ABSTRACT
Mitochondria are multifunctional organelles that not only produce energy for the cell, but are also important for cell signalling, apoptosis and many biosynthetic pathways. In most cell types, they form highly dynamic networks that are constantly remodelled through fission and fusion events, repositioned by motor-dependent transport and degraded when they become dysfunctional. Motor proteins and their tracks are key regulators of mitochondrial homeostasis, and in this Review, we discuss the diverse functions of the three classes of motor proteins associated with mitochondria – the actin-based myosins, as well as the microtubule-based kinesins and dynein. In addition, Miro and TRAK proteins act as adaptors that link kinesin-1 and dynein, as well as myosin of class XIX (MYO19), to mitochondria and coordinate microtubule- and actin-based motor activities. Here, we highlight the roles of motor proteins and motor-linked track dynamics in the transporting and docking of mitochondria, and emphasize their adaptations in specialized cells. Finally, we discuss how motor–cargo complexes mediate changes in mitochondrial morphology through fission and fusion, and how they modulate the turnover of damaged organelles via quality control pathways, such as mitophagy. Understanding the importance of motor proteins for mitochondrial homeostasis will help to elucidate the molecular basis of a number of human diseases.
KEY WORDS: Myosin, Kinesin, Dynein, Actin, Microtubules, Mitochondria
Summary: This Review highlights the diverse functions of myosin, kinesin and dynein motor proteins in the transport and positioning, fission and fusion, and turnover of mitochondria.
Introduction
Mitochondria are the powerhouses of the cell, generating adenosine triphosphate (ATP) by oxidative phosphorylation (OXPHOS). In addition to producing energy, they also play critical roles in the biosynthesis of macromolecules, Ca2+ homeostasis, and signalling in programmed cell death and immunity. Mitochondria are double-membraned organelles of endosymbiotic origin that have an outer mitochondrial membrane (OMM) facing the cytosol and an inner mitochondrial membrane (IMM) marking the boundary of the mitochondrial matrix, which contains the mitochondrial DNA (mtDNA). The intermembrane space (IMS) is sandwiched between the OMM and IMM. Mitochondria can sense stress stimuli, such as nutrient deprivation, and the overall metabolic state of the cell (Chandel, 2015; Giacomello et al., 2020). To maintain organellar homeostasis, mitochondria constantly remodel their network through fission and fusion events, reposition themselves through motor protein- and cytoskeleton-dependent transport, and finally are degraded via several quality control mechanisms (Chan, 2020; Harper et al., 2018; Sleigh et al., 2019). Indeed, defects in mitochondrial homeostasis manifesting as fission–fusion imbalance, impaired transport or reduced clearance of faulty mitochondria by mitophagy increase with age and can lead to a wide range of neurodegenerative disorders (Bakula and Scheibye-Knudsen, 2020; Seo et al., 2010; Sleigh et al., 2019; Tilokani et al., 2018). The overall control of mitochondrial homeostasis involves three types of motor proteins that translocate along cytoskeletal tracks: myosins moving along actin filaments (Box 1), as well as kinesins and dynein moving along microtubules (MTs; Box 2; Vale and Milligan, 2000). The key adaptor complex linking kinesin-1 and dynein to mitochondria consists of Miro1 or Miro2 (referred to collectively as Miro1/2; also known as RHOT1 or RHOT2, respectively) and trafficking kinesin protein 1 or 2 (TRAK1 or TRAK2, referred to collectively as TRAK1/2), and is hereafter referred to as the Miro–TRAK complex (Eberhardt et al., 2020; Melkov and Abdu, 2018). Interestingly, Miro1/2 without TRAK1/2 can also recruit myosin of class XIX (MYO19) to mitochondria (Bocanegra et al., 2020a).
Box 1. Myosin motors translocate along actin filaments.
Actin monomers polymerize to form actin filaments. A host of actin-binding proteins regulate filament dynamics and the assembly into higher-ordered structures, such as stress fibres, lamellipodia, filopodia and actin networks associated with the plasma membrane or intracellular membranes (Rottner et al., 2017). These include: (1) actin nucleators, such as the ARP2/3 complex, formins and tandem WH2 domain-containing proteins, such as mitoSPIRE; (2) actin filament cross-linking or bundling proteins, such as fascin; (3) actin disassembly factors; and (4) monomer-sequestering proteins.
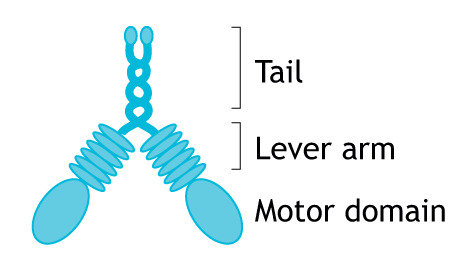
Myosins are a diverse family of molecular motors that undergo directed movement along actin filaments; they form dynamic tethers between cellular membrane compartments and the actin cytoskeleton. In addition, several classes of myosin motors are actively involved in regulating organization of the actin cytoskeleton (Masters et al., 2016). A total of 39 myosins belonging to 12 classes are expressed in humans. All known myosins, regardless of species, have the same basic structure: an N-terminal motor domain, a lever arm and a tail domain (see figure showing the domain organization of myosin V) (Masters et al., 2016). In vitro experiments with myosins indicate that their behaviour broadly falls into three types – transporters, tethers or contractile motors. The lever arm, which typically contains one or more calmodulin- or light-chain-binding IQ motifs, amplifies structural changes in the motor domain into large steps along the actin track. The tail domains either self-assemble to form filaments or contain the majority of binding sites for lipid membranes and cargo adaptor proteins, which recruit the motor to different organelles and vesicular compartments.
Box 2. Microtubule-based kinesin and dynein motor proteins.
MTs are rigid but dynamic polymers, growing and shrinking through the addition or loss of α-tubulin–β-tubulin heterodimers, which provide the tracks for polarized transport (Brouhard and Rice, 2018).
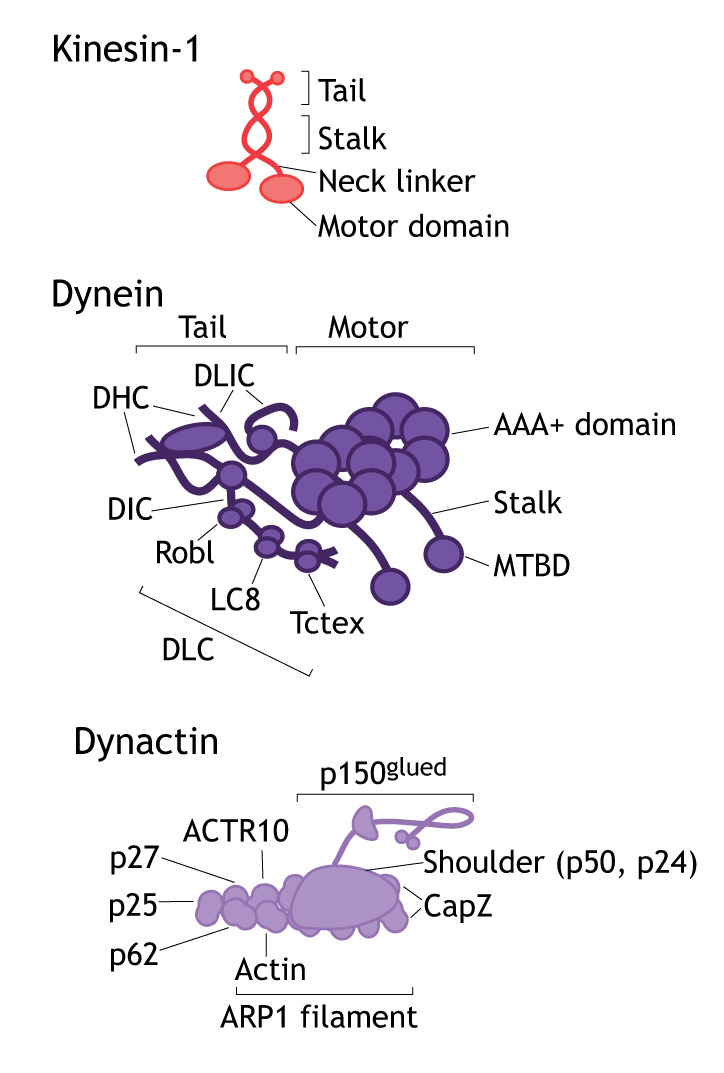
Kinesins are a diverse family of motor proteins that transport cargo unidirectionally along MTs (mostly towards the plus end) and regulate MT network organization (Endow et al., 2010). The kinesin superfamily consists of 15 classes, with over 45 known kinesins expressed in mammalian cells. All kinesins share the same basic domain organization: a conserved head domain that is structurally homologous to the myosin motor domain, followed by the neck linker, a divergent stalk, and a tail domain (Hirokawa et al., 2009; see figure, top). Cargo can either bind directly or through adaptor proteins to the kinesin tail or to one of the kinesin light chains that bind to the kinesin heavy chain (Verhey and Hammond, 2009). Kinesins can be grouped according to the position of their motor domain. Kinesins with motor domains at either the N- or C-terminus drive plus- or minus-end directed transport, respectively, whereas kinesins with the motor domain in the middle of the protein are involved in depolymerizing MTs.
Cytoplasmic dynein 1 (referred to as dynein) drives transport of a wide range of cargos towards the minus-end of MTs (Reck-Peterson et al., 2018). Human dynein is a 1.5 MDa complex comprising the dynein heavy chain (DHC, also known as DYNC1H1), the dynein intermediate chain (DIC), the dynein light intermediate chain (DLIC) and three classes of dynein light chains (DLCs; Robl, LC8 and Tctex) (see figure, middle). The DHC consists of an N-terminal tail domain that is important for their dimerization and that binds to DIC and DLIC, and a linker region connecting the tail domain to the C-terminal motor domain, which comprises a ring of six AAA+ domains, a coiled-coil stalk and a MT-binding domain (MTBD). Dynein motility along MTs is powered by ATP hydrolysis in the AAA+ ring; however, processive movement of dynein requires the co-factor dynactin (see figure, bottom). Dynactin is a 1.1 MDa protein complex with 23 subunits assembled around an actin-like filament (ARP1 filament) and p150glued (also known as DCTN1), which can also bind to MTs (Olenick and Holzbaur, 2019). Cargo attachment to dynein requires long coiled-coil activating adaptors, such as BICD, Hook, and Spindly proteins, which bind to the C-terminus of DLIC and run along the dynactin filament (Olenick and Holzbaur, 2019).
In this Review, we will discuss the importance of motor proteins and motor-linked track dynamics not only for the movement of mitochondria but also for the positioning of these organelles, for the production of force associated with fission and fusion, and for the three-dimensional organization of the cytoskeleton around mitochondria. We will focus on kinesin-1, kinesin-3 and related family members, cytoplasmic dynein-1, MYO19 and myosin V (MYO5), which are involved in mitochondrial transport, as well as non-muscle myosin II (NMII) and myosin VI (MYO6), which have prominent roles in mitochondrial fission and mitophagy, respectively. Finally, we highlight human diseases with mitochondrial phenotypes caused by mutations in motor proteins that open up potential new avenues for treatment (Box 3).
Box 3. Mutations in motor proteins linked to human diseases.
Motor proteins and other members of the machinery important for mitochondrial dynamics that have been linked to human diseases are summarized in Table S1 (see also table 1 in Tilokani et al., 2018). For instance, a mutation in MYH14 (encoding the heavy chain of NMIIC) affects mitochondrial fission and leads to a peripheral neuropathy (Almutawa et al., 2019). Fibroblasts from patients harbouring the mutations K671E or I584L in DYNC1H1, which have been linked to the neuromuscular disorder spinal muscular atrophy lower extremity-predominant 1 (SMA-LED), display fragmented and fewer mitochondria with decreased MFN1 levels (Eschbach et al., 2013). KIF5A mutations causing myoclonus also lead to mitochondrial abnormalities and manifest as atrophy of type I muscle fibres and complex IV deficiency (Duis et al., 2016). Moreover, mutations in the motor adaptor TRAK1 are linked to epileptic encephalopathy early infantile 68; in TRAK1-deficient patient fibroblasts, the mitochondria are irregularly distributed and have reduced motility, decreased membrane potential and a lower respiratory capacity (Barel et al., 2017). Interestingly, Miro1/2 and MYO19 have not been linked to any Mendelian diseases. In many cases where there are disease-causing mutations in motor proteins, it remains to be established whether the motor-linked disruption in mitochondrial homeostasis is also the underlying cause of the disease. Nevertheless, the emerging importance of motor proteins in maintaining mitochondrial homeostasis might open new avenues for treatment of mitochondria-linked diseases, as motor proteins are attractive drug targets.
Mitochondrial motility in neurons – a balance between transport and docking
The dynamic positioning of mitochondria within different cell types and tissues is spatially regulated to fulfil subcellular requirements for local energy production and Ca2+ signalling, which is particularly important in neurons due to their extended morphology (Seager et al., 2020). Mitochondrial movement over long distances in axons or dendrites is dependent on kinesin and dynein motors moving in opposite directions along MTs due to the intrinsic track polarity. Fast axonal transport with velocities between 0.25 and 1 µm/s is driven in the anterograde direction from the cell body to the distal part of the axon and the synapse by plus-end-directed kinesins, whereas retrograde movement depends on the minus-end-directed dynein motor moving back towards the soma (cell body) (Pilling et al., 2006). This bidirectional transport of mitochondria along axonal MTs, which characteristically displays frequent pauses and directional changes, and the docking mechanisms that ensure the correct distribution of mitochondria throughout the neuron, is fundamentally the same in all eukaryotic cells as it uses equivalent motors and machinery. In contrast to the uniform MT polarity in the axon, neuronal dendrites contain MTs of mixed polarity with approximately equal numbers of plus- and minus-ends of MTs directed towards the cell surface; hence, the track orientation determines the direction of transport and the motor involved (Satoh et al., 2008; van Spronsen et al., 2013; Yan et al., 2013). Mitochondrial transport has been extensively studied in neurons (for examples, see recent reviews by Cardanho-Ramos et al., 2020; Mandal and Drerup, 2019), and we will focus here only on the key motors and mitochondrial adaptors (Fig. 1).
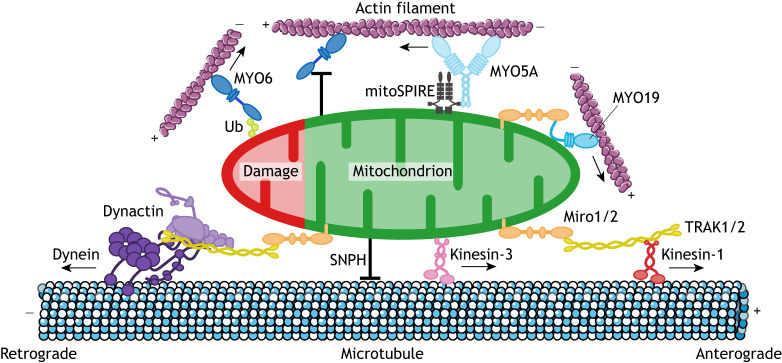
Fig. 1.
Mechanisms of motor protein attachment to mitochondria. Schematic summary of the different classes of myosin, kinesin and dynein motors associated with mitochondria. The MT-based motor proteins, kinesin-1 and the dynein–dynactin complex transport mitochondria towards the plus and minus end of MTs, respectively. They are linked to mitochondria via the adaptor protein TRAK1/2, which binds to the mitochondrially anchored Miro1/2. MYO19 also attaches to mitochondria directly through a lipid-binding region in its tail and by interacting with Miro1/2. Although less well documented than the involvement of kinesin-1 in the movement of mitochondria, kinesin-3 may also play a role in mitochondrial movement towards the plus end of MTs; however, the mechanism of attachment to mitochondria remains unknown. MYO5A is targeted to mitochondria by the mitochondrial isoform of the actin nucleator SPIRE (mitoSPIRE). Syntaphilin (SNPH) is attached to the OMM and acts as an anchor by either directly binding to MTs or to actin filaments via its interaction with MYO6, resulting in stationary mitochondria. During PINK1- and PRKN-mediated mitophagy, MYO6 is recruited to damaged mitochondria by ubiquitin (Ub). Actin filament and microtubule images are adapted from Servier Medical Art (https://smart.servier.com/) under the terms of a CC-BY 3.0 license.
Kinesins mediate anterograde mitochondrial transport
Kinesin-1 (which in mammals has three heavy chain isoforms – the ubiquitously expressed KIF5B, and neuron-specific KIF5A and KIF5C) is the primary motor for anterograde mitochondrial transport (Fig. 1) in axons in Drosophila, mouse and zebrafish (Campbell et al., 2014; Hurd and Saxton, 1996; Karle et al., 2012; Pilling et al., 2006; Tanaka et al., 1998). Kinesin-1 is a highly processive motor that takes over 100 steps before detaching from the MT track in vitro (Taylor and Borisy, 2000). Within the cell, kinesin-1 is activated and the run length increases upon binding to TRAK1, thereby promoting kinesin-1-dependent long-range transport of mitochondria in the crowded cytosol (Henrichs et al., 2020).
In contrast to the well-documented role of kinesin-1, the importance of other classes of kinesins for mitochondrial transport remains controversial. The highly processive kinesin-3 protein KIF1B has been suggested to transport mitochondria in vitro (Nangaku et al., 1994), although no defects in mitochondrial transport are observed in KIF1B-mutant mice (Zhao et al., 2001) or zebrafish (Drerup et al., 2016; Lyons et al., 2009). Furthermore, one publication has described changes in mitochondrial morphology that are linked to the depletion or overexpression of the dominant-negative tail domain of kinesin-like protein 6 (KLP6, also known as KIF28P), a distantly related member of the kinesin-3 family (Tanaka et al., 2011). Although expression of mutant KLP6 also affects mitochondrial distribution in neurites (Tanaka et al., 2011), further work is required to establish the role of KLP6 in mitochondrial transport. Finally, loss of KIF1-binding protein (KIFBP, also known as KBP and KIF1BP), the adaptor protein for both kinesin-3 and KLP6, has been reported to change mitochondrial distribution in some specific cell types and developmental stages (Drerup et al., 2016; Lyons et al., 2008; Wozniak et al., 2005). Since KIFBP alters MT dynamics and mitochondrial biogenesis, it is more likely to play an indirect role in mitochondrial transport by modulating MT tracks (Atherton et al., 2020; Donato et al., 2017; Kevenaar et al., 2016).
Dynein transports mitochondria in the retrograde direction
Dynein is the main motor driving the retrograde transport of axonal mitochondria (Fig. 1) (Drerup et al., 2017). Interestingly, this activity also requires kinesin-1, which delivers newly synthesized dynein from the soma via anterograde slow axonal transport out to the synapse (Twelvetrees et al., 2016). The majority of disease-associated dynein mutations affect overall retrograde dynein-dependent motility, whereas mutations causing loss of function in the dynactin subunit ACTR10 (Box 2) selectively impair binding of dynein to mitochondria in zebrafish (Drerup et al., 2017).
Motor adaptors for mitochondria
Microtubule and myosin motors are bound to the mitochondrial surface by the conserved Miro–TRAK adaptor complex (Fig. 1), which is the key regulator of axonal transport of mitochondria in many different organisms (Schwarz, 2013). TRAK1 and TRAK2, the mammalian homologues of the Drosophila Milton protein, bind directly to Miro proteins, which are anchored to the OMM via a C-terminal transmembrane domain (Fransson et al., 2006; Glater et al., 2006). In mammals, two Miro proteins are expressed, Miro1 and Miro2, and both have two GTPase domains flanking two Ca2+-sensing EF-hand domains (Fransson et al., 2003; Klosowiak et al., 2013; Macaskill et al., 2009). The TRAK family has an extensive N-terminal coiled-coil region similar to huntingtin-associated protein (HAP1) and a C-terminal domain that binds to Miro proteins (Stowers et al., 2002). Whereas TRAK1 predominantly mediates axonal transport by simultaneously interacting with both the cargo-binding tails of kinesin-1 and dynein (Glater et al., 2006; Smith et al., 1995), TRAK2 promotes dendritic transport by forming a complex with dynein in a conformation that interferes with kinesin-1 binding (Brickley and Stephenson, 2011; Loss and Stephenson, 2015; van Spronsen et al., 2013).
Interestingly, Miro proteins not only regulate transport, but also play a role in maintaining mitochondrial morphology and the architecture of the IMM, since Miro1/2 double-knockout (KO) mouse embryonic fibroblasts (MEFs) have a non-uniform arrangement of mitochondrial cristae (Modi et al., 2019).
Finally, in addition to mitochondrial motility, kinesin-1, together with TRAK1/2, also drives the tubulation of mitochondria in the cell periphery. This process requires kinesin-1 motor activity and gives rise to highly dynamic thin tubules that fuse to form lattices and eventually generate an interconnected mitochondrial network (Wang et al., 2015). Indeed, overexpression of TRAK1/2 leads to striking changes in mitochondrial network morphology, inducing the formation of extended, highly dynamic mitochondrial tubules in a MT-dependent manner (Fransson et al., 2006; Koutsopoulos et al., 2010).
Docking and anchoring
For mitochondria to become stationary, their movement is either simply paused (Chung et al., 2016; Wang and Schwarz, 2009) or they are actively anchored to either the actin or MT track. Mitochondrial positioning and docking at locations of nutrient abundance increases the efficiency of ATP generation and is caused by stalling due to TRAK1 glycosylation (O-GlcNAcylation) at high glucose levels (Pekkurnaz et al., 2014). Miro proteins can also act as Ca2+ sensors and induce mitochondrial immobilization by a Ca2+-dependent mechanism, for example, at synaptic sites. Ca2+ binding to the EF-hand motifs of Miro1/2 triggers conformational changes that arrest mitochondrial movement by disrupting interactions between kinesin-1 and MTs, or by leading to dissociation of the Miro–TRAK complex from kinesin-1 (Macaskill et al., 2009; Saotome et al., 2008; Wang and Schwarz, 2009). Elevated Ca2+ levels also activate anchoring proteins that mediate docking of mitochondria, such as syntaphilin (SNPH). Syntaphilin, previously thought to be axonal-specific, is inserted into the OMM with its C-terminal transmembrane domain and directly binds to MTs via its N-terminal MT-binding domain (Kang et al., 2008). It functions as a brake to axonal mitochondrial transport by competing with TRAK1/2 for kinesin-1 binding and, once bound, inhibiting kinesin-1 motor activity (Chen and Sheng, 2013). The dynein light chain (DLC) LC8 (also known as DYNLL1) is thought to enhance SNPH-based anchoring of mitochondria (Chen et al., 2009b).
Mitochondrial positioning and immobilization may require the inactivation of kinesin and dynein motors, but also involves actin-based myosin motor proteins, which may act as short-range transporters or may regulate mitochondrial tethering. Disrupting the actin cytoskeleton with drug treatments has been shown to reduce mitochondrial anchoring in axons (Gutnick et al., 2019), and disruption of the actin- or MT-based cytoskeleton destabilizes mitochondrial tethering in dendrites (Rangaraju et al., 2019). MYO6 (also known as Jaguar in Drosophila) is thought to oppose MT-based mitochondrial movement to facilitate organelle docking in Drosophila neurons, in line with recent findings that the cargo-binding domain of MYO6 directly binds to SNPH (Fig. 1) (Li et al., 2020; Pathak et al., 2010). MYO6 is a unique myosin that moves towards the minus ends of actin filaments, in the opposite direction to all other known myosins (Wells et al., 1999). MYO6 has essential roles in membrane trafficking pathways and mediates the delivery of endosomal membranes to autophagosomes during autophagosome maturation (de Jonge et al., 2019). The direct interaction between MYO6 and SNPH suggests a new role for MYO6 in capturing mitochondria on F-actin and mediating the cytoskeletal switch from MTs to actin, which serves as a platform for anchoring organelles in presynaptic terminals (Li et al., 2020). MYO6-dependent mitochondrial capture is regulated by AMP-activated protein kinase (AMPK)–p21-activated kinase (PAK) signalling pathways and phosphorylation of MYO6 in the motor domain (Li et al., 2020).
A role for MYO5 in mitochondrial docking was observed in Drosophila, where depletion of MYO5 (encoded by didum in Drosophila) causes an increase in anterograde and retrograde axonal mitochondrial motility (Pathak et al., 2010). Myosins of class V (MYO5A, -B and -C) function as a dimer and move processively along actin filaments. Each MYO5 heavy chain binds six calmodulins, giving rise to a long lever arm, which allows this myosin to take large steps of 36 nm and move straight along the helical actin track (Mehta et al., 1999). MYO5A can bind to a mitochondrially-anchored isoform of the actin nucleator and tandem WH2 domain-containing protein SPIRE1 (mitoSPIRE), which cooperates with formins to assemble actin filaments (Fig. 1) (Straub et al., 2020 preprint). These actin–myosin networks are thought to oppose mitochondrial motility by anchoring mitochondria, since loss of mitoSPIRE in MEFs increases mitochondrial motility (Straub et al., 2020 preprint).
In vertebrates, MYO19 is tightly associated with mitochondria and regulates the cellular distribution of this organelle (Fig. 1). Overexpression of full-length MYO19 significantly increases the overall mitochondrial motility over short distances in epithelial cells (Quintero et al., 2009). In neuronal cells, however, MYO19 causes a decrease in mitochondrial movement by inducing track switching from MTs to actin filaments, which results in reduced mitochondrial run length (Quintero et al., 2009). MYO19 displays a slow rate of ADP release linked to a prolonged actin-binding state and high duty ratio in vitro (Lu et al., 2014; Ušaj and Henn, 2017). The very short unique C-terminal tail of MYO19 contains a lipid-binding region of 30–45 amino acids essential for mitochondrial targeting (Hawthorne et al., 2016; Shneyer et al., 2016). In addition, its far C-terminal tail directly interacts with Miro1/2 and competes with TRAK1/2 for binding (Oeding et al., 2018). Thus, Miro proteins not only form a complex with MT motors but also with MYO19, thereby coordinating MT- and actin-based mitochondrial movement. This is confirmed by studies on fibroblasts isolated from Miro1/2 double-KO mice, which not only display reduced retrograde and anterograde mitochondrial movement along MTs, but have also lost MYO19 from mitochondria (López-Domenéch et al., 2018). The N-terminal GTPase domain of Miro1/2 is critical for recruiting and stabilizing MYO19 on the OMM by facilitating its membrane insertion and protecting it from proteasome-mediated degradation (Bocanegra et al., 2020b; López-Domenéch et al., 2018; Oeding et al., 2018). Finally, MYO19 is also able to regulate the precise localization of mitochondria in response to metabolic stimuli, such as glucose starvation or reactive oxygen species (ROS), which activate MYO19 and lead to re-localization of mitochondria into the tips of filopodia (Shneyer et al., 2016, 2017).
As discussed above, the correct distribution of mitochondria is particularly important in highly polarized neuronal cells. In the next section, we highlight other specialized adaptations regulating mitochondrial positioning.
Specialized cell adaptations for mitochondrial positioning
Cardiac cells
Mitochondrial transport in cardiac myoblasts is driven by the same players (kinesin-1, dynein, TRAK1/2) as in neuronal cells and is regulated by similar Ca2+-dependent mechanisms. Indeed, Ca2+ is a key regulator of contractile activity, and elevated cytoplasmic Ca2+ resulting from ionomycin or thapsigargin treatment inhibits mitochondrial transport (Iqbal and Hood, 2014; Yi et al., 2004).
Kinesin-1-mediated mitochondrial transport is also important during cardiac maintenance. Cardiac hypertrophy (cardiomyocyte growth) is an adaptive response aimed at reducing wall stress and maintaining cardiac function (Nakamura and Sadoshima, 2018). Kinesin-1 expression is upregulated in a mouse model of pathological cardiac hypertrophy and in neonatal rat ventricular cardiomyocytes. In these cardiomyocytes, mitochondria are re-localized to the cell periphery accompanied by increased mitochondrial respiration associated with the hypertrophic response, which can be reverted by depletion of kinesin-1 (Tigchelaar et al., 2016). Interestingly, altered mitochondrial morphology and distribution are hallmarks of heart failure (Chen et al., 2009a; Schaper et al., 1991).
The intercellular communication between cardiomyocytes and cardiac myofibroblasts is important for maintaining normal myocardial function and is facilitated by membrane nanotubes (MNTs), which are long, thin membrane-based connections. These connections enable kinesin-1-dependent transport of mitochondria from cell to cell, thus acting as direct highways for the exchange of mitochondria (He et al., 2011; Rustom et al., 2004; Shen et al., 2018; Zhang and Zhang, 2013). Kinesin-1-mediated transport of mitochondria along MTs in MNTs is physiologically important in ischaemic cardiomyopathy for rescuing cardiomyocytes from hypoxia–reoxygenation-induced apoptosis (Shen et al., 2018).
Migrating cells
Migrating cells have a radial MT organization with plus ends facing the cell periphery (Bettencourt-Dias and Glover, 2007). In migrating lymphocytes, the centrosome, the main MT-organizing centre (MTOC), is located in the uropod at the rear or trailing end of the cell, whereas in motile epithelial cells, the centrosome is oriented towards the migration axis, establishing a high-density MT network towards the leading edge (Barker et al., 2016; Melkov and Abdu, 2018).
In lymphocytes or T cells, mitochondria accumulate in the energy-demanding uropod, where they provide ATP for actomyosin II-dependent force generation required for cell movement (Campello et al., 2006; Sanchez-Madrid and del Pozo, 1999). The Miro1–dynein complex is important in controlling lymphocyte and T cell migration by regulating mitochondrial accumulation around the MTOC at the uropod (Morlino et al., 2014). Another example of polarized distribution of mitochondria can be observed in immune cells during the formation of the immunological synapse between T cells and antigen-presenting cells. Mitochondria regulate sustained Ca2+ influx, and kinesin-1 drives anterograde mitochondrial transport to the immunological synapse at the plasma membrane (Quintana et al., 2006, 2007). Miro proteins also regulate mitochondrial recruitment in leukocytes to sites of contact with inflamed endothelial cells (Morlino et al., 2014).
In cancer cells, mitochondrial positioning is crucial for facilitating the energy-expensive processes of dissemination and invasion (da Silva et al., 2014). Mitochondria provide localized energy production by infiltrating the leading edge lamellipodia of migrating epithelial cancer cells in a MT-dependent manner (Cunniff et al., 2013, 2016; McKenzie et al., 2011). Anterior localization of mitochondria correlates with faster cell movement and increased directional persistence, which may account for the ability to invade further and metastasize earlier. Kinesin-1 and Miro1/2 are responsible for transporting mitochondria to the leading edge, since their depletion suppresses mitochondrial re-localization and results in slower cell migration rates as well as reduced tumour cell invasion (Caino et al., 2016; Desai et al., 2013). These findings are supported by observations in Miro1 KO MEFs, which are slower in collective and single-cell migration assays as a result of impaired membrane ruffling, leading-edge protrusion and focal adhesion dynamics (Schuler et al., 2017). Interestingly, downregulation or loss of the SNPH anchor enhances tumour progression in humans, whereas SNPH expression in tumour cells inhibits mitochondrial transport and blocks metastasis in vivo (Caino et al., 2016).
Gametes – oocytes and spermatids
Kinesins and their mitochondrial adaptor proteins are also involved in the spatial redistribution of mitochondria during oocyte maturation and spermatogenesis. Mitochondria in many species (human, Xenopus and Drosophila) enter oocytes from interconnected germ cells to generate the Balbiani body, a non-membrane-bound compartment packed with mitochondria. In Drosophila, oocyte acquisition of mitochondria depends on kinesin-1- and Milton-dependent movement, since mutations in these proteins cause premature and excessive mitochondrial transport into the oocyte (Cox and Spradling, 2003, 2006). During Drosophila oogenesis, the long isoform of the Oskar protein tethers mitochondria to the posterior cortex at the site of primordial germ cell formation through an actin-dependent mechanism requiring the non-muscle tropomyosin, Tropomyosin II; however, at present, it is not known whether any myosin motors are involved in this process (Hurd et al., 2016). During meiotic maturation of mouse oocytes, kinesin-3 is responsible for mitochondrial redistribution near the metaphase plate (Kong et al., 2016).
In Drosophila spermatogenesis, mitochondria undergo dramatic rearrangements, during which they aggregate and fuse to form the spherical Nebenkern (Varuzhanyan and Chan, 2020). Milton has been implicated in anchoring mitochondria of the Nebenkern at the minus end of MTs to the nucleus (Aldridge et al., 2007). Furthermore, in Milton-mutant flies, the unfurling and elongation of mitochondria along MTs is impaired during axonemal growth (Aldridge et al., 2007). Finally, in the testis of some fish species (Larimichthys polyactis and Boleophthalmus pectinirostris), kinesin-2 (specifically KIF3A) is highly expressed and colocalizes with mitochondria in the nuclear periphery during spermiogenesis and in the midpiece of mature sperm (Wang et al., 2019; Zhao et al., 2018), suggesting an active role in mitochondrial positioning during sperm maturation.
Dividing cells
During cell division, mitochondria are divided between the two daughter cells by either active or passive mechanisms. In metaphase, phosphorylation triggers the release of the dynein and kinesin motors from the Miro–TRAK adaptor complex on the mitochondrial surface, causing mitochondria to dissociate from MTs, ensuring symmetrical mitochondrial distribution and inheritance (Chung et al., 2016). Thus, passive inheritance results from the active release of motor proteins from mitochondria. In contrast, at later stages of mitosis, both MT- and actin-based motors play an active role in mitochondrial segregation between the two daughter cells. During anaphase, MYO19 has been shown to tether mitochondria to actin filaments, thereby ensuring equal distribution between the two daughter cells (Rohn et al., 2014). In cells undergoing cytokinesis, mitochondria are linked via kinesin-1 to the growing tips of astral MTs of the mitotic spindle and thereby are transported to the cleavage furrow (Lawrence and Mandato, 2013a; Lawrence et al., 2016; Lawrence and Mandato, 2013b). In addition, Miro1/2-dependent recruitment of the centromeric protein F (CENP-F), a non-motor MT-binding protein that associates with growing MTs, to mitochondria is required for their transport towards the periphery of the two daughter cells in late cytokinesis (Kanfer et al., 2015, 2017). All these processes are likely to involve Miro1/2, which coordinates mitochondrial distribution by MT- and actin-based motors, since Miro1/2 double-KO MEFs display aberrant mitochondrial positioning resulting from asymmetric mitochondrial segregation during mitosis (López-Domenéch et al., 2018).
Mitochondrial dynamics: fission and fusion
Mitochondria undergo regulated fission and fusion events to maintain a healthy, dynamic network (Giacomello et al., 2020; Tilokani et al., 2018). In mammals, the major mediators of fission and fusion are members of the dynamin-related GTPase family and include mitofusins 1 and 2 (MFN1 and MFN2, respectively), optic atrophy 1 (OPA1) and dynamin-related protein 1 (DRP1, also known as DNM1L).
Fission
Mitochondrial fission first involves separation of the IMM followed by OMM division (Chakrabarti et al., 2018; Cho et al., 2017; Labrousse et al., 1999; Lee and Yoon, 2014; Legesse-Miller et al., 2003). Endoplasmic reticulum (ER) tubules wrap around mitochondria, inducing a first constriction at these ER–mitochondria contacts, which mark the sites of mitochondrial fission as well as of active mtDNA replication (Friedman et al., 2011; Korobova et al., 2013; Lewis et al., 2016).
The machinery driving IMM fission in the matrix is poorly understood; however, the process is triggered by a transient rise in mitochondrial matrix Ca2+ that occurs at ER–mitochondria contact sites, which are generated and stabilized by the actin cytoskeleton (Cho et al., 2017). The formin INF2, an actin nucleator embedded in the ER membrane through a prenylated C-terminal CAAX motif, polymerizes actin away from the ER (Fig. 2) (Chhabra et al., 2009). These actin filaments together with NMIIA (which has a heavy chain encoded by MYH9), which forms bipolar mini-filaments, are thought to provide the contractile tension to draw the ER and mitochondria closer together, thereby stimulating ER-to-mitochondrial Ca2+ transfer (Chakrabarti et al., 2018).
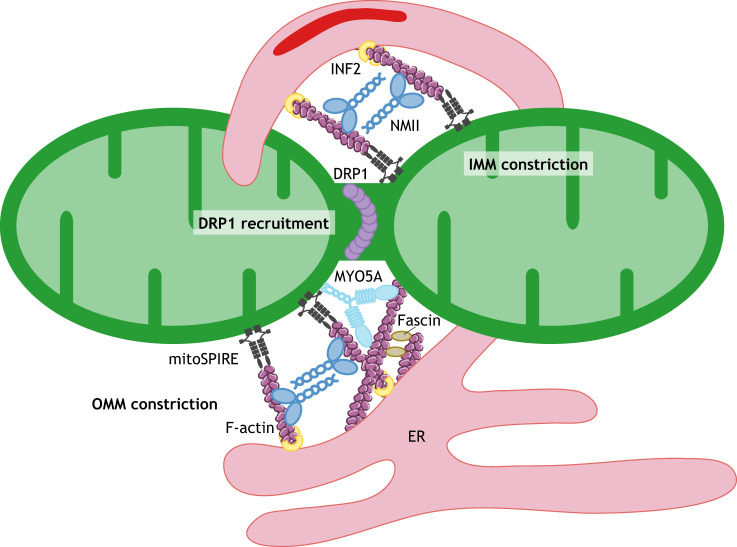
Fig. 2.
Motor proteins and cytoskeletal machinery regulating mitochondrial fission. Under steady-state conditions, the dynamic assembly of actin filaments onto mitochondria occurs at ER–mitochondria contact sites. Here, increased mitochondrial matrix Ca2+ activates the ER-bound formin INF2, resulting in actin polymerization. NMIIA provides the force to draw the ER and mitochondria closer together. At these ER–mitochondria contact sites, Ca2+ is transferred from the ER to mitochondria, triggering IMM constriction through an as-yet-unknown mechanism. Recruitment of the fission factor DRP1 from the cytosol to mitochondria involves several motor proteins (not shown). INF2 cooperates with mitoSPIRE and the actin bundling-protein fascin to drive polymerization of actin filaments on the mitochondrial surface, which target DRP1 to mitochondrial fission sites and stimulate its oligomerization. MYO5A, which interacts with mitoSPIRE, is recruited adjacent to DRP1 on mitochondria and facilitates fission; however, the exact role of MYO5A and actin filaments in this process remains to be established. Finally, NMII accumulates at mitochondrial constriction sites enabling DRP1 oligomerization and, by pulling actin filaments, provides the force for mitochondrial constriction. Actin filament images are adapted from Servier Medical Art (https://smart.servier.com/) under the terms of a CC-BY 3.0 license.
OMM scission requires the dynamic recruitment of DRP1 from the cytosol to tail-anchored proteins that act as DRP1 receptors (Fig. 2), such as the mitochondrial fission factor (MFF) and the mitochondria dynamics proteins 49 and 51 (MiD49 and MiD51, also known as MIEF2 and MIEF1, respectively) (Giacomello et al., 2020). DRP1 oligomerizes into ring-like structures around mitochondria, causing initial constriction of the organelle, before GTP hydrolysis leads to further mitochondrial membrane constriction (Tilokani et al., 2018). The final scission event downstream of DRP1 has recently been proposed to involve Golgi-derived vesicles (Fonseca et al., 2019; Nagashima et al., 2020) and occur at mitochondria–lysosome contact sites (Wong et al., 2018). In addition, several MT-based motors regulate DRP1 recruitment from the cytosol to mitochondria. Kinesin-3 (specifically the KIF1Bβ isoform) drives mitochondrial fission by activating calcineurin, which dephosphorylates DRP1 at Ser637, leading to DRP1 translocation to mitochondria (Li et al., 2016). In contrast, under oxidative stress, kinesin-1 has been shown to enhance DRP1 phosphorylation at Ser616 through JNK activation, which also promotes DRP1 recruitment to mitochondria (Perdiz et al., 2017; Reed et al., 2006). Finally, the dynein–dynactin complex also facilitates DRP1 translocation to mitochondria, since disruption of dynein function by overexpression of the dynactin subunit p50 (also known as DCTN2) in HeLa cells leads to accumulation of DRP1 in the cytosol, resulting in the formation of long, highly branched mitochondria (Varadi et al., 2004). Interestingly, in ageing Dync1h1-mutant mice, mitochondria progressively increase in size and become profoundly dysfunctional in muscle (Eschbach et al., 2013). In contrast to MT-based motors, Miro1/2 negatively regulates DRP1 recruitment to mitochondria, thereby suppressing fission, although the recruitment of fusion-promoting factors may also be affected (Covill-Cooke et al., 2020; Ding et al., 2016; Saotome et al., 2008).
OMM constriction is mediated by myosin motor proteins and the actin cytoskeleton (Fig. 2). INF2, which directly interacts and cooperates with mitoSPIRE and the actin-bundling protein fascin, drives actin polymerization on the mitochondrial surface (Korobova et al., 2013; Lin et al., 2019; Manor et al., 2015; Yang and Svitkina, 2019). These mitochondrial actin filaments not only directly bind DRP1 and target it to mitochondrial fission sites, but also stimulate DRP1 oligomerization (De Vos et al., 2005; Hatch et al., 2016; Ji et al., 2017, 2015; Korobova et al., 2013). In melanoma cell lines, mitoSPIRE binds directly to MYO5A, which recruits this motor to mitochondrial fission sites adjacent to DRP1 puncta; MYO5A recruitment facilitates mitochondrial fission, whereas MYO5A depletion leads to mitochondrial elongation (Araujo et al., 2019 preprint). NMIIA, NMIIB and NMIIC (which have heavy chains encoded by MYH9, MYH10 and MYH14, respectively) also accumulate at mitochondrial constriction sites and enable DRP1 oligomerization, as inhibition of NMII activity leads to increased mitochondrial length and loss of mitochondrial DRP1 (Almutawa et al., 2019; DuBoff et al., 2012; Hatch et al., 2014; Ji et al., 2015; Korobova et al., 2013; Yang and Svitkina, 2019). Furthermore, bipolar NMII filaments can pull on actin filaments to provide the tension force for mitochondrial constriction (Friedman et al., 2011; Osellame et al., 2016). Interestingly, NMIIC with the R94L heavy-chain mutation, which has been linked to peripheral neuropathies, acts as a dominant-negative protein that inhibits fission (Almutawa et al., 2019). In cardiomyocytes after ischaemia–reperfusion injury, inhibition of myosin II with blebbistatin or loss of NMIIA reduces mitochondrial fission and mitophagy (Li et al., 2018).
Finally, dynamic assembly and disassembly of actin filaments onto mitochondrial subpopulations has been observed at ER–mitochondria contact sites; this is blocked by inhibition of actin nucleators, such as the ARP2/3 complex and formins (Moore et al., 2016). This cycling is suggested to act as a steady-state surveillance method to modulate the mitochondrial fission–fusion balance to promote network remodelling and content mixing (Fung et al., 2019; Moore et al., 2016).
Fusion
OMM fusion requires tethering of MFN1 and MFN2 on the OMM between adjacent mitochondria followed by GTP hydrolysis (Tilokani et al., 2018). IMM fusion is mediated by OPA1, which is inserted into the IMM and is cleaved at S1 and S2 sites by two membrane-bound metalloproteases, OMA1 and YME1L1, resulting in at least five OPA1 fragments (two larger L-OPA1 fragments and three shorter S-OPA1 fragments) thought to regulate fusion (Giacomello et al., 2020).
The importance of the cytoskeleton for mitochondrial fusion is less well understood. Inhibiting actin filament polymerization or inducing changes in MT dynamics has little effect on fusion kinetics (Mattenberger et al., 2003). Recent results, however, highlight a potential role of kinesin-3 in IMM fusion, as it binds directly to YME1L1 and its overexpression promotes cleavage of L-OPA1, which leads to mitochondrial fission and fragmentation triggering nerve growth factor (NGF)-induced apoptosis (Ando et al., 2019). However, where binding between kinesin-3 and YME1L1 occurs remains to be established, since kinesin-3 is a cytosolic protein, whereas YME1L1 is present at the IMM.
With their many links to cytoskeletal motors, Miro proteins appear to be important for maintaining the balance between fission and fusion, which controls mitochondrial morphology; however, whether this is a direct effect or due to an indirect effect on transport requires further investigation. Miro proteins may facilitate fusion, since the overexpression of the wild-type protein, or expression of a constitutively active form of the protein, results in mitochondrial enlargement in Drosophila and induces the formation of long thread-like mitochondria (Babic et al., 2015; Ding et al., 2016; Fransson et al., 2006; López-Domenéch et al., 2018; Saotome et al., 2008), whereas in yeast, loss of the Miro homologue Gem1 or its GTPase activity leads to fragmented mitochondria and a collapsed network (Frederick et al., 2004). Miro1/2 and Milton interact with both MFN1 and MFN2, which are also required for mitochondrial transport (Misko et al., 2010). Thus, there is crosstalk and coordination between mitochondrial transport and fusion; however, the dependence of mitochondrial fusion on transport has made it difficult to determine the precise role of Miro proteins and MFNs in mitochondrial fusion. Depletion or mutation of MFN2 reduces the rate of anterograde and retrograde mitochondrial transport in neurons (Misko et al., 2010), while reducing mitochondrial motility by suppression of Miro1/2 inhibits the rate of mitochondrial fusion (Cagalinec et al., 2013).
Mitochondrial turnover and quality control
Several pathways are known to regulate the turnover of whole mitochondria by mitophagy; one of the best studied pathways involves the serine/threonine kinase PINK1 and the E3 ubiquitin ligase Parkin (PRKN), which when mutated cause autosomal recessive Parkinson's disease (Narendra et al., 2008; Ryan et al., 2015). A hallmark of mitochondrial damage is membrane depolarization, leading to the accumulation of PINK1 on the OMM and phosphorylation of ubiquitin attached to OMM proteins (Harper et al., 2018). This, in turn, triggers the recruitment of auto-inhibited PRKN to the OMM, where it binds to phosphorylated ubiquitin. PINK1-dependent phosphorylation leads to full PRKN activation, which then allows PRKN to ubiquitylate OMM proteins including Miro1/2, thereby amplifying the mitophagy signal. Cargo-selective autophagy receptors, such as optineurin (OPTN), Ca2+-binding and coiled-coil domain-containing protein 2 (CALCOCO2, also known as NDP52), and Tax1-binding protein 1 (TAX1BP1), recognize and capture ubiquitylated mitochondria through their ubiquitin-binding domains and simultaneously bind to LC3 proteins (also known as MAP1LC3) to recruit autophagosomal membranes for the formation of mitophagosomes, which ultimately fuse with lysosomes for degradation (Pickles et al., 2018).
Before damaged mitochondria are eliminated by mitophagy, they are transported by dynein towards the cell body/soma (Fig. 3) (Cai et al., 2012; Miller and Sheetz, 2004), and defects in retrograde transport of senescent mitochondria lead to increased autophagy in axonal swellings (Pilling et al., 2006). Enhancing retrograde transport by inducing binding of mitochondria to dynein increases neuronal mitophagy (Zheng et al., 2019), whereas Dync1h1 mutations lead to impaired perinuclear clustering of damaged mitochondria in fibroblasts (Eschbach et al., 2013). Furthermore, the release of the MT anchor SNPH from stressed mitochondria enhances retrograde mitochondrial transport before activation of PRKN-mediated mitophagy, whereas overexpression of SNPH blocks mitophagy (Lin et al., 2017). Compartmental restriction of mitophagy to the soma of stressed neurons (Evans and Holzbaur, 2020) is supported by in vivo data from Drosophila neurons (Devireddy et al., 2015; Sung et al., 2016) and Purkinje cells from mito-QC mice expressing a pH-sensitive fluorescent mitochondrial marker (McWilliams et al., 2016); however, clearance of damaged mitochondria has also been shown to occur locally in distal axons in response to acute stress (Ashrafi et al., 2014).
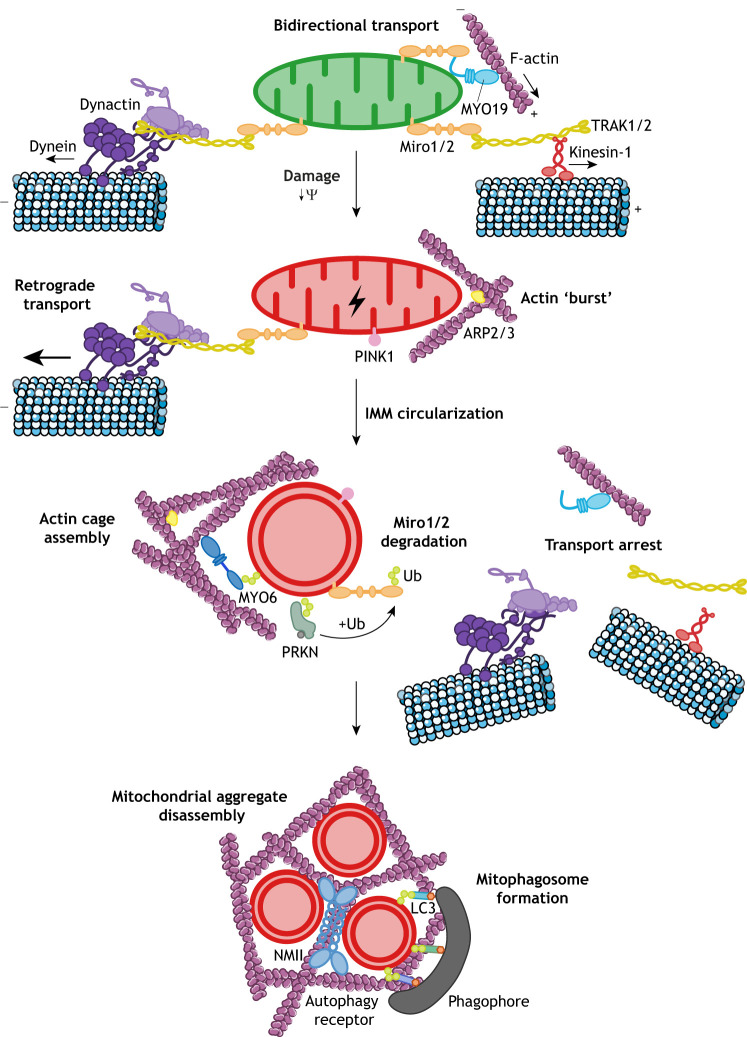
Fig. 3.
Motor proteins and machinery associated with damaged mitochondria during mitophagy. Healthy mitochondria are actively transported by kinesin-1 and dynein motors and positioned by MYO19. Upon mitochondrial damage, loss of mitochondrial membrane potential (Ψ) results in the accumulation of the kinase PINK1 on the OMM. Within 5 min of damage induction, a short ‘burst’ of mitochondrial actin can be observed, which requires the ARP2/3 complex and leads to IMM circularization followed by actin disassembly. The ubiquitin ligase PRKN translocates from the cytosol to the mitochondria where it ubiquitylates (+Ub) Miro proteins, leading to their subsequent proteasomal degradation. This results in release of kinesin-1, dynein–dynactin and MYO19 from mitochondria and the arrest of mitochondrial transport. At ∼2 h after mitochondrial damage, the OMM has fragmented and MYO6 is recruited to PRKN-mediated ubiquitin chains on the mitochondrial surface, where it assembles an actin cage to isolate damaged, fragmented mitochondria, thereby preventing their re-fusion with neighbouring populations. The actin meshwork around mitochondria, together with NMII, may reorganize to disrupt any mitochondrial aggregates, creating small ‘bite-size’ mitochondria, and serve as a structural scaffold for the growing LC3-positive phagophore. Actin filament and microtubule images are adapted from Servier Medical Art (https://smart.servier.com/) under the terms of a CC-BY 3.0 license.
In addition to phosphorylating PRKN and ubiquitin, PINK1 also phosphorylates Miro1/2, thereby facilitating PRKN recruitment to Miro proteins (Shlevkov et al., 2016; Wang et al., 2011). Ubiquitylation of Miro1/2 by PRKN then leads to its subsequent proteasomal degradation (Fig. 3) (Birsa et al., 2014). This results in the release of MYO19 and kinesin from the mitochondrial surface, leading to a reduced motility of damaged mitochondria (Liu et al., 2012; López-Domenéch et al., 2018; Safiulina et al., 2019; Shlevkov et al., 2016; Tsai et al., 2014; Wang et al., 2011; Weihofen et al., 2009). Hence, arresting mitochondrial transport is an early step in the clearance of dysfunctional mitochondria, which potentially isolates them before their elimination by mitophagy (Fig. 3).
Mitochondrial damage not only halts the retrograde dynein-dependent movement of mitochondria, but also leads to distinct F-actin assembly events on mitochondria that are linked to recovery, segregation and the initiation of downstream mitophagy. A rapid, dynamic and transient ‘burst’ in mitochondria-localized F-actin occurs within minutes (∼5 min) of damage and requires ARP2/3 (Fig. 3), but is independent of INF2 and MYO6 (Fung et al., 2019; Kruppa et al., 2018; Li et al., 2015). This leads to circularization of mitochondria, which is their last chance to recover before being committed to clearance by mitophagy. Prolonged depolarization leads to mitochondrial fragmentation and initiates a second wave of MYO6-dependent actin polymerization after several hours, which initiates the formation of actin cages around damaged mitochondria during PRKN-mediated mitophagy (Fu and Lippincott-Schwartz, 2018; Kruppa et al., 2018). MYO6 forms a complex with PRKN and is selectively recruited to damaged, ubiquitylated mitochondria (Kruppa et al., 2018; Sarraf et al., 2013). Signalling downstream of the Rho family GTPase CDC42, as well as actin nucleators, such as the ARP2/3 complex, formins and N-WASP (also known as WASL), are all required for actin cage assembly. These MYO6-induced actin cages serve as a quality control mechanism at the onset of mitophagy by isolating dysfunctional mitochondria, thereby preventing their re-fusion with neighbouring populations and reintegration into the network (Hsieh and Yang, 2019; Kruppa et al., 2018) (Fig. 3).
In recent years, there has been growing evidence that the cytoskeletal machinery plays crucial roles during mitophagy; therefore, the actin filaments assembled by MYO6 on the mitochondrial surface may not only form actin cages, but also have important roles during mitophagy by facilitating phagophore formation and mitophagosome maturation (Kast and Dominguez, 2017; Kruppa and Buss, 2018; Kruppa et al., 2016). Indeed, since MYO6-dependent actin cages restrict the size of mitochondrial fragments, they may create small, ‘bite-size’ mitochondria that are more easily engulfed by the autophagy machinery. The synchronous formation of mitochondrial actin structures near autophagy initiation sites suggests a coordination between a reduction in mitochondrial dimensions and mitophagosome formation (Hsieh and Yang, 2019). This is driven by NMII and F-actin (Fig. 3), which have been implicated in disassembling mitochondrial aggregates into smaller pieces (1.5 h after damage) in preparation for efficient mitophagy into autophagosomes (with a typical diameter of 0.5–1.5 µm) (Hsieh and Yang, 2019; Mizushima et al., 2002).
Furthermore, the actin meshwork around mitochondria may persist after the initial damage and therefore could serve as a structural scaffold for the growing phagophore (Fig. 3) as well as supporting fusion of individual phagophores into a single mitophagosome (Hsieh and Yang, 2019; Kruppa and Buss, 2018; Tang et al., 2011). Mitochondrial actin may also drive autophagosome generation by disrupting mitochondrial aggregates to increase the available surface accessible to phagophores (Hsieh and Yang, 2019). Indeed, blocking the disassembly of mitochondrial aggregates and actin generation with an ARP2/3 inhibitor leads to prolonged clustering and inhibited mitophagosome formation (Hsieh and Yang, 2019).
Conclusions and perspectives
In this Review, we have highlighted a distinct subset of MT- and actin-based motor proteins that are linked to mitochondrial transport, fission, fusion and turnover. Even though we have discussed these key mitochondrial processes in separate sections, they are of course intimately linked and interdependent, and thus studying the precise roles of motors and their adaptors in isolation has been difficult due to their overlapping functions. One important avenue for future research is the regulation of motors and their adaptor proteins on mitochondria in response to changes in cell physiology and metabolism. The balance between fission and fusion, as well as transport and anchoring, are fine-tuned and require the coordination and handover of MT- and actin-based motors. Furthermore, a detailed understanding of motor protein regulation in maintaining mitochondrial homeostasis is crucial to elucidate how defects in these pathways can lead to human diseases.
Supplementary Material
Acknowledgements
We apologize to our colleagues whose work we were unable to cite due to space limitations. We would like to thank Evan Reid, Alex Whitworth, John Kendrick-Jones, Christopher Batters, Janeska de Jonge, Chloe Johnson, Sue Arden, and Alex Holmes for critical reading of the manuscript and helpful suggestions. CIMR is supported by the Wellcome Trust with an equipment grant (093026).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Our work in this area is funded by project grants from the Medical Research Council (MR/S007776/1) and the Biotechnology and Biological Sciences Research Council (BB/R001316/1) to F.B. Open access funding provided by University of Cambridge. Deposited in PMC for immediate release.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.226084.supplemental
References
- Aldridge, A. C., Benson, L., Siegenthaler, M. M., Whigham, B. T., Stowers, R. S. and Hales, K. G. (2007). Roles for Drp1, a dynamin-related protein, and milton, a kinesin-associated protein, in mitochondrial segregation, unfurling, and elongation during Drosophila spermatogenesis. Fly 1, 38-46. 10.4161/fly.3913 [DOI] [PubMed] [Google Scholar]
- Almutawa, W., Smith, C., Sabouny, R., Smit, R. B., Zhao, T., Wong, R., Lee-Glover, L., Desrochers-Goyette, J., Ilamathi, H. S., Care4Rare Canada Consortium et al. (2019). The R941L mutation in MYH14 disrupts mitochondrial fission and associates with peripheral neuropathy. EBioMedicine 45, 379-392. 10.1016/j.ebiom.2019.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando, K., Yokochi, T., Mukai, A., Wei, G., Li, Y., Kramer, S., Ozaki, T., Maehara, Y. and Nakagawara, A. (2019). Tumor suppressor KIF1Bβ regulates mitochondrial apoptosis in collaboration with YME1L1. Mol. Carcinog. 58, 1134-1144. 10.1002/mc.22997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo, J. S., Silva-Junior, R. M. P., Zhang, T., Schiavon, C. R., Chu, Q., Wu, M., Pontes, C. L. S., Souza, A. O., Alberici, L. C., Santos, A. M.et al. (2019). A novel role for Myosin-Va in mitochondrial fission. bioRxiv, 655803. [Google Scholar]
- Ashrafi, G., Schlehe, J. S., LaVoie, M. J. and Schwarz, T. L. (2014). Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J. Cell Biol. 206, 655-670. 10.1083/jcb.201401070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton, J., Hummel, J. J. A., Olieric, N., Locke, J., Peña, A., Rosenfeld, S. S., Steinmetz, M. O., Hoogenraad, C. C. and Moores, C. A. (2020). The mechanism of kinesin inhibition by kinesin-binding protein. eLife 9, e61481. 10.7554/eLife.61481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babic, M., Russo, G. J., Wellington, A. J., Sangston, R. M., Gonzalez, M. and Zinsmaier, K. E. (2015). Miro's N-terminal GTPase domain is required for transport of mitochondria into axons and dendrites. J. Neurosci. 35, 5754-5771. 10.1523/JNEUROSCI.1035-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakula, D. and Scheibye-Knudsen, M. (2020). MitophAging: mitophagy in aging and disease. Front. Cell Dev. Biol. 8, 239. 10.3389/fcell.2020.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barel, O., Malicdan, M. C. V., Ben-Zeev, B., Kandel, J., Pri-Chen, H., Stephen, J., Castro, I. G., Metz, J., Atawa, O., Moshkovitz, S.et al. (2017). Deleterious variants in TRAK1 disrupt mitochondrial movement and cause fatal encephalopathy. Brain 140, 568-581. 10.1093/brain/awx002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, A. R., McIntosh, K. V. and Dawe, H. R. (2016). Centrosome positioning in non-dividing cells. Protoplasma 253, 1007-1021. 10.1007/s00709-015-0883-5 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias, M. and Glover, D. M. (2007). Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 8, 451-463. 10.1038/nrm2180 [DOI] [PubMed] [Google Scholar]
- Birsa, N., Norkett, R., Wauer, T., Mevissen, T. E. T., Wu, H.-C., Foltynie, T., Bhatia, K., Hirst, W. D., Komander, D., Plun-Favreau, H.et al. (2014). Lysine 27 ubiquitination of the mitochondrial transport protein Miro is dependent on serine 65 of the Parkin ubiquitin ligase. J. Biol. Chem. 289, 14569-14582. 10.1074/jbc.M114.563031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocanegra, J. L., Adikes, R. and Quintero, O. A. (2020a). Myosin XIX. Adv. Exp. Med. Biol. 1239, 439-451. 10.1007/978-3-030-38062-5_20 [DOI] [PubMed] [Google Scholar]
- Bocanegra, J. L., Fujita, B. M., Melton, N. R., Cowan, J. M., Schinski, E. L., Tamir, T. Y., Major, M. B. and Quintero, O. A. (2020b). The MyMOMA domain of MYO19 encodes for distinct Miro-dependent and Miro-independent mechanisms of interaction with mitochondrial membranes. Cytoskeleton 77, 149-166. 10.1002/cm.21560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickley, K. and Stephenson, F. A. (2011). Trafficking kinesin protein (TRAK)-mediated transport of mitochondria in axons of hippocampal neurons. J. Biol. Chem. 286, 18079-18092. 10.1074/jbc.M111.236018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard, G. J. and Rice, L. M. (2018). Microtubule dynamics: an interplay of biochemistry and mechanics. Nat. Rev. Mol. Cell Biol. 19, 451-463. 10.1038/s41580-018-0009-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagalinec, M., Safiulina, D., Liiv, M., Liiv, J., Choubey, V., Wareski, P., Veksler, V. and Kaasik, A. (2013). Principles of the mitochondrial fusion and fission cycle in neurons. J. Cell Sci. 126, 2187-2197. 10.1242/jcs.118844 [DOI] [PubMed] [Google Scholar]
- Cai, Q., Zakaria, H. M., Simone, A. and Sheng, Z.-H. (2012). Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr. Biol. 22, 545-552. 10.1016/j.cub.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caino, M. C., Seo, J. H., Aguinaldo, A., Wait, E., Bryant, K. G., Kossenkov, A. V., Hayden, J. E., Vaira, V., Morotti, A., Ferrero, S.et al. (2016). A neuronal network of mitochondrial dynamics regulates metastasis. Nat. Commun. 7, 13730. 10.1038/ncomms13730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, P. D., Shen, K., Sapio, M. R., Glenn, T. D., Talbot, W. S. and Marlow, F. L. (2014). Unique function of Kinesin Kif5A in localization of mitochondria in axons. J. Neurosci. 34, 14717-14732. 10.1523/JNEUROSCI.2770-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campello, S., Lacalle, R. A., Bettella, M., Mañes, S., Scorrano, L. and Viola, A. (2006). Orchestration of lymphocyte chemotaxis by mitochondrial dynamics. J. Exp. Med. 203, 2879-2886. 10.1084/jem.20061877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardanho-Ramos, C., Faria-Pereira, A. and Morais, V. A. (2020). Orchestrating mitochondria in neurons: cytoskeleton as the conductor. Cytoskeleton 77, 65-75. 10.1002/cm.21585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti, R., Ji, W.-K., Stan, R. V., de Juan Sanz, J., Ryan, T. A. and Higgs, H. N. (2018). INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division. J. Cell Biol. 217, 251-268. 10.1083/jcb.201709111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, D. C. (2020). Mitochondrial dynamics and its involvement in disease. Annu. Rev. Pathol. Mech. Dis. 15, 235-259. 10.1146/annurev-pathmechdis-012419-032711 [DOI] [PubMed] [Google Scholar]
- Chandel, N. S. (2015). Evolution of mitochondria as signaling organelles. Cell Metab. 22, 204-206. 10.1016/j.cmet.2015.05.013 [DOI] [PubMed] [Google Scholar]
- Chen, Y. and Sheng, Z.-H. (2013). Kinesin-1-syntaphilin coupling mediates activity-dependent regulation of axonal mitochondrial transport. J. Cell Biol. 202, 351-364. 10.1083/jcb.201302040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., Gong, Q., Stice, J. P. and Knowlton, A. A. (2009a). Mitochondrial OPA1, apoptosis, and heart failure. Cardiovasc. Res. 84, 91-99. 10.1093/cvr/cvp181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.-M., Gerwin, C. and Sheng, Z.-H. (2009b). Dynein light chain LC8 regulates syntaphilin-mediated mitochondrial docking in axons. J. Neurosci. 29, 9429-9438. 10.1523/JNEUROSCI.1472-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra, E. S., Ramabhadran, V., Gerber, S. A. and Higgs, H. N. (2009). INF2 is an endoplasmic reticulum-associated formin protein. J. Cell Sci. 122, 1430-1440. 10.1242/jcs.040691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, B., Cho, H. M., Jo, Y., Kim, H. D., Song, M., Moon, C., Kim, H., Kim, K., Sesaki, H., Rhyu, I. J.et al. (2017). Constriction of the mitochondrial inner compartment is a priming event for mitochondrial division. Nat. Commun. 8, 15754. 10.1038/ncomms15754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, J. Y.-M., Steen, J. A. and Schwarz, T. L. (2016). Phosphorylation-induced motor shedding is required at mitosis for proper distribution and passive inheritance of mitochondria. Cell Rep. 16, 2142-2155. 10.1016/j.celrep.2016.07.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covill-Cooke, C., Toncheva, V. S., Drew, J., Birsa, N., López-Doménech, G. and Kittler, J. T. (2020). Peroxisomal fission is modulated by the mitochondrial Rho-GTPases, Miro1 and Miro2. EMBO Rep. 21, e49865. 10.15252/embr.201949865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, R. T. and Spradling, A. C. (2003). A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development 130, 1579-1590. 10.1242/dev.00365 [DOI] [PubMed] [Google Scholar]
- Cox, R. T. and Spradling, A. C. (2006). Milton controls the early acquisition of mitochondria by Drosophila oocytes. Development 133, 3371-3377. 10.1242/dev.02514 [DOI] [PubMed] [Google Scholar]
- Cunniff, B., Benson, K., Stumpff, J., Newick, K., Held, P., Taatjes, D., Joseph, J., Kalyanaraman, B. and Heintz, N. H. (2013). Mitochondrial-targeted nitroxides disrupt mitochondrial architecture and inhibit expression of peroxiredoxin 3 and FOXM1 in malignant mesothelioma cells. J. Cell. Physiol. 228, 835-845. 10.1002/jcp.24232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunniff, B., McKenzie, A. J., Heintz, N. H. and Howe, A. K. (2016). AMPK activity regulates trafficking of mitochondria to the leading edge during cell migration and matrix invasion. Mol. Biol. Cell 27, 2662-2674. 10.1091/mbc.e16-05-0286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva, A. F., Mariotti, F. R., Máximo, V. and Campello, S. (2014). Mitochondria dynamism: of shape, transport and cell migration. Cell. Mol. Life Sci. 71, 2313-2324. 10.1007/s00018-014-1557-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge, J. J., Batters, C., O'Loughlin, T., Arden, S. D. and Buss, F. (2019). The MYO6 interactome: selective motor-cargo complexes for diverse cellular processes. FEBS Lett. 593, 1494-1507. 10.1002/1873-3468.13486 [DOI] [PubMed] [Google Scholar]
- De Vos, K. J., Allan, V. J., Grierson, A. J. and Sheetz, M. P. (2005). Mitochondrial function and actin regulate dynamin-related protein 1-dependent mitochondrial fission. Curr. Biol. 15, 678-683. 10.1016/j.cub.2005.02.064 [DOI] [PubMed] [Google Scholar]
- Desai, S. P., Bhatia, S. N., Toner, M. and Irimia, D. (2013). Mitochondrial localization and the persistent migration of epithelial cancer cells. Biophys. J. 104, 2077-2088. 10.1016/j.bpj.2013.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devireddy, S., Liu, A., Lampe, T. and Hollenbeck, P. J. (2015). The organization of mitochondrial quality control and life cycle in the nervous system in vivo in the absence of PINK1. J. Neurosci. 35, 9391-9401. 10.1523/JNEUROSCI.1198-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, L., Lei, Y., Han, Y., Li, Y., Ji, X. and Liu, L. (2016). Vimar is a novel regulator of mitochondrial fission through miro. PLoS Genet. 12, e1006359. 10.1371/journal.pgen.1006359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato, V., Bonora, M., Simoneschi, D., Sartini, D., Kudo, Y., Saraf, A., Florens, L., Washburn, M. P., Stadtfeld, M., Pinton, P.et al. (2017). The TDH–GCN5L1–Fbxo15–KBP axis limits mitochondrial biogenesis in mouse embryonic stem cells. Nat. Cell Biol. 19, 341-351. 10.1038/ncb3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drerup, C. M., Lusk, S. and Nechiporuk, A. (2016). Kif1B interacts with KBP to promote axon elongation by localizing a microtubule regulator to growth cones. J. Neurosci. 36, 7014-7026. 10.1523/JNEUROSCI.0054-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drerup, C. M., Herbert, A. L., Monk, K. R. and Nechiporuk, A. V. (2017). Regulation of mitochondria-dynactin interaction and mitochondrial retrograde transport in axons. eLife 6, e22234. 10.7554/eLife.22234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBoff, B., Götz, J. and Feany, M. B. (2012). Tau promotes neurodegeneration via DRP1 mislocalization in vivo. Neuron 75, 618-632. 10.1016/j.neuron.2012.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duis, J., Dean, S., Applegate, C., Harper, A., Xiao, R., He, W., Dollar, J. D., Sun, L. R., Waberski, M. B., Crawford, T. O.et al. (2016). KIF5A mutations cause an infantile onset phenotype including severe myoclonus with evidence of mitochondrial dysfunction. Ann. Neurol. 80, 633-637. 10.1002/ana.24744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt, E. L., Ludlam, A. V., Tan, Z. and Cianfrocco, M. A. (2020). Miro: a molecular switch at the center of mitochondrial regulation. Protein Sci. 29, 1269-1284. 10.1002/pro.3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow, S. A., Kull, F. J. and Liu, H. (2010). Kinesins at a glance. J. Cell Sci. 123, 3420-3424. 10.1242/jcs.064113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschbach, J., Sinniger, J., Bouitbir, J., Fergani, A., Schlagowski, A.-I., Zoll, J., Geny, B., René, F., Larmet, Y., Marion, V.et al. (2013). Dynein mutations associated with hereditary motor neuropathies impair mitochondrial morphology and function with age. Neurobiol. Dis. 58, 220-230. 10.1016/j.nbd.2013.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, C. S. and Holzbaur, E. L. F. (2020). Degradation of engulfed mitochondria is rate-limiting in Optineurin-mediated mitophagy in neurons. eLife 9, e50260. 10.7554/eLife.50260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca, T. B., Sánchez-Guerrero, A., Milosevic, I. and Raimundo, N. (2019). Mitochondrial fission requires DRP1 but not dynamins. Nature 570, E34-E42. 10.1038/s41586-019-1296-y [DOI] [PubMed] [Google Scholar]
- Fransson, A., Ruusala, A. and Aspenström, P. (2003). Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J. Biol. Chem. 278, 6495-6502. 10.1074/jbc.M208609200 [DOI] [PubMed] [Google Scholar]
- Fransson, S., Ruusala, A. and Aspenström, P. (2006). The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem. Biophys. Res. Commun. 344, 500-510. 10.1016/j.bbrc.2006.03.163 [DOI] [PubMed] [Google Scholar]
- Frederick, R. L., McCaffery, J. M., Cunningham, K. W., Okamoto, K. and Shaw, J. M. (2004). Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology via a novel pathway. J. Cell Biol. 167, 87-98. 10.1083/jcb.200405100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, J. R., Lackner, L. L., West, M., DiBenedetto, J. R., Nunnari, J. and Voeltz, G. K. (2011). ER tubules mark sites of mitochondrial division. Science 334, 358-362. 10.1126/science.1207385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, D. and Lippincott-Schwartz, J. (2018). Monitoring the effects of pharmacological reagents on mitochondrial morphology. Curr. Protoc. Cell Biol. 79, e45. 10.1002/cpcb.45 [DOI] [PubMed] [Google Scholar]
- Fung, T. S., Ji, W.-K., Higgs, H. N. and Chakrabarti, R. (2019). Two distinct actin filament populations have effects on mitochondria, with differences in stimuli and assembly factors. J. Cell Sci. 132, jcs234435. 10.1242/jcs.234435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomello, M., Pyakurel, A., Glytsou, C. and Scorrano, L. (2020). The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Biol. 21, 204-224. 10.1038/s41580-020-0210-7 [DOI] [PubMed] [Google Scholar]
- Glater, E. E., Megeath, L. J., Stowers, R. S. and Schwarz, T. L. (2006). Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J. Cell Biol. 173, 545-557. 10.1083/jcb.200601067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutnick, A., Banghart, M. R., West, E. R. and Schwarz, T. L. (2019). The light-sensitive dimerizer zapalog reveals distinct modes of immobilization for axonal mitochondria. Nat. Cell Biol. 21, 768-777. 10.1038/s41556-019-0317-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J. W., Ordureau, A. and Heo, J.-M. (2018). Building and decoding ubiquitin chains for mitophagy. Nat. Rev. Mol. Cell Biol. 19, 93-108. 10.1038/nrm.2017.129 [DOI] [PubMed] [Google Scholar]
- Hatch, A. L., Gurel, P. S. and Higgs, H. N. (2014). Novel roles for actin in mitochondrial fission. J. Cell Sci. 127, 4549-4560. 10.1242/jcs.153791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch, A. L., Ji, W.-K., Merrill, R. A., Strack, S. and Higgs, H. N. (2016). Actin filaments as dynamic reservoirs for Drp1 recruitment. Mol. Biol. Cell 27, 3109-3121. 10.1091/mbc.e16-03-0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne, J. L., Mehta, P. R., Singh, P. P., Wong, N. Q. and Quintero, O. A. (2016). Positively charged residues within the MYO19 MyMOMA domain are essential for proper localization of MYO19 to the mitochondrial outer membrane. Cytoskeleton 73, 286-299. 10.1002/cm.21305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, K., Shi, X., Zhang, X., Dang, S., Ma, X., Liu, F., Xu, M., Lv, Z., Han, D., Fang, X.et al. (2011). Long-distance intercellular connectivity between cardiomyocytes and cardiofibroblasts mediated by membrane nanotubes. Cardiovasc. Res. 92, 39-47. 10.1093/cvr/cvr189 [DOI] [PubMed] [Google Scholar]
- Henrichs, V., Grycova, L., Barinka, C., Nahacka, Z., Neuzil, J., Diez, S., Rohlena, J., Braun, M. and Lansky, Z. (2020). Mitochondria-adaptor TRAK1 promotes kinesin-1 driven transport in crowded environments. Nat. Commun. 11, 3123. 10.1038/s41467-020-16972-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa, N., Noda, Y., Tanaka, Y. and Niwa, S. (2009). Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682-696. 10.1038/nrm2774 [DOI] [PubMed] [Google Scholar]
- Hsieh, C.-W. and Yang, W. Y. (2019). Omegasome-proximal PtdIns(4,5)P2 couples F-actin mediated mitoaggregate disassembly with autophagosome formation during mitophagy. Nat. Commun. 10, 969. 10.1038/s41467-019-08924-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd, D. D. and Saxton, W. M. (1996). Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics 144, 1075-1085. 10.1093/genetics/144.3.1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd, T. R., Herrmann, B., Sauerwald, J., Sanny, J., Grosch, M. and Lehmann, R. (2016). Long oskar controls mitochondrial inheritance in Drosophila melanogaster. Dev. Cell 39, 560-571. 10.1016/j.devcel.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal, S. and Hood, D. A. (2014). Cytoskeletal regulation of mitochondrial movements in myoblasts. Cytoskeleton 71, 564-572. 10.1002/cm.21188 [DOI] [PubMed] [Google Scholar]
- Ji, W.-K., Hatch, A. L., Merrill, R. A., Strack, S. and Higgs, H. N. (2015). Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. eLife 4, e11553. 10.7554/eLife.11553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, W.-K., Chakrabarti, R., Fan, X., Schoenfeld, L., Strack, S. and Higgs, H. N. (2017). Receptor-mediated Drp1 oligomerization on endoplasmic reticulum. J. Cell Biol. 216, 4123-4139. 10.1083/jcb.201610057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfer, G., Courthéoux, T., Peterka, M., Meier, S., Soste, M., Melnik, A., Reis, K., Aspenström, P., Peter, M., Picotti, P.et al. (2015). Mitotic redistribution of the mitochondrial network by Miro and Cenp-F. Nat. Commun. 6, 8015. 10.1038/ncomms9015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfer, G., Peterka, M., Arzhanik, V. K., Drobyshev, A. L., Ataullakhanov, F. I., Volkov, V. A. and Kornmann, B. (2017). CENP-F couples cargo to growing and shortening microtubule ends. Mol. Biol. Cell 28, 2400-2409. 10.1091/mbc.e16-11-0756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J.-S., Tian, J.-H., Pan, P.-Y., Zald, P., Li, C., Deng, C. and Sheng, Z.-H. (2008). Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell 132, 137-148. 10.1016/j.cell.2007.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karle, K. N., Möckel, D., Reid, E. and Schöls, L. (2012). Axonal transport deficit in a KIF5A−/− mouse model. Neurogenetics 13, 169-179. 10.1007/s10048-012-0324-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast, D. J. and Dominguez, R. (2017). The cytoskeleton–autophagy connection. Curr. Biol. 27, R318-R326. 10.1016/j.cub.2017.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevenaar, J. T., Bianchi, S., van Spronsen, M., Olieric, N., Lipka, J., Frias, C. P., Mikhaylova, M., Harterink, M., Keijzer, N., Wulf, P. S.et al. (2016). Kinesin-binding protein controls microtubule dynamics and cargo trafficking by regulating kinesin motor activity. Curr. Biol. 26, 849-861. 10.1016/j.cub.2016.01.048 [DOI] [PubMed] [Google Scholar]
- Klosowiak, J. L., Focia, P. J., Chakravarthy, S., Landahl, E. C., Freymann, D. M. and Rice, S. E. (2013). Structural coupling of the EF hand and C-terminal GTPase domains in the mitochondrial protein Miro. EMBO Rep. 14, 968-974. 10.1038/embor.2013.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, X.-W., Wang, D.-H., Zhou, C.-J., Zhou, H.-X. and Liang, C.-G. (2016). Loss of function of KIF1B impairs oocyte meiotic maturation and early embryonic development in mice. Mol. Reprod. Dev. 83, 1027-1040. 10.1002/mrd.22744 [DOI] [PubMed] [Google Scholar]
- Korobova, F., Ramabhadran, V. and Higgs, H. N. (2013). An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464-467. 10.1126/science.1228360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsopoulos, O. S., Laine, D., Osellame, L., Chudakov, D. M., Parton, R. G., Frazier, A. E. and Ryan, M. T. (2010). Human Miltons associate with mitochondria and induce microtubule-dependent remodeling of mitochondrial networks. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 1803, 564-574. 10.1016/j.bbamcr.2010.03.006 [DOI] [PubMed] [Google Scholar]
- Kruppa, A. J. and Buss, F. (2018). Actin cages isolate damaged mitochondria during mitophagy. Autophagy 14, 1644-1645. 10.1080/15548627.2018.1486152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruppa, A. J., Kendrick-Jones, J. and Buss, F. (2016). Myosins, actin and autophagy. Traffic 17, 878-890. 10.1111/tra.12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruppa, A. J., Kishi-Itakura, C., Masters, T. A., Rorbach, J. E., Grice, G. L., Kendrick-Jones, J., Nathan, J. A., Minczuk, M. and Buss, F. (2018). Myosin VI-dependent actin cages encapsulate parkin-positive damaged mitochondria. Dev. Cell 44, 484-499.e6. 10.1016/j.devcel.2018.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse, A. M., Zappaterra, M. D., Rube, D. A. and van der Bliek, A. M. (1999). C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell 4, 815-826. 10.1016/S1097-2765(00)80391-3 [DOI] [PubMed] [Google Scholar]
- Lawrence, E. and Mandato, C. (2013a). Mitochondrial inheritance is mediated by microtubules in mammalian cell division. Commun. Integr. Biol. 6, e27557. 10.4161/cib.27557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, E. J. and Mandato, C. A. (2013b). Mitochondria localize to the cleavage furrow in mammalian cytokinesis. PLOS ONE 8, e72886. 10.1371/journal.pone.0072886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, E. J., Boucher, E. and Mandato, C. A. (2016). Mitochondria-cytoskeleton associations in mammalian cytokinesis. Cell Div. 11, 3. 10.1186/s13008-016-0015-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. and Yoon, Y. (2014). Transient contraction of mitochondria induces depolarization through the inner membrane dynamin OPA1 protein. J. Biol. Chem. 289, 11862-11872. 10.1074/jbc.M113.533299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legesse-Miller, A., Massol, R. H. and Kirchhausen, T. (2003). Constriction and Dnm1p recruitment are distinct processes in mitochondrial fission. Mol. Biol. Cell 14, 1953-1963. 10.1091/mbc.e02-10-0657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, S. C., Uchiyama, L. F. and Nunnari, J. (2016). ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science 353, aaf5549. 10.1126/science.aaf5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Xu, S., Roelofs, B. A., Boyman, L., Lederer, W. J., Sesaki, H. and Karbowski, M. (2015). Transient assembly of F-actin on the outer mitochondrial membrane contributes to mitochondrial fission. J. Cell Biol. 208, 109-123. 10.1083/jcb.201404050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Fell, S. M., Surova, O., Smedler, E., Wallis, K., Chen, Z. X., Hellman, U., Johnsen, J. I., Martinsson, T., Kenchappa, R. S.et al. (2016). The 1p36 tumor suppressor KIF 1Bβ is required for calcineurin activation, controlling mitochondrial fission and apoptosis. Dev. Cell 36, 164-178. 10.1016/j.devcel.2015.12.029 [DOI] [PubMed] [Google Scholar]
- Li, F., Fan, X., Zhang, Y., Zhang, Y., Ma, X., Kou, J. and Yu, B. (2018). Inhibition of myosin IIA-actin interaction prevents ischemia/reperfusion induced cardiomyocytes apoptosis through modulating PINK1/Parkin pathway and mitochondrial fission. Int. J. Cardiol. 271, 211-218. 10.1016/j.ijcard.2018.04.079 [DOI] [PubMed] [Google Scholar]
- Li, S., Xiong, G.-J., Huang, N. and Sheng, Z.-H. (2020). The cross-talk of energy sensing and mitochondrial anchoring sustains synaptic efficacy by maintaining presynaptic metabolism. Nat. Metab. 2, 1077-1095. 10.1038/s42255-020-00289-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, M.-Y., Cheng, X.-T., Tammineni, P., Xie, Y., Zhou, B., Cai, Q. and Sheng, Z.-H. (2017). Releasing syntaphilin removes stressed mitochondria from axons independent of mitophagy under pathophysiological conditions. Neuron 94, 595-610.e6. 10.1016/j.neuron.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S., Huang, C., Gunda, V., Sun, J., Chellappan, S. P., Li, Z., Izumi, V., Fang, B., Koomen, J., Singh, P. K.et al. (2019). Fascin controls metastatic colonization and mitochondrial oxidative phosphorylation by remodeling mitochondrial actin filaments. Cell Rep. 28, 2824-2836.e8. 10.1016/j.celrep.2019.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S., Sawada, T., Lee, S., Yu, W., Silverio, G., Alapatt, P., Millan, I., Shen, A., Saxton, W., Kanao, T.et al. (2012). Parkinson's disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genet. 8, e1002537. 10.1371/journal.pgen.1002537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Domenéch, G., Covill-Cooke, C., Ivankovic, D., Halff, E. F., Sheehan, D. F., Norkett, R., Birsa, N. and Kittler, J. T. (2018). Miro proteins coordinate microtubule- and actin-dependent mitochondrial transport and distribution. EMBO J. 37, 321-336. 10.15252/embj.201696380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loss, O. and Stephenson, F. A. (2015). Localization of the kinesin adaptor proteins trafficking kinesin proteins 1 and 2 in primary cultures of hippocampal pyramidal and cortical neurons. J. Neurosci. Res. 93, 1056-1066. 10.1002/jnr.23549 [DOI] [PubMed] [Google Scholar]
- Lu, Z., Ma, X.-N., Zhang, H.-M., Ji, H.-H., Ding, H., Zhang, J., Luo, D., Sun, Y. and Li, X.-D. (2014). Mouse myosin-19 is a plus-end-directed, high-duty ratio molecular motor. J. Biol. Chem. 289, 18535-18548. 10.1074/jbc.M114.569087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons, D. A., Naylor, S. G., Mercurio, S., Dominguez, C. and Talbot, W. S. (2008). KBP is essential for axonal structure, outgrowth and maintenance in zebrafish, providing insight into the cellular basis of Goldberg-Shprintzen syndrome. Development 135, 599-608. 10.1242/dev.012377 [DOI] [PubMed] [Google Scholar]
- Lyons, D. A., Naylor, S. G., Scholze, A. and Talbot, W. S. (2009). Kif1b is essential for mRNA localization in oligodendrocytes and development of myelinated axons. Nat. Genet. 41, 854-858. 10.1038/ng.376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaskill, A. F., Rinholm, J. E., Twelvetrees, A. E., Arancibia-Carcamo, I. L., Muir, J., Fransson, A., Aspenstrom, P., Attwell, D. and Kittler, J. T. (2009). Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron 61, 541-555. 10.1016/j.neuron.2009.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, A. and Drerup, C. M. (2019). Axonal transport and mitochondrial function in neurons. Front. Cell. Neurosci. 13, 373. 10.3389/fncel.2019.00373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor, U., Bartholomew, S., Golani, G., Christenson, E., Kozlov, M., Higgs, H., Spudich, J. and Lippincott-Schwartz, J. (2015). A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. eLife 4, e08828. 10.7554/eLife.08828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters, T. A., Kendrick-Jones, J. and Buss, F. (2016). Myosins: domain organisation, motor properties, physiological roles and cellular functions. In The Actin Cytoskeleton (Handbook of Experimental Pharmacology), Vol. 235 (ed. B. Jockusch), pp. 77-122. Springer. 10.1007/164_2016_29 [DOI] [PubMed] [Google Scholar]
- Mattenberger, Y., James, D. I. and Martinou, J.-C. (2003). Fusion of mitochondria in mammalian cells is dependent on the mitochondrial inner membrane potential and independent of microtubules or actin. FEBS Lett. 538, 53-59. 10.1016/S0014-5793(03)00124-8 [DOI] [PubMed] [Google Scholar]
- McKenzie, A. J., Campbell, S. L. and Howe, A. K. (2011). Protein kinase A activity and anchoring are required for ovarian cancer cell migration and invasion. PLOS ONE 6, e26552. 10.1371/journal.pone.0026552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams, T. G., Prescott, A. R., Allen, G. F. G., Tamjar, J., Munson, M. J., Thomson, C., Muqit, M. M. K. and Ganley, I. G. (2016). mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J. Cell Biol. 214, 333-345. 10.1083/jcb.201603039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, A. D., Rock, R. S., Rief, M., Spudich, J. A., Mooseker, M. S. and Cheney, R. E. (1999). Myosin-V is a processive actin-based motor. Nature 400, 590-593. 10.1038/23072 [DOI] [PubMed] [Google Scholar]
- Melkov, A. and Abdu, U. (2018). Regulation of long-distance transport of mitochondria along microtubules. Cell. Mol. Life Sci. 75, 163-176. 10.1007/s00018-017-2590-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, K. E. and Sheetz, M. P. (2004). Axonal mitochondrial transport and potential are correlated. J. Cell Sci. 117, 2791-2804. 10.1242/jcs.01130 [DOI] [PubMed] [Google Scholar]
- Misko, A., Jiang, S., Wegorzewska, I., Milbrandt, J. and Baloh, R. H. (2010). Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. J. Neurosci. 30, 4232-4240. 10.1523/JNEUROSCI.6248-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima, N., Ohsumi, Y. and Yoshimori, T. (2002). Autophagosome formation in mammalian cells. Cell Struct. Funct. 27, 421-429. 10.1247/csf.27.421 [DOI] [PubMed] [Google Scholar]
- Modi, S., López-Doménech, G., Halff, E. F., Covill-Cooke, C., Ivankovic, D., Melandri, D., Arancibia-Cárcamo, I. L., Burden, J. J., Lowe, A. R. and Kittler, J. T. (2019). Miro clusters regulate ER-mitochondria contact sites and link cristae organization to the mitochondrial transport machinery. Nat. Commun. 10, 4399. 10.1038/s41467-019-12382-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, A. S., Wong, Y. C., Simpson, C. L. and Holzbaur, E. L. F. (2016). Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission-fusion balance within mitochondrial networks. Nat. Commun. 7, 12886. 10.1038/ncomms12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlino, G., Barreiro, O., Baixauli, F., Robles-Valero, J., Gonzalez-Granado, J. M., Villa-Bellosta, R., Cuenca, J., Sanchez-Sorzano, C. O., Veiga, E., Martin-Cofreces, N. B.et al. (2014). Miro-1 links mitochondria and microtubule Dynein motors to control lymphocyte migration and polarity. Mol. Cell Biol. 34, 1412-1426. 10.1128/MCB.01177-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima, S., Tábara, L.-C., Tilokani, L., Paupe, V., Anand, H., Pogson, J. H., Zunino, R., McBride, H. M. and Prudent, J. (2020). Golgi-derived PI(4)P-containing vesicles drive late steps of mitochondrial division. Science 367, 1366-1371. 10.1126/science.aax6089 [DOI] [PubMed] [Google Scholar]
- Nakamura, M. and Sadoshima, J. (2018). Mechanisms of physiological and pathological cardiac hypertrophy. Nat. Rev. Cardiol. 15, 387-407. 10.1038/s41569-018-0007-y [DOI] [PubMed] [Google Scholar]
- Nangaku, M., Sato-Yoshitake, R., Okada, Y., Noda, Y., Takemura, R., Yamazaki, H. and Hirokawa, N. (1994). KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell 79, 1209-1220. 10.1016/0092-8674(94)90012-4 [DOI] [PubMed] [Google Scholar]
- Narendra, D., Tanaka, A., Suen, D.-F. and Youle, R. J. (2008). Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795-803. 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeding, S. J., Majstrowicz, K., Hu, X.-P., Schwarz, V., Freitag, A., Honnert, U., Nikolaus, P. and Bähler, M. (2018). Identification of Miro1 and Miro2 as mitochondrial receptors for myosin XIX. J. Cell Sci. 131, jcs219469. 10.1242/jcs.219469 [DOI] [PubMed] [Google Scholar]
- Olenick, M. A. and Holzbaur, E. L. F. (2019). Dynein activators and adaptors at a glance. J. Cell Sci. 132, jcs227132. 10.1242/jcs.227132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osellame, L. D., Singh, A. P., Stroud, D. A., Palmer, C. S., Stojanovski, D., Ramachandran, R. and Ryan, M. T. (2016). Cooperative and independent roles of the Drp1 adaptors Mff, MiD49 and MiD51 in mitochondrial fission. J. Cell Sci. 129, 2170-2181. 10.1242/jcs.185165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak, D., Sepp, K. J. and Hollenbeck, P. J. (2010). Evidence that myosin activity opposes microtubule-based axonal transport of mitochondria. J. Neurosci. 30, 8984-8992. 10.1523/JNEUROSCI.1621-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekkurnaz, G., Trinidad, J. C., Wang, X., Kong, D. and Schwarz, T. L. (2014). Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase. Cell 158, 54-68. 10.1016/j.cell.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiz, D., Lorin, S., Leroy-Gori, I. and Poüs, C. (2017). Stress-induced hyperacetylation of microtubule enhances mitochondrial fission and modulates the phosphorylation of Drp1 at616Ser. Cell. Signal. 39, 32-43. 10.1016/j.cellsig.2017.07.020 [DOI] [PubMed] [Google Scholar]
- Pickles, S., Vigié, P. and Youle, R. J. (2018). Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28, R170-R185. 10.1016/j.cub.2018.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling, A. D., Horiuchi, D., Lively, C. M. and Saxton, W. M. (2006). Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol. Biol. Cell 17, 2057-2068. 10.1091/mbc.e05-06-0526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana, A., Schwarz, E. C., Schwindling, C., Lipp, P., Kaestner, L. and Hoth, M. (2006). Sustained activity of calcium release-activated calcium channels requires translocation of mitochondria to the plasma membrane. J. Biol. Chem. 281, 40302-40309. 10.1074/jbc.M607896200 [DOI] [PubMed] [Google Scholar]
- Quintana, A., Schwindling, C., Wenning, A. S., Becherer, U., Rettig, J., Schwarz, E. C. and Hoth, M. (2007). T cell activation requires mitochondrial translocation to the immunological synapse. Proc. Natl. Acad. Sci. USA 104, 14418-14423. 10.1073/pnas.0703126104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero, O. A., DiVito, M. M., Adikes, R. C., Kortan, M. B., Case, L. B., Lier, A. J., Panaretos, N. S., Slater, S. Q., Rengarajan, M., Feliu, M.et al. (2009). Human Myo19 is a novel myosin that associates with mitochondria. Curr. Biol. 19, 2008-2013. 10.1016/j.cub.2009.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju, V., Lewis, T. L., Jr., Hirabayashi, Y., Bergami, M., Motori, E., Cartoni, R., Kwon, S.-K. and Courchet, J. (2019). Pleiotropic mitochondria: the influence of mitochondria on neuronal development and disease. J. Neurosci. 39, 8200-8208. 10.1523/JNEUROSCI.1157-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson, S. L., Redwine, W. B., Vale, R. D. and Carter, A. P. (2018). The cytoplasmic dynein transport machinery and its many cargoes. Nat. Rev. Mol. Cell Biol. 19, 382-398. 10.1038/s41580-018-0004-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, N. A., Cai, D., Blasius, T. L., Jih, G. T., Meyhofer, E., Gaertig, J. and Verhey, K. J. (2006). Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 16, 2166-2172. 10.1016/j.cub.2006.09.014 [DOI] [PubMed] [Google Scholar]
- Rohn, J. L., Patel, J. V., Neumann, B., Bulkescher, J., McHedlishvili, N., McMullan, R. C., Quintero, O. A., Ellenberg, J. and Baum, B. (2014). Myo19 ensures symmetric partitioning of mitochondria and coupling of mitochondrial segregation to cell division. Curr. Biol. 24, 2598-2605. 10.1016/j.cub.2014.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner, K., Faix, J., Bogdan, S., Linder, S. and Kerkhoff, E. (2017). Actin assembly mechanisms at a glance. J. Cell Sci. 130, 3427-3435. 10.1242/jcs.206433 [DOI] [PubMed] [Google Scholar]
- Rustom, A., Saffrich, R., Markovic, I., Walther, P. and Gerdes, H.-H. (2004). Nanotubular highways for intercellular organelle transport. Science 303, 1007-1010. 10.1126/science.1093133 [DOI] [PubMed] [Google Scholar]
- Ryan, B. J., Hoek, S., Fon, E. A. and Wade-Martins, R. (2015). Mitochondrial dysfunction and mitophagy in Parkinson's: from familial to sporadic disease. Trends Biochem. Sci. 40, 200-210. 10.1016/j.tibs.2015.02.003 [DOI] [PubMed] [Google Scholar]
- Safiulina, D., Kuum, M., Choubey, V., Gogichaishvili, N., Liiv, J., Hickey, M. A., Cagalinec, M., Mandel, M., Zeb, A., Liiv, M.et al. (2019). Miro proteins prime mitochondria for Parkin translocation and mitophagy. EMBO J. 38, e99384. 10.15252/embj.201899384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Madrid, F. and del Pozo, M. A. (1999). Leukocyte polarization in cell migration and immune interactions. EMBO J. 18, 501-511. 10.1093/emboj/18.3.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saotome, M., Safiulina, D., Szabadkai, G., Das, S., Fransson, A., Aspenstrom, P., Rizzuto, R. and Hajnoczky, G. (2008). Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc. Natl. Acad. Sci. USA 105, 20728-20733. 10.1073/pnas.0808953105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf, S. A., Raman, M., Guarani-Pereira, V., Sowa, M. E., Huttlin, E. L., Gygi, S. P. and Harper, J. W. (2013). Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, 372-376. 10.1038/nature12043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, D., Sato, D., Tsuyama, T., Saito, M., Ohkura, H., Rolls, M. M., Ishikawa, F. and Uemura, T. (2008). Spatial control of branching within dendritic arbors by dynein-dependent transport of Rab5-endosomes. Nat. Cell Biol. 10, 1164-1171. 10.1038/ncb1776 [DOI] [PubMed] [Google Scholar]
- Schaper, J., Froede, R., Hein, S., Buck, A., Hashizume, H., Speiser, B., Friedl, A. and Bleese, N. (1991). Impairment of the myocardial ultrastructure and changes of the cytoskeleton in dilated cardiomyopathy. Circulation 83, 504-514. 10.1161/01.CIR.83.2.504 [DOI] [PubMed] [Google Scholar]
- Schuler, M.-H., Lewandowska, A., Caprio, G. D., Skillern, W., Upadhyayula, S., Kirchhausen, T., Shaw, J. M. and Cunniff, B. (2017). Miro1-mediated mitochondrial positioning shapes intracellular energy gradients required for cell migration. Mol. Biol. Cell 28, 2159-2169. 10.1091/mbc.e16-10-0741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, T. L. (2013). Mitochondrial trafficking in neurons. Cold Spring Harb. Perspect. Biol. 5, a011304. 10.1101/cshperspect.a011304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seager, R., Lee, L., Henley, J. M. and Wilkinson, K. A. (2020). Mechanisms and roles of mitochondrial localisation and dynamics in neuronal function. Neuronal Signal. 4, NS20200008. 10.1042/NS20200008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, A. Y., Joseph, A.-M., Dutta, D., Hwang, J. C. Y., Aris, J. P. and Leeuwenburgh, C. (2010). New insights into the role of mitochondria in aging: mitochondrial dynamics and more. J. Cell Sci. 123, 2533-2542. 10.1242/jcs.070490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, J., Zhang, J.-H., Xiao, H., Wu, J.-M., He, K.-M., Lv, Z.-Z., Li, Z.-J., Xu, M. and Zhang, Y.-Y. (2018). Mitochondria are transported along microtubules in membrane nanotubes to rescue distressed cardiomyocytes from apoptosis. Cell Death Dis. 9, 81. 10.1038/s41419-017-0145-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlevkov, E., Kramer, T., Schapansky, J., LaVoie, M. J. and Schwarz, T. L. (2016). Miro phosphorylation sites regulate Parkin recruitment and mitochondrial motility. Proc. Natl. Acad. Sci. USA 113, E6097-E6106. 10.1073/pnas.1612283113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shneyer, B. I., Ušaj, M. and Henn, A. (2016). Myo19 is an outer mitochondrial membrane motor and effector of starvation-induced filopodia. J. Cell Sci. 129, 543-556. 10.1242/jcs.175349 [DOI] [PubMed] [Google Scholar]
- Shneyer, B. I., Ušaj, M., Wiesel-Motiuk, N., Regev, R. and Henn, A. (2017). ROS induced distribution of mitochondria to filopodia by Myo19 depends on a class specific tryptophan in the motor domain. Sci. Rep. 7, 11577. 10.1038/s41598-017-11002-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleigh, J. N., Rossor, A. M., Fellows, A. D., Tosolini, A. P. and Schiavo, G. (2019). Axonal transport and neurological disease. Nat. Rev. Neurol. 15, 691-703. 10.1038/s41582-019-0257-2 [DOI] [PubMed] [Google Scholar]
- Smith, M. G., Simon, V. R., O'Sullivan, H. and Pon, L. A. (1995). Organelle-cytoskeletal interactions: actin mutations inhibit meiosis-dependent mitochondrial rearrangement in the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 6, 1381-1396. 10.1091/mbc.6.10.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers, R. S., Megeath, L. J., Górska-Andrzejak, J., Meinertzhagen, I. A. and Schwarz, T. L. (2002). Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron 36, 1063-1077. 10.1016/S0896-6273(02)01094-2 [DOI] [PubMed] [Google Scholar]
- Straub, F., Welz, T., Alberico, H., Brandão, R. O., Huber, A., Samol-Wolf, A., Brakebusch, C., Woods, D., Kollmar, M., Martin-Gonzalez, J.et al. (2020). The SPIRE1 actin nucleator coordinates actin/myosin functions in the regulation of mitochondrial motility. bioRxiv, 2020.06.19.161109. [Google Scholar]
- Sung, H., Tandarich, L. C., Nguyen, K. and Hollenbeck, P. J. (2016). Compartmentalized regulation of parkin-mediated mitochondrial quality control in the Drosophila nervous system in vivo. J. Neurosci. 36, 7375-7391. 10.1523/JNEUROSCI.0633-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, Y., Kanai, Y., Okada, Y., Nonaka, S., Takeda, S., Harada, A. and Hirokawa, N. (1998). Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell 93, 1147-1158. 10.1016/S0092-8674(00)81459-2 [DOI] [PubMed] [Google Scholar]
- Tanaka, K., Sugiura, Y., Ichishita, R., Mihara, K. and Oka, T. (2011). KLP6: a newly identified kinesin that regulates the morphology and transport of mitochondria in neuronal cells. J. Cell Sci. 124, 2457-2465. 10.1242/jcs.086470 [DOI] [PubMed] [Google Scholar]
- Tang, D., Kang, R., Livesey, K. M., Kroemer, G., Billiar, T. R., Van Houten, B., Zeh, H. J., III and Lotze, M. T. (2011). High-mobility group box 1 is essential for mitochondrial quality control. Cell Metab. 13, 701-711. 10.1016/j.cmet.2011.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, E. W. and Borisy, G. G. (2000). Kinesin processivity. J. Cell Biol. 151, F27-F29. 10.1083/jcb.151.5.F27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigchelaar, W., de Jong, A. M., Bloks, V. W., van Gilst, W. H., de Boer, R. A. and Silljé, H. H. W. (2016). Hypertrophy induced KIF5B controls mitochondrial localization and function in neonatal rat cardiomyocytes. J. Mol. Cell. Cardiol. 97, 70-81. 10.1016/j.yjmcc.2016.04.005 [DOI] [PubMed] [Google Scholar]
- Tilokani, L., Nagashima, S., Paupe, V. and Prudent, J. (2018). Mitochondrial dynamics: overview of molecular mechanisms. Essays Biochem. 62, 341-360. 10.1042/EBC20170104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, P.-I., Course, M. M., Lovas, J. R., Hsieh, C.-H., Babic, M., Zinsmaier, K. E. and Wang, X. (2014). PINK1-mediated phosphorylation of Miro inhibits synaptic growth and protects dopaminergic neurons in Drosophila. Sci. Rep. 4, 6962. 10.1038/srep06962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twelvetrees, A. E., Pernigo, S., Sanger, A., Guedes-Dias, P., Schiavo, G., Steiner, R. A., Dodding, M. P. and Holzbaur, E. L. F. (2016). The dynamic localization of cytoplasmic dynein in neurons is driven by Kinesin-1. Neuron 90, 1000-1015. 10.1016/j.neuron.2016.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ušaj, M. and Henn, A. (2017). Kinetic adaptation of human Myo19 for active mitochondrial transport to growing filopodia tips. Sci. Rep. 7, 11596. 10.1038/s41598-017-11984-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale, R. D. and Milligan, R. A. (2000). The way things move: looking under the hood of molecular motor proteins. Science 288, 88-95. 10.1126/science.288.5463.88 [DOI] [PubMed] [Google Scholar]
- van Spronsen, M., Mikhaylova, M., Lipka, J., Schlager, M. A., van den Heuvel, D. J., Kuijpers, M., Wulf, P. S., Keijzer, N., Demmers, J., Kapitein, L. C.et al. (2013). TRAK/Milton motor-adaptor proteins steer mitochondrial trafficking to axons and dendrites. Neuron 77, 485-502. 10.1016/j.neuron.2012.11.027 [DOI] [PubMed] [Google Scholar]
- Varadi, A., Johnson-Cadwell, L. I., Cirulli, V., Yoon, Y., Allan, V. J. and Rutter, G. A. (2004). Cytoplasmic dynein regulates the subcellular distribution of mitochondria by controlling the recruitment of the fission factor dynamin-related protein-1. J. Cell Sci. 117, 4389-4400. 10.1242/jcs.01299 [DOI] [PubMed] [Google Scholar]
- Varuzhanyan, G. and Chan, D. C. (2020). Mitochondrial dynamics during spermatogenesis. J. Cell Sci. 133, jcs235937. 10.1242/jcs.235937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey, K. J. and Hammond, J. W. (2009). Traffic control: regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 10, 765-777. 10.1038/nrm2782 [DOI] [PubMed] [Google Scholar]
- Wang, X. and Schwarz, T. L. (2009). The mechanism of Ca2+ -dependent regulation of kinesin-mediated mitochondrial motility. Cell 136, 163-174. 10.1016/j.cell.2008.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Winter, D., Ashrafi, G., Schlehe, J., Wong, Y. L., Selkoe, D., Rice, S., Steen, J., LaVoie, M. J. and Schwarz, T. L. (2011). PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147, 893-906. 10.1016/j.cell.2011.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Du, W., Su, Q. P., Zhu, M., Feng, P., Li, Y., Zhou, Y., Mi, N., Zhu, Y., Jiang, D.et al. (2015). Dynamic tubulation of mitochondria drives mitochondrial network formation. Cell Res. 25, 1108-1120. 10.1038/cr.2015.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Gao, X., Zheng, X., Hou, C., Xie, Q., Lou, B. and Zhu, J. (2019). Expression and potential functions of KIF3A/3B to promote nuclear reshaping and tail formation during Larimichthys polyactis spermiogenesis. Dev. Genes Evol. 229, 161-181. 10.1007/s00427-019-00637-5 [DOI] [PubMed] [Google Scholar]
- Weihofen, A., Thomas, K. J., Ostaszewski, B. L., Cookson, M. R. and Selkoe, D. J. (2009). Pink1 forms a multiprotein complex with Miro and Milton, linking Pink1 function to mitochondrial trafficking. Biochemistry 48, 2045-2052. 10.1021/bi8019178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, A. L., Lin, A. W., Chen, L.-Q., Safer, D., Cain, S. M., Hasson, T., Carragher, B. O., Milligan, R. A. and Sweeney, H. L. (1999). Myosin VI is an actin-based motor that moves backwards. Nature 401, 505-508. 10.1038/46835 [DOI] [PubMed] [Google Scholar]
- Wong, Y. C., Ysselstein, D. and Krainc, D. (2018). Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 554, 382-386. 10.1038/nature25486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak, M. J., Melzer, M., Dorner, C., Haring, H.-U. and Lammers, R. (2005). The novel protein KBP regulates mitochondria localization by interaction with a kinesin-like protein. BMC Cell Biol. 6, 35. 10.1186/1471-2121-6-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, J., Chao, D. L., Toba, S., Koyasako, K., Yasunaga, T., Hirotsune, S. and Shen, K. (2013). Kinesin-1 regulates dendrite microtubule polarity in Caenorhabditis elegans. eLife 2, e00133. 10.7554/eLife.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C. and Svitkina, T. M. (2019). Ultrastructure and dynamics of the actin-myosin II cytoskeleton during mitochondrial fission. Nat. Cell Biol. 21, 603-613. 10.1038/s41556-019-0313-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, M., Weaver, D. and Hajnóczky, G. (2004). Control of mitochondrial motility and distribution by the calcium signal: a homeostatic circuit. J. Cell Biol. 167, 661-672. 10.1083/jcb.200406038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. and Zhang, Y. (2013). Membrane nanotubes: novel communication between distant cells. Sci. China Life Sci. 56, 994-999. 10.1007/s11427-013-4548-3 [DOI] [PubMed] [Google Scholar]
- Zhao, C., Takita, J., Tanaka, Y., Setou, M., Nakagawa, T., Takeda, S., Yang, H. W., Terada, S., Nakata, T., Takei, Y.et al. (2001). Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bbeta. Cell 105, 587-597. 10.1016/s0092-8674(01)00363-4 [DOI] [PubMed] [Google Scholar]
- Zhao, Y.-Q., Mu, D.-L., Wang, D., Han, Y.-L., Hou, C.-C. and Zhu, J.-Q. (2018). Analysis of the function of KIF3A and KIF3B in the spermatogenesis in Boleophthalmus pectinirostris. Fish Physiol. Biochem. 44, 769-788. 10.1007/s10695-017-0461-1 [DOI] [PubMed] [Google Scholar]
- Zheng, Y., Zhang, X., Wu, X., Jiang, L., Ahsan, A., Ma, S., Xiao, Z., Han, F., Qin, Z.-H., Hu, W.et al. (2019). Somatic autophagy of axonal mitochondria in ischemic neurons. J. Cell Biol. 218, 1891-1907. 10.1083/jcb.201804101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.