Abstract
Objective
Although attention deficit hyperactivity disorder (ADHD) is a disease with high genetic transition, our knowledge about the mechanism of the disease is limited. In this study, it was aimed to evaluate the levels of miR-132-3p and miR-942-5p that are associated with the dopamine carrier protein gene (DAT1) and dopamine receptor 5 (DRD5) genes, which have been shown to play a role in the development of ADHD.
Methods
According to the Diagnostic and Statistical Manual of Mental Disorders 5th edition, 50 children diagnosed with ADHD and 48 healthy controls were included in the study. Affective Disorders and Schizophrenia Interview Schedule-Now and Lifetime Version-Turkish Adaptation was used to evaluate ADHD and the diagnoses accompanying ADHD. Quantitative Real-Time Polymerase Chain Reaction was used to evaluate miR-132-3p and miR-942-5p expression levels.
Results
It was observed that miR-132-3p level (p = 0.001) was significantly higher with children with ADHD compared to the control group, and the level of miR-942-5p (p = 0.181) was higher in ADHD but did not reach statistically significant level.
Conclusion
In our study, we found that the increase in the miR-132-3p levels of children with ADHD may be a therapeutic target of the disease.
Keywords: Attention deficit hyperactivity disorder, MicroRNA, MiR-132-3p, MiR-942-5p.
INTRODUCTION
Attention deficit hyperactivity disorder (ADHD) is a neurodevelopmental disorder that is characterized by distraction, hyperactivity and impulsivity, and its symptoms continue mostly for life [1]. Genetic factors have been shown to be very important in the etiology of ADHD. In a study with a identical twins, it was found that the inheritance of ADHD was 76% [2]. Some genes have been found associated with ADHD significantly such as dopamine D4 receptor gene (DRD4), dopamine D5 receptor gene (DRD5), dopamine transporter 1 (DAT1), serotonergic receptor (HTR1B), dopamine beta hydroxylase enzyme (DBH), serotonin transporter enzyme (5-HTT) and, synaptosomal-associated protein 25 (SNAP-25) genes [2].
MicroRNAs (miRNAs) are RNA molecules that are encoded with genes, have regulatory properties, and are single-stranded, consisting of approximately 21−23 nucleotides, and do not encode endogenous protein; and they suppress the expression of the target gene [3,4]. It has been shown to have an effect on important biological processes such as neurogenesis, synaptic plasticity, cell death decision, apoptosis and stress response by acting on messenger RNA (mRNA) [5,6]. Although 70% of miRNAs have been shown to be expressed in the brain and most miRNAs are specific to neurons, their role in development and nervous system activity is largely unknown [7,8]. Studies on miR-132-3p have shown that miR-132-3p and miR-134 play a role in the regulation of synaptic plasticity, and miR 132 also increases dentrit sizes and dentritic branching [9]. In a study with transgenic mice, it was found that increased miR-132-3p within the physiological limits increases cognitive capacity, and miR-132-3p overex-pression that exceeds physiological limits distrups learning [10]. In addition to dopamine-related genes, the brain-derived growth factor (BDNF) gene, an important neurotrophin, is also included in the ‘hot gene’ class in ADHD [11]. BDNF protein, which is effective on synaptic plasticity, neuronal survival and differentiation, is critical for the survival and differentiation of dopaminergic neurons in the midbrain [12]. BDNF is also indirectly regulated by miR-132-3p through MeCP2 [13], and a decrease in miR-132-3p level has been shown in Rett Syndrome caused by mutation in the MeCP2 gene [14]. MiRNA-related studies have been performed in many psychiatric diseases such as schizophrenia, bipolar disorder, depression, anxiety disorder, autism, and Tourette Syndrome [15-19]. While these studies are still in their infancy, these small molecules are thought to lead to significant improvements in the diagnosis and treatment of psychiatric diseases.
So far, studies evaluating the relationship between miRNA and ADHD are also limited [20]. In a study by Wu et al., the relationship between galectin-3 and miR let-7d, which leads to down-regulation in tyrosine hydroxylase enzyme, which has an important role in dopamine metabolism, was investigated. It was observed that there was an increase in miR let-7d level in ADHD patients and this increase caused a decrease in serum galectin-3 levels [21]. miR-942-5p is a relatively new detected microRNA that has also been shown to play a role in regulating the Wnt/ß-catenin signal pathway, which plays a role in regulating stem cell functions [22]. It is estimated that mi942 is one of the miRNAs that show different expression in ADHD patients [23]. In this study, it was aimed to evaluate the microRNA-132 (miR-132-3p) and microRNA-942 (miR-942-5p) levels, which are stated to be associated with DAT1 and DRD5 genes respectively in the miRNA database (http://www.mirbase.org/), in newly diagnosed ADHD patients. Our hypothesis was that these miRNAs would be different in ADHD patients compared to controls.
METHODS
Fifty ADHD patients who met the inclusion criteria of the study from the patients diagnosed with ADHD Compound Type according to the Diagnostic and Statistical Manual of Mental Disorders 5th edition (DSM-5) diagnostic criteria between February 2015 and December 2015 in Gaziantep University Faculty of Medicine, Department of Child and Adolescent Psychiatry, and 48 healthy volunteer controls were included. Affective Disorders and Schizophrenia Interview Schedule-Now and Lifetime Version-Turkish Adaptation for ADHD and comorbid diagnoses were evaluated using the semi-structured interview Schedule [24]. Conners Parent Rating Scale (CPRS) were used as a screening diagnostic measurement [25].
Children were excluded from the study if they had mental retardation, autism spectrum disorder, mood disorder, tic disorder, chronic physical disease (diabetes mellitus, asthma, cancer, epilepsy, etc.). In addition, children who did not want to participate in the study and whose mother was not at a sufficient sociocultural level to fill the scales were excluded from the study. Therefore, a written informed consent form was signed for those who wanted to participate in the research. For this study, approval was received from the Medical Ethics Committee of Gaziantep University Medical Faculty (Date: 09.02.2015, No: 2015/50) and our study was funded by Gaziantep University Scientific Research Projects (Project No: TF.15.31). Criteria for inclusion in the research for the group with ADHD; to be diagnosed with ADHD according to DSM-5 diagnostic criteria, not to have been treated for ADHD before, being between the ages of 6−14 and voluntarily accepting to participate in the study. Criteria for inclusion in the research for the control group; not to have any psychiatric and additional chronic medical illness or disease history and voluntarily accepting to participate in the study. The sociodemographic data of the participants included informtion about age, sex, education level, ages of parents, professions and education levels of parents, family structure, family status and income level.
miRNAs were identified by using the following databases: (http://www.targetscan.org), (http://www.mirbase.org/) and (http://mirwalk.umm.uni-heidelberg.de) (Table 1). Plasma was obtained from whole blood samples of ADHD and control groups. Total RNA, including miRNA, was obtained from the patient and control plasma using the miRNeasy Mini Kit (Qiagen, Santa Clarita, CA, USA). The isolated RNA samples were translated into complementary DNA (cDNA) using TaqMan miRNA Reverse Transcription Kit (Life Technologies, Foster City, CA, USA) in 384-well Thermal Cycler (BioEr, Hangzhou, China). Before quantitative Real-Time Polymerase Chain Reaction (qRT-PCR), cDNA samples were amplified using TaqMan PreAmp Master Mix (Life Technologies). The preamplification protocol was performed as follows: 10 minutes at 95°C, 2 minutes at 55°C, 2 minutes at 72°C and 15 seconds at 95°C for 14 cycles as a cycle step, and 4 minutes at 60°C. Amplified cDNAs were kept at −80°C for later investigations. qRT-PCR reactions were performed with a high processing device (BioMark; Fluidigm, San Francisco, CA, USA). Amplified cDNA samples were mixed with TaqMan Universal PCR Master Mix and Sample Loading Reagent and loaded into the sample inputs of the 96.96-chip Dynamic Array (Fluidigm). QRT-PCR reactions were performed in the BioMark Real-Time PCR system following the protocol. Ten minutes at 95°C, 15 seconds at 95°C and 1 minute at 60°C for 30 cycles. In this way, it was aimed to investigate the expression levels of miR-132-3p and miR-942-5p and compare them between the groups. Serum miR-132-3p and miR-942-5p levels were evaluated in Gaziantep University Genetics Laboratory.
Table 1.
miRNA’s target genes and primer sequences
| miRNA’s | miRbase accession number | Sequence | Target gene |
|---|---|---|---|
| hsa-miR-132-3p | MIMAT0000426 | UAACAGUCUACAGCCAUGGUCG | DAT1(SLC6A3) |
| hsa-miR-942-5p | MIMAT0004985 | UCUUCUCUGUUUUGGCCAUGUG | DRD5 |
miRNAs are identified by using the following databases: (http://www.targetscan.org), (http://www.mirbase.org) and (http://mirwalk.umm.uni- heidelberg.de).
Statistical Analysis
NCSS (Number Cruncher Statistical System) 2007 (Kaysville, UT, USA) program was used for statistical analysis. While evaluating the study data, besides descriptive statistical methods (mean, standard deviation, median, frequency, ratio, minimum, maximum), Student’s ttest was used for comparing two groups of parameters that showed normal distribution, and Mann−Whitney Utest for two group comparisons of non-normally distributed parameters. In comparison of qualitative data, Pearson chi-square test, Fisher-Freeman-Halton test, Fisher’s Exact test and Yates ’Continuity Correction test (Yates’ corrected chi-square) were used. Spearman’s Correlation Analysis was also used to evaluate the relationships between parameters. Diagnostic screening tests (sensitivity, specificity, positive estimation value, negative estimation) and ROC curve analysis were used to determine the cut off for the variables. Significance was evaluated at p < 0.01 and p < 0.05 levels.
RESULTS
Sociodemographic data of the ADHD group and the control group included in the study are given in Table 1. When the ADHD group was compared with the control group in terms of age, sex, educational background, income level, number of siblings and family status, there was no difference (p > 0.05) (Table 2). When the ADHD group and the control group were compared according to the CPRS subscale scores, it was seen that the scores of the ADHD group from all subscales were statistically significantly higher than the control group.
Table 2.
Descriptive features by groups
| Variable | Patient group (n = 50) | Control group (n = 48) | pvalue |
|---|---|---|---|
| Age (yr) | 8.45 ± 2.25 | 9.50 ± 2.82 | 0.055 |
| Sex | |||
| Male | 37 (74) | 33 (68.8) | 0.725 |
| Female | 13 (26) | 15 (31.3) | |
| Education status | |||
| Kindergarten | 3 (6) | 3 (6.3) | 0.171 |
| Primary education | 37 (74) | 27 (56.3) | |
| Middle school | 9 (18) | 13 (27.1) | |
| High school | 1 (2) | 5 (10.4) | |
| Number of siblings | |||
| 1 | 5 (10) | 3 (6.3) | 0.785 |
| 2 | 19 (38) | 17 (35.4) | |
| ≥ 3 | 26 (52) | 28 (58.3) | |
| Income status | |||
| Poor | 5 (10) | 1 (2.1) | 0.228 |
| Middle | 39 (78) | 38 (79.2) | |
| High | 6 (12) | 9 (18.8) | |
| Family status | |||
| Married | 47 (94) | 44 (91.7) | 0.438 |
| Divorced | 1 (2) | 0 (0) | |
| Dead | 2 (4) | 4 (8.3) |
Values are presented as mean ± standard deviation or number (%).
When the miR-132-3p and miR-942-5p measurements were compared according to the groups, miR-132-3p measurements of the control group were significantly higher than the patient group (p = 0.001; p < 0.01), while there were no statistically significant differences between the groups in the miR-942-5p measurements (Table 3).
Table 3.
Evaluation of miR-132-3p and miR-942-5p measurements by groups
| miRNA’s | Patient group (n = 50) | Control group (n = 48) | pvalue |
|---|---|---|---|
| miR-132 | 27.7−34 | 28.4−35.2 | 0.001 |
| 30.35 ± 1.27 | 31.43 ± 1.41 | ||
| miR-942 | 26.6−34 | 28−36 | 0.181 |
| 30.79 ± 1.33 | 31.20 ± 1.65 |
Values are presented as Min−Max or mean ± standard deviation.
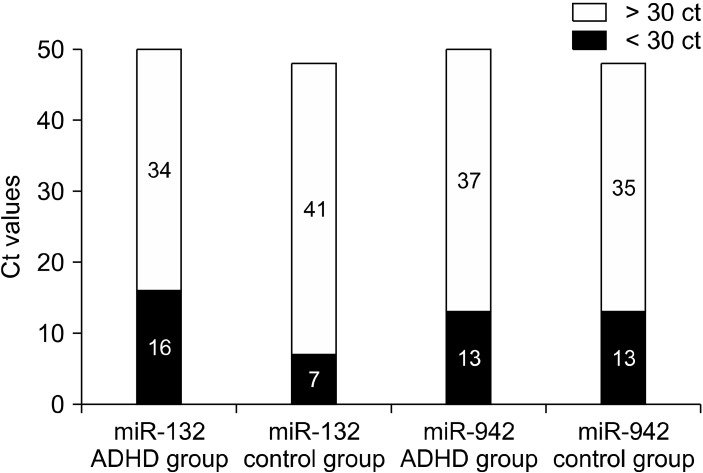
The patient and control group miRNA levels were evaluated in terms of fold changes (Fig. 1). It was seen that miR-132-3p expression increased 2.12 times and miR-942-5p expression increased 1.3 times in the patient group. In the correlation analysis in terms of age; the relationship between the age and the miR-132-3p and miR-942-5p measurements of the patients (with increasing age, miRNA measurement increased) was not statistically significant (Table 4).
Fig. 1.
miR-132-3p and miR-942-5p measurements (high ct values indicate low expression). ADHD, attention deficit hyperactivity disorder.
Table 4.
Characteristics of miR-132-3p and miR-942-5p measurements
| miRNA’s | Fold change (95% confidence interval) | r (p) |
|---|---|---|
| Comparison of cases in terms of miR-132-3p and miR-942-5p Fold Change values | ||
| miR-132-3p | 2.119 (1.34−2.90) | |
| miR-942-5p | 1.3069 (0.78−1.84) | |
| Relationship between ages of cases and miR-132-3p and miR-942-5p measurements | ||
| miR-132 | 0.169 (0.097) | |
| miR-942 | 0.073 (0.478) | |
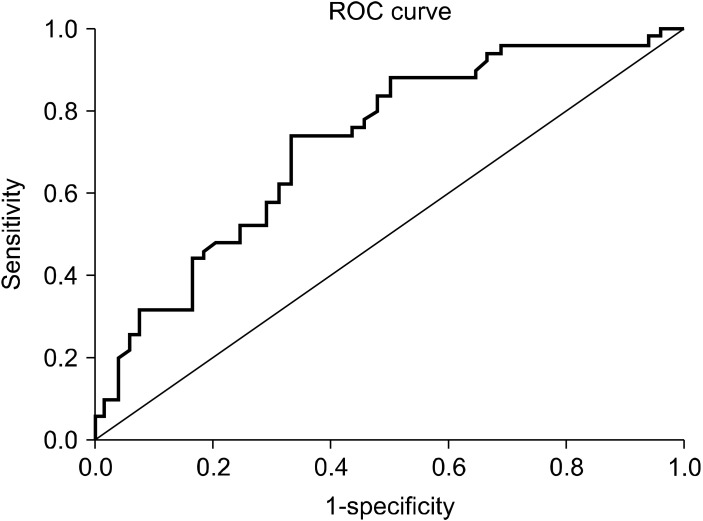
A statistically significant difference was found between miR-132-3p measurements according to the presence of disease. Based on this significance, ROC analysis and diagnostic screening tests were used to determine the cut off point for miR-132-3p (Fig. 2). Depending on the presence of the disease, the cut off point for miR-132-3p was found to be 30.93 and below. A statistically significant relationship was found between ADHD and the cut-off value of miR-132-3p level. The ODDS ratio for miR-132-3p is 5,692 (95% confidence interval: 2,381−13,607). We can evaluate this situation in cases with miR-132-3p measurement of 30.93 and below, with an increased risk of disease by 5.69 times (Table 5).
Fig. 2.
ROC curve for miR-132-3p measurement.
Table 5.
Diagnostic screening tests and ROC curve results for miR-132-3p
| miRNA | Diagnostic scan | ROC curve | pvalue | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Cut off | Sensitivity | Specificity | Positive estimation value | Negative estimation value | Area | 95% confidence interval | |||
| miR-132-3p | ≤ 30.93 | 74.00 | 66.67 | 69.80 | 71.10 | 0.721 | 0.620−0.822 | 0.001 | |
DISCUSSION
In our study, we found that the miR-132-3p level in children with ADHD was significantly higher compared to the control group. Although miR-942-5p expression was higher in ADHD patients compared to the control group, the difference between the groups was not statistically significant.
Few studies evaluating miRNA levels in ADHD have focused on different miRNAs. It has been shown that miRNA let-7d [26] expression, which has been shown to be effective on brain development and morphogenesis, has increased in children with ADHD and this increase is significantly associated with ADHD [21]. In the study conducted by Kandemir et al., it was shown that there was a significant decrease in miR 18a-5p, miR 22-3p, miR 24-3p, miR 106b-5p and miR 107 levels and a significant increase in miR 155a-5p in the ADHD group. In addition, significant results were obtained in the ROC analysis with miR 107, and it was stated that miR 107 can be used as a marker in the diagnosis of the disease [27]. According to the ROC analysis data in our study, it was observed that the positive predictive value was 69.80% and the negative predictive value was 71.10% in cases with a miR-132-3p measurement of 30.93 and below, and the risk of disease was increased by 5.69 times in these cases. In the light of these data, it is thought that miR-132-3p may be a marker for the diagnosis of the disease, but more extensive research is needed in this area. In many neuropsy-chiatric diseases such as schizophrenia, substance use dis-order, major depressive disorder, Rett syndrome, Alzheimer, miR-132-3p levels have been shown to change [28-31].
In studies evaluating the relationship between schizophrenia and ADHD, it was shown that the risk of developing schizophrenia increases in children with a history of ADHD and the frequency of ADHD is higher in children who have a high risk of developing schizophrenia, but the cause of the relationship between schizophrenia and ADHD has not been clearly established [32]. Studies have shown that there is a relationship between miR-132-3p levels in the brain and peripheral blood mononucleer cells [33,34]. In schizophrenia, a significant reduction in miR-132-3p levels was found in the dorsolateral prefrontal cortex (DLPFC) [35]. In another study evaluating miR-132-3p levels in the peripheral blood of schizophrenia patients, a similar result was found and a decrease in miR-132-3p expression in the blood was observed. In this study, in 12 new-onset schizophrenia patients, an increase in miR-132-3p expression was detected after antipsychotic drug treatment and it was pointed out that miR-132-3p may be a potential target in the diagnosis and treatment of the disease [31]. In our study, it was thought that the significantly higher miR-132-3p level of the ADHD group. This may indicate that mi-132, which is high in other psychiatric diseases, can be a potential target in diagnosis and treatment, considering the common etiopathogenesis of psychiatric diseases.
Deletion at the miR-132-3p/212 locus caused morphological changes in the emerging adult hippocampus neurons in the form of a decrease in dentritic growth and a decrease in spine density [6]. In a study with transgenic mice, miR-132-3p overexpression has been shown to cause impairments in the new object recognition memory [36]. While learning process in non-neuronal cells does not increase miR-132-3p, it has been shown that learning process in neuronal cells causes a significant increase in miR-132-3p levels, and increased miR-132-3p within physiological limits increases cognitive capacity. In this study, it was also determined that miR-132-3p overexpression disrupts learning in a level that exceeds physiological limits. According to these data, it was emphasized that miR-132-3p expression should remain within a certain range for learning and memory, miR-132-3p defined as a dynamic regulator of cognitive capacity [10]. Considering these relationships between miR-132-3p and cognitive functions, it is thought that disorders of learning and memory functions that can be seen in children with ADHD may occur due to increased miR-132-3p expression.
BDNF, also indirectly regulated by MiR-132-3p, is a protein that acts on synaptic plasticity, neuronal survival and differentiation [12,13]. A decrease in PFC and hippocampus volumes has been shown in ADHD patients with polymorphism in the BDNF gene [37]. It has been observed that there is an increase in serum BDNF level in children with ADHD and this increase correlates with the severity of careless symptoms [38]. In another study, a decrease in hippocampus BDNF level was found in rats with ADHD and it was found that BDNF level increased after exercise, thus improving spatial learning abilities [39]. In our study, it was thought that increased miR-132-3p expression in children with ADHD might have disrupted the adjustment in BDNF levels and this may be related to the emergence of problems related to attention and learning.
In the literature review, only one ADHD study was found with miR942. In this study, it was stated that miR-942-5p was statistically upregulated in the group with ADHD compared to the other group [23]. This is a result that partially matches our study. Although there was a higher expression level in our study, no statistically significant difference was found. This may be due to the fact that the previous study had a small number of samples. This study included only 9 children with ADHD and 20 age-matched typically developing subjects. However, the minimum number of patients should be 33 for moderate effect size in power analyzes and previous studies.
Limitations of our study; The low sample size, the low number of miRNAs evaluated, the literature on ADHD and miRNA relationship are under development, so the comparison of our findings with previous study findings has not been sufficiently made. As a result; The change in miR-132-3p levels in psychiatric diseases such as ADHD, schizophrenia, MSD, Rett syndrome may indicate the need for miR-132-3p expression to remain within a certain range for normal development. Studies to understand the relationship between miR-132-3p and ADHD are thought to contribute to developments in the diagnosis and treatment process of the disease. Studies in larger samples are needed to understand the role of miRNAs in ADHD.
Acknowledgments
We would like to thank Gaziantep University Scientific Research Center for financing this project. We are also grateful to all the participants and their families for being included in the research.
Footnotes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Author Contributions
Conceptualization: Seyma Coskun, Cem Gokcen, Serdar Oztuzcu. Data acquisition: Seyma Coskun, Mehmet Karadag, Serdar Oztuzcu. Formal analysis: Seyma Coskun, Mehmet Karadag. Writing-original draft: Seyma Coskun, Mehmet Karadag Writing-review&editing: Cem Gokcen. Supervision: Cem Gokcen.
References
- 1.American Psychiatric Association, author. Diagnostic and statistical manual of mental disorders: DSM-5. 5th ed. American Psychiatric Association; Arlington: 2013. [Google Scholar]
- 2.Faraone SV, Perlis RH, Doyle AE, Smoller JW, Goralnick JJ, Holmgren MA, et al. Molecular genetics of attention-deficit/hyperactivity disorder. Biol Psychiatry. 2005;57:1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 5.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Magill ST, Cambronne XA, Luikart BW, Lioy DT, Leighton BH, Westbrook GL, et al. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc Natl Acad Sci U S A. 2010;107:20382–20387. doi: 10.1073/pnas.1015691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- 8.Camkurt MA, Güneş S, Coşkun S, Fındıklı E. Peripheral signatures of psychiatric disorders: microRNAs. Clin Psychopharmacol Neurosci. 2017;15:313–319. doi: 10.9758/cpn.2017.15.4.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, et al. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen KF, Karelina K, Sakamoto K, Wayman GA, Impey S, Obrietan K. miRNA-132: a dynamic regulator of cognitive capacity. Brain Struct Funct. 2013;218:817–831. doi: 10.1007/s00429-012-0431-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Chang SH, Zhang LY, Gao L, Wang J. Molecular genetic studies of ADHD and its candidate genes: a review. Psychiatry Res. 2014;219:10–24. doi: 10.1016/j.psychres.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Hyman C, Hofer M, Barde YA, Juhasz M, Yancopoulos GD, Squinto SP, et al. BDNF is a neurotrophic factor for dopaminergic neurons of the substantia nigra. Nature. 1991;350:230–232. doi: 10.1038/350230a0. [DOI] [PubMed] [Google Scholar]
- 13.Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 14.Wu H, Tao J, Chen PJ, Shahab A, Ge W, Hart RP, et al. Genome-wide analysis reveals methyl-CpG-binding protein 2-dependent regulation of microRNAs in a mouse model of Rett syndrome. Proc Natl Acad Sci U S A. 2010;107:18161–18166. doi: 10.1073/pnas.1005595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abelson JF, Kwan KY, O’Roak BJ, Baek DY, Stillman AA, Morgan TM, et al. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 16.Abu-Elneel K, Liu T, Gazzaniga FS, Nishimura Y, Wall DP, Geschwind DH, et al. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics. 2008;9:153–161. doi: 10.1007/s10048-008-0133-5. [DOI] [PubMed] [Google Scholar]
- 17.Bocchio-Chiavetto L, Maffioletti E, Bettinsoli P, Giovannini C, Bignotti S, Tardito D, et al. Blood microRNA changes in depressed patients during antidepressant treatment. Eur Neuropsychopharmacol. 2013;23:602–611. doi: 10.1016/j.euroneuro.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Rong H, Liu TB, Yang KJ, Yang HC, Wu DH, Liao CP, et al. MicroRNA-134 plasma levels before and after treatment for bipolar mania. J Psychiatr Res. 2011;45:92–95. doi: 10.1016/j.jpsychires.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 19.Camkurt MA, Karababa F, Erdal ME, Bayazıt H, Kandemir SB, Ay ME, et al. Investigation of dysregulation of several microRNAs in peripheral blood of schizophrenia patients. Clin Psychopharmacol Neurosci. 2016;14:256–260. doi: 10.9758/cpn.2016.14.3.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karadag M, Gokcen C, Nacarkahya G, Namiduru D, Dandil F, Calisgan B, et al. Chronotypical characteristics and related miR-142-3p levels of children with attention deficit and hyperactivity disorder. Psychiatry Res. 2019;273:235–239. doi: 10.1016/j.psychres.2018.12.175. [DOI] [PubMed] [Google Scholar]
- 21.Wu LH, Peng M, Yu M, Zhao QL, Li C, Jin YT, et al. Circulating microRNA let-7d in attention-deficit/hyperactivity disorder. Neuromolecular Med. 2015;17:137–146. doi: 10.1007/s12017-015-8345-y. [DOI] [PubMed] [Google Scholar]
- 22.Ge C, Wu S, Wang W, Liu Z, Zhang J, Wang Z, et al. miR-942 promotes cancer stem cell-like traits in esophageal squamous cell carcinoma through activation of Wnt/b-catenin signalling pathway. Oncotarget. 2015;6:10964–10977. doi: 10.18632/oncotarget.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nuzziello N, Craig F, Simone M, Consiglio A, Licciulli F, Margari L, et al. Integrated analysis of microRNA and mRNA expression profiles: an attempt to disentangle the complex interaction network in attention deficit hyperactivity disorder. Brain Sci. 2019;9:288. doi: 10.3390/brainsci9100288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gökler B, Ünal F, Pehlivantürk B, Kültür EÇ, Akdemir D, Taner Y. [Reliability and validity of schedule for affective disorders and schizophrenia for school age children-present and lifetime version-Turkish version (K-SADS-PL-T)] Turk J Child Adolesc Ment Health. 2004;11:109–116. Turkish. [Google Scholar]
- 25.Kaner S, Büyüköztürk Ş, İşeri E, Ak A, Özaydin L. [Conners’ parent rating scale long form-revised: factor structure, reliability and validity studies] Turk J Child Adolesc Ment Health. 2011;18:45–58. Turkish. [Google Scholar]
- 26.Lehmann SM, Krüger C, Park B, Derkow K, Rosenberger K, Baumgart J, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 27.Kandemir H, Erdal ME, Selek S, Ay Öİ, Karababa IF, Kandemir SB, et al. Evaluation of several micro RNA (miRNA) levels in children and adolescents with attention deficit hyperactivity disorder. Neurosci Lett. 2014;580:158–162. doi: 10.1016/j.neulet.2014.07.060. [DOI] [PubMed] [Google Scholar]
- 28.Hollander JA, Im HI, Amelio AL, Kocerha J, Bali P, Lu Q, et al. Striatal microRNA controls cocaine intake through CREB signalling. Nature. 2010;466:197–202. doi: 10.1038/nature09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li YJ, Xu M, Gao ZH, Wang YQ, Yue Z, Zhang YX, et al. Alterations of serum levels of BDNF-related miRNAs in patients with depression. PLoS One. 2013;8:e63648. doi: 10.1371/journal.pone.0063648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pichler S, Gu W, Hartl D, Gasparoni G, Leidinger P, Keller A, et al. The miRNome of Alzheimer’s disease: consistent downregulation of the miR-132/212 cluster. Neurobiol Aging. 2017;50:167.e1–167.e10. doi: 10.1016/j.neurobiolaging.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 31.Yu HC, Wu J, Zhang HX, Zhang GL, Sui J, Tong WW, et al. Alterations of miR-132 are novel diagnostic biomarkers in peripheral blood of schizophrenia patients. Prog Neuropsycho-pharmacol Biol Psychiatry. 2015;63:23–29. doi: 10.1016/j.pnpbp.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 32.Dalsgaard S, Mortensen PB, Frydenberg M, Maibing CM, Nordentoft M, Thomsen PH. Association between attention- deficit hyperactivity disorder in childhood and schizophrenia later in adulthood. Eur Psychiatry. 2014;29:259–263. doi: 10.1016/j.eurpsy.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 33.Lai CY, Lee SY, Scarr E, Yu YH, Lin YT, Liu CM, et al. Aberrant expression of microRNAs as biomarker for schizophrenia: from acute state to partial remission, and from peripheral blood to cortical tissue. Transl Psychiatry. 2016;6:e717. doi: 10.1038/tp.2015.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao T, Zhen XC. Dysregulation of miRNA and its potential therapeutic application in schizophrenia. CNS Neurosci Ther. 2018;24:586–597. doi: 10.1111/cns.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci U S A. 2012;109:3125–3130. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen KF, Sakamoto K, Wayman GA, Impey S, Obrietan K. Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS One. 2010;5:e15497. doi: 10.1371/journal.pone.0015497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, Straub RE, et al. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shim SH, Hwangbo Y, Kwon YJ, Jeong HY, Lee BH, Lee HJ, et al. Increased levels of plasma brain-derived neurotrophic factor (BDNF) in children with attention deficit-hyperactivity disorder (ADHD) Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:1824–1828. doi: 10.1016/j.pnpbp.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 39.Jeong HI, Ji ES, Kim SH, Kim TW, Baek SB, Choi SW. Treadmill exercise improves spatial learning ability by enhancing brain- derived neurotrophic factor expression in the attention-deficit/hyperactivity disorder rats. J Exerc Rehabil. 2014;10:162–167. doi: 10.12965/jer.140111. [DOI] [PMC free article] [PubMed] [Google Scholar]