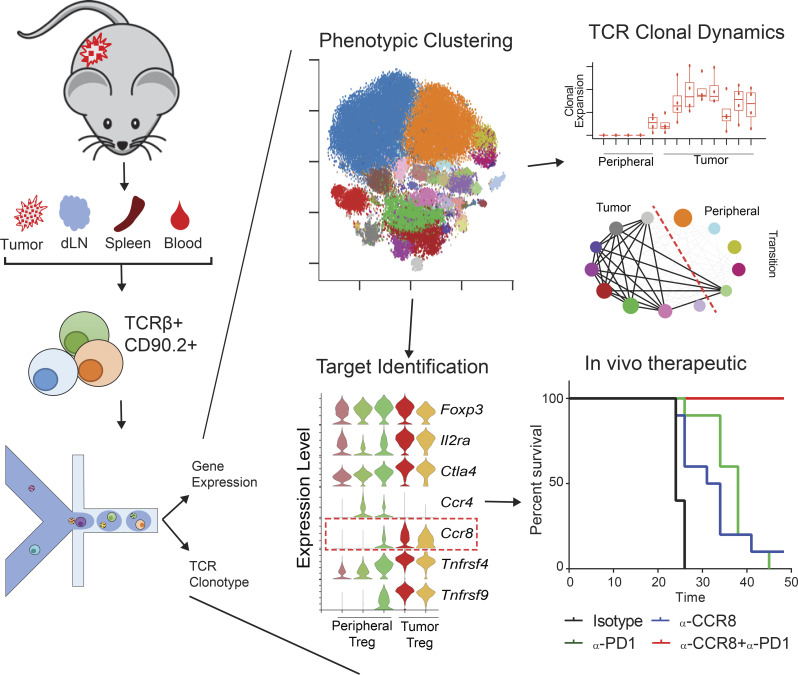
Single cell RNAseq analyses were performed on T cells from MC38 and CT26 tumor models. PCA-based sub-clustering and STARTRAC analyses revealed fine-tuned T cell subsets and dynamic in tumor. CCR8 was identified as a novel target on Tregs to synergize with anti–PD-1 for cancer immunotherapy.
Abstract
Single-cell RNA sequencing is a powerful tool to examine cellular heterogeneity, novel markers and target genes, and therapeutic mechanisms in human cancers and animal models. Here, we analyzed single-cell RNA sequencing data of T cells obtained from multiple mouse tumor models by PCA-based subclustering coupled with TCR tracking using the STARTRAC algorithm. This approach revealed various differentiated T cell subsets and activation states, and a correspondence of T cell subsets between human and mouse tumors. STARTRAC analyses demonstrated peripheral T cell subsets that were developmentally connected with tumor-infiltrating CD8+ cells, CD4+ Th1 cells, and T reg cells. In addition, large amounts of paired TCRα/β sequences enabled us to identify a specific enrichment of paired public TCR clones in tumor. Finally, we identified CCR8 as a tumor-associated T reg cell marker that could preferentially deplete tumor-associated T reg cells. We showed that CCR8-depleting antibody treatment provided therapeutic benefit in CT26 tumors and synergized with anti–PD-1 treatment in MC38 and B16F10 tumor models.
Graphical Abstract
Introduction
Immunotherapies including anti–programmed death 1 (PD-1) and anti–CTLA-4 antibodies have achieved major progress in the treatment of various cancers over the past decade (Sharma and Allison, 2015). Although many patients have benefited from these revolutionary treatments, many cancers respond inadequately due to the ability of cancer cells to develop various escape mechanisms and evade immune surveillance (Sharma et al., 2017). Understanding these immune evasion mechanisms holds the key for the development of the next generation of immunotherapies. T cells play a central role in mediating antitumor immunity (Fridman et al., 2012). Solid tumors are frequently infiltrated with a heterogenous population of T cells as part of the endogenous antitumor response. Cytotoxic T cells inside tumors frequently display an exhausted or dysfunctional phenotype marked by the upregulation of various coinhibitory receptors, such as PD-1, TIM-3, and LAG-3 (Pardoll, 2012; Ahmadzadeh et al., 2009). Checkpoint inhibitor therapies, such as anti–PD-1 antibodies, block these inhibitory pathways and reinvigorate the cytolytic capacity of these T cells (Egen et al., 2020; Huang et al., 2017; Blank et al., 2019). Recent studies further divide exhausted T cells into predysfunctional (progenitor or stem like), early dysfunctional, and late dysfunctional (terminally exhausted) subsets (Im et al., 2016; van der Leun et al., 2020). In addition to exhausted T cells, cytotoxic T cells in tumors and tissues display various other phenotypes, including effector T cells (Temra/TEFF cells), central memory T cells (TCM cells), resident memory T cells, effector memory T cells (TEM cells), and stem cell–like phenotypes (Tsc; Zhang et al., 2018; Yost et al., 2019; Yu et al., 2020; van der Leun et al., 2020). Although some of these CD8+ cells are developmentally linked to exhausted T cells, their precise contributions to the antitumor response are not fully understood.
Conventional CD4+ T cells and CD4+Foxp3+ regulatory T cells (T reg cells) also participate in cancer immunity. While CD4+ T helper type 1 (Th1) and T follicular helper (Tfh) cells promote CD8+ cytotoxicity and antitumor activities, T reg cells suppress antitumor immunity (Borst et al., 2018; Nishikawa and Sakaguchi, 2014). In preclinical models, many therapies, such as anti–CTLA-4 and anti-OX40, achieved therapeutic benefits through the depletion of tumor-infiltrating T reg cells (Egen et al., 2020). Similarly, although not conclusive in human cancers, there are data that suggest that partial depletion of T reg cells in the tumor is one of the mechanisms of action for anti–CTLA-4 checkpoint inhibitors (Simpson et al., 2013; Wei et al., 2017; Sharma et al., 2019); however, anti–CTLA-4 therapy and, indeed, most checkpoint blockade therapies lead to significant adverse effects in cancer patients, including autoimmune reactions (Myers, 2018; Michot et al., 2016). New strategies to manipulate cancer-infiltrating T reg cells should aim to further improve their therapeutic window while minimizing nonspecific inhibition that may lead to adverse effects.
Although the interplay of tumor cells and tumor-infiltrating T cells has been well documented, several key questions regarding the heterogeneity, diverse functionalities, dynamics of T cell clonal expansion, cross-tissue migration, and stimulus-dependent phenotypic transition of tumor-infiltrating T cells remain poorly understood. The advancement of single-cell sequencing technology has allowed an unprecedented and unbiased view of the whole transcriptome of various cell types and the heterogeneity of cellular compositions within a given tissue or organ (Puram et al., 2017; Tirosh et al., 2016; Zhang et al., 2018). Single-cell RNA sequencing (scRNAseq) technology has been a valuable tool with which to dissect the cellular composition of heterogeneous leukocyte populations within the tumor microenvironment of various human cancers (Wu et al., 2020; Guo et al., 2018; Zhang et al., 2018; Deng et al., 2018). It has also provided insights into the cellular impact of therapies and elucidated mechanisms of drug resistance in cancer patients and preclinical animal models (Yost et al., 2019; Zhang et al., 2020).
Another major advantage in single-cell immunology is the ability to simultaneously sequence the transcriptome and the α/β TCR antigen receptors. Because of the highly diverse nature of antigen receptor variable, diversity and joining gene segement (V(D)J) recombination, each unique TCR clonotype represents a unique clonal lineage arising from a single progenitor passed onto daughter cells and gives a direct measurement of clonal expansion. Simultaneous determination of the TCR clonotype and total gene expression at the single-cell level enables a precise categorization of the multitude of phenotypic states of a TCR clone undergoing clonal expansion and enables tracking of these clones across multiple tissues. To best integrate transcriptome and TCR clonotype data, we previously developed a single T cell analysis using the RNA sequencing and TCR tracking (STARTRAC) algorithm for the study of T cell dynamics in human colorectal cancer (CRC) samples (Zhang et al., 2018). This analysis revealed many interesting features of tumor-infiltrating T cell subsets. For example, exhausted CD8+ T cells inside tumors exhibited high clonal expansion and expressed high levels of effector genes, but showed low mobility and appeared to be trapped inside the tumor. Conversely, tumor T reg cells displayed moderate levels of clonal expansion and appeared to be developmentally different from the T reg cells identified in the adjacent normal tissue.
Large numbers of α/β TCR clonotypes from individuals also enable us to identify public TCRs. Public TCR clonotypes have been traditionally defined as epitope-specific TCRs that are frequently observed in multiple individuals (Venturi et al., 2008); however, analysis of mouse naive T cells and preterm neonates has shown that public TCRs may comprise a significant portion of the naive T cell repertoire (Bousso et al., 1998; Carey et al., 2017). These studies have typically relied on identification of either α or β chains within a bulk population; however, it has been shown that the sharing of paired TCRs between individuals does indeed occur, albeit at a lower frequency (Grigaityte et al., 2017 Preprint; Carter et al., 2019). Both recombinational bias and convergent recombination are proposed to contribute to the occurrence of public TCRs between individuals (Venturi et al., 2008; Li et al., 2012). Public TCRs for either α or β chain alone have been used in most of the previous studies that feature the properties of public TCRs. For example, >1% of amino acid sequences encoded for TCRβ CDR3 are shared in two individuals (Robins et al., 2009). Public CD8+ TCRβ CDR3 sequences specific to influenza epitope have been detected in mice, suggesting a convergent selection for best fit immunodominant epitope (Kedzierska et al., 2004). Public TCRβ CDRs on CD4+ T cells were also revealed in EAE models and were implicated as potential drivers of disease progression (Zhao et al., 2016). Similarly, public TCRs have been found in cancers (Le Gal et al., 2005; Derré et al., 2008) and in mouse tumor models (Sainz-Perez et al., 2012). Both recombinational bias and convergent recombination seem to drive the occurrence of public TCRs in cancer (Wang et al., 2017), and immunotherapy has been reported to enhance public TCRα and β clonotypes (Hosoya et al., 2018); however, public TCR single chains may not entirely reflect the full antigen specificity of these cells, especially when tumor antigens are the source of public TCR enrichment. Many public β chain CDR3 sequences have been identified in various human cancers and individuals, having been identified in tumor as well as in peripheral blood (Li et al., 2016). The frequency of shared public CDR3s in blood samples was also similar between healthy individuals and bladder cancer patients (Elhanati et al., 2018). From these data, the contribution of tumor antigens in driving the frequency of these public TCR chains remains unclear.
In this study, we performed principal component analysis (PCA)–based subclustering and applied the STARTRAC method of clonotype tracking to gain a greater understanding of the complexities of T cell subsets in mouse tumors. We conducted paired single-cell TCR and gene expression sequencing with the 10x Genomics Chromium 5′-based system on T cells from mice bearing MC38 tumors at different time points and mice bearing CT26 model—widely used syngeneic models to evaluate various antitumor immune-modulation strategies. We harvested T cells from the tumor, spleen, draining LN, and peripheral blood. We sequenced over 60,000 T cells with complete TCRα and β chains from several MC38 mice, enabling investigation into the presence of public TCRs and revealing the enrichment of public TCR clones inside the tumors. We also combined the transcriptome and clonotype information of all cells using STARTRAC, which revealed a dynamic relationship between CD8+, CD4+, and CD4+Foxp3+ T reg cell subsets that reflects much of what has been seen in human cancers. Our data identified many unique markers for various T cell subsets in mouse tumor models, but also for the first time described the dynamic status of these T cell subsets in the MC38 syngeneic tumor model. We also confirmed these subsets further by scRNAseq data obtained from different time points of the MC38 model and CT26 model. Finally, by comparing the transcriptome of human and mouse T reg cells from cancers, we identified CCR8 as a potential depletion marker for tumor-infiltrating T reg cells. When compared with previously characterized tumor-infiltrating T reg cell markers, such as CTLA-4, CCR4, and OX40, the expression of CCR8 was more restricted in these T reg cells, suggesting that CCR8 was potentially a better target for preferentially depleting T reg cells in tumors. Indeed, an anti-CCR8 depletion antibody, but not ligand-blocking antibody, was able to drive antitumor activity in the CT26 model and provide synergistic therapeutic benefit with anti–PD-1 antibody in MC38 and B16F10 tumor models.
Results
T cell subsets identified in MC38 tumor model based on transcriptome and paired TCR data
To understand T cell clonal expansion, activation and migration dynamics, and developmental trajectories in a mouse tumor, we used the well-established MC38 syngeneic tumor model due to its amenability to immunotherapy. Tumors, spleens, draining LNs, and peripheral blood mononuclear cells were harvested from four tumor-bearing C57BL/6 mice 16 d after tumor implantation. Total T cells from each tissue were purified and processed for scRNAseq and TCR sequencing via drop sequencing using the 10x Genomics Chromium system. Samples were prepared in two separate batches of purified FACS-sorted T cells from two mice, with purified cells from each individual mouse’s tissue run on a separate microfluidic lane.
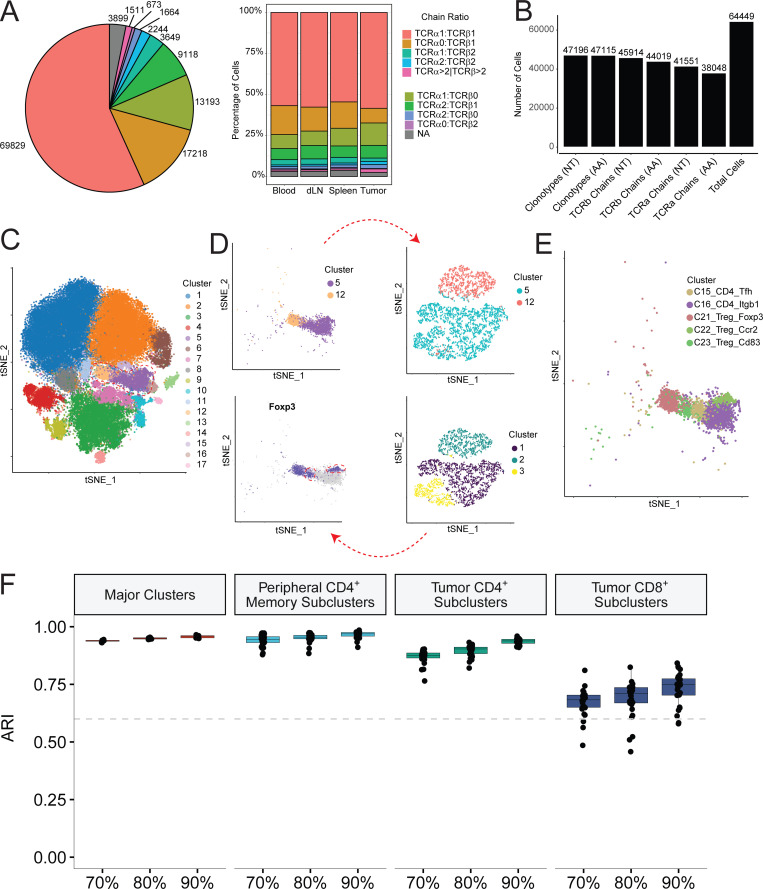
All single-cell datasets were aligned using 10x Genomics Cell Ranger count and vdj pipelines, and postalignment filtering and clustering was performed using Seurat. The aggregated single-cell datasets were filtered based on transcript levels, number of unique genes observed, mitochondrial content, and finally selection of cells with paired single TCRα and β chains. Distinct clonotypes were assigned according to the presence of a unique α and β chain, with each chain being defined as a unique combination of V segment, J segment, and nucleotide-level CDR3. The clonotype composition of all cells revealed that 56% of all clonotypes contained a single paired α and β chain pair. The next largest fractions of cells contained only one chain, either α or β, most likely representing sequencing dropouts. A smaller proportion of cells expressing two copies of either α or β alleles possibly represent either biallelic expression within a single cell or a cell doublet of two single-chain pairs. A small fraction of cells (1–2%) contained more than two copies of any chain allele were likely cell multiplets captured on a single bead (Fig. S1 A). Importantly, these numbers were consistent regardless of the tissue of origin (Fig. S1 A, right).
Figure S1.
Clonotype analysis and clustering methodology. (A) Number of cells with the indicated ratios of TCR α to β chains per clonotype (left). Proportion of cells within each tissue with α:β clonotype ratio indicated. (B) Number of unique paired clonotypes—individual α and individual β chains at both nucleotide and amino acid levels compared with total cells in this study. (C–E) Schematic of iterative clustering approach. Initially assigned clusters in C are selected and examined for heterogeneity of key marker genes and subclustered by PCA-based clustering (D), resulting in final cluster assignments (E). (F) Stability of clusters under different levels of downsampling. Adjusted Rand Index (ARI) was computed for all clusters, peripheral CD4/T reg cell clusters (C16:C23), tumor CD4 effector clusters (C17:C20), and tumor CD8 clusters (C8:C11). dLN, draining LN.
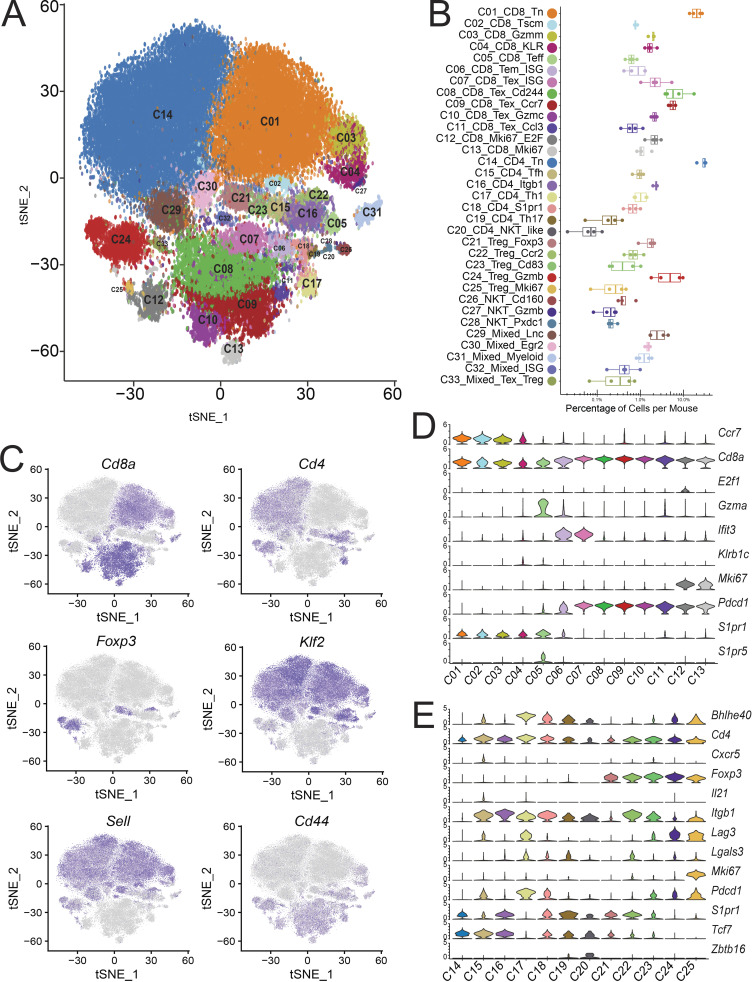
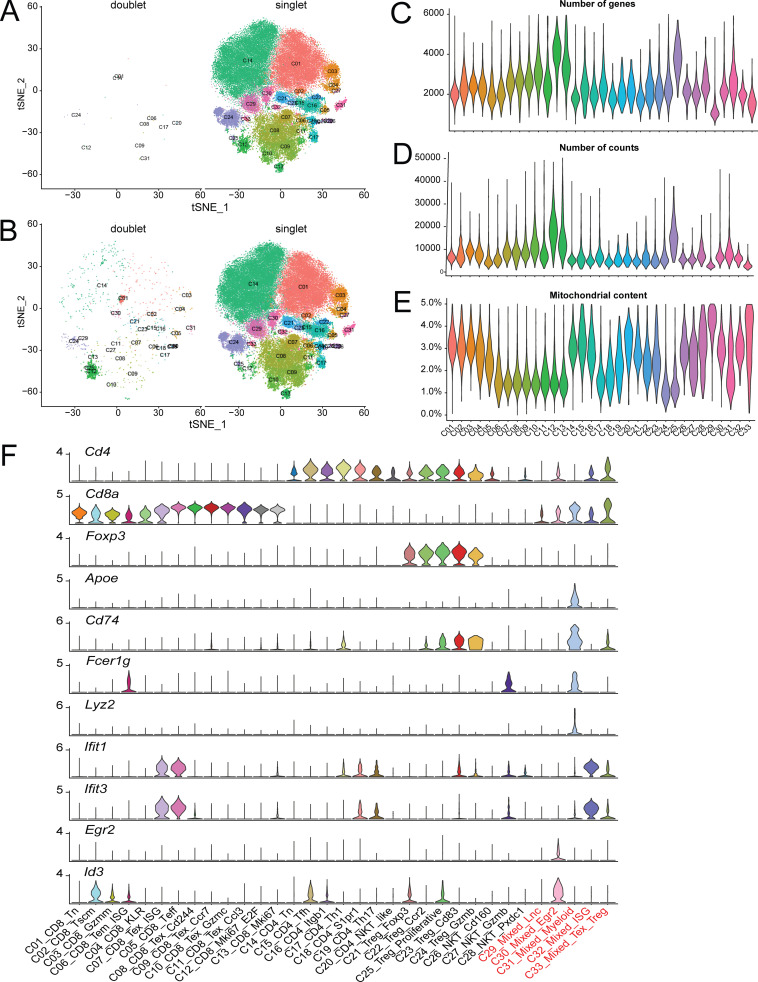
To maintain the highest degree of certainty about clonal identity, we restricted our analyses to cells exhibiting a single α and β chain pair and performed gene expression clustering on this filtered set of cells. This yielded between 1,000 and 6,000 single cells per sample that passed gene expression quality filters and had a clonotype comprised of a productive pair of α and β TCR chains and an aggregate total of 64,449 cells from all samples. 47,196 unique clonotypes with a single α and β pair were found from these cells, indicating that 17,253 cells were clonally expanded (Fig. S1 B). These 64,449 cells were used to perform gene expression–based clustering. We performed batch correction on the datasets as described in the Materials and methods and then graph-based Louvain clustering on all aggregated cells (Stuart et al., 2019), which identified 17 major clusters (Fig. S1 C). However, when we examined these clusters with well-established T cell subset marker genes, such as FOXP3, RORγt, IL-21, and IL-17, etc., we observed that some of these clusters contained biologically meaningful subclusters. For example, in the peripheral T reg cell and memory CD4+ subsets (Fig. S1 C, red circle), FOXP3 clearly marked two additional subsets within the memory cluster of cells (Fig. S1 D, red circles). We therefore further refined the initial major cluster assignments by iteratively examining for heterogeneous substructures and splitting heterogeneous groups by PCA-based clustering. Clusters that displayed at least five significantly differentially expressed genes among its subclusters were assigned as separate clusters, yielding 33 distinct clusters (Fig. 1 A, Table S1, and Table S2). Since this procedure might result in overclustering, we confirmed the reproducibility of these clusters by validating their presence and similar distribution in all four individual mice from two batches of scRNAseq data (Fig. 1 B), and examined their stability by calculating the concordance of the clusters from the down-sampled dataset with that from the full dataset for all major clusters and PCA-based subclusters (Fig. S1 F). In addition, as discussed later, we also verified that many of the subclusters identified through this procedure were consistent with previously defined T cell subsets.
Figure 1.
Gene expression and profiles of T cell subsets. (A) tSNE plot of 64,449 tumor and peripheral-derived T cells derived from four mice, colored as 33 distinct phenotypic clusters. (B) Distribution of the percentage of cells in each cluster from individual animals. (C) tSNE plot of key marker gene expression. (D and E) Violin plots of major population and cluster-specific marker genes in CD8+ (D) and CD4+/T reg cell clusters (E).
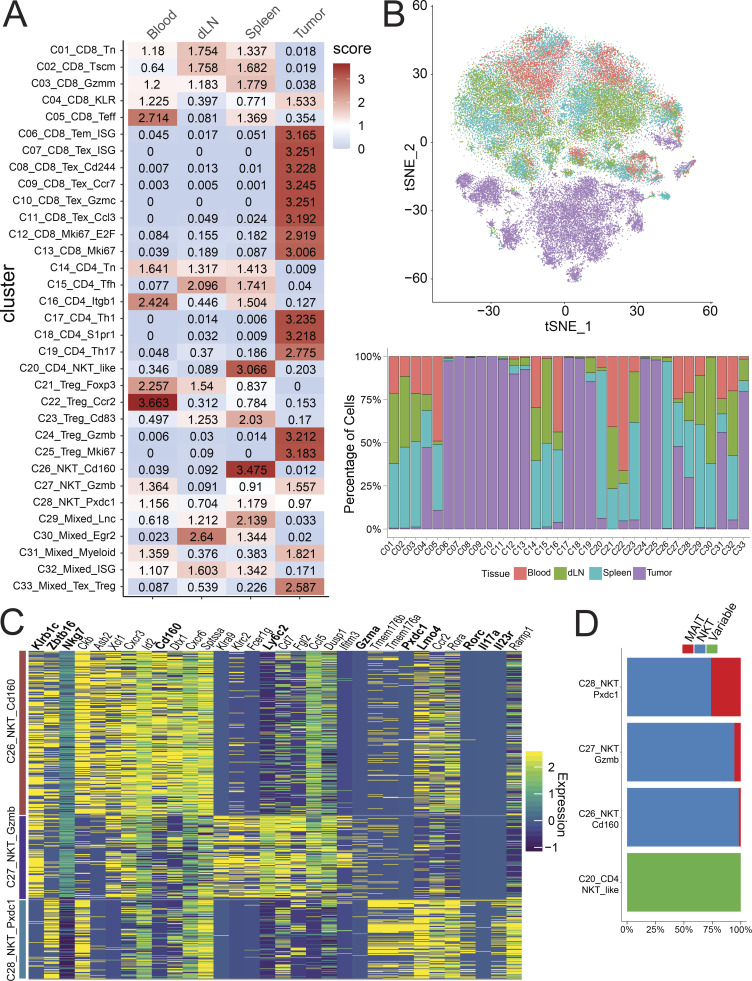
Among these clusters, we identified 13 CD8+ clusters (C01–C13), 7 CD4+Foxp3− clusters (C14–C20), and 5 CD4+Foxp3+ T reg cell clusters (C21–C25) based on key marker gene expression (Fig. 1, C–E) and their tissue of origin (Fig. 2, A and B). We also found five ambiguous clusters that were composed of a mixture of multiple cell types (C29–C33). We first applied the doublet detection methods, Scrublet and DoubletDetection (Wolock et al., 2019; Gayoso et al., 2018), to show that none of the mixed clusters was identified as a collection of experimental doublets (Fig. S2, A and B). We then applied per-cluster inspection of gene counts, read counts, and mitochondrial content for all clusters (Fig. S2, C–E). The results indicated that C29_Mixed_Lnc and C33_Mixed_Texhausted_Treg might be composed of low-quality cells due to their lower number of genes, lower number of read counts, and higher mitochondrial content, while the quality metrics of other mixed clusters did not deviate much from other clusters. However, marker gene analyses indicated that C30, C31, and C32 contained mixed cell types. For example, cluster C31 was marked by the presence of canonical B cell and myeloid markers, such as Cd74 and Lyz2, as well as canonical T cell markers (Fig. S2 F). Similarly, clusters C30 and C32 were heterogeneous T cell clusters comprised of both CD4+ and CD8+ single positive cells. Differential gene expression analysis did not exhibit obvious major phenotypic differences from nearby related clusters, such as naive T cells (Fig. S2 F). These latter two heterogeneous mixed clusters were minimally derived from tumor samples (Fig. 2, A and B), and their TCRs were not clonally expanded (data not shown) and were thus excluded from further analyses.
Figure 2.
Tissue distribution of T cell clusters. (A) Tissue distribution preference of each cluster shown as a STARTRAC observed/expected distribution score. (B) Tissue distribution of cells within each cluster displayed as tSNE (top) and bar graph (bottom). (C) Heatmap of differentially expressed genes in three NKT/MAIT clusters. (D) Proportion of MAIT, NKT, and conventional variable TCR T cells in clusters C20 and C26-C28. dLN, draining LN.
Figure S2.
Quality measurements of single-cell clusters. (A and B) Analysis of putative doublets using Scrublet (A) or DoubletDetection (B) packages. Doublets are shown in the respective left panels and singlets in the right panels. (C–E) Violin plots of unique genes (C), transcripts (D), and mitochondrial transcript percentage (E). (F) Violin plots of key T cell, B cell, and myeloid cell marker gene expression across all clusters.
In addition to conventional T cell subsets, PCA-based subclustering helped to better identify invariant natural killer T cells (iNKT) and mucosal-associated invariant T cells (MAITs; Figs. 1 A and S1 C, C26–C28). These clusters differed from other standard variant clusters by the high expression of canonical NKT genes, such as the lineage-specifying transcription factor Zbtb16 (PLZF) and cell surface markers Klrb1c (NK1.1) and Nkg7 (Fig. 2 C). The semi-invariant T cells in these subclusters were confirmed by their specific TCR α chain use, with 509 iNKT cells that have the specific TRAV11-TRAJ18 α chain and 48 MAITs defined by a TRAV1-TRAJ33 α chain (Garner et al., 2018). These three were distinguished from each other by their gene profile (Fig. 2 C), tissue localization (Fig. 2, A and B), and invariant cell composition (Fig. 2 D and Table S3). C26_NKT_Cd160 was almost solely composed of splenic iNKTs, while C27_NKT_Gzmb and C28_NKT_Pxdc1 showed a broader tissue distribution and included many tumor-derived cells. Of these two clusters, C27 was distinguished by high levels of multiple granzyme molecules and Ly6c2 (Fig. 2 C). Cluster C28 had a higher proportion of MAITs (Fig. 2 D) and expressed higher levels of the Th17 marker genes Il17a, Rorc, Il23r, and Pxdc1, consistent with reported phenotypes of NKT17/MAIT17 (Fig. 2 C; Salou et al., 2019).
Public clonotypes are enriched in tumor-derived and clonally expanded clusters
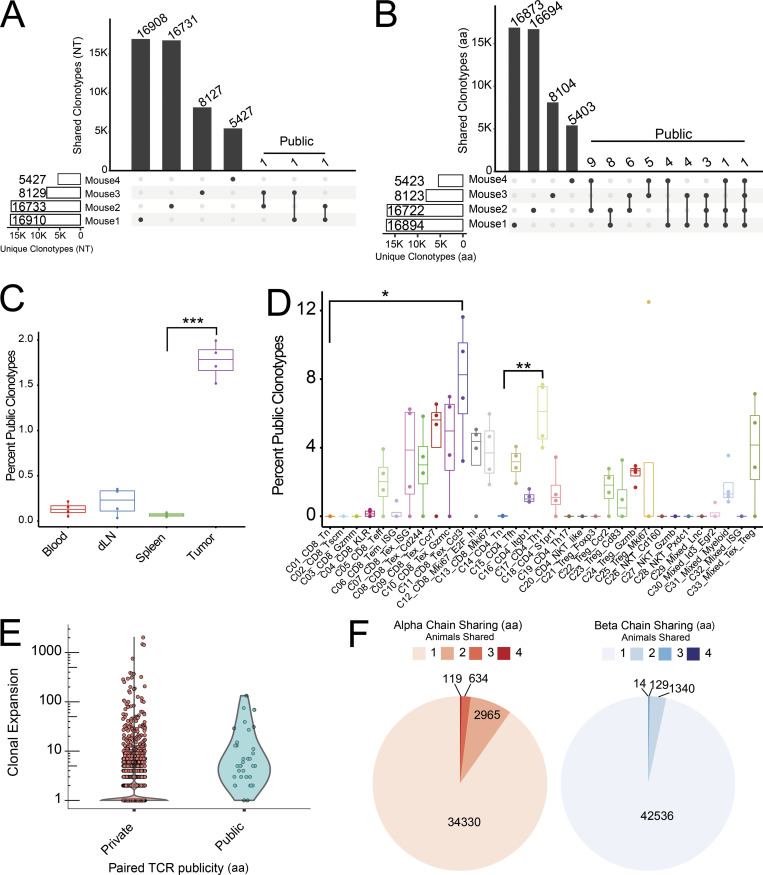
The large number of clonotype sequences from multiple animals allowed us to address the frequency of clonotype sharing across individual animals. Public TCR clonotypes have been traditionally defined as epitope-specific TCRs that are frequently observed in multiple individuals (Venturi et al., 2008); however, analysis of mouse naive T cells and preterm neonates has shown that public TCRs may comprise a significant portion of the naive T cell repertoire (Bousso et al., 1998; Carey et al., 2017). These studies have typically relied on identification of either α or β chains within a bulk population; however, it has been shown that the sharing of paired TCRs between individuals does indeed occur, albeit at a lower frequency (Grigaityte et al., 2017 Preprint; Carter et al., 2019). Here, we used both paired and unpaired α and β chain sequence to address clonotype sharing among animals (public) based on the presence of identical nucleotide or amino acid CDR3 sequences. In our data, the number of unique amino acid–defined clonotypes was slightly lower: 47,115 compared with 47,196 nucleotide-defined clonotypes. The reduced 81 clonotypes was caused by a low level of convergence from a different nucleotide sequence of either one or both TCR chains to identical amino acid sequence (Fig. S1 B and Table S4). Considering α and β paired clonotypes at the nucleotide level revealed that only three clonotypes were shared between two animals with no clonotype shared by all four animals (Fig. 3 A). Loosening the sharing criteria to the amino acid sequence increased the paired public TCR (ppTCR) clonotypes shared by at least two animals to 41 clonotypes, which comprises 0.087% of total unique paired amino acid CDR3 sequences (Fig. 3 B). Phenotypic characteristics of the ppTCR clonotypes revealed that they are mostly restricted to tumor-derived clusters (Fig. 3 C). ppTCRs were identified in various tumor-infiltrating CD8+ subsets and CD4+ subsets, with C11_CD8_Tex_Ccl3 and C17_CD4_Th1 containing the highest percentage of public clonotypes in CD8+ and CD4+ subsets, respectively (Fig. 3 D). These data suggest that common tumor antigens may select for a convergence at the amino acid level of tumor-specific TCR clonotypes. Consistently, as further discussed below, these public clonotypes are markedly clonally expanded, although not among the most highly expanded CD8+ effector clonotypes (Fig. 3 E). In all mice, this expansion was only observed with public clonotypes from tumors, but not those from peripheral lymphoid tissues (data not shown), suggesting that the expansion might be driven by tumor-associated antigens.
Figure 3.
Analysis of public TCRs. (A and B) Numbers of private and public nucleotide (NT; A) and amino acid (AA; B) levels of TCR clonotypes. Clonotypes shared by more than one mouse are marked as public. (C and D) Public aa-level clonotypes as a percentage of all aa clonotypes within each tissue (C) and phenotypic cluster for each mouse (n = 4; D). Paired Student’s t test was applied to determine the significance between clusters and tissues in C and D. ***, P < 0.001; **, P < 0.01; *, P < 0.05. (E) Clonal expansion of tumor T cells between private and public aa-level clonotypes. (F) Sharing of individual TCR chains displayed as proportions. dLN, draining LN.
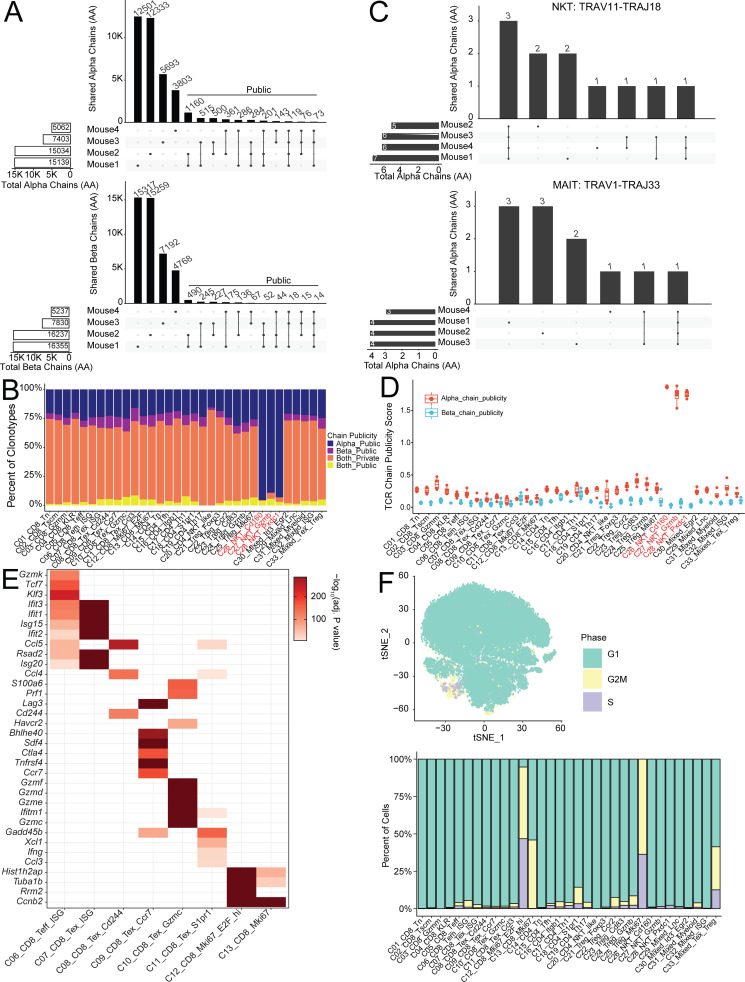
Next, we analyzed the cross-animal publicity of individual α or β chains. Comparison of amino acid chains yielded a higher number of public TCR chains, with 119 α and 14 β amino acid–level chains shared by all four animals (Figs. 3 F and S3 A). As expected, the shared single public chain numbers are much higher than that of ppTCR chains (Fig. S3 B). Invariant T cells defined by the specific Vα-Jα combination discussed above only comprised a few of the total public α chains—three NKT α chains and only one MAIT α chain were shared by all four subjects (Fig. S3 C). Despite the presence of a fixed germline gene segment, these invariant α chains still displayed diverse and subject-specific CDR3s (Fig. S3 C). Interestingly, the paired clonotypes of semi-invariant NKTs and MAITs did not exhibit cross-individual sharing despite having invariant α chains, showing that the β chains in these cells maintain a high degree of individual diversity (data not shown). We further investigated whether public chains were associated with specific phenotypic clusters by measuring the clonotype proportions (Fig. S3 B) and calculating a STARTRAC publicity score for each cluster. This score was derived in two steps: First, we calculated a publicity index for each individual chain based on its clonal expansion and distribution across multiple animals, and then the cluster-level STARTRAC publicity score was defined as the weighted average of all TCR chain publicity indices contained in the cluster (see Materials and methods). A high publicity score for a cluster meant that the public chains in that cluster were highly expanded or constituted multiple paired clonotypes, possibly implying that commonly shared antigens would drive expansion of a particular phenotype. Most clusters had a similar level of chain publicity, with the notable exception of the three invariant clusters, which had high α chain publicity scores and clonotype proportions (Fig. S3, B and D).
Figure S3.
Analysis of public TCR chains, Tex cluster markers and cell cycle states. (A) Sharing of individual amino acid–level TCR chains between multiple animals. (B) Proportion of clonotypes with public α or β amino acid chains in each cluster. (C) Clonotype sharing of invariant chains between animals. (D) Amino acid chain publicity score for each cluster calculated by STARTRAC method (see Materials and methods). (E) Significance of marker gene expression in Fig. 4 B. Adjusted P value of indicated gene (y axis) in specified cluster (x axis) calculated by Wilcoxon rank sum test. (F) Cell-cycle status of cells within each cluster displayed as tSNE (top) and bar graph (bottom).
Tumor-infiltrating CD8+ cells displayed gradual expression profiles and activation states
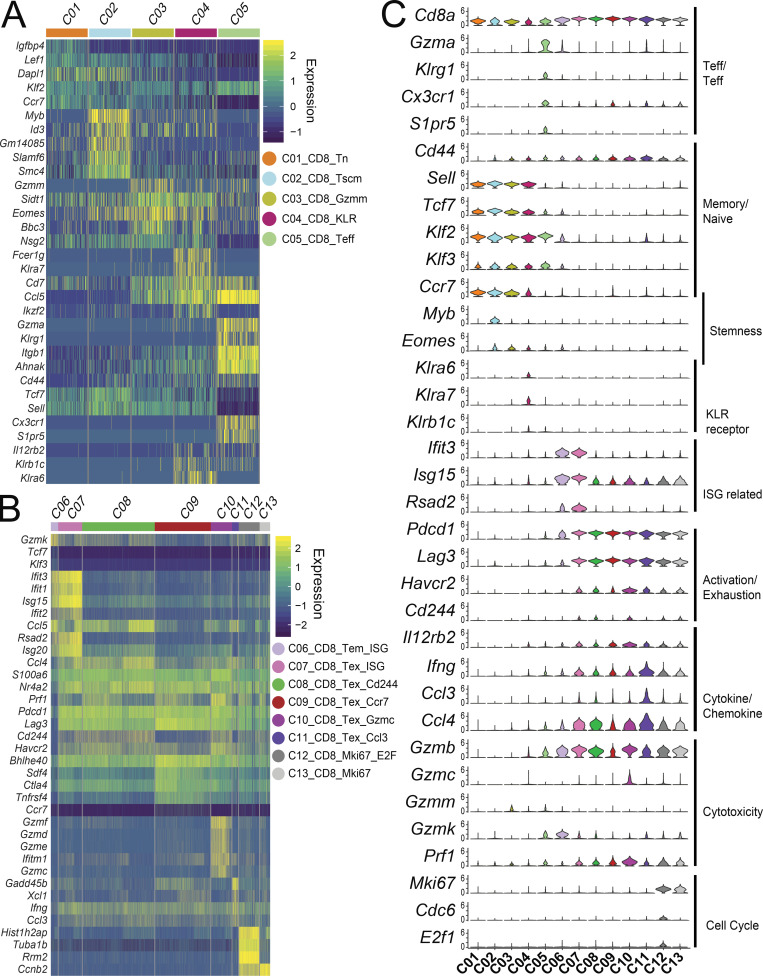
The naive CD4+, CD8+, and T reg cell subsets could be broadly identified by the absence or low expression of Cd44 (Fig. 1 C). The expression of several other marker genes—Tcf7, Lef1, Ccr7, and Sell, encoding L-selectin CD62L—further defined naive subsets and helped to distinguish several memory subsets. Within CD8+ T cells, naive CD8+ cells (C01_CD8_Tn), marked by Cd44−Tcf7+Lef1+ expression (Table S5), comprised the largest cluster and was detected in the peripheral lymphoid tissues, but not in the tumor (Figs. 1 C; and 2, A and B). The Cd44+Sell− C05_CD8_Teff cluster belonged to an effector population, equivalent to human Teff cells or Temra cells, with high expression of Klrg1, Cx3cr1, and S1pr5 (Fig. 4, A and C). There were three CD8+ central memory–like subsets marked as Cd44+Sell+ primarily found in peripheral lymph tissues, including C02_CD8_Tscm, C03_CD8_Gzmm, and C04_CD8_KLR (Fig. 4 A). In addition to typical central memory cell markers, the C02_CD8 cluster had higher expression of Myb, Tcf7, and Eomes (Fig. 4 C). Both Myb and Tcf7 help to maintain the stemness of T cell subsets, implying that the C02_CD8 cluster may be a memory stem cell–like (Tscm) population (Gattinoni et al., 2009). C04_CD8_KLR cells exhibited markers associated with activation by IFN-γ pathway, which included Il12rb2 and several KLR family genes, such as Klrb1c (NK1.1 protein), Klra6, and Klra7 (Fig. 4 C). C03_CD8_Gzmm and C05_CD8_Teff cells resided mainly in peripheral tissues, whereas cluster C04_CD8_KLR is of heterogeneous tissue origin (Fig. 2, A and B).
Figure 4.
Gene expression signatures of CD8+ clusters. (A and B) Heatmaps of peripheral-derived CD8 clusters C01-C05 (A) and tumor-derived CD8+ clusters C06-C13 (B). Genes displayed are either significantly differentially expressed between CD8+ populations or are notable markers of T cell differentiation, function, and exhaustion. Rows represent genes and columns represent individual cells with cluster identities indicated by the color bar on top. (C) Violin plots of key functional and differentiation markers. KLR, killer lectin related gene; ISG, IFN stimulated genes.
Although exhausted or dysfunctional CD8+ T cells are dominant in cancer-infiltrating CD8+ subset (Hashimoto et al., 2018), these cells display a gradient of cell states from predysfunction and early dysfunction to late dysfunction (van der Leun et al., 2020). Because we harvested a large number of single cells, we attempted to capture more granularity of these heterogeneous substructures of the major exhausted subsets by PCA-based subclustering (see Materials and methods; Figs. S1 C and 1 A). Eight CD8+ T cells subsets or states were identified inside the tumor, including C06_CD8_Tem_ISG, C07_CD8_Tex_ISG, C08_CD8_Tex_Cd244, C09_CD8_Tex_Ccr7, C10_CD8_Tex_Gzmc, C11_CD8_Tex_Ccl3, C12_CD8_Mki67_E2F, and C13_ CD8_Mki67 based on statistically significant unique gene signatures (Figs. 4 B and S3 E and Table S5). Among these clusters, C06_CD8_Tem_ISG had moderate expression of exhaustion markers, including Pdcd1 and Lag3 (Fig. 4, B and C). Instead, cells in this cluster expressed genes that are characteristic of effector and memory cells, for example, Gzmk, Klf3, and Tcf7, as well as high levels of IFN-stimulated genes (ISGs), such as Ifit2/Ifit3 and Isg15 (Fig. 4 C), presumably from exposure to the tumor microenvironment. The remaining tumor-infiltrating clusters all expressed high levels of Pdcd1, a marker associated with exhausted or dysfunctional CD8+ T cells (van der Leun et al., 2020). C07_CD8_Tex_ISG, C08_CD8_Tex_Cd244, C10_CD8_Tex_Gzmc, C12_CD8_Tex_Mki67_E2F, and C13_CD8_Tex_Mki67 were similar to the typically defined tumor-infiltrating exhausted T cells and expressed other exhaustion marker genes, Havcr2 and Cd244, in addition to Pdcd1 and Lag3 (Fig. 4 C), but PCA-based subclustering captured subtle different gene expression profiles that may reflect their functional status and exposure to the microenvironment. For example, cluster C10_CD8_Tex_Gzmc expressed a higher and broader range of granzyme molecules and Prf1 than any other clusters (Fig. 4 C), associating with a potential effector function in action. Cluster C07_CD8_Tex_ISG, in contrast, had a higher IFN signature, suggesting an exposure to an IFN microenvironment. In addition, both C12_CD8_Tex_Mki67_E2F and C13_CD8_Tex_Mki67 clusters expressed Mki67 (Fig. 4 C), a marker for cell cycle. C12_CD8_Tex_Mki67_E2F also highly expressed E2f1 and Cdc6, implying that they were in G1/S stage of the cell cycle (Figs. 4 B and S4 F). Finally, both C09_CD8_Tex_Ccr7 and C11_CD8_Tex_Ccl3 clusters had lower expression of Havcr2 and Cd244, suggesting a less exhausted or predysfunctional status (Fig. 4 C). Indeed, C09_CD8_Tex_Ccr7 also had less expression of other effector molecules, but higher expression of Ccr7, a marker gene for the predysfunction or stem precursor of CD8+ Tex cells (Fig. S4 A; Brummelman et al., 2018; Galletti et al., 2020). In contrast, C11_CD8_Tex_Ccl3 had higher expression of Ifng, Ccl3,and Ccl4, and genes associated with NF-κB pathways, categorizing these cells in an early TCR activation stage (Fig. 4 C). Since only fewer effector marker genes, such granzymes, Ifng, Ccl3,and Prf1, were identified to classify different clusters of tumor-infiltrating CD8+ T cells by PCA-based subclustering, these clusters might not represent distinct differentiated subsets as those identified in CD4+ cells. Instead, these genes likely captured different activation states or stimulatory conditions.
Figure S4.
Marker genes for tumor-infiltrating clusters and pseudotime analysis. (A) tSNE plots of cluster-specific marker genes. Specific clusters discussed in text are circled in red. (B) Pseudotime analysis of tumor-infiltrating CD8+ clusters 5–11 (top), CD4+ Th clusters 14–19 (middle), and T reg cell clusters 21:24 (bottom), aggregated together (leftmost plot) or separated by constituent clusters. Axes represent the two major components generated by Monocle2 dimensionality reduction, black nodes denote the branching points of trajectory, and clusters are denoted by color. (C) Pairwise migration between tissue pairs of CD4/T reg cell clusters quantified as a STARTRAC pMigr score. dLN, draining LN.
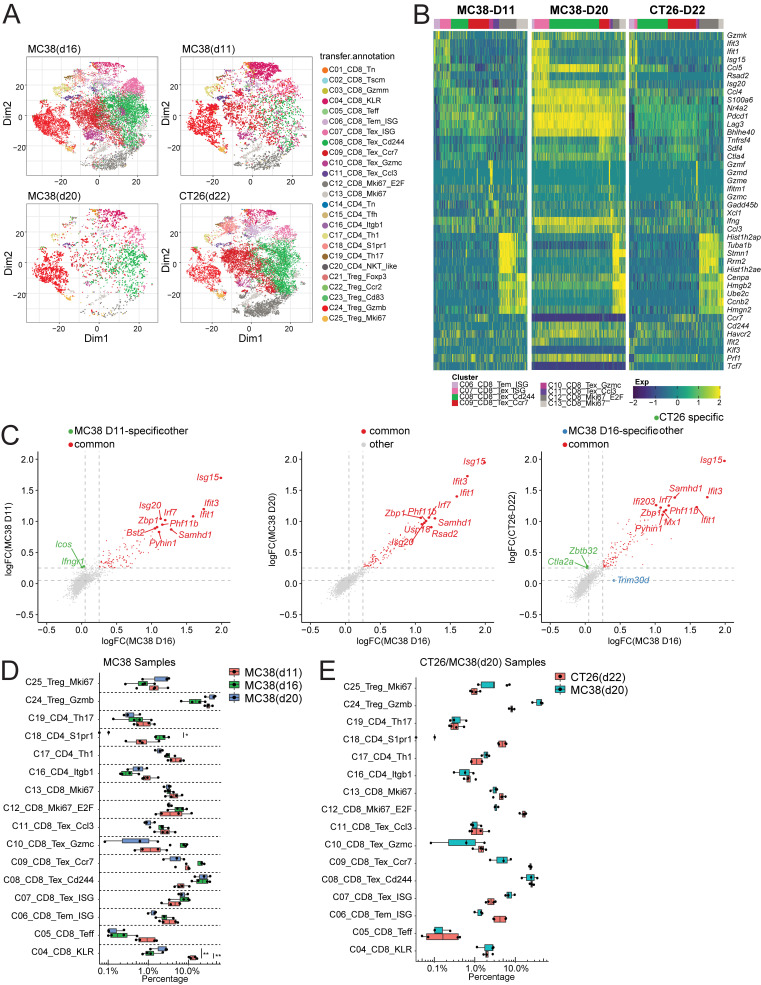
To better understand the developmental relationships among all the T cell subsets in tumor, we performed monocle trajectory analyses (Fig. S4 B). Consistent with our premise, the results did not map clearly the developmental trajectories for most CD8+ Tex subsets (C09-C11), which are overlapped but distinguished from C05_CD8_Teff and C06_Tem_ISG populations (Fig. S4 B), supporting the idea that they were not distinct differentiated subsets. This could be caused by related few distinct marker genes among these Tex subsets or by Monocle2 to force cells along branching trajectories. Interestingly, C07_Tex_ISG resided at one end of the developmental trajectory and was more overlapped with the cluster C06_CD8_Tem_ISG (Fig. S4 B). To confirm that these different exhausted states we identified were not simply due to overclassification, we analyzed additional scRNAseq data from T cells isolated from MC38 tumors at different time points (day 11 and day 20) and from CT26 tumors (Fig. 5 A). By using the Seurat3 pipeline, the same clusters have been identified from these additional datasets (Fig. 5 A). Importantly, the original marker genes identified from the original day 16 data demonstrated similar differential expression patterns and marked the similar clusters in these new datasets (Fig. 5 B). Furthermore, as demonstrated as an example for C07_CD8_Tex-ISG, the marker genes for the same cluster that derived from different datasets showed a high degree of correlation (Fig. 5 C). Similar results were also obtained for other Tex subsets (data not shown). Finally, all these clusters were identified with similar distribution patterns from different time points of MC38 tumors and CT26 tumors. In the MC38 model, there was a higher percentage of C04_CD8_KLR cluster in day 11 than in later time points. There was also a trend toward an increase of C08_CD8_Tex_CD244 in the later time points (Fig. 5, D and E). Taken together, these data support the idea that these different states of Tex cells were broadly present in mouse tumor models.
Figure 5.
Correlation of tumor CD8+ clusters across multiple tumor time points and models. (A) tSNE plot of integrated tumor T cell datasets, split by tumor model and time point indicated at top. (B) Heatmap of key cluster-specific genes in multiple tumor models. (C) Correlation of key marker genes within cluster C07_Tex_ISG between different mouse tumor time points and models. Fold change of genes over other tumor CD8+ clusters are plotted, with top 10 common and top five model-specific genes highlighted. (D and E) Boxplots showing the distribution of percentage of cells within each phenotypic cluster from each indicated tumor type and time point. P values were calculated using unpaired t test. ***, P < 0.001; **, P < 0.01; *, P < 0.05.
Shared TCR uses among tumor CD8+ Tex subsets and peripheral Teff cells revealed by STARTRAC analyses
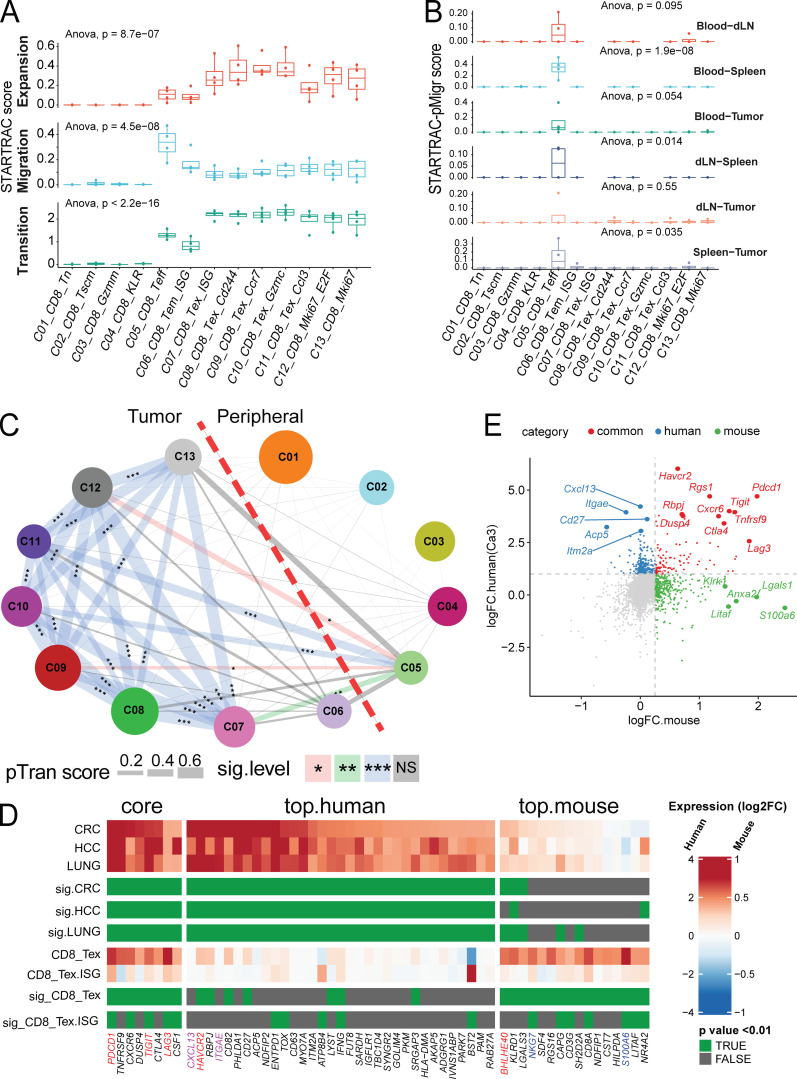
By taking advantage of the higher cell numbers sequenced in this study, we identified more gradual states of the intratumor dysfunctional or exhausted CD8+ subsets than those reported in human cancers (Zhang et al., 2018; Guo et al., 2018; Zheng et al., 2017; Azizi et al., 2018; Tirosh et al., 2016; van der Leun et al., 2020). Although these clusters might capture a more detailed activation status and environmental conditions for intratumoral CD8+ Tex cells, trajectory analyses further supported that these clusters were not developmentally distinct subsets. We therefore used TCR clonal expansion data to examine the relationships among these Tex clusters. Previously, we devised the STARTRAC method to combine both transcriptome data and TCR clonal information to describe various T cell clusters (Zhang et al., 2018). Briefly, this method relies on the premise that cells bearing the same TCRαβ chain are derived from the same progenitor cells and are thus developmentally related. The cross-cluster transition or cross-tissue migration scores can be calculated by the weighted Shannon diversity index for individual clonotypes in a specific cluster or tissue by comparing with multiple other clusters or tissues (Zhang et al., 2018). Consistent with the characteristics of human tumor-infiltrating CD8+ Tex cells (Zhang et al., 2018), all Tex clusters (C7-C13) observed in MC38 tumors displayed higher clonal expansion scores than other CD8+ clusters, especially naive and central memory clusters (Fig. 6 A, top), but relatively lower migration scores than Teff cluster C05_CD8_Temra (Fig. 6 A, middle, and Fig. 6 B). Importantly, these Tex clusters had high STARTRAC transition scores (Fig. 6 A, bottom), indicating that they were developmentally connected, as supported by their significant sharing of common TCRs among each other, further supporting the conclusion from the pseudotime trajectory analyses (Fig. S4 B) and pairwise clonotype transition analysis discussed below (Fig. 6 C).
Figure 6.
STARTRAC analysis of CD8+ clusters. (A) Clonal expansion, cross-tissue migration, and phenotypic transition scores for all CD8+ clusters quantified by STARTRAC for each animal (n = 4). Significance calculated by ANOVA. (B) Pairwise migration between tissue pairs of CD8+ clusters quantified as a STARTRAC pMigr score. (C) Cross-cluster phenotypic transition scores for all combined animals. P values calculated by permutation of clusters to determine significance of pairwise cluster sharing. ***, P < 0.001; **, P < 0.01; *, P < 0.05. (D) Gene signature comparison between human and mouse Tex cells. Mouse CD8+ clusters C08-C11 were combined into CD8_Tex and compared against all other clusters, while human Tex from previous human findings (Guo et al., 2018; Zhang et al., 2018; Zheng et al., 2017) were compared with non-Tex clusters. Gene expression from both species are displayed as log2FC (color bar) with adjusted P value (green). Homologous genes are classified as human or mouse enriched genes based on adjusted P value of < 0.01 and log2FC > 1.5 (human) or P value < 0.01 and log2FC > 0.3 (mouse). Genes enriched in both species are indicated as core. (E) Correlation of key marker genes between aggregated mouse Tex clusters C08-C11 and aggregated human Tex clusters from three human cancers labeled in D as Ca3. Gene expression fold change of genes over other clusters by species are plotted, with common markers highlighted in red, mouse specific in green, and human specific in blue. dLN, draining LN; HCC, hepatocellular carcinoma.
Next, we compared expression profiles of orthologous genes between the human and mouse tumor-infiltrating CD8+ exhausted T cell subsets (Table S6). In this analysis, we combined all mouse Tex subsets and generated combined gene signatures to compare with the expression signature of human tumor-associated CD8+ Tex cells described in previous studies (Zhang et al., 2019; Guo et al., 2018; Zhang et al., 2018). Although many canonical exhaustion markers Pdcd1, Tigit, and Havcr2 showed a broad correlation in both species (Fig. 6, D and E; and Table S6), there did remain a few unique signatures that were distinct to each species. For example, Itgae and Cxcl13 were highly expressed in all human tumor T cells examined, but absent in mouse. Conversely, mouse CD8+ Tex clusters express Nkg7 and S100a6 at higher levels than human counterparts. Furthermore, we found that many other genes, such as Bhlhe40, showed a cross-species correlation (Fig. 6, D and E). Together, these data suggest that CD8+ Tex cells in human cancer and in a mouse tumor model share similar markers and dynamic behaviors.
Within tumor-associated subsets, C06_CD8_Tem_ISG cells were more overlapped with C07_CD8_Tex_ISG, but not with other Tex subsets in the trajectory analysis (Fig. S4 B). They instead showed moderate clonal expansion and slightly higher migration capability than Tex subsets (Fig. 6 A). Notably, cells in this cluster did not have a significant TCR clonotype-based developmental transition score to any Tex subset within the tumor (Fig. 6 C). Importantly, although C07_CD8_Tex_ISG shared a common IFN response signature with C06_CD8_Tem_ISG, STARTRAC analyses suggested that these two subsets had different developmental origins (Fig. 6, A and C). Cells in the C06_CD8_Tem_ISG cluster might not be directly reactive to the tumor and might belong to bystander T cells reported in previous studies (Scheper et al., 2019; Simoni et al., 2018); however, these cells may be exposed to cytokines such as IFN, similar to C07_CD8_Tex_ISG, in the tumor microenvironment that result in upregulation of IFN-responsive family genes such as Ifit and Isg in their transcriptome (Fig. 4, B and C). Together, these data supported the idea that STARTRAC could be used to better understand the developmental and migration relationships among different clusters in supplement to transcriptome and trajectory-based analyses.
Like its human counterpart, the periphery-enriched cluster C05_CD8_Teff was the only subset that had a high expansion score and a high cross-tissue migration score (Fig. 6 A). Although most cells in this cluster migrated between blood and spleen, clonotypes within this cluster were highly shared with tumor CD8+ clusters (Fig. 6 B). The gene signature and dynamic properties of C05_CD8_Teff cluster were consistent with what has been previously described for CD8+ Teff cells in human CRC (Zhang et al., 2018). Interestingly, this cluster displayed high STARTRAC transition score with a majority of the CD8 Tex subclusters, except the cluster C08_CD8_Tex_Cd244 (Fig. 6 C), supporting a developmental connection between intratumor Tex T cells and peripheral effector-like cells.
In summary, the combined transcriptome, trajectory, and STARTRAC analyses revealed more detailed dynamic states of T cell exhaustion in the microenvironment of MC38 tumors.
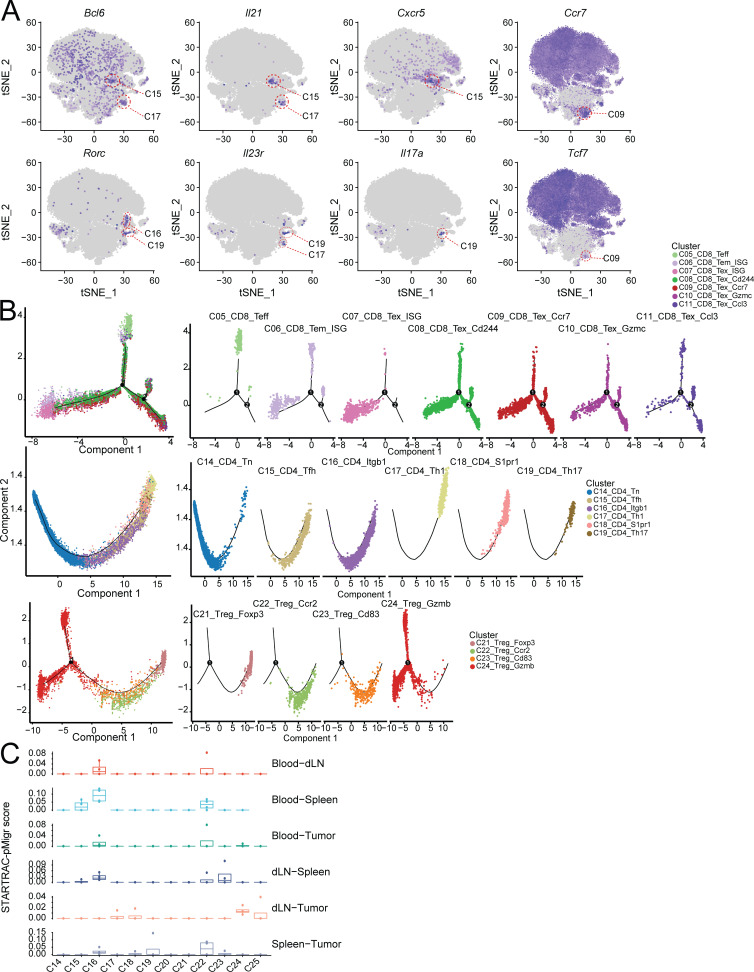
STARTRAC analysis unveiled developmental connection between intratumor Bhlhe40+ Th1-like cells and peripheral Tfh cells
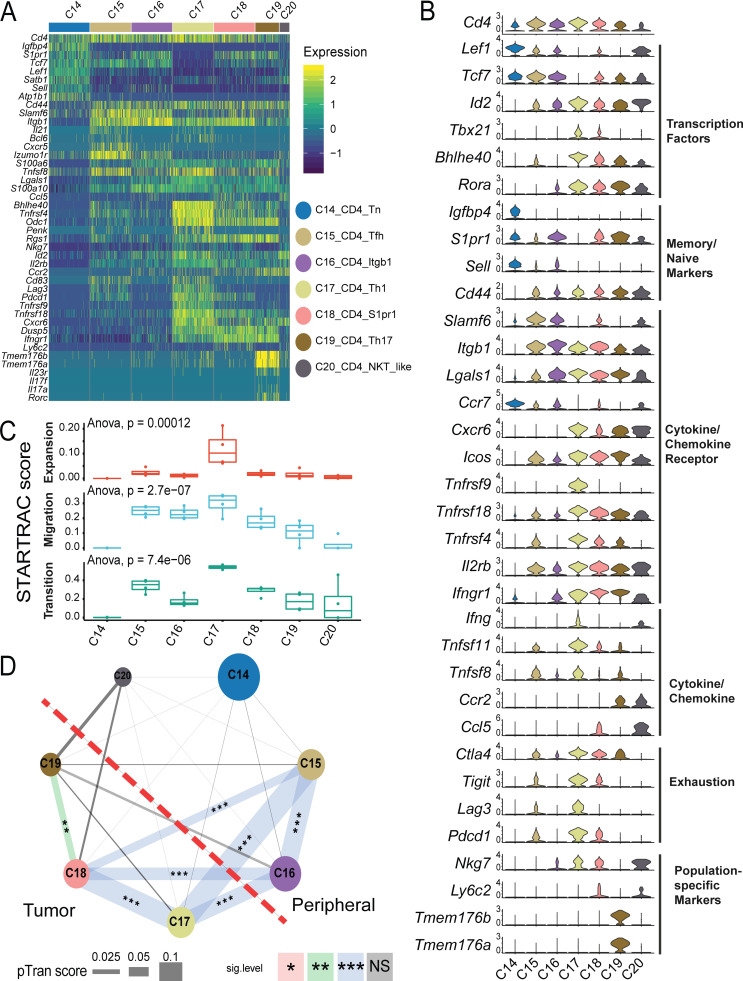
In the case of CD4+ cells, seven conventional CD4+ T helper cell subsets and five T reg cell subsets could be readily distinguished by differential expression of Foxp3 (Fig. 1 A, Fig. S1 C, and Table S7). PCA-based subclustering helped to identified small, well-established T helper cell clusters, such as Th17 and Tfh cells (Fig. 1 A and Fig. S1 C). Among conventional CD4+ clusters, the largest cluster C14_CD4_Tn was composed of peripheral lymphoid tissue–derived naive cells expressing high levels of Tcf7/Lef1 (Fig. 7, A and B). These cells were nearly evenly distributed in the blood, LN, and spleen (Fig. 2, A and B). Two peripherally enriched CD4+ memory subsets, C15_CD4_Tfh and C16_CD4_Itgb1, were identified based on their high expression of Cd44, Slamf6, and Itgb1 (Fig. 7, A and B). The C16_CD4_Itgb1 cluster was preferentially enriched in the blood and spleen, while C15_CD4_Tfh cells had high expression of known Tfh cell genes Il21, Bcl6, and Cxcr5 (Figs. 7 A and S4 A) and were enriched in spleen and draining LNs (Fig. 2, A and B). Their high STARTRAC migration scores implied that cells in both clusters were mobile in the peripheral lymphoid tissues (Fig. 7 C, middle, and Fig. S4 C). Interestingly, there is a high level of TCR clonal sharing between C15 and C16 clusters, indicating their developmental connection (Fig. 7 D). Consistently, both clusters were largely overlapped in the trajectory analyses and preceded the more differentiated Th subsets, such as Th1 and Th17 cells (Fig. S4 B). Neither of these two clusters displayed substantial clonal expansion scores (Fig. 7 C, top). The C20_CD4_NKT_like cluster represents a small number of mostly spleen-resident cells that clustered closely together with the C28_NKT_Pxdc1 cluster (Fig. 1 A) and specifically upregulated the expression of Zbtb16 (Fig. 1 D), but did not express the canonical TRAV11-TRAJ18 α chain (Figs. 1 B and S2 B).
Figure 7.
Gene expression and STARTRC analysis of CD4+ effector clusters. (A) Heatmaps of all CD4+ clusters C14-C20. (B) Violin plots of key marker genes. (C) Clonal expansion, cross-tissue migration, and phenotypic transition scores for CD4+ clusters quantified by STARTRAC for each animal (n = 4). Significance calculated by ANOVA. (D) Cross-cluster phenotypic transition scores between CD4+ clusters for all animals combined. P values calculated by permutation of clusters to determine significance of pairwise cluster sharing. ***, P < 0.001; **, P < 0.01; *, P < 0.05.
Although conventional CD4+ T cells only represented a smaller portion of tumor-infiltrating cells compared with CD8+ T cells, we identified three clusters—C17_CD4_Th1, C18_Cd4_S1pr1, and C19_CD4_Th17—that were specifically enriched in the tumor (Fig. 1, A and B; and Fig. S1 C). The C18_CD4_S1pr1 cluster had higher expression of S1pr1 and Il7r (Fig. 7, A and B), a feature of central memory cells, suggesting that they might have recently migrated into the tumor. In support of this hypothesis, C18_CD4_S1pr1 was developmentally linked with the peripheral C15_CD4_Tfh and C16_CD4_Itgb1 subsets in terms of their STARTRAC pairwise transition scores (Fig. 7, C and D). In addition, consistent with the dynamic features observed in human tumor-infiltrating central memory cells (Zhang et al., 2018), cells in the C18_CD4_S1pr1 cluster had lower clonal expansion, but a relatively high migration score (Figs. 7 C and S4 C). C19_CD4_Th17 were Th17 cells that preferentially expressed Il17a, Il23r, and Rorc (Fig. S4 A). These Th17 cells were not highly clonally expanded (Fig. 7 C, top), suggesting that they were not actively proliferating within the tumor. Interestingly, they were developmentally related to C18_CD4_S1pr1 central memory cells, but not to any peripheral memory subsets, indicating they may be derived from those TCM cells and remain within the tumor as a resident memory subset (Fig. 7 D).
The C17_CD4_Th1 cluster was similar to Bhlhe40+ Th1 cells described in human CRC (Fig. 7, A and B; Zhang et al., 2018, 2020; Yu et al., 2018; Emming et al., 2020). Cells in this cluster expressed higher activation markers, Bhlhe40, Tbx21, and Ifng (Fig. 7 B and Table S7). These cells might be activated by tumor antigens, as supported by their high clonal expansion score relative to all other CD4+ subsets (Fig. 7 C, top). Additionally, the C17_CD4_Th1 cluster had both high migration and global developmental transition scores (Fig. 7 C, middle and bottom). Trajectory analysis placed these cells at the end of the cell differentiation pathway and closely related to C18_CD4_S1pr1 central memory cells (Fig. S4 B). In contrast to Th17 cells, STARTRAC further revealed that these Th1-like cells were developmentally connected to not only C18_CD4_S1pr1 central memory cells, but also to peripheral C15_CD4_Tfh and C16_CD4_Itgb1 subsets (Fig. 7 D), because of the significant sharing of common TCRs, implying that these Th1 cells could be derived from different sources. Recently, the importance of IL-21–producing Tfh cells in cancer immunity has been highlighted in different models (Hollern et al., 2019). Consistently, a portion of these Bhlhe40+ Th1 cells were able to express Il21 (Figs. 7 A and S4 A).
Identification of clonally expanded tumor-associated T reg cells in peripheral lymph organs
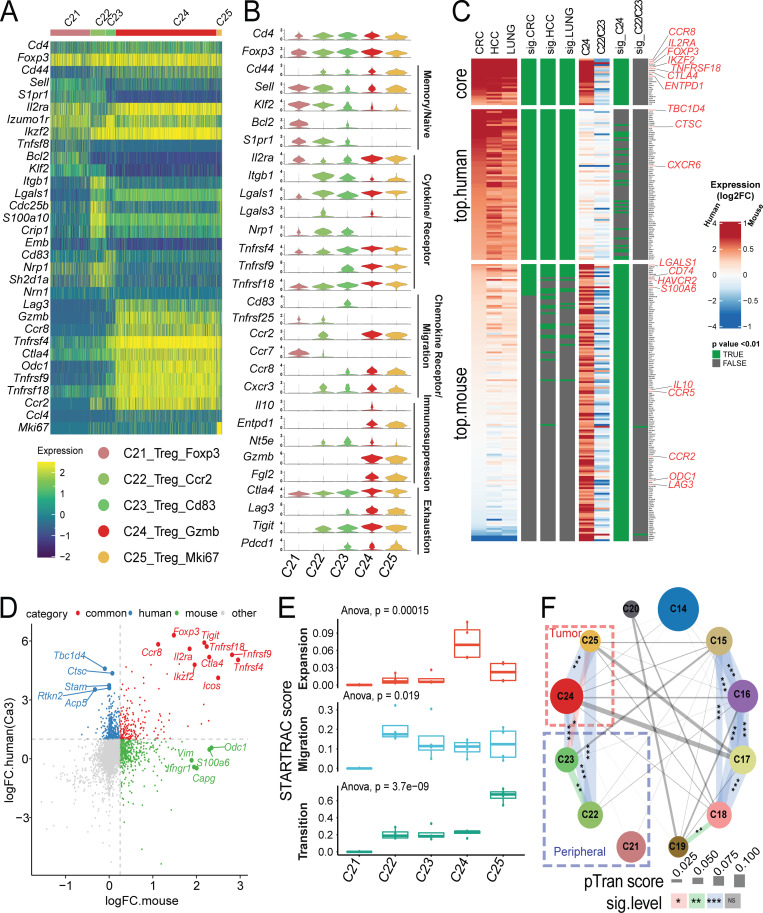
Like in CD4+ and CD8+ cells, PCA-based subclustering identified five T reg cell clusters that exhibited distinct gene expression profiles and tissue distributions (Fig. 8, A and B; and Table S1 and Table S2). The naive T reg cell subset C21_Treg_Foxp3 mainly resided within peripheral tissues and not in the tumor (Fig. 2, A and B). Two memory T reg cell subsets—C22_Treg_Ccr2 and C23_Treg_Cd83—were identified in the periphery as well (Fig. 2 A), but were not identified in previous human studies (Zhang et al., 2018; Guo et al., 2018; Zheng et al., 2017). C22_Treg_Ccr2 was preferentially enriched in blood, while the C23_Treg_Cd83 cluster was mainly enriched in LN and spleen. Consistently, these two clusters could be distinguished by different marker genes that might reflect their physical location. C22_Treg_Ccr2 was marked by high expression of S1pr1, Itgb1, Ccr2, and Lgals3 (Galectin-3) that were related to migration, while C23_Treg_Cd83 had higher expression of Cd83, Lag3, and Pdcd1 that might be caused by priming and activation (Fig. 8, A and B; and Table S8). Two T reg cell clusters—C24_Treg_Gzmb and C25_Treg_Ki67—were tumor-associated T reg cells, distinguished by high Gzmb, Lag3, and Tnfrsf9, while the latter cluster contained proliferating T reg cells (Fig. 8, A and B; and Table S8). We next sought to understand the correlations between tumor T reg cell phenotypes observed in mice and previously published human tumor T reg cells (Zhang et al., 2018; Guo et al., 2018; Zheng et al., 2017). Comparative analysis of T reg cell samples revealed similarities between the expression profiles of mouse tumor T reg cells and human tumor T reg cells. The genes Foxp3, Ikzf2, Il2ra (CD25), and Tnfrsf18 (GITR) showed strong concordance as tumor T reg cell markers across species (Fig. 8, C and D; and Table S9); however, many orthologous genes had divergent expression patterns; CXCR6 was a significant marker of human T reg cells, but was not highly expressed in mouse cells, while Ccr5 and Ccr2 were strongly upregulated in mouse but not in human (Fig. 8 C and Table S9). Other common T reg cell marker genes identified in both human and mouse tumors included Ccr8 and Entpd1.
Figure 8.
Gene expression and STARTRC analysis of Foxp3+ T reg cells. (A) Heatmaps of all Foxp3+ clusters C21-C25. (B) Violin plots of key marker genes. (C) Gene signature comparison between human and mouse T reg cell cells. Mouse T reg cell clusters C24 and the combination of C22/23 clusters, while human T reg cell from previous human studies (Guo et al., 2018; Zhang et al., 2018; Zheng et al., 2017) were compared with non–T reg cell clusters. Gene expression from both species are displayed as log2FC (color bar) with adjusted P value (green). Homologous genes are classified as human or mouse enriched genes based on adjusted P value of < 0.01 and log2FC > 1.5 (human) or P value < 0.01 and log2FC > 0.3 (mouse). Genes enriched in both species are indicated as core. Notable genes are highlighted in red. (D) Correlation of genes between human and mouse-derived tumor T reg cells. Fold change of genes over non–T reg cell clusters are plotted, with top 10 common and top 5 species-specific genes highlighted. Human data are taken as an average of tumor T reg cell clusters shown in C. (E) Clonal expansion, cross-tissue migration, and phenotypic transition scores for T reg cell clusters quantified by STARTRAC for each animal (n = 4). Significance calculated by ANOVA. (F) Cross-cluster phenotypic transition scores between CD4+ and Foxp3+ clusters for all animals combined. P values calculated by permutation of clusters to determine significance of pairwise cluster sharing. ***, P < 0.001; **, P < 0.01; *, P < 0.05. HCC, hepatocellular carcinoma.
TCR-based STARTRAC analyses indicated that only the tumor-infiltrating T reg cells exhibited high clonal expansion in the MC38 model (Fig. 8 E, top), consistent with the observations of T reg cells in human tumors. The mobility of tumor-infiltrating T reg cells was similar to peripheral memory T reg cells (Fig. 8 E, middle). C22_Treg_Ccr2 and C23_Treg_Cd83 were developmentally overlapped and placed between naive T reg cells and tumor-infiltrating T reg cells based on trajectory data (Fig. S4 B). Interestingly, in contrast to the relatively low developmental connectivity to the peripheral T reg cells observed in previous studies for human tumor-infiltrating T reg cells, the tumor-infiltrating T reg cells in the MC38 model are strongly developmentally connected with peripheral memory T reg cell subsets, especially C22_Treg_Ccr2 (Fig. 8 E, bottom, and Fig. 8 F). This discrepancy could be due to the fact that either only a relatively small number of peripheral blood T reg cells was examined in humans or T reg cells from peripheral lymphoid tissues, such as spleen and LN, were not examined in previous human studies (Zhang et al., 2018). In summary, these data support the model that tumor-infiltrating T reg cells in the MC38 model are capable of undergoing clonal expansion in the tumor and that these expanded T reg cell clones can be detected in peripheral lymph tissues.
Antitumor activity of depleting CCR8-expressing tumor-infiltrating T reg cells
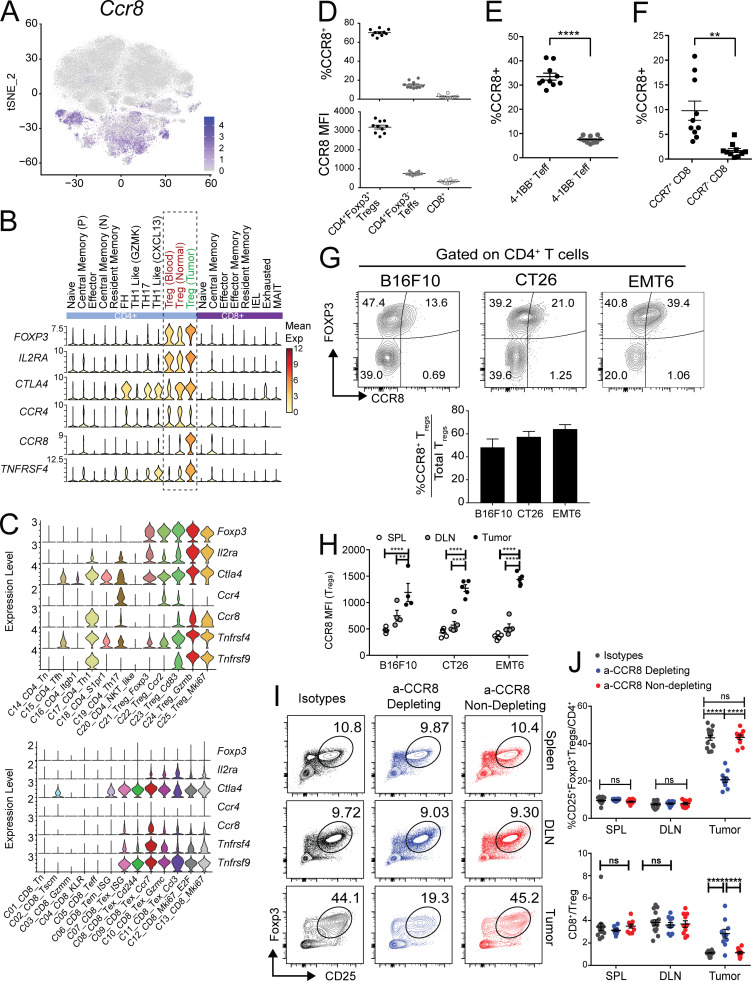
Both tumor-infiltrating CD8+ T cells and T reg cells are essential targets for cancer immunotherapy. Our scRNAseq data in the MC38 and CT26 models provided an opportunity to identify common targets expressed on these cells from both mouse models and human tumors. Targeting T reg cells by anti–CTLA-4 antibody is clinically beneficial but also associated with significant side effects in cancer patients (Tanaka and Sakaguchi, 2017; Nishikawa and Sakaguchi, 2014). We therefore focused on the identification of additional targets commonly expressed in both human and mouse tumor-related T reg cells, but not in bystander T reg cells. Ccr8 was a top gene identified that was preferentially expressed on T reg cells isolated from various human cancer types and in the MC38 and CT26 models (Figs. 8 C and 9, A–C). Importantly, compared with other known T reg cell depletion targets, such as CTLA-4, CD25, OX40 (TNFRSF4), and CCR4 (Onda et al., 2019; Sharma et al., 2019; Sugiyama et al., 2013; Bulliard et al., 2014), CCR8 was more preferentially enriched in CRC-infiltrating T reg cells (Fig. 9 B). Unlike Ctla4, the frequency of Ccr8 expression was much lower in peripheral and normal tissue T reg cells, suggesting that it might be a better T reg cell–depleting target. Similar patterns were also observed in mouse (Fig. 9 C). Although in mouse T cell subsets, Ccr8 expression was broader than that in human (Fig. 9 C), its expression was still better restricted in tumor T reg cells compared with CTLA-4 and OX40 (Fig. 9 C).
Figure 9.
Characterization of CCR8 expression in mouse tumor T reg cells. (A) tSNE plot of Ccr8 expression. (B) Violin plots displaying the cluster-specific expression of Foxp3, Ccr8, and other key T reg cell–related marker genes in human tumors taken from Zhang et al. (2018). (C) Violin plots displaying the cluster-specific expression of Foxp3, Ccr8, and other key T reg cell–related marker genes in mouse. (D) Flow cytometry analysis of CCR8 expression in T cells from MC38 tumors (upper: percentages; lower: mean fluorescence intensity [MFI]). (E) Coexpression of CCR8 and 4-1BB in MC38 tumor-derived CD4 TEFF populations (****, P < 0.0001 paired t test). (F) Coexpression of CCR8 and CCR7 in MC38 tumor CD8 populations (**, P < 0.005 paired t test). (G) Flow cytometry analysis of CCR8+ levels on tumor-infiltrating T reg cells across multiple syngeneic mouse tumor models shown as representative FACS plots (left) and as average bar graphs relative to total T reg cells (right). (H) CCR8 expression on tumor-infiltrating T reg cells compared with expression in peripheral lymphoid organ T reg cells. (*, P = 0.0250; **, P = 0.0021; ****, P < 0.0001; two-way ANOVA with Tukey’s post-hoc analysis.) (I) Representative FACS plots of T reg cell levels in CT26 tumor and peripheral lymphoid tissues after administration of CCR8-specific depleting, nondepleting, and isotype control antibodies. (J) Summary of CD25+/FOXP3+ T reg cell infiltration (left) and changes in CD8/T reg cell ratio in tumors and peripheral lymphoid upon antibody dosing (****, P < 0.0001; two-way ANOVA with Tukey’s post-hoc analysis). DLN, draining LN.
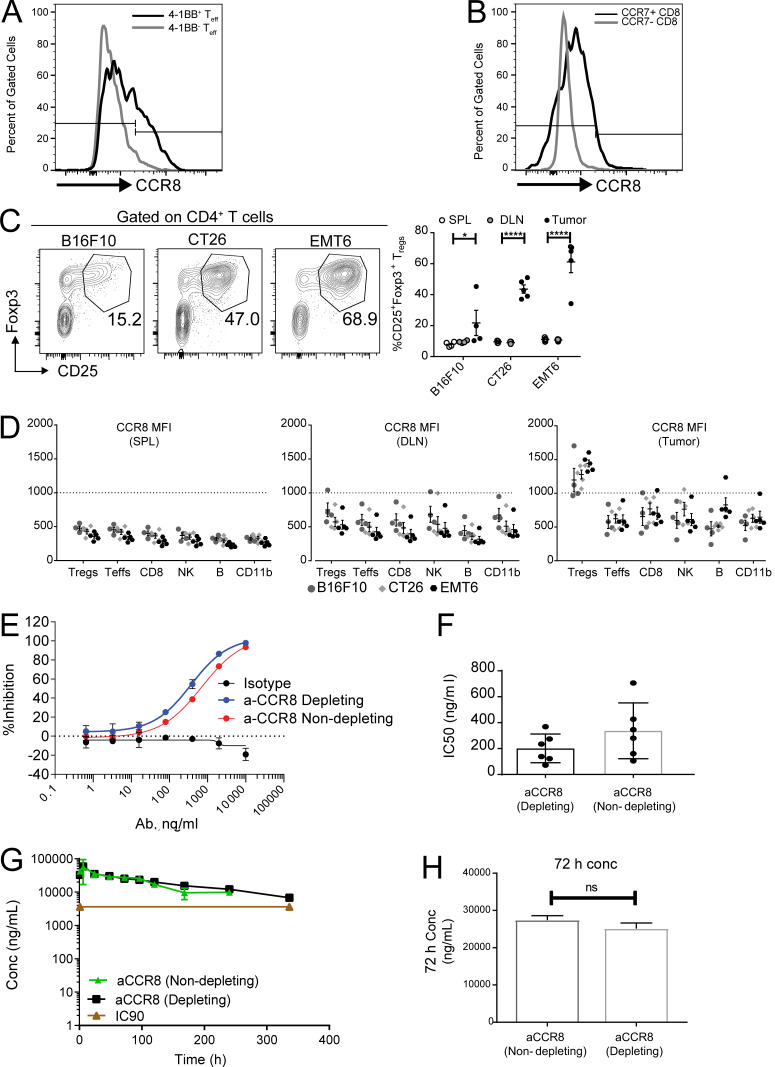
To better understand the protein expression of CCR8 in tumor-infiltrating T reg cells and other T cell subsets, we analyzed the expression of CCR8 on tumor-infiltrating T cells from MC38 model tumors by FACS. While ∼70% of tumor-infiltrating T reg cells were positive for CCR8, <5% of CD8+ cells and ∼10–15% of CD4+FOXP3− cells had CCR8 expression (Fig. 9 D). In addition, the quantity of CCR8 expression on positive cells was significantly higher in T reg cells than in other cell types as measured by mean fluorescence intensity (Fig. 9 D). Within the CD4+FOXP3− population, CCR8+ cells were enriched in 4-1BB+ Th1-like cells (Figs. 9 E and S5 A), and in CD8+ cells the percentage of CCR8+ cells was significantly higher in the CCR7+ population than in the CCR7− population (Figs. 9 F and S5 B), consistent with the observations from scRNAseq data (Fig. 9 C). We next validated whether increased CCR8 protein expression could be detected on tumor-infiltrating T reg cells across several syngeneic mouse tumor models—CT26, B16F10, and EMT6. CD25+Foxp3+ T reg cells comprise ∼15–60% of the total CD4+ T cells infiltrating mouse tumors (Fig. S5 C). Within the total T reg cell compartment, CCR8+ T reg cells constituted ∼50–70% of total tumor T reg cells in these models (Fig. 9 G). Importantly, CCR8 expression was specifically upregulated on tumor-infiltrating T reg cells relative to its expression on T reg cells from peripheral lymphoid organs (spleen and draining LNs), as well as other immune cell types across all tumor models (Figs. 9 H and S5 D). These analyses confirmed enriched CCR8 protein expression on tumor T reg cells across multiple syngeneic mouse tumor models, in alignment with the scRNAseq analysis from the MC38 and CT26 models.
Figure S5.
CCR8 expression in tumor T reg cells and characterization of anti-CCR8 antibodies. (A) Representative histogram plot depicting expression of CCR8 on 4-1BB+ and 4-1BB− CD4+Foxp3− TEFF population in MC38 tumors. (B) Representative histogram plot depicting expression of CCR8 on CCR7+ and CCR7−CD8+ T cells in MC38 tumors. (C) Flow cytometry analysis of tumor-infiltrating T reg cells across multiple mouse tumor models shown as representative FACS plots (left) and frequencies in tumors and peripheral lymphoid organs (right). *, P = 0.0250; ****, P < 0.0001; two-way ANOVA with Tukey’s post-hoc analysis. (D) Flow cytometry analysis of CCR8 expression on immune cell types (Foxp3+ T reg cells, Foxp3− T effectors, CD8+, NKp46+ NK, CD19+ B, and CD11b+) in tumors and peripheral lymphoid tissues across tumor models. (E) In vitro chemotaxis assay evaluating the ability of CCR8-depleting and -nondepleting antibodies to block 100 pM CCL1-induced chemotaxis. (F) Bar graph comparing IC50 values from the chemotaxis assay testing CCR8-depleting and -nondepleting antibodies (n = 6). (G) Pharmacokinetic assessment of serum antibody concentration after single-dose administration of CCR8-depleting and -nondepleting antibodies (10 mg/kg i.p.). (H) Serum concentration at 72 h of animals treated as in G. DLN, draining LN; SPL, spleen; MFI, mean fluorescence intensity.
Owing to previous publications demonstrating that the CT26 tumor model is sensitive to anti–CTLA-4 monotherapy (Selby et al., 2013; Simpson et al., 2013), we decided to first evaluate the antitumor potential of targeting CCR8 as a single agent in the CT26 tumor model. Furthermore, we sought to dissect the relative contribution to antitumor activity of a T reg cell–depletion strategy mediated by anti-CCR8 antibody versus inhibition of T reg cell recruitment to the tumor via CCL1-induced chemotaxis (Iellem et al., 2001). We generated a CCR8-depleting antibody with antibody-dependent cytotoxicity activity (aCCR8 mIgG2a) and a nondepleting version that lacks antibody-dependent cytotoxicity (aCCR8 mIgG1 N297G). Both of these antibodies could block the chemotaxis activity induced by CCL1, the dominant ligand for CCR8, in vitro (Fig. S5 E), while the half-maximal inhibitory potency (IC50) of the depleting antibody trended higher than the nondepleting antibody in multiple experiments (Fig. S5, E and F). To ensure both antibodies could sufficiently block CCL1-induced chemotaxis in vivo, we measured the pharmacokinetic parameters of both antibodies in vivo with 10 mg/kg i.p. dosing. Both antibodies had similar pharmacokinetics and, at 72 h, the nondepleting antibody could still maintain the high serum level at 27.4 μg/ml, which is 7.6-fold higher than the IC90 (3.6 ug/ml) for blocking CCL1-induced chemotaxis. We thus chose the 10-mg/kg i.p. dose every 3 d for all in vivo studies.
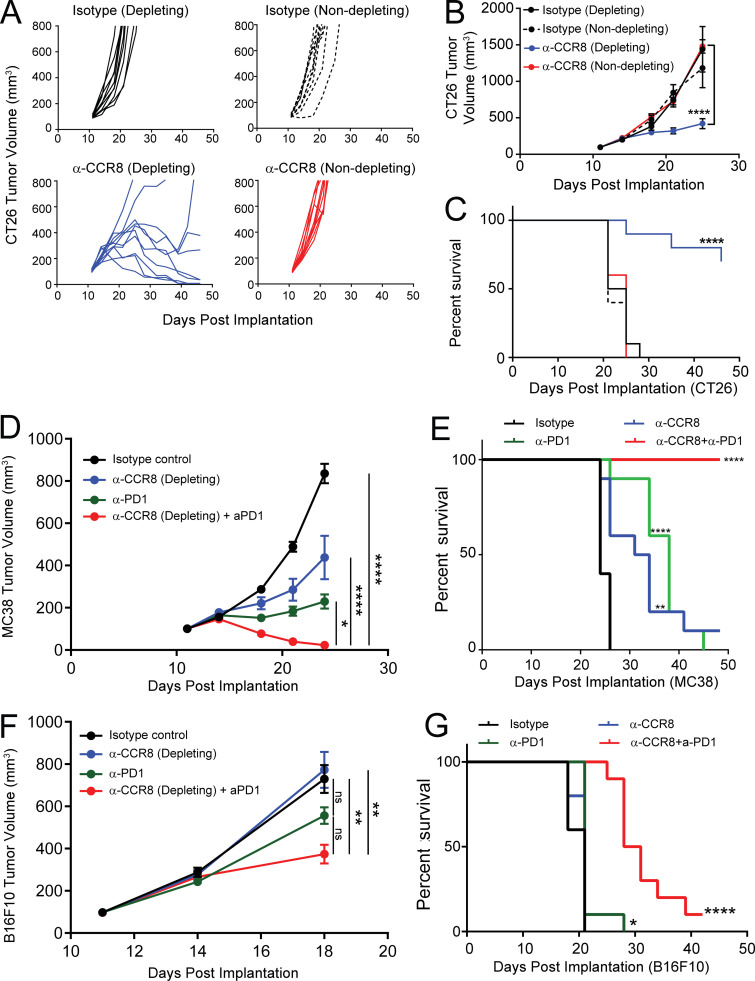
Single-dose administration of these antibodies in CT26 tumor-bearing animals led to a preferential depletion of tumor T reg cells with the CCR8-depleting antibody and a corresponding increase in the CD8/T reg cell ratio in tumors without impacting T reg cells in peripheral tissues (Fig. 9, I and J). Importantly, treatment with the CCR8-nondepleting antibody did not lead to any changes in T reg cell frequencies or the CD8/T reg cell ratio in any of the tissues evaluated (Fig. 9, I and J). Although these data could not exclude the possibility that anti-CCR8 antibody also depleted some small portions of CCR8+ non–T reg cells, dosing therapeutically—day 11 after tumor implantation—with the CCR8-depleting antibody in CT26 tumor-bearing animals demonstrated significantly reduced tumor burden and improved overall survival compared with isotype-treated animals (Fig. 10, A and B), supporting the idea that depletion of CCR8+ T reg cells was sufficient to drive a robust antitumor immune response. In contrast, blockade of CCR8/CCL1 chemotaxis alone did not suffice to drive antitumor activity, since animals treated with the nondepleting CCR8 antibody failed to exhibit any reduction in tumor burden (Fig. 10, A and B). Next, we asked whether anti–CCR8-depleting antibody could synergize with anti–PD-1 therapeutically in mouse tumor models. Since this antibody demonstrated superior efficacy in the CT26 model as a single agent with minimal window for a synergistic effect, we tested anti-CCR8 antibody and anti–PD-1 antibody combination in MC38 and B16F10 tumor models. Anti-CCR8 antibody alone also significantly reduced the tumor progression in the MC38 model (Fig. 10, D and E), although it was less than that in CT26 model, whereas it did not provide therapeutic benefit in the B16F10 model as a single agent (Fig. 10, F and G). In both models, however, anti-CCR8 significantly synergized with anti–PD-1 to slow the tumor progression and prolong the survival of the animals. These preclinical data in mouse models highlight the preferential enrichment of CCR8 expression on tumor T reg cells as an important target to specifically deplete tumor T reg cells without impacting peripheral T reg cells to boost antitumor immunity. Furthermore, it suggests that blockade of the chemotaxis function of CCR8 alone may not suffice for a robust antitumor therapy.
Figure 10.
Antitumor activity of targeting CCR8 expression on tumor-infiltrating T reg cells. (A) CT26 tumor growth curves of animals treated with CCR8-specific depleting and nondepleting antibodies, along with depleting and nondepleting isotype controls (top). (B and C) Averaged tumor growth (B) and overall survival of animals (C) treated as in A. (D) Averaged tumor growth curves of MC38 tumors treated with isotype control, anti-CCR8, anti–PD-1, or combination. (E) Overall survival of MC38 tumor bearing mice treated in D. (F) Averaged tumor growth curves of B16F10 tumors treated with Isotype control, anti-CCR8, anti–PD-1, or combination. (G) Overall survival of B16F10 tumor-bearing mice treated in F (****, P < 0.0001; ***, P < 0.0002; **, P < 0.002; *, P < 0.05; two-way ANOVA with Tukey’s post-hoc for tumor growth inhibition and Mantel-Cox log rank test for survival curve analysis).
Discussion
The explosion of scRNAseq data from various human cancers and preclinical models has enabled a highly granular view of intratumoral immune cells (Zhang et al., 2020; Tirosh et al., 2016; van der Leun et al., 2020; Yu et al., 2020). These data have unveiled potential mechanisms underlying the susceptibility or the resistance of cancers to immunotherapies (Yost et al., 2019; Zhang et al., 2020). scRNAseq data can be used to identify novel cell subsets and states, their potential functions and unique marker genes, and their dynamic behavior over time or in response to treatment. Advanced bioinformatic analyses of these data have also provided insights into developmental trajectories between related cell subsets (Park et al., 2020; Miragaia et al., 2019); however, there remain challenges on how to best interpret scRNAseq data from multiple studies when analyzing transcriptome data. Individual studies usually capture the status of cells at fixed time points due to the limitation of sample collection and cost. Challenges also remain in the comparison of cellular subsets and cellular status across studies, especially across species. In studies of T cells, TCR information provides a rich layer of data with which to address these questions. The STARTRAC method we have previously developed is uniquely suited to integrate transcriptome and TCR clonal information to better describe the dynamics of T cell subsets (Zhang et al., 2018, 2020).
In this study, we sequenced a large number of MC38 tumor and peripheral tissue–derived T cells, capturing both transcriptome and TCR information with the goal of understanding the expansion, migration, and differentiation of tumor-infiltrating CD8+, CD4+, and T reg cell subsets. By using a stepwise cell-partitioning approach in the single-cell transcriptome analysis, we excluded most technical noises while retaining the true biological variations among different cell populations, thereby identifying robust cell subsets across animals and experiment batches. We confirmed our findings with additional scRNAseq analyses from MC38 tumor at different time points and from CT26 tumor. Although cell subsets of different major populations may not be partitioned at a uniform depth, this method performed well at revealing biologically unique subsets—for example, Th1, Th17, and Tfh subsets within memory CD4+ cells; however, this method might result in overclustering and artificial subsets. Thus, the biological significance of certain subsets, such as various CD8+ Tex subsets, needs to be further examined in vivo. We also aimed to understand the commonalities and differences of these subsets in human cancers versus syngeneic mouse tumor models. We found that tumor T cells are comprised of mostly highly clonally expanded CD8+ exhausted cells and moderately clonally expanded T reg cells. In contrast, peripheral tissue–derived T cells are more clonally diverse and have a low degree of clonal expansion. We have identified cells with clonotypes found in multiple tissues. In particular, those that migrate between tumor and any peripheral tissue have distinct gene expression phenotypes from those that are tissue-restricted clonotypes. By comparing TCRs from individual mice, we also identified many public TCR CDR3s. Paired TCRαβ sequences allowed us to characterize the features of ppTCRs inside the tumor. We focused only on ppTCR CDR3s, which more stringently define antigen specificity, and demonstrated that there was a clear enrichment of ppTCR preferentially inside tumors, supporting tumor antigen–driven selection of these TCRs. In the future, it will be intriguing to explore the potential antitumor functions of these ppTCRs.
Combined expression and clonality analysis of T cell subsets by STARTRAC between species provided important insights to translate detailed mechanistic data from animal models to the development of novel therapeutic agents for human cancer (Zhang et al., 2020). By comparing the gene signature of mouse tumor Tex and various human cancer Tex cells (Fig. 6, D and E), we identified many common and species-specific markers. Within all PD-1hi subsets, expression of some key exhaustion markers, such as Pdcd1 and Lag3, were similar, but expression of other markers, including Havcr2 and Cd244, were not as strongly correlated. We also found that the combination of trajectory analysis and STARTRAC may be a validated method by which to fine-tune the corresponding subsets in different studies according to their clonotype-centric behaviors. For example, C08_CD8_Tex_Cd244 was closely related to terminally differentiated Tex cells in human tumors based on STARTRAC migration and transition indices. This approach may help to better interpret data from mouse models with various treatments in the future and enhance translatability.
Augmented IFN-γ–producing Th1 cells are associated with better responses to anti–PD-1 treatments in CRC patients with microsatellite instability status (Llosa et al., 2015; Mlecnik et al., 2016). Interestingly, only the BHLHE40+ Th1-like cells, not EOMES+ Th1, are significantly enriched in CRC patients with microsatellite instability status over microsatellite stable CRC patients (Zhang et al., 2018). Our recent study also demonstrated that an anti-CD40 agonist antibody can synergize with anti–PD-1 to induce remission in MC38 mouse model through the early induction of Bhlhe40+ Th1-like cells (Zhang et al., 2020), supporting the important contribution of these cells in cancer immunotherapies. However, the relationship of these tumor-infiltrating Bhlhe40+ Th1-like cells with other Th cell subsets is not fully understood. In this study, STARTRAC clonotype analysis revealed these Th1 cells are not only developmentally related to intratumoral TCM-like CD4+ cells, but also to peripheral Tfh cells in the LN and spleen. CD4+ Tfh cells express Bcl6, Il21, and Cxcr5, and are defined as germinal center T cells essential for T-dependent humoral immune responses (Crotty, 2019). Recent studies have also supported the potential role of Tfh cells in antitumor immune responses. Tfh cells are associated with longer progression-free survival in head and neck squamous cell carcinoma (Cillo et al., 2020). In addition, in a study of immune checkpoint blockade in a triple-negative breast cancer mouse model, the therapy activated Tfh cells and enhanced the antitumor response through activation of B cells (Hollern et al., 2019). In our study, although Th1-like and Tfh cells were indeed clonally related, the developmental order of this relationship was not clear, and future research is needed to reveal the biological significance of their developmental connection.
Finally, the refined cell subsets and transcriptome data enabled us to identify potential novel therapeutic candidates. T reg cells are important target cells for cancer immunotherapy. Here, by using STARTRAC, we identified two peripheral T reg cell subsets that were developmentally connected with tumor T reg cell populations. Interestingly, CCR8 was preferentially expressed on the C23_Treg_Cd83 subset and on tumor-infiltrating T reg cells. CCR8 has also been identified to be preferentially expressed on T reg cells infiltrating various cancers (Plitas et al., 2016; De Simone et al., 2016; Zheng et al., 2017; Zhang et al., 2018). CCR8 is a chemokine receptor expressed on various leukocytes, especially T cells (Karin, 2018). Human CCR8 has four putative ligands: CCL1, CCL8, CCL16, and CCL18. CCL1 is its key ligand governing T cell homing to skin (Schaerli et al., 2004) and a critical modulator of immunosuppression of autoimmune models (Barsheshet et al., 2017). CCR8 was reported to be differentially expressed on Th2 cells (Zingoni et al., 1998) and on tissue-resident memory T cells in human skin (McCully et al., 2018). In humans there are three subsets of T reg cells in the blood: T reg cell I (CD45RA+FOXP3lo), T reg cell II (CD45RA−FOXP3hi), and T reg cell III (CD45RA−FOXP3lo). T reg cell II expresses the highest CCR8 and shares TCRs with intratumor T reg cells in breast cancer patients (Wang et al., 2019). Thus, the C23_Treg_Cd83 subset from this study is likely the mouse counterpart of the human T reg cell II population.
Based on its expression profile, CCR8 may be a good target to specifically deplete tumor-related T reg cells. Previous studies showed that an anti-CCR8 antibody treatment suppressed CT26 tumor growth when antibody was administered on day 4 after CT26 tumor implantation (Villarreal et al., 2018). This antibody also synergized with Listeria monocytogenes–based tumor vaccines to significantly delay growth of established tumors and to prolong survival (Villarreal et al., 2018). The authors hypothesized that this antibody acts to block ligand-receptor interactions that result in T reg cell depletion inside the tumor; however, it is still possible that the mechanism of action in vivo was through an Fc receptor–mediated direct T reg cell depletion by the anti-CCR8 antibody. In our study, by using an engineered effectorless antibody, we showed that depletion, but not blocking the ligand binding alone, was essential for its anticancer activity. We were able to demonstrate monotherapy effect even in an established CT26 tumor model. Importantly, this depletion antibody could also synergize with anti–PD-1 antibody to treat established tumors in both MC38 and B16F10 models.
In summary, we have successfully used the STARTRAC algorithm in a mouse model to describe the intratumor dynamics of various T cell subsets and revealed a potential novel therapeutic candidate. STARTRAC may serve as a powerful tool with which to elucidate various mechanisms underlining the antitumor T cell response in both mouse and human cancers.
Finally, we have created an interactive portal at http://cancer-pku.cn:3838/mc38startrac/, which can be used as a resource for data exploration and identification of novel regulatory mechanisms for tumor infiltrating T cells.
Materials and methods
Animals and cell lines
MC38, B16F10, EMT6, and CT26 mouse tumor cell lines were acquired from the American Type Culture Collection and cultured in complete RPMI medium consisting of 10% heat-inactivated FBS, penicillin (100 U/ml), streptomycin (100 mg/ml), and 1× Glutamax and were periodically tested for Mycoplasma by IDEXX. Cells were thawed from liquid nitrogen stocks and passaged two to three times before being used for in vivo or in vitro experiments.
6–8-wk-old female C57BL/6 and BALB/C mice were purchased from Charles River Laboratories. All animals were housed at Association for Assessment and Accreditation of Laboratory Care International–accredited facilities (Amgen). Animal procedures were approved by the Amgen Institutional Animal Care and Use Committee and were compliant with the Guide for the Care and Use of Laboratory Animals (eighth edition).
For single-cell studies, mice were injected s.c. in the rear right flank with 3 × 105 MC38 or CT26 cells per site of injection. Tumor growth was monitored until takedown on day 16 for the main experiment, or at days 11 and 20 after implantation for comparative experiments shown in Fig. 5. Day 20 MC38 scRNAseq data were derived from the isotype-treated samples described in Zhang et al. (2020). For experiments represented in Fig. 9, G–J, Fig. 10, and Fig. S5, C and D, mice were injected with 3 × 105 CT26, EMT6 cells, or 2 × 105 B16F10 cells. Animals subject to antibody treatment were injected with 3 × 105 CT26 cells on the day of implantation. Tumors were allowed to grow for 10–11 d, after which animals were assigned (mean tumor volume = 110 mm3) to different treatment groups and dosed with either isotype antibodies (mIgG1 MOPC-21 and mIgG2a C1.18.4; catalogue #BE0083 and #BE0085; BioXCell) or CCR8-depleting and -nondepleting antibodies (rat SA214G2; BioLegend; murinized internally on mIgG1 N297G effectorless and mIgG2a effector-competent backbones). Antibodies were dosed i.p. at 300 µg once every 3 d for a total of three doses (every 3 d × 3). For pharmacodynamic studies, tumors and peripheral lymphoid tissues were harvested 48 h after single antibody dose (300 µg). For efficacy studies, tumor volumes were measured twice per week until survival end point (tumor volume ≥ 800 mm3).
Tissue isolation and cell preparation
Peripheral blood was isolated from anesthetized animals via cardiac puncture and diluted into a solution of room temperature 1× PBS + EDTA (5 mM). Tumor, spleens, and tumor-draining LNs were subsequently collected after sacrifice.
Tumors were minced into 1-mm pieces and resuspended in 10 ml DMEM/F12 supplemented with 200 µg/ml Liberase TL (Roche) and DNase I 5 µg/ml (Sigma-Aldrich), followed by mechanical dissociation using Miltenyi C-tubes and the gentleMACS dissociater with the manufacturer program, hu_tumor_01. Suspensions were incubated at 37°C with shaking at 180 rpm for 20 min, followed by enzyme quenching by addition of 1 ml FBS and an additional two rounds of gentleMACS dissociation using program hu_tumor_02. Tumor suspensions were strained through 70-µm filters. The filter was rinsed with 10 ml MACS buffer (PBS, 2% FBS, 2 mM EDTA). Cells were then pelleted at 1,500 rpm for 5 min at 4°C and resuspend in 5 ml MACS buffer. Cells were counted on ViCell XR (Beckman Coulter) and resuspended to the appropriate concentration for T cell enrichment.
Peripheral blood mononuclear cells were isolated from peripheral blood by density centrifugation as follows: ∼3–4 ml of diluted blood was layered over 3 ml Lympholyte-Mammal (Cederlane) and centrifuged for 20 min at 800 g at room temperature, brakes off. Lymphocytes were washed twice with 40 ml MACS to remove platelets. Spleen and LN were homogenized through 40-µm strainers. Samples were preenriched for total T cells by immunomagnetic negative selection using StemCell Technologies EasySep Mouse T Cell Isolation Kit (19851) following the manufacturer protocols for all three peripheral tissue suspensions, with minor modifications for processing of tumor suspensions. The negative selection antibody cocktail for tumor samples was supplemented with 15 μg/ml anti–CD34-biotin (13–0341-82; eBioscience) to deplete MC38 tumor cells. After labeling, the tumor cocktail suspension was washed once with 5 ml MACS buffer before a 10-min incubation with eBioscience MagniSort Streptavidin Negative Selection Beads (MSNB600274; Invitrogen) at 20 µl beads/100 ml sample. All sample volumes were brought up to 2.5 ml with additional MACS buffer. Samples were then placed on a StemCell Technologies magnet for 10 min and T cells were subsequently collected.
Antibodies and flow cytometry
All enriched mouse single T cell samples were stained for viability (Sytox Red) and with antibodies against CD45, TCRβ, and CD90.2 (eBioscience). Approximately 10e5 viable CD45+TCRβ+CD90.2+ cells were sorted, washed in PBS, counted, and resuspended to a concentration range of 7e5–1e6 cells per ml. For in vivo immune cell profiling in Fig. 7 and Fig. S5, cells were stained with antibodies against CD45 (30-F11; catalogue #103116), TCRβ (H57-597; catalogue #109228), CD4 (GK1.5; catalogue #100438), CD8 (53–6.7; catalogue #100722), CD19 (6D5; catalogue #115546), CD11b (M1/70; catalogue #101226), NKp46 (29A1.4; catalogue #137606), CCR8 (SA214G2; catalogue #150312; all from BioLegend); CD25 (PC61; catalogue #564022; BD Biosciences); CCR7 (4B12; catalogue #562675; BD Biosciences); 4-1BB (17B5; catalogue #25137180; eBioscience); and Foxp3 (FJK-16s; catalogue #11-5773-82; eBioscience).
Chemotaxis assay
Assessment of mouse anti-CCR8 antibodies’ ability to block chemotaxis was done using BW5147.G.1.4 cells, a murine T lymphocyte cell line endogenously expressing CCR8. Testing was done in a 96-well Transwell plate with a 5-µm pore size in complete BW5147.G.1.4 growth medium. Cells were preincubated with test antibodies for 30 min and transferred to the top Transwell chambers in total 50 µl volume and 200,000 cells per well load. Recombinant human CCL1 (R&D Systems) was prepared at suboptimal concentration of 100 pM and added to the bottom Transwell chambers at 100 µl per well. Transwell plates were incubated at 37°C 5% CO2 for 4 h. Ligand suboptimal concentration was established based on the cells’ chemotactic dose-response curve and varied from the effective response concentration (EC50 to EC80) for different experiments. At the end of incubation, the top chambers were removed and 50 µl/well CellTiterGlo reagent (Promega) was added to the bottom chambers with migrated cells. After 10 min of incubation at room temperature, 100 µl of the mix from the bottom chamber was transferred to the black well clear-bottom plates for Luminescence readout (Envision plate reader). The percent inhibition of chemotaxis was calculated using Basal and Max chemotaxis control wells present on each plate. The percent inhibition and IC50 values were calculated using Screener analysis software.
Single-cell sequencing library preparation
∼17,000 cells were loaded onto 10× chromium instrument for single-cell encapsulation with the 5′ VDJ kit (10x Genomics) to recover an estimated 10,000 cells. RT was performed according to the manufacturer’s protocol, followed by emulsion breakage and cleanup of single-cell cDNA. cDNA was preamplified for 12 cycles to generate an enriched library for parallel TCR and total cDNA libraries. 5% of preamplified cDNA was reserved for TCR library generation by two rounds of nested multiplex PCR using nested primers complementary to the mouse Trac, Trbc1, and Trbc2 loci. A single Cβ primer was designed complementary to the identically conserved 5′ ends of Trbc1 and Trbc2, and all primers were restricted to have a similar temperature of 58–60°C and used at a final 1 μM concentration. Primer sequences are as follows: mTRAC_R1: 5′-GCATCACAGGGAACGTCTGA-3′; mTRAC_R2: 5′-AAGTCGGTGAACAGGCAGAG-3′; mTRBC_R1: 5′-TGCTCAGGCAGTAGCTATAATTGCT-3′; and mTRBC_R2: 5′-TTTGATGGCTCAAACAAGGA-3′. 50 ng of enriched total cDNA and 50 ng of enriched mTCR libraries were used to complete the remainder of the single-cell sequencing library preparation according to the 10x Genomics protocol. Final total cDNA and TCR libraries for each tissue were diluted to 20 nM and pooled at equimolar ratios. Pooled total cDNA libraries were sequenced to a depth of 500 million reads per sample on a NovaSeq (26 bp read 1, 98 bp read 2, and 8 bp index i7). Pooled TCR library was sequenced to a depth of 50 million reads per sample on a HiSeq4000 (150 bp paired end read, 8 bp index i7). Fastq files have are available at GEO accession no. GSE168944.
Sequencing alignment and data analysis
Sequences were aligned to the Ensembl mouse reference transcriptome using the 10x Genomics CellRanger pipeline (version 2.1.0). Quality control and transcriptome analysis of the single-cell datasets were performed using the R package Seurat (version 2.3.0; Butler et al., 2018). Cell matrices are available at GEO accession no. GSE168944. Genes detected in less than three cells were excluded. Cells were filtered for quality based on the following criteria: of having a unique molecular identifier (UMI) count of between 2,000 and 60,000, mapping to over 200 unique genes, and the fraction of unique mitochondrial transcripts was less than 5%. The TCR library sequences were processed using the CellRanger vdj function supplied with mouse TCR gene annotations and reference sequence files acquired from the International Immunogenetics Information System database (http://www.IMGT.org) and prepared using the mkvdjref function. Cells were further filtered for the presence of a productive paired single TCR α and β chain.
After quality control, 16,136 genes and 64,449 cells remained and constituted an expression matrix. The expression matrix was first normalized using the Seurat function, LogNormalize, and then scaled to universal mean and variance using the Seurat function, ScaleData, with library size and percentage of mitochondrial reads regressed. Next, data from two different sequencing batches (T01 and T05 as the first batch, and T09 and T10 as the second batch) were aligned using the Seurat function AlignSubspace. Specifically, highly variable genes were selected separately in two batches using the Seurat function FindVariableFeatures, with the default parameter, and the first 30 canonical correlation components were computed on the intersecting highly variable genes and aligned across batches. t-Distributed stochastic neighbor embedding (tSNE) dimensionality reduction and graph-based Louvain clustering were performed on the aligned components using the Seurat functions, RunTSNE and FindClusters. A total of 17 major clusters emerged at the clustering resolution of 0.6. Every cluster was examined iteratively for heterogeneous substructures. Briefly, the expression matrix of the examined major cluster was first extracted, and then a second round of graph-based clustering was applied to <10 principal components identified as meaningful—exhibiting significance of explained variance by the Seurat function JackStraw and no obvious difference between the two batches. Subgroups with more than five genes different from other cells in the same cluster (P < 0.05; average log fold-change > 1) were assigned as separate clusters. The final dataset had 33 clusters. Wilcoxon rank sum test was performed to identify significantly upregulated genes in each cluster using the Seurat FindMarkers function. Cell-cycle analysis of cells was done in Seurat using the CellCycleScoring function and cell-cycle genes described in Nestorowa et al. (2016).
To validate that the T cell subsets identified in the main dataset (MC38 tumor, day 16) were prevalent in mouse models, scRNAseq data of MC38 tumors at different time points (day 11 and day 20) and from CT26 tumors were also obtained. Day 11 mc38 data were obtained from Zhang et al. (2020). For those validation datasets, the Seurat3 pipeline was applied to obtain the same cluster annotation as that in the main dataset. First, the data of the two batches of the main dataset were integrated by calling function FindIntegrationAnchors and IntegrateData with default settings. Then, the integrated main dataset was used as a reference, and the cluster annotations were transferred to each of the validation dataset by calling function FindTransferAnchors and TransferData. For visualization, all datasets were integrated by calling function FindIntegrationAnchors and IntegrateData. The integrated data were then scaled, after which PCA and tSNE analysis were performed.
STARTRAC analysis
STARTRAC analysis of clonal transition, migration, and expansion was done as described in Zhang et al. (2018). Briefly, clonotype expansion was calculated as 1-evenness in each cluster, while migration and transition scores for each cluster were calculated as the weighted sum of the Shannon entropy indices of each clonotype in said cluster based on its distribution across multiple tissues or clusters, respectively. In the case of pairwise migration and transition scores as in Fig. 4, B and C, the Shannon entropy score was restricted to the clonotype distribution across individual pairs of tissues or clusters. The significance of individual STARTRAC scores was determined by random permutation of cluster identities (number of permutations, n = 1,000). Specifically, in each permutation, the cluster identities of cells were randomly sampled without replacement from the original data and reassigned to cells. Then, the STARTRAC score (transition/migration/expansion) was calculated using the permuted data. Significance was obtained by comparing the score of original data to the distribution of the scores of the 1,000 permuted data, and P value was calculated as the number of times the scores of permuted data exceeded the score of original data divided by 1,000. Code for STARTRAC analysis is available at https://github.com/Japrin/STARTRAC. Scores and significance for CD8+ clusters and CD4+/T reg cell clusters were calculated separately. To calculate STARTRAC scores for TCR chain publicity within clusters, first a publicity index for each individual amino acid–level α or β TCR chain was calculated as:
where is the ratio of the number of cells with chain c in mouse m to the total number of cells with TCR chain c across all mice, and The total publicity index for each cluster is then defined as the weighted average of all TCR chain publicity indices within the cluster and is calculated as follows:
in which is the ratio of the number of cells with chain c in cluster cls to the total number of cells in cluster cls. See Table S10, Table S11, and Table S12.
Gene signature analysis
CD8+ Tex and CD4+ T reg cell cluster-specific marker genes that were homologous between human and mouse were compared. First, differential expressed genes were identified using limma by comparing CD8+ Tex (or CD4+ tumor-enriched T reg cell) against all other CD8+ (or CD4+) clusters. Then, for humans, cluster-specific marker genes were defined as those with log2FC > 1.5 and adjusted P value < 0.01 in all three human cancers (hepatocellular carcinoma, non–small-cell lung cancer, and CRC); for mouse, cluster-specific marker genes were those with log2FC > 0.3 and adjusted P value < 0.01. Genes showing cluster specificity in both species are indicated as core, and other cluster-specific marker genes were indicated as top.human or top.mouse.
Online supplemental material
Fig. S1 shows basic clonotype information and illustrates the methodology and stability of the clustering approach taken. Fig. S2 shows quality control metrics and doublet analyses of the single-cell dataset. Fig. S3 shows additional analyses of TCR chain publicity status across multiple animals and within different phenotypic clusters, subset-specific gene expression among CD8+ Tex clusters, and the cell-cycle phase assigned by different clusters. Fig. S4 shows the gene expression levels of key Th subset marker genes; monocle trajectory analysis of CD8+, CD4+ Th, and T reg cell populations; and the STARTRAC pairwise tissue migration scores of CD4+ Th populations. Fig. S5 shows more detailed flow cytometry analysis of CCR8 levels within specific tumor TEFF and CD8+ populations, CCR8+ levels within different mouse tissues and tumor models, and detailed pharmacodynamic characterization of the α-CCR8+ antibodies used in this study. Table S1 is a summary table of cluster attributes, including clonal expansion, tissue localization, and representative marker gene expression. Table S2 lists cell metadata indicating tissue, individual mouse, tSNE coordinates, cell-cycle phase, gene expression cluster, and TCR clonotype. Table S3 contains differential expression values of NKT clusters C20 and C26:28. Table S4 lists cell metadata, as in Table S2, with additional annotation of the V, D, and J segments and TCR publicity status. Table S5 contains differential expression analysis of genes in each CD8+ cluster. Table S6 contains the cross-species comparison of human gene expression and mouse gene expression in CD8+ Tex clusters. Table S7 and Table S8 list differential expression analyses of genes in each CD4+ cluster and each CD4+Foxp3+ T reg cell cluster, respectively. Table S9 contains the cross-species comparison of human and mouse gene expression in CD4+Foxp3+ T reg cell clusters. Table S10, Table S11, and Table S12 contain the cluster-specific STARTRAC clonotype analysis, given as aggregated scores, tissue pairwise migration scores, and cluster pair transition scores, respectively.
Supplementary Material
is a summary table of cluster attributes, including clonal expansion, tissue localization, and representative marker gene expression.
lists cell metadata indicating tissue, individual mouse, tSNE coordinates, cell-cycle phase, gene expression cluster, and TCR clonotype.
provides differential expression values of NKT clusters C20 and C26:28.
lists cell metadata, as in Table S2, with additional annotation of the V, D, and J segments and TCR publicity status.
is a differential expression analysis of genes in each CD8+ cluster compared to all other CD8 clusters as in Table S3.
provides a cross-species comparison of human and mouse CD8 Tex clusters.
shows a differential expression analysis of genes in each CD4+ cluster compared to all other CD4+ clusters as in Table S3.
shows a differential expression analysis of genes in each CD4+Foxp3+ T reg cell cluster compared to all other T reg cell clusters as in Table S3.
provides a comparison of human gene expression and mouse gene expression in CD4+Foxp3+ T reg cell clusters, as in Table S6.
is a STARTRAC analysis of T cell clusters.
is a pairwise STARTRAC analysis of T cell migration.
is a pairwise STARTRAC analysis of T cell cluster transitions.
Acknowledgments
We thank Xiaobin Tang, Charles Tran, and Nathan Deer for assisting with sample preparation and sorting, and Ian Driver for assisting with sequence alignment.
All studies in this manuscript were funded by Amgen research.
Author contributions: W. Ouyang, D. Bhatt, and D. Sawant designed the project and all experiments; W. Ouyang, Z. Zhang, J. DeVoss, and A. Symons supervised the project; D. Bhatt and J.Y. Huang performed single-cell experiments; D. Bhatt, B. Kang, L. Zheng, and W. Ouyang performed single-cell analysis; D. Sawant, K. Perez, Z. Huang, and X. Liu performed and analyzed in vivo experiments; L. Sekirov performed in vitro experiments; D. Wolak, J. DeVoss, P.S. Manzanillo, N. Pierce, A. Symons, and Z. Zhang provided resources, analysis, or reagents; and D. Bhatt, W. Ouyang, B. Kang, D. Sawant, and L. Zheng wrote and edited manuscript with the contributions from all other authors.
References
- Ahmadzadeh, M., Johnson L.A., Heemskerk B., Wunderlich J.R., Dudley M.E., White D.E., and Rosenberg S.A.. 2009. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood. 114:1537–1544. 10.1182/blood-2008-12-195792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi, E., Carr A.J., Plitas G., Cornish A.E., Konopacki C., Prabhakaran S., Nainys J., Wu K., Kiseliovas V., Setty M., et al. 2018. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 174:1293–1308.e36. 10.1016/j.cell.2018.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsheshet, Y., Wildbaum G., Levy E., Vitenshtein A., Akinseye C., Griggs J., Lira S.A., and Karin N.. 2017. CCR8+FOXp3+ Treg cells as master drivers of immune regulation. Proc. Natl. Acad. Sci. USA. 114:6086–6091. 10.1073/pnas.1621280114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank, C.U., Haining W.N., Held W., Hogan P.G., Kallies A., Lugli E., Lynn R.C., Philip M., Rao A., Restifo N.P., et al. 2019. Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 19:665–674. 10.1038/s41577-019-0221-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst, J., Ahrends T., Bąbała N., Melief C.J.M., and Kastenmüller W.. 2018. CD4+ T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 18:635–647. 10.1038/s41577-018-0044-0 [DOI] [PubMed] [Google Scholar]
- Bousso, P., Casrouge A., Altman J.D., Haury M., Kanellopoulos J., Abastado J.P., and Kourilsky P.. 1998. Individual variations in the murine T cell response to a specific peptide reflect variability in naive repertoires. Immunity. 9:169–178. 10.1016/S1074-7613(00)80599-3 [DOI] [PubMed] [Google Scholar]
- Brummelman, J., Mazza E.M.C., Alvisi G., Colombo F.S., Grilli A., Mikulak J., Mavilio D., Alloisio M., Ferrari F., Lopci E., et al. 2018. High-dimensional single cell analysis identifies stem-like cytotoxic CD8+ T cells infiltrating human tumors. J. Exp. Med. 215:2520–2535. 10.1084/jem.20180684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulliard, Y., Jolicoeur R., Zhang J., Dranoff G., Wilson N.S., and Brogdon J.L.. 2014. OX40 engagement depletes intratumoral Tregs via activating FcγRs, leading to antitumor efficacy. Immunol. Cell Biol. 92:475–480. 10.1038/icb.2014.26 [DOI] [PubMed] [Google Scholar]
- Butler, A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey, A.J., Hope J.L., Mueller Y.M., Fike A.J., Kumova O.K., van Zessen D.B.H., Steegers E.A.P., van der Burg M., and Katsikis P.D.. 2017. Public clonotypes and convergent recombination characterize the naïve CD8+ T-cell receptor repertoire of extremely preterm neonates. Front. Immunol. 8:1859. 10.3389/fimmu.2017.01859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, J.A., Preall J.B., Grigaityte K., Goldfless S.J., Jeffery E., Briggs A.W., Vigneault F., and Atwal G.S.. 2019. Single T cell sequencing demonstrates the functional role of αβ TCR pairing in cell lineage and antigen specificity. Front. Immunol. 10:1516. 10.3389/fimmu.2019.01516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cillo, A.R., Kürten C.H.L., Tabib T., Qi Z., Onkar S., Wang T., Liu A., Duvvuri U., Kim S., Soose R.J., et al. 2020. Immune landscape of viral- and carcinogen-driven head and neck cancer. Immunity. 52:183–199.e9. 10.1016/j.immuni.2019.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty, S. 2019. T follicular helper cell biology: A decade of discovery and diseases. Immunity. 50:1132–1148. 10.1016/j.immuni.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone, M., Arrigoni A., Rossetti G., Gruarin P., Ranzani V., Politano C., Bonnal R.J.P., Provasi E., Sarnicola M.L., Panzeri I., et al. 2016. Transcriptional landscape of human tissue lymphocytes unveils uniqueness of tumor-infiltrating T regulatory cells. Immunity. 45:1135–1147. 10.1016/j.immuni.2016.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, M., Gui X., Kim J., Xie L., Chen W., Li Z., He L., Chen Y., Chen H., Luo W., et al. 2018. LILRB4 signalling in leukaemia cells mediates T cell suppression and tumour infiltration. Nature. 562:605–609. 10.1038/s41586-018-0615-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derré, L., Bruyninx M., Baumgaertner P., Ferber M., Schmid D., Leimgruber A., Zoete V., Romero P., Michielin O., Speiser D.E., and Rufer N.. 2008. Distinct sets of αβ TCRs confer similar recognition of tumor antigen NY-ESO-1157-165 by interacting with its central Met/Trp residues. Proc. Natl. Acad. Sci. USA. 105:15010–15015. 10.1073/pnas.0807954105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egen, J.G., Ouyang W., and Wu L.C.. 2020. Human anti-tumor immunity: Insights from immunotherapy clinical trials. Immunity. 52:36–54. 10.1016/j.immuni.2019.12.010 [DOI] [PubMed] [Google Scholar]
- Elhanati, Y., Sethna Z., Callan C.G. Jr., Mora T., and Walczak A.M.. 2018. Predicting the spectrum of TCR repertoire sharing with a data-driven model of recombination. Immunol. Rev. 284:167–179. 10.1111/imr.12665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emming, S., Bianchi N., Polletti S., Balestrieri C., Leoni C., Montagner S., Chirichella M., Delaleu N., Natoli G., and Monticelli S.. 2020. A molecular network regulating the proinflammatory phenotype of human memory T lymphocytes. Nat. Immunol. 21:388–399. 10.1038/s41590-020-0622-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman, W.H., Pagès F., Sautès-Fridman C., and Galon J.. 2012. The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer. 12:298–306. 10.1038/nrc3245 [DOI] [PubMed] [Google Scholar]
- Galletti, G., De Simone G., Mazza E.M.C., Puccio S., Mezzanotte C., Bi T.M., Davydov A.N., Metsger M., Scamardella E., Alvisi G., et al. 2020. Two subsets of stem-like CD8+ memory T cell progenitors with distinct fate commitments in humans. Nat. Immunol. 21:1552–1562. 10.1038/s41590-020-0791-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner, L.C., Klenerman P., and Provine N.M.. 2018. Insights into mucosal-associated invariant T cell biology from studies of invariant natural killer T cells. Front. Immunol. 9:1478. 10.3389/fimmu.2018.01478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni, L., Zhong X.S., Palmer D.C., Ji Y., Hinrichs C.S., Yu Z., Wrzesinski C., Boni A., Cassard L., Garvin L.M., et al. 2009. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat. Med. 15:808–813. 10.1038/nm.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayoso, A., Shor J., Carr A.J., Sharma R., and Pe’er D.. 2018. DoubletDetection (Version v2.4). http://doi.org/. 10.5281/zenodo.2678041 [DOI]
- Grigaityte, K., Carter J.A., Goldfless S.J., Jeffery E.W., Hause R.J., Jiang Y., Koppstein D., Briggs A.W., Church G.M., Vigneault F., et al. 2017. Single-cell sequencing reveals αβ chain pairing shapes the T cell repertoire. bioRxiv. doi: (Preprint posted November 2, 2017). 10.1101/213462 [DOI]
- Guo, X., Zhang Y., Zheng L., Zheng C., Song J., Zhang Q., Kang B., Liu Z., Jin L., Xing R., et al. 2018. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 24:978–985. 10.1038/s41591-018-0045-3 [DOI] [PubMed] [Google Scholar]
- Hashimoto, M., Kamphorst A.O., Im S.J., Kissick H.T., Pillai R.N., Ramalingam S.S., Araki K., and Ahmed R.. 2018. CD8 T cell exhaustion in chronic infection and cancer: Opportunities for interventions. Annu. Rev. Med. 69:301–318. 10.1146/annurev-med-012017-043208 [DOI] [PubMed] [Google Scholar]
- Hollern, D.P., Xu N., Thennavan A., Glodowski C., Garcia-Recio S., Mott K.R., He X., Garay J.P., Carey-Ewend K., Marron D., et al. 2019. B cells and T follicular helper cells mediate response to checkpoint inhibitors in high mutation burden mouse models of breast cancer. Cell. 179:1191–1206.e21. 10.1016/j.cell.2019.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya, T., Sato-Kaneko F., Ahmadi A., Yao S., Lao F., Kitaura K., Matsutani T., Carson D.A., and Hayashi T.. 2018. Induction of oligoclonal CD8 T cell responses against pulmonary metastatic cancer by a phospholipid-conjugated TLR7 agonist. Proc. Natl. Acad. Sci. USA. 115:E6836–E6844. 10.1073/pnas.1803281115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, A.C., Postow M.A., Orlowski R.J., Mick R., Bengsch B., Manne S., Xu W., Harmon S., Giles J.R., Wenz B., et al. 2017. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 545:60–65. 10.1038/nature22079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iellem, A., Mariani M., Lang R., Recalde H., Panina-Bordignon P., Sinigaglia F., and D’Ambrosio D.. 2001. Unique chemotactic response profile and specific expression of chemokine receptors CCR4 and CCR8 by CD4+CD25+ regulatory T cells. J. Exp. Med. 194:847–854. 10.1084/jem.194.6.847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im, S.J., Hashimoto M., Gerner M.Y., Lee J., Kissick H.T., Burger M.C., Shan Q., Hale J.S., Lee J., Nasti T.H., et al. 2016. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 537:417–421. 10.1038/nature19330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin, N. 2018. Chemokines and cancer: new immune checkpoints for cancer therapy. Curr. Opin. Immunol. 51:140–145. 10.1016/j.coi.2018.03.004 [DOI] [PubMed] [Google Scholar]
- Kedzierska, K., Turner S.J., and Doherty P.C.. 2004. Conserved T cell receptor usage in primary and recall responses to an immunodominant influenza virus nucleoprotein epitope. Proc. Natl. Acad. Sci. USA. 101:4942–4947. 10.1073/pnas.0401279101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gal, F.A., Ayyoub M., Dutoit V., Widmer V., Jäger E., Cerottini J.C., Dietrich P.Y., and Valmori D.. 2005. Distinct structural TCR repertoires in naturally occurring versus vaccine-induced CD8+ T-cell responses to the tumor-specific antigen NY-ESO-1. J. Immunother. 28:252–257. 10.1097/01.cji.0000161398.34701.26 [DOI] [PubMed] [Google Scholar]
- Li, H., Ye C., Ji G., and Han J.. 2012. Determinants of public T cell responses. Cell Res. 22:33–42. 10.1038/cr.2012.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., Li T., Pignon J.-C., Wang B., Wang J., Shukla S.A., Dou R., Chen Q., Hodi F.S., Choueiri T.K., et al. 2016. Landscape of tumor-infiltrating T cell repertoire of human cancers. Nat. Genet. 48:725–732. 10.1038/ng.3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llosa, N.J., Cruise M., Tam A., Wicks E.C., Hechenbleikner E.M., Taube J.M., Blosser R.L., Fan H., Wang H., Luber B.S., et al. 2015. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 5:43–51. 10.1158/2159-8290.CD-14-0863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCully, M.L., Ladell K., Andrews R., Jones R.E., Miners K.L., Roger L., Baird D.M., Cameron M.J., Jessop Z.M., Whitaker I.S., et al. 2018. CCR8 expression defines tissue-resident memory T cells in human skin. J. Immunol. 200:1639–1650. 10.4049/jimmunol.1701377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot, J.M., Bigenwald C., Champiat S., Collins M., Carbonnel F., Postel-Vinay S., Berdelou A., Varga A., Bahleda R., Hollebecque A., et al. 2016. Immune-related adverse events with immune checkpoint blockade: a comprehensive review. Eur. J. Cancer. 54:139–148. 10.1016/j.ejca.2015.11.016 [DOI] [PubMed] [Google Scholar]
- Miragaia, R.J., Gomes T., Chomka A., Jardine L., Riedel A., Hegazy A.N., Whibley N., Tucci A., Chen X., Lindeman I., et al. 2019. Single-cell transcriptomics of regulatory T cells reveals trajectories of tissue adaptation. Immunity. 50:493–504.e7. 10.1016/j.immuni.2019.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlecnik, B., Bindea G., Angell H.K., Maby P., Angelova M., Tougeron D., Church S.E., Lafontaine L., Fischer M., Fredriksen T., et al. 2016. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity. 44:698–711. 10.1016/j.immuni.2016.02.025 [DOI] [PubMed] [Google Scholar]
- Myers, G. 2018. Immune-related adverse events of immune checkpoint inhibitors: a brief review. Curr. Oncol. 25:342–347. 10.3747/co.25.4235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestorowa, S., Hamey F.K., Pijuan Sala B., Diamanti E., Shepherd M., Laurenti E., Wilson N.K., Kent D.G., and Göttgens B.. 2016. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 128:e20–e31. 10.1182/blood-2016-05-716480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa, H., and Sakaguchi S.. 2014. Regulatory T cells in cancer immunotherapy. Curr. Opin. Immunol. 27:1–7. 10.1016/j.coi.2013.12.005 [DOI] [PubMed] [Google Scholar]
- Onda, M., Kobayashi K., and Pastan I.. 2019. Depletion of regulatory T cells in tumors with an anti-CD25 immunotoxin induces CD8 T cell-mediated systemic antitumor immunity. Proc. Natl. Acad. Sci. USA. 116:4575–4582. 10.1073/pnas.1820388116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardoll, D.M. 2012. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 12:252–264. 10.1038/nrc3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.E., Botting R.A., Domínguez Conde C., Popescu D.M., Lavaert M., Kunz D.J., Goh I., Stephenson E., Ragazzini R., Tuck E., et al. 2020. A cell atlas of human thymic development defines T cell repertoire formation. Science. 367:eaay3224. 10.1126/science.aay3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plitas, G., Konopacki C., Wu K., Bos P.D., Morrow M., Putintseva E.V., Chudakov D.M., and Rudensky A.Y.. 2016. Regulatory T cells exhibit distinct features in human breast cancer. Immunity. 45:1122–1134. 10.1016/j.immuni.2016.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram, S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S., Rodman C., Luo C.L., Mroz E.A., Emerick K.S., et al. 2017. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 171:1611–1624.e24. 10.1016/j.cell.2017.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins, H.S., Campregher P.V., Srivastava S.K., Wacher A., Turtle C.J., Kahsai O., Riddell S.R., Warren E.H., and Carlson C.S.. 2009. Comprehensive assessment of T-cell receptor β-chain diversity in alphabeta T cells. Blood. 114:4099–4107. 10.1182/blood-2009-04-217604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz-Perez, A., Lim A., Lemercier B., and Leclerc C.. 2012. The T-cell receptor repertoire of tumor-infiltrating regulatory T lymphocytes is skewed toward public sequences. Cancer Res. 72:3557–3569. 10.1158/0008-5472.CAN-12-0277 [DOI] [PubMed] [Google Scholar]
- Salou, M., Legoux F., Gilet J., Darbois A., du Halgouet A., Alonso R., Richer W., Goubet A.G., Daviaud C., Menger L., et al. 2019. A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J. Exp. Med. 216:133–151. 10.1084/jem.20181483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerli, P., Ebert L., Willimann K., Blaser A., Roos R.S., Loetscher P., and Moser B.. 2004. A skin-selective homing mechanism for human immune surveillance T cells. J. Exp. Med. 199:1265–1275. 10.1084/jem.20032177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper, W., Kelderman S., Fanchi L.F., Linnemann C., Bendle G., de Rooij M.A.J., Hirt C., Mezzadra R., Slagter M., Dijkstra K., et al. 2019. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat. Med. 25:89–94. 10.1038/s41591-018-0266-5 [DOI] [PubMed] [Google Scholar]
- Selby, M.J., Engelhardt J.J., Quigley M., Henning K.A., Chen T., Srinivasan M., and Korman A.J.. 2013. Anti-CTLA-4 antibodies of IgG2a isotype enhance antitumor activity through reduction of intratumoral regulatory T cells. Cancer Immunol. Res. 1:32–42. 10.1158/2326-6066.CIR-13-0013 [DOI] [PubMed] [Google Scholar]
- Sharma, P., and Allison J.P.. 2015. The future of immune checkpoint therapy. Science. 348:56–61. 10.1126/science.aaa8172 [DOI] [PubMed] [Google Scholar]
- Sharma, P., Hu-Lieskovan S., Wargo J.A., and Ribas A.. 2017. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 168:707–723. 10.1016/j.cell.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, A., Subudhi S.K., Blando J., Scutti J., Vence L., Wargo J., Allison J.P., Ribas A., and Sharma P.. 2019. Anti-CTLA-4 immunotherapy does not deplete FOXP3+ regulatory T cells (Tregs) in human cancers. Clin. Cancer Res. 25:1233–1238. 10.1158/1078-0432.CCR-18-0762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoni, Y., Becht E., Fehlings M., Loh C.Y., Koo S.-L., Teng K.W.W., Yeong J.P.S., Nahar R., Zhang T., Kared H., et al. 2018. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 557:575–579. 10.1038/s41586-018-0130-2 [DOI] [PubMed] [Google Scholar]
- Simpson, T.R., Li F., Montalvo-Ortiz W., Sepulveda M.A., Bergerhoff K., Arce F., Roddie C., Henry J.Y., Yagita H., Wolchok J.D., et al. 2013. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J. Exp. Med. 210:1695–1710. 10.1084/jem.20130579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., W.M. Mauck, III, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive integration of single-cell data. Cell. 177:1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama, D., Nishikawa H., Maeda Y., Nishioka M., Tanemura A., Katayama I., Ezoe S., Kanakura Y., Sato E., Fukumori Y., et al. 2013. Anti-CCR4 mAb selectively depletes effector-type FoxP3+CD4+ regulatory T cells, evoking antitumor immune responses in humans. Proc. Natl. Acad. Sci. USA. 110:17945–17950. 10.1073/pnas.1316796110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, A., and Sakaguchi S.. 2017. Regulatory T cells in cancer immunotherapy. Cell Res. 27:109–118. 10.1038/cr.2016.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh, I., Izar B., Prakadan S.M., Wadsworth M.H. II, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G., et al. 2016. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 352:189–196. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Leun, A.M., Thommen D.S., and Schumacher T.N.. 2020. CD8+ T cell states in human cancer: insights from single-cell analysis. Nat. Rev. Cancer. 20:218–232. 10.1038/s41568-019-0235-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi, V., Price D.A., Douek D.C., and Davenport M.P.. 2008. The molecular basis for public T-cell responses? Nat. Rev. Immunol. 8:231–238. 10.1038/nri2260 [DOI] [PubMed] [Google Scholar]
- Villarreal, D.O., L’Huillier A., Armington S., Mottershead C., Filippova E.V., Coder B.D., Petit R.G., and Princiotta M.F.. 2018. Targeting CCR8 induces protective antitumor immunity and enhances vaccine-induced responses in colon cancer. Cancer Res. 78:5340–5348. 10.1158/0008-5472.CAN-18-1119 [DOI] [PubMed] [Google Scholar]
- Wang, T., Wang C., Wu J., He C., Zhang W., Liu J., Zhang R., Lv Y., Li Y., Zeng X., et al. 2017. The different T-cell receptor repertoires in breast cancer tumors, draining lymph nodes, and adjacent tissues. Cancer Immunol. Res. 5:148–156. 10.1158/2326-6066.CIR-16-0107 [DOI] [PubMed] [Google Scholar]
- Wang, L., Simons D.L., Lu X., Tu T.Y., Solomon S., Wang R., Rosario A., Avalos C., Schmolze D., Yim J., et al. 2019. Connecting blood and intratumoral Treg cell activity in predicting future relapse in breast cancer. Nat. Immunol. 20:1220–1230. 10.1038/s41590-019-0429-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, S.C., Levine J.H., Cogdill A.P., Zhao Y., Anang N.A.S., Andrews M.C., Sharma P., Wang J., Wargo J.A., Pe’er D., et al. 2017. Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell. 170:1120–1133.e17. 10.1016/j.cell.2017.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolock, S.L., Lopez R., and Klein A.M.. 2019. Scrublet: Computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8:281–291.e9. 10.1016/j.cels.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T.D., Madireddi S., de Almeida P.E., Banchereau R., Chen Y.J.J., Chitre A.S., Chiang E.Y., Iftikhar H., O’Gorman W.E., Au-Yeung A., et al. 2020. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature. 579:274–278. 10.1038/s41586-020-2056-8 [DOI] [PubMed] [Google Scholar]
- Yost, K.E., Satpathy A.T., Wells D.K., Qi Y., Wang C., Kageyama R., McNamara K.L., Granja J.M., Sarin K.Y., Brown R.A., et al. 2019. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 25:1251–1259. 10.1038/s41591-019-0522-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F., Sharma S., Jankovic D., Gurram R.K., Su P., Hu G., Li R., Rieder S., Zhao K., Sun B., and Zhu J.. 2018. The transcription factor Bhlhe40 is a switch of inflammatory versus antiinflammatory Th1 cell fate determination. J. Exp. Med. 215:1813–1821. 10.1084/jem.20170155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X., Zhang L., Chaudhry A., Rapaport A.S., and Ouyang W.. 2020. Unravelling the heterogeneity and dynamic relationships of tumor-infiltrating T cells by single-cell RNA sequencing analysis. J. Leukoc. Biol. 107:917–932. 10.1002/JLB.6MR0320-234R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Yu X., Zheng L., Zhang Y., Li Y., Fang Q., Gao R., Kang B., Zhang Q., Huang J.Y., et al. 2018. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 564:268–272. 10.1038/s41586-018-0694-x [DOI] [PubMed] [Google Scholar]
- Zhang, Q., He Y., Luo N., Patel S.J., Han Y., Gao R., Modak M., Carotta S., Haslinger C., Kind D., et al. 2019. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 179:829–845.e20. 10.1016/j.cell.2019.10.003 [DOI] [PubMed] [Google Scholar]
- Zhang, L., Li Z., Skrzypczynska K.M., Fang Q., Zhang W., O’Brien S.A., He Y., Wang L., Zhang Q., Kim A., et al. 2020. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell. 181:442–459.e29. 10.1016/j.cell.2020.03.048 [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Nguyen P., Ma J., Wu T., Jones L.L., Pei D., Cheng C., and Geiger T.L.. 2016. Preferential use of public TCR during autoimmune encephalomyelitis. J. Immunol. 196:4905–4914. 10.4049/jimmunol.1501029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, C., Zheng L., Yoo J.K., Guo H., Zhang Y., Guo X., Kang B., Hu R., Huang J.Y., Zhang Q., et al. 2017. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 169:1342–1356.e16. 10.1016/j.cell.2017.05.035 [DOI] [PubMed] [Google Scholar]
- Zingoni, A., Soto H., Hedrick J.A., Stoppacciaro A., Storlazzi C.T., Sinigaglia F., D’Ambrosio D., O’Garra A., Robinson D., Rocchi M., et al. 1998. The chemokine receptor CCR8 is preferentially expressed in Th2 but not Th1 cells. J. Immunol. 161:547–551. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is a summary table of cluster attributes, including clonal expansion, tissue localization, and representative marker gene expression.
lists cell metadata indicating tissue, individual mouse, tSNE coordinates, cell-cycle phase, gene expression cluster, and TCR clonotype.
provides differential expression values of NKT clusters C20 and C26:28.
lists cell metadata, as in Table S2, with additional annotation of the V, D, and J segments and TCR publicity status.
is a differential expression analysis of genes in each CD8+ cluster compared to all other CD8 clusters as in Table S3.
provides a cross-species comparison of human and mouse CD8 Tex clusters.
shows a differential expression analysis of genes in each CD4+ cluster compared to all other CD4+ clusters as in Table S3.
shows a differential expression analysis of genes in each CD4+Foxp3+ T reg cell cluster compared to all other T reg cell clusters as in Table S3.
provides a comparison of human gene expression and mouse gene expression in CD4+Foxp3+ T reg cell clusters, as in Table S6.
is a STARTRAC analysis of T cell clusters.
is a pairwise STARTRAC analysis of T cell migration.
is a pairwise STARTRAC analysis of T cell cluster transitions.