Abstract
Pulmonary macrophages play a critical role in the recognition of pathogens, initiation of host defense via inflammation, clearance of pathogens from the airways, and resolution of inflammation. Recently, we have shown a pivotal role for the nuclear factor of activated T-cell cytoplasmic member 3 (NFATc3) transcription factor in modulating pulmonary macrophage function in LPS-induced acute lung injury (ALI) pathogenesis. Although the NFATc proteins are activated primarily by calcineurin-dependent dephosphorylation, here we show that LPS induces posttranslational modification of NFATc3 by polyADP-ribose polymerase 1 (PARP-1)-mediated polyADP-ribosylation. ADP-ribosylated NFATc3 showed increased binding to iNOS and TNFα promoter DNA, thereby increasing downstream gene expression. Inhibitors of PARP-1 decreased LPS-induced NFATc3 ribosylation, target gene promoter binding, and gene expression. LPS increased NFAT luciferase reporter activity in lung macrophages and lung tissue that was inhibited by pretreatment with PARP-1 inhibitors. More importantly, pretreatment of mice with the PARP-1 inhibitor olaparib markedly decreased LPS-induced cytokines, protein extravasation in bronchoalveolar fluid, lung wet-to-dry ratios, and myeloperoxidase activity. Furthermore, PARP-1 inhibitors decreased NF-кB luciferase reporter activity and LPS-induced ALI in NF-кB reporter mice. Thus, our study demonstrates that inhibiting NFATc3 and NF-кB polyADP-ribosylation with PARP-1 inhibitors prevented LPS-induced ALI pathogenesis.
Keywords: Macrophage, PolyADP-ribose polymerase 1, NFATc3, Acute lung injury, Pulmonary edema
Introduction
Nuclear factor of activated T cells (NFATs) are evolutionarily conserved transcription factors that share a common “Rel” homology DNA-binding domain and show active nuclear translocation similar to NF-κB with distinctly different cellular functions [1]. NFATs have been reported to play key roles in regulating T lymphocyte cell functions [1, 2]. Studies from our group and others have identified an important regulatory role for NFATs in innate immune cell functions [3, 4, 5, 6]. The NFAT protein family comprises 5 members, of which the cytoplasmic members NFATc1, NFATc2, NFATc3, and NFATc4 are activated by calcineurin-mediated dephosphorylation, whereas the fifth member, NFAT5, is regulated by changes in osmolarity [7, 8]. We have previously demonstrated that LPS-induced acute lung injury (ALI) pathophysiology is critically dependent on lung macrophages [9, 10]. In addition, LPS-activated NFATc3 upregulated iNOS expression and enhanced macrophage bactericidal activity in mice subjected to cecal ligation and puncture (CLP)-induced polymicrobial abdominal sepsis [3]. Furthermore, LPS-induced iNOS expression was significantly decreased in NFATc3−/− bone marrow-derived macrophages, and subjecting NFATc3−/− mice to CLP-abdominal sepsis resulted in increased bacterial burden in the lungs [3]. Thus, NFATc3−/− mice subjected to CLP-sepsis were expected to show compromised bacterial clearance from the lungs and increased sepsis-induced ALI. In contrast, we have observed that NFATc3-deficient mice subjected to CLP-abdominal sepsis showed decreased ALI, improved alveolar capillary barrier function, increased pulmonary arterial oxygen saturation, and a survival benefit [4]. Similarly, inhibition of NFAT activation by the cell permeable calcineurin peptide inhibitor CP9-ZIZIT mitigated LPS-induced ALI [4]. Thus, decreased severity of lung injury in NFATc3−/− mice during sepsis indicates additional mechanisms of NFATc3 regulation in lung macrophages.
In the current article, we present data that NFATc3 activation in lung- and bone marrow-derived macrophages (BMDMs) is also regulated by posttranslational modification via polyADP-ribose polymerase 1 (PARP-1)-mediated ADP-ribosylation. Protein ADP-ribosylation is a reversible posttranslational modification mediated by the enzymatic addition of polyADP-ribose (PAR) moieties catalyzed by the PARP super family of proteins in the presence of NAD+[11]. PARP-1 cleaves NAD+ into nicotinamide and ADP-ribose during the ribosylation process. The ADP-ribose acceptor proteins include histones, DNA repair enzymes, transcription factors, and PARP-1 itself [12, 13, 14]. While histone ribosylation is associated with chromatin remodeling, the functional significance of NFATc3 ribosylation, in terms of downstream gene regulation, is unknown.
PARP-1 is extensively studied for its role in DNA damage signaling and oxidative stress-induced transcription factor activation, including NF-κB [15, 16, 17]. PolyADP-ribosylated NF-κB showed altered transcriptional activity and effector gene expression levels, including iNOS [17, 18]. PARP-1 activation is implicated in the pathogenesis of several chronic inflammatory pathologies including diabetes, diabetic endothelial dysfunction, arthritis, colitis, dermatitis, and uveitis [19, 20]. Despite the scientific literature and clinical applications for PARP-1 inhibitors in the oncology field, cell-specific molecular mechanisms of PARP-1-associated disease pathologies are unknown. Previous studies have shown that NFATc proteins are ribosylated by PARP-1 in T cells, and ribosylated NFATc2 modulated IL-2/IL-4 expression [21, 22]. Similarly, ADP-ribosylation of NF-κB was correlated with changes in TLR4-mediated NF-κB signaling, including modulation of iNOS [23]. However, the in vivo role of ADP-ribosylated NFAT and NF-κB in LPS-induced ALI has not been established. In the current article, we show that LPS causes NFATc3 ribosylation and that the ribosylated NFATc3 was associated with increased iNOS and TNFα promoter DNA-binding activities and gene expression. Notably, PARP-1-mediated ribosylation of NF-κB and NFATc3 showed an important in vivo role in LPS-induced ALI in mice.
Materials and Methods
Mice
C57BL/6 and NFAT luciferase reporter mice (B6.Cg-Tg(Il2/NFAT-luc)83Rinc/J; JAX stock #006098) were purchased from Jackson Research Laboratories and maintained at the vivarium of the Ohio State University, Columbus, OH, USA. NFAT luciferase reporter mice harbor an IL-2 promoter NFAT consensus sequence conjugated to luciferase reporter [24]. NF-κB reporter mice express Photinus luciferase cDNA under the control of the NF-κB-dependent HIV-1 long terminal repeat promoter, as described previously [25]. Mice aged 8–12 weeks were used in the study. All the animal experiments were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the Ohio State University, protocol no. 2013A00000105-R1.
Cells
BMDMs from C57BL/6/NF-κB reporter mice were isolated, as described earlier [26], and grown in DMEM supplemented with 10% FBS, 50 IU/mL penicillin, and 50 μg/mL streptomycin. As described earlier, total lung leukocytes were isolated by digesting mouse lungs with collagenase/DNase, and macrophages were further purified by adherence [27].
Co-Immunoprecipitation and Western Blot Analysis
Cells were lysed in RIPA lysis buffer (catalog no. 9806; Cell Signaling Technologies) with 1X protease inhibitor cocktail (catalog no. 87786; Thermo Fisher Scientific). Cell lysates containing 300 μg of protein were immunoprecipitated with 10 μg of anti-NFATc3 antibody at 4°C for 16 h and adsorbed onto protein A + G agarose beads at RT for 2 h. Protein A + G beads containing anti-NFATc3 immunoprecipitated proteins were washed once with RIPA buffer and 4 times with 1X PBS. Proteins bound to A + G beads were denatured in Laemmli sample buffer and electrophoresed on 4–20% SDS PAGE gradient gels. Electrophoresed proteins were transferred onto PVDF membranes and immunoblotted with anti-PAR antibodies and HRP-conjugated secondary antibodies. Proteins are detected using a streptavidin-HRP chemiluminescence detection system (catalog no. 34095; Thermo Fisher). Total cellular NFATc3, iNOS, and β-actin protein levels were detected using 4–20% SDS PAGE and by immunoblotting with protein-specific antibodies.
Histone Ribosylation Activity Assay
E. coli LPS (serotype O55:B5 S-form) was purchased from Enzo Life Sciences. PARP-1 inhibitors, olaparib, 3-aminobenzamide, and 1,5-isoquinolinediol, and PARP-2 inhibitor UPF1035 were purchased from Thermo Fisher Scientific. An equal volume of 0.01% DMSO was used as vehicle control in all the experiments. PolyADP-ribosylation activity in macrophages was measured using the Trevigen Universal PARP Assay kit (catalog no. 4677-096-K) that replaced the HAS enzyme with nuclear protein extracts from macrophages stimulated with LPS/PARP inhibitors. Lung macrophages were stimulated with LPS (100 ng/mL) for 0, 1, 4, 16, or 24 h, and total nuclear proteins were used as the enzyme source to measure histone ribosylation. Incorporation of biotinylated PAR onto histone proteins coated onto 96-well plates was measured by colorimetric assay. ADP-ribosylation activity of PARP-1 was expressed as absorbance at 450 nm multiplied by 1,000, and units of PAR incorporated into histone substrate per milligram of nuclear protein were compared between different groups. To determine the effect of inhibitors, lung macrophages were pretreated with PARP-1/2/NF-κB inhibitors for 30 min prior to LPS stimulation for 4 h. PARP-1/2 activity is calculated as units of PAR/mg protein.
NFAT Luciferase Reporter Activity
Lung macrophages stimulated with LPS/PARP-1/PARP-2/NF-κB inhibitors were lysed in Promega reporter lysis buffer (catalog no. E4030), and lung tissue homogenates were prepared in the same buffer using a Qiagen TissueLyser. Luciferase activity was measured by adding 20 μL cell lysate to 100 μL of freshly prepared CycLuc1 (AOBIOUS, catalog no. AOB 1117) luciferin substrate using a Promega GloMax luminometer. Luciferase activity was expressed as relative light units per milligram of protein.
NO Measurement
Nitrite concentration in cell culture supernatants was determined using a Griess reagent kit (G-7921; Molecular Probes, Inc.) according to the manufacturer's protocol. Nitrite concentration in the samples was calculated from a standard curve of sodium nitrite.
Measurement of Cytokines
Cytokine release was analyzed using R&D Systems ELISA kits for mouse TNFα (catalog no. MTA00B) and IL-6 (catalog no. M6000B) following the protocols supplied by the manufacturer.
Electrophoretic Mobility Shift Assay
NFATc3 promoter DNA-binding activity was analyzed by electrophoretic mobility shift assay (EMSA) as described earlier [26]. In brief, nuclear extracts from macrophages stimulated with LPS and PARP inhibitors were incubated with biotin-labeled NFATc3 consensus oligonucleotide sequences from the iNOS and TNFα promoters at 4°C for 30 min. The reaction mix was separated on an 8% native polyacrylamide gel, transferred onto Biodyne A membrane (Pierce, catalog no. 77015), and detected using a streptavidin-HRP chemiluminescence detection system (Pierce, catalog no. 34095).
Chromatin Immunoprecipitation
NFATc3 binding to target gene promoters was analyzed by chromatin immunoprecipitation (ChIP) as described earlier [26]. Cells were stimulated with LPS after pretreatment with PARP-1 inhibitor 1,5-isoquinolinediol or 0.01% DMSO for 30 min and cross-linked with 1% formaldehyde for 10 min at room temperature. The cross-linking was stopped by adding 125 mM glycine for 5 min and washed twice with 1X PBS. Cross-linked chromatin was extracted using the ChromaFlash Chromatin extraction kit (Epigentek, catalog no. P-2001-100), and the chromatin was sheared by using a Virtis Virsonic cell disrupter for 10 cycles of 20 s each with a 10-s gap. The DNA-protein complex was immunoprecipitated using a 1:1 mixture of anti-NFATc3 antibodies (Santa Cruz Biotechnology, Inc, catalog nos. sc-23814 and sc-1152) or pre-immune serum as control for each treatment. The immunoprecipitated DNA fragments were washed, de-cross-linked, purified with phenol:chloroform:isoamyl alcohol, and analyzed by qPCR using iNOS promoter primers flanking − 2342 NFAT consensus sequence (forward primer 5′ CTG TTC CCC TTA GGT GGG 3′; reverse primer 5′ ACA GGA GTG TGT GCA TCT G 3′). One percent of chromatin from each treatment was processed along with immunoprecipitated chromatin and used as input control in qPCR. Binding of NFATc3 to the iNOS promoter was calculated using the SABioscience EpiTecChIP qPCR Data Analysis Template (http://www.sabiosciences.com/chippcrarray_data_analysis.php) and presented as % input values.
LPS-Induced ALI Mouse Models
NFAT or NF-κB luciferase mice were pretreated with 10 mg/kg olaparib by intranasal insufflation and, 1 h later, subjected to LPS-induced ALI by a single dose of intraperitoneal LPS (15 mg/kg, dissolved in pharmaceutical grade saline). A dose of LPS 15 mg/kg was required to induce acute neutrophilic lung inflammation as previously described [10]. Control mice received either 0.01% DMSO in saline or 10 mg/kg olaparib dissolved in 0.01% DMSO in saline. Untreated and LPS only-treated mice were included as controls for cytokine/chemokine measurements. ALI severity was measured as described previously [10]. After 16 h of LPS treatment, mice were euthanized, the right bronchus was ligated, and bronchoalveolar fluid (BALF) was collected from the left lung lobe by instilling 750 μL sterile saline 2 times and aspirating as much as possible. Aspirated BALF was centrifuged at 300 g to separate cells, and the supernatant was used for TNFα, IL-6, KC, and extravasated protein measurements. Right lung lobes from different treatment groups were used for luciferase activity and lung wet-to-dry weight measurements. For macrophage isolation, mice were perfused with 5 mL saline in the right ventricle, and lung tissues were collected and digested with collagenase/DNase to isolate macrophages as previously described [27].
Myeloperoxidase Activity
One of the right lung lobes from right bronchus ligated mice was homogenized and used to assay myeloperoxidase (MPO) activity. MPO activity in lung tissue was assayed using an Abcam MPO assay kit (catalog no. ab111749) according to the manufacturer's instructions. MPO activity is expressed as change in fluorescence units per mg protein.
Bioluminescence Imaging of Mice
Whole body bioluminescence imaging of mice from different treatment groups was performed on an IVIS Lumina II (PerkinElmer Inc.) bioluminescence optical imaging system at the small animal imaging core (SAIC) facility of the Ohio State University Wexner Medical Center. Mice were anesthetized, and the hair from the neck-to-abdomen region was removed 12 h before imaging. These mice were injected with 150 mg/kg luciferin (GoldBio, #LUCNA), and then bioluminescence was measured after determining exposure time. NF-κB or NFAT luciferase activity was expressed as photon values per second per square centimeter of chest area of each mouse.
Statistical Analysis
Data are expressed as mean ± SEM and analyzed using GraphPad Prism 5.0. All in vitro experiments utilized pooled macrophages from 4 to 5 mice; each experiment was repeated 3 times. Representative blots of co-immunoprecipitation and immunoblotting are shown. Each in vivo experimental group consisted of 5 mice, and each experiment was repeated 3 times. Statistical differences between treatments were calculated by ANOVA with Bonferroni correction. p values <0.05 were considered statistically significant, as indicated in individual figure legends.
Results
LPS Induces PolyADP-Ribosylation of NFATc3 and Histones in Macrophages
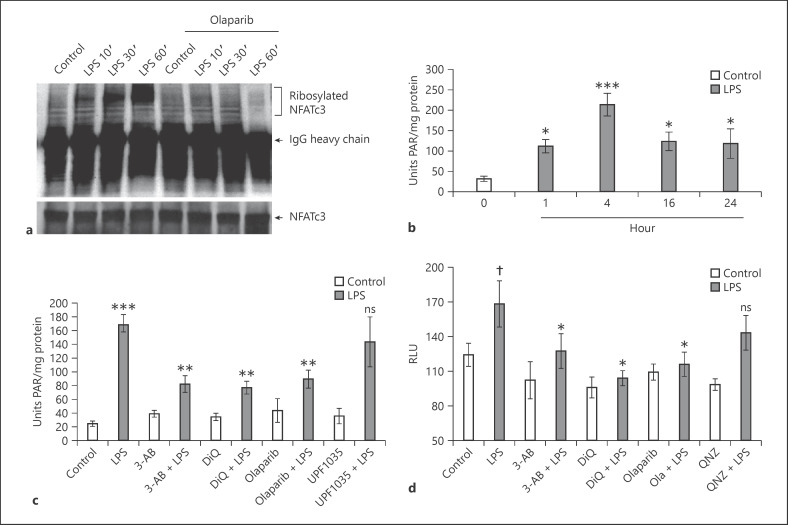
PARP-1 ribosylates different protein substrates, including histones, nuclear proteins, and transcription factors. To determine if LPS activates NFATc3 ribosylation, total cellular proteins from BMDMs were immunoprecipitated with anti-NFATc3 and immunoblotted with PAR antibodies. Levels of ribosylated NFATc3 increased with LPS treatment in a time-dependent manner, whereas 10 μM of PARP-1 inhibitor olaparib significantly decreased NFATc3 ribosylation (Fig. 1a). Next, lung macrophages were stimulated with LPS for 0, 1, 4, 16 and 24 h; total nuclear proteins were used as the enzymatic source to measure histone ribosylation using a Trevigen PARP assay kit. LPS increased histone ribosylation for up to 4 h, which then decreased slightly and remained stable for up to 24 h (Fig. 1b). PARP-1 activity was reduced to ∼50% when cells were pretreated with various PARP-1 inhibitors: 3-aminobenzamide, 1,5-isoquinolinediol, and olaparib (Fig. 1c). In contrast, the PARP-2 inhibitor UPF1035 showed no effect on LPS-induced histone ribosylation activity, thus indicating that the majority of cellular ADP-ribosylation activity is contributed by PARP-1 (Fig. 1c).
Fig. 1.
LPS increases PARP-1-mediated polyADP-ribosylation of NFATc3 and histones. a Mouse BMDMs pretreated with 5 μM olaparib or equal volume of 0.01% DMSO (control) were stimulated with LPS (100 ng/mL) for different time periods. Total cellular proteins were immunoprecipitated with anti-NFATc3 and immunoblotted with PAR and NFATc3 antibody separately. b–cLung macrophages were stimulated with LPS (100 ng/mL) for 0, 1, 4, 16, and 24 h. c Pretreated with PARP-1 and PARP-2 inhibitors separately and then stimulated with LPS for 4 h. Total nuclear proteins were used as an enzyme source to measure histone ribosylation. d Lung macrophages from NFAT luciferase reporter mice were pretreated with PARP-1/NF-κB inhibitors and stimulated with LPS for 4 h. Cell lysates were assayed for luciferase activity. Statistical difference between different treatments was calculated by 1-way ANOVA with Bonferroni correction. bp ≤ 0.001 vehicle + LPS versus vehicle control alone; *p ≤ 0.05 LPS 1, 16, 24 versus 4 h. c ***p ≤ 0.001 vehicle + LPS versus vehicle control alone; **p ≤ 0.05 PARP-1/2 inhibitors + LPS versus vehicle + LPS. d †p ≤ 0.05 vehicle + LPS versus vehicle control alone; *p ≤ 0.05 PARP-1/NF-κB inhibitors + LPS versus vehicle + LPS. PARP-1, polyADP-ribose polymerase 1; NFAT, nuclear factor of activated T cell; NFATc3, nuclear factor of activated T-cell cytoplasmic member 3; PAR, polyADP-ribose; 3-AB, 3-aminobenzamide; DiQ, 1,5-isoquinolinediol; RLU, relative light units; BMDMs, bone marrow-derived macrophages.
NFATc3 Ribosylation Modulates NFATc3 Binding to Target Gene Promoters and Downstream Gene Expression
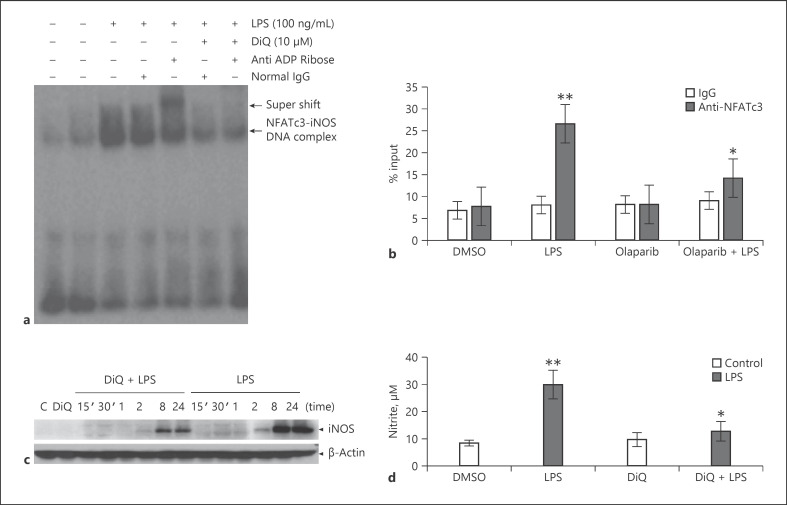
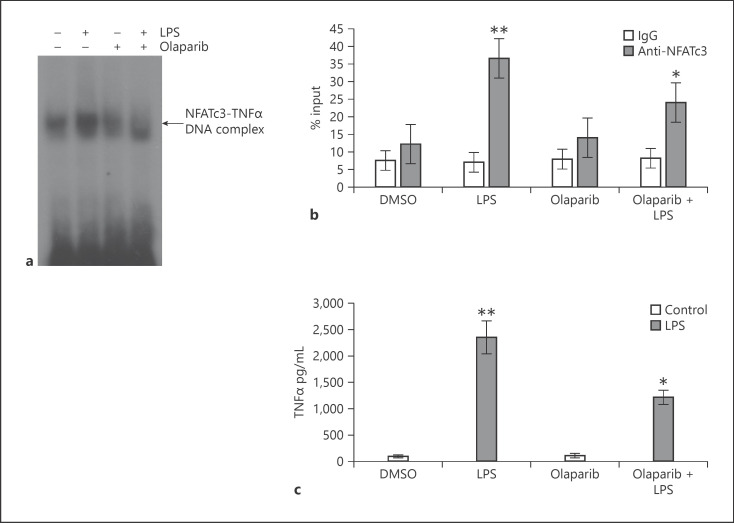
The genomic sequence containing the promoter region of mouse iNOS and TNFα mRNA was downloaded from the National Center for Biotechnology Information (NCBI) database. Potential NFAT-binding sites were identified by analysis of the promoter region using the online software MatInspector [28]. The iNOS gene promoter harbors 3 NFAT-binding elements at −2,409, −2,342, and −754 bp upstream of the transcription start site. In vitro binding of ribosylated NFATc3 to the iNOS gene promoter was observed only at the −2,342 region, which increased with LPS (lane 3 vs. 2, Fig. 2a). Specificity of NAFTc3 binding was confirmed by using an anti-ADP-ribose antibody in EMSA. As shown, the anti-ADP-ribose antibody formed a higher molecular mass complex of iNOS-promoter DNA oligo-NFATc3 protein than normal IgG (lane 5 vs. 4; Fig. 2a). Further, ChIP assay with anti-NFATc3 showed that LPS caused a significant increase in NFATc3 binding to the −2,342 NFAT consensus sequence in iNOS promoter, and pretreatment with olaparib inhibited NFATc3 promoter DNA binding (Fig. 2b). Increase in iNOS promoter DNA binding directly correlated with an increase in iNOS protein and NO levels (Fig. 2c, d). Similarly, LPS treatment showed increased binding of NFAT with TNFα promoter assayed by EMSA and ChIP (Fig. 3a, b). PARP-1 inhibitors decreased both DNA binding and TNFα secretion by macrophages (Fig. 3a–c).
Fig. 2.
NFATc3 ribosylation increases binding of NFATc3 to iNOS gene promoter and gene expression. (a) Nuclear extracts from vehicle and LPS-stimulated BMDMs were incubated with a biotin-labeled NFAT consensus sequence from the iNOS promoter; separate binding reactions with DiQ, normal rabbit IgG, or anti-rabbit ADP-ribose antibody were analyzed by EMSA, and a representative blot is presented. (b) Macrophages were pretreated with vehicle (0.01% DMSO) or DiQ for 30 min and stimulated with LPS for 2 h. Treated cells were cross-linked with 1% formaldehyde, immunoprecipitated with anti-NAFTc3 or pre-immune serum, purified and used to amplify NFATc3 bound DNA on iNOS promoter by qPCR. (c) Expression levels of iNOS in vehicle or PARP-1 inhibitor pretreated cells that were stimulated with LPS for different time periods were determined by immunoblotting with anti-iNOS antibodies; (d) Release of NO into culture medium was measured by Greiss reagent. Statistical difference between different groups was calculated by 1-way ANOVA with Bonferroni correction. **p ≤ 0.01 LPS versus vehicle; *p ≤ 0.05 vehicle + LPS versus PARP-1 inhibitor + LPS. PARP-1, polyADP-ribose polymerase 1; NFATc3, nuclear factor of activated T-cell cytoplasmic member 3; BMDMs, bone marrow-derived macrophages; EMSA, electrophoretic mobility shift assay; DiQ, 1,5-isoquinolinediol; NFAT, nuclear factor of activated T cell.
Fig. 3.
NFATc3 ribosylation increases binding of NFATc3 to TNFα gene promoter and gene expression. (a) Nuclear extracts from vehicle and LPS-stimulated BMDMs were incubated with a biotin-labeled NFAT consensus sequence from the TNFα promoter. Separate binding reactions with vehicle or olaparib were analyzed by EMSA, and a representative blot is presented. (b) Macrophages were pretreated with vehicle or olaparib for 30 min and stimulated with LPS for 2 h. LPS- or inhibitor-treated cells were cross-linked with 1% formaldehyde and immunoprecipitated with anti-NFATc3 or pre-immune serum. NFATc3 cross-linked chromatin was processed and used for PCR to amplify NFATc3-bound DNA on TNFα promoter. (c) Expression levels of TNFα in vehicle or olaparib-pretreated and LPS-stimulated (8 h) cells were determined by ELISA. Statistical differences between different groups were calculated by 1-way ANOVA with Bonferroni correction. **p ≤ 0.01 LPS versus vehicle; *p ≤ 0.05 vehicle + LPS versus olaparib + LPS. NFATc3, nuclear factor of activated T-cell cytoplasmic member 3; BMDMs, bone marrow-derived macrophages; EMSA, electrophoretic mobility shift assay; NFAT, nuclear factor of activated T cell.
Prophylactic Effect of PARP-1 Inhibitors on LPS-Induced ALI
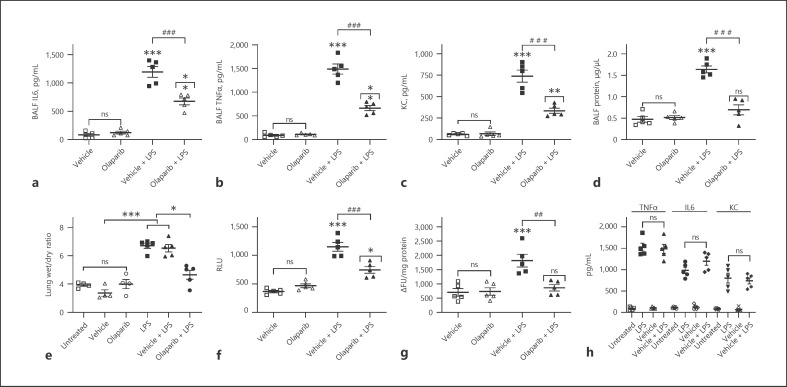
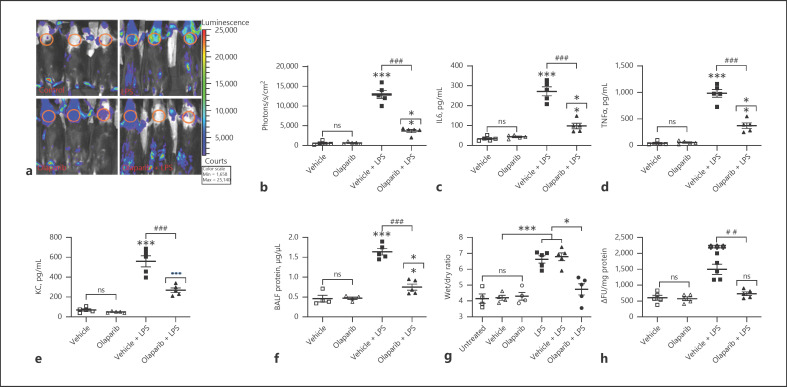
LPS-induced ALI severity was determined in mice pretreated with 10 mg/kg olaparib for 1 h and subjected to a single intraperitoneal dose of LPS (15 mg/kg) for 16 h. LPS-treated NFAT luciferase reporter mice showed increased lung inflammation measured in terms of BALF IL-6, TNFα, and KC levels (Fig. 4a–c). Furthermore, LPS-induced ALI increased pulmonary edema as measured by protein extravasation in BALF, lung wet/dry ratios (Fig. 4d, e), and lung tissue luciferase reporter and MPO activities (Fig. 4f, g). LPS-induced ALI and pulmonary edema were consistently inhibited by pretreatment with olaparib (Fig. 4a–g). In addition, TNFα, IL-6, and KC levels and lung wet-to-dry ratios were not statistically different between untreated and vehicle groups of mice or LPS only and vehicle + LPS groups of mice (Fig. 4h, e). Similarly, NF-κB reporter mice showed increased BALF IL-6, TNFα, and KC levels; protein; lung wet/dry ratios; and tissue luciferase and MPO activities that were abrogated by PARP-1 inhibitor olaparib (Fig. 5c–h). In addition, the lung wet-to-dry ratios were similar in untreated and vehicle-treated NF-κB luciferase reporter mice subjected to LPS-induced ALI (Fig. 5g). Furthermore, NF-κB reporter activity in LPS-challenged mouse lungs increased significantly as measured by the increase in photons per cm2 area of lung area per minute using an IVIS bioluminescence imaging system (Fig. 5a, b). However, NFAT luciferase mice did not show strong bioluminescence signals under the IVIS imaging system, possibly because of the weak reporter activity associated with the IL-2 promoter linked NFAT consensus sequence used in generating NFAT luciferase mice (online suppl. Fig. 1, see www.karger.com/doi/10.1159/000510269). Lung tissue luciferase activity was moderately inducible in NFAT luciferase mice subjected to LPS-induced ALI, which was inhibited by pretreatment with PARP-1 inhibitor (Fig. 4f). In summary, the current study convincingly demonstrates the in vivo role of NFATc3 and NF-κB ribosylation in accentuating ALI pathogenesis in a murine LPS-induced ALI model. More importantly, LPS-induced ALI severity was prophylactically inhibited by pretreatment of NFAT and NF-κB luciferase mice with PARP-1 inhibitors.
Fig. 4.
Prophylactic inhibition of NFAT polyADP-ribosylation by olaparib inhibits LPS-induced ALI and pulmonary edema in NFAT luciferase mice. NFAT reporter mice were untreated (naïve), pretreated with 0.01% DMSO or olaparib (10 mg/kg) for 1 h, followed by intraperitoneal LPS (15 mg/kg) injection. At 16 h after LPS, ALI and pulmonary edema in the treated mice were determined by measuring BALF levels of IL-6 (a), TNFα (b), KC (c), extravasated protein (d), lung wet/dry ratios (e), and lung tissue luciferase (f) and MPO activities (g). Similarly, ALI and pulmonary edema were compared in untreated or 0.01% DMSO-pretreated (1 h) NFAT luciferase subjected to LPS-induced ALI by measuring BALF levels of IL-6, TNFα, and KC (h). The bars represent the mean ± SEM from 5 mice in each group. Statistical difference between different treatments was calculated by 1-way ANOVA with Bonferroni correction. ***p ≤ 0.001 vehicle + LPS or LPS alone versus vehicle or untreated control; ###p ≤ 0.001 vehicle + LPS versus olaparib + LPS; ##p ≤ 0.01 vehicle + LPS versus olaparib + LPS; **p ≤ 0.01 olaparib versus olaparib + LPS; *p ≤ 0.05 olaparib versus olaparib + LPS (f) or untreated or vehicle + LPS versus olaparib + LPS (e). NFAT, nuclear factor of activated T cell; BALF, bronchoalveolar fluid; ALI, acute lung injury; MPO, myeloperoxidase; RLU, relative light units.
Fig. 5.
Prophylactic inhibition of NF-κB polyADP-ribosylation by olaparib inhibits LPS-induced ALI and pulmonary edema in NF-κB luciferase mice. NF-κB reporter mice were untreated, pretreated with 0.01% DMSO or olaparib (10 mg/kg) for 1 h, followed by intraperitoneal LPS (15 mg/kg) i.p. injection. At 16 h after LPS, ALI and pulmonary edema in the treated mice were measured by (a) bioluminescence imaging of the animals using an IVIS Lumina II imaging system. The circled chest area represents the region of interest for comparing the photon readings. b In vivo pulmonary inflammation was measured as photons emitted per second per cm2 mouse lung area. Similarly, parallel groups of NF-κB reporter mice were treated as described in Fig. 4, and BALF levels of IL-6 (c), TNFα (d), and KC (e), BALF protein (f), lung wet/dry ratios (g), and MPO levels in lung tissue were measured (h). Error bars represent mean ± SEM from 5 mice in each group. Statistical difference between different treatments was calculated by 1-way ANOVA with Bonferroni correction. ***p ≤ 0.001 vehicle + LPS or LPS alone versus vehicle or untreated control; ###p ≤ 0.001 vehicle + LPS versus olaparib + LPS; ##p ≤ 0.01 vehicle + LPS versus olaparib + LPS; **p ≤ 0.01 olaparib versus olaparib + LPS; •••p ≤ 0.001 olaparib versus olaparib + LPS; *p ≤ 0.05 untreated or vehicle + LPS versus olaparib + LPS (g). ALI, acute lung injury; BALF, bronchoalveolar fluid; MPO, myeloperoxidase.
Discussion
Studies from different cell types have implicated the transcription factors NF-κB, AP1, CREB, c/EBP, and IRF4/5 in inflammatory signaling pathways [29, 30, 31]. Recent data suggest that NFAT proteins also play critical roles in innate immune regulation [32, 33]. Previous work by our group has demonstrated that macrophage-specific NFATc3 activation plays an important role in ALI pathophysiology [4]. While calcineurin-mediated dephosphorylation is the primary mechanism of activation of cytoplasmic NFATs, other posttranslational modifications such as polyADP-ribosylation, SUMOylation, and acetylation were known to modulate their activity [34, 35, 36]. Similarly to NFATs, cytoplasmic signaling events that control activation of RelA by phosphorylation are extensively studied. Intranuclear or cytoplasmic protein modifications, including acetylation, methylation, SUMOylation, ubiquitination, and nitrosylation also play a major role in the regulation of NF-κB activity in different cell types [37]. More importantly, the protein modifications can vary depending on the nature of the NF-κB-inducing stimulus, and these modifications can have different effects in different cells. For example, studies indicate that in vivo acetylation of RelA at lysine 221 enhances DNA binding and impairs assembly with I-κBα, while acetylation of lysine 310 is required for full transcriptional activity of RelA independent of changes in DNA binding or IκBα binding [38]. Deacetylation of lysine 221 by HDAC3 promotes high-affinity binding of RelA to newly synthesized IκBα proteins and rapid nuclear export of the NF-κB complex [38]. The complexity of posttranslational modifications depending on cell type and stimulation explains the diverse functions of a single protein under normal and pathophysiological conditions. In this regard, we have determined the in vivo role of polyADP-ribosylated NFATc3 in pulmonary macrophages and BMDMs during LPS-induced ALI pathogenesis. We have previously reported that LPS causes dephosphorylation of NFATc3 that translocates to the nucleus of lung macrophages, thereby regulating macrophage inflammatory genes [3, 4]. In the current study, co-immunoprecipitation of ribosylated NFATc3 with anti-ADP ribose strongly suggests that PARP-1 interacts with transcription factor NFATc3 and ribosylates NFATc3 in mouse primary macrophages. We have also demonstrated that ADP-ribosylation of NFATc3 altered its function by increased binding to iNOS and TNFα gene promoters. LPS increased histone ribosylation in parallel with NFATc3, and this was abolished by PARP-1 inhibitors. Both the histone ribosylation and NFAT luciferase reporter activity were unaffected by PARP-2 inhibitor U1035, thus indicating that PARP-1 is the major NFATc3 ribosylating factor.
Studies from different research groups have indicated that LPS-induced ALI is associated with an increase in PARP-1 activation and inhibition of PARP-1 decreased the ALI severity [39, 40]. However, these studies have primarily implicated cellular DNA damage responses, NF-κB-mediated inflammation, and oxidative stress as the causative factors of PARP-1-mediated ALI [39, 40, 41]. In a study by Zerfaoui et al. [23], ADP-ribosylation of RelA/p65 was found to be essential for the interaction of RelA/p65 with a nuclear export protein Crm1 (exportin 1) to promote the nuclear retention of NF-κB, thereby regulating downstream cytokine gene expression in smooth muscle cells. Nuclear translocation of NF-κB decreased with the inhibition of PARP-1 in this study [23]. Interestingly, a coordinated interaction of NFATs and NF-κB is also implicated in the upregulation of cyclooxygenase 2 gene expression in BEAS-2B cells [42]. Our data show that NF-κB inhibitor QNZ had no effect on LPS-induced NFAT luciferase reporter activity, ruling out the cross-talk between NFAT and NF-κB. Different PARP-1 inhibitors decreased both the NFAT luciferase and ribosylation activities. Interestingly, both PARP-1 and NF-κB inhibitors decreased LPS-induced NF-κB luciferase reporter activity. Further studies are required to determine the cross-talk between NFAT and NF-κB in LPS-regulated macrophage gene expression, as the TNFα and iNOS promoters harbor NF-κB and NFAT cis binding elements and are regulated by both the transcription factors [43, 44].
Our recent studies demonstrated that inhibition of NFATc3 activation by calcineurin inhibitor cyclopsorin or NFATc-specific competitive peptide 11-R VIVIT decreased expression of iNOS, leading to compromised bacterial clearence during CLP-mediated abdominal sepsis in NFATc3−/− mice [3]. However, the use of the broad-spectrum antibiotic imipenem before CLP in NFATc3−/− mice significantly improved their survival [4]. Similar to the protective effect observed in NFATc3−/− or cell permeable 11-R VIVIT peptide-treated mice, inhibition of NFATc3 ADP-ribosylation by PARP-1 inhibitor olaparib decreased LPS-induced BALF cytokine/chemokine levels, BALF protein leakage, and lung wet/dry ratios, thus advocating the beneficial effects of PARP-1 inhibition in ameliorating LPS-induced ALI severity. Together, these results demonstrate that ADP-ribosylation mediated by PARP-1 is an additional mechanism to regulate NFATc3-dependent gene transcription. These results also imply that, similar to calcineurin inhibition, targeting PARP-1 by pharmacological inhibitors would be beneficial in modulating macrophage functions in ALI pathophysiology.
Statement of Ethics
All the in vivo experiments using mice were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the Ohio State University (protocol no. 2013A00000105-R1).
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
This work was supported by grants from the National Institutes of Health 5R01HL137224-JWC, American Heart Association 17SDG33410982-MK.
Author Contributions
M.K. and J.W.C. designed the study, analyzed the data, and wrote the manuscript. Y.N., T.S.N., R.R., B.F.R., S.C., J.A.E. and M.N.B. acquired and analyzed the data.
Supplementary Material
Supplementary data
References
- 1.Serfling E, Berberich-Siebelt F, Avots A, Chuvpilo S, Klein-Hessling S, Jha MK, et al. NFAT and NF-kappaB factors-the distant relatives. Int J Biochem Cell Biol. 2004 Jul;36((7)):1166–70. doi: 10.1016/j.biocel.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Hogan PG. Calcium-NFAT transcriptional signalling in T cell activation and T cell exhaustion. Cell Calcium. 2017 May;63:66–9. doi: 10.1016/j.ceca.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ranjan R, Deng J, Chung S, Lee YG, Park GY, Xiao L, et al. The transcription factor nuclear factor of activated T cells c3 modulates the function of macrophages in sepsis. J Innate Immun. 2014;6((6)):754–64. doi: 10.1159/000362647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karpurapu M, Lee YG, Qian Z, Wen J, Ballinger MN, Rusu L, et al. Inhibition of nuclear factor of activated T cells (NFAT) c3 activation attenuates acute lung injury and pulmonary edema in murine models of sepsis. Oncotarget. 2018 Jan 25;9((12)):10606–20. doi: 10.18632/oncotarget.24320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zanoni I, Granucci F. Regulation and dysregulation of innate immunity by NFAT signaling downstream of pattern recognition receptors (PRRs) Eur J Immunol. 2012 Aug;42((8)):1924–31. doi: 10.1002/eji.201242580. [DOI] [PubMed] [Google Scholar]
- 6.Minematsu H, Shin MJ, Celil Aydemir AB, Kim KO, Nizami SA, Chung GJ, et al. Nuclear presence of nuclear factor of activated T cells (NFAT) c3 and c4 is required for toll-like receptor-activated innate inflammatory response of monocytes/macrophages. Cell Signal. 2011 Nov;23((11)):1785–93. doi: 10.1016/j.cellsig.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horsley V, Pavlath GK. NFAT: ubiquitous regulator of cell differentiation and adaptation. J Cell Biol. 2002 Mar 4;156((5)):771–4. doi: 10.1083/jcb.200111073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JH, Kim M, Im YS, Choi W, Byeon SH, Lee HK. NFAT5 induction and its role in hyperosmolar stressed human limbal epithelial cells. Invest Ophthalmol Vis Sci. 2008 May;49((5)):1827–35. doi: 10.1167/iovs.07-1142. [DOI] [PubMed] [Google Scholar]
- 9.Koay MA, Gao X, Washington MK, Parman KS, Sadikot RT, Blackwell TS, et al. Macrophages are necessary for maximal nuclear factor-kappa B activation in response to endotoxin. Am J Respir Cell Mol Biol. 2002 May;26((5)):572–8. doi: 10.1165/ajrcmb.26.5.4748. [DOI] [PubMed] [Google Scholar]
- 10.Karpurapu M, Wang X, Deng J, Park H, Xiao L, Sadikot RT, et al. Functional PU.1 in macrophages has a pivotal role in NF-κB activation and neutrophilic lung inflammation during endotoxemia. Blood. 2011 Nov 10;118((19)):5255–66. doi: 10.1182/blood-2011-03-341123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amé JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26((8)):882–93. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Zamudio R, Ha HC. Histone ADP-ribosylation facilitates gene transcription by directly remodeling nucleosomes. Mol Cell Biol. 2012 Jul;32((13)):2490–502. doi: 10.1128/MCB.06667-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kameoka M, Ota K, Tetsuka T, Tanaka Y, Itaya A, Okamoto T, et al. Evidence for regulation of NF-kappaB by poly(ADP-ribose) polymerase. Biochem J. 2000;346((Pt 3)):641–9. [PMC free article] [PubMed] [Google Scholar]
- 14.Tao Z, Gao P, Liu HW. Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. J Am Chem Soc. 2009 Oct 14;131((40)):14258–60. doi: 10.1021/ja906135d. [DOI] [PubMed] [Google Scholar]
- 15.Pascal JM. The comings and goings of PARP-1 in response to DNA damage. DNA Repair. 2018 Nov;71:177–82. doi: 10.1016/j.dnarep.2018.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langelier MF, Eisemann T, Riccio AA, Pascal JM. PARP family enzymes: regulation and catalysis of the poly(ADP-ribose) posttranslational modification. Curr Opin Struct Biol. 2018 Dec;53:187–98. doi: 10.1016/j.sbi.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang WJ, Alvarez-Gonzalez R. The sequence-specific DNA binding of NF-kappa B is reversibly regulated by the automodification reaction of poly (ADP-ribose) polymerase 1. J Biol Chem. 2001;276((50)):47664–70. doi: 10.1074/jbc.M104666200. [DOI] [PubMed] [Google Scholar]
- 18.Le Page C, Sanceau J, Drapier JC, Wietzerbin J. Inhibitors of ADP-ribosylation impair inducible nitric oxide synthase gene transcription through inhibition of NF kappa B activation. Biochem Biophys Res Commun. 1998 Feb 13;243((2)):451–7. doi: 10.1006/bbrc.1998.8113. [DOI] [PubMed] [Google Scholar]
- 19.Berger NA, Besson VC, Boulares AH, Bürkle A, Chiarugi A, Clark RS, et al. Opportunities for the repurposing of PARP inhibitors for the therapy of non-oncological diseases. Br J Pharmacol. 2018 Jan;175((2)):192–222. doi: 10.1111/bph.13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morales J, Li L, Fattah FJ, Dong Y, Bey EA, Patel M, et al. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit Rev Eukaryot Gene Expr. 2014;24((1)):15–28. doi: 10.1615/critreveukaryotgeneexpr.2013006875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valdor R, Schreiber V, Saenz L, Martínez T, Muñoz-Suano A, Dominguez-Villar M, et al. Regulation of NFAT by poly(ADP-ribose) polymerase activity in T cells. Mol Immunol. 2008 Apr;45((7)):1863–71. doi: 10.1016/j.molimm.2007.10.044. [DOI] [PubMed] [Google Scholar]
- 22.Olabisi OA, Soto-Nieves N, Nieves E, Yang TT, Yang X, Yu RY, et al. Regulation of transcription factor NFAT by ADP-ribosylation. Mol Cell Biol. 2008 May;28((9)):2860–71. doi: 10.1128/MCB.01746-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zerfaoui M, Errami Y, Naura AS, Suzuki Y, Kim H, Ju J, et al. Poly(ADP-ribose) polymerase-1 is a determining factor in Crm1-mediated nuclear export and retention of p65 NF-kappa B upon TLR4 stimulation. J Immunol. 2010 Aug 1;185((3)):1894–902. doi: 10.4049/jimmunol.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rincon M, Flavell RA. Regulation of AP-1 and NFAT transcription factors during thymic selection of T cells. Mol Cell Biol. 1996 Mar;16((3)):1074–84. doi: 10.1128/mcb.16.3.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yull FE, Han W, Jansen ED, Everhart MB, Sadikot RT, Christman JW, et al. Bioluminescent detection of endotoxin effects on HIV-1 LTR-driven transcription in vivo. J Histochem Cytochem. 2003 Jun;51((6)):741–9. doi: 10.1177/002215540305100605. [DOI] [PubMed] [Google Scholar]
- 26.Karpurapu M, Ranjan R, Deng J, Chung S, Lee YG, Xiao L, et al. Krüppel like factor 4 promoter undergoes active demethylation during monocyte/macrophage differentiation. PLoS One. 2014 Apr 2;9((4)):e93362. doi: 10.1371/journal.pone.0093362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ojielo CI, Cooke K, Mancuso P, Standiford TJ, Olkiewicz KM, Clouthier S, et al. Defective phagocytosis and clearance of Pseudomonas aeruginosa in the lung following bone marrow transplantation. J Immunol. 2003 Oct 15;171((8)):4416–24. doi: 10.4049/jimmunol.171.8.4416. [DOI] [PubMed] [Google Scholar]
- 28.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21((13)):2933–42. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 29.Liu T, Zhang L, Joo D, Sun SC. NF-κB signaling in inflammation. Signal Transduct Target Ther. 2017;2:17023. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen AY, Sakamoto KM, Miller LS. The role of the transcription factor CREB in immune function. J Immunol. 2010 Dec 1;185((11)):6413–9. doi: 10.4049/jimmunol.1001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chistiakov DA, Myasoedova VA, Revin VV, Orekhov AN, Bobryshev YV. The impact of interferon-regulatory factors to macrophage differentiation and polarization into M1 and M2. Immunobiology. 2018 Jan;223((1)):101–11. doi: 10.1016/j.imbio.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Fric J, Zelante T, Wong AY, Mertes A, Yu HB, Ricciardi-Castagnoli P. NFAT control of innate immunity. Blood. 2012 Aug 16;120((7)):1380–9. doi: 10.1182/blood-2012-02-404475. [DOI] [PubMed] [Google Scholar]
- 33.Herbst S, Shah A, Mazon Moya M, Marzola V, Jensen B, Reed A, et al. Phagocytosis-dependent activation of a TLR9-BTK-calcineurin-NFAT pathway co-ordinates innate immunity to Aspergillus fumigatus. EMBO Mol Med. 2015 Mar;7((3)):240–58. doi: 10.15252/emmm.201404556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim ET, Kwon KM, Lee MK, Park J, Ahn JH. Sumoylation of a small isoform of NFATc1 is promoted by PIAS proteins and inhibits transactivation activity. Biochem Biophys Res Commun. 2019 Apr 2;513((1)):172–178. doi: 10.1016/j.bbrc.2019.03.171. [DOI] [PubMed] [Google Scholar]
- 35.Vihma H, Timmusk T. Sumoylation regulates the transcriptional activity of different human NFAT isoforms in neurons. Neurosci Lett. 2017 Jul 13;653:302–7. doi: 10.1016/j.neulet.2017.05.074. [DOI] [PubMed] [Google Scholar]
- 36.Meissner JD, Freund R, Krone D, Umeda PK, Chang KC, Gros G, et al. Extracellular signal-regulated kinase 1/2-mediated phosphorylation of p300 enhances myosin heavy chain I/beta gene expression via acetylation of nuclear factor of activated T cells c1. Nucleic Acids Res. 2011 Aug;39((14)):5907–25. doi: 10.1093/nar/gkr162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006 Oct 30;25((51)):6717–30. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 38.Chen Lf L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001 Aug 31;293((5535)):1653–7. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 39.Liaudet L, Pacher P, Mabley JG, Virág L, Soriano FG, Haskó G, et al. Activation of poly(ADP-ribose) polymerase-1 is a central mechanism of lipopolysaccharide-induced acute lung inflammation. Am J Respir Crit Care Med. 2002;165((3)):372–7. doi: 10.1164/ajrccm.165.3.2106050. [DOI] [PubMed] [Google Scholar]
- 40.Kapoor K, Singla E, Sahu B, Naura AS. PARP inhibitor, olaparib ameliorates acute lung and kidney injury upon intratracheal administration of LPS in mice. Mol Cell Biochem. 2015 Feb;400((1–2)):153–62. doi: 10.1007/s11010-014-2271-4. [DOI] [PubMed] [Google Scholar]
- 41.Wang G, Huang X, Li Y, Guo K, Ning P, Zhang Y. PARP-1 inhibitor, DPQ, attenuates LPS-induced acute lung injury through inhibiting NF-κB-mediated inflammatory response. PLoS One. 2013 Nov 21;8((11)):e79757. doi: 10.1371/journal.pone.0079757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai T, Li X, Ding J, Luo W, Li J, Huang C. A cross-talk between NFAT and NF-κB pathways is crucial for nickel-induced COX-2 expression in Beas-2B cells. Curr Cancer Drug Targets. 2011 Jun;11((5)):548–59. doi: 10.2174/156800911795656001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Obasanjo-Blackshire K, Mesquita R, Jabr RI, Molkentin JD, Hart SL, Marber MS, et al. Calcineurin regulates NFAT-dependent iNOS expression and protection of cardiomyocytes: co-operation with Src tyrosine kinase. Cardiovasc Res. 2006 Sep 1;71((4)):672–83. doi: 10.1016/j.cardiores.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 44.Kleinert H, Schwarz PM, Förstermann U. Regulation of the expression of inducible nitric oxide synthase. Biol Chem. 2003 Oct-Nov;384((10–11)):1343–64. doi: 10.1515/BC.2003.152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data