VPS13D mutations result in severe mitochondrial defects. Guillén-Samander et al. show that VPS13D binds VAP in the ER and interacts with Miro on mitochondria and peroxisomes, where it could provide a bridge for lipid transport between these organelles.
Abstract
Mitochondria, which are excluded from the secretory pathway, depend on lipid transport proteins for their lipid supply from the ER, where most lipids are synthesized. In yeast, the outer mitochondrial membrane GTPase Gem1 is an accessory factor of ERMES, an ER–mitochondria tethering complex that contains lipid transport domains and that functions, partially redundantly with Vps13, in lipid transfer between the two organelles. In metazoa, where VPS13, but not ERMES, is present, the Gem1 orthologue Miro was linked to mitochondrial dynamics but not to lipid transport. Here we show that Miro, including its peroxisome-enriched splice variant, recruits the lipid transport protein VPS13D, which in turn binds the ER in a VAP-dependent way and thus could provide a lipid conduit between the ER and mitochondria. These findings reveal a so far missing link between function(s) of Gem1/Miro in yeast and higher eukaryotes, where Miro is a Parkin substrate, with potential implications for Parkinson’s disease pathogenesis.
Introduction
Homeostasis of membranous subcellular organelles relies on appropriate synthesis, metabolism, and distribution of bilayer lipids. Most such lipids are synthesized in the ER, and their delivery to other destinations relies both on vesicular traffic and on protein-mediated lipid transfer. Much of the latter route of delivery occurs at membrane contact sites, where many lipid transport proteins also function as tethers between the two membranes (Prinz, 2014; Saheki and De Camilli, 2017; Wong et al., 2019). For mitochondria, which are excluded from the membrane traffic flow of the secretory pathway, protein-mediated lipid transfer represents the only mechanism for their supply of lipids from the ER (Scharwey et al., 2013). In yeast, ER–mitochondria encounter structure (ERMES), a heterotetrametric protein complex that contains lipid transport modules and tethers the ER to the outer mitochondrial membrane (OMM), was shown to account for some of this transport (Kornmann et al., 2009). Another yeast protein, Vps13, which also localizes at membrane contact sites, albeit not ER–mitochondria contacts in this organism, was shown to have partially overlapping function with ERMES, possibly by participating in an alternative route for the delivery of lipids from the ER to mitochondria via the vacuole (Lang et al., 2015; Park et al., 2016; John Peter et al., 2017). While the ERMES complex is not conserved in metazoans, the single yeast Vps13 has four different homologues in mammals (including in humans; Velayos-Baeza et al., 2004). Additionally, an accessory subunit of ERMES, the OMM GTPase Gem1, is also conserved in higher eukaryotes (Frederick et al., 2004; Kornmann et al., 2011). However, the two mammalian Gem1 orthologues, Miro1 and Miro2, have been associated primarily with mitochondrial dynamics and not with lipid transport (Fransson et al., 2003; Glater et al., 2006; Saotome et al., 2008; MacAskill et al., 2009; Wang and Schwarz, 2009; Nguyen et al., 2014). Notably, a localization of Miro at ER–mitochondria contact sites has also been reported (Kornmann et al., 2009; Modi et al., 2019). Additionally, splice variants of Miro1 (Miro1v2 and Miro1v4) were shown to be enriched at peroxisomes and to be implicated in the dynamics of these organelles as well (Okumoto et al., 2018; Castro et al., 2018; Covill-Cooke et al., 2020).
Recent findings have shown that Vps13 is indeed a lipid transport protein and have shed light on the molecular properties of VPS13 family proteins and on some of their sites of action in animal cells (Kumar et al., 2018; Li et al., 2020). The N-terminal portion of Vps13 folds as an elongated tube with a hydrophobic groove that runs along its entire length, thus suggesting that Vps13 acts as a bridge allowing lipid flow from one bilayer to another at sites of bilayer apposition (Li et al., 2020; Lees and Reinisch, 2020; Ugur et al., 2020). Such structure may account for net transfer of lipids and thus for a role of VPS13 proteins and of its distant relative ATG2 (Kumar et al., 2018), an autophagy protein with a very similar fold (Valverde et al., 2019; Osawa et al., 2019; Maeda et al., 2019), in membrane expansion. Studies of mammalian VPS13A and VPS13C have demonstrated that they not only are localized at membrane contact sites but also tether adjacent membranes (Kumar et al., 2018). Moreover, studies of the mammalian VPS13 protein family have shown paralogue-specific localizations (Seifert et al., 2011; Kumar et al., 2018; Yeshaw et al., 2019; Park and Neiman, 2020). VPS13A populates ER–mitochondria contacts, where it may have taken over some of the functions of ERMES; VPS13C is localized at ER–late endosome/lysosome contacts (Kumar et al., 2018); and VPS13B resides predominantly in the Golgi complex region (Seifert et al., 2011). The localization and potential tethering properties of VPS13D remain unclear. Distinct functions of the four mammalian VPS13 proteins are further demonstrated by the different neurological diseases associated with their loss-of-function mutations, namely chorea acanthocytosis (VPS13A; Rampoldi et al., 2001; Ueno et al., 2001), Cohen syndrome (VPS13B; Kolehmainen et al., 2003), Parkinson’s disease (VPS13C; Lesage et al., 2016; Schormair et al., 2018), and ataxia (VPS13D; Gauthier et al., 2018; Seong et al., 2018).
VPS13D is the only mammalian VPS13 protein proven to be essential for cell and organismal survival (Wang et al., 2015; Blomen et al., 2015; Seong et al., 2018; Anding et al., 2018). While complete absence of each of the other three VPS13 proteins is compatible with life in humans, albeit with developmental (VPS13B) or neurodegenerative (VPS13A and VPS13C) defects, in patients with compound heterozygous VPS13D mutations leading to severe ataxias, at least one of the alleles carries a missense mutation, thus probably encoding an at least partially functioning protein (Dziurdzik et al., 2020; Ugur et al., 2020). Complete absence of the VPS13D orthologue in mice leads to embryonic lethality, and in flies it leads to death at the larval stage (Seong et al., 2018; Anding et al., 2018). Given its physiological importance, investigation of this protein will likely reveal some fundamental aspect of the biology of the VPS13 protein family. Studies in both human cells and flies have suggested a role for VPS13D in mitochondrial function. Enlarged spherical mitochondria have been reported in midgut cells and neurons of VPS13D-knockdown flies, in VPS13D-knockout HeLa cells, and in patient-derived fibroblasts (Seong et al., 2018; Anding et al., 2018; Insolera et al., 2020 Preprint). However, so far, VPS13D has not been visualized in association with mitochondria and was instead reported, on the basis of immunofluorescence of its orthologue in Drosophila melanogaster, to be localized on lysosomes (Anding et al., 2018).
Here, we show that VPS13D binds to mitochondria and peroxisomes via an interaction with the GTPase Miro. It also binds the ER via its interaction with the ER protein vesicle-associated membrane protein (VAP) through a phospho-FFAT motif. These findings suggest that, like its yeast orthologue Gem1, Miro participates in the control of lipid transport between adjacent membranes, thus revealing an evolutionary link between the functions of Miro and Gem1 that had remained elusive until now. Collectively, our results provide new insight into the crosstalk between the ER and other organelles at contact sites and point to an explanation for the mitochondrial phenotypes caused by the lack of VPS13D.
Results
Localization of VPS13D in the Golgi complex and on mitochondria
VPS13D, with 4,388 residues in humans, is the largest protein of the human VPS13 family. Its predicted domain architecture is overall very similar to that of other VPS13 proteins (Velayos-Baeza et al., 2004; Kumar et al., 2018). It comprises a long N-terminal lipid transfer domain that starts with the conserved chorein domain and is predicted to be slightly longer than in other VPS13 paralogues, a β-propeller region (so-called Vps13 adaptor binding [VAB; Bean et al., 2018] or WD40-like region [Kumar et al., 2018]), a Dbl homology (DH)-like domain, and a C-terminal pleckstrin homology (PH) domain. Differently from other human VPS13 proteins, VPS13D also contains a ubiquitin-binding domain (UBA; Fig. 1 A, left; Anding et al., 2018; Ugur et al., 2020). To investigate VPS13D localization in cells, overcoming the absence of antibodies that yield a detectable immunofluorescence signal and the limited expression expected upon transfection of such a large protein, a synthetic codon optimized cDNA sequence encoding VPS13D was generated, which also included a fluorescent tag (Halo or EGFP) after residue 1,576 (hereafter referred to as VPS13D^tag). This position, which is localized in a predicted disordered loop, corresponds to the position where tag insertion did not affect function in other VPS13 proteins (Fig. 1 A, left; Park et al., 2016; Kumar et al., 2018). Expression of VPS13D^EGFP in COS7 cells resulted in a diffuse cytosolic fluorescence with some variable accumulation in the Golgi complex region and a weak enrichment, also variable in intensity from cell to cell but detectable in about half of the cells, around mitochondria and in sparse additional spots (Fig. 1 B). In cells where the localization on mitochondria was detectable, VPS13D was not restricted to a small subset of mitochondria, speaking against a selective binding of VPS13D via its UBA domain to ubiquitinated mitochondria targeted for degradation.
Figure 1.
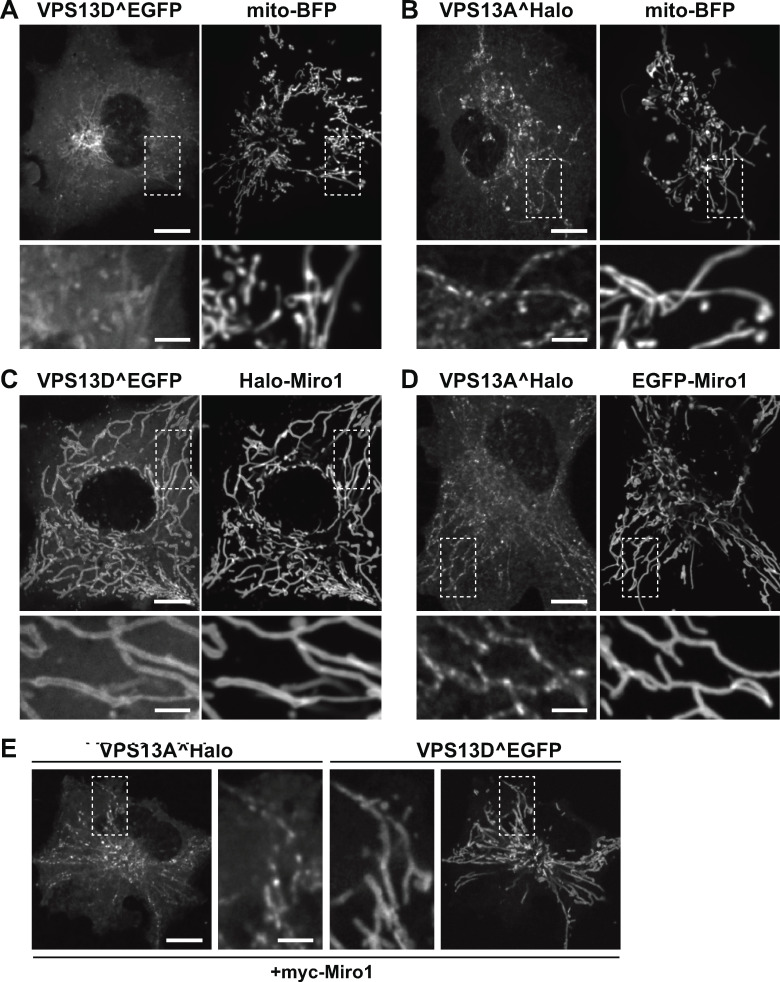
VPS13D recruitment by Miro GTPases. (A) Domain organization of human VPS13D and Miro1. TM, transmembrane. (B) Confocal images of COS7 cell expressing VPS13D^EGFP, showing that this protein decorates mitochondria, as shown by colocalization with mito-BFP (Box 1), in addition to being enriched in the Golgi area, visualized by the trans-Golgi marker ST-Halo (Box 2). (C) COS7 cell coexpressing VPS13D^EGFP and Halo-Miro1, showing dramatic increase in the localization of VPS13D^EGFP to mitochondria produced by Miro overexpression. The region within the white rectangle is shown at higher magnification next to the main field. Fig. 1, B and C, are also shown in Fig. S1, A and C. Scale bars, 10 µm in main panels, 3 µm in insets. (D) Recruitment of VPS13D to mitochondria can also be achieved by coexpression with Miro2. (E) VPS13D^Halo and EGFP-Miro1 colocalize in the cell body and processes of a mouse hippocampal neuron. (F) Optogenetic recruitment of the cytosolic domain of Miro1 to the OMM. Top panels: Schematic representation of the experiment, showing that Venus-iLID-Mito in the OMM recruits mCh-Miro1(ΔTM)-SspB upon blue light excitation. The recruitment of Miro1, in turn, triggers the recruitment of VPS13D. Middle and bottom panels: Confocal images of a COS7 cell show blue light–dependent recruitment and shedding of mCh-Miro1(ΔTM)-SspB and correspondingly of VPS13D^Halo. The illumination was started at time 0 s on a 5-µm2 area shown in the leftmost panel. See also Video 1. Scale bars for D–F, 10 µm. (G) Time course of the recruitment and shedding of VPS13D^Halo and mCh-Miro1(ΔTM)-SSPB to the OMM. Top left panel: Snapshots of an isolated mitochondrion at the peak of recruitment. Top right panel: Example kymographs showing the increase and decrease in fluorescence along the stippled line shown in the left panels. Note that while the mCh-Miro1(ΔTM)-SSPB signal on mitochondria is lower than in the surrounding cytosol before illumination and after recovery, this is not the case for VPS13D^Halo, as a pool of this protein is bound to endogenous Miro. The kymographs start 10 s before illumination. Bottom panel: Graph showing the normalized fluorescence intensity (average ± SEM) along the length of kymographs of 18 independently illuminated mitochondria in 14 different cells. Scale bars, 1 µm. The decay in intensity of VPS13D^Halo and mCh-Miro1(ΔTM)-SspB on mitochondria was fitted to an exponential equation: For VPS13D^Halo, time constant τ = 86.51 s; 95% confidence interval, 77.64, 97.56; for mCh-Miro1(ΔTM)-SspB, time constant τ = 54.11 s; 95% confidence interval, 51.07, 57.54; adjusted R2 of fits = 0.97 and 0.98, respectively.
Overexpression of Miro very strongly enhances the recruitment of VPS13D to mitochondria
Among the OMM proteins that could function as regulated binding partners for VPS13D at the mitochondrial surface, we considered Miro1 and Miro2 (henceforth referred to as Miro) as potential candidates. Miro, which contains two GTPase domains and two EF-hand Ca2+-binding domains exposed to the cytosol (Fig. 1 A, right), has been primarily implicated in the regulation of mitochondrial transport in mammalian cells (Fransson et al., 2003; Glater et al., 2006; Saotome et al., 2008; MacAskill et al., 2009; Wang and Schwarz, 2009; Nguyen et al., 2014). The Miro orthologue in yeast, Gem1, is an accessory subunit of the ERMES complex, whose function in lipid transport between ER and mitochondria is partially redundant with that of yeast Vps13 (Lang et al., 2015; Park et al., 2016). Thus, a potential connection of Miro to a mammalian VPS13 family member seemed plausible. When either Miro1or Miro2 was coexpressed with VPS13D in COS7 cells, a major impact on the localization of VPS13D was observed. This protein was now strikingly concentrated on mitochondria in more than 85% of the cells, where it colocalized with Miro along the entire mitochondrial surface (Fig. 1, C and D; and Fig. S1; see also Fig. 2 B). Minimal residual fluorescence was observed elsewhere in the cell as if Miro had sequestered the entire VPS13D pool on mitochondria. Colocalization of cotransfected VPS13D^Halo and EGFP-Miro1 was also observed in mouse hippocampal neurons, where VPS13D was recruited to mitochondria both in the cell soma and in the neurites (Fig. 1 E).
Figure S1.
Both VPS13D and VPS13A interact with mitochondria, but only the interaction of VPS13D is mediated by Miro. (A and B) Confocal images of COS7 cells coexpressing either VPS13D^EGFP (A) or VPS13A^Halo (B) and the mitochondrial marker mito-BFP, but not Miro. The fluorescence of both VPS13 paralogues decorates mitochondria, with the fluorescence of VPS13A showing the typical discontinuous pattern that was shown (Kumar et al., 2018) to reflect its selective concentration at mitochondria–ER contact sites even without VAP overexpression. (C and D) Upon coexpression with Miro1, the localization of VPS13D^EGFP (C) but not VPS13A^Halo (D) at mitochondria is drastically enhanced. Note that VPS13D^EGFP colocalizes with Miro1 along the entire mitochondrial surface, while the punctate localization of VPS13A^Halo along mitochondria is unaffected by the overexpression of Miro1. (E) COS7 cell coexpressing VPS13A^Halo and VPS13D^EGFP as well as myc-Miro1 (not shown), showing the different localization of the two proteins on mitochondria: VPS13D^EGFP is enriched throughout the entire mitochondrial surface, while VPS13A^Halo localizes only to hot spots. Higher magnifications of the areas enclosed by stippled white rectangles are shown for all fields. Fig. S1, A and C, are also shown in Fig. 1, B and C. Scale bars, 10 µm in the main panels and 3 µm in insets.
Figure 2.
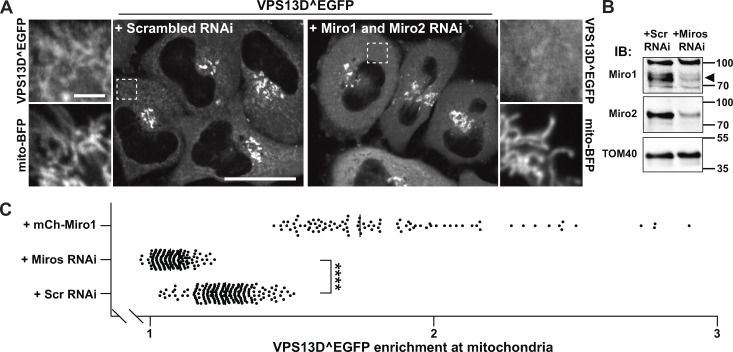
Miro is required for VPS13D recruitment to mitochondria. (A) Confocal images of HeLa cells showing the moderate recruitment of VPS13D^EGFP to mitochondria in control cells (left panel) and the loss of such signal upon knockdown of the two Miro genes via RNAi (right panel). Areas enclosed by stippled white rectangles are shown at higher magnification next to the main field, and they also show the localization of the mitochondrial marker mito-BFP. Scale bars, 30 µm in the main panel and 3 µm in the insets. (B) Immunoblot (IB) of mitochondrial fractions from HeLa cells showing the decrease of Miro1 and Miro2 levels upon RNAi treatment. Sizes in kD are indicated next to the blot. The black arrowhead indicates the Miro1 band. (C) Quantification of VPS13D^EGFP enrichment at mitochondria in control conditions and upon Miro knockdown or overexpression. The signal from mito-BFP was first used to generate a mitochondrial mask and a mask profiling a thin (1-pixel wide) cytosolic area surrounding mitochondria; the intensity from EGFP was then measured within each of these masks, and, for each cell analyzed, the ratio between these two measurements was plotted on the graph. Number of cells analyzed for scrambled (Scr) RNAi, Miro RNAi, and mCh-Miro1 conditions are 204, 204, and 96, respectively. ****, P < 0.0001 (Welch’s corrected ANOVA with Games-Howell's post hoc test).
Miro-dependent acute recruitment of VPS13D to mitochondria
To confirm a role for Miro in VPS13D recruitment to mitochondria, the improved light-induced dimer (iLID) optical dimerization system (Guntas et al., 2015) was used to induce acutely, upon blue light illumination, the association of the cytosolic portion of Miro to the mitochondrial surface. We cotransfected VPS13D^Halo with a mitochondrial anchored bait (iLID fused to a mitochondrial targeting sequence), and the cytosolic portion of Miro1 fused to the cognate prey for iLID (the stringent starvation protein B [SspB] peptide). Blue light illumination induced immediate massive recruitment of the Miro1 construct to mitochondria, which was rapidly followed by a corresponding massive recruitment of VPS13D (Fig. 1, F and G), proving the role of Miro in VPS13D recruitment. Conversely, upon light interruption, both proteins were shed from mitochondria (Fig. 1 G and Video 1). The short latency of the shedding of VPS13D relative to shedding of Miro during the postillumination phase (Fig. 1 G) may reflect the occurrence of stabilizing VPS13D interactions at the mitochondrial membrane, including its interaction with endogenous Miro, once its recruitment has occurred.
Video 1.
Confocal time-lapse imaging of a COS7 cell coexpressing VPS13D^Halo, mCh-Miro1(ΔTM)-SspB, and EGFP-iLID-Mito (not shown), showing recruitment of VPS13D and Miro1 upon blue light irradiation (at 0 s) of the area indicated by the white box. Images were acquired at a rate of 0.5 frames/s for a total of 4 min. Scale bars, 10 µm.
Loss of Miro results in a defect in the targeting of VPS13D to mitochondria
We next examined whether Miro is required for the recruitment to mitochondria of exogenous VPS13D. HeLa cells were transfected with VPS13D^EGFP and siRNAs targeting the two Miro genes or control scrambled siRNAs. As shown by Fig. 2 A and as quantified in Fig. 2 B, in cells treated with Miro siRNAs, where the decrease of both Miro1 and Miro2 was validated by Western blotting (Fig. 2 C), the VPS13D^EGFP signal on mitochondria was significantly reduced, while the robust signal in the Golgi complex, where Miro is not localized, was not affected. We also attempted to analyze the localization of VPS13D^EGFP in cells where one Miro paralogue was knocked out by CRISPR-Cas9 and the other one was knocked down, but the poor viability of these cells prevented a reliable analysis.
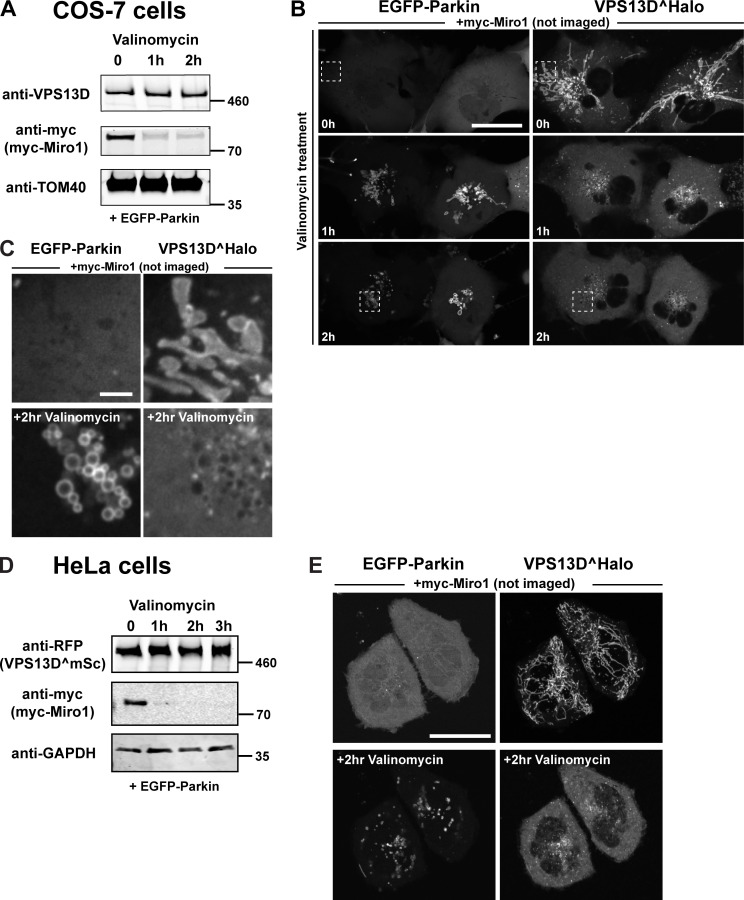
Miro is a key target of the PINK1/Parkin-dependent degradation pathway of mitochondrial proteins leading to mitophagy. Triggering of this pathway with valinomycin, which disrupts mitochondrial membrane potential and promotes the recruitment of Parkin to mitochondria (Narendra et al., 2010; Yamano et al., 2018), results in the rapid degradation of Miro proteins (Wang et al., 2011; Birsa et al., 2014; Shlevkov et al., 2016). Accordingly, 1 h of valinomycin treatment of either COS7 or HeLa cells expressing myc-Miro1 and EGFP-Parkin resulted in a robust decrease in the levels of this protein (Fig. S2, A and D, respectively). While levels of VPS13D, either endogenous (Fig. S2 A) or exogenous (Fig. S2 D), did not change, Miro degradation coincided with the shedding of VPS13D from mitochondria and with an increase in its cytosolic pool (Fig. S2, B–D). While these experiments do not prove the role of Miro in the recruitment of VPS13D to mitochondria, because Parkin-induced degradation is not specific for Miro, they are consistent with the importance of Miro demonstrated by the Miro-knockdown experiments.
Figure S2.
Parkin-mediated Miro degradation correlates with the dissociation of VPS13D from mitochondria. COS7 or HeLa cells were cotransfected with myc-Miro1, VPS13D, and EGFP-Parkin. (A and D) Western blots showing the decrease of myc-Miro1 level upon 10 µM valinomycin treatment of COS7 (A) or HeLa cells (D). Scarlet, a modified RFP, was visualized by anti-RFP antibodies. Sizes in kD are indicated next to each blot. (B, C, and E) Confocal time lapses of COS7 (B and C) or HeLa cells (E) coexpressing EGFP-Parkin, VPS13D^Halo, and myc-Miro1. VPS13D, initially recruited to mitochondria by Miro, is shed from mitochondria and relocated to the cytosol upon treatment with valinomycin, which induces the recruitment of EGFP-Parkin. The boxed regions in B are shown in higher magnification in C. Scale bars, 30 µm in the main panels and 3 µm in insets.
A Miro1 splice variant targeted to peroxisomes recruits VPS13D to peroxisomes
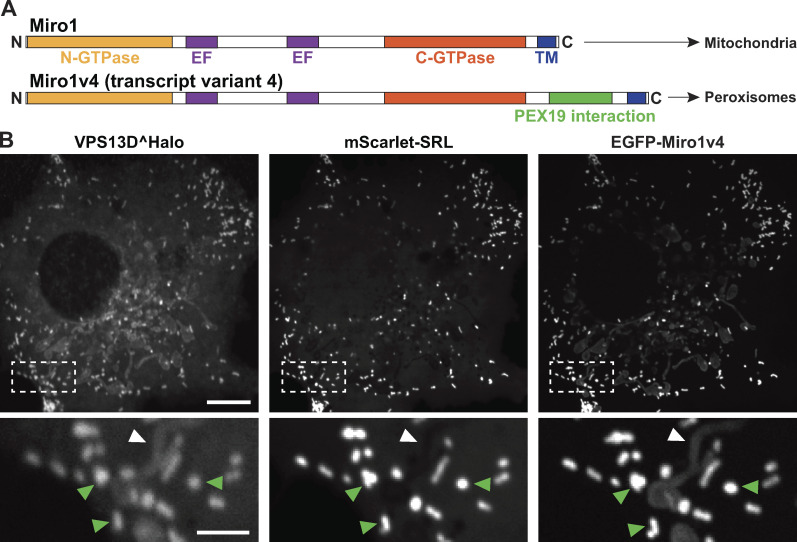
A splice variant of Miro1 including exons 19 and 20 in its C-terminal region (transcript variant 4, referred to as Miro1v4) was shown to localize predominantly at the membrane of peroxisomes, and only weakly on the OMM, via a mechanism that requires its interaction with Pex19 (Fig. 3 A; Okumoto et al., 2018; Covill-Cooke et al., 2020), a cytosolic chaperone that functions as a receptor for a subset of peroxisomal membrane proteins (Jones et al., 2004). If Miro is sufficient to recruit VPS13D to an organelle, one would expect that Miro1v4 may recruit VPS13D to peroxisomes. In fact, upon coexpression with Miro1v4 in COS7 cells, the majority of VPS13D^Halo was recruited to peroxisomes where Miro1v4 was predominantly localized, and only a faint VPS13D^Halo signal was observed on mitochondria (Fig. 3 B).
Figure 3.
A transcript variant of Miro1 preferentially targeted to peroxisomes recruits VPS13D to peroxisomes. (A) Domain organization of two splice variants of Miro1 (Okumoto et al., 2018). TM, transmembrane. In the alternative splice variant 4 (Miro1v4), two extra exons are included that encode 73 additional amino acids in the C-terminal region of Miro1. These amino acids promote the interaction of the protein with the chaperone Pex19, which leads to the predominant insertion of this variant into the peroxisomal membrane, although a pool of this variant still localizes to mitochondria. (B) COS7 cell showing the recruitment of VPS13D^Halo to peroxisomes (visualized by the peroxisomal luminal marker mScarlet-SRL) upon coexpression with transcript variant Miro1v4. The insets show colocalization with the peroxisomal marker (green arrowheads) and a weaker signal for both VPS13D and Miro1v4 on mitochondria (white arrowheads). Scale bars, 10 µm in the main panel and 3 µm in the insets.
Integrity of the GTPase domains and EF-hand regions of Miro are required for the recruitment of VPS13D
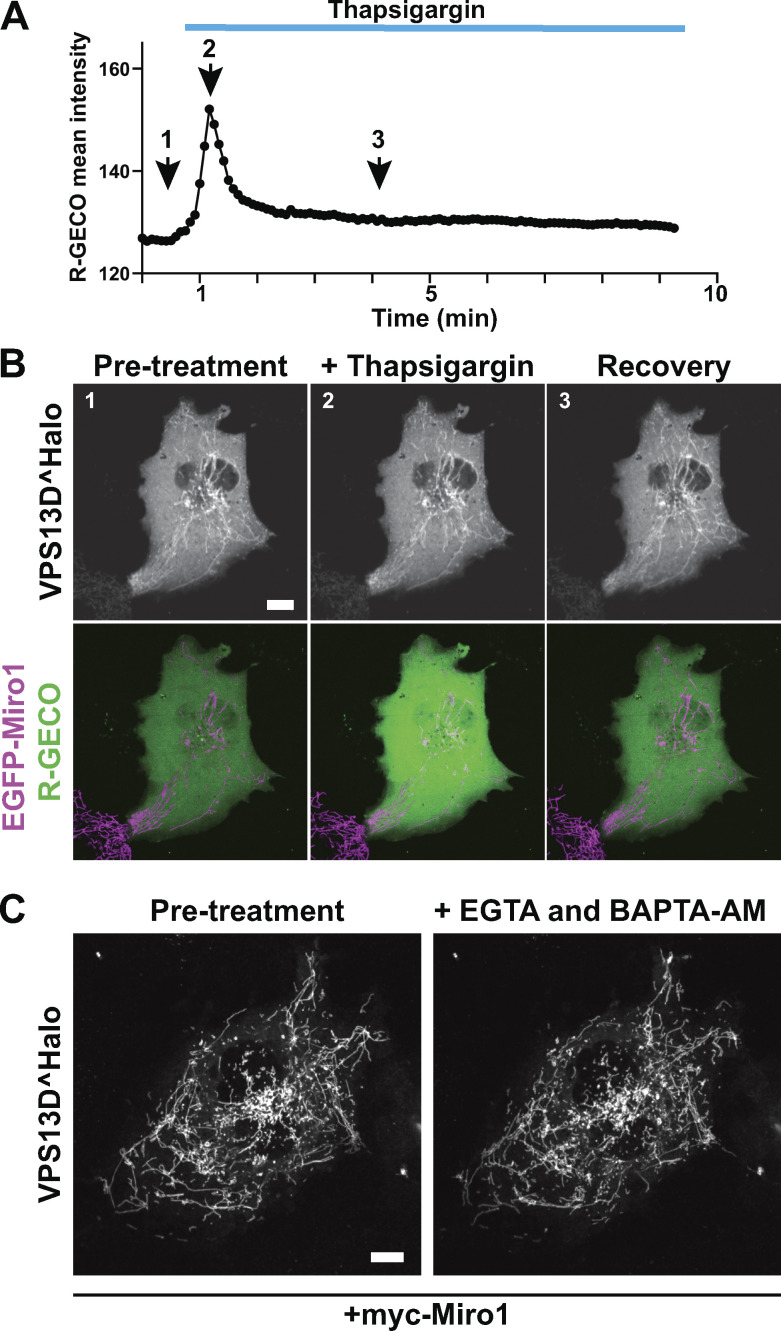
Binding of Miro1 to some of its known other interactors and of Gem1 to ERMES in yeast requires integrity of its GTPase and EF-hand domains (Kornmann et al., 2011; Kanfer et al., 2015; Oeding et al., 2018). When inactivating mutations based on previous studies were introduced in the GTP binding site of the N-terminal GTPase domain (T18N) or in the Ca2+-binding sites of both EF hands (E208K/E328K) of Miro1 (Fig. 4 A; Fransson et al., 2003; Frederick et al., 2004), the corresponding constructs no longer promoted recruitment of VPS13D to mitochondria (Fig. 4 B). In contrast, a mutation in the C-terminal GTPase (S432N) of Miro1 (Fransson et al., 2003; Frederick et al., 2004) did not affect the recruitment of VPS13D (Fig. 4 B). To assess a potential regulatory role of cytosolic Ca2+, we treated cells with the ER Ca2+ pump inhibitor thapsigargin to raise its concentration, but no further increase of VPS13D recruitment to mitochondria in cell overexpression of Miro1 was observed (Fig. S3, A and B). Likewise, incubation of cells in the absence of extracellular Ca2+ and in the presence of EGTA (to chelate residual extracellular Ca2+) and 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid acetoxymethyl ester (BAPTA-AM), to chelate intracellular Ca2+ had no obvious effect on VPS13D recruitment to mitochondria (Fig. S3 C). In light of this insensitivity to cytosolic Ca2+, the possibility that some of the disrupting effects of these mutations reflect regulatory mechanisms or protein misfolding, as previously considered (Kanfer et al., 2015), remains to be determined.
Figure 4.
Recruitment of VPS13D by Miro GTPases requires their intact N-GTPase domain and EF-hand domains and the β-propeller region of VPS13D. (A) Domain organization of human Miro1 and mutations used for the experiments shown in B. TM, transmembrane. (B) Confocal images of COS7 cells showing that the recruitment of VPS13D^Halo by EGFP-Miro1 is impaired by mutations in the GTP binding site of its N-GTPase domain and in the Ca2+ binding sites of both its EF hands, but not by a mutation in the GTP binding site of its C-GTPase domain. Asterisks represent the point mutations in the Miro1 construct. Scale bar, 3 µm. (C) Top: Cartoons showing VPS13D constructs used for the experiments shown below. Bottom: Confocal images showing that removal of the C-terminal half of the protein or of the β-propeller region selectively, but not of the DH-PH domain, abolishes recruitment of VPS13D^EGFP by Halo-Miro1. All of several constructs encoding only the β-propeller region formed small aggregates, possibly due to misfolding. See also Fig. S4. Scale bar, 10 µm.
Figure S3.
Recruitment of VPS13D to mitochondria by Miro is unaffected by changes in cytosolic Ca2+. (A and B) COS7 cell expressing EGFP-Miro1, VPS13D^Halo and the RFP genetically encoded Ca2+ indicator for optical imaging (R-GECO). (A) Cytosolic Ca2+ levels before and after addition of the sarco/endoplasmic reticulum Ca2+-ATPase pump inhibitor thapsigargin. (B) Time-lapse confocal images showing snapshot from the time points indicated in A and demonstrating that the localization of VPS13D^Halo on mitochondria in the presence of overexpressed Miro1 is not affected by the Ca2+ concentration. (C) Time-lapse confocal images showing that the binding of VPS13D^Halo to mitochondria in the presence of coexpressed myc-Miro1 is unaffected by the addition of EGTA and BAPTA-AM to lower intracellular cytosolic Ca2+. Scale bars, 10 µm.
The VAB/β-propeller domain of VPS13D is required for its recruitment by Miro
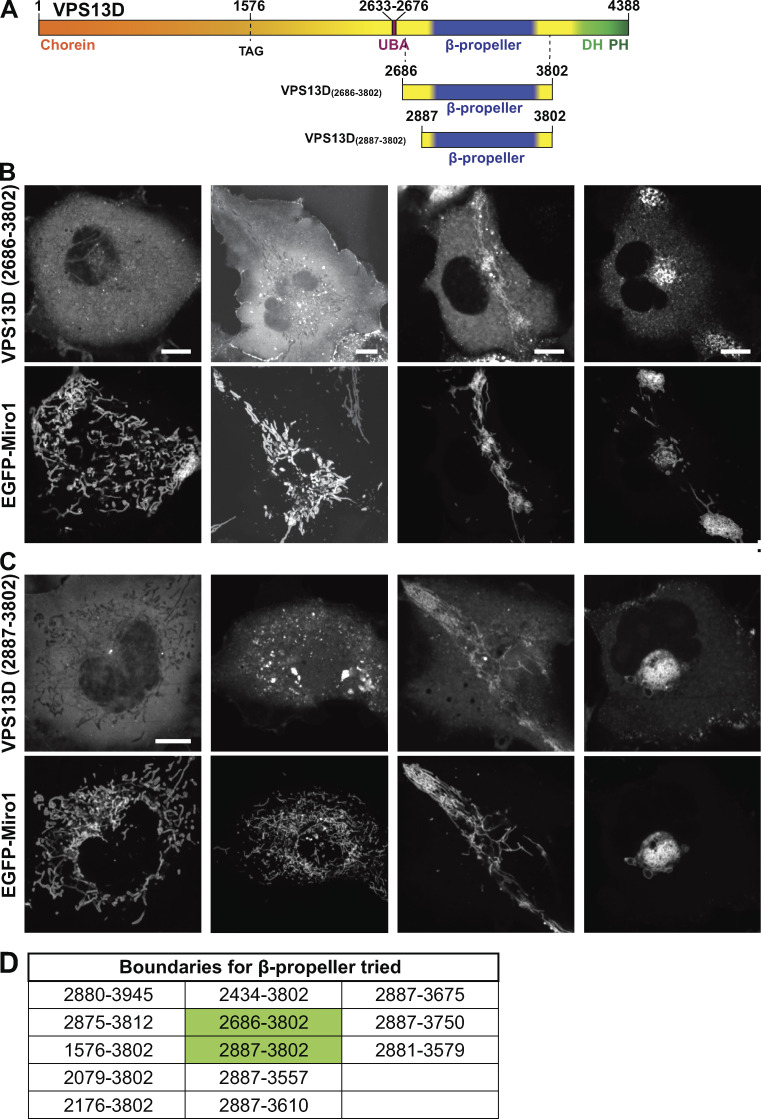
To determine the region of VPS13D required for Miro-dependent recruitment, we coexpressed in COS7 cells Miro with several VPS13D fragments. Removal of the C-terminal region of VPS13D, including both the VAB/β-propeller domain and DH-PH domains, abolished its Miro-dependent recruitment to mitochondria (Fig. 4 C). In contrast, VPS13D constructs lacking the DH-PH domain only were still recruited by Miro, whereas constructs containing the DH-PH domain but not the VAB/β-propeller domain were not (Fig. 4 C), suggesting that presence of the VAB/β-propeller domain was necessary for this recruitment. Efforts to determine whether the VAB/β-propeller domain was sufficient for the recruitment were inconclusive because each of several constructs of this domain that we generated produced heterogeneous subcellular localizations ranging from cytosolic distribution, to small aggregates, to a colocalization with mitochondria (with roughly equal proportions of the three localization patterns). However, in the latter case, the morphology and intracellular distribution of mitochondria was disrupted, often resulting in their massive clustering, as shown by the EGFP-Miro1 signal. These results raise the possibility that constructs of the VAB/β-propeller region alone may not fold correctly (Fig. S4). While all of our data are compatible with a direct interaction between VPS13D and Miro, we cannot rule out an indirect interaction.
Figure S4.
Localization of the constructs comprising the β-propeller region of VPS13D only. (A) Cartoon depicting full-length VPS13D and the VPS13D fragments used in the imaging experiment shown in B and C. (B and C) Confocal images of representative examples of the heterogeneous localization of two mCherry-tagged VPS13D fragments in COS7 cells coexpressing EGFP-Miro1. The first column shows a cytosolic localization; the second column shows the presence of small aggregates; and the third and fourth columns show the colocalization of VPS13D with Miro on mitochondria whose localization is disrupted. Scale bars, 10 µm. (D) List of VPS13D fragments containing the β-propeller region that were tested for Miro-induced recruitment to mitochondria. All of them behaved similarly. The constructs used for fields in B and C are highlighted in green.
VPS13A, a paralogue of VPS13D, also binds mitochondria, but it does so via its C-terminal DH-PH domain whose interactor(s) at the mitochondrial surface remain(s) unknown (Kumar et al., 2018). Accordingly, cotransfection of VPS13A^Halo with EGFP-Miro1 did not result in an increased recruitment of VPS13A^Halo to mitochondria (Fig. S1, C–E). Moreover, when VPS13A^Halo and VPS13D^EGFP were cotransfected in COS7 cells (Fig. S1 E) also overexpressing Miro1, they were both concentrated at mitochondria, but with a different pattern: VPS13A had a punctate distribution previously shown to reflect its restricted localization at ER–mitochondria contact sites (Kumar et al., 2018), while VPS13D decorated the entire mitochondrial surface. This indicates a different mechanism of mitochondrial binding of VPS13A and VPS13D.
VPS13D binds the ER protein VAP via an unconventional FFAT motif
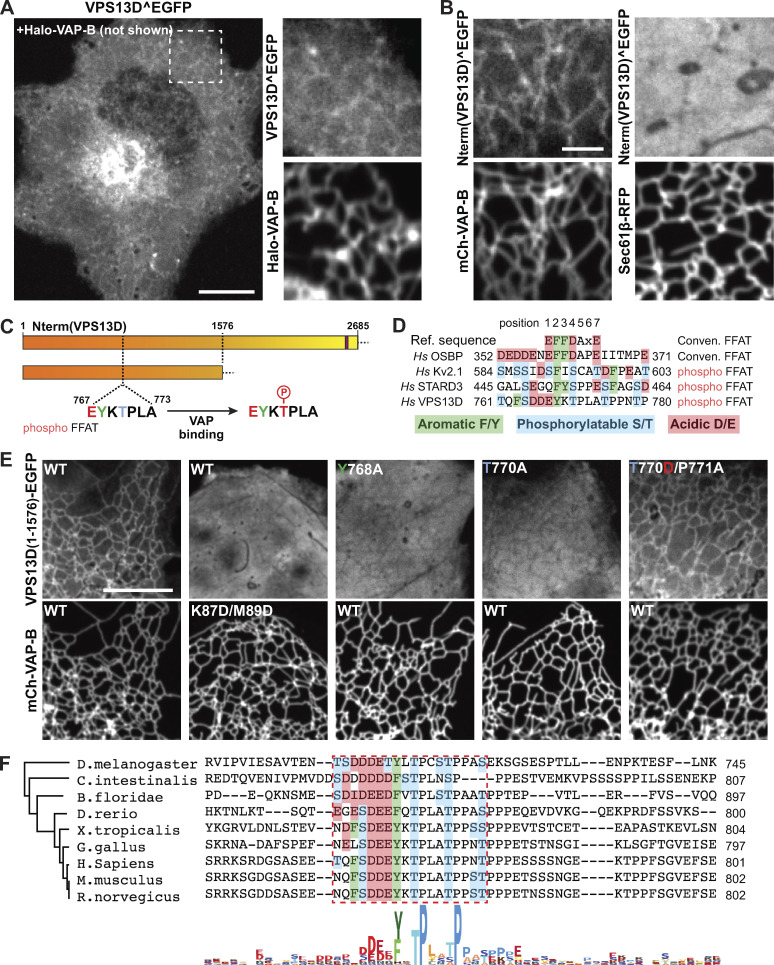
Studies of yeast Vps13 and of mammalian VPS13A and VPS13C have shown that a shared property of these proteins is to bridge organelles to provide a conduit for the flow of lipids between adjacent bilayers. Both VPS13A and VPS13C tether other organelles to the ER via an interaction of their FFAT motifs with the MSP domain of the ER proteins VAP-A and VAP-B (Kumar et al., 2018). While the homogeneous localization of VPS13D on the mitochondrial surface does not support a selective localization of this protein at contact sites with the ER, we considered that an interaction of VPS13D with the ER, and possibly with VAP, may be of low affinity and/or regulated. To explore this possibility, we examined the localization of VPS13D^EGFP in COS7 cells when coexpressed with Halo–VAP-B but without Miro. While no obvious fluorescent signal on the ER was observed when VPS13D^EGFP was expressed alone (Fig. 1 B), a weak ER localization of VPS13D^EGFP was observed in cells also expressing Halo–VAP-B (Fig. 5 A). Most likely, endogenous VAP expression levels are not sufficient to bind a detectable amount of VPS13D and/or the diffuse cytosolic fluorescence of VPS13D^EGFP obscures such localization. Overexpression of VAP-B, but not of a VAP-B construct harboring two mutations (K87D/M89D) that impair FFAT motif binding (Dong et al., 2016), also induced the recruitment to the ER of the VPS13D N-terminal region alone, further validating VAP-dependent binding and indicating that the ER binding site, as in the case of VPS13A and VPS13C, lies somewhere in this region (Fig. 5, B and E).
Figure 5.
VPS13D binds VAP on the ER via a phospho-FFAT in its N-terminal region. (A) Left: Confocal image of a COS7 cell coexpressing Halo–VAP-B and VPS13D^EGFP showing weak recruitment of VPS13D^EGFP to the ER. Scale bar, 10 µm. Right: High-magnification view of Halo–VAP-B and VPS13D^EGFP fluorescence of the field enclosed by a rectangle. (B) VAP-dependent binding of the N-terminal portion of VPS13D to the ER. Left: Confocal images of a COS7 cell coexpressing the N-terminal portion of VPS13D fused to EGFP and mCherry-VAP-B. Right: Confocal images of a COS7 cell coexpressing the N-terminal portion of VPS13D fused to EGFP and Sec61β-RFP, but not VAP. Scale bar, 3 µm. (C) Cartoons showing VPS13D constructs used for the experiments shown in E, indicating the position of the predicted phospho-FFAT motif. (D) Comparison of the conventional FFAT motif with phospho-FFAT motifs, including the one found in VPS13D (Di Mattia et al., 2020). The aromatic residue indicated in green, present in both conventional and phospho-FFAT motifs, is essential for binding to VAP. Acidic amino acid residues in the region contribute to the binding in conventional FFAT motifs, but based on a previous study (Di Mattia et al., 2020), they can be replaced by phosphorylatable residues in phospho-FFAT motifs. (E) Evidence for a phospho-FFAT motif-dependent binding of the N-terminal region of VPS13D to VAP. First column: The N-terminal construct (1–1576) of VPS13D is recruited to the ER upon VAP overexpression. Second column: Two mutations in the MSP domain of VAP that disrupt the FFAT motif binding pocket also disrupt the recruitment of the VPS13D construct. Third column: Mutation to alanine of the aromatic residue of the phospho-FFAT motif disrupts binding. Fourth and fifth columns: No binding occurs when the threonine that corresponds to an aspartate in the conventional FFAT motif is replaced by a nonphosphorylatable alanine, but binding is restored when the threonine is replaced by aspartate, as long as the adjacent proline is also mutated (Di Mattia et al., 2020). Scale bar, 10 µm. (F) Alignment of the region of VPS13D orthologues from different species centered on the amino acid region required for Miro binding in human VPS13D. The alignment shows a high degree of conservation of the key residues of the phospho-FFAT motif among several chordates and also observed in flies. The phylogenetic tree was generated by maximum likelihood.
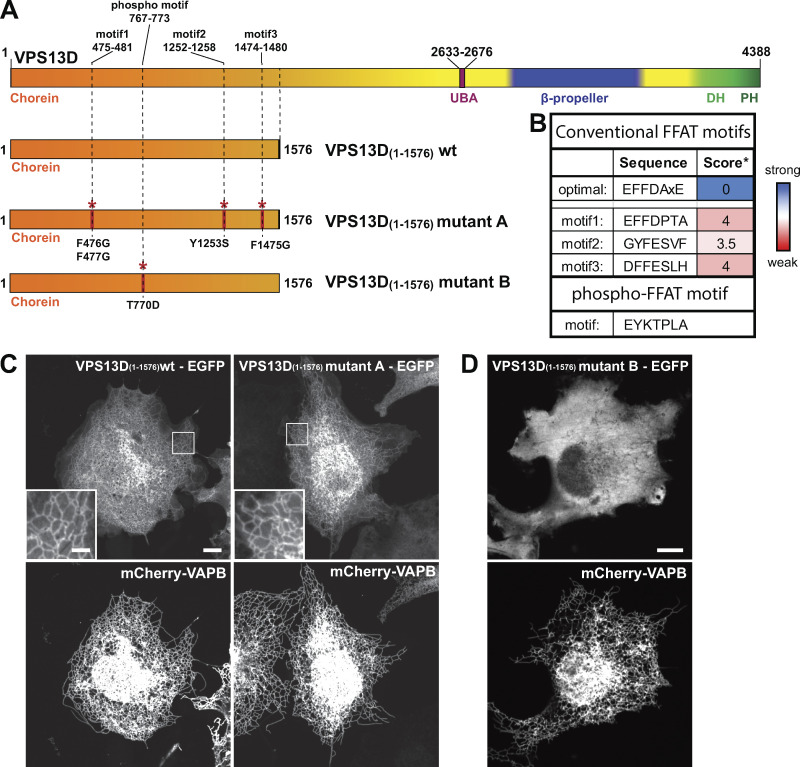
Three conventional FFAT motifs are predicted with low score (Murphy and Levine, 2016; Slee and Levine, 2019) in the N-terminal region of VPS13D, but, surprisingly, the combined mutation of all three sites did not abolish VAP-dependent recruitment to the ER (Fig. S5). Recent studies of VAP binding proteins revealed the occurrence of unconventional VAP binding motifs that bind the FFAT binding pocket of VAP in spite of several differences, including the proposed dependence on phosphorylation rather than on acidic amino acids (hence referred to as phospho-FFAT motifs; Johnson et al., 2018; Kirmiz et al., 2018; Di Mattia et al., 2020). As a phospho-FFAT motif was predicted in the N-terminal region of VPS13D (aa 767–773; Fig. 5, C and D; Di Mattia et al., 2020), we tested its importance. Mutation to alanine of tyrosine (Y768A) at position 2 of the core motif (i.e., a position where there is an absolute requirement for an aromatic amino acid in all FFAT motifs) abolished the recruitment of the N-terminal region of VPS13D to the ER in VAP-B–overexpressing cells (Fig. 5 E, third column). Furthermore, mutation to alanine of the threonine (T770A) of the motif, which was suggested as a phosphorylatable site (Di Mattia et al., 2020), also disrupted the interaction with VAP (Fig. 5 E, fourth column). Conversely, a phosphomimetic mutation of this threonine to aspartate, when combined with the mutation of the adjacent proline to alanine (T770D/P771A), rescued its ability to bind VAP (Fig. 5 E, fifth column). The proline residue immediately downstream of the phosphosite is a shared characteristic among other phospho-FFAT motifs, and mutation of this proline to alanine was shown to be necessary to allow the phosphomimetic construct to bind VAP (Di Mattia et al., 2020). It was suggested by Di Mattia et al. (2020) that the proline imposes a conformational rigidity that “would prevent the aspartate residue from properly mimicking a phosphorylated serine, as it possesses a shorter side chain.” Supporting this idea, the single mutation of the threonine to an aspartate (T770D) was not enough to allow binding of the VPS13D construct to VAP (Fig. S5 D). Alignment of VPS13D protein sequences from different animal species revealed a high conservation of this phospho-FFAT motif, pointing to its physiological importance (Fig. 5 F).
Figure S5.
Mutations in the phospho-FFAT motif of VPS13D but not in conventional FFAT motifs disrupt VAP binding. (A) Cartoon depicting full-length VPS13D and the constructs used for C and D, displaying the localization of the predicted three conventional FFAT motifs and the single phospho-FFAT motif and the mutations that were introduced to disrupt them. (B) Sequence of each of the predicted conventional FFAT and phospho-FFAT motifs. *, Conventional FFAT motif score was calculated using a previously described algorithm; scores 3.5 and 4 are considered weak FFAT motifs (Slee and Levine, 2019). (C) Left: Coexpression of an EGFP-tagged N-terminal fragment of VPS13D (amino acid 1–1576) with the ER protein VAP-B shows robust recruitment of the fragment to the ER. Right: The combined disruption of the three best predicted conventional FFAT motifs as indicated in A did not affect ER recruitment of this VPS13D fragment by VAP-B. (D) The point mutation T770D alone (without the additional mutation of the adjacent proline to alanine; see Fig. 5 E and Di Mattia et al., 2020) abolishes the VAP-dependent recruitment of the N-terminal fragment of VPS13D to the ER. Scale bars, 10 µm.
Binding of VPS13D to VAP and to Miro allows it to function as a bridge between the ER and mitochondria
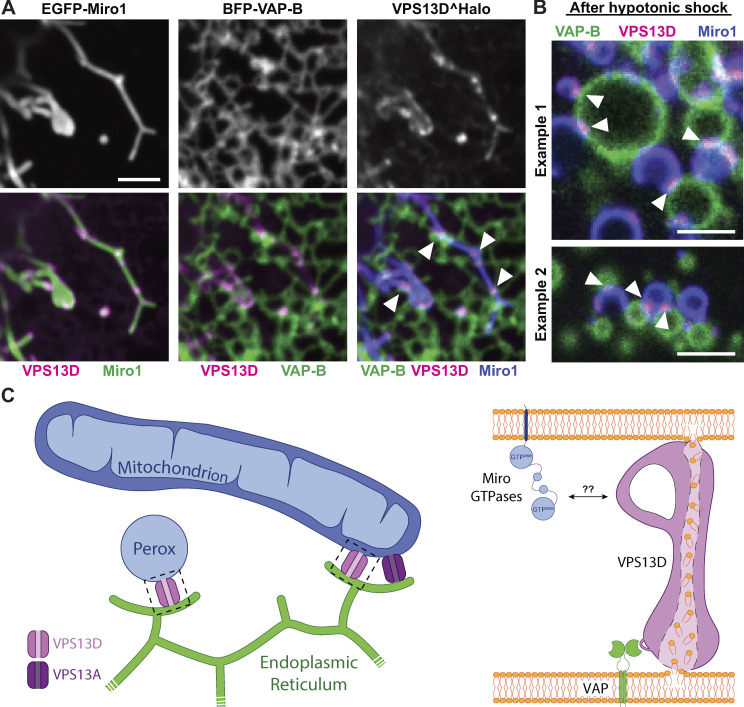
Prompted by the property of the N-terminal region of VPS13D to bind VAP, we examined whether VPS13D can bridge ER to mitochondria. When VPS13D^Halo was coexpressed with both BFP–VAP-B and EGFP-Miro1, the signal of VPS13D^Halo no longer decorated the entire mitochondrial surface homogeneously but was primarily concentrated at hot spots. Such hot spots precisely colocalized with sites of apposition between the mitochondria and the ER, consistent with a localization of VPS13D at contacts between the two organelles (Fig. 6 A). This conclusion was supported by observing VPS13D dynamics during exposure of cells to strong hypotonic solutions (Fig. 6 B and Video 2). Under these conditions, dramatic shape changes and vesiculation of organelles occurs, but tethers between organelles persist (King et al., 2020). In cells expressing all three proteins, VPS13D^Halo fluorescence coalesced at sites where mitochondria- and ER-derived vesicles remained in contact with each other, as expected for a protein that can bridge the two organelles (Fig. 6 B and Video 2).
Figure 6.
VPS13D can tether the ER to mitochondria in a VAP- and Miro-dependent way. (A) COS7 cells coexpressing EGFP-Miro1, BFP–VAP-B, and VPS13D^Halo. Top: Single fluorescence images. Bottom: Merge of the different fluorescence channels as indicated. White arrowheads in the triple merge show that VPS13D^Halo (magenta) is enriched at hot spots where the ER (green) and mitochondria (blue) intersect. (B) Vesiculated ER and mitochondria induced by hypotonic treatment of COS7 cells coexpressing VPS13D^Halo, GFP-Miro1, and BFP–VAP-B remain tethered to each other, and VPS13D^Halo concentrates at these sites (white arrowheads). See also Video 2. Scale bars, 3 µm. (C) Schematic cartoon summarizing key findings of this study. Left: VPS13D can bridge the ER and either mitochondria or peroxisomes. Right: The interaction of VPS13D with the ER is mediated by VAP, and its interaction with either mitochondria or peroxisomes is mediated by Miro. Based on the reported properties of VPS13 family proteins, it is proposed that VPS13D allows flux of lipids between the two tethered bilayers.
Video 2.
Confocal time-lapse imaging of a COS7 cell coexpressing VPS13D^Halo, EGFP-Miro1, and BFP–VAP-B upon hypotonic shock. Mitochondria and ER vesiculate but remain tethered by hot spots of VPS13D. Cells were imaged right after water addition at a rate of 0.5 frames/s for a total of 2 min. Some frames were lost due to focal plane changes. Scale bar, 3 µm.
Discussion
Our findings provide insight into the properties of VPS13D, a VPS13 family member previously shown to have an impact on mitochondrial biology based on loss-of-function studies, but whose site of action remained elusive. We show that VPS13D can bridge mitochondria to the ER and identify proteins on the two organelles responsible for this localization: the protein VAP in the ER membrane and the protein Miro in the OMM (Fig. 6 C). Splice variants of Miro are targeted to peroxisomes (Okumoto et al., 2018; Covill-Cooke et al., 2020), and, accordingly, we have shown that VPS13D can also be recruited to peroxisomes. A membrane-bridging property of VPS13D is consistent with the recently discovered core function of VPS13 proteins: mediating lipid fluxes between bilayers (Kumar et al., 2018; Li et al., 2020). Both mitochondria and peroxisomes, whose functions are closely interrelated, critically depend on nonvesicular lipid transport from the ER for their growth and function (Vance, 1990; Raychaudhuri and Prinz, 2008; Kornmann et al., 2009; Petrungaro and Kornmann, 2019).
An association of VPS13D with mitochondria is also supported by recent proximity labeling–based studies listing this protein among the hits retrieved with outer membrane protein baits (Hung et al., 2017; Liu et al., 2018; Antonicka et al., 2020). Interestingly, these studies also retrieved VPS13A, which we had shown previously to act as a bridge between ER and mitochondria (Kumar et al., 2018), but did not retrieve VPS13B or VPS13C, for which there is no evidence for such a localization (Seifert et al., 2015; Kumar et al., 2018; Ugur et al., 2020). Based on these results, VPS13D may have a partially overlapping role with VPS13A in allowing lipid fluxes between the ER and mitochondria. However, VPS13D likely has a more fundamental function, as it is required for life (Wang et al., 2015; Blomen et al., 2015; Seong et al., 2018), while VPS13A is not. Clearly, the organelle-bridging roles of VPS13A and VPS13D are differentially regulated, as the recruitment of VPS13A to mitochondria is not mediated by Miro (the mechanism of its binding to mitochondria remains unknown) and the FFAT motifs through which the two proteins bind VAP have different properties.
While VPS13A has a conventional FFAT motif (Murphy and Levine, 2016; Kumar et al., 2018), the VAP binding site of VPS13D fits the consensus of the recently defined phospho-FFAT motif (Di Mattia et al., 2020), so called because acidic amino acids of the motif can be replaced by phosphorylatable serines or threonines. In the phospho-FFAT motif of VPS13D, position 2 of the core sequence is occupied by a tyrosine (Y768), which we have shown to be essential for VAP binding as predicted, and position 4 is occupied by a threonine (T770) instead of an acidic amino acid as in conventional FFAT motifs. We have shown that replacement of this threonine with the nonphosphorylatable alanine abolishes binding, while its replacement with the phosphomimetic aspartate (along with the replacement of the neighboring proline with an alanine; see Di Mattia et al., 2020) rescues binding. Additional serines/threonines are present in the proximity of the core phospho-FFAT motif of VPS13D, which may contribute to binding affinity via phosphorylation, as has been shown to happen in other phospho-FFAT motifs (Di Mattia et al., 2020).
The different regulation of the bridging functions of VPS13A and VPS13D is further revealed by the observation that, at least in the cells used in this study, detectable ER binding of VPS13D, but not of VPS13A, requires VAP overexpression, suggesting a lower affinity, likely regulated by phosphorylation. As a result of this difference, in cells that do not overexpress VAP, overexpressed VPS13D localizes along the entire mitochondrial surface, while overexpressed VPS13A accumulates at hot spots, which represent sites of ER–mitochondria contacts (Fig. S1; Kumar et al., 2018).
The recruitment of VPS13D to peroxisomes via a splicing variant of Miro1 points to VPS13D as a critical player in delivering lipids to these organelles from the ER and is consistent with previous data indicating the importance of peroxisome–ER contacts for the expansion of their membranes in fungi (Raychaudhuri and Prinz, 2008; Akşit and van der Klei, 2018; Islinger et al., 2018) and more specifically of Vps13 in Hansenula polymorpha (Akşit, 2018). A new study, also published in this issue, reports a critical role for human VPS13D in peroxisome biogenesis, as absence of VPS13D was associated with partial or complete loss of peroxisomes in human cell lines and in fibroblasts from patients carrying VPS13D mutations (Baldwin et al., 2021). Roles of VPS13D at other cellular sites should also be considered in view of the enrichment of VPS13D^EGFP in the Golgi complex area, a localization that we have not further investigated in this study. Interestingly, another VPS13 paralogue, VPS13B, is selectively enriched in the Golgi complex area (Seifert et al., 2011; 2015), once again suggesting differences, but also partial overlap in the role of distinct VPS13 paralogues.
A partnership between VPS13D and Miro fits with the critical role of both proteins for life (López-Doménech et al., 2018; Seong et al., 2018). Moreover, in cellular models, loss-of-function perturbations of either Miro or VPS13D include similar alterations in mitochondrial size, distribution, and degradation (Nguyen et al., 2014; Anding et al., 2018; Seong et al., 2018; López-Doménech et al., 2018; Insolera et al., 2020 Preprint). The accumulation of round mitochondria is a particularly striking feature of VPS13D- and Miro-deficient cells. Among the many potential scenarios to be considered, one is that the altered mitochondrial morphology in cells lacking VPS13D may result from reduced membrane lipid availability for membrane expansion with an increase of volume-to-surface ratio. Swelling of mitochondria, in turn, may impair the action of the fission machinery. Interestingly, it was reported that, in mammalian cells, expansion of the so-called “mitochondrial derived compartment,” a newly described membranous organelle derived from the OMM, requires Miro and contacts between mitochondria and the ER, consistent with a role of Miro in a process that may involve lipid flux from the ER (English et al., 2020; Schuler et al., 2020 Preprint).
Collectively, our study advances our understanding of pathways of nonvesicular lipid traffic within cells. It adds new evidence for the fundamental importance in cell physiology of proteins of the VPS13/ATG2 superfamily (i.e., proteins thought to act in the bulk delivery of lipids from one bilayer to another to mediate bilayer expansion; Ugur et al., 2020; Lees and Reinisch, 2020). It also reveals an evolutionary link, missing so far, between the function of the Gem1/Miro family in fungi, where Gem1 is an accessory subunit of ERMES (Kornmann et al., 2011), a major lipid transfer complex, and in mammals, where Miro had been primarily implicated in the control of mitochondrial dynamics and/or transport via molecular motors (Fransson et al., 2003; Saotome et al., 2008; Wang and Schwarz, 2009; Nguyen et al., 2014; López-Doménech et al., 2018). While in yeast the single Vps13 can compensate for the lack of the ERMES complex, no link between Gem1 and Vps13 has been observed so far in this organism. But ERMES is not present in metazoa, possibly explaining why the role of Gem1 as a partner of ERMES in lipid transport has evolved to a role of its orthologue Miro as a partner of VPS13D in mammalian cells and possibly other high eukaryotes. It will be of interest in the future to determine how the distinct functions of Miro in the control of mitochondria mobility and lipid transport are coordinated.
Finally, as Miro is a substrate for Parkin (Wang et al., 2011; Birsa et al., 2014; Shlevkov et al., 2016) and is itself a target of Parkinson’s disease–associated mutations (Berenguer-Escuder et al., 2019), our results may also be relevant to pathogenetic mechanisms of Parkinson’s disease.
Materials and methods
DNA plasmids
A plasmid containing codon-optimized cDNA encoding human VPS13D, also including an mScarlet fluorescent protein after amino acid residue 1576 flanked by BamHI restriction enzyme sites, was generated by and purchased from GenScript Biotech. This plasmid was linearized with BamHI and used to clone VPS13D^EGFP and VPS13D^Halo by In-Fusion Cloning (Takara Bio). The general protocol used for Infusion cloning (Takara) was deposited in protocols.io: https://dx.doi.org/10.17504/protocols.io.bvgwn3xe. An N-terminal fragment of VPS13D, Nterm(VPS13D)^EGFP, and a deletion mutant of VPS13D lacking the β-propeller region (VAB/WD40-like domain), VPS13D(Δβ-prop)^EGFP, were generated by PCR amplification and ligated into pEGFP-C1 by In-Fusion Cloning using the EcoRI and AgeI sites. A C-terminally truncated VPS13D, VPS13D(ΔDHPH)^EGFP, was generated using site-directed mutagenesis (QuikChange II XL; Agilent Technologies) by adding an early stop codon replacing the codon encoding amino acid residue 3802. VPS13D constructs including the β-propeller region were generated by PCR amplification of the sequences encoding the different portions indicated in Fig. S4 D and ligation into a pmCherry-C1 backbone by In-Fusion Cloning using EcoRI and KpnI sites. Plasmids containing human Miro1 (variant 1) and human Miro2 were a gift from P. Aspenström (Uppsala University, Uppsala, Sweden; Addgene; 47888 and 47891). EGFP-Miro1, EGFP-Miro2, and mCherry-Miro1 were generated by PCR amplification of the coding sequence of Miro and ligated into pEGFP-C1 (Addgene) or pmCherry-C1 (Addgene) using EcoRI and XhoI sites. Halo-Miro1 was generated by PCR amplification of the coding sequence of Miro from EGFP-Miro1 and the coding sequence of the Halo protein from pHalo-C1 and ligated by In-Fusion Cloning into a pcDNA3.1 backbone. For EGFP-Miro1v4, a double-stranded DNA fragment encoding the amino acid residues from exons 8 and 9 of Miro1 (i.e., the exons missing from Miro variant 1) was ordered from Integrated DNA Technologies (IDT) and ligated into EGFP-Miro1 at residue 580 by In-Fusion Cloning. For the optogenetic experiments, the cytosolic domain of Miro1 (Miro1(ΔTM)) and the iLID binding peptide (SspB, obtained from Addgene; 60415, gift from B. Kuhlman, University of North Carolina, Chapel Hill, NC) were fused into pmCherry-C1 using EcoRI and KpnI, by In-Fusion Cloning (mCh-Miro1(ΔTM)-SSPB). EGFP-Miro1 constructs containing mutations T18N, E208/E328K and S432N as well as VPS13D N-term fragments containing mutations of conventional FFAT motifs (F476/F477G, Y1253S, F1475G) or of the phospho-FFAT motif (Y768A, T770A, T770D/P771A) were generated using site-directed mutagenesis (QuikChange II XL; Agilent Technologies) of EGFP-Miro1. To generate BFP–VAP-B, the VAP-B coding sequence was amplified by PCR from an mCherry–VAP-B plasmid (our laboratory), and the BFP coding sequence was amplified from mitoBFP (Addgene; 49151), and they were assembled by HiFi reaction (New England Biolabs). HALO–VAP-B was obtained by PCR amplification of a VAP-B coding sequence from mCherry-VAP-B and pHALO-C1 using EcoRI and KpnI sites. Sialyltransferase (ST)-Halo was generated by PCR amplification of the ST coding sequence from ST-mRFP (our laboratory) and ligated into pHALO-N1 using EcoRI and KpnI sites. EGFP-Parkin was obtained by PCR amplification of rat Parkin cDNA (kind gift from E. Fon, McGill University, Montreal, QC, Canada) and ligated into pEGFP-C1 using EcoRI and XhoI. Other plasmids used in this study were kind gifts: mito-BFP from G. Voeltz (University of Colorado Boulder, Boulder, CO; Addgene; 49151), Venus-iLID-Mito from B. Kuhlman (Addgene; 60413), and mScarlet-SRL from D. Gadella (University of Amsterdam, Amsterdam, Netherlands; Addgene; 85063). Mutant mCherry-VAP-B (K87D, M89D) was previously generated in our laboratory (Dong et al., 2016).
Antibodies and reagents
Primary antibodies used were as follows: mouse anti-RHOT1 (Miro1, H00055288-M01; Abnova), rabbit anti-Miro2 (ab224089; Abcam), rabbit anti-TOMM40 (18409-1-AP; Proteintech), rabbit anti-VPS13D (ab202285; Abcam), mouse anti-GAPDH (40-1246; Proteus Biosciences Inc.), rabbit anti-GFP (ab290; Abcam), mouse anti-myc (sc-40; Santa Cruz Biotechnology), rabbit anti-RFP (600-401-379; Rockland Inc.). Secondary antibodies used were goat anti-mouse IgG (926-32210; LI-COR Biosciences) and goat anti-rabbit IgG (926-68021; LI-COR Biosciences).
Halo tag ligands were a kind gift from L. Lavis (Janelia Research Campus, Ashburn, VA). Valinomycin was purchased from Sigma-Aldrich and used at 10 µM concentration. RNAis for Miro1 were purchased from Ambion (4390824), and the ones for Miro2 were purchased from IDT (hs.Ri.RHOT2.13). Specific primers were purchased from IDT; for sequences, refer to Table S1.
Cell culture and transfection
COS7 and HeLa cells (American Type Culture Collection) were cultured at 37°C and 5% CO2 in DMEM containing 10% FBS, 1 mM sodium pyruvate, 100 U/ml penicillin, 100 mg/ml streptomycin, and 2 mM L-glutamine (all from Gibco). The general protocol for maintenance and preparation of cells for imaging was deposited in protocols.io: https://dx.doi.org/10.17504/protocols.io.bvgmn3u6. For imaging experiments, cells were seeded on glass-bottomed dishes (MatTek) at a concentration of 75 × 103 cells per dish and transiently transfected after 6 h using FuGene HD (Promega). For Miro1- and Miro2-knockdown experiments, 12 h following the transient transfection with VPS13D and mitoBFP, HeLa and COS7 cells were treated RNAi (30 pmol/gene) using Lipofectamine RNAiMAX (Life Technologies) and imaged 36 h afterward. For experiments where the recruitment of Parkin to mitochondria was triggered, 10 µM final valinomycin (dissolved in DMSO) was added.
Neuronal cultures
Experiments were performed in accordance with the Yale University Institutional Animal Care and Use Committee. Briefly, hippocampi were dissected from P0 mouse brains, and neurons were dissociated by papain treatment and seeded in serum-based medium on poly-D-lysine–coated, glass-bottomed dishes (MatTek), as previously described (Sun et al., 2019). After 6 h, the serum-based medium was removed and replaced with neuronal growth media (Neurobasal A supplemented with B-27 and GlutaMAX; all from Gibco). After 11 d in vitro, cells were transfected using Lipofectamine 2000 (Life Technologies). The detailed protocol for preparation of hippocampal cultures was deposited in protocols.io: https://dx.doi.org/10.17504/protocols.io.bvgkn3uw.
Microscopy
Live-cell imaging
Just before imaging, the growth medium was removed and replaced with live-cell imaging solution (Life Technologies). All live-cell imaging was performed at 37°C and 5% CO2. Spinning-disk confocal microscopy was performed using an Andor Dragonfly system equipped with a plan apochromat objective (63×, 1.4 NA, oil) and a Zyla scientific CMOS camera. For the hypotonic lysis experiments, live-cell imaging solution was replaced with distilled water, and cells were imaged at a rate of 0.5 Hz.
For experiments involving cytosolic Ca2+ changes, COS7 cells were seeded and transfected as explained above. To acutely increase cytosolic Ca2+, 2 µM thapsigargin (Life Technologies) was added to the live-cell imaging solution (Life Technologies), which contains 1.8 mM Ca2+, and cytosolic Ca2+ was monitored by the intensity of the RFP genetically encoded Ca2+ indicator for optical imaging (plasmid was a gift from R. Campbell, University of Alberta, Edmonton, AB, Canada; Addgene catalog no. 45494). To decrease cytosolic Ca2+, 4 mM EGTA and 10 µM BAPTA-AM (Thermo Fisher Scientific) were added to the medium during imaging. Calcium experiments were repeated twice in at least four cells for each case. The general protocol for the imaging experiments described was deposited in protocols.io: https://dx.doi.org/10.17504/protocols.io.bvgmn3u6.
Optogenetic experiments
For whole-cell activation experiments, an Andor Dragonfly system (see above) was used, and recruitment to mitochondria was achieved with a single 200-ms pulse of the 488-nm laser. For localized activation to promote recruitment to a single mitochondrion, an Improvision UltraVIEW VoX system (PerkinElmer), built around a Nikon Ti-E inverted microscope and controlled by Volocity software (Quorum Technologies), was used. Imaging was performed at 37°C with a 63× plan apochromat oil objective (1.45 NA). A built-in photoperturbation unit was used to deliver 488-nm light pulses in a 5-µm2 area. The protocol for the optogenetic experiments was deposited in protocols.io: https://dx.doi.org/10.17504/protocols.io.bvgvn3w6.
Image processing, analysis, and statistics
Fluorescence images were processed using Fiji software (ImageJ; National Institutes of Health). Gaussian Blur filters were applied on some of the images presented. The fluorescent signals in Videos 1 and 2 were corrected for photobleaching using the Bleach Correction function in Fiji.
For the quantification of the optogenetic experiment, kymographs were built by tracing a line across the mitochondria in the unprocessed images, resulting in images such as the examples shown in Fig. 1 G, from which the intensity profile was measured. The fluorescence intensities were normalized to the minimum and maximum values using the following formula:
where
For the analysis of VPS13D mitochondrial recruitment, cells coexpressing VPS13D^EGFP and mitoBFP were imaged and analyzed individually, using an automated macro designed in Fiji. Gaussian Blur and Enhance Local Contrast were applied to the mitoBFP channel to generate an accurate mask of mitochondria. The total amount of VPS13D signal on mitochondria was then obtained by measuring the VPS13D^EGFP intensity within that mask. To obtain a mask covering the cytosolic area surrounding mitochondria, the Dilate function was applied to the mitochondrial mask; specifically, a two-times dilated mitochondrial mask was subtracted from a three-times dilated one. The resulting mask, a 1-pixel-wide mask covering the area lining the profile of mitochondria, was then used to measure VPS13D^EGFP intensity in the cytosol surrounding mitochondria. This second value was used to normalize for expression levels. The ratio between the intensity of VPS13D on mitochondria and the intensity of VPS13D in the cytosol surrounding them was plotted on a graph as the value of VPS13D enrichment at mitochondria. For statistical analysis, GraphPad Prism 8 software was used. The Brown-Forsythe test and Welch’s ANOVA were used to assess significant differences among experimental groups, followed by the Games-Howell multiple comparisons test. The protocol describing the image analysis, as well as the FIJI algorithm to perform the analysis, were deposited in protocols.io: https://dx.doi.org/10.17504/protocols.io.bvgnn3ve.
Online supplemental material
Fig. S1 shows that VPS13D and VPS13A can bind to mitochondria but that only the interaction of VPS13D is mediated by Miro. Fig. S2 shows that triggering of Parkin-mediated mitophagy leads to degradation of Miro and shedding of VPS13D from mitochondria. Fig. S3 shows that recruitment of VPS13D by Miro is unaffected by changes in cytosolic Ca2+. Fig. S4 shows the localization of truncated constructs of VPS13D containing the β-propeller region.Fig. S5 shows that low-score conventional FFAT motifs in the N-terminal region of VPS13D are not involved in its binding to VAP. Video 1 shows acute recruitment of VPS13D to mitochondria upon optogenetic recruitment of Miro. Video 2 shows that VPS13D coalesces at sites of ER–mitochondria tethering upon organelle swelling in response to hypotonic shock. Table S1 shows a list of oligonucleotides used in this study.
Supplementary Material
lists oligonucleotides used in this study.
Acknowledgments
We thank Y. Wu, M. Hammarlund, H. Falahati, and J.H. Park for discussion. We also thank J.Liu, F. Wilson, and A. Dao for outstanding technical assistance.
This work was supported in part by National Institutes of Health grants NS36251 and DA018343, the Kavli Foundation, the Parkinson’s Foundation (PF-RCE-1946 to P. De Camilli), and by the Chan Zuckerberg Initiative Donor-Advised Fund, an advised fund of the Silicon Valley Community Foundation (grant 2020-221912) to P. De Camilli and H. Shen. A. Guillén-Samander was supported in part by a Jung-Stiftung für Wissenschaft und Forschung scholarship. The study was also funded by the joint efforts of the Michael J. Fox Foundation for Parkinson's Research (MJFF) and the Aligning Science Across Parkinson's (ASAP) initiative. MJFF administers the grant ASAP-000580 on behalf of ASAP and itself. For the purpose of open access, the author has applied a CC-BY public copyright license to the Author Accepted Manuscript (AAM) version arising from this submission.
P. De Camilli serves on the scientific advisory board of Casma Therapeutics. The authors declare no further competing financial interests.
Author contributions: A. Guillén-Samander, M. Leonzino, and P. De Camilli conceptualized the project. A. Guillén-Samander, M. Leonzino, and M.G. Hanna, IV designed and performed experiments. A. Guillén-Samander, M. Leonzino, N. Tang, H. Shen, and P. De Camilli analysed data. A. Guillén-Samander, M. Leonzino, and P. De Camilli wrote the original draft of the manuscript, which was then reviewed and edited by all the authors.
References
- Akşit, A. 2018. Peroxisomal Membrane Contact Sites in the Yeast Hansenula Polymorpha. PhD Thesis. University of Groningen, Groningen, Netherlands. Available at: https://research.rug.nl/en/publications/peroxisomal-membrane-contact-sites-in-the-yeast-hansenula-polymor. [Google Scholar]
- Akşit, A., and van der Klei I.J.. 2018. Yeast peroxisomes: How are they formed and how do they grow? Int. J. Biochem. Cell Biol. 105:24–34. 10.1016/j.biocel.2018.09.019 [DOI] [PubMed] [Google Scholar]
- Anding, A.L., Wang C., Chang T.-K., Sliter D.A., Powers C.M., Hofmann K., Youle R.J., and Baehrecke E.H.. 2018. Vps13D encodes a ubiquitin-binding protein that is required for the regulation of mitochondrial size and clearance. Curr. Biol. 28:287–295.e6. 10.1016/j.cub.2017.11.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonicka, H., Lin Z.-Y., Janer A., Aaltonen M.J., Weraarpachai W., Gingras A.-C., and Shoubridge E.A.. 2020. A high-density human mitochondrial proximity interaction network. Cell Metab. 32:479–497.e9. 10.1016/j.cmet.2020.07.017 [DOI] [PubMed] [Google Scholar]
- Baldwin, H., Wang C., Kanfer G., Shah H., Velayos-Baeza A., Dulovic-Mahlow M., Brüggemann N., Anding A., Baehrecke E.H., Maric D., et al. 2021. VPS13D promotes peroxisome biogenesis. J. Cell Biol. 220:e202001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean, B.D.M., Dziurdzik S.K., Kolehmainen K.L., Fowler C.M.S., Kwong W.K., Grad L.I., Davey M., Schluter C., and Conibear E.. 2018. Competitive organelle-specific adaptors recruit Vps13 to membrane contact sites. J. Cell Biol. 217:3593–3607. 10.1083/jcb.201804111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenguer-Escuder, C., Grossmann D., Massart F., Antony P., Burbulla L.F., Glaab E., Imhoff S., Trinh J., Seibler P., Grünewald A., and Krüger R.. 2019. Variants in Miro1 cause alterations of ER-mitochondria contact sites in fibroblasts from Parkinson’s disease patients. J. Clin. Med. 8:2226. 10.3390/jcm8122226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsa, N., Norkett R., Wauer T., Mevissen T.E.T., Wu H.-C., Foltynie T., Bhatia K., Hirst W.D., Komander D., Plun-Favreau H., and Kittler J.T.. 2014. Lysine 27 ubiquitination of the mitochondrial transport protein Miro is dependent on serine 65 of the Parkin ubiquitin ligase. J. Biol. Chem. 289:14569–14582. 10.1074/jbc.M114.563031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen, V.A., Májek P., Jae L.T., Bigenzahn J.W., Nieuwenhuis J., Staring J., Sacco R., van Diemen F.R., Olk N., Stukalov A., et al. 2015. Gene essentiality and synthetic lethality in haploid human cells. Science. 350:1092–1096. 10.1126/science.aac7557 [DOI] [PubMed] [Google Scholar]
- Castro, I.G., Richards D.M., Metz J., Costello J.L., Passmore J.B., Schrader T.A., Gouveia A., Ribeiro D., and Schrader M.. 2018. A role for Mitochondrial Rho GTPase 1 (MIRO1) in motility and membrane dynamics of peroxisomes. Traffic. 19:229–242. 10.1111/tra.12549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covill-Cooke, C., Toncheva V.S., Drew J., Birsa N., López-Doménech G., and Kittler J.T.. 2020. Peroxisomal fission is modulated by the mitochondrial Rho-GTPases, Miro1 and Miro2. EMBO Rep. 21:e49865. 10.15252/embr.201949865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mattia, T., Martinet A., Ikhlef S., McEwen A.G., Nominé Y., Wendling C., Poussin-Courmontagne P., Voilquin L., Eberling P., Ruffenach F., et al. 2020. FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts. EMBO J. 39:e104369. 10.15252/embj.2019104369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, R., Saheki Y., Swarup S., Lucast L., Harper J.W., and De Camilli P.. 2016. Endosome-ER contacts control actin nucleation and retromer function through VAP-dependent regulation of PI4P. Cell. 166:408–423. 10.1016/j.cell.2016.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziurdzik, S.K., Bean B.D.M., Davey M., and Conibear E.. 2020. A VPS13D spastic ataxia mutation disrupts the conserved adaptor-binding site in yeast Vps13. Hum. Mol. Genet. 29:635–648. 10.1093/hmg/ddz318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English, A.M., Schuler M.-H., Xiao T., Kornmann B., Shaw J.M., and Hughes A.L.. 2020. ER-mitochondria contacts promote mitochondrial-derived compartment biogenesis. J. Cell Biol. 219:e202002144. 10.1083/jcb.202002144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson, A., Ruusala A., and Aspenström P.. 2003. Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J. Biol. Chem. 278:6495–6502. 10.1074/jbc.M208609200 [DOI] [PubMed] [Google Scholar]
- Frederick, R.L., McCaffery J.M., Cunningham K.W., Okamoto K., and Shaw J.M.. 2004. Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology via a novel pathway. J. Cell Biol. 167:87–98. 10.1083/jcb.200405100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier, J., Meijer I.A., Lessel D., Mencacci N.E., Krainc D., Hempel M., Tsiakas K., Prokisch H., Rossignol E., Helm M.H., et al. 2018. Recessive mutations in VPS13D cause childhood onset movement disorders. Ann. Neurol. 83:1089–1095. 10.1002/ana.25204 [DOI] [PubMed] [Google Scholar]
- Glater, E.E., Megeath L.J., Stowers R.S., and Schwarz T.L.. 2006. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J. Cell Biol. 173:545–557. 10.1083/jcb.200601067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntas, G., Hallett R.A., Zimmerman S.P., Williams T., Yumerefendi H., Bear J.E., and Kuhlman B.. 2015. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. USA. 112:112–117. 10.1073/pnas.1417910112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, V., Lam S.S., Udeshi N.D., Svinkina T., Guzman G., Mootha V.K., Carr S.A., and Ting A.Y.. 2017. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. eLife. 6:e24463. 10.7554/eLife.24463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insolera, R., Lorincz P., Wishnie A.J., Juhász G., and Collins C.A.. 2020. Mitochondrial fission, integrity and completion of mitophagy require separable functions of Vps13D in Drosophila neurons. bioRxiv. doi:2020.01.21.914523 (Preprint posted December 1, 2020) [DOI] [PMC free article] [PubMed]
- Islinger, M., Voelkl A., Fahimi H.D., and Schrader M.. 2018. The peroxisome: an update on mysteries 2.0. Histochem. Cell Biol. 150:443–471. 10.1007/s00418-018-1722-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John Peter, A.T., Herrmann B., Antunes D., Rapaport D., Dimmer K.S., and Kornmann B.. 2017. Vps13-Mcp1 interact at vacuole-mitochondria interfaces and bypass ER-mitochondria contact sites. J. Cell Biol. 216:3219–3229. 10.1083/jcb.201610055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, B., Leek A.N., Solé L., Maverick E.E., Levine T.P., and Tamkun M.M.. 2018. Kv2 potassium channels form endoplasmic reticulum/plasma membrane junctions via interaction with VAPA and VAPB. Proc. Natl. Acad. Sci. USA. 115:E7331–E7340. 10.1073/pnas.1805757115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.M., Morrell J.C., and Gould S.J.. 2004. PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J. Cell Biol. 164:57–67. 10.1083/jcb.200304111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfer, G., Courthéoux T., Peterka M., Meier S., Soste M., Melnik A., Reis K., Aspenström P., Peter M., Picotti P., and Kornmann B.. 2015. Mitotic redistribution of the mitochondrial network by Miro and Cenp-F. Nat. Commun. 6:8015–8019. 10.1038/ncomms9015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, C., Sengupta P., Seo A.Y., and Lippincott-Schwartz J.. 2020. ER membranes exhibit phase behavior at sites of organelle contact. Proc. Natl. Acad. Sci. USA. 117:7225–7235. 10.1073/pnas.1910854117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmiz, M., Vierra N.C., Palacio S., and Trimmer J.S.. 2018. Identification of VAPA and VAPB as Kv2 channel-interacting proteins defining endoplasmic reticulum-plasma membrane junctions in mammalian brain neurons. J. Neurosci. 38:7562–7584. 10.1523/JNEUROSCI.0893-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolehmainen, J., Black G.C.M., Saarinen A., Chandler K., Clayton-Smith J., Träskelin A.-L., Perveen R., Kivitie-Kallio S., Norio R., Warburg M., et al. 2003. Cohen syndrome is caused by mutations in a novel gene, COH1, encoding a transmembrane protein with a presumed role in vesicle-mediated sorting and intracellular protein transport. Am. J. Hum. Genet. 72:1359–1369. 10.1086/375454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann, B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., and Walter P.. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325:477–481. 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann, B., Osman C., and Walter P.. 2011. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc. Natl. Acad. Sci. USA. 108:14151–14156. 10.1073/pnas.1111314108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, N., Leonzino M., Hancock-Cerutti W., Horenkamp F.A., Li P., Lees J.A., Wheeler H., Reinisch K.M., and De Camilli P.. 2018. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 217:3625–3639. 10.1083/jcb.201807019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, A.B., John Peter A.T., Walter P., and Kornmann B.. 2015. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J. Cell Biol. 210:883–890. 10.1083/jcb.201502105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees, J.A., and Reinisch K.M.. 2020. Inter-organelle lipid transfer: a channel model for Vps13 and chorein-N motif proteins. Curr. Opin. Cell Biol. 65:66–71. 10.1016/j.ceb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage, S., Drouet V., Majounie E., Deramecourt V., Jacoupy M., Nicolas A., Cormier-Dequaire F., Hassoun S.M., Pujol C., Ciura S., et al. International Parkinson’s Disease Genomics Consortium (IPDGC) . 2016. Loss of VPS13C function in autosomal-recessive parkinsonism causes mitochondrial dysfunction and increases PINK1/Parkin-dependent mitophagy. Am. J. Hum. Genet. 98:500–513. 10.1016/j.ajhg.2016.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P., Lees J.A., Lusk C.P., and Reinisch K.M.. 2020. Cryo-EM reconstruction of a VPS13 fragment reveals a long groove to channel lipids between membranes. J. Cell Biol. 219:e202001161. 10.1083/jcb.202001161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Salokas K., Tamene F., Jiu Y., Weldatsadik R.G., Öhman T., and Varjosalo M.. 2018. An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat. Commun. 9:1188. 10.1038/s41467-018-03523-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Doménech, G., Covill-Cooke C., Ivankovic D., Halff E.F., Sheehan D.F., Norkett R., Birsa N., and Kittler J.T.. 2018. Miro proteins coordinate microtubule- and actin-dependent mitochondrial transport and distribution. EMBO J. 37:321–336. 10.15252/embj.201696380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaskill, A.F., Rinholm J.E., Twelvetrees A.E., Arancibia-Carcamo I.L., Muir J., Fransson A., Aspenström P., Attwell D., and Kittler J.T.. 2009. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron. 61:541–555. 10.1016/j.neuron.2009.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, S., Otomo C., and Otomo T.. 2019. The autophagic membrane tether ATG2A transfers lipids between membranes. eLife. 8:e45777. 10.7554/eLife.45777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi, S., López-Doménech G., Halff E.F., Covill-Cooke C., Ivankovic D., Melandri D., Arancibia-Cárcamo I.L., Burden J.J., Lowe A.R., and Kittler J.T.. 2019. Miro clusters regulate ER-mitochondria contact sites and link cristae organization to the mitochondrial transport machinery. Nat. Commun. 10:4399. 10.1038/s41467-019-12382-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, S.E., and Levine T.P.. 2016. VAP, a versatile access point for the endoplasmic reticulum: review and analysis of FFAT-like motifs in the VAPome. Biochim. Biophys. Acta. 1861(8, 8 Pt B):952–961. 10.1016/j.bbalip.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Narendra, D.P., Jin S.M., Tanaka A., Suen D.-F., Gautier C.A., Shen J., Cookson M.R., and Youle R.J.. 2010. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8:e1000298. 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T.T., Oh S.S., Weaver D., Lewandowska A., Maxfield D., Schuler M.-H., Smith N.K., Macfarlane J., Saunders G., Palmer C.A., et al. 2014. Loss of Miro1-directed mitochondrial movement results in a novel murine model for neuron disease. Proc. Natl. Acad. Sci. USA. 111:E3631–E3640. 10.1073/pnas.1402449111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeding, S.J., Majstrowicz K., Hu X.-P., Schwarz V., Freitag A., Honnert U., Nikolaus P., and Bähler M.. 2018. Identification of Miro1 and Miro2 as mitochondrial receptors for myosin XIX. J. Cell Sci. 131:jcs219469. 10.1242/jcs.219469 [DOI] [PubMed] [Google Scholar]
- Okumoto, K., Ono T., Toyama R., Shimomura A., Nagata A., and Fujiki Y.. 2018. New splicing variants of mitochondrial Rho GTPase-1 (Miro1) transport peroxisomes. J. Cell Biol. 217:619–633. 10.1083/jcb.201708122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa, T., Kotani T., Kawaoka T., Hirata E., Suzuki K., Nakatogawa H., Ohsumi Y., and Noda N.N.. 2019. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 26:281–288. 10.1038/s41594-019-0203-4 [DOI] [PubMed] [Google Scholar]
- Park, J.-S., and Neiman A.M.. 2020. XK is a partner for VPS13A: a molecular link between Chorea-Acanthocytosis and McLeod Syndrome. Mol. Biol. Cell. 31:2425–2436. 10.1091/mbc.E19-08-0439-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.-S., Thorsness M.K., Policastro R., McGoldrick L.L., Hollingsworth N.M., Thorsness P.E., and Neiman A.M.. 2016. Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol. Biol. Cell. 27:2435–2449. 10.1091/mbc.e16-02-0112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrungaro, C., and Kornmann B.. 2019. Lipid exchange at ER-mitochondria contact sites: a puzzle falling into place with quite a few pieces missing. Curr. Opin. Cell Biol. 57:71–76. 10.1016/j.ceb.2018.11.005 [DOI] [PubMed] [Google Scholar]
- Prinz, W.A. 2014. Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics. J. Cell Biol. 205:759–769. 10.1083/jcb.201401126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampoldi, L., Dobson-Stone C., Rubio J.P., Danek A., Chalmers R.M., Wood N.W., Verellen C., Ferrer X., Malandrini A., Fabrizi G.M., et al. 2001. A conserved sorting-associated protein is mutant in chorea-acanthocytosis. Nat. Genet. 28:119–120. 10.1038/88821 [DOI] [PubMed] [Google Scholar]
- Raychaudhuri, S., and Prinz W.A.. 2008. Nonvesicular phospholipid transfer between peroxisomes and the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 105:15785–15790. 10.1073/pnas.0808321105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saheki, Y., and De Camilli P.. 2017. Endoplasmic reticulum–plasma membrane contact sites. Annu. Rev. Biochem. 86:659–684. 10.1146/annurev-biochem-061516-044932 [DOI] [PubMed] [Google Scholar]
- Saotome, M., Safiulina D., Szabadkai G., Das S., Fransson A., Aspenström P., Rizzuto R., and Hajnóczky G.. 2008. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc. Natl. Acad. Sci. USA. 105:20728–20733. 10.1073/pnas.0808953105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharwey, M., Tatsuta T., and Langer T.. 2013. Mitochondrial lipid transport at a glance. J. Cell Sci. 126:5317–5323. 10.1242/jcs.134130 [DOI] [PubMed] [Google Scholar]
- Schormair, B., Kemlink D., Mollenhauer B., Fiala O., Machetanz G., Roth J., Berutti R., Strom T.M., Haslinger B., Trenkwalder C., et al. 2018. Diagnostic exome sequencing in early-onset Parkinson’s disease confirms VPS13C as a rare cause of autosomal-recessive Parkinson’s disease. Clin. Genet. 93:603–612. 10.1111/cge.13124 [DOI] [PubMed] [Google Scholar]
- Schuler, M.-H., English A.M., VanderMeer L., Shaw J.M., and Hughes A.L.. 2020. Amino acids promote mitochondrial-derived compartment formation in mammalian cells. bioRxiv. doi:2020.12.23.424218 (Preprint posted December 23, 2020)
- Seifert, W., Kühnisch J., Maritzen T., Horn D., Haucke V., and Hennies H.C.. 2011. Cohen syndrome-associated protein, COH1, is a novel, giant Golgi matrix protein required for Golgi integrity. J. Biol. Chem. 286:37665–37675. 10.1074/jbc.M111.267971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert, W., Kühnisch J., Maritzen T., Lommatzsch S., Hennies H.C., Bachmann S., Horn D., and Haucke V.. 2015. Cohen syndrome-associated protein COH1 physically and functionally interacts with the small GTPase RAB6 at the Golgi complex and directs neurite outgrowth. J. Biol. Chem. 290:3349–3358. 10.1074/jbc.M114.608174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong, E., Insolera R., Dulovic M., Kamsteeg E.-J., Trinh J., Brüggemann N., Sandford E., Li S., Ozel A.B., Li J.Z., et al. 2018. Mutations in VPS13D lead to a new recessive ataxia with spasticity and mitochondrial defects. Ann. Neurol. 83:1075–1088. 10.1002/ana.25220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlevkov, E., Kramer T., Schapansky J., LaVoie M.J., and Schwarz T.L.. 2016. Miro phosphorylation sites regulate Parkin recruitment and mitochondrial motility. Proc. Natl. Acad. Sci. USA. 113:E6097–E6106. 10.1073/pnas.1612283113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slee, J.A., and Levine T.P.. 2019. Systematic prediction of FFAT motifs across eukaryote proteomes identifies nucleolar and eisosome proteins with the predicted capacity to form bridges to the endoplasmic reticulum. Contact (Thousand Oaks). 2:1–21. 10.1177/2515256419883136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, E.W., Guillén-Samander A., Bian X., Wu Y., Cai Y., Messa M., and De Camilli P.. 2019. Lipid transporter TMEM24/C2CD2L is a Ca2+-regulated component of ER–plasma membrane contacts in mammalian neurons. Proc. Natl. Acad. Sci. USA. 116:5775–5784. 10.1073/pnas.1820156116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno, S., Maruki Y., Nakamura M., Tomemori Y., Kamae K., Tanabe H., Yamashita Y., Matsuda S., Kaneko S., and Sano A.. 2001. The gene encoding a newly discovered protein, chorein, is mutated in chorea-acanthocytosis. Nat. Genet. 28:121–122. 10.1038/88825 [DOI] [PubMed] [Google Scholar]
- Ugur, B., Hancock-Cerutti W., Leonzino M., and De Camilli P.. 2020. Role of VPS13, a protein with similarity to ATG2, in physiology and disease. Curr. Opin. Genet. Dev. 65:61–68. 10.1016/j.gde.2020.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde, D.P., Yu S., Boggavarapu V., Kumar N., Lees J.A., Walz T., Reinisch K.M., and Melia T.J.. 2019. ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 218:1787–1798. 10.1083/jcb.201811139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance, J.E. 1990. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265:7248–7256. 10.1016/S0021-9258(19)39106-9 [DOI] [PubMed] [Google Scholar]
- Velayos-Baeza, A., Vettori A., Copley R.R., Dobson-Stone C., and Monaco A.P.. 2004. Analysis of the human VPS13 gene family. Genomics. 84:536–549. 10.1016/j.ygeno.2004.04.012 [DOI] [PubMed] [Google Scholar]
- Wang, X., and Schwarz T.L.. 2009. The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell. 136:163–174. 10.1016/j.cell.2008.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Winter D., Ashrafi G., Schlehe J., Wong Y.L., Selkoe D., Rice S., Steen J., LaVoie M.J., and Schwarz T.L.. 2011. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 147:893–906. 10.1016/j.cell.2011.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T., Birsoy K., Hughes N.W., Krupczak K.M., Post Y., Wei J.J., Lander E.S., and Sabatini D.M.. 2015. Identification and characterization of essential genes in the human genome. Science. 350:1096–1101. 10.1126/science.aac7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, L.H., Gatta A.T., and Levine T.P.. 2019. Lipid transfer proteins: the lipid commute via shuttles, bridges and tubes. Nat. Rev. Mol. Cell Biol. 20:85–101. 10.1038/s41580-018-0071-5 [DOI] [PubMed] [Google Scholar]
- Yamano, K., Wang C., Sarraf S.A., Münch C., Kikuchi R., Noda N.N., Hizukuri Y., Kanemaki M.T., Harper W., Tanaka K., et al. 2018. Endosomal Rab cycles regulate Parkin-mediated mitophagy. eLife. 7:e31326. 10.7554/eLife.31326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeshaw, W.M., van der Zwaag M., Pinto F., Lahaye L.L., Faber A.I., Gómez-Sánchez R., Dolga A.M., Poland C., Monaco A.P., van IJzendoorn S.C., et al. 2019. Human VPS13A is associated with multiple organelles and influences mitochondrial morphology and lipid droplet motility. eLife. 8:e43561. 10.7554/eLife.43561 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists oligonucleotides used in this study.