Rasicci et al. investigate the mechanochemical properties of distinct isoforms of cardiac myosin carrying the K104E regulatory light chain mutation, previously associated with hypertrophic cardiomyopathy. They demonstrate that the mutation does not affect the ATPase or motor properties of human cardiac myosin subfragment-1 (M2β-S1), but the mutation increases the sliding velocity and disrupts RLC incorporation in full-length α-cardiac myosin.
Abstract
Mutations in the cardiac myosin regulatory light chain (RLC, MYL2 gene) are known to cause inherited cardiomyopathies with variable phenotypes. In this study, we investigated the impact of a mutation in the RLC (K104E) that is associated with hypertrophic cardiomyopathy (HCM). Previously in a mouse model of K104E, older animals were found to develop cardiac hypertrophy, fibrosis, and diastolic dysfunction, suggesting a slow development of HCM. However, variable penetrance of the mutation in human populations suggests that the impact of K104E may be subtle. Therefore, we generated human cardiac myosin subfragment-1 (M2β-S1) and exchanged on either the wild type (WT) or K104E human ventricular RLC in order to assess the impact of the mutation on the mechanochemical properties of cardiac myosin. The maximum actin-activated ATPase activity and actin sliding velocities in the in vitro motility assay were similar in M2β-S1 WT and K104E, as were the detachment kinetic parameters, including the rate of ATP-induced dissociation and the ADP release rate constant. We also examined the mechanical performance of α-cardiac myosin extracted from transgenic (Tg) mice expressing human wild type RLC (Tg WT) or mutant RLC (Tg K104E). We found that α-cardiac myosin from Tg K104E animals demonstrated enhanced actin sliding velocities in the motility assay compared with its Tg WT counterpart. Furthermore, the degree of incorporation of the mutant RLC into α-cardiac myosin in the transgenic animals was significantly reduced compared with wild type. Therefore, we conclude that the impact of the K104E mutation depends on either the length or the isoform of the myosin heavy chain backbone and that the mutation may disrupt RLC interactions with the myosin lever arm domain.
Introduction
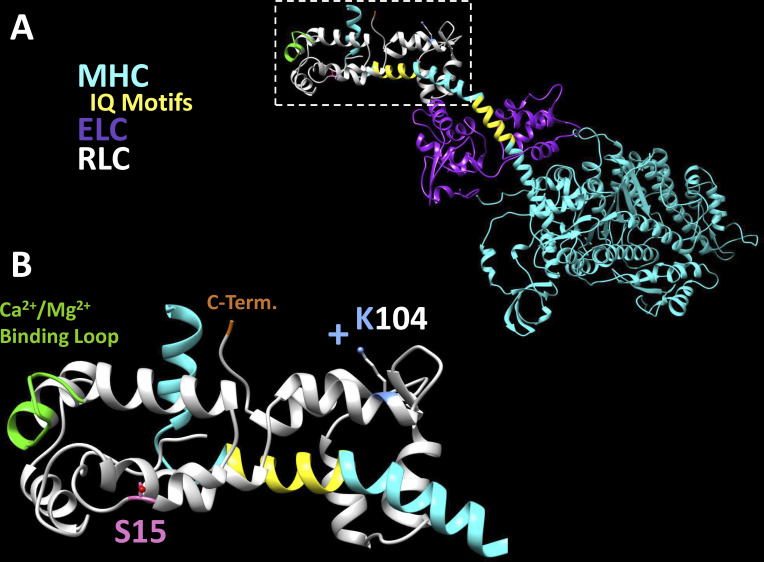
Myosin is a hexameric protein, composed of two heavy chains (MHCs), two essential light chains (ELCs), and two regulatory light chains (RLCs), with one ELC and RLC molecule per MHC. The MHC contains a globular head with nucleotide and actin-binding sites, a lever arm domain with sites for light chain binding, and a tail region that assembles into thick filaments of sarcomeres in striated muscle tissue (Rayment et al., 1993). Cardiac myosin couples the energy from its ATP hydrolysis cycle with a mechanical power stroke when bound to actin-containing thin filaments, culminating in the shortening of the sarcomere and cardiomyocytes (Sweeney and Hammers, 2018). In ventricular heart tissue, humans predominantly express the β-cardiac isoform of MHC, while smaller organisms with a higher metabolic rate (e.g., mice) predominantly express the α-cardiac MHC isoform (Alpert et al., 2002). Myosin molecules may be cleaved at various locations, but the subfragment-1 (S1) of myosin is commonly studied, as it contains the motor domain and light chain-containing lever arm of one MHC, and represents a portion of the molecule fully capable of the mechanochemical adenosine triphosphatase (ATPase) cycle and force generation (Fig. 1 A).
Figure 1.
Structural model of K104 residue in M2β-S1. The location of the K104 residue in the RLC is depicted in the structural model of human M2β-S1 (HBCprestrokeS1 Homology Model, Spudich Lab, Stanford University, Stanford, CA). (A) M2β-S1 in the prestroke state with MHC (cyan), ELC (purple), and RLC (white) bound to the lever arm domain at respective IQ motifs (yellow). (B) Magnified view of the inset of RLC in A. Critical regions of the RLC molecule are highlighted, including phosphorylatable S15 (pink), the divalent cation binding loop (green, residues 37–48), and C terminus D166 (orange). The K104 residue is predicted to point away from the cyan lever arm in this model.
Point mutations in several sarcomeric proteins have been associated with various degrees of hypertrophic cardiomyopathy (HCM), a disease characterized by pronounced left ventricular hypertrophy, diastolic dysfunction, fibrosis, and myofibrillar disarray (Alcalai et al., 2008; Yotti et al., 2019). HCM is the leading cause of sudden cardiac death in young adults, and thus, the genotype–phenotype relationship of HCM-causing mutations is of high clinical interest. While most known HCM mutations are in the MHC (MYH7) or myosin binding protein C (MYBPC3), less studied mutations in other sarcomeric proteins such as the RLC or ELC (MYL2 and MYL3, respectively) of myosin also can drive severe phenotypes of HCM (Alcalai et al., 2008; Yadav et al., 2019b).
The RLC binds an IQ motif in the distal portion of the 8.5-nm α-helix of the myosin lever arm domain, while the ELC binds a more proximal IQ motif of the α-helix (Fig. 1). This long α-helix has been demonstrated to function as a lever arm that swings during the power stroke to generate force when myosin is strongly bound to actin (Houdusse and Sweeney, 2016). Thus, RLC and ELC biochemistry is believed to play a critical role in supporting lever arm structure and function (Sitbon et al., 2020). Importantly, RLC can bind a divalent cation (Mg2+ or Ca2+) between residues 37 and 48 in its first EF-hand, which may contribute to overall cellular Ca2+ homeostasis (Szczesna-Cordary et al., 2004). The N-terminal lobe also is characterized by a phosphorylatable serine residue, S15 in humans (Fig. 1 B). A phosphorylated S15 residue is associated with disruption of the autoinhibitory state of myosin and increased cardiac contractility (Sweeney et al., 1993; Chang et al., 2016). Given these various mechanisms of myosin lever arm regulation, it is intuitive that light chain mutations may perturb lever arm dynamics and myosin mechanochemistry, driving changes that may result in cardiac hypertrophy. To date, at least 17 HCM-associated RLC mutations have been identified, which demonstrate a wide range of clinical phenotypes as well as effects on myosin molecular function (Yadav et al., 2019b).
The K104E RLC mutation was first reported in a Danish population study, in which the proband demonstrated pronounced septal hypertrophy and diastolic filling abnormalities, hallmark characteristics of HCM (Andersen et al., 2001). However, the proband exhibited compound heterozygosity, carrying a second RLC mutation (IVS6-1, a splice site acceptor mutation), effectively complicating the genotype–phenotype relationship. In 2014, the Szczesna-Cordary laboratory generated a transgenic (Tg) mouse model to study the specific effects of the K104E mutation. In their study, Tg K104E mice developed severe HCM only with senescence (>13 mo). The full-length α-cardiac myosin isolated from Tg K104E hearts had increased ATPase activity, decreased load tolerance in loaded in vitro motility assays, and a twofold decrease in RLC phosphorylation, relative to myosin from Tg WT animals. Myofibrils from Tg K104E animals had a decreased ability to generate force, and histological sections demonstrated increased fibrotic lesions with age and increased mitochondrial content. Collectively, the authors suggested that diastolic disturbance and bioenergetic inefficiencies drove the late-onset hypertrophic phenotype in Tg K104E mice (Huang et al., 2014).
To our knowledge, the K104E RLC mutation has not been studied outside of a Tg murine model. Moreover, while some RLC mutations have been exchanged into porcine myosin (β-cardiac myosin), recombinant human β-cardiac myosin has not been used to study the impact of an HCM-associated RLC mutation. Thus, the purpose of this study was to assess the impact of the K104E mutation in human β-cardiac myosin S1 (M2β-S1), which contains the motor domain and light chain–binding regions of MHC. Based on previously reported results in mice, we hypothesized that we would observe significant differences in the mechanochemical properties of the mutated myosin including in vitro actin sliding velocity, steady-state actin-activated ATPase, and transient kinetics of the myosin ATPase cycle. To augment this data, we also extracted full-length α-cardiac myosin from (Tg) mouse hearts, which allowed us to compare the impact of the mutation in α- and β-cardiac myosin. We hypothesized that the full-length α-cardiac myosin extracted from Tg K104E mice would show deficits in mechanochemical properties, similar to previous results (Huang et al., 2014).
Materials and methods
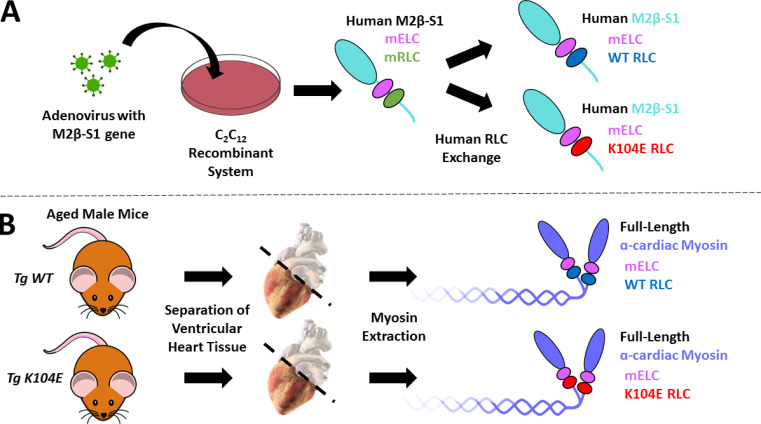
A dual approach of recombinantly expressed M2β-S1 (WT versus mutant, K104E) and full-length α-cardiac myosin extracted from Tg mouse hearts (Tg WT versus mutant, Tg K104E) was employed in the study. Fig. 2 summarizes these constructs including the associated MHC and light chain isoforms, which are referred to throughout the manuscript.
Figure 2.
Generation of myosin constructs. Illustration demonstrating our methodology to generate the various myosin constructs in the study. (A) Human M2β-S1 myosin (cyan) was generated via our C2C12 adenovirus recombinant system. C2C12 myoblasts are murine skeletal muscle cells, and thus the myosin contains the endogenous murine light chains (mELC and mRLC, pink and green, respectively). We performed an RLC exchange in order to study the impact of the human RLC (WT versus K104E, blue and red, respectively) on the mechanochemical properties of M2β-S1. Notably, the constructs still contain the endogenous mELC. (B) Full-length α-cardiac myosin (purple) was extracted from the ventricles of Tg animals expressing either human WT RLC (blue, n = 3) or human K104E RLC (red, n = 3). The image of the virus, petri dish, and mouse are under public domain (CC0). The image of the heart (titled “Heart oblique external”) was illustrated by Patrick J. Lynch; the original image was rotated slightly, and the atria and great vessels were made transparent. The original work and license (CCBY2.5) for the heart image can be found at the following link: https://commons.wikimedia.org/wiki/File:Heart_oblique_external.jpg.
Recombinant β-cardiac myosin expression and purification
A recombinant adenovirus was constructed containing the human M2β-S1 WT with an N-terminal FLAG tag for column purification and C-terminal Avi tag for in vitro motility (Swenson et al., 2017). Differentiating C2C12 cells (murine skeletal myoblasts) were infected with the human M2β-S1 adenovirus, as described previously (Tang et al., 2017; Swenson et al., 2017). The M2β-S1 protein was purified by anti-FLAG affinity chromatography, followed by AmSO4 precipitation, and overnight dialysis against MOPS 20 buffer (10 mM MOPS, pH 7.0, 20 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 1 mM dithiothreitol [DTT]) as previously described (Tang et al., 2017; Swenson et al., 2017). Final protein concentrations were determined by Bradford assays using BSA as a standard. Prior to elution from the FLAG resin, the M2β-S1 constructs were subjected to an RLC exchange, using either human WT RLC or human K104E RLC (see protocol below). Actin was purified (Pardee and Spudich, 1982) and labeled with pyrene iodoacetamide as previously described (Criddle et al., 1985).
RLC expression and exchange
Recombinant M2β-S1 generated in the C2C12 system contains endogenous murine isoforms of RLC and ELC (Swenson et al., 2017), and thus a protocol was used to exchange on human recombinant RLC (either WT RLC or K104E RLC) during purification. Human WT versus K104E RLC constructs with an N-terminal His-tag were inserted into pET15b plasmids, expressed in the Escherichia coli Rossetta BL21 cell line, and purified with cobalt chelating resin centrifugation. Purified RLC was flash-frozen and kept at −80°C until the day of the exchange (Wu et al., 1999). The M2β-S1 was stripped of its endogenous murine RLC and replaced with either human WT RLC or human K104E RLC through a column exchange protocol, as described previously (Nag et al., 2017). Briefly, a stripping buffer containing 20 mM Tris, pH 7.5, 0.5% Triton X-100, 200 mM KCl, 5 mM CDTA, 2 mM ATP, 1 mM DTT, 0.01 mg/ml aprotinin, 0.01 mg/ml leupeptin, and 1 mM PMSF was applied to the M2β-S1 WT construct-containing resin for two 30-min incubation periods. Following the stripping protocol, 1 ml of 20 µM human RLC constructs was incubated on the column for a total of 2 h, with three resuspensions of the resin. The column then was eluted with FLAG peptide (0.2 mg/ml), AmSO4-precipitated, and dialyzed into MOPS 20 buffer overnight.
Prior to AmSO4 precipitation, myosin intended for in vitro motility was biotinylated with BirA in the presence of ATP and biotin (Swenson et al., 2017). SDS-PAGE was performed to assess the quality of protein purification and RLC exchange using NuPage Novex 4–12% Bis-Tris, 1.0 mm, 10- or 12-well precast gradient gels (Invitrogen). To assess efficiency of the RLC exchange, the stoichiometry of M2β-S1 to RLC was calculated by comparing the total concentration of M2β-S1 by Bradford assay with the amount of RLC by Western blot (primary antibody: 6x-His tag monoclonal antibody (Invitrogen); secondary antibody: 680RD donkey α-mouse IgG (LiCor).
Steady-state and transient kinetic measurements
Steady-state actin-activated ATPase
An Applied Photophysics stopped-flow apparatus was used to perform NADH-coupled ATPase assays to measure steady-state actin-activated ATPase of the human M2β-S1 constructs (De La Cruz and Ostap, 2009; Swenson et al., 2017). Briefly, 0.1 µM M2β-S1 was assayed in the presence of varying concentrations of F-actin (0, 5, 10, 20, 40, and 60 µM). After the addition of 1 mM ATP, changes in absorption at 340 nm were measured for 200 s, and the transients were fit to a linear function. A standard curve of known ADP concentrations was performed to determine the ATPase rate at each actin concentration, and the data were fit to a Michaelis–Menten relationship to determine the ATPase parameters.
Transient kinetic analysis (ADP release and ATP-induced dissociation)
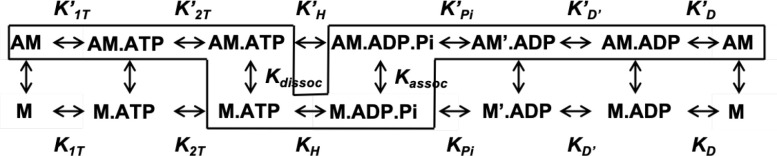
Transient kinetic analyses of key steps in the actomyosin ATPase cycle of M2β-S1 with WT RLC versus K104E were measured with an Applied Photophysics stopped-flow apparatus equipped with an excitation monochrometer (9.3-nm band pass) and appropriate emission filters. The 2′-deoxy-ADP labeled with N-methylanthraniloyl at the 3′-ribose position (mantADP) fluorescence was excited at 290 nm and the emission measured with a 395 nm long-pass filter. The pyrene actin fluorescence was excited at 365 nm and the emission measured with a 395-nm long pass filter. Fluorescence transients were fit with custom software provided with the instrument or GraphPad Prism software. Data were analyzed using the kinetic scheme (Scheme 1) used in recent studies of human cardiac myosin (Swenson et al., 2017; Tang et al., 2019). ATP and ADP, highest purity commercially available, were prepared fresh from powder, and concentrations were determined by absorbance at 259 (ε259 = 15,400 M−1 ⋅ cm–1). mantADP was purchased from Jenna Biosciences.
(Scheme 1).
Extraction of α-cardiac myosin from Tg mice
To compare the impact of the mutation in α- versus β-cardiac myosin, ventricular heart samples of six age-matched (∼19 mo) male B6SJLF1/J Tg mice were obtained from the University of Kentucky (graciously donated by the Szczesna-Cordary laboratory at the University of Miami). All animal procedures and experiments were performed in accordance with institutional guidelines at the University of Miami and the University of Kentucky. Three Tg mice overexpressed the human WT isoform of RLC (Tg WT), while three overexpressed the human K104E RLC mutation (Tg K104E). In vivo and in vitro cardiac functions of these mice have been characterized previously by the Szczesna-Cordary laboratory (Huang et al., 2014). All mice were sacrificed at the University of Kentucky. Ventricular tissue was separated, flash-frozen, and shipped to Penn State College of Medicine, where full-length myosin was extracted, as previously described (Woodward et al., 2015). Inactive myosin molecules (i.e., dead-heads) were removed with an actin spin-down in the presence of ATP. The myosin was resuspended into a final volume of 300–400 µl MOPS 20 buffer, containing additional KCl to 300 mM. Protein concentration was determined by Bradford assay, and the full-length α-cardiac myosin was studied via mass spectrometry (MS) and unloaded in vitro motility assays.
MSs
To determine the levels of RLC expression, 100 µl aliquots of the full-length α-cardiac myosin (2–5 µM) were flash frozen and sent to the University of Vermont on dry ice. The proteins were digested in solution and denatured by the addition of 75 µl of 0.1% RapiGest SF Surfactant (Waters Corporation) and heating (50°C, 45 min). Cysteine residues were reduced and alkylated by adding 5 µl of 0.1 M DTT and heating at 100°C for 10 min and then adding 10.4 µl of 100 µM iodoacetamide and incubating in the dark (22°C, 30 min). Proteins were digested to peptides by adding 25 µl trypsin in 50 mM ammonium bicarbonate (0.2 µm/ml) and heating overnight (37°C). The solution was dried in a speed vacuum. Trypsin was deactivated by adding 100 µl of 7% formic acid/50 mM ammonium bicarbonate (37°C, 1 h) and then dried in a speed vacuum. The RapiGest was cleaved by repeating the previous step with 0.1% trifluoroacetic acid. The peptides were reconstituted in 100 µl 0.1% trifluoroacetic acid and centrifuged. The top 75 µl of the solution was put into an MS analysis vial.
The samples were analyzed by liquid chromatography (LC) MS as described previously (O’Leary et al., 2019). Briefly, a 20-µl aliquot of each sample was injected onto a 100-Å, 1.8-µm, 1 × 150 mm Acquity UPLC HSS T3 column (Waters) attached to an UltiMate 3000 ultra-high pressure LC system (Dionex). The ultra-high pressure LC effluent was directly infused into a Q Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). Data were collected in data-dependent tandem MS mode. Peptides were identified by SEQUEST, and LC peak areas of peptides of interest, those unique to or shared between sequences, were extracted using the Proteome Discoverer 2.2 software package. LC peak areas were imported into Excel for further analysis by mass balance (O’Leary et al., 2019).
Unloaded in vitro motility
The in vitro motility assay was performed on M2β-S1 constructs and full-length α-cardiac myosin extracted from tissue, using established protocols (Kron et al., 1991; Tang et al., 2017). Microscope coverslips were coated with 1% nitrocellulose in amyl acetate (Ladd Research) and applied to a microscope slide with double-sided tape to create a flow cell. For M2β-S1 motility, the coverslip surface was coated with streptavidin (0.1 mg/ml) and blocked with BSA (1 mg/ml) before flowing through biotinylated M2β-S1, with densities from 0.2 µM to 0.8 µM in MOPS 20 buffer. Unlabeled sheared actin (2 µM) followed by ATP (2 mM) was added to ensure blocking of inactive myosin heads (dead-heads). Actin was labeled with phalloidin–Alexa 555 (DsRed filter; excitation/emission, 555/588 nm). To initiate motility, an activation buffer containing 0.35% methylcellulose, an ATP regeneration system (2 mM ATP, 5 mg/ml glucose, 20 U/ml pyruvate kinase, and 2.5 mM phosphoenolpyruvate), and oxygen scavengers (0.1 mg/ml glucose oxidase and 0.018 mg/ml catalase) was added to the flow cell. Temperature (25 ± 1°C) was monitored using a thermocouple meter (Stable Systems International). The slide was visualized promptly with a NIKON TE2000 microscope equipped with a 60×/1.4 NA phase objective and a Perfect Focus System. All images were acquired at 1-s intervals for 2 min using a shutter-controlled CoolSNAP HQ2 cooled charge-coupled device digital camera (Photometrics) binned 2 × 2. For M2β-S1 motility, videos were exported to ImageJ and prepared for automated FAST software motility analysis (Aksel et al., 2015), from which >2,500 actin filaments from one experiment (i.e., slide) per protein preparation (n = 3) at 0.5 µM myosin were compiled for statistical analysis, WT versus K104E. Videos with >20% stuck filaments were excluded from the analysis.
For full-length α-cardiac constructs, the in vitro motility assay was performed on myosin extracted from six Tg male mice (three Tg WT versus three Tg K104E) as stated above, except myosin was attached directly to the nitrocellulose motility surface in the absence of streptavidin, and the activation buffer contained 10 mM DTT. Importantly, due to inherently faster actin sliding velocities of α-cardiac myosin, actin filaments were analyzed manually via MTrackJ plug-in in ImageJ (Meijering et al., 2012), as opposed to FAST motility analysis. 30 fluorescent actin filaments from one experiment per myosin extraction (n = 3) at 0.5 µM myosin were compiled for statistical analysis, Tg WT versus Tg K104E.
Statistics
All experiments compared either M2β-S1 WT versus K104E or full-length α-cardiac Tg WT versus K104E, and thus, differences in each dataset were examined with unpaired Student’s t tests (*, P ≤ 0.05). To avoid statistical analysis of technical replicates in the in vitro motility experiments (i.e., actin filament velocities from the same experiment), the sample means of independent experiments were averaged, and the data have been presented as SuperPlots, as described previously (Lord et al., 2020). Data are reported as mean ± SE, unless otherwise indicated.
Online supplemental material
Fig. S1 shows a representative SDS-PAGE gel demonstrating the quality of M2β-S1 purification and RLC exchange, while Fig. S2 shows a representative Western blot used to quantitate the efficiency of RLC exchange. Fig. S3 demonstrates density dependence of M2β-S1 in the in vitro motility assay. Fig. S4 shows potential biochemical changes in the C-terminal lobe of RLC induced by the K104E mutation in M2α-S1, as predicted by Chimera Modeller (Sali and Blundell, 1993).
Figure S1.
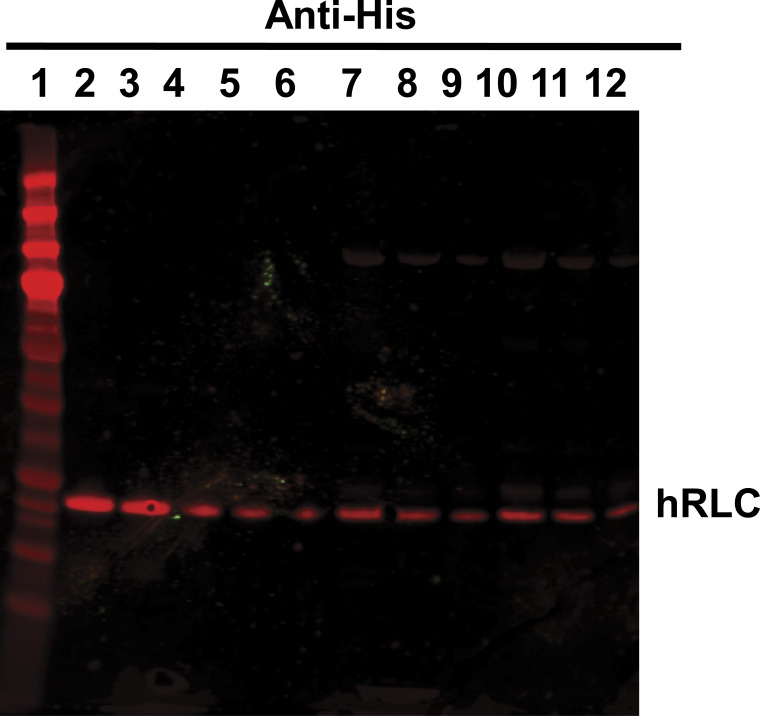
SDS-PAGE of RLC exchange. Representative SDS-PAGE gel of protein preparation run in precast NuPage Novex 4–12% Bis-Tris, 1.0 mm, 10- or 12-well gels, demonstrating quality of M2β-S1 purification and RLC exchange. Lanes were loaded with the following samples: (1) mol wt marker (BioRad Precision Plus Protein Standard: 250, 150, 100, 75, 50, 37, 25, 20, 15, and 10 kD, respectively), (2) stripped endogenous mRLC, (3) M2β-S1 WT human RLC (hRLC), (4) M2β-S1 K104E hRLC, (5) His-tagged hRLC control, and (6) M2β-S1 nonexchanged control. See labels for S1, endogenous murine RLC (mRLC) and ELC (mELC), as well as the exchanged hRLC. Note differential migration between His-tagged hRLC (19,589 D) and mRLC (18,864 D).
Figure S2.
Quantitative Western blot for hRLC exchange efficiency. Representative Western blot used to quantitate human RLC (hRLC) exchange in M2β-S1. Image is a representative Western blot (primary antibody: α-HIS; secondary antintintibody: α-mouse 680 nm) used to calculate the efficiency of the RLC exchange. WT and K104E M2β-S1 were loaded in decreasing amounts and compared with a RLC standard curve. Lanes of the SDS-PAGE gel were loaded with the following samples: (1) mol wt marker (BioRad Precision Plus Protein Standard: 250, 150, 100, 75, 37, 25, 20, 15, and 10 kD, respectively), (2–6) decreasing amounts of His-tagged hRLC standard protein (564, 423, 282, 141, and 70.5 ng WT RLC, respectively), (7–9) decreasing amounts of M2β-S1 WT RLC (20, 10, and 5 µl, respectively), and (10–12) decreasing amounts of M2β-S1 K104E RLC (20, 10, and 5 µl, respectively). Concentration was calculated by gel densitometry and compared with Bradford assay of total M2β-S1 concentration as a measure of exchange efficiency. Blots from three protein preparations were averaged to calculate heavy chain:light chain ratio (± SD): 0.97 ± 0.11 M2β-S1:WT RLC and 0.96 ± 0.11 for M2β-S1 K104E RLC.
Figure S3.
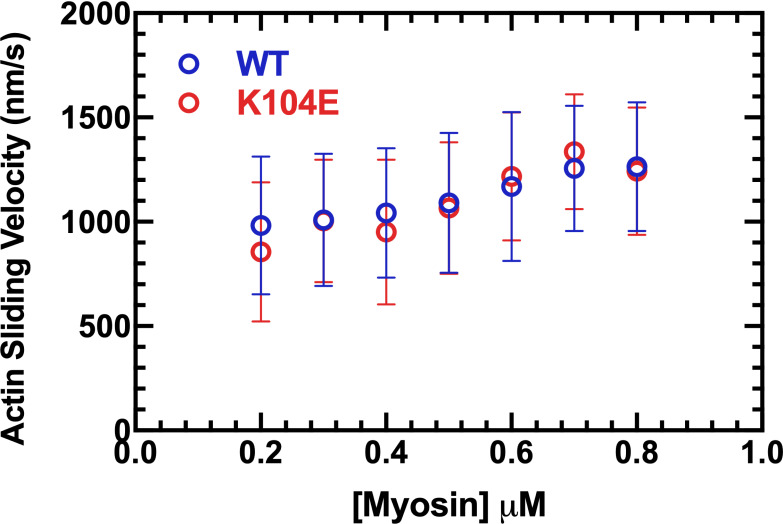
Density dependence of in vitro motility of human β-cardiac myosin. Actin sliding velocities from the in vitro motility assay depicted at increasing densities of biotinylated M2β-S1 adhered to a streptavidin-nitrocellulose–coated glass coverslip. For each density, filament velocities analyzed by FAST tracking software from 3 or 4 videos were compiled between two protein preparations. Videos with >20% stuck filaments were excluded. Error bars represent mean ± SD. Actin sliding velocities are similar between M2β-S1 WT and K104E across all myosin densities, with <13% difference at each density.
Figure S4.
Structural model of K104E mutation in M2α-S1. The effect of the K104E mutation in the RLC is depicted in the structural model of modified M2α-S1 (HBCprestrokeS1 Homology Model, Spudich Lab, Stanford University, Stanford, CA). (A) The murine α-MHC (Myh6, cyan) and the murine RLC (Myl1, purple) were threaded through the model of human cardiac S1 (http://spudlab.stanford.edu/homology-models) using Chimera Modeller (zDOPE scores = −0.47 and −0.71, respectively). The human isoform of K104E (red) was threaded through and overlaid upon the human WT isoform (blue). The IQ motifs (yellow) represent the sites of light chain binding to the lever arm. (B) Magnified view of overlaid WT and K104E RLC from inset in A. The N-terminal lobe of K104E is not predicted to shift in the model (left, nearly perfect overlap), while the C-terminal lobe shifts significantly (right), such that K104E is more closely associated to the lever arm. Yellow dotted lines indicate distance from functional groups of lysine (K104, blue) and glutamate (K104E, red) to nearby carbon of isoleucine, the first residue of the IQ motif, which measure 11.0 Å and 4.5 Å, respectively.
Results
Expression and purification of M2β-S1 with RLC exchange
SDS-PAGE was performed for each preparation to assess the removal of the endogenous murine RLC as well as reconstitution with the His-tagged human RLC isoform (Fig. S1). Differential migration of murine RLC versus His-tagged human RLC allowed qualitative assessment of the RLC exchange. To quantitatively assess the efficiency of RLC exchange, the ratio of M2βS1:RLC was determined from the total myosin concentration by Bradford assay, and the amount of RLC was measured by quantitative Western blotting (Fig. S2). The ratio of M2βS1:RLC was determined for three separate protein preparations and averaged. The average ratio (± SD) of M2β-S1 to RLC was 0.97 ± 0.11 for WT and 0.96 ± 0.11 for K104E, indicative of an efficient RLC exchange in the recombinant system. The amount of ELC before and after exchange was difficult to assess because of similar migration profiles of human His-tagged RLC and an endogenous ELC (Fig. S1).
Actin-activated ATPase
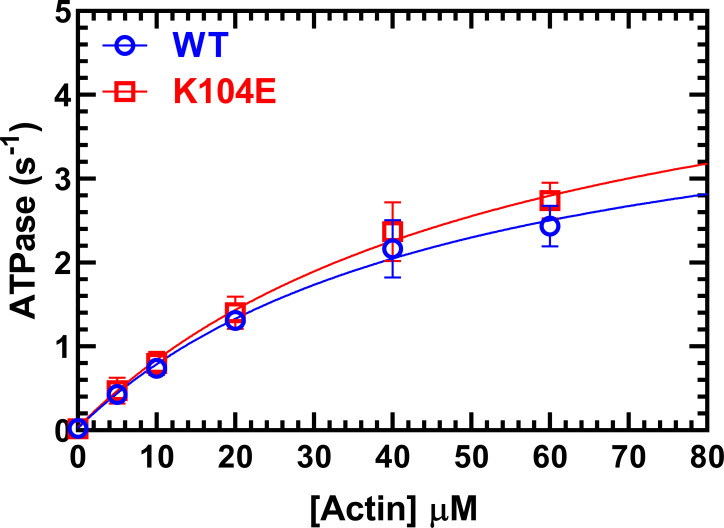
We examined the impact of K104E RLC in M2β-S1 constructs on actin-activated ATPase activity using the steady-state NADH-coupled ATPase assay (Fig. 3 and Table 1). There were no observed differences in steady-state actin-activated ATPase between M2β-S1 WT and K104E. The ATPase rates (± SD) between M2β-S1 with WT versus K104E RLC were similar in the absence of actin (0.02 ± 0.02 versus 0.04 ± 0.04 s–1) as well as the maximal actin-activated ATPase activity (4.5 ± 0.6 versus 5.4 ± 0.8 s–1), respectively. The actin concentration at which the ATPase rate is one half maximal was similar between the WT and K104E (49 ± 11 µM versus 57 ± 14 µM), respectively. The nonexchanged M2β-S1 controls also had similar ATPase parameters (Table 1).
Figure 3.
Actin-activated ATPase. The actin-activated ATPase activity of M2β-S1 WT RLC versus K104E RLC (0.1 µM) was examined as a function of actin concentration in MOPS 20 buffer at 25°C. The ATPase activity is plotted as a function of actin concentration and fit to a hyperbolic relationship to determine maximal ATPase and actin concentration at which ATPase is one-half maximal. The results represent an average of three experiments from three protein preparation (mean ± SD). See Table 1 for a summary of ATPase parameters.
Table 1. Summary of steady-state ATPase results for human M2β-S1 exchanged with K104E versus WT RLC.
| Steady-state ATPase values (± SD), n = 3 | WT | K104E | WT Non-ex |
|---|---|---|---|
| v0 (s–1) | 0.02 ± 0.02 | 0.04 ± 0.04 | 0.02 ± 0.01 |
| kcat (s–1) | 4.5 ± 0.6 | 5.4 ± 0.8 | 5.4 ± 1.3 |
| KATPase (µM) | 49 ± 11 | 57 ± 14 | 53 ± 22 |
KATPase, the actin concentration at which the ATPase rate is one half maximal actin-activated ATPase activity; kcat, maximal actin-activated ATPase activity; v0, absence of actin; Non-ex, non-exchanged myosin with endogenous RLC.
Transient kinetic analysis
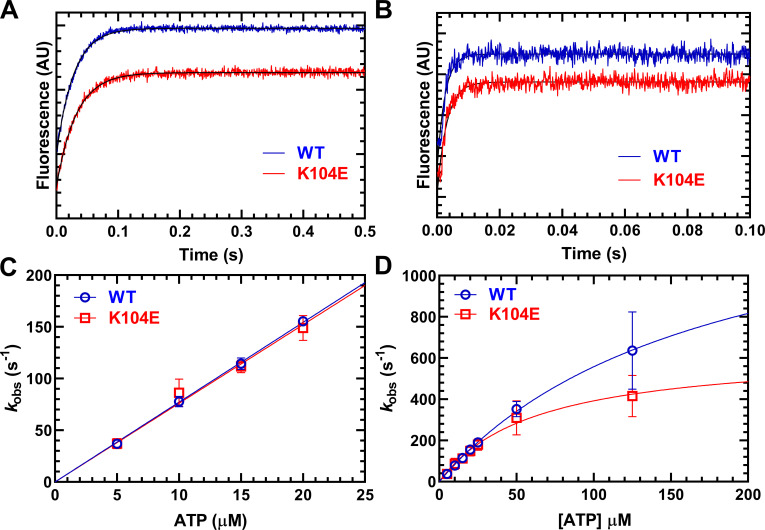
To further examine differences in the ATPase kinetics of WT and K104E M2β-S1, we measured two key steps that are associated with the detachment of myosin from actin (Table 2). The detachment of myosin from actin is limited by the rates of ADP release from actomyosin and ATP binding–induced dissociation from actin. We examined ATP-induced dissociation by mixing a pyrene actomyosin complex with increasing concentrations of ATP (5–125 µM; Fig. 4). Fluorescence transients indicative of unquenching of pyrene actin were fit to a single exponential function at all ATP concentrations (Fig. 4, A and B). At low ATP concentrations (5–20 µM ATP), the observed rate constant was linearly dependent on ATP concentration, which allowed determination of the second-order rate constant for ATP binding. We observed no difference in the second-order rate constant (μM–1⋅s–1) between WT (7.7 ± 0.1 µM–1⋅s–1) and K104E (7.6 ± 0.3 µM–1⋅s–1; Fig. 4 C), or in the maximal rate of ATP-induced dissociation, represented by the maximum value of the hyperbolic fit of the data (K104E = 636 ± 76; WT = 1,527 ± 358 s–1; P = 0.079; Fig. 4 D). We also observed no significant difference in the apparent affinity for ATP, as estimated by the K0.5 (ATP concentration at which kobs is one-half maximal) of the hyperbolic fit, in the mutant (62 ± 14 µM) compared with WT (174 ± 60 µM) M2β-S1.
Table 2. Summary of transient kinetic results for human M2β-S1 exchanged with K104E versus WT RLC.
| Rate/equilibrium constants (± SE) | WT | K104E |
|---|---|---|
| ATP binding (actomyosin)a, n = 3 | ||
| K’1Tk’+2T (µM–1·s–1) | 7.7 ± 0.1 | 7.6 ± 0.3 |
| K0.5 (µM) | 174 ± 60 | 62 ± 14 |
| k’+2T (s–1) | 1,527 ± 358 | 636 ± 76 |
| Actomyosin ADP release (pyrene)a, n = 4 | ||
| k’+D (s–1) | 242.5 ± 17.2 | 284.4 ± 9.9 |
| Actomyosin mantADP releaseb, n = 4 | ||
| k’+D (s–1) | 196.5 ± 15.6 | 157.1 ± 5.8 |
k’+2T, maximal rate of ATP-induced dissociation; K’1Tk’+2T, second-order rate constant for ATP binding. K0.5’, ATP concentration at which the observed rate constant is one-half maximal.
Pyrene actin fluorescence.
mantADP fluorescence.
Figure 4.
ATP-induced dissociation from actin. The experiment was performed by mixing a complex of M2β-S1 and pyrene actin with increasing concentrations of ATP (5–125 µM). (A and B) The fluorescence transients were best fit by a single exponential function at all ATP concentrations. Representative fluorescence transients at 5 µM ATP (A; kobs = 36.0 ± 0.2 and 31.1 ± 0.3 s–1 in WT and K104E, respectively) and 125 µM ATP (B; kobs = 553.6 ± 18.5 and 403.3 ± 12.0 s–1 in WT and K104E, respectively). (C) The second-order rate constant K’1Tk’+2 was determined from the linear-dependence on ATP concentration at low ATP concentrations (5–20 µM). (D) The maximal rate of ATP-induced dissociation (k’+2T) was determined from the hyperbolic fit of kobs as a function of ATP concentration. AU, arbitrary unit.
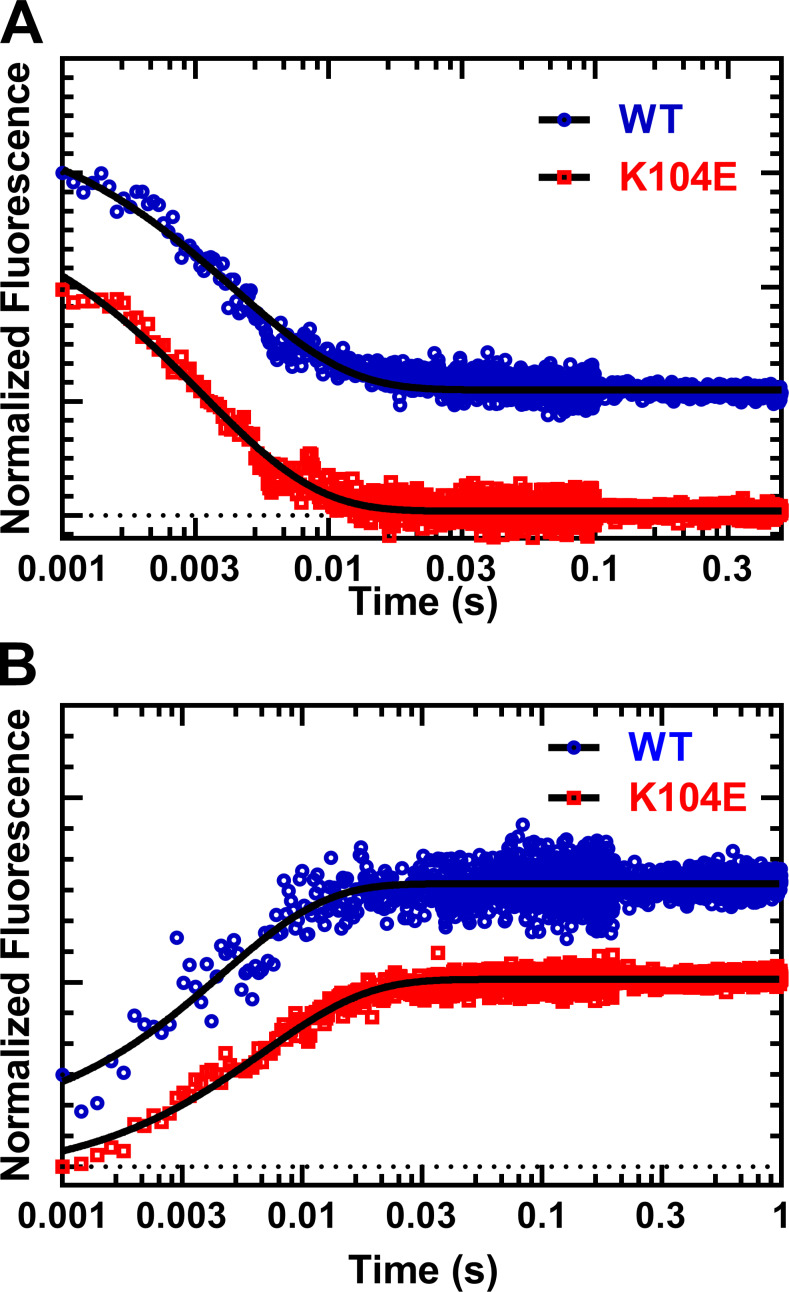
We examined the ADP release rate constant with two experimental methods, directly using mant-labeled ADP and indirectly with pyrene-labeled actin (Fig. 5 and Table 2). The release of mantADP was performed by mixing an actomyosin complex (M2β-S1, F-actin, and mantADP) with 1 mM ATP. The fluorescence transients were fit to a single exponential function, and the mantADP release rate constant was not significantly different in K104E (k’+D = 284.4 ± 9.9) compared with WT (k’+D = 242.5 ± 17.2; Fig. 5 A). The release of unlabeled ADP was performed by mixing a pyrene actomyosin.ADP complex (M2β-S1, pyrene-actin, and ADP) with 2 mM ATP. The rate of pyrene unquenching, which is limited by the rate of ADP release and ATP-induced dissociation in these conditions, was fit to a single exponential function (Fig. 5 B). We found the overall rate of detachment (k’+D) under these conditions was not statistically different (WT = 196.5 ± 15.6; K104E = 157.1 ± 5.8 s–1; P = 0.056). Therefore, we conclude that there is no difference between these myosin constructs in k’+D under physiological conditions of ATP (2 mM) and ADP (50 µM).
Figure 5.
ADP-release from actomyosin. Representative fluorescence transients of ADP release experiments using mantADP or pyrene actin. (A) We monitored mantADP dissociation by mixing acto–M2β-S1 complexed with mantADP with saturating ATP (1 mM), and the fluorescence transients were fit to a single exponential function (WT, kobs = 228.3 ± 3.8 s–1, and K104E, kobs = 297.5 s–1). (B) We examined ADP dissociation by mixing pyrene acto–M2β-S1.ADP with saturating ATP (2 mM), and the fluorescence transients were fit to a single exponential function (WT, kobs = 215.8 ± 8.9 s–1, and K104E, kobs = 145.0 ± 2.5 s–1).
Acting sliding velocities of M2β-S1 versus full-length α-cardiac myosin
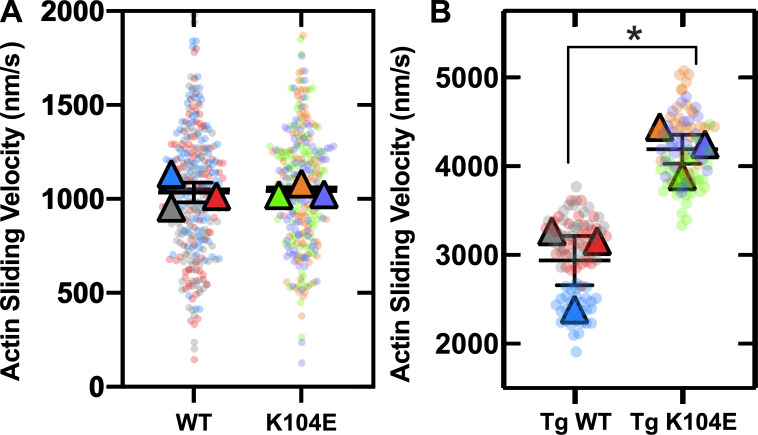
We also investigated the ability of our M2β-S1 constructs (WT versus K104E) to slide actin filaments in the unloaded in vitro motility assay. There were no significant differences in actin sliding velocities in M2β-S1 WT (1,036 ± 53 nm/s) versus K104E (1,043 ± 20 nm/s) at 0.5 µM myosin density across three distinct myosin protein preparations (Fig. 6 A and Table 3). Density dependence of myosin also was examined in both M2β-S1 WT and K104E across two protein preparations. For both myosin constructs, actin sliding velocity slightly increased with myosin density from 0.2 µM to 0.7 µM, and a plateau was observed between the final two concentrations of 0.7 and 0.8 µM (Fig. S3). Sliding velocities were similar between M2β-S1 WT and K104E at all of the studied densities.
Figure 6.
SuperPlots of in vitro motility assay from β-cardiac and α-cardiac myosin. (A) SuperPlot demonstrating 100 randomly sampled actin filament velocities analyzed by FAST (transparent circles, 100 of >2,500 filaments/experiment) and means from three experiments from distinct protein preparations (triangles) at 0.5 µM β-cardiac myosin. Student’s t test of sample means (n = 3) showed no difference (P = 0.8875) between WT (1,035 ± 53) and K104E (1,043 ± 20) in M2β-S1 constructs. (B) SuperPlot demonstrating 90 actin filament velocities analyzed manually using MTrackJ (transparent circles, 30 filaments/experiment) and means from experiments from three distinct ventricular myosin extractions (triangles) per group of Tg animals at 0.5 µM α-cardiac myosin. Student’s t test of sample means (n = 3) demonstrated a significant difference (*, P = 0.0176) between Tg WT (2,939 ± 278) and Tg K104E (4,194 ± 163) animals.
Table 3. Summary of in vitro motility results.
| Actin sliding velocity (± SE) | WT | K104E |
|---|---|---|
| Human M2β-S1, n = 3 experiments from three distinct protein preparations, 0.5 µM density | 1,036 ± 53 | 1,043 ± 20 |
| Mouse M2α-full length, n = 3 experiments from three distinct myosin extractions, 0.5 µM density | 2,939 ± 278 | 4,194 ± 163a |
P < 0.05.
In vitro motility also was employed to study the mechanical properties of full-length α-cardiac myosin, extracted from the ventricles of six Tg male mice (three WT versus three K104E) and attached directly to a nitrocellulose-coated coverslip at 0.5 µM density of myosin. A significant increase in actin sliding velocity (43%) was observed in Tg K104E (4,194 ± 163 nm/s) compared with Tg WT (2,939 ± 278 nm/s) when comparing sample means across three experiments (Fig. 6 B).
Expression and phosphorylation of RLC in Tg mice
Purified myosin samples were analyzed by MS in order to determine the degree of incorporation of the human WT and K104E RLC into myosin. By comparison with myosin purified from WT mouse hearts, 95 ± 2% (SD, n = 3) of the endogenous mouse RLC was replaced with human WT RLC in the Tg WT animals. In contrast, only 54 ± 5% (SD, n = 3) of the endogenous mouse RLC was replaced with human K104E RLC in the Tg K104E animals. The mono-and bis-phosphorylated mouse phosphopeptides (IEGGSpSNVFSMFEQTQIQEFK and IEGGSpSpNVFSMFEQTQIQEFK) and mono-phosphorylated human peptide (AGGANSpNVFSMFEQTQIQEFK) were found in each sample, where p denotes the site of phosphorylation. These sites corresponded to serines 14 and 15 in the mouse sequence and serine 15 in the human sequence. The relative ratio of the phosphopeptide and nonphosphopeptides did not differ (P > 0.2) between samples (Table 4). Note that these relative ratios do not account for the difference in the ionization efficiencies between the peptides and should not be interpreted as absolute phosphorylation levels. These results demonstrate that the human WT RLC incorporates more efficiently into the endogenous mouse myosin than the human K104E RLC.
Table 4. Summary of MS data from α-cardiac myosin extracted from mouse hearts.
| Tg WT RLC | Tg K104E RLC | |
|---|---|---|
| Protein (%) | ||
| Mouse WT RLC | 4.3 ± 1.8 | 46.6±5.1 |
| Human WT RLC | 95.7 ± 1.8 | — |
| Human K104E RLC | — | 53.4 ± 5.1 |
| α-Cardiac myosin (Myh6) | 98.3 ± 0.5 | 97.1 ± 1.7 |
| Peptidea | ||
| IEGGSpSNVFSMFEQTQIQEFK (mRLC, singly phosphorylated) | 10.0 ± 5.6 | 5.1 ± 0.3 |
| IEGGSpSpNVFSMFEQTQIQEFK (mRLC, doubly phosphorylated) | 0.6 ± 0.3 | 0.3 ± 0.1 |
| AGGANSpNVFSMFEQTQIQEFK (hRLC, phosphorylated) | 11.9 ± 10.4 | 3.4 ± 1.2 |
All data are averages ± SD where n = 3. hRLC, human RLC; mRLC, murine RLC.
Data are expressed as the ratio of the phosphopeptide and the nonphosphopeptide. These ratios do not account for the difference in the ionization efficiencies between the peptides and should not be interpreted as absolute phosphorylation levels.
Discussion
This study investigated the impact of an HCM-associated RLC mutation, K104E, on cardiac myosin motor function. We successfully exchanged the mutant RLC onto human M2β-S1 and compared it to its WT counterpart. We demonstrate that the K104E RLC mutation does not alter the intrinsic myosin motor properties of human M2β-S1, including ATPase kinetics and actin filament sliding in the in vitro motility assay. Interestingly, experiments with full-length mouse α-cardiac myosin isolated from Tg mouse hearts that overexpress either human WT RLC or K104E RLC reveal that the mutant produced enhanced unloaded actin sliding velocities, similar to previous results (Huang et al., 2014). The mutation also reduces the ability of human RLC to incorporate into α-cardiac myosin in Tg animals. We conclude that the K104E mutation may alter RLC interactions with the myosin lever arm, and the impact of the mutation may depend on the MHC isoform and/or the interactions with the subfragment-2 (S2) region of the myosin tail.
Impact of K104E on human M2β-S1 motor function
Although several studies have examined the impact of HCM mutations in the RLC, most have used murine α-cardiac myosin, and some have exchanged human RLC onto porcine β-cardiac myosin. In the current study, we used recombinantly expressed human M2β-S1 to investigate the impact of an HCM-associated mutation, highlighting the clinically relevant human myosin isoform. We demonstrate the K104E does not alter the intrinsic motor properties of human M2β-S1 in unloaded conditions. The mutation had no effect on actin-activated ATPase activity (Fig. 3). In addition, actin sliding velocities were very similar in M2β-S1 containing WT or K104E RLC, regardless of myosin surface density (Fig. 6 A and Fig. S3). Since the mutation is not located near the active site, these results may be expected, though long-range allosteric changes in myosin have been reported in the literature (Gunther et al., 2019; Trivedi et al., 2020). Previous work has provided evidence that other RLC mutations can alter the mechanical properties of porcine β-cardiac myosin in the in vitro motility assay in both unloaded (Farman et al., 2014) and loaded conditions (Greenberg et al., 2009; Greenberg et al., 2010; Karabina et al., 2015). The current work highlights that the K104E mutation does not impact the unloaded mechanochemistry of M2β-S1.
We investigated key transient kinetic steps in the ATPase cycle associated with the detachment of myosin from actin, which is an important determinant of shortening velocity. Since the ADP release rate constant is typically slower than the maximum rate of ATP-induced dissociation, ADP release is proposed to limit the overall rate of detachment (Siemankowski et al., 1985). However, if the maximum rate of ATP-induced dissociation is slowed, it also can contribute to the overall detachment rate (Nyitrai et al., 2006). We did not observe a significant difference in the ADP release rate constant or ATP-induced dissociation of β-myosin from actin (Table 2, Fig. 4, and Fig. 5 A). We also found that the overall rate of detachment in the presence of physiological ATP and ADP concentrations was not different between M2β-S1 WT versus K104E (Table 2 and Fig. 5 B). While the detachment rate constants were not significantly different, the results suggest that under other conditions, such as the presence of load, the detachment kinetics may be depressed in K104E, as has been suggested in other RLC mutations such as N47K and R58Q (Greenberg et al., 2010). The ADP release rate constant is known to be altered by the presence of load and is crucial for myosin to adapt its ATPase cycle to different physiological conditions (Holmes et al., 2004). Small changes in ADP-dependent detachment kinetics can have profound effects on the force–velocity properties of cardiac myosin (Greenberg et al., 2014). Thus, future studies in which the detachment kinetics can be examined in the presence of load may provide key insight into the impact of the K104E and other HCM-associated RLC mutations.
Impact of K104E on full-length mouse α-cardiac myosin
Previous work in the Tg mouse model of K104E demonstrated that senescent animals develop HCM (Huang et al., 2014). Our work mirrors the previous study in that we observed a 43% increase in unloaded actin sliding velocities with the Tg K104E mutant compared with Tg WT counterparts (Fig. 6 B; Huang et al., 2014). Previous work in this model also found an increase in the rate of cross-bridge binding to thin filaments and the rate of execution of the power stroke in contracting myofibrils from ex vivo left ventricles of Tg K104E mice (Duggal et al., 2015). Purified myosin from this tissue also revealed a slightly slower second-order ATP-binding constant for Tg K104E compared with Tg WT myosin (Duggal et al., 2015). Collectively, these findings in full-length α-cardiac myosin are contrary to those we observed in M2β-S1, supporting the notion that the effects of the mutation on intrinsic motor properties may be dependent on the length or isoform of the myosin backbone.
We determined the efficiency of RLC incorporation in the full-length myosin purified from Tg mouse hearts, in which K104E RLC had less efficient incorporation (53%) than WT RLC (96%), as per MS (Table 4). This contrasts the previous study that found nearly 100% exchange efficiency of both the mutant and WT RLC, as per gel densitometry of atrial tissue (Huang et al., 2014). Intriguingly, the K104E mutation has been shown to decrease α-helical content in circular dichroism spectroscopy experiments with purified RLC (Huang and Szczesna-Cordary, 2015). Our structural modeling studies suggest that the conformation of the α-helix containing lysine-104 is altered by introducing glutamate at this position, which results in a reorientation of the C-terminal lobe of RLC toward the lever arm helix (Fig. S4; Sali and Blundell, 1993). Thus, structural changes induced by the K104E mutation may weaken the binding affinity of the RLC for the lever arm, explaining the reduced incorporation we observed in the Tg mouse model. We also found that there was no difference in the degree of RLC phosphorylation in the Tg WT and Tg K104E mice, which contradicts the earlier study that found a twofold decrease in phosphorylation levels in Tg K104E mice. Collectively, our work indicates that a 53% incorporation of K104E RLC is sufficient to cause an enhancement in full-length α-cardiac myosin unloaded sliding velocity and that this impact is not dependent on significant changes in RLC phosphorylation levels.
Several potential mechanisms could explain how the mutation enhances unloaded sliding velocity in full-length mouse α-cardiac myosin but does not impact the intrinsic motor properties of human M2β-S1. The α-cardiac myosin isoform is known to have a significantly faster ATPase activity and unloaded sliding velocity (Deacon et al., 2012), and thus the mutation may impact α-cardiac differently than β-cardiac myosin. The ATPase cycle has several key differences, including a 10-fold faster ADP release rate constant in α-cardiac myosin (Deacon et al., 2012). Previous work found that a classic heavy chain HCM mutation, R403Q, has a different impact on α- versus β-cardiac myosin ATPase activity and in vitro motility (Lowey et al., 2008; Lowey et al., 2013). Therefore, α/β differences are likely to contribute to the differential impact of the K104E mutation in these two cardiac myosin isoforms. Interestingly, one could argue that the different ratios of murine versus human isoforms in the α-cardiac myosin may slow the in vitro actin sliding velocity compared with α-cardiac myosin containing its native mouse RLC. However, a previous motility study demonstrated no difference in α-cardiac myosin containing the human versus the endogenous murine RLC (i.e., non-Tg animals; Greenberg et al., 2009).
Another important factor may be the different lengths of the constructs (S1 versus full-length cardiac myosin), and the ability to form the auto-inhibited state or super-relaxed (SRX) state of myosin. It is proposed that in the SRX state, myosin head–head and head–rod interactions that allow the formation of the interacting heads motif (IHM) significantly slow the ATPase activity and prevent interactions with actin (Alamo et al., 2008; Hooijman et al., 2011; McNamara et al., 2015). Thus, since human M2β-S1 lacks the coiled-coil rod, it cannot form inhibitory head–rod interactions. Interestingly, the previous study demonstrated that the ATPase activity of full-length mouse α-cardiac myosin purified from mouse hearts was enhanced twofold in the K104E mutant (Huang et al., 2014). One possible interpretation of these results is that the mutant may destabilize the SRX state and increase the number of myosin heads available to interact with actin, increasing the overall ATPase rate. The K104E RLC mutation is located in a region that could interact with the S2 region of the rod domain, which is known to be important for forming the IHM (Alamo et al., 2017). The change in charge at the 104 position from a positive lysine to a negative glutamate residue is consistent with disruption of the interactions with S2, which carries a net negative charge (Blankenfeldt et al., 2006). K104E also may alter SRX state formation due to disruptions in RLC–lever arm interactions, as RLC structural dynamics were found to be important in forming the IHM of smooth muscle myosin (Vileno et al., 2011). Therefore, future studies that can examine the impact of K104E and other RLC mutations on the formation of the IHM/SRX state could reveal crucial information about the structure–function relationships that mediate this important regulatory state. Intriguingly, previous studies in HCM-associated RLC mutant R58Q demonstrated this mutant stabilizes the SRX state (Kampourakis et al., 2018; Yadav et al., 2019a), contrary to a leading hypothesis in the field that HCM-associated mutations destabilize the SRX state (Trivedi et al., 2018).
We acknowledge that our study does not come without limitations. Our humanized M2β-S1 constructs contain endogenous murine ELC isoforms, native to the C2C12 myoblast expression system. Our previous work demonstrated that Myl1/3 (fast skeletal) and Myl4 (fetal) are both present in these constructs, which are 75% and 79% homologous to human ventricular cardiac ELC (MYL3), respectively (Swenson et al., 2017). Thus, we recognize that these murine ELC isoforms may impact lever arm structure and function in our M2β-S1 study and that fully humanized constructs (human M2β-S1, RLC, and ELC) are most indicative of cardiac myosin function in humans. In addition, we recognize that the study of either full-length α- versus β-cardiac myosin or M2α-S1 versus M2β-S1 would make for direct comparison and would enhance the interpretation of our results. However, it is difficult to produce full-length β-cardiac myosin in the recombinant expression system we used. In addition to various lengths of myosin constructs, we also recognize that HCM-associated mutations should be studied in the presence of load when possible.
Conclusions
In the current study, we present a powerful approach to examine the impact of an RLC mutation associated with inherited heart disease using both recombinantly expressed human cardiac myosin and a Tg mouse model. We found that the K104E RLC mutation, for which there is limited clinical data, does not disrupt the intrinsic motor properties of human M2β-S1. However, the same mutation in full-length α-cardiac myosin demonstrates that the mutation has a dramatic impact on the motor properties of this isoform, which may be attributed to inherent isoform differences, myosin autoregulatory states, and/or disrupted RLC–lever arm interactions. Tg animal models of HCM-associated mutations are irreplaceable, as they leverage the ability to study the effects of a mutant at the molecular, cellular, tissue, and organ levels. However, the differential impact we observed in the two myosin isoforms highlights the importance of evaluating the impact of mutations in human cardiac myosin as well. Overall, we suggest future HCM studies that can utilize both Tg animal models and recombinant expression systems will improve our understanding of mutation-associated mechanisms and disease pathogenesis.
Acknowledgments
Henk L. Granzier served as editor.
We would like to acknowledge Rohini Desetty for her outstanding technical assistance in the Yengo laboratory at Penn State College of Medicine. We also acknowledge C. Kurtis Mann, Hossein Sharifi, and Alexus Rockward at the University of Kentucky for their assistance with the Tg mouse colonies.
This work was supported by National Institutes of Health grants HL127699 (to C.M. Yengo), HL133359 (to K.S. Campbell and J.F. Wenk), HL124041 (to M.J. Previs), and HL143830 (to D. Szczesna-Cordary).
The authors declare no competing financial interests.
Author contributions: C.M. Yengo, D. Szczesna-Cordary, K.S. Campbell, and J.F. Wenk conceived the study and supervised each step of the work. F.H. Moonschi maintained the mouse colony and harvested the heart tissue. D.V. Rasicci and O. Kirkland, Jr. prepared the myosin constructs and performed and analyzed in vitro motility assays. C.M. Yengo designed, performed, and analyzed stopped-flow experiments. N.B. Wood and M.J. Previs performed the MS. D.V. Rasicci wrote the first draft of the paper. All authors contributed to the final version of the manuscript.
Footnotes
This work is part of a special collection on myofilament function and disease.
References
- Aksel, T., Choe Yu E., Sutton S., Ruppel K.M., and Spudich J.A.. 2015. Ensemble force changes that result from human cardiac myosin mutations and a small-molecule effector. Cell Rep. 11:910–920. 10.1016/j.celrep.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamo, L., Wriggers W., Pinto A., Bártoli F., Salazar L., Zhao F.Q., Craig R., and Padrón R.. 2008. Three-dimensional reconstruction of tarantula myosin filaments suggests how phosphorylation may regulate myosin activity. J. Mol. Biol. 384:780–797. 10.1016/j.jmb.2008.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamo, L., Ware J.S., Pinto A., Gillilan R.E., Seidman J.G., Seidman C.E., and Padrón R.. 2017. Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes. eLife. 6:e24634. 10.7554/eLife.24634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcalai, R., Seidman J.G., and Seidman C.E.. 2008. Genetic basis of hypertrophic cardiomyopathy: from bench to the clinics. J. Cardiovasc. Electrophysiol. 19:104–110. [DOI] [PubMed] [Google Scholar]
- Alpert, N.R., Brosseau C., Federico A., Krenz M., Robbins J., and Warshaw D.M.. 2002. Molecular mechanics of mouse cardiac myosin isoforms. Am. J. Physiol. Heart Circ. Physiol. 283:H1446–H1454. 10.1152/ajpheart.00274.2002 [DOI] [PubMed] [Google Scholar]
- Andersen, P.S., Havndrup O., Bundgaard H., Moolman-Smook J.C., Larsen L.A., Mogensen J., Brink P.A., Børglum A.D., Corfield V.A., Kjeldsen K., et al. 2001. Myosin light chain mutations in familial hypertrophic cardiomyopathy: phenotypic presentation and frequency in Danish and South African populations. J. Med. Genet. 38:E43. 10.1136/jmg.38.12.e43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenfeldt, W., Thomä N.H., Wray J.S., Gautel M., and Schlichting I.. 2006. Crystal structures of human cardiac beta-myosin II S2-Delta provide insight into the functional role of the S2 subfragment. Proc. Natl. Acad. Sci. USA. 103:17713–17717. 10.1073/pnas.0606741103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, A.N., Mahajan P., Knapp S., Barton H., Sweeney H.L., Kamm K.E., and Stull J.T.. 2016. Cardiac myosin light chain is phosphorylated by Ca2+/calmodulin-dependent and -independent kinase activities. Proc. Natl. Acad. Sci. USA. 113:E3824–E3833. 10.1073/pnas.1600633113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criddle, A.H., Geeves M.A., and Jeffries T.. 1985. The use of actin labelled with N-(1-pyrenyl)iodoacetamide to study the interaction of actin with myosin subfragments and troponin/tropomyosin. Biochem. J. 232:343–349. 10.1042/bj2320343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz, E.M., and Ostap E.M.. 2009. Kinetic and equilibrium analysis of the myosin ATPase. Methods Enzymol. 455:157–192. 10.1016/S0076-6879(08)04206-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon, J.C., Bloemink M.J., Rezavandi H., Geeves M.A., and Leinwand L.A.. 2012. Identification of functional differences between recombinant human α and β cardiac myosin motors. Cell. Mol. Life Sci. 69:2261–2277. 10.1007/s00018-012-0927-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal, D., Nagwekar J., Rich R., Huang W., Midde K., Fudala R., Das H., Gryczynski I., Szczesna-Cordary D., and Borejdo J.. 2015. Effect of a myosin regulatory light chain mutation K104E on actin-myosin interactions. Am. J. Physiol. Heart Circ. Physiol. 308:H1248–H1257. 10.1152/ajpheart.00834.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farman, G.P., Muthu P., Kazmierczak K., Szczesna-Cordary D., and Moore J.R.. 2014. Impact of familial hypertrophic cardiomyopathy-linked mutations in the NH2 terminus of the RLC on β-myosin cross-bridge mechanics. J Appl Physiol (1985). 117:1471–1477. 10.1152/japplphysiol.00798.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, M.J., Watt J.D., Jones M., Kazmierczak K., Szczesna-Cordary D., and Moore J.R.. 2009. Regulatory light chain mutations associated with cardiomyopathy affect myosin mechanics and kinetics. J. Mol. Cell. Cardiol. 46:108–115. 10.1016/j.yjmcc.2008.09.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, M.J., Kazmierczak K., Szczesna-Cordary D., and Moore J.R.. 2010. Cardiomyopathy-linked myosin regulatory light chain mutations disrupt myosin strain-dependent biochemistry. Proc. Natl. Acad. Sci. USA. 107:17403–17408. 10.1073/pnas.1009619107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, M.J., Shuman H., and Ostap E.M.. 2014. Inherent force-dependent properties of β-cardiac myosin contribute to the force-velocity relationship of cardiac muscle. Biophys. J. 107:L41–L44. 10.1016/j.bpj.2014.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther, L.K., Rohde J.A., Tang W., Walton S.D., Unrath W.C., Trivedi D.V., Muretta J.M., Thomas D.D., and Yengo C.M.. 2019. Converter domain mutations in myosin alter structural kinetics and motor function. J. Biol. Chem. 294:1554–1567. 10.1074/jbc.RA118.006128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, K.C., Trentham D.R., Simmons R., Nyitrai M., and Geeves M.A.. 2004. Adenosine diphosphate and strain sensitivity in myosin motors. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359:1867–1877. 10.1098/rstb.2004.1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooijman, P., Stewart M.A., and Cooke R.. 2011. A new state of cardiac myosin with very slow ATP turnover: a potential cardioprotective mechanism in the heart. Biophys. J. 100:1969–1976. 10.1016/j.bpj.2011.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houdusse, A., and Sweeney H.L.. 2016. How myosin generates force on actin filaments. Trends Biochem. Sci. 41:989–997. 10.1016/j.tibs.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W., and Szczesna-Cordary D.. 2015. Molecular mechanisms of cardiomyopathy phenotypes associated with myosin light chain mutations. J. Muscle Res. Cell Motil. 36:433–445. 10.1007/s10974-015-9423-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W., Liang J., Kazmierczak K., Muthu P., Duggal D., Farman G.P., Sorensen L., Pozios I., Abraham T.P., Moore J.R., et al. 2014. Hypertrophic cardiomyopathy associated Lys104Glu mutation in the myosin regulatory light chain causes diastolic disturbance in mice. J. Mol. Cell. Cardiol. 74:318–329. 10.1016/j.yjmcc.2014.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampourakis, T., Ponnam S., and Irving M.. 2018. Hypertrophic cardiomyopathy mutation R58Q in the myosin regulatory light chain perturbs thick filament-based regulation in cardiac muscle. J. Mol. Cell. Cardiol. 117:72–81. 10.1016/j.yjmcc.2018.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabina, A., Kazmierczak K., Szczesna-Cordary D., and Moore J.R.. 2015. Myosin regulatory light chain phosphorylation enhances cardiac β-myosin in vitro motility under load. Arch. Biochem. Biophys. 580:14–21. 10.1016/j.abb.2015.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron, S.J., Toyoshima Y.Y., Uyeda T.Q., and Spudich J.A.. 1991. Assays for actin sliding movement over myosin-coated surfaces. Methods Enzymol. 196:399–416. 10.1016/0076-6879(91)96035-P [DOI] [PubMed] [Google Scholar]
- Lord, S.J., Velle K.B., Mullins R.D., and Fritz-Laylin L.K.. 2020. SuperPlots: Communicating reproducibility and variability in cell biology. J. Cell Biol. 219:e202001064. 10.1083/jcb.202001064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowey, S., Lesko L.M., Rovner A.S., Hodges A.R., White S.L., Low R.B., Rincon M., Gulick J., and Robbins J.. 2008. Functional effects of the hypertrophic cardiomyopathy R403Q mutation are different in an alpha- or beta-myosin heavy chain backbone. J. Biol. Chem. 283:20579–20589. 10.1074/jbc.M800554200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowey, S., Bretton V., Gulick J., Robbins J., and Trybus K.M.. 2013. Transgenic mouse α- and β-cardiac myosins containing the R403Q mutation show isoform-dependent transient kinetic differences. J. Biol. Chem. 288:14780–14787. 10.1074/jbc.M113.450668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara, J.W., Li A., Dos Remedios C.G., and Cooke R.. 2015. The role of super-relaxed myosin in skeletal and cardiac muscle. Biophys. Rev. 7:5–14. 10.1007/s12551-014-0151-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering, E., Dzyubachyk O., and Smal I.. 2012. Methods for cell and particle tracking. Methods Enzymol. 504:183–200. 10.1016/B978-0-12-391857-4.00009-4 [DOI] [PubMed] [Google Scholar]
- Nag, S., Trivedi D.V., Sarkar S.S., Adhikari A.S., Sunitha M.S., Sutton S., Ruppel K.M., and Spudich J.A.. 2017. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat. Struct. Mol. Biol. 24:525–533. 10.1038/nsmb.3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyitrai, M., Rossi R., Adamek N., Pellegrino M.A., Bottinelli R., and Geeves M.A.. 2006. What limits the velocity of fast-skeletal muscle contraction in mammals? J. Mol. Biol. 355:432–442. 10.1016/j.jmb.2005.10.063 [DOI] [PubMed] [Google Scholar]
- O’Leary, T.S., Snyder J., Sadayappan S., Day S.M., and Previs M.J.. 2019. MYBPC3 truncation mutations enhance actomyosin contractile mechanics in human hypertrophic cardiomyopathy. J. Mol. Cell. Cardiol. 127:165–173. 10.1016/j.yjmcc.2018.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee, J.D., and Spudich J.A.. 1982. Purification of muscle actin. Methods Enzymol. 85(Pt B):164–181. 10.1016/0076-6879(82)85020-9 [DOI] [PubMed] [Google Scholar]
- Rayment, I., Rypniewski W.R., Schmidt-Bäse K., Smith R., Tomchick D.R., Benning M.M., Winkelmann D.A., Wesenberg G., and Holden H.M.. 1993. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 261:50–58. 10.1126/science.8316857 [DOI] [PubMed] [Google Scholar]
- Sali, A., and Blundell T.L.. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815. 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- Siemankowski, R.F., Wiseman M.O., and White H.D.. 1985. ADP dissociation from actomyosin subfragment 1 is sufficiently slow to limit the unloaded shortening velocity in vertebrate muscle. Proc. Natl. Acad. Sci. USA. 82:658–662. 10.1073/pnas.82.3.658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitbon, Y.H., Yadav S., Kazmierczak K., and Szczesna-Cordary D.. 2020. Insights into myosin regulatory and essential light chains: a focus on their roles in cardiac and skeletal muscle function, development and disease. J. Muscle Res. Cell Motil. 41:313–327. 10.1007/s10974-019-09517-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney, H.L., and Hammers D.W.. 2018. Muscle Contraction. Cold Spring Harb. Perspect. Biol. 10:a023200. 10.1101/cshperspect.a023200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney, H.L., Bowman B.F., and Stull J.T.. 1993. Myosin light chain phosphorylation in vertebrate striated muscle: regulation and function. Am. J. Physiol. 264:C1085–C1095. 10.1152/ajpcell.1993.264.5.C1085 [DOI] [PubMed] [Google Scholar]
- Swenson, A.M., Tang W., Blair C.A., Fetrow C.M., Unrath W.C., Previs M.J., Campbell K.S., and Yengo C.M.. 2017. Omecamtiv Mecarbil Enhances the Duty Ratio of Human β-Cardiac Myosin Resulting in Increased Calcium Sensitivity and Slowed Force Development in Cardiac Muscle. J. Biol. Chem. 292:3768–3778. 10.1074/jbc.M116.748780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesna-Cordary, D., Guzman G., Ng S.S., and Zhao J.. 2004. Familial hypertrophic cardiomyopathy-linked alterations in Ca2+ binding of human cardiac myosin regulatory light chain affect cardiac muscle contraction. J. Biol. Chem. 279:3535–3542. 10.1074/jbc.M307092200 [DOI] [PubMed] [Google Scholar]
- Tang, W., Blair C.A., Walton S.D., Málnási-Csizmadia A., Campbell K.S., and Yengo C.M.. 2017. Modulating Beta-Cardiac Myosin Function at the Molecular and Tissue Levels. Front. Physiol. 7:659. 10.3389/fphys.2016.00659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, W., Unrath W.C., Desetty R., and Yengo C.M.. 2019. Dilated cardiomyopathy mutation in the converter domain of human cardiac myosin alters motor activity and response to omecamtiv mecarbil. J. Biol. Chem. 294:17314–17325. 10.1074/jbc.RA119.010217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi, D.V., Adhikari A.S., Sarkar S.S., Ruppel K.M., and Spudich J.A.. 2018. Hypertrophic cardiomyopathy and the myosin mesa: viewing an old disease in a new light. Biophys. Rev. 10:27–48. 10.1007/s12551-017-0274-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi, D.V., Nag S., Spudich A., Ruppel K.M., and Spudich J.A.. 2020. The Myosin Family of Mechanoenzymes: From Mechanisms to Therapeutic Approaches. Annu. Rev. Biochem. 89:667–693. 10.1146/annurev-biochem-011520-105234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vileno, B., Chamoun J., Liang H., Brewer P., Haldeman B.D., Facemyer K.C., Salzameda B., Song L., Li H.C., Cremo C.R., and Fajer P.G.. 2011. Broad disorder and the allosteric mechanism of myosin II regulation by phosphorylation. Proc. Natl. Acad. Sci. USA. 108:8218–8223. 10.1073/pnas.1014137108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward, M., Previs M.J., Mader T.J., and Debold E.P.. 2015. Modifications of myofilament protein phosphorylation and function in response to cardiac arrest induced in a swine model. Front. Physiol. 6:199. 10.3389/fphys.2015.00199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X., Clack B.A., Zhi G., Stull J.T., and Cremo C.R.. 1999. Phosphorylation-dependent structural changes in the regulatory light chain domain of smooth muscle heavy meromyosin. J. Biol. Chem. 274:20328–20335. 10.1074/jbc.274.29.20328 [DOI] [PubMed] [Google Scholar]
- Yadav, S., Kazmierczak K., Liang J., Sitbon Y.H., and Szczesna-Cordary D.. 2019a. Phosphomimetic-mediated in vitro rescue of hypertrophic cardiomyopathy linked to R58Q mutation in myosin regulatory light chain. FEBS J. 286:151–168. 10.1111/febs.14702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, S., Sitbon Y.H., Kazmierczak K., and Szczesna-Cordary D.. 2019b. Hereditary heart disease: pathophysiology, clinical presentation, and animal models of HCM, RCM, and DCM associated with mutations in cardiac myosin light chains. Pflugers Arch. 471:683–699. 10.1007/s00424-019-02257-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yotti, R., Seidman C.E., and Seidman J.G.. 2019. Advances in the Genetic Basis and Pathogenesis of Sarcomere Cardiomyopathies. Annu. Rev. Genomics Hum. Genet. 20:129–153. 10.1146/annurev-genom-083118-015306 [DOI] [PubMed] [Google Scholar]