Abstract
The level of long interspersed nuclear element-1 (LINE-1) methylation, representing the global deoxyribonucleic acid methylation level, could contribute to the prognosis of cancer via the activation of oncogenes. This study was performed to evaluate the prognostic implications of LINE-1 hypomethylation in patients with hepatocellular carcinoma (HCC) and the possible mechanisms related to oncogene activation.
Seventy-seven HCC patients between October 2014 and September 2015 were enrolled in this prospective study. Quantitative pyrosequencing was performed to assess the LINE-1 methylation level of HCC and matched non-HCC tissue samples. The expression of suppression of tumorigenicity 18 was measured by immunohistochemistry and its correlation with LINE-1 methylation levels was examined.
LINE-1 was significantly hypomethylated in the HCC tissue compared with the matched nontumor tissue (64.0 ± 11.6% vs 75.6 ± 4.0%, P < .001). LINE-1 hypomethylation was an independent risk factor for overall survival (hazard ratio = 27.291, P = .032) and disease progression (hazard ratio = 5.298, P = .005). The expression of suppression of tumorigenicity 18 was higher in the hypomethylated LINE-1 HCC tissue than the hypermethylated LINE-1 tumor tissue (P = .030).
LINE-1 hypomethylation may serve as a potential prognostic marker for patients with HCC.
Keywords: hepatocellular carcinoma, long interspersed nuclear element-1, methylation, prognosis, suppression of tumorigenicity 18
1. Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related deaths worldwide and its incidence is increasing.[1] Despite advances in HCC treatment, the prognosis is still unsatisfactory because of advanced disease status at initial diagnosis and high recurrence rates.[2] Predicting the prognosis in HCC patients could help to determine the optimal therapeutic modalities and affect the treatment outcomes, thus indicating the importance of identifying prognostic markers. Although various molecular and biological factors have been studied in HCC patients, reliable prognostic markers have not yet been identified.[3]
Genetic and epigenetic alterations such as mutations in oncogenes and tumor suppressor genes, insertion or deletion of chromosome regions, and deoxyribonucleic acid (DNA) methylation are related to many biological processes and underlie the carcinogenesis of most malignancies.[4,5] Identification of these molecular alterations provides valuable tools for early diagnosis, prognosis prediction, and therapeutic targets.[6,7] Among these, global DNA hypomethylation, the genome-wide reduction of DNA methylation, is one of the distinguishing features found in various types of human cancer and plays a significant role in cancer development and progression.[8–10] The global reduction of DNA methylation induces activation and translocation of retrotransposons, thus facilitating chromosomal instability which is crucial to tumor development.[11,12] Hence, the global DNA methylation level may fulfill the role of a prognostic biomarker for tumors.
Long interspersed nuclear element-1 (LINE-1) retrotransposons, comprising approximately 17% of the human genome, are one of the most fundamental mobile genetic elements. The methylation level of LINE-1 is a good indicator of the global DNA methylation level. LINE-1 is epigenetically suppressed in most differentiated somatic cells by the methylation of a promoter and is often aberrantly hypomethylated and highly expressed in human cancers.[13,14] LINE-1 hypomethylation increases the expression of retrotransposon elements and significantly alters gene function during cancer initiation and progression, and is thus a promising prognostic factor.[15,16] LINE-1 hypomethylation correlates with poor prognosis in various types of human malignancies, including gastric cancer, colon cancer, esophageal squamous cell carcinoma, and ovarian cancer.[17–20] LINE-1 hypomethylation in tumors frequently causes retrotransposition at various loci containing proto-oncogenes and activates the genomic expression associated with tumor invasion or metastasis.[21,22] In colon cancer, LINE-1 hypomethylation activates an alternate transcript of the c-MET proto-oncogenes.[17] In addition, previous reports revealed that tumors with LINE-1 hypomethylation showed increased CDK6 expression and promoted tumor progression in esophageal squamous cell carcinoma and HCC.[23,24] A recent study indicated that the hypomethylation of LINE-1 is associated with the activation of suppression of tumorigenicity 18 (ST18) expression in HCC.[25] However, it was mainly based on experiments using liver cancer cell lines and mouse models, and so the prognostic significance and the molecular mechanisms of LINE-1 methylation in HCC patients remain unclear.
In the present study, we examined the association between LINE-1 promoter hypomethylation and prognosis in HCC patients. In addition, we also assessed the correlation between LINE-1 methylation level and ST18 expression in HCC.
2. Methods
2.1. Patients and follow-up
HCC patients who visited Kyungpook National University Hospital, Daegu, South Korea between October 2014 and September 2015 were consecutively enrolled in this prospective study. HCC tissue and nontumor tissue samples were obtained by liver biopsy from all of these patients before specific treatment for HCC. The clinical data and laboratory investigations of the enrolled patients were examined at the initial diagnosis. Exclusion criteria were age under 20 years, severe or uncontrolled medical illness, malignancy other than HCC, infection, and pregnancy. HCC was diagnosed by histopathology according to the Edmondson-Steiner classification system and staged using the system of the modified International Union Against Cancer.[26,27] Liver cirrhosis was defined as a liver biopsy specimen showing METAVIR stage F4.
During the follow-up period, laboratory tests including alpha-fetoprotein and imaging studies using computed tomography or magnetic resonance imaging were performed every 3 to 6 months to monitor the disease status. Overall survival time was defined as the period from the initial diagnosis to the last follow-up or death. Disease progression was defined according to the modified Response Evaluation Criteria in Solid Tumors criteria, or death. The primary endpoint was overall survival and the secondary endpoint was disease progression.
The study was performed in accordance with the ethical guidelines of the Helsinki Declaration of 1975, as revised in 2013. All patients included in the study provided informed consent, and the study protocol was approved by the Institutional Review Board of Kyungpook National University Hospital (IRB No. KNUH-2014-04-056-001).
2.2. Tissue specimens and DNA methylation analysis
Seventy-seven formalin-fixed, paraffin-embedded human HCC tissue, and 77 corresponding nontumor tissue samples were used for this study. The formalin-fixed, paraffin-embedded tissue specimens were microdissected and analyzed, and DNA was extracted with a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), as per the manufacturer's instructions. DNA was modified with sodium-bisulfite using an EZ Methylation-Gold Kit (Zymo Research, CA), and LINE-1 was amplified using a LINE-1-specific polymerase chain reaction. The primer sequences were forward: 5’-TGG TTA AGG GTT TGG GGA TAT T-3’, reverse: 5’-(Biotin)-AAC ACA ATT CCC AAC CCA C-3’, and sequencing: 5’-GGG TTT TGA ATT TGG TA-3’. Quantitative bisulfite pyrosequencing using the PyroMark Q24 Advanced (Qiagen) was performed to quantify the methylation levels of four CpG sites in the repetitive LINE-1 sequence promoter, as described previously.[17]
2.3. Immunohistochemistry
ST18 expression levels were assessed by an immunohistochemical study. The paraffin blocks containing representative tumor regions were selected after a review of the corresponding hematoxylin and eosin-stained slides. Immunohistochemical staining of the ST18 antigens was performed using an automated immunostainer, Ventana BenchMark XT (Ventana Medical Systems, Tucson, AZ) with an UltraView kit, according to the manufacturer's protocol. Rabbit monoclonal ST18 (1:100, sc-46675, Santa Cruz Biotechnology, Santa Cruz, CA) was applied as the primary antibody, after which the samples were incubated at 37°C for 32 minutes followed by standard Ventana signal amplification, and hematoxylin and a bluing reagent counterstaining, consecutively. After the autostainer process, the slides were mounted and examined by light microscopy. Positive staining was indicated by a prominent brownish pigmentation in the cytoplasm. Negative controls were obtained by omitting the specific primary antibodies of the same species. Positive and negative controls stained appropriately.
2.4. Evaluation of immunohistochemical staining
ST18 immunostaining in a predominantly cytoplasmic pattern was considered as positive; it was stained in most of the tumor cells, although the staining intensity of ST18 staining was diverse in each case. ST18 expression was assessed according to staining and scored from 1 to 3 as follows:
-
1.
weak staining,
-
2.
moderate staining, and
-
3.
strong staining (Fig. 1).
Figure 1.
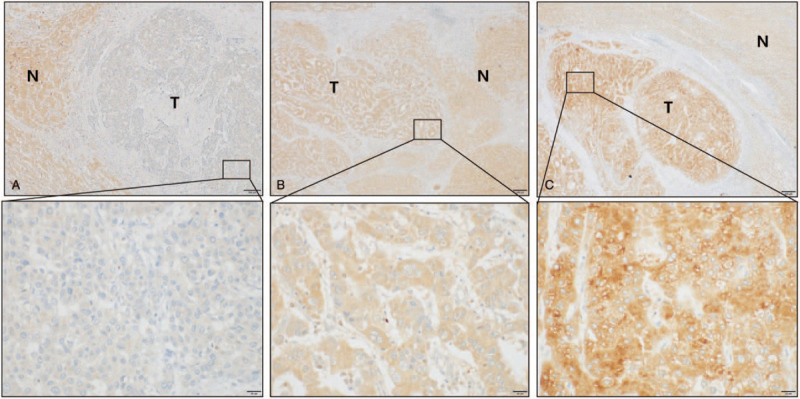
Different expression status of ST18 in hepatic tumors and adjacent nontumor areas detected by immunohistochemistry. (A) Low expression, (B) Moderate expression, (C) High expression. Magnifications: X10; boxed area: 40X. T = tumor region, N = adjacent nontumor area.
The stained samples were evaluated by an expert pathologist blinded to the clinicopathological features or clinical outcome.
2.5. Statistical analysis
Data are presented as median (interquartile range) or n (%) values as appropriate. Categorical variables were compared using a Chi-square test (or Fisher exact test), while continuous variables were compared using the Mann–Whitney test (or the Kruskal–Wallis test). The overall survival and cumulative disease progression rate were estimated using the Kaplan–Meier method, and the differences were compared using a log-rank test. Factors associated with overall survival and disease progression were identified by applying a multivariate Cox proportional-hazards regression test using variables that were significant in the univariate analysis. The hazard ratio (HR) and 95% confidence interval (CI) were also determined. A probability value of P < .05 was considered statistically significant. The statistical analyses were performed using SPSS (version 18.0, PASW Statistics Incorporated, Chicago, IL).
3. Results
3.1. Patients’ characteristics and treatment outcomes of HCC patients
The baseline characteristics of the 77 patients (69 men and 8 women) enrolled in this study are summarized in Table 1. The most common etiology of HCC was chronic hepatitis B (n = 54), followed by alcohol (n = 9), chronic hepatitis C (n = 7), and coinfection of chronic hepatitis B and chronic hepatitis C (n = 2). The tumors were smaller than 5 cm in 42 patients (54.5%), and 37 patients (48.1%) had multiple tumors. Of the 77 patients, 42 had cirrhosis as the underlying liver disease.
Table 1.
Correlations between LINE-1 methylation levels and clinical characteristics in HCC (n = 77).
| All | LINE-1 hypermethylation (methylation ≥64.5%) (n = 39) | LINE-1 hypomethylation (methylation <64.5%) (n = 38) | P value | |
| Demographic variables | ||||
| Age, yr (<60/≥60) | 36/41 (46.8/53.2) | 21/18 (53.8/46.2) | 15/23 (39.5/60.5) | .206 |
| Sex, male/female | 69/8 (89.6/1.4) | 37/2 (94.9/5.1) | 32/6 (84.2/15.8) | .154 |
| Etiology, viral/nonviral | 63/14 (81.8/18.2) | 31/8 (79.5/2.5) | 32/6 (84.2/15.8) | .944 |
| Previous HCC treatment history, no/yes | 60/17 (77.9/22.1) | 33/6 (84.6/15.4) | 27/11 (71.1/28.9) | .151 |
| Tumor variables | ||||
| Size of tumor, <5 cm/≥5 cm | 42/35 (54.5/45.5) | 19/20 (48.7/51.3) | 23/15 (60.5/39.5) | .298 |
| Number of tumor, single/multiple | 40/37 (51.9/48.1) | 22/17 (56.4/43.6) | 18/20 (47.4/52.6) | .427 |
| PVT, no/yes | 57/20 (74.0/26.0) | 28/11 (71.8/28.2) | 29/9 (76.3/23.7) | .651 |
| T stage, 1–2/3–4 | 37/40 (48.1/51.9) | 20/19 (51.3/48.7) | 17/21 (44.7/55.3) | .565 |
| N stage, 0/1 | 75/2 (97.4/2.6) | 37/2 (94.9/5.1) | 38/0 (100.0/0.0) | .494 |
| M stage, 0/1 | 72/5 (93.5/6.5) | 38/1 (97.4/2.6) | 34/4 (89.5/10.5) | .200 |
| Modified UICC stage, I–II/III–IV | 36/41 (46.8/53.2) | 20/19 (51.3/48.7) | 16/22 (42.1/57.9) | .420 |
| Performance status, 0–1/2–3 | 74/3 (96.1/3.9) | 38/1 (97.4/2.6) | 36/2 (94.7/5.3) | .615 |
| Laboratory variables | ||||
| Aspartate aminotransferase, IU/L (<40/≥40) | 40/37 (51.9/48.1) | 21/18 (53.8/46.2) | 19/19 (50.0/50.0) | .736 |
| Alanine aminotransferase, IU/L (<40/≥40) | 53/24 (68.6/31.2) | 27/12 (69.2/30.8) | 26/12 (68.4/31.6) | .939 |
| Alpha-fetoprotein, ng/mL (<20/≥20) | 47/30 (61.0/39.0) | 25/14 (64.1/35.9) | 22/16 (57.9/42.1) | .704 |
| LC, no/yes | 35/42 (45.5/54.5) | 18/21 (46.2/53.8) | 17/21 (44.7/55.3) | .901 |
| Child-Pugh class, A/B | 27/15 (64.3/35.7) | 13/8 (61.9/38.1) | 14/7 (66.7/33.3) | .747 |
| Presence of ascites, no/yes | 71/6 (92.2/7.8) | 34/5 (87.2/12.8) | 37/1 (97.4/2.6) | .2 |
Patients were treated according to the Barcelona Clinic Liver Cancer guidelines. Twenty-two patients underwent hepatic resection, 8 liver transplantation, 19 radiofrequency ablation, 2 chemoembolization, 9 systemic therapy (sorafenib), and 7 patients received the best supportive care including management of pain, nutrition, and psychological support. Among the enrolled patients, the treatment modalities of 10 were unknown because of follow-up loss. Complete remission was achieved in 47 patients (61.0%).
During the study period (median 42.3 months), disease progression was identified in 48 patients (62.3%) at a median of 4.9 months, and 35 patients (45.5%) died at a median of 6.6 months. The most common cause of death was the progression of HCC (n = 29, 82.9%), followed by hepatic failure. The overall survival rates were 79.2%, 68.8%, and 58.4% at 6, 12, and 24 months, respectively.
3.2. LINE-1 methylation levels in HCC and nontumor tissue
LINE-1 methylation levels were examined by pyrosequencing analysis in 77 HCC and matched nontumor tissue. LINE-1 methylation levels in HCC samples were significantly lower than in the matched non-HCC tissue (64.6 ± 11.6% vs 75.6 ± 4.0%, P < .001 by the paired t test) (Fig. 2A). There was a decreasing tendency for LINE-1 methylation from nontumor tissue to HCC tissue (Fig. 2B). There was no significant difference in LINE-1 methylation level between normal (n = 35) and cirrhotic tissue (n = 42) (75.0 ± 5.3% vs 76.2 ± 3.0%, P = .223).
Figure 2.
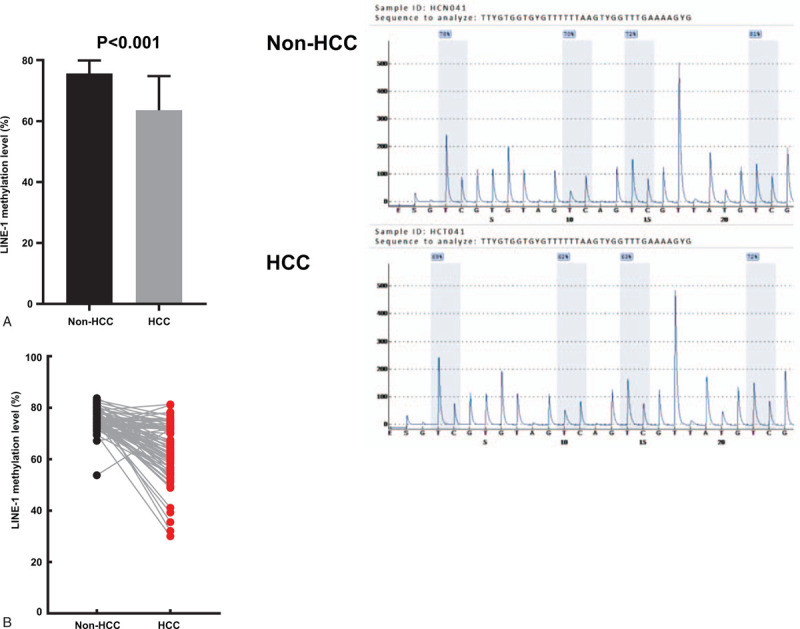
The levels of LINE-1 methylation in HCC tissue and matched nontumor tissue (n = 77). (A) LINE-1 methylation levels were significantly lower in the HCC tissue compared with the nontumor tissue (left, 64.6 ± 11.6% vs 75.6 ± 4.0%, paired t test; P < .001; right, grey vertical boxes in the pyrograms illustrate individual CpG sites analyzed). (B) There was a decreasing tendency for LINE-1 methylation levels from non-tumor tissue to HCC tissue. HCC = hepatocellular carcinoma, LINE-1 = long interspersed nuclear element-1.
3.3. LINE-1 methylation levels and HCC patient outcomes
Dividing the methylation level by the quartile categorical variable (i.e., first quartile cases [Q1; ≥72.21%], second quartile cases [Q2; 64.5–72.21%], third quartile cases [Q3; 56.52–64.5%], and fourth quartile cases [Q4; < 56.52%]), overall survival was higher in Q1 than in Q2, Q3, and Q4 (Fig. 3A). Therefore, we defined the LINE-1 methylation level as follows: Q1 as the “hypermethylation group” and a combination of Q2, Q3, and Q4 as the “hypomethylation group.” The hypomethylation group tended to have shorter overall survival (median 36.2 months vs 44.6 months) and progression-free survival (median 8.1 months vs 19.5 months) compared with the hypermethylation group, although these were not statistically significant (log-rank test: P = .102, P = .075, respectively) (Fig. 3B and C). The overall survival rates at 6, 12, and 24 months were lower in the hypomethylation group (77.6%, 67.2%, and 53.4%, respectively) than in the hypermethylation group (84.2%, 73.7%, and 73.7%, respectively). The rates of disease progression at 6, 12, and 24 months were also higher in the hypomethylation group (43.3%, 57.9%, and 72.6%, respectively) than in the hypermethylation group (17.6%, 35.3%, and 47.1%, respectively).
Figure 3.
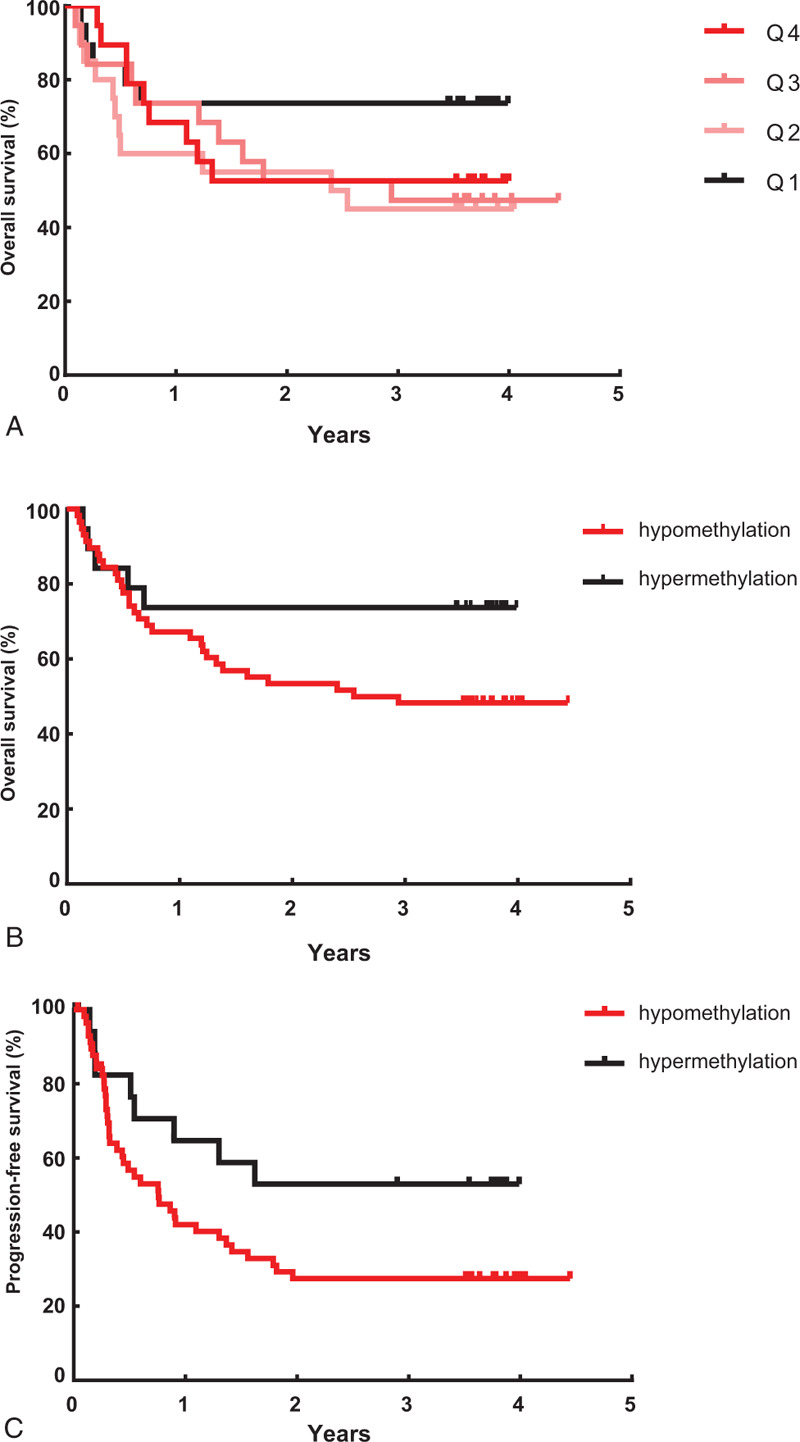
(A) Kaplan–Meier plots of overall survival according to LINE-1 methylation level in 77 hepatocellular carcinoma (HCC) patients divided into quartiles Q1–4: Overall survival was higher in Q1 than in Q2, Q3, and Q4. Q1 represents the “hypermethylation group” and a combination of Q2, Q3, and Q4 represents the “hypomethylation group.” (B) Kaplan–Meier plots of overall survival according to LINE-1 methylation level in 77 HCC patients. The survival rate was lower in patients with LINE-1 hypomethylation (median 36.2 months) than those with LINE-1 hypermethylation (median 44.6 months), although not statistically significant (log-rank test: P = .102). (C) Kaplan–Meier plots of progression-free survival according to LINE-1 methylation level in 77 HCC patients. The progression-free survival rate had a tendency to be lower in patients with LINE-1 hypomethylation (median 8.1 months) than those with LINE-1 hypermethylation (median 19.5 months), which showed borderline statistical significance (log-rank test: P = .075). HCC = hepatocellular carcinoma, LINE-1 = long interspersed nuclear element-1.
In a multivariate analysis, LINE-1 hypomethylation was identified as an independent prognostic factor of overall survival (HR = 27.291, 95% CI = 1.326–561.749, P = .032) and disease progression (HR = 5.298, 95% CI = 1.663–16.873, P = .005) in HCC patients (Table 2). In a subgroup analysis of 41 patients with stage III or IV, LINE-1 hypomethylation was also found to be independently associated with overall survival and disease progression (P = .027, P = .006, respectively) (Table 3).
Table 2.
Multivariate analysis of factors associated with overall survival and disease progression in all HCC patients (n = 77).
| Overall survival | Disease progression | |||||
| Univariate | Multivariate analysis | Univariate | Multivariate analysis | |||
| P value | P value | Hazard ratio (95% CI) | P value | P value | Hazard ratio (95% CI) | |
| Previous HCC treatment history | .02 | .028 | ||||
| Number of tumors (multiple vs single) | .004 | .092 | ||||
| Size of tumor, cm (≥5 vs <5) | <.001 | .003 | 6.459 (1.910–21.844) | <.001 | .004 | 2.958 (1.411–6.200) |
| PVT | <.001 | <.001 | .009 | 3.311 (1.348–8.137) | ||
| N stage | <.001 | .013 | 30.813 (2.066–459.512) | .001 | .022 | 7.934 (1.347–46.737) |
| M stage | .003 | .001 | 13.575 (3.090–59.649) | .001 | .013 | 4.141 (1.345–12.753) |
| Performance status | .004 | .001 | ||||
| CRP, mg/dL | <.001 | .011 | 1.418 (1.084–1.856) | <.001 | .012 | 1.391 (1.075–1.800) |
| Aspartate aminotransferase, IU/L | .001 | .001 | .009 | 1.012 (1.003–1.021) | ||
| Serum albumin, g/dL | <.001 | .022 | ||||
| Alpha-fetoprotein, ng/mL (≥20 vs <20) | .037 | .108 | ||||
| Presence of LC | .048 | .005 | 5.055 (1.612–15.851) | .454 | ||
| Presence of ascites | .692 | .014 | 22.425 (1.874–268.357) | .776 | .055 | |
| LINE-1 hypomethylation | .111 | .032 | 27.291 (1.326–561.749) | .081 | .005 | 5.298 (1.663–16.873) |
Table 3.
Multivariate analysis of factors associated with overall survival and disease progression in TNM stage III–IV (n = 41).
| Overall survival | Disease progression | |||||
| Univariate | Multivariate | Univariate | Multivariate | |||
| P value | P value | Hazard ratio (95% CI) | P value | P value | Hazard ratio (95% CI) | |
| Previous HCC treatment history | .044 | .084 | ||||
| Size of tumor, cm (≥5 vs <5) | .003 | .011 | 3.959 (1.367–11.467) | .007 | .075 | |
| PVT | .007 | .004 | 3.531 (1.487–8.387) | .038 | .021 | 3.185 (1.188–8.535) |
| N stage | .004 | .004 | 17.653 (2.558–121.812) | .006 | <.001 | 56.026 (6.772–463.505) |
| M stage | .202 | .056 | ||||
| Performance status | .016 | .017 | .003 | 11.935 (2.387–59.683) | ||
| CRP, mg/dL | .001 | <.001 | ||||
| Aspartate aminotransferase, IU/L | .033 | .01 | .003 | 1.019 (1.006–1.031) | ||
| Presence of ascites | .014 | .001 | 41.993 (4.853–363.401) | .016 | <.001 | 156.833 (12.930–1902.309) |
| LINE-1 hypomethylation | .404 | .027 | 4.216 (1.179–15.074) | .437 | .006 | 7.519 (1.774–31.864) |
3.4. Association of LINE-1 methylation with the expression of ST18 in HCC
Since recent evidence suggests that ST18 is activated via LINE-1 hypomethylation,[25] we examined ST18 expression using immunohistochemical analysis in 51 HCC patients. The patients were classified into 2 groups based on the median LINE-1 methylation level (64.5%) of the HCC tissue samples: 25 with a LINE-1 methylation level of <64.5% and 26 with a LINE-1 methylation level of ≥64.5%. When the immunohistochemical staining scores were grouped into low expression (score 1–2) and high expression (score 3), 11 of the 25 cases (44%) showed high ST18 expression in hypomethylated LINE-1 patients, whereas only 6 of 26 cases (23.1%) exhibited high ST18 expression in hypermethylated LINE-1 patients (P = .030) (Fig. 4). Hence, elevated ST18 expression was significantly associated with LINE-1 hypomethylation in the HCC patients.
Figure 4.
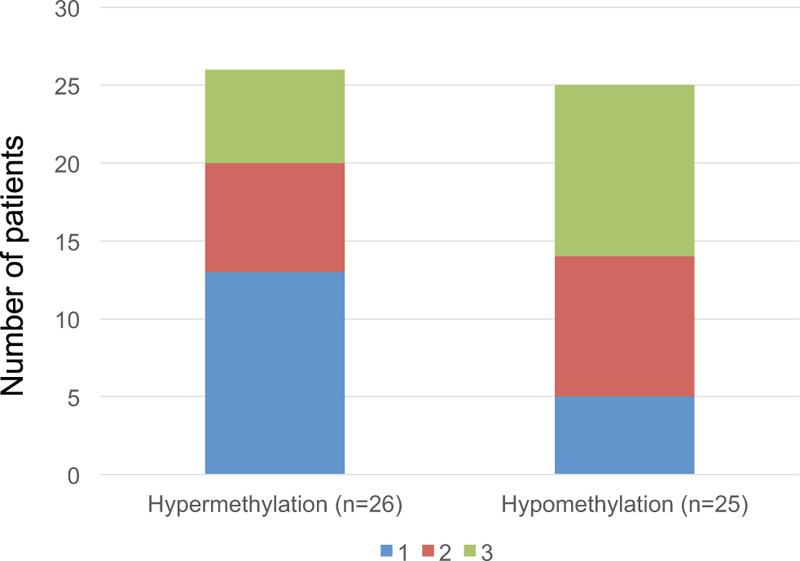
Comparison of ST18 expression between hypermethylated and hypomethylated LINE-1 hepatocellular carcinoma (HCC) tissue by immunohistochemical staining (n = 51). ST18 expression was significantly higher in the hypomethylated LINE-1 HCC tissue than in the hypermethylated LINE-1 HCC tissue (P = .030). HCC = hepatocellular carcinoma, LINE-1 = long interspersed nuclear element-1.
4. Discussion
LINE-1 methylation level is regarded as a useful representative for estimating the global DNA methylation status,[28,29] while aberrant LINE-1 hypomethylation is known as a potential prognostic marker for various types of cancer.[18,19] However, the role of LINE-1 hypomethylation in the pathogenesis of HCC is still not fully understood. In this study, we evaluated the prognostic relevance of LINE-1 hypomethylation in HCC as well as the relationship between ST18 expression and LINE-1 methylation.
In the present study, we demonstrated that the LINE-1 methylation level was significantly lower in HCC tissue compared with nontumor tissue, and LINE-1 hypomethylation was significantly associated with overall survival and disease progression in HCC patients. Moreover, LINE-1 hypomethylation was correlated with elevated ST18 expression.
LINE-1 was significantly hypomethylated in HCC tissue compared with matched nontumor tissue. However, there was no significant difference in LINE-1 methylation level between the nontumorous tissue samples (normal vs cirrhosis), which indicates that hypomethylation of LINE-1 has a crucial role during the hepatocarcinogenesis and is a specific event associated with the development of HCC rather than other pathologic processes in the liver, including chronic hepatitis and cirrhosis.[25,30] In the present study, LINE-1 hypomethylation was associated with poor overall survival and disease progression after adjusting for other factors. LINE-1 methylation status could be used as a reliable prognostic biomarker in HCC patients. Furthermore, the LINE-1 methylation level could affect the management of HCC patients and help to decide the frequency of follow-up examination. Previous studies of LINE-1 hypomethylation in human HCC included patients who had undergone hepatectomy, so only early-stage HCC was examined.[21,24] Our study included HCC patients with various cancer stages and therapeutic modalities. Interestingly, LINE-1 hypomethylation was also revealed as an independent predictor of overall survival and disease progression in patients with advanced-stage HCC (stage III–IV). Thus, LINE-1 hypomethylation could affect the prognosis of not only early-stage but also advanced-stage HCC patients. However, this requires validation with further studies.
LINE-1 comprises highly repetitive sequences that exist throughout the human genome. It contains a CpG island in the 5’-UTR region which is often heavily methylated in normal somatic cells.[29,31] The biological function of LINE-1 hypomethylation has not been thoroughly investigated, although several hypotheses have been suggested to explain the involvement of LINE-1 hypomethylation in tumorigenesis and cancer progression. The hypomethylation of retrotransposons induces genomic instability, a high frequency of DNA-double strand breaks, and reactivation of transposable elements.[9,32,33] By a “copy and paste” mechanism, retrotransposons are replicated and inserted into new genomic regions. Activated retrotransposition disrupts gene structure and coding sequence, dysregulates gene expression, and activates oncogenic pathways during carcinogenesis.[25,34,35] Once the tumor has developed, LINE-1 promoter methylation gradually decreases as the tumor progresses,[36,37] aberrantly activating an additional antisense promoter and downstream proto-oncogenes.[38] Previous findings in HCC patients were consistent with other cancer patients, in which LINE-1 methylation was significantly lower in tumor, and LINE-1 hypomethylation was associated with poor overall survival and disease progression in HCC patients. Moreover, LINE-1 hypomethylation in HCC promoter proceeded to poor outcomes via inducing expression of c-MET and CD133.[21,39]
ST18 is a member of the NZF/MyT1 zinc-finger transcription factor family[40] and it has been proposed to act as either a tumor suppressor or an oncogene in different cancers.[41,42] While ST18 expression is usually repressed in normal liver tissue, the activation of its expression is revealed in human and mouse models of HCC. ST18 has a significant role in liver tumorigenesis and proliferation of tumor cells, suggesting it as a possible oncogene in HCC.[25,43] A previous study demonstrated that inserted LINE-1 induced by LINE-1 hypomethylation interrupted a negative feedback loop and resulted in ST18 activation in liver cancer cell lines and mouse models for HCC.[25] In addition, it was indicated that ST18 expression in HCC was induced by tumor-associated macrophages, and these contributed reciprocally in liver carcinogenesis.[43] However, these studies were performed mainly in vitro. In this study, HCC patients with hypomethylated LINE-1 expressed higher ST18 expression than those with hypermethylated LINE-1. These results support that LINE-1 hypomethylation might affect the development and progression of human HCC through activation of ST18.
There are several limitations in the present study. First, the prognostic role of LINE-1 methylation according to the etiology and treatment modalities could not be evaluated because of the small number of patients who met the conditions. Second, we did not examine the LINE-1 methylation level in dysplastic nodules, regarded as precancerous lesions. Further studies with a large number of patients are required to confirm the potential prognostic value of LINE-1 methylation.
In conclusion, the present study revealed that LINE-1 hypomethylation is a prognostic marker in HCC patients. LINE-1 hypomethylation plays an important role during the development and progression of HCC via activation of ST18 expression.
Author contributions
Conceptualization: Yu Rim Lee, Gyeonghwa Kim, Keun Hur.
Data curation: Yu Rim Lee, Gyeonghwa Kim.
Formal analysis: Yu Rim Lee, Won Young Tak, Soo Young Park.
Funding acquisition: Yu Rim Lee.
Investigation: Young Seok Han, Jae Min Chun, Ja Ryung Han.
Methodology: Gyeonghwa Kim, Hye Won Lee, Keun Hur.
Project administration: Se Young Jang, Young Oh Kweon.
Supervision: Jung Gil Park, Keun Hur.
Writing – original draft: Yu Rim Lee, Gyeonghwa Kim, Hye Won Lee, Keun Hur.
Writing – review & editing: Yu Rim Lee, Gyeonghwa Kim, Hye Won Lee, Keun Hur.
Footnotes
Abbreviations: CI = confidence interval, HCC = hepatocellular carcinoma, HR = hazard ratio, LINE-1 = long interspersed nuclear element-1, ST18 = suppression of tumorigenicity 18.
How to cite this article: Lee YR, Kim G, Lee HW, Tak WY, Park SY, Jang SY, Kweon YO, Park JG, Han YS, Chun JM, Han JR, Hur K. Long interspersed nuclear element-1 hypomethylation is associated with poor outcomes via the activation of ST18 in human hepatocellular carcinoma. Medicine. 2021;100:16(e25552).
YRL and GK contributed equally to this work.
This work was supported by Biomedical Research Institute grant, Kyungpook National University Hospital (2017).
The authors have no conflicts of interest to disclose.
All data generated or analyzed during this study are included in this published article [and its supplementary information files]. The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
HCC = hepatocellular carcinoma, LC = liver cirrhosis, LINE-1 = long interspersed nuclear element-1, PVT = portal vein thrombosis.
CI = confidence interval, CRP = C-reactive protein, HCC = hepatocellular carcinoma, LC = liver cirrhosis, LINE-1 = long interspersed nuclear element-1, PVT = portal vein thrombosis.
CI = confidence interval, CRP = C-reactive protein, HCC = hepatocellular carcinoma, LINE-1 = long interspersed nuclear element-1, PVT = portal vein thrombosis, TNM = tumor-node-metastasis.
References
- [1].Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet (London, England) 2012;379:1245–55. [DOI] [PubMed] [Google Scholar]
- [2].Carr BI. Hepatocellular carcinoma: current management and future trends. Gastroenterology 2004;127: 5 Suppl 1: S218–224. [DOI] [PubMed] [Google Scholar]
- [3].Haruyama Y, Yorita K, Yamaguchi T, et al. High preoperative levels of serum glypican-3 containing N-terminal subunit are associated with poor prognosis in patients with hepatocellular carcinoma after partial hepatectomy. Int J Cancer 2015;137:1643–51. [DOI] [PubMed] [Google Scholar]
- [4].Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell 2012;150:12–27. [DOI] [PubMed] [Google Scholar]
- [5].Herath NI, Leggett BA, MacDonald GA. Review of genetic and epigenetic alterations in hepatocarcinogenesis. J Gastroenterol Hepatol 2006;21(1 Pt 1):15–21. [DOI] [PubMed] [Google Scholar]
- [6].Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer 2006;6:674–87. [DOI] [PubMed] [Google Scholar]
- [7].Feng GS. Conflicting roles of molecules in hepatocarcinogenesis: paradigm or paradox. Cancer cell 2012;21:150–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Weisenberger DJ. Characterizing DNA methylation alterations from The Cancer Genome Atlas. J Clin Investig 2014;124:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Esteller M. Epigenetics in cancer. N Engl J Med 2008;358:1148–59. [DOI] [PubMed] [Google Scholar]
- [10].Berman BP, Weisenberger DJ, Aman JF, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet 2011;44:40–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet 2009;10:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science (New York, N Y ) 2003;300:489–92. [DOI] [PubMed] [Google Scholar]
- [13].Coufal NG, Garcia-Perez JL, Peng GE, et al. L1 retrotransposition in human neural progenitor cells. Nature 2009;460:1127–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet 2011;12:615–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Herceg Z, Paliwal A. Epigenetic mechanisms in hepatocellular carcinoma: how environmental factors influence the epigenome. Mutat Res 2011;727:55–61. [DOI] [PubMed] [Google Scholar]
- [16].Kim MJ, White-Cross JA, Shen L, et al. Hypomethylation of long interspersed nuclear element-1 in hepatocellular carcinomas. Modern Pathol 2009;22:442–9. [DOI] [PubMed] [Google Scholar]
- [17].Hur K, Cejas P, Feliu J, et al. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 2014;63:635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Iwagami S, Baba Y, Watanabe M, et al. LINE-1 hypomethylation is associated with a poor prognosis among patients with curatively resected esophageal squamous cell carcinoma. Ann Surg 2013;257:449–55. [DOI] [PubMed] [Google Scholar]
- [19].Shigaki H, Baba Y, Watanabe M, et al. LINE-1 hypomethylation in gastric cancer, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastric Cancer 2013;16:480–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pattamadilok J, Huapai N, Rattanatanyong P, et al. LINE-1 hypomethylation level as a potential prognostic factor for epithelial ovarian cancer. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society 2008;18:711–7. [DOI] [PubMed] [Google Scholar]
- [21].Zhang C, Xu Y, Zhao J, et al. Elevated expression of the stem cell marker CD133 associated with Line-1 demethylation in hepatocellular carcinoma. Ann Surg Oncol 2011;18:2373–80. [DOI] [PubMed] [Google Scholar]
- [22].Rodriguez J, Frigola J, Vendrell E, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res 2006;66:8462–9468. [DOI] [PubMed] [Google Scholar]
- [23].Baba Y, Watanabe M, Murata A, et al. LINE-1 hypomethylation, DNA copy number alterations, and CDK6 amplification in esophageal squamous cell carcinoma. Clinical Cancer Res 2014;20:1114–24. [DOI] [PubMed] [Google Scholar]
- [24].Harada K, Baba Y, Ishimoto T, et al. LINE-1 methylation level and patient prognosis in a database of 208 hepatocellular carcinomas. Ann Surg Oncol 2015;22:1280–7. [DOI] [PubMed] [Google Scholar]
- [25].Shukla R, Upton KR, Muñoz-Lopez M, et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 2013;153:101–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ueno S, Tanabe G, Nuruki K, et al. Prognostic performance of the new classification of primary liver cancer of Japan (4th edition) for patients with hepatocellular carcinoma: a validation analysis. Hepatol Res 2002;24:395–403. [DOI] [PubMed] [Google Scholar]
- [27].Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer 1954;7:462–503. [DOI] [PubMed] [Google Scholar]
- [28].Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- [29].Kazazian HH, Jr. Mobile elements: drivers of genome evolution. Science (New York, N Y ) 2004;303:1626–32. [DOI] [PubMed] [Google Scholar]
- [30].Gao XD, Qu JH, Chang XJ, et al. Hypomethylation of long interspersed nuclear element-1 promoter is associated with poor outcomes for curative resected hepatocellular carcinoma. Liver Int 2014;34:136–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hoffmann MJ, Schulz WA. Causes and consequences of DNA hypomethylation in human cancer. Biochem Cell Biol = Biochimie et biologie cellulaire 2005;83:296–321. [DOI] [PubMed] [Google Scholar]
- [32].Cruickshanks HA, Tufarelli C. Isolation of cancer-specific chimeric transcripts induced by hypomethylation of the LINE-1 antisense promoter. Genomics 2009;94:397–406. [DOI] [PubMed] [Google Scholar]
- [33].Howard G, Eiges R, Gaudet F, et al. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene 2008;27:404–8. [DOI] [PubMed] [Google Scholar]
- [34].Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Developmental cell 2010;19:698–711. [DOI] [PubMed] [Google Scholar]
- [35].Bjornsson HT, Brown LJ, Fallin MD, et al. Epigenetic specificity of loss of imprinting of the IGF2 gene in Wilms tumors. J Nat Cancer Inst 2007;99:1270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Calvisi DF, Ladu S, Gorden A, et al. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Investig 2007;117:2713–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pogribny IP, Rusyn I. Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer letters 2014;342:223–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Carreira PE, Richardson SR, Faulkner GJ. L1 retrotransposons, cancer stem cells and oncogenesis. FEBS J 2014;281:63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhu C, Utsunomiya T, Ikemoto T, et al. Hypomethylation of long interspersed nuclear element-1 (LINE-1) is associated with poor prognosis via activation of c-MET in hepatocellular carcinoma. Ann Surg Oncol 2014;21: Suppl 4: S729–35. [DOI] [PubMed] [Google Scholar]
- [40].Yee KS, Yu VC. Isolation and characterization of a novel member of the neural zinc finger factor/myelin transcription factor family with transcriptional repression activity. J Biol Chem 1998;273:5366–74. [DOI] [PubMed] [Google Scholar]
- [41].Steinbach D, Schramm A, Eggert A, et al. Identification of a set of seven genes for the monitoring of minimal residual disease in pediatric acute myeloid leukemia. Clin Cancer Res 2006;12:2434–41. [DOI] [PubMed] [Google Scholar]
- [42].Jandrig B, Seitz S, Hinzmann B, et al. ST18 is a breast cancer tumor suppressor gene at human chromosome 8q11.2. Oncogene 2004;23:9295–302. [DOI] [PubMed] [Google Scholar]
- [43].Ravà M, D’Andrea A, Doni M, et al. Mutual epithelium-macrophage dependency in liver carcinogenesis mediated by ST18. Hepatology (Baltimore, Md) 2017;65:1708–19. [DOI] [PMC free article] [PubMed] [Google Scholar]


