and Pittsburgh SCCOR, the first report of sRAGE associations with longitudinal decline in FEV1 in ECLIPSE, and the first report of an integrated protein-SNP analysis of emphysema. These observations confirm the concept that lower sRAGE is a biomarker for the presence of emphysema and airflow obstruction as have been previously reported for COPDGene and ECLIPSE [20, 29]. These associations were highly significant regardless of which sRAGE platform was used, whether plasma or serum was assayed, or how emphysema was measured (quantitative or visual). However, while baseline sRAGE was predictive of progression of emphysema and airflow obstruction in ECLIPSE [20], we were not able to replicate these associations in other cohorts. Until additional cohorts can replicate the ECLIPSE associations with emphysema progression and FEV1 decline, the current consensus should be limited to sRAGE serving best as a blood biomarker of emphysema/airflow obstruction or COPD affection status.
The replication of most associations across four independent cohorts is noteworthy for COPD, as there are few publications which consistently replicate biomarkers across multiple diverse cohorts. The challenge of replicating biomarkers of airflow decline or emphysema progression is not limited to proteomic approaches, but also other omics such as genetics, transcriptomics, and metabolomics. There are many genetic variants associated with lung function and COPD affection status, including the AGER locus which was among the first identified in large general population GWAS [36–39], and rs2070600 was recently included in a 279-SNP genetic risk score (GRS) for COPD based on a UK BioBank GWAS [36]. Since most large GWAS have only evaluated cross-sectional lung function phenotypes, there have been limited discoveries of genetic variation associated with progression of disease [40]. Furthermore, large studies of other longitudinal COPD outcomes such as exacerbations have suffered from inability to replicate significant findings across different populations [41]. The reason for replication difficulties is not completely understood, but likely includes the heterogeneity of COPD study populations, inherent variability in longitudinal spirometric and QCT measurements, as well as potentially fundamental issues such as how to define COPD affection status and how to define progression. For example, COPD affection status is based on a single spirometric measurement based on FEV1/FVC and severity is determined by FEV1% predicted. The former measure can be confounded by age and the latter may be low because full lung function was never achieved in adulthood rather than any actual loss of any lung function during adulthood. Thus, many COPD genes or biomarkers (including sRAGE) may actually be better markers of lung mass (size), density (emphysema), or structural abnormalities rather than airflow obstruction. A study that had multiple sRAGE measurements and quantitative CTs over many years (> 10) would be ideal to address this hypothesis; however, such a large studies does not yet exist.
An important aspect of this study was its use of different sRAGE assay platforms (antibody and aptamer based (SOMAscan) on the identical aliquots from the same blood sample. We demonstrated these platforms correlate with each other regardless of whether they are using antibodies or aptamers, although correlation is not perfect. We also showed that subjects who carried the minor allele for rs2070600 SNP in AGER (the gene that codes for RAGE) had lower measurements of sRAGE on most assay platforms. The rs2070600 SNP codes for a glycine-to-serine at amino acid 82 (G82S). Regardless of genotype, both carriers and non-carriers of the rs2070600 minor allele showed a similar inverse relationship between plasma or serum sRAGE and emphysema severity even though carriers had significantly lower sRAGE. An exception to this observation was the COPDGene QBR assay, which reportedly used a polyclonal detection antibody. There was no difference by genotype for this QBR assay, suggesting that the commonly used monoclonal or single aptamer assays may poorly bind to the G82S isoform due to epitope differences in the antibody or aptamer binding area adjacent to G82S. The G82S variant has an amino acid change adjacent to an important glycosylation site at amino acid N81. Both the G82S isoform and de-glycosylation at N81 decrease binding of RAGE to damage-associated molecular pattern (DAMPs) [42]. Additional molecular work needs to be done to determine whether the G82S isoform glycosylation pattern is sufficient to alter antigenicity of RAGE thereby leading to different binding affinities of monoclonal antibodies or aptamers. Nevertheless, the lower levels of measurement in the G82S carriers suggest that researchers consider adding rs2070600 genotype when modelling sRAGE—clinical phenotype relationships and also underscores the finding that most proteins have some genetic variants associated with their measurements and the gene-biomarker-disease modelling should account for this relationship [43].
Additionally, we were able to evaluate rs2071288, another SNP in the AGER gene, which is associated with circulating levels of sRAGE in non-Hispanic African American populations [33–35]. We confirmed that the minor allele (A) was associated with lower sRAGE with the QBR platform and observed a similar, but statistically non-significant, trend with the SOMAscan platform. This SNP is intronic, located at a splice site in intron 9 and is reported to be associated with diffusing capacity of carbon monoxide and with emphysema severity in COPD patients [19, 35]. This SNP has low MAF in non-Hispanic whites and our non-Hispanic African American population was a small sample size, but our findings demonstrate the importance of conducting biomarker research in ethnically and racially diverse populations to identify ethnic and racial specific gene-by-biomarker interactions.
While this study is novel in that it evaluates sRAGE platform correlations, presents new sRAGE associations with COPD severity, identifies assay specific genetic quantitative trait loci of protein expression (pQTLs), and exhaustively evaluates disease progression from four independent cohorts, there are limitations. Foremost, there is considerable heterogeneity in cohort composition with ECLIPSE having a much larger number of participants with moderate or severe COPD and emphysema. Similarly, each cohort used slightly different CT acquisition protocols [i.e., differences in tube current exposure time product (ma × sec)], which may explain higher and noisier emphysema data. Even though these cohorts are some of the largest with sRAGE and longitudinal data, a lack of association between sRAGE and disease progression in other cohorts may be due to power. In addition, it could be due to selection bias introduced by those with the most rapid decline in FEV1 or progression of emphysema being less likely to follow-up, resulting in results toward the null. The ROC curves for sRAGE, which do not exceed 0.75 for emphysema, suggest that it should not be used as a sole diagnostic test to rule emphysema in or out, rather that it can be used as an enrichment measure to increase or decrease the probability that an individual has emphysema similar to how ventilation perfusion scintigraphy has been used. While we did adjust analyses for important covariates such as BMI, there were some covariates that were not available in most cohorts, but might affect sRAGE measurements (e.g., lipid measurements [44]). Finally, sRAGE is also lower in participants with idiopathic pulmonary fibrosis [45], suggesting that it may be a non-specific marker of loss of lung epithelium (similar to DLco), rather than a specific marker of emphysema.
Conclusion
In conclusion, sRAGE is identified as one of the best blood biomarkers of emphysema and airflow obstruction which makes it a strong candidate as a Drug Development Tool for screening potential clinical trial participants for interventions assessing the impact of treatment on emphysema. Additional larger studies are needed to confirm its role in predicting progression of airflow obstruction or emphysema as well as its value as a surrogate marker for efficacy of interventions. Finally, we note that there are common racially specific pQTL SNPs (rs2070600 in non-Hispanic whites and rs2071288 in non-Hispanic African Americans) and there is potential platform isoform detection specificity (monoclonal (e.g., Quantikine) versus aptamer (SOMAscan) assays versus polyclonal sRAGE assays) which may influence interpretation of sRAGE levels. Therefore, both population genetics and assay platforms should be considered when planning to interpret clinical associations.
Supplementary Information
Additional file 1. Additional methods, figures, tables.
Acknowledgements
ECLIPSE: We thank all the study participants for their willingness to advance knowledge in the field of COPD, study site staff for all the detailed assessments, as well as T Candido, S Cogswell, H Davis, L Holy, N Krowchuk, H Lee, E Phillips, C Storness-Bliss, N Tai, A-T Tran, N Tran, E Wang, and T Yokogawa for technical assistance with the CT analysis and data management.
Abbreviations
- AGEs
Advanced glycation end products
- AGER
Gene encoding RAGE
- ARDS
Acute respiratory distress syndrome
- COPD
Chronic obstructive pulmonary disease
- COPDGene
Genetic Epidemiology of COPD
- CRP
C-reactive protein
- DAMPs
Damage-associated molecular pattern
- ECLIPSE
Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points
- EDTA
Ethylenediaminetetraacetic acid
- ELISA
Enzyme-linked immunosorbent assay
- FEV1
Forced expiratory volume in one second
- FEV1/FVC
Ratio of forced expiratory volume in one second to functional vital capacity
- FEV1% predicted
FEV1 percent predicted: forced expiratory volume in one second/predicted FEV1
- GOLD
Global Initiative for Chronic obstructive Lung Disease
- GRS
Genetic risk score
- GWAS
Genome-wide association study
- LCMS
Liquid chromatography-mass spectrometry
- MAF
Minor allele frequency
- (NF)-kB
Nuclear factor
- PAMPs
Pathogen-associated molecular patterns
- PD15adj
15Th percentile density of lung density
- PRRs
Pattern recognition receptors
- QBR
Quotient Bioresearch
- QCT
Quantitative computed tomography
- RAGE
Receptor of advanced glycation end products
- RBM
Myriad-RBM Myriad-Rules Based Medicine
- ROC
Receiver operating characteristic
- RSV
Respiratory syncytial virus
- SCCOR
Specialized Center for Clinically Oriented Research
- SD
Standard deviation
- SNP
Single nucleotide polymorphism
- SPIROMICS
Subpopulations and Intermediate Outcome Measures in COPD Study
- sRAGE
Soluble advanced glycation end products
- %LAA
Percent low attenuation areas below − 950 Hounsfield Units on inspiratory CT
Authors’ contributions
All authors participated in the writing and made critical revisions to the manuscript. KAP haromonized and analyzed data from the cohorts. JC,DC, DLD, FS, YZ, VEO, WO, JDN, APC, SDP, NHTH, RB, FK, PW, RP, RBG, JH, CMD,JPC,RB, collected clinical data used in these analysis. KAP, JC, KK, DC, MHC, EKS, DLD, FS, YZ, LG, DAL, EH, JDN, PJC, BM, SDP, NHTH, RB, FK, PW, RS, NL, JY, SJ, RT-S, DM, RB contributed to the study design and data interpretation. All authors read and approved the final manuscript.
Funding
COPDGene: The COPDGene study (NCT00608764) is also supported by the COPD Foundation through contributions made to an Industry Advisory Committee comprised of AstraZeneca, Boehringer-Ingelheim, Genentech, GlaxoSmithKline, Novartis, Pfizer, Siemens, and Sunovion. The project described was supported by Awards Number U01 HL089897, Number U01 HL089856, R01 HL137995 and R01 HL129937 from the National Heart, Lung, and Blood Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health. SCCOR: This project was supported by Awards Number P50HL084948 and R21HL129917 from the National Heart, Lung and Blood Institute and Pennsylvania CURE SAP 4100062224. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung and Blood Institutes of Health. SPIROMICS: Was supported by contracts from the NIH/NHLBI (HHSN268200900013C, HHSN268200900014C, HHSN268200900015C, HHSN268200900016C, HHSN268200900017C, HHSN268200900018C, HHSN268200900019C, HHSN268200900020C), grants from the NIH/NHLBI (U01 HL137880 and U24 HL141762), and supplemented by contributions made through the Foundation for the NIH and the COPD Foundation from AstraZeneca/MedImmune; Bayer; Bellerophon Therapeutics; Boehringer-Ingelheim Pharmaceuticals, Inc.; Chiesi Farmaceutici S.p.A.; Forest Research Institute, Inc.; GlaxoSmithKline; Grifols Therapeutics, Inc.; Ikaria, Inc.; Novartis Pharmaceuticals Corporation; Nycomed GmbH; ProterixBio; Regeneron Pharmaceuticals, Inc.; Sanofi; Sunovion; Takeda Pharmaceutical Company; and Theravance Biopharma and Mylan.
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
All participants signed an informed consent. All studies were approved by ethics and review boards (Institutional Review Boards) at all participating centers.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dawn L. DeMeo, Email: redld@channin.harvard.edu
Lucas A. Gillenwater, Email: lagillenwater@gmail.com
Bruce E. Miller, Email: bmiller@copdfoundation.org
Nicholas Locantore, Email: nlocantore@att.net.
Russell P. Bowler, Email: BowlerR@NJHealth.org
References
- 1.Xie J, Mendez JD, Mendez-Valenzuela V, Aguilar-Hernandez MM. Cellular signalling of the receptor for advanced glycation end products (RAGE) Cell Signal. 2013;25(11):2185–2197. doi: 10.1016/j.cellsig.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Selvin E, Halushka MK, Rawlings AM, Hoogeveen RC, Ballantyne CM, Coresh J, Astor BC. sRAGE and risk of diabetes, cardiovascular disease, and death. Diabetes. 2013;62(6):2116–2121. doi: 10.2337/db12-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindsey JB, de Lemos JA, Cipollone F, Ayers CR, Rohatgi A, Morrow DA, Khera A, McGuire DK. Association between circulating soluble receptor for advanced glycation end products and atherosclerosis: observations from the Dallas Heart Study. Diabetes Care. 2009;32(7):1218–1220. doi: 10.2337/dc09-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Mesallamy HO, Hammad LN, El-Mamoun TA, Khalil BM. Role of advanced glycation end product receptors in the pathogenesis of diabetic retinopathy. J Diabetes Complications. 2011;25(3):168–174. doi: 10.1016/j.jdiacomp.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Yonchuk JG, Silverman EK, Bowler RP, Agusti A, Lomas DA, Miller BE, Tal-Singer R, Mayer RJ. Circulating soluble receptor for advanced glycation end products (sRAGE) as a biomarker of emphysema and the RAGE axis in the lung. Am J Respir Crit Care Med. 2015;192(7):785–792. doi: 10.1164/rccm.201501-0137PP. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Wang T, Yuan Z, Cao Y, Zhou Y, He J, Shen Y, Zeng N, Dai L, Wen F, Chen L. Role of receptor for advanced glycation end products in regulating lung fluid balance in lipopolysaccharide-induced acute lung injury and infection-related acute respiratory distress syndrome. Shock. 2018;50(4):472–482. doi: 10.1097/SHK.0000000000001032. [DOI] [PubMed] [Google Scholar]
- 7.Kankova K, Kalousova M, Hertlova M, Krusova D, Olsovsky J, Zima T. Soluble RAGE, diabetic nephropathy and genetic variability in the AGER gene. Arch Physiol Biochem. 2008;114(2):111–119. doi: 10.1080/13813450802033818. [DOI] [PubMed] [Google Scholar]
- 8.Waden JM, Dahlstrom EH, Elonen N, Thorn LM, Waden J, Sandholm N, Forsblom C, Groop PH, FinnDiane Study G Soluble receptor for AGE in diabetic nephropathy and its progression in Finnish individuals with type 1 diabetes. Diabetologia. 2019;62(7):1268–1274. doi: 10.1007/s00125-019-4883-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JK, Park S, Lee MJ, Song YR, Han SH, Kim SG, Kang SW, Choi KH, Kim HJ, Yoo TH. Plasma levels of soluble receptor for advanced glycation end products (sRAGE) and proinflammatory ligand for RAGE (EN-RAGE) are associated with carotid atherosclerosis in patients with peritoneal dialysis. Atherosclerosis. 2012;220(1):208–214. doi: 10.1016/j.atherosclerosis.2011.07.115. [DOI] [PubMed] [Google Scholar]
- 10.Al Rifai M, Schneider AL, Alonso A, Maruthur N, Parrinello CM, Astor BC, Hoogeveen RC, Soliman EZ, Chen LY, Ballantyne CM, Halushka MK, Selvin E. sRAGE, inflammation, and risk of atrial fibrillation: results from the Atherosclerosis Risk in Communities (ARIC) Study. J Diabetes Complications. 2015;29(2):180–185. doi: 10.1016/j.jdiacomp.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian J, Huang K, Krishnan S, Svabek C, Rowe DC, Brewah Y, Sanjuan M, Patera AC, Kolbeck R, Herbst R, Sims GP. RAGE inhibits human respiratory syncytial virus syncytium formation by interfering with F-protein function. J Gen Virol. 2013;94(Pt 8):1691–1700. doi: 10.1099/vir.0.049254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stogsdill MP, Stogsdill JA, Bodine BG, Fredrickson AC, Sefcik TL, Wood TT, Kasteler SD, Reynolds PR. Conditional overexpression of receptors for advanced glycation end-products in the adult murine lung causes airspace enlargement and induces inflammation. Am J Respir Cell Mol Biol. 2013;49(1):128–134. doi: 10.1165/rcmb.2013-0013OC. [DOI] [PubMed] [Google Scholar]
- 13.Sambamurthy N, Leme AS, Oury TD, Shapiro SD. The receptor for advanced glycation end products (RAGE) contributes to the progression of emphysema in mice. PLoS ONE. 2015;10(3):e0118979. doi: 10.1371/journal.pone.0118979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann MA, Drury S, Fu C, Qu W, Taguchi A, Lu Y, Avila C, Kambham N, Bierhaus A, Nawroth P, Neurath MF, Slattery T, Beach D, McClary J, Nagashima M, Morser J, Stern D, Schmidt AM. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97(7):889–901. doi: 10.1016/S0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- 15.Bierhaus A, Schiekofer S, Schwaninger M, Andrassy M, Humpert PM, Chen J, Hong M, Luther T, Henle T, Kloting I, Morcos M, Hofmann M, Tritschler H, Weigle B, Kasper M, Smith M, Perry G, Schmidt AM, Stern DM, Haring HU, Schleicher E, Nawroth PP. Diabetes-associated sustained activation of the transcription factor nuclear factor-kappaB. Diabetes. 2001;50(12):2792–2808. doi: 10.2337/diabetes.50.12.2792. [DOI] [PubMed] [Google Scholar]
- 16.Bopp C, Bierhaus A, Hofer S, Bouchon A, Nawroth PP, Martin E, Weigand MA. Bench-to-bedside review: the inflammation-perpetuating pattern-recognition receptor RAGE as a therapeutic target in sepsis. Crit Care. 2008;12(1):201. doi: 10.1186/cc6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teissier T, Boulanger E. The receptor for advanced glycation end-products (RAGE) is an important pattern recognition receptor (PRR) for inflammaging. Biogerontology. 2019;20(3):279–301. doi: 10.1007/s10522-019-09808-3. [DOI] [PubMed] [Google Scholar]
- 18.Carolan BJ, Hughes G, Morrow J, Hersh CP, O'Neal WK, Rennard S, Pillai SG, Belloni P, Cockayne DA, Comellas AP, Han M, Zemans RL, Kechris K, Bowler RP. The association of plasma biomarkers with computed tomography-assessed emphysema phenotypes. Respir Res. 2014;15:127. doi: 10.1186/s12931-014-0127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng DT, Kim DK, Cockayne DA, Belousov A, Bitter H, Cho MH, Duvoix A, Edwards LD, Lomas DA, Miller BE, Reynaert N, Tal-Singer R, Wouters EF, Agusti A, Fabbri LM, Rames A, Visvanathan S, Rennard SI, Jones P, Parmar H, MacNee W, Wolff G, Silverman EK, Mayer RJ, Pillai SG, Tesra, Investigators E Systemic soluble receptor for advanced glycation endproducts is a biomarker of emphysema and associated with AGER genetic variants in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2013;188(8):948–957. doi: 10.1164/rccm.201302-0247OC. [DOI] [PubMed] [Google Scholar]
- 20.Coxson HO, Dirksen A, Edwards LD, Yates JC, Agusti A, Bakke P, Calverley PM, Celli B, Crim C, Duvoix A, Fauerbach PN, Lomas DA, Macnee W, Mayer RJ, Miller BE, Muller NL, Rennard SI, Silverman EK, Tal-Singer R, Wouters EF, Vestbo J, Evaluation of CLtIPSEI The presence and progression of emphysema in COPD as determined by CT scanning and biomarker expression: a prospective analysis from the ECLIPSE study. Lancet Respiratory Med. 2013;1(2):129–136. doi: 10.1016/S2213-2600(13)70006-7. [DOI] [PubMed] [Google Scholar]
- 21.Iwamoto H, Gao J, Koskela J, Kinnula V, Kobayashi H, Laitinen T, Mazur W. Differences in plasma and sputum biomarkers between COPD and COPD-asthma overlap. Eur Respiratory J. 2014;43(2):421–429. doi: 10.1183/09031936.00024313. [DOI] [PubMed] [Google Scholar]
- 22.Iwamoto H, Gao J, Pulkkinen V, Toljamo T, Nieminen P, Mazur W. Soluble receptor for advanced glycation end-products and progression of airway disease. BMC Pulm Med. 2014;14:68. doi: 10.1186/1471-2466-14-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sukkar MB, Wood LG, Tooze M, Simpson JL, McDonald VM, Gibson PG, Wark PA. Soluble RAGE is deficient in neutrophilic asthma and COPD. Eur Respiratory J. 2012;39(3):721–729. doi: 10.1183/09031936.00022011. [DOI] [PubMed] [Google Scholar]
- 24.Vestbo J, Anderson W, Coxson HO, Crim C, Dawber F, Edwards L, Hagan G, Knobil K, Lomas DA, MacNee W, Silverman EK, Tal-Singer R. Evaluation of COPD longitudinally to identify predictive surrogate End-points (ECLIPSE) Eur Respiratory J. 2008;31(4):869–873. doi: 10.1183/09031936.00111707. [DOI] [PubMed] [Google Scholar]
- 25.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, Crapo JD. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7(1):32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Couper D, LaVange LM, Han M, Barr RG, Bleecker E, Hoffman EA, Kanner R, Kleerup E, Martinez FJ, Woodruff PG, Rennard S, Group SR. Design of the subpopulations and intermediate outcomes in COPD Study (SPIROMICS) Thorax. 2014;69(5):491–494. doi: 10.1136/thoraxjnl-2013-203897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stamm JA, Belloli EA, Zhang Y, Bon J, Sciurba FC, Gladwin MT. Elevated N-terminal pro-brain natriuretic peptide is associated with mortality in tobacco smokers independent of airflow obstruction. PLoS ONE. 2011;6(11):e27416. doi: 10.1371/journal.pone.0027416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ajala O, Zhang Y, Gupta A, Bon J, Sciurba F, Chandra D. Decreased serum TRAIL is associated with increased mortality in smokers with comorbid emphysema and coronary artery disease. Respiratory Med. 2018;145:21–27. doi: 10.1016/j.rmed.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zemans RL, Jacobson S, Keene J, Kechris K, Miller BE, Tal-Singer R, Bowler RP. Multiple biomarkers predict disease severity, progression and mortality in COPD. Respiratory Res. 2017;18(1):117. doi: 10.1186/s12931-017-0597-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun W, Kechris K, Jacobson S, Drummond MB, Hawkins GA, Yang J, Chen TH, Quibrera PM, Anderson W, Barr RG, Basta PV, Bleecker ER, Beaty T, Casaburi R, Castaldi P, Cho MH, Comellas A, Crapo JD, Criner G, Demeo D, Christenson SA, Couper DJ, Curtis JL, Doerschuk CM, Freeman CM, Gouskova NA, Han MK, Hanania NA, Hansel NN, Hersh CP, Hoffman EA, Kaner RJ, Kanner RE, Kleerup EC, Lutz S, Martinez FJ, Meyers DA, Peters SP, Regan EA, Rennard SI, Scholand MB, Silverman EK, Woodruff PG, O'Neal WK, Bowler RP, Group SR, Investigators CO Common genetic polymorphisms influence blood biomarker measurements in COPD. PLoS Genet. 2016;12(8):e1006011. doi: 10.1371/journal.pgen.1006011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klont F, Pouwels SD, Hermans J, van de Merbel NC, Horvatovich P, ten Hacken NHT, Bischoff R. A fully validated liquid chromatography-mass spectrometry method for the quantification of the soluble receptor of advanced glycation end-products (sRAGE) in serum using immunopurification in a 96-well plate format. Talanta. 2018;182:414–421. doi: 10.1016/j.talanta.2018.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Jang Y, Kim JY, Kang SM, Kim JS, Chae JS, Kim OY, Koh SJ, Lee HC, Ahn CW, Song YD, Lee JH. Association of the Gly82Ser polymorphism in the receptor for advanced glycation end products (RAGE) gene with circulating levels of soluble RAGE and inflammatory markers in nondiabetic and nonobese Koreans. Metabolism. 2007;56(2):199–205. doi: 10.1016/j.metabol.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 33.Loomis SJ, Chen Y, Sacks DB, Christenson ES, Christenson RH, Rebholz CM, Selvin E. Cross-sectional analysis of AGE-CML, sRAGE, and esRAGE with diabetes and cardiometabolic risk factors in a community-based cohort. Clin Chem. 2017;63(5):980–989. doi: 10.1373/clinchem.2016.264135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maruthur NM, Li M, Halushka MK, Astor BC, Pankow JS, Boerwinkle E, Coresh J, Selvin E, Kao WH. Genetics of plasma soluble receptor for advanced glycation end-products and cardiovascular outcomes in a community-based population: results from the atherosclerosis risk in communities study. PLoS ONE. 2015;10(6):e0128452. doi: 10.1371/journal.pone.0128452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serveaux-Dancer M, Jabaudon M, Creveaux I, Belville C, Blondonnet R, Gross C, Constantin JM, Blanchon L, Sapin V. Pathological implications of receptor for advanced glycation end-product (AGER) gene polymorphism. Dis Markers. 2019;2019:2067353. doi: 10.1155/2019/2067353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shrine N, Guyatt AL, Erzurumluoglu AM, Jackson VE, Hobbs BD, Melbourne CA, Batini C, Fawcett KA, Song K, Sakornsakolpat P, Li X, Boxall R, Reeve NF, Obeidat M, Zhao JH, Wielscher M, Understanding Society Scientific G. Weiss S, Kentistou KA, Cook JP, Sun BB, Zhou J, Hui J, Karrasch S, Imboden M, Harris SE, Marten J, Enroth S, Kerr SM, Surakka I, Vitart V, Lehtimaki T, Allen RJ, Bakke PS, Beaty TH, Bleecker ER, Bosse Y, Brandsma CA, Chen Z, Crapo JD, Danesh J, DeMeo DL, Dudbridge F, Ewert R, Gieger C, Gulsvik A, Hansell AL, Hao K, Hoffman JD, Hokanson JE, Homuth G, Joshi PK, Joubert P, Langenberg C, Li X, Li L, Lin K, Lind L, Locantore N, Luan J, Mahajan A, Maranville JC, Murray A, Nickle DC, Packer R, Parker MM, Paynton ML, Porteous DJ, Prokopenko D, Qiao D, Rawal R, Runz H, Sayers I, Sin DD, Smith BH, Soler Artigas M, Sparrow D, Tal-Singer R, Timmers P, Van den Berge M, Whittaker JC, Woodruff PG, Yerges-Armstrong LM, Troyanskaya OG, Raitakari OT, Kahonen M, Polasek O, Gyllensten U, Rudan I, Deary IJ, Probst-Hensch NM, Schulz H, James AL, Wilson JF, Stubbe B, Zeggini E, Jarvelin MR, Wareham N, Silverman EK, Hayward C, Morris AP, Butterworth AS, Scott RA, Walters RG, Meyers DA, Cho MH, Strachan DP, Hall IP, Tobin MD, Wain LV. New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat Genet. 2019;51(3):481–493. doi: 10.1038/s41588-018-0321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakornsakolpat P, Prokopenko D, Lamontagne M, Reeve NF, Guyatt AL, Jackson VE, Shrine N, Qiao D, Bartz TM, Kim DK, Lee MK, Latourelle JC, Li X, Morrow JD, Obeidat M, Wyss AB, Bakke P, Barr RG, Beaty TH, Belinsky SA, Brusselle GG, Crapo JD, de Jong K, DeMeo DL, Fingerlin TE, Gharib SA, Gulsvik A, Hall IP, Hokanson JE, Kim WJ, Lomas DA, London SJ, Meyers DA, O'Connor GT, Rennard SI, Schwartz DA, Sliwinski P, Sparrow D, Strachan DP, Tal-Singer R, Tesfaigzi Y, Vestbo J, Vonk JM, Yim JJ, Zhou X, Bosse Y, Manichaikul A, Lahousse L, Silverman EK, Boezen HM, Wain LV, Tobin MD, Hobbs BD, Cho MH, SpiroMeta C, International CGC Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Nat Genet. 2019;51(3):494–505. doi: 10.1038/s41588-018-0342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hobbs BD, de Jong K, Lamontagne M, Bosse Y, Shrine N, Artigas MS, Wain LV, Hall IP, Jackson VE, Wyss AB, London SJ, North KE, Franceschini N, Strachan DP, Beaty TH, Hokanson JE, Crapo JD, Castaldi PJ, Chase RP, Bartz TM, Heckbert SR, Psaty BM, Gharib SA, Zanen P, Lammers JW, Oudkerk M, Groen HJ, Locantore N, Tal-Singer R, Rennard SI, Vestbo J, Timens W, Pare PD, Latourelle JC, Dupuis J, O'Connor GT, Wilk JB, Kim WJ, Lee MK, Oh YM, Vonk JM, de Koning HJ, Leng S, Belinsky SA, Tesfaigzi Y, Manichaikul A, Wang XQ, Rich SS, Barr RG, Sparrow D, Litonjua AA, Bakke P, Gulsvik A, Lahousse L, Brusselle GG, Stricker BH, Uitterlinden AG, Ampleford EJ, Bleecker ER, Woodruff PG, Meyers DA, Qiao D, Lomas DA, Yim JJ, Kim DK, Hawrylkiewicz I, Sliwinski P, Hardin M, Fingerlin TE, Schwartz DA, Postma DS, MacNee W, Tobin MD, Silverman EK, Boezen HM, Cho MH, Investigators CO, Investigators E, LifeLines I, Group SR, International CGNI, Investigators UKB, International CGC Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet. 2017;49(3):426–432. doi: 10.1038/ng.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wain LV, Shrine N, Artigas MS, Erzurumluoglu AM, Noyvert B, Bossini-Castillo L, Obeidat M, Henry AP, Portelli MA, Hall RJ, Billington CK, Rimington TL, Fenech AG, John C, Blake T, Jackson VE, Allen RJ, Prins BP, Understanding Society Scientific G. Campbell A, Porteous DJ, Jarvelin MR, Wielscher M, James AL, Hui J, Wareham NJ, Zhao JH, Wilson JF, Joshi PK, Stubbe B, Rawal R, Schulz H, Imboden M, Probst-Hensch NM, Karrasch S, Gieger C, Deary IJ, Harris SE, Marten J, Rudan I, Enroth S, Gyllensten U, Kerr SM, Polasek O, Kahonen M, Surakka I, Vitart V, Hayward C, Lehtimaki T, Raitakari OT, Evans DM, Henderson AJ, Pennell CE, Wang CA, Sly PD, Wan ES, Busch R, Hobbs BD, Litonjua AA, Sparrow DW, Gulsvik A, Bakke PS, Crapo JD, Beaty TH, Hansel NN, Mathias RA, Ruczinski I, Barnes KC, Bosse Y, Joubert P, van den Berge M, Brandsma CA, Pare PD, Sin DD, Nickle DC, Hao K, Gottesman O, Dewey FE, Bruse SE, Carey DJ, Kirchner HL, Geisinger-Regeneron Discov EHRC. Jonsson S, Thorleifsson G, Jonsdottir I, Gislason T, Stefansson K, Schurmann C, Nadkarni G, Bottinger EP, Loos RJ, Walters RG, Chen Z, Millwood IY, Vaucher J, Kurmi OP, Li L, Hansell AL, Brightling C, Zeggini E, Cho MH, Silverman EK, Sayers I, Trynka G, Morris AP, Strachan DP, Hall IP, Tobin MD. Genome-wide association analyses for lung function and chronic obstructive pulmonary disease identify new loci and potential druggable targets. Nat Genet. 2017;49(3):416–425. doi: 10.1038/ng.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.John C, Soler Artigas M, Hui J, Nielsen SF, Rafaels N, Pare PD, Hansel NN, Shrine N, Kilty I, Malarstig A, Jelinsky SA, Vedel-Krogh S, Barnes K, Hall IP, Beilby J, Musk AW, Nordestgaard BG, James A, Wain LV, Tobin MD. Genetic variants affecting cross-sectional lung function in adults show little or no effect on longitudinal lung function decline. Thorax. 2017;72(5):400–408. doi: 10.1136/thoraxjnl-2016-208448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keene JD, Jacobson S, Kechris K, Kinney GL, Foreman MG, Doerschuk CM, Make BJ, Curtis JL, Rennard SI, Barr RG, Bleecker ER, Kanner RE, Kleerup EC, Hansel NN, Woodruff PG, Han MK, Paine R, Martinez FJ, Bowler RP, O'Neal WK. Biomarkers predictive of exacerbations in the SPIROMICS and COPDGene Cohorts. Am J Respir Crit Care Med. 2017;195(4):473–481. doi: 10.1164/rccm.201607-1330OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osawa M, Yamamoto Y, Munesue S, Murakami N, Sakurai S, Watanabe T, Yonekura H, Uchigata Y, Iwamoto Y, Yamamoto H. De-N-glycosylation or G82S mutation of RAGE sensitizes its interaction with advanced glycation endproducts. Biochim Biophys Acta. 2007;1770(10):1468–1474. doi: 10.1016/j.bbagen.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Yao C, Chen G, Song C, Keefe J, Mendelson M, Huan T, Sun BB, Laser A, Maranville JC, Wu H, Ho JE, Courchesne P, Lyass A, Larson MG, Gieger C, Graumann J, Johnson AD, Danesh J, Runz H, Hwang SJ, Liu C, Butterworth AS, Suhre K, Levy D. Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat Commun. 2018;9(1):3268. doi: 10.1038/s41467-018-05512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNair E, Qureshi M, Prasad K, Pearce C. Atherosclerosis and the hypercholesterolemic AGE-RAGE Axis. Int J Angiol. 2016;25(2):110–116. doi: 10.1055/s-0035-1570754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manichaikul A, Sun L, Borczuk AC, Onengut-Gumuscu S, Farber EA, Mathai SK, Zhang W, Raghu G, Kaufman JD, Hinckley-Stukovsky KD, Kawut SM, Jelic S, Liu W, Fingerlin TE, Schwartz DA, Sell JL, Rich SS, Barr RG, Lederer DJ. Plasma soluble receptor for advanced glycation end products in idiopathic pulmonary fibrosis. Ann Am Thorac Soc. 2017;14(5):628–635. doi: 10.1513/AnnalsATS.201606-485OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Additional methods, figures, tables.
Data Availability Statement
The datasets used during the current study are available from the corresponding author on reasonable request.