VMP1 and TMEM41B, integral membrane proteins of the ER, regulate the formation of autophagosomes, lipid droplets, and lipoproteins. Here, Li, Wang, et al. show that they have phospholipid scramblase activity, which is essential to the normal distribution of cholesterol and phosphatidylserine in mammalian cells.
Abstract
TMEM41B and VMP1 are integral membrane proteins of the endoplasmic reticulum (ER) and regulate the formation of autophagosomes, lipid droplets (LDs), and lipoproteins. Recently, TMEM41B was identified as a crucial host factor for infection by all coronaviruses and flaviviruses. The molecular function of TMEM41B and VMP1, which belong to a large evolutionarily conserved family, remains elusive. Here, we show that TMEM41B and VMP1 are phospholipid scramblases whose deficiency impairs the normal cellular distribution of cholesterol and phosphatidylserine. Their mechanism of action on LD formation is likely to be different from that of seipin. Their role in maintaining cellular phosphatidylserine and cholesterol homeostasis may partially explain their requirement for viral infection. Our results suggest that the proper sorting and distribution of cellular lipids are essential for organelle biogenesis and viral infection.
Introduction
Lipid droplets (LDs) are ER-derived cellular organelles that regulate lipid homeostasis and many other cellular functions. LDs are believed to originate from discreet sites of ER tubules, although the molecular underpinnings of LD biogenesis remain to be elucidated (Gao et al., 2019; Olzmann and Carvalho, 2019; Pol et al., 2014; Santinho et al., 2020; Walther et al., 2017). Both proteins and lipids can regulate LD formation from the ER, and membrane curvature may be a determining factor for LD initiation (Santinho et al., 2020). Seipin, an integral membrane protein of the ER with two transmembrane helices, plays a prominent role in LD biogenesis and growth (Chung et al., 2019; Fei et al., 2008; Szymanski et al., 2007). Recent evidence suggests that seipin and its partner LDAF1 organize LD initiation from the ER in part by facilitating the nucleation of storage lipids such as triacylglycerols (Chung et al., 2019), although other studies have suggested that seipin may directly regulate the synthesis and distribution of phospholipids, including phosphatidic acid (PA), in the ER (Fei et al., 2011; Pagac et al., 2016; Soltysik et al., 2021; Yan et al., 2018). To better understand LD biogenesis, it is crucial to identify and dissect additional ER resident proteins that regulate LD formation.
TMEM41B and VMP1 are integral membrane proteins of the ER and belong to a large, evolutionarily conserved family of proteins that share a VTT (VMP1, TMEM41, and Tvp38) domain of unknown function (Morita et al., 2018; Zhao et al., 2017). Whereas both TMEM41B and VMP1 are required for the formation of autophagosomes, they also regulate many other cellular processes. Recently, TMEM41B, and to a lesser extent VMP1, were identified as essential host factors for infection by all coronaviruses (including severe acute respiratory syndrome coronavirus 2) and flaviviruses (Hoffmann et al., 2021; Schneider et al., 2021). Notably, TMEM41B, but not autophagy per se, is required for flavivirus infection (Hoffmann et al., 2021). Moreover, VMP1, but not other autophagy-related genes, is essential for the assembly and secretion of lipoproteins in the intestine and liver (Morishita et al., 2019). Finally, giant LDs were observed in VMP1- or TMEM41B-deficient cells, reminiscent of the seipin phenotype (Fei et al., 2008; Morita et al., 2018; Pagac et al., 2016; Szymanski et al., 2007; Zhao et al., 2017). Both seipin and VMP1 were shown to regulate cellular calcium concentrations, and both were reported to interact with SERCA physically and functionally (Bi et al., 2014; Zhao et al., 2017). Thus, it seems likely that seipin and VMP1/TMEM41B may cooperate to regulate LD formation. More broadly, it is important to determine the molecular function of VMP1 and TMEM41B because they regulate a diverse range of critical cellular functions.
Results and discussion
TMEM41B/VMP1 and seipin regulate LD biogenesis likely through distinct mechanisms
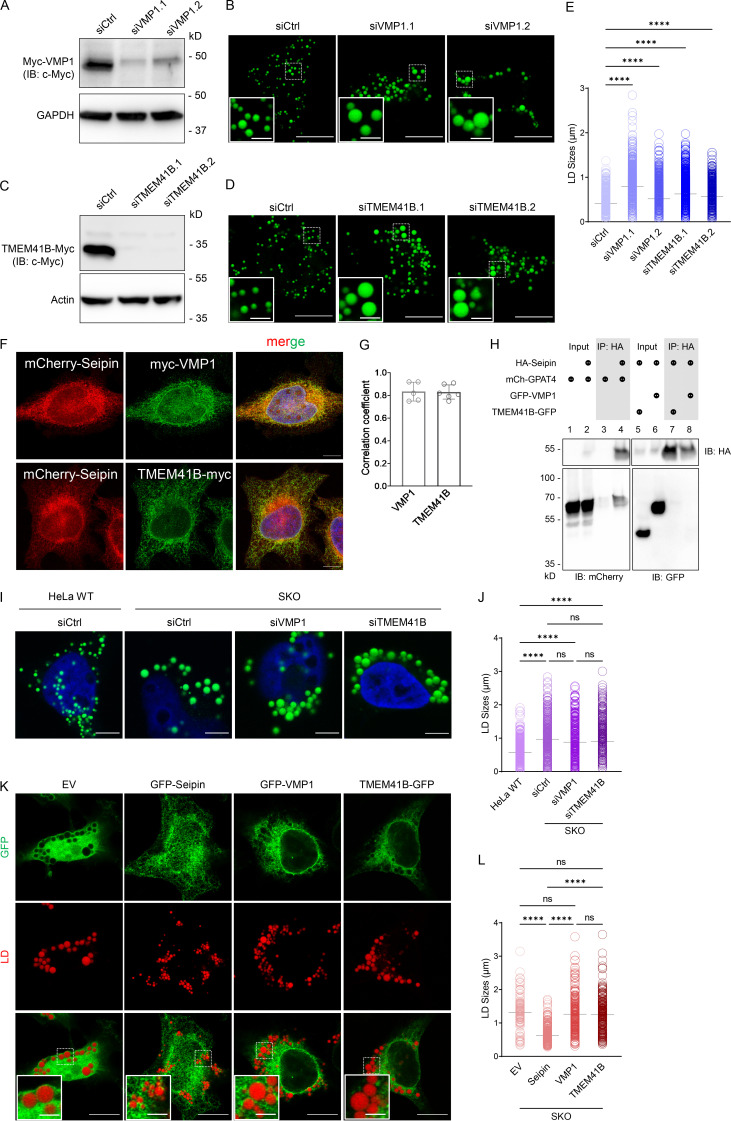
Given the reported links between seipin and VMP1/TMEM41B, we initiated this study to investigate the physical and functional relationship between seipin and VMP1/TMEM41B in LD formation. Knocking down VMP1 or TMEM41B from HeLa cells formed large LDs as reported previously (Fig. 1, A–E; Morita et al., 2018; Zhao et al., 2017). Moreover, both VMP1 and TMEM41B showed near-perfect colocalization with seipin (Fig. 1, F and G). However, glycerol-3-phosphate acyltransferase 4 (GPAT4), but not VMP1 or TMEM41B, coprecipitated with seipin (Fig. 1 H; Pagac et al., 2016), suggesting no detectable physical association between seipin and TMEM41B/VMP1. Overexpressing or knocking down VMP1/TMEM41B had little impact on LD size in seipin knockout (SKO) cells treated with oleate (Fig. 1, I–L). Because seipin deficiency also impairs early LD formation (Chung et al., 2019; Salo et al., 2016; Wang et al., 2016), we further investigated VMP1’s role in LD initiation. We tagged endogenous perilipin 3 (PLIN3) by genome engineering with mCherry as described (Chung et al., 2019). PLIN3 is an endogenous protein that indicates the earliest steps of LD formation, whereas lipophilic dyes, such as LipidTOX/BODIPY, only stain LDs that have acquired enough neutral lipids. In normal cells, PLIN3 rapidly accumulated in small puncta that grew larger, and nearly all PLIN3 puncta became BODIPY positive within 20 min of oleate loading (Fig. S1, A and B). In seipin-deficient SKO cells, there was a sharp increase in the number of PLIN3 puncta within 10 min of oleate treatment (Fig. S1, A and B), but very few BODIPY-positive LDs were detected up to 20 min of oleate treatment (Fig. S1, A and B), indicating a delay in initial LD lipidation as reported (Chung et al., 2019). By contrast, formation of early LDs appeared normal in VMP1-deficient cells (Fig. S1, A and B). Together, these results suggest that seipin and VMP1/TMEM41B likely impact LD formation through distinct mechanisms.
Figure 1.
Seipin and VMP1/TMEM41B regulate LD growth likely through distinct mechanisms. (A and C) Western blot analysis of myc-VMP1 or TMEM41B-myc in HeLa cells treated with control or two different VMP1/TMEM41B siRNAs. Cells were treated with siRNA for 24 h followed by the transfection of myc-VMP1 or TMEM41B-myc cDNA for 24 h. GAPDH/actin was included as loading control. (B and D) Confocal images of oleate-treated (6 h) cells transfected with siRNAs as in A or C and stained with BODIPY. (E) LD diameters measured from images shown in B and D. ****, P < 0.0001 (one-way ANOVA, n = 1,280–2,340 LDs). (F) Confocal images of HeLa cells cotransfected with cDNAs encoding mCherry-tagged seipin and myc-tagged VMP1 or TMEM41B, which were stained with anti-myc antibody and detected by immunofluorescence. (G) Colocalization analysis of mCherry-seipin and myc-tagged VMP1 or TMEM41B in F. Pearson r (correlation coefficient) value is shown (unpaired t test, mean ± SD, n = 5–6 cells). (H) Coimmunoprecipitation of HA-tagged seipin with mCherry-tagged GPAT4 or GFP-tagged VMP1/TMEM41B expressed in HEK293 cells. HA-seipin was pulled down by HA-agarose beads. Immunoblotting of mCherry-GPAT4, GFP-VMP1, and TMEM41B-GFP from both input and immunoprecipitated samples are shown. (I) Confocal images of oleate-treated (6 h) SKO cells transfected with siVMP1 and siTMEM41B for 48 h and stained with BODIPY. HeLa WT cells were treated with control siRNA only. (J) LD diameters measured from images shown in I. ****, P < 0.0001 (one-way ANOVA, n = 335–620 LDs). (K) Confocal images of oleate-treated (16 h) SKO cells transfected with cDNAs encoding GFP empty vector (EV) or GFP-tagged seipin, VMP1, or TMEM41B for 40 h and stained with LipidTOX Deep Red. (L) LD diameters measured from images shown in K. ****, P < 0.0001 (one-way ANOVA, n = 130–485 LDs). Scale bars = 10 µm (inset scale bars = 2 µm) for all images. Ctrl, control; IB, immunoblot; IP, immunoprecipitation; mCh, mCherry.
Figure S1.
VMP1 deficiency does not impact early LD formation. (A) Confocal images of mCherry-tagged endogenous PLIN3 and BODIPY-positive LD puncta in HeLa WT or SKO cells treated with control or VMP1 siRNAs for 24 h followed by starvation in 1% lipoprotein-deficient serum for 16 h. Images were taken at a 2-min interval for up to 18 min after oleate addition (400 µM). (B) Quantification of the numbers of PLIN3- and BODIPY-positive LD puncta shown in A at indicated time points. The value of pairs indicates overlapped PLIN3 and BODIPY puncta. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (two-way ANOVA, mean ± SD, n = 15–20 cells). Scale bars = 10 µm. Ctrl, control.
TMEM41B and VMP1 regulate the cellular distribution of free cholesterol
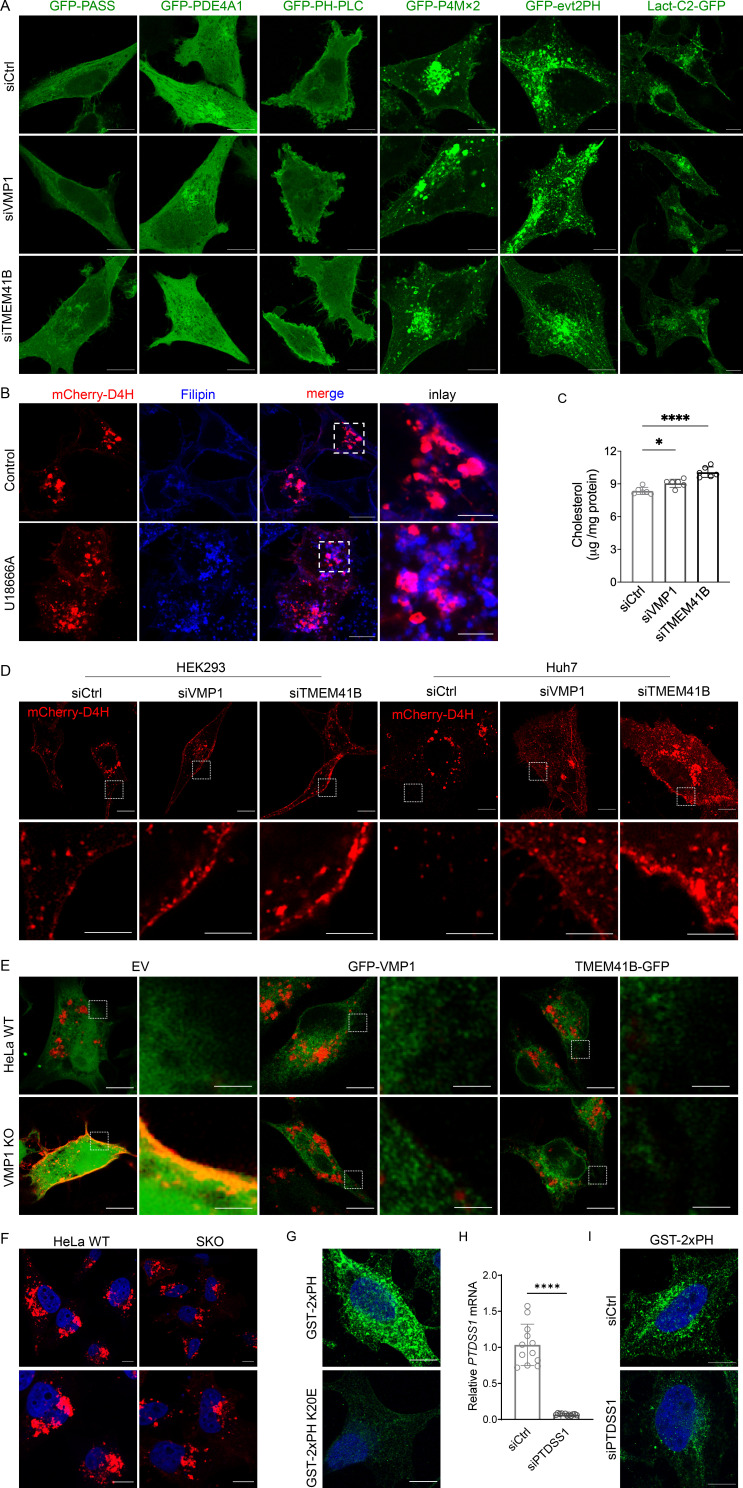
To further understand how VMP1/TMEM41B may regulate LD formation, we focused on their potential role in cellular lipid metabolism, since LD morphology is highly sensitive to the concentration and distribution of lipids (Fei et al., 2011; Gao et al., 2019; Krahmer et al., 2011). We examined the distribution of all available lipid sensors in our laboratory in WT and VMP1/TMEM41B-deficient cells. No change was observed for sensors detecting PA (GFP-PASS, GFP-PDE4A1; Kassas et al., 2017), phosphatidylinositol (4,5)-bisphosphate (GFP-PH-PLC; Várnai and Balla, 1998), phosphatidylinositol (4)-phosphate (GFP-P4Mx2; Hammond et al., 2014), and phosphatidylserine (PS; GFP-evt2PH, Lact-C2-GFP; Uchida et al., 2011; Yeung et al., 2008) between WT and VMP1/TMEM41B knockdown cells (Fig. S2 A). Strikingly, mCherry-D4H, a probe for cholesterol on the cytoplasmic side of cellular membranes (Maekawa and Fairn, 2015; Wang et al., 2019), relocated from internal membranes, most likely lysosomes (Fig. S2 B; Maekawa and Fairn, 2015), to the plasma membrane (PM) in VMP1/TMEM41B-deficient HeLa cells (Fig. 2, A–C). Total cellular free cholesterol moderately but significantly increased in VMP1/TMEM41B-deficient HeLa cells (Fig. S2 C). The redistribution of D4H was also observed in HEK293 and Huh7 cells deficient in VMP1/TMEM41B (Fig. S2 D). Interestingly, the redistribution of D4H in TMEM41B-deficient HeLa cells was rescued by overexpressing VMP1 and vice versa, suggesting that both proteins share a common function (Fig. 2, D–G). We further made VMP-1 knockout (KO) cells by CRISPR-mediated gene targeting as described (Morita et al., 2018). In VMP1 KO cells, the PM distribution of D4H was pronounced, which was rescued upon expressing VMP1 or TMEM41B (Fig. S2 E). As a control, no change in D4H distribution was observed in SKO cells (Fig. S2 F). Finally, we monitored cholesterol with a newly developed probe for cholesterol: GRAM1b (G187L), which is a mutated form of the GRAM domain of GramD1b, a cholesterol transfer protein (Ercan et al., 2021). GRAM1b (G187L) specifically detects the accessible cholesterol pool in the inner leaflet of the PM. Strong GRAM1b (G187L) signal was detected in the PM of TMEM41B/VMP1-deficient cells but not that of WT cells (Fig. 2, H and I), consistent with results from D4H. Together, these results suggest that TMEM41B and VMP1 may regulate the physical properties of cellular membranes; in their absence, more cholesterol becomes accessible on the cytoplasmic leaflet of the PM.
Figure S2.
Supporting data and controls for cholesterol and PS detection. (A) Confocal images of indicated lipid sensors in HeLa cells treated with control, VMP1, or TMEM41B siRNAs. (B) Confocal images of mCherry-D4H in HeLa cells in the absence or presence of U18666A. Cellular cholesterol was stained with filipin. (C) Cholesterol content measured by Amplex Red assay in cells treated with control, VMP1, or TMEM41B siRNAs. Values were normalized to total cellular proteins. *, P < 0.05; ****, P < 0.0001 (one-way ANOVA, mean ± SD, n = 6). (D) Confocal images of mCherry-D4H in HEK293 and Huh7 cells treated with control, VMP1, or TMEM41B siRNAs. Cells were treated with siRNA for 24 h followed by the transfection of mCherry-D4H cDNA for 24 h. Inset images show the labeling of the plasma membrane by mCherry-D4H. (E) Confocal images of coexpressed mCherry-D4H and GFP empty vector (EV), GFP-VMP1, or TMEM41B-GFP in VMP1 KO HeLa cells. Inset images show the difference of the plasma membrane labeling by mCherry-D4H. (F) Confocal images of mCherry-D4H in HeLa WT and SKO cells. (G) PS labeling by GST-2xPH or GST-2xPH K20E (deficient in PS binding) in digitonin-permeabilized HeLa cells. (H) Quantitative RT-PCR analysis of HeLa cells treated with control or PTDSS1 siRNAs for 48 h. ****, P < 0.0001 (unpaired t test, mean ± SD, n = 12). (I) PS labeling by GST-2xPH in digitonin-permeabilized HeLa cells treated with control or PTDSS1 siRNAs for 48 h. Scale bars = 10 µm (inset scale bars in B and D = 4 µm and in E = 2 µm). Ctrl, control.
Figure 2.
Altered cholesterol distribution in VMP1- and TMEM41B-deficient cells. (A and B) Confocal images of mCherry-D4H in HeLa cells treated with control or two different VMP1 or TMEM41B siRNAs. Cells were treated with siRNA for 24 h followed by transfection of mCherry-D4H cDNA for 24 h. Inset images show the labeling of the PM by mCherry-D4H. (C) Relative intensity of mCherry-D4H (PM/cytoplasm) of cells shown in A and B. *, P < 0.05; ****, P < 0.0001 (one-way ANOVA, mean ± SD, n = 17–30 cells). (D) Confocal images of coexpressed mCherry-D4H and GFP empty vector (EV) or TMEM41B-GFP in HeLa cells treated with control or VMP1 siRNA. (E) Relative intensity of mCherry-D4H (PM/cytoplasm) of cells shown in D. ****, P < 0.0001 (one-way ANOVA, mean ± SD, n = 12–29 cells). (F) Confocal images of coexpressed mCherry-D4H and GFP EV or GFP-VMP1 in HeLa cells treated with control or TMEM41B siRNA. (G) Relative intensity of mCherry-D4H (PM/cytoplasm) of cells shown in F. **, P < 0.01; ****, P < 0.0001 (one-way ANOVA, mean ± SD, n = 8–22 cells). (H) Confocal images of GFP-GRAM1b and GFP-GRAM1b (G187L) in HeLa cells treated with control, VMP1, or TMEM41B siRNAs. Inset images show the intensity of the probes associated with the PM. (I) Relative intensity of GFP-GRAM1b and GFP-GRAM1b (G187L) (PM/cytoplasm) of cells shown in H. **, P < 0.01 (one-way ANOVA, mean ± SD, n = 13–49 cells). Scale bars = 10 µm (inset scale bars in A and B = 5 µm; inset scale bar in H = 2 µm). Ctrl, control.
TMEM41B and VMP1 regulate the cellular distribution of PS
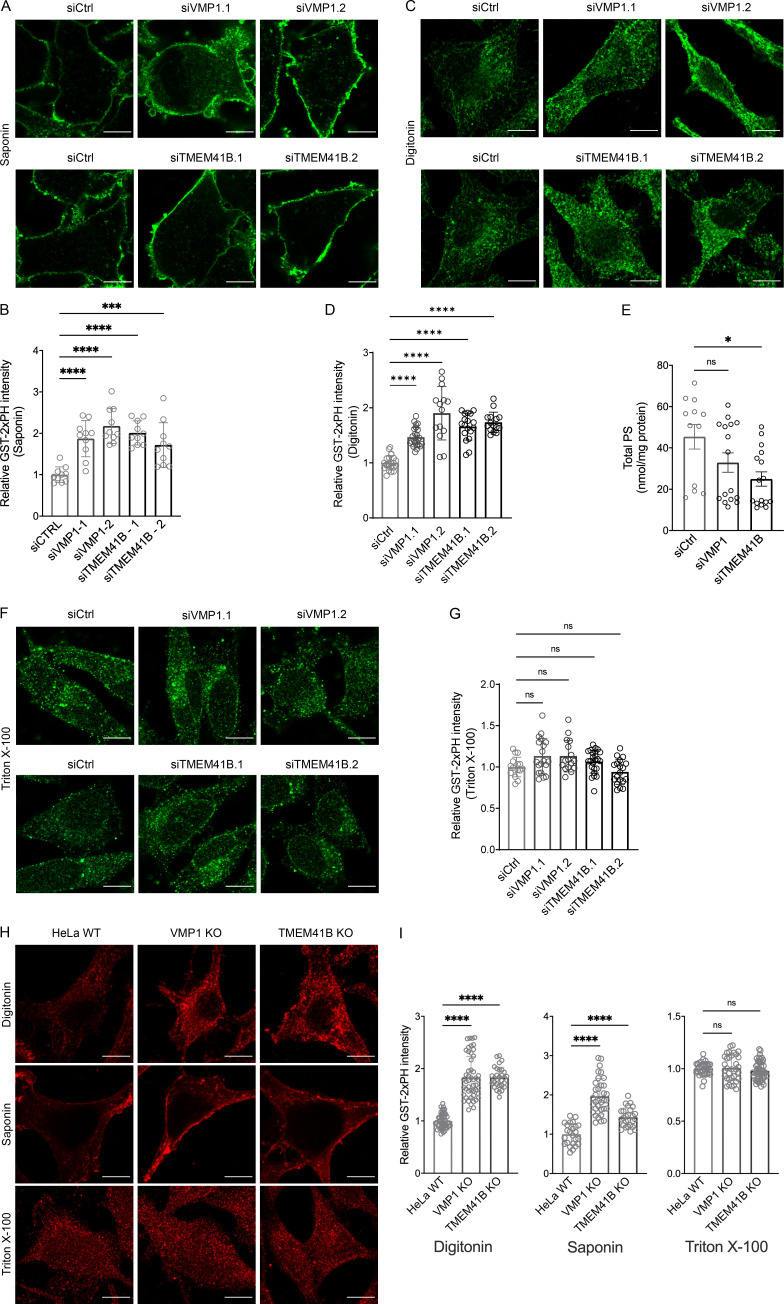
In our miniscreen of lipid sensors, we also observed a significant increase in the intensity of GST-2xPH, a PS sensor, in VMP1/TMEM41B-deficient cells (Fig. 3, A–D; Tsuji et al., 2019). GST-2xPH contains the PH domain of evectin-2 in tandem and has recently been shown to bind PS with much higher specificity and sensitivity than other PS sensors (Tsuji et al., 2019; Uchida et al., 2011). We further tested the specificity of GST-2xPH in our cells. GST-2xPH, but not its mutant form GST-2xPH (K20E), showed intracellular puncta when cells were permeabilized with digitonin (Fig. S2 G). Importantly, the GST-2xPH signal was almost completely lost in cells lacking PTDSS1, a major enzyme for the synthesis of PS (Fig. S2, H and I; Trinh et al., 2020). We detected a significant increase in the intensity of GST-2xPH in TMEM41B- or VMP1-deficient cells when cells were permeabilized with saponin (for detecting the PM; Fig. 3, A and B) and digitonin (for detecting internal membranes; Fig. 3, C and D). The increase in PS/GST-2xPH under these conditions is somewhat confusing because total cellular PS was not increased as assessed by lipidomic analyses (Fig. 3 E). Treating cells with saponin or digitonin permeabilizes only the PM and allows detection of PS on the cytoplasmic side but not on the luminal side of cellular organelles, such as ER and Golgi (Ohsaki et al., 2005; Plutner et al., 1992). Despite increased cytoplasmic PS, luminal PS could be decreased in TMEM41B- or VMP1-deficient cells, leading to little change in overall PS. We therefore treated cells with Triton X-100, which permeabilizes all membranes and allows access to both membrane leaflets by GST-2xPH. No apparent difference in GST-2xPH staining was observed between WT and TMEM41B/VMP1-deficient cells (Fig. 3, F and G). Similar results were also observed in TMEM41B/VMP1 KO cells (Fig. 3, H and I). Thus, it appears that TMEM41B and VMP1 may regulate the balance of PS between two membrane leaflets.
Figure 3.
VMP1/TMEM41B deficiency increases PS in the cytoplasmic leaflet of cellular membranes. (A, C, and F) PS labeling by GST-2xPH in saponin/digitonin/Triton X-100–permeabilized HeLa cells treated with control (Ctrl) or two different VMP1 or TMEM41B siRNAs for 48 h. (B, D, and G) Relative intensity of GST-2xPH (PM/cytoplasm) of cells permeabilized by saponin/digitonin/Triton X-100 shown in A, C, and F. ****, P < 0.0001 (one-way ANOVA, mean ± SD, n = 10–24 cells). ***, P < 0.001. (E) Lipidomic analysis of total PS extracted from HeLa cells treated with Ctrl, VMP1, or TMEM41B siRNAs for 48 h. Data are pooled from two independent experiments and expressed as mean ± SEM. *, P < 0.05 (one-way ANOVA; siCtrl, n = 12; siVMP1 and siTMEM41B, n = 16). (H) PS labeling by GST-2xPH in digitonin/saponin/Triton X-100–permeabilized HeLa WT cells or VMP1/TMEM41B KO cells. (I) Relative intensity of GST-2xPH of cells permeabilized by digitonin/saponin/Triton X-100 shown in H. ****, P < 0.0001 (one-way ANOVA, mean ± SD, n = 24–49 cells). Scale bars = 10 µm for all images.
TMEM41B and VMP1 have phospholipid scramblases activity
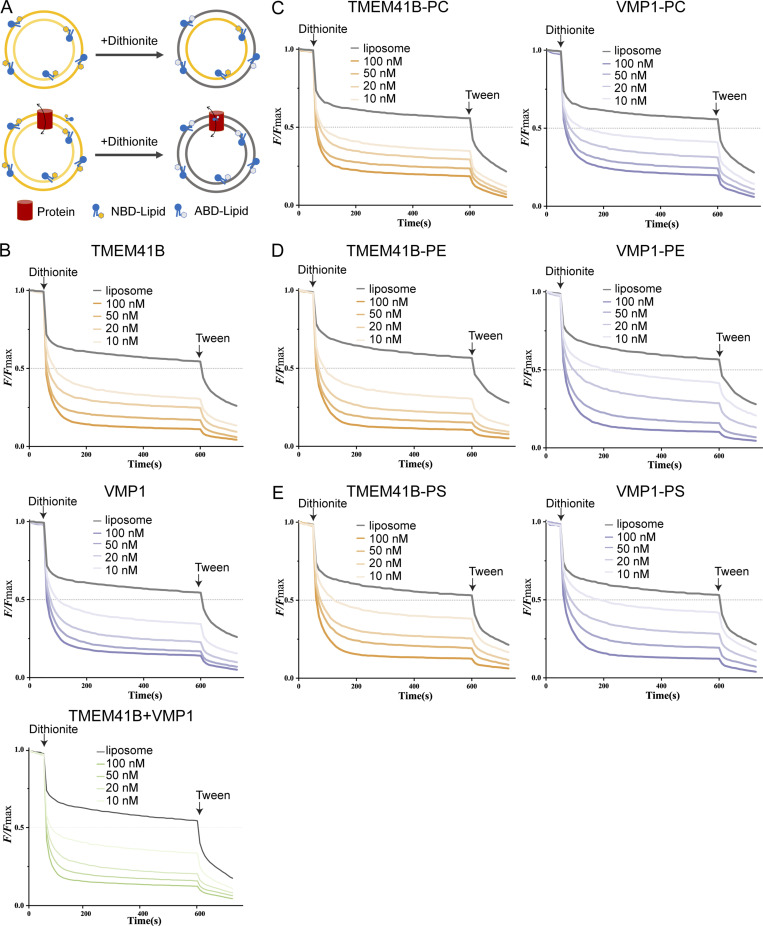
Given their impact on the biogenesis of LDs (cytoplasmic side of the ER) and lipoproteins (luminal side), as well as on the distribution of PS, we wondered whether TMEM41B and VMP1 may have flippase/floppase/scramblase activity. We purified VMP1 and TMEM41B to examine their impact on the movement of phospholipids from the inner to the outer leaflet of liposomes (Fig. S3, A and B). For this purpose, the effect of dithionite, a reducing agent that cannot penetrate lipid bilayers, on liposomes containing fluorescent NBD acyl-labeled phospholipids was examined (Fig. 4 A; Menon et al., 2011). Dithionite is known to reduce and quench the fluorescence of NBD-lipids only within the outer leaflet of a liposome. Adding Tween 20 enables dithionite to access and quench the fluorescence of NBD-lipids within both leaflets of the liposome. Thus, ∼50% of the fluorescence should be lost from NBD-lipid–containing liposomes with dithionite alone, and more than half of the fluorescence should be lost when proteins with scramblase activity are incorporated into the liposomes. Liposomes containing NBD-labeled phosphatidylethanolamine (PE), phosphatidylcholine (PC), and PS with or without purified proteins were treated with dithionite. NBD fluorescence of liposomes without any protein was reduced by ∼50% upon adding dithionite (Fig. S3 C). Opsin, a known scramblase, but not digeranylgeranylglyceryl phosphate synthase (DGGGPase), a lipid-modifying enzyme with nine transmembrane helices, reduced NBD fluorescence by >50% upon addition of dithionite (Fig. S3 C; Menon et al., 2011; Ren et al., 2020). The presence of TMEM41B or VMP1 in liposomes also decreased NBD fluorescence by >50% (Fig. 4 B and Fig. S3 C). The degree of NBD quenching depended on the protein concentration of TMEM41B/VMP1 (Fig. 4 B). The scramblase activity of TMEM41B/VMP1 did not depend on ATP and was not affected by calcium (Fig. S3, D and E). Of note, the scramblase activity of TMEM41B was not impacted by the presence of VMP1 (Fig. 4 B) and vice versa, indicating that TMEM41B’s or VMP1’s scramblase activity does not require the other in vitro. Moreover, no apparent specificity toward the head groups of phospholipids was detected (Figs. 4, C–E). Together, these data indicate that both TMEM41B and VMP1 are scramblases. To corroborate this conclusion, a BSA-based lipid extraction assay was also performed (Vehring et al., 2007). In this assay, fatty acid–free BSA was added to extract and quench the fluorescence of NBD-lipids within the outer leaflets of liposomes (Fig. S3 F). Adding BSA accelerated the decrease of NBD fluorescence in TMEM41B- or VMP1-containing liposomes (Fig. S3 G).
Figure S3.
Protein purification and the scramblase assays. (A) Purification of TMEM41B. The chromatography of gel filtration (Superose 6 Increase 10/300 GL) and corresponding SDS-PAGE of peak fractions are shown. The elution volume is indicated below. (B) Purification of VMP1. The chromatography of gel filtration (Superose 6 Increase 10/300 GL) and corresponding SDS-PAGE of peak fractions are shown. The elution volume is indicated below. (C) Setup of the fluorescence-based scramblase assay. All protein concentrations used here are 100 nM. The time points of adding dithionite and Tween 20 are denoted. Opsin is a known scramblase; DGGGpase is a lipid-modifying enzyme without any known scramblase activity. (D) The rate of scrambling NBD-lipids by TMEM41B (left) and VMP1 (right) is independent of Ca2+. The concentration of the tested protein in this assay was 10 nM. The concentration of Ca2+ is indicated. Data represent the average of three independent measurements using the same batch of NBD-lipid–containing liposomes. (E) The scrambling NBD-lipids rate by TMEM41B (left) and VMP1 (right) is independent of ATP and Mg2+. The concentration of the tested protein in this assay was 10 nM. The concentration of Ca2+ is indicated. Data represent the average of three independent measurements using the same batch of NBD-lipid–containing liposomes. (F) Schematic of the BSA extraction assay. (G) Results of the BSA extraction assay. The concentrations of proteins used in this assay are 100 nM VMP1/TMEM41B and 2 mg/ml BSA. Data represent the average of three independent measurements using the same batch of NBD-lipid–containing liposomes.
Figure 4.
TMEM41B and VMP1 have scramblase activity in vitro. (A) Schematic of the fluorescence-based scrambling assay. Only the NBD-lipids in the outer leaflet of liposome can be bleached into nonfluorescent ABD-lipid by dithionite. (B) Scrambling of NBD-lipids by TMEM41B (top), VMP1 (middle), and TMEM41B-VMP1 complex (bottom) with different concentrations of proteins as indicated. The time points of adding dithionite and Tween 20 are denoted. Data represent the average of three independent measurements using the same batch of NBD-lipid–containing liposomes. (C–E) Substrate preference for TMEM41B (left) and VMP1 (right) against NBD-PC (C), NBD-PE (D), and NBD-PS (E). The concentration of tested protein is indicated. The time points of adding dithionite and Tween 20 are denoted. Data represent the average of three independent measurements using the same batch of NBD-lipid–containing liposomes.
Most phospholipids, including PS, are synthesized on the cytoplasmic face of the ER. Thus, there is an imbalance in newly synthesized glycerophospholipids between the cytoplasmic and luminal leaflets of the ER. ATP-independent scramblases catalyze the trans-bilayer movement of charged phospholipids so as to maintain the stability of the ER bilayer (Pomorski and Menon, 2016). While such scramblases in the ER have historically been highly elusive, recent studies have suggested that TMEM16K/E, and perhaps some G-protein–coupled receptors, are candidate scramblases in the ER (Bushell et al., 2019; Pomorski and Menon, 2016; Suzuki et al., 2010; Tsuji et al., 2019). Here, we show that TMEM41B and VMP1 have scramblase activity, which could be the core function of all members of the evolutionarily conserved DedA family of proteins (Morita et al., 2018). The scramblase activity of TMEM41B/VMP1 appears to be independent of calcium, distinct from the TMEM16 family of proteins (Suzuki et al., 2010; Tsuji et al., 2019).
A general role of TMEM41B/VMP1 in maintaining cellular lipid distribution helps to explain the diverse functions of these proteins. Lipid composition of the ER can greatly impact the biogenesis of ER-derived structures, such as LDs, lipoproteins, and autophagosomes (Gao et al., 2019; Joshi et al., 2017). The entry and assembly of viruses also rely on optimal membrane lipid/cholesterol composition (Lee et al., 2021). The change in cholesterol metabolism in TMEM41B/VMP1-deficient cells is unexpected, but it may partially explain the critical requirement for TMEM41B/VMP1 in viral infection. The level and distribution of cholesterol are well-established regulators of infection by many viruses, possibly by impacting the fusion activity of the glycoproteins/spike proteins of the viruses as recently reported (Lee et al., 2021). Notably, several nonbiased genetic screens for host factors essential for viral infection identified genes/proteins regulating cholesterol homeostasis as top hits (Hoffmann et al., 2021; Schneider et al., 2021; Wang et al., 2021). Thus, among other possibilities, maintaining normal cholesterol distribution may contribute to how TMEM41B/VMP1 facilitates viral replication/infection. Although the link between the scramblase activity of TMEM41B/VMP1 and cholesterol is not apparent and requires further investigation, it is well recognized that phospholipids, especially PS, can impact cellular cholesterol trafficking and distribution (Ercan et al., 2021; Maekawa and Fairn, 2015). Recently, PS was demonstrated to control the transport of low-density lipoprotein–derived cholesterol from lysosomes to the ER (Trinh et al., 2020). A role for the distribution of PS/cholesterol in viral infection is further supported by a recent study that identified OSBPL9 (a PS transporter), PTDSS1 (a major PS synthase), and TMEM30A (cofactor of a PS flippase) as host factors for severe acute respiratory syndrome coronavirus 2 (Baggen et al., 2021). Cholesterol on the plasma membrane was dramatically increased in PTDSS1-deficient cells (Trinh et al., 2020), and cholesterol level/distribution is also likely to change in cells lacking OSBPL9 or TMEM30A. Therefore, the ability of TMEM41B/VMP1 to maintain cellular PS and cholesterol homeostasis may be a major contributing factor to viral infection.
Our data also left open several questions. Because cholesterol sensors (D4H, GRAM1b) are not quantitative, their increased signal at the PM does not necessarily indicate an increase of total cholesterol in the PM. It is possible that the three pools of cholesterol redistributed within the PM in VMP1/TMEM41B-deficient cells (Das et al., 2014), exposing more cholesterol to the sensors. Future work is needed to quantify cholesterol content in all organelles and to determine whether cholesterol trafficking between organelles remains intact in VMP1/TMEM41B-deficient cells. We cannot explain the differences in detecting PS between purified sensors (GST-2xPH) and live sensors (GFP-evt2PH and Lact-C2-GFP). It is possible that using purified sensors in permeabilized cells enables more sensitive detection, since many factors can impact the sensors in live cells (Tsuji et al., 2019). In this regard, purified sensors are commonly used for detecting cholesterol (Saha et al., 2020; Trinh et al., 2020). However, detergents may alter membrane composition/integrity, thereby compromising the accuracy of lipid detection by purified sensors. Moreover, the degree of membrane permeabilization by saponin/digitonin may be impacted by cholesterol. Thus, more accurate detection of PS in its native environment with better techniques, such as freeze-fracture cryo-electron microscopy, is needed in the future. Finally, the impact of VMP1/TMEM41B on the quantity and distribution of other phospholipids (e.g., PC and PE) should be investigated.
In summary, our results unveil a key biochemical function of TMEM41B/VMP1 and highlight the importance of lipid distribution in cell biology and disease. These results should lay the foundation for future investigations into the mechanisms of lipid sorting in the ER, the biogenesis of organelles as well as viral infection.
Materials and methods
Cell culture and transfection
HeLa cells were obtained from ATCC. HEK293 and Huh7 cells were from Invitrogen and Cell Bank Australia, respectively. Monolayers of cells were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin sulfate at 37°C with 5% CO2. Transient plasmid transfection and siRNA transfection were performed using Lipofectamine LTX/PLUS Reagent and Lipofectamine RNAiMAX Reagent (Invitrogen), according to the manufacturer’s instructions.
RNAi and cDNA constructs
siRNAs against VMP1 (SASI_Hs01_0009-2297, SASI_Hs01_0009-2298) and TMEM41B (SASI_Hs01_0017-1330, SASI_Hs01_0017-1331) were obtained from Sigma-Aldrich. Details of the cDNA constructs used in this study were described previously, including myc-VMP1, GFP-VMP1 (myc/GFP-fused vacuole membrane protein 1; Morita et al., 2018); TMEM41B-myc, TMEM41B-GFP (human gene NP_055827.1 tagged with myc/GFP; Morita et al., 2018); GFP-Evt2PH (GFP-tagged tandem fusion of evectin-2 [2×PH]; Uchida et al., 2011); GFP-GRAM1b WT and G187L (GFP-conjugated GRAM domain of GRAMD1b with a single G187L mutation; Ercan et al., 2021); GFP-P4Mx2 (GFP-conjugated tandem fusion of P4M domain consisting 546–647 of Legionella pneumophila SidM; Hammond et al., 2014); GFP-PASS (GFP-fused PA biosensor with superior sensitivity; Kassas et al., 2017); GFP-PDE4A1 (GFP-tagged single exon in the unique N-terminal region of the cAMP-specific phosphodiesterase; Kassas et al., 2017); GFP-PH-PLC (GFP-tagged PH domain and phospholipase C δ, which is known to specifically bind PtdIns(4,5)P2; Várnai and Balla, 1998); GST-2xPH (GST tagged two of the PH domains of evectin-2 in tandem; Tsuji et al., 2019); mCherry-D4H (mCherry-tagged fourth domain of perfringolysin O theta toxin with D434S mutation; Maekawa and Fairn, 2015); Lact-C2-GFP (GFP-conjugated C2 domain of lactadherin; Yeung et al., 2008); mCherry-GPAT4 (mCherry-tagged GPAT4; Pagac et al., 2016); and HA-tagged seipin, GFP-tagged seipin, and mCherry-tagged seipin (HA/GFP/mCherry-fused human seipin; Pagac et al., 2016).
LD staining
Cells were treated with 400 µM oleate for 6 h followed by 15-min fixation with 4% PFA at RT. After three washes with PBS, cells were permeabilized with 0.1% Triton X-100 for 10 min. LDs were stained with 1 µg/ml BODIPY 493 (Invitrogen) in PBS for 15 min or HCS LipidTOX Deep Red Neutral Lipid Stain (1:1,000; Invitrogen) in PBS for 1 h at RT with light avoidance. Cells were mounted on glass slides with ProLong Gold Antifade Mounting Media with DAPI (Invitrogen).
Immunofluorescence
Buffers used in this study included washing buffer (PBS containing 50 mM NH4Cl), buffer A (PBS containing 50 mM NH4Cl and 5% vol/vol NGS), buffer B (PBS containing 5% NGS), and buffer C (0.05% saponin [84510; Sigma-Aldrich] and 5% NGS in PBS). For immunostaining of intracellular PS, cells were harvested with 2% PFA fixation for 15 min at RT. After washing coverslips three times with washing buffer, cells were permeabilized with 20 µM digitonin (D141; Sigma-Aldrich) or 0.1% Triton X-100 in PBS for 5 min. Each coverslip was blocked in buffer A for 45 min after washing with PBS three times followed by 1-h incubation with purified GST-2xPH (60 µg/ml), 1-h incubation with rabbit polyclonal to anti-GST antibody (1:500, G7781; Sigma-Aldrich), and 45-min incubation with secondary antibody (Alexa Fluor 488, 1:500, A21206 [Life Technology]; Alexa Fluor 594, 1:500, A11012 [Invitrogen]). Sufficient washes with PBS were applied between each incubation. The recombinant protein and the antibodies were diluted in buffer B. Cells were then postfixed with 2% PFA for 5 min followed by three rinses with washing buffer. Before mounting on the slides with ProLong Gold Antifade Mounting Media with DAPI, coverslips were rinsed with distilled water once. For immunostaining of PM PS, cells were harvested with 4% PFA fixation for 15 min at RT. After washing coverslips three times with washing buffer, cells were blocked with buffer C for 45 min followed by 1-h incubation with GST-2xPH, 1-h incubation with anti-GST antibody, and 45-min incubation with secondary antibody as described above. The recombinant protein and antibodies were diluted in buffer C. Sufficient washes with PBS were applied between each incubation. Postfixation with 2% PFA for 5 min was applied to the cells followed by three washes with washing buffer. Coverslips were rinsed with distilled water once before mounting on the slides.
Filipin staining
Cells grown on coverslips were harvested with 4% PFA for 15 min at RT followed by three washes with PBS. Fixed cells were then incubated with freshly prepared filipin (F9765; Sigma-Aldrich) at 50 µg/ml in PBS for 1–2 h at RT in the dark. After washing three times with PBS, cells were mounted with ProLong Gold Antifade reagent.
Microscopy and image analysis
Confocal microscopy was performed using a Zeiss LSM 900 confocal microscope equipped with GaAsP detectors and a Zeiss Airyscan 2 detector. A 63×/1.4 or 40×/1.3 oil immersion objective was applied for imaging at RT. The fluorochromes used were Alexa Fluor 488, Alexa Fluor 594, BODIPY FL, DAPI (filipin), EGFP, HCS LipidTOX Deep Red neutral lipid stain, and mCherry. For comparisons of fluorescence intensity, images were taken during a single session at identical excitation and detection settings. To measure the colocalization between GFP-tagged VMP1 or TMEM41B and seipin, the Coloc 2 plugin in Fiji software was used with Threshold regression setting as Costes, point spread function as 3.0, and Costes randomization as 10. The Pearson correlation coefficients (no threshold) within individual cells of interest that expressed both VMP1/TMEM41B and seipin were recorded. The intensity of cellular GST-2xPH was analyzed using Fiji software. The ratio of raw integrated density by area for each cell of interest was acquired and normalized by the mean intensity in control group. The same method was applied to measure intracellular intensity of GST-2xPH (i.e., intensity from cell of interest excluding the PM regions). The difference between cellular and intracellular measurements of the same cell was referred to as the PM intensity of GST-2xPH. This method was also used to quantify the PM and intracellular intensities of mCherry-D4H and GFP-GRAM probes.
To measure temporal colocalization between PLIN3 and BODIPY in either WT HeLa or SKO cells, live cell imaging was performed with 37°C temperature control on the Zeiss Elyra microscope. A Plan-Apochromat 63×/1.4 oil lens was used with 0.2% of 561-nm and 0.06% 488-nm laser excitations. Images were taken with exposure time of 500 ms on an Andor Ultra 897 EMCCD camera (gain at 200) in HiLo mode (angle = 58°). A custom-built script in MATLAB (MathWorks) was used to quantify the colocalization by detecting the spots (droplets) in each channel at every time point and counting the number of green, red, and mixed (colocalizing) pairs. Briefly, the script loads CZI files into MATLAB via OME formats, and a smoothing Gaussian or Laplacian of Gaussian filter is applied, with sigma of 2.5 pixels, to denoise images and isolate droplets from the background. The feature-finding algorithm was applied to detect spots in each channel (Pelletier et al., 2009). Once detected at every time point in images series, a green spot center was checked to see whether it was within a droplet diameter distance of a green spot. If this criterion was fulfilled, these two were assigned to be a pair, and the procedure was repeated until all the spots in both channels were checked. The final count of green, red, and mixed spots per time point was exported into an Excel file for further processing.
Generation of VMP1 KO cells
VMP1 KO and TMEM41B KO HeLa cells were generated by the CRISPR/Cas9 system. Single-guide RNAs were constructed into pSpCas9(BB)-2A-GFP vector (48138; Addgene). DNA oligonucleotide sequence 5′-CTTTTGTATGCCTACTGGAT-3′ and 5′-GTCGCCGAACGATCGCAGTT-3′ (Morita et al., 2018) was chosen as single-guide RNAs to KO VMP1 or TMEM41B accordingly. HeLa WT cells transfected with CRISPR plasmid for 48 h were sorted by FACSMelody Cell Sorter (BD Biosciences). Single cells were sorted into each well of 96-well plates. VMP1 or TMEM41B deficiency was screened by genomic sequencing and Western blot analysis.
Immunoblot analysis and antibodies
Cells were harvested and resuspended in lysis buffer (1% Triton X-100, 100 mM NaCl, 50 mM Tris, pH 7.8) to incubate for 1 h on ice. Cell lysates were clarified by centrifugation, and protein concentration was determined by BCA protein assay kit. After mixing with 2× Laemmli buffer, samples were subjected to 7.5% or 10% SDS-PAGE. Proteins were then transferred to a nitrocellulose membrane (0.45 µm, #1620115; Bio-Rad) followed by blocking with 5% (wt/vol) skim milk in TBS plus Tween 20 for 1 h at RT. Incubation with primary antibody (1:1,000) was performed overnight at 4°C after three washes for 5 min each in TBS plus Tween 20. Primary antibodies included in this study were mouse monoclonal c-myc monoclonal antibody (clone 9E10, 13-2500; Invitrogen), GFP (sc-9996; Santa Cruz Biotechnology), rabbit polyclonal to VMP1 (12929S; Cell Signaling Technology), GAPDH (2118; Cell Signaling Technology), β-actin (4970; Cell Signaling Technology), and HA-tag (3724S; Cell Signaling Technology). Secondary antibodies were peroxidase-conjugated AffiniPure donkey anti-rabbit or donkey anti-mouse IgG (H+L; Jackson ImmunoResearch Laboratories) used at a 1:5,000 dilution at RT for 1 h. The bound antibodies were detected by ECL Western Blotting Detection Reagent (GE Healthcare or Merck Millipore) and visualized with Molecular Imager® ChemiDoc™ XRS+ (Bio-Rad).
Cholesterol measurement
HeLa cells plated in six-well plates were transfected with the indicated siRNA for 48 h. Cellular free cholesterol concentrations were measured using an Amplex™ Red cholesterol assay kit (Invitrogen) according to manufacturer’s instructions. In brief, cells were lysed and incubated with cholesterol oxidase, horseradish peroxidase, and Amplex Red in the absence of cholesterol esterase. The amount of cholesterol was determined indirectly by measuring resorufin absorbance at 560 nm. The values were normalized to the total cellular protein levels, which were determined with a BCA Protein Assay Kit (Thermo Fisher Scientific).
Coimmunoprecipitation
HEK293 cells in 10-cm dishes were cotransfected constructs encoding HA-seipin and GFP-tagged VMP1, TMEM41B, or mCherry-tagged GPAT4 for 24–48 h. Each dish of cells was lysed in 0.75 ml lysis buffer (25 mM Hepes pH 7.4, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% wt/vol n-dodecyl-β-D-maltoside [DDM]) on ice for 30 min. The lysates were cleared by centrifuging at 18,000 ×g for 15 min at 4°C. An equal volume of cell lysates (1∼2 mg of proteins) was mixed with 25 µl HA-agarose beads (#A2095; Sigma-Aldrich). The protein bead mixture was then gently rotated at 4°C for 3 h followed by washing three times with PBS and centrifuging at 10,000 ×g for 3 min at 4°C. Immunoprecipitated proteins were eluted with 30 µl 2× Laemmli sample buffer without β-mercaptoethanol by vortexing at RT for 5 min. The elutes were collected by centrifuging at 18,000 ×g for 5 min at RT and mixed with 30 µl 2× Laemmli sample buffer with β-mercaptoethanol. The resultant samples were subjected to SDS-PAGE and immunoblotting.
Recombinant protein purification
GST-2xPH was transfected in BL21(DE3)-competent cells (EC0114; Thermo Fisher Scientific). When bacterial growth reached an optical density at 600 nm of ∼1, 0.1 mM IPTG was added to induce protein production at 18°C overnight. Cells were then harvested and resuspended in lysis buffer (20 mM Tris-Cl, 100 mM NaCl, 1 mM DTT, 1 mM EDTA, a protease inhibitor cocktail tablet [5892791001; Roche], and 0.35 mg/ml lysozyme, pH 8.0) on ice for 30 min. Lysate was then sonicated on ice with 120 cycles of a 3-s pulse followed by a 10-s break at 10% amplitude output for further lysis. After rocking with 1% Triton X-100 at 4°C for 15 min, lysate was clarified at 17,000 ×g at 4°C for 30 min. The supernatant was incubated with Pierce Glutathione Superflow Agarose (#25237; Thermo Fisher Scientific) at 4°C overnight. GST-2xPH proteins were eluted from beads with eluting buffer (50 mM Tris-Cl, 200 mM NaCl, and 25 mM L-glutathione, pH 8.8). Eluate was dialyzed in dialysis buffer (50 mM Tris-Cl and 0.5 mM DTT, pH 8.0) at 4°C overnight. The purification efficiency was validated by Coomassie blue staining. Concentration of protein was determined by BCA Protein Assay Kit (Sigma-Aldrich).
Cloning and protein purification for scramblase assays
The cDNA of full-length human TMEM41B (UniProt ID: Q5BJD5) was subcloned into pFastBac dual with a C-terminal 2×Strep/FLAG-tag under the control of the polyhedron promoter. VMP1 (UniProt ID: Q96GC9) was cloned under control of the p10 promoter with C-terminal His10-tag. All tags can be removed by TEV protease. The baculoviruses were generated in Sf9 cells using the Bac-to-Bac system (Life Technologies). The cells were infected by the virus and harvested after 48 h. Cells were pelleted by centrifugation at 3,000 ×g for 15 min. The pellets were lysed in 20 mM Tris-HCl pH 8.0, 200 mM NaCl, 0.5 mM Tris(2-carboxyethyl)phosphine (TCEP)-HCl, 10% glycerol, 2 mM PMSF, 1.6 µM aprotinin, 2 µM pepstatin, and 20 µM leupeptin by French Press. Then the lysate was supplemented with 1% DDM and incubated at 4°C for 2 h. The lysate was centrifuged at 10,000 ×g for 50 min at 4°C. The supernatant of VMP1 was incubated with Ni-nitrilotriacetic acid resin at 4°C for 30 min supplemented with 5 mM imidazole pH 8.0. VMP1 was eluted with 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5 mM TCEP-HCl, 250 mM imidazole pH 8.0, 10% glycerol, 0.05% DDM, 0.01% cholesteryl hemisuccinate (CHS), 1 mM PMSF, 0.8 µM aprotinin, 1 µM pepstatin, and 10 µM leupeptin. The supernatant of TMEM41B was incubated with Strep-Tactin Superflow Resin (IBA Lifesciences) at 4°C for 30 min. TMEM41B was eluted with 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5 mM TCEP-HCl, 2.56 mM desthiobiotin, 10% glycerol, 0.05% DDM, 0.01% CHS, 1 mM PMSF, 0.8 µM aprotinin, 1 µM pepstatin, and 10 µM leupeptin. The eluate of target proteins was concentrated to 2 ml and loaded on Superose 6 Increase 10/300 GL column equilibrated with 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5 mM TCEP-HCl, 5% glycerol, 0.03% DDM, and 0.006% CHS. The peak fractions were flash frozen in liquid N2 and stored at −80°C for further experiments. The full-length human opsin (UniProt ID: P03999) and DGGPase (UniProt ID: Q57727) were purified according to previous reports (Menon et al., 2011; Ren et al., 2020).
Scramblase activity assay
POPC (850457P; Avanti) and POPS (840034P; Avanti) were mixed at a molar ratio of 9:1 and then vacuumed overnight at 25°C after being dried with nitrogen gas stream. The lipid film was rehydrated in buffer A (50 mM Hepes, pH 7.4, 100 mM NaCl). Liposomes were made by extruding the lipid solution 21 times through a 400-nm-pore-size membrane and 11 times through a 200-nm-pore-size membrane after 10 freeze–thaw cycles. After destabilized using 0.36% DDM (Anatrace) for 3 h at 25°C, liposomes were incubated with target proteins and 0.4 mol% NBD-lipids (NBD-PS Avanti 810192P, NBD-PC Avanti 810130P, and NBD-PE Avanti 810144P) together or separately for 1 h at 25°C. The Bio-Beads were then added and incubated to remove DDM as reported (Menon et al., 2011). The prepared proteoliposomes were diluted 20-fold using buffer A. The fluorescence signal was monitored using Synergy H1 Microplate Reader (BioTek Instruments) at excitation and emission wavelengths of 470 nm and 530 nm, respectively. 150 mM sodium hydrosulfite (157953; Sigma) was added after the system was equilibrated for 50 s, and 0.1% Tween 20 was supplemented at 600 s. ATP, MgCl2, and CaCl2 were supplemented in the buffer at indicated concentrations. The fatty acid–free BSA (B2064; Sigma) was used in the BSA extraction assay at a final concentration of 2 mg/ml instead of the dithionite. The maximum fluorescence signal data were normalized to 1, and the plots were generated using Origin Pro.
Lipidomics
Lipids were extracted from cell pellets using a modified methyl-tert-butyl ether method (Turner et al., 2018). Methyl tert-butyl ether and methanol (3:1 vol/vol with 0.01% butylated hydroxytoluene) were added to cell pellets alongside an internal standard solution containing 10 µM PS 17:0/17:0. Extracted lipids were analyzed by liquid chromatography-mass spectrometry using a Dionex UltiMate 3000 Pump liquid chromatograph coupled to a Q Exactive Plus mass spectrometer equipped with a heated electrospray ionization source (Thermo Fisher Scientific Australia). Data were acquired in full-scan/data-dependent MS2 mode (full-scan resolution, 70,000 full width at half maximum; maximum ion injection time, 50 ms; scan range mass/charge ratio, 200–1,500), with the 10 most abundant ions being subjected to collision-induced dissociation using an isolation window of 1.5 D and a normalized stepped collision energy of 15/27 eV. Lipids were analyzed using MS-DIAL. Exported aligned data were background subtracted, quantified from internal standards using the statistical package R, and normalized to total proteins. One-way ANOVA with Tukey post hoc analysis was used to identify differences between groups, with statistical significance set at an adjusted P < 0.05.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 9.0.2 for Windows (GraphPad Software) with t test, ordinary one-way ANOVA unless otherwise specified. Data were expressed as mean ± SD.
Online supplemental material
Fig. S1 compares the effects of VMP1 and seipin on early LD formation. Fig. S2 shows supporting data and controls for cholesterol and PS detection. Fig. S3 shows supporting data and controls for scramblase assay.
Supplementary Material
Acknowledgments
We thank Dr. Noboru Mizushima for sending us plasmids encoding TMEM41B and VMP1, Dr. Toyoshi Fujimoto for GST-2xPH, and Dr. Nicolas Vitale for GFP-PASS and GFP-PDE4A1.
H. Yang is supported by project grants from the National Health and Medical Research Council of Australia (grant nos. 1141938, 1141939, and 1144726). S. Qi is supported by National Key Research and Development Program of China grant 2017YFA0506300.
The authors declare no competing financial interests.
Author contributions: Y.E. Li, Y. Wang, X. Du, T. Zhang, H.Y. Mak, S. Hancock, H. McEwen, Y.C. Aw, I. Lukmantara, Y. Yuan, and X. Dong contributed to the investigation, data curation, and formal analysis. Y.E. Li and X. Du also contributed to visualization and writing. A. Don, E. Pandzic, R.M. Whan, and N. Turner contributed to data curation and supervision. S. Qi contributed to the methodology for the scramblase assays, formal analysis, supervision, data curation, funding acquisition, and writing. H. Yang contributed to the conceptualization, formal analysis, project administration, supervision, funding acquisition, and writing.
References
- Baggen, J., Persoons L., Vanstreels E., Jansen S., Van Looveren D., Boeckx B., Geudens V., De Man J., Jochmans D., Wauters J., et al. 2021. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat. Genet. 53:435–444. [DOI] [PubMed] [Google Scholar]
- Bi, J., Wang W., Liu Z., Huang X., Jiang Q., Liu G., Wang Y., and Huang X.. 2014. Seipin promotes adipose tissue fat storage through the ER Ca2+-ATPase SERCA. Cell Metab. 19:861–871. 10.1016/j.cmet.2014.03.028 [DOI] [PubMed] [Google Scholar]
- Bushell, S.R., Pike A.C.W., Falzone M.E., Rorsman N.J.G., Ta C.M., Corey R.A., Newport T.D., Christianson J.C., Scofano L.F., Shintre C.A., et al. 2019. The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K. Nat. Commun. 10:3956. 10.1038/s41467-019-11753-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, J., Wu X., Lambert T.J., Lai Z.W., Walther T.C., and Farese R.V. Jr. 2019. ldaf1 and seipin form a lipid droplet assembly complex. Dev. Cell. 51:551–563.e7. 10.1016/j.devcel.2019.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, A., Brown M.S., Anderson D.D., Goldstein J.L., and Radhakrishnan A.. 2014. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. eLife. 3:e02882. 10.7554/eLife.02882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan, B., Naito T., Koh D.H.Z., Dharmawan D., and Saheki Y.. 2021. Molecular basis of accessible plasma membrane cholesterol recognition by the GRAM domain of GRAMD1b. EMBO J. 40:e106524. 10.15252/embj.2020106524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A.J., Wenk M.R., Parton R.G., and Yang H.. 2008. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180:473–482. 10.1083/jcb.200711136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, W., Shui G., Zhang Y., Krahmer N., Ferguson C., Kapterian T.S., Lin R.C., Dawes I.W., Brown A.J., Li P., et al. 2011. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 7:e1002201. 10.1371/journal.pgen.1002201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M., Huang X., Song B.L., and Yang H.. 2019. The biogenesis of lipid droplets: lipids take center stage. Prog. Lipid Res. 75:100989. 10.1016/j.plipres.2019.100989 [DOI] [PubMed] [Google Scholar]
- Hammond, G.R., Machner M.P., and Balla T.. 2014. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J. Cell Biol. 205:113–126. 10.1083/jcb.201312072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, H.H., Schneider W.M., Rozen-Gagnon K., Miles L.A., Schuster F., Razooky B., Jacobson E., Wu X., Yi S., Rudin C.M., et al. 2021. TMEM41B Is a Pan-flavivirus Host Factor. Cell. 184:133–148.e20. 10.1016/j.cell.2020.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, A.S., Zhang H., and Prinz W.A.. 2017. Organelle biogenesis in the endoplasmic reticulum. Nat. Cell Biol. 19:876–882. 10.1038/ncb3579 [DOI] [PubMed] [Google Scholar]
- Kassas, N., Tanguy E., Thahouly T., Fouillen L., Heintz D., Chasserot-Golaz S., Bader M.F., Grant N.J., and Vitale N.. 2017. Comparative characterization of phosphatidic acid sensors and their localization during frustrated phagocytosis. J. Biol. Chem. 292:4266–4279. 10.1074/jbc.M116.742346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer, N., Guo Y., Wilfling F., Hilger M., Lingrell S., Heger K., Newman H.W., Schmidt-Supprian M., Vance D.E., Mann M., et al. 2011. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 14:504–515. 10.1016/j.cmet.2011.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., Kreutzberger A.J.B., Odongo L., Nelson E.A., Nyenhuis D.A., Kiessling V., Liang B., Cafiso D.S., White J.M., and Tamm L.K.. 2021. Ebola virus glycoprotein interacts with cholesterol to enhance membrane fusion and cell entry. Nat. Struct. Mol. Biol. 28:181–189. 10.1038/s41594-020-00548-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa, M., and Fairn G.D.. 2015. Complementary probes reveal that phosphatidylserine is required for the proper transbilayer distribution of cholesterol. J. Cell Sci. 128:1422–1433. 10.1242/jcs.164715 [DOI] [PubMed] [Google Scholar]
- Menon, I., Huber T., Sanyal S., Banerjee S., Barré P., Canis S., Warren J.D., Hwa J., Sakmar T.P., and Menon A.K.. 2011. Opsin is a phospholipid flippase. Curr. Biol. 21:149–153. 10.1016/j.cub.2010.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita, H., Zhao Y.G., Tamura N., Nishimura T., Kanda Y., Sakamaki Y., Okazaki M., Li D., and Mizushima N.. 2019. A critical role of VMP1 in lipoprotein secretion. eLife. 8:e48834. 10.7554/eLife.48834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, K., Hama Y., Izume T., Tamura N., Ueno T., Yamashita Y., Sakamaki Y., Mimura K., Morishita H., Shihoya W., et al. 2018. Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J. Cell Biol. 217:3817–3828. 10.1083/jcb.201804132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki, Y., Maeda T., and Fujimoto T.. 2005. Fixation and permeabilization protocol is critical for the immunolabeling of lipid droplet proteins. Histochem. Cell Biol. 124:445–452. 10.1007/s00418-005-0061-5 [DOI] [PubMed] [Google Scholar]
- Olzmann, J.A., and Carvalho P.. 2019. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20:137–155. 10.1038/s41580-018-0085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagac, M., Cooper D.E., Qi Y., Lukmantara I.E., Mak H.Y., Wu Z., Tian Y., Liu Z., Lei M., Du X., et al. 2016. SEIPIN regulates lipid droplet expansion and adipocyte development by modulating the activity of glycerol-3-phosphate acyltransferase. Cell Rep. 17:1546–1559. 10.1016/j.celrep.2016.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier, V., Gal N., Fournier P., and Kilfoil M.L.. 2009. Microrheology of microtubule solutions and actin-microtubule composite networks. Phys. Rev. Lett. 102:188303. 10.1103/PhysRevLett.102.188303 [DOI] [PubMed] [Google Scholar]
- Plutner, H., Davidson H.W., Saraste J., and Balch W.E.. 1992. Morphological analysis of protein transport from the ER to Golgi membranes in digitonin-permeabilized cells: role of the P58 containing compartment. J. Cell Biol. 119:1097–1116. 10.1083/jcb.119.5.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol, A., Gross S.P., and Parton R.G.. 2014. Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J. Cell Biol. 204:635–646. 10.1083/jcb.201311051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomorski, T.G., and Menon A.K.. 2016. Lipid somersaults: Uncovering the mechanisms of protein-mediated lipid flipping. Prog. Lipid Res. 64:69–84. 10.1016/j.plipres.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, S., de Kok N.A.W., Gu Y., Yan W., Sun Q., Chen Y., He J., Tian L., Andringa R.L.H., Zhu X., et al. 2020. Structural and functional insights into an archaeal lipid synthase. Cell Rep. 33:108294. 10.1016/j.celrep.2020.108294 [DOI] [PubMed] [Google Scholar]
- Saha, P., Shumate J.L., Caldwell J.G., Elghobashi-Meinhardt N., Lu A., Zhang L., Olsson N.E., Elias J.E., and Pfeffer S.R.. 2020. Inter-domain dynamics drive cholesterol transport by NPC1 and NPC1L1 proteins. eLife. 9:e57089. 10.7554/eLife.57089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo, V.T., Belevich I., Li S., Karhinen L., Vihinen H., Vigouroux C., Magré J., Thiele C., Hölttä-Vuori M., Jokitalo E., and Ikonen E.. 2016. Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 35:2699–2716. 10.15252/embj.201695170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santinho, A., Salo V.T., Chorlay A., Li S., Zhou X., Omrane M., Ikonen E., and Thiam A.R.. 2020. Membrane curvature catalyzes lipid droplet assembly. Curr. Biol. 30:2481–2494.e6. 10.1016/j.cub.2020.04.066 [DOI] [PubMed] [Google Scholar]
- Schneider, W.M., Luna J.M., Hoffmann H.H., Sánchez-Rivera F.J., Leal A.A., Ashbrook A.W., Le Pen J., Ricardo-Lax I., Michailidis E., Peace A., et al. 2021. Genome-Scale Identification of SARS-CoV-2 and Pan-coronavirus Host Factor Networks. Cell. 184:120–132.e14. 10.1016/j.cell.2020.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sołtysik, K., Ohsaki Y., Tatematsu T., Cheng J., Maeda A., Morita S.Y., and Fujimoto T.. 2021. Nuclear lipid droplets form in the inner nuclear membrane in a seipin-independent manner. J. Cell Biol. 220:e202005026. 10.1083/jcb.202005026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, J., Umeda M., Sims P.J., and Nagata S.. 2010. Calcium-dependent phospholipid scrambling by TMEM16F. Nature. 468:834–838. 10.1038/nature09583 [DOI] [PubMed] [Google Scholar]
- Szymanski, K.M., Binns D., Bartz R., Grishin N.V., Li W.P., Agarwal A.K., Garg A., Anderson R.G., and Goodman J.M.. 2007. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA. 104:20890–20895. 10.1073/pnas.0704154104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh, M.N., Brown M.S., Goldstein J.L., Han J., Vale G., McDonald J.G., Seemann J., Mendell J.T., and Lu F.. 2020. Last step in the path of LDL cholesterol from lysosome to plasma membrane to ER is governed by phosphatidylserine. Proc. Natl. Acad. Sci. USA. 117:18521–18529. 10.1073/pnas.2010682117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, T., Cheng J., Tatematsu T., Ebata A., Kamikawa H., Fujita A., Gyobu S., Segawa K., Arai H., Taguchi T., et al. 2019. Predominant localization of phosphatidylserine at the cytoplasmic leaflet of the ER, and its TMEM16K-dependent redistribution. Proc. Natl. Acad. Sci. USA. 116:13368–13373. 10.1073/pnas.1822025116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, N., Lim X.Y., Toop H.D., Osborne B., Brandon A.E., Taylor E.N., Fiveash C.E., Govindaraju H., Teo J.D., McEwen H.P., et al. 2018. A selective inhibitor of ceramide synthase 1 reveals a novel role in fat metabolism. Nat. Commun. 9:3165. 10.1038/s41467-018-05613-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida, Y., Hasegawa J., Chinnapen D., Inoue T., Okazaki S., Kato R., Wakatsuki S., Misaki R., Koike M., Uchiyama Y., et al. 2011. Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes. Proc. Natl. Acad. Sci. USA. 108:15846–15851. 10.1073/pnas.1109101108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várnai, P., and Balla T.. 1998. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 143:501–510. 10.1083/jcb.143.2.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vehring, S., Pakkiri L., Schröer A., Alder-Baerens N., Herrmann A., Menon A.K., and Pomorski T.. 2007. Flip-flop of fluorescently labeled phospholipids in proteoliposomes reconstituted with Saccharomyces cerevisiae microsomal proteins. Eukaryot. Cell. 6:1625–1634. 10.1128/EC.00198-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, T.C., Chung J., and Farese R.V. Jr. 2017. Lipid droplet biogenesis. Annu. Rev. Cell Dev. Biol. 33:491–510. 10.1146/annurev-cellbio-100616-060608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Becuwe M., Housden B.E., Chitraju C., Porras A.J., Graham M.M., Liu X.N., Thiam A.R., Savage D.B., Agarwal A.K., et al. 2016. Seipin is required for converting nascent to mature lipid droplets. eLife. 5:e16582. 10.7554/eLife.16582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Ma Q., Qi Y., Dong J., Du X., Rae J., Wang J., Wu W.F., Brown A.J., Parton R.G., et al. 2019. ORP2 delivers cholesterol to the plasma membrane in exchange for phosphatidylinositol 4, 5-bisphosphate (PI(4,5)P2). Mol. Cell. 73:458–473.e7. 10.1016/j.molcel.2018.11.014 [DOI] [PubMed] [Google Scholar]
- Wang, R., Simoneau C.R., Kulsuptrakul J., Bouhaddou M., Travisano K.A., Hayashi J.M., Carlson-Stevermer J., Zengel J.R., Richards C.M., Fozouni P., et al. 2021. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell. 184:106–119.e14. 10.1016/j.cell.2020.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, R., Qian H., Lukmantara I., Gao M., Du X., Yan N., and Yang H.. 2018. Human SEIPIN binds anionic phospholipids. Dev. Cell. 47:248–256.e4. 10.1016/j.devcel.2018.09.010 [DOI] [PubMed] [Google Scholar]
- Yeung, T., Gilbert G.E., Shi J., Silvius J., Kapus A., and Grinstein S.. 2008. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 319:210–213. 10.1126/science.1152066 [DOI] [PubMed] [Google Scholar]
- Zhao, Y.G., Chen Y., Miao G., Zhao H., Qu W., Li D., Wang Z., Liu N., Li L., Chen S., et al. 2017. The ER-localized transmembrane protein EPG-3/VMP1 regulates SERCA activity to control ER-isolation membrane contacts for autophagosome formation. Mol. Cell. 67:974–989.e6. 10.1016/j.molcel.2017.08.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.