ABSTRACT
The small Rho-family GTPase Cdc42 has long been known to have a role in cell motility and axon growth. The eukaryotic Ccd42 gene is alternatively spliced to generate mRNAs with two different 3′ untranslated regions (UTRs) that encode proteins with distinct C-termini. The C-termini of these Cdc42 proteins include CaaX and CCaX motifs for post-translational prenylation and palmitoylation, respectively. Palmitoyl-Cdc42 protein was previously shown to contribute to dendrite maturation, while the prenyl-Cdc42 protein contributes to axon specification and its mRNA was detected in neurites. Here, we show that the mRNA encoding prenyl-Cdc42 isoform preferentially localizes into PNS axons and this localization selectively increases in vivo during peripheral nervous system (PNS) axon regeneration. Functional studies indicate that prenyl-Cdc42 increases axon length in a manner that requires axonal targeting of its mRNA, which, in turn, needs an intact C-terminal CaaX motif that can drive prenylation of the encoded protein. In contrast, palmitoyl-Cdc42 has no effect on axon growth but selectively increases dendrite length. Together, these data show that alternative splicing of the Cdc42 gene product generates an axon growth promoting, locally synthesized prenyl-Cdc42 protein.
This article has an associated First Person interview with one of the co-first authors of the paper.
KEY WORDS: Alternative splicing, Axon regeneration, Growth cone, mRNA transport, Geranylgeranylation
Summary: Axon regeneration drives selective localization of alternatively spliced Cdc42 mRNA isoform to PNS axons.
INTRODUCTION
Concentrating proteins in subcellular regions is used to create unique functional domains in polarized cells. This is achieved by specifically transporting proteins from their sites of translation to subcellular regions through protein targeting sequences that are used for protein localization (Emanuelsson et al., 2007). Transport of mRNAs to subcellular domains with subsequent localized translation in those domains is also used to spatially regulate the proteome of polarized cells. For some mRNAs, this localized translation imparts unique functions to the cell. As remarkably polarized cells with widely separated and specialized subcellular domains, neurons are a very useful model system to profile the populations of mRNAs that are transported into axons and dendrites, as well as to test functions of proteins synthesized locally in these subcellular compartments (Kar et al., 2018; Terenzio et al., 2017). Indeed, axons can extend to lengths that are more than 1000-fold longer than the diameter of cell body, and localized synthesis of new proteins in distal axons provides a level of autonomy to respond to extracellular stimuli (Sahoo et al., 2018). Both growth-promoting and growth-inhibiting stimuli regulate intra-axonal protein synthesis to support directional growth and branching of axons (Dalla Costa et al., 2020).
RNA profiling studies have shown hundreds to thousands of mRNAs are localized into axons of sensory, cortical, retinal ganglion cell and motor neurons (Kar et al., 2018). These populations include several mRNAs encoding proteins that can impact the axonal cytoskeleton, either directly through generation of cytoskeletal proteins or indirectly by generation of proteins that regulate polymerization, depolymerization, severing or branching of the cytoskeleton. The mRNA encoding the small GTPase RhoA, whose activation has been shown to trigger axon retraction, localizes into and is translated within axons in response to growth inhibitory stimuli (Wu et al., 2005). The mRNA for the small GTPase Rac1 was recently shown to localize into sympathetic axons, with its encoded protein undergoing prenylation in axons (Scott-Solomon and Kuruvilla, 2020). RhoA and Rac belong to the Rho GTPase family, which also includes Cdc42 protein. Rho GTPases regulate cytoskeleton organization, and antagonistic functions among these family members have been reported in neurons (Etienne-Manneville and Hall, 2002). RhoA activation leads to neurite retraction through an increase in actomyosin contractility and actin depolymerization, while Cdc42 and Rac activation lead to neurite extension through promoting actin polymerization (Da Silva et al., 2003; Jalink et al., 1994; Luo, 2000; Luo et al., 1994). The Cdc42 gene undergoes alternative splicing to generate two distinct Cdc42 mRNAs, and recent work points to local translation of the Cdc42 mRNA variant containing 3′ terminal exon 7, rather than exon 6, in neurites of cultured neurons (Ciolli Mattioli et al., 2019). We find that the exon 7 containing-Cdc42 mRNA, which encodes the prenylated Cdc42 isoform [mRNA denoted as prenyl-Cdc42; also known as the ‘placental isoform’ (Wirth et al., 2013)], selectively localizes into axons, and this axonally localized mRNA accounts for the axon growth-promoting effects of Cdc42 protein. In contrast, the Cdc42 mRNA variant encoding the palmitoylated Cdc42 mRNA isoform [mRNA denoted as palm-Cdc42; also known as the ‘brain isoform’ (Wirth et al., 2013)] does not affect axon growth but rather supports extension of dendrites. The contribution of Cdc42 to axonal growth has been known for many years (Hall, 1998), and our data indicate that this is uniquely driven by the prenyl-Cdc42 mRNA isoform through axonal localization of its mRNA and its C-terminal CaaX motif that supports prenylation of its locally translated protein product.
RESULTS
The prenyl-Cdc42 mRNA isoform uniquely localizes into peripheral nervous system axons
The rat Cdc42 gene consists of seven exons that generate two mRNA isoforms by alternative splicing that contain either 3′ terminal exon 6 or 7 (Fig. 1A). The splice variant ending with exon 6 produces palm-Cdc42 mRNA, which generates palmitoylated Cdc42 protein, while the mRNA skipping exon 6 and ending with exon 7 produces the prenyl-Cdc42 mRNA that generates prenylated Cdc42 protein (Wirth et al., 2013). This alternative splicing to generate two Cdc42 mRNA isoforms with unique 3′ untranslated regions (UTRs) and different C-termini is conserved from fish to mammals (Table S1). Ciolli Mattioli et al. (2019) recently reported that the prenyl-Cdc42 mRNA variant, and not the palm-Cdc42 mRNA, can localize into neurites of mouse embryonic stem cell (ESC)-derived and primary cortical neurons, but they did not distinguish between dendrites and axons (Ciolli Mattioli et al., 2019). However, RNA-Seq data for mouse cortical neurons Taliaferro et al. (2016) shows near equivalent proportions of prenyl-Cdc42 and palm-Cdc42 mRNAs in neurites (Table S2). By mining published next-generation sequencing (RNA-Seq) data from axon-only RNA isolates from cultured embryonic mouse sensory and motor neurons, we find that an overall higher proportion of the prenyl-Cdc42 mRNA localizes into axons than the palm-Cdc42 mRNA (Table S2) (Briese et al., 2016; Minis et al., 2014). RNA-seq data from adult mouse dorsal root ganglia (DRG) cultures similarly show much higher proportion of the prenyl-Cdc42 than palm-Cdc42 mRNA in isolated axons (Table S2).
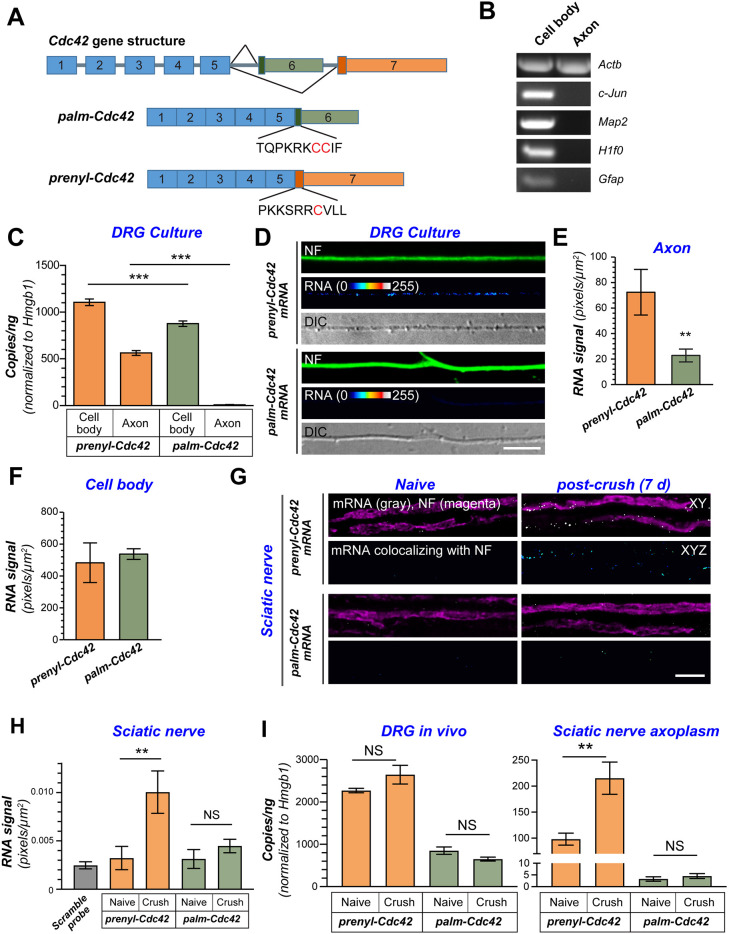
Fig. 1.
Alternatively spliced Cdc42 mRNA isoforms differentially localize to peripheral nervous system axons. (A) Schematic for alternative splicing for Cdc42 mRNAs. The two Cdc42 mRNAs have distinct 3′UTRs from alternative use of exons 6 or 7. These two mRNAs encode proteins with different C-termini. (B) Representative ethidium bromide-stained agarose gel of RT-PCR products for RNA isolated from axonal and cell body compartments of dissociated DRG neurons. (C) RT-ddPCR for prenyl-Cdc42 and palm-Cdc42 isoforms is shown as mean±s.e.m. copy number; Hmgb1 was used for input RNA normalization (N=3 biological replicates). ***P≤0.005 (Student's t-test for the indicated data pairs). (D) Representative exposure-matched smFISH and immunofluorescence (IF) images for Cdc42 mRNA isoforms smFISH and neurofilament (NF) in adult DRG neuron cultures. Scale bars: 10 µm. (E,F) Quantification of smFISH signal intensities shown as mean±s.e.m. pixel intensityfor axons (E) and cell bodies (F) and axons (N≥60 neurons in three independent cultures). **P<0.01 (Kruskal–Wallis test with multiple comparisons). (G,H) Representative FISH and IF images for naïve and 7 day post-crush injured sciatic nerve are shown in G. The upper row of each image set shows the merged confocal xy optical plane; the lower row of each image set shows RNA signal overlapping with NF across individual optical planes that was extracted to a separate channel and projected as xyz images. For scrambled probe signals, see Fig. S1C. Quantification of FISH signals for RNA probe signals overlapping with NF shown in H as mean±s.e.m. (N=15). **P<0.01, NS, non-significant (Kruskal–Wallis test with multiple comparisons). Scale bar: 10 µm. (I) RT-ddPCR for RNA isolated from extruded sciatic nerve axoplasm or L4–L6 DRG lysates shown as mean±s.e.m. mRNA copy number is normalized to Hmgb1 mRNA levels (N=6). **P<0.01; NS, not significant (one-way ANOVA with pair-wise comparison with Tukey post-hoc tests).
To more directly test the possibility for selective axonal localization of prenyl-Cdc42 mRNA, we cultured L4–L6 DRG neurons from adult rat sensory neurons on a porous membrane to allow for isolation of axonal RNA (Zheng et al., 2001). Reverse transcriptase-coupled PCR (RT-PCR) showed that the axonal preparations contained Actb mRNA (encoding β-actin) but not the somatodendritic-restricted Map2 mRNA, cell body-restricted Jun and H1f0 mRNAs, or glial Gfap mRNA (Fig. 1B). In reverse transcription-coupled droplet digital PCR (RT-ddPCR) experiments, more prenyl- than palm-Cdc42 mRNA was detected in the DRG cell body RNA preparations, and prenyl-Cdc42 mRNA was much more abundant than palm-Cdc42 mRNA in the axonal RNA isolates, with only a few copies of palm-Cdc42 mRNA detected in the axonal RNA isolates (Fig. 1C). Consistent with the RT-ddPCR results, the cell bodies of cultured DRG neurons showed stronger signals for prenyl- than palm-Cdc42 mRNA by single-molecule fluorescence in situ hybridization (smFISH; Fig. S1A). Prominent smFISH signals for prenyl-Cdc42 mRNA were seen in the axons of these neurons, but axonal signals for palm-Cdc42 mRNA were comparable to the scrambled probe control (Fig. 1D; Fig. S1B). Quantification showed significantly higher prenyl- than palm-Cdc42 mRNA signals in the DRG axons, with no significant differences in overall levels in the cell bodies (Fig. 1E,F)
With this preferential localization of prenyl-Cdc42 mRNA into axons of cultured adult DRG neurons, we asked whether prenyl-Cdc42 mRNA localizes into axons in vivo. Unexpectedly, smFISH signals for prenyl-Cdc42 mRNA were very low in uninjured sciatic nerve axons (Fig. 1G). However, the axonal prenyl-Cdc42 mRNA signals were quite prominent in the regenerating axons of the seven day post-crush sciatic nerve, with palm-Cdc42 mRNA signals being again comparable to the scrambled probe (Fig. 1G; Fig. S1C). Quantifying the smFISH signals across multiple animals showed a significant increase in axonal prenyl-Cdc42 mRNA in the regenerating axons compared to uninjured axons, while the axonal palm-Cdc42 mRNA signals were not significantly different to the scrambled control (Fig. 1H). There did not appear to be a correlation of prenyl-Cdc42 mRNA signals with axon diameter in the sciatic nerve, suggesting that this differential localization of prenyl- versus palm-Cdc42 mRNAs is not limited to sensory axons.
As a second test for Cdc42 mRNA isoform levels in sciatic nerve axons, we used RT-ddPCR with a sciatic nerve axoplasm preparation that is highly enriched in axonal contents (Hanz et al., 2003). Again, there was a significant increase in prenyl-Cdc42 mRNA in the regenerating nerve axoplasm of the compared to naïve nerve axoplasm with very low signals for palm-Cdc42 mRNA in both (Fig. 1I). Interestingly, DRG levels of prenyl-Cdc42 and palm-Cdc42 mRNA did not significantly change with injury (Fig. 1I), suggesting that the increase in axonal prenyl-Cdc42 mRNA in regenerating axons is driven either by increased transport into or stabilization of the mRNA in axons. Together, these data indicate that elevated axonal levels of prenyl-Cdc42 mRNA is a feature of growing axons.
The prenyl-Cdc42 3′UTR drives axon localization in sensory neurons
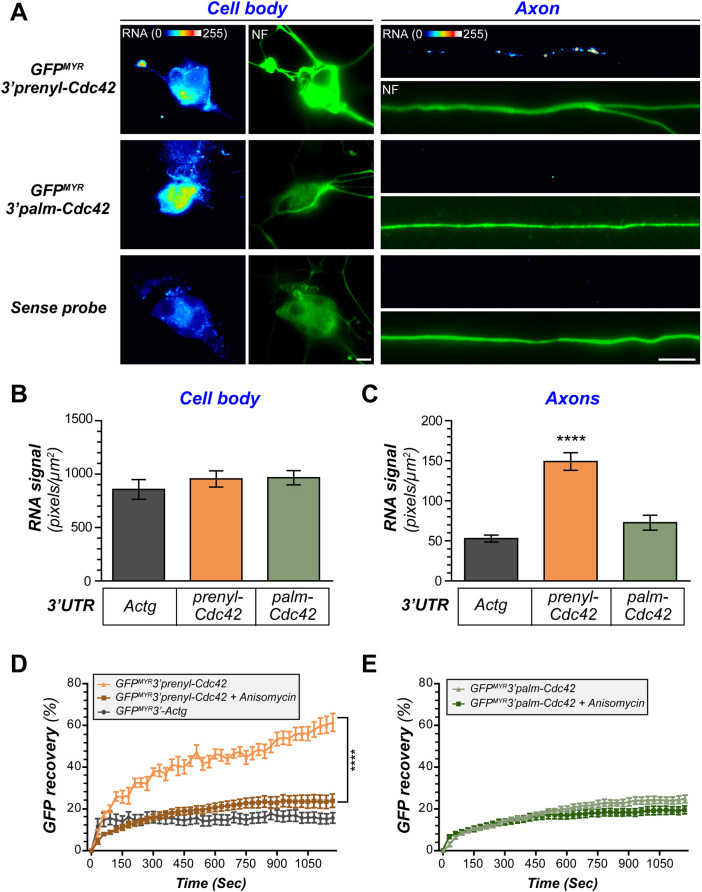
The unique 3′UTR of prenyl-Cdc42 has been suggested to be sufficient for translation of mCherry reporter mRNA in neurites of ESC-derived neurons on the basis of a puromycin-proximity ligation assay and immunoblotting (Ciolli Mattioli et al., 2019). We used myristoylated (MYR) eGFP reporter constructs containing the 3′UTRs of prenyl-Cdc42 versus palm-Cdc42 mRNAs (GFPMYR3′palm-Cdc42 and GFPMYR3′prenyl-Cdc42, respectively) to compare potential axonal localizing activity of the Cdc42 gene exons 6 and 7 in adult DRG cultures. As shown by smFISH for GFP mRNA, only the GFPMYR3′prenyl-Cdc42-transfected neurons showed detectable axonal GFP RNA signals (Fig. 2A–C). Axonal smFISH signals for GFPMYR3′palm-Cdc42 mRNA were not significantly different to those of the GFPMYR mRNA carrying the 3′UTR of the mRNA encoding γ-actin (Actg; GFPMYR3′Actg), which does not localize into DRG axons (Fig. 2B,C) (Willis et al., 2007). The MYR tag of the GFP markedly limits diffusion of the protein synthesized in neurites, so that sites of protein synthesis can be visualized using fluorescence recovery after photobleaching (FRAP) (Aakalu et al., 2001; Yudin et al., 2008). FRAP studies revealed that distal axons of DRGs expressing GFPMYR3′prenyl-Cdc42 mRNA showed significantly higher GFPMYR recovery after photobleaching than those expressing GFPMYR3′palm-Cdc42 (Fig. 2D,E). Pre-incubation with the translation inhibitor anisomycin attenuated the post-bleach fluorescence recovery of GFPMYR3′prenyl-Cdc42, indicating that the axonal recovery seen for this reporter derives from locally synthesized GFP (Fig. 2D). The GFPMYR3′palm-Cdc42 showed more recovery than GFPMYR3′Actg-transfected neurons (Fig. 2E), which may reflect the low axonal levels of palm-Cdc42 mRNA seen in Fig. 1. Taken together, these data show that the 3′UTR from Cdc42 gene exon 7 is sufficient for axonal localization and translation of prenyl-Cdc42 mRNA.
Fig. 2.
The prenyl-Cdc42 mRNA isoform 3′UTR drives axonal mRNA localization and translation in adult sensory neurons. (A) Representative exposure-matched smFISH and immunofluorescence (IF) images for GFP mRNA and NF in DRG neurons transfected with the GFPMYR reporter containing 3′UTRs of prenyl-Cdc42 or palm-Cdc42 mRNAs (GFPMYR-3′prenyl-Cdc42 and GFPMYR3′palm-Cdc42, respectively) are shown. Scale bars: 10 µm. (B,C) Quantification of GFP mRNA signals from smFISH for cell bodies (B) and axons (C) shown as mean±s.e.m. pixel intensity. GFPMYR3′-Actg was used as a negative control (N≥45 neurons in three independent experiments). ***P≤0.001 by Kruskal–Wallis test with multiple comparisons). (D,E) FRAP analyses for distal axons of DRG neurons expressing indicated GFPMYR reporter constructs as in A are shown as mean±s.e.m. percentage normalized average recovery. The GFPMYR3′prenyl-Cdc42 shows significantly greater fluorescence recovery than GFPMYR3′palm-Cdc42 and GFPMYR3′-Actg. Note that translation inhibition with anisomycin prior to photobleaching shows that the GFPMYR3′prenyl-Cdc42 recovery requires protein synthesis (N≥15 neurons over three independent experiments). ****P<0.001 (two-way ANOVA with pair-wise comparison and Tukey post-hoc tests).
Alternative splicing of the Cdc42 gene transcript accounts for the different 3′UTRs of prenyl- and palm-Cdc42 mRNAs. The GFPMYR constructs used above would not be subjected to these splicing events, and retention of proteins at splice sites has been suggested to impact subcellular localization and translation of the endogenous mRNAs (Le Hir et al., 2016). To address the possibility that splicing rather than the RNA sequence determines axonal localization of prenyl-Cdc42 mRNA, we generated Cdc42 ‘mini-gene’ constructs that have GFP cDNA fused to rat Cdc42 exons 4, 5 and 6 or exons 4, 5 and 7 to mimic the splicing events for endogenous palm-Cdc42 and prenyl-Cdc42 mRNAs, respectively (Fig. S2A). Exons 4 and 5 were separated by the endogenous 103 nucleotide (nt) intervening intron; exons 5 and 6 or 5 and 7 were separated by 500 nt intronic sequence downstream of exon 5 plus 500 nt intronic upstream of exon 6 or 7 (i.e. an 1000 nt intronic sequence). RT-PCR of DRGs transfected with these mini-gene constructs showed PCR products at the anticipated sizes of the mature (i.e. spliced) mRNAs for GFP+Cdc42 exons 4-6 and GFP+Cdc42 exons 4-5+7 (Fig. S2B). As shown by smFISH, GFP mRNA levels in cell bodies were comparable in the DRG neurons transfected with the different mini-gene constructs (Fig. S2C). Robust axonal localization was seen for the mini-gene construct containing Cdc42 exon 7 but not for exon 6 (Fig. S2D). Neither mRNAs from the mini-gene containing Cdc42 exon 6 nor the GFP-3′palm-Cdc42 showed axonal levels significantly greater than the GFP control (Fig. S2D). These data further emphasize that sequences within the 3′UTR sequences of Cdc42 exon 7 are necessary and sufficient for axonal localization, which explains the more robust axonal localization of prenyl-Cdc42 compared to palm-Cdc42 mRNA.
Axonally localizing prenyl-Cdc42 mRNA drives axon growth in sensory neurons
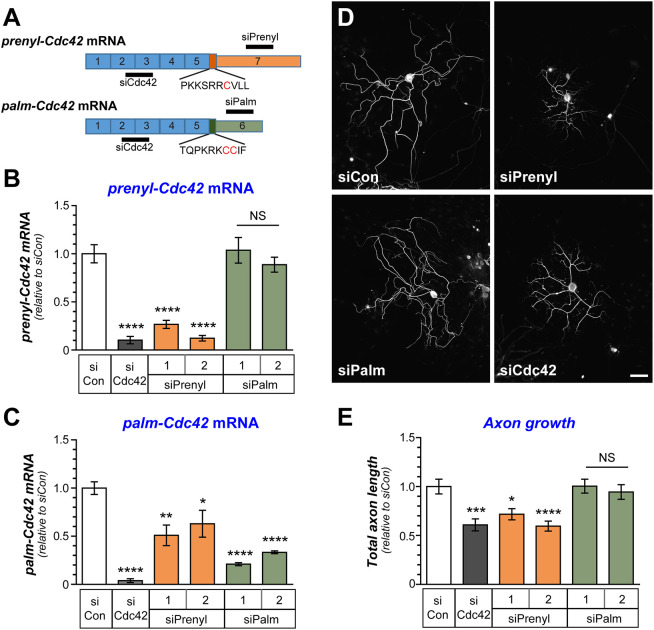
Cdc42 protein has been shown to localize to growth cones and its activity supports neurite outgrowth (Friesland et al., 2013; Matsuura et al., 2004; Myers et al., 2012). Recent work linked the prenyl-Cdc42 protein isoform to axon specification in CNS neurons while the palm-Cdc42 protein isoform was needed for dendritic spine development (Yap et al., 2016). To determine whether axonal localization of prenyl-Cdc42 mRNA contributes to these growth effects of Cdc42 protein, we selectively knocked down prenyl- versus palm-Cdc42 mRNAs or both using isoform-specific or pan-Cdc42 siRNAs (siPrenyl, siPalm and siCdc42; Fig. 3A). siCdc42 depleted both Cdc42 mRNA isoforms and, consistent with a previous report (Chandran et al., 2016), decreased axon growth in the DRG cultures (Fig. 3B–E). siPalm depleted palm-Cdc42 mRNA, but had no appreciable effect on prenyl-Cdc42 mRNA levels nor did it affect axon lengths (Fig. 3B–E). In contrast, siPrenyl significantly decreased axon growth to levels comparable to depletion of both mRNA isoforms using pan-siCdc42 (Fig. 3B–E). Notably despite trying multiple prenyl-Cdc42 targeting siRNAs, siPrenyl depleted prenyl-Cdc42 and palm-Cdc42 mRNAs, albeit with higher efficiency for the prenyl-Cdc42 isoform (≥75% versus 40–50%, respectively; Fig. 3B,C).
Fig. 3.
prenyl-Cdc42 isoform depletion decreases axon growth in adult sensory neurons. (A) Schematic for siRNAs targeting both Cdc42 isoforms (pan-siCdc42) and prenyl-Cdc42 isoform and palm-Cdc42 mRNA isoforms. Two closely spaced siRNAs were designed to target each isoform mRNA (indicated as siPrenyl 1 and 2 and siPalm 1 and 2). (B,C) Cdc42 mRNA levels were assessed by RT-ddPCR using primers selective for the prenyl-Cdc42 (B) and palm-Cdc42 (C) mRNA, where both were normalized to Hmgb1 mRNA. Levels are shown as mean±s.e.m. fold change relative to control siRNA (siCon) (N=3). *P<0.05; **P<0.01; ***P<0.005 (one-way ANOVA with pair-wise comparison with Tukey post-hoc tests). (D,E) Representative images for NF immunofluorescence in rat DRG neuron cultures transfected with siRNAs from A at 48 h post-transfection shown in D. Total axon length/neuron for transfected neurons shown as mean± s.e.m. (N≥90 neurons in three independent experiments). *P<0.05; ***P<0.005 by Kruskal–Wallis test with multiple comparisons). Scale bar: 100 µm.
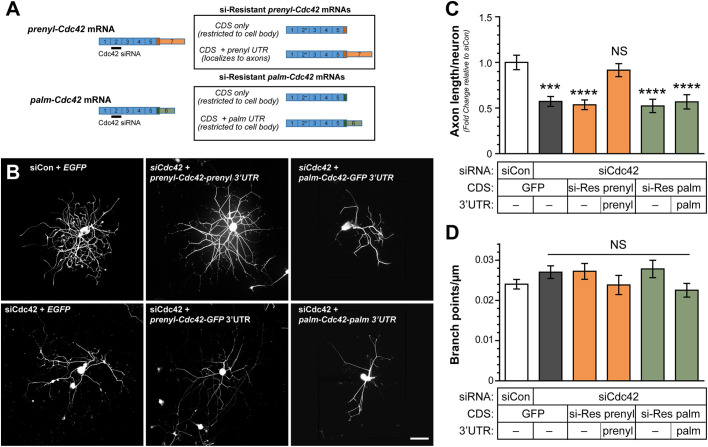
The comparable effect of siCdc42 and siPrenyl on axon outgrowth is consistent with the hypothesis that prenyl- and not palm-Cdc42 mRNA selectively supports axon growth, but we could not rule out contributions of the palm-Cdc42 mRNA isoform to axon growth given that its levels were decreased by both siCdc42 and siPrenyl transfections. Thus, we tested the ability of siRNA-resistant (‘si-Res’) prenyl-Cdc42 and palm-Cdc42 cDNA constructs to rescue the decrease in axonal growth seen after pan-Cdc42 depletion (Fig. 4A). Axon growth was only fully rescued by expressing the si-Res prenyl-Cdc42 mRNA with its axonally localizing 3′UTR (Fig. 4B,C). There was no rescue when the si-Res prenyl-Cdc42 mRNA including the non-localizing 3′UTR of the GFP construct was used (Fig. 4B,C). The si-resistant palm-Cdc42 mRNA similarly did not rescue the axon growth deficit in the siCdc42-transfected DRGs (Fig. 4B,C). Notably, branching of axons was not affected by pan-Cdc42 knockdown or rescue transfections with si-Res prenyl-Cdc42 or palm-Cdc42 constructs (Fig. 4D), indicating that Cdc42 expression contributes to axon elongation and not axon branching. Taken together, these data indicate that the axonally localizing prenyl-Cdc42 mRNA isoform supports axon growth in adult sensory neurons, but not the palm-Cdc42 mRNA isoform.
Fig. 4.
Axonal prenyl-Cdc42 mRNA rescues axon growth reduction by siRNA. (A) Schematic for siRNA-resistant cDNA constructs used in panels B–D. (B) Representative NF immunofluorescence images for rat DRG neurons transfected with siCon or pan-siCdc42 plus si-resistant cDNA constructs from A at 96 h post-transfection. Scale bar: 20 µm. (C,D) Total axon length/neuron for DRG neurons transfected as in B is shown in C as mean±s.e.m. fold change relative to siCon plus GFP transfection. D shows axon branching as mean±s.e.m. branch points/µm (N≥150 neurons in three independent experiments). ***P<0.005 (Kruskal–Wallis test with multiple comparisons).
To determine whether activity of prenyl-Cdc42 protein is needed for axon growth, we tested two small molecules that directly and specifically target Cdc42 activity. ZCL278 and ZCL367 were identified by an in silico screen for chemical structures predicted to bind to residues in Cdc42 protein that interact with the Cdc42-specific guanine nucleotide exchange factor (GEF) intersectin (herein referring to intersectin 1 and intersectin 2) (Friesland et al., 2013). ZCL278 has dual activities in that it decreases Cdc42 activity in vitro and in cultured cells when Cdc42 is ligand activated (Friesland et al., 2013), while it can increase Cdc42 activity when the protein is not ligand activated (Aguilar et al., 2019). Consistent with this, axon growth was significantly increased by 10 and 25 µM ZCL278 treatment, while axon growth was significantly attenuated by 25 µm ZCL367 treatment (Fig. S3A). The growth-promoting effect of ZCL278 in these adult DRG cultures was blocked by transfection with siPrenyl and siCdc42, but DRG neurons transfected with siPalm showed growth promotion by ZCL278 similar to control siRNA-transfected DRG neurons (Fig. S3B). In contrast, axon growth was equally decreased in siCon-, pan-siCdc42-, siPrenyl- and siPalm-transfected neurons (Fig. S3B). Taken together with the siRNA rescue data above, these observations indicate that activity of the prenyl-Cdc42 and not the palm-Cdc42 isoform promotes axon growth.
Differential localization of prenyl-Cdc42 and palm-Cdc42 mRNAs in cortical axons and dendrites
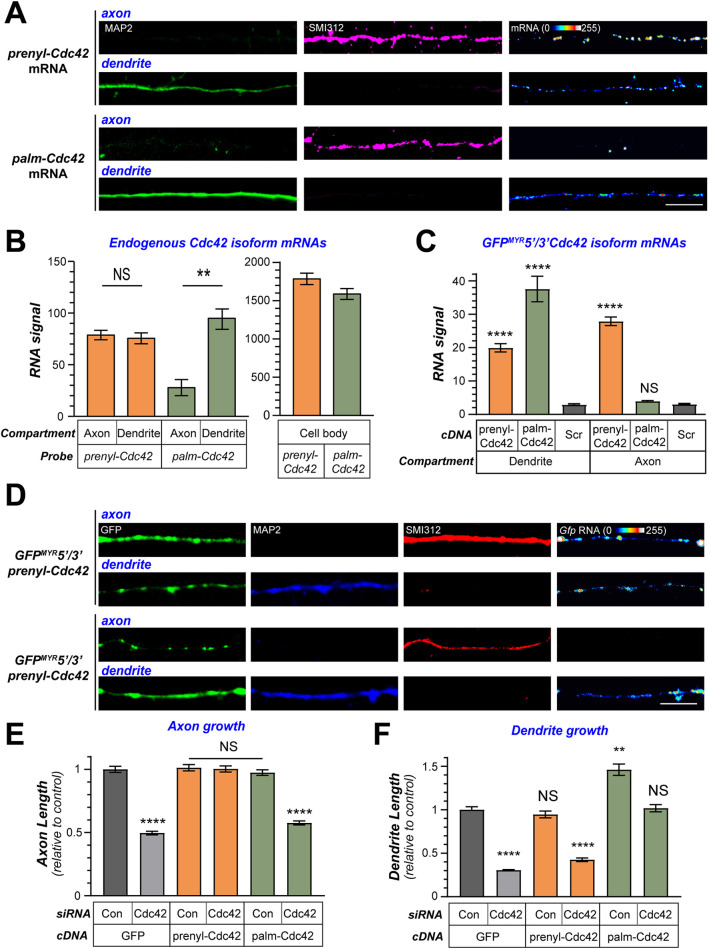
The DRG neurons used in the experiments above only extend neurites with axonal features in culture (Vuppalanchi et al., 2010; Zheng et al., 2001), but in our hands they do not full recapitulate the pseudo-unipolar morphology of DRG neurons in vivo. Thus, the selective axonal localization of prenyl-Cdc42 mRNA could reflect a feature of these uniquely polarized neurons compared to multipolar neurons with both axons and dendrites. Taliaferro et al. (2016) detected palm-Cdc42 mRNA in mouse cortical neurites by RNA-seq analyses (Table S2), so we asked whether the Cdc42 mRNA variants might show differential localization in axons and dendrites in experiments using embryonic rat cortical neurons that show distinct localization of axonal versus dendritic marker proteins by 5 days in vitro (DIV). The palm-Cdc42 mRNA was detected in dendrites and not axons of DIV9 cortical neurons from embryonic day 18 (E18) rat pups by smFISH, while prenyl-Cdc42 mRNA was detected in both axons and dendrites (Fig. 5A,B; Fig. S4A). The dendritic localization of palm-Cdc42 mRNA conflicts with a recent report that palm-Cdc42 mRNA is restricted to the cell body in mouse ESC-derived and DIV21 mouse cortical neurons by Ciolli Mattiolli et al. (2019). Considering the DIV9 cortical neurons used here showed different localization of the prenyl-Cdc42 and palm-Cdc42 mRNAs, we asked whether the 3′UTRs of these mRNAs might be sufficient for localizing GFP mRNA into the axonal or dendritic compartments of the cortical neurons. Consistent with the differential localization of the endogenous prenyl-Cdc42 and palm-Cdc42 mRNAs, GFPMYR5′/3′prenyl-Cdc42 mRNA localized into both axons and dendrites while the GFPMYR5′/3′palm-Cdc42 mRNA only localized into the dendrites (Fig. 5C,D). Thus, only the prenyl-Cdc42 mRNA isoform can localize into axonal compartment, while the palm-Cdc42 mRNA appears to be restricted to the somatodendritic compartment of these cortical neurons.
Fig. 5.
Differential localization and functions for Cdc42 mRNA isoforms in polarized cortical neurons. (A,B) Representative exposure matched smFISH and immunofluorescence (IF) images of DIV9 cortical neuron for prenyl-Cdc42 and palm-Cdc42 mRNAs plus phosphorylated NF (SMI312), as an axonal marker, and MAP2, as a dendritic marker show in A. For RNA signals in corresponding cell bodies, refer to Fig. S1B. Quantification of prenyl-Cdc42 and palm-Cdc42 mRNA signals in axons, dendrites, and cell bodies across multiple cortical neuron culture preparations is shown as mean±s.e.m. pixel intensity in B (n≥60 neurons in three independent transfections). **P<0.01; NS, not significant (Kruskal–Wallis test with multiple comparisons). Scale bars: 10 µm. (C,D) Axon and dendrite localization as determined by smFISH for GFP mRNA is shown for GFPMYR5/3′palm and GFPMYR5/3′prenyl mRNAs in dendrites versus axons. C shows quantitative data and D shows representative images using Map2 as a dendritic marker and SMI312 as an axonal marker (n≥60 neurons in three independent transfections). ****P≤0.001; NS, not significant versus scrambled smFISH probe [Scr] (Kruskal–Wallis test with multiple comparisons). (E,F) Quantification of axon and dendrite length are shown for DIV9 cortical neurons co-transfected with control or pan-Cdc42 siRNAs plus GFP, GFP-prenyl-Cdc42 or GFP-palm-Cdc42 mRNA is shown. See Fig. S4 for representative images and branching data (n≥100 neurons in three independent transfections). **P≤0.01; ****P≤0.001; NS, not significant versus control siRNA plus GFP (Kruskal–Wallis test with multiple comparisons).
Although only differing by 10 amino acids, unique functions for neuronal prenyl- versus palm-Cdc42 protein isoforms have been found in previous studies, with dendritic spine development ascribed to the palm-Cdc42 and axonogenesis ascribed to the prenyl-Cdc42 (Kang et al., 2008; Yap et al., 2016). Our findings in the DRGs are consistent with these previous reports, but do not test the functions for palm-Cdc42 in earlier stages of dendrite growth nor did they distinguish axonal versus dendritic growth functions for the prenyl-Cdc42. With the prenyl-Cdc42 mRNA selectively localizing into the cortical axons, we asked whether the two isoforms might have a differential effect on axon versus dendrite growth. For this, we co-transfected cortical neurons with siCdc42 plus si-resistant GFP–prenyl-Cdc42 or GFP–palm-Cdc42-encoding cDNAs that included the 5′ and 3′ UTRs of the prenyl-Cdc42 and palm-Cdc42 mRNAs. As expected, siCDC42 plus control GFP cDNA-transfected cortical neurons showed decreased axon and dendrite length compared to siCon plus GFP cDNA-transfected neurons, indicating that Cdc42 proteins contribute to both axon and dendrite growth (Fig. 5E,F; Fig. S4B). The axon growth deficit seen with pan-Cdc42 mRNA depletion was completely reversed by cotransfection with GFP-prenyl-Cdc42 but not GFP-palm-Cdc42 cDNA (Fig. 5E; Fig. S4B). In contrast, the decreased dendrite length seen with pan-Cdc42 mRNA depletion was reversed by co-transfection with GFP–palm-Cdc42- but not GFP–prenyl-Cdc42-encoding cDNA (Fig. 5F; Fig. S4B). Branching of axons was not affected by pan-Cdc42 mRNA depletion (Fig. S4C). However, the number of dendrites per neuron and branches from those dendrites were decreased by pan-Cdc42 mRNA depletion (Fig. S4D,E). Surprisingly, co-transfecting pan-Cdc42-depleted cortical neurons with GFP–prenyl-Cdc42-encoding cDNA, but not GFP–palm-Cdc42, rescued both branch points along dendrites as well as number of dendrites/neuron (Fig. S4D,E). Together, these data indicate that prenyl-Cdc42 and palm-Cdc42 serve distinct functions in axonal and dendritic growth. Notably, overexpression of prenyl-Cdc42 mRNA in the control siRNA-transfected cortical neurons significantly increased the number of primary dendrites per neuron; this could reflect a role for prenyl-Cdc42 in initiation of dendrite growth since technical limitations required cDNA transfection at DIV0 and siRNAs were introduced at DIV5.
Axonal growth promotion requires axonal localization of prenyl-Cdc42 mRNA and an intact CaaX motif
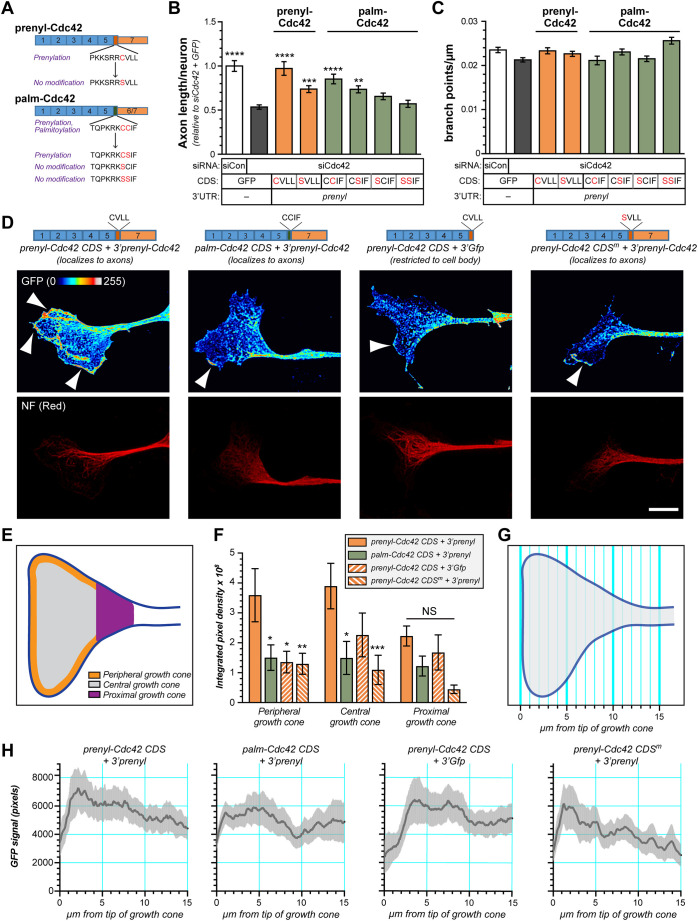
With the distinct subcellular localization for prenyl- versus palm-Cdc42 mRNAs and the axonally localizing prenyl-Cdc42 mRNA selectively rescuing axon growth deficits from pan-Cdc42 depletion, we wondered whether axonal localization of the mRNA or the differential post-translational modifications of locally synthesized CDC42 protein contribute to axon growth promotion. To address this possibility, we targeted si-Res prenyl-Cdc42 and palm-Cdc42 mRNAs into axons of cultured DRGs using the 3′UTR of the prenyl-Cdc42 mRNA. We further mutated the reported prenylation and palmitoylation sites in the C-terminal coding sequence of these constructs. The first 180 amino acids of palm-Cdc42 and prenyl-Cdc42 proteins are identical, with the distinct C-terminal 10 amino acids coming from exons 6 and 7, respectively (Chen et al., 2012). The four C-terminal residues of these two Cdc42 protein isoforms contain CCaX and CaaX motifs for palmitoylation and prenylation, respectively (where C represents cysteine, a is an aliphatic amino acid and X is any amino acid) (Wirth et al., 2013). The cysteine residues in the CaaX and CCaX motifs can be covalently modified by prenylation and palmitoylation, and mutating the cysteine residues to serine has been used to prevent prenylation and palmitoylation of proteins (Nishimura and Linder, 2013). The CCaX motif in the palm-Cdc42 protein product (CCIF) can undergo an initial prenylation at the first cysteine followed by palmitoylation at the second cysteine (Wirth et al., 2013). Thus, we generated siRNA-resistant mutants expressing palm-Cdc42 plus the 3′UTR of prenyl-Cdc42 with the C-terminal CCIF residues of the palm-Cdc42-encoding sequence mutated to CSIF, SCIF and SSIF. For prenyl-CDC42, we generated siRNA-resistant mutants with C-terminal CVLL residues mutated to SVLL (Fig. 6A).
Fig. 6.
The C-terminal CaaX motif is needed for optimal axon growth promotion by axonal prenyl-Cdc42 mRNA. (A) Schematic for si-resistant Cdc42 isoforms with CaaX and CCaX motifs. (B,C) Total axon length/neuron for DRG neurons transfected with pan-siCdc42 plus the indicated si-resistant CaaX and CCaX mutants for Cdc42 isoforms is shown in B as mean±s.e.m. fold change relative to siCon plus eGFP. C shows axon branching as mean±s.e.m. branch points/µm axon length. Note that mRNAs for all Cdc42 mutants were targeted into axons using the 3′UTR from prenyl-Cdc42 (N≥120 neurons over three independent experiments). *P<0.05; ***P<0.005 (Kruskal–Wallis test with multiple comparisons). (D) Representative exposure-matched fluorescence images for GFP (upper row) and NF (lower row) for indicated GFP–Cdc42 expression constructs with mutated C-termini in growth cones (arrowheads) of DRG neuron are shown as maximum xyz projection from deconvolved confocal z stacks. mRNAs for the cDNA constructs were cell body-restricted using 3′UTR of GFP or axonally localizing using 3′UTR of prenyl-Cdc42 mRNAs. Representative immunofluorescence (IF) images in each column are for neurons expressing the indicated Cdc42 cDNA construct. Scale bar: 10 µm. (E,F) Schematic peripheral, central and proximal domains of the growth cone used for GFP–Cdc42 signal quantification across the growth cone shown in E. Quantification of GFP–Cdc42 intensity in growth cone domains defined in E is shown as mean±s.e.m. in F. The protein product of the axonally localizing GFP-prenyl-Cdc42 plus 3′prenyl-Cdc42 shows significantly greater signals in the periphery and central domains compared to protein products of cell body-restricted GFP-Prenyl-Cdc42 plus 3′Gfp and axonally localizing GFP-Palm-Cdc42 plus 3′prenyl-Cdc42 and the GFP-prenyl-Cdc42 SVLL mutant (labeled CDSm) plus 3′prenyl-cdc42 (N≥9 growth cones over three independent experiments). *P<0.05; **P<0.01; ***P<0.001 (two-way ANOVA with Tukey post-hoc). (G,H) Schematic of method for unbiased assessment of GFP–Cdc42 signals across growth cone shown in G. Quantification of growth cone signals for indicated GFP–Cdc42 variants using the method defined in G is shown in H. The distribution of protein encoded by GFP-Cdc42-3′prenyl is significantly different than those from the other prenyl-Cdc42 and palm-Cdc42 constructs to SVLL mutant (N≥11 growth cones over three independent experiments; comparing GFP-Cdc42-3′prenyl curve to others by Kolmogorov–Smirnov test shows P<0.0001 for all and GFP-Cdc42-3′prenyl has D value=0.450 versus GFP-palm-Cdc42 plus 3′prenyl-cdc42, D=0.322 versus GFP-prenyl-Cdc42 CDS plus 3′Gfp, D=0.705 versus the GFP-prenyl-Cdc42 SVLL mutant (labeled CDSm) + 3′prenyl-Cdc42 where lower D values indicate greater differences between curves). CDS, coding sequence.
The axonally targeted, siRNA-resistant palm-Cdc42 mRNA with SCIF and SSIF mutations did not significantly affect axon length in the Cdc42-depleted DRG neurons (Fig. 6B). The axonally targeted siRNA-resistant palm-Cdc42 mRNA with an intact CCIF motif, and to a lesser extent with CSIF, induced partial rescue of axon growth in Cdc42-depleted DRG cultures compared to axonally targeted siRNA-resistant prenyl-Cdc42 with an intact CVLL motif (Fig. 6B). Notably, there was no significant change to axon branching upon expression of the wild-type or of these mutant Cdc42 constructs (Fig. 6C). Taken together, these data suggest that both axonal mRNA localization of prenyl-Cdc42 mRNA and an intact C-terminal CVLL sequence (i.e. a CaaX motif) for prenylation are needed for the full axon growth-promoting effects of the prenyl-Cdc42 isoform, with the palm-Cdc42 isoform only partially supporting axon growth when its mRNA is targeted into axons.
Prenyl-Cdc42 and palm-Cdc42 proteins show distinct localizations in growth cones
Since the axonally targeted palm-Cdc42 mRNA with the intact CCIF motif did not promote axon growth to the same extent as prenyl-Cdc42 mRNA, we asked whether prenyl-Cdc42 and palm-Cdc42 proteins might show different localization in growth cones. We used confocal microscopy to visualize GFP-tagged prenyl-Cdc42 and palm-CDC42 proteins in distal axons of DRG neurons transfected with Cdc42 constructs with or without axonally localizing 3′UTR from Cdc42 exon 7 (Fig. 6D, schematics). The protein product of the axonally targeted GFP-prenyl-Cdc42 mRNA showed fluorescent signals concentrated at the periphery of the growth cone, frequently with localization at or just beneath the cytoplasmic membrane at the leading edge of the growth cone (Fig. 6D). In contrast, signals from axonally targeted translation products for GFP-prenyl-Cdc42 mRNA with the CVLL to SVLL mutation and the axonally targeted translation products for GFP-palm-Cdc42 mRNA rarely gave rise to concentrated signals at the leading edge of the growth cone (Fig. 6D). When the axonal-targeting 3′UTR of GFP-prenyl-Cdc42 was replaced with a cell body-restricted 3′UTR, signals at the leading edge of the growth cone were not nearly as prominent as those seen with the axonally targeted GFP-prenyl-Cdc42 (Fig. 6D). Quantifying the signal intensities for these GFP–prenyl-Cdc42 and GFP–palm-Cdc42 showed significantly higher levels of protein at the growth cone periphery for the axonally targeted GFP-prenyl-Cdc42-3′prenyl-Cdc42 than cell body-restricted GFP-prenyl-Cdc42-3′Gfp or axonally targeted GFP-palm-Cdc42-3′prenyl-Cdc42 (Fig. 6E–H). The protein encoded by GFP-prenyl-Cdc42-3′prenyl-Cdc42 with the SVLL mutation showed growth cone peripheral signals that were intermediate between axonally targeted GFP-prenyl-Cdc42-3′prenyl-Cdc42 and the cell body-restricted GFP-palm-Cdc42-3′Gfp (Fig. 6E–H). These data indicate that axonal localization of prenyl-Cdc42 mRNA and an intact CaaX motif in the encoded protein are needed for targeting Cdc42 protein to the growth cone periphery.
DISCUSSION
Intra-axonally synthesized proteins contribute to axon growth, both during development and after axonal injury, and a few thousand mRNAs are now known to localize into axons (Kar et al., 2018). Our data indicate that the prenyl-Cdc42 mRNA, but not the palm-Cdc42 isoform, is transported into axons where its protein product increases lengths of growing axons. In contrast, both prenyl- and palm-Cdc42 mRNAs localize into dendrites, with the dendritically targeted palm-Cdc42 mRNA increasing dendrite length and the prenyl-Cdc42 mRNA increasing dendrite branching. With roles in actin polymerization and filopodia formation, Cdc42 activity has long been linked to axon growth and cell migration (Aguilar et al., 2017; Hall and Lalli, 2010). The palm-Cdc42 and prenyl-Cdc42 mRNA isoforms are generated by alternative splicing with differential inclusion of exons 6 and 7 in their mRNAs, respectively (Chen et al., 2012). Previous work in cultured CNS neurons pointed to roles for the palm-Cdc42 in dendrite maturation and prenyl-Cdc42 in axon specification, respectively (Yap et al., 2016). Our data emphasize that axonal mRNA targeting by Cdc42 gene exon 7 and an intact CaaX motif, which is needed for prenylation of its protein product, are responsible for the axon growth-promoting functions of Cdc42 protein and its localization to the leading edge of growth cones.
Alternative RNA splicing allows generation of different protein isoforms from a single genes as well as mRNAs with different post-transcriptional regulatory motifs. Subcellular localization of mRNAs is most frequently driven by 3′UTR sequences (Andreassi and Riccio, 2009; Gomes et al., 2014), as we show here for prenyl-Cdc42 mRNA. Kpnb1 (encoding importin β1), Ranbp1, Stat3a and Bdnf mRNA isoforms with long 3′UTRs are selectively transported into neuronal processes (An et al., 2008; Ben-Yaakov et al., 2012; Perry et al., 2012; Yudin et al., 2008). These mRNA variants are generated by alternative poly-adenylation site usage, with an RNA localization motif introduced from the additional 3′UTR sequences. The Kuruvilla laboratory recently showed that an mRNA isoform for another RhoA family member, Rac1, localizes into sympathetic axons via a long 3′UTR similarly generated by alternative poly-adenylation site usage (Scott-Solomon and Kuruvilla, 2020). In contrast, RNA-seq studies from neurites of neural cell lines showed prevalence for distal alternative last exons and additional exon usage over differential poly-adenylation site usage for subcellularly targeted mRNA isoforms (Taliaferro et al., 2016). This prevalence was similarly seen for mRNA isoforms that localize into neurites of primary cortical neurons and axons of embryonic DRG neuron cultures. Moreover, Taliaferro et al. (2016) observed a shift of alternative splicing to favor mRNA isoforms with localizing 3′UTRs during neuronal differentiation of CAD cells. Similarly, we see that inclusion of Cdc42 exon 7 rather than exon 6 selectively generates a Cdc42 mRNA isoform can localize into axons through its 3′UTR, while exon 6-containing Cdc42 mRNA only localizes into dendrites. Despite that, Cdc42 expression is reported to shift from predominantly prenyl-Cdc42 mRNA to predominantly palm-Cdc42 mRNA during brain development (Makeyev et al., 2007), RNA-seq analyses of cultured adult sensory neurons shown here indicate ∼5-fold higher cell body compartment and ∼10-fold higher axonal compartment levels for prenyl-Cdc42 compared with palm-Cdc42 mRNA. The cultured neurons used here continuously extend axons, suggesting that continued use of Cdc42 exon 7 drives axon growth by generating prenyl-Cdc42 mRNA that can localize into axons. Consistent with this notion, axonal prenyl-Cdc42 mRNA levels were much lower in uninjured, non-growing axons of the sciatic nerve than in regenerating axons. Since the sciatic nerve contains sensory and motor axons, the selective localization of prenyl-Cdc42 mRNA into regenerating adult axons also likely applies to injured motor axons.
Neuronal Cdc42 has been shown to contribute to axon growth initiation, elongation and growth cone guidance (Hall and Lalli, 2010). Linking Cdc42 functions to axon growth was determined in part based on overexpression of constitutively-active or dominant-negative mutations of Cdc42 (Banzai et al., 2000; Nishimura et al., 2005). Exogenous overexpression of one Rho GTPase has been shown to compete with endogenous Rho GTPases for binding to RhoGDI1 (also known as ARHGDIA), which decreases levels of endogenous Rho GTPases (Boulter et al., 2010). This complicates interpretation of previous findings for expression of dominant-negative and constitutively-active Cdc42s, and emphasizes the utility of the knockdown, pharmacological manipulations and rescue approaches used here for distinguishing functions of the Cdc42 isoforms. Our data further emphasize that ZCL278 can function as a Cdc42 agonist under some conditions, as shown by Aguilar et al. (2019). Since the growth promotion by ZCL278 was completely abrogated by depletion of prenyl-Cdc42 and not palm-Cdc42 mRNA, activity of prenyl-Cdc42 and not palm-Cdc42 is needed for axon growth promotion. Our data raise the possibility that roles previously ascribed to Cdc42 in axon specification and growth cone guidance derive from the axonally translated prenyl-Cdc42 mRNA. Notably, previous work with overexpression of Cdc42 and mutant Cdc42 proteins did not consider axonal localization of the mRNA (Banzai et al., 2000; Nishimura et al., 2005). The results of experiments with GFP fusion proteins show that Cdc42 proteins derived from the cell body-restricted palm-Cdc42 mRNA and prenyl-Cdc42 mRNA are clearly transported into distal axons, but the proteins show do not show enriched localization at the growth cone periphery as we see for the Cdc42 protein generated from axonally targeted prenyl-Cdc42 mRNA.
With isoform-specific overexpression and knockdown/knockout approaches, previous work showed that prenyl-Cdc42 is required for axon development and palm-Cdc42 is required for maturation of dendritic spines (Yap et al., 2016). This implies that neuronal polarity is determined in part by the different subcellular activities of the two Cdc42 isoforms. Although recent work in mouse ESC-derived and DIV21 primary cortical neurons only detected prenyl-Cdc42 mRNA in neurites (Ciolli Mattioli et al., 2019), RNA-seq data from primary cortical neuron cultures detected palm-Cdc42 mRNA in neurites (Taliaferro et al., 2016) and we see that palm-Cdc42 mRNA selectively localizes into dendrites of DIV9 rat cortical neurons. These discrepancies for palm-Cdc42 mRNA localization may reflect that Ciolli Mattioli et al. (2019) used DIV21 cortical cultures, which is well beyond the DIV9 neurons that we used here. Moreover, the effects of the dendritically targeted palm-Cdc42 mRNA seen here at DIV9 precede the initiation of dendritic spine growth and occur during a period of active dendrite extension (Craig and Banker, 1994). siRNA-based depletion of both Cdc42 mRNA isoforms from hippocampal neuron cultures has also been shown to decrease dendritic protrusions, both long and short protrusions (likely filopodia and spines, respectively), and this was rescued by expression of either prenyl- or palm-Cdc42 in DIV15 cultures (Wirth et al., 2013). It is not clear whether 3′UTRs for either the prenyl-Cdc42 or palm-Cdc42 mRNAs were included in the expression constructs used by Yap et al. (2016) and Wirth et al. (2013). In contrast, cortical neurons used here were transfected at DIV0 with rescue cDNAs encoding axonal and dendritic localizing prenyl- and palm-Cdc42 mRNAs followed by siRNA depletions beginning at DIV5 and analyses at DIV9. Under these conditions, depletion of both Cdc42 mRNA isoforms from cortical neurons decreases dendrite length and branching, with the length deficit selectively rescued by the localizing GFP–palm-Cdc42 and the branching deficit selectively rescued by the localizing GFP–prenyl-Cdc42. It will be of high interest to determine the mechanisms underlying these differential effects of prenyl- and palm-Cdc42 mRNAs on dendrite growth in future studies.
Our studies in the DRG neurons indicate that full axon growth promotion by Cdc42 protein requires both axonal localization of its mRNA and an intact CaaX motif, which is needed for post-translational modification. Covalent addition of lipids to proteins, specifically palmitoylation, prenylation, farnesylation and myristoylation, can alter subcellular localization of those modified proteins (Resh, 2006). For example, farnesylated RhoB protein is targeted to the plasma membrane, but prenylated RhoB protein is targeted to late endosomes (Wherlock et al., 2004). Palm- and prenyl-Cdc42 proteins have also been shown to have different motility in cell membranes, with palm-Cdc42 being more motile than prenyl-Cdc42 (Wirth et al., 2013). Palmitoylated proteins are reported to localize to lipid rafts in cell membranes while prenylated proteins are excluded from the cholesterol-rich rafts, although sequences within the Rho GTPases are known to contribute to their membrane localization beyond the modified CaaX and CCaX motifs (Levental et al., 2010). Thus, membrane localization of the prenyl-Cdc42 and palm-Cdc42 proteins could be indeed be altered by their post-translational modifications. Wirth et al. (2013) showed that the initial cysteine in palm-Cdc42 CCaX motif can also be prenylated, but both GFP–palm-Cdc42 with CCaX and a mutated CAaX C-termini showed faster recovery than GFP–prenyl-Cdc42 at the cytoplasmic membrane of N1E-115 cells after photobleaching. In our hands, the axonally synthesized GFP–prenyl-Cdc42 protein showed significantly increased localization to the periphery of growth cones compared to the GFP–palm-Cdc42 protein encoded by mRNA that included the axonally targeting 3′UTR of prenyl-Cdc42. The only sequence difference between these axonally targeted GFP–prenyl-Cdc42 and GFP–palm-Cdc42 constructs is in the nucleotides encoding the C-terminal most 10 amino acids (Fig. 6A). The GFP–prenyl-CDC42 CVLL to SVLL mutant showed growth cone localization intermediate between the GFP fusion proteins derived from the axonally targeted and cell body-restricted GFP-prenyl-Cdc42 mRNAs, emphasizing functionality of the CaaX motif in the locally synthesized Cdc42. Additionally, pan-Cdc42-depleted DRG neurons that were expressing axonally targeted palm-Cdc42 mRNA with the C-terminal CCIF to CSIF mutation, which previous work has indicated would allow for palmitoylation or prenylation of the encoded protein (Wirth et al., 2013), showed less axon growth rescue than the neurons expressing palm-Cdc42 mRNA encoding CCIF at the protein C-terminus (i.e. native palm-Cdc42 protein). This further emphasize the importance of the CaaX motif in prenyl-Cdc42 and provides further evidence that axonally translated proteins can be locally prenylated, likely increasing membrane localization of or stabilizing the protein, or both.
The prenyl transferases, farnesyl transferase (FT) and geranylgeranyl transferase (GGT), catalyze addition of either farnesyl or geranylgeranyl isoprenoid lipids to the cysteine residue in the CaaX motif. Scott-Solomon and Kuruvilla (2020) very recently showed that sympathetic axons have GGT activity, with nerve growth factor increasing axonal GGT activity; furthermore, knocking out GGT from sympathetic neurons attenuated TrkA signaling through loss of prenylated Rac1 from those axons. In contrast to the developing sympathetic neurons used by Solomon and Kuruvilla, the adult DRG neurons in our studies do not require exogenous trophic support for survival or axon growth. Both prenyl-Cdc42 and RhoA are substrates for GGT and not FT (Wirth et al., 2013). Both CCIF and CSIF motifs included in the axonally targeted palm-Cdc42 mRNA would be substrates for GGT, and hence prenylation (Nishimura and Linder, 2013; Wirth et al., 2013). Thus, the protein product of the palm-Cdc42 mRNA targeted into axons through the prenyl-Cdc42 3′UTR could undergo localized post-translational modifications. Consistent with this, the axonal growth deficit seen with pan-Cdc42 depletion was partially rescued by expression of axonally targeted palm-Cdc42 mRNA with intact CCIF and CSIF but not with the SCIF or SSIF mutations. The growth-promoting effects of prenyl-Cdc42 mRNA were lost when the mRNA was restricted to the soma by replacing the 3′UTR with that of GFP. Together, these observations emphasize that the optimal axon growth promotion of Cdc42 requires that its mRNA is spliced to exclude exon 6 and include exon 7 for axonal mRNA localization and that it includes the CaaX motif in the C-terminus of the locally translated protein.
In summary, our data indicate that the prenyl-Cdc42 isoform specifically drives the axon growth-promoting effects of Cdc42 in adult sensory neurons though axonal translation of its mRNA. An intact CaaX motif for post-translational prenylation of the axonally synthesized Cdc42 protein is essential for optimal axon growth promotion. In contrast, both palm- and prenyl-Cdc42 localize into dendrites, where palm-Cdc42 supports dendrite extension and prenyl-Cdc42 supports dendrite branching over the early stages of dendrite maturation. On first glance, our axon growth data seem to contradict a study that used prenylation inhibitors to link the post-translational modification to slowed axon growth on myelin-associated glycoprotein (MAG), a substrate that inhibits axon growth (Li et al., 2016). In cultured neurons, highly selective FT inhibitors were more effective than GGT inhibitors for increasing axon growth (Li et al., 2016), but the combination of the two synergistically block prenylation (Roberts et al., 2008). RhoA shows altered subcellular localization upon inhibition of GGT (Roberts et al., 2008), and in contrast to the growth-promoting effects of prenyl-Cdc42 shown here, RhoA attenuates axon growth on non-permissive substrates like MAG and the protein is synthesized in axons (Walker et al., 2012; Wu et al., 2005). Previous studies have shown compensatory geranylgeranylation through GGT for some farnesylated GTPases when FT is inhibited (Roberts et al., 2008), which could account for the increased efficacy of FT plus GGT inhibition seen by Li et al. (2016). Interestingly, reducing membrane cholesterol by inhibition of HMG-CoA reductase using Lovastatin was recently shown to decrease axon growth, but this was rescued by supplementing neurons with geranylgeranyl pyrophosphate, which is an eventual product of cholesterol synthesis and is needed for protein prenylation (Shabanzadeh et al., 2021). Based on our observations, depletion of cholesterol-rich lipid rafts combined with loss of geranylgeranylation of prenyl-Cdc42 could indeed account for the decreased axon growth with Lovastatin treatment.
MATERIALS AND METHODS
Animal use and neuron cultures
All animal experiments were conducted under IACUC approved protocols. For DRG cultures, L4–L6 ganglia were isolated from adult male Sprague Dawley rats (175–250 g) and dissociated with collagenase (Invitrogen). Cells were washed and then plated with DMEM/F12 (Cellgro) supplemented with 10% fetal bovine serum (Gemini Bio), 10 µM cytosine arabinoside (Sigma) and N1 supplement (Sigma). Cells were cultured on either glass coverslips or polyethylene-tetrathalate (PET) membrane (1 µm pores; Corning) inserts coated with poly-L-lysine (Sigma) and laminin (Millipore).
For cultures of cortical neurons, cortices from E18 male and female rat pups were harvested in Hibernate E (BrainBits) and dissociated using the Neural Tissue Dissociation kit (Miltenyi Biotec). Briefly, the harvested cortices were incubated in pre-warmed enzyme mix at 37°C for 15 min and triturated using fire-polished glass pipettes and applied to a 40 µm cell strainer. After washing and centrifugation, neurons were seeded onto poly-D-lysine coated glass coverslips at a density of 10,000 cells/coverslip in culture medium containing NbActive-1 medium (BrainBits) supplemented with 100 U/ml of penicillin-streptomycin (Life Technologies), 2 mM L-Glutamine (Life Technologies), and 1× N21 supplement (R&D Systems) and grown at 37°C, 5% CO2 for 7 days.
DRG cultures were transfected with 2–3 µg of plasmid using an Amaxa Nucleofection device (Lonza) with a Basic Neuron SCN Nucleofector kit (Program SCN-8) before plating and maintained for 48–72 h. For depletion experiments, synthetic siRNAs (predesigned from Integrated DNA Tech., 100 nM) were transfected into DRG neurons using DharmaFECT-3 and incubated for 3 DIV. For control, identical amounts of non-targeting siControl pools were used. Cortical cultures were similarly transfected with expression plasmids on DIV 0 using Amaxa Nucleofection, followed by siRNA transfections with DarmaFECT-3 reagent at DIV 5 and DIV 7 and then analyzed at DIV 9. RT-ddPCR was used to test the efficiency of depletion (see below). siRNA sequences were: siPrenyl (#1), 5′-GUGUUGUCAUCAUACUAAAAGCAAU-3′; siPrenyl (#2), 5′-GCAAUGUUUAAAUCAAACUAAAGAU-3′; siPalm (#1), 5′-GUGAUCAGUAGUCACAUUAGACUUG-3′; siPalm (#2), 5′-UCAGUAGUCACAUUAGACUUGUUUA 3′; siCdc42, 5′-UGGUAAAACAUGUCUCCUG-3′.
DRG cultures were exposed to the Cdc42-targeting small molecules, ZCL278 and ZCL367 (1–25 μM; Friesland et al. 2013), after the dissociated cells were adhered to coverslips (3–4 h post plating), and then assayed for axon growth 48 h later. Chemical structures and rationale for selection of ZCL278 and ZCL367 as Cdc42 targeting agents was previously described (Friesland et al., 2013).
Plasmid constructs
All fluorescent reporter constructs for analyses of RNA translation were based on eGFP with an myristoylation element (GFPMYR; originally provided by Dr. Erin Schuman, Max-Plank Inst., Frankfurt) (Aakalu et al., 2001). To isolate the cDNA of rat Cdc42 (NCBI ID: XM_008764286 for prenyl-Cdc42; XM_008764287 for palm-Cdc42), total RNA from rat DRG was reverse transcribed with Superscript II (Invitrogen) and then amplified by PCR using PrimeStar HS polymerase (Takara). Primers for amplifying the prenyl-Cdc42 coding sequence and 3′UTR were engineered to contain terminal NotI and SalI restriction sites. Using these restriction sites, the amplicons were then inserted into pEGFP-C3 (Clontech) vectors. Quikchange site directed mutagenesis kit (Stratagene) was used to introduce point mutations of CaaX or CCaX motifs.
Fluorescence in situ hybridization and immunofluorescence
Myristoylated eGFP was fused with 3′UTRs of Cdc42 isoforms to test their axonal localization. Transfected DRG neurons were fixed 15 min in 4% paraformaldehyde (PFA) and hybridized with DIG-labeled GFP cRNA probes (Roche) as described previously (Merianda et al., 2013). Sheep anti-DIG (1:100, cat. no. 713-175-147, Roche) was used to detect probes with Cy5-conjugated donkey anti-sheep-IgG (1:200, Jackson ImmunoResearch). Endogenous Cdc42 isoforms were detected with 5′ Cy5-labelled Stellaris probes to the exons 6 or 7 (probe sequences available upon request; BioSearch Tech.). Hybridization conditions for the Stellaris probes for cultured neurons and for tissue sections was performed as recently described (Terenzio et al., 2017).
For immunofluorescence, neurons plated on glass coverslips were fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. Cultures were permeabilized with 0.3% Triton X-100 in PBS for 15 min and incubated with primary antibodies for overnight in humidified chambers at 4°C. Chicken anti-NF (1:1000, cat. no. NFM, NFL and NFH, Aves labs), SMI312 mouse anti-phospho-NF (1:500, cat. no. 837904, Biolegend), and chicken anti-MAP2 (1:500; cat. no. ab5392, Abcam) were as primary antibodies; FITC-conjugated donkey anti-chicken and donkey anti-mouse (1:200, Jackson ImmunoResearch) were used as secondary antibodies. For visualization of GFP-tagged Cdc42 proteins in growth cones, rabbit anti-GFP (1:500, cat. no. ab290, Abcam) and chicken anti-NF (1:1000, Aves labs) was used for primary antibodies and FITC-conjugated donkey anti-rabbit-IgG (1:200, Jackson ImmunoResearch) and Cy5-conjugated donkey anti-chicken-IgY (1:200, Jackson ImmunoResearch) were used as secondary antibodies. Coverslips were washed with PBS and then incubated with secondary antibodies for an hour before mounting with Prolong anti-fade mounting solution (Invitrogen).
For analyses of smFISH signals, images were obtained using an epifluorescence microscope equipped with ORCA ER CCD camera (Hamamatsu) using matched acquisition parameters. Pixel intensities were measured from distal axons from more than 50 neurons in three replicate experiments. GFP signals in the growth cone were detected by Leica SP8X confocal microscope with HyD detectors. Z-stack images were post-processed by Huygens deconvolution (Scientific Volume Imaging) integrated into the Leica LASX software (HyVolution). Deconvolved image stacks were projected into single-plane images using the maximum pixel intensities.
‘Stellaris’ probes were used for mRNA detection in sciatic nerve sections as described previously (Spillane et al., 2013). Briefly, sciatic nerve segments were fixed overnight in 2% PFA at 4°C and then cryoprotected overnight in 30% sucrose at 4°C. Next, 25 µm thick cryosections were prepared and stored at −20°C until use. Slides were dried at 37°C for 1 h then brought to room temperature, and all subsequent steps were performed at room temperature unless indicated otherwise. Warmed tissue sections were washed 10 min in PBS once, then 10 min in 20 mM glycine three times. Sections were then incubated three times for 5 min in fresh 0.25 M NaBH4. Sections were quickly rinsed in 0.1 M triethanolamine (TEA), incubated for 10 min in 0.25% acetic anhydride in 0.1 M TEA, washed twice with 2× saline-sodium citrate (SSC) buffer, dehydrated in graded ethanol solutions (70%, 95% and 100% for 3 min each), and delipidated in chloroform for 5 min. After incubation in 100% and 95% ethanol for 3 min each, sections were equilibrated in 2× SSC and then incubated at 37°C in a humidified chamber in hybridization buffer without probes for 5 min before incubating overnight with probe (7 µM) and RT97 anti-NF (1:100) in hybridization buffer (Perry et al., 2016). Sections were then washed twice in 2X SSC plus 10% formamide at 37°C for 30 min and once in 2× SSC for 5 min, permeabilized in PBS plus 1% Triton X-100 (PBST) for 5 min, and then incubated for 1 h in donkey anti-mouse-IgG FITC-conjugated antibody (1:200) in 0.3% Triton X-100 supplemented with 1× blocking buffer (Roche). After washing with PBS for 5 min, sections were post-fixed in buffered 2% PFA for 15 min (Spillane et al., 2013), washed in PBS three times for 5 min, rinsed in DEPC-treated water, and mounted using Prolong Gold Antifade.
RNA isolation and PCR analyses
RNA was isolated from dissociated DRG neurons or cell body/axon compartments collected from insert cultures using RNeasy micro isolation kit (Qiagen). Purified RNAs were quantified with Ribogreen (Invitrogen) and 10-50 ng of RNA were used for reverse transcription with SensiFAST cDNA synthesis kit (Bioline) according to the manufacturer's protocol. To assess the purity of axonal RNA, RT-PCR was performed with primers designed to detect cell body-restricted mRNAs (c-Jun, H1f0, Map2) and glial cell-specific mRNAs (Gfap). Droplet digital PCR (ddPCR) was performed according to the manufacturer's procedure (Bio-Rad) with either Evagreen (Bio-Rad) or Taqman probe assays (Integrated DNA Tech). Hmgb1 mRNA levels were used for normalizing RNA yields across different isolates; we have previously shown that cell body and axonal RNA levels of Hmgb1 mRNA do not change after axotomy (Merianda et al., 2015).
Neuron growth analyses
For analyses of axon growth, neurons grown on glass coverslips or glass bottom 24-well plates were immunostained with neurofilament antibodies for DRGs and SMI312 and MAP2 antibodies for cortical neurons as described above. Entire coverslips or bottoms of the 24 well plates were scanned with ImageXpress Micro system (Molecular Devices) using a 20X objective. Stitched images were assessed for axon and dendrite length and branching using WIS-Neuromath (Rishal et al., 2013). All neurons where neurite arbors could be traced by the software were assessed (≥75 neurons per group/per experiment).
Fluorescence recovery after photobleaching
FRAP analyses was performed with minor modifications (Vuppalanchi et al., 2010). DRG neurons were transfected with GFPMYR3′prenyl-Cdc42 or GFPMYR3′palm-Cdc42, or GFPMYR3′Actg as a negative control. Cells were maintained at 37°C, 5% CO2 during imaging sequences. The 488 nm laser line on Leica SP8X confocal microscope was used to bleach GFP signal (argon laser at 70% power, pulsed every 0.82 s for 80 frames). Pinhole was set to 3 Airy units to ensure full thickness bleaching and acquisition (63×/1.4 NA oil immersion objective) (Yudin et al., 2008). Prior to photobleaching, neurons were imaged every 60 s for 2 min to acquire baseline fluorescence in the region of interest (ROI; 15% laser power, 498–530 nm for GFP). The same excitation and emission parameters were used to assess recovery over 15 min post-bleach with images acquired every 30 s. To determine whether fluorescence recovery in axons due to translation, DRG cultures were treated with 100 µM anisomycin (Sigma) for 30 min prior to photobleaching.
Analysis of RNA-seq data
Raw FASTQ files of GSE51572, GSE66230, and GSE67828 were downloaded from NCBI GEO database and adaptor sequences were trimmed as necessary (Briese et al., 2016; Minis et al., 2014; Taliaferro et al., 2016). Unpublished axonal RNA-seq data from our work (J.L.T., R.K. and G.C.) with cultures of adult C57Bl/6 mouse DRGs was similarly analyzed. Reads were then aligned to mm10 genome using STAR (Dobin et al., 2013) with default parameters. Mapped reads for mRNAs were counted by HTSeq and then normalized by trimmed mean of M values using EdgeR. Normalized counts were used to quantify levels of Cdc42 splice variants in axons, neurites or cell bodies.
Experimental design, image analyses and statistical analyses
All experiments using cultured neurons were designed to account for technical and inter-experimental variations. Consequently, all imaging experiments included at least three technical replicates on each culture (i.e. separate coverslips) and experiments were replicated across at least three separate culture preparations. High-content imaging was used for neurite outgrowth analyses such that at least 75 neurons per coverslip were analyzed. For molecular studies of transfected cultures (i.e. for ddPCR), analyses were performed on at least three separate culture preparations. Positive and negative controls were included as outlined above and in the Results section.
All smFISH/IF performed on tissue sections were imaged by confocal microscopy using a Leica SP8X confocal microscope with HyD detectors and post-processing measures to distinguish axonal signals from non-neuronal signals. Scrambled probes were used to assign image acquisition parameters to limit any nonspecific signal from the probes. A 145×145 μm nerve segment was scanned by taking xyz image stacks with 63× oil-immersion objective (1.4 NA). This xyz plane stack was captured at two locations along each nerve section. The Colocalization Plug-in for NIH ImageJ (https://imagej.nih.gov/ij/plugins/colocalization.html) was used to extract RNA signals from smFISH probes in each optical plane overlapping with axonal markers (NF) in the xy plane (Terenzio et al., 2018). All smFISH signal quantifications for axonal mRNA signals from tissue culture sections were generated by analyses of pixel intensity across each xy plane of the extracted ‘axon only’ channels for the image sequences using ImageJ. These smFISH signal intensities across the individual xy planes were then normalized to the area of NF immunoreactivity in each xy plane and averaged across the image z stack of the tile images (Kalinski et al., 2015). The relative mRNA signal intensity was averaged for all tiles in each biological replicate.
For quantification of signal intensity distribution in growth cones and terminal axon shaft and growth cones were imaged by confocal microscopy using a Leica SP8X confocal microscope with HyD detectors and deconvolved using Huygens Deconvolution software (https://svi.nl/Huygens-Deconvolution). Imaging sequences for terminal axon shaft and growth cones consisted of acquiring a 40–50 μm segment by taking xyz image stacks with 63× oil-immersion objective (1.4 NA) for 23–27 optical planes at z depth of 5.8–6.8 μm (0.25 μm interval between planes). The image stacks were projected using the maximum xyz projection. For unbiased protein distribution profiling across the growth cone, lines of 20 µm width were drawn using the line tool in ImageJ and tracked 15 µm proximally in to the axon shaft starting from the growth cone tip. Surface plots of the tracked growth cones were measured using ImageJ. For peripheral, central and proximal growth cone protein quantification, the segmented line tool in ImageJ was used to manually track these areas and integrated pixel intensities were measured.
Fluorescent intensities in the bleached regions of interest (ROI) were calculated by the Leica LASX software. For normalizing across experiments, the fluorescence intensity value at t=0 min post-bleach from each image sequence was set as 0%. The percentage of fluorescence recovery at each time point after photobleaching was calculated by normalizing relative to the pre-bleach fluorescence intensity for each image sequence was set at 100%.
Quantitative data are reported as mean±s.d. or mean±s.e.m. as indicated in the figure legends. A Student's t-test or one-way ANOVA with pairwise comparisons and Tukey post-hoc was used to test for significance between groups for the RT-ddPCR and FRAP studies as indicated in figure legends. A Kolmogorov–Smirnov test was used to compare for significant differences in GFP–Cdc42 isoform distribution for Fig. 6G,H. Large data sets (e.g. growth experiments and smFISH analyses of RNA levels) did not show a normalized data distribution, so a Kruskal–Wallis test with multiple comparisons was used to determine significance differences between groups as indicated in the figure legends. GraphPad Prism 5 software was used for all statistical analyses.
Supplementary Material
Acknowledgements
The authors thank members of the UofSC SmartState Center in Childhood Neurotherapeutics for guidance in these experiments.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.J.L., M.D.Z., J.L.T.; Methodology: S.J.L., M.D.Z., P.K.S., A.N.K., B.J.A., Q.L.; Validation: S.J.L., P.K.S., A.N.K.; Formal analysis: S.J.L., M.D.Z., P.K.S., A.N.K., R.K., G.C., J.L.T.; Investigation: S.J.L., M.D.Z., P.K.S., A.N.K., P.P., R.K., K.D.L., C.R.M., J.L.T.; Resources: B.J.A., Q.L.; Data curation: S.J.L., R.K., G.C.; Writing - original draft: S.J.L., M.D.Z., J.L.T.; Writing - review & editing: M.D.Z., P.K.S., A.N.K., B.J.A., J.L.T.; Supervision: A.N.K., J.L.T.; Project administration: J.L.T.; Funding acquisition: G.C., J.L.T.
Funding
This work was supported by grants from the National Institutes of Health (NIH; R01-NS089663 to JLT; R01-CA111891 and R15-CA165202 to Q.L.), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (to J.L.T. and G.C.), the Wings for Life - Spinal Cord Research Foundation (WFL-US-09/18 to P.P.), South Carolina Spinal Cord Injury Research Fund (2019 PD-02 to P.K.S.), the Harriet and John Wooten Foundation for Alzheimer's Disease Research (to Q.L.), and ASPIRE award from the University of South Carolina Office of Research (to S.J.L. and A.N.K.). J.L.T. is the incumbent SmartState Chair in Childhood Neurotherapeutics at the University of South Carolina. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.251967.supplemental
Peer review history
The peer review history is available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.251967.reviewer-comments.pdf
References
- Aakalu, G., Smith, W. B., Nguyen, N., Jiang, C. and Schuman, E. M. (2001). Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron 30, 489-502. 10.1016/S0896-6273(01)00295-1 [DOI] [PubMed] [Google Scholar]
- Aguilar, B. J., Zhu, Y. and Lu, Q. (2017). Rho GTPases as therapeutic targets in Alzheimer's disease. Alzheimers Res. Ther. 9, 97. 10.1186/s13195-017-0320-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar, B. J., Zhao, Y., Zhou, H., Huo, S., Chen, Y.-H. and Lu, Q. (2019). Inhibition of Cdc42-intersectin interaction by small molecule ZCL367 impedes cancer cell cycle progression, proliferation, migration, and tumor growth. Cancer Biol. Ther. 20, 740-749. 10.1080/15384047.2018.1564559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, J. J., Gharami, K., Liao, G.-Y., Woo, N. H., Lau, A. G., Vanevski, F., Torre, E. R., Jones, K. R., Feng, Y., Lu, B.et al. (2008). Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell 134, 175-187. 10.1016/j.cell.2008.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassi, C. and Riccio, A. (2009). To localize or not to localize: mRNA fate is in 3'UTR ends. Trends Cell Biol. 19, 465-474. 10.1016/j.tcb.2009.06.001 [DOI] [PubMed] [Google Scholar]
- Banzai, Y., Miki, H., Yamaguchi, H. and Takenawa, T. (2000). Essential role of neural Wiskott-Aldrich syndrome protein in neurite extension in PC12 cells and rat hippocampal primary culture cells. J. Biol. Chem. 275, 11987-11992. 10.1074/jbc.275.16.11987 [DOI] [PubMed] [Google Scholar]
- Ben-Yaakov, K., Dagan, S., Segal-Ruder, Y., Shalem, O., Vuppalanchi, D., Willis, D. E., Yudin, D., Rishal, I., Blesch, A., Pilpel, Y.et al. (2012). Axonal transcription factors signal retrogradely in lesioned peripheral nerve. EMBO J. 31, 1350-1363. 10.1038/emboj.2011.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulter, E., Garcia-Mata, R., Guilluy, C., Dubash, A., Rossi, G., Brennwald, P. J. and Burridge, K. (2010). Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat. Cell Biol. 12, 477-483. 10.1038/ncb2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese, M., Saal, L., Appenzeller, S., Moradi, M., Baluapuri, A. and Sendtner, M. (2016). Whole transcriptome profiling reveals the RNA content of motor axons. Nucleic Acids Res. 44, e33. 10.1093/nar/gkv1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran, V., Coppola, G., Nawabi, H., Omura, T., Versano, R., Huebner, E. A., Zhang, A., Costigan, M., Yekkirala, A., Barrett, L.et al. (2016). A systems-level analysis of the peripheral nerve intrinsic axonal growth program. Neuron 89, 956-970. 10.1016/j.neuron.2016.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Wirth, A. and Ponimaskin, E. (2012). Cdc42: an important regulator of neuronal morphology. Int. J. Biochem. Cell Biol. 44, 447-451. 10.1016/j.biocel.2011.11.022 [DOI] [PubMed] [Google Scholar]
- Ciolli Mattioli, C., Rom, A., Franke, V., Imami, K., Arrey, G., Terne, M., Woehler, A., Akalin, A., Ulitsky, I. and Chekulaeva, M. (2019). Alternative 3′ UTRs direct localization of functionally diverse protein isoforms in neuronal compartments. Nucleic Acids Res. 47, 2560-2573. 10.1093/nar/gky1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, A. and Banker, G. (1994). Neuronal polarity. Ann. Rev. Neurosci. 17, 267-310. 10.1146/annurev.ne.17.030194.001411 [DOI] [PubMed] [Google Scholar]
- Da Silva, J. S., Medina, M., Zuliani, C., Di Nardo, A., Witke, W. and Dotti, C. G. (2003). RhoA/ROCK regulation of neuritogenesis via profilin IIa-mediated control of actin stability. J. Cell Biol. 162, 1267-1279. 10.1083/jcb.200304021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla Costa, I., Buchanan, C., Zdradzinski, M. D., Sahoo, P. K., Smith, T. P., Thames, E., Kar, A. N. and Twiss, J. L. (2020). Functional platforms for organizing axonal mRNA transport and translation. Nat. Rev. Neurosci. 22, 77-91. 10.1038/s41583-020-00407-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., Batut, P., Chaisson, M. and Gingeras, T. R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15-21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed]
- Emanuelsson, O., Brunak, S., von Heijne, G. and Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953-971. 10.1038/nprot.2007.131 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville, S. and Hall, A. (2002). Rho GTPases in cell biology. Nature 420, 629-635. 10.1038/nature01148 [DOI] [PubMed] [Google Scholar]
- Friesland, A., Zhao, Y., Chen, Y. H., Wang, L., Zhou, H. and Lu, Q. (2013). Small molecule targeting Cdc42-intersectin interaction disrupts Golgi organization and suppresses cell motility. Proc. Natl. Acad. Sci. USA 110, 1261-1266. 10.1073/pnas.1116051110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, C., Merianda, T. T., Lee, S. J., Yoo, S. and Twiss, J. L. (2014). Molecular determinants of the axonal mRNA transcriptome. Dev. Neurobiol. 74, 218-232. 10.1002/dneu.22123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A. (1998). Rho GTPases and the actin cytoskeleton. Science 279, 509-514. 10.1126/science.279.5350.509 [DOI] [PubMed] [Google Scholar]
- Hall, A. and Lalli, G. (2010). Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb. Perspect Biol. 2, a001818. 10.1101/cshperspect.a001818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanz, S., Perlson, E., Willis, D., Zheng, J. Q., Massarwa, R., Huerta, J. J., Koltzenburg, M., Kohler, M., van-Minnen, J., Twiss, J. L.et al. (2003). Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron 40, 1095-1104. 10.1016/S0896-6273(03)00770-0 [DOI] [PubMed] [Google Scholar]
- Jalink, K., van Corven, E. J., Hengeveld, T., Morii, N., Narumiya, S. and Moolenaar, W. H. (1994). Inhibition of lysophosphatidate- and thrombin-induced neurite retraction and neuronal cell rounding by ADP ribosylation of the small GTP-binding protein Rho. J. Cell Biol. 126, 801-810. 10.1083/jcb.126.3.801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinski, A. L., Sachdeva, R., Gomes, C., Lee, S. J., Shah, Z., Houle, J. D. and Twiss, J. L. (2015). mRNAs and Protein synthetic machinery localize into regenerating spinal cord axons when they are provided a substrate that supports growth. J. Neurosci. 35, 10357-10370. 10.1523/JNEUROSCI.1249-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, R., Wan, J., Arstikaitis, P., Takahashi, H., Huang, K., Bailey, A. O., Thompson, J. X., Roth, A. F., Drisdel, R. C., Mastro, R.et al. (2008). Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456, 904-909. 10.1038/nature07605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar, A., Lee, S. and Twiss, J. (2018). Expanding axonal transcriptome brings new functions for axonally synthesized proteins in health and disease. Neuroscientist 24, 111-129. 10.1177/1073858417712668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Sauliere, J. and Wang, Z. (2016). The exon junction complex as a node of post-transcriptional networks. Nat. Rev. Mol. Cell Biol. 17, 41-54. 10.1038/nrm.2015.7 [DOI] [PubMed] [Google Scholar]
- Levental, I., Grzybek, M. and Simons, K. (2010). Greasing their way: lipid modifications determine protein association with membrane rafts. Biochem 49, 6305-6316. 10.1021/bi100882y [DOI] [PubMed] [Google Scholar]
- Li, H., Kuwajima, T., Oakley, D., Nikulina, E., Hou, J., Yang, W. S., Lowry, E. R., Lamas, N. J., Amoroso, M. W., Croft, G. F.et al. (2016). Protein prenylation constitutes an endogenous brake on axonal growth. Cell Rep. 16, 545-558. 10.1016/j.celrep.2016.06.013 [DOI] [PubMed] [Google Scholar]
- Luo, L. (2000). Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 1, 173-180. 10.1038/35044547 [DOI] [PubMed] [Google Scholar]
- Luo, L., Liao, Y. J., Jan, L. Y. and Jan, Y. N. (1994). Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Devel 8, 1787-1802. 10.1101/gad.8.15.1787 [DOI] [PubMed] [Google Scholar]
- Makeyev, E. V., Zhang, J., Carrasco, M. A. and Maniatis, T. (2007). The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435-448. 10.1016/j.molcel.2007.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura, R., Tanaka, H. and Go, M. J. (2004). Distinct functions of Rac1 and Cdc42 during axon guidance and growth cone morphogenesis in Drosophila. Europ. J. Neurosci. 19, 21-31. 10.1046/j.1460-9568.2003.03084.x [DOI] [PubMed] [Google Scholar]
- Merianda, T. T., Gomes, C., Yoo, S., Vuppalanchi, D. and Twiss, J. L. (2013). Axonal localization of neuritin/CPG15 mRNA in neuronal populations through distinct 5′ and 3’ UTR elements. J. Neurosci. 33, 13735-13742. 10.1523/JNEUROSCI.0962-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merianda, T. T., Coleman, J., Kim, H. H., Kumar Sahoo, P., Gomes, C., Brito-Vargas, P., Rauvala, H., Blesch, A., Yoo, S. and Twiss, J. L. (2015). Axonal amphoterin mRNA is regulated by translational control and enhances axon outgrowth. J. Neurosci. 35, 5693-5706. 10.1523/JNEUROSCI.3397-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minis, A., Dahary, D., Manor, O., Leshkowitz, D., Pilpel, Y. and Yaron, A. (2014). Subcellular transcriptomics-dissection of the mRNA composition in the axonal compartment of sensory neurons. Dev. Neurobiol. 74, 365-381. 10.1002/dneu.22140 [DOI] [PubMed] [Google Scholar]
- Myers, J. P., Robles, E., Ducharme-Smith, A. and Gomez, T. M. (2012). Focal adhesion kinase modulates Cdc42 activity downstream of positive and negative axon guidance cues. J. Cell Sci. 125, 2918-2929. 10.1242/jcs.100107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, A. and Linder, M. E. (2013). Identification of a novel prenyl and palmitoyl modification at the CaaX motif of Cdc42 that regulates RhoGDI binding. Mol. Cell. Biol. 33, 1417-1429. 10.1128/MCB.01398-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, T., Yamaguchi, T., Kato, K., Yoshizawa, M., Nabeshima, Y., Ohno, S., Hoshino, M. and Kaibuchi, K. (2005). PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat. Cell Biol. 7, 270-277. 10.1038/ncb1227 [DOI] [PubMed] [Google Scholar]
- Perry, R. B., Doron-Mandel, E., Iavnilovitch, E., Rishal, I., Dagan, S. Y., Tsoory, M., Coppola, G., McDonald, M. K., Gomes, C., Geschwind, D. H.et al. (2012). Subcellular knockout of importin beta1 perturbs axonal retrograde signaling. Neuron 75, 294-305. 10.1016/j.neuron.2012.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, R. B., Rishal, I., Doron-Mandel, E., Kalinski, A. L., Medzihradszky, K. F., Terenzio, M., Alber, S., Koley, S., Lin, A., Rozenbaum, M.et al. (2016). Nucleolin-mediated RNA localization regulates neuron growth and cycling cell size. Cell Rep. 16, 1664-1676. 10.1016/j.celrep.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh, M. D. (2006). Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2, 584-590. 10.1038/nchembio834 [DOI] [PubMed] [Google Scholar]
- Rishal, I., Golani, O., Rajman, M., Costa, B., Ben-Yaakov, K., Schoenmann, Z., Yaron, A., Basri, R., Fainzilber, M. and Galun, M. (2013). WIS-NeuroMath enables versatile high throughput analyses of neuronal processes. Dev. Neurobiol. 73, 247-256. 10.1002/dneu.22061 [DOI] [PubMed] [Google Scholar]
- Roberts, P. J., Mitin, N., Keller, P. J., Chenette, E. J., Madigan, J. P., Currin, R. O., Cox, A. D., Wilson, O., Kirschmeier, P. and Der, C. J. (2008). Rho Family GTPase modification and dependence on CAAX motif-signaled posttranslational modification. J. Biol. Chem. 283, 25150-25163. 10.1074/jbc.M800882200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo, P. K., Smith, D. S., Perrone-Bizzozero, N. and Twiss, J. L. (2018). Axonal mRNA transport and translation at a glance. J. Cell Sci. 131, jcs196808. 10.1242/jcs.196808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Solomon, E. and Kuruvilla, R. (2020). Prenylation of axonally translated Rac1 controls NGF-dependent axon growth. Dev. Cell 53, 691-705.e7. 10.1016/j.devcel.2020.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabanzadeh, A. P., Charish, J., Tassew, N. G., Farhani, N., Feng, J., Qin, X., Sugita, S., Mothe, A. J., Walchli, T., Koeberle, P. D.et al. (2021). Cholesterol synthesis inhibition promotes axonal regeneration in the injured central nervous system. Neurobiol. Dis. 150, 105259. 10.1016/j.nbd.2021.105259 [DOI] [PubMed] [Google Scholar]
- Spillane, M., Ketschek, A., Merianda, T. T., Twiss, J. L. and Gallo, G. (2013). Mitochondria coordinate sites of axon branching through localized intra-axonal protein synthesis. Cell Rep. 5, 1564-1575. 10.1016/j.celrep.2013.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliaferro, J. M., Vidaki, M., Oliveira, R., Olson, S., Zhan, L., Saxena, T., Wang, E. T., Graveley, B. R., Gertler, F. B., Swanson, M. S.et al. (2016). Distal alternative last exons localize mRNAs to neural projections. Mol. Cell 61, 821-833. 10.1016/j.molcel.2016.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenzio, M., Schiavo, G. and Fainzilber, M. (2017). Compartmentalized signaling in neurons: from cell biology to neuroscience. Neuron 96, 667-679. 10.1016/j.neuron.2017.10.015 [DOI] [PubMed] [Google Scholar]
- Terenzio, M., Koley, S., Samra, N., Rishal, I., Zhao, Q., Sahoo, P. K., Urisman, A., Marvaldi, L., Oses-Prieto, J. A., Forester, C.et al. (2018). Locally translated mTOR controls axonal local translation in nerve injury. Science 359, 1416-1421. 10.1126/science.aan1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuppalanchi, D., Coleman, J., Yoo, S., Merianda, T. T., Yadhati, A. G., Hossain, J., Blesch, A., Willis, D. E. and Twiss, J. L. (2010). Conserved 3'-untranslated region sequences direct subcellular localization of chaperone protein mRNAs in neurons. J. Biol. Chem. 285, 18025-18038. 10.1074/jbc.M109.061333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, B. A., Ji, S. J. and Jaffrey, S. R. (2012). Intra-axonal translation of RhoA promotes axon growth inhibition by CSPG. J. Neurosci. 32, 14442-14447. 10.1523/JNEUROSCI.0176-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherlock, M., Gampel, A., Futter, C. and Mellor, H. (2004). Farnesyltransferase inhibitors disrupt EGF receptor traffic through modulation of the RhoB GTPase. J. Cell Sci. 117, 3221-3231. 10.1242/jcs.01193 [DOI] [PubMed] [Google Scholar]
- Willis, D. E., van Niekerk, E. A., Sasaki, Y., Mesngon, M., Merianda, T. T., Williams, G. G., Kendall, M., Smith, D. S., Bassell, G. J. and Twiss, J. L. (2007). Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J. Cell Biol. 178, 965-980. 10.1083/jcb.200703209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth, A., Chen-Wacker, C., Wu, Y.-W., Gorinski, N., Filippov, M. A., Pandey, G. and Ponimaskin, E. (2013). Dual lipidation of the brain-specific Cdc42 isoform regulates its functional properties. Biochem. J. 456, 311-322. 10.1042/BJ20130788 [DOI] [PubMed] [Google Scholar]
- Wu, K. Y., Hengst, U., Cox, L. J., Macosko, E. Z., Jeromin, A., Urquhart, E. R. and Jaffrey, S. R. (2005). Local translation of RhoA regulates growth cone collapse. Nature 436, 1020-1024. 10.1038/nature03885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap, K., Xiao, Y., Friedman, B. A., Je, H. S. and Makeyev, E. V. (2016). Polarizing the neuron through sustained co-expression of alternatively spliced isoforms. Cell Rep 15, 1316-1328. 10.1016/j.celrep.2016.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudin, D., Hanz, S., Yoo, S., Iavnilovitch, E., Willis, D., Gradus, T., Vuppalanchi, D., Segal-Ruder, Y., Ben-Yaakov, K., Hieda, M.et al. (2008). Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron 59, 241-252. 10.1016/j.neuron.2008.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J.-Q., Kelly, T. K., Chang, B., Ryazantsev, S., Rajasekaran, A. K., Martin, K. C. and Twiss, J. L. (2001). A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. J. Neurosci. 21, 9291-9303. 10.1523/JNEUROSCI.21-23-09291.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.