Abstract
Background
Treacher Collins syndrome‐1 (TCS1; OMIM# 154500) is a rare autosomal dominant disease that is defined by congenital craniofacial dysplasia. Here, we report four sporadic and one familial case of TCS1 in Chinese patients with clinical features presenting as hypoplasia of the zygomatic complex and mandible, downslanting palpebral fissures, coloboma of the lower eyelids, and conductive hearing loss.
Materials and Methods
Audiological, radiological, and physical examinations were performed. Targeted next‐generation sequencing (NGS) was performed to examine the genetics of this disease in five probands, and Sanger sequencing was used to confirm the identified variants. A literature review discusses the pathogenesis, treatment, and prevention of TCS1.
Results
We identified a novel insertion of c.939_940insA (p.Gly314Argfs*35; NM_001135243.1), a novel deletion of c.1766delC (p.Pro589Leufs*7), two previously reported insertions of c.1999_2000insC (p.Arg667Profs*31) and c.4218_4219insG (p.Ser1407Valfs*23), and one previously reported deletion of c.4369_4373delAAGAA (p.Lys1457Glufs*12) in the TCOF1 gene. All five cases exhibited a degree of interfamilial and intrafamilial phenotypic variability. A review of the literature revealed no clear evidence of a genotype–phenotype correlation in TCS1.
Conclusion
Our results expand the variant spectrum of TCOF1 and highlight that NGS is essential for the diagnosis of TCS and that genetic counseling is beneficial for guiding prevention.
Keywords: conductive hearing loss, craniofacial dysplasia, TCOF1, Treacher Collins syndrome
Treacher Collins syndrome‐1 (TCS1; OMIM# 154500) is a rare autosomal dominant disease that is defined by congenital craniofacial dysplasia. Here, we report four sporadic and one familial case of TCS1 in Chinese patients with clinical features presenting as hypoplasia of the zygomatic complex and mandible, downslanting palpebral fissures, coloboma of the lower eyelids, and conductive hearing loss. We identified a novel insertion of c.939_940insA (p.Gly314Argfs*35; NM_001135243.1), a novel deletion of c.1766delC (p.Pro589Leufs*7), two previously reported insertions of c.1999_2000insC (p.Arg667Profs*31) and c.4218_4219insG (p.Ser1407Valfs*23), and one previously reported deletion of c.4369_4373delAAGAA (p.Lys1457Glufs*12) in the TCOF1 gene.

1. INTRODUCTION
Treacher Collins syndrome‐1 (TCS1; OMIM# 154500) was named to commemorate Edward Treacher Collins, an English ophthalmologist, after he first described the basic components of the disorder in 1900 (Teber et al., 2004). TCS1 is an uncommon autosomal dominant disorder that is defined by congenital craniofacial dysplasia. It is also referred to as mandibulofacial dysostosis or Treacher Collins––Franceschetti syndrome. TCS1 is characterized by conductive hearing loss due to malformation of the external or middle ear, and a unique “bird‐like” facial appearance presenting as hypoplasia of the zygomatic complex and mandible, downslanting palpebral fissures, and coloboma of the lower eyelids. TCS1 clinically overlaps with other disorders such as Goldenhar syndrome (OMIM# 164210), Nagar syndrome (OMIM# 154400), and Miller syndrome (OMIM# 263750). This presents the difficulties when making a differential diagnosis according to phenotype. Therefore, an accurate diagnosis can be confirmed by molecular analysis of the pathogenic gene of TCS1 (Zhang et al., 2013). There are four subtypes of TCS, all with similar phenotypes: TCS1 (OMIM# 154500), TCS2 (OMIM# 613717), TCS3 (OMIM# 248390), and TCS4 (OMIM# 618939). These subtypes are caused by variants in the genes TCOF1, POLR1D, POLR1C, and POLR1B, respectively.
The incidence of TCS is estimated to be about 1 in 50,000 live births (Trainor & Andrews, 2013). Variants in the TCOF1 gene account for over 90% of TCS cases and variants in POLR1D and POLR1C are responsible for approximately 9% of non‐TCOF1‐related TCS cases (Dauwerse et al., 2011; Weiner et al., 2012). To date, there are over 300 variants in the TCOF1 gene have been reported, and approximately 60% of TCS1 is the result of de novo variants (Human Gene Mutation Database®; Li et al., 2019). In most cases, TCS1 is caused by frameshift deletions or duplications (1–41 bp) in the TCOF1 coding region resulting in a premature stop codon (Masotti et al., 2009). Phenotypic variability has been observed in patients with TCS within and among families and there has been no clear evidence of a genotype–phenotype correlation in TCS.
Most cases of TCS have been reported in Western countries. Here, we report four sporadic cases and one familial case of TCS1 in Chinese patients. We identified two novel variants and three previously reported variants in the TCOF1 gene. A review of the current literature indicated that our findings expand the variant spectrum of TCOF1. Our results also indicate that NGS has a vital role in the diagnosis of TCS and, therefore, genetic counseling is beneficial for the prevention of the disease.
2. MATERIALS AND METHODS
2.1. Patients and clinical data
In this study, four sporadic and one familial case clinically diagnosed as TCS were recruited at the Department of Otorhinolaryngology, Head and Neck Surgery, the First Affiliated Hospital of Zhengzhou University. Medical and family histories were collected, and all subjects underwent physical examinations and pure‐tone audiometry (at frequencies from 250 to 8000 Hz) tests. Objective audiometry for pediatric patients consisted of auditory steady‐state response (ASSR), tympanometry, auditory brainstem response (ABR), and distortion product otoacoustic emissions (DPOAEs). All probands underwent temporal bone thin‐section computed tomography (CT). Written informed consent was obtained from all cases (Pan, Lu, et al., 2020). This study was performed with the approval of the Ethics Committee of the Second Affiliated Hospital of Zhengzhou University (reference number 2018008). We have submitted the information of phenotype and variants to the LOVD database (https://databases.lovd.nl/shared/individuals/00305490, 00305493, 00305494, 00305495, and 00305496).
2.2. Targeted next‐generation sequencing (NGS), bioinformatics analyses, and variant interpretation
Peripheral venous blood was collected from the probands, parents, and siblings if available. Genomic DNA was extracted from peripheral blood samples using a GenMagBio Genomic DNA Purification kit (GenMagBio) following the manufacturer's protocol. Targeted NGS, bioinformatics analyses, and variant interpretation were performed as previously described (Pan, Xu, et al., 2020). Variant nomenclature was based on TCOF1 transcript NM_001135243.1.
2.3. Sanger sequencing
The variants in the probands detected by NGS were validated by Sanger sequencing. The co‐segregation of the variants with the disease in family members was also tested by Sanger sequencing. The NCBI Primer‐Blast tool was used to design primers targeting these variants (Data S1). Purified PCR products were sequenced on a SeqStudio Genetic Analyzer (Applied Biosystems/Life Technologies).
2.4. Literature review
A search of the literature was performed spanning from 1955 to 2020 using the NCBI PubMed database with the keywords “Treacher Collins syndrome” or “TCOF1.” We subsequently summarized the natural history and the genotype–phenotype correlation of TCS1.
3. RESULTS
3.1. Clinical features of TCS1 cases
We summarized the clinical features of four sporadic and one familial TCS1 cases in Table 1. All five probands had zygomatic complex hypoplasia, mandibular hypoplasia, downslanting palpebral fissures, coloboma of the lower eyelids, and bilateral conductive hearing loss. Dysplastic auricles and external auditory canals were not observed in proband case 4. Hence, a degree of interfamilial phenotypic variability was observed. In familial case 5, the proband suffered from severe craniofacial deformities and conductive hearing loss, whereas the proband's mother was carrying the same variant but was only mildly affected. Therefore, intrafamilial phenotypic variability was also observed to some extent.
TABLE 1.
A review of clinical characteristics in Chinese patients with TCS1
| Publications | Zygomatic complex hypoplasia | Downslanting palpebral fissures | Lower eyelid coloboma | Micrognathia/retrognathia | Auricle deformity | EAC stenosis/atresia | Middle ear hypoplasia | Conductive hearing loss |
|---|---|---|---|---|---|---|---|---|
|
Li et al. (2012) Proband Spo |
+ | + | NA | + | − | + | + | + |
|
Zhang et al. (2013) Family 1, proband |
+ | + | + | + | + | + | + | + |
|
Zhang et al. (2013) Family 1, Ⅱ−1 |
+ | + | + | + | + | + | − | + |
|
Zhang et al. (2013) Family 1, Ⅱ−3 |
+ | + | + | + | + | + | − | + |
|
Zhang et al. (2013) Family 1, Ⅱ−5 |
+ | + | + | + | + | + | − | + |
|
Zhang et al. (2013) Family 2, proband Spo |
+ | + | + | + | + | + | + | + |
|
Wang et al. (2014) Patient 1 Spo |
+ | + | + | − | − | + | − | + |
|
Wang et al. (2014) Patient 2 Spo |
+ | + | + | + | + | + | − | + |
|
Wang et al. (2014) Patient 3 |
− | + | + | + | + | + | + | + |
|
Chen et al. (2018) F24P |
+ | + | + | + | + | + | + | + |
|
Chen et al. (2018) S15P Spo |
+ | + | + | + | + | + | NA | NA |
|
Chen et al. (2018) S2P Spo |
NA | NA | NA | NA | NA | NA | NA | NA |
|
Chen et al. (2018) S5P Spo |
+ | + | + | + | + | + | + | + |
|
Chen et al. (2018) S28P Spo |
+ | + | + | + | + | + | + | + |
|
Chen et al. (2018) S7P Spo |
NA | + | NA | NA | + | + | NA | NA |
|
Chen et al. (2018) F1P1 |
+ | + | + | + | + | + | + | + |
|
Chen et al. (2018) S6P Spo |
+ | + | + | + | + | + | + | + |
|
Chen et al. (2018) F23P |
+ | + | + | + | + | + | + | + |
|
Chen et al. (2018) S29P Spo |
+ | + | + | + | − | + | + | + |
|
Chen et al. (2018) F4P |
+ | + | − | + | + | + | NA | NA |
|
Chen et al. (2018) S22P Spo |
+ | + | + | + | + | NA | NA | + |
|
Chen et al. (2018) F30P |
− | + | + | − | − | − | + | + |
|
Chen et al. (2018) S25P Spo |
+ | + | + | + | + | + | + | + |
|
Chen et al. (2018) F2P |
+ | + | + | + | + | + | + | + |
|
Chen et al. (2018) S20P Spo |
+ | + | + | + | + | − | + | + |
|
Chen et al. (2018) S21P Spo |
+ | − | + | + | + | − | − | + |
|
Chen et al. (2018) F32P |
+ | + | + | + | + | + | + | + |
|
Chen et al. (2018) F26P |
+ | + | + | + | + | + | + | + |
|
Chen et al. (2018) S31P Spo |
+ | + | + | + | + | + | + | + |
|
Chen et al. (2018) F27P |
+ | + | + | + | + | + | + | + |
|
Chen et al. (2018) S23P Spo |
+ | − | − | + | + | NA | NA | NA |
|
Yan et al. (2018) Proband |
+ | + | + | + | + | + | + | + |
|
Yan et al. (2018) Proband's mother |
+ | + | + | + | − | − | − | + |
|
Li et al. (2019) Case 2, proband |
+ | + | + | + | + | + | NA | + |
|
Li et al. (2019) Case 2, Ⅰ−2 |
− | − | − | − | − | − | NA | + |
|
Li et al. (2019) Case 3 Spo |
+ | + | + | + | + | − | NA | + |
|
Li et al. (2019) Case 4 Spo |
+ | + | + | + | − | − | NA | + |
|
Li et al. (2019) Case 5 Spo |
+ | + | + | + | + | + | NA | + |
|
Fan et al. (2019) Patient 2313 |
+ | + | + | + | − | − | + | + |
|
Fan et al. (2019) Patient 3538 |
+ | + | + | + | − | − | + | + |
|
Fan et al. (2019) Patient 2848 |
+ | + | + | + | + | + | + | + |
|
Fan et al. (2019) Patient 2721 |
+ | + | − | + | + | + | + | + |
|
Fan et al. (2019) Patient 3314 |
+ | + | + | + | − | − | + | + |
|
Fan et al. (2019) Patient 3316 |
+ | + | + | + | − | − | + | + |
|
Fan et al. (2019) Patient 3286 |
+ | + | + | + | − | − | + | + |
|
Fan et al. (2019) Patient 3287 |
+ | + | + | + | − | − | + | + |
|
Fan et al. (2019) Patient 3288 |
+ | + | + | + | − | − | + | + |
|
Fan et al. (2019) Patient 3289 |
+ | + | − | + | − | − | + | + |
|
Fan et al. (2019) Patient 3290 |
+ | + | + | + | − | − | + | + |
|
Fan et al. (2019) Patient 3291 |
+ | + | − | + | − | − | + | + |
|
Fan et al. (2019) Patient 3292 |
+ | + | + | + | + | − | + | + |
|
Fan et al. (2019) Patient 3293 |
− | + | − | + | − | − | + | + |
|
Liu et al. (2020) Fetus |
+ | + | + | + | + | NA | NA | NA |
|
Liu et al. (2020) Father |
+ | + | + | + | NA | − | NA | + |
|
Zhang et al. (2020) Case 1 Spo |
+ | + | NA | + | + | + | NA | + |
|
Zhang et al. (2020) Case 2 Spo |
+ | + | NA | + | − | + | NA | + |
|
Zhang et al. (2020) Case 3 Spo |
+ | + | NA | + | + | + | NA | + |
|
Zhang et al. (2020) Case 4 Spo |
+ | + | NA | + | + | + | NA | + |
|
Zhang et al. (2020) Case 5 Spo |
+ | + | NA | + | + | + | NA | + |
|
Zhang et al. (2020) Case 6 Spo |
+ | + | NA | + | + | + | NA | + |
|
Zhang et al. (2020) Case 7 Spo |
+ | + | NA | + | + | + | NA | + |
|
This study Case 1, proband Spo |
+ | + | + | + | + | + | + | + |
|
This study Case 2, proband Spo |
+ | + | + | + | + | + | + | + |
|
This study Case 3, proband Spo |
+ | + | + | + | + | + | + | + |
|
This study Case 4, proband Spo |
+ | + | + | + | − | − | + | + |
|
This study Case 5, proband |
+ | + | + | + | + | + | + | + |
|
This study Case 5, Ⅱ−5 |
+ | + | + | + | + | − | NA | + |
| Frequency | 61/65 | 63/66 | 50/57 | 62/65 | 45/65 | 41/63 | 39/46 | 61/61 |
Abbreviations: EAC, external auditory canal; NA, Not available; Spo, Sporadic; TCS1, Treacher Collins syndrome‐1.
3.2. Identification of variants
Variants in the TCOF1 gene were detected in the five probands with targeted NGS and then, confirmed in their family members by Sanger sequencing. Two previously reported insertions of c.1999_2000insC (p.Arg667Profs*31; NM_001135243.1) and c.4218_4219insG (p.Ser1407Valfs*23) were detected in case 1 as well as case 2 and further confirmed by Sanger sequencing (Bowman et al., 2012; Masotti et al., 2009). One previously reported de novo deletion of c.4369_4373delAAGAA (p.Lys1457Glufs*12) in the TCOF1 gene was identified in case 3 and this variant has been described as a hotspot variant that is frequently detected in TCS1 cases (Bowman et al., 2012; Chen et al., 2018; Conte et al., 2011; Li et al., 2019; Masotti et al., 2009; Splendore et al., 2000; Zhang et al., 2013, 2020). A novel de novo insertion of c.939_940insA (p.Gly314Argfs*35) was detected in case 4. A novel familial deletion of c.1766delC (p.Pro589Leufs*7) was identified in proband case 5 and in the proband's mother, therefore, it is co‐segregated in the family. Variants identified in this study are summarized in Table 2.
TABLE 2.
Reported pathogenic TCOF1 variants in Chinese TCS1 patients
| Location | Nucleotide | Protein | Previous reports |
|---|---|---|---|
| Exon 2 | c.136C>G | p.Leu46Val | Chen et al. (2018) |
| Exon 2 | c.149A > G | p.Tyr50Cys | Zhang et al. (2020) |
| Exon 2 | c.159G>A | p.Trp53* | Chen et al. (2018) |
| Exon 2‐6 | exons deletion | — | Liu et al. (2020) |
| Intron 2 | c.165‐1G>A | Splice | Yan et al. (2018) |
| Exon 5 | c.384_385delGA | p.Glu128Aspfs*46 | Li et al. (2019) |
| Exon 5 | c.430_431insA | p.Thr144Asnfs*31 | Chen et al. (2018) |
| Exon 5 | c.451delC | p.Leu151Phefs*68 | Chen et al. (2018) |
| Exon 5 | c.489delC | p.Ser164Glnfs*55 | Fan et al. (2019) |
| Exon 6A | c.648delC | p.Ser217Glnfs*2 | Fan et al. (2019) |
| Exon 6A | c.810_811insA | p.Glu271Argfs*3 | Chen et al. (2018) |
| Exon 7 | c.939_940insA | p.Gly314Argfs*35 | This study, Case 4 |
| Exon 8 | c.1142delC | p.Arg383Glyfs*109 | Zhang et al. (2020) |
| Exon 8 | c.1155_1160delAGCTGCinsGGGACTT | p.Ala386Glyfs*35 | Chen et al. (2018) |
| Exon 9 | c.1303_1304insC | P.Gln45Profs*23 | Wang et al. (2014) |
| Exon 9 | c.1307_1344del | p.Val436Glufs*9 | Chen et al. (2018) |
| Exon 9 | c.1393C > T | p.Gln465* | Zhang et al. (2020) |
| Exon 9‐13 | exons deletion | — | Li et al. (2019) |
| Exon 10 | c.1639_1640delAG | p.Ser547Glnfs*2 |
Li et al. (2012) Chen et al. (2018) |
| Exon 10 | c.1658C>G | p.Ser553* | Wang et al. (2014) |
| Exon 11 | c.1719delG | p.Asn574 Thrfs*22 | Zhang et al. (2020) |
| Exon 11 | c.1719_1720insG | p.Asn574Glufs*29 | Chen et al. (2018) |
| Exon 11 | c.1766delC | p.Pro589Leufs*7 | This study, Case 5 |
| Exon 12 | c.1999_2000insC | p.Arg667Profs*31 | This study, Case 1 |
| Exon 12 | c.2103_2106delTGAG | p.Ser701Argfs*9 | Wang et al. (2014) |
| Exon 13 | c.2285_2286delCT | p.Ser762* | Zhang et al. (2020) |
| Intron 14 | c.2478+5G>A | Splice | Fan et al. (2019) |
| Exon 15 | c.2626_2627delGA | p.Asp876Glnfs*2 | Li et al. (2019) |
| Intron 17 | c.3047‐2A>G | Splice | Fan et al. (2019) |
| Exon 20 | c.3386delA | p.Lys1129Serfs*79 | Chen et al. (2018) |
| Exon 20 | c.3496delG | P.Ala1166Profs*42 | Chen et al. (2018) |
| Exon 23 | c.3823delC | p.Arg1275Glyfs*32 | Chen et al. (2018) |
| Exon 23 | c.4129_4130delGT | p.Val1377Phefs*21 | Chen et al. (2018) |
| Exon 23 | c.4218_4219insG | p.Ser1407Valfs*23 | This study, Case 2 |
| Exon 23 | c.4231C>T | p.Gln1411* | Chen et al. (2018) |
| Intron 23 | c.4342+5G>C | Splice | Zhang et al. (2020) |
| Exon 24 | c.4369_4373delAAGAA | p.Lys1457Glufs*12 |
Zhang et al. (2013) Chen et al. (2018) Li et al. (2019) Zhang et al. (2020) This study, Case 3 |
| Exon 24 | c.4420C>T | p.Gln1474* | Zhang et al. (2013) |
Nucleotide numbering is based on GenBank reference sequence NM_001135243.1.
Abbreviation: TCS1, Treacher Collins syndrome‐1.
3.3. Case information
3.3.1. Case 1
The female patient (Ⅱ‐7, Figure 1a) was 50 years old at the time of recruitment. The patient had a unique “bird‐like” facial appearance presenting as hypoplasia of the zygomatic complex and mandible, downslanting palpebral fissures, and coloboma of the lower eyelids (Figure 2a). She also had the auricle deformity and external auditory canal atresia bilaterally. Pure‐tone audiometry showed bilateral conductive hearing loss. Temporal bone thin‐section CT showed craniofacial malformations and deformity of the ossicular chains. Her parents and her six siblings had no symptoms or signs of TCS1. Targeted NGS identified a previously reported insertion c.1999_2000insC (p.Arg667Profs*31) in exon 12 of TCOF1 in the proband (Figure 3a), which may cause the formation of a stop codon 31 amino acids downstream of the insertion (Bowman et al., 2012). We could not confirm whether the variant was de novo because the proband's parents had died. Neither her husband nor her son carried this pathogenic variant by Sanger sequencing (PVS1+PM2+PP3+PP4).
FIGURE 1.

Pedigree of five unrelated families with TCS1. (a) Case 1. (b) Case 2. (c) Case 3. (d) Case 4. (e) Case 5
FIGURE 2.

Facial phenotypes and audiograms of individuals with TCS1. (a) Case 1. (b) Case 2. (c) Case 3. (d) Case 4. (e) The proband of Case 5 and his mother
FIGURE 3.
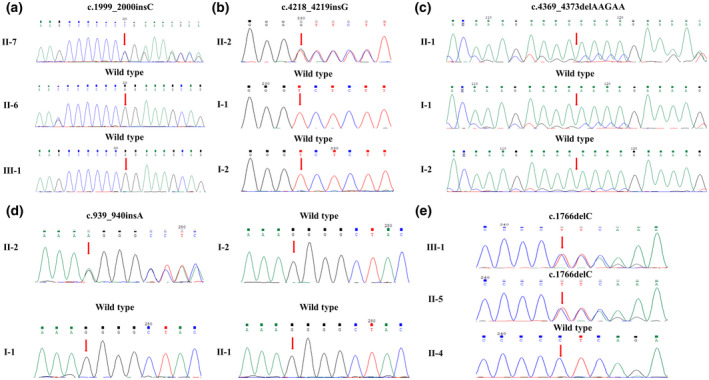
Sanger sequencing of five unrelated families with TCS1. (a) Case 1. (b) Case 2. (c) Case 3. (d) Case 4. (e) Case 5
3.3.2. Case 2
A 12‐year‐old boy (Ⅱ‐2, Figure 1b) was born to non‐consanguineous Chinese parents. Paternity was established through a paternity test. He had noticeable hypoplastic zygomatic complex, mandibular hypoplasia, downslanting palpebral fissures, coloboma of lower eyelids, and hearing loss (Figure 2b). Atresia of external auditory canals was observed and auricle reconstruction surgery of the right ear was performed. Audiological studies revealed approximately 75 dB threshold for bilateral conductive hearing loss. He had been using soft‐band hearing aids for 6 years, which allowed learning needs to be met. Temporal bone CT scans showed bilateral deformity of the ossicular chains. All of the family members had a regular facial appearance and hearing. A previously reported insertion c.4218_4219insG (p.Ser1407Valfs*23) in exon 23 of TCOF1 was detected in the proband by targeted NGS (Figure 3b), which may cause a premature stop codon (Bowman et al., 2012; Masotti et al., 2009). Sanger sequencing analysis confirmed that the variant was absent in the proband's parents, indicative of a de novo variant. This variation interpretation based on ACMG guidelines was classified as pathogenic (PVS1+PM6+PM2+PP3+PP4).
3.3.3. Case 3
A 4‐year‐old boy (Ⅱ‐1, Figure 1c) was born to non‐consanguineous parents. Paternity was established through a paternity test. There was no family history, and the history of pregnancy was uneventful. He had microtia with left external auditory canal atresia and right external auditory canal stenosis. Zygomatic complex hypoplasia, mandibular micrognathia with retrognathia, downslanting palpebral fissures, and coloboma of the lower eyelids were also marked at birth (Figure 2c). DPOAEs were absent in all frequencies on the right side. Average thresholds of ABR (right, 70 dB nHL; left, 80 dB nHL) and the thresholds of bone conduction ABR (right, 20 dB nHL; left, 20 dB nHL) were confirmed. The audiograms of ASSR at 500, 1000, 2000, and 4000 Hz are recorded in Figure 2c. He had been using soft‐band hearing aids and had a normal level of language development. Temporal bone CT scans showed bilateral deformity of the ossicular chains. The proband's parents had no abnormalities or hearing problems. Genetic analyses of the proband showed a previously reported variant c.4369_4373delAAGAA (p.Lys1457Glufs*12) in exon 24 of TCOF1 (Figure 3c), which may result in a premature stop codon causing translation of a truncated protein (Bowman et al., 2012; Chen et al., 2018; Conte et al., 2011; Li et al., 2019; Masotti et al., 2009; Splendore et al., 2000; Zhang et al., 2013, 2020). This variant was not identified by Sanger sequencing in either of his parents, confirmed a de novo status. This variation interpretation based on ACMG guidelines was classified as pathogenic (PVS1+PM6+PM2+PP3+PP4).
3.3.4. Case 4
The proband (II‐2, Figure 1d), a 2‐year‐old infant, was the daughter of non‐consanguineous parents. Paternity was established through a paternity test. The history of exposure to teratogens and the evidence of hereditary diseases were uneventful. At birth, she had mandibular hypoplasia, downslanting palpebral fissures, coloboma of the lower eyelids, and mandibular micrognathia with retrognathia (Figure 2d). Mandibular distraction surgery was performed when the proband was 2 months old. Normal auricles were observed. DPOAEs were absent in all frequencies on both sides. Average thresholds of ABR were recorded (right, 70 dB nHL; left, 65 dB nHL). Bone conduction ABR confirmed conductive hearing loss with thresholds of 25 dB nHL on both sides. The audiograms of ASSR at 500, 1000, 2000, and 4000 Hz are shown in Figure 2d. Temporal bone CT scans showed hypoplastic zygomatic arches, deformity of the ossicular chains but no deformities of external auditory canals. Targeted NGS analyses of the proband identified a novel insertion c.939_940insA (p.Gly314Argfs*35) in exon 7 of the TCOF1 gene (Figure 3d), which may lead to a premature stop codon and result in translation of a truncated 347 amino acid protein predicted by the SWISS‐MODEL program (http://swissmodel.expasy.org). This variant was absent in her unaffected parents or elder sister by Sanger sequencing, indicative of a novel de novo variant. The variant was also not detected in 500 healthy Chinese controls. It has not been reported in the gnomAD database. This variation interpretation based on ACMG guidelines was classified as pathogenic (PVS1+PM6+PM2+PP3+PP4).
3.3.5. Case 5
The proband (Ⅲ‐1, Figure 1e), a 12‐year‐old male, had left external auditory canal atresia, right external auditory canal stenosis, and bilateral auricle deformity that was treated by auricle reconstruction surgery on the right side. In addition, the proband was severely affected with noticeable craniofacial malformations, including hypoplastic zygomatic complex, mandibular hypoplasia, downslanting palpebral fissures, and coloboma of the lower eyelids (Figure 2e). Pure‐tone audiometry revealed approximately 80 dB threshold for bilateral conductive hearing loss. Temporal bone CT scans showed the deformity of the ossicular chains. There was a family history in TCS1 in the proband's mother (Ⅱ‐5, Figure 1e), but she displayed a mild phenotype of craniofacial malformations, cupped protruding ears, and approximately 70 dB threshold for conductive hearing loss. The proband's father had no dysmorphic facial features and normal hearing. Targeted NGS of the proband identified a novel deletion c.1766delC (p.Pro589Leufs*7) in exon 11 of TCOF1, which was predicted to give rise to a premature stop codon causing a truncated 594 amino acid protein (Figure 3e). Sanger sequencing confirmed that the variant was inherited from his mother; his father did not carry the variant. The variant was absent in 500 healthy Chinese controls. It has not been reported in the gnomAD database. This variation interpretation based on ACMG guidelines was classified as pathogenic (PVS1+PM2+PP3+PP4).
4. DISCUSSION
4.1. Mechanism
Treacle, a nucleolar phosphoprotein encoded by TCOF1, plays a vital role in both the formation and migration of craniofacial neural crest cells (Li et al., 2019). Neural crest cells are involved in cartilage bone, connective tissue, and sensory ganglia in the head (Dixon et al., 2006). The lack of treacle can result in the hypoplastic characteristics of TCS1 craniofacial abnormalities by disrupting the formation and proliferation phases of neural crest cells. Tcof1 +/− mice reportedly have craniofacial malformations and may die within 24 hours of birth due to respiratory arrest, which is caused by malformations of skeletal elements in the palatine, premaxilla, maxilla, and nasal (Dixon et al., 2006). Tcof1 +/− mice were used to analyze the developmental mechanisms of conductive hearing loss, and it was indicated that the effective cavitation of the middle ear was closely related to the growth of the auditory bulla, which is a structure derived from neural crest cells and includes all of the components of the middle ear (Richter et al., 2010). Defects in these processes have a profound and adverse effect on hearing. All five variants observed in our study gave rise to premature stop codons in the reading frame, corroborating that the fundamental molecular mechanism of the TCS1 is haploinsufficiency. All five probands had craniofacial malformations and conductive hearing loss.
4.2. Molecular mapping
The linkage of the TCS locus to markers in the chromosomal region 5q32q33.1 was gradually concluded (Dixon et al., 1991, 1992, 1993). The coding sequence of the gene was isolated and named TCOF1, and it was confirmed to encode a low complexity, serine/alanine‐rich protein named treacle (Dixon et al., 1997). This 1,488 amino acid protein of approximately 152 kD has three domains with unique N‐ and C‐termini and a sizeable central repeat domain (So et al., 2004). The LIS1 homology (LisH) putative domain belonging to the N‐terminal is proposed to regulate microtubule dynamics, and the multiple nucleolar localization signals (NLS) belonging to the C‐terminal are essential for locating to the nucleolus (Emes & Ponting, 2001; Marsh et al., 1998). Treacle, a highly phosphorylated nucleolar protein, mainly consists of the central repeat domain (Isaac et al., 2000). The complete exon/intron genomic structure and the coding sequence of the TCOF1 gene were reported to contain 26 exons (Wise et al., 1997). Two additional exons were identified in the TCOF1 gene: exon 6A and exon 16A (So et al., 2004). The sequence for exon 6A transcribes the major TCOF1 isoform (isoform d, NM_001135243.1). Screening for TCOF1 variants in a large cohort of TCS1 patients from different populations, Splendore et al. (2002) confirmed that exons 10, 15, and 16 of the central repeat region and exons 23 and 24 of the NLS domain are mutational hotspots, accounting for more than half of the pathogenic variants in TCS1 cases. We reviewed the literature and summarize the reported pathogenic TCOF1 variants in Chinese TCS1 patients in Table 2. It is concluded that the hotspots of TCOF1 variants associated with TCS1 in the Chinese population are exons 5, 23, and 24. The only frequently recurring variant, c.4369_4373delAAGAA (p.Lys1457Glufs*12) in exon 24, contributes to approximately 12% of Chinese TCS1 cases. In this study, c.939_940insA (p.Gly314Argfs*35) was reported as the first variant in exon 7 in Chinese TCS1 patients.
4.3. Natural history of TCS1
In the natural history of Chinese TCS1 cases, Li et al. (2012) first identified a deletion in the TCOF1 gene in a 5‐year‐old boy with hearing loss and craniofacial malformations. Wang et al. (2014) found two variants in a TCS1 patient, one of which was a silent deletion variant, which emphasized the significance of the clinical and pedigree assessment in interpreting novel sequence changes. Li et al. (2019) described five Chinese TCS cases that exhibited high inter‐ and intra‐familial phenotypic heterogeneity and illustrated that genotype–phenotype variability is extreme in Chinese cases of TCS.
We have reviewed the literature and summarized the clinical characteristics of all reported Chinese TCS1 cases in Table 1. All reported cases had conductive hearing loss, indicating that audiological examinations should be performed regularly for suspected TCS patients and that a greater focus should be placed on the treatment of conductive hearing loss in TCS. An estimated 93% of reported Chinese TCS1 patients have zygomatic complex hypoplasia, mandibular micrognathia with retrognathia, downslanting palpebral fissures, and coloboma of the lower eyelids. Hence, TCS is relatively easy to diagnose by a unique “bird‐like” face. The numbers of patients (shown in Table 1) with malformation of auricles, external auditory canals, and the middle ear are approximately 69%, 65%, and 85%, respectively. These findings reflect the importance of temporal bone CT scanning and the close association between TCS and otorhinolaryngology and plastic surgery. The phenotypic variability in patients with TCS1 is evident within and among families. There has been no distinct evidence of a genotype–phenotype correlation in TCS1.
4.4. Treatment
TCS is a rare congenital disorder defined by craniofacial dysplasia. For newborn patients with TCS, early intervention is required to avoid respiratory failure arising from the narrowing of the airways caused by craniofacial malformations. Intervention includes the clearing and maintaining of the airways, feeding, protecting the eyes, and improving auditory ability and speech development. Operations later in life include aesthetic and functional reconstruction of the face and external ear. Concerning conductive hearing loss, ossicular chain deformities can be surgically corrected if external auditory canals are normal; otherwise, bone conduction hearing aids are often used to improve hearing in these patients (Fan et al., 2019). Ideally, soft‐banded bone conduction hearing aids should be started before 12 months of age to allow for normal development of the central auditory nervous system. Bone conduction hearing device implantation surgery is recommended after the patient is over 6 years of age when the thickness of cranial bones is a minimum of 4 mm. In a study that investigated the hearing rehabilitation effect in six Chinese TCS1 patients, the mean pure‐tone threshold improvements were calculated to be 30 dB for both soft‐band bone conduction hearing aids and implantation of a bone conduction hearing device. The mean speech discrimination scores improved approximately 50% after hearing interventions (Fan et al., 2019). The probands of case 2 and case 3 who wore soft‐band hearing aids experienced a satisfactory improvement in their hearing. Therefore, we recommend that the probands of case 4 and case 5 undergo hearing rehabilitation.
4.5. Prevention
TCS1 has a 50% probability of being inherited due to an autosomal dominant trait. The probands in cases 2–5 and their future spouses are recommended for genetic counseling. The recommendation for probands’ parents to avoid another TCS1 child is preimplantation genetic diagnosis (PGD). Targeted NGS has a vital role in the diagnosis of TCS1 and genetic counseling is beneficial for the prevention of the disease.
At least two professional editors, both native speakers of English, have checked the English in this document. For a certificate, please see: http://www.textcheck.com/certificate/N9upt6.
ETHICS APPROVAL
This study was performed with the approval of the Ethics Committee of the Second Affiliated Hospital of Zhengzhou University (reference number 2018008).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTION
Wei Lu and Wenxue Tang: study design and genetic counseling; Zhaoyu Pan and Hongen Xu: data collection, data analysis, and preparation of the manuscript; Bei Chen: phenotypic analysis; Yongan Tian, Linlin Zhang, and Sen Zhang: sample collection and Sanger sequencing; Danhua Liu: variant interpretation; Huanfei Liu, Ruijun Li, Xinxin Hu, and Jingyuan Guan: next‐generation sequencing. All authors have read and approved the final manuscript.
Funding information
The study is funded by the Collaborative Innovation Project of Zhengzhou (Zhengzhou University) (Grant no. 18XTZX12004) and the Medical Science and Technology Projects in Henan (Grant no. SBGJ2018043) to WT, the Scientific and Technological Research in Henan (Grant no. 192102310383) to WL.
Supporting information
Data S1
ACKNOWLEDGMENTS
The authors thank all subjects for their participation in this study. The Supercomputing Center in Zhengzhou University (Zhengzhou) provided support for data analysis.
Wei Lu and Wenxue Tang contributed equally to this work.
Zhaoyu Pan, Hongen Xu, and Bei Chen contributed equally to this work.
Zhaoyu Pan, Hongen Xu, and Bei Chen are co‐first authors.
Contributor Information
Wenxue Tang, Email: luweimd@hotmail.com, Email: twx@zzu.edu.cn.
Wei Lu, Email: luweimd@hotmail.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
REFERENCES
- Bowman, M. , Oldridge, M. , Archer, C. , O'Rourke, A. , McParland, J. , Brekelmans, R. , Seller, A. , & Lester, T. (2012). Gross deletions in TCOF1 are a cause of Treacher‐Collins‐Franceschetti syndrome. European Journal of Human Genetics, 20(7), 769–777. 10.1038/ejhg.2012.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Guo, L. , Li, C.‐L. , Shan, J. , Xu, H.‐S. , Li, J.‐Y. , Sun, S. , Hao, S.‐J. , Jin, L. , Chai, G. , & Zhang, T.‐Y. (2018). Mutation screening of Chinese Treacher Collins syndrome patients identified novel TCOF1 mutations. Molecular Genetics and Genomics, 293(2), 569–577. 10.1007/s00438-017-1384-3 [DOI] [PubMed] [Google Scholar]
- Conte, C. , D'Apice, M. R. , Rinaldi, F. , Gambardella, S. , Sangiuolo, F. , & Novelli, G. (2011). Novel mutations of TCOF1 gene in European patients with Treacher Collins syndrome. BMC Medical Genetics, 12, 125. 10.1186/1471-2350-12-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauwerse, J. G. , Dixon, J. , Seland, S. , Ruivenkamp, C. A. L. , van Haeringen, A. , Hoefsloot, L. H. , Peters, D. J. M. , Boers, A.‐D. , Daumer‐Haas, C. , Maiwald, R. , Zweier, C. , Kerr, B. , Cobo, A. M. , Toral, J. F. , Hoogeboom, A. J. M. , Lohmann, D. R. , Hehr, U. , Dixon, M. J. , Breuning, M. H. , & Wieczorek, D. (2011). Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nature Genetics, 43(1), 20–22. 10.1038/ng.724 [DOI] [PubMed] [Google Scholar]
- Dixon, J. , Hovanes, K. , Shiang, R. , & Dixon, M. J. (1997). Sequence analysis, identification of evolutionary conserved motifs and expression analysis of murine tcof1 provide further evidence for a potential function for the gene and its human homologue, TCOF1. Human Molecular Genetics, 6(5), 727–737. 10.1093/hmg/6.5.727 [DOI] [PubMed] [Google Scholar]
- Dixon, J. , Jones, N. C. , Sandell, L. L. , Jayasinghe, S. M. , Crane, J. , Rey, J.‐P. , Dixon, M. J. , & Trainor, P. A. (2006). Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proceedings of the National Academy of Sciences of United States of America, 103(36), 13403–13408. 10.1073/pnas.0603730103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, M. J. , Dixon, J. , Houseal, T. , Bhatt, M. , Ward, D. C. , Klinger, K. , & Landes, G. M. (1993). Narrowing the position of the Treacher Collins syndrome locus to a small interval between three new microsatellite markers at 5q32‐33.1. American Journal of Human Genetics, 52(5), 907–914. [PMC free article] [PubMed] [Google Scholar]
- Dixon, M. J. , Dixon, J. , Raskova, D. , Le Beau, M. M. , Williamson, R. , Klinger, K. , & Landes, G. M. (1992). Genetic and physical mapping of the Treacher Collins syndrome locus: refinement of the localization to chromosome 5q32‐33.2. Human Molecular Genetics, 1(4), 249–253. 10.1093/hmg/1.4.249 [DOI] [PubMed] [Google Scholar]
- Dixon, M. J. , Read, A. P. , Donnai, D. , Colley, A. , Dixon, J. , & Williamson, R. (1991). The gene for Treacher Collins syndrome maps to the long arm of chromosome 5. American Journal of Human Genetics, 49(1), 17–22. [PMC free article] [PubMed] [Google Scholar]
- Emes, R. D. , & Ponting, C. P. (2001). A new sequence motif linking lissencephaly, Treacher Collins and oral‐facial‐digital type 1 syndromes, microtubule dynamics and cell migration. Human Molecular Genetics, 10(24), 2813–2820. 10.1093/hmg/10.24.2813 [DOI] [PubMed] [Google Scholar]
- Fan, X. , Wang, Y. , Fan, Y. , Du, H. , Luo, N. , Zhang, S. , & Chen, X. (2019). TCOF1 pathogenic variants identified by Whole‐exome sequencing in Chinese Treacher Collins syndrome families and hearing rehabilitation effect. Orphanet J Rare Dis, 14(1), 178. 10.1186/s13023-019-1136-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac, C. , Marsh, K. L. , Paznekas, W. A. , Dixon, J. , Dixon, M. J. , Jabs, E. W. , & Meier, U. T. (2000). Characterization of the nucleolar gene product, treacle. Treacher Collins Syndrome. Mol Biol Cell, 11(9), 3061–3071. 10.1091/mbc.11.9.3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. B. , Zhang, X. , Li, Z. Y. , Chen, J. , Lu, Y. , Jia, J. J. , Yuan, H. J. , & Han, D. Y. (2012). Clinical and genetic analysis of a patient with Treacher Collins syndrome in TCOF1 gene. Journal of Otolaryngology – Head & Neck Surgery, 26(10), 459–462. 10.13201/j.issn.1001-1781.2012.10.011 [DOI] [PubMed] [Google Scholar]
- Li, X. , Su, Y. U. , Huang, S. , Gao, B. O. , Zhang, D. , Wang, X. , Gao, Q. , Pang, H. , Zhao, Y. , Yuan, Y. , & Dai, P. U. (2019). Genotype‐phenotype variability in Chinese cases of Treacher Collins syndrome. Acta Oto‐Laryngologica, 139(7), 567–575. 10.1080/00016489.2019.1612530 [DOI] [PubMed] [Google Scholar]
- Liu, J. , Lin, P. , Pang, J. , Jia, Z. , Peng, Y. , Xi, H. , & Wang, H. (2020). Identification of a novel gross deletion of TCOF1 in a Chinese prenatal case with Treacher Collins syndrome. Mol Genet Genomic Med, 8(8), e1313. 10.1002/mgg3.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, K. L. , Dixon, J. , & Dixon, M. J. (1998). Mutations in the Treacher Collins syndrome gene lead to mislocalization of the nucleolar protein treacle. Human Molecular Genetics, 7(11), 1795–1800. 10.1093/hmg/7.11.1795 [DOI] [PubMed] [Google Scholar]
- Masotti, C. , Ornelas, C. C. , Splendore‐Gordonos, A. , Moura, R. , Félix, T. M. , Alonso, N. , Camargo, A. A. , & Passos‐Bueno, M. R. (2009). Reduced transcription of TCOF1 in adult cells of Treacher Collins syndrome patients. BMC Medical Genetics, 10, 136. 10.1186/1471-2350-10-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Z. , Lu, W. , Li, X. , Huang, S. , Dai, P. , & Yuan, Y. (2020). Multiple synostoses syndrome: Clinical report and retrospective analysis. American Journal of Medical Genetics. Part A, 182(6), 1438–1448. 10.1002/ajmg.a.61583 [DOI] [PubMed] [Google Scholar]
- Pan, Z. , Xu, H. , Tian, Y. , Liu, D. , Liu, H. , Li, R. , Dou, Q. , Zuo, B. , Zhai, R. , Tang, W. , & Lu, W. (2020). Perrault syndrome: Clinical report and retrospective analysis. Molecular Genetics & Genomic Medicine, 8(10). 10.1002/mgg3.1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, C. A. , Amin, S. , Linden, J. , Dixon, J. , Dixon, M. J. , & Tucker, A. S. (2010). Defects in middle ear cavitation cause conductive hearing loss in the Tcof1 mutant mouse. Human Molecular Genetics, 19(8), 1551–1560. 10.1093/hmg/ddq028 [DOI] [PubMed] [Google Scholar]
- So, R. B. , Gonzales, B. , Henning, D. , Dixon, J. , Dixon, M. J. , & Valdez, B. C. (2004). Another face of the Treacher Collins syndrome (TCOF1) gene: identification of additional exons. Gene, 328, 49–57. 10.1016/j.gene.2003.11.027 [DOI] [PubMed] [Google Scholar]
- Splendore, A. , Jabs, E. W. , & Passos‐Bueno, M. R. (2002). Screening of TCOF1 in patients from different populations: Confirmation of mutational hot spots and identification of a novel missense mutation that suggests an important functional domain in the protein treacle. Journal of Medical Genetics, 39(7), 493–495. 10.1136/jmg.39.7.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splendore, A. , Silva, E. O. , Alonso, L. G. , Richieri‐Costa, A. , Alonso, N. , Rosa, A. , & Passos‐Bueno, M. R. (2000). High mutation detection rate in TCOF1 among Treacher Collins syndrome patients reveals clustering of mutations and 16 novel pathogenic changes. Human Mutation, 16(4), 315–322. [DOI] [PubMed] [Google Scholar]
- Teber, Ö. A. , Gillessen‐Kaesbach, G. , Fischer, S. , Böhringer, S. , Albrecht, B. , Albert, A. , Arslan‐Kirchner, M. , Haan, E. , Hagedorn‐Greiwe, M. , Hammans, C. , Henn, W. , Hinkel, G. K. , König, R. , Kunstmann, E. , Kunze, J. , Neumann, L. M. , Prott, E.‐C. , Rauch, A. , Rott, H.‐D. , … Wieczorek, D. (2004). Genotyping in 46 patients with tentative diagnosis of Treacher Collins syndrome revealed unexpected phenotypic variation. European Journal of Human Genetics, 12(11), 879–890. 10.1038/sj.ejhg.5201260 [DOI] [PubMed] [Google Scholar]
- Trainor, P. A. , & Andrews, B. T. (2013). Facial dysostoses: Etiology, pathogenesis and management. American Journal of Medical Genetics Part C Seminars, 163C(4), 283–294. 10.1002/ajmg.c.31375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Yin, X. J. , Han, T. , Peng, W. , Wu, H. L. , Liu, X. , & Feng, Z. C. (2014). A novel silent deletion, an insertion mutation and a nonsense mutation in the TCOF1 gene found in two Chinese cases of Treacher Collins syndrome. Molecular Genetics and Genomics, 289(6), 1237–1240. 10.1007/s00438-014-0883-8 [DOI] [PubMed] [Google Scholar]
- Weiner, A. M. , Scampoli, N. L. , & Calcaterra, N. B. (2012). Fishing the molecular bases of Treacher Collins syndrome. PLoS One, 7(1), e29574. 10.1371/journal.pone.0029574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise, C. A. , Chiang, L. C. , Paznekas, W. A. , Sharma, M. , Musy, M. M. , Ashley, J. A. , Lovett, M. , & Jabs, E. W. (1997). TCOF1 gene encodes a putative nucleolar phosphoprotein that exhibits mutations in Treacher Collins Syndrome throughout its coding region. Proceedings of the National Academy of Sciences of the United States of America, 94(7), 3110–3115. 10.1073/pnas.94.7.3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Z. , Lu, Y. U. , Wang, Y. , Zhang, X. , Duan, H. , Cheng, J. , Yuan, H. , & Han, D. (2018). Identification of a novel TCOF1 mutation in a Chinese family with Treacher Collins syndrome. Experimental and Therapeutic Medicine, 16(3), 2645–2650. 10.3892/etm.2018.6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , An, L. , Xue, H. , Hao, S. , Yan, Y. , Zhang, Q. , & Ma, X. (2020). Mutation analysis of TCOF1 gene in Chinese Treacher Collins syndrome patients. Journal of Clinical Laboratory Analysis, e23567. 10.1002/jcla.23567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Fan, Y. , Zhang, Y. , Xue, H. , & Chen, X. (2013). A novel mutation in the TCOF1 gene found in two Chinese cases of Treacher Collins syndrome. International Journal of Pediatric Otorhinolaryngology, 77(9), 1410–1415. 10.1016/j.ijporl.2013.05.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.


