PURPOSE
We sought to investigate clinical outcomes of relapsed medulloblastoma and to compare molecular features between patient-matched diagnostic and relapsed tumors.
METHODS
Children and infants enrolled on either SJMB03 (NCT00085202) or SJYC07 (NCT00602667) trials who experienced medulloblastoma relapse were analyzed for clinical outcomes, including anatomic and temporal patterns of relapse and postrelapse survival. A largely independent, paired molecular cohort was analyzed by DNA methylation array and next-generation sequencing.
RESULTS
A total of 72 of 329 (22%) SJMB03 and 52 of 79 (66%) SJYC07 patients experienced relapse with significant representation of Group 3 and wingless tumors. Although most patients exhibited some distal disease (79%), 38% of patients with sonic hedgehog tumors experienced isolated local relapse. Time to relapse and postrelapse survival varied by molecular subgroup with longer latencies for patients with Group 4 tumors. Postrelapse radiation therapy among previously nonirradiated SJYC07 patients was associated with long-term survival. Reirradiation was only temporizing for SJMB03 patients. Among 127 patients with patient-matched tumor pairs, 9 (7%) experienced subsequent nonmedulloblastoma CNS malignancies. Subgroup (96%) and subtype (80%) stabilities were largely maintained among the remainder. Rare subgroup divergence was observed from Group 4 to Group 3 tumors, which is coincident with genetic alterations involving MYC, MYCN, and FBXW7. Subgroup-specific patterns of alteration were identified for driver genes and chromosome arms.
CONCLUSION
Clinical behavior of relapsed medulloblastoma must be contextualized in terms of up-front therapies and molecular classifications. Group 4 tumors exhibit slower biological progression. Utility of radiation at relapse is dependent on patient age and prior treatments. Degree and patterns of molecular conservation at relapse vary by subgroup. Relapse tissue enables verification of molecular targets and identification of occult secondary malignancies.
INTRODUCTION
Medulloblastoma relapse represents a key determinant of cancer-related mortality in the pediatric population.1,2 Multimodal therapy that incorporates maximal surgical resection with craniospinal irradiation (CSI; children older than 3 years) and chemotherapy has driven 5-year overall survival rates to 80%-85% for average-risk disease and 60%-70% for high-risk disease.3,4 Nevertheless, treatment failure and relapse occur in up to one-third of patients and confers abysmal prognosis, with only approximately 10% of patients surviving beyond 5 years postrelapse.5-7 Relapsed disease thus remains extremely refractory to existing therapies, whereas rare survivors often experience serious toxicity and devastating neurocognitive sequalae.8,9 Although no standard approach for treating relapsed medulloblastoma exists, salvage therapies, including conventional and targeted agents, have largely failed to confer durable survival benefit.10,11 A deeper clinical and biological understanding of medulloblastoma recurrence is needed for the design and interpretation of next-generation clinical trials for relapsed disease.
CONTEXT
Key Objective
To determine the clinical outcomes and molecular features of relapsed medulloblastoma.
Knowledge Generated
Time to relapse and postrelapse survival are associated with subgroup with Group 4 tumors exhibiting slower biological progression. Utility of radiation therapy at relapse depends on age and previous therapies. Most relapses exhibit concordant subgroup classifications, except in the cases of occult secondary malignancies or rare divergence from group 4 to group 3. Driver gene and chromosome arm alteration patterns vary according to molecular subgroup.
Relevance
Future trials for relapsed medulloblastoma must be contextualized by molecular subgroup and up-front therapies. Relapse tissue should be acquired and used for confirmation of diagnosis and verification of molecular targets.
Although extensive genomic characterization of medulloblastoma has identified four biologically and clinically distinct subgroups (wingless [WNT], sonic hedgehog [SHH], Group 3, and Group 4), most studies have relied on diagnostic samples that have not been exposed to tumor-directed therapies.12 Comparative molecular studies of diagnostic versus relapsed medulloblastoma have suggested divergent clonal selection, leading to emergence of specific molecular alterations at relapse in the context of subgroup conservation.13-16 However, the generalizability of such findings is diminished by relatively modest cohort sizes and employment of different molecular assays. Additionally, exclusion of secondary CNS malignancies subsequent to medulloblastoma therapy, particularly high-grade gliomas, requires robust classification methods to prevent inadvertent cross-entity comparisons and may represent a key limitation of previous comparative studies.17,18
Multiple recent studies have leveraged DNA methylation profiling to describe additional intertumoral heterogeneity within the core medulloblastoma subgroups.19-21 These subtypes, primarily defined among SHH (α, β, γ, and δ) and Groups 3 and 4 (I-VIII) tumors, have distinctive genetic and clinical features.22,23 Furthermore, subtype-defined risk paradigms are an active area of investigation. However, contextualization of these subtypes in relapsed disease and assessment of conservation at relapse are lacking.
In this study, subgroup-specific clinical behavior of relapsed medulloblastoma is described for patients enrolled on two, multi-institutional, risk-adapted clinical trials. A largely independent, paired molecular cohort composed of patient-matched diagnostic and relapse tumors is investigated to determine the molecular features of medulloblastoma relapse using DNA methylation profiling and next-generation sequencing.
METHODS
Patients and Samples
Study populations are summarized in the Data Supplement (online only). Eligible patients with relapsed medulloblastoma from two multi-institutional, risk-adapted clinical trials, SJMB03 (NCT00085202; Gajjar et al24 and SJYC07 (NCT00602667),23 were included (Appendix Table A1, online only; Data Supplement). Patients with a clinical diagnosis of nonmedulloblastoma subsequent malignancy were excluded. Given the differing eligibility criteria, risk stratifications, and treatment protocols between the trials, patients from each trial were analyzed separately for clinical outcomes based on molecular features garnered from primary tumor specimens. A largely independent, paired molecular cohort of 127 patients with formalin-fixed paraffin-embedded (FFPE) or frozen tissue specimens available from both their histopathologically diagnosed primary medulloblastoma and relapse or subsequent tumors was also assembled (Appendix Table A2, online only; Data Supplement). Comparative molecular analyses were performed among the paired molecular cohort using patient-matched primary and relapsed tumor specimens.
Tumor Molecular Profiling
Tumor specimens were analyzed using Infinium Methylation EPIC or 450K BeadChip arrays (Illumina, San Diego, CA) from either freshly frozen or FFPE tissue (Data Supplement). Medulloblastoma subgroup and subtype predictions were determined using DNA methylation–based classification of CNS tumors (MolecularNeuropathology, Heidelberg, Germany, version 11b4) and trained random forest predictions.18 Genome-wide DNA copy number alterations were inferred from DNA methylation arrays using the Conumee R package.
Next-generation (whole-exome or targeted gene panel) sequencing was performed on tumor samples with sufficient material available. Additional details regarding bioinformatic processing are given in the Appendix Methods (Data Supplement). Genomic datasets included in this study can be freely explored using the online St Jude Cloud pediatric genomic data resource.25
Statistical Analysis
Time-to-event analyses were performed using Kaplan-Meier methods with log-rank tests. Hazard ratios (HRs) with associated 95% CIs and P values were computed using Cox regression. Distributions of categorical variables were compared using Fisher's exact or chi-square test. Multiple testing correction was performed using false discovery rate. Statistical analyses were performed using R version 3.5.1 and are further detailed in the Appendix Methods (Data Supplement).
RESULTS
Incidence of Relapsed Disease
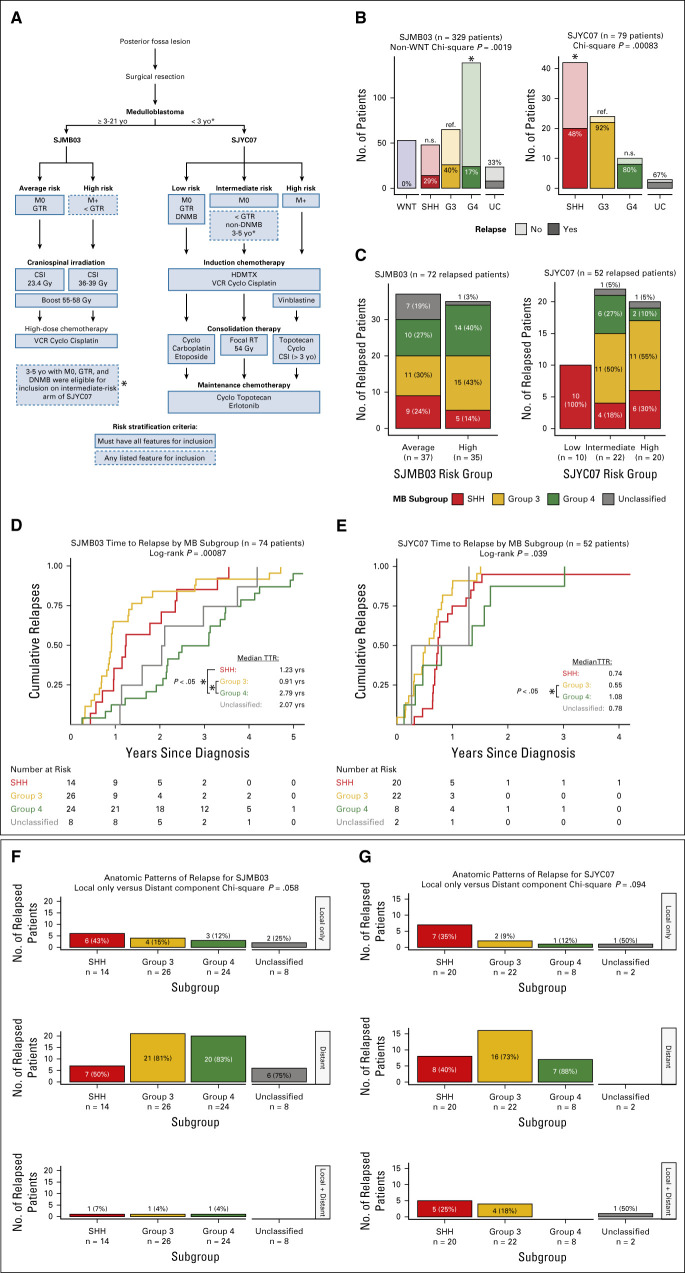
An overview of inclusion criteria, risk stratifications, and treatment protocols for SJMB03 and SJYC07 is shown in Figure 1A. A total of 72 of 329 (22%) patients from SJMB03 and 52 of 79 from SJYC07 (66%) relapsed (Fig 1B). Appendix Table A1 summarizes the demographic and clinical characteristics of relapsed patients from each trial according to molecular subgroup. Notably, WNT subgroup relapsed medulloblastomas were absent in both trials. Distributions of subgroups across clinical trial risk groups for relapsed patients are shown in Figure 1C.
FIG 1.
Relapse patterns for SJMB03 and SJYC07 patients with medulloblastoma. (A) Overview of trial designs and risk stratifications. (B) Apparent relapse rates by medulloblastoma subgroup per trial. (C) Medulloblastoma subgroup distributions per trial risk stratifications. Time to relapse by subgroup for (D) SJMB03 patients and (E) SJYC07 patients. (F) Anatomic patterns of relapse per subgroup for SJMB03 and (G) SJYC07 patients. CSI, craniospinal irradiation; DNMB, desmoplastic nodular medulloblastoma; GTR, gross total resection; HDMTX, high-dose methotrexate; MB, medulloblastoma; RT, radiation therapy; SHH, sonic hedgehog; TTR, time to relapse; UC, unclassified; VCR, vincristine; WNT, wingless.
Subgroup distribution between relapsed and nonrelapsed patients was not uniform in either SJMB03 (P = .0019, chi-square test) or SJYC07 (P = .00083, chi-square test; Fig 1B). Compared with Group 3 tumors, Group 4 tumors had a lower relapse rate in SJMB03 (odds ratio [OR], 0.31; 95% CI, 0.16 to 0.61), whereas SHH tumors had a lower relapse rate in SJYC07 (OR, 0.08; 95% CI, 0.01 to 0.33). The distribution of novel subtypes19,20,22,23 between relapsed and nonrelapsed patients was not uniform for Groups 3 and 4 subtypes in SJMB03 (P < .0001, chi-square test) or SHH subtypes in SJYC07 (P = .0058, chi-square test; Data Supplement). Proportions of relapsed patients between SJMB03 and SJYC07 differed for novel Groups 3 and 4 subtypes IV (P < .0001, Fisher's exact test) and VII (P < .0001, Fisher's exact test).
Time to Relapse
The median time to relapse was 1.64 years (interquartile range [IQR], 0.91-3.03) for SJMB03 and 0.72 year (IQR, 0.47-1.00) for SJYC07 (Data Supplement). For SJMB03, time to relapse varied by molecular subgroup (P = .00087, log-rank test) with a median time to relapse for patients with Group 4 tumor of 2.79 years (IQR, 1.99-3.64) compared with 0.91 year (IQR, 0.68-1.37) for Group 3 (HR, 3.01; 95% CI, 1.67 to 5.41) and 1.23 years (IQR, 0.96-2.34) for patients with SHH (HR, 2.41; 95% CI, 1.20 to 4.81; Fig 1D). For SJYC07, time to relapse also varied by molecular subgroup (P = .039, log-rank test), largely driven by difference between Group 4 and Group 3 tumors (HR, 2.74; 95% CI, 1.14 to 6.60; Fig 1E). Clinical risk group was a significant covariate for time to relapse only among patients with Group 4 tumor in SJMB03 (Data Supplement). Time to relapse according to novel SHH and Groups 3 and 4 subtypes is shown in Data Supplement.
Anatomic Patterns of Relapse
Relapse occurred with a distant component in 57 (79%) SJMB03 patients (Fig 1F) and 41 (79%) SJYC07 patients (Fig 1G). Of patients with SHH tumors, 43% in SJMB03 and 35% in SJYC07 presented with isolated local relapse compared with only 14% (P = .028, Fisher's exact test) and 10% (P = .067, Fisher's exact test) of non-SHH tumors, respectively (Figs 1F and 1G). Distant relapses were significantly associated with shorter time to relapse only for Group 4 tumors in SJMB03 (HR, 5.08; 95% CI, 1.14 to 22.6) and SHH tumors in SJYC07 (HR, 3.48; 95% CI, 1.07 to 11.3; Data Supplement).
Survival After Relapse
At data cutoff, seven relapsed patients (10%) from SJMB03 and 24 (46%) from SJYC07 were alive. The median postrelapse survival (PRS) was 1.14 years (IQR, 0.30-2.36) for SJMB03 and 2.12 (IQR, 0.39-NA) for SJYC07 (Data Supplement). Clinical trial risk group was not significantly associated with PRS for SJMB03 (P = .43, log-rank test) or SJYC07 (P = .099, log-rank test; Data Supplement).
For SJMB03, PRS time varied by molecular subgroup (P = .016, log-rank) with the median PRS time for patients with Group 4 tumors of 2.27 years (IQR, 1.19-3.89) compared with 0.44 year (IQR, 0.29-1.29) for patients with Group 3 tumors (HR, 2.39; 95% CI, 1.29 to 4.42) and 1.06 years (IQR, 0.27-1.88) for patients with SHH tumors (HR, 1.90; 95% CI, 0.94 to 3.83; Fig 2A). For SJYC07, PRS did not vary significantly by molecular subgroup (P = .88, log-rank test; Fig 2B). Time to relapse was significantly associated with PRS for SJMB03 (HR, 0.65; 95% CI, 0.52 to 0.82) but not for SJYC07 (HR, 0.53; 95% CI, 0.23 to 1.23). Notably, the association of time to relapse and PRS for SJMB03 remained significant with subgroup and clinical trial risk group as additional covariates (HR, 0.67; 95% CI, 0.50 to 0.90). PRS according to novel SHH and Groups 3 and 4 subtypes is shown in the Data Supplement.
FIG 2.
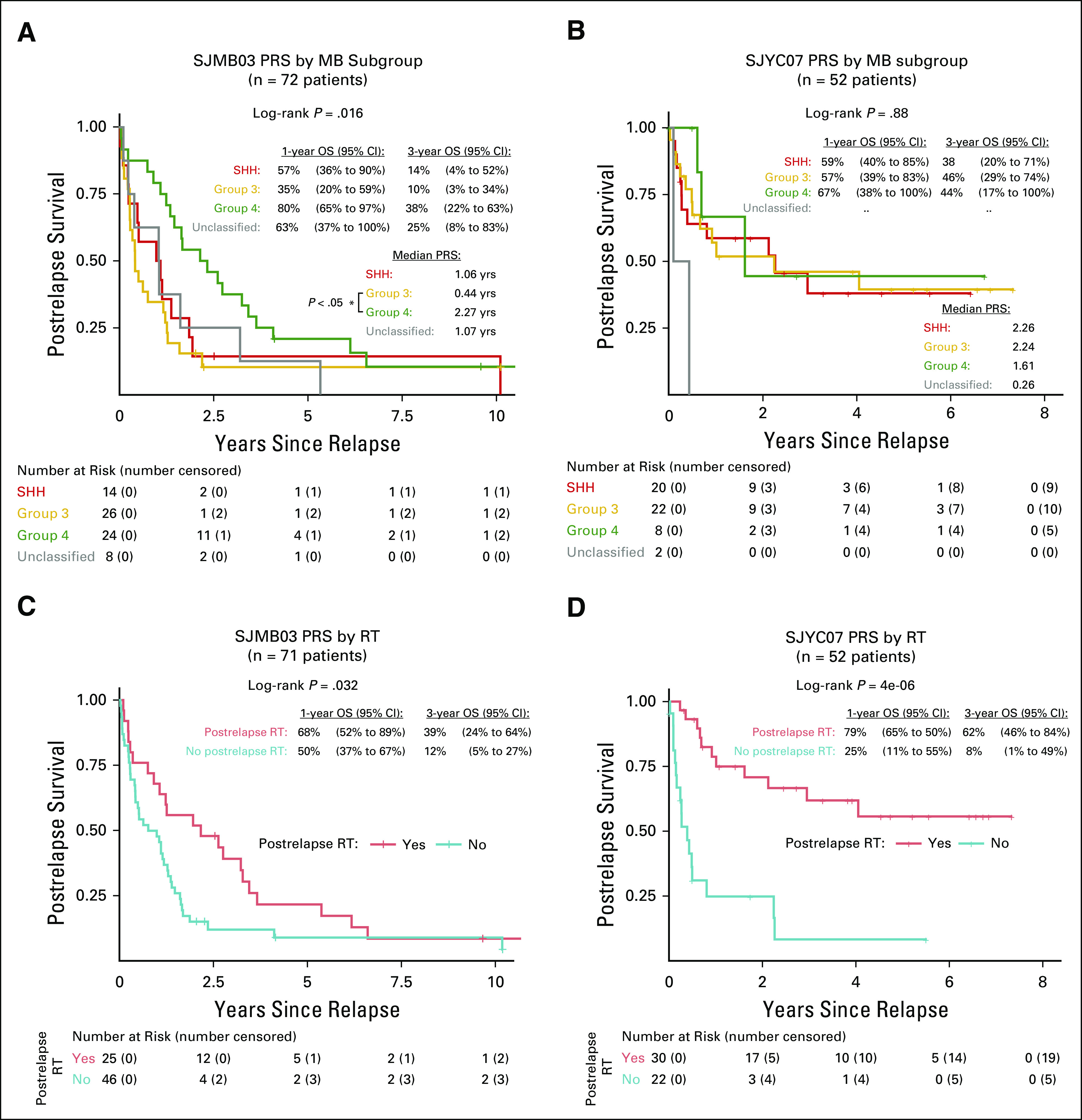
Postrelapse outcomes for SJMB03 and SJYC07 patients with medulloblastoma. PRS by subgroup for (A) SJMB03 and (B) SJYC07 patients. Postrelapse survival by receipt of radiation therapy after relapse for (C) SJMB03 and (D) SJYC07 patients. MB, medulloblastoma; OS, overall survival; PRS, postrelapse survival; RT, radiation therapy; SHH, sonic hedgehog.
Twenty-five (35%) relapsed SJMB03 patients (one patient missing data) and 30 (58%) SJYC07 patients received radiation after relapse. For SJMB03 patients, a transient PRS benefit (P = .032, log-rank test; Fig 2C) was observed with additional radiation after relapse (HR, 0.56; 95% CI, 0.33 to 0.96). For SJYC07 patients, postrelapse CSI (median, 36.0 Gy) was significantly associated with long-term survival (P < .0001, log-rank test; Fig 2D), particularly for patients with SHH (HR, 0.04; 95% CI, 0.01 to 0.37) and Group 3 tumors (HR, 0.27; 95% CI, 0.08 to 0.85) (Data Supplement).
Patient-Matched Molecular Landscapes
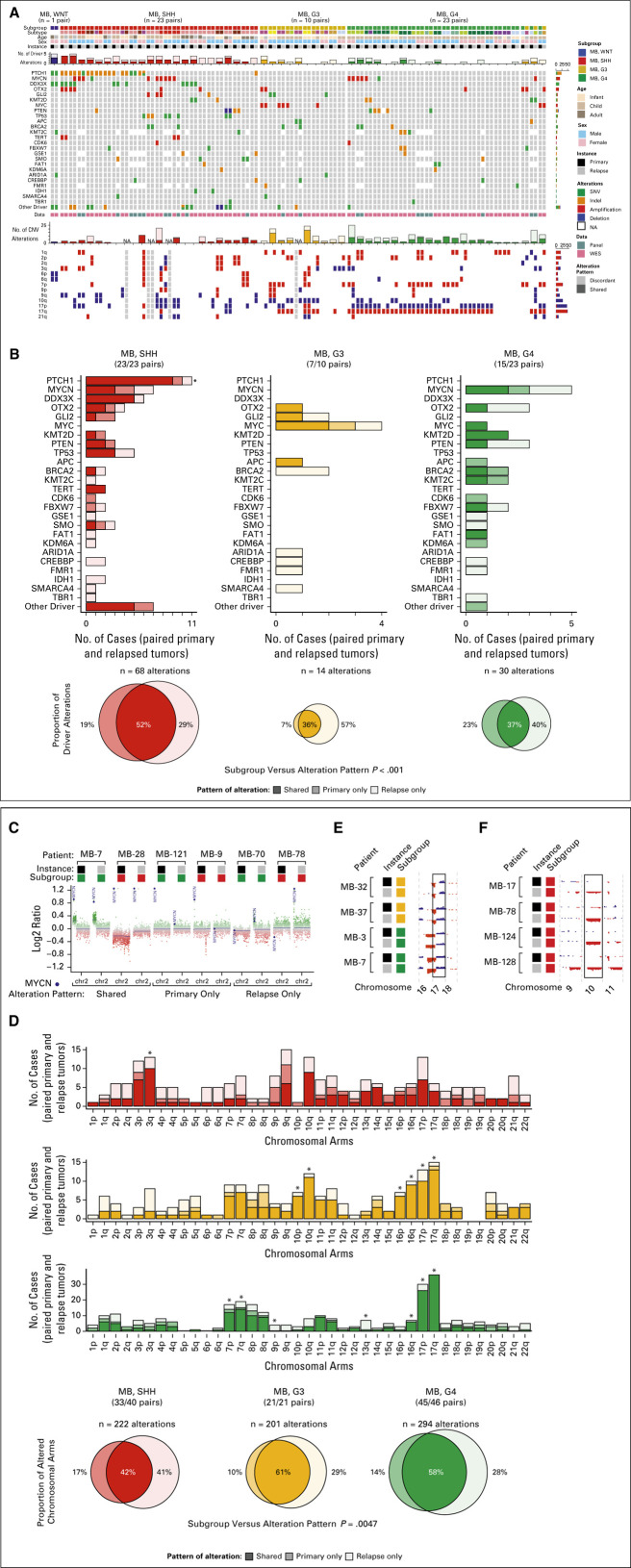
Given the rarity of tissue availability from relapsed medulloblastoma and with only seven patients from each trial with relapse tissue available, a large multi-institutional, paired molecular cohort of 127 patient-matched diagnostic and relapsed tumors was assembled (Fig 3A; Appendix Table A2; Data Supplement). Sufficient tissue for DNA methylation profiling was required for inclusion, and whole-exome or targeted panel sequencing data were available for both patient-matched samples in 50% (64 of 127) of the paired molecular cohort (Fig 3B). DNA methylation classification identified nine relapse cases (7%) as nonmedulloblastoma subsequent tumors within the paired molecular cohort (Fig 3C; Data Supplement).18
FIG 3.
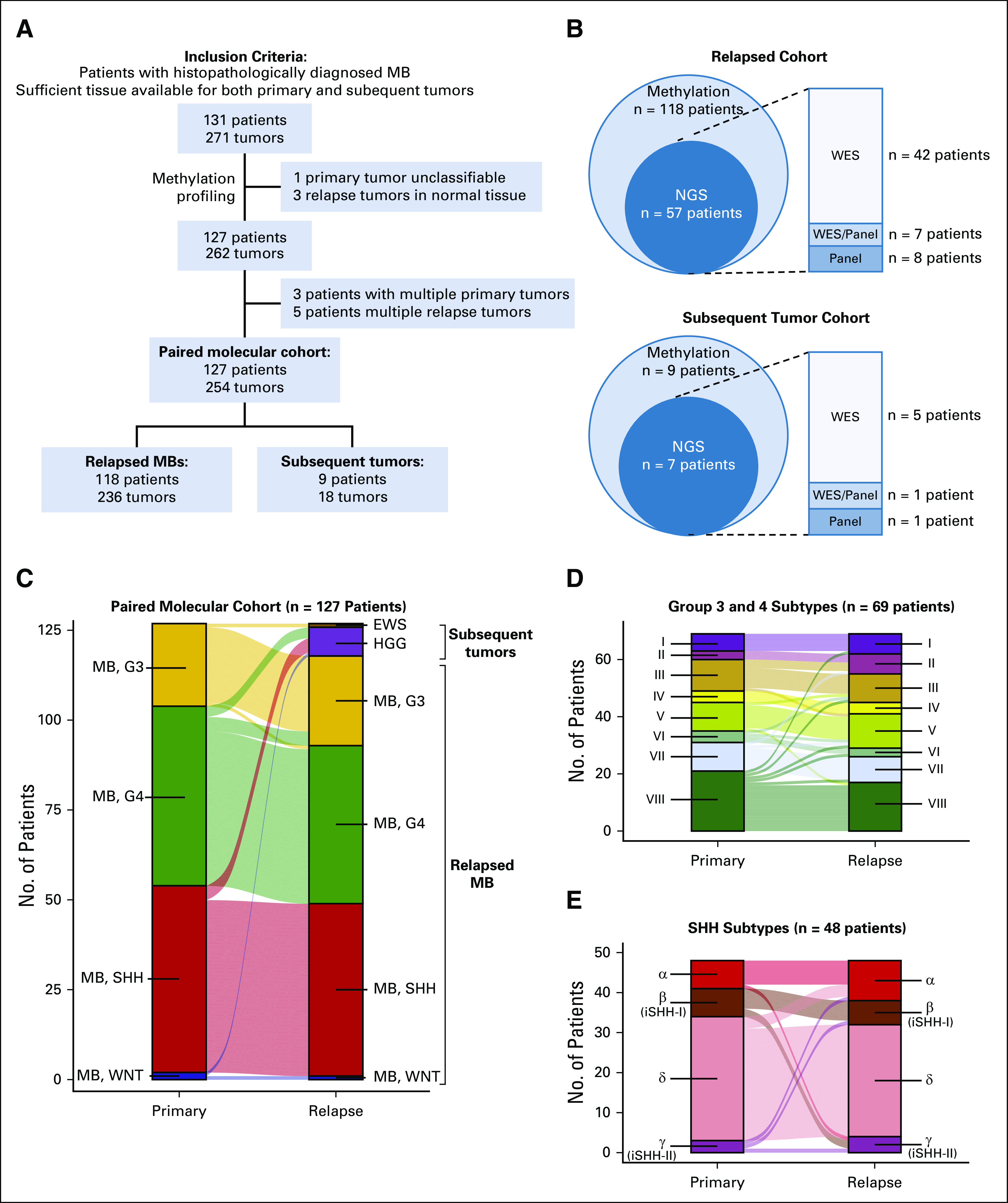
Overview of patient-matched molecular cohort. (A) Flowchart describing assembly of paired molecular cohort composed of patients with truly relapsed medulloblastomas and those with other subsequent CNS malignancies. (B) Methylation and next-generation sequencing data availability for each arm of the paired molecular cohort. (C) Subgroup and entity classification of patient-matched tissue from diagnosis and relapse. (D) Novel Groups 3 and 4 and (E) SHH subtype classifications of patient-matched tissue from diagnosis and relapse. EWS, Ewing sarcoma; HGG, high-grade glioma; iSHH, infant SHH; MB, medulloblastoma; NGS, next-generation sequencing; SHH, sonic hedgehog; WES, whole-exome sequencing; WNT, wingless.
Molecular Subgroups and Subtypes
Among 118 patients with molecularly confirmed relapsed medulloblastoma, diagnostic tumors were classified by methylation as WNT in 1 (1%), SHH in 48 (41%), Group 3 in 22 (19%), and Group 4 in 46 (39%). Medulloblastoma subgroup was conserved at relapse in 113 patients (96%) with divergence observed in a total of five patients between Groups 3 and 4 tumors. Novel molecular subtypes within SHH and Groups 3 and 4 tumors were conserved at relapse in 80% of patients (Figs 3D and 3E).19,20
To assess the degree of molecular conservation between patient-matched diagnostic and relapsed tumors, mutations and copy number alterations of curated medulloblastoma driver genes and chromosomal arms were analyzed (Fig 4A; Data Supplement).19 Subgroup designation for paired patient-matched cases was based on subgroup at diagnosis.
FIG 4.
Molecular landscape of relapsed medulloblastoma. (A) Oncoprint depicting patient characteristics, driver gene alterations, and chromosomal copy number variation for patient-matched tumor pairs with available next-generation sequencing (n = 57 patients). (B) Compartment-specific patterns of driver gene alterations by molecular subgroup. (C) Genomic track of chromosome 2 depicting alteration patterns observed for MYCN. (D) Compartment-specific patterns of chromosome arm copy number variation by molecular subgroup for patient-matched tumor pairs (n = 107 patients). Genomic tracks depicting alteration patterns of (E) chromosome 17 and (F) chromosome 10. CNV, copy number variation; G3, Group 3; G4, Group 4; MB, medulloblastoma; SHH, sonic hedgehog; SNV, single nucleotide variant; WES, whole-exome sequencing; WNT, wingless.
Driver Gene Alteration Patterns
The distribution and pattern of driver gene alterations by subgroup are shown in Figure 4B. Subgroup was associated with a significant difference in conservation pattern of driver gene alterations, largely driven by biases in SHH, which were predominantly conserved (P < .0001, analysis of variance). The median number of discordant driver gene alterations between patient-matched tumors was one with no difference between subgroups (P = .10, Kruskal-Wallis test; Data Supplement). A total of 29% of cases had completely conserved driver gene alterations with no statistical difference between subgroups (P = .29, chi-square test). Within subgroups, the number of discordant copy number variations or mutations was not significantly associated with age at diagnosis, time to relapse, or recurrence pattern (Data Supplement).
The incidence of shared driver gene alterations was 52% in SHH, 36% in Group 3, and 37% in Group 4 tumors (Figs 4A and 4B). Notable shared driver gene alterations included those affecting PTCH1 (10 of 12, 88% shared) and DDX3X (6 of 7, 86%) in SHH tumors. The incidence of relapse-specific driver gene alterations was 29% in SHH, 57% in Group 3, and 40% in Group 4 tumors. Additionally, numerous low incidence relapse-specific alterations, particularly among chromatin modifiers (eg, CREBBP and SMARCA4) and DNA repair machinery (eg, BRCA2), were observed across subgroups. Primary-specific driver gene alterations, such as focal CDK6 amplification in one primary SHH tumor and one primary Group 4 tumor, were rare and comprised the minority in observed patterns of conservation and divergence.
Driver genes with increased odds of alteration in relapsed patients were identified using primary tumor molecular data available for the clinical trial cohorts (Data Supplement). Conservation pattern of such alterations, including TP53 mutations and MYC and MYCN amplifications, was then queried in the paired molecular cohort (Figs 4A and 4B; Data Supplement).
Among relapsed cases in the paired molecular cohort, mutations in TP53 were restricted to SHH tumors (P = .0080, Fisher's exact test) and were relapse-specific in two (40%) and shared in three (60%) patients (Figs 4A and 4B). Predominantly occurring within Group 3 tumors (P = .00049, Fisher's exact test), amplifications of MYC were identified as shared in five (63%, including one Group 4), primary-specific in one (13%), and relapse-specific in two (25%, including one SHH) patients (Data Supplement). Amplifications of MYCN were identified among seven patients with SHH and five patients with Group 4 tumors with 42% occurring as shared, 25% as primary-specific, and 33% as relapse-specific with no statistical difference in alteration pattern between subgroups (P = .77, chi-square test; Figs 4A-4C; Data Supplement). Of 34 driver genes with identified alterations, 15 (44%) are theoretically actionable and occurred in 72% of patients (Data Supplement). Of theoretically actionable alterations, 46% were shared between patient-matched tumors, whereas 33% were relapse-specific and 21% were primary-specific.
Chromosomal Alteration Patterns
Genome-wide distribution and patterns of chromosomal arm alterations were also investigated (Figs 4A, 4D, and 4E, Data Supplement). The incidence of shared chromosome arm alterations was 42% for SHH, 61% for Group 3, and 58% for Group 4 tumors (Fig 4D). Subgroup was associated with a significant difference in the pattern of chromosome arm alterations, largely driven by biases in Group 3 tumors, which were predominantly conserved (P = .0047, analysis of variance). The median number of discordant chromosome arms between patient-matched tumors was two (IQR, 0-5) with no difference between subgroups (P = .10, Kruskal-Wallis test; Data Supplement). Thirty-three percent of patients had completely conserved cytogenetic landscapes with no statistical difference between subgroups (P = .13, chi-square test). Chromosome arms did not exhibit uniform alteration patterns (asterisks in Fig 4D, false discovery adjusted P < .1, chi-square test), as exemplified by highly conserved isochromosome 17q in Group 3 and Group 4 tumors (Fig 4E). Additionally, frequent relapse-specific losses of chromosomes 10q (31%) and 17p (46%) were recognized in SHH tumors (Fig 4F).
Subgroup and Subtype Divergence at Relapse
Subgroup divergence at relapse was observed in five of 118 cases (4%). Four patients diagnosed with high-confidence Group 4 tumors (median classification score, 0.97; IQR, 0.92-0.99) developed relapse tumors that were classified as high-confidence Group 3 tumor (median classification score, 0.92; IQR, 0.88-0.97; Fig 5A). One patient was diagnosed with a Group 3 tumor (classification score, 0.96) and experienced relapse with a Group 4 tumor of intermediate classification score (Group 4 classification score, 0.53; Group 3 classification score, 0.47).
FIG 5.

Subgroup divergence at medulloblastoma relapse. (A) Scatterplot depicting Group 3 and 4 classification scores for patients (n = 5) with divergent subgroup at relapse. (B) Oncoprint depicting alterations among patients with diagnostic group 4 tumors experiencing Group 3 relapse. A patient with primary Group 4 primary tumor (C) who experienced relapse with a Group 3 tumor (D) coincident with focal amplification of MYCN. G3, Group 3; G4, Group 4; MB, medulloblastoma; NGS, next-generation sequencing; SNV, single nucleotide variant.
All cases of Group 4 to Group 3 switching exhibited discordant subtype classifications (Data Supplement). Two cases with primary subtype VII tumors diverged to subtypes II and V at relapse. Other two cases diverged to subtype III from subtypes VI and VIII. Relapse-specific genetic alterations affecting MYC, MYCN, and FBXW7 were also observed in such subgroup-switching cases (Fig 5B). An exemplary case is depicted in Figures 5C and 5D. Notably, the single Group 3 to Group 4 switching case was classified as subtype V at both diagnosis and relapse.
Subtype divergence occurred at similar rates among both SHH (17%) and Groups 3 and 4 tumors (22%; P = .64, Fisher's exact test). No specific pattern of subtype inhomogeneity was identified (Data Supplement; P = .34, Stuart-Maxwell test). Notably, only the presence of MYC amplification or chromosome 2p gain in relapsed tumors was significantly associated with Groups 3 and 4 subtype divergence (P < .05, Fisher's exact test; Fig 4A; Data Supplement).
Nonmedulloblastoma Subsequent Tumors
Nonmedulloblastoma subsequent tumors were identified in nine (7%) patients (Fig 3A). High-grade gliomas accounted for eight (89%) such cases (Fig 6A). The median age of medulloblastoma diagnosis for these patients was 10.4 years (IQR, 9.2-10.8), and seven patients received radiotherapy for medulloblastoma (radiation therapy treatment status for one patient was unknown). The median interval from initial medulloblastoma to diagnosis of subsequent high-grade glioma was 3.77 years (IQR, 2.73-4.38).
FIG 6.
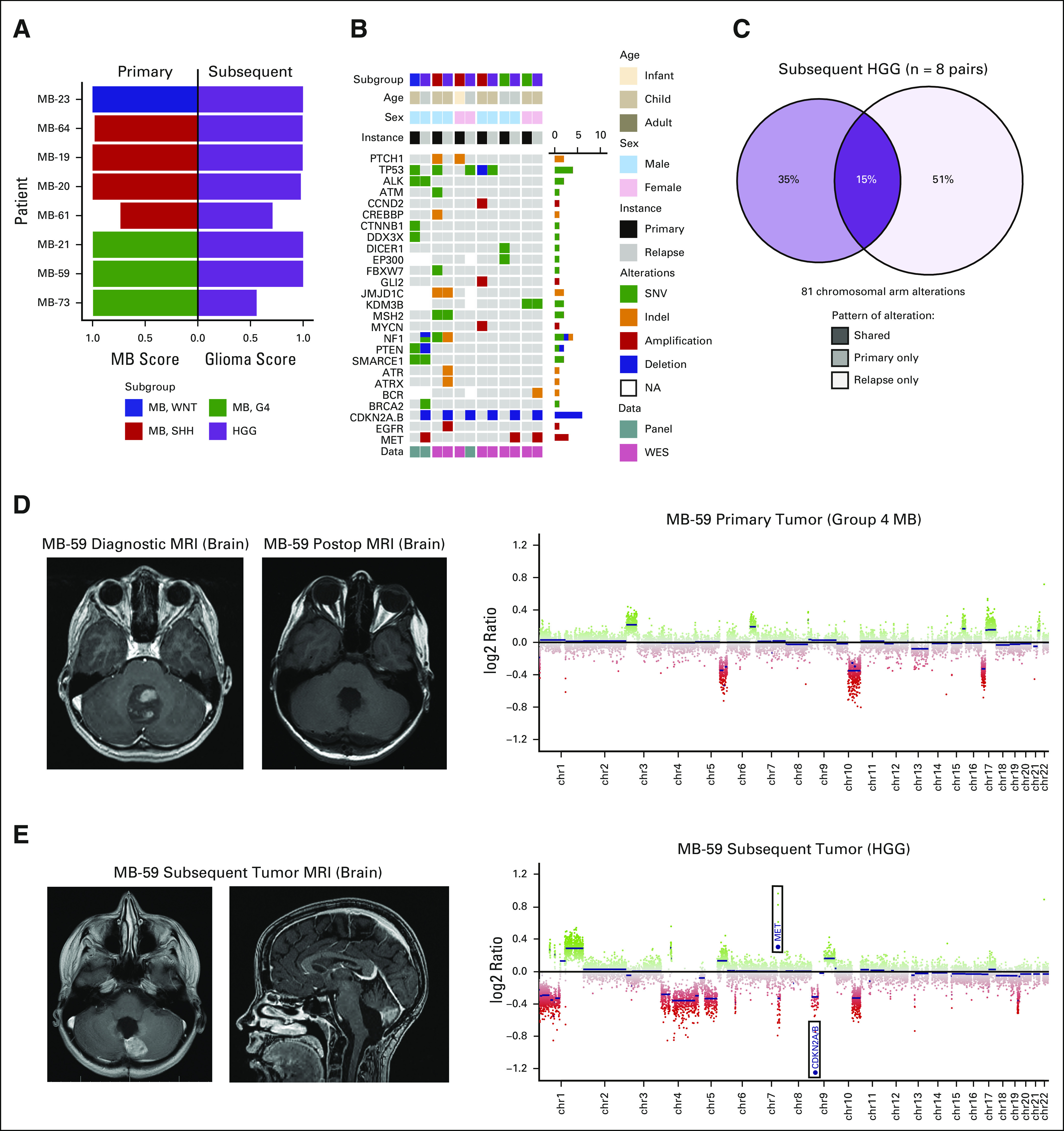
High-grade gliomas subsequent to primary medulloblastoma. (A) Classification scores of primary medulloblastomas and subsequent high-grade gliomas. (B) Oncoprint depicting patient characteristics and driver gene alterations for patient-matched tumor pairs with available next-generation sequencing (n = 6 patients). (C) Compartment-specific patterns of chromosome arm copy number variation for patient-matched tumor pairs (n = 8 patients). A patient with primary Group 4 tumor (D) who experienced subsequent high-grade glioma (E) in the local tumor bed. MRI, magnetic resonance imaging. HGG, high-grade glioma; MB, medulloblastoma; SHH, sonic hedgehog; SNV, single nucleotide variant; WES, whole-exome sequencing; WNT, wingless.
Molecular alterations observed in subsequent tumors are depicted in Figure 6B. Recurrent focal CDKN2A and/or CDKN2B homozygous deletions and MET amplifications were identified almost exclusively among subsequent gliomas (Data Supplement). No strong overlap of chromosome arm alterations was identified between diagnostic medulloblastomas and subsequent high-grade gliomas (Fig 6C). An exemplary case of subsequent high-grade glioma following Group 4 medulloblastoma is depicted in Figures 6D and 6E.
DISCUSSION
Although relapse rates in SJMB03 (22%) and SJYC07 (66%) suggest the critical role of up-front CSI in disease control, the temporal patterns of relapse highlight the slower biological progression of Group 4 tumors. Patients not previously treated with CSI (SJYC07) largely relapsed on or shortly after therapy, irrespective of subgroup. Although the aggressive nature of Group 3 tumors is highlighted in both trials, the different relapse rates observed for tumors assigned to Groups 3 and 4 subtypes IV and VII between trials motivate exploration of tailored treatment strategies driven by molecular subtype. Additionally, the propensity for local relapse among a subset of SHH tumors aligns with previous findings.13 Although our ability to discern subgroup-specific anatomic patterns of relapse might have been limited by cohort size, the high proportion of medulloblastomas with distant relapse highlights the dire need for therapies directed against the metastatic compartment.
Salvage rates of relapsed medulloblastoma are low and highly dependent on previous radiation therapy.11,26 Standardized protocols for salvage therapy are lacking, and studies describing potential survival benefits of various therapies are plagued by small numbers and further confounded by subgroup heterogeneity.27 In this study, radiation therapy after relapse was associated with durable long-term survival in a subset of radiation-naive patients (SJYC07), irrespective of subgroup. However, young patients, particularly infants, will benefit from continuous efforts to spare, delay, or reduce CSI, whereas alternative approaches that mitigate or avoid associated long-term sequelae are still needed.
The finding that SJMB03 patients with Group 4 tumors tend to survive longer postrelapse strongly suggests subgroup determination as a critical component for analyzing patient outcomes within and across trials for relapsed disease. Although subgroup and time to relapse appear to be important factors related to postrelapse survival in our study, additional molecular and clinical features not explicitly explored in the current study, such as underlying mutational signatures and genetic predisposition, may influence biological progression of disease and warrant further investigation.
Although absolute subgroup stability at relapse has been reported in patient-matched tumors profiled using the medulloblastoma-specific expression–based NanoString assay,13 methylation-based classification of the paired molecular cohort presented here suggests that a rare proportion of Group 4 tumors switches subgroup affiliation at relapse to Group 3. These results suggest possible subgroup plasticity between bulk Group 3 and Group 4 tumors and warrant further investigation using laboratory models, particularly to characterize the molecular mechanisms driving subgroup divergence. Given that subgroup switching was a rare observational phenomenon in the current study, genetic drivers underlying this divergence must be explored further and functionally validated. Overall, subtype divergence was more common than subgroup divergence. Associations of subtype divergence with MYC amplification and chromosome 2p gain (containing the MYCN locus) suggest the potential influence of these oncogenic transcription factors on subtype identity. Although no specific molecular feature was identified in cases of divergent subtype classification for SHH tumors, further exploration into intrinsic stability of novel subtypes and potential drivers of such divergence is needed.
The previous literature has described drastic clonal divergence of medulloblastoma at relapse and the emergence of relapse-specific molecular signatures, including MYC, MYCN, and TP53 pathway alterations.13,15 In our restricted analysis of known driver genes, we observed a more variable degree of conservation than suggested previously. Nevertheless, the previous literature leveraged a variety of molecular techniques to query specific lesions, such as iFISH for MYC and MYCN amplifications, which were only used as a validation in a subset of cases here albeit with high concordance.15,28 The incidence of such alterations may be underestimated by methylation array compared to more sensitive methods, such as iFISH. Also, by leveraging robust classification methods that can confidently identify a gamut of CNS tumors, the current study overcomes the appreciable incidence of high-grade gliomas that might have confounded previous studies.
Strides toward implementation of novel therapies, including targeted agents, have largely been based on the molecular landscape of untreated diagnostic tumors. The current study reveals primary-specific alterations as a relative minority, suggesting feasibility of target screening based on diagnostic tumor specimens. Although relatively infrequent, relapse-specific disruptions of DNA repair machinery might represent a viable therapeutic avenue. Similarly, emergence of relapse-specific alterations in chromatin modifiers suggests epigenetic dysregulation as a potentially actionable mechanism underlying therapeutic resistance. Nevertheless, the degree to which additional molecular alterations in other unexplored genes, noncoding regions, or rare subclones may affect target selection must be evaluated.
Key limitations of the current study stem from the scarcity of relapse tumor specimens. The paired molecular cohort is biased toward locally recurrent SHH and nonmetastatic Group 4 tumors with inclusion of older children and adults with SHH medulloblastoma. Assembly of the paired molecular cohort relied heavily on archival FFPE material without matched germline, thereby limiting mutational analysis to tumor-only inferences of previously annotated driver genes and preventing investigation into the role of germline predisposition. Additional molecular features, including gene expression and noncoding variants, should be explored in future studies. The current study of bulk tumor samples may have also under-called subclonal variants (< 10%) and is thus not adequately equipped for analyzing changes in clonal architecture. Single-cell and deep whole-genome sequencing will provide further insights into mechanisms of tumor evolution and oncogenic cascades contributing to relapse.
Given that the majority of patients with medulloblastoma exhibit at least some divergence in molecular features at relapse, targets should be verified at relapse, particularly in next-generation clinical trials. Biopsy may represent a safe and effective alternative to optimize patient selection and confirm targets for experimental therapies.29 Furthermore, definitive molecular diagnosis of relapse should be made given the incidence of other CNS malignancies arising after medulloblastoma therapy, particularly with increasing time from initial diagnosis.17,30 Clinical trials of relapsed medulloblastoma paired with confirmatory molecular profiling are urgently needed to establish standardized protocols for this deadly disease.
APPENDIX
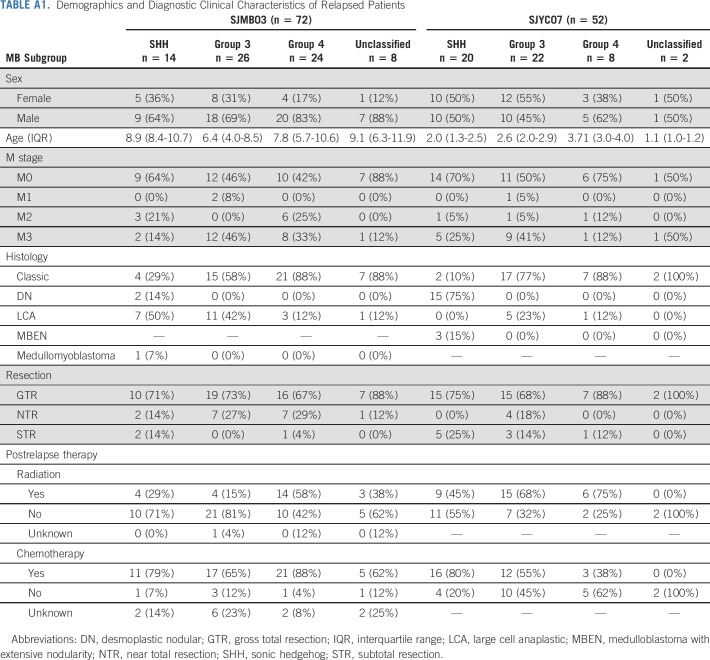
TABLE A1.
Demographics and Diagnostic Clinical Characteristics of Relapsed Patients
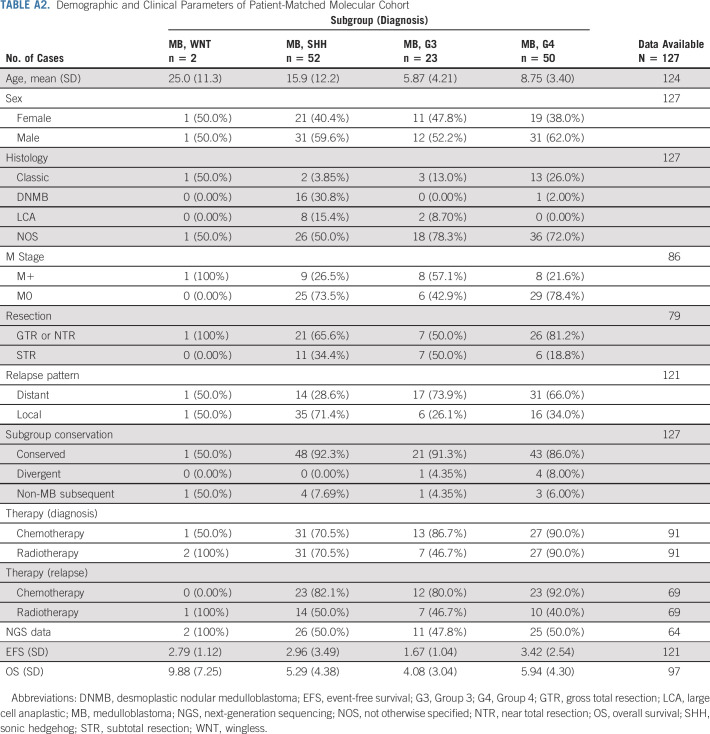
TABLE A2.
Demographic and Clinical Parameters of Patient-Matched Molecular Cohort
DISCLAIMER
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data and had the final responsibility to submit for publication.
SUPPORT
Supported by American Lebanese Syrian Associated Charities, St Jude Children's Research Hospital; Alexander and Margaret Stewart Trust (P.A.N.); American Association for Cancer Research (P.A.N.); St Baldrick's Foundation (P.A.N.); Alex's Lemonade Stand Young Investigator Award (V.R.); C.R. Younger Foundation (V.R.); Canadian Institutes of Health Research (V.R.); Nelina's Hope and Garron Family Cancer Centre (V.R.).
R.K. and K.S.S. contributed equally to this work. D.T.W.J., A.G., V.R., and P.A.N. contributed equally to this work.
AUTHOR CONTRIBUTIONS
Conception and design: Rahul Kumar, Kyle S. Smith, Giles W. Robinson, David T. W. Jones, Amar Gajjar, Vijay Ramaswamy, Paul A. Northcott
Financial support: David T. W. Jones, Amar Gajjar, Vijay Ramaswamy, Paul A. Northcott
Administrative support: Giles W. Robinson, Amar Gajjar, Paul A. Northcott
Provision of study materials or patients: Giles W. Robinson, Brent A. Orr, Murali Chintagumpala, Daniel C. Bowers, Timothy E. Hassall, Jordan R. Hansford, Dong Anh Khuong-Quang, John R. Crawford, Anne E. Bendel, Sridharan Gururangan, Kristin Schroeder, Eric Bouffet, Ute Bartels, Michael J. Fisher, Richard Cohn, Sonia Partap, Stewart J. Kellie, Geoffrey McCowage, Arnold C. Paulino, Stefan Rutkowski, Gudrun Fleischhack, Girish Dhall, Laura J. Klesse, Sarah Leary, Javad Nazarian, Marcel Kool, Pieter Wesseling, Marina Ryzhova, Olga Zheludkova, Andrey V. Golanov, Roger E. McLendon, Roger J. Packer, Christopher Dunham, Juliette Hukin, Maryam Fouladi, Claudia C. Faria, Jose Pimentel, Andrew W. Walter, Nada Jabado, Yoon-Jae Cho, Sebastien Perreault, Sidney E. Croul, Michal Zapotocky, Cynthia Hawkins, Uri Tabori, Michael D. Taylor, Stefan M. Pfister, Paul Klimo Jr, Frederick A. Boop, David W. Ellison, Thomas E. Merchant, Andrey Korshunov, David T. W. Jones, Amar Gajjar, Vijay Ramaswamy
Collection and assembly of data: Rahul Kumar, Kyle S. Smith, Maximilian Deng, Giles W. Robinson, Brent A. Orr, Colt Terhune, Anthony P. Y. Liu, Tong Lin, Catherine A. Billups, Arzu Onar-Thomas, Andrey Korshunov, David T.W. Jones, Amar Gajjar, Vijay Ramaswamy, Paul A. Northcott
Data analysis and interpretation: Rahul Kumar, Kyle S. Smith, Maximilian Deng, Giles W. Robinson, Brent A. Orr, Anthony P. Y. Liu, Tong Lin, Catherine A. Billups, Arzu Onar-Thomas, Andrey Korshunov, David T. W. Jones, Amar Gajjar, Vijay Ramaswamy, Paul A. Northcott
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Clinical Outcomes and Patient-Matched Molecular Composition of Relapsed Medulloblastoma
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Giles W. Robinson
Consulting or Advisory Role: Lilly, Roche/Genentech
Research Funding: Novartis, Genentech/Roche, Novartis
Jordan R. Hansford
Consulting or Advisory Role: Bayer
John R. Crawford
Honoraria: Illumina
Speakers' Bureau: AstraZeneca
Eric Bouffet
Consulting or Advisory Role: Novartis
Research Funding: Roche, Bristol-Myers Squibb
Michael J. Fisher
Honoraria: AstraZeneca
Research Funding: AstraZeneca, Array BioPharma, Exelixis
Travel, Accommodations, Expenses: AstraZeneca, SpringWorks
Sonia Partap
Consulting or Advisory Role: Bayer, Gerson Lehrman Group
Research Funding: Abbvie
Geoffrey McCowage
Consulting or Advisory Role: Biogen
Research Funding: Novartis, Merck
Travel, Accommodations, Expenses: BIOGEN
Arnold C. Paulino
Employment: MD Anderson Cancer Center
Patents, Royalties, Other Intellectual Property: Royalty from Elsevier Inc for book on PET/CT in Radiotherapy Treatment Planning
Travel, Accommodations, Expenses: University of Southern California
Stefan Rutkowski
Consulting or Advisory Role: Bristol-Myers Squibb GmbH & Co KGaA, Germany, Celgene, Roche Pharma AG, Grenzach-Wyhlen
Research Funding: Riemser Pharma GmbH, Greifswald, Germany
Laura J. Klesse
Consulting or Advisory Role: AstraZeneca
Travel, Accommodations, Expenses: Springworks, AstraZeneca
Sarah Leary
Research Funding: Blaze Bioscience
Roger E. McLendon
Stock and Other Ownership Interests: Gilead Sciences, NantKwest
Expert Testimony: Johnson & Johnson
Roger J. Packer
Honoraria: Novartis
Consulting or Advisory Role: Novartis, AstraZeneca
Juliette Hukin
Stock and Other Ownership Interests: Abbvie
Maryam Fouladi
Research Funding: PTC therapeutics, Bayer Schering Pharma
Sebastien Perreault
Leadership: Bayer
Stock and Other Ownership Interests: Novocure
Honoraria: Bayer
Consulting or Advisory Role: Bayer
Speakers' Bureau: Bayer
Expert Testimony: Bayer
Michal Zapotocky
Consulting or Advisory Role: Bayer
Patents, Royalties, Other Intellectual Property: IP for low grade glioma and sarcoma fusion panels as well as medulloblastoma subgrouping panel
Stefan M. Pfister
Research Funding: Lilly, Bayer, Roche, PharmaMar, Pfizer
Patents, Royalties, Other Intellectual Property: Patent on utilizing DNA methylation profiling for tumor classification
Frederick A. Boop
Employment: Semmes Murphey Clinic
David W. Ellison
Patents, Royalties, Other Intellectual Property: Sole inventor of US Patent No. 9,005,907 issued April 14, 2015 “Methods and Compositions for Typing Molecular Subgroups of Medulloblastoma”, 62627291/S88435 1190US.P1 February 2018 “Epigenetic Histone Regulation Mediated by CXorf67” published August 2019 as WO 2019/155387
Thomas E. Merchant
Travel, Accommodations, Expenses: Philips Healthcare
Arzu Onar-Thomas
Consulting or Advisory Role: Roche
Research Funding: Novartis, Apexigen, Pfizer, Celgene, Novartis, Merck, Novocure
Travel, Accommodations, Expenses: Roche
David T. W. Jones
Patents, Royalties, Other Intellectual Property: Patent WO 2013075237 A1, titled “Mutations of histone proteins associated with proliferative disorders”
Amar Gajjar
Consulting or Advisory Role: Roche/Genentech
Research Funding: Genentech, Kazia Pharmaceutical
Vijay Ramaswamy
Honoraria: AstraZeneca
No other potential conflicts of interest were reported.
REFERENCES
- 1.Curtin SC, Minino AM, Anderson RN: Declines in cancer death rates among children and adolescents in the United States, 1999-2014. NCHS Data Brief:1-8, 2016 [PubMed] [Google Scholar]
- 2.Ostrom QT Cioffi G Gittleman H, et al. : CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2012-2016. Neuro Oncol 21:v1-v100, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gajjar A Chintagumpala M Ashley D, et al. : Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): Long-term results from a prospective, multicentre trial. Lancet Oncol 7:813-820, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Tarbell NJ Friedman H Polkinghorn WR, et al. : High-risk medulloblastoma: A pediatric oncology group randomized trial of chemotherapy before or after radiation therapy (POG 9031). J Clin Oncol 31:2936-2941, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnston DL Keene D Strother D, et al. : Survival following tumor recurrence in children with medulloblastoma. J Pediatr Hematol Oncol 40:e159-e163, 2018 [DOI] [PubMed] [Google Scholar]
- 6.Sabel M Fleischhack G Tippelt S, et al. : Relapse patterns and outcome after relapse in standard risk medulloblastoma: A report from the HIT-SIOP-PNET4 study. J Neurooncol 129:515-524, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koschmann C Bloom K Upadhyaya S, et al. : Survival after relapse of medulloblastoma. J Pediatr Hematol Oncol 38:269-273, 2016 [DOI] [PubMed] [Google Scholar]
- 8.Gajjar A Mulhern RK Heideman RL, et al. : Medulloblastoma in very young children: Outcome of definitive craniospinal irradiation following incomplete response to chemotherapy. J Clin Oncol 12:1212-1216, 1994 [DOI] [PubMed] [Google Scholar]
- 9.Valteau-Couanet D Fillipini B Benhamou E, et al. : High-dose busulfan and thiotepa followed by autologous stem cell transplantation (ASCT) in previously irradiated medulloblastoma patients: High toxicity and lack of efficacy. Bone Marrow Transplant 36:939-945, 2005 [DOI] [PubMed] [Google Scholar]
- 10.Grill J Geoerger B Gesner L, et al. : Phase II study of irinotecan in combination with temozolomide (TEMIRI) in children with recurrent or refractory medulloblastoma: A joint ITCC and SIOPE brain tumor study. Neuro Oncol 15:1236-1243, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gururangan S Krauser J Watral MA, et al. : Efficacy of high-dose chemotherapy or standard salvage therapy in patients with recurrent medulloblastoma. Neuro Oncol 10:745-751, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor MD Northcott PA Korshunov A, et al. : Molecular subgroups of medulloblastoma: The current consensus. Acta Neuropathol 123:465-472, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramaswamy V Remke M Bouffet E, et al. : Recurrence patterns across medulloblastoma subgroups: An integrated clinical and molecular analysis. Lancet Oncol 14:1200-1207, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrissy AS Garzia L Shih DJ, et al. : Divergent clonal selection dominates medulloblastoma at recurrence. Nature 529:351-357, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill RM Kuijper S Lindsey JC, et al. : Combined MYC and P53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease. Cancer Cell 27:72-84, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korshunov A Okonechnikov K Sahm F, et al. : Molecular progression of SHH-activated medulloblastomas. Acta Neuropathol 138:327-330, 2019 [DOI] [PubMed] [Google Scholar]
- 17.Salloum R Chen Y Yasui Y, et al. : Late morbidity and mortality among medulloblastoma survivors diagnosed across three decades: A report from the childhood cancer survivor study. J Clin Oncol 37:731-740, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Capper D Jones DTW Sill M, et al. : DNA methylation-based classification of central nervous system tumours. Nature 555:469-474, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Northcott PA Buchhalter I Morrissy AS, et al. : The whole-genome landscape of medulloblastoma subtypes. Nature 547:311-317, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavalli FMG Remke M Rampasek L, et al. : Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 31:737-754.e6, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwalbe EC Lindsey JC Nakjang S, et al. : Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: A cohort study. Lancet Oncol 18:958-971, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma T Schwalbe EC Williamson D, et al. : Second-generation molecular subgrouping of medulloblastoma: An international meta-analysis of group 3 and group 4 subtypes. Acta Neuropathol 138:309-326, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson GW Rudneva VA Buchhalter I, et al. : Risk-adapted therapy for young children with medulloblastoma (SJYC07): Therapeutic and molecular outcomes from a multicentre, phase 2 trial. Lancet Oncol 19:768-784, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gajjar A Robinson GW Smith KS, et al. : Outcomes by clinical and molecular features in children with medulloblastoma treated with risk-adapted therapy: Results of an international phase III trial (SJMB03). J Clin Oncol 39:822-835, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The St Jude Cloud . https://pecan.stjude.cloud/proteinpaint/study/MB-Relapse [Google Scholar]
- 26.Dunkel IJ Gardner SL Garvin JH Jr, et al. : High-dose carboplatin, thiotepa, and etoposide with autologous stem cell rescue for patients with previously irradiated recurrent medulloblastoma. Neuro Oncol 12:297-303, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kameda-Smith MM Wang A Abdulhadi N, et al. : Salvage therapy for childhood medulloblastoma: A single center experience. Can J Neurol Sci 46:403-414, 2019 [DOI] [PubMed] [Google Scholar]
- 28.Pickles JC, Stone TJ, Jacques TS: Methylation-based algorithms for diagnosis: Experience from neuro-oncology. J Pathol 250:510-517, 2020 [DOI] [PubMed] [Google Scholar]
- 29.Balter-Seri J Mor C Shuper A, et al. : Cure of recurrent medulloblastoma: The contribution of surgical resection at relapse. Cancer 79:1241-1247, 1997 [DOI] [PubMed] [Google Scholar]
- 30.Phi JH Park AK Lee S, et al. : Genomic analysis reveals secondary glioblastoma after radiotherapy in a subset of recurrent medulloblastomas. Acta Neuropathol 135:939-953, 2018 [DOI] [PubMed] [Google Scholar]