Abstract
Introduction
Lower gastrointestinal (GI) cancers are a major cause of cancer deaths worldwide. Prognosis improves with earlier diagnosis, and non-invasive biomarkers have the potential to aid with early detection. Substantial investment has been made into the development of biomarkers; however, studies are often carried out in specialist settings and few have been evaluated for low-prevalence populations.
Methods
We aimed to identify novel biomarkers for the detection of lower GI cancers that have the potential to be evaluated for use in primary care. MEDLINE, Embase, Emcare and Web of Science were systematically searched for studies published in English from January 2000 to October 2019. Reference lists of included studies were also assessed. Studies had to report on measures of diagnostic performance for biomarkers (single or in panels) used to detect colorectal or anal cancers. We included all designs and excluded studies with fewer than 50 cases/controls. Data were extracted from published studies on types of biomarkers, populations and outcomes. Narrative synthesis was used, and measures of specificity and sensitivity were meta-analysed where possible.
Results
We identified 142 studies reporting on biomarkers for lower GI cancers, for 24,844 cases and 45,374 controls. A total of 378 unique biomarkers were identified. Heterogeneity of study design, population type and sample source precluded meta-analysis for all markers except methylated septin 9 (mSEPT9) and pyruvate kinase type tumour M2 (TuM2-PK). The estimated sensitivity and specificity of mSEPT9 was 80.6% (95% CI 76.6–84.0%) and 88.0% (95% CI 79.1–93.4%) respectively; TuM2-PK had an estimated sensitivity of 81.6% (95% CI 75.2–86.6%) and specificity of 80.1% (95% CI 76.7–83.0%).
Conclusion
Two novel biomarkers (mSEPT9 and TuM2-PK) were identified from the literature with potential for use in lower-prevalence populations. Further research is needed to validate these biomarkers in primary care for screening and assessment of symptomatic patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12325-021-01645-6.
Keywords: Biomarkers, Clinical practice, Colorectal cancer, Early detection, Lower gastrointestinal cancers, Primary care
Key Summary Points
| To our knowledge, this is the first systematic review to characterise the range of novel biomarkers being investigated for the early detection of lower GI cancers, with a focus on their readiness to progress to further evaluation in low-prevalence populations such as primary care. |
| We identified 378 unique biomarkers from the literature; a meta-analysis of diagnostic accuracy data indicated mSEPT9 and TuM2-PK have potential for further evaluation in low-prevalence populations. |
| We highlight the need for (1) further studies on mSEPT9 and TuM2-PK in low-prevalence populations; (2) better reporting to facilitate translation; (3) more consistency in the use of biomarkers. By doing so, we will be able to progress to a different step in the evaluation process of promising biomarkers, and ultimately ascertain clinical benefits for our intended population. This will require going beyond test performance, investigating implementation (including feasibility and acceptability), safety and cost-effectiveness. |
Digital Features
This article is published with digital features, including a summary slide, to facilitate understanding of the article. To view digital features for this article go to 10.6084/m9.figshare.13664042.
Introduction
Gastrointestinal (GI) cancers account for over 25% of global cancer incidence and 35% of all cancer-related deaths [1]. Lower GI cancers, particularly colorectal cancer (CRC), contribute the most significant proportion with over 1.8 million new cases in 2018 [1]. CRC is the most commonly diagnosed GI cancer and constitutes 1 in 10 cancer cases and deaths [1].
Around 90% of patients with cancer first present with symptoms in primary care, highlighting a key role for primary care providers in the early detection of GI cancers [2, 3]. Diagnosis of GI cancers can prove challenging in the community setting: while gastrointestinal symptoms are commonly encountered, they are usually due to benign or self-limiting conditions and rarely to GI cancer [3]. Initial symptoms are often non-specific, and more specific symptoms usually represent more advanced disease [3].
Increased demand for diagnostic services for lower GI cancer and pressure on waiting times have been seen internationally in countries like Australia, the UK and Canada, where primary care plays a ‘gatekeeping’ role to specialist care. In many countries implementation of faecal occult blood tests (FOBTs) or faecal immunochemical tests (FITs) for CRC screening and diagnostic triage adds further pressure on colonoscopy services. In some healthcare systems, over-screening via colonoscopy is also an issue [4]. New diagnostic approaches are needed to help reduce the burden on specialist care, particularly in the current context of COVID-19 and associated delays in access to cancer diagnostic and treatment services [5].
There is considerable interest in the potential of biomarkers to detect GI cancers [3]. To date, carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9) have played an important role in clinical practice to detect recurrent disease, but their diagnostic performance is inadequate for the early detection of new disease [3, 6, 7]. Substantial investment has been made developing new biomarkers for early detection, but most studies of these tests have occurred in specialist clinical settings [8] where cancer prevalence is higher than in the community settings where they would eventually be applied [9, 10]
The performance characteristics of a diagnostic test are strongly determined by the prevalence and severity of the target disease and of other diseases within the study population [9]. In populations where the prevalence of the target disease is low (e.g. GI cancer in primary care), the corresponding positive predictive values (PPV) are lower than in high-prevalence populations. Tests that are evaluated only in these high-prevalence populations tend to have lower sensitivity and higher specificity when translated to low-prevalence populations [10, 11]. This is known as the ‘spectrum effect’, and has crucial implications for comparing the performance of tests in different populations [9, 10].
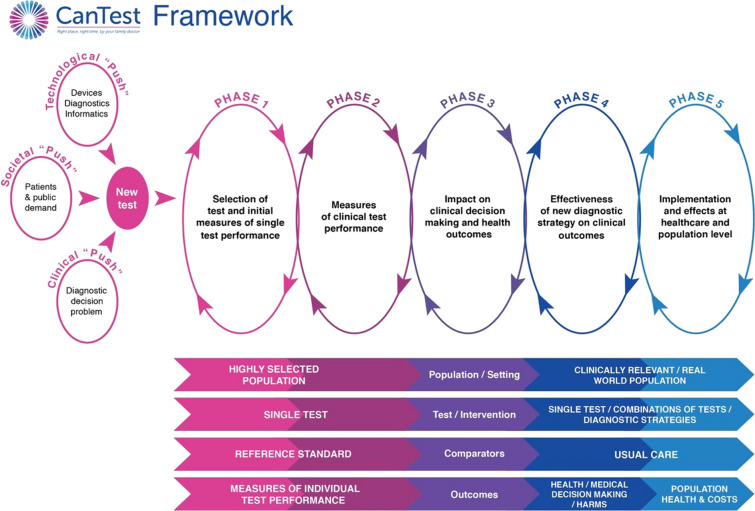
In recognition of these issues, the CanTest Framework was developed (Fig. 1) [10]. This novel framework encompasses a translational pathway for diagnostic tests, from new test discovery to health system implementation in low-prevalence populations [11]. The framework highlights the importance of evaluating clinical performance, implementation, patient safety, quality of care, and cost-effectiveness in the intended setting. It is vital that these elements are investigated alongside test performance in order to ascertain clinical utility and improved outcomes for patients [8].
Fig. 1.
The CanTest framework.
Source: Walter et al. 2019 [10]
This review aimed to systematically identify novel biomarkers for the early detection of lower GI cancers that have measures of diagnostic performance and show sufficient promise to warrant further evaluation in low-prevalence populations.
Methods
Search Strategy and Inclusion/Exclusion Criteria
These have been reported elsewhere [12]. The protocol for this review was registered on PROSPERO (registration ID CRD42020165005) and the Preferred Reporting Items for Systematic Reviews and Meta-Analysis of Diagnostic Test Accuracy Studies (PRISMA-DTA) statement was followed [13]. MEDLINE, Embase, Emcare and Web of Science were electronically searched for primary studies published in English between the 1 January 2000 and 31 October 2019. The search strategy was developed with the assistance of a medical librarian (Appendix 1 in the supplementary material).
Studies eligible for inclusion were situated within phase 2 (i.e. providing measures of diagnostic accuracy beyond discovery, even if carried out in high-prevalence settings) and phase 3 (i.e. examining diagnostic accuracy in intended low-prevalence settings, and providing measures of clinical utility, including feasibility and acceptability) of the CanTest framework [10] (Fig. 1). We included studies which reported measures of diagnostic performance in an independent population (i.e. beyond measures from the initial discovery phase). Studies were excluded if no references to previous research evaluating biomarker performance were available, and if the study provided only one set of performance measures (reflecting discovery phase only). As studies beyond the discovery phase require larger sample sizes [14, 15], we included those which reported data on at least 50 cancer cases and at least one group of homogeneous non-cancer controls (n ≥ 50) with similar clinical characteristics (e.g. healthy, or with non-malignant conditions), as in previous reviews [15, 16].
We included studies on non-invasive biomarkers feasible for use in the community setting: blood (serum and plasma), urine, faecal, salivary or breath samples. Both observational (cohort or case–control, cross-sectional or longitudinal, prospective or retrospective) and experimental designs were eligible for inclusion. Studies undertaken in all recruitment settings were included.
We included studies if they reported on at least one measure of diagnostic performance, namely sensitivity, specificity, PPV, negative predictive value (NPV), false positive, false negative or area under the curve (AUC) for biomarkers used to detect lower GI cancers, including colorectal (colon, rectum, caecum) or anal cancers, in adult populations (mean/median age 18 or older; studies including individuals aged less than 18 were accepted if these were outliers in large samples). Non-specified GI cancers, neuroendocrine cancers, and studies only reporting on familial populations at risk of hereditary cancers were excluded.
Novel biomarkers were considered individually, in combination or as part of a panel. Studies reporting only on a single, established biomarker (CEA, CA19-9, or FIT or FOBT) were excluded [16, 17]. Studies providing measures of diagnostic performance for combinations of established and novel biomarkers were included.
Covidence systematic review software [18] was used to facilitate article screening. Titles and abstracts were screened independently by two reviewers (any two of PD, NC, CS, KM, DB or RB). Full-text articles were also independently evaluated for inclusion by two reviewers (any two of the aforementioned). Reference lists of included studies were manually reviewed by one author to identify additional studies (NC). Full-text articles selected at this stage were also independently assessed by two reviewers (any two of PD, NC, RB or DB). Disagreements were resolved by consensus; when this could not be reached, a senior reviewer was consulted (JE or FMW).
Data Extraction and Analysis
Data extraction was piloted to ensure consistency and was performed independently (by SS, DB, RB, JMG, JO). Information on study characteristics, populations, biomarkers and measures of diagnostic performance were extracted. When studies reported on different phases of biomarker development, only data from the eligible phases were extracted. When studies had more than one eligible phase, data were extracted for all eligible phases. Extracted data were collated and checked for consistency and inaccuracies (PD).
Biomarkers were categorised according to a modified version of the classifications reported by Uttley et al. [15]: microRNAs and other RNAs, autoantibodies and other immunological markers, other proteins (i.e. proteins that did not fit into other categories), metabolic markers, DNA-related markers (protein-coding genes, gene mutations), circulating tumour DNA, DNA methylation markers and other biomarkers. Controls were classified as normal/healthy, having non-malignant conditions or those with adenomas/polyps. Controls described as healthy were coded as such unless studies described underlying conditions. Full details of the control population classification are available in Appendix 2 in the supplementary material.
Quality Assessment and Risk of Bias
Considering the key issue of spectrum bias, studies were classified as either single-gate or two-gate designs. Single-gate studies recruit participants before disease status is known, with a single route of entry, and with the same inclusion criteria. Two-gate studies recruit participants through different routes and use different criteria for cases and controls. This can lead to over-inflated measures of diagnostic performance for example if there is an over-representation of individuals with advanced disease within the study population and comparison with the ‘fittest of the fit’ healthy controls [11]. One author (PD) classified all studies and another (NC) checked the classification. A full description of this classification and how it approaches issues covered by the QUADAS-2 critical appraisal tool [19] is available in Appendix 3 in the supplementary material. Studies included in the meta-analysis were assessed using the QUADAS-2 [19] tool by two reviewers (PD and NC).
Data Synthesis
As significant heterogeneity was anticipated, we used narrative synthesis to summarise the data [20]. An overview of the evidence was developed to describe the key characteristics of the included studies, their populations, biomarkers and outcome measures. Data were examined for similarities that would allow for subgroup analyses, namely the same biomarker, with similar study design and appropriate accuracy performance measures. For meta-analysis to occur, biomarkers had to be investigated in more than two studies, with individual outcome measures provided, similar populations included and a single-gate study design. We focused the analysis on single-gate studies, as this design reduces spectrum bias, and is more likely to provide results that translate for use in low-prevalence populations. Meta-analysis of diagnostic test accuracy was performed using MetaDTA (version 1.43) [21] and RevMan (5.3) [22] software.
For meta-analysis, we used the random effects bivariate binomial model of Chu and Cole fitted as a generalised linear mixed effect model [21]. Sensitivity and specificity were jointly modelled and the estimates from each study were assumed to vary [21]. Hierarchical summary receiver operating characteristic (HSROC) parameters were estimated using the bivariate model parameters. Summary points of sensitivity and specificity were presented alongside forest plots and SROC curves. Heterogeneity and threshold effects were evaluated using the SROC plots and random effects correlation.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Results
A total of 16,597 records were identified in database searches; 9172 were retained after removing duplicates (Fig. 2). During title and abstract screening 8179 records were excluded. After assessing the full text of the remaining 993 records, 731 of them were excluded. Of the remaining studies, 142 are included in this review. The characteristics of included studies are described further in Table 1, and measures of diagnostic performance are described in supplementary Table 1.
Fig. 2.
Study selection
Table 1.
Characteristics of included studies: country, setting and population
| Author (year) | Country (population) | Settinga | Cases and controls | Biomarkers | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases (N) | Controls (N) | Type (N) | ||||||||||||||
| Hosp | Other | CRC | A/P | All | HC | NM | A/P | miRNA | Autoab | DNA | Protein | Metab | ctDNA | Otherb | ||
| Ahlquist et al. (2012) [23] | USA, Canada, Denmark | – | X | 82 | 0 | 960 | 0 | 0 | 0 | – | – | 1 | 1 | – | – | 4 |
| Amiot et al. (2014) [24] | France | – | X | 66 | 0 | 123 | 123 | 0 | 34 | – | – | – | – | – | – | 3 |
| Bagaria et al. (2013) [25] | India | X | – | 50 | 0 | 50 | 50 | 0 | 0 | – | – | – | 2 | – | – | – |
| Broll et al. (2001) [26] | Germany | U | U | 122 | 0 | 65 | 65 | 0 | 0 | – | – | – | 2 | – | – | – |
| Bunger et al. (2012) [27] | Germany | – | X | 83 | 60 | 50 | 50 | 0 | 0 | – | 1 | – | 2 | – | – | – |
| Calistri et al. (2009) [28] | Italy | X | – | 100 | 0 | 100 | 100 | 0 | 0 | 2 | – | – | – | – | – | – |
| Cao et al. (2019a) [29] | China | X | U | 62 | 0 | 59 | 59 | 0 | 0 | – | – | – | – | 1 | – | – |
| Cao et al. (2019b) [30] | China | X | – | 118 | 0 | 85 | 85 | 0 | 0 | – | 1 | – | 2 | 1 | – | – |
| Chan et al. (2010) [31] | Taiwan, China | X | – | 94 | 0 | 54 | 54 | 0 | 0 | – | 5 | – | 1 | – | – | – |
| Chang et al. (2016) [32] | Taiwan, China | U | U | 229 | 0 | 368 | 368 | 0 | 0 | 2 | – | – | – | – | – | – |
| Chang et al. (2011) [33] | China | X | – | 60 | 60 | 57 | 57 | 0 | 0 | – | 5 | – | 1 | – | – | – |
| Chang et al. (2014) [34] | Taiwan, China | U | U | 56 | 0 | 120 | 120 | 0 | 0 | 5 | – | – | – | – | – | – |
| Chao et al. (2013) [35] | Canada, USA | X | X | 316 | 0 | 328 | 328 | 0 | 0 | 7 | – | – | – | – | – | – |
| Chen et al. (2019) [36] | China | X | – | 111 | 0 | 114 | 114 | 0 | 0 | – | – | – | – | – | – | 3 |
| Choi et al. (2018) [37] | Korea | U | U | 69 | 0 | 74 | 74 | 0 | 0 | – | – | – | – | – | – | 11 |
| Church et al. (2014) [38] | USA, Germany | – | X | 53 | 0 | 1457 | 934 | 0 | 523 | – | – | – | – | – | – | 1 |
| Ciarloni et al. (2016) [39] | Switzerland, South Korea | X | – | 52 | 0 | 74 | 74 | 0 | 0 | 27 | – | – | 4 | – | – | – |
| DeVos et al. (2009) [40] | Germany, USA | U | U | 90 | 0 | 155 | 155 | 0 | 0 | – | – | – | – | – | – | 1 |
| Duran-Sanchon et al. (2019) [41] | Spain | – | X | 67 | 0 | 353 | 217 | 0 | 483 | 9 | – | – | – | – | – | – |
| Duvillard et al. (2014) [42] | France | X | – | 224 | 0 | 252 | 0 | 252 | 0 | – | 1 | – | 1 | – | – | – |
| Fan et al. (2017) [43] | Taiwan, China | U | U | 92 | 0 | 100 | 100 | 0 | 0 | – | 4 | – | 1 | – | – | – |
| Fernandes et al. (2005) [44] | Brazil | X | – | 169 | 169 | 100 | 100 | 0 | 0 | – | 1 | – | 1 | – | – | – |
| Flamini et al. (2006) [45] | Italy | X | – | 75 | 57 | 75 | 75 | 0 | 0 | – | – | – | 1 | – | 1 | – |
| Fouad et al. (2017) [46] | Egypt | U | U | 50 | 0 | 50 | 0 | 50 | 0 | 1 | – | – | – | – | – | – |
| Fu et al. (2018) [47] | China | X | – | 98 | 0 | 329 | 253 | 76 | 0 | – | – | – | – | – | – | 1 |
| Fung et al. (2015) [48] | Australia | X | – | 98 | 0 | 99 | 0 | 99 | 0 | – | – | – | 7 | – | – | – |
| Gao et al. (2018) [49] | China | X | – | 279 | 104 | 74 | 0 | 0 | 0 | – | – | – | 3 | – | – | – |
| Groblewska et al. (2010) [50] | Poland | X | U | 91 | 0 | 91 | 91 | 0 | 0 | – | – | – | 4 | – | – | – |
| Grutzmann et al. (2008) [51] | Germany | U | U | 127 | 0 | 369 | 184 | 149 | 0 | – | – | – | – | – | – | 1 |
| Guo et al. (2018) [52] | China | X | – | 107 | 0 | 218 | 120 | 0 | 98 | 5 | – | – | – | – | – | – |
| Han et al. (2019) [53] | South Korea | X | – | 245 | 199 | 245 | 245 | 245 | 0 | – | – | – | – | – | – | 1 |
| Hao et al. (2018) [54] | China | U | U | 186 | 0 | 97 | 97 | 0 | 0 | 2 | – | – | – | – | – | – |
| Hata et al. (2017) [55] | Japan | X | X | 225 | 0 | 916 | 916 | 0 | 0 | – | – | – | – | 1 | – | – |
| Haug et al. (2007) [56] | Germany | – | X | 65 | 0 | 917 | 917 | 0 | 0 | – | – | – | 1 | – | – | – |
| He et al. (2010) [57] | China | X | – | 182 | 0 | 170 | 170 | 0 | 0 | – | – | – | – | – | – | 3 |
| Herreros-Villanueva et al. (2019) [58] | Spain | X | – | 96 | 78 | 101 | 100 | 1 | 0 | 6 | – | – | – | – | – | – |
| Imaoka et al. (2016) [59] | Japan | X | – | 211 | 0 | 57 | 57 | 0 | 0 | 1 | – | – | – | – | – | – |
| Imperiale et al. (2014) [60] | USA, Canada | X | – | 65 | 0 | 9167 | 6274 | 0 | 2893 | 1 | – | – | – | – | – | 2 |
| Jaberie et al. (2019) [61] | Iran | X | U | 113 | 242 | 51 | 50 | 0 | 0 | – | – | – | 2 | – | – | – |
| Jensen et al. (2018) [62] | Denmark | X | – | 143 | 0 | 91 | 91 | 0 | 0 | – | – | – | – | – | 3 | – |
| Jin et al. (2015) [63] | China | X | – | 135 | 0 | 341 | 91 | 0 | 250 | 1 | – | – | – | – | – | – |
| Johnson et al. (2014) [64] | USA | U | U | 101 | 0 | 171 | 94 | 0 | 77 | – | – | – | 1 | – | – | 1 |
| Jones et al. (2016) [65] | USA | U | U | 68 | 0 | 68 | 68 | 0 | 0 | – | 1 | – | 12 | – | – | – |
| Karam et al. (2018) [66] | Egypt | X | U | 65 | 0 | 70 | 70 | 0 | 0 | – | – | – | – | – | – | 1 |
| Karl et al. (2008) [67] | Germany | X | – | 186 | 0 | 252 | 252 | 0 | 0 | – | – | – | 5 | – | – | – |
| Kim et al. (2017) [68] | Korea | X | – | 166 | 60 | 336 | 120 | 81 | 135 | – | – | – | 2 | – | – | – |
| Kim et al. (2015) [69] | Korea | X | – | 139 | 0 | 60 | 60 | 0 | 0 | – | – | – | 2 | – | – | – |
| Koga et al. (2013) [70] | Japan | X | – | 117 | 0 | 107 | 107 | 0 | 0 | 1 | – | – | 1 | – | – | – |
| Lee et al. (2013a) [71] | Korea | X | – | 101 | 0 | 96 | 96 | 0 | 0 | – | – | – | – | – | – | 1 |
| Lee et al. (2013b) [72] | Korea | X | U | 132 | 0 | 228 | 124 | 67 | 37 | – | – | – | 3 | – | – | – |
| Li et al. (2019a) [73] | China | X | – | 147 | 0 | 147 | 147 | 0 | 0 | – | – | – | 1 | – | – | – |
| Li et al. (2019b) [74] | China | X | – | 62 | 60 | 155 | 155 | 0 | 0 | – | – | – | – | – | – | 1 |
| Li et al. (2012) [75] | China | X | – | 70 | 0 | 141 | 141 | 0 | 0 | – | 1 | – | – | – | – | – |
| Liu et al. (2013) [76] | China | X | U | 200 | 0 | 80 | 80 | 0 | 0 | 2 | – | – | – | – | – | – |
| Liu et al. (2016) [77] | China | X | – | 148 | 0 | 320 | 80 | 80 | 160 | 1 | – | – | 1 | – | – | – |
| Liu et al. (2018a) [78] | China | X | – | 130 | 0 | 140 | 90 | 0 | 50 | 2 | – | – | – | – | – | – |
| Liu et al. (2018b) [79] | China | X | – | 50 | 0 | 100 | 50 | 0 | 50 | 2 | – | – | – | – | – | – |
| Lumachi et al. (2012) [80] | Italy | U | U | 102 | 0 | 99 | 0 | 99 | 0 | – | 1 | – | 4 | – | – | – |
| Luo et al. (2019) [81] | China | X | – | 57 | 0 | 192 | 192 | 0 | 0 | 12 | – | – | 1 | – | – | – |
| Marcuello et al. (2019) [82] | Spain | – | X | 59 | 0 | 80 | 80 | 0 | 0 | 6 | – | – | – | – | – | – |
| Marshall et al. (2010) [83] | Canada, USA | X | – | 202 | 0 | 208 | 0 | 0 | 0 | 7 | – | – | – | – | – | – |
| Matsubara et al. (2011) [84] | Japan | X | – | 101 | 0 | 109 | 109 | 0 | 0 | – | 1 | – | 1 | – | – | – |
| Matsushita et al. (2005) [85] | Japan | X | – | 116 | 169 | 83 | 83 | 0 | 0 | – | – | – | – | – | – | 1 |
| Melotte et al. (2015) [86] | Germany | X | – | 66 | 57 | 240 | 240 | 0 | 0 | – | – | – | – | – | – | 2 |
| Meng et al. (2012) [87] | China | – | X | 93 | 0 | 158 | 158 | 0 | 0 | – | – | – | 2 | – | – | – |
| Min et al. (2019) [88] | China | X | – | 58 | 0 | 76 | 0 | 69 | 7 | 4 | – | – | – | – | – | – |
| Mizuno et al. (2003) [89] | Japan | X | – | 100 | 0 | 100 | 100 | 0 | 0 | – | 1 | – | – | – | – | – |
| Mroczko et al. (2010) [90] | Poland | X | – | 75 | 104 | 70 | 70 | 0 | 0 | – | – | – | 4 | – | – | – |
| Mroczko et al. (2006) [91] | Poland | X | – | 76 | 0 | 65 | 65 | 0 | 0 | – | – | – | 3 | – | – | – |
| Mulder et al. (2007) [92] | Netherlands | X | – | 52 | 0 | 63 | 63 | 0 | 0 | – | – | – | 1 | – | – | – |
| Murakoshi et al. (2011) [93] | Japan | X | – | 115 | 0 | 230 | 230 | 0 | 0 | – | 1 | – | 1 | – | – | – |
| Murata et al. (2012) [94] | Japan | X | – | 252 | 199 | 81 | 81 | 0 | 0 | 9 | – | – | – | – | – | – |
| Ng et al. (2017) [95] | Hong Kong | X | – | 117 | 0 | 90 | 90 | 0 | 0 | 2 | – | – | 1 | – | – | – |
| Nielsen et al. (2011) [96] | Denmark | X | – | 294 | 0 | 4202 | 2173 | 1176 | 843 | – | – | – | 2 | – | – | – |
| Ning et al. (2018) [97] | China | X | U | 513 | 0 | 75 | 75 | 0 | 0 | – | – | – | 4 | – | – | – |
| Nishiumi et al. (2012) [98] | Japan | X | – | 59 | 0 | 63 | 63 | 0 | 0 | – | – | – | 2 | 27 | – | – |
| Niu et al. (2012) [99] | China | X | – | 119 | 78 | 397 | 300 | 97 | 0 | – | – | – | – | 1 | – | – |
| Ørntoft et al. (2015) [100] | Denmark | – | X | 128 | 0 | 150 | 150 | 0 | 0 | – | – | – | – | – | – | 1 |
| Palmqvist et al. (2003) [101] | Sweden | – | X | 124 | 0 | 243 | 0 | 0 | 0 | – | – | – | 2 | – | – | |
| Pedersen et al. (2013) [102] | England, Wales, Northern Ireland | – | X | 97 | 242 | 94 | 0 | 0 | 0 | – | 18 | – | – | – | – | – |
| Pedersen et al. (2015) [103] | Australia, Netherlands | X | – | 129 | 0 | 1288 | 450 | 542 | 296 | – | 1 | – | – | 1 | – | – |
| Peng et al. (2017) [104] | China | X | U | 559 | 0 | 559 | 559 | 0 | 0 | – | – | – | 1 | – | – | 2 |
| Pengjun et al. (2013) [105] | China | X | – | 149 | 0 | 69 | 69 | 0 | 0 | – | – | – | 4 | – | – | – |
| Qian et al. (2018) [106] | Germany | X | – | 212 | 0 | 106 | 106 | 0 | 0 | – | – | – | 5 | – | – | – |
| Qu et al. (2019) [107] | China | X | – | 100 | 0 | 100 | 100 | 0 | 0 | 5 | – | – | – | – | – | – |
| Ren et al. (2016) [108] | China | X | – | 422 | 0 | 1019 | 747 | 272 | 0 | – | 1 | – | 1 | – | – | – |
| Rho et al. (2018) [109] | Japan | X | – | 514 | 60 | 386 | 168 | 59 | 159 | – | – | – | 5 | – | – | – |
| Ritchie et al. (2010) [110] | USA, Japan | – | X | 70 | 0 | 70 | 0 | 0 | 0 | – | – | – | – | 3 | – | – |
| Ritchie et al. (2013) [111] | Canada | X | X | 98 | 0 | 964 | 0 | 964 | 0 | – | – | – | – | 1 | – | – |
| Ruffin et al. (2010) [112] | USA | U | U | 69 | 137 | 93 | 70 | 23 | 0 | – | – | – | 1 | 1 | – | – |
| Schneider et al. (2005) [113] | Germany | U | U | 247 | 0 | 53 | 0 | 0 | 0 | – | – | – | 3 | – | – | – |
| Shastri et al. (2008) [114] | Germany | U | U | 55 | 69 | 516 | 498 | 18 | 0 | – | – | – | 1 | – | – | – |
| Shastri et al. (2006) [115] | Germany | U | U | 74 | 0 | 128 | 117 | 11 | 60 | – | – | – | 1 | – | – | – |
| Shi et al. (2019) [116] | China | X | – | 211 | 0 | 103 | 0 | 0 | 103 | – | – | – | 2 | – | – | 1 |
| Sithambaram et al. (2015) [117] | Malaysia | U | U | 100 | 0 | 200 | 200 | 0 | 0 | – | – | – | 1 | – | – | – |
| Song et al. (2017a) [118] | China | X | – | 85 | 0 | 324 | 324 | 0 | 0 | – | – | – | – | – | – | 1 |
| Song et al. (2018a) [119] | Germany | X | – | 465 | 0 | 882 | 610 | 0 | 272 | – | – | – | – | – | – | 1 |
| Song et al. (2017b) [120] | China | X | – | 388 | 0 | 837 | 590 | 0 | 247 | – | – | – | – | – | – | 1 |
| Song et al. (2018b) [121] | China | X | – | 783 | 0 | 794 | 331 | 0 | 463 | – | – | – | 2 | – | – | 1 |
| Stojkovic Lalosevic et al. (2019) [122] | Serbia | – | X | 300 | 0 | 300 | 300 | 0 | 0 | – | – | – | – | – | – | 3 |
| Sun et al. (2019a) [123] | China | X | – | 133 | 0 | 587 | 494 | 11 | 82 | – | – | – | 3 | – | – | 1 |
| Sun et al. (2019b) [124] | China | U | U | 105 | 0 | 102 | 102 | 0 | 0 | – | – | 1 | 1 | – | – | 3 |
| Swellam et al. (2016) [125] | Egypt | U | U | 162 | 0 | 57 | 57 | 0 | 0 | – | – | 5 | – | – | – | |
| Symonds et al. (2016) [126] | Australia | X | – | 66 | 169 | 246 | 246 | 0 | 0 | – | 1 | – | – | 1 | – | – |
| Tagore et al. (2003) [127] | USA | – | X | 52 | 57 | 212 | 113 | 0 | 99 | – | – | 23 | – | – | – | – |
| Taguchi et al. (2015) [128] | USA | U | U | 60 | 0 | 60 | 60 | 0 | 0 | – | – | – | 10 | – | – | – |
| Tang et al. (2011) [129] | China | X | – | 169 | 0 | 139 | 30 | 0 | 109 | – | – | – | – | – | – | 1 |
| Tibble et al. (2001) [130] | UK | U | U | 62 | 0 | 233 | 0 | 233 | 0 | – | – | – | 1 | – | – | – |
| Toiyama et al. (2013) [131] | Japan | X | – | 200 | 104 | 53 | 53 | 0 | 0 | 2 | – | – | – | – | – | – |
| Tomasevic et al. (2016) [132] | Serbia | X | – | 181 | 0 | 191 | 0 | 191 | 0 | – | – | – | 2 | – | – | – |
| Toth et al. (2012) [133] | Hungary | U | U | 92 | 0 | 92 | 92 | 0 | 0 | – | – | – | 1 | – | – | 1 |
| Uchiyama et al. (2018) [134] | Japan | X | – | 56 | 0 | 60 | 60 | 0 | 0 | – | – | – | 5 | – | – | – |
| Vychytilova-Faltejskova et al. (2016) [135] | Czech Republic | – | X | 203 | 199 | 100 | 100 | 0 | 0 | 4 | – | – | – | – | – | – |
| Vychytilova-Faltejskova et al. (2018) [136] | Czech Republic | – | X | 179 | 0 | 100 | 100 | 0 | 0 | 4 | – | – | – | – | – | – |
| Wang et al. (2018) [137] | China | U | U | 96 | 0 | 60 | 60 | 0 | 0 | 1 | – | – | – | – | – | – |
| Wang et al. (2017a) [138] | China | X | – | 91 | 0 | 91 | 91 | 0 | 0 | – | – | – | 10 | – | – | – |
| Wang et al. (2014) [139] | China | X | U | 113 | 0 | 59 | 59 | 0 | 0 | 6 | – | – | 2 | – | – | – |
| Wang et al. (2007) [140] | Taiwan | X | U | 157 | 78 | 80 | 80 | 0 | 0 | 4 | – | – | – | – | – | – |
| Wang et al. (2017b) [141] | China | X | – | 60 | 0 | 50 | 50 | 0 | 0 | 1 | – | – | 2 | – | – | – |
| Wang et al. (2016) [142] | China | X | – | 120 | 0 | 120 | 120 | 0 | 0 | 4 | – | – | 1 | – | – | – |
| Warren et al. (2011) [143] | USA and Russia | X | X | 50 | 242 | 94 | 0 | 0 | 0 | – | – | – | – | – | – | 1 |
| Wilhelmsen et al. (2017) [144] | Denmark | X | – | 512 | 0 | 3320 | 1978 | 1342 | 0 | – | 1 | – | 6 | 1 | – | – |
| Wu et al. (2014) [145] | Hong Kong, China | X | – | 104 | 0 | 109 | 109 | 0 | 0 | 1 | – | – | – | – | – | – |
| Wu et al. (2012) [146] | Hong Kong | X | – | 88 | 0 | 101 | 101 | 0 | 0 | 2 | – | – | – | – | – | – |
| Wu et al. (2017) [147] | China | X | – | 135 | 0 | 140 | 140 | 0 | 0 | – | – | – | 1 | – | – | – |
| Wu et al. (2015) [148] | China | – | X | 100 | 0 | 100 | 100 | 0 | 0 | – | – | 2 | – | – | – | – |
| Wu et al. (2016) [149] | China | X | – | 291 | 0 | 295 | 295 | 0 | 0 | – | – | – | 2 | – | – | 1 |
| Xie et al. (2017) [150] | China | X | – | 132 | 60 | 107 | 0 | 0 | 0 | – | – | – | 2 | – | – | – |
| Xie et al. (2018) [151] | China | X | – | 123 | 0 | 125 | 19 | 106 | 0 | – | – | – | 3 | – | – | 1 |
| Xu et al. (2013) [152] | China | X | X | 87 | 0 | 73 | 73 | 0 | 0 | 2 | 1 | 14 | 1 | – | – | – |
| Yang et al. (2018) [153] | China | U | U | 50 | 0 | 50 | 0 | 0 | 0 | – | – | – | 2 | 1 | – | – |
| Yau et al. (2016) [154] | Hong Kong | U | U | 198 | 0 | 198 | 198 | 0 | 0 | 1 | – | – | – | – | – | – |
| Yuan et al. (2016) [155] | China | X | – | 187 | 0 | 109 | 109 | 0 | 0 | – | – | – | 5 | – | – | 1 |
| Zhang et al. (2015a) [156] | China | X | – | 138 | 60 | 111 | 0 | 46 | 65 | – | – | – | 2 | – | – | – |
| Zhang et al. (2016) [157] | China | X | – | 80 | 0 | 171 | 116 | 55 | 0 | – | – | – | – | 6 | – | – |
| Zhang et al. (2015b) [158] | Japan | U | U | 130 | 0 | 54 | 54 | 0 | 0 | 4 | – | – | – | – | – | – |
| Zhao et al. (2019a) [159] | China | X | – | 117 | 0 | 166 | 166 | 0 | 0 | – | – | – | – | – | – | 2 |
| Zhao et al. (2019b) [160] | China | X | – | 358 | 0 | 286 | 286 | 0 | 0 | – | – | – | 1 | – | – | – |
| Zheng et al. (2014) [161] | China | X | – | 117 | 0 | 175 | 102 | 0 | 73 | 4 | – | – | 1 | – | – | – |
| Zhou et al. (2017) [162] | China | X | – | 242 | 0 | 262 | 262 | 0 | 0 | – | – | – | – | – | – | 1 |
| Zhu et al. (2013) [163] | China | X | U | 269 | 0 | 110 | 110 | 0 | 0 | – | – | – | 5 | – | – | – |
| Zhu et al. (2015) [164] | China | X | – | 70 | 0 | 70 | 70 | 0 | 0 | 1 | – | – | 1 | – | – | – |
autoab autoantibodies and other immunological markers, ctDNA circulating tumour DNA, HC healthy control, Hosp hospital, Ind individual, NM non-malignant, A/P adenomas/polyps, U unclear, UK United Kingdom, USA United States of America
aDue to wide variations in health systems across different countries, hospital setting is a broad definition than can encompass secondary and tertiary care. Other setting refers to biobanks, reference sets, databases or archived samples; general population cohorts or cohorts from population screening programmes; or cohorts from previous trials or observational studies
bOther biomarker type refers to methylation markers, platelets, white blood cells, red blood cells and colonocytes
Characteristics of Included Studies
Most papers (n = 124) recruited patients from a single country. China was the most common country (n = 62), followed by Japan and Germany (both n = 14), and the USA (n = 13). Most studies recruited from single settings with few studies recruiting from at least two different settings (n = 11). The most common recruitment settings were hospitals and other secondary care settings (n = 106). Only one study recruited controls from a primary care setting [57]. All included studies reported on CRC, with six studies specifically referring to colon cancer, and one study specifying rectal and caecum cancer cases separately to colon cancer. Some studies (n = 22) also referred to adenomas or polyps as cases, and five studies also included data on upper GI cancers (e.g. gastric, oesophageal and pancreatic cancers).
Characteristics of Cases and Controls
Overall, the included studies reported on 24,844 cases; the majority were diagnosed with CRC (80.2%) and a minority with adenomas/polyps (19.8%). Most cases had their age reported (79%), either as a range, mean or median. The overall mean age for CRC cases was 61.3 years, and 60.7 years for adenoma/polyp cases. The minimum age for cases was 18 years, while the oldest was 97 years old. The majority (59%) of CRC and adenoma/polyp cases were male. Most studies provided data on tumour staging, mainly using the TNM system (n = 101), though some studies used Dukes’ classification (n = 22), with one study providing data for both. When combining TNM and Dukes’ staging data, over half of the cancers (54%) were diagnosed at early stages (I–II/A + B). Adenomas included as cases were most frequently defined by size, dysplasia, villous component and/or number of adenomas.
The included studies reported on a total of 45,374 controls (31,352 normal/healthy, 6414 with non-malignant conditions and 7608 with adenomas or polyps). A number of studies (n = 37) investigated more than one type of control population. The control populations of most studies (n = 108) were tested to rule out CRC, mainly using colonoscopy (n = 65). The majority of studies (n = 17) with adenomas or polyps as controls included those that were low risk (hyperplastic, non-neoplastic polyps or non-advanced adenomas), though some were high risk (advanced adenomas, those with villous histology or high-grade dysplasia). Age data were extractable for 47.1% of controls. The minimum age for a control was 16 years (healthy control), while the oldest was 99 years old. The majority of both healthy (50.6%) and non-malignant (58%) controls were male.
Types of Biomarkers
Most studies investigated more than one biomarker (79.6%), and these often reported on measures of performance for individual and combinations or panels of biomarkers (45.8%). The commonest sample source was blood (82.4%); these analysed serum (n = 62), plasma (n = 41) or whole blood (n = 14). Faeces was also a common sample source (24.6%); two studies analysed urine, and 13 studies analysed more than one type of sample.
A total of 378 unique biomarkers were identified across the 142 included studies (Appendix 4 in the supplementary material). The commonest biomarkers were microRNAs and other RNAs, followed by proteins, DNA markers, autoantibodies and other immunological markers, and metabolic markers. Proteins were further classified into subcategories, with the most common being novel proteins (Table 2).
Table 2.
Classification of identified biomarkers
| Identified biomarkers (142 studies) | N (%) |
|---|---|
| MicroRNAs and other RNAs | 126 (33.3%) |
| DNA markers (protein coding genes, mutations) | 45 (12.2%) |
| Proteins | 86 (22.8%) |
| Adhesion and matrix proteins | 11 (2.9%) |
| Classic tumour markers | 6 (1.6%) |
| Coagulation and angiogenesis molecules | 6 (1.6%) |
| Cytokines, chemokines and insulin-like growth factors | 15 (4.0%) |
| Hormones | 1 (0.3%) |
| Novel proteins | 39 (10.3%) |
| Not otherwise specified | 8 (2.1%) |
| Autoantibodies and other immunological markers | 44 (11.6%) |
| Metabolic markers | 42 (11.1%) |
| Circulating tumour DNA | 4 (1.1%) |
| DNA methylation | 15 (4.0%) |
| Other biomarkersa | 16 (4.2%) |
aOther biomarkers included platelets, white blood cells, red blood cells and colonocytes
A total of 54 biomarkers were reported in more than one study (Appendix 5 in the supplementary material). Three biomarkers were investigated by more than 10 studies: CA19-9, CEA and mSEPT9 (methylated septin 9). Additionally, six other biomarkers were investigated in five or more studies: tumour pyruvate kinase isoenzyme type M2 (TuM2-PK), microRNA-21 (miR-21), FIT, microRNA-92a (miR-92a), cancer antigen 72-4 (CA72-4) and TIMP metallopeptidase inhibitor 1 (TIMP-1) (see Appendix 5 for references).
Measures of Diagnostic Performance
Individual measures of diagnostic performance (i.e. measures outside of combinations or panels) were available for 35 biomarkers evaluated more than once (Appendix 5 in the supplementary material). Heterogeneity of study design and included populations precluded meta-analysis for the majority of these biomarkers; however, three had individual measures from multiple studies adopting a classic single-gate design: CEA (n = 7 studies), mSEPT9 (n = 4 studies) and TuM2-PK (n = 3 studies). Differences in the sample sources and diagnostic performance measures provided across the studies precluded meta-analysis for any accuracy measures available for CEA, which was included as a comparator to the novel markers. Meta-analysis was performed for the markers mSEPT9 and TuM2-PK.
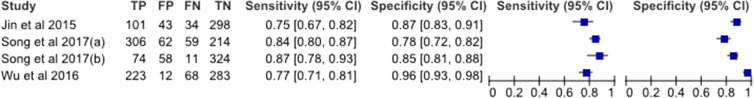
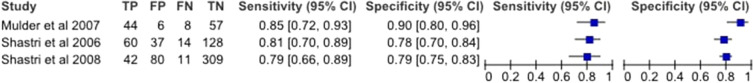
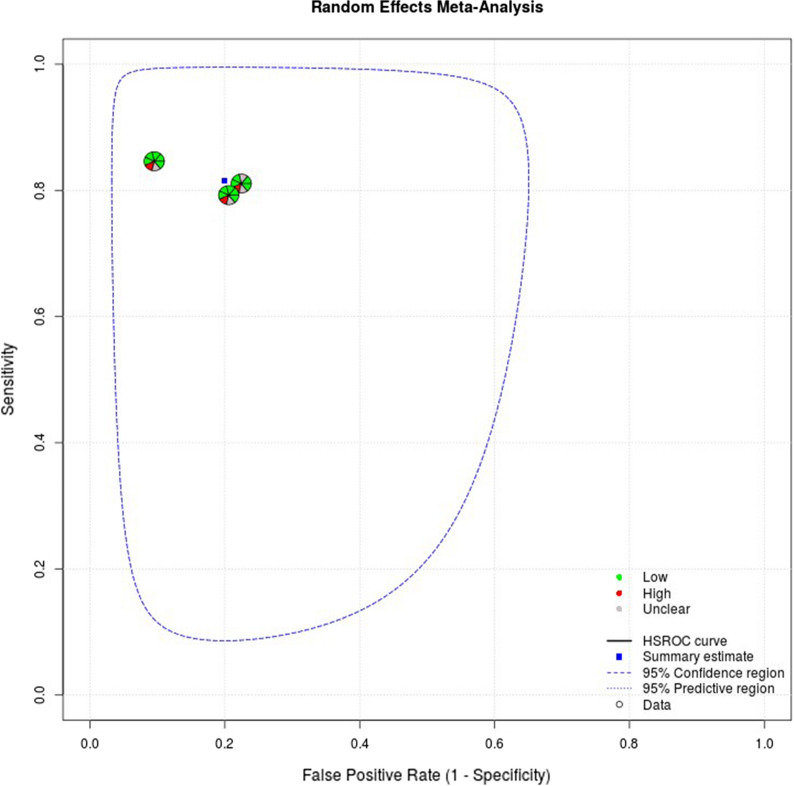
The estimated sensitivity and specificity of mSEPT9 was 80.6% (95% CI 76.6–84.0%) and 88.0% (95% CI 79.1–93.4%), respectively, and the diagnostic odds ratio was 30.3 (95% CI 17.8–51.4). TuM2-PK had an estimated sensitivity of 81.6% (95% CI 75.2–86.6%) and a specificity of 80.1% (95% CI 76.7–83.0%), and a diagnostic odds ratio of 17.8 (95% CI 11.6–27.2). Paired forest plots of the sensitivity and specificity for both mSEPT9 and TuM2-PK are shown in Figs. 3 and 4.
Fig. 3.
Forest plots of sensitivity and specificity for mSEPT9 in plasma
Fig. 4.
Forest plots of sensitivity and specificity for TuM2-PK in stool
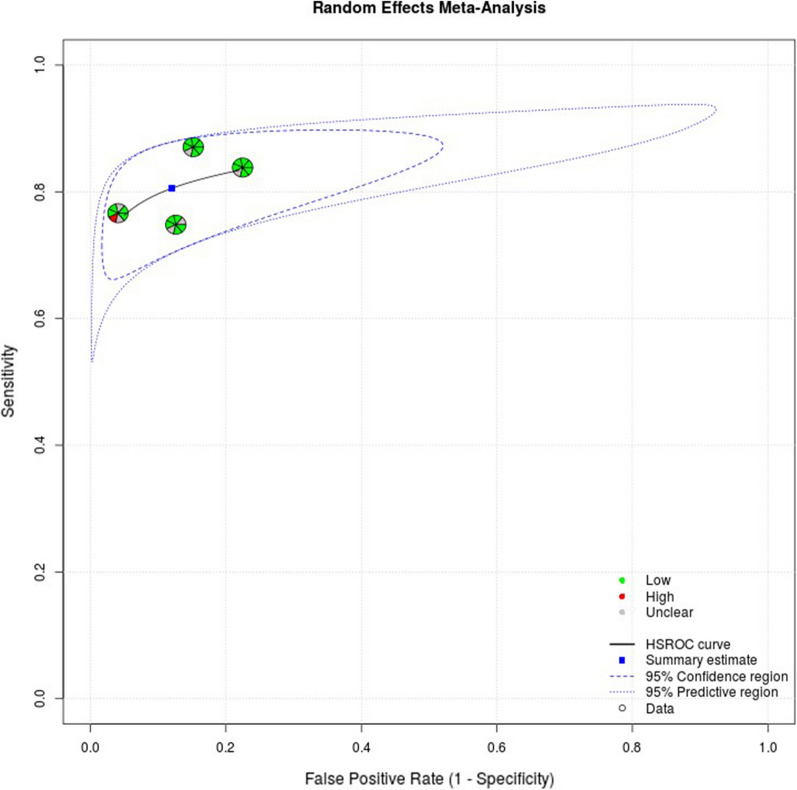
The random effects correlation for mSEPT9 was − 1, indicating a significant threshold effect. Heterogeneity and threshold effect were harder to evaluate statistically for the meta-analysis of TuM2-PK as the low number of included studies impeded accurate fitting of the HSROC curve and generation of a random effects correlation. A cut-off value of 4 U/ml was used for the TuM2-PK assays across all studies. The studies included in the meta-analyses were at low risk of bias across most domains, except for the domains related to patient selection and the index test. Full appraisal data can be found in Appendix 6 in the supplementary material. Summary plots including risk of bias and applicability ratings from QUADAS-2 are shown in Figs. 5 and 6.
Fig. 5.
HSROC curve for mSEPT9 (with risk of bias and applicability ratings)
Fig. 6.
HSROC plot for TuM2-PK (with risk of bias and applicability ratings)
Discussion
This systematic review identified 142 studies reporting on 378 different biomarkers for CRC. The included papers were very heterogeneous, with differences in study design, control populations, sample sources, types of biomarkers, test thresholds and reported performance measures. Meta-analysis of diagnostic accuracy data was only possible for two novel markers: mSEPT9 and TuM2-PK. Both demonstrated high sensitivity, specificity and diagnostic odds ratios in hospital populations.
The most common biomarkers (both individually and in panels) were CEA, CA19-9, mSEPT9 and TuM2-PK. CEA and CA19-9 have a more established role in clinical practice for detecting recurrent disease [3, 7] so it is not surprising that these markers are prevalent throughout the literature. Most of the studies included CEA (42/53) and CA19-9 (20/21) in panels or used them as comparators for novel markers. Meta-analysis was not possible for these studies because of heterogeneity in sample sources and performance measures. Twenty studies reported on the performance of mSEPT9 for CRC detection, mostly as a blood-based biomarker sampled from plasma. While most measures of diagnostic performance were for mSEPT9 as an individual marker, it was also included in panels or combinations across seven studies. Fewer studies reported on the performance of TuM2-PK (nine overall, three included TuM2-PK in panels or combinations). Unlike mSEPT9, TuM2-PK was predominantly sampled from stool, though some studies also reported it as a blood-based biomarker. The studies that evaluated mSEPT9 and TuM2-PK included a number of two-gate studies or hybrid designs, and multiple instances where the study design was unclear. The meta-analyses for mSEPT9 and TuM2-PK included only those with a clear, classic single-gate design [11] to reduce heterogeneity and spectrum bias; consequently, both meta-analyses included a low number of studies, resulting in wide confidence intervals for the diagnostic odds ratios.
The meta-analysis for mSEPT9 synthesised diagnostic performance data on 899 CRC cases. Cancer cases were mostly diagnosed in stages II or III and diagnostic performance data were also provided for adenomas and polyps in most cases. This is important to note, as the diagnostic performance results for early stage cancers are more likely to translate for use in early detection, and the ability for biomarkers also to detect high-risk adenomas, polyps or dysplasia could provide additional clinical utility. Across all studies, the test sensitivity was higher when detecting advanced CRC cases. Conversely, test sensitivity decreased when used to detect either adenomas or degrees of dysplasia. As previously mentioned, several studies evaluated mSEPT9 within diagnostic panels or in combination with other markers. Three studies in particular [66, 151, 153] showed the sensitivity of mSEPT9 to detect CRC increased when combined with more established markers such as FIT and CEA. The results from our review show a slightly higher sensitivity for mSEPT9 in comparison to a recent meta-analysis of 19 studies [165] though it should be noted the analysis from that review included a mixture of study designs and focused on high-risk populations. Our results are comparable to previous analyses which estimated the sensitivity of mSEPT9 as up to 88% [165, 166].
The meta-analysis of diagnostic accuracy data for TuM2-PK as a stool marker included 183 CRC cases. Similarly to mSEPT9, the sensitivity of TuM2-PK was higher for more advanced cancers (Dukes’ stage C and D; stages III and IV) and lower when it was used to detect adenomas, polyps or dysplasia. All three studies included in the meta-analysis compared the diagnostic performance of TuM2-PK to the established stool marker FIT, and demonstrated that FIT was preferable to TuM2-PK as a faecal biomarker for screening populations [94, 116, 117]. Three studies [50, 67, 115] evaluated TuM2-PK as a blood-based biomarker in combination with other markers or in panels, and all found sensitivity to be higher for TuM2-PK in combination with other markers. TuM2-PK may therefore be more promising in blood-based diagnostic panels than as a stand-alone stool marker.
Two-gate and hybrid designs were used widely in the included studies. These types of study designs can lead to over-inflated measures of diagnostic performance due to an over-representation of individuals with advanced disease within the study population [11]. While many studies attempted single-gate designs and recruited participants through one route (usually screening populations where all participants attended for a colonoscopy), the low prevalence of CRC cases meant that extra cases were sourced from alternative routes. This study design issue highlights the importance of large-scale studies and trials that are adequately powered to evaluate diagnostic performance in truly low-prevalence populations.
Several other methodological limitations were identified across the studies. These included the parallel analysis of large numbers of biomarkers during discovery studies; limited external, independent validation of test performance; and selective reporting for validation including alternative analyses and combinations or use of several cut-off points. Insufficient reporting regarding population characteristics and recruitment was also an issue in many studies, with information often provided as supplementary data and with little detail. As a result of the large amount of evidence on biomarker development and evaluation, we believe the field could benefit from a “living systematic review”; this refers to high-quality, up-to-date online summaries of evidence which can be constantly updated as new research becomes available [167].
Although our search was restricted to studies published in English, recent reviews indicate that this has minimal impact on review conclusions [168, 169]. Further limitations of this review include the exclusion of studies that evaluated biomarkers within risk assessment tools or risk prediction models. These studies have strong potential to be used in the community; however, we believe they should be investigated in a separate systematic review. The heterogeneity of the published literature meant we could only conduct meta-analyses on a limited subset of included studies. Nonetheless, we believe the narrative synthesis of additional studies provides a useful summary of the current state of the science in this area. There was insufficient homogeneous data on biomarker panels to report summary estimates of their diagnostic performance. A study from Fung et al. [48] describes ColoSTAT, a novel blood-based diagnostic panel for CRC that includes TuM2-PK with two other biomarkers (IL-8 and DKK-3) and is currently being trialled in Australia. The ColoSTAT panel has reported sensitivity and specificity of 73% and 95%, respectively, for CRC, which is comparable to reported values for FIT (64–73% and 92–95%, respectively [170–173]) for the detection of CRC in screening populations. Previous trials using this panel have been conducted in high-prevalence settings, with two-gate designs. Once further data are available on ColoSTAT and its performance to detect early stage CRC, it may have applicability in low-prevalence settings as an alternative to FIT, either for screening or in symptomatic populations.
Conclusion
There is a large body of evidence on novel biomarkers being developed to aid with the early detection of lower GI cancers. Few of these markers have yet demonstrated their validity or clinical utility, but two show promise for further evaluation, mSEPT9 and TuM2-PK, and could contribute towards the early detection of CRC as part of blood-based diagnostic panels. Further, large-scale studies in low-prevalence populations are required to evaluate their potential role to support diagnostic assessment in primary care and community settings. This review offers a comprehensive overview of the current state of evidence, situates it within a translational framework for diagnostic tests and makes recommendations in order to build the evidence base for the early detection of lower GI cancers in low-prevalence settings.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Funding
This study was supported by the CanTest Collaborative (funded by Cancer Research UK C8640/A23385) of which Fiona M. Walter is Director, Jon Emery is an Associate Director, Mike Messenger is co-investigator, and Natalia Calanzani and Garth Funston are researchers. The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. Paige Druce, Kristi Milley and Jon Emery are supported by the Cancer Australia Primary Care Collaborative Cancer Clinical Trials Group (PC4). Mike Messenger is funded by the NIHR Leeds In Vitro Diagnostic Co-operative (UK). No Rapid Service Fee or Open Access fee was received by the journal for the publication of this article.
Other Assistance
We thank Veronica Phillips, Assistant Librarian, University of Cambridge Medical Library, and Jim Berryman, Liaison Librarian, Brownless Biomedical Library University of Melbourne for expert input when developing the search strategy.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Disclosures
Mike Messenger has previously held research grants with Roche Diagnostics, Everest Detection Ltd, SomaLogic Inc, Abbott Laboratories, Myriad Genetics, Inivata, Oncimmune, Janssen and Siemens Healthineers and has also received paid consultancy from PinPoint Data Science Ltd and Cepheid Inc in the field of cancer diagnostics. Garth Funston is a current member of the Oncology Editorial Board for Advances in Therapy. Paige Druce, Natalia Calanzani, Claudia Snudden, Kristi Milley, Rachel Boscott, Dawnya Behiyat, Javiera Martinez-Gutierrez, Smiji Saji, Jasmeen Oberoi, Jon Emery and Fiona M Walter have nothing to disclose.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
- 1.Arnold M, Abnet CC, Neale RE, et al. Global burden of 5 major types of gastrointestinal cancer. Gastroenterology. 2020;159(1):335–349. doi: 10.1053/j.gastro.2020.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emery JD, Shaw K, Williams B, et al. The role of primary care in early detection and follow-up of cancer. Nat Rev Clin Oncol. 2014;11(1):38–48. doi: 10.1038/nrclinonc.2013.212. [DOI] [PubMed] [Google Scholar]
- 3.Rubin G, Walter F, Emery J, de Wit N. Reimagining the diagnostic pathway for gastrointestinal cancer. Nat Rev Gastroenterol Hepatol. 2018;15(3):181–188. doi: 10.1038/nrgastro.2018.1. [DOI] [PubMed] [Google Scholar]
- 4.Emery JD, Pirotta M, Macrae F, et al. ‘Why don't I need a colonoscopy?’ A novel approach to communicating risks and benefits of colorectal cancer screening. Aust J Gen Pract. 2018;47(6):343–349. doi: 10.31128/AJGP-11-17-4386. [DOI] [PubMed] [Google Scholar]
- 5.Maringe C, Spicer J, Morris M, et al. The impact of the COVID-19 pandemic on cancer deaths due to delays in diagnosis in England, UK: a national, population-based, modelling study. Lancet Oncol. 2020;21(8):1023–1034. doi: 10.1016/S1470-2045(20)30388-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pereira SP, Oldfield L, Ney A, et al. Early detection of pancreatic cancer. Lancet Gastroenterol Hepatol. 2020;5(7):698–710. doi: 10.1016/S2468-1253(19)30416-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall C, Clarke L, Pal A, et al. A review of the role of carcinoembryonic antigen in clinical practice. Ann Coloproctol. 2019;35(6):294–305. doi: 10.3393/ac.2019.11.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ioannidis JPA, Bossuyt PMM. Waste, leaks, and failures in the biomarker pipeline. Clin Chem. 2017;63(5):963–972. doi: 10.1373/clinchem.2016.254649. [DOI] [PubMed] [Google Scholar]
- 9.Usher-Smith JA, Sharp SJ, Griffin SJ. The spectrum effect in tests for risk prediction, screening, and diagnosis. BMJ. 2016;353:i3139. doi: 10.1136/bmj.i3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walter FM, Thompson MJ, Wellwood I, et al. Evaluating diagnostic strategies for early detection of cancer: the CanTest framework. BMC Cancer. 2019;19(1):586. doi: 10.1186/s12885-019-5746-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rutjes AW, Reitsma JB, Vandenbroucke JP, Glas AS, Bossuyt PM. Case-control and two-gate designs in diagnostic accuracy studies. Clin Chem. 2005;51(8):1335–1341. doi: 10.1373/clinchem.2005.048595. [DOI] [PubMed] [Google Scholar]
- 12.Calanzani N, Druce PE, Snudden C, et al. Identifying novel biomarkers ready or evaluation in low-prevalence populations for the early detection of upper gastrointestinal cancers: a systematic review. Adv Ther. 2021;38:793–834. doi: 10.1007/s12325-020-01571-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moher D, Liberati A, Tetzlaff J, Altman DG, The PG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93(14):1054–1061. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- 15.Uttley L, Whiteman BL, Woods HB, Harnan S, Philips ST, Cree IA. Building the evidence base of blood-based biomarkers for early detection of cancer: a rapid systematic mapping review. EBioMedicine. 2016;10:164–173. doi: 10.1016/j.ebiom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vukobrat-Bijedic Z, Husic-Selimovic A, Sofic A, et al. Cancer antigens (CEA and CA 19–9) as markers of advanced stage of colorectal carcinoma. Med Arch. 2013;67(6):397–401. doi: 10.5455/medarh.2013.67.397-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkin C, Bell S, Mirbagheri N. Colorectal cancer screening in Australia. Aust J Gen Pract. 2018;47:859–863. doi: 10.31128/AJGP-01-18-4472. [DOI] [PubMed] [Google Scholar]
- 18.Covidence systematic review software. Melbourne, Australia. VHI.
- 19.Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 20.Popay J, Roberts H, Sowden A, et al., editors. Guidance on the conduct of narrative synthesis in systematic reviews. A product from the ESRC Methods Programme. Version 12006.
- 21.Freeman SC, Kerby CR, Patel A, Cooper NJ, Quinn T, Sutton AJ. Development of an interactive web-based tool to conduct and interrogate meta-analysis of diagnostic test accuracy studies: MetaDTA. BMC Med Res Methodol. 2019;19(1):81. doi: 10.1186/s12874-019-0724-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Review Manager (RevMan) [Computer program]. Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2014.
- 23.Ahlquist DA, Zou H, Domanico M, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012;142(2):248–256. doi: 10.1053/j.gastro.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amiot A, Mansour H, Baumgaertner I, et al. The detection of the methylated Wif-1 gene is more accurate than a fecal occult blood test for colorectal cancer screening. PLoS One. 2014;9(7):e99233. doi: 10.1371/journal.pone.0099233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagaria B, Sood S, Sharma R, Lalwani S. Comparative study of CEA and CA19-9 in esophageal, gastric and colon cancers individually and in combination (ROC curve analysis) Cancer Biol Med. 2013;10(3):148–157. doi: 10.7497/j.issn.2095-3941.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broll R, Erdmann H, Duchrow M, et al. Vascular endothelial growth factor (VEGF)—a valuable serum tumour marker in patients with colorectal cancer? Eur J Surg Oncol. 2001;27(1):37–42. doi: 10.1053/ejso.2000.1052. [DOI] [PubMed] [Google Scholar]
- 27.Bunger S, Haug U, Kelly M, et al. A novel multiplex-protein array for serum diagnostics of colon cancer: a case-control study. BMC Cancer. 2012;12:393. doi: 10.1186/1471-2407-12-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calistri D, Rengucci C, Molinari C, et al. Quantitative fluorescence determination of long-fragment DNA in stool as a marker for the early detection of colorectal cancer. Cell Oncol. 2009;31(1):11–17. doi: 10.3233/CLO-2009-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao H, Wang Q, Gao Z, Xu X, Lu Q, Wu Y. Clinical value of detecting IQGAP3, B7-H4 and cyclooxygenase-2 in the diagnosis and prognostic evaluation of colorectal cancer. Cancer Cell Int. 2019;19:163. doi: 10.1186/s12935-019-0881-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao LL, Liu HQ, Yue ZH, Pei L, Wang H, Jia M. Ferritin is a potential tumor marker for colorectal cancer and modulates histone methylation in colorectal cancer cells. Int J Clin Exp Med. 2019;12(6):7200–7208. [Google Scholar]
- 31.Chan C-C, Fan C-W, Kuo Y-B, et al. Multiple serological biomarkers for colorectal cancer detection. Int J Cancer. 2010;126(7):1683–1690. doi: 10.1002/ijc.24912. [DOI] [PubMed] [Google Scholar]
- 32.Chang PY, Chen CC, Chang YS, et al. MicroRNA-223 and microRNA-92a in stool and plasma samples act as complementary biomarkers to increase colorectal cancer detection. Oncotarget. 2016;7(9):10663–10675. doi: 10.18632/oncotarget.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang W, Wu L, Cao F, et al. Development of autoantibody signatures as biomarkers for early detection of colorectal carcinoma. Clin Cancer Res. 2011;17(17):5715–5724. doi: 10.1158/1078-0432.CCR-11-0199. [DOI] [PubMed] [Google Scholar]
- 34.Chang YT, Huang CS, Yao CT, et al. Gene expression profile of peripheral blood in colorectal cancer. World J Gastroenterol. 2014;20(39):14463–14471. doi: 10.3748/wjg.v20.i39.14463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chao S, Ying J, Liew G, Marshall W, Liew CC, Burakoff R. Blood RNA biomarker panel detects both left- and right-sided colorectal neoplasms: a case–control study. J Exp Clin Cancer Res. 2013;32:44. doi: 10.1186/1756-9966-32-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y, Wang Z, Zhao G, et al. Performance of a novel blood-based early colorectal cancer screening assay in remaining serum after the blood biochemical test. Dis Mark. 2019;2019:5232780. doi: 10.1155/2019/5232780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi J, Maeng HG, Lee SJ, et al. Diagnostic value of peripheral blood immune profiling in colorectal cancer. Ann Surg Treat Res. 2018;94(6):312–321. doi: 10.4174/astr.2018.94.6.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Church TR, Wandell M, Lofton-Day C, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63(2):317–325. doi: 10.1136/gutjnl-2012-304149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ciarloni L, Ehrensberger SH, Imaizumi N, et al. Development and clinical validation of a blood test based on 29-gene expression for early detection of colorectal cancer. Clin Cancer Res. 2016;22(18):4604–4611. doi: 10.1158/1078-0432.CCR-15-2057. [DOI] [PubMed] [Google Scholar]
- 40.deVos T, Tetzner R, Model F, et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem. 2009;55(7):1337–1346. doi: 10.1373/clinchem.2008.115808. [DOI] [PubMed] [Google Scholar]
- 41.Duran-Sanchon S, Moreno L, Auge JM, et al. Identification and validation of microRNA profiles in fecal samples for detection of colorectal cancer. Gastroenterology. 2019;14:14. doi: 10.1053/j.gastro.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 42.Duvillard L, Ortega-Deballon P, Bourredjem A, et al. A case-control study of pre-operative levels of serum neutrophil gelatinase-associated lipocalin and other potential inflammatory markers in colorectal cancer. BMC Cancer. 2014;14:912. doi: 10.1186/1471-2407-14-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan CW, Kuo YB, Lin GP, et al. Development of a multiplexed tumor-associated autoantibody-based blood test for the detection of colorectal cancer. Clin Chim Acta. 2017;475:157–163. doi: 10.1016/j.cca.2017.10.022. [DOI] [PubMed] [Google Scholar]
- 44.Fernandes LC, Kim SB, Matos D. Cytokeratins and carcinoembryonic antigen in diagnosis, staging and prognosis of colorectal adenocarcinoma. World J Gastroenterol. 2005;11(5):645–648. doi: 10.3748/wjg.v11.i5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flamini E, Mercatali L, Nanni O, et al. Free DNA and carcinoembryonic antigen serum levels: an important combination for diagnosis of colorectal cancer. Clin Cancer Res. 2006;12(23):6985–6988. doi: 10.1158/1078-0432.CCR-06-1931. [DOI] [PubMed] [Google Scholar]
- 46.Fouad H, Sabry D, Morsi H, Shehab H, Abuzaid NF. XRCC1 gene polymorphisms and miR-21 expression in patients with colorectal carcinoma. Eurasian J Med. 2017;49(2):132–136. doi: 10.5152/eurasianjmed.2017.17021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu B, Yan P, Zhang S, et al. Cell-free circulating methylated SEPT9 for noninvasive diagnosis and monitoring of colorectal cancer. Dis Mark. 2018;2018:6437104. doi: 10.1155/2018/6437104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fung KY, Tabor B, Buckley MJ, et al. Blood-based protein biomarker panel for the detection of colorectal cancer. PLoS One. 2015;10(3):e0120425. doi: 10.1371/journal.pone.0120425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao L, He SB, Li DC. Effects of miR-16 plus CA19-9 detections on pancreatic cancer diagnostic performance. Clin Lab. 2014;60(1):73–77. doi: 10.7754/clin.lab.2013.121210. [DOI] [PubMed] [Google Scholar]
- 50.Groblewska M, Mroczko B, Gryko M, Kedra B, Szmitkowski M. Matrix metalloproteinase 2 and tissue inhibitor of matrix metalloproteinases 2 in the diagnosis of colorectal adenoma and cancer patients. Folia Histochem Cytobiol. 2010;48(4):564–571. doi: 10.2478/v10042-010-0076-1. [DOI] [PubMed] [Google Scholar]
- 51.Grutzmann R, Molnar B, Pilarsky C, et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS One. 2008;3(11):e3759. doi: 10.1371/journal.pone.0003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo S, Zhang J, Wang B, et al. A 5-serum miRNA panel for the early detection of colorectal cancer. OncoTargets Ther. 2018;11:2603–2614. doi: 10.2147/OTT.S153535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han YD, Oh TJ, Chung TH, et al. Early detection of colorectal cancer based on presence of methylated syndecan-2 (SDC2) in stool DNA. Clin Epigenet. 2019;11(1):51. doi: 10.1186/s13148-019-0642-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hao JP, Ma A. The ratio of miR-21/miR-24 as a promising diagnostic and poor prognosis biomarker in colorectal cancer. Eur Rev Med Pharmacol Sci. 2018;22(24):8649–8656. doi: 10.26355/eurrev_201812_16629. [DOI] [PubMed] [Google Scholar]
- 55.Hata T, Takemasa I, Takahashi H, et al. Downregulation of serum metabolite GTA-446 as a novel potential marker for early detection of colorectal cancer. Br J Cancer. 2017;117(2):227–232. doi: 10.1038/bjc.2017.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haug U, Rothenbacher D, Wente MN, Seiler CM, Stegmaier C, Brenner H. Tumour M2-PK as a stool marker for colorectal cancer: comparative analysis in a large sample of unselected older adults vs colorectal cancer patients. Br J Cancer. 2007;96(9):1329–1334. doi: 10.1038/sj.bjc.6603712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He CZ, Zhang KH, Li Q, Liu XH, Hong Y, Lv NH. Combined use of AFP, CEA, CA125 and CAl9-9 improves the sensitivity for the diagnosis of gastric cancer. BMC Gastroenterol. 2013;13:87. doi: 10.1186/1471-230X-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Herreros-Villanueva M, Duran-Sanchon S, Martin AC, et al. Plasma microRNA signature validation for early detection of colorectal cancer. Clin Transl Gastroenterol. 2019;10(1):e00003. doi: 10.14309/ctg.0000000000000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Imaoka H, Toiyama Y, Fujikawa H, et al. Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann Oncol. 2016;27(10):1879–1886. doi: 10.1093/annonc/mdw279. [DOI] [PubMed] [Google Scholar]
- 60.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370(14):1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 61.Jaberie H, Hosseini SV, Naghibalhossaini F. Evaluation of alpha 1-antitrypsin for the early diagnosis of colorectal cancer. Pathol Oncol Res. 2019;10:10. doi: 10.1007/s12253-019-00679-0. [DOI] [PubMed] [Google Scholar]
- 62.Jensen SO, Orntoft MBW, Ogaard N, et al. Novel DNA methylation biomarkers show high sensitivity and specificity for blood-based detection of colorectal cancer: a clinical biomarker discovery and validation study. Cancer Res. 2018;78(13):2. doi: 10.1186/s13148-019-0757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin P, Kang Q, Wang X, et al. Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm. J Gastroenterol Hepatol. 2015;30(5):830–833. doi: 10.1111/jgh.12855. [DOI] [PubMed] [Google Scholar]
- 64.Johnson DA, Barclay RL, Mergener K, et al. Plasma Septin9 versus fecal immunochemical testing for colorectal cancer screening: a prospective multicenter study. PLoS One. 2014;9(6):e98238. doi: 10.1371/journal.pone.0098238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones JJ, Wilcox BE, Benz RW, et al. A plasma-based protein marker panel for colorectal cancer detection identified by multiplex targeted mass spectrometry. Clin Colorectal Cancer. 2016;15(2):186–194. doi: 10.1016/j.clcc.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karam RA, Zidan HE, Abd Elrahman TM, Badr SA, Amer SA. Study of p16 promoter methylation in Egyptian colorectal cancer patients. J Cell Biochem. 2018;28:28. doi: 10.1002/jcb.28146. [DOI] [PubMed] [Google Scholar]
- 67.Karl J, Wild N, Tacke M, et al. Improved diagnosis of colorectal cancer using a combination of fecal occult blood and novel fecal protein markers. Clin Gastroenterol Hepatol. 2008;6(10):1122–1128. doi: 10.1016/j.cgh.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 68.Kim M, Kim HJ, Lee IK, Oh ST, Han K. Fecal occult blood test/fecal carcinoembriogenic antigen dual rapid test as a useful tool for colorectal cancer screening. Eur Surg Acta Chirurgica Austriaca. 2017;49(3):127–131. [Google Scholar]
- 69.Kim YC, Kim JH, Cheung DY, et al. The usefulness of a novel screening kit for colorectal cancer using the immunochromatographic fecal tumor M2 pyruvate kinase test. Gut Liver. 2015;9(5):641–648. doi: 10.5009/gnl13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koga Y, Yamazaki N, Yamamoto Y, et al. Fecal miR-106a is a useful marker for colorectal cancer patients with false-negative results in immunochemical fecal occult blood test. Cancer Epidemiol Biomark Prev. 2013;22(10):1844–1852. doi: 10.1158/1055-9965.EPI-13-0512. [DOI] [PubMed] [Google Scholar]
- 71.Lee HS, Hwang SM, Kim TS, et al. Circulating methylated septin 9 nucleic acid in the plasma of patients with gastrointestinal cancer in the stomach and colon. Transl Oncol. 2013;6(3):290–296. doi: 10.1593/tlo.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee JH. Clinical usefulness of serum CYFRA 21–1 in patients with colorectal cancer. Nucl Med Mol Imaging. 2013;47(3):181–187. doi: 10.1007/s13139-013-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H, Wang Z, Zhao G, et al. Performance of a MethyLight assay for methylated SFRP2 DNA detection in colorectal cancer tissue and serum. Int J Biol Mark. 2019;34(1):54–59. doi: 10.1177/1724600818820536. [DOI] [PubMed] [Google Scholar]
- 74.Li L, Zhang L, Tian Y, et al. Serum chemokine CXCL7 as a diagnostic biomarker for colorectal cancer. Front Oncol. 2019;9:921. doi: 10.3389/fonc.2019.00921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Y, Jiang T, Zhang J, et al. Elevated serum antibodies against insulin-like growth factor-binding protein-2 allow detecting early-stage cancers: evidences from glioma and colorectal carcinoma studies. Ann Oncol. 2012;23(9):2415–2422. doi: 10.1093/annonc/mds007. [DOI] [PubMed] [Google Scholar]
- 76.Liu GH, Zhou ZG, Chen R, et al. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol. 2013;34(4):2175–2181. doi: 10.1007/s13277-013-0753-8. [DOI] [PubMed] [Google Scholar]
- 77.Liu T, Zhang X, Gao S, et al. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7(51):85551–85563. doi: 10.18632/oncotarget.13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu X, Pan B, Sun L, et al. Circulating exosomal miR-27a and miR-130a act as novel diagnostic and prognostic biomarkers of colorectal cancer. Cancer Epidemiol Biomark Prev. 2018;27(7):746–754. doi: 10.1158/1055-9965.EPI-18-0067. [DOI] [PubMed] [Google Scholar]
- 79.Liu X, Xu T, Hu X, et al. Elevated circulating miR-182 acts as a diagnostic biomarker for early colorectal cancer. Cancer Manag Res. 2018;10:857–865. doi: 10.2147/CMAR.S158016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lumachi F, Marino F, Orlando R, Chiara GB, Basso SM. Simultaneous multianalyte immunoassay measurement of five serum tumor markers in the detection of colorectal cancer. Anticancer Res. 2012;32(3):985–988. [PubMed] [Google Scholar]
- 81.Luo X, Wu Y, Ji M, Zhang S. Combined plasma microRNA and fecal occult blood tests in early detection of colorectal cancer. Clin Lab. 2019;65(5):01. doi: 10.7754/Clin.Lab.2018.180926. [DOI] [PubMed] [Google Scholar]
- 82.Marcuello M, Duran-Sanchon S, Moreno L, et al. Analysis of A 6-Mirna signature in serum from colorectal cancer screening participants as non-invasive biomarkers for advanced adenoma and colorectal cancer detection. Cancers. 2019;11(10):12. doi: 10.3390/cancers11101542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marshall KW, Mohr S, Khettabi FE, et al. A blood-based biomarker panel for stratifying current risk for colorectal cancer. Int J Cancer. 2010;126(5):1177–1186. doi: 10.1002/ijc.24910. [DOI] [PubMed] [Google Scholar]
- 84.Matsubara J, Honda K, Ono M, et al. Identification of adipophilin as a potential plasma biomarker for colorectal cancer using label-free quantitative mass spectrometry and protein microarray. Cancer Epidemiol Biomark Prev. 2011;20(10):2195–2203. doi: 10.1158/1055-9965.EPI-11-0400. [DOI] [PubMed] [Google Scholar]
- 85.Matsushita H, Matsumura Y, Moriya Y, et al. A new method for isolating colonocytes from naturally evacuated feces and its clinical application to colorectal cancer diagnosis. Gastroenterology. 2005;129(6):1918–1927. doi: 10.1053/j.gastro.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 86.Melotte V, Yi JM, Lentjes MH, et al. Spectrin repeat containing nuclear envelope 1 and forkhead box protein E1 are promising markers for the detection of colorectal cancer in blood. Cancer Prev Res. 2015;8(2):157–164. doi: 10.1158/1940-6207.CAPR-14-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meng W, Zhu HH, Xu ZF, et al. Serum M2-pyruvate kinase: a promising non-invasive biomarker for colorectal cancer mass screening. World J Gastrointest Oncol. 2012;4(6):145–151. doi: 10.4251/wjgo.v4.i6.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Min L, Zhu S, Chen L, et al. Evaluation of circulating extracellular vesicles derived miRNAs as biomarkers of early colon cancer: a comparison with plasma total miRNAs. J Extracell Vesicles. 2019;8(Supplement 1):261–262. doi: 10.1080/20013078.2019.1643670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mizuno M, Mizuno M, Iwagaki N, et al. Testing of multiple samples increases the sensitivity of stool decay-accelerating factor test for the detection of colorectal cancer. Am J Gastroenterol. 2003;98(11):2550–2555. doi: 10.1111/j.1572-0241.2003.08672.x. [DOI] [PubMed] [Google Scholar]
- 90.Mroczko B, Groblewska M, Okulczyk B, Kedra B, Szmitkowski M. The diagnostic value of matrix metalloproteinase 9 (MMP-9) and tissue inhibitor of matrix metalloproteinases 1 (TIMP-1) determination in the sera of colorectal adenoma and cancer patients. Int J Colorectal Dis. 2010;25(10):1177–1184. doi: 10.1007/s00384-010-0991-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mroczko B, Groblewska M, Wereszczynska-Siemiatkowska U, Kedra B, Konopko M, Szmitkowski M. The diagnostic value of G-CSF measurement in the sera of colorectal cancer and adenoma patients. Clin Chim Acta. 2006;371(1–2):143–147. doi: 10.1016/j.cca.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 92.Mulder SA, Van Leerdam ME, van Vuuren AJ, et al. Tumor pyruvate kinase isoenzyme type M2 and immunochemical fecal occult blood test: performance in screening for colorectal cancer. Eur J Gastroenterol Hepatol. 2007;19(10):878–882. doi: 10.1097/MEG.0b013e3282cfa49c. [DOI] [PubMed] [Google Scholar]
- 93.Murakoshi Y, Honda K, Sasazuki S, et al. Plasma biomarker discovery and validation for colorectal cancer by quantitative shotgun mass spectrometry and protein microarray. Cancer Sci. 2011;102(3):630–638. doi: 10.1111/j.1349-7006.2010.01818.x. [DOI] [PubMed] [Google Scholar]
- 94.Murata K, Moriya A, Fujita Y, Kakugawa O, Saito MY. Application of miRNA expression analysis on exfoliated colonocytes for diagnosis of colorectal cancer. Gastrointest Cancer Targets Ther. 2012;2:11–18. [Google Scholar]
- 95.Ng L, Wan TM, Man JH, et al. Identification of serum miR-139-3p as a non-invasive biomarker for colorectal cancer. Oncotarget. 2017;8(16):27393–27400. doi: 10.18632/oncotarget.16171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nielsen HJ, Brünner N, Jorgensen LN, et al. Plasma TIMP-1 and CEA in detection of primary colorectal cancer: a prospective, population based study of 4509 high-risk individuals. Scand J Gastroenterol. 2011;46(1):60–69. doi: 10.3109/00365521.2010.513060. [DOI] [PubMed] [Google Scholar]
- 97.Ning S, Wei W, Li J, et al. Clinical significance and diagnostic capacity of serum TK1, CEA, CA 19–9 and CA 72–4 levels in gastric and colorectal cancer patients. J Cancer. 2018;9(3):494–501. doi: 10.7150/jca.21562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nishiumi S, Kobayashi T, Ikeda A, et al. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS One. 2012;7(7):e40459. doi: 10.1371/journal.pone.0040459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Niu R, Jing H, Chen Z, Xu J, Dai J, Yan Z. Differentiating malignant colorectal tumor patients from benign colorectal tumor patients by assaying morning urinary arylsulfatase activity. Asia Pac J Clin Oncol. 2012;8(4):362–367. doi: 10.1111/j.1743-7563.2012.01545.x. [DOI] [PubMed] [Google Scholar]
- 100.Ørntoft MB, Nielsen HJ, Ørntoft TF, Andersen CL. Performance of the colorectal cancer screening marker Sept9 is influenced by age, diabetes and arthritis: a nested case–control study. BMC Cancer. 2015;15:819. doi: 10.1186/s12885-015-1832-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Palmqvist R, Engaras B, Lindmark G, et al. Prediagnostic levels of carcinoembryonic antigen and CA 242 in colorectal cancer: a matched case–control study. Dis Colon Rectum. 2003;46(11):1538–1544. doi: 10.1007/s10350-004-6810-z. [DOI] [PubMed] [Google Scholar]
- 102.Pedersen JW, Gentry-Maharaj A, Fourkala EO, et al. Early detection of cancer in the general population: a blinded case–control study of p53 autoantibodies in colorectal cancer. Br J Cancer. 2013;108(1):107–114. doi: 10.1038/bjc.2012.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pedersen SK, Symonds EL, Baker RT, et al. Evaluation of an assay for methylated BCAT1 and IKZF1 in plasma for detection of colorectal neoplasia. BMC Cancer. 2015;15:654. doi: 10.1186/s12885-015-1674-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Peng HX, Yang L, He BS, et al. Combination of preoperative NLR, PLR and CEA could increase the diagnostic efficacy for I–III stage CRC. J Clin Lab Anal. 2017;31(5):6. doi: 10.1002/jcla.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pengjun Z, Xinyu W, Feng G, et al. Multiplexed cytokine profiling of serum for detection of colorectal cancer. Future Oncol. 2013;9(7):1017–1027. doi: 10.2217/fon.13.71. [DOI] [PubMed] [Google Scholar]
- 106.Qian J, Tikk K, Werner S, et al. Biomarker discovery study of inflammatory proteins for colorectal cancer early detection demonstrated importance of screening setting validation. J Clin Epidemiol. 2018;104:24–34. doi: 10.1016/j.jclinepi.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 107.Qu A, Wang W, Yang Y, et al. A serum piRNA signature as promising non-invasive diagnostic and prognostic biomarkers for colorectal cancer. Cancer Manag Res. 2019;11:3703–3720. doi: 10.2147/CMAR.S193266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ren S, Zhang Z, Xu C, et al. Distribution of IgG galactosylation as a promising biomarker for cancer screening in multiple cancer types. Cell Res. 2016;26(8):963–966. doi: 10.1038/cr.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rho JH, Ladd JJ, Li CI, et al. Protein and glycomic plasma markers for early detection of adenoma and colon cancer. Gut. 2018;67(3):473–484. doi: 10.1136/gutjnl-2016-312794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ritchie SA, Ahiahonu PW, Jayasinghe D, et al. Reduced levels of hydroxylated, polyunsaturated ultra long-chain fatty acids in the serum of colorectal cancer patients: implications for early screening and detection. BMC Med. 2010;8:13. doi: 10.1186/1741-7015-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ritchie SA, Tonita J, Alvi R, et al. Low-serum GTA-446 anti-inflammatory fatty acid levels as a new risk factor for colon cancer. Int J Cancer. 2013;132(2):355–362. doi: 10.1002/ijc.27673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ruffin MT, Normolle DP, Evelegh MJ, et al. Rectal mucosal quantitative galactose oxidase-Schiff reaction as an early detection biomarker for colorectal cancer: comparison to fecal occult stool blood test. Cancer Biomark Sect A Dis Mark. 2010;8(2):109–112. doi: 10.3233/CBM-2011-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schneider J, Bitterlich N, Schulze G. Improved sensitivity in the diagnosis of gastro-intestinal tumors by fuzzy logic-based tumor marker profiles including the tumor M2-PK. Anticancer Res. 2005;25(3A):1507–1515. [PubMed] [Google Scholar]
- 114.Shastri YM, Loitsch S, Hoepffner N, et al. Comparison of an established simple office-based immunological FOBT with fecal tumor pyruvate kinase type M2 (M2-PK) for colorectal cancer screening: prospective multicenter study. Am J Gastroenterol. 2008;103(6):1496–1504. doi: 10.1111/j.1572-0241.2008.01824.x. [DOI] [PubMed] [Google Scholar]
- 115.Shastri YM, Naumann M, Oremek GM, et al. Prospective multicenter evaluation of fecal tumor pyruvate kinase type M2 (M2-PK) as a screening biomarker for colorectal neoplasia. Int J Cancer. 2006;119(11):2651–2656. doi: 10.1002/ijc.22243. [DOI] [PubMed] [Google Scholar]
- 116.Shi C, Xie M, Li L, Li K, Hu BL. The association and diagnostic value of red blood cell distribution width in colorectal cancer. Medicine. 2019;98(19):e15560. doi: 10.1097/MD.0000000000015560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sithambaram S, Hilmi I, Goh KL. The diagnostic accuracy of the M2 pyruvate kinase quick stool test—a rapid office based assay test for the detection of colorectal cancer. PLoS One. 2015;10(7):e0131616. doi: 10.1371/journal.pone.0131616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Song L, Jia J, Yu H, et al. The performance of the mSEPT9 assay is influenced by algorithm, cancer stage and age, but not sex and cancer location. J Cancer Res Clin Oncol. 2017;143(6):1093–1101. doi: 10.1007/s00432-017-2363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Song L, Peng X, Li Y, et al. The SEPT9 gene methylation assay is capable of detecting colorectal adenoma in opportunistic screening. Epigenomics. 2017;9(5):599–610. doi: 10.2217/epi-2016-0146. [DOI] [PubMed] [Google Scholar]
- 120.Song L, Wang J, Wang H, et al. The quantitative profiling of blood mSEPT9 determines the detection performance on colorectal tumors. Epigenomics. 2018;10(12):1569–1583. doi: 10.2217/epi-2017-0154. [DOI] [PubMed] [Google Scholar]
- 121.Song Y, Huang Z, Kang Y, et al. Clinical usefulness and prognostic value of red cell distribution width in colorectal cancer. Biomed Res Int. 2018;2018:9858943. doi: 10.1155/2018/9858943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stojkovic Lalosevic M, Pavlovic Markovic A, Stankovic S, et al. Combined diagnostic efficacy of neutrophil-to-lymphocyte ratio (NLR), platelet-to-lymphocyte ratio (PLR), and mean platelet volume (MPV) as biomarkers of systemic inflammation in the diagnosis of colorectal cancer. Dis Mark. 2019;2019:6036979. doi: 10.1155/2019/6036979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sun J, Fei F, Zhang M, et al. The role of mSEPT9 in screening, diagnosis, and recurrence monitoring of colorectal cancer. BMC Cancer. 2019;19(1):450. doi: 10.1186/s12885-019-5663-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sun M, Liu J, Hu H, et al. A novel panel of stool-based DNA biomarkers for early screening of colorectal neoplasms in a Chinese population. J Cancer Res Clin Oncol. 2019;145(10):2423–2432. doi: 10.1007/s00432-019-02992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Swellam M, Abdelmaksoud MDE, Hassan AK. Clinical significance of NGAL, MMP-9, and VEGF in colorectal cancer patients. Int J Pharm Clin Res. 2016;8(1):117–122. [Google Scholar]
- 126.Symonds EL, Pedersen SK, Baker RT, et al. A blood test for methylated BCAT1 and IKZF1 vs a fecal immunochemical test for detection of colorectal neoplasia. Clin Transl Gastroenterol. 2016;7:e137. doi: 10.1038/ctg.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tagore KS, Lawson MJ, Yucaitis JA, et al. Sensitivity and specificity of a stool DNA multitarget assay panel for the detection of advanced colorectal neoplasia. Clin Colorectal Cancer. 2003;3(1):47–53. doi: 10.3816/CCC.2003.n.011. [DOI] [PubMed] [Google Scholar]
- 128.Taguchi A, Rho JH, Yan Q, et al. MAPRE1 as a plasma biomarker for early-stage colorectal cancer and adenomas. Cancer Prev Res. 2015;8(11):1112–1119. doi: 10.1158/1940-6207.CAPR-15-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tang D, Liu J, Wang DR, Yu HF, Li YK, Zhang JQ. Diagnostic and prognostic value of the methylation status of secreted frizzled-related protein 2 in colorectal cancer. Clin Investig Med. 2011;34(2):E88–E95. doi: 10.25011/cim.v34i1.15105. [DOI] [PubMed] [Google Scholar]
- 130.Tibble J, Sigthorsson G, Foster R, Sherwood R, Fagerhol M, Bjarnason I. Faecal calprotectin and faecal occult blood tests in the diagnosis of colorectal carcinoma and adenoma. Gut. 2001;49(3):402–408. doi: 10.1136/gut.49.3.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Toiyama Y, Takahashi M, Hur K, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105(12):849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tomasevic R, Milosavljevic T, Stojanovic D, et al. Predictive value of carcinoembryonic and carbohydrate antigen 19–9 related to some clinical, endoscopic and histological colorectal cancer characteristics. J Med Biochem. 2016;35(3):324–332. doi: 10.1515/jomb-2016-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Toth K, Sipos F, Kalmar A, et al. Detection of methylated SEPT9 in plasma is a reliable screening method for both left- and right-sided colon cancers. PLoS One. 2012;7(9):e46000. doi: 10.1371/journal.pone.0046000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Uchiyama K, Naito Y, Yagi N, et al. Selected reaction monitoring for colorectal cancer diagnosis using a set of five serum peptides identified by BLOTCHIP®-MS analysis. J Gastroenterol. 2018;53(11):1179–1185. doi: 10.1007/s00535-018-1448-0. [DOI] [PubMed] [Google Scholar]
- 135.Vychytilova-Faltejskova P, Radova L, Sachlova M, et al. Serum-based microRNA signatures in early diagnosis and prognosis prediction of colon cancer. Carcinogenesis. 2016;37(10):941–950. doi: 10.1093/carcin/bgw078. [DOI] [PubMed] [Google Scholar]
- 136.Vychytilova-Faltejskova P, Stitkovcova K, Radova L, et al. Circulating PIWI-interacting RNAs piR-5937 and piR-28876 are promising diagnostic biomarkers of colon cancer. Cancer Epidemiol Biomark Prev. 2018;27(9):1019–1028. doi: 10.1158/1055-9965.EPI-18-0318. [DOI] [PubMed] [Google Scholar]
- 137.Wang DS, Zhong B, Zhang MS, Gao Y. Upregulation of serum miR-103 predicts unfavorable prognosis in patients with colorectal cancer. Eur Rev Med Pharmacol Sci. 2018;22(14):4518–4523. doi: 10.26355/eurrev_201807_15506. [DOI] [PubMed] [Google Scholar]
- 138.Wang H, Luo C, Zhu S, et al. Serum peptidome profiling for the diagnosis of colorectal cancer: discovery and validation in two independent cohorts. Oncotarget. 2017;8(35):59376–59386. doi: 10.18632/oncotarget.19587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang J, Huang SK, Zhao M, et al. Identification of a circulating microRNA signature for colorectal cancer detection. PLoS One. 2014;9(4):e87451. doi: 10.1371/journal.pone.0087451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang JY, Lin SR, Wu DC, et al. Multiple molecular markers as predictors of colorectal cancer in patients with normal perioperative serum carcinoembryonic antigen levels. Clin Cancer Res. 2007;13(8):2406–2413. doi: 10.1158/1078-0432.CCR-06-2054. [DOI] [PubMed] [Google Scholar]
- 141.Wang QJ, Zhang HC, Shen XJ, Ju SQ. Serum microRNA-135a-5p as an auxiliary diagnostic biomarker for colorectal cancer. Ann Clin Biochem. 2017;54(1):76–85. doi: 10.1177/0004563216638108. [DOI] [PubMed] [Google Scholar]
- 142.Wang R, Du L, Yang X, et al. Identification of long noncoding RNAs as potential novel diagnosis and prognosis biomarkers in colorectal cancer. J Cancer Res Clin Oncol. 2016;142(11):2291–2301. doi: 10.1007/s00432-016-2238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Warren JD, Xiong W, Bunker AM, et al. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011;9:133. doi: 10.1186/1741-7015-9-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wilhelmsen M, Christensen IJ, Rasmussen L, et al. Detection of colorectal neoplasia: combination of eight blood-based, cancer-associated protein biomarkers. Int J Cancer. 2017;140(6):1436–1446. doi: 10.1002/ijc.30558. [DOI] [PubMed] [Google Scholar]
- 145.Wu CW, Ng SC, Dong Y, et al. Identification of microRNA-135b in stool as a potential noninvasive biomarker for colorectal cancer and adenoma. Clin Cancer Res. 2014;20(11):2994–3002. doi: 10.1158/1078-0432.CCR-13-1750. [DOI] [PubMed] [Google Scholar]
- 146.Wu CW, Ng SS, Dong YJ, et al. Detection of miR-92a and miR-21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut. 2012;61(5):739–745. doi: 10.1136/gut.2011.239236. [DOI] [PubMed] [Google Scholar]
- 147.Wu HK, Liu C, Fan XX, Wang H, Zhou L. Spalt-like transcription factor 4 as a potential diagnostic and prognostic marker of colorectal cancer. Cancer Biomark Sect A Dis Mark. 2017;20(2):191–198. doi: 10.3233/CBM-170204. [DOI] [PubMed] [Google Scholar]
- 148.Wu Y, Yang L, Zhao J, et al. Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol Cancer. 2015;14:191. doi: 10.1186/s12943-015-0455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wu D, Zhou G, Jin P, et al. Detection of colorectal cancer using a simplified SEPT9 gene methylation assay is a reliable method for opportunistic screening. J Mol Diagn. 2016;18(4):535–545. doi: 10.1016/j.jmoldx.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 150.Xie H, Guo JH, An WM, et al. Diagnostic value evaluation of trefoil factors family 3 for the early detection of colorectal cancer. World J Gastroenterol. 2017;23(12):2159–2167. doi: 10.3748/wjg.v23.i12.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xie L, Jiang X, Li Q, et al. Diagnostic value of methylated septin 9 for colorectal cancer detection. Front Oncol. 2018;8:247. doi: 10.3389/fonc.2018.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xu Y, Xu Q, Yang L, et al. Identification and validation of a blood-based 18-gene expression signature in colorectal cancer. Clin Cancer Res. 2013;19(11):3039–3049. doi: 10.1158/1078-0432.CCR-12-3851. [DOI] [PubMed] [Google Scholar]
- 153.Yang W, Luo Y, Hu S, Li Y, Liu Q. Value of combined detection of serum carcino-embryonic antigen, carbohydrate antigen 19–9 and cyclooxygenase-2 in the diagnosis of colorectal cancer. Oncol Lett. 2018;16(2):1551–1556. doi: 10.3892/ol.2018.8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yau TO, Wu CW, Tang CM, et al. MicroRNA-20a in human faeces as a non-invasive biomarker for colorectal cancer. Oncotarget. 2016;7(2):1559–1568. doi: 10.18632/oncotarget.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yuan P, Cheng XJ, Wu XJ, et al. OSMR and SEPT9: promising biomarkers for detection of colorectal cancer based on blood-based tests. Transl Cancer Res. 2016;5(2):131–139. [Google Scholar]
- 156.Zhang SY, Lin M, Zhang HB. Diagnostic value of carcinoembryonic antigen and carcinoma antigen 19–9 for colorectal carcinoma. Int J Clin Exp Pathol. 2015;8(8):9404–9409. [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang Y, He C, Qiu L, et al. Serum unsaturated free fatty acids: a potential biomarker panel for early-stage detection of colorectal cancer. J Cancer. 2016;7(4):477–483. doi: 10.7150/jca.13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang Y, Suehiro Y, Shindo Y, et al. Long-fragment DNA as a potential marker for stool-based detection of colorectal cancer. Oncol Lett. 2015;9(1):454–458. doi: 10.3892/ol.2014.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhao G, Li H, Yang Z, et al. Multiplex methylated DNA testing in plasma with high sensitivity and specificity for colorectal cancer screening. Cancer Med. 2019;8(12):5619–5628. doi: 10.1002/cam4.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhao XT, Zhang YQ, Deng LL, Wang YM, Li YX, Chen MW. The association between Chinese patients’ elevated omentin-1 levels, their clinicopathological features, and the risk of colorectal cancer. Int J Clin Exp Pathol. 2019;12(6):2264–2274. [PMC free article] [PubMed] [Google Scholar]
- 161.Zheng G, Du L, Yang X, et al. Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br J Cancer. 2014;111(10):1985–1992. doi: 10.1038/bjc.2014.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhou WW, Chu YP, An GY. Significant difference of neutrophil-lymphocyte ratio between colorectal cancer, adenomatous polyp and healthy people. Eur Rev Med Pharmacol Sci. 2017;21(23):5386–5391. doi: 10.26355/eurrev_201712_13924. [DOI] [PubMed] [Google Scholar]
- 163.Zhu D, Wang J, Ren L, et al. Serum proteomic profiling for the early diagnosis of colorectal cancer. J Cell Biochem. 2013;114(2):448–455. doi: 10.1002/jcb.24384. [DOI] [PubMed] [Google Scholar]
- 164.Zhu J, Dong H, Zhang Q, Zhang S. Combined assays for serum carcinoembryonic antigen and microRNA-17-3p offer improved diagnostic potential for stage I/II colon cancer. Mol Clin Oncol. 2015;3(6):1315–1318. doi: 10.3892/mco.2015.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hariharan R, Jenkins M. Utility of the methylated SEPT9 test for the early detection of colorectal cancer: a systematic review and meta-analysis of diagnostic test accuracy. BMJ Open Gastroenterol. 2020;7(1):e000355. doi: 10.1136/bmjgast-2019-000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nian J, Sun X, Ming S, et al. Diagnostic accuracy of methylated SEPT9 for blood-based colorectal cancer detection: a systematic review and meta-analysis. Clin Transl Gastroenterol. 2017;8(1):e216. doi: 10.1038/ctg.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Elliott JH, Turner T, Clavisi O, et al. Living systematic reviews: an emerging opportunity to narrow the evidence-practice gap. PLoS Med. 2014;11(2):e1001603. doi: 10.1371/journal.pmed.1001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jüni P, Holenstein F, Sterne J, et al. Direction and impact of language bias in meta-analyses of controlled trials: empirical study. Int J Epidemiol. 2002;31(1):115–123. doi: 10.1093/ije/31.1.115. [DOI] [PubMed] [Google Scholar]
- 169.Nussbaumer-Streit B, Klerings I, Dobrescu AI, et al. Excluding non-English publications from evidence-syntheses did not change conclusions: a meta-epidemiological study. J Clin Epidemiol. 2020;118:42–54. doi: 10.1016/j.jclinepi.2019.10.011. [DOI] [PubMed] [Google Scholar]
- 170.Wong CK, Fedorak RN, Prosser CI, Stewart ME, van Zanten SV, Sadowski DC. The sensitivity and specificity of guaiac and immunochemical fecal occult blood tests for the detection of advanced colonic adenomas and cancer. Int J Colorectal Dis. 2012;27(12):1657–1664. doi: 10.1007/s00384-012-1518-3. [DOI] [PubMed] [Google Scholar]
- 171.Allison JE, Fraser CG, Halloran SP, Young GP. Comparing fecal immunochemical tests: improved standardization is needed. Gastroenterology. 2012;142(3):422–424. doi: 10.1053/j.gastro.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 172.Allison JE, Sakoda LC, Levin TR, et al. Screening for colorectal neoplasms with new fecal occult blood tests: update on performance characteristics. J Natl Cancer Inst. 2007;99(19):1462–1470. doi: 10.1093/jnci/djm150. [DOI] [PubMed] [Google Scholar]
- 173.Morikawa T, Kato J, Yamaji Y, Wada R, Mitsushima T, Shiratori Y. A comparison of the immunochemical fecal occult blood test and total colonoscopy in the asymptomatic population. Gastroenterology. 2005;129(2):422–428. doi: 10.1016/j.gastro.2005.05.056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.