SUMMARY
Adult T cell leukemia/lymphoma (ATLL) is a frequently incurable disease associated with the human lymphotropic virus type I (HTLV-I). RNAi screening of ATLL lines revealed that their proliferation depends on BATF3 and IRF4, which cooperatively drive ATLL-specific gene expression. HBZ, the only HTLV-I encoded transcription factor that is expressed in all ATLL cases, binds to an ATLL-specific BATF3 super-enhancer and thereby regulates the expression of BATF3 and its downstream targets, including MYC. Inhibitors of bromodomain-and-extra-terminal-domain (BET) chromatin proteins collapsed the transcriptional network directed by HBZ and BATF3, and were consequently toxic for ATLL cell lines, patient samples, and xenografts. Our study demonstrates that the HTLV-I oncogenic retrovirus exploits a regulatory module that can be attacked therapeutically with BET inhibitors.
INTRODUCTION
After neonatal human lymphotropic virus type I (HTLV-I) infection through breast feeding, approximately 5% of HTLV-I carriers eventually develop adult T cell leukemia/lymphoma (ATLL) with a latency of ~50 years, suggesting that acquired genetic and/or epigenetic changes in cellular genes act in concert with HTLV-I to initiate and maintain oncogenic transformation (Matsuoka and Jeang, 2007). ATLL is classified into various clinical subtypes that vary in prognosis (Shimoyama, 1991). Median survival with the aggressive acute and lymphomatous subtypes is short, typically under 1 year. Various chemotherapy regimens have been evaluated, but none extend median survival beyond 1–2 years (Dittus and Sloan, 2017). Azidothymidine (AZT) plus interferon-α, a regimen that targets the HTLV-I retrovirus, may extend survival in patients receiving chemotherapy, but survival is still less than 2 years for most patients (Hodson et al., 2011). Clinical evaluation of mogamulizumab, a monoclonal antibody that recognizes CCR4 on the surface of most ATLL cells, has demonstrated reasonable complete response rates (31%), but again has not improved outcomes appreciably (13-month median overall survival) (Ishida et al., 2012).
A further understanding of ATLL biology may lead to more effective strategies for this often incurable cancer. Frequent somatic genetic changes are acquired by ATLL cells, including mutations targeting the chemokine receptor CCR4, which enhance chemotaxis, and mutations targeting nuclear factor κB (NF-κB) regulators that enhance malignant cell survival (Kataoka et al., 2015; Nakagawa et al., 2014). Despite these frequent somatic alterations, an ongoing role for the HTLV-1 virus is suggested by the persistent expression of HBZ, the only viral protein expressed in all ATLL cases (Matsuoka and Jeang, 2007). HBZ belongs to the bZIP family of transcription factors, but the exact mechanism by which it may promote ATLL malignancy is unknown.
A key transcriptional network in normal T cells is directed by a transcription factor complex involving IRF4 and BATF, which bind to AP1-IRF composite (AICE) DNA motifs (Murphy et al., 2013). The transcriptional complex of IRF4 and BATF plays key roles in the differentiation and function of certain mouse helper T cell subsets by regulating cytokines and other lineage-specifying factors (Ciofani et al., 2012; Glasmacher et al., 2012; Li et al., 2012). A close paralog of BATF, BATF3, is an indispensable transcription factor in a mouse dendritic cell subset (Tussiwand et al., 2012), but also appears to play a redundant role with BATF in the differentiation of TH2 cells (Tussiwand et al., 2012) and can substitute for BATF in Batf knockout T cells (Tussiwand et al., 2012). IRF4 is highly expressed in ATLL (Imaizumi et al., 2001), associated with therapeutic resistance (Ramos et al., 2007) and somatically mutated in 14% of primary ATLL cases (Kataoka et al., 2015), but the functional roles of IRF4 and BATF/BATF3 in ATLL cells have not been elucidated.
Here we addressed the hypothesis that HBZ controls an essential transcriptional network in ATLL cells with the hope that understanding this regulatory network would suggest therapeutic strategies for this often fatal malignancy.
RESULTS
RNA Interference Screening of ATLL Lines
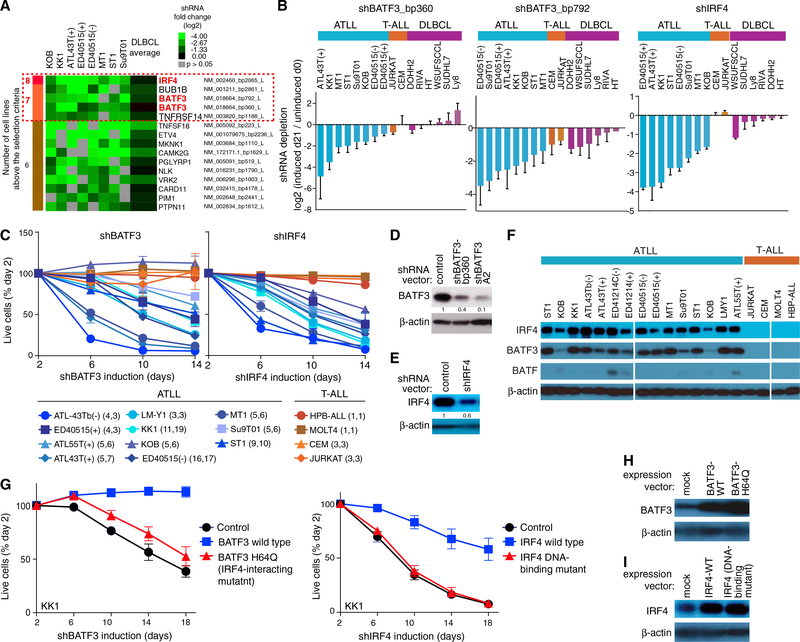
We performed a pooled shRNA screen in eight ATLL cell lines as previously described (Ceribelli et al., 2016; Ngo et al., 2006; Schmitz et al., 2012; Yang et al., 2012), using a library enriched for short hairpin RNAs (shRNAs) targeting lymphoid regulatory factors. Two BATF3 shRNAs (shBATF3_bp360 and shBATF3_bp792) and one IRF4 shRNA (shIRF4) were highly toxic for all ATLL lines, but had little if any effect in other T cell and B cell lines (Figures 1A, 1B, S1A, and S1B; Table S1). To extend these findings, we identified another BATF3 shRNA (shBATF3_A2) with superior knockdown efficiency, which was toxic for all but one ATLL line (Figures 1C and 1D). Induction of shIRF4 was similarly toxic for all 11 ATLL lines tested (Figures 1C, 1E, and S1), whereas knockdown of BATF had no effect (data not shown). By contrast, T cell acute lymphoblastic leukemia (T-ALL) lines were not affected by BATF3 or IRF4 knockdown. Consistent with this selectivity, BATF3 and IRF4, but not BATF, were highly expressed in ATLL lines relative to T-ALL lines (Figure 1F).
Figure 1. BATF3 and IRF4 Are Essential Transcription Factors in ATLL.
(A) Summary figure for shRNA library screening. The shRNAs showing toxicity for ATLL cell lines are listed. The red dashed box indicates the genes above the selection criteria. See STAR Methods for the selection criteria.
(B) Shown are data from shRNA library screening of the indicated cell lines, in which the relative abundance of shBATF3_bp360 (left panel), shBATF3_bp792 (middle panel), and shIRF4 (right panel) is compared at day 0 and day 21 of culture. Error bars represent SEM of quadruplicates.
(C) The indicated cell lines were infected with a retrovirus that expresses shBATF3_A2 or shIRF4 together with GFP. Shown is the fraction of GFP-positive cells over time relative to the GFP-positive fraction on day 2. Error bars represent the SEM of replicates. In parentheses is the number of replicates for shBATF3 followed by number of replicates of shIRF4.
(D) Immunoblot analysis of BATF3 protein in shBATF3_bp360 or shBATF3_A2-transduced KK1. Quantification of BATF3 immunoblot bands, normalized to β-actin and compared with control is shown.
(E) Immunoblot analysis of IRF4 protein in shIRF4-transduced KK1. Quantification of IRF4 immunoblot bands, normalized to β-actin and compared with control is shown.
(F) Immunoblot analysis of BATF3, BATF and IRF4 in the ATLL and T-ALL cell lines.
(G) Wild-type or Q63K BATF3 (left) or wild-type or a DNA-binding mutant IRF4 (right) were retrovirally expressed in KK1 ATLL cells. After puromycin selection of transduced cells, cells were transduced with shBATF3_A2 (left) or shIRF4 (right) and monitored as in (C). Error bars represent the SEM of duplicates.
(H and I) Immunoblot analysis of BATF3 (H) and IRF4 (I) proteins in KK1 ATLL cells that were transduced with the indicated expression vectors. </p/> See also Figure S1 and Table S1.
These observations suggested that IRF4 and BATF3 may cooperate to drive a transcriptional program that is essential for ATLL viability. In support of this hypothesis, a mutant BATF3 isoform that cannot interact with IRF4 (BATF3 Q63K [Tussiwand et al., 2012]) was unable to rescue ATLL cells from shBATF3-mediated toxicity (Figures 1G [left graph] and 1H). Moreover, a mutant IRF4 isoform with impaired DNA-binding ability (Yang et al., 2012) was unable to rescue ATLL cells from shIRF4-mediated toxicity (Figures 1G [right graph] and 1I). By contrast, wild-type BATF3 and IRF4 isoforms were able to rescue ATLL cells from the toxicity of shBATF3 and shIRF4, respectively.
The BATF3/IRF4 Transcriptional Program in ATLL Cells
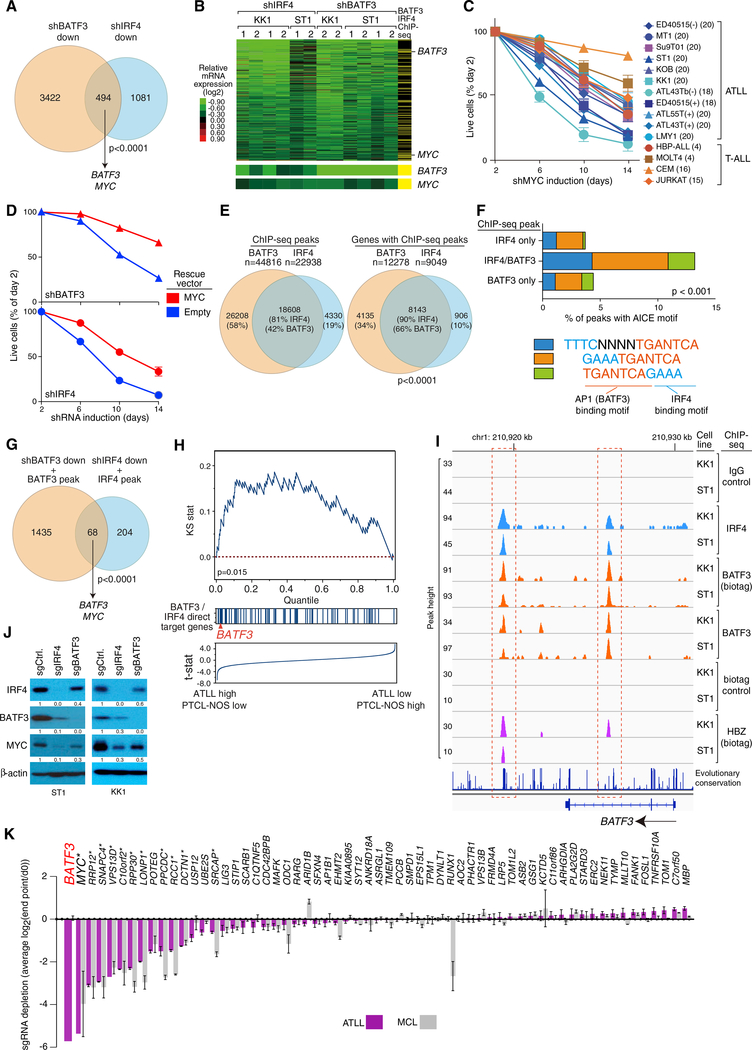
To identify the transcriptional program controlled by BATF3 and IRF4 in ATLL cells, we knocked down each factor and profiled changes in gene expression over time. The gene expression signatures of BATF3 and IRF4 overlapped significantly (p < 0.001), with 494 genes decreasing in expression following knockdown of either factor (Figures 2A and S2A). Analysis of a library of gene expression signatures revealed significant overlap of the BATF3-IRF4 gene expression signature with proliferation-associated and MYC-associated gene expression signatures (Table S2). Accordingly, knockdown of BATF3 or IRF4 caused a pronounced arrest of ATLL cells in the G1 phase of the cell cycle, as did MYC knockdown (Figure S2B). MYC is in the BATF3-IRF4 co-regulated signature (Figure 2B), and knockdown of MYC was toxic for all ATLL lines (Figure 2C). Ectopic expression of MYC partially rescued ATLL cells from the toxicity of BATF3 and IRF4 knockdown, supporting the view that MYC is one of the essential downstream targets of these transcription factors in ATLL (Figure 2D).
Figure 2. Genome-wide Landscape of BATF3/IRF4 Binding.
(A) Venn diagram showing the overlap between genes downregulated by shBATF3 (A2 and bp360) or by shIRF4 in KK1 and ST1 ATLL cell lines. Included are genes with a log2 fold change in mRNA expression of less than −0.3 in more than 3 of 6 shRNA-induced KK1 and ST1 ATLL cell lines.
(B) Heatmap of relative mRNA levels of the 494 genes downregulated by both shBATF3 and shIRF4 in (A), according to the color scale shown. Yellow bars in the far right column indicate genes with ChIP-seq peaks for BATF3 (biotag) and IRF4 within a promoter/gene body window, defined as −15 kb upstream of the transcription start site to +2 kb downstream of the gene body. Log2 fold changes of BATF3 and MYC mRNA expression are shown at the bottom.
(C) MYC addiction in ATLL cell lines. Toxicity assay was done as in Figure 1C. Error bars represent the SEM of replicates. The number of replicates are shown in parentheses.
(D) KK1 cells were transduced with MYC cDNA-expressing vector, and subsequently with vectors co-expressing GFP and either shBATF3_A2 or shIRF4. The GFP-positive cell fraction was monitored as in Figure 1C. Error bars represent the SEM of replicates.
(E) Venn diagrams show overlap of biotag BATF3 ChIP-seq and IRF4 ChIP-seq data with respect to ChIP-seq peaks (left) or genes (right), within a promoter/gene body window (see B).
(F) AICE motif identified within ChIP-seq peaks bound by biotagBATF3, IRF4, or both in KK1 cells and plotted as the fraction of all peaks.
(G) Venn diagram showing the overlap between direct target genes of BATF3 and IRF4 in KK1 and ST1 ATLL cell lines.
(H) Gene set enrichment analysis of BATF3/IRF4 direct target genes in mRNA expression data from primary T cell lymphoma biopsies. Genes were ranked according to the T statistic shown for their relative mRNA expression in ATLL and PTCL-NOS samples. The distribution of direct target genes of BATF3 and IRF4, as defined in the intersection in (G), is shown.
(I) ChIP-seq tracks from KK1 and ST1 cells at the BATF3 locus for IRF4, biotagBATF3, endogenous BATF3, and HBZ. Red dashed boxes indicate regions of IRF4, BATF3 and HBZ binding. Genomic coordinates are based on the NCBI36/hg18 assembly.
(J) Immunoblot analysis of the indicated proteins 6 days after induction of sgRNAs targeting BATF3 and IRF4 in Cas9-expressing ST1 or KK1 cells. Quantification of IRF4, BATF3, and MYC immunoblot bands, normalized to β-actin and compared with shCtrl, is shown.
(K) CRISPR/Cas9 screening for essential BATF3/IRF4 direct target genes in ATLL cell lines. BATF3/IRF4 direct target genes were ranked based on their essentiality, which was quantified as the average log2 fold change in abundance of all sgRNAs corresponding to the indicated genes from day 0 to experimental endpoint (day 28 for ATLL lines; day 21 for MCL lines). Three ATLL cell lines (ST1, KK1, and Su9T01) and two MCL cell lines (Jeko and UPN1) were analyzed. Error bars represent the SEM of replicates. Asterisk indicates core fitness genes in all cell types.
Genome-wide Binding of BATF3 and IRF4 in ATLL Cells
We next used genome-wide chromatin immunoprecipitation sequencing (ChIP-seq) to identify genes that are directly regulated by BATF3 and IRF4. We engineered ATLL cells to express the biotinylation enzyme BirA along with a BATF3 fusion protein containing a “biotag” domain that can be biotinylated by BirA, thereby enabling streptavidin enrichment of BATF3-bound chromatin. We also performed endogenous BATF3 ChIP-seq by using a BATF3 antibody. More than 80% of the genome-binding peaks were shared between the biotag-BATF3 ChIP-seq and endogenous BATF3 ChIP-seq datasets in both KK1 and ST1 cell lines, confirming the specificity of the biotag-BATF3 ChIP-seq peaks (Figure S2C). BATF3 and IRF4 ChIP-seq binding peaks were preferentially localized to enhancer rather than promoter regions (Figure S2D). More than 80% of IRF4 ChIP-seq binding peaks overlapped with biotag-BATF3 peaks in the KK1 and ST1 ATLL lines, and the set of genes with IRF4 and BATF3 ChIP-seq peaks intersected significantly (Figures 2E and S2E; Table S3). In contrast, the IRF4 binding peaks in the diffuse large B cell lymphoma (DLBCL) line HBL1 only overlapped with 18% of the biotag-BATF3 peaks of ST1 ATLL cells, suggesting that BATF3 directs IRF4 to distinct genomic locations in ATLL cells (Figure S2F). This view is also supported by previous reports showing that IRF4 requires interaction with other transcription factors to bind DNA efficiently in many cells, including BATF in T cells and PU.1/SPIB in B cells (Glasmacher et al., 2012; Li et al., 2012; Tussiwand et al., 2012). Analysis of the peaks in ATLL bound by both IRF4 and BATF3 revealed significant enrichment (p < 0.001) for the “AICE” motif (AP-1-IRF composite element) (Glasmacher et al., 2012; Li et al., 2012). The AICE motif consists of an IRF4-binding motif (GAAA) that is near an AP-1-binding motif (TGAnTCA), which can be bound by heterodimers composed of an FOS family member, such as BATF3, and a JUN family member. Three types of AICE motif have been defined, two in which the submotifs are directly adjacent (GAAATGAnTCA or TGAnTCAGAAA) and another in which they are spaced by four base pairs (TTTCnnnnTGAnTCA). All three AICE motifs were observed more often in genomic regions bound by both BATF3 and IRF4 than in regions bound by either factor alone (Figures 2F, S2G, and S2H). These findings suggest that cooperative binding of BATF3 and IRF4 to AICE motifs is likely responsible for their ability to co-regulate genes in ATLL cells (Figure 2B).
By integrating the ChIP-seq and gene expression profiling data, we defined a set of 68 BATF3-IRF4 direct target genes that had overlapping binding peaks for both factors and that decreased in expression following BATF3 or IRF4 knockdown (Figures 2B and 2G). Gene set enrichment analysis using gene expression profiling data from biopsies of various primary T cell lymphoma subtypes (Iqbal et al., 2014) demonstrated that BATF3-IRF4 direct target genes were significantly enriched among genes that are more highly expressed in ATLL than in peripheral T cell lymphoma not otherwise specified (PTCL-NOS) (Figure 2H, Kolmogorov-Smirnov p = 0.015), suggesting that the BATF3 and IRF4 cooperatively regulate transcription in primary ATLL cells. BATF3 is itself a BATF3-IRF4 direct target gene (Figures 2B and 2G) and is in the “leading edge” of genes that are preferentially expressed in ATLL biopsies (Figures 2H and S2I). BATF3 and IRF4 bound together to two regions of the BATF3 locus, one in the second intron and one 3′ of the gene body (Figure 2I). We next developed single guide RNAs (sgRNAs) that could target the endonuclease Cas9 to the coding regions of IRF4 and BATF3, resulting in full inactivation of both proteins in ATLL cells engineered to express Cas9 (Figures 2J and S2J). Inactivation of IRF4 caused a substantial drop in BATF3 protein levels, whereas inactivation of BATF3 caused a modest reduction in IRF4 protein levels (Figure 2J). Knockdown of IRF4 by shRNA also decreased BATF3 protein levels modestly, whereas shBATF3 did not affect IRF4 protein levels (data not shown). Together, these observations suggest that BATF3 works with IRF4 to drive a positive autoregulatory loop for BATF3 expression in ATLL cells. Given the functional importance of MYC in ATLL cells (Figures 2C and 2D), it was also notable that BATF3 and IRF4 bound directly to the MYC promoter region, suggesting that they directly upregulate MYC in ATLL cells (Figures 2G and S2K).
To investigate the essentiality of the 68 BATF3-IRF4 direct target genes, we performed CRISPR/Cas9-mediated screening. A lentiviral sgRNA expression library containing three to four different sgRNAs per gene was used to transduce, in duplicate, three Cas9-expressing ATLL cell lines (KK1, ST1, Su9T01) and, as a control, two non-ATLL cell lines (mantle cell lymphoma [MCL] cell lines Jeko and UPN1). The abundance of each sgRNA was assessed by next-generation sequencing of genomic DNA harvested from the cell populations following puromycin selection for lentiviral integration (time 0) and again after 3–4 weeks in culture. Essential genes are those for which the corresponding sgRNAs were depleted in the final cell pool relative to the starting cell pool. For each gene, essentiality was quantified as an average log2 fold change of all well-measured sgRNAs (Table S4) (see STAR Methods for details). This analysis revealed BATF3 and MYC to be the most essential direct BATF3-IRF4 target genes in ATLL lines, whereas BATF3 was not essential in the control MCL lines (Figure 2K). While the majority of BATF3-IRF4 direct target genes were not essential in either ATLL or MCL cell lines, 11 were essential in both lymphoma subtypes and are known to be core fitness genes in mammalian cells (MYC, RRP12, SNAPC4, VPS13D, C10orf2, RPP30, LONP1, PPCDC, RCC1, DCTN1, and SRCAP) (Hart et al., 2015; Wang et al., 2015). Gene ontology analysis of the 68 BATF3-IRF4 direct target genes showed enrichment for mitochondrial-related gene sets (Table S5), and two of the essential genes have been implicated in mitochondrial function (C10orf2; LONP1), suggesting a role for BATF3 and IRF4 in the regulation of mitochondrial processes in ATLL cells.
HBZ-Driven BATF3 Expression in ATLL Cells
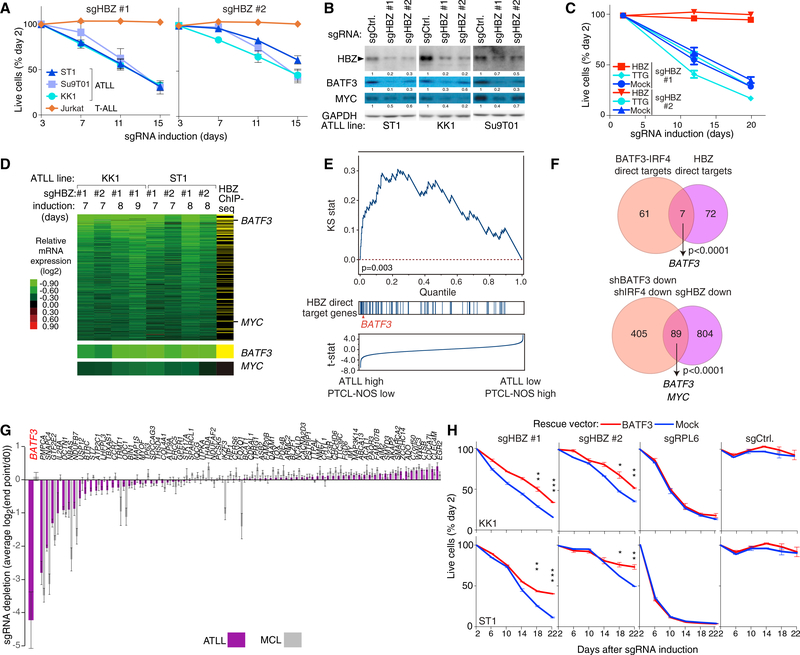
HBZ is unique among HTLV-I viral proteins in being maintained in expression in all ATLL cases, suggesting that it may help maintain the malignant phenotype (Matsuoka and Jeang, 2007). We therefore examined whether HBZ is an essential viral gene in ATLL lines using CRISPR/Cas9-mediated knockout. Induction of two different HBZ sgRNAs decreased HBZ protein levels, resulting in a time-dependent decrease in viable cells (Figures 3A, 3B, and S3A). The toxicity of sgRNA-mediated HBZ inactivation could be prevented by ectopic provision of an HBZ isoform that cannot be targeted by these sgRNAs, demonstrating their specificity (Figures 3C and S3B). Moreover, ectopic induction of TTG-HBZ, in which the start codon (ATG) of HBZ-cDNA was replaced by TTG to prevent protein expression, failed to rescue the sgHBZ-mediated toxicity, indicating that HBZ protein is required for KK1 ATLL cell proliferation.
Figure 3. HBZ Drives BATF3 Expression in ATLL.
(A) The indicated cell lines were infected with a retrovirus that expresses sgHBZ #1 or sgHBZ #2 together with GFP. Shown is the fraction of GFP-positive cells relative to the GFP-positive fraction on day 3. Error bars represent the SEM of replicates (at least three).
(B) Immunoblot analysis of the indicated proteins after 6 days of induction of sgHBZ #1 or sgHBZ #2 in the indicated cell lines. Quantification of HBZ, BATF3 and MYC immunoblot bands, normalized to GAPDH and compared with shCtrl, is shown.
(C) KK1 cells were transduced with retroviruses expressing an sgRNA-resistant HBZ isoform, with retroviruses expressing TTG-HBZ or with empty vector, and were subsequently transduced with retroviruses co-expressing GFP and either sgHBZ #1 or sgHBZ #2. The GFP-positive cell fraction was monitored as in Figure 1C. Error bars represent the SEM of quadruplicates.
(D) Heatmap of relative mRNA expression levels of 894 genes that were downregulated by log2 fold change of less than −0.3 in more than 4 of 8 HBZ-knockout KK1 and ST1 ATLL cells, according to the color scale shown. Yellow bars in the far right column indicate genes with biotagHBZ-ChIP-seq peaks within a promoter region gene window (see Figure 2B legend). Log2 fold changes of BATF3 and MYC mRNA expression are shown at the bottom.
(E) Gene set enrichment analysis of HBZ direct target genes in mRNA expression data from primary T cell lymphoma biopsies. Genes were ranked according to the T statistic shown for their relative mRNA expression in ATLL and PTCL-NOS samples. The distribution of HBZ direct target genes, as defined in (D), is shown.
(F) Venn diagram of 68 direct target genes of BATF3 and IRF4 and 79 direct target genes of HBZ (top) or of genes that are downregulated following knockdown of BATF3 or IRF4 and those downregulated by HBZ inactivation in ATLL (bottom).
(G) HBZ direct target genes were ranked based on their essentiality quantified as average log2 fold change (day 0/endpoint; day 21 in MCL lines, and day 28 in ATLL lines) of all replicates of all corresponding sgRNAs in CRISPR/Cas9-mediated screening. Three ATLL cell lines (ST1, KK1, and Su9T01) or two MCL cell lines (Jeko and UPN1) were analyzed. Error bars represent the SEM of replicates.
(H) ATLL lines were transduced with retroviruses expressing BATF3, and subsequently with retroviruses co-expressing GFP and the indicated shRNAs. The GFP-positive cell fraction was monitored as in Figure 1C. Results are shown as means ±SEM (n = 2). *p < 0.05, **p < 0.01, ***p < 0.001.
We next identified genes that decreased in mRNA abundance upon HBZ inactivation (n = 894), among which were BATF3 and MYC (Figure 3D). To define HBZ direct target genes, we intersected this gene set with the set of genes bound by HBZ, as assessed by ChIP-seq using ATLL cells expressing BirA and a biotag-HBZ fusion protein (Figure 3D, right column). The 79 HBZ direct target genes thus defined were enriched among genes that are more highly expressed ATLL biopsies than in PTCL-NOS biopsies by gene set enrichment analysis (Iqbal et al., 2014) (Figure 3E, Kolmogorov-Smirnov p = 0.003).
Only seven genes were direct targets of both HBZ and BATF3-IRF4, but BATF3 was notably among these (Figure 3F, upper). Indeed, BATF3 mRNA and protein expression decreased following HBZ inactivation (Figures 3B, S3C, and S3D). The set of genes that were downregulated following knockdown of BATF3 or IRF4 overlapped significantly with those that were downregulated following HBZ inactivation (n = 89, p < 0.001; Figure 3F, lower). This shared gene expression program included MYC, which was decreased in both mRNA and protein abundance following HBZ inactivation (Figures 3B, S3C, and S3D). By ChIP-seq analysis, MYC is a direct target of BATF3/IRF4 but not of HBZ (Figure S2K), suggesting that HBZ regulates MYC expression indirectly through BATF3.
Since HBZ has many direct targets in ATLL cells in addition to BATF3, we investigated whether these genes were essential in ATLL cell lines by CRISPR/Cas9-mediated screening, as in Figure 2K. This revealed that BATF3 is the most essential HBZ target gene in ATLL lines, but was not essential in MCL lines (Figure 3G and Table S6). While the majority of HBZ direct target genes were not essential in either ATLL or MCL cell lines, a few were essential in both lymphoma subtypes and are known to be core fitness genes in mammalian cells (Hart et al., 2015; Wang et al., 2015) (e.g., PMPCA, SNAPC4, GTF2E2; Figure 3G and Table S6).
This analysis suggested that BATF3 was one of the most quantitatively important downstream targets of HBZ. Indeed, ectopic expression of BATF3 partially, but consistently, rescued ATLL cells from the toxicity of two HBZ sgRNAs with statistical significance, whereas BATF3 failed to rescue the cells from the toxicity of an sgRNA targeting the ribosomal protein gene RPL6 (Figure 3H). While the incomplete rescue by BATF3 is likely due to loss of one or more core fitness genes in sgHBZ-expressing cells, these data support the view that BATF3 contributes to the oncogenic action of HBZ.
Targeting the HBZ/BATF3/IRF4 Regulatory Network with BET Protein Inhibitors
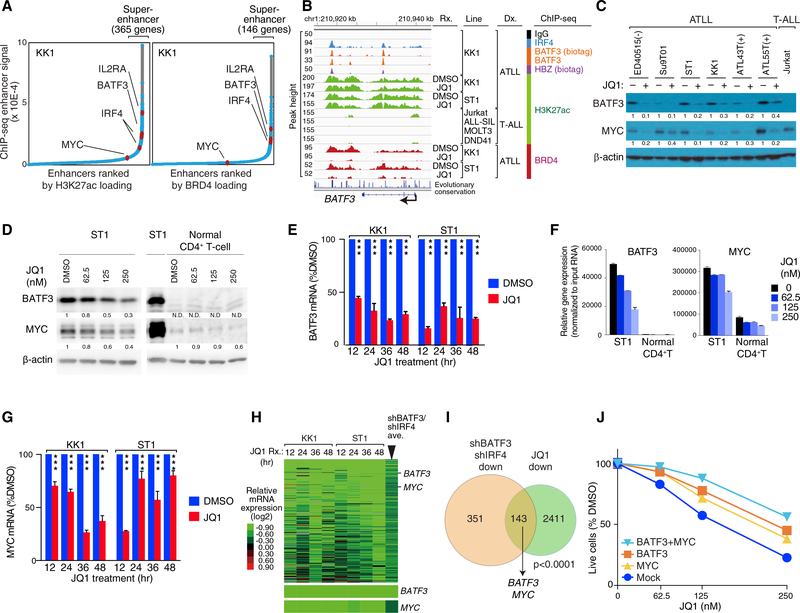
The above considerations suggested that pharmacologic inhibition of the BATF3-IRF4 regulatory network might be a means to attack the HBZ oncogenic program therapeutically. The expression levels of oncogenes and cell lineage-specific genes in cancer cells are often governed by large clusters of regulatory elements known as super-enhancers (Loven et al., 2013; Whyte et al., 2013), which can be pharmacologically disrupted by inhibitors of bromodomain-and-extra-terminal (BET) domain chromatin regulators (Delmore et al., 2011). We defined superenhancers in two complementary ways, one using ChIP-seq of histone H3 lysine 27 acetylation (H3K27ac) and the other using ChIP-seq for BRD4 binding. H3K27ac and BRD4 defined 365 and 146 super-enhancers, respectively, in KK1 ATLL cells (Figure 4A and Table S7), many of which were also observed in ST1 ATLL cells (Figure S4A and Table S7). Both ATLL lines had super-enhancers at the BATF3 locus and at IL2RA, a gene that is characteristically expressed in ATLL. The BATF3 superenhancer spanned roughly 25 kb in KK1 and ST1 ATLL cells, but was not observed in four T-ALL lines, demonstrating its cell type specificity (Figure 4B). The IRF4 locus had a superenhancer in KK1 but not ST1 cells, whereas MYC was not associated with a super-enhancer in either line (Figures S4A–S4C). Importantly, the BATF3 super-enhancer was not observed in various normal human T cell subpopulations (Figure S4D and Table S7), suggesting that the epigenetic status of the BATF3 locus in ATLL may be the result of aberrant gene regulation in these malignant cells.
Figure 4. BET Inhibitors Disrupt the BATF3/IRF4/HBZ Transcriptional Network in ATLL.
(A) Enhancers were ranked based on increasing H3K27ac marks (left panel) and BRD4 loading (right panel) to identify the indicated number of genes with super-enhancers.
(B) ChIP-seq tracks at the BATF3 locus for the indicated proteins and histone marks are shown for the indicated ATLL and T-ALL lines. Also shown is the effect of JQ1-treated KK1 (500 nM) and ST1 (250 nM) cells on H3K27ac and BRD4 ChIP-seq signals compared with the DMSO-treated control cells.
(C) Immunoblot analysis of the indicated proteins following treatment of the indicated cell lines with JQ1 (500 nM).
(D) Immunoblot analysis of BATF3 and MYC protein in JQ1-treated ST1 ATLL cells and normal CD4+ T cells. Quantification of BATF3 and MYC immunoblot bands, normalized to β-actin and compared with DMSO-treated control cells, is shown. N.D., not determined.
(E) qRT-PCR to measure relative mRNA expression levels of BATF3 in the indicated ATLL lines treated with JQ1 (500 nM for KK1 and 250 nM for ST1) or DMSO as in (B) for the indicated times. Error bars represent the SEM of triplicates. ***P < 0.001.
(F) BATF3 and MYC mRNA in JQ1-treated ST1 cells. Normal CD4+ T cell was also studied. mRNA expression levels of the indicated genes were measured and normalized by the amount of input RNA. Error bars represent the SEM of duplicates.
(G) qRT-PCR to measure relative mRNA expression levels of MYC in the indicated ATLL lines treated with JQ1 (500 nM for KK1 and 250 nM for ST1) or DMSO as in (B) for the indicated times. Error bars represent the SEM of triplicates. ***P < 0.001.
(H) Heatmap of mRNA expression changes (log2) after JQ1 treatment relative to DMSO-treated cells according the color scale shown. Genes were chosen that were downregulated by JQ1 and by both shBATF3 and shIRF4. Far right column indicates the average expression values of 12 shBATF3- and shIRF4-transduced ATLL lines. The relative mRNA levels of BATF3 and MYC are shown at the bottom.
(I) Venn diagram of genes downregulated by both shBATF3 and shIRF4 induction and by JQ1 treatment in KK1 and ST1 ATLL lines.
(J) ST1 cells were transduced with retroviral vectors expressing BATF3 along with GFP and/or MYC along with lyt2, and subsequently treated with JQ1. The GFP+/lyt2+ cell fraction was monitored as in Figure 1C. Error bars represent the SEM of duplicates, but they are short and obscured by the symbols.
See also Figures S4 and S5; Table S7.
The small molecule JQ1 prevents the BET protein BRD4 from interacting with chromatin, which is required for the function of super-enhancers (Filippakopoulos et al., 2010). JQ1 treatment reduced BATF3 mRNA and protein levels in all ATLL lines tested (Figures 4C–4F), correlating with eviction of BRD4 from BATF3 super-enhancer (Figure 4B). MYC mRNA and protein expression levels were also decreased by JQ1 treatment without a loss of BRD4 occupancy at the MYC locus (Figures 4C, 4D, 4F, 4G, and S4B), presumably due, in part, to BATF3 downregulation (Figures 4C–4F). Gene expression profiling identified 143 genes that were downregulated by JQ1 treatment of ATLL lines and also by knockdown of BATF3 and IRF4, including BATF3 and MYC (Figures 4H, 4I, and S5A). Ectopic expression of BATF3 and MYC separately and together was able to partially rescue ATLL cells from JQ1-mediated toxicity, suggesting that JQ1 toxicity was due, in part, to downmodulation of the BATF3-dependent transcriptional program (Figure 4J).
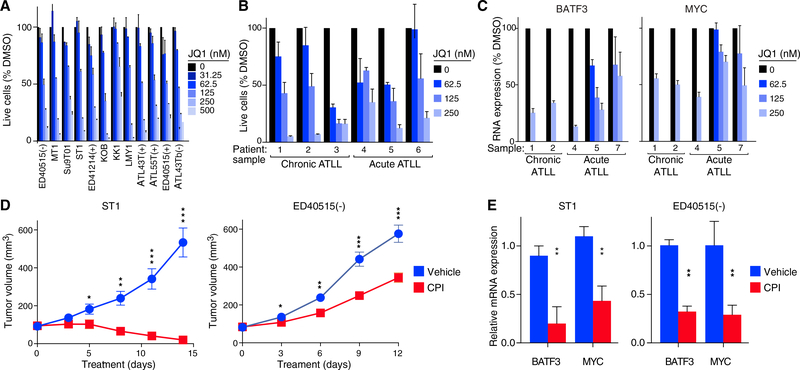
JQ1 treatment was consistently toxic for all ATLL cell lines tested at dose ranges that killed cell line models of T-ALL and DLBCL, which are known to rely on the binding of BET proteins to chromatin (Ceribelli et al., 2014; Loosveld et al., 2014) (Figures 5A and S5B). Moreover, JQ1 reduced the viability of primary ATLL samples and downregulated BATF3 and MYC mRNA levels in a dose-dependent manner (Figures 5B, 5C, and S5C). Samples taken from patients in both the acute and chronic phases of the disease responded equivalently to JQ1 treatment. Finally, we treated mouse xenograft models of ATLL with the BET protein inhibitor CPI-203, a JQ1 analog with superior bioavailability in mice (Ceribelli et al., 2014; King et al., 2013) and with similar toxicity in vitro (Figure S5D) (Ceribelli et al., 2014). In two different xenograft models, we observed significant regression or inhibition of tumor growth along with a reduction in BATF3 and MYC mRNA levels, without evidence of systemic toxicity (Figures 5D, 5E, and S5E).
Figure 5. Toxicity of BET Protein Inhibitors for ATLL Cells In Vitro and In Vivo.
(A) Viable cells measured by the MTS assay for the indicated ATLL cell lines treated with the indicated concentrations of JQ1 for 4 days. Error bars show the SEM of duplicates.
(B) Viable cells were measured ([3H]thymidine incorporation assay) following JQ1 treatment at the indicated concentrations for 6 days (chronic subtype of ATLL) or for 3 days (acute subtype of ATLL). Error bars represent the SEM of triplicates.
(C) BATF3 and MYC mRNA expression quantified by qRT-PCR, normalized to HPRT1 mRNA expression, in primary ATLL cells treated with JQ1 for 24 hr at the indicated concentrations. Error bars represent the SEM of triplicates.
(D) ST1 and ED405151(−) ATLL cells were established as subcutaneous tumors (average 93 mm3 and 83 mm3) in immunodeficient mice, which were then treated daily for 14 days and 12 days, respectively, with CPI-203 (5 mg/kg/injection, twice a day) or vehicle control by intraperitoneal injection. Tumor growth was monitored as a function of tumor volume. Error bars show the SEM of 6 mice (ST1) and 7 mice (ED40515(−)) per group.
(E) BATF3 and MYC mRNA expression quantified by qRT-PCR, normalized to HPRT1 expression, in ST1 and ED40515(−) ATLL xenograft tumors harvested after daily treatment for 4 days (ST1) and 9 days (ED40515(−)) with CPI-203 (5 mg/kg/injection, twice a day) or with vehicle control by intraperitoneal injection. Error bars represent the SEM of triplicates.
*p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S5.
DISCUSSION
While the molecular interactions between IRF4 and BATF/BATF3 are known to control the differentiation of normal T cells, our functional genomic investigations uncovered an essential role for the BATF3/IRF4 transcriptional network in malignant ATLL cells (Figure 6). The activity of BATF3 in ATLL cells is promoted by a positive autoregulatory loop in which BATF3 binds to its own super-enhancer. Genetic ablation of BATF3 or IRF4 in ATLL cells induces a profound cell-cycle arrest, presumably due in part to the fact that the oncogene MYC is a direct BATF3/IRF4 target gene. The expression of BATF3 and BATF3/IRF4 target genes distinguished primary ATLL tumors from PTCL-NOS, demonstrating that the transcription network we describe in ATLL cell lines is a characteristic feature of the disease as whole.
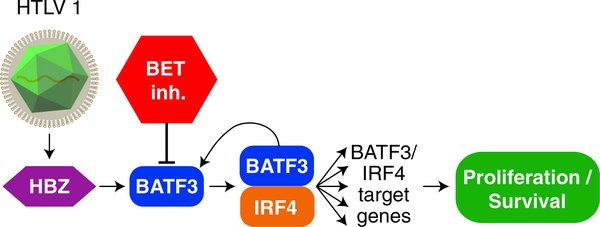
Figure 6.
Schematic of the Malignant HBZ/BATF3/IRF4 Transcriptional Network in ATLL
The viral oncoprotein HBZ, which is consistently expressed in ATLL at all stages of the disease, binds to a super-enhancer region in the BATF3 locus, thereby upregulating BATF3 itself as well as BATF3/IRF4 target genes (Figure 6). While BATF3 plays a subordinate or redundant role to BATF in regulating normal T cell biology, the characteristic expression and function of BATF3 in ATLL appears to be promoted by persistent HBZ expression, rather than reflecting the expression status of BATF3 in a normal T cell subpopulation. This viewpoint is supported by the observation that normal human helper T cell subsets seldom bear the activating H3K27ac epigenetic mark at the BATF3 locus. Genetic inactivation of HBZ was toxic for ATLL lines and downregulated the BATF3/IRF4 network of target genes, including MYC. ChIP-seq analysis coupled with gene expression profiling revealed that HBZ binds and directly upregulates the expression of 79 cellular genes, which are more highly expressed in primary ATLL patient samples than in samples from patients with PTCL-NOS. CRISPR/Cas9 screening of ATLL cell lines revealed that BATF3 is quantitatively the most essential direct HBZ target gene in these cancer cells, and ectopic provision of BATF3 was able to partially reverse the toxicity of HBZ knockdown in ATLL. These observations underscore the central importance of BATF3 and its network of target genes to the oncogenic action of the viral HBZ protein.
Since all ATLL cell lines tested rely on the transcriptional network established by HBZ and BATF3/IRF4, it is conceivable that therapies targeting this network could be effective across a broad range of ATLL cases. BET inhibitors attenuated the expression of BATF3 and its downstream targets, and were toxic for ATLL cell lines and primary samples from both the chronic and acute subtypes of this disease. The toxicity of these agents for ATLL cells could be partially reversed by ectopic provision of BATF3. Thus, even though BET inhibitors are expected to have pleiotropic effects on gene expression, one of the important aspects of their toxicity for ATLL cells is downregulation of BATF3 and its targets, notably including MYC.
We discovered that BET inhibitors were toxic for established ATLL cell lines that model a late stage of the ATLL disease process in which Tax is typically not expressed. Tax expression is lost in more than 60% of advanced-stage ATLL cases due to genomic deletion or epigenetic silencing, presumably in order to evade the host immune system (Matsuoka and Jeang, 2007). A previous study suggested the possible use of the BET inhibitor JQ1 for Tax-expressing T cells (Wu et al., 2013), which represent HTLV-I-infected non-malignant T cells or ATLL cells in the initiation phase of the disease. By contrast, we studied cell line models of late-stage ATLL that harbor many of the previously identified recurrent genetic events that activate oncogenic programs in ATLL (Kataoka et al., 2015; Nakagawa et al., 2014). Using a computational pipeline that can effectively identify cancer gene mutations from RNA-sequencing data (M.N. and L.M.S., unpublished data; Schmitz et al., 2012), we observed that the ST1 ATLL line has a T128M CARD11 mutation that is recurrent in DLBCL (Bohers et al., 2014) and has the potential to activate NF-κB by targeting the CARD11 coiled-coil domain (Lenz et al., 2008). The KK1 ATLL line has a Y338* CCR4 mutation that has the potential to activate phosphoinositide 3-kinase and AKT (Nakagawa et al., 2014), and a S345F PLCG1 mutation that is a mutational hot-spot in ATLL (Kataoka et al., 2015), which has been shown to activate NF-AT and potentially NF-κB in cutaneous T cell lymphoma (Vaque et al., 2014). Both of these ATLL lines were highly sensitive to JQ1 treatment, raising the possibility that BET protein inhibition may be an effective strategy across a wide range of ATLL cases, irrespective of oncogenic abnormalities, due to their ability to extinguish the essential HBZ/BATF3/IRF4 transcriptional program.
Chemotherapy of acute ATLL does not produce lasting remissions for the majority of patients, and although allogeneic bone marrow transplantation can be curative in a subset of patients, it is associated with significant morbidity and mortality (for review see Kato and Akashi, 2015). Treatment with anti-CCR4 antibodies can induce complete but not lasting remissions and can have significant toxicity in some patients (Fuji et al., 2016; Ishida et al., 2012). Faced with the clear need for effective therapies for ATLL, our work supports the clinical evaluation of BET-domain inhibitors as a strategy to extinguish an essential transcriptional network directed by the viral oncoprotein HBZ.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to the Lead Contact Lou Staudt at lstaudt@mail.nih.gov.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human ATLL Samples
Written informed consent was obtained in accordance with the Declaration of Helsinki and the protocol for this study was approved by the Investigational Review Board of the National Cancer Institute (NCI). Peripheral blood mononuclear cells (PBMNCs) were isolated from ATLL patients by Ficoll-Hypaque. Normal human CD4 T-cells were purified from PBMNCs from healthy volunteers by using human CD4+ T Cell Isolation Kit (Miltenyi).
Mice
All animal experiments were approved by the National Cancer Institute Animal Care and Use Committee (NCI ACUC) and were performed in accordance with NCI ACUC guidelines. Female NSG mice and NOD/SCID mice were purchased from Jackson laboratory.
Cell Lines and Cell Culture
The ATLL cell lines were kindly provided by the following researchers: Michiyuki Maeda (Kyoto University; ED40515(+), ED40515(−), ED41214(+), ATL43T(+), ATL43Tb(−), and ATL55T(+)), Yasuaki Yamada (Nagasaki University; ST1, KOB, KK1, LMY1), Tomoko Hata (Nagasaki University; ST1), Naomichi Arima (Kagoshima University; Su9T01). All lines were cultured with RPMI media containing10% fetal calf serum (FCS), penicillin and streptomycin, with IL-2-dependent lines (ED40515(+), ED41214(+), ATL43T(+), ATL55(+), KOB, KK and LMY1) cultured with the addition of human recombinant IL2 (100IU/mL, Hoffmann-La Roche) to this media. Mycoplasma contamination was eradicated by Mycoplasma Removal Agent. All cell lines were tested for unique profiles of polymorphic DNA copy number variants (CNV fingerprint; unpublished protocol from L. Bergsagel). HBZ mRNA and Protein expression were confirmed in all ATLL cell lines (Figure S1). All ATLL cell lines were engineered to express ecotropic retroviral receptors and TET repressor for transduction of retroviral shRNA vectors and expression vectors as previously described(Ngo et al., 2006). For Cas9-mediated gene inactivation, KK1, ST1, Su9T01 and Jurkat cell lines were engineered to express human codon-optimized S. pyogenes Cas9 using the pTO-Cas9-hygro vector (Cas9 from lentiCRISPR v2 ligated into pRCMV/TO-hygro vector) or lentiCas9-Blast. LentiCRISPR v2 and lentiCas9-Blast were gifts from Dr. Feng Zhang (Addgene plasmid # 52961 and # 52962, respectively).
METHOD DETAILS
shRNA Library Screening
shRNA library screening was performed as previously described (Ceribelli et al., 2016; Yang et al., 2012). A pool of 12,513 shRNA expression vectors was retrovirally transduced into ATLL lines. After puromycin selection, shRNA expression was induced by doxycycline (50 ng/ml). Genomic DNA from both uninduced (day 0) and induced cultures (day 21) was harvested. The half hairpin shRNA sequences were amplified by TaKaRa LA Taq Tm Hot Start DNA polymerase (Clontech-TaKaRa) using a Forward primer that hybridizes in the loop region and the Indexed Reverse Primers, which hybridize in the vector region flanking the shRNA. The primers are listed in the Key Resources Table. Depletion of specific shRNAs was quantitatively assessed by next generation sequencing. The screening was done in quadruplicate and the shRNAs which showed toxicity (log2 fold change (day 21 /day 0)) <−1 plus p<0.05) in 7–8/8 of ATLL cell lines were selected. shRNAs showing general toxicity (toxic in ≥ 50% of B-cell lymphoma cell lines (n=6)) were excluded in this analysis.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| goat polyclonal IRF-4 Antibody (M-17) | Santa Cruz Biotechnology | Cat# sc-6059, RRID:AB_2127145 |
| goat polyclonal IRF-4 Antibody (N-18) | Santa Cruz Biotechnology | Cat# sc-11450, RRID:AB_2127144 |
| mouse monoclonal BATF3 Antibody | Abnova Corporation | Cat# H00055509-M04, RRID:AB_10903097 |
| Anti-c-Myc antibody [Y69] | Abcam | Cat# ab32072, RRID:AB_731658 |
| BATF (D7C5) Rabbit mAb #8638 | Cell Signaling | Cell Signaling Technology Cat# 8638S, RRID:AB_11141425 |
| Rabbit-polyclonal-HBZ serum | Arnold et al., 2008 | N/A |
| Polyclonal Sheep-Human BATF3 Antibody | R&D | Cat# AF7437, RRID:AB_11127798 |
| Rabbit polyclonal BRD4 antibody | Bethyl | Cat# A301-985A, RRID:AB_1576498 |
| Rabbit polyclonal anti-Histone H3 (acetyl K27) antibody | Abcam | Cat# ab4729, RRID:AB_2118291 |
| CD8a (Ly-2) MicroBeads | Miltenyi | Order no: 130-117-044 |
| Biological Samples | ||
| Human primary ATLL PBMNC | NIH/NCI | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| JQ1 | Constellation Pharmaceuticals/ Tocris Bioscience | NA/ Cat. No. 4499, Cas No. 1268524-70-4 |
| CPI-203 | Constellation Pharmaceuticals | NA, Cas No. 1446144-04-2 |
| Critical Commercial Assays | ||
| TaqMan assay, HPRT1 | Applied Biosystems | Hs99999909_m1 |
| TaqMan assay, BATF3 | Applied Biosystems | Hs00232744_m1 |
| TaqMan assay, IRF4 | Applied Biosystems | Hs00180031_m1 |
| TaqMan assay, MYC | Applied Biosystems | Hs00153408_m1 |
| Two-color human Agilent 4x44K gene-expression arrays | AGILENT | G4112F |
| LowInput QuickAmp Labeling Kit Two-Color | AGILENT | 5190-2306 |
| NEXTflex™ Illumina ChIP-seq Library Prep Kit | Bioo Scientific | #5143-02 |
| Deposited Data | ||
| ChIP-seq datasets | This study | GSE94732 |
| Gene expression datasets | This study | GSE94409 |
| Microarray data of primary human T-cell lymphoma samples | Gene Expression Omnibus (GEO) | GSE6338 |
| Microarray data of primary human T-cell lymphoma samples | Gene Expression Omnibus (GEO) | GSE14879 |
| Microarray data of primary human T-cell lymphoma samples | Gene Expression Omnibus (GEO) | GSE19069 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Short Read Archive SRA: SRX290664 | |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Short Read Archive SRA: SRX290665 | |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Short Read Archive SRA: SRX290666 | |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1462467 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1462468 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1414734 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1414738 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1056919 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1056920 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1056940 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1056941 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1056942 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1056943 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1056948 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1056949 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1056950 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1056951 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM772987 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1893223 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1893224 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1893225 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1893226 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1893227 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1893228 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1893229 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1893230 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1893231 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1893232 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1893233 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1893234 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1893235 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1893236 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1893237 |
| H3K27ac ChIP-seq data of human helper T-cell subsets | Gene Expression Omnibus (GEO) | GSM1893238 |
| H3K27ac ChIP-seq data of T-ALL cell lines | Short Read Archive SRA: SRR1057274 | |
| H3K27ac ChIP-seq data of T-ALL cell lines | Short Read Archive SRA: SRR969480 | |
| H3K27ac ChIP-seq data of T-ALL cell lines | Gene Expression Omnibus (GEO) | GSM1816978 |
| H3K27ac ChIP-seq data of T-ALL cell lines | Gene Expression Omnibus (GEO) | GSM1816979 |
| H3K27ac ChIP-seq data of T-ALL cell lines | Gene Expression Omnibus (GEO) | GSM1519644 |
| H3K27ac ChIP-seq data of T-ALL cell lines | Gene Expression Omnibus (GEO) | GSM1519645 |
| H3K27ac ChIP-seq data of T-ALL cell lines | Gene Expression Omnibus (GEO) | GSM1003462 |
| H3K27ac ChIP-seq data of T-ALL cell lines | Gene Expression Omnibus (GEO) | GSM1003558 |
| Experimental Models: Cell Lines | ||
| ED40515(+) | Michiyuki Maeda, Kyoto University | N/A |
| ED40515(−) | Michiyuki Maeda, Kyoto University | N/A |
| ED41214(+) | Michiyuki Maeda, Kyoto University | N/A |
| ATL43T(+) | Michiyuki Maeda, Kyoto University | N/A |
| ATL43Tb(−) | Michiyuki Maeda, Kyoto University | N/A |
| ATL55T(+) | Michiyuki Maeda, Kyoto University | N/A |
| ST1 | Yasuaki Yamada and Tomoko Hata, Nagasaki University | N/A |
| KOB | Yasuaki Yamada, Nagasaki University | N/A |
| KK1 | Yasuaki Yamada, Nagasaki University | N/A |
| LM-Y1 | Yasuaki Yamada, Nagasaki University | N/A |
| Su9T01 | Naomichi Arima, Kagoshima University | N/A |
| Experimental Models: Organisms/Strains | ||
| Female NSG mice | Jackson Laboratory | 005557 - NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ |
| Female NOD/SCID mice | Jackson Laboratory | 001303 - NOD.CB17-Prkdcscid/J |
| Oligonucleotides | ||
| shCtrl: CTCTCAACCCTTTAAATCTGA | This paper | N/A |
| shBATF3_bp360: GAGTTGCTGCTCAGAGAAGTC (CDS) | This paper | N/A |
| shBATF3_bp792: CACAGAGCCCTTGTGCAGATC (3’ UTR) | This paper | N/A |
| shBATF3_A2: ACATCCAACAGGCGCCAAACT (3’ UTR) | This paper | N/A |
| shIRF4: GTGCCATTTCTCAGGGAAGTA (3’ UTR) | This paper | N/A |
| shMYC: CGATTCCTTCTAACAGAAATG (3’ UTR) | This paper | N/A |
| sgAAVS1 (control): GGGGCCACTAGGGACAGGAT | This paper | N/A |
| sgBATF3: GAAGGCTGACAAGCTCCATG | This paper | N/A |
| sgIRF4: CAAGCAGGACTACAACCGCG | This paper | N/A |
| sgHBZ #1: TCCTTAGAAGAGGAAAGCCG | This paper | N/A |
| sgHBZ #2: AGAGGAAGCGAAAAAAAGAG | This paper | N/A |
| sgRPL6: GTACTCTCACCTTGCCCCTG | This paper | N/A |
| Forward primer (shRNA library screening): AATGATACGGCGACCACCGTCAGGGCAGTGATGTTGCCCCTCGGAAG | Yang et al., 2012 | N/A |
| Indexed Reverse Primer (shRNA library screening): 3’-GAGCTCTCTAGCTAGATTAAGGCCGCGGATCTCTTCC TGCATACGGCAGAAGACGAAC -5’So13_00_rev | Yang et al., 2012 | N/A |
| Indexed Reverse Primer (shRNA library screening): 3’-GAGTTCTCTAGCTAGATTAAGGCCGCGGATCTCTTCC TGCATACGGCAGAAGACGAAC -5’ So13_01_rev | Yang et al., 2012 | N/A |
| Indexed Reverse Primer (shRNA library screening): 3’-GAGCCCTCTAGCTAGATTAAGGCCGCGGATCTCTTCCTGCATACGGCAGAAGACGAAC -5’ So13_02_rev | Yang et al., 2012 | N/A |
| Indexed Reverse Primer (shRNA library screening): 3’-GAGCTTTCTAGCTAGATTAAGGCCGCGGATCTCTTCCTGCATACGGCAGAAGACGAAC -5’ So13_03_rev | Yang et al., 2012 | N/A |
| Indexed Reverse Primer (shRNA library screening): 3’-GAGCTCCCTAGCTAGATTAAGGCCGCGGATCTCTTCCTGCATACGGCAGAAGACGAAC -5’ So13_04_rev | Yang et al., 2012 | N/A |
| Indexed Reverse Primer (shRNA library screening): 3’-GAGCTCTTTAGCTAGATTAAGGCCGCGGATCTCTTCCTGCATACGGCAGAAGACGAAC -5’ So13_05_rev | Yang et al., 2012 | N/A |
| Indexed Reverse Primer (shRNA library screening): 3’-GAGCTCTCCAGCTAGATTAAGGCCGCGGATCTCTTCCTGCATACGGCAGAAGACGAAC -5’ So13_06_rev | Yang et al., 2012 | N/A |
| Indexed Reverse Primer (shRNA library screening): 3’-GAGCTCTCTGGCTAGATTAAGGCCGCGGATCTCTTCCTGCATACGGCAGAAGACGAAC -5’ So13_11_rev | Yang et al., 2012 | N/A |
| i5 indexed PCR primer (CRISPR-Cas9 screening): D501,AATGATACGGCGACCACCGAGATCTACACTATAGCCTACACTCTTTCCCTACACGACGCTCTTCCGATCTATGCATGCTCTTGTGGAAAGGACGAAACACCG | This paper | N/A |
| i5 indexed PCR primer (CRISPR-Cas9 screening): D502,AATGATACGGCGACCACCGAGATCTACACATAGAGGCACACTCTTTCCCTACACGACGCTCTTCCGATCTTGCATGCAGTCTTGTGGAAAGGACGAAACACCG | This paper | N/A |
| i5 indexed PCR primer (CRISPR-Cas9 screening): D503,AATGATACGGCGACCACCGAGATCTACACCCTATCCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGCATGCATCGTCTTGTGGAAAGGACGAAACACCG | This paper | N/A |
| i5 indexed PCR primer (CRISPR-Cas9 screening): D504,AATGATACGGCGACCACCGAGATCTACACGGCTCTGAACACTCTTTCCCTACACGACGCTCTTCCGATCTCATGCATGACGTCTTGTGGAAAGGACGAAACACCG | This paper | N/A |
| i5 indexed PCR primer (CRISPR-Cas9 screening): D505,AATGATACGGCGACCACCGAGATCTACACAGGCGAAGACACTCTTTCCCTACACGACGCTCTTCCGATCTCGTACGTATACATCTTGTGGAAAGGACGAAACACCG | This paper | N/A |
| i5 indexed PCR primer (CRISPR-Cas9 screening): D506,AATGATACGGCGACCA CCGAGATCTACACTAATCTTAACACTCTTTCCCTACACGACGCTCTTCCGATCTACGTACGTGTATATCTTGTGGAAAGGACGAAACACCG | This paper | N/A |
| i5 indexed PCR primer (CRISPR-Cas9 screening): D507,AATGATACGGCGACCACCGAGATCTACACCAGGACGTACACTCTTTCCCTACACGACGCTCTTCCGATCTTACGTACGCGTGTATCTTGTGGAAAGGACGAAACACCG | This paper | N/A |
| i5 indexed PCR primer (CRISPR-Cas9 screening): D508,AATGATACGGCGACCACCGAGATCTACACGTACTGACACACTCTTTCCCTACACGACGCTCTTCCGATCTGTACGTACACCCGTATCTTGTGGAAAGGACGAAACACCG | This paper | N/A |
| i7 indexed PCR primer (CRISPR-Cas9 screening): D701,CAAGCAGAAGACGGCATACGAGATCGAGTAATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCtctactattctttcccctgcactgt | This paper | N/A |
| i7 indexed PCR primer (CRISPR-Cas9 screening): D702,CAAGCAGAAGACGGCATACGAGATTCTCCGGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCtctactattctttcccctgcactgt | This paper | N/A |
| i7 indexed PCR primer (CRISPR-Cas9 screening): D703,CAAGCAGAAGACGGCATACGAGATAATGAGCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCtctactattctttcccctgcactgt | This paper | N/A |
| i7 indexed PCR primer (CRISPR-Cas9 screening): D704,CAAGCAGAAGACGGCATACGAGATGGAATCTCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCtctactattctttcccctgcactgt | This paper | N/A |
| i7 indexed PCR primer (CRISPR-Cas9 screening): D705,CAAGCAGAAGACGGCATACGAGATTTCTGAATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCtctactattctttcccctgcactgt | This paper | N/A |
| i7 indexed PCR primer (CRISPR-Cas9 screening): D706,CAAGCAGAAGACGGCATACGAGATACGAATTCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCtctactattctttcccctgcactgt | This paper | N/A |
| i7 indexed PCR primer (CRISPR-Cas9 screening): D707,CAAGCAGAAGACGGCATACGAGATAGCTTCAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCtctactattctttcccctgcactgt | This paper | N/A |
| i7 indexed PCR primer (CRISPR-Cas9 screening): D708,CAAGCAGAAGACGGCATACGAGATGCGCATTAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCtctactattctttcccctgcactgt | This paper | N/A |
| i7 indexed PCR primer (CRISPR-Cas9 screening): D709,CAAGCAGAAGACGGCATACGAGATCATAGCCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCtctactattctttcccctgcactgt | This paper | N/A |
| i7 indexed PCR primer (CRISPR-Cas9 screening): D710,CAAGCAGAAGACGGCATACGAGATTTCGCGGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCtctactattctttcccctgcactgt | This paper | N/A |
| i7 indexed PCR primer (CRISPR-Cas9 screening): D711,CAAGCAGAAGACGGCATACGAGATGCGCGAGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCtctactattctttcccctgcactgt | This paper | N/A |
| i7 indexed PCR primer (CRISPR-Cas9 screening): D712,CAAGCAGAAGACGGCATACGAGATCTATCGCTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCtctactattctttcccctgcactgt | This paper | N/A |
| Recombinant DNA | ||
| pTO-Cas9-hygro vector | This paper | N/A |
| LentiCRISPR v2 | A gift from Dr. Feng Zhang (Addgene plasmid) | # 52961 |
| LentiCas9-Blast | A gift from Dr. Feng Zhang (Addgene plasmid) | # 52962 |
| Brunello CRISPR knockout pooled library | A gift from Drs. David Root and John Doench (Addgene plasmid) | #73178 |
| Retroviral shRNA library | Ceribelli et al., 2016; Yang et al., 2012 | N/A |
| pRSMX-PG | Ceribelli et al., 2016; Yang et al., 2012 | N/A |
| pLenti-sg-pgk-PG | This paper | N/A |
| pRCMV/TO-puro | Schmitz et al., 2012 | N/A |
| pRCMV/TO-BATF3-WT-puro | This paper | N/A |
| pRCMV/TO-BATF3- H64Q -puro | This paper | N/A |
| pRCMV/TO-IRF4-WT-puro | Yang et al., 2012 | N/A |
| pRCMV/TO-IRF4-DNA binding mutant-puro | Yang et al., 2012 | N/A |
| pRCMV/TO-flag-BIOTIN-puro | Schmitz et al., 2012 | N/A |
| pRCMV/TO-flag-BIOTIN-HBZ-puro | This paper | N/A |
| pRCMV/TO-flag-BIOTIN-sgRNA resistant-HBZ(SM)-puro | This paper | N/A |
| pRCMV/TO-TTG-HBZ-puro | This paper | N/A |
| BMN-ires-lyt2 | Schmitz et al., 2012 | N/A |
| BMN-MYC-ires-lyt2 | This paper | N/A |
| Software and Algorithms | ||
| Integrative Genome Browser | Broad Institute | http://software.broadinstitute.org/software/igv/ |
| MEME-AME | McLeay and Bailey, 2010 | http://meme-suite.org/doc/ame.html |
| Signature DB | NIH | https://lymphochip.nih.gov/signaturedb/ |
| ImageJ software | NIH | https://imagej.nih.gov/ij/index.html |
| GraphPad Prism 7.0 | Graphpad | www.graphpad.com |
| Cluster 3.0 | de Hoon et al., 2004 | http://bonsai.ims.u-tokyo.ac.jp/mdehoon/software/cluster |
shRNA Toxicity Assay
shRNA toxicity assay was performed as previously described (Ceribelli et al., 2016; Yang et al., 2012). Each shRNA was cloned into the pRSMX-PG vector that enables co-expression of the shRNA and a GFP-puromycin resistance protein fusion, and retrovirally expressed in the target cells. Two days after retroviral transduction, shRNA expression was induced by doxycycline and the fraction of GFP+ shRNA-expressing cells was monitored over time by flow cytometry. The gene targeting sequences of the shRNA vectors used in this study are listed in the Key Resources Table.
sgRNA Toxicity Assay
pLenti-sg-pgk-PG was generated to coexpress a GFP-puromycin resistance protein fusion driven by the pgk promoter and an sgRNA driven by a U6 promoter. Each sgRNA was cloned into pLenti-sg-pgk-PG and lentivirally expressed in Cas9-expressing cells. sgRNA-expressing cells were monitored over time by flow cytometry. The sgRNA target sequences used in this study are listed in the Key Resources Table.
Rescue Experiment for shRNA- or sgRNA-Mediated Toxicity
ATLL cells were first infected with a pRCMV/TO-puro vector (Schmitz et al., 2012) (empty, BATF3-wild type, BATF3-H64Q (Tussiwand et al., 2012), IRF4-wild type, IRF4-DNA binding mutant (Yang et al., 2012), and MYC) or a pRCMV/TO-flag-BIOTIN-puro vector (Schmitz et al., 2012) (empty or sgRNA-resistant HBZ). The sgRNA-resistant HBZ isoform was the SM mutant, in which the entire coding region was mutated to introduce silent mutations at all possible positions, resulting in a 12 bp and 9 bp mismatch with the sgHBZ #1 and sgHBZ #2 target sequences, respectively. For TTG-HBZ, the start codon (ATG) of HBZ-cDNA was replaced by TTG to prevent protein expression, but mRNA of HBZ will be transcribed. Following purification of transduced cells using puromycin, cells were infected with pRSMX-PG vectors co-expressing GFP and the shRNAs of interest, or pLenti-sg-pgk-PG co-expressing GFP and the sgRNAs of interest. The fraction of viable, GFP+ cells was monitored over time by flow cytometry.
Rescue Experiment for JQ1-Mediated Toxicity
ST1 cells were transduced with BMN-ires-lyt2 retroviral expression vector (Schmitz et al., 2012) (empty or MYC). Following purification of transduced cells using lyt2-microbeads (Miltenyi), cells were infected with pRCMV/TO-puroGFP expression vector (empty or BATF3), and subsequently treated with JQ1. The number of the lyt2+/GFP+ cells were counted on flow cytometer by using 123count eBeads (eBioscience).
Q-RT-PCR
Total RNA was extracted from ATLL cell lines with TRIzol (Life Technologies) and subsequently cleaned up using RNeasy mini-columns (Qiagen). Total RNA was reverse-transcribed with random primers and SuperScript III reverse transcriptase (Invitrogen). TaqMan Q-RT-PCR on an ABI7500 instrument (Applied Biosystems) was performed using the resultant cDNA. The TaqMan assays used for this study are listed in the Key Resources Table.
Gene Expression Microarray
Two-color human Agilent 4×44K gene-expression arrays were used, as described by the manufacturer, comparing signal from control cells (Cy3-labelled) and test cells (Cy5-labelled). Array elements were filtered for those meeting confidence thresholds for spot size, architecture, and level above local background. These criteria are a feature of the Agilent gene expression software package for Agilent 4×44k arrays.
Chromatin Immunoprecipitation Sequencing (ChIP-seq)
ChIP was performed as previously described (Ceribelli et al., 2016; Yang et al., 2012) with minor modifications. 20 million exponentially growing cells were collected per sample and cross-linked with 1% formaldehyde for 10 min at room temperature. For H3K27ac and BRD4 ChIP-seq, cells were pre-treated for 24 hr with either DMSO or JQ1 (Constellation Pharmaceuticals; 500 nM for KK1 and 250 nM for ST1) before being cross-linked. The cross-linked cells were resuspended in ice-cold RIPA buffer (10mM Tris-HCl pH 8, 140 mM NaCl, 1 mM EDTA pH 8, 0.5 mM EGTA, 1% Triton X-100, 0.1% SDS and 0.1% sodium deoxycholate) to a final concentration of 4 million cells/ml. DNA was sheared with a Misonix XL sonicator. For each immune precipitation reaction, the chromatins were incubated overnight with 5 μg of normal rabbit IgG (Santa Cruz Biotechnology, sc2027), BATF3 antibody (R&D, AF7437), IRF4 antibody (Santa Cruz Biotechnology, sc6059 and sc11450 (1:1 mix)), BRD4 antibody (Bethyl, Cat No. A301–985A), H3K27ac antibody (Abcam, ab4729). The following day, chromatin/antibody complexes were incubated with Protein G Dynabeads (50 μl; Thermo Fisher Scientific) for 4 hr at 4°C, washed 3 times with RIPA Buffer, once with LiCl Buffer (10 mM Tris-HCl pH8, 250 mM LiCl, 0.5% NP40, 0.5% Sodium Deoxycholate, 1 mM EDTA), once with TE pH 8.0 and finally resuspended in 100 μl TE pH8 containing RNase A (0.2 μg/μl). For biotin-tagged BATF3-ChIP-seq and biotin-tagged HBZ-ChIP-seq, MyOne Streptavidin T1 Dynabeads (Thermo Fisher Scientific) were used for the chromatin of KK1 and ST1 transduced with pRCMV/TO-flag-BIOTIN-BATF3-puro or pRCMV/TO-flag-BIOTIN-HBZ-puro. Reverse crosslinking was performed overnight at 65°C, followed by treatment with 20 μg Proteinase K (Invitrogen) for 2 hr at 50°C. Final DNA purification was performed with QIAquick PCR Purification columns (QIAGEN). ChIP DNA was used to generate ChIP-seq libraries with the NEXTflex™ Illumina ChIP-seq Library Prep Kit (Bioo Scientific) according to manufacturer’s instructions. Sequencing of ChIP-seq samples was performed on a GA2x sequencer (Illumina), with single reads of 36 bp length or on a NextSeq 500 sequencer (Illumina), with single reads of 76 bp length. The sequence tags were aligned to human genome build 36 (hg18) with Bowtie software. Redundant reads were removed and reads uniquely mapping to reference genome were used for further analysis. The final data were visualized using the Integrative Genome Browser (Broad Institute). Promoter peaks were defined as peaks whose apex was located within a -+ 2 Kb window from a representative TSS. Enhancer peaks were defined as peaks was located within peaks of H3K27ac ChIPseq, and removed the peaks defined as Promoter peaks.
MEME Motif Analysis
We selected ChIP-seq peaks with depth of 50 and 100 read counts for ST1 and KK1 line respectively. The sequences were extended to include the region within 80 bp from the middle of each selected peak. MEME-AME software was used for the motif enrichment analysis.
Super-Enhancer Analysis
Super-enhancer was defined as previously described with minor modification (Ceribelli et al., 2016). We first defined BRD4 peaks and H3K27ac marks, and then the peaks and marks within 15 Kbp were merged and ranked by increasing total BRD4 or H3K27ac occupancy. These plots revealed an obvious point at which the BRD4 or H3K27ac loading began to increase rapidly, and enabled us to define super-enhancers. Geometrically, this point was defined as the loading point (x-axis) for which a tangent to the curve with slope 1 could be found. All the defined genomic regions above this point were defined as super-enhancers. A representative RefSeq transcript was associated to a super-enhancer when the super-enhancer region overlapped with the RefSeq gene window (Transcription start site (TSS) −15 kb through gene body +2 kb). In the case that multiple RefSeqs were found within a super-enhancer region, a representative RefSeq was selected based on the highest down modulation of mRNA expression after JQ1 treatment.
CRISPR-Cas9 Screen Analysis
ATLL cell lines (ST1, KK1, Su9T01) and mantle cell lymphoma (MCL) lines were transduced in duplicate with the Brunello CRISPR knockout pooled library (gift of David Root and John Doench; Addgene #73178). Following selection with puromycin (2 μg/mL) for 2–4 days, a cell aliquot was frozen as the day 0 sample. Cells were cultured for an additional 3–4 weeks, and an end point cell aliquot was harvested for genomic DNA extraction by QIAamp DNA blood Maxi kit (QIAGEN). sgRNA sequences were amplified by NEBNext High-Fidelity 2x PCR Master Mix (NEB) from the genomic DNA using the indexed PCR primers with next-generation sequencing adapters compatible with Illumina’s NEXTSeq500. PCR products were size-selected using E-Gel, quantified by Qubit (Thermo Fisher Scientific) and sequenced using NEXTSeq500 (Illumina). Sequenced libraries were de-multiplexed using indexes compatible with the Illumina TrueSeq HT kit. The primers are listed in the Key Resources Table. sgRNAs for HBZ target genes or BATF3/IRF4 target genes were selected that had an average next-generation sequencing count >50 in the day 0 populations. For each sgRNA, log2 fold changes (end point / day 0) were calculated for each replicate in each cell line. For each HBZ target gene or each BATF3/IRF4 target gene, an average log2 fold change was calculated across all replicates of all corresponding sgRNAs.
Signature Enrichment Analysis
A database that contained the lymphoid biology-associated signatures and associated genes (Signature DB; https://lymphochip.nih.gov/signaturedb/) was used. For each signature, the number of signature genes overlapping with a given gene set was identified and a 2-by-2 contingency table with observed values for each association was generated. P-values for signature enrichments were calculated using a Fisher’s exact test.
Cell Cycle Analysis
The cells were fixed with PBS/2% Paraformaldehyde, permeabilized with PBS/1% FBS/0.25% saponin and then stained with PBS/PI (propidium iodide, 50 mg/ml)/RNAse A (500 μg/ml). DNA content was analyzed with a FACScalibur (BD Biosciences).
Immunoblot Analysis
Cells were washed and resuspended in 2X SDS sample buffer and boiled for 5 minutes. Samples were separated on Novex 4–12% Tris-Glycine gel (Invitrogen) and transferred to a PVDF membrane (Millipore, Immobilon-P). Proteins were detected with the following antibodies: IRF4 (Santa Cruz Biotechnology, sc6059 and sc11450 (1:1 mix)), BATF3 (Abnova, 3H1), MYC (Epitomics/Abcam, ab32972), BATF (Cell Signaling, #8638) and Rabbit-HBZ serum (Arnold et al., 2008). Signal intensity was analyzed densitometrically with ImageJ software.
Proliferation Assay
For proliferation assays, a total of 5,000 cells per well was seeded in a final volume of 100 μl with the indicated amount of JQ1 per well in 96-well plates. Two days after, 100 μl of fresh media with the indicated amount of JQ1 was added. Metabolic activity was measured after 4 days of JQ1 treatment by using CellTiter 96® AQueous One Solution Cell Proliferation (MTS) assay (Promega) as described by the manufacturer.
Ex Vivo Cultures of PBMNCs from ATLL Patients
ATLL Patient blood samples were obtained from patients under the care of the Clinical Trials Team, Lymphoid Malignancies Branch, NCI. This study protocol was approved by the Institutional Review Board of the NCI. Informed consent was obtained in writing in accordance with the Declaration of Helsinki. 105 of PBMNCs from chronic and acute ATLL patients were cultured with PRMI/10% FBS/Penicillin-Streptomycin with the indicated concentration of JQ1 in a well of 96 well plate for 6 days and 4 days, respectively. The cells were pulsed with 1 μCi of 3H-thymidine for 6 hours, harvested and counted in a MicroBeta2 plate counter (Perkin Elmer).
Xenograft Experiments
The xenograft tumor model of human ST1 and ED40515(−) ATLL were established by subcutaneous injection of 10 million cells and 6.5 million cells into the right flank of 6 weeks old female NSG mice and NOD/SCID mice, respectively. After the average tumor volume reached approximately 100 mm3, the therapies were started. CPI-203 (Constellation Pharmaceuticals) was dissolved in 10% DMSO+90% 0f 30% 2-Hydroxypropyl-beta-cyclo-dextrin (Sigma, H107) and intraperitoneally injected at 5 mg/kg/injection, twice/day. The tumor growth was monitored by measuring tumor size in two orthogonal dimensions and calculated by using the formula 1⁄2(long dimension) (short dimension). All animal experiments were approved by the National Cancer Institute Animal Care and Use Committee (NCI ACUC) and were performed in accordance with NCI ACUC guidelines.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Analyses
All experiments presented have been repeated at least 2 times and consistent results were obtained. Data were analyzed with one-tailed t-test and Fisher’s exact test. Error bars or p-values are shown to indicate statistical significance. In some Figures, error bars are not visible due to their short heights relative to the size of the symbols. p < 0.05 was considered statistically significant. No statistical methods were used to predetermine sample size. No specific blinding method or randomization was applied for mouse experiments.
DATA AND SOFTWARE AVAILABILITY
Accession Codes
All gene expression datasets produced in this study have been deposited in GEO, under the accession: GSE94409. All ChIP-seq datasets produced in this study have been deposited in SRA, under the accession: GSE94732. GSE6338, GSE14879, GSE19069 were used for analyzing microarray data of primary human T-cell lymphoma samples. SRA: SRR1057274, SRA: SRR969480, GSM1816978, GSM1816979, GSM1519644, GSM1519645, GSM1003462 and GSM1003558 were used for analyzing H3K27ac ChIP-seq data of T-ALL cell lines. SRA: SRX290664, SRA: SRX290665, SRA: SRX290666, GSM1462467, GSM1462468, GSM1414734, GSM1414738, GSM1056919, GSM1056920, GSM1056940, GSM1056941, GSM1056942, GSM1056943, GSM1056948, GSM1056949, GSM1056950, GSM1056951, GSM772987, GSM1893223, GSM1893224, GSM1893225, GSM1893226, GSM1893227, GSM1893228, GSM1893229, GSM1893230, GSM1893231, GSM1893232, GSM1893233, GSM1893234, GSM1893235, GSM1893236, GSM1893237, GSM1893238 were used for analyzing H3K27ac ChIP-seq data of human helper T-cell subsets.
Supplementary Material
Significance.
The HTLV-I retrovirus initiates adult T cell leukemia/lymphoma (ATLL), an often fatal malignancy that responds poorly to chemotherapy. The virally encoded transcription factor HBZ is the only HTLV-I gene that is expressed in all ATLL cases, but its contribution to ATLL malignancy is poorly understood. Using functional genomics, we demonstrate that the BATF3/IRF4 transcriptional complex is a master regulator of ATLL gene expression and proliferation. HBZ directly transactivates BATF3 by binding to its super-enhancer, thereby contributing to ATLL proliferation. Small-molecule BET inhibitors collapse the BATF3/IRF4 transcriptional network and are toxic for ATLL cells, supporting their clinical evaluation in this recalcitrant cancer.
ACKNOWLEDGEMENTS
We thank Y. Yamada, T. Hata, and N. Arima for ATLL cell lines; K.C. Conlon for ATLL patient samples; C. Masison for Tax antibody; and R.N. Bamford, L.P. Perera, R. Schmitz, R.M. Young, D.J. Hodson, and A. Mazzucco for discussions and comments. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. M.N. was supported by Research Fellowship from the Uehara Memorial Foundation. The views presented in this article do not necessarily reflect those of the US Food and Drug Administration.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPORTING CITATIONS
The following references appear in the Supplemental Information: Chen et al., 2008; Li and Green, 2007.
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures and seven tables and can be found with this article online at https://doi.org/10.1016/j.ccell.2018.06.014.
REFERENCES
- Arnold J, Zimmerman B, Li M, Lairmore MD, and Green PL (2008). Human T-cell leukemia virus type-1 antisense-encoded gene, Hbz, promotes T-lymphocyte proliferation. Blood 112, 3788–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohers E, Mareschal S, Bouzelfen A, Marchand V, Ruminy P, Maingonnat C, Menard AL, Etancelin P, Bertrand P, Dubois S, et al. (2014). Targetable activating mutations are very frequent in GCB and ABC diffuse large B-cell lymphoma. Genes Chromosomes Cancer 53, 144–153. [DOI] [PubMed] [Google Scholar]
- Ceribelli M, Hou ZE, Kelly PN, Huang DW, Wright G, Ganapathi K, Evbuomwan MO, Pittaluga S, Shaffer AL, Marcucci G, et al. (2016). A druggable TCF4- and BRD4-dependent transcriptional network sustains malignancy in blastic plasmacytoid dendritic cell neoplasm. Cancer Cell 30, 764–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceribelli M, Kelly PN, Shaffer AL, Wright GW, Xiao W, Yang Y, Mathews Griner LA, Guha R, Shinn P, Keller JM, et al. (2014). Blockade of oncogenic IkappaB kinase activity in diffuse large B-cell lymphoma by bromodomain and extraterminal domain protein inhibitors. Proc. Natl. Acad. Sci. USA 111, 11365–11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Petrus M, Bryant BR, Phuc Nguyen V, Stamer M, Goldman CK, Bamford R, Morris JC, Janik JE, and Waldmann TA (2008). Induction of the IL-9 gene by HTLV-I Tax stimulates the spontaneous proliferation of primary adult T-cell leukemia cells by a paracrine mechanism. Blood 111, 5163–5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkhurst CN, Muratet M, et al. (2012). A validated regulatory network for Th17 cell specification. Cell 151, 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, and Miyano S (2004). Open source clustering software. Bioinformatics 20, 1453–1454. [DOI] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. (2011). BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittus C, and Sloan JM (2017). Adult T-cell leukemia/lymphoma: a problem abroad and at home. Hematol. Oncol. Clin. North Am 31, 255–272. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. (2010). Selective inhibition of BET bromodomains. Nature 468, 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuji S, Inoue Y, Utsunomiya A, Moriuchi Y, Uchimaru K, Choi I, Otsuka E, Henzan H, Kato K, Tomoyose T, et al. (2016). Pretransplantation anti-CCR4 antibody mogamulizumab against adult T-cell leukemia/lymphoma is associated with significantly increased risks of severe and corticosteroid-refractory graft-versus-host disease, nonrelapse mortality, and overall mortality. J. Clin. Oncol 34, 3426–3433. [DOI] [PubMed] [Google Scholar]
- Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, Khan AA, Ciofani M, Spooner CJ, Rutz S, et al. (2012). A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science 338, 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, et al. (2015). High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526. [DOI] [PubMed] [Google Scholar]
- Hodson A, Crichton S, Montoto S, Mir N, Matutes E, Cwynarski K, Kumaran T, Ardeshna KM, Pagliuca A, Taylor GP, and Fields PA (2011). Use of zidovudine and interferon alfa with chemotherapy improves survival in both acute and lymphoma subtypes of adult T-cell leukemia/lymphoma. J. Clin. Oncol 29, 4696–4701. [DOI] [PubMed] [Google Scholar]
- Imaizumi Y, Kohno T, Yamada Y, Ikeda S, Tanaka Y, Tomonaga M, and Matsuyama T (2001). Possible involvement of interferon regulatory factor 4 (IRF4) in a clinical subtype of adult T-cell leukemia. Jpn. J. Cancer Res 92, 1284–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J, Wright G, Wang C, Rosenwald A, Gascoyne RD, Weisenburger DD, Greiner TC, Smith L, Guo S, Wilcox RA, et al. (2014). Gene expression signatures delineate biological and prognostic subgroups in peripheral T-cell lymphoma. Blood 123, 2915–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Joh T, Uike N, Yamamoto K, Utsunomiya A, Yoshida S, Saburi Y, Miyamoto T, Takemoto S, Suzushima H, et al. (2012). Defucosylated anti-CCR4 monoclonal antibody (KW-0761) for relapsed adult T-cell leukemia-lymphoma: a multicenter phase II study. J. Clin. Oncol 30, 837–842. [DOI] [PubMed] [Google Scholar]
- Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga J, Totoki Y, Chiba K, Sato-Otsubo A, Nagae G, et al. (2015). Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat. Genet 47, 1304–1315. [DOI] [PubMed] [Google Scholar]
- Kato K, and Akashi K (2015). Recent advances in therapeutic approaches for adult T-cell leukemia/lymphoma. Viruses 7, 6604–6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King B, Trimarchi T, Reavie L, Xu L, Mullenders J, Ntziachristos P, Aranda-Orgilles B, Perez-Garcia A, Shi J, Vakoc C, et al. (2013). The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell 153, 1552–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, Dave SS, Zhao H, Xu W, Rosenwald A, et al. (2008). Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 319, 1676–1679. [DOI] [PubMed] [Google Scholar]
- Li M, and Green PL (2007). Detection and quantitation of HTLV-1 and HTLV-2 mRNA species by real-time RT-PCR. J. Virol. Methods 142, 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Spolski R, Liao W, Wang L, Murphy TL, Murphy KM, and Leonard WJ (2012). BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature 490, 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loosveld M, Castellano R, Gon S, Goubard A, Crouzet T, Pouyet L, Prebet T, Vey N, Nadel B, Collette Y, and Payet-Bornet D (2014). Therapeutic targeting of c-Myc in T-cell acute lymphoblastic leukemia, T-ALL. Oncotarget 5, 3168–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, and Young RA (2013). Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M, and Jeang KT (2007). Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat. Rev. Cancer 7, 270–280. [DOI] [PubMed] [Google Scholar]
- McLeay RC, and Bailey TL (2010). Motif enrichment analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics 11, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TL, Tussiwand R, and Murphy KM (2013). Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat. Rev. Immunol 13, 499–509. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Schmitz R, Xiao W, Goldman CK, Xu W, Yang Y, Yu X, Waldmann TA, and Staudt LM (2014). Gain-of-function CCR4 mutations in adult T cell leukemia/lymphoma. J. Exp. Med 211, 2497–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, and Staudt LM (2006). A loss-of-function RNA interference screen for molecular targets in cancer. Nature 441, 106–110. [DOI] [PubMed] [Google Scholar]
- Ramos JC, Ruiz P Jr., Ratner L, Reis IM, Brites C, Pedroso C, Byrne GE Jr., Toomey NL, Andela V, Harhaj EW, et al. (2007). IRF-4 and c-Rel expression in antiviral-resistant adult T-cell leukemia/lymphoma. Blood 109, 3060–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, Wright G, Shaffer AL, Hodson DJ, Buras E, et al. (2012). Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 490, 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoyama M (1991). Diagnostic criteria and classification of clinical subtypes of adult T- cell leukaemia-lymphoma. A report from the Lymphoma Study Group (1984–87). Br. J. Haematol 79, 428–437. [DOI] [PubMed] [Google Scholar]
- Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, Kc W, Albring JC, Satpathy AT, Rotondo JA, Edelson BT, Kretzer NM, et al. (2012). Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature 490, 502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaque JP, Gomez-Lopez G, Monsalvez V, Varela I, Martinez N, Perez C, Dominguez O, Grana O, Rodriguez-Peralto JL, Rodriguez-Pinilla SM, et al. (2014). PLCG1 mutations in cutaneous T-cell lymphomas. Blood 123, 2034–2043. [DOI] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, and Sabatini DM (2015). Identification and characterization of essential genes in the human genome. Science 350, 1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, and Young RA (2013). Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Qi J, Bradner JE, Xiao G, and Chen LF (2013). Bromodomain and extraterminal (BET) protein inhibition suppresses human T cell leukemia virus 1 (HTLV-1) Tax protein-mediated tumorigenesis by inhibiting nuclear factor kappaB (NF-kappaB) signaling. J. Biol. Chem 288, 36094–36105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Shaffer AL 3rd, Emre NC, Ceribelli M, Zhang M, Wright G, Xiao W, Powell J, Platig J, Kohlhammer H, et al. (2012). Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell 21, 723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accession Codes
All gene expression datasets produced in this study have been deposited in GEO, under the accession: GSE94409. All ChIP-seq datasets produced in this study have been deposited in SRA, under the accession: GSE94732. GSE6338, GSE14879, GSE19069 were used for analyzing microarray data of primary human T-cell lymphoma samples. SRA: SRR1057274, SRA: SRR969480, GSM1816978, GSM1816979, GSM1519644, GSM1519645, GSM1003462 and GSM1003558 were used for analyzing H3K27ac ChIP-seq data of T-ALL cell lines. SRA: SRX290664, SRA: SRX290665, SRA: SRX290666, GSM1462467, GSM1462468, GSM1414734, GSM1414738, GSM1056919, GSM1056920, GSM1056940, GSM1056941, GSM1056942, GSM1056943, GSM1056948, GSM1056949, GSM1056950, GSM1056951, GSM772987, GSM1893223, GSM1893224, GSM1893225, GSM1893226, GSM1893227, GSM1893228, GSM1893229, GSM1893230, GSM1893231, GSM1893232, GSM1893233, GSM1893234, GSM1893235, GSM1893236, GSM1893237, GSM1893238 were used for analyzing H3K27ac ChIP-seq data of human helper T-cell subsets.