Abstract
Background
Specific diagnostic tests to detect severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) and resulting COVID‐19 disease are not always available and take time to obtain results. Routine laboratory markers such as white blood cell count, measures of anticoagulation, C‐reactive protein (CRP) and procalcitonin, are used to assess the clinical status of a patient. These laboratory tests may be useful for the triage of people with potential COVID‐19 to prioritize them for different levels of treatment, especially in situations where time and resources are limited.
Objectives
To assess the diagnostic accuracy of routine laboratory testing as a triage test to determine if a person has COVID‐19.
Search methods
On 4 May 2020 we undertook electronic searches in the Cochrane COVID‐19 Study Register and the COVID‐19 Living Evidence Database from the University of Bern, which is updated daily with published articles from PubMed and Embase and with preprints from medRxiv and bioRxiv. In addition, we checked repositories of COVID‐19 publications. We did not apply any language restrictions.
Selection criteria
We included both case‐control designs and consecutive series of patients that assessed the diagnostic accuracy of routine laboratory testing as a triage test to determine if a person has COVID‐19. The reference standard could be reverse transcriptase polymerase chain reaction (RT‐PCR) alone; RT‐PCR plus clinical expertise or and imaging; repeated RT‐PCR several days apart or from different samples; WHO and other case definitions; and any other reference standard used by the study authors.
Data collection and analysis
Two review authors independently extracted data from each included study. They also assessed the methodological quality of the studies, using QUADAS‐2. We used the 'NLMIXED' procedure in SAS 9.4 for the hierarchical summary receiver operating characteristic (HSROC) meta‐analyses of tests for which we included four or more studies. To facilitate interpretation of results, for each meta‐analysis we estimated summary sensitivity at the points on the SROC curve that corresponded to the median and interquartile range boundaries of specificities in the included studies.
Main results
We included 21 studies in this review, including 14,126 COVID‐19 patients and 56,585 non‐COVID‐19 patients in total. Studies evaluated a total of 67 different laboratory tests. Although we were interested in the diagnotic accuracy of routine tests for COVID‐19, the included studies used detection of SARS‐CoV‐2 infection through RT‐PCR as reference standard. There was considerable heterogeneity between tests, threshold values and the settings in which they were applied. For some tests a positive result was defined as a decrease compared to normal vaues, for other tests a positive result was defined as an increase, and for some tests both increase and decrease may have indicated test positivity. None of the studies had either low risk of bias on all domains or low concerns for applicability for all domains. Only three of the tests evaluated had a summary sensitivity and specificity over 50%. These were: increase in interleukin‐6, increase in C‐reactive protein and lymphocyte count decrease.
Blood count
Eleven studies evaluated a decrease in white blood cell count, with a median specificity of 93% and a summary sensitivity of 25% (95% CI 8.0% to 27%; very low‐certainty evidence). The 15 studies that evaluated an increase in white blood cell count had a lower median specificity and a lower corresponding sensitivity. Four studies evaluated a decrease in neutrophil count. Their median specificity was 93%, corresponding to a summary sensitivity of 10% (95% CI 1.0% to 56%; low‐certainty evidence). The 11 studies that evaluated an increase in neutrophil count had a lower median specificity and a lower corresponding sensitivity. The summary sensitivity of an increase in neutrophil percentage (4 studies) was 59% (95% CI 1.0% to 100%) at median specificity (38%; very low‐certainty evidence). The summary sensitivity of an increase in monocyte count (4 studies) was 13% (95% CI 6.0% to 26%) at median specificity (73%; very low‐certainty evidence). The summary sensitivity of a decrease in lymphocyte count (13 studies) was 64% (95% CI 28% to 89%) at median specificity (53%; low‐certainty evidence). Four studies that evaluated a decrease in lymphocyte percentage showed a lower median specificity and lower corresponding sensitivity. The summary sensitivity of a decrease in platelets (4 studies) was 19% (95% CI 10% to 32%) at median specificity (88%; low‐certainty evidence).
Liver function tests
The summary sensitivity of an increase in alanine aminotransferase (9 studies) was 12% (95% CI 3% to 34%) at median specificity (92%; low‐certainty evidence). The summary sensitivity of an increase in aspartate aminotransferase (7 studies) was 29% (95% CI 17% to 45%) at median specificity (81%) (low‐certainty evidence). The summary sensitivity of a decrease in albumin (4 studies) was 21% (95% CI 3% to 67%) at median specificity (66%; low‐certainty evidence). The summary sensitivity of an increase in total bilirubin (4 studies) was 12% (95% CI 3.0% to 34%) at median specificity (92%; very low‐certainty evidence).
Markers of inflammation
The summary sensitivity of an increase in CRP (14 studies) was 66% (95% CI 55% to 75%) at median specificity (44%; very low‐certainty evidence). The summary sensitivity of an increase in procalcitonin (6 studies) was 3% (95% CI 1% to 19%) at median specificity (86%; very low‐certainty evidence). The summary sensitivity of an increase in IL‐6 (four studies) was 73% (95% CI 36% to 93%) at median specificity (58%) (very low‐certainty evidence).
Other biomarkers
The summary sensitivity of an increase in creatine kinase (5 studies) was 11% (95% CI 6% to 19%) at median specificity (94%) (low‐certainty evidence). The summary sensitivity of an increase in serum creatinine (four studies) was 7% (95% CI 1% to 37%) at median specificity (91%; low‐certainty evidence). The summary sensitivity of an increase in lactate dehydrogenase (4 studies) was 25% (95% CI 15% to 38%) at median specificity (72%; very low‐certainty evidence).
Authors' conclusions
Although these tests give an indication about the general health status of patients and some tests may be specific indicators for inflammatory processes, none of the tests we investigated are useful for accurately ruling in or ruling out COVID‐19 on their own. Studies were done in specific hospitalized populations, and future studies should consider non‐hospital settings to evaluate how these tests would perform in people with milder symptoms.
Plain language summary
How accurate are routine laboratory tests for diagnosis of COVID‐19?
What are routine laboratory tests?
Routine laboratory tests are blood tests that assess the health status of a patient. Tests include counts of different types of white blood cells (these help the body fight infection), and detection of markers (proteins) that indicate organ damage, and general inflammation. These tests are widely available and in some places they may be the only tests available for diagnosis of COVID‐19.
What did we want to find out?
People with suspected COVID‐19 need to know quickly whether they are infected so that they can self‐isolate, receive treatment, and inform close contacts.
Currently, the standard test for COVID‐19 is usually the RT‐PCR test. In the RT‐PCR, samples from the nose and throat are sent away for testing, usually to a large, central laboratory with specialist equipment. Other tests include imaging tests, like X‐rays, which also require specialist equipment.
We wanted to know whether routine laboratory tests were sufficiently accurate to diagnose COVID‐19 in people with suspected COVID‐19. We also wanted to know whether they were accurate enough to prioritize patients for different levels of treatment.
What did we do?
We searched for studies that assessed the accuracy of routine laboratory tests to diagnose COVID‐19 compared with RT‐PCR or other tests. Studies could be of any design and be set anywhere in the world. Studies could include participants of any age or sex, with suspected COVID‐19, or use samples from people known to have – or not to have ‐ COVID‐19.
What we found
We found 21 studies that looked at 67 different routine laboratory tests for COVID‐19. Most of the studies looked at how accurately these tests diagnosed infection with the virus causing COVID‐19. Four studies included both children and adults, 16 included only adults and one study only children. Seventeen studies were done in China, and one each in Iran, Italy, Taiwan and the USA. All studies took place in hospitals, except one that used samples from a database. Most studies used RT‐PCR to confirm COVID‐19 diagnosis.
Accuracy of tests is most often reported using ‘sensitivity’ and ‘specificity’. Sensitivity is the proportion of people with COVID‐19 correctly detected by the test; specificity is the proportion of people without COVID‐19 who are correctly identified by the test. The nearer sensitivity and specificity are to 100%, the better the test. A test to prioritize people for treatment would require a high sensitivity of more than 80%.
Where four or more studies evaluated a particular test, we pooled their results and analyzed them together. Our analyses showed that only three of the tests had both sensitivity and specificity over 50%. Two of these were markers for general inflammation (increases in interleukin‐6 and C‐reactive protein). The third was for lymphocyte count decrease. Lymphocytes are a type of white blood cell where a low count might indicate infection.
How reliable are the results?
Our confidence in the evidence from this review is low because the studies were different from each other, which made them difficult to compare. For example, some included very sick people, while some included people with hardly any COVID‐19 symptoms. Also, the diagnosis of COVID‐19 was confirmed in different ways: RT‐PCR was sometimes used in combination with other tests.
Who do the results of this review apply to?
Routine laboratory tests can be issued by most healthcare facilities. However, our results are probably not representative of most clinical situations in which these tests are being used. Most studies included very sick people with high rates of COVID‐19 virus infection of between 27% and 76%. In most primary healthcare facilities, this percentage will be lower.
What does this mean?
Routine laboratory tests cannot distinguish between COVID‐19 and other diseases as the cause of infection, inflammation or tissue damage. None of the tests performed well enough to be a standalone diagnostic test for COVID‐19 nor to prioritize patients for treatment. They will mainly be used to provide an overall picture about the health status of the patient. The final COVID‐19 diagnosis has to be made based on other tests.
How up‐to‐date is this review?
We searched all COVID‐19 studies up to 4 May 2020.
Summary of findings
Background
On 30 December 2019, a cluster of patients with pneumonia of unknown origin in Wuhan, China, was publicly reported via ProMED (promedmail.org/promed-posts). In January 2020, it became clear that this was caused by a new coronavirus and that it was spreading to other countries as well. In March 2020, the World Health Organization (WHO) declared the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) and resulting COVID‐19 a worldwide pandemic. This pandemic, in combination with the novelty of the virus, presents important diagnostic challenges.
These challenges range from understanding the value of signs and symptoms in predicting possible infection, assessing whether existing biochemical and imaging tests can identify infection and patients who need critical care, and evaluating whether new diagnostic tests can provide accurate rapid and point‐of care testing, either to identify current infection, rule out infection, identify people in need of care escalation, or to test for past infection and immunity.
This review follows a generic protocol that covers the full series of Cochrane diagnostic test accuracy (DTA) reviews for the diagnosis of COVID‐19 (Deeks 2020b). The Background and Methods sections of this review therefore use some text that was originally published in the protocol, and text that overlaps some of our other reviews (Deeks 2020a; Dinnes 2020; Struyf 2020).
The present review concentrates on the diagnostic accuracy of routine laboratory testing as a triage test to determine if a person has COVID‐19 pneumonia or SARS‐CoV‐2 infection, and to facilitate further testing. In clinical care, routine laboratory markers such as white blood cell count, measures of anticoagulation, C‐reactive protein (CRP) and procalcitonin, are used to assess the health status of a patient. These laboratory markers are also used in patients with COVID‐19 infection and may be useful for triage of people with potential COVID‐19 infection for treatment or more intensive treatment, especially in situations where time and resources are limited.
Target condition being diagnosed
COVID‐19 is the disease caused by infection with SARS‐CoV‐2. The key target condition for this review was current COVID‐19. SARS‐CoV‐2 infection can be asymptomatic (no symptoms); mild or moderate (symptoms such as fever, cough, aches, lethargy but without difficulty breathing at rest); severe (symptoms include breathlessness and increased respiratory rate indicative of pneumonia); or critical (requiring respiratory support due to severe acute respiratory syndrome (SARS) or acute respiratory distress syndrome (ARDS)). People with COVID‐19 pneumonia (severe or critical disease) require distinctive patient management, and it is important to be able to identify these patients.
In this review, we focus on COVID‐19, without making the distinction between mild to moderate and severe disease.
Index test(s)
We collated evidence on all routine biomarker tests reported in the identified studies. These can be classified into:
full blood count, haemoglobin and red blood cells;
coagulation markers;
liver markers, cardiac markers and kidney function markers;
general inflammatory markers; and
metabolic markers.
Clinical pathway
Decisions about patient and isolation pathways for COVID‐19 vary according to health services and settings, available resources, and stages of the epidemic. They will change over time if and when effective treatments and vaccines are identified. The decision points between these pathways vary, but all include points at which knowledge of the accuracy of diagnostic information is needed to be able to inform rational decisions.
Standard workup for individuals suspected of COVID‐19 infection consists of assessing signs and symptoms and a polymerase chain reaction (PCR) test. It is common practice that, when patients enter (either outpatient or admission) the hospital, they will generally have routine laboratory tests done.
Routinely available tests for infection and inflammation may be considered in the investigation of people with possible COVID‐19 infection. For example, many healthcare facilities have access to standard laboratory tests for infection, such as CRP, procalcitonin, measures of anticoagulation, and white blood cell count with leukocyte differentiation. Routine laboratory markers may be used as a triage test, either on their own, or in combination with signs and symptoms. In low‐resource settings, they may sometimes even be the only tests available. In order to function as a triage test or stand‐alone test, a high sensitivity is needed, to prevent infected patients from being sent home or into a general ward with uninfected patients. For a triage test, specificity may be less important, as positive tests will be further investigated. Also, routine laboratory tests may be used to tip the decision to treat the patient as having COVID‐19 or not in case of mixed results from other tests or where a definite diagnosis cannot be made. In that case, knowledge of the sensitivity and specificity in a particular (pre‐tested) patient population may be useful. Routine laboratory tests may also be used in the further diagnostic workup, to predict mild versus severe outcomes, or to monitor treatment response. These aims of testing will not be the focus of this systematic review.
Alternative test(s)
The test that is believed to be most accurate in detecting SARS‐CoV‐2 is reverse transcriptase polymerase chain reaction (RT‐PCR). In many settings, this test will be available, but the results take time before they become available. Although rapid antigen and molecular‐based tests are also available, the value of these rapid tests is still not clear. Antibody tests provide insights into the antibody response, but may also take a few days before the response is detectable and therefore the results are available.
Alternatives to routine laboratory tests may depend on the setting and situation where the tests are done. For example, in primary care, alternatives may consist of signs and symptoms and rapid and point‐of‐care tests. Similarly, point‐of‐care ultrasound may be used, if resources allow. The benefit of routine laboratory tests (and of signs and symptoms) may be as an indication of the severity of a disease: a value further from the reference values may indicate more severe infections.
In emergency departments, chest X‐ray, ultrasound, and computed tomography (CT) are widely used diagnostic imaging tests to identify COVID‐19 pneumonia. Which imaging test is available may depend on the type of hospital and available resources: a tertiary care hospital in a high‐income country may have a mobile CT scan available, while in smaller hospitals only X‐ray and ultrasound are accessible. These imaging tests have the advantage that the condition of the lungs can be assessed visually.
These other tests are all addressed in the other Cochrane DTA reviews in this suite of reviews (Deeks 2020a; Dinnes 2020; McInnes 2020; Struyf 2020).
Rationale
It is essential to understand the accuracy of tests and diagnostic features to identify how they can be used optimally in different settings to develop effective diagnostic and management pathways. New evidence about routine laboratory testing is becoming available quickly. Therefore, we have produced a Cochrane 'living systematic review’ (a systematic review that is continually updated, incorporating relevant new evidence as it becomes available) that will summarize new and existing evidence on the clinical accuracy of routine laboratory markers. Estimates of accuracy from this review will help inform diagnostic, screening, and patient management decisions.
Objectives
To assess the diagnostic accuracy of routine laboratory testing as a triage test to determine if a person has COVID‐19.
Secondary objectives
Where data are available, we investigated the accuracy (either by stratified analysis or meta‐regression) according to a specific measurement or test, days of symptoms, severity of symptoms, reference standard, sample type, study design, and setting.
Methods
Criteria for considering studies for this review
Types of studies
We kept the eligibility criteria broad to include all patient groups and all variations of a test (that is, if patient population was unclear, we included the study).
We included studies of all designs that produce estimates of test accuracy or provide data from which estimates can be computed: cross‐sectional studies, case‐control designs and consecutive series of patients assessing the diagnostic accuracy of routine laboratory testing as a triage test to determine if a person has COVID‐19.
We intended to include studies recruiting only COVID‐19 cases, to estimate sensitivity, or those restricted to people without COVID‐19, to estimate specificity (Deeks 2020a). We decided to deviate from this rule as the added value of such studies for our review is questionable. We included both single‐gate designs, where a single group of participants, often suspected of having the target condition, is recruited, and multi‐gate designs, where people with and without the target condition are recruited separately. We Intended to include studies that based their results on individual patients as well as studies that based their results on samples. We carefully considered the limitations of different study designs, using quality assessment and analysis.
Participants
We included studies recruiting people presenting with suspected SARS‐CoV‐2 infection, studies that recruited people to screen for disease, and studies based on serum banks created from known cases of COVID‐19 and controls.
Studies had to include a minimum of 10 samples or 10 participants.
Index tests
We collected evidence on all routine biomarker tests reported in the identified studies. We interpreted the term 'routine' broadly, considering that some markers will be more routine in some settings or countries than in others. Test positivity could have been defined as an increase in values compared to the normal ranges, or as a decrease compared to normal values.
Target conditions
To be eligible, studies needed to identify at least one of:
current SARS‐CoV‐2 infection;
COVID‐19 pneumonia.
Reference standards
Reverse transcriptase polymerase chain reaction (RT‐PCR) is considered the best available test, although due to rapidly evolving knowledge about the target conditions, multiple reference standards on their own as well as in combination have emerged.
Therefore, we included the following reference standards:
RT‐PCR alone;
RT‐PCR, clinical expertise, and imaging (for example, CT thorax);
repeated RT‐PCR several days apart or from different samples;
plaque reduction neutralization test (PRNT) or enzyme‐linked immunosorbent assay (ELISA);
information available at a subsequent time point;
WHO (Appendix 1), and other case definitions;
any other reference standard used by study authors.
Search methods for identification of studies
Electronic searches
We conducted a single literature search to cover our suite of Cochrane COVID‐19 diagnostic test accuracy (DTA) reviews (Deeks 2020b; McInnes 2020).
We conducted electronic searches using two primary sources. Both of these searches aimed to identify all published articles and preprints related to COVID‐19, and were not restricted to those evaluating tests. Thus, there are no test terms, diagnosis terms, or methodological terms in the searches. Searches were limited to 2019 and 2020, and for this version of the review have been conducted to 4 May 2020.
Cochrane COVID‐19 Study Register searches
We used the Cochrane COVID‐19 Study Register (covid-19.cochrane.org), for searches conducted to 28 March 2020. At that time, the register was populated by searches of PubMed, as well as trials registers at ClinicalTrials.gov and the WHO International Clinical Trials Registry Platform (ICTRP).
Search strategies were designed for maximum sensitivity, to retrieve all human studies on COVID‐19 and with no language limits (Appendix 2).
COVID‐19 Living Evidence Database from the University of Bern
From 28 March 2020, we used the COVID‐19 Living Evidence database from the Institute of Social and Preventive Medicine (ISPM) at the University of Bern (www.ispm.unibe.ch), as the primary source of records for the Cochrane COVID‐19 DTA reviews. This search includes PubMed, Embase, and preprints indexed in bioRxiv and medRxiv databases. The strategies as described on the ISPM website are described here (ispmbern.github.io/covid-19/; Appendix 3).
The decision to focus primarily on the 'Bern' feed was due to the exceptionally large numbers of COVID‐19 studies available only as preprints. The Cochrane COVID‐19 Study Register has undergone a number of iterations since the end of March and we anticipate moving back to the Register as the primary source of records for subsequent review updates.
Searching other resources
We identified Embase records obtained through Martha Knuth for the Centers for Disease Control and Prevention (CDC), Stephen B Thacker CDC Library, COVID‐19 Research Articles Downloadable Database (cdc.gov/library/researchguides/2019novelcoronavirus/researcharticles.html), and de‐duplicated them against the Cochrane COVID‐19 Study Register up to 1 April 2020.
We also checked our search results against two additional repositories of COVID‐19 publications including:
the Evidence for Policy and Practice Information and Co‐ordinating Centre (EPPI‐Centre) 'COVID‐19: Living map of the evidence' (eppi.ioe.ac.uk/COVID19_MAP/covid_map_v4.html);
the Norwegian Institute of Public Health 'NIPH systematic and living map on COVID‐19 evidence' (www.nornesk.no/forskningskart/NIPH_diagnosisMap.html).
Both of these repositories allow their contents to be filtered according to studies potentially relating to diagnosis, and both have agreed to provide us with updates of new diagnosis studies added. For this iteration of the review, we examined all diagnosis studies from either source up to 4 May 2020.
We did not apply any language restrictions.
Data collection and analysis
Selection of studies
First, all retrieved articles were screened by an overall team of screeners who divided the articles over the different rapid DTA reviews. Then, the set of studies possibly involving routine laboratory markers was imported into Covidence. Two review authors screened each title and abstract independent of each other for possible inclusion. In the next step, two review authors independently screened the full text of each possibly relevant article. For articles only available in languages other than English, we used Google Translate and review authors who could read and understand that language. We solved disagreements by discussion. If discussion could not solve the dispute, we consulted a third review author.
Data extraction and management
Two review authors carried out data extraction for each study. We assigned multiple studies with first authors with the same last name to one extractor, so that they could detect preprints from already peer‐reviewed, published articles. We contacted study authors when we needed to check details and obtain missing information. Data were extracted on the country and region, the setting, the time period of the study, funding, and information needed for the Characteristics of included studies tables. Studies may have defined a positive test result as a decrease compared to normal vaues, as an increase compared to normal values, and as both increase and decrease. Where possible, we adapted the two‐by‐two tables in such a way that all studies included in the analyses reported on the same test positivity definition. However, if studies reported both in‐ and decrease as a positive test result, we included both. We resolved disagreements by discussion between the two review authors, and two other review authors checked the results when these were entered into Review Manager 5.4 (Review Manager 2020).
Assessment of methodological quality
QUADAS‐2 assessment
Two review authors independently assessed risk of bias and applicability concerns using the Quality Assessment of Diagnostic Accuracy Studies (QUADAS‐2) tool (Table 3). We resolved disagreements by discussion between three review authors.
1. QUADAS‐2 checklist.
| Index test(s) | Review #1. Laboratory based molecular tests | Review #2. Point‐of‐care tests | Review #3. Antibody tests | Review #4. Signs and symptoms | Review #5. Routine laboratory tests |
| Patients (setting, intended use of index test, presentation, prior testing) | Considered to be the 'gold standard' for acute infection. May have been used with different samples, in different settings, for case‐finding or confirmation of infection in patients with suspected COVID‐19. |
In patients with suspected COVID‐19 or contact tracing. Point‐of‐care: case‐finding in the general population, care homes for elderly people, emergency departments. |
In patients with signs and symptoms suspected of COVID‐19 and for case finding; also in patients with past exposure to SARS‐CoV‐2. | General practice, primary care, emergency care. In patients presenting with suspected COVID‐19. No prior testing. Signs and symptoms often used for triage or referral. |
Mainly meant for situations where a laboratory was close; emergency care, hospital, ICU. COVID triage centres. In patients presenting with suspected COVID‐19. |
| Reference standard and target condition | The focus will be on the diagnosis of COVID‐19 pneumonia or infection with SARS‐CoV‐2. For this protocol, the focus will not be on prognosis. | ||||
| PARTICIPANT SELECTION | |||||
| Was a consecutive or random sample of patients enrolled? | This will be similar for all index tests, target conditions, and populations. YES: if a study explicitly stated that all participants within a certain time frame were included; that this was done consecutively; or that a random selection was done. NO: if it was clear that a different selection procedure was employed; for example, selection based on clinician's preference, or based on institutions. UNCLEAR: if the selection procedure was not clear or not reported. |
||||
| Was a case‐control design avoided? | This will be similar for all index tests, target conditions, and populations. YES: if a study explicitly stated that all participants came from the same group of (suspected) patients. NO: if it was clear that a different selection procedure was employed for the participants depending on their COVID‐19 (pneumonia) status or SARS‐CoV‐2 infection status. UNCLEAR: if the selection procedure was not clear or not reported. |
||||
| Did the study avoid inappropriate exclusions? | Studies may have excluded patients, or selected patients in such a way that they avoided including those who were difficult to diagnosis or likely to be borderline. Although the inclusion and exclusion criteria will be different for the different index tests, inappropriate exclusions and inclusions will be similar for all index tests: for example, only elderly patients excluded, or children (as sampling may be more difficult). This needs to be addressed on a case‐to‐case basis. YES: if a high proportion of eligible patients was included without clear selection. NO: if a high proportion of eligible patients was excluded without providing a reason; if, in a retrospective study, participants without index test or reference standard results were excluded; if exclusion was based on severity assessment postfactum or comorbidities (cardiovascular disease, diabetes, immunosuppression). UNCLEAR: if the exclusion criteria were not reported. |
||||
| Did the study avoid inappropriate inclusions? | Some laboratory studies may have intentionally included groups of patients in whom the accuracy was likely to differ, such as those with particularly low or high viral loads, or who had other diseases, such that the sample over‐represented these groups. This needs to be addressed on a case‐to‐case basis. Artificial spiked samples are a clear example. YES: if samples included were likely to be representative of the spectrum of disease. NO: if the study oversampled patients with particular characteristics likely to affect estimates of accuracy. UNCLEAR: if the exclusion criteria were not reported. |
||||
| Could the selection of patients have introduced bias? | HIGH: if one or more signalling questions were answered with NO, as any deviation from the selection process may lead to bias. LOW: if all signalling questions were answered with YES. UNCLEAR: all other instances. |
||||
| Is there concern that the included patients do not match the review question? | HIGH: if accuracy of RT‐PCR was assessed in a case‐control design; to screen contacts or for stopping contact isolation. Studies done in sample banks and spiked samples. LOW: any other situation: these tests may be used in different settings and for different purposes. UNCLEAR: if a description about the participants was lacking. |
HIGH: if accuracy of tests was assessed in a case‐control design; if not used to diagnose early acute infection; to screen contacts or for stopping contact isolation. Studies done in sample banks and spiked samples. LOW: any other situation: these tests may have been used in different settings and for different purposes. UNCLEAR: if a description about the participants was lacking. |
HIGH: if accuracy of tests was assessed in a case‐control design; when patients were tested too early in the disease phase for detection of past infection. Studies done in sample banks and spiked samples. LOW: any other situation: these tests may be used in different settings and for different purposes. UNCLEAR: if a description about the participants was lacking. |
HIGH: if accuracy of signs and symptoms were assessed in a case‐control design, or in an already highly selected group of participants, or the study was able to only estimate sensitivity or specificity. LOW: any situation where signs and symptoms were the first assessment/test to be done on the included participants. UNCLEAR: if a description about the participants was lacking. |
HIGH: if accuracy of laboratory tests was assessed in a case‐control design, or in an already highly selected group of participants. LOW: any situation where generic laboratory tests were among the first tests to be done on the included participants. UNCLEAR: if a description about the participants was lacking. |
| INDEX TESTS | |||||
| Were the index test results interpreted without knowledge of the results of the reference standard? | This will be similar for all index tests, target conditions, and populations. YES: if blinding was explicitly stated or index test was recorded before the results from the reference standard were available. NO: if it was explicitly stated that the index test results were interpreted with knowledge of the results of the reference standard. UNCLEAR: if blinding was unclearly reported. |
||||
| If a threshold was used, was it prespecified? | This will be similar for all index tests, target conditions, and populations. YES: if the test was dichotomous by nature, or if the threshold was stated in the methods section, or if authors stated that the threshold as recommended by the manufacturer was used. NO: if a receiver operating characteristic curve was drawn or multiple threshold reported in the results section; and the final result was based on one of these thresholds; if fever was not defined beforehand (in review # 4, Signs and symptoms). UNCLEAR: if threshold selection was not clearly reported. |
||||
| Could the conduct or interpretation of the index test have introduced bias? | HIGH: if one or more signalling questions were answered with NO, as even in a laboratory situation knowledge of the reference standard may lead to bias. LOW: if all signalling questions were answered with YES. UNCLEAR: all other instances. |
||||
| Is there concern that the index test, its conduct, or interpretation differ from the review question? | HIGH: if tests were built in‐house. If tests were undertaken in a different setting, or using samples, equipment, or personnel not available in practice. | HIGH: if tests were built in‐house. If tests were undertaken in a different setting, or using samples, equipment or personnel not available in practice. | HIGH: if tests were built in‐house. If tests were undertaken in a different setting, or using samples, equipment. or personnel not available in practice. | This will probably be answered 'LOW' in all cases except when assessments were made in a different setting, or using personnel not available in practice. | This will probably be answered 'LOW' in all cases, except when tests used a threshold that was much higher or lower than in practice, or undertaken in a different setting, or using samples, equipment, or personnel not available in practice. |
| REFERENCE STANDARD | |||||
| Is the reference standard likely to correctly classify the target condition? | In this review, we focused on the target condition COVID‐19 disease. Although we defined acceptable reference standards using a consensus process once the list of reference standards that have been used has been obtained from the eligible studies, Studies of which it is clear that only RT‐PCR was used will be considered high risk of bias. | ||||
| Were the reference standard results interpreted without knowledge of the results of the index test? | YES: if it was explicitly stated that the reference standard results were interpreted without knowledge of the results of the index test, or if the result of the index test was obtained after the reference standard. NO: if it was explicitly stated that the reference standard results were interpreted with knowledge of the results of the index test or if the index test was used to make the final diagnosis. UNCLEAR: if blinding was unclearly reported. |
||||
| Did the definition of the reference standard incorporate results from the index test(s)? | YES: if results from the index test were a component of the reference standard definition. NO: if the reference standard did not incorporate the index standard test. UNCLEAR: if it was unclear whether the results of the index test formed part of the reference standard. |
||||
| Could the conduct or interpretation of the reference standard have introduced bias? | HIGH: if one or more signalling questions were answered with NO. LOW: if all signalling questions were answered with YES. UNCLEAR: all other instances. |
||||
| Is there concern that the target condition as defined by the reference standard does not match the review question? | HIGH: if only RT‐PCR was used (as it measures a different target condition); if alternative diagnosis was highly likely and not excluded (will happen in paediatric cases, where exclusion of other respiratory pathogens is also necessary); if tests used to follow‐up viral load in known test positives. LOW: if above situations were not present. UNCLEAR: if intention for testing was not reported in the study. |
||||
| FLOW AND TIMING | |||||
| Was there an appropriate interval between index test(s) and reference standard? | YES: this will be similar for all index tests, populations for the current infection target conditions: as the situation of a patient, including clinical presentation and disease progress, evolves rapidly and new/ongoing exposure can result in case status change, an appropriate time interval will be within 24 hours. For testing for previous infection, a time interval of at least two weeks is required since resolution of symptoms before the index test was undertaken. NO: if there was more than 24 hours between the index test and the reference standard or if patients were otherwise reported to be assessed with the index versus reference standard test at moments of different severity. UNCLEAR: if the time interval was not reported. |
||||
| Did all patients receive a reference standard? | YES: if all patients received a reference standard (clearly no partial verification). NO: if only (part of) the index test positives or index test negatives received the complete reference standard. UNCLEAR: if it was not reported. |
||||
| Did all patients receive the same reference standard? | YES: if all patients received the same reference standard (clearly no differential verification). NO: if (part of) the index test positives or index test negatives received a different reference standard. UNCLEAR: if it was not reported. |
||||
| Were all patients included in the analysis? | YES: if all included patients were included in the analyses as well. NO: if after the inclusion/exclusion process, patients were removed from the analyses for different reasons: no reference standard done, no index test done, intermediate results of both index test or reference standard, indeterminate results of both index test or reference standard, samples unusable. UNCLEAR: if this was not clear from the reported numbers. |
||||
| Could the patient flow have introduced bias? | HIGH: if one or more signalling questions were answered with NO. LOW: if all signalling questions were answered with YES. UNCLEAR: all other instances. |
||||
| ICU: intensive care unit; RT‐PCR: reverse transcriptase polymerase chain reaction; SARS‐CoV‐2: severe acute respiratory syndrome coronavirus 2; WHO: World Health Organization | |||||
QUADAS‐2 facilitates assessment across four domains: patient selection, index test, reference standard and flow and timing (Whiting 2011). Each domain is assessed in terms of risk of bias and the first three domains are also assessed in terms of concerns regarding applicability. Signalling questions are included to help judge bias. Table 3 shows the definitions used for assessing the methodological quality.
Statistical analysis and data synthesis
Most routine laboratory tests provide test results as continuous measurements. That means that an explicit threshold is needed to provide positive and negative results for estimation of sensitivity and specificity. Some tests indicate disease if the value is decreased relative to the normal ranges, for other tests disease is indicated when the value is increased, and for some tests, both increase and decrease may indicate the presence of disease. For each test in each study, we reported the threshold used in our analyses, and whether an increase or a decrease in value was regarded as a positive test result.
From each study, we included one threshold for each test. If multiple thresholds were reported, we chose the threshold that was most often used in the other studies. We presented the resulting sensitivity and specificity in forest plots. We reported median and interquartile range (IQR) of pre‐test probability of the target condition in 2x2 tables from single‐gate studies.
We considered a meta‐analysis appropriate when four or more studies reported on a particular test. As studies reported mostly different thresholds for the same test, we used the Hierarchical Summary Receiver Operator Curve (HSROC) model for meta‐analyses to estimate summary curves, as recommended by the Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy (Macaskill 2010). Since summary sensitivities and specificities are only clinically interpretable when the studies included in a meta‐analysis use a common cut‐off, we estimated sensitivity at points on the SROC curves corresponding to the median specificity observed in the studies included in the meta‐analysis. The 'Summary of findings' table also reported the estimates for the first and third quartile specificity. Meta‐analyses were undertaken in SAS 9.4, using PROC NLMIXED (SAS 2015).
In resource‐limited situations, or in case SARS‐CoV‐2‐specific tests are not available, routine laboratory tests may be the only tests available. In order to identify the most discriminative test in such a situation, we compared the diagnostic accuracy of biomarkers that had at least a sensitivity of 50% at a minimum specificity of 50% (either median or IQR). We performed these analyses on all studies that evaluated one of these tests (indirect comparison). We performed additional analyses restricted to studies that made head‐to‐head comparisons (i.e. assessed two of the biomarkers in the same participants) when at least four studies were included that enabled these direct comparisons. We made test comparisons by adding a covariate for test type to the HSROC model to assess the effect of test type on the accuracy, cut‐off or shape parameters of the model. In addition, whenever the estimated SROC curves had the same shape, we calculated the relative diagnostic odds ratio (RDOR) as a summary of the relative accuracy of two biomarkers at hand. To assess the statistical significance of differences in test accuracy, we used likelihood ratio tests for comparisons of models with and without covariate terms. If too few primary studies (n < 10) were available for the head‐to‐head comparison, we assumed the shape parameter of the model to be equal for the biomarkers under evaluation.
Investigations of heterogeneity
We investigated sources of heterogeneity if adequate data were available, as listed in the Secondary objectives, either using stratification (where we believed it was inappropriate to combine studies) or through meta‐regression models.
Summary of findings and assessment of the certainty of the evidence
We developed a list of key findings in 'Summary of findings' tables and determined the certainty in the summary estimates for each test and findings, using the GRADE approach (Schünemann 2020a; Schünemann 2020b. Starting at high certainty, we downgraded meta‐analyses by one level when at least half of the studies had high risk of bias on one or more domains; we downgraded for indirectness when at least half of the studies in the meta‐analyses had high concerns regarding applicability on at least one domain; we downgraded for imprecision when fewer people with the target condition were included than would have been needed to achieve the sensitivity estimates listed, with a width of the confidence interval of at most 10 percentage points; and we downgraded for inconsistency when study estimates differed more than 20 percentage points from each other. We did not consider publication bias to be a problem.
Updating
We will undertake the searches of published literature, preprints, and new test approvals weekly, and, dependent on the number of new and important studies found, we will consider updating each review with each search if resources allow.
Results
Results of the search
The overall search for all reviews in this suite was done on 4 May 2020 and resulted in 10,965 records. The first selection resulted in 651 records that were potentially eligible for this review of routine laboratory tests. After title and abstract screening, we excluded 239 records leaving 412 to be assessed on full text (Figure 1). Of these, we removed 17 duplicates and preprints, 31 studies that were not in the scope of the review, 66 studies that did not contain original data and 7 studies that were retracted or otherwise no longer available. Of the remaining 291 studies, 246 studies only considered proven cases of COVID‐19. These reported percentages of proven patients that had an increased or decreased biomarker level. We decided not to extract these data, as only the sensitivity of these markers would be estimable. Furthermore, the aim of these excluded studies was not to assess the accuracy of routine markers for COVID‐19, but just to describe the findings or to assess the accuracy of markers to distinguish between mild and severe disease.
1.
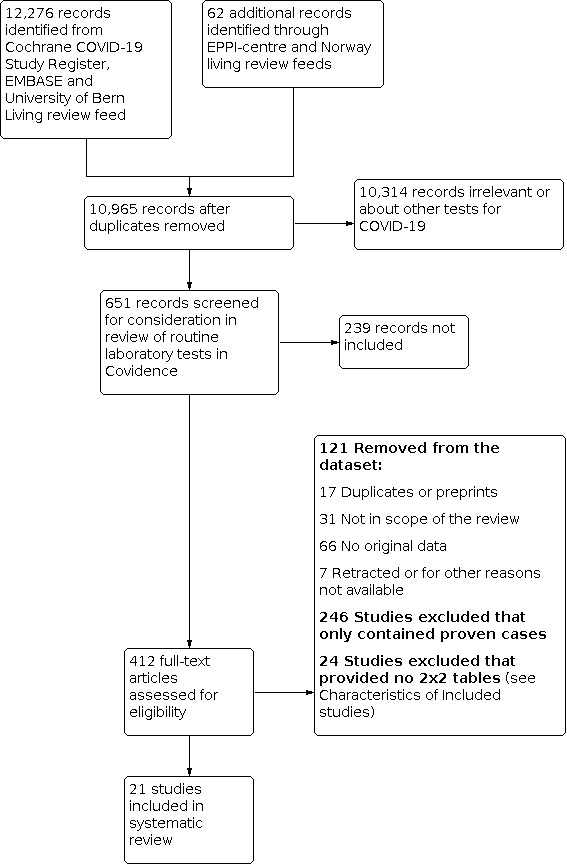
Study flow diagram. Studies were retrieved in a combined search process for all DTA reviews about tests for COVID‐19 and then divided over the different review teams. Due to this process, some preprints only came to light after the data‐extraction phase
The Characteristics of excluded studies table lists the 24 studies that included both patients with and without the target condition, but provided insufficient data to construct 2x2 tables to estimate sensitivity and specificity.
The remaining 21 studies are included in this review.
Included studies
Of the 21 included studies, 14 were single‐gate studies (a study including patients with suspected COVID‐19), six were multiple‐gate studies (including proven COVID‐19 patients and separately one or more groups of non‐COVID‐19 patients). In the remaining study the design was unclear (Characteristics of included studies).
The included studies comprised in total 14,126 COVID‐19 patients and 56,585 people without COVID‐19. They included a total of 67 laboratory tests (Table 4). Four studies included a mix of children and adults, 16 included only adults and one study was only in children. Seventeen studies were done in China, and one each in Iran, Italy, Taiwan and the USA. Nine studies included patients in general hospitals, six studies included patients in emergency departments, three studies included patients in fever clinics, and the remaining three studies included patients in a paediatric hospital, tertiary hospitals, and in veterans affairs databases.
2. List of tests and cut‐off values per study.
| Ai 2020b | Chen 2020c | Feng 2020 | Ferrari 2020 | Hsih 2020 | Li 2020d | Li 2020e | Li 2020f | Li 2020g | Liang 2020 | Liu 2020 | Lu 2020 | Mardani 2020 | Miao 2020 | Pan 2020 | Rentsch 2020 | Yang 2020b | Yang 2020c | Zhang 2020 | Zhao 2020 | Zhu 2020 | |
| a‐HBDH increase | 182 | 182 | |||||||||||||||||||
| ALB decrease | 3.4 | NR | 3.5 | 3 | |||||||||||||||||
| ALP increase | NR | 120 | |||||||||||||||||||
| ALT increase | 50 | 40 | 40 | 40 | NR | 40 | 40 | 50 | 50 | ||||||||||||
| AST increase | 40 | 35 | NR | 40 | 40 | 40 | 40 | ||||||||||||||
| Basophil count increase | 0.1 | NR | |||||||||||||||||||
| Basophil percentage increase | 1 | ||||||||||||||||||||
| Bile acid total | NR | ||||||||||||||||||||
| Bilirubin total increase | 21 | 20.5 | NR | 21 | |||||||||||||||||
| Bilirubin unconjugated | NR | ||||||||||||||||||||
| Corpuscular volume mean decrease | NR | ||||||||||||||||||||
| Corpuscular volume mean increase | NR | ||||||||||||||||||||
| Creatine kinase ‐ increase | 200 | 200 | 185 | 174 | 310 | ||||||||||||||||
| Creatine kinase MB ‐ increase | 24 | 25 | |||||||||||||||||||
| CRP increase | 8 | 11 | 0.8 | 30 | 10 | 4 | 5 | NR | NR | 4 | 34.8 | 10 | 4 | 8 | |||||||
| D‐dimer increase | 0.5 | 0.5 | 0.55 | ||||||||||||||||||
| Direct bilirubin | NR | ||||||||||||||||||||
| eGFR | 15 | ||||||||||||||||||||
| Eosinophil count decrease | 0.02 | NR | |||||||||||||||||||
| Eosinophil count increase | 0.3 | 0.52 | NR | ||||||||||||||||||
| Eosinophil percentage increase | 5 | ||||||||||||||||||||
| Erythrocyte mean corpuscular haemoglobin decrease | NR | ||||||||||||||||||||
| Erythrocyte mean corpuscular haemoglobin increase | NR | ||||||||||||||||||||
| ESR increase | 20 | ||||||||||||||||||||
| Erythrocytemean corpuscular haemoglobin concentrate decrease | NR | ||||||||||||||||||||
| Erythrocytemean corpuscular haemoglobin concentrate increase | NR | ||||||||||||||||||||
| GGT increase | NR | 57 | 45 | ||||||||||||||||||
| GLB decrease | NR | ||||||||||||||||||||
| GLB increase | NR | 30 | |||||||||||||||||||
| HCT decrease | 40 | NR | |||||||||||||||||||
| HCT increase | 52 | ||||||||||||||||||||
| HGB | 13.7 | NR | 10 | ||||||||||||||||||
| Haematuria | NR | ||||||||||||||||||||
| IL‐10 | NR | ||||||||||||||||||||
| IL‐2 | NR | ||||||||||||||||||||
| IL‐4 | NR | ||||||||||||||||||||
| IL‐6 increase | 5.9 | NR | 7 | 7 | |||||||||||||||||
| IL‐8 | NR | ||||||||||||||||||||
| INR increase | 1.25 | 1.15 | |||||||||||||||||||
| LDH increase | 250 | 250 | 245 | 243 | 250 | ||||||||||||||||
| Leukocyturia | NR | ||||||||||||||||||||
| Lymphocyte count decrease | 1.1 | 1.1 | 1 | 1 | 1.1 | 1.1 | 1.1 | 1.1 | NR | 0.8 | 0.8 | 1.1 | 1.1 | ||||||||
| Lymphocyte count increase | 3.2 | 4 | NR | ||||||||||||||||||
| Lymphocyte percentage decrease | 20 | 20 | 23.7 | 20 | |||||||||||||||||
| Lymphocyte percentage increase | 40 | ||||||||||||||||||||
| Monocyte count decrease | 0.1 | NR | |||||||||||||||||||
| Monocyte count increase | 0.6 | 0.8 | 0.6 | NR | |||||||||||||||||
| Monocyte percentage increase | 8 | ||||||||||||||||||||
| Neutrophil count decrease | 1.8 | 2 | 1.8 | NR | |||||||||||||||||
| Neutrophil count increase | 6.3 | 6.3 | 7 | 6.3 | 6.3 | 6.3 | 6.3 | NR | 7 | 4.61 | 6.3 | ||||||||||
| Neutrophil Percentage decrease | 50 | ||||||||||||||||||||
| Neutrophil percentage increase | 75 | 70 | 65.8 | 75 | |||||||||||||||||
| Platelets decreased | 300 | NR | 150 | 100 | |||||||||||||||||
| Platelet mean volume | NR | ||||||||||||||||||||
| pro‐BNP | 450 | ||||||||||||||||||||
| PCT increase | 0.1 | 0.5 | 0.1 | NR | 0.5 | 0.5 | |||||||||||||||
| Protein total | NR | ||||||||||||||||||||
| Proteinuria | 0 | ||||||||||||||||||||
| PT increase | 16 | 15 | |||||||||||||||||||
| RBC decrease | 4.3 | NR | |||||||||||||||||||
| RBC volume distribution increase | 14.5 | NR | |||||||||||||||||||
| s‐CR increase | 73 | 120 | 133 | 115 | |||||||||||||||||
| TNF alpha | NR | ||||||||||||||||||||
| Troponin I | 0.04 | ||||||||||||||||||||
| Urea increase | 7.5 | 8.2 | |||||||||||||||||||
| WBC decrease | 3.5 | 3.5 | 3.6 | 4 | 3.5 | 4 | NR | 4 | 4 | 4 | 3.5 | ||||||||||
| WBC increase | 9.5 | 9.5 | 10 | 10 | 11.2 | 9.5 | 10 | 9.5 | 10 | 10 | NR | 10 | 6.44 | 10 | 9.5 | ||||||
| a‐HBDH: α‐Hydroxybutyrate dehydrogenase; ALB: albumin; ALP: alkaline phosphatase; ALT: alanine aminotransferase; AST: aspartate aminotransferase; CRP: C‐reactive protein; eGFR: estimated glomerular filtration rate; ESR: erythrocyte sedimentation rate; GGT: gamma‐glutamyl transferase; GLB: globulin; HCT: haematocrit; HGB: haemoglobin; IL: interleukin; INR: international normalized ratio; LDH: lactate dehydrogenase; pro‐BNP: pro B‐type natriuretic peptide; PCT: procalcitonin; PT: prothrombin time; RBC: red blood cell; s‐CR: serum creatinine; TNF: tumour necrosis factor; WBC: white blood cell | |||||||||||||||||||||
Thirteen studies used RT‐PCR as reference standard, three studies used other nucleic acid tests, one combined RT‐PCT and chest CT, one used a ‘pharyngeal swab’ (unclear for which test), one combined RT‐PCR, signs and symptoms and chest CT, one used a non‐specific SARS‐CoV‐2 assay, and one based diagnosis on the Diagnosis and Treatment Program of New Coronavirus Pneumonia, China National Health Commission of the People's Republic of China (CDC) case definition (sixth trial version). The target condition was SARS‐CoV‐2 infection in 17 studies, and SARS‐Cov‐2 pneumonia in two studies and COVID‐19 in two other studies.
Eight studies were prepublications and 13 were published in peer reviewed journals.
Methodological quality of included studies
Of the 21 studies, four studies had low or unclear risk of bias on all domains; all other studies had high risk of bias for at least one domain (Figure 2). Six studies had low concerns regarding applicability for all domains. Eleven studies were judged to have a high risk of bias with respect to the patient selection domain, mainly because of including separate groups of cases and non‐cases. Six studies did not describe the order of inclusion of their participants and two did not include a random or consecutive sample. Five studies were case‐control designs and in two studies the design was unclear. We judged risk of bias for patient selection unclear in four studies. We judged three studies as having a high risk of bias regarding the index test. In these studies the index test was either interpreted with knowledge of the reference standard or there was no predefined cut‐off value. Fourteen studies used RT‐PCR as a reference standard for SARS‐CoV‐2 as a target condition, and three used RT‐PCR as a reference standard with COVID‐19 as a target condition. Only four studies reported multiple tests (e.g. RT‐PCR and CT scans) or criteria (e.g. the criteria of the National Health Commission China) as a reference standard for COVID‐19 as a target condition. Flow and timing was unclear in the majority of studies (n = 12), because the time between the reference standard and index test was unclear.
2.
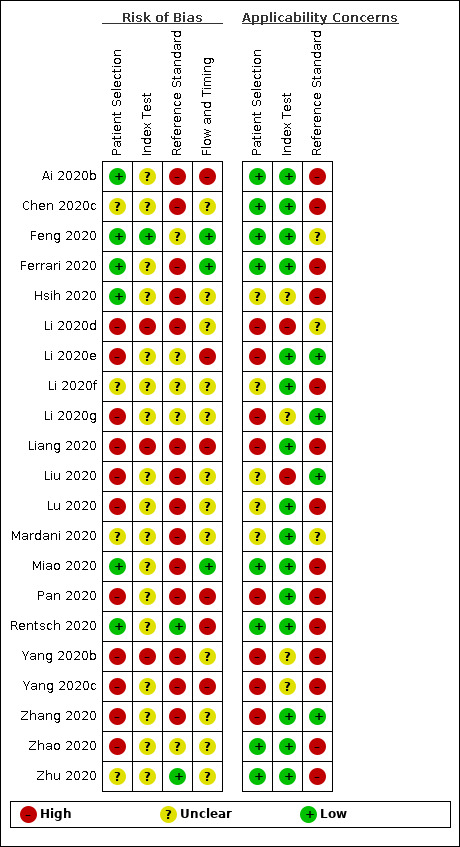
Risk of bias and applicability concerns summary: review authors' judgements about each domain for each included study
None of the studies had low concerns regarding applicability for all domains. As the index test consisted of routine laboratory measurements, these were considered to be low concerns regarding applicability for most studies. In some cases, studies used different cut‐off values, leading to high concerns regarding applicability. As the focus of our review was COVID‐19, we assessed the 14 studies that only used RT‐PCR as a reference standard as high concerns regarding applicability of the reference standard.
Findings
Below we describe the findings for tests assessed in four or more studies: white blood cell count increase and decrease, neutrophil count increase and decrease, monocyte count increase, lymphocyte count decrease, platelets decrease, alanine aminotransferase increase, aspartate aminotransferase increase, albumin decrease, total bilirubin, CRP increase, procalcitonin increase, IL‐6 increase, creatine kinase increase, serum creatinine and lactate dehydrogenase increase. See Table 4 for an overview of tests and cut‐off values per study. Table 1 shows the summary of findings for the individual tests, including sensitivity, specificity and diagnostic odds ratios (DORs). All HSROC curves were close to the non‐informative diagonal, with DORs varying between 0.23 (95% confidence interval (CI) 0.07 to 0.78) and 4.53 (95% CI 1.89 to 10.88). As an indication, a test with a sensitivity of 70% and a specificity of 70% has a DOR of 5.0.
Summary of findings 1. Routine laboratory tests for COVID‐19: single tests.
| Routine laboratory tests for COVID‐19: single tests | |||||||
| Test | Number of studies (number of cases/number of non‐cases) | Median prevalence (IQR) |
Specificity Q1a Mediana Q3a |
Summary sensitivity corresponding with fixed specificity (95% CI) |
Diagnostic odds ratio (95% CI)b | Certainty of the evidencec | Interpretation of the results |
| White blood cell count increase | 15 studies (1262/5318) |
36% (25% to 50%) |
78% | 12% (4.0% to 31%) |
0.35 (0.14 to 0.89) | Very low | WBC count increase is a general marker of inflammation, but most patients with COVID‐19 will be missed at any cut‐off value. Very low‐certainty evidence because of risk of bias, indirectness and inconsistency |
| 85% | 6.0% (2% to 17%) |
||||||
| 92% | 2% (0.0% to 8.0%) |
||||||
| White blood cell count decrease | 11 studies (1211/3900) |
28% (20% to 47%) |
82% | 26% (15% to 40%) |
1.81 (0.90 to 3.67 | Very low | Low WBC is called leukopenia and is a general marker for immune problems. Most patients with COVID‐19 will be missed at any cut‐off value. Very low‐certainty evidence because of risk of bias, indirectness and inconsistency |
| 93% | 25% (8.0% to 27%) |
||||||
| 95% | 22% (5.0% to 26%) |
||||||
| Neutrophil count increase | 11 studies (824/1014) |
36% (25% to 61%) |
66% | 13% (4.0% to 38%) |
0.24 (0.09 to 0.66) | Very low | Neutrophils respond to bacterial infections. An increase may also be caused by other diseases; most patients with COVID‐19 will be missed at any cut‐off value. Very low‐certainty evidence because of risk of bias, indirectness and inconsistency |
| 80% | 4.0% (1.0% to 17%) |
||||||
| 86% | 2.0% (0.0% to 12%) |
||||||
| Neutrophil count decrease | 4 studies (220/514) |
27% (34% to 24%) |
92% | 12% (1.0% to 54%) |
1.29 (0.74 to 2.24) | Low | A decrease in neutrophils is called neutropenia. It is not indicative of COVID‐19, as most patients with COVID‐19 will be missed at any cut‐off value. Low‐certainty evidence because of risk of bias and indirectness |
| 93% | 10% (1.0% to 56%) |
||||||
| 94% | 8.0% (1.0% to 54%) |
||||||
| Neutrophil percentage increase | 4 studies (176/107) |
67% (39% to 74%) |
37% | 62% (1.0 to 100%) |
0.59 (0.13 to 2.61) | Very low | As neutrophils may increase with a general increase of WBCs, the percentage of neutrophils among all WBCs may be given. Most patients without COVID‐19 will still have decreased neutrophil levels. Very low‐certainty evidence because of risk of bias, imprecision and inconsistency |
| 38% | 59% (1.0% to 100%) |
||||||
| 45% | 44% (1.0% to 99%) |
||||||
| Monocyte count Increase | 4 studies (126/332) |
73% (2 studies) |
67% | 14% (6.0% to 30%) |
0.39 (0.17 to 0.86) | Very low | Monocytes are the precursors of macrophages and dendritic cells, the cells that actively catch viruses and bacteria. An increase is called monocytosis and caused by many different inflammatory mechanisms. Most patients with COVID‐19 will be missed at any cut‐off value. Very low‐certainty evidence because of risk of bias, indirectness, imprecision and inconsistency. |
| 73% | 13% (6.0% to 26%) |
||||||
| 80% | 12% (7.0% to 20%) |
||||||
| Lymphocyte count decrease | 13 studies (2752/1066) | 37% (27% to 65%) |
43% | 100% (81% to 100%) |
1.42 (0.93 to 2.17) | Low | Lymphocytes (e.g. T‐cells and B‐cells) play a crucial role in immunity. A decrease (lymphopenia) is not more accurate than tossing a coin. Low‐certainty evidence because of risk of bias and inconsistency |
| 53% | 64% (28% to 89%) |
||||||
| 71% | 0.0% (0.0% to 24%) |
||||||
| Lymphocyte percentage decrease | 4 studies (190/177) |
37% (27% to 65%) |
34% | 70% (0.0% to 100%) |
0.55 (0.08 to 3.73) | Low | A decrease in lymphocyte percentage means that among WBCs the lymphocytes are specifically decreased. This is not indicative for COVID‐19. Low‐certainty evidence because of imprecision and inconsistency |
| 50% | 35% (0.0% to 99%) |
||||||
| 63% | 14% (0.0% to 99%) |
||||||
| Platelets decrease | 4 studies (939/3232) |
76% (38% to 87%) |
83% | 23% (13% to 38%) |
1.68 (1.07 to 2.65) | Very low | A decrease in platelets is called thrombocytopenia and may be caused by various processes. It is not indicative of COVID‐19, as most patients with COVID‐19 will be missed at any cut‐off value. Very low‐certainty evidence because of risk of bias, indirectness and inconsistency |
| 88% | 19% (10% to 32%) |
||||||
| 92% | 16% (7.0% to 31%) |
||||||
| Alanine aminotransferase (ALT) increase | 9 studies (1375/3787) |
42% (34% to 66%) |
85% | 23% (14% to 35%) |
1.29 (0.98 to 1.71) | Low | ALT is an indicator of liver cell damage, but is not specifically indicative for COVID‐19, as most patients with COVID‐19 will be missed at any cut‐off value. Low‐certainty evidence because of risk of bias and indirectness |
| 92% | 12% (3.0% to 34%) |
||||||
| 97% | 4% (0.0% to 41%) |
||||||
| Aspartate aminotransferase (AST) increase | 7 studies (1260/3631) |
53% (29% to 68%) |
79% | 32% (17% to 52%) |
1.63 (1.09 to 2.44) | Low | AST is found in liver, muscles, heart, kidney, brain and red blood cells. It is a marker for liver damage; it is not an indication of COVID‐19, as most patients with COVID‐19 will be missed at any cut‐off value. Low‐certainty evidence because of risk of bias and indirectness |
| 81% | 29% (17% to 45%) |
||||||
| 88% | 17% (8.0% to 33%) |
||||||
| Albumin decrease | 4 studies (799/3273) |
75% (51% to 87%) |
46% | 36% (7.0% to 82%) |
0.51 (0.20 to 1.34) | Low | Hypoalbuminaemia is the term used for low albumin levels and an indication of increased protein loss or decreased protein synthesis (e.g. due to kidney disease, sepsis or severe liver damage). Most patients with COVID‐19 will be missed at any cut‐off value. Low‐certainty evidence because of risk of bias and indirectness |
| 66% | 21% (3.0% to 67%) |
||||||
| 79% | 13% (1.0% to 64%) |
||||||
| Total bilirubin increase | 4 studies (333/438) |
51% (25% to 61%) |
85% | 23% (14% to 35%) |
0.62 (0.15 to 2.61) | Very low | Bilirubin is a breakdown product of haemoglobin. An excess may be an indication that the liver is not capable of removing bilirubin from the blood stream; it is not a specific indication of COVID‐19, as most patients with COVID‐19 will be missed at any cut‐off. Very low‐certainty evidence because of risk of bias, indirectness and inconsistency |
| 92% | 12% (3.0% to 34%) |
||||||
| 97% | 4.0% (0.0% to 41%) |
||||||
| C‐reactive protein (CRP) increase | 14 studies (997/1284) |
51% (28% to 60%) |
23% | 82% (70% to 90%) |
1.50 (0.98 to 2.29) | Very low | CRP levels rise in many different inflammatory situations. It is not a specific indication of COVID‐19, but the majority of cases do seem to have a rise in CRP level, although many patients without COVID‐19 also show a rise in CRP levels. Very low‐certainty evidence because of risk of bias, indirectness and inconsistency |
| 44% | 66% (55% to 75%) |
||||||
| 53% | 58% (45% to 70%) |
||||||
| Procalcitonin increase | 6 studies (607/738) |
38% (31% to 70%) |
66% | 14% (3.0% to 48%) |
0.23 (0.07 to 0.78) | Very low | Procalcitonin levels rise in many different inflammatory situations, especially in bacterial infections. Most patients with COVID‐19 will be missed at any cut‐off value. Very low‐certainty evidence because of risk of bias, indirectness and inconsistency |
| 86% | 3.0% (1.0% to 19%) |
||||||
| 95% | 1.0% (0.0% to 10%) |
||||||
| IL‐6 increase | 4 studies (86/130) |
84% (65% to 94%) |
42% | 83% (47% to 96%) |
4.53 (1.89 to 10.88) | Very low | IL‐6 increases in a various number of conditions and may be linked to a worse prognosis. In this review, it is one of the more sensitive tests. Still, the test by itself cannot rule in or rule out COVID‐19. Very low‐certainty evidence because of risk of bias, imprecision and inconsistency |
| 58% | 73% (36% to 93%) |
||||||
| 74% | 59% (25% to 86%) |
||||||
| Creatine kinase increase | 5 studies (575/498) |
55% (37% to 70%) |
88% | 15% (10% to 22%) |
2.01 (1.01 to 3.98) | Low | Creatine kinase (CK) is an enzyme found in many different tissues in the body. Increased CK is an indication of muscle damage, but most patients with COVID‐19 will be missed at any cut‐off value. Low‐certainty evidence because of risk of bias and indirectness |
| 94% | 11% (6.0% to 19%) |
||||||
| 98% | 7.0% (2.0% to 20%) |
||||||
| Serum creatinine | 4 studies (1005/3311) |
33% (52% to 68%) |
76% | 15% (2.0% to 63%) |
0.70 (0.23 to 2.13) | Low | Serum creatinine is a marker for kidney damage. It is not a specific indication of COVID‐19, as most patients with COVID‐19 will be missed at any cut‐off value. Low‐certainty evidence because of risk of bias and inconsistency |
| 91% | 7% (1.0% to 37%) |
||||||
| 97% | 3% (0.0% to 36%) |
||||||
| Lactate dehydrogenase (LDH) increase | 5 studies (382 cases/431 non‐cases) |
54% (40% to 71%) |
69% | 26% (15% to 42%) |
0.86 (0.52 to 1.45) | Very low | LDH is a marker for general cell and tissue damage. It is not a specific indication of COVID‐19, as most patients with COVID‐19 will be missed at any cut‐off value. Very low‐certainty evidence because of risk of bias, indirectness and inconsistency |
| 72% | 25% (15% to 38%) |
||||||
| 77% | 22% (11% to 40%) |
||||||
|
GRADE Working Group grades of evidence High certainty: we are very confident that the true effect lies close to that of the estimate of the effect. Moderate certainty: we are moderately confident in the effect estimate; the true effect is likely to be close to the estimate of the effect, but there is a possibility that it is substantially different. Low certainty: our confidence in the effect estimate is limited; the true effect may be substantially different from the estimate of the effect. Very low certainty: we have very little confidence in the effect estimate; the true effect is likely to be substantially different from the estimate of effect. ALT: alanine aminotransferase; AST: aspartate aminotransferase; CI: confidence interval; CRP: C‐reactive protein; IL‐6: interleukin‐6; IQR: interquartile range; LDH: lactate dehydrogenase; WBC: white blood cell. Included studies defined a positive test result as an increase or a decrease compared to normal range values, or both. | |||||||
aThe specificity marking the first quartile (Q1) of all specificities of the studies included, the median specificity, and the third quartile (Q3) specificity were used to estimate the corresponding sensitivity estimates from the HSROC model. bA sensitivity and specificity both of 70% would lead to a diagnostic odds ratio of 5.0. cStarting at high certainty of the evidence, the evidence was downgraded by one level when at least half of the studies had high risk of bias on one or more domains; downgraded for indirectness when at least half of the studies in the meta‐analyses had high concerns regarding applicability on at least one domain; downgraded for imprecision when fewer people with the target condition were included then would have been needed to achieve the sensitivity‐estimates listed with a width of the confidence interval of at most 10% points; and downgraded for inconsistency when study estimates differed more than 20% points from each other. Publication bias was not considered to be a problem.
Complete blood count
White blood cell count increase
Fifteen studies (1262 cases/5318 non‐cases) reported on white blood cell count increase (Figure 3). The cut‐off values for an increase in white blood cell count varied from 9.5 x 109 cells/L to 11.2 x 109 cells/L, with the exception of one study that used a cut‐off value of 6.4 x 109 cells/L. The median prevalence of COVID‐19 in the 12 single‐gate studies that reported on white blood cell count increase was 36% (IQR 25% to 50%).
3.
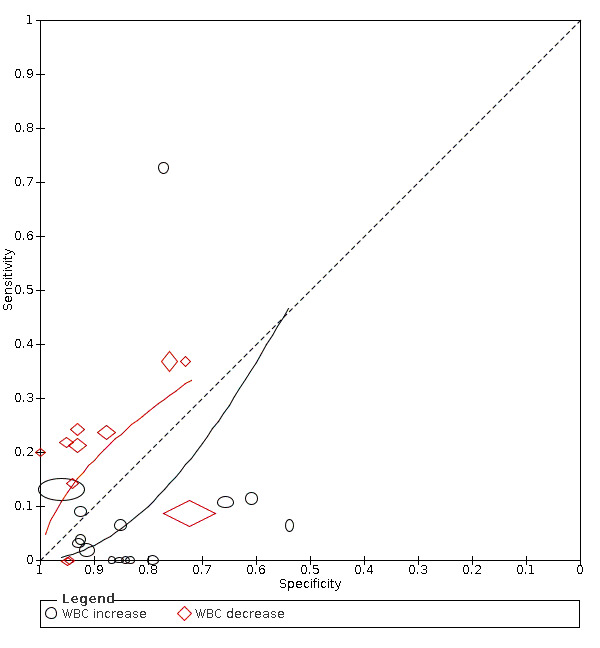
Summary ROC plot of tests. 1: white blood cell count (WBC) increase; 2: WBC decrease
Sensitivity in the 15 included studies ranged from 0% to 73%. Fourteen studies had a sensitivity within the range between 0% and 13% and one study reported a sensitivity of 73%. This outlier also was the only study that used the lower cut‐off of 6.4 x 109 cells/L. Specificity ranged from 54% to 96%.
The median specificity was 85%, with the interquartile range from 78% (Q1) to 92% (Q3). The summary estimate of sensitivity following from the HSROC model and corresponding with a specificity of 78%, was 12% (95% CI 4% to 31%). The summary estimate of sensitivity corresponding with the median specificity of 85%, was 6% (95% CI 2% to 17%) and the summary estimate of sensitivity corresponding with a specificity of 92%, was 2% (95% CI 0% to 8%).
White blood cell count decrease
Eleven studies (1211 cases/3900 non‐cases) reported on white blood cell count decrease (Figure 3). The cut‐off values for a decrease in white blood cell count varied from 3.5 x 109 cells/L to 4.0 x 109 cells/L. The median prevalence of COVID‐19 in the nine single‐gate studies was 28% (IQR 20% to 47%). Sensitivity in the 11 studies ranged from 0% to 37%. Specificity ranged from 72% to 100%.
The median specificity was 93%, with the interquartile range from 82% (Q1) to 95% (Q3). The summary estimates of sensitivity corresponding to these numbers were: 26% (95% CI 15% to 40%) at a specificity of 82%; 25% (95% CI 8% to 27%) at a specificity of 93%; and 22% (95% CI 5% to 26%) at a specificity of 95%.
Neutrophil count increase
Eleven studies (824 cases/1014 non‐cases) reported on neutrophil count (Figure 4). The cut‐off values for an increase in neutrophil count varied from 6.3 x 109 cells/L to 7.0 x 109 cells/L, with the exception of one study that used a cut‐off value of 4.6 x 109 cells/L. The median prevalence of COVID‐19 in the eight single‐gate studies was 36% (IQR 25% to 61%).
4.
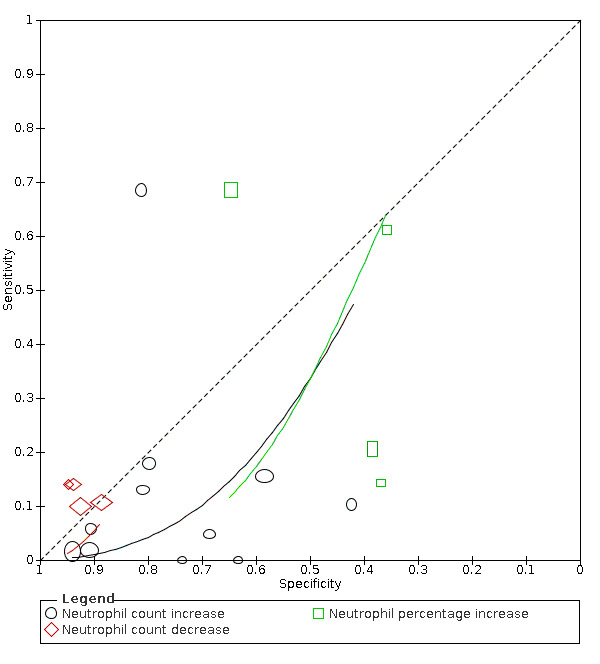
Summary ROC plot of tests: neutrophil count increase, and neutrophil count decrease
Sensitivity ranged from 0% to 68%; in 10 studies the sensitivity ranged between 0% and 18%, one study reported a sensitivity of 68% (this outlier is probably due to the low cut‐off value of 4.6 x 109 cells/L). Specificity ranged from 42% to 94%, with a median of 80% (IQR 66% to 86%).
Meta‐analysis yielded a sensitivity of 13% (95% CI 4% to 38%), 4% (95% CI 1% to 17%) and 2% (95% CI 0% to 12%) at fixed specificity of 66% (Q1), 80% (median) and 86% (Q3), respectively.
Neutrophil count decrease
Four studies (220 cases/514 non‐cases) reported on the accuracy of decrease in neutrophil count (Figure 4). The cut‐off values for a decrease in neutrophil count varied from 1.8*109 cells/L to 2*109 cells/L. The median prevalence of COVID‐19 in the three single‐gate studies was 27% (IQR 34% to 24%). The sensitivity of the four studies ranged from 10% to 14% and specificity ranged from 89% to 95%. Meta‐analysis yielded a sensitivity of 12% (95% CI 1% to 54%), 10% (95% CI 1% to 56%) and 8% (95% CI 1% to 54%) at a fixed specificity of 92% (Q1), 93% (median) and 94% (Q3), respectively.
Neutrophil percentage increase
Four studies (176 cases/107 non‐cases) reported on the accuracy of increase in neutrophil percentage (Figure 4). The cut‐off values for an increase in neutrophil count varied from 65.78% to 75.0%. The median prevalence of COVID‐19 in the three single‐gate studies was 67% (IQR 39% to 74%). The sensitivity of the four studies ranged from 14% to 68% and specificity ranged from 36% to 65%. Meta‐analysis yielded a sensitivity of 62% (95% CI 1% to 100%), 59% (95% CI 1% to 100%) and 44% (95% CI 1% to 99%) at fixed specificity of 37% (Q1), 38% (median) and 45% (Q3), respectively.
Monocyte count increase
Four studies (126 cases/332 non‐cases) reported on monocyte increase (Figure 5). The cut‐off values for an increase in monocyte count varied from 0.00 cells/L to 0.8 cells/L. The median prevalence of COVID‐19 in the two single‐gate studies was 73%. Sensitivity ranged from 10% to 14%; Specificity ranged from 56% to 89%. Meta‐analysis yielded a sensitivity of 14% (95% CI 6% to 30%), 13% (95% CI 6% to 26%) and 12% (95% CI 7% to 20%) at fixed specificity of 67% (Q1), 73% (median) and 80% (Q3), respectively.
5.
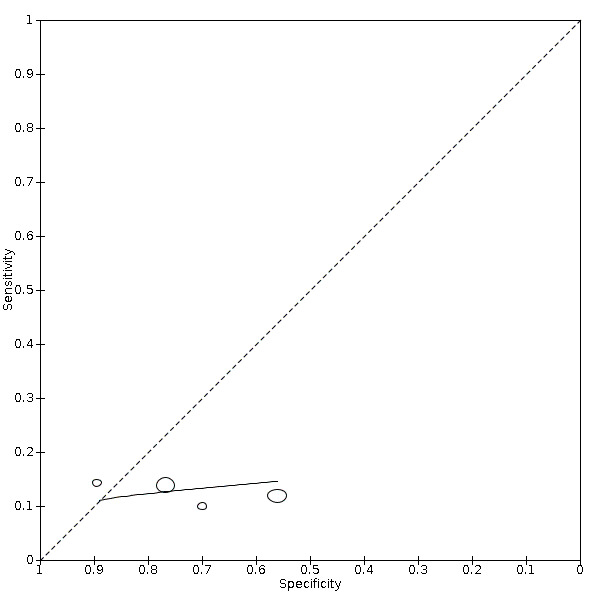
Summary ROC plot of monocyte count increase
Lymphocyte count decrease
Thirteen studies (2752 cases/1066 non‐cases) reported on decrease in lymphocyte count (Figure 6). The cut‐off values for a decrease in lymphocyte count ranged from 8.0*109 cells/L to 1.1*109 cells/L. The median prevalence of COVID‐19 in the 11 single‐gate studies was 37% (27% to 65%), with sensitivity ranging from 0% to 81%, with one outlier of 0% (based on two COVID‐19 cases and specificity from 33% to 89%. Meta‐analysis yielded a sensitivity of 100% (95% CI 81% to 100%), 64% (95% CI 28% to 89%) and 0% (95% CI 0% to 24%) at fixed specificity of 43% (Q1), 53% (median) and 71% (Q3), respectively.
6.
Summary ROC plot of lymphocyte count decrease
Lymphocyte percentage decrease
Four studies (190 cases/177 non‐cases) reported on decrease in lymphocyte percentage (Figure 6). The cut‐off values for a decrease in lymphocyte percentage ranged from 20% to 23.65%. The median prevalence of COVID‐19 in the 11 single‐gate studies was 37% (27% to 65%), with sensitivity ranging from 0% to 79% and specificity from 27% to 65%. Meta‐analysis yielded a sensitivity of 70% (95% CI 0% to 100%), 35% (95% CI 0% to 99%) and 14% (95% CI 0% to 99%) at fixed specificity of 34% (Q1), 50% (median) and 63% (Q3), respectively.
Platelets decrease
Four studies (939 cases/3232 non‐cases) reported on decrease in platelets (Figure 7). The cut‐off values for a decrease in platelets ranged from 0.00 to 300.0 per microlitre. The median prevalence of COVID‐19 in the three single‐gate studies was 76% (38% to 87%), with sensitivity ranging from 13% to 30% and specificity from 71% to 100%. Meta‐analysis yielded a sensitivity of 23% (95% CI 13% to 38%), 19% (95% CI 10% to 32%) and 16% (95% CI 7% to 31%) at fixed specificity of 83% (Q1), 88% (median) and 92% (Q3), respectively.
7.
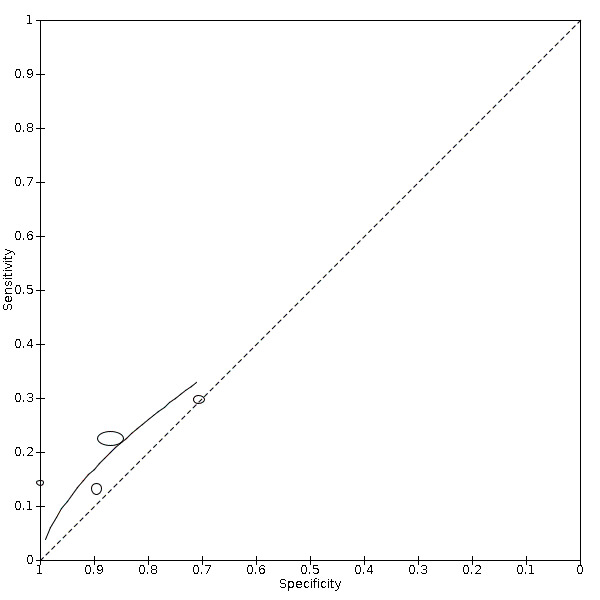
Summary ROC plot of 22 platelets, decreased
Liver function tests
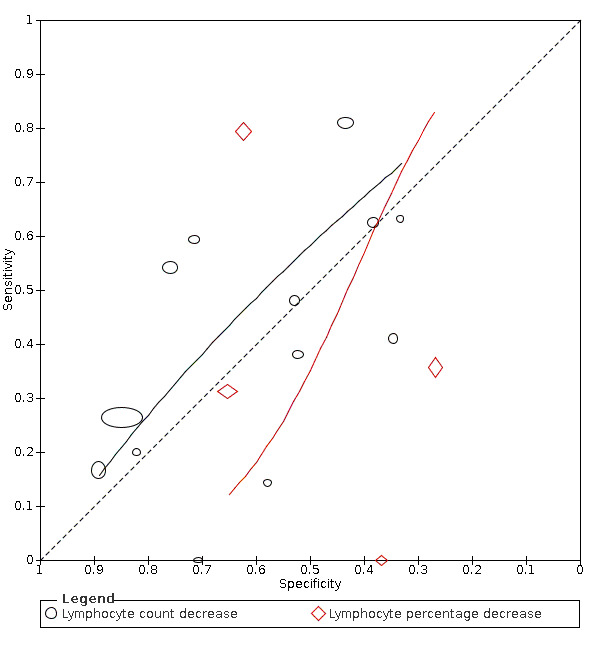
Alanine aminotransferase (ALT) increase
Nine studies (1375 cases/3787 non‐cases) reported on ALT increase (Figure 8). The cut‐off values for in ALT increase varied from 40 U/L to 50 U/L. The median prevalence of COVID‐19 in the seven single‐gate studies was 42% (IQR 34% to 66%). Sensitivity ranged from 10% to 28% and specificity ranged from 74% to 100%. Meta‐analysis yielded a sensitivity of 23% (95% CI 14% to 35%), 12% (95% CI 3% to 34%) and 4% (95% CI 0% to 41%) at fixed specificity of 85% (Q1), 92% (median) and 97% (Q3), respectively.
8.
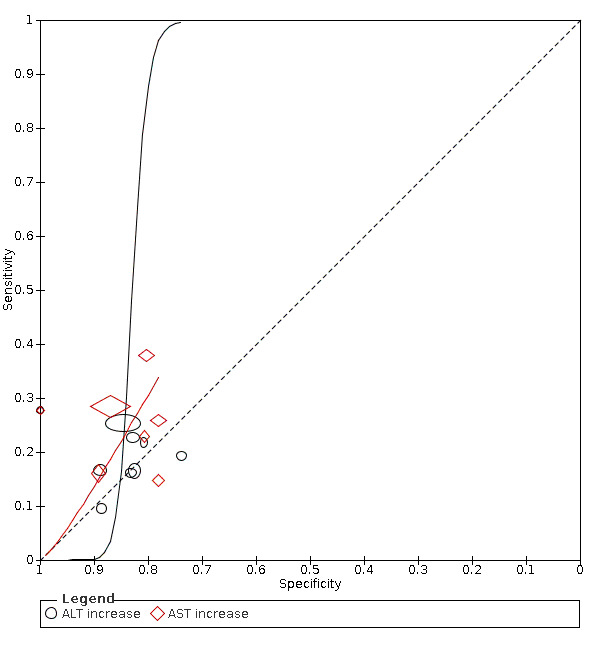
Summary ROC plot of tests: alanine aminotransferase (ALT) increase, aspartate aminotransferase( AST) increase.
Aspartate aminotransferase (AST) increase
Seven studies (1260 cases/3631 non‐cases) reported on AST increase (Figure 8). The cut‐off values of AST increase varied from 35 U/L to 40 U/L. The median prevalence of COVID‐19 in the six single‐gate studies was 53% (IQR 29% to 68%). Sensitivity ranged from 15% to 38%, and specificity from 78% to 100%. Meta‐analysis yielded a sensitivity of 32% (95% CI 17% to 52%), 29% (95% CI 17% to 45%) and 17% (95% CI 8% to 33%) at fixed specificity of 79% (Q1), 81% (median) and 88% (Q3), respectively.
Albumin decrease
Four studies (799 cases/3273 non‐cases) reported on albumin decrease (Figure 9). The cut‐off values of albumin decrease varied from 0 to 3.5 g/L. The median prevalence of COVID‐19 in the three single‐gate studies was 75% (IQR 51% to 87%). Sensitivity ranged from 4% to 55%, and specificity from 16% to 87%. Meta‐analysis yielded a sensitivity of 36% (95% CI 7% to 82%), 21% (95% CI 3% to 67%) and 13% (95% CI 1% to 64%) at fixed specificity of 46% (Q1), 66% (median) and 79% (Q3), respectively.
9.

Summary ROC plot of tests: 30 total bilirubin (TBIL) increase, 36 albumin (ALB) decrease
Total bilirubin increase
Four studies (333 cases/438 non‐cases) reported total bilirubin increase (Figure 9). The cut‐off varied from 0 to 21 µmol/L. The median prevalence of COVID‐19 in the four single‐gate studies was 51% (IQR 25% to 61%). Sensitivity ranged from 3% to 9% and specificity ranged from 77% to 97%. Meta‐analysis yielded a sensitivity of 23% (95% CI 14% to 35%), 12% (95% CI 3% to 34%) and 4% (95% CI 0% to 41%) at fixed specificity of 85% (Q1), 92% (median) and 97% (Q3), respectively.
Markers of inflammation
C‐reactive protein (CRP) increase
Fourteen studies (997 cases/1284 non‐cases) reported on CRP increase (Figure 10). The cut‐off values for an increase in CRP increase varied from 8 mg/L to 34.8 mg/L. The median prevalence of COVID‐19 in the 11 single‐gate studies was 51% (IQR 28% to 60%). Sensitivity ranged from 0% to 95%, with one outlier of 0% (based on two COVID‐19 cases), and the other 13 studies ranging from 31% to 95%. Specificity ranged from 20% to 81%. Meta‐analysis yielded a sensitivity of 82% (95% CI 70% to 90%), 66% (95% CI 55% to 75%) and 58% (95% CI 45% to 70%) at fixed specificity of 23% (Q1), 44% (median) and 53% (Q3), respectively.
10.
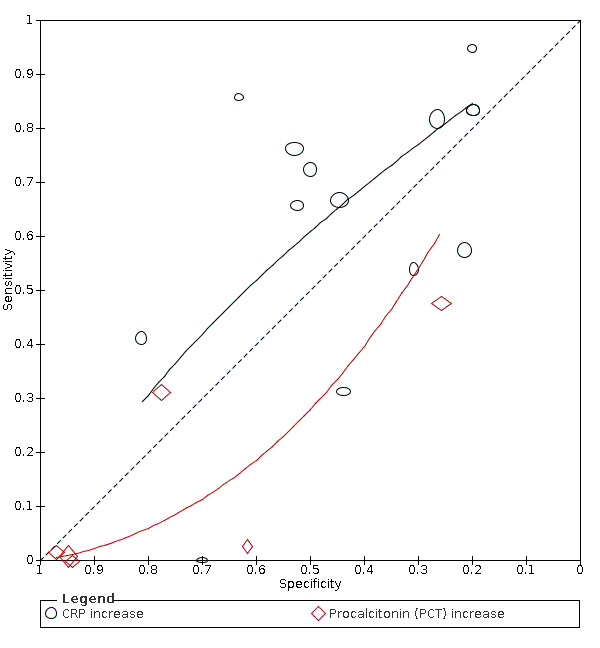
Summary ROC plot of tests: CRP increase and procalcitonin (PCT) increase
Procalcitonin increase
Six studies (607 cases/738 non‐cases) reported on procalcitonin increase (Figure 10). The cut‐off values for an increase in procalcitonin varied from 0.1 ng/mL to 0.5 ng/mL. The median prevalence of COVID‐19 in the five studies was 38% (IQR 31% to 70%). Sensitivity ranged from 0% to 48%. Specificity ranged from 26% to 95%. Meta‐analysis yielded a sensitivity of 14% (95% CI 3% to 48%), 3% (95% CI 1% to 19%) and 1% (95% CI 0% to 10%) at fixed specificity of 66% (Q1), 86% (median) and 95% (Q3), respectively.
IL‐6 increase
Four studies (86 cases/130 non‐cases) reported on IL‐6 increase (Figure 11). The cut‐off values for an increase in IL‐6 varied from 0 to 7 pg/mL. The median prevalence of COVID‐19 in the four studies was 84% (IQR 65% to 94%). Sensitivity ranged from 22% to 86%. Specificity ranged from 27% to 92%. Meta‐analysis yielded a sensitivity of 83% (95% CI 47% to 96%), 73% (95% CI 36% to 93%) and 59% (95% CI 25% to 86%) fixed specificity of 42% (Q1), 58% (median) and 74% (Q3), respectively.
11.
Summary ROC plot of 53 interleukin‐6 (IL‐6) increase. Height and width of the symbols represent the number of cases and non‐cases in the studies
Other tests
Creatine kinase increase
Creatine kinase is a muscle damage marker, which increases upon muscle damage. It is sometimes used as an indicator for cardiac infarction. Five studies (575 cases/498 non‐cases) reported on creatine kinase increase (Figure 12). The cut‐off values for an increase in creatine kinase were between 174 µmol/L and 310 µmol/L. The median prevalence of COVID‐19 in the five single‐gate studies was 55% (IQR 37% to 70%). Meta‐analysis yielded a sensitivity of 15% (95% CI 10% to 22%), 11% (95% CI 6% to 19%) and 7% (95% CI 2% to 20%) at fixed specificity of 88% (Q1), 94% (median) and 98% (Q3), respectively.
12.
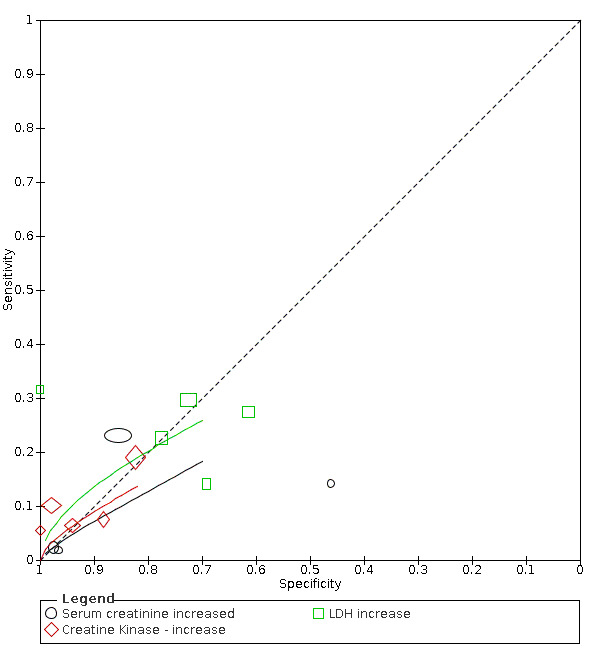
Summary ROC plot of tests: 24 Serum creatinine increased, 25 Creatine kinase ‐ increase, 55 lactate dehydrogenase (LDH) increase
Serum creatinine
Serum creatinine is an indicator of kidney damage. Four studies (1005 cases/3311 non‐cases), all single‐gate design, reported on serum creatinine increase (Figure 12). The cut‐off values for an increase in serum creatinine kinase were between 73 µmol/L and 133 µmol/L. The prevalence in the four studies was 16%, 66%, 38% and 75%. Meta‐analysis yielded a sensitivity of 15% (95% CI 2% to 63%), 7% (95% CI 1% to 37%) and 3% (95% CI 0% to 36%) at fixed specificity of 76% (Q1), 91% (median) and 97% (Q3), respectively.
Lactate dehydrogenase (LDH) increase
LDH is a general marker for tissue damage. Five studies (382 cases/431 non‐cases) reported on LDH increase (Figure 12). The cut‐off values for in LDH increase varied from 243 to 25 U/L. The median prevalence of COVID‐19 in the five single‐gate studies was 54% (IQR 40% to 71%). Sensitivity ranged from 14% to 32% and specificity ranged from 61% to 100%. Meta‐analysis yielded a sensitivity of 26% (95% CI 15% to 42%), 25% (95% CI 15% to 38%) and 22% (95% CI 11% to 40%) at fixed specificity of 69% (Q1), 72% (median) and 77% (Q3), respectively.
Comparisons between tests
For three tests, we found a pair of sensitivity and specificity where both sensitivity and specificity exceeded 50%. These were IL‐6 increase, CRP increase and lymphocyte count decrease. Using all available studies in an indirect comparison (i.e. unrestricted to head‐to‐head studies), we compared the test performance of IL‐6 increase (4 studies), CRP increase (14 studies) and lymphocyte count decrease (13 studies) in one meta‐regression analysis. The shape of the SROC curves significantly differed (P < 0.001). Figure 13 shows the summary ROC curves for the three tests in one Figure (Table 2).
13.
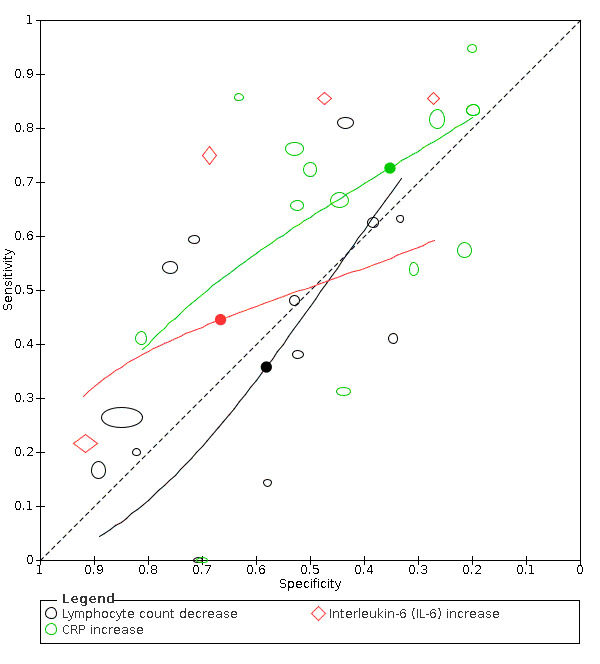
Summary ROC plot of tests: 12 lymphocyte count decrease, 32 CRP increase, 47 interleukin‐6 (IL‐6) increase
Summary of findings 2. Comparisons of routine laboratory tests for COVID‐19 with sensitivity and specificity higher than 50%.
| Comparisons of routine laboratory tests for COVID‐19 with sensitivity and specificity higher than 50% | ||||||||
| Number of studies (number of cases/number of non‐cases) | Fixed specificity |
Summary sensitivity corresponding with fixed specificity (95% CI) |
Interpretation of the results: tests used in a hypothetical cohort of 1000 people tested for COVID‐19, at a pre‐test probability of 5% and 36%a | |||||
| Prevalence | TP | FP | FN | TN | ||||
| Lymphocyte Count Decreaseb | 13 studies (2752/1066) |
53% | 64% (28% to 89%) |
0.05 | 32 | 447 | 18 | 504 |
| 0.36 | 230 | 611 | 130 | 339 | ||||
| C‐reactive protein (CRP) increaseb | 14 studies (997/1284) |
53% | 58% (45% to 70%) |
0.05 | 29 | 447 | 21 | 504 |
| 0.36 | 209 | 611 | 151 | 339 | ||||
| IL‐6 increase at a lower threshold | 4 studies (86/130) |
58% | 73% (36% to 93%) |
0.05 | 37 | 399 | 14 | 551 |
| 0.36 | 263 | 579 | 97 | 371 | ||||
| IL‐6 increase at a higher threshold | 4 studies (86/130) |
74% | 59% (25% to 86%) |
0.05 | 30 | 247 | 21 | 703 |
| 0.36 | 212 | 476 | 148 | 474 | ||||
| CI: confidence interval; FN: false negative; FP: false positive; TN: true negative; TP: true positive. Included studies defined a positive test result as an increase or a decrease compared to normal range values, or both. | ||||||||
aThe median pre‐test probability in the meta‐analyses varied between 27% and 84%, meaning that the included studies are not representative for situations where the prevalence is 5% or lower. The median prevalence over all the single‐gate studies was 36%. bThe direct comparison between lymphocyte count increase and C‐reactive protein (CRP) increase (9 studies) showed that CRP was considerably more accurate than lymphocyte count increase: relative diagnostic odds ratio (DOR) was 2.02 (95% confidence interval 1.47 to 2.78). As the confidence intervals of all the DORs in the indirect comparisons included a non‐informative value (i.e. DOR = 1), a relative DOR of 2 does not mean the alternative is much more informative.
The median specificity in the 19 studies evaluating one or more of the three tests, was 52% (IQR 34% to 67%). Within the specificity interquartile range, sensitivity varied between 6% (95% CI 0% to 49%) and 100% (22% to 100%) for lymphocyte count decrease, between 51% (95% CI 34% to 68%) and 73% (95% CI 64% to 80%) for CRP increase, and between 67% (95% CI 51% to 79%) and 73% (95% CI 45% to 79%) for IL‐6 increase.
Nine studies directly compared CRP increase with lymphocyte count decrease for the detection of COVID‐19. Especially for lymphocyte count decrease, this direct comparison (Figure 14), shows a different picture from the indirect comparisons (Figure 13), or the separate analyses (Figure 6). Despite differences in cut‐offs, the results from most studies were consistent with CRP increase showing higher sensitivity than the lymphocyte count decrease. The RDOR was 2.02 (95% CI 1.47 to 2.78), meaning that the overall accuracy was higher for CRP increase than for lymphocyte count decrease. However, both tests are close to the diagonal line corresponding with an uninformative test.
14.
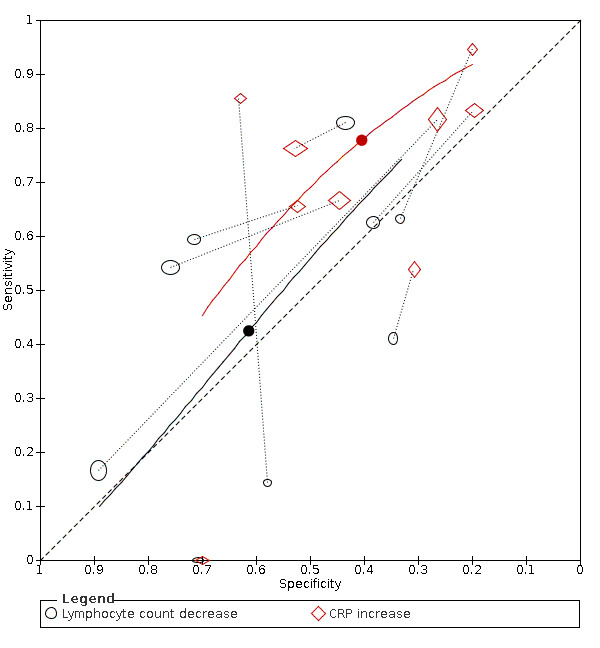
Summary ROC plot of tests: 12 lymphocyte count decrease, 32 CRP increase
Discussion
Summary of main results
We included 21 studies in this review and analyzed the results for 67 different routine laboratory tests, focusing on diagnosing COVID‐19. For 16 tests, we have summarized the results in a meta‐analysis. As the majority of the included studies only reported RT‐PCR as a reference standard, the meta‐analyses may be more applicable to detecting SARS‐CoV‐2 infection than COVID‐19 diseased. Only three tests performed at sensitivity‐specificity combinations where both sensitivity and specificity were above 50%. There was low to very low certainty in the summary estimates of the tests.
The low accuracy of these tests does not render them useless. They are all indicators of the general health status of a patient. They may indicate infection, inflammation, or tissue damage and thus support diagnoses made based on other diseases. However, evidence to date suggests that in sick hospitalized patients, they cannot discriminate between COVID‐19 and other diseases as the cause of infection, inflammation or tissue damage and should preferably not be used as stand‐alone tests for COVID‐19. As a triage test would require a high sensitivity (< 80%), these tests have limited use as triage tests. How these tests would perform in those with milder symptoms cannot be inferred from our data.
In some situations, where resources are very limited, these tests are the only ones at hand when making a diagnosis. In these situations, it may be worthwhile to consider the three tests with a slightly better performance than the others: lymphocyte count decrease, IL‐6 increase and CRP increase. These tests are also available as point‐of‐care tests, although that is not how they were used in the included studies, so any inference should be made with caution.
Of those three, IL‐6 has the highest summary sensitivity at the highest median specificity. Both the median specificity and the boundary of the third quartile were above 50% (58% and 74% respectively). If we chose to use the test at a higher specificity of 74%, then the sensitivity would only be 59% (95% CI 25% to 86%). When testing 1000 people using this cut‐off value, at 5% pre‐test probability, then 29 or 30 out of 50 cases would have a true positive result and be contained or put in quarantine, and 20 or 21 out of 50 cases would be sent home, possibly infectious. It would also mean that of the 950 non‐cases, 247 would be considered to be positive, while they are not. Using the test at a lower cut‐off value to increase sensitivity, would decrease specificity even further.
The median pre‐test probability of all included studies was 36% and most patients were hospitalized. In such a scenario, when testing 1000 people with IL‐6 at a specificity of 74% and a sensitivity of 59%, then 212 out of 360 cases would have a true positive result and be contained or put in quarantine, and 148 out of 360 cases would be sent home or to a non‐COVID‐19 ward, possibly infectious. It would also mean that of the 640 non‐cases, 166 would be considered to be positive, while they are not.
Nine studies directly compared leukocyte count increase and CRP increase. From the meta‐analysis including these two tests, we found that CRP is more accurate than leukocyte count increase, but as explained above, the point estimates do require caution when using the tests as sole markers. Furthermore, we did not assess the quality of the comparisons made in the included studies.
Strengths and weaknesses of the review
We assessed the diagnostic accuracy of a broad spectrum of routine laboratory tests for COVID‐19. Included studies demonstrated considerable heterogeneity in the accuracy of many biomarkers, and used cut‐off values and reference standards that were, in many cases, suboptimally described. The current review included a range of different cut‐off values for most index tests, which we took into account using HSROC analyses and pooling studies with similar cut‐off values for a given laboratory marker.
A limitation is suboptimal reporting that hampered assessment of the QUADAS‐2 flow and timing domain in many studies. In many instances the timing of index test and reference standard was unclear, which could have led to unreliable results concerning the diagnostic abilities of the tests. While most studies used RT‐PCR as reference standard, some used a combination of RT‐PCR and signs and symptoms or other tests. This potentially introduced heterogeneity because of differences in patients marked as cases and controls according to the differences in reference standards.
Some tests of interest, such as d‐dimer or cardiac markers were evaluated in too few studies to meta‐analyse their results.
Applicability of findings to the review question
We retrieved information on multiple index tests. The availability of laboratory tests is dependent on the type of hospital, department and available resources of the place in which the test is to be performed. In order to make the findings suitable for different settings we have included a broad range of biomarkers, and settings. We did not find studies that included participants in a primary care or general population setting. In clinical practice, not a single test, but the results of a combination of tests might be important for diagnosing COVID‐19. These tests can be used for the first triage of patients in case of limited access to diagnostic tests, after which at a later stage further testing can be done. For triage tests, a high sensitivity is important to safely rule out the disease, however all tests had a low sensitivity. Also, the cut‐off values used may differ by clinic and location, this could lead to different treatment decisions if a single patient were tested in different settings. In this review we included all different cut‐off points available in current literature. Lastly, the reference standard in most studies was RT‐PCR only, which means that there are concerns regarding applicability of the results of this review to COVID‐19 as a target condition. However, the reporting of the studies was unclear and sometimes confusing. It may therefore be possible that in the study practice also other criteria were used to assess the diagnosis, but that this was not or insufficiently reported.
Authors' conclusions
Implications for practice.
None of these markers as stand‐alone tests are useful for accurately ruling in or ruling out COVID‐19. As a triage test would require a high sensitivity (< 80%), these tests have limited value as triage tests. Although there is low or very low certainty about the summary estimates in this review, we do not expect that studies with a low risk of bias will show a better performance than the tests included.
Implications for research.
Future studies focusing on the usefulness of routine laboratory tests for COVID‐19 may consider a more representative sample of the population, focus on markers with prespecified, clinically sound cut‐offs and focus on single, but also on the combination of regular blood markers. Furthermore, considering the test results as continuous values may be more informative, as larger deviations from the reference values will have greater impact on the health status of the tested people, and might enable more personalized treatment.
History
Review first published: Issue 11, 2020
Acknowledgements
Members of the Cochrane COVID‐19 Diagnostic Test Accuracy Review Group include:
the project team (Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Leeflang MMG, Spijker R, Hooft L, Van den Bruel A, McInnes MDF, Emperador D, Dittrich S, Cunningham J);
-
the systematic review teams for each review:
Molecular, antigen, and antibody tests (Adriano A, Beese S, Dretzke J, Ferrante di Ruffano L, Harris I, Price M, Taylor‐Phillips S)
Signs and symptoms (Stuyf T, Domen J, Horn S)
Routine laboratory markers (Yang B, Langendam M, Ochodo E, Guleid F, Holtman G, Verbakel J, Wang J, Stegeman I)
Imaging tests (Salameh JP, McGrath TA, van der Pol CB, Frank RA, Prager R, Hare SS, Dennie C, Jenniskens K, Korevaar DA, Cohen JF, van de Wijgert J, Damen JAAG, Wang J);
the wider team of systematic reviewers from University of Birmingham, UK who assisted with title and abstract screening across the entire suite of reviews for the diagnosis of COVID‐19 (Agarwal R, Baldwin S, Berhane S, Herd C, Kristunas C, Quinn L, Scholefield B).
We thank Dr Jane Cunningham (World Health Organization) for participation in technical discussions and comments on the manuscript.
The Cochrane Editorial and Methods Department (EMD) Editorial Service collaborated with Cochrane Infectious Diseases Group (CIDG) on the management of the editorial process. We thank Helen Wakeford (EMD), Anne‐Marie Stephani (EMD) and Deirdre Walshe (CIDG) for editorial checks; Robin Featherstone and Douglas M Salzwedel for comments on the search; Jennifer Hilgart for comments on the abstract; to the peer referees for this review including Jessica Watson and Olabisi A. Oduwole; and Mike Brown and Paul Garner for sign‐off comments. We thank Denise Mitchell for her efforts in copy‐editing this review.
We would like to thank the Cochrane Diagnostic Test Accuracy (DTA) Editorial Team including Sophie Beese and Bella Harris for managing the editorial process; Karen Steingart for methods peer review, Matthew Grainge for providing statistical peer review and acting as Contact Editor for the full review, and to Prof Danielle Van der Windt who was the DTA Contact Editor for the protocol.
The editorial base of the Cochrane Infectious Diseases Group is funded by UK aid from the UK government for the benefit of low‐ and middle‐income countries (project number 300342‐104). The views expressed do not necessarily reflect the UK government’s official policies.
Jonathan Deeks is a United Kingdom National Institute for Health Research (NIHR) Senior Investigator Emeritus. Yemisi Takwoingi is supported by a NIHR Postdoctoral Fellowship. Jonathan Deeks, Jacqueline Dinnes, Yemisi Takwoingi, and Clare Davenport are supported by the NIHR Birmingham Biomedical Research Centre. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health and Social Care.
Appendices
Appendix 1. World Health Organization case definitions
Severe pneumonia
Adolescent or adult: fever or suspected respiratory infection, plus one of the following: respiratory rate > 30 breaths/minute; severe respiratory distress; or oxygen saturation (SpO2) ≤ 93% on room air. Child with cough or difficulty in breathing, plus at least one of the following: central cyanosis or SpO2 < 90%; severe respiratory distress (for example, grunting, very severe chest indrawing); signs of pneumonia with a general danger sign: inability to breastfeed or drink, lethargy or unconsciousness, or convulsions.
Other signs of pneumonia may be present: chest indrawing, fast breathing (in breaths/minute): aged < 2 months: ≥ 60; aged 2 to 11 months: ≥ 50; aged 1 to 5 years: ≥ 40. While the diagnosis is made on clinical grounds; chest imaging may identify or exclude some pulmonary complications.
Acute respiratory distress syndrome (ARDS)
Onset within one week of a known clinical insult or new or worsening respiratory symptoms.
Chest imaging (that is, X‐ray, computer tomography scan, or lung ultrasound): bilateral opacities, not fully explained by volume overload, lobar or lung collapse, or nodules.
Origin of pulmonary infiltrates: respiratory failure not fully explained by cardiac failure or fluid overload. Need objective assessment (for example, echocardiography) to exclude hydrostatic cause of infiltrates/oedema if no risk factor present.
Oxygenation impairment in adults:
mild ARDS: 200 mmHg < ratio of arterial oxygen partial pressure/fractional inspired oxygen (PaO2/FiO2) ≤ 300 mmHg (with positive end‐expiratory pressure (PEEP) or continuous positive airway pressure (CPAP) ≥ 5 cmH2O, or non‐ventilated);
moderate ARDS: 100 mmHg < PaO2/FiO2 ≤ 200 mmHg (with PEEP ≥ 5 cmH2O, or non‐ventilated);
severe ARDS: PaO2/FiO2 ≤ 100 mmHg (with PEEP ≥ 5 cmH2O, or non‐ventilated);
when PaO2 is not available, SpO2/FiO2 ≤ 315 mmHg suggests ARDS (including in non‐ventilated patients).
Oxygenation impairment in children: note OI = Oxygenation Index and OSI = Oxygenation Index using SpO2. Use PaO2‐based metric when available. If PaO2 not available, wean FiO2 to maintain SpO2 ≤ 97% to calculate OSI or SpO2/FiO2 ratio:
bilevel (non‐invasive ventilation or CPAP) ≥ 5 cmH2O via full‐face mask: PaO2/FiO2 ≤ 300 mmHg or SpO2/FiO2 ≤ 264;
mild ARDS (invasively ventilated): 4 ≤ OI < 8 or 5 ≤ OSI < 7.5;
moderate ARDS (invasively ventilated): 8 ≤ OI < 16 or 7.5 ≤ OSI < 12.3;
severe ARDS (invasively ventilated): OI ≥ 16 or OSI ≥ 12.3.
Appendix 2. Cochrane COVID‐19 Study Register searches
| Source | Strategy |
| CT.gov | COVID‐19a |
| WHO ICTRP | Health topic: 2019‐nCov/COVID‐19 |
| PubMed | (("2019 nCoV"[tiab] OR 2019nCoV[tiab] OR "2019 novel coronavirus"[tiab] OR "COVID 19"[tiab] OR COVID19[tiab] OR "new coronavirus"[tiab] OR "novel coronavirus"[tiab] OR "novel corona virus"[tiab] OR "SARS CoV‐2"[tiab] OR (Wuhan[tiab] AND (coronavirus[tiab] OR "corona virus"[tiab])) OR "COVID‐19"[Supplementary Concept] OR "severe acute respiratory syndrome coronavirus 2"[Supplementary Concept]) NOT ("animals"[MeSH Terms] NOT "humans"[MeSH Terms])) NOT (editorial[pt] OR comment[pt] OR letter[pt] OR newspaper article[pt]) |
aAutomatic term mapping links results for 2019‐nCoV, 2019 novel coronavirus, SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Ovid Embase Search
Embase records from the Stephen B. Thacker CDC Library, Covid‐19 Research articles Downloadable database. Records were obtained by the CDC library by searching Embase through Ovid using the following search strategy:
(coronavir* OR corona virus* OR betacoronavir* OR covid19 OR covid 19 OR nCoV OR novel CoV OR CoV 2 OR CoV2 OR sarscov2 OR 2019nCoV OR wuhan virus*).mp. OR ((wuhan OR hubei OR huanan) AND (severe acute respiratory OR pneumonia*) AND outbreak*).mp. OR Coronavirus infection/ OR coronavirinae/ OR exp betacoronavirus/
Limits: 2020‐
OR
(novel coronavir* OR novel corona virus* OR covid19 OR covid 19 OR nCoV OR novel CoV OR CoV 2 OR CoV2 OR sarscov2 OR 2019nCoV OR wuhan virus*).mp. OR ((wuhan OR hubei OR huanan) AND (severe acute respiratory OR pneumonia*) AND outbreak*).mp. OR ((wuhan OR hubei OR huanan) AND (coronavir* OR betacoronavir*)).mp.
Limits: 2019‐
Appendix 3. Living search from the University of Bern
The following information is taken from the university of Bern website (see: ispmbern.github.io/covid-19/living-review/collectingdata.html).
The register is updated daily and CSV file downloads are made available.
1 April 2020
From 1 April 2020, we will retrieve the curated bioRxiv/medRxiv dataset (connect.medrxiv.org/relate/content/181).
26 to 31 March 2020
MEDLINE: (\"Wuhan coronavirus\" [Supplementary Concept] OR \"COVID‐19\" OR \"2019 ncov\"[tiab] OR ((\"novel coronavirus\"[tiab] OR \"new coronavirus\"[tiab]) AND (wuhan[tiab] OR 2019[tiab])) OR 2019‐nCoV[All Fields] OR (wuhan[tiab] AND coronavirus[tiab])))))
Embase: (nCoV or 2019‐nCoV or ((new or novel or wuhan) adj3 coronavirus) or covid19 or covid‐19 or SARS‐CoV‐2).mp.
bioRxiv/medRxiv: ncov or corona or wuhan or COVID or SARS‐CoV‐2
With the kind support of the Public Health & Primary Care Library PHC (www.unibe.ch/university/services/university_library/faculty_libraries/medicine/public_health_amp_primary_care_library_phc/index_eng.html), and following guidance of the Medical Library Association (www.mlanet.org/p/cm/ld/fid=1713).
1 January 2020 to 25 March 2020
MEDLINE: ("Wuhan coronavirus" [Supplementary Concept] OR "COVID‐19" OR "2019 ncov"[tiab] OR (("novel coronavirus"[tiab] OR "new coronavirus"[tiab]) AND (wuhan[tiab] OR 2019[tiab])) OR 2019‐nCoV[All Fields] OR (wuhan[tiab] AND coronavirus[tiab])))))
Embase: ncov OR (wuhan AND corona) OR COVID
bioRxiv/medRxiv: ncov or corona or wuhan or COVID
Data
Presented below are all the data for all of the tests entered into the review.
Tests. Data tables by test.
1. Test.
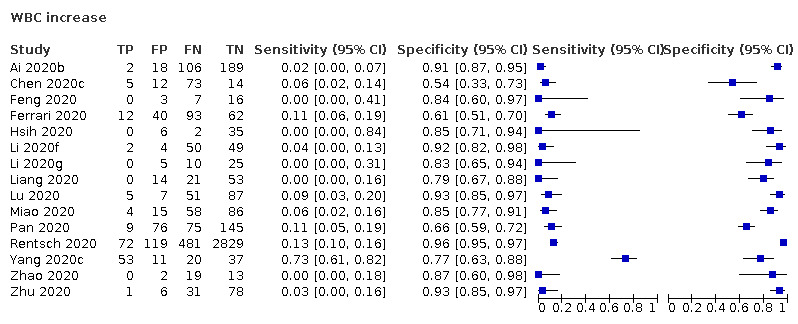
WBC increase
2. Test.
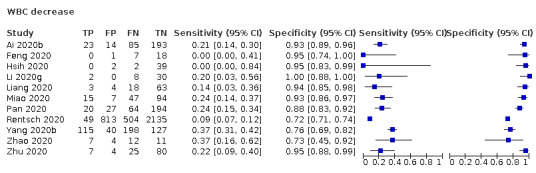
WBC decrease
3. Test.

Leukocyturia
4. Test.
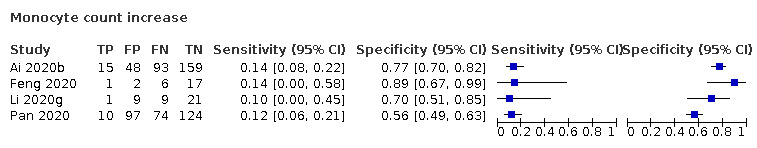
Monocyte count increase
5. Test.

Monocyte count decrease
6. Test.

Monocyte percentage increase
7. Test.
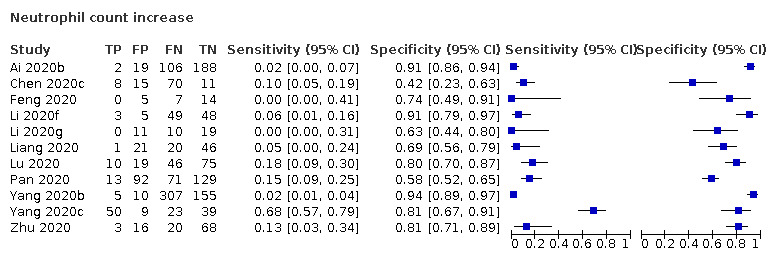
Neutrophil count increase
8. Test.
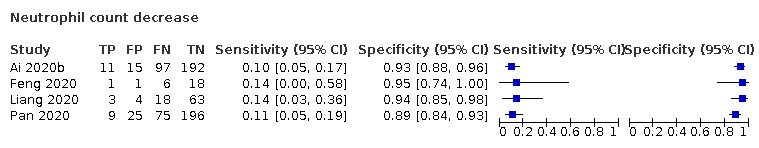
Neutrophil count decrease
9. Test.
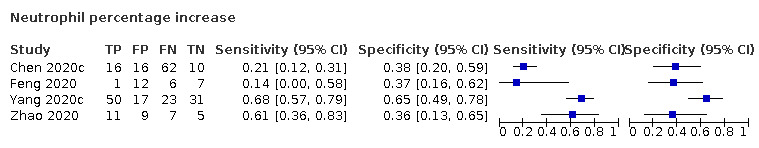
Neutrophil percentage increase
10. Test.

Neutrophil Percentage decrease
11. Test.

Lymphocyte count increase
12. Test.
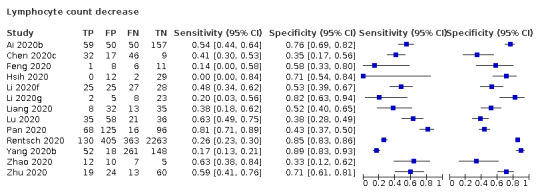
Lymphocyte count decrease
13. Test.

Lymphocyte percentage increase
14. Test.
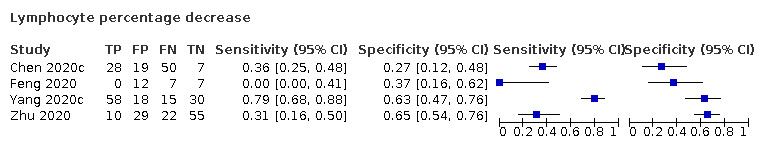
Lymphocyte percentage decrease
15. Test.
Eosinophil count increase
16. Test.
Eosinophil count decrease
17. Test.

Eosinophil percentage increase
18. Test.
Basophil count increase
19. Test.

Basophil percentage increase
20. Test.
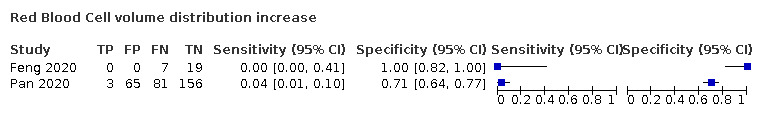
Red Blood Cell volume distribution increase
21. Test.

RBC decrease
22. Test.
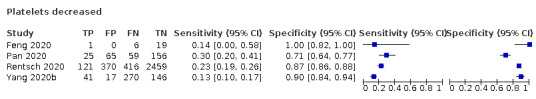
Platelets decreased
23. Test.
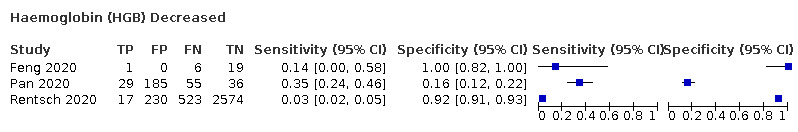
Haemoglobin (HGB) Decreased
24. Test.
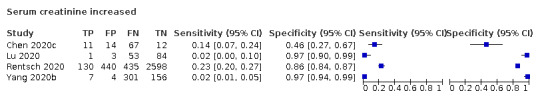
Serum creatinine increased
25. Test.
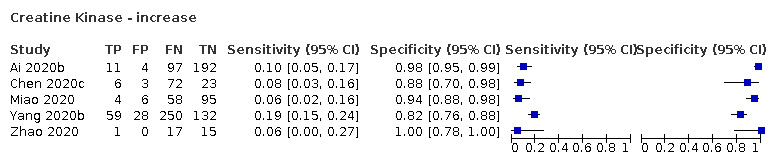
Creatine Kinase ‐ increase
26. Test.
Creatine Kinase MB ‐ increase
27. Test.
Urea increase
28. Test.
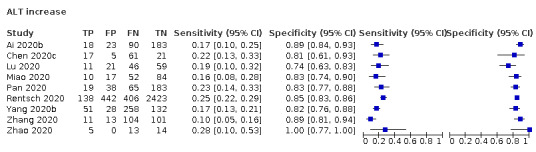
ALT increase
29. Test.
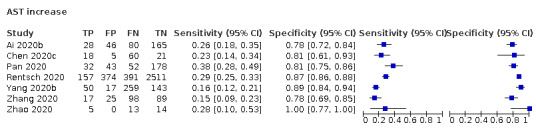
AST increase
30. Test.
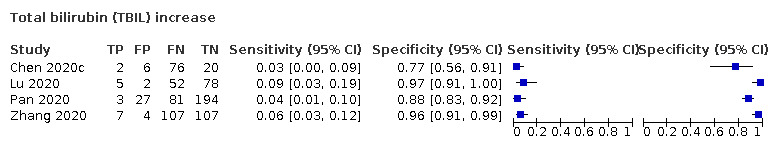
Total bilirubin (TBIL) increase
31. Test.

Erythrocyte Sedimentation Rate (ESR) increase
32. Test.
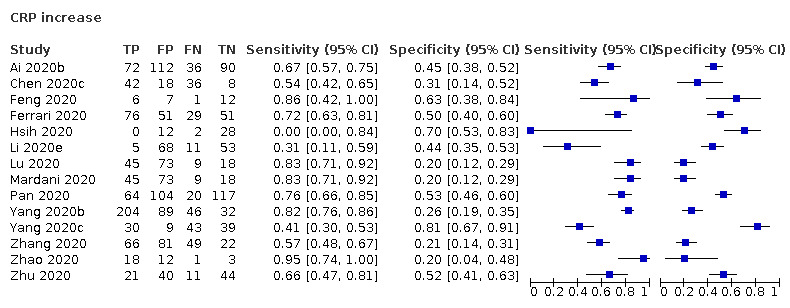
CRP increase
33. Test.

a‐HBDH increased
34. Test.

HCT increased
35. Test.

HCT decreased
36. Test.
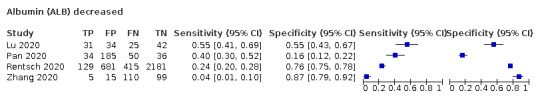
Albumin (ALB) decreased
37. Test.

Globulin (GLB) increase
38. Test.

Globulin (GLB) decrease
39. Test.
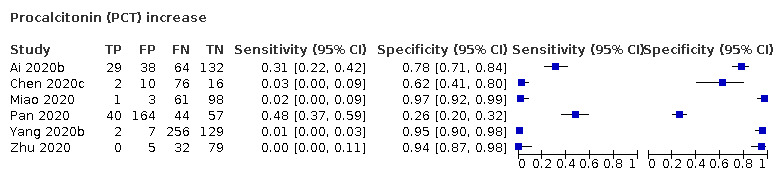
Procalcitonin (PCT) increase
40. Test.

eGFR
41. Test.

Proteinuria
42. Test.
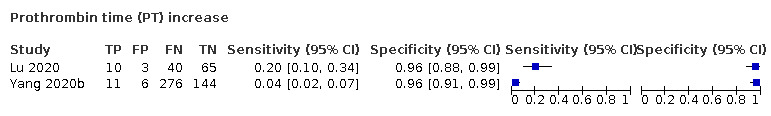
Prothrombin time (PT) increase
43. Test.
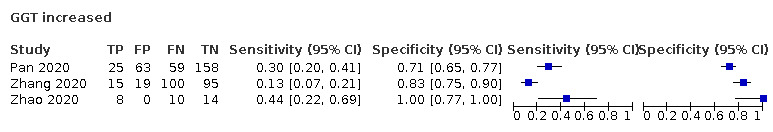
GGT increased
44. Test.
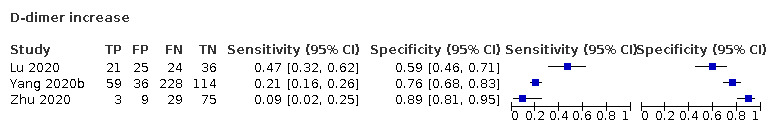
D‐dimer increase
45. Test.

IL‐2
46. Test.

IL‐4
47. Test.
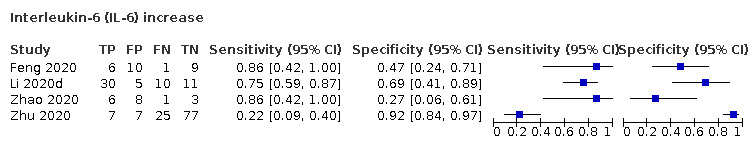
Interleukin‐6 (IL‐6) increase
48. Test.

IL‐8
49. Test.

IL‐10
50. Test.

TNF alpha
51. Test.
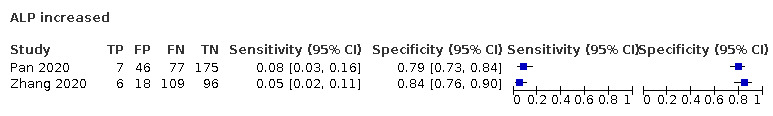
ALP increased
52. Test.

pro‐BNP
53. Test.

Hematuria
54. Test.
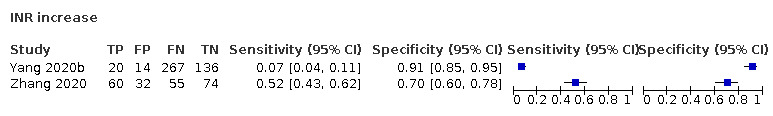
INR increase
55. Test.
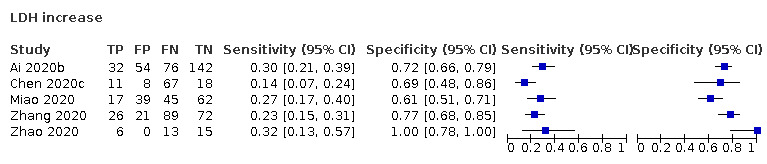
LDH increase
56. Test.

Mean corpuscular volume increase
57. Test.

Mean corpuscular volume decrease
58. Test.

Erythrocyte mean corpuscular hemoglobin increase
59. Test.

Erythrocyte mean corpuscular hemoglobin decrease
60. Test.

Erythrocytemean corpuscular hemoglobin concentrate increase
61. Test.

Erythrocytemean corpuscular hemoglobin concentrate decrease
62. Test.

Mean Platelet Volume
63. Test.

Direct bilirubin
64. Test.

unconjugated bilirubin
65. Test.

Total protein
66. Test.

Total bile acid
67. Test.

Troponin I
Characteristics of studies
Characteristics of included studies [ordered by study ID]
Ai 2020b.
| Study characteristics | |||
| Patient Sampling | Study including patients suspected of having COVID‐19, all suspected patients are classified between COVID‐19 or not COVID‐19 (single gate). Inclusion until February 9, 2020 and follow‐up was until 20 March. Patients were hospitalized in a hospital in China (Xiangyang No.1 People's Hospital). | ||
| Patient characteristics and setting | Setting: hospital, not specified which department
Site: Xiangyang, Hubei province
Country: China
Symptoms and severity: not reported
Demographics: cases: 49% male, age: mean 50.3 years (SD 17.4). non‐cases: 44% male, age: mean 38.8 years (SD 20.1) ‐ both children and adults Exposure history: cases: 75.9% had contact history. Non‐cases: 41.5% had contact history Time since onset of symptoms: not reported |
||
| Index tests | Routine laboratory tests (Table 4) Blood routine examination results were before hospitalization, first enzyme level test results after hospitalization of these 2 groups; person doing the testing not stated. Hospital lab technicians processed samples. Thresholds for positivity or negativity were not reported but we assumed that the same thresholds were used as in Ai 2020b, which was a study on the same 102 participants with COVID‐19. |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR was used to confirm cases. For some cases, RT‐PCR was repeated 5 times before a positive test was confirmed. Sample not reported. Hence target condition was SARS‐CoV‐2 infection. |
||
| Flow and timing | All participants received the RT‐PCR to confirm diagnosis. It is not clear what the time interval between index and reference text is. Missing data for cases: lymphocytes + 1 sample, PCT: 15 missing, ESR: 9 missing. Missing data for controls: ALT: 1 missing, AST: 4 more | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Chen 2020c.
| Study characteristics | |||
| Patient Sampling | Patients suspected of having SARS‐CoV‐2 pneumonia and hospitalized at Chongqing Three Gorges Central Hospital from 26 to 31 January 2020 were included in our study. Suspected = (1) contact with Wuhan or surrounding areas of Wuhan or confirmed patient within 14 days from the onset of the disease; (2) with symptoms of fever or respiratory; (3) with imaging features of COVID‐19 |
||
| Patient characteristics and setting | Setting: hospital, not specified which department
Site: Chongqing Three Gorges Central Hospital
Country: China
Symptoms and severity: cases: 82.1% and 76.9%, respectively of the participants had fever and cough. 10.3% had chest pains and 7.7% had diarrhoea. All the participants had clinical symptoms, such as sputum production, fatigue, shortness of breath, headache, arthralgia and vomiting. Controls: 53.8% and 46.2% had fever and cough respectively
Demographics: 78 COVID‐19 patients and 26 controls. cases median age 45 (range 15‐79) and controls was 61 years; 50% males in both cases and controls Exposure history: 83.3% of COVID‐19 patients admitted exposure to Wuhan (controls 26.9%), among whom 48 participants resided in Wuhan, 3 participants had travelled to Wuhan, and 14 participants had contact with people in Wuhan before the onset of the disease within 14 days Time since onset of symptoms: not reported |
||
| Index tests | Routine laboratory tests (Table 4) Data collection tables were based on electronic medical records. Person doing the testing, sample, timing of testing not stated |
||
| Target condition and reference standard(s) | 2 consecutive positive nucleic acid test result of high‐throughput sequencing or real‐time RT‐PCR assay; upper respiratory throat swab samples 2‐6 times Target condition is SARS‐CoV‐2 infection |
||
| Flow and timing | The time interval between index and reference test is not clear but likely short as all participants were already hospitalized. All participants received the same reference standard. No missed data noticed. | ||
| Comparative | |||
| Notes | Funding: Fundamental Research Funds for the Central Universities (Project No.2020CDJYGRH‐YJ03 to Xianxiang Zhang); Natural Science Foundation of China (Grants No. 81972416, 81672554 and 81472417 to BH) | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Feng 2020.
| Study characteristics | |||
| Patient Sampling | Study including adult patients with suspected infection, all suspected patients are classified between COVID‐19 or not COVID‐19 (single gate). Between 14 January and 9 February All patients admitted to the fever clinic of emergency department of the First Medical Center, Chinese People's Liberation Army General Hospital (PLAGH) in Beijing with the epidemiological history of exposure to COVID‐19 according to WHO interim guidance were enrolled in this study. |
||
| Patient characteristics and setting | Setting: fever clinic of emergency department
Site: First Medical Center, Chinese People's Liberation Army General Hospital (PLAGH) in Beijing
Country: China
Symptoms and severity: all 7 cases had moderate disease as defined by the 6th‐Guidelines‐CNHHC
Demographics: 7 cases and 19 controls. Median age: 39 years in cases and 40 years for controls. Cases were 71.4% male and controls were 63.2% male (adults only) Exposure history: history of sojourn or residence: 57.1% for cases and 21.1% for controls. History of contact with confirmed patient: cases: 28.6% and controls 5.3%. History of contact with person who had fever or respiratory symptoms: cases 14.3% and controls 57.9% Time since onset of symptoms: not reported. Days from illness onset to first admission: median 5 days for cases and 1 day for controls |
||
| Index tests | Routine laboratory tests (Table 4) Lymphocyte count (LYMPH#), CRP and IL‐6 were evaluated on admission. Lymphopenia (< 1.0 × 109/L) was 1 of the 3 diagnostic criteria for S‐COVID‐19‐P according to the 6th‐Guidelines‐CNHHC. Elevated CRP (> 0.8 mg/L) and elevated IL‐6 (> 5.9 pg/mL) were both important infection‐related biomarkers |
||
| Target condition and reference standard(s) | Target condition: S‐COVID‐19‐P COVID‐19 infection was confirmed by real‐time RT‐PCR using the same protocol described previously (Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395(10223): 497‐506.). RT‐PCR detection reagents were provided by the four institutions. Not clear how other criteria were included in the diagnosis. |
||
| Flow and timing | Nothing reported about flow and timing. | ||
| Comparative | |||
| Notes | Funding: the present study was supported by grants from the PLA Science and Technology Project (14CXZ005, AWS15J004, 16BJZ19), National Key R&D Program of China 2019YFF0302300), Construction Project of Key Disciplines in the 13th Five‐Year Plan of the PLA (Traumatic Surgery in the Battlefield, 2019‐126, 2019‐513), Beijing Science and Technology New Star Project (XX2018019/Z181100006218028), the PLA General Hospital Science and technology Project (2019XXJSYX20, 2018XXFC‐20, ZH19016). | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Ferrari 2020.
| Study characteristics | |||
| Patient Sampling | Study including suspected patients, all suspected patients are classified between COVID‐19 or not COVID‐19 (single‐gate, case‐control design). Between 20 February and 20 March 2020 The participants were randomly chosen (alphabetical order) to have a similar number of individuals in the positive (105) and negative (102) rRT‐PCR test groups |
||
| Patient characteristics and setting | Setting: fever clinic of emergency department
Site: San Raffaele Hospital (Milan, Italy) emergency room
Country: Italy
Symptoms and severity: currently Italy has strict directives suggesting an rRT‐PCR test only if patients show ≥ 3 ARS symptoms, review authors assumed that most, if not all, of the individuals enrolled in this study went to the hospital emergency room with fever, cough and fatigue.
Demographics: median age for cases is 61.8 and for controls is 59.2 cases: 70.5% male and controls 52% male (adults only) Exposure history: not stated Time since onset of symptoms: not reported |
||
| Index tests | Routine laboratory tests (Table 4) Blood samples were collected on the same day of the rRT‐PCR test. CRP, AST, ALT, GGT, ALP and LDH were measured on a Roche Cobas 8000 device (Roche Diagnostic, Basel, Switzerland) using either a spectrophotometric assay (AST, ALT and LDH), a colorimetric assay (ALP and GGT) or an immunoturbidimetric assay (CRP). WBC, platelets and the leukocyte formula were measured on Sysmex XE 2100 (Sysmex, Japan). |
||
| Target condition and reference standard(s) | Target condition: SARS‐CoV‐2 infection Reference standard: rRT‐PCR was performed on a Roche Cobas Z480 thermocycler (Roche Diagnostic, Basel, Switzerland) using the Roche‐provided Tib‐Molbiol’s 2019‐nCoV Real‐Time Reverse Transcription PCR Kit. RNA purification was performed using the Roche Magna pure system. Number of samples tested per participant not reported; blinding not reported; no other criteria used. |
||
| Flow and timing | Blood samples were collected on the same day of the rRT‐PCR test; none missing | ||
| Comparative | |||
| Notes | We could not extract 2x2 table because study only reported means and SDs. Study authors contacted; they sent data for 2 tests | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Hsih 2020.
| Study characteristics | |||
| Patient Sampling | Patients admitted to China Medical University Hospital meeting the screening criteria of COVID‐19 reported by Taiwan CDC (travel history to China and presented fever or any respiratory symptoms within 14 days). All eligible patients were included. | ||
| Patient characteristics and setting | Setting: hospital, emergency room Country: Taiwan Symptoms and severity: most common symptoms were fever, nonproductive cough, rhinorrhoea, sore throat, productive cough and dyspnea Demographics: mean age 34 (range 3‐68), female 60% Exposure history: travel to China, contact with people travelling to China, or contact with COVID‐19 patients Time since onset of symptoms: not reported | ||
| Index tests | Index tests (threshold):
For all tests
|
||
| Target condition and reference standard(s) | RT‐PCR (conducted multiple times in each participant; at least upon admission and 24h after admission, and for some participants even every few days). Target condition was SARS‐CoV‐2 infection. Sample: naso‐oropharyngeal specimen, sputum Threshold: not reported |
||
| Flow and timing | Time interval between index test and reference standard: not clearly reported Verification: all participants received the same reference standard Missing data: no missing data or uninterpretable results | ||
| Comparative | |||
| Notes | Funding: this study was supported by a grant, CMUH DMR‐108‐189, from China Medical University Hospital, Taichung, Taiwan. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Li 2020d.
| Study characteristics | |||
| Patient Sampling | Children with confirmed 2019‐nCoV pneumonia (cases) admitted between 24 January and 22 February 2020 and children with RSV pneumonia (controls) admitted between 10 December 2019 and 22 February 2020 in Wuhan Children’s hospital and patients who underwent the detection of peripheral blood lymphocyte subsets were included in the study. Previously healthy children were included in the study, and children receiving chemotherapy, treatment of glucocorticoids or immunosuppressant before the diagnosis of the pneumonia were not included in the study as their immune response to viral infections might be different. | ||
| Patient characteristics and setting | Setting: Wuhan Children's hospital
Site: Wuhan
Country: China
Symptoms and severity: of all children, 3 participants developed severe pneumonia, 1 (2.5%) in cases and 2 (12.5%) in control
Demographics: cases 57% male; controls 62.5% male Age: cases: mean age 5.09 years and controls 1.36 years Exposure history: not stated Time since onset of symptoms: not stated Any other info: |
||
| Index tests | Whole blood Demographic data, clinical manifestations, laboratory findings (including CRP, PCT, Scr, ALT, lymphocyte subsets, cytokines (IL‐2, IL‐4, IL‐6, IL‐10,TNF‐α, IFN‐γ) ) and treatments were recorded from the medical records Cytokines may not be standard in all places, hence unclear concerns regarding applicability. |
||
| Target condition and reference standard(s) | Real‐time RT‐PCR; not reported how often sampled; not reported about blinding. Also, 2019‐nCoV infection was confirmed with RT‐PCR, but unclear how 2019‐nCoV was defined in the first place, before confirming |
||
| Flow and timing | Cases and controls were selected based on detection of peripheral blood lymphocyte subsets. Time interval unclear, but likely before RT‐PCR test | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Unclear | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Li 2020e.
| Study characteristics | |||
| Patient Sampling | Pregnant women who were admitted into the Hubei Provincial Maternal and Child Health Center, during 24 January–29 February 2020. The study also included suspected patients with typical chest CT imaging but negative in RT‐PCR tests. Eleven pregnant women who were tested positive for SARS‐CoV‐2 were classified as laboratory‐confirmed case group, and eighteen with typical chest CT imaging but tested negative in RT‐PCR tests as suspected case group. The control group of pregnant women without pneumonia during hospital stay were randomly selected from the medical records by an investigator (MP), who was not involved in statistical analysis. Only those aged 25‐35 years were selected to match the age range of cases. 121 women admitted during 24 January–11 February 2019 (control 2019 group) |
||
| Patient characteristics and setting | Pregnant women (and therefore high concern regarding applicability) Setting: admission to hospital Site: Hubei Provincial Maternal and Child Health Center Country: China Symptoms and severity: 4 of the cases were admitted with fever for investigation and 8 developed fever after childbirth. None presented other respiratory symptoms on admission nor during hospital stay. 2 of the patients with suspected COVID‐19 pneumonia reported cough, sore throat, dyspnea, diarrhea and vomiting. Demographics: pregnant women Age: confirmed cases: 30.9 years, suspected cases 29.8 years. Control 1:30.1 years and control 2: 29.3 years Exposure history: none of confirmed COVID‐19 patients reported an exposure history. Retrospective analysis of medical records of pregnant women with COVID‐19 pneumonia and pregnant women without COVID‐19 pneumonia. Time since onset of symptoms: not reported |
||
| Index tests | Whole blood. See Table 4 Clinical characteristics, laboratory test results, maternal and neonatal outcomes were collected from medical records and reviewed independently by 2 investigators Index tests were: WBC, lymphocytes, neutrophils, CRP, eosinophils, ALT, AST |
||
| Target condition and reference standard(s) | Cases: RT‐PCR and chest CT Controls: 121 women admitted during 24 January–11 February 2019 (control 2019 group) Target condition: COVID‐19 |
||
| Flow and timing | Blood test results were also retrieved from medical records. 2 case groups underwent blood tests every three days but 2 control groups only taken once | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Li 2020f.
| Study characteristics | |||
| Patient Sampling | Data of this retrospective case‐negative control study were collected from 105 patients first visiting the Fever Clinic of Wuhan Union Hospital from 3‐7 February 2020. | ||
| Patient characteristics and setting | Setting: hospital, emergency department, outpatient setting/fever clinic/COVID triage centre
Site: Wuhan union hospital, Wuhan
Country: China
Symptoms and severity: 59.6% of cases had fever, 38.5% had respiratory symptoms and 1.9% had weakness compared to controls where 52.8% had fever, 47.2% had respiratory symptoms and 0% had weakness.
Demographics: cases 50% male: controls: 56.6% male Age: cases average years 57 years; controls average age 51 years (adults) Exposure history: not stated Time since onset of symptoms: not stated Any other info: |
||
| Index tests | People conducting the test, sample tested were not stated. Tests were conducted at first medical visit. Leukocyte (x 109/L; ref 3.5‐9.5) normal or increased (≤ 3.5); neutrophil (x 109/L; ref 1.8‐6.3) increased; lymphocyte (x 109/L; ref 1.1‐3.2) decreased (< 1.1); monocytes (x 109/L; ref 0.1‐0.6) increased; eosinophil (x 109/L; ref 0.02‐0.52) decreased; hCRP (mg/L; ref < 4) increased. Whole blood (otherwise WBC cannot be assessed) |
||
| Target condition and reference standard(s) | Nasopharyngeal swab specimens of all participants were subject to real time RT‐PCR tests through amplifying ORF1ab gene and N gene of SARS‐CoV‐2 (BioGerm, Shanghai, China) | ||
| Flow and timing | All participants received the same reference test. Index tests were performed at participant's first medical visit. No missing data | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Li 2020g.
| Study characteristics | |||
| Patient Sampling | No inclusion criteria reported, other than patients with suspected COVID‐19 viral pneumonia admitted to the infection department, emergency department, and Jinshan Branch of hospital from 22 January‐17 February 2020. Design was unclear, but study includes COVID‐19 patients and patients with other viral infections. |
||
| Patient characteristics and setting | Setting: hospital, emergency department and infection department
Country: China
Symptoms and severity: unclear
Demographics: 21 male, 19 female Age: adults; median age in diseased 46.5 (IQR 36.5‐64.3), median age in non‐diseased 37.5 (IQR 29.8‐63.2) Exposure history: not reported Time since onset of symptoms: 2 (1.4) days of onset |
||
| Index tests | |||
| Target condition and reference standard(s) | The COVID‐19 group is a confirmed case, that is, the throat swab and blood 2019‐nCOV nucleic acid test are positive. The non‐COVID‐19 group is a suspected case of COVID‐19, tested negative by 2 times of pharyngeal swabs and blood 2019‐nCOV nucleic acid, other viruses (influenza A/B virus or Coxsackie virus or herpes simplex virus or RSV, etc.) positive test, or imaging findings consistent with viral pneumonia | ||
| Flow and timing | No information about flow and timing | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Liang 2020.
| Study characteristics | |||
| Patient Sampling | Based on epidemiological history, clinical and radiological manifestations, cases with possible or probable COVID‐19 were sent for panel discussion. Paediatric patients were not included. | ||
| Patient characteristics and setting | Setting: fever clinic, pre‐screened
Site: Peking University Third Hospital from 21 January‐15 February 2020
Country: China
Symptoms and severity: on presentation, most patients (85.7%) had fever with a mean body temperature of 37.8. Cough (42.9%), expectoration (33.3%), fatigue (57.1%), headache or dizziness (38.1%) were common symptoms. Other symptoms included shortness of breath, myalgia or arthralgia, sore throat, nasal symptoms and diarrhoea.
Demographics: male/female Age: 24‐85 years (median 42.0, range 34.5‐66) Exposure history: imported cases from Wuhan City or Hubei Province 6 (28.6%); known contact with individuals from Wuhan or Hubei 1 (4.8%); known contact with cases of confirmed COVID‐19 5 (23.8%); family aggregation onset 7 (33.3%) Time since onset of symptoms: between 2 and 10 days |
||
| Index tests | Not much information reported. For all index tests, see Table 4 |
||
| Target condition and reference standard(s) | RT‐PCR. Laboratory testing of 2019‐nCoV in throat swabs was performed by both Beijing Centers for Disease Control and Prevention (CDC) and Haidian District CDC. 2019‐nCoV infection was target condition | ||
| Flow and timing | |||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Liu 2020.
| Study characteristics | |||
| Patient Sampling | No sampling method reported, other than these were patients with COVID‐19 in the Renmin Hospital in Wuhan from 31 January‐26 February 2020 | ||
| Patient characteristics and setting | Setting: hospital Site: Renmin Hopsital of Wuhan University Country: China Symptoms and severity: Demographics: cases: 55 male and 57 female; controls: 23 male and 22 female Age: adults; mean age cases subgroups 62‐63 and cases 62 years Exposure history: not stated Time since onset of symptoms: not stated Any other info: |
||
| Index tests | Urine samples, collected from catheters. All collected specimens were tested within 2 h; no blinding reported; no timing reported, no thresholds reported | ||
| Target condition and reference standard(s) | Diagnosis and Treatment Program of New Coronavirus Pneumonia (sixth trial version); no further information on reference standard | ||
| Flow and timing | No information reported | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Unclear | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Lu 2020.
| Study characteristics | |||
| Patient Sampling | Hospitalized patients with confirmed or suspected COVID‐19 and at least one post‐admission evaluation | ||
| Patient characteristics and setting | Setting: hospital
Site: Wuhan Hankou Hospital
Country: China
Symptoms and severity: the most common signs and symptoms at onset of illness were fever (323 (76.5%)), cough (258
(60.4%)), and fatigue (148 (33.4%))
Demographics: median age was 55 years (IQR 39‐66) and 254 (44.0%) were men 6 days from illness onset to admission (IQR 4‐9) |
||
| Index tests | Whole blood Index tests: WBC count, neutrophil count, lymphocyte count, prothrombin time, D‐dimer, ALB, ALT, total BIL, Scr, CRP Blinding not reported |
||
| Target condition and reference standard(s) | Diagnosis was only based on SARS‐CoV‐2 RT‐PCR (no further information provided) | ||
| Flow and timing | Only 199/577 received RT‐PCR Time interval was unclear | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Mardani 2020.
| Study characteristics | |||
| Patient Sampling | Outpatients who presented to Behpooyan ClinicMedical center in Tehran (Iran) from 22 February‐14 March 2020. with suspected COVID‐19 having initial respiratory signs (including sore throat without shortness of breath), fever, cough, muscle ache, and headache were included | ||
| Patient characteristics and setting | Setting: hospital Site: Behpooyan Clinic Medical center in Tehran Country: Iran Symptoms and severity: outpatients with suspected COVID‐19 having initial respiratory signs (including sore throat without shortness of breath), fever, cough, muscle ache, and headache were included Demographics: 200 cases with the mean age of 41.3, SD 14.6 (range: 19‐78) years were studied (0.53% male). 40.2% of cases were in the 30‐49 years age range. Exposure history and time since onset of symptoms: not reported | ||
| Index tests | Only 2x2 table for CRP. Blood samples were collected from each participant. Whole blood | ||
| Target condition and reference standard(s) | RT‐PCR for COVID‐19 using pharyngeal swab samples; no information on blinding | ||
| Flow and timing | Pharyngeal swab was collected on presentation, unclear when blood samples were collected | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Miao 2020.
| Study characteristics | |||
| Patient Sampling | 163 consecutive adult patients with suspected COVID‐19 from three tertiary hospitals in two provinces outside Hubei province | ||
| Patient characteristics and setting | Setting: tertiary hospitals Site: 2 provinces outside Hubei province; fever emergency clinics at Shanghai General Hospital, High‐tech hospital (First Affiliated Hospital of Nanchang University) and People's hospital of Yinchun City from 12 January‐13 February 2020 Country: China Symptoms and severity: suspected of COVID‐19 visiting fever emergency clinics; the most common symptoms on admission were fever (49 (79.0%)), dry cough (37 (59.7%)), fatigue or myalgia (15 (24.2%)) Demographics: 62 cases confirmed and 102 cases unconfirmed. Mean age confirmed group: 43.8 (SD 13.9; range 19‐77); mean age unconfirmed group: 41.3 (SD 14.7; range 19‐81); confirmed group 32 (51.6%) men and non confirmed group 68 (67.3%) men Time since onset of symptoms was 7.0 (3.5‐9.0) days (confirmed group) and 6.0 (4.0‐9.0) days (unconfirmed group). Compared with participants in unconfirmed group, participants in confirmed group had significantly higher proportion of Wuhan residence history, having visited Wuhan, clustering diseases and dry cough |
||
| Index tests | WBC count, PCT, ALT, LDH, creatinine kinase, troponin I. Table 4 | ||
| Target condition and reference standard(s) | RT‐PCR. sample: nasopharyngeal swabs or sputum specimens; the confirmed group was defined as a positive result of at least 1 RT‐PCR test for SARS‐CoV‐2. The unconfirmed group was defined as all results of RT‐PCR tests were negative | ||
| Flow and timing | Time interval not reported; all participants received the same reference standard; no missing data or uninterpretable results | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Pan 2020.
| Study characteristics | |||
| Patient Sampling | COVID‐19 cases: laboratory‐confirmed SARS‐CoV‐2 infection by real‐time RT‐PCR
CAP cases:
Exclusion criteria:
Healthy controls: people who made the physical check‐up in our hospital |
||
| Patient characteristics and setting | Setting: hospital Site: Zhongnan Hospital of Wuhan University Country: China Symptoms and severity: patients with COVID‐19 vs patients with CAP, COVID‐19 patients Demographics: median age 58 (48‐70) M/F: 51/33, CAP patients: median age 71 (56‐86), M/F: 142/79, healthy controls: median age 33 (24‐39) M/F: 68/52 Time since onset of symptoms and exposure history not reported |
||
| Index tests | Hb, lymphocytes, and monocytes, were analyzed. Routine serum biochemical parameters, including ALT, AST, AST/ALT ratio, total BIL, direct BIL, unconjugated BIL, total protein (TP), ALB, GLB, GGT, ALP, and total bile acid (TBA) were measured. | ||
| Target condition and reference standard(s) | Cases: RT‐PCR once, no further specification Hospital controls without pneumonia, patients with CAP: not reported how confirmed |
||
| Flow and timing | No information | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Rentsch 2020.
| Study characteristics | |||
| Patient Sampling | Those tested for COVID‐19 in participants from the Veterans Affairs national Corporate Data Warehouse on members of the VA Birth Cohort from 8 February‐30 March 2020 | ||
| Patient characteristics and setting | Setting: all Country: USA Median age: 65.7 (IQR 60.5‐70.7) 3417 (90.2) male; 372 (9.8) female |
||
| Index tests | See Table 4 Whole blood |
||
| Target condition and reference standard(s) | SARS‐COV‐2 assays. COVID‐19 tests conducted in the VA using text searching of laboratory results 141 containing terms consistent with SARS‐CoV‐2 or COVID‐19. If a participant had more than one test and all were negative we selected first negative, otherwise we used date of first positive. Nearly all tests utilized nasopharyngeal swabs, 1% were from other sources | ||
| Flow and timing | All participants received same ref standard. Missings are participants for whom test are pending (n = 93) or inconclusive (n = 33). Laboratory findings closest to baseline within a year prior or up to 1 week after baseline were used. Baseline was defined as the date of specimen collection for COVID‐19 test unless testing was occurred during hospitalization, in which case it was date of admission. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | No | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | High risk | ||
Yang 2020b.
| Study characteristics | |||
| Patient Sampling | Inclusion criteria of the patients suspected of moderate type novel coronavirus pneumonia for this study are:
Exclusion criteria are:
In this study, the participants suspected of moderate type novel coronavirus pneumonia confirmed with positive nucleic acid tests were designated as the study group and the ones with negative findings as the control group. Duration 31 January‐11 February 2020 |
||
| Patient characteristics and setting | Setting: triaged for admission to the Southeast Hospital of Xiaogan Central Hospital from the fever clinics of Xiaogan Central Hospital, Xiaogan First People’s Hospital and Hubei Aerospace Hospital. From 31 January‐11 February 2020 Country: China Severity: none of the participants were severely or critically ill Demographics: in cases, 51% was male and in controls 48% was male; mean age was 49.2 years +/‐ 13.7 (95% CI 48‐50) Exposure status: more than half were exposed to travellers from Wuhan Time since onset of symptoms: mean 4.6 days from onset of symptoms (+/‐ 2.9); 0.22% died |
||
| Index tests | The data were retrieved from the outpatient and inpatient electronic medical record system (HealthOne, Shenzhen, China), nursing records, laboratory reports and chest CT scans. Laboratory findings: WBC, neutrophils, lymphocytes, Hb, platelets, CRP, PCT, ALT, AST Scr, urea, CK, CK‐MB, pro‐BNP, prothrombin time, INR, D‐Dimer Whole blood; thresholds not reported Some of the routine lab tests were part of the inclusion criteria: normal or decreased WBC count at the early stage, or decreased lymphocyte count |
||
| Target condition and reference standard(s) | Pharyngeal swabs of the suspected participants were collected by a specifically trained nurse and the specimens were delivered to the central lab. The tests were conducted with the novel coronavirus 2019‐nCoV nucleic acid test kit (Shanghai ZJ BioTech, Shanghai, China) using Applied BiosystemsTM 7500 Real‐Time PCR System (Thermo Fisher Scientific, USA) Positive finding of the novel coronavirus nucleic acid test is defined as positive results with both Open reading frame 1ab (ORF 1ab) and Nucleocapsid protein (N) for respiratory specimens examined with real‐time fluorescence PCR. Negative finding of the novel coronavirus nucleic acid test is defined as 2 consecutive tests for respiratory specimens collected with intervals of at least 1 day displaying negative results as examined with real‐time fluorescence PCR |
||
| Flow and timing | Not reported | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Yang 2020c.
| Study characteristics | |||
| Patient Sampling | A consecutive cohort of 73 COVID‐19 and 48 influenza pneumonia patients were retrospectively recruited from 5 independent institutions | ||
| Patient characteristics and setting | COVID: 73 consecutive patients confirmed with SARS‐Cov2, from 5 independent hospitals in 4 Chinese cities, mean age was 41.9, 41 men 32 women Non‐COVID: from 1 January 2015‐30 September 2019, a total of 205 consecutive patients confirmed with influenza pneumonia from Shantou and Meizhou city were recruited. Finally, 48 influenza pneumonia patients (mean age: 40.4 years, range: 0.1‐83 years) were enrolled as controls, including 30 men and 18 women; influenza A = 36, Influenza B = 12 | ||
| Index tests | Total WBC count, lymphocyte count, lymphocyte ratio, neutrophil count, neutrophil ratio and CRP level | ||
| Target condition and reference standard(s) | RT‐PCR for COVID. Influenza controls were confirmed with respiratory pathogen IgM antibody test | ||
| Flow and timing | Time interval, unclear; COVID patients, RT‐PCR and influenza (IgM antibody test), missing data not noticed | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Zhang 2020.
| Study characteristics | |||
| Patient Sampling | COVID‐19 cases: hospitalized patients from Zhongnan Hospital of Wuhan university. COVID‐19 was diagnosed based on criteria issued by the National Health Commission of China.
Controls: CAP hospitalized in Department of Respiratory and Critical Care Medicine between 22 January‐22 February 2019. 5 control patients with chronic Hepatitis B or cirrhosis were excluded |
||
| Patient characteristics and setting | Setting: infectious diseases department hospital; controls in pulmonary and critical care departments Site: Department of Infectious Disease, Zhongnan Hospital of Wuhan University Country: China Demographics: 4 participants < 14 years of age; of the 115 participants in the COVID‐19 group, 49 (42.60%) were male and 66 (57.40%) were female. Mean age at diagnosis was 49.52 ± 17.06 years (IQR, 35‐62; range, 20‐86 years). The CAP group included 55 (48.25%) male participants and 59 (51.75%) female participants, mean age 61.11 ± 18.84 years (IQR, 47‐76; range, 18‐89 years). Severity: 2 patients with chronic Hepatitis B were excluded, and 115 patients were included to COVID‐19 group; from the controls group, four patients with Hepatitis B or cirrhosis were excluded. |
||
| Index tests | Routine laboratory tests: ALT, AST, total BIL, ALP, GGT, LDH, ALB, GLB, INR, CRP | ||
| Target condition and reference standard(s) | COVID‐19 was diagnosed based on criteria issued by the National Health Commission of China; includes RT‐PCR once, Clinical signs and symptoms, chest CT Controls: CAP |
||
| Flow and timing | Not reported | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Unclear | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Zhao 2020.
| Study characteristics | |||
| Patient Sampling | Study recruited 19 COVID‐19 patients and 15 non‐COVID‐19 patients; no further information about selection criteria. Unclear if study was a 2‐gate design or a single‐gate design, but the way the methods and results are described, we assumed a single‐gate design. |
||
| Patient characteristics and setting | 19 COVID‐19 patients and 15 non‐COVID‐19 patients from the Second Affiliated Hospital of Anhui Medical University and Suzhou Municipal Hospital in Anhui province, China were included in this study. The mean age was 48 (IQR 27~56) and 35 (IQR 27~46) in COVID‐19 and non‐COVID‐19 patients, respectively. 8 (42.11%) were female in COVID‐19 patients, and 9 (60%) in non‐COVID‐19 patients. The median duration from exposure to onset is 8 (IQR 6~11) and 5 (IQR 4~11) days in COVID‐19 and non‐COVID‐19 patients, respectively. All participants had a history of exposure to confirmed case of 2019‐nCoV or travel to Hubei before illness | ||
| Index tests | Index tests done: WBC and lymphocyte count, neutrophil count, AST; ALT; LDH; GGT; α‐hydroxybutyric dehydrogenase; CK; CRP and IL‐6. Tests were done on admission (4‐5 days from onset), person doing the testing is not stated. As WBC was assessed, sample must have been whole blood |
||
| Target condition and reference standard(s) | COVID‐19 cases were confirmed to be infected with or without 2019‐nCoV by real‐time RT‐PCR. COVID‐19 was defined to be 2019‐nCoV negative by PCR detection. For non‐COVID‐19 confirmation, we collected a throat swab or sputum sampling every other day. The patient was confirmed as non‐COVID‐19 if 3 consecutive real‐time PCR tests were negative during first 7 days of admission | ||
| Flow and timing | All participants received the same reference test. Test interval is 4‐5 days. No missing data. Index tests were performed at admission. it is not clear when the reference test was done. |
||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Zhu 2020.
| Study characteristics | |||
| Patient Sampling | The inclusion criteria were
The exclusion criteria were
Inclusion period between 24 January 2020 and 20 February 2020 |
||
| Patient characteristics and setting | Setting: hospital, emergency department and infectious diseases satellite hospital Site: The First Affiliated Hospital of University of Science and Technology of China, Hefei Country: China Symptoms and severity: there were 6 (19%) smokers among diagnosed participants and 13 (15%) among negative cases. 7 (22%) diagnosed and 15 (18%) negative cases had hypertension. There were no other commonly found comorbidities in either group. Demographics: median age 40 (IQR 27‐53); 46% male Exposure history: there was no specific exposure history common to all participants with suspected disease: 8 (25%) diagnosed participants had visited Wuhan in the previous 2 weeks and 12 (38%) had been exposed to participants with infection in the previous 2 weeks. In negative cases, these numbers were 7 (20%) and 8 (24%), respectively. None of the participants had a history of exposure to the seafood market in Wuhan. Time since onset of symptoms: median 5 days (IQR 2‐7 days) |
||
| Index tests | Clinical and laboratory data on admission were obtained from detailed medical records, collected in a standardized case report form by 2 experienced emergency doctors. Laboratory tests included a complete blood count, serum biochemistry, IL‐6 test, CK test, LDH test, and tests for the identification of other respiratory pathogens Timing of tests not reported; blinding not reported |
||
| Target condition and reference standard(s) | A nucleic acid amplification test was performed on swab specimens from participants with suspected disease at admission. Participants with a positive diagnosis were admitted to the hospital, while participants with a negative initial result were kept in quarantine and underwent a second nucleic acid test after 24 h; of these, participants with a second negative result on the nucleic acid test were considered to not have an infection and were discharged from the hospital once they tested negative for SARS‐CoV‐2 antigens on 2 consecutive tests. | ||
| Flow and timing | Exact timing of lab tests was not reported. Quote: "Not all patients presented at the same infection stage and some data were missing; thus, data could not be integrated." |
||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
ALB: albumin; ALP: alkaline phosphatase; ALT: alanine aminotransferase; ARS: acute respiratory syndrome; AST: aspartate aminotransferase; BIL: bilirubin; BNP: B‐type natriuretic peptide; CAP: community‐acquired pneumonia; CI: confidence interval; CK: creatine kinase; CK‐MB: creatine kinase (blood); CRP: C‐reactive protein; CT: computed tomography; ESR: erythrocyte sedimentation rate; GGT: γ‐glutamyl transpeptidase; GLB: globulin; Hb: haemoglobin; ICU: intensive care unit; IFN‐y: interferon gamma; IL: interleukin; INR: international normalized ratio; IQR: interquartile range; LDH: lactate hydrogenase; PCR: polymerase chain reaction; PCT: procalcitonin; RNA: ribonucleic acid; (r)RT‐PCR: (rapid) reverse‐transcriptase polymerase chain reaction; RSV: respiratory syncytial virus; Scr: serum creatinine; SD: standard deviation; WBC: white blood cell; WHO: World Health Organization;
Characteristics of excluded studies [ordered by study ID]
| Study | Reason for exclusion |
|---|---|
| Ai 2020a | Insufficient data for 2x2 table |
| Chen 2020a | Insufficient data for 2x2 table |
| Chen 2020b | Insufficient data for 2x2 table + target condition not clear |
| Cheng 2020 | Insufficient data for 2x2 table |
| Giamarellos 2020 | Insufficient data for 2x2 table |
| Han 2020 | Insufficient data for 2x2 table |
| Kurstjens 2020 | Insufficient data for 2x2 table |
| Li 2020a | Insufficient data for 2x2 table + Hospital discharge versus no discharge |
| Li 2020b | Insufficient data for 2x2 table + Mechanical ventilation versus no mechanical ventilation |
| Li 2020c | Insufficient data for 2x2 table + RNA positive versus RNA negative |
| Ling 2020 | Insufficient data for 2x2 table |
| Meng 2020 | Insufficient data for 2x2 table |
| Peng 2020 | Insufficient data for 2x2 table |
| Peng 2020a | Insufficient data for 2x2 table |
| Shi 2020 | Insufficient data for 2x2 table |
| Song 2020 | Insufficient data for 2x2 table |
| Spiezia 2020 | Insufficient data for 2x2 table |
| Sun 2020 | Insufficient data for 2x2 table |
| Tang 2020 | Insufficient data for 2x2 table |
| Wang 2020 | Insufficient data for 2x2 table + diagnostic prediction model |
| Wu 2020 | Insufficient data for 2x2 table + diagnostic artificial intelligence model |
| Xu 2020 | Insufficient data for 2x2 table |
| Yang 2020a | Insufficient data for 2x2 table |
| Yin 2020 | Insufficient data for 2x2 table |
Differences between protocol and review
We deviated from our protocol on some occasions. We intended to include studies that recruited only COVID‐19 cases, to estimate sensitivity or those restricted to people without COVID‐19, to estimate specificity (Deeks 2020a). We decided to deviate from this rule as the added value of such studies for our review is questionable.
We planned to investigate test accuracy, either by stratified analysis or meta‐regression, according to a specific measurement or biomarker, days of symptoms, severity of symptoms, reference standard, sample type, study design, and setting. We decided not to do these analyses in the first version of this review because of the lack of primary studies per subgroup.
We did not specify some details about the analyses in our protocol. We chose to present sensitivity and median interquartile range values for cut‐offs of specificity.
Contributions of authors
Inge Stegeman: Study selection, data‐extraction and quality assessment, first draft of the review and subsequent revisions;
Eleanor A Ochodo: Study selection, data‐extraction and quality assessment, multiple revisions of the review;
Fatuma Guleid: Study selection, data‐extraction and quality assessment, multiple revisions of the review;
Gea A. Holtman: Study selection, data‐extraction and quality assessment, multiple revisions of the review;
Bada Yang: Study selection, data‐extraction and quality assessment, multiple revisions of the review;
Jane Cunningham contributed clinical, methodological and/or technical expertise to drafting the protocol; contributed to multiple revisions of the review;
Clare Davenport contributed clinical, methodological and/or technical expertise to drafting the protocol; contributed to multiple revisions of the review;
Jonathan J Deeks: contributed clinical, methodological and/or technical expertise to drafting the protocol; contributed to multiple revisions of the review and co‐ordinated all contributions to all Cochrane Rapid DTA reviews;
Jacqueline Dinnes contributed clinical, methodological and/or technical expertise to drafting the protocol; did the initial screening titles and abstracts for all reviews; contributed to multiple revisions of the review;
Sabine Dittrich contributed clinical, methodological and/or technical expertise to drafting the protocol; contributed to multiple revisions of the review;
Devy Emperador contributed clinical, methodological and/or technical expertise to drafting the protocol; contributed to multiple revisions of the review;
Lotty Hooft contributed clinical, methodological and/or technical expertise to drafting the protocol; contributed to multiple revisions of the review;
René Spijker contributed clinical, methodological and/or technical expertise to drafting the protocol; co‐ordinated and conducted the study retrieval en initial selection steps; contributed to multiple revisions of the review;
Yemisi Takwoingi contributed clinical, methodological and/or technical expertise to drafting the protocol; supervised the meta‐analyses; contributed to multiple revisions of the review;
Ann Van den Bruel contributed clinical, methodological and/or technical expertise to drafting the protocol; contributed to multiple revisions of the review;
Junfeng Wang translated articles from Chinese to English whenever necessary; retrieved articles in Chinese; extracted data from and assessed quality of Chinese language articles; contributed to revised versions of the review;
Miranda Langendam: Study selection, data‐extraction and quality assessment, multiple revisions of the review;
Jan Verbakel: Study selection, data‐extraction and quality assessment, meta‐analyses; multiple revisions of the review;
Mariska MG Leeflang contributed clinical, methodological and/or technical expertise to drafting the protocol; drafted the QUADAS‐2 criteria; co‐ordinated the review process; overall supervision; drafted all non‐automatic Tables; GRADE assessment; contributed to the first draft and subsequent revisions of the review.
Sources of support
Internal sources
Liverpool School of Tropical Medicine, UK
External sources
-
Foreign, Commonwealth and Development Office (FCDO), UK
Project number: 300342‐104
National Institute for Health Research (NIHR), UK
Declarations of interest
Inge Stegeman: has provided freelance consultancy for approved professional organizations and learned societies (physiotherapists, optometrists, opticians), and has no known conflicts of interest in relation to this review.
Eleanor A Ochodo: none known
Fatuma Guleid: none known.
Gea A. Holtman: none known.
Bada Yang: none known.
Jane Cunningham: none known.
Clare Davenport: none known.
Jonathan J Deeks: none known.
Jacqueline Dinnes: none known.
Sabine Dittrich: is employed by FIND. FIND has several clinical research projects to evaluate multiple new diagnostic tests against published Target Product Profiles that have been defined through consensus processes. These studies are for diagnostic products developed by private sector companies who provide access to know‐how, equipment/reagents, and contribute through unrestricted donations as per FIND policy and external SAC review.
Devy Emperador: is employed by FIND. FIND has several clinical research projects to evaluate multiple new diagnostic tests against published Target Product Profiles that have been defined through consensus processes. These studies are for diagnostic products developed by private sector companies who provide access to know‐how, equipment/reagents, and contribute through unrestricted donations as per FIND policy and external SAC review.
Lotty Hooft: none known.
René Spijker: the Dutch Cochrane Centre (DCC) has received grants for performing commissioned systematic reviews. In no situation, the commissioner had any influence on the results of the work.
Yemisi Takwoingi: none known.
Ann Van den Bruel: none known.
Junfeng Wang: has received consultancy fee from Biomind, an Artificial Intelligence (AI) company providing machine intelligence solutions in medical imaging. The consultancy service was about design of clinical studies, not related to this review. The company had no influence on the results of the work.
Miranda Langendam: none known.
Jan Verbakel: none known.
Mariska MG Leeflang: none known.
New
References
References to studies included in this review
Ai 2020b {published data only}
- Ai J, Gong J, Xing L, He R, Tian F, Wang J, et al. Analysis of factors associated early diagnosis in coronavirus disease 2019 (COVID-19). medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Chen 2020c {published data only}
- Chen X, Yang Y, Huang M, Liu L, Zhang X, Xu J, et al. Differences between COVID-19 and suspected then confirmed SARS-CoV-2-negative pneumonia: a retrospective study from a single center. Journal of Medical Virology 2020 Apr 1 [Epub ahead of print]. [DOI: 10.1002/jmv.25810] [DOI] [PubMed] [Google Scholar]
Feng 2020 {published data only}
- Feng C, Huang Z, Wang L, Chen X, Zhai Y, Zhu F, et al. A novel triage tool of artificial intelligence assisted diagnosis aid system for suspected COVID-19 pneumonia in fever clinics. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Ferrari 2020 {published data only}
- Ferrari D, Motta A, Strollo M, Banfi G, Locatelli M. Routine blood tests as a potential diagnostic tool for COVID-19. Clinical Chemistry and Laboratory Medicine 2020;58(7):1095-9. [DOI] [PubMed] [Google Scholar]
Hsih 2020 {published data only}
- Hsih WH, Cheng MY, Ho MW, Chou CH, Lin PC, Chi CY, et al. Featuring COVID-19 cases via screening symptomatic patients with epidemiologic link during flu season in a medical center of central Taiwan. Journal of Microbiology, Immunology and Infection 2020;53(3):459-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Li 2020d {published data only}
- Li H, Chen K, Liu M, Xu H, Xu Q. The profile of peripheral blood lymphocyte subsets and serum cytokines in children with 2019 novel coronavirus pneumonia. Journal of Infection 2020;8(1):115-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Li 2020e {published data only}
- Li N, Han L, Peng M, Lv Y, Ouyang Y, Liu K, et al. Maternal and neonatal outcomes of pregnant women with COVID-19 pneumonia: a case-control study. Clinical Infectious Diseases 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Li 2020f {published data only}
- Li Q, Ding X, Xia G, Geng Z, Chen F, Wang L, et al. A simple laboratory parameter facilitates early identification of COVID-19 patients. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Li 2020g {published data only}
- Li YX, Wu W, Yang T, Zhou W, Fu YM, Feng QM, et al. Characteristics of peripheral blood leukocyte differential counts in patients with COVID-19. Zhonghua Nei Ke Za Zhi 2020;59(0):E003. [PubMed] [Google Scholar]
Liang 2020 {published data only}
- Liang Y, Liang J, Zhou Q, Li X, Lin F, Deng Z, et al. Prevalence and clinical features of 2019 novel coronavirus disease (COVID-19) in the fever clinic of a teaching hospital in Beijing: a single-center, retrospective study. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Liu 2020 {published data only}
- Liu R, Ma Q, Han H, Su H, Liu F, Wu K, et al. The value of urine biochemical parameters in the prediction of the severity of coronavirus disease 2019. Clinical Chemistry and Laboratory Medicine 2020;58(7):1121-4. [DOI] [PubMed] [Google Scholar]
Lu 2020 {published data only}
- Lu J, Hu S, Fan R, Liu Z, Yin X, Wang Q, et al. ACP risk grade: a simple mortality index for patients with confirmed or suspected severe acute respiratory syndrome coronavirus 2 disease (COVID-19) during the early stage of outbreak in Wuhan, China. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Mardani 2020 {published data only}
- Mardani R, Ahmadi Vasmehjani A, Zali F, Gholami A, Mousavi Nasab SD, Kaghazian H, et al. Laboratory parameters in detection of COVID-19 patients with positive RT-PCR; a diagnostic accuracy study. Archives of Academic Emergency Medicine 2020;8(1):e43. [PMC free article] [PubMed] [Google Scholar]
Miao 2020 {published data only}
- Miao C, Zhuang J, Jin M, Xiong H, Huang P, Zhao Q, et al. A comparative multi-centre study on the clinical and imaging features of comfirmed and uncomfirmed patients with COVID-19. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Pan 2020 {published data only}
- Pan Y, Ye G, Zeng X, Liu G, Zeng X, Jiang X, et al. Can routine laboratory tests discriminate SARS-CoV-2 infected pneumonia from other causes of community acquired pneumonia? Clinical and Translational Medicine 2020;10(1):161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Rentsch 2020 {published data only}
- Rentsch CT, Kidwai-Khan F, Tate JP, Park LS, King JT, Skanderson M, et al. COVID-19 testing, hospital admission, and intensive care among 2,026,227 United States veterans aged 54-75 years. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.09.20059964] [DOI] [Google Scholar]
Yang 2020b {published data only}
- Yang L, Bai Y, Qiu Q, Wang T, Jiang L, Liu X, et al. Early-stage vigilance of novel coronavirus pneumonia. Available at ssrn.com/abstract=3544820 [Preprint] 2020. [DOI: ] [Google Scholar]
Yang 2020c {published data only}
- Yang Z, Lin D, Chen X, Qiu J, Li S, Huang R, et al. Distinguishing COVID-19 from influenza pneumonia in the early stage through CT imaging and clinical features. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhang 2020 {published data only}
- Zhang Y, Zheng L, Liu L, Zhao M, Xiao J, Zhao Q. Liver impairment in COVID-19 patients: a retrospective analysis of 115 cases from a single center in Wuhan city, China. Liver International 2020;40(9):2095-2103. [DOI] [PubMed] [Google Scholar]
Zhao 2020 {published data only}
- Zhao D, Yao F, Wang L, Zheng L, Gao Y, Ye J, et al. A comparative study on the clinical features of COVID-19 pneumonia to other pneumonias. Clinical Infectious Diseases 2020;71(15):756-61. [DOI: 10.1093/cid/ciaa247] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhu 2020 {published data only}
- Zhu W, Xie K, Lu H, Xu L, Zhou S, Fang S. Initial clinical features of suspected coronavirus disease 2019 in two emergency departments outside of Hubei, China. Journal of Medical Virology 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
References to studies excluded from this review
Ai 2020a {published data only}
- Ai JW, Zhang HC, Xu T, Wu J, Zhu M, Yu YQ et al. Optimizing diagnostic strategy for novel coronavirus pneumonia, a multi-center study in Eastern China. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Chen 2020a {published data only}
- Chen X, Ling J, Mo P, Zhang Y, Jiang Q, Ma Z, et al. Restoration of leukomonocyte counts is associated with viral clearance in COVID-19 hospitalized patients. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Chen 2020b {published data only}
- Chen Y, Chen L, Deng Q, Zhang G, Wu K, Ni L, et al. The presence of SARS-CoV-2 RNA in feces of COVID-19 patients. Journal of Medical Virology 2020;92(7). [DOI: ] [DOI] [PubMed] [Google Scholar]
Cheng 2020 {published data only}
- Cheng Z, Lu Y, Cao Q, Qin L, Pan Z, Yan F, et al. Clinical features and chest CT manifestations of coronavirus disease 2019 (COVID-19) in a single-center study in Shanghai, China. American Journal of Roentgenology 2020;215(1):121-6. [DOI: ] [DOI] [PubMed] [Google Scholar]
Giamarellos 2020 {published data only}
- Giamarellos-Bourboulis EJ, Netea MG, Rovina N, Akinosoglou K, Antoniadou A, Antonakos N, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host & Microbe 2020;27(6):992-1000. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Han 2020 {published data only}
- Han H, Yang L, Liu R, Liu F, Wu KL, Li J, et al. Prominent changes in blood coagulation of patients with SARS-CoV-2 infection. Clinical Chemistry and Laboratory Medicine 2020;58(7). [DOI: ] [DOI] [PubMed] [Google Scholar]
Kurstjens 2020 {published data only}
- Kurstjens S, Horst A, Herpers R, Geerits MW, Kluiters-de Hingh YC, Göttgens EL, et al. Rapid identification of SARS-CoV-2-infected patients at the emergency department using routine testing. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PubMed] [Google Scholar]
Li 2020a {published data only}
- Li J, Li S, Cai Y, Liu Q, Li X, Zeng Z, et al. Epidemiological and clinical characteristics of 17 hospitalized patients with 2019 novel coronavirus Infections outside Wuhan, China. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Li 2020b {published data only}
- Li Y, Wang Z, Hui Y, Tong X, Mao X, Huang L, et al. Clinical characteristics of 77 novel coronavirus 2019 infected patients with respiratory failure in the terminal stage in Wuhan. Available at ssrn.com/abstract=3551325 [Preprint] 2020. [DOI: ] [Google Scholar]
Li 2020c {published data only}
- Li YY, Wang WN, Lei Y, Zhang B, Yang J, Hu JW, et al. Comparison of the clinical characteristics between RNA positive and negative patients clinically diagnosed with 2019 novel coronavirus pneumonia. Zhonghua Jie He He Hu Xi Za Zhi 2020;43(5):427-430. [DOI: 10.3760/cma.j.cn112147-20200214-00095 ] [DOI] [PubMed] [Google Scholar]
Ling 2020 {published data only}
- Ling Y, Xu SB, Lin YX, Tian D, Zhu ZQ, Dai FH, et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chinese Medical Journal 2020;133(9):1039-1043. [DOI: 10.1097/CM9.0000000000000774] [DOI] [PMC free article] [PubMed] [Google Scholar]
Meng 2020 {published data only}
- Meng Z, Wang M, Song H, Guo S, Zhou Y, Li W, et al. Development and utilization of an intelligent application for aiding COVID-19 diagnosis. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Peng 2020 {published data only}
- Peng D, Zhang J, Xu Y, Liu Z, Wu P. Clinical analysis and early differential diagnosis of suspected pediatric patients with 2019 novel coronavirus infection. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Peng 2020a {published data only}
- Peng L, Liu KY, Xue F, Miao YF, Tu PA, Zhou C. Improved early recognition of coronavirus disease-2019 (COVID-19): single-center data from a Shanghai screening hospital. Archives of Iranian Medicine 2020;23(4):272-276. [DOI: 10.34172/aim.2020.10] [DOI] [PubMed] [Google Scholar]
Shi 2020 {published data only}
- Shi Y, Tan M, Chen X, Liu Y, Huang J, Ou J, et al. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Song 2020 {published data only}
- Song CY, Xu J, He JQ, Lu YQ. COVID-19 early warning score: a multi-parameter screening tool to identify highly suspected patients. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Spiezia 2020 {published data only}
- Spiezia L, Boscolo A, Poletto F, Cerruti L, Tiberio I, Campello E, et al. COVID-19-Related severe hypercoagulability in patients admitted to intensive care unit for acute respiratory failure. Thrombosis and Haemostasis 2020;120(06):998-1000. [DOI: 10.1055/s-0040-1710018] [DOI] [PMC free article] [PubMed] [Google Scholar]
Sun 2020 {published data only}
- Sun Y, Koh V, Marimuthu K, Ng OT, Young B, Vasoo S, et al. Epidemiological and clinical predictors of COVID-19. Clinical Infectious Diseases 2020;71(15):786-792. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Tang 2020 {published data only}
- Tang X, Du R, Wang R, Cao T, Guan L, Yang C, et al. Comparison of hospitalized patients with acute respiratory distress syndrome caused by COVID-19 and H1N1. Chest 2020;158(1):195-205. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Wang 2020 {published data only}
- Wang Z, Weng J, Li Z, Hou R, Zhou L, Ye H, et al. Development and validation of a diagnostic nomogram to predict COVID-19 pneumonia. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Wu 2020 {published data only}
- Wu J, Zhang P, Zhang L, Meng W, Li J, Tong C, et al. Rapid and accurate identification of COVID-19 infection through machine learning based on clinical available blood test results. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Xu 2020 {published data only}
- Xu Y, Li Y, Zeng Q, Lu Z, Li Y, Wu W, et al. Clinical characteristics of SARS-CoV-2 pneumonia compared to controls in Chinese Han population. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Yang 2020a {published data only}
- Yang Y, Shen C, Li J, Yuan J, Yang M, Wang F, et al. Exuberant elevation of IP-10, MCP-3 and IL-1ra during SARS-CoV-2 infection is associated with disease severity and fatal outcome. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Yin 2020 {published data only}
- Yin S, Huang M, Li D, Tang N. Difference of coagulation features between severe pneumonia induced by SARS-CoV2 and non-SARS-CoV2. Journal of Thrombosis and Thrombolysis 2020. [DOI: 10.1007/s11239-020-02105-8] [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional references
Covidence [Computer program]
- Veritas Health Innovation Covidence. Version accessed before 22 October 2020. Melbourne, Australia: Veritas Health Innovation. Available at covidence.org.
Deeks 2020a
- Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Spijker R, Taylor-Phillips S, et al. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database of Systematic Reviews 2020, Issue 6. Art. No: CD013652. [DOI: 10.1002/14651858.CD013652] [DOI] [PMC free article] [PubMed] [Google Scholar]
Deeks 2020b
- Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Leeflang MMG, Spijker R, et al. Diagnosis of SARS-CoV-2 infection and COVID-19: accuracy of signs and symptoms; molecular, antigen, and antibody tests; and routine laboratory markers. Cochrane Database of Systematic Reviews 2020, Issue 4. Art. No: CD013596. [DOI: 10.1002/14651858.CD013596] [DOI] [Google Scholar]
Dinnes 2020
- Dinnes J, Deeks JJ, Adriano A, Berhane S, Davenport C, Dittrich S, et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database of Systematic Reviews 2020, Issue 8. Art. No: CD013705. [DOI: 10.1002/14651858.CD013705] [DOI] [PMC free article] [PubMed] [Google Scholar]
Macaskill 2010
- Macaskill P, Gatsonis C, Deeks JJ, Harbord RM, Takwoingi Y. Chapter 10: analysing and presenting results. In: Deeks JJ, Bossuyt PM, Gatsonis C editor(s). Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 1.0. The Cochrane Collaboration, 2010. Available from methods.cochrane.org/sdt/handbook-dta-reviews.
McInnes 2020
- Salameh J-P, Leeflang MM, Hooft L, Islam N, McGrath TA, Pol CB, et al. Thoracic imaging tests for the diagnosis of COVID-19. Cochrane Database of Systematic Reviews 2020, Issue 9. Art. No: CD013639. [DOI: 10.1002/14651858.CD013639] [DOI] [PubMed] [Google Scholar]
Review Manager 2020 [Computer program]
- The Cochrane Collaboration Review Manager. Version 5.4. Copenhagen: The Cochrane Collaboration, 2020.
SAS 2015 [Computer program]
- SAS Institute SAS. Version 9.4. Cary, NC, USA: SAS Institute, 2015. Available at www.sas.com.
Schünemann 2020a
- Schünemann HJ, Mustafa RA, Brozek J, Steingart KR, Leeflang M, Murad MH, et al. GRADE guidelines: 21 part 1. Study design, risk of bias, and indirectness in rating the certainty across a body of evidence for test accuracy. Journal of Clinical Epidemiology 2020;122:129-41. [PMID: ] [DOI] [PubMed] [Google Scholar]
Schünemann 2020b
- Schünemann HJ, Mustafa RA, Brozek J, Steingart KR, Leeflang M, Murad MH, et al. GRADE guidelines: 21 part 2. Test accuracy: inconsistency, imprecision, publication bias, and other domains for rating the certainty of evidence and presenting it in evidence profiles and summary of findings tables. Journal of Clinical Epidemiology 2020;122:142-52. [PMID: ] [DOI] [PubMed] [Google Scholar]
Struyf 2020
- Struyf T, Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Leeflang MM, et al. Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19 disease. Cochrane Database of Systematic Reviews 2020, Issue 7. Art. No: CD013665. [DOI: 10.1002/14651858.CD013665] [DOI] [PMC free article] [PubMed] [Google Scholar]
Whiting 2011
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al, QUADAS-2 Group. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine 2011;155(8):529-36. [DOI] [PubMed] [Google Scholar]


