ABSTRACT
Niemann–Pick disease type C (NPC) is a rare, fatal, neurodegenerative lysosomal disease caused by mutations of either NPC1 or NPC2. NPC2 is a soluble lysosomal protein that functions in coordination with NPC1 to efflux cholesterol from the lysosomal compartment. Mutations of either gene result in the accumulation of unesterified cholesterol and other lipids in the late endosome/lysosome, and reduction of cellular cholesterol bioavailability. Zygotic null npc2m/m zebrafish showed significant unesterified cholesterol accumulation at larval stages, a reduction in body size, and motor and balance defects in adulthood. However, the phenotype at embryonic stages was milder than expected, suggesting a possible role of maternal Npc2 in embryonic development. Maternal-zygotic npc2m/m zebrafish exhibited significant developmental defects, including defective otic vesicle development/absent otoliths, abnormal head/brain development, curved/twisted body axes and no circulating blood cells, and died by 72 hpf. RNA-seq analysis conducted on 30 hpf npc2+/m and MZnpc2m/m embryos revealed a significant reduction in the expression of notch3 and other downstream genes in the Notch signaling pathway, suggesting that impaired Notch3 signaling underlies aspects of the developmental defects observed in MZnpc2m/m zebrafish.
KEY WORDS: Zebrafish, Niemann–Pick type C, Niemann–Pick type C2, Npc2, Notch3, Cholesterol
Summary: Generation of maternal zygotic npc2 mutant zebrafish to uncover the role of Npc2 and intracellular cholesterol trafficking during embryonic development.
INTRODUCTION
Intracellular cholesterol trafficking starts with low-density lipoproteins (LDL) entering cells via receptor-mediated endocytosis (Brown and Goldstein, 1986). These LDL-containing endosomes later fuse with lysosomes to enable further processing of LDL. Once inside the lysosomal lumen, cholesterol-ester molecules are released from LDL de-esterified by cholesterol ester acid lipase and transferred to NPC2, a soluble lysosomal luminal protein. The cholesterol is then transferred from NPC2 to NPC1, an endolysosomal transmembrane protein. NPC1 functions to transport cholesterol from the endolysosomal lumen to the external limiting membrane from where it is transported to other subcellular destinations, such as endoplasmic reticulum and mitochondria, for utilization (Cruz et al., 2000; Kwon et al., 2009; Pfeffer, 2019).
Dysfunctional NPC1 or NPC2 protein leads to accumulation of unesterified cholesterol and glycolipids in late endosomes/lysosomes, causing a severe, lethal lysosomal storage disease called Niemann–Pick disease, type C. About 95% of NPC patients have mutations in NPC1 and the remaining cases are associated with mutations in NPC2 (Vanier, 2010). NPC patients exhibit a variety of symptoms, including neonatal jaundice, hepatosplenomegaly, ataxia, tremor, seizures and learning difficulties (Patterson et al., 2013; Vanier, 2010). Infant NPC patients often manifest hepatic dysfunction ranging from prolonged neonatal jaundice to severe cirrhosis and liver failure. Many NPC2 patients have respiratory distress during early infancy (Griese et al., 2010). The onset of neurological manifestations of NPC is variable and ranges from early-infantile to adolescent/adult onset (Mengel et al., 2017). Currently, there are no FDA-approved therapeutics for treatments for NPC. However, intrathecal administration of adrabetadex (VTS-270, Kleptose HPB), a specific 2-hydroxypropyl-β-cyclodextrin (2HPβCD), has been shown to be effective in slowing neurological disease progression in NPC1 patients (Ory et al., 2017), and miglustat, a glycosphingolipid synthesis inhibitor, has been shown to increase survival (Patterson et al., 2020).
Cholesterol biosynthesis, uptake and transport play important roles in embryonic development, including the modification and regulation of proteins involved in Hedgehog signaling (Deshpande et al., 2019; Farese and Herz, 1998; Kinnebrew et al., 2019; Lewis et al., 2001; Xiao et al., 2017). Loss of function in enzymes participating in cholesterol synthesis often results in fetal malformations or even embryonic lethality (Horvat et al., 2011). Cholesterol is also the precursor of steroid hormones, such as glucocorticoids, mineralocorticoids and sex hormones, which also play important roles in embryonic development (Hsu et al., 2006; Miller, 1988). Pregnenolone, a steroid product of cholesterol, has been shown to stabilize microtubules and promote epiboly movement during zebrafish embryonic development (Hsu et al., 2006). Furthermore, pregnenolone was able to rescue the epiboly movement delay in zebrafish embryos injected with morpholino antisense oligonucleotides against npc1 (Schwend et al., 2011), highlighting the diverse roles that cholesterol metabolism might play during the embryonic development.
Previously, we reported development of a genetic zebrafish NPC1 model using CRISPR/Cas9-mediated gene targeting. npc1 null mutant zebrafish manifested both peripheral and central nervous system defects consistent with the clinical issues observed in NPC1 patients, suggesting that the consequences of impaired intracellular cholesterol transport are similar between zebrafish and mammals (Tseng et al., 2018). This prior study supports the use of zebrafish as a model organism in which to study NPC and intracellular cholesterol trafficking. Mice and fruit flies lacking functional NPC2 have been shown to exhibit phenotypes resembling those identified in NPC1 patients and animal models (Huang et al., 2007; Sleat et al., 2004), indicating that both NPC1 and NPC2 act in a common biological pathway. Although mouse, cat, fruit fly and zebrafish models of NPC1 (Huang et al., 2005; Lin et al., 2018; Loftus et al., 1997; Maue et al., 2012; Muñana et al., 1994; Tseng et al., 2018) and mouse and fruit fly models of NPC2 (Huang et al., 2007; Sleat et al., 2004) have been developed, little is known regarding the role of either NPC1 or NPC2 during embryonic development.
Zebrafish are an ideal model organism for studying embryonic development owing to the transparency of embryos, easy breeding, and rapid embryonic development (Driever et al., 1994). However, even though cholesterol plays a major role in embryonic development, zygotic npc1 null mutant zebrafish displayed only very mild abnormalities during early embryonic development. This may be due to the presence of maternal npc1 transcripts that compensate for the lack of functional npc1 embryonic transcripts. Generation of maternal-zygotic (MZ)npc1 mutants was not previously undertaken owing to the unavailability of sexually mature zygotic npc1m/m adult females (Tseng et al., 2018). For this study, we generated zebrafish npc2 null mutants using CRISPR/Cas9-mediated gene targeting. Zygotic npc2 mutant zebrafish exhibited similar defects to those observed in npc1 mutant zebrafish (Lin et al., 2018; Tseng et al., 2018). In contrast to our NPC1 zebrafish model, we were able to obtain sexually mature female zygotic npc2m/m zebrafish and subsequently MZnpc2m/m embryos. MZnpc2 mutants displayed severe embryonic defects, including defective otic vesicle development/lack of otoliths, abnormal head/brain development, absent circulating red blood cells and a curved/twisted body axis. These findings suggest that Npc2 plays a crucial role during early embryonic development in zebrafish. In this paper, we show that the dysmorphic features in MZnpc2 mutants are due to decreased cholesterol bioavailability and appear to be at least partially mediated through the downregulation of Notch3 signaling. Our findings reveal a novel mechanism through which low cholesterol bioavailability negatively regulates embryonic development in zebrafish.
RESULTS
Generation of npc2 null mutant zebrafish
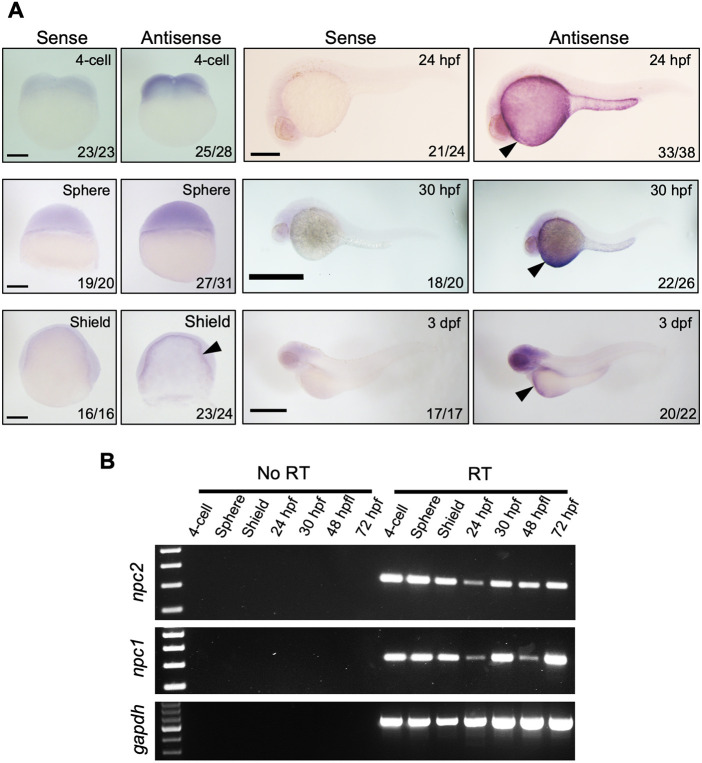
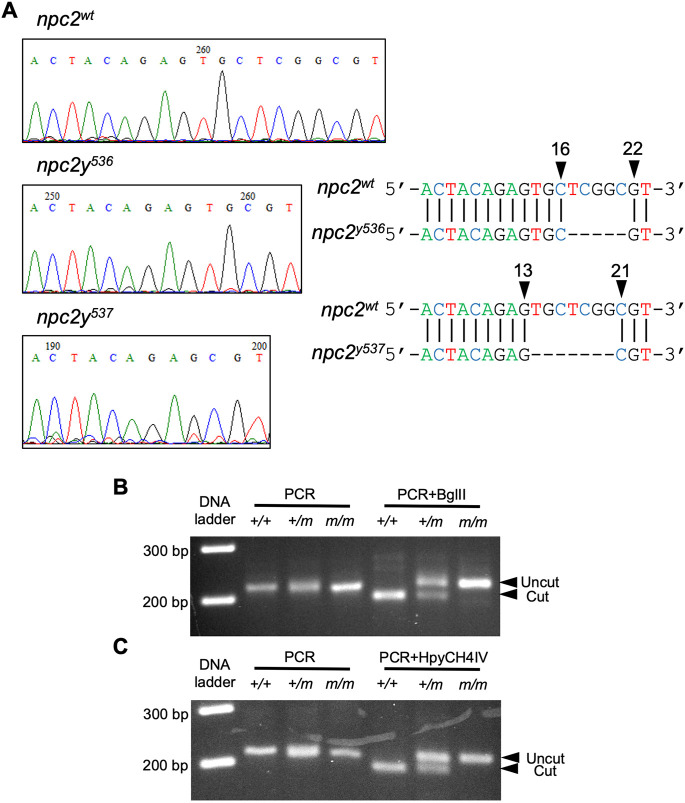
Zebrafish have a single npc2 gene (Ensembl Gene ID: ENSDARG00000090912) located on chromosome 17. The npc2 gene consists of five exons that encode Npc2, a 149 amino acid lysosomal protein. Expression of zebrafish npc2 mRNA was detected as early as the 4-cell stage (Fig. 1A), consistent with maternal expression. npc2 expression is ubiquitous prior to the sphere stage, but after initiation of gastrulation, npc2 expression becomes more prominent in the yolk syncytial layer (Fig. 1A). The expression pattern of npc2 resembles the expression of npc1 (Fig. 1B) (Schwend et al., 2011). This is consistent with the functional interaction of Npc1 and Npc2 to transport cholesterol out of the endolysosomal compartment (Sleat et al., 2004). To understand the function of npc2 in zebrafish, npc2 mutants were generated using CRISPR/Cas9-mediated gene targeting. A single site in exon 2 was chosen as the single-guide RNA (sgRNA) target to induce a DNA double-strand break and error-prone DNA damage repair. Wild-type zebrafish (F0) were injected with a combination of npc2-specific sgRNA and cas9 mRNA at the 1-cell stage to induce mutations in germline cells. Two npc2 mutant alleles harboring frame-shift mutations were identified in the F1 generation. The two mutations are npc2y536 (NM_173224.1: c.17_21del; p.(Leu6Argfs*13) and npc2y537 (NM_173224.1: c.14_20del; p.(Val5Alafs*16) (Fig. 2A). These mutations predict very short, frameshifted and truncated Npc2 peptides. Because we were not able to demonstrate absence of Npc2 protein due to lack of antibodies against zebrafish Npc2 protein, we sequenced npc2 cDNA obtained from livers of three 8 months post-fertilization (mpf) npc2m/m (npc2y537 allele) zebrafish. All sequenced npc2 transcripts contained the expected mutation. These data suggest that the npc2 mutation does not result in alternative splicing that skips over the mutation (Figs S1-S3). PCR fragment length polymorphisms can be used to genotype both npc2y536 and npc2y537 (Fig. 2B,C). The homozygous mutant phenotype was similar for both alleles. The npc2y537 allele was used for subsequent data reported in this paper.
Fig. 1.
RNA expression of npc2 in zebrafish embryos and larvae. (A) mRNA expression of npc2 was characterized by in situ hybridization. Expression was ubiquitous at the 4-cell and sphere stage and became more intense in the yolk syncytial layer (arrowheads) to up to 3 dpf. Number of embryos showing representative staining over total embryos examined are indicated in each image. Scale bars: 200 μm (4-cell, Sphere and Shield); 500 μm (24 hpf, 30 hpf and 3 dpf). (B) RT-PCR assessment of npc2 and npc1 mRNA expression from the 4-cell stage to 72 hpf. gapdh was used as an internal control.
Fig. 2.
Generation of npc2 mutant alleles in zebrafish. (A) Sequencing chromatographs of control npc2, npc2y536 and npc2y537 alleles. Arrowheads indicate the area of the induced deletion and nucleotide positions corresponding to the wild-type npc2 gene sequence. (B) Genotyping of npc2y536 was carried out by using a derived cleaved amplified polymorphic sequence (dCAPS) primer to introduce an artificial BglII restriction enzyme site in the PCR product amplified from the wild-type npc2 allele, followed by BglII digest. The PCR product from control but not npc2y536 was expected to be cut by BglII. (C) A dCAPS primer was used to introduce an artificial HpyCH4IV restriction enzyme site in the PCR product amplified from the control npc2 allele, followed by HpyCH4IV digest. The PCR product from control but not npc2y537 was expected to be cut by HpyCH4IV.
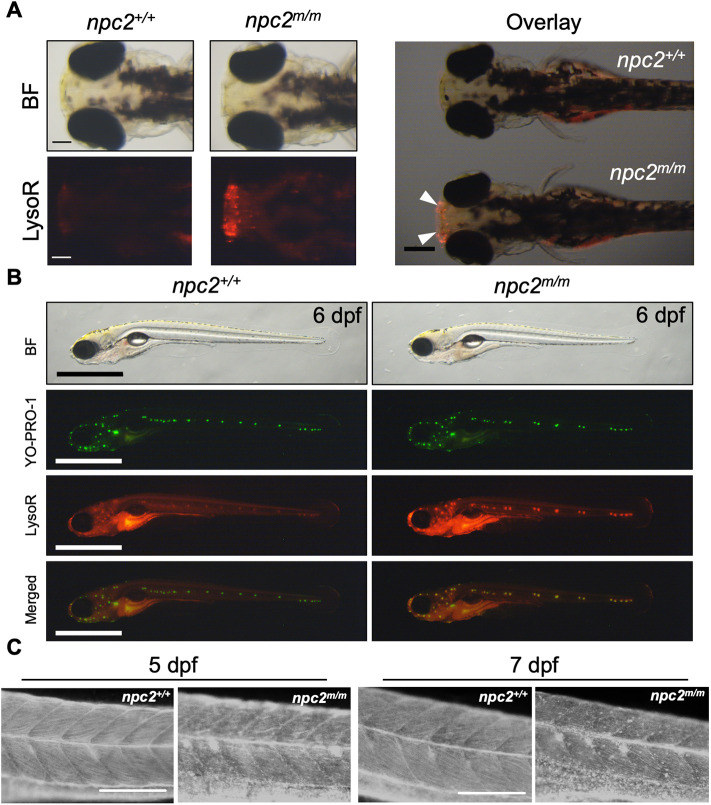
Expansion of the acidic endolysosomal compartment and storage of unesterified cholesterol are the biochemical hallmarks of both NPC1 and NPC2. Similar to what we reported with npc1 zygotic mutant zebrafish (Tseng et al., 2018), npc2m/m larvae showed intense LysoTracker Red staining of the olfactory placode at 3 days post-fertilization (dpf) in comparison with control npc2+/+ larvae (Fig. 3A). LysoTracker Red is a vital stain than labels acidic vesicles (Sugimoto et al., 2001; te Vruchte et al., 2014). Again, like npc1m/m larvae, 6 dpf npc2m/m larvae showed intense LysoTracker Red staining of lateral line neuromast cells (Fig. 3B). Although clearly showing expansion of the lysosomal compartment, the number of lateral line neuromast cells staining with YO-PRO-1 was similar in npc2+/+ and npc2m/m larvae (Fig. 3B, Fig. S4). Filipin is a fluorescent antibiotic that binds to unesterified cholesterol (Cruz et al., 2000). Again, like npc1m/m larvae, we observed increased punctate filipin staining in npc2m/m larvae at 5 and 7 dpf (Fig. 3C). This phenotypic convergence with npc1m/m and npc2m/m zebrafish is consistent with both the coordinated function of these two proteins and the expected NPC lysosomal storage phenotype.
Fig. 3.
npc2m/m larvae displayed intense LysoTracker Red and filipin staining. (A) Live 3 dpf npc2+/+ and npc2m/m larvae were stained with LysoTracker Red (LysoR) for 1 h and destained in egg water for 2 h before images were taken. npc2m/m larva showed intense LysoTracker Red staining of the olfactory placodes (arrowheads). A total of 47 npc2+/+ and 59 npc2m/m larvae were examined in three experiments. All examined larvae displayed the same staining patterns as shown here. Scale bars: 100 μm (individual); 400 μm (overlay). (B) Live double staining of npc2+/+ and npc2m/m larvae with YO-PRO-1 (neuromast cell marker) and LysoTracker Red. npc2m/m larva exhibited intense LysoTracker Red staining along lateral line neuromasts but the overall pattern of neuromast organization was unaffected. A total of 67 npc2+/+ and 75 npc2m/m larvae were examined from three experiments. All larvae displayed the same staining. Scale bars: 1 mm. (C) Filipin-positive puncta were present in 5 and 7 dpf npc2m/m larvae, indicating unesterified cholesterol accumulation. A total of 26 npc2+/+ and 19 npc2m/m 5 dpf larvae and 23 npc2+/+ and 28 npc2m/m 7 dpf larvae were examined in three experiments. All examined larvae displayed the same staining patterns. Scale bars: 200 μm.
Most npc2m/m larvae exhibited a relatively normal gross phenotype at 6 dpf; however, a variable, but small, percentage showed defects in swim bladder inflation and dorsal body curvature (Fig. 4A). Although of normal appearance at 6 dpf, very few npc2m/m fish survived to adulthood when reared with npc2+/+ and npc2+/m siblings. We hypothesized that they were not able to compete sufficiently with siblings for food after introduction to a rotifer diet. Consistent with this, if we sorted the larvae based upon LysoTracker Red-positive staining of the olfactory placode at 3 dpf and separated the npc2m/m larvae from unaffected siblings, most of the npc2m/m zebrafish survived to adulthood. The 10 weeks post-fertilization (10 wpf) adult npc2m/m zebrafish were on average significantly (P<0.001) shorter than either npc2+/+ or npc2+/m zebrafish (Fig. 4B,C). The skeletal structure appeared normal, except for a small number of npc2m/m fish that developed scoliosis (Fig. 4B, Fig. S5). Owing to the difficulty of obtaining accurate length measurements, fish with scoliosis were excluded from the length data. npc2m/m adults survived up to 8 months and died shortly after displaying signs consistent with a neurological defect (Movies 1 and 2). This neurological phenotype was similar to what we observed previously in npc1m/m zebrafish (Tseng et al., 2018) and included inability to maintain an upright swimming posture and vigorous spinning movements.
Fig. 4.
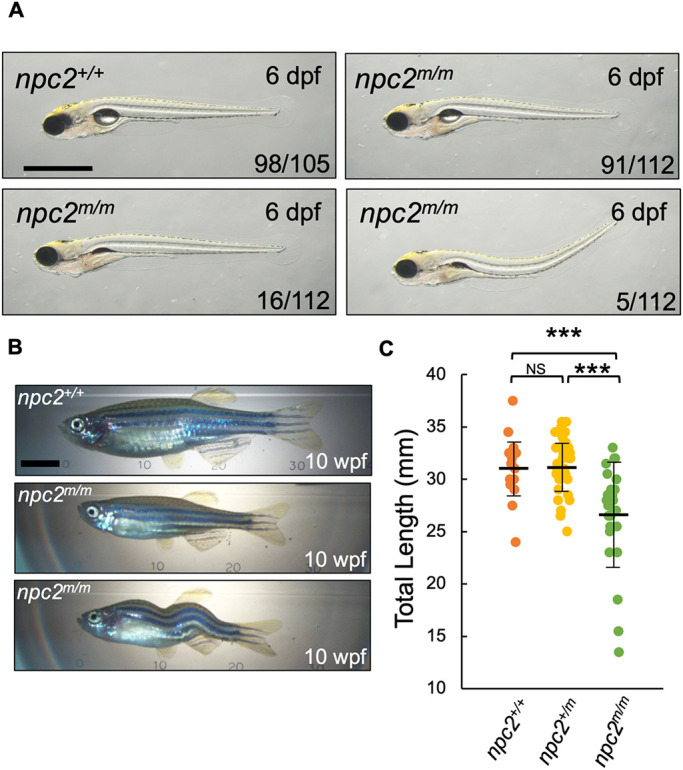
Phenotype of zygotic npc2m/m zebrafish. (A) npc2m/m larvae are mostly identical to npc2+/+ at 6 dpf; however, a few npc2m/m larvae lacked inflated swim bladders or had dorsally curved body axis. A total of 105 npc2+/+ and 112 npc2m/m from five crosses were examined. Numbers of larvae showing representing phenotypes are indicated in each image. Scale bar: 1 mm. (B) Live images of 10 wpf npc2m/m adults showed relatively normal morphology but smaller body size compared with npc2+/+ individuals. Occasionally, npc2m/m adults exhibited scoliosis. Scale bar: 5 mm. (C) Total length of npc2+/+ (31.16±3.01 mm), npc2+/m (31.34±2.68 mm) and npc2m/m (26.62±4.76 mm) adults at 10 wpf. A total of 16 npc2+/+, 32 npc2+/m and 26 npc2m/m adults were examined from two independent crosses. ***P<0.001 (one-way ANOVA and Tukey's test). Total length is presented as mean±s.d. NS, not significant.
MZnpc2m/m embryos displayed severe developmental defects
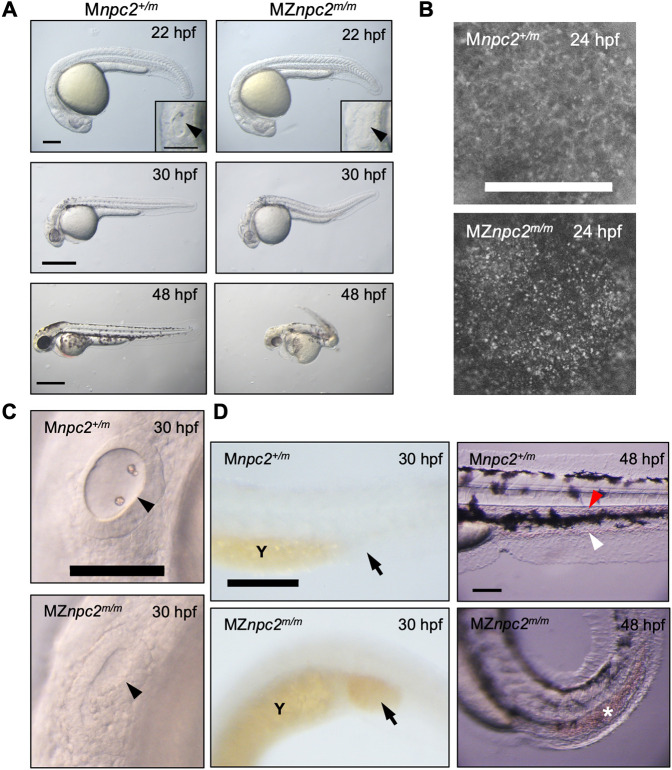
The mild phenotype observed in npc2m/m zygotic larvae could be due to the maternal expression of npc2 (Fig. 1A). Thus, to determine whether Npc2 plays a functional role in early embryonic development of zebrafish, the maternal npc2 transcripts must be depleted. Whereas we were not able to generate sexually mature male npc2m/m fish, we were able to obtain, at a low frequency, fertile female npc2m/m fish. We crossed adult zygotic npc2m/m females with npc2+/m male fish to obtain MZnpc2m/m and maternal (M) npc2+/m embryos. MZnpc2m/m embryos contain no functional npc2 gene product, whereas Mnpc2+/m retain zygotic expression of functional npc2 RNA but no functional maternal npc2 gene product. Mnpc2+/m fish appear normal, suggesting that the zygotic npc2 transcript is sufficient to support normal development. MZnpc2m/m embryos developed normally until 22 hours post-fertilization (hpf). After 22 hpf, we noted an absence of otoliths and by 30 hpf additional developmental abnormalities had arisen, including abnormal head/brain development with characteristic hindbrain reduction, a lack of circulating red blood cells and a curved/twisted body axis (Fig. 5A,C,D, Movies 3 and 4). The abnormal head/brain development with reduced hindbrain area at 30 hpf became more evident at 48 hpf when an enlarged, hollow compartment could be found in the brain (Fig. 5A). The curved/twisted body axis phenotype was observed at 30 hpf when tails randomly curved dorsally, ventrally, left or right and became more severe at 48 hpf when embryos exhibited a twisted body axis with tail pointing toward left or right, and the angle of head and tail was <90° (Fig. 5A). This MZ phenotype was observed for both npc2y536 and npc2y537 alleles (Fig. 5, Fig. S6). MZnpc2m/m embryos not only exhibited a lack of otoliths but the otic vesicle was also smaller and less organized at 30 hpf (Fig. 5C). By electron microscopy, the cells around the lumen of MZnpc2m/m otic vesicles appeared thin with decreased cytoplasm, suggesting that the overall structure of the otic vesicle was abnormal (Fig. S7). The defective otolith formation is likely a result of defective otic vesicle development. These two defects were always associated with each other in our study; thus, we used the lack of otolith phenotype as a surrogate for defective otic vesicle development. Although no circulating blood cells were found in the caudal artery and vein of MZnpc2m/m embryos at 30 hpf (Movies 3 and 4), red blood cells could be seen accumulated in the posterior blood island (Fig. 5D). This phenotype continued to be present at 48 hpf, suggesting that this phenotype was not just due to developmental delay (Fig. 5D). The development of the central nervous system (CNS) in MZnpc2m/m embryos was also severely affected as the RNA expression of nkx2.2a, a marker for CNS development, was greatly reduced at 30 hpf (Fig. S8A). This finding further supports our observation of abnormal head/brain development with reduced hindbrain area in 30 hpf MZnpc2m/m embryos. Axons of Rohon–Beard neurons from the spinal cord were also greatly reduced, consistent with a defect in CNS development (Fig. S8B). By 48 hpf, the defects described above persisted and worsened (Fig. 5A). The MZnpc2m/m embryos died by 3 dpf or soon after. Consistent with the expected cholesterol storage defect, MZnpc2m/m embryos displayed intense filipin-positive puncta throughout the entire body with the strongest signal at the yolk surface, possibly in the yolk syncytial layer at 24 and 30 hpf (Fig. 5B, Figs S9 and S10). Quantification of filipin-positive puncta showed a significant (P<0.0001) increase from a mean number of 91.5±22.9 counts/sample in control embryos to 1028±109.2 counts/sample in MZnpc2m/m embryos. In contrast, supporting the idea that maternal transcripts are protective, filipin-positive puncta were not observed in 30 hpf zygotic npc2m/m embryos (Fig. S10). To verify that the cause of these defects was the lack of both maternal and zygotic functional Npc2, MZnpc2m/m embryos were injected with zebrafish full-length npc2 mRNA at the 1-cell stage. None of the npc2 mRNA-injected MZnpc2m/m embryos showed any of the defects described above, establishing that these malformations present a bona fide maternal-zygotic phenotype (Fig. 6).
Fig. 5.
Developmental malformations in MZnpc2m/m embryos. (A) Live imaging of MZnpc2m/m and Mnpc2+/m embryos. MZnpc2m/m embryos had essentially normal gross morphology except the lack of otoliths at 22 hpf. Otic vesicles are shown in the insets (arrowheads). Older MZnpc2m/m embryos manifested additional malformations, including abnormal head/brain development with reduced hindbrain, no circulating red blood cells, and a curved/twisted body axis at 30 and 48 hpf. All of the embryos from three separate crosses exhibited representing phenotypes as shown in each image (for 22 hpf: Mnpc2+/m n=22, MZnpc2m/m, n=18; for 30 hpf: Mnpc2+/m n=28, MZnpc2m/m n=33; for 48 hpf: Mnpc2+/m n=24, MZnpc2m/m n=21). Scale bars: 200 μm (22 hpf); 500 μm (30 and 48 hpf). (B) Filipin-positive puncta were found on the yolk surface of MZnpc2m/m embryos at 24 hpf. This signal appears to be from the yolk syncytial layer. Mnpc2+/m (n=25) and MZnpc2m/m (n=21) embryos were examined from three experiments. Scale bar: 200 μm. (C) Live imaging of otic vesicles at 30 hpf. Note that Mnpc2+/m embryos (n=28) exhibited two otoliths in the oval-shaped otic vesicle whereas MZnpc2m/m embryos (n=33) displayed a less-organized otic vesicle with no otoliths (arrowhead). Scale bar: 100 μm. (D) Red blood cells stained with O-dianisidine at 30 hpf. Red blood cells accumulated in the posterior blood island (arrow) in the MZnpc2m/m embryo (n=24), indicating that red blood cells were formed but not circulating in these embryos. This accumulation of RBCs was not observed in Mnpc2+/m embryos (n=21). At 48 hpf, circulating blood cells were observed in caudal artery (red arrowhead) and vein (white arrowhead) of the Mnpc2+/m embryos (n=18), whereas blood cells remained sequestered in the posterior blood island in MZnpc2m/m embryos (asterisk) (n=23). Y, yolk stalk extension. Scale bars: 200 μm (30 hpf); 100 μm (48 hpf). All experiments were repeated three times.
Fig. 6.
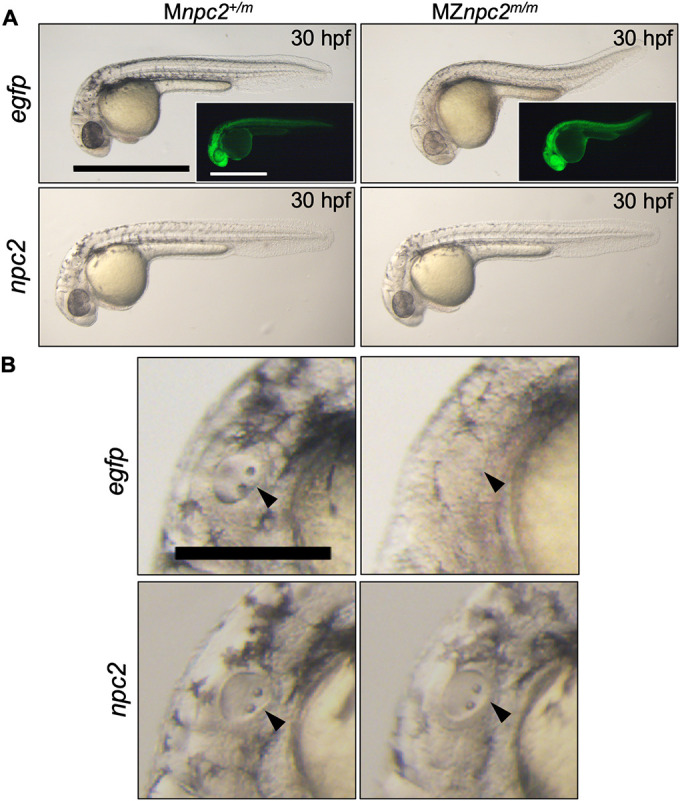
MZnpc2m/m embryonic phenotype was rescued by re-expression of npc2 mRNA. (A) Live imaging of 30 hpf Mnpc2+/m and MZnpc2m/m embryos injected with either 50 pg egfp (Mnpc2+/m n=27, MZnpc2m/m n=31) or npc2 mRNA (Mnpc2+/m n=32, MZnpc2m/m n=37) at the 1-cell stage (n=3 experiments). MZnpc2m/m embryos injected with npc2 mRNA appeared identical to the Mnpc2+/m embryo injected with either egfp or npc2 mRNA. MZnpc2m/m embryos injected with egfp RNA still displayed the defects found in uninjected MZnpc2m/m embryos. EGFP signal is shown in the insets. Scale bars: 500 μm. (B) Magnified images of otic vesicles. The otic vesicle and otoliths were normal in the MZnpc2m/m embryo injected with npc2 mRNA (arrowheads). Scale bar: 250 μm.
Impaired cellular cholesterol homeostasis underlies the early embryonic defects observed in MZnpc2m/m zebrafish
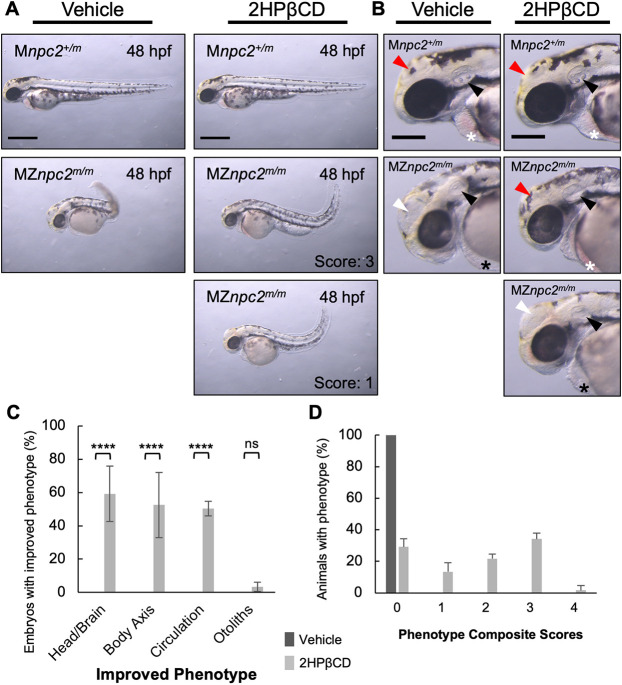
To understand whether the maternal-zygotic npc2 phenotype was a result of decreased cholesterol bioavailability or a novel function of Npc2 unrelated to the lysosomal cholesterol storage defect, we injected both MZnpc2m/m and Znpc2+/m embryos with either vehicle (ddH2O) or 100 pmol of 2-hydroxypropyl-β-cyclodextrin (2HPβCD) at 2.5 hpf. 2HPβCD has been shown to promote redistribution of cholesterol in NPC1 cells and restore intracellular cholesterol homeostasis (Feltes et al., 2020). 2HPβCD treatment significantly improved the NPC1 phenotype in both mouse and cat models of NPC1 and appeared to be effective in a phase 1/2 therapeutic trial (Davidson et al., 2009; Liu et al., 2009; Ory et al., 2017; Vite et al., 2015). In zebrafish, npc1m/m larvae treated with 2HPβCD showed significant reduction of LysoTracker Red staining in lateral line neuromasts, suggesting that 2HPβCD is an applicable potential treatment for NPC1 in different organisms (Tseng et al., 2018). 2HPβCD-injected MZnpc2m/m embryos showed improvement in abnormal head, body axis, and circulation phenotypes compared with vehicle-injected MZnpc2m/m embryos at 30 hpf (Fig. 7, Movies 5 and 6). Nevertheless, almost all 2HPβCD-injected MZnpc2m/m embryos lacked otoliths, a marker of impaired otic vesicle development (Fig. 7B). As part of this study, we were able to obtain one (and only one to date) fertile female zygotic npc1m/m zebrafish. Consistent with their sequential function in moving unesterified cholesterol out of the endolysosomal system, the phenotype of MZnpc1m/m embryos is similar to MZnpc2m/m embryos, including absence of otoliths (Fig. S11). Because our data from MZnpc1m/m embryos is very limited, we treated wild-type zebrafish with U18666A. U18666A inhibits NPC1 function (Lu et al., 2015). Wild-type zebrafish embryos were treated from 4 to 30 hpf with increasing concentrations of U18666A (Fig. 8). In embryos treated with 96 µM, we observed abnormal brain structures, absence of circulating red blood cells, and curved/twisted body axes (Fig. 8A). Furthermore, treating MZnpc2m/m embryos with 96 µM U18666A neither corrected nor exacerbated the defects found in control MZnpc2m/m embryos (Fig. S12), suggesting that there is no synergistic effect between npc1 and npc2 mutations. A dose-dependent effect on otolith size was observed. Otolith diameter was significantly (P<0.0001) decreased in embryos treated with either 24 µM or 48 µM U18666A with mean size decreased by 29.38% and 83.25%, respectively, compared with control (Fig. 8B). Otoliths were not observed in embryos treated with 96 µM U18666A (Fig. 8B). Phenotypic correction by 2HPβCD combined with similar dysmorphic findings when NPC1 function is impaired strongly supports the idea that the MZnpc2m/m embryonic phenotype is secondary to the known cellular cholesterol homeostasis defect and not due to a novel function of NPC2.
Fig. 7.
2HPβCD partially corrects the MZnpc2m/m phenotype. (A) Live imaging of 48 hpf Mnpc2+/m and MZnpc2m/m embryos injected with either vehicle (Mnpc2+/m n=53, MZnpc2m/m n=76) or 100 pmol of 2HPβCD (Mnpc2+/m n=81, MZnpc2m/m n=80) at 2.5 hpf (n=3 experiments). Many 2HPβCD-injected MZnpc2m/m embryos showed milder defects, including more normal head/brain appearance, restored red blood cell circulation, and less-curved body axis than observed in vehicle-injected MZnpc2m/m embryos. Two representative images of 2HPβCD-injected MZnpc2m/m embryos are shown with their corresponding corrected phenotype. Phenotype composite scores of vehicle and 2HPβCD-treated MZnpc2m/m embryos are shown (Score 3: improved head/brain appearance, restored red blood cell circulation, and less-curved body axis; Score 1: less-curved body axis). Individuals exhibiting all four defects (the lack of otoliths, abnormal head/brain development, absent circulating red blood cells and a curved/twisted body axis) received a score of 0, and individuals with none of the four defects received a score 4. Experiments were repeated three times. All Mnpc2+/m embryos injected with either vehicle or 2HPβCD displayed normal phenotype, and all MZnpc2m/m injected with vehicle displayed abnormal phenotypes including all defects described above. Scale bars: 500 μm. (B) Magnifications of the images in A showed that although 2HPβCD was able to partially rescue some defects in MZnpc2m/m embryos, otoliths were still not seen in 2HPβCD-treated MZnpc2m/m embryos despite some of embryos displaying more organized otic vesicles (black arrowheads). Some 2HPβCD-treated MZnpc2m/m embryos displayed improved head/brain appearance (red arrowheads) as they did not exhibit the enlarged hollow compartment (white arrowheads) found in vehicle-treated MZnpc2m/m embryos. Red blood cells were found in the pericardial region of some 2HPβCD-injected MZnpc2m/m embryos (white asterisks) but not in vehicle-injected MZnpc2m/m embryos and some more severe 2HPβCD-injected MZnpc2m/m embryos (black asterisks), consistent with restored circulation. Scale bars: 200 μm. (C) 2HPβCD-injected 48 hpf MZnpc2m/m embryos displayed improved phenotype in head/brain structure, body axis and circulation but the absent otolith phenotype was still largely unchanged. n=76 (vehicle-injected MZnpc2m/m embryos) and n=80 (2HPβCD-injected MZnpc2m/m embryos) (n=3 experiments. ****P<0.0001 (χ2, Fisher's exact test). ns, not significant. (D) Distribution of phenotype composite scores of vehicle and 2HPβCD-treated MZnpc2m/m embryos. Most 2HPβCD-treated MZnpc2m/m embryos showed improvement in one to three individual findings.
Fig. 8.
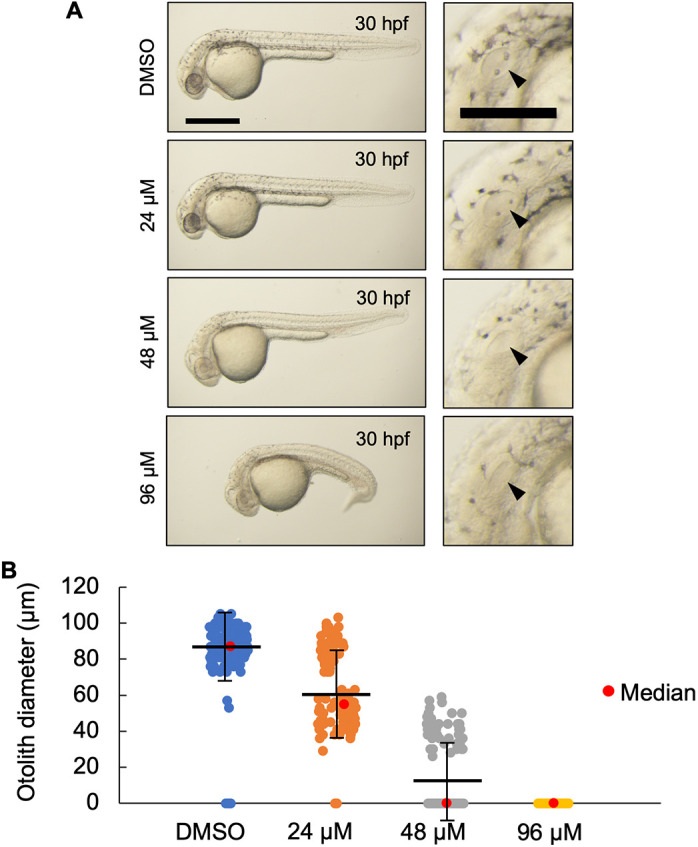
U18666A-treated wild-type zebrafish phenocopied MZnpc2m/m embryos. (A) Live imaging of 30 hpf wild-type embryos treated with the indicated dose of U18666A starting at 4 hpf. U18666A-treated embryos exhibited a dose-dependent phenotype ranging from smaller otoliths at 24 μM to no otoliths, abnormal head, no circulating blood cells, and curved/twisted body axis at 96 μM. Arrowheads indicate the otic vesicle. Scale bars: 500 μm (whole embryo); 200 μm (otic vesicle). (B) Measurement of otolith diameters from DMSO- and U18666A-treated wild-type embryos at 30 hpf. n=118 (DMSO), 118 (24 μM), 114 (48 μM) and 94 (96 μM) (n=2 experiments). Two otoliths per embryo were measured. 0 indicates that otolith was not detectable. P<0.0001 for each treatment group compared with DMSO (Kruskal–Wallis test).
Transcriptome analysis reveals potential pathways underlying the developmental defects observed in MZnpc2m/m embryos
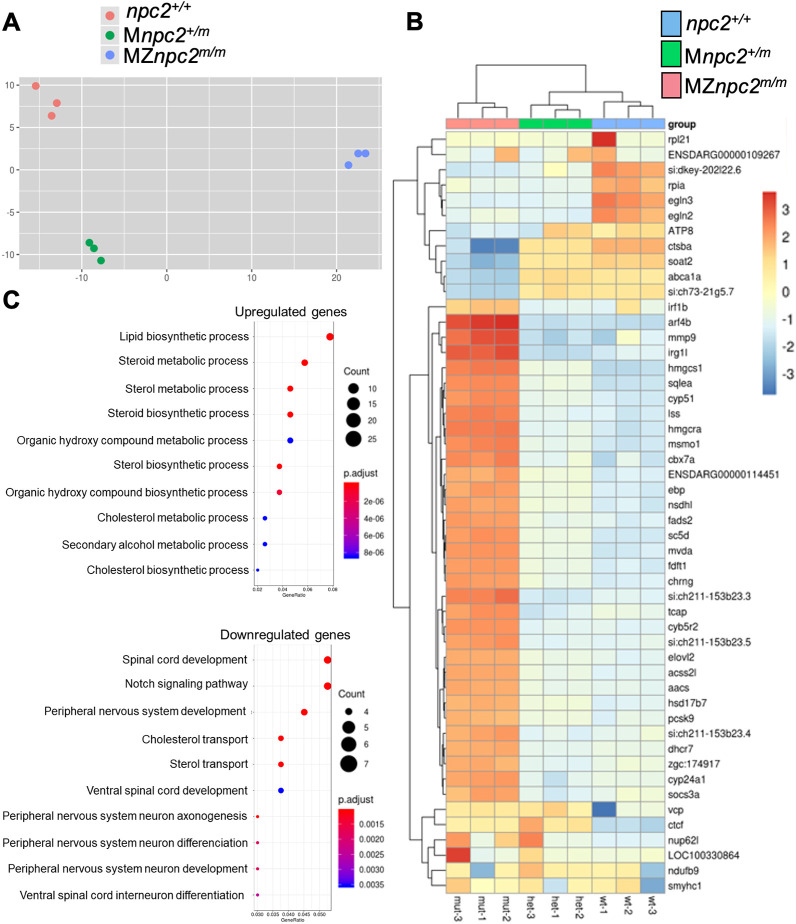
To gain insight into the developmental pathways that are disrupted by impaired cellular cholesterol homeostasis, we compared RNA expression data between 30 hpf MZnpc2m/m, Mnpc2+/m and npc2+/+ zebrafish embryos. Total RNA was extracted from three independent pools of 30 embryos for each genotype. Principal component analysis (PCA) of the RNA expression data showed very good clustering of the independent samples for all three genotypes (Fig. 9A).
Fig. 9.
RNA-seq analysis of MZnpc2m/m and Mnpc2+/m embryos at 30 hpf. (A) Principal component analysis (PCA) of each pool of mRNA from all three genotypes: npc2+/+, Mnpc2+/m and MZnpc2m/m. These data suggested that MZnpc2m/m transcriptomes differed more compared with npc2+/+ and Mnpc2+/m transcriptomes than npc2+/+ differed from Mnpc2+/m (x-axis: 67% of variance; y-axis: 14% of variance). (B) Heatmap of the top 50 genes with the most expression variation. The heatmap plots the deviation from the row mean. (C) Dotplot of GO biological process enrichment in genes differentially expressed between mutant and heterozygote. This represents highly enriched GO biological process terms from the analysis with the color corresponding to level of enrichment (adjusted P-value) and the size of the dots representing the number of genes associated with the GO term.
The heatmap of the top 50 most differentially expressed genes showed similar trends for Mnpc2+/m and npc2+/+ embryos, whereas MZnpc2m/m embryos displayed different gene expression patterns (Fig. 9B). A total of 511 upregulated and 186 downregulated genes were identified in MZnpc2m/m embryos from the differential expression analysis comparing with Mnpc2+/m embryos (Fig. S13, Table S7). For functional enrichment analysis, we compared transcriptomes from MZnpc2m/m and Mnpc2+/m sibling embryos, revealing significant upregulation in genes related to lipid, sterol and cholesterol metabolism and synthesis (Fig. 9C, Table S1) and significant downregulation of genes involved in spinal cord development, Notch signaling and peripheral nervous system development (Fig. 9C, Table S2). As anticipated, owing to decreased cellular cholesterol bioavailability, expression of Srebf2-regulated genes (such as hmgcs1, hmgcra, ebp, nsdhl, sc5d, mvda and dhcr7) involved in endogenous cholesterol synthesis was significantly increased in MZnpc2m/m embryos (Fig. 9B, Table S3) (Eid et al., 2017; Sakakura et al., 2001). The mRNA expression of many genes involved in cholesterol transport was also altered in MZnpc2m/m embryos (Table S4). For example, an important transporter for cholesterol efflux, ABCA1, is downregulated in human NPC1 cells (Choi et al., 2003). The mRNA expression of its zebrafish ortholog abca1a in MZnpc2m/m embryos was also significantly downregulated (Table S4). The RNA-seq analysis data was further validated as otic placode formation and blood vessel endothelial cell differentiation, two biological processes related to the lack of otoliths and absence of circulating red blood cells in MZnpc2m/m embryos, were also identified as significantly downregulated biological processes (Table S5).
Notably, the RNA expression data also showed downregulation of Notch signaling (Fig. 9C, Table S2). Among all the genes related to Notch signaling, notch3 was the only gene at the Notch receptor level that showed significant downregulation of mRNA expression in MZnpc2m/m embryos. Delta-like protein B precursor, dlb, is the only Notch ligand gene for which expression was significantly increased in MZnpc2m/m embryos. Other downregulated genes related to Notch signaling, such as hey2, hey1, her12 and her15.1 act downstream of Notch receptor signaling, so their downregulation is likely secondary to decreased notch3 expression (Table S6). These data suggested that decreased Notch3 signaling might contribute to the dysmorphic findings in MZnpc2m/m embryos, a hypothesis that was strengthened by the fact that notch3 is implicated in many downregulated biological processes, such as spinal cord and peripheral nervous system development, identified in our RNA-seq analysis (Table S2). notch3 mRNA expression by in situ hybridization was reduced in the head area and completely missing from the otic vesicles of 30 hpf MZnpc2m/m embryos, suggesting that decreased notch3 signaling underlies the absent otolith phenotype (Fig. 10). In situ hybridization of msx3, another gene involved in the brain and otic vesicle development and downregulated in our RNA-seq dataset, also showed great reduction of mRNA in the brain, dorsal trunk, and otic vesicles (Fig. 10, Table S5). These in-situ hybridization results validated the data from our RNA-seq analysis, as both notch3 and msx3 were reduced in tissues exhibiting defects in MZnpc2m/m embryos.
Fig. 10.
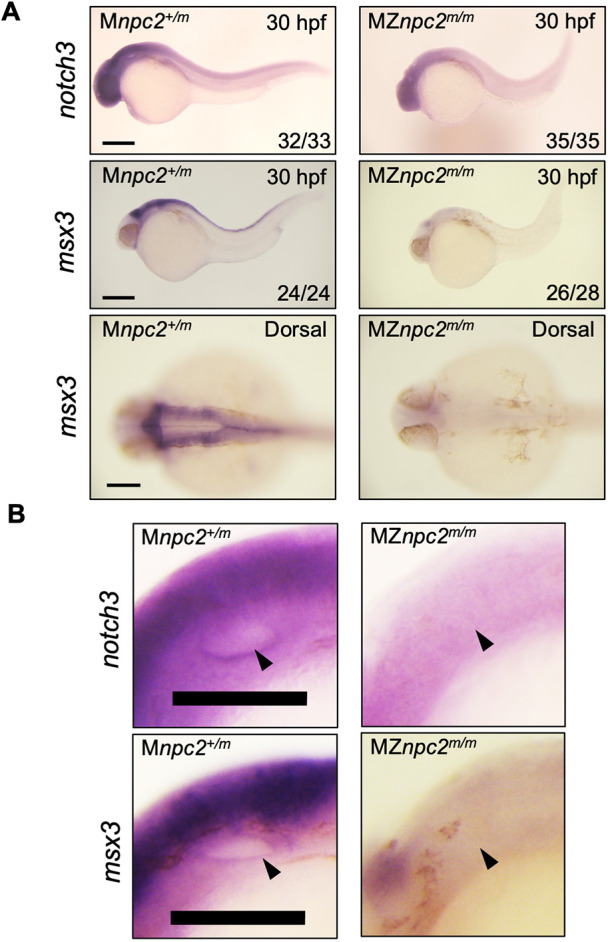
mRNA expression in and adjacent to otic vesicles. (A) Whole-mount RNA in situ hybridization for notch3 and msx3 in 30 hpf Mnpc2+/m and MZnpc2m/m embryos. Expression of both notch3 and msx3 was observed mainly in the head area and was reduced in MZnpc2m/m embryos compared with Mnpc2+/m embryos. The numbers of embryos examined showing the represented staining over the total numbers of embryos examined are indicated in each image (n=3 experiments). Scale bars: 200 μm (30 hpf); 100 μm (dorsal). (B) Magnifications of the otic vesicles shown in A. Both notch3 and msx3 were expressed along the lining of the otic vesicle in Mnpc2+/m embryos. However, their expression was diminished around the otic vesicle in MZnpc2m/m embryos (arrowheads). Scale bars: 100 μm.
To explore directly a potential causative role of Notch3 downregulation for defects found in MZnpc2m/m embryos, Notch3 signaling was enhanced by overexpression of constitutively active Notch3 intracellular domain (N5icd) via microinjection of a plasmid encoding n5icd into MZnpc2m/m and Mnpc2+/m embryos. Consistent with our hypothesis, some MZnpc2m/m embryos injected with n5icd plasmid were almost identical to their control Mnpc2+/m siblings at 30 hpf (Fig. 11). Expression of N5icd in MZnpc2m/m embryos corrected the developmental defects in 8/57 embryos; however, no phenotypic correction was observed in 59 control injections (P=0.0028, χ2, Fisher's exact test). Although the n5icd rescue rate was low, possibly due to transient and mosaic expression of plasmid DNA, this result supports the conclusion that impaired intracellular cholesterol transport due to Npc2 deficiency leads to impaired Notch3 signaling.
Fig. 11.
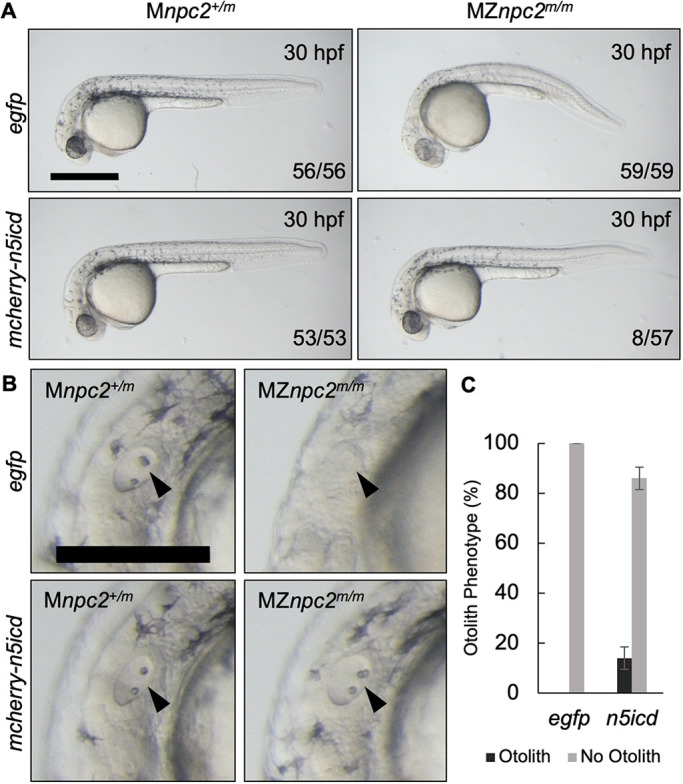
MZnpc2m/m embryos were rescued by re-activation of Notch3 signaling. (A) Live imaging of 30 hpf Mnpc2+/m and MZnpc2m/m embryos injected with either egfp or mcherry-n5icd (notch3 intracellular domain) plasmid DNA at the 1-cell stage. Some of the mcherry-n5icd-injected MZnpc2m/m embryos (13.92±4.43%) displayed a relatively normal phenotype in comparison with egfp-injected MZnpc2m/m embryos. Numbers of embryos examined showing representing staining over total numbers of embryos examined are indicated in each image (n=4 experiments). Scale bar: 500 μm. (B) Magnifications of the images shown in A in the area of the otic vesicle. Both otoliths and otic vesicle were restored in this mcherry-n5icd-injected MZnpc2m/m embryo (arrowheads). Scale bar: 250 μm. (C) Quantification of otolith phenotype recovery in egfp- (n=59) and mecherry-n5icd-injected MZnpc2m/m embryos (n=58). P=0.0028 for ‘no otolith' in n5icd-treated group versus ‘no otolith' in egfp-treated group (χ2, Fisher's exact text).
DISCUSSION
Our npc2 mutant zebrafish model displayed phenotypes similar to those found in npc1 mutant zebrafish (Lin et al., 2018; Tseng et al., 2018), with additional low-penetrant defects such as dorsally curved body axes and a lack of properly inflated swim bladders. The signature biochemical feature of NPC, intracellular unesterified cholesterol accumulation, was found in 5 and 7 dpf npc2m/m larvae throughout the entire body. Similar to what we observed in npc1m/m zebrafish (Tseng et al., 2018), npc2m/m larvae exhibited intense LysoTracker Red-positive staining in olfactory placodes and lateral line neuromasts. Although most npc2m/m larvae appeared to develop normally, very few of them survived to adulthood when raised with npc2+/+ and npc2+/m siblings. This appeared to be due to competition for food by unaffected siblings, as found for npc1m/m zebrafish (Tseng et al., 2018). We used the LysoTracker phenotype to stain and sort npc2m/m larvae away from their npc2+/+ and npc2+/m siblings at 3 dpf and raised them separately. Survival of npc2m/m larvae and juveniles was greatly improved with many surviving to adulthood. Although most of npc2m/m adults were small, we identified a few mature npc2m/m females for breeding. In our previous study, we did not find any mature and fertile npc1m/m individuals (Tseng et al., 2018). Mature npc2m/m males capable of breeding were never found, possibly due to smaller body size.
We postulated that the mild early embryonic phenotypes of both npc1m/m and npc2m/m mutant zebrafish is due to maternal contribution of npc1 and npc2 gene products. To address this issue, we crossed npc2+/m males to npc2m/m females to obtain MZnpc2m/m embryos. In contrast to zygotic npc2m/m embryos, MZnpc2m/m embryos exhibited severe defects, including a lack of otoliths/defective otic vesicle development, abnormal head and brain anatomy, a lack of circulating blood cells and curved/twisted body axes at 30 hpf. This suggested that maternal npc2 gene product plays an important role during early embryonic development. Many maternally expressed genes are important for early embryonic development, as many MZ mutants display early defects that are more severe than those of their zygotic mutant counterparts (Despic and Neugebauer, 2018; Langdon and Mullins, 2011; Pelegri, 2003).
To establish that the MZnpc2m/m phenotype was due to Npc2 disruption, we showed that the phenotype was rescued by embryonic microinjection of npc2 mRNA. Rescue of the MZnpc2m/m phenotype by 2HPβCD treatment further indicated that the MZnpc2m/m phenotype was due to the impaired intracellular cholesterol trafficking common to both NPC1 and NPC2, as opposed to a novel function of Npc2. Further supporting this conclusion, wild-type zebrafish embryos treated with an Npc1 inhibitor, U18666A, exhibited dose-dependent defects consistent with defects observed in MZnpc2m/m embryos at 30 hpf with more than 50% embryos exhibiting small otoliths phenotype at 24 μM and 100% embryos exhibiting no otoliths at 96 μM. This result suggests that similar malformations would be observed in zebrafish if early function of Npc1, including that derived from maternal transcripts, was blocked. Consistent with this conclusion, we were able to obtain MZnpc1m/m embryos from the cross of an npc1+/m male and a single Znpc1m/m female. Although this experiment could not be replicated because we have only found one fertile npc1m/m female, the malformations observed in MZnpc1m/m larvae were similar to those observed in MZnpc2m/m larvae. The defects we found in both MZnpc2m/m and MZnpc1m/m embryos were not reported in zebrafish npc1 morphants, which have an epiboly defect that we did not observe (Schwend et al., 2011). These differences might be attributed to a combination of incomplete knockdown by their morpholino, genetic background effects or morpholino artefact. These data are consistent with the conclusion that decreased cellular cholesterol bioavailability due to impaired transport of cholesterol out of the endolysosomal compartment leads to the malformations common to these interventions. Cholesterol is known to be essential for embryonic development (Farese and Herz, 1998). An alternative explanation would be generalized lysosomal dysfunction due to lysosomal storage. This is unlikely as similar defects have not been reported for other zebrafish models of lysosomal disease, including mucolipidosis type IV (Li et al., 2017), CLN3 (Wager et al., 2016) and Krabbe disease (Zizioli et al., 2014). The ability of HPβCD to prevent the embryonic defects supports the conclusion that these defects result from decreased cellular cholesterol bioavailability as HPβCD is known to restore cellular cholesterol homeostasis in other NPC model systems (Davidson et al., 2009; Feltes et al., 2020; Liu et al., 2009; Vite et al., 2015).
MZnpc2m/m embryos exhibited a variety of defects, including missing otoliths, abnormal head and brain anatomy, no circulating blood cells and curved/twisted body axes at 30 hpf. Among these defects, the lack of otoliths was the first gross defect observed in MZnpc2m/m embryos, as early as 22 hpf, whereas other parts of those embryos looked relatively normal. Zebrafish otoliths are calcified biominerals composed of the calcium carbonate polymorph aragonite and certain organic molecules found in the inner ear. They are involved in zebrafish balance and hearing (Lundberg et al., 2015; Sollner et al., 2003). Although other defects in MZnpc2m/m embryos could be partially rescued by 2HPβCD injection, the lack of otoliths was unchanged. This suggests that mechanisms affecting otolith formation in MZnpc2m/m embryos are more sensitive to the blockage of intracellular cholesterol trafficking than other phenotypes or, alternatively, it could reflect a lack of adequate HPβCD penetration. Evaluation of our RNA-seq data showed that the mRNA expression of genes directly involved in otolith formation, such as oc90 (Lundberg et al., 2015) and otol1a (Lundberg et al., 2015), was not altered significantly in 30 hpf MZnpc2m/m embryos. Although the expression of tecta, a gene required for otolith tethering in zebrafish (Stooke-Vaughan et al., 2015), was significantly decreased in 30 hpf MZnpc2m/m embryos, zebrafish rolling stone (rst) mutants, which have a mutation in tecta, exhibit an otolith-tethering defect at larval stages that is distinct from the formation of otoliths (Stooke-Vaughan et al., 2015). Therefore, it is likely that the lack of otoliths in MZnpc2m/m embryos is due to more general problems with otic vesicle development. Indeed, when examining the otic vesicles of live MZnpc2m/m embryos, we found that the otic vesicles were formed but very unorganized. Transmission electron microscopy revealed that the cells lining the otic vesicles of MZnpc2m/m embryos were very thin and barely contained any cytoplasm. Moreover, the mRNA expression of sox9b, sox3 and msx3, three genes involved in otic vesicle development, was downregulated significantly in MZnpc2m/m embryos at 30 hpf. Taken together, it is likely that the lack of otoliths results from compromised otic vesicle development.
To gain insight into the mechanisms underlying the malformations observed in npc2 mutants, we compared transcriptomes from 30 hpf MZnpc2+/m and MZnpc2m/m embryos. In addition to identifying biological processes associated with sterol/cholesterol metabolism, we observed significant (adjusted P=0.001) alteration in expression of genes involved in Notch signaling, including dab2, nrarpb, notch3, hey2, hey1, her12 and her15.1. notch3 encodes a Notch receptor whereas hey2, hey1, her12 and her15.1 encode downstream effector genes involved in Notch signaling (Gaudet et al., 2011; Matsuda and Chitnis, 2009; Shankaran et al., 2007; Zhong et al., 2001). Based on our expression data, we suspect that impaired Notch signaling is occurring at the level of the Notch receptor rather than due to decreased expression of Notch ligands. The only Notch ligand with altered expression was dlb and its expression was increased, perhaps consistent with a receptor/agonist feedback loop.
Notch3 is involved in many different developmental processes in zebrafish embryos, including maintenance of rhombomere boundaries, oligodendrocyte development and vascular integrity (Ando et al., 2019; Qiu et al., 2009; Wang et al., 2014; Zaucker et al., 2013). Compromised blood vessel development could explain the lack of circulating blood cells in MZnpc2m/m embryos even though erythrocytes were present in the posterior blood island. Abnormal head/brain development in 30 hpf MZnpc2m/m embryos was possibly the consequence of reduced Notch3 signaling strength, as expression of genes involved in nervous system development, such as nkx2.2a, was significantly decreased (Zaucker et al., 2013). Despite an absence of known links to Notch3 signaling, the lack of otoliths and the curved/twisted body axis, together with other defects, in 30 hpf MZnpc2m/m embryos could be rescued by reactivating Notch3 signaling. This suggests that reduced Notch3 signaling underlies the multiple developmental abnormalities observed in MZnpc2m/m embryos.
The connection between decreased cellular cholesterol bioavailability and decreased Notch3 signaling remains to be elucidated. Notch3 is an integral membrane protein. Although we were unable to find specific literature support for Notch3 dependence on cholesterol, perturbation of membrane cholesterol content often alters receptor function (Incardona and Eaton, 2000). Consistent with this idea that Notch3 function is influenced by membrane cholesterol content, Obniski et al. (2018) showed that dietary cholesterol influences intracellular Notch signaling and enteroendocrine cell differentiation in the midgut of adult fruit flies. In addition, activation of the Notch receptor may be sensitive to membrane cholesterol levels. Upon ligand/receptor binding, Notch proteins undergo a series of proteolytic cleavages, liberating only the Notch intracellular domain (ICD), which goes on to enter the nucleus and turn on the transcription of target genes. Among the list of enzymes involved in Notch activation, γ-secretase plays an important role, as it cleaves and releases Notch ICD from the cell membrane (Olsauskas-Kuprys et al., 2013). γ-secretase is a complex consisting of Presenillin, Nicastrin, APH-1 and PEN-2 and is known to be involved in the production of amyloid β-peptides (Wolfe, 2020a,b). Decreased membrane cholesterol reduces γ-secretase activity (Kim et al., 2016). Inhibitors of γ-secretase have been shown to inhibit Notch signaling in different models, including zebrafish (Geling et al., 2002; Olsauskas-Kuprys et al., 2013; Yang et al., 2010). Increased expression of the cholesterol efflux proteins ABCG1 and ABCG4 has been shown to suppress γ-secretase activity, likely secondary to redistribution of membrane cholesterol content (Sano et al., 2016). We found mRNA expression of abcg4b (a zebrafish homolog of human ABCG4) to be increased significantly in 30 hpf MZnpc2m/m embryos. Increased Abcg4b activity could compound the decreased cellular cholesterol bioavailability as a result of impaired lysosomal export of cholesterol. Reduced Notch3 signaling would be further reduced by a self-regulated feedback loop as it also reduces the production of notch3 mRNA transcripts (Fouillade et al., 2013; Liu et al., 2010). The above proposed mechanisms notwithstanding, the details of the cholesterol-Notch link we have uncovered and the specific causes of its breakdown in MZNpc2m/m embryos remain to be elucidated.
In conclusion, we developed a zebrafish model of NPC2 disease in which the zygotic phenotype resembles that of NPC1 zebrafish models (Lin et al., 2018; Tseng et al., 2018). Developmental defects in maternal zygotic mutants appear to be due to decreased cholesterol bioavailability and secondary impairment of Notch3 signaling. Future work will be needed to determine how decreased cellular cholesterol bioavailability impairs Notch3 signaling. This work provides additional evidence of the importance of cholesterol homeostasis in regulation of developmental processes.
MATERIALS AND METHODS
Zebrafish husbandry and generation of mutant lines
Control EKW zebrafish were used to generate npc2 mutant lines by CRISPR/Cas9-mediated gene targeting. The targeting region was located within npc2 exon 2 (5′-GGACTACAGAGTGCTCGGCG-3′) for sgRNA recognition. The corresponding sgRNA (Integrated DNA Technologies) was injected into control zebrafish embryos at the 1-cell stage together with cas9 mRNA. For mutation screening, sgRNA-injected F0 embryos were raised to adulthood and outcrossed to wild-type adults to obtain F1 embryos. PCR and fragment analysis using genomic DNA from 16 randomly selected F1 embryos were performed to identify potential F0 adults carrying mutations (founders). Additional F1 embryos from F0 founders were raised to adulthood and screened for mutations by PCR and fragment analysis. Two npc2 frame-shift mutant alleles were identified during the screening from F1 adults. The mutant alleles are npc2y536 (NM_173224.1: c.17_21del) and npc2y537 (NM_173224.1: c.14_20del). Both alleles were recovered and used for subsequent experiments. The npc1 frame-shift mutant line npc1hg37 was also used to generate MZnpc1 mutants. All zebrafish lines were maintained in the aquatic animal facility at 28.5°C according to the Animal Use Protocol (AUP), approved by IACUC committee of the National Institute of Child and Human Development.
Genotyping
Genomic DNA extraction was carried out as previously described (Tseng et al., 2018). Neither npc2 mutations altered any endogenous restriction enzyme sites; therefore, artificial restriction enzyme sites were introduced via dCAPS PCR to identify mutations. dCAPS PCR primers were designed using the dCAPS Finder 2.0 online tool (Department of Biology, Washington University, St. Louis, MO, USA). For the npc2y536 allele, PCR was carried out using forward primer Znpc2 Ex2-BglII-F (5′-CATTCATTTGAACCATGGACTACAGAGATC-3′) and reverse primer Znpc2 Ex3-R (5′-CTACTTTTCCGTCTACCGAGCCTG-3′); for the npc2y537 allele, PCR was carried out using forward primer Znpc2 Ex2-HpyCH4IV-F (5′-CATTCATTTGAACCATGGACTACAACG-3′) and reverse primer Znpc2 Ex3-R (5′-CTACTTTTCCGTCTACCGAGCCTG-3′). The PCR conditions were 94°C for 2 min, then 35 cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 30 s; followed by 72°C for 5 min and 4°C infinitely for both alleles. PCR products from npc2y536 and npc2y537 were digested with restriction enzyme BglII (R0144, New England BioLabs) and HpyCH4IV (R0619, New England BioLabs), respectively, at 37°C for 8 h. Restriction enzyme digestion products were resolved on 2% agarose gels to identify genotypes.
Live imaging
Larval or adult zebrafish for live imaging were first anesthetized with 0.16 mg/ml tricaine (A5040, MilliporeSigma) in egg/system water. They were mounted on a glass depression slide with 3% methylcellulose solution. All images of live zebrafish were taken using a Leica MZ16F stereomicroscope (Leica Microsystems) and AxioCam HRc CCD camera (Carl Zeiss).
Whole-mount filipin staining and immunofluorescence
Embryos/larvae were fixed in 4% paraformaldehyde in 1× PBS at 4°C overnight. For filipin staining, freshly fixed embryos/larvae were then stained with 0.05 mg/ml Filipin (08707, Polysciences) for 2.5 h at room temperature. For immunofluorescence, fixed embryos were dehydrated and stored in 100% methanol at −20°C prior to the staining. Samples were rehydrated, rinsed, and permeabilized extensively with 1× PBT (0.5% Triton X-100 in 1× PBS) and subsequently blocked in blocking solution (2% bovine serum albumin, 1% DMSO, 0.5% Triton X-100, 0.5% goat serum in 1× PBS) at room temperature for 1 h. Samples were incubated with primary antibody diluted in blocking solution at 4°C overnight. After rinsing larvae a few times with 1× PBT, they were incubated with secondary antibody at 4°C overnight. Stained embryos were rinsed several times with 1× PBT and 1× PBST (0.1% Tween-20 in 1× PBS) to remove excess antibodies. The following antibodies and concentrations were used in experiments: mouse monoclonal anti-acetylated tubulin clone 6-11-B1 (1:1000, T7451, MilliporeSigma), Alexa Fluor 594-conjugated donkey anti-mouse IgG (1:1000, A21203, ThermoFisher Scientific). Filipin-stained images were taken using a Zeiss Observer Z1 inverted compound fluorescence microscope with Calibri.2 LED lighting system and CCD camera (Carl Zeiss). Immunofluorescence images were taken using a Zeiss LSM780 inverted laser scanning confocal microscope (Carl Zeiss). Filipin-positive puncta were quantified by measuring a 384×384 pixels area from each image of the yolk surface using ImageJ (National Institutes of Health, Bethesda, MD, USA).
RT-PCR and cDNA sequencing
Total RNA was extracted from pools of 30-60 embryos at each developmental stage using Trizol (15596018, ThermoFisher Scientific) and RNeasy Mini kit (74104, Qiagen) and 1 μg total RNA from each stage was treated with Turbo DNA-Free kit (AM1907, ThermoFisher Scientific) to remove residual genomic DNA. For reverse transcription, 20 ng DNase-treated total RNA from each stage was used to synthesize cDNA using iScript RT kit (1708890, Bio-Rad) and 1 μl cDNA was used for a 20 μl PCR reaction to amplify PCR amplicons from each sample (‘no RT’ control was also included). PCR was carried out using the following primers: npc2Ex1F2 (5′-TCGTGACAAGGAAACAGGGTGT-3′) and npc2Ex4R2 (5′-CCAGCAAGCACACCATGAAC-3′) for npc2 (predicted PCR product=303 bp); Znpc1Ex9F (5′-CAATGACAACTGCACCATCC-3′) and Znpc1Ex11R (5′-GTGTCATTCAGGTAGTTGGTGAC-3′) for npc1 (predicted PCR product=254 bp); ZgapdhEx3F (5′-CGTCTGGTGACCCGTGCTGC-3′) and ZgapdhEx9R (5′-TGGGGGTGGGGACACGGAAG-3′) for gapdh (predicted PCR product=670 bp). The PCR conditions were: 94°C for 2 min, then 35 cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 30 s; followed by 72°C for 5 min and 4°C infinitely for npc1; 94°C for 2 min, then 35 cycles of 94°C for 30 s, 65°C for 30 s, 72°C for 30 s; followed by 72°C for 5 min and 4°C infinitely for npc2 and gapdh. PCR products were resolved on 2% agarose gel for analysis. For npc2 full-length cDNA sequencing, total RNA was extracted from livers of two npc2+/+ and three npc2m/m 8 mpf adults as previously described and 10 ng total RNA from each sample was used to synthesize cDNA with iScript gDNA Clear cDNA Synthesis Kit (1725035, Bio-Rad). PCR was carried out using 1 μl cDNA from each sample with Platinum SuperFi II DNA Polymerase (12361010, ThermoFisher Scientific) (‘no RT’ control was also included) with a primer pair for amplifying amplicons to include the entire coding region of npc2 [npc2-ex1-2F1: 5′-GACAGAAGGAAGAACCTACATTC-3′; npc2-ex5-R1: 5′-GTAAGCATTTTATCCTGACTTC-3; predicted PCR product size=515 (npc2+) and 508 (npc2y537)]. PCR amplicons were subsequently cloned into pCR4Blunt-TOPO plasmid using Zero Blunt TOPO PCR Cloning Kit for Sequencing (K287520, ThermoFisher Scientific). After transformation, six Escherichia coli colonies from each sample were randomly selected for sequencing. Positive sequencing results were obtained from eight out of 12 colonies from two npc2+/+ zebrafish and 16 out of 18 colonies from three npc2m/m zebrafish. Sequencing results were aligned to wild-type npc2 sequence to confirm their nucleotide composition.
Whole-mount RNA in situ hybridization
Embryos were fixed in 4% paraformaldehyde at 4°C overnight and subsequently dehydrated and stored in 100% methanol at −20°C. RNA in situ hybridization was performed as previously described (Sun et al., 2011) using digoxigenin (DIG)-labeled RNA probes against npc2, notch3 and msx3. Signal was detected using a combination of an anti-DIG AP-conjugated antibody (1:3000, 11093274910, MilliporeSigma) and BM-Purple (11442074001, MilliporeSigma) as the substrate for AP. Embryos were mounted in glycerol and imaged using a Leica MZ16F stereomicroscope (Leica Microsystems) and AxioCam HRc CCD camera (Carl Zeiss). Experiments were repeated three times with three separate pools of wild-type zebrafish embryos or larvae at each stage.
Vital dye staining
LysoTracker Red DND-99 (L7528, ThermoFisher Scientific) and YO-PRO-1 Iodide (Y3603, ThermoFisher Scientific) were used for staining acidic organelles and neuromast cell nuclei. The procedure was executed as previously described (Tseng et al., 2018). The result of staining was examined using a Leica MZ16F stereomicroscope (Leica Microsystems) and AxioCam HRc CCD camera (Carl Zeiss). LysoTracker Red-stained 3 dpf larvae from the intercross of npc2+/m adults were sorted based on presence or absence of the LysoTracker Red-positive signal in the olfactory placodes. The LysoTracker Red staining-sorted larvae were kept and raised separately until 9 weeks old then genotyped and sorted again accordingly.
Electron microscopy
Zebrafish embryos were collected and fixed in 2% glutaraldehyde and 2% paraformaldehyde in 0.2 M sodium cacodylate buffer at room temperature for 2 h. Fixed embryos were then dehydrated and embedded in epoxy resin. Semi-thin sections were cut to confirm the localization of tissues. Ultra-thin sections were cut and viewed using a JOEL JEM-1400 Transmission Electron Microscope (Akishima). All images were taken using an AMT BioSprint-29 camera (AMT Imaging).
Microinjection
For RNA microinjection, egfp and full-length zebrafish npc2 mRNA were first synthesized using mMESSAGE mMACHINE SP6 Transcription Kit (AM1340, ThermoFisher Scientific) following manufacturer's instructions and 50 pg of mRNA was injected into each zebrafish embryo at the 1-cell stage. For DNA microinjection, 5 pg of pCS2+-egfp or pCScherryN5icd (Villefranc et al., 2007) plasmid containing the intracellular domain of zebrafish notch3 cDNA driven by CMV promoter was injected into each zebrafish embryo at the 1-cell stage. For 2-hydroxypropyl-β-cyclodextrin (2HPβCD) injection, 2HPβCD (VTS-270, Mollinckrodt Pharmaceuticals) was dissolved in ddH2O as a 100 mM stock solution. Each 2.5 hpf embryo was injected with 100 pmol of 2HPβCD or ddH2O directly into the yolk. All the injected embryos were examined at different developmental stages and genotyped subsequently.
U18666A treatment
U18666A (66201510MG, MilliporeSigma) was dissolved in DMSO as 9.6 mM stock solution to be used in the treatment. Zebrafish embryos were treated with 1% DMSO (vehicle) or 24, 48 or 96 μM U18666A in 1% DMSO starting at 4 hpf. Embryos were examined at 30 hpf for their phenotype.
Alizarin Red S staining
Adult zebrafish were euthanized by ice water bath and fixed in 10% neutral buffered formalin overnight. Fixed zebrafish were eviscerated and descaled before being bleached to remove surface pigment by incubation in 1% KOH and 3% H2O2. Samples were later incubated in saturated sodium tetraborate solution overnight before being placed in 1% KOH and 1% Alizarin Red S (A5533, MilliporeSigma) overnight. Stained zebrafish were rinsed with tap water and incubated in a series of 1% KOH/glycerol solutions until tissues were clear. Samples were mounted in glycerol and imaged with a Leica MZ16F stereomicroscope (Leica Microsystem) and AxioCam HRc CCD camera (Carl Zeiss).
O-dianisidine staining
To visualize hemoglobin in red blood cells, zebrafish embryos were anesthetized and stained with 0.8 mg/ml o-dianisidine (D9143, MilliporeSigma) in 40% ethanol, 0.01 M sodium acetate pH 5.2 and 2% hydrogen peroxide for 15 min in the dark.
Transcriptome analysis (RNA-seq)
Total RNA was extracted from zebrafish npc2+/+, Mnpc2+/m and MZnpc2m/m embryos at 30 hpf using Trizol (15596018, ThermoFisher Scientific) and RNeasy Mini kit (74104, Qiagen). Mnpc2+/m and MZnpc2m/m embryos were siblings obtained from the same crosses distinguished by their phenotypes whereas npc2+/+ embryos were from the crosses of npc2+/+ adults, which were siblings of Mnpc2+/m and MZnpc2m/m embryos' parents. Thirty embryos of each genotype were pooled together as one pool for RNA extraction. Three separate pools of each genotype were collected for further analysis. RNA concentration and quality were measured using Nanodrop 2000 (ThermoFisher Scientific) and Bioanalyzer (Agilent Biotechnologies). RNA-seq libraries were constructed by fragmenting RNA from each sample, reverse transcribing cDNA, and adding adapters for Illumina sequencing. Each library was then used to generate 30-50 million total sequences for RNA-seq with Illumina HiSeq 2500 (Illumina). RNA-seq data were initially analyzed using MultiQC for quality control (Ewels et al., 2016). Reads were aligned using HISAT2 to the UCSC danRer11 assembly to facilitate visual inspection of the genomic signal. Reads were counted in genes annotated in the Ensembl GRCz11.94 GTF file using featureCounts with default parameters with the exception of -s2 to take advantage of strand-specific counting afforded by the library preparation (Liao et al., 2014). Ensembl chromosome IDs in the GTF file were converted to UCSC chromosome names in the assembly based on the md5sum hash of respective chromosome sequences. Differential expression analysis used the DESeq2 v1.20.0 package in the R statistical programming environment (v3.5.1) (Love et al., 2014). Variance was estimated on all assayed genotypes to improve estimation and then the contrast was performed only on Mnpc2+/m and MZnpc2m/m. Genes were deemed significantly differentially expressed if they had an adjusted P-value<0.1. Note that there are genes shown in the MA plot marked as statistically significant but with relatively low log2 fold change. These are genes that had count outliers as detected by DESeq2 that shrank their log2 fold change accordingly due to the lower information content for those genes. Functional enrichment analysis was performed using gene ontology, KEGG pathway and Reactome annotation databases using the clusterProfiler package by providing the up- and downregulated genes separately to the enrichment routines (Yu et al., 2012).
Statistical analyses
All graphs were plotted as mean±s.d. Differences between experimental groups were analyzed by Student's t-test, χ2, Fisher's exact test, Kruskal–Wallis test and one-way ANOVA followed by Tukey's test. P<0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
We thank staff members from NICHD Molecular Genomics Core for their assistance with RNA-seq. We thank Louis Dye from NICHD Microscopy and Imaging Core for assisting with TEM samples processing and imaging. We also thank Wuhong Pei from NHGRI for providing notch3 and msx3 probes for RNA in situ hybridization. Finally, we thank staff members from Charles River Laboratories for daily zebrafish husbandry in the zebrafish facility at NICHD.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: W.-C.T., B.F., C.A.W., F.D.P.; Methodology: W.-C.T., A.J.J.E., C.-H.T.-M., B.F., R.K.D., C.A.W., F.D.P.; Software: R.K.D.; Formal analysis: W.-C.T., A.J.J.E., C.-H.T.-M., B.F., R.K.D., C.A.W., F.D.P.; Investigation: W.-C.T., A.J.J.E., C.-H.T.-M.; Resources: C.-H.T.-M., B.F.; Data curation: B.F., R.K.D., C.A.W.; Writing - original draft: W.-C.T.; Writing - review & editing: B.F., C.A.W., F.D.P.; Supervision: W.-C.T., C.A.W., F.D.P.; Project administration: B.F., C.A.W., F.D.P.; Funding acquisition: F.D.P.
Funding
This research was funded by the intramural research programs of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (ZIAHD008988). Deposited in PMC for release after 12 months.
Data availability
RNA-seq data have been deposited in Gene Expression Omnibus under accession number GSE161867.
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.194258.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.194258.reviewer-comments.pdf
References
- Ando, K., Wang, W., Peng, D., Chiba, A., Lagendijk, A. K., Barske, L., Crump, J. G., Stainier, D. Y. R., Lendahl, U., Koltowska, K.et al. (2019). Peri-arterial specification of vascular mural cells from naive mesenchyme requires Notch signaling. Development 146, dev165589. 10.1242/dev.165589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, M. S. and Goldstein, J. L. (1986). A receptor-mediated pathway for cholesterol homeostasis. Science 232, 34-47. 10.1126/science.3513311 [DOI] [PubMed] [Google Scholar]
- Choi, H. Y., Karten, B., Chan, T., Vance, J. E., Greer, W. L., Heidenreich, R. A., Garver, W. S. and Francis, G. A. (2003). Impaired ABCA1-dependent lipid efflux and hypoalphalipoproteinemia in human Niemann-Pick type C disease. J. Biol. Chem. 278, 32569-32577. 10.1074/jbc.M304553200 [DOI] [PubMed] [Google Scholar]
- Cruz, J. C., Sugii, S., Yu, C. and Chang, T.-Y. (2000). Role of Niemann-Pick type C1 protein in intracellular trafficking of low density lipoprotein-derived cholesterol. J. Biol. Chem. 275, 4013-4021. 10.1074/jbc.275.6.4013 [DOI] [PubMed] [Google Scholar]
- Davidson, C. D., Ali, N. F., Micsenyi, M. C., Stephney, G., Renault, S., Dobrenis, K., Ory, D. S., Vanier, M. T. and Walkley, S. U. (2009). Chronic cyclodextrin treatment of murine Niemann-Pick C disease ameliorates neuronal cholesterol and glycosphingolipid storage and disease progression. PLoS ONE 4, e6951. 10.1371/journal.pone.0006951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande, I., Liang, J., Hedeen, D., Roberts, K. J., Zhang, Y., Ha, B., Latorraca, N. R., Faust, B., Dror, R. O., Beachy, P. A.et al. (2019). Smoothened stimulation by membrane sterols drives Hedgehog pathway activity. Nature 571, 284-288. 10.1038/s41586-019-1355-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despic, V. and Neugebauer, K. M. (2018). RNA tales - how embryos read and discard messages from mom. J. Cell Sci. 131, jcs201996. 10.1242/jcs.201996 [DOI] [PubMed] [Google Scholar]
- Driever, W., Stemple, D., Schier, A. and Solnica-Krezel, L. (1994). Zebrafish: genetic tools for studying vertebrate development. Trends Genet. 10, 152-159. 10.1016/0168-9525(94)90091-4 [DOI] [PubMed] [Google Scholar]
- Eid, W., Dauner, K., Courtney, K. C., Gagnon, A. M., Parks, R. J., Sorisky, A. and Zha, X. (2017). mTORC1 activates SREBP-2 by suppressing cholesterol trafficking to lysosomes in mammalian cells. Proc. Natl Acad. Sci. USA 114, 7999-8004. 10.1073/pnas.1705304114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels, P., Magnusson, M., Lundin, S. and Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047-3048. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farese, R. V., Jr and Herz, J. (1998). Cholesterol metabolism and embryogenesis. Trends Genet. 14, 115-120. 10.1016/S0168-9525(97)01377-2 [DOI] [PubMed] [Google Scholar]
- Feltes, M. K., Gale, S. E., Moores, S., Ory, D. S. and Schaffer, J. E. (2020). Monitoring the itinerary of lysosomal cholesterol in Niemann-Pick Type C1-deficient cells after cyclodextrin treatment. J. Lipid Res. 61, 403-412. 10.1194/jlr.RA119000571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouillade, C., Baron-Menguy, C., Domenga-Denier, V., Thibault, C., Takamiya, K., Huganir, R. and Joutel, A. (2013). Transcriptome analysis for Notch3 target genes identifies Grip2 as a novel regulator of myogenic response in the cerebrovasculature. Arterioscler. Thromb. Vasc. Biol. 33, 76-86. 10.1161/ATVBAHA.112.251736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet, P., Livstone, M. S., Lewis, S. E. and Thomas, P. D. (2011). Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief. Bioinform. 12, 449-462. 10.1093/bib/bbr042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geling, A., Steiner, H., Willem, M., Bally-Cuif, L. and Haass, C. (2002). A γ-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 3, 688-694. 10.1093/embo-reports/kvf124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griese, M., Brasch, F., Aldana, V. R., Cabrera, M. M., Goelnitz, U., Ikonen, E., Karam, B. J., Liebisch, G., Linder, M. D., Lohse, P.et al. (2010). Respiratory disease in Niemann-Pick type C2 is caused by pulmonary alveolar proteinosis. Clin. Genet. 77, 119-130. 10.1111/j.1399-0004.2009.01325.x [DOI] [PubMed] [Google Scholar]
- Horvat, S., McWhir, J. and Rozman, D. (2011). Defects in cholesterol synthesis genes in mouse and in humans: lessons for drug development and safer treatments. Drug Metab. Rev. 43, 69-90. 10.3109/03602532.2010.540580 [DOI] [PubMed] [Google Scholar]
- Hsu, H.-J., Liang, M.-R., Chen, C.-T. and Chung, B.-C. (2006). Pregnenolone stabilizes microtubules and promotes zebrafish embryonic cell movement. Nature 439, 480-483. 10.1038/nature04436 [DOI] [PubMed] [Google Scholar]
- Huang, X., Suyama, K., Buchanan, J., Zhu, A. J. and Scott, M. P. (2005). A Drosophila model of the Niemann-Pick type C lysosome storage disease: dnpc1a is required for molting and sterol homeostasis. Development 132, 5115-5124. 10.1242/dev.02079 [DOI] [PubMed] [Google Scholar]
- Huang, X., Warren, J. T., Buchanan, J., Gilbert, L. I. and Scott, M. P. (2007). Drosophila Niemann-Pick type C-2 genes control sterol homeostasis and steroid biosynthesis: a model of human neurodegenerative disease. Development 134, 3733-3742. 10.1242/dev.004572 [DOI] [PubMed] [Google Scholar]
- Incardona, J. P. and Eaton, S. (2000). Cholesterol in signal transduction. Curr. Opin. Cell Biol. 12, 193-203. 10.1016/S0955-0674(99)00076-9 [DOI] [PubMed] [Google Scholar]
- Kim, Y., Kim, C., Jang, H. Y. and Mook-Jung, I. (2016). Inhibition of cholesterol biosynthesis reduces γ-secretase activity and amyloid-β generation. J. Alzheimer's Dis. 51, 1057-1068. 10.3233/JAD-150982 [DOI] [PubMed] [Google Scholar]
- Kinnebrew, M., Iverson, E. J., Patel, B. B., Pusapati, G. V., Kong, J. H., Johnson, K. A., Luchetti, G., Eckert, K. M., McDonald, J. G., Covey, D. F.et al. (2019). Cholesterol accessibility at the ciliary membrane controls hedgehog signaling. eLife 8, e50051. 10.7554/eLife.50051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, H. J., Abi-Mosleh, L., Wang, M. L., Deisenhofer, J., Goldstein, J. L., Brown, M. S. and Infante, R. E. (2009). Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell 137, 1213-1224. 10.1016/j.cell.2009.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon, Y. G. and Mullins, M. C. (2011). Maternal and zygotic control of zebrafish dorsoventral axial patterning. Annu. Rev. Genet. 45, 357-377. 10.1146/annurev-genet-110410-132517 [DOI] [PubMed] [Google Scholar]
- Lewis, P. M., Dunn, M. P., McMahon, J. A., Logan, M., Martin, J. F., St-Jacques, B. and McMahon, A. P. (2001). Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell 105, 599-612. 10.1016/S0092-8674(01)00369-5 [DOI] [PubMed] [Google Scholar]
- Li, H., Pei, W., Vergarajauregui, S., Zerfas, P. M., Raben, N., Burgess, S. M. and Puertollano, R. (2017). Novel degenerative and developmental defects in a zebrafish model of mucolipidosis type IV. Hum. Mol. Genet. 26, 2701-2718. 10.1093/hmg/ddx158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y., Smyth, G. K. and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923-930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lin, Y., Cai, X., Wang, G., Ouyang, G. and Cao, H. (2018). Model construction of Niemann-Pick type C disease in zebrafish. Biol. Chem. 399, 903-910. 10.1515/hsz-2018-0118 [DOI] [PubMed] [Google Scholar]
- Liu, B., Turley, S. D., Burns, D. K., Miller, A. M., Repa, J. J. and Dietschy, J. M. (2009). Reversal of defective lysosomal transport in NPC disease ameliorates liver dysfunction and neurodegeneration in the npc1-/- mouse. Proc. Natl. Acad. Sci. USA 106, 2377-2382. 10.1073/pnas.0810895106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Zhang, W., Kennard, S., Caldwell, R. B. and Lilly, B. (2010). Notch3 is critical for proper angiogenesis and mural cell investment. Circ. Res. 107, 860-870. 10.1161/CIRCRESAHA.110.218271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus, S. K., Morris, J. A., Carstea, E. D., Gu, J. Z., Cummings, C., Brown, A., Ellison, J., Ohno, K., Rosenfeld, M. A., Tagle, D. A.et al. (1997). Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science 277, 232-235. 10.1126/science.277.5323.232 [DOI] [PubMed] [Google Scholar]
- Love, M. I., Huber, W. and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, F., Liang, Q., Abi-Mosleh, L., Das, A., De Brabander, J. K., Goldstein, J. L. and Brown, M. S. (2015). Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. eLife 4, e12177. 10.7554/eLife.12177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg, Y. W., Xu, Y., Thiessen, K. D. and Kramer, K. L. (2015). Mechanisms of otoconia and otolith development. Dev. Dyn. 244, 239-253. 10.1002/dvdy.24195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, M. and Chitnis, A. B. (2009). Interaction with Notch determines endocytosis of specific Delta ligands in zebrafish neural tissue. Development 136, 197-206. 10.1242/dev.027938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maue, R. A., Burgess, R. W., Wang, B., Wooley, C. M., Seburn, K. L., Vanier, M. T., Rogers, M. A., Chang, C. C., Chang, T.-Y., Harris, B. T.et al. (2012). A novel mouse model of Niemann-Pick type C disease carrying a D1005G-Npc1 mutation comparable to commonly observed human mutations. Hum. Mol. Genet. 21, 730-750. 10.1093/hmg/ddr505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengel, E., Pineda, M., Hendriksz, C. J., Walterfang, M., Torres, J. V. and Kolb, S. A. (2017). Differences in Niemann-Pick disease Type C symptomatology observed in patients of different ages. Mol. Genet. Metab. 120, 180-189. 10.1016/j.ymgme.2016.12.003 [DOI] [PubMed] [Google Scholar]
- Miller, W. L. (1988). Molecular biology of steroid hormone synthesis. Endocr. Rev. 9, 295-318. 10.1210/edrv-9-3-295 [DOI] [PubMed] [Google Scholar]
- Muñana, K. R., Luttgen, P. J., Thrall, M. A., Mitchell, T. W. and Wenger, D. A. (1994). Neurological manifestations of Niemann-Pick disease type C in cats. J. Vet. Intern. Med. 8, 117-121. 10.1111/j.1939-1676.1994.tb03208.x [DOI] [PubMed] [Google Scholar]
- Obniski, R., Sieber, M. and Spradling, A. C. (2018). Dietary lipids modulate Notch signaling and influence adult intestinal development and metabolism in Drosophila. Dev. Cell 47, 98-111.e115. 10.1016/j.devcel.2018.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsauskas-Kuprys, R., Zlobin, A. and Osipo, C. (2013). Gamma secretase inhibitors of Notch signaling. Onco Targets Ther. 6, 943-955. 10.2147/OTT.S33766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory, D. S., Ottinger, E. A., Farhat, N. Y., King, K. A., Jiang, X., Weissfeld, L., Berry-Kravis, E., Davidson, C. D., Bianconi, S., Keener, L. A.et al. (2017). Intrathecal 2-hydroxypropyl-β-cyclodextrin decreases neurological disease progression in Niemann-Pick disease, type C1: a non-randomised, open-label, phase 1-2 trial. Lancet 390, 1758-1768. 10.1016/S0140-6736(17)31465-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, M. C., Mengel, E., Wijburg, F. A., Muller, A., Schwierin, B., Drevon, H., Vanier, M. T. and Pineda, M. (2013). Disease and patient characteristics in NP-C patients: findings from an international disease registry. Orphanet J. Rare Dis. 8, 12. 10.1186/1750-1172-8-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, M. C., Garver, W. S., Giugliani, R., Imrie, J., Jahnova, H., Meaney, F. J., Nadjar, Y., Vanier, M. T., Moneuse, P., Morand, O.et al. (2020). Long-term survival outcomes of patients with Niemann-Pick disease type C receiving miglustat treatment: a large retrospective observational study. J. Inherit. Metab. Dis. 43, 1060-1069. 10.1002/jimd.12245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegri, F. (2003). Maternal factors in zebrafish development. Dev. Dyn. 228, 535-554. 10.1002/dvdy.10390 [DOI] [PubMed] [Google Scholar]
- Pfeffer, S. R. (2019). NPC intracellular cholesterol transporter 1 (NPC1)-mediated cholesterol export from lysosomes. J. Biol. Chem. 294, 1706-1709. 10.1074/jbc.TM118.004165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, X., Lim, C.-H., Ho, S. H.-K., Lee, K.-H. and Jiang, Y.-J. (2009). Temporal Notch activation through Notch1a and Notch3 is required for maintaining zebrafish rhombomere boundaries. Dev. Genes Evol. 219, 339-351. 10.1007/s00427-009-0296-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakura, Y., Shimano, H., Sone, H., Takahashi, A., Inoue, N., Toyoshima, H., Suzuki, S. and Yamada, N. (2001). Sterol regulatory element-binding proteins induce an entire pathway of cholesterol synthesis. Biochem. Biophys. Res. Commun. 286, 176-183. 10.1006/bbrc.2001.5375 [DOI] [PubMed] [Google Scholar]
- Sano, O., Tsujita, M., Shimizu, Y., Kato, R., Kobayashi, A., Kioka, N., Remaley, A. T., Michikawa, M., Ueda, K. and Matsuo, M. (2016). ABCG1 and ABCG4 suppress γ-secretase activity and amyloid β production. PLoS ONE 11, e0155400. 10.1371/journal.pone.0155400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwend, T., Loucks, E. J., Snyder, D. and Ahlgren, S. C. (2011). Requirement of Npc1 and availability of cholesterol for early embryonic cell movements in zebrafish. J. Lipid Res. 52, 1328-1344. 10.1194/jlr.M012377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaran, S. S., Sieger, D., Schröter, C., Czepe, C., Pauly, M.-C., Laplante, M. A., Becker, T. S., Oates, A. C. and Gajewski, M. (2007). Completing the set of h/E(spl) cyclic genes in zebrafish: her12 and her15 reveal novel modes of expression and contribute to the segmentation clock. Dev. Biol. 304, 615-632. 10.1016/j.ydbio.2007.01.004 [DOI] [PubMed] [Google Scholar]
- Sleat, D. E., Wiseman, J. A., El-Banna, M., Price, S. M., Verot, L., Shen, M. M., Tint, G. S., Vanier, M. T., Walkley, S. U. and Lobel, P. (2004). Genetic evidence for nonredundant functional cooperativity between NPC1 and NPC2 in lipid transport. Proc. Natl. Acad. Sci. USA 101, 5886-5891. 10.1073/pnas.0308456101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner, C., Burghammer, M., Busch-Nentwich, E., Berger, J., Schwarz, H., Riekel, C. and Nicolson, T. (2003). Control of crystal size and lattice formation by starmaker in otolith biomineralization. Science 302, 282-286. 10.1126/science.1088443 [DOI] [PubMed] [Google Scholar]
- Stooke-Vaughan, G. A., Obholzer, N. D., Baxendale, S., Megason, S. G. and Whitfield, T. T. (2015). Otolith tethering in the zebrafish otic vesicle requires Otogelin and alpha-Tectorin. Development 142, 1137-1145. 10.1242/dev.116632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, Y., Ninomiya, H., Ohsaki, Y., Higaki, K., Davies, J. P., Ioannou, Y. A. and Ohno, K. (2001). Accumulation of cholera toxin and GM1 ganglioside in the early endosome of Niemann–Pick C1-deficient cells. Proc. Natl Acad. Sci. USA 98, 12391. 10.1073/pnas.221181998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Wloga, D. and Dougan, S. T. (2011). Embryological manipulations in zebrafish. Methods Mol. Biol. 770, 139-184. 10.1007/978-1-61779-210-6_6 [DOI] [PubMed] [Google Scholar]
- te Vruchte, D., Speak, A. O., Wallom, K. L., Al Eisa, N., Smith, D. A., Hendriksz, C. J., Simmons, L., Lachmann, R. H., Cousins, A., Hartung, R.et al. (2014). Relative acidic compartment volume as a lysosomal storage disorder-associated biomarker. J. Clin. Invest. 124, 1320-1328. 10.1172/JCI72835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng, W.-C., Loeb, H. E., Pei, W., Tsai-Morris, C.-H., Xu, L., Cluzeau, C. V., Wassif, C. A., Feldman, B., Burgess, S. M., Pavan, W. J.et al. (2018). Modeling Niemann-Pick disease type C1 in zebrafish: a robust platform for in vivo screening of candidate therapeutic compounds. Dis. Model. Mech. 11, dmm034165. 10.1242/dmm.034165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanier, M. T. (2010). Niemann-Pick disease type C. Orphanet J. Rare Dis. 5, 16. 10.1186/1750-1172-5-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villefranc, J. A., Amigo, J. and Lawson, N. D. (2007). Gateway compatible vectors for analysis of gene function in the zebrafish. Dev. Dyn. 236, 3077-3087. 10.1002/dvdy.21354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vite, C. H., Bagel, J. H., Swain, G. P., Prociuk, M., Sikora, T. U., Stein, V. M., O'Donnell, P., Ruane, T., Ward, S., Crooks, A.et al. (2015). Intracisternal cyclodextrin prevents cerebellar dysfunction and Purkinje cell death in feline Niemann-Pick type C1 disease. Sci. Transl. Med. 7, 276ra226. 10.1126/scitranslmed.3010101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager, K., Zdebik, A. A., Fu, S., Cooper, J. D., Harvey, R. J. and Russell, C. (2016). Neurodegeneration and Epilepsy in a Zebrafish Model of CLN3 Disease (Batten Disease). PLoS ONE 11, e0157365. 10.1371/journal.pone.0157365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Pan, L., Moens, C. B. and Appel, B. (2014). Notch3 establishes brain vascular integrity by regulating pericyte number. Development 141, 307-317. 10.1242/dev.096107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, M. S. (2020a). Substrate recognition and processing by γ-secretase. Biochim. Biophys. Acta Biomembr. 1862, 183016. 10.1016/j.bbamem.2019.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, M. S. (2020b). Unraveling the complexity of γ-secretase. Semin. Cell Dev. Biol. 105, 3-11. 10.1016/j.semcdb.2020.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, X., Tang, J.-J., Peng, C., Wang, Y., Fu, L., Qiu, Z.-P., Xiong, Y., Yang, L.-F., Cui, H.-W., He, X.-L.et al. (2017). Cholesterol modification of smoothened is required for Hedgehog signaling. Mol. Cell 66, 154-162.e110. 10.1016/j.molcel.2017.02.015 [DOI] [PubMed] [Google Scholar]
- Yang, T., Arslanova, D., Xu, X., Li, Y.-M. and Xia, W. (2010). In vivo manifestation of Notch related phenotypes in zebrafish treated with Alzheimer's amyloid reducing γ-secretase inhibitors. J. Neurochem. 113, 1200-1209. 10.1111/j.1471-4159.2010.06681.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G., Wang, L.-G., Han, Y. and He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A J. Integr. Biol. 16, 284-287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaucker, A., Mercurio, S., Sternheim, N., Talbot, W. S. and Marlow, F. L. (2013). notch3 is essential for oligodendrocyte development and vascular integrity in zebrafish. Dis. Model. Mech. 6, 1246-1259. 10.1242/dmm.012005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, T. P., Childs, S., Leu, J. P. and Fishman, M. C. (2001). Gridlock signalling pathway fashions the first embryonic artery. Nature 414, 216-220. 10.1038/35102599 [DOI] [PubMed] [Google Scholar]
- Zizioli, D., Guarienti, M., Tobia, C., Gariano, G., Borsani, G., Bresciani, R., Ronca, R., Giacopuzzi, E., Preti, A., Gaudenzi, G.et al. (2014). Molecular cloning and knockdown of galactocerebrosidase in zebrafish: new insights into the pathogenesis of Krabbe's disease. Biochim. Biophys. Acta 1842, 665-675. 10.1016/j.bbadis.2014.01.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.