Abstract
Severe fever with thrombocytopenia syndrome virus (SFTSV) and Heartland virus (HRTV) cause viral hemorrhagic fever-like illnesses in humans due to an aberrant host inflammatory response, which contributes to pathogenesis. Here, we established two separate minigenome (MG) systems based on the M-segment of SFTSV and HRTV. Following characterization of both systems for SFTSV and HRTV, we used them as a platform to screen potential compounds that inhibit viral RNA synthesis. We demonstrated that the NF-κB inhibitor, SC75741, reduces viral RNA synthesis of SFTSV and HRTV using our MG platform and validated these results using infectious SFTSV and HRTV. These results may lead to the use of MG systems as potential screening systems for the identification of antiviral compounds and yield novel insights into host-factors that could play role in bandavirus transcription and replication.
Keywords: Bandavirus, Severe fever with thrombocytopenia syndrome, virus, Heartland virus, Antiviral, And minigenome
1. Introduction
Severe fever with thrombocytopenia syndrome (SFTS) virus (SFTSV) and Heartland virus (HRTV) are emerging, tick-borne bandaviruses in the family Phenuiviridae. Bandaviruses have enveloped spherical virions containing tripartite single-stranded RNA with negative-sense and/or ambisense polarity (L, M, and S segments) (Hornak et al., 2016; Mendoza et al., 2019). The L and M segments encode the RNA-dependent RNA polymerase (RdRp; L protein) and surface glycoproteins (Gn and Gc), which are responsible for viral RNA replication/transcription and viral cell attachment/entry, respectively (Hornak et al., 2016; Spiegel et al., 2016). The S segment encodes two proteins: the nucleocapsid protein (N) and NSs in an ambisense manner. N encapsidates the viral genomic RNA (Jiao et al., 2013; Zhou et al., 2013), whereas NSs is known as a virulence factor, which dysregulates host innate immune responses, including the inhibition of interferon (IFN)-mediated antiviral and induction/suppression of the nuclear factor-κB (NF-κB)-mediated inflammatory response (Li et al., 2020; Qu et al., 2012; Sun et al., 2015). NF-κB pathway is major signaling pathway that induces the host inflammatory response leading to the expression of cytokines, chemokines, and other inflammatory mediators (Zhang et al., 2017).
SFTS in humans caused by SFTSV was first identified in 2009 in China, with subsequent cases reported in South Korea, Japan and Taiwan (Yu et al., 2011; Choi et al., 2016; Kim et al., 2018; Takahashi et al., 2014; Kurihara et al., 2016; Kobayashi et al., 2020; Lin et al., 2020). Case fatality rates range from 6 to 30% depending on the country (Kobayashi et al., 2020; Robles et al., 2018). Human-to-human cases of SFTS have been reported in China and zoonotic transmission from companion animals to humans has been described (Bao et al., 2011; Z. Gai et al., 2012a; Liu et al., 2012; Jiang et al., 2015; Jung et al., 2019; Kida et al., 2019; Yoo et al., 2019). HRTV was identified in the United States in 2009 with the two cases of SFTS-like febrile illness in Missouri (McMullan et al., 2012) with at least 50 cases of HRTV-infection reported, including three fatalities (“Statistics & Maps | Heartland virus | CDC,” 2020).
The clinical symptoms and disease course of SFTS and HRTV disease (HRTVD) in humans are characterized by high fever, thrombocytopenia, leukopenia, multiple organ involvement, and in severe and fatal cases, neurological abnormalities and hemorrhagic fever-like manifestations (Yu et al., 2011; McMullan et al., 2012; Z. T. Gai et al., 2012b; Pastula et al., 2014; Kato et al., 2016; Brault et al., 2018; Saijo, 2018). The sustained high serum viral load, uncontrolled systemic inflammatory, and impairment of adaptive immune response are considered to be a prognostic marker associated with fatal outcome (Z. T. Gai et al., 2012b; Deng et al., 2013; Ding et al., 2014; Sun et al., 2016). While experimental vaccines and therapeutic candidates have been reported by several groups (Dong et al., 2019; Kwak et al., 2019; Tani et al., 2016a, 2018; Westover et al., 2017), no vaccines or therapeutics against SFTSV or HRTV have been approved and licensed for use in humans. T-705 (Favipiravir), a nucleoside analog, has demonstrated efficacy in vitro and in type I IFN signaling-deficient rodents with SFTSV or HRTV infections (Tani et al., 2016a, 2018; Westover et al., 2017).
Here, we developed, characterized, and optimized the SFTSV and HRTV minigenome (MG) systems for use in the screening of anti-inflammatory compounds, which also exhibit inhibitory activity on viral RNA transcription and replication processes. Screening potential antiviral compounds in vitro under biosafety-level 2 conditions is advantageous as it provides an avenue for rapid evaluation of therapeutic candidates without the use of infectious virus in a biosafety level-3 facility.
2. Materials and methods
2.1. Chemical compounds
Doxycycline was purchased from Millipore Sigma (cat. No. D9891; Burlington, MA). Tigecycline (cat. no. A10933), T-705 (cat. no. A11590), and SC75741 (cat. no. A14278) were purchased from AdooQ (Irvine, CA). Tetracycline (cat. no. S2574), minocycline (cat. no. S4226), ribavirin (cat. no. S2504), doxorubicin (cat. no. S1208), mesalamine (cat. no. S1681), and wortmannin (cat. no. S278) were purchased from SelleckChem (Houston, TX). GYY4137 (cat. no. 13345), LY294002 (cat. no. 170920), and BAY11-7082 (cat. no. 100102) were purchased from Cayman Chemicals (Ann Arbor, MI). Compounds were resuspended in DMSO upon receipt and stored at −80 °C.
2.2. Cells and viruses
HEK293 (ATCC, CRL-1573), and Vero E6 cells (ATCC, CRL-1586) were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with sodium pyruvate and L-glutamine (Thermo-Fisher Scientific, Waltham, MA) supplied with heat-inactivated fetal bovine serum (FBS; Thermo-Fisher Scientific) and penicillin-streptomycin (Millipore Sigma, Burlington, MA). Huh7 cells (a kind gift from Yoshiharu Matsuura, Osaka University) were maintained in DMEM as described above. THP-1 cells (ATCC, TIB-202) were maintained in RPMI 1640 medium containing L-glutamine (Thermo-Fisher Scientific) supplemented with 10% FBS (Thermo-Fisher Scientific), penicillin-streptomycin (Millipore Sigma), non-essential amino acids, 10 mM HEPES, sodium pyruvate, and β-mercaptoethanol to a final concentration of 0.05 mM.
The SFTSV and HRTV strains used in this study were kindly provided by the Reference and Reagent Laboratory, Arboviral Diseases Branch, Centers for Disease Control and Prevention (CDC); and the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) arthropod-borne virus reference collection at the University of Texas Medical Branch (UTMB). SFTSV YL1 was isolated in 2011 from acute-phase serum from a Chinese patient (UTMB Arbovirus collection; virus pool number TVP 16907) and passaged twice in Huh7 cells prior to titration on Vero E6 cells. HRTV isolate R99207b from a male Tennessee patient in 2013 (CDC Arbovirus reference collection) was passaged 3 times in Huh7 cells.
2.3. Titration of viral stocks
Vero E6 cells were infected with 10-fold serial dilutions of viral stocks. Following adsorption for 1 h at 37 °C, 1.2% carboxymethylcellulose (CMC; Millipore Sigma) was added to the viral inoculum. Cells were incubated for 2 days for SFTSV and 3 days for HRTV following addition of CMC prior to fixation in 10% neutral buffered formalin, and left at 4 °C overnight. Cells were washed thoroughly in PBS (phosphate buffered saline (1x)) followed by fixation using a 1:1 ratio of methanol and acetone, and washed twice with PBS prior to blocking for 1 h at room temperature in PBS containing 5% goat serum (Millipore Sigma), 1% bovine serum albumin (BSA, Millipore Sigma). Primary antibodies for SFTSV and HRTV (UTMB YL1 and MO-4 anti-body; a generous gift from Robert Tesh at UTMB) were added to the cells at a concentration of 1:500 in PBS containing 1% BSA and incubated at 4 °C overnight. Visualization of viral foci was accomplished by using goat anti-mouse AlexaFluor 488 (ThermoFisher Scientific).
2.4. SFTSV and HRTV growth kinetics
THP-1 cells were seeded into 6-well plates at a cell density of 3 × 106 cells/well and treated with 75 nM Vitamin D3 (Millipore Sigma) for 48 h prior to infection to differentiate the cells without skewing them towards an M1 or M2 phenotype (Daigneault et al., 2010). HEK293 (4 × 105/well) and Huh7 (3 × 105/well) cells were seeded one day prior to infection. Cells were infected at an MOI of 0.1 and washed three times with serum-free media following adsorption in order to remove un-bound virus. 3 ml of cell medium containing 2% FBS was added to the cells, and 0.5 ml of supernatant samples were harvested for up to 5 days post-infection (p.i.) and replaced with an equal volume of fresh medium supplemented with 2% FBS. Samples were spun down in a micro-centrifuge for 10 min at 3000 rpm and aliquoted in prior to storage at −80 °C.
2.5. In vitro efficacy of NF-kB inhibitors on SFTSV and HRTV replication
Huh7 cells were seeded in 24-well plate at a cell density of 0.8 × 105 cells per well, and were infected at an MOI of 0.1 for 1 h. Following removal of viral inoculum, cells were washed three times with serum-free media, and replaced with media containing 2% FBS and indicated compounds at concentrations ranging from 0.048 μM to 50 μM (1 well per compound per concentration). Infected cells were treated with indicated compounds for 3 days prior to harvest of cell supernatants for virus titration. Supernatants were spun down at 3000 rpm for 10 min and were stored at −80 °C and titrations were performed in triplicate for each dilution in VeroE6 cells. Inhibitory concentration values at 50% and 90%, IC50 and IC90 respectively, were calculated using nonlinear regression analysis using Prism 8 software (Graphpad). Data are represented as an average of three independent experiments.
2.6. Generation of SFTSV/HRTV minigenome plasmids and helper plasmids
Minigenome (MG) plasmids were generated based on the M-segment of the SFTSV WSQ strain (GenBank accession number HQ419235.1) and HRTV TN strain (KJ740147.1) where the open reading frame (ORF) was replaced with a NanoLuciferase (nLuc) gene. The 3′ and 5’ untranslated regions (UTRs), which are important for initiation of transcription and replication by the viral RNA-dependent RNA polymerase (Ren et al., 2020) were left intact. The human RNA polymerase I (hPolI) MG plasmid possesses hPolI promoter and terminator sequences as previously described (Neumann et al., 1994; Tsuda et al., 2017) and the T7-polymerase (T7-pol) MG plasmid possesses T7 bacteriophage promoter and terminator sequences. Both hPolI and T7-pol MG plasmid systems also contain Hepatitis δ ribozyme (HDV) sequences. Helper plasmids containing the ORFs of the SFTSV/HRTV RNA-dependent RNA polymerase (SFTSV L: HQ419226.1; HRTV L: KJ740148.1) or nucleocapsid (SFTSV S: HQ419239.1; HRTV S: KJ740146.1) protein were generated by cloning the ORFs into a pCAGGs vector via restriction digest and ligation. All plasmids were verified via Sanger Sequencing.
2.7. Rescue, characterization and compound treatment of SFTSV and HRTV MGs
Huh7 or HEK293 cells were seeded at a cell density of 1.6 × 105 cells/well in 12-well plates. 18–24 h post-seeding, cells were transfected using TransIT-LT1 with MG plasmid expressing nLuc, helper plasmids, T7-pol-expressing plasmid, and a firefly luciferase (ffLuc)-expressing plasmid as a transfection control. Cells were transfected with 1000 ng L, 500 ng N, 250 ng MG-nLuc, 100 ng T7-pol, and 10 ng ffLuc control per well. Rescue of the hPolI promoter-dependent MG were similar as described above for the T7-dependent MG without the supplementation of T7-pol. Cells were harvested at indicated time points post-transfection (p.t.) using PLB (Promega), and luciferase activity was assessed by subjecting 20 μl of the cell lysate to the Nano-Dual Luciferase assay (Promega). MG activity was normalized to the polymerase (L) negative control. For compound treatment, cells were scaled up to T75 flasks, transfected with MGs and helper plasmids, and reseeded 23 h p.t.. 6 h post-seeding, cells were treated with indicated compounds for 24 h before equal amounts of the cell lysates were subject to NanoDual Luciferase assay and CellTiter Glo assay.
2.8. Biosafety
All experiments using infectious SFTSV and HRTV were performed in a biosafety level 3 (BSL3) facility at the Mayo Clinic in accordance with approval and guidelines provided by the Mayo Clinic Institutional Biosafety Committee (IBC). Sample inactivation and removal from the facility was performed in accordance to standard operating protocols approved by the IBC.
2.9. Statistical methods
Experiments were performed as three biological replicates and calculation of mean values and standard deviation values was performed by using GraphPad Prism 8 version 8.4.2 (La Jolla, CA). Statistical analyses were performed using GraphPad Prism (La Jolla, CA).
3. Results
3.1. Establishment of MG systems for HRTV and SFTSV
We developed two MG systems based on the T7-pol and hPolI-mediated transcription for both SFTSV and HRTV (Fig. 1A). To assess successful expression of our nLuc reporter gene from MG, we compared the MG activity with or without L helper plasmid transfection at 72 h p.t. in Huh7 (Fig. 1B) (Chen et al., 2017; Tani et al., 2016b). We observed a three to four-log induction of MG activity for both HRTV and SFTSV MGs relative to the negative control, which is similar to the T7-based MG systems established by other groups (Rezelj et al., 2019). Next, we monitored kinetics of MG activities in Huh7 cells and found a gradual increase in MG activity out to 72 h p.t. (Fig. 1C–F, Supplementary Fig. 1). To understand the correlation between the concentration of the RNA transcription/replication complex and MG activity, we performed titrations using the T7-pol MG systems (Supplementary Fig. 2) and found that a reduction in helper plasmid expressing either the L or the N proteins still resulted in similar MG activity in both HEK293 and Huh7 cells at 48 h p.t..
Fig. 1. Bandavirus minigenome (MG) constructs and kinetics.
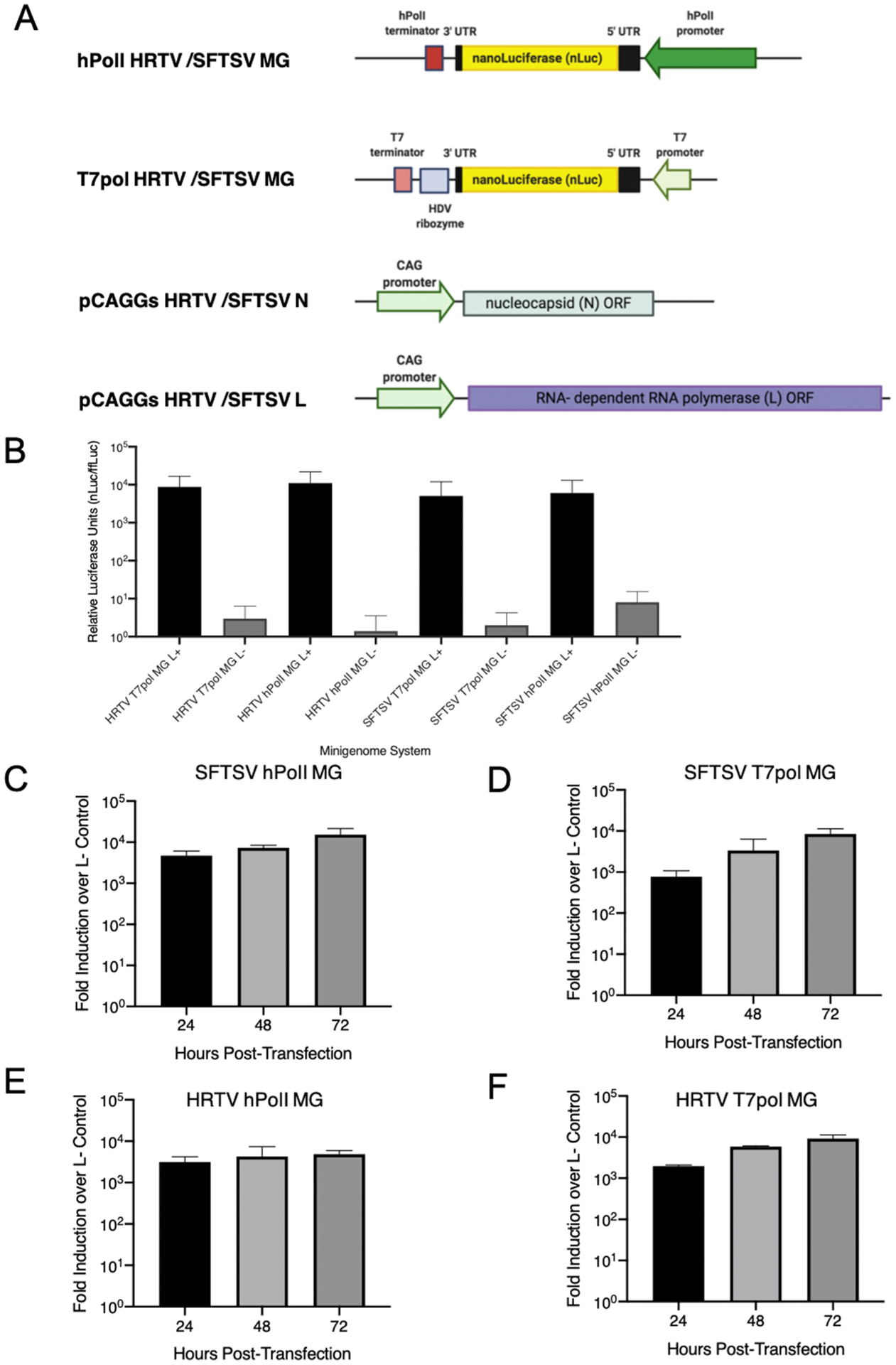
Schematics of the M-segment based MG nLuc plasmids used in this study (T7 Promoter-driven and Human PolI promoter-driven) are shown in (A) in addition to helper plasmids. (B) Comparison between HRTV and SFTSV T7pol promoter and hPolI promoter with and without supplementation of L helper plasmid in Huh7 cells at 72 h post-transfection and kinetics of different MG plasmids for SFTSV and HRTV (C–F). Data are representative of the average of three independent experiments.
3.2. Screening of compounds which inhibit viral RNA synthesis
We assessed the capacity of the established MG systems to serve as a screening platform for compounds that could inhibit viral RNA synthesis. SFTSV and HRTV infection result in an aberrant inflammatory response, thus we selected compounds that had previously been reported to have anti-inflammatory capabilities, as well as antiviral efficacy (Bazhanov et al., 2017; Cameron and Castro, 2001; Dunn et al., 2009; Ehrhardt et al., 2013, 2013; 2013; Furuta et al., 2017; Rothan et al., 2014; Sun et al., 2015; Wu et al., 2015) in Supplementary Table 1.
We found that the NF-κB inhibitors, SC75741 and LY294002 reduced viral RNA synthesis while the other compounds did not at the concentrations tested (Fig. 2, Supplementary Table 1). SC75741 had a half-maximal inhibitory concentration (IC50) value of 0.662 μM for the SFTSV hPolI MG (Fig. 2A, top) and 2.973 μM for the SFTSV T7-Pol MG (Fig. 2B, top). Similarly, SC75741 was effective in reducing the MG activity for both HRTV MG systems: HRTV hPolI MG IC50: 0.4119 μM (Fig. 2C, top).; HRTV T7-Pol MG IC50: 0.333 μM (Fig. 2D, top). Interestingly, LY294002 only reduced SFTSV MG activity for both systems (Fig. 2A and B bottom panels), but not in either HRTV MG system (Fig. 2C and D bottom panels). We noted cell toxicity in our initial MG studies when cells were treated with SC75741 compared to our DMSO vehicle control.
Fig. 2. Efficacy of NF-κB inhibitors in SFTSV and HRTV M-segment hPolI and T7pol MG systems.
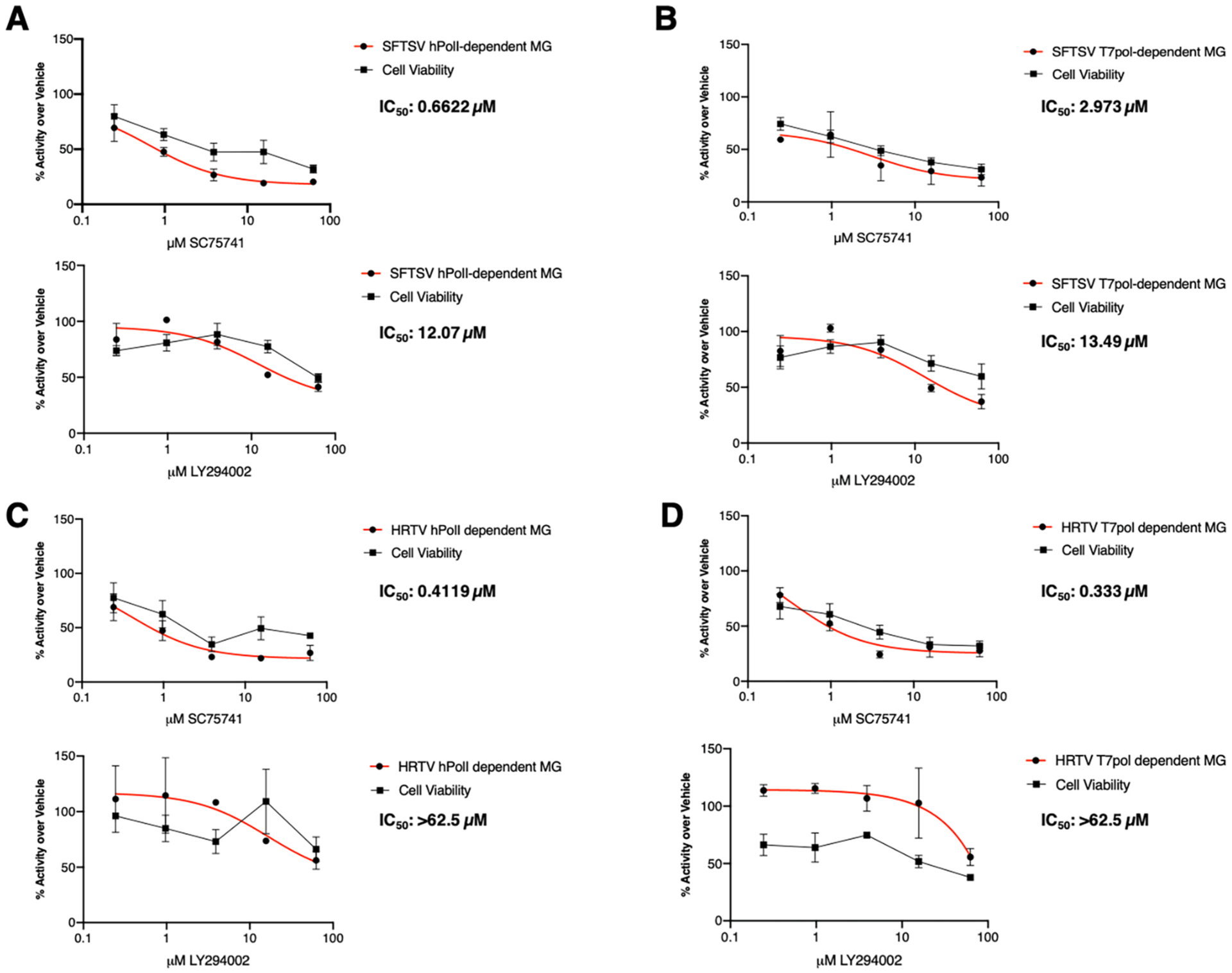
For SFTSV, Huh7 cells were transfected with either SFTSV M-segment Human PolI promoter-driven (A) or SFTSV M-segment based T7 promoter-driven (B) MG systems and treated with SC75741 (top) or LY294002 (bottom) at the indicated concentrations for 24 h. For HRTV, Huh7 cells were transfected with either HRTV M-segment Human PolI promoter-driven (C) or HRTV M-segment based T7 promoter-driven (D) MG systems and treated with SC75741 (top) or LY294002 (bottom) at the indicated concentrations. MG activity was assessed via dual-luciferase assay and cell viability was assessed via CellTiter-Glo assay. Half-maximal inhibitory concentrations (IC50) were calculated using non-linear regression analysis using Prism 8 software (GraphPad). Data are representative of the average of three independent experiments.
T-705, which has shown efficacy in vitro and in vivo against SFTSV and HRTV (Takayama-Ito and Saijo, 2020; Tani et al., 2016b; Westover et al., 2017), interestingly was not efficacious under our initial MG conditions tested (Supplementary Table 1). Under reduced helper plasmid conditions, we observed reduction of MG activity for the SFTSV hPolI MG system with SC75741 and LY294002 (Fig. 3A and B), but only saw a 20% decrease using T-705 at the concentrations tested (Fig. 3C).
Fig. 3. Efficacy of SC75741, LY294002, and T-705 in SFTSV hPolI MG system with reduced helper plasmids.
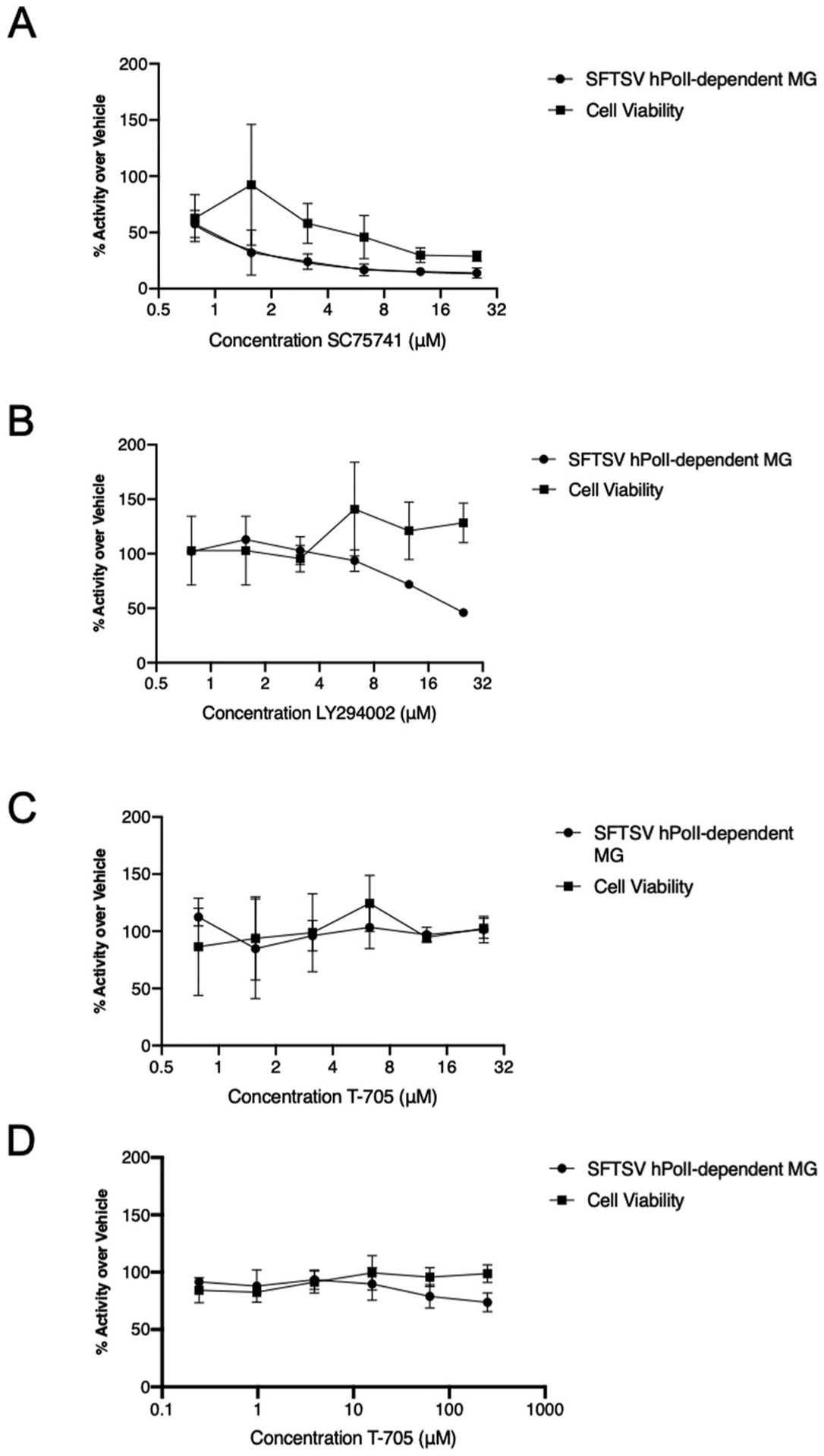
Huh-7 cells transfected with the SFTSV PolI MG plasmid and denoted helper plasmids (125 ng pCAGGs L, 125 ng M-seg MG nLuc, 62.5 ng pCAGGs N, and 10 ng pCAGGs ffLuc) were treated with 2-fold dilution concentrations of SC75741 (A), LY294002 (B), T-705 (C) or 4-fold dilutions of T-705 (D) at 24 h post-transfection. Cell lysates were harvested 24 h post-compound addition and equal amounts of the cell lysates subjected to dual luciferase assay for MG activity and CellTiter-Glo assay for cell viability. Data are representative of the average of three independent experiments.
3.3. Kinetics of SFTSV and HRTV
Although there are no report showing a direct comparison of SFTSV and HRTV growth kinetics, our data suggested MG-driven transcription/replication was higher using the SFTSV plasmids compared to HRTV plasmids, thus we compared growth of SFTSV and HRTV to understand their ability to replicate in different human cell lines. Overall, we observed that SFTSV grew to higher titers than HRTV in HEK293 and Huh7 cells, while both viruses grow to similar titers in THP-1 monocytes (Fig. 4A–C).
Fig. 4. SFTSV and HRTV kinetics in human cell lines.
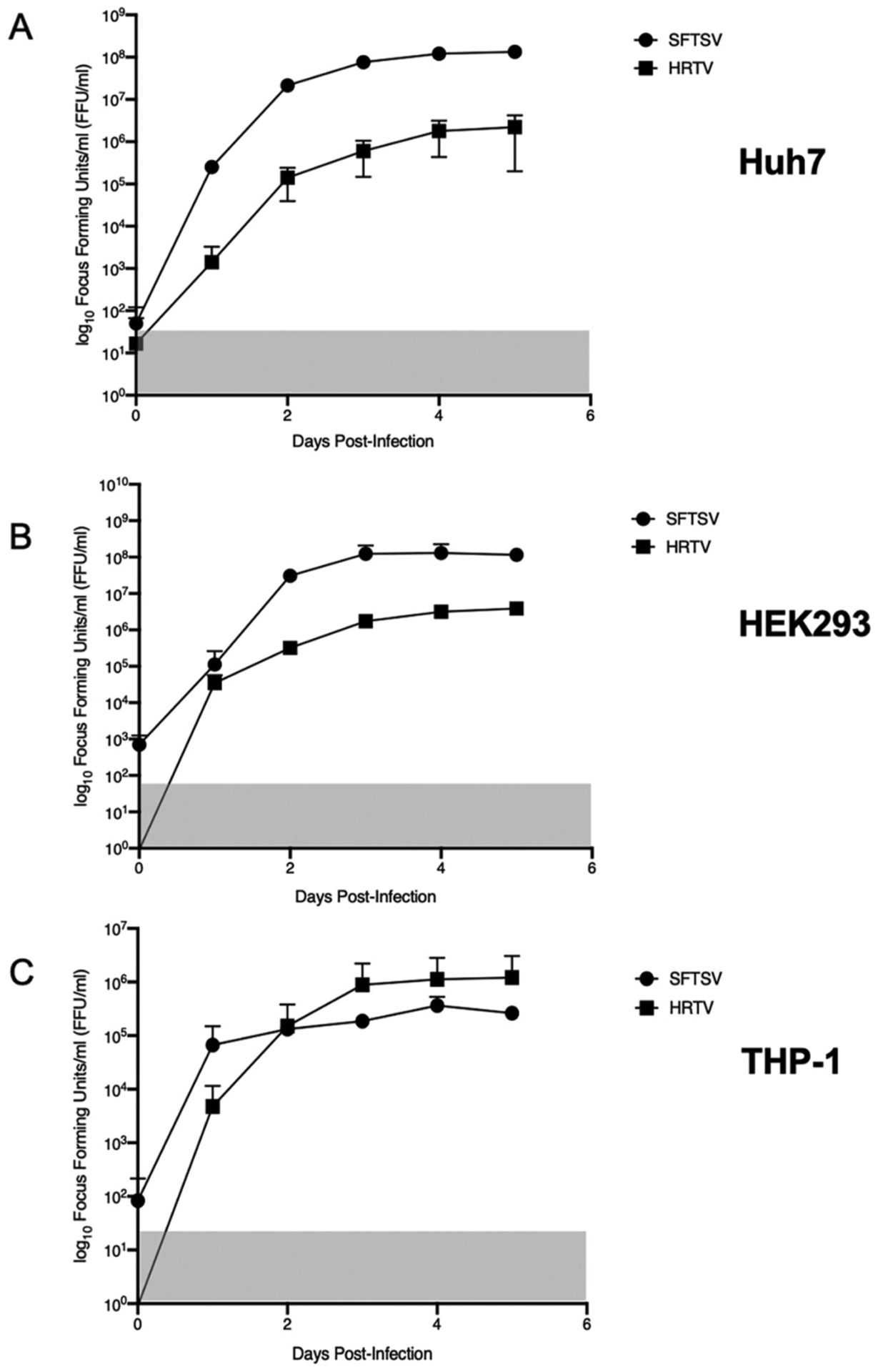
Huh7 (A), HEK293 (B), or THP-1 (C) cells were infected with SFTSV YL1 or HRTV R99207 b at an MOI of 0.1 for 1 h and monitored for 5 days post-infection. Cell supernatants containing virus were titrated on Vero E6 cells. Data are representative of the average of three independent experiments for Huh7, HEK293, and THP-1 cells.
3.4. SC75741 reduces HRTV and SFTSV viral titers in vitro
We assessed antiviral efficacy of SC75741, LY294002, and T-705 in cells infected with either SFTSV or HRTV at an MOI of 0.1 (Figs. 5 and 6, respectively). Similar to previous studies, we observed similar IC50 and IC90 values for T-705 treatment in either SFTSV or HRTV infected Huh7 cells (IC50, IC90 for SFTSV: 2.262 μM, 20.358 μM; IC50, IC90 for HRTV: 3.539 μM, 31.85 μM) (Tani et al., 2016a; Westover et al., 2017) (Fig. 5E and F for SFTSV and Fig. 6E–F). Interestingly, LY294002 treatment mirrored our observations from our MG systems: SFTSV was more sensitive to LY294002 treatment compared to HRTV (IC50, IC90: 12.72 μM, >50 μM for SFTSV vs. 18.86 μM, >50 μM for HRTV) (Fig. 5C, D for SFTSV and Fig. 6C and D for HRTV). SC75741 is a potent inhibitor of both SFTSV and HRTV replication in vitro with an IC90 value for HRTV of 1.85 μM and SFTSV of 2.1 μM (IC50: 0.2337 μM for SFTSV vs. 0.2061 μM for HRTV) (Fig. 5A and B for SFTSV and Fig. 6A and B for HRTV). We observed minimal cytoxicity of SC75741 and LY294002 at lower concentrations tested, however we did visualize cytotoxicity at concentrations above 25 μM in virus-infected Huh7 cells treated with compounds. No toxicity of T-705 treatment at higher concentrations was noted. We performed cytotoxicity assays in uninfected Huh7 cells and saw no significant difference between cells treated with SC75741, LY294002, T-705 or DMSO compared to cells with media alone at concentrations lower than 25 μM for the SC75741 and LY294002 (CytoToxOne Membrane Integrity Assay, Promega; data not shown). Together, these results indicate that the NF-κB inhibitor SC75741 is a novel inhibitor of two pathogenic bandaviruses.
Fig. 5. SC75741 reduces SFTSV viral titers in Huh7 cells.
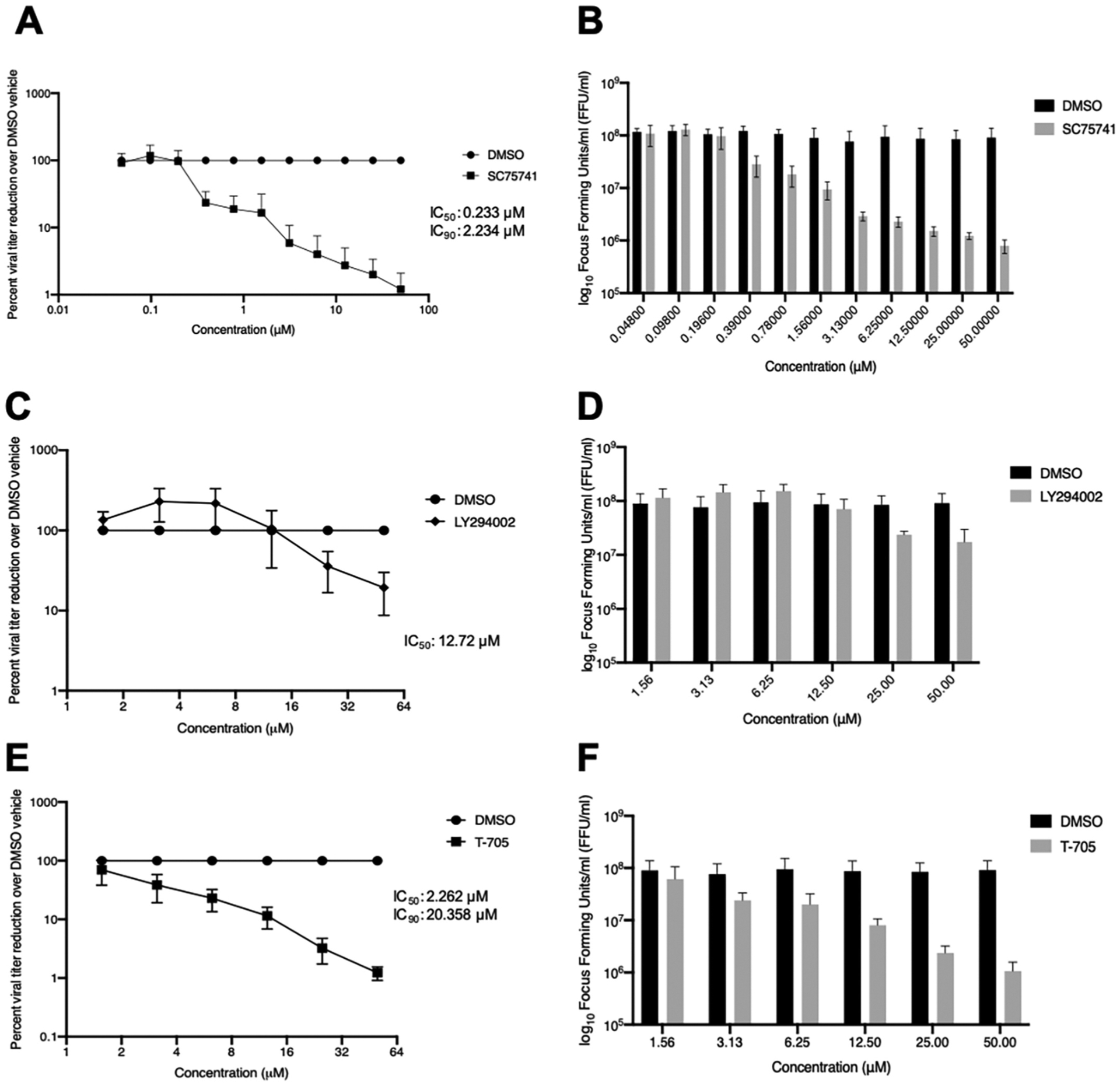
Huh7 cells were infected with SFTSV YL1 strain for 1 h and treated with SC75741 (A,B), LY294002 (C, D), or T-705 (E, F) 1-h post-infection and incubated for three days prior to titration on VeroE6 cells. Data are shown as percent viral titer reduction (A, C, E) or log10 focus forming units/mL (B, D, F). Inhibitory concentration values at 50% and 90%, IC50 and IC90 respectively, were calculated using nonlinear regression analysis using Prism 8 software (Graphpad). Data are represented as an average of three independent experiments.
Fig. 6. SC75741 reduces HRTV viral titers in Huh7 cells.
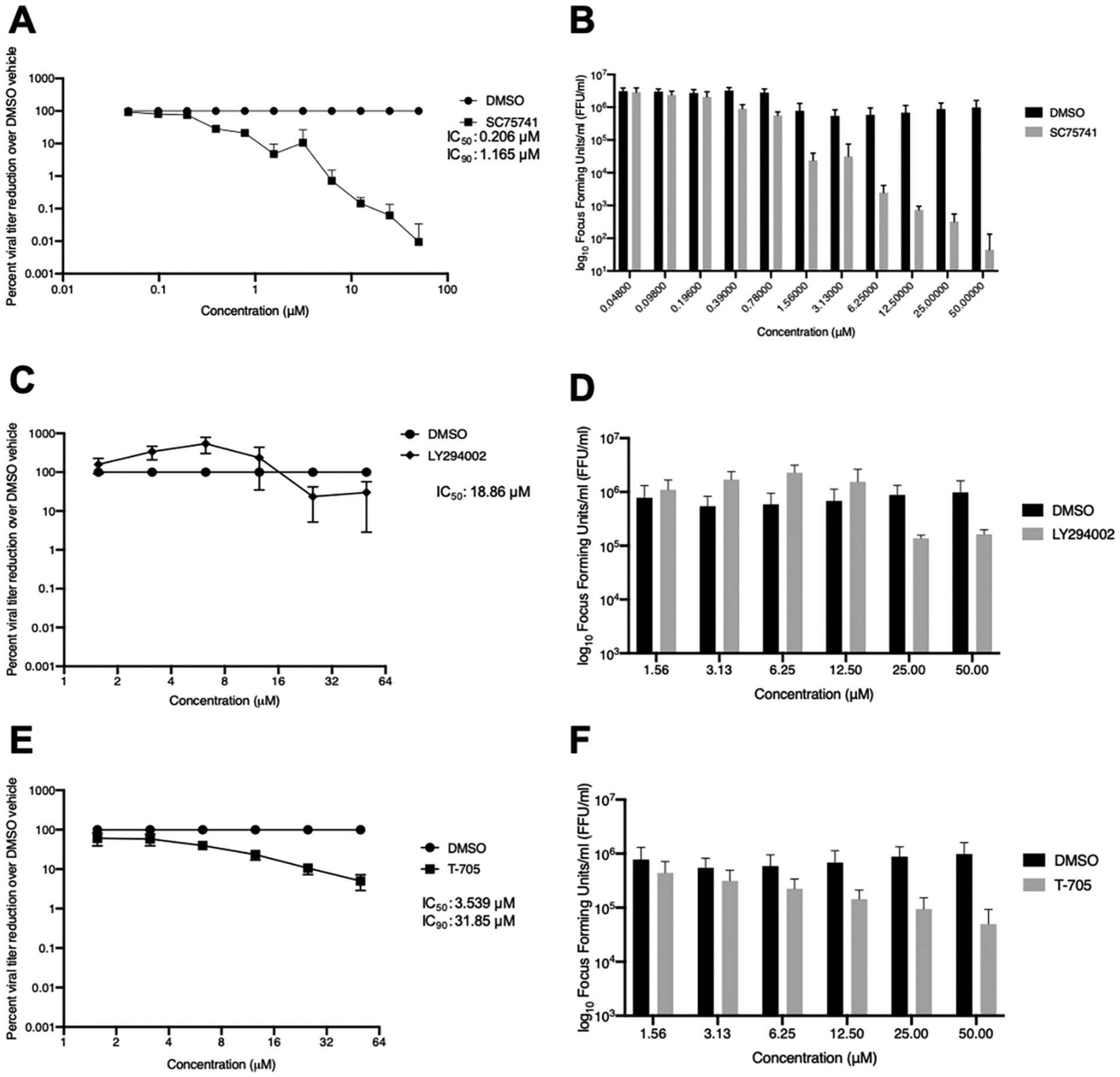
Huh7 cells were infected with HRTV (R99207b) 1 h and treated with SC75741 (A), LY294002 (C), and T-705 (E) 1-h post-infection and incubated for three days prior to titration on VeroE6 cells. Data are shown as percent viral titer reduction (A, C, E) or log10 focus forming units/mL (B, D, F). Inhibitory concentration values at 50% and 90%, IC50 and IC90 respectively, were calculated using nonlinear regression analysis using Prism 8 software (Graphpad). Data are represented as an average of three independent experiments.
4. Discussion
We describe a method for screening novel antiviral compounds against emerging tick-borne bandaviruses based on hPolI MG systems for both SFTSV and HRTV, which is advantageous as it is that it can be used in human cell lines commonly used for SFTSV and HRTV infection in vitro (Ren et al., 2020). The NF-κB inhibitor, SC75741 reduced viral RNA synthesis and replication of SFTSV and HRTV in vitro. SC75741 demonstrated antiviral efficacy against influenza viruses in vitro and in vivo (Ehrhardt et al., 2013, p. 75,741; Haasbach et al., 2013, p. 75,741). Compared to T-705, we observed a remarkable reduction in SFTSV and HRTV viral titers when cells were treated 1 h p.i. with SC75741 (IC90: HRTV: 1.165 μM and SFTSV of 2.234 μM). The SC75741-mediated reduction of SFTSV and HRTV replication/transcription via our MG systems indicates a potential role for NF-kB pathway in viral RNA synthesis of tick-borne bandaviruses, which supports the work of other groups (Qu et al., 2012; Sun et al., 2015) for which the mechanism remains to be identified and will be a future point of study.
While the efficacy of T-705 against SFTSV and HRTV has been widely demonstrated and was also demonstrated within our own study using infectious SFTSV and HRTV (Fig. 5E, F and 6E, 6F) (Tani et al, 2016a, 2018; Westover et al., 2017), we observed a modest reduction in MG activity for SFTSV hPolI MG at only high concentrations of T-705 (Fig. 3C). As we observed, titration of the helper plasmid indeed reduced the resistance barrier of MG activity upon T-705 treatment indicating a threshold of expression of viral replication machinery targeted by T-705 (Fig. 3C and D). A MG system for human metapneumovirus (HMPV) showed similar resistance at lower concentrations of T-705 in 293T cells (Jochmans et al., 2016). T-705 was originally hypothesized to inhibit the active site of the viral RdRp (Furuta et al, 2005, 2013), however more recent studies have suggested T-705 ribofuranosyltriphosphate, which functions as a purine nucleotide analog, induces lethal mutagenesis (Baranovich et al., 2013; Jin et al., 2013; Goldhill et al., 2018; Borrego et al., 2019; Espy et al., 2019; de Ávila et al., 2016; de Avila et al., 2017). The respective contributions of lethal mutagenesis induced by T-705 on the viral genome have yet to be described for SFTSV and HRTV indicating a potential explanation for resistance in a MG system against T-705 (Jochmans et al., 2016). We hypothesize this effect in our system may be due to a saturation of the components of the viral replication complex inhibiting T-705 as it directly acts on the RdRp and therefore cannot lead to an introduction of excess mutations, however further studies are needed to elucidate this mechanism of action.
Notably, we observed differences in growth kinetics in several human cell lines between HRTV and SFTSV, which has not been previously examined. This study will expand the knowledge of not only potential therapeutics for tick-borne bandaviruses, but also the potential screening methods, such as MG systems, for inhibitors of viral RNA synthesis and identification of host-cell factors that contribute to viral transcription/replication. Future studies will focus on identification of bandavirus-specific therapeutics that can reduce the aberrant inflammatory response seen in SFTS and HRTVD that lead to severe pathology seen in patients.
Supplementary Material
Acknowledgements
The authors would like to thank Robert Tesh (University of Texas Medical Branch), Kenneth Plante (University of Texas Medical Branch), the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA), and Brandy Russell (Division of Vector-Borne Infectious Diseases at the Centers for Disease Control and Prevention) for viruses and reagents vital to this study.
Funding
C.A.M. was supported in part by grant T32 AI132165 from the National Institutes of Health.
Footnotes
Declaration of competing interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2020.104993.
References
- Bao C, Guo X, Qi X, Hu J, Zhou M, Varma JK, Cui L, Yang H, Jiao Y, Klena JD, Li L, Tao W, Li X, Chen Y, Zhu Z, Xu K, Shen A, Wu T, Peng H, Li Z, Shan J, Shi Z, Wang H, 2011. A family cluster of infections by a newly recognized bunyavirus in eastern China, 2007: further evidence of person-to-person transmission. Clin. Infect. Dis 53, 1208–1214. 10.1093/cid/cir732. [DOI] [PubMed] [Google Scholar]
- Baranovich T, Wong S-S, Armstrong J, Marjuki H, Webby RJ, Webster RG, Govorkova EA, 2013. T-705 (favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro. J. Virol 87, 3741–3751. 10.1128/JVI.02346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazhanov N, Escaffre O, Freiberg AN, Garofalo RP, Casola A, 2017. Broad-range antiviral activity of hydrogen sulfide against highly pathogenic RNA viruses. Sci. Rep 7, 41029. 10.1038/srep41029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrego B, de Ávila AI, Domingo E, Brun A, 2019. Lethal mutagenesis of rift valley fever virus induced by favipiravir. Antimicrob. Agents Chemother 63 10.1128/AAC.00669-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault AC, Savage HM, Duggal NK, Eisen RJ, Staples JE, 2018. Heartland virus epidemiology, vector association, and disease potential. Viruses 10. 10.3390/v10090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron CE, Castro C, 2001. The mechanism of action of ribavirin: lethal mutagenesis of RNA virus genomes mediated by the viral RNA-dependent RNA polymerase. Curr. Opin. Infect. Dis 14, 757–764. 10.1097/00001432-200112000-00015. [DOI] [PubMed] [Google Scholar]
- Chen X, Ye H, Li S, Jiao B, Wu J, Zeng P, Chen L, 2017. Severe fever with thrombocytopenia syndrome virus inhibits exogenous Type i IFN signaling pathway through its NSs in vitro. PloS One 12, 1–12. 10.1371/journal.pone.0172744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SJ, Park S-W, Bae I-G, Kim S-H, Ryu SY, Kim HA, Jang H-C, Hur J, Jun J-B, Jung Y, Chang H-H, Kim YK, Yi J, Kim K-H, Hwang J-H, Kim Y-S, Jeong HW, Song K-H, Park WB, Kim ES, Oh M, Network for, K.S.C., 2016. Severe fever with thrombocytopenia syndrome in South Korea, 2013–2015. PLoS Neglected Trop. Dis 10, e0005264 10.1371/journal.pntd.0005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigneault M, Preston JA, Marriott HM, Whyte MKB, Dockrell DH, 2010. The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PloS One 5, e8668. 10.1371/journal.pone.0008668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ávila AI, Gallego I, Soria ME, Gregori J, Quer J, Esteban JI, Rice CM, Domingo E, Perales C, 2016. Lethal mutagenesis of hepatitis C virus induced by favipiravir. PloS One 11, e0164691. 10.1371/journal.pone.0164691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Avila AI, Moreno E, Perales C, Domingo E, 2017. Favipiravir can evoke lethal mutagenesis and extinction of foot-and-mouth disease virus. Virus Res. 233, 105–112. 10.1016/j.virusres.2017.03.014. [DOI] [PubMed] [Google Scholar]
- Deng B, Zhou B, Zhang S, Zhu Y, Han L, Geng Y, Jin Z, Liu H, Wang D, Zhao Y, Wen Y, Cui W, Zhou Y, Gu Q, Sun C, Lu X, Wang W, Wang Yu, Li C, Wang Yanli, Yao W, Liu P, 2013. Clinical features and factors associated with severity and fatality among patients with severe fever with thrombocytopenia syndrome bunyavirus infection in Northeast China. PloS One 8 (11). 10.1371/journal.pone.0080802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Guan XH, Kang K, Ding SJ, Huang LY, Xing XS, Sha S, Liu L, Wang XJ, Zhang XM, You AG, Du YH, Zhou H, Vong S, Zhang XD, Feng ZJ, Yang WZ, Li Q, Yin WW, 2014. Risk factors for bunyavirus-associated severe fever with thrombocytopenia syndrome. China. PLoS Neglected Tropical Diseases 8 (10). 10.1371/journal.pntd.0003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Li D, Wen D, Li S, Zhao C, Qi Y, Jangra RK, Wu C, Xia D, Zhang X, Deng F, Chandran K, Zou Z, Yuan F, Zheng A, 2019. Single dose of a rVSV-based vaccine elicits complete protection against severe fever with thrombocytopenia syndrome virus. NPJ Vaccines 4, 5. 10.1038/s41541-018-0096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn EF, Fearns R, Connor JH, 2009. Akt inhibitor Akt-IV blocks virus replication through an Akt-independent mechanism. J. Virol 83, 11665–11672. 10.1128/JVI.01092-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt C, Rückle A, Hrincius ER, Haasbach E, Anhlan D, Ahmann K, Banning C, Reiling SJ, Kühn J, Strobl S, Vitt D, Leban J, Planz O, Ludwig S, 2013. The NF-κB inhibitor SC75741 efficiently blocks influenza virus propagation and confers a high barrier for development of viral resistance. Cell Microbiol. 15, 1198–1211. 10.1111/cmi.12108. [DOI] [PubMed] [Google Scholar]
- Espy N, Nagle E, Pfeffer B, Garcia K, Chitty AJ, Wiley M, Sanchez-Lockhart M, Bavari S, Warren T, Palacios G, 2019. T-705 induces lethal mutagenesis in Ebola and Marburg populations in macaques. Antivir. Res 170, 104529. 10.1016/j.antiviral.2019.06.001. [DOI] [PubMed] [Google Scholar]
- Furuta Y, Gowen BB, Takahashi K, Shiraki K, Smee DF, Barnard DL, 2013. Favipiravir (T-705), a Novel Viral RNA Polymerase Inhibitor. 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed]
- Furuta Y, Komeno T, Nakamura T, 2017. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci 93, 449–463. 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y, Takahashi K, Kuno-Maekawa M, Sangawa H, Uehara S, Kozaki K, Nomura N, Egawa H, Shiraki K, 2005. Mechanism of action of T-705 against influenza virus. Antimicrob. Agents Chemother 49, 981–986. 10.1128/AAC.49.3.981-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai Z, Liang M, Zhang Y, Zhang S, Jin C, Wang S-W, Sun L, Zhou N, Zhang Q, Sun Y, Ding S-J, Li C, Gu W, Zhang F, Wang Y, Bian P, Li X, Wang Z, Song X, Wang X, Xu A, Bi Z, Chen S, Li D, 2012a. Person-to-Person transmission of severe fever with thrombocytopenia syndrome bunyavirus through blood contact. Clin. Infect. Dis 54, 249–252. 10.1093/cid/cir776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai ZT, Zhang Y, Liang MF, Jin C, Zhang S, Zhu CB, Li C, Li XY, Zhang QF, Bian PF, Zhang LH, Wang B, Zhou N, Liu JX, Song XG, Xu A, Bi ZQ, Chen SJ, Li DX, 2012b. Clinical progress and risk factors for death in severe fever with thrombocytopenia syndrome patients. JID (J. Infect. Dis.) 206 (7), 1095–1102. 10.1093/infdis/jis472. [DOI] [PubMed] [Google Scholar]
- Goldhill DH, Te Velthuis AJW, Fletcher RA, Langat P, Zambon M, Lackenby A, Barclay WS, 2018. The mechanism of resistance to favipiravir in influenza. Proc. Natl. Acad. Sci. U.S.A 115, 11613–11618. 10.1073/pnas.1811345115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasbach E, Reiling SJ, Ehrhardt C, Droebner K, Ruckle A, Hrincius ER, Leban J, Strobl S, Vitt D, Ludwig S, Planz O, 2013. The NF-kappaB inhibitor SC75741 protects mice against highly pathogenic avian influenza A virus. Antivir. Res 99, 336–344. 10.1016/j.antiviral.2013.06.008. [DOI] [PubMed] [Google Scholar]
- Hornak KE, Lanchy J-M, Lodmell JS, 2016. RNA encapsidation and packaging in the phleboviruses. Viruses 8. 10.3390/v8070194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang XL, Zhang S, Jiang M, Bi ZQ, Liang MF, Ding SJ, Wang SW, Liu JY, Zhou SQ, Zhang XM, Li DX, Xu AQ, 2015. A cluster of person-to-person transmission cases caused by SFTS virus in Penglai, China. Clin. Microbiol. Infect 21, 274–279. 10.1016/j.cmi.2014.10.006. [DOI] [PubMed] [Google Scholar]
- Jiao L, Ouyang S, Liang M, Niu F, Shaw N, Wu W, Ding W, Jin C, Peng Y, Zhu Y, Zhang F, Wang T, Li C, Zuo X, Luan C-H, Li D, Liu Z-J, 2013. Structure of severe fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential. J. Virol 87, 6829–6839. 10.1128/JVI.00672-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Smith LK, Rajwanshi VK, Kim B, Deval J, 2013. The ambiguous base-pairing and high substrate efficiency of T-705 (Favipiravir) Ribofuranosyl 5’-triphosphate towards influenza A virus polymerase. PloS One 8, e68347. 10.1371/journal.pone.0068347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochmans D, Van Nieuwkoop S, Smits SL, Neyts J, Fouchier RAM, Van Den Hoogen BG, 2016. Antiviral activity of favipiravir (T-705) against a broad range of paramyxoviruses in vitro and against human metapneumovirus in hamsters. Antimicrob. Agents Chemother 60, 4620–4629. 10.1128/AAC.00709-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung IY, Choi W, Kim J, Wang E, Park S-W, Lee W-J, Choi JY, Kim HY, Uh Y, Kim YK, 2019. Nosocomial person-to-person transmission of severe fever with thrombocytopenia syndrome. Clin. Microbiol. Infect 25, 633. 10.1016/j.cmi.2019.01.006 e1–633.e4. [DOI] [PubMed] [Google Scholar]
- Kato H, Yamagishi T, Shimada T, Matsui T, Shimojima M, Saijo M, Oishi K, 2016. Epidemiological and clinical features of severe fever with thrombocytopenia syndrome in Japan, 2013–2014. PloS One 11, 165207. 10.1371/journal.pone.0165207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida K, Matsuoka Y, Shimoda T, Matsuoka H, Yamada H, Saito T, Imataki O, Kadowaki N, Noguchi K, Maeda K, Mochizuki Y, Kishimoto T, 2019. A case of cat-to-human transmission of severe fever with thrombocytopenia syndrome virus. Jpn. J. Infect. Dis 72, 356–358. 10.7883/yoken.JJID.2018.526. [DOI] [PubMed] [Google Scholar]
- Kim YR, Yun Y, Bae SG, Park D, Kim S, Lee JM, Cho N-H, Kim YS, Lee KH, 2018. Severe fever with thrombocytopenia syndrome virus infection, South Korea, 2010. Emerg. Infect. Dis 24, 2103–2105. 10.3201/eid2411.170756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kato H, Yamagishi T, Shimada T, Matsui T, Yoshikawa T, Kurosu T, Shimojima M, Morikawa S, Hasegawa H, Saijo M, Oishi K, SFTS Epidemiological Research Group Japan, 2020. Severe fever with thrombocytopenia syndrome, Japan, 2013–2017. Emerg. Infect. Dis 26, 692–699. 10.3201/eid2604.191011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara S, Satoh A, Yu F, Hayasaka D, Shimojima M, Tashiro M, Saijo T, Takazono T, Imamura Y, Miyazaki T, Tsukamoto M, Yanagihara K, Mukae H, Saijo M, Morita K, Kohno S, Izumikawa K, 2016. The world first two cases of severe fever with thrombocytopenia syndrome: an epidemiological study in Nagasaki, Japan. J. Infect. Chemother 22 (7), 461–465. 10.1016/j.jiac.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Kwak J-E, Kim Y-I, Park S-J, Yu M-A, Kwon H-I, Eo S, Kim T-S, Seok J, Choi W-S, Jeong JH, Lee H, Cho Y, Kwon JA, Jeong M, Maslow JN, Kim Y-E, Jeon H, Kim KK, Shin E-C, Song M-S, Jung JU, Choi YK, Park S-H, 2019. Development of a SFTSV DNA vaccine that confers complete protection against lethal infection in ferrets. Nat. Commun 10, 3836. 10.1038/s41467-019-11815-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Li H, Zhang Y-L, Xin Q-L, Guan Z-Q, Chen X, Zhang X-A, Li X-K, Xiao G-F, Lozach P-Y, Cui J, Liu W, Zhang L-K, Peng K, 2020. SFTSV infection induces BAK/BAX-Dependent mitochondrial DNA release to trigger NLRP3 inflammasome activation. Cell Rep. 30, 4370–4385. 10.1016/j.celrep.2020.02.105 e7. [DOI] [PubMed] [Google Scholar]
- Lin T-L, Ou S-C, Maeda K, Shimoda H, Chan JPW, Tu W-C, Hsu W-L, Chou C-C, 2020. The first discovery of severe fever with thrombocytopenia syndrome virus in Taiwan. Emerg. Microb. Infect 9, 148–151. 10.1080/22221751.2019.1710436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li Q, Hu W, Wu J, Wang Yubi, Mei L, Walker DH, Ren J, Wang Yu, Yu X-J, 2012. Person-to-person transmission of severe fever with thrombocytopenia syndrome virus. Vector Borne Zoonotic Dis. 12, 156–160. 10.1089/vbz.2011.0758. [DOI] [PubMed] [Google Scholar]
- McMullan LK, Folk SM, Kelly AJ, MacNeil A, Goldsmith CS, Metcalfe MG, Batten BC, Albariño CG, Zaki SR, Rollin PE, Nicholson WL, Nichol ST, 2012. A new phlebovirus associated with severe febrile illness in Missouri. N. Engl. J. Med 367, 834–841. 10.1056/nejmoa1203378. [DOI] [PubMed] [Google Scholar]
- Mendoza CA, Ebihara H, Yamaoka S, 2019. Immune modulation and immune-mediated pathogenesis of emerging tickborne banyangviruses. Vaccines 7. 10.3390/vaccines7040125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Zobel A, Hobom G, 1994. RNA polymerase I-mediated expression of influenza viral RNA molecules. Virology 202, 477–479. 10.1006/viro.1994.1365. [DOI] [PubMed] [Google Scholar]
- Pastula DM, Turabelidze G, Yates KF, Jones TF, Lambert AJ, Panella AJ, Kosoy OI, Velez JO, Fisher M, Staples E, Centers for Disease Control and Prevention (CDC), 2014. Notes from the field: Heartland virus disease - United States, 2012–2013. MMWR Morb. Mortal. Wkly. Rep 63, 270–271. [PMC free article] [PubMed] [Google Scholar]
- Qu B, Qi X, Wu X, Liang M, Li C, Cardona CJ, Xu W, Tang F, Li Z, Wu B, Powell K, Wegner M, Li D, Xing Z, 2012. Suppression of the interferon and NF-κB responses by severe fever with thrombocytopenia syndrome virus. J. Virol 86, 8388–8401. 10.1128/jvi.00612-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F, Zhou M, Deng F, Wang H, Ning Y-J, 2020. Combinatorial minigenome systems for emerging banyangviruses reveal viral reassortment potential and importance of a protruding nucleotide in genome “panhandle” for promoter activity and reassortment. Front. Microbiol 11, 599. 10.3389/fmicb.2020.00599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezelj VV, Mottram TJ, Hughes J, Elliott RM, Kohl A, Brennan B, 2019. M segment-based minigenomes and virus-like particle assays as an approach to assess the potential of tick-borne phlebovirus genome reassortment. J. Virol 93 10.1128/JVI.02068-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles NJC, Han HJ, Park S-J, Choi YK, 2018. Epidemiology of severe fever and thrombocytopenia syndrome virus infection and the need for therapeutics for the prevention. Clinical and Experimental Vaccine Research 7, 43–50. 10.7774/cevr.2018.7.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothan HA, Mohamed Z, Paydar M, Rahman NA, Yusof R, 2014. Inhibitory effect of doxycycline against dengue virus replication in vitro. Arch. Virol 159, 711–718. 10.1007/s00705-013-1880-7. [DOI] [PubMed] [Google Scholar]
- Saijo M, 2018. Pathophysiology of severe fever with thrombocytopenia syndrome and development of specific antiviral therapy. J. Infect. Chemother 24, 773–781. 10.1016/j.jiac.2018.07.009. [DOI] [PubMed] [Google Scholar]
- Spiegel M, Plegge T, Pöhlmann S, 2016. The role of phlebovirus glycoproteins in viral entry, assembly and release. Viruses 8, 202. 10.3390/v8070202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statistics & Maps, 2020. Heartland Virus | CDC. WWW Document. https://www.cdc.gov/heartland-virus/statistics/index.html. (Accessed 8 December 2020).
- Sun J, Gong Z, Ling F, Zhang R, Tong Z, Chang Y, Chen E, Liu Q, Lin J, Chen Z, Jiang J, 2016. Factors associated with severe fever with thrombocytopenia syndrome infection and fatal outcome. Sci. Rep 6, 33175. 10.1038/srep33175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Jin C, Zhu L, Liang M, Li C, Cardona CJ, Li D, Xing Z, 2015. Host responses and regulation by NFκB signaling in the liver and liver epithelial cells infected with A novel tick-borne bunyavirus. Sci. Rep 5, 11816. 10.1038/srep11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Maeda K, Suzuki T, Ishido A, Shigeoka T, Tominaga T, Kamei T, Honda M, Ninomiya D, Sakai T, Senba T, Kaneyuki S, Sakaguchi S, Satoh A, Hosokawa T, Kawabe Y, Kurihara S, Izumikawa K, Kohno S, Azuma T, Suemori K, Yasukawa M, Mizutani T, Omatsu T, Katayama Y, Miyahara M, Ijuin M, Doi K, Okuda M, Umeki K, Saito T, Fukushima K, Nakajima K, Yoshikawa T, Tani H, Fukushi S, Fukuma A, Ogata M, Shimojima M, Nakajima N, Nagata N, Katano H, Fukumoto H, Sato Y, Hasegawa H, Yamagishi T, Oishi K, Kurane I, Morikawa S, Saijo M, 2014. The first identification and retrospective study of Severe Fever with Thrombocytopenia Syndrome in Japan. J. Infect. Dis 209, 816–827. 10.1093/infdis/jit603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama-Ito M, Saijo M, 2020. Antiviral drugs against severe fever with thrombocytopenia syndrome virus infection. Front. Microbiol 11, 150. 10.3389/fmicb.2020.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Fukuma A, Fukushi S, Taniguchi S, Yoshikawa T, Iwata-Yoshikawa N, Sato Y, Suzuki T, Nagata N, Hasegawa H, Kawai Y, Uda A, Morikawa S, Shimojima M, Watanabe H, Saijo M, 2016a. Efficacy of T-705 (favipiravir) in the treatment of infections with lethal severe fever with thrombocytopenia syndrome virus. mSphere 1, 1–11. 10.1128/msphere.00061-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Komeno T, Fukuma A, Fukushi S, Taniguchi S, Shimojima M, Uda A, Morikawa S, Nakajima N, Furuta Y, Saijo M, 2018. Therapeutic effects of favipiravir against severe fever with thrombocytopenia syndrome virus infection in a lethal mouse model: dose-efficacy studies upon oral administration. PloS One 13, e0206416. 10.1371/journal.pone.0206416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Shimojima M, Fukushi S, Yoshikawa T, Fukuma A, Taniguchi S, Morikawa S, Saijo M, 2016b. Characterization of glycoprotein-mediated entry of severe fever with thrombocytopenia syndrome virus. J. Virol 90, 5292–5301. 10.1128/JVI.00110-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda Y, Weisend C, Martellaro C, Feldmann F, Haddock E, 2017. Pathogenic analysis of the pandemic 2009 H1N1 influenza A viruses in ferrets. J. Vet. Med. Sci 79, 1453–1460. 10.1292/jvms.16-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westover JB, Rigas JD, Van Wettere AJ, Li R, Hickerson BT, Jung KH, Miao J, Reynolds ES, Conrad BL, Nielson S, Furuta Y, Thangamani S, Wang Z, Gowen BB, 2017. Heartland virus infection in hamsters deficient in type I interferon signaling: protracted disease course ameliorated by favipiravir. Virology 511, 175–183. 10.1016/j.virol.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z-C, Wang X, Wei J-C, Li B-B, Shao D-H, Li Y-M, Liu K, Shi Y-Y, Zhou B, Qiu Y-F, Ma Z-Y, 2015. Antiviral activity of doxycycline against vesicular stomatitis virus in vitro. FEMS Microbiol. Lett 362 10.1093/femsle/fnv195. [DOI] [PubMed] [Google Scholar]
- Yoo JR, Choi JH, Kim YR, Lee KH, Heo ST, 2019. Occupational risk of severe fever with thrombocytopenia syndrome in healthcare workers. Open Forum Infect Dis 6. 10.1093/ofid/ofz210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X-J, Liang M-F, Zhang S-Y, Liu Y, Li J-D, Sun Y-L, Zhang L, Zhang Q-F, Popov VL, Li C, Qu J, Li Q, Zhang Y-P, Hai R, Wu W, Wang Q, Zhan F-X, Wang X-J, Kan B, Wang S-W, Wan K-L, Jing H-Q, Lu J-X, Yin W-W, Zhou H, Guan X-H, Liu J-F, Bi Z-Q, Liu G-H, Ren J, Wang H, Zhao Z, Song J-D, He J-R, Wan T, Zhang J-S, Fu X-P, Sun L-N, Dong X-P, Feng Z-J, Yang W-Z, Hong T, Zhang Y, Walker DH, Wang Y, Li D-X, 2011. Fever with thrombocytopenia associated with a novel bunyavirus in China. N. Engl. J. Med 364, 1523–1532. 10.1056/NEJMoa1010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Lenardo MJ, Baltimore D, 2017. 30 Years of NF-κB: a blossoming of relevance to human pathobiology. Cell 168, 37–57. 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Sun Y, Wang Y, Liu M, Liu C, Wang W, Liu X, Li L, Deng F, Wang H, Guo Y, Lou Z, 2013. The nucleoprotein of severe fever with thrombocytopenia syndrome virus processes a stable hexameric ring to facilitate RNA encapsidation. Protein Cell 4, 445–455. 10.1007/s13238-013-3901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


