Bowman et al. show that in melanocytes, the vSNARE VAMP7 is sorted from endosomes into tubular membrane transport carriers bound for maturing melanosomes in a complex with the tSNARE syntaxin 13 via redundant recognition of each SNARE by an AP-3–BLOC-1 super-complex.
Abstract
Membrane transport carriers fuse with target membranes through engagement of cognate vSNAREs and tSNAREs on each membrane. How vSNAREs are sorted into transport carriers is incompletely understood. Here we show that VAMP7, the vSNARE for fusing endosome-derived tubular transport carriers with maturing melanosomes in melanocytes, is sorted into transport carriers in complex with the tSNARE component STX13. Sorting requires either recognition of VAMP7 by the AP-3δ subunit of AP-3 or of STX13 by the pallidin subunit of BLOC-1, but not both. Consequently, melanocytes expressing both AP-3δ and pallidin variants that cannot bind their respective SNARE proteins are hypopigmented and fail to sort BLOC-1–dependent cargo, STX13, or VAMP7 into transport carriers. However, SNARE binding does not influence BLOC-1 function in generating tubular transport carriers. These data reveal a novel mechanism of vSNARE sorting by recognition of redundant sorting determinants on a SNARE complex by an AP-3–BLOC-1 super-complex.
Introduction
The endolysosomal system requires proper regulation of membrane trafficking for maintenance of organelle identity, cargo distribution, signaling, metabolic control, and organelle biogenesis (Huotari and Helenius, 2011). Membrane-bound vesicular or tubular transport carriers bud from donor organelles and are transported to the appropriate target organelle, to which they then dock and fuse to deliver their contents (Bonifacino and Glick, 2004). Docking and fusion are primarily mediated by specific interactions between SNARE proteins on the membrane carrier and the target membrane (Jahn and Scheller, 2006; Yoon and Munson, 2018). Typically, a vesicular SNARE (or R-SNARE, defined by the central R residue in the SNARE helix) on the vesicle membrane engages a three-helix target SNARE complex (or Qa, Qb, and Qc helices, defined by the central Q residue in the SNARE helix) on the target membrane to form a stable four-helix bundle “trans”-SNARE complex that drives fusion (Jahn and Scheller, 2006), often promoted by Sec1-Munc18 family members and other regulators (Baker and Hughson, 2016; Rizo and Südhof, 2012). In endosomal fusion, the four SNARE helices can be differentially distributed among opposing membranes (Zwilling et al., 2007). A key step in determining the fusion capacity of a transport carrier with its target is the incorporation of the R-SNARE at the time of transport carrier formation. However, our understanding of the mechanisms by which R-SNAREs are sorted into transport carriers is incomplete.
Lysosome-related organelles are specialized, tissue-specific structures that derive from the endolysosomal network. Lysosome-related organelle biogenesis and maturation via membrane transport are well studied for melanosomes, organelles within pigment cells of the skin, hair, and eyes that synthesize and store melanin pigments (Bowman et al., 2019; Sitaram and Marks, 2012). Melanosome precursors segregate from early endosomes (Raposo et al., 2001) and then mature in a process that requires delivery of melanogenic enzymes and transporters by at least three distinct membrane transport pathways, one from the Golgi and two from distinct domains of early endosomes (Bowman et al., 2019; Sitaram and Marks, 2012). The endosome-derived pathways are regulated by the protein complexes adaptor protein-3 (AP-3) and biogenesis of lysosome-related organelles complex (BLOC)-1, -2, and -3, subunits of which are mutated in heritable disorders called the Hermansky-Pudlak syndromes (HPS; Bowman et al., 2019; Di Pietro and Dell’Angelica, 2005). One function of AP-3 is to sort the enzyme tyrosinase (TYR) from early endosomes into clathrin-coated vesicles destined for maturing melanosomes (Huizing et al., 2001; Theos et al., 2005). BLOC-1, in collaboration with actin nucleators and the microtubule motor, KIF13A, is required to generate tubular membrane transport carriers that emerge from vacuolar early endosomes, have features of recycling endosomes, and transiently fuse with melanosomes to deliver essential melanosome cargoes such as TYR-related protein 1 (TYRP1; Delevoye et al., 2016; Delevoye et al., 2009; Dennis et al., 2016; Dennis et al., 2015; Setty et al., 2008; Setty et al., 2007). BLOC-2 helps target the tubular transport carriers to maturing melanosomes (Dennis et al., 2015). Fusion of these carriers and subsequent cargo delivery to melanosomes requires the R-SNARE, vesicle-associated membrane protein 7 (VAMP7; also known as tetanus neurotoxin insensitive VAMP, or TI-VAMP; Dennis et al., 2016; Tamura et al., 2011). VAMP7 is subsequently recycled from melanosomes in distinct tubular carriers (Dennis et al., 2016; Ripoll et al., 2018). Sorting of VAMP7 into melanosome-derived tubules likely requires binding to the scaffolding protein VAMP7- and Rab32/38-interacting protein (VARP; Dennis et al., 2016; Schäfer et al., 2012; Tamura et al., 2009), but how VAMP7 is sorted from endosomes into melanosome-bound tubular carriers is unknown.
VAMP7 is one of several R-SNAREs that possess a longin domain (Daste et al., 2015), which folds back onto the SNARE domain to impede SNARE complex formation (Kent et al., 2012; Martinez-Arca et al., 2003; Schäfer et al., 2012; Vivona et al., 2010). The VAMP7 longin domain facilitates several sorting events, including endocytosis via binding to the clathrin-associated Hrb (also known as AGFG1; Pryor et al., 2008) and likely sorting into transport carriers from endosomes and/or melanosomes by binding to VARP (Burgo et al., 2009; Dennis et al., 2016; Hesketh et al., 2014; Tamura et al., 2009). The VAMP7 longin domain also binds in vitro to AP-3 via the AP-3δ subunit, and AP-3 regulates VAMP7 sorting in several cell types (Chaineau et al., 2009; Kent et al., 2012; Martinez-Arca et al., 2003). AP-3 was proposed to have little impact on the delivery of the BLOC-1–dependent cargo, TYRP1, to melanosomes (Huizing et al., 2001). However, a cohort of AP-3 in cells is bound to BLOC-1 (Di Pietro et al., 2006; Salazar et al., 2006), and TYRP1 cycling through the cell surface is increased in AP-3–deficient melanocytes (Di Pietro et al., 2006). Thus, a role for AP-3 in sorting VAMP7 into BLOC-1–dependent transport carriers is not ruled out.
Another potential mechanism for SNARE sorting is in a “cis”-SNARE complex (i.e., all SNAREs on the same membrane) with partner SNAREs that themselves have sorting potential. For example, endocytic vesicles containing syntaxin 6 are targeted to distinct compartments depending on the absence or presence of a partner SNARE (Koike and Jahn, 2019). Among the known Q-SNAREs that interact with VAMP7 is syntaxin 12/13 (STX13; Chung et al., 2013; Kuster et al., 2015), a Qa SNARE that is present on BLOC-1–dependent transport carriers but is not itself incorporated into melanosomes (Delevoye et al., 2016; Dennis et al., 2016; Dennis et al., 2015) and thus is not likely a part of the fusogenic complex for cargo delivery. STX13 binds in vitro to the pallidin (Pldn; also known as BLOC1S6) subunit of BLOC-1 via its SNARE domain (Ghiani et al., 2010; Huang et al., 1999; Moriyama and Bonifacino, 2002), suggesting a potential direct mechanism for sorting STX13 into BLOC-1–dependent tubules. If this were true, then VAMP7 could be sorted into these tubules by association with STX13.
Here, we tested both models for VAMP7 sorting into BLOC-1–dependent transport carriers in melanocytes by exploiting cell lines lacking AP-3δ, the BLOC-1 Pldn subunit, or both and reconstituting these cells with variants of AP-3δ and/or Pldn that do or do not bind their target SNAREs. We show that binding of either AP-3 to VAMP7 or BLOC-1 to STX13 is both necessary and sufficient to support VAMP7 sorting and melanogenesis, documenting an example of redundancy in a SNARE sorting step. This mechanism of SNARE sorting and regulation provides insight into how SNAREs that localize broadly within the endolysosomal system can maintain specificity in fusion reactions, and how recognition by multiple molecular components can ensure the sorting of an essential SNARE component.
Results
The BLOC-1–dependent cargo TYRP1 is partially missorted in AP-3–deficient melanocytes
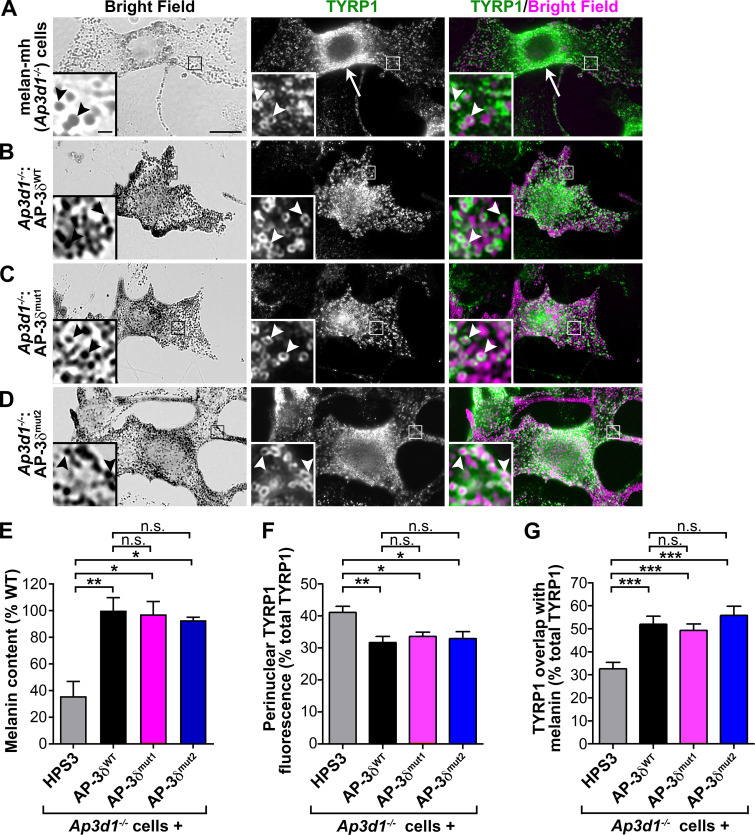
In AP-3–deficient melanocytes from AP3B1−/− HPS type 2 patients or Ap3b1−/− pearl mice, the BLOC-1–independent cargo TYR is largely depleted from melanosomes (Huizing et al., 2001; Theos et al., 2005), but the BLOC-1–dependent cargo TYRP1 was reported to localize normally to melanosomes (Huizing et al., 2001). Consistently, the AP-3δ/σ3 subcomplex binds in a yeast three-hybrid assay to a dileucine-based signal in the cytoplasmic domain of TYR, but not a dileucine-like signal in TYRP1 (Theos et al., 2005). Nevertheless, TYRP1 trafficking through the plasma membrane is increased in Ap3b1−/− melanocytes (Di Pietro et al., 2006), suggesting that AP-3 might indirectly influence BLOC-1–dependent TYRP1 trafficking. To directly test whether AP-3/VAMP-7 binding contributes to this, we used deconvolution immunofluorescence microscopy (dIFM) to quantify TYRP1 localization in AP-3δ–deficient melan-mh melanocytes, derived from mocha (C57BL/6J-Ap3d1mh/mh) mice, and in stably transduced “rescued” melan-mh:AP3δ cells that express WT or mutated forms of chimpanzee AP-3δ. When grown in 20% FBS, melan-mh melanocytes are hypopigmented due to reduced melanin synthesis (Fig. 1 A, left; Fig. S1 A; and Fig. 1 E shows melanin quantification of transduced melan-mh cells expressing AP-3δ variants or a negative control protein, HPS3) as described for AP-3β–deficient melanocytes from pearl mice and HPS2 patients (Huizing et al., 2001; Sitaram et al., 2012; Theos et al., 2005) and consistent with the hypopigmentation of mocha mice (Lane and Deol, 1974). Expression of WT AP-3δ in these cells (melan-mh:AP3δ) increases pigmentation (Fig. 1 B, left; Fig. 1 E; and Fig. S1 D) to levels comparable to those of melanocytes from WT C57BL/6J mice. As described in AP-3β–deficient melanocytes (Huizing et al., 2001; Theos et al., 2005), TYR is largely confined to the perinuclear region and depleted from peripheral melanosomes in melan-mh relative to melan-mh:AP3δ cells (Fig. S1, B and E). However, while a cohort of TYRP1 localizes to melanosomes in melan-mh cells, relative to melan-mh:AP3δ, the degree of TYRP1 overlap with melanosomes is lower (32.6 ± 2.9% versus 51.9 ± 3.5%), and TYRP1 accumulation within the perinuclear region is higher (41.1± 1.9% versus 31.6 ± 2.0%; Fig. 1, A and B, middle and right; Fig. 1, F and G; and Fig. S1, C and F), whereas melanosome distribution is similar in both cell types. The peripheral TYRP1-containing structures in melan-mh cells do not overlap with STX13-containing endosomes (Fig. S2) as they do in BLOC-1–deficient melanocytes (Setty et al., 2007; also note in Fig. S2 that pigment granules in melan-mh cells are not labeled by STX13 in our culture conditions). These data indicate that AP-3 influences the trafficking of TYRP1, a non–AP-3-binding cargo protein that requires both BLOC-1 and VAMP7 for effective delivery to melanosomes.
Figure 1.
VAMP7 binding by AP-3δ is not required for normal pigmentation or cargo localization in melanocytes. (A–D) Untransduced (Ap3d1−/−; A) or retrovirally transduced AP-3δ–deficient melan-mh cells stably expressing WT AP-3δ (AP-3δWT; B) or AP-3δ VAMP7 binding mutants (AP-3δmut1, AP-3δmut2; C and D) were fixed, labeled for TYRP1 and STX13, and analyzed by dIFM and brightfield microscopy. Brightfield images (taken at same gain and exposure) are pseudocolored magenta in the right panels. Boxed regions are magnified fivefold in insets. Arrow, TYRP1 accumulation in the perinuclear region of an untransduced cell; arrowheads, TYRP1 on pigmented melanosomes. Scale bar, 5 µm; inset scale bar, 1 µm. (E) Melanin content in cell lysates quantified by spectrophotometry. Data (mean ± SEM from three separate experiments) are normalized to values from melan-mh cells expressing AP-3δWT. (F and G) Quantification of imaging data; melan-mh cells stably expressing HPS3 are the virally transduced negative control. (F) Quantification of TYRP1 perinuclear signal by dIFM relative to total cellular TYRP1 (percent). Data represent mean ± SEM from 25 (HPS3 control), 20 (AP-3δWT), 26 (AP-3δmut1), and 21 (AP-3δmut2) cells over three separate experiments. (G) Quantification of percentage overlap of TYRP1 by dIFM with melanin by brightfield microscopy. Data represent mean ± SEM from 28 (HPS3 control), 20 (AP-3δWT), 34 (AP-3δmut1), and 20 (AP-3δmut2) cells over three separate experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
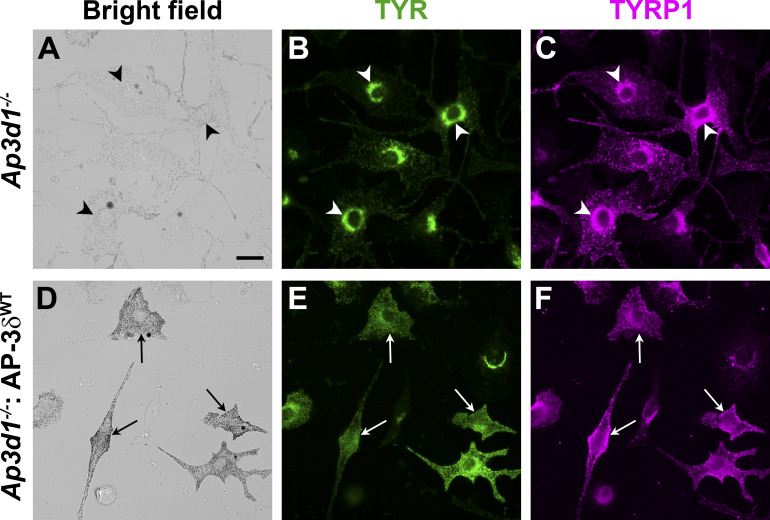
Figure S1.
Disruption of TYR and TYRP1 localization in Ap3d1−/− melan-mh cells. (A–F) Melan-mh cells that were untransduced (A–C) or stably transduced to express WT AP-3δ (melan-mh: AP-3δWT; D–F) were fixed, immunolabeled for TYR and for TYRP1, and analyzed by dIFM and brightfield microscopy. Arrowheads, perinuclear accumulation of both TYR and TYRP1 in hypopigmented melan-mh cells; arrows, peripheral localization of TYR and TYRP1 in darkly pigmented melan-mh: AP-3δWT cells. Scale bar, 20 µm.
Figure S2.
TYRP1 does not accumulate in STX13-containing early endosomes in melanocytes that either lack AP-3 or express an AP-3δ variant that cannot bind VAMP7. (A–D) Melan-mh cells that were untransduced (A) or transduced to express WT AP-3δ (melan-mh: AP-3δWT; B) or the VAMP7 nonbinding variants AP-3δmut1 (C) or AP-3δmut2 (D) were fixed, immunolabeled for TYRP1 and STX13, and analyzed by dIFM. Shown are separate images of TYRP1 (left) and STX13 (middle) labeling; a merged image is shown at right. Boxed regions are magnified sevenfold in the insets. Arrowheads point to TYRP1-labeled structures that lack labeling for STX13. Scale bar, 10 µm; inset scale bar, 1 µm. These images are from the same cells as those shown in Fig. 1, A–D. (E) Quantification (mean ± SEM) of the percentage of structures labeled for TYRP1 that also label for STX13.
AP-3δ binding to VAMP7 is not required for trafficking of melanosome cargoes
AP-3δ binds to the VAMP7 longin domain via the AP-3δ “linker” region (amino acid residues 680–728; Kent et al., 2012; Martinez-Arca et al., 2003). Mutagenesis to serines of either of two pairs of key hydrophobic residues—Ile702 and Val704 (mut1) or Leu709 and Leu713 (mut2)—dramatically reduces AP-3δ binding to VAMP7 in vitro and alters VAMP7 localization in vivo (Kent et al., 2012). To test whether VAMP7 binding to AP-3 was necessary for BLOC-1–dependent cargo localization to melanosomes, we analyzed pigmentation and TYRP1 distribution relative to pigmented melanosomes in Ap3d1−/− melan-mh melanocytes stably expressing a control protein (HPS3) or either WT, mut1, or mut2 variants of AP-3δ. Surprisingly to us, stable expression of either AP-3δWT, AP-3δmut1, or AP-3δmut2, but not HPS3, equally restored WT levels of pigmentation, reduced TYRP1 accumulation in the perinuclear area (33.6 ± 1.4% for mut1, 32.9 ± 2.2% for mut2), and increased TYRP1 overlap with melanin (49.3 ± 2.9% for mut1, 55.8 ± 4.1% for mut2; Fig. 1). Together, these data suggest that the AP-3δ–VAMP7 interaction alone is not required for pigmentation or trafficking of BLOC-1–dependent melanosome cargoes.
STX13 binding to the Pldn subunit of BLOC-1 is not required for melanosome cargo sorting
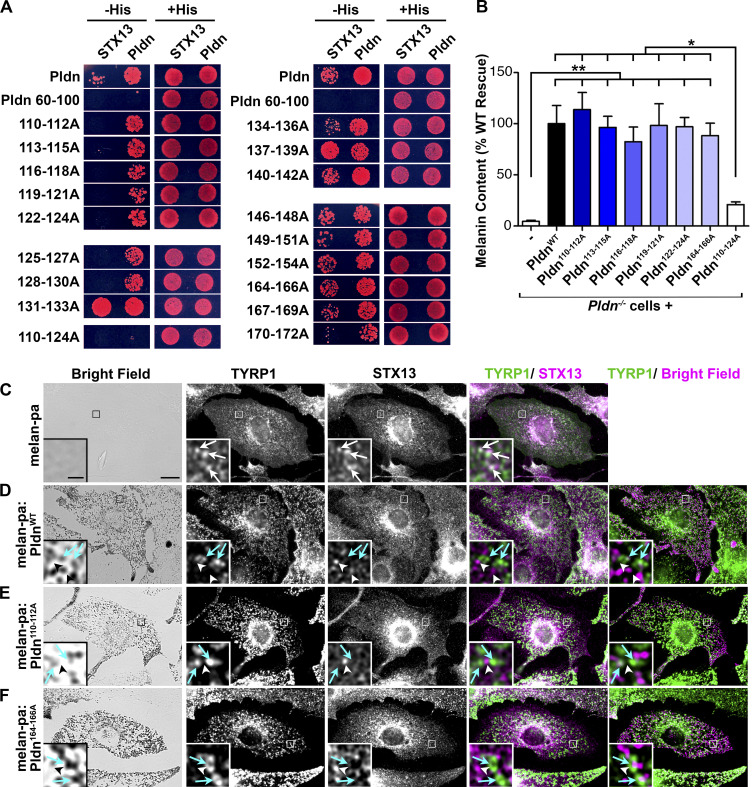
In other cell types, a cohort of VAMP7 associates with STX13 (Chung et al., 2013; Kuster et al., 2015). Thus, VAMP7 could be sorted into endosomal tubules in complex with STX13, which binds to BLOC-1 (Ghiani et al., 2010) via its Pldn subunit (Moriyama and Bonifacino, 2002). To assess BLOC-1 function in SNARE sorting toward melanosomes, we first defined residues within Pldn required for STX13 interaction. A predicted coiled-coil region spanning residues 110–172 of Pldn is necessary and sufficient to bind STX13 in a yeast two-hybrid assay (Moriyama and Bonifacino, 2002). Using this assay, we found that alanine mutagenesis of any three adjacent residues within Pldn residues 110–130 ablated Pldn interaction with STX13 but not a Pldn–Pldn interaction (Fig. 2 A). We then stably expressed a subset of Pldn-STX13 binding mutants in BLOC-1–deficient melan-pa cells, which are immortalized melanocytes derived from pallid (Pldnpa/pa) mice (Dennis et al., 2016; Setty et al., 2007) that do not express Pldn (Huang et al., 1999). Melan-pa cells are severely hypopigmented (Fig. 2, B and C, left panel) and trap TYRP1 in STX13-labeled early endosomes (Setty et al., 2007; Fig. 2 C, arrows). Surprisingly to us, stable expression of any of the triple-alanine Pldn mutants restored pigmentation in melan-pa cells as well as WT Pldn, regardless of whether the mutant bound to STX13 in vitro (Fig. 2 B). Consistently, TYRP1 was equally localized to melanosomes and not to STX13-containing endosomes in melan-pa cells expressing WT Pldn (PldnWT) or Pldn mutants that do (Pldn110-112A, hereafter referred to as Pldnmut1) or do not (Pldn164-166A, as a control) interfere with STX13 binding (Fig. 2, D–F; see also Fig. 5). Together, these data suggest that binding of Pldn to STX13 alone is not required for pigmentation or cargo trafficking to melanosomes.
Figure 2.
STX13 binding by the BLOC-1 subunit Pldn is not required for pigmentation or melanosome cargo localization in melanocytes. (A) Yeast two-hybrid assay for WT or triple alanine scanning mutants of full-length Pldn (fused to the GAL4 activation domain) binding to either the STX13 cytoplasmic domain or full-length Pldn (fused to the GAL4 DNA binding domain). A truncated Pldn mutant harboring residues 60–100 and a full-length mutant with residues 110–124 mutagenized to alanines (110-124A) are negative controls. Representative of at least two experiments showing growth of transformed his3 yeast colonies harboring HIS3 driven by the GAL4 promoter on plates lacking (−His) or containing (+His) histidine; growth on −His plates indicates an interaction. (B–F) WT Pldn (PldnWT) or the indicated mutants were expressed stably in Pldn-deficient melan-pa (Pldn−/−) melanocytes by retroviral transduction. (B) Melanin content in cell lysates was quantified by spectrophotometry. Data (mean ± SEM from two separate experiments done in triplicate) are normalized to the mean value from cells expressing PldnWT. *, P < 0.05; **. P < 0.01; if not indicated, values were not significantly different from cells expressing PldnWT. (C–F) Untransduced melan-pa (Pldn−/−) melanocytes (C) or stable transductants expressing PldnWT (D), a mutant that does not bind STX13 (Pldn110-112A; E) or a mutant that retains STX13 binding (Pldn164-166A; F) were fixed, immunolabeled for TYRP1 and STX13, and analyzed by dIFM and brightfield microscopy. Boxed regions are magnified fivefold in insets. White arrows, TYRP1 colocalized with STX13; cyan arrows, TYRP1 on melanosomes; arrowheads, STX13 on TYRP1-negative compartments. Scale bar, 10 µm; inset scale bar, 2 µm.
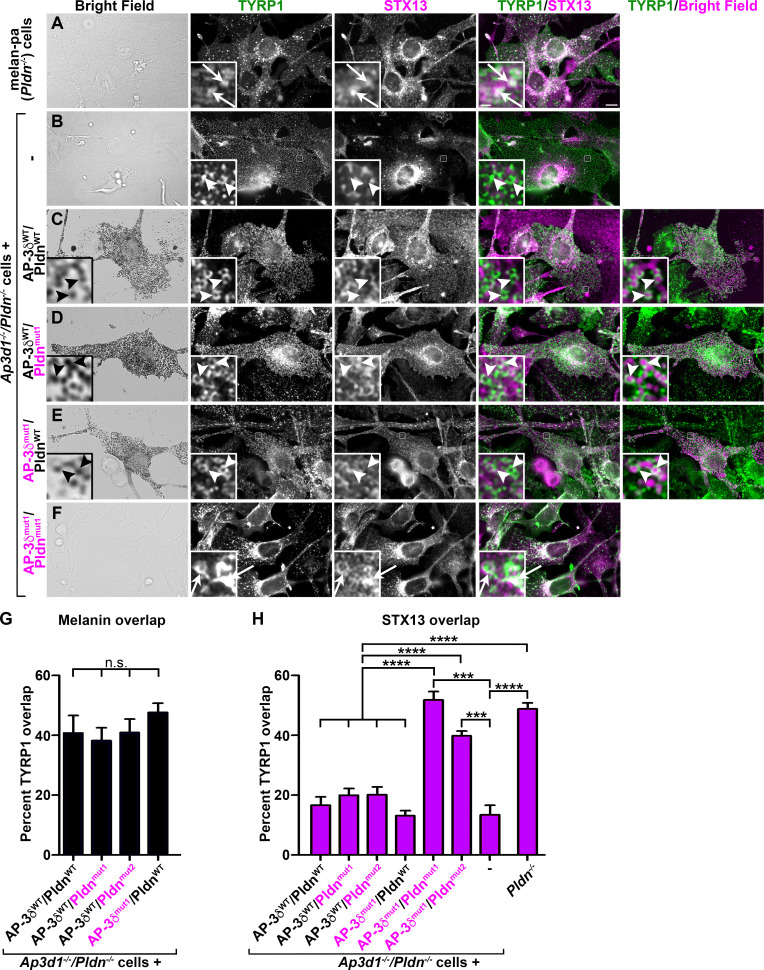
Figure 5.
Binding of either AP-3δ to VAMP7 or Pldn to STX13 is necessary for TYRP1 localization to melanosomes. (A–F) Pldn−/− melan-pa melanocytes (A) or Ap3d1−/−/Pldn−/− melanocytes that were either untransduced (B) or stably transduced to express the indicated WT (black) or SNARE binding mutant (magenta) variants of AP-3δ and Pldn (C–F) were fixed, immunolabeled for STX13 and TYRP1, and analyzed by dIFM and brightfield microscopy. Shown are corresponding brightfield images (left), individual labels for TYRP1 and STX13, and merged images of TYRP1/STX13 and (for pigmented cells) of TYRP1/pigment granules pseudocolored magenta (right). Boxed regions are magnified fivefold in the insets. Arrows, TYRP1/STX13 overlap; arrowheads, TYRP1 label that does not overlap with STX13 (overlaps with pigment granules in C–E). Scale bars, 5 µm; inset scale bars, 1 µm. (G and H) Quantification of percentage overlap of TYRP1 with melanin (G) or STX13 (H) from dIFM/brightfield analyses of Pldn−/− cells and Ap3d1−/−/Pldn−/− cells that were untransduced or express the indicated pairs of WT or SNARE binding mutant forms of AP-3δ and Pldn. Data represent mean ± SEM across 10–22 cells for each sample over three experiments. ***, P < 0.001; ****, P < 0.0001; nonsignificant differences between values are not indicated in H.
Binding of either AP-3δ to VAMP7 or Pldn to STX13 is necessary for pigmentation
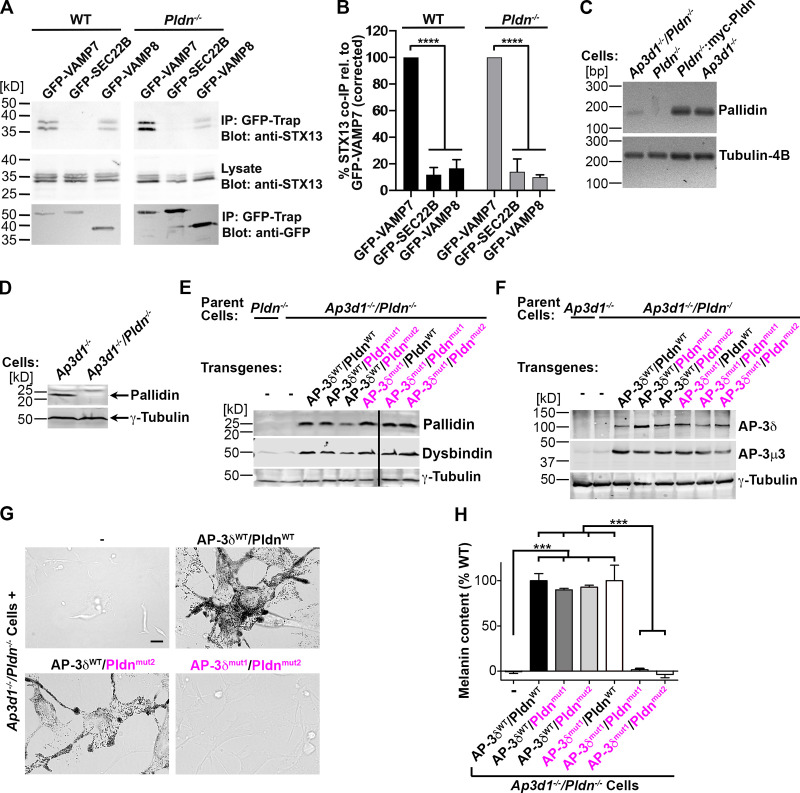
Although neither VAMP7 binding to AP-3 nor STX13 binding to BLOC-1 alone is independently required for BLOC-1–dependent melanosome cargo delivery, we considered whether these two SNARE binding modes are redundant based on the following. (1) The VAMP7 longin domain engages the SNARE domain intramolecularly and AP-3δ intermolecularly using the same interface, suggesting that AP-3 only engages VAMP7 when in complex with other SNAREs and its longin domain is available (Kent et al., 2012). (2) Similarly, BLOC-1 binds effectively to the STX13 SNARE domain only in the absence of the intramolecular inhibitory Habc domain (Ghiani et al., 2010), suggesting that BLOC-1 binds STX13 in a SNARE complex. (3) A cohort of VAMP7 and STX13 interacts within melanocytes independently of BLOC-1, as shown by the selective coprecipitation of both endogenous STX13 and stably expressed myc-tagged STX13 with transiently expressed GFP-VAMP7 but not with GFP-VAMP8 or -SEC22B in both WT and BLOC-1–deficient melanocytes (Fig. 3, A and B). Thus, a SNARE complex containing VAMP7 and STX13 might engage both BLOC-1 and AP-3, a cohort of which associates in cells (Di Pietro et al., 2006; Salazar et al., 2006). We therefore hypothesized that binding of either AP-3 or BLOC-1 to a VAMP7/STX13-containing cis-SNARE complex is sufficient to sort VAMP7 into melanosome-destined tubular carriers.
Figure 3.
Binding of either AP-3δ to VAMP7 or of Pldn to STX13 is required for pigmentation. (A and B) WT melan-Ink4a or Pldn-deficient melan-pa (Pldn−/−) cells stably expressing Myc-STX13 were transiently transfected to express GFP-VAMP7, -VAMP8, or -SEC22B. 1–2 d later, detergent cell lysates were prepared and immunoprecipitated using GFP-Trap agarose, and immunoprecipitates (IP) and 3% of the original cell lysates were fractionated by SDS-PAGE and analyzed by immunoblotting for STX13 or for GFP using HRP-conjugated secondary antibodies and analysis by ECL. (A) Representative images of relevant regions from the blots from a single experiment are shown. The upper band of the STX13 triplet represents Myc-STX13, and the two lower bands represent endogenous STX13. (B) Quantification from multiple experiments of coimmunoprecipitated STX13 band intensity (mean ± SEM) normalized to total STX13 in the lysate, immunoprecipitated GFP-tagged SNARE, and the value of STX13 coimmunoprecipitated (co-IP) with GFP-VAMP7; n = 3 (WT) or 4 (Pldn−/−). (C–H) The Bloc1s6 gene (encoding Pldn) in Ap3d1−/− melanocytes was disrupted by CRISPR/Cas9 mutagenesis to generate Ap3d1−/−/Pldn−/− cells. Data from a single clone and its transduced derivatives (representative of a second clone) are shown below. (C) RT-PCR of RNA from Ap3d1−/−/Pldn−/−, melan-pa (Pldn−/−), melan-pa cells stably expressing myc-tagged human Pldn (Pldn−/−:myc-Pldn), and melan-mh (Ap3d1−/−) cells, amplified for Pldn or tubulin-4B (as a loading control) were fractionated by agarose gel electrophoresis and visualized with SYBR Safe. Left: Positions of DNA markers (bp). Representative of three separate experiments. (D) Immunoblotting of fractionated melan-mh (Ap3d1−/−) and Ap3d1−/−/Pldn−/− whole-cell lysates for Pldn or γ-tubulin as a loading control. Left: Positions of molecular weight markers (kD). Representative of three separate experiments. (E and F) Fractionated whole-cell lysates of untransduced (−) melan-pa (Pldn−/−), melan-mh (Ap3d1−/−), or Ap3d1−/−/Pldn−/− cells or Ap3d1−/−/Pldn−/− cells transduced to express the indicated WT (black) or SNARE binding mutant (magenta) forms of AP-3δ and Pldn were immunoblotted for the BLOC-1 subunits Pldn and dysbindin (E), the AP-3 subunits δ and µ1 (F), or γ-tubulin (E and F). Left: Positions of molecular weight markers (kD). (G) Brightfield microscopy of untransduced Ap3d1−/−/Pldn−/− melanocytes (−) or cells expressing the indicated WT or SNARE binding mutant forms of AP-3δ and Pldn. Scale bar, 10 µm. (H) Melanin content in lysates of untransduced Ap3d1−/−/Pldn−/− cells (−) or cells expressing the indicated WT or SNARE binding mutant forms of AP-3δ and Pldn was assessed by spectrophotometry. Data (mean ± SEM from three separate experiments) are normalized to the mean value from cells expressing AP-3δWT and PldnWT. ***, P < 0.001; ****, P < 0.0001; nonsignificant differences are not indicated.
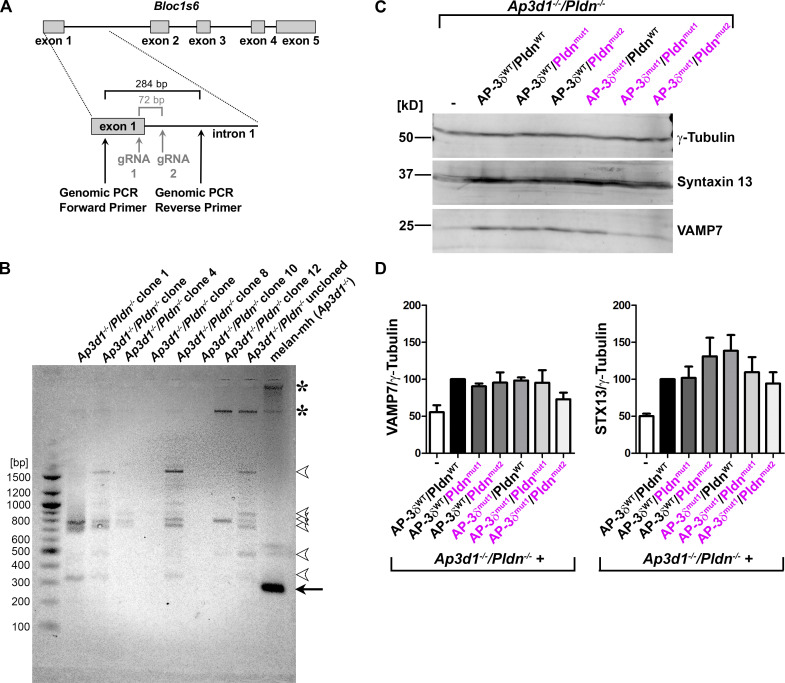
To test this hypothesis, we first generated cells lacking both AP-3δ and Pldn by disrupting Bloc1s6 (encoding Pldn) in Ap3d1−/− melan-mh melanocytes using CRISPR/Cas9 mutagenesis (Fig. S3 A). PCR analysis of genomic DNA showed that a region surrounding the targeted sequence within Bloc1s6 was altered in the selected cell population and all 10 clones tested (Fig. S3 B). By RT-PCR, Pldn mRNA levels were reduced or absent in two of five clones analyzed (Ap3d1−/−/Pldn−/− melanocytes) compared with controls, and they were similar to levels in melan-pa (Pldn−/−) cells (Fig. 3 C). Accordingly, immunoblotting confirmed that Pldn protein was absent in Ap3d1−/−/Pldn−/− cells (Fig. 3 D). We then transduced these cells with paired retroviruses to stably express combinations of WT or VAMP7-binding mutant variants of AP-3δ (AP-3δWT, AP-3δmut1, or AP-3δmut2) and WT or STX13-binding mutant variants of Pldn (PldnWT, Pldnmut1, or the 113-115A mutant [Pldnmut2]). Immunoblotting analyses of whole-cell lysates from selected transduced clones confirmed that they express Pldn and AP-3δ, whereas the parental untransduced Ap3d1−/−/Pldn−/− lines do not (Fig. 3, E and F). Moreover, whereas lysates of the parental line also have reduced levels of the BLOC-1 subunit dysbindin and of AP-3μ3 (due to their instability without BLOC-1 and AP-3 assembly), WT levels of these subunits were restored in each transduced clone (Fig. 3, E and F). Thus, all of the expressed Pldn and AP-3δ subunits incorporate into and restore stability to the respective BLOC-1 and AP-3 complexes. STX13 and VAMP7 levels were modestly but not significantly lower in Ap3d1−/−/Pldn−/−cells than in cells expressing any combination of WT and SNARE binding mutant AP-3δ and Pldn variants (Fig. S3, C and D), suggesting that loss of BLOC-1 and AP-3 expression, but not SNARE binding per se, might partially destabilize these SNAREs.
Figure S3.
The Bloc1s6 gene is disrupted in Ap3d1−/−/Pldn−/− cells. (A) Schematic diagram of the Bloc1s6 gene (top), encoding Pldn, and the CRISPR/Cas9 mutagenesis and screening strategy for Bloc1s6 gene knockout (bottom) in Ap3d1−/− melan-mh cells. Relative positions of gRNAs 1 and 2 within exon 1 and intron 1 are indicated (gray); if used for precise excision, they would be predicted to remove a 72-bp region as indicated. Positions of genomic primers for PCR in B and the size of the predicted fragment from unmodified Bloc1s6 are indicated. (B) Agarose gel analysis of PCR fragments amplified using the primers indicated in A from genomic DNA of melan-mh, the uncloned population of cells surviving selection following CRISPR/Cas9 mutagenesis (Ap3d1−/−/Pldn−/− uncloned), and 7 of 10 selected Ap3d1−/−/Pldn−/− clones. Positions of DNA size standards (bp) are shown at left. Arrow points to the expected 284-bp fragment detected only in parental melan-mh cells. Arrowheads point to faint bands not observed in melan-mh cells that likely represent off-target PCR products. Asterisks, nonspecific bands detected in melan-mh cells. The additional three clones analyzed had banding patterns similar to those of the clones tested here (not shown). (C) Fractionated whole-cell lysates of Ap3d1−/−/Pldn−/− cells that were untransduced or reconstituted with WT (black) or SNARE binding mutants (magenta) of AP-3δ and Pldn were immunoblotted for VAMP7, STX13, and γ-tubulin (loading control). (D) Quantification of STX13 and VAMP7 protein levels (mean ± SEM), normalized to γ-tubulin levels, over seven (STX13) and four (VAMP7) experiments, respectively. None of the values for either STX13 or VAMP7 content among the different cell lines were statistically different by one-way ANOVA.
Using these reconstituted cell lines, we tested whether impaired SNARE binding by AP-3 and BLOC-1 alone or in combination impacts cargo sorting and pigmentation. Like BLOC-1–deficient cells (Setty et al., 2007; Fig. 2, B and C), Ap3d1−/−/Pldn−/− melanocytes are severely hypopigmented, and coexpression of WT forms of AP-3δ and Pldn restored pigmentation (Fig. 3, G and H). Consistent with results of Figs. 1 and 2, coexpression of either AP-3δWT and Pldnmut1 or of AP-3δmut1 and PldnWT in Ap3d1−/−/Pldn−/− melanocytes restored WT pigmentation levels (Fig. 3, G and H). By contrast, coexpression of AP-3δmut1 with either Pldnmut1 or Pldnmut2 failed to restore pigmentation (Fig. 3, G and H). These data suggest that binding of either AP-3δ to VAMP7 or Pldn to STX13 is necessary for pigmentation.
Binding of either AP-3δ to VAMP7 or Pldn to STX13 is necessary for BLOC-1–dependent sorting of VAMP7 and cargo to melanosomes
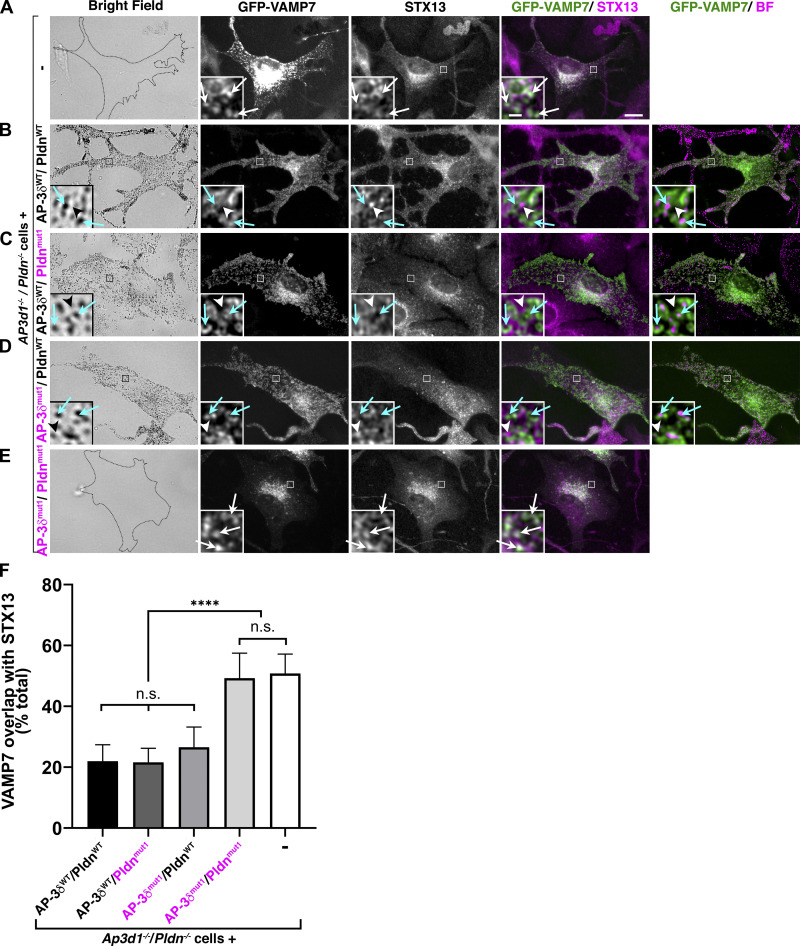
The results above could reflect a need for either AP-3 or BLOC-1 SNARE binding to sort VAMP7 into cargo carriers toward melanosomes or rather an indirect effect on SNARE trafficking. To test whether SNARE binding is needed to sort VAMP7 to melanosomes, we used dIFM to localize transiently expressed GFP-tagged VAMP7 in stably transduced Ap3d1−/−/Pldn−/− melanocytes expressing combinations of WT or SNARE binding mutants of AP-3δ and Pldn. As in BLOC-1–deficient cells (Dennis et al., 2016), GFP-VAMP7 in Ap3d1−/−/Pldn−/− melanocytes was trapped largely in STX13-labeled endosomes (Fig. 4 A, white arrows). Stable expression of AP-3δWT and PldnWT resulted in predominant GFP-VAMP7 steady-state localization to pigmented melanosomes (Fig. 4 B, cyan arrows) and segregation from STX13 (Fig. 4 B, white arrowheads) as in control melanocytes (Fig. 4 F; Dennis et al., 2016). Similarly, expression of either AP-3δWT and Pldnmut1 or of AP-3δmut1 and PldnWT resulted in GFP-VAMP7 localization to melanosomes and reduced overlap with STX13 (Fig. 4, C and D, cyan arrows and white arrowheads; and Fig. 4 F), showing that in melanocytes, unlike in fibroblasts (Kent et al., 2012), VAMP7 localization is not dramatically influenced by loss of its interaction with AP-3δ alone. By contrast, stable expression of both AP-3δmut1 and Pldnmut1 in Ap3d1−/−/Pldn−/− melanocytes did not rescue GFP-VAMP7 localization from STX13-containing endosomes (Fig. 4 E, white arrows, and Fig. 4 F). Thus, binding of either AP-3 to VAMP7 or of BLOC-1 to STX13 is necessary and sufficient for VAMP7 localization to melanosomes.
Figure 4.
Binding of either AP-3δ to VAMP7 or Pldn to STX13 is required for BLOC-1–dependent VAMP7 localization to melanosomes. (A–E) Ap3d1−/−/Pldn−/− melanocytes that were untransduced (-; A) or stably transduced with the indicated WT (black text) or SNARE binding mutant (magenta text) AP-3δ and Pldn variants (B–E) were transiently transfected with GFP-VAMP7, fixed, immunolabeled for STX13, and analyzed by dIFM and brightfield (BF) microscopy. Shown are corresponding BF images (left), individual labels for GFP-VAMP7 and STX13, merged images of GFP-VAMP7 and STX13, and overlap of GFP-VAMP7 with pigment granules (if any) pseudocolored magenta from the BF image (right). Boxed regions are magnified sevenfold in the insets. White arrows, overlap of GFP-VAMP7 with STX13; cyan arrows, overlap of GFP-VAMP7 with pigment granules; white arrowheads, lack of overlap of GFP-VAMP7 with STX13. The cell outline is shown in the BF images in A and E. Scale bar, 10 µm; inset scale bar, 1 µm. (F) Quantification (mean ± SEM) of percentage GFP-VAMP7 overlap with STX13 across 18 cells for each sample over three separate experiments. ****, P < 0.0001; n.s., not significant.
Since VAMP7 is the R-SNARE for fusion of BLOC-1–dependent cargo carriers with melanosomes, we used dIFM to test whether AP-3/VAMP7 or BLOC-1/STX13 binding is required for melanosomal localization of the BLOC-1–dependent cargo, TYRP1. Unlike cells that lack BLOC-1 only (such as Pldn−/− melan-pa), in which TYRP1 and other cargoes are trapped in STX13-labeled endosomes (Setty et al., 2008; Setty et al., 2007; Fig. 5 A, arrows; Fig. 5 H), TYRP1 did not overlap significantly with STX13 in Ap3d1−/−/Pldn−/− melanocytes and instead was distributed broadly throughout the cell and in small puncta (Fig. 5 B, arrowheads; and Fig. 5 H); we speculate that this represents the plasma membrane and clathrin-coated pits, respectively. The data indicate that complete loss of AP-3 exacerbates post-Golgi TYRP1 trafficking defects resulting from loss of BLOC-1. Expression in Ap3d1−/−/Pldn−/− melanocytes of either AP-3δWT and PldnWT, AP-3δWT and Pldnmut1, or AP-3δmut1 and PldnWT restored TYRP1 localization to pigmented melanosomes and maintained minimal overlap with STX13 (Fig. 5, C–E, arrowheads, quantified in Fig. 5, G and H), consistent with the rescue of cargo localization by SNARE binding mutant Pldn or AP-3δ in single Pldn−/− or Ap3d1−/− cells (Figs. 1 and 2). However, expression of either double-mutant combination—AP-3δmut1 and Pldnmut1 or AP-3δmut1 and Pldnmut2—enhanced TYRP1 localization to STX13-containing endosomes (Fig. 5 F, arrows, and Fig. 5 H) as in melan-pa cells (Fig. 5, A and H). Together, these data indicate that binding of either AP-3 to VAMP7 or of BLOC-1 to STX13 is necessary and sufficient for delivery of BLOC-1–dependent cargo to melanosomes.
SNARE binding by either AP-3 or BLOC-1 is required for STX13 sorting into BLOC-1–dependent tubular transport carriers but not for carrier formation
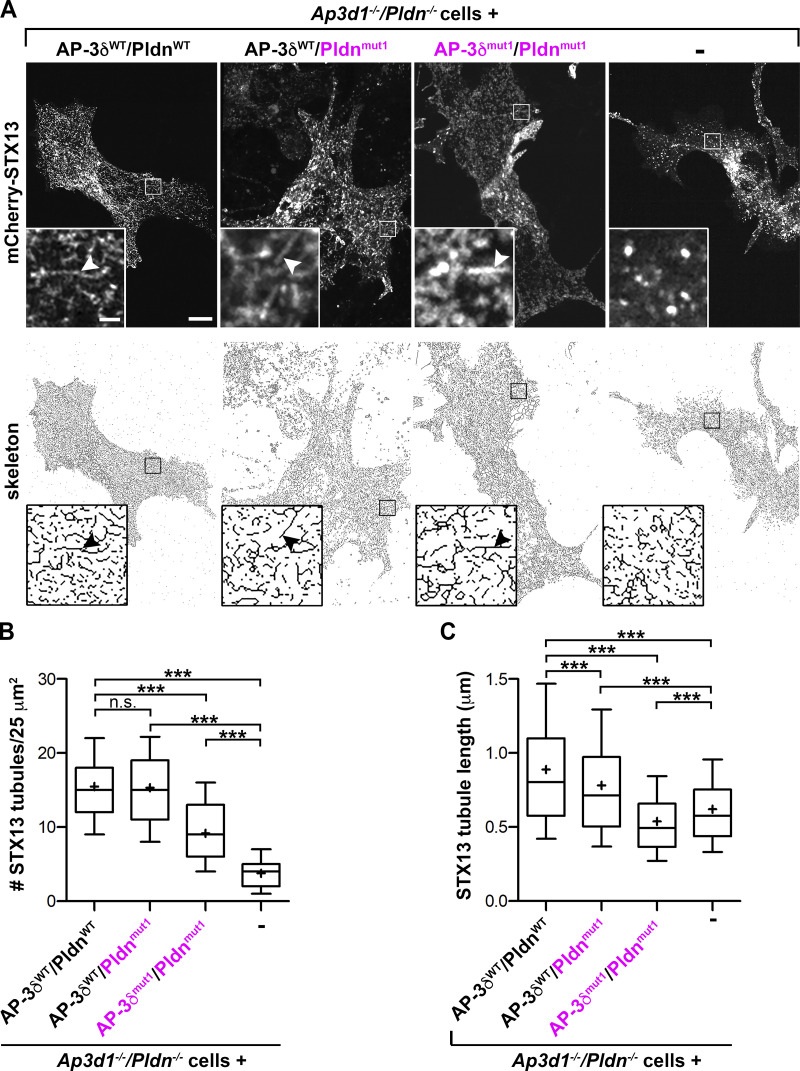
BLOC-1 is required for the formation of tubular transport carriers that deliver cargo from endosomes to maturing melanosomes (Delevoye et al., 2016; Delevoye et al., 2009; Dennis et al., 2016; Dennis et al., 2015). To test whether disruption of both Pldn/STX13 and AP-3δ/VAMP7 binding impacts the formation of tubules and/or STX13 sorting into them, AP3d1−/−/Pldn−/− cells expressing mCherry-STX13, which labels the tubules (Delevoye et al., 2016; Dennis et al., 2016; Dennis et al., 2015), and combinations of WT and SNARE binding mutants of Pldn and AP-3δ were analyzed by live-cell confocal fluorescence imaging. As in WT melanocytes (Delevoye et al., 2016; Dennis et al., 2016; Dennis et al., 2015), numerous long mCherry-STX13–containing tubules were observed in Ap3d1−/−/Pldn−/− melanocytes expressing AP-3δWT and either PldnWT or Pldnmut1 (15.5 ± 0.5 versus 15.3 ± 0.6/25 µm2; Fig. 6 A, arrowheads; quantified in Fig. 6 B), although the average length of tubules in AP-3δWT/Pldnmut1-expressing cells (0.78 ± 0.01 µm) was slightly lower than in AP-3δWT/PldnWT cells (0.89 ± 0.01 µm; Fig. 6 C). As in BLOC-1–deficient melanocytes (Delevoye et al., 2016), mCherry-STX13 in Ap3d1−/−/Pldn−/− cells labeled predominantly punctate structures and vacuoles but very few tubules (3.8 ± 0.3/25 µm2; Fig. 6, A and B) that were significantly shorter (0.62 ± 0.02 µm) than in AP-3δ/Pldn-expressing cells (Fig. 6 C). Importantly, we also observed significantly fewer (9.2 ± 0.4/25 µm2; more than in Ap3d1−/−/Pldn−/− melanocytes) and shorter (0.54 ± 0.01 µm) STX13 tubules in Ap3d1−/−/Pldn−/− melanocytes coexpressing SNARE binding mutants AP-3δmut1 and Pldnmut1 than in cells coexpressing WT AP-3δ and Pldnmut1 (Fig. 6, A–C) or AP-3δmut1 and WT Pldn (not shown). These data suggest that SNARE binding by either AP-3 or BLOC-1 is necessary for either optimal tubule formation or STX13 sorting into the tubules.
Figure 6.
Binding of either AP-3δ to VAMP7 or Pldn to STX13 is required for STX13 sorting into BLOC-1–dependent tubules. Ap3d1−/−/Pldn−/− melanocytes that were either untransduced (-) or stably transduced to express the indicated WT (black) or SNARE binding mutant (magenta) variants of AP-3δ and Pldn were transiently transfected with mCherry-STX13 and analyzed by live-cell spinning disk microscopy for tubule formation. (A) Top panels: Individual frames from representative image streams. Bottom panels: Binary skeletonized images of the top panels. Boxed regions are magnified 6.5-fold in the insets. Arrowheads, STX13-positive tubules. Scale bar, 5 µm; inset scale bar, 1 µm. (B and C) STX13-positive tubules in ten 25-µm2 regions from skeletonized images of each of at least five analyzed cells per condition in each of three separate experiments were analyzed. (B) Tubule number per 25-µm2 region was quantified. (C) Length was quantified for 1,268 tubules in AP-3δWT/PldnWT-expressing cells, 1,247 tubules in AP-3δWT/Pldnmut-expressing cells, 1,190 tubules in AP-3δmut1/Pldnmut1-expressing cells, and 303 tubules in untransduced cells. Data in B and C are represented by Tukey box-and-whisker plots (10th–90th percentiles); mean is denoted as +. ***, P < 0.001.
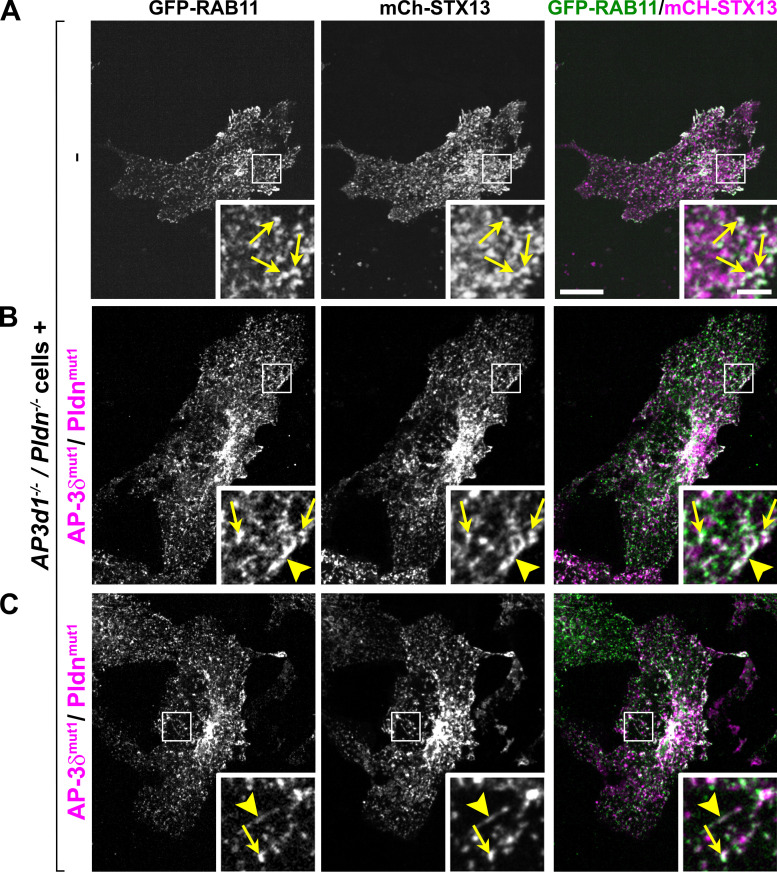
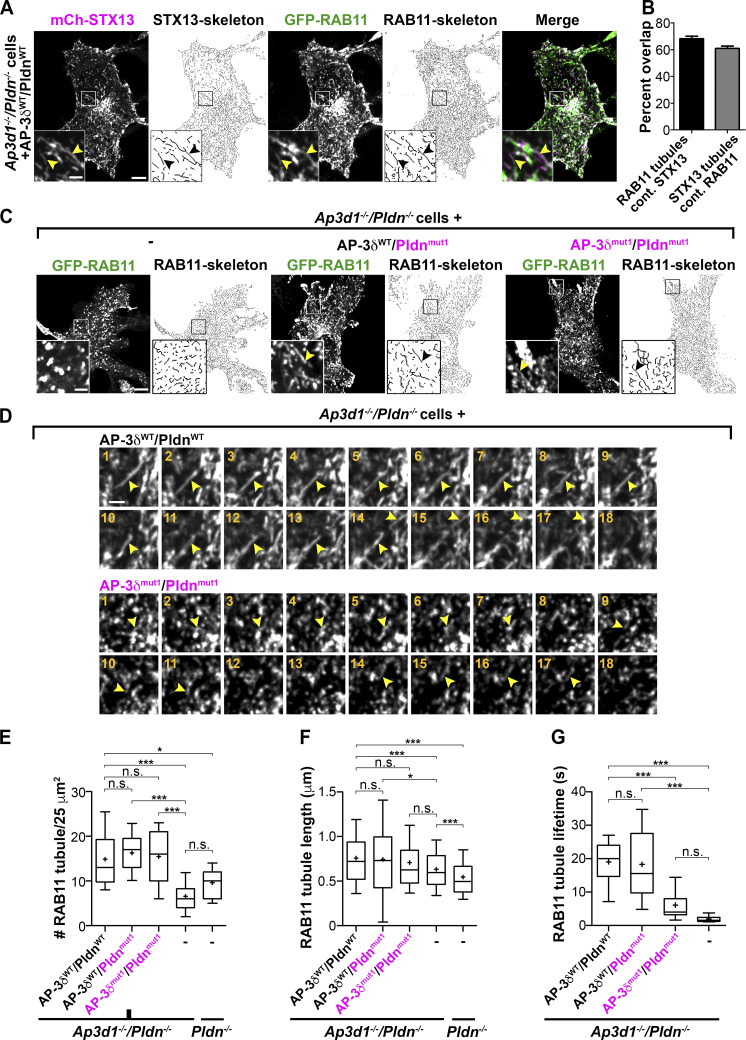
To distinguish between these possibilities, we labeled the BLOC-1–dependent tubules with GFP-RAB11. Consistent with previous data (Delevoye et al., 2014; Dennis et al., 2015), GFP-RAB11 overlapped well with mCherry-STX13–labeled tubules and/or vesicular structures when coexpressed in Ap3d1−/−/Pldn−/− melanocytes that were untransduced (Fig. S4 A) or reconstituted with either AP-3δWT and PldnWT (Fig. 7 A, arrowheads; quantified in Fig. 7 B; see also Video 1) or AP-3δmut1 and Pldnmut1 (Fig. S4, B and C), validating GFP-RAB11 as a marker of recycling endosomes and melanosome-bound tubules. Analyses in Ap3d1−/−/Pldn−/− cells expressing GFP-RAB11 confirmed that the tubules in these cells, as in Pldn−/− melan-pa cells (Delevoye et al., 2016), were reduced in number, length, and lifetime relative to cells reconstituted with WT forms of AP-3δ and Pldn (Fig. 7 C, left two panels; quantified in Fig. 7, E–G). By contrast, both the number and length of GFP-RAB11–labeled tubules were similar in Ap3d1−/−/Pldn−/− cells expressing either AP-3δWT and PldnWT, AP-3δWT and Pldnmut1, or AP-3δmut1 and Pldnmut1 (Fig. 7 C [middle and right panels] and Fig. 7, E and F). Taken together with Fig. 6, we conclude that SNARE binding by AP-3 and BLOC-1 is not required for the formation of endosomal tubules, but is required for sorting STX13 and VAMP7 into them.
Figure S4.
GFP-RAB11 and mCherry-STX13 largely overlap in Ap3d1−/−/Pldn−/− cells or cells expressing SNARE binding mutants of AP-3 and BLOC-1. Ap3d1−/−/Pldn−/− cells that were untransduced (-) or reconstituted with AP-3δmut1 and Pldnmut1 were transiently transfected to express GFP-RAB11 and mCherry-STX13 and analyzed by live-cell spinning disk microscopy. (A) Most endosomal structures in Ap3d1−/−/Pldn−/− cells contain both GFP-RAB11 and mCherry-STX13; most structures were punctate (arrows), and very few endosomal tubules were observed. (B and C) Two examples of GFP-RAB11 and mCherry-STX13 localization in Ap3d1−/−/Pldn−/− cells expressing AP-3δmut1 and Pldnmut1. GFP-RAB11 and mCherry-STX13 colocalize to a subpopulation of endosomal structures, including both punctate structures (arrows) and long tubules (arrowheads). Scale bar, 5 µm. Insets are magnified 3.5-fold. Inset scale bar, 1 µm.
Figure 7.
BLOC-1 function in the formation of endosomal tubules does not require SNARE binding by AP-3 or BLOC-1. (A and B) Ap3d1−/−/Pldn−/− melanocytes expressing AP-3δWT/PldnWT and transiently transfected with mCherry-STX13 and GFP-RAB11 were analyzed by live-cell spinning disk microscopy. (A) Each channel from a frame of a representative image is shown alongside skeletonized tubules; a merged nonskeletonized image is shown at right. Boxed regions are magnified fourfold in the insets. Black or yellow arrowheads, tubules positive for both GFP-Rab11 and mCherry-STX13. Scale bar, 5 µm; inset scale bar, 1 µm. (B) GFP-RAB11–positive tubules per 25-µm2 region (10 per cell) were manually counted from skeletonized images of 40 cells and assessed for mCherry-STX13 overlap. mCherry-STX13–positive tubules were similarly quantified and assessed for GFP-RAB11 overlap. Bars represent the percentage (mean ± SEM) of overlapped tubule number relative to total tubule number. (C–G) Pldn−/− melanocytes or Ap3d1−/−/Pldn−/− melanocytes that were untransduced (−) or transduced to express AP-3δWT/PldnWT, AP-3δWT/Pldnmut1, or AP-3δmut1/Pldnmut1 were transiently transfected with GFP-RAB11 and analyzed by live-cell spinning disk microscopy. (C) A representative frame from an image sequence of the indicated cell type is shown beside the corresponding skeletonized image. Boxed regions are magnified fourfold in the insets. Arrowheads, GFP-RAB11–labeled tubules. Scale bar, 5 µm; inset scale bar, 1 µm. (D) Montages from image streams of Ap3d1−/−/Pldn−/− melanocytes expressing AP-3δWT/PldnWT (top) or AP-3δmut1/Pldnmut1 (bottom) exemplifying tubule lifetimes. Arrowheads, tubules followed through multiple frames. Scale bar, 1 µm. (E–G) GFP-RAB11–labeled tubules in indicated cell samples were quantified as in A for tubule number/25 µm2 (E) and length (F) and also for lifetime (the length of time a given tubule could be detected within the image stream; G). Results are displayed as Tukey box-and-whisker plots (10th–90th percentiles); mean is denoted by +. At least five cells were analyzed for each condition. The numbers of tubules quantified for length and lifetime were 278/37 for AP-3δWT/PldnWT-expressing cells, 326/18 for AP-3δWT/Pldnmut1-expressing cells, 952/31 for AP-3δmut1/Pldnmut1-expressing cells, 322/15 for untransduced Ap3d1−/−/Pldn−/− cells, and 379 (tubule length) for Pldn−/− cells. *, P < 0.05; ***, P < 0.001; n.s, not significant.
Video 1.
GFP-RAB11 labels the majority of mCherry-STX13–positive tubules (related to Fig. 7). Ap3d1−/−/Pldn−/− melanocytes stably expressing AP-3δWT and PldnWT were transiently transfected with mCherry-STX13 and GFP-RAB11 and visualized 48 h later by live-cell spinning disk confocal microscopy in a humidified chamber at 10% CO2 and 37°C. Images were acquired every 0.8 s for 2 min. Grayscale images of GFP-RAB11 and mCherry-STX13 are shown in the left and middle panels of the movie, respectively. The fluorescent channels are merged in the right panel (GFP-RAB11, green; mCherry-STX13, red).
While loss of SNARE binding did not impact the number or length of GFP-RAB11–containing tubules, kinetic analyses of image streams showed that tubule lifetimes were significantly decreased in Ap3d1−/−/Pldn−/− melanocytes expressing AP-3δmut1 and Pldnmut1 relative to cells expressing a WT form of either protein (Fig. 7, D and G). Tubules in cells expressing AP-3δWT and either PldnWT or Pldnmut1 last 19.0 ± 6.9 and 18.3 ± 12.0 s, respectively, with the longest tubules persisting for >30 s (Fig. 7, D and G; and Video 2). By contrast, tubules in cells expressing AP-3δmut1 and Pldnmut1 lasted 6.1 ± 0.9 s (Fig. 7, D and G; and Video 3). These data indicate that incorporation of STX13 and VAMP7 influences tubule lifetime, perhaps reflecting tubule stabilization by SNARE-dependent interactions with the target melanosome membrane.
Video 2.
Tubule formation and duration in Ap3d1−/−/Pldn−/− melanocytes expressing AP-3δWT and PldnWT (related to Fig. 7). Ap3d1−/−/Pldn−/− melanocytes stably expressing AP-3δWT and PldnWT were transiently transfected with GFP-RAB11 and imaged 48 h later by live-cell spinning disk confocal fluorescence microscopy in a humidified chamber at 10% CO2 and 37°C. Images were acquired for 2 min at 1 frame/s. The yellow arrow indicates a GFP-RAB11–positive tubule that persists for at least 15 s.
Video 3.
Tubule formation and duration in Ap3d1−/−/Pldn−/− melanocytes stably expressing AP-3δmut1 and Pldnmut1 (related to Fig. 7). Ap3d1−/−/Pldn−/− melanocytes stably expressing AP-3δmut1/Pldnmut1 were transiently transfected with GFP-RAB11 and imaged 48 h later by live-cell spinning disk confocal fluorescence microscopy in a humidified chamber at 10% CO2 and 37°C. Images were acquired for 2 min at 1 frame/s. The yellow arrow indicates a GFP-RAB11–positive tubule that persists for 5 s.
Discussion
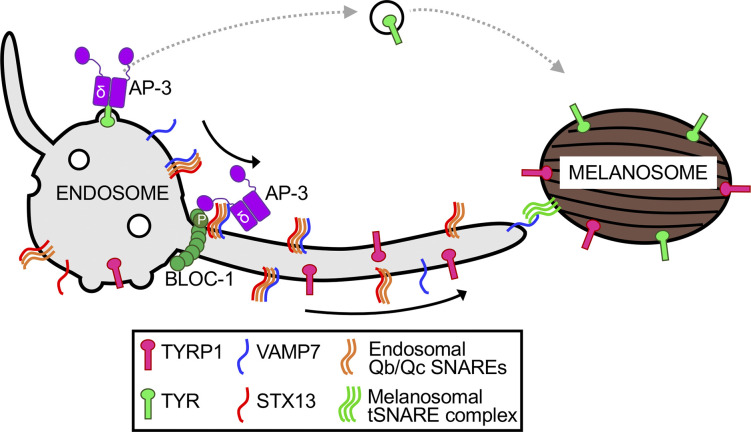
Melanosome maturation in melanocytes requires cargo delivery from endosomes to maturing melanosomes through tubular membrane carriers, which require BLOC-1 for their formation (Delevoye et al., 2016; Delevoye et al., 2009; Dennis et al., 2015; Setty et al., 2007) and VAMP7 for their fusion with melanosome membranes (Dennis et al., 2016). Our data indicate that VAMP7 is sorted into the tubules in a complex with STX13 using a redundant mechanism that requires recognition either of VAMP7 by AP-3 or of STX13 by BLOC-1 (Fig. 8). We show that mutagenesis of either a region of the BLOC-1 subunit Pldn required for STX13 binding or of the VAMP7-binding site on AP-3δ alone does not impact melanosome biogenesis or cargo sorting in melanocytes. However, loss of SNARE binding by both Pldn and AP-3δ blocks VAMP7 localization, melanosome cargo sorting, pigmentation, STX13 entry into the tubules, and tubule stability, but not BLOC-1 function in tubule formation from endosomes. Our data thus support a model in which a preformed STX13–VAMP7 complex on endosomes, likely as part of a cis-SNARE four-helix bundle, is recognized by either component of an AP-3–BLOC-1 super-complex (Di Pietro et al., 2006; Salazar et al., 2006) for entry into the endosome-derived tubules and thus for their ultimate fusion with melanosomes (Fig. 8). This model may have broader implications for SNARE sorting within the endomembrane system.
Figure 8.
AP-3 and BLOC-1 coordinate sorting of VAMP7–STX13 cis-SNARE complexes into melanosome-bound tubule carriers. Model showing VAMP7 sorting from vacuolar early endosomes into tubules as a cis-SNARE complex, with STX13 and unknown Qb and Qc SNARE helices, into tubules by virtue of VAMP7 engagement by AP-3δ (δ) and/or STX13 engagement by Pldn (P) within an AP-3–BLOC-1 super-complex. AP-3 separately sorts TYR from other endosomal domains into melanosome-bound clathrin-coated vesicles. Tubule fusion is mediated by VAMP7 coupling with unknown Qabc SNARE helices on the melanosome membrane after disruption of the VAMP7–STX13 complex. tSNARE, target SNARE.
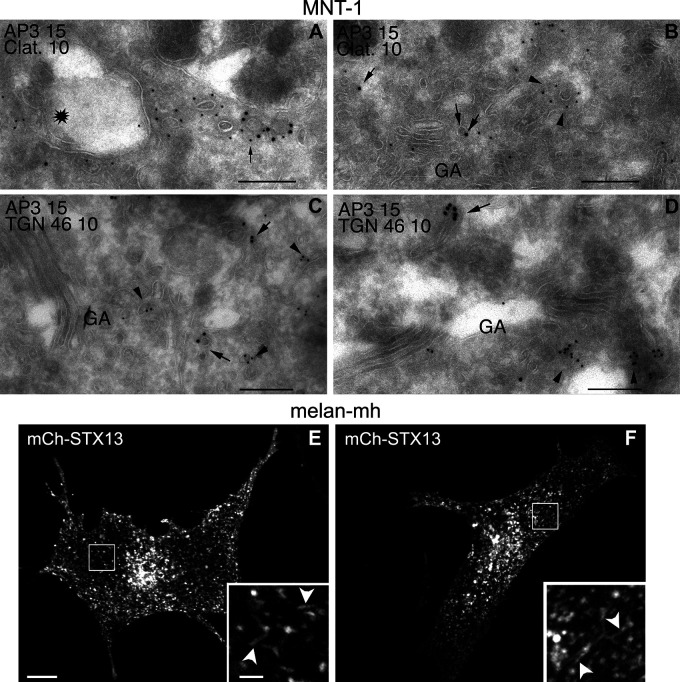
AP-3 sorts TYR into BLOC-1–independent, melanosome-bound vesicles, and AP-3–deficient melanocytes from HPS2 patients and pearl mice are hypopigmented and missort TYR toward endolysosomes (Huizing et al., 2001; Setty et al., 2007; Theos et al., 2005). Although AP-3 does not directly engage the TYRP1 melanosome sorting signal (Theos et al., 2005), our quantitative imaging data reveal a requirement for AP-3 in efficient BLOC-1–dependent trafficking of TYRP1 to melanosomes. In melanocytes, this role is likely mediated by a cohort of AP-3 that physically associates with BLOC-1 (Di Pietro et al., 2006; Salazar et al., 2006), localizes to BLOC-1–associated endosomal tubules (Di Pietro et al., 2006), and is distinct from the cohort (41–46%) associated with clathrin-coated vesicles near peripheral endosomes or melanosomes (Peden et al., 2004; Theos et al., 2005). Indeed, a cohort of AP-3 in melanocytes localizes to tubulovesicular structures that lack a clathrin coat and the vesicular AP-3–dependent cargo TYR (Theos et al., 2005) and that are situated near, but not in, the TGN (Fig. S5, A–D). We speculate that this might reflect the BLOC-1–associated AP-3 pool emerging from endosomes. Distinct and cell type–specific functions for the two AP-3 pools might explain the codependence of AP-3 on BLOC-1 but not clathrin for endosomal sorting in neurons (Newell-Litwa et al., 2009; Zlatic et al., 2013) despite the prominent role of the clathrin-associated pool in sorting lysosomal and melanosomal cargoes (Peden et al., 2004; Theos et al., 2005). Besides VAMP7 sorting, AP-3 must play additional roles in BLOC-1–dependent trafficking, given that TYRP1 distribution in AP-3δ–deficient melanocytes is normalized by expressing an AP-3δ subunit that cannot engage VAMP7. These roles may be indirect via effects on the composition of the early endosomal system (Theos et al., 2005), since a qualitative deficiency in STX13 tubule formation in AP-3–deficient melanocytes is not detected under our culture conditions (Fig. S5, E and F).
Figure S5.
Presence of non–clathrin-associated AP-3 on tubulovesicular structures near, but not in, the TGN in WT melanocytic cells and of STX13-labeled tubules in Ap3d1−/− cells. (A–D) Ultrathin cryosections of the human melanoma cell line, MNT-1, were fixed and immunogold labeled with the anti–AP-3δ antibody SA4 followed by protein A conjugated to 15-nm gold particles (AP3 15) and then antibodies to either the clathrin heavy chain (Clat.; A and B) or the TGN marker TGN46 (C and D) followed by protein A conjugated to 10-nm gold particles (10). Sections were then processed and analyzed by electron microscopy. (A) A section near a vacuolar endosome (star) emphasizing clathrin-coated vesicles, some of which also label for AP-3 (arrow). (B–D) Sections near the Golgi apparatus (GA). (B) Separate tubulovesicular structures near the Golgi are labeled for either clathrin (arrowheads) or AP-3 (arrows), but not both. (C and D) Tubulovesicular structures labeled for AP-3 (arrows) are distinct from TGN structures labeled for TGN46 (arrowheads). (E and F) Ap3d1−/− melan-mh cells transiently transfected to express mCherry-labeled STX13 (mCh-STX13) were analyzed 48 h after transfection by live-cell spinning disk microscopy. Shown are individual frames from two representative cells documenting the presence of long STX13-containing tubules (arrowheads), similar to those observed in WT cells. Boxed regions are magnified fourfold in the insets. Scale bar, 10 µm; inset scale bar, 2 µm.
While STX13 was known to bind BLOC-1 via the Pldn subunit (Ghiani et al., 2010; Huang et al., 1999; Moriyama and Bonifacino, 2002), here we document a function for this interaction. Mutations within a 21-aa region at the beginning of a coiled-coil domain of Pldn impair binding to STX13 but not to another Pldn molecule; we speculate that this region engages an exposed area of the STX13 SNARE domain (Ghiani et al., 2010) in a SNARE complex via a coiled-coil interface. When expressed in Pldn-deficient cells, non–STX13-binding Pldn variants with mutations in this domain were as effective as WT Pldn in supporting melanogenesis and BLOC-1–dependent cargo transport. This and biochemical data indicated that the STX13 binding site is not required for Pldn assembly into BLOC-1 or for BLOC-1 function in an otherwise intact melanocyte. The STX13–Pldn interaction was only functionally required for melanosome maturation upon concomitant disruption of the AP-3δ–VAMP7 interaction. Thus, either the STX13–Pldn or the AP-3δ–VAMP7 interaction, but not both, is necessary for STX13 entry into BLOC-1–dependent tubules, BLOC-1–dependent delivery of VAMP7 and cargo to melanosomes, and consequent melanogenesis.
The simplest interpretation of these data is that AP-3 and BLOC-1 function redundantly in sorting a STX13–VAMP7 complex—we speculate as part of a four-helix cis-SNARE complex—into BLOC-1–dependent tubular transport intermediates (Fig. 8). This model is consistent with observations that (1) AP-3, like Hrb at the plasma membrane (Pryor et al., 2008), engages the VAMP7 longin domain only when VAMP7 is in a SNARE complex (Kent et al., 2012) and (2) the autoinhibitory Habc domain of STX13 impairs interaction of the SNARE domain with BLOC-1 (Ghiani et al., 2010). VAMP7 might be specifically permissive for the Pldn–STX13 interaction, since BLOC-1 did not bind to a SNARE complex containing STX13 and VAMP2 (Ghiani et al., 2010). BLOC-1 might also engage the Qbc SNAREs SNAP-25 (Ilardi et al., 1999; Vites et al., 2004) and SNAP-47 (Ghiani et al., 2010). However, since STX13-binding mutants of Pldn cannot support BLOC-1–dependent trafficking in cells expressing VAMP7-binding mutants of AP-3δ, either Qbc SNARE binding is not sufficient to support STX13/VAMP7 sorting in melanocytes or the STX13–VAMP7 cis-SNARE complex that is sorted by BLOC-1/AP-3 incorporates distinct Qb and Qc helices. An alternative model to explain our data is that the Pldn coiled-coil domain engages a STX13–VAMP7 dimer by replacing one or both of the Qbc SNARE helices, analogous to the clamping mechanism of complexin in blocking trans-SNARE assembly in neurons (Giraudo et al., 2006; Kümmel et al., 2011).
The sorting mechanism for STX13–VAMP7 complexes described here poses an intriguing paradigm for SNARE sorting within the endosomal system. VAMP7 is the predominant R-SNARE for fusing BLOC-1–dependent cargo transport intermediates with melanosomes (Dennis et al., 2016; Jani et al., 2015; Tamura et al., 2011), but it also participates in other endolysosomal fusions (Fader et al., 2009; Martinez-Arca et al., 2003; Moreau et al., 2011; Pols et al., 2013; Pryor et al., 2004; Rao et al., 2004; Wade et al., 2001). We propose that BLOC-1–AP-3–dependent SNARE complex sorting permits VAMP7 entry into newly forming tubular transport carriers while protecting VAMP7 from engaging in nonproductive SNARE complexes during transport. This could provide spatial regulation of VAMP7 sorting and fusion complex formation and could extend to other SNAREs that are distributed among multiple organelles. Indeed, VAMP7 within a SNARE complex at the plasma membrane is sorted into clathrin-coated vesicles by Hrb (Pryor et al., 2008); Vti1b within a SNARE complex is sorted by epsinR (Miller et al., 2007); and STX6-containing vesicles engage distinct tethers on different compartments, depending on whether they also contain STX13 (Koike and Jahn, 2019). We thus speculate that SNARE complex sorting is a general mode of specifying directionality in SNARE complex trafficking. To our knowledge, however, this is the first example in which dual recognition of two components of a SNARE complex function redundantly in sorting.
Our live-cell imaging data using tagged RAB11 showed that SNARE binding by Pldn or AP-3δ was not required for BLOC-1–dependent tubule formation but impacted tubule lifetime. Thus, cargo carrier formation does not require incorporation of the SNAREs that will ultimately mediate carrier fusion with its target, as also observed during COPII-coated vesicle formation (Campbell and Schekman, 1997). The data further suggest that SNARE interactions with the target membrane regulate the stability of BLOC-1–dependent tubules. VAMP7 could dock the tubule ends to melanosomes via trans-SNARE complex formation, SNARE–tether interactions, or both (Gillingham and Munro, 2019; Ungermann and Kümmel, 2019); distinguishing these will require identifying the Q-SNAREs and tether on the melanosome membrane. We propose that in the absence of such VAMP7-dependent interactions, the tubules retract back to the endosome from which they were derived, as appears to occur physiologically upon completion of cargo transfer (Dennis et al., 2015). Moreover, the reduction in tubule lifetime but not steady-state tubule number upon loss of AP-3 and BLOC-1 SNARE binding suggests that more tubules may be generated per unit of time in cells expressing both SNARE binding mutants. This could reflect either a feedback mechanism resulting from the loss of cargo delivery or a faster rate of BLOC-1 cycling to more frequently initiate tubule formation from endosomal vacuoles.
Our results raise questions about the fate of the VAMP7–STX13 complexes within BLOC-1–dependent tubules. STX13 is present throughout the tubules but, unlike VAMP7, is not transferred to melanosomes upon fusion (Dennis et al., 2016; Dennis et al., 2015; STX13 does not accumulate on melanosomes of Ap3d1−/− cells under our conditions as suggested by Jani et al., 2015; Fig. S2). This implies that VAMP7-dependent fusion with melanosomes involves a distinct Qa-SNARE on the melanosome membrane. For VAMP7 to interact with a melanosomal Q-SNARE complex, the VAMP7–STX13 complex must be disassembled before tubule engagement with melanosomes. Although how such disassembly is mediated is not known, it is intriguing that the SNARE recycling factor NSF was identified as a BLOC-1 binding partner (Gokhale et al., 2015). It is possible that timed interactions of BLOC-1, first with AP-3 and subsequently with NSF, might support sorting of the SNARE complexes into the tubules followed by their ultimate dissolution. Defining the SNARE partners both on the endosomal tubules and at the melanosome will allow us to address these questions.
Materials and methods
Reagents
Unless otherwise noted, chemicals were from Sigma-Aldrich, and tissue culture reagents were from Invitrogen. Hygromycin B was from Roche, puromycin was from Takara Bio, and Matrigel was from Becton Dickinson.
Design of Bloc1s6-targeting gRNAs
To generate melanocytes lacking both AP-3δ and Pldn, the Bloc1s6 gene encoding Pldn was targeted by CRISPR/Cas9 mutagenesis in AP-3δ–deficient melan-mh melanocytes (see Cell culture and generation of stable cell lines section below). Two gRNAs targeting exon 1 and intron 1 (Table S1) of Bloc1s6 were designed using CHOPCHOP (https://chopchop.cbu.uib.no/) to delete a 72-bp region spanning the intron–exon border (Fig. S4), as described previously for targeting Bloc1s6 in a mouse alveolar type 2 cell line (Kook et al., 2018). As detailed below, duplex oligonucleotides corresponding to each gRNA bounded by a BsmBI site were uniquely inserted into a lentiviral vector, and melan-mh cells were simultaneously infected with both lentiviruses.
DNA constructs
Complementary oligonucleotides encoding either gRNA 1 or gRNA 2 and bounded with BsmBI sites were phosphorylated using T4 PNK (New England Biolabs) and then annealed by incubation at 37°C for 30 min, 95°C for 5 min, and a ramp-down to 25°C at 5°C per minute. To generate the CRISPR/Cas9 lentiviral gene targeting vectors, pLentiCRISPRv2 (Addgene plasmid 52961; http://n2t.net/addgene:52961; Research Resource Identifier [RRID], Addgene_52961; Sanjana et al., 2014), a gift from Feng Zhang (Massachusetts Institute of Technology, Cambridge, MA) and purchased from Addgene, was digested with BsmBI (New England Biolabs) and then ligated with either of the annealed oligonucleotide duplexes. Proper insertion of gRNAs was confirmed by Sanger sequencing (Table S1). The retroviral vectors pBMN-I-Hygro-myc-Pldn, pBMN-I-Hygro-myc-Muted, and pBMN-I-Hygro-myc-STX13 encoding myc-tagged human Pldn, muted (BLOC1S5), or STX12/13 and hygromycin resistance from an internal ribosome entry site (Dennis et al., 2016; Jani et al., 2015; Setty et al., 2007); the yeast two-hybrid vector pGAD424-myc-Pldn constructs encoding full-length myc-tagged human Pldn fused to the Gal4 activation domain (Moriyama and Bonifacino, 2002); and the yeast two-hybrid vector pGBT9 constructs encoding the complete N-terminal cytosolic domain (aa 1–249) of human STX12/13 or full-length myc-tagged human Pldn fused to the Gal4 DNA binding domain (Moriyama and Bonifacino, 2002) have been described. All yeast two-hybrid vectors were a generous gift from J.S. Bonifacino (National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD). Myc-Pldn triple alanine-scanning mutants were generated by site-directed mutagenesis using two-step amplification as described (Letourneur and Klausner, 1992) using primers listed in Table S2 and cloned into the EcoRI-SalI restriction sites of pGAD424. Selected myc-Pldn constructs were subcloned from the pGAD424 plasmids into the BamHI and XhoI sites of pBMN-I-Hygro. pEGFP-C1-VAMP8 encoding GFP-tagged human VAMP8 was a generous gift of Paul Roche (National Cancer Institute, Bethesda, MD). pEGFP-C1-SEC22B encoding GFP-tagged mouse SEC22B was a generous gift from Thierry Galli (Institute of Psychiatry and Neuroscience, Paris, France; Addgene plasmid 101918; http://n2t.net/addgene:101918; RRID, Addgene_101918; Petkovic et al., 2014). The pBMN-I-Hygro plasmids encoding untagged WT, I702S/V704S (mut1), and L709S/L713S (mut2) variants of chimpanzee AP3D1 (Peden et al., 2004), human HPS3 (Dennis et al., 2015), pmCherry-C1-STX13 encoding mCherry-tagged human STX12/13 (Dennis et al., 2015), pEGFP-C1-RAB11 encoding GFP-tagged human RAB11A (Delevoye et al., 2014), and pEGFP-C1-VAMP7 encoding GFP-tagged rat VAMP7/TiVAMP (Dennis et al., 2016) have been described. In the text and figures, EGFP is shortened to GFP. Plasmids encoding vesicular stomatitis virus G (pMD2.G; Addgene plasmid 12259; http://n2t.net/addgene:12259; RRID, Addgene_12259) and HIV-1 structural components (psPAX2; Addgene plasmid 12260; http://n2t.net/addgene:12260; RRID, Addgene_12260) for lentiviral production were generous gifts from Didier Trono (Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) and were purchased from Addgene. All recombinant plasmids were verified by automated sequencing by the University of Pennsylvania Cell Center or the NAPCore Facility at the Children’s Hospital of Philadelphia Research Institute.
Cell culture and generation of stable cell lines
Immortalized melanocyte cell lines melan-Ink4a derived from C57BL/6J-Ink4a-Arf−/− (Cdkn2a-null) mice (Sviderskaya et al., 2002) and melan-pa1 (referred to here as “melan-pa”) derived from Pldn/BLOC-1–deficient C57BL/6J-Pldnpa/pa pallid mice (Setty et al., 2007) have been described. melan-mh1 (described here as “melan-mh”) and melan-mh2 melanocytes were derived by spontaneous immortalization from the skins of newborn AP-3δ–deficient C57BL/6J-Ap3d1mh/mh mocha mice as described for melan-a (Bennett et al., 1987). All melanocyte cell lines were cultured in RPMI 1640 medium (Invitrogen or Corning) supplemented with 200 nM 12-O-tetradecanoylphorbol-13-acetate and either 10% (melan-pa, melan-Ink4a) or 20% (melan-mh and derived sublines) FBS (Atlanta Biologicals).
Stably modified cell lines and clones lacking both AP-3δ and Pldn (Ap3d1−/−/Pldn−/− cells) were generated by infecting AP-3δ–deficient melan-mh with a 1:1 mixture of recombinant lentiviruses encoding Cas9 and either gRNA 1 or gRNA 2 to target the Pldn gene, followed 2 d later by selection in medium containing 2 µg/ml puromycin. Stable lines and clones of melan-pa expressing myc-tagged Pldn variants, melan-mh expressing AP-3δ variants, or HPS3 as a control, or Ap3d1−/−/Pldn−/− cells expressing both Pldn and AP-3δ variants, were generated by infection with recombinant retroviruses encoding the indicated chains (Ap3d1−/−/Pldn−/− cells were coinfected with a 1:1 mixture of supernatants containing retroviruses expressing either Pldn or AP-3δ) followed 2 d later by selection with medium containing 150 µg/ml Hygromycin B. To obtain monoclonal populations, cells were diluted into a 15-cm dish at 5 cells/ml 1 wk after selection, and colonies were isolated using cloning cylinders. Most experiments were repeated using two clones; only results from one clone are shown in this paper. Stable lines of melan-pa and melan-Ink4a expressing myc-tagged STX13 were generated similarly using recombinant retroviruses and selection with Hygromycin B, and were each used as polyclonal cell populations.
Recombinant lentiviruses were generated following transient transfection of ∼80% confluent HEK 293T cells in a 35-mm well with 2.5 µg pLentiCRISPRv2 encoding gRNA 1 or 2, 0.8 µg pMD2.G, and 1.8 µg psPAX2 with 8 µl Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. Retroviruses were generated following transient transfection of ∼70% confluent Platinum-E cells in 35-mm wells with 4 µg of pBMN plasmid and 8 µl Lipofectamine 2000 (Morita et al., 2000). For both lentivirus and retrovirus preparations, the medium for the transfected cells was replaced with melanocyte culture medium 24 h after transfection, and viral supernatants were harvested 48–60 h later, sterile filtered, and added directly to melanocytes seeded the previous day at 300,000 cells per 35-mm well. For transient transfection with pEGFP-C1-VAMP7, melanocytes were seeded at 5 × 104 on Matrigel-coated 12-mm coverslips in a 24-well plate and transfected 24 h later with Lipofectamine 3000 using 500 ng DNA according to the manufacturer’s instructions. For transient transfections with pEGFP-C1-RAB11 and/or pmCherry-STX13, melanocytes were seeded at 8 × 105 in 35-mm wells and transfected 24 h later with 8 µl Lipofectamine 2000 and 2 µg of plasmid DNA; carrier DNA (pCI plasmid; Clontech) was added as needed to maintain proper DNA-to-Lipofectamine reagent ratios. Cells were transferred 24 h later onto Matrigel-coated 35-mm glass-bottomed dishes (VWR Scientific), and cells with low levels of plasmid expression were imaged 24 h (GFP-VAMP7) or 48 h (mCherry-STX13, GFP-RAB11) after transfection.
Antibodies
mAbs and their sources included mouse anti-TYRP1 (TA99, also known as Mel-5; American Type Culture Collection) and mouse anti-Myc for dIFM (9E10; American Type Culture Collection), mouse anti-Pldn (clone 2G6; a generous gift from Esteban Dell’Angelica, University of California, Los Angeles, CA; Ghiani et al., 2010), mouse anti–AP-3δ (clone SA4; Developmental Studies Hybridoma Bank), rabbit anti-AP3M1 (ab201227; Abcam), mouse anti–γ-tubulin (GTU88; Sigma-Aldrich), mouse anti-VAMP7 (clone 158.2; Synaptic Systems), mouse anti-GFP (NB600-597, Novus Biologicals; or clones 7.1 and 13.1, Roche), and mouse anti-clathrin heavy chain (clone 23; BD Biosciences). Polyclonal antibodies included rabbit anti-STX13 (Prekeris et al., 1998), as previously described (Setty et al., 2007); rabbit anti-VAMP7 TG50, a kind gift from Thierry Galli (Verraes et al., 2018); rabbit αPep7h-msm to the cytoplasmic domain of TYR (Berson et al., 2000); rabbit anti-dysbindin (Starcevic and Dell’Angelica, 2004), a kind gift from Esteban Dell’Angelica; and sheep anti-TGN46 (Serotec). Species-specific secondary antibodies from donkey and conjugated to Alexa Fluor 488, Cy3, Alexa Fluor 594, and Alexa Fluor 647 were used for dIFM or to IRDye-790CW, IRDye-680LT, or HRP for immunoblotting (Jackson ImmunoResearch Laboratories, Inc.).
Yeast two-hybrid assay
The assay was performed essentially as described (Moriyama and Bonifacino, 2002). Briefly, Saccharomyces cerevisiae strain HF7c (Clontech) was cotransformed by the lithium acetate procedure with pGAD424- and pGBT9-based vectors. Double transformants were selected on dropout agar plates lacking Leu and Trp (+His plates). After 4 d, pooled colonies from each transformation were resuspended in water; suspensions were normalized to equivalent ODs (0.1 OD units at 600 nm); and 5 µl were transferred onto +His plates and plates lacking Leu, Trp, and His (−His plates). After 3–4 d, plates were scanned on a flatbed scanner for colony growth analysis. Growth of HF7c double transformants on −His plates indicated an interaction between the Gal4 binding domain and Gal4 activation domain fusion proteins. Each myc-Pldn construct in pGAD424 was tested at least twice against both WT myc-Pldn and STX12/13.
PCR analysis of genomic DNA
Genomic DNA was isolated from cell lines or clones using the DNeasy genomic DNA extraction kit (Qiagen) according to the manufacturer’s instructions. A region of Bloc1s6 encompassing the gRNA target sites and spanning the exon 1–intron 1 border was amplified from genomic DNA to analyze genomic editing (primers in Table S1) using the GoTaq Master Mix system (Promega). PCR products were analyzed on a 1.5% agarose gel, and staining was performed with SYBR Safe DNA Gel Stain (Invitrogen).
RT-PCR analysis
RNA was isolated from melanocytes using the RNeasy Mini RNA extraction kit (Qiagen) according to the manufacturer’s instructions. RNA was then reverse transcribed into cDNA using the SuperScript II First-Strand Synthesis System for RT-PCR (Invitrogen) according to the manufacturer’s instructions using the included random hexamer primers. cDNAs corresponding to Bloc1s6 (encoding Pldn) and Tubb4b (encoding tubulin β-4b) were amplified using the GoTaq Master Mix system (Promega; primers listed in Table S1). Amplified cDNA products were analyzed on a 1.5% agarose gel, and staining was performed with SYBR Safe DNA Gel Stain (Invitrogen).
Immunoblotting
Melanocytes were seeded at 6 × 106 cells in 10-cm dishes and harvested 1–2 d later by rinsing cells once in cold PBS and incubating in PBS + 5 mM EDTA for 3 min. Cells were scraped off of dishes and collected by centrifugation, washed once in cold RPMI + 10% FBS, and washed twice in cold PBS. Cell pellets were resuspended in cold lysis buffer (50 mM Tris, 150 mM NaCl, 0.1% Triton X-100, and 10 mM EDTA, pH 7.2, and protease inhibitor cocktail; Roche) and incubated on ice for 20–30 min with manual inversion every 5 min. Cell lysates were centrifuged for 15–20 min at 13,000 rpm at 4°C, and supernatants were added to an equal volume of SDS-PAGE sample buffer (2× concentrated); boiled for 5 min; loaded onto 12% (BLOC-1 subunits), 15% (VAMP7, and STX13), or 10% (AP-3 subunits) polyacrylamide gels; and subjected to SDS-PAGE. Gels were transferred in transfer buffer (25 mM Tris, 192 mM glycine with 20% [for BLOC-1 subunits, VAMP7, and STX13] or 10% [for AP-3 subunits] methanol) to polyvinyl difluoride membranes (EMD Millipore) for 1 h at 100 V. Membranes were blocked with TBS (20 mM Tris, 140 mM NaCl, pH 7.6) + 5% powdered milk (Lab Scientific BioKEMIX, Inc.) for 1 h, and membranes were incubated in primary antibody and diluted in TBS + 5% milk overnight at 4°C. Membranes were washed once with TBST (TBS + 0.1% Tween 20) and once with TBS and then incubated for 30 min at RT with secondary antibodies conjugated to IRDye-790CW or IRDye-680LT diluted in TBS + 5% milk. Membranes were washed once or twice with TBST and twice with TBS. Membranes were analyzed using a LI-COR Odyssey CLx imager, and images were further processed using ImageJ (National Institutes of Health).
Immunoprecipitation/immunoblotting
BLOC-1–deficient melan-pa and either WT melan-Ink4a or WT melan-pa:HA-Pldn cells, in some cases stably expressing myc-STX13, were seeded at 2–2.5 × 106 cells in 10-cm dishes and transfected 24–48 h later with pEGFP plasmids encoding GFP-tagged VAMP7, VAMP8, or SEC22B using Lipofectamine 3000 and 12 μg of plasmid DNA. Cells were harvested the following day and solubilized in lysis buffer either as described above for immunoblotting or by adding 0.5–1.5 ml of lysis buffer directly to plates that had been washed twice with ice-cold PBS and scraping before incubation on ice and clarification. Samples of cell lysate were preserved, and the rest was added to GFP-Trap agarose or magnetic beads (Chromotek) that had been washed twice with cold lysis buffer and then incubated with rotation at 4°C for 30 min to 1 h; in some experiments, lysates were first precleared by incubation for 1–2 h with Protein A-agarose beads and centrifugation before incubation with GFP-Trap beads. Beads were washed either five times with lysis buffer or twice with 10 mM Tris-HCl/300 mM NaCl/0.1% Triton X-100, pH 8.0, twice with 10 mM Tris-HCl/150 mM NaCl/0.1% Triton X-100, pH 8.0, and once with water. Beads were then incubated with SDS-PAGE sample buffer and heated to 65°C for 10 min. Samples of lysates were brought to 2% SDS and maintained at RT for 5 min in the data shown in Fig. 3 A or at 65°C for 10 min in some experiments. Samples were fractionated by SDS-PAGE on 10% or 12% gels, transferred, blocked, and immunoblotted as above except that nitrocellulose- (Bio-Rad Laboratories) and HRP-tagged secondary antibodies were used (Jackson ImmunoResearch, Inc.), and blots were developed using Amersham ECL Prime or Pierce ECL (Thermo Fisher Scientific) and analyzed using an iBright CL1500 and analysis software (Invitrogen) or a LI-COR C-DiGit. For analysis of immunoprecipitates, the same blots probed for STX13 were washed with TBST and stripped using Restore Western Blot Stripping Buffer (Thermo Fisher Scientific) with incubation at 20°C or 37°C for 20 min, washed once with TBST, and then blocked and reprobed for GFP as above. Band intensities were quantified using either iBright or ImageStudioLite (LI-COR Biosciences) software by selecting a box surrounding the band(s) and subtracting the value of an equal-sized box in a blank area of the same lane. STX13 band intensity in immunoprecipitates for each sample was normalized to the intensities of STX13 in cell lysates and of GFP-tagged SNARE in the immunoprecipitates.
Quantification of melanin content in cell lysates
Melanin content was determined essentially as described (Delevoye et al., 2009). Melanocytes were seeded at 8 × 105 in 6-well plates and harvested 24–48 h later by trypsinization. Cells were counted, washed once with RPMI + 10% FBS and twice with ice-cold PBS, and resuspended in 50 mM Tris-HCl, pH 7.4, 2 mM EDTA, 150 mM NaCl, 1 mM DTT, and 1× cOmplete Protease Inhibitor Cocktail Tablets (Roche). Cells were probe sonicated on ice using the Sonic Dismembrator Model 100 (Fisher Scientific), and sonicates were fractionated into supernatant and pellet fractions by centrifugation at 20,000 × g for 15 min at 4°C. Protein concentrations in supernatants were estimated using the DC colorimetric protein assay (Bio-Rad Laboratories) with a standard curve from differing concentrations of BSA. Melanin-containing pellets were washed in 500 µl 50% ethanol, 50% ether, collected by centrifugation at 20,000 × g, and resuspended in 1 ml of 2 M NaOH, 20% DMSO. Melanin was solubilized by heating at 60°C for 40–60 min and quantified using spectrophotometry to measure OD at 492 nm. OD values were normalized to total protein content in each lysate. Duplicates or triplicates were analyzed for each experimental condition tested in each separate experiment.
Immunofluorescence microscopy and image deconvolution
Imaging was performed essentially as described (Dennis et al., 2016). Melanocytes were seeded onto Matrigel-coated coverslips 24–48 h before fixation. Cells were fixed with 4% PFA, PBS for 15 min, blocked and permeabilized in blocking buffer (PBS, 0.1% BSA, 0.02% saponin), and then labeled with primary antibodies diluted in blocking buffer for 1 h at RT. After a 15-min wash in PBS, cells were incubated with secondary antibodies diluted in blocking buffer for 30 min at RT. Samples were washed for 15 min in PBS, mounted with Prolong Gold Antifade Mountant (Invitrogen), and analyzed by epifluorescence microscopy on a DMI 6000B microscope (Leica Biosystems) equipped with a 63× plan apochromat (Plan Apo) objective (1.4 NA) and a Hamamatsu Photonics ORCA-Flash4.0 scientific complementary metal–oxide–semiconductor (sCMOS) digital camera; for Fig. S1, a 40× Plan Apo objective (1.2 NA) was used. Both fluorescence and brightfield images were captured. Images were acquired as a z-series with 0.19-µm steps and were deconvolved using the blind deconvolution algorithm of Microvolution software (BioVision Technologies) and ImageJ.
Live-cell fluorescence imaging
8 × 105 Melanocytes were seeded into 35-mm wells and 24 h later were transiently transfected with GFP- or mCherry-tagged fusion proteins. 24 h later, cells were reseeded onto Matrigel-coated 35-mm glass-bottomed dishes and imaged 24 h later in Leibovitz’s CO2-independent medium without phenol red (Invitrogen) supplemented with 10% FBS in an environmental chamber set to 37°C with 10% CO2. Confocal fluorescence images were acquired using a DMI8 inverted microscope (Leica Biosystems) equipped with a spinning disc system (VisiScope CSU-W1), a 100× total internal reflection fluorescence objective (1.47 NA), and a Hamamatsu Photonics ORCA-Flash 4.0 sCMOS digital camera. Z-series images covering six to eight 0.19-µm layers were captured every 0.8–1.2 s using VisiView software (Visitron Systems) for a minimum of 2 min and a maximum of 5 min. Images were further manipulated and analyzed using ImageJ and Adobe Photoshop. For Fig. S5, E and F, cells in glass-bottomed dishes were transfected and imaged as described (Dennis et al., 2016) using an Axiovert 200 microscope (Zeiss) equipped with a 63× Plan Apo objective lens (NA 1.4), an UltraVIEW ERS6 spinning-disk confocal scan head (PerkinElmer), an environmental chamber at 37°C, an ORCA-Flash4.0 sCMOS camera (Hamamatsu Photonics), and Volocity (PerkinElmer) software for image acquisition.
Image quantification
Colocalization analyses
The area of overlap between two fluorescently labeled proteins or between fluorescently labeled proteins and pigmented melanosomes was quantified using ImageJ on deconvolved fluorescence images essentially as described (Dennis et al., 2016). Single cells were cropped, and the fluorescence in the perinuclear region was removed. Prior to thresholding, local background was subtracted using a rolling ball radius of 10 pixels. Binary images were created by subtracting background using the subtract operation before applying the Bernsen auto local threshold algorithm in ImageJ. For pigmented melanosomes in the brightfield channel, the binary operation “fill holes” was used to create a more accurate estimate of overlap between TYRP1 (on the limiting membrane of melanosomes) and melanin (in the lumen of melanosomes). The ImageJ Image Calculator plugin was used to multiply two binary images together to create an image representing the area of overlap between the two fluorescent markers or between one fluorescent marker and melanin in the brightfield channel. Structures in the original and overlap binary images larger than 0.2 µm2 were counted using the Analyze Particles plugin, and the ratio of overlap pixels to total fluorescent pixels of the imaging channel of interest was used to generate the percentage overlap between two channels. The results represent values obtained from at least 10 cells from at least 3 individual experiments.
Perinuclear TYRP1 fluorescence
Perinuclear TYRP1 fluorescence was quantified from deconvolved fluorescence images using ImageJ. Local background was subtracted as described above. For each cell, perinuclear fluorescence intensity was obtained by measuring the integrated density of fluorescence in a 5-µm diameter circular region of interest drawn around the nucleus, identified by staining with DAPI (included in the ProLong Gold Antifade Mountant). Total fluorescence intensity was obtained by measuring the integrated density of fluorescence in the entire cell, identified as a region of interest by manual drawing. Perinuclear fluorescence was then divided by total TYRP1 fluorescence to determine the percentage of TYRP1 fluorescence concentrated in the perinuclear region. The results represent values obtained from at least 20 cells over 3 individual experiments.
Tubule number, length, and lifetimes
All analyses were blinded and conducted using ImageJ. Analyses were limited to cells with moderate transgene expression, as determined by maximal signal intensity of peripheral objects in VisiView (average maximal signal ∼250 using standard gain settings). A maximum-intensity projection was generated from four consecutive z-stacks of each analyzed single time frame image, and 10 nonnuclear 25-µm2 regions of each whole cell within the image were randomly chosen and analyzed as described below. Fluorescence images were rendered binary using the Bernsen algorithm of the auto local threshold plugin, followed by the binary skeletonize plugin. These skeletonized 25-µm2 z projections were used for quantification of tubule number, length, and lifetimes. Steady-state tubule number within a single time frame was determined by manual counting of all ten 25-µm2 regions from at least five cells per condition in each of at least three separate experiments. Note that this approach underestimates tubules because not all tubules appear continuous within the z-series. Tubule length was measured using the free-form line tool. Lines were drawn along the entire length of a tubule, and the measure function was used to obtain the length of the line in micrometers. Tubule lifetimes were counted over the entire image sequence by marking the middle of a tubule with the line selection tool and manually counting the number of frames that the tubule persisted. The start of each tubule lifetime was defined as the first frame in which the tubule appeared. Tubules that existed in the first frame of a time-lapse series were excluded because the start of the tubule lifetime could not be determined. The end of a tubule lifetime was defined as the last frame in which the tubule appeared. Tubules that persisted after the last frame of a time-lapse series were excluded from analysis. Note that these approaches underestimate tubule length and lifetime due to potential tubule movement out of the planes of focus and loss of signal along the length of a tubule due to thresholding.
Statistical analysis
Statistical significance was determined using a one-way ANOVA and a Tukey post hoc test. All values are indicated as mean ± SEM, and P > 0.05 was defined as a nonsignificant difference. All statistical analysis was performed using GraphPad Prism software. In most cases, data distribution was assumed to be normal, but this was not formally tested; in Fig. 3 B, the Geisser-Greenhouse correction was applied.
Online supplemental materials
Fig. S1 documents disruption of both TYR and TYRP1 localization in Ap3d1−/− melan-mh melanocytes. Fig. S2 shows that the BLOC-1 cargo, TYRP1, does not accumulate in early endosomes in untreated Ap3d1−/− melanocytes or cells reconstituted with WT or VAMP7 binding–deficient forms of AP-3δ. Fig. S3 illustrates the CRISPR/Cas9 approach for knocking out Bloc1s6, documents successful knockout in Ap3d1−/−/Pldn−/− melanocytes, and shows VAMP7 and STX13 expression levels in cells reconstituted with AP-3δ and Pldn variants. Fig. S4 documents GFP-RAB11 and mCherry-STX13 overlap in Ap3d1−/−/Pldn−/− cells that do or do not express AP-3δ and Pldn variants. Fig. S5 documents non–clathrin-associated AP-3 on tubulovesicular structures near but not in the TGN in WT melanocytic cells and the presence of STX13-containing tubules in Ap3d1−/− melanocytes. Video 1 shows that GFP-RAB11 labels the majority of mCherry-STX13–positive tubules in WT melanocytes. Videos 2 and 3 show GFP-RAB11–containing tubule formation and duration in Ap3d1−/−/Pldn−/− melanocytes expressing AP-3δWT and PldnWT (Video 2) or AP-3δmut1/Pldnmut1 (Video 3). Table S1 lists the oligonucleotide sequences for gRNAs and validation of the CRISPR/Cas9 knockout of Pldn. Table S2 lists the oligonucleotide sequences used for plasmid construction.
Supplementary Material
lists the oligonucleotide sequences for gRNAs and validation of the CRISPR/Cas9 knockout of Pldn.
lists the oligonucleotide sequences used for plasmid construction.
Acknowledgments
We thank J. Bonifacino for reagents and protocols; P. Roche, C. Burd (Yale University, New Haven, CT), E. Dell’Angelica, A. Peden (University of Sheffield, Sheffield, UK), T. Galli, R. Prekeris (University of Colorado, Aurora, CO), D. Trono, and F. Zhang for reagents; and D. Tenza (Institut Curie, Paris, France) for technical assistance.
This work was funded by the following National Institutes of Health grants: National Eye Institute grant no. R01 EY015625 (to M.S. Marks and G. Raposo); National Institute of General Medical Sciences grants no. K12 GM081259 University of Pennsylvania Post-Doctoral Opportunities in Research and Training (supporting S.L. Bowman), T32 GM007229 Training Program in Cell and Molecular Biology (supporting L. Le and A. Sitaram), and R15 GM132810 (to M.K. Dennis); and National Institute of Arthritis and Musculoskeletal and Skin Diseases grant F32 AR062476 (to M.K. Dennis). This work was also funded by Wellcome Trust grants 108429/Z/15/Z (to E.V. Sviderskaya and D.C. Bennett) and 207455/Z/17/Z (to D.J. Owen) and by M.J. Murdock Charitable Trust grant FSU-201811782 (to S.L. Bowman).
The authors declare no competing financial interests.
Author contributions: All authors edited the manuscript. In addition, S.L. Bowman contributed to conceptualization of the project and experimental design, performed most experiments, analyzed most data, assembled most figures, and wrote much of the original draft. L. Le contributed to conceptualization and experimental design, performed experiments, analyzed data, and drafted the associated text for and assembled Fig. 2. Y. Zhu contributed to experimental design, performed experiments, analyzed data, and assembled part of Fig. 4 and associated text. D.C. Harper contributed to experimental design, performed experiments, and analyzed data for Fig. 3, A and B. A. Sitaram contributed to conceptualization and experimental design, analyzed data, and contributed to figure assembly for Fig. 2. A.C. Theos, E.V. Sviderskaya, and D.C. Bennett contributed to conceptualization, experimental design, and validation of the melan-mh cell lines. G. Raposo contributed to conceptualization of part of the project and to experimental design, performed experiments, analyzed data, and assembled Fig. S5. D.J. Owen contributed to conceptualization of the entire project, provided key reagents, and consulted extensively for project administration. M.K. Dennis contributed to project conceptualization, performed experiments and provided data for some figures, analyzed data, supervised some experiments, and contributed to writing the original draft and revisions. M.S. Marks contributed to conceptualization, supervised and administered the project, coordinated among contributors, provided resources, analyzed data, assembled and edited figures, wrote part of the original draft, coordinated the revisions, and coordinated editing by all coauthors.
References
- Baker, R.W., and Hughson F.M.. 2016. Chaperoning SNARE assembly and disassembly. Nat. Rev. Mol. Cell Biol. 17:465–479. 10.1038/nrm.2016.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, D.C., Cooper P.J., and Hart I.R.. 1987. A line of non-tumorigenic mouse melanocytes, syngeneic with the B16 melanoma and requiring a tumour promoter for growth. Int. J. Cancer. 39:414–418. 10.1002/ijc.2910390324 [DOI] [PubMed] [Google Scholar]
- Berson, J.F., Frank D.W., Calvo P.A., Bieler B.M., and Marks M.S.. 2000. A common temperature-sensitive allelic form of human tyrosinase is retained in the endoplasmic reticulum at the nonpermissive temperature. J. Biol. Chem. 275:12281–12289. 10.1074/jbc.275.16.12281 [DOI] [PubMed] [Google Scholar]
- Bonifacino, J.S., and Glick B.S.. 2004. The mechanisms of vesicle budding and fusion. Cell. 116:153–166. 10.1016/S0092-8674(03)01079-1 [DOI] [PubMed] [Google Scholar]
- Bowman, S.L., Bi-Karchin J., Le L., and Marks M.S.. 2019. The road to lysosome-related organelles: insights from Hermansky-Pudlak syndrome and other rare diseases. Traffic. 20:404–435. 10.1111/tra.12646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgo, A., Sotirakis E., Simmler M.C., Verraes A., Chamot C., Simpson J.C., Lanzetti L., Proux-Gillardeaux V., and Galli T.. 2009. Role of Varp, a Rab21 exchange factor and TI-VAMP/VAMP7 partner, in neurite growth. EMBO Rep. 10:1117–1124. 10.1038/embor.2009.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, J.L., and Schekman R.. 1997. Selective packaging of cargo molecules into endoplasmic reticulum-derived COPII vesicles. Proc. Natl. Acad. Sci. USA. 94:837–842. 10.1073/pnas.94.3.837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaineau, M., Danglot L., and Galli T.. 2009. Multiple roles of the vesicular-SNARE TI-VAMP in post-Golgi and endosomal trafficking. FEBS Lett. 583:3817–3826. 10.1016/j.febslet.2009.10.026 [DOI] [PubMed] [Google Scholar]
- Chung, W.Y., Park H.W., Han J.W., Lee M.G., and Kim J.Y.. 2013. WNK4 inhibits plasma membrane targeting of NCC through regulation of syntaxin13 SNARE formation. Cell. Signal. 25:2469–2477. 10.1016/j.cellsig.2013.08.006 [DOI] [PubMed] [Google Scholar]
- Daste, F., Galli T., and Tareste D.. 2015. Structure and function of longin SNAREs. J. Cell Sci. 128:4263–4272. 10.1242/jcs.178574 [DOI] [PubMed] [Google Scholar]
- Delevoye, C., Hurbain I., Tenza D., Sibarita J.-B., Uzan-Gafsou S., Ohno H., Geerts W.J.C., Verkleij A.J., Salamero J., Marks M.S., et al. 2009. AP-1 and KIF13A coordinate endosomal sorting and positioning during melanosome biogenesis. J. Cell Biol. 187:247–264. 10.1083/jcb.200907122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delevoye, C., Miserey-Lenkei S., Montagnac G., Gilles-Marsens F., Paul-Gilloteaux P., Giordano F., Waharte F., Marks M.S., Goud B., and Raposo G.. 2014. Recycling endosome tubule morphogenesis from sorting endosomes requires the kinesin motor KIF13A. Cell Rep. 6:445–454. 10.1016/j.celrep.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delevoye, C., Heiligenstein X., Ripoll L., Gilles-Marsens F., Dennis M.K., Linares R.A., Derman L., Gokhale A., Morel E., Faundez V., et al. 2016. BLOC-1 brings together the actin and microtubule cytoskeletons to generate recycling endosomes. Curr. Biol. 26:1–13. 10.1016/j.cub.2015.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, M.K., Mantegazza A.R., Snir O.L., Tenza D., Acosta-Ruiz A., Delevoye C., Zorger R., Sitaram A., de Jesus-Rojas W., Ravichandran K., et al. 2015. BLOC-2 targets recycling endosomal tubules to melanosomes for cargo delivery. J. Cell Biol. 209:563–577. 10.1083/jcb.201410026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, M.K., Delevoye C., Acosta-Ruiz A., Hurbain I., Romao M., Hesketh G.G., Goff P.S., Sviderskaya E.V., Bennett D.C., Luzio J.P., et al. 2016. BLOC-1 and BLOC-3 regulate VAMP7 cycling to and from melanosomes via distinct tubular transport carriers. J. Cell Biol. 214:293–308. 10.1083/jcb.201605090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro, S.M., and Dell’Angelica E.C.. 2005. The cell biology of Hermansky-Pudlak syndrome: recent advances. Traffic. 6:525–533. 10.1111/j.1600-0854.2005.00299.x [DOI] [PubMed] [Google Scholar]
- Di Pietro, S.M., Falcón-Pérez J.M., Tenza D., Setty S.R.G., Marks M.S., Raposo G., and Dell’Angelica E.C.. 2006. BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol. Biol. Cell. 17:4027–4038. 10.1091/mbc.e06-05-0379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader, C.M., Sánchez D.G., Mestre M.B., and Colombo M.I.. 2009. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim. Biophys. Acta. 1793:1901–1916. 10.1016/j.bbamcr.2009.09.011 [DOI] [PubMed] [Google Scholar]
- Ghiani, C.A., Starcevic M., Rodriguez-Fernandez I.A., Nazarian R., Cheli V.T., Chan L.N., Malvar J.S., de Vellis J., Sabatti C., and Dell’Angelica E.C.. 2010. The dysbindin-containing complex (BLOC-1) in brain: developmental regulation, interaction with SNARE proteins and role in neurite outgrowth. Mol. Psychiatry. 15:115. 10.1038/mp.2009.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham, A.K., and Munro S.. 2019. Transport carrier tethering - how vesicles are captured by organelles. Curr. Opin. Cell Biol. 59:140–146. 10.1016/j.ceb.2019.04.010 [DOI] [PubMed] [Google Scholar]
- Giraudo, C.G., Eng W.S., Melia T.J., and Rothman J.E.. 2006. A clamping mechanism involved in SNARE-dependent exocytosis. Science. 313:676–680. 10.1126/science.1129450 [DOI] [PubMed] [Google Scholar]
- Gokhale, A., Mullin A.P., Zlatic S.A., Easley C.A. IV, Merritt M.E., Raj N., Larimore J., Gordon D.E., Peden A.A., Sanyal S., et al. 2015. The N-ethylmaleimide-sensitive factor and dysbindin interact to modulate synaptic plasticity. J. Neurosci. 35:7643–7653. 10.1523/JNEUROSCI.4724-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh, G.G., Pérez-Dorado I., Jackson L.P., Wartosch L., Schäfer I.B., Gray S.R., McCoy A.J., Zeldin O.B., Garman E.F., Harbour M.E., et al. 2014. VARP is recruited on to endosomes by direct interaction with retromer, where together they function in export to the cell surface. Dev. Cell. 29:591–606. 10.1016/j.devcel.2014.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L., Kuo Y.M., and Gitschier J.. 1999. The pallid gene encodes a novel, syntaxin 13-interacting protein involved in platelet storage pool deficiency. Nat. Genet. 23:329–332. 10.1038/15507 [DOI] [PubMed] [Google Scholar]
- Huizing, M., Sarangarajan R., Strovel E., Zhao Y., Gahl W.A., and Boissy R.E.. 2001. AP-3 mediates tyrosinase but not TRP-1 trafficking in human melanocytes. Mol. Biol. Cell. 12:2075–2085. 10.1091/mbc.12.7.2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari, J., and Helenius A.. 2011. Endosome maturation. EMBO J. 30:3481–3500. 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilardi, J.M., Mochida S., and Sheng Z.H.. 1999. Snapin: a SNARE-associated protein implicated in synaptic transmission. Nat. Neurosci. 2:119–124. 10.1038/5673 [DOI] [PubMed] [Google Scholar]
- Jahn, R., and Scheller R.H.. 2006. SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7:631–643. 10.1038/nrm2002 [DOI] [PubMed] [Google Scholar]
- Jani, R.A., Purushothaman L.K., Rani S., Bergam P., and Setty S.R.G.. 2015. STX13 regulates cargo delivery from recycling endosomes during melanosome biogenesis. J. Cell Sci. 128:3263–3276. 10.1242/jcs.171165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, H.M., Evans P.R., Schäfer I.B., Gray S.R., Sanderson C.M., Luzio J.P., Peden A.A., and Owen D.J.. 2012. Structural basis of the intracellular sorting of the SNARE VAMP7 by the AP3 adaptor complex. Dev. Cell. 22:979–988. 10.1016/j.devcel.2012.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike, S., and Jahn R.. 2019. SNAREs define targeting specificity of trafficking vesicles by combinatorial interaction with tethering factors. Nat. Commun. 10:1608. 10.1038/s41467-019-09617-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kook, S., Qi A., Wang P., Meng S., Gulleman P., Young L.R., and Guttentag S.H.. 2018. Gene-edited MLE-15 cells as a model for the Hermansky-Pudlak syndromes. Am. J. Respir. Cell Mol. Biol. 58:566–574. 10.1165/rcmb.2017-0324MA [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümmel, D., Krishnakumar S.S., Radoff D.T., Li F., Giraudo C.G., Pincet F., Rothman J.E., and Reinisch K.M.. 2011. Complexin cross-links prefusion SNAREs into a zigzag array. Nat. Struct. Mol. Biol. 18:927–933. 10.1038/nsmb.2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuster, A., Nola S., Dingli F., Vacca B., Gauchy C., Beaujouan J.-C., Nunez M., Moncion T., Loew D., Formstecher E., et al. 2015. The Q-Soluble-N-ethylmaleimide-sensitive factor attachment protein receptor (Q-SNARE) SNAP-47 regulates trafficking of selected vesicle-associated membrane proteins (VAMPs). J. Biol. Chem. 290:28056–28069. 10.1074/jbc.M115.666362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, P.W., and Deol M.S.. 1974. Mocha, a new coat color and behavior mutation on chromosome 10 of the mouse. J. Hered. 65:362–364. 10.1093/oxfordjournals.jhered.a108551 [DOI] [PubMed] [Google Scholar]
- Letourneur, F., and Klausner R.D.. 1992. A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell. 69:1143–1157. 10.1016/0092-8674(92)90636-Q [DOI] [PubMed] [Google Scholar]
- Martinez-Arca, S., Rudge R., Vacca M., Raposo G., Camonis J., Proux-Gillardeaux V., Daviet L., Formstecher E., Hamburger A., Filippini F., et al. 2003. A dual mechanism controlling the localization and function of exocytic v-SNAREs. Proc. Natl. Acad. Sci. USA. 100:9011–9016. 10.1073/pnas.1431910100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S.E., Collins B.M., McCoy A.J., Robinson M.S., and Owen D.J.. 2007. A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles. Nature. 450:570–574. 10.1038/nature06353 [DOI] [PubMed] [Google Scholar]
- Moreau, K., Ravikumar B., Renna M., Puri C., and Rubinsztein D.C.. 2011. Autophagosome precursor maturation requires homotypic fusion. Cell. 146:303–317. 10.1016/j.cell.2011.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, S., Kojima T., and Kitamura T.. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063–1066. 10.1038/sj.gt.3301206 [DOI] [PubMed] [Google Scholar]
- Moriyama, K., and Bonifacino J.S.. 2002. Pallidin is a component of a multi-protein complex involved in the biogenesis of lysosome-related organelles. Traffic. 3:666–677. 10.1034/j.1600-0854.2002.30908.x [DOI] [PubMed] [Google Scholar]
- Newell-Litwa, K., Salazar G., Smith Y., and Faundez V.. 2009. Roles of BLOC-1 and adaptor protein-3 complexes in cargo sorting to synaptic vesicles. Mol. Biol. Cell. 20:1441–1453. 10.1091/mbc.e08-05-0456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden, A.A., Oorschot V., Hesser B.A., Austin C.D., Scheller R.H., and Klumperman J.. 2004. Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J. Cell Biol. 164:1065–1076. 10.1083/jcb.200311064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkovic, M., Jemaiel A., Daste F., Specht C.G., Izeddin I., Vorkel D., Verbavatz J.M., Darzacq X., Triller A., Pfenninger K.H., et al. 2014. The SNARE Sec22b has a non-fusogenic function in plasma membrane expansion. Nat. Cell Biol. 16:434–444. 10.1038/ncb2937 [DOI] [PubMed] [Google Scholar]
- Pols, M.S., van Meel E., Oorschot V., ten Brink C., Fukuda M., Swetha M.G., Mayor S., and Klumperman J.. 2013. hVps41 and VAMP7 function in direct TGN to late endosome transport of lysosomal membrane proteins. Nat. Commun. 4:1361. 10.1038/ncomms2360 [DOI] [PubMed] [Google Scholar]
- Prekeris, R., Klumperman J., Chen Y.A., and Scheller R.H.. 1998. Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J. Cell Biol. 143:957–971. 10.1083/jcb.143.4.957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor, P.R., Mullock B.M., Bright N.A., Lindsay M.R., Gray S.R., Richardson S.C., Stewart A., James D.E., Piper R.C., and Luzio J.P.. 2004. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 5:590–595. 10.1038/sj.embor.7400150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor, P.R., Jackson L., Gray S.R., Edeling M.A., Thompson A., Sanderson C.M., Evans P.R., Owen D.J., and Luzio J.P.. 2008. Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell. 134:817–827. 10.1016/j.cell.2008.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, S.K., Huynh C., Proux-Gillardeaux V., Galli T., and Andrews N.W.. 2004. Identification of SNAREs involved in synaptotagmin VII-regulated lysosomal exocytosis. J. Biol. Chem. 279:20471–20479. 10.1074/jbc.M400798200 [DOI] [PubMed] [Google Scholar]
- Raposo, G., Tenza D., Murphy D.M., Berson J.F., and Marks M.S.. 2001. Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells. J. Cell Biol. 152:809–824. 10.1083/jcb.152.4.809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll, L., Heiligenstein X., Hurbain I., Domingues L., Figon F., Petersen K.J., Dennis M.K., Houdusse A., Marks M.S., Raposo G., et al. 2018. Myosin VI and branched actin filaments mediate membrane constriction and fission of melanosomal tubule carriers. J. Cell Biol. 217:2709–2726. 10.1083/jcb.201709055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo, J., and Südhof T.C.. 2012. The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices—guilty as charged? Annu. Rev. Cell Dev. Biol. 28:279–308. 10.1146/annurev-cellbio-101011-155818 [DOI] [PubMed] [Google Scholar]
- Salazar, G., Craige B., Styers M.L., Newell-Litwa K.A., Doucette M.M., Wainer B.H., Falcon-Perez J.M., Dell’Angelica E.C., Peden A.A., Werner E., et al. 2006. BLOC-1 complex deficiency alters the targeting of adaptor protein complex-3 cargoes. Mol. Biol. Cell. 17:4014–4026. 10.1091/mbc.e06-02-0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana, N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer, I.B., Hesketh G.G., Bright N.A., Gray S.R., Pryor P.R., Evans P.R., Luzio J.P., and Owen D.J.. 2012. The binding of Varp to VAMP7 traps VAMP7 in a closed, fusogenically inactive conformation. Nat. Struct. Mol. Biol. 19:1300–1309. 10.1038/nsmb.2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty, S.R.G., Tenza D., Truschel S.T., Chou E., Sviderskaya E.V., Theos A.C., Lamoreux M.L., Di Pietro S.M., Starcevic M., Bennett D.C., et al. 2007. BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol. Biol. Cell. 18:768–780. 10.1091/mbc.e06-12-1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty, S.R.G., Tenza D., Sviderskaya E.V., Bennett D.C., Raposo G., and Marks M.S.. 2008. Cell-specific ATP7A transport sustains copper-dependent tyrosinase activity in melanosomes. Nature. 454:1142–1146. 10.1038/nature07163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaram, A., and Marks M.S.. 2012. Mechanisms of protein delivery to melanosomes in pigment cells. Physiology (Bethesda). 27:85–99. 10.1152/physiol.00043.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaram, A., Dennis M.K., Chaudhuri R., De Jesus-Rojas W., Tenza D., Setty S.R.G., Wood C.S., Sviderskaya E.V., Bennett D.C., Raposo G., et al. 2012. Differential recognition of a dileucine-based sorting signal by AP-1 and AP-3 reveals a requirement for both BLOC-1 and AP-3 in delivery of OCA2 to melanosomes. Mol. Biol. Cell. 23:3178–3192. 10.1091/mbc.e11-06-0509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcevic, M., and Dell’Angelica E.C.. 2004. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J. Biol. Chem. 279:28393–28401. 10.1074/jbc.M402513200 [DOI] [PubMed] [Google Scholar]
- Sviderskaya, E.V., Hill S.P., Evans-Whipp T.J., Chin L., Orlow S.J., Easty D.J., Cheong S.C., Beach D., DePinho R.A., and Bennett D.C.. 2002. p16Ink4a in melanocyte senescence and differentiation. J. Natl. Cancer Inst. 94:446–454. 10.1093/jnci/94.6.446 [DOI] [PubMed] [Google Scholar]
- Tamura, K., Ohbayashi N., Maruta Y., Kanno E., Itoh T., and Fukuda M.. 2009. Varp is a novel Rab32/38-binding protein that regulates Tyrp1 trafficking in melanocytes. Mol. Biol. Cell. 20:2900–2908. 10.1091/mbc.e08-12-1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Ohbayashi N., Ishibashi K., and Fukuda M.. 2011. Structure-function analysis of VPS9-ankyrin-repeat protein (Varp) in the trafficking of tyrosinase-related protein 1 in melanocytes. J. Biol. Chem. 286:7507–7521. 10.1074/jbc.M110.191205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theos, A.C., Tenza D., Martina J.A., Hurbain I., Peden A.A., Sviderskaya E.V., Stewart A., Robinson M.S., Bennett D.C., Cutler D.F., et al. 2005. Functions of adaptor protein (AP)-3 and AP-1 in tyrosinase sorting from endosomes to melanosomes. Mol. Biol. Cell. 16:5356–5372. 10.1091/mbc.e05-07-0626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann, C., and Kümmel D.. 2019. Structure of membrane tethers and their role in fusion. Traffic. 20:479–490. 10.1111/tra.12655 [DOI] [PubMed] [Google Scholar]
- Verraes, A., Cholley B., Galli T., and Nola S.. 2018. Comparative study of commercially available and homemade anti-VAMP7 antibodies using CRISPR/Cas9-depleted HeLa cells and VAMP7 knockout mice. F1000 Res. 7:1649. 10.12688/f1000research.15707.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vites, O., Rhee J.S., Schwarz M., Rosenmund C., and Jahn R.. 2004. Reinvestigation of the role of snapin in neurotransmitter release. J. Biol. Chem. 279:26251–26256. 10.1074/jbc.M404079200 [DOI] [PubMed] [Google Scholar]
- Vivona, S., Liu C.W., Strop P., Rossi V., Filippini F., and Brunger A.T.. 2010. The longin SNARE VAMP7/TI-VAMP adopts a closed conformation. J. Biol. Chem. 285:17965–17973. 10.1074/jbc.M110.120972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, N., Bryant N.J., Connolly L.M., Simpson R.J., Luzio J.P., Piper R.C., and James D.E.. 2001. Syntaxin 7 complexes with mouse Vps10p tail interactor 1b, syntaxin 6, vesicle-associated membrane protein (VAMP)8, and VAMP7 in b16 melanoma cells. J. Biol. Chem. 276:19820–19827. 10.1074/jbc.M010838200 [DOI] [PubMed] [Google Scholar]
- Yoon, T.-Y., and Munson M.. 2018. SNARE complex assembly and disassembly. Curr. Biol. 28:R397–R401. 10.1016/j.cub.2018.01.005 [DOI] [PubMed] [Google Scholar]
- Zlatic, S.A., Grossniklaus E.J., Ryder P.V., Salazar G., Mattheyses A.L., Peden A.A., and Faundez V.. 2013. Chemical-genetic disruption of clathrin function spares adaptor complex 3-dependent endosome vesicle biogenesis. Mol. Biol. Cell. 24:2378–2388. 10.1091/mbc.e12-12-0860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwilling, D., Cypionka A., Pohl W.H., Fasshauer D., Walla P.J., Wahl M.C., and Jahn R.. 2007. Early endosomal SNAREs form a structurally conserved SNARE complex and fuse liposomes with multiple topologies. EMBO J. 26:9–18. 10.1038/sj.emboj.7601467 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the oligonucleotide sequences for gRNAs and validation of the CRISPR/Cas9 knockout of Pldn.
lists the oligonucleotide sequences used for plasmid construction.