The VPS13 proteins (VPS13A–D) are thought to mediate lipid transport between organelles and are linked to distinct neurological disorders in humans. Baldwin et al. found that, in addition to known involvement in mitochondrial morphology, VPS13D is essential for peroxisome biogenesis.
Abstract
The VPS13 gene family consists of VPS13A–D in mammals. Although all four genes have been linked to human diseases, their cellular functions are poorly understood, particularly those of VPS13D. We generated and characterized knockouts of each VPS13 gene in HeLa cells. Among the individual knockouts, only VPS13D-KO cells exhibit abnormal mitochondrial morphology. Additionally, VPS13D loss leads to either partial or complete peroxisome loss in several transformed cell lines and in fibroblasts derived from a VPS13D mutation–carrying patient with recessive spinocerebellar ataxia. Our data show that VPS13D regulates peroxisome biogenesis.
Introduction
Peroxisomes mediate a wide range of metabolic pathways, including the β-oxidation of fatty acids, production of lipid precursors, and amino acid catabolism. Critically, peroxisomes perform essential antioxidant functions that prevent damage by reactive oxygen species. The importance of peroxisomes is highlighted by the clinical presentation of a group of peroxisome biogenesis disorders (PBDs), including the Zellweger syndrome spectrum. These patients exhibit a combination of neurological, liver, and/or adrenal defects to varying levels of severity. 14 identified peroxin (PEX) and nonperoxin genes are involved in mammalian peroxisome biogenesis.
Mammalian peroxisomes proliferate via growth and fission of preexisting peroxisomes or de novo from the ER. Recent work suggests that mitochondria may also serve as a source of de novo peroxisome formation (Sugiura et al., 2017). Interestingly, peroxisomes and mitochondria share fission factors DRP1 (dynamin-1-like protein) and MFF (mitochondrial fission factor). In mammals, peroxisomes are required for the breakdown of very long chain fatty acids (VLCFAs), which are then passed to the mitochondria for further metabolism. The accumulation of VLCFAs in peroxisome-deficient cells causes mitochondrial damage and swelling in Pex19 mutant flies and human skin fibroblasts (Bülow et al., 2018). Mitochondrial fragmentation and swelling were found in mouse cells following PEX5 or PEX3 loss (Tanaka et al., 2019). This interplay is also illustrated by the observation of mitochondrial abnormalities in PBD patients (Salpietro et al., 2015; Hughes et al., 1990; Goldfischer et al., 1973). We recently reported that VPS13D is a novel factor regulating mitochondrial size and fission in both human cells and Drosophila (Anding et al., 2018). Like mitochondria, peroxisomes are highly dynamic, altering their size, number, and cellular distribution in response to external and internal stimuli (Mast and Aitchison, 2018).
Vacuolar protein sorting 13 (VPS13; originally named SOI1) was discovered in Saccharomyces cerevisiae for its role in Golgi trafficking (Brickner and Fuller, 1997). Vps13p, the fifth-largest protein in yeast, has been implicated in sporulation, phospholipid regulation (Park and Neiman, 2012), and mitochondrial integrity (Park et al., 2016). Vps13p has also been linked to mitochondria–ER membrane contact sites. VPS13 dominant mutants suppress the lipid transport defect in ER-mitochondria encounter structure (ERMES) loss-of-function mutant S. cerevisiae cells (Lang et al., 2015). The ERMES complex acts as a physical tether between the ER and the outer mitochondrial membrane. Vps13p’s lipid transport functions bypass the ERMES at alternate membrane contact sites. There is increasing evidence of the VPS13 family’s role in lipid transport–related pathways in humans (Yeshaw et al., 2019; Ramseyer et al., 2018; Park et al., 2015). Kumar et al. (2018) demonstrated the role of human VPS13A and VPS13C in lipid transport at ER contact sites.
Mammals have four VPS13 homologues: VPS13A–D (Velayos-Baeza et al., 2004). Mutations in VPS13A cause the neurodegenerative disease chorea-acanthocytosis (Rampoldi et al., 2001; Ueno et al., 2001). VPS13B is associated with Cohen syndrome (Kolehmainen et al., 2003), a neurodevelopmental disorder that shares features with autism. While fewer studies have examined VPS13B biology, there is evidence that the protein regulates trafficking from the Golgi apparatus and the integrity of the Golgi complex structure (Seifert et al., 2015; Duplomb et al., 2014). Genome-wide association studies have implicated VPS13C in Parkinson’s disease (Lesage et al., 2016). In contrast to VPS13A–C, VPS13D is considered an essential gene in human cells (Hart et al., 2015; Blomen et al., 2015). Complete loss of VPS13D causes embryonic lethality in mice and flies (Seong et al., 2018; Anding et al., 2018).
In humans, VPS13D mutations are linked to movement disorders classified as a subtype of spinocerebellar ataxia, spinocerebellar ataxia autosomal recessive 4 (SCAR4; Gauthier et al., 2018; Seong et al., 2018; Koh et al., 2019). Peroxisomes play an important role in overall nervous system function and are particularly critical for cerebellar development and function through adulthood (De Munter et al., 2015). Cerebellar pathologies are associated with dysfunctions in peroxisomal ether lipid synthesis, α-oxidation, and β-oxidation. However, no peroxisomal phenotype has been reported in VPS13D mutant cerebellar ataxia patients.
We investigated the cellular biology of VPS13A–D knockout (KO) in human cells and found a dual role for VPS13D in mitochondrial morphology and peroxisome biogenesis.
Results
KO and exogenous expression of VPS13A–D genes
Our previous work showed that VPS13D deletion leads to round mitochondria (Anding et al., 2018), a phenotype that is conserved from flies to humans. Two independent VPS13D-KO cell lines were generated by disrupting exon 3 (KO 45) or exon 34 (KO 19) via CRISPR, as previously described (Anding et al., 2018). Both VPS13D-KO lines exhibit normal cell growth and viability (Fig. S1) and have indistinguishable mitochondrial phenotypes: abnormal rounding and fragmentation. The two VPS13D-KO cell lines were considered interchangeable in our experimental design. To investigate if other VPS13 gene family members play a role in mitochondrial morphology, we generated VPS13A-KO, VPS13B-KO, and VPS13C-KO HeLa cells. In contrast to VPS13D-KO cells, there is no observable aberration in the mitochondrial morphology or connectivity in the VPS13A–C KO lines (Fig. 1, A and B). Although some VPS13B-KO cells displayed fragmented mitochondria, the magnitude did not reach substantial significance (Fig. 1 C). Due to the increasing evidence of the presence of VPS13 family proteins at multiple types of membrane contact sites, we assessed the involvement of VPS13A–D in the morphology of other organelles. As shown in Fig. S2, there is no observable difference in the appearance of the Golgi, ER, endosomes, and lysosomes between the four VPS13A–D KO and control HeLa cells, with the exception of the Golgi structure in VPS13B-KO cells, which appears slightly more fragmented. This finding is consistent with a previous report on a VPS13B role in Golgi integrity (Seifert et al., 2011).
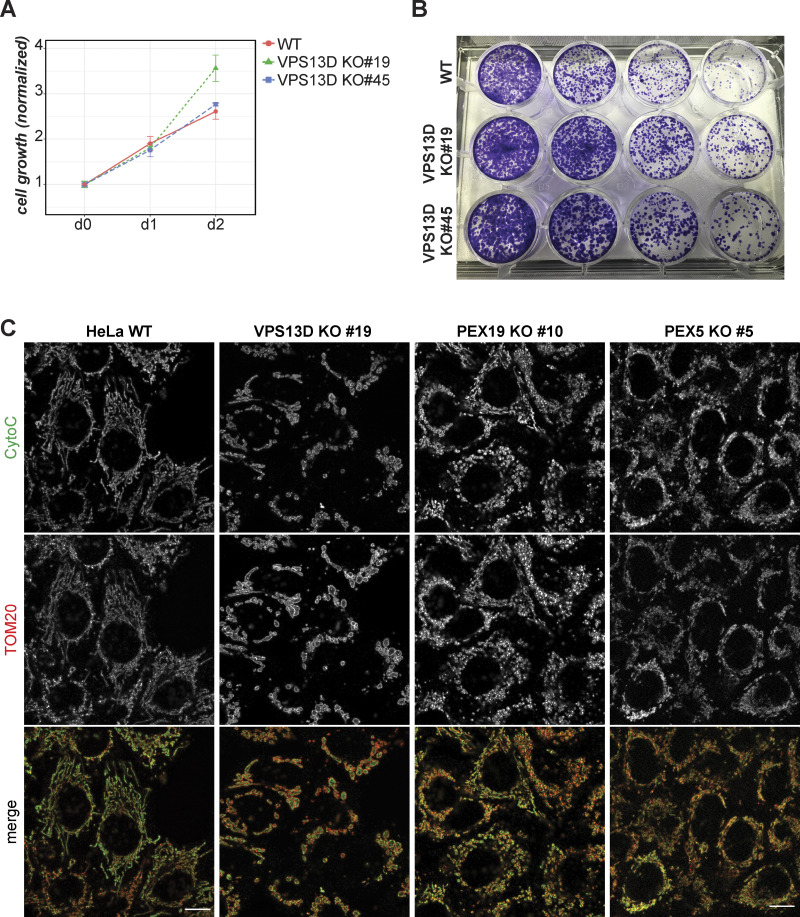
Figure S1.
VPS13D loss does not reduce viability of HeLa population. (A) Cell growth rate of WT, VPS13D KO 19, and VPS13D KO 45 HeLa cells measured by CellTiter-Glo assay. Luminescence normalized to day 0 for each group; n = 5,000 cells/well; 3 wells per group. Error bars show SD among the technical replicates. (B) Clonogenic assay of WT and VPS13D KO 19 and 45 cells. Colonies were fixed and stained for crystal violet 8 d after seeding. Four dilution rates were used (columns from left to right): 4 × 103, 2 × 103, 1 × 103, and 5 × 102. (C) HeLa WT, VPS13D KO 19, PEX5 KO 5, and PEX19 KO 10 cells costained for cytochrome c (green) and TOM20 (red). Scale bars = 10 µm.
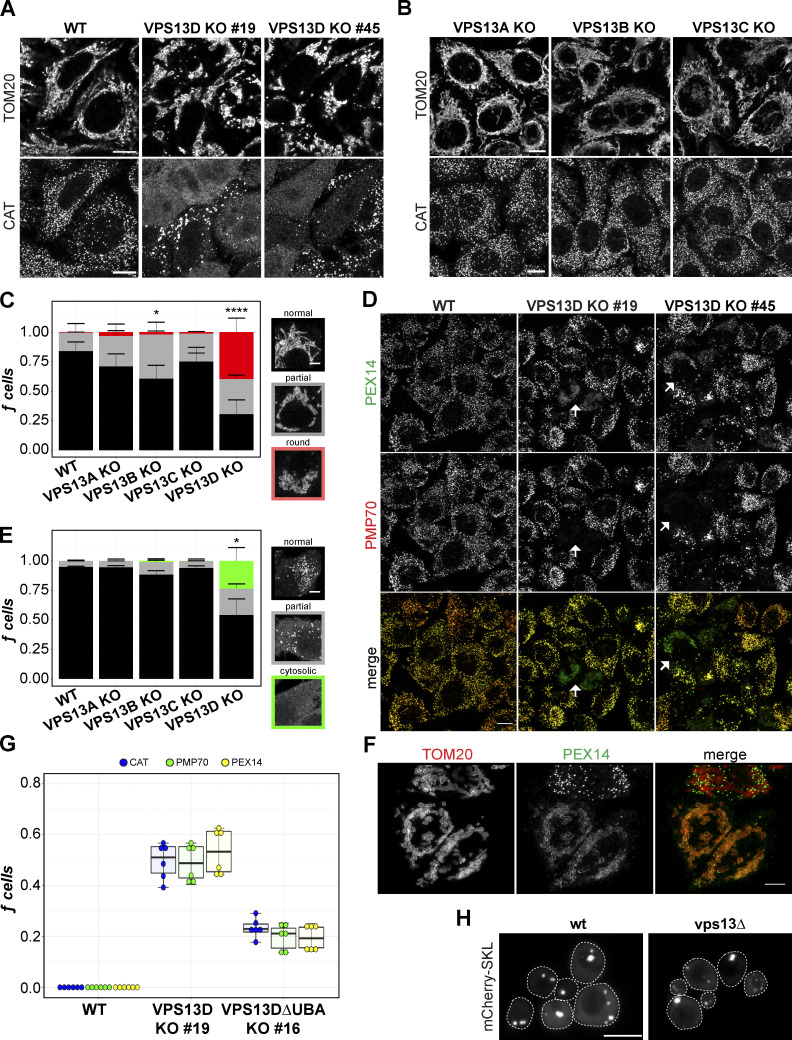
Figure 1.
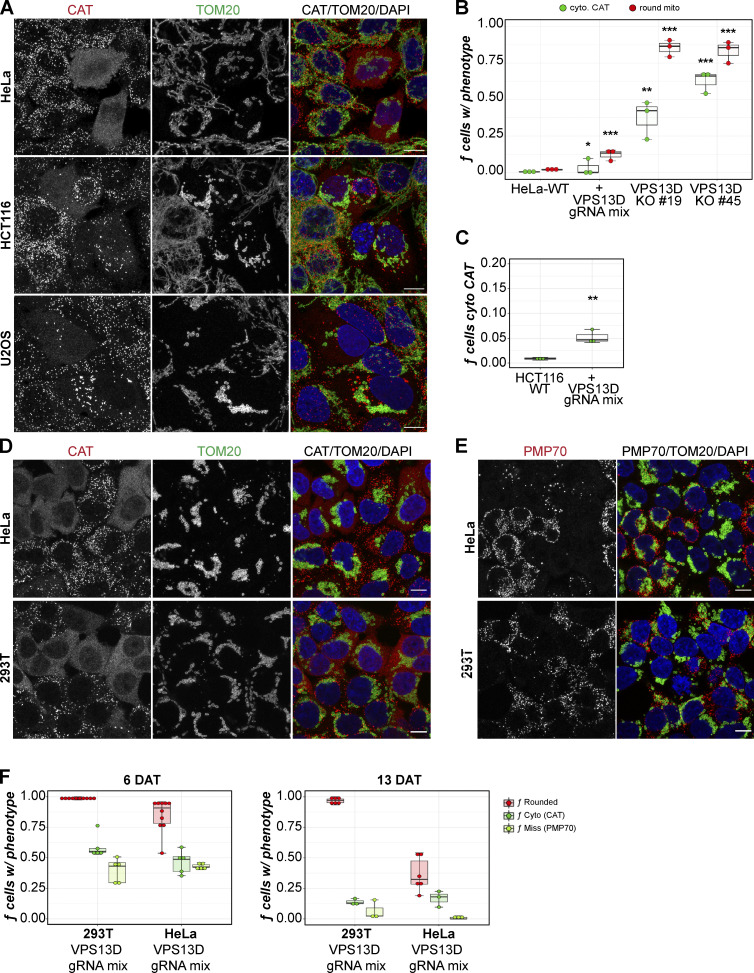
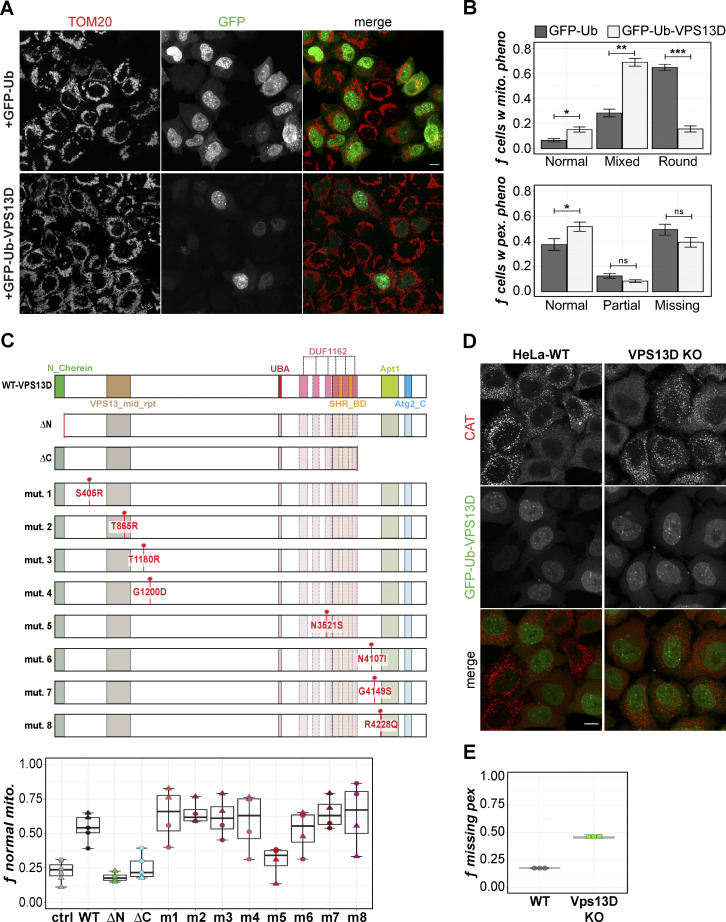
VPS13D, but not VPS13A–C, is required for normal mitochondrial morphology and peroxisome maintenance. (A) HeLa WT and VPS13D KO 19 and KO 45 cells fixed and stained for TOM20 and CAT. (B) VPS13A–C KO cells similarly stained for TOM20 and CAT. Scale bars = 10 µm. (C) The fraction of each cell line’s population manually classified as “rounded,” “partial,” or “normal” mitochondrial morphology. *, P = 0.0128; ****, P = 0.0003. (D) VPS13D-KO and WT cells stained for PEX14 and PMP70. Arrows indicate examples of VPS13D-KO cells with simultaneous nonperoxisomal PEX14 and missing PMP70. Scale bars = 10 µm. (E) Cells classified by peroxisome phenotype (CAT signal) as “cytosolic,” “partial,” or “normal.” *, P = 0.0224 (VPS13D KO versus WT). Kruskal-Wallis and Dunn’s tests for multiple comparisons (versus WT) of normal (mitochondrial/peroxisomal) cells were used in C and E. Each count represents a field of ≥ 100 cells; n counts: WT = 15; VPS13A-KO, VPS13B-KO, VPS13C-KO = 12; VPS13D-KO = 18. Error bars represent SEM. (F) VPS13D KO 45 cells costained for TOM20 and PEX14. Scale bar = 5 µm. (G) Quantification of peroxisomal phenotype in fixed HeLa WT, VPS13D ΔUBA KO 16, and VPS13D KO 19 cells. The fraction of cells missing peroxisomes was determined by cytosolic CAT (blue), negative PMP70 (dark-green), or mislocalized PEX14 (yellow-green) signal. n = 3 counts; ≥100 cells per count. Boxes mark 25th, 50th, and 75th percentiles, and lines show the upper and lower extreme values (Tukey method). (H) vps13Δ and control yeast transiently transfected with peroxisomal marker mCherry-SKL. Cells were outlined (dashed white lines) by hand for visibility. Scale bar = 1 µm.
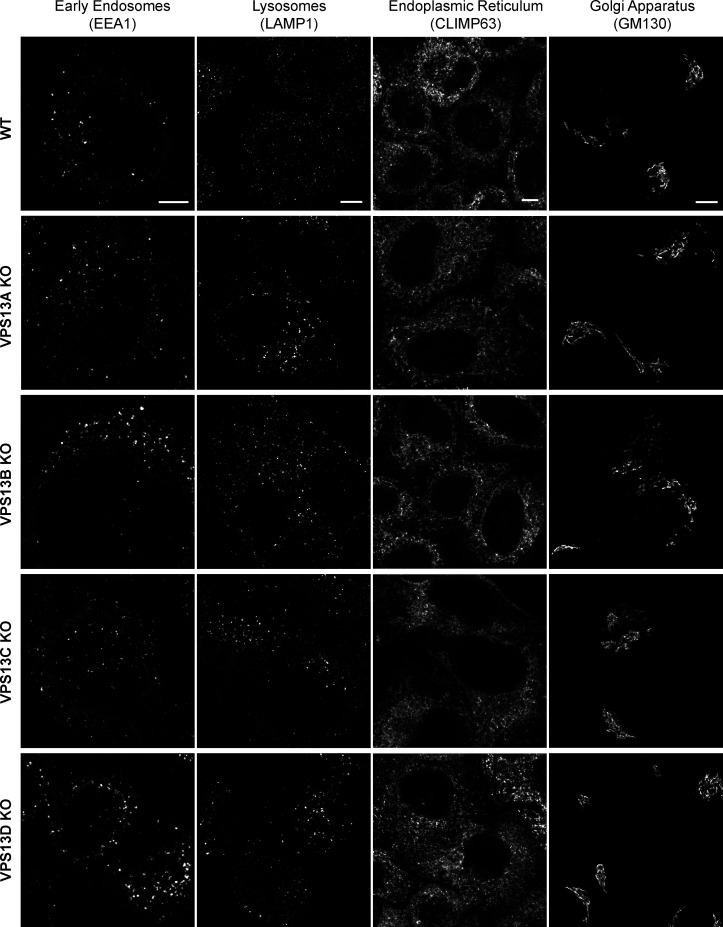
Figure S2.
Other organelles in VPS13A–D KO cells are morphologically normal. WT, VPS13A-KO, VPS13B-KO, VPS13C-KO, and VPS13D KO 45 cells were fixed and immunostained for EEA1, LAMP1, GM130, or CLIMP63. Scale bars = 5 µm.
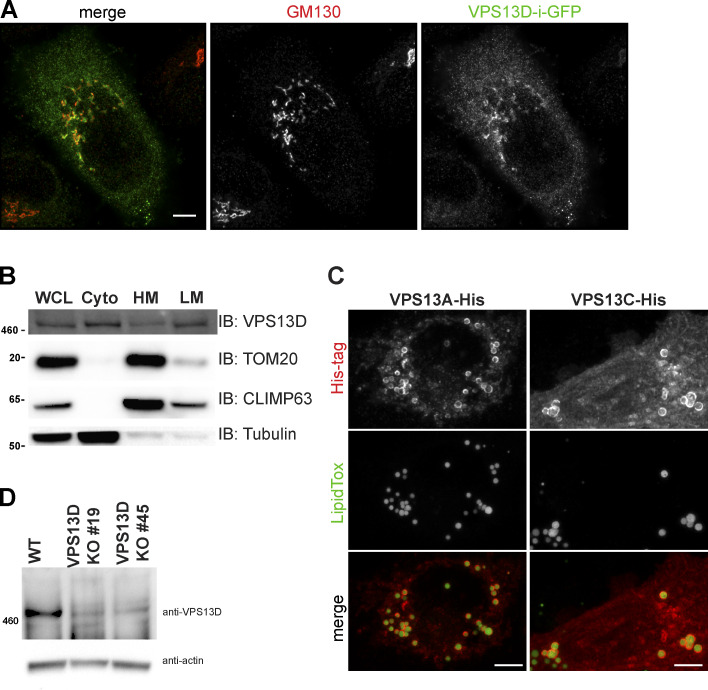
Owing to the lack of suitable antibodies and/or the relatively low endogenous protein expression level in HeLa cells, we were not able to determine the subcellular localization of endogenous VPS13D by immunostaining. To overcome this issue, we generated an internally tagged VPS13D construct, VPS13D-i-GFP, in which GFP is inserted in frame, replacing exon 40. Exon 40 is present in variant 2A (main variant found in the brain) but skipped in variant 1A (main variant in nonneural tissues; Velayos-Baeza et al., 2004). This tagged VPS13D construct has previously been shown to rescue the mitochondrial phenotype in VPS13D-KO cells (Anding et. al., 2018). We found the GFP signal to be imperceptible unless immunostained for GFP. VPS13D-i-GFP immunofluorescence is primarily cytosolic, with a partial concentration of GFP signal on and around the Golgi apparatus (Fig. S3 A). We also performed subcellular fractionation and found that endogenous VPS13D is present in both heavy (mitochondria and ER) and light (peroxisomes/lysosomes/endosomes) membrane fractions (Fig. S3 B). His-tagged VPS13A and VPS13C constructs were also overexpressed in WT and their respective KO cells. Both had a partial cytosolic signal. Exogenously expressed VPS13A and VPS13C clearly encircle the outer membrane of lipid droplets (Fig. S3 C), concurring with recent reports of VPS13A and VPS13C localization to lipid droplets (Yang et al., 2016; Kumar et al., 2018; Yeshaw et al., 2019).
Figure S3.
Exogenous VPS13D preferentially localizes to Golgi, while VPS13A and VPS13C localize to lipid droplets. (A) HeLa cells transiently transfected with VPS13D-i-GFP immunostained for GFP and GM130 (red). (B) Subcellular fractionation Western blot of HeLa WT cells for endogenous VPS13D. WCL, whole cell lysate; Cyto, cytosol; HM, heavy membrane; LM, light membrane. (C) His-tagged VPS13A or VPS13C stained for His-tag (red) and lipid droplets (LipidTox; green). (D) Western blot of VPS13D in HeLa WT, VPS13D KO 19, and VPS13D KO 45 cells with actin loading control. Scale bars in A and C = 5 µm.
VPS13D loss disrupts peroxisome density
Surprisingly, immunofluorescence of peroxisomal markers revealed that at least half of the VPS13D-KO cells exhibit partial or complete peroxisome loss (Fig. 1, A and E). A substantial portion of VPS13D-KO cells express catalase (CAT) in the cytosol, instead of in peroxisomes as in control cells. Peroxisome biogenesis disruption is most commonly associated with a “ghost” peroxisomal phenotype where cells show defective CAT import but have peroxisomal bodies positive for peroxisomal membrane protein 70 (PMP70). PEX3, PEX16, and PEX19 are the only peroxisome biogenesis genes known to be essential for the formation of the peroxisomal membrane structures. We found that PMP70 is completely absent in certain VPS13D-KO cells (Fig. 1 D). PEX14 is a single-transmembrane peroxisomal membrane protein important for the import of certain peroxisomal matrix proteins and normally localized to peroxisomes. In the absence of peroxisomes, several PEX proteins, such as PEX3, PEX12, PEX13, PEX26, and PEX14, mislocalize to mitochondria (Sugiura et al., 2017). Indeed, PEX14 is mistargeted to mitochondria in the population of VPS13D-KO cells lacking PMP70 signal, further confirming the loss of peroxisomes in VPS13D-KO cells (Fig. 1, D and F). The ubiquitin (Ub)-associated binding domain (UBA) is unique to VPS13D in the VPS13 family and has been shown to be required for normal mitochondrial morphology in both flies and HeLa cells (Anding et al., 2018). Cells with the VPS13D UBA domain deleted (VPS13DΔUBA KO 16) lose PMP70-positive peroxisomes and have rounded mitochondria, though at a lower incidence than the complete VPS13D KOs (Fig. 1 G). As with the mitochondria, peroxisomes are also normal in VPS13A–C KO HeLa cells (Fig. 1 B). We did not observe a similar peroxisomal phenotype in S. cerevisiae vps13Δ yeast (Fig. 1 H), revealing a novel function for a VPS13 family protein.
VPS13D loss induces peroxisomal and mitochondrial aberrations in multiple human cell lines
To confirm the peroxisome loss phenotype observed in HeLa cells, we knocked out VPS13D in other mammalian cell lines using distinct VPS13D CRISPR gRNAs. We employed a new strategy that involved using a mixture of three unique VPS13D CRISPR guides, each targeting different exons: 13, 19, and 21. Following selection, the total pool of mixed CRISPR-edited cells was examined for peroxisomal and mitochondrial phenotypes. The pooled VPS13D-KO HeLa cells displayed peroxisomal loss and rounded mitochondria, as in the original individual VPS13D-KO clones (Fig. 2, A and B). VPS13D-deficient HCT116 (colorectal carcinoma) and U2OS (osteosarcoma) cells similarly displayed heterogeneous levels of partial to complete peroxisome loss (Fig. 2, A and B). However, in both HCT116 and HeLa cells, a smaller fraction of cells exhibited the peroxisomal phenotype than did the two individual HeLa VPS13D-KO clones (Fig. 2, B and C). Several attempts at screening individual KO clones in HCT116 and U2OS cells using gRNA targeting exon 19 failed to identify any KO clones. (On the basis of sequencing data, only a few heterozygous clones were obtained.) This led us to speculate that the complete elimination of VPS13D may be lethal, as reported in three different whole-genome CRISPR screens (Hart et al., 2015; Blomen et al., 2015; Wang et al., 2015). There may be a minor residual amount of VPS13D protein in our original VPS13D-KO 19 and 45 lines (Fig. S3 D), perhaps due to unexpected splicing events. The use of three gRNAs greatly enhances the likelihood of completely deleting VPS13D in individual cells. Therefore, we checked the penetrance of the mitochondrial and peroxisomal phenotypes over time after the transient CRISPR transfection into HeLa and HEK293T (human embryonic kidney) cells (Fig. 2, D and E). While the percentage of cells exhibiting a round mitochondrial phenotype decreased in 293T and HeLa cells from 6 d after transfection (DAT) to 13 DAT, a significant number of cells retained the mitochondrial phenotype at 13 DAT (Fig. 2 F). In contrast, the fraction of cells without visible peroxisomes dropped significantly in both 293T and HeLa cells from 6 DAT to 13 DAT.
Figure 2.
VPS13D loss causes mitochondrial and peroxisomal phenotypes in multiple cell lines. (A) Representative images of HeLa, HCT116, and U2OS cells after transfection of VPS13D CRISPR gRNA mix were fixed and stained for CAT (red) and TOM20 (green). (B) Quantification of cells with cytosolic (cyto.) CAT and rounded mitochondria (mito) in HeLa cells with and without VPS13D gRNA mix in parallel with VPS13D KO 19 and 45 cells. One-way ANOVA, Tukey post-hoc multiple comparisons (vs. HeLa-WT): *, P = 0.0320; **, P = 0.0016; ***, P < 0.0005. (C) Quantification of the peroxisomal phenotype (CAT) of HCT116 cells with and without VPS13D gRNA mix. Welch t test: **, P = 0.0045. (D and E) Representative images of HeLa and 293T cells 6 DAT with VPS13D gRNA mix stained for CAT (red) and TOM20 (green; D) or for PMP70 (red) and TOM20 (green; E). (F) Quantification of cells with rounded mitochondria, cytosolic (Cyto) CAT, and missing (Miss) PMP70 signal in 293T and HeLa cells either 6 DAT or 13 DAT with VPS13D gRNA mix. Each dot represents the count from one field of images. Mean cells/field for 6 DAT: n = 116 (HeLa), n = 138 (293T); mean cells/field for 13 DAT: n = 72 (HeLa), n = 86 (293T). Partial peroxisome phenotype cells were excluded from the total count. Box-and-whisker plot whiskers in B, C, and F represent minimum (before lower fence) and maximum (before upper fence) values. Scale bars = 10 µm.
Heterogeneity of peroxisome abundance is stochastic in VPS13D-KO cells
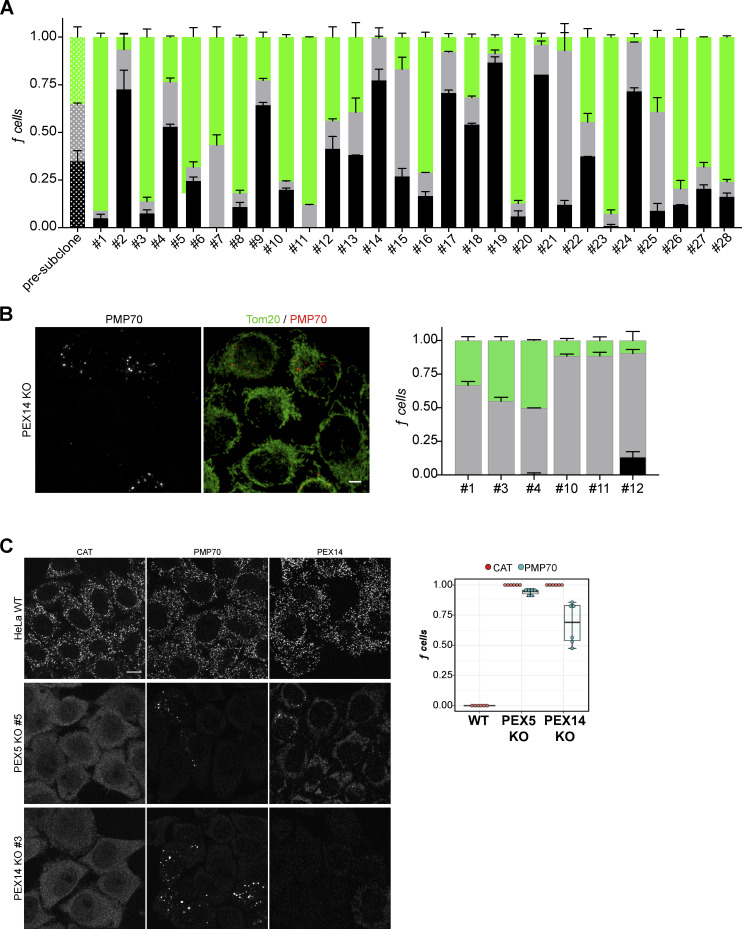
Any given population of VPS13D-KO cells examined displayed a mixture of three distinct peroxisomal phenotypes that we defined as “normal” (no loss), “partial” (≤50% missing), or “missing” (nondetectable). VPS13D-KO cells were subcloned to reestablish single colonies for expansion and phenotypic analysis. Despite several rounds of separate subcloning experiments, we were unable to achieve a homogeneous population of cells with either “all normal” or “all missing” peroxisomes (Fig. S4 A). The stochastic peroxisomal phenotype is not limited to VPS13D-KO cells, as the extent of peroxisome loss also varies among six different PEX14-KO subclone lines (Fig. S4 B). We also identified a varying portion of PEX5-KO cells lacking PMP70-positive structures and mislocalized PEX14, in addition to the expected cytosolic CAT in all PEX5-KO cells (Fig. S4 C).
Figure S4.
Stochastic peroxisome phenotype heterogeneity in Vps13D-KO populations. (A) VPS13D KO 19 cells were subcloned. Selected single colonies were sampled 16 d after subcloning for immunofluorescence (CAT): “normal” (black), “partial” loss (gray), and “missing” peroxisomes or cytosolic CAT (green). For each experiment, n = 6 counts, ≥100 cells per count. (B) Six independent PEX14-KO clones were immunostained with PMP70 and TOM20 and manually classified by peroxisome phenotype: “normal” (black), “partial” (gray), or “missing” (green). One-way ANOVA (fraction of normal peroxisome cells between clones; P = 0.0054; n = 3 counts/clone, ≥100 cells per count. Scale bar = 5 µm. Error bars in A and B represent SEM. (C) Representative images and box plots of the fraction of HeLa WT, PEX5-KO, and PEX14-KO cells with cytosolic CAT (red) or missing PMP70 (blue). Also included are representative images of PEX14 staining. Per group, n = 3 counts/clone, ≥100 cells per count. Boxes mark 25th, 50th, and 75th percentiles, and lines show the upper and lower extreme values (Tukey method). Scale bar = 10 µm.
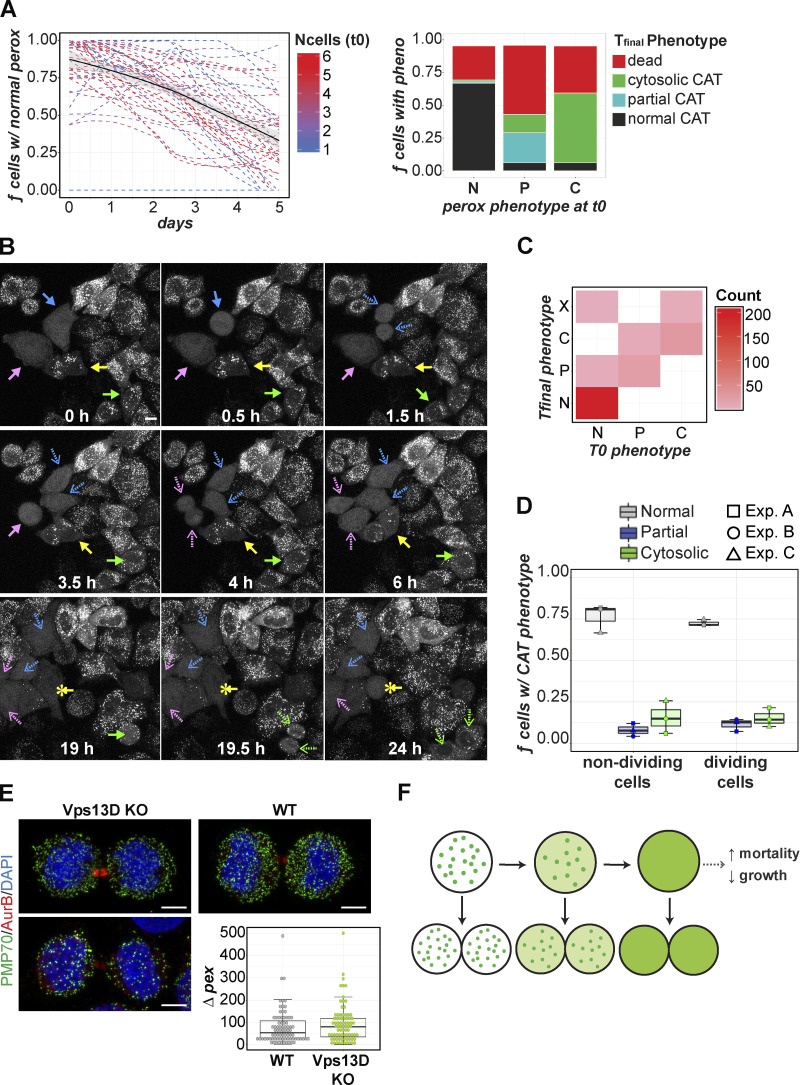
Because of the inability to achieve complete homogeneity in the VPS13D-KO phenotype, we tracked if, and how, the peroxisomal phenotype changed in individual VPS13D-KO cells over time. We generated a VPS13D-KO line stably expressing GFP-CAT in order to observe the peroxisomal phenotype in live cells. A population of GFP-CAT VPS13D-KO cells were subcloned into separate wells on a single plate. Each well was imaged over 5 d at 12-h intervals. Within a well of 1–6 cells, each cell’s phenotype was manually assigned a peroxisomal phenotype (normal, partial depletion, or complete absence) and tracked over the 10 time points. The fraction of cells with normal peroxisome number for each time point per well is displayed in Fig. 3 A (left). The trend line for the average of all the wells analyzed (black) indicates a gradual decrease in the number of cells with a normal number of peroxisomes over time. This indicates a progressive loss of peroxisomes in VPS13D-KO cells. Therefore, we hypothesized that the VPS13D-KO cells without peroxisomes may be more vulnerable to environmental or other stress, leading to slower growth, death, or senescence. We separated individual cells (excluding predecessors or progeny) into groups by their initial peroxisomal phenotype and calculated the fraction of cells exhibiting normal, partial, or missing peroxisome phenotypes, or death, by the final time point (day 5; Fig. 3 A, right). VPS13D-KO cells exhibiting an abnormal peroxisomal phenotype were more likely to die than those with normal peroxisomes within the 5 d. We would expect this difference to be even greater over longer periods of observation. Although cells with fewer peroxisomes die more frequently than those with a normal number of peroxisomes, we did not find a substantial survival or growth defect in the whole population of VPS13D-KO HeLa cells (Fig. S1, A and B). Consistent with this finding, immunostaining detected no cytosolic cytochrome c signal in VPS13D-KO cells or in PEX5-KO or PEX19-KO cells (Fig. S1 C), indicating the lack of basal apoptosis underlying peroxisome loss.
Figure 3.
Peroxisome phenotype changes over time in VPS13D-KO. (A) GFP-CAT stable VPS13D-KO cells were seeded at one to six cells per well. Time-lapse imaging of live cells at 12-h intervals (interrupted by media changes between intervals). Time point 0 (t0) is defined as the first day of imaging (1 d after seeding). Left: Fraction of normal peroxisome (perox) cells (per well). Trend line (dashed) for each well was fitted via LOWESS. Light gray region indicates SE. The lines are colored by total number of cells (Ncells) in the well at t0. Best fit line of total wells modeled via LOWESS is indicated by the black solid line. Right: Individual GFP-CAT VPS13D-KO cells separated into groups by their initial peroxisomal phenotype (normal, partial, missing) and fraction of cells classified as normal (dark gray), partial (turquoise), or missing (green) peroxisomes or dead (red) at final time point (day 5). n = 43 wells; 1–30 unique cells per well. GFP-CAT VPS13D-KO were seeded into chamber wells, and selected fields were imaged continuously overnight. Three independent experiments (detailed in Fig. S5) were performed over different time periods: 8 (experiment A), 12 (experiment B), or 25 (experiment C) h at 900-s (experiment A) or 600-s (experiments B and C) intervals. (B) Select frames from one field of experiment C. Arrows point to cells of interest for tracking with cytosolic CAT/missing peroxisomes (pink and blue) or partial peroxisome phenotype (yellow and green). Dashed arrows (pink, blue, or green) point to the progeny of the same (color-matched) cells at later time points. Note one of the pink daughter cells drifts out of frame at ∼24 h. Yellow asterisks indicate the transformation of partial to cytosolic CAT phenotype within an individual cell (19 h). Scale bar = 10 µm. (C) Heatmap plot correlating a cell’s peroxisomal phenotype (CAT localization) at the initial (0 h) time-point with their (or their progeny’s) final phenotype (8-25 h). N, normal; P, partial; C, cytosolic; X, dead/dying. (D) Cells at the initial time point were classified by whether they divided by the end of the experiment according to initial peroxisomal phenotype: normal (gray), partial (blue), or cytosolic (green) CAT. Each point is one independent experiment.Boxes mark 25th, 50th, and 75th percentiles, and lines show the upper and lower extreme values (Tukey method). n cells per experiment were as follows: A (square) = 91 nondividing, 12 dividing; B (circle) = 47 nondividing, 7 dividing; C (triangle) = 39 nondividing and 79 dividing. No significant difference in peroxisome phenotype prevalence in the dividing versus nondividing groups (Welch’s two-sample t test). (E) Representative images of dividing WT and VPS13D-KO cells stained for PMP70, DAPI, and Aurora B. Two representative images of dividing VPS13D-KO cells that originally had normal (top) or reduced (bottom) peroxisomes are shown. Difference in peroxisome count between two daughter cells (each dot represents one pair of newly divided daughter cells) is shown in the right-hand bottom panel. Both WT and VPS13D-KO: n = 80 images (2 daughter cells per image). Scale bar = 5 µm. (F) Model of VPS13D’s variable effect on peroxisome number. Green represents localization of CAT.
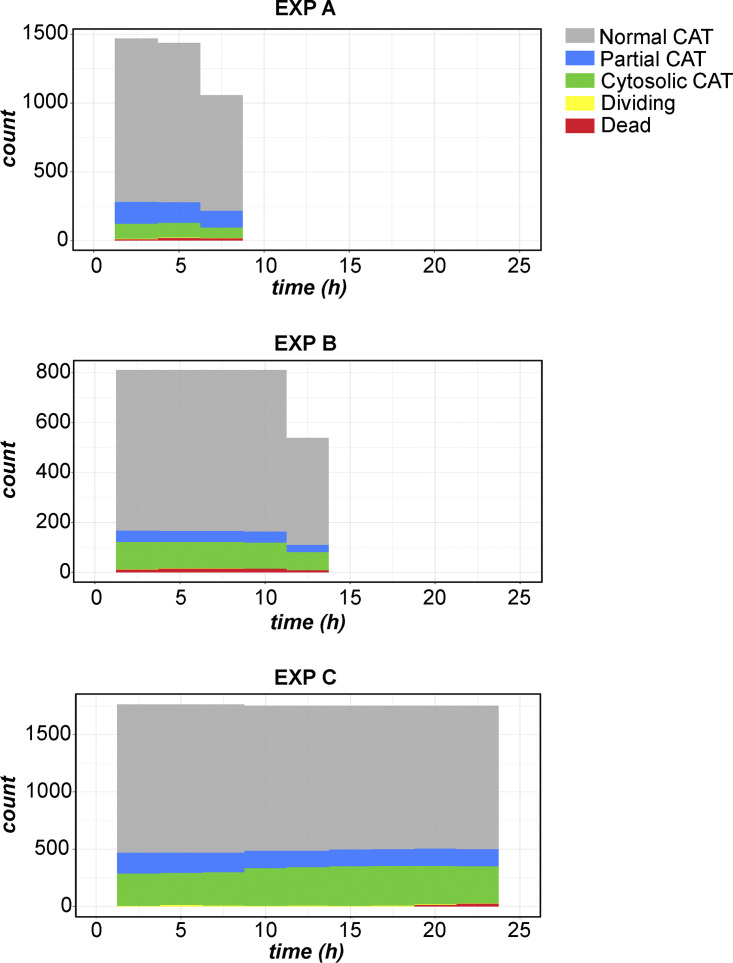
We also performed shorter-term (≤25 h) imaging of live GFP-CAT VPS13D-KO cells at much shorter time intervals, allowing more accurate tracing of individual cells and the timing of their division, death, or phenotypic changes. The analysis of three independent short-term imaging experiments (Fig. S5) confirmed that the peroxisomal phenotype (normal versus cytosolic CAT) has no effect on the VPS13D-KO cell’s ability to divide and survive the division (Fig. 3 B). Comparison of the cell’s initial versus final peroxisomal phenotypes validated the loss, but never the gain of peroxisomes within an individual cell or its progeny (Fig. 3 C). Within each experiment, cells were separated by whether they divided or not during the observed time period. There was no significant difference in the fraction of cells exhibiting either a normal, partial, or cytosolic phenotype between the two groups (Fig. 3 D).
Figure S5.
Histograms of peroxisome phenotypes over time in overnight GFP-CAT VPS13D-KO experiments. Binned histograms showing counts of cells observed as having either normal (gray), partial (blue) missing/cytosolic CAT (green) peroxisomal phenotype, dividing (yellow), or dead (red) at each time point. Experiment A: 900-s intervals, 34 time points (30,600 s final time point), 103 cells at initial time point; experiment B: 600-s intervals, 78 time points (46,200 s), 54 cells at first time point; experiment C: 600-s intervals, 151 time points (90,000 s), 118 initial cells.
VPS13D does not impact peroxisomal segregation during division
One possible etiology of the peroxisome loss in VPS13D-KO cells is the asymmetrical distribution of peroxisomes during cell division. Therefore, we investigated whether VPS13D is important for the normal segregation and roughly equal distribution of peroxisomes during cellular division. Recently divided or dividing daughter VPS13D-KO and control HeLa cells were identified by immunostaining for mitosis factor Aurora B. Using a custom image analysis program, peroxisomes were counted for each daughter cell in a pair. The difference in peroxisome number between two daughter cells was not significantly affected by VPS13D loss (Fig. 3 E). The peroxisome number likely decreases within individual VPS13D-KO cells over time, eventually affecting cell health. Our proposed model of the progressive peroxisomal phenotype change in VPS13D-KO cells is illustrated in Fig. 3 F.
Overexpression of peroxisomal membrane proteins and VPS13D can induce peroxisomal loss in WT cells
We found that GFP tagging on either the N-terminus or C-terminus of VPS13D inactivates VPS13D function (Anding et al., 2018), similar to yeast Vps13p (Lang et al., 2015; Park et al., 2016). To exogenously express untagged VPS13D with a fluorescent marker for transfection, we made a GFP-Ub-VPS13D construct in which VPS13D is transcribed and translated with GFP-Ub but is subsequently cleaved by endogenous deubiquitinases, leaving an untagged VPS13D and GFP-Ub reporter for transfected cells. GFP-Ub-VPS13D overexpression rescues the round mitochondrial phenotype in HeLa VPS13D-KO cells (Fig. 4, A and B).
Figure 4.
Exogenous expression of VPS13D rescues VPS13D-KO mitochondrial phenotype, but not peroxisomal phenotype. (A) VPS13D KO 19 cells were transiently transfected with either GFP-Ub-VPS13D or GFP-Ub (ctrl) for 2 d and stained for TOM20. (B) Proportion of GFP-positive cells manually classified as having either round and fragmented, fragmented only, or normal mitochondria for GFP-Ub-VPS13D and GFP-Ub groups. Welch’s two-sample t test between groups for each phenotype class: *, P = 0.03055; **, P = 0.0007925; ***, P = 0.0001459. The same analysis was performed for peroxisome phenotypes (CAT): *, P = 0.0126. Error bars in A and B represent SEM. (C) Schematic (top panel) of WT VPS13D protein domains, including the β-propeller/DUF1162 regions (light pink). Patient mutations are indicated by red star. Bottom: Quantification of the proportion of VPS13D KO 45 cells with normal mitochondrial morphology following transfection with the corresponding VPS13D constructs. Each point is an independent field of view of two independent experiments, A (circles) and B (triangles). 3–4 images/experiment (ctrl); 2–3 images/experiment (ΔN, ΔC, mutants 1–8); mean number of cells per image: n = 272 (ctrl), n = 81 (nonctrl). (D) HeLa WT and VPS13D-KO cells stably expressing GFP-Ub-VPS13D and stained for CAT. (E) Quantification of peroxisomal phenotype in cells from D; n = 3 counts, ≥100 cells/count. Scale bar = 5 µm. Box-and-whisker plot whiskers in C and E represent minimum (before lower fence) and maximum (before upper fence) values.
Transiently transfecting either an N-terminal (ΔN) or C-terminal (ΔC) truncated form of VPS13D does not rescue mitochondrial morphology (Fig. 4 C), suggesting that both the N-chorein and Atg2_C domains are essential for VPS13D’s function. Symptomatic patients usually carry compound heterozygous VPS13D mutations: one nonsense and one missense (Gauthier et al., 2018; Seong et al., 2018). Among the eight tested point mutations, only R4228Q is a homozygous patient mutation. All of the VPS13D mutants examined, except for N3521S, rescued VPS13D mitochondrial morphology like WT VPS13D (Fig. 4 C). N3521S is the only mutation of the group that is in a DUF1162 domain. The DUF1162 repeats are also referred to as the “Vps13 adaptor binding (VAB) domain,” following the discovery of their role in the organelle-specific recruitment of Vps13p (Bean et al., 2018). The yeast Vps13 mutant N2428S, which corresponds to the N3521S mutation in VPS13D spastic ataxia patients, has been reported to be defective in binding to all three adaptor proteins (Dziurdzik et al., 2020), highlighting the important role of the VAB/DUF1162 domain.
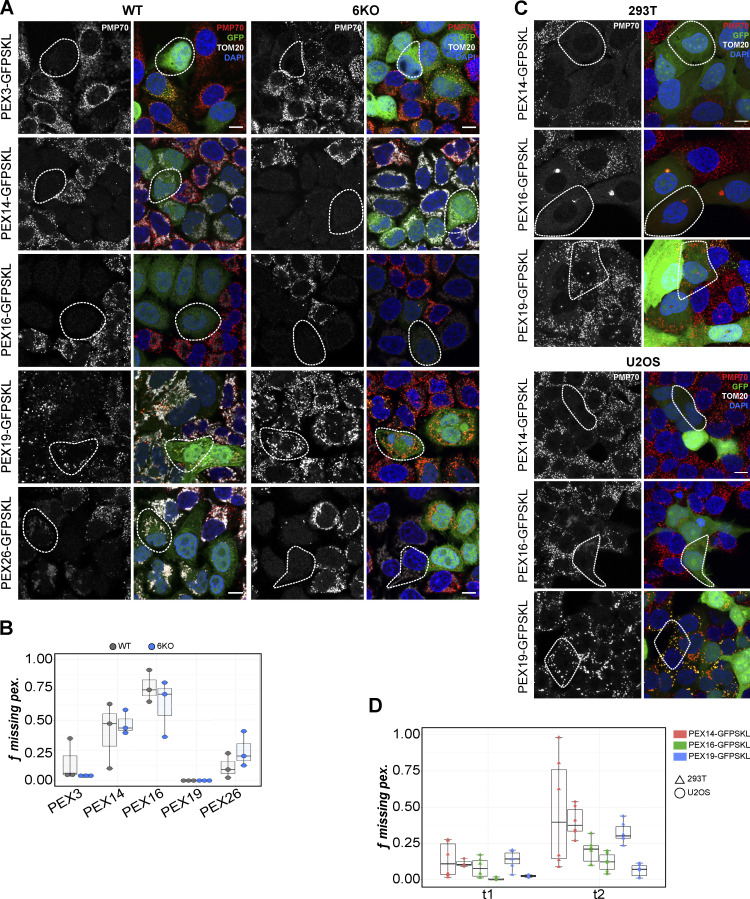
In contrast to mitochondrial morphology, overexpression of the WT GFP-Ub-VPS13D construct only slightly reduced the fraction of VPS13D-KO cells missing peroxisomes (Fig. 4 B). Unexpectedly, overexpression of VPS13D reduces peroxisomes even in WT cells (Fig. 4, D and E). It appears that the peroxisomal phenotype of HeLa cells is affected by too high or too low VPS13D expression. We asked if this phenomenon is unique to VPS13D or whether it is also found in cells overexpressing known peroxisomal proteins, particularly those found in the peroxisomal membrane. We generated viral constructs coexpressing a GFP-SKL and the untagged PEX3, 14, 16, 19, or 26 genes, separated by an internal ribosome entry site (IRES) element to avoid artifacts caused by a fusion tag. HeLa cells stably overexpressing PEX3, PEX14, PEX16, or PEX26 had cytosolic GFP-SKL and fewer PMP70-positive peroxisomal membrane structures (Fig. 5, A and B). In contrast, overexpression of PEX19, localized to the cytosol, had no impact on peroxisome amount (Fig. 5). The observed overexpression-induced disruption of peroxisomes is independent of autophagy, as PEX3, PEX14, PEX16, and PEX26 overexpression still led to peroxisome loss in Atg8 hexa-KO (6KO) cells (Nguyen et al., 2016; Fig. 5, A and B). Overexpression of PEX14 also disrupted peroxisome membrane assembly in U2OS and 293T cells, but not as severely as in HeLa cells (Fig. 5, C and D).
Figure 5.
Overexpression of peroxisomal membrane proteins disrupts peroxisomal biogenesis in WT cells. (A) Representative images of HeLa WT and ATG8 6KO cells stably coexpressing untagged PEX genes and GFP-SKL. Cells were stained for PMP70 (red in merged) and TOM20 (white in merged). The PMP70 channel is isolated as a grayscale image. (B) Box plots quantify the fraction of WT (gray) or 6KO (blue) cells missing PMP70-positive peroxisomesfollowing overexpression of each PEX gene; each dot represents an independent field of view; total cells per group: n = 562 (PEX3, WT), n = 522 (PEX3, 6KO); n = 131 (PEX14, WT), n = 285 (PEX14, 6KO); n = 95 (PEX16, WT), n = 190 (PEX16, 6KO); n = 352 (PEX19, WT), n = 388 (PEX19, 6KO); n = 370 (PEX26, WT), n = 388 (PEX26, 6KO). (C) 293T and U2OS cells transduced with the PEX-IRES-GFPSKL (green) constructs stained with PMP70 and DAPI. (D) Box plots of the fraction of PEX-overexpressing 293T (triangles) and U2OS (circles) cells missing peroxisomes 1 wk (t1) and 2 wk (t2) after transduction. Each point represents an independent field of view; mean number of cells per field: n = 262 (PEX14, 293T), n = 259 (PEX16, 293T), n = 200 (PEX19, 293T); n = 79 (PEX14, U2OS), n = 71 (PEX16, U2OS), n = 70 (PEX19, U2OS). Representative images in D are from the second time point. Exemplifying cells are highlighted by white dashed outlining.Scale bar = 10 µm. For B and D, boxes mark 25th, 50th, and 75th percentiles, and lines show the upper and lower extreme values (Tukey).
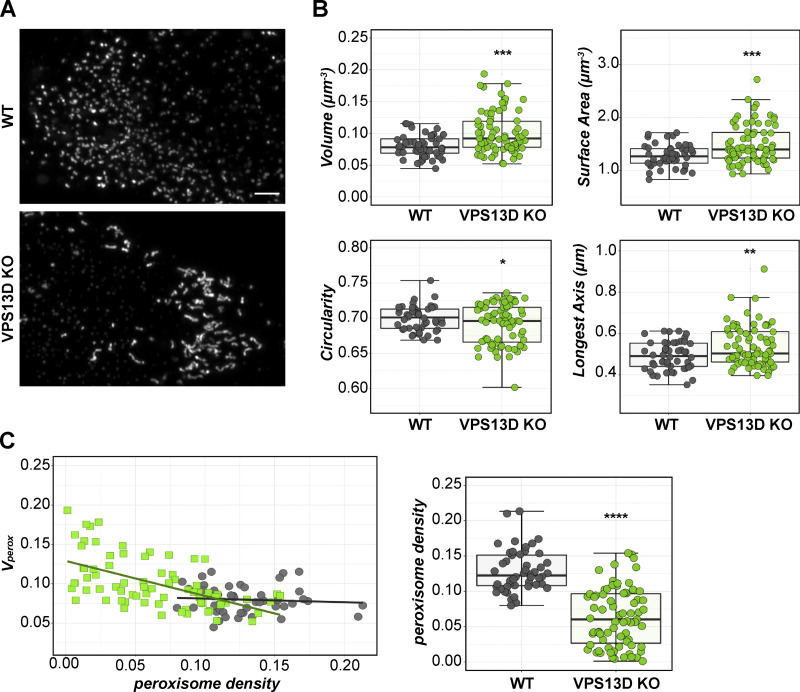
VPS13D-KO cells have morphologically distinct peroxisomes
We noticed a trend between the peroxisome loss in VPS13D-KO cells and the appearance of unusually shaped peroxisomal structures. Most peroxisomes in WT cells appear spherical and consistent in diameter, typical of mammalian cells (∼0.5 µm). In contrast, a subset of VPS13D-KO cells have larger elongated peroxisomes that form in a bent, ellipsoid-like manner (Fig. 6 A). The morphological analysis of peroxisomes visualized by PMP70 revealed a higher level of larger and less circular peroxisomal structure in VPS13D-KO cells (Fig. 6 B). Linear regression analysis shows that increased peroxisome volume is correlated with lower numbers of peroxisomes in VPS13D-KO cells (Fig. 6 C), whereas no such relationship between peroxisome size and number exists in WT cells. Abnormally shaped and elongated peroxisomes have also been observed in cells from patients with PBDs (Bjørgo et al., 2017; Hughes et al., 1990; Ebberink et al., 2010).
Figure 6.
VPS13D-KO cells have elongated peroxisomes correlating with peroxisome loss. (A) Maximum-intensity projections of super-resolution images of WT and VPS13D-KO cells stained for PMP70. (B) Morphology and size measurements of individual peroxisomes in WT or VPS13D-KO cells. Each data point represents the average values of all peroxisomes in an individual cell. Shape factor represents deviation from circularity (circle = 1). Welch’s t test; ****, P < 0.0001 (volume, surface area); **, P = 0.0024 (longest axis); *, P = 0.0358 (circularity). (C) Linear regression analysis: peroxisomal volume versus peroxisome density (number of peroxisomes per cell volume, µm−3) plot (left) inVPS13D-KO (green) or WT (gray) cells. VPS13D-KO slope = −0.5944 to −0.29242; WT slope = −0.2191 to 0.127 (95% CI); test for if slope is significantly nonzero: KO, P < 0.0001; WT, P = 0.5950. Right: Relative peroxisome density (number per cell volume) in WT and VPS13D-KO cells; Welsh’s t test; ****, P < 0.0001. VPS13D-KO 19 and 45 cells included in analysis. WT, n = 48 cells; VPS13D-KO, n = 75 cells. Scale bar = 5 µm. For B and C (right), boxes mark 25th, 50th, and 75th percentiles, and lines show the upper and lower extreme values (Tukey).
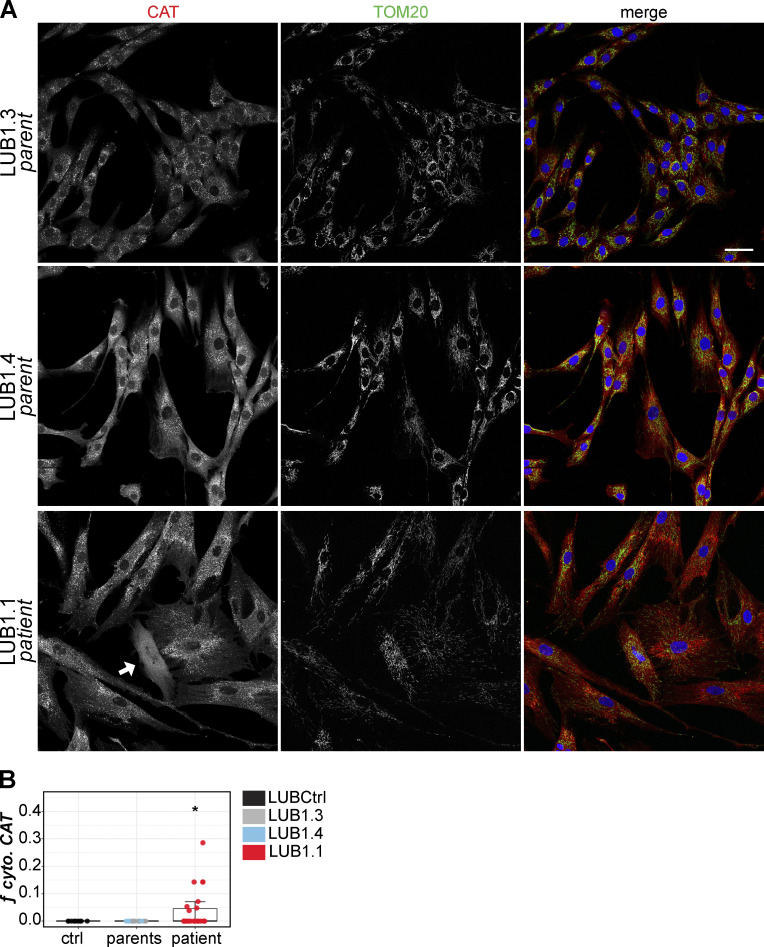
Evidence of peroxisome reduction in VPS13D mutant patient cells
VPS13D dysfunction has recently been linked to a form of spinocerebellar ataxia, SCAR4 (Gauthier et al., 2018; Seong et al., 2018). We examined fibroblasts derived from a VPS13D mutation–carrying patient and the patient’s two heterozygous parents from the German family identified as LUB1. Compound heterozygous mutations in VPS13D were identified by Seong et al. (2018) in the patient and a symptomatic sibling: a maternal nonsense mutation (Tyr1803Ter) and a paternal missense mutation in the highly conserved C-terminal region of the VPS13D gene (Ala4210Val, rs746736545). Both siblings in this family demonstrated adult-onset ataxia and neuropathy. We examined the peroxisomal phenotype of fibroblasts derived from the patient (LUB1.1) and the patient’s parents (LUB1.3, LUB1.4). Some LUB1.1 fibroblasts, but not the asymptomatic parental cells, displayed cytosolic CAT expression indicative of peroxisome loss (Fig. 7 A). We observed this phenomenon in several separate experiments, but the average proportion of cells with cytosolic CAT within a given population is <10% (Fig. 7 B). A mitochondrial morphology phenotype was reported in fibroblasts derived from an unrelated SCAR4 patient (Seong et al., 2018). We failed to identify an obvious mitochondrial phenotype in LUB1.1 cells, but we did not perform a quantitative analysis as did Seong et al.
Figure 7.
VPS13D mutation carrying patient fibroblasts also show peroxisome loss. (A) Fibroblasts from SCAR4 patient (LUB1.1; bottom panel) and the patient’s two heterozygous nonsymptomatic parents (LUB1.3 and LUB1.4; top and middle panels) immunostained for CAT (red) and TOM20 (green); nuclei were visualized by DAPI (blue). Cell with cytosolic CAT is identified with a white arrow. Scale bar = 50 µm. (B) Quantification of cytosolic (cyto.) CAT cells in LUB1.1 patient cells (red) compared with the two unaffected parental lines, LUB1.3 (light blue) and LUB1.4 (gray), or an unrelated control fibroblast line, LUBCtrl (black). Three separate experiments are grouped; each data point represents an independent field of view. Parental and unrelated control data were combined into a single group (“control”) for statistical comparison with LUB1.1 (patient). Welch’s t test; *, P = 0.0154. Per group: n = 26 (patient), n = 22 (control); boxes mark 25th, 50th, and 75th percentiles, and lines show the upper and lower extreme values (Tukey).
Peroxisomal loss in VPS13D-KO cells is independent of pexophagy
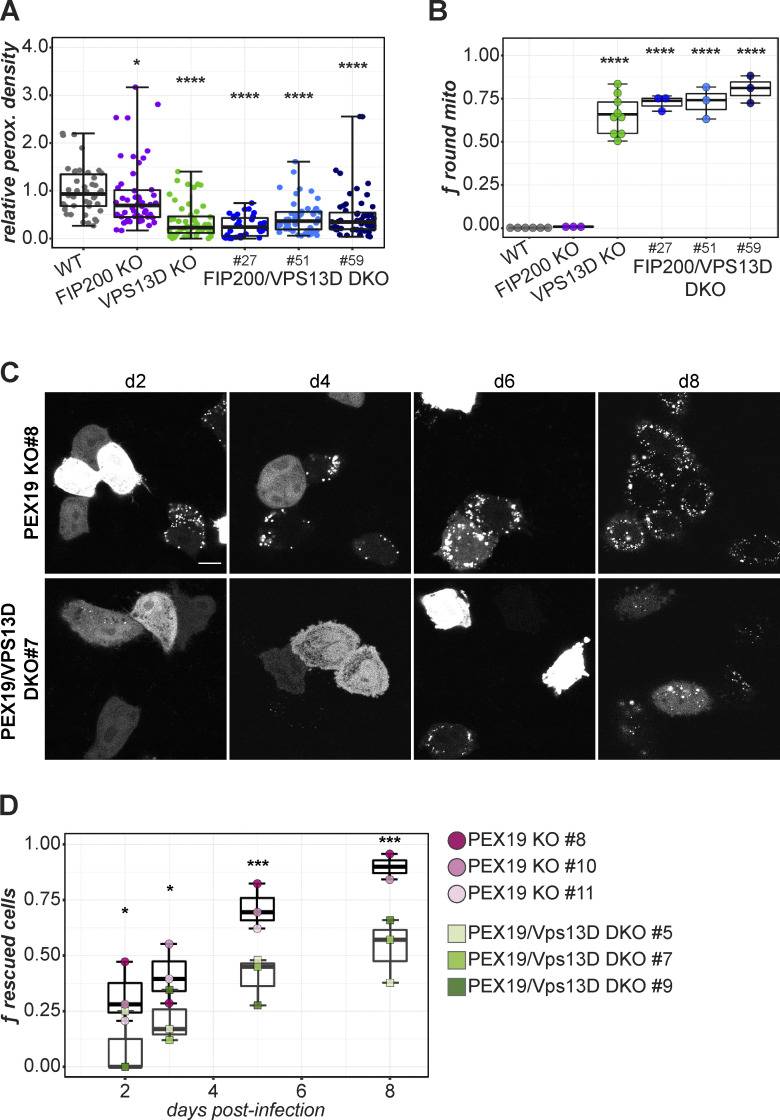
Selective autophagy of peroxisomes, pexophagy, allows the maintenance of peroxisome number in response to cellular needs and environmental cues. To test if aberrantly increased pexophagy may explain the peroxisome loss in VPS13D-KO cells, we knocked out the autophagy gene, FAK family kinase-interacting protein of 200 kD (FIP200), in the VPS13D-KO cells. The FIP200/VPS13D double-KO (DKO) cells allowed us to observe changes in peroxisomal phenotype under long-term autophagy inhibition. There was no observable difference in the proportion of cells lacking peroxisomes in the FIP200/VPS13D-DKO cells compared with VPS13D single-KO cells, and the average peroxisome amounts (relative to cell size) in three independent FIP200/VPS13D-DKO clones are not significantly different from those in VPS13D-KO cells (Fig. 8 A). It is unlikely that pexophagy is stimulated by loss of VPS13D. FIP200 loss also did not affect the mitochondrial morphology aberrations in VPS13D-KO cells (Fig. 8 B).
Figure 8.
PEX19-mediated biogenesis is attenuated by VPS13D loss. (A) Relative peroxisome density in WT, FIP200-KO, VPS13D KO 45, and three FIP200/VPS13D DKO subclones. Kruskal-Wallis and Dunn tests for multiple comparisons versus WT; ****, P < 0.0001. Cells per group: n = 42 (WT), n = 52 (VPS13D-KO), n = 50 (FIP200-KO), n = 36 (FIP200/VPS13D-DKO 27, 51), n = 56 (FIP200/VPS13D-DKO 59). (B) Quantification of round mitochondria in each cell line. Each point represents an independent field of view. One-way ANOVA, Tukey post hoc multiple comparisons test (versus WT); ****, P < 0.0001. Per group number of images (N) and total cells (n): WT, n = 6, n = 1,987; VPS13D-KO, n = 9, n = 3,179; FIP200-KO, n = 3, n = 1,016; FIP200/VPS13D-DKO 27, n = 3, n = 811; DKO 51, n = 3, n = 828; DKO 59, n = 3, n = 872. (C) Representative images of live PEX19-KO or PEX19/VPS13D-DKO cells transduced with PEX19-IRES-GFP-SKL at each time point. Peroxisome presence or absence is indicated by punctate or cytosolic GFP-SKL. Scale bar = 20 µm. (D) Time course of rescued cells (containing peroxisomes) following PEX19 reintroduction. Each dot represents the mean fraction of 6 fields of view per PEX19-KO or PEX19/VPS13D-DKO subclone. Welch’s t test; *, P < 0.05; ***, P < 0.001. Total cells across all time points: n = 252 (PEX19 KO 8), n = 196 (PEX19 KO 10), n = 189 (PEX19 KO 11), n = 226 (PEX19/VPS13D DKO 5), n = 217 (PEX19/VPS13D DKO 7), n = 228 (PEX19/VPS13D DKO 9). Mean number of cells per time point: n = 63 (PEX19 KO 8, PEX19 KO 11), n = 49 (PEX19 KO 10), n = 56 (PEX19/VPS13D DKO 5), n = 54 (PEX19/VPS13D DKO 7), n = 57 (PEX19/VPS13D DKO 9). For A, B, and D, boxes mark 25th, 50th, and 75th percentiles, and lines show the upper and lower extreme values (Tukey).
VPS13D loss attenuates PEX19-dependent peroxisome formation
Of the peroxins, only three are essential for peroxisome biogenesis in mammals: PEX3, PEX16, and PEX19. All three are involved in the import of peroxisomal membrane proteins (Fujiki, et al., 2012). The loss of any of these three genes results in a homogeneous population of cells completely lacking peroxisomes. Each of the KO lines can be rescued within days following replacement of the missing gene (South and Gould 1999; Honsho et al., 2002). If VPS13D is involved in peroxisome biogenesis, the loss of VPS13D would attenuate peroxisome formation in certain essential PEX gene KO cells following rescue with the expression of the respective gene. Peroxisome biogenesis in WT HeLa cells is disrupted by overexpression of peroxisomal membrane proteins PEX3 and PEX16, but not by the cytosolic PEX19 protein. Thus, we used PEX19-based rescue to assay the effect of VPS13D on peroxisome biogenesis. We used CRISPR to knock out PEX19 in WT and VPS13D-KO HeLa cells, and we generated the lentiviral construct pHAGE-PEX19-IRES-GFP-SKL to visualize the peroxisomal phenotype in live PEX19-KO or PEX19/VPS13D-DKO cells. The GFP signal also serves as an indicator of expression of the untagged PEX19 gene. Cells were imaged 2–8 d following PEX19-IRES-GFP-SKL transduction. These images were subject to an automatic quantification using a custom deep learning model designed to classify peroxisome absence in HeLa cells to minimize human-introduced bias while maximizing image analysis efficiency. Following reintroduction of PEX19, a growing proportion of PEX19-KO cells gain peroxisomes (as indicated by a punctate GFP signal; Fig. 8 C). The population of rescued cells increases over time, and, by 8 d after virus addition, >75% of the PEX19-KO cells displayed peroxisomes (Fig. 8 D). In contrast, only ∼50% of the PEX19/VPS13D-DKO cells had peroxisomes at the 8-d time point. Across the observed time points, VPS13D loss significantly attenuated the rescue of peroxisome biogenesis in PEX19-KO cells by PEX19 (Fig. 8 D). The impaired peroxisome rescue is consistent across three independent PEX19/VPS13D-DKO clones. These findings strongly indicate that VPS13D is involved in peroxisome biogenesis.
Discussion
Until recently, there has been little understanding of the function of VPS13D in cellular biology or human pathology since its discovery over a decade ago (Velayos-Baeza et al., 2004). In this study, we report on the role of VPS13D in peroxisome biogenesis. In contrast to the essential peroxisome biogenesis genes (PEX3, PEX16, and PEX19), VPS13D loss results in incomplete and variable peroxisome loss as populations of VPS13D-KO cells, such as PEX14-KO cells, display variable levels of peroxisome depletion. KO of VDAC2 in mouse embryonic fibroblasts also led to a heterogeneous degree of peroxisome absence (Hosoi et al., 2017). Hypomorphic PEX mutants, commonly associated with milder presentations of PBDs, also display peroxisomal mosaicism (Weller et al., 2003; Wanders and Waterham, 2006). In these cases, the severity of the peroxisomal phenotype can be increased with elevated temperature (40°C) or rescued with lower-temperature (30°C) incubation (Gootjes et al., 2004; Berendse et al., 2013; Ratbi et al., 2015). We did not observe in preliminary experiments a significant change in severity of VPS13D-KO peroxisomal phenotype after incubation at 40°C.
VPS13A and VPS13C have been found at various ER–organelle contact sites, including those with mitochondria, lipid droplets, and/or late endosomes (Yang et al., 2016; Kumar et al., 2018; Yeshaw et al., 2019). We also localized exogenous VPS13A and VPS13C to lipid droplets in HeLa cells. VPS13D expression is predominantly cytosolic, with a slight preferential localization proximal to the Golgi apparatus. Subcellular fractionation confirms the presence of VPS13D in membrane fractions. Although this observation is consistent with the Golgi-sorting function first identified for Vps13p in yeast, the connection between Golgi localization of VPS13D and its mitochondrial and peroxisomal functions remains unknown. Supporting our observations, Guillén-Samander et al. (2021) recently also reported that VPS13D is mainly diffuse in the cytosol with some variable accumulation in the Golgi complex as well as weak enrichment around mitochondria. In addition, VPS13D can be recruited to mitochondria and peroxisomes by the outer mitochondrial membrane GTPase, Miro1/2. Interestingly, the VAB/β-propeller domain of VPS13, which contains N3531S patient mutation, is required for Miro1/2 recruitment.
VPS13D is the only VPS13 family member with a UBA, and our data now link the VPS13D UBA domain to peroxisome biogenesis in addition to the previously reported role in mitochondrial morphology (Anding et al., 2018). It remains unclear how the mitochondrial and peroxisomal phenotypes in VPS13D-KO cells may be connected. Peroxisome loss has previously been linked to mitochondrial dysfunction. Swollen, rounded mitochondria were reported in the livers of mice modeling a common PEX1 variant found in Zellweger syndrome patients (Hiebler et al., 2014). Mitochondrial swelling was reported in both a PEX19 Drosophila model and PEX19-null patient fibroblasts. Bülow et al. (2018) posited that the accumulation of free fatty acids (particularly VLCFAs) and the activation of lipolysis due to peroxisome loss directly affect mitochondrial morphology. Yet, we do not find evidence of the severely rounded mitochondria characteristic of VPS13D KO in the PEX-KO HeLa cells completely lacking peroxisomes.
Mitochondria are not the only organelle whose morphology is influenced by VPS13D. Large, elliptical peroxisomes (visualized by either PMP70 or CAT) were found in VPS13D-KO cells. PEX14-KO cells display particularly striking misshaped and long peroxisomes. The finding that the prevalence of abnormally elongated peroxisomes in VPS13D-KO cells is correlated with reduced peroxisomal amount may reflect altered peroxisomal fission in an attempt to compensate for lower peroxisomal biogenesis. Elongated peroxisomes are also observed in fibroblasts derived from patients with mutations in two genes involved in peroxisomal biogenesis: PEX11β and PEX16 (Ebberink et al., 2010, 2012). The difference in peroxisomal morphology in control and VPS13D-KO cells supports a role of VPS13D in peroxisome biogenesis akin to that of other known peroxisomal biogenesis genes.
On the basis of our findings, we conclude that VPS13D affects peroxisome number through its role in peroxisome biogenesis. The specific pathways of peroxisome biogenesis are poorly understood and disputed. At least 14 PEX genes involved in peroxisome biogenesis have been identified in mammals (Tanaka et al., 2019). Likely more are yet to be discovered, for >30 peroxisome biogenesis genes are identified in yeast (Waterham and Ebberink, 2012). While the details remain to be elucidated, there are two widely accepted pathways of peroxisome biogenesis in mammalian cells: fission of preexisting peroxisomes and de novo formation from the ER. The latter was confirmed by the finding that peroxisomes are able to form in PEX3-KO cells that completely lacked any preexisting peroxisomes (Sugiura et al., 2017). In contrast to PEX3 and PEX16, PEX19 is a cytosolic protein and has an insignificant effect on peroxisome number when overexpressed in WT cells. Thus, use of a PEX19 rescue assay allowed us to directly assess VPS13D’s role in peroxisome biogenesis. We found that VPS13D loss significantly attenuates the ability of exogenous PEX19 to rescue peroxisome formation in PEX19-KO cells, identifying VPS13D as a factor in PEX19-mediated peroxisome biogenesis.
Further study is required to elucidate how direct VPS13D acts on peroxisome biogenesis and through which cellular pathway(s). There is mounting evidence of the involvement of VPS13 proteins in interorganelle contact and lipid transport. Vps13p preferentially binds to specific phospholipids (De et al., 2017; Rzepnikowska et al., 2017) and dynamically localizes to multiple membrane contact sites (Lang et al., 2015; John Peter et al., 2017; Bean et al., 2018). Recent studies indicate a similar role for mammalian VPS13A and VPS13C (Park et al., 2015; Kumar et al., 2018; Yeshaw et al., 2019). VPS13D likely contributes to peroxisome biogenesis via interorganelle lipid transport and/or other pathways regulating cellular phospholipids.
Peroxisomes are important in human neurons, as exemplified by the cognitive and neurological defects associated with PBDs (Aubourg and Wanders, 2013; Wanders et al., 2017). They also play an important role in cerebellar development (De Munter et al., 2015). Mutations in genes involved in peroxisomal biogenesis and peroxisomal lipid metabolism are linked to cerebellar defects in PBD patients and ataxia syndromes (Faust, 2003; Pierce et al., 2010; Dick et al., 2011; Buchert et al., 2014). We observed a peroxisomal phenotype in a fibroblast cell line derived from a patient with a VPS13D-linked form of spinocerebellar ataxia (SCAR4). The incidence of cells missing peroxisomes is low and much less penetrant than in the VPS13D-KO HeLa cells. Homozygous VPS13D loss is embryonic lethal, and the patient cell lines stem from partial loss of function of VPS13D, likely explaining the reduced peroxisome phenotype in the patient cells relative to the other VPS13D-KO cell lines we examined. Our finding of a partial peroxisomal defect in the VPS13D mutant patient cells may provide a critical link between VPS13D’s peroxisomal role and the neurological pathology of SCAR4 and similar disorders.
Materials and methods
All materials used in this study are detailed in Table S1.
vps13Δ yeast phenotype
vps13Δ yeast (BY74741 background) were a gift from Aaron M. Neiman (Stony Brook University, Stony Brook NY; Park et al., 2016). The WT BY74741n yeast strain and pRS425-mCherry-SKL plasmid (Joshi et al., 2016) were provided by William Prinz. Cells were grown in yeast extract peptone dextrose medium (1% Bacto yeast extract, 2% Bacto peptone, 2% glucose) at 30°C. Yeast were transformed with mCherry-SKL using standard techniques, then imaged on a Zeiss LSM 880 confocal microscope (100× objective).
Cloning
VPS13A-C Myc-His–tagged constructs were generated by inserting the ORF of the gene (VPS13A variant 1A, VPS13C variant 1A) into pcDNA4-TO-mycHis vectors (Invitrogen) according to the manufacturer’s instructions. Cloning of VPS13D-i-GFP was described previously (Anding et al., 2018). GFP-Ub-VPS13D was generated by inserting VPS13D cDNA (variant 2A) into GFP-Ub-C1 plasmid. GFP-Ub was a gift from Nico Dantuma (Addgene plasmid #11928) at Karolinska Institutet, Stockholm, Sweden (Dantuma et al., 2006). ΔN or ΔC truncated GFP-Ub-VPS13DΔN and GFP-Ub-VPS13DΔC were made as described above for WT GFP-Ub-VPS13D by cloning the last 351–13,092 (ΔN) and first 1–11,868 bp (ΔC) of VPS13D cDNA into GFP-Ub-C1. The retroviral pBMN-GFP-CAT construct was made by inserting CAT cDNA into pBMN-Z vector. All cloning is PCR based via the Gibson assembly cloning method (New England Biolabs). To generate the PEX overexpression lentiviral constructs, Gateway elements were removed from the original pHAGE vector, and new multiple cloning sites were introduced to create a new pHAGE-IRES-puromycin vector. Next, GFP-SKL is PCR amplified (SKL sequence is introduced in the reverse primer) to replace the puromycin maker to make pHAGE-IRES-GFP-SKL. For PEX gene overexpression, PEX3, PEX5, PEX14, PEX16, PEX19, and PEX26 were PCR amplified and cloned into the NheI/SalI sites to make pHAGE-PEX-IRES-GFP-SKL constructs. All plasmids were confirmed by Sanger sequencing. Complete sequence maps of each plasmid are available upon request and will be deposited to Addgene. Primers used for cloning are detailed in Table 1.
Table 1. Primers used for cloning in this study.
| Plasmid cloning | Primer | Sequence |
|---|---|---|
| GFP-Ub-VPS13D | EGFP-Ub forward | 5′-TCTAGATAACTGATCATAATCAGCCATAC-3′ |
| GFP-Ub-VPS13D | EGFPC1 forward | 5′-GATCACTCTCGGCATGGAC-3′ |
| GFP-Ub-VPS13D | EGFPC1 reverse | 5′-CATTTTATGTTTCAGGTTCAGGG-3′ |
| GFP-Ub-VPS13D | Ub-Vps13D forward | 5′-TACTCCGTCTCAGAGGTGGGATGTTGGAAGGCCTTGTAGC-3′ |
| GFP-Ub-VPS13D | Ub-Vps13D reverse | 5′-ATTATGATCAGTTATCTAGATCAGGAGTCCAGCTCCAGCTG-3′ |
| GFP-Ub-VPS13DΔC | Ub-Vps13D-dC reverse | 5′-ATTATGATCAGTTATCTAGATCAAATTGGTGTTCCACCTTG-3′ |
| GFP-Ub-VPS13DΔN | Ub-Vps13D-dN forward | 5′-TACTCCGTCTCAGAGGTGGGATGGAGGAGAAATGGAAGAATGACC-3′ |
| GFP-Ub-VPS13DΔN | Ub-Vps13D-dN reverse | 5′-ATTATGATCAGTTATCTAGATCAGGAGTCCAGCTCCAGCTG-3′ |
| GFP-Ub-VPS13DmutG1200D | Vps13D-G1200D forward | 5′-AGTATAGGTGACACCAAAGTTAATGTCTCAATG-3′ |
| GFP-Ub-VPS13DmutG1200D | Vps13D-G1200D reverse | 5′-ACTTTGGTGTCACCTATACTTGCAGTTG-3′ |
| GFP-Ub-VPS13DmutG4149S | Vps13D-G4149S forward | 5′-CCTTGTAGCCAGCATCCATGGCCTGGCTC-3′ |
| GFP-Ub-VPS13DmutG4149S | Vps13D-G4149S reverse | 5′-CATGGATGCTGGCTACAAGGTGTTCACC-3′ |
| GFP-Ub-VPS13DmutL2900S | Vps13D-L2900S forward | 5′-GGGTGCACTTCGTGGTTTGCCACCCTGAC-3′ |
| GFP-Ub-VPS13DmutL2900S | Vps13D-L2900S reverse | 5′-GCAAACCACGAAGTGCACCCCGTGTGGT-3′ |
| GFP-Ub-VPS13DmutN34107I | Vps13D-N4107I forward | 5′-GGAGTATCAATCTCTGCTGCCAAGTTTGC-3′ |
| GFP-Ub-VPS13DmutN34107I | Vps13D-N4107I reverse | 5′-TTGGCAGCAGAGATTGATACTCCGTGTGTAACATTTC-3′ |
| GFP-Ub-VPS13DmutN3521S | Vps13D-N3521S forward | 5′-CGAATTGACAGCTTTTCTAAGGTCCCGGTTG-3′ |
| GFP-Ub-VPS13DmutN3521S | Vps13D-N3521S reverse | 5′-TTAGAAAAGCTGTCAATTCGGAAAGGAG-3′ |
| GFP-Ub-VPS13DmutR4228Q | Vps13D reverse4228Q forward | 5′-CAGAGGGTTCAGAAACCGCGTTGCTGCAC-3′ |
| GFP-Ub-VPS13DmutR4228Q | Vps13D reverse4228Q reverse | 5′-CGCGGTTTCTGAACCCTCTGTGCTTGAG-3′ |
| GFP-Ub-VPS13DmutS405R | Vps13D_S405R forward | 5′-TTAGTGAACCGTCAGATCCGCTAGCGCTACCGGTCGCCACCAT-3′ |
| GFP-Ub-VPS13DmutS405R | Vps13D_S405R reverse | 5′-AGACATGATCTCGCTTTGTATACGTGTTCATACACGAGGCATC-3′ |
| GFP-Ub-VPS13DmutT1118M | Vps13D-T1118M forward | 5′-AATCCAGAGATGATTGTGGAGCTAATTGG-3′ |
| GFP-Ub-VPS13DmutT1118M | Vps13D-T1118M reverse | 5′-TCCACAATCATCTCTGGATTGAGGATAATATC-3′ |
| GFP-Ub-VPS13DmutT865A | Vps13D-T865A forward | 5′-GATTGATTTATGCTTCAGATCCCAAATATCCAG-3′ |
| GFP-Ub-VPS13DmutT865A | Vps13D-T865A reverse | 5′-GATCTGAAGCATAAATCAATCGACGCTC-3′ |
| pBMNz-GFP-CAT | pBMN-CAT_reverse | 5′-CGCGGTACCGTCGACTGCAGAATTCTCACAGATTTGCCTTCTCCCTTGC-3′ |
| pBMNz-GFP-CAT | pBMN-GFP reverse | 5′-ATAAAATCTTTTATTTTATCGTCGACTTACTTGTACAGCTCGTC-3′ |
| pBMNz-GFP-CAT | pBMNeGFP-CAT_forward | 5′-TACAAGTCCGGACTCAGATCTCGAGGGATGGCTGACAGCCGGGATC-3′ |
| pCDNA-VPS13A/C-mycHis | pcDNA-3xHA-C forward | 5′-ATCCAGCACAGTGGCGGCCGCTACCCATACGATGTTCCTGACTATGCGGGCTATCCCTATGACGTCCCGGACTATGCAGG-3′ |
| pCDNA-VPS13A/C-mycHis | pcDNA-3xHA-C reverse | 5′-GGTTTAAACGGGCCCTCTAGACTAAGCGTAATCTGGAACGTCATATGGATAGGAGCCTGCATAGTCCGGGACGTCATAGG-3′ |
| pCDNA-VPS13C-mycHis | Vps13A forward | 5′-TACTCCGTCTCAGAGGTGGGATGGTTTTCGAGTCGGTGGT-3′ |
| pCDNA-VPS13C-mycHis | Vps13A reverse | 5′-ATTATGATCAGTTATCTAGATCAGAGGCTCGGAGAAGGTTCTC-3′ |
| pCDNA-VPS13C-mycHis | Vps13C forward | 5′-TACTCCGTCTCAGAGGTGGGATGGTGCTGGAGTCGGT-3′ |
| pCDNA-VPS13C-mycHis | Vps13C reverse | 5′-ATTATGATCAGTTATCTAGATTTAAGATGGCAATTGGGGTCTGA-3′ |
| pHAGE-IRES-GFP-SKL | IRES-GFP forward | 5′-TGAAAAACACGATGATAATATGGCCACAACCATGGTGAGCAAGGGCGAG-3′ |
| pHAGE-IRES-GFP-SKL | pHAGE-GFP-SKL reverse | 5′-GTAATCCAGAGGTTGATTAGGATCCCTACAGCTTGGAACCGGACTTGTACAGCTCGTCCATGC-3′ |
| pHAGE-IRES-GFP-SKL | pHAGE-linker forward | 5′-CCTCCATAGAAGACACCGGCGGCCGCGCTAGCCTCGAGTCTAGAGTCGACACGCGTGAATTCCGCCCCCCCCCCCTAACGT-3′ |
| pHAGE-IRES-GFP-SKL | pHAGE-linker reverse | 5′-ACGTTAGGGGGGGGGGGCGGAATTCACGCGTGTCGACTCTAGACTCGAGGCTAGCGCGGCCGCCGGTGTCTTCTATGGAGG-3′ |
| pHAGE-PEX14-IRES-GFPSKL | pHAGE-PEX14 forward | 5′-CATAGAAGACACCGGCGGCCGCGCTAGCCACCATGGCGTCCTCGGAGCAG-3′ |
| pHAGE-PEX14-IRES-GFPSKL | pHAGE-PEX14 reverse | 5′-TTAGGGGGGGGGGGCGGAATTCACGCGTGTCGACCTAGTCCCGCTCACTCTCG-3′ |
| pHAGE-PEX16-IRES-GFPSKL | pHAGE-PEX16 forward | 5′-CATAGAAGACACCGGCGGCCGCGCTAGCCACCATGGAGAAGCTGCGGCTC-3′ |
| pHAGE-PEX16-IRES-GFPSKL | pHAGE-PEX16 reverse | 5′-TTAGGGGGGGGGGGCGGAATTCACGCGTGTCGACTCAGCCCCAACTGTAGAAG-3′ |
| pHAGE-PEX19-IRES-GFPSKL | pHAGE-PEX19 forward | 5′-CATAGAAGACACCGGCGGCCGCGCTAGCCACCATGGCCGCCGCTGAGGAA-3′ |
| pHAGE-PEX19-IRES-GFPSKL | pHAGE-PEX19 reverse | 5′-TTAGGGGGGGGGGGCGGAATTCACGCGTGTCGACTCACATGATCAGACACTGTTCACCAC |
| pHAGE-PEX26-IRES-GFPSKL | pHAGE-PEX26 forward | CATAGAAGACACCGGCGGCCGCGCTAGCCACCATGAAGAGCGATTCTTCGACC-3′ |
| pHAGE-PEX26-IRES-GFPSKL | pHAGE-PEX26 reverse | 5′-TTAGGGGGGGGGGGCGGAATTCACGCGTGTCGACTCAGTCACGGATGCGGAG-3′ |
| pHAGE-PEX3-IRES-GFPSKL | PEX3-IRES reverse | 5′-TTAGGGGGGGGGGGCGGAATTCACGCGTGTCGACTCATTTCTCCAGTTGCTG-3′ |
| pHAGE-PEX3-IRES-GFPSKL | pHAGE-PEX3 forward | 5′-CATAGAAGACACCGGCGGCCGCGCTAGCCACCATGCTGAGGTCTGTATGG-3′ |
Stable cell line generation, transduction, and transfection
Retroviral and lentiviral plasmids were cotransfected in 293T cells with the appropriate helper plasmids and polyethylenimine. Fresh media were added the day after transfection. Viruses were harvested 48 h and 72 h after transfection, then transduced into HeLa cells with 8 µg/ml polybrene. Stable GFP-CAT expressing VPS13D-KO cells were generated by transducing pBMN-GFP-CAT into VPS13D KO 45 cells, then sorting by FACS for optimal GFP expression. Lentiviral (pHAGE) PEX3-26 constructs were similarly stably expressed in WT and Atg8 6KO cells for measurement of peroxisome disruption by PMP overexpression. In the biogenesis assay, cells were used directly following lentiviral PEX19-IRES-GFP-SKL transduction. Transient expression of nonviral plasmids (pcD13A-1A-mH, pcD13C-1A-mH, and VPS13D-i-GFP, GFP-Ub-VPS13D, and GFP-Ub, all CRISPR constructs) into HeLa cells was done using FuGENE HD (at 1:3 ratio of DNA to FuGENE) in Opti-MEM medium. For subcellular localization experiments, all constructs were transfected at 1 µg/100 μl concentrations (total DNA dependent on size of chamber slide wells).
KO cell generation and confirmation
CRISPR sgRNA sequences targeting genes of interest were cloned into pSpCas9(BB)-2A-Puro (PX459) plasmid (Addgene #62988), a gift from Feng Zhang (Broad Institute of Massachusetts Institute of Technology, Cambridge, MA), as described in Ran et al. (2013). Two different CRISPR design strategies were used: double guides targeting introns on the 5′ and 3′ sides of the exon of interest (VPS13A-KO, VPS13B-KO, VPS13C-KO) or a single guide targeting the exon of interest (PEX3-KO, PEX16-KO, PEX19-KO). CRISPR plasmids were transfected into HeLa cells using FuGENE HD following the manufacturer’s protocol (1 µg DNA; 1:3 DNA/FuGENE ratio). Transfected cells were selected with 1 µg/ml puromycin; time of treatment was determined by total death of nontransfected puromycin-treated control cells. See Table 2 for target gRNA sequences and PCR primers.
Table 2. Target gRNA sequences and PCR primers used for KO cell generation in this study.
| Gene | Exon | Target sequence | Forward primer | Reverse primer | |
|---|---|---|---|---|---|
| PEX5 | 6 | 5′-AGGGAACAGCCACCGATCGC-3′ | 5′-GTGTGGCTAAGAGGGTCAGT-3′ | 5′-AAGTTGGTTCGAGAACGTGGAT-3′ | |
| PEX14 | 3 | 5′-TGTAGAAACTTCACTGCCG-3′ | 5′-CTGCCGGGGAGTATGAAGAA-3′ | 5′-GTCATCTTGCCGCTCCATTG-3′ | |
| PEX19 | 4 | 5′-TGCCACTGACCTTCAGGTG-3′ | 5′-TGAAAGGTCACAGTGTTATTGT-3′ | 5′-ACAGCACATCCTTGGAGAGT-3′ | |
| VPS13A | 3 | 5′ | 5′-CTTCTTTTTCTTAAAATAA-3′ | 5′-TGGGGAGAACGTTTTGAAAG-3′ | 5′-TTCAAATCGGCCCCTAGTCA-3′ |
| 3′ | 5′-CCTTCCTGGACATGCAGCC-3′ | ||||
| VPS13B | 8 | 5′ | 5′-TATCTTTCTCCTTTAGTAG-3′ | 5′-GGGTGCTGATCAGGAAATGAAG-3′ | 5′-ACGAAGGAGAGCACAGAAAGGA-3′ |
| 3′ | 5′-CCACTTGAAATCTTTGAAT-3′ | ||||
| VPS13C | 8 | 5′ | 5′-TTGAGTAGTACCTTAGCTA-3′ | 5′-GCTTCCACTGTGAAGGACGTA-3′ | 5′-TGAGACTACAGTCAAGTGACCA-3′ |
| 3′ | 5′-CTTGTGTTAGCCTCGTTTT-3′ | ||||
| VPS13D | 13 | 5′-CATACACGAGGCATCTGCAG-3′ | 5′-GCTCCTCCTGGCGGAATTTA-3′ | 5′-GACATTGGCCCACAGAGAGA-3′ | |
| VPS13D | 19 | 5′-CATACACGAGGCATCTGCAG-3′ | 5′-GCTCCTCCTGGCGGAATTTA-3′ | 5′-ACGTGCTACCCATTGAGACA-3′ | |
| VPS13D | 21 | 5′-AAGAACCTGATGGTGTCTCG-3′ | 5′-CTCCCCTTGCCCTGATTCTC-3′ | 5′-AACCCAGGTTTGCAGTGTGA-3′ | |
| FIP200 | 4–5 | 5' | 5′-ACTATGTAAAAACACCTTAG-3′ | 5′-AGACCTGATAACCAGTTTGAGCAT-3′ | 5′-TGTCAAACTTTTTGCATACTTCCT-3′ |
| 3' | 5'-GTAGTTTTAGGAATAGCAGG-3' | ||||
Single colonies were established by subcloning populations into 96-well plates. Cell lines expanded from the individual colonies are referred to as subclones in this study. When the colonies were large enough, they were sampled to be screened by PCR and ultimately confirmed by sequencing. The VPS13D KO 19 and KO 45 cell lines studied are the same as those generated for Anding et al. (2018). Generation of the VPS13DΔUBA KO line is also described in Anding et al. (2018). FIP200-KO (Vargas et al., 2019), and Atg8 6KO (Nguyen et al., 2016) cells have been described previously. For all (gene)/VPS13D-DKO cell lines, the CRISPR plasmids for the gene of interest were transfected into VPS13D KO 45 cells. All KO cell lines were confirmed with Western blotting and sequencing, with the exception of PEX19 single-KO and DKO lines. The loss of PEX19 was confirmed through phenotype (complete cytosolic expression of CAT in whole cell population).
For “pooled” VPS13D CRISPR experiments, HeLa, HCT116, 293T, and U2OS cells were similarly transfected, each with a mixture of the three gRNA plasmids targeting either exon 13, 19, or 21. After transient transfection overnight, cells were selected with puromycin for 2 d to enrich for transfection events. The total population of cells after selection was used for phenotypic analysis.
Cell culture
Human fibroblast cell lines (LUB1.1, LUB1.3, LUB1.4) were kindly provided by Katja Lohmann and Nobert Brüggemann (University of Lübeck, Lübeck, Germany). All cell lines (HeLa, HEK-293, human fibroblasts) were incubated at 37°C in 5% CO2. HeLa, HEK-293T, and U2OS cell lines were cultured with high-glucose DMEM supplemented with 10% FBS, GlutaMAX, nonessential amino acid (NEAA), Hepes, and sodium pyruvate. HCT116 cells were grown in McCoy’s 5A (modified) media, supplemented with GlutaMAX, NEAA, and 10% FBS. Human fibroblasts were cultured in high-glucose DMEM supplemented with 15% FBS, GlutaMAX, 50 IU/ml penicillin, and 50 µg/ml streptomycin. All fibroblasts used for analysis were between 5 and 10 passages old.
Subcellular fractionation
Cells harvested from a 10-cm dish were washed twice with 5 ml 1× PBS and resuspended in 4.5 ml solution B (20 mM Hepes-KOH, pH 7.6, 220 mM mannitol, 70 mM sucrose, 1 mM EDTA, 0.5 mM PMSF), then transferred to a 15-ml glass homogenizer and homogenized with 30 strokes using a drill-fitted pestle. Cell homogenates were then spun at 800 g for 10 min at 4°C to remove the nuclei, and the supernatant was collected and centrifuged at 10,000 g for 20 min at 4°C. The pellet (mitochondria-rich heavy membrane) was resuspended in 200–500 μl solution B. The cytosolic supernatant was collected and centrifuged again at 100,000 g for 30 min at 4°C to obtain the cytosol fraction, which was concentrated with TCA and finally lysed in lithium dodecyl sulfate loading buffer (Thermo Fisher Scientific). The pellet from the above ultracentrifugation was resuspended in 100–200 μl solution B to obtain the light membrane fraction.
Immunofluorescence
Cells were trypsinized, then seeded at approximately equal confluency in Lab-Tek chamber slides (∼50,000–100,000 cells for two-well slides and ∼25,000–50,000 for four-well slides). Following 16–24 h, cells underwent fixation with 4% PFA (in PBS) and were blocked with buffer (2.5% goat serum, 0.15% Triton X-100–PBS). Cells were incubated with primary antibody at RT for 2 h or at 4°C overnight, then stained with complementary (rabbit or mouse) Alexa Fluor antibody for 1 h at RT. Nuclei were stained with DAPI (1:10,000; PBS). Washes with PBS–Triton X-100 were performed between each step, and cells were stored and imaged in PBS. All immunostained cells were imaged with Zeiss LSM confocal microscope under 63× oil-immersion objective, except for the patient fibroblasts, which were acquired using the 40× oil objective.
Tracking GFP-CAT VPS13D-KO cells
GFP-CAT stable VPS13D-KO 45 cells were seeded into 384-plate wells at 1–10 cells/well. The wells containing 1–6 total cells were then imaged 10 times at 12-h intervals using a Nikon spinning disk confocal microscope under a 20× water objective, incubating at 5% CO2 and 37°C. Multiple fields of the entire well were acquired, then stitched to show the entire well. Acquisition and image stitching were performed in NIS-Elements software. Plates were removed between the 12-h imaging intervals to refresh media. For phenotypic analysis of each well at each time point, individual cells were manually classified by individual cell peroxisomal phenotype (CAT expression/localization): normal number, partial number, or completely missing peroxisomes. In cases in which a cell’s phenotype was unclear, no classification was assigned. Cells visually determined to be dead on the basis of morphology and/or absence in later time points were also counted. Cells classified as unclear or dead were included in the total number of cells per well when calculating the fraction of GFP-CAT VPS13D-KO live cells within an individual well exhibiting a normal peroxisome amount. Trend lines for each well’s fraction of normal cells over time were modeled using the locally weighted scatterplot smoothing (LOWESS) method (fraction of cells with normal peroxisomes [fNorm] vs. time) for display in graphs. Individual GFP-CAT VPS13D-KO cells were also analyzed to compare initial and final peroxisomal phenotypes. Both those found at the initial time points and any of their progeny (appearing at later time points) were separated into groups by their initial (at first time of appearance) peroxisomal phenotype (normal, partial, missing). The fraction of cells classified as either of the three peroxisomal phenotypes or dead at the last time point (5 d) were calculated for each group.
For overnight (short-term) experiments, GFP-CAT VPS13D-KO cells were plated into four-chamber Lab-Tek wells at ∼75% confluency in normal culturing media (described above). The following day, cells were refreshed with the same type of media and imaged with Zeiss LSM 880 confocal microscope with 63× oil objective, incubating at 5% CO2 and 37°C. Per experiment, 10 independent fields of view (2–4 wells of a single chamber slide) were selected as a series of positions in ZEN software (Carl Zeiss). A time series was set up to image each position every 600 (experiments B and C) or 900 (experiment A) s. Z-stacks of five slices at 2-µm intervals were acquired per position. Maximum-intensity projection time series were made for each position for manual classification and figure images. The most visible cells (∼50% per image) identified at the initial time point (first frame) were assigned a unique identifier and applied to the same individual cell or its two progeny across time series. Each unique cell was labeled “cell” for all time points if it never divided or as “parent” if it divided at some point in the time series (predivision) and then “progeny” (daughter cells after division). For analysis of initial phenotypes, cells were grouped as nondividing or dividing if labeled as cell or parent, respectively, at time point 0 (first frame). Final phenotypes were determined by the recorded phenotype of the final cell (or its progeny) at the last visible frame of the series, which occurred at 27,900–30,600 s (A), 46,200 s (B), or 90,000 s (C), depending on the experiment.
Manual quantification of VPS13A–D KO phenotypes
For quantification of mitochondrial and peroxisomal phenotypes, WT and VPS13A–D KO cells stained for translocase of outer membrane 20 (TOM20) and CAT were viewed under a Zeiss confocal microscope (LSM 410). Cells were manually counted by one of three phenotype classifications. Mitochondrial phenotype was defined by observed mitochondrial morphology: “normal” (tubular, connected), “fragmented” (nontubular but not swollen), and “swollen” (fragmented but also round and enlarged). The peroxisomal phenotype counts were classified as “normal” level of peroxisomes per cell, “partial” (peroxisomes present but only in ≤50% of the normal population per cell), and “missing” (no peroxisomes observed, completely cytosolic CAT signal). Each count consists of 100–150 cells viewed and counted from areas within distinct areas of the chamber slide well. Combined mitochondrial phenotype analysis includes data from four independent experiments. Peroxisomal phenotype is a combination of three experiments. (All three were performed in parallel with three of the mitochondrial phenotype experiments.) Each experiment consisted of from three to six technical replicates (fields within slide well) per cell line. VPS13D KO 19 and VPS13D KO 45 were included in both mitochondrial and peroxisomal quantifications. Statistical analysis was performed on the mean values (fraction of “normal” cells) of the technical replicates normalized to the WT group (per experiment). One of the experiments used for both mitochondrial and peroxisomal analyses was blinded. Due to failure to meet the assumption of variance homogeneity and/or normal distribution, measurement of significance for both mitochondrial and peroxisomal analyses used the Kruskal-Wallis rank-sum test and Dunn’s test for pairwise multiple comparisons (with no P-adjustment method applied). Peroxisome phenotype (determined by CAT, PMP70, or PEX14 immunostaining) for VPS13DΔUBA KO and PEX14, PEX5 KO experiments were manually quantified as above.
For the quantification of peroxisome phenotype in human fibroblasts, cells were seeded, fixed, and stained for CAT and DAPI. Stitched images of independent fields of view were acquired for each slide chamber. The number of cytosolic CAT cells was manually determined for each image. Manual classification was blind to which cell type the image belonged. The total number of cells per image was automatically counted using nuclei segmentation of DAPI via R (EBImage package). The fraction of cytosolic CAT to total number of cells (nuclei) was calculated by dividing the former by the latter.
Automatic/deep learning classification of phenotypes
A custom convolutional neural network (CNN) was built and trained using the Keras R interface (Chollet et al., 2017) and Tensorflow (Abadi et. al., 2016) software for the binary classification of peroxisome phenotype (“normal” versus “missing” peroxisomes). Images of individual CAT (immunofluorescence)-stained WT HeLa and PEX19-KO cells were used as examples of “normal” and “missing” phenotypes, respectively, for training the model. For quantification of PMP70 and PEX14 phenotypes, a new CNN was trained via transfer learning from the CAT CNN model with the same model architecture. A separate CNN was built and trained to classify mitochondrial phenotype of HeLa cells into either “normal” or “swollen” on the basis of images of WT and manually selected VPS13D-KO cells, respectively. The probability value of 0 indicates normal phenotype, and the probability value of 1 indicates an abnormal mitochondrial morphology phenotype. Each model was tested against a separate set of manually classified images. A custom code using the R package EBImage (Pau et al., 2010) was used for segmentation of individual cells. For either phenotype classification, each cell analyzed is assigned a probability by the deep learning model between 0 and 1, with 0 representing a normal phenotype and 1 an abnormal (missing peroxisomes or rounded mitochondria) phenotype. The probability values were thresholded near 0 or 1 to quantify “true” normal and abnormal cells, filtering out the intermediate probability values. This method was used to quantify peroxisomal or mitochondrial phenotypes in the following experiments: pooled VPS13D gRNA KO HeLa and non-HeLa cells, peroxisome biogenesis (PEX19 rescue assay), and PEX gene overexpression (in HeLa and non-HeLa cells).
Pexophagy assay
WT, VPS13D KO 45, FIP200 KO, and three FIP200/VPS13D DKO subclones were seeded into two-well Lab-Tek chamber slides. The following day, cells were fixed and immunostained for TOM20 and counterstained with DAPI. 3 × 3 single-plane images were acquired on a Zeiss LSM 880 confocal microscope using a 63× objective, then stitched using ZEN software. These images were subjected to automated classification of mitochondrial phenotype using the mitochondrial deep learning model. The fraction of cells with rounded mitochondria was calculated. A significant difference between group means was found using one-way ANOVA and the Tukey honestly significant difference multiple comparisons test.
Peroxisome biogenesis assay
Three PEX19-KO and three PEX19/VPS13D-DKO subclones were seeded into a six-well plate. The following day, cells were transduced with an equal volume of PEX19-IRES-GFP-SKL lentivirus. Approximately 24 h later, the same cells were seeded into two-well Lab-Tek chamber slides. The first imaging session was the next day (2 d after virus). The same cells were used for subsequent imaging time points. 30 min before imaging, each well was treated with 1 µM deep red anthraquinone 5 (DRAQ5) in fresh DMEM (without phenol red). 3 × 3 single-plane tiled images were acquired (Zeiss LSM 880 confocal microscope, 63× objective, incubation at 5% CO2, 37°C) at six independent (randomly selected) fields of view. Following stitching (ZEN software), images were subjected to analysis using the peroxisomal deep learning model described above. Cells were thresholded by a manually determined minimum GFP intensity (measured by EBImage) to filter out nontransduced cells for analysis.
Peroxisome morphology analysis
WT and VPS13D-KO cells were fixed and stained for PMP70. Z-stacks encompassing the entire cell of interest (step size 0.5 µm) were acquired using the Zeiss LSM 880 Airyscan high-resolution confocal microscope with a 63× objective. Cells were identified and selected within a snapshot image taken of the live scan at 1.8×. The “cropped” cell image was used to automatically determine (ZEN software) the optimal zoom and resolution parameters for acquisition. Final Z-stacks of images were processed using ZEN software’s 3D Airyscan deconvolution algorithm (processing strength automatically determined). Peroxisome morphology was quantified using the Volocity (PerkinElmer) 3D image analysis program. For each image, a manual region of interest was drawn around the cell to segment the cell before peroxisome analysis and to determine the cell volume (automatically quantified by Volocity on the basis of region of interest shape/size and Z-stack range). Peroxisome density was calculated by dividing the number of peroxisomes found by Volocity by the volume (µm3) of the cell. A combination of VPS13D KO 19 and VPS13D KO 45 cells was used in analysis. For each cell analyzed, the peroxisomal parameters (surface area, volume, longest axis, circularity) were averaged. These averaged values were used for statistical analysis (Welch’s t test and linear regression analysis). Linear regression models were made for WT and VPS13D-KO groups, plotting the average volume of peroxisomes versus peroxisome density (per individual cell). A t test was used to determine whether each group’s slope is significantly nonzero.
Peroxisome density quantification
For peroxisome quantification between dividing cells, daughter cells were identified by staining for Aurora B kinase (AIM1). Cells were imaged using Zeiss LSM 880 Airyscan microscope and ZEN software. Images were automatically processed, segmented, and analyzed via R using the EBImage package (Pau et. al., 2010). The automated program involves the creation of a maximum-intensity projection of the image Z-stack, segmentation of daughter cells (via DAPI and background fluorescence outlining cell), then further segmentation and quantification of peroxisomes (PMP70 signal) within each daughter cell. Welch’s t test found no significance between WT and VPS13D-KO groups.
A similar analysis was used in the FIP200-KO experiment, but it was performed in Python using the skimage package (van der Walt et al., 2014). This program allowed 3D segmentation and analysis directly on the Z-stack (no maximum-intensity projections created). Cells with volume <200,000 voxels were discarded from analysis. To calculate peroxisome density, the number of peroxisomes found was divided by the volume of the cell (calculated via the marching cubes algorithm in skimage). These values were then all divided by the average peroxisome density of the WT group to calculate relative peroxisomal density. Kruskal-Wallis and Dunn’s tests for multiple comparisons were used to find significance between the group means.
Image processing and statistical analysis
All figures were created in Adobe Illustrator 2020 (Adobe). All microscopic images shown in the figures were processed and converted to TIFF format in ImageJ/Fiji (Schindelin et al., 2012). Similar adjustments of brightness and contrast for visibility were used across all microscopic images within an experiment. No images underwent nonlinear transformations. In the case when Z-stacks were acquired, maximum-intensity projections were generated in Fiji for figures. The scale bars generated by Fiji based on image metadata (ZEN; Carl Zeiss) were traced in Adobe Illustrator to create a vectorized scale bar to add to images, allowing better visibility and consistency between figures. All statistical analysis was conducted in R for Mac OS (R Core Team, 2017). Graphs were created using the ggplot2 R package (Wickham, 2009). Normality of sample distribution and heterogeneity of variances between groups (for multiple comparisons tests) were tested with the Shapiro-Wilk normality function (Royston, 1982) and the Levene test (Levene, 1960) for equality of variance. P values >0.05 indicate normal distribution or homogeneity of variances and thus are suitable for post hoc analyses requiring normality. In the case where homogeneity of variance was not found, nonparametric multiple comparison tests to compare means were used. Welch’s t test was used for all two-sample comparisons.
Online supplemental material
Fig. S1 shows the results of cell viability and clonogenic growth analysis with cytochrome c immunostaining of the HeLa VPS13D-KO cells. Fig. S2 shows the phenotype of additional organelles in VPS13A–D KO cells. Fig. S3 shows localization of overexpressed VPS13A, VPS13C, and VPS13D; subcellular fractionation of VPS13D localization; and Western blotting for HeLa VPS13D KOs. Fig. S4 shows the heterogeneity of peroxisome phenotype variance in VPS13D-KO cells and PEX14-KO cells and the immunostaining of CAT, PMP70, and PEX14 in PEX5- and PEX14-KO cells. Fig. S5 shows three overnight GFP-CAT VPS13D-KO experiments used for analysis in Fig. 3, detailed via histograms of peroxisomal phenotype versus time. Table S1 lists all materials used in this study, including antibodies, cell lines, vectors and plasmids, and cell culture solutions, chemicals, etc.
Supplementary Material
lists all materials used in this study, including antibodies, cell lines, vectors and plasmids, and cell culture solutions, chemicals, etc.
Acknowledgments
We thank Dr. Katja Lohmann at University of Lübeck for the VPS13D mutant human fibroblasts. We also thank Carolyn Smith and Vincent Schram of the National Institute of Neurological Disorders and Stroke and Eunice Kennedy Shriver National Institute of Child Health and Human Development light imaging microscopy facilities. A large portion of our analysis, including the development of the deep learning models, used the computational resources of the National Institutes of Health High Performance Computing Biowulf cluster.
This work was supported by the National Institute of Neurological Disorders and Stroke Intramural Research Program.
The authors declare no competing financial interests.
Author contributions: H.A. Baldwin and C. Wang performed most of the experimental work and acquired all images. C. Wang generated the transfection and transduction constructs and cell lines with assistance from H.A. Baldwin. H.A. Baldwin performed image and statistical analyses and wrote the related code. All code, particularly related to the image classification CNN model, was adapted and/or written with guidance from G. Kanfer. M. Dulovic-Mahlow and N. Brüggemann provided the human patient fibroblasts and contributed associated discussion and project guidance. H.V. Shah provided experimental resources and assisted in the research and manuscript writing process. D. Maric performed all FACs experiments. The project was inspired by the research on VPS13D’s mitochondrial role by C. Wang, A. Anding, and E. Baehrecke. A. Velayos-Baeza provided critical experimental guidance on working with VPS13D and helpful feedback during the manuscript construction. The manuscript was written by H.A. Baldwin, C. Wang, and, R.J. Youle. All authors contributed to the editing process and provided helpful feedback during the experimental or writing process. W.A. Prinz and R.J. Youle supervised the project.
References
- Abadi, M., Agarwal A., Barham P., Brevdo E., Chen Z., Citro C., Corrado G.S., Davis A., Dean J., Devin M., et al. 2016. TensorFlow: large-scale machine learning on heterogeneous distributed systems. http://tensorflow.org
- Anding, A.L., Wang C., Chang T.K., Sliter D.A., Powers C.M., Hofmann K., Youle R.J., and Baehrecke E.H.. 2018. Vps13D encodes a ubiquitin-binding protein that is required for the regulation of mitochondrial size and clearance. Curr. Biol. 28:287–295.e6. 10.1016/j.cub.2017.11.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubourg, P., and Wanders R.. 2013. Peroxisomal disorders. Handb. Clin. Neurol. 113:1593–1609. 10.1016/B978-0-444-59565-2.00028-9 [DOI] [PubMed] [Google Scholar]
- Bean, B.D.M., Dziurdzik S.K., Kolehmainen K.L., Fowler C.M.S., Kwong W.K., Grad L.I., Davey M., Schluter C., and Conibear E.. 2018. Competitive organelle-specific adaptors recruit Vps13 to membrane contact sites. J. Cell Biol. 217:3593–3607. 10.1083/jcb.201804111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendse, K., Ebberink M.S., Ijlst L., Poll-The B.T., Wanders R.J.A., and Waterham H.R.. 2013. Arginine improves peroxisome functioning in cells from patients with a mild peroxisome biogenesis disorder. Orphanet J. Rare Dis. 8:138. 10.1186/1750-1172-8-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørgo, K., Fjær R., Mørk H.H., Ferdinandusse S., Falkenberg K.D., Waterham H.R., Øye A.M., Sikiric A., Amundsen S.S., Kulseth M.A., et al. 2017. Biochemical and genetic characterization of an unusual mild PEX3-related Zellweger spectrum disorder. Mol. Genet. Metab. 121:325–328. 10.1016/j.ymgme.2017.06.004 [DOI] [PubMed] [Google Scholar]
- Blomen, V.A., Májek P., Jae L.T., Bigenzahn J.W., Nieuwenhuis J., Staring J., Sacco R., Van Diemen F.R., Olk N., Stukalov A., et al. 2015. Gene essentiality and synthetic lethality in haploid human cells. Science. 350:1092–1096. [DOI] [PubMed] [Google Scholar]
- Brickner, J.H., and Fuller R.S.. 1997. SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J. Cell Biol. 139:23–36. 10.1083/jcb.139.1.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchert, R., Tawamie H., Smith C., Uebe S., Innes A.M., Al Hallak B., Ekici A.B., Sticht H., Schwarze B., Lamont R.E., et al. 2014. A peroxisomal disorder of severe intellectual disability, epilepsy, and cataracts due to fatty acyl-CoA reductase 1 deficiency. Am. J. Hum. Genet. 95:602–610. 10.1016/j.ajhg.2014.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bülow, M.H., Wingen C., Senyilmaz D., Gosejacob D., Sociale M., Bauer R., Schulze H., Sandhoff K., Teleman A.A., Hoch M., et al. 2018. Unbalanced lipolysis results in lipotoxicity and mitochondrial damage in peroxisome-deficient Pex19 mutants. Mol. Biol. Cell. 29:396–407. 10.1091/mbc.E17-08-0535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet, F., Allaire J.J., and Falbel D.. 2017. R interface to keras. https://keras.rstudio.com.
- Dantuma, N.P., Groothuis T.A.M., Salomons F.A., and Neefjes J.. 2006. A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J Cell Biol. 173:19–26. 10.1083/jcb.200510071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De, M., Oleskie A.N., Ayyash M., Dutta S., Mancour L., Abazeed M.E., Brace E.J., Skiniotis G., and Fuller R.S.. 2017. The Vps13p-Cdc31p complex is directly required for TGN late endosome transport and TGN homotypic fusion. J. Cell Biol. 216:425–439. 10.1083/jcb.201606078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Munter, S., Verheijden S., Régal L., and Baes M.. 2015. Peroxisomal disorders: a review on cerebellar pathologies. Brain Pathol. 25:663–678. 10.1111/bpa.12290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick, D., Horvath R., and Chinnery P.F.. 2011. AMACR mutations cause late-onset autosomal recessive cerebellar ataxia. Neurology. 76:1768–1770. 10.1212/WNL.0b013e31821a4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplomb, L., Duvet S., Picot D., Jego G., El Chehadeh-Djebbar S., Marle N., Gigot N., Aral B., Carmignac V., Thevenon J., et al. 2014. Cohen syndrome is associated with major glycosylation defects. Hum. Mol. Genet. 23:2391–2399. 10.1093/hmg/ddt630 [DOI] [PubMed] [Google Scholar]
- Dziurdzik, S.K., Bean B.D.M., Davey M., and Conibear E.. 2020. A VPS13D spastic ataxia mutation disrupts the conserved adaptor-binding site in yeast Vps13. Hum. Mol. Genet. 29:635–648. 10.1093/hmg/ddz318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebberink, M.S., Csanyi B., Chong W.K., Denis S., Sharp P., Mooijer P.A.W., Dekker C.J.M., Spooner C., Ngu L.H., De Sousa C., et al. 2010. Identification of an unusual variant peroxisome biogenesis disorder caused by mutations in the PEX16 gene. J. Med. Genet. 47:608–615. 10.1136/jmg.2009.074302 [DOI] [PubMed] [Google Scholar]
- Ebberink, M.S., Koster J., Visser G., Spronsen F., Stolte-Dijkstra I., Smit G.P.A., Fock J.M., Kemp S., Wanders R.J.A., and Waterham H.R.. 2012. A novel defect of peroxisome division due to a homozygous non-sense mutation in the PEX11β gene. J. Med. Genet. 49:307–313. 10.1136/jmedgenet-2012-100778 [DOI] [PubMed] [Google Scholar]
- Faust, P.L. 2003. Abnormal cerebellar histogenesis in PEX2 Zellweger mice reflects multiple neuronal defects induced by peroxisome deficiency. J. Comp. Neurol. 461:394–413. 10.1002/cne.10699 [DOI] [PubMed] [Google Scholar]
- Fujiki, Y., Yagita Y., and Matsuzaki T.. 2012. Peroxisome biogenesis disorders: molecular basis for impaired peroxisomal membrane assembly: in metabolic functions and biogenesis of peroxisomes in health and disease. Biochim. Biophys. Acta. 1822:1337–1342. 10.1016/j.bbadis.2012.06.004 [DOI] [PubMed] [Google Scholar]
- Gauthier, J., Meijer I.A., Lessel D., Mencacci N.E., Krainc D., Hempel M., Tsiakas K., Prokisch H., Rossignol E., Helm M.H., et al. 2018. Recessive mutations in VPS13D cause childhood onset movement disorders. Ann. Neurol. 83:1089–1095. 10.1002/ana.25204 [DOI] [PubMed] [Google Scholar]
- Goldfischer, S., Moore C.L., Johnson A.B., Spiro A.J., Valsamis M.P., Wisniewski H.K., Ritch R.H., Norton W.T., Rapin I., and Gartner L.M.. 1973. Peroxisomal and mitochondrial defects in the cerebro-hepato-renal syndrome. Science. 182:62–64. 10.1126/science.182.4107.62 [DOI] [PubMed] [Google Scholar]
- Gootjes, J., Schmohl F., Mooijer P.A.W., Dekker C., Mandel H., Topcu M., Huemer M., Von Schütz M., Marquardt T., Smeitink J.A., et al. 2004. Identification of the molecular defect in patients with peroxisomal mosaicism using a novel method involving culturing of cells at 40°C: implications for other inborn errors of metabolism. Hum. Mutat. 24:130–139. 10.1002/humu.20062 [DOI] [PubMed] [Google Scholar]
- Guillén-Samander, A., Leonzino M., Hanna M.G. IV, Tang N., Shen H., and De Camilli P.. 2021. VPS13D bridges the ER to mitochondria and peroxisomes via Miro. J. Cell Biol. 10.1083/jcb.202010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart, T., Chandrashekhar M., Aregger M., Steinhart Z., Brown K.R., MacLeod G., Mis M., Zimmermann M., Fradet-Turcotte A., Sun S., et al. 2015. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 163:1515–1526. 10.1016/j.cell.2015.11.015 [DOI] [PubMed] [Google Scholar]
- Hiebler, S., Masuda T., Hacia J.G., Moser A.B., Faust P.L., Liu A., Chowdhury N., Huang N., Lauer A., Bennett J., et al. 2014. The Pex1-G844D mouse: a model for mild human Zellweger spectrum disorder. Mol. Genet. Metab. 111:522–532. 10.1016/j.ymgme.2014.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honsho, M., Hiroshige T., and Fujiki Y.. 2002. The membrane biogenesis peroxin Pex16p. Topogenesis and functional roles in peroxisomal membrane assembly. J. Biol. Chem. 277:44513–44524. 10.1074/jbc.M206139200 [DOI] [PubMed] [Google Scholar]
- Hosoi, K.I., Miyata N., Mukai S., Furuki S., Okumoto K., Cheng E.H., and Fujiki Y.. 2017. The VDAC2-BAK axis regulates peroxisomal membrane permeability. J. Cell Biol. 216:709–722. 10.1083/jcb.201605002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, J.L., Poulos A., Robertson E., Chow C.W., Sheffield L.J., Christodoulou J., and Carter R.F.. 1990. Pathology of hepatic peroxisomes and mitochondria in patients with peroxisomal disorders. Virchows Arch. A Pathol. Anat. Histopathol. 416:255–264. 10.1007/BF01678985 [DOI] [PubMed] [Google Scholar]
- John Peter, A.T., Herrmann B., Antunes D., Rapaport D., Dimmer K.S., and Kornmann B.. 2017. Vps13-Mcp1 interact at vacuole-mitochondria interfaces and bypass ER-mitochondria contact sites. J. Cell Biol. 216:3219–3229. 10.1083/jcb.201610055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, A.S., Huang X., Choudhary V., Levine T.P., Hu J., and Prinz W.A.. 2016. A family of membrane-shaping proteins at ER subdomains regulates pre-peroxisomal vesicle biogenesis. J. Cell Biol. 215:515–529. 10.1083/jcb.201602064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, K., Ichinose Y., Ishiura H., Nan H., Mitsui J., Takahashi J., Sato W., Itoh Y., Hoshino K., Tsuji S., et al. Japan Spastic Paraplegia Research Consotium . 2019. PLA2G6-associated neurodegeneration presenting as a complicated form of hereditary spastic paraplegia. J. Hum. Genet. 64:55–59. 10.1038/s10038-018-0519-7 [DOI] [PubMed] [Google Scholar]
- Kolehmainen, J., Black G.C.M., Saarinen A., Chandler K., Clayton-Smith J., Träskelin A.L., Perveen R., Kivitie-Kallio S., Norio R., Warburg M., et al. 2003. Cohen syndrome is caused by mutations in a novel gene, COH1, encoding a transmembrane protein with a presumed role in vesicle-mediated sorting and intracellular protein transport. Am. J. Hum. Genet. 72:1359–1369. 10.1086/375454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, N., Leonzino M., Hancock-Cerutti W., Horenkamp F.A., Li P., Lees J.A., Wheeler H., Reinisch K.M., and De Camilli P.. 2018. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 217:3625–3639. 10.1083/jcb.201807019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, A.B., John Peter A.T.A.T., Walter P., and Kornmann B.. 2015. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J. Cell Biol. 210:883–890. 10.1083/jcb.201502105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage, S., Drouet V., Majounie E., Deramecourt V., Jacoupy M., Nicolas A., Cormier-Dequaire F., Hassoun S.M., Pujol C., Ciura S., et al. International Parkinson’s Disease Genomics Consortium (IPDGC) . 2016. Loss of VPS13C function in autosomal-recessive parkinsonism causes mitochondrial dysfunction and increases PINK1/Parkin-dependent mitophagy. Am. J. Hum. Genet. 98:500–513. 10.1016/j.ajhg.2016.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene, H. 1960. Intrablock and interblock estimates. In Contributions to Probability and Statistics: Essays in Honor of Harold Hotelling. Olkin I., Ghurye S.G., Hoeffding W., Madow W.G., and Mann H.B., editors. Stanford University Press, Stanford, CA. 278–292. [Google Scholar]
- Mast, F.D., and Aitchison J.D.. 2018. Characterization of peroxisomal regulation networks. Subcell. Biochem. 89:367–382. [DOI] [PubMed] [Google Scholar]
- Nguyen, T.N., Padman B.S., Usher J., Oorschot V., Ramm G., and Lazarou M.. 2016. Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J. Cell Biol. 215:857–874. 10.1083/jcb.201607039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.S., and Neiman A.M.. 2012. VPS13 regulates membrane morphogenesis during sporulation in Saccharomyces cerevisiae. J. Cell Sci. 125:3004–3011. 10.1242/jcs.105114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.S., Halegoua S., Kishida S., and Neiman A.M.. 2015. A conserved function in phosphatidylinositol metabolism for mammalian Vps13 family proteins. PLoS One. 10:e0124836. 10.1371/journal.pone.0124836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.S., Thorsness M.K., Policastro R., McGoldrick L.L., Hollingsworth N.M., Thorsness P.E., and Neiman A.M.. 2016. Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol. Biol. Cell. 27:2435–2449. 10.1091/mbc.e16-02-0112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pau, G., Fuchs F., Sklyar O., Boutros M., and Huber W.. 2010. EBImage—an R package for image processing with applications to cellular phenotypes. Bioinformatics. 26:979–981. 10.1093/bioinformatics/btq046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce, S.B., Walsh T., Chisholm K.M., Lee M.K., Thornton A.M., Fiumara A., Opitz J.M., Levy-Lahad E., Klevit R.E., and King M.C.. 2010. Mutations in the DBP-deficiency protein HSD17B4 cause ovarian dysgenesis, hearing loss, and ataxia of Perrault syndrome. Am. J. Hum. Genet. 87:282–288. 10.1016/j.ajhg.2010.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2017. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.r-project.org/. [Google Scholar]
- Rampoldi, L., Dobson-Stone C., Rubio J.P., Danek A., Chalmers R.M., Wood N.W., Verellen C., Ferrer X., Malandrini A., Fabrizi G.M., et al. 2001. A conserved sorting-associated protein is mutant in chorea-acanthocytosis. Nat. Genet. 28:119–120. 10.1038/88821 [DOI] [PubMed] [Google Scholar]
- Ramseyer, V.D., Kimler V.A., and Granneman J.G.. 2018. Vacuolar protein sorting 13C is a novel lipid droplet protein that inhibits lipolysis in brown adipocytes. Mol. Metab. 7:57–70. 10.1016/j.molmet.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratbi, I., Falkenberg K.D., Sommen M., Al-Sheqaih N., Guaoua S., Vandeweyer G., Urquhart J.E., Chandler K.E., Williams S.G., Roberts N.A., et al. 2015. Heimler syndrome is caused by hypomorphic mutations in the peroxisome-biogenesis genes PEX1 and PEX6. Am. J. Hum. Genet. 97:535–545. 10.1016/j.ajhg.2015.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royston, J.P. 1982. Algorithm AS 181: the W test for normality. Appl. Stat. 31:176–180. 10.2307/2347986 [DOI] [Google Scholar]
- Rzepnikowska, W., Flis K., Muñoz-Braceras S., Menezes R., Escalante R., and Zoladek T.. 2017. Yeast and other lower eukaryotic organisms for studies of Vps13 proteins in health and disease. Traffic. 18:711–719. 10.1111/tra.12523 [DOI] [PubMed] [Google Scholar]
- Salpietro, V., Phadke R., Saggar A., Hargreaves I.P., Yates R., Fokoloros C., Mankad K., Hertecant J., Ruggieri M., McCormick D., et al. 2015. Zellweger syndrome and secondary mitochondrial myopathy. Eur. J. Pediatr. 174:557–563. 10.1007/s00431-014-2431-2 [DOI] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert, W., Kühnisch J., Maritzen T., Horn D., Haucke V., and Hennies H.C.. 2011. Cohen syndrome-associated protein, COH1, is a novel, giant Golgi matrix protein required for Golgi integrity. J. Biol. Chem. 286:37665–37675. 10.1074/jbc.M111.267971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert, W., Kühnisch J., Maritzen T., Lommatzsch S., Hennies H.C., Bachmann S., Horn D., and Haucke V.. 2015. Cohen syndrome-associated protein COH1 physically and functionally interacts with the small GTPase RAB6 at the Golgi complex and directs neurite outgrowth. J. Biol. Chem. 290:3349–3358. 10.1074/jbc.M114.608174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong, E., Insolera R., Dulovic M., Kamsteeg E.-J., Trinh J., Brüggemann N., Sandford E., Li S., Ozel A.B., Li J.Z., et al. 2018. Mutations in VPS13D lead to a new recessive ataxia with spasticity and mitochondrial defects. Ann. Neurol. 83:1075–1088. 10.1002/ana.25220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- South, S.T., and Gould S.J.. 1999. Peroxisome synthesis in the absence of preexisting peroxisomes. J. Cell Biol. 144:255–266. 10.1083/jcb.144.2.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura, A., Mattie S., Prudent J., and McBride H.M.. 2017. Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature. 542:251–254. 10.1038/nature21375 [DOI] [PubMed] [Google Scholar]
- Tanaka, H., Okazaki T., Aoyama S., Yokota M., Koike M., Okada Y., Fujiki Y., and Gotoh Y.. 2019. Peroxisomes control mitochondrial dynamics and the mitochondrion-dependent apoptosis pathway. J. Cell Sci. 132:jcs224766. 10.1242/jcs.224766 [DOI] [PubMed] [Google Scholar]
- Ueno, S., Maruki Y., Nakamura M., Tomemori Y., Kamae K., Tanabe H., Yamashita Y., Matsuda S., Kaneko S., and Sano A.. 2001. The gene encoding a newly discovered protein, chorein, is mutated in chorea-acanthocytosis. Nat. Genet. 28:121–122. 10.1038/88825 [DOI] [PubMed] [Google Scholar]
- van der Walt, S., Schönberger J.L., Nunez-Iglesias J., Boulogne F., Warner J.D., Yager N., Gouillart E., Yu T.. scikit-image contributors. 2014. scikit-image: image processing in Python. PeerJ. 2:e453. [DOI] [PMC free article] [PubMed]
- Vargas, J.N.S., Wang C., Bunker E., Hao L., Maric D., Schiavo G., Randow F., and Youle R.J.. 2019. Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Mol. Cell. 74:347–362.e6. 10.1016/j.molcel.2019.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayos-Baeza, A., Vettori A., Copley R.R., Dobson-Stone C., and Monaco A.P.. 2004. Analysis of the human VPS13 gene family. Genomics. 84:536–549. 10.1016/j.ygeno.2004.04.012 [DOI] [PubMed] [Google Scholar]
- Wanders, R.J.A., and Waterham H.R.. 2006. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem. 75:295–332. 10.1146/annurev.biochem.74.082803.133329 [DOI] [PubMed] [Google Scholar]
- Wanders, R.J.A., Klouwer F.C.C., Ferdinandusse S., Waterham H.R., and Poll-Thé B.T.. 2017. Clinical and laboratory diagnosis of peroxisomal disorders. Methods Mol. Biol. 1595:329–342. 10.1007/978-1-4939-6937-1_30 [DOI] [PubMed] [Google Scholar]
- Wang, T., Birsoy K., Hughes N.W., Krupczak K.M., Post Y., Wei J.J., Lander E.S., and Sabatini D.M.. 2015. Identification and characterization of essential genes in the human genome. Science. 350:1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham, H.R., and Ebberink M.S.. 2012. Genetics and molecular basis of human peroxisome biogenesis disorders. Biochim. Biophys. Acta. 1822:1430–1441. 10.1016/j.bbadis.2012.04.006 [DOI] [PubMed] [Google Scholar]
- Weller, S., Gould S.J., and Valle D.. 2003. Peroxisome biogenesis disorders. Annu. Rev. Genomics Hum. Genet. 4:165–211. 10.1146/annurev.genom.4.070802.110424 [DOI] [PubMed] [Google Scholar]
- Wickham, H. 2009. ggplot2: Elegant Graphics for Data Analysis. Springer, New York. 213 pp. [Google Scholar]
- Yang, R.-Y., Xue H., Yu L., Velayos-Baeza A., Monaco A.P., and Liu F.-T.. 2016. Identification of VPS13C as a galectin-12-binding protein that regulates galectin-12 protein stability and adipogenesis. PLoS One. 11:e0153534. 10.1371/journal.pone.0153534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeshaw, W.M., van der Zwaag M., Pinto F., Lahaye L.L., Faber A.I.E., Gómez-Sánchez R., Dolga A.M., Poland C., Monaco A.P., van IJzendoorn S.C.D., et al. 2019. Human VPS13A is associated with multiple organelles and influences mitochondrial morphology and lipid droplet motility. eLife. 8:e43561. 10.7554/eLife.43561 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists all materials used in this study, including antibodies, cell lines, vectors and plasmids, and cell culture solutions, chemicals, etc.