Abstract
Background
Leiomyosarcoma (LMS), the most common soft tissue sarcoma, exhibits heterogeneous and complex genetic karyotypes with severe chromosomal instability and rearrangement and poor prognosis.
Methods
Clinical variables associated with NKX6-1 were obtained from The Cancer Genome Atlas (TCGA). NKX6-1 mRNA expression was examined in 49 human uterine tissues. The in vitro effects of NXK6-1 in LMS cells were determined by reverse transcriptase PCR, western blotting, colony formation, spheroid formation, and cell viability assays. In vivo tumor growth was evaluated in nude mice.
Results
Using The Cancer Genome Atlas (TCGA) and human uterine tissue datasets, we observed that NKX6-1 expression was associated with poor prognosis and malignant potential in LMS. NKX6-1 enhanced in vitro tumor cell aggressiveness via upregulation of cell proliferation and anchorage-independent growth and promoted in vivo tumor growth. Moreover, overexpression and knockdown of NKX6-1 were associated with upregulation and downregulation, respectively, of stem cell transcription factors, including KLF8, MYC, and CD49F, and affected sphere formation, chemoresistance, NOTCH signaling and Sonic hedgehog (SHH) pathways in human sarcoma cells. Importantly, treatment with an SHH inhibitor (RU-SKI 43) but not a NOTCH inhibitor (DAPT) reduced cell survival in NKX6-1-expressing cancer cells, indicating that an SHH inhibitor could be useful in treating LMS. Finally, using the TCGA dataset, we demonstrated that LMS patients with high expression of NKX6-1 and HHAT, an SHH pathway acyltransferase, had poorer survival outcomes compared to those without.
Conclusions
Our findings indicate that NKX6-1 and HHAT play critical roles in the pathogenesis of LMS and could be promising diagnostic and therapeutic targets for LMS patients.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12929-021-00726-6.
Keywords: Leiomyosarcoma (LMS), NK6 homeobox 1 (NKX6-1), Sonic hedgehog (SHH) signaling, SHH inhibitor, Chemoresistance
Background
Leiomyosarcoma (LMS) is a malignant mesenchymal neoplasm of the smooth muscle lining, occurring most frequently in the uterus, retroperitoneum, extremities, and other primary sites throughout the body [1]. LMS accounts for approximately 24% of soft tissue sarcomas [2] and presents largely as high-grade tumors, which exhibit aggressive behavior and a high propensity for local recurrence and metastasis via hematogenous spread [3]. Uterine LMS accounts for approximately 60% of all uterine sarcomas [4]. LMS is conventionally treated with surgical resection, adjuvant chemotherapy, and radiotherapy [2, 5]. However, systemic treatment for uterine LMS demonstrates only a modest response with approximately 40% metastasis [4, 6] and a 5 year survival rate of approximately 50% in patients with uterus-confined tumors [7]. The median overall survival is less than two years with doxorubicin- or gemcitabine-based therapies [8, 9]. Adjuvant pelvic radiation is not associated with significant improvement in the local control rate [10]. Even with combination regimens, treatment typically fails due to local recurrence and metastasis, resulting in a dismal prognosis. [7]. Such evidence indicates the malignant behavior of LMS and the urgent need for novel treatment options. In addition, the molecular mechanisms of LMS etiology are heterogeneous and are associated with diverse cytogenetic lesions. Consequently, elucidation of LMS pathogenesis is critical, especially considering that no epithelial precursor lesion of origin is known. Since the benefits of chemotherapy and radiotherapy for recurrent and metastatic disease are limited, an in-depth understanding of molecular disease mechanisms may provide a key to new therapeutics.
The NKX family of homeodomain-containing transcription factors is heavily involved in tissue-specific differentiation and development. Accumulating evidence indicates that these transcription factors are associated with a variety of cancers, including sarcomas. More specifically, NKX6-3 depletion results in multiple genetic mutations that drive carcinogenesis of the stomach [11], while NKX2-5 regulates thyroid cell differentiation in thyroid cancer [12]. NKX2-1 mutations have been found in 16% of lung cancers [13] and are associated with lung cancer metastasis [14, 15]. NKX3-1 deletion and promoter hypomethylation have been identified in prostate cancer [16]. The NKX6-1 gene product interacts directly with the oncogenic EWS/ATF-1 chimeric transcription factor in clear cell sarcoma [17]. Moreover, hypermethylation of the NKX6-1 promoter is frequently detected in leukemia, cervical cancer, ovarian cancer and colon cancer [18–21]. Additionally, NKX2-2 was identified as a target of EWS-FLI-1, the fusion protein that is now considered a biomarker for Ewing sarcoma [22]. These results highlighted the critical role of the NKX family in the pathogenesis of cancer development and prompted us to investigate the previously undefined role of the NKX family in the initiation and progression of LMS.
In the present study, we investigated the potential role of the NKX family in LMS using human sarcoma cell lines, The Cancer Genome Atlas datasets, and uterine tissues from 49 patients. Our results demonstrated that NKX6-1 acts through the sonic hedgehog (SHH) pathway to increase cell proliferation, drug resistance, and cancer stemness in vitro and tumorigenicity in vivo. Importantly, inhibition of the SHH pathway significantly inhibits the growth of sarcoma cells with high NKX6-1 expression, indicating possible new treatment options for LMS patients.
Methods
Clinical samples
Tissue samples were collected with the informed consent of patients at the Tri‐Service General Hospital, National Defense Medical Center, Taipei, Taiwan. The samples included 19 LMS, 14 nonmalignant leiomyomas and 16 normal myometrium. These specimens were obtained during surgery, frozen immediately in liquid nitrogen and stored at − 80 °C until analysis. This study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of the Tri-Service General Hospital. All of the patients signed informed consent forms before the study.
Cell lines
Two human uterine sarcoma cell lines (MES-SA and MES-SA/DX5) were obtained from BCRC (Bioresource Collection and Research Center, Taiwan). Cells were cultured at 37 °C and 5% CO2 in McCoy's 5a medium (Invitrogen) supplemented with 10% fetal bovine serum (Biological), 100 U/mL penicillin, and 100 μg/mL streptomycin.
Transfections and NKX6-1 overexpression
Cells were seeded in 24-well plates and were transfected at 50–80% confluence with a NKX6-1 expression vector or with empty vector controls using the liposome-mediated transfection method (Invitrogen). To establish cells with stable expression of NKX6-1, MES-SA cells were transfected with the plasmid of choice (pcDNA3.1/NKX6-1 tag V5) for 2 days and then trypsinized and plated at low density. Stable clones were selected by maintaining cells in medium containing G418 antibiotic (Merck).
RNA interference
Short hairpin RNAs (shRNAs) against human NKX6-1 (TRCN0000017548 and TRCN0000072223) were obtained from the National RNAi Core Facility located at the Institute of Molecular Biology/Genomic Research Center, Academia Sinica, Taiwan. The target sites for the NKX6-1 shRNAs were: 5′‐ CCG GGA AGA CTT TCG AAC AAA CAA ACT CGA GTT TGT TTG TTC GAA AGT CTT CTT TTT‐3′ (TRCN0000017548) and 5′‐ CCG GCC GCT GTA CCC TGC CGC GTA TCT CGA GAT ACG CGG CAG GGT ACA GCG GTT TTT‐3′ (TRCN0000017551). A scrambled shRNA (LacZ) 5′‐TGT TCG CAT TAT CCG AAC CAT‐3′ (TRCN0000072223) was used as a negative control. MES-SA/DX5 cells endogenously expressing NKX6-1 were transfected with different shRNA constructs to evaluate the effects on tumor cells. Stable NKX6-1-knockdown clones were generated by transfecting cells with NKX6-1 or control shRNAs and were selected with puromycin (Invitrogen).
RNA isolation and reverse transcriptase-PCR
Total RNA was extracted from clinical specimens using TRIzol reagent (Invitrogen), and mRNA was isolated from each cell line sample using the Qiagen RNeasy Kit (Qiagen). An additional DNase I digestion step was added to the RNA isolation procedure to remove DNA and was performed according to the manufacturer’s protocol (Qiagen). Five micrograms of total RNA from each sample were subjected to cDNA synthesis using Superscript III reverse transcriptase (Invitrogen) with random hexamer primers (Promega). cDNA was then PCR amplified with primers specific for NKX6-1 and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene using a PCR Master Mix Reagents Kit (RBC Bioscience). After heating at 95 °C for 10 min, PCR was performed in a thermal cycler (Biometra) for 37 cycles, each of which consisted of denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C for 30 s, followed by a final 10 min extension at 72 °C. The PCR products were analyzed by electrophoresis on 2% agarose gels (Bioshop).
Real-time quantitative PCR
Quantitative reverse transcriptase-PCR analysis was performed on a Roche LC480 real-time system. GAPDH was used as an internal control. PCR was performed using a SYBR Green PCR Master Mix Reagent Kit (PE Applied Biosystems). Relative gene expression was determined based on the threshold cycles (Cts) of the genes of interest and the internal control gene. The mRNA levels of the genes of interest are expressed as the ratio of each gene of interest to GAPDH for each sample; mRNA expression was compared between sarcoma cell lines with different constructs. The average Ct value of the GAPDH gene was subtracted from the average Ct value of the gene of interest for each sample. The fold change (2−△Ct) in expression of the target gene was calculated relative to the internal control gene (GAPDH) for each sample. The primers used in this study are shown in Additional file 1: Table S1.
Western blot analysis
Cell pellets were lysed in 10 mM HEPES, 10 mM NaCl, 0.1 mM EDTA, 0.1 mM EGTA, 1% NP-40, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 0.1 mM DTT, 0.1 mM Na3VO4, and protease inhibitors. Protein samples (80 μg of each) were separated by SDS-PAGE (10%) and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). Anti-β-actin antibody was purchased from Abcam (Cambridge, MA), and anti-NKX6-1 antibody was purchased from Cell Signaling (Danvers, MA).
Cell viability assay
Cells were plated at 2000 cells per well in a 96-well plate for 1 day and then treated with chemotherapeutic drugs and the SHH inhibitor RU-SKI 43 (Cayman) for 96 h. Cell viability was measured using an MTS assay kit (CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay, Promega). Briefly, 20 μl/well MTS reagent was added to 80 μl of medium containing cells in each well of a 96-well plate and left for 1 h in a humidified incubator at 37 °C and 5% CO2. For colorimetric analysis, the absorbance at 490 nm was recorded using a microplate reader (Multiskan EX, Thermo). Each condition was repeated at least 4 times. All cells were harvested at the designated times after treatment.
Anchorage-independent growth (AIG)
A 2.5 ml base layer of agar (0.7% agar in culture medium) was allowed to solidify in a six-well flat-bottomed plate before the addition of 2 ml of cell suspensions containing 10,000 cells in 0.5% agar. Colonies were allowed to grow for 14–21 days at 37 °C with 5% CO2 before imaging. Medium was changed 2 times per week. Plates were stained with p-iodonitrotetrazolium violet (INT, Sigma-Aldrich). Colony numbers in the entire dish were counted.
In vivo tumor xenograft model
Four week-old athymic nude mice (BALB/cByJNarl) were purchased from the National Laboratory Animal Center (Taipei, Taiwan). Mice were allowed to acclimate to animal housing for 7 days before study. The protocol for this animal experiment was approved by the Institutional Animal Care and Use Committee of the National Defense Medical Center, Taipei, Taiwan. All animal procedures and animal care were performed according to institutional animal research guidelines. A total of 1 × 106 cells were resuspended in 100 μl PBS and subcutaneously (s.c.) injected into each murine flank using a 1-cc syringe with a 29-gauge needle. N = 3 for each transfectant. At the end of the experiment, all mice were euthanized and tumors were harvested.
Immunohistochemistry
LMS tissues were fixed in 4% paraformaldehyde for 24 h and then placed in 30% sucrose PBS buffer before being embedded at the optimal cutting temperature and frozen. Approximately 5-μm-thick sections were prepared for hematoxylin and eosin (H&E) staining and immunohistochemical analyses. For morphometric lesion analysis, the sections were stained with Mayer’s H&E (Atom Scientific, Manchester, UK). Sections for GLI1 staining were fixed by immersion in ice-cold acetone/methanol (1:1) for 3 min. These sections were incubated in a blocking solution of 3% goat serum in PBS for 1 h at room temperature and then with monoclonal mouse anti-human GLI1 antibody (sc-515751, Santa Cruz) overnight. The sections were then incubated with a horseradish peroxidase-labeled secondary antibody for 1 h at room temperature, and peroxidase activity was visualized using a chromogenic solution of diaminobenzidine at room temperature.
Statistical analysis
A two-tailed Mann–Whitney U test was used to compare data groups for in vitro studies and relative mRNA expression in the different stable transfectants. Standard deviations were used for error bars and various comparisons. Kaplan–Meier analysis, Cox regression and log-rank tests were used to calculate survival and to evaluate differences between overall survival (OS). p-values < 0.05 were considered to be statistically significant. RNA expression profiles (RNA-sequencing data) of LMS cases were obtained from The Cancer Genome Atlas (TCGA). LMS classification was defined by TCGA. In the survival analysis, patients with FPKM > 66th percentile (1/3) were defined as having high expression, while patients with FPKM ≤ 66th percentile (2/3) were defined as having low expression. The results and clinical data were downloaded from the Broad Institute GDAC Firehose (http://gdac.broadinstitute.org/) and used in compliance with TCGA’s data usage policy.
Results
Clinical correlation of the NKX transcription factor family in LMS
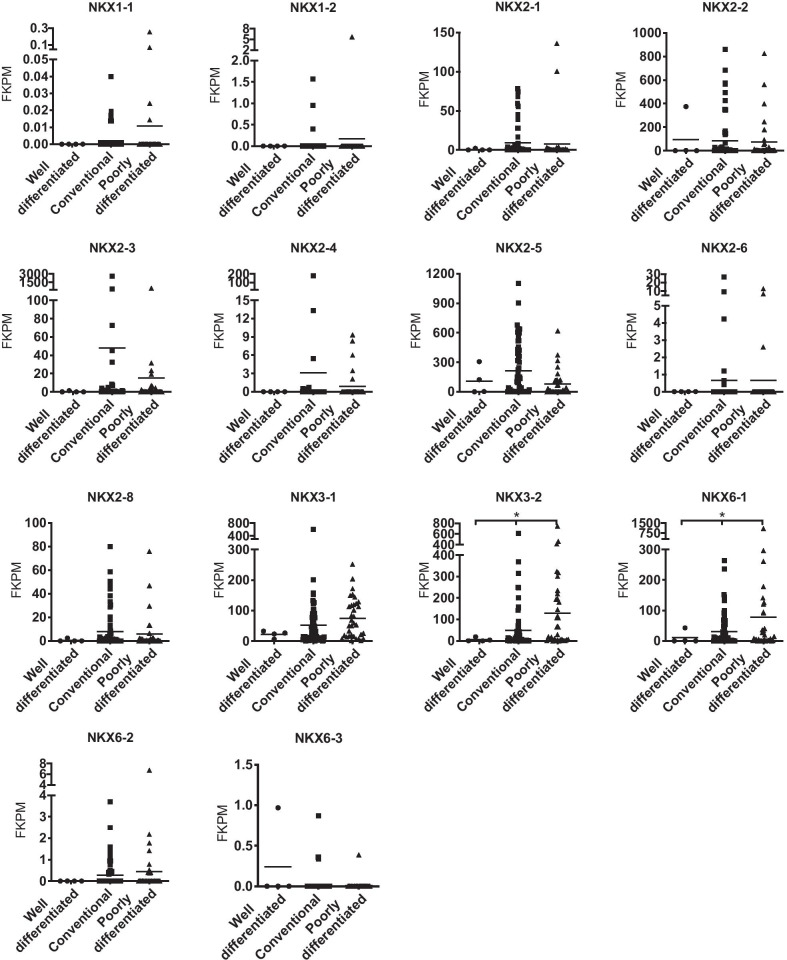
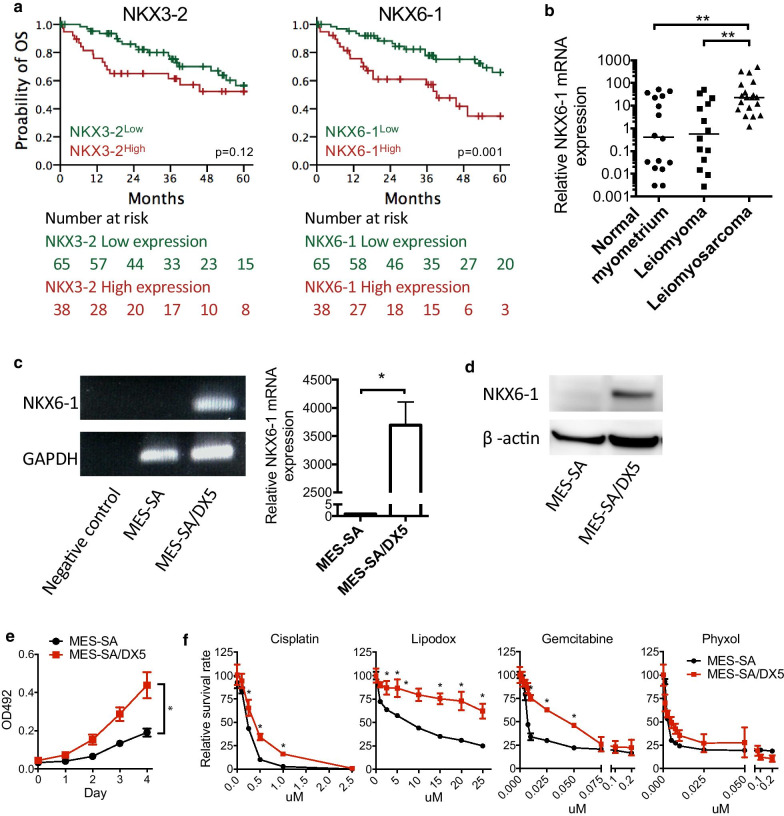
To explore the role of the NKX family in LMS, we analyzed the expression levels of NKX family mRNAs in well-differentiated, conventional, and poorly differentiated LMS cases from TCGA. Among the 14 NKX genes, the mRNA expression levels of NKX3-2 and NXK6-1 were higher in poorly differentiated LMS than in well-differentiated or conventional LMS (Fig. 1). LMS patients with both high and low NKX3-2 mRNA expression levels had similar median 5 year OS durations (NKX3-2high vs. NKX3-2low, > 60 months vs. > 60 months, hazard ratio (HR) = 1.7, 95% confidence interval (CI): 0.9–3.3) and 5 year survival rates (NKX3-2high vs. NKX3-2low, 53 vs. 57%). However, compared to NKX6-1low patients, LMS patients with high NKX6-1 mRNA expression had a worse prognosis, with a median 5 year OS of 39.2 months (NKX6-1high vs. NKX6-1low, 39.2 vs. > 60 months, HR = 2.9, 95%: 1.5–5.7) and a 5 year survival rate of 34% (NKX6-1high vs. NKX6-1low, 34% vs. 66%). (Fig. 2a). We further found that NKX6-1 expression was higher in LMS (n = 19) than in nonmalignant leiomyomas (n = 14) or normal myometrium (n = 16) (Fig. 2b).
Fig. 1.
RNA expression of NKX family members in LMS. The expression levels of 14 NKX family mRNAs in well-differentiated, conventional, and poorly differentiated LMS from TCGA. *p < 0.05
Fig. 2.
NKX6-1 expression is upregulated in LMS tissues and correlates with malignancy. a Kaplan–Meier analysis of NKX3-2 and NKX6-1 in LMS patients from a dataset in The Cancer Genome Atlas (TCGA). b NKX6-1 mRNA levels in 49 human uterine tissues, including normal myometrium (n = 16), leiomyoma (n = 14), and LMS (n = 19). NKX6-1 mRNA c and protein d expression was determined by qRT-PCR and immunoblotting in MES-SA and MES-SA/DX5 cells. The NKX6-1/β-actin protein expression ratios were 0.31 and 1.19, respectively. In vitro growth curves e and analyses of resistance to cisplatin, lipodox, gemcitabine, and phyxol f in the MES-SA parental and MES-SA/DX5 daughter cell lines. *p < 0.05, **p < 0.01
NKX6-1 promotes malignant phenotypes in LMS cell lines
To determine the functional effects of NKX6-1 in LMS, we chose the human sarcoma cell line MES-SA and its chemoresistant daughter cell line MES-SA/DX5 [23]. We observed that NKX6-1 mRNA (Fig. 2c) and protein (Fig. 2d) expression levels were higher in MES-SA/DX5 cells than in MES-SA cells. Interestingly, malignant phenotypes, including proliferation and resistance to cisplatin, lipodox, and gemcitabine, were significantly higher in MES-SA/DX5 cells than in MES-SA cells (Fig. 2e, f).
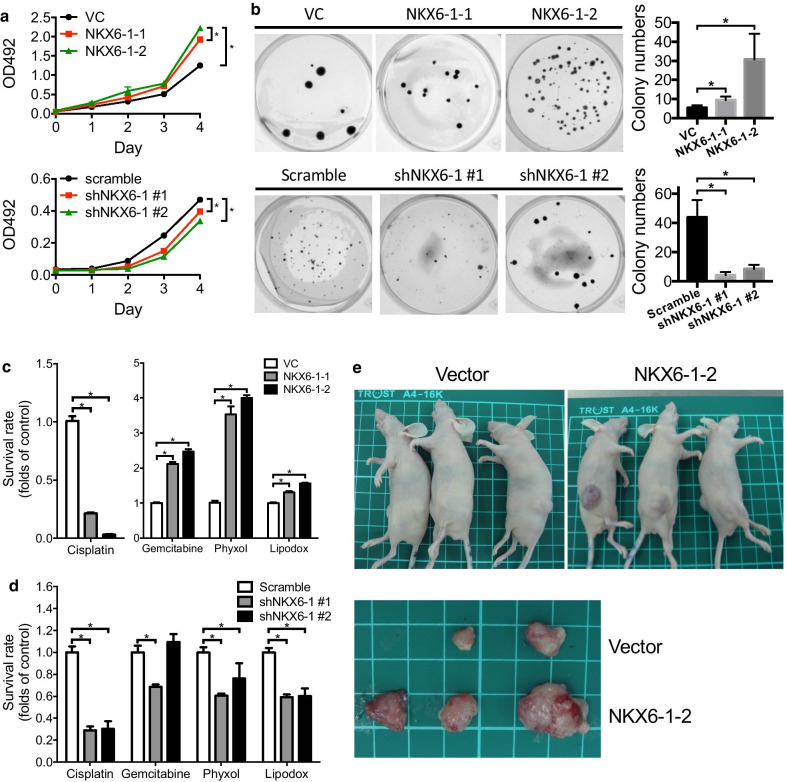
Thereafter, we generated NKX6-1 gain-of-function cells by transfecting full-length NKX6-1 into the MES-SA cell line and loss-of-function cells by transfecting NKX6-1 shRNAs into the MES-SA/DX5 cell line (confirmed by western blotting in Additional file 2: Figure S1). Overexpression of NKX6-1 protein enhanced numerous malignant phenotypes, including proliferation, colony formation, and resistance to gemcitabine, phyxol, and lipodox (Fig. 3a–c). Conversely, knockdown of NKX6-1 in MES-SA/DX5 cells suppressed proliferation, colony formation, and resistance to cisplatin, gemcitabine, phyxol, and lipodox (Fig. 3a, b, d). Tumor xenograft experiments demonstrated that overexpression of NKX6-1 enhanced tumor growth in vivo (Fig. 3e). These results demonstrate that NKX6-1 plays an oncogenic role in LMS.
Fig. 3.
NKX6-1 promotes malignancy in LMS cells. Overexpression of NKX6-1 in MES-SA LMS cells increases proliferation and colony formation. Knockdown of NKX6-1 in MES-SA/DX5 cells, by contrast, decreases proliferation. a A cell proliferation assay was performed using the MTS assay. b Colony formation ability was assessed using the AIG assay. NKX6-1-overexpressing MES-SA cells c and NKX6-1 knockdown MES-SA/DX5 cells d were evaluated for resistance to cisplatin, gemcitabine, phyxol, and doxorubicin. The concentrations of cisplatin, gemcitabine, phyxol, and doxorubicin in c are 0.33, 0.01, 0.005 and 7.5 μM, respectively (the IC50 in MES-SA-VC cells). The concentrations of cisplatin, gemcitabine, phyxol, and doxorubicin in d are 0.5, 0.05, 0.0125 and 30 μM, respectively (the IC50 in MES-SA/DX5-Scramble cells). e In vivo tumor growth of NKX6-1-overexpressing MES-SA cells. *p < 0.05
NKX6-1 enhances stem cell properties and regulates the SHH and NOTCH pathways in LMS cells
Since NKX6-1 was reported to be related to stem cell differentiation [24, 25], we explored the role of NKX6-1 in cancer stemness. Our results demonstrated that overexpression of NKX6-1 in MES-SA cells enhanced sphere formation while knockdown of NKX6-1 in MES-SA/DX5 cells suppressed sphere formation (Fig. 4a, b). Next, we assessed expression of stemness markers, including NESTIN, NANOG, KLF4, KLF8, MYC, OCT4, SOX2, CD44, and CD49F, following the manipulation of NKX6-1 expression in MES-SA and MES-SA/DX5 cells. These results showed that expression of the stemness markers KLF8, MYC, and CD49F was increased in NKX6-1-overexpressing MES-SA cells and reduced after knockdown of NKX6-1 in MES-SA/DX5 cells (Fig. 4c, d). These results suggest that NKX6-1 plays a phenotype-regulatory role in LMS.
Fig. 4.
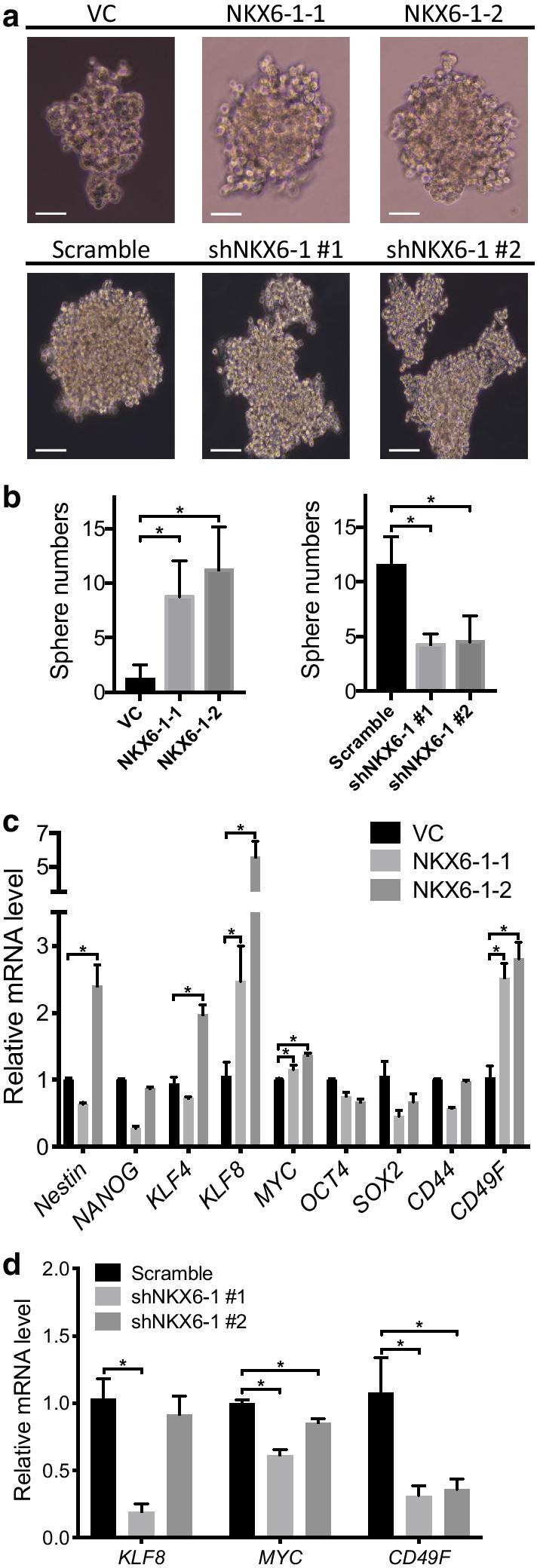
NKX6-1 promotes cancer stemness properties in LMS cells. Cells were seeded in ultralow attachment plates to assess sphere-forming ability. a Representative spheroid images from different NKX6-1 transfectants. NKX6-1-overexpressing MES-SA cells formed spheroids with round morphology (upper), and NKX6-1 knockdown MES-SA/DX5 cells formed clusters of loosely associated cells (lower). The scale bar represents 100 μm. b The number of spheres was quantified using ImageJ software. The total number of spheres increased upon NKX6-1 overexpression in MES-SA cells and decreased upon NKX6-1 knockdown in MES-SA/DX5 cells. The expression of cancer stemness-related genes correlated with sphere formation ability. c qRT-PCR was performed to evaluate stemness markers (including NESTIN, NANOG, KLF4, KLF8, MYC, OCT, SOX2, CD44, and CD49F) in MES-SA cells transfected with either NKX6-1 or vector control. d Expression of KLF8, MYC, and CD49F in MES-SA/DX5 cells transfected with either shNKX6-1 or scrambled control was determined by qRT-PCR. *p < 0.05
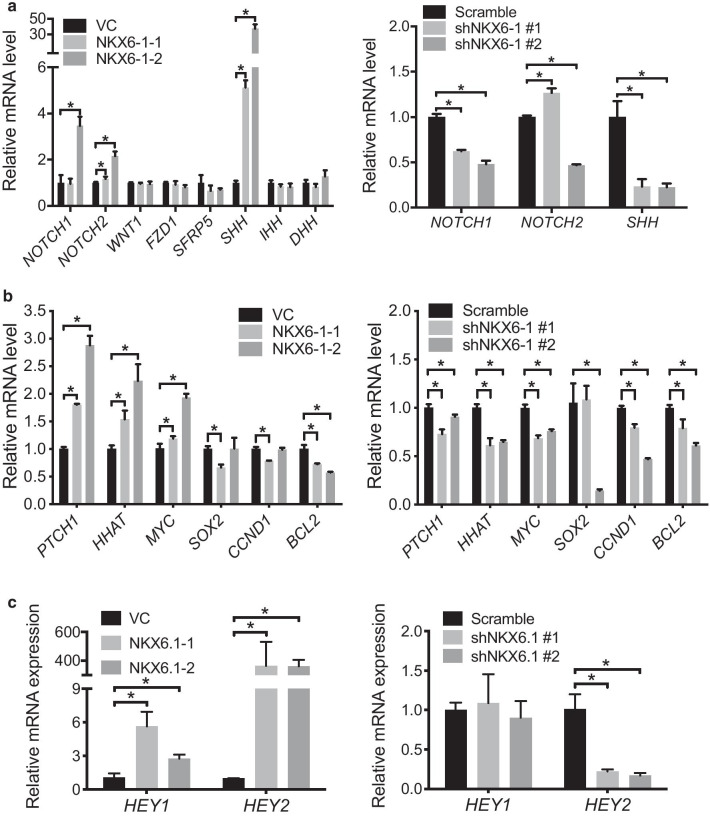
To elucidate the regulatory effects of NKX6-1 on stemness, potential relevant signaling pathways, such as WNT, Hedgehog and Notch, were evaluated [26]. These results demonstrated that the SHH and NOTCH1/2 signaling pathways were upregulated in NKX6-1-overexpressing MES-SA cells and downregulated in NKX6-1-knockdown MES-SA/DX5 cells (Fig. 5a). Genes downstream of SHH signaling, such as PTCH1 and MYC, and SHH-related acyltransferases, such as HHAT, were upregulated upon NKX6-1 overexpression in MES-SA cells. Conversely, PTCH1, HHAT, MYC, SOX2, CCND1 and BCL2 expression levels were decreased after NKX6-1 knockdown in MES-SA/DX5 cells (Fig. 5b). SOX2, CCND1, and BCL2 demonstrated the same trend in opposing treatment conditions, which may be due to crosstalk between signaling pathways. These regulatory pathways, such as PI3K/AKT and SOX2 [27], DNA damage/ATM and CCND1[28], JAK2/ERK and BCL2 [29], may compensate for the effects of NKX6-1, though these compensatory effects should be investigated in future studies. Moreover, the NOTCH signaling downstream genes HEY1 and HEY2 were upregulated in NKX6-1-expressing MES-SA cells, while HEY2 was downregulated in NKX6-1-knockdown MES-SA/DX5 cells (Fig. 5c). Taken together, these data indicate that NKX6-1 promotes stemness phenotypes by regulating the SHH and NOTCH signaling pathways in LMS cells.
Fig. 5.
NKX6-1 mediates the activation of stemness, SHH, and Notch signaling pathways. qRT-PCR analysis of Notch, WNT and SHH signaling. a qRT-PCR for NOTCH1, NOTCH2, WNT1, FZD1, SFRP5, SHH, IHH, and DHH in NKX6-1-overexpressing MES-SA cells. b qRT-PCR for NOTCH1, NOTCH2, and SHH in NKX6-1 knockdown MES-SA/DX5 cells. c Expression of SHH c and NOTCH d downstream genes was determined by qRT-PCR. *p < 0.05
NKX6-1 upregulates SHH signaling and sensitizes LMS cells to an SHH inhibitor
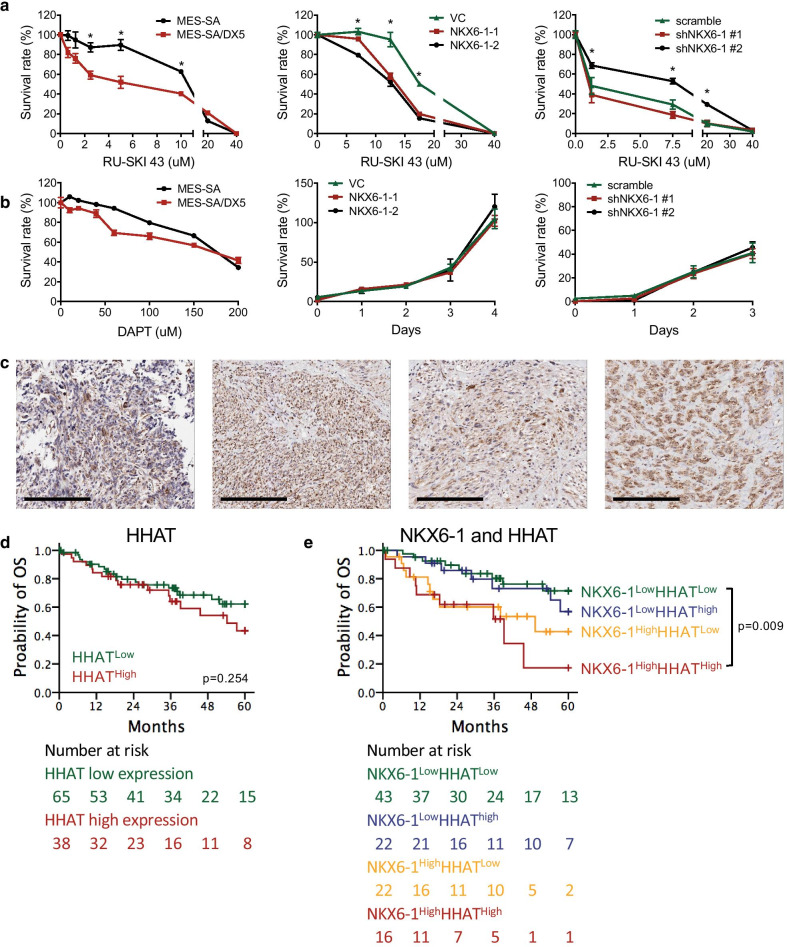
To determine whether the SHH and/or NOTCH pathways could be potential therapeutic targets in NKX6-1-overexpressing LMS cells, we tested the effects of the SHH inhibitor RU-SKI 43 (an HHAT inhibitor) [30] and the NOTCH inhibitor DAPT [31]. We found that LMS cells with higher NKX6-1 expression were more sensitive to RU-SKI 43. MES-SA/DX5 cells were more sensitive than MES-SA cells. NKX6-1-overexpressing MES-SA cells were more sensitive than control cells. NKX6-1 knockdown MES-SA/DX5 clone 1 was more sensitive than control cells, and there was no sensitivity difference between clone 2 and control cells (Fig. 6a). MYC and SOX2 were downregulated in both cell lines after treatment (Additional file 2: Figure S2). Moreover, other gynecological cancer cell lines with higher NKX6-1 expression were more sensitive to RU-SKI 43 (Additional file 2: Figure S3). Treatment with DAPT inhibited cell proliferation in vitro; however, it had no significant effects on the various NKX6-1 transfectants (Fig. 6b), suggesting that NKX6-1 is not involved in the molecular mechanisms targeted by DAPT. Recent studies have shown that the SHH signaling pathway is implicated not only in cancer cells but also in stromal cells [32]. We evaluated GLI1 protein expression in LMS tissue by immunohistochemistry and demonstrated that the expression of GLI1 protein in LMS cancer cells (Fig. 6c and Additional file 2: Figure S4) is consistent with that of a previous study indicating the expression of GLI in LMS tissue [33, 34]. Moreover, the SHH downstream genes BCL-2, c-MYC and CCND1 were expressed in LMS tissues (Additional file 2: Figure S5). All these data suggested that SHH signaling is activated in LMS cells.
Fig. 6.
NKX6-1 activation enhances chemosensitivity to an SHH inhibitor. Dose–response curves (a and b, left panel) and growth curves (b, middle and right panel) of LMS cells and NKX6-1-overexpressing vs. -knockdown cells treated for 96 h with SHH (RU-SKI 43) and NOTCH (DAPT) inhibitors. c Immunohistochemical analysis of GLI1 protein expression in LMS tissue from four patients. The scale bar represents 200 μm. d Kaplan–Meier analysis of overall survival stratified by HHAT expression in LMS patients from the TCGA dataset. e Kaplan–Meier analysis of overall survival stratified by combined NKX6-1 and HHAT expression in LMS patients from the TCGA dataset. *p < 0.05
Since RU-SKI 43 suppresses the SHH acyltransferase HHAT, we assessed HHAT clinical relevance and prognostic markers in LMS patient data from the TCGA database [35]. We found that the expression of HHAT alone was not associated with OS (Fig. 6d). LMS patients with high HHAT mRNA expression had a median 5 year OS of 54.2 vs. > 60 months (HR = 1.5, 95% Cl: 0.8–2.9), and a 5 year survival rate of 45 vs. 61% compared to HHATlow LMS patients. Interestingly, combined assessment of HHAT and NKX6-1 demonstrated that patients with the highest expression of both NKX6-1 and HHAT (i.e., NKX6-1highHHAThigh) had the worst survival outcomes (Fig. 6e), with a median 5 year OS of 39.2 vs. > 60 months (HR = 4.1, 95% CI: 1.6–10.4) and a 5 year survival rate of 26 vs. 71.0% compared to NKX6-1lowHHATlow LMS patients.
Discussion
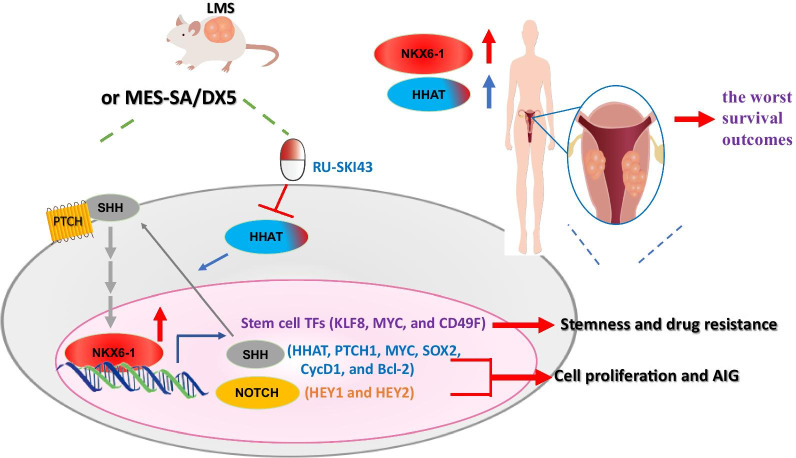
LMS originates from undifferentiated mesenchymal cells and exhibits highly aggressive behavior, characterized by high rates of recurrence and metastasis. Neither chemotherapy nor radiotherapy has been shown to improve overall survival, thus posing a major challenge to adjuvant treatment in LMS patients. Whether NKX6-1 is important in the pathogenesis of LMS and regulates SHH signaling has not been reported. In the present study, we investigated how oncogenic NKX6-1 confers poor prognosis in LMS and how NKX6-1 regulates cancer stem cells through activation of the SHH and NOTCH pathways. Our findings reveal novel molecular insights into LMS and lay a foundation for future diagnostic and therapeutic improvements (Fig. 7).
Fig. 7.
Proposed model of NKX6-1-mediated stemness signaling and cancer stemness properties in LMS. NKX6-1 activates SHH and NOTCH but not WNT, resulting in enhanced malignant phenotypes and poor prognosis in LMS. Inhibition of SHH but not NOTCH inhibits cell growth, suggesting the potential of SHH inhibitors for the treatment of LMS
The relationship between SHH and NKX6-1 is interesting. The SHH pathway plays a crucial role in embryonic vertebral development and tissue homeostasis [36]. In addition, NKX6-1 expression controls fate specification and differentiation in cells surrounding SHH-expressing cells. While a SHH-independent pathway appears to regulate NKX6-1 expression in the foregut [37], SHH signaling is required for NKX6-1 expression in the ventral neural tube and spinal meninges. Previous reports have demonstrated that the NKX gene family is expressed in the medial neural plate above the SHH-expressing axial mesendoderm [38, 39]. Similar to many embryonic signaling pathways in cancer, the SHH signaling pathway plays important roles in promoting oncogenesis and tumor growth and progression. The dysregulated activation of SHH signaling is implicated in several cancers and has been linked to the maintenance of cancer stem-like cells, which are associated with the development of therapeutic resistance [40–45]. Interestingly, our study found that overexpression of NKX6-1 modulates SHH pathway genes and promotes cancer stemness, indicating a novel reciprocal regulatory axis between NKX6-1 and SHH signaling in cancer.
Previous developmental biology studies have shown that Nkx transcription factors act with coactivators or corepressors to regulate tissue-specific gene expression and development. NKX2-5 interacts with HAND2 and GATA4 to promote gene expression, cardiomyocyte differentiation and chamber identity [46]. Nkx2.2 interacts with Grg3 and functions as a transcriptional repressor during islet beta-cell specification in the pancreas [47]. Nkx2.1 interacts with Gata6 to direct pulmonary epithelial differentiation and development [48]. The partners of NKX6-1 should be evaluated in the future.
The different roles of NKX6-1 suggest a bifunctional divergence whereby downstream target genes can be activated or repressed in a cell type-dependent manner [49]. In cervical cancer, NKX6-1 plays a tumor suppressive role by suppressing the epithelial-to-mesenchymal transition and cancer metastasis [50]. However, in other contexts NKX6-1 behaves as an oncogene [51–53]. NKX6-1 upregulates mesenchymal markers, facilitates disease progression and is associated with poor prognosis in patients with primary hepatocellular carcinoma [51]. In breast cancer, NKX6-1 increases IL-6 expression and promotes cell proliferation through an NKX6-1/IL-6 network [52]. Moreover, immunohistochemical staining for NKX6-1 could be an effective marker for pancreatic and duodenal neuroendocrine tumors [53]. NKX6-1 methylation status is an indicator of survival outcome and could inform treatment selection in stage III colon cancer [19]. Our current study indicates that NKX6-1 plays an oncogenic role in LMS.
Through molecular characterization and analyses of LMS patient prognosis in public datasets, we provide evidence to support the oncogenic role of NKX6-1, which serves as a novel biomarker to predict prognosis and guide precision medicine against LMS. A prognostic marker can help clinicians and patients estimate OS and progression-free survival, helping to inform therapeutic decisions. However, few models have been proposed to predict the prognosis of LMS. Cohen et al. observed the association between tumor infiltrating CD8 cytotoxic lymphocytes, PD-L1 staining, expression of mismatch repair-related proteins (MSH2, MLH1, MSH6 and PSM2) and survival in patients with LMS [54]. Studies from Xue et al. suggested that age greater than 60 years, high tumor grade, distant metastasis, tumor size ≥ 5 cm, and lack of surgery were associated with decreased OS and cancer-specific survival [55]. These models were characterized by complexity and clinical impracticality. The results obtained in our study from both cell lines and patient samples provide strong evidence to support the oncogenic role of NKX6-1 in LMS and suggest that NKX6-1 could serve as a novel biomarker and guide the treatment of LMS.
The SHH signaling pathway has been implicated in the progression of cancers. In an essential step for SHH pathway activation, HHAT catalyzes the transfer of the fatty acid palmitate onto SHH-related signaling proteins [56]. Therefore, the potential of HHAT inhibition has been tested in various cancers, including breast cancer [57] and pancreatic ductal adenocarcinoma [58]. Recently, SHH signaling activation was shown to induce undifferentiated soft tissue sarcomas in a mouse model. [59]. Moreover, high expression of SHH signaling pathway proteins has been observed in LMS patients [33]. Based on the present and previous studies, it is important to consider a new therapeutic intervention that targets the SHH pathway to block the progression of this type of malignancy [60]. Here, we have identified a relationship between NKX6-1 expression and SHH signaling activation in cancer cells and we therefore propose that SHH inhibition may represent a feasible strategy for the treatment of NKX6-1high LMS patients.
Primary cilia play important roles in the activation of the Hedgehog pathway. Upon sensing extracellular signals, the components of the hedgehog pathway, including GLI transcription factors, accumulate at primary cilia [61]. In rhabdomyosarcoma and Ewing sarcoma, GLI1 is upregulated and contributes to drug resistance [62]. Our results also demonstrated increased GLI1 expression in LMS tissue. However, cilia may play tumor suppressive or oncogenic roles depending on SHH pathway regulation [63]. Various types of neoplasms, including ovarian, pancreatic, and renal cancers, fail to express cilia and centrioles [64], indicating that the role of cilia dysfunction in the tumorigenic process is complex and deserves further investigation in the future.
Some limitations should be mentioned in our study. First, the role of NKX6-1, SHH, and NOTCH in the in vivo chemoresistance of LMS were not evaluated in the current study. Moreover, the inconsistent effects of NKX6-1 overexpression and knockdown during cisplatin treatment in human sarcoma cells require further investigation. The current study did not use omics-level approaches to analyze the putative genes regulated by NKX6-1; this should be evaluated in a future study. Additionally, manipulation of NKX6-1 exhibits inconsistent effects on the expression of NOTCH1 and NOTCH2. Although inhibition of NOTCH signaling has modest effects on LMS prognosis with either partial response or stable disease in approximately 40% of patients [65, 66], our unpublished results demonstrate that NOTCH inhibition displays NKX6-1-independent chemosensitizing effects in LMS. Further studies should be conducted to uncover the mechanisms by which NOTCH signaling promotes stemness and to determine how we can best incorporate them into LMS therapeutics. Even with these limitations, our studies reveal novel roles of NKX6-1 and SHH signaling in the pathogenesis of LMS, which opens new avenues for the treatment of LMS.
Conclusions
Although large-scale studies are needed, our study demonstrated that NKX6-1 expression regulates the cancer stemness phenotype through activation of the SHH pathway and is correlated with poor prognosis in LMS. Along with the diagnostic potential of detecting NKX6-1 in cancer tissues, our findings suggest that SHH inhibitors could be applied to treat LMS.
Supplementary Information
Additional file 1: Table S1. Primers used in the present study.
Additional file 2: Figure S1. NKX6-1 protein expression in the different stable transfectants. Figure S2. Effects of RU-SKI 43 treatment on SHH downstream genes. Figure S3. Increased NKX6-1 expression correlates with increased RU-SKI 43 resistance in LMS cell lines. Figure S4. GLI1 protein expression in LMS. Figure S5. IHC analysis of BCL-2, c-MYC and Cyclin D1 expression in LMS.
Acknowledgements
We thank Dr. Chin-Sheng Lin (Division of Cardiology, Tri-Service General Hospital, National Defense Medical Center, Taipei, Taiwan) for manuscript editing. We thank Ms. Hui-Yin Su (Division of Cardiology, Tri-Service General Hospital, National Defense Medical Center, Taipei, Taiwan) for helpful artwork.
Abbreviations
- BCL2
B-cell lymphoma 2
- CCND1
Cyclin D1
- DHH
Desert hedgehog
- FZD1
Frizzled-1
- HHAT
Hedgehog Acyltransferase
- IHH
Indian hedgehog
- MYC
MYC Proto-Oncogene, BHLH Transcription Factor
- NKX6-1
NK6 Homeobox 1
- NOTCH1
Notch Receptor 1
- NOTCH2
Notch Receptor 2
- PTCH1
Patched 1
- SFRP5
Secreted Frizzled Related Protein 5
- SHH
Sonic hedgehog
- SOX2
SRY-Box Transcription Factor 2
Authors' contributions
CC Wu and HC Lai initiated the project. PH Su and HC Lai designed the study. YC Weng, LY Chen, CC Wang and PH Su performed the experiments. RL Huang performed the bioinformatics analysis. PH Su, CC Wu and HC Lai wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was found by grants from the Tri-Service General Hospital, Taipei, Taiwan (Grant number: TSGH-C106-075, TSGH-C105-093, TSGH-C104-097, TSGH-C103-098, to CC Wu) and Ministry of Science and Technology (Grant number: NSC100-2314-B-016-028, to CC Wu).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
The protocol of clinical samples collection was approved This study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of the Tri-Service General Hospital. All of the patients signed informed consent forms before study.
The protocol of animal experiment was approved by the Institutional Animal Care and Use Committee (IACUC) of the National Defense Medical Center, Taipei, Taiwan.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gladdy RA, Qin LX, Moraco N, Agaram NP, Brennan MF, Singer S. Predictors of survival and recurrence in primary leiomyosarcoma. Ann Surg Oncol. 2013;20(6):1851–1857. doi: 10.1245/s10434-013-2876-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toro JR, Travis LB, Wu HJ, Zhu K, Fletcher CD, Devesa SS. Incidence patterns of soft tissue sarcomas, regardless of primary site, in the surveillance, epidemiology and end results program, 1978–2001: an analysis of 26,758 cases. Int J Cancer. 2006;119(12):2922–2930. doi: 10.1002/ijc.22239. [DOI] [PubMed] [Google Scholar]
- 3.Abeler VM, Royne O, Thoresen S, Danielsen HE, Nesland JM, Kristensen GB. Uterine sarcomas in Norway. A histopathological and prognostic survey of a total population from 1970 to 2000 including 419 patients. Histopathology. 2009;54(3):355–364. doi: 10.1111/j.1365-2559.2009.03231.x. [DOI] [PubMed] [Google Scholar]
- 4.Huang CY, Chen CA, Chen YL, Chiang CJ, Hsu TH, Lin MC, Lai MS, Chen CJ, You SL, Cheng WF. Nationwide surveillance in uterine cancer: survival analysis and the importance of birth cohort: 30 year population-based registry in Taiwan. PLoS ONE. 2012;7(12):e51372. doi: 10.1371/journal.pone.0051372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinha S, Peach AH. Diagnosis and management of soft tissue sarcoma. BMJ. 2010;341:c7170. doi: 10.1136/bmj.c7170. [DOI] [PubMed] [Google Scholar]
- 6.Iasonos A, Keung EZ, Zivanovic O, Mancari R, Peiretti M, Nucci M, George S, Colombo N, Carinelli S, Hensley ML, Raut CP. External validation of a prognostic nomogram for overall survival in women with uterine leiomyosarcoma. Cancer. 2013;119(10):1816–1822. doi: 10.1002/cncr.27971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gadducci A, Landoni F, Sartori E, Zola P, Maggino T, Lissoni A, Bazzurini L, Arisio R, Romagnolo C, Cristofani R. Uterine leiomyosarcoma: analysis of treatment failures and survival. Gynecol Oncol. 1996;62(1):25–32. doi: 10.1006/gyno.1996.0185. [DOI] [PubMed] [Google Scholar]
- 8.Hensley ML, Blessing JA, Mannel R, Rose PG. Fixed-dose rate gemcitabine plus docetaxel as first-line therapy for metastatic uterine leiomyosarcoma: a Gynecologic Oncology Group phase II trial. Gynecol Oncol. 2008;109(3):329–334. doi: 10.1016/j.ygyno.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raut CP, Nucci MR, Wang Q, Manola J, Bertagnolli MM, Demetri GD, Morgan JA, Muto MG, Fletcher CD, George S. Predictive value of FIGO and AJCC staging systems in patients with uterine leiomyosarcoma. Eur J Cancer. 2009;45(16):2818–2824. doi: 10.1016/j.ejca.2009.06.030. [DOI] [PubMed] [Google Scholar]
- 10.Friedman CF, Hensley ML. Options for adjuvant therapy for uterine leiomyosarcoma. Curr Treat Options Oncol. 2018;19(2):7. doi: 10.1007/s11864-018-0526-0. [DOI] [PubMed] [Google Scholar]
- 11.Yoon JH, Kim O, Eun JW, Choi SS, Ashktorab H, Smoot DT, Nam SW, Park WS. Multiple genetic mutations caused by NKX6.3 depletion contribute to gastric tumorigenesis. Sci Rep. 2018;8(1):17609. doi: 10.1038/s41598-018-35733-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penha RCC, Buexm LA, Rodrigues FR, de Castro TP, Santos MCS, Fortunato RS, Carvalho DP, Cardoso-Weide LC, Ferreira ACF. NKX2.5 is expressed in papillary thyroid carcinomas and regulates differentiation in thyroid cells. BMC Cancer. 2018;18(1):498. doi: 10.1186/s12885-018-4399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L, Lin M, Ruan WJ, Dong LL, Chen EG, Wu XH, Ying KJ. Nkx2-1: a novel tumor biomarker of lung cancer. J Zhejiang Univ Sci B. 2012;13(11):855–866. doi: 10.1631/jzus.B1100382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tagne JB, Gupta S, Gower AC, Shen SS, Varma S, Lakshminarayanan M, Cao Y, Spira A, Volkert TL, Ramirez MI. Genome-wide analyses of Nkx2-1 binding to transcriptional target genes uncover novel regulatory patterns conserved in lung development and tumors. PLoS ONE. 2012;7(1):e29907. doi: 10.1371/journal.pone.0029907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM, Hubbard DD, DuPage MJ, Whittaker CA, Hoersch S, Yoon S, Crowley D, Bronson RT, Chiang DY, Meyerson M, Jacks T. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473(7345):101–104. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen MM, Abate-Shen C. Roles of the Nkx3.1 homeobox gene in prostate organogenesis and carcinogenesis. Dev Dyn. 2003;228(4):767–778. doi: 10.1002/dvdy.10397. [DOI] [PubMed] [Google Scholar]
- 17.Jishage M, Fujino T, Yamazaki Y, Kuroda H, Nakamura T. Identification of target genes for EWS/ATF-1 chimeric transcription factor. Oncogene. 2003;22(1):41–49. doi: 10.1038/sj.onc.1206074. [DOI] [PubMed] [Google Scholar]
- 18.Chang CC, Huang RL, Wang HC, Liao YP, Yu MH, Lai HC. High methylation rate of LMX1A, NKX6-1, PAX1, PTPRR, SOX1, and ZNF582 genes in cervical adenocarcinoma. Int J Gynecol Cancer. 2014;24(2):201–209. doi: 10.1097/IGC.0000000000000054. [DOI] [PubMed] [Google Scholar]
- 19.Chang SY, Kuo CC, Wu CC, Hsiao CW, Hu JM, Hsu CH, Chou YC, Shih YL, Lin YW. NKX6.1 hypermethylation predicts the outcome of stage II colorectal cancer patients undergoing chemotherapy. Genes Chromosomes Cancer. 2018;57(5):268–277. doi: 10.1002/gcc.22529. [DOI] [PubMed] [Google Scholar]
- 20.Lai HC, Lin YW, Huang TH, Yan P, Huang RL, Wang HC, Liu J, Chan MW, Chu TY, Sun CA, Chang CC, Yu MH. Identification of novel DNA methylation markers in cervical cancer. Int J Cancer. 2008;123(1):161–167. doi: 10.1002/ijc.23519. [DOI] [PubMed] [Google Scholar]
- 21.Taylor KH, Pena-Hernandez KE, Davis JW, Arthur GL, Duff DJ, Shi H, Rahmatpanah FB, Sjahputera O, Caldwell CW. Large-scale CpG methylation analysis identifies novel candidate genes and reveals methylation hotspots in acute lymphoblastic leukemia. Cancer Res. 2007;67(6):2617–2625. doi: 10.1158/0008-5472.CAN-06-3993. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida A, Sekine S, Tsuta K, Fukayama M, Furuta K, Tsuda H. NKX2.2 is a useful immunohistochemical marker for Ewing sarcoma. Am J Surg Pathol. 2012;36(7):993–999. doi: 10.1097/PAS.0b013e31824ee43c. [DOI] [PubMed] [Google Scholar]
- 23.Roomi MW, Kalinovsky T, Roomi NW, Rath M, Niedzwiecki A. Anticancer effects of a specific mixture of nutrients in the multidrug-resistant human uterine sarcoma MES-SA/Dx5 and the drug-sensitive MES-SA cell lines. Oncol Rep. 2012;27(1):17–27. doi: 10.3892/or.2011.1471. [DOI] [PubMed] [Google Scholar]
- 24.Gupta SK, Wesolowska-Andersen A, Ringgaard AK, Jaiswal H, Song L, Hastoy B, Ingvorsen C, Taheri-Ghahfarokhi A, Magnusson B, Maresca M, Jensen RR, Beer NL, Fels JJ, Grunnet LG, Thomas MK, Gloyn AL, Hicks R, McCarthy MI, Hansson M, Honore C. NKX6.1 induced pluripotent stem cell reporter lines for isolation and analysis of functionally relevant neuronal and pancreas populations. Stem Cell Res. 2018;29:220–231. doi: 10.1016/j.scr.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Tzatzalos E, Kwan KY, Grumet M, Cai L. Transcriptional regulation of notch1 expression by Nkx6.1 in neural stem/progenitor cells during ventral spinal cord development. Sci Rep. 2016;6:38665. doi: 10.1038/srep38665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, Yang SX, Ivy SP. Targeting Notch, hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol. 2015;12(8):445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh MH, Choe JH, Gadhvi J, Kim YJ, Arguez MA, Palmer M, Gerold H, Nowak C, Do H, Mazambani S, Knighton JK, Cha M, Goodwin J, Kang MK, Jeong JY, Lee SY, Faubert B, Xuan Z, Abel ED, Scafoglio C, Shackelford DB, Minna JD, Singh PK, Shulaev V, Bleris L, Hoyt K, Kim J, Inoue M, DeBerardinis RJ, Kim TH, Kim JW. p63 and SOX2 dictate glucose reliance and metabolic vulnerabilities in squamous cell carcinomas. Cell Rep. 2019;28(7):1860–1878.e1869. doi: 10.1016/j.celrep.2019.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jirawatnotai S, Hu Y, Michowski W, Elias JE, Becks L, Bienvenu F, Zagozdzon A, Goswami T, Wang YE, Clark AB, Kunkel TA, van Harn T, Xia B, Correll M, Quackenbush J, Livingston DM, Gygi SP, Sicinski P. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature. 2011;474(7350):230–234. doi: 10.1038/nature10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waibel M, Solomon VS, Knight DA, Ralli RA, Kim SK, Banks KM, Vidacs E, Virely C, Sia KC, Bracken LS, Collins-Underwood R, Drenberg C, Ramsey LB, Meyer SC, Takiguchi M, Dickins RA, Levine R, Ghysdael J, Dawson MA, Lock RB, Mullighan CG, Johnstone RW. Combined targeting of JAK2 and Bcl-2/Bcl-xL to cure mutant JAK2-driven malignancies and overcome acquired resistance to JAK2 inhibitors. Cell Rep. 2013;5(4):1047–1059. doi: 10.1016/j.celrep.2013.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrova E, Rios-Esteves J, Ouerfelli O, Glickman JF, Resh MD. Inhibitors of hedgehog acyltransferase block sonic hedgehog signaling. Nat Chem Biol. 2013;9(4):247–249. doi: 10.1038/nchembio.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang M, Ma X, Wang J, Wang L, Wang Y. Pretreatment with the gamma-secretase inhibitor DAPT sensitizes drug-resistant ovarian cancer cells to cisplatin by downregulation of Notch signaling. Int J Oncol. 2014;44(4):1401–1409. doi: 10.3892/ijo.2014.2301. [DOI] [PubMed] [Google Scholar]
- 32.Jeng KS, Chang CF, Lin SS. Sonic hedgehog signaling in organogenesis, tumors, and tumor microenvironments. Int J Mol Sci. 2020;21(3):758. doi: 10.3390/ijms21030758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia N, Bozzini N, Baiocchi G, da Cunha IW, Maciel GA, Soares JM, Soares FA, Baracat EC, Carvalho KC. May sonic hedgehog proteins be markers for malignancy in uterine smooth muscle tumors? Hum Pathol. 2016;50:43–50. doi: 10.1016/j.humpath.2015.08.026. [DOI] [PubMed] [Google Scholar]
- 34.Ragazzini P, Gamberi G, Pazzaglia L, Serra M, Magagnoli G, Ponticelli F, Ferrari C, Ghinelli C, Alberghini M, Bertoni F, Picci P, Benassi MS. Amplification of CDK4, MDM2, SAS and GLI genes in leiomyosarcoma, alveolar and embryonal rhabdomyosarcoma. Histol Histopathol. 2004;19(2):401–411. doi: 10.14670/HH-19.401. [DOI] [PubMed] [Google Scholar]
- 35.Omberg L, Ellrott K, Yuan Y, Kandoth C, Wong C, Kellen MR, Friend SH, Stuart J, Liang H, Margolin AA. Enabling transparent and collaborative computational analysis of 12 tumor types within the cancer genome atlas. Nat Genet. 2013;45(10):1121–1126. doi: 10.1038/ng.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma Y, Zhang P, Wang F, Yang J, Yang Z, Qin H. The relationship between early embryo development and tumourigenesis. J Cell Mol Med. 2010;14(12):2697–2701. doi: 10.1111/j.1582-4934.2010.01191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai J, Xu X, Yin H, Wu R, Modderman G, Chen Y, Jensen J, Hui CC, Qiu M. Evidence for the differential regulation of Nkx-6.1 expression in the ventral spinal cord and foregut by Shh-dependent and -independent mechanisms. Genesis. 2000;27(1):6–11. doi: 10.1002/1526-968X(200005)27:1<6::AID-GENE20>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 38.Qiu M, Shimamura K, Sussel L, Chen S, Rubenstein JL. Control of anteroposterior and dorsoventral domains of Nkx-6.1 gene expression relative to other Nkx genes during vertebrate CNS development. Mech Dev. 1998;72(1–2):77–88. doi: 10.1016/S0925-4773(98)00018-5. [DOI] [PubMed] [Google Scholar]
- 39.Shimamura K, Hartigan DJ, Martinez S, Puelles L, Rubenstein JL. Longitudinal organization of the anterior neural plate and neural tube. Development. 1995;121(12):3923–3933. doi: 10.1242/dev.121.12.3923. [DOI] [PubMed] [Google Scholar]
- 40.Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432(7015):324–331. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- 41.Chung SI, Moon H, Ju HL, Cho KJ, Kim DY, Han KH, Eun JW, Nam SW, Ribback S, Dombrowski F, Calvisi DF, Ro SW. Hepatic expression of Sonic Hedgehog induces liver fibrosis and promotes hepatocarcinogenesis in a transgenic mouse model. J Hepatol. 2016;64(3):618–627. doi: 10.1016/j.jhep.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 42.Cochrane CR, Szczepny A, Watkins DN, Cain JE. Hedgehog signaling in the maintenance of cancer stem cells. Cancers (Basel) 2015;7(3):1554–1585. doi: 10.3390/cancers7030851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noman AS, Uddin M, Chowdhury AA, Nayeem MJ, Raihan Z, Rashid MI, Azad AK, Rahman ML, Barua D, Sultana A, Shirin A, Ferdous J, Parag RR, Rahman SM, Mahmud MR, Jerin CS, Jahan N, Siddiqua A, Ara T, Sabur EB, Alam SS, Baidya S, Akther S, Rahman MZ, Banu T, Murugan AK, Sabri S, Islam SMS, Karakas B, Aboussekhra A, Yeger H, Farhat WA, Islam SS. Serum sonic hedgehog (SHH) and interleukin-(IL-6) as dual prognostic biomarkers in progressive metastatic breast cancer. Sci Rep. 2017;7(1):1796. doi: 10.1038/s41598-017-01268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riobo-Del Galdo NA, Lara Montero A, Wertheimer EV. Role of hedgehog signaling in breast cancer: pathogenesis and therapeutics. Cells. 2019;8(4):375. doi: 10.3390/cells8040375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu R, Gires O, Zhu L, Liu J, Li J, Yang H, Ju G, Huang J, Ge W, Chen Y, Lu Z, Wang H. TSPAN8 promotes cancer cell stemness via activation of sonic Hedgehog signaling. Nat Commun. 2019;10(1):2863. doi: 10.1038/s41467-019-10739-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCulley DJ, Black BL. Transcription factor pathways and congenital heart disease. Curr Top Dev Biol. 2012;100:253–277. doi: 10.1016/B978-0-12-387786-4.00008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doyle MJ, Loomis ZL, Sussel L. Nkx2.2-repressor activity is sufficient to specify alpha-cells and a small number of beta-cells in the pancreatic islet. Development. 2007;134(3):515–523. doi: 10.1242/dev.02763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Rath N, Hannenhalli S, Wang Z, Cappola T, Kimura S, Atochina-Vasserman E, Lu MM, Beers MF, Morrisey EE. GATA and Nkx factors synergistically regulate tissue-specific gene expression and development in vivo. Development. 2007;134(1):189–198. doi: 10.1242/dev.02720. [DOI] [PubMed] [Google Scholar]
- 49.Syu LJ, Uhler J, Zhang H, Mellerick DM. The Drosophila Nkx6 homeodomain protein has both activation and repression domains and can activate target gene expression. Brain Res. 2009;1266:8–17. doi: 10.1016/j.brainres.2009.01.068. [DOI] [PubMed] [Google Scholar]
- 50.Li HJ, Yu PN, Huang KY, Su HY, Hsiao TH, Chang CP, Yu MH, Lin YW. NKX6.1 functions as a metastatic suppressor through epigenetic regulation of the epithelial-mesenchymal transition. Oncogene. 2016;35(17):2266–2278. doi: 10.1038/onc.2015.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang LL, Zhang Y, Zhang JX, He LJ, Lai YR, Liao YJ, Tian XP, Deng HX, Liang YJ, Kung HF, Xie D, Zhu SL. Overexpression of NKX6.1 is closely associated with progressive features and predicts unfavorable prognosis in human primary hepatocellular carcinoma. Tumour Biol. 2015;36(6):4405–4415. doi: 10.1007/s13277-015-3080-4. [DOI] [PubMed] [Google Scholar]
- 52.Li W, Itou J, Tanaka S, Nishimura T, Sato F, Toi M. A homeobox protein, NKX6.1, up-regulates interleukin-6 expression for cell growth in basal-like breast cancer cells. Exp Cell Res. 2016;343(2):177–189. doi: 10.1016/j.yexcr.2016.03.023. [DOI] [PubMed] [Google Scholar]
- 53.Tseng IC, Yeh MM, Yang CY, Jeng YM. NKX6-1 is a novel immunohistochemical marker for pancreatic and duodenal neuroendocrine tumors. Am J Surg Pathol. 2015;39(6):850–857. doi: 10.1097/PAS.0000000000000435. [DOI] [PubMed] [Google Scholar]
- 54.Cohen JE, Eleyan F, Zick A, Peretz T, Katz D. Intratumoral immune-biomarkers and mismatch repair status in leiyomyosarcoma -potential predictive markers for adjuvant treatment: a pilot study. Oncotarget. 2018;9(56):30847–30854. doi: 10.18632/oncotarget.25747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xue M, Chen G, Dai J, Hu J. Development and validation of a prognostic nomogram for extremity soft tissue leiomyosarcoma. Front Oncol. 2019;9:346. doi: 10.3389/fonc.2019.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodgers UR, Lanyon-Hogg T, Masumoto N, Ritzefeld M, Burke R, Blagg J, Magee AI, Tate EW. Characterization of hedgehog acyltransferase inhibitors identifies a small molecule probe for hedgehog signaling by cancer cells. ACS Chem Biol. 2016;11(12):3256–3262. doi: 10.1021/acschembio.6b00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matevossian A, Resh MD. Hedgehog Acyltransferase as a target in estrogen receptor positive, HER2 amplified, and tamoxifen resistant breast cancer cells. Mol Cancer. 2015;14:72. doi: 10.1186/s12943-015-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petrova E, Matevossian A, Resh MD. Hedgehog acyltransferase as a target in pancreatic ductal adenocarcinoma. Oncogene. 2015;34(2):263–268. doi: 10.1038/onc.2013.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fleming JT, Brignola E, Chen L, Guo Y, Zhao S, Wang Q, Li B, Correa H, Ermilov AN, Dlugosz AA, Chiang C. Insight into the etiology of undifferentiated soft tissue sarcomas from a novel mouse model. Mol Cancer Res. 2019;17(5):1024–1035. doi: 10.1158/1541-7786.MCR-18-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bhateja P, Cherian M, Majumder S, Ramaswamy B. The hedgehog signaling pathway: a viable target in breast cancer? Cancers (Basel) 2019;11(8):1126. doi: 10.3390/cancers11081126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bangs F, Anderson KV. Primary cilia and mammalian hedgehog signaling. Cold Spring Harb Perspect Biol. 2017 doi: 10.1101/cshperspect.a028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoon JW, Lamm M, Chandler C, Iannaccone P, Walterhouse D. Up-regulation of GLI1 in vincristine-resistant rhabdomyosarcoma and Ewing sarcoma. BMC Cancer. 2020;20(1):511. doi: 10.1186/s12885-020-06985-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fabbri L, Bost F, Mazure NM. Primary cilium in cancer hallmarks. Int J Mol Sci. 2019;20(6):1336. doi: 10.3390/ijms20061336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Higgins M, Obaidi I, McMorrow T. Primary cilia and their role in cancer. Oncol Lett. 2019;17(3):3041–3047. doi: 10.3892/ol.2019.9942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Notch Inhibitor Shows Modest Efficacy. Cancer Discov 7(2):OF3, 2017. [DOI] [PubMed]
- 66.Mir O, Azaro A, Merchan J, Chugh R, Trent J, Rodon J, Ohnmacht U, Diener JT, Smith C, Yuen E, Oakley GJ, 3rd, Le Cesne A, Soria JC, Benhadji KA, Massard C. Notch pathway inhibition with LY3039478 in soft tissue sarcoma and gastrointestinal stromal tumours. Eur J Cancer. 2018;103:88–97. doi: 10.1016/j.ejca.2018.08.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Primers used in the present study.
Additional file 2: Figure S1. NKX6-1 protein expression in the different stable transfectants. Figure S2. Effects of RU-SKI 43 treatment on SHH downstream genes. Figure S3. Increased NKX6-1 expression correlates with increased RU-SKI 43 resistance in LMS cell lines. Figure S4. GLI1 protein expression in LMS. Figure S5. IHC analysis of BCL-2, c-MYC and Cyclin D1 expression in LMS.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.