ABSTRACT
The recognition and disposal of misfolded proteins is essential for the maintenance of cellular homeostasis. Perturbations in the pathways that promote degradation of aberrant proteins contribute to a variety of protein aggregation disorders broadly termed proteinopathies. The AAA-ATPase p97 (also known as VCP), in combination with adaptor proteins, functions to identify ubiquitylated proteins and target them for degradation by the proteasome or through autophagy. Mutations in p97 cause multi-system proteinopathies; however, the precise defects underlying these disorders are unclear. Here, we systematically investigate the role of p97 and its adaptors in the process of formation of aggresomes, membrane-less structures containing ubiquitylated proteins that arise upon proteasome inhibition. We demonstrate that p97 mediates aggresome formation and clearance, and identify a novel role for the adaptor UBXN1 in the process of aggresome formation. UBXN1 is recruited to aggresomes, and UBXN1-knockout cells are unable to form aggresomes. Loss of p97–UBXN1 results in increased Huntingtin polyQ inclusion bodies both in mammalian cells and in a C. elegans model of Huntington's disease. Together, our results identify evolutionarily conserved roles for p97–UBXN1 in the disposal of protein aggregates.
KEY WORDS: Ubiquitin, Aggresome, Aggregate, PolyQ, Inclusion body, Proteasome
Highlighted Article: The p97 AAA-ATPase and adaptor UBXN1 recognize ubiquitylated, misfolded proteins and deliver them to the aggresome for degradation. Loss of p97-UBXN1 inhibits aggresome formation and disposal of Huntingtin inclusion bodies.
INTRODUCTION
The process of protein translation, folding and targeting to subcellular compartments is intrinsically error-prone, and is therefore continuously and closely monitored by various quality control mechanisms (Chen et al., 2011; Harper and Bennett, 2016; Tyedmers et al., 2010; Wolff et al., 2014). Proper folding and sorting of proteins can be affected by transcription and translation errors, as well as thermal stress or pH fluctuations. As a result, an estimated 30% of newly synthesized proteins are misfolded (Harper and Bennett, 2016; Kim et al., 2011). Such proteins are engaged by chaperone systems to rescue functionality; however, should this approach fail, these proteins are rapidly and efficiently cleared by the ubiquitin-proteasome system (UPS) and autophagy (Tyedmers et al., 2010). These protein quality control mechanisms ensure that protein aggregates do not accumulate in normal cells. However, aberrant accumulation of protein aggregates that are frequently ubiquitylated is a hallmark of several human disorders, such as amyloidosis, diabetes and neurodegeneration, suggesting that failure of these systems to prevent or clear the protein aggregates may have deleterious consequences (Olzscha et al., 2011; Walther et al., 2015).
A variety of cellular stressors, such as proteasome impairment as examined in this study, result in a substantial increase and aggregation of misfolded, ubiquitylated proteins. These small protein aggregates are dynamically recruited to a structure known as the aggresome (Johnston et al., 1998; Kaganovich et al., 2008; Kopito, 2000). Aggresomes are membrane-less, juxta-nuclear structures encased in intermediate filament vimentin cages. In the prevailing model, ubiquitylated proteins are recognized by histone deacetylase 6 (HDAC6) and delivered by retrograde transport via the dynein motor to the microtubule organizing center (Kawaguchi et al., 2003). Aggresomes are not mere storage sites for protein aggregates, but actively recruit chaperones, E3 ligases such as tripartite motif-containing 50 (TRIM50), the 26S proteasome, autophagy receptors, such as p62 (also known as SQSTM1) and neighbor of BRCA1 gene 1 (NBR1), and lysosomes to rescue functional, or degrade non-functional proteins (Fusco et al., 2012; Kopito, 2000; Olzmann et al., 2007). Thus, aggresomes are generally believed to be cytoprotective structures that contribute to cell survival by sequestering misfolded, aggregation-prone proteins, and alleviating proteotoxic stress (Arrasate et al., 2004). However, their long-term persistence is detrimental to cell viability.
Numerous studies have reported the recruitment of the AAA-ATPase p97 (also known as VCP) to aggresomes and other disease-causing protein aggregates (Boyault et al., 2006; Ju et al., 2008; Kawaguchi et al., 2003; Song et al., 2008; Tiwari et al., 2016). p97 is a ubiquitin-selective unfoldase that processes proteins for degradation by the 26S proteasome (Bodnar and Rapoport, 2017b; Meyer et al., 2012). The functionality of p97 is best understood in its role in ER-associated degradation (ERAD), wherein misfolded proteins in the ER lumen are retro-translocated by p97 in an ATP-dependent manner to facilitate proteasomal degradation (Bodnar and Rapoport, 2017a). p97 is now appreciated to regulate processes such as extraction of ubiquitylated proteins from chromatin, ribosome-associated quality control and the cell cycle, and to contribute to the selective autophagy of organelles such as mitochondria (Meyer et al., 2012; Raman et al., 2011, 2015). Numerous dedicated p97 adaptor proteins mediate specificity in p97 recognition of ubiquitylated substrates. The largest family of adaptors contains 13 members with conserved ubiquitin-X domains (UBXDs) that bind to the N-terminus of p97 (Hanzelmann and Schindelin, 2017). Other adaptors include heterodimeric ubiquitin fusion degradation 1 (UFD1) and nuclear protein localization 4 (NPL4; also known as NPLOC4), as well as proteins that associate with the C-terminus of p97 via short linear motifs. Apart from a handful of adaptors, including the UFD1–NPL4 dimer, that have well-defined roles in ERAD, many adaptors remain largely unstudied and their specific roles in p97-regulated processes remain to be elucidated.
Mutations in p97 have been identified in individuals with a variety of aggregation-based disorders collectively termed multi-system proteinopathies (MSPs). These include amyotrophic lateral sclerosis (ALS), inclusion body myopathy (IBM), Paget's disease (PD) and frontotemporal dementia (FTD), among others (Hubbers et al., 2007; Meyer and Weihl, 2014; Weihl et al., 2007). Approximately 20 of the 50 disease-associated mutations that have been identified to date span the N-terminus, linker and first ATPase domain. This region is utilized for interaction with UBXD adaptors, and some reports suggest that these mutations impact the balance of UBXD adaptors associated with p97 and may underlie disease phenotypes (Ritz et al., 2011; Zhang et al., 2015). Disease-relevant mutations in p97 impact virtually all known p97 functions, making it difficult to identify a single pathway impacted in disease. However, one striking feature common to these disorders is the accumulation and persistence of ubiquitin-positive insoluble aggregates in patient tissue.
Previous studies investigating the relationship between p97 and aggresomes have reached conflicting conclusions, with some studies supporting a role for p97 in aggresome formation (Ju et al., 2008; Wojcik et al., 2004), while others suggesting a role in clearance (Boyault et al., 2006; Kobayashi et al., 2007). Furthermore, the role of p97-associated adaptors has not been examined. Understanding the role of specific p97 complexes and the mechanisms by which they recognize, sequester and clear aggregates may spur the development of agents that mitigate aggregation-based disorders. We sought to address the role of p97 and the UBXD family of adaptors in the formation and subsequent clearance of aggresomes. We find that in cells with impaired proteasomal function, p97 is required for both the formation and clearance of ubiquitylated substrates at aggresomes. Importantly, we provide evidence that the p97 UBXD adaptor UBXN1 is uniquely required for the formation of aggresomes. The failure to form and clear aggresomes via p97 complexes negatively impacts cell viability. We extend these results to demonstrate that loss of the p97-UBXN1 complex results in an increase in aggregates caused by CAG (glutamine, Q) expansions (polyQ) in the Huntingtin gene both in mammalian cells and in a C. elegans model of Huntington's disease (HD).
Our studies have identified an evolutionarily conserved p97-dependent pathway that regulates the identification and disposal of ubiquitylated, misfolded aggregates.
RESULTS
p97 is required for aggresome formation and clearance
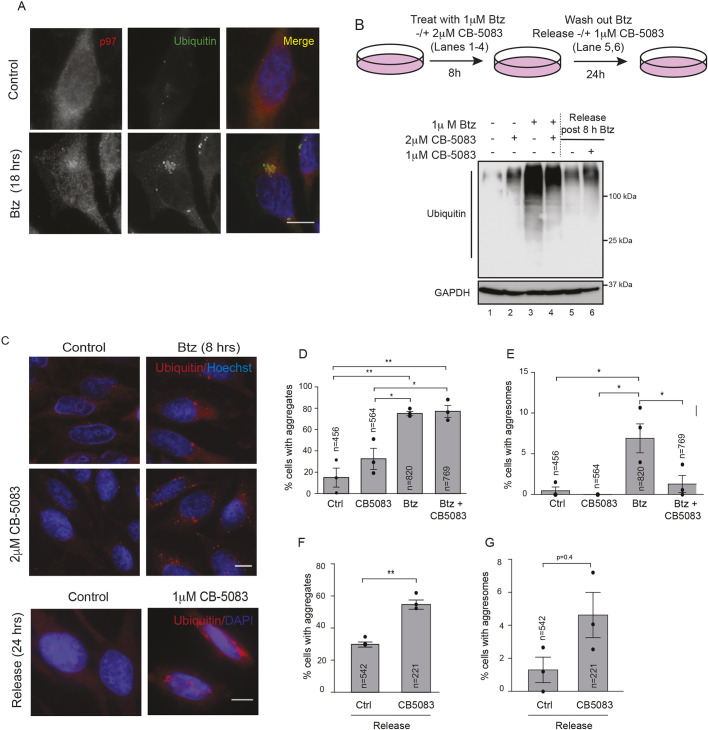
It has been previously reported that p97 colocalizes with ubiquitin-positive aggresomes as well as aggregates caused by over-expression of disease-causing mutant proteins. However, whether p97 escorts ubiquitylated substrates to aggresomes or aids in disassembly and clearance of protein aggregates is presently unclear (Boyault et al., 2006; Ju et al., 2008; Kobayashi et al., 2007; Wojcik et al., 2004). We treated cells with the proteasome inhibitor Bortezomib (Btz) in a time-course study and visualized aggregates by staining for ubiquitin. We observe aggresomes forming in the perinuclear region as early as 8 h and, by 18 h, a single large aggresome is apparent (Fig. S1A,B). We use 18 h of Btz treatment in all following studies except in instances where we co-treated cells with the p97 inhibitor CB-5083 (Fig. 1). In these experiments, aggresome formation was visualized at 8 h as incubation with both Btz and CB-5083 for 18 h resulted in significant cell death. Consistent with previous studies, we find that p97 is recruited to a ubiquitin-positive, perinuclear aggresome (Fig. 1A). We also observe ubiquitin staining in nuclear structures resembling the nucleolus (Fig. S1A, 18 h). Although previous studies have reported the redistribution of ubiquitylated proteins to the nucleolus upon proteasome impairment, the significance of this observation is presently unclear and nuclear aggregates were not included in our analysis (Mattsson et al., 2001). To provide quantitative data on cellular aggregates, we developed a facile ImageJ macro called AggreCount that allows for rapid and unbiased image analysis (Klickstein et al., 2020). AggreCount reports several metrics: (1) total number of cellular aggregates, (2) percentage of cells with aggregates, (3) aggregates per cell, (4) area of aggregates, and (5) localization of aggregates (cytosol, perinuclear or nuclear). Aggresomes are identified based on (1) perinuclear localization and (2) a minimum area cut-off that was empirically determined as the mean perinuclear aggregate area in Btz-treated cells for the 8 or 18 h time point (Fig. S1B). To investigate the role of p97 in aggresome formation we used a two-pronged approach, utilizing shRNA-mediated depletion of p97, as well as acute inhibition with CB-5083, an ATP-competitive p97 specific inhibitor (Zhou et al. 2015). We created stable, doxycycline-inducible shRNA HeLa Flp-in TRex cell lines (denoted sh-p97) that allowed us to achieve transient, partial depletion of p97 without overtly impacting cell viability (Fig. S1C). Two independent sh-p97 cell lines were treated with doxycycline (to deplete p97) and Btz to promote aggresome formation, followed by release into drug-free medium for a further 24 h to monitor clearance. We imaged ubiquitin-positive aggregates by immunofluorescence and analyzed aggregates using AggreCount (Fig. S1D,E). We found that long-term p97 depletion results in an increase in the percentage of cells with aggresomes and that the aggresomes were larger (mean±s.e.m. perinuclear aggregate area of 6.78±0.85 µm2), when compared to wild-type cells (perinuclear aggregate area of 4.2±0.15 µm2) (Fig. S1D,E). Notably, whereas wild-type cells cleared aggresomes upon removal of Btz, ubiquitin-positive aggresomes and aggregates persisted in a significant fraction of p97-depleted cells (Fig. S1D,E). In the second approach, we acutely inhibited p97 using CB-5083 concurrently with proteasome inhibition (for 8 h) to determine the impact on aggresome formation and clearance (Fig. 1B–G). Surprisingly, concurrent CB-5083 and Btz treatment resulted in a decrease in the number of cells with aggresomes compared to that seen upon Btz treatment alone even though the percentage of cells with aggregates was comparable between the two samples (Fig. 1B–E). We then visualized clearance of aggresomes in the presence of CB-5083 (1 µM of CB-5083 was the highest tolerated dose in release experiments due to significant cell death at higher concentrations of the drug) and found an increase in the persistence of ubiquitin conjugates (Fig. 1B, compare lanes 5 and 6). This was confirmed in imaging studies, where we found an increase in aggregates and, to a lesser extent, aggresomes, relative to what is found in untreated cells (Fig. 1C,F,G). We measured cell viability by quantifying cellular ATP levels in cells treated with Btz and/or CB-5083 during aggresome formation and release conditions. We observed no significant loss of cell viability when cells were treated with Btz in the presence or absence of CB-5083 for 8 h (Fig. S1F). However, the presence of CB-5083 during release severely impacted cell viability correlating with the persistence of aggregates under these conditions (Fig. S1F). We conclude that long-term depletion of p97 (∼72 h in shRNA-based studies) results in the accumulation of various p97 substrates that are recruited to aggresomes, increasing the size of and percentage of cells with aggresomes. However acute treatment (8 h in CB-5083 studies) leads to a direct inhibition of processes that recruit ubiquitylated substrates to aggresomes. Our studies suggest that the duration of p97 inhibition or depletion can have distinct outcomes that should be taken into account when designing studies to interfere with p97 function.
Fig. 1.
p97 is required for aggresome formation and clearance. (A) HeLa Flp-in T-Rex cells were treated with 1 µM Btz for 18 h. Cells were stained for p97, ubiquitin (FK2) and nuclei (Hoechst dye). (B) Hela Flp-in T-Rex cells were treated with 1 µM Btz, 2 µM CB-5083 or both for 8 h. Cells were released into drug-free medium or medium containing 1 µM CB-5083 for 24 h. Cell lysates were probed for ubiquitin. (C) Cells were treated as in B and imaged for ubiquitin-positive aggregates and aggresomes. (D) The percentage of cells with cellular aggregates (encompassing cytosolic and perinuclear aggregates) was quantified using AggreCount for images as in C. (E) The percentage of cells with perinuclear aggresomes (minimum size cutoff 2 µm2) was quantified using AggreCount for the images as in C. (F) The percentage of cells with cellular aggregates (encompassing cytosolic and perinuclear aggregates) in the release samples were quantified using AggreCount for images as in C. (G) The percentage of cells with perinuclear aggresomes in release samples were quantified using AggreCount for the images as in C. The black dot represents the mean from each biological replicate. The indicated number of cells (n) analyzed from all three independent biological replicates is shown for each condition. Graphs show the mean±s.e.m. *P≤0.05, **P≤0.01 as determined by one-way ANOVA with Bonferroni corrections (D,E) or an unpaired Student's t-test (F,G). Scale bars: 10 µm.
The p97 adaptors UBXN1 and NPL4 are localized to aggresomes
Given the significant role of p97 in aggresome formation and clearance, we next investigated the role of p97 adaptors in this process. We focused on the UBXD family, as these are the largest family of dedicated p97 adaptors, and many contain ubiquitin-associated domains (UBAs) for interaction with ubiquitin chains on substrates. Notably, p97 mutations known to cause MSPs have been reported to alter UBXD adaptor binding (Ritz et al., 2011; Zhang et al., 2015). We also tested the involvement of the UFD1–NPL4 dimer as it participates in most known p97-dependent processes. We first screened for UBXD adaptors that localized to the aggresome upon proteasome inhibition using antibodies to endogenous proteins. Of the nine adaptors tested (seven UBXD proteins, UFD1 and NPL4), we found that UBXN1 robustly localized to the aggresome (mean±s.d. Manders’ co-efficient of localization of 0.9±0.087; Fig. 2A; Figs S1A and S2A). Consistent with previous reports, we also observed recruitment of UFD1 and NPL4 to aggresomes (Manders’ co-efficient of localizations of UFD1, 0.842±0.065, and NPL4, 0.870±0.073; Fig. 2B) (Nowis et al., 2006). In contrast, the UBXD adaptor p47 (NSFL1C) was not present at aggresomes (Fig. 2B). In time-course studies, we find that UBXN1 is recruited as early as 6 h post-Btz treatment to nascent aggresomes as soon as ubiquitin staining is visible (Fig. S1A). We verified that UBXN1-positive structures were aggresomes by staining with canonical aggresome markers, such as Proteostat (a dye that detects aggregated proteins; Shen et al., 2011), HDAC6 and the 20S proteasome (Fig. 2C). In agreement with the dependence of aggresome formation on the microtubule network, nocodazole treatment caused the dispersal of aggresomes and numerous UBXN1- and ubiquitin-positive foci were observed instead (Fig. 2D). UBXN1 localization to aggresomes was not limited to HeLa cells but was observed in multiple cell types including the neuroblastoma cell line SH-SY5Y and the multiple myeloma cell line MM1.S (Fig. S2B,C). We next asked whether the localization of UBXN1 to ubiquitin-positive aggregates was limited to those formed as a result of proteasome inhibition or whether UBXN1 recognized a wider array of protein aggregates induced by distinct mechanisms. We found UBXN1 did not colocalize with stress granules formed as a result of sodium arsenite treatment (Fig. 2E). Unexpectedly, we observed the recruitment of another p97 adaptor, p47 to stress granules; the significance of this is presently under investigation (Fig. 2E). UBXN1 is not recruited to P-bodies, which are sites of mRNA decay (Fig. 2F). However, UBXN1 colocalization was observed with aggresome-like induced structures (ALIS), which contain defective ribosoal products that arise from treatment with translational poisons such as puromycin (Fig. S2D) (Ganji et al., 2018). Consistent with previous reports, we find that aggresome formation and polyubiquitin accumulation can be reversed by inhibiting protein translation with cycloheximide (Fig. S2E,F) (Nawrocki et al., 2005). Taken together, we have identified UBXN1 as an aggresome-localized protein that likely recognizes specific types of misfolded, ubiquitylated proteins.
Fig. 2.
The p97 adaptors UBXN1 and NPL4 are localized to aggresomes. (A) HeLa Flp-in T-Rex cells were treated with 1 µM Btz for 18 h and cells were stained for UBXN1 and ubiquitin. Colocalization was determined through the Manders’ overlap coefficient for 25 cells in three replicate experiments. (B) NPL4, UFD1 and p47 localization to aggresomes labeled with ubiquitin in cells treated with 1 µM Btz for 18 h. Colocalization was determined through the Manders’ overlap coefficient for 25 cells in three replicate experiments. (C) GFP–UBXN1 colocalizes with the aggresome markers Proteostat, HDAC6 and 20S proteasomes in cells treated with 1 µM Btz for 18 h. (D) Microtubules are required for GFP–UBXN1 localization to aggresomes. Nocodazole co-incubation in Btz-treated (1 µM Btz for 18 h) cells prevents aggresome formation. Lower panel, the number of aggresomes was quantified. (E) Hela Flp-in T-Rex cells were treated with 0.1 mM sodium arsenite for 2 h. Cells were stained for stress granule marker G3BP1 and p97 adaptors (UBXN1 or p47, used here as a positive control. (F) Stable mCherry–Dcp1a cells (labeling P bodies) were stained with UBXN1. The indicated number of cells was analyzed from the three independent biological replicates. Manders’ coefficients are given as mean±s.e.m. For D, the box represents the tenth to 90th percentiles, and the median is indicated. The whiskers show the s.e.m. ***P≤0.001 as determined by unpaired Student's t-test. Scale bars: 10 µm.
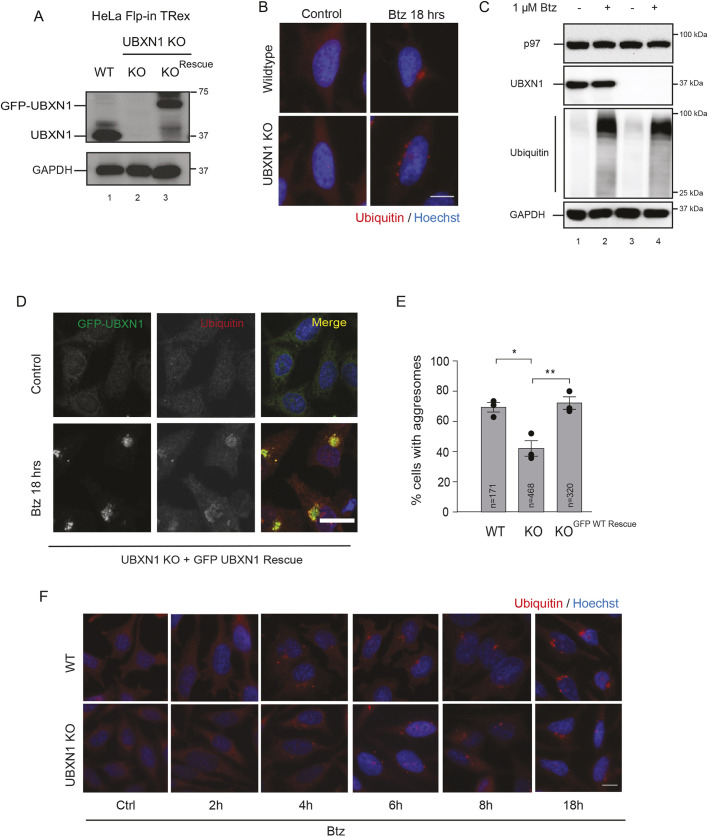
UBXN1 is required for the formation of aggresomes
To determine the role of UBXN1 in aggresome formation, we used the UBXN1-knockout (KO) HeLa Flp-in TRex cell lines created using CRISPR/Cas9 gene-editing that we published previously (Fig. 3A) (Ganji et al., 2018). We assessed aggresome formation in response to Btz treatment in two separate UBXN1 KO clonal lines. We found that loss of UBXN1 resulted in the loss of a single perinuclear aggresome and an increase in aggregates throughout the cytosol (Fig. 3B,E; Fig. S3A). We additionally verified the UBXN1 KO phenotype by transient depletion of UBXN1 with two separate siRNAs and similarly observed the loss of aggresomes (Fig. S3B). Notably, there was no change in total ubiquitin conjugates between wild-type and UBXN1 KO cells (Fig. 3C). To rule out off-target effects, we introduced doxycycline-inducible GFP–UBXN1 into the single FRT site in the UBXN1 KO cell line. Induction of GFP–UBXN1 reinstated aggresome formation as well as GFP–UBXN1 localization to these structures in response to Btz treatment (Fig. 3A,D,E). We examined the possibility of rapid onset and dissolution of aggresomes in KO cells that were missed in our endpoint assay, and performed time-course studies in the UBXN1 KO cells treated with Btz. However, we were unable to observe aggresomes at all time points tested, ruling out temporal differences in aggresome formation in UBXN1 KO cells (Fig. 3F).
Fig. 3.
UBXN1 is required for the formation of aggresomes. (A) Immunoblot showing loss of UBXN1 in CRISPR/Cas9 generated knockout cells and re-expression of wild-type GFP–UBXN1 by doxycycline induction. (B) Wild-type (WT) and UBXN1 KO HeLa Flp-in TRex cell lines were treated with 1 µM Btz for 18 h. Cells were stained for ubiquitin. (C) Cell lysates from WT and KO cells treated with 1 µM Btz for 18 h were probed for ubiquitin. (D) GFP–UBXN1 expression in UBXN1 KO cells was induced by doxycycline. Cells were treated with Btz and stained for ubiquitin. Expression of GFP–UBXN1 reinstated aggresome formation in UBXN1 KO cells. (E) Quantification of data in B and D. (F) WT and UBXN1 KO lines were treated with 1 µM Btz for the indicated times and stained for ubiquitin. The black dot represents the mean from each biological replicate. The indicated number of cells (n) analyzed from all three independent biological replicates is shown for each condition. Graphs show the mean±s.e.m. *P≤0.05, **P≤0.01, as determined by one-way ANOVA with Bonferroni correction. Scale bars: 10 µm.
Although we did not observe other UBXD adaptors recruited to the aggresome, we nevertheless asked whether other adaptors were required for the formation of aggresomes. We created a p47 KO cell line and a NPL4 knockdown cell line using CRISPR/Cas9 gene-editing in HeLa Flp-in TRex cells (Fig. 4A). We used siRNA-mediated depletion of p47 and NPL4 to confirm our findings and to additionally examine the roles of UFD1 and FAF1 (Fig. S4A,B). In addition to UBXN1, both NPL4 and UFD1 (but not p47 or FAF1) were required for aggresome formation (Fig. 4A–C; Fig. S4A,B). These findings are in agreement with prior studies that identified a role for the UFD1–NPL4 dimer in aggresome formation and are not all together surprising given the importance of UFD1–NPL4 in p97 activity. We fractionated cells to isolate detergent insoluble proteins. Notably, the level of soluble and insoluble ubiquitin conjugates was comparable between wild-type, UBXN1 KO and NPL4 KD cell lines indicating that the total levels of ubiquitylated substrates does not change, only their ability to get recruited to aggresomes (Fig. S4C). This was additionally verified by staining these cells with Proteostat. Despite the inability to form aggresomes, the smaller aggregates in UBXN1 KO and NPL4 KD cells were Proteostat positive, indicating the presence of insoluble aggregated proteins (Fig. 4D). To determine whether UBXN1 and UFD1–NPL4 functioned together on a single p97 hexamer or whether they were part of exclusive p97 complexes we performed two-step affinity purifications in cells transfected with FLAG–NPL4 and Myc–UBXN1. In the first step, FLAG–NPL4-associated complexes were affinity purified via the FLAG epitope, and bound proteins were eluted with FLAG peptide. Endogenous p97 in the eluate was purified with a p97 antibody and the presence of Myc–UBXN1 was determined by immunoblotting. As seen in Fig. S4D, a small proportion of UBXN1 was bound to the same p97 hexamer as NLP4, and the interaction was modestly stimulated by Btz treatment (lanes 7, 8, 11 and 12). We co-depleted UBXN1 and UFD1–NPL4 to observe the effect on aggresome formation. However, depletion of UBXN1 and UFD1 or NPL4 in (by dual siRNA transfection) in combination with Btz resulted in cell death precluding further analysis. Next, we asked whether either UBXN1 or NPL4 were required for the recruitment of p97 to ubiquitin-positive aggregates; however, we were unable to see loss of p97 recruitment in single knockdown cells and toxicity in double-knockdown samples prevented further analysis. Collectively, our studies have uncovered a novel role for UBXN1 in mediating aggresome formation.
Fig. 4.
UBXN1 and NPL4 are required for aggresome formation. (A) Immunoblot validation of CRISPR/Cas9 gene-edited knockout (p47; KO) or knockdown (NPL4; KD) cell lines. WT, wild type. (B) Aggresome formation in CRISPR/Cas9 gene-edited KO cell lines for the indicated UBXD adaptors. (C) Quantification of data in B. (D) Aggregates in UBXN1 KO and NPL4 KD cells contain insoluble aggregated proteins as observed by staining with Proteostat. The black dot represents the mean from each biological replicate. The indicated number of cells (n) analyzed from all three independent biological replicates is shown for each condition. Graphs show the mean±s.e.m. *P≤0.05, **P≤0.01 as determined by one-way ANOVA with Bonferroni correction. Scale bars: 10 µm.
p97 association is required for the role of UBXN1 in aggresome formation
Our results thus far suggest that p97 and its adaptors UBXN1 and UFD1–NPL4 are required for aggresome formation. Since the UFD1–NPL4 dimer functions in many p97-dependent processes, we focus on the role of UBXN1 in aggresome formation for the remainder of the study.
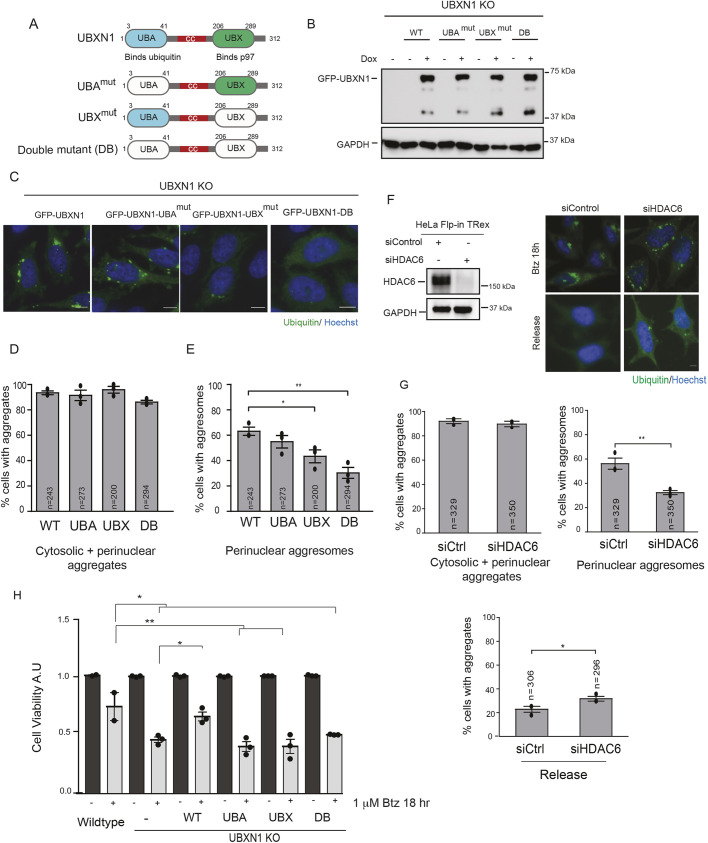
UBXN1 contains an N-terminal UBA domain that associates with ubiquitin and a C-terminal ubiquitin-X (UBX) domain that interacts with p97, separated by a coiled-coil region (Fig. 5A). We asked whether the association with ubiquitin and/or p97 was required for aggresome formation. We mutated conserved residues within the UBA domain (Met13 and Phe15 to Ala, UBAmut) that mediate association with the hydrophobic isoleucine patch on ubiquitin, or the UBX domain (Phe265Pro266Arg267 truncated to Ala-Gly, UBXmut) that associates with the N-terminus of p97. A double mutant with both domains mutated (DB) was also created (Fig. 5A). GFP-tagged mutants were introduced into the FRT site within the UBXN1 KO cell line and equal levels of induction of GFP-tagged proteins was determined by immunoblotting (Fig. 5A,B). We verified that the mutants were unable to bind ubiquitin or p97 using co-immunoprecipitation assays in HEK293T cells expressing Myc-tagged UBXN1 constructs (wild-type, UBAmut or UBXmut) and probing for endogenous ubiquitin or p97. As predicted, the UBXN1 UBAmut had a decreased ability to bind ubiquitin conjugates and UBXN1 UBXmut was deficient in p97 binding (Fig. S5A,B). We next induced expression of GFP–UBXN1 wild-type, UBAmut, UBXmut or the DB mutant in UBXN1 KO cells and assessed aggresome formation (based on perinuclear localization and size). Re-expression of wild-type UBXN1 rescued aggresome formation as before. Both the UBAmut- and UBXmut-expressing cells had defects in aggresome formation compared to cells expressing wild-type GFP–UBXN1, with a greater defect in the UBXmut (Fig. 5C-E, Fig. S5C). However, the DB cells had the most severe phenotype, with less than 40% of cells forming aggresomes. Notably, when we measured the fraction of cells that contained both cytosolic and perinuclear aggregates, it was comparable between wild-type and mutants, indicating that the aggregate burden was similar in all cases, ruling out increased clearance in the mutant expressing cells (Fig. 5D). These results suggest that association of UBXN1 with ubiquitin and p97 is an important requirement for aggresome formation.
Fig. 5.
Analysis of domains within UBXN1 that are necessary for aggresome formation. (A) Domain organization of UBXN1 showing N-terminal ubiquitin associated domain (UBA), coiled coil domain (cc) and C-terminal ubiquitin X domain (UBX). (B) Expression of GFP–UBXN1, GFP–UBXN1-UBAmut, GFP–UBXN1-UBXmut and the double mutant (DB) in the UBXN1 KO cell line induced by the addition of doxycycline for 72 h. (C) UBXN1 KO cells expressing GFP–UBXN1 wild-type (WT), GFP–UBXN1-UBAmut, GFP–UBXN1-UBXmut and GFP–UBXN1-DB were treated with 1 µM Btz for 18 h and stained for ubiquitin. Re-expression of WT UBXN1 but not the single mutants or double mutant rescued aggresome formation. The UBAmut has smaller aggresomes but did not reach significance. (D) The percentage of cells with aggregates (encompassing cytosolic and perinuclear aggregates) were quantified for images as in C. (E) The percentage of cells with perinuclear aggresomes were quantified for the images as in C. (F) The depletion of HDAC6 inhibits aggresome formation and clearance in HeLa Flp-in TRex cells treated with 1 µM Btz for 18 h. Left panel shows knockdown of HDAC6. (G) Quantification of images in F. (H) WT, UBXN1 KO (and indicated rescue lines) and NPL4 cell lines were plated in triplicate into 96-well plates and treated with 1 µM Btz for 18 h. Cell viability was measured and normalized to the value of untreated controls for each cell line. The black dot represents the mean from each biological replicate. The indicated number of cells (n) analyzed from all three independent biological replicates is shown for each condition. Graphs show the mean±s.e.m. for panels in D, E and G and mean±s.d. of panel H. *P≤0.05, **P≤0.01 as determined by one-way ANOVA with Bonferroni correction (D,E) or Dunnett's multiple comparison test (H), or unpaired Student's t-test (G). Scale bars: 10 µm.
Our studies indicate that UBXN1 acts early in the recognition of ubiquitylated substrates to mediate aggresome formation in collaboration with p97. Given that p97-depleted or inhibited cells were also unable to clear aggregates upon Btz removal, we asked whether the same was true for the aggregates observed in cells lacking UBXN1. Surprisingly, we found that cells lacking UBXN1 or NPL4 were capable of clearing aggregates upon Btz removal (Fig. S5D). This finding was further substantiated in cell viability studies, which found no difference in the viability of wild-type, UBXN1 KO and NPL4 KD cells during release conditions (Fig. S5E). One possibility is that p97 utilizes distinct partners for the formation and clearance of aggresomes. p97 has been reported to associate with HDAC6 in prior studies, and HDAC6 overexpression enhances clearance of aggregates in cells expressing p97 disease mutants (Ju et al., 2008). In agreement with previous studies, we find that p97 and HDAC6 associated with each other when transiently overexpressed in cells (Fig. S5F) (Boyault et al., 2006). The depletion of HDAC6 resulted in the loss of aggresomes as expected (Fig. 5F,G). Notably, HDAC6-depleted cells were less able to clear aggregates upon Btz removal (Fig. 5F,G).
To determine whether deregulated aggresome dynamics resulting from UBXN1 or NPL4 loss of function impacted cell survival in the face of proteasome impairment, we measured cell viability in these cells. We found that loss of UBXN1 and NPL4 sensitized cells to Btz treatment compared to what was seen in wild-type cells (Fig. S5E). Importantly, re-expression of wild-type UBXN1 but not UBAmut, UBXmut or DB rescued survival (Fig. 5H). Taken together, our data suggest that p97–UBXN1 is required for the formation of aggregates, but that aggresome clearance is mediated via other p97 adaptors or interactors, possibly HDAC6. Both these events are independently important for alleviating proteotoxic stress.
Loss of p97 and UBXN1 leads to an increase in Huntingtin polyQ aggregates
We were interested in determining whether UBXN1 has roles in the recognition of disease-relevant aggregates. Expansion of a CAG tract in the first exon of the huntingtin (HTT) gene beyond a threshold of ∼35–40 repeats causes Huntington's disease (HD) and results in a mutant HTT protein containing an expanded polyglutamine (polyQ) segment (MacDonald et al., 1993). Expression of exon 1 HTT fragments longer than 40 residues leads to the formation of insoluble amyloid-like aggregates of HTT, known as inclusion bodies (that share features with aggresomes) and contributes to neurodegeneration (Scherzinger et al., 1997, 1999; Waelter et al., 2001). We used a previously published, doxycycline-inducible HTT polyQ91-mCherry U2OS cell line to investigate the role of UBXN1 in inclusion body formation (Bersuker et al., 2016). Treatment with doxycycline led to the appearance of ubiquitin and mCherry–Q91-positive aggregates in ∼1–2% of cells, consistent with previous studies (Bersuker et al., 2016; Ramdzan et al., 2012). A recent study suggested that the ubiquitylation of HTT inclusion bodies is not a pre-requisite for their formation. We used the ubiquitin E1 inhibitor TAK-273 at a dose that depleted endogenous ubiquitin conjugates in both untreated and Btz-treated cells (Fig. S6A) and indeed found that Q91 inclusion bodies still formed but were not ubiquitylated (Fig. S6B). However, in cells treated with TAK-273, there were significantly more Q91 inclusion bodies compared to untreated cells, to a similar extent to what was seen with Btz treatment (Fig. S6C). Unlike the role of p97 in aggresome formation (Fig. 1), we found that inhibition of p97 using CB-5083 resulted in an increase in Q91 inclusion bodies (Fig. S6C). Thus, while the formation of HTT inclusion bodies does not require ubiquitylation, it is dependent on the ubiquitin-proteasome system and p97 for clearance.
We observed recruitment of UBXN1 to Q91 aggregates where it formed a ring-like structure around the aggregate, similar to ubiquitin (Fig. 6A). We tested the impact of UBXN1 depletion on Q91 aggregate formation using siRNA transfection followed by microscopy. Surprisingly, loss of UBXN1 resulted in an increase in the number of polyQ inclusion bodies in cells in a manner comparable to proteasome or p97 inhibition (Fig. 6B; Fig. S6C,D). This was unexpected given the role of UBXN1 in aggresome formation. The high fluorescence intensity of Q91 aggregates necessitated lower capture settings during image acquisition that prevented us from quantifying smaller, less-intense aggregates. To address this shortcoming, we used the flow cytometry-based pulse shape analysis (PulSA) method, which has been used to analyze particles of different sizes including polyQ inclusion bodies (Bersuker et al., 2016; Ramdzan et al., 2012). PulSA uses standard pulse width and height in the fluorescence channel. The formation of HTT-containing aggregates leads to a decrease in pulse width and an increase in pulse height, allowing for the visualization of smaller aggregates and larger inclusion bodies. As we had observed from imaging studies, PulSA analysis of p97- or UBXN1-depleted cells indicated an ∼2.5-fold increase in polyQ inclusion bodies compared to control samples (Fig. S6E). PulSA also enabled us to measure less intense aggregates that were missed in the imaging studies. We found an increase in less intense Q91–mCherry aggregates in cells depleted of UBXN1 or p97, suggesting that loss of this complex promotes the transition of Q91 oligomers into aggregates (Fig. 6C).
Fig. 6.
Role of p97 and UBXN1 in Huntingtin polyQ aggregation. (A) U2OS cells were treated with doxycycline to induce the expression of HTT Q91–mCherry. Cells were fixed and stained for ubiquitin and UBXN1 to demonstrate colocalization with HTT Q91–mCherry. (B) UBXN1 was transiently depleted with siRNAs in U2OS HTT Q91-mCherry cells and imaged for inclusion body formation. The percentage of cells with HTT Q91–mCherry inclusion bodies was quantified. (C) PuLSA analysis of HTT Q91-mCherry aggregates in UBXN1- and p97-depleted cells. (D,E) Representative fluorescent images of L4 larvae stage wild-type, cdc-48.1(tm544) (D), ubxn-1(tm2759) and ubxn-4(ok3343) worms. (E) Loss-of-function mutants expressing polyQ40::YFP in body wall muscle. Images were taken of worms precisely age-matched at the L4.4 vulva developmental stage. Bottom panels in each show quantification of visible fluorescent aggregates in L4 larvae animals expressing polyQ40::YFP in wild-type, cdc-48.1, ubxn-1 and ubxn-4 mutant animals. Quantification was only performed on worms at the L4.4 vulva development stage based on vulva morphology. The black dot represents the mean from each biological replicate. The indicated number of cells (n) analyzed from all three independent biological replicates is shown for each condition. Graphs show mean±s.e.m. *P≤0.05, **P≤0.01, ****P≤0.0001 as determined by one-way ANOVA with Tukey post-hoc testing (B) or Dunnett's test (E), and unpaired Student's t-test (D). Scale bar: 10 µm. Scale bars: 10 μm (B); 100 μm (D).
We next asked whether p97–UBXN1 regulated polyQ aggregation in vivo. We used transgenic worms expressing YFP–polyQ40 under the control of the unc-54 myosin heavy chain promoter to direct expression in the body wall muscle cells (mQ40) (Morley et al., 2002). Previous studies have demonstrated that mQ40 marks a threshold at which the reporter begins to aggregate in worms (Morley et al., 2002). There are two isoforms of p97 (cdc-48.1 and cdc-48.2) with redundant functions in worms. In addition, six UBXD adaptors (including ubxn-1) and ufd-1 and npl-4 are conserved. We crossed cdc-48.1, ubxn-1 and ubxn-4 (worm ortholog of mammalian UBXD2) mutant worms with the mQ40 strain. To accurately quantify aggregates in control and mutant strains, worms were age-matched at the L4.4 vulval development stage, and aggregates were quantified by microscopy. We found that control mQ40 worms typically had on average 20 aggregates; however, cdc-48.1 and ubxn-1 mutant worms had a twofold increase in mQ40 aggregates, in agreement with our findings in mammalian cells (Fig. 6D,E). This finding is in agreement with a previous study where the overexpression of either cdc-48 isoform in mQ40-expressing worms partially suppressed Q40 aggregation (Yamanaka et al., 2004). Surprisingly, ubxn-4 mutant worms also displayed an increase in mQ40 aggregates, indicating that additional adaptors may be utilized for HTT aggregate clearance in worms (see Discussion).
In summary, we have identified an evolutionarily conserved role for p97–UBXN1 in the recognition, formation and clearance of ubiquitylated protein aggregates.
DISCUSSION
Cellular proteostasis networks are complex, yet flexible to accommodate the need for thousands of proteins to perform various cellular functions while minimizing protein misfolding and aggregation. Defects in this large network result in MSPs, and several recent reports suggest that the age-related decline in the quantity, capacity and efficiency of these systems may be an underlying cause of these disorders (Walther et al., 2015). Whether aggregates observed in human diseases are causative of disease-relevant phenotypes or if they represent a cytoprotective structure meant to sequester and shield aggregates is controversial. However, at least in the case of the polyQ inclusions observed in Huntingtin's disease, the soluble microaggregates are believed to be more toxic than the inclusion body (Arrasate et al., 2004; Bauerlein et al., 2017; Takahashi et al., 2008).
In this study, we have identified a novel role for the p97 adaptor UBXN1 in the recognition of specific types of cellular aggregates. UBXN1 is the only UBXD adaptor that robustly localizes to aggresomes, and cells lacking UBXN1 have a diminished ability to form a single aggresome but instead display dispersed microaggregates. In particular, the total ubiquitin burden (measured by immunoblotting total cellular ubiquitin conjugates, as well as by quantifying cytosolic and perinuclear aggregates through imaging) remains comparable in wild-type and UBXN1 KO cells. Although p97 has been implicated in a large number of ubiquitin-dependent events, few of its numerous adaptors have been linked to these processes. Indeed, apart from the UFD1–NPL4 dimer, which functions in most p97 processes (due to dual roles in enhancing ubiquitylated substrate binding to p97 as well as stimulating the unfoldase activity of p97) many of its adaptors including UBXN1 remain poorly studied (Blythe et al., 2017; Bodnar and Rapoport, 2017b). A single UFD1–NPL4 dimer is believed to associate with the p97 hexamer, and at least in the case of FAF1 and UBXD7, UFD1–NPL4 association with the p97 hexamer is a pre-requisite for binding (Hanzelmann et al., 2011). Our studies suggest that a fraction of the cellular pool of UBXN1 and the UFD1–NPL4 dimer associates with the same p97 hexamer. The presence of dual adaptors on p97 may increase the capacity of a single hexamer in capturing multiple substrates or may enhance the avidity of substrate interaction by providing multiple ubiquitin-binding domains on the same molecule. Further studies are required to identify the cargo and molecular architecture of the p97 aggresome targeted complex. Neither UBXN1 nor NPL4 are required for aggresome clearance following Btz removal, and this suggests that p97 may switch adaptors for clearance purposes. In support of this, we and others have found that p97 interacts with HDAC6, which is required for aggresome clearance and may collaborate with p97 in this process. Identifying sites of interaction between these molecules and testing interaction-deficient mutants in aggresome formation will provide conclusive evidence for the role of a p97–HDAC6 complex in aggresome clearance. What factors may dictate adaptor choice during these distinct phases and how assembly of the p97 complex is regulated remains to be determined.
Surprisingly, depleting p97 protein levels or inhibiting its catalytic activity have distinct outcomes in terms of aggresome formation. We find that long-term depletion of p97 leads to an increase in cells with aggresomes that are larger than control cells. In contrast, acute p97 inhibition with CB-5083 leads to loss of aggresomes. Previous studies using ubiquitin remnant capture proteomics suggest that although many p97 and 26S proteasome targets are shared, p97 additionally processes ubiquitylated substrates that are not proteasomal targets (Gendron et al., 2016). Thus, the long-term accumulation of these substrates may contribute to the enhanced aggresome phenotype. In contrast, both depletion and inhibition of p97 lead to defects in the clearance of aggresomes.
We find that depletion of p97 and UBXN1 increases polyQ-containing aggregates in mammalian cells in vitro and in a C. elegans model of HD. Using PuLSA, we show that depletion of p97 and UBXN1 in mammalian cells leads to an increase polyQ inclusion bodies, which is distinct from the role of p97–UBXN1 in aggresome formation. What factors could account for these different phenotypes? PolyQ inclusion bodies share several features with aggresomes; they are ubiquitylated, frequently (although not exclusively) form near the perinuclear area, are dependent on microtubules for their formatio, and recruit similar protein quality control components (Muchowski et al., 2002). However, there may be distinctions that influence what proteins are recruited to each structure and in the discrete steps that culminate in their removal. Aggresomes are composed of a diverse array of ubiquitylated proteasomal substrates, whereas polyQ inclusion bodies are seeded by misfolded HTT and eventually trap other cellular components. UBXN1 may implement aggresome formation via association with a distinct cohort of proteins (both ubiquitylated substrates and regulators). However, any increase in smaller polyQ aggregates in UBXN1-depleted cells may enable direct seeding of larger inclusion bodies. We have previously shown that loss of UBXN1 impairs the clearance of ALIS, whereas UBXN1 appears to have no role in aggresome clearance, further underscoring distinctions in how different aggregates are handled by the same protein (Ganji et al., 2018). In worms, ubxn-1 does not appear to be the sole p97 adaptor in the recognition of inclusion bodies, we find that ubxn-4 mutant worms also have increased polyQ aggregates. Mammalian UBXD2 (the ortholog of ubxn-4) is a component of the ERAD system and the increased polyQ aggregation that we observe in ubxn-4 may be due to inhibition of ERAD and the accumulation of misfolded proteins that exacerbates polyQ aggregation. Further studies are warranted to explore the role of other p97 adaptors in polyQ aggregation.
How does UBXN1 recognize cargo destined for the aggresome? One possibility is the recognition of ubiquitin chains on substrates by the UBA domain in UBXN1. Our studies with UBA and UBX mutants suggest that both ubiquitin binding and p97 recruitment are critical for aggresome formation. However, it is likely that UBXN1 associates with other regulators for aggresome formation to access dynein motors that transport cargo to the perinuclear area. Consistent with this, depolymerization of the microtubule cytoskeleton, caused dispersal of UBXN1 and ubiquitin-positive aggregates. Future studies will involve identification of UBXN1 interactors induced by proteasome stress. Recently, components of the linear ubiquitin chain assembly complex (LUBAC) E3 ligase have been shown to be recruited to and modify HTT polyQ inclusion bodies with linear ubiquitin chains (van Well et al., 2019). LUBAC recruitment was dependent on p97, and both p97 recruitment and LUBAC-mediated linear ubiquitylation of HTT polyQ aggregates were required for efficient clearance. What other protein quality control components associate with UBXN1 and the types of ubiquitin linkages are recognized by UBXN1 and will be a topic of future studies.
Aggresomes were identified more than 20 years ago, yet the full complement of proteins that are recruited to aggresomes remains incomplete (Johnston et al., 1998). It is unclear what distinguishes different types of protein aggregates from one another and how specific components of protein quality control pathways are appropriately engaged. For example, aggresomes have long been known to be encased in intermediate filament vimentin cages. However, it was only recently that it was demonstrated that vimentin specified the recruitment of proteasomes to aggresomes and is an important determinant in how aggregates are triaged as adult neural stem cells exit quiescence (Morrow et al., 2020). Given the prevalence of protein aggregates in numerous neurodegenerative diseases, a complete understanding of the principles that govern aggresome formation and clearance will aid in our understanding of disease-relevant aggregates. Our study demonstrates that the process of aggresome formation and clearance are distinct events mediated by multiple proteins. Given the role of p97 mutations in human disorders related to aggregate clearance, it will be of interest to determine whether UBXN1 regulates their formation and whether targeting this event may have therapeutic benefit.
MATERIALS AND METHODS
Cell lines and culture
HEK-293T (ATCC), HeLa Flp-in TRex (a kind gift from Brian Raught, University of Toronto, Canada), SHSY-5Y (a kind gift from Wade Harper, Harvard Medical School, USA), and U2OS HTT Q91-mCherry (a kind gift from Ron Kopito, Stanford University, USA) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin and streptomycin. Multiple myeloma MM1.S cells (a gift from Filemon Dela Cruz, Memorial Sloan Kettering, USA), were cultured in RPMI-1640 containing 10% FBS and 100 U/ml penicillin and streptomycin. Cells were tested for mycoplasma contamination on a routine basis. Cells were maintained in a humidified, 5% CO2 atmosphere at 37°C.
Antibodies, chemicals and reagents
The rabbit anti-p97 antibody used for immunofluorescence (10736-AP), and UBXN1 (16135-1-AP), UFD1 (10615-1-AP), NPL4 (11638-1-AP), FAF1 (10271-1-AP), UBXD1 (14706-1-AP), UBXD2 (21052-1-AP), UBXD3 (26062-1-AP), UBXD5 (13109-1-AP), p47 (15620-1-AP) antibodies are from Proteintech. Rabbit anti-p97 antibody for immunopurification and immunoblotting is from Bethyl (A300-589A), mouse FLAG-M2 antibody was from Sigma-Aldrich; mouse ubiquitin (FK2) antibody used for immunofluorescence was from EMD Millipore; ubiquitin antibody (clone P4D1) for immunoblotting, GAPDH (sc-47724), and mouse Myc antibody were from Santa Cruz Biotechnology (sc-40). Rabbit anti-HDAC6 antibody (clone D2E5) was from Cell Signaling Technologies (7558), 20S proteasome antibody (MCP257) was from EMD Millipore. For immunofluorescence studies antibodies were used at 1:100 dilution; for immunoblotting, antibodies were used at 1:1000 dilution. Bortezomib and TAK273 were from Selleckchem, CB-5083 from Cayman Chemicals, puromycin, doxycycline, cycloheximide and nocodazole were from Sigma-Aldrich. Anti-FLAG magnetic beads were from Sigma-Aldrich, and Myc magnetic beads were from Pierce.
siRNA transfections and generation of CRISPR/Cas9 knockout cell lines
pTRIPZ sh-p97 hairpin vectors were purchased from Open Biosystems (clone D1: V2THS_111071 Clone D5: V2THS_201657). For the generation of shp97 stable cell lines, lentivirus was produced and packaged in HEK293T cells reported previously and subsequently used to infect HeLa Flp-in TRex cells. Stable cell lines were selected with 1 µg/ml puromycin (Raman et al., 2015). The depletion of p97 was achieved by treatment with 4 µg/ml of doxycycline for the 24–72 h.
siRNAs targeting p97 (15975124), UFD1 (4392420), NPL4 (4392420), HDAC6 (4427038) were purchased from Ambion. siRNAs targeting UBXN1 (D-008652-01, D-008652-02), FAF1 (D-009106-03), p47 (D-017222-01) were purchased from or Dharmacon. For siRNA transfections, cells were trypsinized and reverse transfected with 30 nM siRNA by using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer's protocol in six-well plates. These cells were then trypsinized and plated on 12-well plates with glass coverslips where appropriate. In general, cells were harvested at 60 h post transfection.
The CRISPR/Cas9 gene-editing system was used to generate knockout cell lines in HeLa Flp-in T-REX cells. The guide sequences for UBXN1 (5′-GCCGTCCCAGGATATGTCCAA-3′), p47 (5′-AGATCATCCACCAGCTCGTT-3′), and NPL4 (5′-GATCCGCTTCACTCCATCCG-3′) were cloned into the pX459 vector carrying hSpCas9 and transiently transfected into HeLa cells by using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer's protocol. At 36 h post transfection, the cells were pulsed briefly with 1 µg/ml puromycin for a further 24 h. The surviving cells were then serially diluted to achieve 1 cell/well in 96-well plates for clonal selection. The knockout of the gene of interest was verified by immunoblotting. HTT Q91–mCherry expression was induced by treating cells with 1 µg/ml of doxycycline for 48 h.
Cloning and generation of inducible rescue lines
UBA, UBX point mutants and the double mutant (DB) in UBXN1 were created by overlap PCR and Gibson cloning into pDON223 (Thermo Fisher Scientific, Gibson). Gateway recombination (Thermo Fisher Scientific) was used to recombine into either the pcDNA FRT/TO-GFP or the pDEST N-Myc vector. All clones were verified by Sanger sequencing. GFP-UBXN1 rescue cell lines were generated by transfecting pcDNA FRT/TO-GFP-UBXN1, UBAmut, UBXmut and DB mutants along with pOG-44 (1:9 ratio) into UBXN1 KO cells by using Lipofectamine 3000. Stable integrant cells were treated with 200 µg/ml hygromycin for 7-10 days until visible colonies were observed. GFP-UBXN1 wildtype expression was induced with 1 µg/ml doxycycline, UBAmut and UBXmut were induced with 1.5 µg/ml doxycycline and DB was induced with 1 µg/ml doxycycline to achieve equal levels of expression for 72 h.
Cell lysis, transfections and immunoprecipitation
Cells were lysed in mammalian cell lysis buffer [MCLB; 50 mM Tris-HCl pH 6.8, 150 mM NaCl, 0.5% Nonidet P-40, Halt protease inhibitors (Pierce), and 1 mM DTT]. To recover total protein (soluble and insoluble), SDS was added to a final concentration of 1% (Fig. 1B; Figs S2F and S6A). Cells were incubated at 4°C for 10 min and then centrifuged at 18,800 g for 15 min. The supernatant was removed, and the protein concentration was estimated using the DCA assay (Bio-Rad). To recover insoluble protein, the pellet from the MCLB lysis was resuspended in MCLB containing 1% SDS, vortexed for 10 min, boiled for 5 min and total protein was estimated using the DCA assay.
Myc or FLAG magnetic beads in a 1:1 slurry in mammalian cell lysis buffer was added to samples and rotated at 4°C overnight. A magnetic stand (Thermo Fisher) was used to wash the beads three times with lysis buffer. Beads were resuspended in 25 µl SDS sample buffer, boiled briefly, and processed for SDS-PAGE and immunoblotting. FLAG elutions were performed two times with the 250 µg/ml 3×FLAG peptide (ApexBio) for 30 min at room temperature. Endogenous p97 was further immunoprecipitated (Bethyl) at 4°C overnight. Beads were washed three times in lysis buffer and resuspended in Laemmli buffer and boiled briefly.
Immunofluorescence and microscopy
Cells were grown on coverslips (no. 1.5) in a 12-well plate treated with 1 µM bortezomib (Btz), 2 µM TAK243, 2 µM CB5083 (or 1 µM CB5083 for release experiments), 100 µg/ml cycloheximide or 5 µg/ml nocodazole (for the last 4 h of Btz treatment) for the indicated times. Cells were washed briefly in phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde in PBS at room temperature for 15 min or ice-cold methanol for 10 min at −20°C. Cells were washed in PBS then blocked in 2% bovine serum albumin (BSA) with 0.3% Triton X-100 in PBS for 1 h. The coverslips were incubated with the indicated antibodies in a humidified chamber overnight at 4°C. Coverslips were washed and incubated with the appropriate Alexa Fluor-conjugated secondary antibodies (Molecular Probes) for 1 h in the dark at room temperature. Cells were washed with PBS, and nuclei were stained with Hoechst 33342 dye and mounted onto slides. All images were collected using a Nikon A1R scan head with a spectral detector and resonant scanners on a Ti-E motorized inverted microscope equipped with a 60× Plan Apo 1.4 NA objective lens. The indicated fluorophores were excited with either a 405-nm, 488-nm or 594-nm laser line. Images were analyzed by using FIJI (https://imagej.net/Fiji). Colocalization analysis was performed by using the Coloc2 and JACoP plug-ins in FIJI.
Cell viability assay
Cell viability was measured using Cell Titer-Glo (Promega) according to the manufacturer's instructions. Briefly, 4000 cells for each cell line and condition in the appropriate figures were plated in triplicate in a 96-well plate. GFP–UBXN1 mutant expression was induced with the indicated concentration of doxycycline. Cells were treated after 24 h (in Fig. S1F) or 48 h (Fig. S5E) with Btz or CB-5083 at the indicated concentrations and times. At 30 min prior to the end of the incubation, the plates were equilibrated to room temperature. An equal volume of Cell Titer Glo assay reagent was added to each well. Cell lysis was induced by vigorous shaking at room temperature for 2 min. Luminescence was captured using a spectrophotometer and readings were blank subtracted and normalized to the control untreated condition for each cell line. Three biological replicates were performed.
PuLSA analysis of HTT Q91-mCherry aggregates
U2OS HTT Q91–mCherry cells were trypsinized and reverse transfected with 20 nM siRNA by using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer's protocol in six-well plates. Expression of HTT Q91–mCherry was induced with 1 μg /ml dox 8 h after siRNA transfection, and cells were incubated for an additional 48 h before harvesting and analysis. For PulSA flow cytometric analysis, cells were analyzed on a flow cytometer (LSR II; BD) equipped with 535-nm lasers. Measurements for mCherry peak width, peak height, and total intensity was collected. Flow cytometry analysis software (Flow Jo; Tree Star) was used to analyze the fraction of cells with Inclusion bodies (IBs) by PulSA. Cells with IBs were identified and gated using a mCherry peak width versus peak height scatter plot. The percentage of cells was plotted against total mCherry fluorescence intensity. 10,000 cells were analyzed for each of the three independent experiments.
Fluorescence microscopy and quantification of aggregates in C. elegans
The following strains were used in this study: N2 (Bristol), rmIs133 [unc-54p::Q40::YFP] (polyQ40::YFP expressed under a muscle-specific promoter) (Morley et al., 2002), cdc-48.1(tm544), ubxn-1(tm2759) and ubxn-4(ok3343). All strains were maintained at 20°C as described previously (Brenner, 1974).
Representative fluorescent images of polyQ40::YFP in L4 larval stage C. elegans were acquired using a Carl Zeiss Axiovert M1 microscope equipped with a 5× objective and a YFP filter (Zeiss). Images were collected with an Orca-ER charged coupled device (CCD) camera (Hamamatsu) and Metamorph (version 7.1) software. Exposure settings and gain were adjusted to fill the 12-bit dynamic range without saturation and were identical for all images. Larval-stage 4 (L4) animals were immobilized in a droplet of M9 containing 2.3 mM levamisole (Tetramisole, Sigma-Aldrich) and placed on a 2% agarose pad containing 3.4 mM levamisole. To quantify fluorescent puncta, worms were precisely age-matched at the L4.4 vulva development stage based on vulva morphology using a 100× objective. Then the number of polyQ40::YFP aggregates were visually counted by a researcher who was blind to the experimental conditions with microscope using the 5× objective and recorded. At least three to five animals were measured for each genotype across three independent experiments and statistical analyses were performed using an unpaired two-tailed Student's t-test or one-way ANOVA followed by Dunnett's multiple comparisons test.
Aggresome quantification
Aggresomes were quantified using AggreCount (Klickstein et al., 2020), an ImageJ macro we developed for the unbiased analysis of cellular aggregates. AggreCount can analyze hundreds of cells and images in a matter of minutes and provides quantification of number of aggregates, size of aggregates, and their cellular distribution (cytosolic, perinuclear, and nuclear) at single-cell resolution. We used a 2 µm2 minimum size cut-off for aggresomes formed after 8 h of Btz treatment and a 4 µm2 minimum size cut-off for aggresomes formed after 18 h of Btz treatment based on the mean size of the largest perinuclear aggregate from hundreds of analyzed cells (see quantification in Fig. S1B).
Statistical analysis
Several hundred cells (indicated in the figures) were counted for each of three biological replicates. Means±s.d. or s.e.m. for triplicate measurements were calculated. For image analysis-based quantification, each black dot in the figures represents the mean of at least 50–100 cells for each biological replicate. The error bars represent the standard error of the three means. The total number of cells (n) analyzed across all three biological replicates is denoted for each condition. Statistical significance was calculated by using an unpaired two-tailed Student's t-test or one-way ANOVA with the indicated post-hoc testing.
Acknowledgements
We are grateful to Rakesh Ganji for creating the knock-out cell lines, members of the Raman lab, Shireen Sarraf, and Karl Munger for critical reading of the manuscript. We thank the flow cytometry core and the Center for Neuroscience Imaging Core at Tufts, Caenorhabditis Genetics Center (funded by the National Institutes of Health Office of Research Infrastructure Programs Grant P40ODD010440) and Shohei Mitani (National Bioresource Project) for C. elegans strains.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.M., M.R.; Methodology: S.M., J.A.K., P.J., M.R.; Software: J.A.K.; Validation: S.M., J.A.K.; Formal analysis: S.M., J.A.K., B.O.B., M.R.; Investigation: S.M., B.O.B., M.R.; Resources: J.A.K.; Data curation: S.M., J.A.K., B.O.B.; Writing - original draft: S.M.; Writing - review & editing: S.M., M.R.; Visualization: S.M., J.A.K., B.O.B., M.R.; Supervision: P.J., M.R.; Project administration: M.R.; Funding acquisition: P.J., M.R.
Funding
This work is supported in part by the American Cancer Society Research Scholar grant (RSG-19-022-01-CSM) to M.R., National Institutes of Health grant (R01-GM127557) to M.R., (R21NS101534) to P.J., and a Tufts University Russo Family Award to M.R and P.J. B.O was supported in part by National Institutes of Health (2R25GM066567) as a Post-Baccalaureate Research Program (PREP) trainee. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.254201.supplemental
References
- Arrasate, M., Mitra, S., Schweitzer, E. S., Segal, M. R. and Finkbeiner, S. (2004). Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805-810. 10.1038/nature02998 [DOI] [PubMed] [Google Scholar]
- Bauerlein, F. J. B., Saha, I., Mishra, A., Kalemanov, M., Martinez-Sanchez, A., Klein, R., Dudanova, I., Hipp, M. S., Hartl, F. U., Baumeister, W.et al. (2017). In Situ architecture and cellular interactions of PolyQ inclusions. Cell 171, 179. 10.1016/j.cell.2017.08.009 [DOI] [PubMed] [Google Scholar]
- Bersuker, K., Brandeis, M. and Kopito, R. R. (2016). Protein misfolding specifies recruitment to cytoplasmic inclusion bodies. J Cell Biol. 213, 229-241. 10.1083/jcb.201511024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe, E. E., Olson, K. C., Chau, V. and Deshaies, R. J. (2017). Ubiquitin- and ATP-dependent unfoldase activity of P97/VCP*NPLOC4*UFD1L is enhanced by a mutation that causes multisystem proteinopathy. Proc. Natl. Acad. Sci. USA 114, E4380-e4388. 10.1073/pnas.1706205114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar, N. and Rapoport, T. (2017a). Toward an understanding of the Cdc48/p97 ATPase. F1000Research 6, 1318. 10.12688/f1000research.11683.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar, N. O. and Rapoport, T. A. (2017b). Molecular mechanism of substrate processing by the Cdc48 ATPase complex. Cell 169, 722-735.e729. 10.1016/j.cell.2017.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyault, C., Gilquin, B., Zhang, Y., Rybin, V., Garman, E., Meyer-Klaucke, W., Matthias, P., Muller, C. W. and Khochbin, S. (2006). HDAC6-p97/VCP controlled polyubiquitin chain turnover. EMBO J. 25, 3357-3366. 10.1038/sj.emboj.7601210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. 10.1093/genetics/77.1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B., Retzlaff, M., Roos, T. and Frydman, J. (2011). Cellular strategies of protein quality control. Cold Spring Harb. Perspect. Biol. 3, a004374. 10.1101/cshperspect.a004374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco, C., Micale, L., Egorov, M., Monti, M., D'Addetta, E. V., Augello, B., Cozzolino, F., Calcagni, A., Fontana, A., Polishchuk, R. S.et al. (2012). The E3-ubiquitin ligase TRIM50 interacts with HDAC6 and p62, and promotes the sequestration and clearance of ubiquitinated proteins into the aggresome. PLoS ONE 7, e40440. 10.1371/journal.pone.0040440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganji, R., Mukkavalli, S., Somanji, F. and Raman, M. (2018). The VCP-UBXN1 complex mediates triage of ubiquitylated cytosolic proteins bound to the BAG6 complex. Mol. Cell Biol. 38, e00154-e00118. 10.1128/MCB.00154-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron, J. M., Webb, K., Yang, B., Rising, L., Zuzow, N. and Bennett, E. J. (2016). Using the ubiquitin-modified proteome to monitor distinct and spatially restricted protein homeostasis dysfunction. Mol. Cell Proteomics 15, 2576-2593. 10.1074/mcp.M116.058420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzelmann, P., Buchberger, A. and Schindelin, H. (2011). Hierarchical binding of cofactors to the AAA ATPase p97. Structure (London, England : 1993) 19, 833-843. 10.1016/j.str.2011.03.018 [DOI] [PubMed] [Google Scholar]
- Hanzelmann, P. and Schindelin, H. (2017). The interplay of cofactor interactions and post-translational modifications in the regulation of the AAA+ ATPase p97. Front. Mol. Biosci. 4, 21. 10.3389/fmolb.2017.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J. W. and Bennett, E. J. (2016). Proteome complexity and the forces that drive proteome imbalance. Nature 537, 328-338. 10.1038/nature19947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbers, C. U., Clemen, C. S., Kesper, K., Boddrich, A., Hofmann, A., Kamarainen, O., Tolksdorf, K., Stumpf, M., Reichelt, J., Roth, U.et al. (2007). Pathological consequences of VCP mutations on human striated muscle. Brain 130, 381-393. 10.1093/brain/awl238 [DOI] [PubMed] [Google Scholar]
- Johnston, J. A., Ward, C. L. and Kopito, R. R. (1998). Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143, 1883-1898. 10.1083/jcb.143.7.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju, J. S., Miller, S. E., Hanson, P. I. and Weihl, C. C. (2008). Impaired protein aggregate handling and clearance underlie the pathogenesis of p97/VCP-associated disease. J. Biol. Chem. 283, 30289-30299. 10.1074/jbc.M805517200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich, D., Kopito, R. and Frydman, J. (2008). Misfolded proteins partition between two distinct quality control compartments. Nature 454, 1088-1095. 10.1038/nature07195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi, Y., Kovacs, J. J., McLaurin, A., Vance, J. M., Ito, A. and Yao, T. P. (2003). The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115, 727-738. 10.1016/S0092-8674(03)00939-5 [DOI] [PubMed] [Google Scholar]
- Kim, W., Bennett, E. J., Huttlin, E. L., Guo, A., Li, J., Possemato, A., Sowa, M. E., Rad, R., Rush, J., Comb, M. J.et al. (2011). Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325-340. 10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klickstein, J. A., Mukkavalli, S. and Raman, M. (2020). AggreCount: an unbiased image analysis tool for identifying and quantifying cellular aggregates in a spatially defined manner. J. Biol. Chem. 295, 17672-17683. 10.1074/jbc.RA120.015398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T., Manno, A. and Kakizuka, A. (2007). Involvement of valosin-containing protein (VCP)/p97 in the formation and clearance of abnormal protein aggregates. Genes Cells 12, 889-901. 10.1111/j.1365-2443.2007.01099.x [DOI] [PubMed] [Google Scholar]
- Kopito, R. R. (2000). Aggresomes, inclusion bodies and protein aggregation. Trends in cell biology 10, 524-530. 10.1016/S0962-8924(00)01852-3 [DOI] [PubMed] [Google Scholar]
- MacDonald, M. E., Barnes, G., Srinidhi, J., Duyao, M. P., Ambrose, C. M., Myers, R. H., Gray, J., Conneally, P. M., Young, A. and Penney, J. (1993). Gametic but not somatic instability of CAG repeat length in Huntington's disease. J. Med. Genet. 30, 982-986. 10.1136/jmg.30.12.982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson, K., Pokrovskaja, K., Kiss, C., Klein, G. and Szekely, L. (2001). Proteins associated with the promyelocytic leukemia gene product (PML)-containing nuclear body move to the nucleolus upon inhibition of proteasome-dependent protein degradation. Proc Natl Acad Sci USA 98, 1012-1017. 10.1073/pnas.98.3.1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, H., Bug, M. and Bremer, S. (2012). Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 14, 117-123. 10.1038/ncb2407 [DOI] [PubMed] [Google Scholar]
- Meyer, H. and Weihl, C. C. (2014). The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J. Cell Sci. 127, 3877-3883. 10.1242/jcs.093831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley, J. F., Brignull, H. R., Weyers, J. J. and Morimoto, R. I. (2002). The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci USA 99, 10417-10422. 10.1073/pnas.152161099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow, C. S., Porter, T. J., Xu, N., Arndt, Z. P., Ako-Asare, K., Heo, H. J., Thompson, E. A. N. and Moore, D. L. (2020). Vimentin Coordinates Protein Turnover at the Aggresome during Neural Stem Cell Quiescence Exit. Cell Stem Cell 26, 558-568.e559. 10.1016/j.stem.2020.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski, P. J., Ning, K., D'Souza-Schorey, C. and Fields, S. (2002). Requirement of an intact microtubule cytoskeleton for aggregation and inclusion body formation by a mutant huntingtin fragment. Proc. Natl. Acad. Sci. USA 99, 727-732. 10.1073/pnas.022628699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki, S. T., Carew, J. S., Dunner, K., Jr, Boise, L. H., Chiao, P. J., Huang, P., Abbruzzese, J. L. and McConkey, D. J. (2005). Bortezomib inhibits PKR-like endoplasmic reticulum (ER) kinase and induces apoptosis via ER stress in human pancreatic cancer cells. Cancer Res. 65, 11510-11519. 10.1158/0008-5472.CAN-05-2394 [DOI] [PubMed] [Google Scholar]
- Nowis, D., McConnell, E. and Wojcik, C. (2006). Destabilization of the VCP-Ufd1-Npl4 complex is associated with decreased levels of ERAD substrates. Exp. Cell Res. 312, 2921-2932. 10.1016/j.yexcr.2006.05.013 [DOI] [PubMed] [Google Scholar]
- Olzmann, J. A., Li, L., Chudaev, M. V., Chen, J., Perez, F. A., Palmiter, R. D. and Chin, L. S. (2007). Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J. Cell Biol. 178, 1025-1038. 10.1083/jcb.200611128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzscha, H., Schermann, S. M., Woerner, A. C., Pinkert, S., Hecht, M. H., Tartaglia, G. G., Vendruscolo, M., Hayer-Hartl, M., Hartl, F. U. and Vabulas, R. M. (2011). Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144, 67-78. 10.1016/j.cell.2010.11.050 [DOI] [PubMed] [Google Scholar]
- Raman, M., Havens, C. G., Walter, J. C. and Harper, J. W. (2011). A genome-wide screen identifies p97 as an essential regulator of DNA damage-dependent CDT1 destruction. Mol. Cell 44, 72-84. 10.1016/j.molcel.2011.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman, M., Sergeev, M., Garnaas, M., Lydeard, J. R., Huttlin, E. L., Goessling, W., Shah, J. V. and Harper, J. W. (2015). Systematic proteomics of the VCP-UBXD adaptor network identifies a role for UBXN10 in regulating ciliogenesis. Nat. Cell Biol. 17, 1356-1369. 10.1038/ncb3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdzan, Y. M., Polling, S., Chia, C. P., Ng, I. H., Ormsby, A. R., Croft, N. P., Purcell, A. W., Bogoyevitch, M. A., Ng, D. C., Gleeson, P. A.et al. (2012). Tracking protein aggregation and mislocalization in cells with flow cytometry. Nat. Methods 9, 467-470. 10.1038/nmeth.1930 [DOI] [PubMed] [Google Scholar]
- Ritz, D., Vuk, M., Kirchner, P., Bug, M., Schütz, S., Hayer, A., Bremer, S., Lusk, C., Baloh, R. H., Lee, H.et al. (2011). Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat. Cell Biol. 13, 1116-1123. 10.1038/ncb2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzinger, E., Lurz, R., Turmaine, M., Mangiarini, L., Hollenbach, B., Hasenbank, R., Bates, G. P., Davies, S. W., Lehrach, H. and Wanker, E. E. (1997). Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90, 549-558. 10.1016/S0092-8674(00)80514-0 [DOI] [PubMed] [Google Scholar]
- Scherzinger, E., Sittler, A., Schweiger, K., Heiser, V., Lurz, R., Hasenbank, R., Bates, G. P., Lehrach, H. and Wanker, E. E. (1999). Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: implications for Huntington's disease pathology. Proc Natl Acad Sci USA 96, 4604-4609. 10.1073/pnas.96.8.4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, D., Coleman, J., Chan, E., Nicholson, T. P., Dai, L., Sheppard, P. W. and Patton, W. F. (2011). Novel cell- and tissue-based assays for detecting misfolded and aggregated protein accumulation within aggresomes and inclusion bodies. Cell Biochem. Biophys. 60, 173-185. 10.1007/s12013-010-9138-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, C., Xiao, Z., Nagashima, K., Li, C. C., Lockett, S. J., Dai, R. M., Cho, E. H., Conrads, T. P., Veenstra, T. D., Colburn, N. H.et al. (2008). The heavy metal cadmium induces valosin-containing protein (VCP)-mediated aggresome formation. Toxicol Appl. Pharmacol 228, 351-363. 10.1016/j.taap.2007.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, T., Kikuchi, S., Katada, S., Nagai, Y., Nishizawa, M. and Onodera, O. (2008). Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum. Mol. Genet. 17, 345-356. 10.1093/hmg/ddm311 [DOI] [PubMed] [Google Scholar]
- Tiwari, A., Copeland, C. A., Han, B., Hanson, C. A., Raghunathan, K. and Kenworthy, A. K. (2016). Caveolin-1 is an aggresome-inducing protein. Sci. Rep. 6, 38681. 10.1038/srep38681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers, J., Mogk, A. and Bukau, B. (2010). Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Biol. 11, 777-788. 10.1038/nrm2993 [DOI] [PubMed] [Google Scholar]
- van Well, E. M., Bader, V., Patra, M., Sanchez-Vicente, A., Meschede, J., Furthmann, N., Schnack, C., Blusch, A., Longworth, J., Petrasch-Parwez, E.et al. (2019). A protein quality control pathway regulated by linear ubiquitination. EMBO J. 38, e100730. 10.15252/embj.2018100730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waelter, S., Boeddrich, A., Lurz, R., Scherzinger, E., Lueder, G., Lehrach, H. and Wanker, E. E. (2001). Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell 12, 1393-1407. 10.1091/mbc.12.5.1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, D. M., Kasturi, P., Zheng, M., Pinkert, S., Vecchi, G., Ciryam, P., Morimoto, R. I., Dobson, C. M., Vendruscolo, M., Mann, M.et al. (2015). Widespread proteome remodeling and aggregation in aging C. elegans. Cell 161, 919-932. 10.1016/j.cell.2015.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihl, C. C., Miller, S. E., Hanson, P. I. and Pestronk, A. (2007). Transgenic expression of inclusion body myopathy associated mutant p97/VCP causes weakness and ubiquitinated protein inclusions in mice. Hum. Mol. Genet. 16, 919-928. 10.1093/hmg/ddm037 [DOI] [PubMed] [Google Scholar]
- Wojcik, C., Yano, M. and DeMartino, G. N. (2004). RNA interference of valosin-containing protein (VCP/p97) reveals multiple cellular roles linked to ubiquitin/proteasome-dependent proteolysis. J. Cell Sci. 117, 281-292. 10.1242/jcs.00841 [DOI] [PubMed] [Google Scholar]
- Wolff, S., Weissman, J. S. and Dillin, A. (2014). Differential scales of protein quality control. Cell 157, 52-64. 10.1016/j.cell.2014.03.007 [DOI] [PubMed] [Google Scholar]
- Yamanaka, K., Okubo, Y., Suzaki, T. and Ogura, T. (2004). Analysis of the two p97/VCP/Cdc48p proteins of Caenorhabditis elegans and their suppression of polyglutamine-induced protein aggregation. J. Struct. Biol. 146, 242-250. 10.1016/j.jsb.2003.11.017 [DOI] [PubMed] [Google Scholar]
- Zhang, X., Gui, L., Zhang, X., Bulfer, S. L., Sanghez, V., Wong, D. E., Lee, Y., Lehmann, L., Lee, J. S., Shih, P. Y.et al. (2015). Altered cofactor regulation with disease-associated p97/VCP mutations. Proc. Natl. Acad. Sci. USA 112, E1705-E1714. 10.1073/pnas.1418820112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H.-J., Wang, J., Yao, B., Wong, S., Djakovic, B., Kumar, B., Rice, J., Valle, E., Soriano, F., Menon, M.-K.et al. (2015). Discovery of a first-in-class, potent, selective, and orally bioavailable inhibitor of the p97 AAA ATPase (CB-5083). J. Med. Chem. 58, 9480-9497. 10.1021/acs.jmedchem.5b01346 [DOI] [PubMed] [Google Scholar]